Abstract
A panoramic analysis of chemokines, pro-inflammatory/regulatory cytokines, and growth factors was performed in serum samples from patients with acute DENV infection (n=317) by a high-throughput microbeads array. Most soluble mediators analyzed were increased in DENV patients regardless of the DENV serotype. The substantial increase (≥10-fold) of CXCL10, IL-6, and IFN-γ, and decreased levels of PDGF (<0.4-fold) was universally identified in all DENV serotypes. Of note, increased levels of CXCL8, CCL4, and IL-12 (≥3-9-fold) were selectively observed in DENV2 as compared to DENV1 and DENV4. Heatmap and biomarker signatures further illustrated the massive release of soluble mediators observed in DENV patients, confirming the marked increase of several soluble mediators in DENV2. Integrative correlation matrices and networks showed that DENV infection exhibited higher connectivity among soluble mediators. Of note, DENV2 displayed a more complex network, with higher connectivity involving a higher number of soluble mediators. The timeline kinetics (Day 0-1, D2, D3, D4-6) analysis additionally demonstrated differences among DENV serotypes. While DENV1 triggers a progressive increase of soluble mediators towards D3 and with a decline at D4-6, DENV2 and DENV4 develop with a progressive increase towards D4-6 with an early plateau observed in DENV4. Overall, our results provided a comprehensive overview of the immune response elicited by DENV infection, revealing that infection with distinct DENV serotypes causes distinct profiles, rhythms, and dynamic network connectivity of soluble mediators. Altogether, these findings may provide novel insights to understand the pathogenesis of acute infection with distinct DENV serotypes.
Introduction
Dengue virus infection (DENV) is a mosquito-borne disease caused by one of the four dengue serotypes (DENV-1, DENV-2, DENV-3, and DENV-4) that belongs to the Flaviviridae family. DENV serotypes circulate concomitantly in different regions worldwide, covering approximately 100 countries in tropical and subtropical areas of the globe with expanding geographical range and incidence (1, 2). The precise incidence of DENV is difficult to determine but it is estimated that the annual number of cases ranges from 284 to 528 million worldwide (3) depending on the year and intensity of outbreaks.
DENV infection induces a spectrum of clinical manifestations, ranging from asymptomatic to life-threatening hemorrhagic fever/shock syndrome. The pathogenesis of DENV involves a complex interplay of viral and host factors, including viral serotype, host age, genetic background, and immunological status (4–7).
Previous studies have suggested that distinct serotypes may cause more severe disease. It has been shown that DENV-2 infection is associated with higher rates of severe clinical forms of DENV infections (8). It has been considered that different serotypes may vary in their ability to infect target cells, triggering distinct immune response profiles that, in turn, impact their capacity to cause severe clinical forms (9, 10). However, the precise mechanisms explaining the association between higher disease severity and distinct viral serotypes are still unclear.
Among the host factors, the immune responses are critical for the outcome of DENV infection. High levels of serum soluble mediators have been detected during DENV infection (11–16). The upregulated levels of chemokines and cytokines may contribute to both protective and immunopathological mechanisms during DENV infection (17). Moreover, higher plasma levels of pro-inflammatory mediators have been found in patients with severe dengue (18). It is believed that inflammatory cytokine storm and other soluble mediators can act on the endothelium, and alter normal fluid barrier functions, leading to increased plasma leakage (19, 20). However, much remains to be elucidated about the immunopathogenesis of acute DENV infections in humans, particularly focusing on the impact of distinct DENV serotypes during acute infection.
In the present study, a panoramic analysis of several chemokines, pro-inflammatory/regulatory cytokines, and growth factors was performed in serum samples from patients with acute DENV infection with distinct serotypes (DENV-1, DENV-2, and DENV-4) using a high-throughput microbeads array. Overall, our findings demonstrate that the profile of circulating soluble mediators differs substantially during acute DENV infection according to distinct serotypes. Selective profiles elicited in each response suggest the participation of distinct serotype-associated immune pathways that may ultimately represent a potential target for future therapeutic intervention.
Material and Methods
Subjects and Samples
This is a cross-sectional study submitted and approved by the Ethical Committee at Instituto René Rachou/Fundação Oswaldo Cruz (FIOCRUZ-Minas) – Belo Horizonte, Minas Gerais, Brazil (CAAE 41591015.2.0000.5091), and Fundação de Medicina Tropical Dr. Heitor Vieira Dourado, Manaus, Amazonas, Brazil (CAAE 80866017.8.0000.0005). A total of 317 patients with acute DENV infection, enrolled from January 2010 to December 2019, were selected for this investigation as a non-probabilistic convenience sample, comprising 153 males and 164 females, aging from 6 months to 74 years old (median = 27 years). Serum samples from DENV patients were obtained from biorepositories maintained at Instituto Evandro Chagas, Ananindeua, Pará, Brazil, and Fundação de Medicina Tropical Dr. Heitor Vieira Dourado, Manaus, Amazonas, Brazil. Demographic, clinical, and laboratory records were obtained from the compulsory notification form for DENV infection available at the Information System for Notifiable Diseases (SINAN) of the Ministry of Health of Brazil. Laboratory diagnosis of DENV infection was carried out by serological test (MAC-ELISA), molecular tests (RT-qPCR), or viral isolation in cell culture. Differential diagnosis with other acute febrile viral infections (ZIKV and CHIKV) was also carried out, and cases of co-infection were excluded from the present investigation. DENV serotype diagnosis was performed in 269 DENV patients by qPCR or indirect immunofluorescence test, allowing further classification of patients into three subgroups, referred to as DENV1 (n=116), DENV2 (n=52), and DENV4 (n=101). No DENV3 infection was detected amongst the patients enrolled in the present investigation. Days after symptom onset was available for 313 DENV patients and was used to cluster them into subgroups for a cross-sectional analysis at four consecutive time points (Days = D), including D0-1, D2, D3, and D4-6.
As a reference control group, a total of 319 age-matching healthy subjects were included, comprising 148 males and 171 females, aging from 1 to 75 years old (median = 20 years). The control group was composed of a non-probabilistic convenience sampling from biorepositories maintained at Grupo Integrado de Pesquisas em Biomarcadores, Instituto René Rachou, Fundação Oswaldo Cruz (FIOCRUZ-Minas), Belo Horizonte, MG, Brazil and at Fundação Hospitalar de Hematologia e Hemoterapia do Amazonas (HEMOAM), Manaus, Amazonas, Brazil.
All participants enrolled in the present investigation signed the informed consent form under the Declaration of Helsinki and Resolution 466/2012 of the Brazilian National Health Council for research involving human subjects.
DENV Diagnosis
DENV diagnosis was performed by serological test, molecular tests, or viral isolation in cell culture, as described in detail below.
Serological diagnosis was performed by in-house IgM capture enzyme-linked immunosorbent assay for DENV (MAC-ELISA) as previously described (21). Briefly, flat-bottom 96-well plates were sensitized with goat anti-human IgM and blocked with 4% bovine serum albumin (BSA) solution. Plates were washed and incubated with 50 µL/well of serum samples for 2 hours. Following washing steps, 50 µL of soluble DENV antigen were added to each well, and plates were incubated at 4°C overnight. Plates were washed and 25 µL/well of anti-flavivirus monoclonal antibody (6B6C, Jackson Immunoresearch Laboratories, PA, USA) conjugated with horseradish peroxidase were added. Plates were washed and incubated with 100 µL/well of ABTS substrate (2.2’-azino-di-(3-ethyl-benzthiazoline sulfonate) and hydrogen peroxide. After incubation for 30 min at room temperature (RT), plates were read in an ELISA reader at 405 nm and positivity was assessed according to the results observed for negative control samples.
Molecular diagnosis was performed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) as described (22). Briefly, purified RNA was extracted from 200 μL of serum samples using the ReliaPrep™ Viral TNA Miniprep System (Promega, WI, USA) and then reverse transcribed to cDNA using SuperScript™ II Reverse Transcriptase (Invitrogen, USA). Experiment batches were carried out according to the manufacturer’s instructions. cDNA was used as a template for the qPCR employing the GoTaq® qPCR Master Mix Kit (Promega, WI, USA) for the first screening of Flavivirus using the following primers: mFU-1 (5’-TACAACATGATGGGAAAGCGAGAGAAAAA-3’), and CFD2 (5’-GTGTCCCAGCCGGCGGTGTCATCAGC-3’) (22). DENV molecular diagnosis was performed according to previous studies (23) using D1 (5’-TCAATATGCTGAAACGCGCGAGAAACCG-3’), and D2 (5’-TTGCACCAACAGTCAATGTCTTCAGGTTC-3’) primers.
DENV viral isolation was carried out in continuous C6/36 cell line cultures in vitro as described previously (24).
DENV Serotype Diagnosis
Differential diagnosis of DENV serotypes (DENV1-4) was performed as previously described (23). Samples that tested positive for DENV by RT-qPCR were submitted to a second round of amplification using 10 μL of the first reaction with a mix of DENV serotype-specific reverse primers (TS1 = 5’-CGTCTCAGTGATCCGGGGG-3’; TS2 = 5’-CGCCACAAGGGCCATGAACAG-3’; TS3 = 5’-TAACATCATCATGAGACAGAGC-3’ and TS4 = 5’-CTCTGTTGTCTTAAACAAGAGA-3’), using the same cycling parameters as the first reaction.
DENV samples isolated by cell culture in vitro were further employed for differential DENV serotype diagnosis by indirect immunofluorescence using polyclonal antibodies to DENV serotypes (Biomanguinhos, FIOCRUZ, RJ, Brazil) as previously described (25).
Quantification of Serum Soluble Mediators
Serum chemokines (CXCL8, CCL11, CCL3, CCL4, CCL2, CCL5, and CXCL10), pro-inflammatory cytokines (IL-1β, IL-6, TNF-α, IL-12, IFN-γ, IL-15, and IL-17), regulatory cytokines (IL-1Ra, IL-4, IL-5, IL-9, IL-10, and IL-13) and growth factors (FGF-basic, PDGF, VEGF, G-CSF, GM-CSF, IL-7, and IL-2) were quantified by a high-throughput microbeads array (Bio-Plex Pro™ Human Cytokine 27-plex Assay, Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions. Data acquisition was carried out on a Luminex 200 System using the Bioplex Manager Software. Final concentrations of serum soluble mediators were estimated by 5-parameter logistic regression according to the standard curve inserted on each experimental batch and the results expressed in pg/mL.
Statistical Analysis
Data analysis was performed using GraphPad Prism v.9.1.1 software (San Diego, CA, USA). The Shapiro-Wilk normality test was conducted to assess data normality distribution. Considering the non-parametric distribution of the results, comparative analysis between DENV vs NI was performed by Mann-Whitney test. Multiple comparative analysis among subgroups according to DENV serotypes and days after symptom onset was carried out by Kruskal-Wallis test, followed by Dunn’s post-test. In all cases, significance was considered at p < 0.05.
Heatmap constructs were assembled using the heatmap.2 function in the R and gplots package (Project for Statistical Computing Version 3.0.1). The analysis was performed using customized functions available at Bioconductor packages, considering a Row Z-score, scaled from −2 to +2. Signatures of serum soluble mediators were constructed for NI, DENV, and DENV subgroups by first converting the serum levels of soluble mediators, originally expressed as continuous variables (pg/mL) into categorical data (proportion, %) using the global 3rd tercile values of each soluble mediator as the cut-off to calculate the proportion of subjects above the cut-off edges. The following cut-offs were employed: CXCL8 = 5.68; CCL11 = 34.67; CCL3 = 3.64; CCL4 = 18.86; CCL2 = 51.61; CCL5 = 953.95; CXCL10 = 537.07; IL-1β = 0.45; IL-6 = 2.43; TNF-α = 16.26; IL-12 = 1.13; IFN-γ = 25.50; IL-15 = 102.75; IL-17 = 3.80; IL-1Ra = 303.94; IL-4 = 1.33; IL-5 = 5.90; IL-9 = 10.00; IL-10 = 11.12; IL-13 = 2.29; FGF-basic = 3.69; PDGF = 357.22; VEGF = 137.43; G-CSF = 34.62; GM-CSF = 1.61; IL-7 = 15.36; and IL-2 = 1.75. Thereafter, the proportions of subjects with increased levels of serum soluble mediators (above the 3rd tercile) were calculated and the ascendant signatures built to underscore those molecules with proportion higher than 50%.
Correlation analysis was employed to construct integrative correlation matrices and assemble integrative serum soluble mediator networks. Pearson and Spearman correlation tests were used to obtain the significant “r” scores at p<0.05. The Cytoscape software platform (available at https://cytoscape.org) was employed to construct the complex networks with eccentric layout, with nodes representing the serum soluble mediators. Connecting edges illustrate weak/moderate (“r” scores between |0.1 to 0.66|) and strong correlations (“r” scores ≥ |0.67|) between pairs of attributes. Attributes without strong correlations were distributed in the network periphery. Serum soluble mediators presenting at least 5 strong correlations were assembled in the center with the number of serum soluble mediators and connections between them used to define the central connectivity.
The magnitude of change in the serum levels of soluble mediators was calculated as the proportion ratio according to the median values observed in non-infected healthy controls. Venn diagram analysis (http://bioinformatics.psb.ugent.be/webtools/Venn/) was carried out online to identify common and exclusive soluble mediators with altered levels observed in DENV serotypes.
Results
Overall Profile of Serum Soluble Mediators During Acute DENV Infection
The panoramic analysis of chemokines (CXCL8, CCL11, CCL3, CCL4, CCL2, CCL5, CXCL10), pro-inflammatory cytokines (IL-1β, IL-6, TNF-α, IL-12, IFN-γ, IL-15, IL-17), regulatory cytokines (IL-1Ra, IL-4, IL-5, IL-9, IL-10, IL-13), and growth factors (FGF-basic, PDGF, VEGF, G-CSF, GM-CSF, IL-7, and IL-2) was performed in serum samples from patients with acute DENV infection as compared to the non-infected healthy controls (NI) and the results are shown in Figure 1. Overall, data analysis demonstrated that most chemokines, cytokines, and growth factors were increased in DENV patients as compared to NI. Conversely, lower levels of IL-17, IL-4, and PDGF were observed in DENV patients as compared to NI. No significant differences were observed in the serum levels of CCL11 (Figure 1).
Figure 1
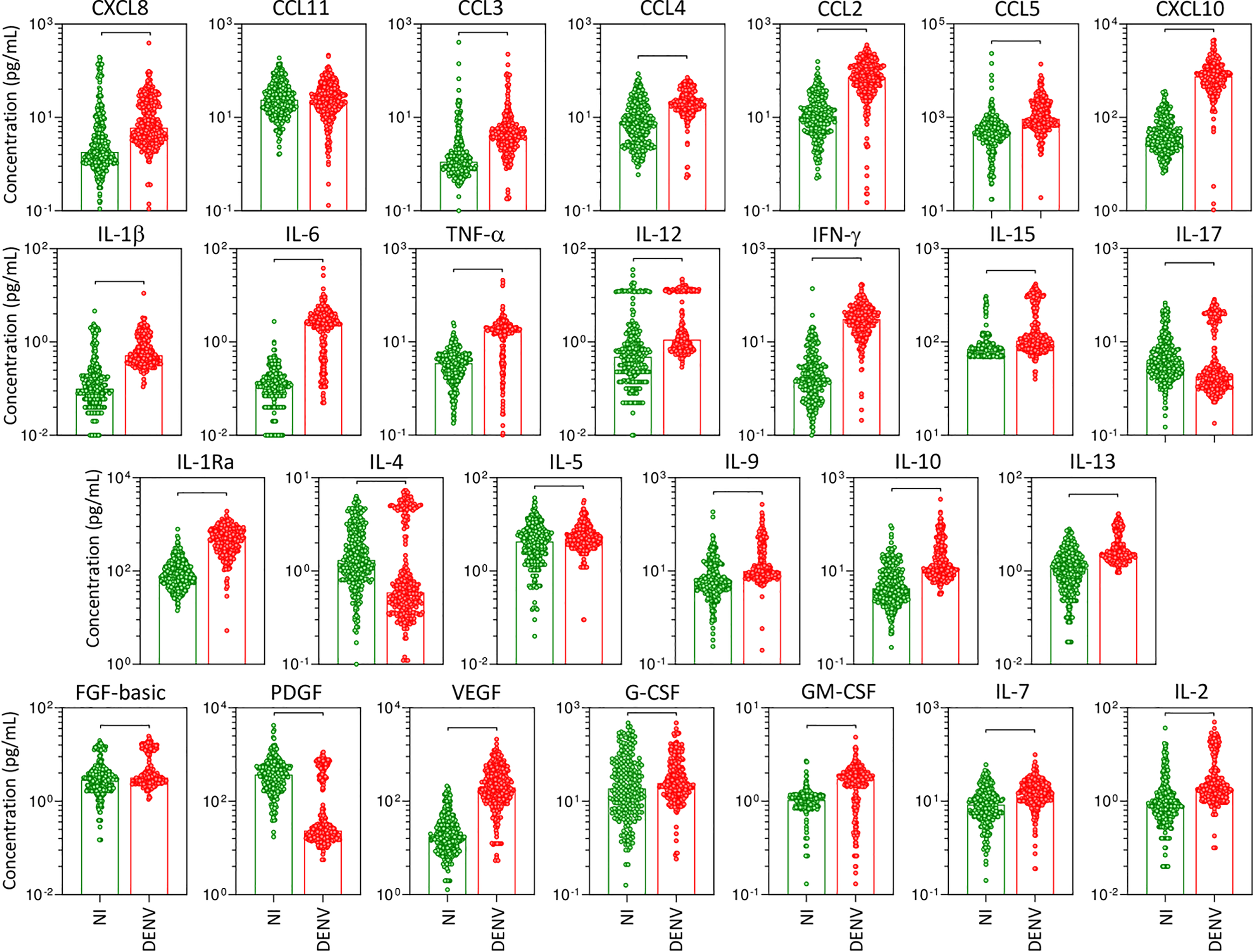
Overall profile of serum soluble mediators during acute DENV infection. The levels of chemokines (CXCL8, CCL11, CCL3, CCL4, CCL2, CCL5, CXCL10), pro-inflammatory cytokines (IL-1β, IL-6, TNF-α, IL-12, IFN-γ, IL-15, IL-17), regulatory cytokines (IL-1Ra, IL-4, IL-5, IL-9, IL-10, IL-13), and growth factors (FGF-basic, PDGF, VEGF, G-CSF, GM-CSF, IL-7, and IL-2) were measured in serum samples from patients with acute DENV infection – “DENV” ( n=317), represented by red dots/bars on the right side of each panel, as compared to the reference non-infected healthy controls – “NI” (
n=317), represented by red dots/bars on the right side of each panel, as compared to the reference non-infected healthy controls – “NI” ( n=319), represented by green dots/bars on the left side of each panel. Quantification of serum soluble mediators was carried out by a high-throughput microbeads array as described in the Material and Methods section. The results are presented as the scattering of individual values expressed in pg/mL over bar plots highlighting the median on each bar. Significant differences at p <0.05 are indicated by connecting lines.
n=319), represented by green dots/bars on the left side of each panel. Quantification of serum soluble mediators was carried out by a high-throughput microbeads array as described in the Material and Methods section. The results are presented as the scattering of individual values expressed in pg/mL over bar plots highlighting the median on each bar. Significant differences at p <0.05 are indicated by connecting lines.
Panoramic Overview of Serum Soluble Mediators During Acute Infection With Distinct DENV Serotypes
The overall snapshot of chemokines, pro-inflammatory cytokines, regulatory cytokines, and growth factors was performed in serum samples from patients with acute DENV infection, further classified according to the DENV serotypes as DENV1, DENV2, and DENV4, as compared to NI (Figure 2). Data analysis demonstrated that in general, regardless of the DENV serotype, the levels of the soluble factors were increased as compared to NI. Of note was the superior increase of several soluble mediators observed in DENV2 as compared to DENV1 and DENV4, including CXCL8, CCL4, IL-1β, IL-12, IL-15, IL-17, IL-4, IL-9, IL-13, FGF-basic, PDGF, IL-7, and IL-2. Conversely, lower levels of CXCL10, IL-6, VEGF, and GM-CSF were observed in DENV2 patients as compared to DENV1 and DENV4. Several changes were observed between DENV1 and DENV4, including increased levels of IL-1β, TNF-α, IL-12, IL-15, IL-5, IL-9, FGF-basic, GM-CSF, and IL-2 observed in DENV1, while increased levels of CCL11 and IL-1Ra along lower levels of IL-17 and PDGF were observed in DENV4 (Figure 2).
Figure 2
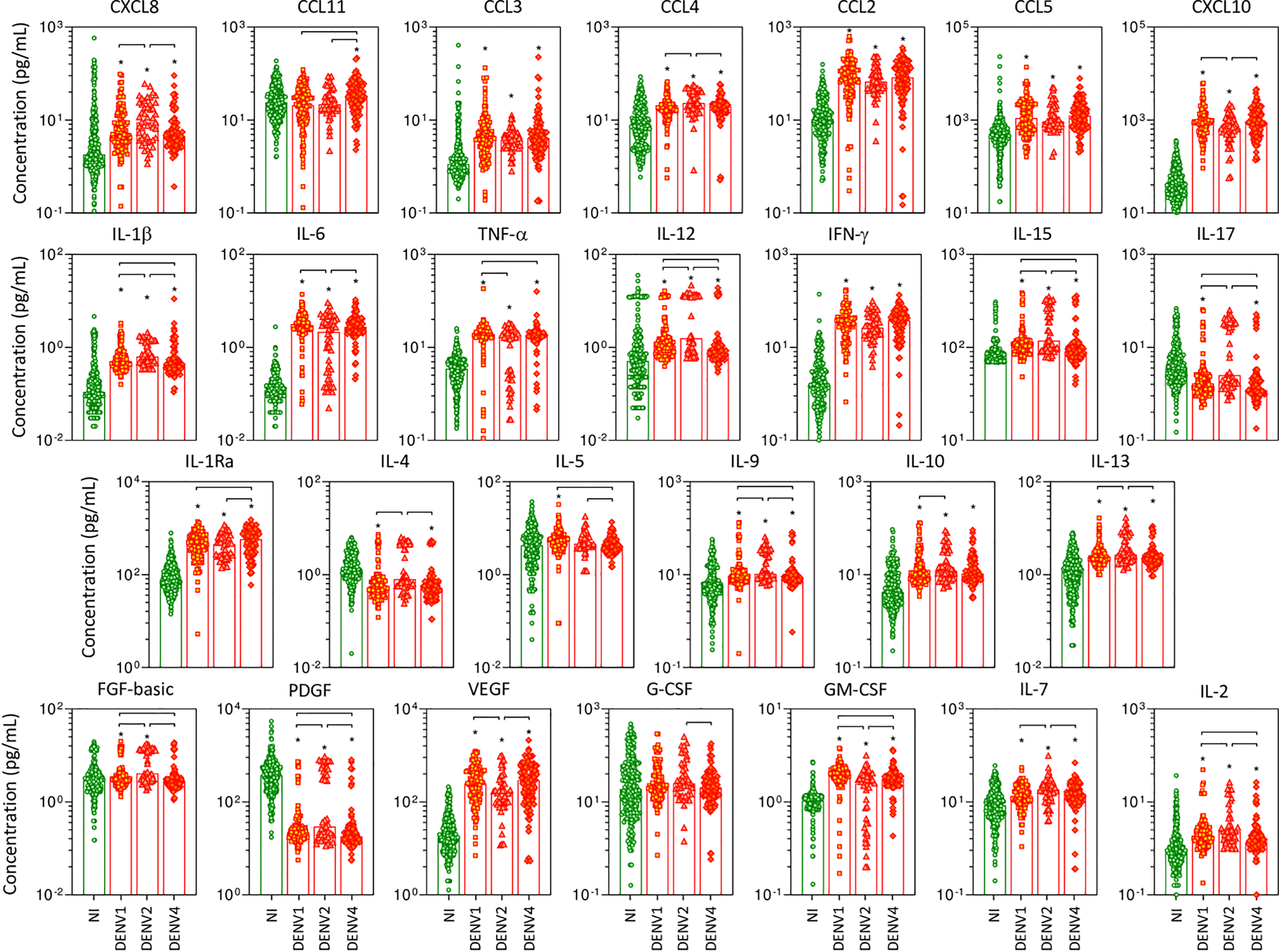
Panoramic overview of serum soluble mediators during acute infection with distinct DENV serotypes. The levels of chemokines (CXCL8, CCL11, CCL3, CCL4, CCL2, CCL5, CXCL10), pro-inflammatory cytokines (IL-1β, IL-6, TNF-α, IL-12, IFN-γ, IL-15, IL-17), regulatory cytokines (IL-1Ra, IL-4, IL-5, IL-9, IL-10, IL-13), and growth factors (FGF-basic, PDGF, VEGF, G-CSF, GM-CSF, IL-7, and IL-2) were measured in serum samples from patients with acute infection with distinct DENV serotypes (n=269), referred as: “DENV1” ( n=116), “DENV2” (
n=116), “DENV2” ( n=52), and “DENV4” (
n=52), and “DENV4” ( n=101), represented by symbols/bars on the right side of each panel, as compared to the reference non-infected healthy controls – “NI” (
n=101), represented by symbols/bars on the right side of each panel, as compared to the reference non-infected healthy controls – “NI” ( n=319), represented by green dots/bars on the left side of each panel. Quantification of serum soluble mediators was carried out by high-throughput microbeads array as described in the Material and Methods section. The results are presented as the scattering of individual values expressed in pg/mL over bar plots highlighting the median on each bar. Significant differences at p <0.05 are indicated by an asterisk (*), and connecting lines for comparisons with non-infected healthy controls and among DENV serotypes, respectively.
n=319), represented by green dots/bars on the left side of each panel. Quantification of serum soluble mediators was carried out by high-throughput microbeads array as described in the Material and Methods section. The results are presented as the scattering of individual values expressed in pg/mL over bar plots highlighting the median on each bar. Significant differences at p <0.05 are indicated by an asterisk (*), and connecting lines for comparisons with non-infected healthy controls and among DENV serotypes, respectively.
To further assess the magnitude of changes in serum levels of soluble mediators in patients with acute infection with distinct DENV serotypes, the fold change ratio was calculated for each sample according to the median values observed in NI (Supplementary Figure 1). Data analysis demonstrated that PDGF is universally identified with decreased levels (fold change <0.4) in all DENV serotypes as compared to NI. On the other hand, the levels of CXCL10, IL-6, and IFN-γ are substantially increased (fold change ≥10), regardless of the DENV serotypes. The analysis of soluble mediators with noteworthy increased levels (fold change ≥3-9) revealed that while CCL3, CCL2, IL-1β, TNF-α, and IL1-Ra were commonly observed in all DENV serotypes, CXCL8, CCL4, and IL-12 were elevated only in patients with DENV2 infections (Supplementary Figure 1).
Heatmap Analysis and Signatures of Serum Soluble Mediators During Acute DENV Infection
Aiming at characterizing the global outlook of serum soluble mediators during acute DENV infection, heatmap constructs and soluble mediator signatures were assembled, and the data are presented in Figure 3. Overall, the heatmaps illustrate the massive release of most serum soluble mediators in DENV patients as compared to NI. Moreover, the heatmap constructs further confirmed the marked increase of several soluble mediators in DENV2 patients as compared to DENV1 and DENV4 (Figure 3A). The analysis of serum soluble mediator signatures assembled with categorical data further corroborates these findings. Using a general cut-off at the 50th percentile, higher release of serum soluble mediators was observed in DENV (CCL3, CCL2, CCL5, CXCL10, IL-1β, IL-6, TNF-α, IFN-γ, IL-1Ra, VEGF, and GM-CSF), and lower PDGF as compared to NI. Moreover, the results also showed that DENV2 (CXCL8, CCL3, CCL4, CCL2, CXCL10, IL-1β, TNF-α, IL-12, IFN-γ, IL-15, IL-1Ra, IL-9, IL-10, IL-13, FGF-basic, VEGF, IL-7, and IL-2) displayed higher upregulation of serum soluble mediators as compared to DENV1 (CCL3, CCL2, CCL5, CXCL10, IL-1β, IL-6, TNF-α, IFN-γ, IL-15, IL-1Ra, VEGF, GM-CSF and IL-2), and DENV4 (CCL3, CCL4, CCL2, CCL5, CXCL10, IL-6, TNF-α, IFN-γ, IL-1Ra, VEGF, GM-CSF) (Figure 3B).
Figure 3
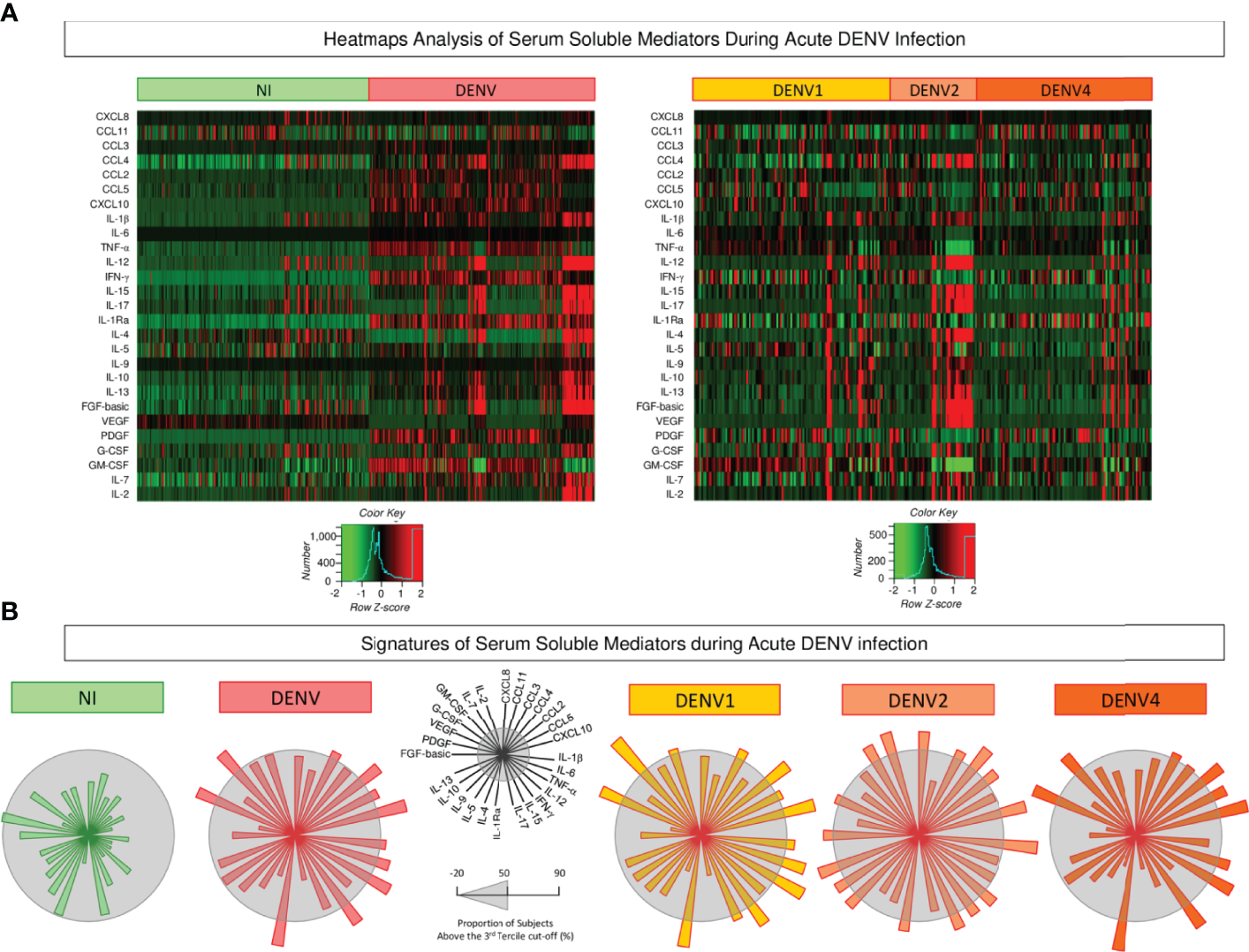
Heatmap analysis and signatures of serum soluble mediators during acute DENV infection. (A) Heatmap constructs of soluble mediators defined to cluster profiles of chemokines (CXCL8, CCL11, CCL3, CCL4, CCL2, CCL5, CXCL10), pro-inflammatory cytokines (IL-1β, IL-6, TNF-α, IL-12, IFN-γ, IL-15, IL-17), regulatory cytokines (IL-1Ra, IL-4, IL-5, IL-9, IL-10, IL-13), and growth factors (FGF-basic, PDGF, VEGF, G-CSF, GM-CSF, IL-7, and IL-2) observed in serum samples from patients with acute DENV infection – “DENV” ( n=319), further classified according to the DENV serotypes (n=269) into: DENV1 (
n=319), further classified according to the DENV serotypes (n=269) into: DENV1 ( n=116), DENV2 (
n=116), DENV2 ( n=52), DENV4 (
n=52), DENV4 ( n=101) as compared to the reference non-infected healthy controls – “NI” (
n=101) as compared to the reference non-infected healthy controls – “NI” ( n=319). The soluble mediators were measured by a high-throughput microbeads array as described in the Material and Methods section. The individual levels of serum soluble mediators (attributes), expressed in pg/mL, were used to calculate the Z-score for each attribute and further generation of heatmap constructs. The Row Z-score scaled from −2 to 2 is illustrated as defined in the color key gradients provided in the Figure. (B) Overall signature of chemokines, pro-inflammatory cytokines, regulatory cytokines, and growth factors assembled for patients with acute DENV infection – “DENV”, further classified according to the DENV serotypes as: DENV1, DENV2, and DENV4 as compared to the reference non-infected healthy controls – “NI”. Signatures of serum soluble mediators were built as described in Material and Methods, by first converting the serum levels of soluble mediators, originally expressed as continuous variables (pg/mL) into categorical data (percentual, %) using the upper 3rd tertile values of each serum mediator as the cut-off to identify the proportion of subjects above the cut-off edges. The final data are shown in a radar chart, with each axis representing one serum mediator. The 50th percentile (gray zone) was used to underscore the serum soluble mediators with increased proportion (≥ 50%) in each study group.
n=319). The soluble mediators were measured by a high-throughput microbeads array as described in the Material and Methods section. The individual levels of serum soluble mediators (attributes), expressed in pg/mL, were used to calculate the Z-score for each attribute and further generation of heatmap constructs. The Row Z-score scaled from −2 to 2 is illustrated as defined in the color key gradients provided in the Figure. (B) Overall signature of chemokines, pro-inflammatory cytokines, regulatory cytokines, and growth factors assembled for patients with acute DENV infection – “DENV”, further classified according to the DENV serotypes as: DENV1, DENV2, and DENV4 as compared to the reference non-infected healthy controls – “NI”. Signatures of serum soluble mediators were built as described in Material and Methods, by first converting the serum levels of soluble mediators, originally expressed as continuous variables (pg/mL) into categorical data (percentual, %) using the upper 3rd tertile values of each serum mediator as the cut-off to identify the proportion of subjects above the cut-off edges. The final data are shown in a radar chart, with each axis representing one serum mediator. The 50th percentile (gray zone) was used to underscore the serum soluble mediators with increased proportion (≥ 50%) in each study group.
Integrative Correlation Matrices and Networks of Serum Soluble Mediators During Acute DENV Infection
To better understand the interrelationship between distinct serum soluble mediators during acute DENV infection, integrative correlation analysis was carried out and comprehensive matrices were created to assemble networks and calculate the neighborhood connectivity (Figure 4). Connectivity power networks were assembled using an eccentric layout based on the profile of significant correlation indices. The analysis of central connectivity scores, defined for serum soluble mediators with at least 5 strong correlations, revealed that acute DENV infection exhibited higher connectivity (n=120) involving more serum soluble mediators (n=13) as compared to NI (n= 103 and 10, respectively). In addition, the analysis of networks triggered by distinct DENV serotypes demonstrated that DENV2 displayed a more complex network, with higher central connectivity (n=180) enrolling more serum soluble mediators (n=17) as compared to DENV1 (n=98 and 12) and DENV4 (n=74 and 9) (Figure 4).
Figure 4
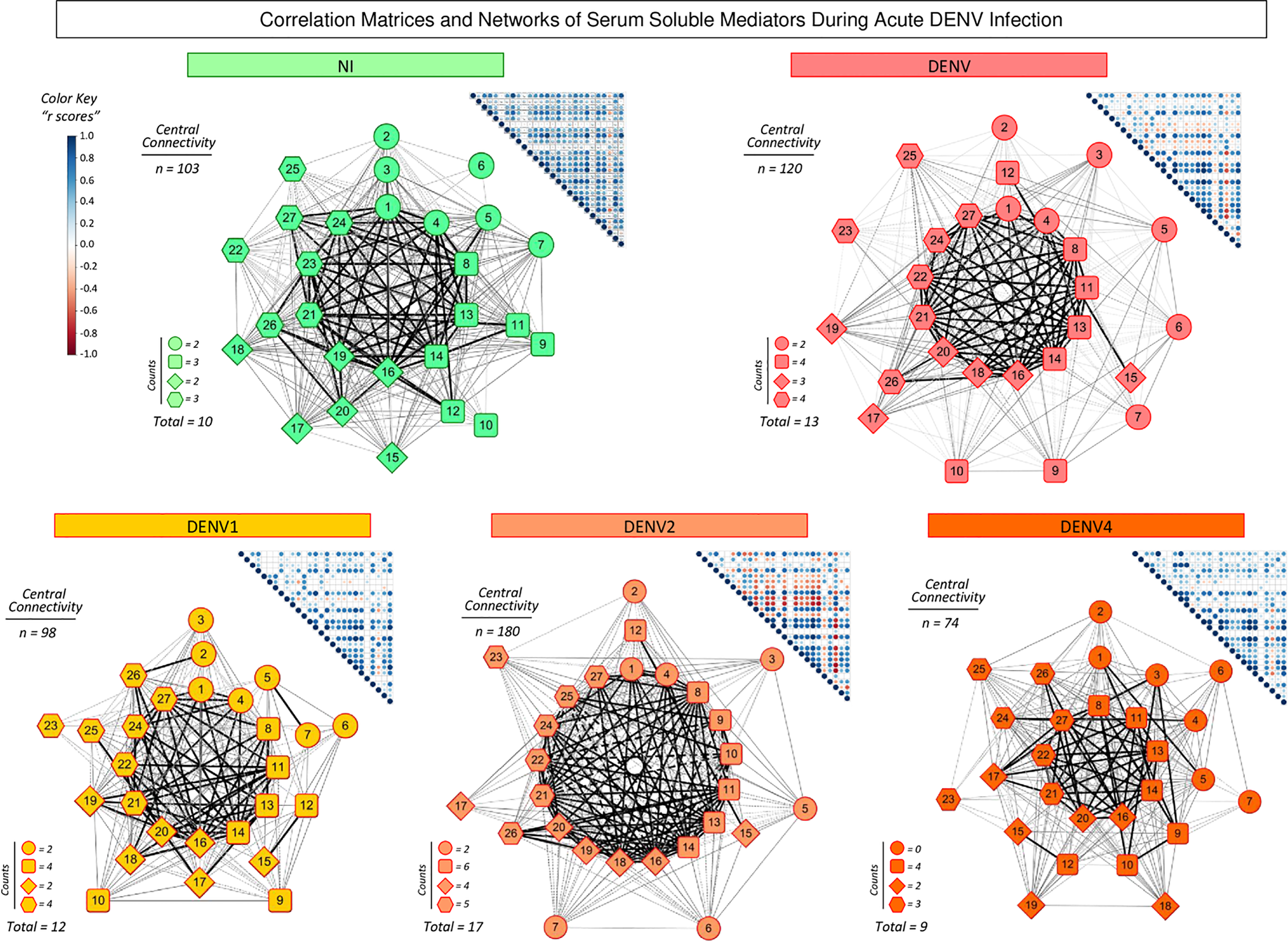
Integrative correlation matrices and networks of serum soluble mediators during acute DENV infection. Comprehensive correlation matrices were assembled based on the Pearson and Spearman “r” scores between chemokines, pro-inflammatory cytokines, regulatory cytokines and growth factors measured in serum samples from patients with acute DENV infection – “DENV” ( n=319), further classified according to the DENV serotypes (n=269) into: DENV1 (
n=319), further classified according to the DENV serotypes (n=269) into: DENV1 ( n=116), DENV2 (
n=116), DENV2 ( n=52), DENV4 (
n=52), DENV4 ( n=101) as compared to the reference non-infected healthy controls – “NI” (
n=101) as compared to the reference non-infected healthy controls – “NI” ( n=319). The soluble mediators were measured by a high-throughput microbeads array as described in the Material and Methods section. Panoramic correlation overviews are shown as triangle template matrices with each square intersection representing the correlation “r” score between pairs of attributes. The “r” scores of significant correlations (p< 0.05) are represented by circles of proportional sizes, scaled from −1 to +1 with a gradient for negative (red dots) and positive (blue dots) color keys. The white squares represent non-significant correlations. Networks were built using an eccentric layout based on significant correlations, with nodes representing the chemokines (
n=319). The soluble mediators were measured by a high-throughput microbeads array as described in the Material and Methods section. Panoramic correlation overviews are shown as triangle template matrices with each square intersection representing the correlation “r” score between pairs of attributes. The “r” scores of significant correlations (p< 0.05) are represented by circles of proportional sizes, scaled from −1 to +1 with a gradient for negative (red dots) and positive (blue dots) color keys. The white squares represent non-significant correlations. Networks were built using an eccentric layout based on significant correlations, with nodes representing the chemokines (  1=CXCL8; 2=CCL11; 3=CCL3; 4=CCL4; 5=CCL2; 6=CCL5 and 7=CXCL10), pro-inflammatory cytokines (8=IL-1β; 9=IL-6; 10=TNF-α; 11=IL-12; 12=IFN-γ; 13=IL-15 and 14=IL-17), regulatory cytokines (
1=CXCL8; 2=CCL11; 3=CCL3; 4=CCL4; 5=CCL2; 6=CCL5 and 7=CXCL10), pro-inflammatory cytokines (8=IL-1β; 9=IL-6; 10=TNF-α; 11=IL-12; 12=IFN-γ; 13=IL-15 and 14=IL-17), regulatory cytokines ( 15=IL-1Ra; 16=IL-4; 17=IL-5; 18=IL-9; 19=IL-10 and 20=IL-13), and growth factors (
15=IL-1Ra; 16=IL-4; 17=IL-5; 18=IL-9; 19=IL-10 and 20=IL-13), and growth factors ( 21=FGF-basic; 22=PDGF; 23=VEGF; 24=G-CSF; 25=GM-CSF; 26=IL-7; and 27=IL-2). Connecting edges illustrate weak/moderate (|0.1 to 0.66|, gradient gray thin lines) and strong “r” scores (≥ |0.67|, thick black lines) between pairs of attributes. Negative correlations are underscored by dashed lines. Attributes without strong correlations are distributed in the network periphery. Serum soluble mediators presenting at least 5 strong correlations are assembled in the center with the number of each category (
21=FGF-basic; 22=PDGF; 23=VEGF; 24=G-CSF; 25=GM-CSF; 26=IL-7; and 27=IL-2). Connecting edges illustrate weak/moderate (|0.1 to 0.66|, gradient gray thin lines) and strong “r” scores (≥ |0.67|, thick black lines) between pairs of attributes. Negative correlations are underscored by dashed lines. Attributes without strong correlations are distributed in the network periphery. Serum soluble mediators presenting at least 5 strong correlations are assembled in the center with the number of each category ( ,
,  ,
,  and
and  ), and connections between them (central connectivity) provided in the Figure.
), and connections between them (central connectivity) provided in the Figure.
Timeline Kinetics of Serum Soluble Mediators During Acute DENV Infection
The kinetics of serum soluble mediators in patients with acute DENV infection was carried out by cross-sectional analysis at four consecutive time points (Days = D), including D0-1, D2, D3, and D4-6 after symptom onset. The data are shown in Figure 5. Data analysis revealed several serum soluble mediators with a progressive increase along the timeline kinetics towards D4-6, including CXCL8, CCL3, CCL4, IL-1β, IL-12, IL-15, IL-17, IL-4, IL-5, IL-9, IL-10, IL-13, FGF-basic, PDGF, G-CSF, IL-7, and IL-2. On the other hand, a range of serum soluble mediators displayed a progressive decline towards D4-6, as observed for CCL11, CCL2, CCL5, CXCL10, IL-6, TNF-α, IFN-γ, IL-1Ra, VEGF, and GM-CSF (Figure 5).
Figure 5
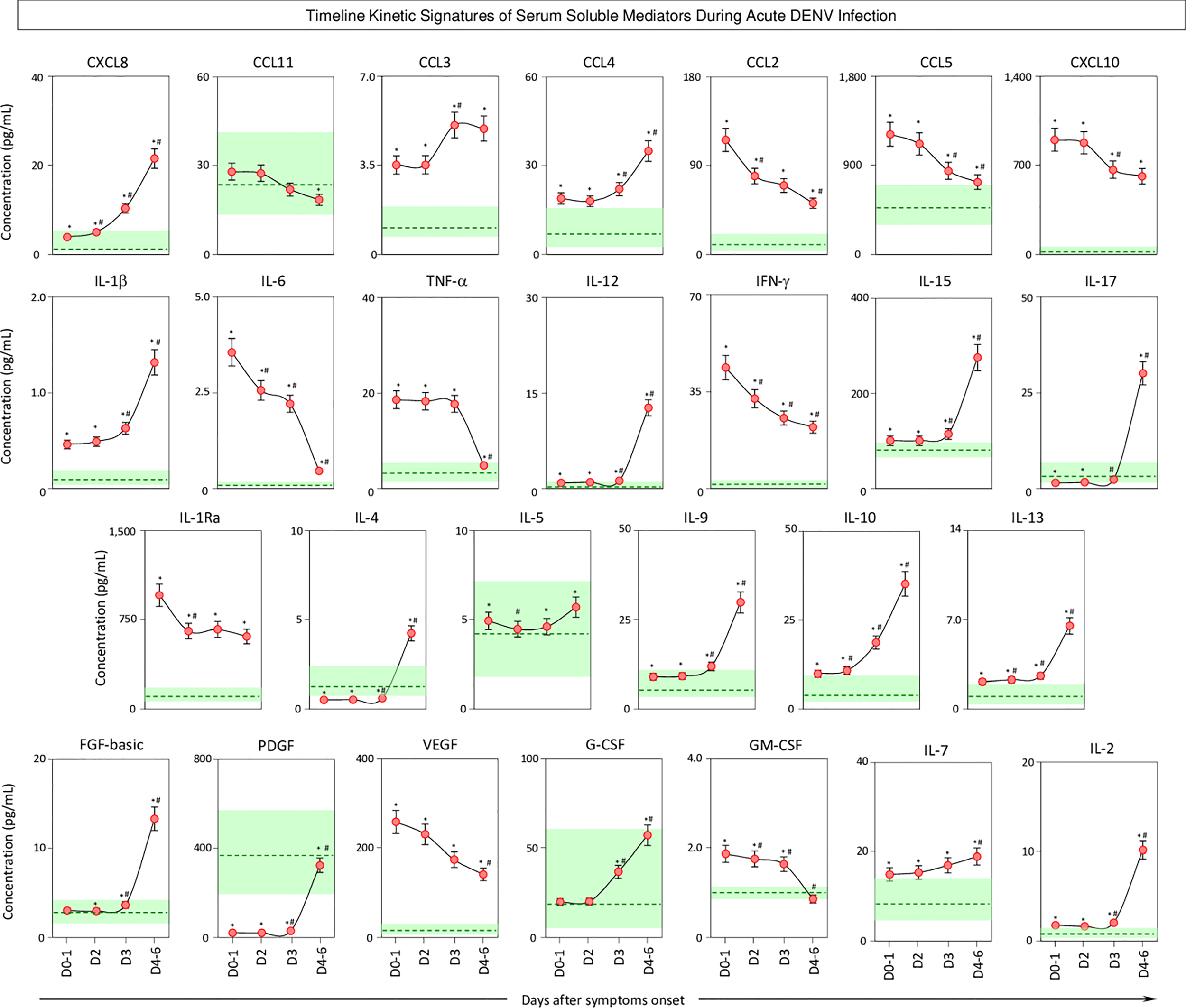
Timeline kinetics of serum soluble mediators during acute DENV infection. The dynamic kinetics of chemokines (CXCL8, CCL11, CCL3, CCL4, CCL2, CCL5, CXCL10), pro-inflammatory cytokines (IL-1β, IL-6, TNF-α, IL-12, IFN-γ, IL-15, IL-17), regulatory cytokines (IL-1Ra, IL-4, IL-5, IL-9, IL-10, IL-13), and growth factors (FGF-basic, PDGF, VEGF, G-CSF, GM-CSF, IL-2, and IL-7) was characterized in serum samples from patients with acute DENV infection ( n=313) by cross-sectional analysis at four consecutive time points (Days = D), including: D0-1 (n=89), D2 (n=100), D3 (n=71), and D4-6 (n=53) after symptom onset. Measurements were carried out by a high-throughput microbeads array as described in the Material and Methods section. The results are presented as a line chart of median values (± 10% of median) at each time point along with the timeline kinetics. The green zone represents the reference interquartile range (25th-75th) observed for non-infected healthy controls “NI” (n=319). Significant differences at p<0.05 are identified by asterisks (*) for comparisons with “NI” and (#) for comparisons with the immediately preceding time-point.
n=313) by cross-sectional analysis at four consecutive time points (Days = D), including: D0-1 (n=89), D2 (n=100), D3 (n=71), and D4-6 (n=53) after symptom onset. Measurements were carried out by a high-throughput microbeads array as described in the Material and Methods section. The results are presented as a line chart of median values (± 10% of median) at each time point along with the timeline kinetics. The green zone represents the reference interquartile range (25th-75th) observed for non-infected healthy controls “NI” (n=319). Significant differences at p<0.05 are identified by asterisks (*) for comparisons with “NI” and (#) for comparisons with the immediately preceding time-point.
Timeline Kinetics Signatures of Serum Soluble Mediators During Acute Infection With Distinct DENV Serotypes
To further characterize the dynamic changes of soluble mediators in acute DENV1, 2, and 4 infection, chemokines, growth factors, and pro-inflammatory and regulatory cytokines were quantified at four consecutive time points (D0-1, D2, D3, and D4-6) after symptom onset (Figure 6). DENV1 infection showed a progressive increase of soluble mediators, with a peak at D3 and a decline towards D4-6. DENV2 infection showed a progressive increase of soluble mediators towards D4-6. Interestingly, DENV4 infection showed an early plateau followed by a late upregulation at D3 towards D4-6.
Figure 6

Timeline kinetics signatures of serum soluble mediators during acute infection with distinct DENV serotypes. Overall signature of chemokines (CXCL8, CCL11, CCL3, CCL4, CCL2, CCL5, CXCL10), pro-inflammatory cytokines (IL-1β, IL-6, TNF-α, IL-12, IFN-γ, IL-15, IL-17), regulatory cytokines (IL-1Ra, IL-4, IL-5, IL-9, IL-10, IL-13), and growth factors (FGF-basic, PDGF, VEGF, G-CSF, GM-CSF, IL-7 and IL-2) were assembled for patients with acute infection with distinct DENV serotypes (n=265), referred to as: DENV1 ( n=114), DENV2 (
n=114), DENV2 ( n=52), and DENV4 (
n=52), and DENV4 ( n=99) by cross-sectional analysis at four consecutive time points (Days = D), including: D0-1 (n=42; 17; 26), D2 (n=40; 18; 36), D3 (n=17; 11; 30) and D4-6 (n=15; 6; 7) after symptom onset. The serum soluble mediators were measured by a high-throughput microbeads array as described in Material and Methods. Signatures of serum soluble mediators were built by first converting the serum levels of soluble mediators, originally expressed as continuous variables (pg/mL) into categorical data (percentual, %) using the upper 3rd tertile values of each serum mediator as the cut-off to identify the proportion of subjects above the cut-off edges. The final data are shown as line charts displaying the serum soluble mediator signature at each time point. Column diagrams are provided below to underscore the upregulated soluble serum mediators along the timeline kinetics. The 50th percentile was used to underscore the serum soluble mediators with an increased proportion (≥ 50%) in each study group.
n=99) by cross-sectional analysis at four consecutive time points (Days = D), including: D0-1 (n=42; 17; 26), D2 (n=40; 18; 36), D3 (n=17; 11; 30) and D4-6 (n=15; 6; 7) after symptom onset. The serum soluble mediators were measured by a high-throughput microbeads array as described in Material and Methods. Signatures of serum soluble mediators were built by first converting the serum levels of soluble mediators, originally expressed as continuous variables (pg/mL) into categorical data (percentual, %) using the upper 3rd tertile values of each serum mediator as the cut-off to identify the proportion of subjects above the cut-off edges. The final data are shown as line charts displaying the serum soluble mediator signature at each time point. Column diagrams are provided below to underscore the upregulated soluble serum mediators along the timeline kinetics. The 50th percentile was used to underscore the serum soluble mediators with an increased proportion (≥ 50%) in each study group.
Discussion
The pathogenesis of DENV infection involves a complex interaction between viral and host features. Risk factors for severe disease may include viral serotype, host age, genetic background, and immunological profiles (4–6). In this sense, distinct DENV serotypes leading to diverse immune responses (9) may impact the disease outcome towards severe clinical forms (10). Despite the relevance of the DENV serotype association with higher rates of severe clinical forms of DENV infections (3, 8), the precise mechanism and the specific elements with which the immune response participates in this phenomenon are not completely elucidated.
From an immunological point of view, previous studies have already demonstrated that acute DENV infection elicits a massive release of several immunological mediators, comprising a high level of chemokines and pro-inflammatory cytokines (18, 24). It is strongly believed that soluble mediators play a key role in orchestrating the immune response against viral patterns. Therefore, there is a consensus that the storm of chemokines, cytokines, and growth factors may directly shape the immunopathogenesis of acute DENV infection (26–28). However, distinct studies report conflicting results, about the increased levels of soluble mediators (11–16). These discrepancies among different studies are probably associated with the study design including the time of sampling, the laboratory methods, and instruments employed to measure soluble mediators, and the DENV serotype causing infection.
Aiming at overcoming these drawbacks, the present investigation encompassed a detailed analysis of serum soluble mediators using a sensitive high-throughput microbeads array and took into consideration the DENV serotype causing infection as well as the time for symptom onset.
Our results demonstrate that distinct patterns and characteristic behavior of serum soluble mediators occurred during acute DENV infection, with DENV serotype-associated particularities as characterized by distinct profiles, rhythms, and dynamic network connectivity. The three serum soluble mediators CXCL10, IL-6, and IFN-γ were underscored as universal, regardless of the DENV serotype, with a substantial increase of more than 10 times the control samples. At the opposite extreme, PDGF appeared as the least expressed soluble mediator, with less than 0.4 times the control, regardless of the serotype. PDGF was originally found as a constituent of platelets and the overall universal decrease in this biomarker may be associated with the canonical thrombocytopenia observed in Dengue patients.
The chemokine system appears to play a role during DENV infection. It has been shown that CXCL10 production improves host resistance, controlling the viral replication during DENV infection (29). It has also been shown that IL-6, produced by macrophages and activated endothelial cells, is a major mediator of fever and acute-phase reactions, and is produced by macrophages and activated endothelial cells. Furthermore, IL-6 mediates changes in coagulation and fibrinolysis by inducing the expression of tissue factors by endothelial cells (30). IL-6 production has been linked to several systemic changes in an acute inflammatory response (31), and the overproduction of IL-6 mediated by different DENV serotypes plays an important role in the pathogenesis triggered by the dengue virus (32). Moreover, increased serum levels of IL-6 have been reported in patients with severe DENV infections (33). In agreement with our findings, previous studies have already shown that IFN-γ is highly produced during DENV infection. IFN-γ is a pro-inflammatory cytokine that controls the production of nitric oxide and has an important antiviral role (34). Increased levels of IFN-γ are directly associated with protection against fever and high viremia, and with higher survival rates in patients with hemorrhagic signs (29, 34, 35). In agreement with that, IFN-γ-deficient mice are more susceptible to DENV2 and DENV3 infection (36, 37). It has been previously reported that lower levels of PDGF are observed in thrombocytopenic DENV patients, suggesting the relevant role of this growth factor in the pathogenesis of DENV (38). Overall, together with previous reports, our data suggest that the profile of CXCL10, IL-6, IFN-γ and PDGF may represent a useful set of biomarkers to monitor DENV outcome caused by distinct serotypes with a putative prognostic purpose. It is important to mention that other febrile illness may also lead to similar profile of serum soluble mediators as observed during the acute DENV infection (39, 40). In this sense, we have already reported that patients with acute ZIKV and CHIKV infection presented enhanced levels of serum soluble mediators including CXCL10 and IL-6. Future studies are still required to further address this issue, comparing a wide range of soluble mediators in distinct acute febrile illnesses in addition to the comparison with healthy controls.
Of note, our data have demonstrated that the profile of circulating soluble mediators differs substantially during acute DENV infection with distinct serotypes. Specifically, CXCL8, CCL4, and IL-12 were selectively increased in DENV2 as compared to DENV1 and DENV4. It has been shown that together with IL-6, CXCL8 mediates the derangement of coagulation and fibrinolysis acts and synergistically in the upregulation of tissue factors by endothelial cells (30). Clinical studies in endemic areas have described a correlation between DENV disease outcome and the levels of CCL4 which are intimately related to hypotension, thrombocytopenia, and hemorrhagic shock (14, 41). In general, the studies demonstrated that IL-12 seems to play a protective role during DENV-2 infection, preceding IFN-γ production (36). It has been previously suggested that lower levels of IL-12 are observed in patients with severe hemorrhagic DENV infection (42).
Clearly, there is a substantial redundancy between soluble mediator functions (i.e., the lack of one may be compensated by another with overlapping activities), and it has been previously demonstrated that the same soluble mediator may play distinct roles depending on the immunological microenvironment. In this sense, intending to perform an integrative analysis of chemokines, cytokines, and growth factors, we have constructed a comprehensive landscape of networks to illustrate the panoramic interplay between them. Our findings showed that DENV infection exhibited higher connectivity between more soluble mediators with DENV2 displaying a more complex network.
Regardless of the substantial number of patients enrolled, the present study has some limitations, such as: lack of previous studies on soluble mediators of DENV infection with distinct serotypes for comparative analysis; observational design of multiple comparisons without corrections for co-morbidities or other confounding variables; and lack of information or limited access to clinical records to allow data analysis according to disease severity and outcome to recovery or death. It is well known that secondary infection, regardless of DENV serotype, is associated with more severe disease as compared to primary infection. Moreover, although we have applied distinct laboratory methods to diagnose acute DENV infection, ascertaining whether the cases represent the primary or secondary dengue episode was not possible. Furthermore, the qPCR data were available for a small number of patients which did not allow an accurate and representative measurement of viremia neither to carry out a comparative analysis between viremia and levels of serum soluble mediators. Additional studies are currently in progress to access the impact of aging on the immunological profile of patients infected with distinct DENV serotypes.
Dengue is a disease that has annual epidemic peaks at different intensities and is related to different DENV serotypes. The reintroduction of DENV-4 in 2010 in Brazil, and the establishment of multiple co-circulating serotypes in different regions of the country are factors that may justify the non-predominance of DENV-3, given that in previous years, this serotype was predominant (43).
Altogether, these findings may provide novel insights to subsidize the understanding of DENV pathogenesis caused by distinct serotypes, highlighting useful biomarkers for future applications to predict severe disease outcomes and establish therapeutic interventions.
Funding
The study was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq. Funding was also obtained from Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM/PPP-CNPq, EDITAL N. 016/2014), Ministério da Saúde do Brasil (Chamada Pública n° 01/2012, Convênio # 776823/2012) and INCT para Febres Hemorrágicas Virais (INCT-FHV - 573739/2008-0).
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by Ethical Committee at Instituto René Rachou/Fundação Oswaldo Cruz (FIOCRUZ-Minas) – Belo Horizonte, Minas Gerais, Brazil (CAAE 41591015.2.0000.5091), and Fundação de Medicina Tropical Dr. Heitor Vieira Dourado, Manaus, Amazonas, Brazil (CAAE 80866017.8.0000.0005). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
Designing research study: MB, LR, PV, OM-F, MA, MSF, and LM. Advisory Committee: KB, VP-M, AC-A, JC-d-R, CG, and AC. Funding Acquisition: AT-C, MB, LR, PV, and OM-F. Conducting experiments and acquiring data: MC-d-S, PS, AC-A, VR, FM, JS-A, IC-R, VS, ESF, EM, ECSF, EVPS, BR, ÉBS, and MNOF. Analyzing data: MC-d-S, PS, AC-A, JB-d-S, MG, LRA, OM-F, MA, and MSF. Writing and reviewing the manuscript: MC-d-S, PS, VP-M, MR, JC-d-R, LRVA, CG, MB, AC, PV, OM-F, MA, MSF, and LM. All authors contributed to the article and approved the submitted version.
Acknowledgments
The study was carried out by students and professors enrolled at Post-graduate Programs: Epidemiologia e Vigilância em Saúde (PPGEVS/IEC), Ciências da Saúde (IRR/FIOCRUZ), and Ciências da Saúde (PPGCIS/UFAM), all supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). The authors thank the Program for Technological Development in Tools for Health-RPT-FIOCRUZ for using the flow cytometry facilities. The authors thank Dayane Andriotti Otta for the technical support. JC-d-R, AT-C, LRVA, OM-F, and MA received PQ fellowships from CNPq. AT-C and OM-F are research fellows from FAPEAM (PVN-II, PRÓ-ESTADO Program #005/2019). EM received fellowship support from FAPEAM/POSGRAD. FM received fellowship support from CNPq/INCT. PF-d-CV received PQ fellowship support from CNPq (310295/2021-1).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.892990/full#supplementary-material
References
1
World Health Organization (2021) . Dengue and Severe Dengue . Available at: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (Accessed January 08, 2022).
2
Rico-Hesse R . Molecular Evolution and Distribution of Dengue Viruses Type 1 and 2 in Nature. Virology (1990) 174(2):479–93. doi: 10.1016/0042-6822(90)90102-w
3
Harapan H Michie A Sasmono RT Imrie A . Dengue: A Minireview. Viruses (2020) 12(8):829. doi: 10.3390/v12080829
4
Guzman MG Halstead SB Artsob H Buchy P Farrar J Gubler DJ et al . Dengue: A Continuing Global Threat. Nat Rev Microbiol (2010) 8(12 Suppl):S7–16. doi: 10.1038/nrmicro2460
5
Srikiatkhachorn A . Plasma Leakage in Dengue Haemorrhagic Fever. Thromb Haemost (2009) 102(6):1042–9. doi: 10.1160/TH09-03-0208
6
Clyde K Kyle JL Harris E . Recent Advances in Deciphering Viral and Host Determinants of Dengue Virus Replication and Pathogenesis. J Virol (2006) 80(23):11418–31. doi: 10.1128/JVI.01257-06
7
Guzman MG Gubler DJ Izquierdo A Martinez E Halstead SB . Dengue Infection. Nat Rev Dis Primers (2016) 2:16055. doi: 10.1038/nrdp.2016.55
8
Rico-Hesse R Harrison LM Salas RA Tovar D Nisalak A Ramos C et al . Origins of Dengue Type 2 Viruses Associated With Increased Pathogenicity in the Americas. Virology (1997) 230(2):244–51. doi: 10.1006/viro.1997.8504
9
Simmons CP Farrar JJ van Vinh C Wills B . Dengue. N Engl J Med (2012) 366(15):1423–32. doi: 10.1056/NEJMra1110265
10
Holmes EC Twiddy SS . The Origin, Emergence and Evolutionary Genetics of Dengue Virus. Infect Genet Evol (2003) 3(1):19–28. doi: 10.1016/s1567-1348(03)00004-2
11
Azeredo EL Zagne SM Alvarenga AR Nogueira RM Kubelka CF de Oliveira-Pinto LM . Activated Peripheral Lymphocytes With Increased Expression of Cell Adhesion Molecules and Cytotoxic Markers Are Associated With Dengue Fever Disease. Mem Inst Oswaldo Cruz (2006) 101(4):437–49. doi: 10.1590/s0074-02762006000400016
12
Friberg H Beaumier CM Park S Pazoles P Endy TP Mathew A et al . Protective Versus Pathologic Pre-Exposure Cytokine Profiles in Dengue Virus Infection. PloS Negl Trop Dis (2018) 12(12):e0006975. doi: 10.1371/journal.pntd.0006975
13
de-Oliveira-Pinto LM Gandini M Freitas LP Siqueira MM Marinho CF Setúbal S et al . Profile of Circulating Levels of IL-1ra, CXCL10/IP-10, CCL4/MIP-1β and CCL2/MCP-1 in Dengue Fever and Parvovirosis. Mem Inst Oswaldo Cruz (2012) 107(1):48–56. doi: 10.1590/s0074-02762012000100007
14
Bozza FA Cruz OG Zagne SM Azeredo EL Nogueira RM Assis EF et al . Multiplex Cytokine Profile From Dengue Patients: MIP-1beta and IFN-Gamma as Predictive Factors for Severity. BMC Infect Dis (2008) 8:86. doi: 10.1186/1471-2334-8-86
15
de-Oliveira-Pinto LM Marinho CF Povoa TF de Azeredo EL de Souza LA Barbosa LD et al . Regulation of Inflammatory Chemokine Receptors on Blood T Cells Associated to the Circulating Versus Liver Chemokines in Dengue Fever. PLos One (2012) 7(7):e38527. doi: 10.1371/journal.pone.0038527
16
Badolato-Corrêa J Sánchez-Arcila JC Alves de Souza TM Santos Barbosa L Conrado Guerra Nunes P da Rocha Queiroz Lima M et al . Human T Cell Responses to Dengue and Zika Virus Infection Compared to Dengue/Zika Coinfection. Immun Inflammation Dis (2018) 6(2):194–206. doi: 10.1002/iid3.203
17
Costa VV Fagundes CT Souza DG Teixeira MM . Inflammatory and Innate Immune Responses in Dengue Infection: Protection Versus Disease Induction. Am J Pathol (2013) 182(6):1950–61. doi: 10.1016/j.ajpath.2013.02.027
18
Martina BE Koraka P Osterhaus AD . Dengue Virus Pathogenesis: An Integrated View. Clin Microbiol Rev (2009) 22(4):564–81. doi: 10.1128/CMR.00035-09
19
Gröger M Pasteiner W Ignatyev G Matt U Knapp S Atrasheuskaya A et al . Peptide Bbeta (15-42) Preserves Endothelial Barrier Function in Shock. PLos One (2009) 4(4):e5391. doi: 10.1371/journal.pone.0005391
20
Luplertlop N Missé D Bray D Deleuze V Gonzalez JP Leardkamolkarn V et al . Dengue-Virus-Infected Dendritic Cells Trigger Vascular Leakage Through Metalloproteinase Overproduction. EMBO Rep (2006) 7(11):1176–81. doi: 10.1038/sj.embor.7400814
21
Kuno G Gómez I Gubler DJ . Detecting Artificial Anti-Dengue IgM Immune Complexes Using an Enzyme-Linked Immunosorbent Assay. Am J Trop Med Hyg (1987) 36(1):153–9. doi: 10.4269/ajtmh.1987.36.153
22
Chao DY Davis BS Chang GJ . Development of Multiplex Real-Time Reverse Transcriptase PCR Assays for Detecting Eight Medically Important Flaviviruses in Mosquitoes. J Clin Microbiol (2007) 45(2):584–9. doi: 10.1128/JCM.00842-06
23
Lanciotti RS Calisher CH Gubler DJ Chang GJ Vorndam AV . Rapid Detection and Typing of Dengue Viruses From Clinical Samples by Using Reverse Transcriptase-Polymerase Chain Reaction. J Clin Microbiol (1992) 30(3):545–51. doi: 10.1128/jcm.30.3.545-551.1992
24
Igarashi A . Isolation of a Singh’s Aedes Albopictus Cell Clone Sensitive to Dengue and Chikungunya Viruses. J Gen Virol (1978) 40(3):531–44. doi: 10.1099/0022-1317-40-3-531
25
Gubler DJ Kuno G Sather GE Velez M Oliver A . Mosquito Cell Cultures and Specific Monoclonal Antibodies in Surveillance for Dengue Viruses. Am J Trop Med Hyg (1984) 33(1):158–65. doi: 10.4269/ajtmh.1984.33.158
26
Chaturvedi UC Elbishbishi EA Agarwal R Mustafa AS . Cytotoxic Factor-Autoantibodies: Possible Role in the Pathogenesis of Dengue Haemorrhagic Fever. FEMS Immunol Med Microbiol (2001) 30(3):181–6. doi: 10.1111/j.1574-695X.2001.tb01568.x
27
Malavige GN Ogg GS . Pathogenesis of Vascular Leak in Dengue Virus Infection. Immunology (2017) 151(3):261–9. doi: 10.1111/imm.12748
28
Srikiatkhachorn A Mathew A Rothman AL . Immune-Mediated Cytokine Storm and Its Role in Severe Dengue. Semin Immunopathol (2017) 39(5):563–74. doi: 10.1007/s00281-017-0625-1
29
Chen JP Lu HL Lai SL Campanella GS Sung JM Lu MY et al . Dengue Virus Induces Expression of CXC Chemokine Ligand 10/IFN-Gamma-Inducible Protein 10, Which Competitively Inhibits Viral Binding to Cell Surface Heparan Sulfate. J Immunol (2006) 177(5):3185–92. doi: 10.4049/jimmunol.177.5.3185
30
Shen BQ Lee DY Cortopassi KM Damico LA Zioncheck TF . Vascular Endothelial Growth Factor KDR Receptor Signaling Potentiates Tumor Necrosis Factor-Induced Tissue Factor Expression in Endothelial Cells. J Biol Chem (2001) 276(7):5281–6. doi: 10.1074/jbc.M007969200
31
Akira S Taga T Kishimoto T . Interleukin-6 in Biology and Medicine. Adv Immunol (1993) 54:1–78. doi: 10.1016/s0065-2776(08)60532-5
32
Lee YR Su CY Chow NH Lai WW Lei HY Chang CL et al . Dengue Viruses can Infect Human Primary Lung Epithelia as Well as Lung Carcinoma Cells and Can Also Induce the Secretion of IL-6 and RANTES. Virus Res (2007) 26(1-2):216–25. doi: 10.1016/j.virusres.2007.03.003
33
Marianneau P Steffan AM Royer C Drouet MT Jaeck D Kirn A et al . Infection of Primary Cultures of Human Kupffer Cells by Dengue Virus: No Viral Progeny Synthesis, But Cytokine Production is Evident. J Virol (1999) 73(6):5201–6. doi: 10.1128/JVI.73.6.5201-5206.1999
34
Charnsilpa W Takhampunya R Endy TP Mammen MP Jr Libraty DH Ubol S . Nitric Oxide Radical Suppresses Replication of Wild-Type Dengue 2 Viruses In Vitro. J Med Virol (2005) 77(1):89–95. doi: 10.1002/jmv.20418
35
Gunther VJ Putnak R Eckels KH Mammen MP Scherer JM Lyons A et al . A Human Challenge Model for Dengue Infection Reveals a Possible Protective Role for Sustained Interferon Gamma Levels During the Acute Phase of Illness. Vaccine (2011) 29(22):3895–904. doi: 10.1016/j.vaccine.2011.03.038
36
Fagundes CT Costa VV Cisalpino D Amaral FA Souza PR Souza RS et al . IFN-γ Production Depends on IL-12 and IL-18 Combined Action and Mediates Host Resistance to Dengue Virus Infection in a Nitric Oxide-Dependent Manner. PloS Negl Trop Dis (2011) 5(12):e1449. doi: 10.1371/journal.pntd.0001449
37
Costa VV Fagundes CT Valadão DF Cisalpino D Dias AC Silveira KD et al . A Model of DENV-3 Infection That Recapitulates Severe Disease and Highlights the Importance of IFN-γ in Host Resistance to Infection. PloS Negl Trop Dis (2012) 6(5):e1663. doi: 10.1371/journal.pntd.0001663
38
Barros TAC . Avaliação do Perfil De Mediadores Séricos E Proteínas Intraplaquetarias Em Relação a Plaquetopenia Em Pacientes Infectados Pelo Vírus Dengue [Tese]. Rio de Janeiro: Instituto Osvaldo Cruz (2015).
39
Rothman AL . Immunity to Dengue Virus: A Tale of Original Antigenic Sin and Tropical Cytokine Storms. Nat Rev Immunol (2011) 11(8):532–43. doi: 10.1038/nri3014
40
Naveca FG Pontes GS Chang AY Silva GAVD Nascimento VAD Monteiro DCDS et al . Analysis of the Immunological Biomarker Profile During Acute Zika Virus Infection Reveals the Overexpression of CXCL10, a Chemokine Linked to Neuronal Damage. Mem Inst Oswaldo Cruz (2018) 113(6):e170542. doi: 10.1590/0074-02760170542
41
Jacob-Nascimento LC Carvalho CX Silva MMO Kikuti M Anjos RO Fradico JRB et al . Acute-Phase Levels of CXCL8 as Risk Factor for Chronic Arthralgia Following Chikungunya Virus Infection. Front Immunol (2021) 12:744183. doi: 10.3389/fimmu.2021.744183
42
Pacsa AS Agarwal R Elbishbishi EA Chaturvedi UC Nagar R Mustafa AS . Role of Interleukin-12 in Patients With Dengue Hemorrhagic Fever. FEMS Immunol Med Microbiol (2000) 28(2):151–5. doi: 10.1111/j.1574-695X.2000.tb01470
43
Fares RC Souza KP Añez G Rios M . Epidemiological Scenario of Dengue in Brazil. BioMed Res Int (2015) 2015:321873. doi: 10.1155/2015/321873
Summary
Keywords
dengue infection, dengue virus, immune response, DENV serotypes, chemokines, cytokines
Citation
Coutinho-da-Silva MS, Sucupira PHF, Bicalho KA, Campi-Azevedo AC, Brito-de-Sousa JP, Peruhype-Magalhães V, Rios M, Teixeira-Carvalho A, Coelho-dos-Reis JGA, Antonelli LRdV, Rezende VBd, Melo FLRd, Garcia CC, Silva-Andrade JC, Costa-Rocha IAd, Bastos MdS, Rocha LAd, Silva VA, Ferreira EdS, Marinho EPM, Costa AG, Gomes MdS, Amaral LR, Furtado ECdS, Silva EVPd, Ramos BA, Santos ÉBd, Freitas MNO, Vasconcelos PFdC, Martins-Filho OA, Araújo MSS, Ferreira MS and Martins LC (2022) Serum Soluble Mediator Profiles and Networks During Acute Infection With Distinct DENV Serotypes. Front. Immunol. 13:892990. doi: 10.3389/fimmu.2022.892990
Received
09 March 2022
Accepted
22 April 2022
Published
31 May 2022
Volume
13 - 2022
Edited by
James Edward Pease, Imperial College London, United Kingdom
Reviewed by
Joel V. Chua, University of Maryland, United States; Henry Puerta-Guardo, Universidad Autónoma de Yucatán, Mexico
Updates
Copyright
© 2022 Coutinho-da-Silva, Sucupira, Bicalho, Campi-Azevedo, Brito-de-Sousa, Peruhype-Magalhães, Rios, Teixeira-Carvalho, Coelho-dos-Reis, Antonelli, Rezende, Melo, Garcia, Silva-Andrade, Costa-Rocha, Bastos, Rocha, Silva, Ferreira, Marinho, Costa, Gomes, Amaral, Furtado, Silva, Ramos, Santos, Freitas, Vasconcelos, Martins-Filho, Araújo, Ferreira and Martins.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olindo Assis Martins-Filho, olindo.filho@fiocruz.br; Márcio Sobreira Silva Araújo, marcio.sobreira@fiocruz.br
†These authors have contributed equally to this work and share first authorship
‡These authors share senior authorship
This article was submitted to Cytokines and Soluble Mediators in Immunity, a section of the journal Frontiers in Immunology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.