- 1Ninghai Institute of Mariculture Breeding and Seed Industry, Zhejiang Wanli University, Ningbo, China
- 2Zhejiang Key Laboratory of Aquatic Germplasm Resource, Zhejiang Wanli University, Ningbo, China
- 3Laboratory for Marine Fisheries Science and Food Production Processes, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China
Family I84 protease inhibitors represent a novel family in the MEROPS peptidase database and are likely unique for molluscan host defense. Two Family I84 members, scSI-1 and scSI-2, were reported from the razor clam Sinonovacula constricta in a previous research. In the present study, 12 additional genes, named scSI-3 to scSI-14, were identified via genome wide sequence analyses. Among them, 10 genes were predicted to have a signal sequence, but one (scSI-7) was not. Besides, one sequence (scSI-14) was likely to encode a prematurely terminated peptide. The predicted mature peptides shared characteristics including 12 conserved cysteine residues, isoelectric points of 4.98 to 6.11, and molecular weights of 7.1 to 9.3 kDa with previously reported family members. Four motifs were characterized in 13 predicted mature peptides (with exception of scSI-14), which shared two to four conserved cysteine residues, are possibly to form two functional domain comprised 6 cysteine residues, respectively. At genomic level, all the 14 razor clam Family I84 genes were organized into 3 exons and 2 introns; 13 of them clustered in 3 regions of 100 kb on 3 separate chromosomes, suggesting tandem duplications of related genes. The promoter region of all the 14 genes was predicted to share some transcription factor binding sites, in particular those responsive to pathological and physiological stimuli, but no shared motifs were identified. Analyses also revealed differences in expression patterns among the genes. One gene in a tandem duplicated gene pairs usually showed a higher expression level than the other whereas non-tandem duplicated genes exhibited a higher degree of correlation in expression level. In addition, 8 of the 14 genes demonstrated higher level of expression in Vibrio tolerant clams than in non-tolerant clams following challenges with Vibrio parahaemolyticus. These results generated important information about the evolution of Family I84 protease inhibitors in S. constricta.
Introduction
Protease inhibitors are a special group of molecules ubiquitously present in all organisms and one of their functions is believed to play a role in host immunity (1, 2). The involvement of protease inhibitors in molluscan host defense are indicated by the findings of homologous genes of several known protease inhibitors, such as the tissue inhibitor of metalloprotease and Kazal-type serine protease inhibitor in the Pacific oyster Crassostrea gigas (3), the bay scallop Argopecten irradians (4), and the Manila clam Ruditapes philippinarum (5). More direct evidence for protease inhibitors’ functioning in the immunity of mollusk species, however, comes from a series of research on the Family I84 protease inhibitors in the MEROPS peptidase and peptidase inhibitor classification system (http://www.ebi.ac.uk/merops).
The Family I84 in MEROPS was first created to host cvSI-1 and cvSI-2 from the eastern oyster Crassostrea virginica, the first protease inhibitors to be purified and characterized in a molluscan species (6–8). More family members have been identified later at the gene sequence level in several other mollusks including oysters (C. gigas), mussels (Mytilus galloprovincialis), clams (Sinonovacula constricta and Meretrix meretrix), and abalones (Haliotis discus hannai) (9–11). A more recent research indicates that Family I84 is likely a multigenic family present exclusively in the Phylum Mollusca and the family has expanded significantly in certain molluscan lineages (11, 12).
Family I84 genes encode peptides that have a signal peptide at the N-terminus. Mature molecules have a molecular weight of about 8 kDa and consist of around 70 amino acid residues including 12 conserved cysteines that are speculated to form 6 intramolecular disulfide bonds (7, 10, 11, 13). They selectively inhibit the enzyme activity of serine type of proteases in particular that in the subtilisin family (6, 7). Several features of Family I84 protease inhibitors indicate their role in mollusk immunity. Purified cvSI-1, for example, inactivates the virulence protease of Perkinsus marinus, the protozoan parasite of eastern oysters and inhibits the parasite’s propagation in vitro (6, 7). At the same time, levels of plasma subtilisin inhibitory activity and related gene expression are significantly higher in oyster populations selected for increased resistance or oyster species inheritably resistant to P. marinus than in oysters highly sensitive to the parasite (13). In addition, the expression of Family I84 genes in oysters, clams, and mussels are detected to be up-regulated by injection of bacterial and protozoan pathogens and pathogen associated molecular patterns (PAMPs) (9, 11, 14, 15).
The razor clam S. constricta is a representative species of bivalve mollusks inhabiting on the muddy intertidal zone. Two Family I84 genes, scSI-1 and scSI-2, are identified from the clam through cDNA cloning and sequencing (9). The encoded peptides are predicted to share critical molecular characteristics of the Family I84 protease inhibitors. The clam plasma is also detected to contain the signature protease inhibitory activity of the Family I84 protease inhibitors. Besides, the expressions of the 2 genes are upregulated the clams following challenges by Vibrio harveyi and extreme physiochemical factors, suggesting their function in host defense. In addition to scSI-1 and scSI-2, searches into transcriptomic sequence databased have also revealed possibilities of the presence of more Family I84 members in the genome of S. constricta. However, the entire repertoire of the family in the razor clam remained unknown.
The availability of the genome sequencing data for S. constricta has made it possible to study a gene family via genome wide identification (16). This kind of genomic studies in combination with gene expression profiling at the transcriptomic level can generate important information about functions and evolution of the gene family of interest (17, 18). Thus, we aimed in the present research at: (1) determining total number, structure and chromosome distribution of the Family I84 genes in the S. constricta genome; (2) analyzing the phylogenetic relationship between the identified S. constricta genes with known Family I84 members; (3) decoding the mechanisms of molecular evolution of Family I84 genes in S. constricta; and (4) profiling expressions of Family I84 genes in S. constricta at different developmental stages and following bacteria challenges. Results of the research will provide important information about the molecular and functional evolution of Family I84 protease inhibitors in mollusks.
Materials and Methods
Family I84 Gene Search and Analysis
BLAST search into the S. constricta genome assembly at the GenBank (accession number GCA_009762815.1) was performed in May 2021 using the tBLASTn program with the 12 Family I84 members from 5 mollusk species (cvSI-1, cvSI-2 and cvSI-3 from C. virginica; scSI-1 and scSI-2 from S. constricta; mgSI-1, mgSI-2 and mgSI-3 from M. galloprovincialis; mmSI-1 and mmSI-2 from M. meretrix; hdhSI-1 and hdhSI-2 from Haliotis discus hannai) as queries (9, 11, 12). Sequence hits with E<0.01 were downloaded and checked for the presence of a complete open reading frame (from the initial ATG to the stop codon). Partial sequences were complemented to their full length by comparison with transcriptome assembly (16).
The retrieved sequences were verified by PCR amplification of the predicted open reading frame (ORF). The PCR amplifications were done using cDNA synthesized from the digestive gland RNAs as templates and primers listed in Supplementary Table 1. For sequences that the PCR using cDNA failed to generate an expected amplicon, the PCR verification was carried out using the genomic DNA from the gill tissues as the templates. Tissue samples for RNA and genomic DNA preparations were collected from healthy adult clams maintained in seawater at 20°C and salinity at 20 psu. Total RNAs and genomic DNA were extracted from related tissues respectively using an RNA isolation kit (Omega, USA) and a DNA extraction kit for marine animals (Tiangen, Beijing, China) according to the manufacturer’s instructions. DNA and RNA concentrations were determined using a micro-spectrophotometer (Nanodrop, Allsheng) and the integrity was checked by electrophoresis in 1.2% agarose gel. The cDNA templates were synthesized using the Prime Script™ RT kit (Takara, Japan).
ORFs were predicted with the ORF Finder at NCBI (https://www.ncbi.nlm.nih.gov/orffinder/). Multiple sequence alignments and sequence percent identity calculations were done using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Aligned sequences were visualized using Jalview v2.11.1.0 (19). Signal sequences were predicted using SignalP 5.0 (www.cbs.dtu.dk/services/SignalP/) and SecretomeP 2.0. Molecular weight (MW) and theoretical isoelectric point (pI) were calculated using Protparam at ExPASy (https://web.expasy.org/protparam/).
Phylogenetic Tree Construction
To determine the evolutionary origin of razor clam Family I84 genes, mature peptides sequence of all Family I84 members in razor clam and 15 other identified members in oysters, mussel, clams and abalone were used to construct a phylogenetic tree. Multiple sequence alignment were conducted with Muscle method (20); the best evolutionary model were predicted and model WAG + Gamma + Invariant showed the best parameters and then maximum likelihood (ML) phylogenetic tree was constructed using MEGA6 with a bootstrap of 1,000 replicates (21).
Motif, Gene Structure, Chromosomal Distribution and Duplication Analysis
The de novo motif identification was carried out in mature peptides using the MEME suite (https://meme-suite.org/meme/tools/meme) with the default parameter. Gene structure including length and number of exons and introns were determined by BLAST the full length of the candidate genes on the genome assembly. General feature format (gff) files containing these genes’ annotation information were then generated. Gene structure and motif patterns were visualized via Toolbox for Biologists (TBtools) v1.05 (22). Chromosomal distributions of the Family I84 genes were visualized using the TBtools.
Gene duplication was determined based on two criteria: >70% coverage of the longer sequence and >70% similarity in a pairwise alignment of 2 sequences without signal peptides (23, 24). Duplicated genes separated by five or fewer genes in a 100-kb chromosomal fragment were considered tandem duplicated genes (24).
Positive Selection and Site Conservation Detection
Simple Ka/Ks Calculator in TBtools were used to calculate the number of substitutions per synonymous site (KS) and nonsynonymous site (KA) for duplicated gene pairs (22). CodeML method in EasyCodeml v1.4 was used to test whether the single sites were under positive selection (25). Briefly, nucleotide sequences of Family I84 genes in razor clam were aligned with codon-based method in MEGA, seven codon substitution models encompassing M0 (one-ratio), M1a (nearly neutral), M2a (positive selection), M3 (discrete), M7 (beta), M8 (beta and ω >1) and M8a (beta and ω = 1) were tested. Four likelihood ratios (M0 vs M3, M1a vs M2a, M7 vs M8 and M8a vs M8) were performed and compared using a likelihood ratio test (LRT).
The codon usage frequencies of TGT or TGC for cysteine were calculated on the CDS of all protein coding genes on the genome (GenBank assembly accession: GCA_009762815.1) and the Family I84 genes in S. constricta using the “cusp” tool in the mEMBOSS package (26). The codon usage bias for the twelve conserved cysteine were carried out by Chi-square test assuming a binomial distribution.
Promoter Sequence Analysis
The 2kb sequence upstream of the translation initiation site was extracted as a candidate promoter region according to the Family I84 gene structure and gff file. Multiple sequence alignment for promoter sequences were conducted with Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/) and identities among sequences were calculated and compared. The sequence were then analyzed using simple MEME Wrapper in TBtools v1.05 with default parameters for motif identifications (22). Potential transcription factors and their binding sites were predicted using Alibaba 2.1 program in the GeneXplain online tool with the pairSIM being set at 64 and the min matrix conservation at 80% (http://gene-regulation.com/pub/programs/alibaba2/index.html).
Transcriptome Assembly and Gene Expression Profiling
Transcriptomic sequence data of 66 samples covering S. constricta at developmental stages, under variable challenges and populations differed in resistance to V. parahaemolyticus were retrieved from the NCBI database (Supplementary Table 2). The developmental stages included egg, four cells, blastulae, gastrulae, trochophore, D-shaped larvae, umbo larvae and juvenile. Environmental stressors for the clam challenges included ammonia nitrogen and temperature. Sequence data for clam populations different in V. parahaemolyticus resistance were generated (27). The retrieved RNA-seq reads were mapped to the S. constricta genome using Hisat (V2.1.0) (28), and assembled using StringTie (29). Gene expressions were normalized and reported in Transcripts Per Kilobase of exon model per Million mapped reads (TPM) (30). The correlation in expression levels between Family I84 genes was calculated in the transcriptomic sequence data of V. parahaemolyticus resistant and sensitive clams, 15 individuals each, after the bacterial injection using Pearson analysis with the corrplot package in RStudio software.
Results
Family I84 Gene Repertoire in the Clam Genome
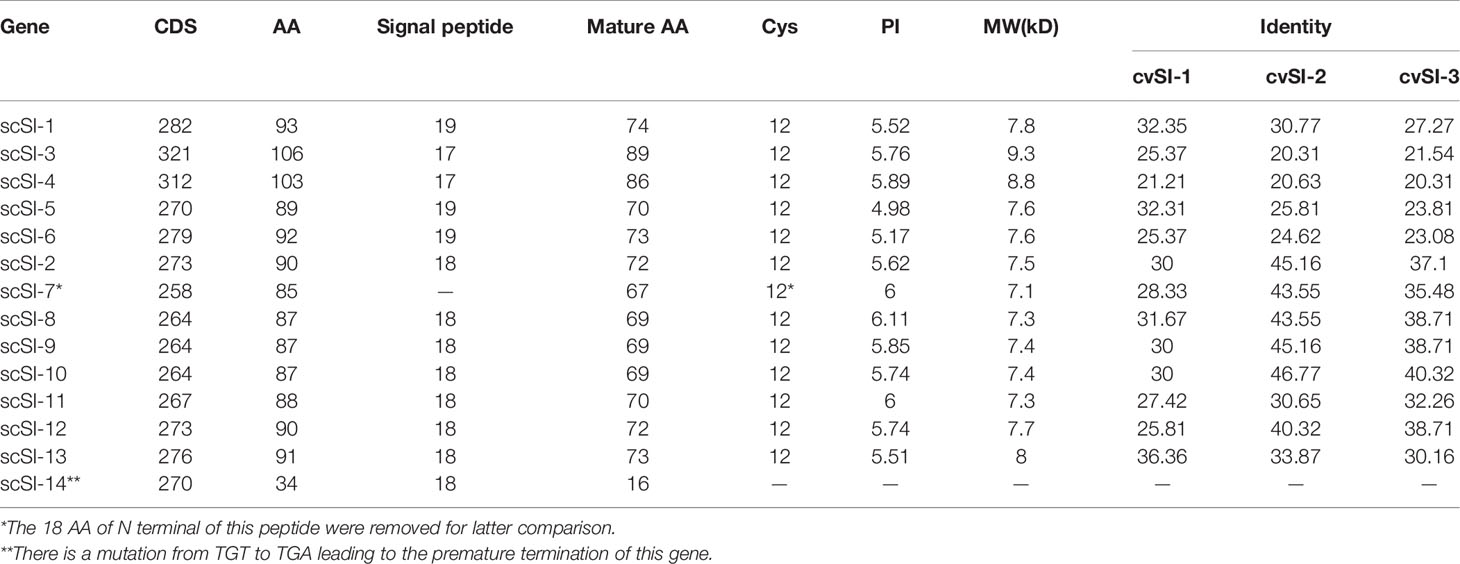
Twelve new Family I84 protease inhibitor genes in addition to scSI-1 and scSI-2 were identified in the S. constricta genome and named scSI-3 to scSI-14, making the razor clam Family I84 gene repertoire being composed of 14 genes. The 12 newly identified genes encompassed an ORF ranging from 258 to 321 bp that encoded peptides from 85 to 106 amino acids. ScSI-14 encoded a much shorter peptide (34 amino acids) than the other members due to a mutation from TGT to TGA leading to premature termination (Table 1; Supplementary Figure 1). All genes but scSI-7 were predicted to have a signal peptide of 17 to 19 amino acids and a mature peptide length ranging from 69 to 89 amino acids. The 12 conserved cysteine were identified in all mature peptides, but the array was highly variable as CX5-8CX4-5CX7-9CX4CTCX6-23CX5-7CX3-6CX5-8CX4CXC (Figure 1). Mature peptides had a predicted PI of 4.98~6.11 and molecular weight of 7.1~ 9.3 kDa. The highest and lowest identities of these members with Family I84 type molecule (cvSI2) were 46.77% and 20.31%, respectively (Table 1), and the sequence identity among the 14 razor clam members ranged from 24.14% to 87.21% (Supplementary Table 3). All 14 genes excluding scSI-7 and scSI-14 of the razor clam Family I84 repertoire were amplified in PCR with cDNA templates. A 1658 bp and a 2596 bp fragments encompassing the predicted ORF of scSI-7 and scSI-14 were amplified in PCR reactions with genomic DNA templates.
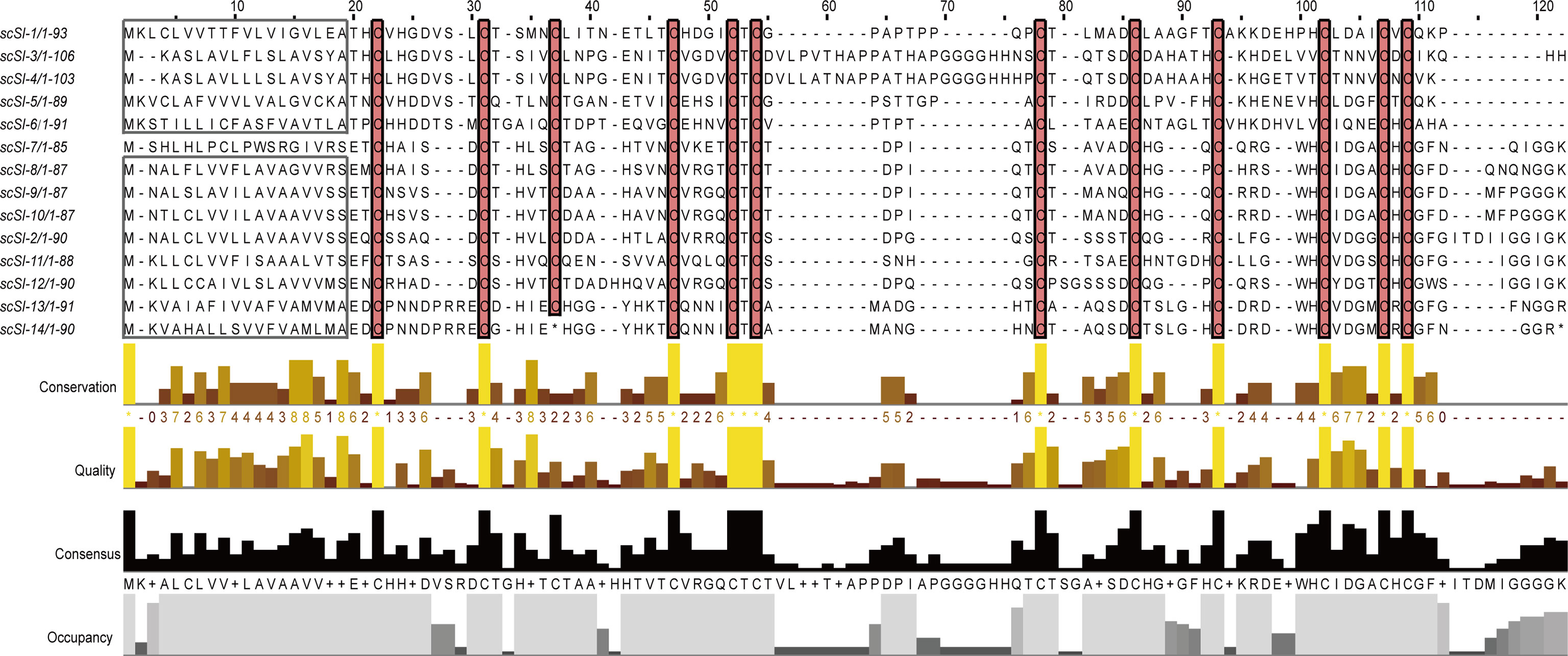
Figure 1 Peptide sequence alignments of the fourteen Family I84 genes from razor clam, two formally characterized members scSI-1 and scSI-2 were included. Predicted signal peptide are framed, conserved cysteine residues are highlighted. Conservation and consensus were shown of the residues. * means the stop codon of peptide.
Phylogenetic Tree
An ML phylogenetic tree containing 28 members’ mature peptide of Family I84 protein sequences was constructed (Figure 2). These sequences were grouped into two major nodes including 23 and 5 members, respectively; however, the bootstrap values poorly supported the two nodes all the time. Sequences from the same or closely related species tended to cluster together and the first node included one cluster which comprised scSI-1, scSI-3, scSI-4, scSI-5 and scSI-6. The other node contained four clusters and they included 7 genes (scSI-2, scSI-7, scSI-8, scSI-9, scSI-10, scSI-11 and scSI-12), 3 genes (cvSI-3, cgSI-5 and cgSI-19), 2 genes (mgSI-2 and mgSI-3), and 2 genes (cvSI-1 and cgSI-6), respectively.
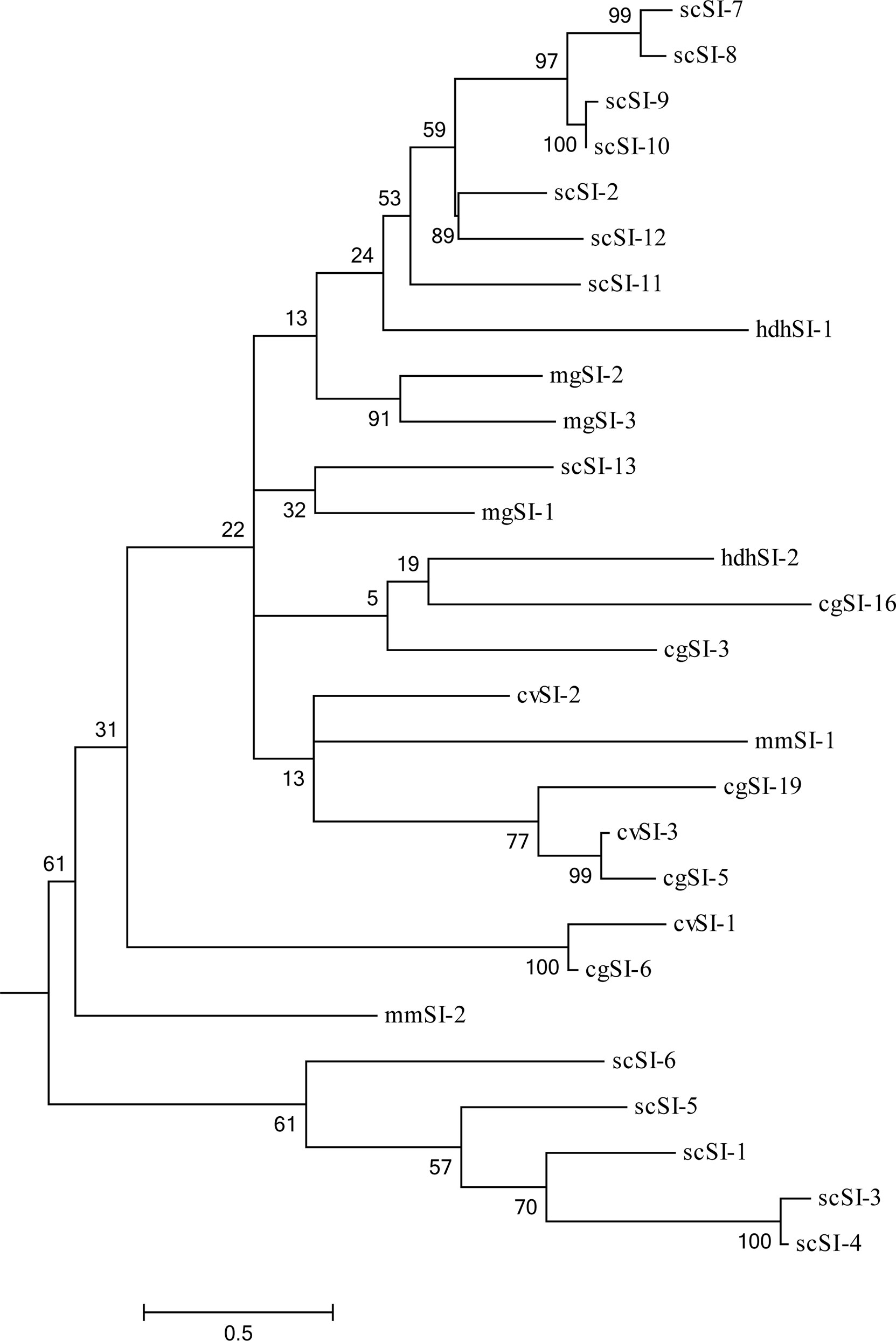
Figure 2 Maximum Likelihood (ML) phylogenetic tree containing 28 members of Family I84 protein sequences without signal peptide. The accession numbers were AAZ41364.1, BAH20735.1 and AQM52281.1 for cvSI-1, cvSI-2 and cvSI-3 in Crassostrea virginica; UBF40522.1, UBF40523.1, UBF40524.1 for mgSI-1, mgSI-2, and mgSI-3 in mussel Mytilus galloprovincialis; ATY73923.1, ATY73924.1 for scSI-1, scSI-2 in clam Sinonovacula constricta; UBF40520.1, UBF40521.1 for mmSI-1, mmSI-2 in clam Meretrix meretrix; QPM65700.1, QPM65701.1 for hdhSI-1, hdhSI-2 of abalone, Haliotis discus hannai used in this phylogeny analysis, the other 11 sequences from clam S.constricta were acquired in the present study. Numbers beside the internal branches indicate bootstrap values based on 1000 replications, the 0.5 scale indicates the genetic distance.
Motifs and Gene Structure
A total of 4 motifs were characterized in 13 mature peptides (Figure 3A). Among them, motif 1 and motif 2 each covered four conserved cysteine residues displaying in arrays of C-X5-C-X4-C-H-C and C-X7-C-X4-C-T-C respectively. Motif 3 and motif 4, however, covered different numbers of conserved cysteines with the former containing 2 and the latter 3 in an array of C-X5-C and C-X4-C-D-C, respectively. Most sequences comprised three motifs, of which seven genes (scSI-2, scSI-7, scSI-8, scSI-9, scSI-10, scSI-11, scSI-12 and scSI-13) contained motif 1, 2 and 3; four genes (scSI-1, scSI-3, scSI-4 and scSI-5) contained motif 2, 3 and 4, but scSI-6 contained only one (motif 2), which were shared by all razor clam Family I84 members.
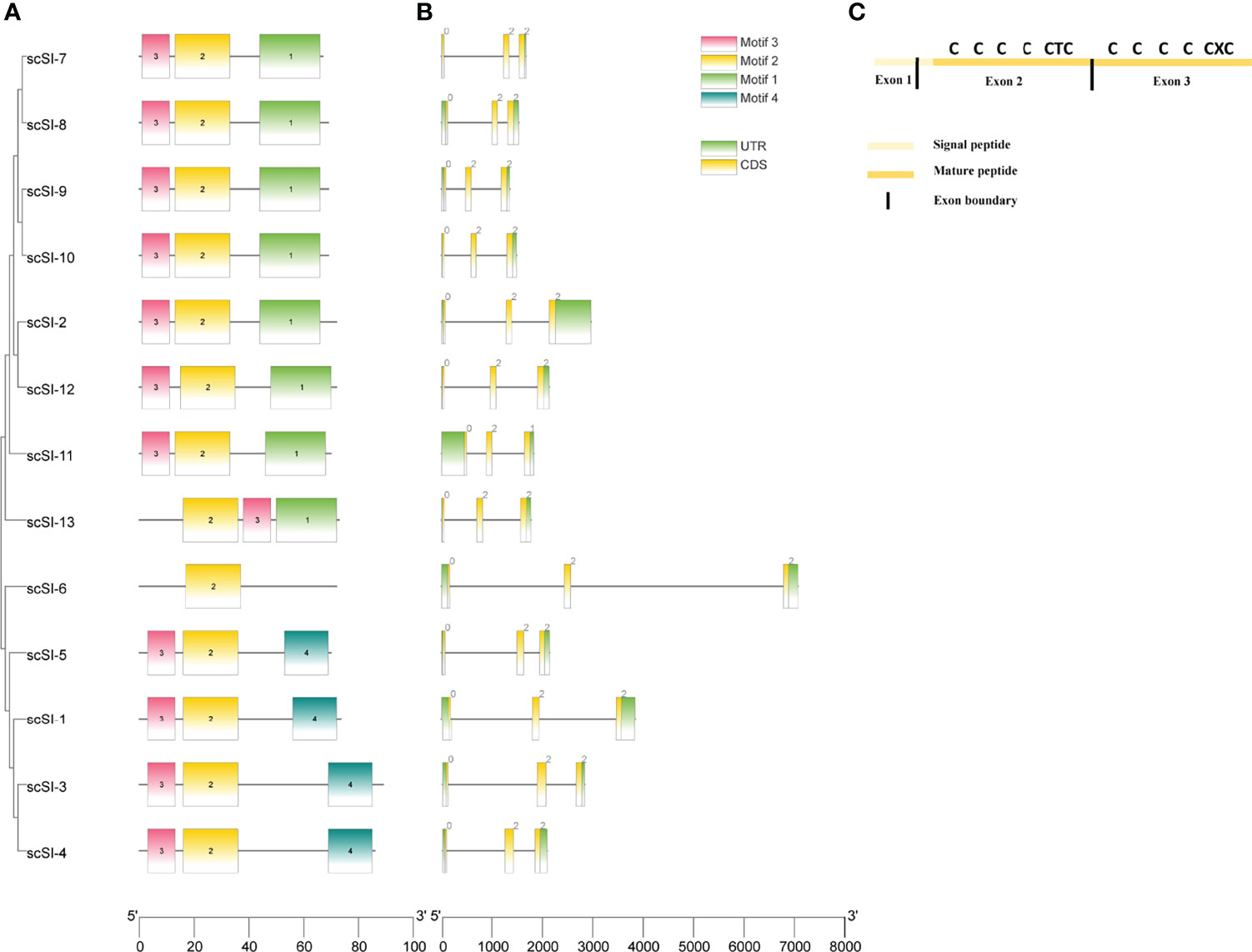
Figure 3 Mature polypeptide motifs and gene structure for all Family I84 genes excluding the scSI-14. (A) Polypeptide motifs of these gene members, different color blocks are motifs and the 1,2,3,4 in the block represent the number of motif. Motif 1: QCQRGWHCIDGACHCGFDINGGG, Motif 2: CDAGHTVTCVRGQCTCTDPIQ, Motif 3: TCGDVSDCTHV, Motif 4: KHDEEVHCLDN VCDCQK; (B) Gene structure of all the I84 family genes and 0, 1, 2 in the block are the phase for translation; (C) Cysteine pattern and their distribution on exons for all of the members.
All genes consisted of three exons and two introns, phase of exons for gene translation were 0, 2 and 2, with exception for scSI-11 (0, 2 and 1); their gene length spanned from 1485 bp (scSI-10) to 7077 bp (scSI-6) on the genome (Figure 3B). The 3 exons coded for different parts of the whole peptide, with first exon for part of the signal peptide, the second exon for the remaining part of signal peptide and the N-terminal segment coving the first to sixth conserved cysteine, and the third exon for the C-terminal segment covering the 6 remaining conserved cysteine residues (Figure 3C).
Gene Chromosomal Location and Tandem Duplication
The 14 Family I84 genes of S. constricta were mapped on three chromosomes- chromosome 6, chromosome 12 and chromosome 16 (Figure 4). For the 8 genes in chromosome 16, 7 (scSI-2, scSI-7, scSI-8, scSI-9, scSI-10, scSI-11 and scSI-12) were localized in a 52 kb genome region, and the other (scSI-6) was about 5M bp away from the 7 gene cluster. On the other hand, four genes (scSI-1 and scSI-3, scSI-4, and scSI-5) clustered in a 23 kb genome region on chromosome 12, and 2 genes (scSI-13 and scSI-14) in a 95 kb region on chromosome 6.
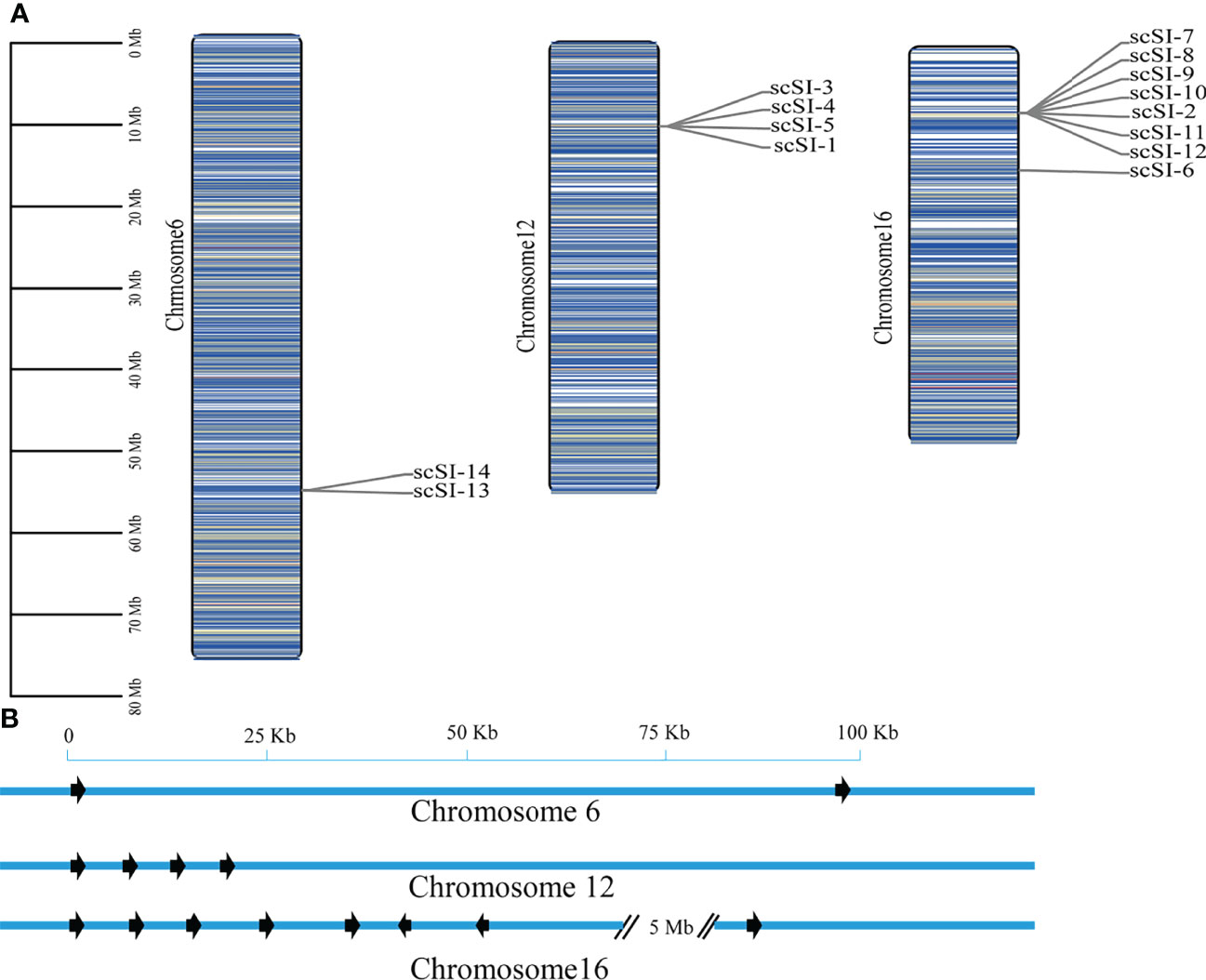
Figure 4 Chromosome distribution of all the Family I84 genes in razor clam, (A) Global view of these genes on three chromosomes, the darker of the bar, the higher density for genes around it; (B) Regional appearance of gene clusters on chromosomes, the arrow are transcriptional direction of genes.
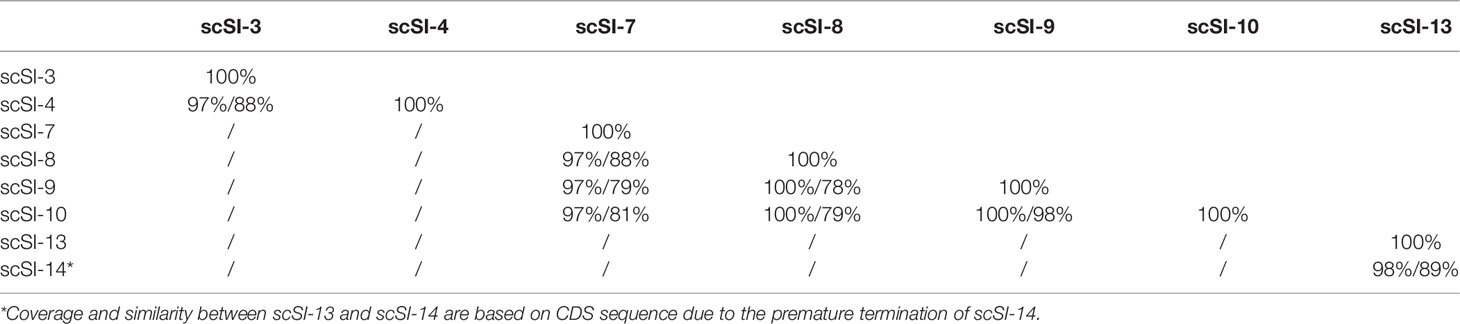
Gene duplications were detected for the razor clam Family I84 genes and based on the defined criteria, 8 genes (57.1%) in three clusters were determined to be tandem duplicated (Table 2). Three tandem duplicated gene clusters included scSI-3 and scSI-4; scSI-7, scSI-8, scSI-9 and scSI-10; scSI-14 and scSI-13, respectively.
Positive Selection and Site Conservation
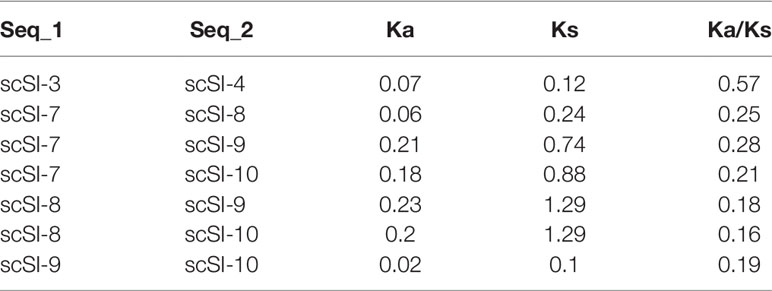
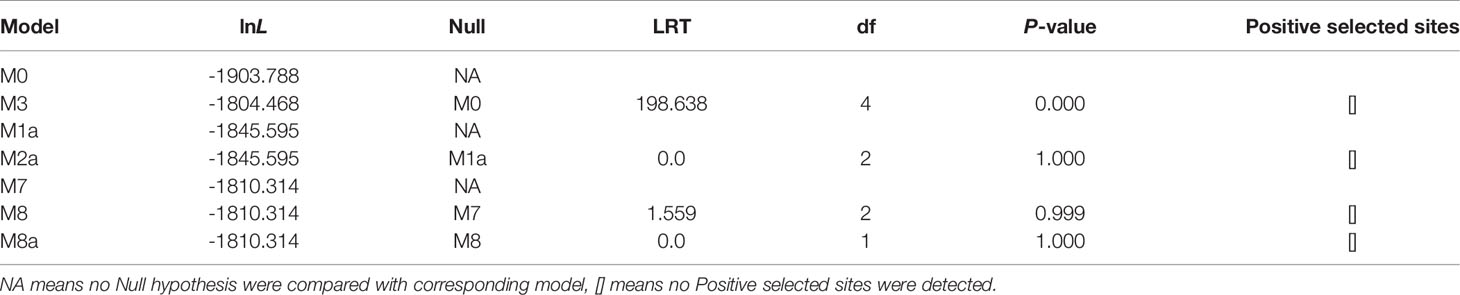
For all the duplicated gene pairs, Ka/Ks for the coding sequence (excluding signal peptide) ranged from 0.16 (scSI-8 with scSI-10) to 0.57 (scSI-3 with scSI-4) (Table 3). Site model analyses did not detect positive selection sites in the razor clam Family I84 genes, however, LRT P-value between M0 and M3 models was lower than 0.000 (Table 4).
The codon usage frequency of cysteine was 0.646 for TGT and 0.354 for TGC in all protein coding genes from the S. constricta genome. Five of the twelve conserved cysteines were significantly deviated from the average (Figure 5). Specially, for the fifth and sixth cysteine, codon preference was TGC, forming the most conserved region (C-T-C) of Family I84 genes.
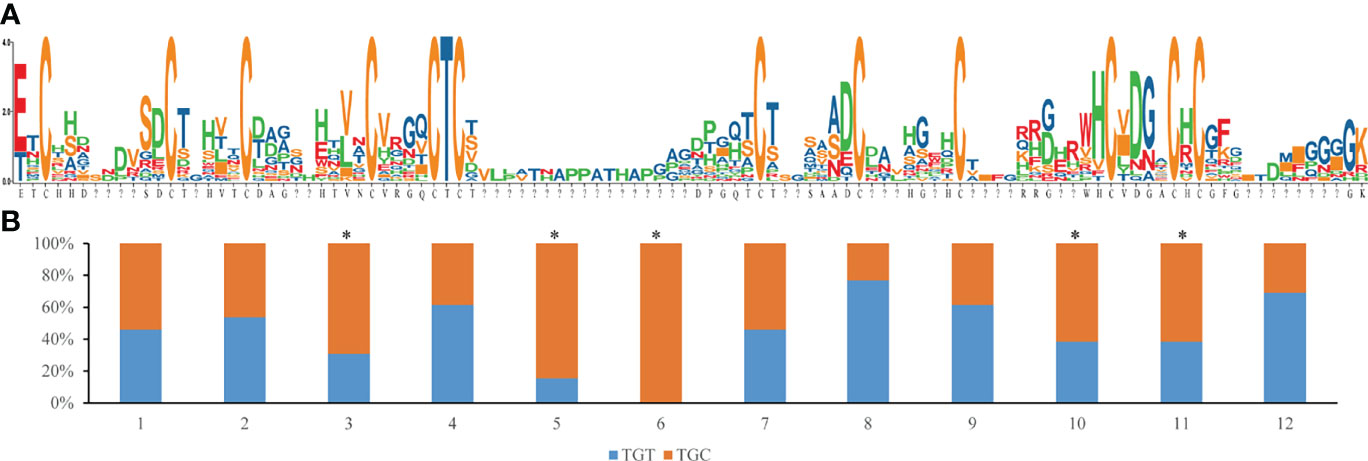
Figure 5 Mature peptide sequence variability and conservation of cysteine residues, (A) Sequence logo of 13 mature peptide, the larger of the letter, the higher proportion of the amino acid in the position,”?” means the position are not shared for all of the sequence; (B) Codon usage percentage of TGT or TGC for 12 conserved cysteine residues, the asterisk means that the ratio in that residue are deviated from the average of the whole genome codon usage frequency (0.646 for TGT and 0.354 for TGC).
Promoter Sequence Feature
The highest and lowest identities among 14 Family I84 members’ promoter sequences were 51.73% and 29.32%, respectively (Supplementary Table 4). No motifs were shared among all razor clam Family I84 members; however one motif was shared among scSI-3 with scSI-4 and another motif was shared by scSI-2, scSI-5, scSI-6, scSI-7, scSI-8, scSI-9 and scSI-10 (Supplementary Figure 2). In total, 14 Family I84 members had binding sites for 14~29 transcription factors, which may functioned in response to physiological and pathological stimuli (Supplementary Table 5). Briefly, 13 razor clam Family I84 genes (92.9%) had one to seven Oct (Octamer-binding protein) binding sites; 7 razor clam Family I84 genes (50%) had one or more IRF (Interferon regulatory factor) binding sites. C/EBP (CCAAT/enhancer binding protein) and HNF (Hepatocyte nuclear factor) were found in 14 and 13 family I84 genes, respectively. Two to five CREB (Cyclic AMP-responsive element-binding protein 1) binding sites existed in 5 Family I84 genes. Altogether, the transcription factors binding sites analysis indicated that razor clam Family I84 genes could involve in different immune and inflammatory responses.
Gene Expression Patterns
The Family I84 genes were not detected in the developmental stages before D-shaped larvae except for scSI-2, scSI-4, scSI-6, scSI-9 and scSI-13. However, all genes except for scSI-7 and scSI-14 were expressed in the umbo larvae and juveniles, and the expression levels increased gradually (Figure 6A).
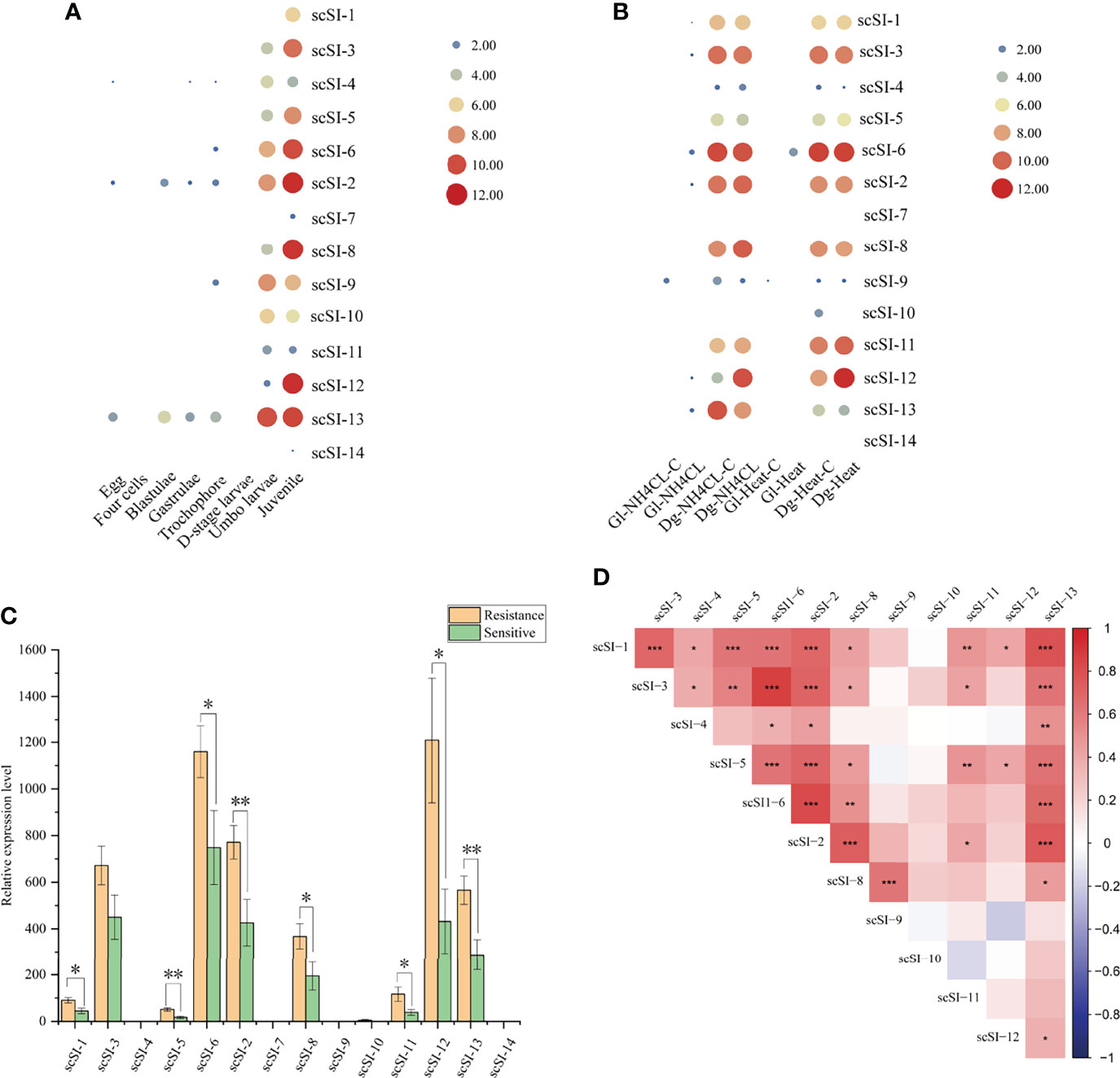
Figure 6 Gene expression patterns and co-expression of Family I84 genes. (A) Expression patterns of genes at different development stages of egg, four cells, blastulae, gastrulae, trochophore, D-stage larvae, umbo larvae and juvenile after log2 transformation; (B) Transcription level of Family I84 genes under the stress of ammonia nitrogen (180 mg/L with 0 mg/L as control) in gill (Gl-NH4CL, Gl-NH4CL-C) and digestive gland (Dg-NH4CL, Dg-NH4CL-C); high temperature (32°C with 22°C as control) in gill (Gl-heat, Gl-heat-C) and digestive gland (Dg-heat, Dg-heat-C) after log2 transformation; (C) Gene expression level for resistant and sensitive groups (n = 15 per group) of razor clam juveniles after V. parahaemolyticus infection, one and two asterisk represent the significant level lower than 0.05 and 0.01, respectively; (D) Co-expression analysis of I84 family genes in resistant and sensitive groups (n = 30 in total) of juveniles under V. parahaemolyticus infection, the darker of the block, the much stronger correlation are between them, *, ** and *** means the p valve for correlation coefficient lower than 0.05, 0.01 and 0.001, respectively.
Under the stress of ammonia nitrogen (180 mg/L with 0 mg/L as control) and high temperature (32°C with 22°C as control), Family I84 genes were hardly expressed in gills, but were much higher in the digestive gland under control or experiment group. ScSI-1, scSI-2, scSI-3, scSI-6 scSI-8, scSI-11 and scSI-12 showed higher expression levels than scSI-4, scSI-7, scSI-9, scSI-10 and scSI-14, but there was no significant difference between the stress group and the control group (Figure 6B).
For resistant and sensitive groups of razor clam juveniles after V. parahaemolyticus infection, expression levels of scSI-4, scSI-7, scSI-9, scSI-10 and scSI-14 were much lower than the other nine Family I84 members (Figure 6C). Significantly higher gene expression was observed in the resistant group than in the sensitive group for scSI-1, scSI-5, scSI-6, scSI-2, scSI-8, scSI-11, scSI-12 and scSI-13.
Correlation coefficient of gene expression of tandem duplicated Family I84 gene pairs (scSI-3 with scSI-4, scSI-8 with scSI-9, scSI-9 with scSI-10) were lower than 0.4 and not significantly different, while correlation coefficient between scSI-8 with scSI-9 were 0.63 (p<0.001). For non-tandem duplicated genes, a high correlation was observed between scSI-3 and scSI-6 (r=0.87, p<0.001), scSI-2 and scSI-6 (r=0.80, p<0.001), scSI-1 and scSI-13 (r=0.77, p<0.001), scSI-2 and scSI-13 (r=0.74, p<0.001) and scSI-2 with scSI-8 (r=0.71, p<0.001), respectively (Figure 6D).
Discussion
The Family I84 in the MEROPS peptidase database consists of novel and mollusk-specific serine protease inhibitors (6, 7, 13). Two Family I84 genes, scSI-1 and scSI-2, have been cloned and their expressions are upregulated in S. constricta following challenges with Vibrio harveyi (9). A better understanding of the Family I84 in the razor clam is now achieved after the identification of 12 additional genes in the present study. The analyses of the entire Family I84 gene repertoire at the genome and transcriptomic levels have generated important information about the function and evolution of Family I84 protease inhibitors.
Family I84 Protease Inhibitors Are Diverse Yet Conserved in Certain Characteristics
The identification of the S. constricta Family I84 gene repertoire reveals the extreme variability of the family members in overall amino acid sequence. The 14-razor clam Family I84 genes encode the peptides that share a sequence identity between each other varying from 24.14% to 87.21%, reflecting the molecular diversity of the Family I84 protease inhibitors within a same species. At the same time, the razor clam Family I84 members also show great degree of difference in amino acid sequences with their homologs in other species (e.g., 20.3%-46.8% identities with the family type member cvSI-2), indicating the molecular variability between the family orthologs in different species. Although molecular diversity seems to be a defining feature in protease inhibitors, the degree of diverseness of the Family I84 is still greater than most other protease inhibitors families including those from molluscan species (e.g., the kazal-type protease inhibitors from clam and scallop (4, 5, 31). In addition, the razor clam Family I84 appears to consist of 2 nonfunctionalized genes, scSI-7 that lacks signal sequence and scSI-14 that encodes a premature peptide, further increasing the family’s diversity. The extremity of Family I84 protease inhibitors in molecular diversity likely reflects the active evolution, which in turn suggests the functional significance, of the mollusk-specific protein family. It should be noted that the diverseness of Family I84 molecules as indicated by the range of sequence identity between intraspecific family members seems to be greater in S. constricta than in several other species such as C. virginica, C. gigas, and M. galloprovincialis (12). This difference in identity among intraspecific family sequences between the razor clam and other species may stem at the lack of recognition of the entire gene repertoire in the latter. Genome wide identification of the Family I84 genes in more species should provide valuable information.
While the Family I84 protease inhibitors are greatly diverse in the overall amino acid sequences, they are conserved in gene structure and cysteine residues. Similar to the cvSI-1 gene in eastern oyster, all the razor clam Family I84 genes consist of three exons and two type 1 introns (32). In addition, most mature peptides contained 12 conserved cysteines with a relatively conserved arrangement in the sequence. Intriguingly, the 12 cysteines are constantly encoded in the second and third exons with 6 in each, suggesting 2 functional modules being encoded by 2 separate exons. However, unlike the Kazal type protease inhibitors where 2-12 repeated modules with a similar 6-cysteines array form a “multiheaded” protease inhibitor (33), the 2 possible modules in Family I84 protease inhibitors differ from each other in the cysteine arrangement patterns and thus likely differ functionally as well (5, 31, 34). It is speculated that the 12 conserved cysteine residues in the Family I84 members form intramolecular disulfide bridges (Xue et al, 2009). Therefore, the conservation of cysteines in the family members appears to be necessary for the proteins to maintain a functional conformation.
Family I84 Genes Expanded via Tandem Duplication
Gene distribution in the genome provide important information about the evolution of Family I84 genes. It is notable that 13 of the 14 razor clam Family I84 genes clustered in 3 regions of 100 kb on 3 separate chromosomes, and 8 genes meet the criteria of tandem duplication. The phenomenon that multiple genes present closely in the genome is also reported in the Pacific oyster Family I84 genes (12). Therefore, Family I84 may have undergone expansion via tandem duplication after speciation of bivalves. Interestingly, tandem duplications have been observed in expansion of several host immunity related gene families including antimicrobial peptides and toll-like receptors (17, 35, 36). Unlike the cysteine-rich antimicrobial polypeptide-big defensins where the species associated monophyletic topology on a phylogenetic tree indicates an exclusive species-specific expansion via tandem duplication; however, we find in the present research that the Family I84 genes are not always grouped in accordance with the species. A similar phenomenon has been reported in an EST contig based research (12). Some genes of the Family I84 may thus have duplicated before the speciation of bivalves and the speculation appears to conform to the distribution of the razor clam genes in 3 different chromosomes.
Positive Selection Was Not Detected in Duplicated Genes
Positive selection is believed to play an important role in driving the diversification of the duplicated genes during the evolution of some protease inhibitors (31, 37, 38). Yu etal. (39) reported a Ka/Ks ratio of 1.4 in the eastern oyster cvSI-1 gene. We calculated a Ka/Ks ratio of 1.3 for scSI-1 in a razor clam population with 150 individuals (data not shown). These research results seem to be indicative for the involvement of positive selection in the evolution of at least some Family I84 genes. However, no positively selected sites were detected in razor clam Family I84 genes in the present research; the significantly lower than 0.001 p-value between M0 and M3 models could only illustrate that selection pressure of different sites are not constant (25). The reason for failing to observe positive selection of Family I84 genes from razor clam, are possibly that these genes have been duplicated several times, as shown by the sequence identity and Ka/Ks for duplicated gene pairs, thus, diversifying time may be insufficient to accumulate enough nonsynonymous mutations in these paralogs (40). However, it cannot be concluded that the evolution of the Family I84 gene does not involve positive selection, on the contrary, the possibility of the gene family evolving through positive selection still exists and identification of more Family I84 orthologs in various mollusk species are required in future studies.
Gene Expression Variations Suggested Functional Divergence
It is noteworthy that expression of Family I84 genes, especially duplicated genes, diversified significantly. Generally, only one gene has a high expression level within tandem duplicated gene pairs, and gene expression between non-tandem duplicated genes has a high correlation. The difference between these genes could reach up to 100-fold when we excluded the above mentioned low expression genes. It had also been observed in eastern oyster, where cvSI-1 and cvSI-2 expression levels are nearly 1000 times that of cvSI-3 (12). ScSI-7 and scSI-14 were barely expressed in the analyzed transcriptome, we speculate that loss of the signal peptide of scSI-7 and premature termination of scSI-14 peptide, may drive their loss of function and silencing compared with their tandem duplicated partner. That is also detected in immune-related multi-gene families, some duplicated genes may have undergo frame-shift or pseudogenization during evolution (17, 36). Similarly, a number of TLR genes expanded in the Pacific oyster genome, but expression patterns of tandem-linked TLR genes in the same scaffold varied dramatically during oyster herpes virus (OsHV-1) infection (17). Their expression variation maybe evolutionarily critical for the functional divergence of Family I84 genes.
Family I84 genes expression increased in response to pathogenic bacteria and parasite; Vibrio challenge led to the elevation of expression for 8 of 14 Family I84 genes in the resistance group compared to the sensitive group. CvSI-1 was the only gene that was highly upregulated (day 15 post challenge) in hemocytes of adult oysters challenged with the pathogen of larval shellfish-V. tubiashii, even if it does not cause mortality in adult oysters (14). Meanwhile, expression of cvSI-1 and cvSI-2 were upregulated by injection of R. crassostreae, the bacterial pathogen of the Roseovarius oyster disease (ROD) in eastern oysters (41). In addition, the expression patterns of the Family I84 genes are inducible by salinity stresses and pathogen-associated molecular pattern stimulations (9, 11). These observation give emphasis to their involvement in function in host defense of related species.
The specific expression of Family I84 genes in the digestive gland should be strictly regulated due to they are hardly expressed in gills. Although no motifs were shared among promoter regions of Family I84 members, it was found that the cis-regulatory motifs of duplicated genes tend to diversify in yeast and mussel (36, 42). On the contrary, several transcription factor binding sites were shared among Family I84 genes, most of which are in response to physiological and pathological stimuli. In the future, functional identification of transcription factors is crucial for the regulation of target gene expression for their diversified function and evolution.
In conclusion, the full repertoire of Family I84 genes shared same gene structure, mature peptide of these member compromise 12 conserved cysteine residues with codon frequency preservation of 5 cysteine. Loss of a predicted signal peptide for scSI-7 and a premature termination for the scSI-14 increased their molecular diversity and may drive their silence compared to previous Family I84 genes. Remarkably, most razor clam Family I84 genes should experience lineage specific expansion by tandem duplication. Differential expression arise for tandem duplicated gene pairs, while high correlation of gene expression observed between non-tandem duplicated genes. These results provides a comprehensive understanding for the evolution of Family I84 protease inhibitor and their function in host defense.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
QX and ZL conceived and designed the study. SL performed gene identification and the statistical analyses. YL conducted transcriptome assembly and gene expression analysis. JL and JM participate in sequence verification. SL and QX wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by National Natural Science Foundation of China (32002428, 32073010), “3315” Innovative Team of Ningbo City, the Fundamental Research Funds for Zhejiang Province Universities and China Agriculture Research System of MOF and MARA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors express their sincere gratitude to Professor Fangluan Gao from Fujian Agriculture and Forestry University, Fuzhou, P. R. China, for his help on using EasyCodeml software.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.907274/full#supplementary-material
References
1. Armstrong PB. The Contribution of Proteinase Inhibitors to Immune Defense. Trends Immunol (2001) 22(1):47–52. doi: 10.1016/S1471-4906(00)01803-2
2. Rubio T, Oyanedel D, Labreuche Y, Toulza E, Luo X, Bruto M, et al. Species-Specific Mechanisms of Cytotoxicity Toward Immune Cells Determine the Successful Outcome of Vibrio Infections. Proc Natl Acad Sci USA (2019) 116(28):14238–47. doi: 10.1073/pnas.1905747116
3. Montagnani C, Le Roux F, Berthe F, Escoubas J-M. Cg-TIMP, an Inducible Tissue Inhibitor of Metalloproteinase From the Pacific Oyster Crassostrea Gigas With a Potential Role in Wound Healing and Defense Mechanisms. FEBS Lett (2001) 500(1):64–70. doi: 10.1016/S0014-5793(01)02559-5
4. Zhu L, Song L, Chang Y, Xu W, Wu L. Molecular Cloning, Characterization and Expression of a Novel Serine Proteinase Inhibitor Gene in Bay Scallops (Argopecten Irradians, Lamarck 1819). Fish. Shellfish. Immunol (2006) 20(3):320–31. doi: 10.1016/j.fsi.2005.05.009
5. Yu Q, Yang D, Wang Q, Zhang Y, Cong M, Wu H, et al. Molecular Characterization, Expression and Functional Analysis of Two Kazal-Type Serine Protease Inhibitors From Venerupis Philippinarum. Fish. Shellfish. Immunol (2017) 70:156–63. doi: 10.1016/j.fsi.2017.09.018
6. Xue QG, Waldrop GL, Schey KL, Itoh N, Ogawa M, Cooper RK, et al. A Novel Slow-Tight Binding Serine Protease Inhibitor From Eastern Oyster (Crassostrea Virginica) Plasma Inhibits Perkinsin, the Major Extracellular Protease of the Oyster Protozoan Parasite Perkinsus Marinus. Comp Biochem Physiol Part B: Biochem Mol Biol (2006) 145(1):16–26. doi: 10.1016/j.cbpb.2006.05.010
7. Xue Q, Itoh N, Schey KL, Cooper RK, La Peyre JF. Evidence Indicating the Existence of a Novel Family of Serine Protease Inhibitors That may be Involved in Marine Invertebrate Immunity. Fish. Shellfish. Immunol (2009) 27(2):250–9. doi: 10.1016/j.fsi.2009.05.006
8. Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS Database of Proteolytic Enzymes, Their Substrates and Inhibitors in 2017 and a Comparison With Peptidases in the PANTHER Database. Nucleic Acids Res (2017) 46(D1):D624–32. doi: 10.1093/nar/gkx1134
9. Wang X, Xue Q, Mao X, Dong Y, Li C, Lin Z. Two I84 Family Protease Inhibitors From Chinese Razor Clams Sinonovacula Constricta Expressed in Response to Environmental Challenges. Fish. Shellfish. Immunol (2018) 75:149–57. doi: 10.1016/j.fsi.2018.02.016
10. Xue Q. Pathogen Proteases and Host Protease Inhibitors in Molluscan Infectious Diseases. J Invertebrate. Pathol (2019) 166:107214. doi: 10.1016/j.jip.2019.107214
11. Lu J, Liu S, Ruan Z, Ye J, Wu Q, Wang S, et al. Family I84 Protease Inhibitors Likely Constitute a Mollusca-Specific Protein Family Functioning in Host Defense. Dis Aquat Org. (2021) 145:89–100. doi: 10.3354/dao03602
12. Xue Q, Beguel JP, Gauthier J, La Peyre J. Identification of cvSI-3 and Evidence for the Wide Distribution and Active Evolution of the I84 Family of Protease Inhibitors in Mollusks. Fish. Shellfish. Immunol (2017) 62:332–40. doi: 10.1016/j.fsi.2017.01.040
13. La Peyre JF, Xue QG, Itoh N, Li Y, Cooper RK. Serine Protease Inhibitor cvSI-1 Potential Role in the Eastern Oyster Host Defense Against the Protozoan Parasite Perkinsus Marinus. Dev Comp Immunol (2010) 34(1):84–92. doi: 10.1016/j.dci.2009.08.007
14. Nikapitiya C, McDowell IC, Villamil L, Muñoz P, Sohn S, Gomez-Chiarri M. Identification of Potential General Markers of Disease Resistance in American Oysters, Crassostrea Virginica Through Gene Expression Studies. Fish. Shellfish. Immunol (2014) 41(1):27–36. doi: 10.1016/j.fsi.2014.06.015
15. Gutiérrez-Rivera JN, Arcos-Ortega GF, Luna-González A, Rodríguez-Jaramillo MC, Arechiga-Carvajal ET, Vázquez-Juárez R. Differential Expression of Serine Protease Inhibitors 1 and 2 in Crassostrea Corteziensis and C. Virginica Infected With Perkinsus Marinus. Dis Aquat Org. (2015) 112(3):185–97. doi: 10.3354/dao02808
16. Dong Y, Zeng Q, Ren J, Yao H, Lv L, He L, et al. The Chromosome-Level Genome Assembly and Comprehensive Transcriptomes of the Razor Clam (Sinonovacula Constricta). Front Genet (2020) 11(664). doi: 10.3389/fgene.2020.00664
17. Zhang L, Li L, Guo X, Litman GW, Dishaw LJ, Zhang G. Massive Expansion and Functional Divergence of Innate Immune Genes in a Protostome. Sci Rep (2015) 5:8693. doi: 10.1038/srep08693
18. Gerdol M, Greco S, Pallavicini A. Extensive Tandem Duplication Events Drive the Expansion of the C1q-Domain-Containing Gene Family in Bivalves. Mar. Drugs (2019) 17(10):583. doi: 10.3390/md17100583
19. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics (2009) 25(9):1189–91. doi: 10.1093/bioinformatics/btp033
20. Edgar RC. MUSCLE: Multiple Sequence Alignment With High Accuracy and High Throughput. Nucleic Acids Res (2004) 32(5):1792–7. doi: 10.1093/nar/gkh340
21. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol (2013) 30(12):2725–9. doi: 10.1093/molbev/mst197
22. Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant (2020) 13(8):1194–202. doi: 10.1016/j.molp.2020.06.009
23. Gu Z, Cavalcanti A, Chen F-C, Bouman P, Li W-H. Extent of Gene Duplication in the Genomes of Drosophila, Nematode, and Yeast. Mol Biol Evol (2002) 19(3):256–62. doi: 10.1093/oxfordjournals.molbev.a004079
24. Zhao P, Wang D, Wang R, Kong N, Zhang C, Yang C, et al. Genome-Wide Analysis of the Potato Hsp20 Gene Family: Identification, Genomic Organization and Expression Profiles in Response to Heat Stress. BMC Genomics (2018) 19(1):61. doi: 10.1186/s12864-018-4443-1
25. Gao F, Chen C, Arab DA, Du Z, He Y, Ho SYW. EasyCodeML: A Visual Tool for Analysis of Selection Using CodeML. Ecol Evol (2019) 9(7):3891–8. doi: 10.1002/ece3.5015
26. Rice P, Longden I, Bleasby A. "EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet (2000) 16(6):276–7. doi: 10.1016/S0168-9525(00)02024-2
27. Zhao X, Wan J, Fu J, Shao Y, Lv Z, Li C. Identification of SNPs Associated With Disease Resistance in Juveniles of Sinonovacula Constricta Using RNA-Seq and High-Resolution Melting Analysis. Aquaculture (2021) 544:737109. doi: 10.1016/j.aquaculture.2021.737109
28. Kim D, Langmead B, Salzberg SL. HISAT: A Fast Spliced Aligner With Low Memory Requirements. Nat Methods (2015) 12:357. doi: 10.1038/nmeth.3317
29. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie Enables Improved Reconstruction of a Transcriptome From RNA-Seq Reads. Nat Biotechnol (2015) 33(3):290–5. doi: 10.1038/nbt.3122
30. Wagner GP, Kin K, Lynch VJ. Measurement of mRNA Abundance Using RNA-Seq Data: RPKM Measure is Inconsistent Among Samples. Theory Biosci (2012) 131(4):281–5. doi: 10.1007/s12064-012-0162-3
31. Christeller JT. Evolutionary Mechanisms Acting on Proteinase Inhibitor Variability. FEBS J (2005) 272(22):5710–22. doi: 10.1111/j.1742-4658.2005.04975.x
32. He Y, Yu H, Bao Z, Zhang Q, Guo X. Mutation in Promoter Region of a Serine Protease Inhibitor Confers Perkinsus Marinus Resistance in the Eastern Oyster (Crassostrea Virginica). Fish. Shellfish. Immunol (2012) 33(2):411–7. doi: 10.1016/j.fsi.2012.05.028
33. Laskowski M Jr., Kato I. Protein Inhibitors of Proteinases. Annu Rev Biochem (1980) 49:593–626. doi: 10.1146/annurev.bi.49.070180.003113
34. Wang B, Zhao J, Song L, Zhang H, Wang L, Li C, et al. Molecular Cloning and Expression of a Novel Kazal-Type Serine Proteinase Inhibitor Gene From Zhikong Scallop Chlamys Farreri, and the Inhibitory Activity of its Recombinant Domain. Fish. Shellfish. Immunol (2008) 24(5):629–37. doi: 10.1016/j.fsi.2008.01.017
35. Gerdol M, Schmitt P, Venier P, Rocha G, Rosa RD, Destoumieux-Garzon D. Functional Insights From the Evolutionary Diversification of Big Defensins. Front Immunol (2020) 11:758. doi: 10.3389/fimmu.2020.00758
36. Rey-Campos M, Novoa B, Pallavicini A, Gerdol M, Figueras A. Comparative Genomics Reveals a Significant Sequence Variability of Myticin Genes in Mytilus Galloprovincialis. Biomolecules (2020) 10:943. doi: 10.3390/biom10060943
37. Laskowski M Jr., Kato I, Kohr WJ, Park SJ, Tashiro M, Whatley HE. Positive Darwinian Selection in Evolution of Protein Inhibitors of Serine Proteinases. Cold Spring Harb Symp. Quant. Biol (1987) 52:545–53. doi: 10.1101/SQB.1987.052.01.062
38. Jongsma MA, Beekwilder J. Co-Evolution of Insect Proteases and Plant Protease Inhibitors. Curr Protein Pept Sci (2011) 12(5):437–47. doi: 10.2174/138920311796391115
39. Yu H, He Y, Wang X, Zhang Q, Bao Z, Guo X. Polymorphism in a Serine Protease Inhibitor Gene and its Association With Disease Resistance in the Eastern Oyster (Crassostrea Virginica Gmelin). Fish. Shellfish. Immunol (2011) 30(3):757–62. doi: 10.1016/j.fsi.2010.12.015
40. Lynch M, Conery JS. “The Evolutionary Fate and Consequences of Duplicate Genes.” Science (2000) 290(5494): 1151–5. doi: 10.1126/science.290.5494.1151
41. McDowell IC, Nikapitiya C, Aguiar D, Lane CE, Istrail S, Gomez-Chiarri M. Transcriptome of American Oysters, Crassostrea Virginica, in Response to Bacterial Challenge: Insights Into Potential Mechanisms of Disease Resistance. PloS One (2014) 9(8):e105097. doi: 10.1371/journal.pone.0105097
Keywords: family I84 protease inhibitor, genome-wide identification, duplication, differentiation, host defense, mollusk
Citation: Liu S, Liu Y, Lu J, Mao J, Lin Z and Xue Q (2022) Genome Wide Identification and Expression Profiling Indicate Expansion of Family I84 Protease Inhibitor via Gene Tandem Duplication and Divergence in Razor Clam Sinonovacula constricta. Front. Immunol. 13:907274. doi: 10.3389/fimmu.2022.907274
Received: 29 March 2022; Accepted: 09 May 2022;
Published: 01 June 2022.
Edited by:
Jun Li, Lake Superior State University, United StatesCopyright © 2022 Liu, Liu, Lu, Mao, Lin and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinggang Xue, cXh1ZUB6d3UuZWR1LmNu
 Sheng Liu
Sheng Liu Youli Liu1,2
Youli Liu1,2 Qinggang Xue
Qinggang Xue