- 1College of Veterinary Medicine, Northeast Agricultural University, Harbin, China
- 2Division of Viral Biologic Testing(I), China Institute of Veterinary Drug Control, Beijing, China
- 3China Ministry of Agriculture Key Laboratory of Animal Pathogen Biology, Northeastern Science Inspection Station, Harbin, China
Dendritic cells (DCs) are professional antigen-presenting cells that can recognize, capture, and process antigens. Fusing molecules targeting DCs with antigens can effectively improve the efficiency with which antigens are recognized and captured by DCs. This targeting strategy can be used for vaccine development to effectively improve the efficiency of antigen recognition and capture by DCs. The targeting sequence of porcine cytotoxic T-lymphocyte associated protein 4 (CTLA4), which binds porcine DCs, was identified in this study. Recombinant Lactobacillus reuteri (L. reuteri) expressing CTLA4-6aa (LYPPPY) and CTLA4-87aa fused to the porcine epidemic diarrhea virus (PEDV) protective antigen core neutralizing epitope (COE) were used to evaluate the ability of the two targeting motifs to bind the B7 molecule on DCs. Our results demonstrate that CTLA4-6aa could bind porcine DCs, and recombinant Lactobacillus expressing the CTLA4-6aa captured by porcine DCs was more efficient than those expressing CTLA4-87aa. In addition, the expression of DC markers, toll-like receptors, and cytokines was significantly higher in the 6aa-COE/L. reuteri-stimulated porcine DCs compared to DCs treated with 87aa-COE/L. reuteri (p<0.01) and recombinant Lactobacillus expressing CTLA4-6aa enhanced the ability of porcine DCs to activate T-cell proliferation. Our analysis of the protein structure revealed that CTLA4-87aa contains intramolecular hydrogen bonds, which may have weakened the intermolecular force between the residues on porcine CTLA4 and that on B7. In conclusion, recombinant Lactobacillus expressing CTLA4-6aa were more efficiently captured by porcine DCs and had a stronger ability to promote DC maturation and enhance T-cell proliferation. The LYPPPY motif is the optimal sequence for binding to porcine DCs. Piglets immunized with recombinant Lactobacillus showed that recombinant Lactobacillus expressing CTLA4-6aa induced significant levels of anti-PEDV-specific IgG and IgA antibody responses. Our study may promote research on DC-targeting strategies to enhance the effectiveness of porcine vaccines.
1 Introduction
Dendritic cells (DCs) are key regulators of T- and B-cell immunity. They can recognize, capture, process, and present antigens to T-cells, which play a critical role in the initiation and regulation of immune responses (1, 2). Moreover, DCs are the “bridge” cells that connect non-specific and specific immune responses and initiate immune responses (3). Consequently, DCs are usually targeted for the delivery of antigens (4). Using DC-targeting molecules enables the delivery of antigens to DCs specifically, which can improve the efficiency of antigens recognized and captured by these cells, leading to stronger T-cell responses (5, 6). Previous studies screened and identified human and chicken DC-targeting peptides using a phage display peptide library (7, 8). Subsequently, oral mucosal vaccines targeting DCs were proposed (9). However, inadequate research on porcine DC-targeting peptides has hampered the development of porcine vaccines.
DC surface receptors can bind specific ligands. Here, we propose a strategy that is based on the binding of natural ligands to the receptors on DCs (10). Increasing evidence indicates that specific ligands can facilitate the targeted delivery of antigens to DCs by interacting with the surface receptors on DCs (11, 12).
Moreover, screening and identifying the ligands that bind DC surface receptors and determining the key binding sites are effective strategies for confirming porcine DC peptides. Cytotoxic T-lymphocyte associated protein 4 (CTLA4) is mainly expressed on the surface of activated T-cells and binds to B7 on DCs (13, 14). CTLA4 consists of extracellular, transmembrane, and intracellular domains, and the extracellular domain plays an indispensable role in binding B7 (15). Many studies have used CTLA4 as a ligand for delivering antigens to DCs. Fusing the extracellular domain of CTLA4 with antigens can improve the efficiency with which antigens are recognized and captured by DCs (16–19). Peach et al. (1995) analyzed the amino acids in the extracellular domains of CTLA4 in humans, mice, cattle, sheep, dogs, pigs, cats, and rabbits and suggested that the extracellular domain of CTLA4 has a highly conserved MYPPPY motif (position 99-104, methionine (Met), Tyr, Pro, Pro, Pro, Tyr). Furthermore, the authors speculated that the conserved MYPPPY motif may be the main binding site in the extracellular domain of the molecule (20). However, the highly conserved MYPPPY motif, which is involved in B7 binding is not fully conserved in the porcine CTLA4 sequence; Met at position 99 is replaced by leucine (Leu) (21). In addition, Metzler et al. (1997) speculated that the regions flanking the MYPPPY motif may be equally important for binding DCs (22). Consequently, although it may be important to ascertain the role of the LYPPPY motif in porcine CTLA4 binding to porcine DCs, no formal studies have been performed to verify these speculations.
To explore the effects of antigen delivery with the help of DC-targeting molecules, we used Lactobacillus reuteri (L. reuteri) expressing antigens that were fused to DC-targeting molecules. Lactobacillus have become the most promising live bacterial vectors due to their probiotic effects (23, 24). Lactobacillus-live vector vaccines are prepared using genetic engineering, which enables the delivery of foreign antigens to the host mucosal system and supports the induction of a subsequent immune response (25, 26). Lactobacillus, serving as vectors that express and deliver antigens, play an important role in inducing intestinal mucosal immunity; however, several problems regarding their use as bacterial vectors remain unresolved. Compared with other expression vectors (such as E. coli, yeast, and insect expression vectors), Lactobacillus expression vectors produce lower amounts of antigen (27). An insufficient number of antigens makes it difficult to stimulate the body’s immune response. Thus, to enhance their immunogenicity and increase the efficiency with which Lactobacillus are recognized by DCs, it is common practice to include sequences of DC-targeting molecules in the recombinant Lactobacillus during vector construction. To date, several studies have analyzed and corroborated the feasibility of using a DC-targeted strategy for Lactobacillus vaccines (28–30). Accordingly, lactic acid bacteria have been widely employed as a delivery model to analyze the targeting functions of DC-targeting molecules.
Here, we aimed to elucidate the effectiveness of CTLA4-6aa (LYPPPY) as a DC-targeting peptide. Additionally, recombinant Lactobacilli expressing CTLA4-6aa and CTLA4-87aa fused to the porcine epidemic diarrhea virus (PEDV) protective antigen COE were used to evaluate the ability of the two targeting motifs to bind the B7 molecule on porcine DCs. Our study suggests that the targeting of DCs could be a potential strategy for the development of porcine vaccines.
2 Materials and methods
2.1 Bacteria and plasmids
Swine-derived L. reuteri was isolated from 3-month-old pig jejunum and cultured in de Man, Rogosa, and Sharpe medium (MRS; Hopebol, Qingdao, China) without shaking. The constitutive expression plasmid pPG-T7g10-PPT was constructed in our laboratory and contained the HCE strong constitutive promoter, T7g10 transcriptional enhancer, pgsA anchor from Bacillus subtilis for stabilizing the heterologous protein on the cell membrane (surface-displaying), and rrnBT1T2 terminator. The pMD19T-COE recombinant plasmid, recombinant plasmid that was inserted into the COE antigen was constructed and preserved in our laboratory. The core neutralizing epitope (COE) gene was obtained from the PEDV LJB/15 strain, which was isolated and identified from clinical samples in 2017 in our laboratory.
Animals
Nine antibody-seronegative, healthy One-month-old piglets were purchased from the Acheng Experimental Practice Base of Northeast Agricultural University (Harbin, China). Animal studies were performed according to the regulations of the Animal Experiment Ethics Committee of the Northeast Agricultural University, China (review number: NEAUEC20210337).
2.2 Peptide sequence
The LYPPPY motif (6aa) and the negative control peptide (YPLPYP, rearranging the sequence of 6aa) were synthesized by Jinsiri Biotechnology (Nanjing, Jiangsu) and purified by high-performance liquid chromatography (purity>95%). The N-terminus was labeled with fluorescein isothiocyanate (FITC).
2.3 Isolation and identification of porcine monocyte derived dendritic cells (MoDCs)
Blood collected from the precaval vein of 5–8-week-old swine was diluted with sterile phosphate-buffered saline (PBS), loaded onto an equal volume Histopaque-1077 (Sigma-Aldrich, St. Louis, MO, USA), and centrifuged at 800 × g for 25 min. The middle white mist-like cell layers were harvested and centrifuged at 600 × g for 15 min. The cell pellets were resuspended in red blood cell lysis buffer (Beyotime, Shanghai, China), rested for 5 min at 37°C, and thereafter centrifuged at 600 × g for 6 min. Cell pellets were washed twice with PBS and centrifuged at 600 × g for 6 min. Peripheral blood mononuclear cells (PBMCs) were obtained as mentioned above and seeded at 106 cells mL-1 in 6-well plates containing pre-warmed Roswell Park Memorial Institute-1,640 (RPMI-1640) medium, and supplemented with 1 U ml-1 penicillin and streptomycin, and 10% fetal calf serum (Gibco, Grand Island, NY, USA) for 6 days at 37°C with 5% CO2. Half of the medium was replaced with pre-warmed complete medium every 2 days, and non-adherent cells were removed on day 2 and day 4. Approximately 20 ng mL-1 recombinant porcine granulocyte-macrophage colony-stimulating factor (GM-CSF) (BD Biosciences, San Jose, CA, USA) and 20 ng mL-1 interleukin (IL-4) (BD Biosciences, San Jose, CA, USA) were added to the medium. Morphological changes in immature porcine MoDCs were observed using optical microscopy.
Immature porcine MoDCs were harvested on day 7 and incubated with CD172a labeled by phycoerythrin (PE) and MHC II labeled by fluorescein isothiocyanate (FITC) for 30 min. The cells were washed three times with sterile PBS, resuspended in 500 µL sterile PBS, and analyzed using flow cytometry and fluorescence microscopy.
2.4 Determining the binding ability of the peptide
On day 6 of MoDC cultivation, complete medium was gently aspirated using pipettes and washed twice with PBS. Porcine MoDCs and intestinal epithelial cells (IPEC) were incubated with FITC-labeled 6aa peptide (LYPPPY) and FITC-labeled negative control peptide (YPLPYP) for 30 min at 4°C. After being washed three times with PBS, the MoDCs were fixed with 4% paraformaldehyde for 5 min at 37°C and observed under a fluorescence microscope (Bio-Rad, CA, USA). Meanwhile, MoDCs that were incubated with the two peptides were collected in Eppendorf tubes, resuspended in 500 µL sterile PBS, and subjected to flow cytometry (BD Biosciences, San Jose, CA, USA).
2.5 Construction of recombinant strains
Primers were designed according to the swine-derived CTLA4 genes, the complete coding sequence published in GenBank (Number NM214149.1), and synthesized by Comate Bioscience (Jilin, Co., Ltd.). The primers used are listed in Table 1. The 6aa-COE gene was amplified using forward and reverse primers (6aa-COE-F/6aa-COE-R) with the plasmid pMD19T-COE as the template. PCR amplification was performed as follows: 95°C for 5 min; 30 cycles at 98°C for 10 s, 55°C for 30 s, 68°C for 30 s, and 68°C for 10 min of final extension. The PCR products were confirmed by DNA sequencing. RNA was extracted from porcine lymph nodes, complementary DNA was obtained by reverse transcription, and the 87aa gene was amplified using forward and reverse primers (87aa-F/87aa-R) with complementary DNA as the template. PCR amplification was performed as follows: 95°C for 5 min; 30 cycles of 98°C for 10 s, 57°C for 15 s, 68°C for 30 s, and 68°C for 10 min of final extension. The PCR products were confirmed by DNA sequencing. Thereafter, the 6aa-COE and 87aa genes were cloned into the expression plasmid pPG-T7g10-PPT to generate pPG-6aa-COE and pPG-87aa-COE. The flag tag was inserted into the 6aa-COE fusion gene, and a recombinant expression plasmid was constructed as shown in Figure S1. To construct the recombinant Lactobacillus strain, recombinant plasmids were electrotransferred into reuteri and verified by PCR, double digestion, and sequencing.
2.6 Protein expression and identification
Expression of the target proteins was analyzed by western blotting. The recombinant Lactobacillus were broken up by ultrasound and electrophoresis using 10% SDS-PAGE. Mouse anti-FLAG monoclonal antibody (diluted 1:1,000; Abcam, Cambridge, MA, USA) was used as the primary antibody, and horseradish peroxidase-conjugated goat anti-mouse IgG (diluted 1:5,000; ZSGB Biotech, Beijing, China) was used as the secondary antibody. Furthermore, 6aa-COE/L. reuteri and 87aa-COE/L. reuteri were cultured overnight at 37°C in MRS broth containing chloramphenicol (10 μg mL-1). The bacteria were pelleted by centrifugation at 1500 × g for 5 min, washed three times with sterile PBS, and resuspended in sterile PBS. The suspended bacterial pellet was sequentially incubated with anti-FLAG tag mouse monoclonal antibody (diluted 1:1,000) and FITC-conjugated anti-mouse IgG secondary antibodies (diluted 1:250). Fluorescence of the target protein expressed on the surface of Lactobacillus was measured by fluorescence microscopy.
2.7 Analysis of the MoDCs targeting ability of recombinant Lactobacillus
2.7.1 Cultivation of recombinant Lactobacillus
Recombinant Lactobacillus were cultured overnight at 37°C in MRS broth at a ratio of 1:50. The bacterial pellet was collected by centrifugation at 1500 × g for 5 min, washed three times with sterile PBS, and resuspended in RPMI-1,640. The recombinant Lactobacillus was used to stimulate porcine MoDCs.
2.7.2 Field-emission scanning electron microscope
MoDCs were cultured in 6-well plates with cell crawling for 6 days. The immature porcine MoDCs were incubated with 6aa-COE/L. reuteri, 87aa-COE/L. reuteri, COE/L. reuteri, and vector/L. reuteri were incubated in 5% CO2 at 37°C for 120 min. Lactobacillus that did not adhere were washed with sterile PBS. The samples were then processed using field-emission scanning electron microscopy.
2.7.3 Flow cytometry
The four strains of cultured recombinant Lactobacillus were cultivated according to the above steps, resuspended in 500 µL PBS, loaded with an equal volume of carboxyfluorescein succinimidyl ester (CFSE; eBioscience, San Diego, CA, USA), and incubated for 30 min at 4°C. Bacteria were washed three times with sterile PBS. Then the labeled recombinant Lactobacillus were added to immature porcine MoDCs, incubated at 37°C for 30 min. After washing thrice with sterile PBS, the cells were subjected to flow cytometry. Immature porcine MoDCs were used as negative controls.
2.7.4 Flat colony counting method
Four strains of cultured recombinant Lactobacillus were incubated with immature porcine MoDCs at 37°C for 30 min and washed three times with sterile PBS. Immature MoDCs were then collected in an Eppendorf tube for lysis. After centrifugation at 1500 × g for 5 min, the precipitate was collected and resuspended in 100 µL PBS. Thereafter, the precipitate was applied to the MRS broth plates with a spreading rod and cultured overnight at 37°C. After cultivation, the colonies were picked and incubated in MRS broth medium overnight at 37°C, PCR was used to identify the 6aa-COE gene in the recombinant plasmid of 6aa-COE/L. reuteri, 87aa gene in the recombinant plasmid 87aa-COE/L. reuteri, COE gene in the recombinant plasmid of COE/L. reuteri, and part of the vector gene in the recombinant plasmid of vector/L. reuteri. The colonies were counted and all measurements were performed in triplicates.
2.8 Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of surface molecules, cytokines, and TLR mRNA expression in porcine MoDCs
The MoDCs were incubated with 6aa-COE/L. reuteri, 87aa-COE/L. reuteri, COE/L. reuteri, vector/L. reuteri, and lipopolysaccharide (LPS) at 37°C for 12 h. Total RNA was extracted from non-stimulated MoDCs and MoDCs incubated with LPS and recombinant Lactobacillus using an RNAprep pure Micro kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. The complementary DNA template for qRT-PCR was obtained by reverse transcription and amplification was performed as follows: 42°C for 2 h, 72°C for 10 min, and rested on ice for 5 min. Primers for the qRT-PCR analysis were designed using Primer 5 (Table 2). The SYBR Green PCR Master Mix (Roche, Shanghai, China) was used to perform qRT-PCR. Amplification and detection of specific products were performed using the 7500 Fast Real-Time PCR System (Applied Biosystems) with the following cycle profile: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. β-actin was used as an internal control. Relative mRNA expression was calculated using the formula delta Ct (ΔΔCt) = [Ct (stimulated-MoDCs, target gene)-Ct (stimulated-MoDCs, β-actin)] - [Ct (non-stimulated-MoDCs, target gene)-Ct (non-stimulated-MoDCs, β-actin)]. The results were calculated by 2-ΔΔCt.
2.9 Evaluation of mixed leukocyte reaction (MLR) in MoDCs
MLR was evaluated using an automated ELISA reader (Bio-Tech Instruments, USA) and CCK-8 cell counting kit (Beyotime, China). Preparation of reaction cells: PBMCs were obtained from piglet peripheral blood by density gradient centrifugation, resuspended in RPMI-1640 basal culture medium, and washed thrice. The cell concentration was adjusted to approximately 2 × 106 mL-1 using RPMI-1640 complete culture medium. For preparation of stimulating cells immature MoDCs were incubated with LPS or 6aa-COE/L. reuteri, 87aa-COE/L. reuteri, COE/L. reuteri, or vector/L. reuteri and treated with 25 µg/mL final mass concentration mitomycin C, incubated at 37°C for 1 h, resuspended in RPMI-1640 basal culture medium, and washed three times. In a 96-well plate, T-lymphocytes were co-cultured with recombinant Lactobacillus-incubated DCs (1 × 105 cells/well) at ratios of 1:1, 1:10, or 1:100 (DCs: lymphocytes). Negative control wells were used for MoDCs and T-cells, and blank control wells were used for RPMI 1640 culture solution. Three replicates per well were incubated at 37°C in a 5% CO2 incubator for 72 h. Finally, CCK-8 was added to the 96-well plates, and OD450 values were read on an ELISA reader. The stimulation index (SI) was calculated using the following formula: SI = (ODsample-ODstimulator cells only)/(ODresponder cells only-ODblank control).
2.10 Detection of cytokines in Mo-DCs by ELISA
To detect the secretion of cytokines, supernatants were obtained from immature or mature MoDCs (5× 105 in 6-well plates) that had been treated with recombinant Lactobacillus at a concentration of 10 TCID50 cell−1 or LPS (2 μg mL−1) for 24 h. Levels of secreted IL-10, IL-12, and IFN-γ (R&D Systems, USA) were determined using commercial ELISA kits according to the manufacturer’s recommendations and calculated based on the curve generated using the cytokine standards.
2.11 Structural analysis of the protein
The crystal structure of porcine CTLA4 needed to be determined. The 3D structural model of porcine CTLA4 protein was generated from the crystal structure of human CTLA4/B7 protein complex (PDB code: 1I8L) template using Phyre2 (https://www.sbg.bio.ic.ac.uk/phyre2). Sequence alignment of porcine CTLA4 and human CTLA4 was performed using NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The binding interface between porcine CTLA4-6aa, CTLA4-87aa, and B7 molecules was analyzed based on the human CTLA4/B7 complex protein model using the PyMOL software (31).
2.12 Immunization and specimen collection
One-month-old specific pathogen-free (SPF) piglets were housed under SPF conditions with free access to a standard chow diet and water. Before oral administration, the recombinant Lactobacillus was cultured overnight in MRS medium, washed with PBS, and resuspended in PBS at a final concentration of 1 × 1010 CFU/mL. The piglets were randomly divided into three groups, with three piglets per group.
The first group was orally administered with 5 mL of recombinant Lactobacillus COE/L. Reuteri (1 × 1010 CFU/mL). The second group was orally administered with 5 mL of recombinant Lactobacillus 6aa-COE/L. Reuteri (1 × 1010 CFU/mL). The third group, which served as an unimmunized control, was inoculated with 5 mL of orally administered PBS. The piglets were immunized orally once a day for three consecutive days on days 1, 2, and 3. Serum, nasal, and anal swab samples were obtained on days 0, 7, 14, 21, 28, 35, 42, and 47 after immunization and were stored at −20°C until use.
2.13 ELISA immunoassay
The levels of anti-COE-specific IgG in the sera and SIgA in the sera, nasal cavity, and feces of the piglets used in this study were determined using ELISA. Briefly, polystyrene microtiter plates were coated with PEDV and incubated overnight at 4°C. After washing thrice with PBST (PBS containing 1% Tween-20), the plates were blocked with 5% skim milk at 37°C for 2 h. After washing thrice with PBST, the collected samples, prepared in triplicate, were diluted with PBS and incubated at 37°C for 1 h. Then, a conjugated goat anti-pig IgG or IgA antibody (Abcam, Cambridge, UK) was added to each well (1:5000) and incubated for 1 h at 37°C. The color was developed using tetramethylbenzidine (TMB; Sigma, Ronkonkoma, NY, USA) as a substrate, and absorbance was measured at 490 nm.
2.14 Statistical analysis
The experimental data were statistically analyzed using GraphPad Prism (version 8.0.2) statistical software. Data were presented as mean ± standard deviation (SD) and analyzed using two-way ANOVA with multiple comparison (LSD) tests using SPSS. Different letters (a vs. b, a vs. c, and b vs. c) indicate significant differences (p < 0.01) at the same time point.
3 Results
3.1 Cells generated by culturing porcine PBMCs with recombinant porcine GM-CSF and IL-4 have the morphology and phenotype of typical DCs
Porcine PBMCs were cultured in the presence of recombinant porcine granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-4. Changes in cell morphology were observed on days 1, 2, 4, and 6 using an optical microscope. As shown in Figure S2A, the isolated monocytes were round in shape and cell aggregates gradually appeared on day 2. As these cell aggregates grew, the surfaces of some cells displayed a veiled and dendritic appearance on day 4, and most cells displayed a typical immature DC appearance on day 6. Fluorescence microscopy and flow cytometry were used to analyze the phenotypes of immature porcine MoDCs. A total of 94.8% of cells stained positive for anti-porcine MHC class II, and 77.1% of cells stained positive for anti-porcine CD172a (Figure S2B). Immature porcine MoDCs expressed high levels of MHC class II molecules and CD172a (Figure S2C). This suggests that GM-CSF and IL-4 treatment facilitated the differentiation of PBMCs into immature porcine MoDCs.
3.2 CTLA4-6aa binds to porcine DCs
The ability of CTLA4-6aa to bind to porcine MoDCs was determined using fluorescence microscopy. We observed that the green fluorescence of porcine MoDCs incubated with FITC-modified CTLA4-6aa was higher than that of MoDCs incubated with the FITC-modified negative control (NC) and no green fluorescence from IPECs incubated with FITC-modified CTLA4-6aa was observed (Figure 1A). For accurate quantitative analysis, flow cytometry was used to evaluate the aforementioned binding ability. The results showed that the positive rate of porcine MoDCs incubated with CTLA4-6aa-FITC was 47.6%, which was significantly higher than that of MoDCs from the NC-FITC and control groups (Figure 1B). These results indicate that CTLA4-6aa can bind porcine DCs.
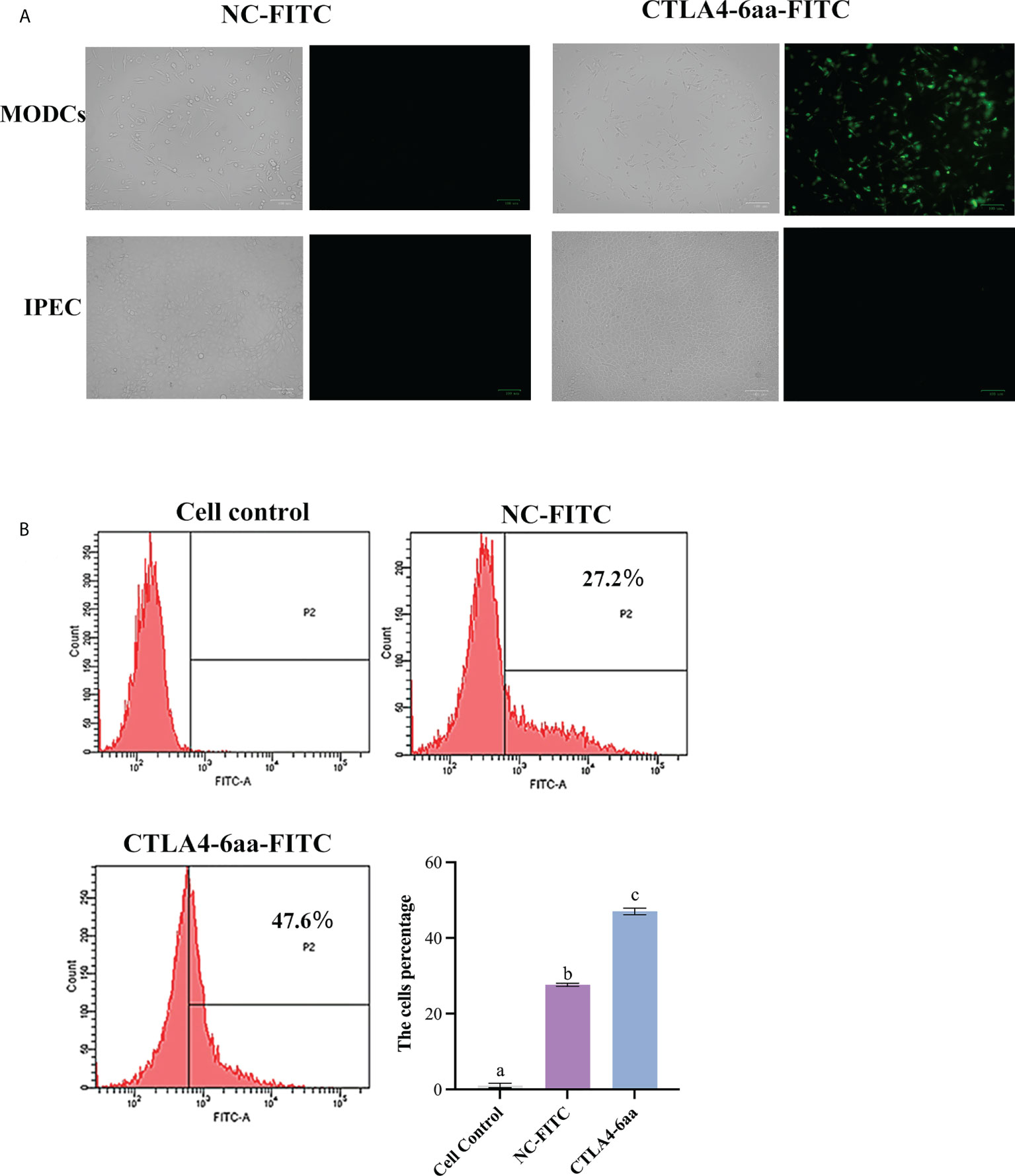
Figure 1 Ability of CTLA4-6aa-FITC to bind porcine monocyte derived dendritic cells (MoDCs) was analyzed by fluorescence microscopy and flow cytometry. Fluorescence microscopy: CTLA4-6aa-FITC is shown in green. Scale bar = 100 µm (A). Flow cytometry: CTLA4-6aa-FITC counts and binding of NC-FITC (as negative control) to porcine MoDCs, and the mean fluorescence intensities of FITC-labeled peptides binding to DCs (B). Different letters (a vs. b, a vs. c, b vs. c) indicate significant differences (p < 0.01) at the same time point.
3.3 Recombinant Lactobacillus expressing partial porcine CTLA4 motifs and PEDV protective antigen
To evaluate the ability of partial porcine CTLA4 motifs and antigens to bind DCs, recombinant Lactobacillus expressing CTLA4-6aa or CTLA4-87aa with PEDV protective antigen were constructed in this study. Expression of the recombinant proteins, namely, 6aa-COE and 87aa-COE, were confirmed by western blotting. As shown in Figure 2A, the fusion proteins 6aa-COE (left, 66 kDa) and 87aa-COE (right, 71 kDa) were detected in the immunoblots. The protein size was consistent with the predicted molecular weight (the protein size of the polyglutamate synthase A (PgsA) anchor was approximately 45 kDa, 6aa-COE was approximately 21 kDa, 87aa-COE was approximately 26 kDa). The NC vector/L. reuteri did not produce the corresponding immunoreactive bands. An indirect immunofluorescence assay was used to determine protein expression in recombinant Lactobacillus. Figure 2B shows that green fluorescence was detected in 6aa-COE/L. reuteri and 87aa-COE/L. reuteri compared to vector/L. reuteri. Therefore, the recombinant proteins were expressed in the Lactobacillus.
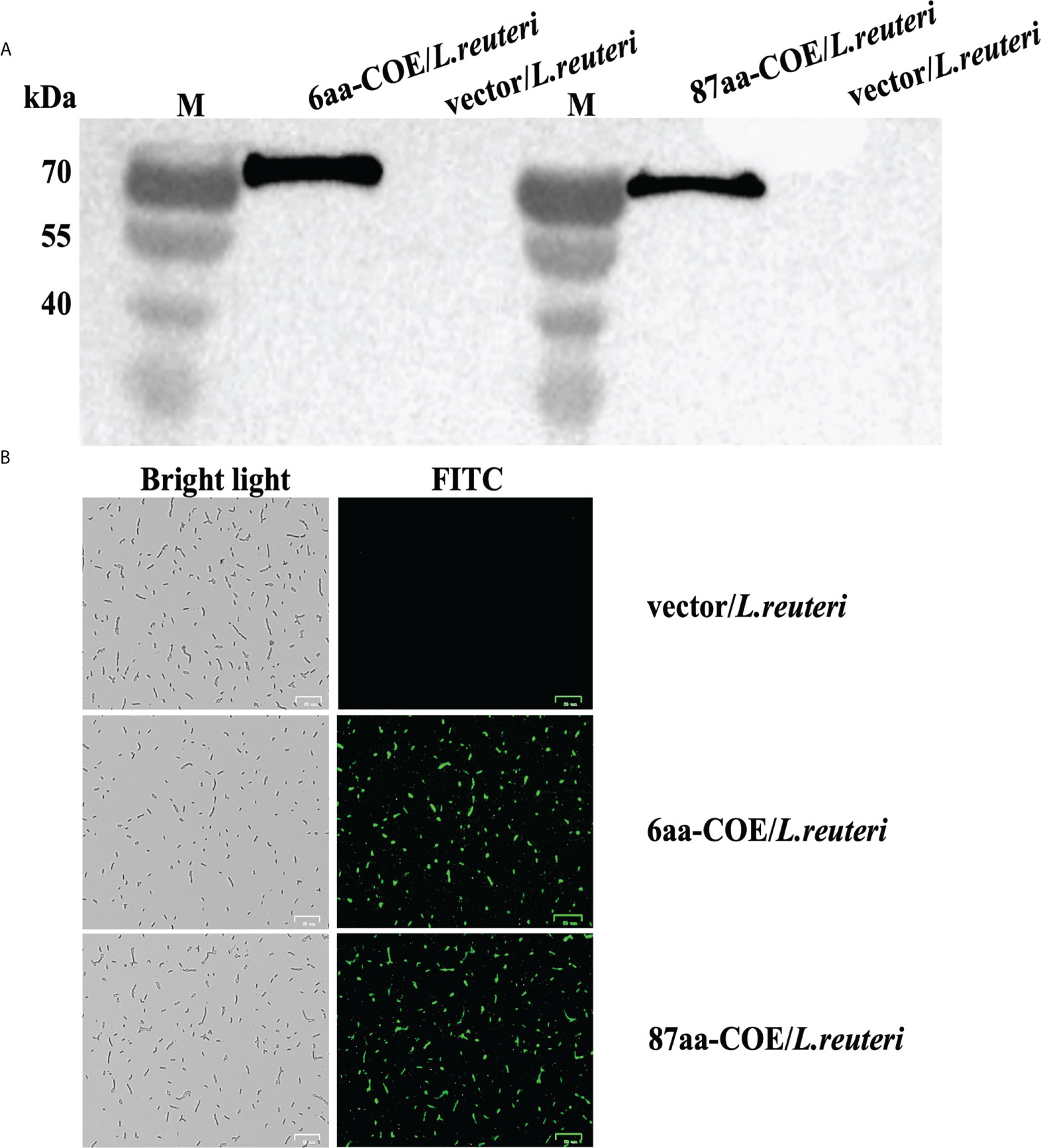
Figure 2 Western blotting analysis for identifying the expression of recombinant proteins 6aa-COE and 87aa-COE in Lactobacillus (Mouse anti-Flag monoclonal antibody was used as the primary antibody) (A). Identification of recombinant proteins expressed in Lactobacillus through fluorescence microscopy (B).
3.4 Recombinant Lactobacillus expressing CTLA4-6aa recognized and captured by porcine MoDCs with increased efficiency
Field-emission scanning electron microscopy was used to evaluate the efficiency with which recombinant Lactobacillus were captured and recognized by porcine MoDCs. The results showed that while 6aa-COE/L. reuteri, 87aa/L. reuteri, and COE/L. reuteri were recognized and captured by porcine MoDCs, vector/L. reuteri could not be captured by porcine MoDCs. Moreover, 6aa-COE/L. reuteri was more efficiently recognized and captured by porcine MoDCs than 87aa-COE/L. reuteri and COE/L. reuteri. There was no difference between the 87aa-COE/L. reuteri and COE/L. reuteri groups with regards to the efficiency of capture and recognition (Figure 3A).
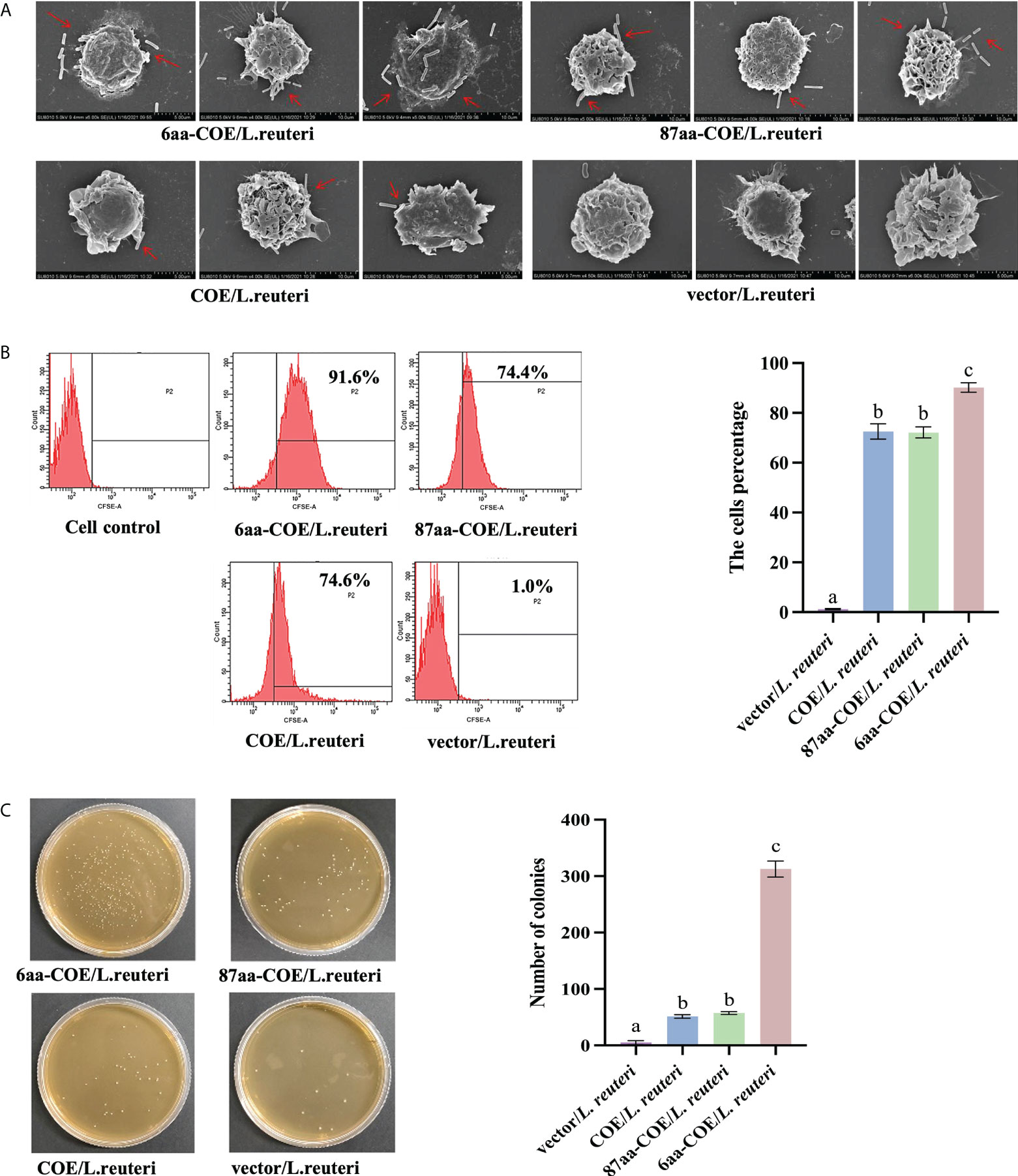
Figure 3 Scanning electron micrographs demonstrate the recombinant Lactobacilli recognized and captured by porcine monocyte derived dendritic cells (MoDCs) (A). Flow cytometry results of recombinant Lactobacillus recognized and captured by porcine MoDCs, and the mean fluorescence intensities of recombinant Lactobacillus recognized and captured by porcine MoDCs (B). Plate count assay for measuring the ability of recombinant Lactobacillus to target porcine MoDCs (C). Different letters (a vs. b, a vs. c, b vs. c) indicate significant differences (p < 0.01) at the same time point.
For a more accurate quantitative analysis, flow cytometry and flat colony counting were used to assess the efficiency with which recombinant Lactobacillus was captured and recognized by porcine MoDCs. Flow cytometry showed that counts of 6aa-COE/L. reuteri (captured by porcine MoDCs) were higher than that of the 87aa-COE/L. reuteri and COE/L. reuteri groups (p<0.01). The counts of 87aa-COE/L. reuteri and COE/L. reuteri groups (p>0.05) did not differ significantly (Figure 3B). The results of the flat colony counting were consistent with those obtained by flow cytometry (Figure 3C). Together, these results indicated that recombinant Lactobacillus expressing CTLA4-6aa was recognized and captured by porcine MoDCs with increased efficiency. Therefore, CTLA4-6aa, not CTLA4-87aa, expression was able to enhance the capture and recognition of recombinant Lactobacillus by porcine MoDCs.
3.5 Recombinant Lactobacillus expressing CTLA4-6aa up-regulate the mRNA expression of markers, cytokines, and TLRs in DCs
DC maturation is critical for priming the immune response. It is characterized by an increase in the expression of DC markers and TLRs, and increased secretion of cytokines. The results of qRT-PCR showed that the levels of DCs markers (CD40, CD80, and CD86), TLRs (TLR-2, 6, and 9), and cytokines (IL-10, IL-12, and IFN-γ) were significantly higher in 6aa-COE/L. reuteri-stimulated porcine DCs compared to DCs treated with 87aa-COE/L. reuteri (p<0.01). Expression of these markers did not differ markedly between the 87aa-COE/L. reuteri and COE/L. reuteri groups (Figures 4A–C). These results suggest that recombinant Lactobacillus expressing CTLA4-6aa may promote DC maturation.
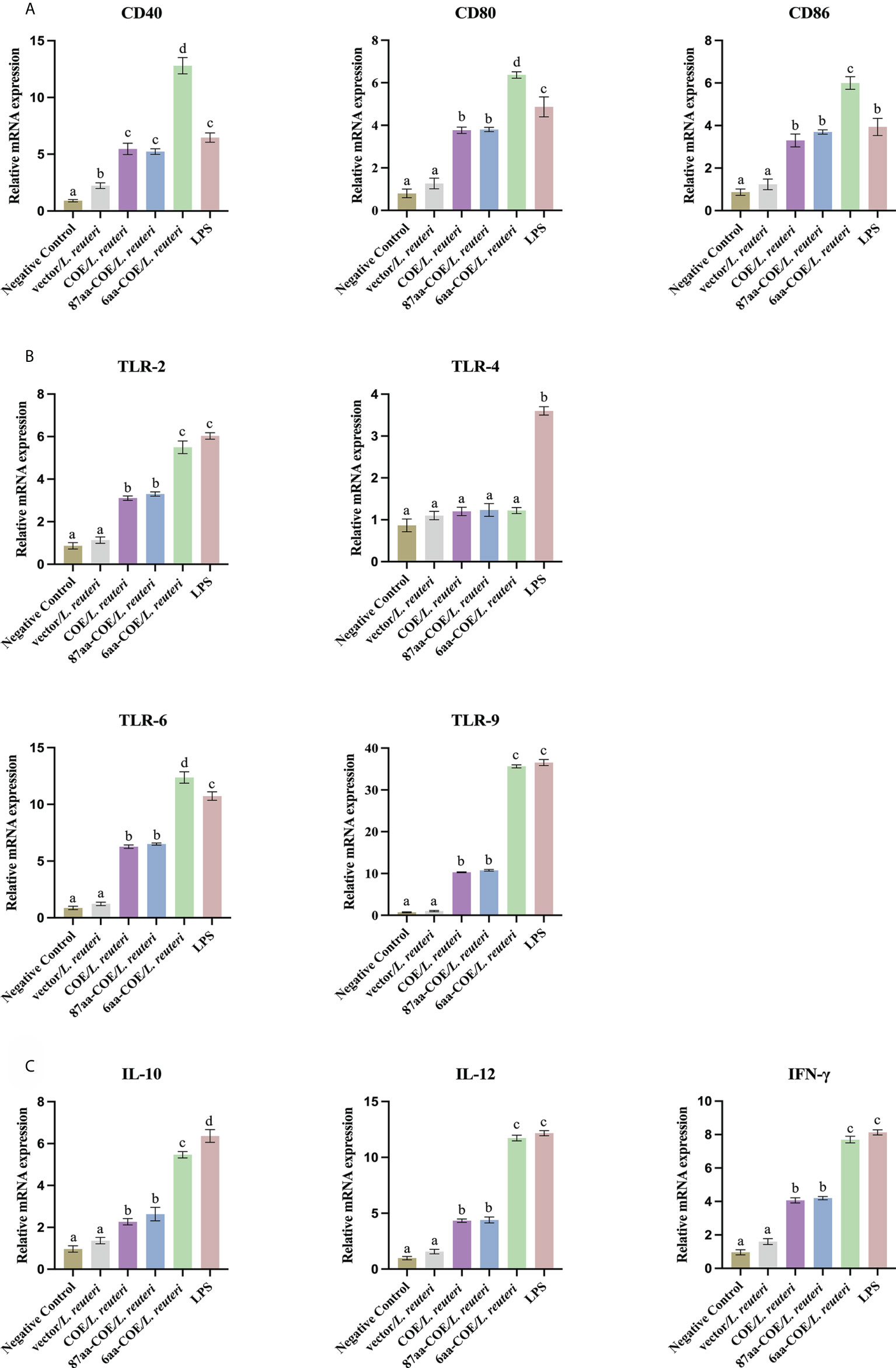
Figure 4 Expression of surface molecules, cytokines, and toll-like receptors (TLRs) by porcine monocyte derived dendritic cells (MoDCs) was assessed by relative quantitative real-time polymerase chain reaction (qRT-PCR). The expression of surface molecules on porcine MoDCs after stimulation with four recombinant Lactobacillus and lipopolysaccharide (LPS) was assessed by relative qRT-PCR (A). Expression of cytokines by porcine MoDCs stimulated with four recombinant Lactobacillus and LPS was assessed by relative qRT-PCR (B). Toll-like receptors expression by porcine MoDCs stimulated by four recombinant Lactobacillus and LPS was assessed by relative qRT-PCR (C). Different letters (a vs. b, a vs. c, b vs. c) indicate significant differences (p < 0.01) at the same time point.
3.6 Fusion of CTLA4-6aa with antigens promotes DC-mediated T-cell differentiation into Th1 cells
To assess whether the fusion of CTLA4-6aa with antigens affects antigen presentation ability, we prepared single-cell suspensions of lymphocytes from piglets and performed cell proliferation tests. The results showed that 6aa-COE/L. reuteri resulted in significantly higher levels of T-cell proliferative responses than COE/L. reuteri (p<0.01) (Figure 5A). This result implies that the fusion of CTLA4-6aa with antigens enhanced the ability of porcine DCs to activate T-cell proliferation.
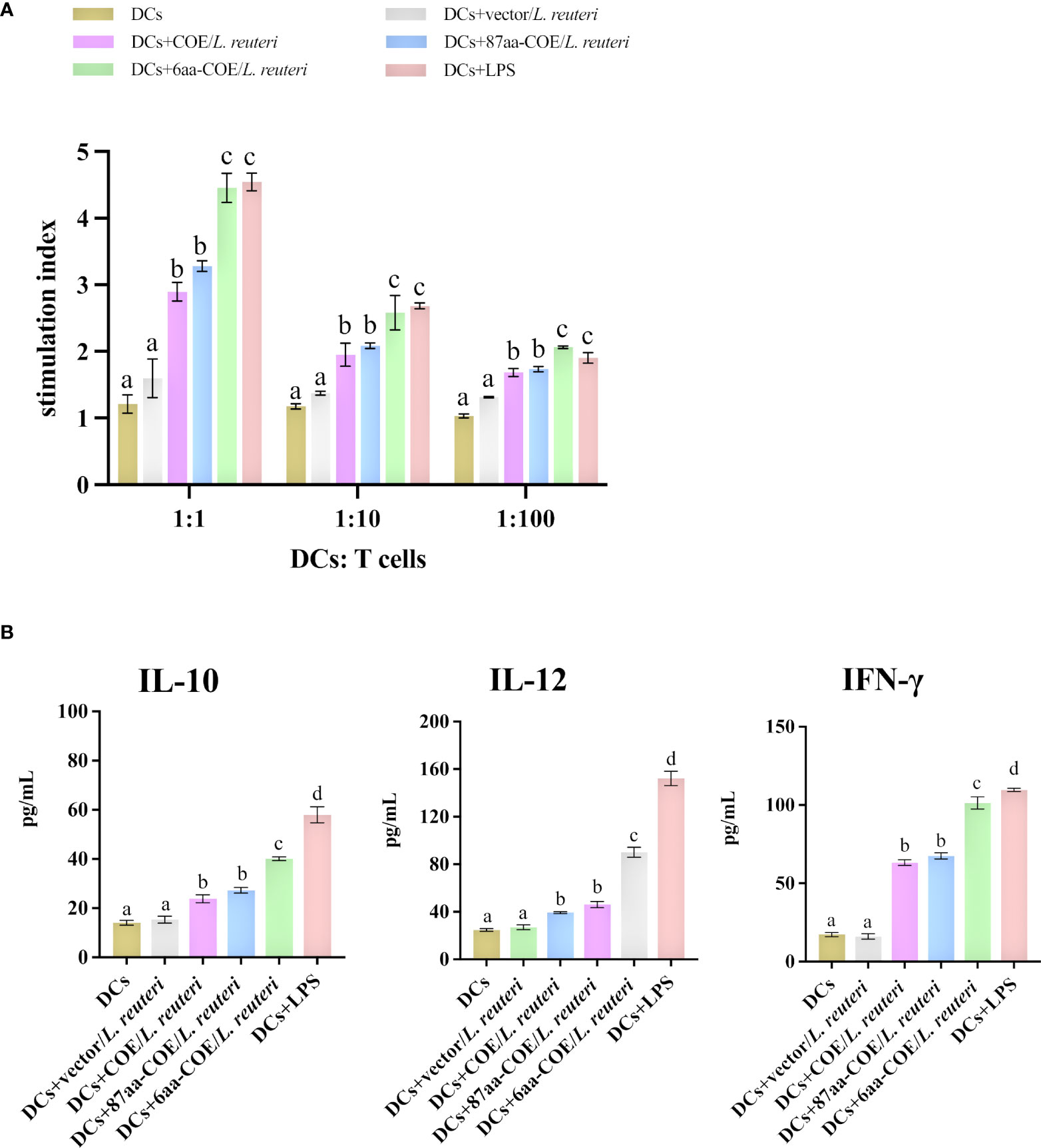
Figure 5 Analysis of MoDCs treated with four recombinant Lactobacillus and LPS skew T-cells toward different effector T-cell profiles. (A) MoDCs treated with recombinant Lactobacillus and LPS stimulate the proliferation of T-lymphocytes in mixed lymphocyte reaction (MLR). Responder cells were added in ratios of 1:1, 1:10, or 1:100 and co-cultured with the stimulated cells for 72 h. Proliferation is expressed as the Stimulation Index (SI) calculated with the formula: SI = (ODsample − ODstimulator cells only)/(ODresponder cells only − ODblank control). All experiments were performed in triplicate at a minimum. Data are presented as mean ± SEM (n = 6 per group) (A). MoDCs were stimulated with recombinant Lactobacillus and LPS for 12 h and then co-cultured with allogenic T-cells at a ratio of 1:1. After 72 h, culture supernatants were collected and analyzed for cytokines by ELISA (B). Different letters (a vs. b, a vs. c, b vs. c) indicate significant differences (p < 0.01) at the same time point.
To further determine the type of T-cell differentiation that is mediated by MoDCs stimulated with recombinant Lactobacillus, we measured the levels of IFN-γ, IL-12, and IL-10 using ELISA kits. Figure 5B shows that 6aa-COE/L. reuteri induced the secretion of a significant number of cytokines (IFN-γ, IL-12, and IL-10) by DCs and T-cells (p<0.01). In addition, the secretion of IFN-γ and IL-12 was higher than that of IL-10 by MoDCs induced by recombinant Lactobacillus. From the above results, it can be concluded that, with the participation of CTLA4-6aa, recombinant Lactobacillus stimulates MoDCs to mediate the differentiation of more T-cells into Th1 cells.
3.7 Forming intramolecular hydrogen bonds between CTLA4-87aa residues reduce binding to the B7 molecule on DCs
The porcine CTLA4 protein structure was simulated using Phyre2 based on the crystal structure of the human CTLA4/B7 complex protein model. As shown in Figure 6A, structural homology searches using Phyre2 suggested that the C chain of the human CTLA4/B7 complex is most similar to the porcine CTLA4 extracellular region, with MYPPPY serving as the CTLA4/B7 binding interface. In addition, the porcine CTLA4 sequence showed 87.6% homology to the human CTLA4 sequence.
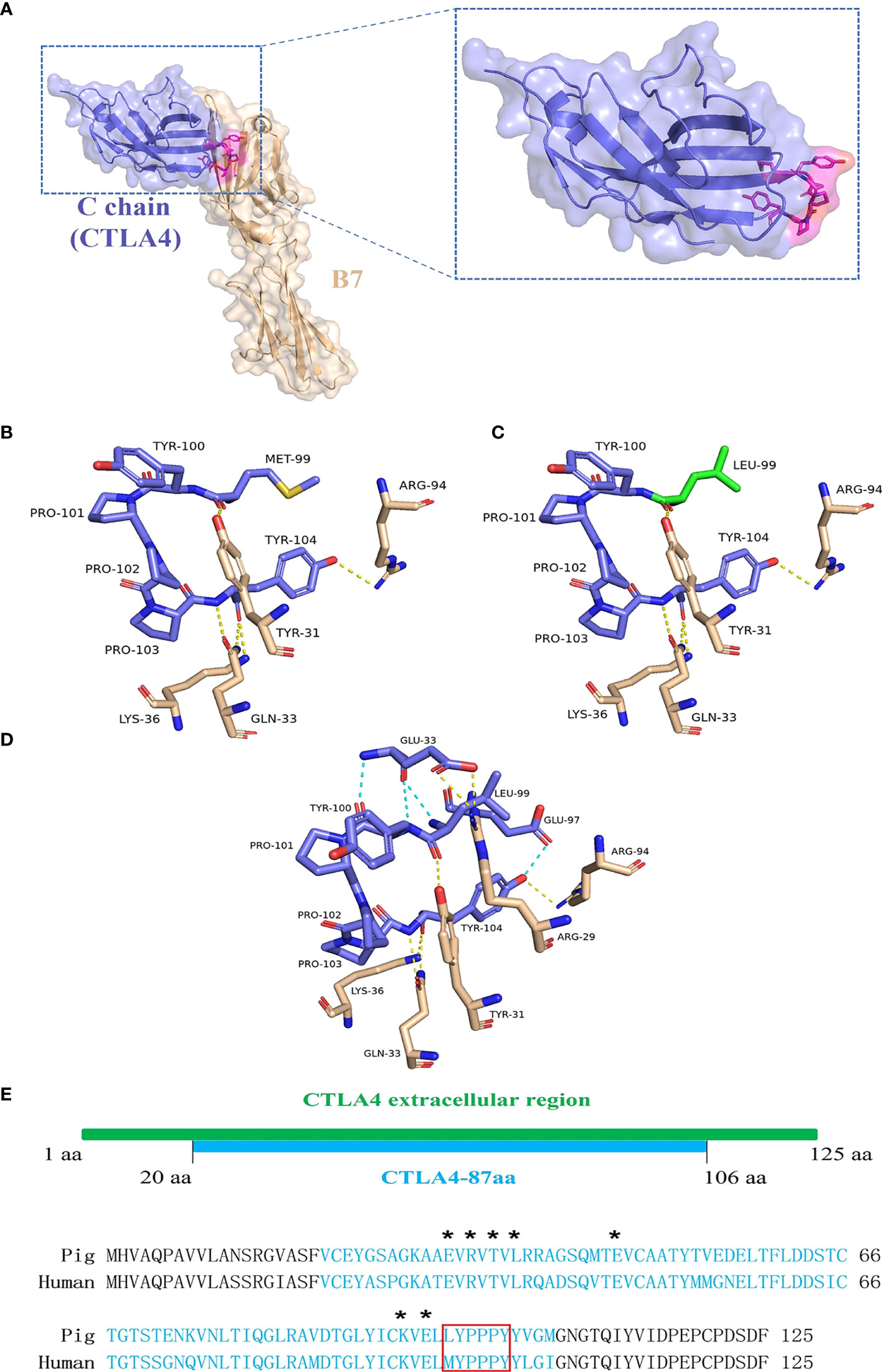
Figure 6 Analysis of porcine CTLA4 protein structure and binding interface. The porcine protein structure was generated from the crystal structure of the human CTLA4/B7 complex’s C chain (Protein Data Bank accession code 1I8L) templated by the Phyre2. (A) Ribbon diagram of the human CTLA4/B7 complex showing CTLA4 (purple), B7 (wheat), and MYPPPY motif (magenta). (B) Structure of the human MYPPPY motif bound to B7 (shown in sticks, colored by atom type). The yellow dotted line indicates hydrogen bonds between the residues. (C) Structure of porcine CTLA4-6aa (LYPPPY) bound to B7 (shown in sticks, colored by atom type). The green sticks represent the residue Leu 99, which was generated by mutating Met 99 and using the PyMOL software. (D) Structure of porcine CTLA4-87aa bound to B7 (shown in sticks, colored by atom type). The intramolecular hydrogen bonds (cyan dotted line) are shown. (E) Sequence alignment of porcine and human CTLA4 extracellular regions. Porcine CTLA4-87aa sequences are labeled in cyan. Asterisks denote the Glu 33, Arg 35, Thr 37, Leu 39, Glu 48, Lys 95, and Glu 97 residues. The LYPPPY and MYPPPY motifs are boxed in red.
The CTLA4/B7 binding interface was analyzed using PyMOL software and was based on the human CTLA4/B7 complex protein model. As shown in Figure 6B, the binding interface of the human MYPPPY motif interacted with B7 via five hydrogen bonds between the Met 99 and Tyr 104 residues on human CTLA4. Since the Met at position 99 was replaced by Leu in the case of the porcine CTLA4 sequence, residue conversion was performed using PyMOL software, and Leu substitutions were selected for 3D structure analysis. The connections between the replaced residues and changes in hydrogen bonds are depicted in Figures 6C and Table 3. The hydrogen bond between Leu 99 on porcine CTLA4 and Tyr 31 on B7 remained unchanged. For the porcine CTLA4-87aa protein, some residues (Glu 33, Arg 35, Thr 37, Leu 39, Glu 48, Lys 95, and Glu 97) speculated by Metzler et al. (1997) may show the importance of DCs binding, which is conserved in both human and porcine sequences (Figure 6E). Notably, as shown in Figure 6D and Table 4, intramolecular hydrogen bonds were formed between Glu 33 and Leu 99, Glu 33 and Tyr 100, and Glu 97 and Tyr 104 residues in the CTLA4-87aa protein, whereas intermolecular hydrogen bonds were formed between Glu 33 and Arg 29 residues. These results indicate that the intramolecular hydrogen bonds in CTLA4-87aa may weaken the intermolecular forces between porcine CTLA4 and B7 residues, thereby negatively affecting the binding of CTLA4 and B7.

Table 3 Analysis of the changes in hydrogen bonds following non-synonymous mutations in the Leu 99 residue of (3D protein structure) porcine CTLA4.
3.8 IgG and SIgA levels induced by recombinant Lactobacillus reuteri via oral immunization
Anti-PEDV-specific IgG was assessed to evaluate the ability of recombinant Lactobacillus strains to induce humoral immune responses in piglets. Serum was collected from piglets between days 0 and 49 post-immunization (collected every seven days), and serum-specific IgG responses were measured using ELISA. As shown in Figure 7, piglets in the 6aa-COE/L. reuteri group showed the highest specific IgG levels. From the fourteenth day after vaccination, a significant level of anti-PEDV-specific IgG antibody was induced in piglets that were orally administered 6aa-COE/L. reuteri (p<0.01) than COE/L. reuteri groups. The level of anti-PEDV-specific IgG antibody was the highest 28 days post-vaccination, significantly higher than in the other groups. This result shows that 6aa-COE/L. reuteri can elicit specific systemic immunity.
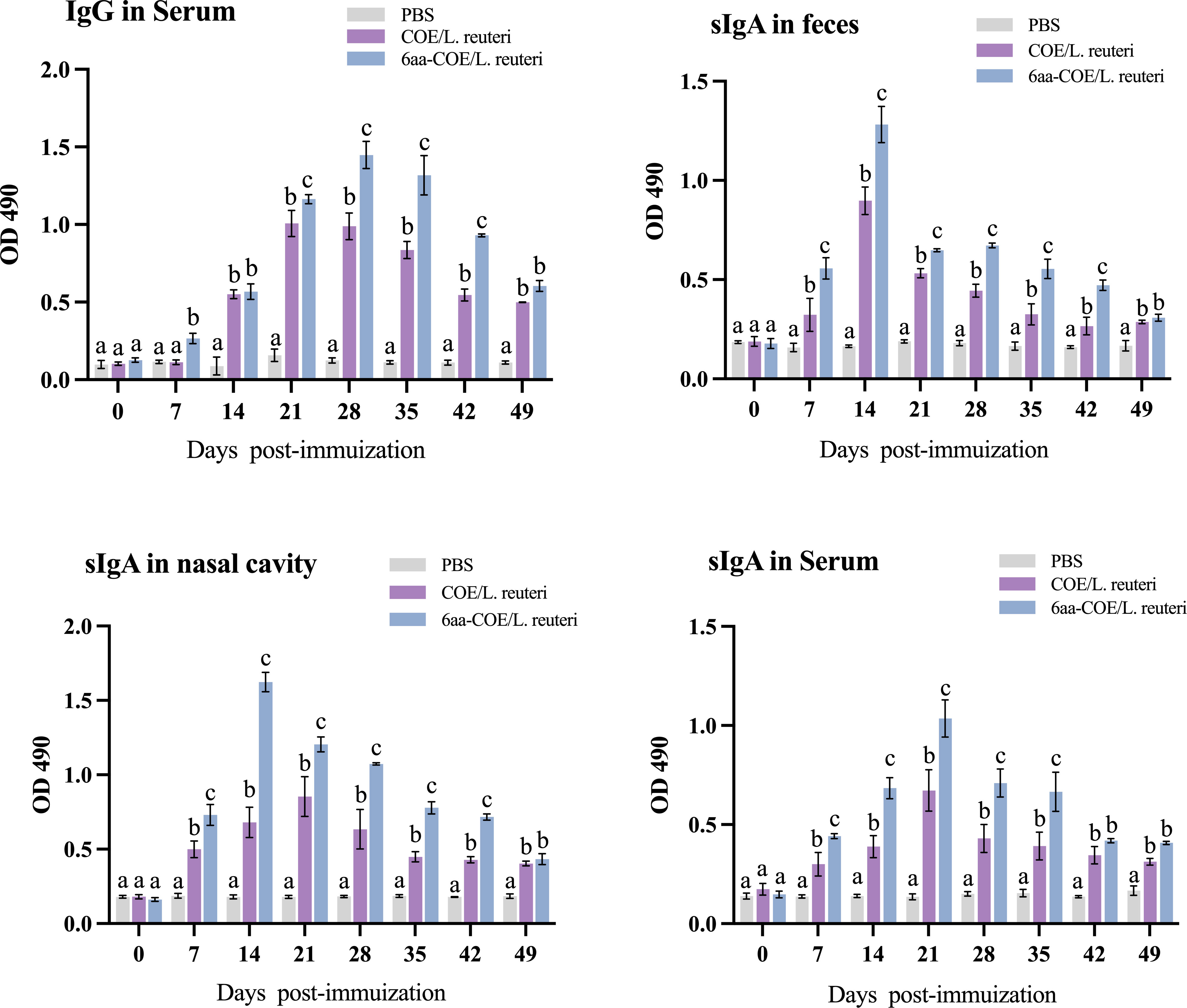
Figure 7 Specific anti-PEDV IgG and sIgA antibody levels in piglets orally immunized with the PBS, recombinant Lactobacillus 6aa-COE/L. reuteri and COE/L. reuteri. Measurement of a specific anti-PEDV IgG antibody in the anti-sera from immunized piglets by ELISA using PEDV as the coating antigen. Measurement of specific anti-PEDV SIgA antibody levels in the feces, nasal cavity, and serum by ELISA using PEDV as the coating antigen. Different letters (a vs. b, a vs. c, b vs. c) indicate significant differences (p < 0.01) at the same time point.
The intestinal mucosa cell-mediated immune response was further evaluated by measuring the specific anti-PEDV SIgA antibody in the nasal cavity, feces, and sera collected from immunized piglets post-immunization using ELISA. Mucosal SIgA levels increased after oral immunization with COE/L. reuteri and 6aa-COE/L. reuteri in the nasal cavity, feces, and sera (p<0.01) than those in the control groups of piglets orally immunized with PBS (Figure 7). Anti-PEDV SIgA in the feces was detected as early as the seventh-day post-immunization with COE/L. reuteri and 6aa-COE/L. reuteri. In addition, the level of SIgA antibody was induced by 6aa-COE/L. reuteri via oral immunization which was significantly higher than that induced by the COE/L. reuteri.
4 Discussion
The molecules on the surface of DCs recognize antigens and enable antigen uptake to facilitate the induction of strong immune responses (32). Therefore, the efficiency of antigen capture by DCs can be improved by the expression of molecules that directly target these cells. CTLA4 can specifically bind B7 molecules on the surface of DCs. Furthermore, the extracellular region of CTLA4 has been identified as the main domain that binds B7 (15). A few key motifs may be crucial for mediating the interaction between the receptor and its ligands. A previous study hypothesized that a peptide motif (MYPPPY) in the extracellular region of human CTLA4 may play an important role in binding the B7 molecule on DCs, but no formal studies have been performed to verify the above speculations (20). Based on these aforementioned study, we identified the key motifs in porcine CTLA4 that are important for binding the B7 molecule to porcine DCs. Thereafter, Lactobacillus expressing these key motifs in combination with the antigen were used to analyze the efficiency of antigen capture and recognition by DCs and monitor their effect on DC function. Our study may promote the targeting of DCs as a potential strategy for the development of porcine vaccines.
For this DC-targeting strategy mentioned above, the expansion of DCs for in vitro studies is necessary because they are rare in body tissues, and in mammals many DCs are generated from blood or bone marrow-derived DCs (33, 34). At present, the isolation of monocytes from peripheral blood is a simple and convenient procedure. Consequently, we generated and used MoDCs in our study. Specifically, PBMCs were cultured in the presence of recombinant porcine GM-CSF and IL-4 for 6 days, and the cultured cells had the morphology of typical DCs. A majority of the cultured cells express MHC class II and CD172a, which are necessary signal transduction molecules for the recognition and capture of antigens by DCs (35, 36).
DC-targeting strategies have become the focus of research, as DCs play a significant role in the induction and regulation of immune responses. The strategy described in our study can be used to develop DC-targeting vaccines to increase antigen availability for DCs (37). To increase the bioavailability of immunogens in pigs, a porcine DC-targeting peptide was identified in this study. Our study indicated that the LYPPPY motif can bind porcine MoDCs and may serve as a suitable alternative for developing targeted vaccines in porcine. In addition, to evaluate the ability of the LYPPPY motif in the extracellular domain of porcine CTLA4 to target DCs, we analyzed the amino acids flanking the LYPPPY motif. Thereafter, recombinant Lactobacillus vaccines with CTLA4-6aa and CTLA4-87aa were constructed to evaluate whether the targeted sequences could direct antigens to DCs.
Choosing suitable antigen delivery vectors is key to evaluating the efficiency of DCs targeting. Lactobacillus in the intestinal tract that can express and deliver antigens (38). They are good vectors substances for studying antigens directly on DCs (39). In particular, Lactobacillus have probiotic properties and can resist gastric acid bile salts and colonize the intestine (40). In this study, a Lactobacillus expression vector (pPG-T7g10-PPT) was used to express DC-targeting sequences and antigens. This vector contains the polyglutamate synthase A (PgsA) anchor protein, which is a transmembrane protein that can be fused with the N-terminus of target proteins such that the target proteins are expressed on the bacterial surface (41). To evaluate the effectiveness of the DC-targeting sequences, recombinant Lactobacillus, namely 6aa-COE/L. reuteri and 87aa-COE/L. reuteri were constructed. This study showed that while recombinant Lactobacillus expressing CTLA4-6aa could efficiently target antigens to DCs, those expressing CTLA4-87aa had no obvious targeting effect compared to the recombinant Lactobacillus expressing COE only. Additionally, we analyzed the protein structure in our study. Analysis of the human CTLA4/B7 complex suggested that the CTLA4 and B7 ligand-binding sites were distal to the binding interface, providing the potential for B7 to bind CTLA4 (42). Notably, it is known that the 99MYPPPY104 motif in human CTLA4 is essential for its interaction with B7. In case of porcine CTLA4, Met 99 in the 99MYPPPY104 motif was replaced by Leu 99. The 3D structural simulation revealed that residue substitutions did not alter the interaction and stability of the CTLA4 protein structure. Therefore, it is plausible that the structure of the human CTLA4/B7 complex could serve as a reasonable model for studying the binding interface of porcine CTLA4. Our analysis using PyMOL software suggested that porcine CTLA4 binds to B7 via the 99LYPPPY104 motif, with Leu 99 and Tyr 104 being the key residues that bind B7. Notably, the unusual cis-trans-cis backbone conformation of the 101Pro-Pro-Pro103 sequence in the 99LYPPPY104 motif of CTLA4 contributes to the considerable geometric complementarity between CTLA4 and B7 (43). Together with the other experimental results in this study, these results show that CTLA4-6aa is a crucial site for binding B7. The binding interface between CTLA4-87aa and B7 was analyzed using a protein structure model. In case of the CTLA4-87aa sequence, the conserved residues (Glu 33, Arg 35, Thr 37, Leu 39, Glu 48, Lys 95, and Glu 97) and two hydrogen bonds between the Glu 33 residue on CTLA4 and the Arg 29 residue on B7 are likely to contribute to the specificity of binding (44). However, intramolecular hydrogen bonds were formed in the CTLA4-87aa protein between the Glu 33 and Leu 99, Glu 33 and Tyr 100, and Glu 97 and Tyr 104 residues. The formation of intramolecular hydrogen bonds may weaken the intermolecular forces between the residues on porcine CTLA4 and those on B7. This negatively affects the interaction between CTLA4 and B7.
Antigens are captured and processed by DCs before being presented to immune cells. DCs gradually mature, with increased expression of surface molecules and TLRs, and augmented cytokine secretion are characteristics of matured DCs (45). TLRs on the surface of DCs can also mediate their maturation and play an important role in initiating innate immune responses (46). Moreover, DCs secrete cytokines that perform different functions and facilitate immune response regulation (47). They have a critical effect on the differentiation of naïve T-cells into T-helper type 1 and type 2 cells (48, 49). Although IL-12 and IFN-γ are pro-inflammatory cytokines, IL-10 is an anti-inflammatory cytokine. The balance between both, the pro- and anti-inflammatory cytokines can regulate the immune response and exert anti-infection and anti-tumor functions (50). The results showed that the expressions of DCs surface molecules (CD40, CD80, and CD86), TLRs (TLR-2, 6, and 9), and cytokines (IL-10, IL-12, and IFN-γ) in 6aa-COE/L. reuteri-stimulated porcine DCs were significantly higher than that in DCs treated with 87aa-COE/L. reuteri. Notably, recombinant Lactobacilli had no effect on TLR-4 expression because TLR-4 can mainly recognize lipopolysaccharides from gram-negative bacteria and its expression may not be affected by gram-positive bacteria (51). A previous study reported DC-targeted peptide-conjugated antigen immunization to preferentially induce the expansion and proliferation of CD4+ T-cells (52). Our results align with previous studies in which recombinant Lactobacillus 6aa-COE/L. reuteri significantly induced MoDCs to stimulate T-cell proliferation at a DC/T ratio of 1:1. The balance between the Th1 and Th2 responses was evaluated based on the secretion levels of these cytokines (IL-10, IL-12, and IFN-γ) (53). In this study, the levels of IL-12 and IFN-γ secreted by recombinant Lactobacillus 6aa-COE/L. reuteri-stimulated MoDCs were higher than those stimulated by IL-10, suggesting that CTLA4-6aa is beneficial to recombinant Lactobacillus to stimulate DCs to mediate the cellular immune response and differentiate T-cells into Th1 type cells.
An effective oral vaccine can induce both systemic and mucosal immune responses. Therefore, we evaluated the immune response of piglets induced by recombinant Lactobacillus via oral immunization. Serum-derived IgG can improve the body’s immune defense by reducing pathogens’ ability to cross the intestinal mucosa, thereby effectively blocking the systemic transmission of pathogens to the organism (54). sIgA plays an important role in the intestinal mucosal immune response and is a major immunoglobulin in mucosal immune responses that protects against pathogen invasion at mucosal sites (55). In this study, we detected high levels of antigen-specific mucosal sIgA in the feces, nasal cavity, and serum of piglets orally immunized with 6aa-COE/L. reuteri and anti-PEDV-specific IgG and sIgA levels significantly increased in the 6aa-COE/L reuteri group after immunization than the PBS and the recombinant Lactobacillus COE/L. reuteri group. The SIgA and IgG contents tended to increase with time. The level of SIgA began to decrease on the twenty-first day after immunization, but the IgG from the blood showed a downward trend on the thirty-fifth day after immunization, indicating that the mucosal immunity induced by recombinant Lactobacillus occurred earlier than humoral immunity. Still, the humoral immunity produced lasted longer, which was confirmed in our previous studies (56, 57). In the present study, we demonstrated oral vaccination with recombinant Lactobacillus 6aa-COE/L. reuteri strongly induces systemic and mucosal immune responses against PEDV. These results indicated that recombinant Lactobacilli expressing CTLA4-6aa have a strong ability to promote DCs maturation and initiate immune responses.
Taken together, this study confirmed that recombinant Lactobacillus expressing CTLA4-6aa was more efficiently captured by porcine DCs and had an increased ability to promote DCs maturation. Additionally, we confirmed that the LYPPPY motif of porcine CTLA4 is the key sequence for its interaction with B7 in porcine DCs. Our results showed that antigen targeting of DCs using Lactobacillus is a promising strategy for the development of porcine vaccines.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by the committee on the Ethics of Animal Experiments of Northeast Agricultural University, Harbin, China.
Author contributions
TX and NW designed and conducted research experiments. TX and NW analyzed data and wrote the paper. YT, CG, and JH contributed to isolation and validation of porcine MoDCs. YG, XW, JL, HZ, WC, ZS, YJ, XQ, LT, LW and YL performed research. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 31772779 and 31972718).
Acknowledgments
We would like to thank Editage (https://www.editage.cn/) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.926279/full#supplementary-material
Figure S1 | Schematic diagram showing the construction of recombinant plasmids. ①6aa-COE fragment, cleaved with SacI and ApaI, was inserted into constitutive expression vector pPG-T7g10-PPT, and recombinant plasmid pPG-T7g10-6aa- COE was obtained by blunt end ligation.② 6aa gene in pPG-T7g10-6aa-COE was disrupted by HindIII and XhoI, 87aa fragment, cleaved with HindIII and XhoI, was inserted into constitutive expression vector pPG-T7g10-PPT-COE, and recombinant plasmid pPG-T7g10-87aa- COE was obtained by blunt end ligation.
Figure S2 | Morphology of peripheral blood mononuclear cells (PBMCs) cultured for 6 days in the presence of recombinant porcine granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin (IL-4). Scale bar 100 µm (A). Fluorescence microscope: FITC-labeled MHC class II is shown in green. Phycoerythrin-labeled CD172a is shown in red. The 4’,6-diamidino-2-phenylindole stained nucleus is shown in blue. Scale bar = 100 µm (B). Flow cytometry: counts of FITC-labeled MHC class II and phycoerythrin-labeled CD172a expressed on immature porcine monocyte derived dendritic cells (MoDCs), and the mean fluorescence intensities of FITC-labeled MHC class II and phycoerythrin-labeled CD172a expressed on immature porcine MoDCs (C). Different letters (a vs. b, a vs. c, b vs. c) indicate significant differences (p < 0.01) at the same time point.
References
1. Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, et al. Immunobiology of dendritic cells. Annu Rev Immunol (2000) 18:767–811. doi: 10.1146/annurev.immunol.18.1.767
2. Eisenbarth SC. Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol (2019) 19:89–103. doi: 10.1038/s41577-018-0088-1
3. Balan S, Saxena M, Bhardwaj N. Dendritic cell subsets and locations. Int Rev Cell Mol Biol (2019) 348:1–68. doi: 10.1016/bs.ircmb.2019.07.004
4. Cohn L, Delamarre L. Dendritic cell-targeted vaccines. Front Immunol (2014) 5:255. doi: 10.3389/fimmu.2014.00255
5. Engering A, Geijtenbeek TB, van Vliet SJ, Wijers M, van Liempt E, Demaurex N, et al. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J Immunol (2002) 168:2118–26. doi: 10.4049/jimmunol.168.5.2118
6. Figdor CG, van Kooyk Y, Adema GJ. C-type lectin receptors on dendritic cells and langerhans cells. Nat Rev Immunol (2002) 2:77–84. doi: 10.1038/nri723
7. Curiel TJ, Morris C, Brumlik M, Landry SJ, Finstad K, Nelson A, et al. Peptides identified through phage display direct immunogenic antigen to dendritic cells. J Immunol (2004) 172:7425–31. doi: 10.4049/jimmunol.172.12.7425
8. Ma S, Qiao X, Xu Y, Wang L, Zhou H, Jiang Y, et al. Screening and identification of a chicken dendritic cell binding peptide by using a phage display library. Front Immunol (2019) 10:1853. doi: 10.3389/fimmu.2019.01853
9. Owen JL, Sahay B, Mohamadzadeh M. New generation of oral mucosal vaccines targeting dendritic cells. Curr Opin Chem Biol (2013) 17:918–24. doi: 10.1016/j.cbpa.2013.06.013
10. Tacken PJ, Torensma R, Figdor CG. Targeting antigens to dendritic cells. vivo. Immunobiol (2006) 211:599–608. doi: 10.1016/j.imbio.2006.05.021
11. Chiang CY, Wu CC, Chen YJ, Liu SJ, Leng CH, Chen HW. Delivery of antigen to CD8+ dendritic cells by fusing antigen with formyl peptide receptor-like 1 inhibitor protein induces antitumor immunity. Front Immunol (2019) 10:1839. doi: 10.3389/fimmu.2019.01839
12. Apostólico JS, Lunardelli VA, Yamamoto MM, Souza HF, Cunha-Neto E, Boscardin SB, et al. Dendritic cell targeting effectively boosts T cell responses elicited by an HIV multiepitope DNA vaccine. Front Immunol (2017) 8:101. doi: 10.3389/fimmu.2017.00101
13. Teft WA, Kirchhof MG, Madrenas J. A molecular perspective of CTLA4 function. Annu Rev Immunol (2006) 24:65–97. doi: 10.1146/annurev.immunol.24.021605.090535
14. Stamper CC, Zhang Y, Tobin JF, Erbe DV, Ikemizu S, Davis SJ, et al. Crystal structure of the B7-1/CTLA4 complex that inhibits human immune responses. Nature (2001) 410:608–11. doi: 10.1038/35069118
15. Morton PA, Fu XT, Stewart JA, Giacoletto KS, White SL, Leysath CE, et al. Differential effects of CTLA4 substitutions on the binding of human CD80 (B7-1) and CD86 (B7-2). J Immunol (1996) 156:1047–54. doi: 10.1084/jem.183.2.705
16. Deliyannis G, Boyle JS, Brady JL, Brown LE, Lew AM. A fusion DNA vaccine that targets antigen-presenting cells increases protection from viral challenge. Proc Natl Acad Sci U.S.A. (2000) 97:6676–80. doi: 10.1073/pnas.120162497
17. Lu M, Isogawa M, Xu Y, Hilken G. Immunization with the gene expressing woodchuck hepatitis virus nucleocapsid protein fused to cytotoxic-t-lymphocyte-associated antigen 4 leads to enhanced specific immune responses in mice and woodchucks. J Virol (2005) 79:6368–76. doi: 10.1128/JVI.79.10.6368-6376.2005
18. Rohrbach F, Weth R, Kursar M, Sloots A, Mittrücker H-W, Wels WS. Targeted delivery of the ErbB2/HER2 tumor antigen to professional APCs results in effective antitumor immunity. J Immunol (2005) 174:5481–9. doi: 10.4049/jimmunol.174.9.5481
19. Gerstmayer B, Pessara U, Wels W. Construction and expression in the yeast pichia pastoris of functionally active soluble forms of the human costimulatory molecules B7-1 and B7-2 and the B7 counter-receptor CTLA4. FEBS Lett (1997) 407:63–8. doi: 10.1016/S0014-5793(97)00294-9
20. Peach RJ, Bajorath J, Brady W, Leytze G, Greene J, Naemura J, et al. Complementarity determining region 1 (CDR1)- and CDR3-analogous regions in CTLA4 and CD28 determine the binding to B7-1. J Exp Med (1994) 180:2049–58. doi: 10.1084/jem.180.6.2049
21. Vaughan AN, Malde P, Rogers NJ, Jackson IM, Lechler RI, Dorling A. Porcine CTLA4-ig lacks a MYPPPY motif, binds inefficiently to human B7 and specifically suppresses human CD4+ T cell responses costimulated by pig but not human B7. J Immunol (2000) 165:3175–81. doi: 10.4049/jimmunol.165.6.3175
22. Metzler WJ, Bajorath J, Fenderson W, Shaw SY, Constantine KL, Naemura J, et al. Solution structure of human CTLA4 and delineation of a CD80/CD86 binding site conserved in CD28. Nat Struct Biol (1997) 4:527–31. doi: 10.1038/nsb0797-527
23. Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, et al. xAn update on the use and investigation of probiotics in health and disease. Gut (2013) 62:787–96. doi: 10.1136/gutjnl-2012-302504
24. Klaenhammer TR, Barrangou R, Buck BL, Azcarate-Peril MA, Altermann E. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol Rev (2005) 29:393–409. doi: 10.1016/j.femsre.2005.04.007
25. Coelho-Rocha ND, de Castro CP, de Jesus LCL, Leclercq SY, de Cicco Sandes SH, Nunes AC, et al. Microencapsulation of lactic acid bacteria improves the gastrointestinal delivery and in situ expression of recombinant fluorescent protein. Front Microbiol (2018) 9:2398. doi: 10.3389/fmicb.2018.02398
26. Del Rio B, Redruello B, Fernandez M, Martin MC, Ladero V, Alvarez MA. Lactic acid bacteria as a live delivery system for the in situ production of nanobodies in the human gastrointestinal tract. Front Microbiol (2019) 9:3179. doi: 10.3389/fmicb.2018.03179
27. Bermúdez-Humarán LG, Kharrat P, Chatel JM, Langella P. Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb Cell Fact (2011) 10:S4. doi: 10.1186/1475-2859-10-S1-S4
28. Mohamadzadeh M, Duong T, Sandwick SJ, Hoover T, Klaenhammer TR. Dendritic cell targeting of bacillus anthracis protective antigen expressed by lactobacillus acidophilus protects mice from lethal challenge. Proc Natl Acad Sci U.S.A. (2009) 106:4331–6. doi: 10.1073/pnas.0900029106
29. Kuczkowska K, Copland A, Øverland L, Mathiesen G, Tran AC, Paul MJ, et al. Inactivated Lactobacillus plantarum carrying a surface-displayed ag85b-esat-6 fusion antigen as a booster vaccine against mycobacterium tuberculosis infection. Front Immunol (2019) 10:1588. doi: 10.3389/fimmu.2019.01588
30. Niu H, Xing JH, Zou BS, Shi CW, Huang HB, Jiang YL, et al. Immune evaluation of recombinant Lactobacillus plantarum with surface display of HA1-DCpep in mice. Front Immunol (2021) 12:800965. doi: 10.3389/FIMMU.2021.800965
31. Rigsby RE, Parker AB. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ (2016) 44(5):433–7. doi: 10.1002/bmb.20966
32. Macri C, Dumont C, Johnston AP, Mintern JD. Targeting dendritic cells: a promising strategy to improve vaccine effectiveness. Clin Transl Immunol (2016) 5:e66. doi: 10.1038/cti.2016.6
33. Barroeta Seijas AB, Simonetti S, Vitale S, Runci D, Quinci AC, Soriani A, et al. GM-CSF inhibits c-kit and SCF expression by bone marrow-derived dendritic cells. Front Immunol (2017) 8:147. doi: 10.3389/fimmu.2017.00147
34. Nedumpun T, Ritprajak P, Suradhat S. Generation of potent porcine monocyte-derived dendritic cells (MoDCs) by modified culture protocol. Vet Immunol Immunopathol (2016) 182:63–8. doi: 10.1016/j.vetimm.2016.10.002
35. Cho KJ, Roche PA. Regulation of MHC class II-peptide complex expression by ubiquitination. Front Immunol (2013) 4:369. doi: 10.3389/fimmu.2013.00369
36. Hartmann SB, Mohanty S, Skovgaard K, Brogaard L, Flagstad FB, Emnéus J, et al. Investigating the role of surface materials and three dimensional architecture on in vitro differentiation of porcine monocyte-derived dendritic cells. PloS One (2016) 11:e0158503. doi: 10.1371/journal.pone.0158503
37. Zaneti AB, Yamamoto MM, Sulczewski FB, Almeida BDS, Souza HFS, Ferreira NS, et al. Dendritic cell targeting using a DNA vaccine induces specific antibodies and CD4+ T cells to the dengue virus envelope protein domain III. Front Immunol (2019) 10:59. doi: 10.3389/fimmu.2019.00059
38. Wang M, Fu T, Hao J, Li L, Tian M, Jin N, et al. A recombinant Lactobacillus plantarum strain expressing the spike protein of SARS-CoV-2. Int J Biol Macromol (2020) 160:736–40. doi: 10.1016/j.ijbiomac.2020.05.239
39. Mercenier A, Müller-Alouf H, Grangette C. Lactic acid bacteria as live vaccines. Curr Issu Mol Biol Curr Issues Mol Biol (2000) 2:17–25. doi: 10.21775/cimb.002.017
40. Yang X, Guo Y, Chen C, Shao B, Zhao L, Zhou Q, et al. Interaction between intestinal microbiota and tumour immunity in the tumour microenvironment. Immunology (2021) 164(3):476–93. doi: 10.1111/imm.13397
41. Yang WT, Yang GL, Yang X, Shonyela SM, Zhao L, Jiang YL, et al. Recombinant lactobacillus plantarum expressing HA2 antigen elicits protective immunity against H9N2 avian influenza virus in chickens. Appl Microbiol Biotechnol (2017) 101(23–24):8475–84. doi: 10.1007/s00253-017-8600-2
42. Schwartz JC, Zhang X, Fedorov AA, Nathenson SG, Almo SC. Structural basis for co-stimulation by the human CTLA-4/B7-2 complex. Nature (2001) 410(6828):604–8. doi: 10.1038/35069112
43. Schwartz JC, Zhang X, Nathenson SG, Almo SC. Structural mechanisms of costimulation. Nat Immunol (2002) 3(5):427–34. doi: 10.1038/ni0502-427
44. Stamper CC, Zhang Y, Tobin JF, Erbe DV, Ikemizu S, Davis SJ, et al. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature (2001) 410(6828):608–11. doi: 10.1038/35069118
45. Liu X, Xia X, Wang X, Zhou J, Sung LA, Long J, et al. Tropomodulin1 expression increases upon maturation in dendritic cells and promotes their maturation and immune functions. Front Immunol (2021) 11:587441. doi: 10.3389/fimmu.2020.587441
46. Ullah MO, Sweet MJ, Mansell A, Kellie S, Kobe B. TRIF-dependent TLR signaling, its functions in host defense and inflammation, and its potential as a therapeutic target. J Leukoc Biol (2016) 100(1):27–45. doi: 10.1189/jlb.2RI1115-531R
47. Dalod M, Chelbi R, Malissen B, Lawrence T. Dendritic cell maturation: functional specialization through signaling specificity and transcriptional programming. EMBO J (2014) 33(10):1104–16. doi: 10.1002/embj.201488027
48. Teng MW, Bowman EP, McElwee JJ, Smyth MJ, Casanova JL, Cooper AM, et al. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med (2015) 21(7):719–29. doi: 10.1038/nm.3895
49. Lee AJ, Ashkar AA. The dual nature of type I and type II interferons. Front Immunol (2018) 9:2061. doi: 10.3389/fimmu.2018.02061
50. Jung HJ, Park SH, Cho KM, Jung KI, Cho D, Kim TS. Threonyl-tRNA synthetase promotes T helper type 1 cell responses by inducing dendritic cell maturation and IL-12 production via an NF-κB pathway. Front Immunol (2020) 11:571959. doi: 10.3389/fimmu.2020.571959
51. Noreen M, Arshad M. Association of TLR1, TLR2, TLR4, TLR6, and TIRAP polymorphisms with disease susceptibility. Immunol Res (2015) 62(2):234–52. doi: 10.1007/s12026-015-8640-6
52. Carter RW, Thompson C, Reid DM, Wong SY, Tough DF. Preferential induction of CD4+ T cell responses through in vivo targeting of antigen to dendritic cell-associated c-type lectin-1. J Immunol (2006) 177:2276–84. doi: 10.4049/jimmunol.177.4.2276
53. Chen CY, Liu HJ, Tsai CP, Chung CY, Shih YS, Chang PC, et al. Baculovirus as an avian influenza vaccine vector: differential immune responses elicited by different vector forms. Vaccine (2010) 28:7644–51. doi: 10.1016/j.vaccine.2010.09.048
54. Holmgren J, Czerkinsky C. Mucosal immunity and vaccines. Nat Med (2005) 11(4):45–53. doi: 10.1038/nm1213
55. Mantis NJ, Rol N, Corthesy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol (2011) 4:603–11. doi: 10.1038/mi.2011.41
56. Gao X, Ma Y, Wang Z, Bai J, Jia S, Feng B, et al. Oral immunization of mice with a probiotic lactobacillus casei constitutively expressing the alpha-toxoid induces protective immunity against clostridium perfringens alpha-toxin. Virulence (2019) 10:166–79. doi: 10.1080/21505594.2019.1582975
Keywords: porcine dendritic cells, CTLA4, targeting, recombinant lactobacillus, fusion expression
Citation: Xia T, Wang N, Tang Y, Gao Y, Gao C, Hao J, Jiang Y, Wang X, Shan Z, Li J, Zhou H, Cui W, Qiao X, Tang L, Wang L and Li Y (2022) Delivery of antigen to porcine dendritic cells by fusing antigen with porcine dendritic cells targeting peptide. Front. Immunol. 13:926279. doi: 10.3389/fimmu.2022.926279
Received: 12 May 2022; Accepted: 18 August 2022;
Published: 08 September 2022.
Edited by:
Urban Švajger, Blood Transfusion Centre of Slovenia, SloveniaReviewed by:
Joseph M. Reynolds, Rosalind Franklin University of Medicine and Science, United StatesMilica M. Perisic Nanut, Institut Jožef Stefan (IJS), Slovenia
Copyright © 2022 Xia, Wang, Tang, Gao, Gao, Hao, Jiang, Wang, Shan, Li, Zhou, Cui, Qiao, Tang, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yijing Li, eWlqaW5nbGlAMTYzLmNvbQ==; Li Wang, d2FuZ2xpY2F1QDE2My5jb20=
†These authors have contributed equally to this work
 Tian Xia1†
Tian Xia1† Xiaona Wang
Xiaona Wang Xinyuan Qiao
Xinyuan Qiao Li Wang
Li Wang

