- 1Department of Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States
- 2Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States
- 3Department of Pediatrics, Children’s Hospital Los Angeles, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States
Colorectal cancer (CRC) is one of the leading causes of cancer-related death in the world. Besides genetic causes, colonic inflammation is one of the major risk factors for CRC development, which is synergistically regulated by multiple components, including innate and adaptive immune cells, cytokine signaling, and microbiota. The complex interaction between CRC and the gut microbiome has emerged as an important area of current CRC research. Metagenomic profiling has identified a number of prominent CRC-associated bacteria that are enriched in CRC patients, linking the microbiota composition to colitis and cancer development. Some microbiota species have been reported to promote colitis and CRC development in preclinical models, while a few others are identified as immune modulators to induce potent protective immunity against colitis and CRC. Mechanistically, microbiota regulates the activation of different immune cell populations, inflammation, and CRC via crosstalk between innate and adaptive immune signaling pathways, including nuclear factor kappa B (NF-κB), type I interferon, and inflammasome. In this review, we provide an overview of the potential interactions between gut microbiota and host immunity and how their crosstalk could synergistically regulate inflammation and CRC, thus highlighting the potential roles and mechanisms of gut microbiota in the development of microbiota-based therapies to prevent or alleviate colitis and CRC.
1 Introduction
Colorectal cancer (CRC) is one of the most common cancers and a major health burden in the world, which accounts for about 10% of all new cancer cases globally and becomes the second leading cause of cancer-related death (1–3). About half of the human population will develop at least one benign colonic adenomatous polyp during their lifetime, with ~3% of these cases developing into CRC (4). Besides genetic alterations, colonic inflammation is a major environmental risk factor for CRC development, as the individuals diagnosed with ulcerative colitis (UC) and Crohn’s disease (CD), the most common types of inflammatory bowel diseases (IBDs), have markedly increased risk of developing CRC (5–7). Although immune cells, cytokines, and microbiota components contribute to colitis and CRC in a context-dependent manner (8, 9), the precise mechanisms remain largely unclear.
The human gut is a complex ecosystem composed of 1013-1014 bacteria, and these microbiota components play critical roles in controlling digestion and benefiting many aspects of human health. Some probiotics have been exploited as food supplements to support the health of the immune and digestive system, and even as novel therapies for disease treatment. Numerous studies have demonstrated that the microbiota composition is significantly altered in both IBD and CRC patients, compared with healthy people. Recent metagenomic profilings have identified multiple prominent CRC-associated bacteria enriched in patients, including the components in genera Fusobacterium, Peptostreptococcus, Porphyromonas, Prevotella, Parvimonas, Bacteroides, and Gemella (10). As most studies link mixed microbiota composition to disease progression, some species have been reported to promote colitis and CRC (11–14); however, only a few bacterial species or strains are identified as immune modulators that induce potent protective immunity. In this review, we provide an overview of the crosstalk between gut microbiota and host immunity and how it regulates inflammation and CRC development. Importantly, microbiota has been implicated in cancer immunotherapy. Due to the lack of clinical evidence on the functions of viruses and fungi, we will mainly focus on bacteria species, as the major component of microbiota.
2 Genetic risk factors in CRC development
CRC is a heterogeneous disease associated with a number of genetic mutations (15), with 10%-20% of all patients possessing a positive family history (16). Besides traditional methods, genome-wide sequencing analyses have been performed to depict the genomic landscape and transcriptome profile, thus allowing the establishment of key promoting and suppressing alterations in CRC development (17).
2.1 Genetic alterations in epithelial cells
Most CRC cases arise from an aberrant crypt on the colonic epithelium, which gradually develops into an adenoma, and ultimately adenocarcinoma (18). The genetic alterations in colon epithelial cells, as the primary site for CRC development, play critical roles in the disease transformation (19). To date, several hundred driver genes have been identified to promote neoplastic transformation by intragenic mutations, with other mutations being passengers that are not associated with selective growth advantages (20). It has been widely accepted that mutations in three major gene clusters, APC, KRAS, and TP53, are sufficient to initiate CRC (21, 22), while BRAF, PIK3CA, and SMAD4, which are also listed as the most frequently mutated genes in CRC (23), are identified as important drivers to promote the progression (24).
Although similar sets of oncogenes are involved, the genetic alterations between sporadic and colitis-associated CRC (CAC) are different in timing and frequency. In sporadic CRC, loss of APC function is a key event to initiate an adenoma, followed by the activation of KRAS, COX-2, and other factors; whereas the abnormality of TP53 usually occurs in the late stage of disease progression and drug resistance (25, 26). On the contrary, in CAC that arises from flat dysplastic mucosa, TP53 mutation is frequently detected in inflamed tissues and is an important step in early cancer development. APC mutation and Wnt dysfunction are relatively infrequently in CAC and occur in the late stage (25, 26).
Interestingly, the pathogenesis and molecular characteristics of CRC also depend on the anatomical locations of the tumors (27), particularly between the proximal (right-sided) and distal (left-sided) colons. Right-sided colon cancer is characterized by the alterations in BRAF, KRAS, and PIK3CA, and has a higher rate of deficient mismatch repair; whereas instability pathway-related APC and TP53 mutations are more frequently observed in left-sided colon cancer, which has a better response to both chemotherapies and targeted therapies, thus showing better prognosis in patients (28, 29). Meanwhile, rectal cancer is featured by the mutations in all three major genes (APC, KRAS, and TP53) and HER2 amplification, and has a lower rate of deficient mismatch repair (29).
2.2 Immune cell-related genetic alterations in CRC development
Besides alterations in colon epithelia as the primary foci, aberrant changes in the immune microenvironment also have profound impacts on the initiation and progression of CRC. One major evidence is that the risk of CRC development is increased in IBD (inflammatory bowel disease) patients (5, 6). It has been demonstrated that the age at diagnosis of IBD-associated CRC is 15-20 years earlier compared to sporadic cancers (30, 31), and CRC accounts for approximately 10%-15% of all deaths in IBD patients (32).
Given the critical roles of the host immune cells and cytokines in controlling CRC development, genetic alterations on immune-related genes or in immune cells, in addition to colon epithelial cells, are equally important for CRC pathogenesis. For instance, IL-10 is an immunoregulatory cytokine that plays a central role in controlling intestinal inflammation (33). Mice deficient in either IL-10 or its receptor develop spontaneous colitis, and became one of the most widely used animal models for studying IBD pathogenesis (34, 35). Furthermore, deficiency in ubiquitin ligase Itch leads to spontaneous colitis and increased susceptibility to CRC through the release of RORγt degradation and excessive production of IL-17A (36). A study shows that the deficiency of Fam64a in mice decreases Th17 cells and ameliorates colitis and CRC (37). These studies indicate that modulation of Th17 cell-related genes has a significant impact on CRC development, although the pro-inflammatory or suppressive roles of these Th17 cells are not validated in detail.
We recently showed that the myeloid-specific deletion of Tak1 (Tak1flox/flox;Lyz2-Cre+/+) renders complete resistance of mice to DSS-induced acute colitis and AOM/DSS-induced CRC (38). Notably, gut microbiota compositions are completely altered in Tak1flox/flox;Lyz2-Cre+/+ mice, compared to wild-type mice. Among them, Odoribacter splanchnicus is markedly accumulated and synergistically cooperated with IL-1β/IL-6 signaling pathways to induce and expand Th17 cells in the intestine. Depletion of Th17 cells by crossing Tak1flox/flox;Lyz2-Cre+/+ mice with either Rag1-/- or Rorc-/- mice abolishes the protection against colitis and CRC (38).
In summary, the genetic alterations of key driver genes in both epithelial cells and immune cells, together with other driver and passenger factors, are crucial in controlling the carcinogenesis and progression of CRC.
3 Gut microbiota in CRC development
In healthy people, colonocytes and their metabolism maintain the anaerobic condition and a homeostatic community of commensal bacteria in the gut, which help consume dietary fiber and produce short-chain fatty acids (SCFA) that are beneficial to the host (39). The shift in colonocytes and their metabolism, due to disease, diet, or other damage, will lead to disordered host-commensal symbiosis and dysbiotic microbiota (39). It has been reported that CRC patients have reduced bacterial diversity and richness than healthy people (40, 41). Whereas Firmicutes, Bacteroidetes, and Proteobacteria are the most dominant phyla in the human large bowel (42), Fusobacterium, Peptostreptococcus, Porphyromonas, Prevotella, Parvimonas, Bacteroides, and Gemella have been indicated as the most prominent CRC-associated bacteria (10), based on metagenomic sequencing analyses between CRC patients and healthy donors.
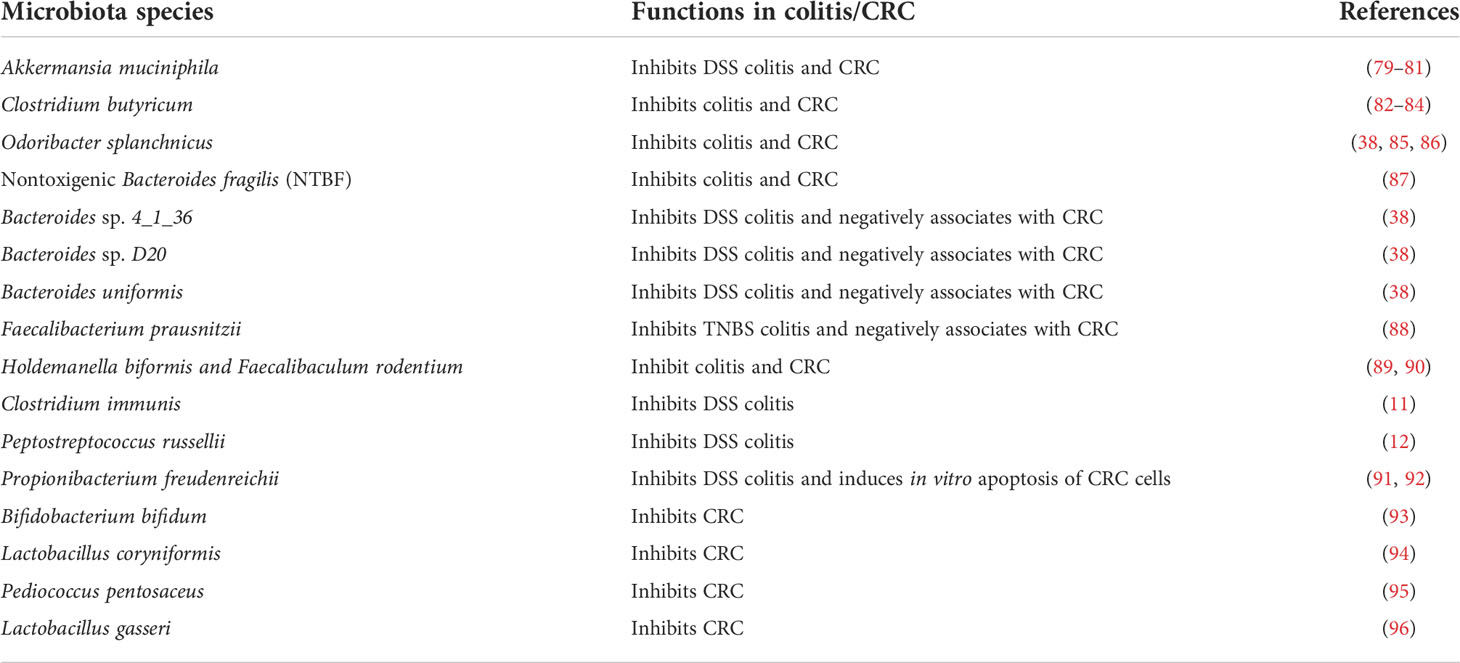
Meanwhile, commensal microbiota has been implicated in modulating colitis, CRC, and cancer immunotherapy (43–47). Depending on specific composition, commensal bacteria may exhibit either promoting or suppressing functions in colitis and CRC development (48–52). Long-term antibiotic use in early-to-middle adulthood is associated with an increased risk of colorectal adenoma (53). Similarly, in a mouse colitis model, depletion of microbiota exacerbated tissue damage and shortened survival (54), which is associated with compromised immunity due to the lack of bacterial stimulation. On the other hand, administration of a common antimicrobial additive, Triclosan, alters mouse gut microbiota, increases the severity of colitis symptoms, and promotes colitis-associated CRC in mouse disease models (55). Several CRC-associated bacteria have been identified in cancer patients and animal models (56); however, the understanding of specific bacterial species or strains that induce and modulate anti-tumor immunity is still limited. Below we list the currently identified promoting (Table 1) and inhibiting (Table 2) microbiota species in colitis and CRC development.
3.1 Cancer-promoting microbiota species
3.1.1 Fusobacterium nucleatum
Fusobacterium nucleatum is an anaerobic oral commensal. As a pro-inflammatory species associated with human colitis (97), it has been widely reported to be positively associated with human CRC (98, 99). Accompanied by bacterial dysbiosis in the gut, an infection with this bacteria is prevalent in human colorectal carcinoma (98). In different clinical reports, Fusobacterium nucleatum has 8.6% and 13% of colonization in CRC tissues, and is associated with increased microsatellite instability (MSI) and impaired immune responses (100, 101). Furthermore, Fusobacterium nucleatum is implicated in accelerating CRC in both human patients and animal models, and is found within metastatic CRC cells in patient biopsies (63, 102). In specific, it adheres to, invades, and induces E-cadherin/β-catenin signaling-mediated oncogenic and inflammatory responses to stimulate CRC carcinogenesis (103–105). CRC cell-resident Fusobacterium nucleatum also promotes the secretion of pro-inflammatory cytokines IL-8 and CXCL1, which in turn stimulate the migration and invasion of both infected and noninfected tumor cells (106). In addition, Fusobacterium nucleatum promotes chemotherapy resistance of CRC through TLR4- and MYD88-mediated innate immune signaling and autophagy pathway (64).
3.1.2 Escherichia coli
Escherichia coli is a Gram-negative facultative anaerobic bacterium in the Enterobacteriaceae family, and the adherent-invasive Escherichia coli species have been associated with human inflammatory bowel disease (IBD) and CRC (42, 107, 108). Under host inflammatory conditions, the mono-colonization of Escherichia coli promotes colitis in Il10−/− mice and invasive carcinoma in azoxymethane (AOM)-treated Il10−/− mice (60, 61). Mechanistically, Escherichia coli produces the genotoxin colibactin through the non-ribosomal peptide synthetase (NRPS)-polyketide synthase (PKS) hybrid gene cluster (109). Colibactin further alkylates DNA and induces double-strand breaks, aneuploidy, and improper division of colonic epithelial cells (60, 109). In this case, the PKS+ Escherichia coli enhances tumorigenesis in preclinical CRC models and is enriched in human CRC tissues. Using optical imaging tools, the massive infiltration of inflammatory cells is also observed in PKS+ Escherichia coli-infected colon tumors, compared with the uninfected group (110).
3.1.3 Enterotoxigenic Bacteroides fragilis
The anaerobic Gram-negative Enterotoxigenic Bacteroides fragilis (ETBF) is a long-studied human GI pathogen that causes diarrhea and GI inflammation (111, 112). In preclinical models, ETBF potentiates colorectal carcinogenesis in ApcMin/+ mice through STAT3 activation and Th17 cell-dependent colitis (57). Furthermore, ETBF and Escherichia coli are detected in biofilms coating human CRCs and precancerous colonic adenomas (58). Tumor-prone mice co-colonized with Escherichia coli and ETBF show increased IL-17A level in the colon and DNA damage in colonic epithelia, with faster tumor onset and greater mortality, compared to mice with either bacterial strain alone (58). Similar to Escherichia coli, ETBF could produce a metalloprotease toxin BFT (Bacteroides fragilis enterotoxin), which has the proteolytic activity to damage the intestinal mucosa and induces a pro-carcinogenic signaling cascade to trigger myeloid-cell-dependent colon tumorigenesis (59, 113).
3.1.4 Campylobacter jejuni
Campylobacter jejuni is a Gram-negative microaerophilic bacterium that has been considered one of the most widespread infectious diseases in developed countries (114). It produces a genotoxin, cytolethal distending toxin (CDT), which has DNAse activity and leads to DNA double-strand breaks (115). The infection of this species is associated with IBD development in human patients (116), and induces colitis in mouse models (65). In germ-free ApcMin/+ mice, Campylobacter jejuni infection promotes colorectal tumorigenesis through the action of CDT (66). As a result, CDT mutation and rapamycin treatment could similarly diminish the tumorigenic capability of Campylobacter jejuni (66).
3.1.5 Enterococcus faecalis
Enterococcus faecalis is a Gram-positive facultative anaerobic bacterium that naturally inhabits the human gastrointestinal tract, and the spread of this bacterium to other organs or tissues can cause severe infection (117). Unlike most other bacteria, this species produces reactive oxygen species (ROS), such as extracellular superoxide (118), thus leading to DNA damage, chromosomal instability, generation of aneuploidy or tetraploidy, and eventually transformation and tumorigenesis of colonic epithelial cells (119, 120). More than half of patients with Enterococcus faecalis infective endocarditis (EFIE) of an unidentifiable source are found to have CRC (121). In Il10−/− mice, Enterococcus faecalis promotes colitis development and colorectal tumorigenesis (67, 68).
3.1.6 Streptococcus bovis
Streptococcus bovis (also known as Streptococcus gallolyticus) is a facultative anaerobic Gram-positive bacterium that serves as a causative agent of septicemia and infective endocarditis (IE) in elderly and immunocompromised people (122). Clinical studies have demonstrated a strong association between invasive infections of Streptococcus bovis and colon neoplasia (123, 124). Consistently, both in vivo and in vitro studies validate the pro-inflammatory and CRC-promoting functions of multiple Streptococcus bovis strains (69, 70, 125, 126). Since Streptococcus bovis is still a normal intestinal tract inhabitant, it may have both passenger and driver functions in CRC tumorigenesis.
3.1.7 Peptostreptococcus anaerobius
Peptostreptococcus anaerobius is an anaerobic Gram-positive bacterium selectively enriched in fecal and mucosal microbiota of CRC patients (71). Although it can produce tryptophan metabolite indoleacrylic acid, which may attenuate inflammatory response and improve barrier function (12), the direct transfer of Peptostreptococcus anaerobius into AOM-treated mice significantly increases colon dysplasia (71). Similarly, in ApcMin/+ mice, it promotes spontaneous CRC development (72). Mechanistically, Peptostreptococcus anaerobius selectively adheres to CRC cells, rather than normal colonic epithelial cells, through a surface protein PCWBR2 (putative cell wall binding repeat 2). PCWBR2 stimulates CRC cells to produce pro-inflammatory cytokines, which in turn mediate the local expansion of tumor-supportive MDSCs, TAMs, and TANs (72).
3.1.8 Other potential cancer-promoting microbiota species
Besides the microbiota species that have been validated in either preclinical or clinical functional studies, several other species also show strong correlations with colitis and CRC development and may serve as potential targets for future investigation on CRC management.
Helicobacter pylori is a Gram-negative capnophile that can grow in both microaerobic and aerobic conditions. It selectively colonizes the gastric epithelia, and is considered one of the most prevalent bacterial pathogens in humans. Helicobacter pylori induces chronic gastritis and is associated with more than 90% of gastric cancers (GC) cases (73, 127, 128), making it a class I carcinogen for GC. Although many reports show that chronic infection of Helicobacter pylori is associated with a moderately increased risk of CRC (74, 129, 130), direct evidence from functional studies is lacking.
Mycobacterium avium is a microaerobic Gram-positive mycobacterium that is commonly grouped with Mycobacterium intracellulare during infection, collectively referred to as Mycobacterium avium complex (MAC). Mycobacterium avium subspecies paratuberculosis (MAP) has long been proposed as a cause of IBD (131, 132); it is increased in IBD patients (75) and can be observed in colon tissues of sporadic CRC patients (76). However, direct functional study of this species is lacking.
Bilophila wadsworthia is an anaerobic Gram-negative saccharolytic bacillus that is a major member of sulfidogenic bacteria in human gut (77). It can produce a genotoxin, hydrogen sulfide, which triggers inflammation and hyperproliferation (Yazici et al., 2017). Sulfidogenic bacteria, including Bilophila wadsworthia, have a race-dependent association with CRC incidence and is expanded in the population with a higher risk of CRC development (Yazici et al., 2017). In genetically susceptible Il10-/- mice, diet-induced blooming of Bilophila wadsworthia promotes the pro-inflammatory Th1 immune response and an increased incidence of colitis (Devkota et al., 2012). The direct administration of Bilophila wadsworthia into specific-pathogen-free (SPF) mice results in systemic inflammation, with reduced body weight and fat mass, apparent hepatosplenomegaly, and elevated serum inflammatory factors (77). However, how Bilophila wadsworthia mediated inflammation may impact tumorigenesis is currently unclear.
3.2 Cancer-inhibiting microbiota species
3.2.1 Akkermansia muciniphila
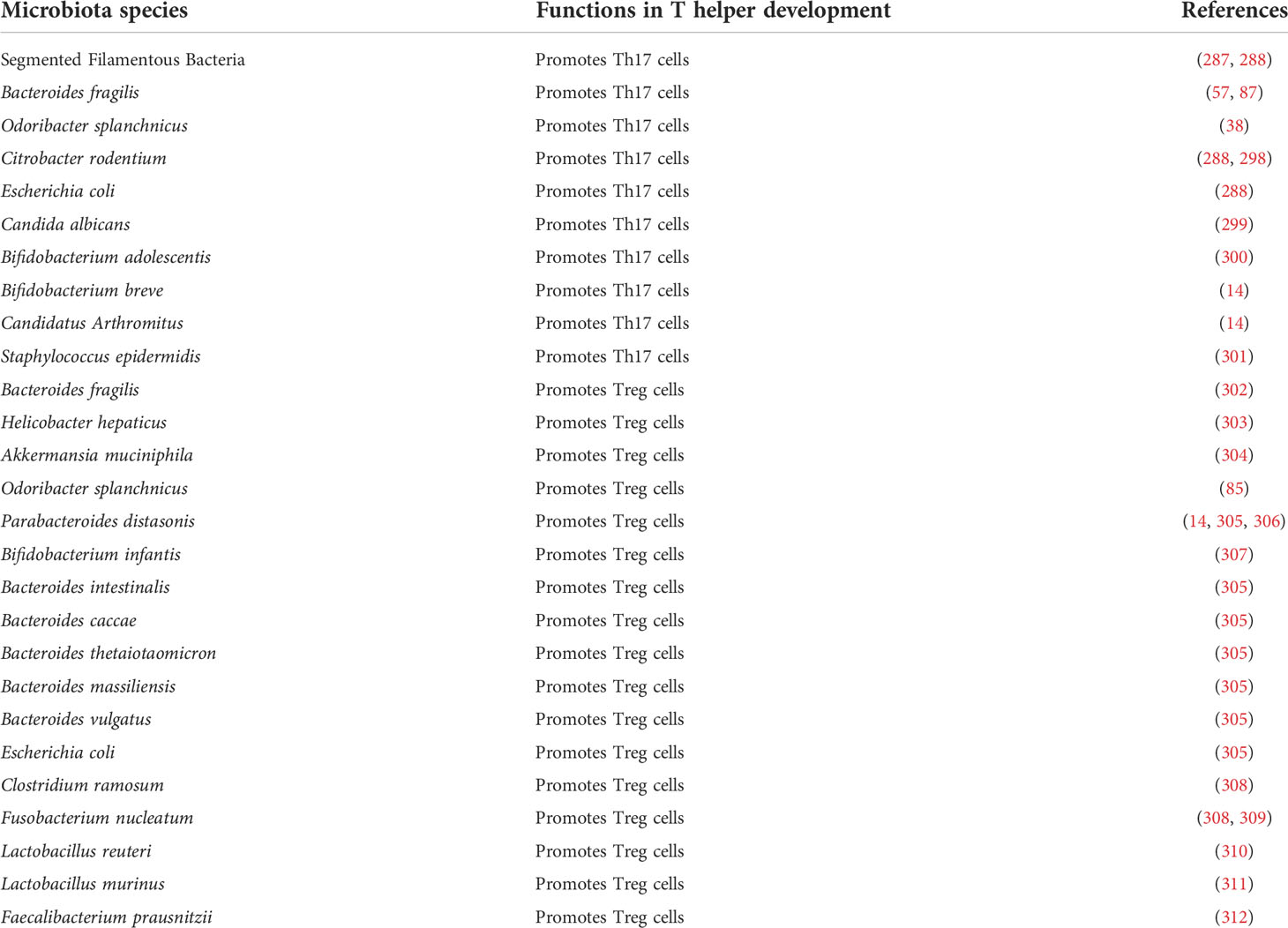
Akkermansia muciniphila is a strictly anaerobic Gram-negative bacterium that resides in the mucus layer and plays a mucin-degrading function in the human intestine (133). It interplays with the intestinal epithelium for nutrition management and controls diet-induced obesity through improved metabolic profiles (134, 135). In preclinical models, Akkermansia muciniphila is positively associated with the induction of CRC in mouse recipients of human fecal transplant (136), and is significantly reduced in CRC-resistant Tak1flox/flox;Lyz2-Cre+/+ mice (38). In humans, its abundance is decreased in most colitis patients but increased in CRC patients (137). Further studies found that Akkermansia muciniphila preferentially expands and colonizes sites of damaged murine mucosa in response to local environmental cues (79), which probably explains the pattern of its distribution and abundance. Notably, Akkermansia muciniphila stimulates the proliferation and migration of enterocytes adjacent to the colonic wounds, through FPR1 (formyl peptide receptor 1) and NOX1 (NADPH Oxidase 1)-mediated redox signaling in epithelial cells, thus enhancing the repair of mucosal wounds and protecting mice from chemically induced colitis (79). A similar protective role is observed in another DSS-induced colitis model, with an improved microbial community (80). In AOM/DSS-induced CAC model, Akkermansia muciniphila treatment could blunt carcinogenesis by enhancing cytotoxic CD8+ T cells (81).
3.2.2 Clostridium butyricum
Clostridium butyricum is a strictly anaerobic Gram-positive butyrate-producing bacillus that is a dietary probiotic for healthy people and an effective approach to IBD treatment (138). In DSS colitis model, Clostridium butyricum directly triggers TLR2/MyD88-dependent IL-10 production by intestinal macrophages in inflamed mucosa to prevent colitis development, and this prevention can be negated in macrophage-specific IL-10-deficient mice (82). In ApcMin/+ mice, Clostridium butyricum inhibits intestinal tumor development by decreasing β-catenin expression in Wnt signaling and modulating gut microbiota (83). Similarly, in AOM/DSS model, Clostridium butyricum regulates gut microbiota composition and reduces CRC development by inhibiting the NF-κB pathway and promoting apoptosis (84).
3.2.3 Odoribacter splanchnicus
Odoribacter splanchnicus, a strictly anaerobic Gram-negative bacterium, is a common member of human intestinal microbiota. Although it is enriched in colorectal adenoma and CRC patients (139), recent studies have identified this species as a CRC-inhibiting and -preventive bacterium. In a preclinical model, treatment with wild-mice microbiota renders normal laboratory mice resistant to CRC, and Odoribacter ranks among the top increased genera after microbiota reconstruction (13). Strikingly, a recent report has characterized Odoribacter splanchnicus as a critical species to protect the host from colitis and CRC (38). This species is highly abundant in a CRC-resistant mouse model (Tak1flox/flox;Lyz2-Cre+/+). Oral transfer of Odoribacter splanchnicus into wild-type (WT) mice induces development of immune-suppressive intestinal Th17 cells, and confers resistance against colitis and CRC (38), probably via increased productions of IL-17A and IL-22 (140, 141). Similar results are observed in a separate report, in which Odoribacter splanchnicus colonization leads to an increase in Foxp3+/RORγt+ Treg cells, induction of IL-10, and production of SCFA, thus reducing colitis in mouse models (85). Furthermore, treatment of Odoribacter splanchnicus supernatant in colon cancer cell lines induces an anti-tumor activity with enhanced apoptosis, and peri-tumoral injection of supernatant significantly decreases CRC formation (86).
3.2.4 Inhibitory Bacteroides species
Bacteroides species are anaerobic Gram-negative bacilli that are normally mutualistic, making up the most substantial portion of commensal microbiota. Some species, such as ETBF, are reported to promote colitis and CRC, while some others have been identified as anti-tumor players. In both WT and CRC mouse models, treatment of nontoxigenic Bacteroides fragilis (NTBF) reduces bacteria-driven chronic colitis and tumor development (87). Recently, Bacteroides sp. 4_1_36, Bacteroides sp. D20, and Bacteroides uniformis are found to accumulate in a CRC-resistant mouse model and significantly inhibit the development of DSS-induced colitis (38). While Bacteroides sp. 4_1_36 and Bacteroides sp. D20 are less reported, Bacteroides uniformis has a reduced abundance in CRC patients (142) and is reported to improve immunological dysfunction and enhance the gut barrier through the production of butyrate and gamma-aminobutyric acid (143, 144).
3.2.5 Faecalibacterium prausnitzii
Faecalibacterium prausnitzii, a Gram-positive anaerobic bacterium, is one of the most abundant and important commensal bacteria in human intestine (145). As a key butyrate producer, the abundance of this bacterium is negatively associated with colon tumorigenesis in multiple scenarios (146–148). In IBD patients, the reduction of Faecalibacterium prausnitzii is associated with a higher risk of postoperative recurrence (88). Furthermore, in TNBS (2,4,6-trinitrobenzenesulphonic acid)-induced mouse colitis model, both live Faecalibacterium prausnitzii and its supernatant exhibit anti-inflammatory effects and markedly ameliorate colitis severity and dysbiosis (88). Mechanistically, metabolites from this species block NF-κB activation in colon epithelial cells and switch the cytokine profile (decreased IFN-γ and IL-12, increased IL-8 and IL-10) (88). However, direct evidence from functional study is in need to determine the role of this species in CRC.
3.2.6 Other potential cancer-inhibiting microbiota species
Holdemanella biformis (formerly Eubacterium biformis) is a Gram-positive obligately anaerobic bacterium that can release both short chain fatty acids (SCFAs) and long chain fatty acids (LCFAs). The abundance of this species and its family Erysipelotrichaceae are reduced in human patients with colon adenomas (89). Although Holdemanella biformis is not able to colonize or survive in the mouse intestine, its mouse homologue, Faecalibaculum rodentium, exhibits anti-tumorigenic function in both ApcMin/+ and AOM/DSS models (89). Mechanistically, both species produce SCFAs that control protein acetylation and tumor cell proliferation by inhibiting calcineurin and NFATc3 activation (89). Holdemanella biformis can also produce 3-hydroxyoctadecaenoic acid (C18-3OH), a LCFA that ameliorates the progression of DSS-induced colitis (90).
Clostridium immunis, an anaerobic Gram-positive bacterium, is a relatively new species identified in the Lachnospiraceae family (11). Lachnospiraceae is dramatically increased in a CRC-resistant mouse model and decreased in Crohn’s disease patients (38, 149), and negatively correlates with CRC development in mouse recipients of human fecal transplant (136). Administration of Clostridium immunis protects formerly colitis-prone mice from DSS-induced colitis (11). Further functional and clinical studies are needed to evaluate the potential of this species as a candidate to control CRC development.
Peptostreptococcus russellii is a Gram-positive anaerobic bacterium that naturally exists in healthy people. It has an enhanced growth rate in the presence of mucin and is thus identified as a “mucin utilizer” (12), which predicts the potential of being a health-associated commensal, such as the CRC-inhibiting Akkermansia muciniphila (133). Oral gavage of Peptostreptococcus russellii protects mice from DSS-induced colitis, with significantly ameliorated body mass and histopathological score (12). It also promotes goblet cell differentiation in colon and the expression of goblet cell-specific secreted protein MUC2. Mechanistically, Peptostreptococcus russellii encodes the phenyllactate gene cluster and produces tryptophan metabolite indoleacrylic acid, which promotes intestinal epithelial barrier function and mitigates inflammatory responses (12).
Propionibacterium freudenreichii is a Gram-positive aerotolerant anaerobe that selectively stimulates the growth of probiotic Bifidobacteria through its component DHNA (1.4-Dihydroxy-2-naphthoic acid) (91). In DSS colitis model, the treatment of DHNA shows both preventive and therapeutic effects in disease amelioration (91). A further study shows that this species induces intrinsic apoptosis of CRC cells via the production of SCFA (propionate and acetate), and thus enhances cytotoxic activity of TRAIL (TNF-Related Apoptosis-Inducing Ligand)-based therapy in CRC (92). Besides, Propionibacterium freudenreichii treatment in healthy people decreases the activity of beta-glucosidase (150), a bacterial enzyme that contributes to CRC development by generating carcinogens. However, direct evidence regarding its function in CRC is lacking.
Recently, the functions of some established natural and engineered probiotics have been investigated in AOM/DSS-induced mouse CRC model. Bifidobacterium bifidum treatment increases the abundance of CRC-inhibiting microbiota and the production of beneficial metabolites, thus protecting mice from tumorigenesis (93). Lactobacillus coryniformis ameliorates CRC by alleviating inflammation, intestinal microenvironment, and intestinal barrier damage (94). Pediococcus pentosaceus inhibits tumor growth in xenograft, and exhibits polyp regression and recovered taxonomic diversity in CRC mice (95). Lactobacillus gasseri, accompanied by other prebiotics, reduces the CRC risk via the regulation of inflammation, carcinogenesis, and compositional change of gut microbiota (96).. In addition, Lactobacillus and Bifidobacterium are the most reported probiotics that exert anti-biofilm activity (151). Strikingly, they can form “probiotic biofilms” to fight against other “pathogenic biofilms” (151, 152). Although most Lactobacillus strains show an anti-inflammatory effect in vitro, only Lactobacillus fermentum NA4 displays a protective effect in vivo (153), suggesting that the beneficial probiotic properties are strain-dependent.
3.3 Metabolic products of microbiota on host immunity and CRC
Besides direct interaction, gut microbiota also produces a diverse metabolite repertoire to trigger specific immune responses that may harm or benefit the host indirectly.
3.3.1 Short-chain fatty acids
SCFAs, mainly consisting of acetate, propionate, and butyrate, are a group of organic acids produced by the anaerobic microbial community from carbohydrate fermentation of undigested dietary fiber (154). High fiber diet promotes SCFA production and suppresses CRC development (155), whereas the removal of dietary carbohydrates alters microbiota and results in susceptibility to infectious colitis (156). In general, SCFAs exhibit potential anti-carcinogenic effects in CRC development, with a decreased gut abundance in CRC and adenoma patients (157), consistent with the reduction of butyrate-producing bacteria (158). Loss of FFAR2 (free-fatty acid receptor 2), a SCFA receptor, promotes colon tumorigenesis in mice by reducing gut barrier integrity, over-activating DCs, and promoting CD8+ T cell exhaustion (159).
Butyrate and propionate, but not acetate, have a histone deacetylase (HDAC)-inhibiting activity and regulate NF-kB and Wnt signaling in colon epithelial cells. These two SCFAs support basal crypt proliferation in healthy tissues and maintain colonic homeostasis, but inhibit cell growth and induce apoptosis in CRC cell lines (160). Through its receptor GPR109A, butyrate also promotes IL-18 production in intestinal epithelial cells (161, 162), which is a protective cytokine in CRC mouse model (163).
In innate immune cells, butyrate functions intracellularly as a histone deacetylase inhibitor to downregulate IL-6 (164). It inhibits LPS-induced pro-inflammatory mediators in both macrophages and dendritic cells (164, 165). In DSS colitis model, butyrate attenuates the intestinal inflammation by enhancing the M2 macrophage polarization (166). In addition, butyrate and other SCFAs can promote Treg cell generation in mice and ameliorate T cell induced colitis (167, 168).
On the other hand, butyrate may show pro-tumorigenic roles in a context-dependent manner. It inhibits intestinal stem/progenitor proliferation (169), which may suppress advanced cancer but delay tissue damage repair at the early stages of CRC. Butyrate also induces the production of reactive oxygen species (ROS), which may have pro- or anti-tumorigenic functions in different models (170, 171).
3.3.2 Polyamine
Polyamines, such as putrescine, spermidine, spermine, and cadaverine, are aliphatic amines derived from amino acid metabolism in both host tissues and commensal microbiota. They bind to negatively charged macromolecules (DNA, RNA, protein) and regulate a series of cancer-related physiological processes, including cell proliferation, differentiation, apoptosis, angiogenesis, and immune response, etc. (172).
Polyamines are generally considered detrimental metabolites in CRC development. Activated KRAS significantly increases the uptake of polyamines by colon cancer cells (173). Consistently, both polyamines and the key enzyme for polyamine biosynthesis, ornithine decarboxylase (ODC), are dramatically increased in CRC tissues (160), while ODC inhibitor alpha-difluoromethylornithine (DFMO) exhibits promising effects in colon adenoma patients (174). The polyamine catabolic enzyme SMO (spermine oxidase) contributes to ETBF-induced colon tumorigenesis (175), while spermidine directly impacts the colibactin production from PKS+ Escherichia coli and is required for genotoxic activity (176). Furthermore, SSAT (spermidine/spermine N1-acetyltransferase)-mediated depletion of polyamines inhibits CRC progression and metastasis through the suppression of AKT, GSK3β, and β-catenin signaling (177). Besides, polyamines can regulate T cell activation and macrophage polarization, thus play an important role in CRC microenvironment (160).
CRC patients have an altered microbiota that is closely associated with a higher abundance of polyamines (178). Bacteria biofilm formation in CRC patients is associated with increased cancer cell proliferation and enhanced polyamine metabolism (179, 180), which can be reduced by antibiotic treatment. Furthermore, CRC-associated microbiota not only has an enhanced capacity for converting amino acids into polyamines via putrefaction and fermentation pathways (181), but also upregulates polyamine production in host cells (154).
3.3.3 Secondary bile acids
Bile acids (BAs) are synthesized in the liver, stored in the gallbladder, and mostly reabsorbed by ileal epithelial cells during lipid absorption. The small number of unabsorbed BAs are converted into secondary BAs by the microbiota, and become detrimental metabolites to the intestine by contributing to neonatal necrotizing enteritis, IBD, and CRC (182). In the African American population who has a higher risk of CRC, high-fiber low-fat diet suppresses secondary BAs synthesis, resulting in the reduction of CRC biomarkers (183). In CRC mouse model, Apc founding mutation leads to a decreased expression of bile acid apical transporter gene Slc10A2, reduced BA reabsorption, and increased secondary BAs, which strongly enhance the gut colonization of CRC-promoting Streptococcus gallolyticus (184).
Metaproteomic analysis in stools from CRC patients identifies a heightened oxidative metabolic microenvironment with increased concentrations of DNA-damaging BAs, especially deoxycholic acid (DCA) (185). DCA inhibits gut epithelial cell proliferation via the activation of BA receptor FXR (farnesoid X receptor), resulting in the inhibition of wound healing and impaired gut barrier function (186). It also activates the beta-catenin signaling pathway and increases proliferation and invasiveness of CRC cells (187). In ApcMin/+ mice, DCA treatment promotes tumorigenesis with a disrupted intestinal mucosal barrier, activated NLRP3 inflammasome, and increased production of inflammatory cytokines (188). Lithocholic acid (LCA), another typical BA, promotes proliferation and invasiveness of CRC cells (189, 190). Both DCA and LCA are reported to induce cancer stemness in colonic epithelial cells (191).
3.3.4 Other cancer-regulating metabolites
Microbial metabolites from healthy colons are reported to inhibit colon tumorigenesis (192). In specific, Lactobacillus reuteri and its metabolite reuterin, which are reduced in mouse and human CRC, could decrease tumor growth and prolong mouse survival by inducing protein oxidation and inhibiting ribosomal biogenesis (192).
Malic acid is a speculated anti-tumor agent produced by Odoribacter splanchnicus, based on the gas chromatography-mass spectrometry (GC/MS) analysis of the bacteria supernatant, which induces the apoptosis of colon cancer cells (86).
Hydrogen Sulfide (H2S), a toxic gas that can be physiologically produced in the large intestine by commensal microbiota, shows both beneficial and deleterious effects on the intestinal mucosa in a dose- and context-dependent manner (193). In particular, H2S ranges from 0.2 mM to 2.0 mM in the mammalian large intestine content and fecal materials, the latter representing the approximate concentration as in the rectum (193). This concentration is critical for the growth of some beneficial microbes, such as Lactobacillus (194).
Beneficial AhR ligands: The aryl hydrocarbon receptor (AhR), a ligand-dependent transcription factor with diverse functions in inflammation, detoxification, and homeostasis (195), has been identified as a tumor suppressor in mouse CRC models (196, 197). Ligand-activation of AhR is required for the maintenance of intestinal immune homeostasis and control of inflammation (198, 199). Several microbial tryptophan catabolites, such as indole-3-acetic acid (IAA) and indolepropionic acid (IPA), are natural AhR ligands that can influence the intestinal epithelial barrier (200). Furthermore, indole treatment leads to the repression of inflammation in CRC cell lines, human duodenum-derived organoids, and mouse models (201).
In summary, CRC-regulating microbiota species mainly function through: (1) directly adhering to epithelial cells for oncogenic or anti-tumor signaling activation; (2) producing detrimental (such as toxins) or beneficial (such as SCFAs) metabolites; and (3) inducing tumor-associated or -inhibiting immune cell populations (Figure 1).
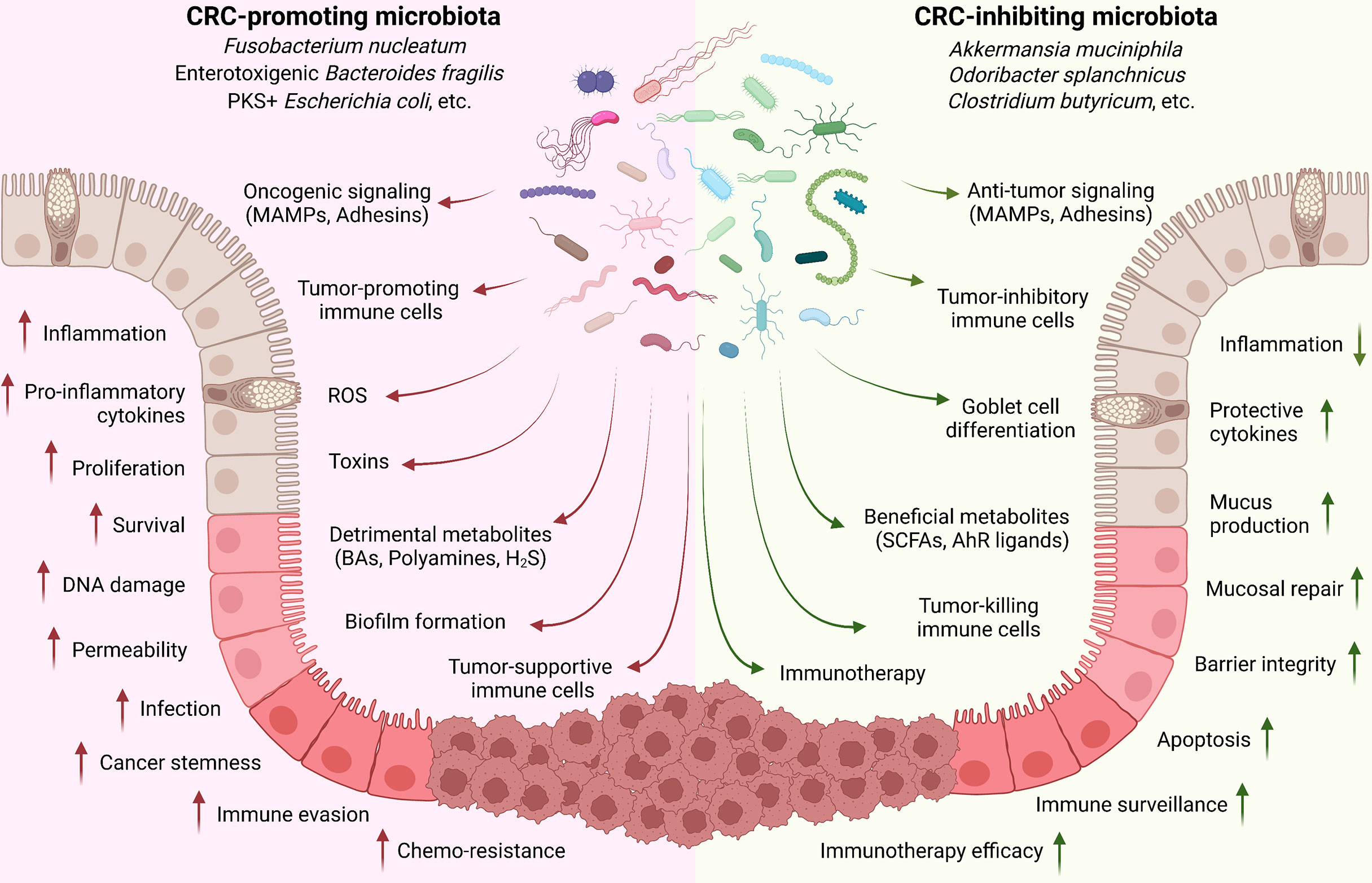
Figure 1 Implication of gut microbiota in CRC development. Commensal microbiota plays critical roles in controlling CRC development. Cancer-promoting microbiota directly adheres to the epithelial cells through MAMPs and adhesins for oncogenic signaling activation (such as Wnt/β-catenin); produce toxins and detrimental metabolites (such as secondary BAs, polyamines, and H2S); and induce tumor-associated immune cell populations (such as TAM, TAN, MDSC, and Treg) to regulate the inflammation, tissue damage, cell proliferation and survival, immune evasion, and drug resistance. On the contrary, CRC-inhibiting microbiota can directly trigger the anti-tumor signaling activation in epithelial cells; produce beneficial metabolites (such as SCFAs and AhR ligands); and stimulate tumor-preventing and -killing immune cells (such as CD8, Th1, Th17, and ILC3).
4 Interplay of microbiota and host immune system in regulating CRC development
As key components of the tumor microenvironment, various immune cell populations, particularly tumor-infiltrating immune cells, play critical roles in mediating promotion or inhibition of CRC development. Even in patients after radiation therapy, repopulation of tumor-infiltrating immune cells could be observed after the initial depletion (202). Microbiota communicates with the immune system through various mechanisms, such as Toll-like receptor (TLR) signaling and inflammasome sensing, and regulates inflammation and cancer development through nuclear factor kappa B (NF-κB), type I interferon, and inflammasome pathways (51, 203–205). Local immune system interacts with gut microbiota to control immune responses, tissue damage, and cancer development (11–14).
4.1 Innate immune system
Innate immune system is the first line of host defense against pathogens that provides microorganism recognition and plays a critical role in mediating inflammation and cancer development under the stimulation of specific microbiota components (203).
4.1.1 Innate immune signaling
Toll-like receptors (TLRs) are well-defined pattern recognition receptors responsible for pathogen recognition and induction of innate immune responses (204). The densely populated microbiota in the intestinal tract could generate various molecules that can be recognized by TLRs, which leads to NF-κB signaling activation and transcription of multiple cytokines (206). Multiple microbial taxa, particularly some pathogenic bacteria, have been reported to activate TLRs, including PSA-producing Bacteroides fragilis, flagellin-producing SFB, Yersinia enterocolitica, Salmonella enterica, Helicobacter hepaticus, Citrobacter rodentium, and LPS-producing Serratia marcescens and Escherichia coli (206, 207). In steady-state, constant recognition of microbiota by TLR4 and TLR1/2 could lead to IL-6, IL-10, and TGF-β production, which is critical for the integrity of intestinal epithelial cells barrier by promoting the expression of tight junction proteins (ZO-1, claudin-1, occludin) and maintaining their proliferation (206). However, aberrant TLR signaling activation in immune cells beneath the IECs could lead to the release of pro-inflammatory cytokines, resulting in acute or chronic intestinal inflammation (204). Therefore, stringent and precise regulation of TLR signaling pathways is essential to maintaining immune balance in the host (204). Particularly, several negative regulators, such as NLRX1, NLRC5, NLRP11, and LRRC25 (208–212), have been identified to control TLR-induced NF-κB signaling pathways at multiple levels, which might be critical for maintaining the delicate balance between bacterial composition, the mucosal immune system, and the intact epithelial barrier.
DNA and RNA sensors: Microbial antigens and potential pathogens are sensed by the host germline-encoded pattern recognition receptors (PRRs) that recognize specific pathogen-associated molecular patterns (PAMPs). As the genetic material, microbial nucleic acids have been identified as the major target for innate immune recognition (213). PRRs that sense intracellular pathogen-derived nucleic acids could mainly divide into three sets, including endosomally localized transmembrane TLRs that sense microbial DNA and RNA in the endolysosomes (214), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) that detect pathogen-derived RNA in the cytosol, and cyclic GMP-AMP (cGAMP) synthase (cGAS) and absent in melanoma 2 (AIM2) (215). Type-I interferon (IFN-I) signaling is a major immune signaling initiated by the nucleic acid sensors upon detecting invading pathogens. Double-stranded RNA of one major commensal species, lactic acid bacteria (LAB) is shown to trigger TLR3-mediated interferon-β production by DCs in the gut, which is beneficial in the protection from infection and colitis (216). Besides, LAB could also induce the production of IFN-I through cGAS and RLRs (215). The initiation of IFN-I system by the gut microbiota is shown to be mediated by tonic activation of the cGAS-STING signaling, which is also crucial for innate resistance to DNA and RNA viruses. Further study suggests that activation of cGAS-STING signaling is triggered by membrane vesicle-mediated dispatch of bacterial DNA (217), and is required to link DNA sensing to immune responses (218). Meanwhile, IFN-I signaling may also show a detrimental role in the course of infection with intracellular bacteria. Deficiency of IFN-I signaling through either the genetic deficiency of IFN or IFNAR results increases resistance to oral infection of Salmonella typhimurium (219, 220). Therefore, activation of IFN-I signaling mediated by DNA and RNA sensors must be under tight control to maintain a steady intestinal mucosal state in response to pathogen infection and colitis. In the past decade, multiple important regulators have been identified in controlling the IFN-I signaling, such as NLRC5, USP3, USP38, TRIM14, and LRRC25 (209, 212, 221–224). These negative regulators may have potential benefits in maintaining the homeostasis of the gut mucosal system during infections, thus regulating colitis and CRC development.
4.1.2 Innate immune cells
Neutrophils: Regulated by TGF-β and IFN-β signaling, neutrophils may have both tumor-suppressive and -supportive functions (225). Besides the canonical role in mediating the cell phagocytosis and enhancing cytotoxicity, the tumor-associated neutrophils (TANs) can also secrete immunoregulatory and angiogenic factors (225). In CRC, neutrophils are increased with tumor progression (226), and suppress the activity of tumor-infiltrating T cells through the activation of TGFβ (227). Although conflicting results are observed regarding the correlation between neutrophils and survival of CRC patients (228, 229), the high ratio of neutrophils to CD8 T cells is associated with a poor prognosis (230, 231).
Macrophages: In specific conditions, macrophages may differentiate into two distinct types: pro-inflammatory M1 and anti-inflammatory M2 macrophages (232). While total macrophages are increased in CRC with tumor progression (226), M2-type TAMs (tumor-associated macrophages) could promote tumor invasion and angiogenesis, and impair the anti-tumor capacity of T cells (233, 234). Although some studies find that high levels of macrophages are associated with improved prognosis in CRC patients (235, 236), the correlation is opposite in metastatic CRC (237), particularly in elderly patients (202).
MDSCs (myeloid-derived suppressor cells): Characterized by the ability to inhibit both innate and adaptive immune responses, MDSCs are a heterogeneous population of myeloid cells that typically express the common myeloid markers (such as CD33 and CD11b) but lack markers of mature myeloid cells (such as HLA-DR) (238). In both CRC patients and animal models, MDSCs are massively accumulated in the blood, lymph nodes, bone marrow, and tumor sites, particularly in the late stage of cancer (239, 240). An increased MDSC level is correlated with advanced tumor stage and metastasis in CRC patients (241, 242), as well as a shorter survival on chemotherapy (243).
Innate lymphoid cells (ILCs), mainly consisting of natural killer (NK) cells, ILC1, ILC2, and ILC3, are considered the innate counterpart of the T lymphocytes (244). Intestinal ILCs play important roles in controlling epithelial protection, metabolic homeostasis, and development of adaptive immune responses (245–247), and show both pro- and anti-tumor functions in balancing CRC development (248). NK cells have been known for their anti-tumor effects for decades, via targeting NCR (natural cytotoxicity receptor) ligands on CRC stem cells and cancer-initiating cells (249). However, developed tumor cells may evade this process by reducing the expression of NCR ligands and upregulating MHC (major histocompatibility complex) class I to suppress NK cell activation (249). Similar to Th1 cells, ILC1s express the transcription factor T-bet and produce IFN-γ and cytotoxic molecules in response to IL-12 and IL-15 for anti-tumor immunity (250). But ILC1-induced inflammation may have negative effects on colitis-associated CRC. ILC2s are rare in the adult human intestine, but are increased in IBD patients (251). Triggered by tumor-derived IL-33, the frequency of IL-13+ ILC2s also increases in colorectal tumors (252). Through the production of IL-4, IL-5, and IL-13, ILC2s exhibit context-dependent roles in CRC development (248). ILC3s are frequently accumulated in the intestine and activated by IL-23 for differentiation and production of IL-17A and IL-22 (253). Due to the context-dependent functions of these cytokines (140, 254–263), ILC3s may also present both pro- and anti-tumor functions in CRC development. Notably, the ILCs temper the expansion of bacterial species and protect the gut epithelium in early life (264). After the maturation of adaptive immune system, transient activation of ILC3s by microbial colonization can be extinguished by CD4 T cells (264), indicating that innate and adaptive lymphocytes operate sequentially and in distinct ways during normal development to establish steady-state commensalism and tissue homeostasis. Meanwhile, the loss of ILCs, which express MHC class II for microbial antigen presentation, is associated with dysregulated adaptive immune cell responses against commensal bacteria (246).
The commensal microbiota can regulate the innate immune system through multiple approaches. It may release microorganism-associated molecular patterns (MAMPs), such as flagellin, elongation factor-Tu (EF-Tu), and lipopolysaccharides (LPS), that can be recognized by PRRs on innate immune cells for direct manipulation of functions (265). It may also indirectly influence the innate immune system by metabolites or triggered productions from colon epithelial cells for manipulating the expansion and recruitment of these cells. Several CRC-associated examples are listed.
Prevotella intermedia, which is associated with a higher risk of developing CRC (266), evades innate immune control by disabling and killing tissue-infiltrating neutrophils in endodontic infection (267). Lactobacillus rhamnosus triggers the anti-inflammatory effects in macrophages and suppresses TNF production through granulocyte-colony stimulating factor (G-CSF)-induced inhibition of c-Jun-N-terminal kinases (JNKs) (268). Bifidobacterium lactis attenuates macrophage senescence and induces M2 macrophage polarization (269). Importantly, certain Lactobacilli and Bifidobacteria species are able to produce butyrate, which in turn control the programming of macrophage for an anti-inflammatory phenotype (270, 271). Clostridium butyricum triggers IL-10 production from intestinal macrophages via the TLR2/MyD88 signaling pathway and prevents mice from DSS-induced colitis (82). Fusobacterium nucleatum administration in mice triggers the increase in tumor-infiltrated immunosuppressive myeloid cells, including MDSCs, TAMs, TANs, and dendritic cells (63). It can also act in a cytokine-independent manner and directly inhibit the cytotoxicity of NK cells against tumors (272). ETBF and Peptostreptococcus anaerobius are also reported to trigger the secretion of chemokines that recruit immunosuppressive MDSCs, TAMs, and TANs (59, 72). In addition, Clostridia bacteria are reported to modulate the balance of retinoic acid and retinyl esters in intestinal epithelial cells, which further regulates the development of IL-22-producing ILC3s (273, 274).
4.2 Adaptive immune system
CD8 T cells are the most potent cytolytic population. Triggered by CRC-derived modulators (such as IL-18) (275), CD8 T cells produce pro-inflammatory cytokines (such as IFN-γ) and cytotoxic molecules for cancer cell clearance (276). Numerous studies have demonstrated the positive association between tumor-infiltrating CD8 T cells with the patients’ prognosis and survival (202, 277). Whereas CD8 cells accumulated at the tumor margin have no effect on survival (278), the ratio of CD8 T cells and Treg cells is a critical determinant of prognosis (279).
CD4 T cells: Different CD4 T helper (Th) cells have distinct roles in regulating CRC development. The cytotoxic Th1 cells are similar to cytolytic CD8 T cells in terms of functions and molecular productions (276), and are positively associated with prolonged survival of CRC patients (280). Treg population plays an immune-suppressive function on multiple immune cell populations through the key cytokines IL-10 and TGF-β (281, 282), but its association with CRC prognosis is controversial (283). Treg infiltration is low in healthy colon, significantly increased in early-stage CRC, but decreased in metastatic cancer (284); and stromal Treg infiltration is 5 times higher than epithelial infiltration in CRC. Further studies find intra-tumoral Treg cells, but not stromal, are associated with increased disease-free survival (285, 286), indicating that the roles of Treg cells may be related to their distribution and immune microenvironment.
Th17 cells are constitutively present in the intestinal lamina propria (LP) due to the activation by microbial flora, such as SFB (segmented filamentous bacteria) (253, 287, 288). Although exhibiting a pro-inflammatory role in autoimmune diseases and host defense against bacteria and fungi (289, 290), Th17 cells in the intestine have an immune-suppressive function (291). In CRC patients, Th17 cells are increased in the tumor and peripheral blood compared with healthy people (292); and the high amount of Th17 cells is associated with tumor progression and a poor prognosis (280, 293). However, the presence of intraepithelial, but not stromal Th17 cells, positively correlates with improved survival (294). The context-dependent function of Th17 population is probably related to the following mechanisms: (1) CRC types. Th17 cells are pathogenic in sporadic CRC models (57, 254–256) but inhibit most CAC models (257–259). (2) Disease stages. Th17 cells may act through altered signaling pathways (such as STAT3) and show distinct roles between the intact epithelial cells in the cancer-initiating stage and the developed tumor cells (295). (3) Differentiation strategies. With the presence of IL-23 or serum amyloid A proteins, Th17 cells could acquire a pathogenic pro-inflammatory phenotype, compared with non-pathogenic Th17 cells induced by IL-6 and TGF-β (296, 297).
The commensal microbiota may regulate the adaptive immune system through multiple approaches. One major aspect is to stimulate and direct the differentiation of several T cell populations in the intestine, particularly Th17 and Treg cells (Table 3). Although dispensable for the induction of peripheral Treg cells (313), microbiota tightly controls the development of gut Treg cells. Bacteroides fragilis directs Treg development in the gut through its unique immunomodulatory molecule, polysaccharide A (PSA), which mediates the conversion of CD4+ T cells into Foxp3+ Treg cells that produce IL-10 (302). Further studies illustrate that the antigenic peptides derived from Akkermansia muciniphila, Helicobacter hepaticus, and several other species induce differentiation of Treg cells in colon and ameliorate intestinal inflammation (303, 304). Similarly, Th17 cell differentiation is mainly directed by specific microbiota strains (49), and some bacteria species have been identified, such as SFB and Bacteroides fragilis (57, 87, 287, 314). Recently, Odoribacter splanchnicus has been reported to induce the development of immune-suppressive intestinal Th17 cells (38) and Foxp3+/RORγt+ regulatory T cells (85), both of which limit colitis development in mouse models. Furthermore, microbiota regulates the antigen recognition and tumor-killing function of cytotoxic T cells, such as CD8 and Th1 cells, thus controlling the efficacy of cancer immunotherapy (45, 46, 315, 316).
In summary, as crucial aspects of the tumor microenvironment, immune cells interplay with the gut microbiota to mediate immune cell functions and control inflammation, anti-tumor immunity, and disease progression (Figure 2). Besides the key innate and adaptive immune populations discussed above, functions of other immune cells in CRC development have been previously reviewed in detail (160, 317, 318).
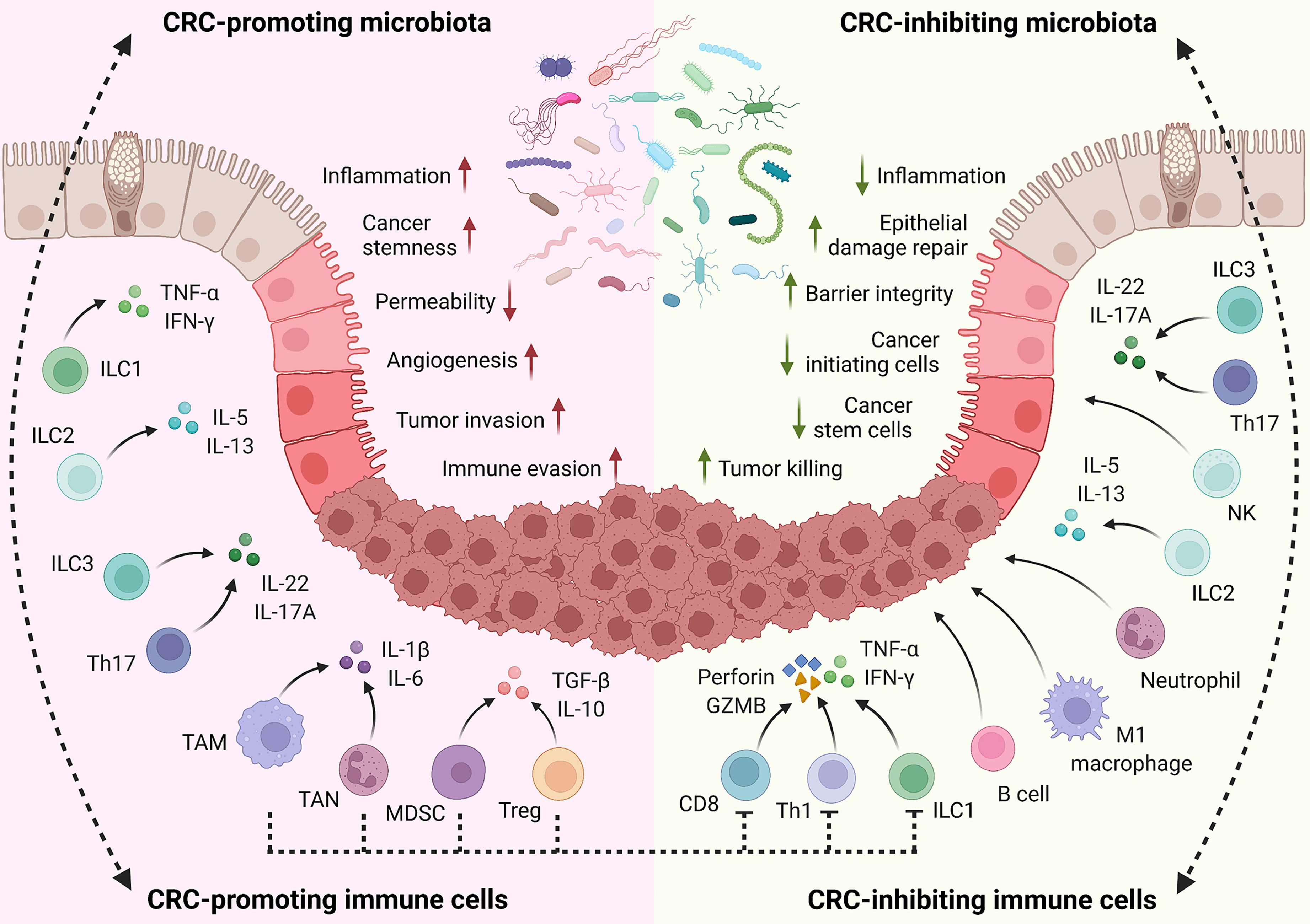
Figure 2 Role of host immune system on CRC. In the host intestine, local immune cells are directly or indirectly stimulated by gut microbiota for activation, proliferation, and differentiation. Meanwhile, the microbiota composition is exquisitely modulated by the immune system. This interplay is important for maintaining homeostasis and plays critical roles in regulating inflammation responses, tissue damage, and CRC development. Through cytokine production and other mechanisms, CRC-promoting immune cells facilitate inflammation, tissue damage, cell proliferation, angiogenesis, tumor invasion, and immune evasion. On the contrary, CRC-inhibiting immune cells enhance epithelial barrier integrity, suppress local inflammation, and eliminate cancer-initiating cells and developed tumors through cytokines, cytotoxic molecules, and other mechanisms. Notably, some immune populations play context-dependent functions in CRC development, based on the disease types, stages, and microenvironment.
5 Gut microbiota in CRC immunotherapy
Microbiota has been identified as a key modulator of cancer immunotherapy (319, 320). Early studies have established the roles of gut microbiota in supporting the CpG-oligonucleotide immunotherapy and the anti-tumor immune responses to cyclophosphamide (CTX) chemotherapy (321, 322). Following research further identified multiple microbiota taxa in enhancing immunomodulatory therapies and controlling tumor-killing efficacy of cytotoxic T cells. Bifidobacterium promotes anti-tumor efficacy of anti-PD-L1 therapy by enhancing CD8+ T cell priming and accumulation in the tumor microenvironment (45). Similarly, Bacteroides fragilis enhances the efficacy of anti-CTLA-4 therapy by triggering a Th1 response and promoting dendritic cell maturation (46). Furthermore, microbiota components from immunotherapy-responding patients lead to improved tumor control, augmented T cell responses, and greater efficacy of immunotherapy in animal models (323, 324). Particularly, Akkermansia muciniphila restores the efficacy of PD-1 blockade in non-responders by recruiting CCR9+CXCR3+CD4+ T cells (325). During CTX treatment, the translocation of Enterococcus hirae from the intestine to secondary lymphoid organs stimulates IFN-producing CD8+ T cells and increases the intratumoral CD8/Treg ratio (315). In addition to the single species, a combination of 11 bacterial strains is reported to improve the anti-tumor efficacy of checkpoint inhibitors with increased tumor antigen-specific CD8+/IFN-γ+ T cells (316). Microbiota-derived metabolites, such as inosine, could also modulate the response to cancer immunotherapy (326).
Besides traditional and targeted therapies, immunotherapy in CRC treatment has not been widely utilized. Classic vaccination strategies and chimeric antigen receptor (CAR) T cells have shown great clinical benefits, but are accompanied by severe toxicity (327–330). Meanwhile, with the discovery of new druggable immune checkpoints (331), checkpoint blockade therapies have shown good responses in several types of CRC, particularly when combined with chemotherapy (43, 332, 333), but have not been widely investigated. In this case, microbiota-related therapies, particularly with previously identified or even FDA-approved probiotics, may provide novel strategies for CRC treatment. Development of microbiota-related therapies may include: (1) specific elimination of detrimental species by antibiotics or targeted bacteriophage therapy; (2) neutralization of harmful metabolites such as bacterial toxins; (3) supplementary administration of anti-tumorigenic species; (4) fecal transplantation of whole microbiota from healthy donors (particularly for antibiotics-treated patients); (5) diet-driven transition of microbiota or metabolites. While promising results are obtained from increasing clinical trials (334–337), more is needed to validate the efficacy and safety of these strategies. Due to the complexity of microbiota and the numerous effects one species may have on host biology, prudent consideration of any therapeutic approach is necessary based on the host microbiota profile, disease stage, and status of the immune system.
6 Gut microbiota and lifestyle risk factors in CRC development
6.1 Diet
Dietary factors play important roles in modulating the gut microbiota, which in turn regulates colon inflammation, genotoxic metabolite production, and eventually CRC development (338). On the other hand, gut microbiota composition can influence the physiological effects of dietary components. For example, trimethylamine-N-oxide (TMAO) is a microbiota-dependent metabolite from protein, in particular red meat (339), and an elevated TMAO level is associated with a higher risk of CRC (340). While Firmicutes species may contribute to TMAO production (341), Eubacterium limosum has the potential to metabolize TMA precursors and reduce TMAO level in gut (342). In addition, dietary carbohydrates, as main fuel sources of the body, have great impacts on the gut microbiota composition and microbiota-related diseases (343). It has been widely reported that the dietary fibers contribute to the reduction of CRC risk (344), mainly through the enhanced production of microbial metabolite SCFAs (345). In this case, specific gut microbiota species, such as Faecalibacterium prausnitzii, Eubacterium rectale, Roseburia faecis, and Eubacterium halli, play important roles in the dietary fiber fermentation and SCFA production (346). The abundances of these bacteria are consistently reduced in the gut microbiota of colorectal adenoma patients (347).
6.2 Obesity
Lipid metabolism plays an essential role in health management, weight control, and risks to cancers and other infectious diseases (348, 349). Obesity has been reported as an important risk factor of CRC that contributes to approximately 5% of incident cases (348), and gut microbiota is one of the leading factors accompanying and pathogenetically contributing to obesity and its metabolic associates, such as diabetes and cardiovascular diseases (350). The gut microbiota in obese people represents a decreased diversity in phyla and an increased ratio of Firmicutes : Bacteroidetes (351), whereas bariatric surgery can reverse these microbial abnormalities (352), associated with changes on dietary habits and macronutrients consumption. In animal models, high-fat diet diminishes the beneficial gut microbiota, such as Actinobacteria, Bifidobacterium, Lactobacillus, and Akkermansia (353–356), thus induces gut inflammation, barrier impairment, and an increased risk of CRC development. On the contrary, several well-established probiotics, including Lactobacillus acidophilus, Bifidobacterium lactis, and Akkermansia muciniphila, show an anti-obesity effect (357). The fecal microbiota transplantation from healthy donors, as a promising approach for the treatment of obesity (358), may also be used to control the risk of CRC development.
6.3 Alcohol
Alcohol consumption, particularly chronic and moderate to heavy alcohol intake, has been recognized as an important risk factor for CRC, and is closely related to the metastasis and poor prognosis in CRC patients (359). The metabolism of alcohol is actively modulated by the gut microbiota, which regulates ethanol conversion into its metabolites that exert carcinogenic effects in the colon (360). For example, Enterobacteriaceae Ruminococcus, and Bifidobacterium mediate the production of carcinogenic acetaldehyde from ethanol (361, 362), which accumulates in the colon and greatly contribute to CRC development as ethanol consumption increases. Furthermore, the gut microbiota in alcoholic people is diminished in dominant obligate anaerobes (such as Bacteroides and Bifidobacterium) and enriched in Streptococcus (363), which in turn contribute to CRC development according to their cancer-inhibiting or -promoting functions as discussed in Section 3.
6.4 Tobacco
Cigarette smoking has long been identified as a risk factor for CRC development, which is attributed to the synergistic effect of multiple carcinogens, including nicotine, aldehydes, polycyclic aromatic hydrocarbons, heavy metals, volatile organic compounds, and toxic gases (364). The long-term exposure to tobacco smoke induces gut microbial dysbiosis and altered metabolites, and promotes CRC development (365). In specific, cigarette smoke toxicants induce the increases of Helicobacter, Streptococci, Firmicutes, Peptococcaceae; as well as the loss of Bacteroidetes, Lachnospiraceae, and Lactobacillaceae (366). Some representative species of these bacterial taxa have been discussed in Section 3 for their CRC-promoting and -inhibiting functions. The increased Firmicutes : Bacteroidetes ratio in tobacco users is also associated with obesity, microbial metabolites, and CRC development.
7 Conclusions
In the past decade, microbiota has been identified as a critical regulator in maintaining homeostasis, while its imbalance triggers numerous pathological conditions, including CRC. Microbiota may regulate CRC development in multiple approaches: directly by tissue invasion, indirectly by producing metabolites, or by triggering host immune responses. Alteration in the microbiota composition is frequently observed in multiple diseases, while the identification of functional species and strains is limited. In this review, we provide an extensive overview of CRC-regulating microbiota species and how they crosstalk with local enterocytes and the host immune system in controlling disease development, thus offering new insights into our understanding and the development of microbiota-based therapies. Based on their functions, different microbiota species may serve as probiotic supplements or therapeutic targets in the prevention and better treatment of colitis and CRC. The profiling of gut microbiota and metabolites may also serve as novel diagnostic markers to evaluate the CRC risk and prognosis in healthy people and cancer patients.
Author contributions
R-FW supervised the entire project. CX and R-FW designed and wrote the manuscript. YD, TD, KN, JC, and HYW assisted in specific sections and manuscript editing. All authors contributed to the article and approved the submitted version.
Funding
This work was in part supported by grants from the NCI, NIH (R01CA101795, R01CA246547, and U54CA210181), Department of Defense (DoD) CDMRP BCRP (BC151081) and LCRP (LC200368) to R-FW.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol (2021) 14(10):101174. doi: 10.1016/j.tranon.2021.101174
2. Cheng Y, Ling Z, Li L. The intestinal microbiota and colorectal cancer. Front Immunol (2020) 11:615056. doi: 10.3389/fimmu.2020.615056
3. Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol (2019) 16(11):690–704. doi: 10.1038/s41575-019-0209-8
4. Kwong LN, Dove WF. APC and its modifiers in colon cancer. Adv Exp Med Biol (2009) 656:85–106. doi: 10.1007/978-1-4419-1145-2_8
5. Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature (2007) 448(7152):427–34. doi: 10.1038/nature06005
6. Jess T, Rungoe C, Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol (2012) 10(6):639–45. doi: 10.1016/j.cgh.2012.01.010
7. Freeman HJ. Colorectal cancer risk in crohn's disease. World J Gastroenterol (2008) 14(12):1810–1. doi: 10.3748/wjg.14.1810
8. Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol (2014) 14(5):329–42. doi: 10.1038/nri3661
9. West NR, McCuaig S, Franchini F, Powrie F. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol (2015) 15(10):615–29. doi: 10.1038/nri3896
10. Ternes D, Karta J, Tsenkova M, Wilmes P, Haan S, Letellier E. Microbiome in colorectal cancer: How to get from meta-omics to mechanism? Trends Microbiol (2020) 28(5):401–23. doi: 10.1016/j.tim.2020.01.001
11. Surana NK, Kasper DL. Moving beyond microbiome-wide associations to causal microbe identification. Nature (2017) 552(7684):244–7. doi: 10.1038/nature25019
12. Wlodarska M, Luo C, Kolde R, d'Hennezel E, Annand JW, Heim CE, et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe (2017) 22(1):25–37.e6. doi: 10.1016/j.chom.2017.06.007
13. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell (2017) 171(5):1015–28.e13. doi: 10.1016/j.cell.2017.09.016
14. Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell (2017) 168(5):928–43 e11. doi: 10.1016/j.cell.2017.01.022
15. Zygulska AL, Pierzchalski P. Novel diagnostic biomarkers in colorectal cancer. Int J Mol Sci (2022) 23(2):852. doi: 10.3390/ijms23020852
16. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet (2019) 394(10207):1467–80. doi: 10.1016/S0140-6736(19)32319-0
17. Kim JC, Bodmer WF. Genomic landscape of colorectal carcinogenesis. J Cancer Res Clin Oncol (2022) 148(3):533–45. doi: 10.1007/s00432-021-03888-w
18. Lee SJ, Yun CC. Colorectal cancer cells - proliferation, survival and invasion by lysophosphatidic acid. Int J Biochem Cell Biol (2010) 42(12):1907–10. doi: 10.1016/j.biocel.2010.09.021
19. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell (1990) 61(5):759–67. doi: 10.1016/0092-8674(90)90186-I
20. Martinez-Jimenez F, Muinos F, Sentis I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer (2020) 20(10):555–72. doi: 10.1038/s41568-020-0290-x
21. Almeida-Lousada H, Mestre A, Ramalhete S, Price AJ, de Mello RA, Marreiros AD, et al. Screening for colorectal cancer leading into a new decade: The "Roaring '20s" for epigenetic biomarkers? Curr Oncol (2021) 28(6):4874–93. doi: 10.3390/curroncol28060411
22. Li J, Ma X, Chakravarti D, Shalapour S, DePinho RA. Genetic and biological hallmarks of colorectal cancer. Genes Dev (2021) 35(11-12):787–820. doi: 10.1101/gad.348226.120
23. Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature (2012) 487(7407):330–7.10.1038/nature11252
24. Huang D, Sun W, Zhou Y, Li P, Chen F, Chen H, et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev (2018) 37(1):173–87. doi: 10.1007/s10555-017-9726-5
25. Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology (2011) 140(6):1807–16. doi: 10.1053/j.gastro.2011.01.057
26. Rhodes JM, Campbell BJ. Inflammation and colorectal cancer: IBD-associated and sporadic cancer compared. Trends Mol Med (2002) 8(1):10–6. doi: 10.1016/S1471-4914(01)02194-3
27. Sumransub N, Vantanasiri K, Prakash A, Lou E. Advances and new frontiers for immunotherapy in colorectal cancer: Setting the stage for neoadjuvant success? Mol Ther Oncol (2021) 22:1–12. doi: 10.1016/j.omto.2021.05.001
28. Baran B, Mert Ozupek N, Yerli Tetik N, Acar E, Bekcioglu O, Baskin Y. Difference between left-sided and right-sided colorectal cancer: A focused review of literature. Gastroenterol Res (2018) 11(4):264–73. doi: 10.14740/gr1062w
29. Salem ME, Weinberg BA, Xiu J, El-Deiry WS, Hwang JJ, Gatalica Z, et al. Comparative molecular analyses of left-sided colon, right-sided colon, and rectal cancers. Oncotarget (2017) 8(49):86356–68. doi: 10.18632/oncotarget.21169
30. Lakatos L, Mester G, Erdelyi Z, David G, Pandur T, Balogh M, et al. Risk factors for ulcerative colitis-associated colorectal cancer in a Hungarian cohort of patients with ulcerative colitis: results of a population-based study. Inflamm Bowel Dis (2006) 12(3):205–11. doi: 10.1097/01.MIB.0000217770.21261.ce
31. Fuszek P, Horvath HC, Speer G, Papp J, Haller P, Fischer S, et al. Location and age at onset of colorectal cancer in Hungarian patients between 1993 and 2004. the high number of advanced cases supports the need for a colorectal cancer screening program in Hungary. Anticancer Res (2006) 26(1B):527–31.
32. Munkholm P. Review article: the incidence and prevalence of colorectal cancer in inflammatory bowel disease. Aliment Pharmacol Ther (2003) 18 Suppl 2:1–5. doi: 10.1046/j.1365-2036.18.s2.2.x
33. Shouval DS, Ouahed J, Biswas A, Goettel JA, Horwitz BH, Klein C, et al. Interleukin 10 receptor signaling: master regulator of intestinal mucosal homeostasis in mice and humans. Adv Immunol (2014) 122:177–210. doi: 10.1016/B978-0-12-800267-4.00005-5
34. Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell (1993) 75(2):263–74. doi: 10.1016/0092-8674(93)80068-P
35. Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, Ryan AM, et al. The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J Exp Med (1998) 187(4):571–8. doi: 10.1084/jem.187.4.571
36. Kathania M, Khare P, Zeng M, Cantarel B, Zhang H, Ueno H, et al. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-gammat ubiquitination. Nat Immunol (2016) 17(8):997–1004. doi: 10.1038/ni.3488
37. Xu ZS, Zhang HX, Li WW, Ran Y, Liu TT, Xiong MG, et al. FAM64A positively regulates STAT3 activity to promote Th17 differentiation and colitis-associated carcinogenesis. Proc Natl Acad Sci USA (2019) 116(21):10447–52. doi: 10.1073/pnas.1814336116
38. Xing C, Wang M, Ajibade AA, Tan P, Fu C, Chen L, et al. Microbiota regulate innate immune signaling and protective immunity against cancer. Cell Host Microbe (2021) 29(6):959–74.e7. doi: 10.1016/j.chom.2021.03.016
39. Litvak Y, Byndloss MX, Baumler AJ. Colonocyte metabolism shapes the gut microbiota. Science (2018) 362(6418):eaat9076. doi: 10.1126/science.aat9076
40. Ai D, Pan H, Li X, Gao Y, Liu G, Xia LC. Identifying gut microbiota associated with colorectal cancer using a zero-inflated lognormal model. Front Microbiol (2019) 10:826. doi: 10.3389/fmicb.2019.00826
41. Saffarian A, Mulet C, Regnault B, Amiot A, Tran-Van-Nhieu J, Ravel J, et al. Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. mBio (2019) 10(4):e01315-19. doi: 10.1128/mBio.01315-19
42. Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes (2010) 1(3):138–47. doi: 10.4161/gmic.1.3.12360
43. Wang RF, Wang HY. Immune targets and neoantigens for cancer immunotherapy and precision medicine. Cell Res (2017) 27(1):11–37. doi: 10.1038/cr.2016.155
44. Spranger S, Dai D, Horton B, Gajewski TF. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell (2017) 31(5):711–23.e4. doi: 10.1016/j.ccell.2017.04.003
45. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
46. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
47. Sears CL, Pardoll DM. The intestinal microbiome influences checkpoint blockade. Nat Med (2018) 24(3):254–5. doi: 10.1038/nm.4511
48. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (2012) 336(6086):1268–73. doi: 10.1126/science.1223490
49. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature (2016) 535(7610):75–84. doi: 10.1038/nature18848
50. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature (2016) 535(7610):65–74. doi: 10.1038/nature18847
51. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol (2016) 16(6):341–52. doi: 10.1038/nri.2016.42
52. Zitvogel L, Ayyoub M, Routy B, Kroemer G. Microbiome and anticancer immunosurveillance. Cell (2016) 165(2):276–87. doi: 10.1016/j.cell.2016.03.001
53. Cao Y, Wu K, Mehta R, Drew DA, Song M, Lochhead P, et al. Long-term use of antibiotics and risk of colorectal adenoma. Gut (2018) 67(4):672–8. doi: 10.1136/gutjnl-2016-313413
54. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell (2004) 118(2):229–41. doi: 10.1016/j.cell.2004.07.002
55. Yang H, Wang W, Romano KA, Gu M, Sanidad KZ, Kim D, et al. A common antimicrobial additive increases colonic inflammation and colitis-associated colon tumorigenesis in mice. Sci Trans Med (2018) 10(443):eaan4116. doi: 10.1126/scitranslmed.aan4116
56. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell (2014) 157(1):121–41. doi: 10.1016/j.cell.2014.03.011
57. Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med (2009) 15(9):1016–22. doi: 10.1038/nm.2015
58. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science (2018) 359(6375):592–7. doi: 10.1126/science.aah3648
59. Chung L, Orberg ET, Geis AL, Chan JL, Fu K, DeStefano Shields CE, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe (2018) 23(3):421. doi: 10.1016/j.chom.2018.02.004
60. Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science (2012) 338(6103):120–3. doi: 10.1126/science.1224820
61. Arthur JC, Gharaibeh RZ, Muhlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun (2014) 5:4724. doi: 10.1038/ncomms5724
62. Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, et al. Precision editing of the gut microbiota ameliorates colitis. Nature (2018) 553(7687):208–11. doi: 10.1038/nature25172
63. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
64. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell (2017) 170(3):548–63.e16. doi: 10.1016/j.cell.2017.07.008
65. Sun X, Threadgill D, Jobin C. Campylobacter jejuni induces colitis through activation of mammalian target of rapamycin signaling. Gastroenterology (2012) 142(1):86–95.e5. doi: 10.1053/j.gastro.2011.09.042
66. He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut (2019) 68(2):289–300. doi: 10.1136/gutjnl-2018-317200
67. Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology (2005) 128(4):891–906. doi: 10.1053/j.gastro.2005.02.009
68. Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am J Pathol (2002) 160(6):2253–7. doi: 10.1016/S0002-9440(10)61172-8
69. Ellmerich S, Scholler M, Duranton B, Gosse F, Galluser M, Klein JP, et al. Promotion of intestinal carcinogenesis by streptococcus bovis. Carcinogenesis (2000) 21(4):753–6. doi: 10.1093/carcin/21.4.753
70. Kumar R, Herold JL, Schady D, Davis J, Kopetz S, Martinez-Moczygemba M, et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PloS Pathog (2017) 13(7):e1006440. doi: 10.1371/journal.ppat.1006440
71. Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology (2017) 152(6):1419–33.e5. doi: 10.1053/j.gastro.2017.01.009
72. Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol (2019) 4(12):2319–30. doi: 10.1038/s41564-019-0541-3
73. Schulz C, Schutte K, Mayerle J, Malfertheiner P. The role of the gastric bacterial microbiome in gastric cancer: Helicobacter pylori and beyond. Therap Adv Gastroenterol (2019) 12:1756284819894062. doi: 10.1177/1756284819894062
74. Fujimori S, Kishida T, Kobayashi T, Sekita Y, Seo T, Nagata K, et al. Helicobacter pylori infection increases the risk of colorectal adenoma and adenocarcinoma, especially in women. J Gastroenterol (2005) 40(9):887–93. doi: 10.1007/s00535-005-1649-1
75. Feller M, Huwiler K, Stephan R, Altpeter E, Shang A, Furrer H, et al. Mycobacterium avium subspecies paratuberculosis and crohn's disease: a systematic review and meta-analysis. Lancet Infect Dis (2007) 7(9):607–13. doi: 10.1016/S1473-3099(07)70211-6
76. Jeyanathan M, Boutros-Tadros O, Radhi J, Semret M, Bitton A, Behr MA. Visualization of mycobacterium avium in crohn's tissue by oil-immersion microscopy. Microbes Infect / Institut Pasteur (2007) 9(14-15):1567–73. doi: 10.1016/j.micinf.2007.09.001
77. Feng Z, Long W, Hao B, Ding D, Ma X, Zhao L, et al. A human stool-derived bilophila wadsworthia strain caused systemic inflammation in specific-pathogen-free mice. Gut Pathog (2017) 9:59. doi: 10.1186/s13099-017-0208-7
78. Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature (2012) 487(7405):104–8. doi: 10.1038/nature11225
79. Alam A, Leoni G, Quiros M, Wu H, Desai C, Nishio H, et al. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat Microbiol (2016) 1:15021. doi: 10.1038/nmicrobiol.2015.21
80. Bian X, Wu W, Yang L, Lv L, Wang Q, Li Y, et al. Administration of akkermansia muciniphila ameliorates dextran sulfate sodium-induced ulcerative colitis in mice. Front Microbiol (2019) 10:2259. doi: 10.3389/fmicb.2019.02259
81. Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C, et al. A purified membrane protein from akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8(+) T cells in mice. Gut (2020) 69(11):1988–97. doi: 10.1136/gutjnl-2019-320105
82. Hayashi A, Sato T, Kamada N, Mikami Y, Matsuoka K, Hisamatsu T, et al. A single strain of clostridium butyricum induces intestinal IL-10-producing macrophages to suppress acute experimental colitis in mice. Cell Host Microbe (2013) 13(6):711–22. doi: 10.1016/j.chom.2013.05.013
83. Chen D, Jin D, Huang S, Wu J, Xu M, Liu T, et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating wnt signaling and gut microbiota. Cancer Lett (2020) 469:456–67. doi: 10.1016/j.canlet.2019.11.019
84. Liu M, Xie W, Wan X, Deng T. Clostridium butyricum modulates gut microbiota and reduces colitis associated colon cancer in mice. Int Immunopharmacol (2020) 88:106862. doi: 10.1016/j.intimp.2020.106862
85. Lima SF, Gogokhia L, Viladomiu M, Chou L, Putzel G, Jin WB, et al. Transferable immunoglobulin a-coated odoribacter splanchnicus in responders to fecal microbiota transplantation for ulcerative colitis limits colonic inflammation. Gastroenterology (2022) 162(1):166–78. doi: 10.1053/j.gastro.2021.09.061
86. Oh BS, Choi WJ, Kim JS, Ryu SW, Yu SY, Lee JS, et al. Cell-free supernatant of odoribacter splanchnicus isolated from human feces exhibits anti-colorectal cancer activity. Front Microbiol (2021) 12:736343. doi: 10.3389/fmicb.2021.736343
87. Chan JL, Wu S, Geis AL, Chan GV, Gomes TAM, Beck SE, et al. Non-toxigenic bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide a. Mucosal Immunol (2019) 12(1):164–77. doi: 10.1038/s41385-018-0085-5
88. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of crohn disease patients. Proc Natl Acad Sci United States Am (2008) 105(43):16731–6. doi: 10.1073/pnas.0804812105
89. Zagato E, Pozzi C, Bertocchi A, Schioppa T, Saccheri F, Guglietta S, et al. Endogenous murine microbiota member faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat Microbiol (2020) 5(3):511–24. doi: 10.1038/s41564-019-0649-5
90. Pujo J, Petitfils C, Le Faouder P, Eeckhaut V, Payros G, Maurel S, et al. Bacteria-derived long chain fatty acid exhibits anti-inflammatory properties in colitis. Gut (2021) 70(6):1088–97. doi: 10.1136/gutjnl-2020-321173
91. Okada Y, Tsuzuki Y, Miyazaki J, Matsuzaki K, Hokari R, Komoto S, et al. Propionibacterium freudenreichii component 1.4-dihydroxy-2-naphthoic acid (DHNA) attenuates dextran sodium sulphate induced colitis by modulation of bacterial flora and lymphocyte homing. Gut (2006) 55(5):681–8. doi: 10.1136/gut.2005.070490
92. Cousin FJ, Jouan-Lanhouet S, Theret N, Brenner C, Jouan E, Le Moigne-Muller G, et al. The probiotic propionibacterium freudenreichii as a new adjuvant for TRAIL-based therapy in colorectal cancer. Oncotarget (2016) 7(6):7161–78. doi: 10.18632/oncotarget.6881
93. Wang Q, Wang K, Wu W, Lv L, Bian X, Yang L, et al. Administration of bifidobacterium bifidum CGMCC 15068 modulates gut microbiota and metabolome in azoxymethane (AOM)/dextran sulphate sodium (DSS)-induced colitis-associated colon cancer (CAC) in mice. Appl Microbiol Biotechnol (2020) 104(13):5915–28. doi: 10.1007/s00253-020-10621-z
94. Wang T, Zhang L, Wang P, Liu Y, Wang G, Shan Y, et al. Lactobacillus coryniformis MXJ32 administration ameliorates azoxymethane/dextran sulfate sodium-induced colitis-associated colorectal cancer via reshaping intestinal microenvironment and alleviating inflammatory response. Eur J Nutr (2022) 61(1):85–99. doi: 10.1007/s00394-021-02627-8
95. Chung Y, Ryu Y, An BC, Yoon YS, Choi O, Kim TY, et al. A synthetic probiotic engineered for colorectal cancer therapy modulates gut microbiota. Microbiome (2021) 9(1):122. doi: 10.1186/s40168-021-01071-4
96. Oh NS, Lee JY, Kim YT, Kim SH, Lee JH. Cancer-protective effect of a synbiotic combination between lactobacillus gasseri 505 and a cudrania tricuspidata leaf extract on colitis-associated colorectal cancer. Gut Microbes (2020) 12(1):1785803. doi: 10.1080/19490976.2020.1785803
97. Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Induction of experimental ulcerative colitis by fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut (2003) 52(1):79–83. doi: 10.1136/gut.52.1.79
98. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res (2012) 22(2):299–306. doi: 10.1101/gr.126516.111
99. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of fusobacterium with colorectal carcinoma. Genome Res (2012) 22(2):292–8. doi: 10.1101/gr.126573.111
100. Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol (2015) 1(5):653–61. doi: 10.1001/jamaoncol.2015.1377
101. Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, et al. Association of fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol (2016) 22(2):557–66. doi: 10.3748/wjg.v22.i2.557
102. Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol (2019) 17(3):156–66. doi: 10.1038/s41579-018-0129-6
103. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating e-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
104. Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-GalNAc. Cell Host Microbe (2016) 20(2):215–25. doi: 10.1016/j.chom.2016.07.006
105. Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun (2011) 79(7):2597–607. doi: 10.1128/IAI.05118-11
106. Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umana A, Zhang Y, et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal (2020) 13(641):eaba9157. doi: 10.1126/scisignal.aba9157
107. Swidsinski A, Khilkin M, Kerjaschki D, Schreiber S, Ortner M, Weber J, et al. Association between intraepithelial escherichia coli and colorectal cancer. Gastroenterology (1998) 115(2):281–6. doi: 10.1016/S0016-5085(98)70194-5
108. Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced escherichia coli adherence and invasion in crohn's disease and colon cancer. Gastroenterology (2004) 127(1):80–93. doi: 10.1053/j.gastro.2004.03.054
109. Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nat Chem (2015) 7(5):411–7. doi: 10.1038/nchem.2221
110. Veziant J, Gagniere J, Jouberton E, Bonnin V, Sauvanet P, Pezet D, et al. Association of colorectal cancer with pathogenic escherichia coli: Focus on mechanisms using optical imaging. World J Clin Oncol (2016) 7(3):293–301. doi: 10.5306/wjco.v7.i3.293
111. Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis (2011) 203(3):306–11. doi: 10.1093/jinfdis/jiq061
112. Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, et al. The bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis (2015) 60(2):208–15. doi: 10.1093/cid/ciu787
113. Obiso RJ Jr., Lyerly DM, Van Tassell RL, Wilkins TD. Proteolytic activity of the bacteroides fragilis enterotoxin causes fluid secretion and intestinal damage in vivo. Infect Immun (1995) 63(10):3820–6. doi: 10.1128/iai.63.10.3820-3826.1995
114. Kaakoush NO, Castano-Rodriguez N, Mitchell HM, Man SM. Global epidemiology of campylobacter infection. Clin Microbiol Rev (2015) 28(3):687–720. doi: 10.1128/CMR.00006-15
115. Lara-Tejero M, Galan JE. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science (2000) 290(5490):354–7. doi: 10.1126/science.290.5490.354
116. Gradel KO, Nielsen HL, Schonheyder HC, Ejlertsen T, Kristensen B, Nielsen H. Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology (2009) 137(2):495–501. doi: 10.1053/j.gastro.2009.04.001
117. Athan E, Cabiltes I, Coghill S, Bowe SJ. Silent but deadly: patients with enterococcal bacteraemia should be assessed for colorectal neoplasia. Med J Aust (2019) 210(2):86. doi: 10.5694/mja2.12027
118. Huycke MM, Moore D, Joyce W, Wise P, Shepard L, Kotake Y, et al. Extracellular superoxide production by enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol Microbiol (2001) 42(3):729–40. doi: 10.1046/j.1365-2958.2001.02638.x
119. Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis (2002) 23(3):529–36. doi: 10.1093/carcin/23.3.529
120. Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res (2008) 68(23):9909–17. doi: 10.1158/0008-5472.CAN-08-1551
121. Pericas JM, Corredoira J, Moreno A, Garcia-Pais MJ, Falces C, Rabunal R, et al. Relationship between enterococcus faecalis infective endocarditis and colorectal neoplasm: Preliminary results from a cohort of 154 patients. Rev Esp Cardiol (Engl Ed) (2017) 70(6):451–8. doi: 10.1016/j.rec.2016.10.013
122. Pasquereau-Kotula E, Martins M, Aymeric L, Dramsi S. Significance of streptococcus gallolyticus subsp. gallolyticus association with colorectal cancer. Front Microbiol (2018) 9:614. doi: 10.3389/fmicb.2018.00614
123. Boleij A, van Gelder MM, Swinkels DW, Tjalsma H. Clinical importance of streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis (2011) 53(9):870–8. doi: 10.1093/cid/cir609
124. Klein RS, Recco RA, Catalano MT, Edberg SC, Casey JI, Steigbigel NH. Association of streptococcus bovis with carcinoma of the colon. New Engl J Med (1977) 297(15):800–2. doi: 10.1056/NEJM197710132971503
125. Biarc J, Nguyen IS, Pini A, Gosse F, Richert S, Thierse D, et al. Carcinogenic properties of proteins with pro-inflammatory activity from streptococcus infantarius (formerly s.bovis). Carcinogenesis (2004) 25(8):1477–84. doi: 10.1093/carcin/bgh091
126. Kumar R, Herold JL, Taylor J, Xu J, Xu Y. Variations among streptococcus gallolyticus subsp. gallolyticus strains in connection with colorectal cancer. Sci Rep (2018) 8(1):1514. doi: 10.1038/s41598-018-19941-7
127. Abreu MT, Peek RM Jr. Gastrointestinal malignancy and the microbiome. Gastroenterology (2014) 146(6):1534–46.e3. doi: 10.1053/j.gastro.2014.01.001
129. Hartwich A, Konturek SJ, Pierzchalski P, Zuchowicz M, Labza H, Konturek PC, et al. Helicobacter pylori infection, gastrin, cyclooxygenase-2, and apoptosis in colorectal cancer. Int J Colorectal Dis (2001) 16(4):202–10. doi: 10.1007/s003840100288
130. Engin AB, Karahalil B, Engin A, Karakaya AE. Oxidative stress, helicobacter pylori, and OGG1 Ser326Cys, XPC Lys939Gln, and XPD Lys751Gln polymorphisms in a Turkish population with colorectal carcinoma. Genet Test Mol Biomark (2010) 14(4):559–64. doi: 10.1089/gtmb.2009.0195
131. Kuenstner JT, Naser S, Chamberlin W, Borody T, Graham DY, McNees A, et al. The consensus from the mycobacterium avium ssp. paratuberculosis (MAP) conference 2017. Front Public Health (2017) 5:208. doi: 10.3389/fpubh.2017.00208
132. Pierce ES. Ulcerative colitis and crohn's disease: is mycobacterium avium subspecies paratuberculosis the common villain? Gut Pathog (2010) 2(1):21. doi: 10.1186/1757-4749-2-21
133. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol (2004) 54(Pt 5):1469–76. doi: 10.1099/ijs.0.02873-0
134. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci United States Am (2013) 110(22):9066–71. doi: 10.1073/pnas.1219451110
135. Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sorensen SJ, et al. Early life treatment with vancomycin propagates akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia (2012) 55(8):2285–94. doi: 10.1007/s00125-012-2564-7
136. Baxter NT, Zackular JP, Chen GY, Schloss PD. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome (2014) 2:20. doi: 10.1186/2049-2618-2-20
137. Gu ZY, Pei WL, Zhang Y, Zhu J, Li L, Zhang Z. Akkermansia muciniphila in inflammatory bowel disease and colorectal cancer. Chin Med J (2021) 134(23):2841–3. doi: 10.1097/CM9.0000000000001829
138. Sun YY, Li M, Li YY, Li LX, Zhai WZ, Wang P, et al. The effect of clostridium butyricum on symptoms and fecal microbiota in diarrhea-dominant irritable bowel syndrome: a randomized, double-blind, placebo-controlled trial. Sci Rep (2018) 8(1):2964. doi: 10.1038/s41598-018-21241-z
139. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun (2015) 6:6528. doi: 10.1038/ncomms7528
140. Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest (2008) 118(2):534–44. doi: 10.1172/JCI33194
141. Lee JS, Tato CM, Joyce-Shaikh B, Gulan F, Cayatte C, Chen Y, et al. Interleukin-23-Independent IL-17 production regulates intestinal epithelial permeability. Immunity (2015) 43(4):727–38. doi: 10.1016/j.immuni.2015.09.003
142. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J (2012) 6(2):320–9. doi: 10.1038/ismej.2011.109
143. Benitez-Paez A, Gomez Del Pulgar EM, Sanz Y. The glycolytic versatility of bacteroides uniformis CECT 7771 and its genome response to oligo and polysaccharides. Front Cell Infect Microbiol (2017) 7:383. doi: 10.3389/fcimb.2017.00383
144. Gauffin Cano P, Santacruz A, Moya A, Sanz Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PloS One (2012) 7(7):e41079. doi: 10.1371/journal.pone.0041079
145. Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J (2017) 11(4):841–52. doi: 10.1038/ismej.2016.176
146. Liang S, Mao Y, Liao M, Xu Y, Chen Y, Huang X, et al. Gut microbiome associated with APC gene mutation in patients with intestinal adenomatous polyps. Int J Biol Sci (2020) 16(1):135–46. doi: 10.7150/ijbs.37399
147. Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun (2015) 6:8727. doi: 10.1038/ncomms9727
148. Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, desulfovibrio and enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol (2008) 23(8 Pt 1):1298–303. doi: 10.1111/j.1440-1746.2008.05490.x
149. Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset crohn's disease. Cell Host Microbe (2014) 15(3):382–92. doi: 10.1016/j.chom.2014.02.005
150. Hatakka K, Holma R, El-Nezami H, Suomalainen T, Kuisma M, Saxelin M, et al. The influence of lactobacillus rhamnosus LC705 together with propionibacterium freudenreichii ssp. shermanii JS on potentially carcinogenic bacterial activity in human colon. Int J Food Microbiol (2008) 128(2):406–10. doi: 10.1016/j.ijfoodmicro.2008.09.010
151. Barzegari A, Kheyrolahzadeh K, Hosseiniyan Khatibi SM, Sharifi S, Memar MY, Zununi Vahed S. The battle of probiotics and their derivatives against biofilms. Infect Drug Resist (2020) 13:659–72. doi: 10.2147/IDR.S232982
152. Speranza B, Liso A, Russo V, Corbo MR. Evaluation of the potential of biofilm formation of bifidobacterium longum subsp. infantis and lactobacillus reuteri as competitive biocontrol agents against pathogenic and food spoilage bacteria. Microorganisms (2020) 8(2):177. doi: 10.3390/microorganisms8020177
153. Aoudia N, Rieu A, Briandet R, Deschamps J, Chluba J, Jego G, et al. Biofilms of lactobacillus plantarum and lactobacillus fermentum: Effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol (2016) 53(Pt A):51–9. doi: 10.1016/j.fm.2015.04.009
154. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol (2014) 12(10):661–72. doi: 10.1038/nrmicro3344
155. Bishehsari F, Engen PA, Preite NZ, Tuncil YE, Naqib A, Shaikh M, et al. Dietary fiber treatment corrects the composition of gut microbiota, promotes SCFA production, and suppresses colon carcinogenesis. Genes (Basel) (2018) 9(2):102. doi: 10.3390/genes9020102
156. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell (2016) 167(5):1339–53.e21. doi: 10.1016/j.cell.2016.10.043
157. Ohigashi S, Sudo K, Kobayashi D, Takahashi O, Takahashi T, Asahara T, et al. Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Dig Dis Sci (2013) 58(6):1717–26. doi: 10.1007/s10620-012-2526-4
158. Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol (2013) 66(2):462–70. doi: 10.1007/s00248-013-0245-9
159. Lavoie S, Chun E, Bae S, Brennan CA, Gallini Comeau CA, Lang JK, et al. Expression of free fatty acid receptor 2 by dendritic cells prevents their expression of interleukin 27 and is required for maintenance of mucosal barrier and immune response against colorectal tumors in mice. Gastroenterology (2020) 158(5):1359–72.e9. doi: 10.1053/j.gastro.2019.12.027
160. Hanus M, Parada-Venegas D, Landskron G, Wielandt AM, Hurtado C, Alvarez K, et al. Immune system, microbiota, and microbial metabolites: The unresolved triad in colorectal cancer microenvironment. Front Immunol (2021) 12:612826. doi: 10.3389/fimmu.2021.612826
161. Kalina U, Koyama N, Hosoda T, Nuernberger H, Sato K, Hoelzer D, et al. Enhanced production of IL-18 in butyrate-treated intestinal epithelium by stimulation of the proximal promoter region. Eur J Immunol (2002) 32(9):2635–43. doi: 10.1002/1521-4141(200209)32:9<2635::AID-IMMU2635>3.0.CO;2-N
162. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity (2014) 40(1):128–39. doi: 10.1016/j.immuni.2013.12.007
163. Salcedo R, Worschech A, Cardone M, Jones Y, Gyulai Z, Dai RM, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med (2010) 207(8):1625–36. doi: 10.1084/jem.20100199
164. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci United States Am (2014) 111(6):2247–52. doi: 10.1073/pnas.1322269111
165. Nastasi C, Candela M, Bonefeld CM, Geisler C, Hansen M, Krejsgaard T, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci Rep (2015) 5:16148. doi: 10.1038/srep16148
166. Ji J, Shu D, Zheng M, Wang J, Luo C, Wang Y, et al. Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Sci Rep (2016) 6:24838. doi: 10.1038/srep24838
167. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature (2013) 504(7480):451–5. doi: 10.1038/nature12726
168. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science (2013) 341(6145):569–73. doi: 10.1126/science.1241165
169. Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell (2016) 165(7):1708–20. doi: 10.1016/j.cell.2016.05.018
170. Irrazabal T, Thakur BK, Kang M, Malaise Y, Streutker C, Wong EOY, et al. Limiting oxidative DNA damage reduces microbe-induced colitis-associated colorectal cancer. Nat Commun (2020) 11(1):1802. doi: 10.1038/s41467-020-15549-6
171. Wang W, Fang D, Zhang H, Xue J, Wangchuk D, Du J, et al. Sodium butyrate selectively kills cancer cells and inhibits migration in colorectal cancer by targeting thioredoxin-1. Onco Targets Ther (2020) 13:4691–704. doi: 10.2147/OTT.S235575
172. Casero RA Jr., Murray Stewart T, Pegg AE. Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer (2018) 18(11):681–95. doi: 10.1038/s41568-018-0050-3
173. Roy UK, Rial NS, Kachel KL, Gerner EW. Activated K-RAS increases polyamine uptake in human colon cancer cells through modulation of caveolar endocytosis. Mol Carcinog (2008) 47(7):538–53. doi: 10.1002/mc.20414
174. Raj KP, Zell JA, Rock CL, McLaren CE, Zoumas-Morse C, Gerner EW, et al. Role of dietary polyamines in a phase III clinical trial of difluoromethylornithine (DFMO) and sulindac for prevention of sporadic colorectal adenomas. Br J Cancer (2013) 108(3):512–8. doi: 10.1038/bjc.2013.15
175. Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, et al. Polyamine catabolism contributes to enterotoxigenic bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci United States Am (2011) 108(37):15354–9. doi: 10.1073/pnas.1010203108
176. Chagneau CV, Garcie C, Bossuet-Greif N, Tronnet S, Brachmann AO, Piel J, et al. The polyamine spermidine modulates the production of the bacterial genotoxin colibactin. mSphere (2019) 4(5). doi: 10.1128/mSphere.00414-19
177. Wang C, Ruan P, Zhao Y, Li X, Wang J, Wu X, et al. Spermidine/spermine N1-acetyltransferase regulates cell growth and metastasis via AKT/beta-catenin signaling pathways in hepatocellular and colorectal carcinoma cells. Oncotarget (2017) 8(1):1092–109. doi: 10.18632/oncotarget.13582
178. Yang Y, Misra BB, Liang L, Bi D, Weng W, Wu W, et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics (2019) 9(14):4101–14. doi: 10.7150/thno.35186
179. Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci USA (2014) 111(51):18321–6. doi: 10.1073/pnas.1406199111
180. Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab (2015) 21(6):891–7. doi: 10.1016/j.cmet.2015.04.011
181. Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med (2019) 25(4):667–78. doi: 10.1038/s41591-019-0405-7
182. Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol (2020) 38:23–48. doi: 10.1146/annurev-immunol-070119-115104
183. O'Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, et al. Fat, fibre and cancer risk in African americans and rural africans. Nat Commun (2015) 6:6342. doi: 10.1038/ncomms7342
184. Aymeric L, Donnadieu F, Mulet C, du Merle L, Nigro G, Saffarian A, et al. Colorectal cancer specific conditions promote streptococcus gallolyticus gut colonization. Proc Natl Acad Sci USA (2018) 115(2):E283–E91. doi: 10.1073/pnas.1715112115
185. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med (2019) 25(6):968–76. doi: 10.1038/s41591-019-0458-7
186. Mroz MS, Lajczak NK, Goggins BJ, Keely S, Keely SJ. The bile acids, deoxycholic acid and ursodeoxycholic acid, regulate colonic epithelial wound healing. Am J Physiol Gastrointestinal Liver Physiol (2018) 314(3):G378–G87. doi: 10.1152/ajpgi.00435.2016
187. Pai R, Tarnawski AS, Tran T. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell (2004) 15(5):2156–63. doi: 10.1091/mbc.e03-12-0894
188. Liu L, Dong W, Wang S, Zhang Y, Liu T, Xie R, et al. Deoxycholic acid disrupts the intestinal mucosal barrier and promotes intestinal tumorigenesis. Food Funct (2018) 9(11):5588–97. doi: 10.1039/C8FO01143E
189. Baek MK, Park JS, Park JH, Kim MH, Kim HD, Bae WK, et al. Lithocholic acid upregulates uPAR and cell invasiveness via MAPK and AP-1 signaling in colon cancer cells. Cancer Lett (2010) 290(1):123–8. doi: 10.1016/j.canlet.2009.08.030
190. Nguyen TT, Lian S, Ung TT, Xia Y, Han JY, Jung YD. Lithocholic acid stimulates IL-8 expression in human colorectal cancer cells Via activation of Erk1/2 MAPK and suppression of STAT3 activity. J Cell Biochem (2017) 118(9):2958–67. doi: 10.1002/jcb.25955
191. Farhana L, Nangia-Makker P, Arbit E, Shango K, Sarkar S, Mahmud H, et al. Bile acid: a potential inducer of colon cancer stem cells. Stem Cell Res Ther (2016) 7(1):181. doi: 10.1186/s13287-016-0439-4
192. Bell HN, Rebernick RJ, Goyert J, Singhal R, Kuljanin M, Kerk SA, et al. Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell (2022) 40(2):185–200.e6. doi: 10.1016/j.ccell.2021.12.001
193. Blachier F, Davila AM, Mimoun S, Benetti PH, Atanasiu C, Andriamihaja M, et al. Luminal sulfide and large intestine mucosa: friend or foe? Amino Acids (2010) 39(2):335–47. doi: 10.1007/s00726-009-0445-2
194. Kushkevych I, Kotrsova V, Dordevic D, Bunkova L, Vitezova M, Amedei A. Hydrogen sulfide effects on the survival of lactobacilli with emphasis on the development of inflammatory bowel diseases. Biomolecules (2019) 9(12):752. doi: 10.3390/biom9120752
195. Xie G, Raufman JP. Role of the aryl hydrocarbon receptor in colon neoplasia. Cancers (Basel) (2015) 7(3):1436–46. doi: 10.3390/cancers7030847
196. Ikuta T, Kobayashi Y, Kitazawa M, Shiizaki K, Itano N, Noda T, et al. ASC-associated inflammation promotes cecal tumorigenesis in aryl hydrocarbon receptor-deficient mice. Carcinogenesis (2013) 34(7):1620–7. doi: 10.1093/carcin/bgt083
197. Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, et al. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci United States Am (2009) 106(32):13481–6. doi: 10.1073/pnas.0902132106
198. Monteleone I, MacDonald TT, Pallone F, Monteleone G. The aryl hydrocarbon receptor in inflammatory bowel disease: linking the environment to disease pathogenesis. Curr Opin Gastroenterol (2012) 28(4):310–3. doi: 10.1097/MOG.0b013e328352ad69
199. Furumatsu K, Nishiumi S, Kawano Y, Ooi M, Yoshie T, Shiomi Y, et al. A role of the aryl hydrocarbon receptor in attenuation of colitis. Dig Dis Sci (2011) 56(9):2532–44. doi: 10.1007/s10620-011-1643-9
200. Rossi T, Vergara D, Fanini F, Maffia M, Bravaccini S, Pirini F. Microbiota-derived metabolites in tumor progression and metastasis. Int J Mol Sci (2020) 21(16):5786. doi: 10.3390/ijms21165786
201. Dvorak Z, Kopp F, Costello CM, Kemp JS, Li H, Vrzalova A, et al. Targeting the pregnane X receptor using microbial metabolite mimicry. EMBO Mol Med (2020) 12(4):e11621. doi: 10.15252/emmm.201911621
202. Mezheyeuski A, Micke P, Martin-Bernabe A, Backman M, Hrynchyk I, Hammarstrom K, et al. The immune landscape of colorectal cancer. Cancers (Basel) (2021) 13(21):5545. doi: 10.3390/cancers13215545
203. Alexander KL, Targan SR, Elson CO 3rd. Microbiota activation and regulation of innate and adaptive immunity. Immunol Rev (2014) 260(1):206–20. doi: 10.1111/imr.12180
204. Duan T, Du Y, Xing C, Wang HY, Wang RF. Toll-like receptor signaling and its role in cell-mediated immunity. Front Immunol (2022) 13:812774. doi: 10.3389/fimmu.2022.812774
205. Ma Q, Xing C, Long W, Wang HY, Liu Q, Wang RF. Impact of microbiota on central nervous system and neurological diseases: the gut-brain axis. J Neuroinflamm (2019) 16(1):53. doi: 10.1186/s12974-019-1434-3
206. Semin I, Ninnemann J, Bondareva M, Gimaev I, Kruglov AA. Interplay between microbiota, toll-like receptors and cytokines for the maintenance of epithelial barrier integrity. Front Med (Lausanne) (2021) 8:644333. doi: 10.3389/fmed.2021.644333
207. Valentini M, Piermattei A, Di Sante G, Migliara G, Delogu G, Ria F. Immunomodulation by gut microbiota: role of toll-like receptor expressed by T cells. J Immunol Res (2014) 2014:586939. doi: 10.1155/2014/586939
208. Xia XJ, Cui J, Wang HLY, Zhu L, Matsueda S, Wang QF, et al. NLRX1 negatively regulates TLR-induced NF-kappa b signaling by targeting TRAF6 and IKK. Immunity (2011) 34(6):843–53. doi: 10.1016/j.immuni.2011.02.022
209. Cui J, Zhu L, Xia XJ, Wang HY, Legras X, Hong J, et al. NLRC5 negatively regulates the NF-kappa b and type I interferon signaling pathways. Cell (2010) 141(3):483–96. doi: 10.1016/j.cell.2010.03.040
210. Wu C, Su Z, Lin M, Ou J, Zhao W, Cui J, et al. NLRP11 attenuates toll-like receptor signalling by targeting TRAF6 for degradation via the ubiquitin ligase RNF19A. Nat Commun (2017) 8(1):1977–. doi: 10.1038/s41467-017-02073-3
211. Feng Y, Duan T, Du Y, Jin S, Wang M, Cui J, et al. LRRC25 functions as an inhibitor of NF-κB signaling pathway by promoting p65/RelA for autophagic degradation. Sci Rep-Uk (2017) 7(1):13448. doi: 10.1038/s41598-017-12573-3
212. Du Y, Duan T, Feng Y, Liu Q, Lin M, Cui J, et al. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J (2018) 37(3):351–66. doi: 10.15252/embj.201796781
213. Tan XJ, Sun LJ, Chen JQ, Chen ZJJ. Detection of microbial infections through innate immune sensing of nucleic acids. Annu Rev Microbiol (2018) 72:447–78. doi: 10.1146/annurev-micro-102215-095605
214. Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell (2020) 180(6):1044–66. doi: 10.1016/j.cell.2020.02.041
215. Wirusanti NI, Baldridge MT, Harris VC. Microbiota regulation of viral infections through interferon signaling. Trends Microbiol (2022) (8):778–92. doi: 10.1016/j.tim.2022.01.007
216. Kawashima T, Kosaka A, Yan HM, Guo ZJ, Uchiyama R, Fukui R, et al. Double-stranded RNA of intestinal commensal but not pathogenic bacteria triggers production of protective interferon-beta. Immunity (2013) 38(6):1187–97. doi: 10.1016/j.immuni.2013.02.024
217. Erttmann SF, Swacha P, Aung KM, Brindefalk B, Jiang H, Hartlova A, et al. The gut microbiota prime systemic antiviral immunity via the cGAS-STING-IFN-I axis. Immunity (2022) 55(5):847–61.e10. doi: 10.1016/j.immuni.2022.04.006
218. Du Y, Luo Y, Hu Z, Lu J, Liu X, Xing C, et al. Activation of cGAS-STING by lethal malaria N67C dictates immunity and mortality through induction of CD11b(+) Ly6C(hi) proinflammatory monocytes. Adv Sci (Weinh) (2022):e2103701. doi: 10.1002/advs.202103701
219. Perkins DJ, Rajaiah R, Tennant SM, Ramachandran G, Higginson EE, Dyson TN, et al. Salmonella typhimurium Co-opts the host type I IFN system to restrict macrophage innate immune transcriptional responses selectively. J Immunol (2015) 195(5):2461–71. doi: 10.4049/jimmunol.1500105
220. Deriu E, Boxx GM, He X, Pan C, Benavidez SD, Cen L, et al. Influenza virus affects intestinal microbiota and secondary salmonella infection in the gut through type I interferons. PloS Pathog (2016) 12(5):e1005572. doi: 10.1371/journal.ppat.1005572
221. Cui J, Song Y, Li Y, Zhu Q, Tan P, Qin Y, et al. USP3 inhibits type I interferon signaling by deubiquitinating RIG-i-like receptors. Cell Res (2014) 24(4):400–16. doi: 10.1038/cr.2013.170
222. Lin M, Zhao Z, Yang Z, Meng Q, Tan P, Xie W, et al. USP38 inhibits type I interferon signaling by editing TBK1 ubiquitination through NLRP4 signalosome. Mol Cell (2016) 64(2):267–81. doi: 10.1016/j.molcel.2016.08.029
223. Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, et al. TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol Cell (2016) 64(1):105–19. doi: 10.1016/j.molcel.2016.08.025
224. Tan P, He L, Cui J, Qian C, Cao X, Lin M, et al. Assembly of the WHIP-TRIM14-PPP6C mitochondrial complex promotes RIG-I-Mediated antiviral signaling. Mol Cell (2017) 68(2):293–307.e5. doi: 10.1016/j.molcel.2017.09.035
225. Mizuno R, Kawada K, Itatani Y, Ogawa R, Kiyasu Y, Sakai Y. The role of tumor-associated neutrophils in colorectal cancer. Int J Mol Sci (2019) 20(3):529. doi: 10.3390/ijms20030529
226. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity (2013) 39(4):782–95. doi: 10.1016/j.immuni.2013.10.003
227. Germann M, Zangger N, Sauvain MO, Sempoux C, Bowler AD, Wirapati P, et al. Neutrophils suppress tumor-infiltrating T cells in colon cancer via matrix metalloproteinase-mediated activation of TGFbeta. EMBO Mol Med (2020) 12(1):e10681. doi: 10.15252/emmm.201910681
228. Governa V, Trella E, Mele V, Tornillo L, Amicarella F, Cremonesi E, et al. The interplay between neutrophils and CD8(+) T cells improves survival in human colorectal cancer. Clin Cancer Res (2017) 23(14):3847–58. doi: 10.1158/1078-0432.CCR-16-2047
229. Wikberg ML, Ling A, Li X, Oberg A, Edin S, Palmqvist R. Neutrophil infiltration is a favorable prognostic factor in early stages of colon cancer. Hum Pathol (2017) 68:193–202. doi: 10.1016/j.humpath.2017.08.028
230. Haram A, Boland MR, Kelly ME, Bolger JC, Waldron RM, Kerin MJ. The prognostic value of neutrophil-to-lymphocyte ratio in colorectal cancer: A systematic review. J Surg Oncol (2017) 115(4):470–9. doi: 10.1002/jso.24523
231. Mazaki J, Katsumata K, Kasahara K, Tago T, Wada T, Kuwabara H, et al. Neutrophil-to-lymphocyte ratio is a prognostic factor for colon cancer: a propensity score analysis. BMC Cancer (2020) 20(1):922. doi: 10.1186/s12885-020-07429-5
232. Wang H, Tian T, Zhang J. Tumor-associated macrophages (TAMs) in colorectal cancer (CRC): From mechanism to therapy and prognosis. Int J Mol Sci (2021) 22(16):8470. doi: 10.3390/ijms22168470
233. Lim SY, Gordon-Weeks A, Allen D, Kersemans V, Beech J, Smart S, et al. Cd11b(+) myeloid cells support hepatic metastasis through down-regulation of angiopoietin-like 7 in cancer cells. Hepatology (2015) 62(2):521–33. doi: 10.1002/hep.27838
234. Yomoda T, Sudo T, Kawahara A, Shigaki T, Shimomura S, Tajiri K, et al. The immunoscore is a superior prognostic tool in stages II and III colorectal cancer and is significantly correlated with programmed death-ligand 1 (PD-L1) expression on tumor-infiltrating mononuclear cells. Ann Surg Oncol (2019) 26(2):415–24. doi: 10.1245/s10434-018-07110-z
235. Forssell J, Oberg A, Henriksson ML, Stenling R, Jung A, Palmqvist R. High macrophage infiltration along the tumor front correlates with improved survival in colon cancer. Clin Cancer Res (2007) 13(5):1472–9. doi: 10.1158/1078-0432.CCR-06-2073
236. Nagorsen D, Voigt S, Berg E, Stein H, Thiel E, Loddenkemper C. Tumor-infiltrating macrophages and dendritic cells in human colorectal cancer: relation to local regulatory T cells, systemic T-cell response against tumor-associated antigens and survival. J Transl Med (2007) 5:62. doi: 10.1186/1479-5876-5-62
237. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science (2011) 331(6024):1565–70. doi: 10.1126/science.1203486
238. Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol (2012) 12(4):253–68. doi: 10.1038/nri3175
239. Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest (2015) 125(9):3356–64. doi: 10.1172/JCI80005
240. OuYang LY, Wu XJ, Ye SB, Zhang RX, Li ZL, Liao W, et al. Tumor-induced myeloid-derived suppressor cells promote tumor progression through oxidative metabolism in human colorectal cancer. J Transl Med (2015) 13:47. doi: 10.1186/s12967-015-0410-7
241. Zhang B, Wang Z, Wu L, Zhang M, Li W, Ding J, et al. Circulating and tumor-infiltrating myeloid-derived suppressor cells in patients with colorectal carcinoma. PloS One (2013) 8(2):e57114. doi: 10.1371/journal.pone.0057114
242. Sun HL, Zhou X, Xue YF, Wang K, Shen YF, Mao JJ, et al. Increased frequency and clinical significance of myeloid-derived suppressor cells in human colorectal carcinoma. World J Gastroenterol (2012) 18(25):3303–9. doi: 10.3748/wjg.v18.i25.3303
243. Tada K, Kitano S, Shoji H, Nishimura T, Shimada Y, Nagashima K, et al. Pretreatment immune status correlates with progression-free survival in chemotherapy-treated metastatic colorectal cancer patients. Cancer Immunol Res (2016) 4(7):592–9. doi: 10.1158/2326-6066.CIR-15-0298
244. Jacquelot N, Seillet C, Vivier E, Belz GT. Innate lymphoid cells and cancer. Nat Immunol (2022) 23(3):371–9. doi: 10.1038/s41590-022-01127-z
245. Chen H, Sun L, Feng L, Yin Y, Zhang W. Role of innate lymphoid cells in obesity and insulin resistance. Front Endocrinol (Lausanne) (2022) 13:855197. doi: 10.3389/fendo.2022.855197
246. Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature (2013) 498(7452):113–7. doi: 10.1038/nature12240
247. Eberl G, Di Santo JP, Vivier E. The brave new world of innate lymphoid cells. Nat Immunol (2015) 16(1):1–5. doi: 10.1038/ni.3059
248. Huang Q, Cao W, Mielke LA, Seillet C, Belz GT, Jacquelot N. Innate lymphoid cells in colorectal cancers: A double-edged sword. Front Immunol (2019) 10:3080. doi: 10.3389/fimmu.2019.03080
249. Tallerico R, Todaro M, Di Franco S, Maccalli C, Garofalo C, Sottile R, et al. Human NK cells selective targeting of colon cancer-initiating cells: a role for natural cytotoxicity receptors and MHC class I molecules. J Immunol (2013) 190(5):2381–90. doi: 10.4049/jimmunol.1201542
250. Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity (2013) 38(4):769–81. doi: 10.1016/j.immuni.2013.02.010
251. Forkel M, van Tol S, Hoog C, Michaelsson J, Almer S, Mjosberg J. Distinct alterations in the composition of mucosal innate lymphoid cells in newly diagnosed and established crohn's disease and ulcerative colitis. J Crohns Colitis (2019) 13(1):67–78. doi: 10.1093/ecco-jcc/jjy119
252. Ercolano G, Gomez-Cadena A, Dumauthioz N, Vanoni G, Kreutzfeldt M, Wyss T, et al. PPAR drives IL-33-dependent ILC2 pro-tumoral functions. Nat Commun (2021) 12(1):2538. doi: 10.1038/s41467-021-22764-2
253. Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid a to promote local effector Th17 responses. Cell (2015) 163(2):381–93. doi: 10.1016/j.cell.2015.08.061
254. Chae WJ, Gibson TF, Zelterman D, Hao L, Henegariu O, Bothwell AL. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc Natl Acad Sci USA (2010) 107(12):5540–4. doi: 10.1073/pnas.0912675107
255. Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature (2012) 491(7423):254–8. doi: 10.1038/nature11465
256. Wang K, Kim MK, Di Caro G, Wong J, Shalapour S, Wan J, et al. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity (2014) 41(6):1052–63. doi: 10.1016/j.immuni.2014.11.009
257. O'Connor W Jr., Kamanaka M, Booth CJ, Town T, Nakae S, Iwakura Y, et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol (2009) 10(6):603–9. doi: 10.1038/ni.1736
258. Ogawa A, Andoh A, Araki Y, Bamba T, Fujiyama Y. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clin Immunol (2004) 110(1):55–62. doi: 10.1016/j.clim.2003.09.013
259. Yang XO, Chang SH, Park H, Nurieva R, Shah B, Acero L, et al. Regulation of inflammatory responses by IL-17F. J Exp Med (2008) 205(5):1063–75. doi: 10.1084/jem.20071978
260. Rutz S, Wang X, Ouyang W. The IL-20 subfamily of cytokines–from host defence to tissue homeostasis. Nat Rev Immunol (2014) 14(12):783–95. doi: 10.1038/nri3766
261. Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity (2008) 29(6):947–57. doi: 10.1016/j.immuni.2008.11.003
262. Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature (2012) 491(7423):259–63. doi: 10.1038/nature11535
263. Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity (2014) 40(5):772–84. doi: 10.1016/j.immuni.2014.03.010
264. Mao K, Baptista AP, Tamoutounour S, Zhuang L, Bouladoux N, Martins AJ, et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature (2018) 554(7691):255–9. doi: 10.1038/nature25437
265. Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science (2021) 371(6536):eabc4552. doi: 10.1126/science.abc4552
266. Yang Y, Cai Q, Shu XO, Steinwandel MD, Blot WJ, Zheng W, et al. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int J Cancer J Int Cancer (2019) 144(10):2381–9. doi: 10.1002/ijc.31941
267. Matsui A, Jin JO, Johnston CD, Yamazaki H, Houri-Haddad Y, Rittling SR. Pathogenic bacterial species associated with endodontic infection evade innate immune control by disabling neutrophils. Infect Immun (2014) 82(10):4068–79. doi: 10.1128/IAI.02256-14
268. Kim SO, Sheikh HI, Ha SD, Martins A, Reid G. G-CSF-mediated inhibition of JNK is a key mechanism for lactobacillus rhamnosus-induced suppression of TNF production in macrophages. Cell Microbiol (2006) 8(12):1958–71. doi: 10.1111/j.1462-5822.2006.00763.x
269. Zhang DY, Pan ZY, Yu XK, Chen YF, Gao CH, Yang YT, et al. Bifidobacterium lactis BB-12 attenuates macrophage aging induced by d-galactose and promotes M2 macrophage polarization. J Immunol Res (2019) 2019:4657928. doi: 10.1155/2019/4657928
270. Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity (2019) 50(2):432–45.e7. doi: 10.1016/j.immuni.2018.12.018
271. Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front Microbiol (2016) 7:979. doi: 10.3389/fmicb.2016.00979
272. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity (2015) 42(2):344–55. doi: 10.1016/j.immuni.2015.01.010
273. Grizotte-Lake M, Zhong G, Duncan K, Kirkwood J, Iyer N, Smolenski I, et al. Commensals suppress intestinal epithelial cell retinoic acid synthesis to regulate interleukin-22 activity and prevent microbial dysbiosis. Immunity (2018) 49(6):1103–15.e6. doi: 10.1016/j.immuni.2018.11.018
274. Mielke LA, Jones SA, Raverdeau M, Higgs R, Stefanska A, Groom JR, et al. Retinoic acid expression associates with enhanced IL-22 production by gammadelta T cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med (2013) 210(6):1117–24. doi: 10.1084/jem.20121588
275. Mutala LB, Deleine C, Karakachoff M, Dansette D, Ducoin K, Oger R, et al. The caspase-1/IL-18 axis of the inflammasome in tumor cells: A modulator of the Th1/Tc1 response of tumor-infiltrating T lymphocytes in colorectal cancer. Cancers (Basel) (2021) 13(2):189. doi: 10.3390/cancers13020189
276. Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu Rev Immunol (2002) 20:323–70. doi: 10.1146/annurev.immunol.20.100201.131730
277. Galon J, Fridman WH, Pages F. The adaptive immunologic microenvironment in colorectal cancer: a novel perspective. Cancer Res (2007) 67(5):1883–6. doi: 10.1158/0008-5472.CAN-06-4806
278. Naito Y, Saito K, Shiiba K, Ohuchi A, Saigenji K, Nagura H, et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res (1998) 58(16):3491–4.
279. Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA (2005) 102(51):18538–43. doi: 10.1073/pnas.0509182102
280. Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res (2011) 71(4):1263–71. doi: 10.1158/0008-5472.CAN-10-2907
281. Saraiva M, O'Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol (2010) 10(3):170–81. doi: 10.1038/nri2711
282. Huber S, Schramm C, Lehr HA, Mann A, Schmitt S, Becker C, et al. Cutting edge: TGF-beta signaling is required for the in vivo expansion and immunosuppressive capacity of regulatory CD4+CD25+ T cells. J Immunol (2004) 173(11):6526–31. doi: 10.4049/jimmunol.173.11.6526
283. Olguin JE, Medina-Andrade I, Rodriguez T, Rodriguez-Sosa M, Terrazas LI. Relevance of regulatory T cells during colorectal cancer development. Cancers (Basel) (2020) 12(7):1888. doi: 10.3390/cancers12071888
284. Loddenkemper C, Schernus M, Noutsias M, Stein H, Thiel E, Nagorsen D. In situ analysis of FOXP3+ regulatory T cells in human colorectal cancer. J Transl Med (2006) 4:52. doi: 10.1186/1479-5876-4-52
285. Yeong J, Thike AA, Lim JC, Lee B, Li H, Wong SC, et al. Higher densities of Foxp3(+) regulatory T cells are associated with better prognosis in triple-negative breast cancer. Breast Cancer Res Treat (2017) 163(1):21–35. doi: 10.1007/s10549-017-4161-4
286. Ward-Hartstonge KA, McCall JL, McCulloch TR, Kamps AK, Girardin A, Cretney E, et al. Inclusion of BLIMP-1(+) effector regulatory T cells improves the immunoscore in a cohort of new Zealand colorectal cancer patients: a pilot study. Cancer Immunol Immunother CII (2017) 66(4):515–22. doi: 10.1007/s00262-016-1951-1
287. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell (2009) 139(3):485–98. doi: 10.1016/j.cell.2009.09.033
288. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell (2015) 163(2):367–80. doi: 10.1016/j.cell.2015.08.058
289. Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol (2008) 8(5):337–48. doi: 10.1038/nri2295
290. Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol (2009) 27:485–517. doi: 10.1146/annurev.immunol.021908.132710
291. Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, et al. Control of TH17 cells occurs in the small intestine. Nature (2011) 475(7357):514–8. doi: 10.1038/nature10228
292. Limagne E, Euvrard R, Thibaudin M, Rebe C, Derangere V, Chevriaux A, et al. Accumulation of MDSC and Th17 cells in patients with metastatic colorectal cancer predicts the efficacy of a FOLFOX-bevacizumab drug treatment regimen. Cancer Res (2016) 76(18):5241–52. doi: 10.1158/0008-5472.CAN-15-3164
293. Lee JY, Seo EH, Oh CS, Paik JH, Hwang DY, Lee SH, et al. Impact of circulating T helper 1 and 17 cells in the blood on regional lymph node invasion in colorectal cancer. J Cancer (2017) 8(7):1249–54. doi: 10.7150/jca.18230
294. Amicarella F, Muraro MG, Hirt C, Cremonesi E, Padovan E, Mele V, et al. Dual role of tumour-infiltrating T helper 17 cells in human colorectal cancer. Gut (2017) 66(4):692–704. doi: 10.1136/gutjnl-2015-310016
295. De Simone V, Pallone F, Monteleone G, Stolfi C. Role of T17 cytokines in the control of colorectal cancer. Oncoimmunology (2013) 2(12):e26617. doi: 10.4161/onci.26617
296. Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, et al. Serum amyloid a proteins induce pathogenic Th17 cells and promote inflammatory disease. Cell (2020) 180(1):79–91.e16. doi: 10.1016/j.cell.2019.11.026
297. Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature (2010) 467(7318):967–71. doi: 10.1038/nature09447
298. Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature (2006) 441(7090):231–4. doi: 10.1038/nature04754
299. Conti HR, Gaffen SL. Host responses to candida albicans: Th17 cells and mucosal candidiasis. Microbes Infect / Institut Pasteur (2010) 12(7):518–27. doi: 10.1016/j.micinf.2010.03.013
300. Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci USA (2016) 113(50):E8141–E50. doi: 10.1073/pnas.1617460113
301. Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature (2015) 520(7545):104–8. doi: 10.1038/nature14052
302. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA (2010) 107(27):12204–9. doi: 10.1073/pnas.0909122107
303. Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, et al. C-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature (2018) 554(7692):373–7. doi: 10.1038/nature25500
304. Kuczma MP, Szurek EA, Cebula A, Chassaing B, Jung YJ, Kang SM, et al. Commensal epitopes drive differentiation of colonic tregs. Sci Adv (2020) 6(16):eaaz3186. doi: 10.1126/sciadv.aaz3186
305. Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Trans Med (2014) 6(220):220ra11. doi: 10.1126/scitranslmed.3008051
306. Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature (2011) 478(7368):250–4. doi: 10.1038/nature10434
307. O'Mahony C, Scully P, O'Mahony D, Murphy S, O'Brien F, Lyons A, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PloS Pathog (2008) 4(8):e1000112. doi: 10.1371/journal.ppat.1000112
308. Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, et al. MUCOSAL IMMUNOLOGY. individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science (2015) 349(6251):993–7. doi: 10.1126/science.aaa9420
309. Jia YP, Wang K, Zhang ZJ, Tong YN, Han D, Hu CY, et al. TLR2/TLR4 activation induces tregs and suppresses intestinal inflammation caused by fusobacterium nucleatum in vivo. PloS One (2017) 12(10):e0186179. doi: 10.1371/journal.pone.0186179
310. Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am J Respir Crit Care Med (2009) 179(3):186–93. doi: 10.1164/rccm.200806-951OC
311. Tang C, Kamiya T, Liu Y, Kadoki M, Kakuta S, Oshima K, et al. Inhibition of dectin-1 signaling ameliorates colitis by inducing lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe (2015) 18(2):183–97. doi: 10.1016/j.chom.2015.07.003
312. Sarrabayrouse G, Bossard C, Chauvin JM, Jarry A, Meurette G, Quevrain E, et al. CD4CD8alphaalpha lymphocytes, a novel human regulatory T cell subset induced by colonic bacteria and deficient in patients with inflammatory bowel disease. PloS Biol (2014) 12(4):e1001833. doi: 10.1371/journal.pbio.1001833
313. Wiechers C, Zou M, Galvez E, Beckstette M, Ebel M, Strowig T, et al. The microbiota is dispensable for the early stages of peripheral regulatory T cell induction within mesenteric lymph nodes. Cell Mol Immunol (2021) 18(5):1211–21. doi: 10.1038/s41423-021-00647-2
314. Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature (2014) 510(7503):152–6. doi: 10.1038/nature13279
315. Daillere R, Vetizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, et al. Enterococcus hirae and barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity (2016) 45(4):931–43. doi: 10.1016/j.immuni.2016.09.009
316. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature (2019) 565(7741):600–5. doi: 10.1038/s41586-019-0878-z
317. Guo L, Wang C, Qiu X, Pu X, Chang P. Colorectal cancer immune infiltrates: Significance in patient prognosis and immunotherapeutic efficacy. Front Immunol (2020) 11:1052. doi: 10.3389/fimmu.2020.01052
318. Briukhovetska D, Dorr J, Endres S, Libby P, Dinarello CA, Kobold S. Interleukins in cancer: from biology to therapy. Nat Rev Cancer (2021) 21(8):481–99. doi: 10.1038/s41568-021-00363-z
319. McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol (2019) 20(2):e77–91. doi: 10.1016/S1470-2045(18)30952-5
320. Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science (2018) 359(6382):1366–70. doi: 10.1126/science.aar6918
321. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science (2013) 342(6161):967–70. doi: 10.1126/science.1240527
322. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science (2013) 342(6161):971–6. doi: 10.1126/science.1240537
323. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
324. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (2018) 359(6371):104–8. doi: 10.1126/science.aao3290
325. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
326. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science (2020) 369(6510):1481–9. doi: 10.1126/science.abc3421
327. Sahin U, Tureci O. Personalized vaccines for cancer immunotherapy. Science (2018) 359(6382):1355–60. doi: 10.1126/science.aar7112
328. Sasada T, Kibe S, Akagi Y, Itoh K. Personalized peptide vaccination for advanced colorectal cancer. Oncoimmunology (2015) 4(5):e1005512. doi: 10.1080/2162402X.2015.1005512
329. Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, et al. T-Cell transfer therapy targeting mutant KRAS in cancer. New Engl J Med (2016) 375(23):2255–62. doi: 10.1056/NEJMoa1609279
330. Parkhurst MR, Yang JC, Langan RC, Dudley ME, Nathan DA, Feldman SA, et al. T Cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther (2011) 19(3):620–6. doi: 10.1038/mt.2010.272
331. Eugene J, Jouand N, Ducoin K, Dansette D, Oger R, Deleine C, et al. The inhibitory receptor CD94/NKG2A on CD8(+) tumor-infiltrating lymphocytes in colorectal cancer: a promising new druggable immune checkpoint in the context of HLAE/beta2m overexpression. Modern Pathol (2020) 33(3):468–82. doi: 10.1038/s41379-019-0322-9
332. Bendell JC, Powderly JD, Lieu CH, Eckhardt SG, Hurwitz H, Hochster HS, et al. Safety and efficacy of MPDL3280A (anti-PDL1) in combination with bevacizumab (bev) and/or FOLFOX in patients (pts) with metastatic colorectal cancer (mCRC). J Clin Oncol (2015) 33. doi: 10.1200/jco.2015.33.3_suppl.704
333. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. New Engl J Med (2015) 372(26):2509–20. doi: 10.1056/NEJMoa1500596
334. Golkhalkhali B, Rajandram R, Paliany AS, Ho GF, Wan Ishak WZ, Johari CS, et al. Strain-specific probiotic (microbial cell preparation) and omega-3 fatty acid in modulating quality of life and inflammatory markers in colorectal cancer patients: a randomized controlled trial. Asia Pac J Clin Oncol (2018) 14(3):179–91. doi: 10.1111/ajco.12758
335. Sun L, Yan Y, Chen D, Yang Y. Quxie capsule modulating gut microbiome and its association with T cell regulation in patients with metastatic colorectal cancer: Result from a randomized controlled clinical trial. Integr Cancer Ther (2020) 19:1534735420969820. doi: 10.1177/1534735420969820
336. Gianotti L, Morelli L, Galbiati F, Rocchetti S, Coppola S, Beneduce A, et al. A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World J Gastroenterol (2010) 16(2):167–75. doi: 10.3748/wjg.v16.i2.167
337. Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgard L, et al. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol (2017) 4(1):e000145. doi: 10.1136/bmjgast-2017-000145
338. Appunni S, Rubens M, Ramamoorthy V, Tonse R, Saxena A, McGranaghan P, et al. Emerging evidence on the effects of dietary factors on the gut microbiome in colorectal cancer. Front Nutr (2021) 8:718389. doi: 10.3389/fnut.2021.718389
339. Chan CWH, Law BMH, Waye MMY, Chan JYW, So WKW, Chow KM. Trimethylamine-n-oxide as one hypothetical link for the relationship between intestinal microbiota and cancer - where we are and where shall we go? J Cancer (2019) 10(23):5874–82. doi: 10.7150/jca.31737
340. Xu R, Wang Q, Li L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine n-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genomics (2015) 16 Suppl 7:S4. doi: 10.1186/1471-2164-16-S7-S4
341. Cho CE, Taesuwan S, Malysheva OV, Bender E, Tulchinsky NF, Yan J, et al. Trimethylamine-n-oxide (TMAO) response to animal source foods varies among healthy young men and is influenced by their gut microbiota composition: A randomized controlled trial. Mol Nutr Food Res (2017) 61(1):1600324. doi: 10.1002/mnfr.201770016
342. Kountz DJ, Behrman EJ, Zhang L, Krzycki JA. MtcB, a member of the MttB superfamily from the human gut acetogen eubacterium limosum, is a cobalamin-dependent carnitine demethylase. J Biol Chem (2020) 295(34):11971–81. doi: 10.1074/jbc.RA120.012934
343. Loke YL, Chew MT, Ngeow YF, Lim WWD, Peh SC. Colon carcinogenesis: The interplay between diet and gut microbiota. Front Cell Infect Microbiol (2020) 10:603086. doi: 10.3389/fcimb.2020.603086
344. Gianfredi V, Salvatori T, Villarini M, Moretti M, Nucci D, Realdon S. Is dietary fibre truly protective against colon cancer? a systematic review and meta-analysis. Int J Food Sci Nutr (2018) 69(8):904–15. doi: 10.1080/09637486.2018.1446917
345. Wu X, Wu Y, He L, Wu L, Wang X, Liu Z. Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. J Cancer (2018) 9(14):2510–7. doi: 10.7150/jca.25324
346. Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio (2019) 10(1):e02566-18. doi: 10.1128/mBio.02566-18
347. Chen HM, Yu YN, Wang JL, Lin YW, Kong X, Yang CQ, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am J Clin Nutr (2013) 97(5):1044–52. doi: 10.3945/ajcn.112.046607
348. Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the united states. CA Cancer J Clin (2018) 68(1):31–54. doi: 10.3322/caac.21440
349. Chu J, Xing C, Du Y, Duan T, Liu S, Zhang P, et al. Pharmacological inhibition of fatty acid synthesis blocks SARS-CoV-2 replication. Nat Metab (2021) 3(11):1466–75. doi: 10.1038/s42255-021-00479-4
350. Marzullo P, Bettini S, Menafra D, Aprano S, Muscogiuri G, Barrea L, et al. Spot-light on microbiota in obesity and cancer. Int J Obes (Lond) (2021) 45(11):2291–9. doi: 10.1038/s41366-021-00866-7
351. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature (2009) 457(7228):480–4. doi: 10.1038/nature07540
352. Tremaroli V, Karlsson F, Werling M, Stahlman M, Kovatcheva-Datchary P, Olbers T, et al. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab (2015) 22(2):228–38. doi: 10.1016/j.cmet.2015.07.009
353. Kim SJ, Kim SE, Kim AR, Kang S, Park MY, Sung MK. Dietary fat intake and age modulate the composition of the gut microbiota and colonic inflammation in C57BL/6J mice. BMC Microbiol (2019) 19(1):193. doi: 10.1186/s12866-019-1557-9
354. Kim KA, Gu W, Lee IA, Joh EH, Kim DH. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PloS One (2012) 7(10):e47713. doi: 10.1371/journal.pone.0047713
355. Lecomte V, Kaakoush NO, Maloney CA, Raipuria M, Huinao KD, Mitchell HM, et al. Changes in gut microbiota in rats fed a high fat diet correlate with obesity-associated metabolic parameters. PloS One (2015) 10(5):e0126931. doi: 10.1371/journal.pone.0126931
356. He C, Cheng D, Peng C, Li Y, Zhu Y, Lu N. High-fat diet induces dysbiosis of gastric microbiota prior to gut microbiota in association with metabolic disorders in mice. Front Microbiol (2018) 9:639. doi: 10.3389/fmicb.2018.00639
357. Gomes AC, de Sousa RG, Botelho PB, Gomes TL, Prada PO, Mota JF. The additional effects of a probiotic mix on abdominal adiposity and antioxidant status: A double-blind, randomized trial. Obes (Silver Spring) (2017) 25(1):30–8. doi: 10.1002/oby.21671
358. Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology (2012) 143(4):913–6.e7. doi: 10.1053/j.gastro.2012.06.031
359. Bishehsari F, Moossavi S, Engen PA, Liu X, Zhang Y. Abnormal food timing promotes alcohol-associated dysbiosis and colon carcinogenesis pathways. Front Oncol (2020) 10:1029. doi: 10.3389/fonc.2020.01029
360. Rossi M, Jahanzaib Anwar M, Usman A, Keshavarzian A, Bishehsari F. Colorectal cancer and alcohol consumption-populations to molecules. Cancers (Basel) (2018) 10(2):38. doi: 10.3390/cancers10020038
361. Elamin EE, Masclee AA, Dekker J, Jonkers DM. Ethanol metabolism and its effects on the intestinal epithelial barrier. Nutr Rev (2013) 71(7):483–99. doi: 10.1111/nure.12027
362. Tsuruya A, Kuwahara A, Saito Y, Yamaguchi H, Tenma N, Inai M, et al. Major anaerobic bacteria responsible for the production of carcinogenic acetaldehyde from ethanol in the colon and rectum. Alcohol Alcohol (2016) 51(4):395–401. doi: 10.1093/alcalc/agv135
363. Tsuruya A, Kuwahara A, Saito Y, Yamaguchi H, Tsubo T, Suga S, et al. Ecophysiological consequences of alcoholism on human gut microbiota: implications for ethanol-related pathogenesis of colon cancer. Sci Rep (2016) 6:27923. doi: 10.1038/srep27923
364. Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer (2003) 3(10):733–44. doi: 10.1038/nrc1190
365. Bai X, Wei H, Liu W, Coker OO, Gou H, Liu C, et al. Cigarette smoke promotes colorectal cancer through modulation of gut microbiota and related metabolites. Gut (2022). doi: 10.1136/gutjnl-2021-325021
Keywords: colorectal cancer, microbiota, innate immunity, adaptive immunity, colitis, metabolites, immune signaling, immunotherapy
Citation: Xing C, Du Y, Duan T, Nim K, Chu J, Wang HY and Wang R-F (2022) Interaction between microbiota and immunity and its implication in colorectal cancer. Front. Immunol. 13:963819. doi: 10.3389/fimmu.2022.963819
Received: 07 June 2022; Accepted: 08 July 2022;
Published: 29 July 2022.
Edited by:
Markus Germann, University of Basel, SwitzerlandReviewed by:
Moisés Laparra, IMDEA Food Institute, SpainGislaine Curty, National Cancer Institute (INCA), Brazil
Copyright © 2022 Xing, Du, Duan, Nim, Chu, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong-Fu Wang, cm9uZ2Z1d2FAdXNjLmVkdQ==
 Changsheng Xing
Changsheng Xing Yang Du
Yang Du Tianhao Duan
Tianhao Duan Kelly Nim1
Kelly Nim1 Junjun Chu
Junjun Chu Helen Y. Wang
Helen Y. Wang Rong-Fu Wang
Rong-Fu Wang