- 1Institute of Biopharmaceutical and Health Engineering, Tsinghua Shenzhen International Graduate School, Tsinghua University, Shenzhen, China
- 2Precision Medicine and Healthcare Research Centre, Tsinghua-Berkeley Shenzhen Institute, Tsinghua University, Shenzhen, China
- 3Programme in Emerging Infectious Diseases, Duke-National University of Singapore Medical School, Singapore, Singapore
- 4Centre of Excellence for Research in AIDS (CERiA), University of Malaya, Kuala Lumpur, Malaysia
- 5Department of Medicine, University of Malaya, Kuala Lumpur, Malaysia
- 6Center for Infectious Medicine, Department of Medicine, Karolinska Institutet, Stockholm, Sweden
- 7Peter Doherty Institute for Infection and Immunity, Melbourne University, Victoria, Australia
MAIT cells are persistently depleted and functionally exhausted in HIV-1-infected patients despite long-term combination antiretroviral therapy (cART). IL-7 treatment supports MAIT cell reconstitution in vivo HIV-1-infected individuals and rescues their functionality in vitro. Single-nucleotide polymorphisms (SNPs) of the IL-7RA gene modulate the levels of soluble(s)IL-7Rα (sCD127) levels and influence bioavailability of circulating IL-7. Here we evaluate the potential influence of IL-7RA polymorphisms on MAIT cell numbers and function in healthy control (HC) subjects and HIV-1-infected individuals on long-term cART. Our findings indicate that IL-7RA haplotype 2 (H2*T), defined as T-allele carriers at the tagging SNP rs6897932, affects the size of the peripheral blood MAIT cell pool, as well as their production of cytokines and cytolytic effector proteins in response to bacterial stimulation. H2*T carriers had lower sIL-7Rα levels and higher MAIT cell frequency with enhanced functionality linked to higher expression of MAIT cell-associated transcription factors. Despite an average of 7 years on suppressive cART, MAIT cell levels and function in HIV-1-infected individuals were still significantly lower than those of HC. Notably, we observed a significant correlation between MAIT cell levels and cART duration only in HIV-1-infected individuals carrying IL-7RA haplotype 2. Interestingly, treatment with sIL-7Rα in vitro suppressed IL-7-dependent MAIT cell proliferation and function following cognate stimulations. These observations suggest that sIL-7Rα levels may influence MAIT cell numbers and function in vivo by limiting IL-7 bioavailability to MAIT cells. Collectively, these observations suggest that IL-7RA polymorphisms may play a significant role in MAIT cell biology and influence MAIT cells recovery in HIV-1 infection. The potential links between IL7RA polymorphisms, MAIT cell immunobiology, and HIV-1 infection warrant further studies going forward.
Introduction
Mucosa-associated invariant T (MAIT) cells are innate-like T cells and abundant in blood, the liver, and mucosal tissues (1). Classical MAIT cells in humans express an invariant T cell receptor (TCR) Vα7.2 coupled with restricted Jα 12/20/33 segments (2–4). In contrast to conventional T cells which recognize peptide fragments presented by the classical major histocompatibility complex (MHC) molecules, MAIT cells are activated by pyrimidine ligands bound to the non-polymorphic MHC-Ib related protein (MR1) (4–6). The TCR Vα7.2, together with high expression of CD161, IL-18 receptor α subunit (IL-18Rα), or recently used MR1 tetramers identify the MAIT cell population in humans (6–8). Although initially identified within the double negative (DN) T cell pool, peripheral blood MAIT cells predominantly express CD8 (9, 10). MAIT cells detect bacterial infection through metabolites of riboflavin biosynthetic pathway, such as 5-OP-RU, presented on MR1 (5). Additionally, MAIT cells can be activated in a TCR-independent manner by cytokines including Interleukin-1β (IL-1β), IL-12, IL-18, and Interferon-α (IFNα) (11, 12). In humans, MAIT cells display antimicrobial responsiveness in the intestinal mucosa pre-natally, prior to exposure to established commensal microflora (13).
Viruses do not directly give rise to MR1-presented riboflavin metabolite antigens. However, due to their high sensitivity to TCR-independent signals, mainly to IL-12 and IL-18, MAIT cells can produce IFNγ in response to different viruses, including influenza A, SARS-CoV-2, HBV/HDV, and HIV-1 (14–21). We and others have reported the loss of MAIT cells in circulation during chronic HIV-1 infection (19, 22–24). Residual MAIT cells from chronically HIV-1-infected patients are highly activated and show functional impairment in response to Escherichia coli stimulation (19, 23, 25). However, during the first weeks of HIV-1 infection, absolute counts of intestinal and circulating MAIT cells are elevated, indicating the proliferation of MAIT cells at the early phase of acute HIV-1 infection (24, 26). Various mechanisms of MAIT cell loss during chronic HIV-1 infection were recently reviewed by Han et al. (25), including cytokine dysregulation (27), persistent activation by microbial metabolites from microbial translocation (28, 29), MAIT cell death due to apoptosis and pyroptosis (30), and homing to inflamed tissues (23, 25). Unfortunately, combination antiretroviral therapy (cART) is ineffective in restoring the circulating MAIT cell pool (19, 24).
MAIT cells express high levels of IL-7Rα (CD127) (31, 32), and IL-7 promotes the activation of circulating MAIT cells, arms cytotoxic capacity, and enhances cytokine production (32). Furthermore, IL-7 amplifies the activation of MAIT cells via TCR or non-TCR signals (31, 32). In addition, IL-7 induces high levels of IL-18Rα on MAIT cells and increases their expression of the transcription factors promyelocytic leukemia zinc finger (PLZF), retinoic acid receptor-related orphan receptor (ROR)γt, T-box transcription factor 21 (TBX21, also known as T-bet), and eomesodermin (Eomes) (32–34). In disease settings, plasma IL-7 levels positively correlate with MAIT cell frequencies in HIV-1-infected patients (32, 35). IL-7 treatment expands the MAIT cell compartment in HIV-1-infected patients in vivo and rescues the antimicrobial effector function of MAIT cells of HIV-1-infected patients in response to E. coli stimulation in vitro (32, 36). In rhesus macaques with chronic SHIV infection, IL-7 alone enhances the proliferation but not degranulation of MAIT cells (33). In mice, pretreatment with IL-7 improves survival in Streptococcus pneumoniae infection (34).
IL-7 is a homeostatic cytokine required for both the production and survival of naïve and memory T cells and is critical for immune recovery in lymphopenic states (37, 38). IL-7 exerts its biological effect by binding to its membrane-bound receptor, mIL-7Rα on T-cells (39). There is also a soluble form of this receptor (sIL-7Rα), which is thought to modulate IL-7 availability (40). In vitro studies found that levels of sIL-7Rα in culture affected IL-7-mediated functions including T cell survival and proliferation (41). This suggests that sIL-7Rα may influence IL-7 mediated function including its capacity to drive immune recovery in HIV-1-infected individuals.
Four common haplotypes of the IL7RA gene (haplotypes 1, 2, 3 and 4) have been previously defined and shown to be associated with sIL-7Rα levels (42, 43). Haplotype 2 (tagged by carriage of T allele in rs6897932) is associated with reduced sIL7Rα levels in plasma. This single nucleotide polymorphism (SNP) influences the amount of soluble and membrane bound IL-7Rα isoforms produced by disrupting an exon splicing silencer (44). Studies assessing the association between rs6897932 and immune reconstitution have reported both positive and negative associations. In a predominantly Caucasian Australian cohort of HIV-1-infected people, there was a significant association between the IL7RA haplotype 2 (H2*T) and faster CD4+ T cell recovery in those receiving suppressive ART (45). In a replicate study in Uganda, the IL7RA H2*T had an inverse association with CD4+ T cell recovery compared to that found in Caucasians, and was associated with poorer CD4+ T cell recovery (46). Subsequently, the differential influences of IL7RA haplotypes on HIV-1 disease progression in Caucasians and Africans have been confirmed (47, 48). The reason for the different associations seen in the various ethnic populations despite a consistent influence of IL7RA haplotype 2 on low sIL-7Rα levels is currently unclear. T-allele carriers of the rs6897932 SNP have also been associated with reduced susceptibility to multiple autoimmune diseases including multiple sclerosis, systemic lupus erythematosus (49), chronic inflammatory arthropathies (50), sarcoidosis (51) and type 1 diabetes (52) in HIV-1-uninfected individuals. Despite the wide ranging effects of this SNP in multiple immune-mediated diseases, there is a notable lack of studies systematically assessing its immunologic effects.
Although the effects of IL-7 on MAIT cells have been clear for some time, it is unknown whether IL7RA polymorphisms affect MAIT cell phenotype and function in health and during HIV-1 infection. In this study, we investigate the association of IL7RA haplotypes with MAIT cell levels and effector function in health and in chronic HIV-1 infection with long-term cART-mediated viral suppression. Our findings indicate that IL7RA H2*T affects the size of the peripheral MAIT cell pool, as well as their production of cytokines and cytolytic effector proteins in response to bacterial stimulation. Furthermore, presence of IL7RA H2*T positively associates with circulating MAIT cell levels following the long-term suppressive ART in chronically HIV-1-infected individuals.
Results
IL7RA haplotype 2 is associated with higher MAIT cell levels in healthy individuals
IL-7 treatment supports increased MAIT cell numbers in vivo in humans and IL7RA SNPs influence IL-7 uptake and downstream biological activity (36, 53). Thus, the relationship between IL7RA SNPs (Supplementary Table 1) and MAIT cell numbers in circulation of healthy individuals (Supplementary Table 2) was initially determined. Peripheral blood (PB) MAIT cells were identified by the combined staining of TCR Vα7.2 with high expression of CD161 (Figure 1A). The vast majority of Vα7.2+ CD161hi MAIT cells also stained positive with MR1-5-OP-RU tetramer and expressed the CD8α co-receptor (Figure 1A), consistent with previously published studies (10, 54). Healthy individuals carrying IL7RA H2*T, defined as T-allele carriers (heterozygous or homozygous) at the tagging SNP rs6897932 (Supplementary Table 1), had higher levels of MAIT cells in circulation than those with non-haplotype 2 (non-H2*T) polymorphisms (Figure 1B). There was no significant age differences between the haplotype groups when stratified by sex (Supplementary Figure 1A). Furthermore, MAIT cell numbers were comparable between females and males when stratified by haplotypes (Supplementary Figure 1B). Correlative analyses also showed that there were no associations between age and MAIT cell numbers as a whole or stratified either by sex or haplotypes (Supplementary Table 3). Therefore, the difference in MAIT cell frequency between the IL7RA haplotypes was probably not influenced by differences in age or sex between the groups. There was no difference in the representation of CD8+ and DN MAIT cells between the IL7RA haplotypes as a whole or when stratified by sex (Figures 1C, D). Similarly, no differences were observed in IL-7Rα surface expression (mIL-7Rα/CD127) by MAIT cells (Figure 1E) and the plasma levels of IL-7 from individuals carrying different IL7RA haplotypes (Figure 1F), but those with H2*T haplotypes had significantly lower sIL-7Rα in their plasma (Figure 1G), consistent with previous studies (42, 44). Notably, there was a significant inverse correlation between MAIT cell frequency and plasma levels of sIL-7Rα in this cohort of healthy individuals (Figure 1H). However, there was no correlation between age and plasma levels of sIL-7Rα as a whole or stratified by haplotype or sex (Supplementary Table 3). Altogether, these results suggest that IL7RA polymorphisms associates with MAIT cell levels in healthy adults.
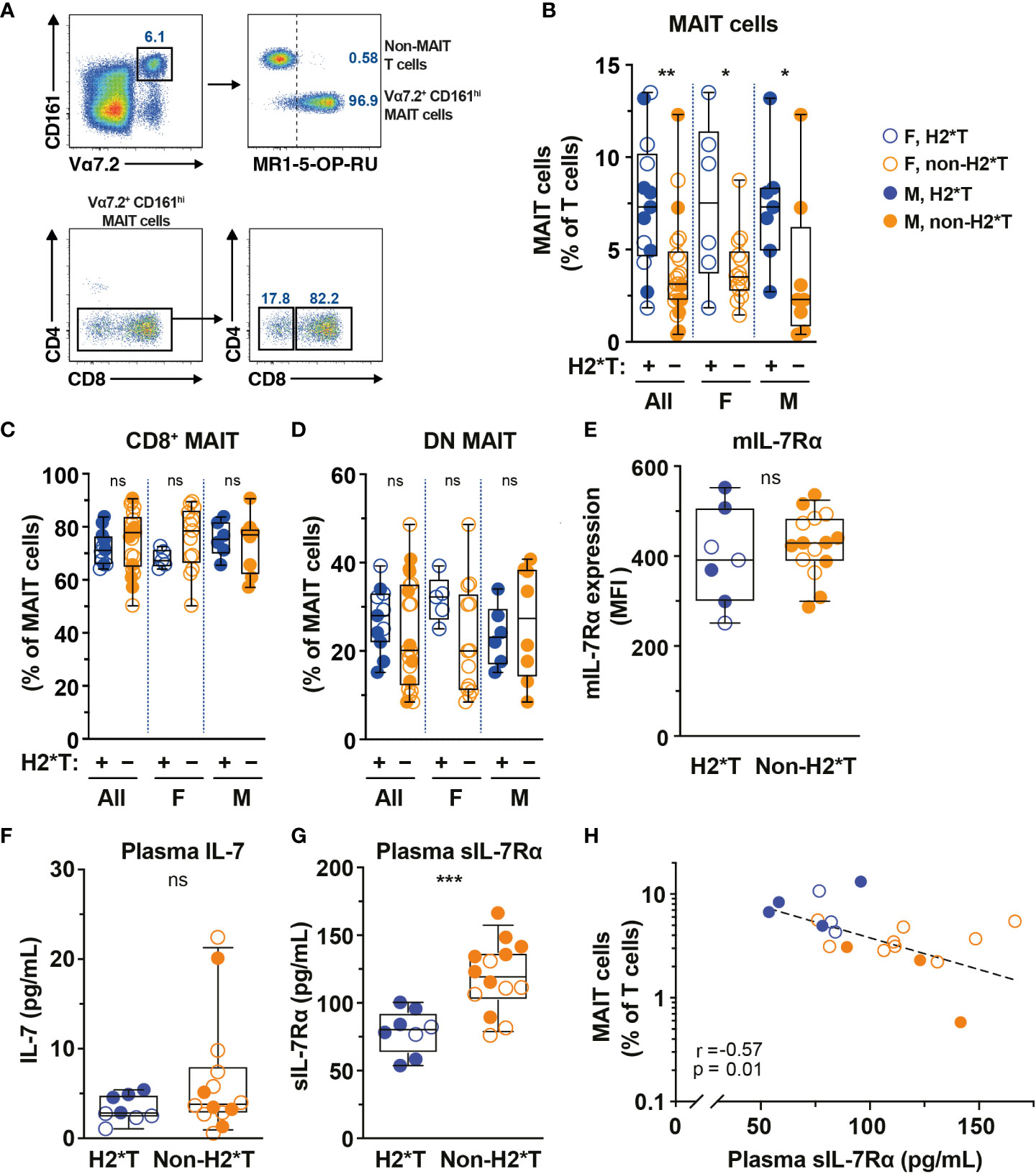
Figure 1 MAIT cell levels are higher in HIV-1-uninfected individuals carrying the IL7RA haplotype 2. (A) Representative flow cytometry gating strategy to identify MAIT cells by TCR Vα7.2 and CD161 co-expression on T cells (Vα7.2+ CD161hi MAIT cells), as well as the CD4 and CD8 co-receptor expression. MR1-5-OP-RU tetramer staining was used to confirm the identity and MR1-restriction of Vα7.2+ CD161hi MAIT cells. (B–E) MAIT cell levels (B), the distribution of CD8+ (C) and CD4- CD8- (DN; D) MAIT cell subsets, and membrane-bound (m)IL-7Rα I levels by MAIT cells in HIV-1-uninfected individuals bearing the different IL-7Rα haplotypes. (F, G) Plasma levels of IL-7 (F) and soluble (s)IL-7Rα (G) in HIV-1-uninfected individuals bearing the different IL-7Rα haplotypes. (H) Spearman’s correlation between circulating MAIT cell levels and plasma sIL-7Rα. Box and whisker plots show all data points (N=7-13 for haplotype 2 individuals and N=14-22 for non-haplotype 2), median, and the interquartile range. Statistical significance was determined using Mann-Whitney’s test (B–F) or unpaired t-test (G). Correlations were assessed using the Spearman rank correlation (H). *** p<0.001, ** p<0.01,* p<0.05. ns, not significant. F, female; M, male; H2*T, IL7RA haplotype 2.
IL7RA SNPs are associated with MAIT cell expression of transcription factors and function in healthy individuals
MAIT cell treatment with IL-7 in vitro leads to enhanced expression of transcription factors (TFs) and augmented MAIT cell antimicrobial function, such as increased production of IFNγ and TNF (31, 32). Therefore, whether IL7RA polymorphisms affect expression of critical TFs and MAIT cell function was next evaluated. In line with higher MAIT cell frequency in individuals carrying H2*T, MAIT cells in these individuals expressed higher levels of PLZF (Figure 2A). RORγt expression also appeared to be higher in MAIT cells from H2*T individuals (Figure 2B). IL-7 downstream biological activity is associated with increased cellular survival partly via the anti-apoptotic protein Bcl-2 (55). However, Bcl-2 expression levels in MAIT cells from individuals carrying the H2*T and non-H2*T haplotypes were comparable (Figure 2C). We were unable to stratify these differences based on sex due to limited sample size. To assess for antimicrobial effector function, MAIT cells were activated using an assay that was previously developed by our group (56, 57). Following stimulation with formaldehyde-fixed E. coli for 24 h, MAIT cells from healthy adults with the H2*T haplotype had increased levels of IFNγ and TNF, and appeared to also be more bifunctional (Figures 2D–G). Such higher expression levels of cytokines were still evident in females after stratification by sex, whereas there was a trend of increasing cytokine production by MAIT cells from male H2*T IL7RA carriers. There was no correlation between age and cytokine expression by MAIT cells stratified by either sex or haplotype (Supplementary Table 3). These findings suggest that IL7RA polymorphism may affect MAIT cell functional activity in vivo through modulation of TF levels.
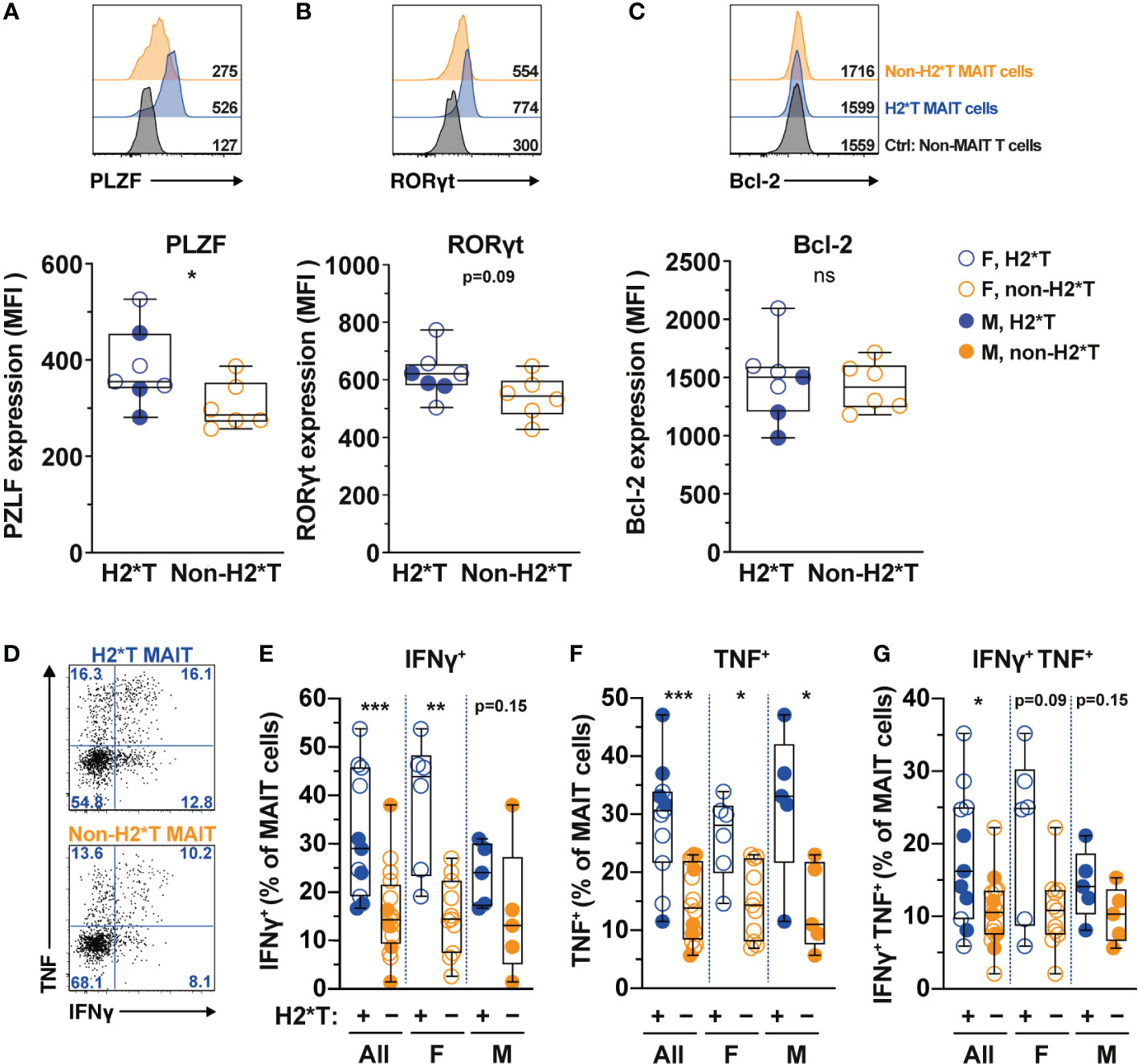
Figure 2 MAIT cells from HIV-1-uninfected individuals carrying IL7RA haplotype 2 express higher levels of MAIT cells-associated transcription factors and effector function. (A–C) Representative histograms for PLZF (A), RORγt (B), and Bcl-2 (C) levels in Vα7.2+ CD161hi MAIT cells in HIV-1-uninfected individuals bearing the different IL-7Rα haplotypes. Non-MAIT T cells represent all other non-MAIT, CD3+ T cells (D–G) Representative flow cytometry gating strategy (D) to enumerate IFNγ (E), TNF (F), and dual IFNγ-TNF (G) production by MAIT cells following 24 h stimulation with formaldehyde-fixed E coli (bacterial dose 1 CFU/cell). Box and whisker plots show all data points (N=7-11 for haplotype 2 individuals and N=6-16 for non-haplotype 2), median, and the interquartile range. Statistical significance was determined using Mann-Whitney’s test. *** p<0.001, ** p<0.01,* p<0.05. ns, not significant. F, female; M, male; H2*T, IL7RA haplotype 2.
MAIT cell levels and function are significantly lower in HIV-1-infected individuals on long-term suppressive ART
The frequency and function of MAIT cells are impaired in chronic untreated HIV-1-infected patients and do not fully recover following long-term cART (19, 22, 25). In agreement with previous studies (19, 22, 58), circulating MAIT cell levels from HIV-1-infected individuals with long-term cART (Supplementary Table 4) were still significantly lower than those from uninfected adult males from the same HC cohort (Figure 3A). The persistent depletion of MAIT cells from the circulation of long-term treated HIV-1-infected individuals was still observed in IL-7RA haplotype 2 carriers but not in non-haplotype 2 carriers (Figure 3A). Since there was no association between MAIT cell levels and age stratified by haplotype in uninfected and HIV-1-infected cohorts in the current study (Supplementary Tables 3, 5, respectively), the differences seen in MAIT cell levels may be less likely due to age differences between the HC and HIV-1-infected cohorts. Similarly, no differences in the proportion of CD8+ and DN MAIT cell subpopulations, as well as in the expression of IL-7Rα by MAIT cells, were noted in healthy controls as compared with those of HIV-1-infected patients (Figures 3B-D). There was no difference in plasma levels of IL-7 and sIL-7Rα between healthy controls and HIV-1-infected patients bearing H2*T and non-H2*T haplotypes (Figures 3E, F). In addition, HIV-1 infection was associated with poor IFNγ and TNF production by MAIT cells following E. coli stimulation despite long-term cART (Figures 3G–J) in line with previous observations (19). This impairment was more obvious in HIV-1-infected individuals carrying the non-H2*T haplotypes (Figures 3G–J). The low sample size for HIV-1-infected individuals carrying the non-H2*T haplotype may explain why there was no significant difference in IFNγ expression found when compared to healthy controls carrying the same IL7RA haplotype. Taken together, these results indicate that MAIT cell dysfunction in chronic HIV-1 infection persists despite long-term viral suppression when compared to healthy controls. This dysfunction may occur regardless of the IL7RA haplotypes.
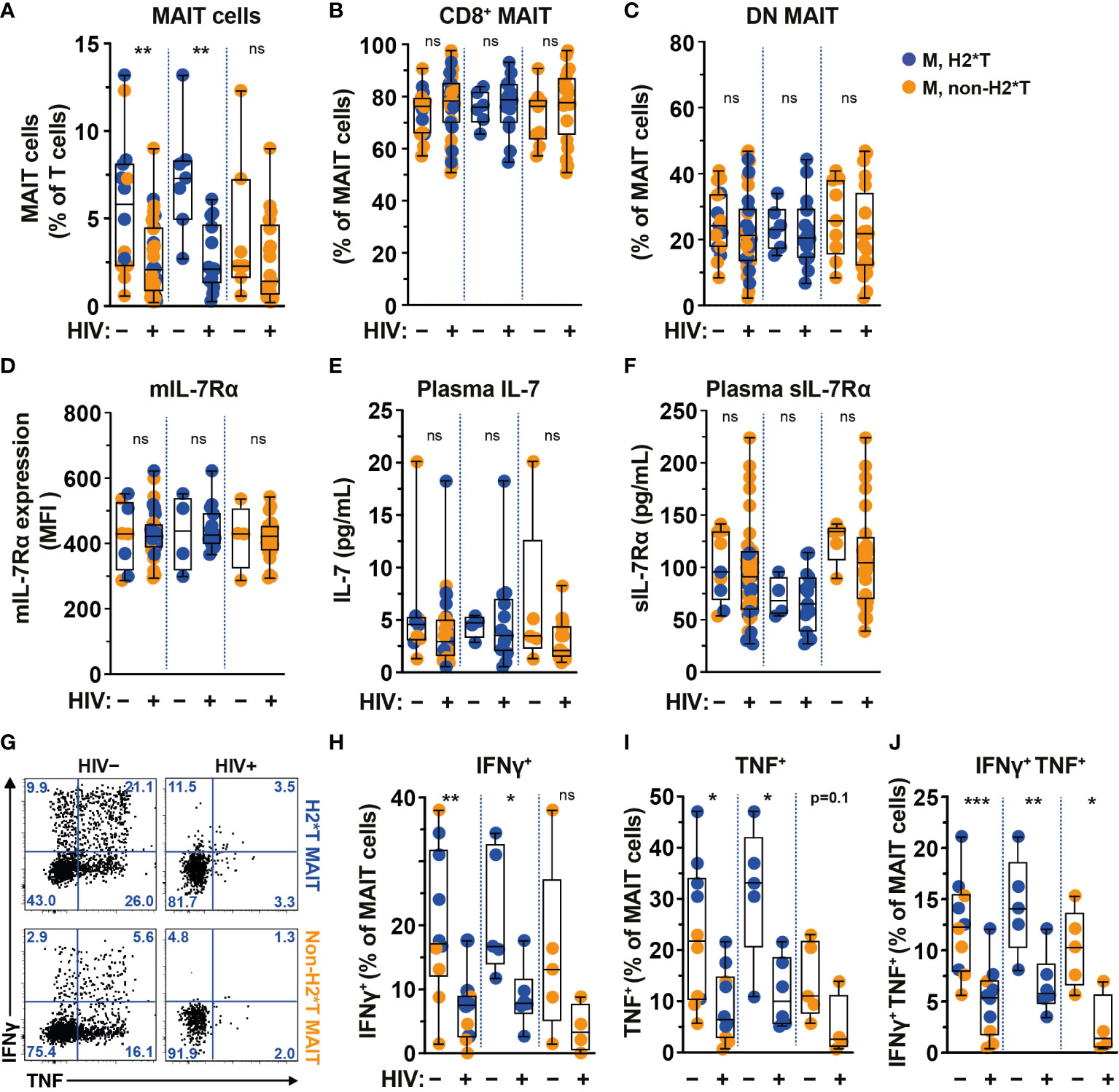
Figure 3 Comparison of MAIT cells in HIV-1-uninfected and -infected individuals with long-term viral suppression and their relationship with the IL7RA SNP. (A–E) Vα7.2+ CD161hi MAIT cell levels (A), the distribution of CD8+ (B) and CD4- CD8- (DN; C) MAIT cell subsets, and membrane-bound (m)IL-7Rα (D) levels by MAIT cells in HIV-1-uninfected and HIV-1-infected individuals carrying the different IL7RA haplotypes. (E, F) Plasma levels of IL-7 (E) and soluble (s)IL-7Rα (F) in HIV-1-uninfected and HIV-1-infected individuals individuals bearing the different IL7RA haplotypes. (G–J) Representative flow cytometry gating strategy (G) to enumerate IFNγ (H), TNF (I), and dual IFNγ-TNF (J) production by MAIT cells following 24 h stimulation with formaldehyde-fixed E coli (bacterial dose 1 CFU/cell). Box and whisker plots show all data points (A–F: HIV-1-uninfected N=8-14, N=4-7 for both haplotype 2 and non-haplotype 2; HIV-1-infected N=34-39, N=14-15 for haplotype 2 and N=20-24 for non-haplotype 2; H–J: HIV-1-uninfected N=10, N=5 for both haplotype 2 and non-haplotype 2; HIV-1-infected N=10, N=6 for haplotype 2 and N=4 for non-haplotype 2), median, and the interquartile range. Statistical significance was determined using Mann-Whitney’s test. *** p<0.001,** p<0.01,* p<0.05. ns, not significant. M, male; H2*T, IL7RA haplotype 2. Male HC dataset were retrieved from HC cohort depicted in Figure 1.
IL7RA haplotypes associate with MAIT cell function and recovery in HIV-1-infected patients on ART
Based on the findings that MAIT cell dysfunction persists in HIV-1-infected patients receiving long-term cART, we next revisited the dataset from the same HIV-1-infected cohort and evaluated the degree of impairment between patients carrying the H2*T and non-H2*T haplotypes. No significant differences in clinical parameters were noted between the HIV-1-infected patient groups carrying IL7RA haplotypes H2*T and non-H2*T, including age, pre-treatment CD4 counts and viral loads, duration of ART (Supplementary Table 4). Interestingly, there was comparable MAIT cell frequency in virally-suppressed chronic HIV-1-infected patients with H2*T and non-H2*T haplotypes (Figure 4A). Additionally, haplotypes H2*T and non-H2*T did not appear to influence MAIT cell major subset distribution within the HIV-1-infected patient group (Figures 4B, C). Although comparable levels mIL-7Rα and plasma IL-7 were observed between the two groups (Figures 4D, E), individuals carrying non-H2*T haplotype had a higher level of plasma sIL-7Rα (Figure 4F), consistent with the observation in healthy individuals (Figure 1G). Moreover, there was no association between age and plasma sIL-7Rα levels as a whole or stratified by haplotype in this HIV-1-infected cohort (Supplementary Table 5). In addition, HIV-1-infected individuals with H2*T haplotype had better MAIT cell functionality following bacterial stimulation when compared to those carrying other haplotypes (Figures 4G–I). There was also no correlation between age and cytokine expression by MAIT cells following E. coli stimulation as a whole or stratified by haplotype in the current HIV-1-infected cohort, although the sample size was small (Supplementary Table 5). Interestingly, despite that there was no difference in MAIT cell levels between group with the different IL7RA haplotypes, duration of ART positively correlated with MAIT cell levels in individuals bearing the H2*T haplotype (Figure 4J).
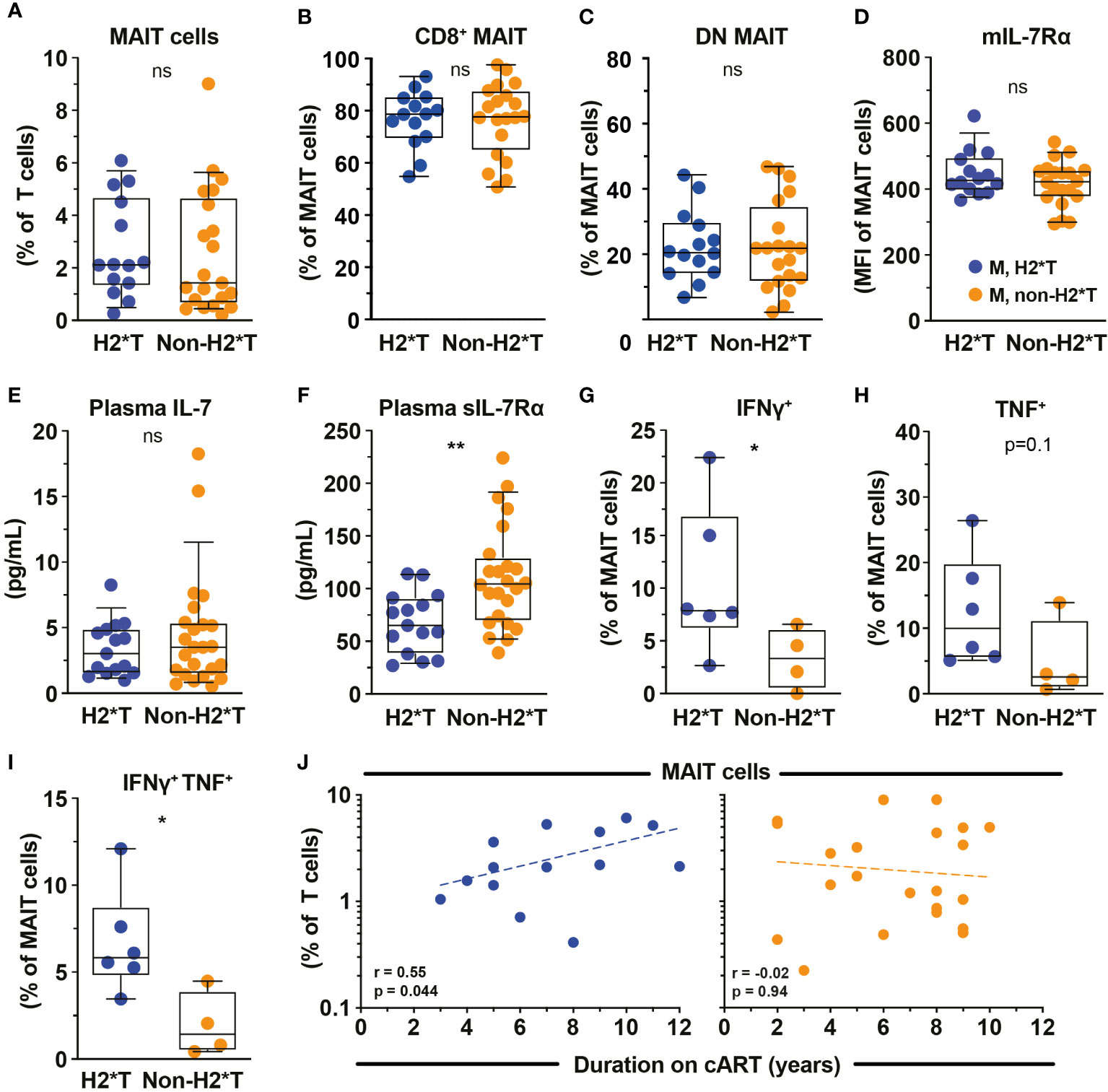
Figure 4 MAIT cell levels and functions in HIV-1-infected individuals with long-term viral suppression and their relationship with the IL-7Rα SNP. Vα7.2+ CD161hi MAIT cell levels (A), the distribution of CD8+ (B) and CD4- CD8- (DN; C) MAIT cell subsets, and membrane-bound (m)IL-7Rα (D) by MAIT cells in HIV-1-infected individuals bearing the different IL7RA haplotypes. (E, F) Plasma levels of IL-7 (E) and soluble (s)IL-7Rα (F) in HIV-1-infected individuals bearing the different IL-7RA haplotypes. (G–I) The IFNγ (G), TNF (H), and dual IFNγ-TNF (I) production by MAIT cells following 24 h stimulation with formaldehyde-fixed E coli (bacterial dose 1 CFU/cell). (J) Spearman’s correlation between circulating MAIT cell levels and duration of cART in individuals bearing haplotype 2 (N=14) and non-haplotype 2 (N=22). Box and whisker plots show all data points (A–E: N=14-15 for haplotype 2 and N=20-24 for non-haplotype 2; G–I: N=6 for haplotype 2 and N=4 for non-haplotype 2), median, and the interquartile range. Statistical significance was determined using Mann-Whitney’s test. ** p<0.01,* p<0.05. ns, not significant. M, male; H2*T, IL7RA haplotype 2. HIV-1-infected dataset were retrieved from HIV-1-infected cohort depicted in Figure 3.
Soluble IL-7Rα inhibits IL-7-mediated MAIT cell proliferation and effector function in vitro
Levels of sIL-7Rα affects IL-7-mediated functions including T cell survival and proliferation (41). Given plasma sIL-7Rα levels negatively correlate with MAIT cell frequency (Figure 1H), we next explored the impact of sIL-7Rα treatment in vitro on IL-7-mediated MAIT cell proliferation and function. Downregulation of surface expression of mIL-7Rα is a surrogate marker of MAIT cell activation following IL-7 treatment in vitro (32). By using this approach, initial experiments showed that sIL-7Rα mitigated the downregulation of mIL-7Rα on MAIT cells following IL-7 treatment in vitro (Figure 5A), suggesting that sIL-7Rα reduces IL-7 binding to mIL-7Rα on MAIT cells. Indeed, sIL-7Rα treatment in vitro reduced MAIT cell proliferation following stimulation with low dose of the prototypical MR1 ligand 5-OP-RU supplemented with low dose IL-7 (Figs. 5B-5G). This was shown by lower levels of Ki67 expression, lower MAIT cell numbers, as well as reduced proliferation and expansion indices calculated from CellTrace Violet (CTV) dilution assay (Figures 5B–G), performed as previously described (56, 59). Such inhibition appeared to be dose-dependent and remained apparent at sIL-7Rα:IL-7 molar ratio > 100 (Figure 5H). The inhibitory effect of sIL-7Rα on MAIT cell proliferation did not occur when IL-2 substituted IL-7 (Supplementary Figures 2A–G), indicating that sIL-7Rα-mediated impairment is specific to IL-7-dependent activity on MAIT cells. Interestingly, sIL-7Rα also inhibited IL-7-mediated enhancement on resting MAIT cell IFNγ and TNF production and Ki-67 expression following E. coli stimulation (Figures 5I–K). This inhibition was also dose-dependent as MAIT cell cytokine production and Ki-67 expression remained reduced at sIL-7Rα:IL-7 molar ratio > 100 (Figure 5L).
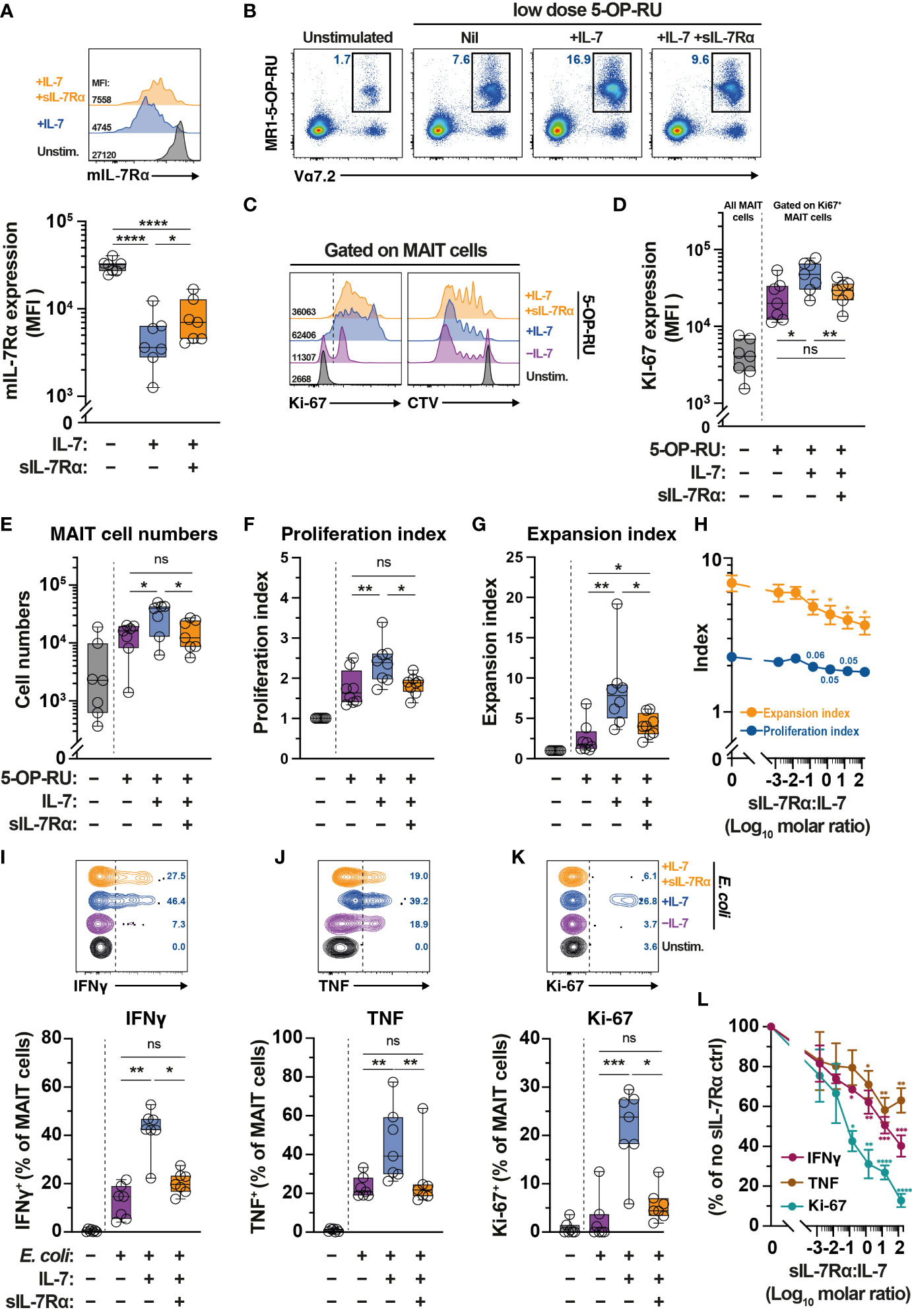
Figure 5 Soluble IL-7Rα inhibits IL-7-mediated MAIT cell proliferation and effector function. (A) Expression levels of mIL-7Rα on MR1-5-OP-RU tetramer+ Vα7.2+ MAIT cells following 24 h treatment of PBMC with IL-7 alone or together with sIL-7Rα (molar ratio 1:1) (N=7). (B–G) MR1-5-OP-RU tetramer+ Vα7.2+ MAIT cell (B) proliferation as determined by Ki-67 expression levels on Ki-67+ MAIT cells (C; left panel, D), absolute MAIT cell numbers (E), and proliferation (F) and expansion (G) indices calculated using CTV-dilution (C; right panel) following 5-days treatment of PBMC with 5-OP-RU, 5-OP-RU+IL-7, or 5-OP-RU+IL-7+sIL-7Rα (sIL-7Rα/IL-7molar ratio 1:1) (N=7-8). (H) Dose-dependent effect of sIL-7Rα treatment on MAIT cells proliferation and expansion indices following 5-days treatment with IL-7- and 5-OP-RU (N=6-7). (I-K) MAIT cell expression of IFNγ (I), TNF (J), and Ki-67 (K) after 24 h treatment of PBMC with IL-7 alone or together with sIL-7Rα (molar ratio 1:1) followed by a further 24 h stimulation with formaldehyde-fixed E coli (bacterial dose 0.1 CFU/cell) (N=7). (L) Dose-dependent effect of sIL-7Rα treatment on MAIT cells cytokine and Ki-67 expression following treatment with E coli with or without IL-7 priming as in (I–K) (N=6-7). Expression levels for (L) were normalised to nil sIL-7Rα controls to minimise inherent donor-to-donor variability, with statistical tests were performed on raw data. Box and whisker plots show all data points, median, and the interquartile range, whereas data presented as line graphs with error bars represent the mean and standard error. Statistical significance was determined using the Friedman test followed by Dunn’s post-hoc test (E, I, J, K), or repeated measure one-way ANOVA with Tukey’s post-hoc test (A, D, F,G), or mixed-effect analysis with Dunnett’s post-hoc test (H, L). **** p<0.0001, *** p<0.001, ** p<0.01, * p<0.05, ns, not significant.
Discussion
In the present study, we report an association between IL7RA polymorphisms and MAIT cell levels and functional activity in healthy individuals and HIV-1-infected individuals with long-term viral suppression. Specifically, we show that IL7RA haplotype 2 positively impacts the frequency and functional activity of MAIT cells in healthy individuals. This IL7RA haplotype is correlated with decreased plasma sIL-7Rα concentration both in health and chronic HIV-1 infection, which is linked with higher MAIT cell levels in healthy controls. Importantly, MAIT cell levels correlate with duration on cART in HIV-1-infected individuals carrying IL7RA haplotype 2, suggesting favorable MAIT cell reconstitution in these individuals. Such MAIT cells also appear to display better functionality upon bacterial stimulation. This observation suggests that IL7RA haplotype 2 may be beneficial for MAIT cell reconstitution in vivo. Finally, in vitro experiments support the notion that the positive effects of IL7RA haplotype 2 on MAIT cell number and function is associated with lower sIL-7Rα concentration, as sIL-7Rα treatment in vitro effectively inhibits IL-7-dependent MAIT cell proliferation and effector function. Altogether, these observations suggest favorable effects of the IL7RA haplotype 2 on MAIT cell reconstitution in virally suppressed chronic HIV-1 infection.
How IL7RA polymorphisms may affect MAIT cell biology in health and during HIV-1-infection is unclear. It is possible that decreased production of sIL-7Rα isoform in IL7RA haplotype 2-carrying healthy and HIV-1-infected individuals may diminish the interference with IL-7 binding to its membrane-bound receptors on T cells (42, 53). However, the effects of sIL-7Rα in shaping IL-7 bioavailability are complex. Several reports demonstrated that sIL-7Rα serves as a reservoir of IL-7 and hence increases its half-life due to relatively low affinity (53, 60). Lundström et al. provided a model for sIL-7Rα-mediated IL-7 regulation, proposing that sIL-7Rα/rhIL-7 with a very high molar ratio (>5000) may increase the bioactivity of IL-7 by stabilizing its reservoir (53). Furthermore, in vivo experiments showed that IL7−/− mice had transiently increased IL-7 levels following administration of rhIL-7 with sIL-7Rα versus rhIL-7 alone (53). In the current study, we saw the plasma levels of sIL-7Rα were significantly lower in haplotype 2 individuals in both healthy and HIV-1-infected individuals, consistent with previous studies (42, 44). Moreover, we found that MAIT cell levels in IL7RA haplotype 2 carriers with chronic HIV-1 infection correlated with duration on cART, whereas non-haplotype 2 carriers did not show this correlation. This observation is in line with better recovery from lymphopenia for haplotype 2 carriers on cART (42). HIV-1-infected IL7RA haplotype 2 carriers also had partial recovery of the cytokine production following E. coli stimulation. Given the stimulatory effects of IL-7 on MAIT cells (31, 32, 61), all these together suggest that IL7RA haplotype 2 may influence MAIT cell homeostasis via enhanced bioactivity of IL-7 due to low plasma sIL-7Rα.
Indeed, our data indicate dose-dependent inhibition of IL-7-mediated MAIT cell proliferation and effector function by sIL-7Rα in vitro. Firstly, sIL-7Rα mitigated the downregulation of mIL-7Rα following low dose IL-7, indicating reduced binding of IL-7 to mIL-7Rα on MAIT cells. Furthermore, sIL-7Rα-mediated inhibition was seen when MAIT cells were stimulated with low dose of the prototypical antigen 5-OP-RU in the presence of low dose of IL-7, but not when in the presence of low dose of a similar γc cytokine IL-2. This indicates the specificity of sIL-7Rα-mediated inhibition on IL-7. Finally, sIL-7Rα negates IL-7-dependent enhancement of MAIT cell effector function following E. coli stimulation. Particularly, IFNγ and TNF levels by IL-7-primed MAIT cells were comparable to those of unprimed MAIT cells following E. coli stimulation when co-administered with sIL-7Rα. Collectively, our in vitro data support the notion that favorable effects of IL7RA haplotype 2 on MAIT cell numbers and function in both HC and HIV-1-infected patients is at least partially a consequence of enhanced bioavailability of IL-7 to MAIT cells due to lower sIL-7Rα levels in vivo. Our in vitro study used much lower sIL-7Rα/rhIL-7 molar ratios (0.015 to 150) and showed inhibitory effect of sIL-7Rα on IL-7 bioactivity, whereas a previous study that used a very high sIL-7Rα/rhIL-7 molar ratio of 5000 showed an enhancement of IL-7 bioactivity (53). Therefore, because of the complexity of IL-7/IL-7Rα biology, it will be important to further examine the influence of these receptor polymorphisms on IL-7 utilization and MAIT cell biology and the mechanisms behind these effects in both HC and HIV-1-infected individuals going forward.
The positive effects of haplotype 2 on MAIT cells may additionally occur through the modulation of critical TFs for MAIT cell functions. Our data indicate that the IL7RA haplotype 2 appears to support higher expression of PLZF and RORγt by peripheral MAIT cells in healthy individuals. These TFs are essential for both TCR-dependent and -independent MAIT cell development and cytokine production (3). Therefore, upregulation of PLZF and RORγt may potentially lead to higher frequency and function of MAIT cells in haplotype 2 individuals. Tang et al. observed upregulation of anti-apoptotic molecule Bcl-2 in intrasinusoidal MAIT cells upon IL-7 stimulation (31). Of note, sIL-7Rα inhibits IL-7–induced proliferation and Bcl-2 expression in peripheral CD8+ T cells (41, 53). Thus, lower sIL-7Rα and increased IL-7 utilisation by MAIT cells in haplotype 2 individuals may potentially lead to enhanced survival of MAIT cells via increased Bcl-2 activity. However, in the current study we did not observe any changes in Bcl-2 expression in the setting of lower plasma sIL-7Rα levels in haplotype 2 individuals.
Our study has several important limitations. Firstly, our sample size was relatively small and there was some differences with regard to age and sex distribution between the healthy and HIV-1-infected cohorts. Nevertheless, our correlative analyses suggest that these differences do not seem to significantly associate with the MAIT cell levels in either HC or HIV-1-infected individuals. Indeed, the differences in MAIT cell numbers and function in individuals carrying different IL7RA haplotypes hold when stratified by sex in HC cohort. Unfortunately, the low number of male donors from either haplotypes in HC cohort does not allow us to make firm conclusion on the influence of IL7RA haplotypes on MAIT cell biology in adult males. We also used only the dataset from male HC for the comparison with the all-male HIV-1-infected cohort, thereby removing biological sex as a confounding factor in these analyses. Secondly, the potential influence of other confounders cannot be ruled out due to the relatively small sample size. In future studies, it will be important to recruit a larger cohort with matching healthy controls, as well as longitudinal cohorts from the HIV-1-infected individuals to confirm better MAIT cell recovery of haplotype 2 carriers. Moreover, further in vitro studies are warranted to tease out the potential mechanisms by which IL-7 and IL-7RA haplotypes influence MAIT cell biology. Complex interaction between MAIT cell biology, IL-7RA polymorphisms, and HIV-1 infection should be explored in-depth going forward. Understanding intricate utilization of IL-7 in reconstituting MAIT cells and enhancing their functions may ultimately benefit the host defense during HIV-1 infection.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Edwin Leeansyah (https://edwin.leeansyah@sz.tsinghua.edu.cn)
Materials availability
This study did not generate new unique reagents. The MR1 ligand 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) was kindly provided by Drs. David P. Fairlie and Jeffrey Y. W. Mak (University of Queensland, Australia) (62, 63).
Data and code availability
No data sets or code were generated or analysed in this study. All raw data associated with this study are available upon request from the Lead Contact. All software is commercially available.
Materials and methods
Ethics statement
Human samples were collected after informed written consent was obtained from all donors in accordance with study protocols conforming to the provisions of the Declaration of Helsinki. All participants (HIV-1-infected and healthy volunteers) provided written informed consent and this study was approved by the University Malaya Medical Centre’s ethics review board (MEC Ref: 896.32) and the Tsinghua Shenzhen International Graduate School Ethics Committee (Ref: 202171).
HIV-1-infected and uninfected cohorts
HIV-1-infected (n=39) individuals previously genotyped for the IL-7Rα SNPs were identified from a previous cohort established to explore biological factors associated with immune recovery (64). Briefly, all patients were recruited at the Infectious Diseases Unit (IDU) in University Malaya Medical Centre (UMMC) during their routine follow-up. Patients who fulfilled the following inclusion criteria were enrolled into the study: 1) Men or women aged at least 18 years, 2) first antiretroviral regimen composed of cART defined by at least three antiretroviral drugs, 3) CD4 T-cell count <500 cells/µl at the start of cART and 4) patients achieving HIV-1 RNA levels ≤50 copies/ml within 12 months following commencement of cART. Demographic and clinical parameters including age at cART initiation, gender, ethnicity, history of AIDS defining illnesses, antiretroviral prescribed and CD4 T cell count and HIV RNA measurements were obtained from patients medical records. HIV-1-negative controls (n=35) were recruited amongst healthy laboratory staff volunteers. All samples were processed using identical protocols within 4 hours of venipuncture.
Blood processing
Peripheral blood mononuclear cells (PBMCs) were isolated by standard Ficoll-Histopaque density gradient separation (Axis-Shield Poc As™). Isolated PBMCs were cryopreserved in liquid nitrogen until further required.
Flow cytometry analysis
Anti-Bcl-2 PE, CD3-FITC, CD8-APC, CD127-BV510, IFNγ-APC, RORγt-BV421, TNF-PE were from BD Biosciences. Anti-CD4-PerCP, CD8-BV510, CD127-PECy7, CD161-BV421, -FITC, and -PE, IFNγ-APC, Ki-67-AF488 and -BV510, TNF-BV421, and Vα7.2 PECy7 were from Biolegend. Anti-PLZF APC was from R&D Systems. CellTrace Violet (CTV) cell proliferation kit, Live/Dead Aqua and Near Infrared fixable cell stain were from Life Technologies. MR1-5-OP-RU tetramer -APC, -BV421, and -PE were from NIH Tetramer Core Facility, Emory University. Staining with the MR1 5-OP-RU tetramers was performed for 40 min at room temperature (RT) (6) before proceeding to the surface and intracellular staining with other mAbs. Cell surface and intracellular staining for TFs, cytokines, and cytotoxic molecules were performed as previously described (10). Samples were acquired on an FACS Canto II (BD Biosciences) equipped with 405, 488, and 633 nm lasers or CytoFLEX (Beckman Coulter) equipped with 405, 488, and 638 nm lasers. Data including the compensation platform were analysed using FlowJo v.10 (BD Biosciences).
Preparation of MAIT cell antigen 5-OP-RU
The MAIT cell antigen 5-OP-RU was synthesized according to a previously published procedure (62, 63). It is stable in DMSO solutions but converts rapidly to much less active lumazine in aqueous media, the exposure time to which should be minimized as much as possible to maximize activity. All 5-OP-RU working solutions were diluted from the DMSO stocks with appropriate culture medium immediately (within 5 min) prior to the proliferation assay.
Preparation of sIL-7Rα/IL-7 or sIL-7Rα/IL-2 mixture
Recombinant human IL-7 (R&D System) or IL-2 (PeproTech) at 2.5 ng/mL in MAIT cell Expansion Medium (see below) were incubated with sIL-7Rα (Biolegend) at varying amounts corresponding to the indicated molar ratios for 1 h at 37°C. PBMCs were then cultured in these media for further MAIT cell functional and proliferation assay as indicated.
MAIT cells activation and proliferation assays
MAIT cells within bulk PBMCs, identified as Vα7.2+ CD161hi CD3+ T cells, were activated using formaldehyde-fixed E. coli at the microbial dose of 1 for 24 h as previously described (65). Monensin (Golgi Stop, BD Biosciences) was added for the last 6 h of incubation. To determine the effect of sIL-7Rα on IL-7-dependent MAIT cell activation, cryopreserved PBMCs were thawed and rested for 48 h in a 37°C/5%CO2 incubator in ImmunoCult-XF T cell expansion medium supplemented with 8% (v/v) xeno-free CTS Immune Cell Serum Replacement (ThermoFisher Scientific), 50 μg/mL gentamicin (ThermoFisher Scientific), and 100 μg/mL normocin (Invivogen) (collectively: Expansion Medium). PBMCs were subsequently supplemented with IL-7 alone or in combination with sIL-7Rα as indicated for a further 24 h prior to analyses. In selected experiments, PBMCs were further stimulated with formaldehyde-fixed E. coli at the microbial dose of 0.1 for another 24 h.
For MAIT cell proliferation assay, MAIT cells were expanded as previously described (59, 66) with modifications. Briefly, cryopreserved PBMCs were thawed and stained using CTV cell proliferation dye as previously described (67), and rested for 48h in a 37°C/5%CO2 incubator in Expansion Medium. Subsequently, PBMCs were cultured with 100 pM 5-OP-RU with or without 2.5 ng/mL IL-7 or IL-2 alone, or with varying amounts of sIL-7Rα as indicated. No further media changes and cytokine supplementations were performed. Expansion and proliferation indices of MAIT cells, identified as MR1-OP-RU tetramer+ Vα7.2+ CD3+ T cells, were determined based on CTV dilution at day 5 post-stimulation using the FlowJo v. 10 software. Absolute MAIT cell numbers were calculated using Precision Count Beads (BioLegend) according to the manufacturer’s instruction.
DNA extraction and IL-7Rα genotyping
DNA was extracted from neutrophils using DNeasy blood & tissue kit according to the manufacturer instruction (QIAGEN, Germany). 200ng of DNA from each patient was used to genotype the IL-7Rα gene. Six specific loci in both coding and non-coding regions of IL-7Rα gene (rs11567762, rs1494555, rs3194051, rs3822731, rs6897932 and rs987106) were genotyped using MassARRAY system through specific primer extension reactions coupled with matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) for multiplexed genotyping (Sequenom MassARRAY iPLEX platform) performed at Australian Genome Research Facility (http://www.agrf.org.au/Genotyping-Platforms.html) or at China Sangon Biotech (https://www.sangon.com/zh/services_snp.html).
IL-7Rα haplotype determination
Haplotypes of IL-7Rα were determined by genotyping six SNPs (haplotype-tagging SNP) of the IL-7Rα gene which collectively defined the four common haplotypes of the IL-7Rα gene as previously described (43). Briefly the rs1494555 SNP (T/C) tags haplotype 1, rs6897932 SNP (C/T) tags haplotype2, rs3822731 SNP (T/C) tags haplotype 3 and rs3194051 SNP (A/G) tags haplotype 4.
Measurement of IL-7
IL-7 levels in plasma samples were measured using Quantikine HS ELISA kit following the manufacturer instruction (R&D Systems). This assay was performed on patient samples in the kinetics study. Briefly, plasma samples were diluted in 1 in 2 using the diluent provided. All samples were assayed in duplicate. Pre- and post-cART samples from the same patient were assayed in the same plate. The inter-assay and intra-assay CV for this assay were 16.3% (n=6) and 9.33% (n=6), respectively. The standard concentrations for this assay ranged from 0.25-16 pg/ml.
Measurement of soluble IL-7Rα (sIL-7Rα)
sIL-7Rα levels in plasma samples were measured using an in-house ELISA method as previously described (42). Human IL-7Rα/Fc chimera extracellular domain immunoglobulin (R&D Systems) was used as standards and the concentration ranged from 0.625-40 ng/ml. Samples, standards, detection antibody and streptavidin-HRP were diluted using 1% BSA in PBS. The inter-assay and intra-assay CV precision for this assay were 20% (n=6) and 2.4% (n=6), respectively. All samples were assayed in duplicate.
Statistical analysis
Statistical analyses were performed using Prism software v.9.4 (GraphPad). Data sets were first assessed for data normality distribution. Box and whisker plots show median, the 10th to 90th percentile, and the interquartile range. Statistically significant differences between samples were determined as appropriate using the unpaired t-test or Mann-Whitney’s test for unpaired samples. The Friedman test, one-way analysis of variance (ANOVA), or mixed-effects analysis followed by the appropriate post-hoc test as indicated was used to detect differences across multiple samples. Correlations were assessed using the Spearman rank correlation. Two-sided p-values < 0.05 were considered significant.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by The University Malaya Medical Centre’s ethics review board (MEC Ref: 896.32). The patients/participants provided their written informed consent to participate in this study.
Author contributions
FH, MG, YZ, NZ, DH, AH, EL performed the experiments. FH, YZ, WS, EL analysed the data. EL designed the experiments. WS, LH, ZL, PQ, PL, AK, L-FW, JS, SL performed critical logistics and/or provided critical reagents. EL conceived the study. RR, EL managed the study. FH, RR, EL wrote the paper. WS, JKS revised the draft, which was approved by all authors. All authors contributed to the article and approved the submitted version.
Funding
The research was supported by grants from Swedish Research Council Grant 2015-00174, Marie Skłodowska Curie Actions, Cofund, Project INCA 600398, the Jonas Söderquist Foundation for Virology and Immunology, the Petrus and Augusta Hedlund Foundation, Tsinghua Shenzhen International Graduate School Research Startup Funds (No. 01030100004), and the Shenzhen Pengcheng Peacock Program (to EL). The establishment of the clinical cohort and genotyping was supported by the High Impact Research grant (HIR/MOHE; H-20001-E000001) to RR, SL, and AK.
Acknowledgments
The MR1 tetramer technology was developed jointly by Dr. James McCluskey, Dr. Jamie Rossjohn, and Dr. David Fairlie; and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne. The MR1 ligand 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) was kindly provided by Drs. David P. Fairlie and Jeffrey Y. W. Mak (University of Queensland, Australia) (62, 63).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.985385/full#supplementary-material
Supplementary Figure 1 | Age distribution and MAIT cell levels in HIV-1-uninfected cohort. (A) Age distribution stratified by sex in HC carrying the IL7RA haplotype 2 vs non-haplotype 2. (B) MAIT cell levels in female and male participants of the healthy cohorts stratified by IL7RA haplotype. (B) Box and whisker plots show all data points, median, and the interquartile range. Statistical significance was determined using Mann-Whitney’s test. ns, not significant.
Supplementary Figure 2 | Soluble IL-7Rα does not inhibit IL-2-mediated MAIT cell proliferation. (A–G) MR1-5-OP-RU tetramer+ Vα7.2+ MAIT cell (A) proliferation as determined by Ki-67 expression levels on Ki-67+ MAIT cells (B; left panel, C), absolute MAIT cell numbers (D), and proliferation (E) and expansion (F) indices calculated using CTV-dilution (B; right panel) following 5-days treatment of PBMC with 5-OP-RU, 5-OP-RU+IL-2, or 5-OP-RU+IL-2+sIL-7Rα (sIL-7Rα/IL-2 molar ratio 1:1) (N=5). (G) Dose-dependent effect of sIL-7Rα treatment on MAIT cells proliferation and expansion indices following 5-days treatment with IL-2 and 5-OP-RU (N=5). Expression levels for (G) were normalised to nil sIL-7Rα controls to minimise inherent donor-to-donor variability, with statistical tests were performed on raw data. Box and whisker plots show all data points, median, and the interquartile range, whereas data presented as line graphs with error bars represent the mean and standard error. Statistical significance was determined using the Friedman test followed by Dunn’s post-hoc test (C, D), or repeated measure one-way ANOVA with Tukey’s post-hoc test (E, F, G). ** p<0.01, * p<0.05, ns, not significant.
References
1. Provine NM, Klenerman P. Mait cells in health and disease. Annu Rev Immunol (2020) 38:203–28. doi: 10.1146/annurev-immunol-080719-015428
2. Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, et al. An invariant T cell receptor alpha chain defines a novel tap-independent major histocompatibility complex class ib-restricted Alpha/Beta T cell subpopulation in mammals. J Exp Med (1999) 189(12):1907–21. doi: 10.1084/jem.189.12.1907
3. Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of mait cells. Nat Immunol (2019) 20(9):1110–28. doi: 10.1038/s41590-019-0444-8
4. Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by Mr1. Nature (2003) 422(6928):164–9. doi: 10.1038/nature01433
5. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. Mr1 presents microbial vitamin b metabolites to mait cells. Nature (2012) 491(7426):717–23. doi: 10.1038/nature11605
6. Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-Cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature (2014) 509(7500):361–5. doi: 10.1038/nature13160
7. Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise development of mait cells in mouse and human. PloS Biol (2009) 7(3):e54. doi: 10.1371/journal.pbio.1000054
8. Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, et al. Human mait cells are xenobiotic-resistant, tissue-targeted, Cd161hi il-17-Secreting T cells. Blood (2011) 117(4):1250–9. doi: 10.1182/blood-2010-08-303339
9. Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (Tcr) expression by human peripheral blood Cd4-8- Alpha/Beta T cells demonstrates preferential use of several V beta genes and an invariant tcr alpha chain. J Exp Med (1993) 178(1):1–16. doi: 10.1084/jem.178.1.1
10. Dias J, Boulouis C, Gorin JB, van den Biggelaar R, Lal KG, Gibbs A, et al. The Cd4(-)Cd8(-) mait cell subpopulation is a functionally distinct subset developmentally related to the main Cd8(+) mait cell pool. Proc Natl Acad Sci U.S.A. (2018) 115(49):E11513–E22. doi: 10.1073/pnas.1812273115
11. Pavlovic M, Gross C, Chili C, Secher T, Treiner E. Mait cells display a specific response to type 1 ifn underlying the adjuvant effect of Tlr7/8 ligands. Front Immunol (2020) 11:2097. doi: 10.3389/fimmu.2020.02097
12. Lamichhane R, Schneider M, de la Harpe SM, Harrop TWR, Hannaway RF, Dearden PK, et al. Tcr- or cytokine-activated Cd8(+) mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep (2019) 28(12):3061–76.e5. doi: 10.1016/j.celrep.2019.08.054
13. Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal mait-cell development. Nat Commun (2014) 5:3143. doi: 10.1038/ncomms4143
14. Loh L, Wang Z, Sant S, Koutsakos M, Jegaskanda S, Corbett AJ, et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity Via il-18-Dependent activation. Proc Natl Acad Sci USA (2016) 113(36):10133–8. doi: 10.1073/pnas.1610750113
15. van Wilgenburg B, Scherwitzl I, Hutchinson EC, Leng T, Kurioka A, Kulicke C, et al. Mait cells are activated during human viral infections. Nat Commun (2016) 7:11653. doi: 10.1038/ncomms11653
16. van Wilgenburg B, Loh L, Chen Z, Pediongco TJ, Wang H, Shi M, et al. Mait cells contribute to protection against lethal influenza infection in vivo. Nat Commun (2018) 9(1):4706. doi: 10.1038/s41467-018-07207-9
17. Flament H, Rouland M, Beaudoin L, Toubal A, Bertrand L, Lebourgeois S, et al. Outcome of sars-Cov-2 infection is linked to mait cell activation and cytotoxicity. Nat Immunol (2021) 22(3):322–35. doi: 10.1038/s41590-021-00870-z
18. Dias J, Hengst J, Parrot T, Leeansyah E, Lunemann S, Malone DFG, et al. Chronic hepatitis delta virus infection leads to functional impairment and severe loss of mait cells. J Hepatol (2019) 71(2):301–12. doi: 10.1016/j.jhep.2019.04.009
19. Leeansyah E, Ganesh A, Quigley MF, Sonnerborg A, Andersson J, Hunt PW, et al. Activation, exhaustion, and persistent decline of the antimicrobial Mr1-restricted mait-cell population in chronic hiv-1 infection. Blood (2013) 121(7):1124–35. doi: 10.1182/blood-2012-07-445429
20. Parrot T, Gorin JB, Ponzetta A, Maleki KT, Kammann T, Emgård J, et al. Mait cell activation and dynamics associated with covid-19 disease severity. Sci Immunol (2020) 5(51):1–11. doi: 10.1126/sciimmunol.abe1670
21. Jouan Y, Guillon A, Gonzalez L, Perez Y, Boisseau C, Ehrmann S, et al. Phenotypical and functional alteration of unconventional T cells in severe covid-19 patients. J Exp Med (2020) 217(12):1–9. doi: 10.1084/jem.20200872
22. Cosgrove C, Ussher JE, Rauch A, Gärtner K, Kurioka A, Hühn MH, et al. Early and nonreversible decrease of Cd161++ /Mait cells in hiv infection. Blood (2013) 121(6):951–61. doi: 10.1182/blood-2012-06-436436
23. Fernandez CS, Amarasena T, Kelleher AD, Rossjohn J, McCluskey J, Godfrey DI, et al. Mait cells are depleted early but retain functional cytokine expression in hiv infection. Immunol Cell Biol (2015) 93(2):177–88. doi: 10.1038/icb.2014.91
24. Greathead L, Metcalf R, Gazzard B, Gotch F, Steel A, Kelleher P. Cd8+/Cd161++ mucosal-associated invariant T-cell levels in the colon are restored on long-term antiretroviral therapy and correlate with Cd8+ T-cell immune activation. Aids (2014) 28(11):1690–2. doi: 10.1097/qad.0000000000000351
25. Han F, Zheng Y, Ho A, Ma S, Sandberg JK, Leeansyah E. Mait cell loss and reconstitution in hiv-1 disease. Crit Rev Immunol (2021) 41(5):69–82. doi: 10.1615/CritRevImmunol.2022042906
26. Lal KG, Kim D, Costanzo MC, Creegan M, Leeansyah E, Dias J, et al. Dynamic mait cell response with progressively enhanced innateness during acute hiv-1 infection. Nat Commun (2020) 11(1):272. doi: 10.1038/s41467-019-13975-9
27. Tang X, Zhang S, Peng Q, Ling L, Shi H, Liu Y, et al. Sustained ifn-I stimulation impairs mait cell responses to bacteria by inducing il-10 during chronic hiv-1 infection. Sci Adv (2020) 6(8):eaaz0374. doi: 10.1126/sciadv.aaz0374
28. Trivedi S, Afroz T, Bennett MS, Angell K, Barros F, Nell RA, et al. Diverse mucosal-associated invariant tcr usage in hiv infection. ImmunoHorizons (2021) 5(5):360–9. doi: 10.4049/immunohorizons.2100026
29. Merlini E, Cerrone M, van Wilgenburg B, Swadling L, Cannizzo ES, d'Arminio Monforte A, et al. Association between impaired Vα7.2+Cd161++Cd8+ (Mait) and Vα7.2+Cd161-Cd8+ T-cell populations and gut dysbiosis in chronically hiv- and/or hcv-infected patients. Front Microbiol (2019) 10:1972. doi: 10.3389/fmicb.2019.01972
30. Xia P, Xing XD, Yang CX, Liao XJ, Liu FH, Huang HH, et al. Activation-induced pyroptosis contributes to the loss of mait cells in chronic hiv-1 infected patients. Mil Med Res (2022) 9(1):24. doi: 10.1186/s40779-022-00384-1
31. Tang XZ, Jo J, Tan AT, Sandalova E, Chia A, Tan KC, et al. Il-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J Immunol (2013) 190(7):3142–52. doi: 10.4049/jimmunol.1203218
32. Leeansyah E, Svard J, Dias J, Buggert M, Nystrom J, Quigley MF, et al. Arming of mait cell cytolytic antimicrobial activity is induced by il-7 and defective in hiv-1 infection. PloS Pathog (2015) 11(8):e1005072. doi: 10.1371/journal.ppat.1005072
33. Murugesan A, Ibegbu C, Styles TM, Jones AT, Shanmugasundaram U, Reddy PBJ, et al. Functional mait cells are associated with reduced simian-human immunodeficiency virus infection. Front Immunol (2019) 10:3053. doi: 10.3389/fimmu.2019.03053
34. Hassane M, Jouan Y, Creusat F, Soulard D, Boisseau C, Gonzalez L, et al. Interleukin-7 protects against bacterial respiratory infection by promoting il-17a-Producing innate T-cell response. Mucosal Immunol (2020) 13(1):128–39. doi: 10.1038/s41385-019-0212-y
35. Beq S, Delfraissy JF, Theze J. Interleukin-7 (Il-7): Immune function, involvement in the pathogenesis of hiv infection and therapeutic potential. Eur Cytokine Netw (2004) 15(4):279–89.
36. Sortino O, Richards E, Dias J, Leeansyah E, Sandberg JK, Sereti I. Il-7 treatment supports Cd8+ mucosa-associated invariant T-cell restoration in hiv-1-Infected patients on antiretroviral therapy. Aids (2018) 32(6):825–8. doi: 10.1097/qad.0000000000001760
37. Schluns KS, Kieper WC, Jameson SC, Lefrançois L. Interleukin-7 mediates the homeostasis of naïve and memory Cd8 T cells in vivo. Nat Immunol (2000) 1(5):426–32. doi: 10.1038/80868
38. Levy Y, Lacabaratz C, Weiss L, Viard JP, Goujard C, Lelièvre JD, et al. Enhanced T cell recovery in hiv-1-Infected adults through il-7 treatment. J Clin Invest (2009) 119(4):997–1007. doi: 10.1172/jci38052
39. Sasson SC, Zaunders JJ, Kelleher AD. The il-7/Il-7 receptor axis: Understanding its central role in T-cell homeostasis and the challenges facing its utilization as a novel therapy. Curr Drug Targets (2006) 7(12):1571–82. doi: 10.2174/138945006779025365
40. Rose T, Lambotte O, Pallier C, Delfraissy JF, Colle JH. Identification and biochemical characterization of human plasma soluble il-7r: Lower concentrations in hiv-1-Infected patients. J Immunol (2009) 182(12):7389–97. doi: 10.4049/jimmunol.0900190
41. Crawley AM, Faucher S, Angel JB. Soluble il-7r alpha (Scd127) inhibits il-7 activity and is increased in hiv infection. J Immunol (2010) 184(9):4679–87. doi: 10.4049/jimmunol.0903758
42. Hoe E, McKay FC, Schibeci SD, Gandhi K, Heard RN, Stewart GJ, et al. Functionally significant differences in expression of disease-associated il-7 receptor alpha haplotypes in Cd4 T cells and dendritic cells. J Immunol (2010) 184(5):2512–7. doi: 10.4049/jimmunol.0902900
43. Teutsch SM, Booth DR, Bennetts BH, Heard RN, Stewart GJ. Identification of 11 novel and common single nucleotide polymorphisms in the interleukin-7 receptor-alpha gene and their associations with multiple sclerosis. Eur J Hum Genet EJHG (2003) 11(7):509–15. doi: 10.1038/sj.ejhg.5200994
44. Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, Prokop A, et al. Interleukin 7 receptor alpha chain (Il7r) shows allelic and functional association with multiple sclerosis. Nat Genet (2007) 39(9):1083–91. doi: 10.1038/ng2103
45. Rajasuriar R, Booth D, Solomon A, Chua K, Spelman T, Gouillou M, et al. Biological determinants of immune reconstitution in hiv-infected patients receiving antiretroviral therapy: The role of interleukin 7 and interleukin 7 receptor α and microbial translocation. J Infect Dis (2010) 202(8):1254–64. doi: 10.1086/656369
46. Rajasuriar R, Booth DR, Gouillou M, Spelman T, James I, Solomon A, et al. The role of snps in the α-chain of the il-7r gene in Cd4+ T-cell recovery in hiv-infected African patients receiving suppressive cart. Genes Immun (2012) 13(1):83–93. doi: 10.1038/gene.2011.65
47. Limou S, Melica G, Coulonges C, Lelièvre JD, Do H, McGinn S, et al. Identification of Il7ra risk alleles for rapid progression during hiv-1 infection: A comprehensive study in the griv cohort. Curr HIV Res (2012) 10(2):143–50. doi: 10.2174/157016212799937209
48. Hartling HJ, Thørner LW, Erikstrup C, Zinyama R, Kallestrup P, Gomo E, et al. Polymorphisms in the interleukin-7 receptor α gene and mortality in untreated hiv-infected individuals. Aids (2013) 27(10):1615–20. doi: 10.1097/QAD.0b013e3283606c2d
49. Wang XS, Wen PF, Zhang M, Hu LF, Ni J, Qiu LJ, et al. Interleukin-7 receptor single nucleotide polymorphism Rs6897932 (C/T) and the susceptibility to systemic lupus erythematosus. Inflammation (2014) 37(2):615–20. doi: 10.1007/s10753-013-9777-x
50. O'Doherty C, Alloza I, Rooney M, Vandenbroeck K. Il7ra polymorphisms and chronic inflammatory arthropathies. Tissue Antigens (2009) 74(5):429–31. doi: 10.1111/j.1399-0039.2009.01342.x
51. Heron M, Grutters JC, van Moorsel CH, Ruven HJ, Huizinga TW, van der Helm-van Mil AH, et al. Variation in Il7r predisposes to sarcoid inflammation. Genes Immun (2009) 10(7):647–53. doi: 10.1038/gene.2009.55
52. Santiago JL, Alizadeh BZ, Martínez A, Espino L, de la Calle H, Fernández-Arquero M, et al. Study of the association between the capsl-Il7r locus and type 1 diabetes. Diabetologia (2008) 51(9):1653–8. doi: 10.1007/s00125-008-1070-4
53. Lundström W, Highfill S, Walsh ST, Beq S, Morse E, Kockum I, et al. Soluble Il7rα potentiates il-7 bioactivity and promotes autoimmunity. Proc Natl Acad Sci USA (2013) 110(19):E1761–70. doi: 10.1073/pnas.1222303110
54. Gherardin NA, Souter MN, Koay H-F, Mangas KM, Seemann T, Stinear TP, et al. Human blood mait cell subsets defined using Mr1 tetramers. Immunol Cell Biol (2018) 96(5):507–25. doi: 10.1111/imcb.12021
55. Koenen P, Heinzel S, Carrington EM, Happo L, Alexander WS, Zhang JG, et al. Mutually exclusive regulation of T cell survival by il-7r and antigen receptor-induced signals. Nat Commun (2013) 4:1735. doi: 10.1038/ncomms2719
56. Dias J, Sobkowiak MJ, Sandberg JK, Leeansyah E. Human mait-cell responses to escherichia coli: Activation, cytokine production, proliferation, and cytotoxicity. J Leukoc Biol (2016) 100(1):233–40. doi: 10.1189/jlb.4TA0815-391RR
57. Dias J, Leeansyah E, Sandberg JK. Multiple layers of heterogeneity and subset diversity in human mait cell responses to distinct microorganisms and to innate cytokines. Proc Natl Acad Sci U.S.A. (2017) 114(27):E5434–43. doi: 10.1073/pnas.1705759114
58. Freeman ML, Morris SR, Lederman MM. Cd161 expression on mucosa-associated invariant T cells is reduced in hiv-infected subjects undergoing antiretroviral therapy who do not recover Cd4(+) T cells. Pathog Immun (2017) 2(3):335–51. doi: 10.20411/pai.v2i3.136
59. Leeansyah E, Hey YY, Sia WR, Ng JHJ, Gulam MY, Boulouis C, et al. Mr1-restricted T cells with mait-like characteristics are functionally conserved in the pteropid bat pteropus alecto. iScience (2020) 23(12):101876. doi: 10.1016/j.isci.2020.101876
60. Steele AK, Carrasco-Medina L, Sodora DL, Crawley AM. Increased soluble il-7 receptor concentrations associate with improved il-7 therapy outcomes in siv-infected art-treated rhesus macaques. PloS One (2017) 12(12):e0188427. doi: 10.1371/journal.pone.0188427
61. Hubrack S, Al-Nesf MA, Agrebi N, Raynaud C, Khattab MA, Thomas M, et al. In vitro interleukin-7 treatment partially rescues mait cell dysfunction caused by sars-Cov-2 infection. Sci Rep (2021) 11(1):14090. doi: 10.1038/s41598-021-93536-7
62. Mak JYW, Xu W, Reid RC, Corbett AJ, Meehan BS, Wang H, et al. Stabilizing short-lived schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nat Commun (2017) 8(1):14599. doi: 10.1038/ncomms14599
63. Mak JYW, Liu L, Fairlie DP. Chemical modulators of mucosal associated invariant T cells. Acc Chem Res (2021) 54(17):3462–75. doi: 10.1021/acs.accounts.1c00359
64. Yap SH, Abdullah NK, McStea M, Takayama K, Chong ML, Crisci E, et al. Hiv/Human herpesvirus Co-infections: Impact on tryptophan-kynurenine pathway and immune reconstitution. PloS One (2017) 12(10):e0186000. doi: 10.1371/journal.pone.0186000
65. Dias J, Sandberg JK, Leeansyah E. Extensive phenotypic analysis, transcription factor profiling, and effector cytokine production of human mait cells by flow cytometry. Methods Mol Biol (Clifton NJ) (2017) 1514:241–56. doi: 10.1007/978-1-4939-6548-9_17
66. Boulouis C, Sia WR, Gulam MY, Teo JQM, Png YT, Phan TK, et al. Human mait cell cytolytic effector proteins synergize to overcome carbapenem resistance in escherichia coli. PloS Biol (2020) 18(6):e3000644. doi: 10.1371/journal.pbio.3000644
Keywords: MAIT cells, HIV-1, IL-7, CD127 (IL7Ra) gene, antiretroviral (ARV) therapy
Citation: Han F, Gulam MY, Zheng Y, Zulhaimi NS, Sia WR, He D, Ho A, Hadadi L, Liu Z, Qin P, Lobie PE, Kamarulzaman A, Wang L-F, Sandberg JK, Lewin SR, Rajasuriar R and Leeansyah E (2022) IL7RA single nucleotide polymorphisms are associated with the size and function of the MAIT cell population in treated HIV-1 infection. Front. Immunol. 13:985385. doi: 10.3389/fimmu.2022.985385
Received: 03 July 2022; Accepted: 04 October 2022;
Published: 20 October 2022.
Edited by:
James E. Ussher, University of Otago, New ZealandReviewed by:
Vijayakumar Velu, Emory University, United StatesChris Bjørnsen Willberg, Medimab Biotherapuetics, United Kingdom
Melanie Harriff, Oregon Health & Science University, United States
Copyright © 2022 Han, Gulam, Zheng, Zulhaimi, Sia, He, Ho, Hadadi, Liu, Qin, Lobie, Kamarulzaman, Wang, Sandberg, Lewin, Rajasuriar and Leeansyah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edwin Leeansyah, ZWR3aW4ubGVlYW5zeWFoQHN6LnRzaW5naHVhLmVkdS5jbg==
†These authors have contributed equally to this work
 Fei Han
Fei Han Muhammad Yaaseen Gulam3†
Muhammad Yaaseen Gulam3† Peiwu Qin
Peiwu Qin Peter E. Lobie
Peter E. Lobie Lin-Fa Wang
Lin-Fa Wang Johan K. Sandberg
Johan K. Sandberg Sharon R. Lewin
Sharon R. Lewin Reena Rajasuriar
Reena Rajasuriar Edwin Leeansyah
Edwin Leeansyah