- 1Department of Laboratory Medicine, Linkou Chang Gung Memorial Hospital, Taoyuan, Taiwan
- 2Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan
- 3Division of Hematology-Oncology, Department of Internal Medicine, Linkou Chang Gung Memorial Hospital, Taoyuan, Taiwan
- 4Department of Life Science, Fu Jen Catholic University, New Taipei City, Taiwan
- 5Graduate Institute of Biomedical Sciences, College of Medicine, Chang Gung University, Taoyuan, Taiwan
In addition to the classical human leukocyte antigen (HLA) genes, the outcomes of post-hematopoietic stem cell transplantation (HSCT) are associated with human leukocyte antigen (HLA)-related genes and non-HLA genes involved in immune regulation. HLA-G gene plays an important role in immune tolerance, assisting immune escape of tumor cells, and decrease of transplant rejection. In this study, we explored the association of genetic variants at the 3’-untranslated region (3’-UTR) and 5’-upstream regulatory region (5’-URR) of HLA-G gene with the adverse outcomes of patients with leukemia receiving HSCT. The genomic DNAs of 164 patients who had acute leukemia and received HSCT were collected for analysis. Nine single nucleotide polymorphisms (SNPs) and six haplotypes in the 3’-UTR and 27 SNPs and 6 haplotypes in the 5’-URR were selected to investigate their relationship with the development of adverse outcomes for patients receiving HSCT, including mortality, relapse, and graft-versus-host disease. Our results revealed that two SNPs (rs371194629 and rs9380142) and one haplotype (UTR-3) located in the 3’-UTR and two SNPs (rs3823321 and rs1736934) and one haplotype (G0104a) located in the 5’-URR of HLA-G were associated with the occurrence of chronic GVHD or development of any forms of GVHD. No SNP was found to associate with the occurrence of mortality and relapse for patients receiving HSCT. These SNPs and haplotypes may play important roles in regulating immune tolerance of allografts post-HSCT that can be used to predict the risk of poor outcomes after receiving HSCT and giving preventive treatment to patients on time.
Introduction
Hematopoietic stem cell transplantation (HSCT) is a treatment of patients with blood disorders or cancer (1). Autologous or allogeneic hematopoietic stem cells are delivered into patients to achieve the purpose of immunotherapy or support patients to receive high-dose radiotherapy or chemotherapy. In allogeneic transplantation, human leukocyte antigen (HLA)-matched stem cells must be used to avoid transplantation rejection. However, there are still cases of transplant failure even using HLA-matched stem cells (2, 3). It is speculated that other factors in addition to HLA may contribute to the occurrence of adverse outcomes such as disease relapse, mortality, and graft-versus-host disease (GVHD) for patients post-HSCT (4, 5).
HLA-A, HLA-B, and HLA-C are the human major histocompatibility complex (MHC) class I proteins which are well documented to associate with the outcomes of HSCT (6). An additional non-classical MHC class Ib gene HLA-G is located on human chromosome 6p21.3, adjacent to the HLA-A locus. HLA-G gene has 8 exons separated by 7 introns. Alternative splicing of HLA-G mRNA selectively produces 7 mature mRNA subtypes, encoding 7 protein isoforms which include 4 membrane-bound proteins (HLA-G1, G2, G3, and G4) and 3 soluble proteins (HLA-G5, G6, and G7) (7). HLA-G1 and HLA-G5 combine with beta-2-microglobulin together to form HLA-G complexes which are the main immunomodulatory subtype with biological functions in the HLA-G family (8, 9). HLA-G has been shown recently to play important roles in immune tolerance of organ transplants (10), immune escape of tumor cells and viruses (11), and fetus-maternal tolerance leading to protection of fetus from attack by the maternal immune system (7, 12). The study by Boukouaci et al. further demonstrated that HLA-G is induced in vivo during allogeneic transplantation leading to immune tolerance and reducing the risk of transplant rejection and GVHD (13). In ex vivo study, HLA-G expression on the surface of CD4+ T cells inhibits the proliferation of allogeneic-specific T cells when mixing with the allogenic lymphocytes (14). These studies indicate that HLA-G is highly related to immune reaction and tolerance.
The 3’-untranslated region (3’-UTR) and the 5’-upstream regulatory region (5’-URR) of HLA-G gene are highly polymorphic. Genetic variations in these regions are known to regulate the levels of HLA-G gene expression and post-transcriptional mRNA stability (15). In addition to the genetic variants of single nucleotide polymorphisms (SNPs) and insertion/deletion (INDEL), 6 haplotypes in the 3’-UTR and 6 haplotypes in the 5’-URR of the HLA-G gene have been defined (16). Genetic polymorphisms and haplotypes of HLA-G have been reported to associate with the risk and prognosis of cancer, organ transplantation, viral and parasitic infections, pregnancy complications, the clinical outcome of patients with beta-thalassemia receiving HSCT, and the susceptibility of autoimmune diseases (17–21). Nevertheless, whether there is an association between HLA-G genetic polymorphism and haplotypes with the risk of adverse outcomes for patients with leukemia receiving HSCT is still not clear.
The aim of this study was to investigate the effects of genotype and haplotypes in the 3’-UTR and 5’-URR of HLA-G gene on the mortality, the risk of disease relapse, and the development of GVHD for patients with acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL) after receiving HSCT. The significance of the findings in this study was discussed.
Methods and materials
Patients and HLA typing
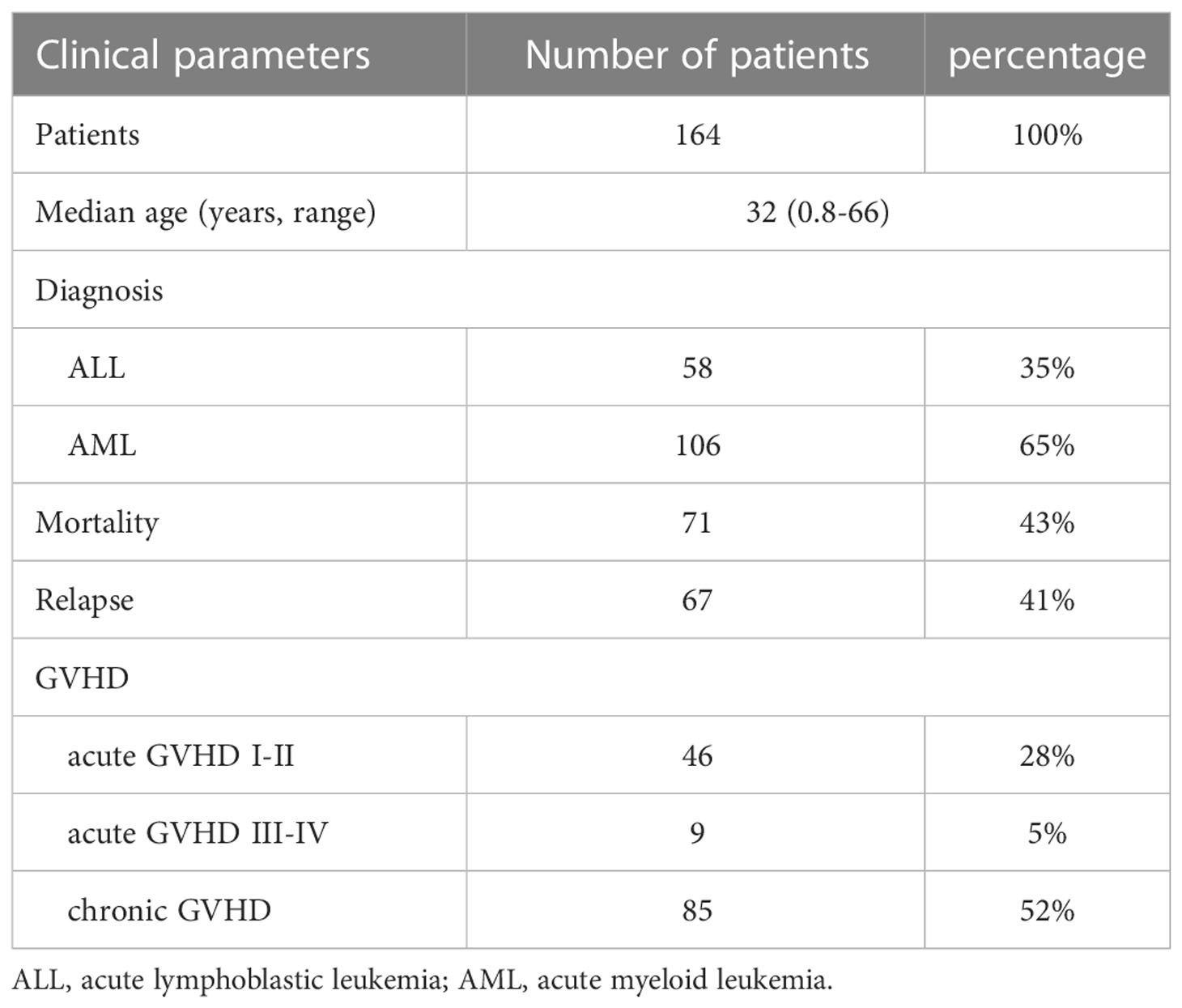
The study has been approved by the Institutional Review Board (IRB) of Chang Gung Memorial Hospital (CGMH) and was performed according to the ethical regulations and requirements. The approved ID was 201304949B0 and 202100302B0. Because HLA-G was mainly expressed in the saliva, peritoneal fluid, plasma, thymus, semen, cerebrospinal fluid but not in the immune cells (7), and only the HLA haplotypes of the recipients but not donors were associated with the outcomes after HSCT (18, 22), the analysis focused on the genetic components of the recipients. The inclusion criteria of participants were male and female adults (over 20 years old) who suffered from AML or ALL and underwent HSCT. Exclude the patients who had other hematological malignancies. A total of 164 patients receiving HSCT was enrolled in this study, in which 106 patients were diagnosed as AML and 58 patients were ALL. The clinical characteristics of the 164 patients are shown in Table 1. All recipients of unrelated HSCT signed informed consents and had fully matched HLA with the donors as revealed by using the SeCore kit (Thermo Fisher, Waltham, MA) for high-resolution HLA typing. The ambiguous alleles of the SeCore typing were resolved by using the MicroSSP Allele Specific Tying Tray with sequence-specific primers (Thermo Fisher, Waltham, MA).
Blood collection and genomic DNA extraction
The specimens for analysis were the remnant DNA for short tandem repeats (STR) analysis obtained from the Department of Laboratory Medicine. Briefly, peripheral blood (3 ml) was collected from patients in the blood collection tube with EDTA as the anticoagulant. The buffy coat was collected for genomic DNA extraction by using QIAamp DNA Blood Mini Kit according to the instruction of the manufacturer (Qiagen, Valencia, California, USA). The OD260/280 of the template genomic DNA had to be in the range of 1.8-2.0 with a minimal concentration of 100 ng/μl for genotyping and haplotyping by PCR.
Selection of genotypes and haplotypes
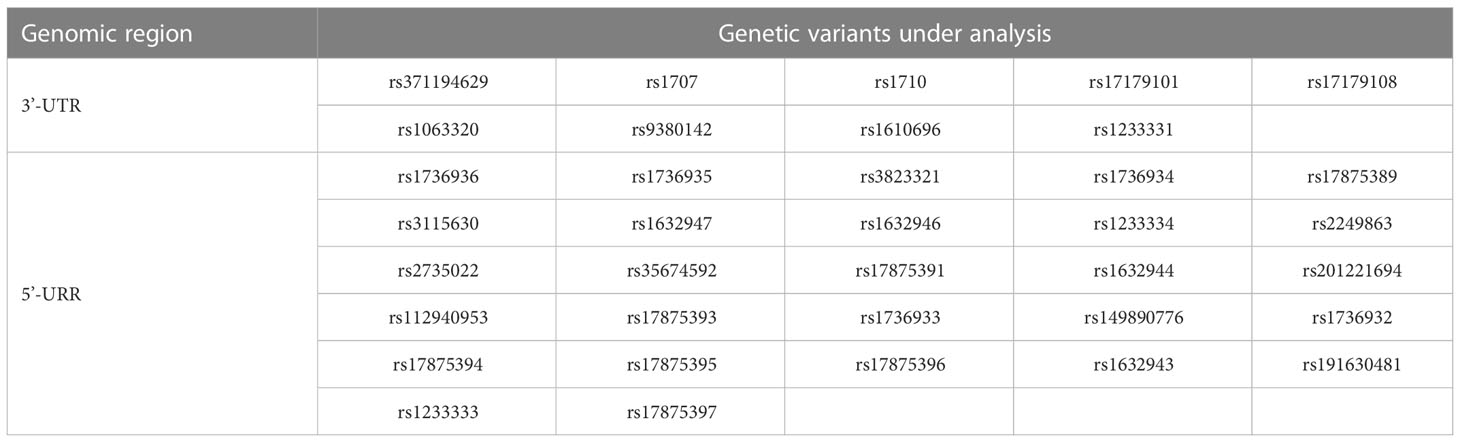
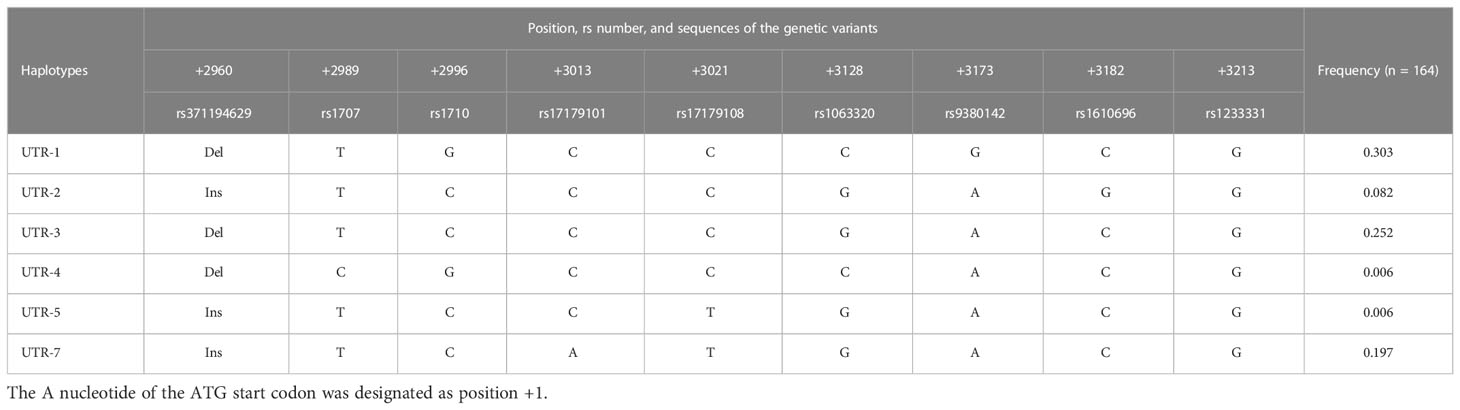
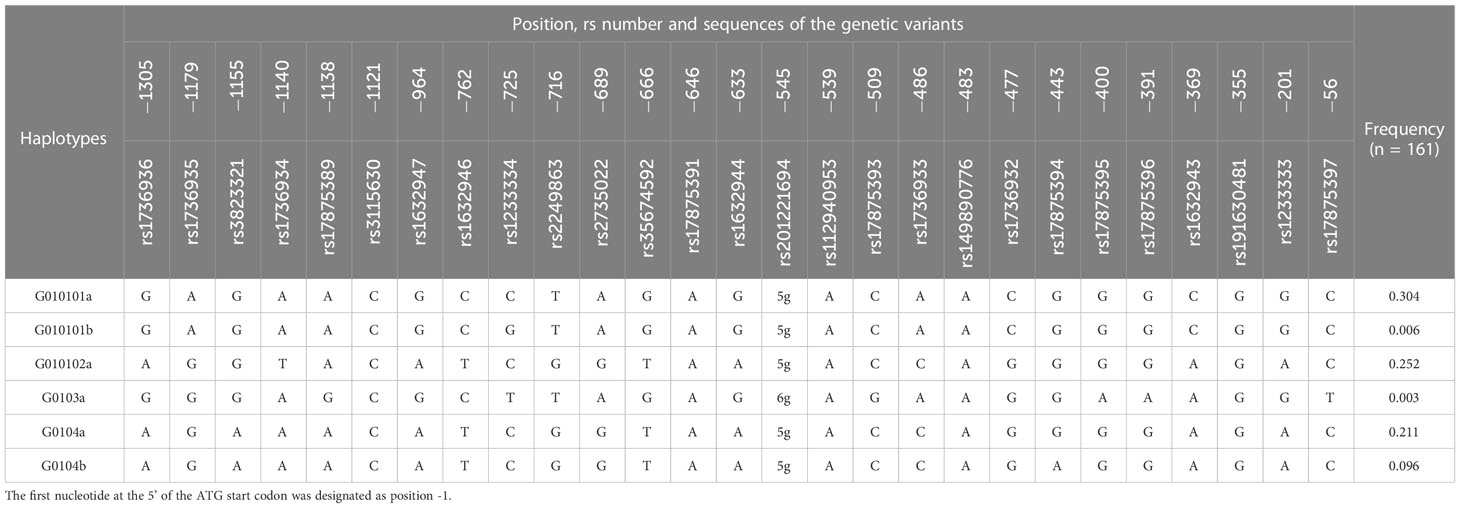
Based on previous studies for the association of HLA-G genetic polymorphisms with the clinical outcomes of various diseases and organ transplantation (17–21, 23), the recipient genotypes and haplotypes in the 3’-UTR and 5’-URR of HLA-G gene were selected for analysis. For 3’-UTR, a total of 9 genetic variants including 8 SNPs and the 14-bp INDEL polymorphism (rs371194629) (Table 2), and 6 haplotypes (Table 3) which were classified according to the sequences of the 9 genetic variants (24) were selected for analysis. For 5’-URR, a total of 27 SNPs (Table 2) and 6 haplotypes (Table 4) which were classified according to the DNA sequences of the 27 SNPs were selected for analysis.
PCR and sequencing
Primers (Table 5) were designed to amplify a 366-bp DNA fragment covering the 3’-UTR of HLA-G gene that contained the sequences of the selected 9 genetic variants and the 6 haplotypes. Primes (Table 5) were also designed to amplify a 1402-bp DNA fragment covering the 5’-URR of HLA-G gene that contained the sequences of the selected 27 SNPs and 6 haplotypes. The 1500 bp upstream of the start codon (ATG) was considered as the promoter region in the 5’-URR as described in most studies (25). PCR amplification was performed in a reaction mixture (25 μl) containing 0.2 mM of dNTP, 1.5 mM of MgCl2, 5 pmol of forward and reversed primers, 1 unit of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), and 200 ng of genomic DNA. PCR was initiated at 94°C for 3 min, followed by 30 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 90 sec, with a final extension step at 72°C for 10 min. The PCR products were verified by fractionating on a 2% agarose gel. The PCR amplicons were sequenced by using the Big Dye Terminator Cycle Sequencing kit (Thermo Fisher, Waltham, Massachusetts, USA) followed by analysis using the ABI PRISM genetic analyzer (Thermo Fisher, Waltham, Massachusetts, USA). Because of either insufficient genomic DNA or PCR failure, not every donor had complete data available.
Definition of the outcomes post-HSCT
The definition of outcomes post-HSCT has been described previously (26). Briefly, mortality was referred to the state of patients who died in the duration of study. Relapse was defined as recurrence of malignancy based on the overall evaluation of one or more of the following test results: bone marrow morphology, minimal residual disease by either flow cytometry, cytogenetics, imaging results, or STR analysis with the presence of > 5% recipient STR alleles in the chimeric test (27). GVHD was considered as acute GVHD (aGVHD) and chronic GVHD (cGVHD) when it occurs within 100 days after transplantation and more than 100 days after transplantation or continually for more than 100 days without remission, respectively (5). aGVHD was divided into four grades (Grade I, II, III, and IV) according to the clinical characteristics of organs (26) Grades I–II were defined as mild GVHD, Grades III–IV were defined as severe GVHD, and patients without aGVHD or cGVHD during the study period were defined as no GVHD. Patients with any forms of GVHD was defined when patients were present with either aGVHD or cGVHD.
Statistical analysis
Patients were grouped as case or control for statistical analyses of the association between the genotype and haplotype of patients and the effectiveness of HSCT. For mortality analysis, patients who died and lived at the end of this study were grouped as case and control, respectively. For relapse analysis, patients who relapsed and did not relapse at the end of this study were grouped as the case and control, respectively. For GVHD analysis, patients who developed stages I-II aGVHD, stages III-IV of aGVHD, cGVHD, and any forms of GVHD were considered as case and patients without development of GVHD were considered as control.
The PLINK software was used for genotyping analysis (http://pngu.mgh.harvard.edu/~purcell/plink/index.shtml). The associations of genotypes and haplotypes in the 3’-UTR and 5’-URR of HLA-G gene with the outcomes of patients post-HSCT (i.e. Grade I-II aGVHD, Grade III-IV aGVHD, cGVHD, any forms of GVHD, mortality, and relapse) were evaluated by analysis of genotype groups (AA, Aa, and aa) using various genetic models including additive, homozygous, heterozygous, dominant and recessive models and logistic regression analysis as described previously (26). Briefly, the allele with higher frequency was referred to major allele “A” and the other was minor allele “a”. For analysis using the additive model, “AA”, “Aa”, and “aa” were compared to each other (AA vs. Aa vs. aa). For homozygous model, the “AA” was compared to “aa”. For the heterozygous model, the “AA” was compared to “Aa”. For the dominant model, the collective genotypes (“Aa” + “aa”) were compared to a reference genotype “AA”. For the recessive model, “aa” is compared to a collective (“AA” + “Aa”) reference genotype.
The haplotypes with frequency less than 1% were excluded from the analysis. The dosage of a specific haplotype was first defined as 2, 1, and 0 when the indicated haplotype was homozygous, heterozygous, and not present in the recipient genomic DNA, respectively. Analysis was performed by either heterozygous or homozygous haplotype models. For heterozygous haplotype model, the occurrence of adverse outcomes for patients with two copies of the same haplotypes were compared with patients with one copy of the haplotype. For homozygous haplotype model, the occurrence of adverse outcomes for patients with two copies of the same haplotypes were compared with patients without the specific haplotype.
The SPSS 17.0 software was used for statistical analysis. A p < 0.05 was considered as statistical significance for all tests. Odds ratio (OR) and 95% confidence interval (CI) were also provided.
Results
Association of genetic polymorphisms in the 3’-UTR and 5’-URR of HLA-G gene with the risk of adverse outcomes for patients with leukemia post-HSCT
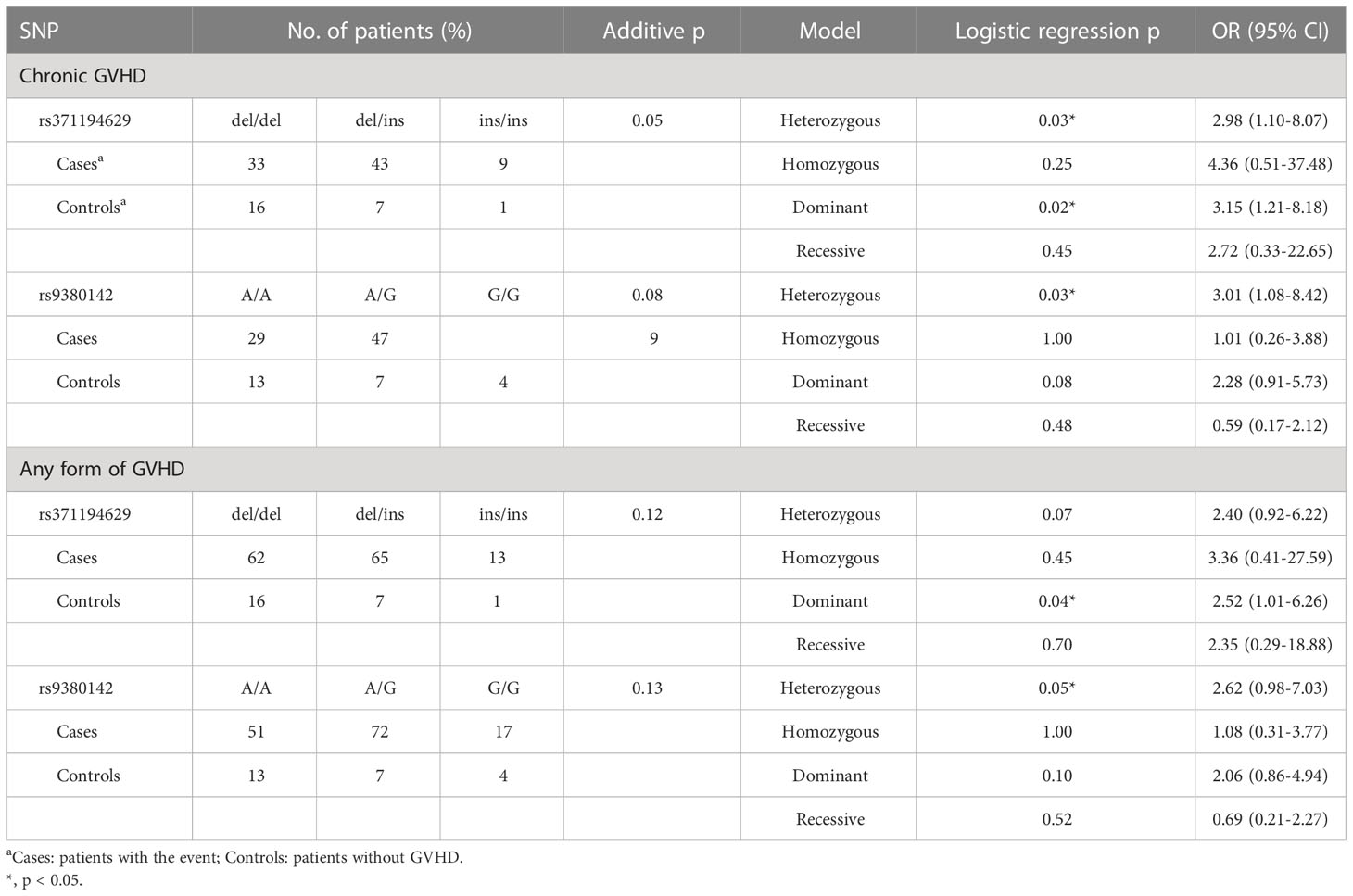
A total of 9 genetic polymorphisms in the 3’-UTR of HLA-G gene were analyzed to determine the association of these genotypes with the risks of adverse outcomes post-HSCT, including mortality, disease relapse, and GVHD. The complete genotype and allele frequency datasets for all recipients are summarized in the Supplementary Tables S1-S4. The genetic polymorphisms that were significantly associated with the risk of adverse outcomes are shown in Tables 6–9. Our data revealed that genetic polymorphisms in the 3’-UTR of HLA-G gene were only associated with the risks for development of GVHD, but not the mortality and disease relapse post- HSCT (Table 6; Supplementary Table S1). By analysis using the heterozygous model, patients with del/ins genotype of the 14-bp INDEL (rs371194629) were associated with an increased risk of cGVHD (OR = 2.98, 95% CI = 1.10 – 8.07, p = 0.03) when compared to patients with del/del genotype. By analysis using the dominant model, patients with the del/ins and ins/ins genotypes of the 14-bp INDEL were associated with an increased risk of cGVHD (OR = 3.15, 95% CI = 1.21 – 8.18, p = 0.02) or any form of GVHD (OR = 2.52, 95% CI = 1.01 – 6.26, p = 0.04) when compared to patients with del/del genotype. As revealed by analysis using the heterozygous model, patients with A/G genotype of rs9380142 had a higher risk to develop cGVHD (OR = 3.01, 95% CI = 1.08-8.42, p = 0.03) or any form of GVHD (OR = 2.62, 95% CI = 0.98 – 7.03, p = 0.05) when compared to patients with A/A genotype.
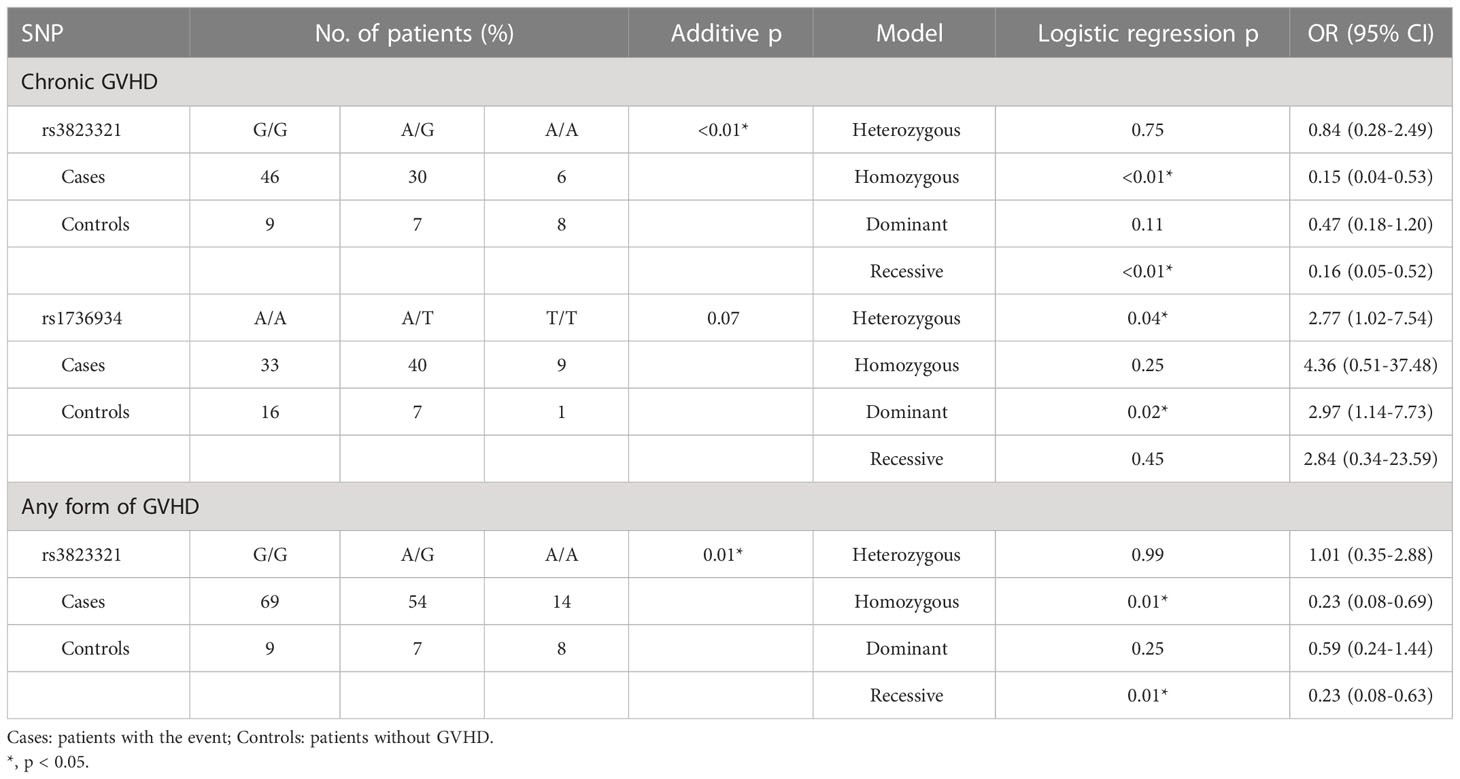
The association of 27 genetic polymorphisms in 5’-URR of HLA-G gene with post-HSCT adverse outcomes of patients were also investigated. Because of the insufficient amount of genomic DNA for PCR, the samples of three patients including one patient with ALL (the patient had mortality and chronic GVHD, but without relapse) and two patients with AML (both patients had mortality and chronic GHVD, and one had relapse) failed to be tested at 5’-URR. No association of these polymorphisms with mortality and relapse was found (Table 7). As revealed by analysis using the additive model, the genotype frequency (G/G vs. A/G vs. A/A) of rs3823321 was significantly different between cGVHD and no GVHD (p < 0.01) as well as between any forms of GVHD and no GVHD (p < 0.01). By analysis using the homozygous model, the A/A genotype of rs3823321 had 0.15 (95% CI = 0.04 - 0.53, p < 0.01) and 0.23 times (95% CI = 0.08 - 0.69, p = 0.01) lower risk for development of cGVHD and any forms of GVHD, respectively, when compared to the G/G genotype. By analysis using recessive model, the A/A genotype of rs3823321 was also associated with a lower risk for development of cGVHD (OR = 0.16, 95% CI = 0.05 – 0.52, p < 0.01) and any forms of GVHD (OR = 0.23, 95% CI = 0.08 – 0.63, p = 0.01). Analysis by heterozygous model revealed that patients with A/A genotype of rs1736934 had an increased risk of cGVHD when compared to patients with T/A genotype (A/A vs. A/T; OR = 2.77, 95% CI = 1.02 - 7.54, p = 0.04). As revealed by analysis with dominant model, patients with T/T and A/T genotypes of rs1736934 had 2.97 times higher risk for development of cGVHD (A/A vs. A/T + T/T; 95% CI =1.14-7.73, p = 0.02) when compared to patients with A/A genotype.
Association of 3’-UTR and 5’-URR haplotypes of HLA-G gene with the risk for post-HSCT adverse outcomes of patients with leukemia
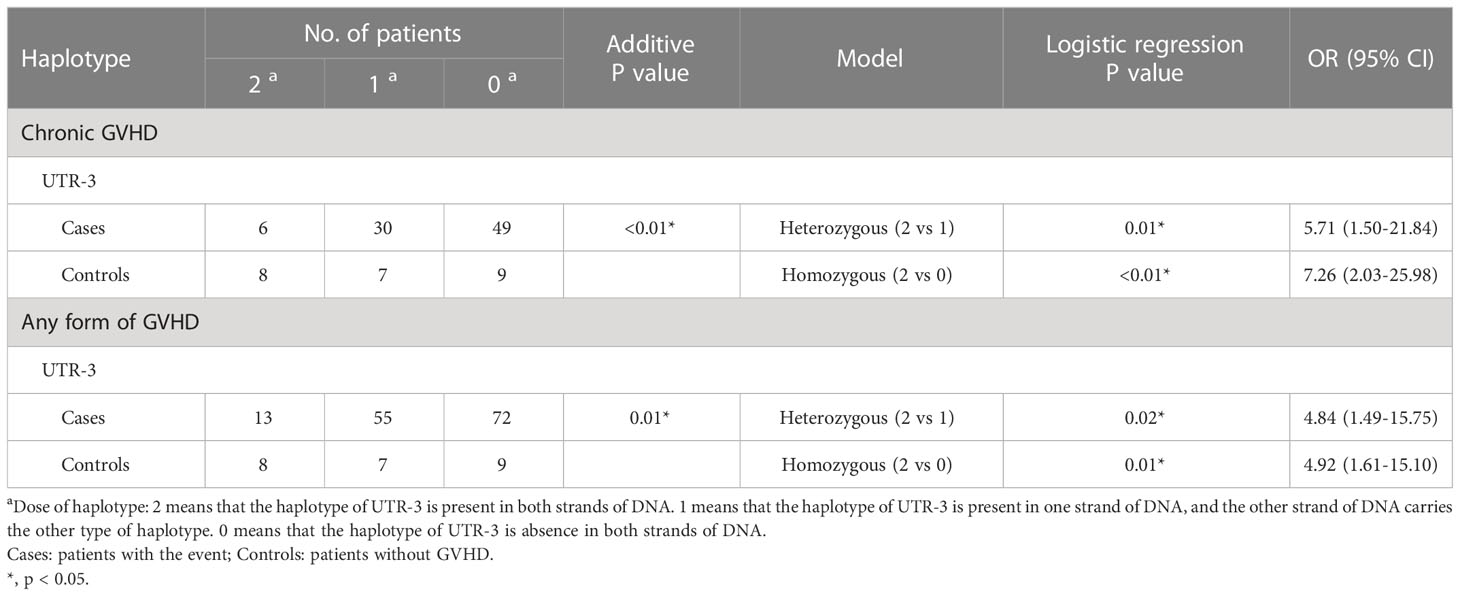
The association of haplotypes in 3’-UTR with the adverse outcomes of patients receiving HSCT was further analyzed. Six different haplotypes including UTR-1, UTR-2, UTR-3, UTR-4, UTR-5, and UTR-7 were classified according to the sequence combination of 8 SNPs (rs371194629, rs1707, rs1710, rs17179101, rs17179108, rs1063320, rs9380142, rs1610696, and rs1233331) located in 3’-UTR. Among patients under analysis, the frequency of UTR-1 (30.3%) was the highest followed by UTR-3 (25.2%) and UTR-7 (19.7%) (Table 3).
During haplotype analysis (Table 8; Supplementary Table S3), the dosage of a haplotype was defined as 2, 1, and 0 when the indicated haplotype was homozygous, heterozygous, and not present in the recipient genomic DNA, respectively. As revealed by additive analysis, the dosage of UTR-3 haplotype was reversely associated with the development of cGVHD (p < 0.01) and any forms of GVHD (p = 0.01). Patients with heterozygous UTR-3 and without UTR-3 had 5.71 times (95% CI = 1.50 - 21.84, p = 0.01), and 7.26 times higher risk (95% CI = 2.03 - 25.98, p < 0.01) for development of cGVHD when compared to patients with homozygous UTR-3, respectively. In addition, patients with heterozygous UTR-3 and without UTR-3 had 4.84 times (95% CI = 1.49 - 15.75, p = 0.02) and 4.92 times (95% CI = 1.61 - 15.10, p = 0.01) higher risk for development of any forms of GVHD when compared to patients with homozygous UTR-3, respectively.
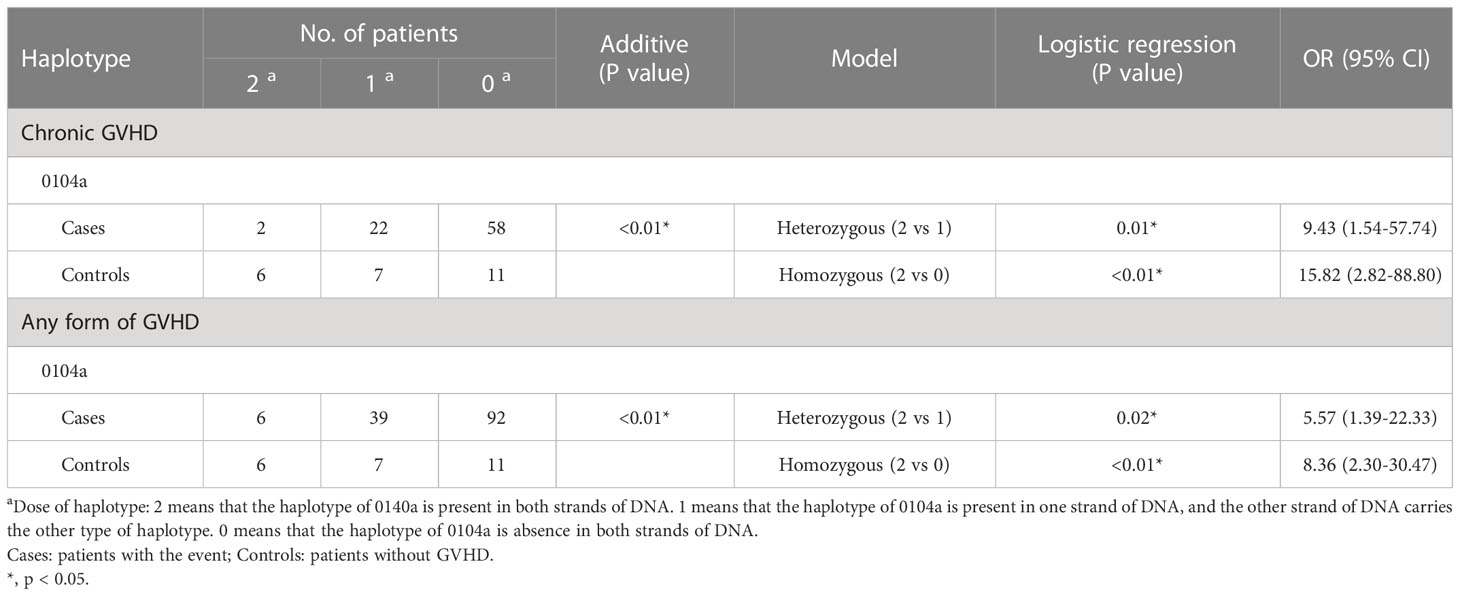
The association of haplotypes in 5’-URR with the adverse outcomes of patients receiving HSCT was also analyzed (Table 9; Supplementary Table S4). Patients enrolled in this study had 6 different haplotypes including 010101a, 010101b, 010102a, 0103a, 0104a, and 0104b. These haplotypes were classified according to the sequence combination of the 27 SNPs located in the 5’-URR including rs1736936, rs1736935, rs3823321, rs1736934, rs17875389, rs3115630, rs1632947, rs1632946, rs1233334, rs2249863, rs2735022, rs35674592, rs17875391, rs1632944, rs201221694, rs112940953, rs17875393, rs1736933, rs149890776, rs1736932, rs17875394, rs17875395, rs17875396, rs1632943, rs191630481, rs1233333, and rs17875397. Among the patients under analysis, the frequency of 010101a (30.4%) was the highest followed by 010102a (25.2%) and 0104a (21.1%).
As revealed by additive analysis, the dosage of 0104a haplotype was reversely associated with the development of cGVHD (p < 0.01) and any forms of GVHD (p < 0.01) post-HSCT. Patients with heterozygous 0104a and without 0104a had 9.43 times (95% CI = 1.54 - 57.74, p = 0.01) and 15.82 times (95% CI = 2.82 - 88.80, p < 0.01) higher risk for development of cGVHD when compared to patients with homozygous 0104a, respectively. Patients with heterozygous 0104a and without 0140a had 5.57 times (95% CI = 1.39 - 22.33, p = 0.02) and 8.36 times (95% CI = 2.30 - 30.47, p < 0.01) higher risk for development of any forms of GVHD when compared to patients with homozygous 0104a, respectively.
Discussion
Genetic polymorphisms and haplotypes of HLA-G have been reported to associate with various diseases (9). The genetic polymorphisms and haplotypes in the 3’-UTR and 5’-URR of HLA-G gene were selected for analysis because genetic variants in these regions have been shown to associate with clinical outcomes of various diseases and organ transplantation (17–21, 23). In this study, we investigated whether the SNPs and haplotypes in the 3’-UTR and 5’-URR of HLA-G are associated with the occurrence of adverse outcomes (mortality, relapse, and GVHD) for patients with AML and ALL. Our data revealed that 2 SNPs and 1 haplotype located in the 3’-UTR and 2 SNPs and 1 haplotype located in the 5’-URR of HLA-G were associated with the occurrence of cGVHD or any forms of GVHD. No SNPs was associated with mortality and relapse. This study provides new insights on the selection of donor/recipient pair when performing HSCT for therapy of patients with AML and ALL.
There are soluble and membrane-bound forms of HLA-G. Through alternative splicing, three major soluble forms of HLA-G (HLA-G5, -G6, and -G7) are expressed in the body fluid such as saliva, plasma, and cerebrospinal fluid (7). HLA-G is also expressed as the membrane-bound form in the placenta trophoblast cells, thymus, peripheral blood mononuclear cells, cornea, and mesenchymal stem cells (7). Soluble and membrane-bound HLA-G interacts with different types of receptors on immune cells and causes negative intracellular signaling leading to a decrease in the inflammatory response of the immune system. Previous study by Boukouaci et al. has demonstrated that patients with low expression of HLA-G have an increased risk of severe aGVHD after HSCT (13). The concentration of the soluble HLA-G in patients after HSCT was also related to the risk and grade of aGVHD (28, 29). Because genetic polymorphism in the untranslated or promoter region may regulate the levels of HLA-G gene expression and alter the status of immune response and the magnitude of immune tolerance, the SNPs and haplotypes locate in the 3’-UTR and 5’-URR were selected in this study to define whether these SNPs and haplotypes are associated with the mortality, the risk of relapse, and the development of GVHD for patients receiving HSCT.
According to the findings of this study, genetic polymorphisms of rs371194629 and rs9380142, and the UTR-3 haplotype located in the 3’-UTR were associated with the risk for the development of cGVHD and any forms of GVHD. These polymorphisms have been shown to involve in the regulation of HLA-G expression (30). Higher HLA-G expression usually causes suppression of immune response and leads to a better immune tolerance. Accordingly, the 14-bp INDEL fragment (5’-ATTTGTTCATGCCT-3’) of rs371194629 is associated with the splicing and the stability of the mRNA, because it contains an AUUUG domain that putatively elicits an AU-pentamer-like effect leading to a decrease in mRNA stability. Hence, the DEL allele provides a higher stability of the mRNA, associated with a high expression of HLA-G (31). The A/G allele of rs9380142 is also implicated in the stability of HLA-G mRNA. The A allele at this position modifies an AU-rich motif and decreases the stability of the corresponding mRNA, while the G allele at this position is associated with an increase in the production of HLA-G (32). Moreover, the UTR-3 haplotype had a 14-bp deletion in rs371194629, an A allele in rs9380142, and a G allele in rs1063320. The presence of a G allele in rs1063320 of the UTR-3 haplotype increases the binding of miR-148a, miR148b and miR-152 to this region and causes degradation of the primary transcript and suppression of its translation, thereby decreasing the production of HLA-G protein. In contrast, the presence of C allele at this position causes a decrease in affinity to the aforementioned miRNA, and an increase in the mRNA availability and the production of HLA-G (15).
Similar to our findings, the association of rs371194629 with the development of GVHD and other post-transplant complications have been reported (33, 34). La Nasa et al. found that in a cohort of 53 thalassemia patients transplanted from an unrelated donor, patients who are homozygous for the 14-bp deletion have a higher risk to develop aGVHD than patients who are homozygous for the 14-bp insertion (34). On the other hand, patients with A/G genotype of rs9380142 had an increased risk for occurrence of cGVHD and any forms of GVHD. Martelli-Palomino et al. demonstrated that individuals with A/G genotype of rs9380142 had higher soluble levels of HLA-G protein when compared to individuals with other genotypes, although significance was not reached (22). In haplotype analysis, we found that UTR-3 was a protective factor for GVHD development. UTR-3 had a 14-bp deletion in rs371194629 and an A allele in rs9380142. The haplotypes of 3’-UTR were associated with the level of soluble HLA-G (22). Poras et al. confirmed that 3’-UTR haplotypes alter the expression of HLA-G by luciferase reporter assay, although the effect is moderate (35). The study by Shivakumar et al. showed that the allele with 14-bp deletion in rs371194629 was associated with reduced IL-6 gene expression and the risk of schizophrenia (36). We have noted that these findings are not always consistent with the hypothesis that patients with higher HLA-G expression tend to have a better immune tolerance leading to a lower risk for the occurrence of cGVHD or any forms of GVHD. The reasons for the discrepancy are not clear. Because the incidence for the polymorphisms is different among different ethnical population and gender (37), these factors may be considered as variates in future analysis. Alternatively, the complexity of HLA system and different analytical methods among different studies may contribute to the inconsistency among the studies. It is also likely that the polymorphism in HLA-G 3’-UTR may associate with GVHD through regulating other factors, such as altering IL-6 gene expression, rather than directly altering the expression of HLA-G (36). Further study is required to investigate whether affecting the levels of HLA-G expression is the major effects of HLA-G polymorphisms on immune tolerance and the occurrence of GVHD.
We further demonstrated in this study that the genotypes of rs3823321 and rs1736934 and the G0104a haplotype located at the 5’-URR of HLA-G gene are associated with the risk for occurrence of cGVHD and development of any forms of GVHD. Patients with G/G genotype of rs3823321 is associated with higher risk for cGVHD as well as development of any forms of GVHD, while T-allele of rs1736934 is associated with higher risk for cGVHD. Notably, G0104a haplotype had the A-allele at rs3823321 and rs1736934. The roles of 5’-URR in the regulation of HLA-G gene expression and in immune tolerance and disease development are not yet well studies. Previous study showed that rs3823321 G was a binding site for SOX5 and progesterone receptor, and rs1736934 T is a binding site for POU2F1 (38). These transcription factors have been shown to play a role in regulating HLA-G expression (38). Progesterone has also been shown to enhance HLA-G expression (39, 40). With the A allele in G0104a haplotype, the binding of these transcription factors to HLA-G promoter may be impaired in patients with G0104a haplotype leading to lower HLA-G expression when compared to non-G0104a haplotypes. This may explain why patients with G0104a are associated with lower risk for development of any forms of GVHD and cGVHD. Nevertheless, the underlying mechanisms remain to be elucidated and require further investigation.
In conclusion, this study showed that six genotypes and haplotypes in 3’-UTR and 5’-URR of HLA-G gene were associated with the risk for development of any forms of GVHD and cGVHD in patients post-HSCT. The findings of this study are applicable to predict the risk of adverse outcomes post-HSCT that allow physicians to prescribe preventive plan or treatment for patients on time. Moreover, effective prevention of GVHD is depended on the induction of peripheral immune tolerance. The role of HLA-G gene polymorphism in the regulation of immune tolerance and the development of GVHD post-HSCT is worthy to investigate further.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in Supplementary Table 5.
Ethics statement
The studies involving human participants were reviewed and approved by Institutional Review Board (IRB) of Chang Gung Memorial Hospital. The patients/participants provided their written informed consent to participate in this study.
Author contributions
D-PC conceived and designed the experiments and reviewed the final draft. P-NW contributed the materials. A-LH, D-PC, and C-PT analyzed and interpreted data. D-PC, F-PH, W-TL, and C-PT wrote draft of the manuscript. W-TW performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants to D-PC from the Chang Gung Memorial Hospital (CMRPG3L0771-2), and to C-PT from the Chang Gung Memorial Hospital (CMRPD1M0071-2, CMRPD1L0281-2, CORPD1K0031, and BMRP466) and the Ministry of Science and Technology (109-2320-B-182-031-MY3). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
The excellent consulting assistance and sample resources from Molecular Diagnosis Laboratory of Chang Gung Memorial Hospital are gratefully acknowledged.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1093514/full#supplementary-material
References
1. Yee GC, McGuire TG. Allogeneic bone marrow transplantation in the treatment of hematologic diseases. Clin Pharm (1985) 4:149–60.
2. Chen DP, Chang SW, Jaing TH, Tseng CP, Chen SH, Wang WT. Effect of HLA mismatching at HLA-a, -b, and -DRB1 for umbilical cord blood transplantation in Taiwan. Clin Chim Acta (2016) 462:162–5. doi: 10.1016/j.cca.2016.09.021
3. Atsuta Y, Suzuki R, Nagamura-Inoue T, Taniguchi S, Takahashi S, Kai S, et al. Disease-specific analyses of unrelated cord blood transplantation compared with unrelated bone marrow transplantation in adult patients with acute leukemia. Blood (2009) 113:1631–8. doi: 10.1182/blood-2008-03-147041
4. Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood (2007) 110:4576–83. doi: 10.1182/blood-2007-06-097386
5. Justiz Vaillant AA, Modi P, Mohammadi O. Graft versus host disease. In: StatPearls. Treasure Island (FL: StatPearls Publishing (2022). Available at: https://pubmed.ncbi.nlm.nih.gov/30855823/.
6. Petersdorf EW, Hansen JA, Martin PJ, Woolfrey A, Malkki M, Gooley T, et al. Major-histocompatibility-complex class I alleles and antigens in hematopoietic-cell transplantation. N Engl J Med (2001) 345:1794–800. doi: 10.1056/NEJMoa011826
7. Li P, Wang N, Zhang Y, Wang C, Du L. HLA-G/sHLA-G and HLA-G-bearing extracellular vesicles in cancers: Potential role as biomarkers. Front Immunol (2021) 12:791535. doi: 10.3389/fimmu.2021.791535
8. Rouas-Freiss N, Moreau P, LeMaoult J, Papp B, Tronik-Le Roux D, Carosella ED. Role of the HLA-G immune checkpoint molecule in pregnancy. Hum Immunol (2021) 82:353–61. doi: 10.1016/j.humimm.2021.01.003
9. Arnaiz-Villena A, Juarez I, Suarez-Trujillo F, López-Nares A, Vaquero C, Palacio-Gruber J, et al. HLA-G: function, polymorphisms and pathology. Int J Immunogenet (2021) 48:172–92. doi: 10.1111/iji.12513
10. Xu HH, Lin A, Yan WH. HLA-G-mediated immunological tolerance and autoimmunity. J Transl Autoimmun (2022) 1:265–95. doi: 10.1016/B978-0-12-822564-6.00021-5
11. Liu L, Wang L, Zhao L, He C, Wang G. The role of HLA-G in tumor escape: manipulating the phenotype and function of immune cells. Front Oncol (2020) 10:597468. doi: 10.3389/fonc.2020.597468
12. Meyer D, Thomson G. How selection shapes variation of the human major histocompatibility complex: A review. Ann Hum Genet (2001) 65:1–26. doi: 10.1046/j.1469-1809.2001.6510001.x
13. Boukouaci W, Busson M, Fortier C, Amokrane K, de Latour RP, Robin M, et al. Association of HLA-G low expressor genotype with severe acute graft-versus-host disease after sibling bone marrow transplantation. Front Immunol (2011) 2:74. doi: 10.3389/fimmu.2011.00074
14. Bu X, Zhong J, Li W, Cai S, Gao Y, Ping B. Immunomodulating functions of human leukocyte antigen-G and its role in graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Ann Hematol (2021) 100:1391–400. doi: 10.1007/s00277-021-04486-z
15. Donadi EA, Castelli EC, Arnaiz-Villena A, Roger M, Rey D, Moreau P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci (2011) 68:369–95. doi: 10.1007/s00018-010-0580-7
16. Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res (2014) 2014:734068. doi: 10.1155/2014/734068
17. Krijgsman D, Roelands J, Hendrickx W, Bedognetti D, Kuppen PJK. HLA-G: a new immune checkpoint in cancer? Int J Mol Sci (2020) 21:4528. doi: 10.3390/ijms21124528
18. de Almeida BS, Muniz YCN, Prompt AH, Castelli EC, Mendes-Junior CT, Donadi EA. Genetic association between HLA-G 14-bp polymorphism and diseases: A systematic review and meta-analysis. Hum Immunol (2018) 79:724–35. doi: 10.1016/j.humimm.2018.08.003
19. Sizzano F, Testi M, Zito L, Crocchiolo R, Troiano M, Mazzi B, et al. Genotypes and haplotypes in the 3' untranslated region of the HLA-G gene and their association with clinical outcome of hematopoietic stem cell transplantation for beta-thalassemia. Tissue Antigens (2012) 79:326–32. doi: 10.1111/j.1399-0039.2012.01862.x
20. Gautam S, Kumar U, Mishra R, Dada R. HLA-G 3'UTR polymorphisms & response to a yoga-based lifestyle intervention in rheumatoid arthritis: A randomized controlled trial. Indian J Med Res (2022) 155:253–63. doi: 10.4103/ijmr.IJMR_3196_20
21. Amoriello R, Rizzo R, Mariottini A, Bortolotti D, Gentili V, Bonechi E, et al. Investigating serum sHLA-G cooperation with MRI activity and disease-modifying treatment outcome in relapsing-remitting multiple sclerosis. Front Neurol (2022) 13:872396. doi: 10.3389/fneur.2022.872396
22. Martelli-Palomino G, Pancotto JA, Muniz YC, Mendes-Junior CT, Castelli EC, Massaro JD, et al. Polymorphic sites at the 3' untranslated region of the HLA-G gene are associated with differential HLA-G soluble levels in the Brazilian and French population. PloS One (2013) 8:e71742. doi: 10.1371/journal.pone.0071742
23. Konishi A, Abe M, Yamaoka M, Satake A, Ito T, Nomura S. Analysis of HLA haplotype and clinical factors during hematopoietic stem cell transplantation. Transpl Immunol (2021) 66:101376. doi: 10.1016/j.trim.2021.101376
24. Castelli EC, Mendes-Junior CT, Veiga-Castelli LC, Roger M, Moreau P, Donadi EA. A comprehensive study of polymorphic sites along the HLA-G gene: Implication for gene regulation and evolution. Mol Biol Evol (2011) 28:3069–86. doi: 10.1093/molbev/msr138
25. Castelli EC, Ramalho J, Porto IO, Lima TH, Felício LP, Sabbagh A, et al. Insights into HLA-G genetics provided by worldwide haplotype diversity. Front Immunol (2014) 5:476. doi: 10.3389/fimmu.2014.00476
26. Chen DP, Chang SW, Wang PN, Lin WT, Hsu FP, Wang WT, et al. The association between single-nucleotide polymorphisms of co-stimulatory genes within non-HLA region and the prognosis of leukemia patients with hematopoietic stem cell transplantation. Front Immunol (2021) 12:730507. doi: 10.3389/fimmu.2021.730507
27. Chen DP, Tsao KC, Wang PN, Tseng CP, Sun CF. Quantitative analysis of chimerism after allogeneic peripheral blood stem cell transplantation. Chang Gung Med J (2002) 25:734–42.
28. Liu H, Chen Y, Xuan L, Wu X, Zhang Y, Fan Z, et al. Soluble human leukocyte antigen G molecule expression in allogeneic hematopoietic stem cell transplantation: good predictor of acute graft-versus-host disease. Acta Haematol (2013) 130:160–8. doi: 10.1159/000350488
29. Kanga U, Mourya M, Seth T, Kumar L, Mahapatra M, Mishra P, et al. Immunobiology of HLA-G in sibling related hematopoietic stem cell transplantation. Hum Immunol (2014) 75:100. doi: 10.1016/j.humimm.2014.08.135
30. Martín-Villa JM, Vaquero-Yuste C, Molina-Alejandre M, Juarez I, Suárez-Trujillo F, López-Nares A, et al. HLA-G: Too much or too little? role in cancer and autoimmune disease. Front Immunol (2022) 13:796054. doi: 10.3389/fimmu.2022.796054
31. Chen XY, Yan WH, Lin A, Xu HH, Zhang JG, Wang XX. The 14 bp deletion polymorphisms in HLA-G gene play an important role in the expression of soluble HLA-G in plasma. Tissue Antigens (2008) 72(4):335–41. doi: 10.1111/j.1399-0039.2008.01107.x
32. Yie SM, Li LH, Xiao R, Librach CL. A single base-pair mutation in the 3'-untranslated region of HLA-G mRNA is associated with pre-eclampsia. Mol Hum Reprod (2008) 14(11):649–53. doi: 10.1093/molehr/gan059
33. Le Maux A, Noël G, Birebent B, Grosset JM, Vu N, De Guibert S, et al. Soluble human leucocyte antigen-G molecules in peripheral blood haematopoietic stem cell transplantation: A specific role to prevent acute graft-versus-host disease and a link with regulatory T cells. Clin Exp Immunol (2008) 152:50–6. doi: 10.1111/j.1365-2249.2008.03598.x
34. La Nasa G, Littera R, Locatelli F, Lai S, Alba F, Caocci G, et al. The human leucocyte antigen-G 14-basepair polymorphism correlates with graft-versus-host disease in unrelated bone marrow transplantation for thalassaemia. Br J Haematol (2007) 139:284–8. doi: 10.1111/j.1365-2141.2007.06779.x
35. Poras I, Yaghi L, Martelli-Palomino G, Mendes-Junior CT, Muniz YC, Cagnin NF, et al. Haplotypes of the HLA-G 3' untranslated region respond to endogenous factors of HLA-g+ and HLA-g- cell lines differentially. PloS One (2017) 12:e0169032. doi: 10.1371/journal.pone.0169032
36. Shivakumar V, Debnath M, Venugopal D, Rajasekaran A, Kalmady SV, Subbanna M, et al. Influence of correlation between HLA-G polymorphism and interleukin-6 (IL6) gene expression on the risk of schizophrenia. Cytokine (2018) 107:59–64. doi: 10.1016/j.cyto.2017.11.016
37. Medeiros FS, Martins AES, Gomes RG, de Oliveira SAV, Welkovic S, Maruza M, et al. Variation sites at the HLA-G 3' untranslated region confer differential susceptibility to HIV/HPV Co-infection and aneuploidy in cervical cell. PloS One (2018) 13:e0204679. doi: 10.1371/journal.pone.0204679
38. Ribeyre C, Carlini F, René C, Jordier F, Picard C, Chiaroni J, et al. HLA-G haplotypes are differentially associated with asthmatic features. Front Immunol (2018) 9:278. doi: 10.3389/fimmu.2018.00278
39. Nilsson LL, Hviid TVF. HLA class ib-receptor interactions during embryo implantation and early pregnancy. Hum Reprod Update (2022) 28:435–54. doi: 10.1093/humupd/dmac007
40. Papúchová H, Saxtorph MH, Hallager T, Jepsen IE, Eriksen JO, Persson G, et al. Endometrial HLA-f expression is influenced by genotypes and correlates differently with immune cell infiltration in IVF and recurrent implantation failure patients. Hum Reprod (2022) 37:1816–34. doi: 10.1093/humrep/deac118
Keywords: hematopoietic stem cell transplantation (HSCT), single nucleotide polymorphism (SNP), human leukocyte antigen-G (HLA-G), graft versus host disease (GVHD), haplotype
Citation: Chen D-P, Wang P-N, Hour A-L, Lin W-T, Hsu F-P, Wang W-T and Tseng C-P (2023) The association between genetic variants at 3’-UTR and 5’-URR of HLA-G gene and the clinical outcomes of patients with leukemia receiving hematopoietic stem cell transplantation. Front. Immunol. 14:1093514. doi: 10.3389/fimmu.2023.1093514
Received: 09 November 2022; Accepted: 02 February 2023;
Published: 23 February 2023.
Edited by:
Leonardo M. R. Ferreira, Medical University of South Carolina, United StatesReviewed by:
Sina Naserian, Hôpital Paul Brousse (INSERM), FranceVivian Weiwen Xue, Shenzhen University, China
Copyright © 2023 Chen, Wang, Hour, Lin, Hsu, Wang and Tseng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ching-Ping Tseng, Y3RzZW5nQG1haWwuY2d1LmVkdS50dw==
 Ding-Ping Chen1,2
Ding-Ping Chen1,2 Ai-Ling Hour
Ai-Ling Hour Fang-Ping Hsu
Fang-Ping Hsu Ching-Ping Tseng
Ching-Ping Tseng