- 1Department of Veterans Affairs, Tennessee Valley Healthcare Service, Nashville, TN, United States
- 2Department of Pathology, Microbiology and Immunology, The Vanderbilt Institute for Infection, Immunology and Inflammation and Vanderbilt Center for Immunology, Vanderbilt University Medical Center, Nashville, TN, United States
- 3Division of Animal Sciences, University of Missouri, Columbia, MO, United States
The large majority of lymphocytes belong to the adaptive immune system, which are made up of B2 B cells and the αβ T cells; these are the effectors in an adaptive immune response. A multitudinous group of lymphoid lineage cells does not fit the conventional lymphocyte paradigm; it is the unconventional lymphocytes. Unconventional lymphocytes—here called innate/innate-like lymphocytes, include those that express rearranged antigen receptor genes and those that do not. Even though the innate/innate-like lymphocytes express rearranged, adaptive antigen-specific receptors, they behave like innate immune cells, which allows them to integrate sensory signals from the innate immune system and relay that umwelt to downstream innate and adaptive effector responses. Here, we review natural killer T cells and mucosal-associated invariant T cells—two prototypic innate-like T lymphocytes, which sense their local environment and relay that umwelt to downstream innate and adaptive effector cells to actuate an appropriate host response that confers immunity to infectious agents.
Introduction: ‘For a secret offence, a secret revenge’
This subtitle ‘For a secret offence, a secret revenge’ (see Box 1) exemplifies the metaphorical descriptions of fin-de-siècle—turn of the 19th century, scientific discoveries written for the benefit of the general public; this style, quite common then and in the early 20th (3, 5), remains in textbooks and lectures in pathology, microbiology, and immunology. By that time, many—Antony van Leeuwenhoek (6), Robert Hooke (7), Theodor Schwann (8), and Matthias Schleiden (9), had independently peered down the microscope, developing the ‘cell theory’—the cell as the fundamental unit of life. Now entered Rudolf Virchow (10) who espoused ‘omnis cellula e cellula’—every living cell derives from another cell, the melodic phrase coined by François-Vincent Raspail (11)—from observations of leukocythemia—leukemic cells in the blood of a 50-year-old woman and formed the cellular basis of disease (10, 12). Robert Koch and Louis Jean Pasteur independently developed the microbial basis of infectious disease (13), and Élie Metchnikoff (previously Ilya Ilyich Mechnikov) whose astute observations of cells swarming toward the splinter prick in the starfish larva and their attempts to eat it, voraciously gnawing at it—that is termed phagocytosis, birthed cellular immunology (5, 14, 15), while from the opposing and warring Paul Ehrlich school originated humoral immunity (15–17).
BOX 1 Fin-de-siècle—a turn-of-the-19th-century metaphorical description of the defense system as a warring system of the body that restores balance when tipped over by an infection.
The subtitle ‘For a secret offence, a secret revenge’ owes to the title of one of the fables in ‘Vacation Stories: Five Science Fiction Tales’ written by the 1906 Nobel Laurate Santiago Ramón y Cajal, published originally in the Spanish language assuming the alias of ‘Dr. Bacteria’. These fables were written for Cajal’s scientific friends. Famed for the ‘neuron doctrine’ and precise and beautiful drawings of the nervous system (1), Cajal is less known for his artistic and literary works because much of these cultural contributions were poorly recorded and archived. Cajal “wrote a collection of twelve fables or semi-philosophical, pseudoscientific tales that [I]” he “never dared take to press, both for the oddness of their ideas and the laxity and carelessness of their style (2).” Fortunately, the collection of five science fiction works have survived Cajal and time in ‘Vacation Stories’; the remaining seven “sleep the slumber … far deeper than the so-called sleep of slumber[.]” not as “failed artistic works” as Cajal’s Preface would make the reader to believe (2) but rather because those manuscripts were never found (3, 4).
Viewed against this historic backdrop, ‘for a secret offence, a secret revenge’ refers to the body’s elegant defense system working against agents that cause infectious diseases—the battles raged between immune cells and bacteria. The immune system is generally described as a warring system that oftentimes wins battles yet may lose a war: the morbidity and mortality caused by severe acute respiratory syndrome coronavirus 2 infection is a sorry reminder of the perils of the warring immune system. While it is a warring system indeed, it does not attack indiscriminately. The immune system has learnt over eons to coexist with billions and zillions of bacteria and other microbes in a symbiotic habit.
Amid kämpfe únd schláchten with microbes and other forms of external (irritants and allergens) and internal (mutant cells and metabolic toxicants) dangers, in complex multicellular metazoans arose a sensing-and-actuating system—the immune system. In vertebrates, the initial response to aforementioned dangers is actuated by the older innate immune system. In vertebrates, the innate immune system, which arose in early metazoan faunas—the simple invertebrates, is made entirely of the myeloid lineage of hematopoietic cells such as macrophages, dendritic cells and mast cells in tissues and by monocytes, neutrophils, basophils, and eosinophils patrolling the blood and, on demand, tissues as well. As the innate immune system responds to danger, it alerts the adaptive immune system, which kicks into full gear should the innate immune response not restore the host’s altered milieu intérieur (homeostasis) to its original state—or close to it. The adaptive system is slow in acting and is made entirely of lymphoid lineage cells. These cells sense alterations in the homeostatic state with the use of antigen-specific receptors encoded by somatically rearranged gene segments, clonally expressed by B and T lymphocytes—the B-cell receptor (BCR) and αβ T-cell receptor (TCR). Such B and T lymphocytes together constitute the conventional lymphocytes. The clonal expression of BCR and TCR requires the priming of the adaptive immune system by either immunization with antigen or natural infection for the clonal expansion of the low-frequency antigen-specific lymphocytes to clear infections and to protect against infectious diseases. This requirement for priming distinguishes the adaptive immune system from the innate, which reacts quickly, without the need for prepriming.
Circa 1973, a non-B, non-T—the ‘null’ killer lymphocyte, which could kill tumor cells without prior priming of the immune system, was discovered. Now called natural killer (NK) cells, their discovery alerted to lymphocytes that behave like the cells of the innate immune system and featured the quiet annunciation of unconventional lymphocytes (18). Next, a decade later, the start of the year 1983 unveiled with the discovery of B lymphocyte subsets: one that secreted natural antibodies (B1a) and the other that produced antibodies to bacterial polysaccharides and T lymphocyte–independent antigens (B1b) in addition to the conventional B2 B cells of the adaptive immune system (19). Then in ca. 1986 came the discovery of γδ T cells, which express the γδ TCR genes—a kin to the αβ TCR (20). The ensuing decades announced the discovery of many more unconventional lymphocytes (Figure 1): e.g., natural killer T (NKT) cells, mucosal-associated invariant T (MAIT) cells, mouse CD8αα intraepithelial T lymphocytes, mouse H-2M3-restricted T cells, mouse/human H-2Qa1/HLA-E-restricted T cells, and human group 1 CD1-restricted T cells as well as lymphoid tissue inducer cells and innate lymphoid cells [reviewed in refs (21, 22).]. This collection of unconventional T lymphocytes we here call innate/innate-like effector lymphocytes.
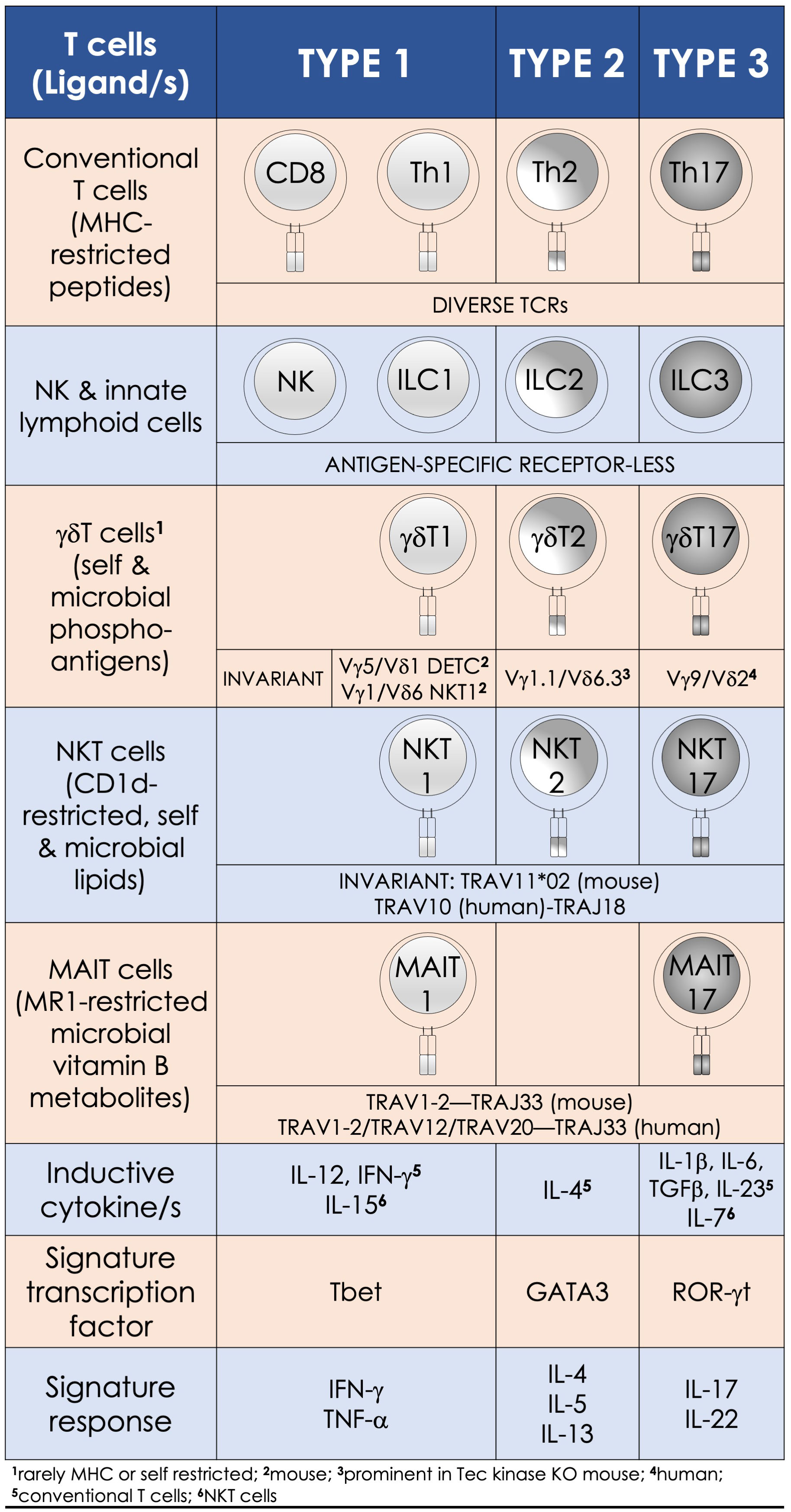
Figure 1 Innate-like effector lymphocyte functions mirror type 1, type 2, and type 3 effector cells. Natural killer T (NKT), mucosal-associated invariant T (MAIT), and γδT cells are characterized by semi-invariant T-cell receptor (TCR) expression by contrast to conventional T cells express a diverse TCR (IMGT nomenclature) repertoire. By contrast, innate lymphoid cells and NK cells do not express rearranged antigen receptors. Type 1 effectors include the cytotoxic NK and CD8+ T cells and T helper (Th) 1 cells, as well as NKT1, MAIT1, and γδT1 cells. They require IL-12 for induction, which is bolstered by IFN-γ. T-bet and the related eomesodermin transcription factors control the differentiation of type 1 effector cells, which are essential for immunity against intracellular pathogens. Type 2 effector cells include Th2, NKT2, and γδT2 cells. These cells are activated by IL-4 and require GATA3 for their effector differentiation. Their physiologic functions—e.g., parasite expulsion, and pathologic—e.g., airway hypersensitivity, are mediated by IL-4, IL-5, and IL-13 secretions. RORγt—the lineage specific transcription factor program type 3 effectors, which include Th17 and NKT17, MAIT17, and γδT17 cells. Lineage-specific inducive factors include IL-6, TGF-β, IL-1β, IL-23, and IL-7. Type 3 effector cells secrete IL-17 and IL-22 upon activation, by which they mediate tissue repair and confer immunity to extracellular bacteria and fungi.
The multitudinous innate/innate-like effector lymphocytes share several common features. In addition to being of lymphoid origin, they act quickly as they display a memory phenotype similar to antigen-experienced conventional lymphocytes yet, unalike conventional lymphocytes, retain no memories of past pathogen encounters. After development, innate/innate-like effector lymphocytes become home to secondary lymphoid and/or nonlymphoid tissues. They are stationed at barrier sites where the microbial consortia are known to congregate (19, 23, 24). As discussed below in the “Hygiene Hypothesis” section, products from these consortia facilitate the development and/or maturation of a subset of innate/innate-like effector lymphocytes (25–29). Those innate-like lymphocytes that express rearranged BCRs or αβ/γδ TCRs recognize their cognate ligands by germ-line encoded portions of the antigen-specific receptors using an innate recognition logic (30–34). Innate/innate-like lymphocytes react to self- and nonself-ligands: Some recognize H-2Qa1/HLA-E-restricted self and/or microbial peptides, H-2M3-restricted N-formylated mitochondrial/microbial peptides, group I and group II CD1-restricted lipids—e.g., αβ and γδ T cells and NKT cells, or major histocompatibility complex (MHC)–related 1 (MR1)-restricted metabolites—e.g., MAIT cells. Others recognize ligands directly without the need for MHC/non-MHC restricted presentation—e.g., intact proteins, small molecules/phosphometabolites—e.g., γδ T cells, or phospholipids—e.g., B1a cells and γδ T cells. Further, inflammatory cytokines alone—e.g., type I interferons (IFNs) or interleukin (IL)-12 and IL-18 by themselves—without the need for antigenic or agonistic ligands, can activate innate-like T lymphocytes. Innate/innate-like effector lymphocytes are quick responders; they can act as quickly as cells of innate immune system or faster [reviewed in refs (21, 22)]. This feature in several innate/innate-like effector lymphocytes is ingrained during development by a genome regulatory network under the control of a promyelocytic leukemia zinc finger transcription factor (encoded by Zbtb16; reviewed in ref (35, 36)]. Activated innate/innate-like effector lymphocytes secrete a wide variety of cytokines and chemokines with which they can steer downstream type I, II, and III immune responses (Figure 2). Thereby, they integrate sensory output/s received from the innate immune system to provide context to downstream innate and adaptive immune responses (35, 42). Here we review how NKT cells and MAIT cells—two prototypic innate-like T lymphocytes, sense their local environment and relay that umwelt to downstream innate and adaptive effector cells to actuate an appropriate response that confers protection from infectious diseases.
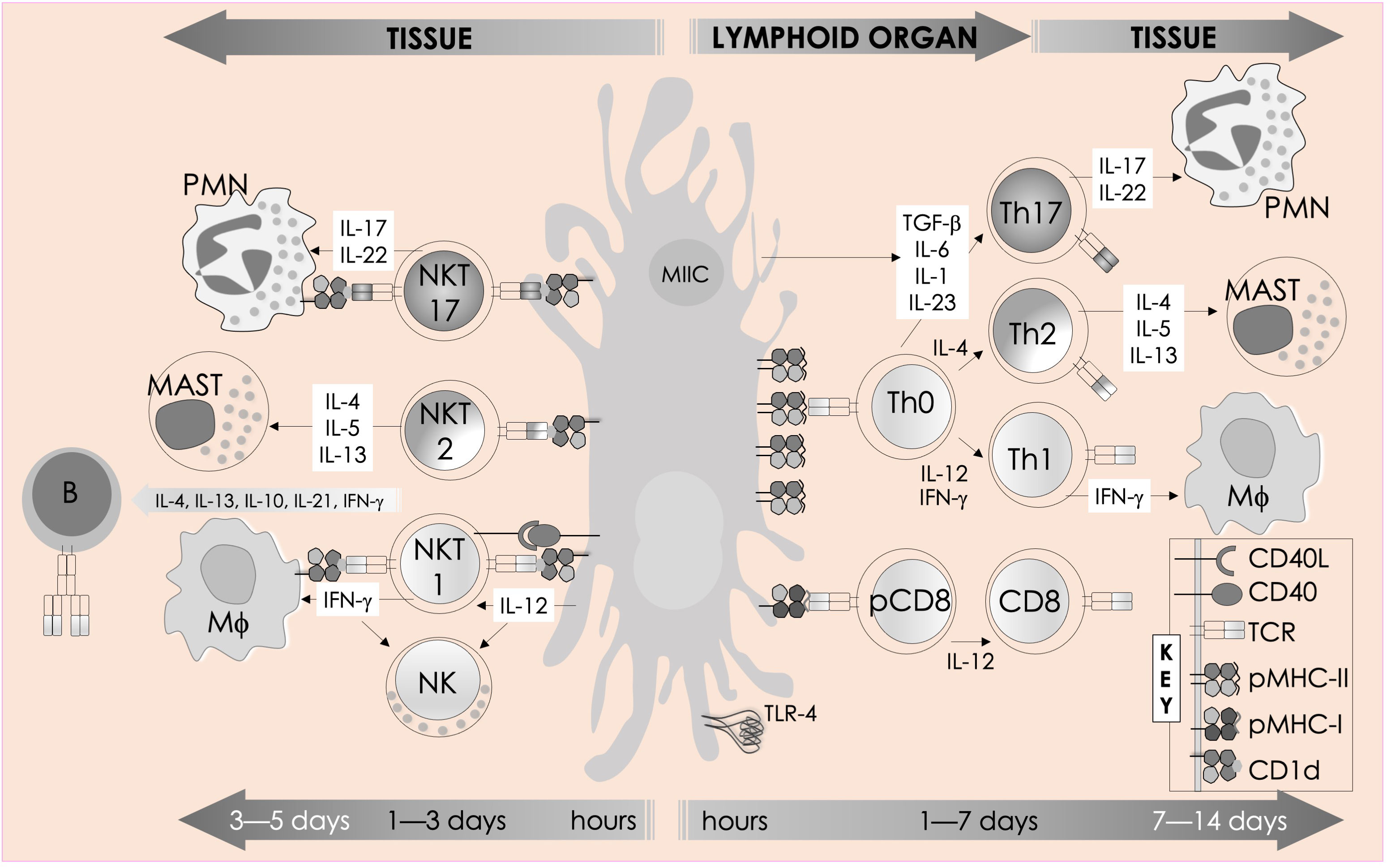
Figure 2 Immune functions of mouse NKT cells. NKT cell activation is initiated by semi-invariant NKT cell receptor interactions with cognate antigen and bolstered by costimulatory interactions between CD28 and CD40 and their cognate ligands CD80/86 (B7.1/7.2) and CD40L, respectively. The resulting activated NKT cells crosstalk with members of the innate and the adaptive immune systems by deploying cytokine and chemokine messengers. Upon activation in vivo, NKT cells rapidly secrete a variety of cytokines and chemokines, which influence the polarization of CD4+ T cells toward Th1 or Th2 cells as well as the differentiation of precursor CD8+ T cells to effector lymphocytes, and B cells to antibody-secreting plasma cells. Some of these mediators facilitate the recruitment, activation, and differentiation of macrophages and DCs, which results in the production of interleukin (IL)-12 and possibly other factors. IL-12, in turn, stimulates NK cells to secrete IFN-γ. Thus, activated NKT cells have the potential to enhance as well as temper the immune response. This schematic rendition is an adaptation of past reviews (35, 37–41) and works cited in the text.
Natural killer T and mucosal-associated invariant T cells—two peas in a pod
There are multiple types of NKT and MAIT cells that are distinguished by their αβ TCR usage and, consequently, the ligands they recognize (21). The focus in this review is on NKT and MAIT cells that express an invariant TCR α-chain: semi-invariant NKT cells begotten from the rearrangement of TRAV11*02 (mouse Vα14i) or TRAV10 (human Vα24i) to TRAJ18 and MAIT cells from TRAV1-2 (mouse and human Vα9i and human TRAV12/TRAV20) to TRAJ33 rearrangement [reviewed in refs (35, 43–45)]. A curious feature of these rearrangements is not only the conserved TRAV to TRAJ usage but also that this rearrangement results in conserved residues that make up the CDR3α (complementarity determining region 3α) loop of the TCR α-chain. Furthermore, invariant Vα14i α-chain pairs with TRBV13-2*01 (Vβ8.2), TRBV29*02 (Vβ7), or TRBV1 (Vβ2) β-chain to form a functional mouse semi-invariant NKT cell TCR. Additionally, the Vα24i α-chain pairs with the mouse TRBV13-2*01 orthologue—TRBV25-1 (Vβ11) to form a functional human semi–invariant NKT cell TCR. Akin to the semi-invariant NKT cells, MAIT cells pair with a limited set of β-chains to form a functional MAIT cell TCR. The conserved nature of the functional NKT and MAIT cell TCRs allow them to recognize their respective ligands—CD1d+lipid/s and MR1+vitamin metabolites, respectively, by means of conserved interactions—i.e., with an innate-like recognition logic (reviewed elsewhere: refs (29, 30, 34)].
In a similar vein, the pig semi-invariant NKT cells use the pTRAV10 TCR Vα gene segment, which is highly homologous to segments encoding human TRAV10, mouse TRAV11, and rat TRAV14S1—the canonical Vα segments used by the semi-invariant NKT cells in these species. The best alignments for pTRAJ18*01 were TRAJ18, TRAJ18, and TRAJ18, the Jα18 gene segments used by the human, rat, and mouse invariant α-chain, respectively. pTRBV25 is most similar to human TRBV25-1 (Vβ11), mouse TRBV13-2*01 (Vβ8.2), and rat Vβ8.2—the canonical Vβ segments used by the semi-invariant NKT cells in these species (46).
NKT cell functions are controlled by a variety of lipid agonists presented by CD1d molecules. These agonists include glycosphingolipids such as α-galactosylceramide (αGalCer) and α-glucosyldiacylglycerols and related compounds—both of host/self and microbial origins (see Table 1 and references therein). MAIT cell functions are controlled by metabolites in the riboflavin biosynthesis pathway when presented by MR1 (43, 44, 59–62). One such MAIT cell agonist is a derivative of vitamin B2 metabolite 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU), which is synthesized by both symbiotic and pathogenic bacteria (43, 44, 59, 60). Consequently, infections with bacteria- harboring mutations in the rib gene/s prevent MAIT cell activation, which in some infections can prove fatal (62).
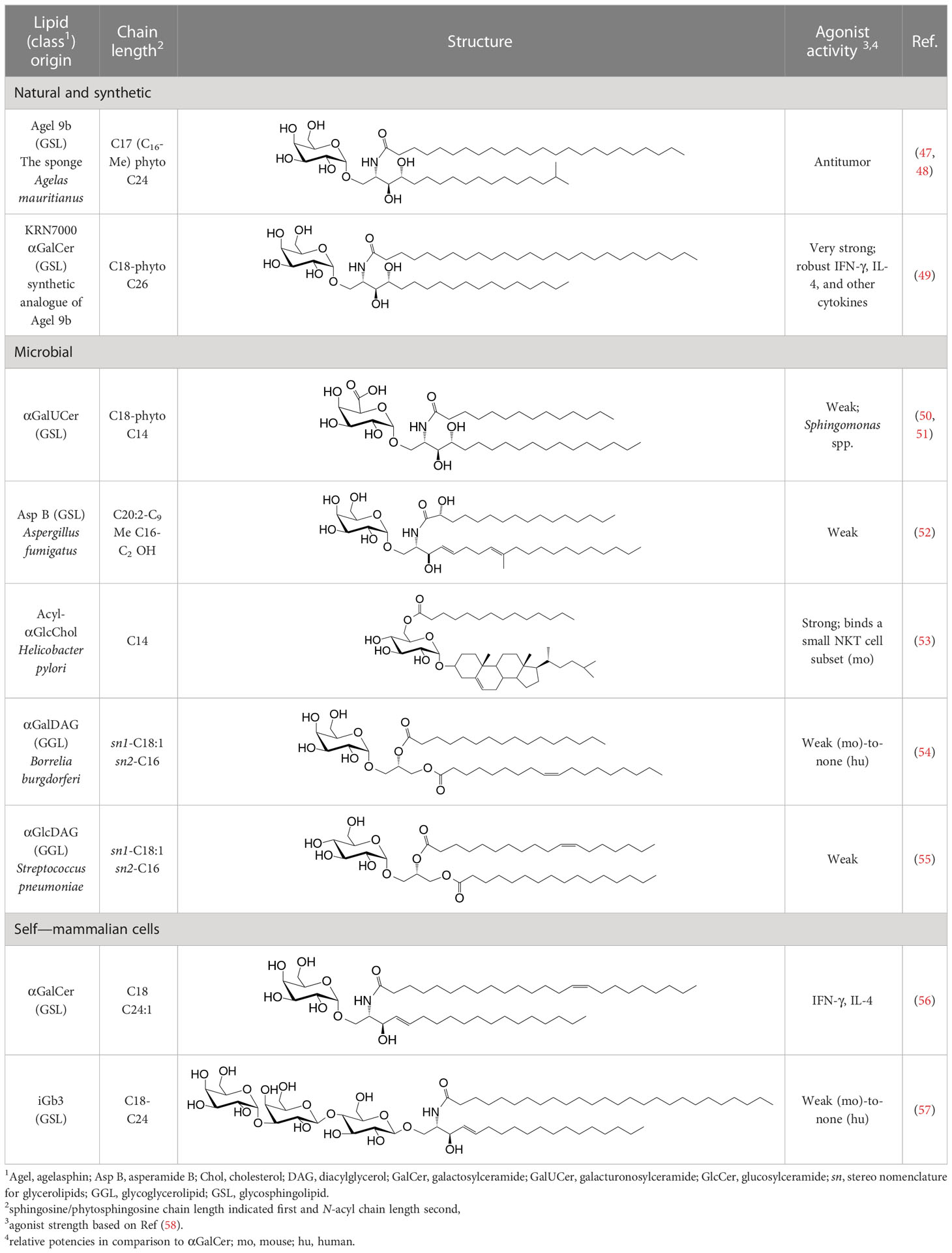
Table 1 Natural, synthetic, microbial, and self natural killer T (NKT) cell agonists: structures and properties.
By the last fin-de-siècle, the roles for NKT cells were implicated in steering immune responses to pathogens: to bacteria—Salmonella choleraesuis, Listeria monocytogenes, Mycobacterium bovis, and M. tuberculosis; to viruses—hepatitis B virus and lymphocytic choriomeningitis virus; to parasites—Plasmodium spp., Leishmania major, and Schistosoma mansonii; and to worms—Nippostrongylus brasiliensis [refs (63–80); see also Supplemental Table 1]. How NKT cells were activated by these pathogens was not understood. At that time, the only known NKT cell agonist was αGalCer (49, 81, 82). αGalCer (KRN7000) was isolated from the marine sponge—Agelas mauritianus, whose potent antitumor activity is mediated by NKT cells (47–49, 83) (see Box 2). In the ensuing two decades, much has been learnt about how NKT and MAIT cells control immune responses to infections with bacteria and viruses, many of which do not biosynthesize agonistic ligands. There are three distinct ways to activate NKT and MAIT cells (Figure 3): the first is termed TCR agonist–dependent direct activation. In this mode, the presentation of the agonist αGalCer by CD1d or 5-OP-RU by MR1 activates NKT or MAIT cells, respectively (Tables 1–3), to initiate their effector function/s (reviewed in ref (35). The second mode is termed TCR agonist–dependent and cytokine-assisted activation. Weak ligands—e.g., α-galacturonosylceramide (αGalUCer) biosynthesized by Sphingomonas spp (50, 51, 87, 111), αGalCer-like asperamide B by Aspergillus fumigatus (52), α-glycosyldiacylglycerols from Borrelia burgdorferi and Streptococcus pneumoniae (54, 55), or self αGalCer or isogloboside 3 (iGb3) induced by certain bacterial infections or sterile inflammation [ref (37, 56, 158–160); for structures, see Table 1]—that poorly activate NKT or MAIT cells require an immune push. That push is provided by inflammatory cytokines produced by the activation of DCs—e.g., IL-1β, IL-12, IL-18, or type I IFNs (96, 103, 116, 131, 155, 161). Hence, the context of infection can influence the activation of NKT and MAIT cells. The third mode of activation occurs in a manner independent of TCR stimulation but is reliant on cytokine/s alone. This mode of NKT and MAIT cell activation is termed TCR-independent inflammatory cytokine–induced activation. Bacteria that do not biosynthesize agonistic lipids but contain microbial pattern recognition receptor ligands such as lipopolysaccharide result in a TCR-independent inflammatory cytokine response from myeloid cells. These inflammatory cytokines can activate NKT cells. This mode of NKT and MAIT cell activation plays a protective role during infectious diseases, especially caused by virus infections (156, 157, 162–166).
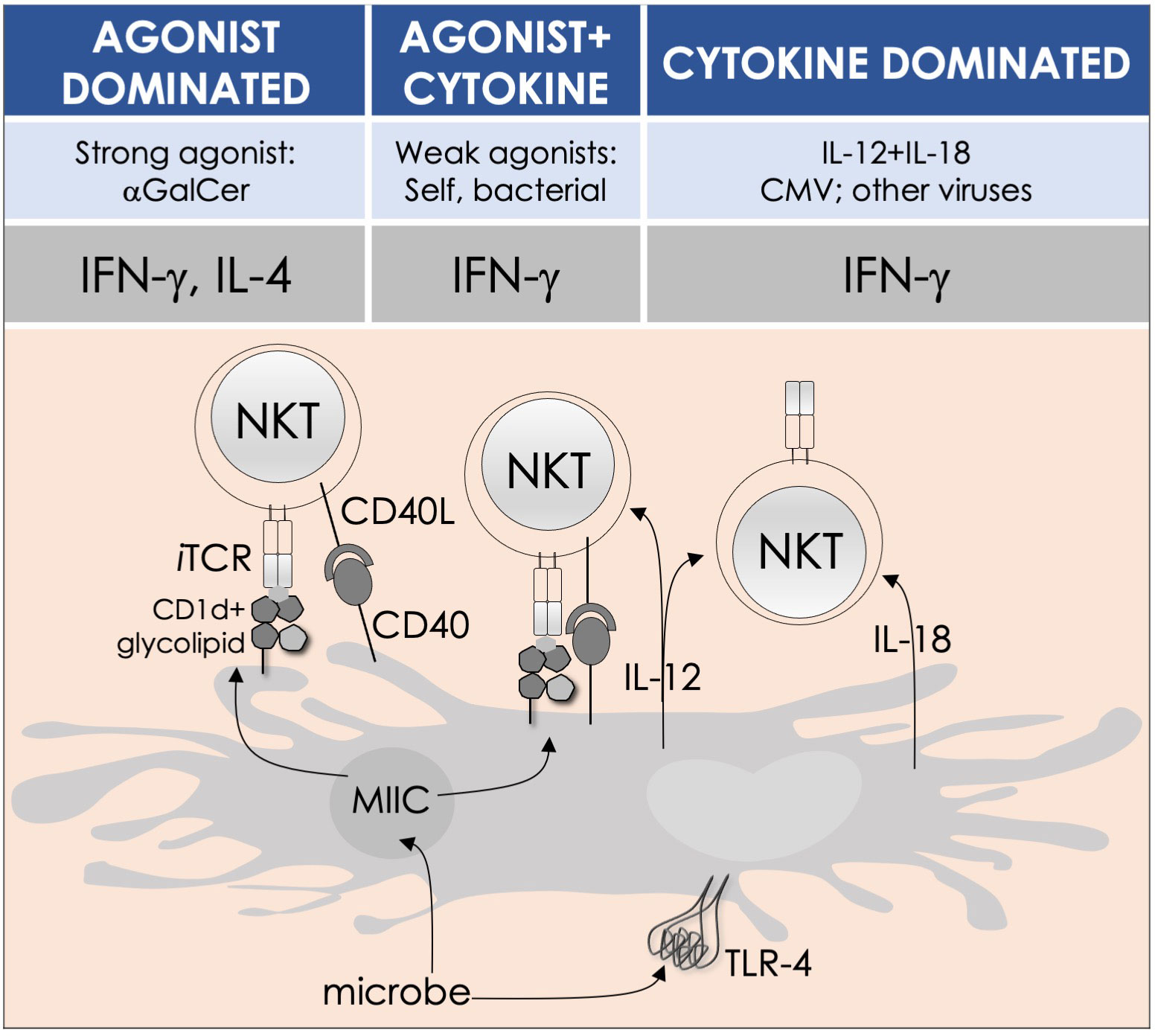
Figure 3 Modes of NKT and MAIT cell activation by microbes. Potent agonists—such as αGalCer, directly activate NKT cells, without the need for a second signal, in a TCR signaling–dominated fashion (left panel). Alternatively, microbes containing TLR ligands such as LPS activate NKT cells by inducing IL-12 production by DCs, which amplifies weak responses elicited upon the recognition of CD1d bound with self-glycolipids by the NKT cell TCR. Several endogenous lipid agonists have been identified and characterized (see Table 1). Some microbes—such as Sphingomonas capsulata and Borrelia burgdorferi—synthesize α-anomeric glycolipids for their cell walls. These glycolipids, when presented by CD1d, weakly activate NKT cells directly. In the presence of a second signal—generally a proinflammatory cytokine such as IL-12, such weak agonists strongly activate NKT cells (middle panel). By contrast, the mode of MAIT cell activation appears to be agonist concentration dependent: microbes that produce high levels of 5-OP-RU—a product of ribD-controlled catalytic activity, directly activate MAIT cells, while those that produce low levels of 5-OP-RU require a cytokine boost. Unlike conventional T cells, cytokines alone can activate both NKT and MAIT cells. Such cytokines, which include a combination of IL-12 and IL-18, activate NKT cells in a TCR-independent manner (right panel). This diagram renders the different strategies for NKT cell activation; they apply to MAIT cells as well. Similarities and differences, if any, are described in the text. Adapted from past reviews (35, 37, 38, 41) and works cited in the text.
BOX 2 A tale of α-galactosylceramides and its biosynthesis.
αGalCer/KRN7000 was first isolated from the marine sponge—Agelas mauritianus. As mammalian symbionts—e.g., Bacteroides fragilis, biosynthesize αGalCer-related compounds (26, 28, 84), it remains open whether the αGalCer was isolated from A. mauritianus or was derived from bacteria living in a symbiotic relationship with those sponges (85, 86). Bacteroidetes and α-Proteobacteria are the residents of sponges, members of which are known to biosynthesize α-anomeric glycosphingolipids that activate NKT cells (26, 28, 50, 51, 87). Of note, however, αGalCer was isolated from an Agelas-related marine sponge species—Axinella corrugata whose symbionts include α-Proteobacteria (88, 89). Nonetheless, current evidence suggests that the A. corrugata αGalCer was derived from the sponge itself and not its symbionts (88, 90). Resolving the source of αGalCer can yield insights into the biosynthesis of αGalCer in mammals (56). One possible route to the biosynthesis of αGalCer and αGlcCer might be the CGT1 (β-galactosylceramide synthase) and CGS (β-glucosylceramide synthase) themselves, which may have an α-linkage retention property. The two hexosylceramide synthases use α-linked uridyldiphosphate-charged sugar donors to form β-linked monohexosylceramides by catalyzing α to β mutarotation prior to the condensation reaction. The potential presence of αGlcCer/αGalCer in the absence of α-hexosylceramide synthase genes within mouse and human genomes poses a quandary, however (56, 91). Biochemical evidence suggests that hexosylceramide synthases may contain α-linkage retention activity, which retains the α-linkage of the charged sugar donor to generate α-linked monohexosylceramides (92–95). This α-anomer retaining activity may explain the synthesis of α-anomeric glycosphingolipids in sponges and mammals, and, potentially, in bacterial species discussed in the text that biosynthesize such lipids.
Once activated, NKT cells produce a variety of cytokines and chemokines that steer downstream innate and adaptive immune responses. This response includes type I, II, and III cytokines, which are secreted by NKT1, NKT2, and NKT17 cells, respectively. Corresponding MAIT1 and MAIT17 cells and attendant cytokine responses are similarly described. The three subsets emerge under the transcriptional activity of factors similar to those established in conventional CD4+ T cells (Figures 1, 2). Broadly, akin to conventional CD4+ T cells, NKT and MAIT cells play roles in immunity to infections and tumors and in autoimmune and allergic reactions. These features of NKT and MAIT cells are reviewed in detail elsewhere (35, 38). In addition to the three NKT cell subsets, NKT10 cells—which secrete IL-10—play regulatory functions in conjunction with T regulatory cells. NKTfh cells—which provide cognate and noncognate help to conventional B cells to secrete antibodies—may control immunity to human pathogens such as Borrelia hermsii, S. pneumoniae, and P. falciparum (167–169). These features of NKT and MAIT cells are reviewed in detail elsewhere (35),
Human NKT cell responses are as diverse as the mouse NKT cells (170). Two functional subsets were recognized that were segregated by the lack of CD4 or CD8 coreceptor expression (NKT1) or by CD4 expression (NKT2). Human NKT1 cells produce IFN-γ and TNF-α and, when activated under the influence of inflammatory cytokines, upregulate NKG2D and perforin expression priming them for cytotoxic response against infected cells and cancer cells (171, 172). Akin to mouse, the human NKT2 subset, which produces IL-4 and IL-13 and their accumulation in the lungs, may underlie the pathology in chronic asthmatic patients (173). Activated human NKT cells also produce IL-17 (170), which may reflect the existence of an NKT17 subset in humans. Further, NKT17 and MAIT17 subsets are present in higher frequency when compared to NKT1 and MAIT1 subsets in liver perfusates, which produce IL-17 and IFN-γ, respectively (174). Human NKT and MAIT subsets have some semblance to mouse NKT and MAIT subsets, but further studies are necessary to understand how similar they are in the two species.
The evolutionary origins of NKT and MAIT cell subsets have not been traced yet. Both NKT and MAIT cells arose as eutherian innovations approximately 125 million years ago in an ancestor after the therian mammals split to metatherians and eutherians—the true placental mammals (35, 175, 176). Among mammals other than the mouse and human, the development and function of NKT cells in pigs—Sus scrofa (var. domesticus)—are intensely studied. Pig NKT cell subsets were recently described using the single-cell RNA sequencing analysis of more than 11,000 differentiating thymic NKT cells (177). The vast majority of porcine NKT thymocytes resemble mouse NKT2 cells. Surprisingly, these pig NKT2-like cells do not differentiate into NKT1 or NKT17 subsets. Instead, some develop into a population enriched for interferon-stimulated genes that simultaneously maintain an NKT2-like gene profile, as well as two very rare subsets, designated iNKT-swine (sw)1 and iNKT-sw2. iNKT-sw1 and iNKT-sw2 cells are most similar to two minor populations of innate-like CD8αα T cells present in pig thymocytes, sharing the expression of FCGR3A, ZNF683, NKG7, and MHC class II–encoding genes. They also downregulate tissue emigration genes, suggesting that both are long-term thymus residents. Similar thymus-resident populations of MAIT cells, γδ T cells, and CD8αα T cells have been described before and have been speculated to modulate thymocyte differentiation to respond to peripheral perturbations, such as infection (24, 174, 178, 179). Interestingly, iNKT-sw2 cells are enriched for CD244 and CXCR6, which are upregulated on a newly discovered population of NKT cells found in mice and humans that are highly cytotoxic and protect mice from melanoma metastasis and influenza infection (180).
Although peripheral pig NKT cells can be stimulated nonspecifically to secrete IFN-γ and IL-17 (181, 182), thymus-resident pig NKT cells appear to produce little if any IFN-γ, IL-4, or IL-17 under steady-state conditions (177). One explanation for the surprisingly undifferentiated state of pig NKT thymocytes is that they emerge from the thymus in a functionally immature state and undergo further differentiation in the periphery. Since human NKT thymocytes do not also produce IFN-γ or IL-4 under steady-state conditions, it is possible that the diversity of NKT thymocyte subsets observed in mice is unusual and that it is more normal for species with the NKT-CD1d system to express fewer and/or less differentiated NKT thymocytes.
In comparison to NKT cells, relatively little is known about porcine MAIT cells. However, MAIT cell TRAV1-TRAJ33 TCRα sequences have been cloned from pig blood and tissues and found to pair with a limited number of TCR β-chains (183). It was further shown that pig MAIT cells can be CD4posCD8pos, CD4posCD8neg, and CD4negCD8pos T cells and express transcripts for the MAIT cell–associated surface molecules IL-18Rα, IL-7Rα, CCR9, CCR5, and/or CXCR6 and the transcription factors PLZF and T-bet or RORγt.
Collectively, current evidence indicate that pig NKT and MAIT cells have characteristics similar to their human and mouse counterparts. Nonetheless, several key lineage-defining differences in mouse and pig NKT cell subsets point toward the acquisition of species-specific innate/innate-like T cell adaptations, perhaps for different pathogens or may reflect the different niches in which the two species evolved and the symbiotic microbes they live with. Hence, the species-specific developmental aspects should be considered, especially in the light of ecology and evolution, when assessing the suitability of mice and pigs as biomedical models for innate/innate-like T cell research.
The hygiene hypothesis: yes, you may pick your nose and eat it
This subtitle was motivated by a burgeoning field of rhinotillexis—yes, nose picking, a new area of scientific enquiry. Beneath this otherwise aversive and socially inept and unacceptable behavior, yet innate to primates, may lie a means to the periodic reinforcement of disease tolerance [see Box 3; ref (184)].
BOX 3 Rhinotillexis—a new, burgeoning field of scientific enquiry.
It is so new and burgeoning that the National Public Radio felt compelled to interview Dr. Anne-Claire Fabre—a pioneer in the field at the Naturhistorisches Museum in Bern, Switzerland, on the matter (npr.org/2022/11/15/1136423436/researchers-dig-into-why-nose-picking-is-a-common-behavior). It is so new that the word rhinotillexis is neither in the Oxford English Dictionary nor the Merriam-Webster American English Dictionary yet but has appeared in Wikipedia, the free encyclopedia, however. Unless careful, excessive rhinotillexis may cause self-induced ethmoidectomy, especially if one suffers from rhinotillexomania (see en.wikipedia.org/wiki/Nose-picking). Rhinotillexis is not peculiar of repulsive men or their man cubs, but it is a primate thing [(184) and references therein]. Self-vaccination, per oral distribution of nasal microflora, and dental hygiene are a few proposed immunologic attributes of rhinotillexis (185–187).
The pervasive presence of microbes, flourishing at every nook and cranny of the earth and on the surfaces and the insides of metazoans, make them a formidable friend and foe. Hence, on being birthed unto a dirty world, to gain fitness, metazoans found ways to befriend and tame microbes, especially the beneficial, and ward off unfriendly ones over eons of evolution. Symbiosis emerged, lending to fitness in both directions—in the metazoan hosts and their microbial partners. So much so, symbiosis has led to the coevolution of the hosts with their microbiota, or vice versa, to the point of codependence, wherein the immune system evolved to manage the microbial consortium from going ‘wild’ and, reciprocally, the diversity of the consortium and its biosynthetic products control the immune system from going ‘rouge’. Thus, the hygiene hypothesis postulates that early life exposure to a full range of diverse microbes (and worms) promotes the development and maturation of an immune system—which reacts in a balanced measure to prevent disease whether incited by external (infections and allergens) or internal (autoinflammation) agencies of inflammation (188).
For example, under sterile, germ-free conditions, the immune system of the laboratory mouse develops and matures poorly, rendering them susceptible to infectious diseases and autoimmune disorders such as colitis (189–196). Conversely, the equilibration of the gut microbiome of the laboratory mouse to that of the ‘dirty’ pet store mouse by cohousing the two, altered, in the former, the immune cell composition at the barrier sites, resistance to infection, and T-cell differentiation in response to virus infection (197). A similar equilibration of the gut microbiome of a laboratory mouse raised under germ-free conditions by the transfer of the gut microbiota from a feral relative of the laboratory mouse and its maintenance over several generations by breeding increased disease tolerance and fitness. Inflammatory responses in such mice to a lethal influenza virus challenge was highly tempered and so was mutagen- and inflammation-induced tumorigenesis (198). All of these altered immune features acquired by the laboratory mouse reflected those of the pet store or feral mouse and those of the adult human (197, 198). The ability to approximate the human immune system in the laboratory mouse by the transfer of the microbiome indigenous of a feral mouse may facilitate and enhance preclinical vaccine development and testing (198–201). Furthermore, the role of the microbiota in the maturation of T cells may explain the intriguing finding that, at steady state—in the absence of an infection—DC emigrees from the barrier epithelium of nonlymphoid tissues stochastically prime and program resting, naïve CD8+ T cells within the local draining lymph nodes for tissue residency (202).
After development in the thymus, NKT and MAIT cells emigrate and home to lymphoid and nonlymphoid tissues, presumably to patrol and maintain the integrity of the tissue borders. The NKT and MAIT cell content at these borders varies by tissues and the mouse strain. Their tissue distribution and functions are best studied in the mouse; only a bit is known of their distribution in the human body (24, 172, 203–205). In mice, thymic NKT cell development, after commitment to this lineage and positive selection, progresses from stage 0 to stage 1 to stage 3—the mature NK1.1pos NKT cells, known to consist largely of NKT1 cells. Of these, CD24neg CCR7pos stage 1/2 NKT cells emigrate from the thymus and seed both the lymphoid and non-lymphoid tissues, where they undergo further maturation, largely driven by the local cytokine milieu (206–208), and perhaps the microbiota.
Verily, the early life exposure of NKT and MAIT cells to the host microbiota has profound, lifelong effect/s on these innate-like lymphocytes (25, 27, 161). Their development itself is dependent on positive selection by agonistic ligands—αGalCer in the case of NKT cells and 5-OP-RU in the case of MAIT cells [reviewed in refs (44, 203), and references therein]. The origins of these agonists are less clearly defined. Because CD4posCD8pos thymocytes activate Vα14i NKT cell hybridomas, it is thought that an NKT cell agonist/s may be of self origin. Thus, β-galactosylceramide synthase (CGT)-deficient thymocytes foster NKT cell development; hence, βGalCer or its derivatives are less likely the thymic NKT cell agonist. Conversely, β-glucosylceramide synthase (CGT-1)–deficient thymocytes poorly activate Vα14i NKT cell hybridomas and conditional CGT1-deficient thymocytes to not promote NKT cell development (57, 209). As βGlcCer itself does not activate Vα14i NKT cell hybridomas, a βGlcCer derivative—iGb3 or a self αGlcCer (Table 1)—is a potential NKT cell–activating self-agonist. While iGb3 synthase deficiency does not alter NKT cell development and function and no known mammalian enzyme/s synthesize α-anomeric glucosylceramide or galactosylceramide, how these agonists are biosynthesized is unclear [see Box 2 for details, see ref (35)]. Alternatively, as several gut symbionts common to many mammals biosynthesize α-anomeric glycosylceramides, their transport by lipid transfer proteins such as apolipoprotein E (210) could potentially deliver the agonist/s to the thymus. This is less likely because NKT cells develop in germ-free mice, but they are not without defects (25, 27, 211).
In a similar vein, mammalian cells do not biosynthesize vitamin B2, whose precursor is a precursor to the MAIT cell agonist 5-OP-RU (33, 59, 147, 212), but rather acquire it from symbionts (161, 203, 213). Consequently, MAIT cells develop poorly in germ-free mice bred under sterile conditions (205, 214). By contrast, NKT cells develop in such mice as noted above. It appears as though NKT cells and MAIT cells compete for niche such that, mice, which have more NKT cells than humans, have a low frequency of MAIT cells. Reciprocally, humans have a high frequency of MAIT cells but are low in NKT cell frequency (205, 214).
NKT cell numbers in the intestinal mucosa are controlled by the neonatal colonization of bacterial symbionts. NKT cells accumulate in significant numbers within the intestinal mucosa, lungs, and liver but not the thymus or spleen of germ-free mice (25, 27). The increased NKT cell number observed in germ-free mouse intestinal mucosa perhaps owes to increased levels of CXCL16—the ligand of CXCR6, the levels of which are controlled by the gut microbiota (25, 215). Moreover, NKT cells developing in germ-free mice do not mature and are hyporesponsive to the glycolipid agonist αGalCer (27). Colonization with NKT cell agonist–bearing bacteria—e.g., Sphingomonas yanoikuyae, during early life but not in adulthood restored NKT cell maturation and normoresponsiveness to αGalCer (27). Nevertheless, αGalCer compounds synthesized by different bacterial symbionts—e.g., Bacteriodes fragilis and S. yanoikuyae (see Table 1), appear to exert differential effects on developing NKT cells (26, 28, 84); why this is awaits resolution.
Early-life microbial ecology has implications for health. Thus, consistent with increased NKT cell frequency in the gut and lungs, germ-free mice are overly sensitive to oxazolone/dextran sodium sulfate–induced inflammatory colitis and airway hypersensitivity (25, 27, 215). This disease phenotype is reversed by early-life exposure to B. fragilis–derived glycosphingolipid(s) (28). Whether the normal development and functions of human NKT cells require interactions with the gut microbiota awaits discovery. So also, whether the microbiota—known to vary between individuals of different genetic, ethnic, and geographic backgrounds (216)—controls human peripheral NKT cell frequency, which varies tremendously between individuals—from undetectable to 5%—remains unknown.
Unlike the gut, which hosts swarms of thousands of microbial species, it is generally assumed that the internal organs not exposed to the outside—such as the liver, heart, and brain—are sterile, devoid of resident microbes. Counter to this assumption, a recent study found mouse and human liver hosts its own, unique microbial consortium distinct from the gut as it was enriched in Proteobacteria (217). This microbiome was seeded from the gut microbiota in a selective manner that depended on the sex of the mouse and the local environment. Moreover, the local immune response was dependent on the liver microbiome, which was influenced by Bacteroidetes species. The hepatic microbiome controlled antigen-presenting cell maturation and adaptive immunity through the mediation of NKT cells (217). Bacteroidetes species biosynthesize αGalCer (26, 28, 84), which activate NKT cells to secrete CCL5 chemokine, in turn, recruiting immune cells to the liver and their activation, expansion, and function (217). Hence, local tissue microbiomes influence local immunity in an NKT cell—dependent mechanism.
NKT cell homeostasis described above requires intestinal microbial lipid presentation by CD11c+ DCs and macrophages (218). Reciprocally, NKT cells appear to control the bacterial composition of the gut microbiota. Consequently, dysbiosis and disruption in intestinal homeostasis ensue in mice deficient in NKT cells—CD1d-/- (218–221) or Jα18-/- (222–224) mice —or mice that lack CD1d expression by DCs, which thereby are unable to present intestinal lipids to activate local (218), intestinal mucosal NKT cells. This dysbiosis and altered intestinal homeostasis are consistent with alterations in the IgA repertoire (223, 225) and the induction and function of regulatory T cells within the gut (192, 222), which are observed in these mice as well (218, 223).
By contrast to the above reports, a recent study found that there are no differences in the composition of the gut microbial consortium in CD1d−/− mice (226). Similarly, no differences were observed in the consortium in Vα14 transgenic mice, which carry high numbers of NKT cells—largely the IL-4 producing NKT2 subset (227, 228). While NKT cell activation by peroral delivery of αGalCer minimally, yet consistently, altered the diversity of the consortium, this effect was only transient. However, the shift in microbiota composition was comparable to the natural drift found in the colony. Critically, this report noted that the natural drift in the microbial composition of individual vivarium over time and, perhaps, the differences in the microbial composition between vivaria, but not NKT cells, had significant influence on the composition of the mouse gut microbial consortium even at steady state (226). Because this is a report from a single center, whether mouse and human NKT cells have an impact on the microbial consortium of the gut will require a concerted, multicenter study.
Mouse and human skin abound with MAIT cells. MAIT cell frequency varies between individuals (229). MAIT cell frequency is similar in genetically identical mice housed in the same cage but varied between those housed in distinct cages. This suggested that the microbiota may have a role in determining the frequency. Studies in germ-free mice revealed that MAIT cells depended on early-life exposure to gut microbial consortium (45, 161, 203, 213). Hence, germ-free mice failed to develop MAIT cells that localize to barrier tissues—such as the skin, when exposed to microbes later in life.
The development of mouse MAIT cells in the thymus is dependent on the presentation of a by-product of riboflavin biosynthesis—5-OP-RU (33, 44, 59, 147, 203, 212). Even though flavonoids are essential, mammalian cells are riboflavin auxotrophs. They depend on external sources of riboflavin, which is biosynthesized by several bacteria and fungi—both symbionts and pathobionts, as well as plants. The microbial origin of riboflavin and biosynthetic metabolites explains the intimate dependence of MAIT cell development on the gut microbiota. 5-OP-RU is biosynthesized in a ribD-dependent manner by the gut, and potentially the skin as well, transported to the thymus, and made available to MR1-expressing cells for assembly and display at the cell surface (213). The mechanism by which 5-OP-RU is transported to the thymus and how cells capture it to make available in the ER lumen for assembly with MR1 are poorly, if at all, understood (203).
Thymic MAIT cell emigrees home to barrier tissues. Their numbers at the barrier tissues depend on the local concentration of microbial derivatives, which is emulated by the painting of skin with varying concentrations of 5-OP-RU (161, 213). In the skin, they surveil the dermal—epidermal interface. Cutaneous-resident cells are the MAIT17 subset; their homeostasis is IL-23 dependent, and they respond to skin commensals upon MR1-ligand recognition in an IL-1- and IL-18-dependent manner. These MAIT17 cells are genetically programmed for tissue repair and, hence, contribute to normal skin physiology (161). Given the intimacies of NKT and MAIT cells with the symbiotic consortium, one might wonder what roles innate-like effector lymphocytes might have in precipitating erythema toxicum neonatorum—which is perhaps an innate immune response to skin microbiont/s that may have penetrated the newborn infant (230).
When van Leeuwenhoek peered down his microscope, curious what might live on his teeth, and perhaps his gums, little did he know he would find many ‘little animals’. In his Letter 39 to the Royal Society, he claimed,
“For my part I judge, from myself (howbeit I clean my mouth …), that all people living in our United Netherlands are not as many as the living animals that I carry in my own mouth this very day: for I noticed one of my back teeth, up against the gum, was coated with the said matter for about the width of a horse-hair, where, to all appearance, it had not been scoured by the salt for a few days; and there were such an enormous number of living animalcules here, that I imagined I could see a good number of ‘em in a quantity of this material that was no bigger than a hundredth part of a sand-grain” (from a collection of surviving van Leeuwenhoek letters, translated and compiled in ref (6). [see letter 39: Phil. Trans. XIV (231) 568, 1684)].
What those ‘little animals’ or ‘animalcules’ on man’s teeth meant remained cloaked for over two centuries. Elie Metchnikoff had a hunch to which, later in his career and life, he laid, to an obsession, much attention to prolong his life, in futility notwithstanding (232). The foregoing advances, which awaited next-generation ‘omics’ technologies and platforms, vindicates Metchnikoff’s hunch on beneficial and harmful gut microbes and lends support to the physiologic functions of early-life exposure to a diverse array of microbes—and, hence, the hygiene hypothesis.
Kämpfe únd schláchten of natural killer T and mucosal-associated invariant T cells with pathogens
NKT cells and MAIT cells perform specialized roles during infections to confer immunity to the host as they struggle (kampf) with and battle (schlacht) pathogens (see Tables 2, 3 and Supplementary Tables 1, 2). While both possess the phenotype of activated T cells, their induction differs from conventional T cells in that they can be triggered during pathogen infections through invariant receptors and cytokine signals in much the same fashion as innate cells. This results in the rapid secretion of multiple cytokines that are released with similar kinetics to innate cell-derived cytokines—i.e., minutes to hours after stimulation. Accordingly, NKT and MAIT cells can influence the behavior of cells in the innate branch of the immune response while also shaping downstream adaptive immune responses. Over the past decades, it has become clear that the innate properties of NKT and MAIT cells are shared by a wide variety of MHC class I–like restricted innate-like αβ T cells with invariant TCRs that are widespread among jawed vertebrates [reviewed in Ref (233)]. These types of lymphocytes are specialized to allow the recognition of common or particular pathogens with relatively few T cells (231). A good example is Xenopus laevis (African clawed frog) tadpoles, which are able to survive in antigen-rich waters using 15,000–20,000 T cells exhibiting limited TCR diversity (234).
As regard the role of NKT cells in immunity, mice deficient in CD1d or TRAJ18 that lack invariant NKT cells have shown that these cells play nonredundant roles in several models of infectious disease (235); NKT cell–deficient mice are more susceptible to several bacteria species (Table 2 and Supplemental Table 1), including S. pneumoniae (97, 236), Borrelia burgdorferi (113), Sphingomonas spp. (50, 51), Pseudomonas spp. (102), Chlamydia pneumoniae (109), and M. tuberculosis (73). They also exhibit greater susceptibility to fungal infections with Cryptococcus neoformans (117) and Aspergillus fumigatus (116); viral infections with herpes simplex virus (124, 237), hepatitis B virus (80, 136), and influenza A virus (131, 132, 238); and protozoan parasite infections with Plasmodium spp (76) and L. donovani (239). A wide array of microbes and microbial products can stimulate NKT cells, either by direct TCR activation, cytokine-mediated activation, or a combination of both and induce them to express activation markers and cytokines, which have diverse effects on other immune cells and the course of an infection (see Tables 1, 2 and references therein). Indeed, microbially activated NKT cells typically secrete a narrower range of cytokines than αGalCer-stimulated NKT cells, which are usually predominated by IFN-γ. This is consistent with the paradigm that the microbial activation of NKT cells is mediated, to a large extent, through innate cytokines such as IL-12 and IL-18, with weak or no TCR stimulation (240). In some infections, NKT17 cells play a significant role. NKT17 cells in a granulocyte–monocyte colony-stimulating factor (CSF2)–dependent manner plays a protective role against S. pneumoniae infection of mouse lungs (236). While Csf2-deficient NKT cells are impaired in αGalCer-induced cytokine secretion and the transactivation of downstream innate and adaptive immune responses (241), anti-CSF2 blocking experiments confirm the role of NKT17 cell–derived CSF2 in immunity against S. pneumoniae (236). Moreover, NKT cells activated by microbes do not usually undergo systemic expansion in vivo even when they contain NKT cell antigens. However, NKT cells have been found to congregate at the sites of infection in mice infected with lymphocytic choriomeningitis virus (79), malaria parasites (119), and C. neoformans (117). They have also been shown to expand in the lungs and draining lymph nodes of pigs infected with influenza and in the peripheral blood, draining lymph nodes, and lungs of pigs infected with African swine fever virus (182). An intriguing aspect of NKT cell biology is that these cells are programmed to undergo apoptosis and/or become functionally anergic after stimulation (242–244). This reduces the risk of a cytokine storm or chronic inflammation arising from the large efflux of proinflammatory cytokines that activated NKT cells produce. Usually, the degree of NKT cell deletion/dysfunction corresponds with the strength of activation, with some microbes such as the lymphocytic choriomeningitis virus capable of rendering NKT cells anergic for up to 3 months after infection (79, 245). Nevertheless, the overactivation of NKT cells does occur in some mouse models of infection, especially in tissues where NKT cells are found at high concentrations, such as the liver in mice (80, 246, 247).
Among the lessons learnt from studying NKT cells in mice is that genetic background can strongly influence the immunomodulatory activities of NKT cells. For example, the same αGalCer analog treatment protocols cause divergent effects on disease between different mouse strains in the mouse models of autoimmune diabetes (248), experimental autoimmune encephalomyelitis (249), collagen-induced arthritis (250, 251), and systemic lupus erythematosus (252). Such differing outcomes are probably related to the diverse concentrations and functional phenotypes of NKT cells that exist among inbred mouse strains. For instance, in a survey of 38 inbred mouse strains, NKT cells as a percentage of αβ T cells ranged from 3.2% to 0.01% in peripheral blood, 4.12% to 0.02% in the spleen, and 9.39% to 0.02% in the thymus (253). The proportion of CD4+ to CD4-CD8- double- negative NKT cells showed similar profound strain variation. Functional differences have been ascribed to these subsets, with the CD4+ subset exerting immunological tolerance in several disease models.
Humans present comparable levels of heterogeneity in NKT cell frequency and cytokine secretion profiles (171, 172, 254–258), which may result in distinct NKT cell responses to microbial infections that vary between individuals. However, whether NKT cells play nonredundant roles in human infectious diseases is largely unknown. Infection with the human immunodeficiency virus, dengue virus, and M. tuberculosis have been linked to reduced NKT cell responses to subsequent αGalCer stimulation (259–261). While these results suggest that at least some of the findings from mouse NKT cell studies apply to human infections, there is little evidence to indicate that humans with unusually high or low NKT cell concentrations or effector responses have altered susceptibility to microbial infections. Moreover, assessing this relationship is complicated by the fact that circulating NKT cells are often a poor reflection of NKT cells in organs and tissues (253, 254). In due course, questions about the translatability of mouse model studies may be partly addressed using CD1d knockout pigs as pig and human immune systems share many similarities, and pigs can be infected with a wide range of human pathogens (262–266).
MAIT cells are activated by microbial species that have an intact riboflavin pathway (Table 3). Accordingly, mice deficient in MAIT cells have an impaired ability to clear 5-OP-RU-producing bacteria, such as Francisella tularensis (151, 267), M. bovis bacillus Calmette-Guérin (268), M. abscesses (147), and Legionella longbeachae (144). Furthermore, TRAV1-TRAJ33 TCR-transgenic mice that express high concentrations of MAIT cells are more resistant to disease in a mouse model of M. tuberculosis infection (147). The mechanisms underlying MAIT cell antimicrobial immunity are not fully understood (see Supplemental Table 2). However, MAIT cells can lyse infected cells through perforin and granzymes (269, 270). They also secrete a variety of effector cytokines, such as IFN-γ, TNF-α, GM-CSF, and IL-17, which potentiate bacterial killing through myeloid cell activation (44, 196, 203, 205, 271, 272).
In addition to TCR-mediated activation, MAIT cells can respond to microbial infections through a variety of cytokine receptors that these cells express, including receptors for IL-1, IL-7, IL-12, IL-15, IL-18, and IL-23 (203, 271). This capacity for TCR-independent stimulation enables MAIT cells to participate in immune responses against viruses that do not produce 5-A-RU derivatives. For instance, in a mouse model of lethal influenza virus infection, MR1-deficient mice had a significantly higher mortality rate than MR1-intact mice (157). Similar results have been reported for both CD1d and TRAJ18 knockout mice demonstrating that NKT cells also play a nonredundant role in influenza virus infections (131, 132, 238). However, while NKT cells were found to be important for inhibiting virus replication, MR1-deficient mice had a similar virus load to MR1-intact mice. Moreover, TCR-dependent stimulation was found to be indispensable and dispensable for NKT cells and MAIT cells, respectively, to control influenza virus infections (132, 157). These results suggest that there exists significant overlap as well as cell type–specific differences in the antiviral activity of NKT cells and MAIT cells.
The role of MAIT cells in human antimicrobial responses remains largely uncertain. However, their high abundance in humans suggests that they may play a more prominent role in host defense and tissue homeostasis than they do in mice. MAIT cell deficiencies have not been directly associated with susceptibility to a particular pathogen in humans. Nevertheless, the frequency of MAIT cells has been found to decrease in the blood of humans infected with various types of bacteria. In some cases, this was accompanied by an increase in MAIT cell frequency at the site of infection (203, 272), suggesting that circulating MAIT cells migrate from circulation to the infection site.
In addition to their contribution to antimicrobial immunity, MAIT cells play a role in wound healing, including repairing host tissues damaged by immune cells during pathogen clearance (203, 272). Activated MAIT cells express a variety of tissue repair factors, including TGF-α, amphiregulin, vascular endothelial growth factor A, IL-5, IL-13, and IL-22 (155, 273, 274). MAIT cells in barrier tissues of the lung and skin are particularly enriched for tissue repair genes, and MAIT cell–mediated wound healing has been demonstrated in punch biopsy and Staphylococcus epidermis infection models of skin damage (161). Together, these findings indicate that MAIT cells play Janus-like opposing roles during infection, on the one hand promoting cytotoxic and proinflammatory responses that destroy infected cells while also restoring tissue integrity after the resolution of the infection.
Stymied by microbial stealth
Unsurprisingly, pathogens have devised ways to stymie CD1d-restricted antigen presentation. Most evade intracellular CD1d trafficking. For example, the modulator of immune recognition (MIR)-1 and MIR-2 proteins of Kaposi sarcoma–associated herpesvirus (KSHV) are ubiquitin ligases. The two KSHV proteins ubiquitinylate the cytoplasmic tail of human CD1d, forcing the endocytosis of surface CD1d and, thereby, reducing cell-surface CD1d expression (275). The human immunodeficiency virus 1-encoded Nef protein mirrors the effects of MIR-1 and MIR-2 proteins to reduce CD1d expression perhaps by increased endocytosis coupled with the inhibition of the return transport of CD1d to the cell surface (276, 277). Similarly, in herpes simplex virus 1 (HSV-1)–infected cells, CD1d molecules accumulate in the MHC class II–enriched compartment due to a defect in CD1d recycling from endosomal compartments back to the cell surface (278). HSV-1 also inhibits the upregulation of cell surface MR1 via the US3 gene product to evade MAIT cell recognition (279). Vaccinia virus and vesicular stomatitis virus also abrogate CD1d antigen presentation, likely by impeding the intracellular trafficking of CD1d molecules induced by mitogen-activated protein kinase signaling (280). Some bacteria have also devised strategies to evade CD1d-restricted antigen presentation. Notably, the infection of monocytes by the human pathogen M. tuberculosis results in reduced CD1d mRNA expression, indicating the transcriptional control of Cd1d expression by a mycobacterial product (281).
While pathogens evade NKT cell activation by way of interference with intracellular CD1d trafficking and, thereby, antigen presentation, pathogens induce MAIT cell dysfunction to evade MAIT cell response. To that end, patients with S. pneumoniae–induced sepsis show significantly reduced but more active and dysfunctional MAIT cell responses compared to healthy donors or paired 90-day samples (139). The hyperactive MAIT cells stir up a pathological cytokine storm thought to be responsible for mortality (141). Furthermore, the hyperactive MAIT cell response poorly induces the differentiation of inflammatory monocytes to dendritic cells during pulmonary infection (139). Similarly, studies of C. difficile pathology indicate that these bacteria potently activate MAIT cells in a combined TCR- and cytokine-dependent manner inducing a pathological cytokine storm. The resultant runaway inflammation perhaps enables C. difficile to overcome cellular barriers to potentiate C. difficile–induced antibiotic-associated colitis (138). In a similar vein, gastric H. pylori infections elicit a hyperactive MAIT cell response, promoting an increased recruitment of inflammatory immune cells to the gastric mucosa exacerbating H. pylori gastritis (145). Thus, while some pathogens evade NKT cell recognition, the effects on MAIT cells focus on inducing MAIT cell hyperactivation and dysfunction as a means of potentiating bacterial pathogenicity.
Sic parvis magna—greatness from small things come
Some 50 years ago, Ivan Riott and John Playfair and their respective groups, independently and a year or so apart, described a small subset of lymphocytes that were neither B nor T cells yet killed tumor cells without prior priming. While no small discovery in and of itself, it was a small beginning considering the numerous unconventional lymphocytes that were discovered in the ensuing decades. Unbeknownst, the discovery of NK cells had silently annunciated the existence of a grander system of cells whose constituents played critical roles in immunity to infectious diseases and cancer, as well as in precipitating autoimmune disorders and allergic reactions. Multitudinous, they are yet cluster together by several common phenotypic and functional features. Their purpose is to process and integrate signals received from the innate immune response to convey that umwelt to downstream innate and adaptive effector responses. In this manner, they appear to function in between, at the edges of the innate and adaptive immune systems. Hence, innate/innate-like effector lymphocytes are called in-betweeners—or, alternatively, Latinate edge, and a ‘limbic immune system’ arises, perchance. In this proposal for a triumvirate immune system, we do not insinuate that the ‘limbic immune system’ is an evolutionary transition between the innate and adaptive systems because the independently acting modules that make up this system arise at different times in evolution, repurposing loosely common genome regulatory circuits to accomplish a common task. The ‘limbic immune system’ functions to integrate information relayed by the innate sensory immune system about the local tissue environment and to provide context to downstream effector innate and adaptive immune responses. The multiple modules add robustness and evolvability to this limbic system to keep abreast of the ever-changing environment and the quick-evolving microbes, especially of those members of an otherwise symbiont community that turn pathobiont without much notice.
Author contributions
SJ, GO and JD wrote and edited the MS. All authors contributed to the article and approved the submitted version.
Funding
VA Research Career Scientist IK6 BX004595; VA Merit Awards: I01 BX001444 and BX001610; NIH grants: R01 AI137082, HD092286 and AI158477; and U.S. Department of Agriculture Grant: 2021-67015.
Acknowledgments
SJ is supported by Research Career Scientist (IK6 BX004595) and Merit (I01 BX001444 and BX001610) Awards from the VA, and an NIH grant (R01 AI137082). GO is supported by the Vanderbilt University Medical Scientist Training Program. JD is supported U.S. Department of Agriculture Grant 2021-67015, the National Institutes of Health Grant HD092286 and AI 158477.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1117825/full#supplementary-material
References
1. Newman E, Araque A, Dubinsky J. The beautiful brain: The drawings of Santiago ramon y cajal. New York: Abrams (2017). p. 208.
2. Otis L. Translation of Santiago Ramon y Cajal’s vacarion stories: Five science fiction tales. Urbana: University of Illinois Press (2001). p. 245.
3. Otis L. Membranes: Metaphors of invasion in ninteenth-century literature, science, and politics. Baltimore, MD: JOhns Hopkins University Press (1999). p. 204.
4. Craigie E, Cano J. Translation of Santiago Ramon y Cajal’s recollections of my life. Massachusetts: MIT Press (1989). p. 638.
5. Tauber A, Chernyak L. Metchnikoff and the origins of immunology: From metaphor to theory. New York: Oxford University Press (1991).
6. Dobell C. Antony van Leeuwenhoek and his “Little animals”. New York: Harcourt, Brace and Company (1932). p. 435.
7. Hooke R. Observ. XVIII. of the schcmatisme or texture of cork, and of the cells and pores of some other such frothy bodies. In: Brouncker P, editor. Micrographia, or some physiological descriptions of minute bodies, made by magnifying glasses, with observations and inquiries thereupon. London: The Royal Society (1665). p. 112–6.
8. Schwann T. Microscopical researches into the accordance in the structure and growth of animals and plants. Smith H, editor. London: The Sydenham Society (1843). p. 228.
9. Schleiden M. Contributions to phytogenesis. In: Schwann T, editor. Reprinted in Microscopical researches into the accordance in the structure and growth of animals and plants. London: The Sydenham Society (1843). p. 229—268.
10. Virchow R. Cellular pathology: as based upon physiological and pathological histology. 2nd ed. London: John Churchill (1860). p 511 E. t. f. G. b. F. Chance.
14. Metchnikoff E. Immunity in infective diseases. 2nd ed. Cambridge: The University Press, Cambridge (1905). p 591 Translated by F. Binnie.
15. Silverstein A. A history of immunology. Amsterdam: Academic Press—an imprint of Elsevier (2009). p. 530.
16. Mazumdar P. The template theory of antibody formation and the chemical synthesis of the twenties. In: Mazumdar P, editor. Immunology 1930–1980: Essays on the history of immunology. Toronto: Wall & Thompson (1989). p. 13—32.
18. Greenberg AH. The origins of the NK cell, or a Canadian in king ivan's court. Clin Invest Med (1994) 17:626–31.
19. Hayakawa K, Hardy RR, Parks DR, Herzenberg LA. The "Ly-1 b" cell subpopulation in normal immunodefective, and autoimmune mice. J Exp Med (1983) 157:202–18. doi: 10.1084/jem.157.1.202
20. Brenner MB, McLean J, Dialynas DP, Strominger JL, Smith JA, Owen FL, et al. Identification of a putative second T-cell receptor. Nature (1986) 322:145–9. doi: 10.1038/322145a0
21. Van Kaer L, Postoak JL, Song W, Wu L. Innate and innate-like effector lymphocytes in health and disease. J Immunol (2022) 209:199–207. doi: 10.4049/jimmunol.2200074
22. Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol (2015) 16:1114–23. doi: 10.1038/ni.3298
23. Lee YJ, Wang H, Starrett GJ, Phuong V, Jameson SC, Hogquist KA, et al. Tissue-specific distribution of iNKT cells impacts their cytokine response. Immunity (2015) 43:566–78. doi: 10.1016/j.immuni.2015.06.025
24. Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, et al. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med (2019) 216:133–51. doi: 10.1084/jem.20181483
25. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science (2012) 336:489–93. doi: 10.1126/science.1219328
26. Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M, et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PloS Biol (2013) 11:e1001610.
27. Wingender G, Stepniak D, Krebs P, Lin L, McBride S, Wei B, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology (2012) 143:418–28. doi: 10.1053/j.gastro.2012.04.017
28. An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell (2014) 156:123–33. doi: 10.1016/j.cell.2013.11.042
29. Scott-Browne JP, Matsuda JL, Mallevaey T, White J, Borg NA, McCluskey J, et al. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol (2007) 8:1105–13. doi: 10.1038/ni1510
30. Wun KS, Borg NA, Kjer-Nielsen L, Beddoe T, Koh R, Richardson SK, et al. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J Exp Med (2008) 205:939–49. doi: 10.1084/jem.20072141
31. Godfrey DI, Rossjohn J, McCluskey J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity (2008) 28:304–14. doi: 10.1016/j.immuni.2008.02.004
32. Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature (2007) 448:44–9. doi: 10.1038/nature05907
33. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin b metabolites to MAIT cells. Nature (2012) 491:717–23. doi: 10.1038/nature11605
34. Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, et al. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med (2012) 209:761–74. doi: 10.1084/jem.20112095
35. Joyce S, Okoye GD, Van Kaer L. Natural killer T lymphocytes integrate innate sensory information and relay context to effector immune responses. Crit Rev Immunol (2021) 41:55–88. doi: 10.1615/CritRevImmunol.2021040076
36. Morgan RC, Kee BL. Genomic and transcriptional mechanisms governing innate-like T lymphocyte development. J Immunol (2022) 209:208–16. doi: 10.4049/jimmunol.2200141
37. Florence WC, Bhat RK, Joyce S. CD1d-restricted glycolipid antigens: presentation principles, recognition logic and functional consequences. Expert Rev Mol Med (2008) 10:e20. doi: 10.1017/S1462399408000732
38. Kumar A, Suryadevara N, Hill TM, Bezbradica JS, Van Kaer L, Joyce S, et al. Natural killer T cells: An ecological evolutionary developmental biology perspective. Front Immunol (2017) 8:1858. doi: 10.3389/fimmu.2017.01858
39. Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol (2013) 13:101–17. doi: 10.1038/nri3369
40. Van Kaer L. α-galactosylceramide therapy for autoimmune diseases: prospects and obstacles. Nat Rev Immunol (2005) 5:31–42. doi: 10.1038/nri1531
41. Hill TM, Bezbradica JS, Van Kaer L, Joyce S. CD1d-restricted natural killer T cells. In: eLS. John Wiley & Sons, Ltd (2001). doi: 10.1002/9780470015902.a0020180.pub2
42. Joyce S. Life in-between: Bridging innate and adaptive immunity. J Immunol (2022) 209:193–5. doi: 10.4049/jimmunol.2290012
43. Mayassi T, Barreiro LB, Rossjohn J, Jabri B. A multilayered immune system through the lens of unconventional T cells. Nature (2021) 595:501–10. doi: 10.1038/s41586-021-03578-0
44. Pellicci DG, Koay HF, Berzins SP. Thymic development of unconventional T cells: how NKT cells, MAIT cells and gammadelta T cells emerge. Nat Rev Immunol (2020) 20:756–70. doi: 10.1038/s41577-020-0345-y
45. Constantinides MG, Belkaid Y. Early-life imprinting of unconventional T cells and tissue homeostasis. Science (2021) 374:eabf0095. doi: 10.1126/science.abf0095
46. Yang G, Artiaga BL, Lomelino CL, Jayaprakash AD, Sachidanandam R, McKenna R, et al. Next generation sequencing of the pig alphabeta TCR repertoire identifies the porcine invariant NKT cell receptor. J Immunol (2019) 202:1981–91. doi: 10.4049/jimmunol.1801171
47. Morita M, Motoki K, Akimoto K, Natori T, Sakai T, Sawa E, et al. Structure-activity relationship of alpha-galactosylceramides against B16-bearing mice. J Med Chem (1995) 38:2176–87. doi: 10.1021/jm00012a018
48. Natori T, Koezuka Y, Higa T. Agelasphins, novel a-galactosylceramides from the marine sponge Agelas mauritianus. Tetrahedron Lett (1993) 34:5591–2. doi: 10.1016/S0040-4039(00)73889-5
49. Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science (1997) 278:1626–9. doi: 10.1126/science.278.5343.1626
50. Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature (2005) 434:520–5. doi: 10.1038/nature03407
51. Mattner J, Debord KL, Ismail N, Goff RD, Cantu C 3rd, Zhou D, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature (2005) 434:525–9. doi: 10.1038/nature03408
52. Albacker LA, Chaudhary V, Chang YJ, Kim HY, Chuang YT, Pichavant M, et al. Invariant natural killer T cells recognize a fungal glycosphingolipid that can induce airway hyperreactivity. Nat Med (2013) 19:1297–304. doi: 10.1038/nm.3321
53. Chang YJ, Kim HY, Albacker LA, Lee HH, Baumgarth N, Akira S, et al. Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. J Clin Invest (2011) 121:57–69. doi: 10.1172/JCI44845
54. Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol (2006) 7:978–86. doi: 10.1038/ni1380
55. Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, et al. Invariant natural killer T cells recognize glycolipids from pathogenic gram-positive bacteria. Nat Immunol (2011) 12:966–74. doi: 10.1038/ni.2096
56. Kain L, Webb B, Anderson BL, Deng S, Holt M, Costanzo A, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian alpha-linked glycosylceramides. Immunity (2014) 41:543–54. doi: 10.1016/j.immuni.2014.08.017
57. Zhou D, Mattner J, Cantu C 3rd, Schrantz N, Yin N, Gao Y, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science (2004) 306:1786–9. doi: 10.1126/science.1103440
58. Joyce S, Girardi E, Zajonc DM. NKT cell ligand recognition logic: molecular basis for a synaptic duet and transmission of inflammatory effectors. J Immunol (2011) 187:1081–9. doi: 10.4049/jimmunol.1001910
59. Eckle SB, Corbett AJ, Keller AN, Chen Z, Godfrey DI, Liu L, et al. Recognition of vitamin b precursors and byproducts by mucosal associated invariant T cells. J Biol Chem (2015) 290:30204–11. doi: 10.1074/jbc.R115.685990
60. Lopez-Sagaseta J, Dulberger CL, Crooks JE, Parks CD, Luoma AM, McFedries A, et al. The molecular basis for mucosal-associated invariant T cell recognition of MR1 proteins. Proc Natl Acad Sci U.S.A. (2013) 110:E1771–1778. doi: 10.1073/pnas.1222678110
61. McWilliam HE, Eckle SB, Theodossis A, Liu L, Chen Z, Wubben JM, et al. The intracellular pathway for the presentation of vitamin b-related antigens by the antigen-presenting molecule MR1. Nat Immunol (2016) 17:531–7. doi: 10.1038/ni.3416
62. Lim HJ, Wubben JM, Garcia CP, Cruz-Gomez S, Deng J, Mak JYW, et al. A specialized tyrosine-based endocytosis signal in MR1 controls antigen presentation to MAIT cells. J Cell Biol (2022) 221:e202110125. doi: 10.1083/jcb.202110125
63. Matsuzaki G, Li XY, Kadena T, Song F, Hiromatsu K, Yoshida H, et al. Early appearance of T cell receptor alpha beta + CD4- CD8- T cells with a skewed variable region repertoire after infection with listeria monocytogenes. Eur J Immunol (1995) 25:1985–91. doi: 10.1002/eji.1830250728
64. Naiki Y, Nishimura H, Kawano T, Tanaka Y, Itohara S, Taniguchi M, et al. Regulatory role of peritoneal NK1.1+ alpha beta T cells in IL-12 production during salmonella infection. J Immunol (1999) 163:2057–63. doi: 10.4049/jimmunol.163.4.2057
65. Emoto M, Emoto Y, Kaufmann SH. Interleukin-4-producing CD4+ NK1.1+ TCR alpha/beta intermediate liver lymphocytes are down-regulated by listeria monocytogenes. Eur J Immunol (1995) 25:3321–5. doi: 10.1002/eji.1830251218
66. Emoto Y, Emoto M, Kaufmann SH. Transient control of interleukin-4-producing natural killer T cells in the livers of listeria monocytogenes-infected mice by interleukin-12. Infect Immun (1997) 65:5003–9. doi: 10.1128/iai.65.12.5003-5009.1997
67. Brown DR, Fowell DJ, Corry DB, Wynn TA, Moskowitz NH, Cheever AW, et al. Beta 2-microglobulin-dependent NK1.1+ T cells are not essential for T helper cell 2 immune responses. J Exp Med (1996) 184:1295–304. doi: 10.1084/jem.184.4.1295
68. Emoto M, Emoto Y, Buchwalow IB, Kaufmann SH. Induction of IFN-gamma-producing CD4+ natural killer T cells by mycobacterium bovis bacillus calmette guerin. Eur J Immunol (1999) 29:650–9. doi: 10.1002/(SICI)1521-4141(199902)29:02<650::AID-IMMU650>3.0.CO;2-M
69. Denkers EY, Gazzinelli RT, Martin D, Sher A. Emergence of NK1.1+ cells as effectors of IFN-gamma dependent immunity to toxoplasma gondii in MHC class I-deficient mice. J Exp Med (1993) 178:1465–72. doi: 10.1084/jem.178.5.1465
70. Denkers EY, Scharton-Kersten T, Barbieri S, Caspar P, Sher A. A role for CD4+ NK1.1+ T lymphocytes as major histocompatibility complex class II independent helper cells in the generation of CD8+ effector function against intracellular infection. J Exp Med (1996) 184:131–9. doi: 10.1084/jem.184.1.131
71. Scharton-Kersten T, Afonso LC, Wysocka M, Trinchieri G, Scott P. IL-12 is required for natural killer cell activation and subsequent T helper 1 cell development in experimental leishmaniasis. J Immunol (1995) 154:5320–30. doi: 10.4049/jimmunol.154.10.5320
72. Behar SM, Dascher CC, Grusby MJ, Wang CR, Brenner MB. Susceptibility of mice deficient in CD1D or TAP1 to infection with mycobacterium tuberculosis. J Exp Med (1999) 189:1973–80. doi: 10.1084/jem.189.12.1973
73. Sousa AO, Mazzaccaro RJ, Russell RG, Lee FK, Turner OC, Hong S, et al. Relative contributions of distinct MHC class I-dependent cell populations in protection to tuberculosis infection in mice. Proc Natl Acad Sci U.S.A. (2000) 97:4204–8.
74. Apostolou I, Takahama Y, Belmant C, Kawano T, Huerre M, Marchal G, et al. Murine natural killer T(NKT) cells [correction of natural killer cells] contribute to the granulomatous reaction caused by mycobacterial cell walls. Proc Natl Acad Sci U.S.A. (1999) 96:5141–6.
75. Pied S, Roland J, Louise A, Voegtle D, Soulard V, Mazier D, et al. Liver CD4-CD8- NK1.1+ TCR alpha beta intermediate cells increase during experimental malaria infection and are able to exhibit inhibitory activity against the parasite liver stage in vitro. J Immunol (2000) 164:1463–9. doi: 10.4049/jimmunol.164.3.1463
76. Gonzalez-Aseguinolaza G, de Oliveira C, Tomaska M, Hong S, Bruna-Romero O, Nakayama T, et al. Alpha -galactosylceramide-activated valpha 14 natural killer T cells mediate protection against murine malaria. Proc Natl Acad Sci U.S.A. (2000) 97:8461–6.
77. Schofield L, McConville MJ, Hansen D, Campbell AS, Fraser-Reid B, Grusby MJ, et al. CD1d-restricted immunoglobulin G formation to GPI-anchored antigens mediated by NKT cells. Science (1999) 283:225–9. doi: 10.1126/science.283.5399.225
78. Joyce S. CD1d and natural T cells: how their properties jump-start the immune system. Cell Mol Life Sci (2001) 58:442–69. doi: 10.1007/PL00000869
79. Hobbs JA, Cho S, Roberts TJ, Sriram V, Zhang J, Xu M, et al. Selective loss of natural killer T cells by apoptosis following infection with lymphocytic choriomeningitis virus. J Virol (2001) 75:10746–54. doi: 10.1128/JVI.75.22.10746-10754.2001
80. Kakimi K, Guidotti LG, Koezuka Y, Chisari FV. Natural killer T cell activation inhibits hepatitis b virus replication in vivo. J Exp Med (2000) 192:921–30. doi: 10.1084/jem.192.7.921
81. Brossay L, Naidenko O, Burdin N, Matsuda J, Sakai T, Kronenberg M, et al. Structural requirements for galactosylceramide recognition by CD1-restricted NK T cells. J Immunol (1998) 161:5124–8. doi: 10.4049/jimmunol.161.10.5124
82. Burdin N, Brossay L, Koezuka Y, Smiley S T, Grusby M J, Gui M, et al. Selective ability of mouse CD1 to present glycolipids: alpha-galactosylceramide specifically stimulates V alpha 14+ NK T lymphocytes. J Immunol (1998) 161:3271–81. doi: 10.4049/jimmunol.161.7.3271
83. Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, et al. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science (1997) 278:1623–6. doi: 10.1126/science.278.5343.1623
84. Oh SF, Jung DJ, Choi E. Gut microbiota-derived unconventional T cell ligands: Contribution to host immune modulation. Immunohorizons (2022) 6:476–87. doi: 10.4049/immunohorizons.2200006
85. Enticknap JJ, Kelly M, Peraud O, Hill RT. Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl Environ Microbiol (2006) 72:3724–32. doi: 10.1128/AEM.72.5.3724-3732.2006
86. Li Z, He L, Miao X. Cultivable bacterial community from south China Sea sponge as revealed by DGGE fingerprinting and 16S rDNA phylogenetic analysis. Curr Microbiol (2007) 55:465–72. doi: 10.1007/s00284-007-9035-2
87. Long X, Deng S, Mattner J, Zang Z, Zhou D, McNary N, et al. Synthesis and evaluation of stimulatory properties of sphingomonadaceae glycolipids. Nat Chem Biol (2007) 3:559–64. doi: 10.1038/nchembio.2007.19
88. Costantino V, Costantino V, Fattorusso E, Imperatore C, Mangoni A, Freigang S, et al. Corrugoside, a new immunostimulatory alpha-galactoglycosphingolipid from the marine sponge axinella corrugata. Bioorg Med Chem (2008) 16:2077–85. doi: 10.1016/j.bmc.2007.10.098
89. White JR, White JR, Patel J, Ottesen A, Arce G, Blackwelder P, et al. Pyrosequencing of bacterial symbionts within axinella corrugata sponges: diversity and seasonal variability. PloS One (2012) 7:e38204. doi: 10.1371/journal.pone.0038204
90. Laroche M, Imperatore C, Grozdanov L, Costantino V, Mangoni A, Hentschel U, et al. Cellular localisation of secondary metabolites isolated from the Caribbean sponge plakortis simplex. Mar Biol (2007) 151:1365–73. doi: 10.1007/s00227-006-0572-1
91. Brennan PJ, Cheng TY, Pellicci DG, Watts GFM, Veerapen N, Young DC, et al. Activation of iNKT cells by a distinct constituent of the endogenous glucosylceramide fraction. Proc Natl Acad Sci U.S.A. (2014) 111:13433–8.
92. Lairson LL, Chiu CPC, Ly HD, He S, Wakarchuk WW, Strynadka NCJ, et al. Intermediate trapping on a mutant retaining alpha-galactosyltransferase identifies an unexpected aspartate residue. J Biol Chem (2004) 279:28339–44. doi: 10.1074/jbc.M400451200
93. Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem (2008) 77:521–55. doi: 10.1146/annurev.biochem.76.061005.092322
94. Lee SS, Hong SY, Errey JC, Izumi A, Davies GJ, Davis BG. Mechanistic evidence for a front-side, SNi-type reaction in a retaining glycosyltransferase. Nat Chem Biol (2011) 7:631–8. doi: 10.1038/nchembio.628
95. Soya N, Fang Y, Palcic MM, Klassen JS. Trapping and characterization of covalent intermediates of mutant retaining glycosyltransferases. Glycobiology (2011) 21:547–52. doi: 10.1093/glycob/cwq190
96. Brigl M, Tatituri R V, Watts G F, Bhowruth V, Leadbetter E A, Barton N, et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med (2011) 208:1163–77. doi: 10.1084/jem.20102555
97. Kawakami K, Yamamoto N, Kinjo Y, Miyagi K, Nakasone C, Uezu K, et al. Critical role of Valpha14+ natural killer T cells in the innate phase of host protection against streptococcus pneumoniae infection. Eur J Immunol (2003) 33:3322–30. doi: 10.1002/eji.200324254
98. Kwiecinski J, Rhost S, Löfbom L, Blomqvist M, Månsson JE, Cardell SL, et al. Sulfatide attenuates experimental staphylococcus aureus sepsis through a CD1d-dependent pathway. Infection Immun (2013) 81:1114–20. doi: 10.1128/IAI.01334-12
99. Kim S, Lalani S, Parekh VV, Vincent TL, Wu L, Van Kaer L. Impact of bacteria on the phenotype, functions, and therapeutic activities of invariant NKT cells in mice. J Clin Invest (2008) 118:2301–15. doi: 10.1172/JCI33071
100. Arrunategui-Correa V, Lenz L, Kim HS. CD1d-independent regulation of NKT cell migration and cytokine production upon listeria monocytogenes infection. Cell Immunol (2004) 232:38–48. doi: 10.1016/j.cellimm.2005.01.009
101. Emoto M, Yoshizawa I, Emoto Y, Miamoto M, Hurwitz R, Kaufmann SH, et al. Rapid development of a gamma interferon-secreting glycolipid/CD1d-specific Valpha14+ NK1.1- T-cell subset after bacterial infection. Infect Immun (2006) 74:5903–13. doi: 10.1128/IAI.00311-06
102. Nieuwenhuis EE, Matsumoto T, Exley M, Schleipman RA, Glickman J, Bailey DT, et al. CD1d-dependent macrophage-mediated clearance of pseudomonas aeruginosa from lung. Nat Med (2002) 8:588–93. doi: 10.1038/nm0602-588
103. Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol (2003) 4:1230–7. doi: 10.1038/ni1002
104. Berntman E, Rolf J, Johansson C, Anderson P, Cardell SL. The role of CD1d-restricted NK T lymphocytes in the immune response to oral infection with salmonella typhimurium. Eur J Immunol (2005) 35:2100–9. doi: 10.1002/eji.200425846
105. Ito Y, Vela JL, Matsumura F, Hoshino H, Tyznik A, Lee H, et al. Helicobacter pylori cholesteryl alpha-glucosides contribute to its pathogenicity and immune response by natural killer T cells. PloS One (2013) 8:e78191.
106. Bilenki L, Wang S, Yang J, Fan Y, Joyee AG, Yang X, et al. NK T cell activation promotes chlamydia trachomatis infection in vivo. J Immunol (2005) 175:3197–206. doi: 10.4049/jimmunol.175.5.3197
107. Habbeddine M, Verbeke P, Delarbre C, Moutier R, Prieto S, Ojcius DM, et al. CD1d-restricted NKT cells modulate placental and uterine leukocyte populations during chlamydial infection in mice. Microbes Infection (2013) 15:928–38. doi: 10.1016/j.micinf.2013.08.006
108. Peng Y, Zhao L, Shekhar S, Liu L, Wang H, Chen Q, et al. The glycolipid exoantigen derived from chlamydia muridarum activates invariant natural killer T cells. Cell Mol Immunol (2012) 9:361–6. doi: 10.1038/cmi.2012.19
109. Joyee AG, Qiu H, Wang S, Fan Y, Bilenki L, Yang X, et al. Distinct NKT cell subsets are induced by different chlamydia species leading to differential adaptive immunity and host resistance to the infections. J Immunol (2007) 178:1048–58. doi: 10.4049/jimmunol.178.2.1048
110. Hill TM, Gilchuk P, Cicek BB, Osina MA, Boyd KL, Durrant DM, et al. Border patrol gone awry: Lung NKT cell activation by francisella tularensis exacerbates tularemia-like disease. PloS Pathog (2015) 11:e1004975. doi: 10.1371/journal.ppat.1004975
111. Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol (2005) 35:1692–701. doi: 10.1002/eji.200526157
112. Mattner J, Savage PB, Leung P, Oertelt SS, Wang V, Trivedi O, et al. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe (2008) 3:304–15. doi: 10.1016/j.chom.2008.03.009
113. Kumar H, Belperron A, Barthold SW, Bockenstedt LK. CD1d deficiency impairs murine host defense against the spirochete, borrelia burgdorferi. J Immunol (Cutting Edge) (2000) 165:4797–801. doi: 10.4049/jimmunol.165.9.4797
114. Tupin E, Benhnia MR, Kinjo Y, Patsey R, Lena CJ, Haller MC, et al. NKT cells prevent chronic joint inflammation after infection with borrelia burgdorferi. Proc Natl Acad Sci U.S.A. (2008) 105:19863–8.
115. Sada-Ovalle I, Chiba A, Gonzales A, Brenner MB, Behar SM. Innate invariant NKT cells recognize mycobacterium tuberculosis-infected macrophages, produce interferon-gamma, and kill intracellular bacteria. PloS Pathog (2008) 4:e1000239.
116. Cohen NR, Tatituri RV, Rivera A, Watts GF, Kim EY, Chiba A, et al. Innate recognition of cell wall beta-glucans drives invariant natural killer T cell responses against fungi. Cell Host Microbe (2011) 10:437–50. doi: 10.1016/j.chom.2011.09.011
117. Kawakami K, Wang X, Shofuda T, Sumimoto H, Tupesis J, Fitzgerald E, et al. Monocyte chemoattractant protein-1-dependent increase of V alpha 14 NKT cells in lungs and their roles in Th1 response and host defense in cryptococcal infection. J Immunol (2001) 167:6525–32. doi: 10.4049/jimmunol.167.11.6525
118. Hansen DS, Siomos MA, Buckingham L, Scalzo AA, Schofield L. Regulation of murine cerebral malaria pathogenesis by CD1d-restricted NKT cells and the natural killer complex. Immunity (2003) 18:391–402. doi: 10.1016/S1074-7613(03)00052-9
119. Soulard V, Roland J, Sellier C, Gruner AC, Leite-de-Moraes M, Franetich JF, et al. Primary infection of C57BL/6 mice with plasmodium yoelii induces a heterogeneous response of NKT cells. Infect Immun (2007) 75:2511–22. doi: 10.1128/IAI.01818-06
120. Roberts TJ, Lin Y, Spence PM, Van Kaer L, Brutkiewicz RR. CD1d1-dependent control of the magnitude of an acute antiviral immune response. J Immunol (2004) 172:3454–61. doi: 10.4049/jimmunol.172.6.3454
121. Spence PM, Sriram V, Van Kaer L, Hobbs JA, Brutkiewicz RR. Generation of cellular immunity to lymphocytic choriomeningitis virus is independent of CD1d1 expression. Immunology (2001) 104:168–74. doi: 10.1046/j.1365-2567.2001.01302.x
122. Amprey JL, Im JS, Turco SJ, Murray HW, Illarionov PA, Besra GS, et al. A subset of liver NK T cells is activated during leishmania donovani infection by CD1d-bound lipophosphoglycan. J Exp Med (2004) 200:895–904. doi: 10.1084/jem.20040704
123. Lotter H, Gonzalez-Roldan N, Lindner B, Winau F, Isibasi A, Moreno-Lafont M, et al. Natural killer T cells activated by a lipopeptidophosphoglycan from entamoeba histolytica are critically important to control amebic liver abscess. PloS Pathog (2009) 5:e1000434. doi: 10.1371/journal.ppat.1000434
124. Grubor-Bauk B, Simmons A, Mayrhofer G, Speck PG. Impaired clearance of herpes simplex virus type 1 from mice lacking CD1d or NKT cells expressing the semivariant V alpha 14-J alpha 281 TCR. J Immunol (2003) 170:1430–4. doi: 10.4049/jimmunol.170.3.1430
125. Cornish AL, Keating R, Kyparissoudis K, Smyth MJ, Carbone FR, Godfrey DI, et al. NKT cells are not critical for HSV-1 disease resolution. Immunol Cell Biol (2006) 84:13–9. doi: 10.1111/j.1440-1711.2005.01396.x
126. Rao P, Wen X, Lo JH, Kim S, Li X, Chen S, et al. Herpes simplex virus 1 specifically targets human CD1d antigen presentation to enhance its pathogenicity. J Virol (2018) 92:e01490-18. doi: 10.1128/JVI.01490-18
127. Ashkar AA, Rosenthal KL. Interleukin-15 and natural killer and NKT cells play a critical role in innate protection against genital herpes simplex virus type 2 infection. J Virol (2003) 77:10168–71. doi: 10.1128/JVI.77.18.10168-10171.2003
128. Kim EY, Battaile JT, Patel AC, You Y, Agapov E, Grayson MH, et al. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nat Med (2008) 14:633–40. doi: 10.1038/nm1770
129. Johnson TR, Hong S, Van Kaer L, Koezuka Y, Graham BS. NK T cells contribute to expansion of CD8(+) T cells and amplification of antiviral immune responses to respiratory syncytial virus. J Virol (2002) 76:4294–303. doi: 10.1128/JVI.76.9.4294-4303.2002
130. Rey-Jurado E, Bohmwald K, Galvez NMS, Becerra D, Porcelli SA, Carreno LJ, et al. Contribution of NKT cells to the immune response and pathogenesis triggered by respiratory viruses. Virulence (2020) 11:580–93. doi: 10.1080/21505594.2020.1770492
131. Paget C, Ivanov S, Fontaine J, Blanc F, Pichavant M, Renneson J, et al. Potential role of invariant NKT cells in the control of pulmonary inflammation and CD8+ T cell response during acute influenza a virus H3N2 pneumonia. J Immunol (2011) 186:5590–602. doi: 10.4049/jimmunol.1002348
132. De Santo C, Salio M, Masri SH, Lee LY, Dong T, Speak AO, et al. Invariant NKT cells reduce the immunosuppressive activity of influenza a virus-induced myeloid-derived suppressor cells in mice and humans. J Clin Invest (2008) 118:4036–48. doi: 10.1172/JCI36264
133. Paget C, Ivanov S, Fontaine J, Renneson J, Blanc F, Pichavant M, et al. Interleukin-22 is produced by invariant natural killer T lymphocytes during influenza a virus infection: potential role in protection against lung epithelial damages. J Biol Chem (2012) 287:8816–29. doi: 10.1074/jbc.M111.304758
134. Ho LP, Denney L, Luhn K, Teoh D, Clelland C, McMichael AJ, et al. Activation of invariant NKT cells enhances the innate immune response and improves the disease course in influenza a virus infection. Eur J Immunol (2008) 38:1913–22. doi: 10.1002/eji.200738017
135. Kok WL, Denney L, Benam K, Cole S, Clelland C, McMichael AJ, et al. Pivotal advance: Invariant NKT cells reduce accumulation of inflammatory monocytes in the lungs and decrease immune-pathology during severe influenza a virus infection. J Leukoc Biol (2012) 91:357–68. doi: 10.1189/jlb.0411184
136. Zeissig S, Murata K, Sweet L, Publicover J, Hu Z, Kaser A, et al. Hepatitis b virus-induced lipid alterations contribute to natural killer T cell-dependent protective immunity. Nat Med (2012) 18:1060–8. doi: 10.1038/nm.2811
137. Bedel R, Matsuda JL, Brigl M, White J, Kappler J, Marrack P, et al. Lower TCR repertoire diversity in Traj18-deficient mice. Nat Immunol (2012) 13:705–6. doi: 10.1038/ni.2347
138. Marquardt I, Jakob J, Scheibel J, Hofmann JD, Klawonn F, Neumann-Schaal M, et al. Clostridioides difficile toxin CDT induces cytotoxic responses in human mucosal-associated invariant T (MAIT) cells. Front Microbiol (2021) 12:752549. doi: 10.3389/fmicb.2021.752549
139. Shaler CR, Choi J, Rudak PT, Memarnejadian A, Szabo PA, Tun-Abraham ME, et al. MAIT cells launch a rapid, robust and distinct hyperinflammatory response to bacterial superantigens and quickly acquire an anergic phenotype that impedes their cognate antimicrobial function: Defining a novel mechanism of superantigen-induced immunopathology and immunosuppression. PloS Biol (2017) 15:e2001930. doi: 10.1371/journal.pbio.2001930
140. Jochems SP, de Ruiter K, Solorzano C, Voskamp A, Mitsi E, Nikolaou E, et al. Innate and adaptive nasal mucosal immune responses following experimental human pneumococcal colonization. J Clin Invest (2019) 129:4523–38. doi: 10.1172/JCI128865
141. Emgard J, Bergsten H, McCormick JK, Barrantes I, Skrede S, Sandberg JK, et al. MAIT cells are major contributors to the cytokine response in group a streptococcal toxic shock syndrome. Proc Natl Acad Sci U.S.A. (2019) 116:25923–31.
142. Georgel P, Radosavljevic M, Macquin C, Bahram S. The non-conventional MHC class I MR1 molecule controls infection by klebsiella pneumoniae in mice. Mol Immunol (2011) 48:769–75. doi: 10.1016/j.molimm.2010.12.002
143. Smith DJ, Hill GR, Bell SC, Reid DW. Reduced mucosal associated invariant T-cells are associated with increased disease severity and pseudomonas aeruginosa infection in cystic fibrosis. PloS One (2014) 9:e109891. doi: 10.1371/journal.pone.0109891
144. Wang H, D'Souza C, Lim XY, Kostenko L, Pediongco TJ, Eckle SBG, et al. MAIT cells protect against pulmonary legionella longbeachae infection. Nat Commun (2018) 9:3350. doi: 10.1038/s41467-018-05202-8
145. D'Souza C, Pediongco T, Wang H, Scheerlinck JY, Kostenko L, Esterbauer R, et al. Mucosal-associated invariant T cells augment immunopathology and gastritis in chronic helicobacter pylori infection. J Immunol (2018) 200:1901–16. doi: 10.4049/jimmunol.1701512
146. Booth JS, Salerno-Goncalves R, Blanchard TG, Patil SA, Kader HA, Safta AM, et al. Mucosal-associated invariant T cells in the human gastric mucosa and blood: Role in helicobacter pylori infection. Front Immunol (2015) 6:466. doi: 10.3389/fimmu.2015.00466
147. Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol (2010) 11:701–8. doi: 10.1038/ni.1890
148. Salerno-Goncalves R, Rezwan T, Luo D, Tettelin H, Sztein MB. B cells control mucosal-associated invariant T cell responses to salmonella enterica serovar typhi infection through the CD85j HLA-G receptor. Front Immunol (2021) 12:728685. doi: 10.3389/fimmu.2021.728685
149. Howson LJ, Napolitani G, Shepherd D, Ghadbane H, Kurupati P, Preciado-Llanes L, et al. MAIT cell clonal expansion and TCR repertoire shaping in human volunteers challenged with salmonella paratyphi a. Nat Commun (2018) 9:253. doi: 10.1038/s41467-017-02540-x
150. Zhao Z, Wang H, Shi M, Zhu T, Pediongco T, Lim XY, et al. Francisella tularensis induces Th1 like MAIT cells conferring protection against systemic and local infection. Nat Commun (2021) 12:4355. doi: 10.1038/s41467-021-24570-2
151. Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci USA (2013) 110:E3119–3128. doi: 10.1073/pnas.1302799110
152. Dey RJ, Dey B, Harriff M, Canfield ET, Lewinsohn DM, Bishai WR, et al. Augmentation of the riboflavin-biosynthetic pathway enhances mucosa-associated invariant T (MAIT) cell activation and diminishes mycobacterium tuberculosis virulence. mBio (2022) 13:e0386521. doi: 10.1128/mbio.03865-21
153. Paquin-Proulx D, Avelino-Silva VI, Santos BAN, Silveira Barsotti N, Siroma F, Fernandes Ramos J, et al. MAIT cells are activated in acute dengue virus infection and after in vitro zika virus infection. PloS Negl Trop Dis (2018) 12:e0006154. doi: 10.1371/journal.pntd.0006154
154. Phetsouphanh C, Phalora P, Hackstein CP, Thornhill J, Munier CML, Meyerowitz J, et al. Human MAIT cells respond to and suppress HIV-1. Elife (2021) 10:e50324. doi: 10.7554/eLife.50324
155. Lamichhane R, Galvin H, Hannaway RF, de la Harpe SM, Munro F, Tyndall JD, et al. Type I interferons are important co-stimulatory signals during T cell receptor mediated human MAIT cell activation. Eur J Immunol (2020) 50:178–91. doi: 10.1002/eji.201948279
156. Loh L, Wang Z, Sant S, Koutsakos M, Jegaskanda S, Corbett AJ, et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18-dependent activation. Proc Natl Acad Sci U.S.A. (2016) 113:10133–8.
157. van Wilgenburg B, Loh L, Chen Z, Pediongco TJ, Wang H, Shi M, et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat Commun (2018) 9:4706. doi: 10.1038/s41467-018-07207-9
158. Darmoise A, Teneberg S, Bouzonville L, Brady RO, Beck M, Kaufmann SH, et al. Lysosomal alpha-galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity (2010) 33:216–28. doi: 10.1016/j.immuni.2010.08.003
159. Bedard M, Shrestha D, Priestman DA, Wang Y, Schneider F, Matute JD, et al. Sterile activation of invariant natural killer T cells by ER-stressed antigen-presenting cells. Proc Natl Acad Sci U.S.A. (2019) 116:23671–81.
160. Govindarajan S, Verheugen E, Venken K, Gaublomme D, Maelegheer M, Cloots E, et al. ER stress in antigen-presenting cells promotes NKT cell activation through endogenous neutral lipids. EMBO Rep (2020) 21:e48927. doi: 10.15252/embr.201948927
161. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science (2019) 366. doi: 10.1126/science.aax6624
162. Nagarajan NA, Kronenberg M. Invariant NKT cells amplify the innate immune response to lipopolysaccharide. J Immunol (2007) 178:2706–13. doi: 10.4049/jimmunol.178.5.2706
163. Tyznik AJ, Tupin E, Nagarajan NA, Her MJ, Benedict CA, Kronenberg M, et al. The mechanism of invariant NKT cell responses to viral danger signals. J Immunol (Cutting Edge) (2008) 181:4452–6. doi: 10.4049/jimmunol.181.7.4452
164. Wesley JD, Tessmer MS, Chaukos D, Brossay L. NK cell-like behavior of Valpha14i NK T cells during MCMV infection. PloS Pathog (2008) 4:e1000106.
165. van Wilgenburg B, Scherwitzl I, Hutchinson EC, Leng T, Kurioka A, Kulicke C, et al. MAIT cells are activated during human viral infections. Nat Commun (2016) 7:11653. doi: 10.1038/ncomms11653
166. Lamichhane R, Munro F, Harrop TWR, de la Harpe SM, Dearden PK, Vernall AJ, et al. Human liver-derived MAIT cells differ from blood MAIT cells in their metabolism and response to TCR-independent activation. Eur J Immunol (2021) 51:879–92. doi: 10.1002/eji.202048830
167. Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A, et al. Identification of bcl-6-dependent follicular helper NKT cells that provide cognate help for b cell responses. Nat Immunol (2011) 13:35–43. doi: 10.1038/ni.2166
168. King IL, Fortier A, Tighe M, Dibble J, Watts GF, Veerapen N, et al. Invariant natural killer T cells direct b cell responses to cognate lipid antigen in an IL-21-dependent manner. Nat Immunol (2012) 13:44–50. doi: 10.1038/ni.2172
169. Tonti E, Fedeli M, Napolitano A, Iannacone M, von Andrian UH, Guidotti LG, et al. Follicular helper NKT cells induce limited b cell responses and germinal center formation in the absence of CD4(+) T cell help. J Immunol (2012) 188:3217–22. doi: 10.4049/jimmunol.1103501
170. Chan AC, Leeansyah E, Cochrane A, d'Udekem d'Acoz Y, Mittag D, Harrison LC, et al. Ex-vivo analysis of human natural killer T cells demonstrates heterogeneity between tissues and within established CD4(+) and CD4(-) subsets. Clin Exp Immunol (2013) 172:129–37. doi: 10.1111/cei.12045
171. Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med (2002) 195:625–36. doi: 10.1084/jem.20011786
172. Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human V(alpha)24 natural killer T cells. J Exp Med (2002) 195:637–41. doi: 10.1084/jem.20011908
173. Akbari O, Faul JL, Hoyte EG, Berry GJ, Wahlström J, Kronenberg M, et al. CD4+ invariant T-Cell–receptor+ natural killer T cells in bronchial asthma. New Engl J Med (2006) 354:1117–29. doi: 10.1056/NEJMoa053614
174. Lee M, Lee E, Han SK, Choi YH, Kwon DI, Choi H, et al. Single-cell RNA sequencing identifies shared differentiation paths of mouse thymic innate T cells. Nat Commun (2020) 11:4367. doi: 10.1038/s41467-020-18155-8
175. Boudinot P, Mondot S, Jouneau L, Teyton L, Lefranc MP, Lantz O. Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc Natl Acad Sci U.S.A. (2016) 113:E2983–2992. doi: 10.1073/pnas.1600674113
176. Harly C, Robert J, Legoux F, Lantz O. Gammadelta T, NKT, and MAIT cells during evolution: Redundancy or specialized functions? J Immunol (2022) 209:217–25. doi: 10.4049/jimmunol.2200105
177. Gu W, Madrid DMC, Joyce S, Driver JP. A single-cell analysis of thymopoiesis and thymic iNKT cell development in pigs. Cell Rep (2022) 40:111050. doi: 10.1016/j.celrep.2022.111050
178. Fan X, Rudensky AY. Hallmarks of tissue-resident lymphocytes. Cell (2016) 164:1198–211. doi: 10.1016/j.cell.2016.02.048
179. Verstichel G, Vermijlen D, Martens L, Goetgeluk G, Brouwer M, Thiault N, et al. The checkpoint for agonist selection precedes conventional selection in human thymus. Sci Immunol (2017) 2:eaah4232. doi: 10.1126/sciimmunol.aah4232
180. Cui G, Shimba A, Jin J, Ogawa T, Muramoto Y, Miyachi H, et al. A circulating subset of iNKT cells mediates antitumor and antiviral immunity. Sci Immunol (2022) 7:eabj8760. doi: 10.1126/sciimmunol.abj8760
181. Thierry A, Robin A, Giraud S, Minouflet S, Barra A, Bridoux F, et al. Identification of invariant natural killer T cells in porcine peripheral blood. Vet Immunol Immunopathol (2012) 149:272–9. doi: 10.1016/j.vetimm.2012.06.023
182. Schafer A, Huhr J, Schwaiger T, Dorhoi A, Mettenleiter TC, Blome S, et al. Porcine invariant natural killer T cells: Functional profiling and dynamics in steady state and viral infections. Front Immunol (2019) 10:1380. doi: 10.3389/fimmu.2019.01380
183. Xiao X, Li K, Ma X, Liu B, He X, Yang S, et al. Mucosal-associated invariant T cells expressing the TRAV1-TRAJ33 chain are present in pigs. Front Immunol (2019) 10:2070. doi: 10.3389/fimmu.2019.02070
184. Fabre A-C, Portela Miguez R, Wall CE, Peckre, Ehmke LR E, Boistel R. A review of nose picking in primates with new evidence of its occurrence in daubentonia madagascariensis. J Zool (2023) 319:91–8. doi: 10.1111/jzo.13034
185. MacClancy J, Henry C, Macbeth H. Consuming the inedible: Neglected dimensions of food choice. New York: Berghahn (2009). p. 258.
186. Wertheim HF, van Kleef M, Vos MC, Ott A, Verbrugh HA, Fokkens W. Nose picking and nasal carriage of staphylococcus aureus. Infect Control Hosp Epidemiol (2006) 27:863–7. doi: 10.1086/506401
187. Frenkel ES, Ribbeck K. Salivary mucins protect surfaces from colonization by cariogenic bacteria. Appl Environ Microbiol (2015) 81:332–8. doi: 10.1128/AEM.02573-14
188. Strachan DP. Hay fever, hygiene, and household size. BMJ (1989) 299:1259–60. doi: 10.1136/bmj.299.6710.1259
189. Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe (2008) 4:337–49. doi: 10.1016/j.chom.2008.09.009
190. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
191. Ivanov II, Littman DR. Modulation of immune homeostasis by commensal bacteria. Curr Opin Microbiol (2011) 14:106–14. doi: 10.1016/j.mib.2010.12.003
192. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (2012) 336:1268–73. doi: 10.1126/science.1223490
193. Hand TW, Vujkovic-Cvijin I, Ridaura VK, Belkaid Y. Linking the microbiota, chronic disease, and the immune system. Trends Endocrinol Metab (2016) 27:831–43. doi: 10.1016/j.tem.2016.08.003
194. Mao K, Baptista AP, Tamoutounour S, Zhuang L, Bouladoux N, Martins AJ, et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature (2018) 554:255–9. doi: 10.1038/nature25437
195. Fitzpatrick Z, Frazer G, Ferro A, Clare S, Bouladoux N, Ferdinand J, et al. Gut-educated IgA plasma cells defend the meningeal venous sinuses. Nature (2020) 587:472–6. doi: 10.1038/s41586-020-2886-4
196. Ansaldo E, Farley TK, Belkaid Y. Control of immunity by the microbiota. Annu Rev Immunol (2021) 39:449–79. doi: 10.1146/annurev-immunol-093019-112348
197. Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature (2016) 532:512–6. doi: 10.1038/nature17655
198. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell (2017) 171:1015–1028.e1013. doi: 10.1016/j.cell.2017.09.016
199. Reese TA, Bi K, Kambal A, Filali-Mouhim A, Beura LK, Burger MC, et al. Sequential infection with common pathogens promotes human-like immune gene expression and altered vaccine response. Cell Host Microbe (2016) 19:713–9. doi: 10.1016/j.chom.2016.04.003
200. Fiege JK, Block KE, Pierson MJ, Nanda H, Shepherd FK, Mickelson CK, et al. Mice with diverse microbial exposure histories as a model for preclinical vaccine testing. Cell Host Microbe (2021) 29:1815–1827 e1816. doi: 10.1016/j.chom.2021.10.001
201. Fay EJ, Balla KM, Roach SN, Shepherd FK, Putri DS, Wiggen TD, et al. Natural rodent model of viral transmission reveals biological features of virus population dynamics. J Exp Med (2022) 219:e20211220. doi: 10.1084/jem.20211220
202. Mani V, Bromley SK, Aijo T, Mora-Buch R, Carrizosa E, Warner RD, et al. Migratory DCs activate TGF-beta to precondition naive CD8(+) T cells for tissue-resident memory fate. Science (2019) 366:eaav5728. doi: 10.1126/science.aav5728
203. Legoux F, Salou M, Lantz O, Cell Development MAIT. And functions: the microbial connection. Immunity (2020) 53:710–23. doi: 10.1016/j.immuni.2020.09.009
204. Crosby CM, Kronenberg M. Tissue-specific functions of invariant natural killer T cells. Nat Rev Immunol (2018) 18:559–74. doi: 10.1038/s41577-018-0034-2
205. Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol (2019) 20:1110–28. doi: 10.1038/s41590-019-0444-8
206. Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol (2010) 11:197–206. doi: 10.1038/ni.1841
207. Wang H, Hogquist KA. CCR7 defines a precursor for murine iNKT cells in thymus and periphery. Elife (2018) 7:e34793. doi: 10.7554/eLife.34793
208. Bortoluzzi S, Dashtsoodol N, Engleitner T, Drees C, Helmrath S, Mir J, et al. Brief homogeneous TCR signals instruct common iNKT progenitors whose effector diversification is characterized by subsequent cytokine signaling. Immunity (2021) 54:2497–2513 e2499. doi: 10.1016/j.immuni.2021.09.003
209. Stanic AK, De Silva AD, Park JJ, Sriram V, Ichikawa S, Hirabyashi Y, et al. Defective presentation of the CD1d1-restricted natural Va14Ja18 NKT lymphocyte antigen caused by beta-d-glucosylceramide synthase deficiency. Proc Natl Acad Sci U.S.A. (2003) 100:1849–54.
210. Elzen Pvd, Garg S, León L, Brigl M, Leadbetter EA, Gumperz JE, et al. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature (2005) 437:906–10. doi: 10.1038/nature04001
211. Park SH, Benlagha K, Lee D, Balish E, Bendelac A. Unaltered phenotype, tissue distribution and function of Valpha14(+) NKT cells in germ-free mice. Eur J Immunol (2000) 30:620–5. doi: 10.1002/1521-4141(200002)30:2<620::AID-IMMU620>3.0.CO;2-4
212. Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PloS Biol (2010) 8:e1000407. doi: 10.1371/journal.pbio.1000407
213. Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science (2019) 366:494–9. doi: 10.1126/science.aaw2719
214. Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol (2016) 17:1300–11. doi: 10.1038/ni.3565
215. Zeissig S, Blumberg RS. Commensal microbiota and NKT cells in the control of inflammatory diseases at mucosal surfaces. Curr Opin Immunol (2013) 25:690–6. doi: 10.1016/j.coi.2013.09.012
216. Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486:207–14. doi: 10.1038/nature11234
217. Leinwand JC, Paul B, Chen R, Xu F, Sierra MA, Paluru MM, et al. Intrahepatic microbes govern liver immunity by programming NKT cells. J Clin Invest (2022) 132:e151725. doi: 10.1172/JCI151725
218. Saez de Guinoa J, Jimeno R, Gaya M, Kipling D, Garzon MJ, Dunn-Walters D, et al. CD1d-mediated lipid presentation by CD11c(+) cells regulates intestinal homeostasis. EMBO J (2018) 37:e97537. doi: 10.15252/embj.201797537
219. Nieuwenhuis EE, Matsumoto T, Lindenbergh D, Willemsen R, Kaser A, Simons-Oosterhuis Y, et al. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J Clin Invest (2009) 119:1241–50. doi: 10.1172/JCI36509
220. Selvanantham T, Lin Q, Guo CX, Surendra A, Fieve S, Escalante NK, et al. NKT cell-deficient mice harbor an altered microbiota that fuels intestinal inflammation during chemically induced colitis. J Immunol (2016) 197:4464–72. doi: 10.4049/jimmunol.1601410
221. Maricic I, Marrero I, Eguchi A, Nakamura R, Johnson CD, Dasgupta S, et al. Differential activation of hepatic invariant NKT cell subsets plays a key role in progression of nonalcoholic steatohepatitis. J Immunol (2018) 201:3017–35. doi: 10.4049/jimmunol.1800614
222. Pillai AB, George TI, Dutt S, Strober S. Host natural killer T cells induce an interleukin-4-dependent expansion of donor CD4+CD25+Foxp3+ T regulatory cells that protects against graft-versus-host disease. Blood (2009) 113:4458–67. doi: 10.1182/blood-2008-06-165506
223. de Aguiar CF, Castoldi A, Amano MT, Ignacio A, Terra FF, Cruz M, et al. Fecal IgA levels and gut microbiota composition are regulated by invariant natural killer T cells. Inflammation Bowel Dis (2020) 26:697–708. doi: 10.1093/ibd/izz300
224. Shen S, Prame Kumar K, Stanley D, Moore RJ, Van TTH, Wen SW, et al. Invariant natural killer T cells shape the gut microbiota and regulate neutrophil recruitment and function during intestinal inflammation. Front Immunol (2018) 9:999. doi: 10.3389/fimmu.2018.00999
225. Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol (2004) 4:478–85. doi: 10.1038/nri1373
226. Lin Q, Kuypers M, Liu Z, Copeland JK, Chan D, Robertson SJ, et al. Invariant natural killer T cells minimally influence gut microbiota composition in mice. Gut Microbes (2022) 14:2104087. doi: 10.1080/19490976.2022.2104087
227. Bendelac A, Hunziker RD, Lantz O. Increased interleukin 4 and immunoglobulin e production in transgenic mice overexpressing NK1 T cells. J Exp Med (1996) 184:1285–93. doi: 10.1084/jem.184.4.1285
228. Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity (2007) 27:751–62. doi: 10.1016/j.immuni.2007.08.020
229. Youssef GB, Tourret M, Salou MM, Ghazarian L, Houdouin V, Mondot S, et al. Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. J Exp Med (2018) 215:459–79. doi: 10.1084/jem.20171739
230. Marchini G, Nelson A, Edner J, Lonne-Rahm S, Stavreus-Evers A, Hultenby K. Erythema toxicum neonatorum is an innate immune response to commensal microbes penetrated into the skin of the newborn infant. Pediatr Res (2005) 58:613–6. doi: 10.1203/01.pdr.0000176836.27156.32
231. Edholm ES, Banach M, Robert J. Evolution of innate-like T cells and their selection by MHC class I-like molecules. Immunogenetics (2016) 68:525–36. doi: 10.1007/s00251-016-0929-7
232. Metchnikoff E. The prolongation of life: Optimistic studies. Chalmers Mitchell P, editor. New York: Andesite Press (2017).
233. Kaufman J. Unfinished business: Evolution of the MHC and the adaptive immune system of jawed vertebrates. Annu Rev Immunol (2018) 36:383–409. doi: 10.1146/annurev-immunol-051116-052450
234. Edholm ES, Albertorio Saez LM, Gill AL, Gill SR, Grayfer L, Haynes N, et al. Nonclassical MHC class I-dependent invariant T cells are evolutionarily conserved and prominent from early development in amphibians. Proc Natl Acad Sci U.S.A. (2013) 110:14342–7.
235. Mori L, Lepore M, De Libero G. The immunology of CD1- and MR1-restricted T cells. Annu Rev Immunol (2016) 34:479–510. doi: 10.1146/annurev-immunol-032414-112008
236. Murray MP, Crosby CM, Marcovecchio P, Hartmann N, Chandra S, Zhao M, et al. Stimulation of a subset of natural killer T cells by CD103(+) DC is required for GM-CSF and protection from pneumococcal infection. Cell Rep (2022) 38:110209. doi: 10.1016/j.celrep.2021.110209
237. Grubor-Bauk B, Arthur JL, Mayrhofer G. Importance of NKT cells in resistance to herpes simplex virus, fate of virus-infected neurons, and level of latency in mice. J Virol (2008) 82:11073–83. doi: 10.1128/JVI.00205-08
238. Ishikawa H, Tanaka K, Kutsukake E, Fukui T, Sasaki H, Hata A, et al. IFN-gamma production downstream of NKT cell activation in mice infected with influenza virus enhances the cytolytic activities of both NK cells and viral antigen-specific CD8+ T cells. Virology (2010) 407:325–32. doi: 10.1016/j.virol.2010.08.030
239. Karmakar S, Bhaumik SK, Paul J, De T. TLR4 and NKT cell synergy in immunotherapy against visceral leishmaniasis. PloS Pathog (2012) 8:e1002646. doi: 10.1371/journal.ppat.1002646
240. Van Kaer L, Parekh VV, Wu L. The response of CD1d-restricted invariant NKT cells to microbial pathogens and their products. Front Immunol (2015) 6:226.
241. Bezbradica JS, Gordy LE, Stanic AK, Dragovic S, Hill T, Hawiger J, et al. Granulocyte-macrophage colony-stimulating factor regulates effector differentiation of invariant natural killer T cells during thymic ontogeny. Immunity (2006) 25:487–97. doi: 10.1016/j.immuni.2006.06.017
242. Parekh VV, Wilson MT, Olivares-Villagomez D, Singh AK, Wu L, Wang CR, et al. Glycolipid antigen induces long-term natural killer T cell anergy in mice. J Clin Invest (2005) 115:2572–83. doi: 10.1172/JCI24762
243. Sullivan BA, Kronenberg M. Activation or anergy: NKT cells are stunned by alpha-galactosylceramide. J Clin Invest (2005) 115:2328–9. doi: 10.1172/JCI26297
244. Uldrich AP, Crowe NY, Kyparissoudis K, Pellicci DG, Zhan Y, Lew AM, et al. NKT cell stimulation with glycolipid antigen in vivo: costimulation-dependent expansion, bim-dependent contraction, and hyporesponsiveness to further antigenic challenge. J Immunol (2005) 175:3092–101. doi: 10.4049/jimmunol.175.5.3092
245. Lin Y, Roberts TJ, Wang CR, Cho S, Brutkiewicz RR. Long-term loss of canonical NKT cells following an acute virus infection. Eur J Immunol (2005) 35:879–89. doi: 10.1002/eji.200425495
246. Osman Y, Kawamura T, Naito T, Takeda K, Van Kaer L, Okumura K, et al. Activation of hepatic NKT cells and subsequent liver injury following administration of alpha-galactosylceramide. Eur J Immunol (2000) 30:1919–28. doi: 10.1002/1521-4141(200007)30:7<1919::AID-IMMU1919>3.0.CO;2-3
247. Takeda K, Hayakawa Y, Van Kaer L, Matsuda H, Yagita H, Okumura K, et al. Critical contribution of liver natural killer T cells to a murine model of hepatitis. Proc Natl Acad Sci U.S.A. (2000) 97:5498–503.
248. Driver JP, Scheuplein F, Chen YG, Grier AE, Wilson SB, Serreze DV, et al. Invariant natural killer T-cell control of type 1 diabetes: a dendritic cell genetic decision of a silver bullet or Russian roulette. Diabetes (2010) 59:423–32. doi: 10.2337/db09-1116
249. Singh AK, Wilson MT, Hong S, Olivares-Villagomez D, Du C, Stanic AK, et al. Natural killer T cell activation protects mice against experimental autoimmune encephalomyelitis. J Exp Med (2001) 194:1801–11. doi: 10.1084/jem.194.12.1801
250. Chiba A, Oki S, Miyamoto K, Hashimoto H, Yamamura T, Miyake S, et al. Suppression of collagen-induced arthritis by natural killer T cell activation with OCH, a sphingosine-truncated analog of alpha-galactosylceramide. Arthritis Rheum (2004) 50:305–13. doi: 10.1002/art.11489
251. Coppieters K, Van Beneden K, Jacques P, Dewint P, Vervloet A, Vander Cruyssen B, et al. A single early activation of invariant NK T cells confers long-term protection against collagen-induced arthritis in a ligand-specific manner. J Immunol (2007) 179:2300–9. doi: 10.4049/jimmunol.179.4.2300
252. Singh AK, Yang JQ, Parekh VV, Wei J, Wang CR, Joyce S, et al. The natural killer T cell ligand alpha-galactosylceramide prevents or promotes pristane-induced lupus in mice. Eur J Immunol (2005) 35:1143–54. doi: 10.1002/eji.200425861
253. Chen YG, Tsaih SW, Serreze DV. Genetic control of murine invariant natural killer T-cell development dynamically differs dependent on the examined tissue type. Genes Immun (2012) 13:164–74. doi: 10.1038/gene.2011.68
254. Berzins SP, Cochrane AD, Pellicci DG, Smyth MJ, Godfrey DI. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur J Immunol (2005) 35:1399–407. doi: 10.1002/eji.200425958
255. Kis J, Engelmann P, Farkas K, Richman G, Eck S, Lolley J, et al. Reduced CD4+ subset and Th1 bias of the human iNKT cells in type 1 diabetes mellitus. J Leukoc Biol (2007) 81:654–62. doi: 10.1189/jlb.1106654
256. Montoya CJ, Pollard D, Martinson J, Kumari K, Wasserfall C, Mulder CB, et al. Characterization of human invariant natural killer T subsets in health and disease using a novel invariant natural killer T cell-clonotypic monoclonal antibody, 6B11. Immunology (2007) 122:1–14. doi: 10.1111/j.1365-2567.2007.02647.x
257. Chan AC, Serwecinska L, Cochrane A, Harrison LC, Godfrey DI, Berzins SP, et al. Immune characterization of an individual with an exceptionally high natural killer T cell frequency and her immediate family. Clin Exp Immunol (2009) 156:238–45. doi: 10.1111/j.1365-2249.2009.03888.x
258. Bernin H, Fehling H, Marggraff C, Tannich E, Lotter H. The cytokine profile of human NKT cells and PBMCs is dependent on donor sex and stimulus. Med Microbiol Immunol (2016) 205:321–32. doi: 10.1007/s00430-016-0449-y
259. Moll M, Kuylenstierna C, Gonzalez VD, Andersson SK, Bosnjak L, Sonnerborg A, et al. Severe functional impairment and elevated PD-1 expression in CD1d-restricted NKT cells retained during chronic HIV-1 infection. Eur J Immunol (2009) 39:902–11. doi: 10.1002/eji.200838780
260. Kee SJ, Kwon YS, Park YW, Cho YN, Lee SJ, Kim TJ, et al. Dysfunction of natural killer T cells in patients with active mycobacterium tuberculosis infection. Infect Immun (2012) 80:2100–8. doi: 10.1128/IAI.06018-11
261. Matangkasombut P, Chan-In W, Opasawaschai A, Pongchaikul P, Tangthawornchaikul N, Vasanawathana S, et al. Invariant NKT cell response to dengue virus infection in human. PloS Negl Trop Dis (2014) 8:e2955. doi: 10.1371/journal.pntd.0002955
262. Yang G, Artiaga BL, Hackmann TJ, Samuel MS, Walters EM, Salek-Ardakani S, et al. Targeted disruption of CD1d prevents NKT cell development in pigs. Mamm Genome (2015) 26:264–70. doi: 10.1007/s00335-015-9564-0
263. Bertho N, Meurens F. The pig as a medical model for acquired respiratory diseases and dysfunctions: An immunological perspective. Mol Immunol (2021) 135:254–67. doi: 10.1016/j.molimm.2021.03.014
264. Kaser T. Swine as biomedical animal model for T-cell research-success and potential for transmittable and non-transmittable human diseases. Mol Immunol (2021) 135:95–115. doi: 10.1016/j.molimm.2021.04.004
265. Sinkora M, Butler JE. Progress in the use of swine in developmental immunology of b and T lymphocytes. Dev Comp Immunol (2016) 58:1–17. doi: 10.1016/j.dci.2015.12.003
266. Yang G, Richt JA, Driver JP. Harnessing invariant NKT cells to improve influenza vaccines: A pig perspective. Int J Mol Sci (2017) 19:68–84. doi: 10.3390/ijms19010068
267. Meierovics AI, Cowley SC. MAIT cells promote inflammatory monocyte differentiation into dendritic cells during pulmonary intracellular infection. J Exp Med (2016) 213:2793–809. doi: 10.1084/jem.20160637
268. Chua WJ, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH, et al. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun (2012) 80:3256–67. doi: 10.1128/IAI.00279-12
269. Le Bourhis L, Dusseaux M, Bohineust A, Bessoles S, Martin E, Premel V, et al. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PloS Pathog (2013) 9:e1003681. doi: 10.1371/journal.ppat.1003681
270. Kurioka A, Ussher JE, Cosgrove C, Clough C, Fergusson JR, Smith K, et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol (2015) 8:429–40. doi: 10.1038/mi.2014.81
271. Franciszkiewicz K, Salou M, Legoux F, Zhou Q, Cui Y, Bessoles S, et al. MHC class I-related molecule, MR1, and mucosal-associated invariant T cells. Immunol Rev (2016) 272:120–38. doi: 10.1111/imr.12423
272. Salou M, Franciszkiewicz K, Lantz O. MAIT cells in infectious diseases. Curr Opin Immunol (2017) 48:7–14. doi: 10.1016/j.coi.2017.07.009
273. Leng T, Akther HD, Hackstein CP, Powell K, King T, Friedrich M, et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep (2019) 28:3077–91:e3075. doi: 10.1016/j.celrep.2019.08.050
274. Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, et al. Activation and In vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep (2019) 28:3249–3262 e3245. doi: 10.1016/j.celrep.2019.07.039
275. Sanchez DJ, Gumperz JE, Ganem D. Regulation of CD1d expression and function by a herpesvirus infection. J Clin Invest (2005) 115:1369–78. doi: 10.1172/JCI200524041
276. Cho S, Knox KS, Kohli LM, He JJ, Exley MA, Wilson SB, et al. Impaired cell surface expression of human CD1d by the formation of an HIV-1 Nef/CD1d complex. Virology (2005) 337:242–52. doi: 10.1016/j.virol.2005.04.020
277. Chen N, McCarthy C, Drakesmith H, Li D, Cerundolo V, McMichael AJ, et al. HIV-1 down-regulates the expression of CD1d via nef. Eur J Immunol (2006) 36:278–86. doi: 10.1002/eji.200535487
278. Yuan W, Dasgupta A, Cresswell P. Herpes simplex virus evades natural killer T cell recognition by suppressing CD1d recycling. Nat Immunol (2006) 7:835–42. doi: 10.1038/ni1364
279. McSharry BP, Samer C, McWilliam HEG, Ashley CL, Yee MB, Steain M, et al. Virus-mediated suppression of the antigen presentation molecule MR1. Cell Rep (2020) 30:2948–2962 e2944. doi: 10.1016/j.celrep.2020.02.017
280. Liu J, Gallo RM, Khan MA, Iyer AK, Kratzke IM, Brutkiewicz RR, et al. JNK2 modulates the CD1d-dependent and -independent activation of iNKT cells. Eur J Immunol (2019) 49:255–65. doi: 10.1002/eji.201847755
Keywords: NKT (natural killer T) cell, MAIT (mucosal-associated invariant T) cell, innate-like effector lymphocyte, symbionts, pathobiont
Citation: Joyce S, Okoye GD and Driver JP (2023) Die Kämpfe únd schláchten—the struggles and battles of innate-like effector T lymphocytes with microbes. Front. Immunol. 14:1117825. doi: 10.3389/fimmu.2023.1117825
Received: 06 December 2022; Accepted: 22 March 2023;
Published: 24 April 2023.
Edited by:
Fu-Dong Shi, Tianjin Medical University General Hospital, ChinaReviewed by:
Seokmann Hong, Sejong University, Republic of KoreaKazuya Iwabuchi, Kitasato University School of Medicine, Japan
Copyright © 2023 Joyce, Okoye and Driver. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John P. Driver, ZHJpdmVyanBAbWlzc291cmkuZWR1; Sebastian Joyce, c2ViYXN0aWFuLmpveWNlQHZ1bWMub3Jn
 Sebastian Joyce
Sebastian Joyce Gosife Donald Okoye
Gosife Donald Okoye John P. Driver
John P. Driver