Abstract
Purpose:
Myocardial injury is a common complication in patients with endotoxaemia/sepsis, especially in children. Moreover, it develops through an unclear pathophysiological mechanism, and effective therapies are lacking. Recently, RNA modification, particularly N6-methyladenosine (m6A) modification, has been found to be involved in various physiological processes and to play important roles in many diseases. However, the role of m6A modification in endotoxaemia/sepsis-induced myocardial injury is still in its infancy. Therefore, we attempted to construct the m6A modification map of myocardial injury in a rat model treated by lipopolysaccharide (LPS) and explore the role of m6A modification in LPS-induced myocardial injury.
Method:
Myocardial injury adolescent rat model was constructed by intraperitoneal injection of LPS. m6A RNA Methylation Quantification Kit was used to detect overall level of m6A modification in rat cardiac tissue. m6A-specific methylated RNA immunoprecipitation followed by high-throughput sequencing (MeRIP-seq) and RNA sequencing (RNA-seq) were conducted to identify the altered m6A-modified genes and differentially expressed genes in cardiac tissue of rats treated by LPS and control rats (6 versus. 6). Bioinformatics was used to analyze the functions of differentially m6A modified genes, differentially expressed genes, and genes with both differential m6A modification and differential expression. qPCR was used to detect expression of m6A modification related enzymes.
Result:
We found that the overall level of m6A modification in cardiac tissue of the LPS group was up-regulated compared with that of the control group. MeRIP-seq and RNA-seq results showed that genes with differential m6A modification, genes with differential expression and genes with both differential m6A modification and differential expression were closely associated with inflammatory responses and apoptosis. In addition, we found that m6A-related enzymes (Mettl16, Rbm15, Fto, Ythdc2 and Hnrnpg) were differentially expressed in the LPS group versus. the control group.
Conclusion:
m6A modification is involved in the pathogenesis process of LPS-induced myocardial injury, possibly through the regulation of inflammatory response and apoptosis-related pathways. These results provide valuable information regarding the potential pathogenic mechanisms underlying LPS-induced myocardial injury.
1 Introduction
Sepsis is a life-threatening organ dysfunction caused by dysregulated host responses to infection (1). Approximately 48.9 million people worldwide are diagnosed with sepsis each year, and sepsis led to ~11 million deaths in 2017 (2). Sepsis is often accompanied by multiple-organ dysfunction, involving conditions such as acute kidney injury, acute lung injury, and myocardial injury. Notably, epidemiological studies have found that the incidence of myocardial injury was as high as 70.2% in children (3), and sepsis-induced myocardial injury was closely related to mortality (4). In addition, endotoxic shock is a subtype of sepsis and is often accompanied by myocardial injury (5, 6). However, the pathophysiological mechanism of endotoxaemia/sepsis-induced myocardial injury is still unclear, and no specific drug can be targeted to treat endotoxaemia/sepsis-induced myocardial injury. Our team has found that myocardial apoptosis, inflammatory disorders (6), mitochondrial dysfunction, oxidative stress (7) and autophagy (8) were involved in the occurrence and development of endotoxaemia/sepsis-induced myocardial injury. Epigenetic modifications have also been implicated (6), but many aspects of the occurrence and development of endotoxaemia/sepsis-induced myocardial injury remain to be discovered.
m6A modification is the most frequent internal modification of eukaryotic mRNAs, and such modifications influence diverse fundamental cellular functions, such as pre-mRNA splicing, nuclear transport, stability, translation, and microRNA biogenesis (9). Recently, m6A modification has been found to be associated with the occurrence and development of many diseases, such as cancer (9), cardiovascular diseases (10). Moreover, recent studies showed that m6A modification played an important regulatory role in mediating inflammation responses, which was to be associated with the occurrence and development of endotoxaemia/sepsis-induced myocardial injury. For example, m6A methyltransferase Mettl3 could regulate inflammatory responses through IGF2BP1/HDAC4 dependent manner in LPS-induced cardiomyocytes (11). However, as a widespread RNA modification, m6A modification sites and regulatory mechanisms are still worthy of further study.
Here, considering endotoxin can trigger all of the cardinal feature of sepsis on its own and likely is a modulating factor during the syndrome (5), we conducted myocardial injury rat model of LPS-induced endotoxic shock and performed MeRIP-seq and RNA-seq to identify differentially m6A-modified RNAs and differentially expressed RNAs involved in LPS-induced myocardial injury. Then, understanding the functions that these differentially m6A-modified RNAs may be involved in regulation through bioinformatics. Combined analysis of MeRIP-seq and RNA-seq helped us identify genes that may be regulated by m6A at RNA levels. Detection of m6A-related enzymes helped us understand which enzymes might play important functions by regulating m6A modification.
2 Materials and methods
2.1 Animals
The study was approved by the Ethics Committee of the Shengjing Hospital of China Medical University (Shenyang, China; approval number 2022PS854K). This study conformed to the relevant ethical standards. The rats used in this study (Beijing HFK Bioscience Co., Ltd.; Beijing, China) had a Wistar genetic background. The rats were adolescent, male, and pathogen-free; weighed approximately 160-180 g; and were housed in cages at 24°C with a 12h alternating light/dark cycle and free access to water and food. An adolescent rat model of endotoxic shock was generated by intraperitoneal (i.p.) injection of LPS, as we described previously (6). Briefly, rats were anesthetized with 20% urethane (1g/kg, i.p.). The left femoral artery was cannulated (Biopac MP150; Biopac Systems, Goleta, CA, USA) to continuously measure the mean arterial pressure (MAP). After the MAP stabilized, LPS (L-2880; Sigma-Aldrich, St. Louis, MO, USA) was administered i.p. (20 mg/kg, 10 mg LPS dissolved in 1 ml of 0.9% saline). 2 hours after LPS injection, MAP began to decline, and the model was considered successful when MAP decreased by 20-25 percent. The control group was injected with an equal volume of 0.9% normal saline solution. The preliminary experiment found that the death rate of rats was relatively high 24 hours after injection of LPS, but could basically be stabilized to 12 hours. In order to obtain the pathological changes in the late stage of LPS-induced myocardial injury, we selected 12 hours after injection of LPS to conduct this study. Twelve hours post-LPS or normal saline injection, euthanasia was performed, and the left ventricle was excised and placed in an RNase-free centrifuge tube, quickly placed in liquid nitrogen, and stored at -80°C. Blood samples were collected from the abdominal aorta at 12h post-LPS or saline administration and centrifuged (3,000 rpm, 10 min) after incubation at room temperature (15°C-25°C) for 15 min. Supernatants were collected and stored at -80°C for enzyme-linked immunosorbent assay (ELISA) analysis. Six pairs of left ventricles with endotoxaemia or controls were selected for MeRIP-seq and RNA-seq, and the remaining samples were saved for validation experiments.
2.2 Hematoxylin and eosin staining
Rat heart samples were fixed in 4% paraformaldehyde for 48 h, dehydrated, permeabilized, and embedded in paraffin. Paraffin-embedded tissues were sectioned (0.35 µm) and stained with H&E. Pathological changes were observed microscopically.
2.3 Echocardiography
Twelve hours after the LPS injection, echocardiography was performed. A special ultrasound instrument (Vinno Technology, China) for small animals was used to obtain echocardiographic images. Two-dimensional guided M-mode measurements of the left ventricular (LV) internal diameter were obtained from the long-axis view at the level of the papillary muscles. The interventricular septal thickness at diastole (IVSd), left ventricular intra-diameter at diastole (LVIDd), left ventricular end-diastolic posterior wall thickness (LVPWd), interventricular septal thickness at systole (IVSs), left ventricular intra-diameter at systole (LVIDs), left ventricular end-systolic posterior wall thickness (LVPWs), left ventricular end-systolic volume (LVESV), left ventricular end-diastolic volume (LVEDV), left ventricular ejection fraction (LVEF), stroke volume (SV), left ventricular fractional shortening (LVFS), heart rate (HR) and cardiac output (CO) were recorded.
2.4 ELISA-based analysis of rat serum
Cardiac troponin I (c-TNI), creatine kinase myocardial band (CK-MB) and interleukin (IL)-6 levels in blood supernatants from both groups were determined via ELISA analysis. IL-6 ELISA kits were purchased from Wanlei Biological Company (Shenyang, China), whereas CK-MB and c-TNI ELISA kits were purchased from Elabscience Biotechnology Co., Ltd. (Wuhan, China). The biomarker concentrations in each sample were estimated based on optical density values at 450 nm and a standard curve.
2.5 Measurement of total m6A levels
Total RNA was isolated from heart tissue using the TRIzol Reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA). The RNA quantity and purity of each sample were determined using Nanodrop 2000 (Thermo Scientific, USA). m6A RNA Methylation Quantification Kit (ab185912, Abcam, Shanghai, China) was used to detect the total m6A levels in 200ng RNA by measuring absorbance at 450nm.
2.6 MeRIP-seq and RNA-seq analyses
RNA extraction and purity measurement are the same as 2.5. RNA-integrity numbers were assessed using a Bioanalyzer 2100 (Agilent, CA, USA) and exceeded 7.0, as confirmed by denaturing agarose-gel electrophoresis. Poly (A) RNA was purified from 50μg total RNA using Dynabeads Oligo (dT)25-61005 (Thermo Fisher, CA, USA) and two rounds of purification. Then, the poly(A) RNA was fragmented using a Magnesium RNA Fragmentation Module (NEB, USA) at 86°C for 7min. The RNA fragments were incubated for 2h at 4°C with an m6A-specific antibody (Synaptic Systems, Germany) in IP buffer (50mM Tris-HCl, 750mM NaCl, and 0.5% Igepal CA-630). Both the input samples (without IP) and the m6A-IP samples were used to construct RNA-seq libraries with the NEBNext Ultra RNA Library Prep Kit for Illumina. Finally, we performed 2×150bp paired-end sequencing (PE150) on an Illumina Novaseq 6000 instrument (LC-Bio Technology Co., Ltd., Hangzhou, China).
2.7 Bioinformatics analysis
Fastp software (12) (default parameters) was used to remove reads containing adaptors, low-quality bases, and undetermined bases. The quality of the IP and input sequences was verified using FastQC and RseQC. Filtered high-quality data were mapped to the Rattus norvegicus genome (Ensemble release 101) using HISAT2 (13). Peak calling and differential-peak analysis were performed using the exomePeak2 package (14), and peaks were annotated using ANNOVAR. The peaks in each group with a fold change (FC) of ≥2 was considered significant. HOMER (15) was used for de novo motif finding, followed by motif localization with respect to the peak summit. Then, StringTie (16) was used to determine the expression levels of all mRNAs from the input libraries using the following relationship: (total exon fragments/mapped reads [millions] × exon length [kB]).
2.8 Protein–protein-interaction network
Genes with differential expression or m6A modification sites were imported into the STRING database (17), and the interaction relationships between the imported genes were determined. A PPI network was constructed by importing the data into Cytoscape 3.8.2 (18)and analyzing the data using a network analyzer. The genes imported with combined interaction scores of >0.4 were selected to construct the PPI network.
2.9 MeRIP assays
MeRIP assays were performed using the m6A RNA Enrichment Kit (Epigentek, USA). Target m6A-containing fragments were immunoprecipitated using a bead-bound m6A capture antibody, and RNA sequences at both ends of the m6A-containing regions were cleaved using Cleavage Enzyme Mix. The enriched RNA was released, purified, and eluted. Quantitative reverse transcriptase-PCR (qRT-PCR) analysis was performed following MeRIP to quantify changes in target-gene m6A modification. Primers (Table S1) were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).
2.10 Real-time RT-PCR analysis
RNA was reverse transcribed to complementary DNA using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara Bio, Being, China), with a reaction time of 15min at 37°C, followed by denaturation for 5s at 85°C. A 7500 Real-Time PCR System (Applied Biosystems, USA) was used for PCR. The primers used are listed in Table S2.
2.11 Western blot
Frozen ventricular tissue samples were homogenized in RIPA buffer (Beyotime Biotechnology, China) containing protein enzyme inhibitors, and protein concentration was quantified using a BCA Protein Assay Kit (Beyotime Biotechnology, China). The lysates were separated on 8–15% SDS-PAGE gels and then electro-transferred to 0.45μm polyvinylidene difluoride membranes (Millipore, USA). After blocking with 5% nonfat milk in Tris-buffered saline at room temperature for 2h, the membranes were incubated for overnight at 4°C with primary antibodies. The primary antibody included IL-6 (Wanlei, China), IL-1β (Affinity, China), IL-18 (Affinity, China) and Tubulin (Affinity, China). After washing three times for 10min per wash, the membranes were incubated with an HRP-conjugated secondary antibody for 2h at room temperature. Subsequently, the blots were imaged using a Bio-Rad imaging system (Bio-Rad, USA) and protein expression levels were determined using ImageJ software.
2.12 Statistical analysis
All data were obtained from three or more independent experiments and are presented as mean ± standard deviation. Statistical analyses were conducted using GraphPad Prism (version 8.0). Student’s t-test for unpaired data was used to compare two groups. Statistical significance was set at P < 0.05. Peaks and transcripts with |FC| ≥1.5 and P < 0.05 between control group and LPS group were considered differential peaks and transcripts.
3 Results
3.1 Myocardial injury induced by LPS was linked to changes in global m6A modification level
In this study, we conducted a rat model of endotoxic shock by intraperitoneal injection of LPS. The MAP in rats decreased by 25-30% 2h after LPS administration (Supplementary Figure 1). Firstly, we assessed cardiac function using echocardiograph. As shown in Table 1, the IVSd, LVIDd, LVIDs, LVEDV, LVESV, SV, HR and CO were deceased significantly in the LPS group while the LVPWd and LVSs were increased significantly in the LPS group. However, no difference in LVPWs, EF and FS were observed between two groups. Then, we detected the pathological changes in the cardiac tissue founding that myocardial cells showed greater disorder and inflammatory cell infiltration in the LPS group than in the control group (H&E staining; Figure 1A). Strikingly, serum c-TNI and CK-MB (markers of injury to cardiomyocytes) were significantly elevated in the LPS group (Figure 1B, C), as was the serum inflammatory biomarker, IL-6 (Figure 1D). We also analyzed the protein expression levels of inflammatory cytokines (IL-6, IL-18, IL-1β) in the heart using western blot. The protein expression levels of IL-6, IL-18, and IL-1β were upregulated in the LPS group compared to the control group (Figure 1E), suggesting inflammatory injury of cardiac tissue in the rat model induced by LPS.
Table 1
| Control | LPS | |
|---|---|---|
| IVSd (mm) | 379.300 ± 52.520 | 303.400 ± 33.230* |
| LVIDd (mm) | 6.395 ± 0.247 | 3.563 ± 0.433*** |
| LVPWd (mm) | 1.313 ± 0.228 | 2.423 ± 0.301*** |
| IVSs (mm) | 2.863 ± 0.197 | 3.328± 0.339* |
| LVIDs (mm) | 1.615 ± 0.240 | 0.937 ± 0.209*** |
| LVPWs (mm) | 3.337 ± 0.239 | 3.522 ± 0.300 |
| LVEDV (ml) | 0.603 ± 0.063 | 0.118 ± 0.042*** |
| LVESV (ml) | 0.012 ± 0.004 | 0.002 ± 0.004** |
| LVEF (%) | 98.000 ± 0.859 | 97.890 ± 0.912 |
| SV (ml) | 0.592 ± 0.063 | 0.117 ± 0.043*** |
| LVFS (%) | 74.760 ± 3.484 | 73.800 ± 4.191 |
| HR (time/min) | 379.300 ± 52.520 | 303.400 ± 33.230* |
| CO (L/min) | 0.223 ± 0.037 | 0.036 ± 0.015*** |
The echocardiographic data of rats in groups.
n=6 per group; * p < 0.05 compared to the control; **p < 0.01 compared to the control; ***p < 0.001 compared to the control; IVSd, interventricular septal thickness at diastole; LVIDd, left ventricular intra-diameter at diastole; LVPWd, left ventricular end-diastolic posterior wall thickness; IVSs, interventricular septal thickness at systole; LVIDs, left ventricular intra-diameter at systole; LVPWs, left ventricular end-systolic posterior wall thickness; LVESV, left ventricular end-systolic volume; LVEDV, left ventricular end-diastolic volume; LVEF, left ventricular ejection fraction; SV, stroke volume; LVFS, left ventricular fractional shortening; HR, heart rate; CO, cardiac output.
Figure 1

Basic characteristics of animal models between LPS group and control group. (A) Gross morphology of cardiac tissue stained with HE (200×) (n=6 in each group); Red arrow, disordered cardiac cells; White arrow, infiltration of inflammatory cells. (B, C) Serum cardiac enzyme c-TNI and CK-MB (D) Serum inflammatory biomarker IL-6 were detected by ELISA (n=6 in each group). (E) Protein expression levels of inflammatory indicators (IL-6, IL-18, IL-1β) were detected by western blot (n=6 in each group). (F) Quantification of m6A levels in cardiac tissue of LPS group and control group (n=6 in each group). HE, hematoxylin and eosin; c-TNI, cardiac troponin I; CK-MB, creatine kinase MB form; *p < 0.05 compared to the control; **p < 0.01 compared to the control; ***p < 0.001 compared to the control.
In addition, higher total m6A modification levels were found in the LPS group than that in the control group (Figure 1F). Thus, we performed transcriptome-wide MeRIP-seq and RNA-Seq to generate an m6A modification map in cardiac tissue of rats with LPS-induced myocardial injury.
3.2 Differential m6A modification patterns in cardiac tissue of rats treated by LPS and control rats
Next, we compared the m6A peaks between LPS group and control group. We identified 14,360 common m6A peaks, representing 9,840 genes in both groups using exomePeak2 (Figure 2A). In addition, 4,497 unique peaks and 3,589 unique genes appeared in the LPS group, whereas 5,162 unique peaks and 4,102 unique genes appeared in the control group (Figure 2B). Chromosome 1 showed the largest number of m6A modification sites with 2,272 and 2,355 m6A peaks in the LPS group and the control group, respectively, followed by chromosome 10 with 1,613 and 1,662 m6A peaks in the LPS group and the control group, respectively (Supplementary Figure 2A). Most genes had one to three m6A modification sites (Supplementary Figure 2B).
Figure 2
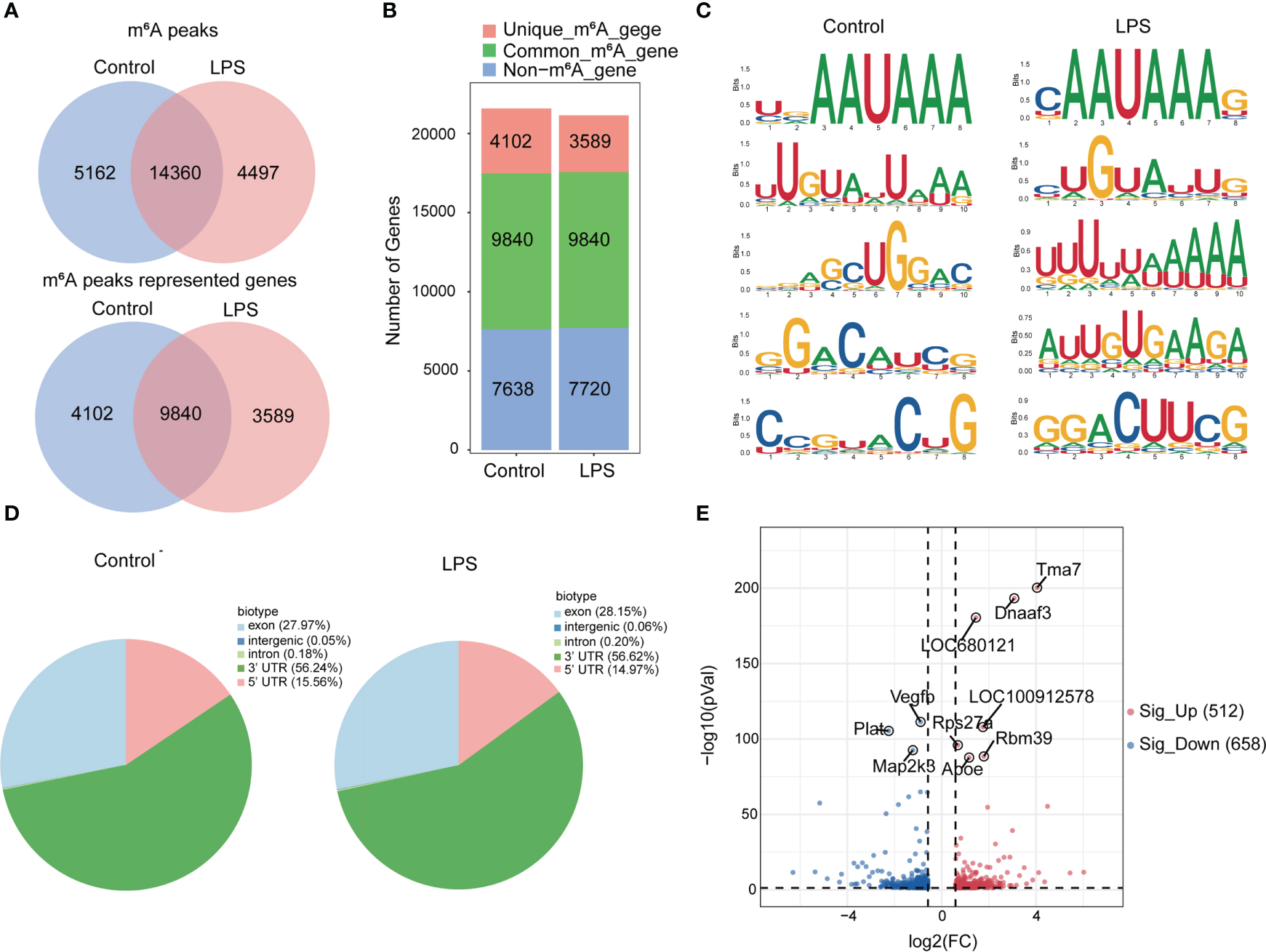
Transcriptome-wide MeRIP-seq and analysis of m6A peaks. (A) Overlap of m6A peaks in the LPS group and the control group. Numbers of LPS-unique, control-unique, and common m6A peaks are shown as Venn diagram as well as m6A peaks representing genes in the two groups. (B) Summary of genes with m6A modification identified in MeRIP-seq. (C) Top 5 m6A modification motifs enriched from all identified m6A peaks in the two groups. (D) Proportion of m6A peaks distributed in the indicated regions in the control group and the LPS group. (E) Volcano map: identification of 512 hyper-m6A modification and 658 hypo-m6A modification peaks in the LPS group compared to the control group (|FC|>=1.5, P<0.05). n=6 in each group; FC, fold change.
In addition, we identified the top five consensus motifs for the m6A peaks in the LPS group and the control group (Figure 2C). RRACH motif was identified in both groups, where R = G or A; A = m6A, and H = U, A, or C, as described (19). The m6A peaks were predominantly distributed in coding sequences and the 3’ untranslated region (UTR) (Supplementary Figure 2C). Finally, m6A peaks were analyzed in whole transcriptome data and divided into exon, intergenic, intron, 3’UTR, 5’UTR regions, based on their location in RNA transcripts. The peaks were mainly distributed in 3’UTR, exon and 5’UTR and different distribution patterns were observed between the two groups. The LPS group showed slightly more m6A peaks in the exon (27.97% versus. 28.15%) and 3′UTR (56.24% versus. 56.62%) and slightly less m6A peaks in the 5′UTR (15.56% versus. 14.97%) than the control group (Figure 2D).
Furthermore, we found 14,360 m6A peaks in the LPS group and the control group, 5,740 of which were differentially modification (P < 0.05). Among them, 512 hypermethylated and 658 hypomethylated m6A modification sites (|FC| ≥ 1.5, P < 0.05) were found in the LPS group versus. the control group. The 10 genes with the most significant differences were identified (Figure 2E). Table 2 showed the top 20 differentially upregulated and downregulated m6A peaks.
Table 2
| Chr | Peak_length | Strand | DiffMod Log2FC |
P-value | Annotation | Transcript ID | Gene Name |
|---|---|---|---|---|---|---|---|
| chr1 | 551 | – | 6.02 | 0.00 | exonic | ENSRNOT00000044195 | LOC100911951 |
| chr7 | 1067 | – | 5.44 | 0.00 | exonic | ENSRNOT00000086062 | Col2a1 |
| chr1 | 466 | + | 4.48 | 0.00 | UTR3 | ENSRNOT00000028222 | Polr2i |
| chr10 | 2933 | + | 4.36 | 0.00 | exonic | ENSRNOT00000089404 | AC117889 |
| chr2 | 1476 | – | 4.11 | 0.00 | UTR3 | ENSRNOT00000009813 | Fgb |
| chr8 | 4687 | – | 4.04 | 0.00 | UTR3 | ENSRNOT00000075819 | Tma7 |
| chr5 | 401 | + | 3.86 | 0.00 | UTR3 | ENSRNOT00000037547 | MGC94199 |
| chr1 | 101 | + | 3.60 | 0.00 | exonic | ENSRNOT00000017948 | Mrgprf |
| chr9 | 376 | – | 3.50 | 0.00 | UTR3 | ENSRNOT00000016237 | LOC100365697 |
| chr9 | 449 | + | 3.30 | 0.00 | UTR5 | ENSRNOT00000051841 | Atn1 |
| chr3 | 201 | + | 3.17 | 0.00 | UTR3 | ENSRNOT00000022265 | RGD1561517 |
| chr1 | 727 | + | 3.07 | 0.00 | UTR3 | ENSRNOT00000058900 | Dnaaf3 |
| chr1 | 3661 | + | 3.00 | 0.00 | exonic | ENSRNOT00000016883 | Cyp2e1 |
| chr18 | 226 | + | 2.95 | 0.00 | exonic | ENSRNOT00000027015 | Pcdhga7 |
| chr8 | 9941 | + | 2.89 | 0.00 | UTR3 | ENSRNOT00000047613 | Faim |
| chr14 | 376 | + | 2.88 | 0.00 | UTR3 | ENSRNOT00000033437 | Gpr75 |
| chr17 | 2440 | – | 2.78 | 0.00 | exonic | ENSRNOT00000010836 | LOC100912163 |
| chr10 | 373 | + | 2.64 | 0.00 | UTR3 | ENSRNOT00000091610 | Gspt1 |
| chr13 | 376 | + | 2.62 | 0.00 | UTR3 | ENSRNOT00000004104 | Gpr161 |
| chr13 | 376 | – | 2.61 | 0.00 | UTR3 | ENSRNOT00000004204 | LOC100911825 |
| chr20 | 101 | – | -6.31 | 0.00 | exonic | ENSRNOT00000073474 | RT1-DMb |
| chr20 | 426 | – | -5.37 | 0.00 | UTR3 | ENSRNOT00000086240 | RT1-DMb |
| chr7 | 2975 | – | -5.17 | 0.00 | UTR5 | ENSRNOT00000021523 | Calcoco1 |
| chr1 | 319 | + | -4.85 | 0.00 | exonic | ENSRNOT00000064178 | LOC100911727 |
| chr12 | 101 | + | -4.33 | 0.00 | exonic | ENSRNOT00000001479 | Fbxl18 |
| chr11 | 251 | – | -4.25 | 0.00 | exonic | ENSRNOT00000029842 | LOC100911374 |
| chr1 | 101 | + | -3.82 | 0.00 | UTR5 | ENSRNOT00000074301 | Zfp260 |
| chr20 | 3286 | – | -3.72 | 0.00 | exonic | ENSRNOT00000073474 | RT1-DMb |
| chr20 | 126 | + | -3.68 | 0.00 | UTR3 | ENSRNOT00000000468 | Slc35f1 |
| chr1 | 7568 | – | -3.59 | 0.00 | exonic | ENSRNOT00000017718 | Lnpep |
| chr12 | 5279 | + | -3.56 | 0.00 | exonic | ENSRNOT00000070868 | Gatc |
| chr15 | 301 | + | -3.36 | 0.00 | UTR3 | ENSRNOT00000074912 | LOC108348161 |
| chr20 | 226 | – | -3.32 | 0.00 | UTR3 | ENSRNOT00000070886 | RT1-DMa |
| chr10 | 176 | – | -3.20 | 0.00 | UTR3 | ENSRNOT00000073706 | LOC100912585 |
| chr1 | 126 | – | -3.08 | 0.00 | UTR3 | ENSRNOT00000075570 | Itprip |
| chr10 | 611 | – | -3.07 | 0.00 | exonic | ENSRNOT00000013281 | Exoc7 |
| chr14 | 401 | – | -2.93 | 0.00 | UTR3 | ENSRNOT00000075415 | Pigg |
| chr18 | 1176 | + | -2.89 | 0.00 | UTR3 | ENSRNOT00000073388 | LOC100910979 |
| chr9 | 6739 | – | -2.71 | 0.00 | UTR5 | ENSRNOT00000019848 | Tmbim1 |
| chr11 | 101 | – | -2.60 | 0.01 | exonic | ENSRNOT00000002713 | Tmprss2 |
Top 20 differentially up- and down-regulated m6A peaks according to the fold change in the LPS group compared to the control group.
3.3 Systematic functional analysis of genes with differential m6A modification
To determine the possible functions of the differentially m6A-methylated genes involved in regulation, we performed Gene Ontology (GO) (20), Kyoto Encyclopedia of Genes and Genomes (KEGG) (21), and PPI network analyses.
Most genes with differential m6A modification were related to apoptosis and inflammatory responses by GO analysis (Figures 3A, D). Furthermore, KEGG pathway analysis of genes with differentially m6A modification revealed enrichment for terms related to (i) inflammatory responses, such as Staphylococcus aureus infection, Fc gamma R-mediated phagocytosis, complement and coagulation cascades, Th17 cell differentiation, chemokine signaling pathway, IL17 signaling pathway and antigen processing and presentation; (ii) important mediators of apoptosis, such as Hippo signaling pathway, AGE-RAGE signaling pathway in diabetic complications, MAPK signaling pathway and PI3K/Akt signaling pathway (Figures 3B, E).
Figure 3
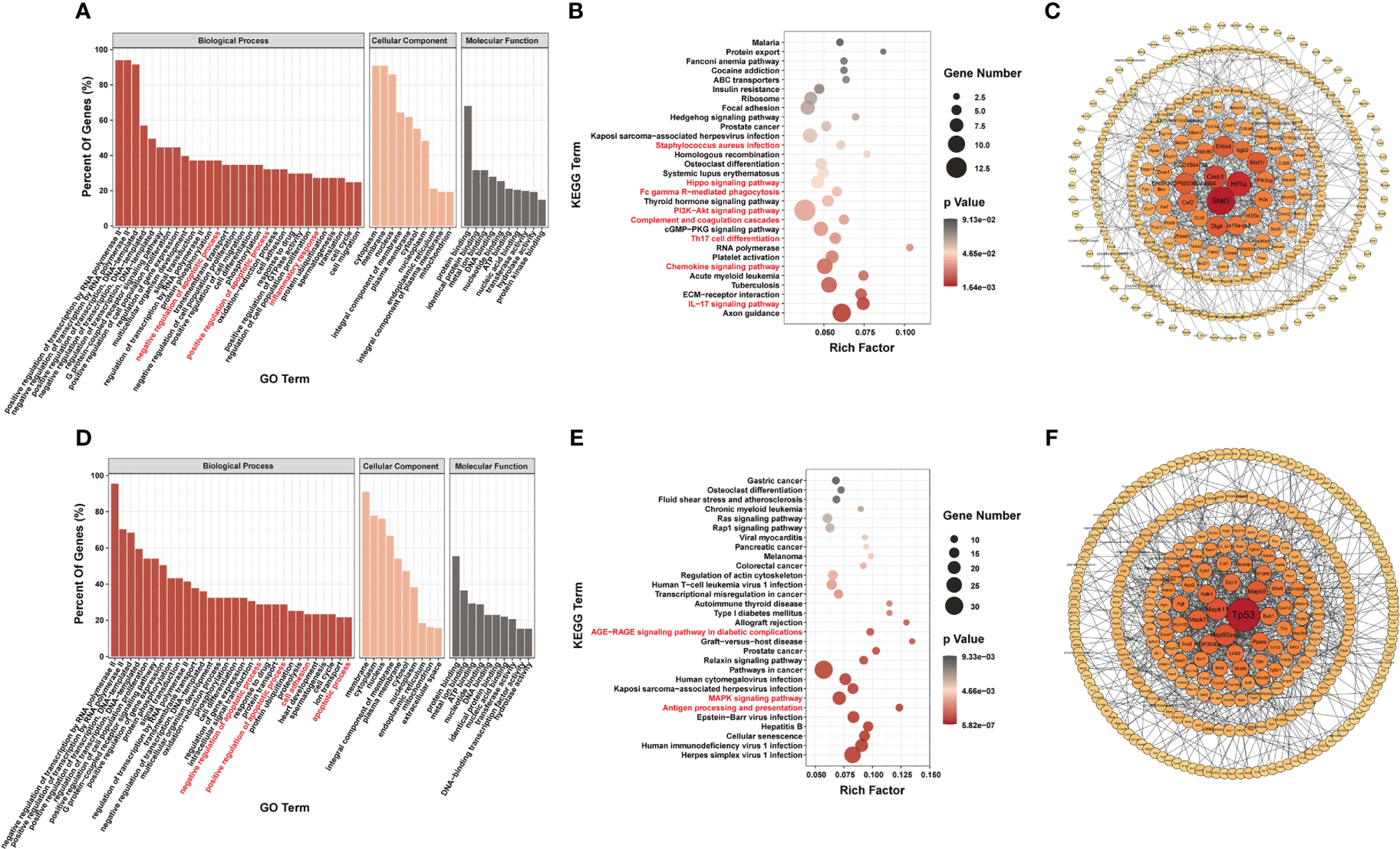
Systematic functional analysis of genes with differentially m6A modification. (A–C) Systematic functional analysis for genes with up-regulated m6A modification; (A) Major gene ontology terms significantly enriched; (B) Top 30 significantly enriched pathways; (C) PPI network. (D–F) Systematic functional analysis for genes with down-regulated m6A modification; (D) Major gene ontology terms significantly enriched; (E) Top 30 significantly enriched pathways; (F) PPI network.
We then performed PPI-network analysis of genes with differential m6A modification using Cytoscape (18). Stat3, Hif1a, and Creb1 were the most central genes with upregulated m6A modification, whereas Tp53, Mapk11, and Hsp90aa1 were the most central genes with downregulated m6A modification (Figures 3C, F). These central proteins are particularly associated with inflammatory responses and apoptosis.
3.4 mRNA-expression differences in cardiac tissue of rats treated by LPS and control rats
RNA-seq data showed that 1,836 genes were significantly upregulated and that 2,467 genes were significantly downregulated in the LPS group compared to the control group (|FC| ≥1.5, P < 0.05). Figure 4A shows the 10 most upregulated and downregulated genes. In addition, principal component analysis showed that samples from the LPS group and control group clustered separately within each group (Supplementary Figure 3A). The heatmap of Pearson correlation coefficient matrix also revealed good intragroup correlations in both groups (Supplementary Figure 3B). Moreover, GO analysis suggested that differentially expressed genes were associated with inflammatory responses and apoptosis (Figure 4B). KEGG pathway analysis revealed enrichment for terms related to (i) inflammatory responses terms, such as the chemokine signaling pathway, Toll-like receptor signaling pathway, Th17 cell differentiation, natural killer cell-mediated cytotoxicity, NOD-like receptor signaling pathway, cytokine-cytokine receptor interaction and TNF signaling pathway; (ii) apoptosis terms such as PI3K-Akt signaling pathway and MAPK signaling pathway (Figure 4C). Furthermore, PPI-network analysis demonstrated that the central proteins were Stat1, Tnf, and IL-6 among the upregulated genes (which are critical for inflammatory responses), and Erbb3, Igf1, and Pik3r3 among the downregulated genes (which are vital for involvement of the PI3k-Akt signaling pathway) (Figures 4D, E).
Figure 4
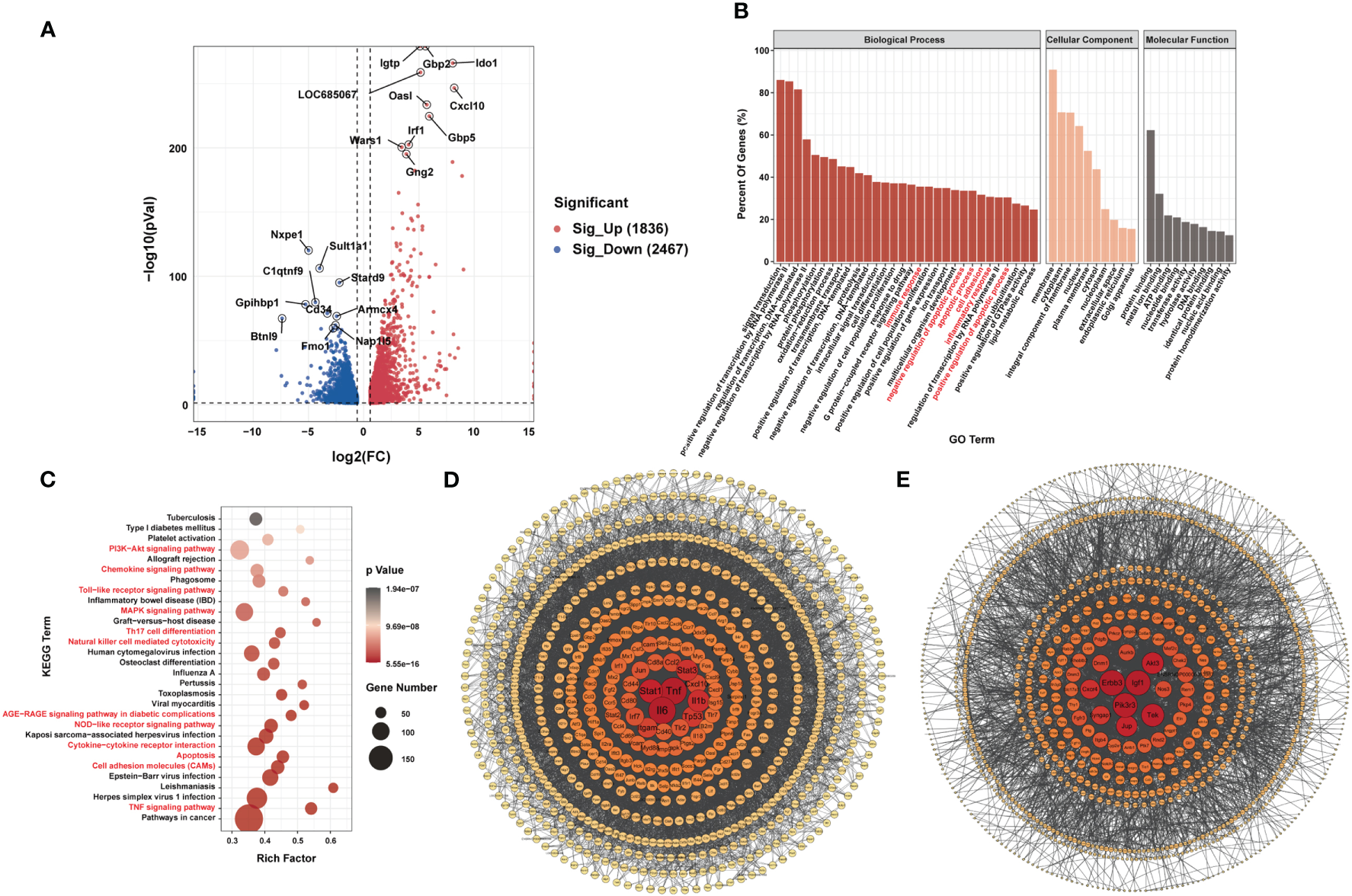
Differences in mRNA Expression in cardiac tissue of the LPS group and the control group. (A) Volcano map of differential expression genes (|FC|>=1.5, P<0.05). (B) Major gene ontology terms significantly enriched. (C) Top 30 significantly enriched pathways. (D) PPI network for up-regulated genes. (E) PPI network for down-regulated genes. n = 6 in each group.
3.5 Conjoint analysis of MeRIP-seq and RNA-seq data
Seventy-two genes with m6A hypermodification and 103 genes with m6A hypomodification were upregulated. In addition, 137 genes with m6A hypermodification and 133 genes with m6A hypomodification were downregulated. Notably, 4 upregulated genes had both m6A upregulated and downregulated modification at different sites, and 9 downregulated genes had both m6A upregulated and downregulated modification sites (Figure 5A).
Figure 5
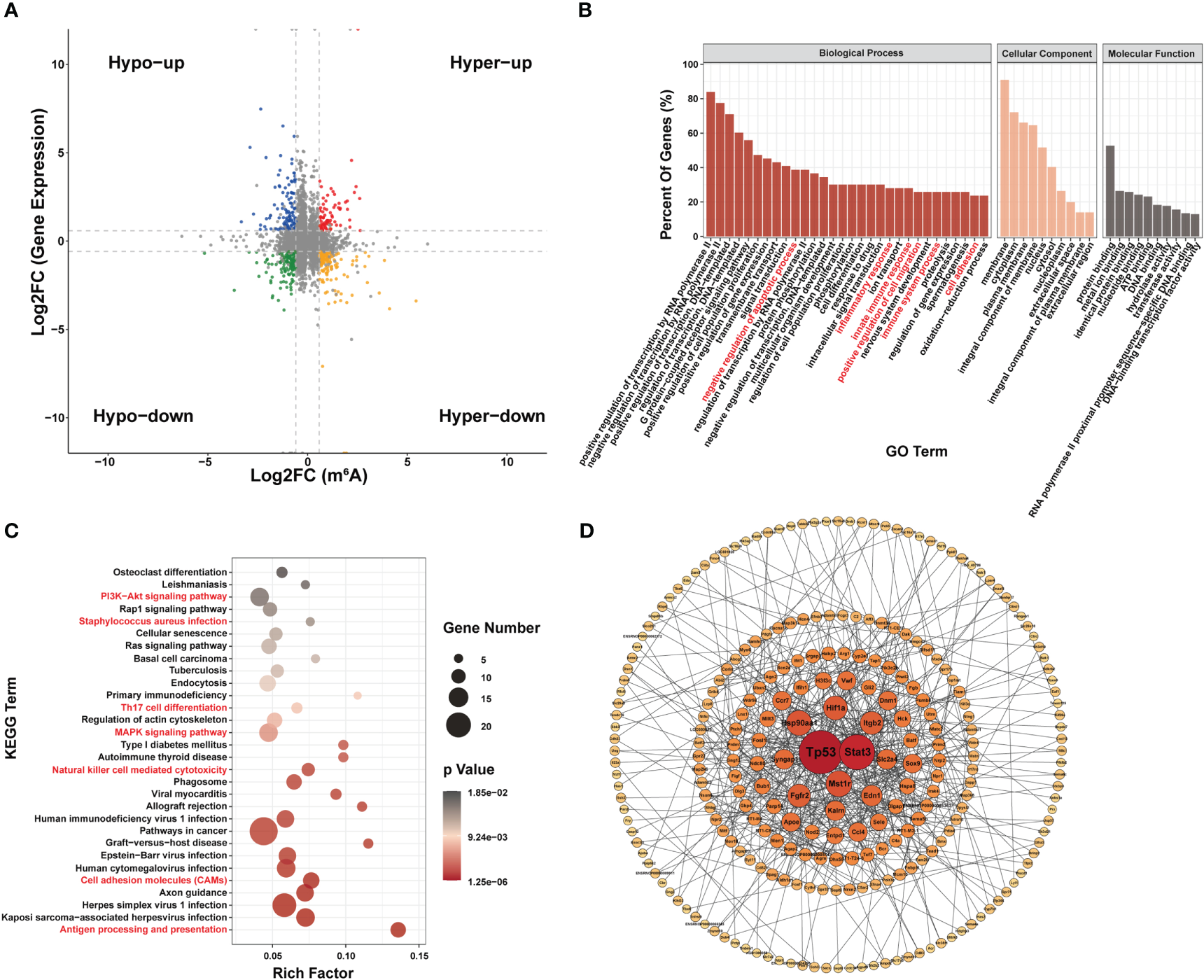
Conjoint analysis of MeRIP-seq and RNA-seq data. (A) Four-quadrant diagram of m6A modification and RNA expression, divided into hypo m6A modification with gene up regulation (Hypo-up), hyper m6A modification with gene up regulation (Hyper-up), hypo m6A modification with gene down regulation (Hypo-down) and hyper m6A modification with gene down regulation (Hyper-down). (B–D) Function analysis of gene with both differential m6A modification and differential expression; (B) Major gene ontology terms significantly enriched; (C) Top 30 significantly enriched pathways; (D) PPI network.
GO analysis suggested that genes with both differential m6A modification and differential expression were related to inflammatory responses and apoptosis (Figure 5B). KEGG terms related to (i) certain immune pathways, such as Staphylococcus aureus sepsis, Th17 cell differentiation, natural killer cell-mediated cytotoxicity, cell adhesion molecules and antigen processing and presentation and (ii) several pathways involved in apoptosis, such as PI3K-Akt signaling pathway and MAPK signaling pathway, were significantly enriched (Figure 5C). In addition, Tp53, stat3, Mst1r, and Hsp90aa1 were the most central proteins in the PPI network associated with immune response and apoptosis (Figure 5D).
Lasso screening identified Utrn, Stat3, Stard9, Pcdh12, LOC108348047, and Dnmt3a as key m6A-hypermodified genes (Figure 6A), whereas Tnfrsf9, Sele, Pcdh12, Myrip, Gng2, Gng12, Bdkrb2, Batf, and Agrn were identified as key m6A-hypomodified genes in the LPS group compared to the control group (Figure 6B). Furthermore, we analyzed the m6A modification sites with the Integrative Genomics Viewer (IGV) (22) (Figure 6C), verified the m6A modification and gene expression levels of leucine-rich repeat kinase 2 (Lrrk2), selectin E (Sele), TNF receptor superfamily member 9 (Tnfrsf9), bradykinin receptor B2 (Bdkrb2), and heat shock protein 90 alpha family class A member 1 (Hsp90aa1) in the LPS group and the control group (Figures 6D, E), which showed that they had the same expression tendencies, consistent with our sequencing data.
Figure 6
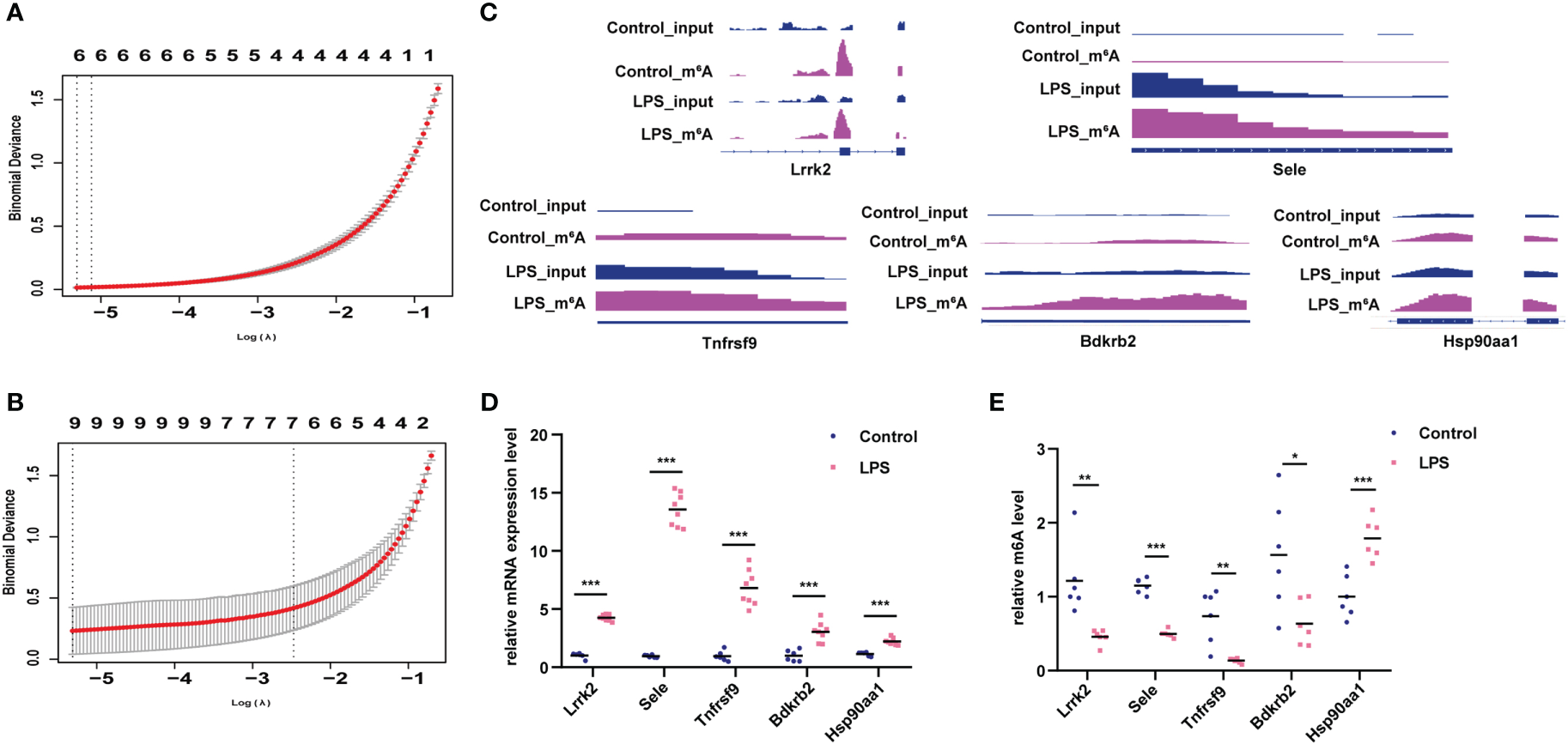
Verification of genes with differential m6A modification and differential expression. (A) Lasso analysis for genes with hyper-m6A modification. (B) Lasso analysis for genes with hypo-m6A modification. (C) m6A peak and gene expression visualization using IGV in the LPS group and control group. (D, E) Verification of m6A modification and gene expression levels of Lrrk2, Sele, Tnfrsf9, Bdkrb2, Hsp90aa1 in the LPS group compared to the control group. n = 6 in each group; *p < 0.05 compared to the control; **p < 0.01 compared to the control. ***p < 0.001 compared to control.
3.6 Changes in m6A-related enzymes in cardiac tissue of rats treated by LPS and control rats
We examined the expression of m6A methyltransferase (“writers”), including Mettl3, Mettl14, Wtap, Mettl16, Rbm15, Rbm15b, Zc3h13 and Virma and found that Mettl16 was upregulated whereas Rbm15 was significantly downregulated in the LPS group versus. the control group (Figure 7A). In addition, among the m6A demethylases (“erasers”) studied (Fto and Alkbh5), Fto was significantly downregulated in the LPS group (Figure 7B). When studying m6A-recognition factors (“readers”), we discovered that the YTH domain family protein, Ythdc2, was significantly upregulated in the LPS group versus. the control group and that heterogeneous nuclear ribonucleoprotein G (Hnrnpg) was downregulated (Figure 7C).
Figure 7
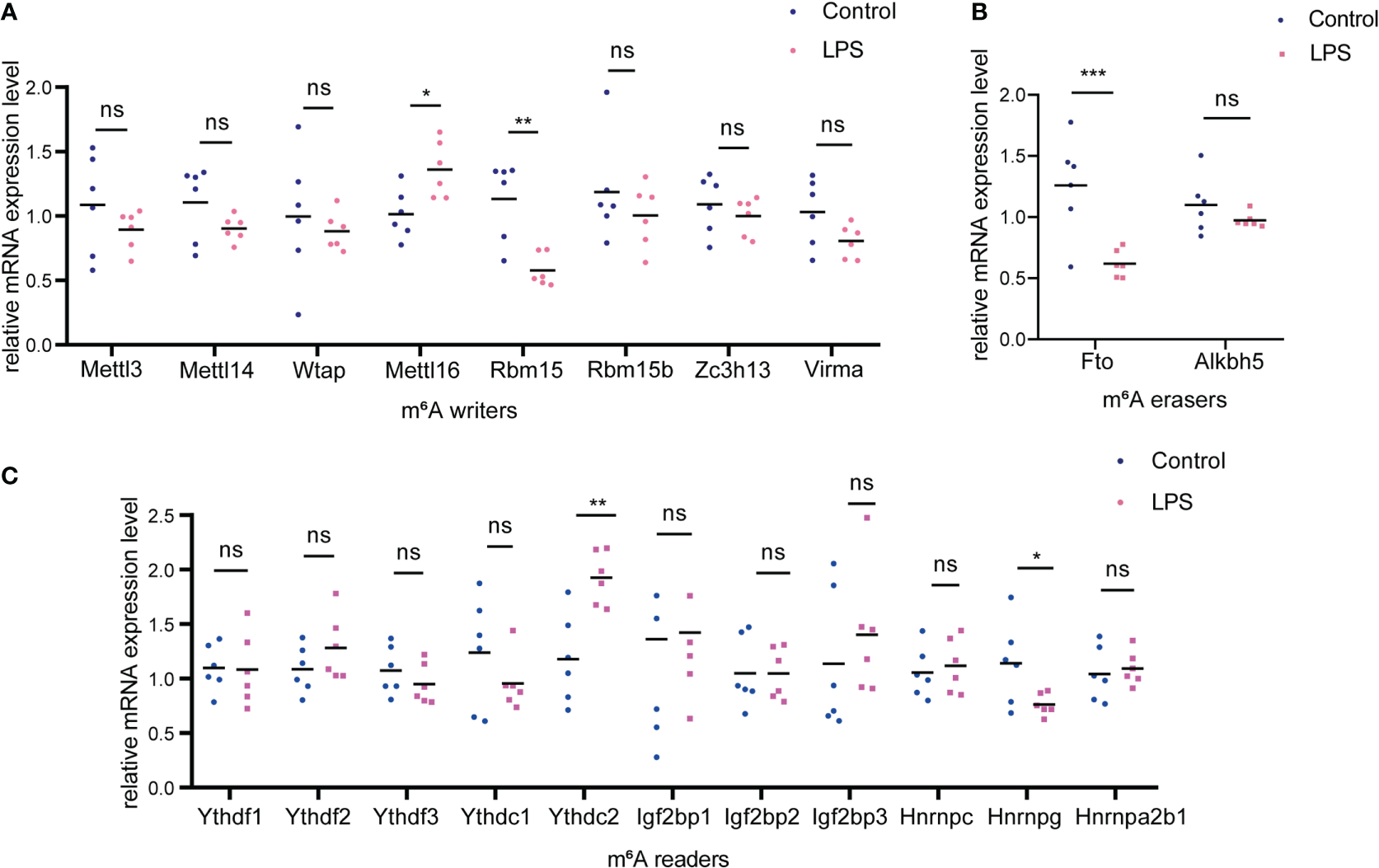
Changes of m6A-related enzymes in cardiac tissue of the LPS group compared to the control group. (A) mRNA expression levels of m6A methyltransferases, also named as “m6A writers” in the LPS group and the control group. (B) mRNA expression levels of m6A demethylases, also named as “erasers”; (C) mRNA expression levels of m6A recognition factors, called as “readers”. n=6 in each group; *p < 0.05 compared to the control; **p < 0.01 compared to the control. ***p < 0.001 compared to control. ns, no significance.
4 Discussion
The main features of endotoxaemia/sepsis-induced myocardial injury are ventricular dilatation, reduced ventricular contractility, and right and left ventricular dysfunction with a reduced response to volume infusion (23). However, no targeted treatments are available for this condition. Additionally, the molecular mechanisms underlying endotoxaemia/sepsis-induced myocardial injury remain unclear. Thus, it is necessary to explore the potential pathophysiological mechanisms underlying endotoxaemia/sepsis-induced myocardial injury. m6A modification is a widespread mRNA modification in eukaryotes that participates in post-transcriptional gene regulation (9). Regulators of m6A modification include m6A writers, erasers, and readers. m6A writers include Mettl3, Mettl14, Wtap, Rbm15/15b, Virma (Kiaa1429), and Zc3h13, which function together with m6A methyltransferase or cooperatively regulate m6A modification. Notably, dynamic m6A methylation can be reversed by nuclear m6A erasers, including Fto and Alkbh5 (24). In addition, m6A readers can regulate mRNA splicing, nuclear export, decay/degradation, translation, and stability by passing the m6A signal (9). The dysregulation of m6A modification and m6A-related enzymes have been found to be associated with cardiac diseases, such as myocardial infarction (10), heart failure (19) and cardiac hypertrophy (25), but the role of m6A modification in endotoxaemia/sepsis-induced myocardial injury is relatively unexplored.
In this study, we conducted the rat model of LPS-induced endotoxic shock. Strikingly, we did not find changes in ejection fraction and shortening fraction of echocardiography results. We speculated that the possible reasons were: 1. Myocardial injury was not serious enough; 2. Ejection fraction and shortening fraction were affected by reduced afterload (26, 27), resulting from vascular paralysis existed in our model. Therefore, the ejection fraction and shortening fraction may remain unchanged even with reduced cardiac systolic function. Some studies have proposed that the diagnosis of endotoxaemia/sepsis-induced myocardial injury based on EF values may underestimate its incidence and lead to increased mortality (26). In addition, the reductions in LVID, LVEDV, cardiac output, and stroke volume in this study may also be secondary to reduced preload. To investigate whether cardiac systolic dysfunction existed in the rat model of LPS induced endotoxic shock, we detected cardiac systolic function by an invasive hemodynamic study in previous studies (6, 7, 28). The MAP began decreasing 2 h after LPS injection, accompanied by significant decreases in the heart rate, peak rate of left ventricular pressure rise, and left ventricular peak rate of pressure decay, as well as a prolonged relaxation time constant. These studies showed that our model existed reduced cardiac systolic function and was stable. In this study, although we did not conduct invasive hemodynamics study, we did examine the decreased MAP as an indicator of endotoxic shock.
Moreover, as highly sensitive and specific markers of myocardial damage, clinical studies have proved that troponin (cTn) including c-TNI and c-TNT were correlated with a greater degree of left ventricular dysfunction, illness severity, and mortality of sepsis patients (23, 29, 30). CK-MB was also biochemical markers of myocyte necrosis (31). We found that the markers of myocardial injury (c-TNI and CK-MB) of serum were up-regulated significantly, indicating the presence of some extent of myocardial injury in the LPS group. In addition, inflammatory markers including IL-6, IL-18 and IL1β of cardiac tissue were also up-regulated significantly, indicating inflammatory injury in the LPS group. Furthermore, we have found increased cardiomyocyte apoptosis with LPS challenge by TUNEL staining in previous studies (6, 32). Therefore, there was myocardial injury in the LPS-induced endotoxic shock rats constructed in this study.
We performed MeRIP-seq and RNA-seq in cardiac tissue of rats treated by LPS and control rats. Bioinformatics analysis revealed that differentially expressed RNAs were mainly concentrated in inflammatory responses and apoptotic pathways. For example, the Hippo signaling pathway, AGE-RAGE signaling pathway in diabetic complications, MAPK signaling pathway and PI3K/Akt signaling pathway are closely associated with apoptosis (33–36). Our team have found infiltration of inflammatory cells and cardiomyocytes apoptosis existed in the cardiac tissue of rats with LPS-induced myocardial injury in previous study (7, 32). Other studies have also shown that inflammation and apoptosis are important pathophysiological processes of LPS-induced myocardial injury (37, 38). For example, TNFα, IL-6, IL1β and other inflammatory factors can directly or indirectly cause myocardial injury (39, 40). Chinese herbal preparations may alleviate LPS-induced myocardial injury by inhibiting myocardial apoptosis (41). These studies suggested the important role of apoptosis and inflammation in LPS-induced myocardial injury. Strikingly, we identified 512 hypermethylated and 658 hypomethylated m6A peaks in the LPS group compared to the control group by MeRIP-seq. GO and KEGG analyses showed that genes with differential m6A modification were also significantly associated with inflammatory responses terms and apoptosis terms, indicating potential important role in LPS-induced myocardial injury. In addition, Dubey et al. demonstrated that increased m6A-RNA methylation and Fto suppression was associated with myocardial inflammation and dysfunction during endotoxaemia in mice (42) and Shen et al. reported that Mettl3 knockdown repressed the inflammatory damage of LPS-induced cardiomyocytes by regulating m6A modification on HDAC4 mRNA (11). These studies indicated that m6A modification was involved in molecular regulation of inflammation in LPS-induced myocardial injury. In this study, we explored the differential m6A modification sites in cardiac tissue of rats with LPS-induced myocardial injury and control rats using deep sequencing, providing more potential regulation mechanisms and therapeutic targets. For example, we found that Stat3, Hif1a and Creb1 were the most interacting genes among the up-regulated m6A modified genes, while Tp53, Mapk11 and Hsp90aa1 were the most interacting genes among the down-regulated m6A modified genes by PPI network analysis. Tp53, Creb1, Mapk11 and Hif1a have been reported to be closely related to apoptosis (43–46), while Stat3 and Hsp90aa1 are classical molecules of inflammatory response (47, 48). Our sequencing results found that these genes were modified by m6A and what role does m6A play in these genes deserve further investigation in LPS-induced myocardial injury. Moreover, other apoptotic and inflammatory pathways in the MeRIP-seq results are also worthy of further exploration, including PI3K-Akt signaling pathway, Hippo signal pathway, AGE-RAGE signaling pathway in diabetes complications, MAPK signaling pathway, etc (33–36). These studies and results suggested that m6A modification may play an important role in LPS-induced myocardial injury through regulation of apoptosis and inflammatory response.
Notably, m6A methylation participates in the post-transcriptional regulation of genes and may regulate RNA-expression levels. Therefore, we performed combined analysis of MeRIP-seq and RNA-seq to identify potential genes that may be regulated by m6A modification at RNA level. Enrichment results of genes with both differential m6A modification and differential expression showed that inflammatory responses and apoptosis were significantly enriched. Moreover, Lasso analysis was used to identify key genes that may be regulated by m6A modification (49). These genes may play a more important role in LPS-induced myocardial injury and deserve further attention. Furthermore, we performed MeRIP-qPCR and qPCR to verify the m6A modification and RNA expression levels of Lrrk2, Sele, Tnfrsf9, Bdkrb2, and Hsp90aa1 in the LPS group and the control group. Lrrk2 is closely associated with the MAPK pathway and participates in many inflammatory diseases (50, 51). Recently, Liu et al. reported that Lrrk2 deficiency protected against cardiac remodeling under pressure overload (52). Sele, as an adhesion molecular, was reported to be involved in occurrence and development of various inflammatory diseases (53). Tnfrsf9 is involved in T cell and natural killer cell activation and cytokine production and plays a critical role in LPS-induced septic shock (54). Bdkrb2, a receptor for bradykinin which act as mediators of pain and inflammation, can activate a phosphatidylinositol-calcium second messenger system (55). Hsp90aa1, an inducible molecular chaperone that functions as a homodimer, can bind LPS and mediates LPS-induced inflammatory response, including Tnf secretion by monocytes (48). In addition, GO analysis revealed that Tnfrsf9, Bdkrb2, and Hsp90aa1 were significantly enriched in the apoptosis pathway. These genes with both differential expression and differential m6A modification provide a new idea for molecular regulation mechanism of LPS-induced myocardial injury. Furthermore, we found Mettl16, Rbm15, Fto, Ythdc2, and Hnrnpg were differentially expressed which may participate in regulation genes with differentially m6A modification in cardiac tissue of rats with LPS-induced myocardial injury compared to control rats, providing a new perspective for further exploring the regulation mechanism of m6A modification.
5 Conclusion
In this study, we established m6A modification map in cardiac tissue of rats with LPS-induced myocardial injury and found that genes with differential expression and differential m6A modification were closely related to inflammation and apoptosis, which enriched the pathophysiological processes of LPS-induced myocardial injury and provided new ideas for more researchers. This study also has some limitations, such as the lack of deeper mechanistic research, the lack of exploration of m6A modification changes at early stage of endotoxaemia and MeRIP-seq including only mRNA but not non-coding RNAs. More research is needed to explore these aspects in the future.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJNA911952 (SRA).
Ethics statement
The animal study was reviewed and approved by the Ethics Committee of the Shengjing Hospital of China Medical University (Shenyang, China; approval number 2022PS854K).
Author contributions
C-FL performed study concept and design. WW performed development of methodology and writing. T-NZ and NY performed review and revision of the paper. RW, Y-JW, B-LZ, and HY provided acquisition, analysis and interpretation of data, and statistical analysis. All authors read and approved the final paper.
Funding
This work was supported by the National Natural Science Foundation of China (81971810 to C-LF), Liaoning Province Science and Technology Major Special Project (No.2020JH1/10300001 to C-LF), Shenyang Science and Technology Plan Project (20-205-4-002 to C-LF).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1122317/full#supplementary-material
Supplementary Figure 1The MAP changes in rats after LPS administration. The rats were divided into control group and LPS group, and the changes of blood pressure over time were measured by femoral artery intubation, including 0h, 2h, 6h, and 12h after injection of LPS or normal saline. n=6 in each group; ***p < 0.001 compared to control.
Supplementary Figure 2Basic characteristics of m6A peaks in the LPS group and the control group. (A) Number of m6A peaks in each chromosome in both groups. (B) Number of m6A peaks in each gene in both groups. (C) Distribution of m6A peaks in 5’UTR, CDS and 3’UTR in both groups.
Supplementary Figure 3Correlation analysis of RNA-seq in the LPS group and the control group. (A) Principal component analysis (PCA) of RNA-seq in both groups. (B) Pearson correlation heatmap of RNA-seq in in both groups.
References
1
Singer M Deutschman CS Seymour CW Shankar-Hari M Annane D Bauer M et al . The third international consensus definitions for sepsis and septic shock (Sepsis-3). Jama (2016) 315:801–10. doi: 10.1001/jama.2016.0287
2
Rudd KE Johnson SC Agesa KM Shackelford KA Tsoi D Kievlan DR et al . Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the global burden of disease study. Lancet (London England) (2020) 395:200–11. doi: 10.1016/s0140-6736(19)32989-7
3
Weiss SL Fitzgerald JC Pappachan J Wheeler D Jaramillo-Bustamante JC Salloo A et al . Global epidemiology of pediatric severe sepsis: the sepsis prevalence, outcomes, and therapies study. Am J Respir Crit Care Med (2015) 191:1147–57. doi: 10.1164/rccm.201412-2323OC
4
Beesley SJ Weber G Sarge T Nikravan S Grissom CK Lanspa MJ et al . Septic cardiomyopathy. Crit Care Med (2018) 46:625–34. doi: 10.1097/ccm.0000000000002851
5
Kellum JA Formeck CL Kernan KF Gómez H Carcillo JA . Subtypes and mimics of sepsis. Crit Care Clinics (2022) 38:195–211. doi: 10.1016/j.ccc.2021.11.013
6
Zhang TN Goodwin JE Liu B Li D Wen R Yang N et al . Characterization of long noncoding RNA and mRNA profiles in sepsis-induced myocardial depression. Mol Ther Nucleic Acids (2019) 17:852–66. doi: 10.1016/j.omtn.2019.07.020
7
Zhang T Liu CF Zhang TN Wen R Song WL . Overexpression of peroxisome proliferator-activated receptor γ coactivator 1-α protects cardiomyocytes from lipopolysaccharide-induced mitochondrial damage and apoptosis. Inflammation (2020) 43:1806–20. doi: 10.1007/s10753-020-01255-4
8
Zhang B Liu C Yang N Wang X . A comparative study of changes of autophagy in rat models of CLP versus LPS induced sepsis. Exp Ther Med (2017) 14:2194–200. doi: 10.3892/etm.2017.4758
9
Deng LJ Deng WQ Fan SR Chen MF Qi M Lyu WY et al . m6A modification: recent advances, anticancer targeted drug discovery and beyond. Mol Cancer (2022) 21:52. doi: 10.1186/s12943-022-01510-2
10
Mathiyalagan P Adamiak M Mayourian J Sassi Y Liang Y Agarwal N et al . FTO-dependent N(6)-methyladenosine regulates cardiac function during remodeling and repair. Circulation (2019) 139:518–32. doi: 10.1161/circulationaha.118.033794
11
Shen H Xie K Li M Yang Q Wang X . N(6)-methyladenosine (m(6)A) methyltransferase METTL3 regulates sepsis-induced myocardial injury through IGF2BP1/HDAC4 dependent manner. Cell Death Discovery (2022) 8:322. doi: 10.1038/s41420-022-01099-x
12
Chen S Zhou Y Chen Y Gu J . Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinf (Oxford England) (2018) 34:i884–i90. doi: 10.1093/bioinformatics/bty560
13
Kim D Langmead B Salzberg SL . HISAT: a fast spliced aligner with low memory requirements. Nat Methods (2015) 12:357–60. doi: 10.1038/nmeth.3317
14
Meng J Lu Z Liu H Zhang L Zhang S Chen Y et al . A protocol for RNA methylation differential analysis with MeRIP-seq data and exomePeak R/Bioconductor package. Methods (San Diego Calif.) (2014) 69:274–81. doi: 10.1016/j.ymeth.2014.06.008
15
Heinz S Benner C Spann N Bertolino E Lin YC Laslo P et al . Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and b cell identities. Mol Cell (2010) 38:576–89. doi: 10.1016/j.molcel.2010.05.004
16
Pertea M Pertea GM Antonescu CM Chang TC Mendell JT Salzberg SL . StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol (2015) 33:290–5. doi: 10.1038/nbt.3122
17
Szklarczyk D Gable AL Nastou KC Lyon D Kirsch R Pyysalo S et al . The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res (2021) 49:D605–d12. doi: 10.1093/nar/gkaa1074
18
Shannon P Markiel A Ozier O Baliga NS Wang JT Ramage D et al . Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res (2003) 13:2498–504. doi: 10.1101/gr.1239303
19
Berulava T Buchholz E Elerdashvili V Pena T Islam MR Lbik D et al . Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart failure (2020) 22:54–66. doi: 10.1002/ejhf.1672
20
The Gene Ontology Consortium . The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res (2019) 47:D330–d8. doi: 10.1093/nar/gky1055
21
Kanehisa M Furumichi M Tanabe M Sato Y Morishima K . KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res (2017) 45:D353–d61. doi: 10.1093/nar/gkw1092
22
Thorvaldsdóttir H Robinson JT Mesirov JP . Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Briefings Bioinf (2013) 14:178–92. doi: 10.1093/bib/bbs017
23
Martin L Derwall M Al Zoubi S Zechendorf E Reuter DA Thiemermann C et al . The septic heart: current understanding of molecular mechanisms and clinical implications. Chest (2019) 155:427–37. doi: 10.1016/j.chest.2018.08.1037
24
Jiang X Liu B Nie Z Duan L Xiong Q Jin Z et al . The role of m6A modification in the biological functions and diseases. Signal transduction targeted Ther (2021) 6:74. doi: 10.1038/s41392-020-00450-x
25
Dorn LE Lasman L Chen J Xu X Hund TJ Medvedovic M et al . The N(6)-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation (2019) 139:533–45. doi: 10.1161/circulationaha.118.036146
26
L'Heureux M Sternberg M Brath L Turlington J Kashiouris MG . Sepsis-induced cardiomyopathy: a comprehensive review. Curr Cardiol Rep (2020) 22:35. doi: 10.1007/s11886-020-01277-2
27
Robotham JL Takata M Berman M Harasawa Y . Ejection fraction revisited. Anesthesiology (1991) 74:172–83. doi: 10.1097/00000542-199101000-00026
28
Yang N Shi XL Zhang BL Rong J Zhang TN Xu W et al . The trend of β3-adrenergic receptor in the development of septic myocardial depression: a lipopolysaccharide-induced rat septic shock model. Cardiology (2018) 139:234–44. doi: 10.1159/000487126
29
Masson S Caironi P Fanizza C Carrer S Caricato A Fassini P et al . Sequential n-terminal pro-B-Type natriuretic peptide and high-sensitivity cardiac troponin measurements during albumin replacement in patients with severe sepsis or septic shock. Crit Care Med (2016) 44:707–16. doi: 10.1097/ccm.0000000000001473
30
Bessière F Khenifer S Dubourg J Durieu I Lega JC . Prognostic value of troponins in sepsis: a meta-analysis. Intensive Care Med (2013) 39:1181–9. doi: 10.1007/s00134-013-2902-3
31
Yilmaz A Yalta K Turgut OO Yilmaz MB Ozyol A Kendirlioglu O et al . Clinical importance of elevated CK-MB and troponin I levels in congestive heart failure. Adv Ther (2006) 23:1060–7. doi: 10.1007/bf02850226
32
Zhu XG Zhang TN Wen R Liu CF . Overexpression of miR-150-5p alleviates apoptosis in sepsis-induced myocardial depression. BioMed Res Int (2020) 2020:3023186. doi: 10.1155/2020/3023186
33
Xie J Wang Y Ai D Yao L Jiang H . The role of the Hippo pathway in heart disease. FEBS J (2021) 289:5819–33. doi: 10.1111/febs.16092
34
Papaconstantinou J . The role of signaling pathways of inflammation and oxidative stress in development of senescence and aging phenotypes in cardiovascular disease. Cells (2019) 8:1383. doi: 10.3390/cells8111383
35
Zhang Q Wang L Wang S Cheng H Xu L Pei G et al . Signaling pathways and targeted therapy for myocardial infarction. Signal transduction targeted Ther (2022) 7:78. doi: 10.1038/s41392-022-00925-z
36
Walkowski B Kleibert M Majka M Wojciechowska M . Insight into the role of the PI3K/Akt pathway in ischemic injury and post-infarct left ventricular remodeling in normal and diabetic heart. Cells (2022) 11:1553. doi: 10.3390/cells11091553
37
Li N Zhou H Wu H Wu Q Duan M Deng W et al . STING-IRF3 contributes to lipopolysaccharide-induced cardiac dysfunction, inflammation, apoptosis and pyroptosis by activating NLRP3. Redox Biol (2019) 24:101215. doi: 10.1016/j.redox.2019.101215
38
Chen L Liu P Feng X Ma C . Salidroside suppressing LPS-induced myocardial injury by inhibiting ROS-mediated PI3K/Akt/mTOR pathway in vitro and in vivo. J Cell Mol Med (2017) 21:3178–89. doi: 10.1111/jcmm.12871
39
Poveda-Jaramillo R . Heart dysfunction in sepsis. J cardiothoracic Vasc Anesth (2021) 35:298–309. doi: 10.1053/j.jvca.2020.07.026
40
Busch K Kny M Huang N Klassert TE Stock M Hahn A et al . Inhibition of the NLRP3/IL-1β axis protects against sepsis-induced cardiomyopathy. J cachexia sarcopenia Muscle (2021) 12:1653–68. doi: 10.1002/jcsm.12763
41
Xu P Zhang WQ Xie J Wen YS Zhang GX Lu SQ . Shenfu injection prevents sepsis-induced myocardial injury by inhibiting mitochondrial apoptosis. J ethnopharmacol (2020) 261:113068. doi: 10.1016/j.jep.2020.113068
42
Dubey PK Patil M Singh S Dubey S Ahuja P Verma SK et al . Increased m6A-RNA methylation and FTO suppression is associated with myocardial inflammation and dysfunction during endotoxemia in mice. Mol Cell Biochem (2022) 477:129–41. doi: 10.1007/s11010-021-04267-2
43
Aubrey BJ Kelly GL Janic A Herold MJ Strasser A . How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death differentiation (2018) 25:104–13. doi: 10.1038/cdd.2017.169
44
Niu RZ Wang LQ Yang W Sun LZ Tao J Sun H et al . MicroRNA-582-5p targeting Creb1 modulates apoptosis in cardiomyocytes hypoxia/reperfusion-induced injury. Immunity Inflammation Dis (2022) 10:e708. doi: 10.1002/iid3.708
45
Zhao X Liu L Li R Wei X Luan W Liu P et al . Hypoxia-inducible factor 1-α (HIF-1α) induces apoptosis of human uterosacral ligament fibroblasts through the death receptor and mitochondrial pathways. Med Sci monitor Int Med J Exp Clin Res (2018) 24:8722–33. doi: 10.12659/msm.913384
46
Chun Yang X Hui Zhao D Bond Lau W Qiang Liu K Yu Tian J Chao Cheng Z et al . lncRNA ENSMUST00000134285 increases MAPK11 activity, regulating aging-related myocardial apoptosis. journals gerontol Ser A Biol Sci Med Sci (2018) 73:1010–7. doi: 10.1093/gerona/gly020
47
Hillmer EJ Zhang H Li HS Watowich SS . STAT3 signaling in immunity. Cytokine Growth factor Rev (2016) 31:1–15. doi: 10.1016/j.cytogfr.2016.05.001
48
Triantafilou K Triantafilou M Dedrick RL . A CD14-independent LPS receptor cluster. Nat Immunol (2001) 2:338–45. doi: 10.1038/86342
49
Zhang Y Xia R Lv M Li Z Jin L Chen X et al . Machine-learning algorithm-based prediction of diagnostic gene biomarkers related to immune infiltration in patients with chronic obstructive pulmonary disease. Front Immunol (2022) 13:740513. doi: 10.3389/fimmu.2022.740513
50
Li T Ning B Kong L Dai B He X Thomas JM et al . A LRRK2 GTP binding inhibitor, 68, reduces LPS-induced signaling events and TNF-α release in human lymphoblasts. Cells (2021) 10:480. doi: 10.3390/cells10020480
51
Cao J Zhuang Y Zhang J Zhang Z Yuan S Zhang P et al . Leucine-rich repeat kinase 2 aggravates secondary brain injury induced by intracerebral hemorrhage in rats by regulating the P38 MAPK/Drosha pathway. Neurobiol Dis (2018) 119:53–64. doi: 10.1016/j.nbd.2018.07.024
52
Liu Y Hao C Zhang W Liu Y Guo S Li R et al . Leucine-rich repeat kinase-2 deficiency protected against cardiac remodelling in mice via regulating autophagy formation and degradation. J advanced Res (2022) 37:107–17. doi: 10.1016/j.jare.2021.07.004
53
Meng ZD Wang XL Du TP Wang Y Qin GF Zhao HB et al . Effects of RNA interference-mediated e-selectin gene silencing on cell adhesion molecule expression and cell-cell adhesion in vascular endothelial cells in mice with immunologic contact urticaria. J Cell Biochem (2019) 120:115–25. doi: 10.1002/jcb.27150
54
Vinay DS Choi BK Bae JS Kim WY Gebhardt BM Kwon BS . CD137-deficient mice have reduced NK/NKT cell numbers and function, are resistant to lipopolysaccharide-induced shock syndromes, and have lower IL-4 responses. J Immunol (Baltimore Md. 1950) (2004) 173:4218–29. doi: 10.4049/jimmunol.173.6.4218
55
Chen Z Deddish PA Minshall RD Becker RP Erdös EG Tan F . Human ACE and bradykinin B2 receptors form a complex at the plasma membrane. FASEB J (2006) 20:2261–70. doi: 10.1096/fj.06-6113com
Summary
Keywords
m6A, cardiac, inflammation, apoptosis, endotoxaemia
Citation
Wang W, Zhang T-N, Yang N, Wen R, Wang Y-J, Zhang B-L, Yang Y-H and Liu C-F (2023) Transcriptome-wide identification of altered RNA m6A profiles in cardiac tissue of rats with LPS-induced myocardial injury. Front. Immunol. 14:1122317. doi: 10.3389/fimmu.2023.1122317
Received
12 December 2022
Accepted
05 May 2023
Published
19 May 2023
Volume
14 - 2023
Edited by
Jianmin Chen, Queen Mary University of London, United Kingdom
Reviewed by
Jing Liu, University of Illinois Chicago, United States; Xingming Ma, Xihua University, China
Updates
Copyright
© 2023 Wang, Zhang, Yang, Wen, Wang, Zhang, Yang and Liu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun-Feng Liu, zhliu258@hotmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.