- 1Department of Animal Science, College of Agriculture and Natural Resources, University of Tehran, Karaj, Iran
- 2Faculty of Agricultural Sciences and Engineering, University of Tehran, Karaj, Iran
- 3Dietary Supplements and Probiotic Research Center, Alborz University of Medical Sciences, Karaj, Iran
- 4Department of Physiology and Pharmacology, School of Medicine, Alborz University of Medical Sciences, Karaj, Iran
- 5School of Pharmacy, Shahid Beheshty University of Medical Sciences, Tehran, Iran
- 6Department of Biochemistry, Faculty of Advanced Sciences and Technology, Tehran Medical Sciences, Islamic Azad University, Tehran, Iran
- 7Department of Pediatric, School of Medicine, Amir al momenin Hospital, Zabol University of Medical Sciences, Zabol, Iran
- 8Zoology Department, Faculty of Science, Mansoura University, Mansoura, Egypt
- 9Virology Department, Faculty of Veterinary Medicine, University of Tehran, Tehran, Iran
- 10Department of Dental Surgery, Mashhad University of Medical Sciences, Mashhad, Iran
- 11Khorasan Covid-19 Scientific Committee, Mashhad, Iran
Coronavirus disease 2019 (COVID-19) is a severe respiratory disease caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that affects the lower and upper respiratory tract in humans. SARS-CoV-2 infection is associated with the induction of a cascade of uncontrolled inflammatory responses in the host, ultimately leading to hyperinflammation or cytokine storm. Indeed, cytokine storm is a hallmark of SARS-CoV-2 immunopathogenesis, directly related to the severity of the disease and mortality in COVID-19 patients. Considering the lack of any definitive treatment for COVID-19, targeting key inflammatory factors to regulate the inflammatory response in COVID-19 patients could be a fundamental step to developing effective therapeutic strategies against SARS-CoV-2 infection. Currently, in addition to well-defined metabolic actions, especially lipid metabolism and glucose utilization, there is growing evidence of a central role of the ligand-dependent nuclear receptors and peroxisome proliferator-activated receptors (PPARs) including PPARα, PPARβ/δ, and PPARγ in the control of inflammatory signals in various human inflammatory diseases. This makes them attractive targets for developing therapeutic approaches to control/suppress the hyperinflammatory response in patients with severe COVID-19. In this review, we (1) investigate the anti-inflammatory mechanisms mediated by PPARs and their ligands during SARS-CoV-2 infection, and (2) on the basis of the recent literature, highlight the importance of PPAR subtypes for the development of promising therapeutic approaches against the cytokine storm in severe COVID-19 patients.
1 Introduction
Coronavirus disease 2019 (COVID-19) is an infectious and severe respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 is a positive sense single-stranded RNA beta-coronavirus that infects the lower and upper respiratory tract and has recently affected millions of people worldwide (1–3). Primary symptoms of COVID-19 include fever, cough, pneumonia, and shortness of breath, and histological pictures of this disease are characterized by mononuclear inflammatory cells, severe pneumocyte hyperplasia, interstitial thickening, hyaline membrane formation, and prominent alveolar damage with eosinophilic exudates (4, 5).
During COVID-19, a cascade of inflammatory pathways is activated, leading to massive cytokine release from the host immune system in response to SARS-CoV-2 infection (6, 7). In this regard, the vast increase in the secretion of circulating proinflammatory cytokines such as tumor necrosis factors (TNFs), interleukins (ILs), chemokines, and interferons (IFNs) leads to the exacerbation of the host inflammatory response to the pathogen. This exacerbation in the host’s inflammatory response increases the severity of the disease (8–10). This hyperinflammation or imbalanced inflammation during SARS-CoV-2 infection is called “cytokine storm”, which is one of the main hallmarks of the deterioration of the COVID-19 immunopathogenesis and triggers acute respiratory distress syndrome (ARDS), multi-organ failure (MOF), acute lung injury (ALI), decreased lung function, and finally, death of the host (11, 12). In this context, in recent years, various clinical and omics-based studies have investigated the molecular mechanisms behind the SARS-CoV-2 infection in different disease stages and different tissues (13–17). Surprisingly, most of these studies have observed the activation of inflammatory mechanisms and hyperinflammation in COVID-19 patients. Therefore, developing effective therapeutic strategies by targeting critical factors in regulating host inflammatory response can provide a potential and promising solution for the survival of COVID-19 patients, especially the prevention of cytokine storms (18–20). The peroxisome proliferator-activated receptors (PPARs) are a subgroup of ligand-activated transcription factors and members of the nuclear receptor superfamily that play a crucial role in regulating energy balance, carbohydrate and lipid metabolism, cell growth, and differentiation (21, 22). PPARs can regulate the transcriptional activity of target genes by two different mechanisms (1): binding to the promoter region of target genes with DNA sequences known as peroxisome proliferator response elements (PPREs) as a ligand-dependent transcription factor, and (2) controlling gene expression through association with PPRE-independent activator proteins (21, 23). Several previous reports highlighted the core role of PPARs in many human diseases, such as different types of cancer (24, 25), atherosclerosis (26), and type 2 diabetes (27, 28).
Interestingly, in addition to the central roles of PPARs in regulating energy homeostasis, such as fatty-acid metabolism and glucose utilization, growing evidence suggests that members of the nuclear receptor superfamily, such as PPARs, also have significant regulatory effects on inflammatory processes (29). Indeed, extensive research has proven that PPARs have potential anti-inflammatory effects during inflammation-related disease (30, 31). In this regard, previous literature, based on available evidence, has suggested that subtypes of PPARs exert their anti-inflammatory effects and subsequently control the host’s inflammatory response through different mechanisms such as successful competition with other inflammatory transcription factors for the recruitment of essential and shared co-activator proteins, inhibition of binding of inflammatory transcription factors such as AP-1, nuclear factor-κB (NF-κB), NFAT, and STATs to their response elements through direct physical protein-protein interaction, blocking MAPK-induced signaling cascades, preventing the clearance of proinflammatory genes co-repressors, and upregulation in the expression of anti-inflammatory genes (32). For example, it has been discussed that the activation of PPARs during inflammatory bowel disease (IBD) leads to the suppression of the main pathways of inflammation, such as NF-κB signaling. Subsequently, the activation of PPARs inhibits the production of proinflammatory cytokines such as TNF-α, IL6, and IL1B. Therefore, it was concluded that anti-inflammatory responses induced by the activation of PPARs might restore the physio-pathological imbalance associated with this disorder (21).
The interference of viral infections such as SARS-CoV-2 in the PPARs signaling is a completely new issue and interest in this area has been very motivated by the COVID-19 pandemic. Emerging studies show that SARS-CoV-2, by modulating PPAR subtypes, leads to metabolic changes (especially lipid metabolism) and exacerbation of pulmonary inflammation in lung epithelial cells of COVID-19 patients (33). Therefore, these findings have suggested that the use of agonists of PPARs with the aim of their activation may be a useful therapeutic strategy to reverse the inflammatory and metabolic changes caused by SARS-CoV-2 infection (34). In this regard, it has been reported that several natural ligands of PPARs, such as turmeric, docosahexaenoic acid (DHA), and eicosapentaenoic acid (EPA), lead to a decrease in the production of proinflammatory cytokines through interaction with PPARs and then induction of their activity (35, 36) For example, a very recent study has identified possible mechanisms by which the PPARα agonist palmitoylethanolamide (PEA) antagonizes the NF-κB signaling pathway and subsequently reduces the production of TNF-α, IL1B, and other inflammatory mediators such as inducible nitric oxide synthase (iNOS) and COX2 through selective activation of PPARα in cultured murine alveolar macrophages during SARS-CoV-2 infection (37). Moreover, it has been suggested that synthetic agonists of PPARγ, such as thiazolidinediones (TZDs), like pioglitazone, are anti-inflammatory drugs with ameliorative effects on severe viral pneumonia-like COVID-19 (38). On the other hand, by integrating different transcriptome datasets with computational network-based systems biology methods, promising therapeutic targets, including PPARα and PPARγ, have been identified for the modulation of inflammatory processes caused by COVID-19 (39, 40). Therefore, PPARs and their ligands have crucial therapeutic potential with key immunomodulatory effects on inflammatory mechanisms and cytokine/chemokine production during infectious and inflammation-related diseases such as the COVID-19 pandemic.
Nevertheless, considering the importance of immunopathology’s role of the inflammatory response in COVID-19 patients and the role of PPARs in controlling inflammation, in this review, we (1) provide a summary of general information about PPARs such as subtypes, structure, tissue expression, and function (2), investigate the molecular mechanisms of the exacerbation of the host inflammatory response during COVID-19 (3), describe the anti-inflammatory mechanisms mediated by PPARs, and (4) discuss the anti-inflammatory roles of PPAR subtypes during COVID-19 pandemic on the basis of the recent clinical and omics-based literature.
2 PPARs: Subtypes, structure, tissue expression, and function
2.1 PPAR subtypes and structure
The PPARs are ligand-dependent/activated transcription factors, members of the nuclear- hormone-receptor superfamily (including the receptors for thyroid hormone, vitamin D, ecdysone, retinoic acids, and some orphan receptors) that transduce a wide range of signals, including environmental, nutritional, and inflammatory events, to a set of cellular responses at the transcriptional gene level; they were named due to their joint property in increasing the number and activity of peroxisomes (41–43). So far, three isoforms of PPARs, namely, PPARα (NR1C1), PPARβ/δ (NR1C2), and PPARγ (NR1C3), have been identified in vertebrates (including human, mouse, rat, hamster, and Xenopus), which are encoded by distinct genes on different chromosomes. They have shown a high degree of sequence and structural homology (Figure 1A) but different tissue distribution, ligand specificity, and regulatory activities (44–46).
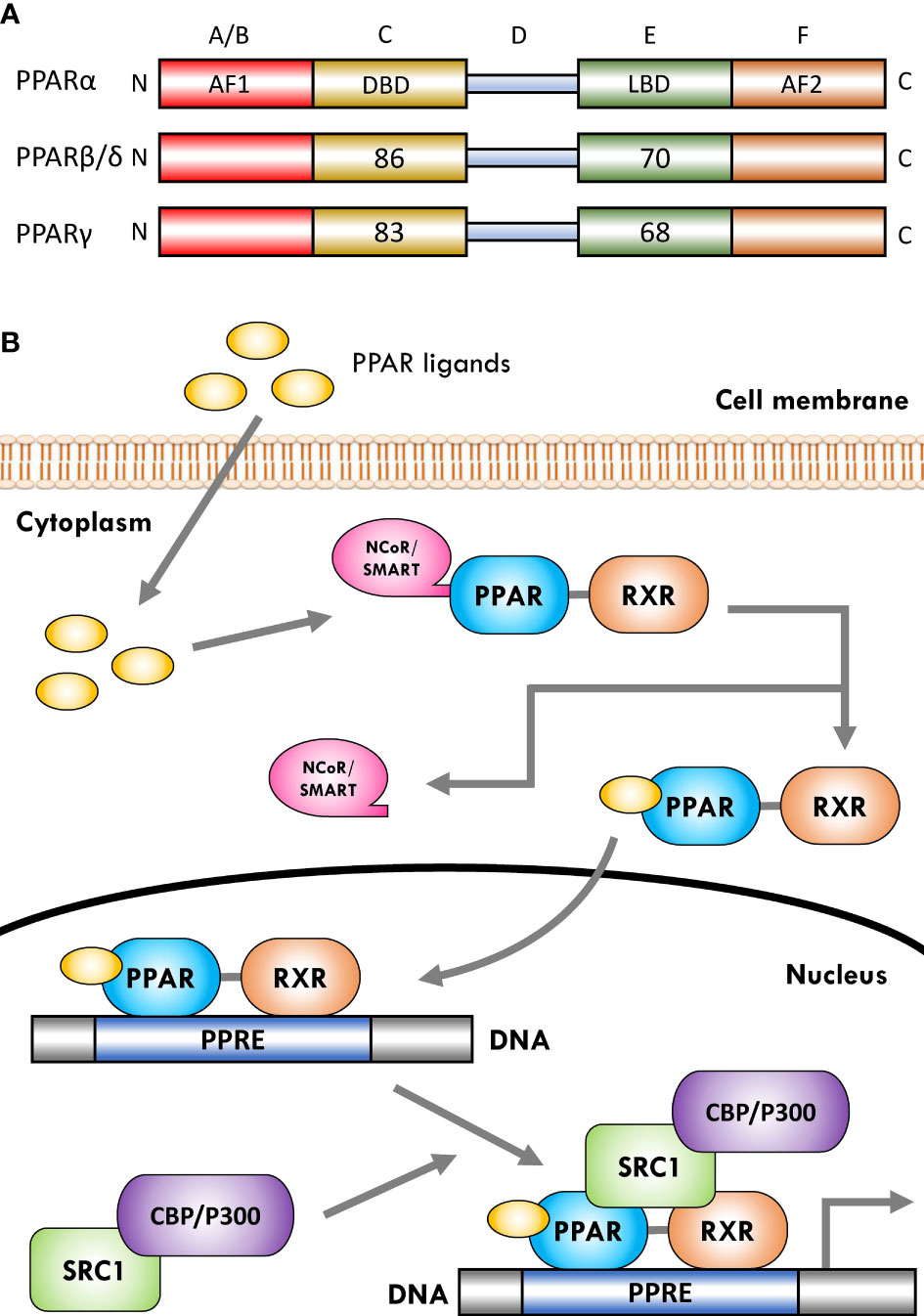
Figure 1 Schematic representation of peroxisome proliferator-activated receptor (PPAR) structure and ligand-induced activation. (A) The PPARs are composed of five distinct regions or domains (1): the ligand-independent activation domain of AF1 located in the N-terminus (amino-terminal A/B domain), which is responsible for receptor phosphorylation (2); the highly conserved DNA-binding domain (DBD) in the C region that contains two zinc finger motifs responsible for receptor binding to DNA targets on the peroxisome proliferator hormone response elements (PPREs) of PPARs target genes (3); a variable hinge region in the D domain that is the docking site for co-factors (4); a moderately conserved ligand-binding domain (LBD) in the E region that is responsible for the ligand specificity and activation, as well as for dimerization of the receptor with the 9-cis-retinoic acid receptor (RXR) (5); an AF2 ligand-dependent activation domain in the C-terminus (carboxyl-terminal in F domain) that is crucial for the recruitment of PPAR co-activators. Numbers shown in C and E regions indicate the percentage amino-acid identity of DBD and LBD of human PPARβ/δ and PPARγ compared to human PPARα. (B) Several co-activator or co-repressor factors affect the activity of PPARs, which can stimulate or inhibit the function of the receptor, respectively. When PPARs are in a non-ligand-bound state in solution (inactive mode), all three PPAR isoforms can bind transcription co-repressors in a DNA-independent manner. These co-repressors, such as nuclear receptor co-repressor/silencing mediators for retinoid and thyroid hormone receptors (NCoR/SMRT), suppress gene transcription by interacting with histone deacetylases (HDACs). The binding of ligands to the PPAR–RXR heterodimer causes the exchange of co-repressors with co-activators, thereby converting PPARs from an inactive state to an active state. Receptor activation generally occurs after agonist binding to the LBD. Following ligand binding and initiation of receptor phosphorylation, PPARs dissociate the co-repressor complex. Then, the ligand–heterodimer (ligand–PPAR–RXR) complex binds to the target DNA promoter through a PPRE. Next, in order to allow the transcriptional machinery to gain access to the promoter region, PPARs bind specific co-activator complexes, such as steroid receptor activator 1 (SRC1) and cAMP response element-binding (CREB)-binding protein (CBP)/p300, which have acetyltransferase activity. Subsequently, they regulate the transcription of various genes that play a key role in various physiological processes.
2.2 Tissue distribution and function
In recent years, various in vitro and in vivo studies have reported that all isoforms of PPARs primarily regulate lipid and glucose metabolism and have additional regulatory roles in cell proliferation and differentiation, vascular homeostasis and atherosclerosis, cancer, and the immune system (38, 47). In addition to the mentioned activities, it is thought that the activation of PPAR subtypes reduces the expression of proinflammatory cytokines and inflammatory cell functions, exerting significant anti-inflammatory properties (48). PPARα is the first known PPAR that was initially cloned from a mouse liver complementary DNA library as a nuclear receptor that mediates the effects of an endogenous group and xenobiotic compounds known as peroxisome proliferators (PPs) (31, 49). This subtype of PPARs is highly expressed in metabolically active tissues such as the liver, heart, skeletal muscles, intestinal mucosa, and brown adipose tissue (50, 51). PPARα is mainly involved in the carbohydrate metabolism and catabolism of fatty acids and their oxidation, such that its activation reduces lipid levels (52–55). Additionally, it has been well highlighted that PPARα increases the expression of IκB, which is a factor that suppresses the nuclear translocation and transcriptional activity of NF-κB, thereby interfering with NF-κB signaling and the inflammatory response (48). Besides, increasing evidence has demonstrated that the anti-inflammatory properties of PPARα are manifested by a decrease in the secretion of several key downstream inflammatory factors such as NF-κB–driven cytokines (TNF-α, IL1B, and IL6), COX2, IL8, IL12, IL2, VCAM1, TLR4, MCP1, STAT3, AP-1, and IL18 (56, 57). Moreover, it has been reported that the activation of PPARα leads to the upregulation of important anti-inflammatory factors such as IL1 receptor antagonist (IL1ra) (58) and vanin-1 (59). Furthermore, PPARα can interfere with angiogenic responses that are critical during chronic inflammation by targeting endothelial vascular endothelial growth factor receptor-2 (VEGFR-2) signaling, thereby controlling the inflammatory response (60).
PPARγ is the most widely studied PPAR isoform, which is expressed in white and brown adipose tissue, large intestine, and immune cells such as macrophages, the pancreas, and the spleen, and it plays a key role in a series of biochemical processes, including insulin sensitivity, inducing tumor cell differentiation and apoptosis, adipogenesis, lipoprotein metabolism, energy balance, reducing blood fat and blood pressure, and lipid biosynthesis (53, 61–63). Activation of PPARγ increases fat storage by increasing adipocyte differentiation and enhancing the transcription of genes important for lipogenesis (64, 65). Moreover, this subtype has been proposed as a potential therapeutic target for different types of cancer due to its various anti-tumor properties (66, 67). In terms of regulating inflammation, recent literature has reported that PPARγ prevent the inflammatory cascades caused by NF-κB activation and the production of proinflammatory cytokines such as TNF-α, IL1B, IFN-γ, IL2, iNOS, IL18, reactive oxygen species (ROS), and IL6 through the inhibition of NF-κB transactivation (38, 68–70). On the other hand, PPARγ exerts its protective effects by targeting major inflammatory factors such as STAT1, AP-1, PI3K, intercellular adhesion molecule (ICAM1), and matrix metallopeptidase 9 (MMP9) and inhibiting their activity to prevent destructive inflammatory damage (71). In addition, PPARγ regulates the expression of several essential inflammatory target genes such as MCP1/CCL2, endothelin-1, and adiponectin (APN) (72). Interestingly, a recent study well demonstrated that PPARγ inhibits dysregulated inflammatory responses by suppressing NLRP3 inflammasome activation as well as decreasing maturation of caspase-1 and IL1B (73).
The PPARβ/δ is the third subtype of PPARs, which has not been as intensely studied as PPARα and PPARγ; it consists of 441 amino acids with a molecular weight of 49.9 kDa. This isoform is expressed in almost all tissues. It is especially abundant in the liver, intestine, kidney, abdominal adipose tissue, and skeletal muscle, all of which are involved in lipid metabolism. Indeed, the PPARβ/δ isoform participates in fatty-acid oxidation, mainly in skeletal and cardiac muscles, and regulates blood cholesterol and glucose concentration (47, 74, 75). However, complete information on the exact role of PPARβ/δ in the regulation of inflammation is still not available, and more research is needed to deeply dissect the relationship between PPARβ/δ and inflammation or inflammatory response. In some contexts, PPARβ/δ has been shown to have anti-inflammatory functions. For example, it was demonstrated that activation of PPARβ/δ reduces the expression of inflammation-associated NF-κB and STAT1-targeted genes including TNF-α, MCP1, IL6, CXCL8, CCL2, CXCR2, and CXCL1 (76–79). Taken together, all three PPAR subtypes have distinct yet overlapping roles in regulating metabolic function and inflammation. Further details on the tissue distribution, function, and natural and synthetic ligands of PPARα, PPARβ/δ, and PPARγ are provided in Table 1.
2.3 Mechanism of PPAR activation
The activation of PPARs by ligands is associated with structural changes in the receptor, including dissociation from co-repressor complexes and association with appropriate transcriptional co-activators, binding to DNA, and acquiring transactivation/transrepression capabilities (31). Moreover, promotion of many biochemical mechanisms of PPARs requires that the receptor is part of a heterodimeric complex with another nuclear receptor, the 9-cis-retinoic acid receptor (RXR; NR2B) (21, 89). Therefore, after activation with ligands/agonists, the PPAR–RXR heterodimers are transported to the nucleus and bind to specific DNA sequences consisting of a direct repeat of DNA recognition motif AGGTCA separated by one or two nucleotides (DR-1 or DR-2 response elements), thereby stimulating/repressing the transcription of target genes (Figure 1B) (89, 90). This sequence is called the peroxisome proliferator response element (PPRE) and is located in the promoter regions of PPAR-regulated target genes (91). Furthermore, after binding the ligand-activated PPAR–RXR complex to the target DNA through PPARE, this complex binds to specific co-activator complexes such as CREB-binding protein (CBP)/p300 and steroid receptor co-activator 1 (SRC1), which have histone acetyltransferase activity and facilitate the remodeling of chromatin structure (92–95). In this regard, previous studies have reported that the binding of co-activator complexes to the ligand-activated, PPRE-associated PPAR–RXR complex can disrupt nucleosomes and induce transcriptional regulatory changes in the chromatin structure near the regulatory regions of PPAR target genes (Figure 1B) (57, 96, 97).
3 COVID-19 and cytokine storm
SARS-CoV-2, which affects the lower and upper respiratory tract, invades host cells through angiotensin-converting enzyme 2 (ACE2) receptors (98, 99) and causes a wide range of clinical manifestations from mild forms such as fever, cough, and myalgia to moderate forms with pneumonia and local inflammation symptoms requiring hospitalization, to severe/critical forms with fatal outcomes (100). Upon cellular entry of SARS-CoV-2 via its ACE2 receptor, viral genomic single-stranded RNA or other RNA compositions (double-stranded RNA) can be recognized as pathogen-associated molecule patterns (PAMPs) by innate immune and epithelial cells through the activation of pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs), retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), and NOD-like receptors (NLRs) (101, 102). Following sensitization of PRRs, downstream key inflammation-related transcription factors such as NF-κB, activator protein-1 (AP-1), and IFN regulatory factors (IRFs) are activated and promote the transcription of proinflammatory cytokines, chemokines, and IFNs such as IL1β, IL18, IL6, IL12, TNF-α, IL8, IL2, IL7, IL17, CCL3, CCL5, CXCL8, CXCL10, and IFN-γ (103–108). Moreover, proinflammatory cytokines such as IL6, TNF-α, and IFN-γ, in turn, activate JAK/STAT, NF-κB, and mitogen-activated protein kinase (MAPK) signaling by binding to their receptors on immune cells to induce further production of proinflammatory cytokines and subsequently form positive feedback to initiate the cytokine storm (Figure 2) (102).
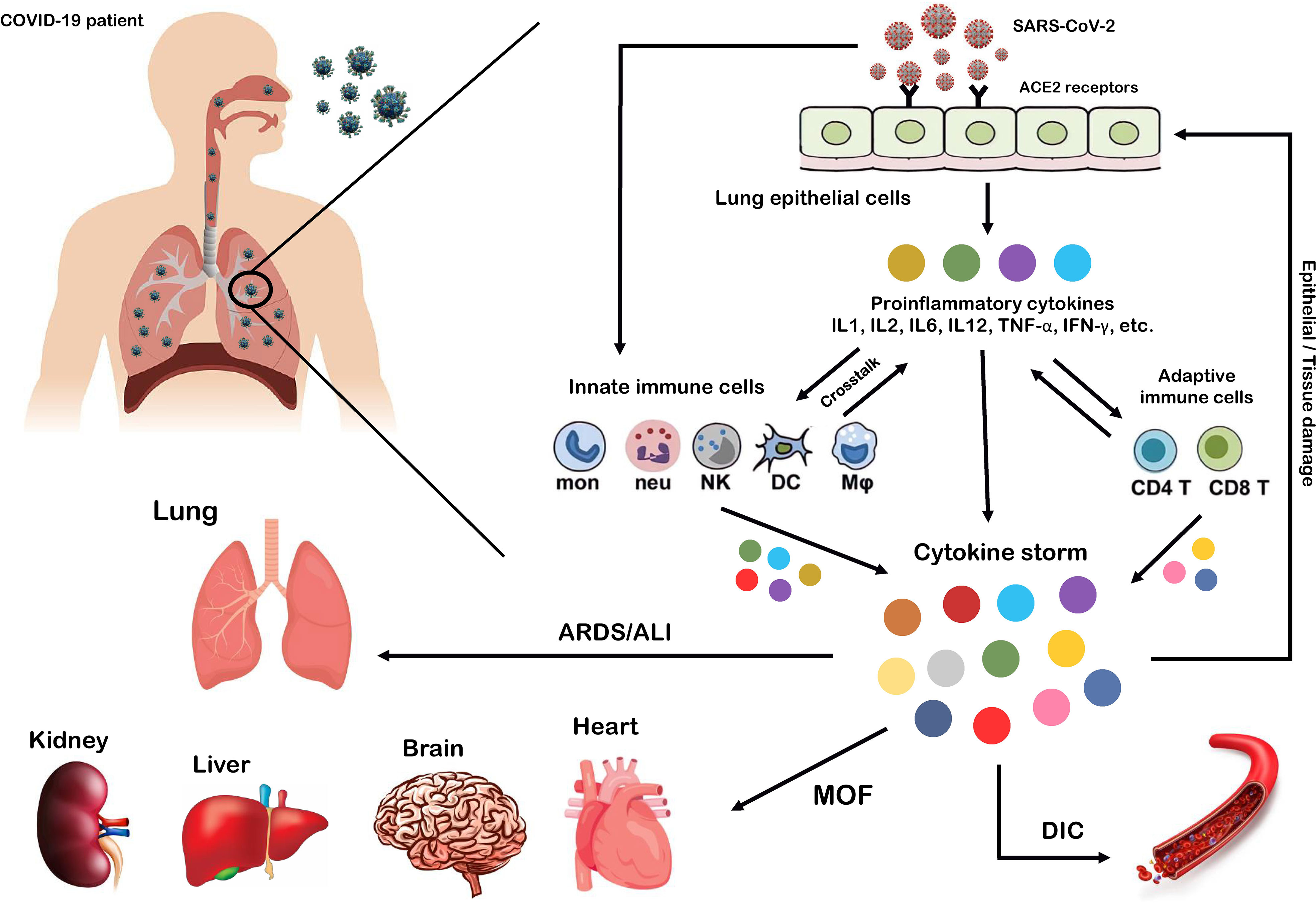
Figure 2 Cytokine storm as the hallmark of COVID-19 immunopathogenesis. Following the entry of SARS-CoV-2 into lung epithelial and immune cells via angiotensin-converting enzyme 2 (ACE2) receptors, a cascade of downstream signaling pathways is activated, ultimately leading to the massive release of proinflammatory cytokines and chemokines and tissue damage. Moreover, these proinflammatory cytokines lead to the recruitment of more innate immune cells, including neutrophils, macrophages, natural killer (NK) cells, monocytes, and dendritic cells (DCs) and active adaptive immune cells, including CD4+ and CD8+ T cells, to the site of infection, in order to induce the production of circulating cytokines. As a result, the crosstalk between epithelial and immune cells in vast cytokine release causes hyperinflammation and cytokine storm, which leads to a wide range of clinical manifestations from mild to severe/critical forms with a fatal outcome. Some of these fatal consequences include macrophage activation syndrome (MAS), hemophagocytic lymphohistiocytosis (HLH), capillary leak syndrome (CLS), thrombosis, disseminated intravascular coagulation (DIC), acute respiratory distress syndrome (ARDS), multi-organ failure (MOF), and acute lung injury (ALI).
Exacerbation of the local inflammatory response and increased secretion of proinflammatory cytokines and chemokines by resident immune and respiratory epithelial cells leads to more recruitment of innate and adaptive immune cells such as macrophages, neutrophils, dendritic cells (DCs), natural killer (NK) cells, monocytes, and CD4+ and CD8+ T cells to the site of infection to produce more persistent inflammatory cytokines (102). Indeed, growing evidence suggests that the crosstalk between epithelial cells and immune cells in COVID-19 produces high levels of proinflammatory cytokines that trigger an uncontrollable inflammatory response, hyperinflammation, or imbalanced inflammation (known as “cytokine storm”) with severe complications and poor outcomes (109, 110). In this regard, extensive studies have recently reported that high circulating levels of proinflammatory cytokines (IFN-α, IFN-γ, IL1β, IL6, IL12, IL18, IL33, TNF-α, TGF-β, IL1RA, IL7, IL8, IL9, VEGFA, etc.) and chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10, etc.) have been identified in patients with severe COVID-19 (105, 111–114). Furthermore, it has been highlighted that cytokine storm is one of the main features of ARDS, ALI, tissue damage, and MOF, which are the major causes of COVID-19 severity and death of patients (106, 111, 115). Therefore, we believe that any intervention approach to target the critical inflammatory factors during SARS-CoV-2 infection could be a fundamental step in developing therapeutic strategies to control hyperinflammation, combat the cytokine storm, and reduce COVID-19 severity.
4 Anti-inflammatory mechanisms mediated by PPARs
In the last decade, many studies have concluded that PPARs, in addition to being critical players in glucose and lipid metabolism, play an essential role in controlling various types of the inflammatory response (57, 116, 117). Indeed, the inflammatory role of PPARs was highlighted when a previous study showed that PPARα knockdown was directly associated with increased levels of proinflammatory cytokines (118). In agreement with this study, a recent study showed that, in addition to PPARα, the knockdown of PPARγ also leads to increased serum levels of IL6, IL1β, and TNF-α during lipopolysaccharide (LPS) stimulation (119). Furthermore, in an animal model, Huang et al. (120) showed that increased PPARγ expression levels prevented pulmonary inflammation and were directly associated with the recovery of influenza virus-infected animals (120). Moreover, several previous studies have shown that PPARα and PPARγ activation lead to reduced inflammation in polymicrobial sepsis (121) and HIV infection (122). In addition to these findings, in recent years, the central role of PPARs to control inflammation and reduce the levels of proinflammatory cytokines has been reviewed in many inflammatory disorders, including lung inflammatory diseases (32), IBD (21), and hepatic inflammation (116). These results indicate that PPARs suppress the transcription of main active inflammatory transcription factors, including NF-κB, AP-1, nuclear factor of activated T cells (NFAT), and signal transducers and activators of transcription (STATs), through an agonist-dependent mechanism (123). Among the various mechanisms PPARs use to repress many distinct transcriptions factor families, the most likely include four main mechanisms in which ligand-activated PPAR–RXR complexes suppress the activity of many inflammatory factors.
The first mechanism is the successful competition of PPARs to limit the amount of essential and shared co-activator proteins (such as CBP/P300) in a cell. As a result of this successful competition, these co-activators are not available for other transcription factors (31, 124). Therefore, the activities of other transcription factors (such as NF-κB) that use the same co-activators are repressed in these situations of co-activator competition. On the other hand, the second mechanism involves direct physical association between PPARs and other transcription factors without the mediation of co-activators. During the second mechanism, known as “cross-coupling” or “mutual receptor antagonism”, ligand-activated PPAR–RXR heterodimers form a new complex with other transcription factors, such as AP-1, NF-κB, NFAT, and STATs through physical protein–protein interactions, thereby preventing transcription factor binding to its response element and also inhibiting their ability to induce the transcription of proinflammatory genes such as IL6, IL1β, and TNF-α (Figure 3) (46). For instance, agonist-activated PPARα and PPARγ negatively regulate the inflammatory gene response through bidirectionally blocking NF-κB and AP-1 signaling pathways via physical interaction with NF-κB p65 (38, 125). Moreover, PPARs can suppress the expression of NF-κB through the upregulation of inhibitors of NF-κB (IκBs) (126, 127). The third mechanism also involves blocking MAPK-inducted signaling cascades by ligand-activated PPAR–RXR heterodimers through inhibition of MAPK phosphorylation and activation (128, 129). Lastly, preventing the clearance of co-repressors whose removal is required for the transcriptional activation of AP-1 and NF-κB target proinflammatory genes is the fourth mechanism of inflammation suppression by PPARs (130, 131). Moreover, another anti-inflammatory effect of PPARs is their agonistic effect with other anti-inflammatory factors. Previous studies have shown that a significant increase in the expression level of PPARs is associated with an increase in the expression of anti-inflammatory factors such as IL10 (132–134). Several human and animal models have reported that PPARs and their ligands downregulate the expression of many chemokines such as CCL2, -4, -7, -12, -17, and -19, CXCL1, -9, and -10, and leukocyte adhesion molecules such as VCAM1, ICAM1, and endothelin-1. This downmodulation inhibits leukocyte recruitment to the site of inflammation and reduces the crosstalk between immune cells and other resident cells for cytokine production (135–139).
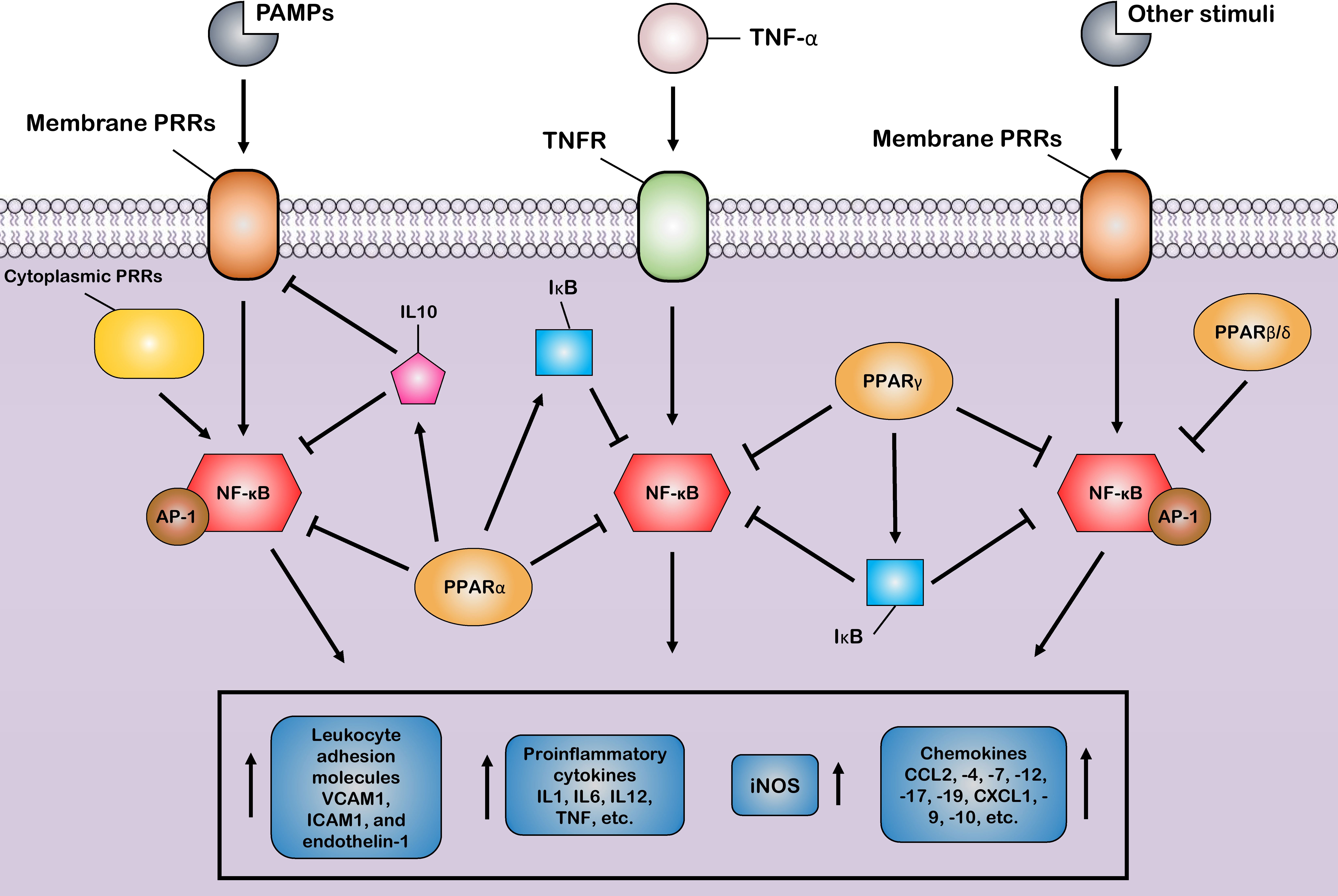
Figure 3 Control of the host inflammatory response mediated by PPARs. Most of the anti-inflammatory properties of PPARs are characterized by the suppression of key inflammatory transcription factors such as nuclear factor-κB (NF-κB) and activator protein-1 (AP-1) via different suppressive mechanisms, as induction of the production of anti-inflammatory cytokines. Through these mechanisms, PPARs block the expression of various inflammatory genes and, thus, reduce the production of many proinflammatory cytokines, chemokines, and other proinflammatory signal mediators, such as inducible nitric oxide synthase (iNOS). Additionally, the PPARs prevent the recruitment of leukocytes to the site of inflammation by inhibiting the production of cell adhesion molecules.
5 Control of inflammation by PPARs during SARS-CoV-2 infection
The recent emergence of COVID-19 in the past years and its rapid worldwide spread have led to extensive clinical studies investigating the molecular regulatory mechanisms behind this severe disease. Intriguingly, at this time, many of these extensive clinical studies reported the influential role of PPAR subtypes, especially PPARγ and their ligands, in controlling the host inflammatory response during SARS-CoV-2 infection. Meanwhile, in previous clinical trials, a decrease in the expression of PPAR subtypes and an increase in the serum level of proinflammatory cytokines have been observed in inflammatory lungs of patients with severe COVID-19 (112, 113, 140). Additionally, in agreement with the results of clinical studies, several recent transcriptomics studies using microarray, RNA-sequencing (RNA-seq), and single-cell RNA-seq techniques have reported downregulation of PPARs in various tissues including whole blood, lung epithelial cells, bronchoalveolar lavage fluids (BALFs), and peripheral blood mononuclear cells (PBMCs) in the SARS-CoV-2-infected individuals (141–145). Following these findings, the results of previous proteomics and metabolomics studies also indicate the interference of SARS-CoV-2 infection in PPAR signaling (146, 147). Surprisingly, Keikha et al. (148) recently demonstrated that a set of miRNAs, including mir-27b, were upregulated during SARS-CoV-2 infection. They also reported that mir-27b has a significant negative correlation with its main target, i.e., PPARγ, and the increase in its expression during SARS-CoV-2 infection directly leads to the downregulation of PPARγ, thus playing a key role in the exacerbation of the inflammatory response in COVID-19 patients (148). One of the main immunopathogenesis strategies of SARS-CoV-2 infection has been suggested to interfere with PPAR signaling to exacerbate the inflammatory response (149). In other words, previous studies have reported that the decrease in the expression of PPARs including PPARγ, PPARα, and PPARβ/δ during SARS-CoV-2 infection is associated with the increased secretion of proinflammatory cytokines such as IL6, IL1β, and TNF-α, as well as cytokine storm; thus, it has a positive correlation with ARDS and ALI in COVID-19 patients (150, 151).
Moreover, a previous study concluded that SARS-CoV-2 suppresses PPAR expression in the lungs and abrogates one of the main anti-inflammatory cores for NF-κB activity, thereby exerting a hyperinflammatory response in patients with severe COVID-19 (152). Moreover, several recent studies also reported that the reduction in PPARγ and PPARα is directly related to acute pulmonary inflammation in COVID-19 and the shift of the disease from mild to severe and, finally, death (33, 153). Additionally, it has been highlighted that suppressing the expression of PPAR subtypes, especially PPARγ, leads to increased susceptibility to SARS-CoV-2 infection (154). Interestingly, the decrease in PPARγ expression during SARS-CoV-2 infection, in addition to being positively related to the occurrence of hyperinflammation, also leads to insulin resistance in COVID-19 patients (155). Besides, it has recently been reviewed that over-activation of the canonical WNT/β-catenin pathway in response to SARS-CoV-2 infection leads to inhibition of PPARγ expression in an opposing interplay (156). Furthermore, COVID-19 has more negative clinical consequences for obese people because clinical trials indicate that the serum levels of PPARγ are lower in obese people. Therefore, the probability of cytokine storm during SARS-CoV-2 infection is higher in these people (157). Furthermore, another study proved that alcohol consumption is directly related to systematic inflammation in COVID-19 patients because ethanol (EtOH) exacerbates the activation of proinflammatory cytokines, including IL6, IL1B, IFN, and TNF-α and inflammation-related transcription factors, including HIF1-α, JUN, NF-κB, and STATs via induction of PPAR–RXR inactivation (158).
Additionally, there is accumulating evidence that the T-helper 2 (Th2) inflammatory response phenotype can induce protective effects against the COVID-19 immunopathogenesis due to the increased secretion and release of Th2 anti-inflammatory cytokines such as IL10, IL4, and IL13 and recruitment of the eosinophils to the site of inflammation (159). Following these results, it has been well reviewed that cytokines associated with Th2 inflammatory response such as IL4 and IL13 inhibit the secretion of several proinflammatory cytokines such as IL6, IL1B, IL1α, IL12, and TNF-α, which play a central role in the pathogenesis of COVID-19 and hyperinflammation (160). Moreover, the anti-inflammatory M2 macrophages is activated by IL4 and IL13, which modulate inflammatory responses by producing anti-inflammatory cytokines, such as IL10 (161). Strikingly, recent results suggest that Th2 responses which driven by IL4, IL5, and IL13 dramatically reduce ACE2 in the respiratory tract and are associated with better clinical outcomes with COVID-19 (162, 163). Therefore, it has been hypothesized that the Th2 inflammatory response may exert potential protective effects against COVID-19 (164). Surprisingly, previous studies in several human inflammatory diseases indicate that both PPARα and PPARγ and their ligands increase the expression levels of anti-inflammatory markers associated with the Th2 inflammation such as IL13, IL4, IL10, and GATA3, thereby limiting the dysregulation of inflammation (165–167). Therefore, based on these findings, it can be concluded that PPARs can induce different anti-inflammatory mechanisms during SARS-CoV-2 infection through a synergistic effect with Th2 inflammatory responses.
Several reports suggest that PPARs play an important role in controlling the inflammatory response during COVID-19 by inducing the inactivation of the key inflammatory transcription factors, especially NF-κB (168). In this regard, it has been suggested that activation of PPARγ during COVID-19 can reduce the circulating levels of TNF-α, IL-1, and IL-6 in the innate immune cells such as macrophages and monocytes through interaction with NF-κB (169). Moreover, PPARγ acts as a negative regulator of cytotoxic T-cell activation and suppresses the production of cytokines by these adaptive immune cells (170). Following these studies, the recent emerging literature has also reported that activation of PPARα, PPARβ/δ, and PPARγ is inversely related to pulmonary fibrosis caused by chronic inflammation in COVID-19 patients (147, 171–173). It has been also hypothesized that exercise may prevent untoward systemic consequences of SARS-CoV-2 infection including inflammation and metabolic dysfunctions such as lipotoxicity by having a positive effect on PPARα (33). The anti-inflammatory role of PPARβ/δ during COVID-19 has been less studied than the other two types of PPARs. However, a few reports have indicated that PPARβ/δ suppressing transcription factors involved in the inflammatory response, including NF-κB and AP-1 (31), reduce the expression levels of GDF-15 (one of the inflammatory biomarkers of COVID-19 severity) in a negative feedback manner (174).
Furthermore, PPARβ/δ and PPARγ have been shown to play a central role in the macrophage polarization toward an anti-inflammatory M2 phenotype during COVID-19 (175). On the other hand, the previous literature has shown that PPARα and PPARγ prevent the apoptosis of inflammatory cells by inducing anti-apoptotic factors of the BCL-2 family, thereby preventing the spread of cytokines and chemokines in the intercellular space (176). Notably, the decrease in the expression levels of PPARs in the early stages of SARS-CoV-2 infection and the increase in their expression during the treatment/recovery period indicate the opposite/inverse relationship of these receptors with the severity of the disease (38, 177).
Intriguingly, PPARα and PPARγ have been proposed as effective adjuvants for the development of COVID-19 vaccines because these receptors through an increase in the population of regulatory T-cells via upregulation in FOXP3 mRNA expression (a transcriptional factor for the function and differentiation of regulatory T-cells) (1): stimulate memory T-cells (2), upregulate the γδ type of T-cells, and (3) prolong B-cell memory and improve the secondary antibody response and thus can induce long-term memory (176, 178). However, the inverse relationship between regulatory T-cells and chronic inflammation has been revealed by previous research (175, 179), and the anti-inflammatory properties of these cells are well established (180, 181). Therefore, it can be predicted that the increase in the population of regulatory T-cells due to the activation of PPARs can play a potential dual-role by stimulating and strengthening long-term memory and exerting significant anti-inflammatory properties during SARS-CoV-2 infection.
Recent advances in high-throughput transcriptome-based technologies and the integration of these techniques with computational network-based algorithms of systems biology have provided an excellent opportunity to identify altered gene regulatory networks under infected conditions, activated pathways, potential therapeutic/diagnostic/prognostic targets, and understanding the complex molecular mechanisms underlying infectious disease at the systemic level (182). In our previous work, we integrated and analyzed the RNA-seq data from PBMCs of healthy individuals and COVID-19 patients with computational network-based methods of systems biology in order to identify potential therapeutic targets and candidate gene modules underlying COVID-19 and develop promising therapeutic strategies for COVID-19 (39). As a result, we identified nine candidate co-expressed gene modules and 290 hub-high traffic genes with the highest betweenness centrality (BC) score directly related to SARS-CoV-2 pathogenesis (39). Indeed, the genes with the highest BC score have the highest rate of “information transfer” in their respective modules with critical biological functions, which are known as “high traffic” genes and can be potential therapeutic, diagnostic, and prognostic targets for COVID-19 therapy (39). We observed that PPARα is among the hub-high traffic genes in one of the key modules with anti-inflammatory function, indicating the crucial anti-inflammatory role of this PPAR subtype during SARS-CoV-2 infection (39). In another study, Auwul et al. (40) integrated various transcriptomic data with computational systems biology and machine learning algorithms and identified 52 common drug targets, including PPARγ, for COVID-19 treatment (40).
Moreover, further studies using pharmacological network approaches have identified PPARα and PPARγ as promising drug/therapeutic targets to control inflammation caused by host–SARS-CoV-2 interactions (183, 184). On the other hand, recently, a study introduced glycyrrhetinic acid as an essential drug against cytokine storm in COVID-19 patients (185). During this study, using protein–protein interaction (PPI) network and molecular docking techniques, it was well established that glycyrrhetinic acid activates or represses 84 core genes to counter the cytokine storm during COVID-19 using multiple strategies (185). As an important result of this study, one of these glycyrrhetinic acid strategies to deal with the cytokine storm was to target PPARγ, PPARα, and PPARβ/δ for activation (185). Figure 4 shows that the PPI structure of the candidate modules identified by these studies that contained critical therapeutic targets, including PPARα, PPARβ/δ, and PPARγ for COVID-19 therapy.
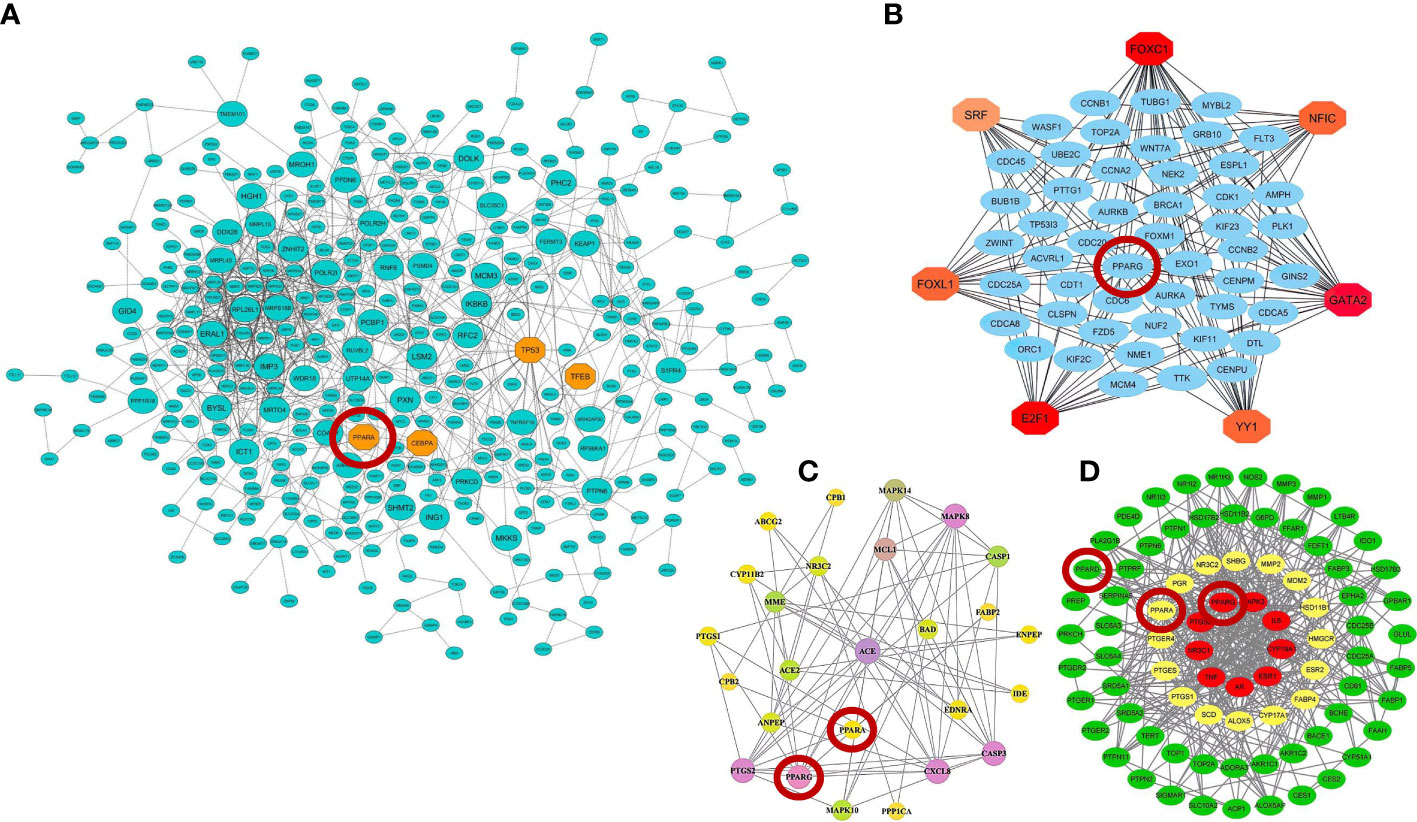
Figure 4 Protein–protein interaction (PPI) networks of therapeutic candidate modules for COVID-19 therapy obtained by (A) our group (39) (B) Auwul et al. (40), (C) Oh et al. (183), and (D) Li et al. (185). These modules had the most biological associations with the immunopathogenesis of COVID-19. Large circles represent hub-high traffic genes. PPAR genes as potential therapeutic targets for COVID-19 pandemic are highlighted by red circles in these PPI networks.
Interestingly, the use of PPARs agonists to activate them to repress the inflammatory processes during COVID-19 has recently attracted much attention. In this regard, it has been demonstrated that PPAR activation through synthetic and nutritional compounds can be an efficient management program to overcome the cytokine storm and prevent the deleterious inflammatory effects after coronavirus infection (38, 186). Moreover, a recent study suggested using synthetic and natural ligands of PPARs in order to target NF-κB transcriptional activity and reduce inflammatory response as an attractive strategy for managing the nutrition of COVID-19 patients (187). Following these results, the recent literature also reported that several natural and synthetic PPARγ agonists suppress NF-κB activity through PPARγ activation, leading to reduced levels of proinflammatory cytokines such as IL1β, IL6, TNF-α, IL18, IFN-γ, IL8, and IL12 (38). It has been well established that PPARα activation using oleoylethanolamide, in addition to suppressing TLR4-mediated NF-κB signaling cascade and reducing proinflammatory cytokines such as COX2, IL6, CRP, IL1β, TNF-α, and iNOS, is also associated with increased levels of anti-inflammatory factors such as IL10 (56, 188–190). Additionally, it has recently been shown that fenofibrate is a PPARα agonist with anti-inflammatory, anti-oxidant, and anti-thrombotic properties that exerts broad anti-inflammatory effects such as inhibition of iNOS, repression of COX2 and MMP9, activation of inhibitory kappa B (IκB), and release of adiponectin through the activating of PPARα during SARS-CoV-2 infection (150). Furthermore, previous studies reported that fenofibrate inhibits viral replication in lung epithelial cells by reversing the metabolic changes caused by SARS-CoV-2 (175). On the other hand, natural astaxanthin (ASX), an important PPARγ agonist, has a clinically proven safety profile with anti-oxidant, anti-inflammatory, and immunomodulatory properties. Interestingly, mounting evidence from clinical studies suggests that this PPAR agonist prevents the ARDS/ALI in COVID-19 patients by downregulating NF-κB and JAK/STAT signaling and then reducing TNF-α, IL1β, and IL6 levels. Moreover, ASX caused a change in the inflammatory response of Th1 cells to Th2, leading to a shift from proinflammatory cytokine secretion to anti-inflammatory cytokine secretion (191). ASX also exerts an anti-oxidant effect and prevents oxidative damage through (1) inhibition of NLRP3 inflammasome and HIF1-α, and (2) suppression of plasma CRP, iNOS, COX2, PGE2, and ICAM1, respectively. Accordingly, ASX-mediated activation of PPARγ has been proposed as an effective therapeutic strategy to control host inflammatory and immune responses, antagonize the cytokine storm, and prevent deleterious inflammatory effects following COVID-19 (191). Moreover, troglitazone, an insulin-sensitizing drug that is prescribed for treating type 2 diabetes mellitus (192), is a synthetic PPARγ agonist that interferes with NF-κB activity and exerts its anti-inflammatory effects through the activation of PPARγ (38). Interestingly, this drug has been introduced as one of the most suitable options for applying anti-inflammatory effects for COVID-19-inducted hyperinflammation (38). In addition to troglitazone, pioglitazone (a synthetic agonist of PPARγ, see Table 1) is another member of the thiazolidinedione (TZD) family that has significant anti-inflammatory effects and has been suggested by Carboni et al. (193) as a support drug for the reduction in many inflammatory parameters in COVID-19 patients (193). Additionally, pioglitazone can reduce SARS-CoV-2 RNA synthesis and replication through potential inhibition of 3-chymotrypsin-like protease (3CL-Pro) (194). Moreover, it has recently been suggested that activation of PPARγ by agonists such as cannabidiol reduces cytokine secretion, pulmonary inflammation, and fibrosis in the lung of the patients during SARS-CoV-2 infection (195). Intriguingly, zinc supplementation has also been reported to have potential health benefits for managing inflammation in COVID-19 by suppressing the expression of many cytokines and adhesion molecules through increasing the expression of PPARα (196). Furthermore, natural compounds such as gamma-oryzanol (the main bioactive constituent from rice bran and germ) have been introduced as a possible adjunctive therapy to prevent the cytokine storm in COVID-19 patients, as this compound positively increases the expression of PPARγ in adipose tissue and as a result reduces the levels of inflammatory cytokines including TNF-α, IL6, and MCP1 (157). To the best of our knowledge, no information is yet available on the role of PPARβ/δ agonists during SARS-CoV-2 infection. Therefore, future research should investigate the anti-inflammatory effects of natural and synthetic agonists of PPARβ/δ during the COVID-19 pandemic. However, looking at the previous literature on similar inflammatory lung diseases in humans, it can be concluded that PPARβ/δ agonists have significant anti-inflammatory effects during lung infection (197). Conversely, using existing natural and synthetic ligands of PPARs may have limitations or challenges. For instance, recent data show that using natural and synthetic ligands of PPARs is highly dose-dependent and can interact with non-PPAR targets due to the complexities of the drug–target complex (94). Moreover, it has been reported that some ligands of PPARs can exert selective agonistic or antagonistic regulatory effects depending on the cell context (191, 198, 199). Additionally, it has been highlighted that using synthetic agonists of PPARs can lead to serious clinical complications, including bone fracture, heart failure, cardiovascular risk, liver failure, gastrointestinal bleeding, and liver and kidney toxicity (200).
These findings highlight the potential role of PPAR subtypes (particularly PPARγ) and their ligands with anti-inflammatory effects during the COVID-19 pandemic, which can be promising candidates for inhibiting key inflammatory factors (especially NF-κB and AP-1), thus regulating inflammation during SARS-CoV-2 infection. Therefore, any intervention methods aimed at activating/upregulating/overexpressing of PPAR subtypes could be a promising therapeutic strategy to reduce the hyperinflammatory response in COVID-19 patients and prevent the cytokine storm.
6 Conclusions and future prospects
COVID-19 is an emerging global health threat caused by SARS-CoV-2 infection with severe inflammatory complications. Treatment of severely ill patients is an important healthcare issue. Despite developing various vaccines for disease prevention, there is still no definitive treatment solution for COVID-19 patients. The massive cytokine secretion caused by the exacerbation of the host inflammatory system in response to SARS-CoV-2 infection is known as a “cytokine storm”, which is directly related to the progression of the disease from mild to severe. In recent years, among previous efforts, it has been suggested that one of the most effective strategies for improving the survival of COVID-19 patients and reducing the severity of the disease is to control the hyperinflammatory response and interfere with cytokine storm. Cytokine storm has been one of the main characteristics of disease severity, decreased lung function, ARDS, ALI, and MOF, and ultimately, the death of COVID-19 patients. Therefore, paying attention to anti-inflammatory factors and examining their response during SARS-CoV-2 infection can provide the basic solution to deal with COVID-19-inducted cytokine storm.
PPARs are ligand-dependent transcription factors belonging to the nuclear receptor superfamily, which are the main regulators of lipid and glucose metabolism. This transcription factor family consists of three subtypes: PPARα, PPARβ/δ, and PPARγ. In the last decade, it has been well established that these subtypes, in addition to their central role in metabolism and energy balance, play important roles in cell proliferation, differentiation, the immune cell system, and inflammation. Concerning inflammation and inflammation-related disease, PPARs play an important anti-inflammatory role as critical inhibitors of the host inflammatory response through adverse regulatory effects on active inflammatory transcription factors such as NF-κB, AP-1, NFAT, and STATs. Currently, extensive clinical and omics studies indicate downregulation in the expression of PPARs in response to SARS-CoV-2 infection, which has been proposed as one of the main causes of SARS-CoV-2 immunopathogenesis to exacerbate the host inflammatory response.
On the other hand, it has been highlighted that the activation of PPARs through natural and synthetic ligands is associated with the reduction of hyperinflammatory response, prevention of cytokine storm, and reduction in disease severity in COVID-19 patients. Therefore, this makes them attractive and practical targets for developing novel therapeutic strategies against COVID-19 and cytokine storm. However, the previous literature has indicated that using existing natural and synthetic ligands of PPARs may lead to severe clinical complications.
Therefore, considering the anti-inflammatory importance of PPARs to control the hyperinflammatory response during COVID-19, further research should deeply investigate the individual or collective effects of PPAR subtypes to inhibit cytokine storms during SARS-CoV-2 infection. Moreover, considering the side-effects and challenges of using existing natural and synthetic ligands to activate PPARs, further exploration of the underlying mechanisms is needed to establish new pathways of PPARs activation without causing severe clinical side-effects.
Author contributions
AH and AB conceived the ideas. AH, AB, AS, and BK designed the study. AH, AB, AS, BT-F, and SI determined the methodology. AH, AB, BK, FG, AS, AA, and MA validated and interpreted the results. AH, AB, BT-F, AS, MA, FG, and SI investigated and collected resources. AH, AB, and BK wrote and prepared the original draft. AH, AB, AS, AA, and BK reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors thank all the teams who contributed to and provided technical assistance during this study. We also thank the reviewers whose critical comments helped in improving the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ACE2, Angiotensin-converting enzyme 2; ALI, Acute lung injury; AP-1, Activator protein-1; ARDS, Acute respiratory distress syndrome; BALFs, bronchoalveolar lavage fluids; COVID-19, Coronavirus disease 2019; IBD, Inflammatory bowel disease; IFNs, Interferons; IκBs, Inhibitors of NF-κB; ILs, Interleukins; MOF, Multi-organ failure; NF-κB, Nuclear factor-κB; PAMPs, Pathogen associated molecular patterns; PBMCs, Peripheral blood mononuclear cells; PRRs, Pattern recognition receptors; PPARs, Peroxisome proliferator-activated receptors; PPI, Protein–protein interaction; PPREs, Peroxisome proliferator response elements; RXR, 9-cis-retinoic acid receptor; SARS-CoV-2, Severe acute respiratory syndrome corona virus 2; TNFs, Tumor necrose factors.
References
1. Ahamad MM, Aktar S, Rashed-Al-Mahfuz M, Uddin S, Liò P, Xu H, et al. A machine learning model to identify early stage symptoms of sars-Cov-2 infected patients. Expert Syst Appl (2020) 160:113661. doi: 10.1016/j.eswa.2020.113661
2. Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, et al. Clinical characteristics of coronavirus disease 2019 in China. New Engl J Med (2020) 382(18):1708–20. doi: 10.1056/NEJMoa2002032
3. Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A, et al. World health organization declares global emergency: A review of the 2019 novel coronavirus (Covid-19). Int J Surg (2020) 76:71–6. doi: 10.1016/j.ijsu.2020.02.034
4. Tian S, Hu W, Niu L, Liu H, Xu H, Xiao S-Y. Pulmonary pathology of early-phase 2019 novel coronavirus (Covid-19) pneumonia in two patients with lung cancer. J Thorac Oncol (2020) 15(5):700–4. doi: 10.1016/j.jtho.2020.02.010
5. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in wuhan, China: A descriptive study. Lancet (2020) 395(10223):507–13. doi: 10.1016/S0140-6736(20)30211-7
6. McGonagle D, Sharif K, O’Regan A, Bridgewood C. The role of cytokines including interleukin-6 in covid-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmun Rev (2020) 19(6):102537. doi: 10.1016/j.autrev.2020.102537
7. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of covid-19: Immunity, inflammation and intervention. Nat Rev Immunol (2020) 20(6):363–74. doi: 10.1038/s41577-020-0311-8
8. Cao X. Covid-19: Immunopathology and its implications for therapy. Nat Rev Immunol (2020) 20(5):269–70. doi: 10.1038/s41577-020-0308-3
9. Merad M, Martin JC. Pathological inflammation in patients with covid-19: A key role for monocytes and macrophages. Nat Rev Immunol (2020) 20(6):355–62. doi: 10.1038/s41577-020-0331-4
10. Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The covid-19 cytokine storm; what we know so far. Front Immunol (2020) 11:1446. doi: 10.3389/fimmu.2020.01446
11. Ahmed F. A network-based analysis reveals the mechanism underlying vitamin d in suppressing cytokine storm and virus in sars-Cov-2 infection. Front Immunol (2020) 11:590459. doi: 10.3389/fimmu.2020.590459
12. Rehan M, Ahmed F, Howladar SM, Refai MY, Baeissa HM, Zughaibi TA, et al. A computational approach identified andrographolide as a potential drug for suppressing covid-19-Induced cytokine storm. Front Immunol (2021) 12:648250. doi: 10.3389/fimmu.2021.648250
13. Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in covid-19 patients. Emerging Microbes Infections (2020) 9(1):761–70. doi: 10.1080/22221751.2020.1747363
14. Ren X, Wen W, Fan X, Hou W, Su B, Cai P, et al. Covid-19 immune features revealed by a Large-scale single-cell transcriptome atlas. Cell (2021) 184(7):1895–913.e19. doi: 10.1016/j.cell.2021.01.053
15. Yang AC, Kern F, Losada PM, Agam MR, Maat CA, Schmartz GP, et al. Dysregulation of brain and choroid plexus cell types in severe covid-19. Nature (2021) 595(7868):565–71. doi: 10.1038/s41586-021-03710-0
16. McClain MT, Constantine FJ, Henao R, Liu Y, Tsalik EL, Burke TW, et al. Dysregulated transcriptional responses to sars-Cov-2 in the periphery. Nat Commun (2021) 12(1):1079. doi: 10.1038/s41467-021-21289-y
17. Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, et al. Systems biological assessment of immunity to mild versus severe covid-19 infection in humans. Science (2020) 369(6508):1210–20. doi: 10.1126/science.abc6261
18. Hariharan A, Hakeem AR, Radhakrishnan S, Reddy MS, Rela M. The role and therapeutic potential of nf-Kappa-B pathway in severe covid-19 patients. Inflammopharmacology (2021) 29(1):91–100. doi: 10.1007/s10787-020-00773-9
19. Kircheis R, Haasbach E, Lueftenegger D, Heyken WT, Ocker M, Planz O. Nf-Kb pathway as a potential target for treatment of critical stage covid-19 patients. Front Immunol (2020) 11:598444. doi: 10.3389/fimmu.2020.598444
20. Sun X, Wang T, Cai D, Hu Z, Ja C, Liao H, et al. Cytokine storm intervention in the early stages of covid-19 pneumonia. Cytokine Growth Factor Rev (2020) 53:38–42. doi: 10.1016/j.cytogfr.2020.04.002
21. Decara J, Rivera P, López-Gambero AJ, Serrano A, Pavón FJ, Baixeras E, et al. Peroxisome proliferator-activated receptors: Experimental targeting for the treatment of inflammatory bowel diseases. Front Pharmacol (2020) 11:730. doi: 10.3389/fphar.2020.00730
22. Li J, Liu Y-P. The roles of ppars in human diseases. Nucleosides Nucleotides Nucleic Acids (2018) 37(7):361–82. doi: 10.1080/15257770.2018.1475673
23. Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W. From molecular action to physiological outputs: Peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog Lipid Res (2006) 45(2):120–59. doi: 10.1016/j.plipres.2005.12.002
24. Gonzalez FJ, Shah YM. Pparα: Mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology (2008) 246(1):2–8. doi: 10.1016/j.tox.2007.09.030
25. You M, Jin J, Liu Q, Xu Q, Shi J, Hou Y. Pparα promotes cancer cell Glut1 transcription repression. J Cell Biochem (2017) 118(6):1556–62. doi: 10.1002/jcb.25817
26. Hennuyer N, Duplan I, Paquet C, Vanhoutte J, Woitrain E, Touche V, et al. The novel selective pparα modulator (Spparmα) pemafibrate improves dyslipidemia, enhances reverse cholesterol transport and decreases inflammation and atherosclerosis. Atherosclerosis (2016) 249:200–8. doi: 10.1016/j.atherosclerosis.2016.03.003
27. Andrulionyte L, Kuulasmaa T, Chiasson J-L, Laakso M, Group ftS-NS. Single nucleotide polymorphisms of the peroxisome proliferator–activated receptor-A gene (Ppara) influence the conversion from impaired glucose tolerance to type 2 diabetes: The stop-niddm trial. Diabetes (2007) 56(4):1181–6. doi: 10.2337/db06-1110
28. Holness MJ, Samsuddin S, Sugden MC. The role of ppars in modulating cardiac metabolism in diabetes. Pharmacol Res (2009) 60(3):185–94. doi: 10.1016/j.phrs.2009.04.006
29. Ramon S, Bancos S, Thatcher TH, Murant TI, Moshkani S, Sahler JM, et al. Peroxisome proliferator-activated receptor Γ b Cell-Specific–deficient mice have an impaired antibody response. J Immunol (2012) 189(10):4740–7. doi: 10.4049/jimmunol.1200956
30. Choi J-M, Bothwell ALM. The nuclear receptor ppars as important regulators of T-cell functions and autoimmune diseases. Molecules Cells (2012) 33(3):217–22. doi: 10.1007/s10059-012-2297-y
31. Daynes RA, Jones DC. Emerging roles of ppars in inflammation and immunity. Nat Rev Immunol (2002) 2(10):748–59. doi: 10.1038/nri912
32. Becker J, Delayre-Orthez C, Frossard N, Pons F. Regulation of inflammation by ppars: A future approach to treat lung inflammatory diseases? Fundam Clin Pharmacol (2006) 20(5):429–47. doi: 10.1111/j.1472-8206.2006.00425.x
33. Heffernan KS, Ranadive SM, Jae SY. Exercise as medicine for covid-19: On ppar with emerging pharmacotherapy. Med Hypotheses (2020) 143:110197. doi: 10.1016/j.mehy.2020.110197
34. Fantacuzzi M, Amoroso R, Ammazzalorso A. Ppar ligands induce antiviral effects targeting perturbed lipid metabolism during sars-Cov-2, hcv, and hcmv infection. Biology (2022) 11(1):114. doi: 10.3390/biology11010114
35. Calder PC. N-3 fatty acids, inflammation and immunity: New mechanisms to explain old actions. Proc Nutr Soc (2013) 72(3):326–36. doi: 10.1017/S0029665113001031
36. Jacob A, Wu R, Zhou M, Wang P. Mechanism of the anti-inflammatory effect of curcumin: Ppar-Γ activation. PPAR Res (2007) 2007:089369. doi: 10.1155/2007/89369
37. Del Re A, Corpetti C, Pesce M, Seguella L, Steardo L, Palenca I, et al. Ultramicronized palmitoylethanolamide inhibits Nlrp3 inflammasome expression and pro-inflammatory response activated by sars-Cov-2 spike protein in cultured murine alveolar macrophages. Metabolites (2021) 11(9):592. doi: 10.3390/metabo11090592
38. Ciavarella C, Motta I, Valente S, Pasquinelli G. Pharmacological (or synthetic) and nutritional agonists of ppar-Γ as candidates for cytokine storm modulation in covid-19 disease. Molecules (2020) 25(9):2076. doi: 10.3390/molecules25092076
39. Hasankhani A, Bahrami A, Sheybani N, Aria B, Hemati B, Fatehi F, et al. Differential Co-expression network analysis reveals key hub-high traffic genes as potential therapeutic targets for covid-19 pandemic. Front Immunol (2021) 12:789317. doi: 10.3389/fimmu.2021.789317
40. Auwul MR, Rahman MR, Gov E, Shahjaman M, Moni MA. Bioinformatics and machine learning approach identifies potential drug targets and pathways in covid-19. Briefings Bioinf (2021) 22(5). doi: 10.1093/bib/bbab120
41. Hihi AK, Michalik L, Wahli W. Ppars: Transcriptional effectors of fatty acids and their derivatives. Cell Mol Life Sci (2002) 59(5):790–8. doi: 10.1007/s00018-002-8467-x
42. Kota BP, Huang TH-W, Roufogalis BD. An overview on biological mechanisms of ppars. Pharmacol Res (2005) 51(2):85–94. doi: 10.1016/j.phrs.2004.07.012
43. Wang Y-X. Ppars: Diverse regulators in energy metabolism and metabolic diseases. Cell Res (2010) 20(2):124–37. doi: 10.1038/cr.2010.13
44. Wagner N, Wagner K-D. Ppars and angiogenesis–implications in pathology. Int J Mol Sci (2020) 21(16):5723. doi: 10.3390/ijms21165723
45. Wahli W, Michalik L. Ppars at the crossroads of lipid signaling and inflammation. Trends Endocrinol Metab (2012) 23(7):351–63. doi: 10.1016/j.tem.2012.05.001
46. Ricote M, Glass CK. Ppars and molecular mechanisms of transrepression. Biochim Biophys Acta (BBA) - Mol Cell Biol Lipids (2007) 1771(8):926–35. doi: 10.1016/j.bbalip.2007.02.013
47. Grygiel-Górniak B. Peroxisome proliferator-activated receptors and their ligands: Nutritional and clinical implications - a review. Nutr J (2014) 13(1):17. doi: 10.1186/1475-2891-13-17
48. Crossland H, Constantin-Teodosiu D, Greenhaff PL. The regulatory roles of ppars in skeletal muscle fuel metabolism and inflammation: Impact of ppar agonism on muscle in chronic disease, contraction and sepsis. Int J Mol Sci (2021) 22(18):9775. doi: 10.3390/ijms22189775
49. Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature (1990) 347(6294):645–50. doi: 10.1038/347645a0
50. Yousefnia S, Momenzadeh S, Seyed Forootan F, Ghaedi K, Nasr Esfahani MH. The influence of peroxisome proliferator-activated receptor Γ (Pparγ) ligands on cancer cell tumorigenicity. Gene (2018) 649:14–22. doi: 10.1016/j.gene.2018.01.018
51. Basilotta R, Lanza M, Casili G, Chisari G, Munao S, Colarossi L, et al. Potential therapeutic effects of ppar ligands in glioblastoma. Cells (2022) 11(4):621. doi: 10.3390/cells11040621
52. Chinetti G, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors (Ppars): Nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflammation Res (2000) 49(10):497–505. doi: 10.1007/s000110050622
53. Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors A and Γ. Proc Natl Acad Sci (1997) 94(9):4318–23. doi: 10.1073/pnas.94.9.4318
54. Neschen S, Morino K, Dong J, Wang-Fischer Y, Cline GW, Romanelli AJ, et al. N-3 fatty acids preserve insulin sensitivity in vivo in a peroxisome proliferator–activated Receptor-A–dependent manner. Diabetes (2007) 56(4):1034–41. doi: 10.2337/db06-1206
55. Tan Y, Wang M, Yang K, Chi T, Liao Z, Wei P. Ppar-A modulators as current and potential cancer treatments. Front Oncol (2021) 11:599995. doi: 10.3389/fonc.2021.599995
56. Yang L, Guo H, Li Y, Meng X, Yan L, Dan Z, et al. Oleoylethanolamide exerts anti-inflammatory effects on lps-induced thp-1 cells by enhancing pparα signaling and inhibiting the nf-Kb and Erk1/2/Ap-1/Stat3 pathways. Sci Rep (2016) 6(1):34611. doi: 10.1038/srep34611
57. Bougarne N, Weyers B, Desmet SJ, Deckers J, Ray DW, Staels B, et al. Molecular actions of pparα in lipid metabolism and inflammation. Endocr Rev (2018) 39(5):760–802. doi: 10.1210/er.2018-00064
58. Stienstra R, Mandard S, Tan NS, Wahli W, Trautwein C, Richardson TA, et al. The interleukin-1 receptor antagonist is a direct target gene of pparα in liver. J Hepatol (2007) 46(5):869–77. doi: 10.1016/j.jhep.2006.11.019
59. Mandard S, Müller M, Kersten S. Peroxisome proliferator-activated receptor A target genes. Cell Mol Life Sci CMLS (2004) 61(4):393–416. doi: 10.1007/s00018-003-3216-3
60. Meissner M, Stein M, Urbich C, Reisinger K, Suske G, Staels B, et al. Pparα activators inhibit vascular endothelial growth factor receptor-2 expression by repressing Sp1-dependent DNA binding and transactivation. Circ Res (2004) 94(3):324–32. doi: 10.1161/01.RES.0000113781.08139.81
61. Medina-Gomez G, Gray SL, Yetukuri L, Shimomura K, Virtue S, Campbell M, et al. Ppar gamma 2 prevents lipotoxicity by controlling adipose tissue expandability and peripheral lipid metabolism. PloS Genet (2007) 3(4):e64. doi: 10.1371/journal.pgen.0030064
62. Lehrke M, Lazar MA. The many faces of pparγ. Cell (2005) 123(6):993–9. doi: 10.1016/j.cell.2005.11.026
63. Cataldi S, Costa V, Ciccodicola A, Aprile M. Pparγ and diabetes: Beyond the genome and towards personalized medicine. Curr Diabetes Rep (2021) 21(6):18. doi: 10.1007/s11892-021-01385-5
64. Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr Rev (1999) 20(5):649–88. doi: 10.1210/edrv.20.5.0380
65. Jamwal S, Blackburn JK, Elsworth JD. Pparγ/Pgc1α signaling as a potential therapeutic target for mitochondrial biogenesis in neurodegenerative disorders. Pharmacol Ther (2021) 219:107705. doi: 10.1016/j.pharmthera.2020.107705
66. Xu WP, Zhang X, Xie WF. Differentiation therapy for solid tumors. J Digestive Dis (2014) 15(4):159–65. doi: 10.1111/1751-2980.12122
67. Cui L, Li Z, Xu F, Tian Y, Chen T, Li J, et al. Antitumor effects of astaxanthin on esophageal squamous cell carcinoma by up-regulation of pparγ. Nutr Cancer Via (2022) 74(4):1399–410. doi: 10.1080/01635581.2021.1952449
68. Wu L, Guo C, Wu J. Therapeutic potential of pparγ natural agonists in liver diseases. J Cell Mol Med (2020) 24(5):2736–48. doi: 10.1111/jcmm.15028
69. Choi M-J, Lee E-J, Park J-S, Kim S-N, Park E-M, Kim H-S. Anti-inflammatory mechanism of galangin in lipopolysaccharide-stimulated microglia: Critical role of ppar-Γ signaling pathway. Biochem Pharmacol (2017) 144:120–31. doi: 10.1016/j.bcp.2017.07.021
70. Luzi L, Radaelli MG. Influenza and obesity: Its odd relationship and the lessons for covid-19 pandemic. Acta Diabetologica (2020) 57(6):759–64. doi: 10.1007/s00592-020-01522-8
71. Caioni G, Viscido A, d’Angelo M, Panella G, Castelli V, Merola C, et al. Inflammatory bowel disease: New insights into the interplay between environmental factors and pparγ. Int J Mol Sci (2021) 22(3). doi: 10.3390/ijms22030985
72. Kökény G, Calvier L, Hansmann G. Pparγ and tgfβ–major regulators of metabolism, inflammation, and fibrosis in the lungs and kidneys. Int J Mol Sci (2021) 22(19):10431. doi: 10.3390/ijms221910431
73. Yang C-C, Wu C-H, Lin T-C, Cheng Y-N, Chang C-S, Lee K-T, et al. Inhibitory effect of pparγ on Nlrp3 inflammasome activation. Theranostics (2021) 11(5):2424. doi: 10.7150/thno.46873
74. Luquet S, Lopez-Soriano J, Holst D, Gaudel C, Jehl-Pietri C, Fredenrich A, et al. Roles of peroxisome proliferator-activated receptor delta (Pparδ) in the control of fatty acid catabolism. a new target for the treatment of metabolic syndrome. Biochimie (2004) 86(11):833–7. doi: 10.1016/j.biochi.2004.09.024
75. Stephen RL, Gustafsson MCU, Jarvis M, Tatoud R, Marshall BR, Knight D, et al. Activation of peroxisome proliferator-activated receptor Δ stimulates the proliferation of human breast and prostate cancer cell lines. Cancer Res (2004) 64(9):3162–70. doi: 10.1158/0008-5472.CAN-03-2760
76. Lee MY, Choi R, Kim HM, Cho EJ, Kim BH, Choi YS, et al. Peroxisome proliferator-activated receptor Δ agonist attenuates hepatic steatosis by anti-inflammatory mechanism. Exp Mol Med (2012) 44(10):578–85. doi: 10.3858/emm.2012.44.10.066
77. Liu Y, Colby JK, Zuo X, Jaoude J, Wei D, Shureiqi I. The role of ppar-Δ in metabolism, inflammation, and cancer: Many characters of a critical transcription factor. Int J Mol Sci (2018) 19(11):3339. doi: 10.3390/ijms19113339
78. Jung TW, Lee SH, Kim H-C, Bang JS, Abd El-Aty AM, Hacımüftüoğlu A, et al. Metrnl attenuates lipid-induced inflammation and insulin resistance Via ampk or pparδ-dependent pathways in skeletal muscle of mice. Exp Mol Med (2018) 50(9):1–11. doi: 10.1038/s12276-018-0147-5
79. Malm T, Mariani M, Donovan LJ, Neilson L, Landreth GE. Activation of the nuclear receptor pparδ is neuroprotective in a transgenic mouse model of alzheimer’s disease through inhibition of inflammation. J Neuroinflamm (2015) 12(1):7. doi: 10.1186/s12974-014-0229-9
80. Delerive P, Furman C, Teissier E, Fruchart J-C, Duriez P, Staels B. Oxidized phospholipids activate pparα in a phospholipase A2-dependent manner. FEBS Lett (2000) 471(1):34–8. doi: 10.1016/S0014-5793(00)01364-8
81. Wagner K-D, Wagner N. Peroxisome proliferator-activated receptor Beta/Delta (Pparβ/Δ) acts as regulator of metabolism linked to multiple cellular functions. Pharmacol Ther (2010) 125(3):423–35. doi: 10.1016/j.pharmthera.2009.12.001
82. Chen W, Wang L-L, Liu H-Y, Long L, Li S. Peroxisome proliferator-activated receptor Δ-agonist, Gw501516, ameliorates insulin resistance, improves dyslipidaemia in monosodium l-glutamate metabolic syndrome mice. Basic Clin Pharmacol Toxicol (2008) 103(3):240–6. doi: 10.1111/j.1742-7843.2008.00268.x
83. Yu B-C, Chang C-K, Ou H-Y, Cheng K-C, Cheng J-T. Decrease of peroxisome proliferator-activated receptor delta expression in cardiomyopathy of streptozotocin-induced diabetic rats. Cardiovasc Res (2008) 80(1):78–87. doi: 10.1093/cvr/cvn172
84. Henke BR. Peroxisome proliferator-activated receptor A/Γ dual agonists for the treatment of type 2 diabetes. J Medicinal Chem (2004) 47(17):4118–27. doi: 10.1021/jm030631e
85. Egerod FL, Brünner N, Svendsen JE, Oleksiewicz MB. Pparα and pparγ are Co-expressed, functional and show positive interactions in the rat urinary bladder urothelium. J Appl Toxicol (2010) 30(2):151–62. doi: 10.1002/jat.1481
86. Schulman IG. Nuclear receptors as drug targets for metabolic disease. Advanced Drug Delivery Rev (2010) 62(13):1307–15. doi: 10.1016/j.addr.2010.07.002
87. Sheu S-H, Kaya T, Waxman DJ, Vajda S. Exploring the binding site structure of the pparγ ligand-binding domain by computational solvent mapping. Biochemistry (2005) 44(4):1193–209. doi: 10.1021/bi048032c
88. Sauer S. Ligands for the nuclear peroxisome proliferator-activated receptor gamma. Trends Pharmacol Sci (2015) 36(10):688–704. doi: 10.1016/j.tips.2015.06.010
89. Poulsen LLC, Siersbæk M, Mandrup S. Ppars: Fatty acid sensors controlling metabolism. Semin Cell Dev Biol (2012) 23(6):631–9. doi: 10.1016/j.semcdb.2012.01.003
90. Brunmeir R, Xu F. Functional regulation of ppars through post-translational modifications. Int J Mol Sci (2018) 19(6):1738. doi: 10.3390/ijms19061738
91. Tzeng J, Byun J, Park JY, Yamamoto T, Schesing K, Tian B, et al. An ideal ppar response element bound to and activated by pparα. PloS One (2015) 10(8):e0134996. doi: 10.1371/journal.pone.0134996
92. Ahmed W, Ziouzenkova O, Brown J, Devchand P, Francis S, Kadakia M, et al. Ppars and their metabolic modulation: New mechanisms for transcriptional regulation? J Internal Med (2007) 262(2):184–98. doi: 10.1111/j.1365-2796.2007.01825.x
93. Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, et al. Ligand binding and Co-activator assembly of the peroxisome proliferator-activated receptor-Γ. Nature (1998) 395(6698):137–43. doi: 10.1038/25931
94. Wright MB, Bortolini M, Tadayyon M, Bopst M. Minireview: Challenges and opportunities in development of ppar agonists. Mol Endocrinol (2014) 28(11):1756–68. doi: 10.1210/me.2013-1427
95. Zhang X, Young HA. Ppar and immune system–what do we know? Int Immunopharmacol (2002) 2(8):1029–44. doi: 10.1016/S1567-5769(02)00057-7
96. Berger JP, Akiyama TE, Meinke PT. Ppars: Therapeutic targets for metabolic disease. Trends Pharmacol Sci (2005) 26(5):244–51. doi: 10.1016/j.tips.2005.03.003
97. Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev (1999) 9(2):140–7. doi: 10.1016/S0959-437X(99)80021-5
98. Wan Y, Shang J, Graham R, Baric Ralph S, Li F. Receptor recognition by the novel coronavirus from wuhan: An analysis based on decade-long structural studies of sars coronavirus. J Virol (2020) 94(7):e00127–20. doi: 10.1128/JVI.00127-20
99. Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H. Tissue distribution of Ace2 protein, the functional receptor for sars coronavirus. a first step in understanding sars pathogenesis. J Pathol (2004) 203(2):631–7. doi: 10.1002/path.1570
100. Fu L, Wang B, Yuan T, Chen X, Ao Y, Fitzpatrick T, et al. Clinical characteristics of coronavirus disease 2019 (Covid-19) in China: A systematic review and meta-analysis. J Infection (2020) 80(6):656–65. doi: 10.1016/j.jinf.2020.03.041
101. Jensen S, Thomsen Allan R. Sensing of rna viruses: A review of innate immune receptors involved in recognizing rna virus invasion. J Virol (2012) 86(6):2900–10. doi: 10.1128/JVI.05738-11
102. Yang L, Xie X, Tu Z, Fu J, Xu D, Zhou Y. The signal pathways and treatment of cytokine storm in covid-19. Signal Transduction Targeted Ther (2021) 6(1):255. doi: 10.1038/s41392-021-00679-0
103. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe covid-19 patients. Science (2020) 369(6504):718–24. doi: 10.1126/science.abc6027
104. Ronit A, Berg RMG, Bay JT, Haugaard AK, Ahlström MG, Burgdorf KS, et al. Compartmental immunophenotyping in covid-19 Ards: A case series. J Allergy Clin Immunol (2021) 147(1):81–91. doi: 10.1016/j.jaci.2020.09.009
105. Nile SH, Nile A, Qiu J, Li L, Jia X, Kai G. Covid-19: Pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev (2020) 53:66–70. doi: 10.1016/j.cytogfr.2020.05.002
106. Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of covid-19. J Pharm Anal (2020) 10(2):102–8. doi: 10.1016/j.jpha.2020.03.001
107. Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, et al. Longitudinal analyses reveal immunological misfiring in severe covid-19. Nature (2020) 584(7821):463–9. doi: 10.1038/s41586-020-2588-y
108. Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, Brüggen M-C, et al. Immune response to sars-Cov-2 and mechanisms of immunopathological changes in covid-19. Allergy (2020) 75(7):1564–81. doi: 10.1111/all.14364
109. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. Covid-19: Consider cytokine storm syndromes and immunosuppression. Lancet (2020) 395(10229):1033–4. doi: 10.1016/S0140-6736(20)30628-0
110. Henderson LA, Canna SW, Schulert GS, Volpi S, Lee PY, Kernan KF, et al. On the alert for cytokine storm: Immunopathology in covid-19. Arthritis Rheumatol (2020) 72(7):1059–63. doi: 10.1002/art.41285
111. Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (Covid-19) outbreak. J Autoimmun (2020) 109:102433. doi: 10.1016/j.jaut.2020.102433
112. Han H, Ma Q, Li C, Liu R, Zhao L, Wang W, et al. Profiling serum cytokines in covid-19 patients reveals il-6 and il-10 are disease severity predictors. Emerging Microbes Infections (2020) 9(1):1123–30. doi: 10.1080/22221751.2020.1770129
113. Liu Y, Zhang C, Huang F, Yang Y, Wang F, Yuan J, et al. Elevated plasma levels of selective cytokines in covid-19 patients reflect viral load and lung injury. Natl Sci Rev (2020) 7(6):1003–11. doi: 10.1093/nsr/nwaa037
114. Xu Z-S, Shu T, Kang L, Wu D, Zhou X, Liao B-W, et al. Temporal profiling of plasma cytokines, chemokines and growth factors from mild, severe and fatal covid-19 patients. Signal Transduction Targeted Ther (2020) 5(1):100. doi: 10.1038/s41392-020-0211-1
115. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet (2020) 395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5
116. Chinetti G, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors and inflammation: From basic science to clinical applications. Int J Obes (2003) 27(3):S41–S5. doi: 10.1038/sj.ijo.0802499
117. Martin H. Role of ppar-gamma in inflammation. prospects for therapeutic intervention by food components. Mutat Research/Fundamental Mol Mech Mutagenesis (2009) 669(1):1–7. doi: 10.1016/j.mrfmmm.2009.06.009
118. Delayre-Orthez C, Becker J, Guenon I, Lagente V, Auwerx J, Frossard N, et al. Pparα downregulates airway inflammation induced by lipopolysaccharide in the mouse. Respir Res (2005) 6(1):91. doi: 10.1186/1465-9921-6-91
119. Heming M, Gran S, Jauch S-L, Fischer-Riepe L, Russo A, Klotz L, et al. Peroxisome proliferator-activated receptor-Γ modulates the response of macrophages to lipopolysaccharide and glucocorticoids. Front Immunol (2018) 9:893. doi: 10.3389/fimmu.2018.00893
120. Huang S, Goplen NP, Zhu B, Cheon IS, Son Y, Wang Z, et al. Macrophage ppar-Γ suppresses long-term lung fibrotic sequelae following acute influenza infection. PloS One (2019) 14(10):e0223430. doi: 10.1371/journal.pone.0223430
121. Kaplan J, Nowell M, Chima R, Zingarelli B. Pioglitazone reduces inflammation through inhibition of nf-Kb in polymicrobial sepsis. Innate Immun (2013) 20(5):519–28. doi: 10.1177/1753425913501565
122. Huang W, Rha GB, Han M-J, Eum SY, András IE, Zhong Y, et al. Pparα and pparγ effectively protect against hiv-induced inflammatory responses in brain endothelial cells. J Neurochem (2008) 107(2):497–509. doi: 10.1111/j.1471-4159.2008.05626.x
123. Neri T, Armani C, Pegoli A, Cordazzo C, Carmazzi Y, Brunelleschi S, et al. Role of nf-Kb and ppar-Γ in lung inflammation induced by monocyte-derived microparticles. Eur Respir J (2011) 37(6):1494–502. doi: 10.1183/09031936.00023310
124. Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, et al. A cbp integrator complex mediates transcriptional activation and ap-1 inhibition by nuclear receptors. Cell (1996) 85(3):403–14. doi: 10.1016/S0092-8674(00)81118-6
125. Delerive P, De Bosscher K, Besnard S, Vanden Berghe W, Peters JM, Gonzalez FJ, et al. Peroxisome proliferator-activated receptor A negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors nf-Kb and ap-1. J Biol Chem (1999) 274(45):32048–54. doi: 10.1074/jbc.274.45.32048
126. Vanden Berghe W, Vermeulen L, Delerive P, De Bosscher K, Staels B, Haegeman G. A paradigm for gene regulation: Inflammation, nf-Kb and ppar. In: Peroxisomal disorders and regulation of genes. Boston, MA: Springer US (2003).
127. Kleemann R, Verschuren L, de Rooij B-J, Lindeman J, de Maat MM, Szalai AJ, et al. Evidence for anti-inflammatory activity of statins and pparα activators in human c-reactive protein transgenic mice in vivo and in cultured human hepatocytes in vitro. Blood (2004) 103(11):4188–94. doi: 10.1182/blood-2003-11-3791
128. Desreumaux P, Dubuquoy L, Nutten S, Peuchmaur M, Englaro W, Schoonjans K, et al. Attenuation of colon inflammation through activators of the retinoid X receptor (Rxr)/Peroxisome proliferator–activated receptor Γ (Pparγ) heterodimer: A basis for new therapeutic strategies. J Exp Med (2001) 193(7):827–38. doi: 10.1084/jem.193.7.827
129. Goetze S, Kintscher U, Kim S, Meehan WP, Kaneshiro K, Collins AR, et al. Peroxisome proliferator–activated receptor-Γ ligands inhibit nuclear but not cytosolic extracellular signal–regulated Kinase/Mitogen–activated protein kinase–regulated steps in vascular smooth muscle cell migration. J Cardiovasc Pharmacol (2001) 38(6):909–21. doi: 10.1097/00005344-200112000-00013
130. Ghisletti S, Huang W, Ogawa S, Pascual G, Lin M-E, Willson TM, et al. Parallel sumoylation-dependent pathways mediate gene- and signal-specific transrepression by lxrs and pparγ. Mol Cell (2007) 25(1):57–70. doi: 10.1016/j.molcel.2006.11.022
131. Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, et al. A sumoylation-dependent pathway mediates transrepression of inflammatory response genes by ppar-Γ. Nature (2005) 437(7059):759–63. doi: 10.1038/nature03988
132. Ferreira AE, Sisti F, Sônego F, Wang S, Filgueiras LR, Brandt S, et al. Ppar-Γ/Il-10 axis inhibits Myd88 expression and ameliorates murine polymicrobial sepsis. J Immunol (2014) 192(5):2357–65. doi: 10.4049/jimmunol.1302375
133. Xu Z, Wang G, Zhu Y, Liu R, Song J, Ni Y, et al. Ppar-Γ agonist ameliorates liver pathology accompanied by increasing regulatory b and T cells in high-Fat-Diet mice. Obesity (2017) 25(3):581–90. doi: 10.1002/oby.21769
134. Are A, Aronsson L, Wang S, Greicius G, Lee YK, Gustafsson J-Å, et al. Enterococcus faecalis from newborn babies regulate endogenous pparγ activity and il-10 levels in colonic epithelial cells. Proc Natl Acad Sci (2008) 105(6):1943–8. doi: 10.1073/pnas.0711734105
135. Marx N, Sukhova GK, Collins T, Libby P, Plutzky J. Pparα activators inhibit cytokine-induced vascular cell adhesion molecule-1 expression in human endothelial cells. Circulation (1999) 99(24):3125–31. doi: 10.1161/01.CIR.99.24.3125
136. Kurebayashi S, Xu X, Ishii S, Shiraishi M, Kouhara H, Kasayama S. A novel thiazolidinedione mcc-555 down-regulates tumor necrosis factor-A-Induced expression of vascular cell adhesion molecule-1 in vascular endothelial cells. Atherosclerosis (2005) 182(1):71–7. doi: 10.1016/j.atherosclerosis.2005.02.004
137. Ahmed W, Orasanu G, Nehra V, Asatryan L, Rader DJ, Ziouzenkova O, et al. High-density lipoprotein hydrolysis by endothelial lipase activates pparα. Circ Res (2006) 98(4):490–8. doi: 10.1161/01.RES.0000205846.46812.be
138. Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell (2005) 122(5):707–21. doi: 10.1016/j.cell.2005.06.029
139. Lee JW, Bajwa PJ, Carson MJ, Jeske DR, Cong Y, Elson CO, et al. Fenofibrate represses interleukin-17 and interferon-Γ expression and improves colitis in Interleukin-10–deficient mice. Gastroenterology (2007) 133(1):108–23. doi: 10.1053/j.gastro.2007.03.113
140. Prasad K, AlOmar SY, Almuqri EA, Rudayni HA, Kumar V. Genomics-guided identification of potential modulators of sars-Cov-2 entry proteases, Tmprss2 and cathepsins B/L. PloS One (2021) 16(8):e0256141. doi: 10.1371/journal.pone.0256141
141. Desterke C, Turhan AG, Bennaceur-Griscelli A, Griscelli F. Hla-dependent heterogeneity and macrophage immunoproteasome activation during lung covid-19 disease. J Trans Med (2021) 19(1):290. doi: 10.1186/s12967-021-02965-5
142. Jackson H, Rivero Calle I, Broderick C, Habgood-Coote D, D’Souza G, Nichols S, et al. Characterisation of the blood rna host response underpinning severity in covid-19 patients. Sci Rep (2022) 12(1):12216. doi: 10.1038/s41598-022-15547-2
143. Vlasov I, Panteleeva A, Usenko T, Nikolaev M, Izumchenko A, Gavrilova E, et al. Transcriptomic profiles reveal downregulation of low-density lipoprotein particle receptor pathway activity in patients surviving severe covid-19. Cells (2021) 10(12):3495. doi: 10.3390/cells10123495
144. Nain Z, Barman SK, Sheam MM, Syed SB, Samad A, Quinn JMW, et al. Transcriptomic studies revealed pathophysiological impact of covid-19 to predominant health conditions. Briefings Bioinf (2021) 22(6):bbab197. doi: 10.1093/bib/bbab197
145. Desterke C, Turhan AG, Bennaceur-Griscelli A, Griscelli F. Pparγ cistrome repression during activation of lung monocyte-macrophages in severe covid-19. iScience (2020) 23(10):101611. doi: 10.1016/j.isci.2020.101611
146. Costanzo M, Caterino M, Fedele R, Cevenini A, Pontillo M, Barra L, et al. Covidomics: The proteomic and metabolomic signatures of covid-19. Int J Mol Sci (2022) 23(5):2414. doi: 10.3390/ijms23052414
147. Yang J, Chen C, Chen W, Huang L, Fu Z, Ye K, et al. Proteomics and metabonomics analyses of covid-19 complications in patients with pulmonary fibrosis. Sci Rep (2021) 11(1):14601. doi: 10.1038/s41598-021-94256-8
148. Keikha R, Hashemi-Shahri SM, Jebali A. The mirna neuroinflammatory biomarkers in covid-19 patients with different severity of illness. Neurología (2021). doi: 10.1016/j.nrl.2021.06.005
149. Batiha GE-S, Gari A, Elshony N, Shaheen HM, Abubakar MB, Adeyemi SB, et al. Hypertension and its management in covid-19 patients: The assorted view. Int J Cardiol Cardiovasc Risk Prev (2021) 11:200121. doi: 10.1016/j.ijcrp.2021.200121
150. Alkhayyat SS, Al-kuraishy HM, Al-Gareeb AI, El-Bouseary MM, AboKamer AM, Batiha GE-S, et al. Fenofibrate for covid-19 and related complications as an approach to improve treatment outcomes: The missed key for holy grail. Inflammation Res (2022) 71:1159–67. doi: 10.1007/s00011-022-01615-w
151. Jha NK, Sharma C, Hashiesh HM, Arunachalam S, Meeran MN, Javed H, et al. B-caryophyllene, a natural dietary Cb2 receptor selective cannabinoid can be a candidate to target the trinity of infection, immunity, and inflammation in covid-19. Front Pharmacol (2021) 12:590201. doi: 10.3389/fphar.2021.590201
152. O’Carroll SM, O’Neill LAJ. Targeting immunometabolism to treat covid-19. Immunother Adv (2021) 1(1). doi: 10.1093/immadv/ltab013
153. Ghasemnejad-Berenji M. Immunomodulatory and anti-inflammatory potential of crocin in covid-19 treatment. J Food Biochem (2021) 45(5):e13718. doi: 10.1111/jfbc.13718
154. Ayres JS. A metabolic handbook for the covid-19 pandemic. Nat Metab (2020) 2(7):572–85. doi: 10.1038/s42255-020-0237-2
155. Mahmudpour M, Vahdat K, Keshavarz M, Nabipour I. The covid-19-Diabetes mellitus molecular tetrahedron. Mol Biol Rep (2022) 49(5):4013–24. doi: 10.1007/s11033-021-07109-y
156. Vallée A, Lecarpentier Y, Vallée J-N. Interplay of opposing effects of the Wnt/B;-catenin pathway and pparγ and implications for sars-Cov2 treatment. Front Immunol (2021) 12:666693. doi: 10.3389/fimmu.2021.666693
157. Francisqueti-Ferron FV, Garcia JL, Ferron AJT, Nakandakare- Maia ET, Gregolin CS, JPdC S, et al. Gamma-oryzanol as a potential modulator of oxidative stress and inflammation Via ppar-y in adipose tissue: A hypothetical therapeutic for cytokine storm in covid-19? Mol Cell Endocrinol (2021) 520:111095. doi: 10.1016/j.mce.2020.111095
158. Huang W, Zhou H, Hodgkinson C, Montero A, Goldman D, Chang SL. Network meta-analysis on the mechanisms underlying alcohol augmentation of covid-19 pathologies. Alcohol Clin Exp Res (2021) 45(4):675–88. doi: 10.1111/acer.14573
159. Adir Y, Saliba W, Beurnier A, Humbert M. Asthma and covid-19: An update. Eur Respir Rev (2021) 30(162):210152. doi: 10.1183/16000617.0152-2021
160. Ramakrishnan RK, Al Heialy S, Hamid Q. Implications of preexisting asthma on covid-19 pathogenesis. Am J Physiol-Lung Cell Mol Physiol (2021) 320(5):L880–L91. doi: 10.1152/ajplung.00547.2020
161. Farruggia C, Kim M-B, Bae M, Lee Y, Pham TX, Yang Y, et al. Astaxanthin exerts anti-inflammatory and antioxidant effects in macrophages in Nrf2-dependent and independent manners. J Nutr Biochem (2018) 62:202–9. doi: 10.1016/j.jnutbio.2018.09.005
162. Steinman JB, Lum FM, Ho PP-K, Kaminski N, Steinman L. Reduced development of covid-19 in children reveals molecular checkpoints gating pathogenesis illuminating potential therapeutics. Proc Natl Acad Sci (2020) 117(40):24620–6. doi: 10.1073/pnas.2012358117
163. Ferastraoaru D, Hudes G, Jerschow E, Jariwala S, Karagic M, de Vos G, et al. Eosinophilia in asthma patients is protective against severe covid-19 illness. J Allergy Clin Immunol: In Pract (2021) 9(3):1152–62.e3. doi: 10.1016/j.jaip.2020.12.045
164. Franceschini L, Macchiarelli R, Rentini S, Biviano I, Farsi A. Eosinophilic esophagitis: Is the Th2 inflammation protective against the severe form of covid-19? Eur J Gastroenterol Hepatol (2020) 32(12):1583. doi: 10.1097/meg.0000000000001909
165. Kostadinova R, Wahli W, Michalik L. Ppars in diseases: Control mechanisms of inflammation. Curr Medicinal Chem (2005) 12(25):2995–3009. doi: 10.2174/092986705774462905
166. Saubermann LJ, Nakajima A, Wada K, Zhao S, Terauchi Y, Kadowaki T, et al. Peroxisome proliferator-activated receptor gamma agonist ligands stimulate a Th2 cytokine response and prevent acute colitis. Inflammatory Bowel Dis (2002) 8(5):330–9. doi: 10.1097/00054725-200209000-00004
167. Chen T, Tibbitt CA, Feng X, Stark JM, Rohrbeck L, Rausch L, et al. Ppar-Γ promotes type 2 immune responses in allergy and nematode infection. Sci Immunol (2017) 2(9):eaal5196. doi: 10.1126/sciimmunol.aal5196
168. Jiang Y, Zhao T, Zhou X, Xiang Y, Gutierrez-Castrellon P, Ma X. Inflammatory pathways in covid-19: Mechanism and therapeutic interventions. MedComm (2022) 3(3):e154. doi: 10.1002/mco2.154
169. Mukherjee JJ, Gangopadhyay KK, Ray S. Management of diabetes in patients with covid-19. Lancet Diabetes Endocrinol (2020) 8(8):666. doi: 10.1016/S2213-8587(20)30226-6
170. Shirazi J, Donzanti MJ, Nelson KM, Zurakowski R, Fromen CA, Gleghorn JP. Significant unresolved questions and opportunities for bioengineering in understanding and treating covid-19 disease progression. Cell Mol Bioengineering (2020) 13(4):259–84. doi: 10.1007/s12195-020-00637-w
171. Kheirollahi V, Wasnick RM, Biasin V, Vazquez-Armendariz AI, Chu X, Moiseenko A, et al. Metformin induces lipogenic differentiation in myofibroblasts to reverse lung fibrosis. Nat Commun (2019) 10(1):2987. doi: 10.1038/s41467-019-10839-0
172. Bargagli E, Refini RM, d’Alessandro M, Bergantini L, Cameli P, Vantaggiato L, et al. Metabolic dysregulation in idiopathic pulmonary fibrosis. Int J Mol Sci (2020) 21(16):5663. doi: 10.3390/ijms21165663
173. Landi C, Bargagli E, Carleo A, Bianchi L, Gagliardi A, Prasse A, et al. A system biology study of balf from patients affected by idiopathic pulmonary fibrosis (Ipf) and healthy controls. Proteomics – Clin Appl (2014) 8(11-12):932–50. doi: 10.1002/prca.201400001
174. Ahmed DS, Isnard S, Berini C, Lin J, Routy J-P, Royston L. Coping with stress: The mitokine gdf-15 as a biomarker of covid-19 severity. Front Immunol (2022) 13:820350. doi: 10.3389/fimmu.2022.820350
175. Batabyal R, Freishtat N, Hill E, Rehman M, Freishtat R, Koutroulis I. Metabolic dysfunction and immunometabolism in covid-19 pathophysiology and therapeutics. Int J Obes (2021) 45(6):1163–9. doi: 10.1038/s41366-021-00804-7
176. AbdelMassih AF, Menshawey R, Ismail JH, Husseiny RJ, Husseiny YM, Yacoub S, et al. Ppar agonists as effective adjuvants for covid-19 vaccines, by modifying immunogenetics: A review of literature. J Genet Eng Biotechnol (2021) 19(1):82. doi: 10.1186/s43141-021-00179-2
177. Li Z, Peng M, Chen P, Liu C, Hu A, Zhang Y, et al. Imatinib and methazolamide ameliorate covid-19-Induced metabolic complications Via elevating Ace2 enzymatic activity and inhibiting viral entry. Cell Metab (2022) 34(3):424–40.e7. doi: 10.1016/j.cmet.2022.01.008
178. AbdelMassih A, El Shershaby M, Gaber H, Habib M, Gamal N, Husseiny R, et al. Should we vaccinate the better seroconverters or the most vulnerable? game changing insights for covid-19 vaccine prioritization policies. Egyptian Pediatr Assoc Gazette (2021) 69(1):39. doi: 10.1186/s43054-021-00086-8
179. Lumeng CN, Liu J, Geletka L, Delaney C, Delproposto J, Desai A, et al. Aging is associated with an increase in T cells and inflammatory macrophages in visceral adipose tissue. J Immunol (2011) 187(12):6208–16. doi: 10.4049/jimmunol.1102188
180. Caza T, Landas S. Functional and phenotypic plasticity of Cd4+ T cell subsets. BioMed Res Int (2015) 2015:521957. doi: 10.1155/2015/521957
181. Meftahi GH, Jangravi Z, Sahraei H, Bahari Z. The possible pathophysiology mechanism of cytokine storm in elderly adults with covid-19 infection: The contribution of “Inflame-aging”. Inflammation Res (2020) 69(9):825–39. doi: 10.1007/s00011-020-01372-8
182. Durmuş S, Çakır T, Özgür A, Guthke R. A review on computational systems biology of pathogen–host interactions. Front Microbiol (2015) 6:235. doi: 10.3389/fmicb.2015.00235
183. Oh KK, Adnan M, Cho DH. Network pharmacology approach to decipher signaling pathways associated with target proteins of nsaids against covid-19. Sci Rep (2021) 11(1):9606. doi: 10.1038/s41598-021-88313-5
184. Afroz S, Fairuz S, Joty JA, Uddin MN, Rahman MA. Virtual screening of functional foods and dissecting their roles in modulating gene functions to support post covid-19 complications. J Food Biochem (2021) 45(12):e13961. doi: 10.1111/jfbc.13961
185. Li H, You J, Yang X, Wei Y, Zheng L, Zhao Y, et al. Glycyrrhetinic acid: A potential drug for the treatment of covid-19 cytokine storm. Phytomedicine (2022) 102:154153. doi: 10.1016/j.phymed.2022.154153
186. Shirazi FM, Banerji S, Nakhaee S, Mehrpour O. Effect of angiotensin ii blockers on the prognosis of covid-19: A toxicological view. Eur J Clin Microbiol Infect Dis (2020) 39(10):2001–2. doi: 10.1007/s10096-020-03932-6
187. Fernández-Quintela A, Milton-Laskibar I, Trepiana J, Gómez-Zorita S, Kajarabille N, Léniz A, et al. Key aspects in nutritional management of covid-19 patients. J Clin Med (2020) 9(8):2589. doi: 10.3390/jcm9082589
188. Di Renzo L, Gualtieri P, Pivari F, Soldati L, Attinà A, Leggeri C, et al. Covid-19: Is there a role for immunonutrition in obese patient? J Trans Med (2020) 18(1):415. doi: 10.1186/s12967-020-02594-4
189. Ghaffari S, Roshanravan N, Tutunchi H, Ostadrahimi A, Pouraghaei M, Kafil B. Oleoylethanolamide, a bioactive lipid amide, as a promising treatment strategy for Coronavirus/Covid-19. Arch Med Res (2020) 51(5):464–7. doi: 10.1016/j.arcmed.2020.04.006
190. Akbari N, Ostadrahimi A, Tutunchi H, Pourmoradian S, Farrin N, Najafipour F, et al. Possible therapeutic effects of boron citrate and oleoylethanolamide supplementation in patients with covid-19: A pilot randomized, double-blind, clinical trial. J Trace Elements Med Biol (2022) 71:126945. doi: 10.1016/j.jtemb.2022.126945
191. Talukdar J, Bhadra B, Dattaroy T, Nagle V, Dasgupta S. Potential of natural astaxanthin in alleviating the risk of cytokine storm in covid-19. Biomed Pharmacother (2020) 132:110886. doi: 10.1016/j.biopha.2020.110886
192. Petersen KF, Krssak M, Inzucchi S, Cline GW, Dufour S, Shulman GI. Mechanism of troglitazone action in type 2 diabetes. Diabetes (2000) 49(5):827–31. doi: 10.2337/diabetes.49.5.827
193. Carboni E, Carta AR, Carboni E. Can pioglitazone be potentially useful therapeutically in treating patients with covid-19? Med Hypotheses (2020) 140:109776. doi: 10.1016/j.mehy.2020.109776
194. Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for sars-Cov-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B (2020) 10(5):766–88. doi: 10.1016/j.apsb.2020.02.008
195. Esposito G, Pesce M, Seguella L, Sanseverino W, Lu J, Corpetti C, et al. The potential of cannabidiol in the covid-19 pandemic. Br J Pharmacol (2020) 177(21):4967–70. doi: 10.1111/bph.15157
196. Oyagbemi AA, Ajibade TO, Aboua YG, Gbadamosi IT, Adedapo ADA, Aro AO, et al. Potential health benefits of zinc supplementation for the management of covid-19 pandemic. J Food Biochem (2021) 45(2):e13604. doi: 10.1111/jfbc.13604
197. Belvisi MG, Mitchell JA. Targeting ppar receptors in the airway for the treatment of inflammatory lung disease. Br J Pharmacol (2009) 158(4):994–1003. doi: 10.1111/j.1476-5381.2009.00373.x
198. Choi C-I. Astaxanthin as a peroxisome proliferator-activated receptor (Ppar) modulator: Its therapeutic implications. Mar Drugs (2019) 17(4):242. doi: 10.3390/md17040242
199. Inoue M, Tanabe H, Matsumoto A, Takagi M, Umegaki K, Amagaya S, et al. Astaxanthin functions differently as a selective peroxisome proliferator-activated receptor Γ modulator in adipocytes and macrophages. Biochem Pharmacol (2012) 84(5):692–700. doi: 10.1016/j.bcp.2012.05.021
Keywords: SARS-CoV-2, cytokine storm, PPARs, hyperinflammation, COVID-19 therapy,
Citation: Hasankhani A, Bahrami A, Tavakoli-Far B, Iranshahi S, Ghaemi F, Akbarizadeh MR, Amin AH, Abedi Kiasari B and Mohammadzadeh Shabestari A (2023) The role of peroxisome proliferator-activated receptors in the modulation of hyperinflammation induced by SARS-CoV-2 infection: A perspective for COVID-19 therapy. Front. Immunol. 14:1127358. doi: 10.3389/fimmu.2023.1127358
Received: 19 December 2022; Accepted: 08 February 2023;
Published: 17 February 2023.
Edited by:
Slaven Crnkovic, Medical University of Graz, AustriaReviewed by:
Mohammad Asad, Albert Einstein College of Medicine, United StatesAlessandra Ammazzalorso, University “G. d’Annunzio” of Chieti-Pescara, Italy
Copyright © 2023 Hasankhani, Bahrami, Tavakoli-Far, Iranshahi, Ghaemi, Akbarizadeh, Amin, Abedi Kiasari and Mohammadzadeh Shabestari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aliakbar Hasankhani, QS5oYXNhbmtoYW5pNzRAdXQuYWMuaXI=; Bahman Abedi Kiasari, YWJlZGlraWFzYXJpLmJAdXQuYWMuaXI=; Alireza Mohammadzadeh Shabestari, RHJzaGFiZXN0YXJpM0BnbWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
 Aliakbar Hasankhani
Aliakbar Hasankhani Abolfazl Bahrami
Abolfazl Bahrami Bahareh Tavakoli-Far3,4
Bahareh Tavakoli-Far3,4