- 1Institute of Human Genetics, Polish Academy of Sciences, Poznan, Poland
- 2Optegra Eye Health Care Clinic in Poznan, Poznan, Poland
- 3Chair of Ophthalmology and Optometry, Poznan University of Medical Sciences, Poznan, Poland
- 4Chair and Department of Genetics and Pharmaceutical Microbiology, Poznan University of Medical Sciences, Poznan, Poland
Background: Keratoconus (KTCN) is the most common corneal ectasia resulting in a conical shape of the cornea. Here, genomic variation in the corneal epithelium (CE) across the keratoconic cone surface in patients with KTCN and its relevance in the functioning of the immune system were assessed.
Methods: Samples from four unrelated adolescent patients with KTCN and two control individuals were obtained during the CXL and PRK procedures, respectively. Three topographic regions, central, middle, and peripheral, were separated towards the whole-genome sequencing (WGS) study embracing a total of 18 experimental samples. The coding and non-coding sequence variation, including structural variation, was assessed and then evaluated together with the previously reported transcriptomic outcomes for the same CE samples and full-thickness corneas.
Results: First, pathway enrichment analysis of genes with identified coding variants pointed to “Antigen presentation” and “Interferon alpha/beta signaling” as the most overrepresented pathways, indicating the involvement of inflammatory responses in KTCN. Both coding and non-coding sequence variants were found in genes (or in their close proximity) linked to the previously revealed KTCN-specific cellular components, namely, “Actin cytoskeleton”, “Extracellular matrix”, “Collagen-containing extracellular matrix”, “Focal adhesion”, “Hippo signaling pathway”, and “Wnt signaling” pathways. No genomic heterogeneity across the corneal surface was found comparing the assessed topographic regions. Thirty-five chromosomal regions enriched in both coding and non-coding KTCN-specific sequence variants were revealed, with a most representative 5q locus previously recognized as involved in KTCN.
Conclusion: The identified genomic features indicate the involvement of innate and adaptive immune system responses in KTCN pathogenesis.
1 Introduction
Keratoconus (KTCN) is the most common corneal ectasia, characterized by progressive stromal thinning and the presence of the keratoconic cone (1). The structural changes, which often manifest asymmetrically, lead to a loss of visual acuity as an effect of irregular astigmatism (2). The first symptoms usually occur in adolescence, and the disease progresses through the third or fourth decade of life (1). Keratoconus affects both genders, but the reason of its higher occurrence in male patients remains unclear (3). The occurrence of KTCN may vary depending on the ethnicity of patients (1), with an estimated prevalence of 1.38 per 1,000 in a general population (4).
Advanced KTCN remains one of the most common indications for full-thickness (penetrating keratoplasty) and partial-thickness (deep anterior lamellar keratoplasty) corneal transplantation in developed countries (5). In the face of the organ shortage crisis in worldwide transplantation, there is a need for other therapies that inhibit the progression of KTCN to an advanced stage. A corneal collagen cross-linking (CXL) is an effective alternative used to stabilize the progression of KTCN and often improves the quality of patients’ vision (6).
Histopathological abnormalities are present in all corneal layers of the KTCN cornea (7), with diverse manifestations in central and peripheral zones being an effect of different biomechanical tension across corneal curvature in KTCN (8). Changes in collagen fibers and other elements of the extracellular matrix (ECM) form stromal irregularities (8) contributing to the KTCN cone formation and also affect the corneal epithelium (CE). The resulting rebuilt epithelial structure is characterized by a central thinning surrounded by a thickened annulus, named doughnut pattern (9). In the morphological observations of the CE (including confocal and immunofluorescence microscopy), the decreased cell density and their elongated shape, disruptions in basal integrity, increased number of apoptotic cells, and depositions of iron particles have been previously described (7, 10, 11). Moreover, some molecular proteomic and transcriptomic findings showed disruption of elements of Wnt signaling (e.g., WNT10A) (12), cell–cell communications (e.g., CDH13) (13), pro-inflammatory cytokines (e.g., IL6) (14), matrix metalloproteinases (e.g., MMP9) (15), and innate immune system (e.g., TLR2 and TLR4) (16).
The available reports regarding molecular and environmental findings in KTCN studies (17–22) indicate the multifactorial nature of the disease. The behavioral, environmental, and socioeconomic factors supposedly induce disease manifestation/progression in genetically predisposed individuals. In clinical studies, KTCN was found to be related to chronic eye rubbing, allergy, atopy, asthma, and UV exposure (18).
Historically, KTCN has been described as a non-inflammatory disease (2), but in the last decade, this aspect has been widely studied and questioned by ophthalmologists. However, the currently valid Global Consensus on Keratoconus and Ectatic Diseases (23) has not addressed this issue.
Previously published transcriptomic and proteomic findings derived a hint for the influence of inflammatory factors in KTCN as the increased levels of inflammatory cytokines (as IL6, TNFα, and MMP9) were found in patients’ tear fluid and serum samples and suggested to contribute to the thinning and weakening of the corneal tissue in KTCN (17, 24–26).
So far, the genetic aspects regarding the functioning of the immune system in the KTCN have not been demonstrated. Numerous genetic findings, including familial inheritance (27, 28), a concordance between monozygotic twins in contrast to dizygotic twins (29), and the occurrence of syndromic KTCN (30), imply the presence of a genetic component for the disease development. The previous results obtained by our research group confirmed the postulated genetic heterogeneity between patients and the involvement of numerous genetic factors in the pathogenesis of KTCN (19, 31–33). Besides the identification of the 13q32 (31), 5q31.1-q35.3 (32), 2q13-q14.3, and 20p13-p12.2 (33) KTCN loci, the numerous sequence variants in VSX1, TGFBI, DOCK9, STK24, IPO5, SKP1 and IL17B candidate genes (19, 32) have been demonstrated. So far, whole-genome sequencing (WGS) has not been applied in KTCN research. As only WGS provides an overview of the entire human genome, and it is a suitable method for causative variant discovery in genetically heterogeneous diseases, this approach is a natural next step in investigating the pathogenesis of KTCN. We hypothesize that variants in non-coding regions of the genome complement the previously identified features specific to the KTCN exome.
The multilevel structural and functional changes in cornea, including the varied morphology and various intensities of inflammatory-causing internal biomechanical tension and external environmental stimuli (e.g., eye rubbing) acting on particular zones of CE, suggest the diverse molecular features across corneal curvature. These premises support the designation and separation of three topographic regions of the CE, namely, central, middle, and peripheral. Here, we assessed the genomic diversity across keratoconic cone surface implementing a unique study design embracing the three topographic regions of the CE of adolescent patients with KTCN.
2 Materials and methods
2.1 Ophthalmic examination and patients’ inclusion and exclusion criteria
Unrelated adolescent patients with KTCN and the youngest available controls (the non-KTCN individuals with mild myopia) were involved in this study. The study protocol was approved by the Bioethics Committee at Poznan University of Medical Sciences, Poznan, Poland. The possible consequences of the study were explained, and informed consent was obtained from all participants, according to the Declaration of Helsinki.
Each individual underwent a complete ophthalmological examination, including the assessments of both uncorrected (UCVA) and best-corrected visual acuity (BCVA), intraocular pressure (IOP), corneal tomography with rotating Scheimpflug camera WaveLight® Oculyzer™ II (Alcon, TX, USA), epithelial thickness mapping [Spectral-Domain Optical Coherence Tomography (SD-OCT) device, Zeiss Cirrus 5000, Carl Zeiss Meditec, Dublin, California, USA], and slit-lamp and dilated funduscopic examination. A questionnaire comprising the behavioral, environmental, and socioeconomic aspects, including eye rubbing, use of contact lenses, atopy, UV exposure, smoking, reading habits, time spent in front of a screen, hormone intake, education level, and place of living, was completed by each participant.
The inclusion and exclusion criteria for adolescents with KTCN, and control individuals are described in detail in Supplementary Methods 1.1.
2.2 CXL and PRK procedures
CXL in patients with KTCN was performed in accordance with the standard Dresden Protocol (34), while the PRK was performed as a refractive error correction procedure (35) in control individuals as described in Supplementary Methods 1.2, 1.3 respectively.
2.3 Material collection and sample preparation
Stamps towards the nose and eyebrow were made on the CE before the excision in the CXL/PRK procedures to enable correct tissue orientation during cutting and separation of the topographic regions. The obtained tissues were submersed in an RNA stabilization solution (RNAlater; Qiagen, Hilden, Germany) immediately after excision and stored at −80°C until further proceeding.
The procedure of designation of the particular topographic region is shown in Figures 1, S1. The details of the sample preparation are described in Supplementary Methods 1.4.
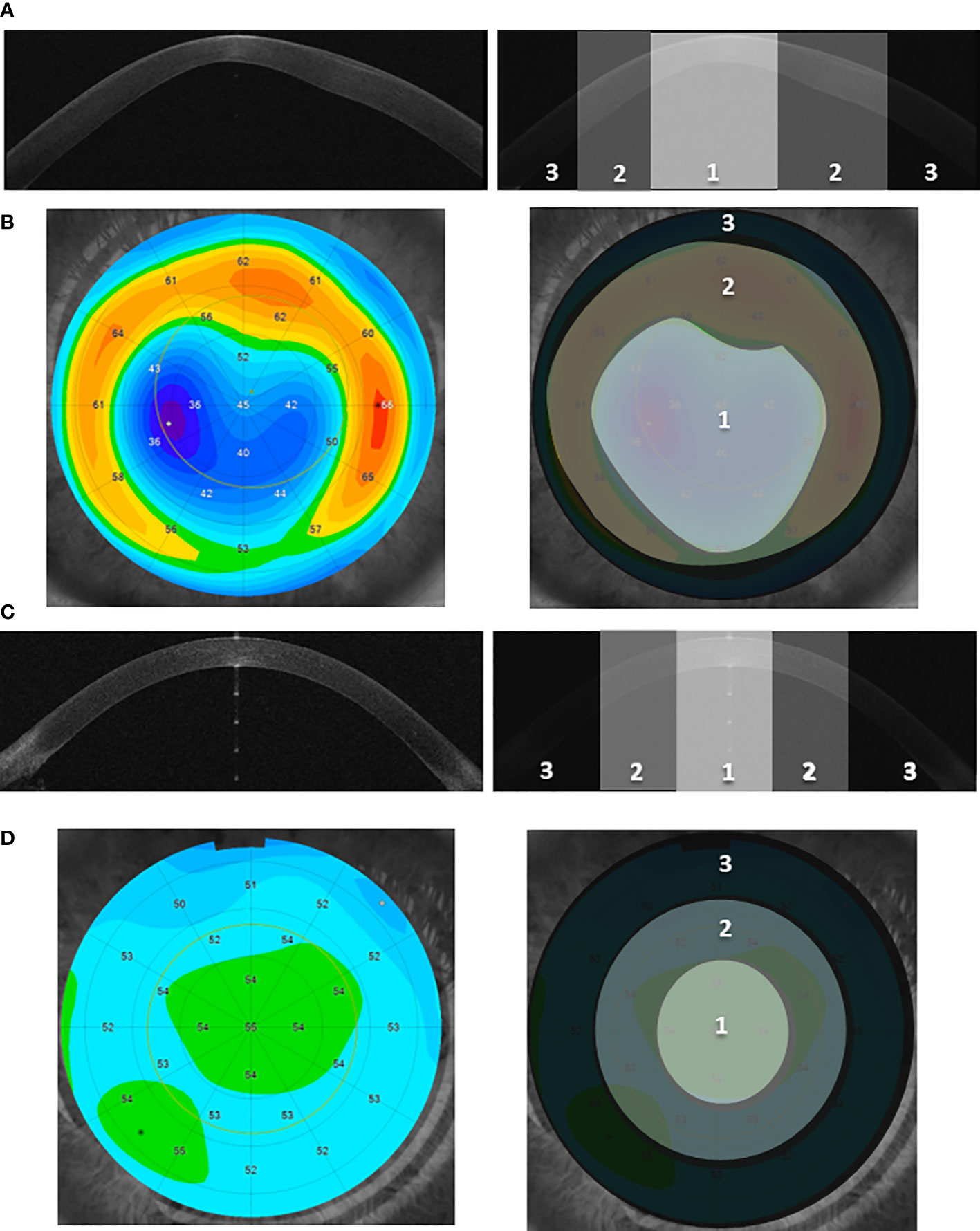
Figure 1 The designation of the three corneal topographic regions. (A, C) Examples of the cross-sectional image of the anterior segment of the eye (Anterior Segment Optical Coherence Tomography, AS-OCT) for the KTCN adolescent (A) and control individual (C), respectively. Three distinct topographic regions (1—central, 2—middle, and 3—peripheral) are indicated. The KTCN-specific thinning at the 1—central region is visible. (B, D) Example of the epithelial thickness mapping for the KTCN adolescent (B) and control individual (D), respectively. Representative images were obtained using MS-39 AS-OCT (CSO, Firenze, Italy). The topographic regions: 1—central, 2—middle, and 3—peripheral, were designated based on the steepness of epithelial thickness and corneal tomography with a rotating Scheimpflug camera (not shown). The designated CE topographic regions were separated and processed towards the DNA extraction.
2.4 DNA extraction
Separated CE samples were transferred from the microscope slides to the lysis solution (Norgen Biotek, Thorold, ON, Canada). Total RNA, DNA, and proteins were extracted and purified according to the instructions of the RNA/DNA/Protein Purification Plus Micro Kit (Norgen Biotek). The quantity of DNA samples was measured by Qubit dsDNA HS Assay Kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) and quality was assessed by 1% gel electrophoresis.
2.5 Library preparation, sequencing, and WGS data analyses
WGS of the 18 CE samples was performed with a TruSeq Nano DNA HT Library Prep Kit (Illumina, San Diego, CA, USA) and the HiSeqX platform (Illumina) with mean coverage depth 30× at CloudHealth Genomics (Shanghai, China) as previously described (36). WGS data processing is summarized in the Supplementary Methods 1.5.
The following subset of sequence variants were analyzed: high impact variants (nonsense mutations/stop gain, stop lost, frameshifts, splice site mutations, etc.), missense variants (missense mutations considered deleterious by either SIFT or PolyPhen), and variants in regulatory regions [overlapping promoters, promoter flanking regions, enhancers, CCCTC-binding factor binding sites, transcription factor (TF) binding sites, or open chromatin regions, based on the Ensembl Regulatory Build for fibroblasts], classified as single-nucleotide variant (SNV)/deletion/insertion/indel/sequence alteration (substitution).
Motif analysis of variants located in regulatory elements (REs) was executed using the web-based FABIAN-variant application (37), and SNVs were additionally verified using MotifbreakR (38).
A set of impacted genes (high impact/missense/regulatory regions variants) was assessed for each patient after removing variants identified in control patients and variants with a MAF_AF (Maximum observed allele frequency in 1000 Genomes, ESP, and gnomAD) ≥0.1 (1000 Genomes Project phase three, ESP6500SI-V2, gnomAD v2.1). The non-redundant set of impacted genes in KTCN patients was analyzed using ConsensusPathDB (39), ShinyGO (40), STRING tool (41), and Reactome database (42). In pathway enrichment and gene ontology analyses, the p-value ≤ 0.05 and/or FDR ≤ 0.05 were the cutoffs.
The identification of genomic regions significantly enriched in genes with KTCN-specific sequence variants, compared to the density of genes in the background, was performed using ShinyGO (40). A “sliding window” is applied to scan the genome and multiple hypergeometric tests to determine whether the evaluated genes are significantly overrepresented within the analyzed window with a size of 6 Mbp and the FDR ≤ 0.05 as a cutoff.
The Integrative Genomics Viewer (IGV) (43) was used for visual exploration of genomic data.
To determine if there is any difference between the genomic characteristics of the particular CE topographic regions and full-thickness corneas, we compared the data obtained for the CE samples with our previously generated molecular data for the same population (Polish Caucasians) (19, 32, 44, 45) and with other published data on candidate genes and gene pathways (31, 46–51).
All experimental samples were analyzed separately and at no step of the analysis (including the analysis of raw next generation sequencing reads) were data from the three regions of one individual combined. In the Results section, the variant(s) referred as recognized in the KTCN patient were present in all three topographic regions.
2.6 Data integration
Simultaneously, the transcriptomic and proteomic assessments as well as morphological evaluation in the same CE samples were performed, as described in detail elsewhere (13). All the data of the mentioned investigation were considered in final interpretation of the study outcomes.
3 Results
3.1 Characteristics of patients and DNA samples
Four unrelated adolescent patients with KTCN (one female/three male patients) and two control individuals (two male individuals) were involved in this study. There was no history of KTCN and genetic diseases (including autoimmune diseases) in families of ascertained patients and controls. Clinical characteristics of the examined individuals and the eyes subjected to the surgery are presented in Table 1, while the information collected for both eyes in the studied individuals is compiled in S1 Table. Selected behavioral, environmental, and socioeconomic aspects evaluated in the questionnaire are presented in S2 Table.
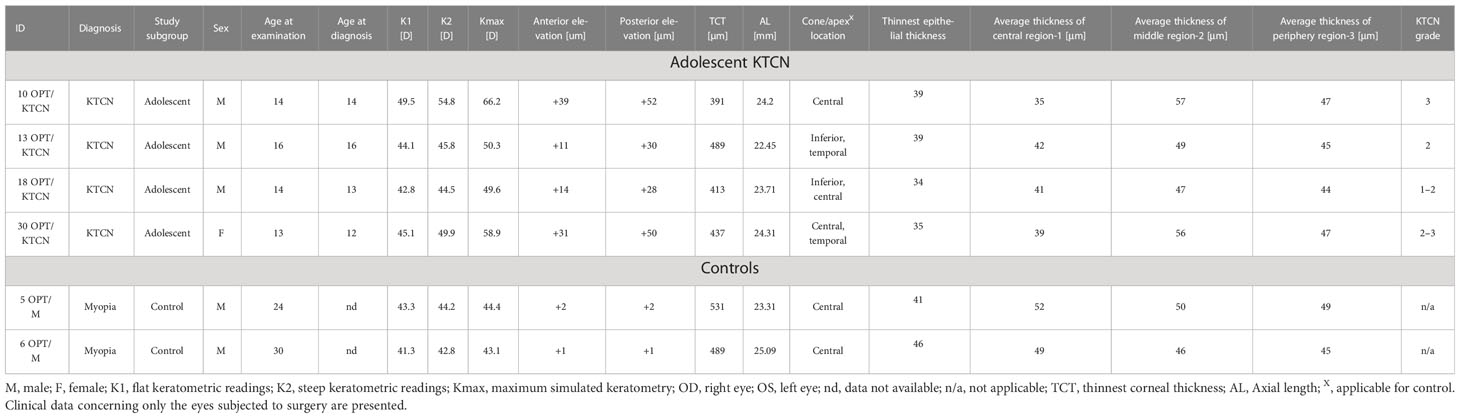
Table 1 The clinical characteristics of the ascertained adolescent patients with KTCN and control individuals (non-KTCN mild myopia patients).
A total of 18 experimental samples of CE (three topographic regions in each of the six ascertained individuals) were collected and used in the WGS experiments, in accordance with a study scheme (Figures 1, S1). The results of quantity and quality control of the 18 DNA samples and quality control of WGS reads are presented in S3 Table.
3.2 Genes shaping the inflammatory responses
Analyzing the coding sequence, a total of 646 variants were identified in KTCN patients, including high impact and missense variants (classified as SNVs, deletions, and insertions). In aspects of molecular function, biological process, and their cellular location, genes with identified variants contributed to ECM structural constituent, collagen-containing ECM, and cytoskeleton organization (S2 Figure and S4 Table). What is important, the pathway enrichment analysis comprising all genes with recognized sequence variation revealed the “Antigen presentation” and “Interferon alpha/beta signaling” as the most overrepresented Reactome pathways (FDR < 0.001, S5 Table). In STRING analysis, embracing the same set of genes with the 646 variants, the significant enrichment of 63 clusters, including network of interferon alpha/beta signaling, was recognized (S3 Figure and S6 Table).
Seventeen out of 646 variants were found as recurring in patients with KTCN, whereas the remaining 158, 159, 187, and 165 variants identified in patients 10 OPT/KTCN, 13 OPT/KTCN, 18 OPT/KTCN, and 30 OPT/KTCN, respectively, were unique for the evaluated individuals. The recurrent coding variants were as follows: rs687485 in KIR2DL; rs643861 and rs652641 in KIR3DL1; rs878913005, rs878949904, and rs1269243287 in MUC5AC; rs767573621 and rs752917826 in TRAV19; rs17229 in TRBV12-5; rs2257251 in NANOGP8; rs1227351091 in NBPF19; rs148235978 in NFE2L3; rs151080920 in TSC22D2; rs140771568 in UCMA; rs28575804 (A>G) in UGT2B25P; rs1563130264 in AC011005.1; and rs879192097 in AL354822.1 (S7 Table).
Furthermore, analyzing the high-impact and missense variants identified in at least two patients with KTCN, the “Graft-versus-host disease” and “Antigen processing and presentation” were found to be the most enriched pathways (Figure 2A and S8 Table).
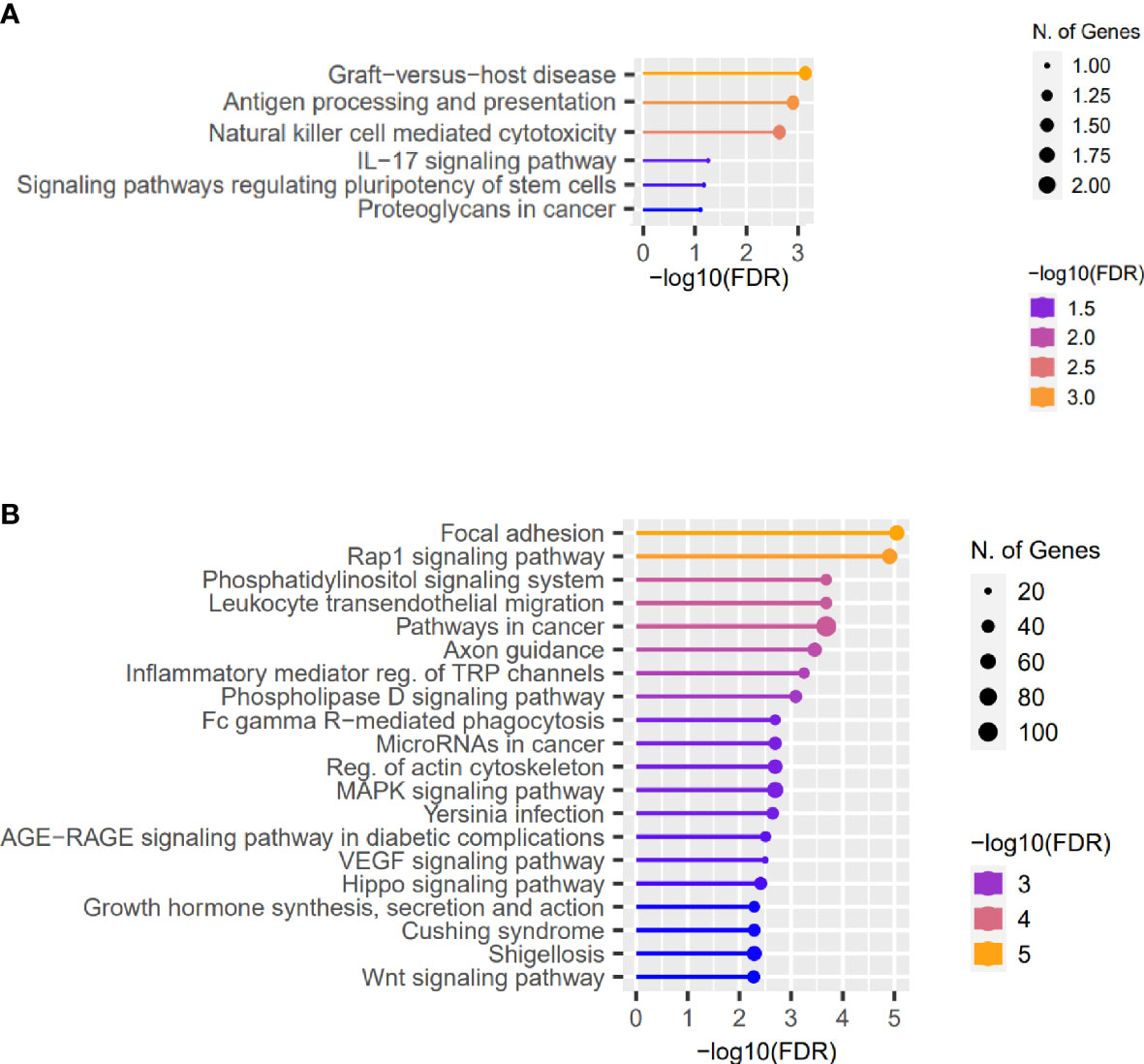
Figure 2 Representative results of the pathway enrichment analysis. Representative results of the pathway enrichment analysis of (A) genes with the same coding sequence variants identified in more than one KTCN patient, pointing to the involvement of innate and adaptive immune system responses in KTCN pathogenesis and (B) genes located close to the variants in REs. On x axes, −log(FDR) values of enriched pathways are presented, and on y axes, the KEGG pathways are presented. The size of the bubbles indicates the number of genes attributed to the particular pathway and present in our set.
3.3 KCTN-specific non-coding sequence features
Excluding variants recognized in control individuals and variants with MAF>0.1, 15,213 sequence variants in REs in KTCN patients, classified as SNVs, deletions, insertions, indels, and substitutions, were identified.
Next, using the FABIAN-variant application that predicts the effects of DNA variant located in RE on transcription factor binding sites (TFBSs), we narrowed down the number of variants and genes for further evaluation to 7,886 and 5,709, respectively. We assessed genes in which close proximity in at least one sequence variant in RE per KTCN patient was recognized as potentially forming or abolishing TFBS. Pathway enrichment analysis of these genes disclosed “Focal adhesion”, “Rap1 signaling pathway”, and also “Hippo signaling pathway”, “Wnt signaling”, and “TGF-beta signaling” pathways (Figure 2B and S9 Table).
3.4 Coexistence of coding and non-coding sequence variants in genes influencing the KTCN-specific pathways
Both coding and non-coding sequence variants of KTCN patients were found in the genes/genes’ close proximity linked to the revealed KTCN-specific cellular components, namely, “Actin cytoskeleton”, “Extracellular matrix”, and “Collagen-containing extracellular matrix”, and KTCN-specific pathways such as “Focal adhesion”, “Hippo signaling pathway”, and “Wnt signaling” (S4 Figure and S10 Table). Genes with coding and non-coding sequence variants, attributed to biological pathways from Reactome database, are listed in Table 2, and in detail in S11 Table.

Table 2 Chosen WGS results, list of genes with high impact, missense, and variants in regulatory elements attributed to the selected biological pathways.
To determine if the evaluated genome regions are significantly enriched, genes with both coding and non-coding sequence variants were analyzed and 35 enriched regions were revealed, as presented in Figure 3, with majority of these enriched regions at chromosomes 5q (13 regions), 9q (9 regions), and 16q (5 regions).
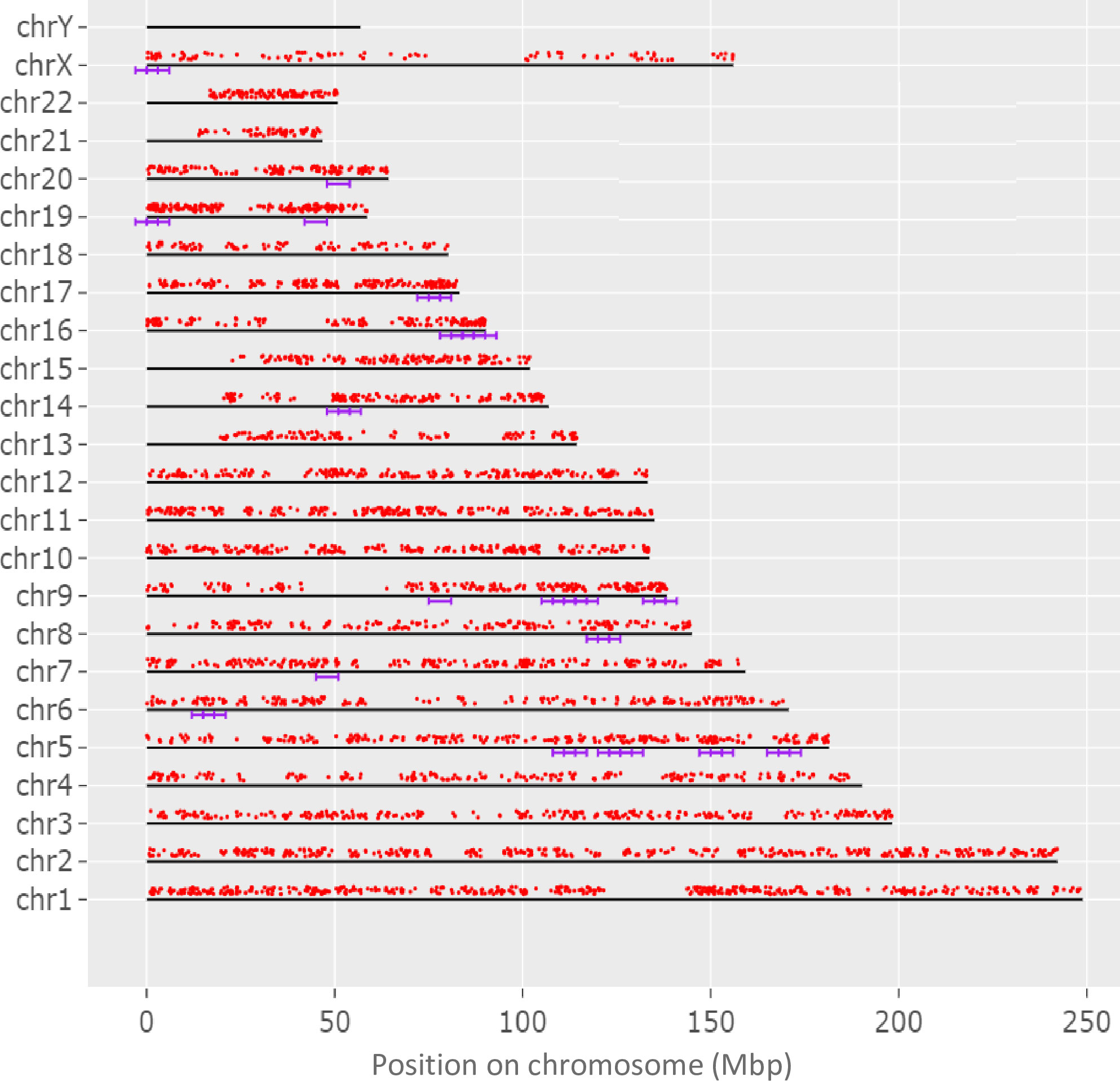
Figure 3 The display of genes, in which both coding and non-coding variants unique for adolescent patients with KTCN were identified, across chromosomes. Genes are represented by red dots. The violet lines indicate 35 regions statistically enriched. The genome was scanned with a sliding window in of size 6 Mbp. Within each window, the hypergeometric tests were used to determine if genes of both coding and non-coding variants were significantly overrepresented. The used FDR cutoff was ≤0.05.
3.5 Genomic heterogeneity across corneal surface and structural variation
All identified coding-sequence variants were assessed for differences between topographic regions, separately for each adolescent patient with KTCN. No genomic heterogeneity across corneal surface was found. Although identified sequence variants in KIR3DL1 (in two KTCN patients), UMAD1 (in two KTCN patients), and GOLGA6A2 (in one KTCN patient) genes have met quality criteria of reads in the applied bioinformatic algorithms and differentiated the topographic CE regions, the BAM files visualized with the IGV showed a variation in one of 26–43 reads only in these listed genes (the total number of reads for a particular genome site varied), indicating uncertain results. Expanding the same analysis for control samples (but applying the criterion of MAF ≤ 0.1), the additional coding sequence variants in 15 genes (AC018682.2, AC092384.1, AC092490.1, AC117481.1, AC118281.1, ANKRD36BP2, CA15P2, CRACD, HLA-DRB5, KRTAP2-2, LINC00634, NBEAP1, OR2T35, SAMD1, UGT2B25P, andZNF571) were recognized as potentially differentiating the corneal surface, but none of these variants was confirmed by the BAM files.
Assessing structural variation, deletions longer than 50 bp were characterized in detail. The length of the deletions and their density in genome are presented in S5 Figure. Most of the deletions were localized in non-coding regions of the genome and in gene introns. We did not observe a difference in the number of deletions comparing patients with KTCN and control individuals, assessing the previously published KTCN-specific loci (S12 Table) (52). Nevertheless, upstream and downstream regions of genes involved in the innate immune system (ITGAM, PRKCE, COL11A2, STK10, ADGRG3, IGHV2-70, IGHG4, MAP2K1, IGHG2, GSDMD, MLC1, MYO9B, IGHV1-69, DEFB131A, KIR2DS4, NKIRAS2, RNF38, and RPS6KA5); the adaptive immune system (TRAC, INPP5D, IGHV2-70, ASB3, PRKG1, TAP2, HMCN2, KIR3DL2, KIR3DL1, IGHV1-69, KIR2DL4, RASGRP3), cytokine signaling (ITGAM, HNRNPA2B1, NUP155, CAMK2A, INPP5D, IGHG4, MAP2K1, CRLF2, GSDMD, NKIRAS2, BOLA2, and RPS6KA5); ECM including collagen biosynthesis, formation, and trimerization (ITGA9, FBLN1, LOXL4, COL6A6, ITGAM, ADAMTS14, P4HA3, COL11A2, COL27A1, and COL26A1); and others were found to contain deletions (51–5,098 bp in size) in patients with KTCN in contrast to control individuals (MAF > 0.1 was an exclusion criterion).
3.6 Influence of the presence of RE variants on gene expression
We evaluated the influence of the presence of identified RE variants on expression of genes localized in close proximity, based on data of RNA-seq performed in the same CE samples, and full-thickness corneas (45). No substantial features were recognized in the assessment of the CE. However, 722 of the previously recognized differentially expressed genes (DEGs) in full-thickness KTCN corneas were re-recognized as the gene containing variants in REs. Then, in the pathway enrichment analysis of the re-recognized genes and transcripts, the ECM pathway as the most overrepresented was revealed (Figure 4 and S13 Table).

Figure 4 Representative results of the pathway enrichment analysis of 722 genes previously recognized as differentially expressed (DEGs) in full-thickness KTCN corneas and re-recognized as containing variants in the REs localized in close proximity. On the x axis, −log(FDR) values of enriched pathways are presented, and on the y axis, the KEGG pathways are presented. The size of bubbles indicated the number of genes attributed to the particular pathway and present in our set. The Reactome database was chosen for the definition of pathways.
4 Discussion
The complex background of KTCN, in addition to environmental factors, includes heterogeneous genetic characteristics. The multiple candidate genes and variants identified using different molecular techniques indicate that this heterogeneity requires the introduction of high-throughput investigation methods, which may be more effective in unraveling the KTCN-specific determinants.
Each individual genome differs from the reference human genome by an average of 3.5 million SNPs, but only a minority of those variations contributes to the phenotypic differences and disease predisposition (53). Only WGS can provide an overview of the entire human genome, allowing the global analysis and the full set of variants to be assessed (54–56). The majority of SNVs and indels is identifiable by both WGS and exome sequencing (ES), but still approximately 3% of coding variants is missed in the ES approach (54). Therefore, WGS is more efficient than ES for detecting coding sequence variants, besides the undoubted benefit of identification of non-coding sequence variation. To date, WGS has not been applied in KTCN research. Since WGS is an effective method of detecting mutations in genetically heterogeneous diseases, including ophthalmic diseases (57), and is more efficient than ES in the identification of sequence variation, this technique is crucial in deciphering the genetic aspects of KTCN.
Here, we found that the genetic features of patients with KTCN correspond with those previously reported regarding sequence variants in components of ECM (19, 50). Also, we identified other sequence variants in genes previously reported in KTCN research in the same population (19), including COL17A1 [playing a role in the integrity of hemidesmosome and the attachment of basal keratinocytes (58), whose expression was decreased in corneal epithelial cells in single-cell RNA-seq analysis of KTCN corneas (59)]; FREM2 [protein required for maintenance of the integrity of the skin epithelium (60) and eye morphogenesis (60)]; SSX2IP [which may connect the nectin–afadin and E-cadherin–catenin system through alpha-actinin and may be involved in the organization of the actin cytoskeleton (61)]; MINK1 [mediating stimulation of the stress-activated protein kinase MAPK14/p38 MAPK downstream of the Raf/ERK pathway (62)]; CAMSAP1 [Calmodulin-regulated spectrin-associated protein 1; key microtubule-organizing protein (63)]; and RGS12 [Regulator of G-protein signaling 12 (64)]. Moreover, we identified variants previously reported in EML6 (50), which is an element of cytoskeletal network and microtubules in cultured keratocytes (50, 65); MAGI1 (50), which regulates cell–cell and cell–matrix adhesion (66); and COL5A1 (48, 49), whose knockout in corneal stroma in mouse showed severe dysfunctional regulation of fibrillogenesis resulting in decreased fibril number and a disorganized lamellae structure with decreased stroma thickness (67).
Also, we have revealed variants in genes involved in keratin-related pathways (namely, the “keratinization” pathway from the Reactome database), such as EVPL, PCSK6, KRTAP10-7, KRT35, FLG, and KRT33B. As keratins participate in forming intermediate filaments (IF), which provide mechanical support, and form desmosomes between cells and hemi-desmosomes with basement membranes for epithelium integrity (68), this result may be of special interest regarding changes in biomechanics of keratoconic cornea.
Importantly, non-coding variants are now considered to be potentially pathogenic (69, 70). Sequence variation in the form of SNVs or indels in the regulatory regions can either generate or abolish TF binding sites, modulating the transcriptional activity of genes and regulating chromatin state (71). The disruption of the TF binding site and its effect on the disease phenotype have already been reported in many cancer types such as T‐cell acute lymphoblastic leukemia (72) and esophageal and gastric cancers (73). However, while great advances in predicting the effects of coding variants have been made, the assessment of non-coding variants remains challenging. In particular, the immense number of variants revealed per patient sample impedes the analysis and biological interpretation. In our investigation, all variants located in REs, including promoters, promoter flanking regions, enhancers, CCCTC-binding factor binding sites, TFBS, or open chromatin regions of fibroblasts, were evaluated in silico towards functional effect (TFBS gain/lost) based on the motif analysis. The obtained list of RE variants showed unity on the pathway level with previously reported variants, namely, focal adhesion (19), Wnt signaling (19), and TGF-beta signaling (74). Moreover, the LAMC2, COL17A1, SDK1, PECAM1, TJP2, MINK1, NEIL3, EVPL, and PCSK6 genes were revealed to have unique variants for patients with KTCN in both coding and non-coding genome sequence, which again were found as involved in the ECM, cell–cell communications, cytoskeleton organization, and keratinization pathways. To further evaluate the effect of variants located in REs, the functional approaches such as gene reporter assay are needed, but firstly transcriptome data from adequate biological material is necessary to be integrated with the genomic data. Therefore, we evaluated the data overlap between DEGs in full-thickness KTCN corneas (45) and variants recognized in the REs. We found the involvement of RE variants in numerous elements of ECM and that fact supports our assumption concerning the role of RE sequence variation in KTCN pathogenesis.
Although single variants could influence the KTCN phenotype in some families (75), our genomic data (both coding and non-coding sequence variation) suggest that KTCN largely occurs through the interplay of multiple variants that interfere with major physiological processes in the cornea. The presented connection between coding and non-coding sequence variants in the gene ontology (“Actin cytoskeleton”, “Extracellular matrix”, and “Collagen-containing extracellular matrix”) and in the pathway assessments (“Focal adhesion”, “Hippo signaling pathway”, and “Wnt signaling”) corroborates this assumption, as well as the identified 13 regions enriched in variants in the 5q locus that had previously been found to be specific to KTCN in distinct populations (32, 46, 47).
It has been known that DNA structural variation modifies the interactions between REs and their target genes (71). Here, the majority of the deletions were found to be located in non-coding and intron regions. Overall, assessing the previously published KTCN-specific loci, we did not observe a difference in the number/density of deletions comparing patients with KTCN and control individuals. However, numerous genes involved in the innate immune system; adaptive immune system; ECM including collagen biosynthesis, formation, and trimerization; and others were found to contain deletions in size of 51–5,098 bp in patients with KTCN in contrast to control individuals. The extent to which this finding is important should be clarified in further research on this aspect. To date, deletions >50 bp in size themselves and their effect on the KTCN phenotype were only once evaluated, in an Ecuadorian family with KTCN (31), but that study did not reveal any copy number variation on evaluated 13q32 in the tested individuals.
We have anticipated that the various intensities of both internal biomechanical tension and external environmental stimuli (e.g., eye rubbing) acting on various topographic regions of CE could result in diverse molecular features across corneal curvature. Our study performed in the designated three topographic regions of the CE, central, middle, and peripheral, enabled the conclusion that there is no genomic heterogeneity across the corneal surface in KTCN. However, in a comprehensive morphological, transcriptomic, and proteomic evaluation performed in the same corneal samples, we identified the variability shaping the distinct features of CE zones (13). Continuing the topographic aspects of the CE, we have considered features of mosaicism to be involved in the process of remodeling of the KTCN CE. To date, based on the results of exome and/or Sanger sequencing performed in matched KTCN cornea–blood pairs, we found no evidence of inter-tissue variants in human KTCN corneas, since all variants possibly related to KTCN were identified in both corneal tissue and peripheral blood of patients (19). Again, here we did not identify features of mosaicism.
In our opinion, the most important findings of this study are those concerning the inflammation in KTCN. Nowadays, the inflammatory aspects in KTCN are widely discussed. The multiple analyses of the tear cytokine and protease profiles of patients with KTCN have shown a persistent inflammation (17, 76). Previously, we identified numerous, randomly distributed variants in genes that encoded inflammatory molecules (IL1A and IL1B) and one variant (c.214+242C>T in the IL1RN) presented in all KTCN individuals (33). Here, the enrichment in variants of genes involved in the innate (neutrophil degranulation) and adaptive (Class I MHC antigen processing and presentation) immune system was revealed. In our study, TRAV19 and TRVB12-5 genes were recognized with three recurring variants (one as a stop gain). Because these genes encode variable regions of T-cell receptors, it could indirectly influence the process of antigen processing (77). Importantly, in a single-cell study of full corneas, the upregulation of the T-cell receptor signaling pathway has been recognized (59). Identified recurring missense variants in KIR2DL1 and KIR3DL1 could influence the activity of the immune system, since they are members of the highly conserved killer cell immunoglobulin-like receptors (KIRs) family (78), which are transmembrane glycoproteins expressed by natural killer (NK) cells and subsets of T cells (79). It was previously reported that the variant in KIR3DL1 has affected the NK cell function (80). NK cells activate neutrophils and induce both their infiltration to the inflamed sites and degranulation (81). Also, NK cells were reported to participate in acute immune response in dry eye syndrome and trigger Th-17 response, manifested by an increase of IL17 (82). Moreover, the involvement of NK cells in the corneal wound healing has been confirmed in a murine model (83). The evidence for migration of cells into corneal limbus and central stroma after a 2-mm epithelial abrasion of the mouse cornea and altered inflammatory reaction after antibody-induced depletion of NK cells were presented (83). Previously, as elements of innate immune responses, the expression of TLR2 and TLR4 has been studied by flow cytometry in corneal epithelial cells and blood samples of KTCN patients and their relatives (16, 84, 85), and significant upregulation has been shown, indicating a potential of the TLR2 and TLR4 proteins to be the KTCN biomarkers. However, no sequence variants in the TLRs were recognized here as well as in our current gene and protein expression assessment (13). Instead, we recognized sequence variants of other innate immune system elements (e.g., MUC5AC, as mucins consist the first-line defense). Moreover, the increased proportions of activated NK cells as well as neutrophils and γδT cells have been reported in KTCN studies (86). Also in many studies of tear fluid collected in KTCN patients, the higher levels of interleukins (IL1β, IL6, and IL17A), interferons (IFNα/β/γ), and other pro-inflammatory factors (TNFα, TGFβ1, LIF, granzyme-B, perforin, and MMP2) were recognized (14, 24, 86–88). Besides published transcriptomic and proteomic findings, the genomic aspects regarding disruption of immune responses have not been demonstrated in KTCN research. Also, the here-identified single high-impact sequence variants (CEP290, MMRN1, and FN1) and missense variants (PECAM1, VWF, CD109, HRG, TLN1, HPSE, EPO, SIGLEC10, DOCK8, ERBB3, and AP3B1) could contribute to molecular dysregulation of wound healing, followed by inflammation, although we have not experimentally determined their impact.
Therefore, we conclude here that the disturbance of the immune responses in KTCN affects various elements of corneal hemostasis, including the antigen presentation and neutrophil degranulation processes, and possibly results in thinning and weakening of the KTCN corneas.
Summarizing, applied WGS allowed for identification and confirmation of numerous genomic features in KTCN. In future studies, WGS data should be integrated with other genome-wide analyses data, including chromatin accessibility (Assay for Transposase Accessible Chromatin with sequencing, ATAC-Seq), transcriptomics, and proteomic data, in order to obtain the most reliable research results. In this study, owing to a limited number of included patients, we intently did not focus on particular sequence variations but rather on the sets of impacted genes and the pathways in which those genes are involved. Additionally, we decided to evaluate adolescent patients with KTCN only, assuming the more pronounced genetic background of the disease. The recruitment of the control group was especially challenging, as nowadays the photorefractive keratectomy (PRK) procedure, during which the CE is removed, is rarely performed and is replaced with the newer techniques of vision correction without CE removal. The youngest possible patients with minimal vision impairment undergoing the PRK procedure were ascertained into the control group to minimize potential bias. The PRK procedure is performed in adults, and this fact also results in the difference in age between the compared groups of patients. Although only four patients and two controls were involved, 18 samples were tested in the WGS experiments and data obtained from those 18 samples were carefully curated. In addition, the sequence variant interpretation was performed using additional allele frequency data derived from the reference databases (1000 Genomes, ESP and gnomAD). Owing to the small size of the compared study groups, we were unable to subdivide these groups into subgroups of patients, taking into account the presence of an allergic disease or dry eye syndrome.
The multiple coding and non-coding sequence variants were found in genes (or in their close proximity) contributing to the previously discussed KTCN-specific cellular components, and newly presented aspects of innate and adaptive immune system responses, pointing to the involvement of inflammatory aspects in KTCN. Therefore, we conclude that variants in non-coding regions of the genome have complemented the previously identified features specific to the KTCN exome.
Additionally, the 35 chromosomal regions shown enriched in both coding and non-coding KTCN-specific sequence variants confirmed the heterogenous background of KTCN with a limited number of common elements.
We did not find the diversity in genetic features of the assessed topographic regions of CE. This lack of genomic heterogeneity across the corneal surface and no evidence of inter-tissue variants in human KTCN corneas suggest other molecular mechanisms of KTCN-specific changes in CE.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/clinvar/ under the accession number SUB13057607.
Ethics statement
The studies involving human participants were reviewed and approved by Bioethics Committee at Poznan University of Medical Sciences, Poznan, Poland. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
KJ: participated in research design, sample collection, and patient surveys; performed sample preparations towards WGS experiments; participated in bioinformatics analyses; figures and tables preparation; participated in the genomic and clinical data interpretation; and wrote the manuscript. MM-K: performed recruitment of the patients and clinical evaluation; facilitated sample collection; performed the clinical data interpretation; participated in the genomic data interpretation; and manuscript writing. MK: performed setup of bioinformatics pipeline; bioinformatics analyses of the genomic data; and preparation of figures and tables. JAK: participated in the genomic data interpretation. MG: research design; secured research funding; participated in the genomic data interpretation; and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Science Centre in Poland (grant number: 2018/31/B/NZ5/03280 and 2021/41/B/NZ5/02245).
Acknowledgments
We are grateful to the director and the team of the Optegra Eye Health Care Clinic in Poznan, Poland, for enabling biological sampling and assisting in patient recruitment and clinical evaluation. Special thanks to Dr. Pawel Stankiewicz for his assistance in the WGS.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1197054/full#supplementary-material
Abbreviations
BCVA, Best-Corrected Visual Acuity; CE, Corneal Epithelium; CXL, Corneal Collagen Cross-linking; DEG, Differentially Expressed Gene; ECM, Extracellular Matrix; ES, Exome Sequencing; FDR, False Discovery Rate; IF, Intermediate Filaments; IGV, Integrative Genomics Viewer; IL, Interleukin; IOP, Intraocular Pressure; KIR, Killer cell Immunoglobulin-like Receptor; KTCN, Keratoconus; MAF, Maximum Allele Frequency; NK, Natural Killer cells; PRK, Photorefractive Keratectomy; RE, Regulatory Element; SD-OCT Spectral-Domain Optical Coherence Tomography; SNP, Single-Nucleotide Polymorphism; SNV, Single-Nucleotide Variant; TF, Transcription Factor; TFBS, Transcription Factor Binding Site; UCVA, Uncorrected Visual Acuity; WGS, Whole-Genome Sequencing.
References
1. Ferrari G, Rama P. The keratoconus enigma: a review with emphasis on pathogenesis. Ocular Surface (2020) 18:363–73. doi: 10.1016/j.jtos.2020.03.006
2. Rabinowitz YS. Keratoconus. Surv Ophthalmol (1998) 42:297–319. doi: 10.1016/s0039-6257(97)00119-7
3. Fink BA, Wagner H, Steger-May K, Rosenstiel C, Roediger T, McMahon TT, et al. Differences in keratoconus as a function of gender. Am J Ophthalmol (2005) 140:459–68. doi: 10.1016/j.ajo.2005.03.078
4. Hashemi H, Heydarian S, Hooshmand E, Saatchi M, Yekta A, Aghamirsalim M, et al. The prevalence and risk factors for keratoconus: a systematic review and meta-analysis. Cornea (2020) 39:263–70. doi: 10.1097/ICO.0000000000002150
5. Gain P, Jullienne R, He Z, Aldossary M, Acquart S, Cognasse F, et al. Global survey of corneal transplantation and eye banking. JAMA Ophthalmol (2016) 134:167–73. doi: 10.1001/jamaophthalmol.2015.4776
6. O’Brart DPS, Kwong TQ, Patel P, McDonald RJ, O’Brart NA. Long-term follow-up of riboflavin/ultraviolet a (370 nm) corneal collagen cross-linking to halt the progression of keratoconus. Br J Ophthalmol (2013) 97:433–7. doi: 10.1136/bjophthalmol-2012-302556
7. Naderan M, Jahanrad A, Balali S. Histopathologic findings of keratoconus corneas underwent penetrating keratoplasty according to topographic measurements and keratoconus severity. Int J Ophthalmol (2017) 10:1640–6. doi: 10.18240/ijo.2017.11.02
8. Scarcelli G, Besner S, Pineda R, Kalout P, Yun SH. In vivo biomechanical mapping of normal and keratoconus corneas. JAMA Ophthalmol (2015) 133:480–2. doi: 10.1001/jamaophthalmol.2014.5641
9. Silverman RH, Urs R, RoyChoudhury A, Archer TJ, Gobbe M, Reinstein DZ. Epithelial remodeling as basis for machine-based identification of keratoconus. Invest Ophthalmol Vis Sci (2014) 55:1580. doi: 10.1167/iovs.13-12578
10. Khaled ML, Helwa I, Drewry M, Seremwe M, Estes A, Liu Y. Molecular and histopathological changes associated with keratoconus. BioMed Res Int (2017) 2017:7803029. doi: 10.1155/2017/7803029
11. Sherwin T, Brookes NH. Morphological changes in keratoconus: pathology or pathogenesis. Clin Exp Ophthalmol (2004) 32:211–7. doi: 10.1111/j.1442-9071.2004.00805.x
12. Foster JW, Parikh RN, Wang J, Bower KS, Matthaei M, Chakravarti S, et al. Transcriptomic and immunohistochemical analysis of progressive keratoconus reveal altered WNT10A in epithelium and bowman’s layer. Invest Ophthalmol Vis Sci (2021) 62:16. doi: 10.1167/iovs.62.6.16
13. Jaskiewicz K, Maleszka-Kurpiel M, Matuszewska E, Kabza M, Rydzanicz M, Malinowski R, et al. The impaired wound healing process is a major factor in remodeling of the corneal epithelium in adult and adolescent patients with keratoconus. Invest Ophthalmol Vis Sci (2023) 64:22. doi: 10.1167/iovs.64.2.22
14. Shetty R, Sathyanarayanamoorthy A, Ramachandra RA, Arora V, Ghosh A, Srivatsa PR, et al. Attenuation of lysyl oxidase and collagen gene expression in keratoconus patient corneal epithelium corresponds to disease severity. Mol Vis (2015) 21:12–25.
15. Yam GH-F, Fuest M, Zhou L, Liu Y-C, Deng L, Chan AS-Y, et al. Differential epithelial and stromal protein profiles in cone and non-cone regions of keratoconus corneas. Sci Rep (2019) 9:2965. doi: 10.1038/s41598-019-39182-6
16. Malfeito M, Regueiro U, Pérez-Mato M, Campos F, Sobrino T, Lema I. Innate immunity biomarkers for early detection of keratoconus. Ocular Immunol Inflammation (2019) 27:942–8. doi: 10.1080/09273948.2018.1511813
17. Balasubramanian SA, Pye DC, Willcox MDP. Effects of eye rubbing on the levels of protease, protease activity and cytokines in tears: relevance in keratoconus. Clin Exp Optom (2013) 96:214–8. doi: 10.1111/cxo.12038
18. Gordon-Shaag A, Millodot M, Shneor E, Liu Y. The genetic and environmental factors for keratoconus. BioMed Res Int (2015) 2015:795738. doi: 10.1155/2015/795738
19. Karolak JA, Gambin T, Rydzanicz M, Polakowski P, Ploski R, Szaflik JP, et al. Accumulation of sequence variants in genes of wnt signaling and focal adhesion pathways in human corneas further explains their involvement in keratoconus. PeerJ (2020) 8:e8982. doi: 10.7717/peerj.8982
20. Nowak-Malczewska DM, Karolak JA, Swierkowska J, Jaworska MM, Kulinska KI, Polakowski P, et al. Changes in nuclear gene expression related to mitochondrial function affect extracellular matrix, collagens, and focal adhesion in keratoconus. Transl Vis Sci Technol (2021) 10:6. doi: 10.1167/tvst.10.11.6
21. Özalp O, Atalay E, Yıldırım N. Prevalence and risk factors for keratoconus in a university-based population in Turkey. J Cataract Refract Surg (2021) 47:1524–9. doi: 10.1097/j.jcrs.0000000000000669
22. Sahebjada S, Chan E, Xie J, Snibson GR, Daniell M, Baird PN. Risk factors and association with severity of keratoconus: the Australian study of keratoconus. Int Ophthalmol (2021) 41:891–9. doi: 10.1007/s10792-020-01644-6
23. Gomes JAP, Tan D, Rapuano CJ, Belin MW, Ambrósio R, Guell JL, et al. Group of panelists for the global Delphi panel of keratoconus and ectatic diseases. Global consensus keratoconus ectatic diseases. Cornea (2015) 34:359–69. doi: 10.1097/ICO.0000000000000408
24. Lema I, Sobrino T, Durán JA, Brea D, Díez-Feijoo E. Subclinical keratoconus and inflammatory molecules from tears. Br J Ophthalmol (2009) 93:820–4. doi: 10.1136/bjo.2008.144253
25. Lema I, Durán JA. Inflammatory molecules in the tears of patients with keratoconus. Ophthalmology (2005) 112:654–9. doi: 10.1016/j.ophtha.2004.11.050
26. López-López M, Regueiro U, Bravo SB, Chantada-Vázquez MDP, Varela-Fernández R, Ávila-Gómez P, et al. Tear proteomics in keratoconus: a quantitative SWATH-MS analysis. Invest Ophthalmol Vis Sci (2021) 62:30. doi: 10.1167/iovs.62.10.30
27. Hutchings H, Ginisty H, Le Gallo M, Levy D, Stoësser F, Rouland JF, et al. Identification of a new locus for isolated familial keratoconus at 2p24. J Med Genet (2005) 42:88–94. doi: 10.1136/jmg.2004.022103
28. Brancati F. A locus for autosomal dominant keratoconus maps to human chromosome 3p14-q13. J Med Genet (2004) 41:188–92. doi: 10.1136/jmg.2003.012872
29. Bechara SJ, Waring GO, Insler MS. Keratoconus in two pairs of identical twins. Cornea (1996) 15:90–3.
30. Elder MJ. Leber congenital amaurosis and its association with keratoconus and keratoglobus. J Pediatr Ophthalmol Strabismus (1994) 31:38–40. doi: 10.3928/0191-3913-19940101-08
31. Gajecka M, Radhakrishna U, Winters D, Nath SK, Rydzanicz M, Ratnamala U, et al. Localization of a gene for keratoconus to a 5.6-Mb interval on 13q32. Invest Ophthalmol Vis Sci (2009) 50:1531–9. doi: 10.1167/iovs.08-2173
32. Karolak JA, Gambin T, Pitarque JA, Molinari A, Jhangiani S, Stankiewicz P, et al. Variants in SKP1, PROB1, and IL17B genes at keratoconus 5q31.1-q35.3 susceptibility locus identified by whole-exome sequencing. Eur J Hum Genet (2017) 25:73–8. doi: 10.1038/ejhg.2016.130
33. Nowak DM, Karolak JA, Kubiak J, Gut M, Pitarque JA, Molinari A, et al. Substitution at IL1RN and deletion at SLC4A11 segregating with phenotype in familial keratoconus. Invest Ophthalmol Vis Sci (2013) 54:2207. doi: 10.1167/iovs.13-11592
34. Wollensak G, Spoerl E, Seiler T. Riboflavin/ultraviolet-a-induced collagen crosslinking for the treatment of keratoconus. Am J Ophthalmol (2003) 135:620–7. doi: 10.1016/s0002-9394(02)02220-1
35. Ganesh S, Brar S, Patel U. Comparison of ReLEx SMILE and PRK in terms of visual and refractive outcomes for the correction of low myopia. Int Ophthalmol (2018) 38:1147–54. doi: 10.1007/s10792-017-0575-6
36. Karolak JA, Vincent M, Deutsch G, Gambin T, Cogné B, Pichon O, et al. Complex compound inheritance of lethal lung developmental disorders due to disruption of the TBX-FGF pathway. Am J Hum Genet (2019) 104:213–28. doi: 10.1016/j.ajhg.2018.12.010
37. Steinhaus R, Robinson PN, Seelow D. FABIAN-variant: predicting the effects of DNA variants on transcription factor binding. Nucleic Acids Res (2022) 50:W322–9. doi: 10.1093/nar/gkac393
38. Coetzee SG, Coetzee GA, Hazelett DJ. motifbreakR : an R/Bioconductor package for predicting variant effects at transcription factor binding sites: fig. 1. Bioinformatics (2015), 31(23):3847–9. doi: 10.1093/bioinformatics/btv470
39. Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc (2016) 11:1889–907. doi: 10.1038/nprot.2016.117
40. Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics (2020) 36:2628–9. doi: 10.1093/bioinformatics/btz931
41. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res (2021) 49:D605–12. doi: 10.1093/nar/gkaa1074
42. Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, et al. The reactome pathway knowledgebase. Nucleic Acids Res (2022) 50:D687–92. doi: 10.1093/nar/gkab1028
43. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol (2011) 29:24–6. doi: 10.1038/nbt.1754
44. Kabza M, Karolak JA, Rydzanicz M, Udziela M, Gasperowicz P, Ploski R, et al. Multiple differentially methylated regions specific to keratoconus explain known keratoconus linkage loci. Invest Ophthalmol Vis Sci (2019) 60:1501. doi: 10.1167/iovs.18-25916
45. Kabza M, Karolak JA, Rydzanicz M, Szcześniak MW, Nowak DM, Ginter-Matuszewska B, et al. Collagen synthesis disruption and downregulation of core elements of TGF-β, hippo, and wnt pathways in keratoconus corneas. Eur J Hum Genet (2017) 25:582–90. doi: 10.1038/ejhg.2017.4
46. Bisceglia L, De Bonis P, Pizzicoli C, Fischetti L, Laborante A, Di Perna M, et al. Linkage analysis in keratoconus: replication of locus 5q21.2 and identification of other suggestive loci. Invest Ophthalmol Vis Sci (2009) 50:1081–6. doi: 10.1167/iovs.08-2382
47. Bykhovskaya Y, Li X, Taylor KD, Haritunians T, Rotter JI, Rabinowitz YS. Linkage analysis of high-density SNPs confirms keratoconus locus at 5q chromosomal region. Ophthalmic Genet (2016) 37(1):109–10. doi: 10.3109/13816810.2014.889172
48. Li X, Bykhovskaya Y, Haritunians T, Siscovick D, Aldave A, Szczotka-Flynn L, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet (2012) 21:421–9. doi: 10.1093/hmg/ddr460
49. Lu Y, Vitart V, Burdon KP, Khor CC, Bykhovskaya Y, Mirshahi A, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet (2013) 45:155–63. doi: 10.1038/ng.2506
50. Shinde V, Sobreira N, Wohler ES, Maiti G, Hu N, Silvestri G, et al. Pathogenic alleles in microtubule, secretory granule and extracellular matrix-related genes in familial keratoconus. Hum Mol Genet (2021) 30:658–71. doi: 10.1093/hmg/ddab075
51. Xu X, Zhang X, Cui Y, Yang H, Ping X, Wu J, et al. Three novel variants identified within ECM-related genes in Chinese han keratoconus patients. Sci Rep (2020) 10:5844. doi: 10.1038/s41598-020-62572-0
52. Karolak JA, Gajecka M. Genomic strategies to understand causes of keratoconus. Mol Genet Genomics (2017) 292:251–69. doi: 10.1007/s00438-016-1283-z
53. Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med (2012) 63:35–61. doi: 10.1146/annurev-med-051010-162644
54. Belkadi A, Bolze A, Itan Y, Cobat A, Vincent QB, Antipenko A, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci U.S.A. (2015) 112:5473–8. doi: 10.1073/pnas.1418631112
55. Esser D, Holze N, Haag J, Schreiber S, Krüger S, Warneke V, et al. Interpreting whole genome and exome sequencing data of individual gastric cancer samples. BMC Genomics (2017) 18:517. doi: 10.1186/s12864-017-3895-z
56. Sun Y, Ruivenkamp CAL, Hoffer MJV, Vrijenhoek T, Kriek M, van Asperen CJ, et al. Next-generation diagnostics: gene panel, exome, or whole genome? Hum Mutat (2015) 36:648–55. doi: 10.1002/humu.22783
57. González-Del Pozo M, Fernández-Suárez E, Bravo-Gil N, Méndez-Vidal C, Martín-Sánchez M, Rodríguez-de la Rúa E, et al. A comprehensive WGS-based pipeline for the identification of new candidate genes in inherited retinal dystrophies. NPJ Genom Med (2022) 7:17. doi: 10.1038/s41525-022-00286-0
58. Löffek S, Hurskainen T, Jackow J, Sigloch FC, Schilling O, Tasanen K, et al. Transmembrane collagen XVII modulates integrin dependent keratinocyte migration via PI3K/Rac1 signaling. PloS One (2014) 9:e87263. doi: 10.1371/journal.pone.0087263
59. Collin J, Queen R, Zerti D, Bojic S, Dorgau B, Moyse N, et al. A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocular Surface (2021) 21:279–98. doi: 10.1016/j.jtos.2021.03.010
60. Zhang X, Wang D, Dongye M, Zhu Y, Chen C, Wang R, et al. Loss-of-function mutations in FREM2 disrupt eye morphogenesis. Exp Eye Res (2019) 181:302–12. doi: 10.1016/j.exer.2019.02.013
61. Reis AH, Xiang B, Ossipova O, Itoh K, Sokol SY. Identification of the centrosomal maturation factor SSX2IP as a wtip-binding partner by targeted proximity biotinylation. PloS One (2021) 16:e0259068. doi: 10.1371/journal.pone.0259068
62. Nicke B, Bastien J, Khanna SJ, Warne PH, Cowling V, Cook SJ, et al. Involvement of MINK, a Ste20 family kinase, in ras oncogene-induced growth arrest in human ovarian surface epithelial cells. Mol Cell (2005) 20:673–85. doi: 10.1016/j.molcel.2005.10.038
63. Zhou Z, Xu H, Li Y, Yang M, Zhang R, Shiraishi A, et al. CAMSAP1 breaks the homeostatic microtubule network to instruct neuronal polarity. Proc Natl Acad Sci U.S.A. (2020) 117:22193–203. doi: 10.1073/pnas.1913177117
64. Schroer AB, Mohamed JS, Willard MD, Setola V, Oestreich E, Siderovski DP. A role for regulator of G protein signaling-12 (RGS12) in the balance between myoblast proliferation and differentiation. PloS One (2019) 14:e0216167. doi: 10.1371/journal.pone.0216167
65. Yin H, Hou X, Zhang T, Shi L. Su y-q. participation of EML6 in the regulation of oocyte meiotic progression in mice. J BioMed Res (2019) 34:44–53. doi: 10.7555/JBR.33.20190014
66. Wörthmüller J, Rüegg C. MAGI1, a scaffold protein with tumor suppressive and vascular functions. Cells (2021) 10:1494. doi: 10.3390/cells10061494
67. Sun M, Chen S, Adams SM, Florer JB, Liu H, Kao WW-Y, et al. Collagen V is a dominant regulator of collagen fibrillogenesis: dysfunctional regulation of structure and function in a corneal-stroma-specific Col5a1-null mouse model. J Cell Sci (2011) 124:4096–105. doi: 10.1242/jcs.091363
68. Kao WW-Y. Keratin expression by corneal and limbal stem cells during development. Exp Eye Res (2020) 200:108206. doi: 10.1016/j.exer.2020.108206
69. Johnston JJ, Williamson KA, Chou CM, Sapp JC, Ansari M, Chapman HM, et al. NAA10 polyadenylation signal variants cause syndromic microphthalmia. J Med Genet (2019) 56:444–52. doi: 10.1136/jmedgenet-2018-105836
70. Wright CF, Quaife NM, Ramos-Hernández L, Danecek P, Ferla MP, Samocha KE, et al. Non-coding region variants upstream of MEF2C cause severe developmental disorder through three distinct loss-of-function mechanisms. Am J Hum Genet (2021) 108:1083–94. doi: 10.1016/j.ajhg.2021.04.025
71. Diederichs S, Bartsch L, Berkmann JC, Fröse K, Heitmann J, Hoppe C, et al. The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med (2016) 8:442–57. doi: 10.15252/emmm.201506055
72. Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. Oncogene regulation. an oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science (2014) 346:1373–7. doi: 10.1126/science.1259037
73. Malik MA, Zargar SA, Mittal B. A six-nucleotide deletion polymorphism in the casp8 promoter is associated with reduced risk of esophageal and gastric cancers in Kashmir valley. Indian J Hum Genet (2011) 17:152–6. doi: 10.4103/0971-6866.92090
74. Lin Q, Zheng L, Shen Z. A novel variant in TGFBI causes keratoconus in a two-generation Chinese family. Ophthalmic Genet (2022) 43:159–63. doi: 10.1080/13816810.2021.2015788
75. Bisceglia L, Ciaschetti M, De Bonis P, Campo PAP, Pizzicoli C, Scala C, et al. VSX1 mutational analysis in a series of Italian patients affected by keratoconus: detection of a novel mutation. Invest Ophthalmol Vis Sci (2005) 46:39–45. doi: 10.1167/iovs.04-0533
76. Acera A, Vecino E, Rodríguez-Agirretxe I, Aloria K, Arizmendi JM, Morales C, et al. Changes in tear protein profile in keratoconus disease. Eye (Lond) (2011) 25:1225–33. doi: 10.1038/eye.2011.105
77. Parkhurst M, Robbins P, Rosenberg S. Isolation of T cell receptors specifically reactive with mutated tumor associated antigens. J immunotherapy Cancer (2014) 2:P33:2051–1426-2-S3-P33. doi: 10.1186/2051-1426-2-S3-P33
78. Sambrook JG, Bashirova A, Andersen H, Piatak M, Vernikos GS, Coggill P, et al. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics (2006) 7:209. doi: 10.1186/1471-2164-7-209
79. Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology (2011) 132:315–25. doi: 10.1111/j.1365-2567.2010.03398.x
80. Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol (2005) 175:5222–9. doi: 10.4049/jimmunol.175.8.5222
81. Jensen KN, Omarsdottir SY, Reinhardsdottir MS, Hardardottir I, Freysdottir J. Docosahexaenoic acid modulates NK cell effects on neutrophils and their crosstalk. Front Immunol (2020) 11:570380. doi: 10.3389/fimmu.2020.570380
82. Zhang X, Volpe EA, Gandhi NB, Schaumburg CS, Siemasko KF, Pangelinan SB, et al. NK cells promote Th-17 mediated corneal barrier disruption in dry eye. PloS One (2012) 7:e36822. doi: 10.1371/journal.pone.0036822
83. Liu Q, Smith CW, Zhang W, Burns AR, Li Z. NK cells modulate the inflammatory response to corneal epithelial abrasion and thereby support wound healing. Am J Pathol (2012) 181:452–62. doi: 10.1016/j.ajpath.2012.04.010
84. Regueiro U, López-López M, Hervella P, Sobrino T, Lema I. Corneal and conjunctival alteration of innate immune expression in first-degree relatives of keratoconus patients. Graefes Arch Clin Exp Ophthalmol (2021) 259:459–67. doi: 10.1007/s00417-020-04929-9
85. Sobrino T, Regueiro U, Malfeito M, Vieites-Prado A, Pérez-Mato M, Campos F, et al. Higher expression of toll-like receptors 2 and 4 in blood cells of keratoconus patiens. Sci Rep (2017) 7:12975. doi: 10.1038/s41598-017-13525-7
86. D’Souza S, Nair AP, Sahu GR, Vaidya T, Shetty R, Khamar P, et al. Keratoconus patients exhibit a distinct ocular surface immune cell and inflammatory profile. Sci Rep (2021) 11:20891. doi: 10.1038/s41598-021-99805-9
87. Chaerkady R, Shao H, Scott S-G, Pandey A, Jun AS, Chakravarti S. The keratoconus corneal proteome: loss of epithelial integrity and stromal degeneration. J Proteomics (2013) 87:122–31. doi: 10.1016/j.jprot.2013.05.023
Keywords: cornea, corneal epithelium, inflammation, non-coding sequence variation, whole genome, WGS, keratoconus
Citation: Jaskiewicz K, Maleszka-Kurpiel M, Kabza M, Karolak JA and Gajecka M (2023) Sequence variants contributing to dysregulated inflammatory responses across keratoconic cone surface in adolescent patients with keratoconus. Front. Immunol. 14:1197054. doi: 10.3389/fimmu.2023.1197054
Received: 30 March 2023; Accepted: 09 June 2023;
Published: 06 July 2023.
Edited by:
Cheng-Rong Yu, National Eye Institute (NIH), United StatesReviewed by:
Susana Bravo, Sanitary Research Institute Foundation of Santiago de Compostela, SpainUxía Regueiro, Health Research Institute of Santiago de Compostela (IDIS), Spain
Copyright © 2023 Jaskiewicz, Maleszka-Kurpiel, Kabza, Karolak and Gajecka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marzena Gajecka, Z2FtYXJAbWFuLnBvem5hbi5wbA==
 Katarzyna Jaskiewicz1
Katarzyna Jaskiewicz1 Justyna A. Karolak
Justyna A. Karolak Marzena Gajecka
Marzena Gajecka