- 1State Key Laboratory for Animal Disease Control and Prevention, College of Veterinary Medicine, Lanzhou University, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
- 2Department of Basic and Diagnostic Sciences, College of Veterinary Science, Mekelle University, Mekelle, Tigray, Ethiopia
In the family of histone-deacetylases, histone deacetylase 6 (HDAC6) stands out. The cytoplasmic class IIb histone deacetylase (HDAC) family is essential for many cellular functions. It plays a crucial and debatable regulatory role in innate antiviral immunity. This review summarises the current state of our understanding of HDAC6’s structure and function in light of the three mechanisms by which it controls DNA and RNA virus infection: cytoskeleton regulation, host innate immune response, and autophagy degradation of host or viral proteins. In addition, we summed up how HDAC6 inhibitors are used to treat a wide range of diseases, and how its upstream signaling plays a role in the antiviral mechanism. Together, the findings of this review highlight HDAC6’s importance as a new therapeutic target in antiviral immunity, innate immune response, and some diseases, all of which offer promising new avenues for the development of drugs targeting the immune response.
Introduction
Viruses are invaders that get inside the cells of many kinds of organisms, from single-celled microbes to plants and mammals. Virus infection is a prevalent cause of human sickness or death worldwide and a major source of hospital admissions in children, adults and the elderly, significantly impacting healthcare systems and human health (1, 2). The coordination of antiviral immune responses in the innate immune system, which is the first line of defense against viral infection, is required (3, 4). The innate immune system is a universal and ancient host defense against infection that recognizes nonspecific pathogens (5, 6). Most innate immune cells have molecular receptors that are genetically conserved. The immune system uses these innate immune receptors, also known as pattern recognition receptors (PRRs), including retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), toll-like receptors (TLRs), nucleotide oligomerization domain-like receptors (NLRs), c-type lectin receptors (CLRs), and others (7). PRRs are used to identify structurally conserved molecules known as pathogen-associated molecular patterns (PAMPs), which are shared by a wide range of microbial species. Even though PAMPs from viruses, bacteria, fungi, and parasites have a wide variety of chemical structures, the immune response brought on by innate immunity cells is quick (8). Infection with a virus or an immunostimulant mimic, such as poly (I:C), can trigger the synthesis of interferon-β (IFN-β), a vital element of innate antiviral immunity (9).
HDACs are acetylation erasers from lysine residues that play critical roles in many biological processes, including immunity, cell cycle progression, apoptosis, and their repressive effects on gene transcription (10–12). HDACs have been classified into four classes in humans. Up to this point, four distinct classes of HDACs have been identified: class I (HDAC1, HDAC2, HDAC3, and HDAC8), class II (quite far subdivided into IIa comprising of HDAC4, HDAC5, HDAC7, HDAC9 and IIb consisting of HDAC6, HDAC10), class III (Sirtuins 1-7) and class IV (HDAC11) (13, 14). Except for class III, eleven of the 18 HDACs rely on Zn2+ for deacetylation (15). Class II HDACs are known to often move between the nucleus and cytoplasm, whereas those of class IIb HDACs are mostly localized in the cytoplasm (16). Immune cells react to invaders’ single- or double-stranded DNA or RNA in a general manner during this viral invasion. HDACs modulate this response by enhancing or weakening the results to alter genetic signatures. The JAK/STAT pathway, which is the most crucial signaling pathway against viral infection, is activated by type-I interferon (IFN-I) (17). It is noteworthy that HDAC3 interacts with forkhead box K1 to control signaling STAT1/2 transcription, which supports macrophages’ antiviral innate immunity (18). To control the expression of the interferon-stimulated gene (ISG), the released IFN cytokines first bind to IFN-I receptors and activate the JAK/STAT signaling cascade (19). HDAC4, a class II member, has anti-influenza A virus (IAV) characteristics and is a component of the host’s innate antiviral response (20). Besides, HDAC1 is crucial for the replication of the IAV, suggesting that it could be a target for innate immunity-based antiviral protection (21). HDAC6 is essential for antiviral innate immune responses (22–24). HDAC6 was first discovered due to its similarity to the Saccharomyces cerevisiae histone deacetylase HDA1 (25, 26). Moreover, HDAC6 is a multisubstrate enzyme that controls several cellular processes, including those that lead to cancer, neurological illnesses and inflammatory disorders. However, it is still unclear how HDAC6 is related to the antiviral signaling network and whether it is essential for antiviral immunity in animals under physiological circumstances. In particular, data show that IFN-β gene activation necessitates HDAC activity and that HDAC6 also serves as a coactivator of interferon regulatory factor 3 (IRF3)-dependent transcription (19). The failure of IFN-β production in cells is linked to the inhibition of antiviral responses (19). As a result of these factors, HDAC6 has emerged as a topic of intense interest among researchers and has become a desirable therapeutic target.
To conclude, we highlight new studies that show HDAC6’s multi-functionality in protein structure, viral infection, innate immune response, upstream signaling, and the application of HDAC6 inhibitors in associated diseases.
HDAC6’s structure and function
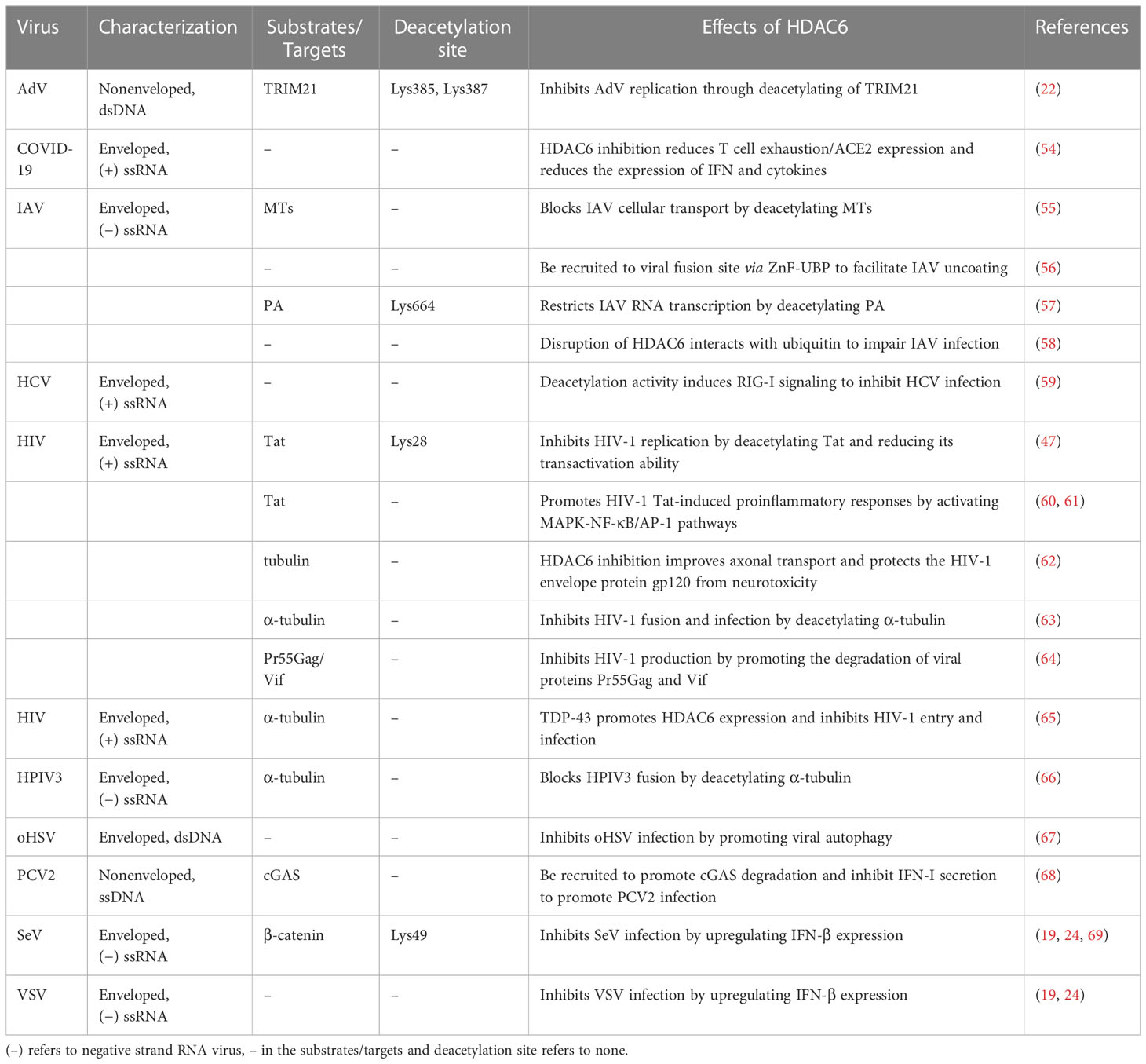
The HDAC6 isoenzyme has two deacetylase domains. This enzyme is a cytoplasmic class II histone deacetylase involved in a variety of cellular functions such as immunological synapse formation, misfolded protein degradation, migration, and cell-cell contact (27, 28). With 1,215 amino acids, HDAC6 protein is the largest HDAC protein found in people. It is unique and different because it is made up of five domains. The N-terminal (1-87 aa) is made up of a nuclear-localized signal (NLS, 14-59 aa) and a nuclear export signal (NES, 67-76 aa), which together control HDAC6’s shift from the nucleus to the cytoplasm. Also, research shows that the N-terminal of HDAC6 is needed for binding and effective acetylation of tubulin (29); it has two highly conserved catalytic domains, the first of which is called catalytic domain 1 (CD1, 88-447 aa) and has been shown to have deacetylase activity (30) and ubiquitin E3 ligase activity (31); the second CD2 (482-800 aa) exhibits broader substrate specificity as a deacetylated domain with catalytic activity (32); a cytoplasmic retention signal, Ser-Glu-containing tetrapeptide (SE14, 884-1022 aa), SE14 has the ability to control how HDAC6 interacts with other proteins (33). Another NES (1049-1058 aa) and a zinc-finger ubiquitin-specific protease (ZnF-UBP domain, also known as BUZ, 1131-1192 aa) recruit ubiquitin protein to stimulate aggresome formation (Figure 1) (34–36). The HDAC6 gene has 21923 base pairs and is on chromosome X p11.22-23. It is found in large amounts in the testis, spermatogenic cells, germ cell tissues, liver, heart, muscle, spleen, and kidney, among other normal human tissues and organs. (Figure 2) (Data from https://www.ncbi.nlm.nih.gov/gene/10013) (25, 37, 38).
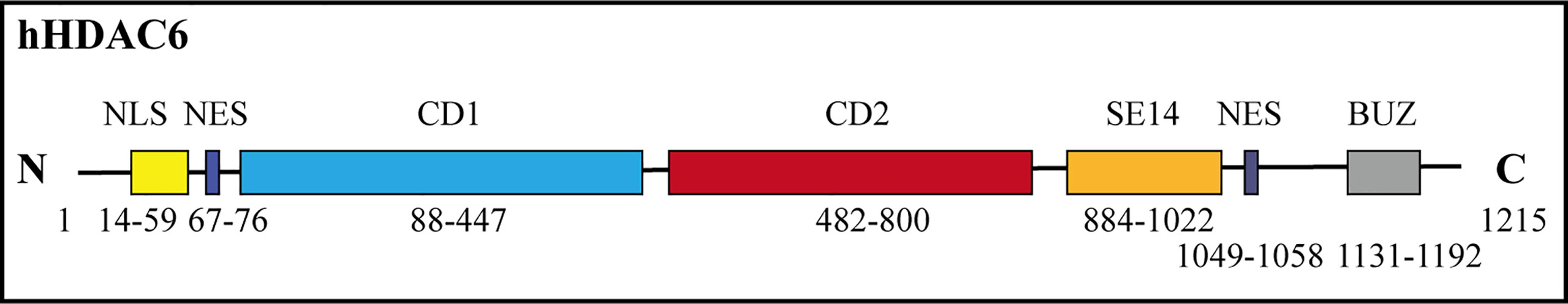
Figure 1 Human HDAC6’s functional domains and schematic depiction. HDAC6 has both catalytic activity and two tandem deacetylase domains (CD1 and CD2). Two nuclear export signal (NES) prevents the protein from building up in the nucleus, and the Ser-Glu-containing tetrapeptide (SE14) region guarantees persistent anchoring of the enzyme in the cytoplasm. In the nucleus, HDAC6 is translocated by the nuclear localization signal (NLS). The C terminal contains a ZnF-UBP.
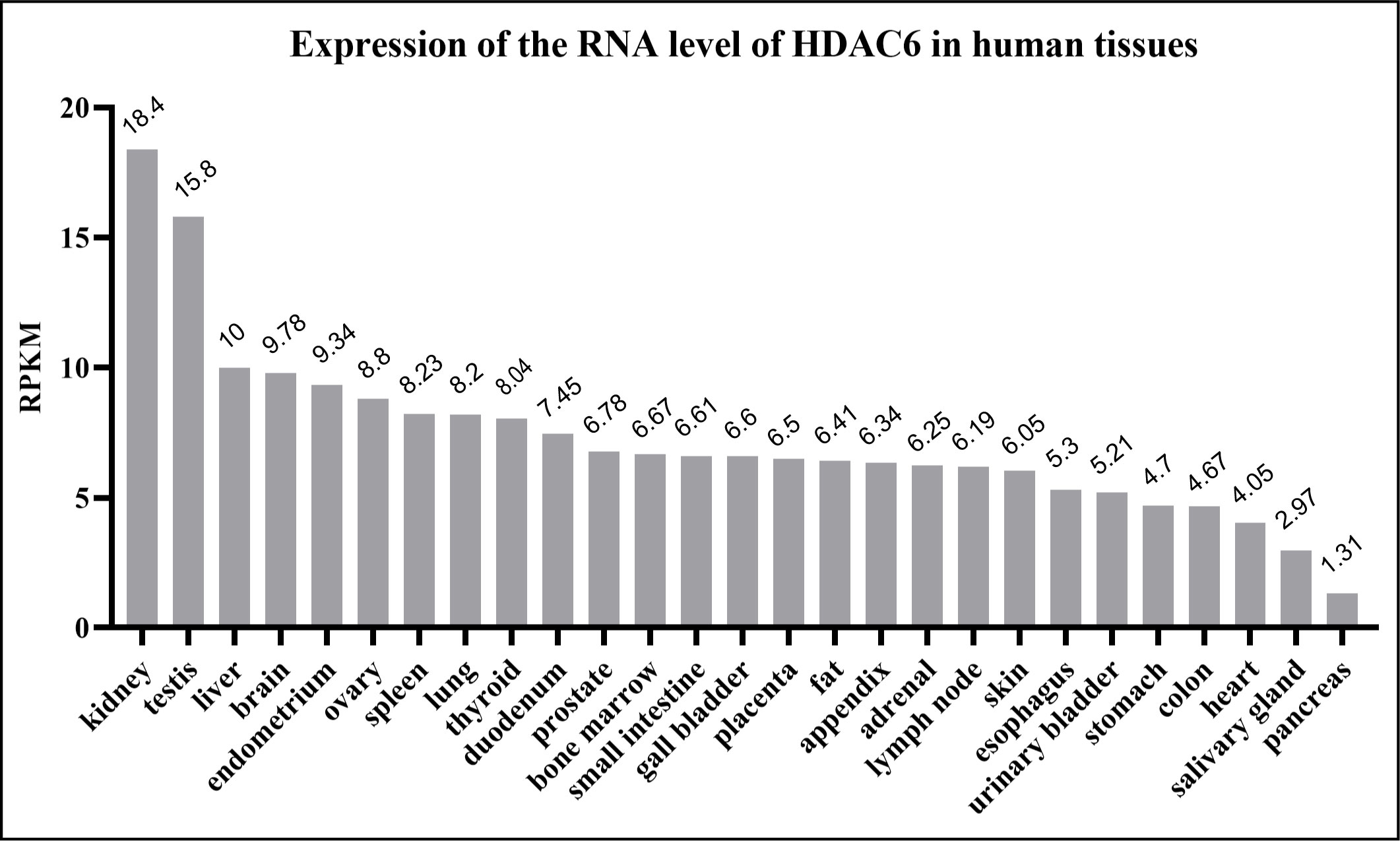
Figure 2 Human HDAC6 RNA expression level in normal tissues. The data from National Center for Biotechnology Information. HDAC6 is abundant in the kidney, testis, liver and brain. RPKM means reads per kilobase of exon model per million mapped reads.
Also, HDAC family members, especially the cytoplasmic protein HDAC6, are very important in controlling the acetylation of non-histone (34). A proteomic analysis found at least 3,600 acetylation sites in 1,750 nuclear and non-nuclear proteins. This shows that acetylation and deacetylation play an important role in processes in both the nucleus and the cytoplasm (39). Many substrates and proteins that work with them are found in the cytoplasm, which makes sense since that is where HDAC6 was found to be concentrated. HDAC6 has been shown to interact with a number of proteins that are not histones. These include α-tubulin (40, 41), heat shock protein 90 (42), cortactin (43), peroxiredoxins (44), β-catenin (45), Ku-70 (46), Tat (47), survivin (48), extracellular signal-regulated kinase-1 (ERK-1) (49, 50), heat shock factor-1 (51), myosin heavy chain 9 (52), heat shock cognate protein 70 (52), dnaJ homolog subfamily A member 1 (52), Miro-1 (53), tripartite motif-containing protein 21 (TRIM21) (22). Table 1 is a summary of the many HDAC6 substrates, what they do, and what viral infections they are linked to. A proteomics study found that 107 proteins in the livers of HDAC6 knockout mice could be HDAC6 deacetylating substrates (52).
HDAC6’s tubulin deacetylase activity was initially reported by Hubbert et al. in both in vivo and in vitro settings (40). In the end, it was found that the activity of HDAC6 recombinant mutants is controlled by mutations in certain catalytic domains. Their analysis showed that in vitro deacetylase activity was only encountered in the CD2 domain (35). In contrast to other HDACs, HDAC6 has a Cys/His-rich C-terminal region that interacts with polyubiquitin in a novel way while preserving the deacetylase activity of the protein (37). It has been reported that HDAC6 inhibitors regulate viral infection, tumorigenesis and disease development. Specific HDAC6 inhibitors are known so far including Tubacin, Tubastatin A, ACY-1215, ACY-24, Nexturastat A and others (34). HDAC6 inhibitors are gradually being used as a novel strategy for antiviral and antitumor drug development.
Upstream antiviral signaling promotes HDAC6
The host antiviral response depends on understanding HDAC6’s upstream signaling molecules. Transactive response DNA-binding protein 43 kDa (TDP-43), a nuclear RNA binding protein, is important in RNA processing (70, 71). TDP-43 particularly binds HDAC6 RNA, according to current research (65). TDP-43 stabilises the antiviral enzyme HDAC6 and increases its mRNA and protein levels, the study found (65). The upregulation of MT acetylation and the downregulation of the antiviral HDAC6 enzyme in response to low TDP-43 levels make target cells more vulnerable to human immunodeficiency virus (HIV) infection (65). High TDP-43 levels, on the other hand, change the cellular level of the HDAC6 antiviral factor, preventing HIV-1 envelope complex (Env) fusion and infection regardless of viral Env tropism and strain (65). The acetylation of α-tubulin lysine-40 amino acids is considered a sign of microtubules (MTs) stability (72), and as early as 2022, researchers found that HDAC6 can deacetylate α-tubulin (73). Rabies is caused by the Rabies virus, which has a high fatality rate and offers a significant health risk to humans. It was shown that expressing the rabies virus M protein alone significantly increased HDAC6 expression, leading to a significant decrease in its substrate, acetylated-tubulin, and ultimately MTs depolymerization (74) (Figure 3A). MTs depolymerization increases viral RNA production substantially (74). Inhibiting HDCA6 deacetylase activity drastically reduced rabies virus RNA synthesis, indicating that HDAC6 plays an essential role in viral infection. The protein kinase C (PKC) family is a type of serine/threonine kinase that is activated by phospholipids and calcium (75). PKCα is a member of the PKC family that is implicated in cancers, viral infection, and other signaling pathways (75, 76). Although IRF3 activation controls interferon synthesis, a recent study has demonstrated that IRF3 also works in conjunction with the protein β-catenin (69). A previous study has shown that HDAC6 activates β-catenin by deacetylation (69). However, following viral infection, how β-catenin was activated to affect downstream signaling pathways is still elusive. Zhu et al. (24) demonstrated a signaling pathway involving sendai virus (SeV) infection activated PKCα, which increases the phosphorylation of HDAC6, which then deacetylates β-catenin and promotes its translocation to the nucleus, promotes the phosphorylation of IRF3 and the production of IFN-β, which eventually lead to the inhibition of SeV replication (Figure 3B).
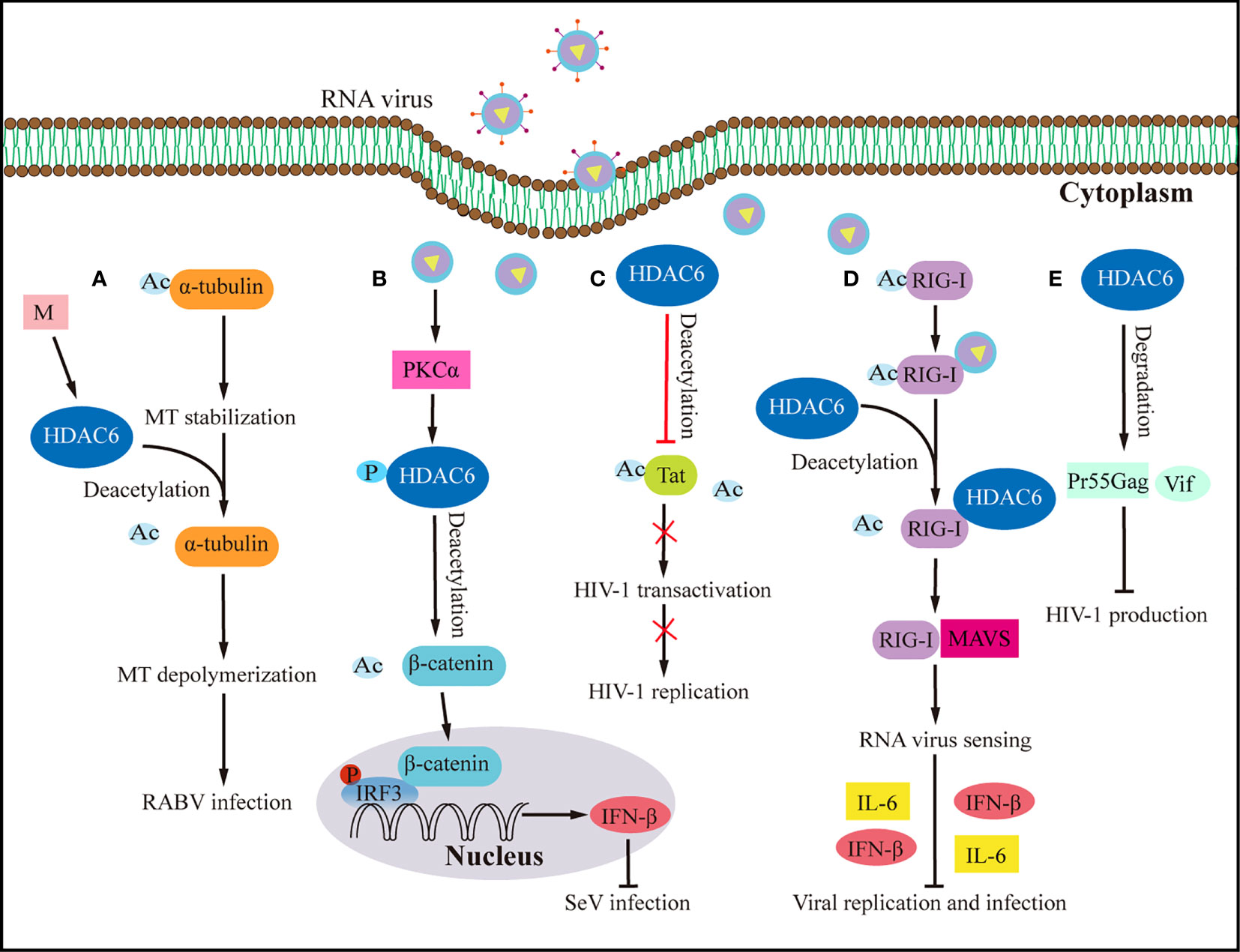
Figure 3 Effects of HDAC6 on RNA virus infection and host antiviral immunity. (A) By upregulating HDAC6, the M protein of the rabies virus induces MTs depolymerization to facilitate viral RNA synthesis. (B) HDAC6 deacetylation of β-catenin promotes nuclear translocation and IRF3-mediated IFN-β transcription, thereby inhibiting the infection by SeV. (C) HDAC6 prevents HIV-1 replication by deacetylating Tat and decreasing its ability to transactivate. (D) By interacting with RIG-I and promoting RIG-I deacetylation and sensing of RNA viruses, HDAC6 accelerates the transmission of the RLR signaling pathway when it is overexpressed, which increases the production of IFN-β and inflammatory proteins to prevent viral infection. (E) HDAC6 prevents the production of HIV-1 by promoting the degradation of viral proteins Pr55Gag and Vif. Ac, acetyl; M, matrix protein of rabies virus; P, phosphorylation; RABV, rabies virus.
HDAC6 mediates viral infection as well as host innate immunity
HDAC6 regulates both DNA and RNA viral replication, with recent research concentrating on influenza viruses among RNA viruses. HDAC6 mainly confronts virus invasion by controlling host cytoskeleton MTs to dynamically control virus transport, fusion, or uncoating. HDAC6 can also control viral infection by modifying the human immune system and autophagy. HDAC6’s upstream signaling pathway and its downstream target in anti-viral signaling, regrettably, remain largely unknown. Recent research on HDAC6 activity emphasizes its importance in the innate immune response to viral infection. Studies on HDAC6’s function in the infection of viruses have mostly focused on three aspects: (a) controlling viral transport, fusion, and viral component release by modulating plasma membrane dynamics and the cytoskeleton (55, 56, 62, 63, 74, 77); (b) influencing host cells’ antiviral immune response (19, 23, 69); (c) tuning the breakdown of viral or host proteins in host cells through autophagy (64, 68, 78).
HDAC6 regulates viral infection by changing the cytoskeleton and dynamics of plasma membranes
Through its effect on the cytoskeleton, HDAC6 controls viral transcription, replication, entry, and the movement of viral parts. This can be good or bad for a viral infection (55, 74). MTs have been known for a long time to be important in how the cytoskeleton moves during viral infection (79). It has been shown that viruses not only change MT and MT-related proteins to help them infect cells, but they also cause specific post-translational modifications (PTM) to help them spread through cells and cause side effects of how they spread (80). Acetylation of lysine is a reversible PTM that affects many cellular processes, such as chromatin remodeling, signaling, RNA splicing, gene expression, the cell cycle, protein stability, and protein transport (81). It happens at the lysine-amino ends, which are watched over by both histone acetyltransferases (HATs) (82) and HDACs (83). Some HATs and HDACs have been found to be very important in controlling IFN-I production and response (19, 24, 84). In 2002 and 2003, researchers found that HDAC6-mediated deacetylation was linked to the stability of MTs dynamics in vivo and that HDAC6 deacetylated both tubulin and MTs (41, 73). The cytoskeleton controls how the host cell membrane moves, which is how animal viruses get in (85). The host cytoskeleton is made up of microfilaments, MTs, and intermediate filaments (86, 87). These filaments guide a number of processes, such as reshaping plasma membranes, capturing and moving cargo, and arranging organelles in space, which are important for cell shape, polarity, movement, or division. Many viruses use the MT-based transport system of the host cell to move around inside the cell (88, 89).
HDAC6 is a major regulator of the invasive pathway of viruses. It affects how MTs move and how T regulatory cells work (40, 90), and it is a key part of how IAV gets infected (57). Seasonal outbreaks of IAV continue to kill a lot of people and make a lot of people sick all over the world. IAVs from birds can spread to humans and change enough to cause a pandemic (91). There is still a chance of a future IAV pandemic because new bird IAV strains (like H10N8, H6N1, H7N9, H9N2, H1N1, and H5N6) keep showing up in humans. IAV has already caused a pandemic this century (92–97). IAV attacks the airway epithelium of a person’s respiratory system to start the infection, which then leads to the flu, an acute fever-related respiratory illness. It has been shown that acetylated MTs help IAV nonmembrane components move (55). However, the deacetylation of MTs by HDAC6 prevents the trafficking of IAV components (55). HDAC6 is an anti-IAV host factor that works by using acetylated MTs as its substrate to inhibit the movement of viral components toward the plasma membrane of the host cell (55) (Figure 4A). However, when in IAV infection, the virus can take advantage of a variety of host characteristics to aid in its reproduction. For example, to aid uncoating and infection, IAV can use the aggresome-processing machinery driven by HDAC6 (56). IAV hijacks host HDAC6 to employ the host’s aggresome pathway to uncoat itself during viral entry (56). The ubiquitin-binding domain, but not the deacetylase activity, was needed for HDAC6 to be brought to viral fusion sites and for viruses to uncoat and infect cells (56). A further study showed that HDAC6 interacts with MT motor protein cytoplasmic dynein and its activator dynactin to bind unanchored ubiquitin chains that mimic unfolded proteins in the capsid, assisting in its transit to the aggresome (56) (Figure 4B). But new research has discovered that interfering with the interaction of HDAC6 and ubiquitin impairs IAV and Zika virus infection (58). It suggests that designed ankyrin repeat proteins (DARPins), which can block the ZnF pocket where ubiquitin engages, also inhibit aggresomes and stress granules (SGs) formation (58) (Figure 4C). The effects of acetylation and deacetylation on viral proteins and human immune pathway proteins impact viral replication. Other RNA viruses, like the HIV-1, use the MTs for their own purposes, just like IAV does. For instance, MTs were found to be crucial for HDAC6 to interact with Tat and for HDAC6 to deacetylate Tat and stop HIV-1 transactivation (47) (Figure 3C). HDAC6 reduction promotes axonal transport and avoids HIV-1 envelope protein gp120 neurotoxicity (62). HDAC6-mediated deacetylation of α-tubulin, for example, inhibits HIV-1 fusion and infection (63). Recent research has demonstrated that the suppression of HDAC6 greatly increases the amount of acetylation of α-tubulin, which enhances the fusion and reproduction of human parainfluenza virus type 3 (HPIV3) (66). TANK-binding kinase 1 (TBK1), an IKK-related serine/threonine kinase, plays a big role in antiviral immunity (98–101). Recent research shows that HDAC6 controls the phosphorylation of TBK1 and Akt in macrophages by poly (I:C) (23). Reports show that when double-stranded RNA (dsRNA) binds to TLR3, it turns on TBK1 and starts signaling transduction that leads to the production of IFN-I (98–101). The cell signaling molecule Akt also interacts with MT (102, 103). It has been observed that Akt regulates glycogen synthase kinase-3β (GSK-3β) activity and interleukin-10 (IL-10) expression (104, 105). GSK-3β has a pivotal role in controlling TBK1 activity (106). In the presence of a virus, GSK-3β interacts to TBK1, leading to the phosphorylation of TBK1 (106). Numerous studies indicate that MTs and GSK-3β have intimate interactions (107–109). Poly (I:C) induced IFN-β expression in macrophages was elevated after HDAC6 deletion, which influenced IFN-I and IL-10 production via enhancing TBK1 activity and removing GSK-3β inhibitory regulation (23). The results of the experiments with HDAC6 knockout mice highlight the effect of HDAC6 deletion on the immune system, confirming the importance of HDAC6 in viral infection. HDAC6 knockout mice exhibited abnormalities in bone homeostasis and immunological function (38). The number of IFN-I and IL-6 produced by these mice is also lower, making them more vulnerable to lethal RNA virus infection (110).
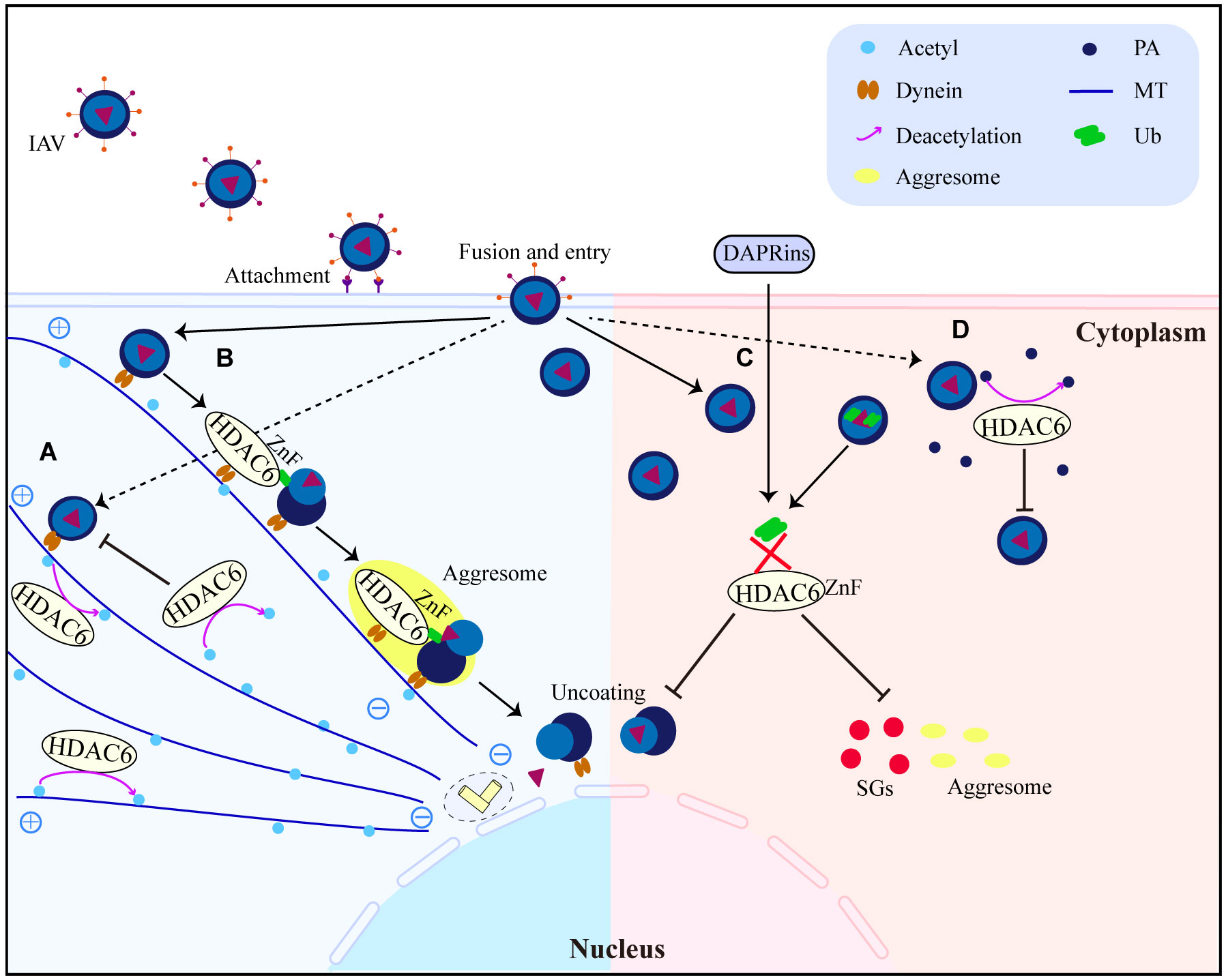
Figure 4 HDAC6 promotes or inhibits IAV replication and infection interestingly. (A) HDAC6 inhibits IAV release by downregulating IAV viral component transport by deacetylating MTs. (B) HDAC6 interacts with dynein, IAV can promote virus uncoating by utilizing the HDAC6-Znf-dependent aggresome formation mechanism. (C) DARPins block the binding of HDAC6-ZnF to Ub, inhibit the production of downstream SGs and impair IAV infection. (D) Through the destabilization of PA, HDAC6 functions as a negative regulator of IAV infection. HDAC6 binds to and deacetylates PA, promoting its proteasomal degradation. PA, polymerase acidic protein; MT, microtubule; Ub, ubiquitin.
Generally, these results highlight the complexity of the relationship between hosts and invaders by showing that HDAC6 can have both positive and negative effects on viral replication and indicating that effective HDAC6-targeting techniques may fundamentally vary between various viral species.
HDAC6 influences the host’s antiviral immunological response
HDAC6 acts as an anti-IAV host factor by deacetylating polymerase acidic protein (PA), which reduces IAV RNA polymerase activity (Figure 4D) (57). A recent study found that HDAC6 mutant mice have a higher proclivity to produce IAV infection (111), emphasising the importance of HDAC6 in anti-IAV infection. In embryonic stem cells and animals, HDAC6 overexpression enhances viral resistance (112). According to the latest research, TRIM21 is a substrate of HDAC6, and acetylation regulates its function (22). HDAC6 interacts with TRIM21 via the PRYSPRY motif and deacetylates it at lysines 385 and 387, facilitating homodimerization (22). Enhanced TRIM21 acetylation caused by HDAC6 inhibition, together with the suppression of TRIM21 dimerization and ubiquitination caused by hyperacetylation, prevents TRIM21 from adhering to the antibody-bound adenovirus (AdV) type 5 complex and being destroyed by the ubiquitin-proteasome pathway (22). HDAC6 depletion or inhibition promotes virus accumulation in cells, indicating a reduced ability for virus intracellular neutralization mediated by antibodies (22), which confirmed that HDAC6 is a new activator of TRIM21-mediated anti-viral innate immune response. Furthermore, HDAC6 deletion suppressed poly (I:C)-induced Akt activation, as evidenced by decreased phosphorylation of Akt at Ser473 near the carboxy terminus, implies that HDAC6 might control the degree to which macrophages activate Akt during viral infection (23). The findings imply that HDAC6 may modulate macrophage innate immunological responses to viruses.
RLRs, which include RIG-I and MDA5, are critical in innate immune responses to viral infection (113–117). RLRs in the cytosol detected invading RNA viruses (118). MDA5 and RIG-I detect different dsRNAs, MDA5 recognizes poly(I:C), and promoting production of IFNs in response to paramyxoviruses, influenza virus, and RIG-I recognizes in vitro transcribed dsRNAs and picornavirus (119). RIG-I containing CARDs and DExD/H box helicase domain and inducing production of IFNs in case of infection with dsRNA (113). RIG-I is required for viral RNA recognition and activation of downstream signaling pathways (110), such as the SeV (120), IAV and vesicular stomatitis virus (VSV) (121). RIG-I has piqued the interest of researchers as a viral sensor, able to distinguish 5’-triphosphate-containing dsRNA from a range of viruses or short dsRNA molecules (113, 114, 122). Additionally, overexpression of HDAC6 but not the catalytically inactive mutant boosted RLR-mediated RNA virus replication decrease (110). Binding of K63-linked polyubiquitin chains to RIG-I and start assembly of mitochondrial anti-viral-signaling protein (MAVS; also known as VISA, CARDIF, and IPS-1) activates TBK1 and IKK through a K63 polyubiquitin-dependent mechanism (123). Subsequently, transcription factors IRF3 and nuclear factor kappa B (NF-κB) are activated, and type I interferons and pro-inflammatory cytokines are turned on (123). HDAC6 regulates deacetylation of the RIG-I C-terminal domain, which limits the protein’s capacity to recognize viral RNA (110). HDAC6 acts as a deacetylase that enhances RIG-I activation and innate antiviral defense in order to identify and reduce hepatitis C virus (HCV) and other RNA virus infections (59). HDAC6 temporarily bound to RIG-I in the presence of viral RNAs, deacetylating lysine 909 (K909) to improve RIG-I’s ability to detect viral RNAs (Figure 3D) (110). Acetyl-mimicking mutants of RIG-I do not induce virus-induced assembly of active homo-oligomers, whereas deacetylation of RIG-I promotes the oligomerization of RIG-I and binding to the ligand MAVS (Figure 3D) (59). Reduced HDAC6 expression led to compromised defense against RNA viruses but not DNA viruses (110). Reducing or eliminating endogenous HDAC6 in immune cells increased viral multiplication and lowered IFN-I and pro-inflammatory cytokine production in response to RNA viruses (110). IFN-I responses are essential for the host innate immunity to protect against infections (124, 125). When cytoplasmic RIG-I-like receptors detect viral RNA, they send signals to the transcription factors IRF3 and NF-κB, which in turn induce IFN-I transcription (126). MAVS binds to RIG-I upon virus recognition and stimulates the production of ISGs by activating NF-κB and other interferon regulatory factors (127, 128). In order to offer antiviral defense and regulate immunity to infection, this mechanism leads to the development of MAVS, which in turn activates NF-κB and IRF3 signaling pathways and triggers the production of IFN-I and a large number of antiviral and immunology-related genes (129). In response to viral infection, RIG-I or TLR3 signaling activated IRF3, a critical transcription factor for IFN induction (130, 131). It has been demonstrated that HDACs participate in the TLR signaling pathway of the innate immune system. For instance, HDAC6 facilitates ubiquitin-dependent myeloid differentiation primary response 88 (MyD88) aggregation, which hinders TLR4 responses and prevents a viable MyD88 signaling platform from emerging (132). In addition, the intracellular bacterium Listeria monocytogenes activates HDAC6 to regulate innate immune and autophagy responses to TLR-mediated signals (133). Ablation of HDAC6 appears to decrease NF-κB activation in response to TLR stimulation and permit faulty autophagy, resulting in impaired bacterial clearance. HDAC6 is linked to the TLR adaptor protein MyD88 (133).
In conclusion, these findings imply that enhancing HDAC6 responses may protect against viral infection and effectively demonstrate that HDAC6 may improve the host IFN-I response to obstruct viral pathogenesis. HDAC6, a host factor, acts as an antiviral target by modulating viral transport, uncoating, replication, and infectivity, as well as IFN-I expression.
HDAC6 controls viral infection by degrading host or viral proteins
In addition to controlling RNA viral infection, HDAC6 affects the host’s innate immune response following DNA virus infection. Porcine circoviruses (PCV) are non-enveloped tiny viruses with single-stranded circular DNA genomes (1.76 kb) that belong to the Circoviridae family (134, 135). PCV2 is a type of PCV that promotes cyclic GMP-AMP synthase (cGAS) phosphorylation at S278 in the early stages of infection by activating phosphatidylinositol 3-kinase (PI3K)/Akt signaling, which directly silences cGAS catalytic activity (68). Then, by the activity of the HDAC6 deacetylase, cGAS phosphorylation at the S278 site can stimulate k48-linked polyubiquitination at the K389 site, which can be exploited as a signal and facilitate the translocation of k48-ubiquitinated cGAS from the cytosol to the autolysosome (68). According to the results, PCV2 inhibits IFN-I induction to enhance DNA virus infections by targeting cGAS (68). As such, TRIM14 recruits USP14 to cleave the K48-linked ubiquitin chains of cGAS at K414 during herpes simplex virus type 1 (HSV-1) infection (136). This inhibits the p62-mediated autophagic degradation of cGAS and increases the activation of IFN-I signaling (136). ISG15 association with HDAC6 and p62 and it was activated by type I IFN signaling, which enhances the activity of selective autophagy, resulting in the effective clearance of ubiquitin and ISG15-tagged unwanted proteins and pathogens via p62 and HDAC6 (137). ISG15-mediated protein conjugation may therefore be the IFN system that permits the cell to utilize autophagy as an innate antiviral defense (137).
To inhibit HIV-1 production, HDAC6 promotes aggresome/autophagic degradation of the viral polyprotein Pr55Gag (64). By targeting the HIV proteins Pr55Gag and viral infectivity factor (Vif), HDAC6 functions as an anti-HIV-1 limiting factor, reducing viral proliferation and infection (64) (Figure 3E). SGs are dynamic structures that can be targeted for autophagic clearance, which is known as granulophagy (138). Aside from that, granulophagy may be an approach used by viruses to suppress antiviral immune responses (78). SGs mentioned above can serve as an antiviral immune complex and play a positive role in IFN-I response, while HDAC6 is also a component of SGs (78). In recent years, studies have found that coxsackie virus A16 infection leads to granulophagy, degradation of SGs, and selective autophagy inhibits IFN-I response (78). The findings demonstrate that viral replication and proliferation can be prevented by limiting granulophagy. Furthermore, HDAC6 inhibits oncolytic herpes simplex virus (oHSV) replication in glioma cells by aiding oHSV endocytic entry and subsequent fusion to lysosomes, hence steering incoming virions to autophagy/xenophagy rather than viral multiplication in the nucleus (67). HDAC6, a ubiquitin-binding deacetylase that targets protein aggregates and damaged mitochondria, is a key component in basic autophagy (139). HDAC6 activates autophagy by activating a cortactin-dependent actin-remodeling mechanism, which then assembles an F-actin network that promotes autophagosome-lysosome fusion and substrate degradation (139). HDAC6 is involved in the regulation of autophagy, whether as an antiviral factor to promote viral protein degradation or as a component of an antiviral complex to be degraded by viruses, which provides new ideas for host antiviral strategies.
Diseases involving HDAC6
More attention has been paid to HDAC6’s many roles in pathology and physiology because it could be used to treat a wide range of diseases. Recent studies and reports have shown that selective HDAC6 inhibition is a good way to fight neurodegenerative diseases like Alzheimer, Huntington, and Parkinson (140). The cells of the innate immune system are the most important line of defense against pathogens and cancer cells. HDAC6 is closely linked to a number of diseases that are linked to cancerous tumors (141). As well as cancers like lung cancer, breast cancer, and ovarian cancer (28). In the case of hepatocellular carcinoma (HCC), pro-inflammatory cytokines increase the amount of HDAC6 in the body. This can cause more cells to grow by stopping p53 from doing its job as a transcription factor and causing it to be broken down (142). Hepatitis B virus (HBV), as a stronger inducer of HCC, chronic infection of HBV increased risk of HCC (143, 144). According to research, the protein expression levels of HDAC6 in HBV patients’ serum and liver decreased significantly after antiviral treatment (145). It has been reported that HBV internalization is sparked by the host-entry cofactor epidermal growth factor receptor (146). A deficiency of HDAC6 promotes epidermal growth factor receptor endocytic trafficking and degradation (147). In addition, the study found that HDAC6 can promote human papillomavirus (HPV) positive cervical cancer by upregulating Wnt5a (148). In the small-cell lung cancer xenograft model, HDAC6 was found to be a possible treatment target when used with JQ1 (149). More research needs to be done to find out if HDAC6 overexpression mice cause tumors or if they are more or less likely to get tumors. The innate immune response is different and ends with the production of cytokines (150). A macrophage releases ILs, cytokines that give instructions to other immune cells when it encounters a virus, bacteria, or other pathogens (151, 152). The innate immune response is a quick, nonspecific reaction that is controlled by a number of molecules. HDACs are important for controlling the size and strength of the response, which makes it possible to make just the right amount of inflammatory cytokines and ILs (153). Early-onset familial Parkinson’s disease is caused by changes in the protein Parkin, which is a ubiquitin ligase (154). Parkin increases mitophagy by accelerating mitochondrial ubiquitination, which attracts ubiquitin-binding autophagic components, HDAC6 and p62, resulting in mitochondrial clearance (154). Tubulin deacetylation is caused by the enzymatic activity of HDAC6 (40), and blocking tubulin deacetylation is effective in avoiding neurodegeneration in animal models of Alzheimer’s and Huntington’s disease (155, 156).
Through the transcriptional activation of their respective genes by a transcription factor NF-κB, which is a crucial component of the immune response, these cytokines are frequently produced by a variety of innate immune cells, HDAC6 has a role in HIV-1 Tat-induced pro-inflammatory gene expression by regulating the mitogen-activated protein kinase NF-κB/AP-1 pathways and it is a biological target for HIV-1 Tat-mediated neuroinflammation (60, 61). Interestingly, HDAC6 is also involved in the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) pandemic that is currently underway (coronavirus disease 2019; COVID-19) (157). HDAC6 inhibition has the potential to be used to minimize the morbidity associated with severe COVID-19 via modulating the innate and adaptive immune systems (54). HDAC6 inhibition, which blocks IFN-I synthesis and its downstream effects in airway epithelial cells and immune cells, may be able to mitigate the negative effects generated in severely ill COVID-19 patients by late or extended activation of the IFN-I pathway (54). These results support the use of HDAC inhibitors as part of an epigenetic therapeutic strategy in the treatment of severe COVID-19 (54).
HDAC6 may be a risk factor for tuberculosis (TB) and a novel host-directed anti-TB therapeutic target (158). HDAC6 also affects allergic skin inflammation. By directly modulating sirtuin 1 expression, HDAC6-negative MiR-9 prevented atopic dermatitis (159). HDAC6 and CXCL13 influence cellular connections and miR-9 and SIRT1 expression (159). HDAC6 regulates inflammation. HDAC6 reduces NACHT, LRR, and PYD domains-containing protein (NLRP3) inflammasome activity by interacting with ubiquitinated NLRP3 (160). HDAC6 knockout mice develop normally, but their immune systems are weakened (38). HDAC6 promotes monocyte/macrophage infiltration during inflammation and suppresses T cell IL-17 production, providing new insights into its role in the immune system (161, 162). A recent study demonstrated that administering an HDAC6 degrader to LPS-induced mice reduced the activation of the NLRP3 inflammasome, confirming for the first time that HDAC6 proteolysis targeting chimera may be a potential treatment for NLRP3-related illnesses (163).
The application of HDAC6 inhibitors
Tubacin, as a selective inhibitor of HDAC6, plays a positive role in anti-viral and anti-tumor diseases (164–166). It is worth noting that “tubacin”, which prevents α-tubulin deacetylation in mammalian cells, was discovered for the first time using an omnifarious, chemical genetic screen of 7,392 small molecules (167). In the early years, as an HDAC6 selective inhibitor, tubacin induces apoptosis in epstein-barr virus (EBV)-burkitt lymphoma cells and kills EBV lymphoblastoid cells by producing reactive oxygen species (165). A few years ago, researchers found that tubacin reduces Japanese encephalitis virus replication by reducing viral RNA synthesis (164). The role of inhibiting viral replication was also demonstrated by HDAC inhibitors including suberoylanilide hydroxamic acid (SAHA) (168), which improves the susceptibility of uninfected CD4+ T cells to HIV by boosting the kinetics and efficiency of postentry viral processes (169). On the other hand, HDAC6 knockout mice can live and grow normally, tubulin hyperacetylation does not interfere with normal mammalian development, indicating that HDAC6 inhibitors could have less negative effects (38), as opposed to the inhibition of specific class I HDACs and other HDACs. These results encourage more research into the efficacy of HDAC6 inhibitors in a variety of diseases, including hematological malignancies (170, 171), neurodegenerative diseases (140) and cancers (28). Moreover, specific inhibition of HDAC6 normalizes B cell activation and germinal centre development in a model of systemic lupus erythematosus (172). These tests will provide the basic science foundation for moving forward with clinical trials of an inhibitor that selectively targets HDAC6. There is great potential for HDAC6 as a target molecule in clinical drug research, as more than a dozen HDAC6s are already in use for the treatment of various disorders.
Conclusions
This review summarizes the function of HDAC6 in viral infection, host antiviral immunity and immune-related disorders. HDAC6 is a cytoplasmic class II histone deacetylase involved in various cellular functions, including immunological synapse formation, misfolded protein degradation, migration, and cell-cell contact. HDAC6 has two highly conserved catalytic domains: catalytic domain 1 (CD1) and CD2, which exhibit broader substrate specificity as a deacetylated domain with catalytic activity. Another NES (1049-1058 aa) and a ZnF-UBP domain recruit ubiquitin protein to stimulate aggresome formation. The HDAC6 gene has 21923 base pairs and is on chromosome X p11.22-23. It is found in large amounts in the testis, spermatogenic cells, germ cell tissues, liver, heart, muscle, spleen, and kidney, among other normal human tissues and organs. HDAC6 functions as a host antiviral factor to prevent the invasion and infection of invaders, while the virus will utilize some HDAC6 properties to escape the host’s defensive response and aid in the virus’s intracellular cycle completion. HDAC6 can reduce IAV release by deacetylating MTs, inhibit viral transcription by inhibiting the IAV RNA polymerase PA subunit, and assist viral uncoating via the aggregation formation mechanism of HDAC6’s ZnF domain. Blocking HDAC6 binding of ZnF to ubiquitin can decrease aggregation formation, reducing IAV uncoating. HDAC6 is involved in the process of a range of viral infections and can be exploited as a potential therapeutic target for many disorders. Therefore, the review’s findings show that HDAC6 is an attractive and new potential therapeutic target for the development of immune-enhancing medicines in the areas of antiviral immunity, innate immune response, and certain diseases.
Author contributions
MQ and XW conceived and drafted the manuscript. MQ, YS, AW and PC discussed the concepts of the manuscript. MQ drew the figures. MQ, XY, XW, HZ and YS approved the version to be submitted. All authors contributed to the article and approved the submitted version.
Funding
This work was sponsored by the Major Science and Technology Project of Gansu Province (21ZD3NA001, 22ZD6NA001), the Key Development and Research Foundation of Gansu (Grant no. 21YF5WA153) and National Natural Science Foundation of China (Grant no. 32202779).
Acknowledgments
We sincerely regret that we were unable to cite certain scholars’ related work in this study due to limited space. We appreciate the editor’s and reviewers’ helpful comments and discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
References
1. Shi T, McAllister DA, O’Brien KL, Simoes EA, Madhi SA, Gessner BD, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet (2017) 390(10098):946–58. doi: 10.1016/s0140-6736(17)30938-8
2. Gower E, Estes C, Blach S, Shearer KR, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol (2014) 61(1 Suppl):S45–57. doi: 10.1016/j.jhep.2014.07.027
3. Laing KJ, Ouwendijk WJD, Koelle DM, Verjans GMGM. Immunobiology of Varicella-Zoster virus infection. J Infect Dis (2018) 218(suppl_2):S68–74. doi: 10.1093/infdis/jiy403
4. Hosseini A, Hashemi V, Shomali N, Asghari F, Gharibi T, Akbari M, et al. Innate and adaptive immune responses against coronavirus. BioMed Pharmacother (2020) 132:110859. doi: 10.1016/j.biopha.2020.110859
5. O’Neill LA, Golenbock D, Bowie. AG. The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol (2013) 13(6):453–60. doi: 10.1038/nri3446
6. Janeway CA Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol (2002) 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359
7. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell (2010) 140(6):805–20. doi: 10.1016/j.cell.2010.01.022
8. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity (2011) 34(5):637–50. doi: 10.1016/j.immuni.2011.05.006
9. Matsumoto M, Seya T. TLR3: interferon induction by double-stranded RNA including poly(I:C). Adv Drug Delivery Rev (2008) 60(7):805–12. doi: 10.1016/j.addr.2007.11.005
10. Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol (2007) 1(1):19–25. doi: 10.1016/j.molonc.2007.01.001
11. Lemoine M, Younes A. Histone deacetylase inhibitors in the treatment of lymphoma. Discovery Med (2010) 10(54):462–70.
12. Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol (2014) 6(4):a018713. doi: 10.1101/cshperspect.a018713
13. Wang XX, Wan RZ, Liu. ZP. Recent advances in the discovery of potent and selective HDAC6 inhibitors. Eur J Med Chem (2018) 143:1406–18. doi: 10.1016/j.ejmech.2017.10.040
14. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature (2000) 403(6771):795–800. doi: 10.1038/35001622
15. Zhang Y, Yan J, Yao. TP. Discovery of a fluorescent probe with HDAC6 selective inhibition. Eur J Med Chem (2017) 141:596–602. doi: 10.1016/j.ejmech.2017.10.022
16. Kumar S, Attrish D, Srivastava A, Banerjee J, Tripathi M, Chandra PS, et al. Non-histone substrates of histone deacetylases as potential therapeutic targets in epilepsy. Expert Opin Ther Targets (2021) 25(1):75–85. doi: 10.1080/14728222.2021.1860016
17. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol (2014) 14(1):36–49. doi: 10.1038/nri3581
18. Yang LP, Chen SC, Zhao Q, Pan CH, Peng LN, Han Y, et al. Histone deacetylase 3 contributes to the antiviral innate immunity of macrophages by interacting with FOXK1 to regulate STAT1/2 transcription. Cell Rep (2022) 38(4):110302. doi: 10.1016/j.celrep.2022.110302
19. Nusinzon I, Horvath CM. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol Cell Biol (2006) 26(8):3106–13. doi: 10.1128/mcb.26.8.3106-3113.2006
20. Galvin HD, Husain M. Influenza A virus-induced host caspase and viral PA-X antagonize the antiviral host factor, histone deacetylase 4. J Biol Chem (2019) 294(52):20207–21. doi: 10.1074/jbc.RA119.010650
21. Chen L, Wang CM, Luo J, Su W, Li M, Zhao N, et al. Histone deacetylase 1 plays an acetylation-independent role in influenza A virus replication. Front Immunol (2017) 8:1757. doi: 10.3389/fimmu.2017.01757
22. Xie SB, Zhang LL, Dong D, Ge R, He Q, Fan C, et al. HDAC6 regulates antibody-dependent intracellular neutralization of viruses via deacetylation of TRIM21. J Biol Chem (2020) 295(42):14343–51. doi: 10.1074/jbc.RA119.011006
23. Wang Y, Wang K, Fu. J. HDAC6 mediates poly (I:C)-induced TBK1 and Akt phosphorylation in macrophages. Front Immunol (2020) 11:1776. doi: 10.3389/fimmu.2020.01776
24. Zhu JZ, Coyne CB, Sarkar. SN. PKC alpha regulates Sendai virus-mediated interferon induction through HDAC6 and beta-catenin. EMBO J (2011) 30(23):4838–49. doi: 10.1038/emboj.2011.351
25. Grozinger CM, Hassig CA, Schreiber. SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci U.S.A. (1999) 96(9):4868–73. doi: 10.1073/pnas.96.9.4868
26. Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J Biol Chem (1999) 274(4):2440–5. doi: 10.1074/jbc.274.4.2440
27. Chen S, Owens GC, Makarenkova H, Edelman DB. HDAC6 regulates mitochondrial transport in hippocampal neurons. PloS One (2010) 5(5):e10848. doi: 10.1371/journal.pone.0010848
28. Li Y, Shin D, Kwon. SH. Histone deacetylase 6 plays a role as a distinct regulator of diverse cellular processes. FEBS J (2013) 280(3):775–93. doi: 10.1111/febs.12079
29. Ustinova K, Novakova Z, Saito M, Meleshin M, Mikesova J, Kutil Z, et al. The disordered N-terminus of HDAC6 is a microtubule-binding domain critical for efficient tubulin deacetylation. J Biol Chem (2020) 295(9):2614–28. doi: 10.1074/jbc.RA119.011243
30. Saito M, Hess D, Eglinger J, Fritsch AW, Kreysing M, Weinert BT, et al. Acetylation of intrinsically disordered regions regulates phase separation. Nat Chem Biol (2019) 15(1):51–61. doi: 10.1038/s41589-018-0180-7
31. Zhang M, Xiang S, Joo HY, Wang L, Williams KA, Liu W, et al. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSalpha. Mol Cell (2014) 55(1):31–46. doi: 10.1016/j.molcel.2014.04.028
32. Hai Y, Christianson DW. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol (2016) 12(9):741. doi: 10.1038/nchembio.2134
33. Balmik AA, Chidambaram H, Dangi A, Marelli UK, Chinnathambi S. HDAC6 ZnF UBP as the modifier of Tau structure and function. Biochemistry (2020) 59(48):4546–62. doi: 10.1021/acs.biochem.0c00585
34. Pulya S, Amin SA, Adhikari N, Biswas S, Jha T, Ghosh B. HDAC6 as privileged target in drug discovery: A perspective. Pharmacol Res (2021) 163:105274. doi: 10.1016/j.phrs.2020.105274
35. Zou H, Wu Y, Navre M, Sang BC. Characterization of the two catalytic domains in histone deacetylase 6. Biochem Biophys Res Commun (2006) 341(1):45–50. doi: 10.1016/j.bbrc.2005.12.144
36. Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell (2003) 115(6):727–38. doi: 10.1016/s0092-8674(03)00939-5
37. Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, et al. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol Cell Biol (2001) 21(23):8035–44. doi: 10.1128/MCB.21.23.8035-8044.2001
38. Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol (2008) 28(5):1688–701. doi: 10.1128/MCB.01154-06
39. Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science (2009) 325(5942):834–40. doi: 10.1126/science.1175371
40. Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, et al. HDAC6 is a microtubule-associated deacetylase. Nature (2002) 417(6887):455–8. doi: 10.1038/417455a
41. Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J (2003) 22(5):1168–79. doi: 10.1093/emboj/cdg115
42. Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell (2005) 18(5):601–7. doi: 10.1016/j.molcel.2005.04.021
43. Zhang X, Yuan Z, Zhang Y, Yong S, Burgos AS, Koomen J, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell (2007) 27(2):197–213. doi: 10.1016/j.molcel.2007.05.033
44. Parmigiani RB, Xu WS, Venta-Perez G, Bromage HE, Yaneva M, Tempst P, et al. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc Natl Acad Sci U.S.A. (2008) 105(28):9633–8. doi: 10.1073/pnas.0803749105
45. Li Y, Zhang X, Polakiewicz RD, Yao TP, Comb MJ. HDAC6 is required for epidermal growth factor-induced beta-catenin nuclear localization. J Biol Chem (2008) 283(19):12686–90. doi: 10.1074/jbc.C700185200
46. Subramanian C, Jarzembowski JA, Opipari AW Jr, Castle VP, Kwok RPS. HDAC6 deacetylates Ku70 and regulates Ku70-Bax binding in neuroblastoma. Neoplasia (2011) 13(8):726–34. doi: 10.1593/neo.11558
47. Huo L, Li D, Sun X, Shi XJ, Karna P, Yang W, et al. Regulation of Tat acetylation and transactivation activity by the microtubule-associated deacetylase HDAC6. J Biol Chem (2011) 286(11):9280–6. doi: 10.1074/jbc.M110.208884
48. Riolo MT, Cooper ZA, Holloway MP, Cheng Y, Bianchi C, Yakirevich E, et al. Histone deacetylase 6 (HDAC6) deacetylates survivin for its nuclear export in breast cancer. J Biol Chem (2012) 287(14):10885–93. doi: 10.1074/jbc.M111.308791
49. Williams KA, Zhang M, Xiang S, Hu C, Wu JY, Zhang SP, et al. Extracellular signal-regulated kinase (ERK) phosphorylates histone deacetylase 6 (HDAC6) at serine 1035 to stimulate cell migration. J Biol Chem (2013) 288(46):33156–70. doi: 10.1074/jbc.M113.472506
50. Wu JY, Xiang S, Zhang M, Fang B, Huang H, Kwon OK, et al. Histone deacetylase 6 (HDAC6) deacetylates extracellular signal-regulated kinase 1 (ERK1) and thereby stimulates ERK1 activity. J Biol Chem (2018) 293(6):1976–93. doi: 10.1074/jbc.M117.795955
51. Pernet L, Faure V, Gilquin B, Guérin SD, Khochbin S, Vourc’h C, et al. HDAC6-ubiquitin interaction controls the duration of HSF1 activation after heat shock. Mol Biol Cell (2014) 25(25):4187–94. doi: 10.1091/mbc.E14-06-1032
52. Zhang L, Liu S, Liu N, Zhang Y, Liu M, Li DW, et al. Proteomic identification and functional characterization of MYH9, Hsc70, and DNAJA1 as novel substrates of HDAC6 deacetylase activity. Protein Cell (2015) 6(1):42–54. doi: 10.1007/s13238-014-0102-8
53. Kalinski AL, Kar AN, Craver J, Tosolini AP, Sleigh JN, Lee SJ, et al. Deacetylation of Miro1 by HDAC6 blocks mitochondrial transport and mediates axon growth inhibition. J Cell Biol (2019) 218(6):1871–90. doi: 10.1083/jcb.201702187
54. Ripamonti C, Spadotto V, Pozzi P, Stevenazzi A, Vergani B, Marchini M, et al. HDAC inhibition as potential therapeutic strategy to restore the deregulated immune response in severe COVID-19. Front Immunol (2022) 13:841716. doi: 10.3389/fimmu.2022.841716
55. Husain M, Cheung CY. Histone deacetylase 6 inhibits influenza A virus release by downregulating the trafficking of viral components to the plasma membrane via its substrate, acetylated microtubules. J Virol (2014) 88(19):11229–39. doi: 10.1128/JVI.00727-14
56. Banerjee I, Miyake Y, Nobs SP, Schneider C, Horvath P, Kopf M, et al. Influenza A virus uses the aggresome processing machinery for host cell entry. Science (2014) 346(6208):473–7. doi: 10.1126/science.1257037
57. Chen H, Qian YJ, Chen X, Ruan ZY, Ye YT, Chen HJ, et al. HDAC6 restricts influenza A virus by deacetylation of the RNA polymerase PA subunit. J Virol (2019) 93(4):e01896-18. doi: 10.1128/jvi.01896-18
58. Wang L, Moreira EA, Kempf G, Miyake Y, Esteves BO, Fahmi A, et al. Disrupting the HDAC6-ubiquitin interaction impairs infection by influenza and Zika virus and cellular stress pathways. Cell Rep (2022) 39(4):110736. doi: 10.1016/j.celrep.2022.110736
59. Liu HM, Jiang FG, Loo YM, Hsu SZ, Hsiang TY, Marcotrigiano J, et al. Regulation of retinoic acid inducible gene-I (RIG-I) activation by the histone deacetylase 6. Ebiomedicine (2016) 9:195–206. doi: 10.1016/j.ebiom.2016.06.015
60. Youn GS, Ju SM, Choi SY, Park J. HDAC6 mediates HIV-1 tat-induced proinflammatory responses by regulating MAPK-NF-kappaB/AP-1 pathways in astrocytes. Glia (2015) 63(11):1953–65. doi: 10.1002/glia.22865
61. Youn GS, Cho H, Kim D, Choi SY, Park J. Crosstalk between HDAC6 and Nox2-based NADPH oxidase mediates HIV-1 Tat-induced pro-inflammatory responses in astrocytes. Redox Biol (2017) 12:978–86. doi: 10.1016/j.redox.2017.05.001
62. Wenzel ED, Speidell A, Flowers SA, Wu CB, Avdoshina V, Mocchetti I. Histone deacetylase 6 inhibition rescues axonal transport impairments and prevents the neurotoxicity of HIV-1 envelope protein gp120. Cell Death Dis (2019) 10:674. doi: 10.1038/s41419-019-1920-7
63. Valenzuela-Fernandez A, Alvarez S, Gordon-Alonso M, Barrero M, Ursa A, Cabrero R. Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol Biol Cell (2005) 16(11):5445–54. doi: 10.1091/mbc.e05-04-0354
64. Marrero-Hernandez S, Marquez-Arce D, Cabrera-Rodriguez R, Estévez-Herrera J, Pérez-Yanes S, Barroso-González J, et al. HIV-1 nef targets HDAC6 to assure viral production and virus infection. Front Microbiol (2019) 10:2437. doi: 10.3389/fmicb.2019.02437
65. Cabrera-Rodriguez R, Perez-Yanes S, Montelongo R, Lorenzo-Salazar JM, Estévez-Herrera J, García-Luis J, et al. Transactive response DNA-binding protein (TARDBP/TDP-43) regulates cell permissivity to HIV-1 infection by acting on HDAC6. Int J Mol Sci (2022) 23(11):6180. doi: 10.3390/ijms23116180
66. Zhang S, Jiang Y, Cheng Q, Zhong Y, Qin YL, Chen MZ. Inclusion body fusion of human parainfluenza virus type 3 regulated by acetylated alpha-tubulin enhances viral replication. J Virol (2017) 91(3):e01802-16. doi: 10.1128/JVI.01802-16
67. Nakashima H, Kaufmann JK, Wang PY, Nguyen T, Speranza MC, Kasai K, et al. Histone deacetylase 6 inhibition enhances oncolytic viral replication in glioma. J Clin Invest (2015) 125(11):4269–80. doi: 10.1172/jci80713
68. Wang Z, Chen J, Wu X, Ma D, Zhang XH, Li RZ, et al. PCV2 targets cGAS to inhibit type I interferon induction to promote other DNA virus infection. PloS Pathog (2021) 17(9):e1009940. doi: 10.1371/journal.ppat.1009940
69. Chattopadhyay S, Fensterl V, Zhang Y, Veleeparambil S, Wetzel JL, Sen GC. Inhibition of viral pathogenesis and promotion of the septic shock response to bacterial infection by IRF-3 are regulated by the acetylation and phosphorylation of its coactivators. Mbio (2013) 4(2):e00636-12. doi: 10.1128/mBio.00636-12
70. Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci (2011) 14(4):459–68. doi: 10.1038/nn.2779
71. Brown AL, Wilkins OG, Keuss MJ, Hill SE, Zanovello M, Lee WC, et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature (2022) 603(7899):131–7. doi: 10.1038/s41586-022-04436-3
72. Perdiz D, Mackeh R, Pous C, Baillet A. The ins and outs of tubulin acetylation: more than just a post-translational modification? Cell Signal (2011) 23(5):763–71. doi: 10.1016/j.cellsig.2010.10.014
73. Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J (2002) 21(24):6820–31. doi: 10.1093/emboj/cdf682
74. Zan J, Liu S, Sun DN, Mo KK, Yan Y, Liu J, et al. Rabies virus infection induces microtubule depolymerization to facilitate viral RNA synthesis by upregulating HDAC6. Front Cell Infect Microbiol (2017) 7:146. doi: 10.3389/fcimb.2017.00146
75. Kawano T, Inokuchi J, Eto M, Murata M, Kang JH. Protein kinase C (PKC) isozymes as diagnostic and prognostic biomarkers and therapeutic targets for cancer. Cancers (Basel) (2022) 14(21):5425. doi: 10.3390/cancers14215425
76. Ghashghaeinia M, Dreischer P, Wieder T, Köberlec M. Coronavirus disease 2019 (COVID-19), human erythrocytes and the PKC-alpha/-beta inhibitor chelerythrine -possible therapeutic implication. Cell Cycle (2020) 19(24):3399–405. doi: 10.1080/15384101.2020.1859197
77. Hornikova L, Brustikova K, Ryabchenko B, Zhernov L, Fraiberk M, Mariničová Z, et al. The major capsid protein, VP1, of the mouse polyomavirus stimulates the activity of Tubulin Acetyltransferase 1 by microtubule stabilization. Viruses (2020) 12(2):227. doi: 10.3390/v12020227
78. Zheng Y, Zhu G, Tang Y, Yan J, Han S, Yin J, et al. HDAC6, A novel cargo for autophagic clearance of stress granules, mediates the repression of the type I interferon response during Coxsackievirus A16 infection. Front Microbiol (2020) 11:78. doi: 10.3389/fmicb.2020.00078
79. Hornikova L, Brustikova K, Forstova. J. Microtubules in polyomavirus infection. Viruses (2020) 12(1):121. doi: 10.3390/v12010121
80. Requena S, Sanchez-Madrid F, Martin-Cofreces. NB. Post-translational modifications and stabilization of microtubules regulate transport of viral factors during infections. Biochem Soc Trans (2021) 49(4):1735–48. doi: 10.1042/bst20210017
81. Zhang J, Sprung R, Pei J, Tan XH, Kim S, Zhu H. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics (2009) 8(2):215–25. doi: 10.1074/mcp.M800187-MCP200
82. Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Curr Opin Genet Dev (2001) 11(2):155–61. doi: 10.1016/s0959-437x(00)00173-8
83. Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp Cell Res (2001) 262(2):75–83. doi: 10.1006/excr.2000.5080
84. Suh HS, Choi S, Khattar P, Choi N, Lee SC. Histone deacetylase inhibitors suppress the expression of inflammatory and innate immune response genes in human microglia and astrocytes. J Neuroimmune Pharmacol (2010) 5(4):521–32. doi: 10.1007/s11481-010-9192-0
85. Jaqaman K, Grinstein S. Regulation from within: the cytoskeleton in transmembrane signaling. Trends Cell Biol (2012) 22(10):515–26. doi: 10.1016/j.tcb.2012.07.006
86. Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol (2011) 12(12):773–86. doi: 10.1038/nrm3227
87. Rottner K, Faix J, Bogdan S, Linder S, Kerkhoff S. Actin assembly mechanisms at a glance. J Cell Sci (2017) 130(20):3427–35. doi: 10.1242/jcs.206433
88. Ploubidou A, Way M. Viral transport and the cytoskeleton. Curr Opin Cell Biol (2001) 13(1):97–105. doi: 10.1016/s0955-0674(00)00180-0
89. Ward BM. The taking of the cytoskeleton one two three: how viruses utilize the cytoskeleton during egress. Virology (2011) 411(2):244–50. doi: 10.1016/j.virol.2010.12.024
90. de Zoeten EF, Wang L, Butler K, Beier UH, Akimova T, Sai H, et al. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol Cell Biol (2011) 31(10):2066–78. doi: 10.1128/MCB.05155-11
91. Long JS, Mistry B, Haslam SM, Barclay WS. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol (2019) 17(2):67–81. doi: 10.1038/s41579-018-0115-z
92. Chen H, Yuan H, Gao R, Zhang JX, Wang DY, Xiong Y, et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet (2014) 383(9918):714–21. doi: 10.1016/S0140-6736(14)60111-2
93. Wei SH, Yang JR, Wu HS, Chang MC, Lin JS, Lin CY, et al. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Respir Med (2013) 1(10):771–8. doi: 10.1016/S2213-2600(13)70221-2
94. Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med (2013) 368(20):1888–97. doi: 10.1056/NEJMoa1304459
95. Um S, Siegers JY, Sar B, Chin S, Patel S, Bunnary S, et al. Human infection with avian influenza A(H9N2) virus, Cambodia, February 2021. Emerg Infect Dis (2021) 27(10):2742–5. doi: 10.3201/eid2710.211039
96. Parys A, Vandoorn E, King J, Graaf A, Pohlmann A, Beer M, et al. Human infection with eurasian avian-like swine influenza A(H1N1) virus, the Netherlands, September 2019. Emerg Infect Dis (2021) 27(3):939–43. doi: 10.3201/eid2703.201863
97. Li J, Fang Y, Qiu X, Yu X, Cheng S, li N, et al. Human infection with avian-origin H5N6 influenza a virus after exposure to slaughtered poultry. Emerg Microbes Infect (2022) 11(1):807–10. doi: 10.1080/22221751.2022.2048971
98. Wang D, Huo H, Werid GM, Werid GM, Ibrahim YM, Tang L, et al. TBK1 mediates innate antiviral immune response against duck enteritis virus. Viruses (2022) 14(5):1008. doi: 10.3390/v14051008
99. Tang JL, Yang Q, Xu CH, Zhao H, Liu CY, Zhou Y, et al. Histone deacetylase 3 promotes innate antiviral immunity through deacetylation of TBK1. Protein Cell (2021) 12(4):261–78. doi: 10.1007/s13238-020-00751-5
100. Louis C, Burns C, Wicks. I. TANK-binding kinase 1-dependent responses in health and autoimmunity. Front Immunol (2018) 9:434. doi: 10.3389/fimmu.2018.00434
101. Zhao W. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett (2013) 587(6):542–8. doi: 10.1016/j.febslet.2013.01.052
102. Shah N, Kumar S, Zaman N, Pan CC, Bloodworth JC, Lei W, et al. TAK1 activation of alpha-TAT1 and microtubule hyperacetylation control AKT signaling and cell growth. Nat Commun (2018) 9(1):1696. doi: 10.1038/s41467-018-04121-y
103. Giustiniani J, Daire V, Cantaloube I, Durand G, Poüs C, Perdiz D, et al. Tubulin acetylation favors Hsp90 recruitment to microtubules and stimulates the signaling function of the Hsp90 clients Akt/PKB and p53. Cell Signal (2009) 21(4):529–39. doi: 10.1016/j.cellsig.2008.12.004
104. Ha SD, Ng D, Pelech SL, Kim SO. Critical role of the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase-3 signaling pathway in recovery from anthrax lethal toxin-induced cell cycle arrest and MEK cleavage in macrophages. J Biol Chem (2007) 282(50):36230–9. doi: 10.1074/jbc.M707622200
105. Martin M, Schifferle RE, Cuesta N, Vogel SN, Katz J, Michalek SM, et al. Role of the phosphatidylinositol 3 kinase-Akt pathway in the regulation of IL-10 and IL-12 by Porphyromonas gingivalis lipopolysaccharide. J Immunol (2003) 171(2):717–25. doi: 10.4049/jimmunol.171.2.717
106. Lei CQ, Zhong B, Zhang Y, Zhang J, Wang S, Shu HB. Glycogen synthase kinase 3beta regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity (2010) 33(6):878–89. doi: 10.1016/j.immuni.2010.11.021
107. Namekata K, Harada C, Guo X, Kimura A, Kittaka D, Watanabe H, et al. Dock3 stimulates axonal outgrowth via GSK-3beta-mediated microtubule assembly. J Neurosci (2012) 32(1):264–74. doi: 10.1523/JNEUROSCI.4884-11.2012
108. Zhou FQ, Snider WD. Cell biology. GSK-3beta and microtubule assembly in axons. Science (2005) 308(5719):211–4. doi: 10.1126/science.1110301
109. Drechsler H, Tan AN, Liakopoulos. D. Yeast GSK-3 kinase regulates astral microtubule function through phosphorylation of the microtubule-stabilizing kinesin Kip2. J Cell Sci (2015) 128(21):3910–21. doi: 10.1242/jcs.166686
110. Choi SJ, Lee HC, Kim JH, Park SY, Kim TH, Lee WK, et al. HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I. EMBO J (2016) 35(4):429–42. doi: 10.15252/embj.201592586
111. Zanin M, DeBeauchamp J, Vangala G, Webby RJ, Husain M. Histone deacetylase 6 knockout mice exhibit higher susceptibility to influenza A virus infection. Viruses-Basel (2020) 12(7):728. doi: 10.3390/v12070728
112. Wang D, Meng Q, Huo L, Yang M, Wang L, Chen X, et al. Overexpression of Hdac6 enhances resistance to virus infection in embryonic stem cells and in mice. Protein Cell (2015) 6(2):152–6. doi: 10.1007/s13238-014-0120-6
113. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol (2004) 5(7):730–7. doi: 10.1038/ni1087
114. Akira S, Uematsu S, Takeuchi. O. Pathogen recognition and innate immunity. Cell (2006) 124(4):783–801. doi: 10.1016/j.cell.2006.02.015
115. Rehwinkel J, Tan CP, Goubau D, Schulz L, Pichlmair A, Bier K, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell (2010) 140(3):397–408. doi: 10.1016/j.cell.2010.01.020
116. Nikonov A, Molder T, Sikut R, Kiiver K, Männik A, Toots U, et al. RIG-I and MDA-5 detection of viral RNA-dependent RNA polymerase activity restricts positive-strand RNA virus replication. PloS Pathog (2013) 9(9):e1003610. doi: 10.1371/journal.ppat.1003610
117. Jorgensen SE, Christiansen M, Ryo LB, Gad HH, Gjedsted J, Staeheli P, et al. Defective RNA sensing by RIG-I in severe influenza virus infection. Clin Exp Immunol (2018) 192(3):366–76. doi: 10.1111/cei.13120
118. Bruns AM, Horvath CM. Antiviral RNA recognition and assembly by RLR family innate immune sensors. Cytokine Growth Factor Rev (2014) 25(5):507–12. doi: 10.1016/j.cytogfr.2014.07.006
119. Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature (2006) 441(7089):101–5. doi: 10.1038/nature04734
120. Hausmann S, Marq JB, Tapparel C, Kolakofsky D, Garcin D. RIG-I and dsRNA-induced IFNbeta activation. PloS One (2008) 3(12):e3965. doi: 10.1371/journal.pone.0003965
121. Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol (2008) 20(1):17–22. doi: 10.1016/j.coi.2008.01.002
122. Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity (2007) 27(3):370–83. doi: 10.1016/j.immuni.2007.08.012
123. Tan X, Sun L, Chen J, Chen ZJ. Detection of microbial infections through innate immune sensing of nucleic acids. Annu Rev Microbiol (2018) 72:447–78. doi: 10.1146/annurev-micro-102215-095605
124. Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol (2006) 8(6):907–22. doi: 10.1111/j.1462-5822.2006.00716.x
125. McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol (2015) 15(2):87–103. doi: 10.1038/nri3787
126. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature (2008) 455(7213):674–8. doi: 10.1038/nature07317
127. Dixit E, Boulant S, Zhang Y, Lee A S.Y., Odendall C, Shum B, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell (2010) 141(4):668–81. doi: 10.1016/j.cell.2010.04.018
128. Horner SM, Liu HM, Park HS, Briley J, Gale M Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U.S.A. (2011) 108(35):14590–5. doi: 10.1073/pnas.1110133108
129. Loo YM, Gale C.OMMAJr. M. Immune signaling by RIG-I-like receptors. Immunity (2011) 34(5):680–92. doi: 10.1016/j.immuni.2011.05.003
130. Brennan K, Bowie AG. Activation of host pattern recognition receptors by viruses. Curr Opin Microbiol (2010) 13(4):503–7. doi: 10.1016/j.mib.2010.05.007
131. Arpaia N, Barton GM. Toll-like receptors: key players in antiviral immunity. Curr Opin Virol (2011) 1(6):447–54. doi: 10.1016/j.coviro.2011.10.006
132. Into T, Inomata M, Niida S, Murakami Y, Shibata KI. Regulation of MyD88 aggregation and the MyD88-dependent signaling pathway by sequestosome 1 and histone deacetylase 6. J Biol Chem (2010) 285(46):35759–69. doi: 10.1074/jbc.M110.126904
133. Moreno-Gonzalo O, Ramirez-Huesca M, Blas-Rus N, Cibrián D, Saiz ML, Jorge I, et al. HDAC6 controls innate immune and autophagy responses to TLR-mediated signalling by the intracellular bacteria Listeria monocytogenes. PloS Pathog (2017) 13(12):e1006799. doi: 10.1371/journal.ppat.1006799
134. Tischer HGI, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature (1982) 295:64–6. doi: 10.1038/295064a0
135. Misinzo G, Delputte PL, Meerts P, Lefebvre DJ, Nauwynck HJ. Porcine circovirus 2 uses heparan sulfate and chondroitin sulfate B glycosaminoglycans as receptors for its attachment to host cells. J Virol (2006) 80(7):3487–94. doi: 10.1128/JVI.80.7.3487-3494.2006
136. Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, et al. TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor p62 to Promote Innate Immune Responses. Mol Cell (2016) 64(1):105–19. doi: 10.1016/j.molcel.2016.08.025
137. Nakashima H, Nguyen T, Goins WF, Chiocca EA. Interferon-stimulated gene 15 (ISG15) and ISG15-linked proteins can associate with members of the selective autophagic process, histone deacetylase 6 (HDAC6) and SQSTM1/p62. J Biol Chem (2015) 290(3):1485–95. doi: 10.1074/jbc.M114.593871
138. Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol (2016) 26(9):668–79. doi: 10.1016/j.tcb.2016.05.004
139. Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J (2010) 29(5):969–80. doi: 10.1038/emboj.2009.405
140. Simoes-Pires C, Zwick V, Nurisso A, Schenker E, Carrupt PA, Cuendet M. HDAC6 as a target for neurodegenerative diseases: what makes it different from the other HDACs? Mol Neurodegener (2013) 8:7. doi: 10.1186/1750-1326-8-7
141. Li T, Zhang C, Hassan S, Liu X, Song F, Chen K, et al. Histone deacetylase 6 in cancer. J Hematol Oncol (2018) 11(1):111. doi: 10.1186/s13045-018-0654-9
142. Ding G, Liu HD, Huang Q, Liang HX, Ding ZH, Liao ZJ, et al. HDAC6 promotes hepatocellular carcinoma progression by inhibiting P53 transcriptional activity. FEBS Lett (2013) 587(7):880–6. doi: 10.1016/j.febslet.2013.02.001
143. Lebosse F, Zoulim F. [Hepatitis B vaccine and liver cancer]. Bull Cancer (2021) 108(1):90–101. doi: 10.1016/j.bulcan.2020.10.014
144. Chen JG, Zhang SW. Liver cancer epidemic in China: past, present and future. Semin Cancer Biol (2011) 21(1):59–69. doi: 10.1016/j.semcancer.2010.11.002
145. Zhang D, Wang Y, Zhang HY, Jiao FZ, Zhang WB, Wang LW, et al. Histone deacetylases and acetylated histone H3 are involved in the process of hepatitis B virus DNA replication. Life Sci (2019) 223:1–8. doi: 10.1016/j.lfs.2019.03.010
146. Iwamoto M, Saso W, Sugiyama R, Ishii K, Ohki M, Nagamori S, et al. Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. Proc Natl Acad Sci U.S.A. (2019) 116(17):8487–92. doi: 10.1073/pnas.1811064116
147. Gao YS, Hubbert CC, Yao TP. The microtubule-associated histone deacetylase 6 (HDAC6) regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation. J Biol Chem (2010) 285(15):11219–26. doi: 10.1074/jbc.M109.042754
148. Shao Y, Zhu F, Zhu S, Bai L. HDAC6 suppresses microRNA-199a transcription and augments HPV-positive cervical cancer progression through Wnt5a upregulation. Int J Biochem Cell Biol (2021) 136:106000. doi: 10.1016/j.biocel.2021.106000
149. Liu Y, Li Y, Liu S, Adeegbe DO, Christensen CL, Quinn MM, et al. NK cells mediate synergistic antitumor effects of combined inhibition of HDAC6 and BET in a SCLC preclinical model. Cancer Res (2018) 78(13):3709–17. doi: 10.1158/0008-5472.Can-18-0161
150. Diacovich L, Gorvel JP. Bacterial manipulation of innate immunity to promote infection. Nat Rev Microbiol (2010) 8(2):117–28. doi: 10.1038/nrmicro2295
151. Radtke AL, O’Riordan MX. Intracellular innate resistance to bacterial pathogens. Cell Microbiol (2006) 8(11):1720–9. doi: 10.1111/j.1462-5822.2006.00795.x
152. Weiss G, Schaible UE. Macrophage defense mechanisms against intracellular bacteria. Immunol Rev (2015) 264(1):182–203. doi: 10.1111/imr.12266
153. Vancurova I, Uddin MM, Zou Y, Vancura A. Combination therapies targeting HDAC and IKK in solid tumors. Trends Pharmacol Sci (2018) 39(3):295–306. doi: 10.1016/j.tips.2017.11.008
154. Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol (2010) 189(4):671–9. doi: 10.1083/jcb.201001039
155. Zhang F, Su B, Wang C, Siedlak SL, Mondragon-Rodriguez S, Lee HG, et al. Posttranslational modifications of alpha-tubulin in alzheimer disease. Transl Neurodegener (2015) 4:9. doi: 10.1186/s40035-015-0030-4
156. Dompierre JP, Godin JD, Charrin BC, Cordelières FP, King SJ, Humbert S, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci (2007) 27(13):3571–83. doi: 10.1523/JNEUROSCI.0037-07.2007
157. Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta BioMed (2020) 91(1):157–60. doi: 10.23750/abm.v91i1.9397
158. Zhang FZ, Yu SS, Chai QY, Wang J, Wu T, Liu R, et al. HDAC6 contributes to human resistance against Mycobacterium tuberculosis infection via mediating innate immune responses. FASEB J (2021) 35(11):e22009. doi: 10.1096/fj.202100614R
159. Kwon Y, Choi Y, Kim M, Jeong MS, Jung H, Jeoung D. HDAC6 and CXCL13 mediate atopic dermatitis by regulating cellular interactions and expression levels of miR-9 and SIRT1. Front Pharmacol (2021) 12:691279. doi: 10.3389/fphar.2021.691279
160. Hwang I, Lee E, Jeon SA, Yu JW. Histone deacetylase 6 negatively regulates NLRP3 inflammasome activation. Biochem Biophys Res Commun (2015) 467(4):973–8. doi: 10.1016/j.bbrc.2015.10.033
161. Yan B, Xie S, Liu Y, Liu W, Li D, Luo HR, et al. Histone deacetylase 6 modulates macrophage infiltration during inflammation. Theranostics (2018) 8(11):2927–38. doi: 10.7150/thno.25317
162. Yan B, Liu Y, Bai H, Chen M, Xie SB, Li D, et al. HDAC6 regulates IL-17 expression in T lymphocytes: implications for HDAC6-targeted therapies. Theranostics (2017) 7(4):1002–9. doi: 10.7150/thno.17615
163. Cao ZX, Gu ZC, Lin SX, Chen D, Wang J, Zhao Y, et al. Attenuation of NLRP3 inflammasome activation by indirubin-derived PROTAC targeting HDAC6. ACS Chem Biol (2021) 16(12):2746–51. doi: 10.1021/acschembio.1c00681
164. Lu CY, Chang YC, Hua CH, Chuang C, Huang SH, Kung SH, et al. Tubacin, an HDAC6 selective inhibitor, reduces the replication of the Japanese encephalitis virus via the decrease of viral RNA synthesis. Int J Mol Sci (2017) 18(5):954. doi: 10.3390/ijms18050954
165. Kawada J, Zou P, Mazitschek R, Bradner JE, Cohen JI. Tubacin kills Epstein-Barr virus (EBV)-Burkitt lymphoma cells by inducing reactive oxygen species and EBV lymphoblastoid cells by inducing apoptosis. J Biol Chem (2009) 284(25):17102–9. doi: 10.1074/jbc.M809090200
166. Chao OS, Chang TC, Di Bella MA, Alessandro R, Anzanello F, Rappa G, et al. The HDAC6 inhibitor tubacin induces release of CD133(+) extracellular vesicles from cancer cells. J Cell Biochem (2017) 118(12):4414–24. doi: 10.1002/jcb.26095
167. Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci U.S.A. (2003) 100(8):4389–94. doi: 10.1073/pnas.0430973100
168. Saha B, Parks RJ. Histone deacetylase inhibitor suberoylanilide hydroxamic acid suppresses human adenovirus gene expression and replication. J Virol (2019) 93(12):e00088-19. doi: 10.1128/JVI.00088-19
169. Lucera MB, Tilton CA, Mao H, Dobrowolski C, Tabler CO, Haqqani AA, et al. The histone deacetylase inhibitor vorinostat (SAHA) increases the susceptibility of uninfected CD4+ T cells to HIV by increasing the kinetics and efficiency of postentry viral events. J Virol (2014) 88(18):10803–12. doi: 10.1128/JVI.00320-14
170. Zhou R, Fang S, Zhang M, Zhang Q, Hu J, Wang M, et al. Design, synthesis, and bioactivity evaluation of novel Bcl-2/HDAC dual-target inhibitors for the treatment of multiple myeloma. Bioorg Med Chem Lett (2019) 29(3):349–52. doi: 10.1016/j.bmcl.2018.12.052
171. Zhang L, Zhang Z, Li C, Zhu T, Gao J, Zhou H, et al. S100A11 promotes liver steatosis via FOXO1-mediated autophagy and lipogenesis. Cell Mol Gastroenterol Hepatol (2021) 11(3):697–724. doi: 10.1016/j.jcmgh.2020.10.006
Keywords: HDAC6, viral infection, innate immunity, autophagy, diseases
Citation: Qu M, Zhang H, Cheng P, Wubshet AK, Yin X, Wang X and Sun Y (2023) Histone deacetylase 6’s function in viral infection, innate immunity, and disease: latest advances. Front. Immunol. 14:1216548. doi: 10.3389/fimmu.2023.1216548
Received: 04 May 2023; Accepted: 14 July 2023;
Published: 11 August 2023.
Edited by:
Fuminori Tokunaga, Osaka Metropolitan University, JapanReviewed by:
Qingjian Ou, Tongji University, ChinaMairaj Ahmed Ansari, Jamia Hamdard University, India
Copyright © 2023 Qu, Zhang, Cheng, Wubshet, Yin, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuefeng Sun, c3VueXVlZmVuZ0BjYWFzLmNu; Xiangwei Wang, d2FuZ3hpYW5nd2VpQGNhYXMuY24=
 Min Qu
Min Qu Huijun Zhang1
Huijun Zhang1 Ashenafi Kiros Wubshet
Ashenafi Kiros Wubshet Xiangping Yin
Xiangping Yin Xiangwei Wang
Xiangwei Wang Yuefeng Sun
Yuefeng Sun