- Institute of Clinical Anatomy and Cell Analysis, University of Tübingen, Tübingen, Germany
Introduction: Wnt-signaling is a key regulator of stem cell homeostasis, extensively studied in the intestinal crypt and other metazoan tissues. Yet, there is hardly any data available on the presence of Wnt-signaling components in the adult enteric nervous system (ENS) in vivo.
Methods: Therefore, we employed RNAscope HiPlex-assay, a novel and more sensitive in situ hybridization technology. By amplifying target specific signals, this technique enables the detection of low abundance, tightly regulated RNA content as is the case for Wnt-signaling components. Additionally, we compared our data to previously published physiological single cell RNA and RiboTag-based RNA sequencing analyses of enteric gliosis using data-mining approaches.
Results: Our descriptive analysis shows that several components of the multidi-mensional regulatory network of the Wnt-signaling pathway are present in the murine ENS. The transport and secretion protein for Wnt-ligands Wntless as well as canonical (Wnt3a and Wnt2b) and non-canonical Wnt-ligands (Wnt5a, Wnt7a, Wnt8b and Wnt11) are detectable within submucosal and myenteric plexus. Further, corresponding Frizzled receptors (Fzd1, Fzd3, Fzd6, and Fzd7) and regulatory signaling mediators like R-Spondin/DKK ligands are present in the ENS of the small and large intestine. Further, data mining approaches revealed, that several Wnt-related molecules are expressed by enteric glial cell clusters and are dynamically regulated during the inflammatory manifestation of enteric gliosis.
Discussion: Our results suggest, that canonical and non-canonical Wnt-signaling has a much broader impact on the mature ENS and its cellular homeostasis in health and inflammation, than previously anticipated.
Introduction
The enteric nervous system (ENS) represents the intrinsic innervation of the gut and arises from migrating vagal neural crest cells originating from the neural tube. These precursor cells proliferate and differentiate into multiple subtypes of enteric neurons and glial cells. In the adult ENS of mammals, enteric neurons and glia are organized in two predominant networks of interconnected plexus (submucosal and myenteric), that are wired into microcircuits enabling gastrointestinal function independent of central nervous system control (1, 2). However, the ENS is still immature at birth and underlies considerable refinement including changes in its neurochemical coding as well as synaptic wiring to exert its proper function (3). Particularly the submucosal plexus develops late during embryogenesis and groups of neurons forming ganglia are not found before birth in the murine colon (4). In addition, a notable number of submucosal and myenteric progenitors undergo enteric neurogenesis within the first postnatal days in rodents (5, 6).
Unlike the early postnatal days, enteric neurogenesis in the adult ENS is controversially discussed and arguably a rare event in the submucosal and myenteric plexus in vivo (6–8). Although many in vitro studies (9–11) demonstrated a regenerative potential of postnatal-derived ENS-progenitor cells of various rodent species and human patients, only models of chemical denervation (12) and pathological settings like colitis (13) resulted in remodeling processes of the ENS in vivo. Still, the ever-changing environment of the gut, like constant physical stimuli by different motility patterns (14, 15), microbiome (16) or immunological interactions (17) suggest that cellular ENS-homeostasis might be continuously regulated by cell-cell-communication systems to maintain the steady state of the adult ENS.
In this context, enteric glial cells have gained increasing attention over the past years, as regulatory interaction partners in the reciprocal communication with enteric neurons (18), immune cells (19) and epithelial stem cells (20). Moreover, glial cell communication pathways involved in phenotypic plasticity induced by gastrointestinal disorders or systemic diseases are mostly unknown (21, 22).
During embryogenesis, Wnt-signaling induces and specifies neural crest formation and is involved in regulating enteric neural cell migration, guidance, and growth of enteric neuronal projections in the developing gut (23, 24). Further, Wnt-signaling is a key regulator of epithelial adult stem cell niches in different metazoan tissues including the gut epithelium (25), and the localization of Wnt-signaling components within the mucosa is well characterized (26). Intriguingly, a recent study by Baghdadi et al. highlighted the presence of enteric glial cells in the Lamina propria mucosae, which express several Wnt-signaling ligands to promote the self-renewal capacity of intestinal epithelial stem cells and thereby influence epithelial regeneration and barrier function in mice (20). Nevertheless, the presence of Wnt-signaling components in large parts of the adult ENS – especially in the submucosal and myenteric plexus in vivo has been largely neglected so far, and the functional evaluation is restricted to a handful of studies (27–29). Interestingly, the myenteric plexus is integrated into a network of myogenic cells, that are known to actively secrete Wnt-signaling molecules (30), conceivably shaping the cellular homeostasis and cytoarchitecture of the adult ganglionated plexus. Intriguingly, recently published transcriptome profiling data of the widespread used in vitro model system of purified ENS-progenitors, suggested, that mesenchymal cells indeed produced ligands belonging to the Wnt-signaling pathway (31). In particular, some of these ligands and corresponding receptors have roles in proliferation, differentiation, or maintenance of neural stem and progenitor cells (32, 33). Furthermore, regulatory Wnt-signaling co-activators like R-Spondins (34)- and Wnt-antagonists like Dickkopf (35)-ligands share heparin-binding properties, that enable interaction with the extracellular matrix component heparin sulfate proteoglycans. Since secretory heparin sulfate proteoglycans, such as collagen 18a and agrin, were shown to have an influence on proper ENS development in chicken (36), it is plausible that Wnt-components also interact with the extracellular matrix, assuming a profound influence on ENS-homeostasis.
To clarify the presence of Wnt-signaling components in vivo, we mapped the expression profile of these molecules in the adult murine small and large intestine. We take the advantage of a novel, sensitive in situ hybridization technology, commercially available as RNAScope HiPlex-assay. By amplify target-specific signals, this technique enables the detection of low abundance, tightly regulated RNA content as it is the case for the Wnt-signaling pathway. Further, to refine the cell type specific localization and contextualise our findings, we compared our RNAscope-based expression atlas to a recently published single cell RNA sequencing dataset of the juvenile (20 days of age) mouse ENS (37). Moreover, to assess the relevance of Wnt-signaling under pathophysiological conditions, we re-evaluated a RiboTag-based bulk RNA sequencing dataset of myenteric glial cells in acute inflammation (22) especially focusing on Wnt-signaling related targets. Thus, in this study, we extended the existing knowledge about the presence of Wnt-signaling components in the gastrointestinal tract by specifically focusing on the enteric nervous system. Our study suggests, that Wnt-signaling has a much broader impact on the adult ENS homeostasis in health and inflammation, than anticipated previously.
Materials and methods
Animals
Animals were handled and kept in accordance with the guidelines, regulating the handling of animals for scientific purposes (TierVersV, Notification number AT 01/19 M), which conform to international guidelines. C57BL/6J were housed in standard cages with standard pathogen-free breeding and a standard 12-hour light/dark cycle at 22 ± 2°C and 60 ± 5% humidity. Germ-free food and water were available ad libitum. For in situ hybridization experiments, small and large intestine samples from male C57BL/6J mice (postnatal day 60) were used.
In situ hybridization: fixed frozen tissue sample preparation, pre-treatment, RNAscope® HiPlex Assay and detection
We applied the RNAscope® HiPlex Assay and HiPlex Assay v2 (ACB biotechne, Wiesbaden-Nordenstadt Germany) according to the manufacturer’s description to detect expression of relevant Wnt-ligands and receptor mRNAs in small and large intestine samples.
Sample preparation
The intestine bundle was removed from the abdomen, transferred to and rapidly dissected in 4% (w/v) phosphate buffered p-formaldehyde (Merck KGaA, Darmstadt, Germany). Adherent mesenteria were dissected to unfold intestine. Afterwards, small, and large intestine were separated and for better handling cut into 2 cm long samples.
Fixed frozen tissue sample preparation and pre-treatment
Before embedding, prepared tissue samples were fixed with 4% (w/v) phosphate buffered p-formaldehyde (Merck KGaA, Darmstadt, Germany) at 4°C for one hour and rinsed 3 times with RNAse-free phosphate-buffered saline (PBS). Afterwards, fixed samples were stored overnight at 4°C in 30% (w/v) sucrose solution (AppliChem, Darmstadt, Germany). Next, samples were frozen in isopentane-nitrogen cooled TissueTek® (Sakura, Staufen, Germany) in such a way that, three transversal small and large intestine samples were on one section. Blocks were stored at -80°C until further processing. Prepared cryosections of 12 µm thickness were washed with PBS and dried for 30 minutes at 60°C, following post-fixation with 4% (w/v) phosphate buffered p-formaldehyde for 15 minutes at 4°C. Then, sections were dehydrated in an ascending alcohol series: each step for 5 minutes in 50% (v/v), 70% (v/v) and twice in 100% (v/v) EtOH at room temperature. Target retrieval was performed in mild-boiling (98 – 102°C) 1x RNAscope® Target Retrieval Reagent solution for 5 minutes, followed by one washing step in distilled water and one in 100% (v/v) EtOH. Sections were dried completely at 60°C for 5 minutes, before the hydrophobic barrier was drawn. One the next day, RNAscope® Protease III treatment was applied for 5 minutes at 40°C. Before continuing with the RNAscope® HiPlex Assay, sections were washed once with distilled water.
RNAscope® HiPlex assay
Prior probe hybridization, pre-warmed RNAscope® HiPlex 50x probe stocks were diluted with RNAscope® HiPlex Diluent according to the manufacturer’s description and cooled to room temperature prior to use. Probe hybridization was carried out at 40°C for 2 hours. Afterwards, sections were washed twice for two minutes at room temperature with 1x RNAscope® Wash Buffer. Next, three amplification rounds were carried out, while sections were treated successively first with the RNAscope® HiPlex Amp1, second the RNAscope® HiPlex Amp2 and third with the RNAscope® HiPlex Amp3 for 30 minutes at 40°C. After each amplification round, sections were washed twice with 1x RNAscope® Wash Buffer for 2 minutes at room temperature.
For detection, sections were treated 15 minutes with the corresponding RNAscope® HiPlex Fluor Solution at 40°C, washed twice with 1x RNAscope® Wash Buffer for 2 minutes at room temperature, counterstained with RNAscope® DAPI for 1 minute and mounted with ProLong Gold Antifade Mountant (Thermo Fisher Scientific, MA, USA). For each experiment a positive and a negative control was performed to evaluate signal strength as well as background staining. Target probe signal was evaluated according to the manufacturer’s description. Commercially designed, generated, and evaluated target probes and their corresponding probe channel used in this study are listed in Tables 1, 2.
Microscopy
Images were acquired using a Zeiss Axio Imager.Z1 fluorescence microscope with Apotome module with 358, 488, 543, 647 nm for excitation and appropriate filter sets. Images were acquired using ZEN software. For in situ hybridization exposure time for DAPI was 150 ms and for the RNAscope HiPlex Fluoro solutions 5000 ms with the 63-objective (Plan-Apochromat 63x/1.40 Oil DIC M27). RNAscope demonstrated a clear punctate staining, whereby each RNA dot derives from a single mRNA molecule.
Hybridization probes targeting housekeeping genes, including GAPDH (Glycerin-aldehyd-3-phosphat-Dehydrogenase), HPRT (Hypoxanthine phosphoribosyl trans-ferase 1) and UBC (Polyubiquitin-C), were used as positive controls for the hybridization procedure in each independent preparation. Samples treated with dapB probes were used as negative controls to assess background signals (Supplementary Figures 1A, B for negative control).
We applied a semiquantitative evaluation, whereby less than five dots correspond to no expression. Low expression is marked by single mRNA-transcripts with less signal intensity, that are clearly recognizable from the tissue background. mRNA transcripts displaying a strong signal intensity, still with clearly distinguishable single dots were defined as medium expression. High expression is characterized by a high density of no separable single dots. The distal jejunum and ileum of the small intestine as well as the proximal half of the large intestine was included in our study, whereby we evaluated the following regions: epithelium (w/o crypts), epithelium (crypts), Tunica muscularis, as well as submucosal and myenteric ganglia. In the small intestine the category epithelium (w/o crypts) covers the area from the tip of the villus surface down to the crypt-villus junction. In the large intestine the epithelium (w/o crypts) category is defined by the complete epithelial surface excluding the crypts. The category epithelium (crypts) includes the entire crypts from the crypt-villus junction down to the crypt bottom in both small and large intestine. If applicable more precise localization was described in the text.
Results
In this study we employed the RNAscope HiPlex-assay, a novel and more sensitive in situ hybridization technology to investigate components of the Wnt signaling network particularly focusing on the ENS of adult P60 mice. This method uses a special amplification procedure that enables the detection of single mRNA transcripts, making it especially useful for the evaluation of low abundance transcripts of tightly regulated signaling cascades. Further, we compared our data to previously published physiological single cell RNA (37) and RiboTag-based RNA (22) sequencing analysis of acute inflammation using data-mining approaches.
In vivo expression of Wnt-ligands in mouse intestine
Overall, we found a broad expression of Wnt-ligand mRNAs in enteric ganglia, with a majority of probes generating clearly detectable signals in the small (16 out of 19) and large intestine (13 out of 19). Additionally, we detected the expression of the multi-pass transmembrane protein Wntless/Evi, which is essential for intracellular Wnt-shuttling and Wnt-secretion of lipidated Wnt-ligands (38, 39) in the submucosal and myenteric plexus of the small intestine (Figure 1). In contrast to the small intestine, Wntless/Evi expression was less abundant and restricted to some cells within enteric ganglia of the colon (Figure 2). In the small intestine, we found the expression of Wnt1-, Wnt2-, Wnt2b-, Wnt3-, Wnt3a-, Wnt4-, Wnt5a-, Wnt5b-, Wnt6-, Wnt7b-, Wnt8a-, Wnt8b-, Wnt9a-, Wnt10a-, Wnt10b-, Wnt11-, and Wnt16-transcripts in the submucosal (Figures 1B, C - SubG) and myenteric (Figures 1B, C – MyG) plexus. The signal intensity was comparable for all detectable transcript, excluding Wnt4-, Wnt5a-, and Wnt8b-mRNAs, which exhibited considerably lower, yet clearly distinguishable, signal levels (Figure 1B). Interestingly, some Wnt-ligands, such as Wnt1, Wnt2, and Wnt2b, showed a preferential expression in myenteric neuronal somata as compared to glial cell bodies and surrounding ganglionic neuropil (Supplementary Table 1). In contrast, most other transcripts did not exhibit a clear localization to a specific cell type (Figure 1C). Moreover, Wnt7a-, Wnt9b-, and Wnt10b-transcripts were not detectable in the ganglia neither in the submucosal nor in the myenteric plexus (Supplementary Figure 1C, Supplementary Table 1).
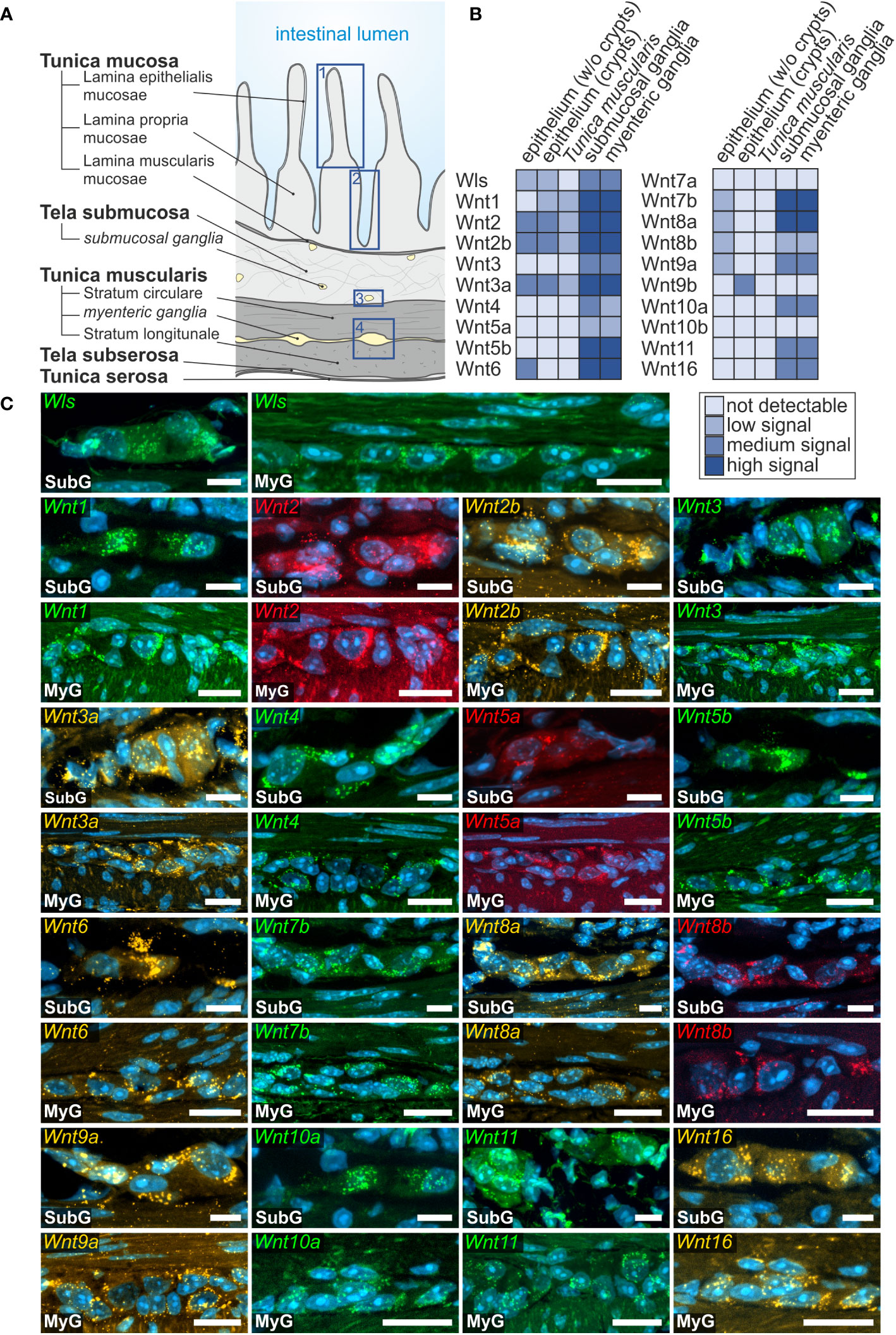
Figure 1 Wnt-ligands expressed in the murine small intestine. (A): Schematic drawing of the gut wall with the histological landmarks. Numbered rectangles (dark blue) indicate the regions examined: 1: epithelium (without crypts); 2: epithelium (only crypt region); 3: submucosal ganglia; 4: myenteric ganglia with surrounding musculature. (B, C): In situ hybridization experiments showed Wls- as well as Wnt1-, Wnt2-, Wnt2b-, Wnt3-, Wnt3a-, Wnt4-, Wnt5a-, Wnt5b-, Wnt6-, Wnt7b-, Wnt8a-, Wnt8b-, Wnt9a-, Wnt10a-, Wnt11- and Wnt16-mRNAs detection (colors as indicated) and the nuclear marker DAPI (blue) within submucosal (SubG) and myenteric plexus (MyG) of murine small intestine with different expression levels. Scale bars: 40 µm.
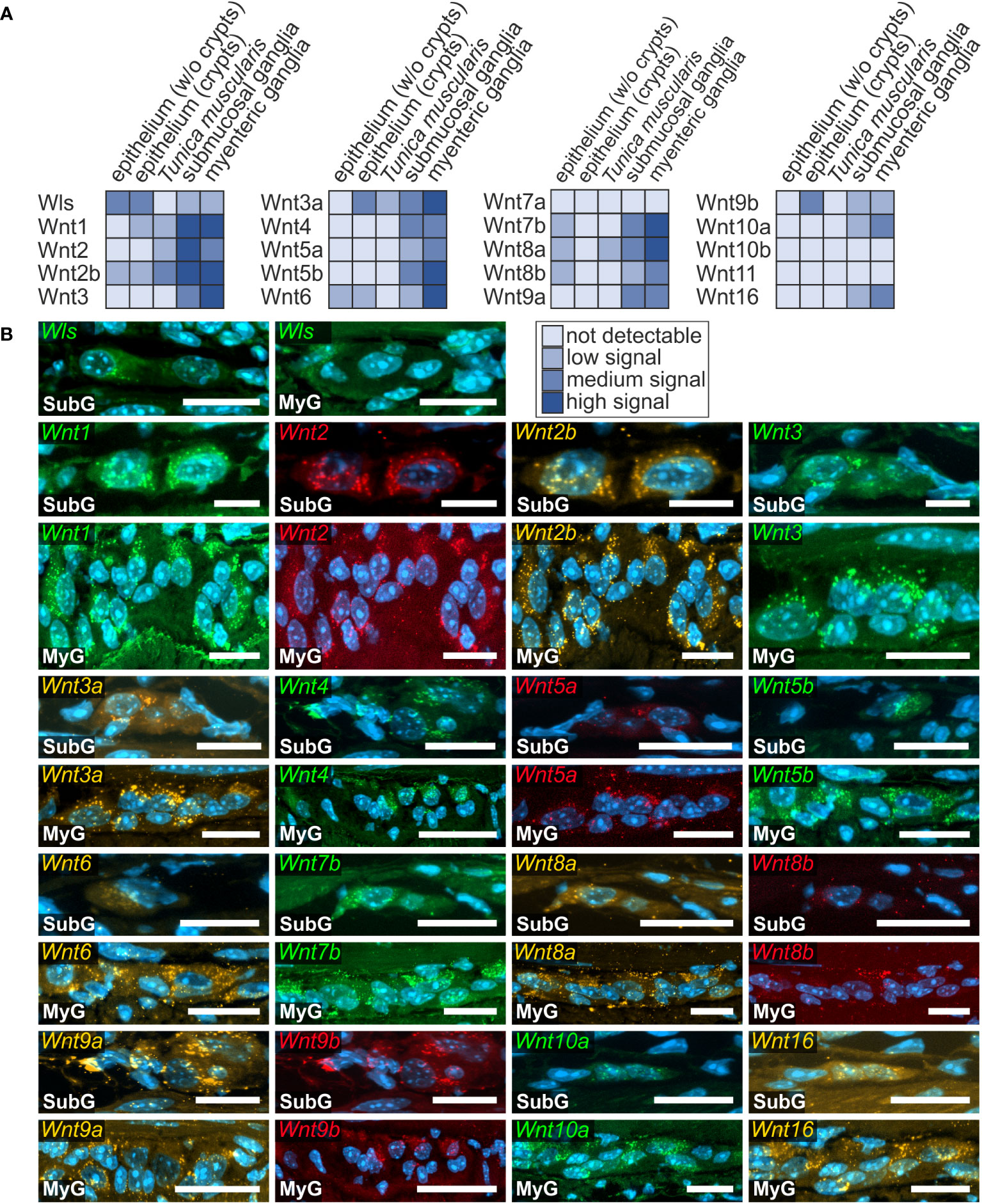
Figure 2 Presence of Wnt-ligand-transcripts in the murine large intestine. (A, B): Wls- as well as Wnt1-, Wnt2-, Wnt2b-, Wnt3-, Wnt3a-, Wnt4-, Wnt5a-, Wnt5b-, Wnt6-, Wnt7b-, Wnt8a-, Wnt8b-, Wnt9a-, Wnt9b-, Wnt10a- and Wnt16-transcripts (colors as indicated) were found within submucosal (SubG) and myenteric plexus (MyG) of murine large intestine with different expression levels. Cell nuclei are stained for the nuclear marker DAPI (blue). Scale bars: SubG 20 µm and MyG 40 µm.
To put our results into a wider context with previously published RNA expression profiles, we compared our RNAscope-based expression atlas (Supplementary Table 1) of the small intestine to a preexisting single cell RNA sequencing dataset of the juvenile murine myenteric plexus (38). Interestingly, there we found that Wnt2-, Wnt2b, Wnt3 and Wnt16 transcripts were only detectable in the myenteric neuronal cell cluster, whereas Wnt5b, Wnt7b and Wnt10a mRNAs were observed in the glial cell cluster. Further, detectable mRNA expression of Wls, Wnt1, Wnt4, Wnt5a, Wnt6, Wnt9a, as well as Wnt11 was found within both, neuronal and glial cell clusters. Wnt3a-, Wnt7a-, Wnt8a-, Wnt8b-, Wnt9b- and Wnt10b-transcripts were not found in the neuronal nor glial clusters (Supplementary Table 2).
In the large intestine, we found the expression of Wnt1-, Wnt2-, Wnt2b-, Wnt3-, Wnt3a-, Wnt4-, Wnt5a-, Wnt5b-, Wnt6-, Wnt7b-, Wnt8a-, Wnt8b-, Wnt9a-, Wnt9b-, Wnt10a-, and Wnt16-transcripts throughout myenteric ganglia, however, with variable intensities (Figure 2A). While Wnt5a, Wnt8b, and Wnt9b were low, yet clearly abundant, the signal intensity of the remaining detectable Wnt-ligands was considerably stronger (Figure 2B). In addition, Wnt1-, Wnt2-, Wnt2b-, Wnt3-, Wnt3a-, Wnt4-, Wnt5a-, Wnt5b-, Wnt6-, Wnt7b-, Wnt8a-, Wnt8b-, Wnt9a-, Wnt9b-, Wnt10a-, and Wnt16-transcripts were also found in submucosal plexus, though we found Wnt5b- and Wnt6 mRNAs only found single submucosal neurons (Figure 2B). Moreover, Wnt2 expression was noticeably more intense in the submucosal than in the myenteric ganglia. In contrast to the small intestine, Wnt7a-, Wnt10b-, and Wnt11-mRNAs were not detectable in the ganglia of the large intestine (Supplementary Figure 1C, Supplementary Table 1). Yet, like the findings in the small intestine, Wnt1, Wnt2, and Wnt2b exhibited a neuronal localization, both in submucosal and myenteric ganglia. Additionally, the expression of Wnt4 appeared to be restricted to a subset of neuronal somata in the myenteric plexus (Figure 2B).
Several Wnts-ligands were also expressed in other compartments of the intestines, including the epithelium, Lamina propria mucosae as well as in the smooth musculature of the Tunica muscularis of small (Supplementary Figure 2) and large intestine (Supplementary Figure 3). Since there is extensive data on the expression of Wnt signaling components in the intestinal epithelium and crypts, we used these compartments as internal positive controls, further validating our findings in the ENS. In the small intestine, transcripts of Wnt2 and the closely related homolog Wnt2b were found in the epithelium (Supplementary Figures 2A-i-ii, B-i-ii, arrowheads), and cells of the Lamina propria mucosae (Supplementary Figures 2A-i-ii, B-i-ii, arrow), as well as within the Tunica muscularis (Supplementary Figures 2A-iii, B-iii, arrowheads).
In contrast, in the large intestine Wnt2 and Wnt2b were less abundant. Wnt2 was hardly expressed in the majority of epithelial cells (Supplementary Figures 3A-i, arrowheads) and exhibited a low expression profile throughout the Lamina propria mucosae (Supplementary Figures 3A-ii, arrowheads) and Tunica muscularis (Supplementary Figures 3A-iii, arrowheads). Wnt2b showed slightly higher expression levels, both in the epithelium as well as the Tunica muscularis (Supplementary Figures 3B-i–B-iii, arrowhead).
Furthermore, Wnt3a-transcripts were strongly expressed throughout the Tunica mucosa of the small intestine (Supplementary Figures 2C-i-ii, arrowhead), but was restricted to the crypt bottom in the large intestine (Supplementary Figure 3C). Wnt6-expression was found in the small intestine throughout the villus but was undetectable in the crypt bottom (Supplementary Figure 2D). In the large intestine, however, Wnt6 was expressed in the epithelium (Supplementary Figures 3D-i, arrowhead), including the crypt bottom (Supplementary Figures 3D-ii, arrowhead). Wnt7b-, Wnt8a-, and Wnt8b-transcripts were detectable with low expression rates in a subset of superficial epithelial cells (Supplementary Figures 3E, F-i, G, arrowhead). However, we did not detect any expression of these transcripts in the epithelial cells of the crypts, both in small and large intestine. Interestingly, we found a population of cells in the Lamina propria mucosae with high expression profiles of Wnt7b und Wnt8a, that lacked a detectable signal for Wnt8b, both in the small intestine (Supplementary Figures 2E, F arrow) and colon (Supplementary Figures 3E, F-i arrows). Moreover, Wnt8a was also expressed in the Tunica muscularis in the large intestine (Supplementary Figures 3F-ii, arrowhead). Finally, Wnt9a was located throughout the epithelium of the small intestine (Supplementary Figure 2H, arrowhead), whereas Wnt9b-transcripts were found at the crypt bottom of small and large intestine only (Supplementary Figures 2I, 3H, arrowhead).
Wntless, the transport and secretion protein for Wnt-ligands, was found within the Lamina propria mucosae and occasionally in epithelial cells of small and large intestine (Supplementary Figures 2J, 3I). Beside their expression within the enteric nervous system, neither Wnt1-, Wnt3-, Wnt4-, Wnt5a- and Wnt5b- and Wnt10a-transcripts, nor Wnt1-, Wnt3-, Wnt5a- and Wnt5b-, Wnt10a- and Wnt16-mRNAs were detectable within small and large intestine.
Wnt-receptor expression in murine small and large intestine
Next, we examined the expression of Frizzled receptors 1-10 as well as Lrp-receptors 5 and 6 to identify Wnt-responsive cells within the enteric nervous system. As shown in Figure 3, Fzd- and Lrp-transcripts were found within submucosal (Figures 3A, B - SubG) and myenteric ganglia (Figures 3A, B – MyG) of the small intestine and were also widely distributed within submucosal (Figures 4A, B – SubG) and myenteric (Figures 4A, B – MyG) ganglia of the large intestine. In the small intestine, the expression intensity was comparable for all transcripts, excluding Fzd1-, and Fzd10-mRNAs, which exhibited considerably lower, but clearly distinguishable signal levels in myenteric, but particularly in submucosal ganglia. Interestingly, some Fzd-receptors, such as Fzd2, Fzd3, Fzd6, and Fzd7 showed a clearly recognizable expression in myenteric neuronal somata. In contrast, most other transcripts did not exhibit a clear localization to a specific cell type (Figure 3, Supplementary Table 1). Compared to the small intestine, expression signals of Fzd-transcripts were considerably stronger in the large intestine. Additionally, like the findings in the small intestine, Fzd2, Fzd3, Fzd6, and Fzd7 displayed a neuronal localization, both in submucosal (Figure 4B – SubG) and myenteric (Figure 4B – MyG) ganglia. The transcripts of the co-receptors Lrp5 and Lrp6 exhibited highly intense signal levels throughout the myenteric and submucosal ganglia of the small intestine (Figure 3) and the colon (Figure 4). Again, comparing our results to scRNA-seq atlas of the juvenile murine myenteric plexus (38), we found a considerable mRNA expression of the Fzd1-10 and Lrp5 and Lrp6 transcripts within neuronal and glial cell cluster (Supplementary Table 2), which is partly in line with our own findings.
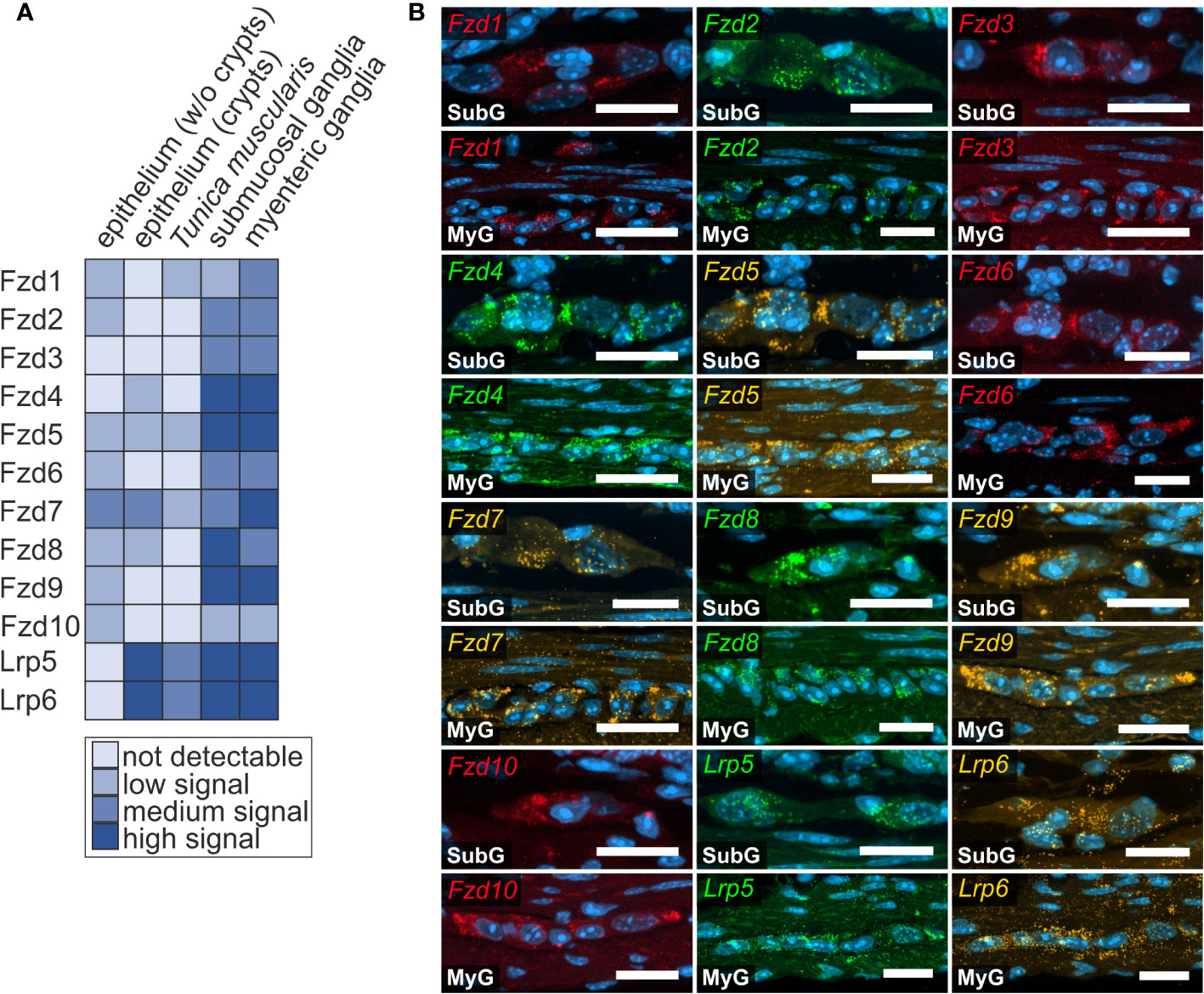
Figure 3 Frizzled-mRNAs in murine small intestine. (A, B): In situ hybridization experiments showed Fzd1-, Fzd2-, Fzd3-, Fzd4-, Fzd5-, Fzd6-, Fzd7-, Fzd8-, Fzd9-, Fzd10-, Lrp5- and Lrp6-transcripts detection (colors as indicated) and the nuclear marker DAPI (blue) within submucosal (SubG) and myenteric plexus (MyG) of murine small intestine with different expression levels. Scale bars: 40 µm.
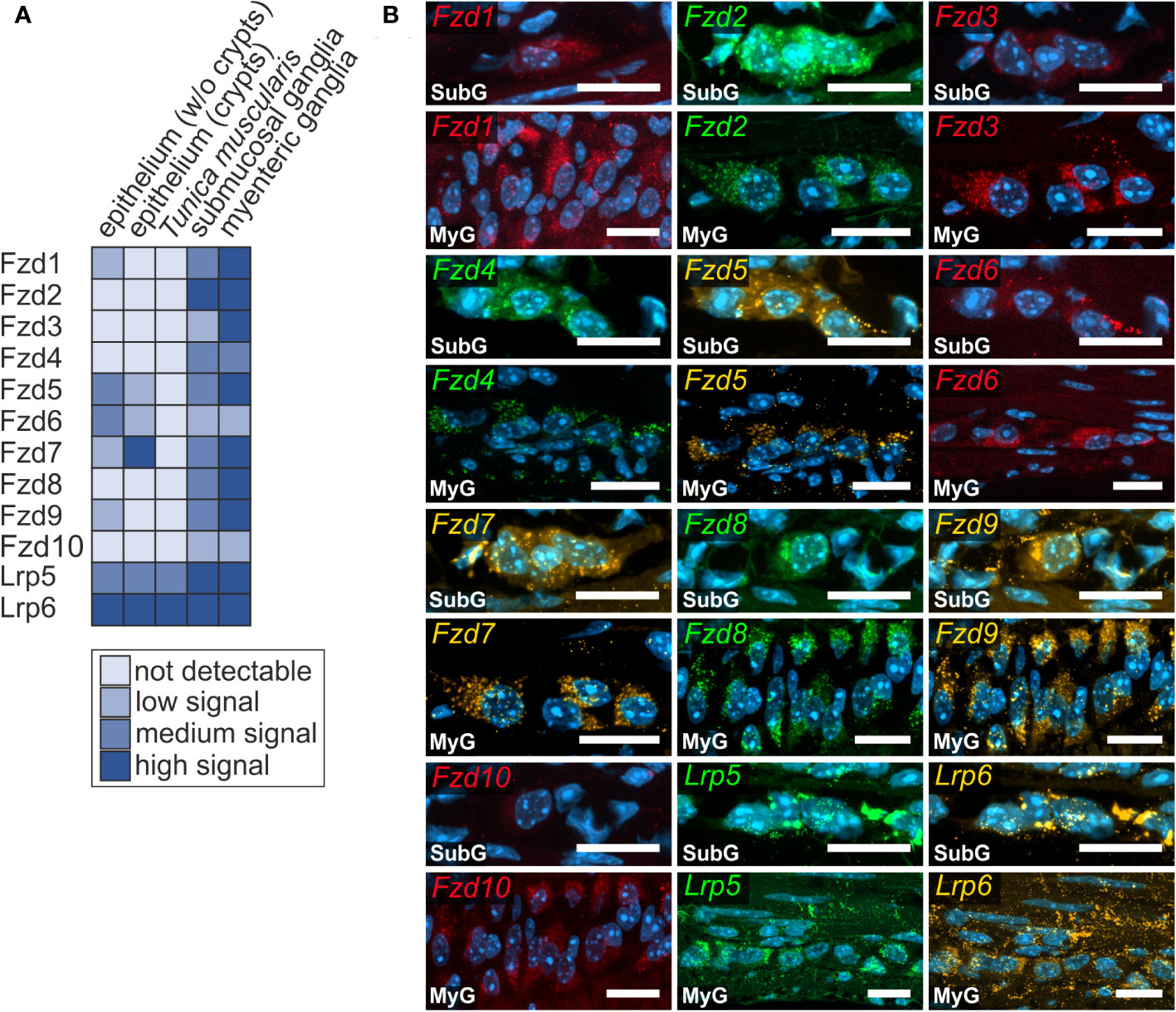
Figure 4 Frizzled-mRNAs are detectable in murine large intestine. (A, B): Fzd1-, Fzd2-, Fzd3-, Fzd4-, Fzd5-, Fzd6-, Fzd7-, Fzd8-, Fzd9-, Fzd10-, Lrp5- and Lrp6-transcripts were detected (colors as indicated) within submucosal (SubG) and myenteric plexus (MyG) of murine large intestine with different intensity levels. Cell nuclei are stained with the nuclear marker DAPI (blue). Scale bars: SubG 20 µm and MyG 40 µm.
We further looked for the expression of Fzd- and Lrp-transcripts in other layers of the gut wall. We found a low hybridization signal for Fzd1 throughout the epithelium of small (Supplementary Figure 4A) and large intestine (Supplementary Figure 5A). Interestingly, we detected a medium to high expression of Fzd2-, Fzd5-, Fzd7-, Fzd8-, and Fzd9-transcripts in a subset of cells in the Lamina propria mucosae of the small intestine (Supplementary Figures 4B, D-i, F, G, H, arrow), whereas in the large intestine only Fzd9 was detectable at a comparable location (Supplementary Figure 5E, arrow). Occasionally, we also found Fzd10-transcritps localized within cells of the Lamina propria mucosae (Supplementary Figure 4I, arrow) as well as in a subset of epithelial cells of the small intestine (Supplementary Figure 4I, arrowhead). In the epithelium, there was a clearly visible concentration of cells expressing Fzd4, Fzd5, Fzd6, and Fzd7 in the small intestinal crypt bottom. While Fzd7 exhibited a partly similar expression at the crypt bottom of the colon, Fzd5 and Fzd6-expressing cells were absent throughout the crypt and rather located at the luminal surface (Supplementary Figures 5B, C, arrowhead). Except for the ENS, Fzd3 was not detectable in other parts of the gut wall both in the large and small intestine.
In contrast to Fzd-receptors, Lrp-receptors showed a stronger expression signal throughout all intestinal layers examined. Intriguingly, Lrp5 was found within epithelial cells of the crypt bottom in the small intestine (Supplementary Figures 4J-i), whereas in the large intestine it was located in the cells of the Lamina propria mucosae surrounding the crypts (Supplementary Figures 5F-i-ii). Lrp6, on the other hand, exhibited an intense expression both, in the epithelial cells of the crypt bottom as well as the Lamina propria cells throughout the small intestine and colon (Supplementary Figures 4K-i-ii, 5G-i-iii).
Expression of the secreted Wnt-antagonist Dickkopf in murine small and large intestine
As a classical functional counterpart of activating Wnt-signals, we screened for classical secreted Wnt-antagonists, namely Dickkopf-ligands 1-4 (Dkk1, Dkk2, Dkk3, Dkk4), and the relevant receptors Kremen 1-2 (Krm1, Krm2) and Low-density lipoprotein receptor-related protein 5 and 6 (Lrp5 and Lrp6), (see also Figures 3, 4). Dkk-ligands and corresponding receptors were detectable within the submucosal and myenteric plexus of the small (Figures 5A, B) and large intestine (Figures 5C, D). In the small intestine, Dkk-ligands showed variable expression levels within the myenteric plexus, in that Dkk2- and Dkk3-transcripts exhibited a more abundant and stronger expression than Dkk1- and Dkk4-mRNAs. Moreover, we observed that Dkk4-signals were weaker than Dkk1-transcripts (Figure 5B), and nearly absent in submucosal ganglia. This expression pattern was comparable in the small and large intestine (Figure 5D). Interestingly, Krm1- and Krm2-transcripts showed a higher expression intensity in myenteric ganglia of the large intestine than in small intestinal ganglia. Noteworthy, Dkk-ligands and their receptors did not exhibit a detectable preference to a specific ganglionic cell type (Figures 5B, D as well as Supplementary Table 1).
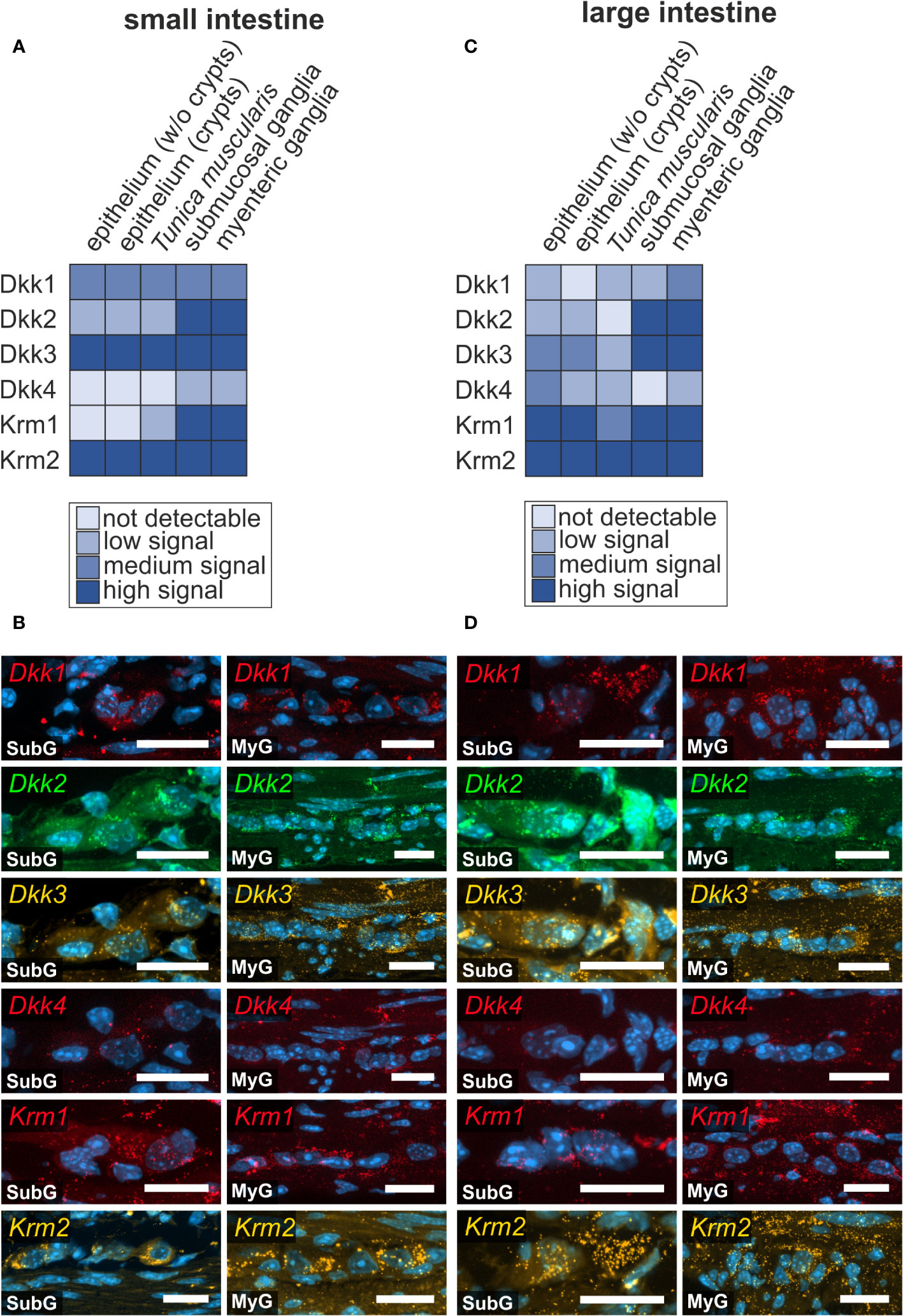
Figure 5 Prominent Wnt-signaling antagonists are expressed in the murine intestine. (A, B): Dkk1, Dkk2, Dkk3, Dkk4, Krm1 and Krm2 mRNAs were detected with in situ hybridization experiments (colors as indicated) and the nuclear marker DAPI (blue) within submucosal (SubG) and myenteric plexus (MyG) of murine small intestine with different signal intensities. Scale bars: 40 µm. (C, D): Dkk1, Dkk2, Dkk3, Dkk4, Krm1 and Krm2 transcripts (colors as indicated) and the nuclear marker DAPI (blue) were also detectable within myenteric plexus of murine large intestine Scale bars: SubG 20 µm and MyG 40 µm.
Similar results were obtained from single cell RNA sequencing. In detail, mRNA expression of Dkk1-, Dkk2-, Dkk3-, Krm1, as well as Krm2 was found within the neuronal and glial cell clusters. In contrast to our dataset, Dkk4 transcripts were not found in the neuronal and glial cluster (Supplementary Table 2).
Dkk- and Krm-expression was also observed within other layers of the intestinal wall of small intestine (Supplementary Figure 6) and large intestine (Supplementary Figure 7). In the small intestine, Dkk1- and Dkk3-transcripts were expressed throughout the villus including the epithelium, Lamina propria mucosae (Supplementary Figures 6A-i, 5C-i), and within the intestinal crypts (Supplementary Figures 6A-ii, 5B-ii).
Interestingly, Dkk1-expression was considerably lower in the crypt bottom compared to more superficial epithelial cells (Supplementary Figures 6A-ii, asterisk). Additionally, Dkk1-, Dkk3-, and Krm2-transcripts were examined within smooth muscle cells of the Tunica muscularis (Supplementary Figures 6A-iii, C-iii, D-iii). In the large intestine, Dkk1- and Dkk3-transcrips were expressed within the epithelium (Supplementary Figures 6A-i, C-i). Additionally, Dkk3 exhibited an expression down to the base of the crypts, in the Lamina propria mucosae below the crypt region and within the Lamina muscularis mucosae (Supplementary Figures 7C-ii). Dkk2 was weakly detectable in epithelial cells, at the crypt bottom as well as in smooth muscle cells of the Tunica muscularis of the small intestine (Supplementary Figures 6B-iii).
In the large intestine, Dkk2-mRNAs were mostly present within the superficial cells of the epithelium and less so in the crypts (Supplementary Figures 7B-i-ii). Moreover, weak expression of Dkk4 was found in intestinal crypts of the small intestine (Supplementary Figures 6E-i), surrounding the crypt bottom. In contrast, in the large intestine Dkk4-mRNAs were mainly detected in epithelial cells outside the crypts (Supplementary Figures 7E-i) and in the Lamina muscularis mucosae (Supplementary Figures 7E-ii). Additionally, Dkk4-transcripts were also present within the Tunica muscularis in both gut regions (Supplementary Figures 6E-ii, 7E-iii).
In the small intestine, Krm1-transcripts were mainly localized within crypts, and in the Tela submucosa and Tunica muscularis (Supplementary Figures 6F-i-ii). In the large intestine, Krm1 was slightly expressed within some epithelial cells, however, predominantly localized at the crypt bottom, in the Lamina propria mucosae surrounding the crypts, as well as weakly within the Tunica muscularis (Supplementary Figures 7D-i-ii). Krm2-transcripts showed a broader presence than Krm1-transcripts, including the expression, throughout the villus and Lamina propria mucosae, the crypt bottom, as well as within the Tunica muscularis of the small intestine (Supplementary Figures 6D-i-iii). In large intestine, Krm2-expression was less intense, but also present within the epithelium including the crypt bottom and within the Tunica muscularis (Supplementary Figures 7F-i-iii).
Expression of the Wnt-agonist R-Spondin in mouse intestine
In cells with ongoing Wnt-signaling, R-Spondin proteins interact with their cognate receptors LGR4, LGR5, and LGR6 to inhibit ligase activity that would otherwise degrade Frizzled receptors, as part of a negative feedback mechanism. This in turn leads to accumulation of Frizzled receptors on the plasma membrane, thus sensitizing target cells towards Wnt ligands (40, 41). Here, we examined the expression of R-Spondin-ligands 1-4 (Rspo1, Rspo2, Rspo3, Rspo4), and the relevant receptors Lgr4, Lgr5, and Lgr6 in the murine ENS. We found that all RSPO-ligands were expressed in the submucosal as well as the myenteric plexus of the small (Figures 6A, B) and large intestine (Figures 6C, D). Yet, Rspo2-, Rspo3-, Rspo4-transcripts exhibited a more abundant and stronger expression than Rspo1 (Figures 6B, D).
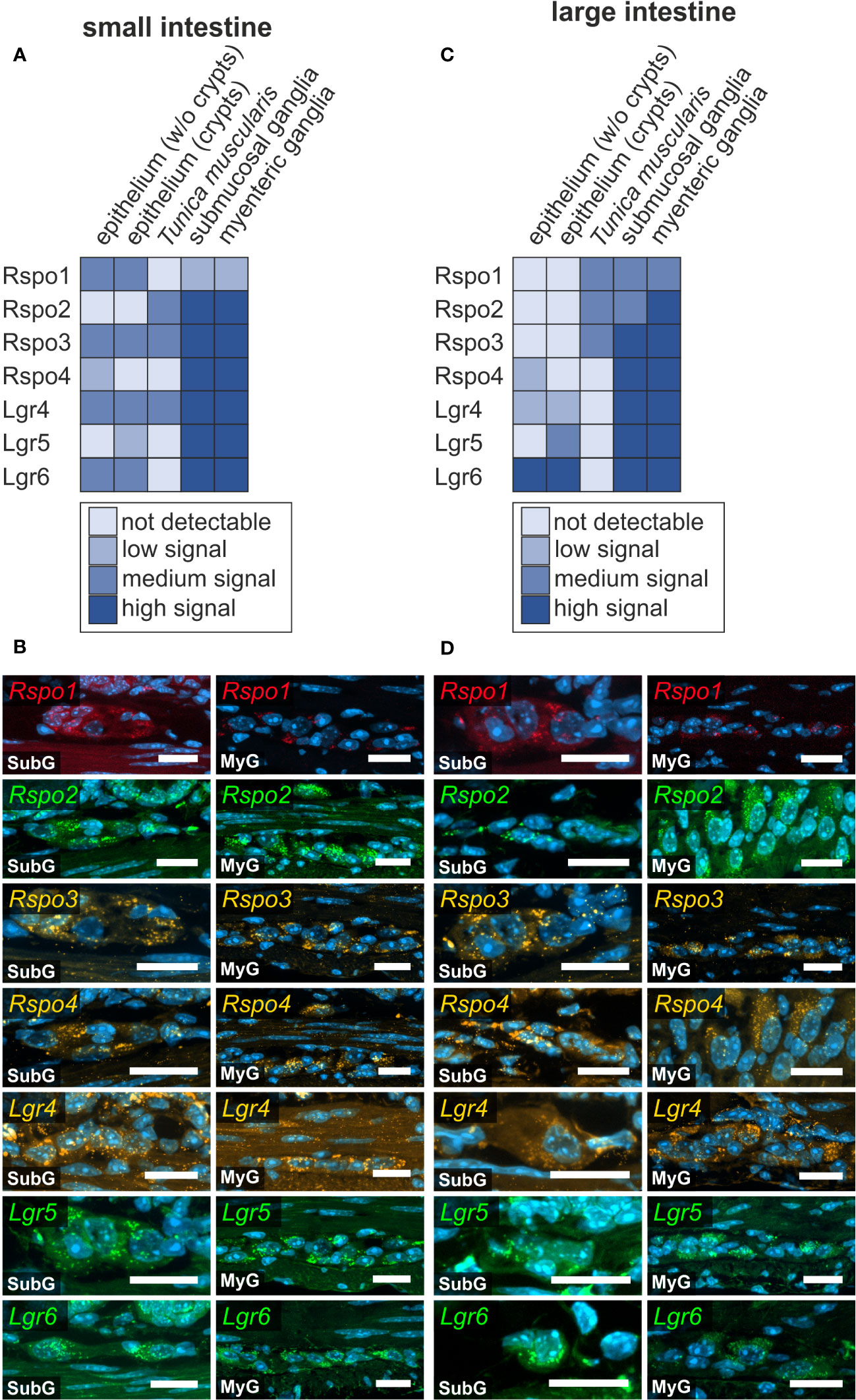
Figure 6 R-spondin-ligands and Lgr-receptors are present in the murine small and large intestine. (A–D): In situ hybridization experiments showed Rspo1-, Rspo2-, Rspo3-, Rspo4-, Lgr4-, Lgr5-, and Lgr6-mRNA detection (colors as indicated) and the nuclear marker DAPI (blue) within submucosal (SubG) and myenteric plexus (MyG) of murine small (A, B) and large intestine (C, D) with different signal levels. Scale bars: 20 µm.
Moreover, Lgr4, Lgr5, and Lgr6 were strongly expressed in both enteric plexus of the small and large intestine. Intriguingly, whereas Lgr5 and Lgr6-transcripts displayed a clear and distinguishable localization within neuronal perikarya compared to glial cell bodies or the ganglionic neuropil, Lgr4-transcripts showed a rather homogeneous expression throughout the different cells of the ganglia (Figures 6A, B). Furthermore, a similar expression pattern was observed in submucosal and myenteric ganglia of the large intestine (Figures 6C, D, Supplementary Table 1). Similar results were obtained from the single cell RNA sequencing dataset, however Rspo4 transcripts were not found in the glial clusters (Supplementary Table 2).
R-Spondins and Lgr-receptors were also found in other layers of the gastrointestinal wall. We detected a considerable expression of Rspo1 in within the epithelium of the small intestine (Supplementary Figure 8A), however, with no expression in the large intestine. Rspo2-expression was not detectable in small and large intestine samples. Moreover, Rspo3-mRNAs were found in the small intestine particularly within the Lamina propria mucosae (Supplementary Figure 8B, arrowhead) and in epithelial cells at the crypt bottom (Supplementary Figure 8B, arrow). Yet, Rspo3 was not detectable in the large intestine. Rspo4 was expressed within cells of the Lamina propria mucosae (Supplementary Figure 8C, arrow) and with lower expression intensity within the epithelium of the small intestine (Supplementary Figure 8C, arrowhead). In the large intestine, Rspo4 was restricted in its expression to the epithelium especially outside the crypts (Supplementary Figure 9A, arrowhead).
In the small intestine, a high expression of the corresponding receptor Lgr4 was observed throughout the entire epithelium (Supplementary Figures 8D-i, arrowhead) and within the Lamina propria mucosae (Supplementary Figures 8D-i, arrow). In the large intestine, Lgr4-transcripts were found within the epithelium (Supplementary Figures 9B-i), as well as throughout the crypts (Supplementary Figures 9B-ii, arrow). However, we found a population of cells at the bottom of the crypt that clearly lacked the expression of Lgr4 (Supplementary Figures 9B-ii, arrowhead). In contrast, Lgr5-mRNA was restricted to a subpopulation of epithelial cells at the crypt bottom as well as to cells of the Lamina propria mucosae in close vicinity to the crypt bottoms both, within the small (Supplementary Figure 8E, arrowhead) and large intestine (Supplementary Figure 9C, arrowhead). Lgr6 was occasionally found in cells of the epithelium (Supplementary Figures 8F-i, arrowhead), and in a subset of cells located at the crypt bottom (Supplementary Figures 8F-ii, arrow), as well as within the Lamina propria mucosae of the small intestine (Supplementary Figures 8F-i, asterisks). In the large intestine, Lgr6-transcripts were found in epithelial cells (Supplementary Figures 9D-i-ii, arrowhead), within the Lamina propria mucosae (Supplementary Figures 9D-ii, asterisk), however not at the crypt bottom (Supplementary Figures 9D-ii, arrow).
To further contextualise the presence of Wnt-signaling related mRNAs under pathophysiological conditions, we re-evaluated a recently published glia-specific RiboTag-based RNA sequencing dataset of acute inflammation in a mouse model of post-operative ileus (21).
Myenteric glial cells express Wnt-signaling related mRNAs in a model of post-operative ileus
A recently published study by Leuven and Schneider et al. describes enteric glial plasticity during acute intestinal inflammation in a post-operative ileus (POI) mouse model. In brief, they induced enteric glial reactivity, by a standardized intestinal manipulation (IM) procedure leading to POI. POI progression was classified into an early/immediate phase (3h after intestinal manipulation), inflammatory/manifestation phase (24h after intestinal manipulation), and recovery/resolution phase (72h after intestinal manipulation). With an established RiboTag-based approach (42), they selectively isolated transcribed mRNAs of hemagglutinin-labeled ribosomes of enteric glial cells of the Tunica muscularis of Sox10iCreERT2/Rpl22HA/+ mice within the relevant POI stages. We re-examined the resulted RNA-Seq dataset (22), thereby focusing on Wnt-signaling related mRNAs and found, that in total 31 Wnt-signaling related mRNAs were differently regulated during the designated timepoints. Fzd2-, Krm1-, Rspo3-, Wnt5b- and Wnt6-mRNA transcripts reached statistical significance during the early phase, 3h post IM (see Supplementary Table 3). However, their expression level normalized to the naive control population or were not detectable after 6h post IM. During the inflammatory phase, 24h post IM, Dkk2 and Fzd7 transcripts were upregulated, whereas Dkk3 and Sfrp1 (Secreted frizzled related protein 1) transcripts were down regulated. After 72h post IM, Dkk2, Sfrp1, Fzd2 and Wnt5a mRNAs expression levels were still significantly regulated.
In summary, by combining our in situ hybridization expression atlas with the enteric gliosis data shed light on a broader, arguably highly dynamic impact of canonical and non-canonical Wnt-signaling on the mature ENS and its cellular homeostasis in health and inflammation.
Discussion
Histological evaluation of small lipid-modified morphogens, such as Wnt-molecules, poses considerable experimental challenges, one of which is the lack of reliable antibodies targeting Wnt-molecules in mammalian tissues (43). To cope with this problem, detecting mRNAs is an effective alternative, which also allows for a more specific identification of Wnt-secreting cells. Here, we applied the RNAscope HiPlex-assay, that uses a special amplification procedure to enable the detection of single mRNA transcripts, making it especially useful for the evaluation of tightly regulated signaling cascades with low abundant mRNA molecules at the cellular level (44). In this study, we present an overview of the expression pattern of 45 relevant Wnt-signaling components within the enteric nervous system, thereby closing a gap of knowledge about the role of Wnt-signaling in tissue homeostasis of the gastrointestinal tract. Our results are partly in concert with and thus substantiate previously published single cell RNA-sequencing analyses of the juvenile murine ENS of the small intestine (37), thereby adding a valuable special component to big data gene expression studies. Nevertheless, we want to point out that cross-interpretation between different studies should always be carried out carefully taking diverse experimental approaches (e.g., sample acquisition, cell isolation, age of animal) and varying sensitivity in the detection of target sequences into account.
Source of Wnt-ligands in the ENS
Prior to secretion, newly synthesized Wnt-ligands undergo various posttranslational modifications in the endoplasmic reticulum (ER) of Wnt-producing cells. These modifications are indispensable for both, intracellular trafficking by the transmembrane protein Wntless (Wls) via the Golgi apparatus to the plasma membrane (i.e., Wnt-ligand secretion), and Wnt-receptor recognition by the target cell. After Wnt-ligand secretion, Wls is endocytosed and recycled to fulfil its trafficking function again (39). Wls indirectly plays an important role for ENS-development, as it is expressed during early embryogenesis at the mid-hindbrain boundary and mediates the secretion of Wnt1, which in turn induces neural crest formation (45). Furthermore, in humans loss of Wls is associated with severe multiorgan defects (46) and epithelial colorectal cancer (47), whereby the latter underlines its importance in tissue homeostasis.
We observed that Wls-transcripts are expressed throughout the cell bodies of submucosal and myenteric plexus cells in the small intestine and were less abundant in the large intestine. Together with previous reports (47, 48), our data indicate that the Wnt-producing cells are thus present within the adult enteric nervous system, suggesting an underlying, yet to be uncovered role in postnatal ENS tissue homeostasis.
Moreover, we found that the mRNA of Wnt ligands, such as Wnt1, Wnt2, or Wnt2b was primarily located in enteric perikarya rather than the neuropil or other intraganglionic cells, strongly indicating that not only glial cells but also enteric neurons themselves could serve as sources of Wnt ligands in the ENS. The expression of Wnt1 mRNA by adult enteric neurons is particularly intriguing as Cre-lox mediated fluorescent reporter mouse strains widely used in the field make use of a Cre-recombinase under the control of a Wnt1-promotor (e.g., 129S4.Cg-E2f1Tg(Wnt1-cre)2Sor/J, The Jackson Laboratory, strain #022137). If crossbred with mice carrying floxed fluorescent reporter proteins, these strains are particularly useful in visualizing the ENS, as all neural crest derivatives (i.e., the ENS in the gut) initiate fluorescent protein expression during Wnt1-dependent neural crest induction. The re-occurrence of Wnt1-mRNA in adult enteric neurons, however, suggest that also Cre-recombinases are expressed at adult stages. It is therefore conceivable that mature enteric neurons would go through repetitive recombination events, if the floxed parent strain uses LoxP-sites that are oriented in opposite direction to each other in order to introduce an inversion of the floxed target gene sequence (see also (49) for recent review on Cre-LoxP strategies). Therefore, beyond its physiological implications, our data point out methodological caveats when planning reporter and/or knock-out experiments using Wnt1-Cre dependent mouse models.
Transport of Wnt-ligands and the extracellular matrix of the ENS
As a result of the posttranslational modifications, Wnt-ligands are hydrophobic and are thus unlikely to diffuse freely within the aqueous extracellular milieu to reach the target cell (for more details on posttranslational modifications, see the recent review by Mehta et al. (50). Yet, the underlying mechanism by which Wnt-ligands are dispersed is hardly characterized in mammals compared to studies in Drosophila (51) and Xenopus (52). Intriguingly, there are implications suggesting that posttranslational modifications (53) enable interactions with extracellular matrix components like heparin sulfate proteoglycans (HSPGs), which are also present on the cell-surface of Wnt-receiving cells (54). This lead to a diffusion model, where Wnt-ligands are transported in a bucket brigade manner by repeated association and dissociation with HSPGs on cell membranes [see also (55)]. Still, there is hardly any data available on secretion and transport processes of Wnt-ligands in the adult enteric nervous system. Nonetheless, immunohistochemical analysis of two heparin sulfate proteoglycans, namely collagen 18 and agrin, in chicken (36) as well as in rat and guniea pig (56) showed a strong expression surrounding enteric ganglia, thus, rendering HSPG-mediated extracellular/extra-ganglionic shuttling of Wnt-ligands plausible. In addition, extracellular matrix interaction of Wnt-ligands secreted from surrounding glia cells (20), myofibroblasts, pacemaker cells, fibroblasts, smooth muscle cells or macrophages (57) might influence adult ENS-homeostasis, as well. In this context, especially the role of resident macrophages of the muscularis externa has been recently shown to have a profound impact on early postnatal development of the ENS (17).
Intracellular Wnt signaling in the target cell
After reaching a target cell, Wnt-ligands activate either β-catenin-dependent, canonical (classical agonists are Wnt1, Wnt3a or Wnt8a) or β-catenin-independent, non-canonical (Wnt4, Wnt5a or Wnt11) signaling cascades. However, it is noteworthy that the signaling output not only depends on the particular Wnt-family member, but is largely influenced by the cellular context and the combination of receptors expressed by the target cell (58). Thus, three core pathways have emerged as the main transducers of Wnt-signaling: (1) The Wnt/βcatenin signaling is of particular importance for cell proliferation. It is triggered by interaction between the canonical Wnt-ligands and a transmembrane receptor complex that is formed by the seven-pass transmembrane receptors of the Frizzled family (Fzd1-10), and the co-receptor low-density lipoprotein receptor-related protein 5 or 6 (LRP5/6) (59).
Among the non-canonical pathways (2), the Wnt/Planar cell polarity (PCP) pathway, employs a signaling cascade involving the small GTPases Rho and Rac and c-Jun N-terminal kinase prominently regulating remodeling of the cytoskeleton and cell differentiation processes (60). In contrast, the non-canonical (3) Wnt/calcium pathway triggers the release of intracellular Ca2+ and the activation of Ca2+-dependent kinases thereby controlling cell motility (61). In this regard our studies demonstrated that the canonical Wnt signaling pathway is inactivated between the transition from proliferation to differentiation in the in vitro model system of postnatally-derived murine ENS-progenitors (62). Moreover, pharmacological activation of canonical Wnt-signaling increases the proliferative capacity of postnatal ENS-progenitors from rodent models and human infants in vitro and successively leads to a higher yield of newly generated neurons (28). It is conceivable, that canonical Wnt signaling, together with the non-canonical Wnt pathways, shapes the cellular homeostasis and cytoarchitecture of the adult ganglionated plexus in vivo, as well.
Fzd-Wnt interactions in the ENS
Despite the apodictic categorization of Wnt-ligands as canonical and non-canonical agonists, mapping a conclusive Fzd-Wnt interactome still is a challenge for the mammalian system, due to high cross-reactivity of individual Wnt-ligands with Fzd subtypes (63). The 10 mammalian Fzd-receptors can be divided into four discrete classes of receptors based on their structural homology. Hereby, Fzd-receptors within each class share between 50% (Fzd3 and Fzd6) and 97% (Fzd1, Fzd2 and Fzd7) amino acid identity (64, 65). In mammals, Frizzled receptors have been implicated in a variety of developmental processes of the central nervous system including neural tube closure (66), axonal outgrowth and guidance of major fiber tracts (67–69), or mediating neurogenic processes in the developing neural tube (70). However, in the adult, misexpression of Fzd receptors is often associated with development and progression of diverse cancers underlining the importance of Wnt-signaling in tissue homeostasis [reviewed in detail by Phesse et al. (71)].
Particularly, Fzd7 is known to activate non-canonical as well as canonical Wnt-signaling in various tissues during development, homeostasis, and disease in several species including humans (72). For instance, the interaction of Fzd7 with Wnt6 induces epithelialisation of the somite’s via canonical signaling cascade (73), and its interplay with Wnt11 orchestrates neural crest cell migration via non-canonical PCP and Ca++ signaling (74). In the adult intestine, Fzd7 is known to be expressed by epithelial stem cells (26) and Flanagan et al. observed an impaired capacity of these cells to regenerate the intestinal epithelium of Fzd7-knockout mice (75). Additionally, the latter study reported on the binding capacity of Fzd7 for the canonical Wnt-ligands Wnt3a as well as Wnt2b (75). These ligands are indispensable for ongoing canonical Wnt-signaling at the stem cell niche at the crypt base (76) and thus are essential to maintain the pool of cycling intestinal stem cells (77, 78). Hence, ectopic expression of Fzd7 has a profound role in tumorigenesis and is critical for the survival, invasion, and metastatic capabilities of colorectal cancer (79, 80). Comparable to the observations by Flanagan et al., we detected Fzd7-expressing cells within the crypts of small and large intestine. In addition, we found a prominent expression of the Fzd7-mRNA within submucosal and myenteric ganglia of adult mice. Interestingly, Wnt3a and Wnt2b were also expressed within submucosal and myenteric plexus cells.
Beyond that, in the small intestine, we detected the expression of supposedly non-canonical Wnt-ligands Wnt5a, Wnt7a, Wnt8b and Wnt11 within the ENS, whereby the latter was not expressed in large intestine samples. It should be mentioned, that although Wnt5a is an important agonist of non-canonical Wnt/Ca++-signaling (81), it also activates canonical Wnt-signaling in the presence of the Lrp5- and Fzd4-receptors (81, 82). Interestingly, in the current study we found Fzd4-expression in submucosal and myenteric plexus of small and large intestine. This is in accordance with previous studies showing, that the Wnt-receptor Fzd4 is expressed on protein level in the human colonic myenteric plexus in Hirschsprung patients (83) and moreover is useful to purify ENS-progenitor cells (27). Further, some Wnt-ligands have been shown to play an important role in the development of the avian intestine. Nagy and colleagues showed, that Wnt11 is expressed in the ceca during hindgut ENS formation and highlighted its importance as an activator of non-canonical Wnt signaling in regulating neural crest cell differentiation (24). Furthermore, data gathered from transcriptome profiling of the widely used in vitro model system of postnatally derived and purified murine ENS-progenitors, suggests that mesenchymal cells express neural stem cell supportive ligands including Wnt5A and Wnt11. Intriguingly, corresponding receptors such as Fzd1, Fzd3, Fzd6, and Fzd7 were expressed by neural cells in the same study (31).
Fine tuning of Wnt-signaling activity by Dickkopf and R-Spondin ligands
Due to its pivotal role in regulating proliferation and differentiation in various organs, Wnt-signaling is tightly regulated. This is achieved not only through the divergent expression of ligands and receptors with partially overlapping function, but also through the secretion of various co-activators and inhibitors that allow for precise fine-tuning of Wnt-activity (84). The prominent group of Wnt-signaling antagonists namely Dickkopf-related proteins are an evolutionary conserved gene family of four glycoproteins (DKK1-4) (85). These proteins are involved in morphogenesis in diverse tissue types in vertebrates (85), and further control a variety of cellular/context-dependent functions, like cell proliferation (86) and differentiation processes (87–89), cell survival and programmed cell death (90). Particularly, DKK1, 3 and 4 interfere with canonical Wnt-signaling by binding LRP5/6 and forming a tertiary complex with their respective receptors Kremen 1 and 2 (Krm1-2), which induce the endocytosis of the essential LRP5/6 receptors (91). Our group has recently published results about the ambivalent function of the Wnt-antagonist DKK1 on murine and human ENS-progenitor cells (29), suggesting a multidimensional regulatory network of Wnt-signaling components in the enteric nervous system. In the current study, we further substantiated our previous findings on DKK expression in the ENS by showing the expression of DKK-ligands and Kremen-receptors in the submucosal and myenteric plexus of the murine small and large intestine.
The regulatory network of Wnt-signaling is completed by the R-spondin protein family (roof plate–specific spondin), which comprises four members (RSPO1-4). These are thought to augment ongoing Wnt-signaling and were first described as secreted activators of canonical Wnt-signaling, and potent mitogens within the stem cell niche of the intestinal epithelium (92, 93). Within this stem-cell niche R-Spondins actively cooperate with Wnt-ligands, such as Wnt3a, Wnt6, and Wnt9 expressed by Paneth cells or the surrounding mesenchyme (94). By binding to the Leucine-rich repeat-containing G-protein coupled receptors 4-6 (LGR4-6), degradation of Frizzled-receptors is inhibited, thus increasing the sensitivity of these target-cells for Wnt-activation (95), enabling the continuous regeneration of the intestinal epithelium (40, 41). Moreover, the corresponding signaling receptor of R-Spondins Lgr5, is an established stem cell marker in adult epithelial stem cell niches (96, 97). Interestingly, unlike in epithelial stem cell niches, Yu and colleagues reported about LGR5-expression in terminally differentiated neurons, but not in the stem cell population of the olfactory bulb (98). Moreover, LGR5- and RSPOs-expression was observed during the WNT-dependent maturation processes in cerebellar granule neurons (99). In this context, we found several R-Spondin ligands as well as corresponding receptors expressed within submucosal and myenteric ganglia of small and large intestine. Especially, LGR5 and LGR6 exhibited a clear localization within enteric neurons. Moreover, intriguingly, the study by Stavely et al. reported R-Spondin 1 and 3 expression in mesenchymal cells and corresponding LGR4 receptor expression by neural cells in vitro (31), thus emphasizing a yet undiscovered role of Wnt-RSPO-LGR signaling in postnatal homeostasis.
Just like Wnt-ligands, Dickkopf- as well as R-Spondin-ligands share posttranslational modifications, that enable interactions with heparin sulfate proteoglycans thereby facilitating extracellular transportation (34, 35, 100). Interestingly, mechanical signals mediated via the extracellular matrix have been reported to influence Wnt-ligand and Wnt-antagonist secretion (101, 102) as well as Fzd-receptor expression (103), resulting in pro-proliferative or pro-differentiation signals. As the adult ENS is constantly motile, changes in the physical property of the surrounding extracellular matrix initiate, not only reflex activity by mechanosensitive enteric neurons (104), but might also influence homeostatic processes in the postnatal ENS in vivo, conceivably by Wnt-signaling components linked to the extracellular matrix.
Wnt-signaling and intestinal immunity
The enteric nervous system and the gut-resident immune system are in constant reciprocal interaction to maintain and modulate intestinal immunity. Conversely, the intestinal immune system influences maintenance of ENS integrity in health and disease and thus shapes ENS structure and function (17). Yet, the pathways of this neuro-immune interaction are still poorly elucidated, and little is known about respective impact of direct neuron-to-immune cell contacts and indirect interactions via intermediary cells, such as enteric glial cells [reviewed in detail by Seguella and Gulbransen (105)].
Although there is hardly any data available on the ENS-derived Wnt-signaling niche, recently published studies suggest, that Wnt-signaling might enable bidirectional communication between enteric glial cells, epithelial cells and immune cells in intestinal immunity. As mentioned above, Baghdadi et al. showed that Wnt ligands, particularly Wnt2, -4, -5a, and -6, are secreted by a subpopulation of type III glial cells in the Lamina propria mucosae and thereby influence epithelial regeneration and barrier function in mice. Interestingly, their results indicate that enteric glial-derived Wnts are dispensable for proper crypt homeostasis under healthy conditions arguably due to functional compensation by Wnts from other stromal sources. However, Wnt ligands from GFAP positive enteric glial cells were important for the high regenerative capacity necessary for the recovery from acute and chronic colitis (20). Further, in their recent study Leuven and Schneider et al. highlighted the plasticity of enteric glial cells during acute inflammation using a post-operative ileus (POI) mouse model (22). By glial specific RiboTag based RNA sequencing, they discovered differentially regulated gene expression profiles during the early/immediate phase (3h after surgery), inflammatory/manifestation phase (24h after surgery), and recovery/resolution phase (72h after surgery). Enteric glial cells switched to an acute immunoreactive phenotype, thereby expressing chemotactic factors, and shaping their microenvironment. At the disease peak, 24h after intestinal manipulation, glial cell reactivity was hallmarked by a migratory and highly proliferative phenotype before declining to a resolution state. Intriguingly, the study showed increased expression of the proliferation-marker Ki67 in a glial subpopulation. Additionally, revisiting their datasets, we found several Wnt-related mRNAs regulated. Interestingly, the presence of the proliferative gilal phenotype coincidences with the downregulation of contra-proliferative Wnt-signals like Dkk ligands or Sfrp1 mRNA. This could indicate a link between enteric gliosis/acute inflammation and our previous works on Wnt-dependent cell-expansion in enteric neural progenitor cells in vitro (28, 29). Additionally, we found that pro-inflammatory Wnt5a expression was steadily decreasing over all tested timepoints reaching statistical significance at 72h post-surgery along with a detectable, yet not significant, expression of its inhibitor Sfrp5 in the early phase. Since the Wnt5a/sFRP5 system is involved in the fine tuning of inflammatory responses in different tissues (106–108), these results might indicate that the enteric glial cells counteracts/limit inflammatory reactions via the Wnt-signaling pathway.
Concluding remarks
Our high-throughput study demonstrated that, a plethora of Wnt-signaling components are expressed within the ENS, indicating a yet to be elucidated functional role of WNTs in ENS-homeostasis. Given the importance of the extracellular matrix for transportation as well as its supposed regulatory influence on secretion of Wnt-signaling components, suggests that further studies are needed to elucidate how morphogens are transported within the ENS. We are aware, that our study does not address the functional role of expressed mRNAs carried out by their translated proteins. Additionally, we want to emphasize, that a detailed cellular mapping of the gene expression patterns of selected genes using combined immunohistological analyses are needed to uncover the missing cell-type specific localization. Especially the lack of differentiability between immune cells like intraganglionic macrophages or leukocytes would be helpful to further stratify the immunological functions of Wnt-signaling related molecules.
However, this is beyond the scope of this work and up to future studies. Still, the presented data, bridges the existing gap of knowledge about the presences of Wnt-signaling components in the so far neglected ENS in vivo. Finally, together with recently published in vitro studies (28, 29), it highlights the importance of the multidimensional regulatory network of Wnt-signaling in the postnatal enteric nervous system that needs to be discovered.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by Regierungspräsidium Tübingen, Landratsamt Tübingen, Veterinäramt, Tübingen, Germany. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. BH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Visualization, Writing – review & editing. PN: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The project was supported by a grant from the German Research Foundation (DFG, Grant number: 438504601).
Acknowledgments
We would like to thank Adrian Krestel for is help in the animal facility as well as Lothar Just and Andreas Mack for their helpful comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1302488/full#supplementary-material
Supplementary Figure 1 | Positive and negative controls. (A): Representative micrographs show mRNA expression of the housekeeping genes GAPDH and HPRT in Tunica mucosa, Tunica submucosa and Tunica muscularis within murine small intestine (i) and large intestine (ii). In contrast, UBC expression was detected within the villi and the Tunica muscularis of small intestine (i+ii), as well as within the Tunica muscularis (iii) of the large intestine. Of note, all three housekeeping genes were expressed within the enteric nervous system. Scale bars: 40 µm. (B): shows a representative negative control of murine large intestine. Scale bar: 100 µm. (C): micrographs depict the non-expressed Wnt-ligands Wnt7a, Wnt10b and Wnt11 in murine small intestine and Wnt7a, Wnt9b and Wnt10b in large intestine. Scale bars: 40µm.
Supplementary Figure 2 | mRNA expression of Wntless and Wnt-ligands within Tunica mucosa and Tunica muscularis of murine small intestine. Wnt2 (A), Wnt2b (B), Wnt3a (C), Wnt6 (D), Wnt7b (E), Wnt8a (F), Wnt8b (G), Wnt9a (H), Wnt9b (I) and Wls (J) transcripts (colors as indicated) are found within different compartments of the intestinal wall (i-iii) of murine small intestine with different expression levels. Cell nuclei are stained for the nuclear marker DAPI (blue). Arrow as well as arrowheads point to positive cells. Scale bars: 40 µm.
Supplementary Figure 3 | Presence of Wntless and Wnt-ligand transcripts within Tunica mucosa and Tunica muscularis of murine large intestine. Wnt2 (A), Wnt2b (B), Wnt3a (C), Wnt6 (D), Wnt7b (E), Wnt8a (F), Wnt8b (G), Wnt9a (H) and Wls (J) transcripts (colors as indicated) are expressed within the intestinal wall (i-iii) of murine large intestine with different intensity levels. Cell nuclei are stained for the nuclear marker DAPI (blue). Arrow as well as arrowheads point to positive cells. Scale bars: 40 µm.
Supplementary Figure 4 | Frizzled and Lrp receptors are expressed in different compartments of the murine small intestine. Detection of Fzd1 (A), Fzd2 (B), Fzd4 (C), Fzd5 (D), Fzd6 (E), Fzd7 (F), Fzd8 (G), Fzd9 (H), Fzd10 (I), Lrp5 (J) and Lrp6 (K) transcripts (colors as indicated) and the nuclear marker DAPI (blue) in different compartments (i-iii) of the small intestine. Arrow as well as arrowheads point to positive cells. Scale bars: 40 µm.
Supplementary Figure 5 | Frizzled- and Lrp-receptor mRNAs are present within the murine large intestine. In situ hybridization experiments detected Fzd1 (A), Fzd5 (B), Fzd6 (C), Fzd7 (D), Fzd9 (E), Lrp5 (F) and Lrp6 (G) transcripts (colors as indicated) and the nuclear marker DAPI (blue) in different compartments (i-iii) of the murine large intestine. Scale bars: 40 µm.
Supplementary Figure 6 | The Wnt antagonist Dickkopf and corresponding receptors Kremen are expressed in the murine small intestine. Micrographs depict expression of Dkk1 (A), Dkk2 (B), Dkk3 (C), Krm2 (D), Dkk4 (E) and Krm1 (F), (colors as indicated) and the nuclear marker DAPI (blue) within the gastrointestinal wall (i-iii) of the murine small intestine. Asterisk indicates no expression at crypt bottom. Scale bars: 40 µm.
Supplementary Figure 7 | Prominent Wnt-signaling antagonists are expressed in the murine intestine. Representative images show detection of Dkk1 (A), Dkk2 (B), Dkk3 (C), Krm1 (D), Dkk4 (E) and Krm2 (F), (colors as indicated) and the nuclear marker DAPI (blue) within the gastrointestinal wall (i-iii) of the murine large intestine. Scale bars: 40 µm.
Supplementary Figure 8 | R-spondin-ligands and Lgr-receptors are present in the murine small intestine. In situ hybridization experiments showed Rspo1- (A), Rspo3- (B), Rspo4- (C), Lgr4- (D), Lgr5- (E), and Lgr6 (F)-mRNA detection (colors as indicated) and the nuclear marker DAPI (blue) within different compartments of the murine small intestine. Arrows, arrowheads, and asterisk indicate respectively for the corresponding transcript expressing or non-expressing cells. Scale bars: 40 µm.
Supplementary Figure 9 | R-spondin-ligands and Lgr-receptors are present in the murine large intestine. Representative images depict the expression of Rspo4- (A), Lgr4- (B), Lgr5- (C), and Lgr6 (D)-mRNAs and the nuclear marker DAPI (blue) within the gastrointestinal wall of the large intestine. Arrows, arrowheads, and asterisk indicate respectively for the corresponding transcript expressing or non-expressing cells. Scale bars: 40 µm.
References
1. Nagy N, Goldstein AM. Enteric nervous system development: A crest cell’s journey from neural tube to colon. Semin Cell Dev Biol (2017) 66:94–106. doi: 10.1016/j.semcdb.2017.01.006
2. Hao MM, Young HM. Development of enteric neuron diversity. J Cell Mol Med (2009) 13(7):1193–210. doi: 10.1111/j.1582-4934.2009.00813.x
3. Parathan P, Wang Y, Leembruggen AJL, Bornstein JC, Foong JPP. The enteric nervous system undergoes significant chemical and synaptic maturation during adolescence in mice. Dev Biol (2020) 458(1):75–87. doi: 10.1016/j.ydbio.2019.10.011
4. McKeown SJ, Chow CW, Young HM. Development of the submucous plexus in the large intestine of the mouse. Cell Tissue Res (2001) 303(2):301–5. doi: 10.1007/s004410000303
5. Bergner AJ, Stamp LA, Gonsalvez DG, Allison MB, Olson DP, Myers MG Jr., et al. Birthdating of myenteric neuron subtypes in the small intestine of the mouse. J Comp Neurol (2014) 522(3):514–27. doi: 10.1002/cne.23423
6. Pham TD, Gershon MD, Rothman TP. Time of origin of neurons in the murine enteric nervous system: Sequence in relation to phenotype. J Comp Neurol (1991) 314(4):789–98. doi: 10.1002/cne.903140411
7. Boesmans W, Nash A, Tasnády KR, Yang W, Stamp LA, Hao MM. Development, diversity, and neurogenic capacity of enteric glia. Front Cell Dev Biol (2021) 9:775102. doi: 10.3389/fcell.2021.775102
8. Virtanen H, Garton DR, Andressoo J-O. Myenteric neurons do not replicate in small intestine under normal physiological conditions in adult mouse. Cell Mol Gastroenterol Hepatol (2022) 14(1):27–34. doi: 10.1016/j.jcmgh.2022.04.001
9. Metzger M, Bareiss PM, Danker T, Wagner S, Hennenlotter J, Guenther E, et al. Expansion and differentiation of neural progenitors derived from the human adult enteric nervous system. Gastroenterology (2009) 137(6):2063–73.e4. doi: 10.1053/j.gastro.2009.06.038
10. Metzger M, Caldwell C, Barlow AJ, Burns AJ, Thapar N. Enteric nervous system stem cells derived from human gut mucosa for the treatment of aganglionic gut disorders. Gastroenterology (2009) 136(7):2214–25.e1-3. doi: 10.1053/j.gastro.2009.02.048
11. Almond S, Lindley RM, Kenny SE, Connell MG, Edgar DH. Characterisation and transplantation of enteric nervous system progenitor cells. Gut (2007) 56(4):489–96. doi: 10.1136/gut.2006.094565
12. Laranjeira C, Sandgren K, Kessaris N, Richardson W, Potocnik A, Vanden Berghe P, et al. Glial cells in the mouse enteric nervous system can undergo neurogenesis in response to injury. J Clin Invest (2011) 121(9):3412–24. doi: 10.1172/JCI58200
13. Belkind-Gerson J, Graham HK, Reynolds J, Hotta R, Nagy N, Cheng L, et al. Colitis promotes neuronal differentiation of Sox2+ and PLP1+ enteric cells. Sci Rep (2017) 7(1):2525. doi: 10.1038/s41598-017-02890-y
14. Chevalier NR, Ammouche Y, Gomis A, Langlois L, Guilbert T, Bourdoncle P, et al. A neural crest cell isotropic-to-nematic phase transition in the developing mammalian gut. Commun Biol (2021) 4(1):770. doi: 10.1038/s42003-021-02333-5
15. Li Z, Hao MM, Van den Haute C, Baekelandt V, Boesmans W, Vanden Berghe P. Regional complexity in enteric neuron wiring reflects diversity of motility patterns in the mouse large intestine. eLife (2019) 8:e42914. doi: 10.7554/eLife.42914.044
16. Margolis KG, Cryan JF, Mayer EA. The microbiota-gut-brain axis: from motility to mood. Gastroenterology (2021) 160(5):1486–501. doi: 10.1053/j.gastro.2020.10.066
17. Viola MF, Chavero-Pieres M, Modave E, Delfini M, Stakenborg N, Estévez MC, et al. Dedicated macrophages organize and maintain the enteric nervous system. Nature (2023) 618(7966):818–26. doi: 10.1038/s41586-023-06200-7
18. McClain JL, Fried DE, Gulbransen BD. Agonist-evoked Ca(2+) signaling in enteric glia drives neural programs that regulate intestinal motility in mice. Cell Mol Gastroenterol hepatol (2015) 1(6):631–45. doi: 10.1016/j.jcmgh.2015.08.004
19. Delvalle NM, Dharshika C, Morales-Soto W, Fried DE, Gaudette L, Gulbransen BD. Communication between enteric neurons, glia, and nociceptors underlies the effects of tachykinins on neuroinflammation. Cell Mol Gastroenterol hepatol (2018) 6(3):321–44. doi: 10.1016/j.jcmgh.2018.05.009
20. Baghdadi MB, Ayyaz A, Coquenlorge S, Chu B, Kumar S, Streutker C, et al. Enteric glial cell heterogeneity regulates intestinal stem cell niches. Cell Stem Cell (2022) 29(1):86–100.e6. doi: 10.1016/j.stem.2021.10.004
21. Clairembault T, Kamphuis W, Leclair-Visonneau L, Rolli-Derkinderen M, Coron E, Neunlist M, et al. Enteric GFAP expression and phosphorylation in Parkinson’s disease. J Neurochem (2014) 130(6):805–15. doi: 10.1111/jnc.12742
22. Leven P, Schneider R, Schneider L, Mallesh S, Vanden Berghe P, Sasse P, et al. β-adrenergic signaling triggers enteric glial reactivity and acute enteric gliosis during surgery. J Neuroinflamm (2023) 20(1):255. doi: 10.1186/s12974-023-02937-0
23. Sasselli V, Boesmans W, Vanden Berghe P, Tissir F, Goffinet AM, Pachnis V. Planar cell polarity genes control the connectivity of enteric neurons. J Clin Invest (2013) 123(4):1763–72. doi: 10.1172/JCI66759
24. Nagy N, Kovacs T, Stavely R, Halasy V, Soos A, Szocs E, et al. Avian ceca are indispensable for hindgut enteric nervous system development. Development (2021) 148(22). doi: 10.1242/dev.199825
25. Clevers H, Loh KM, Nusse R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science (2014) 346(6205):1248012. doi: 10.1126/science.1248012
26. Gregorieff A, Pinto D, Begthel H, Destrée O, Kielman M, Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology (2005) 129(2):626–38. doi: 10.1016/j.gastro.2005.06.007
27. Neckel PH, Scharr M, Seid K, Nothelfer K, Fuchs J, Obermayr F, et al. Wnt receptor frizzled-4 as a marker for isolation of enteric neural progenitors in human children. Cells (2019) 8(8):792. doi: 10.3390/cells8080792
28. Zhang Y, Seid K, Obermayr F, Just L, Neckel PH. Activation of wnt signaling increases numbers of enteric neurons derived from neonatal mouse and human progenitor cells. Gastroenterology (2017) 153(1):154–65.e9. doi: 10.1053/j.gastro.2017.03.019
29. Scharr M, Scherer S, Hirt B, Neckel PH. Dickkopf1 induces enteric neurogenesis and gliogenesis in vitro if apoptosis is evaded. Commun Biol (2023) 6(1):808. doi: 10.1038/s42003-023-05072-x
30. Valenta T, Degirmenci B, Moor AE, Herr P, Zimmerli D, Moor MB, et al. Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep (2016) 15(5):911–8. doi: 10.1016/j.celrep.2016.03.088
31. Stavely R, Bhave S, Ho WLN, Ahmed M, Pan W, Rahman AA, et al. Enteric mesenchymal cells support the growth of postnatal enteric neural stem cells. Stem Cells (2021) 39(9):1236–52. doi: 10.1002/stem.3388
32. Bengoa-Vergniory N, Kypta RM. Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cell Mol Life Sci (2015) 72(21):4157–72. doi: 10.1007/s00018-015-2028-6
33. Ille F, Sommer L. Wnt signaling: multiple functions in neural development. Cell Mol Life Sci CMLS. (2005) 62(10):1100–8. doi: 10.1007/s00018-005-4552-2
34. Ohkawara B, Glinka A, Niehrs C. Rspo3 binds syndecan 4 and induces wnt/PCP signaling via clathrin-mediated endocytosis to promote morphogenesis. Dev Cell (2011) 20(3):303–14. doi: 10.1016/j.devcel.2011.01.006
35. Fedi P, Bafico A, Soria AN, Burgess WH, Miki T, Bottaro DP, et al. Isolation and biochemical characterization of the human dkk-1 homologue, a novel inhibitor of mammalian wnt signaling*. J Biol Chem (1999) 274(27):19465–72. doi: 10.1074/jbc.274.27.19465
36. Nagy N, Barad C, Hotta R, Bhave S, Arciero E, Dora D, et al. Collagen 18 and agrin are secreted by neural crest cells to remodel their microenvironment and regulate their migration during enteric nervous system development. Dev (Cambridge England) (2018) 145(9). doi: 10.1242/dev.160317
37. Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, et al. Molecular architecture of the mouse nervous system. Cell (2018) 174(4):999–1014.e22. doi: 10.1016/j.cell.2018.06.021
38. Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature (2003) 423(6938):448–52. doi: 10.1038/nature01611
39. Yu J, Chia J, Canning CA, Jones CM, Bard FA, Virshup DM. WLS retrograde transport to the endoplasmic reticulum during Wnt secretion. Dev Cell (2014) 29(3):277–91. doi: 10.1016/j.devcel.2014.03.016
40. Koo B-K, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature (2012) 488(7413):665–9. doi: 10.1038/nature11308
41. Hao H-X, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature (2012) 485(7397):195–200. doi: 10.1038/nature11019
42. Leven P, Schneider R, Siemens KD, Jackson WS, Wehner S. Application of a RiboTag-based approach to generate and analyze mRNA from enteric neural cells. Neurogastroenterol Motil (2022) 34(7):e14309. doi: 10.1111/nmo.14309
43. Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods (2008) 5(10):877–9. doi: 10.1038/nmeth.1253
44. Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn (2012) 14(1):22–9. doi: 10.1016/j.jmoldx.2011.08.002
45. Fu J, Jiang M, Mirando AJ, Yu H-MI, Hsu W. Reciprocal regulation of Wnt and Gpr177/mouse Wntless is required for embryonic axis formation. Proc Natl Acad Sci (2009) 106(44):18598–603. doi: 10.1073/pnas.0904894106
46. Chai G, Szenker-Ravi E, Chung C, Li Z, Wang L, Khatoo M, et al. A human pleiotropic multiorgan condition caused by deficient wnt secretion. N Engl J Med (2021) 385(14):1292–301. doi: 10.1056/NEJMoa2033911
47. Voloshanenko O, Erdmann G, Dubash TD, Augustin I, Metzig M, Moffa G, et al. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat Commun (2013) 4(1):2610. doi: 10.1038/ncomms3610
48. Wang L-T, Wang S-J, Hsu S-H. Functional characterization of mammalian Wntless homolog in mammalian system. Kaohsiung J Med Sci (2012) 28(7):355–61. doi: 10.1016/j.kjms.2012.02.001
49. Tian X, Zhou B. Strategies for site-specific recombination with high efficiency and precise spatiotemporal resolution. J Biol Chem (2021) 296:100509. doi: 10.1016/j.jbc.2021.100509
50. Mehta S, Hingole S, Chaudhary V. The emerging mechanisms of wnt secretion and signaling in development. Front Cell Dev Biol (2021) 9. doi: 10.3389/fcell.2021.714746
51. Kamimura K, Ueno K, Nakagawa J, Hamada R, Saitoe M, Maeda N. Perlecan regulates bidirectional Wnt signaling at the Drosophila neuromuscular junction. J Cell Biol (2013) 200(2):219–33. doi: 10.1083/jcb.201207036
52. Mii Y, Takada S. Heparan sulfate proteoglycan clustering in wnt signaling and dispersal. Front Cell Dev Biol (2020) 8:631. doi: 10.3389/fcell.2020.00631
53. Smolich BD, McMahon JA, McMahon AP, Papkoff J. Wnt family proteins are secreted and associated with the cell surface. Mol Biol Cell (1993) 4(12):1267–75. doi: 10.1091/mbc.4.12.1267
54. Mii Y, Yamamoto T, Takada R, Mizumoto S, Matsuyama M, Yamada S, et al. Roles of two types of heparan sulfate clusters in Wnt distribution and signaling in Xenopus. Nat Commun (2017) 8(1):1973. doi: 10.1038/s41467-017-02076-0
55. Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol (2009) 1(3):a002493. doi: 10.1101/cshperspect.a002493
56. Bannerman PG, Mirsky R, Jessen KR, Timpl R, Duance VC. Light microscopic immunolocalization of laminin, type IV collagen, nidogen, heparan sulphate proteoglycan and fibronectin in the enteric nervous system of rat and Guinea pig. J Neurocytol (1986) 15(6):733–43. doi: 10.1007/BF01625191
57. Cosin-Roger J, Ortiz-Masià MD, BarraChina MD. Macrophages as an emerging source of wnt ligands: relevance in mucosal integrity. Front Immunol (2019) 10. doi: 10.3389/fimmu.2019.02297
58. Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin JP, Davenport AP, et al. International Union of Pharmacology. XLVI. G protein-coupled receptor list. Pharmacol Rev (2005) 57(2):279–88. doi: 10.1124/pr.57.2.5
59. Niehrs C, Shen J. Regulation of lrp6 phosphorylation. Cell Mol Life Sci (2010) 67(15):2551–62. doi: 10.1007/s00018-010-0329-3
60. Veeman MT, Axelrod JD, Moon RT. A second canon: functions and mechanisms of β-catenin-independent wnt signaling. Dev Cell (2003) 5(3):367–77. doi: 10.1016/S1534-5807(03)00266-1
61. De A. Wnt/Ca2+ signaling pathway: a brief overview. Acta Biochim Biophys Sin (2011) 43(10):745–56. doi: 10.1093/abbs/gmr079
62. Neckel PH, Mohr R, Zhang Y, Hirt B, Just L. Comparative microarray analysis of proliferating and differentiating murine ENS progenitor cells. Stem Cells Int (2016) 2016:9695827. doi: 10.1155/2016/9695827
63. Hsieh J-C, Rattner A, Smallwood PM, Nathans J. Biochemical characterization of Wnt-Frizzled interactions using a soluble, biologically active vertebrate Wnt protein. Proc Natl Acad Sci (1999) 96(7):3546–51. doi: 10.1073/pnas.96.7.3546
64. Sagara N, Toda G, Hirai M, Terada M, Katoh M. Molecular cloning, differential expression, and chromosomal localization of human frizzled-1, frizzled-2, and frizzled-7. Biochem Biophys Res Commun (1998) 252(1):117–22. doi: 10.1006/bbrc.1998.9607
65. Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol (2003) 63(6):1256–72. doi: 10.1124/mol.63.6.1256
66. Yu H, Smallwood PM, Wang Y, Vidaltamayo R, Reed R, Nathans J. Frizzled 1 and frizzled 2 genes function in palate, ventricular septum and neural tube closure: general implications for tissue fusion processes. Development (2010) 137(21):3707–17. doi: 10.1242/dev.052001
67. Yanshu W, Nini G, Jeremy N. The role of frizzled3 and frizzled6 in neural tube closure and in the planar polarity of inner-ear sensory hair cells. J Neurosci (2006) 26(8):2147. doi: 10.1523/JNEUROSCI.22-19-08563.2002
68. Lyuksyutova AI, Lu C-C, Milanesio N, King LA, Guo N, Wang Y, et al. Anterior-posterior guidance of commissural axons by wnt-frizzled signaling. Science (2003) 302(5652):1984–8. doi: 10.1126/science.1089610
69. Yanshu W, Nupur T, Philip MS, Jennifer PM, Jeremy N. <em<Frizzled-3</em< Is required for the development of major fiber tracts in the rostral CNS. J Neurosci (2002) 22(19):8563. doi: 10.1523/JNEUROSCI.4698-05.2005
70. Alrefaei AF, Münsterberg AE, Wheeler GN. FZD10 regulates cell proliferation and mediates Wnt1 induced neurogenesis in the developing spinal cord. PloS One (2020) 15(6):e0219721. doi: 10.1371/journal.pone.0219721
71. Phesse T, Flanagan D, Vincan E. Frizzled7: A promising achilles’ Heel for targeting the wnt receptor complex to treat cancer. Cancers (Basel) (2016) 8(5). doi: 10.3390/cancers8050050
72. Huang H-C, Klein PS. The Frizzled family: receptors for multiple signal transduction pathways. Genome Biol (2004) 5(7):234. doi: 10.1186/gb-2004-5-7-234
73. Linker C, Lesbros C, Gros J, Burrus LW, Rawls A, Marcelle C. beta-Catenin-dependent Wnt signalling controls the epithelial organisation of somites through the activation of paraxis. Development (2005) 132(17):3895–905. doi: 10.1242/dev.01961
74. De Calisto J, Araya C, Marchant L, Riaz CF, Mayor R. Essential role of non-canonical Wnt signalling in neural crest migration. Development (2005) 132(11):2587–97. doi: 10.1242/dev.01857
75. Flanagan DJ, Phesse TJ, Barker N, Schwab RH, Amin N, Malaterre J, et al. Frizzled7 functions as a Wnt receptor in intestinal epithelial Lgr5(+) stem cells. Stem Cell Rep (2015) 4(5):759–67. doi: 10.1016/j.stemcr.2015.03.003
76. Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet (1998) 19(4):379–83. doi: 10.1038/1270
77. Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature (2011) 469(7330):415–8. doi: 10.1038/nature09637
78. Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology (2012) 143(6):1518–29.e7. doi: 10.1053/j.gastro.2012.08.031
79. Vincan E, Barker N. The upstream components of the Wnt signalling pathway in the dynamic EMT and MET associated with colorectal cancer progression. Clin Exp Metastasis (2008) 25(6):657–63. doi: 10.1007/s10585-008-9156-4
80. Ueno K, Hazama S, Mitomori S, Nishioka M, Suehiro Y, Hirata H, et al. Down-regulation of frizzled-7 expression decreases survival, invasion and metastatic capabilities of colon cancer cells. Br J Cancer (2009) 101(8):1374–81. doi: 10.1038/sj.bjc.6605307
81. Ishitani T, Kishida S, Hyodo-Miura J, Ueno N, Yasuda J, Waterman M, et al. The TAK1-NLK mitogen-activated protein kinase cascade functions in the wnt-5a/ca2+ Pathway to antagonize wnt/β-catenin signaling. Mol Cell Biol (2003) 23(1):131–9. doi: 10.1128/MCB.23.1.131-139.2003
82. Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PloS Biol (2006) 4(4):e115. doi: 10.1371/journal.pbio.0040115
83. Nothelfer K, Obermayr F, Belz N, Reinartz E, Bareiss PM, Bühring H-J, et al. Expression of the wnt receptor frizzled-4 in the human enteric nervous system of infants. Stem Cells Int (2016) 2016:9076823. doi: 10.1155/2016/9076823
84. Cruciat CM, Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb Perspect Biol (2013) 5(3):a015081. doi: 10.1101/cshperspect.a015081
85. Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene (2006) 25(57):7469–81. doi: 10.1038/sj.onc.1210054
86. Solberg N, Machon O, Krauss S. Effect of canonical Wnt inhibition in the neurogenic cortex, hippocampus, and premigratory dentate gyrus progenitor pool. Dev dynamics (2008) 237(7):1799–811. doi: 10.1002/dvdy.21586
87. Verani R, Cappuccio I, Spinsanti P, Gradini R, Caruso A, Magnotti MC, et al. Expression of the Wnt inhibitor Dickkopf-1 is required for the induction of neural markers in mouse embryonic stem cells differentiating in response to retinoic acid. J neurochem (2007) 100(1):242–50. doi: 10.1111/j.1471-4159.2006.04207.x
88. Kunke D, Bryja V, Mygland L, Arenas E, Krauss S. Inhibition of canonical Wnt signaling promotes gliogenesis in P0-NSCs. Biochem Biophys Res Commun (2009) 386(4):628–33. doi: 10.1016/j.bbrc.2009.06.084
89. Ribeiro D, Ellwanger K, Glagow D, Theofilopoulos S, Corsini NS, Martin-Villalba A, et al. Dkk1 regulates ventral midbrain dopaminergic differentiation and morphogenesis. PloS One (2011) 6(2):e15786. doi: 10.1371/journal.pone.0015786
90. Causeret F, Sumia I, Pierani A. Kremen1 and Dickkopf1 control cell survival in a Wnt-independent manner. Cell Death Differ (2016) 23(2):323–32. doi: 10.1038/cdd.2015.100
91. Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature (2002) 417(6889):664–7. doi: 10.1038/nature756
92. Kim KA, Zhao J, Andarmani S, Kakitani M, Oshima T, Binnerts ME, et al. R-Spondin proteins: a novel link to beta-catenin activation. Cell Cycle (2006) 5(1):23–6. doi: 10.4161/cc.5.1.2305
93. Kazanskaya O, Glinka A, del Barco Barrantes I, Stannek P, Niehrs C, Wu W. R-Spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev Cell (2004) 7(4):525–34. doi: 10.1016/j.devcel.2004.07.019
94. de Lau W, Barker N, Low TY, Koo B-K, Li VSW, Teunissen H, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature (2011) 476(7360):293–7. doi: 10.1038/nature10337
95. de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev (2014) 28(4):305–16. doi: 10.1101/gad.235473.113
96. van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol (2005) 7(4):381–6. doi: 10.1038/ncb1240
97. Yan KS, Janda CY, Chang J, Zheng GXY, Larkin KA, Luca VC, et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature (2017) 545(7653):238–42. doi: 10.1038/nature22313
98. Yu Y, Moberly AH, Bhattarai JP, Duan C, Zheng Q, Li F, et al. The stem cell marker lgr5 defines a subset of postmitotic neurons in the olfactory bulb. J Neurosci (2017) 37(39):9403–14. doi: 10.1523/JNEUROSCI.0500-17.2017
99. Miller TE, Wang J, Sukhdeo K, Horbinski C, Tesar PJ, Wechsler-Reya RJ, et al. Lgr5 marks post-mitotic, lineage restricted cerebellar granule neurons during postnatal development. PloS One (2014) 9(12):e114433. doi: 10.1371/journal.pone.0114433
100. Dubey R, van Kerkhof P, Jordens I, Malinauskas T, Pusapati GV, McKenna JK, et al. R-spondins engage heparan sulfate proteoglycans to potentiate WNT signaling. Elife (2020) 9. doi: 10.7554/eLife.54469
101. Przybyla L, Lakins Johnathon N, Weaver Valerie M. Tissue mechanics orchestrate wnt-dependent human embryonic stem cell differentiation. Cell Stem Cell (2016) 19(4):462–75. doi: 10.1016/j.stem.2016.06.018
102. Barbolina MV, Burkhalter RJ, Stack MS. Diverse mechanisms for activation of Wnt signalling in the ovarian tumour microenvironment. Biochem J (2011) 437(1):1–12. doi: 10.1042/BJ20110112
103. Du J, Zu Y, Li J, Du S, Xu Y, Zhang L, et al. Extracellular matrix stiffness dictates Wnt expression through integrin pathway. Sci Rep (2016) 6(1):20395. doi: 10.1038/srep20395
104. Mazzuoli-Weber G, Schemann M. Mechanosensitivity in the enteric nervous system. Front Cell Neurosci (2015) 9:408. doi: 10.3389/fncel.2015.00408
105. Seguella L, Gulbransen BD. Enteric glial biology, intercellular signalling and roles in gastrointestinal disease. Nat Rev Gastroenterol Hepatol (2021) 18(8):571–87. doi: 10.1038/s41575-021-00423-7
106. Schulte DM, Kragelund D, Müller N, Hagen I, Elke G, Titz A, et al. The wingless-related integration site-5a/secreted frizzled-related protein-5 system is dysregulated in human sepsis. Clin Exp Immunol (2015) 180(1):90–7. doi: 10.1111/cei.12484
107. Huang Y, Xue Q, Chang J, Wang X, Miao C. Wnt5a: A promising therapeutic target for inflammation, especially rheumatoid arthritis. Cytokine (2023) 172:156381. doi: 10.1016/j.cyto.2023.156381
Keywords: in situ hybridization, canonical Wnt signaling, non-canonical Wnt signaling, Dickkopf, R-Spondin, enteric nervous system, RNA sequencing, enteric gliosis
Citation: Scharr M, Hirt B and Neckel PH (2024) Spatial gene expression profile of Wnt-signaling components in the murine enteric nervous system. Front. Immunol. 15:1302488. doi: 10.3389/fimmu.2024.1302488
Received: 26 September 2023; Accepted: 02 January 2024;
Published: 18 January 2024.
Edited by:
Reiner Schneider, University Hospital Bonn, GermanyReviewed by:
Ulrika Marklund, Karolinska Institutet (KI), SwedenNandor Nagy, Semmelweis University, Hungary
Copyright © 2024 Scharr, Hirt and Neckel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter H. Neckel, cGV0ZXIubmVja2VsQHVuaS10dWViaW5nZW4uZGU=
 Melanie Scharr
Melanie Scharr Bernhard Hirt
Bernhard Hirt Peter H. Neckel
Peter H. Neckel