- 1Department of General Surgery, Clinical Laboratory of Integrative Medicine, The First Affiliated Hospital of Dalian Medical University, Dalian, China
- 2Department of Cardiology, The Second Hospital of Dalian Medical University, Dalian, China
- 3Institute (College) of Integrative Medicine, Dalian Medical University, Dalian, China
Overcoming the immunosuppressive tumor microenvironment and identifying widely used immunosuppressants with minimal side effects are two major challenges currently hampering cancer immunotherapy. Regulatory T cells (Tregs) are present in almost all cancer tissues and play an important role in preserving autoimmune tolerance and tissue homeostasis. The tumor inflammatory microenvironment causes the reprogramming of Tregs, resulting in the conversion of Tregs to immunosuppressive phenotypes. This process ultimately facilitates tumor immune escape or tumor progression. However, current systemic Treg depletion therapies may lead to severe autoimmune toxicity. Therefore, it is crucial to understand the mechanism of Treg reprogramming and develop immunotherapies that selectively target Tregs within tumors. This article provides a comprehensive review of the potential mechanisms involved in Treg cell reprogramming and explores the application of Treg cell immunotherapy. The interference with reprogramming pathways has shown promise in reducing the number of tumor-associated Tregs or impairing their function during immunotherapy, thereby improving anti-tumor immune responses. Furthermore, a deeper understanding of the mechanisms that drive Treg cell reprogramming could reveal new molecular targets for future treatments.
1 Introduction
Immune checkpoint blockade therapy has shown significant advancements in cancer treatment. However, it is important to recognize that this treatment can only cure a limited proportion of patients, while the majority may not benefit from it and could potentially experience autoimmune diseases and unforeseen toxicities (1, 2). To address these challenges, we can focus on two key aspects: overcoming the immunosuppressive tumor microenvironment and improving local-specific antitumor immunity (3–5). One potential target to achieve these goals is Tregs. Additionally, cancer is strongly influenced by the presence of immune cells and the levels of inflammatory cytokines. In tissues experiencing inflammation, they are also important in avoiding severe tissue injury (6, 7). However, immune cell malfunctions can cause inflammation to advance into cancer.
Regulatory T cells (Tregs) are recognized as a specific subset of CD4+ T cells. Foxp3 is a unique transcription factor (8). According to their origin, we can divide them into thymic Tregs (tTregs)/natural Tregs (nTregs) and peripheral Tregs (pTregs)/inducible Tregs (iTregs) (9). Abul K Abbas et al. proposed a unified classification of Tregs into tTregs and pTregs (10). The tTregs are produced by the thymus and are tissue-intrinsic Tregs. After being stimulated by self-antigens, they are transported to the periphery and exhibit inhibitory activity against self-antigens (11, 12). pTregs are differentiated from naïve T cells or conventional T cells in peripheral tissues, or induced by TGF-β in vitro. pTregs play a role in preventing autoimmunity caused by foreign antigens (13). According to the function and phenotype of Tregs, they are divided into naïve Tregs (or resting Tregs), effector Tregs (eTregs), and non-Tregs. Naïve Tregs manifested as CD4+FOXP3low CD25hiTregs, eTregs manifested as CD4+FOXP3hi CD25hiTregs, and non-Tregs manifested as CD4+FOXP3low Tregs (14). Under steady-state conditions, naïve Tregs have weak immunosuppressive functions and eTregs have strong immunosuppressive functions. Non-Tregs have no suppressive function and secrete pro-inflammatory factors (14). In the inflammatory environment, nonTregs are also known as “exTreg” or “exFoxp3” cells (15).
In the tumor microenvironment (TME), naïve Tregs and exTregs will be converted into tumor-infiltrating Tregs (TI-Tregs) under various stimuli. TI-Tregs have the same phenotype as eTregs (14). This process is called Tregs reprogramming. Infiltration of reprogrammed Tregs into tumors is associated with a negative prognosis (16). Thus, targeting the reprogramming process of Tregs and paying attention to their altered state in both inflammation and cancer could potentially provide new perspectives for cancer immunotherapy. Therefore, this article aims to deeply explore the mechanism of reprogramming exTregs into immunosuppressive Tregs.
Recent research has shown that specific signaling molecules found within the tumor microenvironment, along with factors generated by immune cells and tumor cells, have the potential to initiate the reprogramming of exTregs (17, 18). In the hypoxic, low-glucose, and high-lactic acid conditions of the tumor microenvironment, exTregs undergo metabolic adaptability through reprogramming, gradually transitioning towards an immunosuppressive phenotype (19–21). Furthermore, Foxp3, the most crucial transcription factor regulating Tregs, undergoes post-translational modification involving methylation, acetylation, glycosylation, phosphorylation, and ubiquitination. The function and reprogramming of Tregs rely heavily on these regulatory mechanisms (9, 22, 23).
This work aims to present a summary of the mechanism of reprogramming exTregs into eTregs, from the following three aspects: (1) signaling molecules and intracellular signaling pathways in the tumor microenvironment; (2) metabolic and energy pathways including glycolysis, amino acid metabolism, fatty acid oxidation (FAO) and mevalonate pathways, mitochondrial oxidative phosphorylation and complexes (OXPHOS), and metabolite pathways in TME; (3) the modification of Foxp3 expression at the transcriptional level, such as methylation, acetylation, glycosylation, phosphorylation, and ubiquitination of Foxp3, affects the immunosuppressive function of Tregs.
2 Immunosuppressive immune cells within TME
Targeting specific immune cells, especially Tregs, and inhibiting their immunosuppressive functions is a crucial approach in current immunotherapy. Additionally, other immune cells such as regulatory B cells (Bregs), regulatory dendritic cells (DCregs), myeloid-derived suppressor cells (MDSCs), and tumor-associated macrophages (TAMs) also possess immunosuppressive capabilities (24). Bregs, which constitute only 10% of circulating B cells in healthy individuals, exert their suppressive effects on autoreactive B cells through the secretion of soluble molecules and the expression of inhibitory molecules. However, due to the lack of a specific transcription factor defining Bregs, there is currently limited research in this area (25). DCregs, which are subpopulations of DCs in central and peripheral lymph, exhibit lower levels of MHC and costimulatory molecules on their surface. DCregs exert immunosuppressive functions mainly by inducing the inactivation of autoreactive T cells and Tregs differentiation, and increasing the levels of inhibitory molecules and cytokines such as IL-10 (24, 26). Similarly, MDSCs, known for their immunosuppressive function, inhibit T cell responses, induce Tregs, and differentiate into TAMs (27, 28). TAMs have low expression of inhibitory molecules such as MHCII and PD-1, thereby exerting immunosuppressive effects and promoting tumor growth (29, 30).
3 The reprogramming mechanism of exTregs to eTregs
Under normal circumstances, Tregs exhibit stability. However, in specific conditions like an inflammatory tumor microenvironment, certain Tregs undergo reprogramming (31). In most cancers, there is infiltration of eTregs, while in some cancers, both eTregs and exTregs are present (32). This review focuses on elucidating the mechanism of exTregs reprogramming into eTregs from three distinct perspectives.
3.1 Signaling molecules and pathways
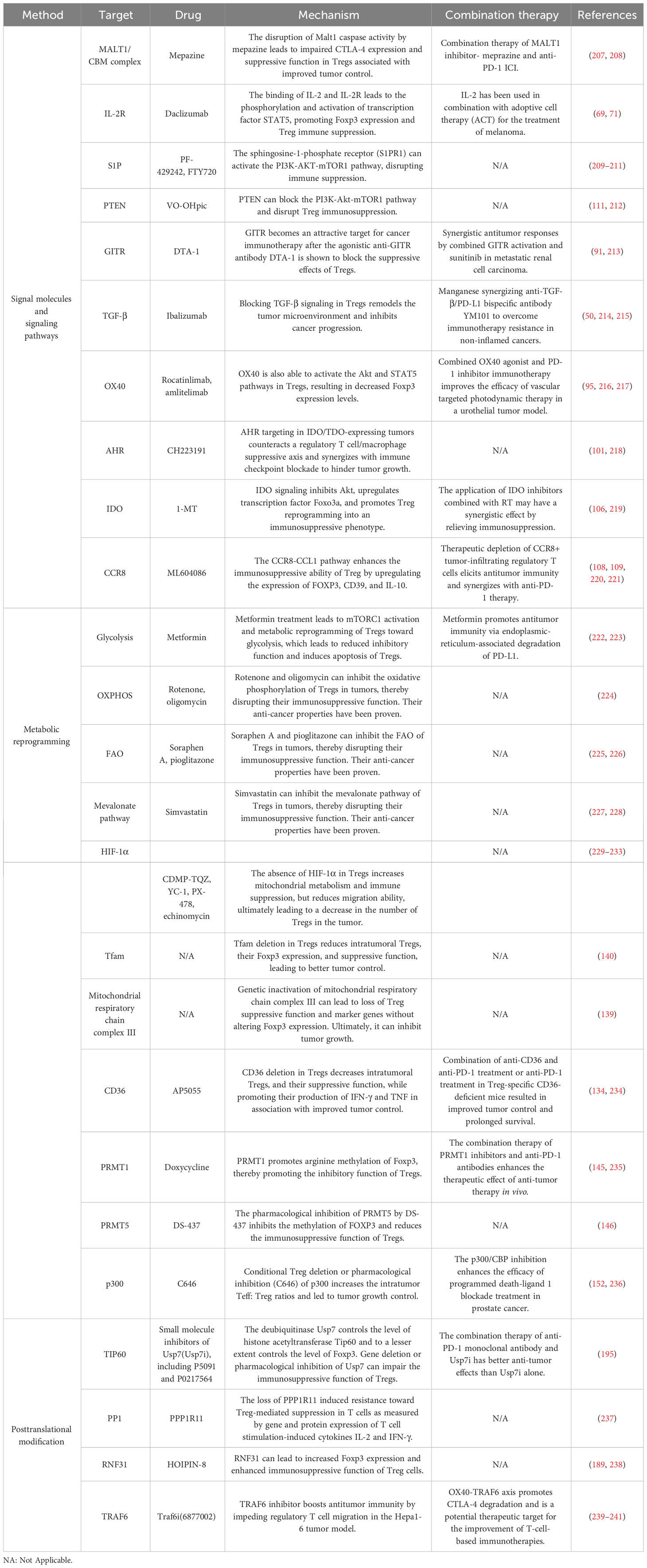
Recent research has demonstrated that certain signaling molecules within TME can cause exTregs to undergo reprogramming (Figure 1). This reprogramming process can lead to a transition of exTregs from their typical immunoregulatory state to a state that promotes tumor growth.
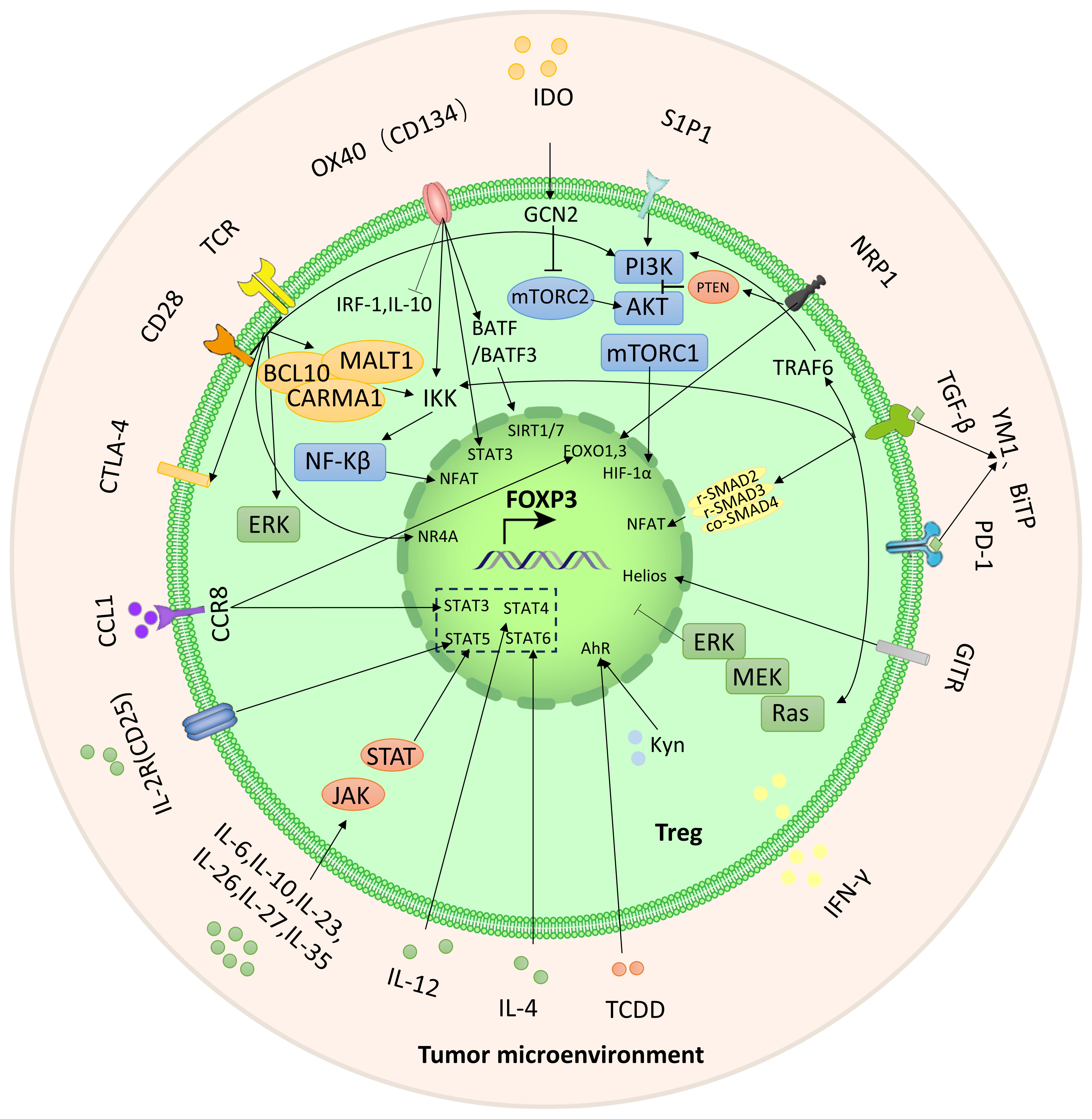
Figure 1 The crucial signals and pathways for the reprogramming of Tregs. Reprogramming of Tregs from inflammatory to immunosuppressive activity is achieved through these signaling molecules and pathways (as highlighted by the lines and arrows in the figure).
3.1.1 TCR/CD28
The leukocyte surface differentiation antigen 28 (CD28) costimulatory receptor forms microclusters with the T cell receptor (TCR) and plays a dual role in the reprogramming of Tregs (33). The TCR-CD28 microclusters trigger various cytoplasmic signaling pathways, such as the induction of the CARMA1-BCL10-MALT1 (CBM) complex. The CBM complex further stimulates IKK-mediated NF-κB activation, which subsequently activates the transcription factor NFAT and promotes immunosuppression (34–37). In addition, TCR/CD28 costimulation can induce cytotoxic T-lymphocyte antigen-4 (CTLA-4). The high expression of CTLA-4 on Tregs supports its immunosuppressive activity, so anti-CTLA-4 antibody has been developed as an immune checkpoint inhibitor (38, 39).
Notably, TCR/CD28 can also subvert immunosuppression, primarily through the phosphatidylinositol 3-kinase (PI3K)-AKT-mammalian target of the rapamycin (mTOR) pathway. It is also crucial for metabolism. In addition to TCR/CD28, sphingosine-1-phosphate receptor 1 (S1PR1) and interleukin-2 receptor (IL-2R) can also activate the PI3K-AKT-mTOR1 signaling pathway (40, 41). AKT can inhibit the activity of transcription factors FOXO1 and FOXO3 and inhibit the expression of Foxp3 (42). mTORC1 can bind with the transcription factor known as hypoxia-inducible factor (HIF-1α). Additionally, HIF-1α exerts inhibitory effects on the expression of Foxp3 (43, 44). Studies by Sharma et al. have shown that neuropilin-1 (Nrp1) in Tregs can inhibit this pathway through phosphatase and tensin homolog (PTEN), and lead to increased nuclear translocation of FOXO1 and FOXO3a resulting in increased Foxp3 expression and enhanced immunosuppression (45, 46). In addition, TCR/CD28 costimulation can also affect the expression of Foxp3 through the nuclear receptor Nr4a, extracellular signal-regulated kinase (ERK), Ca2+, and protein kinases PKA and PKC (47, 48).
3.1.2 Transforming growth factor beta
TGF-β exerts its influence on Treg function through both Smad-dependent and Smad-independent pathways, which may have different effects in different contexts. Smad protein serves as a downstream effector transcription factor in the TGF-β signaling pathway. R-Smad is activated when activated TGF-β interacts with its receptor. R-smad2 and r-smad3 form a complex with co-smad4 and bind to the CNS1 region of Foxp3 through the transcription factor NFAT, thereby promoting Foxp3 expression (49, 50).
Apart from the Smad-dependent signaling pathway, the activated TGF-β receptor (TGFβR) can recruit TAK1 (TGF-β-activated kinase 1) to further activate the NF-κB pathway and enhance Foxp3 expression (51). Moreover, TGF-β stimulates the PI3K-AKT-mTOR pathway through the interaction of activated TGFβRI and TGFβRII with p85 (the regulatory unit of PI3K), then inhibits Foxp3 expression (52). Nrp1-activated PTEN can block the PI3K-AKT-mTOR pathway. Moreover, TGFβR assembles the ShcA-Grb2-SOS complex by phosphorylating ShcA and activates the Ras-MEK-ERK pathway, and the MEK-ERK-dependent pathway can inhibit Foxp3 expression (53). Through the TRAF6-mediated pathway, TGF-β induces ubiquitination of p85, thereby initiating the activation of AKT-mTOR and inhibiting Foxp3 expression (50).
Disruption of TGF-β signaling underlies inflammatory diseases (54). In the early stages of cancer, TGF-β induces apoptosis in precancerous cells and inhibits tumor cell proliferation. As tumors progress, tumor-derived TGF-β can trigger tumorigenic and pro-metastatic responses in cancer cells and stroma, including the formation of an immunosuppressive tumor microenvironment (55, 56). At this time, TGF-β can affect a variety of immune cells, including inhibiting the proliferation and function of T cells, inducing the differentiation of naïve T cells into regulatory T cells, etc (54). At present, great progress has been made in immunotherapy targeting TGF-β-mediated immunosuppression, including TGF-β mRNA-directed agents, ligand traps, antibodies, fusion proteins and small-molecule kinase inhibitors of TGFβRs (57). Mouse model studies have shown a strong synergistic effect between TGF-β pathway inhibitors and ICIs (58). In particular, the anti-PD-L1/TGF-βR bispecific antibodies YM1 and BiTP can effectively inhibit the effect of TGF-β-Smad, have better anticancer effects than antibodies that antagonize TGF-β alone, and can also restore the response to PD- L1 drug resistance (59–62).
3.1.3 IL family and transcription factor STAT family
As a family of cytokines, the interleukin (IL) family is important in TME. According to the homology of cytokines, the IL family can be divided into IL-1, IL-2, IL-6, IL-10, IL-12, IL-17 families, etc. IL-33 and IL-36, which are part of the IL-1 family, play a role in the activation of Tregs (63, 64). IL-36γ signals through the IL-36R, MyD88, and NFκBp50 in CD4+ T cells, effectively inhibiting the development of Foxp3-expressing Tregs (64). The STAT family, a transcriptional activator, could control the expression of Foxp3. A significant correlation between IL family and STAT family has been observed (65–67). IL-2 is crucial for Treg function. The combination of IL-2 and IL-2R phosphorylates and activates the transcription factor STAT5, which in turn increases Foxp3 expression (68–70). CD25, a component of IL-2R, can be targeted by daclizumab to effectively reduce the expression of Foxp3 and increase the secretion of IFN-γ (71, 72). This phenomenon could potentially occur via the IFN-γ-mediated polarization of Tregs towards Th1-like effector T cells (Teffs) (73). Additionally, IL-2 is also required for CD8+ T cells to remain activated and continue to exhibit cytotoxic effects. Tregs, which have high IL-2R expression, impede the activation of Teffs and induce apoptosis of these cells, thereby mediating immune suppression (74, 75). Furthermore, IL-4 activates STAT6 and inhibits the transcription of Foxp3, while IL-12 has been found to regulate STAT4 and inhibit the expression of Foxp3 (76–79).
It is worth noting that IL-6, IL-10, IL-23, IL-26, IL-27, and IL-35 can regulate STAT3 and participate in the JAK-STAT pathway, which in turn affects the expression of Foxp3 (80–85). This suggests that STAT3 can be activated by various cytokines, leading to an impact on Tregs. The effects of certain factors on Tregs can be the opposite, such as the pro-inflammatory properties of IL-6 and the anti-inflammatory properties of IL-10. This suggests that STAT3 may play different roles in Tregs (80). Among these factors, IL-26 is a constituent of the IL-10 family, while IL-23, IL-27, and IL-35 are part of the IL-12 family. The influence of IL-27 on the promotion or suppression of Tregs is influenced by multiple factors and requires further investigation (84).
3.1.4 Glucocorticoid-induced tumor necrosis factor receptor
Glucocorticoid-induced tumor necrosis factor receptor (GITR), a TNFR, acts as a costimulatory molecule and has high expression levels on Tregs. Antibodies targeting GITR have shown promising antitumor effects. These antibodies not only reduce the number of Tregs but also disrupt the reprogramming of Tregs and their immunosuppressive function (86–88). In the inflammatory tumor microenvironment, the transcription factor Helios plays an important role in maintaining the stability of Tregs (89, 90). Treatment with anti-GITR drugs decreases Helios expression, consequently downregulating Foxp3 expression, as well as reducing IL-10 levels and increasing IFN-γ levels (91–93).
3.1.5 OX40
In addition to TCR signaling, the TNF receptor OX40 can also activate IκB kinase β (IKKβ), causing c-Rel and RelA to be translocated to the nucleus. This activation participates in the NF-kB pathway and regulates Foxp3 expression (51, 94). Furthermore, OX40 can activate the Akt and Stat5 pathways, causing temporary proliferation of Tregs and reducing Foxp3 expression levels. However, this will lead to a relative deficiency of IL-2. The use of IL-2 agonists can rescue this situation, enabling Tregs to exert their immunosuppressive function (95). OX40 can also up regulate the expression of BATF and BATF3 in CD4+T cells, and inhibit the expression of Foxp3 by histone deacetylases SIRT1/7 (96). In addition, under the action of OX40 agonist, the expression of transcription factor interferon regulatory factor 1(IRF1) and the production of IL-10 of Tregs were inhibited, inhibiting the immunosuppressive function of Tregs (97, 98).
3.1.6 Aryl hydrocarbon receptor
2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), an environmental pollutant, exhibits a strong affinity for the AhR (99). AhR serves as a crucial transcription factor that regulates genes associated with inflammation and subsequently influences Tregs. According to recent scientific investigations, it has been uncovered that TCDD possesses the capability to stimulate AhR. Consequently, AhR forms a connection with the DRE sequence situated within the Foxp3 promoter, effectively augmenting the expression of Foxp3 (99, 100). L-kynurenine (Kyn), a tryptophan metabolite of indoleamine 2,3-dioxygenase (IDO), interacts with AhR to enhance the expression of Foxp3 (101).
3.1.7 Cdc42 GTP enzyme
Cdc42 is a Rho family GTPase. Recent studies have shown that inhibiting Cdc42 GTPase or its direct downstream effector WASP can disrupt the reprogramming of Tregs and stimulate anti-tumor immunity. In terms of mechanism, inhibition of Cdc42 and its direct downstream effector WASP works by promoting the non-canonical signaling cascade GATA-binding protein 3 (GATA3)-carbonic anhydrase I (CAI). CAI-mediated pH changes induce anti-tumor T cell immunity (102).
3.1.8 IDO signaling
Under inflammatory conditions, dendritic cells (DCs) signal IDO, which reduces tryptophan concentrations near Tregs. The decline in tryptophan levels triggers the activation of GCN2 kinase, which hinders the functioning of the mTORC2 complex and impedes its phosphorylation of Akt at Ser473 (45, 103). The signaling of Akt in Tregs leads to the impairment of their immunosuppressive function (104–106). IDO signaling inhibits Akt, upregulates transcription factor FOXO3a, and promotes Treg reprogramming into an immunosuppressive phenotype.
3.1.9 Chemokine receptor 8
The CC chemokine CCL1 receptor CCR8 is selectively expressed on Tregs in TME and is associated with poor tumor prognosis (107). The CCR8-CCL1 pathway can increase STAT3 expression and FOXO1 entry into the nucleus, thereby increasing Foxp3 expression. In addition, CCR8-CCL1 can also increase the expression of CD39 and IL-10, jointly promoting Treg reprogramming and enhancing immunosuppressive ability (108, 109).
3.2 Metabolic and energy pathways
Metabolic and energy pathways play a crucial role in the survival and functioning of cells. Similar to other immune cells, Tregs in TME are activated and regulate their metabolic pathways to ensure biosynthesis and energy metabolism for their survival (Figure 2). The metabolic characteristics of the TME are often characterized by hypoxia, low glucose levels, and increased lactate production. Under conditions of inflammation, exTregs signal via the TLR1/2 pathway to enhance glycolysis by triggering the expression of Glut1 in a manner that relies on mTORC1. Simultaneously inhibiting Foxp3 expression and promoting glycolysis by affecting mTORC2 and cMyc. At the same time, the fatty acid oxidation pathway is inhibited. After exTregs are reprogrammed into eTregs with immunosuppressive functions, Foxp3 controls the metabolic activities of Treg cells by promoting FAO while restricting glycolysis by inhibiting the signaling pathways of c-Myc and mTORC2. Additionally, amino acid metabolism, mevalonate metabolism, and oxidative phosphorylation are improved (18–21).
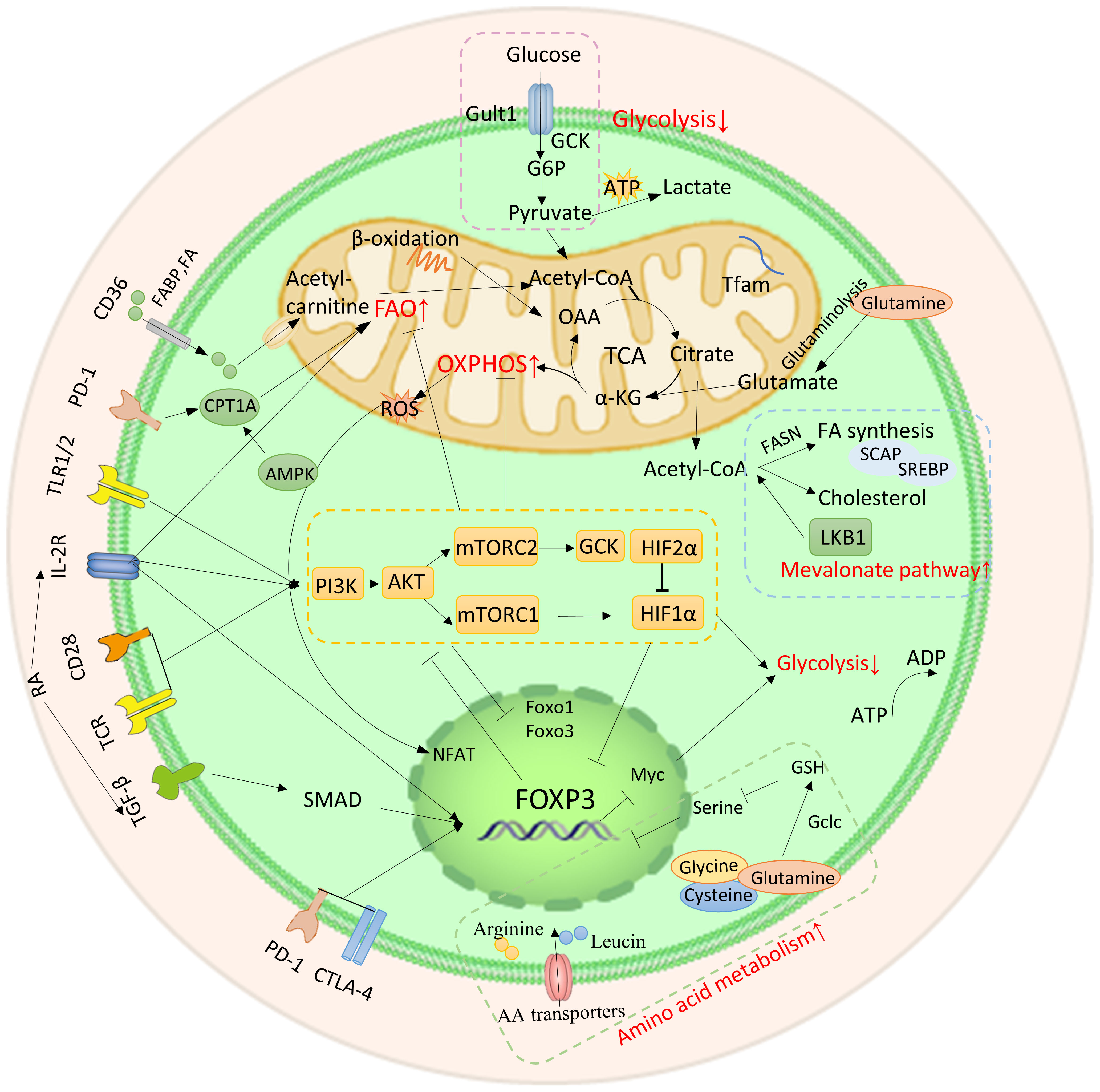
Figure 2 Metabolic and energy pathways reprogrammed by Tregs. After exTregs are reprogrammed into eTregs with immunosuppressive functions, Foxp3 regulates Treg cell metabolism by promoting FAO and inhibiting glycolysis. Meanwhile, amino acid metabolism, mevalonate metabolism and OXPHOS are promoted.
3.2.1 PI3K-Akt-mTORC1 axis
The PI3K-Akt-mTORC1 axis plays an important role in both signaling and metabolism. It is important to highlight that there exists a negative feedback regulatory mechanism between the PI3K-Akt-mTORC1 axis and Foxp3. This pathway can suppress the expression of Foxp3 through transcription factors. Conversely, increased expression of Foxp3 directly inhibits the pathway, resulting in reduced glycolysis and increased OXPHOS and FAO. The Tregs reprogramming process is regulated by the crucial PI3K-Akt-mTOR pathway, which includes important molecules such as AMPK, PTEN, CD28, and HIF-1α (21). Inhibiting PI3K and mTORC1, PTEN and AMPK serve as upstream regulators to enhance Foxp3 expression, thereby playing a significant role in the modulation of Tregs reprogramming (110, 111). The loss of the phosphatase PTEN results in a defect in reprogramming, causing immunosuppressive Tregs to transform into pro-inflammatory Tregs. CD28 promotes the activation of the PI3K-Akt-mTOR pathway (112). Additionally, HIF-2α inhibits HIF-1α and reduces the inhibition of Foxp3 expression by HIF-1α (113).
The mTOR is an important eukaryotic cell signal. Its expression has an impact on Treg formation, proliferation, and activity (114). It also plays an important role in autoimmune disorders and cancer. Sirolimus, often known as rapamycin, is a first-generation mTOR inhibitor. In autoimmune diseases, Treg depletion, mislocalization, or impaired function can be observed (115, 116). This can be improved by rapamycin (117). In contrast, the anticancer effect of rapamycin exceeds its immunosuppressive effect on tumor development (118). A plausible hypothesis is that rapamycin modifies the metabolic and protein synthesis of cancer cells to a greater extent than Tregs’ function (119–121). It’s also important to remember that rapamycin-mediated mTOR blockage may be particularly effective in certain forms of cancer that are preceded by autoimmune inflammation, such as mTOR-dependent cirrhosis that precedes hepatocarcinogenesis in patients with transaldolase deficiency (122, 123).
3.2.2 Glycolysis
Under inflammatory conditions, TLR1/2 receptors on the surface of exTregs activate the PI3K-Akt-mTORC1 pathway, induce the expression of glucose transporter 1 (Glut1), and promote glycolysis (124). After the surface receptor CD28 is activated, it upregulates glucokinase (GCK) through the mTORC2 pathway and promotes glycolysis (125). IL-2R inhibits Foxp3 expression by inhibiting the transcription factors FOXO1 and FOXO3 through Akt (126–128). Nevertheless, the glycolysis inhibition in question arises when Foxp3 effectively dampens the PI3K-Akt-mTORC1 axis via feedback regulation (124). In parallel, the expression of the vital transcription factor Myc is suppressed by Foxp3, leading to the inhibition of glycolysis (129). After reprogramming of exTregs, Foxp3 expression is upregulated and the glycolysis pathway is inhibited.
3.2.3 Amino acid metabolism
Treg reprogramming is largely dependent on amino acid metabolism. For instance, research conducted by Hao Shi et al. showed that arginine and leucine interact with the GTPases Rag A/B and Rheb 1/2. These proteins are necessary to maintain the adaptability of mitochondria and lysosomes, as well as for the expression of Treg suppressor gene signatures (130). Additionally, glutamine, glycine, and cysteine stimulate the expression of Foxp3 and enhance the immunosuppressive ability of Tregs through the activation of glutamate cysteine ligase (Gclc). At the same time, they inhibit serine, which negatively controls Treg function (131).
3.2.4 Fatty acid oxidation and mevalonate metabolism
FAO is essential in promoting the reprogramming of Tregs and improving their immunosuppressive functions. The activation of IL-2R triggers the JAK-AMPK pathway, leading to the activation of FAO (132, 133). Simultaneously, PD-1 increases FAO by upregulating the expression of CPT1A and inducing the production of endogenous lipids. Moreover, the accumulation of free fatty acids (FFA) in TME activates fatty acid-binding protein (FABP) and CD36, thereby further promoting FAO. Notably, loss of CD36 weakens immunosuppressive function (21, 134).
The activation of the mevalonate pathway can be facilitated by liver kinase B1 (LKB1). LKB1 activates the mevalonate pathway by either upregulating the mevalonate gene or regulating intracellular cholesterol homeostasis (135). The mevalonate pathway metabolite geranylgeranyl pyrophosphate (GGPP) enhances STAT5 phosphorylation through IL-2 signaling, which is crucial for reprogramming Tregs to acquire immunosuppressive capabilities (135). Notably, the expression of mevalonate metabolic enzymes, as well as cholesterol synthesis and protein geranylgeranylation, is regulated by SREBP (sterol regulatory element binding protein)/SCAP (SREBP cleavage activating protein) signaling, thereby further supporting the mevalonate pathway (21, 136).
3.2.5 Mitochondrial OXPHOS and mitochondrial complexes
The TME is rich in ROS, which is produced during mitochondrial OXPHOS. These ROS can activate NF-kB in Tregs and enhance the expression of the transcription factor NFAT. NFAT then binds with the non-coding sequence 2 (CNS2) of the upstream enhancer of the Foxp3 gene, leading to increased expression of Foxp3 and contributing to the reprogramming of Tregs (137). Furthermore, Foxp3 has the ability to enhance mitochondrial OXPHOS through the upregulation of genes and proteins involved in the electron transport system within mitochondria (138).
Mitochondrial electron transport chain (ETC) complexes, which are involved in OXPHOS, have been found to have an impact on Tregs function. The mitochondrial respiratory chain consists of five complexes. Alessia Angelin et al. conducted a study that revealed that mitochondrial complex I (also known as CI or NADH) could decrease the immunosuppressive function of Tregs (129). Similarly, Samuel E Weinberg et al. discovered that Tregs lacking complex III still maintained stable expression of Foxp3. However, there was a decrease in the expression of genes linked to Treg function (139). Additionally, in glucose-deprived TME, Tregs deficient in mitochondrial transcription factor A (Tfam) showed a decline in the expression of Foxp3, potentially attributed to increased methylation within the TSDR region of the Foxp3 site (140).
3.2.6 Tumor microenvironment metabolites
Metabolites in the TME also affect Tregs function. As an active derivative of vitamin A, retinoic acid (RA) enhances the expression of Foxp3 by stimulating IL-2 to activate either the downstream JAK/STAT5 pathway or the downstream SMAD signaling pathway via TGF-βR (141, 142). However, there is some controversy surrounding this mechanism. In a different study, it has been observed that the absence of endogenous RA signaling actually enhances the suppressive ability and metabolic adaptation of Tregs. This enhancement was achieved through the stimulation of STAT5 and mTORC1 signaling (143).
3.3 Post-translational modification of Foxp3
Proteins’ posttranslational modifications (PTMs) are important for linking cellular signaling to functional characteristics. PTM refers to the enzymatic process that alters proteins after their synthesis. The Foxp3 protein consists of three functionally significant domains: the N-terminal domain, the zinc finger and leucine zipper regions, and the C-terminal forkhead domain. The stability of Foxp3 expression and Tregs’ immunosuppressive role are facilitated by methylation, acetylation, and glycosylation of the protein (Figure 3). On the other hand, the effects of phosphorylation and ubiquitination are more complex, having both positive and negative impacts (9, 22, 23).
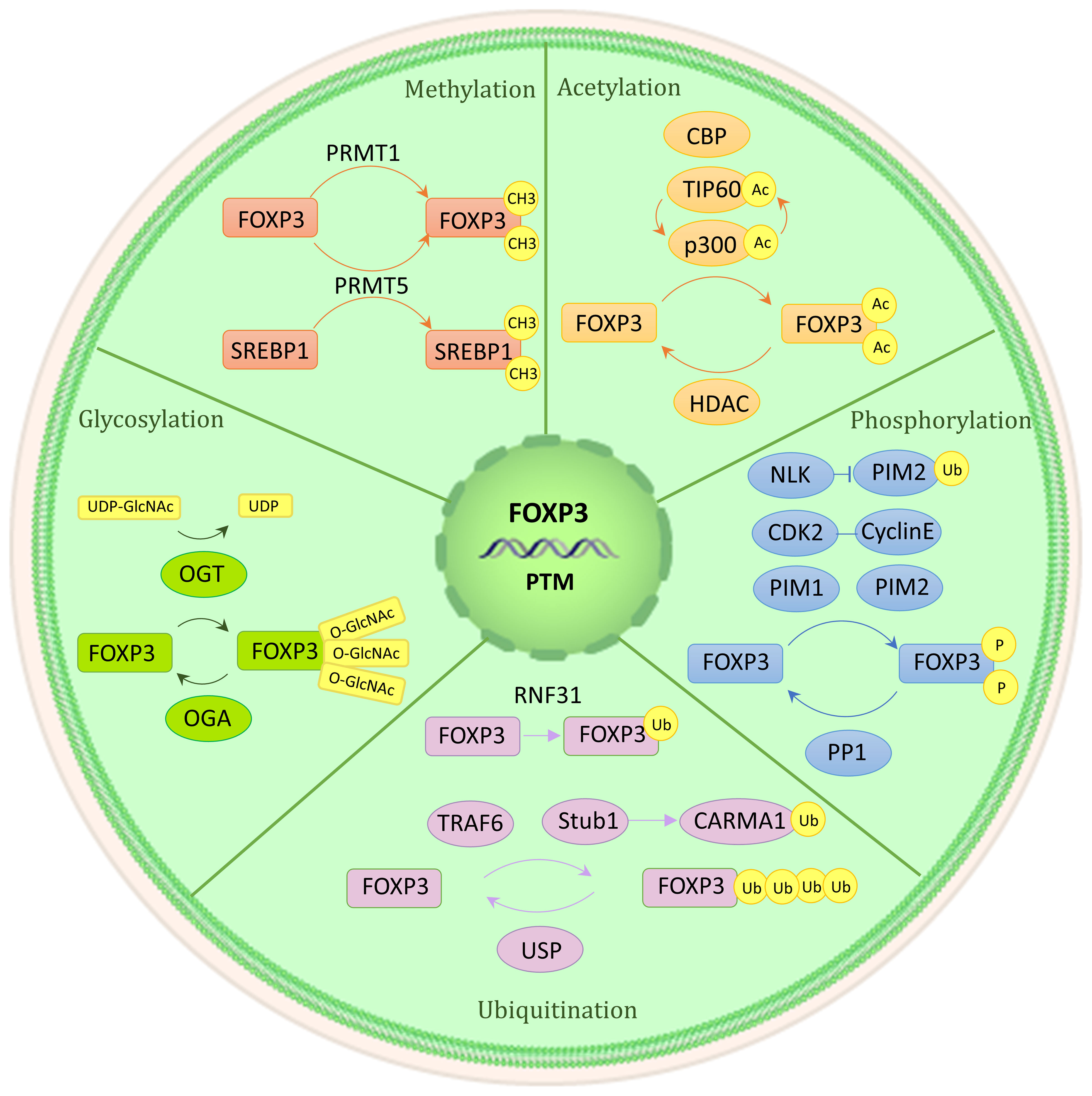
Figure 3 Post-translational modification of Foxp3. Numerous regulatory factors can post-translational modify Foxp3. A brief illustration is presented here, outlining their basic mechanism. Methylation (PRMT1 and PRMT5), acetylation and deacetylation (CBP, p300, TIP60, HDAC), phosphorylation (CDK2, PIM1, PIM2, NLK, PP1), ubiquitination, deubiquitination, and SUMO (RNF31, STUB1, TRAF6, USP1), glycosylation (OGT, OGA).
3.3.1 Methylation
Protein methylation is the enzymatic transfer of a methyl group to certain residues of a protein, such as lysine or arginine. Additional residues that can undergo methylation include histidine, cysteine, and asparagine (144). The methylation of the Foxp3 is significantly impacted by the arginine methyltransferase (PRMT) family members, including PRMT1 and PRMT5 (22). Specifically, PRMT1 adds asymmetric dimethylarginine to Arg-48 and Arg-51 of Foxp3. The immunosuppressive ability of Tregs is greatly reduced when the methylation of these two sites is inhibited (145). Similarly, symmetric dimethylarginines on Foxp3’s Arg-27, Arg-51, and Arg-146 are catalyzed by PRMT5. Among these modifications, methylation silencing on Arg-51 can restrict the inhibitory function (146), potentially because RPMT5 regulates the production of the IL-2R’s γ chain (CD132) (147). Furthermore, PRMT5 can also methylate SREBP1, which can regulate cholesterol biosynthesis, as mentioned previously (148).
3.3.2 Acetylation and deacetylation
Acetylation is a PTM primarily observed on histones and other cellular proteins. Acetyl groups are added to proteins through histone acetyltransferase (HAT), now referred to as acetyltransferase (KAT), which enhances Foxp3 expression and immunosuppression. Conversely, histone deacetylase (HDAC), now known as lysine deacetylase (KDAC), catalyzes the opposite reaction, negatively regulating Foxp3 expression and impairing the immunosuppressive function of Tregs (22, 149). The coordination of Treg formation, function, and reprogramming is greatly aided by these enzymes.
The main HATs involved in Treg acetylation are p300, CBP, and TIP60. Acetylation of Foxp3 by p300 increases the levels of Foxp3 and prevents its degradation by proteases (150). Three acetylation sites, including K31, K262, and K267, have been found using mass spectrometry investigation (151). The loss of p300 leads to a decrease in the immunosuppressive ability of Tregs and an enhancement of anti-tumor immunity (152, 153). CBP, a paralog of p300, also has a significant function in controlling the activity of Treg in specific inflammatory conditions (154). TIP60 and p300 collaborate to control the functioning of Foxp3. p300 forms an interaction with TIP60 and facilitates the autoacetylation of TIP60 at K327. Such adjustment elevates the robustness of the TIP60 protein and accelerates the acetylation process of Foxp3. Similarly, TIP60 also amplifies p300 acetylation and the activity of HAT. When TIP60 and p300 are both present (e.g., at K179 and K227 sites), acetylation of Foxp3 becomes more pronounced (155). Furthermore, if the forkhead domain of Foxp3 undergoes mutation, it disrupts the binding between Foxp3 and TIP60, leading to a reduction of Tregs’ function. Still, this can be reversed by allosteric modifiers that strengthen the interaction between Foxp3 and TIP60 (156, 157).
There are four distinct categories into which HDACs can be classified. HDACs of class I, class II, and class IV function through the dependence on zinc ions (Zn2+). Conversely, sirtuins, known as class III HDACs, make use of nicotinamide adenine dinucleotide (NAD) as a coenzyme (158). For the execution of their role, Class I HDAC1 and HDAC2 attach themselves to the N-terminal section of Foxp3 (159). HDAC3 knockout mice have been found to have significantly impaired immunosuppressive function (160). Class II HDACs include HDAC6,7,9,10. The lack of HDAC6 enhances the expression of numerous genes associated with Tregs, such as IL-10, Lag3, and STAT3, thereby greatly enhancing the suppressive capability of Tregs (161, 162). In the TCR signaling scenario, the separation of HDAC9 from Foxp3 disrupts STAT5 acetylation, which subsequently hinders the functioning of Foxp3 (163–165). On the other hand, HDAC10 facilitates the deacetylation of Foxp3 at Lys-31 (166). The specific mechanism behind these processes is still unknown, though. Class III SIRT1 is a NAD-dependent enzyme that negatively regulates TIP60 acetylation (167, 168). This regulatory process can be disrupted by mammalian sterile 20-like kinase 1 (MST1) (150, 169). Additionally, Class IV Zn2+-dependent HDAC11 can co-associate with Foxp3 and contribute to its deacetylation (165, 170).
3.3.3 Phosphorylation
Protein kinases engage in phosphorylation to covalently attach a phosphate group to a protein’s serine (S), threonine (T), or tyrosine (Y) residue. This process is reversible. Among them, cyclin-dependent kinase (CDK2), proto-oncogene serine/threonine protein kinase (PIM1, PIM2) and protein phosphatase 1 (PP1) negatively regulate Foxp3 expression. Kinase NLK positively regulates Foxp3 expression (22).
CDK2, PIM1, and PIM2 each inhibit Treg immunosuppressive function through distinct mechanisms. Four cyclin-dependent kinase motifs (Ser/Thr-Pro) are present in the N-terminal repressor domain of Foxp3. Cyclin E interacts with CDK2 and phosphorylates Foxp3 at these sites, resulting in decreased stability of Foxp3 protein and reduced suppressive function of Tregs (171, 172). In contrast, PIM1 and PIM2 have no effect on the decrease in stability of Foxp3. Instead, they impact its activity of binding to DNA and interacting with other co-factors. The crucial role of DNA binding for Foxp3 is fulfilled by its forkhead domain (FHD). PIM1 phosphorylates Foxp3 at Ser-422, which is situated in the C-terminal region of FHD (173). Foxp3’s interaction with co-factors such as HDAC7, TIP60, and Eos depends on its N-terminal region. PIM2 phosphorylates Ser-33 and Ser-41 within the N-terminal region of Foxp3 (174, 175). Additionally, lymphocyte-specific protein tyrosine kinase (LCK) phosphorylates Foxp3 at Tyr-342, which inhibits cell invasion. However, the functional impact of LCK phosphorylation on Foxp3 remains unclear (176).
How PP1 affects Foxp3 expression and Treg function remains a topic of debate (9, 23, 177). According to Hong Nie et al.’s study, during inflammatory circumstances, PP1 dephosphorylated Foxp3 at Ser-418 of FHD, leading to a reduction in Foxp3 expression (178). On the other hand, Nemo-like kinase (NLK) was found to promote Foxp3 expression. NLK phosphorylates Foxp3 and prevents its degradation by the ubiquitin ligase Stub1 (179, 180).
3.3.4 Ubiquitination, deubiquitination, and small ubiquitin-like modifier
Ubiquitination, a process mediated by a sequence of enzymes, denotes the covalent attachment of ubiquitin to target proteins. This intricate procedure necessitates the synergies of three distinct ubiquitin enzymes: E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin ligase. Substrate proteins can undergo monoubiquitination or polyubiquitination. Monoubiquitination takes place when a solitary ubiquitin molecule binds to a lone Lys residue of the target protein, whereas polyubiquitination entails the concurrent labeling of various Lys residues of the target protein by a solitary ubiquitin molecule (181–183). The three enzymes responsible for ubiquitinating Foxp3 are RNF31, Stub1, and TRAF6, each serving different functions.
RNF31, a crucial E3 ligase found in the LUBAC complex (linear assembly complex of ubiquitin chains), holds a vital function in facilitating the monoubiquitination of Foxp3. This particular protein actively participates in numerous immune signaling pathways and carries substantial implications in Tregs (184–188). In addition to regulating TCR signaling, RNF31 also facilitates Foxp3 monoubiquitination, resulting in heightened Foxp3 expression and improved immunosuppressive function in Tregs (189).
Stub1 and TRAF6 are involved in the polyubiquitination of Foxp3. Stub1 mediates K48 polyubiquitination at multiple sites, leading to the degradation of Foxp3 and a reduction in Treg suppressive activity (180). Additionally, Stub1 can also mediate K27 polyubiquitination on CARMA1 in the CBM complex, so aiding in the NF-κB’s activation (190). On the other hand, TRAF6, a cyclic E3 ligase, is responsible for mediating inflammation-related signaling (191). K63 polyubiquitination on Foxp3 at Lys-262 is mediated by TRAF6, but the exact mechanism of its impact on Foxp3 still requires further investigation (192).
Deubiquitinase (DUB) can reverse the reversible process of ubiquitination. DUB removes ubiquitin from substrate proteins (193). Several proteins, including USP7, USP21, USP22, and USP44, have been discovered to directly interact with Foxp3. USP7 prevents the ubiquitin-dependent degradation of Foxp3 protein by removing the K48 polyubiquitination tag. Additionally, it promotes the interaction of TIP60 with Foxp3, thereby preserving the level of expression of Foxp3 and the immunosuppressive ability of Treg (194). Similarly, USP21, USP22, and USP44 also deubiquitinate Foxp3, preventing its degradation (195–199).
Small ubiquitin-like modifiers are small molecules that can be covalently bound to other proteins through a cascade reaction mediated by enzymes. This process is known as SUMO. SUMO does not involve the proteolysis of targeted proteins but rather plays a crucial part in regulating diverse biological processes. It is important to note that the SUMO process is reversible. For instance, ROS induced by TCR signaling controls the protein stability of the deubiquitinating enzyme SENP3, thereby mediating the deubiquitinating modification of the Foxp3 transcription factor BACH2 and its activity in maintaining the immunosuppressive function of Tregs (200).
3.3.5 Glycosylation
Glycosylation is a type of PTM that involves attaching glycans to proteins. Observations demonstrate a direct link between the suppressive activity of Tregs and glycosylation, indicating a positive correlation (201, 202). O-GlcNAc glycosylation modification specifically refers to the addition of a single monosaccharide modification to the serine and/or threonine residues of a protein. O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) are enzymes that catalyze this modification in opposite directions. There are several O-GlcNAcylation locations in Foxp3, which facilitate the expression of Foxp3 by regulating the IL-2/STAT5 pathway following glycosylation (203). It is significant to highlight that the glycosylation of c-Rel, another transcription factor, has been demonstrated to diminish its interaction with Foxp3 and inhibit Foxp3 expression (204). Moreover, O-GlcNAcylation could oppose ubiquitination, resulting in the enhancement of Foxp3 protein stability (205).
4 Immunotherapy targeting Treg reprogramming
With the advancement of cancer immunotherapy and the in-depth exploration of the TME, cancer treatment has experienced a revolution. In TME, Treg is a major therapeutic target. There are currently seven immunotherapies targeting Tregs, such as (1) depleting TI-Tregs, (2) preventing the migration of Tregs into tumors, (3) sensitizing TI-Tregs to inhibitory receptor blockade, (4) targeting costimulatory signals on TI-Tregs, (5) targeting Treg cytokine secretion, (6) altering Treg stability, (7) disrupting exTreg reprogramming (206). As shown in Table 1, we summarized potential targets that may act on Tregs reprogramming.
5 Conclusion
After extensive research, there has been a better understanding of Tregs. In the inflammatory microenvironment, Tregs are influenced by signaling molecules, metabolic conditions, and Foxp3 post-translational modifications. As a result, they undergo reprogramming and transform into immunosuppressive Tregs, which enhances immunosuppressive ability. Tregs are now a prominent target in the realm of cancer immunotherapy. By influencing the reprogramming of Tregs, it is possible to effectively reduce their immunosuppressive function, mitigate the negative effects of Treg reprogramming on immunotherapy in inflammatory conditions, and partially prevent the severe autoimmune consequences that may arise from Treg depletion therapy.
Existing technology and approaches are making great efforts to address the inadequacies of various immunotherapies. Considering the current negative consequences of Treg depletion-induced autoimmunity, there are a number of ways to effectively induce tumor immunity by targeting specific Tregs without triggering severe autoimmune reactions. The first one is to selectively target effector Treg cells in tumor tissues, thereby retaining naïve Tregs in other tissues that are needed to prevent autoimmunity. The second aims to adjust the extent and duration of Treg depletion. The third method is to inject Treg-depleting antibodies directly into the tumor tissue. In addition, when Tregs depletion is used in combination with other immunotherapies, Tregs should be depleted or their suppressive activity should be weakened before other treatments (such as immunosuppressants and vaccination) (242).
In targeted Tregs reprogramming, multi-omics approaches combined with novel computational tools can be applied to target various signals and molecules on Tregs (243–245). It enables a better understanding of the gene regulatory networks regulating Treg function, as well as the identification of additional biomarkers that define Tregs function in the TME. This could go a long way toward new therapies targeting Tregs without affecting immune homeostasis in various cancers (206).
The drawback of targeted post-translational modifications is that they are difficult to selectively target because they are present in both cancerous and normal cells on a large scale. The issue with metabolic reprogramming that targets Tregs is the same (21). Furthermore, targeting post-translational modifications is more common in hematological malignancies and lacks application in solid tumors. In recent studies, proteolytic targeting chimeras (PROTACs) have been used as an innovative technology to target parts of proteins that could not be targeted before. They affect Tregs function by connecting target protein ligands and E3 ubiquitin ligase ligands, promoting rapid ubiquitination and consequent degradation of target proteins (246–248).
However, there are still some issues that we need to explore further. (1) Are there other mechanisms affecting Tregs reprogramming? (2) Can tumor-specific Treg-targeted therapy be achieved without affecting tissue Tregs? (3) How much impact do different inflammation and cancer types have on Tregs? We believe that in the future, with continued in-depth research on Tregs, significant progress will be made in the field of tumor immunotherapy, benefiting millions of cancer patients.
Author contributions
JL: Conceptualization, Writing – original draft, Writing – review & editing. BZ: Conceptualization, Writing – original draft, Writing – review & editing. GZ: Conceptualization, Writing – original draft. DS: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Morad G, Helmink BA, Sharma P, Wargo JA. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell (2021) 184(21):5309–37. doi: 10.1016/j.cell.2021.09.020
2. Johnson DB, Nebhan CA, Moslehi JJ, Balko JM. Immune-checkpoint inhibitors: Long-term implications of toxicity. Nat Rev Clin Oncol (2022) 19(4):254–67. doi: 10.1038/s41571-022-00600-w
3. June CH, Warshauer JT, Bluestone JA. Is autoimmunity the achilles' Heel of cancer immunotherapy? Nat Med (2017) 23(5):540–7. doi: 10.1038/nm.4321
4. Kubli SP, Berger T, Araujo DV, Siu LL, Mak TW. Beyond immune checkpoint blockade: Emerging immunological strategies. Nat Rev Drug Discovery (2021) 20(12):899–919. doi: 10.1038/s41573-021-00155-y
5. Zhang B, Huang B, Zhang X, Li S, Zhu J, Chen X, et al. Panoptosis-related molecular subtype and prognostic model associated with the immune microenvironment and individualized therapy in pancreatic cancer. Front Oncol (2023) 13:1217654. doi: 10.3389/fonc.2023.1217654
6. Cao Q, Zhang Q, Chen YQ, Fan AD, Zhang XL. Risk factors for the development of hepatocellular carcinoma in chengdu: A prospective cohort study. Eur Rev Med Pharmacol Sci (2022) 26(24):9447–56. doi: 10.26355/eurrev_202212_30696
7. Liu R, Nikolajczyk BS. Tissue immune cells fuel obesity-associated inflammation in adipose tissue and beyond. Front Immunol (2019) 10:1587. doi: 10.3389/fimmu.2019.01587
8. Ohkura N, Sakaguchi S. Transcriptional and epigenetic basis of treg cell development and function: Its genetic anomalies or variations in autoimmune diseases. Cell Res (2020) 30(6):465–74. doi: 10.1038/s41422-020-0324-7
9. Dong Y, Yang C, Pan F. Post-translational regulations of foxp3 in treg cells and their therapeutic applications. Front Immunol (2021) 12:626172. doi: 10.3389/fimmu.2021.626172
10. Abbas AK, Benoist C, Bluestone JA, Campbell DJ, Ghosh S, Hori S, et al. Regulatory T cells: Recommendations to simplify the nomenclature. Nat Immunol (2013) 14(4):307–8. doi: 10.1038/ni.2554
11. Nishikawa H, Koyama S. Mechanisms of regulatory T cell infiltration in tumors: Implications for innovative immune precision therapies. J immunotherapy Cancer (2021) 9(7):e002591. doi: 10.1136/jitc-2021-002591
12. Cinier J, Hubert M, Besson L, Di Roio A, Rodriguez C, Lombardi V, et al. Recruitment and expansion of tregs cells in the tumor environment-how to target them? Cancers (2021) 13(8):1850. doi: 10.3390/cancers13081850
13. Shevach EM, Thornton AM. Ttregs, ptregs, and itregs: Similarities and differences. Immunol Rev (2014) 259(1):88–102. doi: 10.1111/imr.12160
14. Tanaka A, Sakaguchi S. Targeting treg cells in cancer immunotherapy. Eur J Immunol (2019) 49(8):1140–6. doi: 10.1002/eji.201847659
15. Saxena V, Lakhan R, Iyyathurai J, Bromberg JS. Mechanisms of extreg induction. Eur J Immunol (2021) 51(8):1956–67. doi: 10.1002/eji.202049123
16. Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin Cancer Biol (2015) 35 Suppl:S185–s98. doi: 10.1016/j.semcancer.2015.03.004
17. Li C, Jiang P, Wei S, Xu X, Wang J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol Cancer (2020) 19(1):116. doi: 10.1186/s12943-020-01234-1
18. Moreno Ayala MA, Li Z, DuPage M. Treg programming and therapeutic reprogramming in cancer. Immunology (2019) 157(3):198–209. doi: 10.1111/imm.13058
19. Wang YA, Li XL, Mo YZ, Fan CM, Tang L, Xiong F, et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol Cancer (2018) 17(1):168. doi: 10.1186/s12943-018-0913-y
20. Georgiev P, Charbonnier LM, Chatila TA. Regulatory T cells: The many faces of foxp3. J Clin Immunol (2019) 39(7):623–40. doi: 10.1007/s10875-019-00684-7
21. Yan Y, Huang L, Liu Y, Yi M, Chu Q, Jiao D, et al. Metabolic profiles of regulatory T cells and their adaptations to the tumor microenvironment: Implications for antitumor immunity. J Hematol Oncol (2022) 15(1):104. doi: 10.1186/s13045-022-01322-3
22. Deng G, Song X, Fujimoto S, Piccirillo CA, Nagai Y, Greene MI. Foxp3 post-translational modifications and treg suppressive activity. Front Immunol (2019) 10:2486. doi: 10.3389/fimmu.2019.02486
23. Lu L, Barbi J, Pan F. The regulation of immune tolerance by foxp3. Nat Rev Immunol (2017) 17(11):703–17. doi: 10.1038/nri.2017.75
24. Ghobadinezhad F, Ebrahimi N, Mozaffari F, Moradi N, Beiranvand S, Pournazari M, et al. The emerging role of regulatory cell-based therapy in autoimmune disease. Front Immunol (2022) 13:1075813. doi: 10.3389/fimmu.2022.1075813
25. Catalán D, Mansilla MA, Ferrier A, Soto L, Oleinika K, Aguillón JC, et al. Immunosuppressive mechanisms of regulatory B cells. Front Immunol (2021) 12:611795. doi: 10.3389/fimmu.2021.611795
26. Manicassamy S, Pulendran B. Dendritic cell control of tolerogenic responses. Immunol Rev (2011) 241(1):206–27. doi: 10.1111/j.1600-065X.2011.01015.x
27. Groth C, Hu X, Weber R, Fleming V, Altevogt P, Utikal J, et al. Immunosuppression mediated by myeloid-derived suppressor cells (Mdscs) during tumour progression. Br J Cancer (2019) 120(1):16–25. doi: 10.1038/s41416-018-0333-1
28. Wu Y, Yi M, Niu M, Mei Q, Wu K. Myeloid-derived suppressor cells: An emerging target for anticancer immunotherapy. Mol Cancer (2022) 21(1):184. doi: 10.1186/s12943-022-01657-y
29. Christofides A, Strauss L, Yeo A, Cao C, Charest A, Boussiotis VA. The complex role of tumor-infiltrating macrophages. Nat Immunol (2022) 23(8):1148–56. doi: 10.1038/s41590-022-01267-2
30. Chen Y, Song Y, Du W, Gong L, Chang H, Zou Z. Tumor-associated macrophages: An accomplice in solid tumor progression. J Biomed Sci (2019) 26(1):78. doi: 10.1186/s12929-019-0568-z
31. Sakaguchi S, Vignali DA, Rudensky AY, Niec RE, Waldmann H. The plasticity and stability of regulatory T cells. Nat Rev Immunol (2013) 13(6):461–7. doi: 10.1038/nri3464
32. Tay C, Tanaka A, Sakaguchi S. Tumor-infiltrating regulatory T cells as targets of cancer immunotherapy. Cancer Cell (2023) 41(3):450–65. doi: 10.1016/j.ccell.2023.02.014
33. Yokosuka T, Saito T. Dynamic regulation of T-cell costimulation through tcr-cd28 microclusters. Immunol Rev (2009) 229(1):27–40. doi: 10.1111/j.1600-065X.2009.00779.x
34. Klein T, Fung SY, Renner F, Blank MA, Dufour A, Kang S, et al. The paracaspase malt1 cleaves hoil1 reducing linear ubiquitination by lubac to dampen lymphocyte nf-Κb signalling. Nat Commun (2015) 6:8777. doi: 10.1038/ncomms9777
35. Sun SC, Maggirwar SB, Harhaj EW, Uhlik M. Binding of C-rel to stat5 target sequences in htlv-I-transformed T cells. Oncogene (1999) 18(7):1401–9. doi: 10.1038/sj.onc.1202430
36. Rao A, Luo C, Hogan PG. Transcription factors of the nfat family: Regulation and function. Annu Rev Immunol (1997) 15:707–47. doi: 10.1146/annurev.immunol.15.1.707
37. Liu Q, Chen Y, Auger-Messier M, Molkentin JD. Interaction between nfκb and nfat coordinates cardiac hypertrophy and pathological remodeling. Circ Res (2012) 110(8):1077–86. doi: 10.1161/circresaha.111.260729
38. Zappasodi R, Serganova I, Cohen IJ, Maeda M, Shindo M, Senbabaoglu Y, et al. Ctla-4 blockade drives loss of T(Reg) stability in glycolysis-low tumours. Nature (2021) 591(7851):652–8. doi: 10.1038/s41586-021-03326-4
39. Watanabe T, Ishino T, Ueda Y, Nagasaki J, Sadahira T, Dansako H, et al. Activated ctla-4-independent immunosuppression of treg cells disturbs ctla-4 blockade-mediated antitumor immunity. Cancer Sci (2023) 114(5):1859–70. doi: 10.1111/cas.15756
40. Liu W, Zhou X, Zeng K, Nie C, Huang J, Zhu L, et al. Study on the action mechanism of buyang huanwu decoction against ischemic stroke based on S1p/S1pr1/pi3k/akt signaling pathway. J Ethnopharmacol (2023) 312:116471. doi: 10.1016/j.jep.2023.116471
41. Zeiser R, Negrin RS. Interleukin-2 receptor downstream events in regulatory T cells: Implications for the choice of immunosuppressive drug therapy. Cell Cycle (Georgetown Tex) (2008) 7(4):458–62. doi: 10.4161/cc.7.4.5454
42. Ohkura N, Sakaguchi S. Foxo1 and foxo3 help foxp3. Immunity (2010) 33(6):835–7. doi: 10.1016/j.immuni.2010.12.004
43. Alcántara-Hernández M, Torres-Zárate C, Pérez-Montesinos G, Jurado-Santacruz F, Domínguez-Gómez MA, Peniche-Castellanos A, et al. Overexpression of hypoxia-inducible factor 1 alpha impacts foxp3 levels in mycosis fungoides–cutaneous T-cell lymphoma: Clinical implications. Int J Cancer (2014) 134(9):2136–45. doi: 10.1002/ijc.28546
44. Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, et al. Control of T(H)17/T(Reg) balance by hypoxia-inducible factor 1. Cell (2011) 146(5):772–84. doi: 10.1016/j.cell.2011.07.033
45. Sharma MD, Shinde R, McGaha TL, Huang L, Holmgaard RB, Wolchok JD, et al. The pten pathway in tregs is a critical driver of the suppressive tumor microenvironment. Sci Adv (2015) 1(10):e1500845. doi: 10.1126/sciadv.1500845
46. Overacre AE, Vignali DA. T(Reg) stability: To be or not to be. Curr Opin Immunol (2016) 39:39–43. doi: 10.1016/j.coi.2015.12.009
47. Sekiya T, Kashiwagi I, Yoshida R, Fukaya T, Morita R, Kimura A, et al. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat Immunol (2013) 14(3):230–7. doi: 10.1038/ni.2520
48. Takahashi R, Yoshimura A. Socs1 and regulation of regulatory T cells plasticity. J Immunol Res (2014) 2014:943149. doi: 10.1155/2014/943149
49. Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and nfat cooperate to induce foxp3 expression through its enhancer. Nat Immunol (2008) 9(2):194–202. doi: 10.1038/ni1549
50. Wang J, Zhao X, Wan YY. Intricacies of tgf-Β Signaling in treg and th17 cell biology. Cell Mol Immunol (2023) 20(9):1002–22. doi: 10.1038/s41423-023-01036-7
51. Hövelmeyer N, Schmidt-Supprian M, Ohnmacht C. Nf-Κb in control of regulatory T cell development, identity, and function. J Mol Med (Berlin Germany) (2022) 100(7):985–95. doi: 10.1007/s00109-022-02215-1
52. Yi JY, Shin I, Arteaga CL. Type I transforming growth factor beta receptor binds to and activates phosphatidylinositol 3-kinase. J Biol Chem (2005) 280(11):10870–6. doi: 10.1074/jbc.M413223200
53. Guo J, Zhang J, Zhang X, Zhang Z, Wei X, Zhou X. Constitutive activation of mek1 promotes treg cell instability in vivo. J Biol Chem (2014) 289(51):35139–48. doi: 10.1074/jbc.M114.589192
54. Derynck R, Turley SJ, Akhurst RJ. Tgfβ Biology in cancer progression and immunotherapy. Nat Rev Clin Oncol (2021) 18(1):9–34. doi: 10.1038/s41571-020-0403-1
55. Batlle E, Massagué J. Transforming growth factor-Β Signaling in immunity and cancer. Immunity (2019) 50(4):924–40. doi: 10.1016/j.immuni.2019.03.024
56. David CJ, Massagué J. Contextual determinants of tgfβ Action in development, immunity and cancer. Nat Rev Mol Cell Biol (2018) 19(7):419–35. doi: 10.1038/s41580-018-0007-0
57. Tauriello DVF, Sancho E, Batlle E. Overcoming tgfβ-mediated immune evasion in cancer. Nat Rev Cancer (2022) 22(1):25–44. doi: 10.1038/s41568-021-00413-6
58. Gulley JL, Schlom J, Barcellos-Hoff MH, Wang XJ, Seoane J, Audhuy F, et al. Dual inhibition of tgf-Β and pd-L1: A novel approach to cancer treatment. Mol Oncol (2022) 16(11):2117–34. doi: 10.1002/1878-0261.13146
59. Yi M, Wu Y, Niu M, Zhu S, Zhang J, Yan Y, et al. Anti-tgf-Β/pd-L1 bispecific antibody promotes T cell infiltration and exhibits enhanced antitumor activity in triple-negative breast cancer. J Immunother Cancer (2022) 10(12):e005543. doi: 10.1136/jitc-2022-005543
60. Li T, Wang X, Niu M, Wang M, Zhou J, Wu K, et al. Bispecific antibody targeting tgf-Β and pd-L1 for synergistic cancer immunotherapy. Front Immunol (2023) 14:1196970. doi: 10.3389/fimmu.2023.1196970
61. Yi M, Zhang J, Li A, Niu M, Yan Y, Jiao Y, et al. The construction, expression, and enhanced anti-tumor activity of ym101: A bispecific antibody simultaneously targeting tgf-Β and pd-L1. J Hematol Oncol (2021) 14(1):27. doi: 10.1186/s13045-021-01045-x
62. Cheng B, Ding K, Chen P, Ji J, Luo T, Guo X, et al. Anti-pd-L1/tgf-Βr fusion protein (Shr-1701) overcomes disrupted lymphocyte recovery-induced resistance to pd-1/pd-L1 inhibitors in lung cancer. Cancer Commun (London England) (2022) 42(1):17–36. doi: 10.1002/cac2.12244
63. Faustino LD, Griffith JW, Rahimi RA, Nepal K, Hamilos DL, Cho JL, et al. Interleukin-33 activates regulatory T cells to suppress innate Γδ T cell responses in the lung. Nat Immunol (2020) 21(11):1371–83. doi: 10.1038/s41590-020-0785-3
64. Harusato A, Abo H, Ngo VL, Yi SW, Mitsutake K, Osuka S, et al. Il-36γ Signaling controls the induced regulatory T cell-th9 cell balance via nfκb activation and stat transcription factors. Mucosal Immunol (2017) 10(6):1455–67. doi: 10.1038/mi.2017.21
65. Laurence A, Amarnath S, Mariotti J, Kim YC, Foley J, Eckhaus M, et al. Stat3 transcription factor promotes instability of ntreg cells and limits generation of itreg cells during acute murine graft-versus-host disease. Immunity (2012) 37(2):209–22. doi: 10.1016/j.immuni.2012.05.027
66. Xu J, Yang Y, Qiu G, Lal G, Yin N, Wu Z, et al. Stat4 is critical for the balance between th17 cells and regulatory T cells in colitis. J Immunol (Baltimore Md 1950) (2011) 186(11):6597–606. doi: 10.4049/jimmunol.1004074
67. Contaldi E, Magistrelli L, Milner AV, Cosentino M, Marino F, Comi C. Expression of transcription factors in cd4 + T cells as potential biomarkers of motor complications in parkinson's disease. J Parkinson's Dis (2021) 11(2):507–14. doi: 10.3233/jpd-202417
68. Harris F, Berdugo YA, Tree T. Il-2-based approaches to treg enhancement. Clin Exp Immunol (2023) 211(2):149–63. doi: 10.1093/cei/uxac105
69. Ross SH, Cantrell DA. Signaling and function of interleukin-2 in T lymphocytes. Annu Rev Immunol (2018) 36:411–33. doi: 10.1146/annurev-immunol-042617-053352
70. Jeffery HC, Jeffery LE, Lutz P, Corrigan M, Webb GJ, Hirschfield GM, et al. Low-dose interleukin-2 promotes stat-5 phosphorylation, T(Reg) survival and ctla-4-dependent function in autoimmune liver diseases. Clin Exp Immunol (2017) 188(3):394–411. doi: 10.1111/cei.12940
71. Rech AJ, Mick R, Martin S, Recio A, Aqui NA, Powell DJ Jr., et al. Cd25 blockade depletes and selectively reprograms regulatory T cells in concert with immunotherapy in cancer patients. Sci Trans Med (2012) 4(134):134ra62. doi: 10.1126/scitranslmed.3003330
72. Jones DM, Read KA, Oestreich KJ. Dynamic roles for il-2-stat5 signaling in effector and regulatory cd4(+) T cell populations. J Immunol (Baltimore Md 1950) (2020) 205(7):1721–30. doi: 10.4049/jimmunol.2000612
73. Zagorulya M, Yim L, Morgan DM, Edwards A, Torres-Mejia E, Momin N, et al. Tissue-Specific Abundance of Interferon-Gamma Drives Regulatory T cells to Restrain Dc1-Mediated Priming of Cytotoxic T cells against Lung Cancer. Immunity (2023) 56(2):386–405.e10. doi: 10.1016/j.immuni.2023.01.010
74. Chinen T, Kannan AK, Levine AG, Fan X, Klein U, Zheng Y, et al. An essential role for the il-2 receptor in T(Reg) cell function. Nat Immunol (2016) 17(11):1322–33. doi: 10.1038/ni.3540
75. Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. Cd4+Cd25+Foxp3+ Regulatory T cells induce cytokine deprivation-mediated apoptosis of effector cd4+ T cells. Nat Immunol (2007) 8(12):1353–62. doi: 10.1038/ni1536
76. Chu KH, Lin SY, Chiang BL. Stat6 pathway is critical for the induction and function of regulatory T cells induced by mucosal B cells. Front Immunol (2020) 11:615868. doi: 10.3389/fimmu.2020.615868
77. Chapoval S, Dasgupta P, Dorsey NJ, Keegan AD. Regulation of the T helper cell type 2 (Th2)/T regulatory cell (Treg) balance by il-4 and stat6. J Leukoc Biol (2010) 87(6):1011–8. doi: 10.1189/jlb.1209772
78. Afzali B, Lombardi G, Lechler RI, Lord GM. The role of T helper 17 (Th17) and regulatory T cells (Treg) in human organ transplantation and autoimmune disease. Clin Exp Immunol (2007) 148(1):32–46. doi: 10.1111/j.1365-2249.2007.03356.x
79. O'Malley JT, Sehra S, Thieu VT, Yu Q, Chang HC, Stritesky GL, et al. Signal transducer and activator of transcription 4 limits the development of adaptive regulatory T cells. Immunology (2009) 127(4):587–95. doi: 10.1111/j.1365-2567.2008.03037.x
80. Schmetterer KG, Pickl WF. The il-10/stat3 axis: Contributions to immune tolerance by thymus and peripherally derived regulatory T-cells. Eur J Immunol (2017) 47(8):1256–65. doi: 10.1002/eji.201646710
81. Short WD, Steen E, Kaul A, Wang X, Olutoye OO 2nd, Vangapandu HV, et al. Il-10 promotes endothelial progenitor cell infiltration and wound healing via stat3. FASEB J Off Publ Fed Am Societies Exp Biol (2022) 36(7):e22298. doi: 10.1096/fj.201901024RR
82. Kortylewski M, Xin H, Kujawski M, Lee H, Liu Y, Harris T, et al. Regulation of the il-23 and il-12 balance by stat3 signaling in the tumor microenvironment. Cancer Cell (2009) 15(2):114–23. doi: 10.1016/j.ccr.2008.12.018
83. Truong AD, Hong Y, Hoang CT, Lee J, Hong YH. Chicken il-26 regulates immune responses through the jak/stat and nf-Κb signaling pathways. Dev Comp Immunol (2017) 73:10–20. doi: 10.1016/j.dci.2017.03.001
84. Meka RR, Venkatesha SH, Dudics S, Acharya B, Moudgil KD. Il-27-induced modulation of autoimmunity and its therapeutic potential. Autoimmun Rev (2015) 14(12):1131–41. doi: 10.1016/j.autrev.2015.08.001
85. Cai Z, Zhang S, Wu P, Ren Q, Wei P, Hong M, et al. A novel potential target of il-35-regulated jak/stat signaling pathway in lupus nephritis. Clin Trans Med (2021) 11(2):e309. doi: 10.1002/ctm2.309
86. Shimizu J, Yamazaki S, Takahashi T, Ishida Y, Sakaguchi S. Stimulation of cd25(+)Cd4(+) regulatory T cells through gitr breaks immunological self-tolerance. Nat Immunol (2002) 3(2):135–42. doi: 10.1038/ni759
87. Cohen AD, Schaer DA, Liu C, Li Y, Hirschhorn-Cymmerman D, Kim SC, et al. Agonist anti-gitr monoclonal antibody induces melanoma tumor immunity in mice by altering regulatory T cell stability and intra-tumor accumulation. PloS One (2010) 5(5):e10436. doi: 10.1371/journal.pone.0010436
88. Amoozgar Z, Kloepper J, Ren J, Tay RE, Kazer SW, Kiner E, et al. Targeting treg cells with gitr activation alleviates resistance to immunotherapy in murine glioblastomas. Nat Commun (2021) 12(1):2582. doi: 10.1038/s41467-021-22885-8
89. Yates K, Bi K, Haining WN, Cantor H, Kim HJ. Comparative transcriptome analysis reveals distinct genetic modules associated with helios expression in intratumoral regulatory T cells. Proc Natl Acad Sci USA (2018) 115(9):2162–7. doi: 10.1073/pnas.1720447115
90. Lam AJ, Uday P, Gillies JK, Levings MK. Helios is a marker, not a driver, of human treg stability. Eur J Immunol (2022) 52(1):75–84. doi: 10.1002/eji.202149318
91. Schaer DA, Budhu S, Liu C, Bryson C, Malandro N, Cohen A, et al. Gitr pathway activation abrogates tumor immune suppression through loss of regulatory T cell lineage stability. Cancer Immunol Res (2013) 1(5):320–31. doi: 10.1158/2326-6066.Cir-13-0086
92. Nakagawa H, Sido JM, Reyes EE, Kiers V, Cantor H, Kim HJ. Instability of helios-deficient tregs is associated with conversion to a T-effector phenotype and enhanced antitumor immunity. Proc Natl Acad Sci USA (2016) 113(22):6248–53. doi: 10.1073/pnas.1604765113
93. Zabransky DJ, Nirschl CJ, Durham NM, Park BV, Ceccato CM, Bruno TC, et al. Phenotypic and functional properties of helios+ Regulatory T cells. PloS One (2012) 7(3):e34547. doi: 10.1371/journal.pone.0034547
94. Song J, So T, Croft M. Activation of nf-kappab1 by ox40 contributes to antigen-driven T cell expansion and survival. J Immunol (Baltimore Md 1950) (2008) 180(11):7240–8. doi: 10.4049/jimmunol.180.11.7240
95. Xiao X, Gong W, Demirci G, Liu W, Spoerl S, Chu X, et al. New insights on ox40 in the control of T cell immunity and immune tolerance in vivo. J Immunol (Baltimore Md 1950) (2012) 188(2):892–901. doi: 10.4049/jimmunol.1101373
96. Zhang X, Xiao X, Lan P, Li J, Dou Y, Chen W, et al. Ox40 costimulation inhibits foxp3 expression and treg induction via batf3-dependent and independent mechanisms. Cell Rep (2018) 24(3):607–18. doi: 10.1016/j.celrep.2018.06.052
97. Burocchi A, Pittoni P, Gorzanelli A, Colombo MP, Piconese S. Intratumor ox40 stimulation inhibits irf1 expression and il-10 production by treg cells while enhancing cd40l expression by effector memory T cells. Eur J Immunol (2011) 41(12):3615–26. doi: 10.1002/eji.201141700
98. Piconese S, Valzasina B, Colombo MP. Ox40 triggering blocks suppression by regulatory T cells and facilitates tumor rejection. J Exp Med (2008) 205(4):825–39. doi: 10.1084/jem.20071341
99. Marshall NB, Kerkvliet NI. Dioxin and immune regulation: Emerging role of aryl hydrocarbon receptor in the generation of regulatory T cells. Ann New York Acad Sci (2010) 1183:25–37. doi: 10.1111/j.1749-6632.2009.05125.x
100. Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, et al. Control of T(Reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature (2008) 453(7191):65–71. doi: 10.1038/nature06880
101. Campesato LF, Budhu S, Tchaicha J, Weng CH, Gigoux M, Cohen IJ, et al. Blockade of the ahr restricts a treg-macrophage suppressive axis induced by L-kynurenine. Nat Commun (2020) 11(1):4011. doi: 10.1038/s41467-020-17750-z
102. Kalim KW, Yang JQ, Wunderlich M, Modur V, Nguyen P, Li Y, et al. Targeting of cdc42 gtpase in regulatory T cells unleashes antitumor T-cell immunity. J immunotherapy Cancer (2022) 10(11):e004806. doi: 10.1136/jitc-2022-004806
103. Hou DY, Muller AJ, Sharma MD, DuHadaway J, Banerjee T, Johnson M, et al. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res (2007) 67(2):792–801. doi: 10.1158/0008-5472.Can-06-2925
104. Ali K, Soond DR, Pineiro R, Hagemann T, Pearce W, Lim EL, et al. Inactivation of pi(3)K P110δ Breaks regulatory T-cell-mediated immune tolerance to cancer. Nature (2014) 510(7505):407–11. doi: 10.1038/nature13444
105. Crellin NK, Garcia RV, Levings MK. Altered activation of akt is required for the suppressive function of human cd4+Cd25+ T regulatory cells. Blood (2007) 109(5):2014–22. doi: 10.1182/blood-2006-07-035279
106. Munn DH, Mellor AL. Ido in the tumor microenvironment: Inflammation, counter-regulation, and tolerance. Trends Immunol (2016) 37(3):193–207. doi: 10.1016/j.it.2016.01.002
107. Cadilha BL, Benmebarek MR, Dorman K, Oner A, Lorenzini T, Obeck H, et al. Combined tumor-directed recruitment and protection from immune suppression enable car T cell efficacy in solid tumors. Sci Adv (2021) 7(24):eabi5781. doi: 10.1126/sciadv.abi5781
108. Barsheshet Y, Wildbaum G, Levy E, Vitenshtein A, Akinseye C, Griggs J, et al. Ccr8(+)Foxp3(+) T(Reg) cells as master drivers of immune regulation. Proc Natl Acad Sci United States America (2017) 114(23):6086–91. doi: 10.1073/pnas.1621280114
109. Wang T, Zhou Q, Zeng H, Zhang H, Liu Z, Shao J, et al. Ccr8 blockade primes anti-tumor immunity through intratumoral regulatory T cells destabilization in muscle-invasive bladder cancer. Cancer Immunol Immunother CII (2020) 69(9):1855–67. doi: 10.1007/s00262-020-02583-y
110. Yang YX, Yuan Y, Xia B. Cinnamtannin D1 ameliorates dss-induced colitis by preventing th17/treg imbalance through activation of the ampk/mtor pathway. Allergol Immunopathol (2022) 50(5):153–61. doi: 10.15586/aei.v50i5.654
111. He H, Chen Q, Fan H, Leng XY, Zhu F, Gao F, et al. Extracellular Vesicles produced by bone Marrow Mesenchymal Stem Cells Overexpressing Programmed Death-Ligand 1 Ameliorate Dextran Sodium Sulfate-Induced Ulcerative Colitis in Rats by Regulating Th17/Treg Cell Balance through Pten/Pi3k/Akt/Mtor Axis. J Gastroenterol Hepatol (2022) 37(12):2243–54. doi: 10.1111/jgh.15987
112. Zhang H, Watanabe R, Berry GJ, Nadler SG, Goronzy JJ, Weyand CM. Cd28 Signaling Controls Metabolic fitness of Pathogenic T Cells in Medium and Large vessel vasculitis. J Am Coll Cardiol (2019) 73(14):1811–23. doi: 10.1016/j.jacc.2019.01.049
113. Hsu TS, Lin YL, Wang YA, Mo ST, Chi PY, Lai AC, et al. Hif-2α Is indispensable for regulatory T cell function. Nat Commun (2020) 11(1):5005. doi: 10.1038/s41467-020-18731-y
114. Lai ZW, Kelly R, Winans T, Marchena I, Shadakshari A, Yu J, et al. Sirolimus in Patients with Clinically Active Systemic Lupus Erythematosus Resistant to, or Intolerant of, Conventional Medications: A Single-Arm, Open-Label, Phase 1/2 Trial. Lancet (London England) (2018) 391(10126):1186–96. doi: 10.1016/s0140-6736(18)30485-9
115. Danese S, Rutella S. The janus face of cd4+Cd25+ Regulatory T cells in cancer and autoimmunity. Curr Med Chem (2007) 14(6):649–66. doi: 10.2174/092986707780059599
116. Carbone F, De Rosa V, Carrieri PB, Montella S, Bruzzese D, Porcellini A, et al. Regulatory T cell proliferative potential is impaired in human autoimmune disease. Nat Med (2014) 20(1):69–74. doi: 10.1038/nm.3411
117. Procaccini C, De Rosa V, Galgani M, Abanni L, Calì G, Porcellini A, et al. An oscillatory switch in mtor kinase activity sets regulatory T cell responsiveness. Immunity (2010) 33(6):929–41. doi: 10.1016/j.immuni.2010.11.024
118. Law BK. Rapamycin: an anti-cancer immunosuppressant? Crit Rev Oncol Hematol (2005) 56(1):47–60. doi: 10.1016/j.critrevonc.2004.09.009
119. Wang Y, Huang T, Gu J, Lu L. Targeting the metabolism of tumor-infiltrating regulatory T cells. Trends Immunol (2023) 44(8):598–612. doi: 10.1016/j.it.2023.06.001
120. Li J, Kim SG, Blenis J. Rapamycin: One drug, many effects. Cell Metab (2014) 19(3):373–9. doi: 10.1016/j.cmet.2014.01.001
121. Yecies JL, Manning BD. Transcriptional control of cellular metabolism by mtor signaling. Cancer Res (2011) 71(8):2815–20. doi: 10.1158/0008-5472.Can-10-4158
122. Winans T, Oaks Z, Choudhary G, Patel A, Huang N, Faludi T, et al. Mtor-dependent loss of pon1 secretion and antiphospholipid autoantibody production underlie autoimmunity-mediated cirrhosis in transaldolase deficiency. J Autoimmun (2023) 140:103112. doi: 10.1016/j.jaut.2023.103112
123. Oaks Z, Patel A, Huang N, Choudhary G, Winans T, Faludi T, et al. Cytosolic aldose metabolism contributes to progression from cirrhosis to hepatocarcinogenesis. Nat Metab (2023) 5(1):41–60. doi: 10.1038/s42255-022-00711-9
124. Gerriets VA, Kishton RJ, Johnson MO, Cohen S, Siska PJ, Nichols AG, et al. Foxp3 and toll-like receptor signaling balance T(Reg) cell anabolic metabolism for suppression. Nat Immunol (2016) 17(12):1459–66. doi: 10.1038/ni.3577
125. Kishore M, Cheung KCP, Fu H, Bonacina F, Wang G, Coe D, et al. Regulatory T cell migration is dependent on glucokinase-mediated glycolysis. Immunity (2017) 47(5):875–89.e10. doi: 10.1016/j.immuni.2017.10.017
126. Newton RH, Turka LA. Regulation of T cell homeostasis and responses by pten. Front Immunol (2012) 3:151. doi: 10.3389/fimmu.2012.00151
127. Ouyang W, Liao W, Luo CT, Yin N, Huse M, Kim MV, et al. Novel foxo1-dependent transcriptional programs control T(Reg) cell function. Nature (2012) 491(7425):554–9. doi: 10.1038/nature11581
128. Kim ME, Kim DH, Lee JS. Foxo transcription factors: applicability as a novel immune cell regulators and therapeutic targets in oxidative stress-related diseases. Int J Mol Sci (2022) 23(19):11877. doi: 10.3390/ijms231911877
129. Angelin A, Gil-de-Gómez L, Dahiya S, Jiao J, Guo L, Levine MH, et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab (2017) 25(6):1282–93.e7. doi: 10.1016/j.cmet.2016.12.018
130. Shi H, Chapman NM, Wen J, Guy C, Long L, Dhungana Y, et al. Amino acids license kinase mtorc1 activity and treg cell function via small G proteins rag and rheb. Immunity (2019) 51(6):1012–27.e7. doi: 10.1016/j.immuni.2019.10.001
131. Kurniawan H, FranChina DG, Guerra L, Bonetti L, Baguet LS, Grusdat M, et al. Glutathione restricts serine metabolism to preserve regulatory T cell function. Cell Metab (2020) 31(5):920–36.e7. doi: 10.1016/j.cmet.2020.03.004
132. Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, et al. Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory cd4+ T cell subsets. J Immunol (Baltimore Md 1950) (2011) 186(6):3299–303. doi: 10.4049/jimmunol.1003613
133. Miao Y, Zhang C, Yang L, Zeng X, Hu Y, Xue X, et al. The Activation of Pparγ Enhances Treg Responses through up-Regulating Cd36/Cpt1-Mediated Fatty Acid Oxidation and Subsequent N-Glycan Branching of Tβrii/Il-2rα. Cell Commun Signal CCS (2022) 20(1):48. doi: 10.1186/s12964-022-00849-9
134. Wang H, Franco F, Tsui YC, Xie X, Trefny MP, Zappasodi R, et al. Cd36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat Immunol (2020) 21(3):298–308. doi: 10.1038/s41590-019-0589-5
135. Timilshina M, You Z, Lacher SM, Acharya S, Jiang L, Kang Y, et al. Activation of mevalonate pathway via lkb1 is essential for stability of T(Reg) cells. Cell Rep (2019) 27(10):2948–61.e7. doi: 10.1016/j.celrep.2019.05.020
136. Lim SA, Wei J, Nguyen TM, Shi H, Su W, Palacios G, et al. Lipid signalling enforces functional specialization of T(Reg) cells in tumours. Nature (2021) 591(7849):306–11. doi: 10.1038/s41586-021-03235-6
137. Ligtenberg MA, Çınar Ö, Holmdahl R, Mougiakakos D, Kiessling R. Methylcholanthrene-induced sarcomas develop independently from nox2-derived ros. PloS One (2015) 10(6):e0129786. doi: 10.1371/journal.pone.0129786
138. Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity (2013) 38(2):225–36. doi: 10.1016/j.immuni.2012.10.020
139. Weinberg SE, Singer BD, Steinert EM, Martinez CA, Mehta MM, Martínez-Reyes I, et al. Mitochondrial complex iii is essential for suppressive function of regulatory T cells. Nature (2019) 565(7740):495–9. doi: 10.1038/s41586-018-0846-z
140. Fu Z, Ye J, Dean JW, Bostick JW, Weinberg SE, Xiong L, et al. Requirement of mitochondrial transcription factor a in tissue-resident regulatory T cell maintenance and function. Cell Rep (2019) 28(1):159–71.e4. doi: 10.1016/j.celrep.2019.06.024
141. Liu ZM, Wang KP, Ma J, Guo Zheng S. The role of all-trans retinoic acid in the biology of foxp3+ Regulatory T cells. Cell Mol Immunol (2015) 12(5):553–7. doi: 10.1038/cmi.2014.133
142. Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, et al. Il-2 regulates foxp3 expression in human cd4+Cd25+ Regulatory T cells through a stat-dependent mechanism and induces the expansion of these cells in vivo. Blood (2006) 108(5):1571–9. doi: 10.1182/blood-2006-02-004747
143. Thangavelu G, Andrejeva G, Bolivar-Wagers S, Jin S, Zaiken MC, Loschi M, et al. Retinoic acid signaling acts as a rheostat to balance treg function. Cell Mol Immunol (2022) 19(7):820–33. doi: 10.1038/s41423-022-00869-y
144. Schubert HL, Blumenthal RM, Cheng X. 1 protein methyltransferases: their distribution among the five structural classes of adomet-dependent methyltransferases. Enzymes (2006) 24:3–28. doi: 10.1016/s1874-6047(06)80003-x
145. Kagoya Y, Saijo H, Matsunaga Y, Guo T, Saso K, Anczurowski M, et al. Arginine methylation of foxp3 is crucial for the suppressive function of regulatory T cells. J Autoimmun (2019) 97:10–21. doi: 10.1016/j.jaut.2018.09.011
146. Nagai Y, Ji MQ, Zhu F, Xiao Y, Tanaka Y, Kambayashi T, et al. Prmt5 associates with the foxp3 homomer and when disabled enhances targeted P185(Erbb2/neu) tumor immunotherapy. Front Immunol (2019) 10:174. doi: 10.3389/fimmu.2019.00174
147. Tanaka Y, Nagai Y, Okumura M, Greene MI, Kambayashi T. Prmt5 is required for T cell survival and proliferation by maintaining cytokine signaling. Front Immunol (2020) 11:621. doi: 10.3389/fimmu.2020.00621
148. Webb LM, Sengupta S, Edell C, Piedra-Quintero ZL, Amici SA, Miranda JN, et al. Protein arginine methyltransferase 5 promotes cholesterol biosynthesis-mediated th17 responses and autoimmunity. J Clin Invest (2020) 130(4):1683–98. doi: 10.1172/jci131254
149. Morabito F, Voso MT, Hohaus S, Gentile M, Vigna E, Recchia AG, et al. Panobinostat for the treatment of acute myelogenous leukemia. Expert Opin investigational Drugs (2016) 25(9):1117–31. doi: 10.1080/13543784.2016.1216971
150. van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YY, Beekman JM, van Beekum O, et al. Regulation of treg functionality by acetylation-mediated foxp3 protein stabilization. Blood (2010) 115(5):965–74. doi: 10.1182/blood-2009-02-207118
151. Kwon HS, Lim HW, Wu J, Schnölzer M, Verdin E, Ott M. Three novel acetylation sites in the foxp3 transcription factor regulate the suppressive activity of regulatory T cells. J Immunol (Baltimore Md 1950) (2012) 188(6):2712–21. doi: 10.4049/jimmunol.1100903
152. Liu Y, Wang L, Predina J, Han R, Beier UH, Wang LC, et al. Inhibition of P300 impairs foxp3⁺ T regulatory cell function and promotes antitumor immunity. Nat Med (2013) 19(9):1173–7. doi: 10.1038/nm.3286
153. Lau OD, Kundu TK, Soccio RE, Ait-Si-Ali S, Khalil EM, Vassilev A, et al. Hats off: selective synthetic inhibitors of the histone acetyltransferases P300 and pcaf. Mol Cell (2000) 5(3):589–95. doi: 10.1016/s1097-2765(00)80452-9
154. Liu Y, Wang L, Han R, Beier UH, Akimova T, Bhatti T, et al. Two histone/protein acetyltransferases, cbp and P300, are indispensable for foxp3+ T-regulatory cell development and function. Mol Cell Biol (2014) 34(21):3993–4007. doi: 10.1128/mcb.00919-14
155. Du T, Nagai Y, Xiao Y, Greene MI, Zhang H. Lysosome-dependent P300/foxp3 degradation and limits treg cell functions and enhances targeted therapy against cancers. Exp Mol Pathol (2013) 95(1):38–45. doi: 10.1016/j.yexmp.2013.04.003
156. Bin Dhuban K, d'Hennezel E, Nagai Y, Xiao Y, Shao S, Istomine R, et al. Suppression by human foxp3(+) regulatory T cells requires foxp3-tip60 interactions. Sci Immunol (2017) 2(12):eaai9297. doi: 10.1126/sciimmunol.aai9297
157. Deng G, Song X, Greene MI. Foxp3 in T(Reg) cell biology: A molecular and structural perspective. Clin Exp Immunol (2020) 199(3):255–62. doi: 10.1111/cei.13357
158. Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol (2010) 6(3):238–43. doi: 10.1038/nchembio.313
159. Li B, Samanta A, Song X, Iacono KT, Brennan P, Chatila TA, et al. Foxp3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human xlaad/ipex autoimmune disease. Int Immunol (2007) 19(7):825–35. doi: 10.1093/intimm/dxm043
160. Wang L, Liu Y, Han R, Beier UH, Bhatti TR, Akimova T, et al. Foxp3+ Regulatory T cell development and function require histone/protein deacetylase 3. J Clin Invest (2015) 125(3):1111–23. doi: 10.1172/jci77088
161. de Zoeten EF, Wang L, Butler K, Beier UH, Akimova T, Sai H, et al. Histone deacetylase 6 and heat shock protein 90 control the functions of foxp3(+) T-regulatory cells. Mol Cell Biol (2011) 31(10):2066–78. doi: 10.1128/mcb.05155-11
162. Sadlon TJ, Wilkinson BG, Pederson S, Brown CY, Bresatz S, Gargett T, et al. Genome-wide identification of human foxp3 target genes in natural regulatory T cells. J Immunol (Baltimore Md 1950) (2010) 185(2):1071–81. doi: 10.4049/jimmunol.1000082
163. Li B, Samanta A, Song X, Iacono KT, Bembas K, Tao R, et al. Foxp3 interactions with histone acetyltransferase and class ii histone deacetylases are required for repression. Proc Natl Acad Sci United States America (2007) 104(11):4571–6. doi: 10.1073/pnas.0700298104
164. Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med (2007) 13(11):1299–307. doi: 10.1038/nm1652
165. Beier UH, Wang L, Han R, Akimova T, Liu Y, Hancock WW. Histone deacetylases 6 and 9 and sirtuin-1 control foxp3+ Regulatory T cell function through shared and isoform-specific mechanisms. Sci Signaling (2012) 5(229):ra45. doi: 10.1126/scisignal.2002873
166. Dahiya S, Beier UH, Wang L, Han R, Jiao J, Akimova T, et al. Hdac10 deletion promotes foxp3(+) T-regulatory cell function. Sci Rep (2020) 10(1):424. doi: 10.1038/s41598-019-57294-x
167. Wang J, Chen J. Sirt1 regulates autoacetylation and histone acetyltransferase activity of tip60. J Biol Chem (2010) 285(15):11458–64. doi: 10.1074/jbc.M109.087585
168. Yi J, Huang X, Yang Y, Zhu WG, Gu W, Luo J. Regulation of histone acetyltransferase tip60 function by histone deacetylase 3. J Biol Chem (2014) 289(49):33878–86. doi: 10.1074/jbc.M114.575266
169. Li J, Du X, Shi H, Deng K, Chi H, Tao W. Mammalian sterile 20-like kinase 1 (Mst1) enhances the stability of forkhead box P3 (Foxp3) and the function of regulatory T cells by modulating foxp3 acetylation. J Biol Chem (2015) 290(52):30762–70. doi: 10.1074/jbc.M115.668442
170. Huang J, Wang L, Dahiya S, Beier UH, Han R, Samanta A, et al. Histone/protein deacetylase 11 targeting promotes foxp3+ Treg function. Sci Rep (2017) 7(1):8626. doi: 10.1038/s41598-017-09211-3
171. Morawski PA, Mehra P, Chen C, Bhatti T, Wells AD. Foxp3 protein stability is regulated by cyclin-dependent kinase 2. J Biol Chem (2013) 288(34):24494–502. doi: 10.1074/jbc.M113.467704
172. Chunder N, Wang L, Chen C, Hancock WW, Wells AD. Cyclin-dependent kinase 2 controls peripheral immune tolerance. J Immunol (Baltimore Md 1950) (2012) 189(12):5659–66. doi: 10.4049/jimmunol.1202313
173. Li Z, Lin F, Zhuo C, Deng G, Chen Z, Yin S, et al. Pim1 Kinase Phosphorylates the Human Transcription Factor Foxp3 at Serine 422 to Negatively Regulate Its Activity under Inflammation. J Biol Chem (2014) 289(39):26872–81. doi: 10.1074/jbc.M114.586651
174. Deng G, Nagai Y, Xiao Y, Li Z, Dai S, Ohtani T, et al. Pim-2 kinase influences regulatory T cell function and stability by mediating foxp3 protein N-terminal phosphorylation. J Biol Chem (2015) 290(33):20211–20. doi: 10.1074/jbc.M115.638221
175. Basu S, Golovina T, Mikheeva T, June CH, Riley JL. Cutting edge: foxp3-mediated induction of pim 2 allows human T regulatory cells to preferentially expand in rapamycin. J Immunol (Baltimore Md 1950) (2008) 180(9):5794–8. doi: 10.4049/jimmunol.180.9.5794
176. Nakahira K, Morita A, Kim NS, Yanagihara I. Phosphorylation of foxp3 by lck downregulates mmp9 expression and represses cell invasion. PloS One (2013) 8(10):e77099. doi: 10.1371/journal.pone.0077099
177. Riaz F, Huang Z, Pan F. Targeting post-translational modifications of foxp3: A new paradigm for regulatory T cell-specific therapy. Front Immunol (2023) 14:1280741. doi: 10.3389/fimmu.2023.1280741
178. Nie H, Zheng Y, Li R, Guo TB, He D, Fang L, et al. Phosphorylation of foxp3 controls regulatory T cell function and is inhibited by tnf-Α in rheumatoid arthritis. Nat Med (2013) 19(3):322–8. doi: 10.1038/nm.3085
179. Fleskens V, Minutti CM, Wu X, Wei P, Pals C, McCrae J, et al. Nemo-like kinase drives foxp3 stability and is critical for maintenance of immune tolerance by regulatory T cells. Cell Rep (2019) 26(13):3600–12.e6. doi: 10.1016/j.celrep.2019.02.087
180. Chen Z, Barbi J, Bu S, Yang HY, Li Z, Gao Y, et al. The ubiquitin ligase stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor foxp3. Immunity (2013) 39(2):272–85. doi: 10.1016/j.immuni.2013.08.006
181. Nakagawa T, Nakayama K. Protein monoubiquitylation: Targets and diverse functions. Genes to Cells devoted to Mol Cell Mech (2015) 20(7):543–62. doi: 10.1111/gtc.12250
182. Sadowski M, Suryadinata R, Tan AR, Roesley SN, Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life (2012) 64(2):136–42. doi: 10.1002/iub.589
183. Martinez-Forero I, Rouzaut A, Palazon A, Dubrot J, Melero I. Lysine 63 polyubiquitination in immunotherapy and in cancer-promoting inflammation. Clin Cancer Res an Off J Am Assoc Cancer Res (2009) 15(22):6751–7. doi: 10.1158/1078-0432.Ccr-09-1225
184. Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. Lubac synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep (2012) 13(9):840–6. doi: 10.1038/embor.2012.105
185. Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature (2011) 471(7340):591–6. doi: 10.1038/nature09816
186. Zinngrebe J, Rieser E, Taraborrelli L, Peltzer N, Hartwig T, Ren H, et al. –lubac Deficiency Perturbs Tlr3 Signaling to Cause Immunodeficiency and Autoinflammation. J Exp Med (2016) 213(12):2671–89. doi: 10.1084/jem.20160041
187. Damgaard RB, Nachbur U, Yabal M, Wong WW, Fiil BK, Kastirr M, et al. The ubiquitin ligase xiap recruits lubac for nod2 signaling in inflammation and innate immunity. Mol Cell (2012) 46(6):746–58. doi: 10.1016/j.molcel.2012.04.014
188. Rodgers MA, Bowman JW, Fujita H, Orazio N, Shi M, Liang Q, et al. The linear ubiquitin assembly complex (Lubac) is essential for nlrp3 inflammasome activation. J Exp Med (2014) 211(7):1333–47. doi: 10.1084/jem.20132486
189. Zhu F, Yi G, Liu X, Zhu F, Zhao A, Wang A, et al. Ring finger protein 31-mediated atypical ubiquitination stabilizes forkhead box P3 and thereby stimulates regulatory T-cell function. J Biol Chem (2018) 293(52):20099–111. doi: 10.1074/jbc.RA118.005802
190. Wang S, Li Y, Hu YH, Song R, Gao Y, Liu HY, et al. Stub1 is essential for T-cell activation by ubiquitinating carma1. Eur J Immunol (2013) 43(4):1034–41. doi: 10.1002/eji.201242554
191. Kobayashi T, Walsh PT, Walsh MC, Speirs KM, Chiffoleau E, King CG, et al. Traf6 is a critical factor for dendritic cell maturation and development. Immunity (2003) 19(3):353–63. doi: 10.1016/s1074-7613(03)00230-9
192. Ni X, Kou W, Gu J, Wei P, Wu X, Peng H, et al. Traf6 directs foxp3 localization and facilitates regulatory T-cell function through K63-linked ubiquitination. EMBO J (2019) 38(9):e99766. doi: 10.15252/embj.201899766
193. Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell (2005) 123(5):773–86. doi: 10.1016/j.cell.2005.11.007
194. van Loosdregt J, Fleskens V, Fu J, Brenkman AB, Bekker CP, Pals CE, et al. Stabilization of the transcription factor foxp3 by the deubiquitinase usp7 increases treg-cell-suppressive capacity. Immunity (2013) 39(2):259–71. doi: 10.1016/j.immuni.2013.05.018
195. Wang L, Kumar S, Dahiya S, Wang F, Wu J, Newick K, et al. Ubiquitin-specific protease-7 inhibition impairs tip60-dependent foxp3+ T-regulatory cell function and promotes antitumor immunity. EBioMedicine (2016) 13:99–112. doi: 10.1016/j.ebiom.2016.10.018
196. Li Y, Lu Y, Wang S, Han Z, Zhu F, Ni Y, et al. Usp21 prevents the generation of T-helper-1-like treg cells. Nat Commun (2016) 7:13559. doi: 10.1038/ncomms13559
197. Zhang J, Chen C, Hou X, Gao Y, Lin F, Yang J, et al. Identification of the E3 deubiquitinase ubiquitin-specific peptidase 21 (Usp21) as a positive regulator of the transcription factor gata3. J Biol Chem (2013) 288(13):9373–82. doi: 10.1074/jbc.M112.374744
198. Cortez JT, Montauti E, Shifrut E, Gatchalian J, Zhang Y, Shaked O, et al. Crispr screen in regulatory T cells reveals modulators of foxp3. Nature (2020) 582(7812):416–20. doi: 10.1038/s41586-020-2246-4
199. Yang J, Wei P, Barbi J, Huang Q, Yang E, Bai Y, et al. The deubiquitinase usp44 promotes treg function during inflammation by preventing foxp3 degradation. EMBO Rep (2020) 21(9):e50308. doi: 10.15252/embr.202050308
200. Yu X, Lao Y, Teng XL, Li S, Zhou Y, Wang F, et al. Senp3 maintains the stability and function of regulatory T cells via bach2 desumoylation. Nat Commun (2018) 9(1):3157. doi: 10.1038/s41467-018-05676-6
201. Chang YH, Weng CL, Lin KI. O-glcnacylation and its role in the immune system. J Biomed Sci (2020) 27(1):57. doi: 10.1186/s12929-020-00648-9
202. Cabral J, Hanley SA, Gerlach JQ, O'Leary N, Cunningham S, Ritter T, et al. Distinctive surface glycosylation patterns associated with mouse and human cd4(+) regulatory T cells and their suppressive function. Front Immunol (2017) 8:987. doi: 10.3389/fimmu.2017.00987
203. Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature (2007) 446(7139):1017–22. doi: 10.1038/nature05815
204. de Jesus TJ, Tomalka JA, Centore JT, Staback Rodriguez FD, Agarwal RA, Liu AR, et al. Negative regulation of foxp3 expression by C-rel O-glcnacylation. Glycobiology (2021) 31(7):812–26. doi: 10.1093/glycob/cwab001
205. Liu B, Salgado OC, Singh S, Hippen KL, Maynard JC, Burlingame AL, et al. The lineage stability and suppressive program of regulatory T cells require protein O-glcnacylation. Nat Commun (2019) 10(1):354. doi: 10.1038/s41467-019-08300-3
206. Shan F, Somasundaram A, Bruno TC, Workman CJ, Vignali DAA. Therapeutic targeting of regulatory T cells in cancer. Trends Cancer (2022) 8(11):944–61. doi: 10.1016/j.trecan.2022.06.008
207. Di Pilato M, Kim EY, Cadilha BL, Prüßmann JN, Nasrallah MN, Seruggia D, et al. Targeting the cbm complex causes T(Reg) cells to prime tumours for immune checkpoint therapy. Nature (2019) 570(7759):112–6. doi: 10.1038/s41586-019-1215-2
208. Di Pilato M, Gao Y, Sun Y, Fu A, Grass C, Seeholzer T, et al. Translational studies using the malt1 inhibitor (S)-mepazine to induce treg fragility and potentiate immune checkpoint therapy in cancer. J immunotherapy Precis Oncol (2023) 6(2):61–73. doi: 10.36401/jipo-22-18
209. Wang TB, Geng M, Jin H, Tang AG, Sun H, Zhou LZ, et al. Srebp1 site 1 protease inhibitor pf-429242 suppresses renal cell carcinoma cell growth. Cell Death Dis (2021) 12(8):717. doi: 10.1038/s41419-021-03999-9
210. Tsai HC, Han MH. Sphingosine-1-phosphate (S1p) and S1p signaling pathway: Therapeutic targets in autoimmunity and inflammation. Drugs (2016) 76(11):1067–79. doi: 10.1007/s40265-016-0603-2
211. Liu G, Yang K, Burns S, Shrestha S, Chi H. The S1p(1)-mtor axis directs the reciprocal differentiation of T(H)1 and T(Reg) cells. Nat Immunol (2010) 11(11):1047–56. doi: 10.1038/ni.1939
212. Augello G, Puleio R, Emma MR, Cusimano A, Loria GR, McCubrey JA, et al. A pten inhibitor displays preclinical activity against hepatocarcinoma cells. Cell Cycle (Georgetown Tex) (2016) 15(4):573–83. doi: 10.1080/15384101.2016.1138183
213. Yu N, Fu S, Xu Z, Liu Y, Hao J, Zhang A, et al. Synergistic antitumor responses by combined gitr activation and sunitinib in metastatic renal cell carcinoma. Int J Cancer (2016) 138(2):451–62. doi: 10.1002/ijc.29713
214. Li S, Liu M, Do MH, Chou C, Stamatiades EG, Nixon BG, et al. Cancer immunotherapy via targeted tgf-Β Signalling blockade in T(H) cells. Nature (2020) 587(7832):121–5. doi: 10.1038/s41586-020-2850-3
215. Yi M, Niu M, Zhang J, Li S, Zhu S, Yan Y, et al. Combine and conquer: Manganese synergizing anti-tgf-Β/pd-L1 bispecific antibody ym101 to overcome immunotherapy resistance in non-inflamed cancers. J Hematol Oncol (2021) 14(1):146. doi: 10.1186/s13045-021-01155-6
216. Lé AM, Torres T. Ox40-ox40l inhibition for the treatment of atopic dermatitis-focus on rocatinlimab and amlitelimab. Pharmaceutics (2022) 14(12):2753. doi: 10.3390/pharmaceutics14122753
217. Alvim RG, Georgala P, Nogueira L, Somma AJ, Nagar K, Thomas J, et al. Combined ox40 agonist and pd-1 inhibitor immunotherapy improves the efficacy of vascular targeted photodynamic therapy in a urothelial tumor model. Molecules (Basel Switzerland) (2021) 26(12):3744. doi: 10.3390/molecules26123744
218. Leja-Szpak A, Góralska M, Link-Lenczowski P, Czech U, Nawrot-Porąbka K, Bonior J, et al. The opposite effect of L-kynurenine and ahr inhibitor ch223191 on apoptotic protein expression in pancreatic carcinoma cells (Panc-1). Anti-cancer Agents medicinal Chem (2019) 19(17):2079–90. doi: 10.2174/1871520619666190415165212
219. Liu M, Li Z, Yao W, Zeng X, Wang L, Cheng J, et al. Ido inhibitor synergized with radiotherapy to delay tumor growth by reversing T cell exhaustion. Mol Med Rep (2020) 21(1):445–53. doi: 10.3892/mmr.2019.10816
220. Van Damme H, Dombrecht B, Kiss M, Roose H, Allen E, Van Overmeire E, et al. Therapeutic depletion of ccr8(+) tumor-infiltrating regulatory T cells elicits antitumor immunity and synergizes with anti-pd-1 therapy. J immunotherapy Cancer (2021) 9(2):e001749. doi: 10.1136/jitc-2020-001749
221. Wang L, Jenkins TJ, Dai M, Yin W, Pulido JC, Lamantia-Martin E, et al. Antagonism of chemokine receptor ccr8 is ineffective in a primate model of asthma. Thorax (2013) 68(6):506–12. doi: 10.1136/thoraxjnl-2012-203012
222. Cha JH, Yang WH, Xia W, Wei Y, Chan LC, Lim SO, et al. Metformin promotes antitumor immunity via endoplasmic-reticulum-associated degradation of pd-L1. Mol Cell (2018) 71(4):606–20.e7. doi: 10.1016/j.molcel.2018.07.030
223. Kunisada Y, Eikawa S, Tomonobu N, Domae S, Uehara T, Hori S, et al. Attenuation of cd4(+)Cd25(+) regulatory T cells in the tumor microenvironment by metformin, a type 2 diabetes drug. EBioMedicine (2017) 25:154–64. doi: 10.1016/j.ebiom.2017.10.009
224. Li X, Tian R, Wang L, Xu C, Wu H, Liu L, et al. Oligomycin a promotes radioresistance in ht29 colorectal cancer cells and its mechanisms. Zhong nan da xue xue bao Yi xue ban = J Cent South Univ Med Sci (2021) 46(2):113–20. doi: 10.11817/j.issn.1672-7347.2021.200063
225. Beckers A, Organe S, Timmermans L, Scheys K, Peeters A, Brusselmans K, et al. Chemical inhibition of acetyl-coa carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res (2007) 67(17):8180–7. doi: 10.1158/0008-5472.Can-07-0389
226. Levin D, Bell S, Sund R, Hartikainen SA, Tuomilehto J, Pukkala E, et al. Pioglitazone and bladder cancer risk: A multipopulation pooled, cumulative exposure analysis. Diabetologia (2015) 58(3):493–504. doi: 10.1007/s00125-014-3456-9
227. Yao X, Xie R, Cao Y, Tang J, Men Y, Peng H, et al. Simvastatin induced ferroptosis for triple-negative breast cancer therapy. J nanobiotechnology (2021) 19(1):311. doi: 10.1186/s12951-021-01058-1
228. Duarte JA, de Barros ALB, Leite EA. The potential use of simvastatin for cancer treatment: A review. Biomedicine pharmacotherapy = Biomedecine pharmacotherapie (2021) 141:111858. doi: 10.1016/j.biopha.2021.111858
229. Miska J, Lee-Chang C, Rashidi A, Muroski ME, Chang AL, Lopez-Rosas A, et al. Hif-1α Is a metabolic switch between glycolytic-driven migration and oxidative phosphorylation-driven immunosuppression of tregs in glioblastoma. Cell Rep (2019) 27(1):226–37.e4. doi: 10.1016/j.celrep.2019.03.029
230. Chu PC, Wu YC, Chen CY, Hung YS, Chang CS. Novel hif-1α Inhibitor cdmp-tqz for cancer therapy. Future medicinal Chem (2021) 13(12):1057–72. doi: 10.4155/fmc-2020-0307
231. Ouyang C, Zhang J, Lei X, Xie Z, Liu X, Li Y, et al. Advances in antitumor research of hif-1α Inhibitor yc-1 and its derivatives. Bioorganic Chem (2023) 133:106400. doi: 10.1016/j.bioorg.2023.106400
232. Lee K, Kim HM. A novel approach to cancer therapy using px-478 as a hif-1α Inhibitor. Arch pharmacal Res (2011) 34(10):1583–5. doi: 10.1007/s12272-011-1021-3
233. Bailey CM, Liu Y, Peng G, Zhang H, He M, Sun D, et al. Liposomal formulation of hif-1α Inhibitor echinomycin eliminates established metastases of triple-negative breast cancer. Nanomedicine nanotechnology biology Med (2020) 29:102278. doi: 10.1016/j.nano.2020.102278
234. Rekhi UR, Catunda RQ, Alexiou M, Sharma M, Fong A, Febbraio M. Impact of a cd36 inhibitor on porphyromonas gingivalis mediated atherosclerosis. Arch Oral Biol (2021) 126:105129. doi: 10.1016/j.archoralbio.2021.105129
235. Liu J, Bu X, Chu C, Dai X, Asara JM, Sicinski P, et al. Prmt1 mediated methylation of cgas suppresses anti-tumor immunity. Nat Commun (2023) 14(1):2806. doi: 10.1038/s41467-023-38443-3
236. Liu J, He D, Cheng L, Huang C, Zhang Y, Rao X, et al. P300/cbp inhibition enhances the efficacy of programmed death-ligand 1 blockade treatment in prostate cancer. Oncogene (2020) 39(19):3939–51. doi: 10.1038/s41388-020-1270-z
237. Joshi RN, Fernandes SJ, Shang MM, Kiani NA, Gomez-Cabrero D, Tegnér J, et al. Phosphatase inhibitor ppp1r11 modulates resistance of human T cells toward treg-mediated suppression of cytokine expression. J leukocyte Biol (2019) 106(2):413–30. doi: 10.1002/jlb.2a0618-228r
238. Zhang Z, Kong X, Ligtenberg MA, van Hal-van Veen SE, Visser NL, de Bruijn B, et al. Rnf31 inhibition sensitizes tumors to bystander killing by innate and adaptive immune cells. Cell Rep Med (2022) 3(6):100655. doi: 10.1016/j.xcrm.2022.100655
239. Wu X, Xue R, Peng H, Gan X, Lu X, Yan W, et al. Traf6 inhibitor boosts antitumor immunity by impeding regulatory T cell migration in hepa1-6 tumor model. Int Immunopharmacol (2019) 77:105965. doi: 10.1016/j.intimp.2019.105965
240. Yu J, Cui J, Zhang X, Xu H, Chen Z, Li Y, et al. The ox40-traf6 axis promotes ctla-4 degradation to augment antitumor cd8(+) T-cell immunity. Cell Mol Immunol (2023) 20(12):1445–56. doi: 10.1038/s41423-023-01093-y
241. Bishop RT, Marino S, Carrasco G, Li B, Allen RJ, Sparatore A, et al. Combined administration of a small-molecule inhibitor of traf6 and docetaxel reduces breast cancer skeletal metastasis and osteolysis. Cancer Lett (2020) 488:27–39. doi: 10.1016/j.canlet.2020.05.021
242. Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res (2017) 27(1):109–18. doi: 10.1038/cr.2016.151
243. Zhang B, Sun J, Guan H, Guo H, Huang B, Chen X, et al. Integrated single-cell and bulk rna sequencing revealed the molecular characteristics and prognostic roles of neutrophils in pancreatic cancer. Aging (2023) 15(18):9718–42. doi: 10.18632/aging.205044
244. Zhang B, Liu J, Li H, Huang B, Zhang B, Song B, et al. Integrated multi-omics identified the novel intratumor microbiome-derived subtypes and signature to predict the outcome, tumor microenvironment heterogeneity, and immunotherapy response for pancreatic cancer patients. Front Pharmacol (2023) 14:1244752. doi: 10.3389/fphar.2023.1244752
245. Pan C, Deng D, Wei T, Wu Z, Zhang B, Yuan Q, et al. Metabolomics study identified bile acids as potential biomarkers for gastric cancer: A case control study. Front Endocrinol (2022) 13:1039786. doi: 10.3389/fendo.2022.1039786
246. Békés M, Langley DR, Crews CM. Protac targeted protein degraders: The past is prologue. Nat Rev Drug Discovery (2022) 21(3):181–200. doi: 10.1038/s41573-021-00371-6
247. Sakamoto KM, Kim KB, Verma R, Ransick A, Stein B, Crews CM, et al. Development of protacs to target cancer-promoting proteins for ubiquitination and degradation. Mol Cell Proteomics MCP (2003) 2(12):1350–8. doi: 10.1074/mcp.T300009-MCP200
Keywords: regulatory T cells, reprogramming, tumor immunity, Foxp3, metabolize, posttranslational modification, cancer therapy
Citation: Liu J, Zhang B, Zhang G and Shang D (2024) Reprogramming of regulatory T cells in inflammatory tumor microenvironment: can it become immunotherapy turning point? Front. Immunol. 15:1345838. doi: 10.3389/fimmu.2024.1345838
Received: 28 November 2023; Accepted: 29 January 2024;
Published: 21 February 2024.
Edited by:
Peng Liu, Sun Yat-sen University, ChinaReviewed by:
Andras Perl, Upstate Medical University, United StatesMing Yi, Zhejiang University, China
Michal Kuczma, Georgia State University, United States
Yongwei Zheng, Guangzhou Bio-Gene Technology Co., Ltd., China
Copyright © 2024 Liu, Zhang, Zhang and Shang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Shang, c2hhbmdkb25nQGRtdS5lZHUuY24=
†These authors have contributed equally to this work
 Jinming Liu
Jinming Liu Biao Zhang
Biao Zhang Guolin Zhang2
Guolin Zhang2 Dong Shang
Dong Shang