- 1Department of Medical Laboratory Sciences, College of Medicine and Health Sciences, Wollo University, Dessie, Ethiopia
- 2Department of Medical Laboratory Science, College of Health Sciences, Woldia University, Woldia, Ethiopia
- 3Department of Pediatric and Child Health Nursing, School of Nursing and Midwifery, College of Medicine and Health Sciences, Wollo University, Dessie, Ethiopia
Introduction: Inflammatory bowel disease (IBD) poses a growing global burden, necessitating the discovery of reliable biomarkers for early diagnosis. The clinical significance of dysregulated expression of long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) in diagnosing IBD has not been well established. Thus, our study aimed to investigate the diagnostic value of lncRNAs and circRNAs for IBD based on currently available studies.
Methods: A comprehensive search was carried out in diverse electronic databases, such as PubMed, Embase, Scopus, Science Direct and Wiley Online Library to retrieve articles published until October 30, 2023. Stata 17.0 software was employed to determine pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic ratio (DOR), and area under the curve (AUC). Heterogeneity, subgroup analysis, and meta-regression were explored, and publication bias was assessed using Deeks’ funnel plot. Fagan’s nomogram and likelihood ratio scattergram were employed to evaluate the clinical validity.
Result: A total of 11 articles encompassing 21 studies which involved 1239 IBD patients and 985 healthy controls were investigated. The findings revealed lncRNAs exhibit high level of pooled sensitivity 0.94 (95% CI: 0.87-0.97) and specificity 0.99 (95% CI: 0.89-1.00), along with PLR, NLR, DOR, and AUC values of 64.25 (95% CI: 7.39-558.66), 0.06 (95% CI: 0.03-0.13), 1055.25 (95% CI: 70.61-15770.77), and 0.99 (95% CI: 0.97-0.99), respectively. Conversely, CircRNAs showed moderate accuracy in IBD diagnosis, with sensitivity of 0.68 (95% CI: 0.61-0.73), specificity of 0.73 (95% CI: 0.65-0.79), PLR of 2.47 (95% CI: 1.94-3.16), NLR of 0.45 (95% CI: 0.38-0.53), DOR of 5.54 (95% CI: 3.88-7.93), and AUC value of 0.75 (95% CI: 0.71-0.79). Moreover, findings from subgroup analysis depicted heightened diagnostic efficacy when employing lncRNA H19 and a large sample size (≥100), with notable efficacy in diagnosing both ulcerative colitis (UC) and Crohn’s disease (CD).
Conclusion: LncRNAs exhibit high diagnostic accuracy in distinguishing patients with IBD from healthy controls signifying their possible use as potential biomarkers, while circRNAs showed moderate diagnostic accuracy. Nevertheless, to validate our findings and confirm the clinical utility of lncRNAs and circRNAs in IBD diagnosis, a large pool of prospective and multi-center studies should be undertaken.
Systematic review registration: https://www.crd.york.ac.uk/PROSPERO, identifier CRD42023491840.
Introduction
Inflammatory bowel disease is primarily composed of ulcerative colitis (UC), which is confined to the mucosa of the colon, and Crohn’s disease (CD), which can impact any part of the gastrointestinal tract (GIT) from the mouth to the anus. These are two phenotypes within the GIT that share a persistent state of inflammation but vary in symptoms, the location of the disease, and histopathological features (1–3). The pathogenesis of IBD remains unclear, with various factors potentially interacting to contribute to its development. These factors include genetic predisposition, microbial infection, and environmental influences (4). Genetic susceptibility, environmental factors (such as diet, smoking, and microbial exposure), the presence of pathogenic factors such as abnormal gut microbiota, and dysregulated immune responses including cytokines (such as TNFα) and immune cells with exaggerated inflammatory responses contribute to chronic inflammation in the GIT with repeated cycles of relapse and remission, leading to the characteristic symptoms of IBD (5). Currently, IBD treatments include anti-inflammatory drugs (amino salicylates and corticosteroids), immunomodulators (thiopurines, methotrexate, and newer biologic agents (e.g., anti-TNF drugs like infliximab), biological therapies (monoclonal antibodies targeting specific inflammatory pathways such as anti-integrins and interleukins), and surgery (6). However, these are not entirely curative treatments for IBD. While these treatments can effectively induce and maintain remission, challenges exist, including variable response rates, side effects, and the need for personalized treatment approaches. Additionally, the chronic nature of IBD necessitates long-term management strategies (7, 8).
Between 1990 and 2017, there was a substantial increase in the prevalence of IBD, rising from 3.7 million to over 6.8 million. This marked an 85.1% global rise in IBD cases during that timeframe. Furthermore, across these years, the prevalence rate consistently demonstrated a significant predominance in females over males, with females constituting 57% and males making up 43% (9). IBD exerts profound effects on patients, impacting their quality of life, mental health, work productivity, and the utilization of healthcare resources (10, 11). Additionally, the number of deaths related to IBD increased by 67.0% from 1990 to 2017, rising from 23,000 to 38,000 (9).
The complexity and variability of symptoms associated with IBD make it challenging to establish a singular “gold standard” test for diagnosis, severity assessment, or treatment response evaluation. Physicians adopt a comprehensive approach, considering clinical symptoms, laboratory indices, radiological investigations, endoscopy, and histological examination of tissue specimens to thoroughly assess disease activity and formulate appropriate treatment strategies (12, 13). Extensive studies over the past decades have focused on laboratory indices for IBD, leading to the integration of specific biomarkers into clinical practice (14). Recent progress in biomarker discovery for IBD emphasizes the integration of cutting-edge proteomic techniques through innovations such as liquid chromatography-mass spectrometry (LC-MS), electrospray ionization (ESI) and tandem mass spectrometry (MS) that allow distinct protein signatures associated with IBD subtypes, employing high-throughput proteomic analyses (15, 16). While the integration of proteomic data with genomics and transcriptomics hold promises for accurate diagnosis and personalized treatment, challenges such as standardization, reproducibility, and validation persist (17). Currently, blood and fecal biomarkers such as anti-neutrophil cytoplasm antibody, anti-laminaribioside carbohydrate antibody, C-reactive protein, fecal calprotectin, and anti-Saccharomyces cerevisiae antibody are employed in the management of IBD. However, their utility is limited in the initial diagnosis of IBD due to their low specificity or sensitivity (18, 19). Nevertheless, despite these advancements, there is still no ideal biomarker possessing all the necessary qualities for the precise diagnosis of IBD, differentiation between IBD subtypes, or effective monitoring of disease activity (20). Therefore, discovering ideal diagnostic biomarkers for IBD is crucial. Optimal biomarkers are expected to be non-invasive, sensitive, disease-specific, easy to perform, and cost-effective (14).
Non-coding RNAs (ncRNAs), including long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs), constitute a diverse set of transcripts that are not translated into proteins. Since their discovery, ncRNAs have emerged as significant regulators of various biological functions across different cell types and tissues, and their dysregulation has been associated with disease (21). Noncoding RNAs, exert regulatory control through diverse mechanisms. LncRNAs influence gene expression at transcriptional and post-transcriptional levels by interacting with chromatin and RNA binding proteins, while miRNAs primarily act post-transcriptionally by binding to the 3’ untranslated region (UTR) of target mRNAs, and hinder translation initiation by preventing ribosome binding to target mRNAs. CircRNAs function as competitive endogenous RNAs (ceRNAs) by acting as miRNA sponges. Collectively, these ncRNAs form intricate regulatory networks, and employ diverse mechanisms to regulate biological functions, from influencing chromatin dynamics to fine-tuning gene expression at the post-transcriptional level, impacting cellular processes crucial for understanding diseases like IBD and exploring diagnostic potential (22). Recent literature has uncovered evolving associations between IBD and noncoding RNAs, recognizing them as pivotal regulators of gene expression at both transcriptional and post-transcriptional levels (23). While certain noncoding RNAs play a protective role by preserving gut microbiota homeostasis (24) and controlling intestinal inflammation, the majority are implicated in the pathogenesis of IBD through disruptions in autophagy, the intestinal barrier, and immune homeostasis (25).
Multiple studies have emphasized the notable contribution of dysregulated lncRNAs and circRNAs in diagnosing IBD, indicating that both types of RNAs serve as effective advanced diagnostic biomarkers with increased sensitivity and specificity for IBD diagnosis (26–31). Besides, there are reports highlighting distinct patterns in the relative expression levels of lncRNAs and circRNAs among CD and UC patients (32–35). More importantly, the expression profiling of both parameters in individuals with IBD has produced inconsistent results. Given these varied insights, a comprehensive analysis is imperative to evaluate the applicability of these biomarkers as diagnostic tools for IBD. Therefore, this systematic review and meta-analysis aimed to assess the overall diagnostic accuracy of lncRNAs and circRNAs for IBD.
Methods
Study protocol
This study adheres to the commendations specified in the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) guideline (36) as indicated in Supplementary File 1. The study protocol was officially registered in the Prospective Register of Systematic Reviews (PROSPERO) with the registration identification number of CRD42023491840.
Literature search strategy and data sources
A systematic and thorough search of the existing literature on the diagnostic efficacy of lncRNAs and circRNAs for IBD was conducted by two independent researchers (MAB and EA) using diverse electronic bibliographic databases, such as PubMed, Embase, Scopus, Science Direct and Wiley Online Library. Furthermore, a manual search on Google was executed to uncover any pertinent studies possibly overlooked in the electronic database searches, by scrutinizing the bibliographies of the identified studies. The conclusive search took place on October 30, 2023. The search strategy incorporated Medical Subject Heading (MeSH) terms and keywords, such as “long noncoding RNA”, “long non-coding RNA”, “long non coding RNA”, “long ncRNA”, “long ncRNAs”, “ncRNAs, long”, “lncRNA”, “lnc RNA”, “ncRNA”, “RNA, long noncoding”, “noncoding RNA, long”, “circular RNA”, “circular RNAs”, “RNA, circular”, “circRNA”, “circRNAs”, “diagnos*”, “inflammatory bowel disease*”, “Crohn*”, “ulcerative colitis”. Boolean operators (“OR” and “AND”) were used as necessary in the advanced search databases. The detailed search strategy is available in the Supplementary File 2.
Eligibility criteria
This review focused on specific categories of research, specifically observational studies (including cross-sectional, case-control and cohort studies) published until October 30, 2023, which investigated the potential of lncRNA and circRNA as a diagnostic marker for differentiating between IBD patients and healthy individuals. Furthermore, the selected studies were required to provide crucial information such as sensitivity, specificity, and sample sizes, facilitating the computation of key diagnostic metrics like true positives (TP), false positives (FP), false negatives (FN), and true negatives (TN). Conversely, the review excluded various types of articles, such as review articles, case reports, narrative reviews, conference abstracts, editorials, commentaries, letters to the editor, and author replies. Additionally, studies lacking human subjects or essential data for calculating TP, FP, TN, and FN were excluded. These defined inclusion and exclusion criteria were applied to guide the study selection process for the meta-analysis.
Study selection and data extraction
The literature search results were imported into EndNote 20 software (Clarivate Analytics USA) and duplicate were removed. Following this, a comprehensive screening process was conducted for each chosen article involving the assessment of the title, abstract, and full text by two independent reviewers (MAB and EA), adhering to pre-established eligibility criteria. In instances where discrepancies or disagreements arose between the two reviewers, a discussion ensued, and a third reviewer (AG) was brought in as necessary to make final determinations regarding the inclusion of articles in the review.
Vital data from the eligible studies were extracted into an Excel spreadsheet using a preconceived data abstraction form. Three researchers (MAB, EA and MT) extracted data including first author name, publication year, study country, extracted lncRNA or circRNA type, lncRNA or circRNA expression pattern, specimen type, internal reference control, sample sizes, sex and age for both IBD patients and healthy individuals, diagnostic methods, and cut-off values. Moreover, diagnostic parameters like sensitivity, specificity, and the area under the curve (AUC) were extracted. The three reviewers meticulously reviewed and verified their extraction results. Any disparities between the data extractors were resolved through discussion and consensus, involving a third reviewer (ST). This process ensured the integrity of the collected data.
Quality assessment
Two independent evaluators (ZM and ST) carried out a thorough assessment of the methodological and substantive quality of eligible studies using the modified Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool, implemented with the support of Review Manager (RevMan) 5.3 software (37). The QUADAS-2 tool consists of four domains: patient selection, index test, reference standard, and flow and timing. The process involved appraising the clinical relevance of selected patients, the performance of the index test, and the adequacy of the reference standard. The overall risk of bias for the comparison can then be assessed by considering the risk of bias for each domain. The resulting risk of bias was then categorized as low (L), high (H), or unclear (U) based on the outcomes of this evaluation.
Statistical analysis
Stata version 17.0 software (Stata Corp., College Station, TX) was used for analyzing extracted data. To gauge and assess heterogeneity among the studies, the Cochrane Q test and I2 statistics were employed. Significant heterogeneity was identified when the I2 test statistic values exceeded 50%, and the p-value was less than 0.05. The extracted data from each eligible study were transformed into diagnostic parameters, encompassing true positives (TP), false positives (FP), false negatives (FN), and true negatives (TN). These parameters were utilized to compute pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under the curve (AUC) through a random-effects model. The overall diagnostic accuracy of lncRNA and circRNA in diagnosing IBD was assessed using AUC and DOR from the summary receiver characteristic curve (SROC). The presence of a threshold effect was established by analyzing the Spearman correlation coefficient and visually inspecting the SROC curve. A p-value of less than 0.05 derived from the Spearman correlation coefficient, combined with the absence of the characteristic “shoulder-arm” shape in the SROC curve, indicated the presence of a threshold effect. Subgroup analyses and meta-regression analyses were conducted to explore the primary sources of heterogeneity. The presence of publication bias was assessed using Deeks’ funnel plot asymmetry, where a p-value greater than 0.10 indicated the absence of publication bias. Furthermore, the clinical utility of lncRNAs and circRNAs in distinguishing IBD patients from healthy individuals was assessed deploying the Fagan plot and likelihood ratio scattergram.
Results
Selection of studies
Upon conducting the primary search of the databases and other sources, a total of 434 records were retrieved. Following the removal of duplicates, the remaining 124 articles were screened based on review of title and abstract, and 82 were removed. After conducting an in-depth review, a total of 42 articles were deemed suitable for full-text analysis, and were thoroughly evaluated against the eligibility criteria, resulting in the exclusion of 31 studies for various reasons. Ultimately, after this rigorous process, 21 studies from 11 sources (31, 32, 38–46) were found to be potentially eligible for inclusion in the meta-analysis (Figure 1).
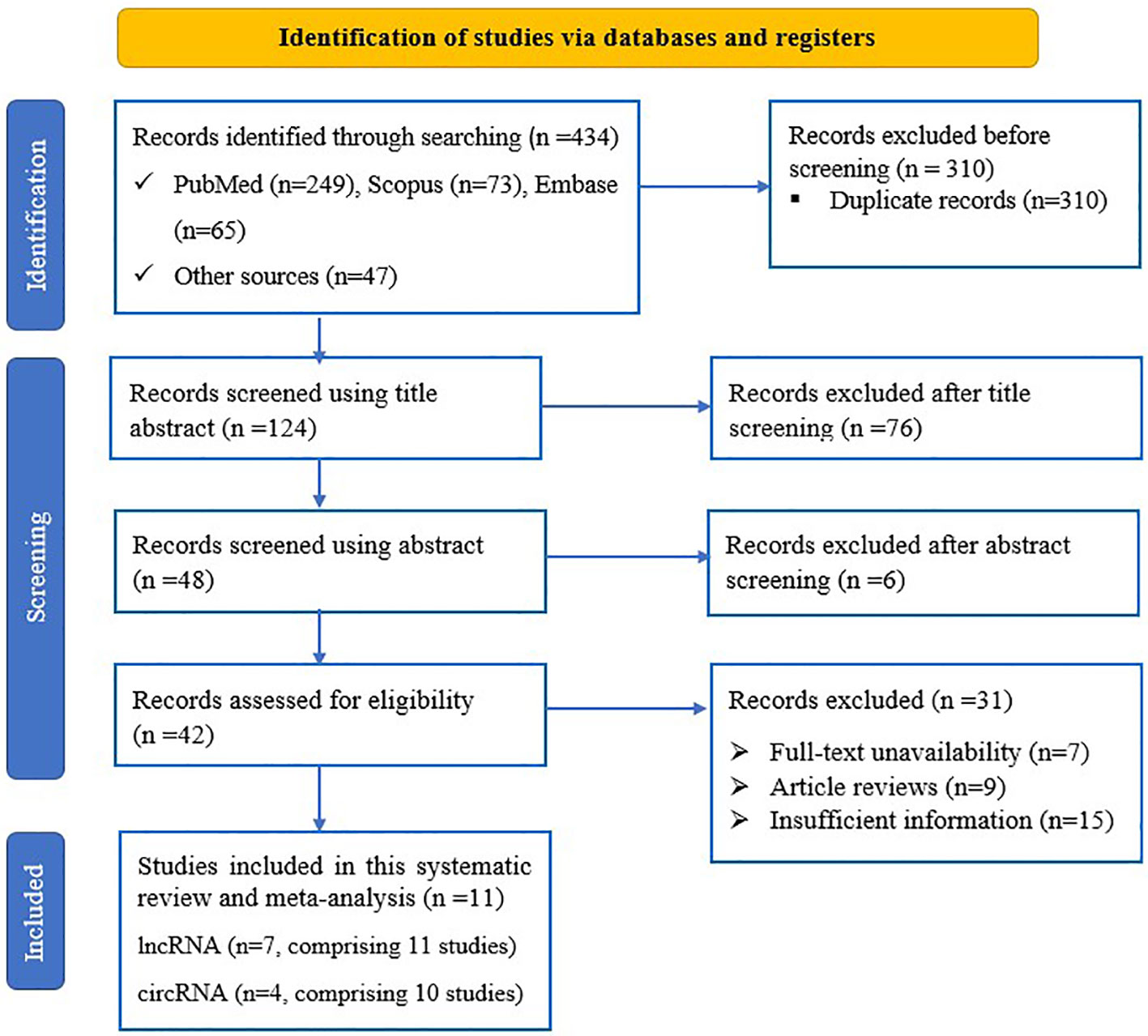
Figure 1 PRISMA flow diagram of eligible study selection process for the systematic review and meta-analysis.
Characteristics of included studies
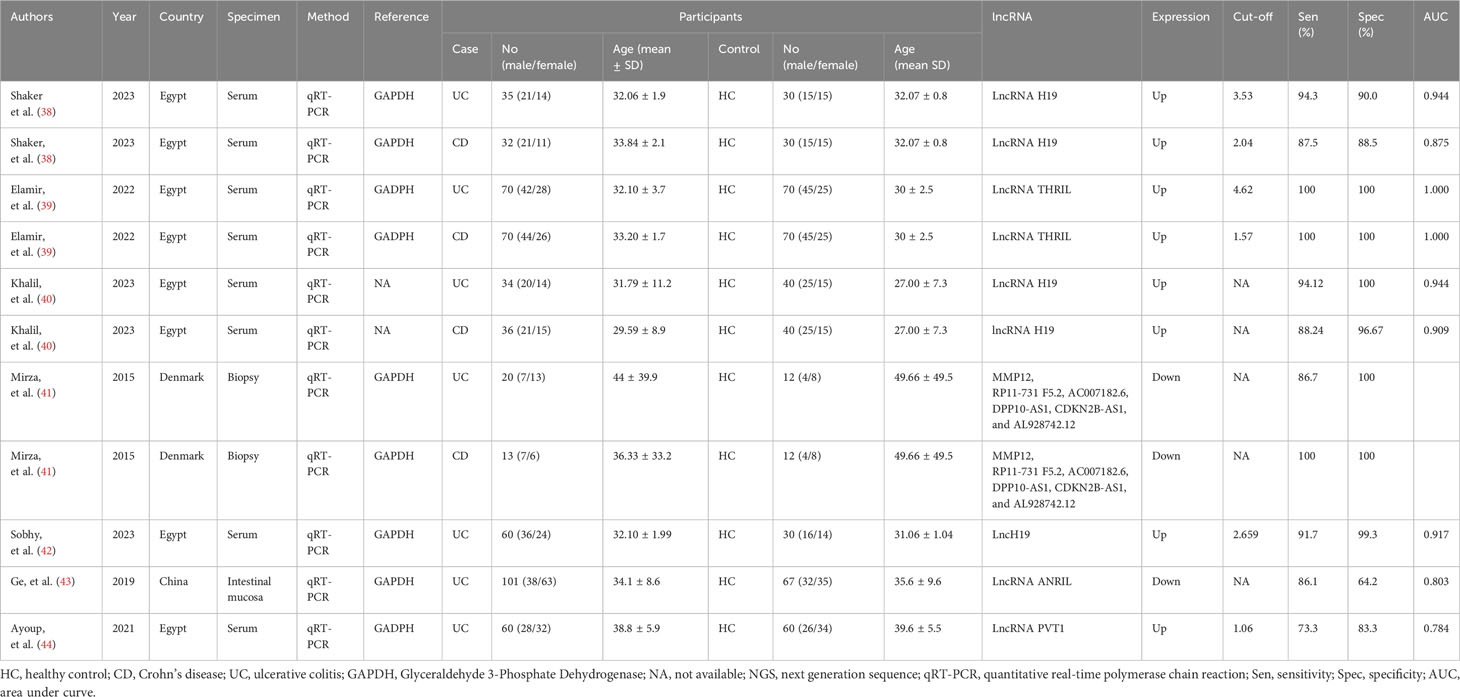
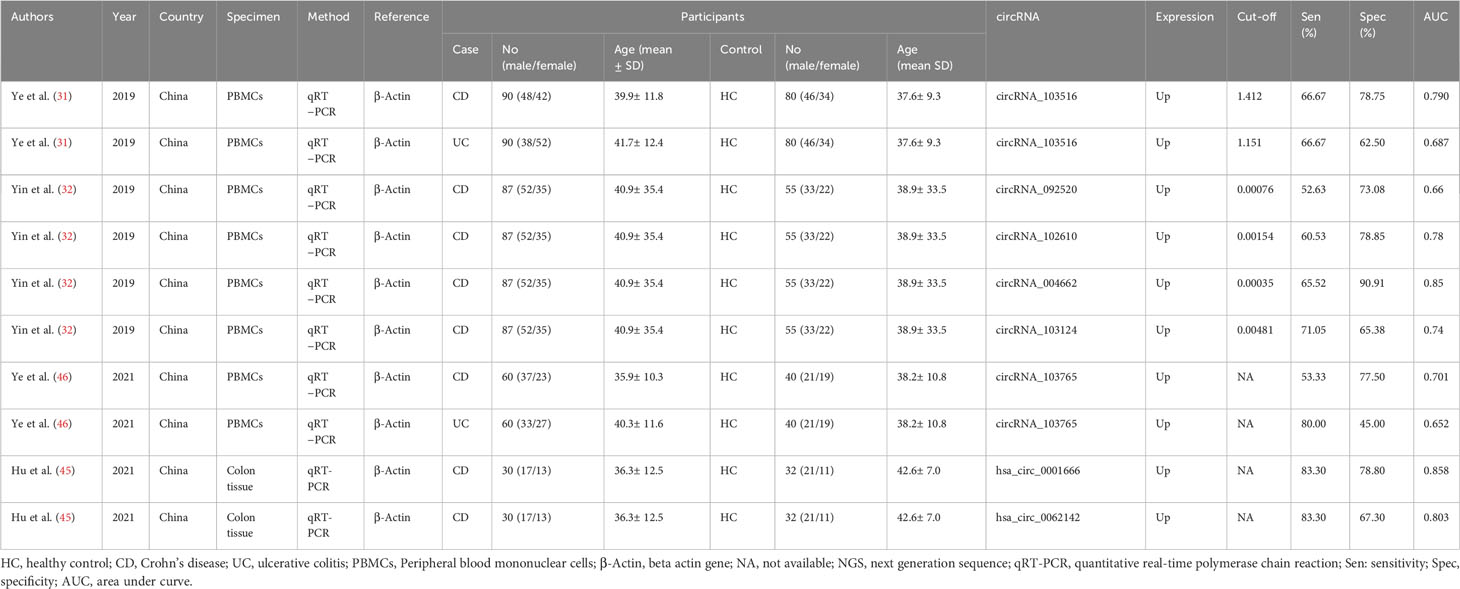
This systematic review and meta-analysis included data on lncRNAs from a total of 11 studies, involving 531 individuals diagnosed with IBD and 461 healthy controls whereas 10 studies were included regarding circRNAs encompassing a total of 708 IBD patients and 524 healthy controls. Majority of the included studies were from China, followed by Egypt, and used serum sample for lncRNA and circRNA detection. All studies deployed quantitative real-time PCR (qRT-PCR) for assessing lncRNA and circRNA expression. Concerning lncRNA profiling, 9 studies focused on individual lncRNA, while 2 studies explored combined lncRNAs. Among the findings, eight studies reported an upregulation in lncRNA expression, whereas three studies reported a downregulation. Among ten lncRNA, three lncRNAs (lncRNA H19, lncRNA THRIL, lncRNA PVT1) were reported to be upregulated. One lncRNA (lncRNA ANRIL) was downregulated in serum specimens of IBD patients (Table 1A). On the other hand, all ten studies focused on individual circRNA, reported upregulation of circRNA expression, and used β-Actin as an internal reference (Table 1B).
Quality assessment of studies included in meta-analysis
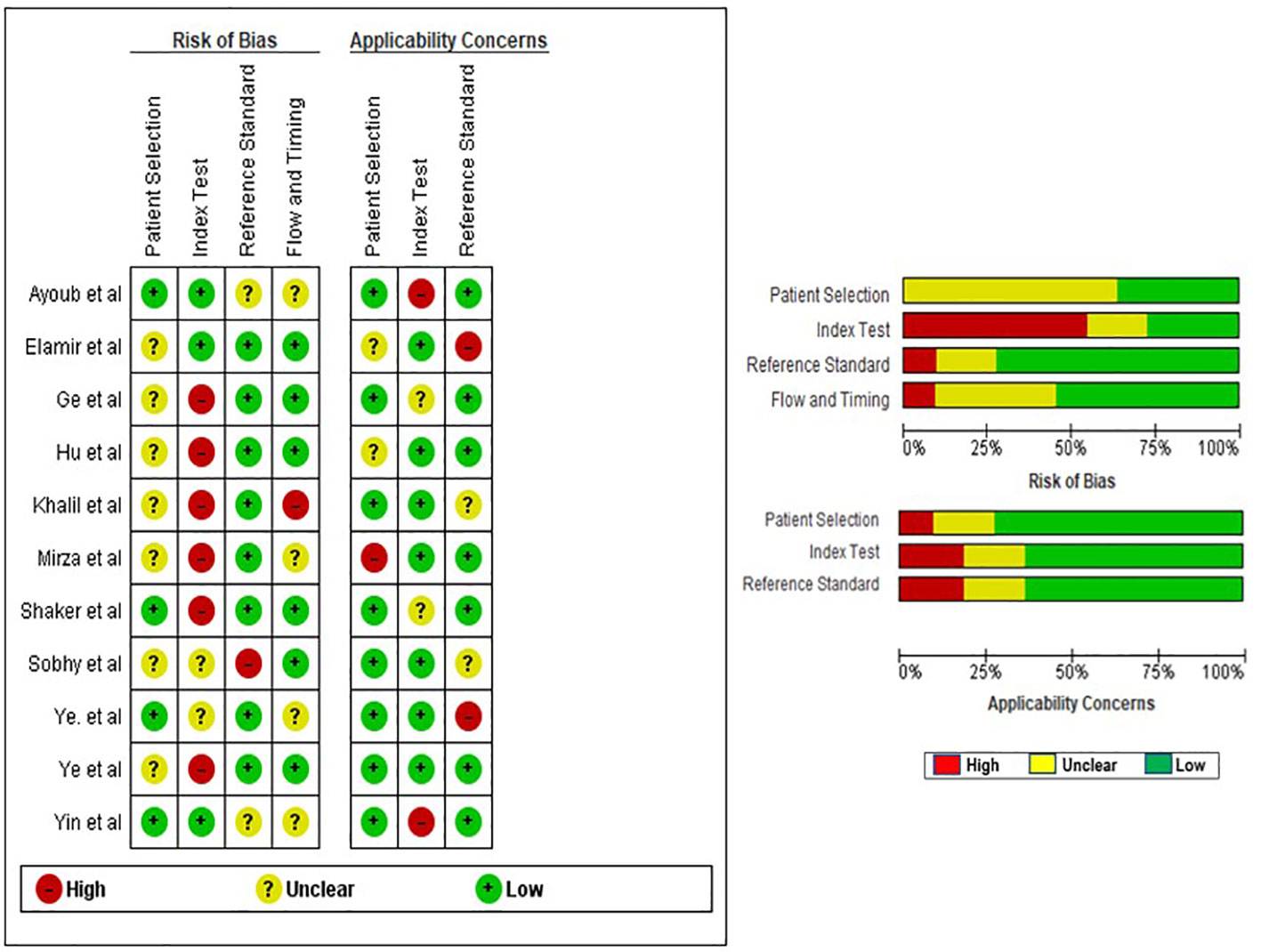
The evaluation of the quality of the eligible studies was carried out through the utilization of the QUADAS-2 tool. Due to the crucial role of patient selection in ensuring experimental integrity, the data incorporated into this meta-analysis predominantly came from well-validated groups. In general, the studies included demonstrated satisfactory and qualifying methodological standards. A comprehensive breakdown of the criteria used in the quality assessment is clearly shown in Figure 2.
Overall diagnostic accuracy of lnc and circular RNA in diagnosing IBD
The presence of a threshold effect of heterogeneity was evaluated using both the Spearman correlation coefficient and the SROC curve. The findings from the Spearman correlation coefficient depicting a rho value of -0.33 and a p-value of 0.34, along with the absence of the characteristic “shoulder-arm” shape in the SROC curve, indicated that there is no evidence of a threshold effect of heterogeneity. Moreover, the I2 values for sensitivity, specificity, PLR, NLR, and DOR for lncRNA markers were 91.69%, 96.85%, 93.47%, 92.78%, and 100%, respectively. Conversely, the I2 values for sensitivity, specificity, PLR, NLR, and DOR for circRNA markers were 66.11%, 73.11%, 39.82%, 47.22%, and 98.44%, respectively. In both cases, given that the I2 results surpass 50% and the p-values for all parameters are below 0.001, it strongly indicates the existence of significant non-threshold effect heterogeneity in this study. Consequently, a random-effects model was utilized for the meta-analysis.
The findings revealed that both lncRNAs and circRNAs demonstrated strong diagnostic potential for detecting IBD. The combined sensitivity and specificity for lncRNA markers were 0.94 (95% CI: 0.87-0.97) and 0.99 (95% CI: 0.89-1.00), respectively (Figure 3A). Similarly, the combined sensitivity and specificity for circRNA markers were 0.68 (95% CI: 0.61-0.73) and 0.73 (95% CI: 0.65-0.79), respectively (Figure 3B). Additionally, the pooled PLR and NLR were 64.25 (95% CI: 7.39-558.66) and 0.06 (95% CI: 0.03-0.13), respectively for lncRNA, and 2.47 (95% CI: 1.94-3.16) and 0.45 (95% CI: 0.38-0.53), respectively for circRNA. Furthermore, the DOR for lncRNA and circRNA markers were 1055.25 (95% CI: 70.61-15770.77) and 5.54 (95% CI: 3.88-7.93). In assessing diagnostic accuracy of lncRNA and circRNA, the SROC curve was generated, resulting in an AUC of 0.99 (95% CI: 0.97-0.99) and 0.75 (95% CI: 0.71-0.79), respectively (Figure 4). These results indicate that both lncRNAs and circRNAs exhibit high diagnostic accuracy in identifying IBD, as an AUC greater than 0.7 is indicative of a strong predictive capability.
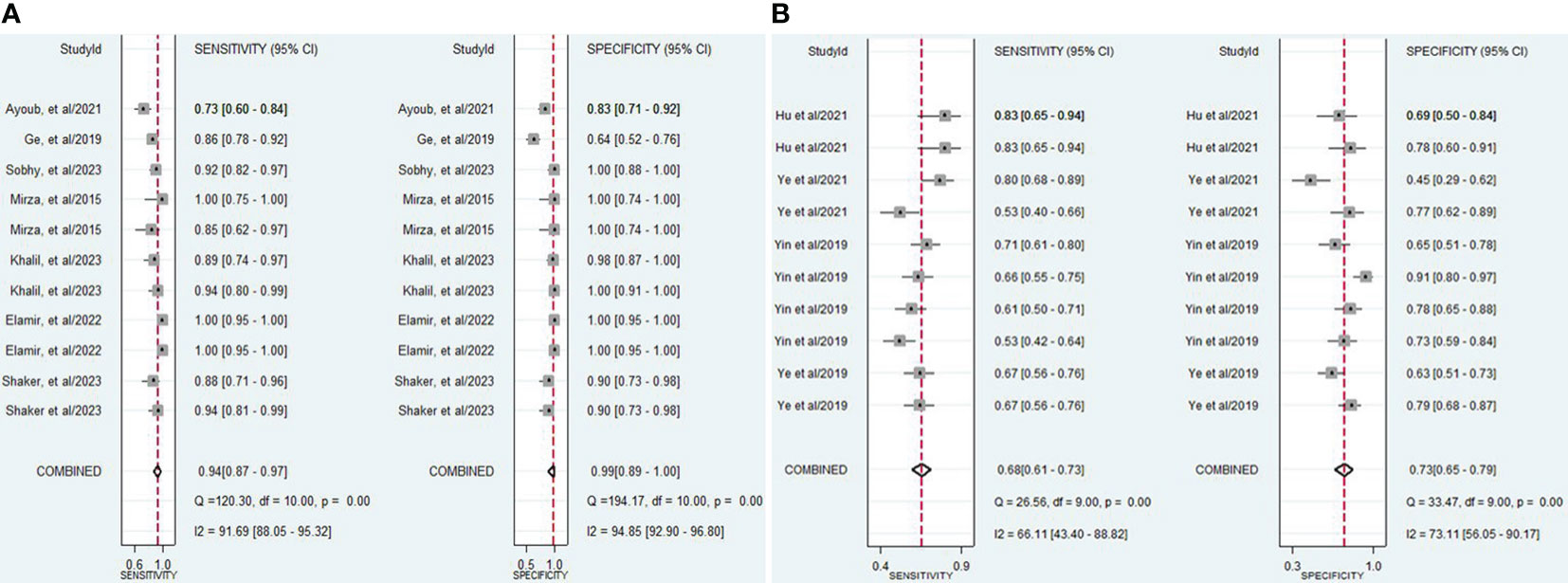
Figure 3 (A) Forest plot of pooled sensitivity and specificity of lncRNAs in diagnosing of IBD. (B) Forest plot of pooled sensitivity and specificity of circRNAs in diagnosing of IBD.
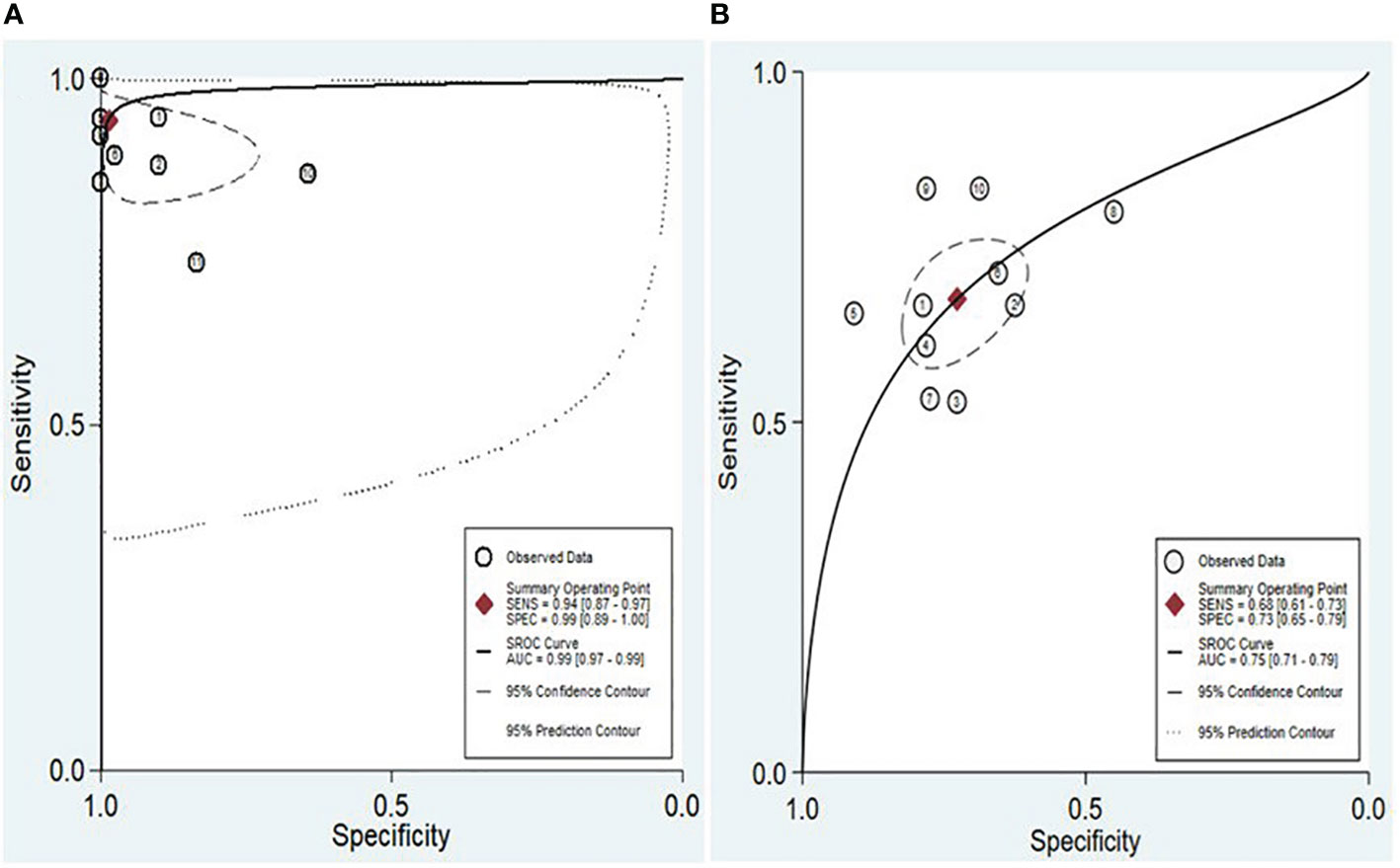
Figure 4 SROC and the 95% confidence contour and 95% prediction contour for lncRNAs (A) and circRNAs (B).
Clinical applicability of lncRNA and circRNA for diagnosing IBD
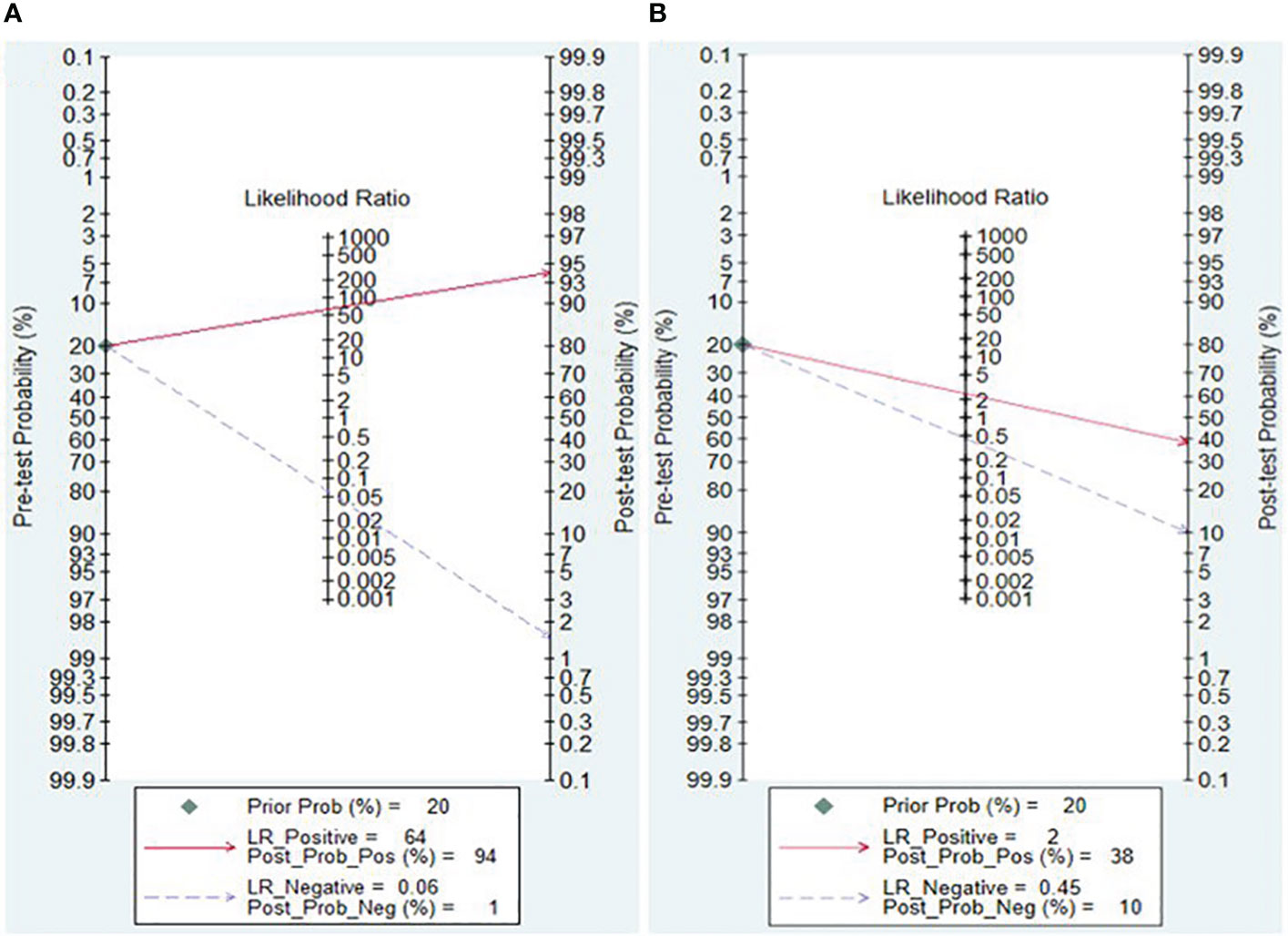
The Fagan nomogram and likelihood ratio scattergram were employed to evaluate the clinical value of lncRNAs and circRNAs in diagnosing IBD. The Fagan’s nomogram showed promising outcomes, revealing post-test probabilities with PLR and NLR values of 0.94 and 0.01 for lncRNAs and 0.38 and 0.1 for circRNA, respectively, under a pre-test probability set at 20% (Figures 5A, B). Furthermore, a scattergram depicting the likelihood ratios (PLR and NLR) was generated to assess the clinical applicability of lncRNAs and circRNA in IBD diagnosis. The results indicated that studies conducted by Khalil et al. (lncRNA H19), Mirza et al. (MMP12, RP11-731 F5.2, AC007182.6, DPP10-AS1, CDKN2B-AS1, and AL928742.12), and Sobby et al. (lncRNA H19) laid over on the left upper quadrant (PLR > 10 and an NLR < 0.1), indicating that the markers could be used for both exclusion and confirmation of IBD (Figure 6A). On the other hand, all eligible studies for circRNAs were positioned in the right lower quadrant (PLR < 10 and an NLR < 1), indicating that these markers are not suitable for confirming or excluding IBD (Figure 6B).
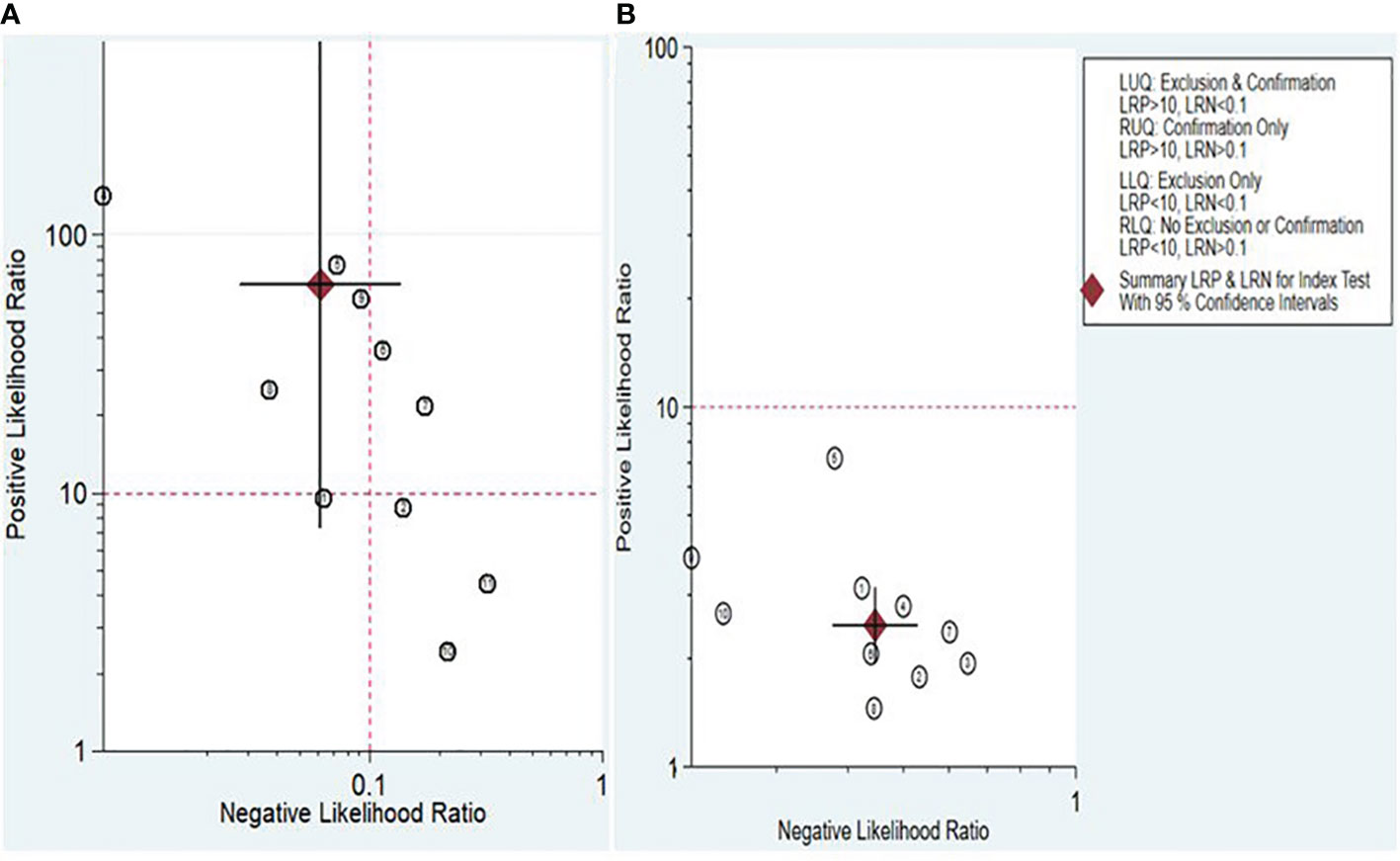
Figure 6 Scattergram assessing the clinical applicability of lncRNAs (A) and circRNAs (B) for diagnosing IBD.
Subgroup analysis and meta-regression
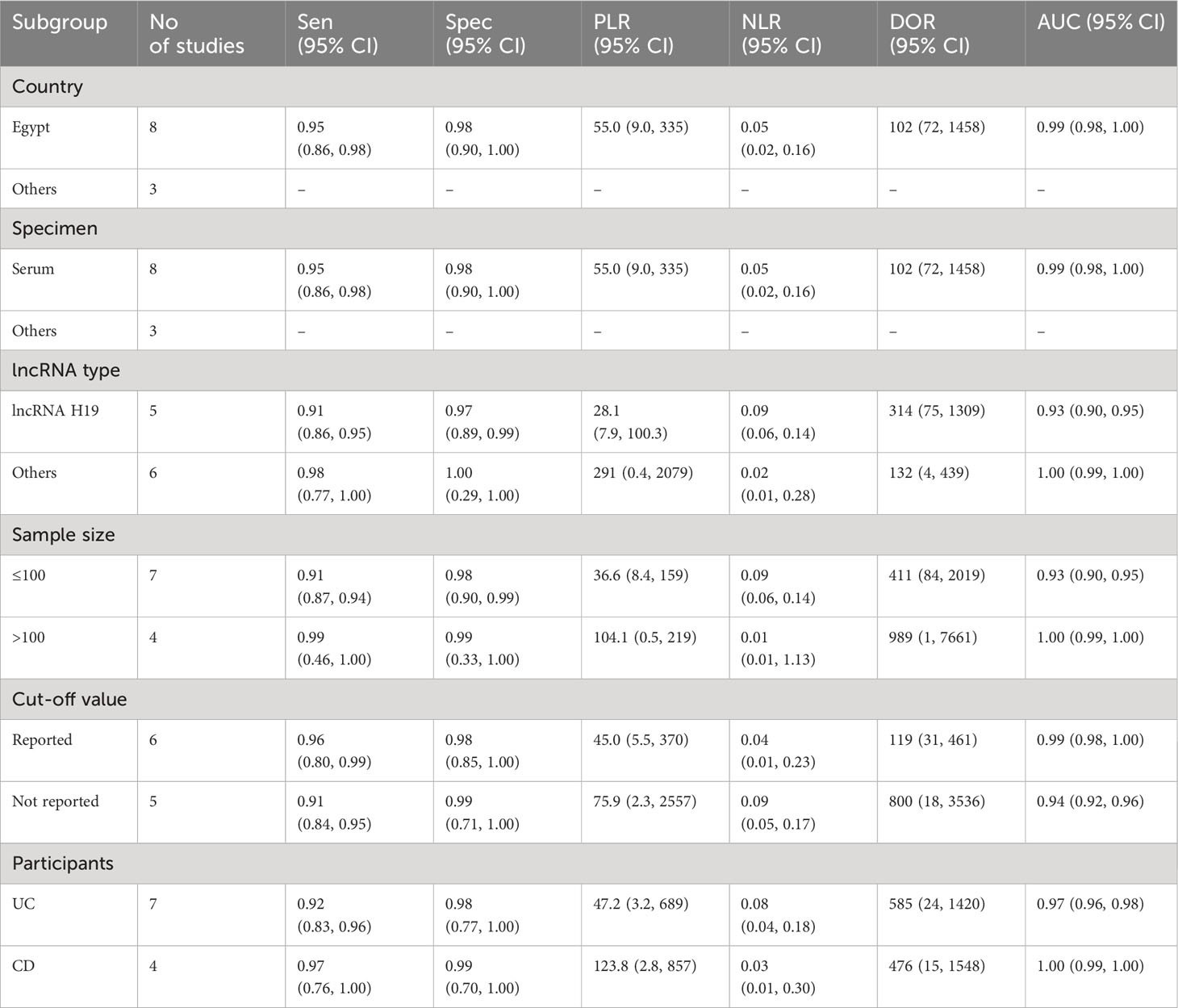
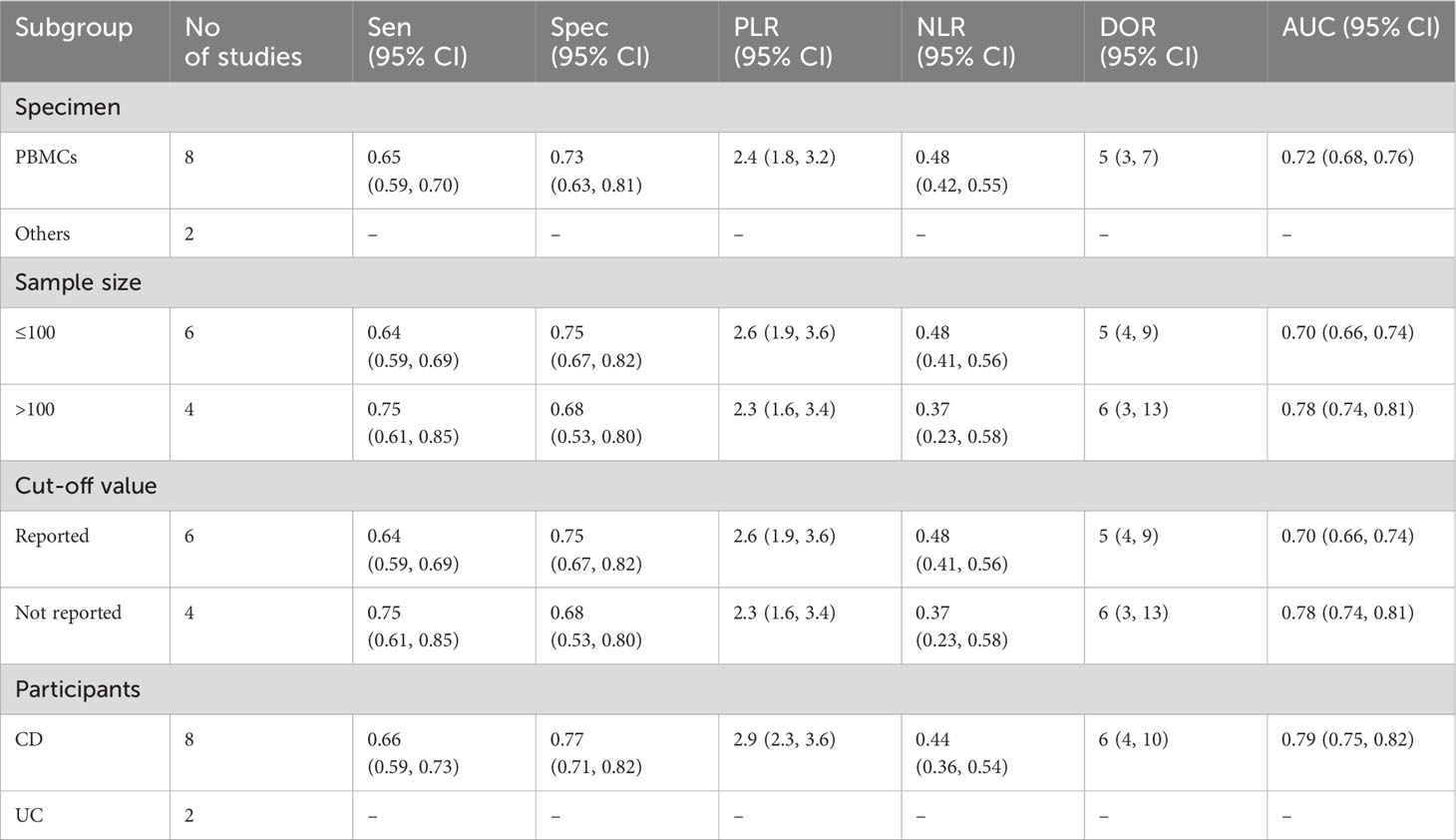
Due to the presence of substantial heterogeneity (I2 > 50% and P < 0.05) across all diagnostic performance parameters, such as sensitivity, specificity, PLR, NLR, DOR, and AUC, meta-regression and subgroup analyses were carried out. The purpose of these analyses was to delve into the sources of heterogeneity among the studies, examining various study characteristics. These characteristics include country, biological specimen, regulation pattern, lncRNA profiling, sample size, internal reference, cut-off value establishment, and the classification of IBD patients.
In the subgroup analysis, lncRNA H9 showed a relatively lower diagnostic performance for IBD compared to other lncRNAs. Notably, lncRNA H9 demonstrated lower sensitivity (0.91, 95% CI: 0.86-0.95), specificity (0.97, 95% CI: 0.89-0.99), PLR (28.1, 95% CI: 7.9-100.3), NLR (0.09, 95% CI: 0.06-0.14), DOR (314, 95% CI: 75-1309), and AUC (0.93, 95% CI: 0.90-0.95) compared to other lncRNAs (sensitivity: 0.98, 95% CI: 0.77-1.00; specificity: 1.00, 95% CI: 0.29-1.00; PLR: 291, 95% CI: 0.4-2079; NLR: 0.02, 95% CI: 0.01-0.28; DOR: 132, 95% CI: 4-439; AUC: 1.00, 95% CI: 0.99-1.00).
In terms of sample size, lncRNAs exhibited the highest overall diagnostic accuracy when the sample size was >100, with a sensitivity 0.99 (95% CI: 0.46-1.00), specificity 0.99 (95% CI: 0.33-1.00), PLR 104.1 (95% CI: 0.5-219), NLR 0.01 (95% CI: 0.01-1.13), DOR 989 (955 CI: 1-7661), and AUC of 1.00 (95% CI: 0.99-1.00), demonstrating superior diagnostic performance compared to instances where the sample size was ≤100. Furthermore, lncRNAs demonstrated notable efficacy in the diagnosis of both UC and CD with minimal differences in parameters. In case of the CD, the diagnostic parameters were as follows: sensitivity of 0.97 (95% CI: 0.76-1.00), specificity of 0.99 (95% CI:0.70-1.00), PLR of 123.8 (95% CI: 2.8-857), NLR of 0.03 (95% CI: 0.01-0.30), DOR of 476 (95% CI: 15-1548), and an AUC of 1.00 (95% CI: 0.99-1.00) (Table 2A). On the other hand, circRNAs exhibited moderate overall diagnostic accuracy when the sample size >100 with sensitivity 0.75 (95% CI: 0.61-0.85), Specificity 0.68 (95% CI: 0.53-0.80), PLR 2.3 (95% CI: 1.6-3.4), NLR 0.37 (95% CI: 0.23-0.58), DOR 6 (95% CI: 3-13), and 0.78 (95% CI: 0.74-0.81) (Table 2B).
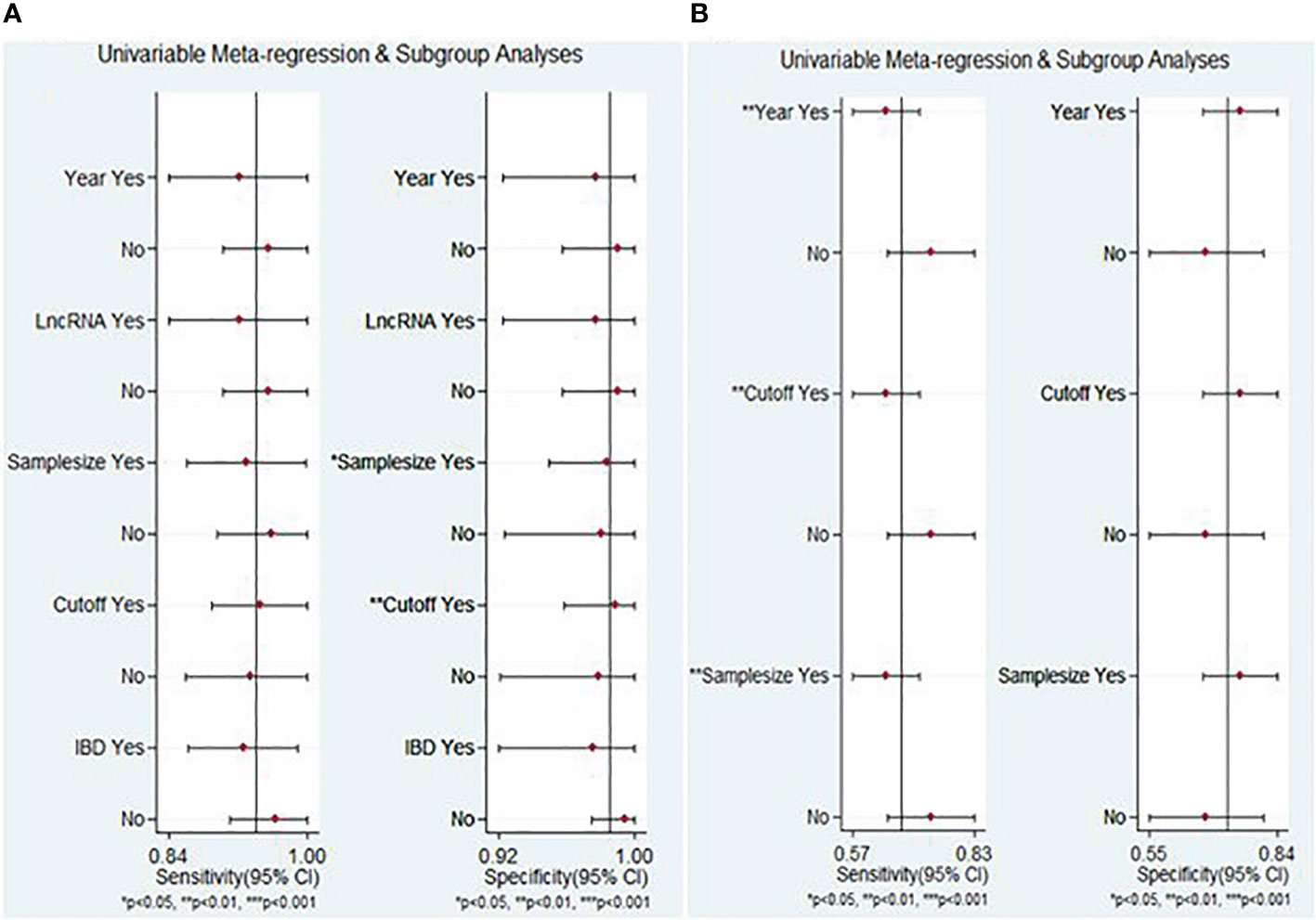
The meta-regression analysis results of lncRNA markers indicated that factors contributing to heterogeneity for specificity encompassed sample size and cut-off value determination (P < 0.05) (Figure 7A). Conversely, the publication year, cut-off value determination, and sample size were identified as the sources of heterogeneity of circRNA markers, particularly for sensitivity (P < 0.05) (Figure 7B).
Sensitivity analysis and publication bias
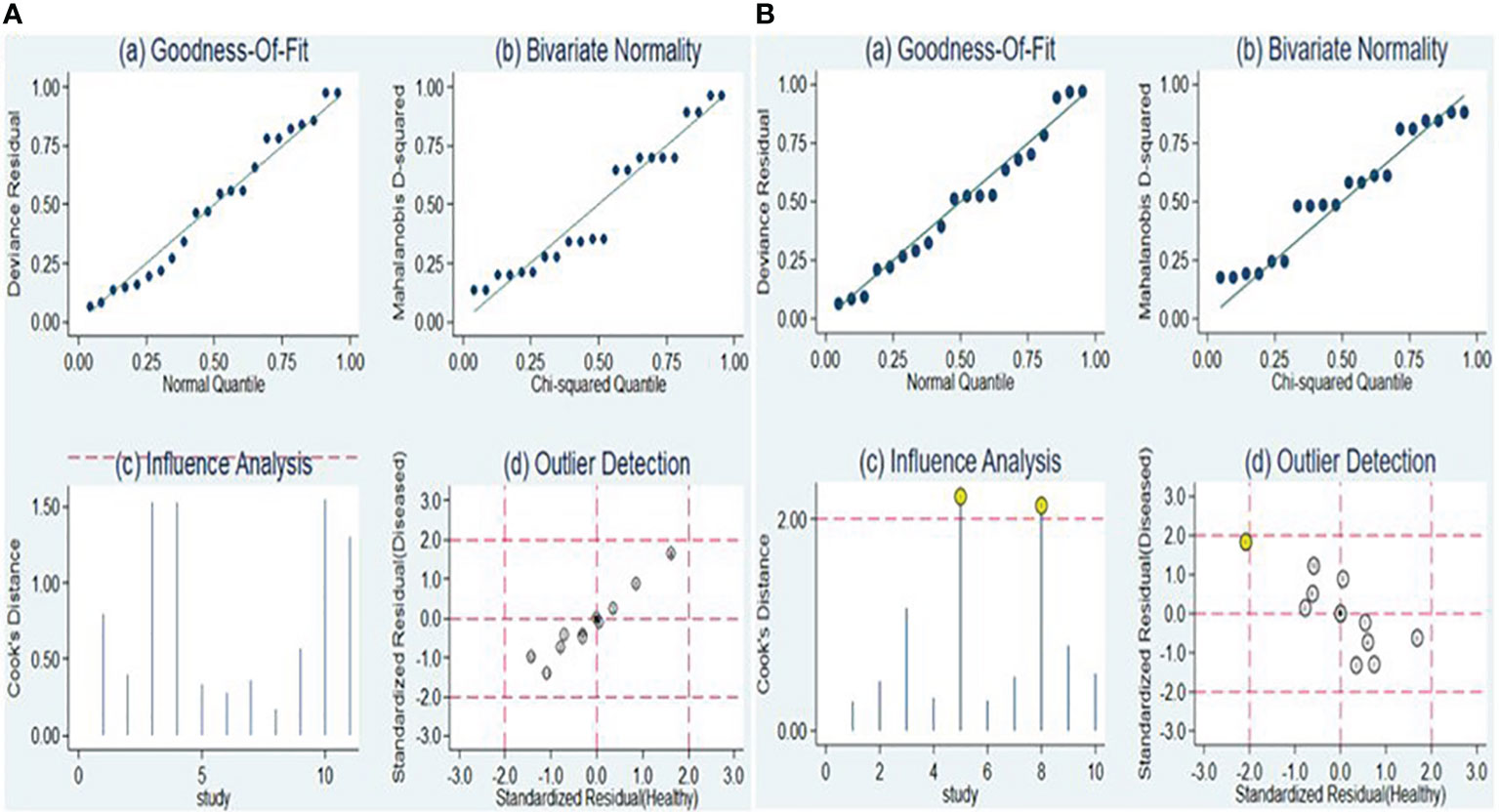
The sensitivity analysis is illustrated in Figures 8A, B. Examination of goodness-of-fit and bivariate normality indicated the strength and reliability of the bivariate mixed-effects model for conducting meta-analysis [Figure 8A (a and b), Figure 8B (a and b)]. Outlier identification for lncRNA markers revealed no outliers were seen indicting the findings were reliable [Figure 8A (d)]. On the other hand, the identification of outliers in the case of circRNA pointed to potential outlier in the form of a study conducted by Ye et al. (circRNA-103765) [Figure 8B (d)]. Upon removing this outlier, we observed no substantial alterations in the overall sensitivity (0.66, 95% CI: 0.60-0.72), specificity (0.75, 95% CI: 0.69-0.80), PLR (2.6, 95% CI: 2.1-3.4), NLR (0.45, 95% CI: 0.38-0.54), DOR (6, 95% CI: 4-8), and AUC (0.76, 95% CI: 0.72-0.80). This suggests that the sensitivity of the studies included was consistently low, and the results became more resilient and trustworthy.
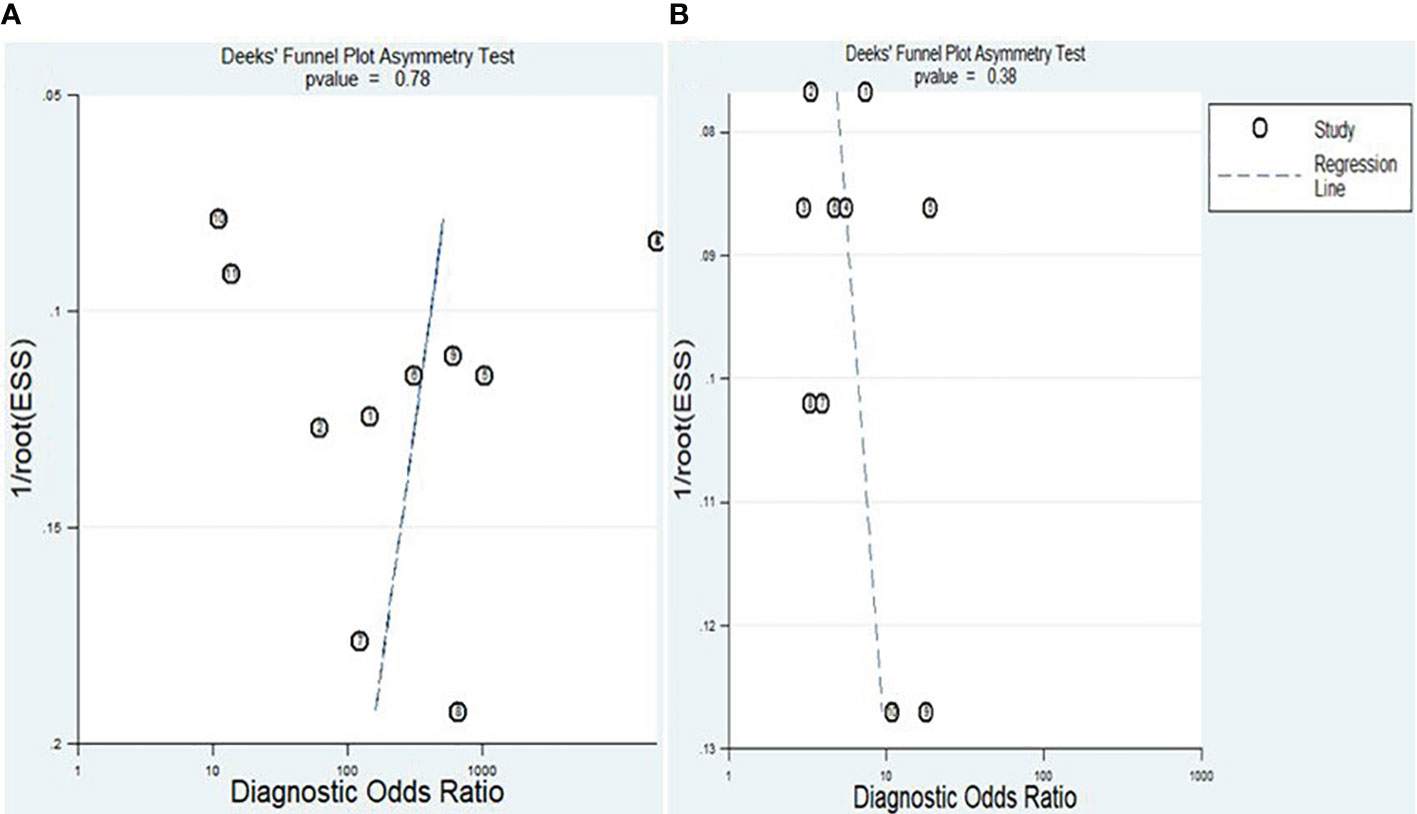
The Deeks’ funnel plot asymmetry test was conducted to assess the presence of publication bias. The obtained P-values of 0.78 and 0.38 for lncRNAs and circRNA, respectively, suggest that there was no apparent indication of publication bias within the included studies (Figures 9A, B).
Discussion
The burden of IBD is raising globally (47). Achieving an early diagnosis of IBD presents a formidable challenge, primarily due to the frequent misinterpretation of its symptoms as indicative of common infections (48, 49). Moreover, limited availability of diagnostic modalities may further delay prompt identification of IBD cases (50, 51). Diagnosis of IBD predominantly depends on clinical evaluation, blood tests (such as complete blood count, c-reactive protein, and erythrocyte sedimentation rate), endoscopic procedures (colonoscopy, sigmoidoscopy, and upper endoscopy), radiological imaging (X-rays, computed tomography (CT) scans), and biopsies (52–54). However, these techniques have several limitation and challenges including the invasiveness of procedures, the potential for false-negative or false-positive results, and the inability to consistently distinguish between CD and UC, necessitating a multifaceted approach for accurate diagnosis and classification (55). Thus, to embark a more accurate and timely detection of IBD, the investigation of novel diagnostic biomarkers has recently become vital, enabling early intervention and improved patient outcomes. In this regard, lncRNAs and circRNAs have gained substantial attention attributable to their diverse roles in the regulation of gene expression and pathophysiology of inflammatory processes (56, 57). LncRNAs and circRNAs with dysregulated expression play a role in diverse complex diseases, including cancer (58–62), neurodegenerative diseases (63), autoimmune diseases (64), cardiovascular diseases (65–67), inflammations and various infections (67, 68).
LncRNAs and circRNAs exhibit typical features that render them promising diagnostic biomarkers for IBD. These features include the noncoding nature of both lncRNAs and circRNAs that allow them to actively involve in gene expression regulation (69, 70), and stability in various body fluids making them readily accessible for noninvasive detection (71, 72). Besides, these biomarkers are resistant to enzymatic degradation (73).
Both lncRNAs and circRNAs are not sufficiently investigated in the context of IBD (74). While few systematic reviews have incorporated specific lncRNAs and circRNAs that exhibit altered expression in individuals with IBD compared to controls (23, 75, 76), none of these reviews have presented information on the diagnostic efficacy of these biomarkers. Thus, this study appears to be the first meta-analysis to investigate the utility of lncRNAs and circRNAs in the diagnosis of IBD. This systematic review and meta-analysis combined findings of existing studies to comprehensively assess the diagnostic potential of lncRNAs and circRNAs for the early detection and stratification of IBD patients.
In this study, a total of 11 articles encompassing 21 studies which involved 1239 IBD patients and 985 healthy controls were investigated. The findings of this meta-analysis enrolling four distinct lncRNAs and one lncRNA panel from a total of 11 studies depicted that lncRNAs could be used as potential biomarker in diagnosing IBD, with a high level of pooled sensitivity 0.94 (95% CI: 0.87-0.97) and specificity 0.99 (95% CI: 0.89-1.00), along with PLR, NLR, DOR, and AUC values of 64.25 (95% CI: 7.39-558.66), 0.06 (95% CI: 0.03-0.13), 1055.25 (95% CI: 70.61-15770.77), and 0.99 (95% CI: 0.97-0.99), respectively. On the other hand, the overall findings of our analysis enrolling eight distinct circRNAs from 10 studies showed that the pooled sensitivity, specificity, PLR, NLR, DOR, and AUC of circRNAs in the diagnosis of IBD were 0.68 (95% CI: 0.61-0.73), 0.73 (95% CI: 0.65-0.79), 2.47 (95% CI: 1.94-3.16), 0.45 (95% CI: 0.38-0.53), 5.54 (95% CI: 3.88-7.93), and 0.75 (95% CI: 0.71-0.79), respectively. The findings revealed that circRNAs illustrate a moderate level of pooled sensitivity and specificity in serving as a diagnostic marker of IBD.
The PLR value of 64.25 suggests that the likelihood of detecting positive lncRNAs in patients with IBD is around 64 times higher compared to healthy controls. Conversely, the NLR, at 0.06, implies that cases with negative test results have an approximately 6% chance of developing IBD. The DOR serves as an indicator of the discriminatory performance of a test (77), and a DOR value exceeding 1 signifies a more effective diagnostic test. In this context, the DOR of 1055.25 underscores the extreme capability of lncRNAs to effectively and efficiently discriminate between patients with IBD and their healthy counterparts. Furthermore, the AUC value serves as a reliable indicator within the assessment system. An ideal test, exhibiting perfect and flawless discrimination, would achieve an AUC of 1.0 (78). As the AUC value of a test approaches 1.0, there is a corresponding increase in the overall efficacy of the test (79). In this study, it was observed that lncRNA holds potential for differentiating IBD patients from healthy controls, as evidenced by a huge AUC value of 0.99. This proximity to 1.0 implies a substantial capacity of lncRNAs to discern effectively between individuals with IBD and those without the condition, signifying lncRNAs as an optimal test.
On the other hand, in the case of circRNAs, the probability of positive circRNA determination in IBD patients is approximately 2.47 times higher (PLR values of 2.47) compared to healthy controls, with NLR of 0.45 denoting that cases partaking negative test results have around 45% chance of developing IBD. The DOR index of 5.54 showed the relative ability of circRNAs to discriminate between patients with IBD and healthy controls. Additionally, an AUC value of 0.75 is indicative of a relative ability of cirRNAs to distinguish IBD patients from health controls. However, such findings of circRNAs and the interpretations drawn from circRNA-related investigations in this meta-analysis may not be conclusive as the number of included studies is limited. To establish more robust conclusions, it is advisable to validate these findings through a large number of studies.
In the subgroup analysis, lncRNA H9 showed a relatively high diagnostic performance (AUC: 0.93, 95% CI: 0.90-0.95) for IBD compared to other lncRNAs and lncRNA clusters. This may be due to the specific expression patterns and biological roles of lncRNA H9 in the context of IBD. However, due to the limited number of studies reporting performances of lncRNA panel and individual lncRNAs including lncRNA THRIL, lncRNA ANRIL and lncRNA PVT1, we could not able to assess their combined diagnostic performances. Consequently, our current evidence showed that lncRNA H9 may be a better and more suitable diagnostic marker for IBD.
Our finding also revealed a substantial difference in diagnostic ability of lncRNAs between the studies that had sample size >100 participants and ≤100 participants. Accordingly, studies with sample size >100 (AUC: 1.00 (95% CI: 0.99-1.00) are found to have more diagnostic performance seamlessly becoming very close to the ideal test compared with studies with sample size ≤100 (AUC: 0.93, 95% CI: 0.90-0.95). Similarly, our study showed a significant difference in diagnostic ability of circRNA between the groups with a sample size > 100 (AUC: 0.78) and ≤ 100 (AUC: 0.70). The possible justification for such discrepancy could be differences in statistical power and sampling bias that arise as a result of variances in sample size. However, large-scale studies deploying large sample size will be paramount to confirm and validate these findings.
In fact, both PLR and AUC are vital diagnostic measures with significant clinical implications. Clinically, a higher PLR enhances the utility of a diagnostic test by indicating that a positive result strongly supports the presence of the condition. It aids in patient stratification, contributing to more accurate identification of individuals who truly have the disease. A higher AUC, a key metric in ROC analysis, signifies the test’s superior ability to differentiate between individuals with and without the condition. Clinicians use the AUC to assess diagnostic accuracy, guide decisions on test suitability for patient care, and determine optimal cutoff values for clinical application (78). However, there are potential sources of bias and limitations in calculating PLR and AUC measures, including study population, verification, choice of different threshold values for defining a positive test result, variations in disease prevalence in different populations, reference standard related bias, publication bias, inclusion of small sample size, and heterogeneity in study designs, patient populations, or test methodologies (80).
Additionally, lncRNAs demonstrated notable efficacy in the diagnosis of both UC and CD with minimal differences in parameters and high AUC values (AUC: 0.97 for UC and AUC: 1.00 for CD). This indicates the possibility of using lncRNA biomarkers for all forms of IBD and their ability to diagnose both UC and CD with high sensitivity and specificity. Such findings are indicative of the applicability of lncRNAs to diagnose IBD and their possible wide spread use.
Facilitating clinical decision-making stands as the paramount value of biomarkers. Likelihood ratios offer valuable insights to clinicians, furnishing information on the probability that a patient with a positive or negative test truly has or does not have IBD (81). In evaluating the clinical utility of lncRNAs for diagnosing IBD, this study condensed and assessed PLR and NLR. A PLR > 10 and NLR < 0.1 signify a high level of diagnostic accuracy. Notably, this study showed that lncRNA H19 exhibited high diagnostic accuracy and clinical applicability. Consequently, lncRNA H19 emerges as a promising candidate deserving of further research in the realm of lncRNAs.
In this meta-analysis, the Fagan’s nomogram for lncRNAs, assuming a pre-test probability set at 20%, yielded post-test probabilities with PLR and NLR values of 0.94 and 0.01, respectively. This finding illustrates potential outcomes wherein samples testing positive for the presence of lncRNAs indicate a 94% probability of IBD development in patients, while the post-test probability of the disease decreases to 1% when samples tested negative for lncRNAs. Accordingly, lncRNAs exhibit a discernible diagnostic potential in distinguishing patients with IBD from healthy controls, and can be used as a viable screening method for IBD.
This meta-analysis revealed that lncRNAs exhibit enormously higher diagnostic performance, characterized by markedly higher sensitivity and specificity compared to circRNAs, which demonstrated moderate sensitivity and specificity values. Investigations of dysregulated expressions of lncRNAs and circRNAs are vital tools in enabling the early diagnosis of IBD and improving overall health outcomes, establishing them as promising diagnostic markers. These biomarkers are beneficial than the conventional methods, which are often invasive and exhibit lower diagnostic efficacy. This advantage is mainly attributed to the easy accessibility of peripheral blood, tissue samples, and body fluids, the consistent expression of biomarkers in these samples (82), their tissue-specific characteristics, and reliability of the qRT-PCR technique in detecting circulating biomarkers (26).
This study has several limitations. Firstly, limited number of eligible studies were used for assessing the diagnostic performance of circRNA which could have constrained and hindered the evaluation. Secondly, the data extracted from the eligible studies were relatively limited due to variations in the cutoff values of the biomarkers, which could possibly lead to potential heterogeneity. Thirdly, the eligible studies originated from limited number of countries, potentially compromising the appropriateness and generalizability of the biomarkers’ diagnostic performance to IBD patients globally. Fourthly, the statistical power of our meta-analysis could be constrained as a result of the small sample size pertained by most of the included studies. Fifthly, the unavailability of a substantial number of similar lncRNA and circRNA biomarkers prevented the pooling of results, making it challenging to identify specific single biomarkers or panels as the optimal diagnostic tools for IBD. Lastly, potential biases in the included studies related with selection, verification, reference standard usage, inappropriate index test dependency, variable diseases prevalence or characteristics, and publication related bias may impact our overall analysis. While these limitations may have impacted our meta-analysis findings, we anticipate that this study will offer baseline data for forthcoming studies. As a result, our results should be interpreted with caution, and we recommend future researchers to confirm and validate our findings through more extensive studies with larger sample sizes, following a more standardized approach.
In conclusion, our study evidenced that lncRNAs have high diagnostic accuracy in distinguishing patients with IBD from healthy controls, while circRNAs showed moderate diagnostic accuracy. This suggests that lncRNAs could serve as effective non-invasive biomarkers for IBD. Notably, both lncRNA H19 and lncRNAs in studies with a large sample size (>100) demonstrate higher diagnostic potential in the diagnosis of IBD. Additionally, lncRNAs displayed notable efficacy in the diagnosis of both UC and CD, with minimal differences in parameters. However, to validate our findings and confirm the clinical utility of lncRNAs and circRNAs in IBD diagnosis, a comprehensive array of prospective, multi-center studies, encompassing both individual and combined lncRNA and circRNA assays, should be undertaken. Furthermore, to mitigate these limitations, future research endeavors should focus on broadening the pool of eligible studies, ensuring diverse global representation, minimizing potential biases, standardizing cutoff values, and incorporating larger sample sizes. Additionally, exploration of a broader array of lncRNAs and circRNAs biomarkers from noninvasive specimens while ensuring consistent methodologies, and promoting transparent reporting practices are imperative to enhance the robustness and generalizability of future research outcomes and facilitate the translation of these biomarkers into clinically meaningful diagnostic tools for IBD.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
MB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. ST: Data curation, Formal analysis, Methodology, Software, Supervision, Writing – review & editing. MT: Data curation, Formal analysis, Investigation, Software, Validation, Writing – review & editing. AG: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – review & editing. AS: Data curation, Formal analysis, Investigation, Software, Supervision, Visualization, Writing – review & editing. ZM: Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing – review & editing. MW: Formal analysis, Methodology, Software, Supervision, Validation, Visualization, Writing – review & editing. SG: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Writing – review & editing. HD: Formal analysis, Investigation, Methodology, Resources, Software, Writing – review & editing. MA: Data curation, Formal analysis, Software, Validation, Visualization, Writing – review & editing. EA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1362437/full#supplementary-material
References
1. Saez A, Herrero-Fernandez B, Gomez-Bris R, Sánchez-Martinez H, Gonzalez-Granado JM. Pathophysiology of inflammatory bowel disease: innate immune system. Int J Mol Sci. (2023) 24:1526. doi: 10.3390/ijms24021526
2. Sairenji T, Collins KL, Evans DV. An update on inflammatory bowel disease. Primary Care: Clinics Office Pract. (2017) 44:673–92. doi: 10.1016/j.pop.2017.07.010
3. Kelsen JR, Russo P, Sullivan KE. Early-onset inflammatory bowel disease. Immunol Allergy Clinics. (2019) 39:63–79. doi: 10.1016/j.iac.2018.08.008
4. Guan Q. A comprehensive review and update on the pathogenesis of inflammatory bowel disease. J Immunol Res. (2019) 2019:1–16. doi: 10.1155/2019/7247238
5. Lee SH, eun Kwon J, Cho M-L. Immunological pathogenesis of inflammatory bowel disease. Intestinal Res. (2018) 16:26–42. doi: 10.5217/ir.2018.16.1.26
6. Cai Z, Wang S, Li J. Treatment of inflammatory bowel disease: a comprehensive review. Front Med. (2021) 8:765474. doi: 10.3389/fmed.2021.765474
7. West J, Tan K, Devi J, Macrae F, Christensen B, Segal JP. Benefits and challenges of treat-to-target in inflammatory bowel disease. J Clin Med. (2023) 12:6292. doi: 10.3390/jcm12196292
8. Clinton JW, Cross RK. Personalized treatment for Crohn’s disease: current approaches and future directions. Clin Exp Gastroenterol. (2023) 16:249–76. doi: 10.2147/CEG.S360248
9. Alatab S, Sepanlou SG, Ikuta K, Vahedi H, Bisignano C, Safiri S, et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. (2020) 5:17–30. doi: 10.1016/S2468-1253(19)30333-4
10. Parra RS, Chebli JM, Amarante HM, Flores C, Parente JM, Ramos O, et al. Quality of life, work productivity impairment and healthcare resources in inflammatory bowel diseases in Brazil. World J Gastroenterol. (2019) 25:5862. doi: 10.3748/wjg.v25.i38.5862
11. Sciberras M, Karmiris K, Nascimento C, Tabone T, Nikolaou P, Theodoropoulou A, et al. Mental health, work presenteeism, and exercise in inflammatory bowel disease. J Crohn's Colitis. (2022) 16:1197–201. doi: 10.1093/ecco-jcc/jjac037
12. Sandberg K, Yarger E, Saeed S. Updates in diagnosis and management of inflammatory bowel disease. Curr Problems Pediatr Adolesc Health Care. (2020) 50:100785. doi: 10.1016/j.cppeds.2020.100785
13. Gomollón F, Dignass A, Annese V, Tilg H, Van Assche G, Lindsay JO, et al. 3rd European evidence-based consensus on the diagnosis and management of Crohn’s disease 2016: part 1: diagnosis and medical management. J Crohn's Colitis. (2017) 11:3–25. doi: 10.1093/ecco-jcc/jjw168
14. Vermeire S, Van Assche G, Rutgeerts P. Laboratory markers in IBD: useful, magic, or unnecessary toys? Gut. (2006) 55:426–31. doi: 10.1136/gut.2005.069476
15. Bennike T, Birkelund S, Stensballe A, Andersen V. Biomarkers in inflammatory bowel diseases: current status and proteomics identification strategies. World J Gastroenterol: WJG. (2014) 20:3231. doi: 10.3748/wjg.v20.i12.3231
16. Tontini GE, Vecchi M, Pastorelli L, Neurath MF, Neumann H. Differential diagnosis in inflammatory bowel disease colitis: state of the art and future perspectives. World J Gastroenterol: WJG. (2015) 21:21. doi: 10.3748/wjg.v21.i1.21
17. Fiocchi C. Omics and multi-omics in IBD: no integration, no breakthroughs. Int J Mol Sci. (2023) 24:14912. doi: 10.3390/ijms241914912
18. Zhou G, Song Y, Yang W, Guo Y, Fang L, Chen Y, et al. ASCA, ANCA, ALCA and many more: are they useful in the diagnosis of inflammatory bowel disease? Digestive Dis. (2016) 34:90–7. doi: 10.1159/000442934
19. Dragoni G, Innocenti T, Galli A. Biomarkers of inflammation in inflammatory bowel disease: how long before abandoning single-marker approaches? Digestive Dis. (2021) 39:190–203. doi: 10.1159/000511641
20. Chen P, Zhou G, Lin J, Li L, Zeng Z, Chen M, et al. Serum biomarkers for inflammatory bowel disease. Front Med. (2020) 7:123. doi: 10.3389/fmed.2020.00123
21. Nemeth K, Bayraktar R, Ferracin M, Calin GA. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat Rev Genet. (2023) 25:1–22. doi: 10.1038/s41576-023-00662-1
22. Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. (2011) 90:430–40. doi: 10.1093/cvr/cvr097
23. Yarani R, Mirza AH, Kaur S, Pociot F. The emerging role of lncRNAs in inflammatory bowel disease. Exp Mol Med. (2018) 50:1–14. doi: 10.1038/s12276-018-0188-9
24. Ma H, Hu T, Tao W, Tong J, Han Z, Herndler-Brandstetter D, et al. A lncRNA from an inflammatory bowel disease risk locus maintains intestinal host-commensal homeostasis. Cell Res. (2023) 33:1–17. doi: 10.1038/s41422-023-00790-7
25. Alfaifi J, Germain A, Heba A-C, Arnone D, Gailly L, Ndiaye NC, et al. Deep dive into microRNAs in inflammatory bowel disease. Inflammatory Bowel Dis. (2023) 29:986–99. doi: 10.1093/ibd/izac250
26. Jabandziev P, Bohosova J, Pinkasova T, Kunovsky L, Slaby O, Goel A. The emerging role of noncoding RNAs in pediatric inflammatory bowel disease. Inflammatory Bowel Dis. (2020) 26:985–93. doi: 10.1093/ibd/izaa009
27. Zhu M, Xie J. LncRNA MALAT1 promotes ulcerative colitis by upregulating lncRNA ANRIL. Digestive Dis Sci. (2020) 65:3191–6. doi: 10.1007/s10620-020-06093-w
28. Wang S, Hou Y, Chen W, Wang J, Xie W, Zhang X, et al. KIF9−AS1, LINC01272 and DIO3OS lncRNAs as novel biomarkers for inflammatory bowel disease. Mol Med Rep. (2018) 17:2195–202. doi: 10.3892/mmr.2017.8118
29. Xia H, Li S, He Y, Ren Q, Qin S. Long non-coding RNA ANRIL serves as a potential marker of disease risk, inflammation, and disease activity of pediatric inflammatory bowel disease. Clinics Res Hepatol Gastroenterol. (2022) 46:101895. doi: 10.1016/j.clinre.2022.101895
30. Rui Q, Lu C, Song S, Zhang L. Identification and validation of key long non-coding RNAs using co-expression network analysis in Crohn's disease. Ann Palliative Med. (2021) 10:9627–39. doi: 10.21037/apm-21-1952
31. Ye Y-L, Yin J, Hu T, Zhang L-P, Wu L-Y, Pang Z. Increased circulating circular RNA_103516 is a novel biomarker for inflammatory bowel disease in adult patients. World J Gastroenterol. (2019) 25:6273. doi: 10.3748/wjg.v25.i41.6273
32. Yin J, Hu T, Xu L, Li P, Li M, Ye Y, et al. Circular RNA expression profile in peripheral blood mononuclear cells from Crohn disease patients. Medicine. (2019) 98:1–9. doi: 10.1097/MD.0000000000016072
33. Xu Y, Xu X, Ocansey DKW, Cao H, Qiu W, Tu Q, et al. CircRNAs as promising biomarkers of inflammatory bowel disease and its associated-colorectal cancer. Am J Trans Res. (2021) 13:1580.
34. Lucafò M, Pugnetti L, Bramuzzo M, Curci D, Di Silvestre A, Marcuzzi A, et al. Long non-coding RNA GAS5 and intestinal MMP2 and MMP9 expression: a translational study in pediatric patients with IBD. Int J Mol Sci. (2019) 20:5280. doi: 10.3390/ijms20215280
35. Ding G, Ming Y, Zhang Y. lncRNA Mirt2 is downregulated in ulcerative colitis and regulates IL-22 expression and apoptosis in colonic epithelial cells. Gastroenterol Res Pract. (2019) 2019:1–5. doi: 10.1155/2019/8154692
36. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Int J Surg. (2021) 88:105906. doi: 10.1016/j.ijsu.2021.105906
37. Yang B, Mallett S, Takwoingi Y, Davenport CF, Hyde CJ, Whiting PF, et al. QUADAS-C: a tool for assessing risk of bias in comparative diagnostic accuracy studies. Ann Internal Med. (2021) 174:1592–9. doi: 10.7326/M21-2234
38. Shaker OG, Safa A, Khairy A, Abozeid NF. Serum long noncoding RNA H19/micro RNA-675-5p axis as a probable diagnostic biomarker in inflammatory bowel disease. Mol Biol Rep. (2023) 50:9029–36. doi: 10.1007/s11033-023-08777-8
39. Elamir A, Shaker O, Kamal M, Khalefa A, Abdelwahed M, Abd El Reheem F, et al. Expression profile of serum LncRNA THRIL and MiR-125b in inflammatory bowel disease. PloS One. (2022) 17:e0275267. doi: 10.1371/journal.pone.0275267
40. Khalil EH, Shaker OG, Hasona NA. lncRNA H-19 and miR-200a implication and frequency of lncRNA H-19 rs2170425 SNP in ulcerative colitis and Crohn’s disease. Comp Clin Pathol. (2023) 32:1–7. doi: 10.1007/s00580-023-03465-2
41. Mirza AH, Berthelsen CH, Seemann SE, Pan X, Frederiksen KS, Vilien M, et al. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. (2015) 7:1–22. doi: 10.1186/s13073-015-0162-2
42. Sobhy R, Shaker O, Khairy A, Gaber DA. Expression of long non coding RNA H19& miR-675 in colorectal cancer and ulcerative colitis patients. Mol Biol Rep. (2023) 50(11):1–8.
43. Ge Q, Dong Y, Lin G, Cao Y. Long noncoding RNA antisense noncoding RNA in the INK4 locus correlates with risk, severity, inflammation and infliximab efficacy in Crohn's disease. Am J Med Sci. (2019) 357:134–42. doi: 10.1016/j.amjms.2018.10.016
44. Ayoup SE. Expression profile of long non coding RNA PVT1 in patients with ulcerative colitis. Egyptian J Med Microbiol. (2021) 30:135–41. doi: 10.51429/EJMM30318
45. Hu Y, Zhu Y, Liu G, Yao X, Yan X, Yang Y, et al. Expression profiles of circular RNAs in colon biopsies from Crohn's disease patients by microarray analysis. J Clin Lab Anal. (2021) 35:e23788. doi: 10.1002/jcla.23788
46. Ye Y, Zhang L, Hu T, Yin J, Xu L, Pang Z, et al. CircRNA_103765 acts as a proinflammatory factor via sponging miR-30 family in Crohn’s disease. Sci Rep. (2021) 11:565. doi: 10.1038/s41598-020-80663-w
47. Wang R, Li Z, Liu S, Zhang D. Global, regional and national burden of inflammatory bowel disease in 204 countries and territories from 1990 to 2019: a systematic analysis based on the Global Burden of Disease Study 2019. BMJ Open. (2023) 13:e065186. doi: 10.1136/bmjopen-2022-065186
48. Watermeyer G, Katsidzira L, Nsokolo B, Isaac Alatise O, Duduyemi BM, Kassianides C, et al. Challenges in the diagnosis and management of IBD: a sub-Saharan African perspective. Ther Adv Gastroenterol. (2023) 16:17562848231184986. doi: 10.1177/17562848231184986
49. Dara N, Nemati S, Teimourian S, Imanzadeh F, Hosseini A, Tajalli S, et al. Diagnostic challenges in the early onset of inflammatory bowel disease: a case report. Int J Mol Cell Med. (2018) 7:251. doi: 10.22088/IJMCM.BUMS.7.4.251
50. Li Y, Qian J-m. The challenge of inflammatory bowel disease diagnosis in Asia. Inflammatory Intestinal Dis. (2017) 1:159–64. doi: 10.1159/000448384
51. Cross E, Saunders B, Farmer AD, Prior JA. Diagnostic delay in adult inflammatory bowel disease: A systematic review. Indian J Gastroenterol. (2023) 42:40–52. doi: 10.1007/s12664-022-01303-x
52. Maaser C, Sturm A, Vavricka SR, Kucharzik T, Fiorino G, Annese V, et al. ECCO-ESGAR Guideline for Diagnostic Assessment in Inflammatory Bowel Disease Part 1. Initial diagnosis, monitoring of known IBD, detections of complications. J Of Crohn's And Colitis. (2019) 13:144–64. doi: 10.1093/ecco-jcc/jjy113
53. Pasternak G, Chrzanowski G, Aebisher D, Myśliwiec A, Dynarowicz K, Bartusik-Aebisher D, et al. Crohn’s disease: basic characteristics of the disease, diagnostic methods, the role of biomarkers, and analysis of metalloproteinases: A review. Life. (2023) 13:2062. doi: 10.3390/life13102062
54. Noiseux I, Veilleux S, Bitton A, Kohen R, Vachon L, White Guay B, et al. Inflammatory bowel disease patient perceptions of diagnostic and monitoring tests and procedures. BMC Gastroenterol. (2019) 19:1–11. doi: 10.1186/s12876-019-0946-8
55. Naganuma M, Hosoe N, Kanai T, Ogata H. Recent trends in diagnostic techniques for inflammatory bowel disease. Korean J Internal Med. (2015) 30:271. doi: 10.3904/kjim.2015.30.3.271
56. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. (2014) 15:7–21. doi: 10.1038/nrg3606
57. Ghafouri-Fard S, Shoorei H, Sabernia T, Hussen BM, Taheri M, Pourmoshtagh H. Circular RNAs and inflammation: Epigenetic regulators with diagnostic role. Pathol-Res Pract. (2023) 251:154912. doi: 10.1016/j.prp.2023.154912
58. Gu Y, Zhao H, Zheng L, Zhou C, Han Y, Wu A, et al. Non-coding RNAs and colitis-associated cancer: Mechanisms and clinical applications. Clin Trans Med. (2023) 13:e1253. doi: 10.1002/ctm2.1253
59. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. (2022) 19:188–206. doi: 10.1038/s41571-021-00585-y
60. Zeng C, Tang Y, Jiang Y, Zuo Z, Tao H. Long noncoding RNAs as biomarkers for the diagnosis of hepatocellular carcinoma: A meta-analysis. Pathol-Res Pract. (2021) 224:153546. doi: 10.1016/j.prp.2021.153546
61. Li Y, Zeng X, He J, Gui Y, Zhao S, Chen H, et al. Circular RNA as a biomarker for cancer: A systematic meta−analysis. Oncol Lett. (2018) 16:4078–84. doi: 10.3892/ol.2018.9125
62. Jiang X, Zhang G, Wu J, Lin S, Liu Y, Ma Y, et al. Long noncoding RNA serve as a potential predictive biomarker for breast cancer: a meta-analysis. BioMed Res Int. (2020) 2020:1–15. doi: 10.1155/2020/9045786
63. Ruffo P, Strafella C, Cascella R, Caputo V, Conforti FL, Andò S, et al. Deregulation of ncRNA in neurodegenerative disease: Focus on circRNA, lncRNA and miRNA in amyotrophic lateral sclerosis. Front Genet. (2021) 12:784996. doi: 10.3389/fgene.2021.784996
64. Lodde V, Murgia G, Simula ER, Steri M, Floris M, Idda ML. Long noncoding RNAs and circular RNAs in autoimmune diseases. Biomolecules. (2020) 10:1044. doi: 10.3390/biom10071044
65. Das S, Shah R, Dimmeler S, Freedman JE, Holley C, Lee J-M, et al. Noncoding RNAs in cardiovascular disease: current knowledge, tools and technologies for investigation, and future directions: a scientific statement from the american heart association. Circulation: Genomic Precis Med. (2020) 13:e000062. doi: 10.1161/HCG.0000000000000062
66. Liao Y, Wang R, Wen F. Diagnostic and prognostic value of long noncoding RNAs in sepsis: a systematic review and meta-analysis. Expert Rev Mol Diagnostics. (2022) 22:821–31. doi: 10.1080/14737159.2022.2125801
67. Qi L, Yan Y, Chen B, Cao J, Liang G, Xu P, et al. Research progress of circRNA as a biomarker of sepsis: a narrative review. Ann Trans Med. (2021) 9:1–6. doi: 10.21037/atm-21-1247
68. Zhong X, Guo Q, Zhao J, Li Y, Li X, Ren M, et al. Diagnostic significance of long non-coding RNAs expression in tuberculosis patients: A systematic review and meta-analysis. Medicine. (2022) 101:1–8. doi: 10.1097/MD.0000000000028879
69. Srijyothi L, Ponne S, Prathama T, Ashok C, Baluchamy S. Roles of non-coding RNAs in transcriptional regulation. Transcriptional Post-transcriptional Regul. (2018) 55. doi: 10.5772/intechopen.76125
70. Li Z, Zhao W, Wang M, Zhou X. The role of long noncoding RNAs in gene expression regulation. Gene Expression Profiling Cancer. (2019), 1–17. doi: 10.5772/intechopen.81773
71. Wang Y-M, Trinh MP, Zheng Y, Guo K, Jimenez LA, Zhong W. Analysis of circulating non-coding RNAs in a non-invasive and cost-effective manner. TrAC Trends Analytical Chem. (2019) 117:242–62. doi: 10.1016/j.trac.2019.07.001
72. Pardini B, Sabo AA, Birolo G, Calin GA. Noncoding RNAs in extracellular fluids as cancer biomarkers: the new frontier of liquid biopsies. Cancers. (2019) 11:1170. doi: 10.3390/cancers11081170
73. Niderla-Bielińska J, Jankowska-Steifer E, Włodarski P. Non-coding RNAs and human diseases: current status and future perspectives. Int J Mol Sci. (2023) 24:11679. doi: 10.3390/ijms241411679
74. Triantaphyllopoulos KA. Long non-coding RNAs and their “discrete” contribution to IBD and Johne’s disease—what stands out in the current picture? A comprehensive review. Int J Mol Sci. (2023) 24:13566. doi: 10.3390/ijms241713566
75. Lin L, Zhou G, Chen P, Wang Y, Han J, Chen M, et al. Which long noncoding RNAs and circular RNAs contribute to inflammatory bowel disease? Cell Death Dis. (2020) 11:456. doi: 10.1038/s41419-020-2657-z
76. Lun J, Guo J, Yu M, Zhang H, Fang J. Circular RNAs in inflammatory bowel disease. Front Immunol. (2023) 14:1307985. doi: 10.3389/fimmu.2023.1307985
77. Dhamnetiya D, Jha RP, Shalini S, Bhattacharyya K. How to analyze the diagnostic performance of a new test? Explained with illustrations. J Lab Physicians. (2021) 14:090–8. doi: 10.1055/s-0041-1734019
78. Çorbacıoğlu ŞK, Aksel G. Receiver operating characteristic curve analysis in diagnostic accuracy studies: A guide to interpreting the area under the curve value. Turkish J Emergency Med. (2023) 23:195. doi: 10.4103/tjem.tjem_182_23
79. Mandrekar JN. Simple statistical measures for diagnostic accuracy assessment. J Thorac Oncol. (2010) 5:763–4. doi: 10.1097/JTO.0b013e3181dab122
80. Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. Curr Pediatr. (2022) 21:209–28. doi: 10.15690/vsp.v21i3.2427
81. Ranganathan P, Aggarwal R. Understanding the properties of diagnostic tests–Part 2: Likelihood ratios. Perspect Clin Res. (2018) 9:99. doi: 10.4103/picr.PICR_41_18
Keywords: lncRNAs, circRNAs, diagnostic biomarkers, inflammatory bowel disease, ulcerative colitis, Crohn’s disease, meta-analysis
Citation: Belete MA, Tadesse S, Tilahun M, Gedefie A, Shibabaw A, Mulatie Z, Wudu MA, Gebremichael S, Debash H, Alebachew M and Alemayehu E (2024) Long noncoding RNAs and circular RNAs as potential diagnostic biomarkers of inflammatory bowel diseases: a systematic review and meta-analysis. Front. Immunol. 15:1362437. doi: 10.3389/fimmu.2024.1362437
Received: 28 December 2023; Accepted: 27 February 2024;
Published: 08 March 2024.
Edited by:
Jiao Li, Sichuan University, ChinaReviewed by:
Cheng Ye, Zhejiang Chinese Medical University, ChinaKiesha Wilson, University of South Carolina, United States
Copyright © 2024 Belete, Tadesse, Tilahun, Gedefie, Shibabaw, Mulatie, Wudu, Gebremichael, Debash, Alebachew and Alemayehu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melaku Ashagrie Belete, bWVsYWt1YXNoYWdyaWVAZ21haWwuY29t
†These authors have contributed equally to this work and share first authorship
 Melaku Ashagrie Belete
Melaku Ashagrie Belete Selamyhun Tadesse2
Selamyhun Tadesse2 Mihret Tilahun
Mihret Tilahun Alemu Gedefie
Alemu Gedefie Agumas Shibabaw
Agumas Shibabaw Muluken Amare Wudu
Muluken Amare Wudu Saba Gebremichael
Saba Gebremichael Ermiyas Alemayehu
Ermiyas Alemayehu