Abstract
Outer membrane vesicles (OMVs) released by Helicobacter pylori (H.pylori) can enter the blood circulation of the host and cause extra-gastric lesions such as atherosclerosis and hyperemesis gravidarum. This study aimed to investigate the effect of OMVs released by H.pylori on the development of thymic T cells in offspring mice and its underlying mechanisms. Through experimental observations, we found that H.pylori OMVs were able to cross the placental barrier, leading to a decrease in the number of CD3+ and CD4+ T cells in the peripheral blood of the offspring mice and a decrease in the response of T cells to H.pylori stimulation. After stimulation with OMVs in T cell positive selection experiments, the expression levels of CHMP5, IKK-β, and NF-κB are up-regulated, and the release of cytokines IL-7, IL-2, IL-4, and IFN-γ is simultaneously increased, whereas in T cell negative selection experiments, the expression of JNK is up-regulated, and the expression of CHMP5 and Bcl-2 is down-regulated in E15-16 fetal thymus organ culture. These results indicate that transmission of H pylori OMVs from mother to fetus might be related to the development of central tolerance in offspring T cells. The underlying mechanism may involve an interaction between the OMVs-stimulated pathway and the TCR pathway, although further research is needed to confirm this hypothesis. The study highlights the importance of preventing H.pylori infection during pregnancy and suggests that the effect of centrally tolerated antigens needs to be considered in vaccine design to maximize prevention.
1 Introduction
Helicobacter pylori (H pylori) is a gram-negative bacterium that can colonize the stomach for a long time (1–3). About half of the world’s population is infected with H pylori (4, 5), and nearly half of pregnant women are also infected (6, 7). Severe H pylori infection in pregnant women has been reported to lead to abortion and developmental malformations (8, 9). However, the effect of H pylori infection during pregnancy on fetal development remains poorly understood. This gap in knowledge may be due to the perception that bacteria cannot pass through the placental barrier.
The outer membrane vesicles (OMVs) released by H pylori play a crucial role in its pathogenesis (10–12). OMVs are key mediators of bacterial-host interactions, capable of delivering bacterial proteins and other molecules into host cells, thereby affecting the host’s immune response and cellular function (13). Recent studies have shown that OMVs can enter the host’s circulatory system and may cross the placental barrier (14). This raises concerns about the potential effects of H pylori infection during pregnancy on the fetus.
Although the placental barrier provides a degree of protection to the fetus from harmful substances, it can be compromised by certain pathogens (15, 16). These pathogens can cross the placental barrier and infect the fetus, leading to various pregnancy complications. Therefore, understanding the integrity of the placental barrier is essential to protect the fetus from infection. However, relatively little research has been conducted on whether H pylori OMVs can cross the placental barrier and affect fetal health. This area of research is important for understanding the maternal-fetal transmission of H pylori infections and their potential impact on offspring health.
Our central hypothesis is that H pylori OMVs can cross the placental barrier and affect fetal immune system development, particularly the development of thymic T cells. The key research questions we aim to address are: (1) How do H pylori OMVs cross the placental barrier? (2) What are the effects of H pylori OMVs on the development of thymic T cells in offspring? (3) What are the underlying mechanisms by which OMVs influence T cell development?
To address these questions, we examined the effects of H pylori OMVs on the development of thymic T cells in offspring mice. Specifically, we assessed the impact of OMVs on the number of CD3+ and CD4+CD8- T cells in the peripheral blood of offspring mice, analyzed T cell responses to H pylori stimulation, described changes in molecular and cytokine expression in positive T cell selection assays, and discussed changes in molecular expression in negative T cell selection assays. We hope to provide new insights into the effects of H pylori infection on fetal immune system development during pregnancy.
The significance of this study is to reveal the impact of maternal-fetal transmission of H pylori OMVs on the central tolerance of offspring T cells (Figure 1), elucidate the mechanism of interaction between the OMVs-stimulated pathway and the TCR pathway. We highlight the importance of screening for H.pylori infection, especially during pregnancy, and increasing awareness among women of child-bearing age. While it may be challenging to completely prevent H.pylori infection, these results emphasize the need for proactive measures to identify and manage the infection during this critical period, as well as considerations regarding centrally tolerated antigens in vaccine design, and potential approaches to maximize the effectiveness of prevention.
Figure 1

Schematic representation of H pylori OMVs mediating central tolerance of T cells from C57BL/6J mice offspring via maternal-fetal transmission.
2 Methods and experimental models
2.1 H.pylori culture and OMVs extraction and purification
According to previous studies, we made appropriate modifications (17). The H.pylori SS1 (purchased from BioVector NTCC Inc., cat.no. Hp SS1) used was resuscitated in Columbia solid medium containing 10% defibrinated sheep blood and transferred to brain heart infusion (BHI) medium (containing 10mg/L vancomycin, 2.5mg/L amphotericin B, 5mg/L trimethoprim, and 2.5IU/L polymyxin B, all antibiotics were purchased from Sigma-Aldrich, Germany). After microaerobic culture (85% N2, 10% CO2, 5% O2) at 37°C for 3-5 days under shaking at 120rpm/min, the supernatant was collected by centrifugation at 2000g for 10 min at 4°C, filtered with 0.45 and 0.22μm membranes, respectively, and then centrifuged with 100kDa (Millipore, USA) ultrafiltration tube at 3000g for 10min at 4°C. The pure OMVs were then extracted through an exclusion column kit (Invitrogen exosome spin column MW3000, USA) according to the manufacturer’s instructions (18). Protein concentrations of OMV preparations were quantified using the BCA Protein Assay Kit (Pierce Thermo Scientific, Rockford, IL, USA).
2.2 Identification of OMVs
Using the NanoSight® NS300 Nanoparticle Tracking Analysis (NTA) device, extracted OMV samples were diluted at a ratio of 1:100 and measured to obtain information on the size, and distribution of the particles in the samples (18).In addition, the samples were dropped onto FCF300-Cu grids (Sigma-Aldrich, Germany), fixed with 1% glutaraldehyde for 5min, dried, observed in a transmission electron microscope, and photographed (Hitachi HT7800, Japan) (19). The samples were then mixed with loading buffer and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) at 95°C for 10min. After being rapidly stained with Coomassie Brilliant blue (R-250), the samples were identified by gel digestion and mass spectrometry.
2.3 Protein extraction and analysis of H.pylori and OMVs
The bacteria were collected by centrifugation (5000r/min, 4°C), washed 3 times with precooled PBS, lysed with protein lysis buffer containing protease inhibitors and nucleases, mixed, and incubated on ice for 30min. The suspension was treated by sonication (3s sonication, 9s interval, cooling by ice bath) until the cells had broken. The precipitate was then discarded by centrifugation (12000r/min, 4°C), and the supernatant was transferred to Eppendorf tubes. Protein was quantified by BCA method.
The extracted H.pylori whole-cell protein and OMVs samples were mixed with loading buffer, respectively, subjected to SDS-PAGE at 95°C for 10min in a water bath, and then transferred to a polyvinylidene difluoride (PVDF) membrane. The membranes were blocked with 5% skim milk powder in TBST for 1h and then incubated overnight at 4°C with the primary antibody (a polyclonal antibody obtained from cows immunized with inactivated whole SS1 bacteria by our research group). After a wash step, the membranes were incubated with the secondary antibody [goat anti-bovine IgG (H+L) HRP, Invitrogen, 1:500 dilution] for 1h at room temperature. Finally, an imaging system (Amersham Imager 600, GE Healthcare, Britain) was used to observe the membranes and to take pictures. The stained bands were subsequently dug out and identified by Coomassie Brilliant blue tape in-gel digestion and mass spectrometry.
2.4 Cell culture of BMDCs and H.pylori antigen extracted by BMDCs
Bone marrow (BM) was washed from the femur and tibia of mice. After purification, the cell concentration was adjusted to 1×106 cells/mL. The cells were plated onto 100mm bacterial culture dishes with RPMI 1640 complete medium containing 10% FBS and GM-CSF (20ng/mL, Abcam, ab9742). Another 10mL fresh medium containing 20ng/mL GM-CSF was added to the culture dish. On day 6 and day 8, the old medium was collected and centrifuged, and the cells were resuspended in fresh medium containing 20 ng/mL GM-CSF, before being returned to their original dish. Cells were harvested at day 10 and identified by flow cytometry (20, 21).
Mature BMDCs (1×107 cells/mL) were incubated with PBS, H.pylori, and H.pylori OMVs for 5 days at 1000 r/min, and the cells were collected by centrifugation. Subsequently, 4mL weak acid extract [0.131mol/L citric acid, 0.066mol/L disodium hydrogen phosphate (Sigma-Aldrich, USA)] was added, and the pH was adjusted to 3.3. The supernatant was shaken gently at room temperature for 5min, aspirated by centrifugation, and then intercepted by centrifugation in a 3kDa (Millipore, USA) ultrafiltration tube at 3000g for 10min at 4°C, and then analyzed by mass spectrometry (22).
2.5 Mass spectrometry analysis
Sample preparation for mass spectrometry began with extraction of the target protein bands from the gel and washing with MilliQ water, followed by treatment with a decolorizing solution of ammonium bicarbonate and acetonitrile for 20 min at 37°C to remove the pigmentation until the color disappeared. The samples were then dehydrated, vacuum dried, and treated with DTT reduction and IAA alkylation. Afterwards, the micelles were eluted using a solution of ammonium bicarbonate and acetonitrile and fixed by vacuum drying. After addition of trypsin buffer solution, the samples were incubated at 4°C for 30 min, followed by overnight digestion at 37°C. The digestion reaction was terminated with 0.1% trichloroacetic acid and the digest was collected by centrifugation. Trypsin-digested peptides were analyzed using a Q Exactive HF-X mass spectrometer and an EASY-nLC 1200-Q Exactive HF-X system, and NCBI databases were searched via the MASCOT search engine (Matrixscience.com) for protein identification.
2.6 Mice experiments
All animal experimental protocols were implemented after approval by the Animal Ethics Committee of the First Hospital of Hebei Medical University, and all animals reached the SPF level that was assured as being non-infected by H.pylori. C57BL/6J mice were housed in a pathogen-free room (12 h day/night, temperature 21 ± 2°C, humidity 55 ± 10%) and fed AD libitum. The H.pylori infection model was established as follows (23): freshly cultured H.pylori was collected, and the concentration of bacterial solution was adjusted to 1×109 CFU/mL. Three to four weeks old female mice were selected. In order to achieve H.pylori colonization, 0.1 mL of 50% alcohol was given to each mouse by gavage one day before inoculation. (18). H.pylori was given by gavage, 0.2mL each time, once every other day, 3 times. All animals were fasted and water-deprived for 12h before feeding and for an additional 4h after feeding. The mice were caged (vaginal plug dates were designed as E1), and the peripheral blood of the offspring mice was collected to analyze the changes of peripheral blood T cell subsets in the offspring mice when they were 6-8 weeks old.
An OMVs model for tail vein injection was established: OMVs were resuspended in sterile PBS, incubated with 5μM DiI (1,1’-dioctadecyl-3,3,3’,3’-tetramethylindocarbocyanine perchlorate, Invitrogen, USA) for 30min, then washed three times to remove impurities such as free DiI and lipoproteins, and resuspended in PBS (24, 25). Pregnant E18-19 mice were injected with DiI-labeled H.pylori OMVs (0.2mL, 20μg/mL) via the tail vein, and Cy5-IgG was injected as a positive control. Offspring were born, and the fluorescence signal of the offspring was detected within 24h by using the FX Pro system (Bruker, USA). Only pregnant mice with 3-6 fetuses were included in this study.
2.7 Positive-selection experiments and negative-selection experiments
The positive and negative selection experiments were appropriately adapted according to the guidance of the literature (26, 27). The experimental steps were as follows: first, pregnant mice (embryonic day E15-E16) were executed and their abdomens were wiped with 70% ethanol. Next, the abdominal wall was incised to expose the uterus, which was carefully incised with scissors and forceps to remove the embryos and to strip the placenta and fetal membranes. The removed embryos were then placed in petri dishes containing RF10-H medium.
For positive and negative selection experiments, thymic lobes were isolated from embryos and transferred to Nucleopore polycarbonate filters (Costar). Thymic lobes were cultured at 37°C with 5% CO2 using RP10 medium (RPMI1640 medium supplemented with 10% v/v heat-inactivated FCS, 2 mM glutamine, 10 mM HEPES, 0.5 mg/mL folate, 0.2 mg/mL glucose, 100 U/mL penicillin, and 100 μg/mL streptomycin).
For positive selection experiments, fetal thymus organ culture (FTOC) was established using thymic lobes from E15-E16 embryos. H pylori-OMVs at a concentration of 0.4 μg/mL were added at the start of the culture process, and the culture was maintained for 24 hours. For the negative selection experiments, FTOC was also established using thymic lobes from E15-E16 embryos. After 3 days of incubation, the thymic lobes were transferred to a medium containing 0.4 μg/mL of H pylori OMVs, and incubation was continued for an additional 24 hours.
Finally, thymocytes that underwent positive and negative selection were collected separately. After gently hand-damaging the thymus blades in 1.5 mL Eppendorf tubes, they were analyzed by flow cytometry. These steps were designed to simulate and study the development and differentiation process of thymocytes under different selection pressures.
2.8 Flow cytometry
E15-E16 pregnant mice (9-week-old, 30-33g) were used. After the fetal mice thymic lobes had been dissected, one half of each lobe was cultured for 4 days (control group), and three groups of duplicate wells were set up. The other half of each of the thymic lobes was cultured for 3 days and then stimulated with H.pylori OMVs for 1 day (experimental group), and 3 groups of duplicate wells were set up. On the fourth day, fetal thymus CD4 and CD8 were analyzed by flow cytometry.
The peripheral blood of 6- to 8-week-old offspring mice was collected, with 4 mice per group. To each 100 μL of whole blood, 30 μL of heparin sodium solution was added to prepare anticoagulated blood. Antibodies CD3 (PE, no.100206), CD4 (FITC, no.100510), and CD8 (PerCP/Cyanine5.5, no.100734) were added to the anticoagulated blood. Antibodies were purchased from BioLegend. The cells were incubated in the dark for 30min. Subsequently, 2mL red blood cell lysate was added to the cells and mixed gently. The reaction was carried out at room temperature in the dark for 10min. The samples were centrifuged at 1500rpm for 5min, and the supernatant from each was discarded. After being mixed with 2mL PBS, the cells were centrifuged at 1500rpm for 5min, the supernatant was discarded, and the cell precipitate was resuspended in 500uL of PBS and examined by flow cytometry.
All experiments included the addition of a mice Fc receptor blocker to perform blocking operations to avoid non-specific staining. The results were analyzed using Attune NxT software (Invitrogen, USA), and the gate strategies are shown in Supplementary Figure.
2.9 Mixed lymphocyte reaction
To detect cell proliferation (21), we first co-incubated mature bone marrow-derived dendritic cells (BMDCs, 1×105/well/100uL) with H pylori for 5 days. At the same time, peripheral blood samples were collected from the offspring of mice injected with H pylori OMVs via tail vein in late gestation (E18-E19). Monocytes were isolated from these samples using isodensity gradient centrifugation and B lymphocytes were cleared by CD19 magnetic beads (Meteni, Germany). Subsequently, purified BMDCs were co-cultured with T cells at a ratio of 1:10 for 5 days. At the end of the culture, cell culture supernatants were collected for subsequent cytokine assays.
2.10 Cellular immunofluorescence
The cell source is the same as that of the mixed lymphocyte reaction experiment. Cells were fixed with 4% paraformaldehyde (PFA) for 15min at room temperature, permeabilized with 0.3% Triton X-100, blocked with 2% goat serum for 1h, and incubated with rabbit anti-Ki-67 (Invitrogen, MA5-14520 at 1:250 dilution) overnight at 4°C. The cells were subsequently incubated with Alexa Fluor 555-conjugated secondary antibody goat anti-rabbit IgG (Invitrogen, A32732, 1:200 dilution) 1 h. After a washing step, coverslips were carefully placed over the cells and sealed using an anti-fluorescence quencher. Images were acquired by confocal microscopy (Zeiss, Oberkochen, Germany) for fluorescence imaging.
2.11 Western blot analysis
The gels and PVDF membranes were cut according to the size of the membrane transfer mold, and the proteins were transferred to the PVDF membranes (Millipore, USA) in a Bio-Rad transfer tank. The PVDF membranes were removed and immersed in blocking solution (3% bovine serum albumin) for 1h at room temperature with shaking. Goat anti-CHMP5 antibody (GeneTex, GTX106692, 1:500 dilution), goat anti-NF-κB antibody (Abcam, ab32536, 1:1000 dilution), and goat anti-IKK-β antibody (Abcam, ab124957, 1:1000 dilution) were added to the membranes, with β-actin being used as control. The membranes were incubated at room temperature for 1h with shaking, after which time they were removed and washed 4 times with PBS for 10min each. They were then immersed in the secondary antibody solution (goat anti-rabbit IgG Dylight 800, diluted 1:5000), incubated for 1h with slow shaking, and washed 4 times with PBS for 10min each. The target protein spots were detected by an ECL colorimetric kit. Image J software was used for analysis and processing.
2.12 Real-time fluorescent quantitative polymerase chain reaction
Total RNA was extracted from tissues according to the Trizol (Invitrogen 15596-026) instructions (28), and its integrity was confirmed by agarose gel electrophoresis. RNA concentration and purity were determined by a Nano Drop 1000 spectrophotometer (DeNovix, USA). cDNA was synthesized by a cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA), followed by a Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific, Waltham, MA) to amplify the genes of interest. The primer sequences for qRT-PCR are shown in Table 1.
Table 1
| Gene assayed | Primer sequence (5'–3') |
|---|---|
| CHMP5 | 5'-GATGAGAGAGGGTCCTGCTAAG-3' 5'-CAGGTTGTCTCGCTGTTGCTCA-3' |
| JNK | 5'-CGCCTTATGTGGTGACTCGCTA-3' 5'-TCCTGGAAAGAGGATTTTGTGGC-3 ' |
| Bcl-2 | 5'-CCTGTGGATGACTGAGTACCTG-3 ' 5' -AGCCAGGAGAAATCAAACAGAGAGG-3 ' |
| GAPDH | 5'-CATCACTGCCACCCAGAAGACTG-3' 5'-ATGCCAGTGAGCTTCCCGTTCAG-3' |
The primer sets used for the detection of murine genes by qRT-PCR analysis.
2.13 Enzyme-linked immunosorbent assay
Cytokines were detected with IL-7, IL-2, IL-4, and IFN-γ ELISA kits, purchased from R&D (USA), according to the manufacturer’s instructions (29). First, the capture antibody was encapsulated in a 96-well plate and incubated at 37°C overnight. Subsequently, the plates were closed for one hour, samples and standards were added and incubated for 2 hours, followed by the addition of the detection antibody and substrate and incubation for another 2 hours. Afterwards, the affinity hormone-HRP coupling was added and incubated for 30 minutes. Finally, 2,2’-azathiazoline [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt solution was added and the optical density was measured at 405 nm. Between each step, the substrate was washed thoroughly using wash buffer to remove unbound substrate. The experimental procedure for each group was repeated three times.
2.14 Statistical analysis
Data are presented as mean±standard deviation (SD). GraphPad Prism 9.0 was used for statistical analysis. The Shapiro - Wilk test was employed to assess the data distribution. The results indicated that the data followed a normal distribution (P > 0.05). Student’s t test was used for paired pairs, the two-tailed t test was used for unpaired pairs, and a one-way analysis of variance (ANOVA) and Tukey’s multiple comparison test were used for multiple group comparisons. Statistical results were expressed as *P < 0.05, ** P < 0.01, *** P < 0.001.
3 Results
3.1 Extraction and identification of H.pylori OMVs
H.pylori OMVs were isolated from H.pylori culture supernatant and identified. Transmission electron microscopy (TEM) showed that the isolate contained spherical vesicular structures (Figure 2A). Nanoparticle tracking analysis (NTA) results showed that the particle size distribution of OMVs ranged from 100 to 150 nm, which was the typical particle size of OMVs produced by Gram-negative bacteria (Figure 2B). The extracts were subjected to SDS-PAGE, gel staining, band collection (Figure 2C), and in-gel enzymatic digestion followed by proteomic analysis; 343 H.pylori proteins were identified, including outer membrane protein (OMP), vacuolating cytotoxin A (VacA), flagellin A (FlaA), and urease (Ure) etc (Table 2).
Figure 2
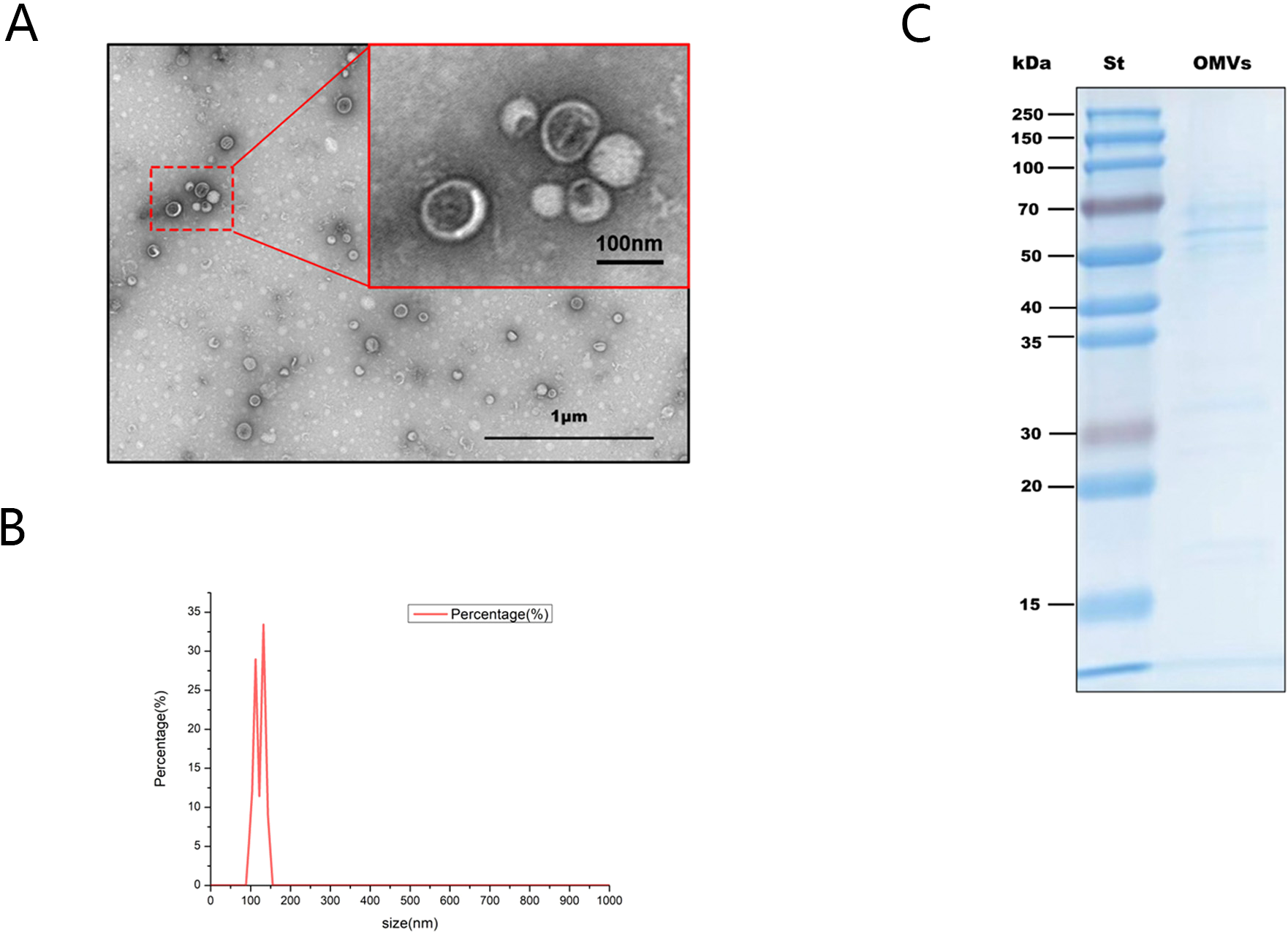
Identification of H.pylori OMVs. (A), Typical spherical vesicle-like morphological structures with diameters between 20 and 400nm were observed by TEM. Scale bars:1 micr/on (black box) and 100 nm (red box). (B), Nanoparticle tracking analysis shows the average concentration (vesicles/mL) of OMV of a specific size (nm). (C), Coomassie blue staining results of H.pylori OMVs after SDS-PAGE electrophoresis. St indicates the molecular marker standard (kDa, left).
Table 2
| Accession | Description | Coverage [%] | Peptides | PSMs | Unique Peptides | Protein Groups | AAs | MW [kDa] | calc. pI | Peptides (by Search Engine): Mascot | Abundance: F14: Sample |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A0A1U9ITG9 | Outer membrane protein HopQ / Omp27 OS=Helicobacter pylori SS1 OX=102617 GN=hopQ PE=4 SV=1 | 43 | 19 | 30 | 8 | 1 | 638 | 69.4 | 9.07 | 19 | 12909955.75 |
| A0A1U9IVK7 | Vacuolating cytotoxin autotransporter OS=Helicobacter pylori SS1 OX=102617 GN=vacA PE=4 SV=1 | 35 | 30 | 33 | 26 | 1 | 1312 | 141.9 | 8.75 | 30 | 225306407.5 |
| A0A1U9IUB7 | Flagellin OS=Helicobacter pylori SS1 OX=102617 GN=flaA PE=3 SV=1 | 61 | 23 | 59 | 23 | 1 | 510 | 53.2 | 6.43 | 23 | 162104462.9375 |
| A0A1U9IS01 | Urease subunit beta OS=Helicobacter pylori SS1 OX=102617 GN=ureB PE=3 SV=1 | 77 | 36 | 153 | 36 | 1 | 569 | 61.6 | 6.01 | 5517 | 2135599392.65625 |
Proteomic analysis of H pylori OMVs by LC-ESI-MS/MS. This table briefly summarizes H pylori SS1proteins identified by mass spectrometry.
3.2 H.pylori OMVs can be transmitted across the placental barrier to the embryo
An appropriate dose of H.pylori OMVs was determined by a dose-effect experiment. E18-19 pregnant mice were injected with 160, 80, 40, 20, or 10μg/mL H.pylori OMVs (0.2mL) via the tail vein. When the concentration was higher than 40μg/mL, the pregnant mice showed premature delivery, vaginal bleeding, and abortion (Figure 3A); when the injection concentration was 20μg/mL, the pregnant mice had no vaginal bleeding, and the offspring mice survived for more than one year and were able to reproduce. Therefore, the 0.2mL(20μg/mL) dose was used as the experimental injection dose.
Figure 3

H.pylori OMVs is able to cross the placental barrier. (A), When the concentration of H.pylori OMVs was greater than 40 μg/mL, vaginal bleeding and abortion were observed in E18-19 pregnant mice injected via the tail vein. (B), Flow chart of DiI-OMVs administered via the tail vein to E18-E19 pregnant mice. (C), Views after FX PRO (Bruker) imaging. Hyperfluorescence signal in fetal mice (upper) via tail vein injection of IgG-Cy5. DiI-OMVs were injected via the tail vein, and high fluorescence signal was observed in the offspring (lower) (right, scale bar). (D), Ex vivo fluorescence imaging of mice placenta in DiI-OMVs groups (right, scale bar).
To evaluate whether H.pylori OMVs were able to pass through the placental barrier to the embryo. H.pylori OMVs were labeled with DiI and injected into E18-19 pregnant mice via the tail vein. Strong fluorescence signals were observed in the offspring mice, indicating that H.pylori OMVs had passed through the placental barrier. Similarly, strong autofluorescent signals were detected in isolated placentas of the DiI-OMVs group but not in offspring mice from the H.pylori OMVs group (Figures 3B-D).
3.3 H.pylori OMVs and H.pylori maternal bacteria have common antigens
To analyze the antigenic components carried by the OMVs, the lysates of H.pylori and of H.pylori OMVs were subjected to SDS-PAGE and Western blot analysis. We compared the protein composition of H.pylori OMVs with that of H.pylori. The results showed that H.pylori and their OMVs had high gray values at 25-35kDa and 15-20kDa. The gels were cut at the corresponding positions and analyzed after in-gel digestion (Figure 4A), We found 1000 proteins in the 25-35kDa band of H.pylori whole bacteria and 581 proteins in H.pylori OMVs, of which 573 proteins were shared by H.pylori and their OMVs. We detected 971 proteins in H.pylori whole bacteria at the 15-20kDa band and 292 proteins in H.pylori OMVs, of which 287 proteins were shared by H.pylori and their OMVs (Figure 4B).
Figure 4
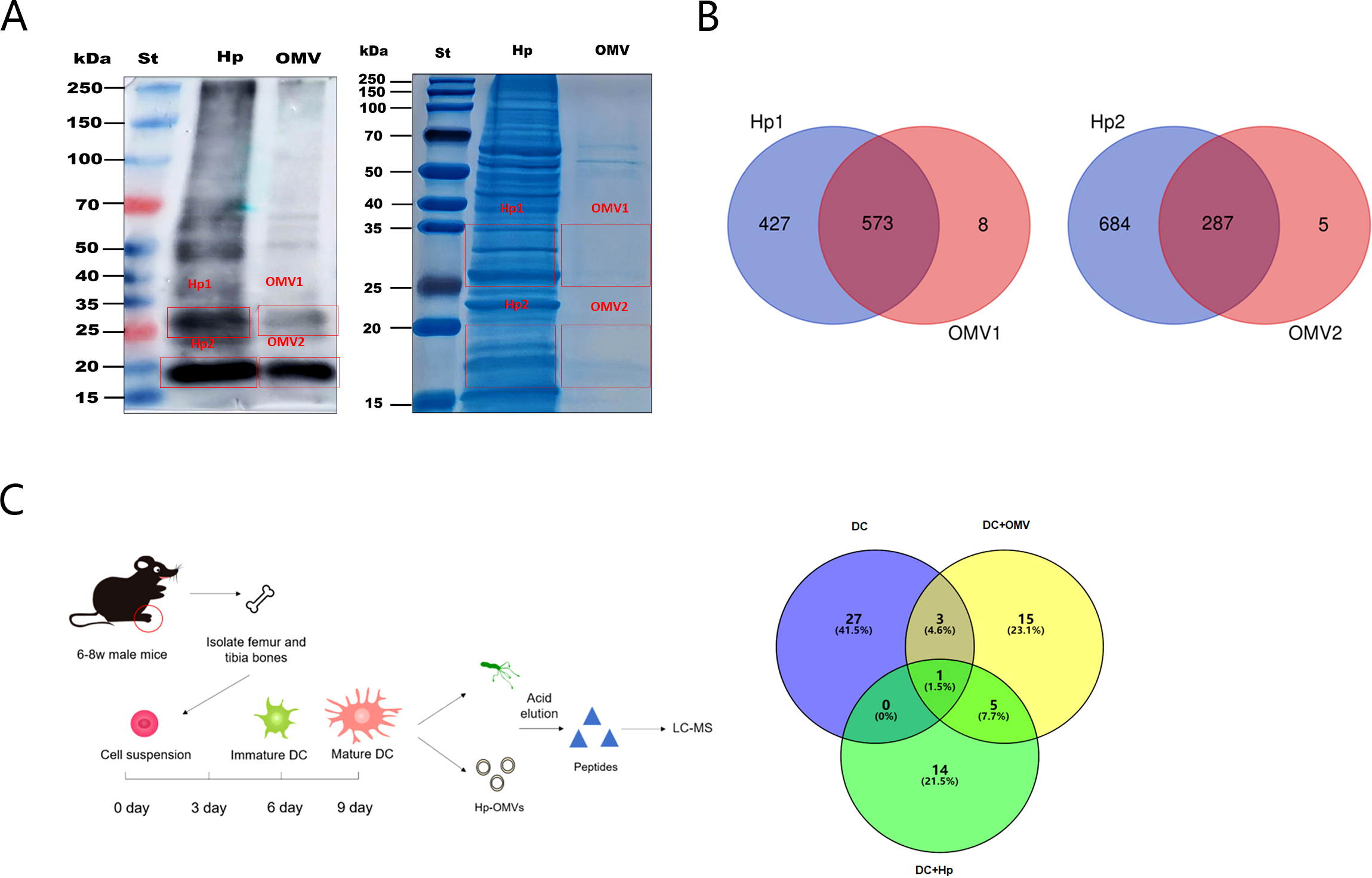
H.pylori OMVs carry the same antigen as H.pylori. (A), Immunoblotting plots of H.pylori and of H.pylori OMVs show high gray values at both 25-35kDa and 15-20kDa. (B), Gel stained with Coomassie brilliant blue at the corresponding positions. Venn diagram shows the amount of protein detected in whole bacteria and OMVs. (C), Mature BMDCs were cultured and stimulated with H.pylori or with H.pylori OMVs. MHC peptides were eluted with weak acid and identified by mass spectrometry. Venn diagram showing antigenic peptides extracted from OMVs and H.pylori by BMDCs.
Subsequently, we co-cultured the induced mature bone-marrow-derived dendritic cells (BMDCs) with H.pylori or their OMVs for 5 days (Figure 4C). The MHC-binding antigenic peptides were then eluted under weak acid conditions, and the eluate was used for peptide identification by liquid chromatography-mass spectrometry (LC-MS/MS). The results showed that H.pylori and their OMVs shared six common antigenic peptides (Table 3).
Table 3
| Accession | Description | Coverage [%] | Peptides | PSMs | Unique Peptides | Protein Groups | AAs | MW [kDa] | calc. pI | Peptides (by Search Engine): Mascot | Abundance: F7: Sample |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A0A1Y3DZ56 | Uncharacterized protein OS=Helicobacter pylori SS1 OX=102617 GN=X568_06790 PE=4 SV=1 | 9 | 1 | 2 | 1 | 1 | 66 | 7.7 | 9.94 | 1 | |
| A0A1U9IVK7 | IS607 resolvase OrfA OS=Helicobacter pylori SS1 OX=102617 GN=HPYLSS1_00302 PE=4 SV=1 | 3 | 1 | 1 | 1 | 1 | 217 | 25.1 | 9.03 | 1 | 13230317 |
| A0A1Y3E1L4 | Plasmid stabilization protein OS=Helicobacter pylori SS1 OX=102617 GN=X568_02515 PE=4 SV=1 | 1 | 1 | 1 | 1 | 1 | 543 | 61.9 | 9.16 | 1 | 6487167 |
| A0A1U9IS95 | Protein RecA OS=Helicobacter pylori SS1 OX=102617 GN=recA PE=3 SV=1 | 2 | 1 | 1 | 1 | 1 | 347 | 37.6 | 5.66 | 1 | |
| A0A1U9IT63 | GMP reductase OS=Helicobacter pylori SS1 OX=102617 GN=guaC PE=3 SV=1 | 2 | 1 | 1 | 1 | 1 | 325 | 35.9 | 8.53 | 1 | |
| A0A1U9ITD1 | Protein translocase subunit SecA OS=Helicobacter pylori SS1 OX=102617 GN=secA PE=3 SV=1 | 1 | 1 | 1 | 1 | 1 | 865 | 99 | 5.91 | 1 | 200291.171875 |
BMDCs extracted the same antigenic peptides from H pylori-OMVs and H pylori.
3.4 H.pylori OMVs can induce central tolerance of progeny T cells
We first examined the effect of H.pylori OMVs stimulation on T cell subsets in fetal mice thymus cultured in vitro. Compared with the control group, CD4+CD8- T cells in the H.pylori OMVs stimulation group decreased significantly, whereas CD4-CD8+, CD4+CD8+, and CD4-CD8- T cells did not change significantly (Figure 5A).
Figure 5
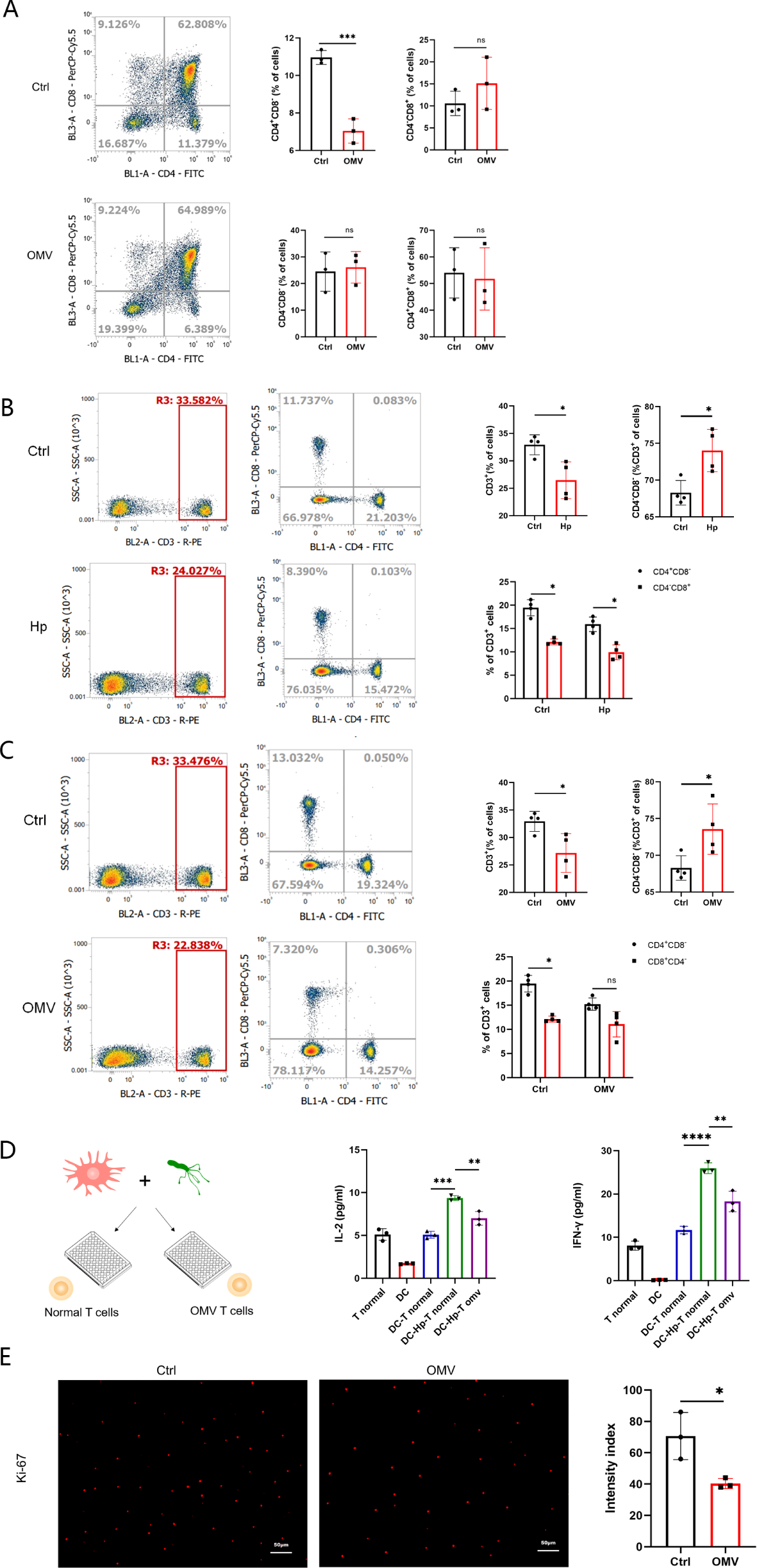
Induction by H.pylori OMVs of changes in T cell subsets in fetal mice thymus and peripheral blood in offspring mice. (A), T cell subsets (CD4+CD8-, CD4-CD8+, CD4+CD8+, CD4-CD8-) in the thymus of fetal mice were detected by flow cytometry; results are expressed as mean ± SD and statistically tested by unpaired two-tailed t tests. (B), T cell subsets (CD3+, CD3+CD4+CD8-, CD3+CD4-CD8+, CD3+CD4-CD8-) in peripheral blood of offspring mice born to mice experiencing H.pylori infection during pregnancy were detected by flow cytometry, 4 mice/group (n=4); results are expressed as mean ± SD, and statistical analysis was performed by unpaired two-tailed t test. (C), Flow cytometry was used to detect the peripheral blood T cell subsets (CD3+, CD3+CD4+CD8-, CD3+CD4-CD8+, CD3+CD4-CD8-) in the offspring of E18-E19 pregnant mice injected with H.pylori OMVs via the tail vein, 4 mice/group (n=4); results are expressed as mean ± SD, and statistical analysis was performed by unpaired two-tailed t test. (D), Expression of IL-2 and IFN-γ in the supernatants of mixed lymphocyte reaction cells was measured by ELISA, repeated three times; results are expressed as mean ± SD and statistically tested by unpaired two-tailed t test. (E), Cells stained with Ki-67 were observed by fluorescence microscopy (×10 eyepiece, ×20 objective), scale: images were randomly collected three times and analyzed in a blind manner; results were expressed as mean ± SD and statistically tested by unpaired two-tailed t test. *P< 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, no significance..
Next, we investigated the changes of T cells in the offspring of pregnant mice infected with H.pylori. The results indicated abnormal variations in the T cell subpopulations of peripheral blood in the offspring mice. Specifically, there was a significant reduction in CD3+, CD3+CD4+CD8-, and CD3+CD4-CD8+ T cells, while CD3+CD4-CD8- T cells increased significantly (Figure 5B).
Then, we determined the changes of T cells in offspring of mice injected with H.pylori OMVs via the tail vein during pregnancy (E18-19). The results showed that the peripheral blood T cell subsets of offspring mice had abnormal changes, and the CD3+; and CD3+CD4+CD8- T cells were significantly reduce, while CD3+CD4-CD8- T cells increased significantly (Figure 5C). Subsequently, the ability of CD3+CD4+CD8+ T cells from the offspring mice to respond to H.pylori stimulation was verified. The production of IL-2 and IFN-γ by T cells from the offspring of the above model mice was shown to be significantly lower than that of the normal group (Figure 5D), and the proliferation ability of T cells was significantly lower than that of the control group (Figure 5E).
3.5 Molecular mechanism responsible for effects of H.pylori OMVs on thymic development in fetal mice
To explore the molecular mechanism responsible for the effects of H.pylori OMVs on thymus development, we tested positive selection and negative selection phase. In the positive selection phase, the expression levels of CHMP5, IKK-β, and NF-κB proteins, as detected by Western blotting, in the thymus tissues of E15-16 fetal mice were shown to be up-regulated (Figure 6A). ELISA revealed that the levels of IL-7, IL-2, IL-4 and IFN-γ in the supernatants were increased (Figure 6B). In the negative selection phase, the gene expression levels of CHMP5, JNK, and Bcl-2 in the thymus tissues of the 3-day stimulation group were examined by qRT-PCR: JNK expression was found to be significantly up-regulated, whereas CHMP5 and Bcl-2 expression was significantly down-regulated (Figure 6C). The above results thus showed that thymic stromal cells and related TCR and MHC signaling pathways were activated by H.pylori OMVs, and that the expression level of CHMP5 was changed in a bidirectional manner (Figure 7).
Figure 6
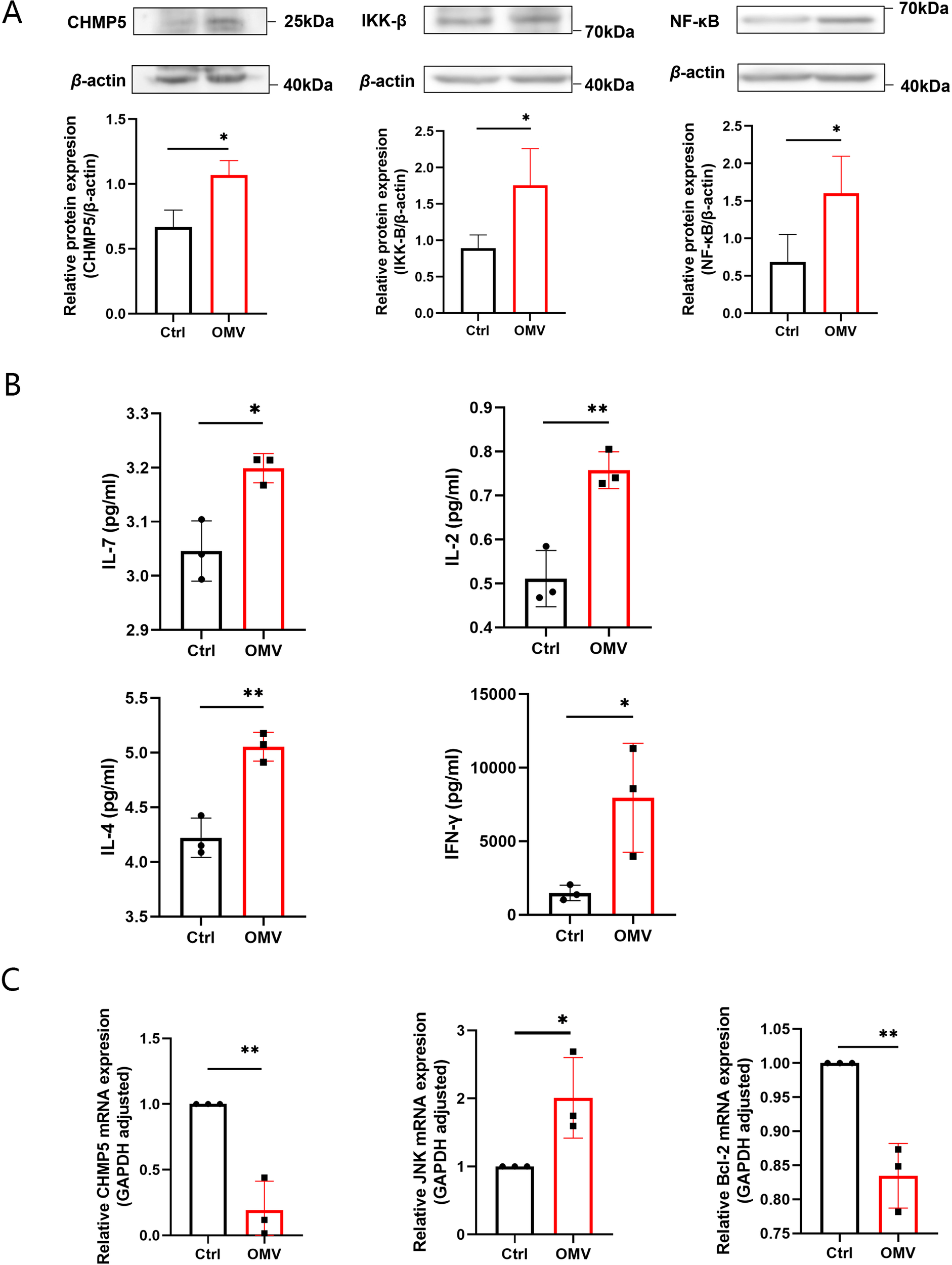
Induction by H.pylori OMVs of changes in signaling molecules related to T cell development in fetal mice thymus and their relationships. On day 0, the thymus of E15-16 fetal mice was stimulated with H.pylori OMVs: (A), Western blot analysis showing the expression of CHMP5, IKK-β, and NF-κB; (B), the expression levels of cytokines IL-7, IL-2, IL-4, and IFN-γ as determined by ELISA. (C), After stimulation with H.pylori OMVs in the thymus of E15-16 fetal mice on day 3 of culture, qRT-PCR showing the expression of JNK, CHMP5 and Bcl-2 mRNA. Each independent experiment was repeated three times; results are presented as mean ± SD. Statistical tests were performed using an unpaired two-tailed t test. *P< 0.05, **P < 0.01.
Figure 7
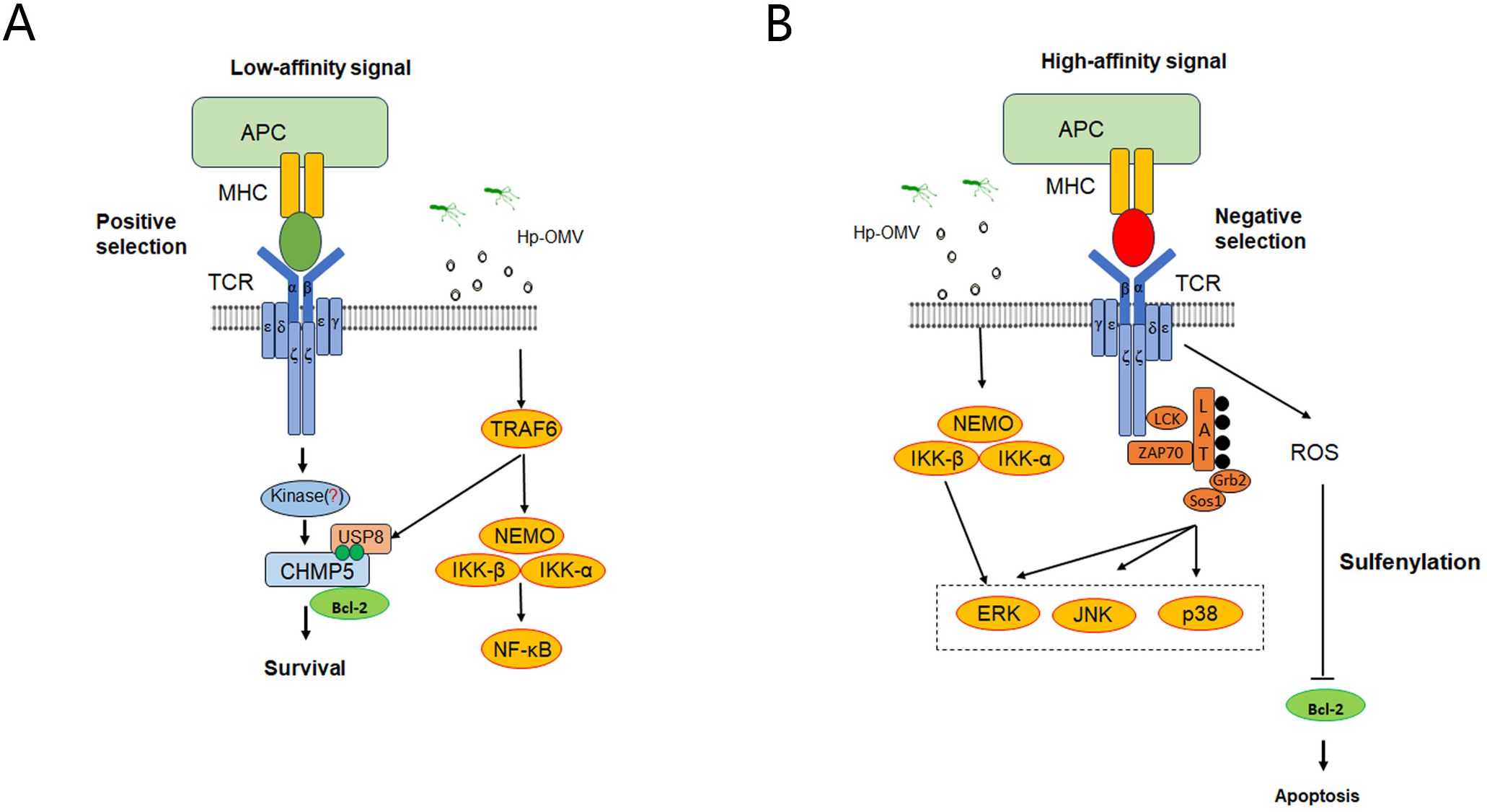
Possible mechanism of central tolerance of thymic T cells induced by H.pylori OMVs. (A), During the positive selection phase, crosstalk might occur between the OMV activation pathway and the MHC pathway, which might regulate the stability of CHMP5 protein and promote the development of positively selected T cells. (B), During the negative selection phase, crosstalk might occur between the OMV activation pathway and the TCR signaling pathway to promote T cell apoptosis. *P < 0.05, ** P < 0.01, *** P < 0.001, ns: no significance.
4 Discussion
In recent years, H.pylori OMVs have attracted much attention in the field of medical research (30–33). As an important class of microbial components, OMVs play a key role in bacteria-host interactions (34). In addition to its colonization and pathogenic role in the gastrointestinal tract, the H.pylori OMVs are able to cross a variety of biological barriers, including the gastric mucosa and the blood circulatory system (11, 33, 35). The background of this study builds on these findings and further explores the potential impact of H pylori OMVs during pregnancy, specifically whether they are able to cross placental barriers and affect the immune development of the offspring. This study provides the first direct evidence that H pylori OMVs are able to cross the placental barrier. In addition, the transfer of OMVs from mother to fetus is associated with T cell development.
It has been found that infection with H pylori during pregnancy leads to significant changes in T cell subsets in the offspring (36), specifically demonstrated by a decrease in the number of CD3+ T cells, which may affect the immune response and the establishment of tolerance in the offspring (37). Additionally, the increase in CD3+CD4-CD8- T cells may be related to the immune response activated by maternal infection with H pylori or exposure to OMVs. These inflammatory signals could be transmitted to the fetus through the placenta, thereby affecting the development of its immune system. The elevation of the CD3+CD4-CD8- T cells population might reflect an adaptive response of the fetal immune system to maternal infection, such as maintaining immune homeostasis through Th cell subsets. Although these CD3+ T cells, lacking both CD4+ and CD8+ coreceptors, account for only 3-5% of peripheral blood T lymphocytes, they possess both innate and adaptive immune functions (38). They play a crucial role in immune surveillance, infection resistance, and immune regulation. Abnormalities in their numbers may either weaken the effective defense against pathogens or increase the risk of autoimmunity due to long-term immune activation, potentially forming a synergistic effect with the Th2 bias immune characteristics of the neonatal period (39, 40). H pylori infection during pregnancy may affect T cell maturation and function through antigens transmitted by OMVs, thereby influencing the long-term immune health of the offspring. The reduction in the number and function of CD4+ T cells could have profound effects on the offspring’s immune function, including decreased defense against pathogens, diminished vaccination efficacy, and increased susceptibility to autoimmune and allergic diseases (41, 42).Understanding these nuances is essential for optimizing vaccine strategies and managing infectious diseases in neonates and infants (43).
Given the observed alterations in the T cell subsets of offspring, we hypothesize that H.pylori OMVs may act as a vector for the transmission of maternal antigens to the offspring, thereby impacting the development and function of offspring T cells. H.pylori OMVs possess the capability to traverse the placental barrier and deliver H.pylori antigens to the fetus. These antigens may then be processed and presented within the fetal thymus, influencing both positive and negative T cell selection processes. This sequence of events may ultimately lead to the establishment of central tolerance to H.pylori within the offspring T cells, subsequently modulating their immune response to H.pylori. This hypothesis is consistent with the well - characterized properties of H.pylori OMVs. For instance, H.pylori OMVs are known to carry a diverse array of bacterial proteins and toxins, and they play a crucial role in mediating bacterial-host interactions. Their unique ability to encapsulate and transport various biomolecules, along with their involvement in intercellular communication between bacteria and host cells, provides a mechanistic basis for the proposed role of H.pylori OMVs in the transfer of maternal antigens and the subsequent impact on offspring T cell development.
Our experimental results reveal that H pylori OMVs can penetrate the placental barrier. By injecting OMVs via the tail vein into pregnant mice at 18-19 days of gestation, it was found that OMVs were able to enter the offspring, confirming the possibility of OMVs crossing the placental barrier. In addition, the study observed changes in T-cell subsets in the offspring, suggesting that OMVs may play a role in the fetus, affecting its immune cell development and function. The placental penetration ability of OMVs may have an important impact on the immune development of the offspring. Placental barrier penetration may result in the exposure of the offspring to maternal H pylori antigens during the embryonic period, thereby affecting their T-cell development. This early antigenic exposure may lead to the development of central tolerance to H pylori in the offspring’s T cells, affecting their immune response to H pylori (44).
The study investigates the impact of H pylori OMVs on T cell development in the thymus. Continuous exposure to OMVs can lead to cross-presentation of H pylori antigen peptides by MHC class I molecules on thymic dendritic cells, affecting CD8+ T cell negative selection and potentially causing non-specific thymic damage if not cleared by antigen-presenting cells (36). Using embryonic thymus organ cultures, we found that OMVs upregulate IKK-β, CHMP5, and NF-κB during positive selection, promoting T cell survival through pathways involving NF-κB and CHMP5-stabilized Bcl-2. OMVs also increase cytokine expression (e.g., IL-2, IL-4, IL-7, IFN-γ), supporting T cell maturation. In negative selection, OMVs upregulate JNK while downregulating Bcl-2 and CHMP5, indicating crosstalk between OMV-activated pathways (e.g., TLRs) and TCR signaling. Further studies are needed to clarify the mechanisms, particularly the kinase responsible for CHMP5 phosphorylation (45).
The central tolerance induced by OMVs has profound implications for the design of H pylori vaccines. Central tolerance is a critical mechanism by which the immune system eliminates self-reactive T cells during development to prevent autoimmune diseases. However, antigens delivered by H pylori OMVs can induce central tolerance, thereby reducing vaccine efficacy. For instance, OMVs can activate TLR pathways and induce the production of immunosuppressive cytokines such as IL-10, limiting inflammatory responses and promoting bacterial survival (46). These immune-modulating mechanisms may dampen the immunogenicity of vaccines, especially if central tolerance is established during fetal development, rendering the immune system less responsive to H pylori antigens later in life.
To address this challenge, several vaccine design strategies should be considered. First, it is essential to choose antigens that are less likely to induce central tolerance. For example, epitope-based vaccines designed through immunoinformatics have demonstrated robust immunogenicity, capable of inducing strong cellular and humoral immune responses (47). Second, leveraging the natural immunogenic properties of OMVs, such as engineering OMVs to carry specific antigenic epitopes, can enhance antigen presentation efficiency and bypass central tolerance mechanisms (48). Additionally, combining novel adjuvants (e.g., OMVs with removed endotoxins) (49) or delivery systems (e.g., lipid nanoparticles) (50) can further enhance vaccine immunogenicity and safety.
Future research should focus on elucidating the mechanisms by which OMVs induce central tolerance and exploring strategies to overcome this challenge in vaccine design. Specifically, further investigation is needed to understand how OMVs modulate T cell selection and central tolerance via TLR pathways and TCR signaling. Moreover, developing novel vaccine platforms based on OMVs, such as glycoengineered OMVs or OMVs incorporating antigens from multiple pathogens, may provide innovative solutions for vaccine design. After validating the efficacy and safety of these vaccines in animal models, advancing them to clinical trials will be essential for developing effective vaccines against H pylori and other pathogens. In summary, by unraveling the immune-modulating mechanisms of OMVs and integrating modern vaccine technologies, it is possible to develop more effective vaccines that overcome the limitations imposed by central tolerance.
5 Conclusions
Our study indicates that H pylori OMVs can cross the placental barrier, inducing central tolerance in offspring T cells to the bacteria. This finding reveals a significant pathogenic mechanism and underscores the need for vaccine strategies to consider central tolerance antigens. However, the specific mechanisms of OMVs influence on T cell tolerance require further investigation, acknowledging the limitations in fully elucidating the impact of OMVs on immune development. Future research should address these gaps to enhance our understanding and develop effective H pylori vaccines.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by the Animal Ethics Committee of the First Hospital of Hebei Medical. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YW: Formal analysis, Writing – original draft, Writing – review & editing. LZ: Investigation, Writing – review & editing. XZ: Software, Writing – review & editing. HQ: Data curation, Writing – review & editing. DH: Methodology, Writing – original draft. ZS: Formal analysis, Project administration, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by grants from Hebei Province Clinical Medicine Excellent Talents Training Program (No.LS201908).
Acknowledgments
We thank Xiangjian Zhang and Rong Chen (Hebei Collaborative Innovation Center for Cardio-cerebrovascular Disease, Hebei Key Laboratory of Vascular Homeostasis, The Second Hospital of Hebei Medical University, Shijiazhuang, China), for technical assistance and we thank Chenming Zhou (Department of Electron Microscopy Laboratory Center, Hebei Medical University, Shijiazhuang, China), for technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1522842/full#supplementary-material
References
1
Malfertheiner P Camargo MC El-Omar E Liou JM Peek R Schulz C et al . Helicobacter pylori infection. Nat Rev Dis Primers. (2023) 9:19. doi: 10.1038/s41572-023-00431-8
2
Elshenawi Y Hu S Hathroubi S . Biofilm of helicobacter pylori: life cycle, features, and treatment options. Antibiotics (Basel). (2023) 12:1260. doi: 10.3390/antibiotics12081260
3
Wang L Cao ZM Zhang LL Dai XC Liu ZJ Zeng YX et al . Helicobacter pylori and autoimmune diseases: involving multiple systems. Front Immunol. (2022) 13:833424. doi: 10.3389/fimmu.2022.833424
4
Sousa C Ferreira R Azevedo NF Oleastro M Azeredo J Figueiredo C et al . Helicobacter pylori infection: from standard to alternative treatment strategies. Crit Rev Microbiol. (2022) 48:376–96. doi: 10.1080/1040841X.2021.1975643
5
Chen YC Malfertheiner P Yu HT Kuo CL Chang YY Meng FT et al . Global Prevalence Of Helicobacter Pylori Infection And Incidence Of Gastric Cancer Between 1980 And 2022. Gastroenterology. (2024) 166:605–19. doi: 10.1053/j.gastro.2023.12.022
6
Abdella B Ibrahim M Tadesse I Hassen K Tesfa M . Association between helicobacter pylori infection and occurrence of anemia among pregnant women attending antenatal care in kulito health center, halaba zone, south Ethiopia, 2018. . Anemia. (2020) 2020:6574358. doi: 10.1155/2020/6574358
7
Merino JS Araneda L Lincoñir-Campos P Parra C Sáez K García A . Dynamics of helicobacter pylori infection in infants during the first six months of life. Enferm Infecc Microbiol Clin (Engl Ed). (2019) 37:109–11. doi: 10.1016/j.eimc.2018.03.016
8
Feng Y Huang Q Luo M Wei J Gao T Chu D et al . The association between helicobacter pylori and gastrointestinal disorders during pregnancy: A multicenter retrospective study. Helicobacter. (2024) 29:e13032. doi: 10.1111/hel.13032
9
Grooten IJ Den Hollander WJ Roseboom TJ Kuipers EJ Jaddoe VW Gaillard R et al . Helicobacter pylori infection: A predictor of vomiting severity in pregnancy and adverse birth outcome. Am J Obstet Gynecol. (2017) 216:512.E1–.E9. doi: 10.1016/j.ajog.2017.01.042
10
González MF Díaz P Sandoval-Bórquez A Herrera D Quest AFG . Helicobacter pylori outer membrane vesicles and extracellular vesicles from helicobacter pylori-Infected cells in gastric disease development. Int J Mol Sci. (2021) 22:4823. doi: 10.3390/ijms22094823
11
Xie J Cools L Van Imschoot G Van Wonterghem E Pauwels MJ Vlaeminck I et al . Helicobacter pylori-derived outer membrane vesicles contribute to alzheimer’s disease pathogenesis via C3-C3ar signalling. . J Extracell Vesicles. (2023) 12:E12306. doi: 10.1002/jev2.12306
12
Wang N Zhou F Chen C Luo H Guo J Wang W et al . Role of outer membrane vesicles from helicobacter pylori in atherosclerosis. Front Cell Dev Biol. (2021) 9:673993. doi: 10.3389/fcell.2021.673993
13
Eletto D Mentucci F Voli A Petrella A Porta A Tosco A . Helicobacter pylori pathogen-associated molecular patterns: friends or foes? Int J Mol Sci. (2022) 23:3531. doi: 10.3390/ijms23073531
14
Lara B Loureiro I Gliosca L Castagnola L Merech F Gallino L et al . Porphyromonas gingivalis outer membrane vesicles shape trophoblast cell metabolism impairing functions associated to adverse pregnancy outcome. J Cell Physiol. (2023) 238:2679–91. doi: 10.1002/jcp.v238.11
15
Yue H Tian Y Zhu H Wu X Xu P Ji X et al . Fetal origin of abnormal glucose tolerance in adult offspring induced by maternal bisphenol A analogs exposure. Environ Sci Technol. (2024) 58:10910–9. doi: 10.1021/acs.est.3c09238
16
Hoo R Ruiz-Morales ER Kelava I Rawat M Mazzeo CI Tuck E et al . Acute response to pathogens in the early human placenta at single-cell resolution. Cell Syst. (2024) 15:425–44.E9. doi: 10.1016/j.cels.2024.04.002
17
Hu W Zhang L Li MX Shen J Liu XD Xiao ZG et al . Vitamin D3 activates the autolysosomal degradation function against helicobacter pylori through the pdia3 receptor in gastric epithelial cells. Autophagy. (2019) 15:707–25. doi: 10.1080/15548627.2018.1557835
18
Palacios E Lobos-González L Guerrero S Kogan MJ Shao B Heinecke JW et al . Helicobacter pylori outer membrane vesicles induce astrocyte reactivity through nuclear factor-κappa B activation and cause neuronal damage in vivo in A murine model. J Neuroinflamm. (2023) 20:66. doi: 10.1186/s12974-023-02728-7
19
Grande R Di Marcantonio MC Robuffo I Pompilio A Celia C Di Marzio L et al . Helicobacter pylori atcc 43629/nctc 11639 outer membrane vesicles (Omvs) from biofilm and planktonic phase associated with extracellular dna (Edna). Front Microbiol. (2015) 6:1369. doi: 10.3389/fmicb.2015.01369
20
Vangala P Murphy R Quinodoz SA Gellatly K McDonel P Guttman M et al . High-resolution mapping of multiway enhancer-promoter interactions regulating pathogen detection. Mol Cell. (2020) 80:359–73.E8. doi: 10.1016/j.molcel.2020.09.005
21
Riegel K Schlöder J Sobczak M Jonuleit H Thiede B Schild H et al . Raf kinases are stabilized and required for dendritic cell differentiation and function. Cell Death Differ. (2020) 27:1300–15. doi: 10.1038/s41418-019-0416-4
22
Mbiribindi B Mukherjee S Wellington D Das J Khakoo SI . Spatial clustering of receptors and signaling molecules regulates nk cell response to peptide repertoire changes. Front Immunol. (2019) 10:605. doi: 10.3389/fimmu.2019.00605
23
Lee A O’rourke J De Ungria MC Robertson B Daskalopoulos G Dixon MF . A standardized mouse model of helicobacter pylori infection: introducing the sydney strain. Gastroenterology. (1997) 112:1386–97. doi: 10.1016/S0016-5085(97)70155-0
24
Hu YB Yan C Mu L Mi YL Zhao H Hu H et al . Exosomal wnt-induced dedifferentiation of colorectal cancer cells contributes to chemotherapy resistance. Oncogene. (2019) 38:1951–65. doi: 10.1038/s41388-018-0557-9
25
Schaack B Mercier C Katby M Hannani D Vollaire J Robert JS et al . Rapid biodistribution of fluorescent outer-membrane vesicles from the intestine to distant organs via the blood in mice. Int J Mol Sci. (2024) 25:1821. doi: 10.3390/ijms25031821
26
Teixeiro E Daniels MA . Fetal thymic organ culture and negative selection. Methods Mol Biol. (2023) 2580:293–302. doi: 10.1007/978-1-0716-2740-2_18
27
Khanom US Ohigashi I Fujimori S Kondo K Takada K Takahama Y . Tcr affinity for in vivo peptide-induced thymic positive selection fine-tunes tcr responsiveness of peripheral cd8(+) T cells. J Immunol. (2019) 203:881–7. doi: 10.4049/jimmunol.1900097
28
Dawicki-Mckenna JM Felix AJ Waxman EA Cheng C Amado DA Ranum PT et al . Mapping ptbp2 binding in human brain identifies syngap1 as A target for therapeutic splice switching. Nat Commun. (2023) 14:2628. doi: 10.1038/s41467-023-38273-3
29
Xu J Wang P Li Z Li Z Han D Wen M et al . Irf3-binding lncrna-isir strengthens interferon production in viral infection and autoinflammation. . Cell Rep. (2021) 37:109926. doi: 10.1016/j.celrep.2021.109926
30
Das S Halder P Banerjee S Mukhopadhyay AK Dutta S Koley H . Establishment of an intragastric surgical model using C57bl/6 mice to study the vaccine efficacy of omv-based immunogens against helicobacter pylori. Biol Open. (2024) 11:bio.060282. doi: 10.1242/bio.060282
31
Melo J Cavadas B Pereira L Figueiredo C Leite M . Transcriptomic remodeling of gastric cells by helicobacter pylori outer membrane vesicles. Helicobacter. (2024) 29:E13031. doi: 10.1111/hel.13031
32
Ahmed AAQ Besio R Xiao L Forlino A . Outer membrane vesicles (Omvs) as biomedical tools and their relevance as immune-Modulating agents against H. Pylori infections: current status and future prospects. Int J Mol Sci. (2023) 24:8542. doi: 10.3390/ijms24108542
33
Qiang L Hu J Tian M Li Y Ren C Deng Y et al . Extracellular vesicles from helicobacter pylori-infected cells and helicobacter pylori outer membrane vesicles in atherosclerosis. Helicobacter. (2022) 27:E12877. doi: 10.1111/hel.12877
34
Magaña G Harvey C Taggart CC Rodgers AM . Bacterial outer membrane vesicles: role in pathogenesis and host-cell interactions. Antibiotics (Basel). (2023) 13:32. doi: 10.3390/antibiotics13010032
35
Chmiela M Walczak N Rudnicka K . Helicobacter pylori outer membrane vesicles involvement in the infection development and helicobacter pylori-related diseases. J BioMed Sci. (2018) 25:78. doi: 10.1186/s12929-018-0480-y
36
Kyburz A Fallegger A Zhang X Altobelli A Artola-Boran M Borbet T et al . Transmaternal helicobacter pylori exposure reduces allergic airway inflammation in offspring through regulatory T cells. J Allergy Clin Immunol. (2019) 143:1496–512.E11. doi: 10.1016/j.jaci.2018.07.046
37
Rossi G Romagnoli S Lauretti L Pancotto L Taccini E Rappuoli R et al . Helicobacter pylori infection negatively influences pregnancy outcome in A mouse model. Helicobacter. (2004) 9:152–7. doi: 10.1111/j.1083-4389.2004.00212.x
38
Wu Z Zheng Y Sheng J Han Y Yang Y Pan H et al . Cd3(+)Cd4(-)Cd8(-) (Double-negative) T cells in inflammation, immune disorders and cancer. Front Immunol. (2022) 13:816005. doi: 10.3389/fimmu.2022.816005
39
Zaghouani H Hoeman CM Adkins B . Neonatal immunity: faulty T-helpers and the shortcomings of dendritic cells. Trends Immunol. (2009) 30:585–91. doi: 10.1016/j.it.2009.09.002
40
Pichichero ME . Challenges in vaccination of neonates, infants and young children. Vaccine. (2014) 32:3886–94. doi: 10.1016/j.vaccine.2014.05.008
41
Kosanovich JL Eichinger KM Lipp MA Gidwani SV Brahmbhatt D Yondola MA et al . Exacerbated lung inflammation following secondary rsv exposure is cd4+ T cell-dependent and is not mitigated in infant balb/C mice born to pref-vaccinated dams. Front Immunol. (2023) 14:1206026. doi: 10.3389/fimmu.2023.1206026
42
Lacorcia M Bhattacharjee S Laubhahn K Alhamdan F Ram M Muschaweckh A et al . Fetomaternal immune cross talk modifies T-cell priming through sustained changes to dc function F. J Allergy Clin Immunol. (2021) 148:843–57.E6. doi: 10.1016/j.jaci.2021.02.031
43
Zemlin M Härtel C . Neonatal immunology. Int J Mol Sci. (2024) 25:9395. doi: 10.3390/ijms25179395
44
Van Wijck Y De Kleijn S John-Schuster G Mertens TCJ Hiemstra PS Müller A et al . Therapeutic application of an extract of helicobacter pylori ameliorates the development of allergic airway disease. J Immunol. (2018) 200:1570–9. doi: 10.4049/jimmunol.1700987
45
Morris GP Hedrick SM . A rheostat tuning thymic selection. Nat Immunol. (2017) 18:713–4. doi: 10.1038/ni.3778
46
Behrouzi A Vaziri F Riazi Rad F Amanzadeh A Fateh A Moshiri A et al . Comparative study of pathogenic and non-pathogenic escherichia coli outer membrane vesicles and prediction of host-interactions with tlr signaling pathways. BMC Res Notes. (2018) 11:539. doi: 10.1186/s13104-018-3648-3
47
Khan MS Khan IM Ahmad SU Rahman I Khan MZ Khan MSZ et al . Immunoinformatics design of B and T-cell epitope-based sars-cov-2 peptide vaccination. Front Immunol. (2022) 13:1001430. doi: 10.3389/fimmu.2022.1001430
48
Jin M Huo D Sun J Hu J Liu S Zhan M et al . Enhancing immune responses of esc-based taa cancer vaccines with A novel omv delivery system. J Nanobiotechnology. (2024) 22:15. doi: 10.1186/s12951-023-02273-8
49
Van De Waterbeemd B Streefland M van der Ley P Zomer B van Dijken H et al . Improved omv vaccine against neisseria meningitidis using genetically engineered strains and A detergent-free purification process. Vaccine. (2010) 28:4810–6. doi: 10.1016/j.vaccine.2010.04.082
50
Li Y Ma X Yue Y Zhang K Cheng K Feng Q et al . Rapid surface display of mrna antigens by bacteria-derived outer membrane vesicles for A personalized tumor vaccine. Adv Mater. (2022) 34:E2109984. doi: 10.1002/adma.202109984
Summary
Keywords
Helicobacter pylori (H.pylori), outer membrane vesicles (OMVs), pregnancy, T cells, central tolerance
Citation
Wei Y, Zhou L, Zhao X, Qiu H, Hu D and Shi Z (2025) Helicobacter pylori outer membrane vesicles mediate central tolerance in C57BL/6J mice offspring T cells via maternal-fetal transmission. Front. Immunol. 16:1522842. doi: 10.3389/fimmu.2025.1522842
Received
05 November 2024
Accepted
26 March 2025
Published
15 April 2025
Volume
16 - 2025
Edited by
Yang Zhang, Brigham and Women’s Hospital and Harvard Medical School, United States
Reviewed by
Jing Luo, Brigham and Women’s Hospital and Harvard Medical School, United States
He Pan, University of Electronic Science and Technology of China, China
Updates
Copyright
© 2025 Wei, Zhou, Zhao, Qiu, Hu and Shi.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongli Shi, 16301050@hebmu.edu.cn; Dailun Hu, 17300574@hebmu.edu.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.