- 1Cardiovascular Pathology, Department of Cardiac, Thoracic and Vascular Sciences and Public Health, University of Padova, Padova, Italy
- 2Department of Surgery, Oncology and Gastroenterology, Oncology Section, University of Padova, Padova, Italy
- 3Immunology and Molecular Oncology Diagnostics, Veneto Institute of Oncology (IOV) - Istituto di Ricovero e Cura a Carattere Scientifico, Padova, Italy
- 4Unit of Biostatistics, Epidemiology and Public Health, Department of Cardiac, Thoracic, Vascular Sciences, and Public Health, University of Padova, Padova, Italy
- 5Division of Cardiac Surgery, Department of Cardiac, Thoracic and Vascular Sciences and Public Health, University of Padova, Padova, Italy
- 6Cardiology Unit, Department of Cardiac, Thoracic, Vascular Sciences and Public Health, University of Padova, Padova, Italy
- 7Department of Molecular Medicine, University of Padova, Padova, Italy
Background: Cytomegalovirus (CMV) infection is a relevant threat to heart-transplanted patients during the first year after surgery, leading to increased morbidity and, in some cases, mortality. This proof-of-concept study aims to assess the transcriptomic profile of CMV infection in cardiac transplanted patients as a new diagnostic approach to discriminate infection and Acute Cellular Rejection (ACR) on EMB specimens.
Methods: We performed a microarray-based messenger RNA (mRNA) and micro-RNA (miRNA) profiling. We analyzed three patient groups in the setting of CMV viremia and inflammatory infiltrate: a control group (n=5), an ACR group (n=5), and an infection group (n=6). Differentially expressed mRNA and miRNA were further investigated through bioinformatic pathway analysis.
Results: Focusing on infection vs rejection comparison, we investigated the role of the 18 differentially expressed mRNAs and the 12 miRNAs with the most significative p-value (gene level fold change, FC <-2 or >2, p-value <0.05). Based on the bioinformatic analysis, we explored the regulatory effects of these miRNAs on the mRNA pathways independently identified in the same samples. The results showed that two genes, IL7R and GZMK (-38.63 and -3.15 FC, respectively), and two miRNAs, mir-93-5p and mir-345-5p (-2.63 and -2.18 FC, respectively), are differentially expressed in infection and can be exploited to differentiate CMV-positive from ACR-positive EMB specimens, reaching an AUC of 0.87 and an accuracy of 91% at cross-validation.
Conclusions: We have identified a distinctive combined molecular profile of mRNAs and miRNAs for infection in post-cardiac transplant follow-up. Based on IL7R, GZMK, mir-93-5p, and mir-345-5p we suggest a novel possible workflow to distinguish infection, where those markers are downregulated, from rejection, where they are overexpressed, on EMB specimens. This analysis showed good accuracy and promising predictive performance. The future combined analysis of these genes and these miRNAs through user-friendly techniques, such as quantitative PCR, could reduce turn-around time and improve our diagnostic power for distinguishing CMV infection from ACR in EMB specimens.
1 Introduction
Cytomegalovirus (CMV) is the most clinically relevant post-transplant infectious agent. CMV is a member of the beta-Herpesviridae family that, in the normal population, latently infects 50-90% of individuals, but normally takes an asymptomatic course (1). Differently, in cardiac transplanted patients, CMV infection impacts morbidity and mortality (2, 3), and some studies showed that CMV infection is a risk factor for Antibody Mediated Rejection (AMR) and Cardiac Allograft Vasculopathy (CAV) (4–7).
The cardiac transplanted patients can be stratified according to the donor/recipient (D/R) serological status. The highest risk is associated with the mismatch between donor-positive and recipient-negative (D+/R-). Based on the approach chosen by each transplant center, these patients may undergo pre-emptive therapy (based on the antiviral administration for early asymptomatic CMV viremia detected by surveillance testing) or antiviral prophylaxis (2, 8, 9).
Routine EMB tissue exhamination represents a crucial procedure to determine CMV replication on the graft and thus intervene with a proper antiviral therapy and taper the immunosuppression regimen accordingly. However, the discrimination between Acute Cellular Rejection (ACR) and the inflammation process triggered by CMV infection itself is quite cryptic. In case of suspected CMV infection, both nucleic acid and immunohistochemical tests should be performed on EMB specimens, e.g. Immunohistochemical staining on Formalin Fixed Paraffine Embedded (FFPE) EMB slices and Polymerase Chain Reaction with the nucleic acid extracted from EMB tissue. Those tests are designed to detect viral components, such as the CMV genome or proteins, in tissue samples. Still, these tests are prone to false results due to CMV activation’s patchy nature in tissue, and the EMB procedure sampling errors.
Seeking novel biomarkers associated with rejection monitoring has been investigated over years in tissue and liquid biopsy (10–13). Transcriptomic studies opened new horizons on cardiac rejection allograft monitoring, showing that Rejection Associated Transcript analyses are similar through solid organ transplants and can differentiate among Cellular Rejection, Antibody-Mediated Rejection, and no-rejecting EMBs (11, 14, 15). Unfortunately, most of these studies focused mainly on inflammation but not on the infection. This limitation urges the need for new approaches that could help the pathologist in characterizing the allograft’s status, differentiating the inflammation itself from the virus induced inflammation pattern.
In this proof-of-concept study, we investigate the transcriptomic profile of CMV-positive patients at mRNA and miRNA levels to determine new candidate biomarkers to distinguish inflammatory infiltrate caused by infection from that due to rejection on FFPE EMB specimens. For this purpose, we analyzed the mRNA and miRNA profile of FFPE EMB through high-density Clariom S Affymetrix GeneChip arrays (Thermofisher Scientific, USA) platform. We demonstrated that combining different biomarkers can significantly improve CMV infection detection and discriminate it from ACR during EMB pathological assessment.
2 Materials and methods
All the details about the Study design, patient selection, EMBs histological evaluation, RNA extraction protocol, Microarray-based mRNA and miRNA analysis protocols, bioinformatics, and statistical analysis are provided in the Extended Methods in Supplementary Materials.
3 Results
3.1 Study population characteristics
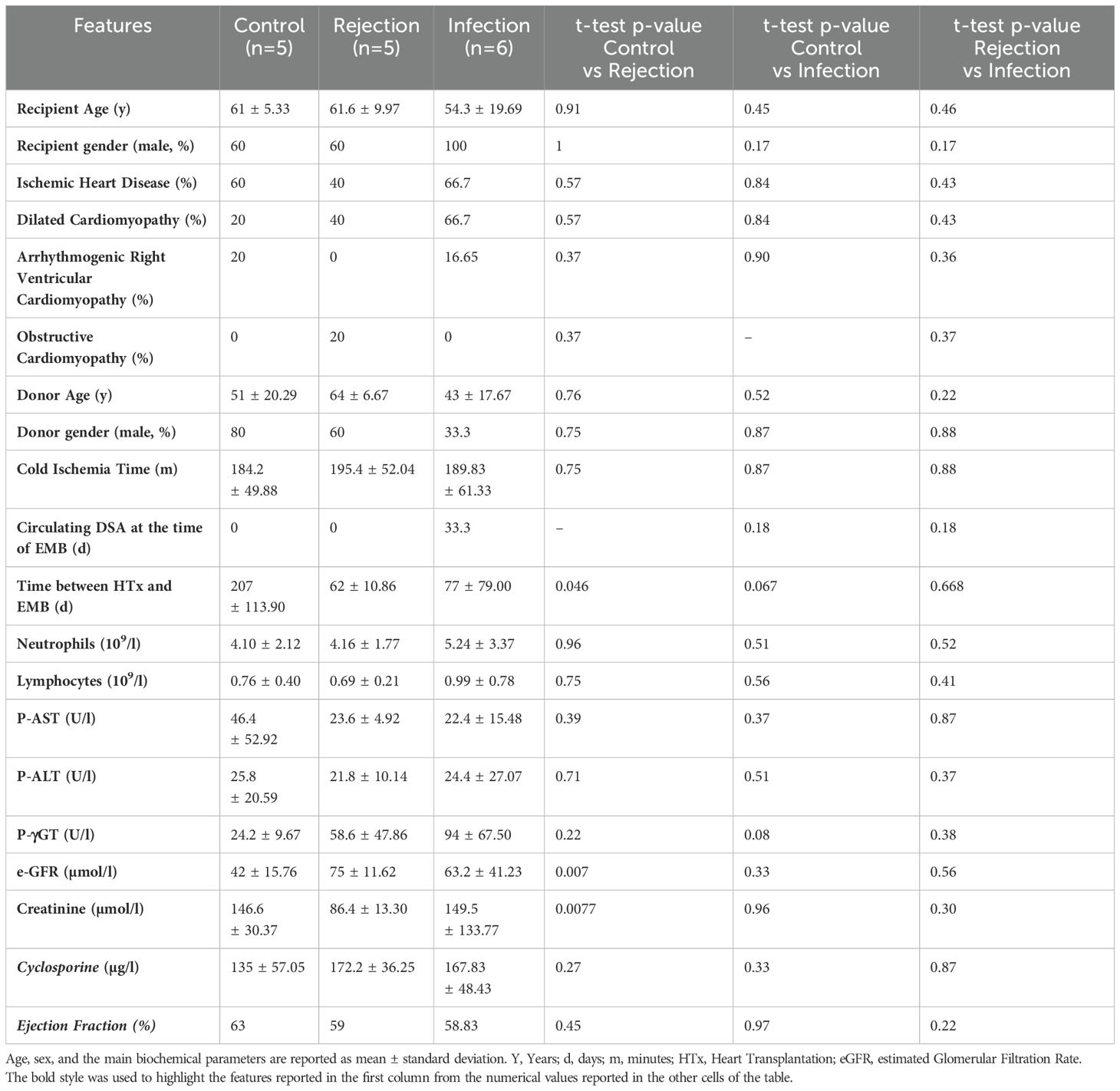
The patients enrolled in this study were divided into three groups according to the ACR grade and the CMV viremia. Recipients’ and Donors’ characteristics are shown in Table 1. Among selected patients, 68.75% were male, and the mean age was 61. Ischemic Heart Disease was the most common diagnosis leading to transplant. These patients were homogenous for Circulating DSA at the time of EMB, lymphocyte count, hepatic functionality, and Ejection Fraction. The control group showed slightly worse renal functionality and a higher time interval between heart transplant and EMB.
3.2 Microarray transcriptomic analysis of FFPE EMBs
We analyzed the transcriptomic profile of our samples through a microarray-based platform. The bioinformatic analysis compared the expression levels of detected mRNAs in the Controls, the Rejection group, and the Infection group.
Notably, 293 genes were differentially expressed between Control and Infection groups (82 up-regulated and 211 down-regulated), 407 genes between Control and Rejection groups (126 up-regulated and 281 down-regulated), and 18 genes between Infection and Rejection groups (10 up-regulated and 8 down-regulated) (p-value <0.05).
Since we aimed to define a possible molecular signature to distinguish infection from rejection at the tissue level, we focused our attention on the differentially expressed genes (DEG) that emerged in the comparison between these two study groups. In particular, PHLDB2, HP1BP3, HMGCS2, UBC, LDB3, ALPK2, MYH7B, ACADVL, HSPB6, and TBX20 resulted to be overexpressed in Infection compared to Rejection, while PDCL2, OCRL, TAOK3, PLAC8, GZMK, MNDA, IL7R, P2RY14 were downregulated in the same comparison (p<0.05) (Figure 1A). Afterward, we investigated the role of all these DEGs. We consulted several databases, such as the GeneCards database and WikiPathways TAC tool, focusing on their cellular functions and the biological processes they are involved in. This analysis revealed that these DEGs are mainly involved in cardiomyocyte metabolism (e.g. HMGCS2, ACADVL), IL-7 signaling and regulatory circuits of the STAT3 signaling (e.g. IL7R), DNA replication (e.g. HP1BP3) and cardiac progenitor differentiation pathways (e.g. PHLDB2, LDB3, ALPK2, TBX20) (Supplementary Table S1).
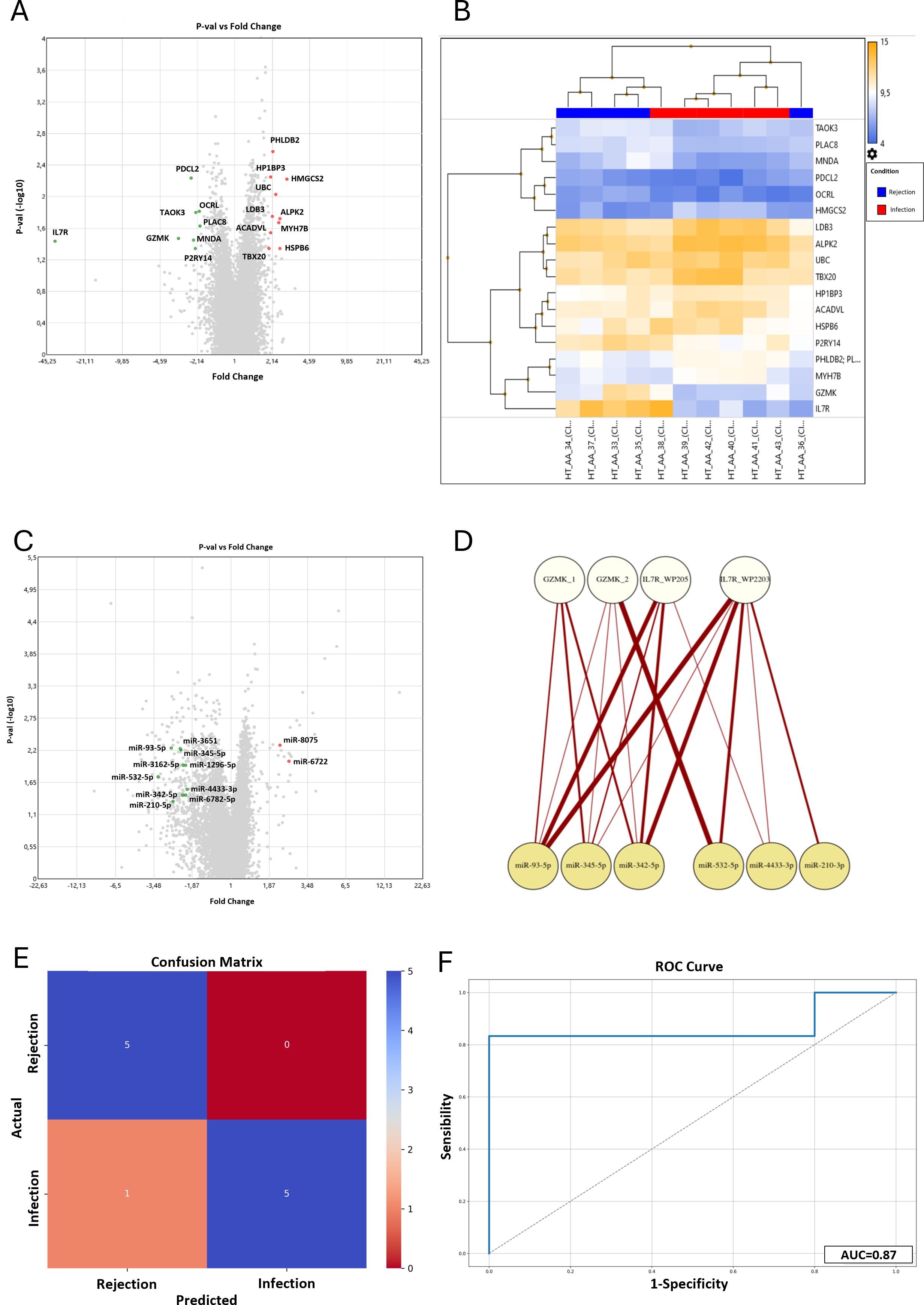
Figure 1. Results of the microarray mRNA analysis in the Infection vs Rejection group. (A) The Volcano Plot reported the differentially expressed genes between the Infection and Rejection groups. On the horizontal axis is reported the fold-change value (fold-change <-2 and >2), while the vertical axis reported the p-values (<0.05) of the same genes. (B) The Hierarchical Clustering reported the fluorescence level of each DEG (higher level in orange and lower level in blue). In the upper part of the graph is reported the clustering of the patients in the two study groups, Rejection in blue and Infection in red. (C) Results of the microarray miRNAs analysis in the Infection vs Rejection group. The Volcano Plot reported the differentially expressed miRNAs between the Infection and Rejection groups. On the horizontal axis is reported the fold-change value (fold-change <-2 and >2) while the vertical axis reports the p-values (<0.05) of the same miRNAs. (D) The bipartite graph between GZMK and IL7R pathways and miRNAs target genes, obtained by igraph library in R. The upper white nodes layer represents the Cytoscape pathways selected: GZMK glucose metabolic (GZMK_1), GZMK type I immune response (GZMK_2), IL7R WP205 - IL-7 signaling pathway (IL7R_WP205), and L7R WP2203 - Thymic stromal lymphopoietin signaling pathway (IL7R_WP2203). The lower yellow node layer represents the infection-related miRNAs. (E) Confusion Matrix. The panel shows the performance of the Support Vector Machine model (SVM) to classify infection and rejection at cross-validation. The y-axis represents the actual enrolled group (5 samples for rejection, and 6 samples for the infection group). The x-axis represents the samples classified by the SVM. (F) ROC curve analysis: Area Under the Curve (AUC) at cross-validation for infection and rejection discrimination is reported.
As shown in the Volcano Plot (Figure 1A), all the significantly differently expressed genes are quite homogeneous in terms of fold-change values, except for GZMK and IL7R. In detail, IL7R resulted downregulated in the Infection group compared to the Rejection (-38.63 fold-change; 12.69 (log2) Avg in the Rejection group vs 7.41 (log2) Avg in the Infection group). IL7R is highly expressed in conventional mature T-cells, except for regulatory T-cells that show a low level of IL7R on their surface (16, 17). Once activated, IL7R induces proliferative and anti-apoptotic signals mainly by activating several cellular pathways, such as JAK/STAT, PI3K/Akt, and MAPK/ERK pathways (18, 19). In addition, IL7R has been reported to activate chronic inflammation (20), autoimmunity (21), and allograft rejection in rodent models (22, 23). Likewise, GZMK is downregulated in the infection group (-3.15 fold-change; 9.27 (log2) Avg in Rejection group vs 7.62 (log2) Avg in Infection group). GZMK is a member of serine proteases that preferentially cleave after basic residues (24, 25), and it is expressed in different immune cell types, such as NK cells, cytotoxic T cells (26), and macrophages (27). GZMK is also reported to be involved in several pathologic conditions, such as cardiovascular diseases (28), vasculitis (29), and endothelial activation, promoting the release of proinflammatory cytokines from endothelial cells (30).
The heatmap reported in Figure 1B confirms that the DEG values are homogenous within each group, in terms of the expression level. Moreover, the hierarchical clustering correctly classifies nine out of eleven samples (82%), in agreement with histology and CMV serology. Only two samples were assigned to the wrong group: sample #36, selected as Infection but classified as Rejection by hierarchical clustering, and sample #38, selected as Rejection but classified as Infection by hierarchical clustering. In detail, the misclassification of these samples is linked to the values of GZMK and IL7R genes. In our cohort, the expression levels of these two genes were generally high in the Rejection group and low in the Infection group. Sample #36 showed a low level of expression of IL7R and GZMK genes, and the system classified it as “Infection” despite it being negative for the CMV serology test. On the other hand, sample #38 showed a high level of expression for IL7R and GZMK genes, and the system classified it as “Rejection” even though the CMV serology positivity. This misclassification suggests that those genes are crucial in differentiating Infections, with low expression, from Rejections, with high expression.
3.3 Microarray miRNAs analysis of FFPE EMB
The same total RNA extracted and analyzed for mRNA profiling was evaluated for miRNA profiling. The Clariom-S miRNA 4.0 chip tested the expression level of 2578 different miRNAs for each patient, and we performed the same comparisons conducted for the mRNA analysis (gene level fold-change <-2 or >2, p-value<0.05). The statistical evaluation of the raw data revealed that 513 miRNAs were differentially expressed between the Control and Rejection groups, with 477 miRNAs upregulated and 36 downregulated, while 111 (62 upregulated and 49 downregulated) between the Control and Infection groups. The comparison between the Infection and the Rejection groups showed that 386 miRNAs were differentially expressed (32 upregulated and 354 downregulated). Among these 386 miRNAs, 29 emerged differentially expressed only in the latter comparison. We further filtered out those results, focusing on the miRNA with the highest p-value. Hence we selected twelve hsa-miRNAs that showed the highest p-value in infection versus vs rejection comparison: miR-8075 (fold-change= 2.23; p-value= 0.0052), miR-93-5p (fold-change= -2.63; p-value= 0.0058), miR-3651 (fold-change= -2.26; p-value= 0.0059), miR-345-5p (fold-change= -2.25; p-value= 0.0061), miR-6722 (fold-change= 2.57; p-value= 0.0098), miR-1296-5p (fold-change= -2.08; p-value= 0.0112), miR-3162-5p (fold-change= -2.15; p-value= 0.0114); miR-532-5p (fold-change= -3.26; p-value= 0.0177), miR-4433-3p (fold-change= -2.02; p-value= 0.0293), miR-6782-5p (fold-change= -2.08; p-value= 0.0365), miR-342-5p (fold-change= -2.18; p-value=0.0371), and miR-210-3p (fold-change= -2.54; p-value= 0.0469) (Figure 1C).
Once determined our miRNAs of interest, we explored their function through a literature review. The screening of PubTator, GeneCards, and PubMed databases revealed that these miRNAs are mainly involved in PI3K/Akt, STAT, and HIF1α pathways, regulating cell proliferation and apoptosis, along with mitochondria stress response.
3.4 Analysis of mRNA and miRNA interactions in CMV infection
Once we identified the mRNAs and miRNAs differentially expressed in the Infection vs Rejection groups, we further investigated in silico the possible interactions among mRNAs and the target genes of miRNAs.
We consulted TAC and Microcosm databases to define the target genes of the selected miRNAs. Through a symmetric matrix (Supplementary Figure S2), we highlighted the number of shared genes among each couple of entries. In detail, it showed that only miR-93-5p targets some of the mRNAs identified as DEG in the Infection group.
Since we focused on the miRNAs shared target genes, we saw that the most relevant miRNAs were miR-93-5p, miR-345-5p, miR-532-5p, miR-342-5p, and miR-210-3p. Indeed, these miRNAs shared a higher number of target genes: miR-93-5p shared 101 target genes with miR-345-5p, 100 target genes with miR-532-5p, 78 target genes with miR-342-5p, and 66 target genes with miR-210-3p (Supplementary Figure S2A). Next, we represented these connections with an undirected weighted graph: the thickness of the edges is proportional to the number of targets shared between two miRNAs (Supplementary Figure S2B). Once again, the miRNAs that were more relevant were miR-93-5p, miR-345-5p, miR-532-5p, miR-342-5p, and miR-210-3p.
Interestingly, miR-93-5p targets some mRNAs identified in infection vs rejection: ACADVL, ALPK2, OCRL, PDCL2, and UBC. Using the EnrichR tool, we saw that these genes are mainly involved in fatty acid metabolism pathways (Supplementary Figures S2C, D).
3.5 The interplay among miRNA targets and GZMK and IL7R pathways
The analysis of mRNAs revealed that GZMK and IL7R are crucial for distinguishing infection from rejection. Thus, we investigated the interplay between the gene pathways and our selected miRNAs. Consulting the Cytoscape database, we chose the two most significant pathways for GZMK and IL7R based on the p-value associated. The genes reported to be involved in these pathways were matched with the miRNAs’ target genes to identify the shared ones, and the results are shown in (See Supplementary Materials, Supplementary Table S2). Notably, only seven miRNAs were shown to target some genes involved in GZMK and IL7R pathways, and their interplay is graphically represented by the bipartite graph reported in Figure 1D. Once again, miR-93-5p shared more target genes with the pathways analyzed, especially with IL7R pathways, such as MAPK1, JAK1, MYC, PIK3R2, STAT3, and CRLF2. MiR-345-5p and miR-342-5p target several genes in GZMK and IL7R pathways, such as ADHFE1, PTK2B, IRF1, PIK3R2, CRLF2, MAP2K2, MAPK3, LCK.
Thus, miR-93-5p and miR-345-5p showed the strongest connections with IL7R and GZMK pathways. Both of them were up-regulated in the Rejection group: mir-93-5p showed 5.98 Avg (log2) in the Rejection group vs 4.59 Avg (log2) in the Infection group; mir-342-5p showed 2.18 Avg (log2) in the Rejection group vs 1.6 Avg (log2) in the Infection group. Li et al. demonstrated that the miR-93 family takes part in PTEN regulation (31). PTEN is a tumor suppressor mutated in many cancers that antagonizes the PI3K/Akt pathway, modulating cell cycle progression and cell survival (32, 33). Furthermore, several studies demonstrated that this miRNA plays an important role in enhancing endothelial activities (34) and regulating integrin-β8 expression (35). Additionally, miR-345-5p showed several shared target genes with other miRNAs. This miRNA is reported to play a role in the regulation of HIF1α, and consequently, modulation of TGFβ/Smad2/Smad3 signaling (36), linked to ischemic damage response. Liu and colleagues reported that miR-345-5p can downregulate the TLR4/NF-κB pathway, altering inflammatory response and apoptosis (37).
Taken together, all these results showed that miR-93-5p and miR-345-5p are the best-ranked candidates to distinguish infection from rejection since they are down-regulated in CMV-positive patients and up-regulated in the Rejection group.
3.6 Performance assessment of the combined molecular markers
We are aware that our study population is small, and this leads to a reduced statistical power. This was confirmed by the Post Hoc Power analysis, which revealed that our cohort is associated with a value of (1-β) of 0.19, and, consequently, with a high risk of false negative results.
However, once defined our new panel of combined markers, we assessed the accuracy of this novel approach to distinguish infection from rejection. We implemented a classifier based on the identified markers using a Support Vector Machine with a polynomial kernel (KSVM). We then performed stratified cross-validation to evaluate its performance. As shown in the Confusion Matrix (Figure 1E), based on the level of expression of GZMK, IL7R, mir-93-5p, and mir-345-5p, the model correctly classified as “rejection” all the real rejection samples. On the other hand, the model correctly classified five out of six infection samples (83,3%), with only one misclassification. This performance is associated with an Infection Precision of 0.83, with an overall accuracy of 91% (F1 score of 0.91). Subsequently, the sensitivity and specificity analysis revealed that the combination of GZMK, IL7R, mir-93-5p, and mir-345-5p reached good results, with an Area Under the Curve (AUC) of 87% (Figure 1F).
In summary, these analyses showed that, although the power analysis highlighted a limited statistical power due to the small sample size, the model based on the combined marker panel exhibited a promising predictive performance.
4 Discussion
Heart-transplanted patients represent a fragile population. After surgery, they need to be strictly monitored to contrast possible rejection episodes. The primary defense against rejection is immunosuppressive therapy, which patients must take for their entire lives. Unfortunately, these treatments make them more susceptible to bacterial and viral infections. Cytomegalovirus is the most clinically relevant infection in cardiac transplanted patients during the first year of follow-up (1). CMV de novo infection or reactivation needs to be treated properly (2, 8). Thus, correct diagnosis is crucial to define the appropriate therapeutic protocol. With this study, we aimed to explore the role of mRNA and miRNA as new biomarkers to improve EMB interpretation and distinguish rejection from infection-related inflammatory infiltrate on graft tissue.
Current molecular tests to assess CMV infection on EMB specimens need improvement. Our study suggests that IL7R and GZMK are decisive in the discrimination between an activate inflammatory status, due to rejection and infection, and control stable patients. These genes were downregulated in the Infection group (-38.63 Fold-change for IL7R and -3.15 Fold-change for GZMK), compared to the Rejection group, suggesting an alteration in the regulatory mechanisms of their pathways. Many groups showed that IL7R (also called CD127) is downregulated in viral infection characterized by a latent phase (38–41). Interestingly, immune activation and CD4 lymphopenia have been associated with decreased IL7R expression (41). These results perfectly fit with the conditions observed in CMV-positive transplanted patients, who are exposed to a viral-induced stimulation in an impaired immunocompetence condition. Conversely, only a few and contrasting studies have been published about GZMK expression in viral infection (42). Of relevance, Verschoor and colleagues showed that GZMK expression decreased in some specific subsets of T cells in CMV-positive patients, and in older patients, who are known to be characterized by a fragile and altered immune response (43). Focusing on the connection between the pathways of GZMK and IL7R and the miRNA dysregulated in our comparison, we identified two key miRNAs that are up-regulated in the Rejection group: miR-93-5p (5.98 Avg (log2) in Rejection vs 4.59 Avg (log2) in Infection) and miR-345-5p (2.18 Avg (log2) in Rejection vs 1.6 Avg (log2) in Infection). These elements represent an important starting point for investigating new CMV diagnosis biomarkers. We could combine gene expression analysis (IL7R and GZMK) to assess the inflammatory status of the graft, and on the other hand, the assessment of miR-93-5p and miR-345-5p could be helpful to discriminate infection, where their levels of expression are decreased, from rejection. In clinical practice, the possible application of this novel and promising approach could be divided into two phases. First of all, thanks to IL7R and GZMK testing, we could perform a screening process and discriminate between stable and unstable patients, who show IL7R and GZMK increased expression levels compared to non-rejecting patients. Secondly, through mir-93-5p and mir-345-5p analysis, we could improve our capacity to distinguish infection, where those miRNAs show a lower level of expression, from rejection, where they are overexpressed. In this ideal new workflow, combining different biomarkers should be decisive in discriminating between infection and rejection. Indeed, inflammatory response is a dynamic process, and using a double level of investigation could improve our ability to identify infection-associated inflammatory infiltrate on tissue samples.
Over the past 30 years, multiple solid molecular and epidemiological evidences have connected CMV infection and the exacerbation of acute and chronic allograft rejection in solid organ transplants, especially heart transplants (4, 7, 44–47). The association between CMV and CAV, CAD, and vascular dysfunction has been deeply investigated at the molecular level, demonstrating the primary role of CMV in inducing focal inflammation, triggering atherosclerosis, and microvascular damage. These results were also confirmed in human hand transplantation and facial vascularized composite allotransplantation (48–50).
Additionally, this novel paradigm could be effective in improving CMV diagnosis and overcoming the limitations of molecular tests currently in use. Indeed, tests based on the detection of the viral genome are prone to false negative results, due to the focal reactivation of CMV and to sampling error of EMB. The application of PCR or immunohistochemistry analysis to detect the viral genomes or proteins may yield false-negative results when the EMBs fail to sample regions of focal CMV infection. Conversely, PCR may also produce false-positive findings by detecting viral particles present on tissue samples as a result of blood contamination rather than true myocardial viral localization. The analysis of genes that test the immune response to the virus could overcome these limits. Finally, thanks to this proof-of-concept study, we can optimistically foresee a wide clinical application of our promising results with cost-effective and user-friendly techniques, such as quantitative PCR (qPCR). Indeed, we identified four molecular candidate markers that could be easily analyzed on EMB specimen extract in all clinical laboratories, with no need for new platforms and minimal equipment costs.
Our study has some limitations. Unfortunately, our enrolled population is quite small, and this is due to the strict inclusion criteria applied in the selection phase. We aimed to evaluate the transcriptomic profile of the EMB in a setting of CMV infection without any other viral or bacterial coinfections, significantly reducing the number of eligible patients. Despite the modest population size, we obtained a huge amount of data thanks to the high-throughput transcriptomic platform employed, without any bias in transcript selection. Our cross-validation results demonstrated good performance, supporting the robustness of our findings despite the relatively low expected values. To address the current limitations, further validation is warranted in an independent cohort, ideally comprising a larger and more heterogeneous patient population. This will enhance the generalizability of our preliminary results to real-world clinical practice and help mitigate the risk of false negatives.
In conclusion, we explored the possibility of identifying new biomarker candidates in post-transplantation infection diagnosis. Our study is a proof-of-concept that infection and rejection are characterized by different transcriptomic profiles, both at mRNA and miRNA levels, shedding new light on the understanding of the pathophysiologic mechanisms that underlie these insidious conditions. Combining CMV-related gene analysis with rejection-related miRNA assessment could further improve our “resolution power” in distinguishing CMV infection from ACR, which can show an overlapping phenotype on histology. These novel and interesting results open up new possible innovative, and promising approaches for CMV infection diagnosis with the use of cost-effective qPCR applications, reduced turnaround time, and large-scale clinical applicability of our results.
Data availability statement
The microarray data have been deposited on Zenodo and they are available at this doi: 10.5281/zenodo.15295266. See the link: https://doi.org/10.5281/zenodo.15295266.
Ethics statement
The studies involving humans were approved by Ethics Committee of Azienda Ospedaliera di Padova. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
IB: Data curation, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. DP: Data curation, Investigation, Software, Visualization, Writing – review & editing. CC: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing. AG: Methodology, Visualization, Writing – review & editing. ER: Methodology, Writing – review & editing. LV: Data curation, Software, Writing – review & editing. SAM: Methodology, Writing – review & editing. CT: Resources, Writing – review & editing. NP: Resources, Writing – review & editing. GT: Resources, Writing – review & editing. FT: Resources, Writing – review & editing. CB: Resources, Writing – review & editing. GG: Resources, Writing – review & editing. SM: Methodology, Resources, Writing – review & editing. DA: Data curation, Supervision, Validation, Writing – review & editing. DG: Data curation, Supervision, Writing – review & editing. AA: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. MF: Conceptualization, Funding acquisition, Investigation, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by University of Padova grants number BIRD191573 and BIRD170215.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1581151/full#supplementary-material
References
1. Potena L, Solidoro P, Patrucco F, and Borgese L. Treatment and prevention of cytomegalovirus infection in heart and lung transplantation: an update. Expert Opin Pharmacother. (2016) 17:1611–22. doi: 10.1080/14656566.2016.1199684
2. Kotton CN, Kumar D, Caliendo AM, Huprikar S, Chou S, Danziger-Isakov L, et al. The third international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation. (2018) 102:900–31. doi: 10.1097/TP.0000000000002191
3. Mendez-Eirin E, Paniagua-Martín MJ, Marzoa-Rivas R, Barge-Caballero E, Grille-Cancela Z, Cañizares A, et al. Cumulative incidence of cytomegalovirus infection and disease after heart transplantation in the last decade: effect of preemptive therapy. Transplant Proc. (2012) 44:2660–2. doi: 10.1016/j.transproceed.2012.09.035
4. Saldan A, Mengoli C, Sgarabotto D, Fedrigo M, Angelini A, Feltrin G, et al. Human cytomegalovirus and Epstein–Barr virus infections occurring early after transplantation are risk factors for antibody-mediated rejection in heart transplant recipients. Front Immunol. (2023) 14:1171197. doi: 10.3389/fimmu.2023.1171197
5. Valantine HA. The role of viruses in cardiac allograft vasculopathy. Am J Transpl. (2003) 4:169–77. doi: 10.1046/j.1600-6143.2003.00316.x
6. Delgado JF, Manito N, Almenar L, Crespo-Leiro M, Roig E, Segovia J, et al. Risk factors associated with cytomegalovirus infection in heart transplant patients: a prospective, epidemiological study. Transpl Infect Dis. (2011) 13:136–44. doi: 10.1111/j.1399-3062.2010.00573.x
7. Potena L and Valantine HA. Cytomegalovirus-associated allograft rejection in heart transplant patients. Curr Opin Infect Dis. (2007) 20:425–31. doi: 10.1097/QCO.0b013e328259c33b
8. Razonable RR and Humar A. Cytomegalovirus in solid organ transplant recipients—Guidelines of the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transpl. (2019) 33:1–23. doi: 10.1111/ctr.13512
9. Kalil AC, Levitsky J, Lyden E, Stoner J, and Freifeld AG. Meta-analysis: the efficacy of strategies to prevent organ disease by cytomegalovirus in solid organ transplant recipients. Ann Intern Med. (2005) 143:870. doi: 10.7326/0003-4819-143-12-200512200-00005
10. Crespo-Leiro MG, Stypmann J, Schulz U, Zuckermann A, Mohacsi P, Bara C, et al. Clinical usefulness of gene-expression profile to rule out acute rejection after heart transplantation: CARGO II. Eur Heart J. (2016) 37:2591–601. doi: 10.1093/eurheartj/ehv682
11. Halloran PF and Madill-Thomsen KS. The molecular microscope diagnostic system: assessment of rejection and injury in heart transplant biopsies. Transplantation. (2023) 107:27–44. doi: 10.1097/TP.0000000000004323
12. Agbor-Enoh S, Shah P, Tunc I, Hsu S, Russell S, Feller E, et al. Cell-free DNA to detect heart allograft acute rejection. Circulation. (2021) 143:1184–97. doi: 10.1161/CIRCULATIONAHA.120.049098
13. Goldberg JF, Truby LK, Agbor-Enoh S, Jackson AM, deFilippi CR, Khush KK, et al. Selection and interpretation of molecular diagnostics in heart transplantation. Circulation. (2023) 148:679–94. doi: 10.1161/CIRCULATIONAHA.123.062847
14. Halloran PF, Potena L, Van Huyen JPD, Bruneval P, Leone O, Kim DH, et al. Building a tissue-based molecular diagnostic system in heart transplant rejection: The heart Molecular Microscope Diagnostic (MMDx) System. J Heart Lung Transpl. (2017) 36:1192–200. doi: 10.1016/j.healun.2017.05.029
15. Loupy A, Lefaucheur C, Vernerey D, Chang J, Hidalgo LG, Beuscart T, et al. Molecular microscope strategy to improve risk stratification in early antibody-mediated kidney allograft rejection. J Am Soc Nephrol. (2014) 25:2267–77. doi: 10.1681/ASN.2013111149
16. Seddiki N, Santner-nanan B, Martinson J, Zaunders J, Sasson S, Landay A, et al. Expression of interleukin (IL) -2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med. (2010) 203:1693–700. doi: 10.1084/jem.20060468
17. Michel L, Soulillou JP, Laplaud DA, Michel L, Berthelot L, Pettré S, et al. Patients with relapsing-remitting multiple sclerosis have normal Treg function when cells expressing IL-7 receptor α -chain are excluded from the analysis. J Clin Invest. (2008) 118:3411–9. doi: 10.1172/JCI35365
18. MacKall CL, Fry TJ, and Gress RE. Harnessing the biology of IL-7 for therapeutic application. Nat Rev Immunol. (2011) 11:330–42. doi: 10.1038/nri2970
19. Carrette F and Surh CD. IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin Immunol. (2012) 24:209–17. doi: 10.1016/j.smim.2012.04.010
20. Belarif L, Mary C, Jacquemont L, Mai HL, Danger R, Hervouet J, et al. IL-7 receptor blockade blunts antigen-speci fi c memory T cell responses and chronic in fl ammation in primates. Nat Commun. (2018) 9:4483. doi: 10.1038/s41467-018-06804-y
21. Dooms H. Interleukin-7: Fuel for the autoimmune attack. J Autoimmun. (2013) 45:40–8. doi: 10.1016/j.jaut.2013.06.007
22. Piotti G, Ma J, Adams E, Cobbold S, and Waldmann H. Guiding postablative lymphocyte reconstitution as a route toward transplantation tolerance. Am J Transpl. (2014) 14:1678–89. doi: 10.1111/ajt.12756
23. Mai H, Boeffard F, Longis J, Danger R, and Martinet B. IL-7 receptor blockade following T cell depletion promotes long-term allograft survival. J Clin Invest. (2014) 124:1723–33. doi: 10.1172/JCI66287
24. Bovenschen N, Quadir R, Berg ALVD, Brenkman AB, Vandenberghe I, Devreese B, et al. Granzyme K displays highly restricted substrate specificity that only partially overlaps with granzyme A *. J Biol Chem. (2009) 284:3504–12. doi: 10.1074/jbc.M806716200
25. Plasman K, Demol H, Bird PI, Gevaert K, and Damme PV. Substrate specificities of the granzyme tryptases A and K. J Proteome Res. (2014) 13:6067–77. doi: 10.1021/pr500968d
26. Chowdhury D and Lieberman J. Death by a thousand cuts: granzymes pathways of programmed cell death. Annu Rev Immunol. (2008) 26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404
27. Turner CT, Zeglinski MR, Richardson KC, Zhao H, Shen Y, Papp A, et al. Granzyme K expressed by classically activated macrophages contributes to inflammation and impaired remodeling. J Invest Dermatol. (2019) 139:930–9. doi: 10.1016/j.jid.2018.09.031
28. Zeglinski MR and Granville DJ. Granzymes in cardiovascular injury and disease. Cell Signal. (2020) 76:109804. doi: 10.1016/j.cellsig.2020.109804
29. Poli A, Michel T, Thérésine M, Andrès E, Hentges F, and Zimmer J. CD56 bright natural killer (NK) cells: an important NK cell subset. Immunology. (2009) 126:458–65. doi: 10.1111/j.1365-2567.2008.03027.x
30. Sharma M, Merkulova Y, Raithatha S, Parkinson LG, and Shen Y. Extracellular granzyme K mediates endothelial activation through the cleavage of protease-activated receptor-1. FEBS J. (2016) 283:1734–47. doi: 10.1111/febs.2016.283.issue-9
31. Li N, Miao Y, Shan Y, Liu B, Li Y, Zhao L, et al. Mir-106b and mir-93 regulate cell progression by suppression of pten via pi3k/akt pathway in breast cancer. Cell Death Dis. (2017) 8:e2796–11. doi: 10.1038/cddis.2017.119
32. Myers MP, Pass I, Batty IH, van der Kaay J, Stolarov JP, Hemmings BA, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci U S A. (1998) 95:13513–8. doi: 10.1073/pnas.95.23.13513
33. Tamura M, Gu J, Matsumoto K, Aota S, Parson R, and Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. (1998) 280:1614–7. doi: 10.1126/science.280.5369.1614
34. Fang L, Du WW, Yang W, Rutnam ZJ, Peng C, Li H, et al. MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle. (2012) 11:4352–65. doi: 10.4161/cc.22670
35. Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW, et al. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene. (2011) 30:806–21. doi: 10.1038/onc.2010.465
36. Wang P, Fang Y, Qiu J, Zhou Y, Wang Z, and Jiang C. miR-345–5p curbs hepatic stellate cell activation and liver fibrosis progression by suppressing hypoxia-inducible factor-1alpha expression. Toxicol Lett. (2022) 370:42–52. doi: 10.1016/j.toxlet.2022.09.008
37. Liu J, Jiang Y, Han M, Jiang L, Liang D, Li S, et al. MicroRNA-345-5p acts as an anti-inflammatory regulator in experimental allergic rhinitis via the TLR4/NF-κB pathway. Int Immunopharmacol. (2020) 86. doi: 10.1016/j.intimp.2020.106522
38. Crawley AM and Angel JB. Expression of γ-chain cytokine receptors on CD8+ T cells in HIV infection with a focus on IL-7Rα (CD127). Immunol Cell Biol. (2012) 90:379–87. doi: 10.1038/icb.2011.66
39. Golden-Mason L, Burton JR, Castelblanco N, Klarquist J, Benlloch S, Wang C, et al. Loss of IL-7 receptor alpha-chain (CD127) expression in acute HCV infection associated with viral persistence. Hepatology. (2006) 44:1098–109. doi: 10.1002/hep.21365
40. Boutboul F, Puthier D, Appay V, Pelle O, Ait-Mohand H, Combadière B, et al. Modulation of interleukin-7 receptor expression characterizes differentiation of CD8 T cells specific for HIV, EBV and CMV. AIDS. (2005) 19:1981–6. doi: 10.1097/01.aids.0000191919.24185.46
41. Koesters SA, Alimonti JB, Wachihi C, Matu L, Anzala O, Kimani J, et al. IL-7Rα expression on CD4+ T lymphocytes decreases with HIV disease progression and inversely correlates with immune activation. Eur J Immunol. (2006) 36:336–44. doi: 10.1002/eji.200535111
42. Bouwman AC, Van Daalen KR, Crnko S, Ten Broeke T, and Bovenschen N. Intracellular and extracellular roles of granzyme K. Front Immunol. (2021) 12:677707. doi: 10.3389/fimmu.2021.677707
43. Verschoor CP, Picard E, Andrew MK, Haynes L, Loeb M, Pawelec G, et al. NK- and T-cell granzyme B and K expression correlates with age, CMV infection and influenza vaccine-induced antibody titres in older adults. Front Aging. (2023) 3:1098200. doi: 10.3389/fragi.2022.1098200
44. Potena L, Holweg CTJ, Chin C, Luikart H, Weisshaar D, Narasimhan B, et al. Acute rejection and cardiac allograft vascular disease is reduced by suppression of subclinical cytomegalovirus infection. Transplantation. (2006) 82:398–405. doi: 10.1097/01.tp.0000229039.87735.76
45. Chang A, Musk M, Lavender M, Wrobel J, Yaw M, Lawrence S, et al. Cytomegalovirus viremia in lung transplantation during and after prophylaxis. Transpl Infect Dis. (2019) 21:e13069. doi: 10.1111/tid.2019.21.issue-3
46. Korneffel K, Mitro G, Buschor K, Rees M, and Ortiz J. Low dose valganciclovir as cytomegalovirus prophylaxis in post-renal transplant recipients induced with alemtuzumab: A single-center study. Transpl Immunol. (2019) 56:101226. doi: 10.1016/j.trim.2019.101226
47. Singh N, Winston DJ, Razonable RR, Lyon GM, Silveira FP, Wagener MM, et al. Effect of preemptive therapy vs antiviral prophylaxis on cytomegalovirus disease in seronegative liver transplant recipients with seropositive donors: A randomized clinical trial. JAMA J Am Med Assoc. (2020) 323:1378–87. doi: 10.1001/jama.2020.3138
48. Schneeberger S, LucChina S, Lanzetta M, Brandacher G, Bösmüller C, Steurer W, et al. Cytomegalovirus-related complications in human hand transplantation. Transplantation. (2005) 80:441–7. doi: 10.1097/01.tp.0000168454.68139.0a
49. Kauke-Navarro M, Panayi AC, Formica R, Marty F, Parikh N, Foroutanjazi S, et al. Cytomegalovirus-related complications and management in facial vascularized composite allotransplantation: an international multicenter retrospective cohort study. Transplantation. (2022) 106:2031–43. doi: 10.1097/TP.0000000000004132
Keywords: cytomegalovirus, heart transplant, rejection, transcriptomic profiling mRNA, miRNA, biomarkers
Citation: Barison I, Perazzolo D, Castellani C, Giarraputo A, Rossi E, Vedovelli L, Minuzzo SA, Tessari C, Pradegan N, Toscano G, Tona F, Basso C, Gerosa G, Mandruzzato S, Abate D, Gregori D, Angelini A and Fedrigo M (2025) Transcriptomic profiling of cytomegalovirus infection in cardiac transplantation: proof-of-concept for a new strategy in tissue markers application. Front. Immunol. 16:1581151. doi: 10.3389/fimmu.2025.1581151
Received: 21 February 2025; Accepted: 09 April 2025;
Published: 16 May 2025.
Edited by:
Martin Kauke-Navarro, Yale-New Haven Hospital, United StatesReviewed by:
Javier Carbone, Gregorio Marañón Hospital, SpainZhendong Ding, Central South University, China
Copyright © 2025 Barison, Perazzolo, Castellani, Giarraputo, Rossi, Vedovelli, Minuzzo, Tessari, Pradegan, Toscano, Tona, Basso, Gerosa, Mandruzzato, Abate, Gregori, Angelini and Fedrigo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Annalisa Angelini, YW5uYWxpc2EuYW5nZWxpbmlAdW5pcGQuaXQ=
 Ilaria Barison
Ilaria Barison Diego Perazzolo
Diego Perazzolo Chiara Castellani
Chiara Castellani Alessia Giarraputo1
Alessia Giarraputo1 Elisabetta Rossi
Elisabetta Rossi Luca Vedovelli
Luca Vedovelli Chiara Tessari
Chiara Tessari Giuseppe Toscano
Giuseppe Toscano Francesco Tona
Francesco Tona Cristina Basso
Cristina Basso Gino Gerosa
Gino Gerosa Davide Abate
Davide Abate Dario Gregori
Dario Gregori Annalisa Angelini
Annalisa Angelini