- 1Department of Radiation Oncology, First Affiliated Hospital of Guangxi Medical University, Nanning, China
- 2Department of Otorhinolaryngology and Head and Neck Surgery, First Affiliated Hospital of Guangxi Medical University, Nanning, China
- 3Guangxi Key Laboratory of Early Prevention and Treatment for Regional High Frequency Tumor, Guangxi Medical University, Nanning, China
The gut microbiota has been increasingly recognized as a critical player in maintaining human health and influencing disease development. The tumor microenvironment (TME) is pivotal in tumor development and progression, comprising immune cells, stromal elements, extracellular matrix components, and cytokines. Recent studies have highlighted the promising potential of gut microbiota-derived metabolites (e.g., short-chain fatty acids, bile acids, polyamines, and tryptophan derivatives) to reshape the TME in various ways, generating significant interest for the development of novel therapeutic strategies. Beyond their established effects on traditional cancer treatments, emerging evidence suggests that microbiome-based interventions can substantially enhance cancer immunotherapy. However, the variable role of gut microbiota in modulating therapeutic responses complicates the prediction of clinical outcomes. Therefore, understanding the crosstalk between the gut microbiota and the TME is crucial and holds promise for the development of personalized and comprehensive cancer management strategies. This review aims to summarize the reciprocal regulatory mechanisms between gut microbiota-derived metabolites and the TME, and to explore how these interactions can be leveraged to improve cancer immunotherapy.
1 Introduction
The gut microbiota is a complex ecosystem of microorganisms in the gastrointestinal tract, importantly contributing to the maintenance of health and, when disrupted, to the development of disease (1). The tumor microenvironment (TME), as a dynamic ecosystem, encompasses the intricate cellular and acellular surroundings in which tumor cells proliferate, invade, and metastasize, comprising various components (e.g., immune cells, stromal elements, extracellular matrix components, and cytokines) that closely interact with the tumor cells (2). The crosstalk between the gut microbiota and the TME is increasingly recognized as an important factor in modulating cancer development, progression, and treatment response (3).
Evidence has accumulated to suggest different metabolites produced by the gut microbiota, including short-chain fatty acids (SCFAs), bile acids, polyamines, and tryptophan derivatives (4), as important mediators facilitating the communication between the gut microbiota and the TME. Specifically, SCFAs, as key metabolites of gut microbiota, are produced via the fermentation of undigested dietary fiber by specific intestinal anaerobic microbial communities and exhibit the highest concentration within the gut, primarily consisting of acetate, propionate, and butyrate (5). Bile acids are primarily synthesized in the liver from cholesterol to modulate host physiology and immune functions, and further interact with the gut microbiota to undergo various biotransformation, generating secondary bile acids (e.g., deoxycholic acid [DCA], lithocholic acid [LCA]) (6, 7). Furthermore, polyamines, including spermine, spermidine and putrescine, derive mainly from dietary protein, which serves as the major source of intestinal polyamines (8), and tryptophan derivatives are described as several indole-derivatives produced by gut microflora through catabolism of dietary tryptophan in the colon (9).
It has been shown that the TME interacts with different microbial metabolites to modulate tumorigenesis, immune evasion, and therapeutic responses (3), whereas microbial metabolites are known to modulate critical pathways in the TME such as immune cell differentiation, cytokine secretion, and tumor cell behavior (10). Conversely, tumor-associated inflammation and metabolic reprogramming within the TME can also influence the composition of the gut microbiota and alter metabolite synthesis (11). Understanding this reciprocal interplay between the gut microbiota and the TME shows therefore significant potential for targeting microbial metabolites to reshape the TME and improve cancer outcomes (12).
Cancer immunotherapy has rapidly evolved, offering transformative treatment options for patients; however, significant challenges, such as immune resistance and immune-related adverse events (irAEs), continue to limit its clinical efficacy and broader application (13). The manifestations of irAEs range from mild side effects to life-threatening complications, depending on factors such as the affected organ, tumor histology, and individual patient characteristics (13). IrAEs often affect the gut, skin, liver, and lungs, compromising treatment adherence and patient quality of life (14). Utilizing microbiota-host interactions to develop innovative strategies, such as fecal microbiota transplantation (FMT), pro- and prebiotics, and dietary interventions, with the aim of enhancing the efficacy of immunotherapy while reducing its side effects, is gaining momentum in cancer research (15). FMT is an innovative approach to restoring gut microbial homeostasis by transferring fecal matter from a healthy donor to a recipient (16). To reach the full potential of such strategies, a deeper understanding of specific microbial metabolites is essential for refining strategies for microbial modulation and identifying reliable biomarkers to guide personalized therapeutic interventions.
This review aims to synthesize current knowledge on the crosstalk between the gut microbiota and the TME, with a focus on summarizing the roles of gut microbiota-derived metabolites and how their interactions with the host TME may enhance cancer immunotherapy.
2 Gut microbiota-derived metabolites and modulation of the TME
Different gut microbiota-derived metabolites may play different regulatory roles in modulating the TME (15). SCFAs have been shown to significantly influence the TME by modulating the interactions between immune cells and the production of cytokines (17, 18). Bile acids and their metabolites could on the other hand influence the TME through regulating diverse immune cells (19). Polyamines may exhibit similar functional properties (8). Moreover, indole metabolites derived from tryptophan metabolism have demonstrated multifaceted roles within the TME, showcasing potential utility in both chemotherapy and immunotherapy (20).
2.1 T cells
T cells are integral to the evolvement and modulation of the TME (21). T cells engage in dynamic and context-dependent interactions within the TME, where T cells are tightly regulated by TME-derived signals (e.g., cytokines, metabolic stress, checkpoint molecules), ultimately dictating the efficacy of anti-tumor immunity or facilitating tumor immune evasion (22, 23). SCFAs help to shape T cell differentiation into either effector or regulatory phenotypes (24). CD8+ T cells function as core effector cells that mediate immune responses, acting as the primary target for various immunotherapeutic strategies (25). Specifically, SCFAs have been shown to enhance the functions of CD8+ T cell through inhibiting histone deacetylase (HDAC) and upregulating effector molecules, contributing to anti-tumor immune responses, particularly in colorectal cancer and gastric cancer (26–29). In addition to CD8+ T cells, SCFAs also exhibit diverse effects on other subsets of T cells. CD4+T cells exhibit an adaptive response to the immune microenvironment, ensuring the initiation of the optimal immune strategy in response to different types of immune challenges (30). Stimulated by specific environmental conditions, they differentiate into various cell subsets, such as Th1, Th2, Th17, and Treg cells, each assuming distinct roles in the immune response (31). Butyrate is known to attenuate CD4+ T cell activation by simultaneously inhibiting HDAC and G protein-coupled receptor 43 (GPR43) signaling, effectively suppressing the proliferation of Th1, Th17, and Th22 cells (32, 33). SCFAs have also been shown to promote regulatory T cells (Tregs), contributing to the maintenance of intestinal homeostasis and alleviation of certain pathological processes, such as abdominal aortic aneurysm (34–36). Moreover, propionate has been shown to inhibit IL-17 production by the γδ T cells during the inflammatory and tumorigenic processes (37).
Bile acids are also natural modulators of Th17/Treg balance. Lithocholic acid derivatives, such as 3-oxoLCA and isoalloLCA, could exhibit reciprocal effects by inhibiting Th17/Treg differentiation and enhancing Treg generation (38). Further, deoxycholic acid (DCA) may negatively influence the function of CD8+ T cells through suppressing the Ca2+-nuclear factor of activated T cells (NFAT)2 signaling, thereby facilitating immune evasion in colorectal cancer (39).
In addition to SCFAs and bile acids, other microbial metabolites can also modulate T cell responses. Polyamine metabolism is essential in T cell differentiation, e.g., spermidine has been shown to promote Treg differentiation and attenuating Th17 responses (40). Similarly, ornithine decarboxylase-dependent polyamine production is crucial for maintaining the fidelity of CD4+ T cells (41). Finally, tryptophan derivatives have been suggested to affect the number of Treg cells and induce apoptosis in Th1/Th17 cells (42, 43).
In summary, metabolites derived from the gut microbiota can intricately regulate T cell responses through epigenetic, metabolic, and receptor-mediated mechanisms, presenting significant opportunities for therapeutic interventions in cancer. Future studies should explore tissue-specific effects, dose-dependent outcomes, and translational applicability of these metabolite-based therapies.
2.2 B cells
B cells exert multifaceted roles that collectively shape anti-tumor immunity and correlate with prognostic outcomes, encompassing antigen presentation, antibody production, organization of tertiary lymphoid structures, and regulation via immunosuppressive B regulatory cells (Bregs) (44). Gut microbiota-derived metabolites can influence B cell responses through multiple metabolic and signaling pathways. The enhancing effect of SCFAs on B cell antibody production essentially works by reshaping the energy metabolism pathway of B cells and indirectly consolidating the intestinal immune barrier (45). SCFAs enhance antibody production in B cells by increasing levels of intracellular acetyl-CoA and subsequently stimulating oxidative phosphorylation, glycolysis, and fatty acid synthesis, thus bolstering intestinal and systemic immunity (46). SCFAs can also function as epigenetic regulators of B cell differentiation and activity, influencing both the homeostatic and pathogen-specific antibody responses (47). For instance, butyrate has been shown to promote the differentiation of IL-10-producing (IL-10+) Bregs, a process associated with the inhibition of HDAC3 activity and the reduction of mitochondrial oxidative stress (48). Furthermore, butyrate can enhance the immunosuppressive capabilities of Bregs, important for maintaining immune tolerance (49).
Nonetheless, it is important to note that the immunomodulatory effects of SCFAs on B cells are dose dependent. Low levels of butyrate and propionate have been shown to moderately enhance class-switch DNA recombination in B cells whereas higher levels can inhibit activation-induced cytidine deaminase and Blimp1 expression, ultimately suppressing class-switch DNA recombination (CSR) and plasma cell differentiation (50). Careful clarification of such nuanced, dose-dependent effects is therefore important for effectively harnessing SCFAs in the therapeutic modulation of B cell responses.
2.3 Macrophages
Macrophages are pivotal components of the TME, and their polarization states are intricately regulated by metabolites derived from the gut microbiota. M1 macrophages exhibit tumoricidal activity and reinvigorates cytotoxic T-cell responses, whereas M2 macrophages foster immune evasion and tumor progression (51, 52). Different metabolites exert distinct and multifaceted effects on macrophages. For instance, SCFAs have been shown to modulate the dynamic balance of M1/M2 macrophages by suppressing M1 macrophage polarization and promoting M2 macrophage polarization, thereby participating in tumor-related pathological processes (53–55). Interestingly, B.thetaiotaomicron-derived acetic acid was proved to improve the polarization of M1 macrophages and further promotes the function of cytotoxic CD8+ T cells, ultimately inhibiting the growth of hepatocellular carcinoma tumors (56).
Other microbial metabolites also influence macrophage function. Trimethylamine N-oxide (TMAO), a metabolite produced by the gut microbiota, was shown to promote M1 macrophage polarization via NOD-like receptor protein 3 (NLRP3) inflammasome activation (57) and enhance the cytotoxic capacity of M1 macrophages against tumor cells (58). Moreover, recent studies have suggested that indole-3-acetic acid, a tryptophan-derived metabolite, promotes the IL-35 production in macrophages and other immune cells, subsequently alleviating intestinal inflammation and suppressing tumorigenesis (59).
These findings collectively highlight the complex and context-specific nature of microbial metabolite-mediated regulation of macrophages. Further research is needed to elucidate tissue-specific mechanisms, enabling more effective therapeutic modulations of macrophages in cancer.
2.4 Other immune cells
In addition to T cells, B cells, and macrophages, gut microbiota-derived metabolites also modulate the functions of other immune cell types that are important to the immunological landscape of the TME.
For instance, dendritic cells (DCs) uniquely orchestrate antitumor responses through their specialized capacity for cross-presenting tumor antigens to naïve T cells (60). SCFAs regulate the expression of genes related to inflammation and immune-cell recruitment through HDAC inhibition, resulting in particularly strong modulatory effects in DCs and enhanced anti-inflammatory activity (61). Moreover, SCFAs promote dendrite elongation in DCs, assisting antigen uptake and key processes for effective T cell activation (62).Secondary bile acids have also been shown to inhibit DC activity through inhibiting nuclear factor κB (NF-κB)-mediated activation via the TGR5-cAMP-PKA axis (63).
Myeloid-derived suppressor cells (MDSCs) represent a heterogeneous population of pathologically responded neutrophils and monocytes, which exhibit a strong correlation with unfavorable clinical prognoses in cancer and immune responses (64–67). Butyrate has been shown to induce epigenetic and metabolic reprogramming in MDSCs, promoting their expansion and immunosuppressive capacity (68). In contrast, bile acid has been shown to recruit MDSCs and help mitigate excessive immunosuppression, via the cancer-associated fibroblast-CCL3/CCR1 axis (69).
Natural killer (NK) cells, as key innate effectors in anti-tumor immunity (70), are also modulated by SCFAs. Recent studies have shown that SCFAs can enhance the proliferation and function of NK cells by promoting the release of NK-derived extracellular vesicles and reducing the levels of anti-inflammatory cytokine IL-10, suggesting that SCFAs can contribute the anti-tumor NK cell responses (71). Finally, high levels of SCFAs have been shown to impair the migration and antiviral defense of neutrophils against human immunodeficiency virus, with potentially age- and sex-dependent regulatory characteristics (72). Moreover, butyrate and propionate can induce apoptosis and degranulation in basophils to modify basophil-mediated immune responses (73).
Collectively, these findings emphasize the important role of metabolites derived from the gut microbiota in regulating a wide array of immune cell types within the TME. Further research is warranted to delineate the specific molecular mechanisms by which these metabolites exert such function under different pathological conditions. Ultimately, these insights could guide the development of microbiota-targeted therapies aimed at reshaping the immune landscape in cancer (Table 1).
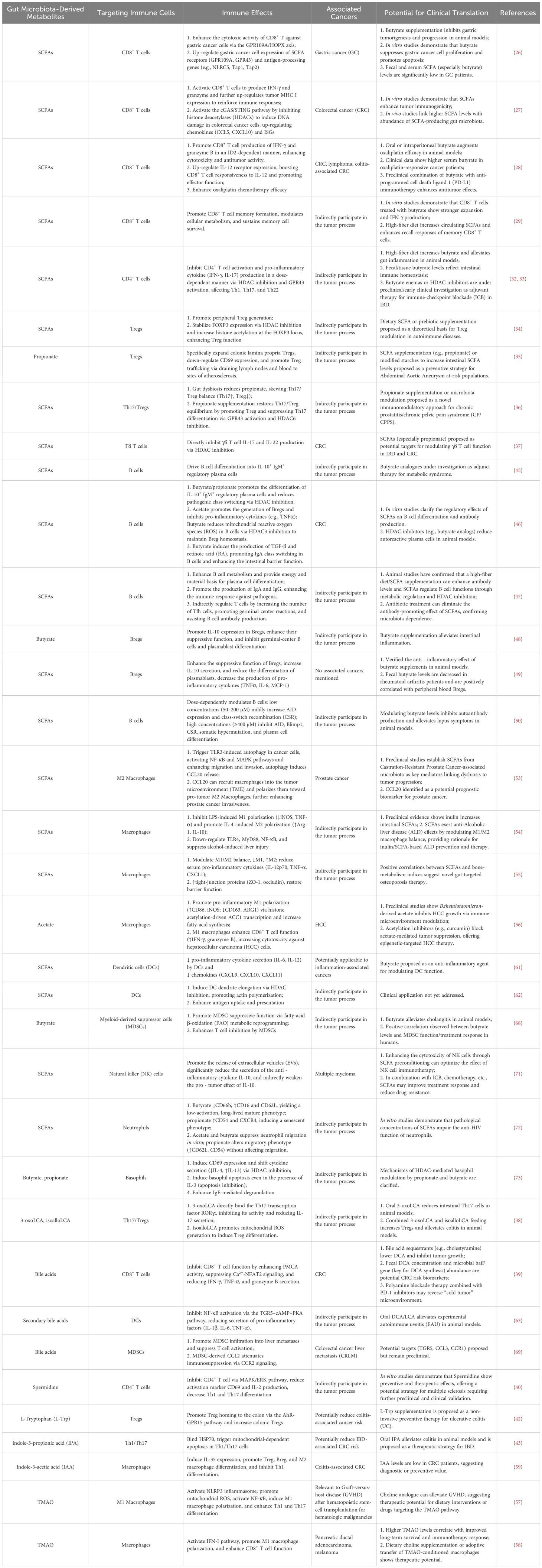
Table 1. Crosstalk of major gut microbiota-derived metabolites and the tumor immune microenvironment: clinical translational potential.
3 Gut microbiota-derived metabolites and cancer immunotherapy
The dynamic interplay between the gut microbiota and the immune system forms the foundation for how gut microbiota-derived metabolites influence immune functions and disease outcomes. Leveraging this interaction offers a promising strategy to enhance immune responses and alleviate immunological disorders. This section examines the translational implications of host-microbiota crosstalk in improving the efficacy of cancer immunotherapy.
3.1 Immune checkpoint blockade therapy
ICB therapy has revolutionized cancer immunotherapy by targeting inhibitory pathways, such as programmed cell death protein 1 (PD-1)/programmed cell death ligand 1 (PD-L1) and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), which regulate immune system homeostasis under physiological conditions while tumors exploit to escape immune surveillance (74, 75). Through blocking these checkpoints, ICB reactivates T cell-mediated anti-tumor responses (76). Emerging evidence has indicated that the gut microbiota significantly influences the efficacy of ICB therapy, whereas the microbial diversity and composition of the gut microbiota contribute importantly to treatment outcomes (77). For instance, melanoma patients responding to anti-PD-1 therapy have been shown to exhibit higher microbial diversity and an enrichment of specific bacterial taxa, compared with non-responders, in the gut microbiota (78).
Microbial metabolites can also modulate ICB therapy. Phenylacetylglutamine (PAGln) has been shown to negatively correlate with ICB efficacy (79), whereas TMAO was shown to synergize with immune checkpoint inhibitors to reduce tumor burden and improve survival in a pancreatic ductal adenocarcinoma model (58). The role of microbial metabolites in immunotherapy is not necessarily monolithic. For example, tryptophan metabolites have been shown to exert dual roles, namely that they enhance ICB efficacy through modulating tumor-associated macrophages but also promote tumor progression via IL4I1-mediated AhR activation (80, 81). A similarly complex picture has been noted for SCFAs. For instance, high levels of butyrate have been suggested to impair anti-CTLA-4 therapy by increasing the frequencies of Tregs and reducing tumor-specific T cell infiltration (82).
Regardless, existing studies suggest a potentially central role of the gut microbiota and its derived metabolites in modulating the efficacy of ICB therapy. Improved understanding of the interactions between different microbial metabolites and the TME helps to develop personalized strategies to enhance therapeutic responses (83).
3.2 Gut microbiota-derived metabolites and adverse events of immunotherapy
Enhancing the efficacy of ICB therapy is utmost important; however, mitigating irAEs is equally critical. The gut microbiota and its derived metabolites have been implicated to modulate the severity of irAEs, particularly in the gastrointestinal tract (84, 85). The gut-liver axis further exemplifies how microbiota-mediated immune regulation can influence systemic toxicity profiles (86).
Specific microbial metabolites have been linked to the susceptibility of irAEs. For instance, menaquinone has been suggested as a potential modulator of adverse immune responses (87) whereas butyrate has been shown to reinforce intestinal barrier integrity and ameliorate immune checkpoint inhibitors (ICIs)-induced colitis (88). Indole-3-carboxaldehyde, a tryptophan metabolite, may exert similar regulatory effects as butyrate (89). To better identify strategies to prevent or alleviate irAEs, in-depth characterization of key microbiota-immune crosstalk pathways is needed.
3.3 Fecal microbiota transplantation
FMT has been shown to reprogram the gut microbiota and the TME among immunotherapy-refractory patients (68, 90) and to restore anti-PD-1 sensitivity among patients with refractory melanoma and other malignancies (91–94). Moreover, FMT has been shown to increase the production of SCFAs and facilitate the infiltration and activation of immune cells to the TME, thereby improving therapeutic efficacy (95). The potential of FMT has also been suggested in hepatocellular carcinoma, particularly in managing intrahepatic metastases (96). Although the potential of FMT as an add-on therapeutic strategy for immunotherapy of diverse cancer types is clear, challenges exist regarding donor screening, protocol standardization, and potential side effects (16, 97). Rigorously designed clinical trials and preclinical models are needed to illuminate the trade-off between benefits and potential harms (90).
3.4 Probiotics and prebiotics
Probiotics and prebiotics represent targeted strategies to modulate the composition and function of the gut microbiota (98). As a vital supplementary treatment method, probiotics have been proved to restore the microbial imbalance caused by cancer treatment, thereby alleviating gastrointestinal adverse reactions and stimulating the immune system to fight against tumor cells (99, 100). Clostridium butyricum, for instance, can suppress colorectal cancer associated with colitis and enhance efficacy of ICB therapy (101–104). Prebiotics, which are selectively utilized by host microorganisms (e.g., glucans and fructans), support the colonization and functions of probiotics and enhance the production of SCFAs (105). For example, pectin has been shown to selectively enrich SCFA-producing taxa (e.g., Bifidobacterium and Lactobacillus), contributing to an immunostimulatory TME (106). Together, prebiotics and probiotics modulate the gut microbiota to promote host health, with overlapping mechanisms such as immune regulation and gut barrier improvement (107). However, as various factors (e.g., strain specificity, host health status, and diet) could influence outcomes of pro- or prebiotics use, individualized approaches and therapeutic guidelines are urgently needed (98, 108). Precision probiotics, tailored to specific microbiome phenotypes, may optimize therapeutic efficacy by promoting the growth of beneficial metabolite-producing microbes (109). Clinical validation and standardized guidelines are therefore essential for the integration of such interventions to personalized oncology (110).
3.5 Dietary interventions
Dietary interventions targeting the gut microbiota have emerged as a non-invasive strategy to improve the immune status and support cancer immunotherapy (111). For example, high dietary cholesterol has been revealed to result in non-alcoholic fatty liver disease-related hepatocellular carcinoma (NAFLD-HCC) through dysbiosis of gut microbiota and metabolites and anticholesterol treatment has significant potential in preventing cancer (112). Furthermore, a high-fiber diet lays a solid immune foundation for strengthening the intestinal immune barrier and enhancing T cell activation to improve responses to anti-PD-1 therapy, promoting the proliferation of gut bacteria that produce SCFAs and increases endogenous SCFA levels (113, 114), especially propionates have been proved to alleviate lipid dysmetabolism and enhance immune homeostasis (115–117). Other microbial metabolites derived from dietary components also exhibit immunomodulatory properties. Polyamines (e.g., spermidine) derived from whole grains and fermented foods help to modulate T cell differentiation and contribute to gut immunity (118). Moreover, appropriate reduction in daily protein intake can enhance the enrichment of beneficial gut bacteria and modulate host health status through microbial-derived metabolites (119).
However, inter-individual microbiome variability and varying adherence to dietary interventions might influence efficacy (120). Successful clinical use of personalized dietary interventions will require a deeper phenotyping of individual microbiota profiles and a validation through rigorously designed clinical trials. Notably, given their relatively minimal side effects, the significant potential of dietary interventions in tumor immunotherapy represents a promising avenue for further exploration.
3.6 Emerging biomarkers for cancer immunotherapy
As the targeted modulation of the gut microbiota has emerged as an innovative therapy for cancer, the information encoded within the compositional and metabolic profiles of the gut microbiota is increasingly being harnessed to develop novel biomarkers for the prediction of risk and prognosis of cancer, indicating another important clinical utility of the gut microbiota (121). Intestinal microbiota exhibits a dynamic and real-time correlation with tumor progression and therapeutic interventions, enabling a more comprehensive and timely assessment of treatment efficacy compared to traditional biomarkers (122). Specifically, there appears to be notable heterogeneity between tumor types. Decreased abundance in specific probiotic species has been linked to a dysbiotic state associated with poor outcomes of colorectal cancer (123).
Gut microbiota metabolites also show potential for non-invasive screening and treatment response prediction (124). Reduced levels and decreased abundance of SCFA-producing bacteria have been shown to be correlated with risk markers in non-small cell lung cancer (84, 125), while secondary bile acids (e.g., deoxycholic acid) with elevated levels and increased abundance of related metabolizing bacteria can act as diagnostic markers in CRC patients (126). Moreover, tryptophan metabolites, particularly indoxyl sulfate (IS), appear to serve as key predictors for differentiating ruptured from unruptured intracranial aneurysms (127).
Nevertheless, owing to the individual variations caused by factors such as diet and antibiotic use, as well as the lack of standardized detection technologies, further verifying the reliability of microbial biomarkers are crucial for fully realizing clinical transformation (123).
4 Conclusions and future perspectives
The gut microbiota plays a pivotal role in modulating the immune responses within the TME and shaping the efficacy of cancer therapies, especially immunotherapy. Investigating the therapeutic potential of gut microbiota-derived metabolites is an emerging frontier in precision oncology, presenting new opportunities to improve clinical outcomes of cancer patients. The convergence of microbiology, immunology, and oncology will facilitate a holistic paradigm shift in cancer care (128) (Figure 1).
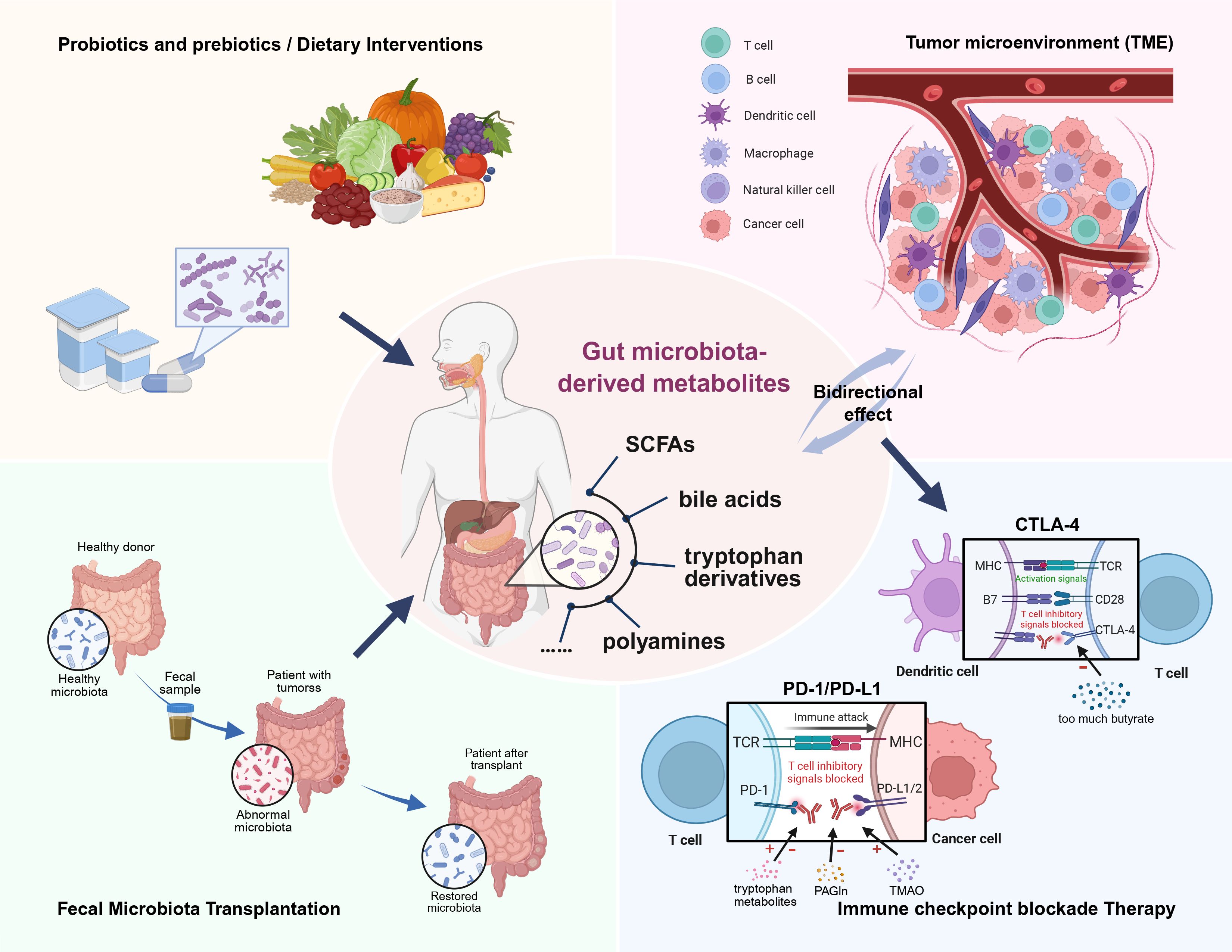
Figure 1. Gut microbiota-derived metabolites play a crucial role in modulating the tumor microenvironment (TME) and influencing the efficacy of cancer therapies. This review highlights the ability of various metabolites to mediate immune responses within the TME. Deciphering and harnessing this crosstalk holds significant promise for advancing cancer immunotherapy, particularly in supporting tailored immune checkpoint blockade (ICB) therapies that target specific molecules. Additionally, targeted fecal microbiota transplantation (FMT), along with other supportive measures such as probiotics, prebiotics, and dietary interventions, may help restore gut microbial homeostasis and its associated metabolic profiles, ultimately enhancing cancer therapy outcomes.
Continued research is clearly needed to best translate bench-side discoveries into clinical applications (129). Innovative technologies and personalized strategies, such as AI-based identification of immunomodulatory gene targets (130), microbiota‐targeted nanomedicine via genetic engineering (131), and development of novel postbiotics or metabolite supplementation (132), could all potentially help improve the efficacy of cancer immunotherapy. Moreover, advanced metabolomics approaches – such as untargeted metabolomics or stable-isotope tracing – should also be leveraged to uncover additional microbiota-derived metabolites of relevance to the TME and efficacy of immunotherapy (133). In addition to the identification of novel metabolites, integrative use of advanced metagenomics and metatranscriptomics techniques can also help identify microbial genes and pathways critical for immune modulation (134, 135). Translational studies should on the other hand expand to include robust, well-powered clinical trials that evaluate different microbiota-targeted therapies such as engineered probiotics, synthetic microbial consortia, and postbiotic supplementation across diverse patient populations (136, 137). Finally, integrating microbiome interventions with emerging cancer therapies – such as CAR-T cells and cancer vaccines – also represents a promising new frontier (138).
Despite significant advancement, several challenges remain. The mechanisms by which microbial metabolites influence immune responses within the TME need further exploration, and their long-term health effects must be thoroughly evaluated. The complexity of host-microbiota interactions necessitates a comprehensive, systems-level research approach. Moreover, population-specific variability underscores the need for large-scale, diverse clinical studies. Personalized therapeutic strategies tailored to individual microbiota profiles could lead to substantial improvements in cancer care. Expanding clinical trial cohorts and ensuring adequate statistical power are essential for generalizing findings and implementing microbiota-based interventions across diverse populations.
Author contributions
XH: Writing – review & editing, Writing – original draft. BL: Formal Analysis, Writing – original draft. YQL: Writing – original draft. YSL: Writing – review & editing. TH: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Youth Foundation of China (82202939 to TH), Guangxi Youth Science Foundation (2025GXNSFBA069088 to TH), Guangxi Science and Technology Program (AD25069077 to TH), the First-class discipline innovation-driven talent program of Guangxi Medical University (2024, to TH), the Medical Excellence Award of Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University (2024, to TH) and General Program of the Guangxi Natural Science Foundation of China [2025GXNSFAA069605 to Liang (YSL)].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1649438/full#supplementary-material
References
1. Adak A and Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. (2019) 76:473–93. doi: 10.1007/s00018-018-2943-4
2. de Visser KE and Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell. (2023) 41:374–403. doi: 10.1016/j.ccell.2023.02.016
3. Luu M, Schütz B, Lauth M, and Visekruna A. The impact of gut microbiota-derived metabolites on the tumor immune microenvironment. Cancers (Basel). (2023) 15(5):1588. doi: 10.3390/cancers15051588
4. Agus A, Clément K, and Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. (2021) 70:1174–82. doi: 10.1136/gutjnl-2020-323071
5. Mann ER, Lam YK, and Uhlig HH. Short-chain fatty acids: linking diet, the microbiome and immunity. Nat Rev Immunol. (2024) 24:577–95. doi: 10.1038/s41577-024-01014-8
6. Fiorucci S, Marchianò S, Urbani G, Di Giorgio C, Distrutti E, Zampella A, et al. Immunology of bile acids regulated receptors. Prog Lipid Res. (2024) 95:101291. doi: 10.1016/j.plipres.2024.101291
7. Ridlon JM and Gaskins HR. Another renaissance for bile acid gastrointestinal microbiology. Nat Rev Gastroenterol Hepatol. (2024) 21:348–64. doi: 10.1038/s41575-024-00896-2
8. Holbert CE, Cullen MT, Casero RA Jr., and Stewart TM. Polyamines in cancer: integrating organismal metabolism and antitumour immunity. Nat Rev Cancer. (2022) 22:467–80. doi: 10.1038/s41568-022-00473-2
9. Roager HM and Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. (2018) 9:3294. doi: 10.1038/s41467-018-05470-4
10. Rossi T, Vergara D, Fanini F, Maffia M, Bravaccini S, and Pirini F. Microbiota-derived metabolites in tumor progression and metastasis. Int J Mol Sci. (2020) 21(16):5786. doi: 10.3390/ijms21165786
11. Nobels A, van Marcke C, Jordan BF, Van Hul M, and Cani PD. The gut microbiome and cancer: from tumorigenesis to therapy. Nat Metab. (2025) 7:895–917. doi: 10.1038/s42255-025-01287-w
12. Xiao Y and Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther. (2021) 221:107753. doi: 10.1016/j.pharmthera.2020.107753
13. Blum SM, Rouhani SJ, and Sullivan RJ. Effects of immune-related adverse events (irAEs) and their treatment on antitumor immune responses. Immunol Rev. (2023) 318:167–78. doi: 10.1111/imr.13262
14. Ramos-Casals M, Brahmer JR, Callahan MK, Flores-Chávez A, Keegan N, Khamashta MA, et al. Immune-related adverse events of checkpoint inhibitors. Nat Rev Dis Primers. (2020) 6:38. doi: 10.1038/s41572-020-0160-6
15. Yang Q, Wang B, Zheng Q, Li H, Meng X, Zhou F, et al. A review of gut microbiota-derived metabolites in tumor progression and cancer therapy. Adv Sci (Weinh). (2023) 10:e2207366. doi: 10.1002/advs.202207366
16. Jamal R, Messaoudene M, de Figuieredo M, and Routy B. Future indications and clinical management for fecal microbiota transplantation (FMT) in immuno-oncology. Semin Immunol. (2023) 67:101754. doi: 10.1016/j.smim.2023.101754
17. Dong Y, Zhang K, Wei J, Ding Y, Wang X, Hou H, et al. Gut microbiota-derived short-chain fatty acids regulate gastrointestinal tumor immunity: a novel therapeutic strategy? Front Immunol. (2023) 14:1158200. doi: 10.3389/fimmu.2023.1158200
18. Kalyanaraman B, Cheng G, and Hardy M. The role of short-chain fatty acids in cancer prevention and cancer treatment. Arch Biochem Biophys. (2024) 761:110172. doi: 10.1016/j.abb.2024.110172
19. Su X, Gao Y, and Yang R. Gut microbiota derived bile acid metabolites maintain the homeostasis of gut and systemic immunity. Front Immunol. (2023) 14:1127743. doi: 10.3389/fimmu.2023.1127743
20. Jia D, Kuang Z, and Wang L. The role of microbial indole metabolites in tumor. Gut Microbes. (2024) 16:2409209. doi: 10.1080/19490976.2024.2409209
21. Kishton RJ, Sukumar M, and Restifo NP. Metabolic regulation of T cell longevity and function in tumor immunotherapy. Cell Metab. (2017) 26:94–109. doi: 10.1016/j.cmet.2017.06.016
22. Edwards M and Brockmann L. Microbiota-dependent modulation of intestinal anti-inflammatory CD4(+) T cell responses. Semin Immunopathol. (2025) 47:23. doi: 10.1007/s00281-025-01049-6
23. Park J, Hsueh PC, Li Z, and Ho PC. Microenvironment-driven metabolic adaptations guiding CD8(+) T cell anti-tumor immunity. Immunity. (2023) 56:32–42. doi: 10.1016/j.immuni.2022.12.008
24. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. (2015) 8:80–93. doi: 10.1038/mi.2014.44
25. Kersten K, Hu KH, Combes AJ, Samad B, Harwin T, Ray A, et al. Spatiotemporal co-dependency between macrophages and exhausted CD8(+) T cells in cancer. Cancer Cell. (2022) 40:624–38.e9. doi: 10.1016/j.ccell.2022.05.004
26. Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8(+) T cells. Immunity. (2019) 51:285–97.e5. doi: 10.1016/j.immuni.2019.06.002
27. He Y, Fu L, Li Y, Wang W, Gong M, Zhang J, et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metab. (2021) 33:988–1000.e7. doi: 10.1016/j.cmet.2021.03.002
28. Mowat C, Dhatt J, Bhatti I, Hamie A, and Baker K. Short chain fatty acids prime colorectal cancer cells to activate antitumor immunity. Front Immunol. (2023) 14:1190810. doi: 10.3389/fimmu.2023.1190810
29. Yu X, Ou J, Wang L, Li Z, Ren Y, Xie L, et al. Gut microbiota modulate CD8(+) T cell immunity in gastric cancer through Butyrate/GPR109A/HOPX. Gut Microbes. (2024) 16:2307542. doi: 10.1080/19490976.2024.2307542
30. Speiser DE, Chijioke O, Schaeuble K, and Münz C. CD4(+) T cells in cancer. Nat Cancer. (2023) 4:317–29. doi: 10.1038/s43018-023-00521-2
31. Ruterbusch M, Pruner KB, Shehata L, and Pepper M. In vivo CD4(+) T cell differentiation and function: revisiting the Th1/Th2 paradigm. Annu Rev Immunol. (2020) 38:705–25. doi: 10.1146/annurev-immunol-103019-085803
32. Kibbie JJ, Dillon SM, Thompson TA, Purba CM, McCarter MD, and Wilson CC. Butyrate directly decreases human gut lamina propria CD4 T cell function through histone deacetylase (HDAC) inhibition and GPR43 signaling. Immunobiology. (2021) 226:152126. doi: 10.1016/j.imbio.2021.152126
33. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504:446–50. doi: 10.1038/nature12721
34. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–5. doi: 10.1038/nature12726
35. Yang F, Xia N, Guo S, Zhang J, Liao Y, Tang T, et al. Propionate alleviates abdominal aortic aneurysm by modulating colonic regulatory T-cell expansion and recirculation. JACC Basic Transl Sci. (2022) 7:934–47. doi: 10.1016/j.jacbts.2022.05.001
36. Du HX, Yue SY, Niu D, Liu C, Zhang LG, Chen J, et al. Gut microflora modulates Th17/Treg cell differentiation in experimental autoimmune prostatitis via the short-chain fatty acid propionate. Front Immunol. (2022) 13:915218. doi: 10.3389/fimmu.2022.915218
37. Dupraz L, Magniez A, Rolhion N, Richard ML, Da Costa G, Touch S, et al. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal γδ T cells. Cell Rep. (2021) 36:109332. doi: 10.1016/j.celrep.2021.109332
38. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature. (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
39. Cong J, Liu P, Han Z, Ying W, Li C, Yang Y, et al. Bile acids modified by the intestinal microbiota promote colorectal cancer growth by suppressing CD8(+) T cell effector functions. Immunity. (2024) 57:876–89.e11. doi: 10.1016/j.immuni.2024.02.014
40. Zheng R, Kong M, Wang S, He B, and Xie X. Spermine alleviates experimental autoimmune encephalomyelitis via regulating T cell activation and differentiation. Int Immunopharmacol. (2022) 107:108702. doi: 10.1016/j.intimp.2022.108702
41. Puleston DJ, Baixauli F, Sanin DE, Edwards-Hicks J, Villa M, Kabat AM, et al. Polyamine metabolism is a central determinant of helper T cell lineage fidelity. Cell. (2021) 184:4186–202.e20. doi: 10.1016/j.cell.2021.06.007
42. Van NT, Zhang K, Wigmore RM, Kennedy AI, DaSilva CR, Huang J, et al. Dietary L-Tryptophan consumption determines the number of colonic regulatory T cells and susceptibility to colitis via GPR15. Nat Commun. (2023) 14:7363. doi: 10.1038/s41467-023-43211-4
43. Gao H, Sun M, Li A, Gu Q, Kang D, Feng Z, et al. Microbiota-derived IPA alleviates intestinal mucosal inflammation through upregulating Th1/Th17 cell apoptosis in inflammatory bowel disease. Gut Microbes. (2025) 17:2467235. doi: 10.1080/19490976.2025.2467235
44. Laumont CM and Nelson BH. B cells in the tumor microenvironment: Multi-faceted organizers, regulators, and effectors of anti-tumor immunity. Cancer Cell. (2023) 41:466–89. doi: 10.1016/j.ccell.2023.02.017
45. Kim M, Qie Y, Park J, and Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. (2016) 20:202–14. doi: 10.1016/j.chom.2016.07.001
46. Qu S, Gao Y, Ma J, and Yan Q. Microbiota-derived short-chain fatty acids functions in the biology of B lymphocytes: From differentiation to antibody formation. BioMed Pharmacother. (2023) 168:115773. doi: 10.1016/j.biopha.2023.115773
47. Kim CH. B cell-helping functions of gut microbial metabolites. Microb Cell. (2016) 3:529–31. doi: 10.15698/mic2016.10.536
48. Föh B, Buhre JS, Lunding HB, Moreno-Fernandez ME, König P, Sina C, et al. Microbial metabolite butyrate promotes induction of IL-10+IgM+ plasma cells. PloS One. (2022) 17:e0266071. doi: 10.1371/journal.pone.0266071
49. Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells. Cell Metab. (2020) 31:837–51.e10. doi: 10.1016/j.cmet.2020.03.003
50. Sanchez HN, Moroney JB, Gan H, Shen T, Im JL, Li T, et al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun. (2020) 11:60. doi: 10.1038/s41467-019-13603-6
51. Vitale I, Manic G, Coussens LM, Kroemer G, and Galluzzi L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. (2019) 30:36–50. doi: 10.1016/j.cmet.2019.06.001
52. Chen D, Zhang X, Li Z, and Zhu B. Metabolic regulatory crosstalk between tumor microenvironment and tumor-associated macrophages. Theranostics. (2021) 11:1016–30. doi: 10.7150/thno.51777
53. Liu Y, Zhou Q, Ye F, Yang C, and Jiang H. Gut microbiota-derived short-chain fatty acids promote prostate cancer progression via inducing cancer cell autophagy and M2 macrophage polarization. Neoplasia. (2023) 43:100928. doi: 10.1016/j.neo.2023.100928
54. Wang Z, Zhang X, Zhu L, Yang X, He F, Wang T, et al. Inulin alleviates inflammation of alcoholic liver disease via SCFAs-inducing suppression of M1 and facilitation of M2 macrophages in mice. Int Immunopharmacol. (2020) 78:106062. doi: 10.1016/j.intimp.2019.106062
55. Chen Y, Yang C, Deng Z, Xiang T, Ni Q, Xu J, et al. Gut microbially produced tryptophan metabolite melatonin ameliorates osteoporosis via modulating SCFA and TMAO metabolism. J Pineal Res. (2024) 76:e12954. doi: 10.1111/jpi.12954
56. Ma H, Yang L, Liang Y, Liu F, Hu J, Zhang R, et al. B. thetaiotaomicron-derived acetic acid modulate immune microenvironment and tumor growth in hepatocellular carcinoma. Gut Microbes. (2024) 16:2297846. doi: 10.1080/19490976.2023.2297846
57. Wu K, Yuan Y, Yu H, Dai X, Wang S, Sun Z, et al. The gut microbial metabolite trimethylamine N-oxide aggravates GVHD by inducing M1 macrophage polarization in mice. Blood. (2020) 136:501–15. doi: 10.1182/blood.2019003990
58. Mirji G, Worth A, Bhat SA, El Sayed M, Kannan T, Goldman AR, et al. The microbiome-derived metabolite TMAO drives immune activation and boosts responses to immune checkpoint blockade in pancreatic cancer. Sci Immunol. (2022) 7:eabn0704. doi: 10.1126/sciimmunol.abn0704
59. Wang J, Hao Y, Yang Y, Zhang Y, Xu C, and Yang R. Gut microbiota derived indole-3-acetic acid ameliorates precancerous inflammatory intestinal milieu to inhibit tumorigenesis through IL-35. J Immunother Cancer. (2025) 13(4):e011155. doi: 10.1136/jitc-2024-011155
60. Møller SH, Wang L, and Ho PC. Metabolic programming in dendritic cells tailors immune responses and homeostasis. Cell Mol Immunol. (2022) 19:370–83. doi: 10.1038/s41423-021-00753-1
61. Nastasi C, Candela M, Bonefeld CM, Geisler C, Hansen M, Krejsgaard T, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci Rep. (2015) 5:16148. doi: 10.1038/srep16148
62. Inamoto T, Furuta K, Han C, Uneme M, Kano T, Ishikawa K, et al. Short-chain fatty acids stimulate dendrite elongation in dendritic cells by inhibiting histone deacetylase. FEBS J. (2023) 290:5794–810. doi: 10.1111/febs.16945
63. Hu J, Wang C, Huang X, Yi S, Pan S, Zhang Y, et al. Gut microbiota-mediated secondary bile acids regulate dendritic cells to attenuate autoimmune uveitis through TGR5 signaling. Cell Rep. (2021) 36:109726. doi: 10.1016/j.celrep.2021.109726
64. Gabrilovich DI and Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. (2009) 9:162–74. doi: 10.1038/nri2506
65. Lasser SA, Ozbay Kurt FG, Arkhypov I, Utikal J, and Umansky V. Myeloid-derived suppressor cells in cancer and cancer therapy. Nat Rev Clin Oncol. (2024) 21:147–64. doi: 10.1038/s41571-023-00846-y
66. Gabrilovich DI. Myeloid-derived suppressor cells. Cancer Immunol Res. (2017) 5:3–8. doi: 10.1158/2326-6066.CIR-16-0297
67. Veglia F, Sanseviero E, and Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. (2021) 21:485–98. doi: 10.1038/s41577-020-00490-y
68. Wang R, Li B, Huang B, Li Y, Liu Q, Lyu Z, et al. Gut microbiota-derived butyrate induces epigenetic and metabolic reprogramming in myeloid-derived suppressor cells to alleviate primary biliary cholangitis. Gastroenterology. (2024) 167:733–49.e3. doi: 10.1053/j.gastro.2024.05.014
69. Li C, Xing X, Li M, Liu Y, Huang S, Zhu T, et al. Bile acids produced by gut microbiota activate TGR5 to promote colorectal liver metastasis progression by inducing MDSCs infiltration in liver. Int Immunopharmacol. (2025) 158:114829. doi: 10.1016/j.intimp.2025.114829
70. Wu SY, Fu T, Jiang YZ, and Shao ZM. Natural killer cells in cancer biology and therapy. Mol Cancer. (2020) 19:120. doi: 10.1186/s12943-020-01238-x
71. Pérez M, Buey B, Corral P, Giraldos D, and Latorre E. Microbiota-derived short-chain fatty acids boost antitumoral natural killer cell activity. J Clin Med. (2024) 13(13):3885. doi: 10.3390/jcm13133885
72. Carrillo-Salinas FJ, Parthasarathy S, Moreno de Lara L, Borchers A, Ochsenbauer C, Panda A, et al. Short-chain fatty acids impair neutrophil antiviral function in an age-dependent manner. Cells. (2022) 11(16):2515. doi: 10.3390/cells11162515
73. Shi Y, Xu M, Pan S, Gao S, Ren J, Bai R, et al. Induction of the apoptosis, degranulation and IL-13 production of human basophils by butyrate and propionate via suppression of histone deacetylation. Immunology. (2021) 164:292–304. doi: 10.1111/imm.13370
74. Zhou CB, Zhou YL, and Fang JY. Gut microbiota in cancer immune response and immunotherapy. Trends Cancer. (2021) 7:647–60. doi: 10.1016/j.trecan.2021.01.010
75. Morad G, Helmink BA, Sharma P, and Wargo JA. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell. (2021) 184:5309–37. doi: 10.1016/j.cell.2021.09.020
76. Zhang C, Hu Y, and Shi C. Targeting natural killer cells for tumor immunotherapy. Front Immunol. (2020) 11:60. doi: 10.3389/fimmu.2020.00060
77. Peng Z, Cheng S, Kou Y, Wang Z, Jin R, Hu H, et al. The gut microbiome is associated with clinical response to anti-PD-1/PD-L1 immunotherapy in gastrointestinal cancer. Cancer Immunol Res. (2020) 8:1251–61. doi: 10.1158/2326-6066.CIR-19-1014
78. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. (2018) 359:97–103. doi: 10.1126/science.aan4236
79. Zhu X, Hu M, Huang X, Li L, Lin X, Shao X, et al. Interplay between gut microbial communities and metabolites modulates pan-cancer immunotherapy responses. Cell Metab. (2025) 37:806–23.e6. doi: 10.1016/j.cmet.2024.12.013
80. Sadik A, Somarribas Patterson LF, Öztürk S, Mohapatra SR, Panitz V, Secker PF, et al. IL4I1 is a metabolic immune checkpoint that activates the AHR and promotes tumor progression. Cell. (2020) 182:1252–70.e34. doi: 10.1016/j.cell.2020.07.038
81. Hezaveh K, Shinde RS, Klötgen A, Halaby MJ, Lamorte S, Ciudad MT, et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity. (2022) 55:324–40.e8. doi: 10.1016/j.immuni.2022.01.006
82. Coutzac C, Jouniaux JM, Paci A, Schmidt J, Mallardo D, Seck A, et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun. (2020) 11:2168. doi: 10.1038/s41467-020-16079-x
83. Yoo JY, Groer M, Dutra SVO, Sarkar A, and McSkimming DI. Gut microbiota and immune system interactions. Microorganisms. (2020) 8(2):e001383. doi: 10.3390/microorganisms8101587
84. Malczewski AB, Navarro S, Coward JI, and Ketheesan N. Microbiome-derived metabolome as a potential predictor of response to cancer immunotherapy. J Immunother Cancer. (2020) 8(16):2515. doi: 10.1136/jitc-2020-001383
85. Dora D, Bokhari SMZ, Aloss K, Takacs P, Desnoix JZ, Szklenárik G, et al. Implication of the gut microbiome and microbial-derived metabolites in immune-related adverse events: emergence of novel biomarkers for cancer immunotherapy. Int J Mol Sci. (2023) 24(3):2769. doi: 10.3390/ijms24032769
86. Kamiya T and Ohtani N. The role of immune cells in the liver tumor microenvironment: an involvement of gut microbiota-derived factors. Int Immunol. (2022) 34:467–74. doi: 10.1093/intimm/dxac020
87. Hu M, Lin X, Sun T, Shao X, Huang X, Du W, et al. Gut microbiome for predicting immune checkpoint blockade-associated adverse events. Genome Med. (2024) 16:16. doi: 10.1186/s13073-024-01285-9
88. Chang AE, Golob JL, Schmidt TM, Peltier DC, Lao CD, and Tewari M. Targeting the gut microbiome to mitigate immunotherapy-induced colitis in cancer. Trends Cancer. (2021) 7:583–93. doi: 10.1016/j.trecan.2021.02.005
89. Renga G, Nunzi E, Pariano M, Puccetti M, Bellet MM, Pieraccini G, et al. Optimizing therapeutic outcomes of immune checkpoint blockade by a microbial tryptophan metabolite. J Immunother Cancer. (2022) 10(3):e003725. doi: 10.1136/jitc-2021-003725
90. Karimi M, Shirsalimi N, Hashempour Z, Salehi Omran H, Sedighi E, Beigi F, et al. Safety and efficacy of fecal microbiota transplantation (FMT) as a modern adjuvant therapy in various diseases and disorders: a comprehensive literature review. Front Immunol. (2024) 15:1439176. doi: 10.3389/fimmu.2024.1439176
91. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. (2021) 371:595–602. doi: 10.1126/science.abf3363
92. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. (2021) 371:602–9. doi: 10.1126/science.abb5920
93. Hayase E and Jenq RR. Role of the intestinal microbiome and microbial-derived metabolites in immune checkpoint blockade immunotherapy of cancer. Genome Med. (2021) 13:107. doi: 10.1186/s13073-021-00923-w
94. Jiang K, Wang Q, Chen XL, Wang X, Gu X, Feng S, et al. Nanodelivery optimization of IDO1 inhibitors in tumor immunotherapy: challenges and strategies. Int J Nanomed. (2024) 19:8847–82. doi: 10.2147/IJN.S458086
95. Ren S, Feng L, Liu H, Mao Y, and Yu Z. Gut microbiome affects the response to immunotherapy in non-small cell lung cancer. Thorac Cancer. (2024) 15:1149–63. doi: 10.1111/1759-7714.15303
96. Deng Z, Mei S, Ouyang Z, Wang R, Wang L, Zou B, et al. Dysregulation of gut microbiota stimulates NETs-driven HCC intrahepatic metastasis: therapeutic implications of healthy faecal microbiota transplantation. Gut Microbes. (2025) 17:2476561. doi: 10.1080/19490976.2025.2476561
97. Bokoliya SC, Dorsett Y, Panier H, and Zhou Y. Procedures for fecal microbiota transplantation in murine microbiome studies. Front Cell Infect Microbiol. (2021) 11:711055. doi: 10.3389/fcimb.2021.711055
98. Reid G, Gadir AA, and Dhir R. Probiotics: reiterating what they are and what they are not. Front Microbiol. (2019) 10:424. doi: 10.3389/fmicb.2019.00424
99. Suez J, Zmora N, Segal E, and Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. (2019) 25:716–29. doi: 10.1038/s41591-019-0439-x
100. Legesse Bedada T, Feto TK, Awoke KS, Garedew AD, Yifat FT, and Birri DJ. Probiotics for cancer alternative prevention and treatment. BioMed Pharmacother. (2020) 129:110409. doi: 10.1016/j.biopha.2020.110409
101. Liu M, Xie W, Wan X, and Deng T. Clostridium butyricum modulates gut microbiota and reduces colitis associated colon cancer in mice. Int Immunopharmacol. (2020) 88:106862. doi: 10.1016/j.intimp.2020.106862
102. Tomita Y, Ikeda T, Sakata S, Saruwatari K, Sato R, Iyama S, et al. Association of probiotic clostridium butyricum therapy with survival and response to immune checkpoint blockade in patients with lung cancer. Cancer Immunol Res. (2020) 8:1236–42. doi: 10.1158/2326-6066.CIR-20-0051
103. Dikeocha IJ, Al-Kabsi AM, Eid EEM, Hussin S, and Alshawsh MA. Probiotics supplementation in patients with colorectal cancer: a systematic review of randomized controlled trials. Nutr Rev. (2021) 80:22–49. doi: 10.1093/nutrit/nuab006
104. Lee SY, Lee DY, Kang JH, Kim JH, Jeong JW, Kim HW, et al. Relationship between gut microbiota and colorectal cancer: Probiotics as a potential strategy for prevention. Food Res Int. (2022) 156:111327. doi: 10.1016/j.foodres.2022.111327
105. Sanders ME, Merenstein DJ, Reid G, Gibson GR, and Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. (2019) 16:605–16. doi: 10.1038/s41575-019-0173-3
106. Zhang SL, Mao YQ, Zhang ZY, Li ZM, Kong CY, Chen HL, et al. Pectin supplement significantly enhanced the anti-PD-1 efficacy in tumor-bearing mice humanized with gut microbiota from patients with colorectal cancer. Theranostics. (2021) 11:4155–70. doi: 10.7150/thno.54476
107. Samanta S. Potential impacts of prebiotics and probiotics on cancer prevention. Anticancer Agents Med Chem. (2022) 22:605–28. doi: 10.2174/1871520621999201210220442
108. Shi N, Li N, Duan X, and Niu H. Interaction between the gut microbiome and mucosal immune system. Mil Med Res. (2017) 4:14. doi: 10.1186/s40779-017-0122-9
109. Veiga P, Suez J, Derrien M, and Elinav E. Moving from probiotics to precision probiotics. Nat Microbiol. (2020) 5:878–80. doi: 10.1038/s41564-020-0721-1
110. Cunningham M, Azcarate-Peril MA, Barnard A, Benoit V, Grimaldi R, Guyonnet D, et al. Shaping the future of probiotics and prebiotics. Trends Microbiol. (2021) 29:667–85. doi: 10.1016/j.tim.2021.01.003
111. Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, et al. Gut-microbiota-targeted diets modulate human immune status. Cell. (2021) 184:4137–53.e14. doi: 10.1016/j.cell.2021.06.019
112. Zhang X, Coker OO, Chu ES, Fu K, Lau HCH, Wang YX, et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut. (2021) 70:761–74. doi: 10.1136/gutjnl-2019-319664
113. Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science. (2021) 374:1632–40. doi: 10.1126/science.aaz7015
114. Mukhopadhya I and Louis P. Gut microbiota-derived short-chain fatty acids and their role in human health and disease. Nat Rev Microbiol. (2025) 23(10):635–51. doi: 10.1038/s41579-025-01183-w
115. Song B, Zhong YZ, Zheng CB, Li FN, Duan YH, and Deng JP. Propionate alleviates high-fat diet-induced lipid dysmetabolism by modulating gut microbiota in mice. J Appl Microbiol. (2019) 127:1546–55. doi: 10.1111/jam.14389
116. Mandaliya DK, Patel S, and Seshadri S. The combinatorial effect of acetate and propionate on high-fat diet induced diabetic inflammation or metaflammation and T cell polarization. Inflammation. (2021) 44:68–79. doi: 10.1007/s10753-020-01309-7
117. Haase S, Mäurer J, Duscha A, Lee DH, Balogh A, Gold R, et al. Propionic acid rescues high-fat diet enhanced immunopathology in autoimmunity via effects on Th17 responses. Front Immunol. (2021) 12:701626. doi: 10.3389/fimmu.2021.701626
118. Carriche GM, Almeida L, Stüve P, Velasquez L, Dhillon-LaBrooy A, Roy U, et al. Regulating T-cell differentiation through the polyamine spermidine. J Allergy Clin Immunol. (2021) 147:335–48.e11. doi: 10.1016/j.jaci.2020.04.037
119. Fan L, Xia Y, Wang Y, Han D, Liu Y, Li J, et al. Gut microbiota bridges dietary nutrients and host immunity. Sci China Life Sci. (2023) 66:2466–514. doi: 10.1007/s11427-023-2346-1
120. Feng W, Liu J, Cheng H, Zhang D, Tan Y, and Peng C. Dietary compounds in modulation of gut microbiota-derived metabolites. Front Nutr. (2022) 9:939571. doi: 10.3389/fnut.2022.939571
121. Yang Y, Misra BB, Liang L, Bi D, Weng W, Wu W, et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. (2019) 9:4101–14. doi: 10.7150/thno.35186
122. Xie Y and Liu F. The role of the gut microbiota in tumor, immunity, and immunotherapy. Front Immunol. (2024) 15:1410928. doi: 10.3389/fimmu.2024.1410928
123. Kang X, Liu C, Ding Y, Ni Y, Ji F, Lau HCH, et al. Roseburia intestinalis generated butyrate boosts anti-PD-1 efficacy in colorectal cancer by activating cytotoxic CD8(+) T cells. Gut. (2023) 72:2112–22. doi: 10.1136/gutjnl-2023-330291
124. Qu R, Zhang Y, Ma Y, Zhou X, Sun L, Jiang C, et al. Role of the gut microbiota and its metabolites in tumorigenesis or development of colorectal cancer. Adv Sci (Weinh). (2023) 10:e2205563. doi: 10.1002/advs.202205563
125. Zhu X, Li K, Liu G, Wu R, Zhang Y, Wang S, et al. Microbial metabolite butyrate promotes anti-PD-1 antitumor efficacy by modulating T cell receptor signaling of cytotoxic CD8 T cell. Gut Microbes. (2023) 15:2249143. doi: 10.1080/19490976.2023.2249143
126. Wang Z, Dan W, Zhang N, Fang J, and Yang Y. Colorectal cancer and gut microbiota studies in China. Gut Microbes. (2023) 15:2236364. doi: 10.1080/19490976.2023.2236364
127. Sun H, Sun K, Tian H, Chen X, Su S, Tu Y, et al. Integrated metagenomic and metabolomic analysis reveals distinctive stage-specific gut-microbiome-derived metabolites in intracranial aneurysms. Gut. (2024) 73:1662–74. doi: 10.1136/gutjnl-2024-332245
128. Schluter J, Peled JU, Taylor BP, Markey KA, Smith M, Taur Y, et al. The gut microbiota is associated with immune cell dynamics in humans. Nature. (2020) 588:303–7. doi: 10.1038/s41586-020-2971-8
129. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. (2019) 7(1):14. doi: 10.3390/microorganisms7010014
130. Luo C, Zhang R, Guo R, Wu L, Xue T, He Y, et al. Integrated computational analysis identifies therapeutic targets with dual action in cancer cells and T cells. Immunity. (2025) 58:745–65.e9. doi: 10.1016/j.immuni.2025.02.007
131. Wei Y, Shen F, Song H, Zhao R, Feng W, Pan Y, et al. The challenge and opportunity of gut microbiota-targeted nanomedicine for colorectal cancer therapy. Imeta. (2024) 3:e213. doi: 10.1002/imt2.213
132. Cho YS, Han K, Xu J, and Moon JJ. Novel strategies for modulating the gut microbiome for cancer therapy. Adv Drug Delivery Rev. (2024) 210:115332. doi: 10.1016/j.addr.2024.115332
133. Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, Jarmusch AK, and Dorrestein PC. Mass spectrometry-based metabolomics in microbiome investigations. Nat Rev Microbiol. (2022) 20:143–60. doi: 10.1038/s41579-021-00621-9
134. Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, Asraf O, Martino C, Nejman D, et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. (2022) 185:3789–806.e17. doi: 10.1016/j.cell.2022.09.005
135. Ojala T, Häkkinen AE, Kankuri E, and Kankainen M. Current concepts, advances, and challenges in deciphering the human microbiota with metatranscriptomics. Trends Genet. (2023) 39:686–702. doi: 10.1016/j.tig.2023.05.004
136. Rozera T, Pasolli E, Segata N, and Ianiro G. Machine learning and artificial intelligence in the multi-omics approach to gut microbiota. Gastroenterology. (2025) 169(3):487–501. doi: 10.1053/j.gastro.2025.02.035
137. Murali SK and Mansell TJ. Next generation probiotics: Engineering live biotherapeutics. Biotechnol Adv. (2024) 72:108336. doi: 10.1016/j.biotechadv.2024.108336
Keywords: gut microbiota, tumor microenvironment, gut microbiota-derived metabolites, cancer immunotherapy, immune cells, crosstalk
Citation: Hu X, Li B, Li Y, Liang Y and Huang T (2025) Communication between gut microbiota-derived metabolites and the tumor microenvironment. Front. Immunol. 16:1649438. doi: 10.3389/fimmu.2025.1649438
Received: 18 June 2025; Accepted: 30 September 2025;
Published: 15 October 2025.
Edited by:
Teresa Zelante, University of Perugia, ItalyReviewed by:
Maria Gazouli, National and Kapodistrian University of Athens, GreeceCopyright © 2025 Hu, Li, Li, Liang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tingting Huang, dGluZ3RpbmdodWFuZzE5ODZAZ21haWwuY29t; Yushan Liang, eXVzaGFubGlhbmczM0BvdXRsb29rLmNvbQ==
†These authors have contributed equally to this work
 Xinyi Hu
Xinyi Hu Bo Li
Bo Li Yuanqing Li
Yuanqing Li Yushan Liang
Yushan Liang Tingting Huang
Tingting Huang