- 1Medical School, The University of Western Australia, Nedlands, WA, Australia
- 2Wal-yan Respiratory Research Centre, The Kids Research Institute Australia, Perth, WA, Australia
- 3Australian Research Council Centre of Excellence in Plant Energy Biology, School of Molecular Sciences, The University of Western Australia, Perth, WA, Australia
- 4European Virus Bioinformatics Center, Friedrich-Schiller-Universitat Jena, Jena, Germany
- 5Center for Child Health Research, The University of Western Australia, Nedlands, WA, Australia
- 6RAN-Biolinks Canada Ltd., Toronto, ON, Canada
- 7Larsson-Rosenquist Foundation Centre for Immunology and Breastfeeding, School of Medicine, The University of Western Australia, Perth, WA, Australia
- 8Immunology and Breastfeeding Team, The Kids Research Institute Australia, Perth, WA, Australia
- 9Department of Pediatrics, Centre for Child Health Research, The University of Western Australia, Nedlands, WA, Australia
- 10Department of Respiratory and Sleep Medicine, Perth Children’s Hospital, Perth, WA, Australia
- 11Canadian Center for Vaccinology, Dalhousie University, IWK Health Centre and the Nova Scotia Health Authority, Halifax, NS, Canada
- 12Department of Pediatrics, Dalhousie University, Halifax, NS, Canada
- 13Department of Microbiology and Immunology, Dalhousie University, Halifax, NS, Canada
Type 1 interferons (T1IFNs) are typically expressed in low concentrations under homeostatic conditions, but upon pathogenic insult or perturbation of the pathway, these critical immune signaling molecules can become either protectors from or drivers of pathology. While essential for initiating antiviral defense and modulating inflammation, dysregulation of T1IFN signaling can contribute to immunopathology, making it and its associated pathways prime targets for immune evasion and disruption by pathogens. This review focuses on the changes in T1IFN signaling across the lifespan, with particular emphasis on the role of the Stimulator of Interferon Genes (STING) pathway in autoimmune and infectious disease susceptibility, especially in the context of viral infections. Aging is associated with diminished T1IFN responsiveness, partially resulting from chronic stimulation of the STING pathway, which contributes to increased susceptibility and impaired viral clearance. Conversely, neonates and young children also show increased vulnerability to certain viral infections, but whether this is driven by T1IFN differences or another mechanism remains incompletely understood. Despite growing interest in T1IFN-based immunotherapies, pediatric and elderly populations remain underrepresented in clinical trials. Here, we advocate for a deeper molecular and systems understanding of how the interferon response evolves across the human lifespan, to inform age-tailored therapeutic approaches and more inclusive study designs, thereby improving outcomes in both the youngest and oldest patients.
1 Introduction
Interferons (IFN) are a class of cytokines that ‘interfere’ with viral replication. There are currently three known IFN families, each defined by its use of distinct signaling receptors. Type 1 Interferons (T1IFN) were the first to be characterized and are key effectors and modulators of both innate and adaptive immunity, with broad effects (1). T1IFNs are highly conserved throughout mammalian evolution, and the human genome contains 17 functional genes encoding 16 proteins, including tissue-specific IFN-ϵ and -κ, the ubiquitous IFN-β, and 12 IFN-α subtypes (2).
T1IFN and their downstream Interferon Stimulated Genes (ISGs) play a unique role in controlling inflammation with both pro- and anti-inflammatory effects. This dual nature makes them highly relevant not just in pathogen-mediated disease, but also in autoimmune and autoinflammatory conditions (3). Due to their broad biological activity, dysregulation of the T1IFN pathway can have harmful consequences (4, 5). Given the diverse and numerous roles of T1IFN in immune function, comprehending these is essential to enhance treatment strategies and elucidate underlying pathologies during infections. Furthermore, emerging evidence suggests that the T1IFN response varies across life stages, potentially influencing disease susceptibility, severity, and treatment in both the young and the elderly. In this review, we outline key age-related differences in T1IFN signaling, with particular focus on antiviral responses mediated by the cyclic GMP-AMP synthase (cGAS) – Stimulator of Interferon Genes (STING) pathway, underscoring the need for a deeper understanding of age-specific immunity to improve infection outcomes.
2 Type 1 interferon signaling in health and disease
T1IFNs are constitutively expressed at very low concentrations in healthy individuals and are essential for immune homeostasis (6). This basal signaling contributes to the development and maintenance of the hematopoietic system, supporting the proliferation and differentiation of immune cells under steady-state conditions (7, 8). Additionally, low-level T1IFN activity also facilitates immune surveillance, regulates tissue integrity, and primes host defenses in the absence of infection (9, 10). However, dysregulation of T1IFN signaling, through genetic factors or triggered by endogenous stimuli, leads to chronic, sterile inflammation. This activation is a hallmark of several autoinflammatory and autoimmune conditions, including systemic lupus erythematosus (SLE), Aicardi-Goutières syndrome, and STING-associated vasculopathy with onset in infancy (SAVI), where disrupted IFN signaling drives pathological immune activation and tissue damage (3, 11, 12). During a pathogenic insult, T1IFNs transition from homeostatic regulators to powerful antimicrobial agents.
During infection, T1IFNs are canonically induced by several pathogen recognition receptors across cellular and tissue compartments in response to diverse stimuli such as bacterial lipopolysaccharide and virion proteins, and intracellular and extracellular detection of nucleotides (Figure 1A) (13–16). The STING pathway is a dedicated induction pathway for T1IFN, primarily activated by cytosolic DNA and double-stranded RNA (dsRNA) via the cGAS-Cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) complex (9). Binding of the cGAS sensor to free nucleotides in the cytoplasm produces the intermediary cGAMP, initiating a complex signaling cascade (as shown in Figure 1) that culminates in the production of T1IFNs and ISGs (17–19). Secreted T1IFNs act in both autocrine and paracrine manners through the Interferon Alpha Receptor 1/Interferon Alpha Receptor 2 (IFNAR1/2) complex, leading to the production of ISGs (Figure 1D). These ISGs contribute to diverse effects, including antiviral function, regulation of antigen presentation, and metabolic modulation (20–24). With their vital role in host defense, T1IFNs are delicately balanced so as to avoid the immunopathology that comes with dysregulated signaling. This duality is evident during infection, where the nature, timing, and context of T1IFN signaling shape the course and outcome of disease.
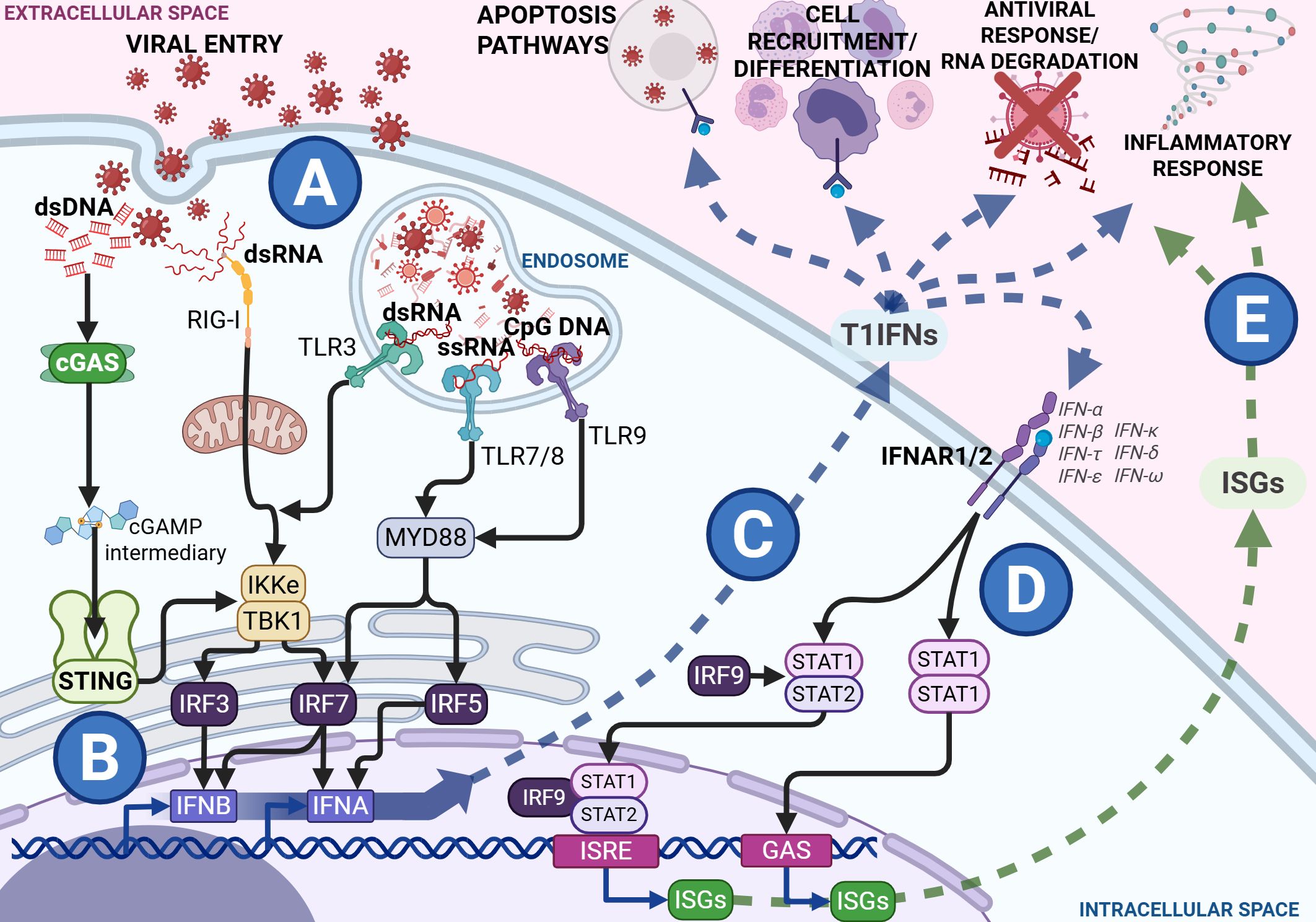
Figure 1. Type 1 interferon (T1IFN) signaling during viral infection with black arrows showing signal transduction within the cytosol from Pathogen Recognition Receptors (PRRs) and the Interferon Alpha Receptor (IFNAR) heterodimer complex to the transcription factors and response elements, the blue arrow showing the release and direct effects of T1IFNs and the green pathway showing the release and effects of Interferon Stimulated Genes (ISGs). During a viral infection and viral entry, viral components are detected through various PRRs in the cytosol and the endosome (A). Cytosolic sensing works through the Stimulator of Interferon Genes (STING) and Retinoic acid-Inducible Gene I (RIG-I) pathways, while endosomal sensing functions involve several Toll-Like Receptor (TLR) pathways. These signal through the endoplasmic reticulum to release Interferon Regulatory Factors (IRFs). Binding of IRFs to promoter regions induces the transcription of T1IFNs, such as IFNA and IFNB (B). These T1IFNs are then released into the extracellular space where they exert antiviral effects such as signaling infected cells to die, recruitment and differentiation of immune cells, RNA degradation, and control of inflammatory response (C). All T1IFNs signal through the same IFNAR heterodimer complex, initiating a Signal Transducer and Activator of Transcription (STAT) cascade, ultimately resulting in transcription of various Interferon ISGs through Interferon-Sensitive Response Elements (ISRE) and Gamma-Activated Sites (GAS) promoter regions (D). These ISGs are released into the extracellular space alongside the T1IFNs, further perpetuating the antiviral and inflammatory responses (E). Figures created in BioRender. Hartnell, L. (2025) https://BioRender.com/ua8nw9b.
3 Type 1 interferons during infection
The immunological impact of T1IFNs during infection is defined not only by their induction pathways but also by pathogen-specific evasion strategies. Signaling through cGAS-STING has been shown to be essential for T1IFN responses to DNA-containing pathogens, with genetic disruption altogether preventing such responses to bacteria (14), as well as DNA viruses and retroviruses. Bacterial induction of cGAS-STING-IFN is not fully elucidated, as, unlike viruses, successful bacteria do not release their genetic material into the cytoplasm of host cells. However, it has been proposed that bacterial induction of T1IFN relies on mistakes in bacterial infection processes, such as the accidental secretion of genetic material or bacterial cyclic dinucleotides (CDNs) along with toxins (25).
The consequences of T1IFN induction in bacterial infection are context-dependent and can be paradoxical (26). While T1IFNs induce bacterial autophagy by host dendritic cells, they also increase cell motility, facilitating cell-to-cell spread and bacterial escape (27). In contrast, during viral infection, the cGAS-STING pathway quickly senses viral nucleotides released into the cytoplasm during the infection process, inducing T1IFN and its downstream effectors (as seen in Figure 1). Interference in viral infection by ISGs occurs at most stages of the viral replication cycle, including preventing viral entry, protein and mRNA synthesis, and assembly (28). Additionally, IFNs and ISGs contribute to wider immune defense by mediating cell-cell interactions and recruitment via altering cell receptor and chemoattractant expression and promoting antigen presentation (Figures 1C, E) (28–30). The timing, magnitude, and duration of the T1IFN response is critical for optimal function in host defense. T1IFN signaling is highly beneficial in the host response mechanisms to acute viral infection but becomes detrimental in prolonged or chronic infection, such as in human immunodeficiency virus (HIV) infection, where it induces a negative feedback loop that reduces the immune response over time, promoting a proviral state (30–32). This exemplifies the finely tuned balance of STING-IFN signaling that is integral to the proper functioning of immune responses, and how pathogens can utilize its mechanisms to perpetuate infection.
4 Evasion and disruption of STING-IFN signaling by pathogens
Given the importance of T1IFNs to immune response and pathogen clearance, many infectious agents disrupt related pathways to either evade detection or improve escape (33, 34). These various strategies not only perpetuate infectious agents within the body but also increase immune-mediated tissue damage (35, 36). Many bacteria induce T1IFNs, especially intracellular bacteria, like Listeria, which stimulate the cGAS-STING pathway (14). Bacterial strategies for the disruption of T1IFN responses typically rely on taking advantage of inflammation and IFN-induced apoptosis of essential immune effector cells (3, 37).
Numerous viruses express viral proteins that prevent signal transduction through the STING-IFN pathway, such as the papain-like protease of coronaviruses (38–40). Non-structural proteins in Respiratory Syncytial Virus (RSV) suppress T1IFN responses, and higher T1IFN levels are associated with better outcomes from the infection (41). Beyond the damage caused by invading microbes, the host’s response to pathogens can lead to hyperinflammation and other pathologies with severe outcomes. Disrupting or delaying the T1IFN response can result in a poorly timed influx of inflammatory cells that perpetuates pathological inflammation and a pro-viral state that is unable to resolve successfully (3, 42, 43). Therefore, understanding the dynamic interplay between pathogens and the host’s immune response is crucial to improving the understanding of infection progression and outcomes.
5 Type 1 interferon signaling in early life
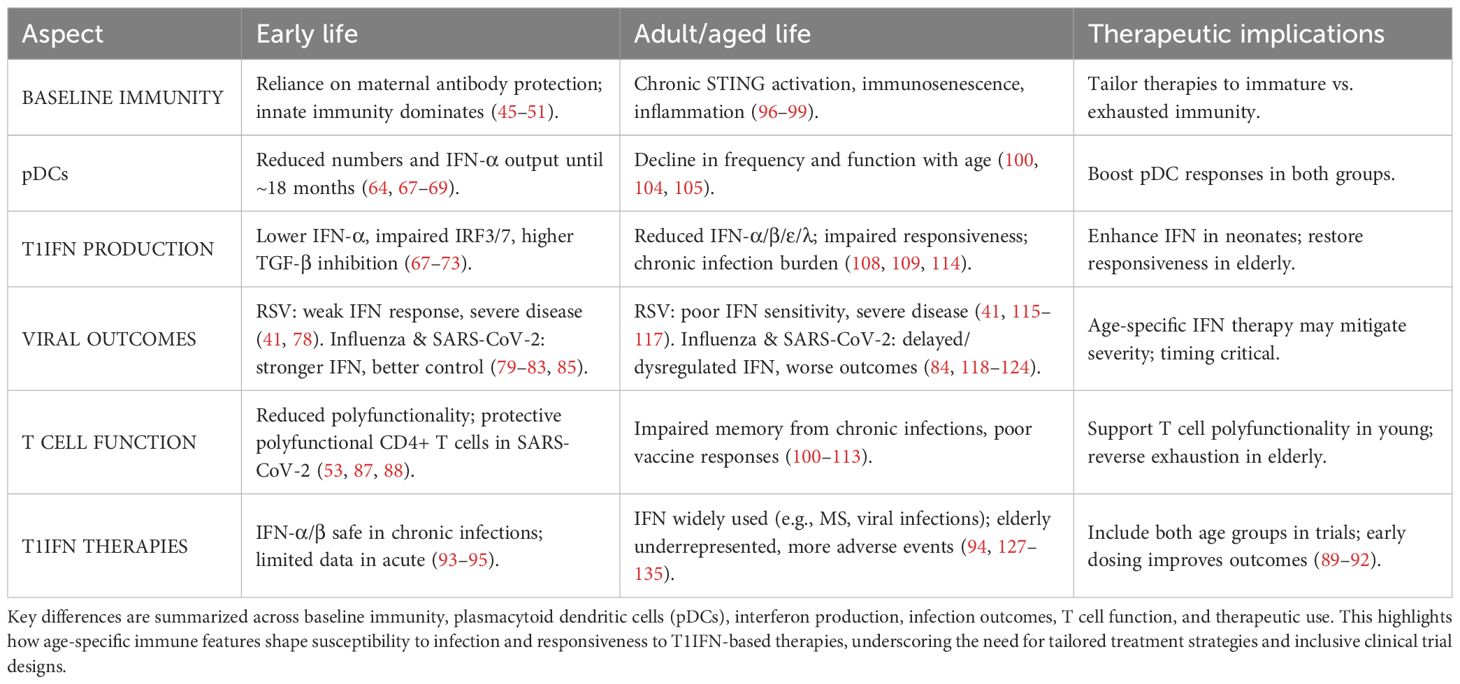
Compared to their adult counterparts, the current understanding of the T1IFN response to infection in early life is limited. In 2019, infectious diseases accounted for up to 49% of deaths in children under five years of age worldwide, with lower respiratory infections responsible for 13.9% of these deaths (44). The immune systems of neonates and children rely heavily on innate immunity due to the naïveté of the adaptive system (45–48). Infant immunity also relies on vertically transmitted antibodies and antimicrobials through their mother’s placenta and breastmilk. Nevertheless, these protective substances wane six months post-birth for placental antibodies and diminish with the cessation of breastfeeding for other immune factors (49–51).
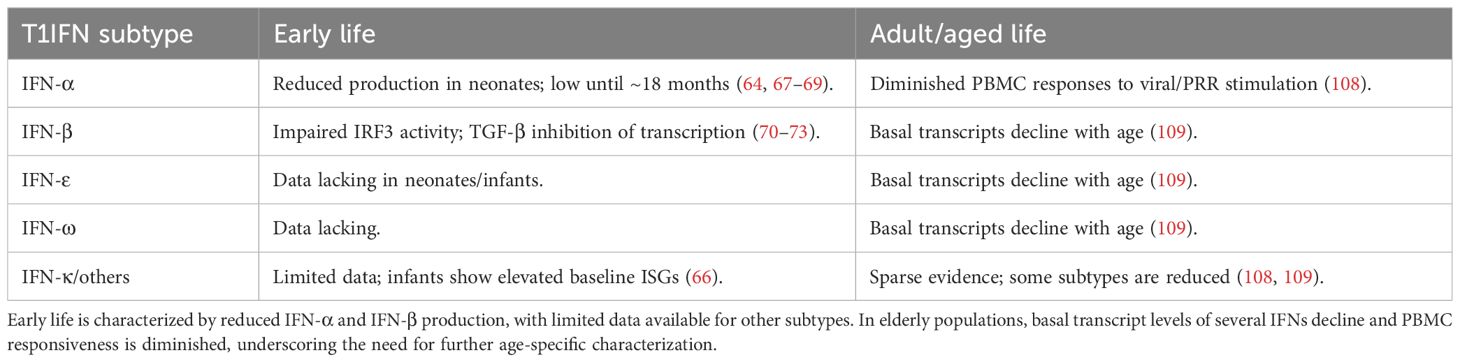
Particularly in neonates, numerous aspects of immune responses appear counterintuitive to infection clearance. Immune cell populations in children differ from those in adults and change dynamically as individuals age, impacting both the capacity and coordination of immune response, with implications for systemic immunity (52–54). At the cellular level, neonatal innate cells produce higher quantities of single cytokines such as interleukins IL-1β, IL-6, IL-23, and IL-10 compared to adults but have diminished simultaneous multi-cytokine responses (55, 56). While almost all immune cell types produce T1IFN, plasmacytoid dendritic cells (pDCs) are the major drivers of T1IFN production during infection (57–60). These specialized dendritic cells have the capacity to produce substantial amounts of T1IFN rapidly and can infiltrate other tissue types such as the airway mucosa (61). Their potent T1IFN production and ability to migrate into infected tissue make them crucial to host immune responses despite representing a small fraction of peripheral blood mononuclear cells (PBMC) (62, 63). Neonatal mononuclear cells produce significantly lower levels of IFN-α than adults, which is less effective at activating pDCs, and while production increases after two months, it remains lower until about 18 months of age (Table 1) (64, 65). Multi-omics profiling of infant PBMCs reveals an elevated baseline of antiviral ISGs, suggesting a partially primed innate state (66). Ex vivo studies of cord blood pDCs also show decreased activity of interferon regulatory factors IRF3 and IRF7, with reduced IRF3 activation observed in cord blood mononuclear cells, and decreased production of the intermediary cGAMP molecule (Figure 2A) (67–69). IRF3 is a key regulator in IFN-β production and Transforming Growth Factor β (TGF-β), by preventing IRF3 phosphorylation and subsequent nuclear translocation, inhibits IFN-β transcription (70–72). Interestingly, Okamoto et al. found that pediatric populations have higher levels of circulating TGF, suggesting one possible mechanism for decreased childhood T1IFN responses (73). These differences in innate T1IFN responses may contribute to observed differences in susceptibility to and severity of infection in children (Table 1).
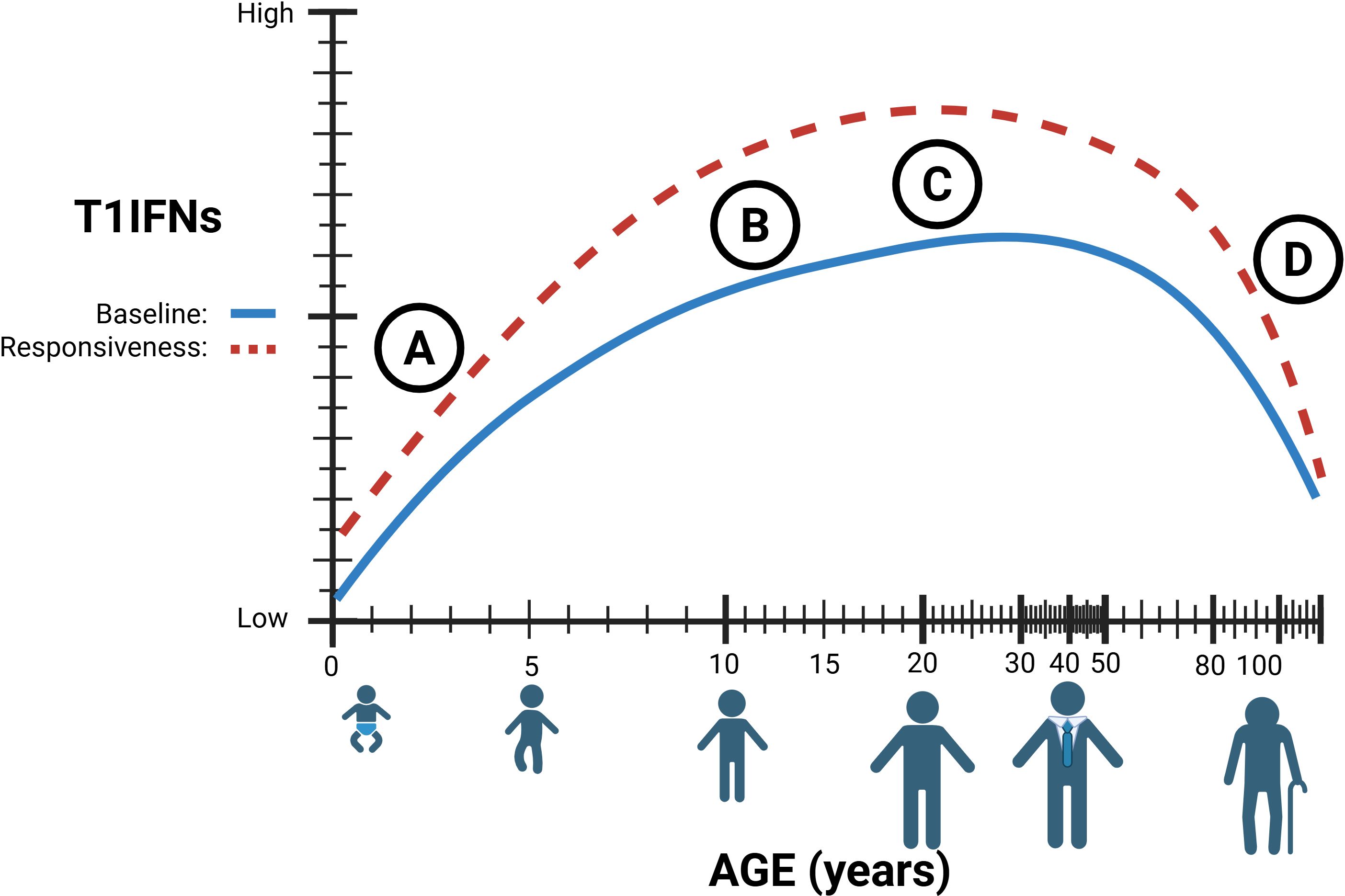
Figure 2. (A) Type 1 interferon (T1IFN) signaling in early life is believed to be diminished resulting from immature and decreased numbers of immune cells. (B) T1IFN signaling increases after puberty, with hormones significantly affecting T1IFN signaling pathways. (C) The true immunological ‘peak’ of T1IFN is still unknown. (D) Lower baseline T1IFNs through chronic STING stimulation, combined with lower circulating T1IFN-producing cells results in decreased sensitivity and responsiveness in elderly individuals. Figures created in BioRender. Hartnell, L. (2025) https://BioRender.com/x7l3f99.
Children exhibit higher infection rates compared to adults, particularly with respiratory viruses, alongside increased rates of coinfection; however, most children experience more favorable health outcomes (74–77). This may be attributed to differences in first-line defense involving T1IFNs. Compared to adults, neonatal and infant infection with RSV is associated with less circulating pDCs, reduced T1IFN responses, and Retinoic acid-Inducible Gene I (RIG-I) signaling (41). More severe RSV infections in infants correlate with lower nasal IFN levels, bronchiolitis, and respiratory failure (78). Contrary to RSV responses, children have a stronger innate immune response to influenza than adults, driven by increased T1IFNs (79). In children, impaired IRF production and T1IFN signaling during influenza infection are associated with susceptibility, and increased T1IFN appears to be protective from severe disease (80, 81). Similar to influenza, pediatric infections with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) are associated with stronger innate IFN responses than adults (82, 83). Children, particularly in the upper airway mucosa, exhibit higher pre-infection levels of T1IFN, allowing for more efficient early viral control and limiting systemic spread (84, 85). Additionally, in vitro co-culture of pediatric nasal epithelial and immune cells shows enhanced epithelial-immune crosstalk, resulting in more rapid and higher magnitude T1IFN induction than adults (86). In early life, T-cells are generally less polyfunctional, producing fewer cytokines simultaneously (53). Polyfunctionality of T cells can influence the course of SARS-CoV-2 infection and, in children, the presence of polyfunctional SARS-CoV-2-specific CD4+ T cells correlates with seroconversion, suggesting a role in the development of lasting immunity (87, 88). This apparent divergence in adult and pediatric responses underscores that the two immune systems function differently. Understanding these differences is crucial for enhancing prevention strategies, therapeutic interventions, and overall health outcomes.
6 Therapeutic use of type 1 interferon in early life
The immune system develops at varying rates from infancy to old age and relies heavily on the accumulation of immunity through repeated exposures. As a result, there is no defined age at which an individual is considered immunologically “mature” (Figure 2) (52). This highlights a need for age-specific treatment approaches and a deeper understanding of the baseline immune environment in children.
Clinical trials of T1IFNs therapy in adults for SARS-CoV-2 infection have shown significant success in reducing viral load, disease severity, and transmission, particularly when administered before peak viral load (89–92). While most trials exclude both young children and the elderly, some studies have demonstrated the effectiveness of T1IFN in reducing viral load in pediatric patients (93). Both IFN-α and IFN-β have been proven safe and effective for treating chronic viral infections in children; however, further research is needed before they can be widely applied for acute infections (94, 95). Given the critical role of timing in IFN therapy, more research is necessary to understand differences in viral response kinetics between adults and children.
7 Type 1 interferon signaling in adult life
Given the role of STING in sensing and responding to damaged DNA, it is highly relevant in understanding health and disease in aging populations. Aging increases the risk and prevalence of cellular and genomic damage (96). Accumulated damage results in the release of self-DNA into the cytoplasm and stimulation of the STING-IFN pathway (97), which has been implicated in age-related chronic inflammation, immunosenescence, and cancer (98, 99). These factors, combined with differences in circulating immune cell populations, lead to an increased susceptibility of aged individuals to disease (100–103). Notably, the T1IFN-producing pDCs decrease in the elderly, and their capacity to produce T1IFNs diminishes, reducing their capacity to mount a response (Figure 2) (100, 104, 105). Additionally, key pathogen recognition pathways that induce T1IFN are decreased in later life, such as RIG-I and numerous TLR pathways, across multiple cell types (106, 107). In short, elderly populations exhibit chronic STING activation, leading to reduced T1IFN production and sensitivity, which results in dysregulated cell differentiation and recruitment during immune responses to infection.
While T1IFNs (including IFN-α, IFN-β, IFN-ϵ, and IFN-λ) are critical for antiviral immunity, the extent to which they change with age varies by subtype, and such characterization remains incomplete (as summarized in Table 1). Ex vivo stimulation of PBMCs from older adults shows delayed and diminished production of IFN-α in response to PRR agonists, including those targeting STING and RIG-I pathways (108). Transcriptomic analysis of upper from SARS-CoV-2-negative individuals shows that basal mRNA transcripts of IFN-ϵ and IFN-λ decline with increasing age, in addition to IFN-α and IFN-β, although to a lesser extent (Figure 2D) (109). Notably, evidence for IFN-κ and other subtypes in the aging immune system remains sparse, highlighting a critical knowledge gap and an area for future investigation across all ages (Table 1).
Deficiencies in T1IFN signaling in aged individuals have been associated with poorer vaccine responses and increased viral load during infection (110, 111). This combination of factors contributes to an overarching susceptibility to viral infections during later life and poorer outcomes from these infections (112). Repeated and chronic infections in elderly populations exacerbate this vulnerability, resulting in a functionally exhausted pDC population that is less able to respond to T1IFN stimulation (113). Among these, human cytomegalovirus (HCMV) is particularly influential. Lifelong HCMV latency is characterized by sustained immune stimulation, chronic STING pathway activation, and dysfunctional memory T cells that can accelerate immunosenescence and impair T1IFN responsiveness (114). The cumulative burden of chronic infections such as HCMV not only reshapes immune cell compartments but also leaves older adults less able to respond effectively to acute viral threats.
These differences in immunity may help to explain disproportionate susceptibility and severity of numerous viruses. RSV is typically more severe in both the young and the elderly, partly because it suppresses T1IFN responses (41, 115). In the elderly, poor RSV outcomes result from multiple factors, including reduced IFN sensitivity, immune exhaustion, and defective immune memory, culminating in an inability to produce a sufficient response to viral assault (116, 117). In geriatric influenza infections, PBMC-derived production of IFN-α is decreased, but the individual capacity of pDCs to produce the cytokine remains intact (118). However, localized expression of ISGs in geriatric airway epithelial cells is skewed to favor an inflammatory phenotype, which impairs viral clearance (119). In addition to RSV and influenza, older adults are also more likely to have poorer outcomes of SARS-CoV-2 infection than their younger counterparts (120). Disease severity of SARS-CoV-2 has been linked to the timing and magnitude of the T1IFN response (84, 121, 122). Aging-related dysfunction of T1IFN, and its role in inflammation and immunosenescence, contributes to increased risk of cytokine storm and adverse outcomes of SARS-CoV-2 infection (123, 124). Taken together, STING-mediated chronic inflammation, lower populations of T1IFN-producing cells, and reduced T1IFN sensitivity become a potentially disastrous combination for older adults with viral infections. To tackle this, intervention strategies that modulate activation along the STING-T1IFN pathway present promising avenues for reversing immunosenescence and restoring T1IFN responsiveness in elderly populations (125, 126). This rationale underpins the growing interest in T1IFN-based therapies in adult and elderly populations.
8 Therapeutic use of type I interferon in adult life
Given its inherent antiviral properties, T1IFN has the potential to complement standard treatments for viral infections. IFN-β is already an established and effective therapy for reducing relapse frequency and disease progression in multiple sclerosis (127). Additionally, IFNs and related modulators are widely used as both frontline and alternative therapies for viral infections, in acute and chronic stages, often in combination with standard care (94, 128–132). A 2022 scoping review on interferon-based therapies for human respiratory viral infections found that 66% of trials evaluated IFN-α as the primary intervention, with rhinovirus (40% of trials) and SARS-CoV-2 (29% of trials) being the most studied pathogens (133). Elderly populations, however, have historically been underrepresented in clinical trials involving IFN-based treatments. Older participants are also more likely to experience higher rates of adverse events and study withdrawal compared to younger participants (134). Despite these challenges, IFN treatment remains a promising option for novel viral infections such as SARS-CoV-2, particularly in elderly populations, where age is a significant risk factor for severe outcomes (135). Given the observed age-related differences in immune response and infection susceptibility, it is essential to ensure broader representation across all age groups in clinical trials, including individuals at both ends of the age spectrum.
9 Discussion
Interferon signaling during infection differs between pediatric, adult, and geriatric populations, influencing disease susceptibility as well as progression to morbidity and mortality. At one end of the lifespan, the early-life environment balances the cost-benefit of growth processes against the need to fight infections. In neonates, immune cells are less polyfunctional, and interferon production is diminished throughout the first 18 months of life. In contrast, children have stronger innate T1IFN responses, resulting in better outcomes than adults despite experiencing higher infection rates. These patterns are observed in both common and severe viral infections. At the other end of the spectrum, the aging immune system is characterized and driven by chronic inflammation, diminished cell populations, and immune exhaustion. Within this context, chronic stimulation of the STING pathway, resulting from accumulated genome damage, along with decreased T1IFN production and sensitivity, perpetuates susceptibility and poor outcomes to disease in the elderly, particularly in cases of respiratory virus infection.
In a world increasingly at risk from novel pathogens such as SARS-CoV-2, T1IFNs offer a valuable treatment option due to their dual role in regulating innate immune responses and exerting antiviral effects. T1IFNs not only restrict pathogen replication but also modulate immune responses to mitigate or prevent the immunopathology associated with severe infections. However, applying this therapy across age groups requires a deeper understanding of age-related differences in T1IFN pathway function and regulation.
This review has outlined age-related differences in the T1IFN pathway, regulated by the cGAS-STING pathway, and response to viral infection (as summarized in Tables 1 and 2). Although significant progress has been made in the development of T1IFN-based therapies, few studies have included or considered pediatric populations. Identifying age-specific T1IFN response characteristics, kinetics, and magnitude shift could help optimize treatment protocols, improving infection management and reducing treatment burdens. Additionally, a deeper understanding of the developing innate interferon response could inform targeted and age-specific treatments for both pediatric and geriatric viral infections, such as RSV, and provide a basis for host-focused therapies to combat future novel pathogens. Ultimately, age-related differences in the innate immune response warrant greater attention, including explicit consideration in the design of clinical trials.
10 Future directions
Together, these findings underscore the importance of contextualizing T1IFN biology across the lifespan. Building on this foundation, future work should leverage emerging technologies and therapeutic strategies to address these age-related differences. Advancing our understanding of T1IFN biology will require integrative approaches that go beyond single-dimensional measurements of cytokine production. The emergence of multi-omics profiling i.e., combining transcriptomics, proteomics, metabolomics, and epigenomics, offers the potential to characterize age-specific signatures of STING activation and T1IFN responsiveness at a higher resolution than ever before. Combining this with systems-level computational modelling could identify predictive biomarkers of infection trajectory, antiviral protection, or immunopathology, as well as regulatory nodes that can be leveraged for therapeutic intervention. In parallel, there is an urgent need to expand translational studies into underrepresented pediatric and geriatric populations. Age-tailored modulation of the STING-T1IFN axis may help restore responsiveness in the elderly or temper hyperresponsiveness in the neonate, ultimately reducing infection-related morbidity at both ends of the lifespan. Incorporating these perspectives into clinical trial design will not only refine therapeutic strategies for existing pathogens but also enhance preparedness against future viral threats.
Author contributions
LH: Writing – original draft, Writing – review & editing, Conceptualization. PA-R: Writing – review & editing, Supervision, Conceptualization. SM: Conceptualization, Supervision, Writing – review & editing. RB-O: Supervision, Writing – review & editing. VV: Writing – review & editing, Supervision. SS: Supervision, Writing – review & editing. TK: Supervision, Writing – review & editing, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. LH is supported by an Australian Government Research Training Program Scholarship at The University of Western Australia.
Conflict of interest
Author RB-O was employed by RAN-Biolinks Canada Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Crow MK and Ronnblom L. Type I interferons in host defence and inflammatory diseases. Lupus Sci Med. (2019) 6:e000336. doi: 10.1136/lupus-2019-000336
2. Wittling MC, Cahalan SR, Levenson EA, and Rabin RL. Shared and unique features of human interferon-beta and interferon-alpha subtypes. Front Immunol. (2020) 11:605673. doi: 10.3389/fimmu.2020.605673
3. Ji L, Li T, Chen H, Yang Y, Lu E, Liu J, et al. The crucial regulatory role of type I interferon in inflammatory diseases. Cell Biosci. (2023) 13:230. doi: 10.1186/s13578-023-01188-z
4. Crow YJ and Stetson DB. The type I interferonopathies: 10 years on. Nat Rev Immunol. (2022) 22:471–83. doi: 10.1038/s41577-021-00633-9
5. Domizio JD, Gulen MF, Saidoune F, Thacker VV, Yatim A, Sharma K, et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature. (2022) 603:145–51. doi: 10.1038/s41586-022-04421-w
6. Gough DJ, Messina NL, Clarke CJ, Johnstone RW, and Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. (2012) 36:166–74. doi: 10.1016/j.immuni.2012.01.011
7. Demerdash Y, Kain B, Essers MAG, and King KY. Yin and Yang: The dual effects of interferons on hematopoiesis. Exp Hematol. (2021) 96:1–12. doi: 10.1016/j.exphem.2021.02.002
8. Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. (2009) 458:904–8. doi: 10.1038/nature07815
9. Li Y, Wilson HL, and Kiss-Toth E. Regulating STING in health and disease. J Inflammation (Lond). (2017) 14:11. doi: 10.1186/s12950-017-0159-2
10. Lima-Junior DS, Krishnamurthy SR, Bouladoux N, Collins N, Han SJ, Chen EY, et al. Endogenous retroviruses promote homeostatic and inflammatory responses to the microbiota. Cell. (2021) 184:3794–811 e19. doi: 10.1016/j.cell.2021.05.020
11. Chasset F, Dayer JM, and Chizzolini C. Type I interferons in systemic autoimmune diseases: distinguishing between afferent and efferent functions for precision medicine and individualized treatment. Front Pharmacol. (2021) 12:633821. doi: 10.3389/fphar.2021.633821
12. Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Sanchez GAM, et al. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. (2014) 371:507–18. doi: 10.1056/NEJMoa1312625
13. Bender AT, Tzvetkov E, Pereira A, Wu Y, Kasar S, Przetak MM, et al. TLR7 and TLR8 differentially activate the IRF and NF-kappaB pathways in specific cell types to promote inflammation. Immunohorizons. (2020) 4:93–107. doi: 10.4049/immunohorizons.2000002
14. Chen Q, Sun L, and Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. (2016) 17:1142–9. doi: 10.1038/ni.3558
15. Chen Y, Lin J, Zhao Y, Ma X, and Yi H. Toll-like receptor 3 (TLR3) regulation mechanisms and roles in antiviral innate immune responses. J Zhejiang Univ Sci B. (2021) 22:609–32. doi: 10.1631/jzus.B2000808
16. Rehwinkel J and Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. (2020) 20:537–51. doi: 10.1038/s41577-020-0288-3
17. Ishikawa H, Ma Z, and Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. (2009) 461:788–92. doi: 10.1038/nature08476
18. Ivashkiv LB and Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. (2014) 14:36–49. doi: 10.1038/nri3581
19. Lemos H, Huang L, McGaha TL, and Mellor AL. Cytosolic DNA sensing via the stimulator of interferon genes adaptor: Yin and Yang of immune responses to DNA. Eur J Immunol. (2014) 44:2847–53. doi: 10.1002/eji.201344407
20. Schoggins JW and Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. (2011) 1:519–25. doi: 10.1016/j.coviro.2011.10.008
21. Gessani S, Conti L, Del Corno M, and Belardelli F. Type I interferons as regulators of human antigen presenting cell functions. Toxins (Basel). (2014) 6:1696–723. doi: 10.3390/toxins6061696
22. Bai J and Liu F. The cGAS-cGAMP-STING pathway: A molecular link between immunity and metabolism. Diabetes. (2019) 68:1099–108. doi: 10.2337/dbi18-0052
23. Yang E and Li MMH. All about the RNA: interferon-stimulated genes that interfere with viral RNA processes. Front Immunol. (2020) 11:605024. doi: 10.3389/fimmu.2020.605024
24. Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. (2013) 41:D1040–6. doi: 10.1093/nar/gks1215
25. Kagan JC. Infection infidelities drive innate immunity. Science. (2023) 379:333–5. doi: 10.1126/science.ade9733
26. Boxx GM and Cheng G. The roles of type I interferon in bacterial infection. Cell Host Microbe. (2016) 19:760–9. doi: 10.1016/j.chom.2016.05.016
27. Guimaraes ES, Marinho FV, de Queiroz N, Antunes MM, and Oliveira SC. Impact of STING inflammatory signaling during intracellular bacterial infections. Cells. (2021) 11:74. doi: 10.3390/cells11010074
28. Schoggins JW. Interferon-stimulated genes: what do they all do? Annu Rev Virol. (2019) 6:567–84. doi: 10.1146/annurev-virology-092818-015756
29. Padovan E, Spagnoli GC, Ferrantini M, and Heberer M. IFN-alpha2a induces IP-10/CXCL10 and MIG/CXCL9 production in monocyte-derived dendritic cells and enhances their capacity to attract and stimulate CD8+ effector T cells. J Leukoc Biol. (2002) 71:669–76. doi: 10.1189/jlb.71.4.669
30. Sedaghat AR, German J, Teslovich TM, CoFrancesco J Jr., Jie CC, Talbot CC Jr., et al. Chronic CD4+ T-cell activation and depletion in human immunodeficiency virus type 1 infection: type I interferon-mediated disruption of T-cell dynamics. J Virol. (2008) 82:1870–83. doi: 10.1128/JVI.02228-07
31. Wang Y, Swiecki M, Cella M, Alber G, Schreiber RD, Gilfillan S, et al. Timing and magnitude of type I interferon responses by distinct sensors impact CD8 T cell exhaustion and chronic viral infection. Cell Host Microbe. (2012) 11:631–42. doi: 10.1016/j.chom.2012.05.003
32. Soper A, Kimura I, Nagaoka S, Konno Y, Yamamoto K, Koyanagi Y, et al. Type I interferon responses by HIV-1 infection: association with disease progression and control. Front Immunol. (2017) 8:1823. doi: 10.3389/fimmu.2017.01823
33. Ahn J and Barber GN. STING signaling and host defense against microbial infection. Exp Mol Med. (2019) 51:1–10. doi: 10.1038/s12276-019-0333-0
34. Liu N, Pang X, Zhang H, and Ji P. The cGAS-STING pathway in bacterial infection and bacterial immunity. Front Immunol. (2021) 12:814709. doi: 10.3389/fimmu.2021.814709
35. Medzhitov R. Origin and physiological roles of inflammation. Nature. (2008) 454:428–35. doi: 10.1038/nature07201
36. Bhattacharyya S. Inflammation during virus infection: swings and roundabouts. In: Dynamics of immune activation in viral diseases Singapore: Springer (2019). p. 43–59.
37. Kovarik P, Castiglia V, Ivin M, and Ebner F. Type I interferons in bacterial infections: A balancing act. Front Immunol. (2016) 7:652. doi: 10.3389/fimmu.2016.00652
38. Cao D, Duan L, Huang B, Xiong Y, Zhang G, and Huang H. The SARS-CoV-2 papain-like protease suppresses type I interferon responses by deubiquitinating STING. Sci Signal. (2023) 16:eadd0082. doi: 10.1126/scisignal.add0082
39. Chen X, Yang X, Zheng Y, Yang Y, Xing Y, and Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. (2014) 5:369–81. doi: 10.1007/s13238-014-0026-3
40. Sun L, Xing Y, Chen X, Zheng Y, Yang Y, Nichols DB, et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PloS One. (2012) 7:e30802. doi: 10.1371/journal.pone.0030802
41. Hijano DR, Vu LD, Kauvar LM, Tripp RA, Polack FP, and Cormier SA. Role of type I interferon (IFN) in the respiratory syncytial virus (RSV) immune response and disease severity. Front Immunol. (2019) 10:566. doi: 10.3389/fimmu.2019.00566
42. Bellgrau D and Modiano JF. The cytokine storm-An appropriate, over-reactive response to SARS-CoV-2 or the wrong immune pathway? Scand J Immunol. (2021) 93:e12979. doi: 10.1111/sji.12979
43. Murira A and Lamarre A. Type-I interferon responses: from friend to foe in the battle against chronic viral infection. Front Immunol. (2016) 7:609. doi: 10.3389/fimmu.2016.00609
44. Perin J, Mulick A, Yeung D, Villavicencio F, Lopez G, Strong KL, et al. Global, regional, and national causes of under-5 mortality in 2000-19: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet Child Adolesc Health. (2022) 6:106–15. doi: 10.1016/S2352-4642(21)00311-4
45. Yu JC, Khodadadi H, Malik A, Davidson B, Salles E, Bhatia J, et al. Innate immunity of neonates and infants. Front Immunol. (2018) 9:1759. doi: 10.3389/fimmu.2018.01759
46. Basha S, Surendran N, and Pichichero M. Immune responses in neonates. Expert Rev Clin Immunol. (2014) 10:1171–84. doi: 10.1586/1744666X.2014.942288
47. Coates BM, Staricha KL, Wiese KM, and Ridge KM. Influenza A virus infection, innate immunity, and childhood. JAMA Pediatr. (2015) 169:956–63. doi: 10.1001/jamapediatrics.2015.1387
48. Wynn JL and Levy O. Role of innate host defenses in susceptibility to early-onset neonatal sepsis. Clin Perinatol. (2010) 37:307–37. doi: 10.1016/j.clp.2010.04.001
49. Lokossou GAG, Kouakanou L, Schumacher A, and Zenclussen AC. Human breast milk: from food to active immune response with disease protection in infants and mothers. Front Immunol. (2022) 13:849012. doi: 10.3389/fimmu.2022.849012
50. Lyons KE, Ryan CA, Dempsey EM, Ross RP, and Stanton C. Breast milk, a source of beneficial microbes and associated benefits for infant health. Nutrients. (2020) 12:4. doi: 10.3390/nu12041039
51. Pierzynowska K, Wolinski J, Westrom B, and Pierzynowski SG. Maternal immunoglobulins in infants-are they more than just a form of passive immunity? Front Immunol. (2020) 11:855. doi: 10.3389/fimmu.2020.00855
52. Simon AK, Hollander GA, and McMichael A. Evolution of the immune system in humans from infancy to old age. Proc Biol Sci. (2015) 282:20143085. doi: 10.1098/rspb.2014.3085
53. Trück J and van der Burg M. Development of adaptive immune cells and receptor repertoires from infancy to adulthood. Curr Opin Syst Biol. (2020) 24:51–5. doi: 10.1016/j.coisb.2020.10.004
54. Schreurs R, Sagebiel AF, Steinert FL, Highton AJ, Klarenbeek PL, Drewniak A, et al. Intestinal CD8(+) T cell responses are abundantly induced early in human development but show impaired cytotoxic effector capacities. Mucosal Immunol. (2021) 14:605–14. doi: 10.1038/s41385-021-00382-x
55. Kollmann TR, Crabtree J, Rein-Weston A, Blimkie D, Thommai F, Wang XY, et al. Neonatal innate TLR-mediated responses are distinct from those of adults. J Immunol. (2009) 183:7150–60. doi: 10.4049/jimmunol.0901481
56. Georgountzou A and Papadopoulos NG. Postnatal innate immune development: from birth to adulthood. Front Immunol. (2017) 8:957. doi: 10.3389/fimmu.2017.00957
57. Hertzog PJ and Williams BR. Fine tuning type I interferon responses. Cytokine Growth Factor Rev. (2013) 24:217–25. doi: 10.1016/j.cytogfr.2013.04.002
58. Hervas-Stubbs S, Perez-Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A, and Melero I. Direct effects of type I interferons on cells of the immune system. Clin Cancer Res. (2011) 17:2619–27. doi: 10.1158/1078-0432.CCR-10-1114
59. Lande R and Gilliet M. Plasmacytoid dendritic cells: key players in the initiation and regulation of immune responses. Ann N Y Acad Sci. (2010) 1183:89–103. doi: 10.1111/j.1749-6632.2009.05152.x
60. Hillyer P, Mane VP, Schramm LM, Puig M, Verthelyi D, Chen A, et al. Expression profiles of human interferon-alpha and interferon-lambda subtypes are ligand- and cell-dependent. Immunol Cell Biol. (2012) 90:774–83. doi: 10.1038/icb.2011.109
61. Bencze D, Fekete T, and Pazmandi K. Type I interferon production of plasmacytoid dendritic cells under control. Int J Mol Sci. (2021) 22:4190. doi: 10.3390/ijms22084190
62. Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. (1999) 284:1835–7. doi: 10.1126/science.284.5421.1835
63. Li G, Cheng L, and Su L. Phenotypic and functional study of human plasmacytoid dendritic cells. Curr Protoc. (2021) 1:e50. doi: 10.1002/cpz1.50
64. Vosters O, Lombard C, Andre F, Sana G, Sokal EM, and Smets F. The interferon-alpha and interleukin-10 responses in neonates differ from adults, and their production remains partial throughout the first 18 months of life. Clin Exp Immunol. (2010) 162:494–9. doi: 10.1111/j.1365-2249.2010.04267.x
65. Lau-Kilby AW, Turfkruyer M, Kehl M, Yang L, Buchholz UJ, Hickey K, et al. Type I IFN ineffectively activates neonatal dendritic cells limiting respiratory antiviral T-cell responses. Mucosal Immunol. (2020) 13:371–80. doi: 10.1038/s41385-019-0234-5
66. Wimmers F, Burrell AR, Feng Y, Zheng H, Arunachalam PS, Hu M, et al. Multi-omics analysis of mucosal and systemic immunity to SARS-CoV-2 after birth. Cell. (2023) 186(21):4632–51.e23. doi: 10.1016/j.cell.2023.08.044
67. Aksoy E, Albarani V, Nguyen M, Laes JF, Ruelle JL, De Wit D, et al. Interferon regulatory factor 3-dependent responses to lipopolysaccharide are selectively blunted in cord blood cells. Blood. (2007) 109:2887–93. doi: 10.1182/blood-2006-06-027862
68. Danis B, George TC, Goriely S, Dutta B, Renneson J, Gatto L, et al. Interferon regulatory factor 7-mediated responses are defective in cord blood plasmacytoid dendritic cells. Eur J Immunol. (2008) 38:507–17. doi: 10.1002/eji.200737760
69. Wang ZS, Liu YL, Mi N, and Duan DY. Intracellular DNA sensing pathway of cGAS-cGAMP is decreased in human newborns and young children. Mol Immunol. (2017) 87:76–85. doi: 10.1016/j.molimm.2017.04.007
70. Thomas BJ, Lindsay M, Dagher H, Freezer NJ, Li D, Ghildyal R, et al. Transforming growth factor-beta enhances rhinovirus infection by diminishing early innate responses. Am J Respir Cell Mol Biol. (2009) 41:339–47. doi: 10.1165/rcmb.2008-0316OC
71. Xu P, Bailey-Bucktrout S, Xi Y, Xu D, Du D, Zhang Q, et al. Innate antiviral host defense attenuates TGF-beta function through IRF3-mediated suppression of Smad signaling. Mol Cell. (2014) 56:723–37. doi: 10.1016/j.molcel.2014.11.027
72. Grunwell JR, Yeligar SM, Stephenson S, Ping XD, Gauthier TW, Fitzpatrick AM, et al. TGF-beta1 suppresses the type I IFN response and induces mitochondrial dysfunction in alveolar macrophages. J Immunol. (2018) 200:2115–28. doi: 10.4049/jimmunol.1701325
73. Okamoto Y, Gotoh Y, Uemura O, Tanaka S, Ando T, and Nishida M. Age-dependent decrease in serum transforming growth factor (TGF)-beta 1 in healthy Japanese individuals; population study of serum TGF-beta 1 level in Japanese. Dis Markers. (2005) 21:71–4. doi: 10.1155/2005/381215
74. Prendergast AJ, Klenerman P, and Goulder PJ. The impact of differential antiviral immunity in children and adults. Nat Rev Immunol. (2012) 12:636–48. doi: 10.1038/nri3277
75. Hwang JK, Na JY, Kim J, Oh JW, Kim YJ, and Choi YJ. Age-specific characteristics of adult and pediatric respiratory viral infections: A retrospective single-center study. J Clin Med. (2022) 11:3197. doi: 10.3390/jcm11113197
76. Glynn JR and Moss PAH. Systematic analysis of infectious disease outcomes by age shows lowest severity in school-age children. Sci Data. (2020) 7:329. doi: 10.1038/s41597-020-00668-y
77. Mandelia Y, Procop GW, Richter SS, Worley S, Liu W, and Esper F. Dynamics and predisposition of respiratory viral co-infections in children and adults. Clin Microbiol Infect. (2021) 27:631 e1– e6. doi: 10.1016/j.cmi.2020.05.042
78. Thwaites RS, Coates M, Ito K, Ghazaly M, Feather C, Abdulla F, et al. Reduced nasal viral load and IFN responses in infants with respiratory syncytial virus bronchiolitis and respiratory failure. Am J Respir Crit Care Med. (2018) 198:1074–84. doi: 10.1164/rccm.201712-2567OC
79. Oshansky CM, Gartland AJ, Wong SS, Jeevan T, Wang D, Roddam PL, et al. Mucosal immune responses predict clinical outcomes during influenza infection independently of age and viral load. Am J Respir Crit Care Med. (2014) 189:449–62. doi: 10.1164/rccm.201309-1616OC
80. Principi N and Esposito S. Severe influenza in children: incidence and risk factors. Expert Rev Anti Infect Ther. (2016) 14:961–8. doi: 10.1080/14787210.2016.1227701
81. Scotta MC, MaChado DG, Oliveira SG, de Moura A, Estorgato GR, de Souza APD, et al. Evaluation of nasal levels of interferon and clinical severity of influenza in children. J Clin Virology. (2019) 114:37–42. doi: 10.1016/j.jcv.2019.02.003
82. Pierce CA, Preston-Hurlburt P, Dai Y, Aschner CB, Cheshenko N, Galen B, et al. Immune responses to SARS-CoV-2 infection in hospitalized pediatric and adult patients. Sci Transl Med. (2020) 12:eabd5487. doi: 10.1126/scitranslmed.abd5487
83. Yoshida M, Worlock KB, Huang N, Lindeboom RGH, Butler CR, Kumasaka N, et al. Local and systemic responses to SARS-CoV-2 infection in children and adults. Nature. (2022) 602:321–7. doi: 10.1038/s41586-021-04345-x
84. Vono M, Huttner A, Lemeille S, Martinez-Murillo P, Meyer B, Baggio S, et al. Robust innate responses to SARS-CoV-2 in children resolve faster than in adults without compromising adaptive immunity. Cell Rep. (2021) 37:109773. doi: 10.1016/j.celrep.2021.109773
85. Loske J, Rohmel J, Lukassen S, Stricker S, Magalhaes VG, Liebig J, et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat Biotechnol. (2022) 40:319–24. doi: 10.1038/s41587-021-01037-9
86. Magalhaes VG, Lukassen S, Drechsler M, Loske J, Burkart SS, Wust S, et al. Immune-epithelial cell cross-talk enhances antiviral responsiveness to SARS-CoV-2 in children. EMBO Rep. (2023) 24:e57912. doi: 10.15252/embr.202357912
87. Adam L, Rosenbaum P, Quentric P, Parizot C, Bonduelle O, Guillou N, et al. CD8+PD-L1+CXCR3+ polyfunctional T cell abundances are associated with survival in critical SARS-CoV-2-infected patients. JCI Insight. (2021) 6:e151571. doi: 10.1172/jci.insight.151571
88. Benede N, Tincho MB, Walters A, Subbiah V, Ngomti A, Baguma R, et al. Distinct T cell polyfunctional profile in SARS-CoV-2 seronegative children associated with endemic human coronavirus cross-reactivity. iScience. (2024) 27:108728. doi: 10.1016/j.isci.2023.108728
89. Castro-Rodriguez JA, Fish EN, Montgomery ST, Kollmann TR, Iturriaga C, Shannon C, et al. Interferon beta-1a ring prophylaxis to reduce household transmission of SARS-CoV-2: a cluster randomised clinical trial. EClinicalMedicine. (2023) 62:102082. doi: 10.1016/j.eclinm.2023.102082
90. Kamyshnyi A, Koval H, Kobevko O, Buchynskyi M, Oksenych V, Kainov D, et al. Therapeutic effectiveness of interferon-alpha2b against COVID-19 with community-acquired pneumonia: the ukrainian experience. Int J Mol Sci. (2023) 24:6887. doi: 10.3390/ijms24086887
91. Monk PD, Marsden RJ, Tear VJ, Brookes J, Batten TN, Mankowski M, et al. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med. (2021) 9:196–206. doi: 10.1016/S2213-2600(20)30511-7
92. Zhou Q, MacArthur MR, He X, Wei X, Zarin P, Hanna BS, et al. Interferon-alpha2b treatment for COVID-19 is associated with improvements in lung abnormalities. Viruses. (2020) 13:44. doi: 10.3390/v13010044
93. Zhou J, Chen X, Lu Y, Wang L, and Yu H. Interferon-alpha-2b nasal spray for treating SARS-coV-2 omicron variant-infected children. J Clin Immunol. (2023) 43:862–4. doi: 10.1007/s10875-023-01452-4
94. Druyts E, Thorlund K, Wu P, Kanters S, Yaya S, Cooper CL, et al. Efficacy and safety of pegylated interferon alfa-2a or alfa-2b plus ribavirin for the treatment of chronic hepatitis C in children and adolescents: a systematic review and meta-analysis. Clin Infect Dis. (2013) 56:961–7. doi: 10.1093/cid/cis1031
95. Hu Y, Ye Y, Ye L, Wang X, and Yu H. Efficacy and safety of interferon alpha therapy in children with chronic hepatitis B: A long-term follow-up cohort study from China. Med (Baltimore). (2019) 98:e16683. doi: 10.1097/MD.0000000000016683
96. Lopez-Otin C, Blasco MA, Partridge L, Serrano M, and Kroemer G. The hallmarks of aging. Cell. (2013) 153:1194–217. doi: 10.1016/j.cell.2013.05.039
97. Miller KN, Victorelli SG, Salmonowicz H, Dasgupta N, Liu T, Passos JF, et al. Cytoplasmic DNA: sources, sensing, and role in aging and disease. Cell. (2021) 184:5506–26. doi: 10.1016/j.cell.2021.09.034
98. Li T and Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med. (2018) 215:1287–99. doi: 10.1084/jem.20180139
99. Schmitz CRR, Maurmann RM, Guma F, Bauer ME, and Barbe-Tuana FM. cGAS-STING pathway as a potential trigger of immunosenescence and inflammaging. Front Immunol. (2023) 14:1132653. doi: 10.3389/fimmu.2023.1132653
100. Jing Y, Shaheen E, Drake RR, Chen N, Gravenstein S, and Deng Y. Aging is associated with a numerical and functional decline in plasmacytoid dendritic cells, whereas myeloid dendritic cells are relatively unaltered in human peripheral blood. Hum Immunol. (2009) 70:777–84. doi: 10.1016/j.humimm.2009.07.005
101. Ventura MT, Casciaro M, Gangemi S, and Buquicchio R. Immunosenescence in aging: between immune cells depletion and cytokines up-regulation. Clin Mol Allergy. (2017) 15:21. doi: 10.1186/s12948-017-0077-0
102. Oh SJ, Lee JK, and Shin OS. Aging and the immune system: the impact of immunosenescence on viral infection, immunity and vaccine immunogenicity. Immune Netw. (2019) 19:e37. doi: 10.4110/in.2019.19.e37
103. Thin KA, Cross A, Angsuwatcharakon P, Mutirangura A, Puttipanyalears C, and Edwards SW. Changes in immune cell subtypes during ageing. Arch Gerontol Geriatr. (2024) 122:105376. doi: 10.1016/j.archger.2024.105376
104. Agrawal A. Mechanisms and implications of age-associated impaired innate interferon secretion by dendritic cells: a mini-review. Gerontology. (2013) 59:421–6. doi: 10.1159/000350536
105. Qian F, Wang X, Zhang L, Lin A, Zhao H, Fikrig E, et al. Impaired interferon signaling in dendritic cells from older donors infected in vitro with West Nile virus. J Infect Dis. (2011) 203:1415–24. doi: 10.1093/infdis/jir048
106. Kollmann TR, Levy O, Montgomery RR, and Goriely S. Innate immune function by Toll-like receptors: distinct responses in newborns and the elderly. Immunity. (2012) 37:771–83. doi: 10.1016/j.immuni.2012.10.014
107. Molony RD, Nguyen JT, Kong Y, Montgomery RR, Shaw AC, and Iwasaki A. Aging impairs both primary and secondary RIG-I signaling for interferon induction in human monocytes. Sci Signal. (2017) 10:eaan2392. doi: 10.1126/scisignal.aan2392
108. Connors J, Taramangalam B, Cusimano G, Bell MR, Matt SM, Runner K, et al. Aging alters antiviral signaling pathways resulting in functional impairment in innate immunity in response to pattern recognition receptor agonists. Geroscience. (2022) 44:2555–72. doi: 10.1007/s11357-022-00612-5
109. Pierangeli A, Gentile M, Oliveto G, Frasca F, Sorrentino L, Matera L, et al. Comparison by age of the local interferon response to SARS-coV-2 suggests a role for IFN-epsilon and -omega. Front Immunol. (2022) 13:873232. doi: 10.3389/fimmu.2022.873232
110. Lauf T, Hader A, Hornung F, Reisser Y, Nietzsche S, Schanz F, et al. Age-related STING suppression in macrophages contributes to increased viral load during influenza a virus infection. Immun Ageing. (2024) 21:80. doi: 10.1186/s12979-024-00482-9
111. Panda A, Qian F, Mohanty S, van Duin D, Newman FK, Zhang L, et al. Age-associated decrease in TLR function in primary human dendritic cells predicts influenza vaccine response. J Immunol. (2010) 184:2518–27. doi: 10.4049/jimmunol.0901022
112. Feng E, Balint E, Poznanski SM, Ashkar AA, and Loeb M. Aging and interferons: impacts on inflammation and viral disease outcomes. Cells. (2021) 10:708. doi: 10.3390/cells10030708
113. Macal M, Jo Y, Dallari S, Chang AY, Dai J, Swaminathan S, et al. Self-renewal and toll-like receptor signaling sustain exhausted plasmacytoid dendritic cells during chronic viral infection. Immunity. (2018) 48:730–44 e5. doi: 10.1016/j.immuni.2018.03.020
114. Bouezzedine F, Baba RE, Morot-Bizot S, Diab-Assaf M, and Herbein G. Cytomegalovirus at the crossroads of immunosenescence and oncogenesis. Explor Immunol. (2023) 3:17–27. doi: 10.37349/ei.2023.00086
115. Stephens LM and Varga SM. Function and modulation of type I interferons during respiratory syncytial virus infection. Vaccines (Basel). (2020) 8:177. doi: 10.3390/vaccines8020177
116. Cherukuri A, Patton K, Gasser RA Jr., Zuo F, Woo J, Esser MT, et al. Adults 65 years old and older have reduced numbers of functional memory T cells to respiratory syncytial virus fusion protein. Clin Vaccine Immunol. (2013) 20:239–47. doi: 10.1128/CVI.00580-12
117. Looney RJ, Falsey AR, Walsh E, and Campbell D. Effect of aging on cytokine production in response to respiratory syncytial virus infection. J Infect Dis. (2002) 185:682–5. doi: 10.1086/339008
118. Canaday DH, Amponsah NA, Jones L, Tisch DJ, Hornick TR, and Ramachandra L. Influenza-induced production of interferon-alpha is defective in geriatric individuals. J Clin Immunol. (2010) 30:373–83. doi: 10.1007/s10875-010-9374-9
119. Chason KD, Jaspers I, Parker J, Sellers S, Brighton LE, Hunsucker SA, et al. Age-associated changes in the respiratory epithelial response to influenza infection. J Gerontol A Biol Sci Med Sci. (2018) 73:1643–50. doi: 10.1093/gerona/gly126
120. Fericean RM, Rosca O, Citu C, Manolescu D, Bloanca V, Toma AO, et al. COVID-19 clinical features and outcomes in elderly patients during six pandemic waves. J Clin Med. (2022) 11:6083. doi: 10.3390/jcm11226803
121. Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, et al. Imbalanced host response to SARS-coV-2 drives development of COVID-19. Cell. (2020) 181:1036–45 e9. doi: 10.1016/j.cell.2020.04.026
122. Lopez L, Sang PC, Tian Y, and Sang Y. Dysregulated interferon response underlying severe COVID-19. Viruses. (2020) 12:1433. doi: 10.3390/v12121433
123. Meftahi GH, Jangravi Z, Sahraei H, and Bahari Z. The possible pathophysiology mechanism of cytokine storm in elderly adults with COVID-19 infection: the contribution of “inflame-aging. Inflammation Res. (2020) 69:825–39. doi: 10.1007/s00011-020-01372-8
124. Tizazu AM, Mengist HM, and Demeke G. Aging, inflammaging and immunosenescence as risk factors of severe COVID-19. Immun Ageing. (2022) 19:53. doi: 10.1186/s12979-022-00309-5
125. Jin X, Wang W, Zhao X, Jiang W, Shao Q, Chen Z, et al. The battle between the innate immune cGAS-STING signaling pathway and human herpesvirus infection. Front Immunol. (2023) 14:1235590. doi: 10.3389/fimmu.2023.1235590
126. Zhang S, Zheng R, Pan Y, and Sun H. Potential therapeutic value of the STING inhibitors. Molecules. (2023) 28:3127. doi: 10.3390/molecules28073127
127. Group TIMSS. Interferon beta-1b is effective in relapsing-remitting multiple sclerosis. I. Clinical results of a multicenter, randomized, double-blind, placebo-controlled trial. The IFNB Multiple Sclerosis Study Group. Neurology. (1993) 43:655–61. doi: 10.1212/WNL.43.4.655
128. Antonelli G, Scagnolari C, Moschella F, and Proietti E. Twenty-five years of type I interferon-based treatment: a critical analysis of its therapeutic use. Cytokine Growth Factor Rev. (2015) 26:121–31. doi: 10.1016/j.cytogfr.2014.12.006
129. Terrault NA, Bzowej NH, Chang KM, Hwang JP, Jonas MM, Murad MH, et al. AASLD guidelines for treatment of chronic hepatitis B. Hepatology. (2016) 63:261–83. doi: 10.1002/hep.28156
130. Thompson AJ and Holmes JA. Treating hepatitis C - what’s new? Aust Prescr. (2015) 38:191–7. doi: 10.18773/austprescr.2015.068
131. Feld JJ and Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. (2005) 436:967–72. doi: 10.1038/nature04082
132. Jaeckel E, Cornberg M, Wedemeyer H, Santantonio T, Mayer J, Zankel M, et al. Treatment of acute hepatitis C with interferon alfa-2b. N Engl J Med. (2001) 345:1452–7. doi: 10.1056/NEJMoa011232
133. Mesic A, Jackson EK, Lalika M, Koelle DM, and Patel RC. Interferon-based agents for current and future viral respiratory infections: A scoping literature review of human studies. PloS Glob Public Health. (2022) 2:e0000231. doi: 10.1371/journal.pgph.0000231
134. Sherigar JM, Gayam V, Khan A, Mukhtar O, Arefiev Y, Khalid M, et al. Clinical efficacy and tolerability of direct-acting antivirals in elderly patients with chronic hepatitis C. Eur J Gastroenterol Hepatol. (2017) 29:767–76. doi: 10.1097/MEG.0000000000000871
Keywords: interferon, type 1 interferon, STING, age-related, pediatric, viral infection, SARS-CoV-2, COVID-19
Citation: Hartnell L, Agudelo-Romero P, Montgomery ST, Ben-Othman R, Verhasselt V, Stick SM and Kollmann TR (2025) What goes up must come down: dynamics of type 1 interferon signaling across the lifespan. Front. Immunol. 16:1654604. doi: 10.3389/fimmu.2025.1654604
Received: 26 June 2025; Accepted: 29 September 2025;
Published: 16 October 2025.
Edited by:
Huiwen Zheng, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaReviewed by:
Fumiyuki Hattori, Kansai Medical University, JapanDavide Proietto, University of Ferrara, Italy
Copyright © 2025 Hartnell, Agudelo-Romero, Montgomery, Ben-Othman, Verhasselt, Stick and Kollmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucy Hartnell, bHVjeS5oYXJ0bmVsbEB0aGVraWRzLm9yZy5hdQ==
 Lucy Hartnell
Lucy Hartnell Patricia Agudelo-Romero
Patricia Agudelo-Romero Samuel T. Montgomery
Samuel T. Montgomery Rym Ben-Othman
Rym Ben-Othman Valerie Verhasselt
Valerie Verhasselt Stephen M. Stick
Stephen M. Stick Tobias R. Kollmann11,12,13
Tobias R. Kollmann11,12,13