- 1Lydia Becker Institute of Immunology and Inflammation, Faculty of Biology, Medicine and Health, University of Manchester, Manchester, United Kingdom
- 2Wellcome Centre for Cell-Matrix Research, Lydia Becker Institute of Immunology and Inflammation, Faculty of Biology, Medicine and Health, Manchester Academic Health Science Centre, University of Manchester, Manchester, United Kingdom
- 3Geoffrey Jefferson Brain Research Centre, Manchester Academic Health Science Centre, Northern Care Alliance NHS Group, University of Manchester, Manchester, United Kingdom
- 4Division of Immunology, Immunity to Infection and Respiratory Medicine Faculty of Biology, Medicine and Health, University of Manchester, Manchester, United Kingdom
- 5The Walter and Eliza Hall Institute of Medical Research, Parkville, VIC, Australia
- 6GlaxoSmithKline, Stevenage, United Kingdom
- 7Medical Research Council Centre for Medical Mycology at the University of Exeter, Faculty of Health and Life Sciences, Exeter, United Kingdom
- 8Institute of Immunology and Infection Research, School of Biological Sciences, University of Edinburgh, Edinburgh, United Kingdom
Introduction: Although human lung macrophages are heterogenous and play key roles during health and disease, the mechanisms that govern their activation and function are unclear, particularly in type 2 settings. Our understanding of how human lung macrophages respond to inflammatory signals have predominantly relied on cell lines or peripheral blood derived cells, which have a limited capacity to reflect the complexity of tissue macrophage responses.
Methods: We isolated macrophages from resected human lung tissue and stimulated them ex vivo under type 2 (IL-4, IL-13, or IL-4 + IL-13) or type 1 (IFNγ + LPS) conditions.
Results: Human lung macrophages stimulated with IL-4/13, alone or in combination, significantly upregulated expression of the chemokines CCL17, CCL18 and CCL22, along with the transglutaminase TGM2 and the lipoxygenase ALOX15. This type 2 activation profile was distinct from LPS + IFNγ activated human lung macrophages, which upregulated IL6, IL8, IL1B, TNFα and CHI3L1 (YKL-40). Further, type 2 activated human lung macrophage products showed differential metabolic reliance for their induction, with IL-4/13 induced CCL22 being glycolytically controlled, while ALOX15 was regulated by fatty acid oxidation.
Discussion: These data clarify hallmarks of human lung macrophage activation and polarisation in addition to revealing novel metabolic regulation of type 2 markers.
Introduction
Lung macrophages are highly specialised, playing a central role in maintaining health and restricting disease (1). Their development, metabolic responsiveness and function are shaped by the unique metabolically challenging lung environment since its available metabolic substrates are tightly controlled (2, 3). For instance, the airways are abundant in lipid-rich surfactant while having one of the lowest glucose environments in the body (4, 5).
Metabolism governs how macrophages respond to type 1 and type 2 cytokines, with the general principle being that glycolysis tends to be more typical of type 1 macrophages, while lipid metabolism is more associated with type 2 macrophages (6). However, the emphasis of work on human lung tissue macrophage immunometabolism has thus far focused on LPS responsiveness, rather than on type 2 stimuli such as IL-4 and IL-13 (7).
Given limitations on collecting primary human tissue cells (8), our understanding of human lung macrophage function and immunometabolism during disease predominantly comes from studies using human peripheral blood monocyte derived macrophages (MDMs) in vitro, or is extrapolated from murine in vivo models (9–11). However, as the tissue environment plays a role in shaping cellular fate, metabolic responsiveness, and function (1), it is unlikely that MDMs accurately represent lung macrophage function and immunometabolism (6). Similarly, the direct translatability of observations in murine lung macrophages to humans is unclear, with previously defined differences being apparent (12, 13). Such differences likely contribute to why so many drug development programmes fail in translation from preclinical models (14, 15).
There also remains a need to define robust markers to identify human tissue macrophages in different activation states, particularly in the context of type 2 inflammation (1, 13). Flow cytometric identification of lung type 2 macrophage activation markers is challenging, since human lung macrophages are highly auto-fluorescent and some commonly used murine markers of type 2 associated macrophages (e.g. Chil3) have no direct human homologs (16, 17) or are differentially regulated in humans (e.g. Arginase 1) (18).
Here, we demonstrate that CCL17, CCL22, ALOX15, CCL18 and TGM2 represent reliable markers of human lung macrophage type 2 activation, and that expression of these markers show distinct metabolic regulation to their type 1 counterparts.
Materials and methods
Sex as a biological variable
In this study, sex was not considered as a biological variable.
Human subjects and samples
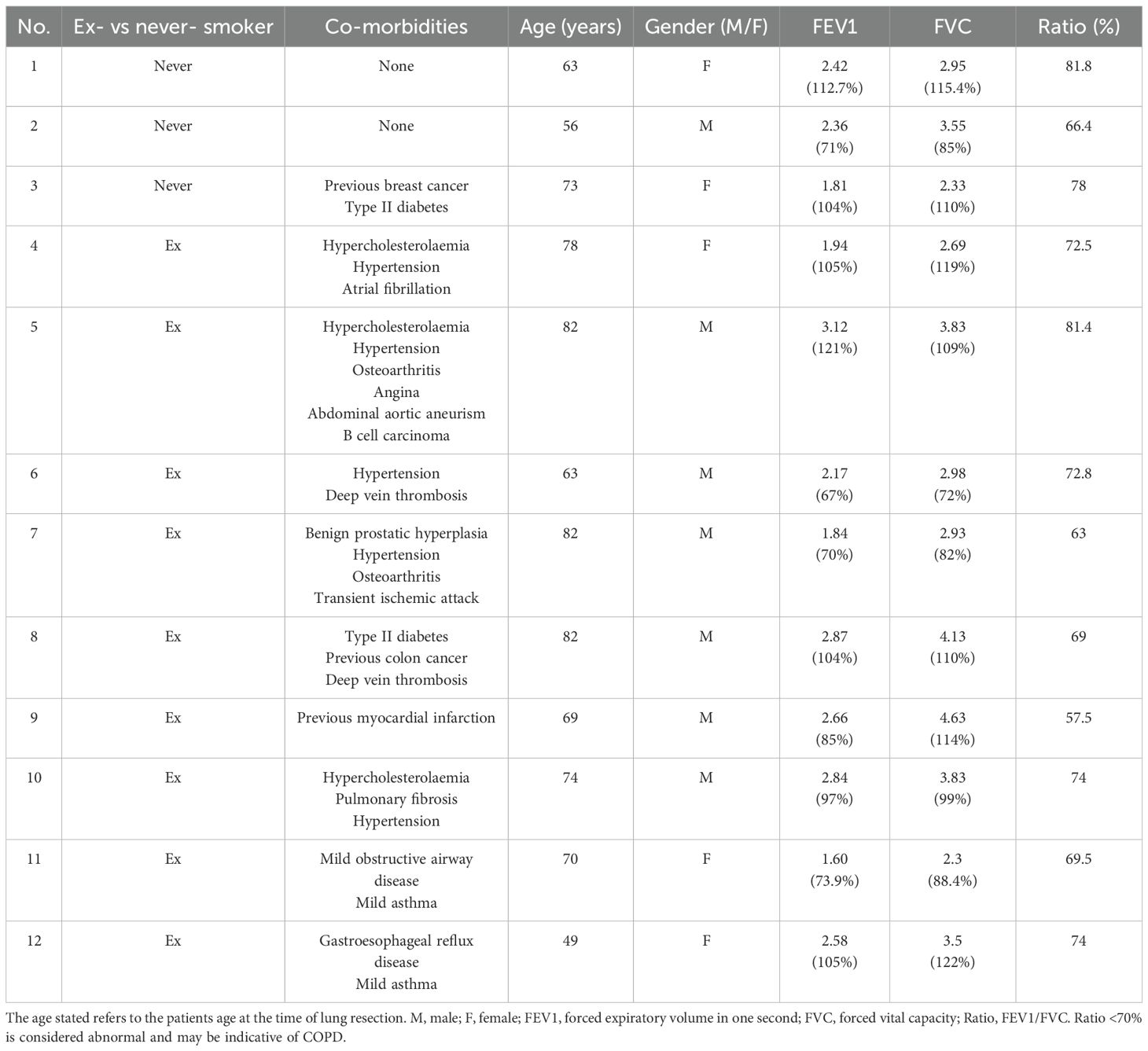
All adult human donors gave written informed consent in accordance with the principles expressed in the Declaration of Helsinki and were recruited into the Manchester, Allergy, Respiratory and Thoracic Surgery (ManARTS) Biobank (located in the Northwest Lung Research Centre at the University Hospital of South Manchester NHS Foundation, Wythenshawe). The research protocol was approved by the National Research Ethics Service (NRES) Committee Northwest – Haydock, ethics approval 15/NW/0409). The ManARTS Biobank collected surgical lung samples from lung cancer patients that had undergone surgical lung tumor resection. The regions of lung marginal tissue samples collected for the experiments described in this study were taken by a histopathologist at a set distance from the lung tumor (>6 cm) during resection and thus deemed non-cancerous for research purposes. Lung resection sample exclusion criteria for this study included patients with Rheumatoid Arthritis, a known history of peripheral arterial disease, sarcoidosis, ankylosing spondylitis, Crohn’s disease, autoimmune diseases (e.g. inflammatory bowel disease, multiple sclerosis and type 1 diabetes), conditions requiring immunomodulatory therapy (e.g. Methotrexate and Azathioprine), a history of tuberculosis and chronic obstructive pulmonary disease (COPD) or emphysema diagnosis. A total of twelve donors aged between 49 and 82 years old were used for the experiments detailed in this study. Their patient demographic and clinical information is listed in Table 1 along with the patient’s medication history in Table 2.
Study approval
The study was approved under the National Research Ethics Service Committee; North West – Haydock ethics reference 20/NW/0302. Informed consent was obtained from all donors included in this study.
Human lung macrophage isolation
Human lung macrophages were isolated from lung tissue resection samples using perfusion and plastic adherence (19). The airways of the resected lung tissue were perfused with sterile 1X PBS using a syringe attached to a 21-gauge needle, 40 mm in length (BD Microlance) until the lung tissue appeared anemic in colour. The perfusate was then centrifuged at 400 x g for 10 min at 4 °C (Heraeus Megafuge 40R) and pelleted cells resuspended in sterile 10 ml Roswell Park Memorial Institute (RPMI)-1640 media before being slowly overlaid onto 10 ml Ficoll-Paque Plus (GE Healthcare) using a Pasteur pipette. The cells and Ficoll were then centrifuged at 400 x g with reduced acceleration and without brakes for 30 min at RT. After centrifugation, the peripheral blood mononuclear cell (PBMC) layer was collected and washed with 20 ml RPMI-1640 media. The isolated cells were centrifuged at 400 x g for 10 min at 4 °C before the pelleted cells were resuspended in prewarmed RPMI-1640 media supplemented with 2 mM L-Glutamine (Sigma-Aldrich), 100 U/ml penicillin (Sigma-Aldrich), 100 μg/ml streptomycin (Sigma-Aldrich) and 10% foetal calf serum (FCS) until manually counted based on size and morphology using a haemocytometer, as described previously (20).
Flow cytometry
Single cell suspension was washed with PBS and stained with Zombie ultraviolet (UV) dye (BioLegend) diluted 1:2000. Cells were washed with FACS buffer (PBS containing 2% FBS and 2 mM EDTA) to sequester unbound Zombie UV dye prior to blocking with 5 μg/ml Human FC block (Fc1; BD Pharmingen). Surface staining was subsequently performed for 30 minutes at 4 °C in FACS buffer using CD45 (HI30), CD64 (10.1) and CD163 (GHI/61) all BioLegend antibodies diluted at 1:200. Cells were fixed and permeabilised using eBioscience fixation/permeabilisation buffer for 45 minutes at 4 °C. Stained cells were acquired using the 7-laser Bigfoot cell sorter (Thermo Fisher Scientific), with spectral signatures for individual fluorochromes established with single stained cells to enable spectral unmixing. Samples were analysed using FlowJo version 10.10 Software (Tree Star, Inc).
In vitro cell culture
Human lung macrophage culture with type 2 and type 1 stimuli, and metabolic inhibitors
Cells were resuspended in RPMI-1640 media containing 2 mM L-Glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 10% FCS at a density of 4 x 105 cells/ml. Next, 40,000 cells in 100 μl were plated per well in a 96-well tissue culture treated, non-pyrogenic, polystyrene flat-bottomed culture plate with lid (Corning) and allowed to adhere for 1 h at 37 °C in a humidified environment of 5% CO2. The total cell counts were based on large cells to ensure enough macrophages were plated and allowed to adhere. Consistent with prior studies focusing on human lung macrophages (13, 21–23), CD64 and CD163 were used to identify human lung macrophages. Using this approach, the average purity of macrophages (CD45+CD163+CD64+) after adherence, as determined by flow cytometry (described above), being ~74.5% (see Supplementary Figure 1). After incubation and, adherence, cells were washed three times with 200 μl 1X PBS per well before 100 µl of fresh supplemented RPMI-1640 media was added per well. Next, either 50 μl supplemented RPMI-1640 media alone or containing 4 mM 2-Deoxy-D-glucose (2-DG) (RPMI containing 1 mM final concentration) (Sigma-Aldrich) or 50 μl 800 µM etomoxir (Eto) (200 µM final concentration) (Sigma-Aldrich) was added per well for 30 min at 37 °C, 5% CO2. After incubation, cells were stimulated with either 50 μl supplemented RPMI-1640 media alone or containing 50 μl 80 ng/ml human rIL-4 (20 ng/ml final concentration) (PeproTech) or 80 ng/ml human rIL-13 (20 ng/ml final concentration) (PeproTech) or 80 ng/ml LPS (20 ng/ml final concentration) (Sigma-Aldrich) plus 80 ng/ml IFNγ (20 ng/ml final concentration) (PeproTech) for 48 h at 37 °C, 5% CO2. After 48 h supernatants were harvested and stored at -80 °C for future secreted protein analysis whilst cells were lysed in 150 μl RLT buffer with 1.5 μl 2-ME and stored at -80 °C prior to RNA extraction.
ELISA
To quantify protein concentrations in human lung macrophage culture supernatants, ELISAs with paired antibodies and recombinant standards were performed (see Supplementary Table 1). Capture antibodies were coated onto 96 well flat bottom non tissue culture plates (NUNC-Immuno Plate, Thermo Fisher Scientific) in 50 µl of 1X PBS overnight at 4 °C. Plates were then washed four times with 200 µl wash buffer (1X PBS containing 0.05% Tween-20) before being blocked using 50 µl 1% bovine serum albumin (BSA) (Sigma-Aldrich) diluted with 1X PBS for 90 min at 37 °C. After blocking, supernatants in duplicate or triplicate at the required dilution along with doubling dilutions of recombinant protein standards in a volume of 50 µl wash buffer were added. Samples were then incubated overnight at 4 °C. Plates were then washed four times with 200 µl wash buffer before secondary detection antibodies were added in 50 µl wash buffer and allowed to bind for 1 h at 37 °C. Plates were washed a further four times with 200 µl wash buffer before streptavidin-horseradish peroxidase (R&D Systems) diluted 1:200 in wash buffer was added to plates in 50 µl and incubated at 37 °C for 1 h. After incubation, plates were washed eight times with 200 µl wash buffer before 100 µl of colorimetric substrate of peroxidase, 3,3’,5,5’-Tetramethylbenzidine (TMB) (Sigma-Aldrich) was added to each well. Following development, the reaction was stopped by addition of 0.18 M sulfuric acid (Sigma-Aldrich). Plates were read at 450 nm, with 570 nm as the reference wavelength, using a plate reader (Tecan, Infinite M200 PRO).
RNA extraction and qPCR
Cells post culture were lysed with 150 µl RLT lysis buffer containing 1.5 μl 2-ME before RNA was isolated using RNeasy Micro kits (Qiagen) according to the manufacturer’s instructions. After RNA extraction, cDNA was generated from sorted cells using SuperScript-III (Thermo Fisher Scientific) and Oligo-dT (12–18 primer, Thermo Fisher Scientific). Relative quantification of genes of interest was performed by qPCR analysis with a QuantStudio 12K Flex Real-Time PCR instrument (Thermo Fisher Scientific) using QuantStudio software (Thermo Fisher Scientific) and SYBR Green master mix (Thermo Fisher Scientific). Five serial 1:4 dilutions of a positive control sample of pooled cDNA were used to create standard curves. Gene expression (Ct values) was normalised to Hypoxanthine Phosphoribosyltransferase (HPRT). Human primer sequences were checked using the NCBI Basic Local Alignment Search Tool (BLAST) and ordered from Sigma-Aldrich (Germany). Primer pairs for sequences with length between 80–150 were selected (Supplementary Table 2).
Statistics
Statistical analysis was performed using R (version 4.3) in RStudio (version 2023.09.1 + 494). Pairwise comparisons statistical comparisons were calculated by Wilcoxon two-sample tests using the rstatix package (version 0.7.2). P values from Wilcoxon comparisons were adjusted for multiple testing using the Holm method. All P values are reported as the Holm-adjusted P value. An adjusted P value less than 0.05 was considered significant. Heatmaps were produced by calculating the geometric mean for each group/condition and plotted using the with tile colours scaled by row. All graphs were produced using the ggplot2 package (version 3.4.1).
Results
Human lung alveolar macrophages display distinct polarisation phenotypes following stimulation
To define changes in gene expression and protein secretion in human lung macrophages during polarisation, we isolated macrophages by perfusion of human lung tissue obtained from patients undergoing surgical tissue resection with no underlying autoimmune diseases or COPD diagnosis. Macrophages were cultured ex vivo under type 2 (IL-4 and/or IL-13) or type 1 (LPS + IFNγ) polarising conditions for 48 h, RNA isolated to quantify gene expression and supernatants collected for measurement of secreted proteins. Macrophages showed distinct profiles of gene expression (Figure 1A). Type 2 polarisation resulted in significantly increased expression of CCL17 (Figure 1B), CCL22 (Figure 1C), TGM2 and ALOX15, along with a trend for elevated CCL18. In contrast, type 1 stimulation significantly increased expression of IL6, TNFα, IL8, IL1B and CHI3L1 (Figure 1A).
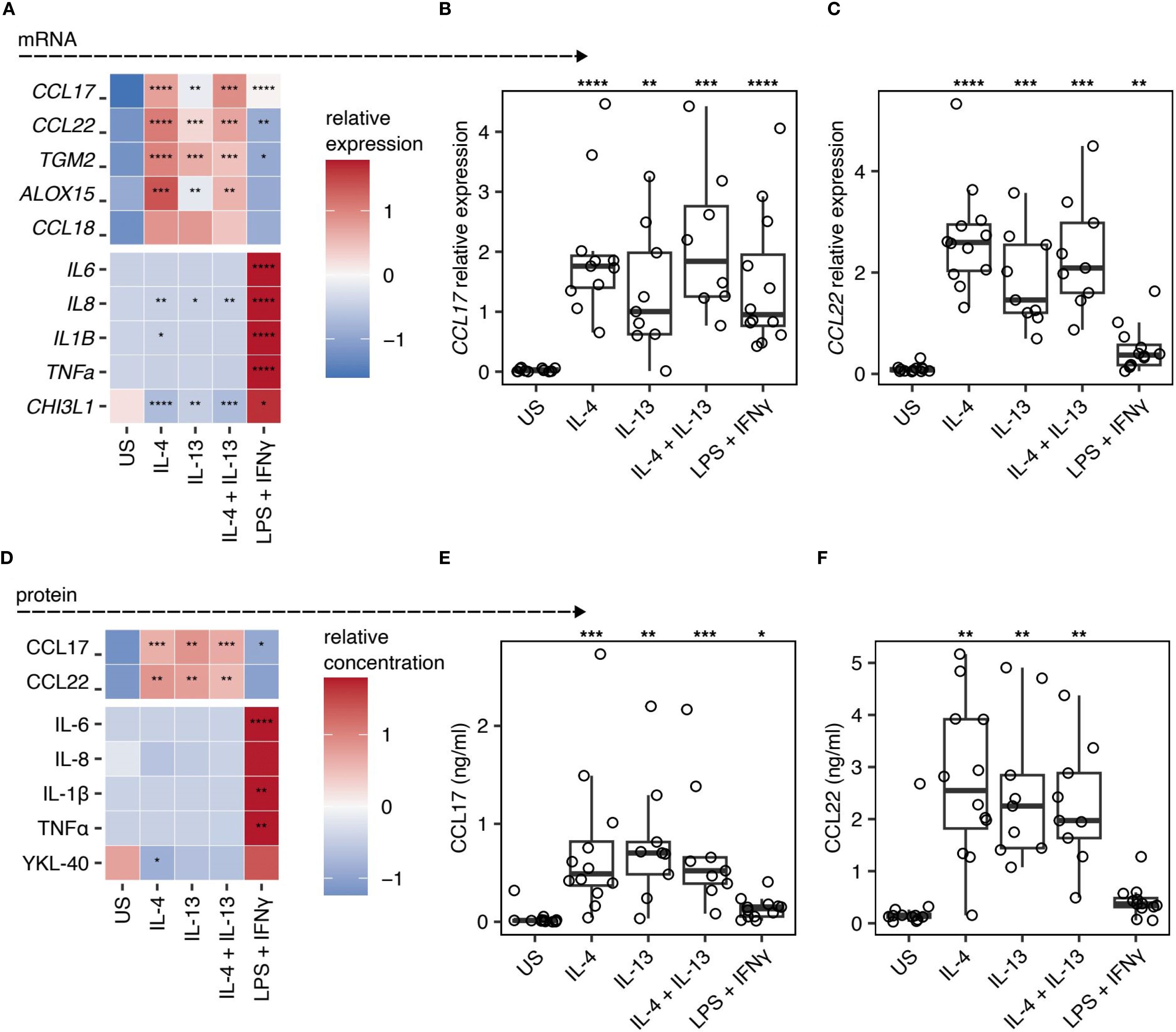
Figure 1. CCL17 and CCL22 is a marker of IL-4 and IL-13 stimulated ex vivo lung macrophages. (A-C) Quantification of mRNA expression in human lung macrophages following stimulation with IL-4, IL-13, IL-4 + IL-13, or LPS + IFNγ. Relative gene expression was normalised to HPRT using the 2-ΔCT method. US indicates unstimulated controls. (A) Heatmap representation of relative gene expression scaled by row. (B&C) Scatter plots showing the individual sample gene expression values for (B) CCL17 and (C) CCL22. Points represent individual patients. Box plots indicate the median and IQ range, whiskers indicate 1.5x the quartile limit. (D-F) Quantification of protein secretion into the supernatant of macrophages stimulated with IL-4, IL-13, IL-4 + IL-13, or LPS + IFNγ, measured by ELISA. (D) Heatmap showing the relative quantification of different proteins scaled to the row value. (E&F) Scatter plots showing the protein concentration of (E) CCL17 and (F) CCL22. Boxes indicate the median and IQ range, whiskers indicate 1.5x the quartile limit. Statistical values were calculated using Wilcoxon two-sample tests and adjusted for multiple testing using the Holm method. * indicates adjust P < 0.05, ** indicates adjust P < 0.01, *** indicates adjust P < 0.001, **** indicates adjust P < 0.0001.
Consistent with our mRNA expression data, we observed significant increases in CCL17 and CCL22 in supernatants of type 2 polarised macrophage cultures, but not type 1 polarised macrophages, which instead demonstrated elevated levels of IL-6, IL-8, IL-1β, TNFα, and YKL-40 (the product of CHI3L1) (Figures 1D–F). Despite being underpowered to perform statistics, we observed a trend for higher CCL17 protein in the supernatants of IL-4 and/or IL-13 stimulated lung macrophages isolated from our two asthmatic donors relative to the non-asthmatic donors (Supplementary Figure 2). All other macrophage responses to IL-4, IL-13, and combined IL-4/IL-13 stimulation fell within the range of responses seen in donors without asthma. Nevertheless, for the first time, we show that CCL17 represents a robust marker of IL-4 and IL-13 stimulated human lung macrophages, in addition to revealing CCL18 and TGM2 as being associated with IL-4 or IL-13 activated human lung macrophages.
Type 2 polarisation of human lung macrophages is metabolically dependent
In addition to increasing clarity surrounding type 2 human tissue macrophage markers, we have also addressed how these cells are governed metabolically. To determine whether human lung macrophages display specific metabolic dependence during type 2/type 1 polarisation, we added either 2-DG (a competitive inhibitor of glucose metabolism which blocks the glycolytic pathway (24)), or Eto (an inhibitor of fatty acid oxidation (FAO) (25)) (Figure 2A and Supplementary Figure 3).
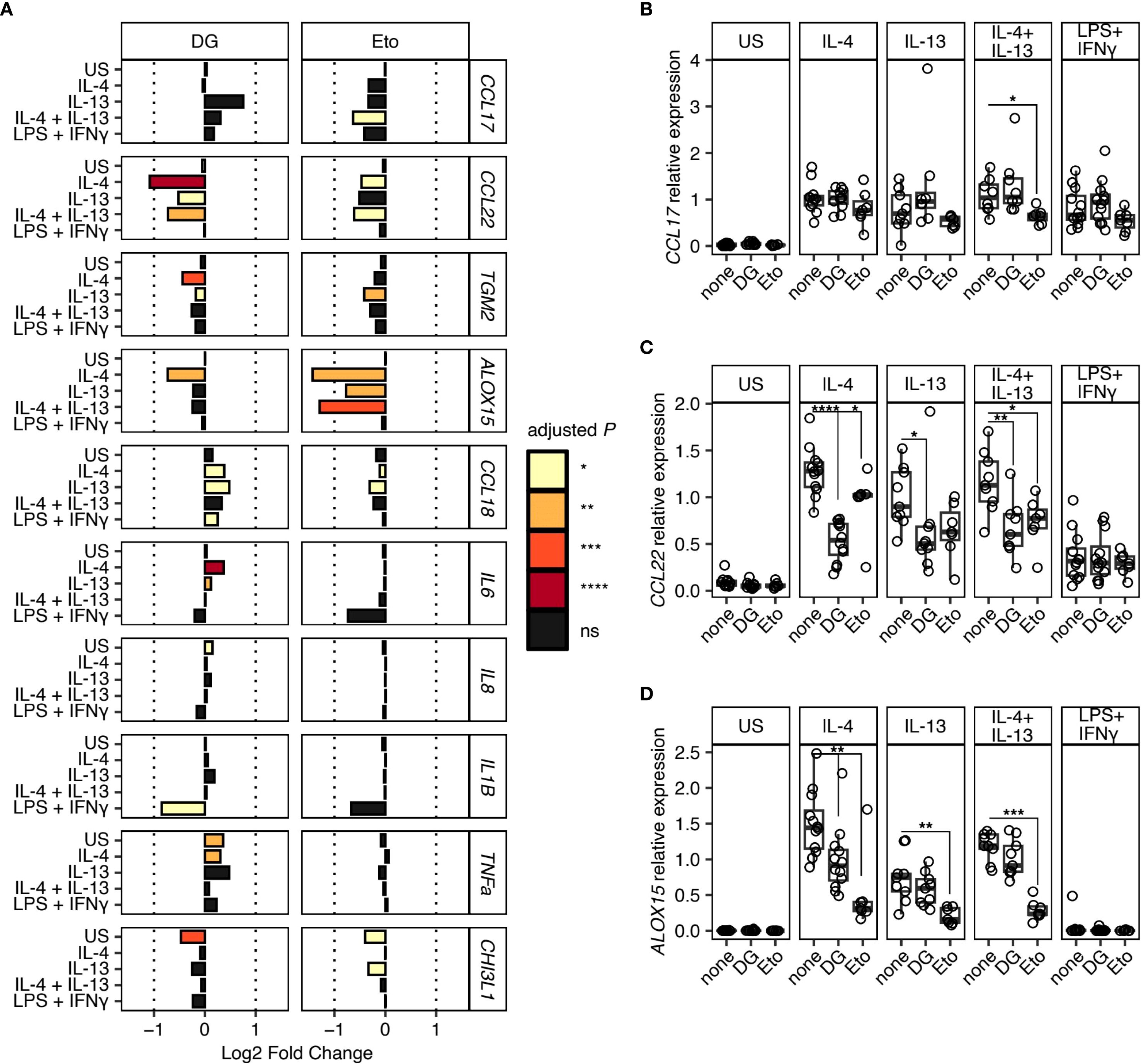
Figure 2. Metabolic dependence in gene expression during type 2 macrophage polarisation. Macrophages isolated from resected human lung tissue were stimulated with IL-4, IL-13, IL-4 + IL-13, or LPS + IFNγ and cultured in the presence of metabolic inhibitors, 2-Deoxy-D-glucose (2-DG) or Etomoxir (Eto). Gene expression was quantified by qPCR normalised against HPRT using the 2-ΔCT method. US indicates unstimulated controls. (A) Bar chart representing Log2 Fold Change in gene expression for each gene and stimulation condition. Fold change is expressed relative to the matched culture condition without metabolic inhibitor. Bar colour indicates the level of statistical significance. (B-D) Scatter plots showing relative expression of (B) CCL17, (C) CCL22, and (D) ALOX15 under different stimulation conditions with (2-DG & Eto) or without (none) metabolic inhibitors. Boxes indicate the median and IQ range, whiskers indicate 1.5x the quartile limit. Statistical values were calculated using Wilcoxon two-sample tests and adjusted for multiple testing using the Holm method. * indicates adjust P < 0.05, ** indicates adjust P < 0.01, *** indicates adjust P < 0.001, **** indicates adjust P < 0.0001.
Culture of human lung macrophages with 2-DG resulted in a significant reduction in the expression of CCL22 during IL-4 and IL-13 induced type 2 polarisation (Figures 2A, C), which strongly contrasted the limited impact of 2-DG on CCL17 expression (Figure 2B). Secretion of CCL22 into culture supernatants was also impaired under these conditions, although this only reached statistical significance following IL-4 stimulation (Supplementary Figure 3). Additionally, following culture with 2-DG, a small but significant reduction in ALOX15 expression was observed in the IL-4 stimulation condition only (Figures 2A, D). In contrast, Eto caused a significant reduction in ALOX15 expression under all type 2 polarisation conditions, with smaller reductions in CCL17, CCL22, TGM2 and CCL18 expression, which only reach statistical significance under specific type 2 stimulation conditions (Figures 2A, B). We also found that, despite displaying a robust LPS and IFNγ response (as evidenced by upregulated IL-6, IL-8, IL-1β, TNFα and YKL-40), addition of the glycolytic inhibitor 2-DG had no significant effect on human lung macrophage expression (Figure 2A) or secretion (Supplementary Figure 3) of these markers. Notably, like 2-DG, there was also a strong trend for reduced expression of IL1B along with IL6 mRNA following addition of Eto (Figure 2A), although the reverse was seen in terms of secretion (Supplementary Figure 3).
Together, these data promote key roles for both glycolysis and FAO in the polarisation of macrophages in a type 2 context, in a marker-specific manner. Specifically, whilst there was a lack of reliance on glycolysis for expression of CCL17 by type 2 activated human lung macrophages, there was a shared reliance on glycolysis and FAO for CCL22 expression (Figures 2A, C). Together, these data support a model in which expression of different genes stimulated by type 2 cytokines are dependent on distinct metabolic pathways.
Discussion
Despite murine models being widely used to study activation and polarisation of macrophages, clear markers for identification of type 2 (IL-4 and IL-13) activated human macrophages are lacking. This is mainly since translation of murine data to humans can be problematic, as many macrophage type 2 markers identified in mice (e.g., Chil3/Ym1) ( (16) have no direct human homologue, or such markers are differentially regulated in humans (e.g., Arginase 1) (17, 18), or the gene exists in humans but not mice (e.g., CCL18) (17, 18). These fundamental differences between mouse and human macrophages highlight the requirement for more markers common to both species to be identified, to enable more meaningful translational studies into macrophages during type 2 inflammatory diseases. At the same time, much of what we currently know about macrophage activation and function is reliant on use of model murine or human macrophages in vitro, such as BMDMs or MDMs, rather than those isolated ex vivo from relevant tissues.
Our work builds upon and extends previous limited human lung tissue macrophage studies to comprehensively identify a suite of markers - CCL17, CCL22, TGM2, ALOX15 and CCL18 - as features of type 2 (IL-4 and IL-13) activation of human pulmonary macrophages ex vivo. Of note, while both these cytokines are broadly classified as ‘type 2’ stimuli and share some receptor components, they signal through distinct receptors (IL-4Rα and IL-13Rα1) and may elicit heterogeneous macrophage responses (26). Nevertheless, here we demonstrate for the first time that CCL17 represents a robust marker of IL-4 and IL-13 stimulated human lung macrophages and confirm previous suggestions that CCL22 may also represent a type 2 human lung macrophage marker (27). Together, our data are in line with previous studies using IL-4 and IL-13 polarised human MDMs (28, 29) and murine BMDMs (30), that suggested that CCL17 (Figures 1B, E) and CCL22 (Figures 1C, F) represent type 2 macrophage markers common to both species. Of note, while CCL17 is strongly induced by IL-4 and IL-13, its expression is not entirely exclusive to type 2 stimulation but remains a robust marker in this context. For instance, CCL17 expression is also observed in several human type 1 inflammatory diseases, including ulcerative colitis, Crohn’s disease (31) and rheumatoid arthritis (32), emphasising that CCL17 can be present in other inflammatory contexts. From our data, it appears that CCL22 is a much more selective marker of type 2 immunity in human lung macrophages. Importantly, CCL22 is co-expressed alongside CCL17 during type 2 stimulation, making the combined expression of both chemokines a hallmark feature of type 2 immunity. Co-expression of CCL17 and CCL22 has been reported in human type 2 diseases such as in the bronchoalveolar fluid of asthmatics (33) and serum of atopic dermatitis patients (34). Our data extend these previous studies by revealing that macrophages represent a cellular source of these two chemokines in the human lung.
We included two donors in our study that had mild asthma. Despite a trend for CCL17 protein being higher in these two participants, all other macrophage responses to IL-4, IL-13, and combined IL-4/IL-13 stimulation fell within the range of responses seen in donors without asthma and did not skew the overall dataset, justifying their inclusion (Supplementary Figure 2).
Other type 2 activation markers we validated for human lung macrophages included ALOX15, CCL18 and TGM2, confirming ALOX15 as a consistent marker of type 2 activated human lung macrophages (27), while revealing CCL18 and TGM2 as being associated with IL-4 or IL-13 activated human lung macrophages (Figure 1A). This supports previous suggestions that isolated human BAL fluid macrophages (35) and human MDMs can express CCL18 in response to IL-4 or IL-13 (29), with the latter also upregulating TGM2 (28, 36, 37). Importantly, our finding that CCL18 is produced by type 2 stimulated human lung macrophages may be clinically relevant, since this chemokine is elevated in serum and BALF of idiopathic pulmonary fibrosis (IPF) (38) where is has been identified as an important biomarker to predict mortality (39). Further, CCL18 has also been found to be increased in bronchoalveolar lavage fluid and serum from allergic asthmatics (40).
In addition to increasing clarity surrounding type 2 human tissue macrophage markers, we have also addressed how these cells are governed metabolically. Of note, human lung macrophage expression of the type 2 marker CCL22 following IL-4 and IL-13 stimulation was strikingly reduced upon addition of the glycolytic inhibitor 2-DG (Figure 2C), which strongly contrasted the limited impact of 2-DG on CCL17 expression (Figure 2B). This reflects the fact that, despite CCL17 and CCL22 being known to share the same G-protein coupled receptor CCR4 (41, 42), they are distinct in terms of their structure (41, 43), receptor binding affinity (44), downstream signalling (45) and expression profiles (46–48). Further, this direct association between glycolytic metabolism and CCL22 production supports previous studies which have reported that diet can influence circulating levels of CCL22 and that its production in response to IL-4 by murine BMDMs is reliant on glutamine (49, 50). Our data also demonstrate for the first time that IL-4 induced ALOX15 mRNA expression by human lung macrophages is controlled by glycolysis and FAO (Figure 2D). Broadly, these data extend previous observations using murine BMDMs and imply that human lung macrophage responsiveness to IL-4 and IL-13 is regulated by both glycolysis and FAO (51–53).
It has been previously reported that FAO is dispensable for human MDM IL-4 responsiveness but not murine MDMs (9, 54). Consistent with this, we demonstrate that human lung macrophage expression of CCL17, CCL22, TGM2 and CCL18 in response to IL-4 and IL-13 was not significantly affected by addition of the FAO inhibitor Eto (Figure 2A). Notably, we did observe a trend for reduced CCL17 and CCL22 expression in this context, indicating a potential role for FAO (Figures 2B, C). Our results also demonstrate that expression of the type 2 marker ALOX15 by human lung macrophages cultured with IL-4 and IL-13 was significantly reduced following addition of Eto (Figure 2C), indicating a role for FAO. Looking at the type 2 markers on an individual basis, it is striking that they appear to be distinctly metabolically programmed, with this level of fine tuning likely to be relevant in the in vivo context.
Similarly to human MDMs (55), we have found that YKL-40, the human equivalent of the type 2 murine macrophage marker Ym1 (17), was upregulated by human lung macrophages in response to LPS, rather than IL-4 or IL-13 (Figure 1D). However, a role for YKL-40 (CHI3L1) during type 2 inflammatory disease cannot be ruled out as it has been reported to be elevated in the serum of asthmatics and associated with increased asthma severity (56, 57).
Our data confirm previous studies using human MDMs (28), human airway macrophages (7) and lung macrophages (58, 59) that IL-6, IL-8, IL-1β and TNFα represent type 1 activation markers (Figures 1A, D). We found that, despite displaying a robust LPS and IFNγ response (as evidenced by upregulated IL-6, IL-8, IL-1β, TNFα and YKL-40), addition of the glycolytic inhibitor 2-DG had no significant effect on human lung macrophage expression (Figure 2A) or secretion (Supplementary Figure 3) of these markers. Although not significant, our data suggests that glycolysis may play a role in regulating IL-1β expression and YKL-40 secretion in type 1 stimulated human lung macrophages, since there was a strong trend for reduced expression of these markers following addition of 2-DG (Figure 2A and Supplementary Figure 3). Whilst the lack of significant impact of 2-DG on these type 1 markers somewhat contradicts murine studies showing that glycolysis controls murine BMDMs responding to LPS (60, 61), it may indicate that glycolysis may be dispensable for LPS activated human MDMs. Consistent with this, one of the few immunometabolism studies to use human BAL fluid macrophages found that they failed to display metabolic reprogramming towards glycolysis after LPS stimulation (7), contrary to murine BMDMs (54, 62) and human MDMs (51). Notably, like 2-DG, there was also a strong trend for reduced expression of IL1B along with IL6 mRNA following addition of Eto (Figure 2A), although the reverse was seen in terms of secretion (Supplementary Figure 3).
It must be noted that a limitation of our study relates to the concentrations of metabolic inhibitors used, specifically Eto and 2-DG. Eto concentrations above 5 µM can lead to off-target effects, including inhibition of mitochondrial complex I (63) and disruption of Coenzyme A metabolism (64). However, we employed a concentration of 200 µM to ensure robust inhibition of FAO, consistent with prior studies in murine alveolar macrophages where this dose effectively blocked FAO-dependent pathways (65). Similarly, 2-DG was used at a concentration previously shown to inhibit glycolysis in murine BMDMs (52, 65, 66) and murine alveolar macrophages (65). Specifically, in these studies, 2-DG (1 mM) treatment inhibiting oxidative phosphorylation, as determined using a Seahorse analyser (52, 65) and significantly reduced Glut1, a molecule in the glycolysis signalling pathway during type 1 (LPS induced) macrophage differentiation (67). It has also been shown that 1 mM 2-DG treatment inhibits glucose consumption and lactate production in BMDMs (66). Similarly, it has been shown using the murine macrophage cell line RAW264.7, that glucose consumption significantly decreases after 1 mM 2-DG treatment (68). Further, our choice of inhibitor concentration was constrained, since the limited number of cells we could isolate from human lung tissue samples precluded full dose-titration experiments. While these concentrations allowed us to interrogate the metabolic dependencies of macrophage activation, we recognise the potential for off-target effects. Thus, future studies employing inhibitor dose-titration, Seahorse, SCENITH (69) or genetic approaches such as siRNA knockdown of key metabolic enzymes will be essential to build upon our results and further delineate the roles of glycolysis and FAO in regulating human macrophage function.
We and others have previously highlighted the use of tissue macrophages for metabolic profiling, noting that BMDMs and MDMs in vitro behave differently and exhibit different metabolic phenotypes to murine or human tissue macrophages ex vivo (61, 65, 70). Building on these earlier observations, our current data further demonstrate the importance of using human tissue macrophages to address paradigms in metabolic requirements for activation and function that have previously been constructed using MDMs or murine macrophages. We show that human tissue macrophage metabolic programming is not as simple as once thought, and requires further delineation, as glycolysis and FAO likely play complementary roles in specific aspects of both type 1 and type 2 human macrophage polarisation. This supports a recent study which showed glycolysis supports cytokine production and functional activation in human airway macrophages polarised with IFNγ, while in MDMs, it is more crucial for upregulating activation markers with IFN-γ but less so for cytokine secretion, and both cell types show reduced glycolytic dependence under IL-4 polarisation (70). Together, our data highlights the critical role of the tissue environment in controlling human macrophage responsiveness to cytokines, a principle that should be considered when interpreting data generated in murine studies or using model human macrophages, given that impaired macrophage metabolism is associated with many human diseases (71–73).
Lung macrophage heterogeneity is complex, consisting of both alveolar and interstitial subsets (74–76), and we and others have previously demonstrated that murine alveolar macrophages show differential metabolism and responsiveness to polarising stimuli than interstitial macrophages (65, 77). Thus, we anticipate that IL-4 and IL-13 induced CCL17 and CCL22 expression by human alveolar and interstitial macrophages may differ, along with their metabolic profiles. This underscores the need for future studies to unequivocally distinguish human airway alveolar macrophages from tissue interstitial macrophages, to be able to address such possibilities experimentally. Although notably, resolving human macrophage population-specific responses remains a challenge since well-defined markers are still lacking and autofluorescence can complicate analysis (1, 78).
In summary, our data show that CCL17 and CCL22 along with TGM2, ALOX15 and CCL18, represent reliable markers that can be used to identify and distinguish type 2 (IL-4 and IL-13) from type 1 (LPS and IFNγ) activated human lung macrophages. Further, these novel data increase our understanding of how metabolism governs pulmonary macrophage responsiveness and activation, since we reveal that type 2 macrophage markers (including CCL22 and ALOX15) are significantly regulated by glycolysis and FAO, respectively. Our data emphasise the importance for studies on human tissue macrophages ex vivo, to reduce reliance on in vitro assays and murine models that may fail to represent the human lung environment. We suggest that CCL17 and CCL22 represent useful human type 2 tissue macrophage markers that may provide potential therapeutic targets for future treatments targeting such mediators or the metabolic pathways that control them in pulmonary type 2 associated inflammatory lung disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving humans were approved by National Research Ethics Service Committee; North West - Haydock. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
AR: Writing – review & editing, Methodology, Writing – original draft, Data curation, Investigation, Conceptualization. AC: Data curation, Methodology, Investigation, Writing – review & editing. SC: Formal analysis, Writing – review & editing, Visualization. JH: Writing – review & editing. DD: Writing – review & editing. AS: Resources, Writing – review & editing. LB: Writing – review & editing. MF: Writing – review & editing. PC: Writing – review & editing. AM: Writing – review & editing, Supervision, Conceptualization, Writing – original draft, Funding acquisition, Resources.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. AR was supported by a MRC CASE studentship (with GSK) (MR/N013751/1). AM was supported by funding from GSK, the Lydia Becker Institute and the MRC (MR/W018748/1). PCC was funded from the MRC Centre for Medical Mycology at the University of Exeter (MR/N006364/2 and MR/V033417/1), the NIHR Exeter Biomedical Research Centre (NIHR203320) and a Wellcome Trust Sir Henry Dale Fellowship (218550/Z/19/Z). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Acknowledgments
This report is independent research supported by the North West Lung Centre Charity at Manchester University NHS Foundation Trust. The authors would like to acknowledge the Manchester Allergy, Respiratory and Thoracic Surgery Biobank and the North West Lung Centre Charity for supporting this project. In addition, we would like to thank the study participants for their contribution.
Conflict of interest
MF: employee of GSK and hold stock or stock options.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor SF declared a past co-authorship with the author AS.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the North West Lung Centre Charity or the Department of Health.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1654717/full#supplementary-material
References
1. Bain CC and MacDonald AS. The impact of the lung environment on macrophage development, activation and function: diversity in the face of adversity. Mucosal Immunol. (2022) 15:223–34. doi: 10.1038/s41385-021-00480-w
2. Burgy O, Loriod S, Beltramo G, and Bonniaud P. Extracellular lipids in the lung and their role in pulmonary fibrosis. Cells. (2022) 11(7):1209. doi: 10.3390/cells11071209
3. Rajesh R, Atallah R, and Bärnthaler T. Dysregulation of metabolic pathways in pulmonary fibrosis. Pharmacol Ther. (2023) 246:108436. doi: 10.1016/j.pharmthera.2023.108436
4. Gill SK, Hui K, Farne H, Garnett JP, Baines DL, Moore LS, et al. Increased airway glucose increases airway bacterial load in hyperglycaemia. Sci Rep. (2016) 6:27636. doi: 10.1038/srep27636
5. Baker EH and Baines DL. Airway glucose homeostasis: A new target in the prevention and treatment of pulmonary infection. Chest. (2018) 153:507–14. doi: 10.1016/j.chest.2017.05.031
6. Van den Bossche J, O'Neill LA, and Menon D. Macrophage immunometabolism: where are we (Going)? Trends Immunol. (2017) 38:395–406. doi: 10.1016/j.it.2017.03.001
7. Lavrich KS, Speen AM, Ghio AJ, Bromberg PA, Samet JM, and Alexis NE. Macrophages from the upper and lower human respiratory tract are metabolically distinct. Am J Physiol Lung Cell Mol Physiol. (2018) 315:L752–64. doi: 10.1152/ajplung.00208.2018
8. Lawrence E, Sims J, Gander A, Garibaldi JM, Fuller B, Davidson B, et al. The barriers and motivators to using human tissues for research: the views of UK-based biomedical researchers. Biopreserv Biobank. (2020) 18:266–73. doi: 10.1089/bio.2019.0138
9. Namgaladze D and Brüne B. Fatty acid oxidation is dispensable for human macrophage IL-4-induced polarization. Biochim Biophys Acta. (2014) 1841:1329–35. doi: 10.1016/j.bbalip.2014.06.007
10. Van den Bossche J, Baardman J, Otto NA, van der Velden S, Neele AE, van den Berg SM, et al. Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Rep. (2016) 17:684–96. doi: 10.1016/j.celrep.2016.09.008
11. Vijayan V, Pradhan P, Braud L, Fuchs HR, Gueler F, Motterlini R, et al. Human and murine macrophages exhibit differential metabolic responses to lipopolysaccharide - A divergent role for glycolysis. Redox Biol. (2019) 22:101147. doi: 10.1016/j.redox.2019.101147
12. Byrne AJ, Powell JE, O'Sullivan BJ, Ogger PP, Hoffland A, Cook J, et al. Dynamics of human monocytes and airway macrophages during healthy aging and after transplant. J Exp Med. (2020) 217(3):e20191236. doi: 10.1084/jem.20191236
13. Leach SM, Gibbings SL, Tewari AD, Atif SM, Vestal B, Danhorn T, et al. Human and mouse transcriptome profiling identifies cross-species homology in pulmonary and lymph node mononuclear phagocytes. Cell Rep. (2020) 33:108337. doi: 10.1016/j.celrep.2020.108337
14. Fabre K, Berridge B, Proctor WR, Ralston S, Will Y, Baran SW, et al. Introduction to a manuscript series on the characterization and use of microphysiological systems (MPS) in pharmaceutical safety and ADME applications. Lab Chip. (2020) 20:1049–57. doi: 10.1039/C9LC01168D
15. Ingber DE. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat Rev Genet. (2022) 23:467–91. doi: 10.1038/s41576-022-00466-9
16. Raes G, De Baetselier P, Noël W, Beschin A, Brombacher F, and Hassanzadeh Gh G. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J Leukoc Biol. (2002) 71:597–602. doi: 10.1189/jlb.71.4.597
17. Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH, Scotton C, Locati M, et al. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J Immunol. (2005) 174:6561; author reply 6561–2. doi: 10.4049/jimmunol.174.11.6561
18. Hieshima K, Imai T, Baba M, Shoudai K, Ishizuka K, Nakagawa T, et al. A novel human CC chemokine PARC that is most homologous to macrophage-inflammatory protein-1 alpha/LD78 alpha and chemotactic for T lymphocytes, but not for monocytes. J Immunol. (1997) 159:1140–9. doi: 10.4049/jimmunol.159.3.1140
19. Cohn ZA. The isolation and cultivation of mononuclear phagocytes. Methods Enzymol. (1974) 32:758–65. doi: 10.1016/0076-6879(74)32079-4
20. Connolly E, Morgan DJ, Franklin M, Simpson A, Shah R, Brand OJ, et al. Neurturin regulates the lung-resident macrophage inflammatory response to viral infection. Life Sci Alliance. (2020) 3(12):E202000780. doi: 10.26508/lsa.202000780
21. Evans BJ, Haskard DO, Sempowksi G, and Landis RC. Evolution of the macrophage CD163 phenotype and cytokine profiles in a human model of resolving inflammation. Int J Inflam. (2013) 2013:780502. doi: 10.1155/2013/780502
22. Bharat A, Bhorade SM, Morales-Nebreda L, McQuattie-Pimentel AC, Soberanes S, Ridge K, et al. Flow cytometry reveals similarities between lung macrophages in humans and mice. Am J Respir Cell Mol Biol. (2016) 54:147–9. doi: 10.1165/rcmb.2015-0147LE
23. Desch AN, Gibbings SL, Goyal R, Kolde R, Bednarek J, Bruno T, et al. Flow cytometric analysis of mononuclear phagocytes in nondiseased human lung and lung-draining lymph nodes. Am J Respir Crit Care Med. (2016) 193:614–26. doi: 10.1164/rccm.201507-1376OC
24. Pajak B, Siwiak E, Sołtyka M, Priebe A, Zieliński R, Fokt I, et al. 2-deoxy-d-glucose and its analogs: from diagnostic to therapeutic agents. Int J Mol Sci. (2019) 21(1):234. doi: 10.3390/ijms21010234
25. Ma Y, Wang W, Devarakonda T, Zhou H, Wang XY, Salloum FN, et al. Functional analysis of molecular and pharmacological modulators of mitochondrial fatty acid oxidation. Sci Rep. (2020) 10:1450. doi: 10.1038/s41598-020-58334-7
26. Junttila IS. Tuning the cytokine responses: an update on interleukin (IL)-4 and IL-13 receptor complexes. Front Immunol. (2018) 9:888. doi: 10.3389/fimmu.2018.00888
27. Abrial C, Grassin-Delyle S, Salvator H, Brollo M, Naline E, and Devillier P. 15-Lipoxygenases regulate the production of chemokines in human lung macrophages. Br J Pharmacol. (2015) 172:4319–30. doi: 10.1111/bph.13210
28. Unuvar Purcu D, Korkmaz A, Gunalp S, Helvaci DG, Erdal Y, Dogan Y, et al. Effect of stimulation time on the expression of human macrophage polarization markers. PloS One. (2022) 17:e0265196. doi: 10.1371/journal.pone.0265196
29. Scott TE, Lewis CV, Zhu M, Wang C, Samuel CS, Drummond GR, et al. IL-4 and IL-13 induce equivalent expression of traditional M2 markers and modulation of reactive oxygen species in human macrophages. Sci Rep. (2023) 13:19589. doi: 10.1038/s41598-023-46237-2
30. Czimmerer Z, Halasz L, Daniel B, Varga Z, Bene K, Domokos A, et al. The epigenetic state of IL-4-polarized macrophages enables inflammatory cistromic expansion and extended synergistic response to TLR ligands. Immunity. (2022) 55:2006–2026.e6. doi: 10.1016/j.immuni.2022.10.004
31. Jugde F, Alizadeh M, Boissier C, Chantry D, Siproudhis L, Corbinais S, et al. Quantitation of chemokines (MDC, TARC) expression in mucosa from Crohn's disease and ulcerative colitis. Eur Cytokine Netw. (2001) 12:468–77.
32. Radstake TR, van der Voort R, ten Brummelhuis M, de Waal Malefijt M, Looman M, Figdor CG, et al. Increased expression of CCL18, CCL19, and CCL17 by dendritic cells from patients with rheumatoid arthritis, and regulation by Fc gamma receptors. Ann Rheum Dis. (2005) 64:359–67. doi: 10.1136/ard.2003.017566
33. Pilette C, Francis JN, Till SJ, and Durham SR. CCR4 ligands are up-regulated in the airways of atopic asthmatics after segmental allergen challenge. Eur Respir J. (2004) 23:876–84. doi: 10.1183/09031936.04.00102504
34. Shimada Y, Takehara K, and Sato S. Both Th2 and Th1 chemokines (TARC/CCL17, MDC/CCL22, and Mig/CXCL9) are elevated in sera from patients with atopic dermatitis. J Dermatol Sci. (2004) 34:201–8. doi: 10.1016/j.jdermsci.2004.01.001
35. Pechkovsky DV, Prasse A, Kollert F, Engel KM, Dentler J, Luttmann W, et al. Alternatively activated alveolar macrophages in pulmonary fibrosis-mediator production and intracellular signal transduction. Clin Immunol. (2010) 137:89–101. doi: 10.1016/j.clim.2010.06.017
36. Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. (2013) 121:e57–69. doi: 10.1182/blood-2012-06-436212
37. Yamaguchi M, Zacharia J, Laidlaw TM, and Balestrieri B. PLA2G5 regulates transglutaminase activity of human IL-4-activated M2 macrophages through PGE2 generation. J Leukoc Biol. (2016) 100:131–41. doi: 10.1189/jlb.3A0815-372R
38. Cai M, Bonella F, He X, Sixt SU, Sarria R, Guzman J, et al. CCL18 in serum, BAL fluid and alveolar macrophage culture supernatant in interstitial lung diseases. Respir Med. (2013) 107(9):1444–52.
39. Prasse A, Probst C, Bargagli E, Zissel G, Toews GB, Flaherty KR, et al. “Serum CC-chemokine ligand 18 concentration predicts outcome in idiopathic pulmonary fibrosis”, Am J Respir Crit Care Med. (2009) 179(8):717–23.
40. de Nadaï P, Charbonnier AS, Chenivesse C, Sénéchal S, Fournier C, Gilet J, et al. Involvement of CCL18 in allergic asthma. J Immunol. (2006) 176(10):6286–93.
41. Imai T, Baba M, Nishimura M, Kakizaki M, Takagi S, and Yoshie O. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J Biol Chem. (1997) 272:15036–42. doi: 10.1074/jbc.272.23.15036
42. Imai T, Chantry D, Raport CJ, Wood CL, Nishimura M, Godiska R, et al. Macrophage-derived chemokine is a functional ligand for the CC chemokine receptor 4. J Biol Chem. (1998) 273:1764–8. doi: 10.1074/jbc.273.3.1764
43. Imai T, Yoshida T, Baba M, Nishimura M, Kakizaki M, and Yoshie O. Molecular cloning of a novel T cell-directed CC chemokine expressed in thymus by signal sequence trap using Epstein-Barr virus vector. J Biol Chem. (1996) 271:21514–21. doi: 10.1074/jbc.271.35.21514
44. Mariani M, Lang R, Binda E, Panina-Bordignon P, and D'Ambrosio D. Dominance of CCL22 over CCL17 in induction of chemokine receptor CCR4 desensitization and internalization on human Th2 cells. Eur J Immunol. (2004) 34:231–40. doi: 10.1002/eji.200324429
45. Ajram L, Begg M, Slack R, Cryan J, Hall D, Hodgson S, et al. Internalization of the chemokine receptor CCR4 can be evoked by orthosteric and allosteric receptor antagonists. Eur J Pharmacol. (2014) 729:75–85. doi: 10.1016/j.ejphar.2014.02.007
46. Campbell JJ, Haraldsen G, Pan J, Rottman J, Qin S, Ponath P, et al. The chemokine receptor CCR4 in vascular recognition by cutaneous but not intestinal memory T cells. Nature. (1999) 400:776–80. doi: 10.1038/23495
47. Horikawa T, Nakayama T, Hikita I, Yamada H, Fujisawa R, Bito T, et al. IFN-gamma-inducible expression of thymus and activation-regulated chemokine/CCL17 and macrophage-derived chemokine/CCL22 in epidermal keratinocytes and their roles in atopic dermatitis. Int Immunol. (2002) 14:767–73. doi: 10.1093/intimm/dxf044
48. Zheng X, Nakamura K, Furukawa H, Nishibu A, Takahashi M, Tojo M, et al. Demonstration of TARC and CCR4 mRNA expression and distribution using in situ RT-PCR in the lesional skin of atopic dermatitis. J Dermatol. (2003) 30:26–32. doi: 10.1111/j.1346-8138.2003.tb00329.x
49. Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. (2015) 42:419–30. doi: 10.1016/j.immuni.2015.02.005
50. Petersen PS, Wolf RM, Lei X, Peterson JM, and Wong GW. Immunomodulatory roles of CTRP3 in endotoxemia and metabolic stress. Physiol Rep. (2016) 4(5):e12735. doi: 10.14814/phy2.12735
51. Huang SC, Everts B, Ivanova Y, O'Sullivan D, Nascimento M, Smith AM, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. (2014) 15:846–55. doi: 10.1038/ni.2956
52. Tan Z, Xie N, Cui H, Moellering DR, Abraham E, Thannickal VJ, et al. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J Immunol. (2015) 194:6082–9. doi: 10.4049/jimmunol.1402469
53. Covarrubias AJ, Aksoylar HI, Yu J, Snyder NW, Worth AJ, Iyer SS, et al. Akt-mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. Elife. (2016) 5:E11612. doi: 10.7554/eLife.11612
54. Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. (2006) 4:13–24. doi: 10.1016/j.cmet.2006.05.011
55. Bonneh-Barkay D, Bissel SJ, Kofler J, Starkey A, Wang G, and Wiley CA. Astrocyte and macrophage regulation of YKL-40 expression and cellular response in neuroinflammation. Brain Pathol. (2012) 22(4):530–46.
56. Lee CG and Elias JA. Role of breast regression protein-39/YKL-40 in asthma and allergic responses. Allergy Asthma Immunol Res. (2010) 2:20–7. doi: 10.4168/aair.2010.2.1.20
57. Tang H, Fang Z, Sun Y, Li B, Shi Z, Chen J, et al. YKL-40 in asthmatic patients, and its correlations with exacerbation, eosinophils and immunoglobulin E. Eur Respir J. (2010) 35:757–60. doi: 10.1183/09031936.00034409
58. Victoni T, Salvator H, Abrial C, Brollo M, Porto LCS, Lagente V, et al. Human lung and monocyte-derived macrophages differ with regard to the effects of β. Respir Res. (2017) 18:126. doi: 10.1186/s12931-017-0613-y
59. Ferrara AL, Galdiero MR, Fiorelli A, Cristinziano L, Granata F, Marone G, et al. Macrophage-polarizing stimuli differentially modulate the inflammatory profile induced by the secreted phospholipase A. Cytokine. (2021) 138:155378. doi: 10.1016/j.cyto.2020.155378
60. Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. (2013) 496:238–42. doi: 10.1038/nature11986
61. Woods PS, Kimmig LM, Meliton AY, Sun KA, Tian Y, O'Leary EM, et al. Tissue-resident alveolar macrophages do not rely on glycolysis for LPS-induced inflammation. Am J Respir Cell Mol Biol. (2020) 62:243–55. doi: 10.1165/rcmb.2019-0244OC
62. Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. (2015) 21:65–80. doi: 10.1016/j.cmet.2014.12.005
63. O'Connor RS, Guo L, Ghassemi S, Snyder NW, Worth AJ, Weng L, et al. The CPT1a inhibitor, etomoxir induces severe oxidative stress at commonly used concentrations. Sci Rep. (2018) 8:6289. doi: 10.1038/s41598-018-24676-6
64. Divakaruni AS, Hsieh WY, Minarrieta L, Duong TN, Kim KKO, Desousa BR, et al. Etomoxir inhibits macrophage polarization by disrupting coA homeostasis. Cell Metab. (2018) 28:490–503.e7. doi: 10.1016/j.cmet.2018.06.001
65. Svedberg FR, Brown SL, Krauss MZ, Campbell L, Sharpe C, Clausen M, et al. The lung environment controls alveolar macrophage metabolism and responsiveness in type 2 inflammation. Nat Immunol. (2019) 20:571–80. doi: 10.1038/s41590-019-0352-y
66. Zhao Q, Chu Z, Zhu L, Yang T, Wang P, Liu F, et al. 2-Deoxy-d-Glucose Treatment Decreases Anti-inflammatory M2 Macrophage Polarization in Mice with Tumor and Allergic Airway Inflammation. Front Immunol. (2017) 8:637.
67. Yu Q, Wang Y, Dong L, He Y, Liu R, Yang Q, et al. Regulations of Glycolytic Activities on Macrophages Functions in Tumor and Infectious Inflammation. Front Cell Infect Microbiol. (2020) 10:287.
68. Kaushik N, Lee SJ, Choi TG, Baik KY, Uhm HS, Kim CH, et al. Non-thermal plasma with 2-deoxy-D-glucose synergistically induces cell death by targeting glycolysis in blood cancer cells. Sci Rep. (2015) 5:8726.
69. Argüello RJ, Combes AJ, Char R, Gigan JP, Baaziz AI, Bousiquot E, et al. SCENITH: A flow cytometry-based method to functionally profile energy metabolism with single-cell resolution. Cell Metab. (2020) 32:1063–1075.e7. doi: 10.1016/j.cmet.2020.11.007
70. Cox DJ, Connolly SA, Ó Maoldomhnaigh C, Brugman AAI, Sandby Thomas O, Duffin E, et al. Human airway macrophages are metabolically reprogrammed by IFN-γ resulting in glycolysis-dependent functional plasticity. Elife. (2024) 13:RP98449. doi: 10.7554/eLife.98449
71. Uranga RM and Keller JN. The complex interactions between obesity, metabolism and the brain. Front Neurosci. (2019) 13:513. doi: 10.3389/fnins.2019.00513
72. Frezza C. Metabolism and cancer: the future is now. Br J Cancer. (2020) 122:133–5. doi: 10.1038/s41416-019-0667-3
73. Sun Y, Ma C, Sun H, Wang H, Peng W, Zhou Z, et al. Metabolism: A novel shared link between diabetes mellitus and alzheimer's disease. J Diabetes Res. (2020) 2020:4981814. doi: 10.1155/2020/4981814
74. Chakarov S, Lim HY, Tan L, Lim SY, See P, Lum J, et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science. (2019) 363(6432):Eaau0964. doi: 10.1126/science.aau0964
75. Schyns J, Bai Q, Ruscitti C, Radermecker C, De Schepper S, Chakarov S, et al. Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat Commun. (2019) 10:3964. doi: 10.1038/s41467-019-11843-0
76. Li X, Kolling FW, Aridgides D, Mellinger D, Ashare A, and Jakubzick CV. ScRNA-seq expression of. Life Sci Alliance. (2022) 5(11):E202201458. doi: 10.26508/lsa.202201458
77. Pisu D, Huang L, Grenier JK, and Russell DG. Dual RNA-seq of mtb-infected macrophages in vivo reveals ontologically distinct host-pathogen interactions. Cell Rep. (2020) 30:335–350.e4. doi: 10.1016/j.celrep.2019.12.033
Keywords: macrophages, lung, tissue, metabolism, humans
Citation: Ridley AJL, Curle AJ, Colombo SAP, Hughes JJ, Dyer DP, Simpson A, Booty LM, Feeney M, Cook PC and MacDonald AS (2025) The chemokines CCL22 and CCL17 are a defining feature of type 2 stimulated human lung macrophages and exhibit different metabolic dependencies. Front. Immunol. 16:1654717. doi: 10.3389/fimmu.2025.1654717
Received: 26 June 2025; Accepted: 15 September 2025;
Published: 13 October 2025.
Edited by:
Susetta Finotto, Universitätsklinikum Erlangen, GermanyCopyright © 2025 Ridley, Curle, Colombo, Hughes, Dyer, Simpson, Booty, Feeney, Cook and MacDonald. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew S. MacDonald, QW5kcmV3Lk1hY0RvbmFsZEBlZC5hYy51aw==
 Amanda J. L. Ridley
Amanda J. L. Ridley Annabel J. Curle1
Annabel J. Curle1 Stefano A. P. Colombo
Stefano A. P. Colombo Douglas P. Dyer
Douglas P. Dyer Maria Feeney
Maria Feeney Peter C. Cook
Peter C. Cook Andrew S. MacDonald
Andrew S. MacDonald