- 1Assisted Reproductive Technology Center, The Affiliated Foshan Women and Children Hospital, Guangdong Medical University, Foshan, China
- 2Hunan Provincial Key Laboratory of the Research and Development of Novel Pharmaceutical Preparations, Changsha Medical University, Changsha, China
- 3Department of Obstetrics and Gynecology, Guangdong Provincial Key Laboratory of Major Obstetric Diseases; Guangdong Provincial Clinical Research Center for Obstetrics and Gynecology; Guangdong-Hong Kong-Macao Greater Bay Area Higher Education Joint Laboratory of Maternal-Fetal Medicine; The Third Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
- 4Department of Obstetrics and Gynecology, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, Guangdong, China
- 5Department of Obstetrics and Gynecology, Center for Reproductive Medicine; Guangdong Provincial Key Laboratory of Major Obstetric Diseases; Guangdong Provincial Clinical Research Center for Obstetrics and Gynecology; Guangdong-Hong Kong-Macao Greater Bay Area Higher Education Joint Laboratory of Maternal-Fetal Medicine; The Third Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
- 6Department of Obstetrics, First Affiliated Hospital of Jinan University, Guangzhou, China
- 7Department of Obstetrics and Gynecology, Guangzhou Women and Children’s Medical Center, Guangzhou Medical, Guangzhou, China
- 8Department of Obstetrics and Gynecology, Guangdong Provincial Key Laboratory of Major Obstetric Diseases, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
Background: Recurrent spontaneous abortion (RSA) significantly impacts women’s health, yet the underlying biological mechanisms remain poorly defined. Understanding the molecular contributors to RSA is crucial for developing targeted interventions.
Objective: This study aims to investigate the causal relationships between plasma proteins and RSA, focusing on the identification of potential therapeutic targets through multi-omic approaches.
Methods: We utilized two-sample Mendelian randomization (MR) analyses integrating genome-wide association study (GWAS) data for both plasma proteins and RSA. Proteomic data were sourced from the UK Biobank-Plasma Proteome Project and deCODE Health Study. We further validated our findings through both bulk and single-cell RNA sequencing of clinical specimens, alongside quantitative real-time polymerase chain reaction and immunohistochemistry. A phenome-wide association study was also conducted to assess the safety and broader implications of identified targets.
Results: Our analyses identified the interleukin 6 receptor (IL6R) as a key candidate, with elevated plasma levels correlating with increased RSA risk. Furthermore, IL6R was found to be upregulated in RSA-related endometrial and decidual tissues. The phenome-wide association study provided insights into potential side effects and additional therapeutic indications for IL6R.
Conclusion: IL6R upregulation is mechanistically implicated in the pathogenesis of RSA, establishing it as a validated causal biomarker and a potentially actionable therapeutic target. This study not only highlights the role of IL6R in RSA but also supports its development into a therapeutic strategy with a comprehensive safety profile.
Introduction
Recurrent spontaneous abortion (RSA), characterized by two or more consecutive miscarriages before 24 weeks of gestation with the same sexual partner, represents a significant challenge to women’s reproductive health (1, 2). Its incidence is estimated at 1% to 5% in the general population (1). RSA has a multifactorial etiology, including chromosomal abnormalities, uterine anatomical defects, autoimmune conditions, and endometrial dysfunction (3). Despite extensive investigation, the underlying biological mechanisms remain poorly understood, limiting effective clinical interventions. RSA not only increases the risk of adverse outcomes in subsequent pregnancies but also elevates the likelihood of long-term metabolic and cardiovascular complications, thereby substantially affecting women’s physical and psychological well-being (4–7).
Recently, plasma proteins are vital in physiological and pathological processes and are increasingly investigated in the context of diseases such as diabetes (8) and ankylosing spondylitis (9). Abnormal plasma protein levels, such as elevated antiphospholipid antibodies, are associated with a higher risk of RSA (10). Proteins, as downstream products of metabolic activities and modifiable entities through dietary or pharmacological means, act as vital biomarkers and prospective therapeutic targets for various disorders (11). Employing proteins intermediate phenotypes might provide crucial understanding of the mechanisms connecting plasma proteins with RSA. Nonetheless, results from observational research are frequently obscured by environmental influences, which constrain the ability to make strong causal determinations. Mendelian randomization (MR) offers a methodological advantage by using genetic variants as instrumental variables (IVs) to infer causality. By leveraging the random allocation of alleles during meiosis, akin to randomization in clinical trials, MR minimizes confounding and reverse causation, offering more robust evidence of causal relationships.
Increasing evidence underscores the importance of endometrial microenvironment remodeling during days 19–21 of the menstrual cycle, corresponding to the implantation window (LH + 7 to +9), in supporting successful implantation and pregnancy (12). This phase is marked by a highly receptive state, conducive to embryo implantation, both biochemically and structurally (13). Transcriptomic analyses during this window reveal dynamic shifts in immune tolerance, vascular remodeling, and steroid hormone responsiveness, with aberrations in these processes strongly implicated in RSA pathogenesis (1, 13–16). However, it remains unclear whether these microenvironmental changes are causally driven by systemic biomarker alterations or reflect localized endometrial dysfunction. To address this gap, we propose an integrative causal inference framework that combines population-level protein quantitative trait locus (pQTL) data with transcriptomic profiling of the endometrium during the implantation window. This approach employs MR to differentiate between systemic and tissue-specific causal pathways, while simultaneously identifying druggable targets supported by both genetic and functional evidence.
Ultimately, this study aims to advance our understanding of RSA pathogenesis and help in formulating targeted clinical strategies.
Materials and methods
Study design
Comprehensive genome-wide association study (GWAS) summary data were derived from the original investigations wherein all participants had provided informed consent. We adhered to the STROBE-MR guidelines to ensure transparent and comprehensive reporting of our MR study (17), which included providing details on the study design, data sources, and the assumptions underlying the MR analysis. Initially, we employed two independent two-sample MR analyses to assess the causal effects of plasma proteins from the UK Biobank and Icelandic cohorts on RSA. Subsequently, proteins displaying concordant causal effects (P < 0.05 in both cohorts) were intersected with differentially expressed genes (DEGs) identified from RSA case-control RNA sequencing (RNA-seq) data using Ensembl gene ID matching. Consequently, the interleukin 6 receptor (IL6R) was identified as a unique candidate protein. Single-cell RNA-seq (scRNA-seq) analysis further examined the expression of IL6R in both the RSA group and the control group. Finally, a phenome-wide association study (PheWAS) was conducted to comprehensively evaluate the role of IL6R across diverse phenotypes, to evaluate the safety implications and therapeutic potential of targeting IL6R for RSA treatment, as well as to explore its potential as a therapeutic target. Figure 1 depicts a flowchart that systematically outlines the research process employed in this study.
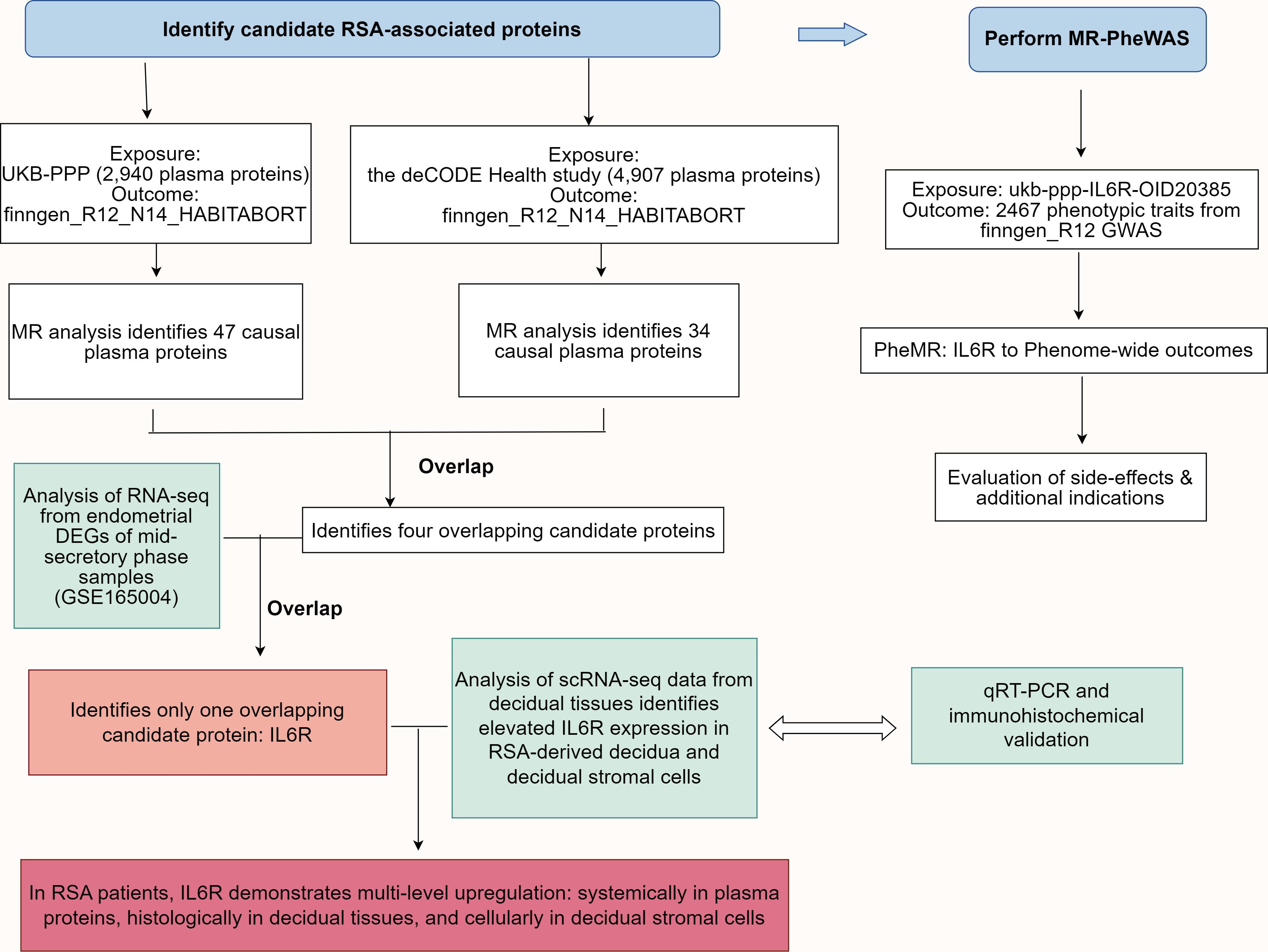
Figure 1. Schematic representation of the research design. MR analysis of plasma proteomics data from the UK Biobank Protein Plasma Project (UKB-PPP) and the deCODE Health Study identified 47 and 34 causal plasma proteins associated with RSA risk, respectively. The genetic data for RSA were obtained from the FinnGen R12_N14_HABITABORT dataset. Four candidate proteins were identified by overlapping the two datasets, with IL6R being the only candidate protein that overlapped with DEGs from mid-secretory phase endometrial RNA-seq data of RSA patients and controls (GSE165004). Reanalysis of single-cell RNA-seq data from the decidual tissues identified elevated IL6R expression in RSA-derived decidua and stromal cells. The IL6R expression was significantly upregulated at both the plasma protein and decidua mRNA levels in RSA patients. Finally, MR-PheWAS was performed using the ukb-ppp-IL6R-OID20385 dataset as the exposure variable and 2467 phenotypic traits from the finngen_R12 GWAS dataset as the outcome to evaluate the potential side effects and additional indications for IL6R as a drug target for RSA. DEGs, Differentially expressed genes; SNP, Single nucleotide polymorphism; IL6R, interleukin 6 receptor; UKB-PPP, UK Biobank Pharma Proteomics Project; RSA, Recurrent spontaneous abortion; RNA-seq, RNA sequencing; MR, Mendelian randomization; MR-PheWAS, MR-Phenome-wide association study; GWAS, genome-wide association study.
Data sources for RSA
Summary statistics for RSA from a GWAS were acquired from FinnGen_R12 (https://r12.risteys.finngen.fi/). The dataset (GWAS ID: finngen_R12_N14_HABITABORT), released in 2024, investigates habitual abortion, commonly known as RSA. This study encompassed 130,971 individuals of Finnish descent, comprising 811 cases and 130,160 control participants, with 20088611 SNPs meeting the quality control parameters.
Data sources for plasma pQTLs
We identified index cis-SNPs associated with the plasma protein levels at a genome-wide significance threshold (P < 5 × 10-8) to serve as IVs, utilizing data from two extensive GWASs: the deCODE Health study (18) and the UK Biobank Pharma Proteomics Project (UKB-PPP) (19). The UKB-PPP performed proteomic profiling on blood plasma samples from 54,219 participants using the Olink platform (19). Cis-SNPs were characterized as single nucleotide polymorphisms (SNPs) located within 1 megabase of the gene that encodes the corresponding protein, with linkage disequilibrium (LD) assessed employing the European reference panel provided by the 1000 Genomes. For 2,940 proteins, a comprehensive mapping of pQTLs was performed, with the genomic location of pQTLs frequently being near the gene associated with the specific protein. In practical terms, a pQTL located close to its associated gene is called a “cis-pQTL,” assuming that it affects the gene through direct interaction. In the two-sample MR analysis, index cis-SNPs corresponding to 1,728 plasma proteins were derived from the deCODE Health study. This study involved the measurement of 4,907 aptamers in a cohort of 35,559 Icelandic individuals by using the SomaScan platform (18). Similarly, we selected index cis-SNPs as IVs for 1980 proteins from the UKB-PPP (19). Our findings highlight the overlapping proteins identified as genetic instruments from two separate analyses so as to evaluate the consistency of results obtained from different proteomic profiling platforms.
Selection of IVs
In MR studies, IVs must adhere to the following three fundamental assumptions: (i) the relevance assumption, which posits that the IV exhibits a robust association with the exposure, typically indicated by an F-value of >10 to denote a strong correlation; (ii) the exclusivity assumption, which states that the IV should not influence the outcome; (iii) the independence assumption, which asserts that the IV must remain unassociated with any other confounding factors (20).
The instrumental variable selection protocol implemented the following three sequential filters (1): cis-pQTLs were operationally defined as genetic variants positioned within a proximal 2 Mb genomic window (spanning ±1 Mb from transcriptional initiation sites) (2); genome-wide significance threshold (P < 5 ×10-8) was applied to ensure high-confidence protein-SNP associations (3); LS pruning (r² < 0.001) was conducted to eliminate correlated variants, thereby preserving genetic instrument independence (4). The clumping window size for MR analyses of phenome-wide analysis was set to 10,000 kb. In contrast, a clumping window size of 1,000 kb was employed for proteome-wide MR (5); F-value calculated as (β2/se2) >10 to exclude weak IV bias (6); SNPs with a minor allele frequency (MAF) of 0.01 or less were not included (21, 22).
MR analysis
The inverse-variance weighted (IVW) method is recognized as the most reliable technique for evaluating all IVs (23). Consequently, this approach was adopted as the primary method for analysis in the present study. Statistically significant outcomes were defined as those with P < 0.05. However, it is important to note that the conclusions drawn from the IVW method may be biased in the presence of a horizontal pleiotropy (23). To address this potential limitation, the analysis was further supported by employing MR–Egger regression and the weighted median method, both of which aimed to bolster the reliability of the findings. MR-Egger regression is known to produce trustworthy estimates, although it may be affected by outliers (24). To investigate the presence of a broader horizontal pleiotropy, we utilized the MR-PRESSO approach (25). The possibility of a directional pleiotropy was assessed by applying the intercept test from MR-Egger regression and a significant non-zero intercept (P < 0.05), indicating a systematic directional pleiotropy (26). Moreover, we conducted a leave-one-out sensitivity analysis to determine whether the IVW estimate was influenced by a single SNP (27). Cochran’s Q test was applied to validate the presence of multiple variants, with heterogeneity acknowledged at a significance level of P < 0.05 (28). All analyses were conducted by using the TwoSampleMR package (version 0.6.8), the MR package (version 0.8.0), and the MR Pleiotropy Residual Sum and Outlier (MR-PRESSO) package (version 1.0) within the R software environment (version 4.4.2; https://www.R-project.org).
RNA-seq data analyses
The endometrial transcriptomic profiles analyzed in this investigation were derived from the publicly accessible Gene Expression Omnibus (GEO) repository with the accession number GSE165004 (12). This dataset, generated using the Illumina HiSeq 4000 system (platform identifier GPL16699), originally comprised endometrial tissue specimens from three distinct cohorts: 24 RSA cases, 24 individuals diagnosed with idiopathic infertility, and 24 reproductively competent females serving as reference subjects. Our analytical cohort was selectively restricted to 24 cases that met the diagnostic criteria for RSA and 24 demographically matched healthy controls with proven fertility. Endometrial samples were obtained during days 19–21 of the menstrual cycle. DEGs were screened using the GEO2R. Then, Gene Ontology (GO) and Kyoto Encyclopedia of Genes, Genomes (KEGG) analyses were conducted using the R statistical software (version 4.2.1) with the clusterProfiler package (v4.4.4) and ggplot2 package (3.4.4) (29). For the visualization of GO and KEGG results in bar charts, a more stringent selection criterion was applied, specifically |log2 fold change| > 0.585 and Padj < 0.05. Conversely, the chord diagram was generated using DEGs selected with the broader criterion of Padj < 0.05 only. The chord diagram not only quantitatively displayed changes in the gene expression but also highlighted the intensity of interactions among gene groups through the use of color and connection strength.
scRNA-seq data reanalysis
The scRNA-seq data of first-trimester decidual tissues (6 RSA cases and 5 normal controls) from our previous investigation (30) were subjected to a secondary analysis to map the IL6R expression landscapes. The expression patterns of IL6R in decidua tissues and decidual stromal cells (DSCs), the main cell type in decidual tissues, between normal and RSA groups were visualized through violin plots generated by geom_violin and ggboxplot function performed using ggpubr v0.6.0. By employing the netVisual_chord method from CellChat, we could effectively visualize the communication patterns of Interleukin 6 signaling among various cell types. This approach can help illustrate how Interleukin 6 regulates signaling across senders, receivers, mediators, and influencers, thereby revealing its key role and the complexity of interactions within the cellular network.
MR-PheWAS
PheWAS, commonly referred to as reverse GWAS, is a methodological approach used to examine associations between SNPs or phenotypes and a wide range of traits across the entire phenome (31). This method is particularly useful for assessing any potential side effects associated with the drug targets (31). The research utilized plasma proteins displaying positive MR results (IL6R, ukb-ppp-IL6R-OID20385), indicating exposure. The Finnish database (version R12), which includes 2467 phenotypic traits, provided the outcome data. To address the false-discovery rate in multiple comparisons, significance was set at an FDR-adjusted P < 0.05. SNPs associated with outcomes were excluded if their P value was below the Bonferroni-corrected threshold (P < 0.05/N. SNPs) in the MR-PheWAS analysis.
Sample collection and ethics statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Medical Ethics Committee of The Third Affiliated Hospital of Guangzhou Medical University (Approval No. 202401). Written informed consent was obtained from all participants prior to tissue collection. The RSA cohort comprised women with two or more consecutive unexplained pregnancy losses following confirmed normal (both parental and fetal) karyotypes, excluding individuals with uterine anomalies, endocrine/metabolic disorders, autoimmune conditions, or infections. Control subjects were gestational age-matched women undergoing voluntary termination of confirmed viable intrauterine pregnancies without history of pregnancy losses. Demographic characteristics of the participants providing decidual specimens for molecular analyses, are comprehensively detailed in Supplementary Table S1. All GWAS datasets, RNA-seq and scRNA-seq data included in this analysis underwent ethical review and were approved by their respective institutional ethics boards, with written informed consent obtained from all participants.
Quantitative real-time polymerase chain reaction
RNA extraction and qRT-PCR were conducted using RNAiso Plus and PrimeScript RT Reagent Kit for total RNA extraction and reverse transcription (TaKaRa, Japan). Gene expression was measured using TBGreen Premix Ex Taq II on a QuantStudio 3 Real-Time PCR System (Applied Biosystems, USA). Primer sequences are in Supplementary Table S2, and mRNA expression was normalized to the RNA18SN1 housekeeping gene.
Immunohistochemistry
Paraffin-embedded endometrial specimens from controls undergoing elective termination and RSA patients, underwent standardized immunohistochemical processing as described previously (32). Following formalin fixation and paraffin embedding, 5 μm sections were mounted on charged slides. Antigen retrieval was performed using citrate buffer (pH 6.0) before incubation with rabbit polyclonal anti-IL6R antibody (Proteintech; 1:200 dilution) overnight at 4 °C. Detection employed secondary antibodies (Cell Signaling Technology; 1:500) with 60-minute room temperature incubation, counterstained with DAPI nuclear marker (Sigma-Aldrich; 1 μg/mL).
Statistical analysis
Image processing and figure composition were conducted using Adobe Photoshop (Version 7.0). Quantitative data from qRT-PCR and immunohistochemistry were analyzed through IBM SPSS (Version 21.0) and GraphPad Prism (Version 8.0) platforms, employing independent two-sample t-tests and nonparametric tests for group comparisons. Continuous variables are presented as mean ± standard deviation, with statistical significance defined as P < 0.05 (two-tailed probability threshold).
Results
Proteome-wide MR intersected with RNA-seq data identifies one plasma protein, IL6R, as a putative therapeutic target for RSA
Using two-sample MR we analyzed plasma proteomic data from two large cohorts: the Icelandic deCODE study (4,907 proteins; n = 35,559) and the UK Biobank-PPP (2,940 proteins; n = 54,219). In total, 34 proteins from the deCODE dataset and 47 from the UKB-PPP dataset were significantly associated with RSA risk (IVW P < 0.05, Supplementary Tables S3-S6). Intersection analysis identified four plasma proteins consistently associated with RSA across both cohorts: IL6R (UKB-PPP OR = 1.29, 95% CI: 1.11–1.51; deCODE OR = 1.13, 95% CI: 1.00–1.28), contactin associated protein 2 (CNTNAP2) (UKB-PPP OR = 1.28, 95% CI: 1.018–1.60; deCODE OR = 1.41, 95% CI: 1.11–1.80), Delta-like canonical Notch ligand 1(DLL1) (UKB-PPP OR = 0.60, 95% CI: 0.36–0.99; deCODE OR = 0.35, 95% CI: 0.14–0.84), and neuronal pentraxin-1 (NPTX1) (UKB-PPP OR = 0.71, 95% CI: 0.56–0.90; deCODE OR = 0.69, 95% CI: 0.49–0.98) (Figure 2).
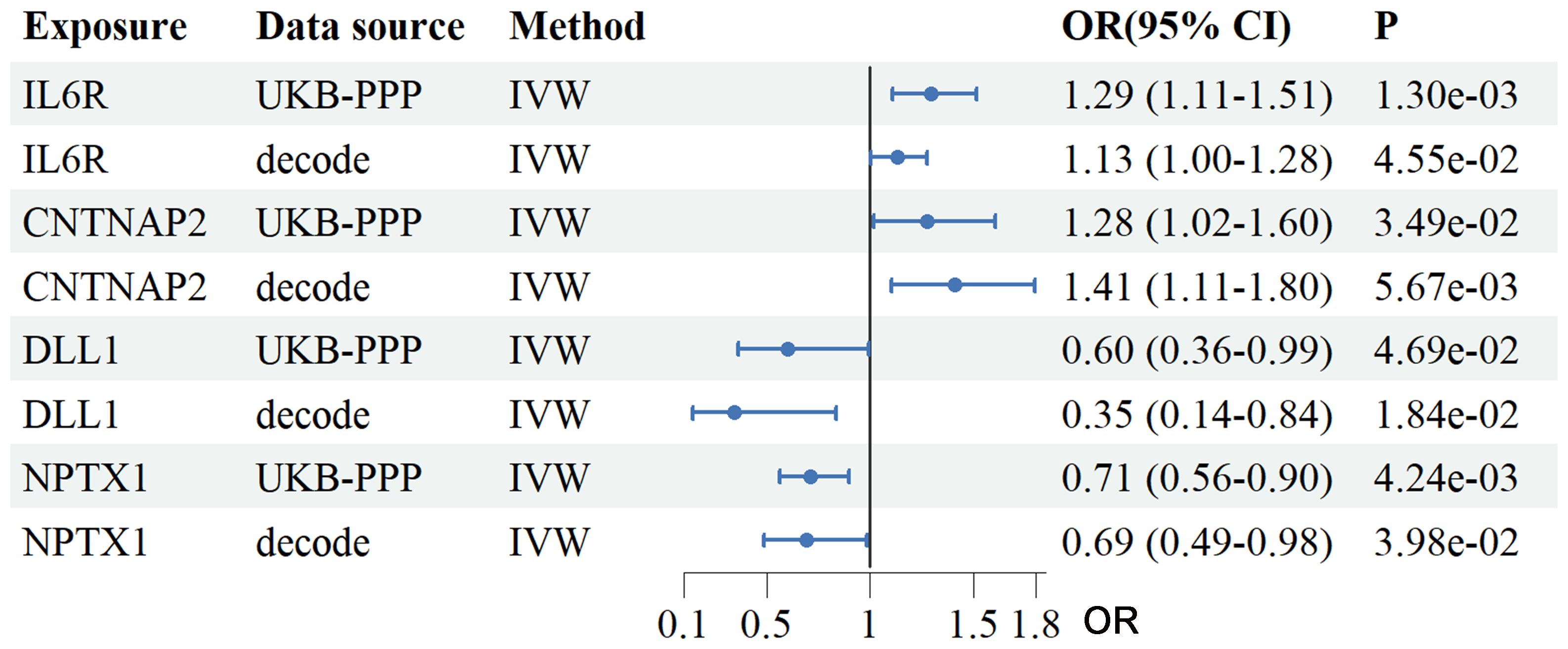
Figure 2. Analysis of the association between plasma proteins and the risk of RSA using the IVW method. The forest plot depicts the causal effects of plasma proteins on RSA risk based on a two-sample MR analysis using data from UKB-PPP and the Icelandic cohort. The four overlapping proteins identified were IL6R, CNTNAP2, DLL1 and NPTX1. OR and 95% CI were estimated using the IVW method. UKB-PPP, UK Biobank Prospective Proteomic Study; deCODE, Icelandic cohort; IVW, Inverse variance weighted; OR, Odds ratios; CI, confidence intervals.
To further refine our identification of tissue-specific therapeutic targets, we integrated mid-secretory-phase endometrial RNA-seq data (GSE165004), identifying 359 differentially expressed genes (DEGs) (Supplementary Table S7), Among these, IL6R was the only candidate that overlapped with the four MR-identified plasma proteins associated with RSA risk. It demonstrated bidirectional consistency: elevated plasma levels were associated with increased disease risk, and there was concomitant upregulation in RSA endometrial tissues (Figures 3A, B). Consequently, IL6R was established as the prioritized therapeutic candidate.
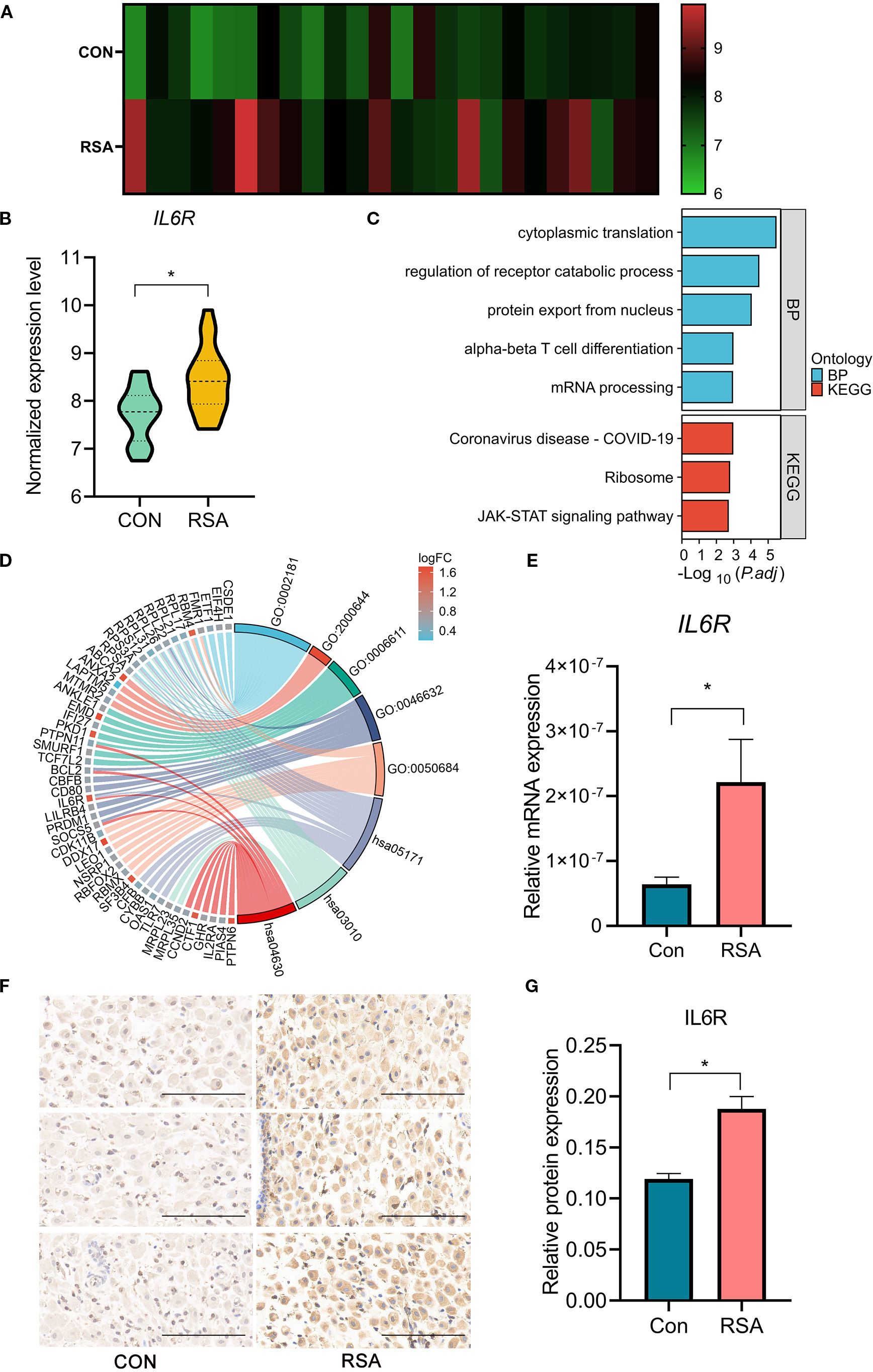
Figure 3. Comparative analysis of IL6R expression and pathway interactions in endometrial and decidual tissues of control group and RSA group. (A) Heatmap of IL6R expression in the mid-secretory endometria from RSA cases (n=24) compared to controls (n=24). Each row represents a sample, and color intensity indicates expression levels, with scale provided. (B) RNA-seq analysis of IL6R expression in mid-secretory endometrium derived from RSA group (n=24) compared to control group (n=24). *P < 0.05. (C) Bar chart displaying GO and KEGG pathway enrichment of DEGs in mid-secretory endometria derived from RSA cases compared to controls. (D) Chord Diagram of pathway-gene associations visualizes associations between DEGs and enriched biological processes and pathways. Ribbon widths indicate the strength of gene-pathway interactions, with significant links between IL6R, JAK-STAT signaling, and α-β T cell differentiation. (E) IL6R mRNA levels in deciduas from women with RSA and those with normal early pregnancies were analyzed using quantitative real-time polymerase chain reaction. The results were normalized to RNA18SN1 expression and are presented as the mean ± SD.*P < 0.05. n=18. (F, G) immunohistochemistry (IHC) staining and analysis of IL6R expression in early pregnancies deciduas from control group and RSA group. Scale bars,100μm. n=13. IL6R, interleukin 6 receptor; CON, Control; RSA, recurrent spontaneous abortion. GO,0002181, cytoplasmic translation; GO,2000644, regulation of receptor catabolic process; GO,0006611, protein export from nucleus; GO,0046632, alpha-beta T cell differentiation; GO,0050684, regulation of mRNA processing; hsa05171, Coronavirus disease- COVID-19; hsa03010, Ribosome; hsa04630, JAK-STAT signaling pathway.
Further transcriptomic analysis of endometrial tissues from RSA cases and controls identified a subset of DEGs enriched in biological pathways critical for immune regulation and cellular signaling. GO and KEGG analyses underscored significant enrichment in pathways such as α-β T cell differentiation and JAK-STAT signaling (Figure 3C). Notably, IL6R emerged as a pivotal gene within the α-β T cell differentiation pathway, suggesting its crucial role in modulating adaptive immune responses. A chord diagram further reinforced these associations, emphasizing the quantitative and functional interplay between IL6R and JAK-STAT signaling pathway (Figure 3D).
Furthermore, IL6R mRNA level was significantly upregulated in RSA decidua at the mRNA level (Figure 3E). In addition, immunohistochemistry further confirmed the significant increase in IL6R protein level in RSA decidua (Figures 3F, G).
Leveraging our published decidual scRNA-seq atlas, comprising first-trimester samples from five controls and six patients with RSA (30), we observed significantly elevated IL6R mRNA expression in the RSA cohort (Figures 4A, B). Furthermore, the communication model of IL6R among different cellular subtypes serving as senders, receivers, mediators, and influencers was disrupted, providing valuable insights into the molecular mechanisms underlying RSA pathogenesis (Figures 4C, D). Specifically, IL6 pathway crosstalk between DSCs and other cell populations was markedly enhanced in RSA, suggesting aberrant immune communication contributes to disease pathogenesis (Figures 4E, F).
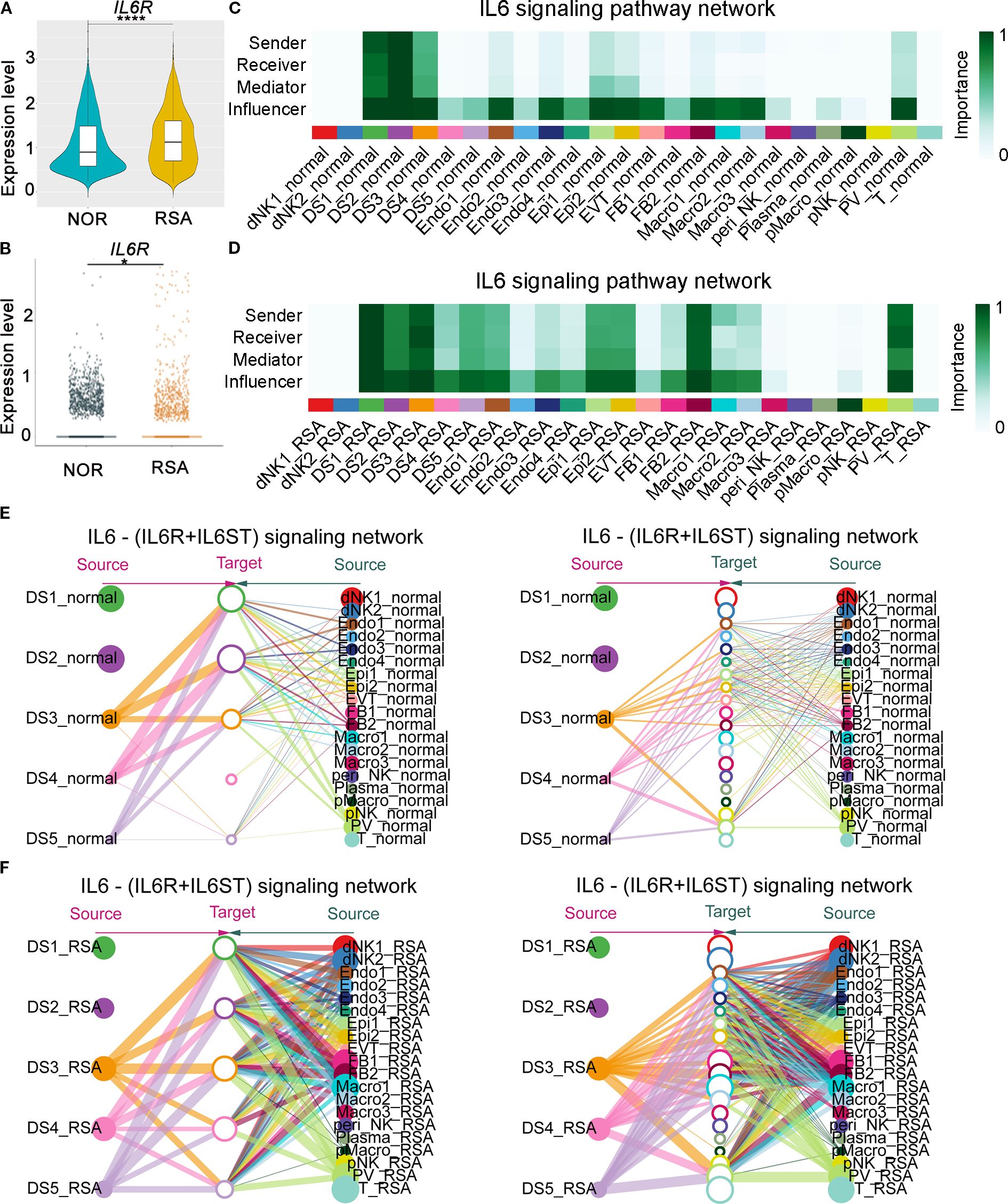
Figure 4. Abnormal upregulation of decidual IL6R mRNA in RSA patients was associated with IL6-signaling pathway dysregulation and compromised intercellular communication at the maternal-fetal interface. (A) Comparative single-cell RNA-seq (scRNA-seq) profiling of the IL6R transcript levels in decidual tissues from RSA patients (n = 6) versus gestational age-matched healthy controls (n = 5). The IL6R mRNA expression demonstrated a significant elevation in the RSA cohort (****P < 0.0001). (B) DSCs-specific analysis of scRNA-seq data demonstrated significantly elevated IL6R mRNA levels within DSCs populations from RSA patients (n = 6) when compared to gestational age-matched healthy controls (n = 5). (C) Analysis of scRNA-seq data revealed how different cell subtypes serve as senders, receivers, mediators, or influencers in the IL6-signaling pathway in the control group. The importance score (bar height) represents the communication probability (range 0–1), quantifying the relative strength of IL-6 ligand–receptor interactions between "sender" and "receiver" cell clusters. Higher values indicate stronger inferred communication. (D) Analysis of scRNA-seq data revealed how different cell subtypes serve as senders, receivers, mediators, or influencers in the IL6-signaling pathway in the RSA group. (E) Analysis of scRNA-seq data indicated the crosstalk between DSCs and the other subtypes in the control group, with a focus on the IL6-signaling pathway. Solid circles on the left represent different DS cell subpopulations, hollow circles in the center represent target cell subpopulations, and solid circles on the right represent other distinct cell subpopulations. The size of each circle is proportional to the number of cells within the corresponding subpopulation. Lines connecting nodes represent inferred significant ligand–receptor interactions, indicating potential cellular communication. The width of each line denotes the relative strength of the interaction. (F) Analysis of scRNA-seq data indicated the crosstalk between DSCs and other cellular subtypes in the RSA group, focusing on the IL6-signaling pathway. DS1-DS5, Decidual stromal cells; dNK1-dNK2, Decidual natural killer cells; Endo1-Endo4, Endometrial cells; Epi1-Epi2, Epithelial cells; FB1-FB2, Fibroblast cells; Macro1-Macro3, Macrophages; peri_NK, Peripheral blood natural killer cells; Plasma, Plasma cells; pMacro, placental macrophages; pNK, Placental natural killer cells; TPV, Trophoblast blood vessels; T, T cells.
Collectively, these findings demonstrate that IL6R is upregulated at both systemic and local tissue levels in RSA, marked by increased plasma protein levels and elevated expression in decidual and stromal cells.
Phenome-wide evaluation of IL6R targeting
A comprehensive two-sample MR-PheWAS was performed using data from the FinnGen cohort, encompassing 784 clinical diagnoses, 17 disease categories, and 2,469 quantitative phenotypes (https://r12.finngen.fi/) (Supplementary Table S8). This analysis evaluated the effect of genetically predicted plasma IL6R concentrations on disease risk, with effect estimates representing changes in disease odds per standard deviation increase in IL6R levels. Associations were classified as detrimental if consistent with increased RSA risk, or protective if opposed. Sensitivity analyses using alternative MR methods confirmed the robustness of the observed associations (Supplementary Table S9). Detailed MR estimates with corresponding confidence intervals for all examined phenotypes are presented in the supplementary materials (Supplementary Table S9). At the Bonferroni-corrected significance threshold (P ≤ 6.38 × 10−5, α = 0.05/784), 20 significant associations were identified (Figure 5; Supplementary Table S10), including 8 potentially protective relationships (40%; Supplementary Table S10). Notably, IL6R exhibited pleiotropic protective effects against several conditions (P ≤ 6.38 × 10−5, α = 0.05/784), including benign neoplasm of the stomach, alcoholic gastritis, and multiple cardiovascular diseases (e.g., ischemic heart disease, coronary artery bypass grafting, aortic aneurysm, calcific aortic valve stenosis, major coronary events, coronary atherosclerosis, myocardial infarction, and valvular heart disease excluding rheumatic fever). Protective associations were also observed for hypothyroidism. Conversely, elevated IL6R levels were associated with increased risk of several respiratory system disorders (P ≤ 6.38 × 10−5, α = 0.05/784), such as asthma, chronic obstructive pulmonary disease (COPD), chronic lower respiratory diseases, nontoxic diffuse goiter, malignant neoplasm of breast, cholelithiasis, infections of the skin and subcutaneous tissue, and acute lymphadenitis (Supplementary Table S10). These findings underscore the broad physiological impact of IL6R modulation, highlighting both the therapeutic potential and the possible adverse effects of IL6R-targeted strategies in the treatment of RSA.
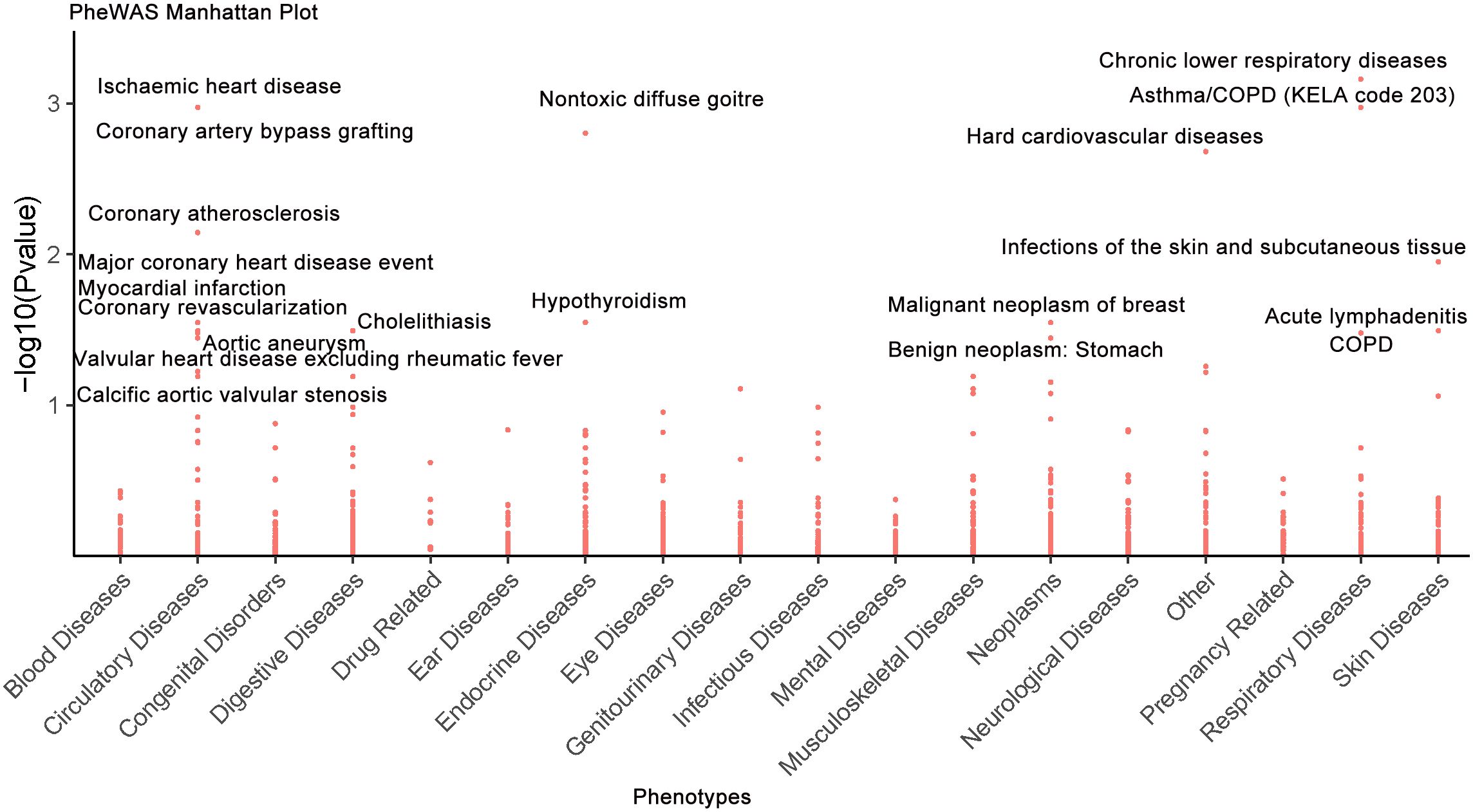
Figure 5. Phenome-wide causal estimates for IL6R perturbation are displayed, as derived from MR analyses of Finnish database clinical traits. The -log10(P-values) are plotted along the ordinate, with 20 labels applied to associations surpassing predefined statistical significance criteria (FDR < 0.05).
Discussion
This study utilized an integrative MR framework to identify causal therapeutic plasma protein targets in RSA, highlighting IL6R as a central pathogenic factor.
Our study identified four plasma proteins with causal associations with RSA risk: IL6R and CNTNAP2 as pathogenic contributors, and DLL1 and NPTX1 as protective factors.
Our multi-omics approach underlines the pivotal role of IL6R in RSA pathogenesis through elevated protein levels and transcriptomic upregulation in reproductive tissues. IL6R’s central position in immune modulation links α-β T cell differentiation (33), and JAK-STAT mediated signal transduction, which is essential for decidualization (34, 35), with implications for cytokine storm mechanisms similar to severe COVID-19 (36).
scRNA-seq analysis revealed IL6R signaling upregulation in decidual stromal and immune cells, which may suggest a potential disruption in immune tolerance that is critical for early pregnancy (37). Specifically, impaired crosstalk between DSCs and uterine natural killer (uNK) cells via IL6R may compromise key immunomodulatory functions critical for placental development, as uNK-derived IL6 family cytokines are known to regulate trophoblast invasion and spiral artery remodeling (38). Moreover, emerging evidence suggests that elevated IL6 levels in specific immune cell clusters (39), may hinder M2 macrophage polarization. This disruption may contribute to the etiopathogenesis of RSA by impairing fetal-maternal immune tolerance.
Mechanistically, IL6R mediates inflammatory responses via its heterodimeric structure, influencing both membrane-bound and soluble pathways (36). Mechanistically, IL6R mediates inflammatory responses primarily through two distinct pathways: classical cis-signaling and trans-signaling (40). In classical cis-signaling, membrane-bound IL6R (mIL6R) on the cell surface binds IL6 and recruits the glycoprotein 130 (gp130) co-receptor, activating intracellular signaling cascades such as JAK-STAT pathways in cells expressing mIL6R. In contrast, trans-signaling involves the soluble form of IL6R (sIL6R), which is generated by proteolytic shedding or alternative splicing. The sIL6R-IL6 complex can bind to gp130 on cells that lack mIL6R, thereby broadening the cellular responsiveness to IL6 and contributing to chronic inflammation. The clinical tractability of IL6R, demonstrated in autoimmune diseases through inhibitors like tocilizumab and sarilumab (41, 42), underscores its therapeutic potential in RSA-related inflammation.
While CNTNAP2 is well known for its role in neural development (43), our MR analyses also implicated CNTNAP2 in RSA susceptibility, supported by array comparative genomic hybridization analysis showing significant 7q34q36.3 duplications encompassing CNTNAP2 in RSA-linked pathogenic CNVs (44). The protective effects of DLL1 appear to be mediated through its involvement in endometrial remodeling via Notch2 signaling essential for decidualization (45). This decidualization process is critical for successful implantation and pregnancy maintenance (32), and increasing evidence implicates impaired decidualization in RSA pathogenesis (30, 32, 46–48). NPTX1 plays a role in uterine biology, potentially affecting uterine receptivity and stability (49). Furthermore, treatment of human endometrial stromal cells with synthetic progestins under hypoxic conditions significantly upregulated NPTX1 secretion (50), indicating its potential role in promoting decidualization.
Our MR-PheWAS analysis revealed IL6R’s significant causal associations with diverse phenotypes (P ≤ 6.38 × 10−5, α = 0.05/784), including reduced risks of COPD and asthma, which are consistent with prior studies (51). Protective trends were also observed for gastritis (P ≤ 6.38 × 10−5, α = 0.05/784), aligning with previously published research (42). This supports the need for careful consideration of IL6R’s pleiotropic effects in therapy development.
This study has several strengths. First, utilizing MR minimizes confounding and reverse causation through genetic instruments. Second, MR acts as a “natural randomized controlled trial”, providing robust evidence without the ethical concerns. Last, integration of large-scale GWAS and proteomic datasets enhances statistical power and reliability.
Several methodological limitations should be considered when interpreting our findings. First, results may lack generalizability due to European-biased cohorts. Second, Unmeasured gene-environment interactions could modify risk estimates (52).Third, residual horizontal pleiotropy remains possible despite sensitivity analyses. Additionally, although our study emphasizes the role of IL6R in RSA, mechanistic insights into its molecular mechanisms and downstream signaling pathways, particularly its mediation of crosstalk between uNK cells and DSCs, remain insufficiently characterized. Future studies involving IL6R inhibitor trial, functional assays and crosstalk between uNK cells and DSCs, are required to validate its mechanistic pathways in RSA. Additionally, the absence of clinical validation may limit the translation of our research findings into clinical practice, potentially impacting the perceived reliability of the therapeutic target. Future research should focus on conducting clinical trials to validate the efficacy and safety of interventions based on these findings. Last, exploring the molecular mechanisms in more diverse populations could enhance the generalizability of the results and provide deeper insights into the biological underpinnings. In summary, while our findings provide significant evidence pointing to IL6R as a key factor in RSA and propose it as a therapeutic target, the generalizability of these results requires cautious interpretation. Further studies involving diverse populations, comprehensive environmental data, functional assays, IL6R inhibitor-related experiment, and clinical validation are essential to confirm and extend the applicability of these findings to broader RSA patient groups and to develop effective, personalized treatment strategies.
Conclusion
This study advances the understanding of RSA by integrating genetic, proteomic, and transcriptomic analyses to uncover the causal role of IL6R as a key pathogenic factor. This was consistently validated across various biological levels, showing upregulation in plasma, decidual tissues, and decidual stromal cells of RSA patients. Additionally, a MR-PheWAS approach was used to explore the broader implications of targeting IL6R, assessing potential side effects and other therapeutic applications. Our results highlight IL6R as a vital therapeutic target for RSA, suggesting new directions for translational research and clinical strategi.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by the Medical Ethics Committee of The Third Affiliated Hospital of Guangzhou Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
WM: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. ZG: Formal analysis, Investigation, Resources, Visualization, Writing – original draft. QL: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft. MC: Conceptualization, Methodology, Writing – review & editing. JZ: Investigation, Writing – review & editing. XL: Funding acquisition, Methodology, Writing – review & editing. HYL: Data curation, Investigation, Methodology, Writing – review & editing. YL: Data curation, Methodology, Writing – review & editing. HL: Funding acquisition, Investigation, Resources, Writing – review & editing. JZ: Investigation, Writing – review & editing. MX: Investigation, Writing – review & editing. JL: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing. LD: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing. GS: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Support for this research was provided by the Opening Fund of NHC Key Laboratory of Chronobiology (Sichuan University) (NHCC-2024-03 to GS), the High-Level Talents Startup Fund from Affiliated Foshan Maternal and Child Health Hospital, Guangdong Medical University (GCCRCKYJF to WM), National Key R&D Program of China (2022YFC2702504 to JL), the National Natural Science Foundation of China (82371659 to JL, 82371674 to LD), Key-Area Research and Development Program of Guangdong Province (2023B1111020006 to JL), the Research Foundation of Guangzhou Women and Children’s Medical Center for Clinical Doctor (2023BS022 to XL), the Science and Technology Program of Guangzhou, China (2024A04J4735 XL and 2024A04J3776 to HL).
Acknowledgments
The authors express their gratitude to the investigators and participants for their valuable contributions and making the summary statistics publicly accessible.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1659251/full#supplementary-material
References
1. Genovese HG and McQueen DB. The prevalence of sporadic and recurrent pregnancy loss. Fertil Steril. (2023) 120:934–6. doi: 10.1016/j.fertnstert.2023.08.954
2. Bender AR, Christiansen OB, Elson J, Kolte AM, Lewis S, Middeldorp S, et al. ESHRE guideline: recurrent pregnancy loss: an update in 2022. Hum Reprod Open. (2023) 2023:d2. doi: 10.1093/hropen/hoad002
3. Dimitriadis E, Menkhorst E, Saito S, Kutteh WH, and Brosens JJ. Recurrent pregnancy loss. Nat Rev Dis Primers. (2020) 6:98. doi: 10.1038/s41572-020-00228-z
4. Bortoletto P, Lucas ES, Melo P, Gallos ID, Devall AJ, Bourne T, et al. Miscarriage syndrome: Linking early pregnancy loss to obstetric and age-related disorders. EBIOMEDICINE. (2022) 81:104134. doi: 10.1016/j.ebiom.2022.104134
5. Coomarasamy A, Dhillon-Smith RK, Papadopoulou A, Al-Memar M, Brewin J, Abrahams VM, et al. Recurrent miscarriage: evidence to accelerate action. Lancet. (2021) 397:1675–82. doi: 10.1016/S0140-6736(21)00681-4
6. Quenby S, Gallos ID, Dhillon-Smith RK, Podesek M, Stephenson MD, Fisher J, et al. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet. (2021) 397:1658–67. doi: 10.1016/S0140-6736(21)00682-6
7. Rai R, Regan L, and Recurrent miscarriage. LANCET. (2006) 368:601–11. doi: 10.1016/S0140-6736(06)69204-0
8. Yuan S, Xu F, Li X, Chen J, Zheng J, Mantzoros CS, et al. Plasma proteins and onset of type 2 diabetes and diabetic complications: Proteome-wide Mendelian randomization and colocalization analyses. Cell Rep Med. (2023) 4:101174. doi: 10.1016/j.xcrm.2023.101174
9. Zhao W, Fang P, Lai C, Xu X, Wang Y, Liu H, et al. Proteome-wide mendelian randomization identifies therapeutic targets for ankylosing spondylitis. Front Immunol. (2024) 15:1366736. doi: 10.3389/fimmu.2024.1366736
10. Santos T, Ieque AL, de Carvalho HC, Sell AM, Lonardoni M, Demarchi IG, et al. Antiphospholipid syndrome and recurrent miscarriage: A systematic review and meta-analysis. J Reprod Immunol. (2017) 123:78–87. doi: 10.1016/j.jri.2017.09.007
11. Huang X and Zhao JV. Exploring the pathways linking fasting insulin to coronary artery disease: a proteome-wide Mendelian randomization study. BMC Med. (2025) 23:321. doi: 10.1186/s12916-025-04127-6
12. Keleş ID, Günel T, Özgör BY, Ülgen E, Gümüşoğlu E, Hosseini MK, et al. Gene pathway analysis of the endometrium at the start of the window of implantation in women with unexplained infertility and unexplained recurrent pregnancy loss: is unexplained recurrent pregnancy loss a subset of unexplained infertility? Hum Fertil (Camb). (2023) 26:1129–41. doi: 10.1080/14647273.2022.2143299
13. Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A, et al. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med. (2020) 26:1644–53. doi: 10.1038/s41591-020-1040-z
14. Alecsandru D, Klimczak AM, Garcia VJ, Pirtea P, and Franasiak JM. Immunologic causes and thrombophilia in recurrent pregnancy loss. Fertil Steril. (2021) 115:561–6. doi: 10.1016/j.fertnstert.2021.01.017
15. Rasmark RE, Hellgren M, Hjertberg R, Blomqvist L, Matthiesen L, Henic E, et al. Treatment efficacy for idiopathic recurrent pregnancy loss - a systematic review and meta-analyses. Acta Obstet Gynecol Scand. (2018) 97:921–41. doi: 10.1111/aogs.13352
16. Ali S, Majid S, Niamat AM, Taing S, El-Serehy HA, and Al-Misned FA. Evaluation of etiology and pregnancy outcome in recurrent miscarriage patients. Saudi J Biol Sci. (2020) 27:2809–17. doi: 10.1016/j.sjbs.2020.06.049
17. Sanderson E, Glymour MM, Holmes MV, Kang H, Morrison J, Munafò MR, et al. Mendelian randomization. Nat Rev Methods Primers. (2022) 2. doi: 10.1038/s43586-021-00092-5
18. Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet. (2021) 53:1712–21. doi: 10.1038/s41588-021-00978-w
19. Sun BB, Chiou J, Traylor M, Benner C, Hsu YH, Richardson TG, et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. (2023) 622:329–38. doi: 10.1038/s41586-023-06592-6
20. Skrivankova VW, Richmond RC, Woolf B, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. Jama. (2021) 326:1614–21. doi: 10.1001/jama.2021.18236
21. Schmidt AF, Finan C, Gordillo-Marañón M, Asselbergs FW, Freitag DF, Patel RS, et al. Genetic drug target validation using Mendelian randomisation. Nat Commun. (2020) 11:3255. doi: 10.1038/s41467-020-16969-0
22. Lv X, Hu Z, Liang F, Liu S, Gong H, Du J, et al. Causal relationship between ischemic stroke and its subtypes and frozen shoulder: a two-sample Mendelian randomization analysis. Front Neurol. (2023) 14:1178051. doi: 10.3389/fneur.2023.1178051
23. Lin Z, Deng Y, and Pan W. Combining the strengths of inverse-variance weighting and Egger regression in mendelian randomization using a mixture of regressions model. PloS Genet. (2021) 17:e1009922. doi: 10.1371/journal.pgen.1009922
24. Bowden J, Davey SG, Haycock PC, and Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
25. Verbanck M, Chen CY, Neale B, and Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
26. Burgess S and Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
27. Hemani G, Bowden J, and Davey SG. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. (2018) 27:R195–208. doi: 10.1093/hmg/ddy163
28. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. (2018). doi: 10.7554/eLife.34408
29. Yu G, Wang LG, Han Y, and He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
30. Du L, Deng W, Zeng S, Xu P, Huang L, Liang Y, et al. Single-cell transcriptome analysis reveals defective decidua stromal niche attributes to recurrent spontaneous abortion. Cell Prolif. (2021):e13125. doi: 10.1111/cpr.13125
31. Denny JC, Bastarache L, and Roden DM. Phenome-wide association studies as a tool to advance precision medicine. Annu Rev Genomics Hum Genet. (2016) 17:353–73. doi: 10.1146/annurev-genom-090314-024956
32. Ma W, Cao M, Bi S, Du L, Chen J, Wang H, et al. MAX deficiency impairs human endometrial decidualization through down-regulating OSR2 in women with recurrent spontaneous abortion. Cell Tissue Res. (2022) 388:453–69. doi: 10.1007/s00441-022-03579-z
33. Wang W, Sung N, Gilman-Sachs A, and Kwak-Kim J. T Helper (Th) cell profiles in pregnancy and recurrent pregnancy losses: Th1/Th2/Th9/Th17/Th22/Tfh cells. Front Immunol. (2020) 11:2025. doi: 10.3389/fimmu.2020.02025
34. Rosario GX, Hondo E, Jeong JW, Mutalif R, Ye X, Yee LX, et al. The LIF-mediated molecular signature regulating murine embryo implantation. Biol Reprod. (2014) 91:66. doi: 10.1095/biolreprod.114.118513
35. Suman P, Malhotra SS, and Gupta SK. LIF-STAT signaling and trophoblast biology. Jakstat. (2013) 2:e25155. doi: 10.4161/jkst.25155
36. Rodrigues F, Da SR, Santos E, de Brito M, Da SA, de Meira LM, et al. Association of polymorphisms of IL-6 pathway genes (IL6, IL6R and IL6ST) with COVID-19 severity in an amazonian population. Viruses. (2023) 15. doi: 10.3390/v15051197
37. Robertson SA, Prins JR, Sharkey DJ, and Moldenhauer LM. Seminal fluid and the generation of regulatory T cells for embryo implantation. Am J Reprod Immunol. (2013) 69:315–30. doi: 10.1111/aji.12107
38. Wallace AE, Host AJ, Whitley GS, and Cartwright JE. Decidual natural killer cell interactions with trophoblasts are impaired in pregnancies at increased risk of preeclampsia. Am J Pathol. (2013) 183:1853–61. doi: 10.1016/j.ajpath.2013.08.023
39. Zhao Y, Sun J, and Jin L. The N6-methyladenosine regulator ALKBH5 mediated stromal cell-macrophage interaction via VEGF signaling to promote recurrent spontaneous abortion: a bioinformatic and in vitro study. Int J Mol Sci. (2022) 23:15819. doi: 10.3390/ijms232415819
40. Garbers C and Rose-John S. Dissecting interleukin-6 classic- and trans-signaling in inflammation and cancer. Methods Mol Biol. (2018) 1725:127–40. doi: 10.1007/978-1-4939-7568-6_11
41. Choe JY, Lee GH, and Kim SK. Radiographic bone damage in chronic gout is negatively associated with the inflammatory cytokines soluble interleukin 6 receptor and osteoprotegerin. J Rheumatol. (2011) 38:485–91. doi: 10.3899/jrheum.100727
42. Fu C, Wang L, and Cai W. IL6 receptor inhibitors: exploring the therapeutic potential across multiple diseases through drug target Mendelian randomization. Front Immunol. (2024) 15:1452849. doi: 10.3389/fimmu.2024.1452849
43. Papale LA, Madrid A, Zhang Q, Chen K, Sak L, Keleş S, et al. Gene by environment interaction mouse model reveals a functional role for 5-hydroxymethylcytosine in neurodevelopmental disorders. Genome Res. (2022) 32:266–79. doi: 10.1101/gr.276137.121
44. Gajjar K, Patel A, Patel B, Chettiar S, and Jhala D. Array comparative genomic hybridization analysis of products of conception in recurrent pregnancy loss for specific anomalies detected by USG. Reprod Fertil. (2023) 4. doi: 10.1530/RAF-22-0092
45. Otti GR, Saleh L, Velicky P, Fiala C, Pollheimer J, and Knöfler M. Notch2 controls prolactin and insulin-like growth factor binding protein-1 expression in decidualizing human stromal cells of early pregnancy. PloS One. (2014) 9:e112723. doi: 10.1371/journal.pone.0112723
46. Jiang Y, Li J, Li G, Liu S, Lin X, He Y, et al. Osteoprotegerin interacts with syndecan-1 to promote human endometrial stromal decidualization by decreasing Akt phosphorylation. Hum Reprod. (2020) 35:2439–53. doi: 10.1093/humrep/deaa233
47. Larsen EC, Christiansen OB, Kolte AM, and Macklon N. New insights into mechanisms behind miscarriage. BMC Med. (2013) 11:154. doi: 10.1186/1741-7015-11-154
48. Liu M, Deng W, Tang L, Liu M, Bao H, Guo C, et al. Menin directs regionalized decidual transformation through epigenetically setting PTX3 to balance FGF and BMP signaling. Nat Commun. (2022) 13:1006. doi: 10.1038/s41467-022-28657-2
49. Krsteski J, Gorenjak M, But I, Pakiž M, and Potočnik U. Dysregulation of synaptic signaling genes is involved in biology of uterine leiomyoma. Genes (Basel). (2021) 12:1179. doi: 10.3390/genes12081179
50. Guzeloglu-Kayisli O, Basar M, Shapiro JP, Semerci N, Huang JS, Schatz F, et al. Long-acting progestin-only contraceptives enhance human endometrial stromal cell expressed neuronal pentraxin-1 and reactive oxygen species to promote endothelial cell apoptosis. J Clin Endocrinol Metab. (2014) 99:E1957–66. doi: 10.1210/jc.2014-1770
51. Wu D, Gong Z, Hao X, and Liu L. Genetic perturbation of IL-6 receptor signaling pathway and risk of multiple respiratory diseases. . J Transl Med. (2024) 22:581. doi: 10.1186/s12967-024-05366-6
Keywords: Mendelian randomization, recurrent spontaneous abortion, plasma proteins, interleukin 6 receptor, phenome-wide association study
Citation: Ma W, Gu Z, Lin Q, Cao M, Zhong J, Li X, Li H, Lin Y, Lin H, Xu M, Liu J, Du L and Song G (2025) Integrative genetic and multi-omics analysis reveals the interleukin-6 receptor’s role in recurrent spontaneous abortion. Front. Immunol. 16:1659251. doi: 10.3389/fimmu.2025.1659251
Received: 03 July 2025; Accepted: 16 September 2025;
Published: 02 October 2025.
Edited by:
Danuta Radzioch, McGill University, CanadaReviewed by:
Abdelgadir Elamin Eltom, Gulf Medical University, United Arab EmiratesYujia Luo, Zhejiang University, China
Copyright © 2025 Ma, Gu, Lin, Cao, Zhong, Li, Li, Lin, Lin, Xu, Liu, Du and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ge Song, c29uZ2dlcHBAMTI2LmNvbQ==; Lili Du, bGlsaWR1Z3lzeUBnemhtdS5lZHUuY24=; Jianqiao Liu, bGl1anFzc3pAZ3pobXUuZWR1LmNu
†These authors have contributed equally to this work
 Weixu Ma
Weixu Ma Zhongjia Gu3†
Zhongjia Gu3† Qiwang Lin
Qiwang Lin Mingzhu Cao
Mingzhu Cao Hongyan Li
Hongyan Li Yue Lin
Yue Lin Han Lin
Han Lin Jianqiao Liu
Jianqiao Liu Lili Du
Lili Du Ge Song
Ge Song