- 1Yibin Institute of Traditional Chinese Medicine, Yibin, Sichuan, China
- 2Yibin Hospital of Traditional Chinese Medicine, Yibin, Sichuan, China
Epithelial ovarian cancer (EOC) remains a lethal epithelial malignancy. Immune-checkpoint inhibitors have entered management for recurrent/metastatic disease; yet durable benefit is confined to a subset, reflecting TGF-β–conditioned stromal barriers and organised T-cell exclusion. In this review we summarise advances from single-cell RNA and ATAC profiling and spatial transcriptomics that resolve fibroblast, tumour and immune programmes linked to TGF-β signalling, and appraise translational opportunities spanning selective pathway modulation, checkpoint combinations and spatial biomarkers. We also discuss enduring challenges—including site-specific heterogeneity across adnexal, omental and peritoneal niches, limited assay standardisation and a scarcity of predictive metrics—that temper implementation. By integrating TGF-β–informed readouts (e.g., INHBA+ cancer-associated fibroblast burden, periostin/fibronectin indices, MHC-I status and CD8–tumour distances) with PD-1–based regimens and TGF-β-axis agents (ALK5 inhibitors, Activin A neutralisation, NOX4-directed reprogramming), emerging strategies aim to restore antigen presentation, improve lymphocyte access and remodel tumour–stroma interfaces. Our synthesis provides an appraisal of the evolving landscape of TGF-β–informed precision immuno-oncology in ovarian cancer and outlines pragmatic standards and avenues for clinical translation. We hope these insights will assist researchers and clinicians as they endeavour to implement more effective, individualised regimens.
1 Introduction
Epithelial ovarian cancer remains the most lethal gynecologic malignancy, with high-grade serous ovarian carcinoma (HGSOC) accounting for the majority of deaths and displaying pronounced genomic instability and tissue-site heterogeneity that complicate immune control (1–3). Single-cell and spatially resolved studies demonstrate that immune activation and suppression can segregate across intraperitoneal niches in HGSOC, with microenvironmental context shaping recognition and escape (4–7). Immunotherapy with immune-checkpoint inhibitors has produced limited and variable benefit in unselected ovarian cancer populations, underscoring the need to resolve mechanisms of immune failure at cellular and spatial resolution.
Transforming growth factor-β1/β2/β3 from tumor cells, CAFs and Tregs drive exclusion via TGFBR1/ALK5-SMAD2/3 plus non-SMAD (p38/ERK/PI3K) arms, while CAF-derived Activin A (INHBA) engages ACVR1B/ACVR2 to phenocopy these suppressive effects and blunt PD-(L)1 responses (8–11). Foundational work in urothelial and colorectal cancer showed that TGF-β–dependent stromal activation confines effector T cells to peritumoral territories and that dual blockade of TGF-β and PD-(L)1 can restore intratumoral T-cell access (12–15). Consistent with these principles, pan-cancer analyses link high TGF-β activity to immune-excluded phenotypes and resistance to checkpoint inhibition.
Evidence specific to ovarian cancer supports a TGF-β–conditioned, stromal-dominated immune low-response state. Integrated digital pathology and transcriptomics identified TGF-β–driven loss of antigen presentation and fibroblast activation as mediators of T-cell exclusion in ovarian tumors, with reduced MHC-I on cancer cells and desmoplastic barriers that hinder infiltration (16–18). Single-cell and spatial profiling of HGSOC further resolve site-specific immune ecosystems, revealing that tumors with copy-number–driven evolution can exhibit elevated TGF-β signaling alongside naïve or memory-skewed T-cell compartments and limited effector access. Spatial atlases also document marked heterogeneity of tumor-infiltrating T cells and their neighborhood relationships with stromal and malignant cells, providing a structural substrate for immune exclusion (19–21). Within the stromal compartment, immunomodulatory cancer-associated fibroblast subsets, including INHBA+ CAFs that enforce SMAD2-dependent PD-L1 expression and regulatory T-cell differentiation, exemplify TGF-β–linked suppressive circuits in advanced ovarian cancer. Preclinical work in HGSOC models shows that concurrent targeting of TGF-β and PD-L1 can enhance antitumor immunity, consistent with a causal role for TGF-β in therapeutic nonresponse.
Single-cell RNA sequencing, single-cell chromatin and spatial transcriptomic technologies now permit direct quantification of TGF-β pathway activity, fibroblast and extracellular-matrix programs, and ligand–receptor interactions that organize T-cell exclusion in ovarian cancer (22–24). By integrating these modalities, it is feasible to define reproducible immune low-response phenotypes, map their stromal drivers, and derive composite biomarkers and testable interventions. The purpose of this review is to synthesize single-cell and spatial transcriptomic evidence on TGF-β–driven T-cell exclusion in ovarian cancer, delineate mechanistic links between signaling and stromal remodeling, and outline diagnostic and therapeutic implications for risk stratification and treatment design.
2 Single-cell and spatial phenotype of TGF-β–conditioned immune low-response in ovarian cancer
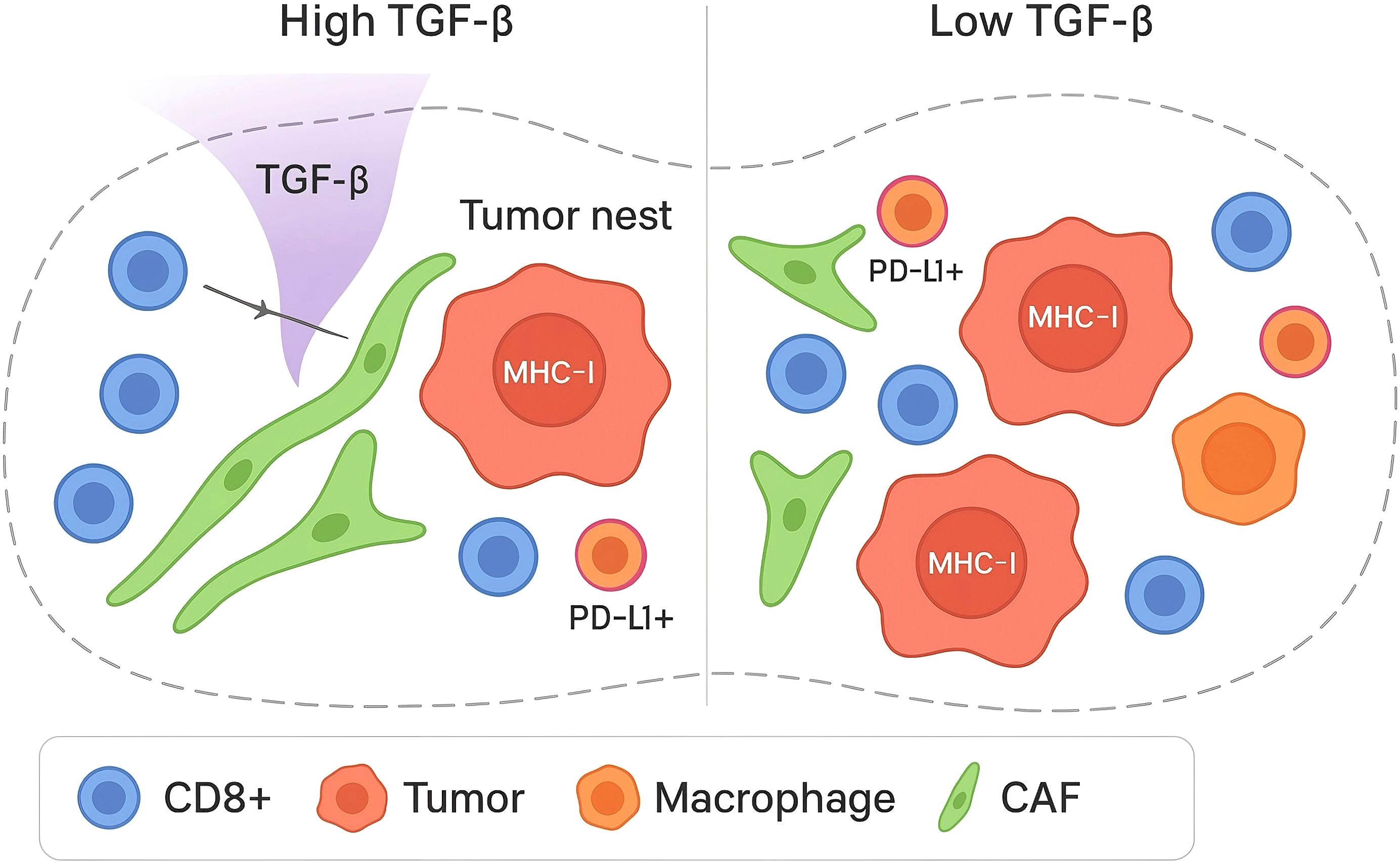
Single-cell and spatial studies in high-grade serous ovarian carcinoma (HGSOC) converge on a reproducible immune–stromal state in which transforming growth factor-β (TGF-β) signaling coincides with peritumoral confinement of effector T cells, reduced antigen presentation, and desmoplastic remodeling (25–30).
In HGSOC, multi-site single-cell and multiplex imaging analyses show that anatomical location and mutational processes stratify immune phenotypes (30–32). Tumors bearing fold-back inversions exhibit elevated TGF-β pathway activity with immune-excluded architectures populated by naïve/stem-like and memory-skewed T-cell compartments, whereas homologous-recombination-deficient tumors display more differentiated dysfunctional CD8+ states (33, , 12). These patterns are quantified by nearest-neighbor distance (centroid-to-centroid μm after nuclei segmentation; k-d tree), tumor–stroma interface length (contiguous boundary μm by skeletonization), and CAF ‘corridor’ width (fibronectin/α-SMA–positive bands via binary morphology). As shown in Figure 1, these site and genotype-linked patterns are quantified by nearest-neighbor distances between CD8+ T cells, PD-L1+ cancer cells, and PD-L1+ macrophages, indicating reduced effector proximity in TGF-β–high contexts.
Single-cell atlases further resolve T-cell heterogeneity across ovarian and omental foci. Ovarian lesions often show ‘cold’ states with Tregs and dysfunctional T cells, while omentum harbors bystanders; canonical markers/niches include TCF1+ stem-like CD8 (TCF7, SLAMF6) perivascular/TLS-adjacent, terminally exhausted CD8 (PD-1, TOX, TIM-3) at margins, bystander CD8 (CD39-) in omentum, and Tregs (FOXP3, CTLA-4, TIGIT) in CAF-rich rims (34–37). These features are consistent with a TGF-β–conditioned, stromal-dominated immune low-response.
Fibroblast programs are central to this phenotype. A TGF-β–driven cancer-associated fibroblast (CAF) subset identified by scRNA-seq adversely associates with outcome and expresses TGF-β pathway and EMT-linked effectors (38–40). Complementing this, INHBA+ (Activin A–producing) CAFs enforce SMAD2-dependent PD-L1 expression and promote regulatory T-cell differentiation, providing a direct cellular mechanism for immunosuppression within advanced ovarian tumors.
Operational markers used in this review to define the TGF-β–conditioned immune low-response state are shown in Table 1. Spatial proteogenomic profiling in ovarian cancer supports these single-cell inferences: immune-excluded regions are enriched for Tregs and fibronectin-rich stroma, whereas diffuse, tumor-proximal immune niches exhibit higher PD-L1/IDO1 and activated lymphocyte markers (41–43). These observations align with a model in which TGF-β-conditioned fibroblast matrices and checkpoint-ligand geography jointly restrict productive cytotoxic engagement.
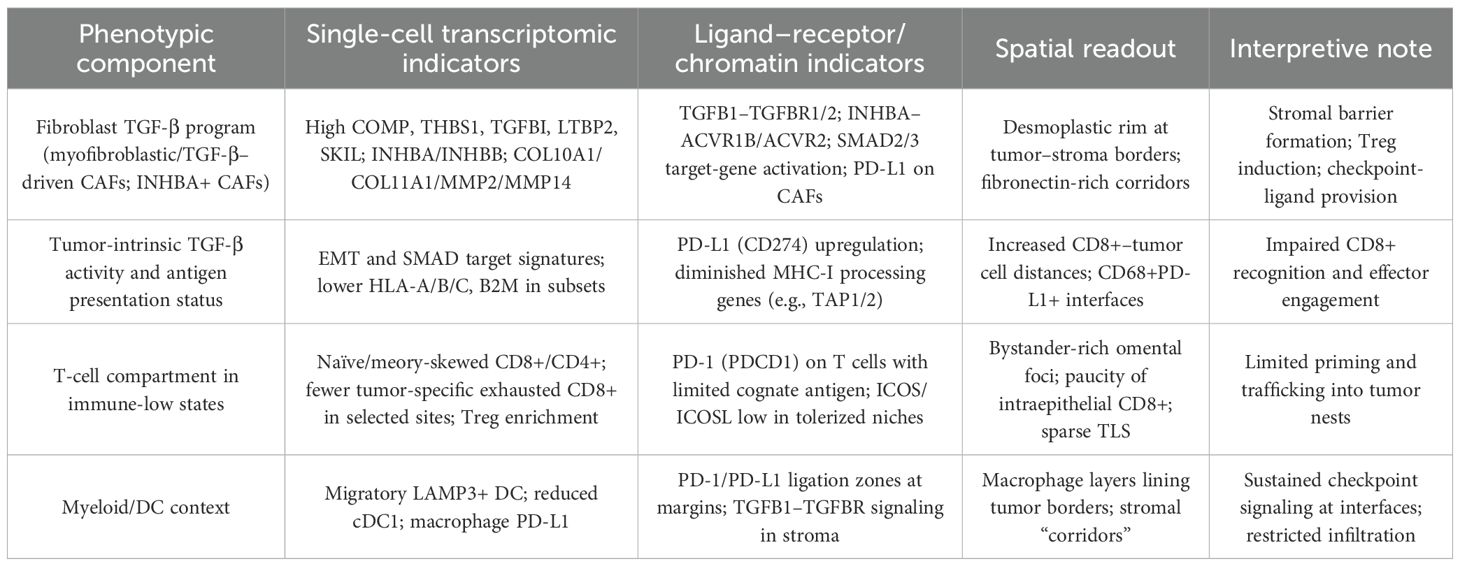
Table 1. Operational features and readouts of a TGF-β–conditioned immune low-response state in ovarian cancer.
These single-cell and spatial criteria delineate an ovarian cancer ecosystem in which TGF-β–responsive fibroblast matrices, altered antigen presentation, and checkpoint-dominated contact zones converge to produce immune exclusion; this phenotype maps onto the broader TGF-β barrier framework defined in other indications and provides tractable readouts for risk stratification and therapeutic testing.
3 Mechanistic links between TGF-β signaling, stromal remodeling, and T-cell exclusion
Transforming growth factor-β orchestrates a fibroblast-centered program that remodels the extracellular matrix and establishes spatial barriers to effector T-cell access. In multiple solid tumors, stromal TGF-β activity correlates with immune-excluded architectures, and experimental inhibition of TGF-β restores intratumoral T-cell penetration when combined with PD-(L)1 blockade, indicating that TGF-β–dependent stromal activation is a proximal cause rather than an epiphenomenon of exclusion (44–46). Mechanistically, TGF-β/SMAD signaling in cancer-associated fibroblasts (CAFs) induces contractile myofibroblastic states and upregulates matrix constituents and modulators—collagens, fibronectin, versican, thrombospondins, latent TGF-β–binding proteins—together with crosslinking and alignment programs that increase stiffness and reduce interstitial porosity, thereby constraining lymphocyte trafficking (47–49). Ovarian tumor stroma exemplifies these dynamics: TGF-β1–induced periostin in activated fibroblasts promotes desmoplastic remodeling and malignant cell motility, reinforcing matrix-rich interfaces at tumor borders that are unfavorable to T-cell ingress (50–52). These observations align with pan-cancer ECM signatures linked to TGF-β and poor ICI outcomes; chemokine circuits (CXCL12–CXCR4, CCL2–CCR2, TGF-β–induced CXCLs) cooperate with aligned collagen/fibronectin to confine cells peritumorally.
In ovarian cancer, TGF-β shapes immunoregulation beyond its effects on physical barriers. Integrated digital pathology and transcriptomics show that TGF-β correlates with diminished tumor-cell antigen presentation and fibroblast activation in T-cell–excluded tumors, indicating concurrent defects in recognition and access (53–55). Spatial proteogenomic profiling further demonstrates that fibronectin-rich stromal territories with regulatory T-cell enrichment co-localize with immune-excluded niches, whereas areas with diffuse tumor–immune proximity display higher antigen-presentation markers and checkpoint expression, consistent with segregation of suppressive matrix from effective cytotoxic engagement (56–58). At the cellular level, INHBA+ CAFs release bioactive Activin A via furin-mediated prodomain cleavage; ACVR1B/ACVR2→SMAD2 signaling in CAFs induces PD-L1 and Treg programs, sustaining exclusion even when T cells reach the margin (59–61). Tumor-derived TGF-β1 promotes CAF differentiation and metastatic competence, supplying ligand to maintain these stromal and immunosuppressive circuits. Taken together, these data delineate a convergent mechanism whereby TGF-β–responsive fibroblast programs generate an ECM-defined barrier, reduce antigen visibility, and install local checkpoint ligation, yielding a T-cell–excluded, immune low-response state.
4 Diagnostic trajectory and risk stratification from single-cell and spatial readouts
Diagnostic evaluation of immune low-response ovarian cancer should progress from discovery-grade single-cell and spatial assays to deployable, site-aware risk stratification that quantifies TGF-β–conditioned stromal programs, antigen-presentation deficits, and the geometry of tumor–immune contacts. In high-grade serous ovarian carcinoma (HGSOC), spatial transcriptomics shows that discrete malignant subclones occupy distinct neighborhoods and engage defined stromal and immune partners, indicating that clone-specific ligand–receptor circuits partly encode the degree and pattern of lymphocyte access; these features are directly measurable in tissue and link to outcome-relevant biology (62–65). Integrative multi-omic mapping across 160 tumor sites further demonstrates that mutational processes and anatomic location co-determine immune states, with fold-back inversion–bearing tumors exhibiting elevated TGF-β signaling, T-cell exclusion, and naïve/memory-skewed T-cell compartments—an axis that plausibly marks a TGF-β–high risk group (66–68). In parallel, spatial proteogenomic profiling in ovarian cancer separates diffuse tumor–immune interdigitation from focal immune niches; the former co-localizes with higher PD-L1/IDO1 and other immunotherapy targets, while focalized macrophage-rich niches (CD163high) associate with preliminarily worse outcomes, supporting the use of neighborhood metrics rather than bulk density alone for risk definition (69–71). These observations provide a rationale to define a composite “TGF-β–conditioned exclusion” classifier that integrates four orthogonal readouts: a stromal/CAF activity index, an antigen-presentation index, a spatial interaction index, and a contextual genetic index.
For the stromal/CAF activity index, single-cell–derived markers of TGF-β–responsive fibroblasts can be translated to practical surrogates. INHBA+ cancer-associated fibroblasts drive SMAD2-dependent PD-L1 expression and promote regulatory T-cell differentiation in advanced ovarian cancer, nominating INHBA protein/RNA and CAF-PD-L1 as tissue surrogates of TGF-β superfamily–linked immune suppression (72–74). Periostin-rich matrices—induced through integrin/NF-κB and TGF-β2 signaling—track with macrophage recruitment and fibroblast activation in ovarian cancer and can serve as desmoplastic sentinels measurable on archival formalin-fixed tissue (75–78). For the antigen-presentation index, HR-deficient contexts show enhanced immunosurveillance, whereas HR-proficient tumors display compartmentalization; incorporate MHC-I surrogates with HRD and note: MHC-I loss via B2M truncation, HLA LOH, or IFN–JAK/STAT defects; readouts—IHC (HLA-A/B/C; B2M 0–3+ rubric) and copy-number flags (79–81). The spatial interaction index should quantify nearest-neighbor distances and interface lengths between CD8+ T cells, PD-L1+ tumor/myeloid cells, and CAF corridors, because diffuse tumor–immune mixing versus focal or peritumoral restriction carries distinct therapeutic implications in ovarian cancer (82–84). The contextual genetic index should register fold-back inversions and other copy-number–driven processes that associate with high TGF-β activity and immune exclusion, as these events stratify immunologic phenotypes across intraperitoneal sites.
Assay implementation can follow a tiered path compatible with routine specimens. Discovery-level single-cell and spatial transcriptomic platforms define cell states, ligands, and receptor topologies; these can be down-translated to validated multiplex protein imaging on formalin-fixed sections. High-dimensional imaging methods such as multiplexed ion-beam imaging by time-of-flight (MIBI-TOF) have demonstrated reproducible, quantitative annotation of clinically relevant cell states in archival tissues and provide a route to standardize spatial scoring rules across centers (85–87). For institutions without high-plex capacity, constrained surrogate panels can approximate the composite score by combining INHBA/α-SMA/fibronectin/periostin with PD-L1, HLA-I components, and pan-T-cell markers, quantified with pre-specified adjacency metrics. Spatial risk assignments should be site-aware, because adnexal, omental, and peritoneal foci exhibit different immune architectures under the same patient-level genotype, and because subclones within a lesion can preferentially associate with fibroblasts or CXCL9+ macrophages. Where available, radiogenomic bridges that correlate spatial transcriptomic phenotypes with computed tomography features can facilitate non-invasive stratification and longitudinal monitoring.
In terms of clinical use, the composite classifier should separate at least two actionable risk states. A TGF-β–dominant, CAF-rich, immune-excluded state—scored by high INHBA/periostin/fibronectin, low MHC-I, long CD8+–tumor distances, macrophage/CAF border interfaces, and FBI-like genomic context—would be predicted to benefit from strategies that decompress or reprogram stroma and restore access, including consideration of TGF-β pathway targeting layered onto PD-(L)1 where feasible. The biological basis rests on studies in other solid tumors in which stromal TGF-β blockade restored intratumoral T-cell access and synergized with PD-(L)1 inhibition, supporting the face validity of this state as a barrier phenotype (88–90). A comparatively inflamed state—characterized by diffuse tumor–immune contact, TLS/B-cell aggregates, and intact antigen presentation—could be triaged toward checkpoint-based regimens or trials emphasizing antigen-presentation and costimulation, with ovarian data showing that TLS and diffuse tumor–immune interactions track with favorable immune targets and improved prognostic signals (26, 91, 92). Prospective validation should predefine analytic thresholds, ensure inter-assay concordance between discovery and surrogate panels, and embed multi-site sampling to avoid misclassification by local ecology; however, the current body of single-cell and spatial evidence already delineates measurable features that can be operationalized to forecast T-cell access, checkpoint-ligand geography, and TGF-β–linked stromal risk in ovarian cancer.
5 Therapeutic strategies and future directions
Therapeutic development for a TGF-β–conditioned, immune-excluded state in ovarian cancer should prioritize combinations that restore intratumoral CD8+ T-cell access while minimizing pathway-wide toxicities. Convergent preclinical work demonstrates that stromal TGF-β activity enforces peritumoral confinement of effector T cells and that simultaneous inhibition of TGF-β and PD-(L)1 converts exclusion into productive antitumor immunity; these data provide a mechanistic basis for layered regimens in TGF-β–high ovarian tumors identified by single-cell and spatial criteria (93–95). However, the pleiotropic roles of TGF-β mandate selective approaches. Strategies that confine pathway blockade to dominant immunoregulatory sources or nodes are attractive—for example, antibody targeting of GARP: TGF-β1 complexes to restrict neutralization to regulatory T-cell–derived ligand, or context-adapted ALK5 inhibition administered in intermittent schedules to mitigate toxicity—both supported by translational and early clinical literature.
Ovarian-specific stromal targets emerging from single-cell and spatial profiling nominate tractable entry points. INHBA+ (Activin A–producing) cancer-associated fibroblasts upregulate PD-L1 via SMAD2-dependent signaling and drive regulatory T-cell differentiation; neutralization of Activin A attenuates disease and remodels the immune–stromal compartment in ovarian models, indicating a rational partner for PD-(L)1 or costimulatory strategies in INHBA-high states (96–98). Periostin-rich matrices induced through integrin/NF-κB and TGF-β2 signaling associate with macrophage recruitment, fibroblast activation, and metastatic competence in epithelial ovarian cancer, supporting periostin or upstream integrin blockade as stroma-decompressing adjuncts in exclusion phenotypes. Because CAF-driven mechanics and chemokine circuits are central to immune geography, pharmacologic reprogramming rather than indiscriminate depletion is preferred; inhibition of NOX4, a TGF-β–linked driver of myofibroblastic states, overcomes CAF-mediated CD8+ T-cell exclusion and potentiates checkpoint efficacy across models, justifying evaluation in ovarian desmoplastic contexts (99–101). Beyond ligand- or matrix-focused interventions, clinically advanced TGF-β receptor I (ALK5) inhibitors offer near-term feasibility for combination regimens. Vactosertib has shown signals of activity in combination with pembrolizumab in microsatellite-stable colorectal cancer and favorable safety in hematologic malignancy when paired with an immunomodulatory backbone, motivating disease-adapted trials in ovarian cancer cohorts molecularly enriched for TGF-β–conditioned exclusion.
Implementation should be explicitly biomarker-driven and site-aware. As outlined by spatial and single-cell evidence, risk assignment can integrate a stromal/CAF activity index (e.g., INHBA, periostin, fibronectin), an antigen-presentation index, and quantitative interaction metrics (nearest-neighbor distances and interface lengths among CD8+ T cells, PD-L1+ tumor/myeloid populations, and CAF corridors). Pharmacodynamic endpoints: ≥20–30% shortening of CD8+–tumor distances, ≥30% reduction of continuous macrophage/CAF–tumor interfaces, and ≥1-grade HLA-I upshift with emergence of tumor–immune interdigitation; biopsy at baseline and ~2–4 weeks on-treatment. Standardized multiplex tissue imaging permits these readouts on archival formalin-fixed sections; MIBI-TOF has demonstrated reproducible, quantitative annotation of clinically relevant cell states and can anchor cross-center harmonization of spatial metrics (102–105). Given the heterogeneity of adnexal, omental, and peritoneal ecosystems, protocols should mandate multi-site sampling and predefine adjudication rules when spatial phenotypes diverge within a patient. In inflamed tumors with B-/T-cell aggregates, preserve/induce TLS: emergence and maintenance require CXCL13 and LTα/β; dose stromal modulation intermittently and tissue-sparing to avoid TLS disruption, pairing with DC/costimulatory support.
Future studies should prospectively test a tiered combination schema aligned to spatially measured biology. In a TGF-β–dominant, CAF-rich exclusion state, a backbone of PD-(L)1 with a TGF-β–axis agent selected to the dominant source (e.g., Activin A neutralization in INHBA-high CAF contexts or ALK5 inhibition with intermittent dosing) can be layered with CAF reprogrammers such as NOX4 inhibitors; in comparatively inflamed, TLS-rich states, emphasis can shift toward antigen-presentation and costimulation with stromal restraint. Trial designs should incorporate adaptive stopping rules tied to on-treatment engagement of the intended axis and include safety guardrails informed by the historical toxicity profile of TGF-β inhibitors (cutaneous events, gastrointestinal symptoms, and rare cardiotoxicity), with dosing schedules and patient selection optimized to minimize non-target immunologic perturbation. These principles convert single-cell and spatial readouts into actionable therapeutic logic: deconstrain access when T cells are present but excluded, restore recognition when antigen visibility is limited, and preserve organized immune niches when they emerge under therapy.
Author contributions
JH: Writing – original draft. JT: Writing – original draft. YZ: Writing – original draft. HL: Writing – original draft. WF: Writing – original draft, Writing – review & editing. YX: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Hu Y, Recouvreux MS, Haro M, Taylan E, Taylor-Harding B, Walts AE, et al. INHBA (+) cancer-associated fibroblasts generate an immunosuppressive tumor microenvironment in ovarian cancer. NPJ Precis Oncol. (2024) 8:35. doi: 10.1038/s41698-024-00523-y
2. Kasikova L, Rakova J, Hensler M, Lanickova T, Tomankova J, Pasulka J, et al. Tertiary lymphoid structures and B cells determine clinically relevant T cell phenotypes in ovarian cancer. Nat Commun. (2024) 15:2528. doi: 10.1038/s41467-024-46873-w
3. Kment J, Newsted D, Young S, Vermeulen MC, Laight BJ, Greer PA, et al. Blockade of TGF-β and PD-L1 by bintrafusp alfa promotes survival in preclinical ovarian cancer models by promoting T effector and NK cell responses. Br J Cancer. (2024) 130:2003–15. doi: 10.1038/s41416-024-02677-9
4. Jackson JW, Frederick Streich Jr C, Pal A, Coricor G, Boston C, Brueckner CT, et al. An antibody that inhibits TGF-β1 release from latent extracellular matrix complexes attenuates the progression of renal fibrosis. Sci Signaling. (2024) 17:eadn6052. doi: 10.1126/scisignal.adn6052
5. Li A, Chang Y, Song NJ, Wu X, Chung D, Riesenberg BP, et al. Selective targeting of GARP-LTGFβ axis in the tumor microenvironment augments PD-1 blockade via enhancing CD8+ T cell antitumor immunity. J Immunotherapy Cancer. (2022) 10:e005433. doi: 10.1136/jitc-2022-005433
6. Alabi FA, Okpalanwaka IF, Oyegbesan A, Okoyeocha E, Oladejo M, and Imodoye SO. Transforming growth factor-beta signaling in cancer: therapeutic implications, challenges, and pathways to progress. MedComm–Oncology. (2025) 4:e70027. doi: 10.1002/mog2.70027
7. Walsh LA and Quail DF. Decoding the tumor microenvironment with spatial technologies. Nat Immunol. (2023) 24:1982–93. doi: 10.1038/s41590-023-01678-9
8. Olbrecht S, Busschaert P, Qian J, Vanderstichele A, Loverix L, Van Gorp T, et al. High-grade serous tubo-ovarian cancer refined with single-cell RNA sequencing: specific cell subtypes influence survival and determine molecular subtype classification. Genome Med. (2021) 13:111. doi: 10.1186/s13073-021-00922-x
9. Regner MJ, Wisniewska K, Garcia-Recio S, Thennavan A, Mendez-Giraldez R, Malladi VS, et al. A multi-omic single-cell landscape of human gynecologic Malignancies. Mol Cell. (2021) 81:4924–4941. e10. doi: 10.1016/j.molcel.2021.10.013
10. Croft W, Pounds R, Jeevan D, Singh K, Balega J, Sundar S, et al. The chromatin landscape of high-grade serous ovarian cancer metastasis identifies regulatory drivers in post-chemotherapy residual tumour cells. Commun Biol. (2024) 7:1211. doi: 10.1038/s42003-024-06909-9
11. Lin SC, Liao YC, Chen PM, Yang YY, Wang YH, Tung SL, et al. Periostin promotes ovarian cancer metastasis by enhancing M2 macrophages and cancer-associated fibroblasts via integrin-mediated NF-κB and TGF-β2 signaling. J Biomed Sci. (2022) 29:109. doi: 10.1186/s12929-022-00888-x
12. Evans ET, Page EF, Choi AS, Shonibare Z, Kahn AG, Arend RC, et al. Activin levels correlate with lymphocytic infiltration in epithelial ovarian cancer. Cancer Med. (2024) 13:e7368. doi: 10.1002/cam4.7368
13. Chung SW, Xie Y, and Suk JS. Overcoming physical stromal barriers to cancer immunotherapy. Drug delivery Trans Res. (2021) 11:2430–47. doi: 10.1007/s13346-021-01036-y
14. Peng D, Fu M, Wang M, Wei Y, and Wei X. Targeting TGF-β signal transduction for fibrosis and cancer therapy. Mol Cancer. (2022) 21:104. doi: 10.1186/s12943-022-01569-x
15. Ford K, Hanley CJ, Mellone M, Szyndralewiez C, Heitz F, Wiesel P, et al. NOX4 inhibition potentiates immunotherapy by overcoming cancer-associated fibroblast-mediated CD8 T-cell exclusion from tumors. Cancer Res. (2020) 80:1846–60. doi: 10.1158/0008-5472.CAN-19-3158
16. Hsieh WC, Budiarto BR, Wang YF, Lin CY, Gwo MC, So DK, et al. Spatial multi-omics analyses of the tumor immune microenvironment. J Biomed Sci. (2022) 29:96. doi: 10.1186/s12929-022-00879-y
17. Li X, Xu H, Du Z, Cao Q, and Liu X. Advances in the study of tertiary lymphoid structures in the immunotherapy of breast cancer. Front Oncol. (2024) 14:1382701. doi: 10.3389/fonc.2024.1382701
18. Garsed DW, Pandey A, Fereday S, Kennedy CJ, Takahashi K, Alsop K, et al. The genomic and immune landscape of long-term survivors of high-grade serous ovarian cancer. Nat Genet. (2022) 54:1853–64. doi: 10.1038/s41588-022-01230-9
19. Guo J, Han X, Li J, Li Z, Yi J, Gao Y, et al. Single-cell transcriptomics in ovarian cancer identify a metastasis-associated cell cluster overexpressed RAB13. J Trans Med. (2023) 21:254. doi: 10.1186/s12967-023-04094-7
20. Taylor MJ, Lukowski JK, and Anderton CR. Spatially resolved mass spectrometry at the single cell: recent innovations in proteomics and metabolomics. J Am Soc Mass Spectrometry. (2021) 32:872–94. doi: 10.1021/jasms.0c00439
21. De Oliveira Macena Y, Cezar MEN, Lira CBF, De Oliveira LBDM, Almeida TN, Costa ADAV, et al. The roles of periostin derived from cancer-associated fibroblasts in tumor progression and treatment response. Cancer Metastasis Rev. (2025) 44:11. doi: 10.1007/s10555-024-10233-3
22. Kelly MR, Wisniewska K, Regner MJ, Lewis MW, Perreault AA, Davis ES, et al. A multi-omic dissection of super-enhancer driven oncogenic gene expression programs in ovarian cancer. Nat Commun. (2022) 13:4247. doi: 10.1038/s41467-022-31919-8
23. Liu R, Liu J, Cao Q, Chu Y, Chi H, Zhang J, et al. Identification of crucial genes through WGCNA in the progression of gastric cancer. J Cancer. (2024) 15:3284. doi: 10.7150/jca.95757
24. Zhao Q, Shao H, and Zhang T. Single-cell RNA sequencing in ovarian cancer: revealing new perspectives in the tumor microenvironment. Am J Trans Res. (2024) 16:3338. doi: 10.62347/SMSG9047
25. Gulley JL, Schlom J, Barcellos-Hoff MH, Wang XJ, Seoane J, Audhuy F, et al. Dual inhibition of TGF-β and PD-L1: a novel approach to cancer treatment. Mol Oncol. (2022) 16:2117–34. doi: 10.1002/1878-0261.13146
26. Pinjusic K, Bulliard M, Rothé B, Ansaryan S, Liu YC, Ginefra P, et al. Stepwise release of Activin-A from its inhibitory prodomain is modulated by cysteines and requires furin coexpression to promote melanoma growth. Commun Biol. (2024) 7:1383. doi: 10.1038/s42003-024-07053-0
27. Peng G, Chi H, Gao X, Zhang J, Song G, Xie X, et al. Identification and validation of neurotrophic factor-related genes signature in HNSCC to predict survival and immune landscapes. Front Genet. (2022) 13:1010044. doi: 10.3389/fgene.2022.1010044
28. Gong S, Wang S, and Shao M. NADPH oxidase 4: a potential therapeutic target of Malignancy. Front Cell Dev Biol. (2022) 10:884412. doi: 10.3389/fcell.2022.884412
29. Liu CC, Bosse M, Kong A, Kagel A, Kinders R, Hewitt SM, et al. Reproducible, high-dimensional imaging in archival human tissue by multiplexed ion beam imaging by time-of-flight (MIBI-TOF). Lab Invest. (2022) 102:762–70. doi: 10.1038/s41374-022-00778-8
30. Lanickova T, Hensler M, Kasikova L, Vosahlikova S, Angelidou A, Pasulka J, et al. Chemotherapy drives tertiary lymphoid structures that correlate with ICI-responsive TCF1+ CD8+ T cells in metastatic ovarian cancer. Clin Cancer Res. (2025) 31:164–80. doi: 10.1158/1078-0432.CCR-24-1594
31. Denisenko E, de Kock L, Tan A, Beasley AB, Beilin M, Jones ME, et al. Spatial transcriptomics reveals discrete tumour microenvironments and autocrine loops within ovarian cancer subclones. Nat Commun. (2024) 15:2860. doi: 10.1038/s41467-024-47271-y
32. Yeh CY, Aguirre K, Laveroni O, Kim S, Wang A, Liang B, et al. Mapping spatial organization and genetic cell-state regulators to target immune evasion in ovarian cancer. Nat Immunol. (2024) 25:1943–58. doi: 10.1038/s41590-024-01943-5
33. Kader T, Lin JR, Hug CB, Coy S, Chen YA, de Bruijn I, et al. Multimodal spatial profiling reveals immune suppression and microenvironment remodeling in fallopian tube precursors to high-grade serous ovarian carcinoma. Cancer Discov. (2025) 15:1180–202. doi: 10.1158/2159-8290.CD-24-1366
34. Kim TW, Lee KW, Ahn JB, Lee J, Ryu J, Oh B, et al. 618P Efficacy and safety of vactosertib and pembrolizumab combination in patients with previously treated microsatellite sta ble metastatic colorectal cancer. Ann Oncol. (2023) 34:S443.
35. Yap TA, Choudhury AD, Hamilton E, Rosen LS, Stratton KL, Gordon MS, et al. PF-06952229, a selective TGF-β-R1 inhibitor: preclinical development and a first-in-human, phase I, dose-escalation study in advanced solid tumors. ESMO Open. (2024) 9:103653. doi: 10.1016/j.esmoop.2024.103653
36. Liu Y, Xiao H, Zeng H, and Xiang Y. Beyond tumor-associated macrophages involved in spheroid formation and dissemination: Novel insights for ovarian cancer therapy. Int J Oncol. (2024) 65:117. doi: 10.3892/ijo.2024.5705
37. Schweizer L, Kenny HA, Krishnan R, Kelliher L, Bilecz AJ, Heide J, et al. Spatial proteo-transcriptomic profiling reveals the molecular landscape of borderline ovarian tumors and their invasive progression. Cancer Cell. (2024) S1535-6108.
38. Chiaro J, Peltonen K, Õunap K, Bailey A, Feola S, Wojciechowski S, et al. Proteogenomic approach to immunopeptidomics of ovarian tumors identifies shared peptide vaccine candidates. NPJ Vaccines. (2025) 10:195. doi: 10.1038/s41541-025-01234-6
39. Li H, Guo L, Su K, Li C, Jiang Y, Wang P, et al. Construction and validation of TACE therapeutic efficacy by ALR score and nomogram: a large, multicenter study. J Hepatocellular Carcinoma. (2023) 25:1009–17. doi: 10.2147/JHC.S414926
40. Luo S, Germain PL, Robinson MD, and von Meyenn F. Benchmarking computational methods for single-cell chromatin data analysis. Genome Biol. (2024) 25:225. doi: 10.1186/s13059-024-03356-x
41. Wang X, Jin Z, Shi Y, and Xi R. Detecting copy-number alterations from single-cell chromatin sequencing data by AtaCNA. Cell Rep Methods. (2025) 5:100939. doi: 10.1016/j.crmeth.2024.100939
42. Coscia F, Nielsen AB, Weigert M, Watters K, Javellana M, Anglesio M, et al. A proteogenomic view of synchronous endometrioid endometrial and ovarian cancer. Clin Cancer Res. (2025) 31:2230–40. doi: 10.1158/1078-0432.CCR-24-1763
43. Zheng S, Wang W, Shen L, Yao Y, Xia W, Ni C, et al. Tumor battlefield within inflamed, excluded or desert immune phenotypes: the mechanisms and strategies. Exp Hematol Oncol. (2024) 13:80. doi: 10.1186/s40164-024-00543-1
44. Karami Z, Mortezaee K, and Majidpoor J. Dual anti-PD-(L) 1/TGF-β inhibitors in cancer immunotherapy–updated. Int Immunopharmacol. (2023) 122:110648. doi: 10.1016/j.intimp.2023.110648
45. He S, Su L, Hu H, Liu H, Xiong J, Gong X, et al. Immunoregulatory functions and therapeutic potential of natural killer cell-derived extracellular vesicles in chronic diseases. Front Immunol. (2024) 14:1328094. doi: 10.3389/fimmu.2023.1328094
46. Rubin MA, Bristow RG, Thienger PD, Dive C, and Imielinski M. Impact of lineage plasticity to and from a neuroendocrine phenotype on progression and response in prostate and lung cancers. Mol Cell. (2020) 80:562–77. doi: 10.1016/j.molcel.2020.10.033
47. Burdett NL, Willis MO, Pandey A, Twomey L, Alaei S, Australian Ovarian Cancer Study Group, et al. Timing of whole genome duplication is associated with tumor-specific MHC-II depletion in serous ovarian cancer. Nat Commun. (2024) 15:6069. doi: 10.1038/s41467-024-50137-y
48. Sun G and Liu Y. Tertiary lymphoid structures in ovarian cancer. Front Immunol. (2024) 15:1465516. doi: 10.3389/fimmu.2024.1465516
49. Tan B, Khattak A, Felip E, Kelly K, Rich P, Wang D, et al. Bintrafusp alfa, a bifunctional fusion protein targeting TGF-β and PD-L1, in patients with esophageal adenocarcinoma: results from a phase 1 cohort. Targeted Oncol. (2021) 16:435–46. doi: 10.1007/s11523-021-00809-2
50. Amengual J, Gonzalez-Sanchez E, Yáñez-Bartolome M, Sererols-Viñas L, Ravichandra A, Guiton C, et al. NADPH oxidase 1/4 dual inhibition impairs transforming growth factor-beta protumorigenic effects in cholangiocarcinoma cancer-associated fibroblasts. Signal Transduction Targeted Ther. (2025) 10:257. doi: 10.1038/s41392-025-02347-z
51. Liu Y, Ye SY, He S, Chi DM, Wang XZ, Wen YF, et al. Single-cell and spatial transcriptome analyses reveal tertiary lymphoid structures linked to tumour progression and immunotherapy response in nasopharyngeal carcinoma. Nat Commun. (2024) 15:7713. doi: 10.1038/s41467-024-52153-4
52. Xie Y, Peng H, Hu Y, Jia K, Yuan J, Liu D, et al. Immune microenvironment spatial landscapes of tertiary lymphoid structures in gastric cancer. BMC Med. (2025) 23:59. doi: 10.1186/s12916-025-03889-3
53. Gao VR, Yang R, Das A, Luo R, Luo H, McNally DR, et al. ChromaFold predicts the 3D contact map from single-cell chromatin accessibility. Nat Commun. (2024) 15:9432. doi: 10.1038/s41467-024-53628-0
54. Zhang K, Hocker JD, Miller M, Hou X, Chiou J, Poirion OB, et al. A single-cell atlas of chromatin accessibility in the human genome. Cell. (2021) 184:5985–6001. e19. doi: 10.1016/j.cell.2021.10.024
55. Lowe MM, Cohen JN, Moss MI, Clancy S, Adler JP, Yates AE, et al. Tertiary lymphoid structures sustain cutaneous B cell activity in hidradenitis suppurativa. JCI Insight. (2024) 9:e169870.
56. Ju HY, Youn SY, Kang J, Whang MY, Choi YJ, Han MR, et al. Integrated analysis of spatial transcriptomics and CT phenotypes for unveiling the novel molecular characteristics of recurrent and non-recurrent high-grade serous ovarian cancer. biomark Res. (2024) 12:80. doi: 10.1186/s40364-024-00632-7
57. Liu W, Xia L, Peng Y, Cao Q, Xu K, Luo H, et al. Unraveling the significance of cuproptosis in hepatocellular carcinoma heterogeneity and tumor microenvironment through integrated single-cell sequencing and machine learning approaches. Discover Oncol. (2025) 16:900. doi: 10.1007/s12672-025-02696-9
58. Carstens JL, Krishnan SN, Rao A, Sorace AG, Seeley EH, Ferri-Borgogno S, et al. Spatial multiplexing and omics. Nat Rev Methods Primers. (2024) 4:54. doi: 10.1038/s43586-024-00330-6
59. Stur E, Peng F, Teng PN, Bayraktar E, Hu M, Corvigno S, et al. The dynamic immune behavior of primary and metastatic ovarian carcinoma. NPJ Precis Oncol. (2025) 9:120. doi: 10.1038/s41698-025-00818-8
60. Vázquez-García I, Uhlitz F, Ceglia N, Lim JLP, Wu M, Mohibullah N, et al. Ovarian cancer mutational processes drive site-specific immune evasion. Nature. (2022) 612:778–86. doi: 10.1038/s41586-022-05496-1
61. Mateiou C, Lokhande L, Diep LH, Knulst M, Carlsson E, Ek S, et al. Spatial tumor immune microenvironment phenotypes in ovarian cancer. NPJ Precis Oncol. (2024) 8:148. doi: 10.1038/s41698-024-00640-8
62. Gendrau-Sanclemente N, Figueras A, Gracova K, Lahiguera Á, Alsina-Sanchís E, Marín-Jiménez JA, et al. Ovarian cancer relies on the PDGFRβ–fibronectin axis for tumorsphere formation and metastatic spread. Mol Oncol. (2024) 18:136–55.
63. Huang Z, Byrd O, Tan S, Hu K, Knight B, Lo G, et al. Periostin facilitates ovarian cancer recurrence by enhancing cancer stemness. Sci Rep. (2023) 13:21382. doi: 10.1038/s41598-023-48485-8
64. You Y, Chen Y, Li J, Zhang Q, Zhang Y, Yang P, et al. Physical activity mitigates the influence of blood cadmium on memory function: a cross-sectional analysis in US elderly population. Environ Sci pollut Res. (2023) 30:68809–20. doi: 10.1007/s11356-023-27053-7
65. Byrne A, Le D, Sereti K, Menon H, Vaidya S, Patel N, et al. Single-cell long-read targeted sequencing reveals transcriptional variation in ovarian cancer. Nat Commun. (2024) 15:6916. doi: 10.1038/s41467-024-51252-6
66. Van Kleunen LB, Ahmadian M, Post MD, Wolsky RJ, Rickert C, Jordan KR, et al. The spatial structure of the tumor immune microenvironment can explain and predict patient response in high-grade serous carcinoma. Cancer Immunol Res. (2024) 12:1492–507. doi: 10.1158/2326-6066.CIR-23-1109
67. Bollhagen A and Bodenmiller B. Highly multiplexed tissue imaging in precision oncology and translational cancer research. Cancer Discov. (2024) 14:2071–88. doi: 10.1158/2159-8290.CD-23-1165
68. Samorodnitsky S, Campbell K, Ribas A, and Wu MC. A SPatial Omnibus Test (SPOT) for spatial proteomic data. Bioinformatics. (2024) 40:btae425. doi: 10.1093/bioinformatics/btae425
69. Feng Y, Ma W, Zang Y, Guo Y, Li Y, Zhang Y, et al. Spatially organized tumor-stroma boundary determines the efficacy of immunotherapy in colorectal cancer patients. Nat Commun. (2024) 15:10259. doi: 10.1038/s41467-024-54710-3
70. Chap BS, Rayroux N, Grimm AJ, Ghisoni E, and Dangaj Laniti D. Crosstalk of T cells within the ovarian cancer microenvironment. Trends Cancer. (2024) 10:1116–30. doi: 10.1016/j.trecan.2024.09.001
71. Barrett RL and Puré E. Cancer-associated fibroblasts and their influence on tumor immunity and immunotherapy. Elife. (2020) 9:e57243. doi: 10.7554/eLife.57243
72. Li Z, Sun C, and Qin Z. Metabolic reprogramming of cancer-associated fibroblasts and its effect on cancer cell reprogramming. Theranostics. (2021) 11:8322. doi: 10.7150/thno.62378
73. Huang J, Zhang L, Wan D, Zhou L, Zheng S, Lin S, et al. Extracellular matrix and its therapeutic potential for cancer treatment. Signal transduction targeted Ther. (2021) 6:153. doi: 10.1038/s41392-021-00544-0
74. Mai Z, Lin Y, Lin P, Zhao X, and Cui L. Modulating extracellular matrix stiffness: a strategic approach to boost cancer immunotherapy. Cell Death Dis. (2024) 15:307. doi: 10.1038/s41419-024-06697-4
75. Di Mauro F and Arbore G. Spatial dissection of the immune landscape of solid tumors to advance precision medicine. Cancer Immunol Res. (2024) 12:800–13. doi: 10.1158/2326-6066.CIR-23-0699
76. Omar M, Fanelli GN, Socciarelli F, Ullanat V, Puchala SR, Wen J, et al. Antibody-based multiplex image analysis: standard analytical workflows and tools for pathologists. Lab Invest. (2025) 104220:307.
77. Lukowski JK, Cho BK, Calderon AZ, Dianati B, Stumpo K, Snyder S, et al. Advances in spatial multi-omics: A review of multi-modal mass spectrometry imaging and laser capture microdissection-LCMS integration. Proteomics. (2025) 7:e202400378. doi: 10.1002/pmic.202400378
78. Hu Q, Zhu Y, Mei J, Liu Y, and Zhou G. Extracellular matrix dynamics in tumor immunoregulation: from tumor microenvironment to immunotherapy. J Hematol Oncol. (2025) 18:65. doi: 10.1186/s13045-025-01717-y
79. Lin P, Lin Y, Chen X, Zhao X, and Cui L. Decoding MHC loss: Molecular mechanisms and implications for immune resistance in cancer. Clin Trans Med. (2025) 15:e70403. doi: 10.1002/ctm2.70403
80. Kubo T, Asano S, Sasaki K, Murata K, Kanaseki T, Tsukahara T, et al. Assessment of cancer cell-expressed HLA class I molecules and their immunopathological implications. HLA. (2024) 103:e15472. doi: 10.1111/tan.15472
81. Egan D, Glennon K, Treacy A, Fabre A, McCormack J, Hill S, et al. A temporal model of tumor-immune dynamics during the metastatic progression of high-grade serous ovarian cancer. NPJ Precis Oncol. (2025) 9:188. doi: 10.1038/s41698-025-00973-y
82. Mantovani A, Marchesi F, Di Mitri D, and Garlanda C. Macrophage diversity in cancer dissemination and metastasis. Cell Mol Immunol. (2024) 21:1201–14. doi: 10.1038/s41423-024-01216-z
83. Xia S, Chen L, Yu M, Li J, Chen J, Xu F, et al. Genetic and therapeutic heterogeneity shape the baseline and longitudinal immune ecosystem of ovarian clear cell carcinoma. J Immunotherapy Cancer. (2024) 12:e010069. doi: 10.1136/jitc-2024-010069
84. Chi J, Gao Q, and Liu D. Tissue-resident macrophages in cancer: friend or foe? Cancer Med. (2024) 13:e70387.
85. Zhu C, Xu Z, Zhang T, Qian L, Xiao W, Wei H, et al. Updates of pathogenesis, diagnostic and therapeutic perspectives for ovarian clear cell carcinoma. J Cancer. (2021) 12:2295. doi: 10.7150/jca.53395
86. Ma Q, Kang R, Xu R, Guan Y, Chang S, Li S, et al. Crosstalk between stromal, immune, and ovarian cancer cells in lipid-rich tumor microenvironment exhibits proliferative features. Front Immunol. (2025) 16:1614815. doi: 10.3389/fimmu.2025.1614815
87. Bareham B, Dibble M, and Parsons M. Defining and modeling dynamic spatial heterogeneity within tumor microenvironments. Curr Opin Cell Biol. (2024) 90:102422. doi: 10.1016/j.ceb.2024.102422
88. Tufail M, Jiang CH, and Li N. Immune evasion in cancer: mechanisms and cutting-edge therapeutic approaches. Signal Transduction Targeted Ther. (2025) 10:227. doi: 10.1038/s41392-025-02280-1
89. Castiglioni A, Yang Y, Williams K, Gogineni A, Lane RS, Wang AW, et al. Combined PD-L1/TGFβ blockade allows expansion and differentiation of stem cell-like CD8 T cells in immune excluded tumors. Nat Commun. (2023) 14:4703. doi: 10.1038/s41467-023-40398-4
90. Nan L, Qin Y, Huang X, Pan M, Wang X, Lv Y, et al. A bifunctional anti-PD-1/TGF-β fusion antibody restores antitumour immunity and remodels the tumour microenvironment. Int J Mol Sci. (2025) 26:7567. doi: 10.3390/ijms26157567
91. Chowdhury S, Kennedy JJ, Ivey RG, Murillo OD, Hosseini N, Song X, et al. Proteogenomic analysis of chemo-refractory high-grade serous ovarian cancer. Cell. (2023) 186:3476–3498. e35. doi: 10.1016/j.cell.2023.07.004
92. Ferreira GA, Thomé CH, Izumi C, Grassi ML, Lanfredi GP, Smolka M, et al. Proteomic analysis of exosomes secreted during the epithelial-mesenchymal transition and potential biomarkers of mesenchymal high-grade serous ovarian carcinoma. J Ovarian Res. (2023) 16:232. doi: 10.1186/s13048-023-01304-0
93. Bell ATF, Mitchell JT, Kiemen AL, Lyman M, Fujikura K, Lee JW, et al. PanIN and CAF transitions in pancreatic carcinogenesis revealed with spatial data integration. Cell Syst. (2024) 15:753–769. e5. doi: 10.1016/j.cels.2024.07.001
94. Liu CC, Greenwald NF, Kong A, McCaffrey EF, Leow KX, Mrdjen D, et al. Robust phenotyping of highly multiplexed tissue imaging data using pixel-level clustering. Nat Commun. (2023) 14:4618. doi: 10.1038/s41467-023-40068-5
95. Fan L, Liu Y, Zhou H, Feng Y, Jiang G, Hou G, et al. Spatially resolved proteomics surveys the chemo-refractory proteins related to high-grade serous ovarian cancer. Clin Trans Med. (2025) 15:e70422. doi: 10.1002/ctm2.70422
96. Wu Y, Yan Y, Guo Y, Niu M, Zhou B, Zhang J, et al. Anti-TGF-β/PD-L1 bispecific antibody synergizes with radiotherapy to enhance antitumor immunity and mitigate radiation-induced pulmonary fibrosis. J Hematol Oncol. (2025) 18:24. doi: 10.1186/s13045-025-01678-2
97. Yi M, Wu Y, Niu M, Zhu S, Zhang J, Yan Y, et al. Anti-TGF-β/PD-L1 bispecific antibody promotes T cell infiltration and exhibits enhanced antitumor activity in triple-negative breast cancer. J Immunotherapy Cancer. (2022) 10:e005543. doi: 10.1136/jitc-2022-005543
98. Mortaheb S, Pezeshki PS, and Rezaei N. Bispecific therapeutics: a state-of-the-art review on the combination of immune checkpoint inhibition with costimulatory and non-checkpoint targeted therapy. Expert Opin Biol Ther. (2024) 24:1335–51. doi: 10.1080/14712598.2024.2426636
99. Zhang H, Lu KH, Ebbini M, Huang P, Lu H, Li L, et al. Mass spectrometry imaging for spatially resolved multi-omics molecular mapping. NPJ Imaging. (2024) 2:20. doi: 10.1038/s44303-024-00025-3
100. Tapia-Galisteo A, Sánchez-Rodríguez I, Narbona J, Iglesias-Hernández P, Aragón-García S, Jiménez-Reinoso A, et al. Combination of T cell-redirecting strategies with a bispecific antibody blocking TGF-β and PD-L1 enhances antitumor responses. Oncoimmunology. (2024) 13:2338558. doi: 10.1080/2162402X.2024.2338558
101. Lind H, Gameiro SR, Jochems C, Donahue RN, Strauss J, Gulley JL, et al. Dual targeting of TGF-β and PD-L1 via a bifunctional anti-PD-L1/TGF-βRII agent: status of preclinical and clinical advances. J immunotherapy Cancer. (2020) 8:e000433. doi: 10.1136/jitc-2019-000433
102. Paz-Ares L, Kim TM, Vicente D, Felip E, Lee DH, Lee KH, et al. Bintrafusp alfa, a bifunctional fusion protein targeting TGF-β and PD-L1, in second-line treatment of patients with NSCLC: results from an expansion cohort of a phase 1 trial. J Thorac Oncol. (2020) 15:1210–22. doi: 10.1016/j.jtho.2020.03.003
103. Grogg J, Vernet R, Charrier E, Urwyler M, Von Rohr O, Saingier V, et al. Engineering a versatile and retrievable cell macroencapsulation device for the delivery of therapeutic proteins. Iscience. (2023) 26:2338558. doi: 10.1016/j.isci.2023.107372
104. Harms PW, Frankel TL, Moutafi M, Rao A, Rimm DL, Taube JM, et al. Multiplex immunohistochemistry and immunofluorescence: a practical update for pathologists. Modern Pathol. (2023) 36:100197. doi: 10.1016/j.modpat.2023.100197
105. Su K, Wang F, Li X, Chi H, Zhang J, He K, et al. Effect of external beam radiation therapy versus transcatheter arterial chemoembolization for non-diffuse hepatocellular carcinoma (≥ 5 cm): a multicenter experience over a ten-year period. Front Immunol. (2023) 14:1265959. doi: 10.3389/fimmu.2023.1265959
Keywords: ovarian cancer, TGF-β signalling, T-cell exclusion, single-cell RNA/ATAC, spatial transcriptomics
Citation: He J, Tao J, Zhou Y, Li H, Feng W and Xu Y (2025) TGF-β–driven T-cell exclusion in ovarian cancer: single-cell and spatial transcriptomic views of immune low-response states. Front. Immunol. 16:1698088. doi: 10.3389/fimmu.2025.1698088
Received: 03 September 2025; Accepted: 03 October 2025;
Published: 17 October 2025.
Edited by:
Shangke Huang, Southwest Medical University, ChinaReviewed by:
Zhiheng Lin, Shanghai University of Traditional Chinese Medicine, ChinaCopyright © 2025 He, Tao, Zhou, Li, Feng and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenqi Feng, bW9vbmx5MTk4MUAxNjMuY29t; Yongqiang Xu, eHUxMzkwOTA5MjkyM0AxNjMuY29t
 Jiang He1
Jiang He1 Jun Tao
Jun Tao Wenqi Feng
Wenqi Feng