Abstract
Background:
In recent years, researchers have identified numerous potential biomarkers and therapeutic targets applicable to cancer immunotherapy, among which the role of tumor-internal microorganisms in the tumor microenvironment has been explored. However, this field is still in its early stages of development, facing limitations such as the unclear mechanisms of interaction between tumor-internal microorganisms and host immunity, as well as significant variations in microbial profiles among different tumor types and patients.
Objective:
This study aims to explore the research hotspots and development trends of tumor-internal microorganisms through bibliometric methods and to construct a systematic knowledge map.
Methods:
This study retrieved publications related to tumor-internal microorganisms from the Web of Science Core Collection (WOSCC) prior to December 22, 2025. Subsequently, the selected literature was analyzed using VOSviewer (v.1.6.20), CiteSpace (v.6.4.1R), and SCImago Graphica. In addition, we integrated PubMed data to assess status and trends in preclinical and clinical studies of intratumoral microbiota interventions for anti-tumor therapy efficacy.
Results:
From the Web of Science database, we retrieved 1,278 relevant articles. Since 2012, the number of papers published on the intratumoral microbiota has shown an overall upward trend. China and the United States are the two major countries in this field. Keyword analysis shows that “tumor microbiome,” “gut microbiome,” “cancer,” and “Fusobacterium nucleatum” are frequent terms. 11 keyword groups have been identified, among which “tumor immunotherapy” and “immune microenvironment” form two important groups. A total of 69 preclinical and clinical studies has intervened in intratumoral microbiota and affected anti-tumor treatment outcomes. Among them, 25 studies involving Fusobacterium nucleatum account for a large proportion. However, most of these studies are still at the basic or preclinical stage, and clinical translation evidence is limited.
1 Introduction
In recent years, research on the tumor microenvironment (TME) has deepened, and in addition to the traditional focus on tumor cells, immune cells, and stromal cells, the intratumoral microbiome (ITM) has gradually emerged as a new direction in oncology research (1–3). Since its systematic reporting in 2020 as being present in cancer cells and immune cells (4), numerous studies have discovered that intratumoral microbes not only exhibit high heterogeneity (5) and low biomass characteristics (6, 7) within tumor tissues but may also play significant roles in tumorigenesis, progression, recurrence, and prognosis by influencing processes (8–10)such as tumor inflammatory responses (11), immune evasion (12), genomic stability (13), and treatment resistance (14). Furthermore, significant differences in the composition of intratumoral microbiota exist among patients of different tumor origins, subtypes (15, 16), ethnicities (17), and lifestyles (18), suggesting that intratumoral microbes hold promise as potential biomarkers and therapeutic targets in cancer diagnosis and treatment.
In terms of detection technology, the development of new methods such as 2bRAD-M sequencing, 16S rRNA sequencing, and spatial transcriptomics has significantly enhanced the detection and functional analysis capabilities of low-abundance microbes, providing powerful tools for elucidating the biological significance of intratumoral microbiota (19–21). In clinical application explorations, existing research indicates that intratumoral microbes can modulate the efficacy of immunotherapy, targeted therapy, chemotherapy, and radiotherapy (22–25). Some studies have even combined antimicrobial therapy with nanocarrier technology to improve microbe-related immune suppression (26, 27). However, this field still faces numerous challenges, including high risks of sample contamination, lack of standardized detection methods, insufficient mechanistic studies, and the complexity of interactions among different microbial communities, which to some extent limits the reproducibility of research results and their clinical translatability.
Against this backdrop, it is essential to systematically review and quantitatively analyze the knowledge structure, hot topics, cutting-edge trends, and academic collaboration networks in the field of intratumoral microbiota research. Bibliometrics, as a method that combines quantitative statistics and visual analysis, can reveal the development history, core scholars, major research institutions, key literature, and future directions of specific research fields, and has been widely applied in the analysis of research hotspots in oncology and microbiology (28). Therefore, this study aims to conduct a systematic analysis of literature related to intratumoral microbiota based on the internationally recognized database Web of Science, utilizing bibliometric and visualization techniques, with the goal of mapping the overall research landscape of this field, identifying its core research hotspots and potential cutting-edge developments, and providing references for subsequent basic research, clinical translation, and interdisciplinary collaboration.
2 Materials and methods
2.1 Sources of literature and search strategy
In this study, the literature search mainly came from two databases: Web of Science and PubMed, covering relevant research from the establishment of the database until December 22, 2025. We conducted a systematic search for the subject term “intratumoral microbiota,” with specific search strategies outlined in the “Supplementary File 1.” Through the search in Web of Science, we initially identified 2,835 records. After screening, a total of 1,278 documents were included, forming the bibliometric dataset used for visual analysis of annual publication volume, co-authorship, co-citation, and keyword mapping, with the specific search process detailed in Figure 1A and “Supplementary File 1.” In addition, 1,867 records were retrieved from the PubMed database. After removing duplicates in comparison with the 1,278 documents included from Web of Science, the total number of documents from both Web of Science and PubMed was 2,951. Subsequently, we excluded 2,886 documents that did not meet the research criteria, resulting in a final number of 69 documents that met the inclusion criteria. Detailed information on the search process can be found in Figure 1B and the “Supplementary File 1.” These 69 documents will be used for subsequent analysis of trends in preclinical research and clinical trial studies. The retrieval and screening process is shown in Figure 1:
Figure 1

Flow chart of literature retrieval. (A) Flowchart of the Web of Science search process, with search results incorporated into software for visualizing bibliometric analysis. (B) Flowchart of the Web of Science and PubMed search processes, with search results analyzed for the status and trends of preclinical and clinical trial research. The flowchart was created using the PRISMA 2020 flow diagram for updated systematic reviews, available at: https://www.prisma-statement.org/prisma-2020-flow-diagram.
2.2 Inclusion and exclusion criteria
Eligible studies include original research and review articles that directly investigate the relationship between intratumoral microbiota and tumors. Exclusion criteria encompass non-research materials (such as editorials, book chapters, and conference abstracts), duplicate entries, retracted publications, and studies with insufficient or incomplete data. Additionally, studies unrelated to intratumoral microbiota were also manually excluded. Two researchers independently conducted the screening, data extraction, and verification, resolving any discrepancies through team discussions. The final dataset includes complete records of Full Record.
The literature on preclinical and clinical studies of intratumoral microbiota interventions for anti-tumor therapy efficacy applied stringent inclusion and exclusion criteria. Inclusion criteria: 1. Research type: Clinical Study, Clinical Trial, Clinical Trial, Phase I, Clinical Trial, Phase II, Clinical Trial, Phase III, Clinical Trial, Phase IV, Controlled Clinical Trial, Pragmatic Clinical Trial, Randomized Controlled Trial. 2. PICO principles: P (Participant): Tumor patients, tumor cells, or experimental animals; no species restrictions. I (Intervention): Interventions targeting naturally occurring intratumoral microbiota to influence anti-tumor treatment efficacy. C (Comparison): Blank control group or placebo group. O (Outcome): Tumor treatment efficacy. Exclusion criteria: 1. A control group was not established. 2. The interventions employed in the study, or the outcomes observed, did not clearly involve the intratumoral microbiota. 3. Not related to Anti-tumor therapy.
2.3 Data analysis and visualization
This study employed four software tools for bibliometric and visualization analysis, including Microsoft Excel 2022, VOSviewer (v.1.6.20), CiteSpace (v6.4.R1), and SCImago Graphica.
2.3.1 VOSviewer (v.1.6.20)
It was used to generate network diagrams of country/regional cooperation (Figure 3A), institutional cooperation network (Figure 4B), and author cooperation network (Figure 5). The analysis adopted the “total count” method built into the software. To focus on core participants, the minimum publication volume threshold was set as follows: authors ≥ 6, countries/regions ≥ 8, institutions ≥ 11. In the network diagrams, the size of the nodes is proportional to the number of publications, the color represents different clusters, and the thickness of the lines reflects the intensity of cooperation.
Figure 2
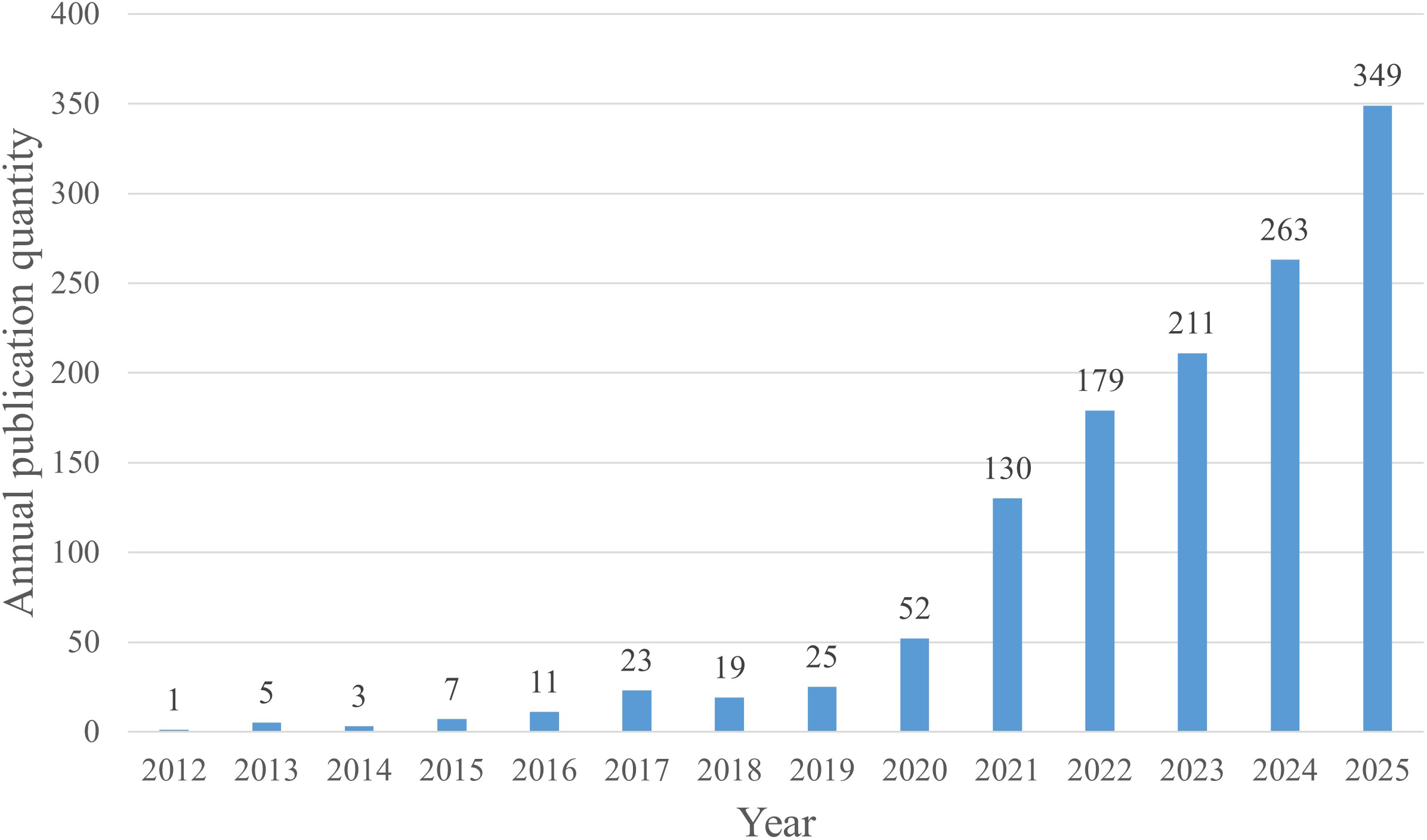
Overview of intratumoral microorganism–related publications by volume. Annual publication numbers from 2012 to 2025.
Figure 3

Participation of countries/regions in intratumoral microbiome research. (A) A network diagram assessing global collaboration. (B) A world map of intratumoral microbiomes. The numerical values correspond to the publication volume of each country, the text denotes the names of the countries, the size of the circles reflects the proportion of publication volume, and the lines indicate collaborations between different countries.
Figure 4

Visualization of institutions related to intratumoral microbiome research: institutional co-occurrence network view.
2.3.2 CiteSpace (v6.4.R1)
CiteSpace (v6.4.R1) was used for institution cooperation network analysis (Figure 4A), keyword co-occurrence (Figure 6), clustering (Figure 7), emergent (Figure 8), and timeline evolution (Figure 9) analysis. The parameters were set as follows: time slice set to 1 year, analysis range up to 2025. The node types included institutions, authors, and keywords. The network was optimized by using the Pathfinder, Pruning sliced network, and Pruning the merged network algorithms. The main network parameters were: g-index (k=25), node number (N)=272, edge number (E)=418, network density (Density)=0.0113. The settings for emergent word detection were γ (1.0), minimum duration = 1. The specific details of keyword merging can be found in the appendix “Supplementary materials.”.
Figure 5
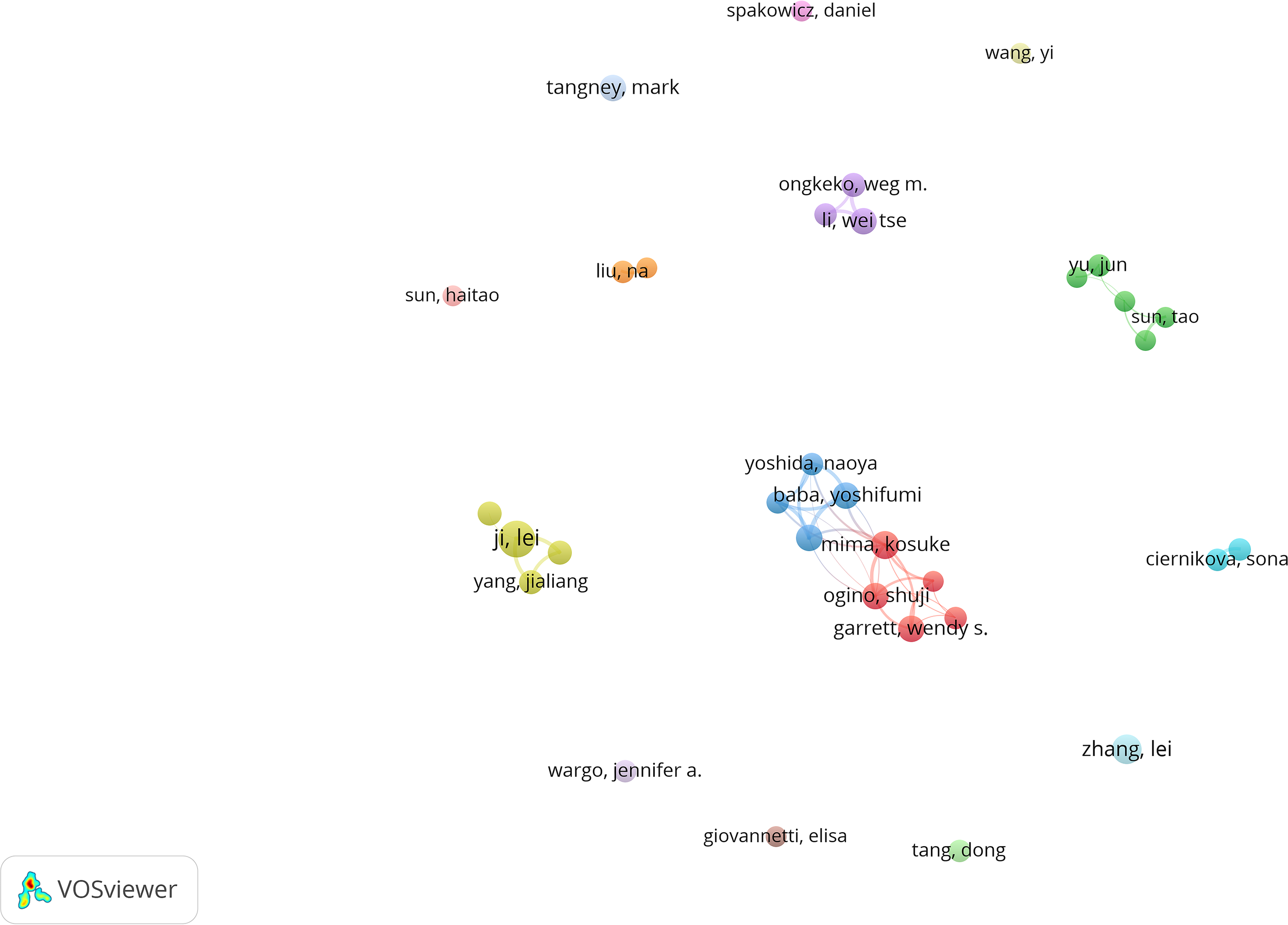
Visual analysis of publishing authors. The cooperation network of co-occurring authors visualized using VOSviewer.
Figure 6
A representation of the keyword mapping focused on intratumoral microbiomes. The shade of the node reflects the frequency of occurrence.
Figure 7
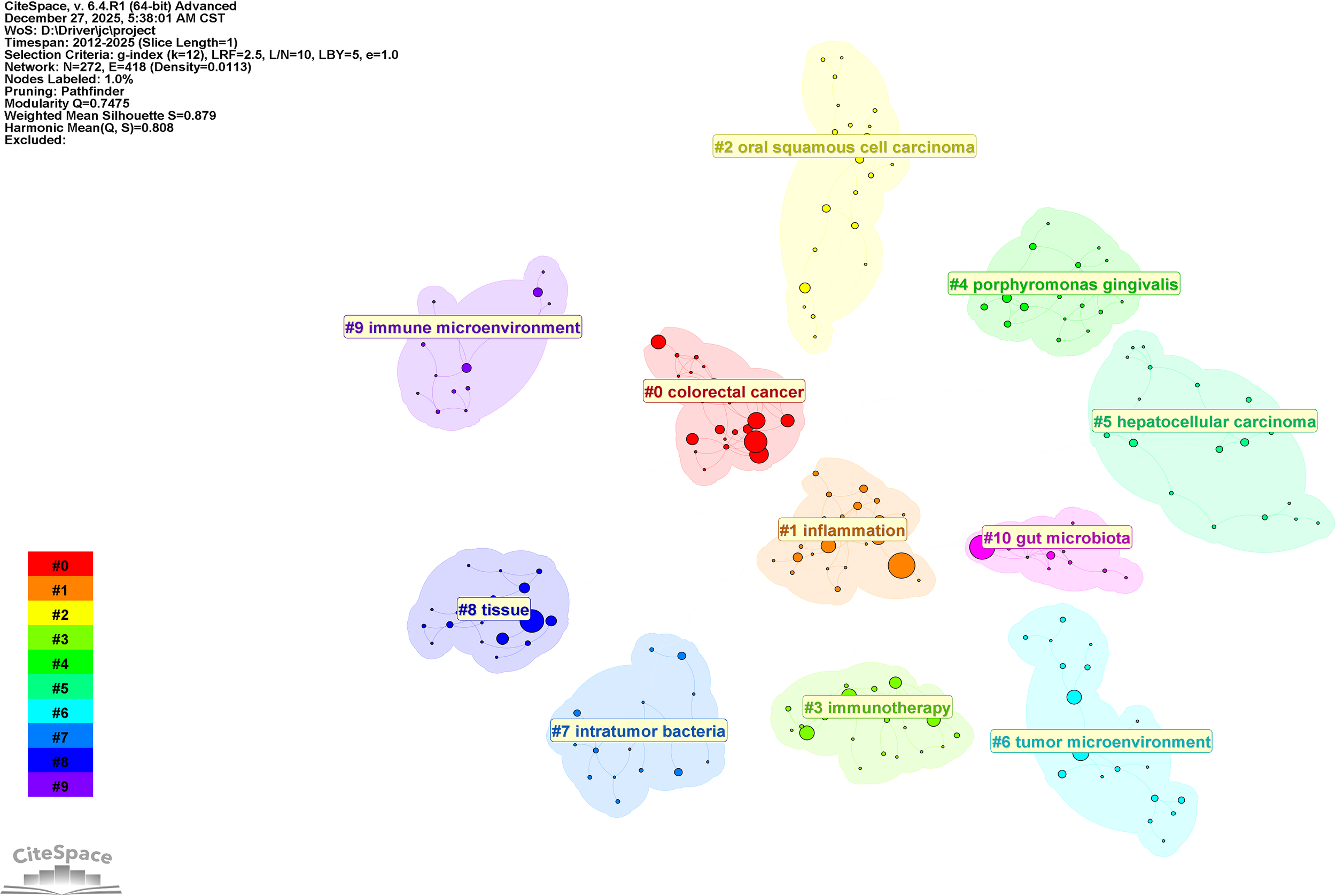
Network map of keyword clustering for intratumoral microbiomes.
Figure 8

The top 15 keywords with the strongest citation burst. The blue bars represent the time periods during which the keywords appeared, while the red bars indicate the intervals in which citation bursts were observed, denoting the starting year, ending year, and duration of the bursts.
2.3.3 SCImago Graphica
SCImago Graphica was used to convert the publication volume and cooperation data of countries/regions into geographic space visualization graphics (Figure 3B). Firstly, the country cooperation matrix data (in CSV format) was exported from VOSviewer, and then the results were imported into SCImago Graphica for visualization analysis. The size of the circles in the figure is proportional to the publication volume, the numbers correspond to the publication volume of the countries, and the lines represent the cooperative relationships between countries/regions. The thickness of the lines represents the number of collaborations.
2.3.4 Microsoft Excel 2022
Microsoft Excel 2022 was used for basic data organization and the drawing of descriptive statistical charts, namely the annual publication volume trend chart (Figure 2).
Figure 9
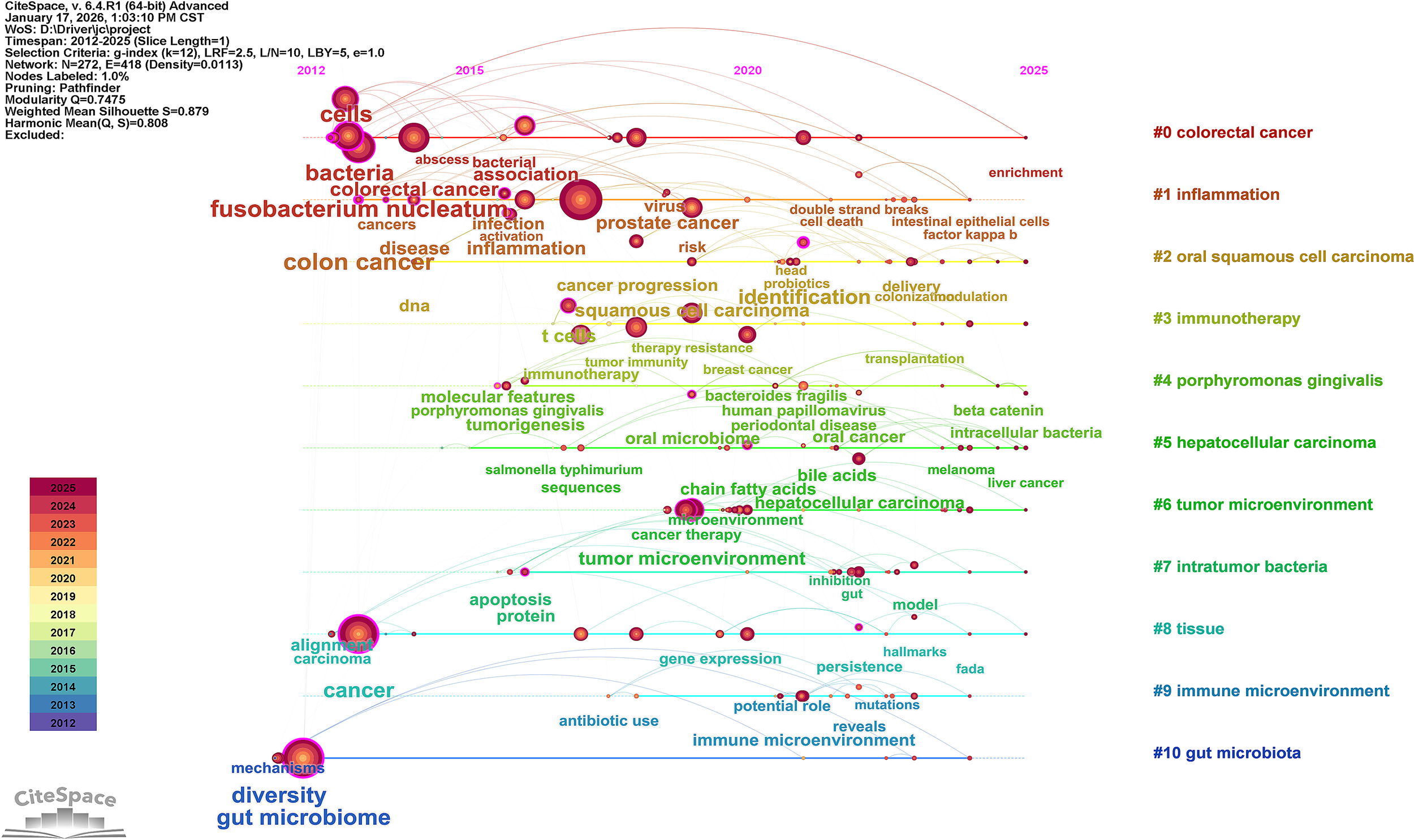
Timeline of the keyword clustering network map for intratumoral microbiomes.
3 Result
3.1 Literature search results
This study incorporated 1,278 eligible documents from the Web of Science for visual analysis. The Web of Science records show that the earliest study addressing the relationship between intratumoral microbiomes and tumors was published in 2012. From 2012 through 2025, annual publication counts in this field have generally increased, with a dip in 2018 and subsequent year-on-year growth from 2019 onward, accompanied by a rapid rise after 2020 (Figure 2), indicating that this field has attracted sustained and rapidly growing research interest in recent years. Notably, in 2020, Nejman et al. proposed that different cancer types exhibit distinct intratumoral microbiome compositions and that most intratumoral bacteria reside within cancer and immune cells (4), a finding that coincided with a surge in publications thereafter. This development has prompted many researchers to shift their focus from merely detecting microbiomes in tumor tissues to conducting in vivo and in vitro experiments on microbiomes within tumors and immune cells. In 2022, a Nature study combined 10x Visium spatial transcriptomics with GeoMx digital spatial analysis, integrating in situ spatial analysis with single-cell RNA sequencing (29), and this work revealed interactions between the host and microbiome at spatial and cellular levels. These technological advances have propelled multi-omics studies of intratumoral microbiomes, further shifting the research emphasis from simply characterizing microbiome composition to exploring function and potential mechanisms. It is important to note that the publication count for 2025 is based on data available through December 22, 2025, and may not fully reflect the final total for that year (Figure 2).
3.2 Distribution and collaboration among countries/regions.
Figure 3 presents a map of publication volume and co-occurrence, depicting the countries and regions engaged in intratumoral microbiome research. The bibliometric analysis employed the Full Counting method available in VOSviewer software. Since the inception of the Web of Science database, a total of 69 countries and regions have contributed to this research field. Table 1 lists the top ten countries and regions ranked by the number of published papers, with China leading at 637 publications, followed by the United States (330 papers), Italy (68 papers), Germany (54 papers), Japan (51 papers), and the United Kingdom (42 papers). In terms of citation frequency, the United States ranks first with a total of 26,772 citations, surpassing China’s 13,592 citations. France, Spain, and Italy also exhibit notable citation performance despite their relatively limited publication volumes. The analysis of national collaboration networks indicates that a close collaborative structure has formed globally, centered around a few high-output countries. Among them, China and the United States constitute dual hubs of the network, with the strongest collaborative link between the two. Additionally, the United States maintains a stable cooperative relationship with major European countries such as Germany and the United Kingdom, while there is also active regional collaboration among European countries, resulting in a tight-knit sub-network. Overall, the field of intratumoral microbiome research demonstrates a broadly international collaborative landscape.
Table 1
| Rank | Country | Documents | Citations |
|---|---|---|---|
| 1 | China | 637 | 13592 |
| 2 | USA | 330 | 26772 |
| 3 | Italy | 68 | 3670 |
| 4 | Germany | 54 | 2392 |
| 5 | Japan | 51 | 2322 |
| 6 | United Kingdom | 42 | 2025 |
| 7 | South Korea | 34 | 895 |
| 8 | Spain | 31 | 2504 |
| 9 | Australia | 29 | 1662 |
| 10 | France | 29 | 2804 |
Top 10 countries/regions by publication output.
3.3 Analysis of collaborative relationships among publishing institutions
The VOSviewer parameters were set as Method (Linlog/modularity) and a minimum number of documents from an institution: 11. The obtained results were retrieved from 1,935 institutions, and 47 met the thresholds. As shown in Figure 4B, six clusters were formed, each comprising institutions that collaborate closely. The first cluster includes the following institutions: Capital Medical University; Central South University; Chinese Academy of Medical Sciences; Chinese Academy of Medical Sciences & Peking Union Medical College; Huazhong University of Science and Technology; Nanjing Medical University; Peking University; Shanghai Jiao Tong University; Soochow University; Southern Medical University; Tongji University; Wuhan University; Zhejiang Chinese Medical University; Zhejiang University; and Zhengzhou University. The second cluster consists of the following institutions: Chinese Academy of Sciences; Mayo Clinic; Ohio State University; Qingdao University; Shandong University; Sichuan University; Sun Yat-sen University; Xi’an Jiaotong University; and Yangzhou University. The third cluster includes the following institutions: Baylor College of Medicine; Cleveland Clinic; Johns Hopkins University; Nanchang University; National Cancer Institute; University of California San Diego; University of Maryland; University of Texas MD Anderson Cancer Center; and Weizmann Institute of Science. The fourth cluster comprises the following institutions: Brigham and Women’s Hospital; Broad Institute of MIT and Harvard; Dana-Farber Cancer Institute; Harvard Medical School; Harvard T.H. Chan School of Public Health; Kumamoto University; and Massachusetts General Hospital. The fifth cluster includes the following institutions: Fujian Medical University; Geneis Beijing Co., Ltd.; Harbin Medical University; and Qingdao Geneis Institute of Big Data and Precision Medicine. The sixth cluster consists of China Medical University; Fudan University; and the University of Hong Kong. Figure 4A displays a visualization map of the CiteSpace network of institutions. We can observe that the connections between institutions are relatively close, with the outer purple and larger nodes representing significant participating institutions in intratumoral microbiome research. Table 2 lists the publication volume and betweenness centrality of institutions, with Sun Yat-sen University ranking first in both metrics (46 papers and 0.41, respectively), indicating that it serves as a key bridge in the inter-institutional collaboration network.
Table 2
| Rank | Organization | Documents | Centrality |
|---|---|---|---|
| 1 | Sun Yat Sen University | 46 | 0.41 |
| 2 | Chinese Academy of Sciences | 45 | 0.24 |
| 3 | University of Texas System | 42 | 0.18 |
| 4 | Harvard University | 40 | 0.01 |
| 5 | Chinese Academy of Medical Sciences - Peking Union Medical College | 39 | 0.02 |
| 6 | Zhejiang University | 39 | 0.05 |
| 7 | Shanghai Jiao Tong University | 38 | 0 |
| 8 | Nanjing Medical University | 36 | 0.09 |
| 9 | Harvard University Medical Affiliates | 33 | 0.03 |
| 10 | Peking Union Medical College | 30 | 0.15 |
Top 10 institutions in terms of publications.
3.4 Author analysis
Figure 5 illustrates the authors’ contributions and connections to the study of the intratumoral microbiome. Table 3 presents the top ten most prolific and influential authors in this research field. The results indicate that research in this area is not concentrated within a single institution or author. In terms of publication volume, Ji Lei ranks first with 16 papers, demonstrating his sustained productivity in intratumoral microbiome research. His studies primarily focus on the associations between intratumoral microbiomes in gastrointestinal tumors and clinical outcomes, treatment responses, and immune-related indicators. Furthermore, Mima, Kosuke, Baba, Hideo, and Baba, Yoshifumi are also among the top authors in terms of publication volume and citation frequency, all maintaining close collaborative ties with Kumamoto University in Japan, forming a persistent and stable research cluster in this field. Notably, Baba, Hideo’s research focuses on the tissue detection of intratumoral Fusobacterium nucleatum and its clinical association with patient prognosis; Mima, Kosuke systematically analyzes the relationship between intratumoral microbiomes and the characteristics of the tumor immune microenvironment, advancing the direction from simple prognostic associations to immune phenotype analysis. Baba, Yoshifumi’s core contribution lies in the establishment of a systematic research framework centered on Fusobacterium nucleatum, integrating clinical outcomes, immune microenvironment, epigenetic changes, metabolic states, and molecular mechanisms. Although Garrett, Wendy S., and Bullman, Susan have published fewer articles, they rank first and third in citation frequency. Bullman, Susan’s work relies on a large prospective cohort and multidimensional data integration, focusing on revealing the heterogeneous contributions of intratumoral microbiomes in different subtypes of colorectal cancer, tumor locations, autophagy status, and treatment responses, particularly emphasizing the potential application value of intratumoral microbiomes in tumor.
Table 3
| Rank | Author | Number of publications | Author | Citations |
|---|---|---|---|---|
| 1 | Ji, Lei | 16 | Garrett, Wendy s. | 2492 |
| 2 | Zhang, Lei | 11 | Wargo, Jennifer a. | 1703 |
| 3 | Mima, Kosuke | 10 | Bullman, Susan | 1421 |
| 4 | Baba, Hideo | 9 | Mima, Kosuke | 1299 |
| 5 | Baba, Yoshifumi | 9 | Ogino, Shuji | 905 |
| 6 | Garrett, Wendy s. | 9 | Chan, Andrew t. | 8281 |
| 7 | Li, Wei tse | 9 | Baba, Hideo | 765 |
| 8 | Ogino, Shuji | 9 | Baba, Yoshifumi | 765 |
| 9 | Tangney, Mark | 9 | Yoshida, Naoya | 723 |
| 10 | Ongkeko, Weg m. | 8 | Tangney, Mark | 643 |
Top 10 authors by number of publications and citations.
3.5 Keyword analysis
Keywords serve as concise indicators of the central theme of research papers, capturing the essence of research questions and significant findings. They provide a brief overview of the research topic and its contributions to the academic field. Analyzing these keywords can identify key research hotspots, emerging trends, and areas of interest within the academic community. Figure 6 visually illustrates the most used keywords, aiding in the identification of focal areas and overall trends in the study of the intratumoral microbiome. Additionally, Table 4 organizes and lists the top ten keywords based on their frequency and betweenness centrality. Terms such as “ tumor microbiome,” “gut microbiome,” “cancer,” “Fusobacterium nucleatum,” and “colorectal cancer” hold significant positions, underscoring their relevance in ongoing discussions.
Table 4
| Rank | Keyword | Counts | Centrality | Rank | Keywords | Counts | Centrality |
|---|---|---|---|---|---|---|---|
| 1 | tumor microbiome | 392 | 0.01 | 1 | colon cancer | 27 | 0.35 |
| 2 | gut microbiome | 353 | 0.25 | 2 | cancer | 318 | 0.29 |
| 3 | cancer | 318 | 0.29 | 3 | gut microbiome | 353 | 0.25 |
| 4 | Fusobacterium nucleatum | 269 | 0.19 | 4 | butyrate | 11 | 0.25 |
| 5 | colorectal cancer | 244 | 0.07 | 5 | bacteria | 179 | 0.24 |
| 6 | bacteria | 179 | 0.24 | 6 | identification | 26 | 0.23 |
| 7 | cells | 150 | 0.12 | 7 | helicobacter pylori | 72 | 0.21 |
| 8 | intratumor microbiome | 137 | 0.02 | 8 | health | 46 | 0.2 |
| 9 | tumor microenvironment | 129 | 0.13 | 9 | Fusobacterium nucleatum | 269 | 0.19 |
| 10 | tumor immunity | 117 | 0.02 | 10 | association | 95 | 0.16 |
The top ten keywords with the highest frequency (left) and the top ten keywords with the highest betweeness centrality (right).
Figure 7 illustrates the clustering of keywords, revealing 11 distinct clusters. Clustering employs the LLR clustering labeling method, resulting in Q = 0.7475 and S = 0.879. Table 5 presents the cluster labels and the primary keywords for each cluster, including#0 colorectal cancer; #1 inflammation; #2 oral squamous cell carcinoma; #3 immunotherapy; #4 porphyromonas gingivalis; #5 hepatocellular carcinoma; #6 tumor microenvironment; #7 intratumor bacteria; #8 tissue; #9 immune microenvironment; #10 gut microbiota. The analysis based on keyword clustering indicates that research related to the intratumoral microbiome can be categorized into several relatively focused research groups. Each cluster exhibits a certain degree of internal consistency in keyword composition; however, some overlap exists between different clusters, suggesting a strong interrelation among research themes in this field. One cluster features high-frequency keywords such as tumor microenvironment, tumor microbiota, and intratumoral microbiota (#6), co-occurring with terms like toll-like receptor, host microbial interactions, and breast cancer, indicating that this cluster primarily consists of keywords describing the local tumor microenvironment and its associated microbial elements. Another cluster centers around core keywords like” colorectal cancer “ and” fusobacterium nucleatum” (#0, #1, and #4), appearing alongside terms such as tumor microbiome, tumorigenesis, and poor prognosis, reflecting that this line of research largely revolves around tumor-associated microbiota and their biological characteristics within the context of specific cancer types. Additionally, some clusters focus on keywords related to “ immunotherapy”, “ immune microenvironment”, and “ immune checkpoint inhibitors “ (#3 and #9), demonstrating a notable co-occurrence between the study of intratumoral microbiota and themes related to tumor immunotherapy. Furthermore, there are clusters concerning intratumor bacteria (#7) and gut microbiota (#10), which respectively examine the presence of bacteria within tumor tissues and the relationship between gut microbiota and tumor immunotherapy research. Overall, the results of the keyword clustering reflect the diversity of current research on intratumoral microbiota in terms of subjects and content; however, the different themes do not form completely isolated structures but rather exhibit a degree of intersection and overlap.
Table 5
| #0 | colorectal cancer | colorectal cancer; pancreatic cancer; immune response; bioinformatics tools; bacteria-fungi interaction; Fusobacterium nucleatum; tumor microbiome; bacteria; cancer; persistence |
| #1 | inflammation | tumor microbiome; Fusobacterium nucleatum; bacteria; risk; cancer; pancreatic cancer; colorectal cancer; immune response; bioinformatics tools; bacteria-fungi interaction |
| #2 | oral squamous cell carcinoma | oral squamous cell carcinoma; associated macrophage; hepatocellular carcinoma; preclinical mechanisms; anti-cancer therapy; tumor microenvironment; tumor microbiome; host-microbe interaction; cancer biology; oral bacteria |
| #3 | immunotherapy | breast cancer; tumor microenvironment; intratumoral microbiome; immune cell; metabolic heterogeneity; immunotherapy; resistance; cancer; microbiome; efficacy |
| #4 | porphyromonas gingivalis | colorectal cancer; tumorigenesis; molecular features; association; familial adenomatous polyposis; Fusobacterium nucleatum; pancreatic cancer; intratumoral microbiota; cervical cancer; poor prognosis |
| #5 | hepatocellular carcinoma | hepatocellular carcinoma; metabolic pathway; fatty acid; lipid synthesis; bacterial theranostics; intratumoral microbiota; Fusobacterium nucleatum; pancreatic cancer; identification; osteomyelitis |
| #6 | tumor microenvironment | tumor microenvironment; toll-like receptor; tumor microbiota; targeting therapy; host microbial interactions; intratumoral microbiota; tumor microenvironment; breast cancer; spatial multi-omics; targeting therapy |
| #7 | intratumor bacteria | tumor microbiome; Fusobacterium nucleatum; tumor microenvironment; tumor-associated macrophages; colorectal cancer; intratumor bacteria; tumor metabolism; ellagic acid; targeted therapy; trastuzumab resistance |
| #8 | tissue | cancer; Fusobacterium nucleatum; tumor microbiome; bacteria; persistence; gut microbiota; lung cancer; intratumoral microbiota; human papillomavirus; penile cancer |
| #9 | immune microenvironment | immune microenvironment; therapeutic target; immune checkpoint inhibitor; mammary carcinoma; host microbial interactions; Fusobacterium nucleatum; cells; reveals; mutations; survival |
| #10 | gut microbiota | gut microbiota; immune checkpoint inhibitors; immune-related adverse events; skin microbiota; tumor microbiome; gut microbiome; cancer immunotherapy; microbiome modulation; dietary interventions; candida albicans |
The names of the eight clusters along with the primary keywords for each cluster.
The purpose of keyword burst analysis is to identify emerging concepts that have gained frequent citations during a specified period, thereby highlighting emerging research hotspots or frontiers within a particular field. This analysis enables researchers to anticipate future research directions and trends by monitoring the frequency and timing of keyword occurrences (Figure 8). The findings are based on a chronologically compiled and presented analysis of keyword appearances, with CiteSpace configured using the following parameters: γ (1.0), minimum duration = 1, as shown in Figure 8.
The keyword “colon cancer” is particularly prominent, exhibiting a burst strength of 4.64, reflecting significant attention between 2013 and 2019. Most keyword bursts are concentrated between 2016 and 2021. Notable early burst keywords include “colon cancer” (2013-2019) and “disease” (2014-2019). Both keywords began showing significant increases in citations in 2013 and 2014, respectively, and peaked in 2019. This trend reflects researchers’ early and sustained focus on the mechanistic progression of colon cancer. Furthermore, the keywords “molecular features” (2016-2022) and “bacterial” (2016-2022) have exhibited continuous bursts since 2016, indicating a deepening exploration of intratumoral microbiota as molecular marker features in tumors. The keyword “Kappa B” began attracting attention in 2023, reflecting a growing emphasis on the role of this key transcription factor in inflammatory and oncogenic processes. Meanwhile, the keyword “binding” shows a significant burst between 2024 and 2025, suggesting that exploring the close integration between microbiology and cancer biology, including research on immunotherapy targets, may represent an emerging frontier in current research.
The timeline view (Figure 9) illustrates the temporal evolution of research related to tumor-associated microbiota from 2012 to 2025, showing a gradual shift in research focus across different thematic clusters. From 2012 to 2015, research activity was primarily concentrated on gastrointestinal cancer topics. Keywords that emerged early in the timeline included “cancer”, “ gut microbiome”, “ gastric cancer”, “colon cancer”, “diversity”, “colorectal cancer”, “bacteria”, “cells”, and “disease”, indicating that early studies mainly focused on the association between intratumoral microbiota and gastrointestinal cancers. During this period, “Fusobacterium nucleatum” was a prominent keyword, reflecting its central role in research on microbiota associated with gastrointestinal tumors. Meanwhile, the early attention to “colon cancer “ and “bacteria “ demonstrated initial explorations into the role of microorganisms in the tumor microenvironment of colorectal cancer. In short, this stage was characterized by exploration and associative research themes.
Between 2015 and 2020, the timeline shows a shift toward more specific and complex interactions between microbiota and cancer processes. Keywords related to microorganisms that emerged included “molecular features”, “Porphyromonas gingivalis”, “Salmonella typhimurium”, “Escherichia coli”, “tumor microbiome”, “oral microbiome”, “Bacteroides fragilis”, and “virus”, while cancer-related keywords included “cancer therapy”, “cancer progression”, “T cells”, “ tumor immunity”, “cancer progression”, and “ tumor microenvironment”. This suggests that research from 2015 to 2020 was relatively focused on the interactions between specific bacterial species and cancer processes. Additionally, the appearance of keywords such as “protein, “ microbial metabolites, and “ gene expression indicated a growing interest in the mechanisms underlying tumor progression.
From 2020 to 2025, more keywords related to tumor-host interactions and tumor therapy frequently appeared on the timeline, including “ identification”, “factor kappa B”, “clinical translation”, “chemoradiotherapy”, “persistence”, “immune microenvironment”, “ inhibition”, and “ risk factors”. These keywords were primarily distributed within clusters related to tumor therapy themes, showing a trend toward integrating microbiota research with therapeutic strategies and immune responses, highlighting clinical translation.
3.6 Analysis of research trends and characteristics
To gain a comprehensive understanding of the developmental trends of tumor-associated microbiomes and their implications for cancer treatment, this study performed a summary analysis of preclinical and clinical trials regarding the application of intratumoral microorganisms in therapeutic interventions, as illustrated in Table 6, which includes a total of 69 studies labeled from *1 to *69. The abbreviations for tumor sites in the table are as follows: Bca (breast cancer), NPC (nasopharyngeal carcinoma), Lc (lung cancer), CRC (colorectal cancer), ESCA (esophageal carcinoma), PDAC (pancreatic cancer), MEL (melanoma), and PCa (prostate cancer), CC (Cervical cancer), OSCC (oral squamous cell carcinoma), SCC (squamous cell carcinoma). The studies are primarily categorized into five types: Animal testing (62 studies), Nano Research (19 studies), Engineered bacteria (3 studies), RCT (2 studies), Phase I Clinical Trial (1 studies) and In vitro experiments (2 studies). Given that both Nano Research and Engineered bacteria involve Animal testing, the total number of studies exceeds 69. Post-intervention primary outcomes include Endocrine and targeted therapy (11 studies), Chemotherapy (20 studies), Intratumoral bacterial clearance (17 studies), and Immunotherapy (21 studies). When considered in conjunction with the results in the table, we find that tumor immunotherapy constitutes a substantial proportion of the studies, which is consistent with the results of the keyword clustering. The symbols “+” and “-” denote whether the reported effects of intratumoral bacteria on tumor treatment were positive or negative. Analysis reveals that most studies reported negative outcomes (21 studies with “+” and 48 studies with “-”), indicating that the presence of intratumoral bacteria is often detrimental to the tumor treatment process, which necessitates interventions targeting these bacteria to enhance anti-tumor therapies. Notably, research on Fusobacterium nucleatum is the most prevalent, with 25 studies, which aligns with the co-occurrence and clustering results of keywords, thereby suggesting that Fusobacterium nucleatum is currently a focal point in the study of intratumoral microbiota.
Table 6
| Endocrine and targeted therapy | |||||
|---|---|---|---|---|---|
| Sequence Number | Year | Tumor site | Classification of interventional studies | Types of microorganisms | Effect |
| *1 (30) | 2025 | Bca | Animal testing | Firmicutes Lactobacillus | + |
| *2 (31) | 2023 | CRC and Bca | Animal testing | Proteus mirabilis (A‐gyo) and Rhodopseudomonas palustris (UN‐gyo) | + |
| *3 (32) | 2017 | RCC, CRC, READ | Animal testing | non-pathogenic E. coli strain MG1655 | + |
| *4 (33) | 2025 | Bca | Animal testing | Pseudomonas aeruginosa 3oc | – |
| *5 (34) | 2024 | NPC | Animal testing | Lactobacillus plantarum | + |
| *6 (35) | 2024 | Bca | Animal testing | Tumor-colonizing microorganisms | – |
| *7 (36) | 2024 | Animal testing | Tumor-colonizing microorganisms | + | |
| *8 (37) | 2023 | Lc | Animal testing | Akkermansia muciniphila | + |
| *9 (38) | 2022 | CRC, Lc and Bca | Nano Research, Animal testing | Intratumoral Microbiome | + |
| *10 (39) | 2022 | Bca | Animal testing | Helicobacter hepaticus | – |
| *11 (40) | 2020 | OSCC | Animal testing | Porphyromonas gingivalis, Treponema denticola, and Fusobacterium nucleatum | – |
| Chemotherapy | |||||
| *12 (41) | 2024 | CRC | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *13 (42) | 2024 | Bca | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *14 (43) | 2023 | CRC | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *15 (44) | 2023 | CRC | Nano Research, Animal testing | Intratumoral Microbiome | – |
| *16 (45) | 2025 | Bca | Nano Research, Animal testing | Intratumoral Microbiome | – |
| *17 (46) | 2024 | CRC | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *18 (47) | 2022 | PDAC | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *19 (48) | 2023 | PDAC, Bca | Engineered bacteria, Animal testing | HAase-expressing Salmonella | + |
| *20 (49) | 2022 | CRC | Animal testing | Fusobacterium nucleatum | – |
| *21 (50) | 2023 | CRC | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *22 (51) | 2022 | ESCA | Animal testing | Fusobacterium nucleatum | – |
| *23 (52) | 2025 | CRC | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *24 (53) | 2024 | Animal testing | Pseudomonas aeruginosa | – | |
| *25 (54) | 2024 | CRC | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *26 (55) | 2024 | Animal testing | Pseudomonas aeruginosa | – | |
| *27 (56) | 2019 | Animal testing | Intratumoral Microbiome | – | |
| *28 (45) | 2025 | Bca | Nano Research, Animal testing | Intratumoral Microbiome | + |
| *29 (57) | 2023 | CC | Animal testing | Lactobacillus iners | – |
| *30 (58) | 2017 | Animal testing | Bacteria containing the long CDD variant | – | |
| *31 (59) | 2023 | Lc | In vitro experiments | Staphylococcus aureus | – |
| Intratumoral bacterial clearance | |||||
| *32 (60) | 2021 | CRC | RCT | Fusobacterium nucleatum | – |
| *33 (61) | 2025 | CRC | Animal testing | Intratumoral Microbiome | – |
| *34 (44) | 2023 | Nano Research, Animal testing | Intratumoral Microbiome | – | |
| *35 (26) | 2024 | Bca | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *36 (62) | 2025 | Animal testing | Fusobacterium nucleatum, Streptococcus sanguinis, Enterococcus faecalis, and Staphylococcus xylosus | – | |
| *37 (63) | 2022 | Engineered bacteria, Animal testing | Intratumoral Microbiome | – | |
| *38 (64) | 2025 | Nano Research, Animal testing | Intratumoral Microbiome | – | |
| *39 (65) | 2017 | RCT | Fusobacterium nucleatum | – | |
| *40 (66) | 2023 | CRC | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *41 (67) | 2021 | Bca | RCT | Intratumoral Microbiome | – |
| *42 (68) | 2025 | Animal testing | Intratumoral Microbiome | – | |
| *43 (69) | 2024 | PDAC | Animal testing | Fusobacterium nucleatum | – |
| *44 (54) | 2024 | CRC | Animal testing | Fusobacterium nucleatum | – |
| *45 (70) | 2022 | Bca | Animal testing | Streptococcus, Lactobacillus, Staphylococcus, and Enterococcus | – |
| *46 (71) | 2025 | Animal testing | Escherichia coli | – | |
| *47 (72) | 2025 | Bca | Nano Research | Fusobacterium nucleatum | – |
| *48 (73) | 2023 | CRC | Animal testing | Fusobacterium nucleatum | – |
| Immunotherapy | |||||
| *49 (74) | 2022 | PDAC | Animal testing | Fungus | – |
| *50 (75) | 2023 | MEL | Animal testing | Lactobacillus reuteri | + |
| *51 (76) | 2021 | Phase I Clinical Trial | mycobacteria | + | |
| *52 (77) | 2024 | CRC | RCT, Animal testing | Escherichia coli Nissle 1917 | + |
| *53 (78) | 2024 | PDAC | Animal testing | Proteobacteria | + |
| *54 (79) | 2024 | CRC | Animal testing | Intratumoral Microbiome | + |
| *55 (80) | 2023 | ESCA | Animal testing | Fusobacterium nucleatum | – |
| *56 (27) | 2023 | PDAC | Nano Research, Animal testing | Fusobacterium nucleatum | – |
| *57 (81) | 2025 | Animal testing | B. parabrevis | + | |
| *58 (82) | 2020 | Animal testing | Bifidobacteria | + | |
| *59 (83) | 2024 | CRC | Animal testing | Fusobacterium nucleatum | + |
| *60 (84) | 2023 | ESCA | Animal testing | Streptococcus | + |
| *61 (85) | 2022 | SCC | In vitro experiments | Fusobacterium | – |
| *62 (86) | 2024 | Bca | Nano Research, Animal testing | Intratumoral Microbiome | + |
| *63 (87) | 2025 | Bca | Animal testing | Stenotrophomonas maltophilia | + |
| *64 (88) | 2022 | CRC | Engineered bacteria, Animal testing | Fusobacterium nucleatum, Escherichia coli, Staphylococcus aureus, Bifidobacterium pseudolongum | + |
| *65 (89) | 2018 | PDAC | Animal testing | Intratumoral Microbiome | – |
| *66 (90) | 2022 | OSCC | Animal testing | Peptostreptococcus | – |
| *67 (91) | 2021 | Nano Research, Animal testing | Intratumoral Microbiome | – | |
| *68 (74) | 2022 | Animal testing | Fungus | – | |
| *69 (92) | 2025 | Bca | Animal testing | Fusobacterium nucleatum | – |
Current preclinical research and clinical trials of intratumoral microorganisms in tumor treatment interventions.
4 Discussion
This study, based on bibliometric analysis, systematically mapped the knowledge structure and evolutionary trajectory of research on intratumoral microbiota, and examined its potential value in tumor initiation, progression, and clinical applications. The results show that research on intratumoral microbiota began to emerge in 2012 and entered a phase of rapid development after 2020. This is in agreement with the results reported by Xiao et al (93). This shift indicates that intratumoral microbiota is gradually becoming a frontier field at the intersection of oncology and immunology. Next, we will discuss the results of the search and analysis conducted using Web of Science and PubMed.
4.1 Evolution and hotspots of research themes
According to our timeline results, research on the intratumoral microbiome has evolved from the characterization of cancer-associated microbial communities and extensive studies in colorectal cancer to the investigation of specific microbial species and their mechanisms, ultimately progressing to immune modulation and therapeutic interventions. Among them, the themes related to intratumoral microorganisms, immunotherapy, and the immune microenvironment have gradually assumed significant prominence in recent years, as evidenced by keyword co-occurrence and temporal evolution analyses. Early studies primarily utilized 16S rRNA sequencing to observe microbial distribution, revealing associations between microbiota such as Helicobacter pylori and Fusobacterium nucleatum with gastric cancer and colorectal cancer (94, 95). The advent of multi-omics technologies (96) and later developments in spatial transcriptomics have enabled researchers to trace the spatial co-localization relationships between microbiota and host cells at both single-cell and subcellular scales. Furthermore, this reveals the intricate interactions between complex microbial communities and the cancer process. In recent years, the integration of immunotherapy, outer membrane vesicle (OMV) coated nanotechnology (27), electronic antibacterial therapy (EAT) (69), and nanocarrier technology (44) has further emphasized the potential value of tumor-associated microorganisms as therapeutic targets and supportive factors. In summary, this evolutionary trend indicates that research on tumor-associated microorganisms is progressively transitioning from correlation validation to functional mechanism elucidation and ultimately to the exploration of clinical applications.
4.2 The application of tumor-associated microorganisms in cancer therapy
Through keyword clustering analysis, we found that themes related to tumor immunotherapy exhibit a high degree of clustering in the research on intratumoral microorganisms, suggesting that this field is gradually becoming one of the important application directions of current research. The results of the keyword co-occurrence analysis indicate that “immunity” has a count of 117 and a centrality of 0.02. Furthermore, the results of the intervention subset reveal that “cancer immunotherapy” is one of the therapeutic approaches with significant clinical translational trends, which accounts for a considerable proportion of the studies on intratumoral microorganisms and tumor treatment. Based on our findings, three key points regarding cancer treatment are worthy of discussion, including functional heterogeneity, diversity of intervention strategies, and the synergistic regulation of representative target bacteria and overall microecology. Firstly, tumor-associated microbiota may exert dual effects—either promoting or inhibiting cancer treatment. As shown in Table 5, different microbiota demonstrates certain variations in immunotherapy, endocrine therapy, targeted therapy, and chemotherapy. On the one hand, the studies summarized in the table indicate that certain naturally occurring intratumoral microorganisms or tumor-associated commensals, including Lactobacillus, Akkermansia muciniphila, and Bifidobacterium, are associated with enhanced CD8+ T-cell infiltration, induction of immunogenic cell death, and modulation of the tumor immune microenvironment, thereby potentiating the efficacy of immune checkpoint inhibitors, chemotherapy, or targeted therapies. On the other hand, several studies employed engineered or exogenously administered bacteria (e.g., Salmonella or Escherichia coli Nissle 1917) as therapeutic tools. Although these bacteria are not intrinsic components of the intratumoral microbiota, they can exert antitumor effects by selectively eliminating deleterious intratumoral microorganisms, reshaping the tumor microenvironment, and/or directly enhancing antitumor immune responses. Together, this evidence suggests that tumor-associated microbiota constitutes an important regulatory factor influencing responses to cancer treatment, rather than simply being classified as “harmful” or “beneficial”. Furthermore, unlike the non-specific bactericidal effects of traditional antibiotics, recent years have seen a substantial focus on the precise modulation of intra-tumoral microbiota rather than their complete eradication. As shown in Table 5, emerging strategies such as nanodrug delivery systems, biomimetic nanomaterials, outer membrane vesicle coatings, photothermal/photodynamic therapy, and electroporation antibacterial treatments have been widely used to target tumor-associated microbiota. These methods have achieved the goal of “specifically eliminating pathogenic bacteria while preserving or reshaping beneficial microbiota” in various tumor models, and they can synergize with chemotherapy, immunotherapy, or targeted therapy. Additionally, engineered bacteria can serve not only as antibacterial tools but also as drug delivery carriers or immune activation platforms, further expanding the therapeutic dimensions of tumor microbiota intervention. However, most current studies remain at the animal experiment or early clinical stages, and their long-term safety, stability of microbiota remodeling, and optimal administration routes still require further validation. Moreover, as indicated by the distribution of interventional studies included in the table, current research often focuses on several representative microorganisms in practical applications. Among them, Fusobacterium nucleatum has become one of the most used model target bacteria in nanodrug, antibacterial vaccine, and combination therapy studies due to its stable colonization in various gastrointestinal tumors and breast cancer, its relatively clear pathogenic mechanisms, and the presence of defined molecular targets. However, the table also shows that Escherichia coli, Pseudomonas aeruginosa, Streptococcus, Proteobacteria, and intra-tumoral fungal communities play important roles in regulating treatment sensitivity and the immune microenvironment. An increasing number of studies are beginning to focus on the combined effects of multi-species coexistence and their interactions within tumors on treatment responses, attempting to achieve systematic improvement in therapeutic outcomes by eliminating key pathogenic bacteria, modulating functional microbiota modules, or overall remodeling of the tumor microecological network. Overall, existing evidence indicates that tumor-associated microbiota has become a key regulatory factor influencing cancer treatment responses and prognosis. Future research needs to advance from correlational descriptions to elucidating mechanisms and establishing causal relationships, and through multi-omics integration and clinical translational studies, explore safer, more precise, and scalable tumor microbiota intervention strategies, thereby providing a new theoretical basis and therapeutic targets for personalized cancer treatment.
4.3 Tumor colonization by gut microbiota
During the literature screening, keyword co-occurrence analysis, and clustering analysis, we observed that “gut microbiota” emerged as a high-frequency keyword and simultaneously exhibited a significant betweenness centrality (0.25) in the study of intratumoral microbiota, indicating that the gut microbiome has become an important and indispensable topic of association in this research field. Upon reviewing relevant literature for further analysis, we discovered that current research involving gut microbiota and intra-tumoral microbiota predominantly focuses on describing key microbial populations that have been repeatedly reported in recent gut microbiome studies, which are also detected in the tumor microenvironment, such as Lactobacillus, Clostridium, Bifidobacterium, and Akkermansia muciniphila (37, 82, 97, 98). These studies, to some extent, expand our understanding of the sources and distribution of tumor-associated microbiota; however, they primarily remain phenomenological in nature. In recent years, numerous studies have begun to investigate the potential functions of these microorganisms within the microenvironment. Yet, systematic research supporting the overall changes and interaction patterns between the gut microbiome and the intra-tumoral microbiome is still lacking. This is particularly evident in the intervention aspect, as the number of studies directly assessing the impact of rapidly developing fecal microbiota transplantation (FMT) methods from gut microbiome research on intra-tumoral microbiota remains limited. This phenomenon reflects a significant methodological bottleneck faced by the field, specifically concerning whether and under what conditions gut-derived microorganisms can effectively colonize and persist stably at tumor sites. Notably, recent studies on “gut microbiota colonization of tumors” have provided new perspectives on this issue. Some research has observed the accumulation of gut-derived microorganisms within tumor tissues (82, 99). Additionally, mechanisms linking host molecular characteristics with the enrichment of specific microorganisms within tumors have been reported. Shi et al. found that KRAS mutations can actively promote the intratumoral colonization of toxin-producing Bacteroides fragilis in colorectal cancer by modulating the miRNA3655/SURF6/IRF7/IFNβ axis (references) (100). This shift indicates that research interest is transitioning from “whether intra-tumoral microbiota exist” to “which factors determine their colonization and stability.” Based on existing literature and keyword co-occurrence analysis results, this study posits that research surrounding the relationship between gut microbiota, fecal microbiota transplantation, and intra-tumoral colonization is forming a new interdisciplinary hotspot. This also leads to a noteworthy research direction: whether it is possible to enhance the colonization potential of beneficial microorganisms at tumor sites by modulating host-related molecular characteristics or microenvironment conditions, thereby providing new strategies for the precise regulation of the tumor microenvironment. The increasing attention to this direction suggests its potential significance in future tumor microbiota research and adjunct strategies for immunotherapy.
4.4 Tumor-associated microorganisms and their metabolites in relation to tumor immunity
The sustained burst of the keyword “molecular features” (2016-2022) indicates that research on the intratumoral microbiota has entered a phase of deep mechanistic exploration during this period. Combined with the results from the timeline chart, both tumor-associated microorganisms themselves and their metabolites, serving as molecular features, are currently one of the hot topics in the field of tumor-associated microbiota research. While the keyword “binding” is projected to show a significant burst between 2024 and 2025, its meaning is relatively broad, encompassing areas such as the integration of multi-omics data and the binding of therapeutic targets to drugs. Based on this, we attempt to discuss the feasibility of combining metabolites with immunotherapy as a future research direction. Short-chain fatty acids, particularly butyrate, have been repeatedly reported as key immunomodulatory metabolites (101–103). For example, butyrate derived from tumor-associated can enhance the T cell receptor (TCR) signaling of cytotoxic CD8+ T cells, thereby improving their effector function and enhancing the anti-tumor efficacy of immune checkpoint inhibitors such as anti-PD-1 (104). Such studies provide a more mechanistic explanation for the association between “microbiota—immunotherapy response” from the perspective of immunometabolism, suggesting that metabolites may serve as critical mediating factors in regulating sensitivity to immunotherapy. In contrast to immune-enhancing metabolites, certain microbially-associated metabolic products have been confirmed to participate in shaping an immunosuppressive tumor microenvironment. The colonization and metabolic activity of symbiotic microorganisms, particularly anaerobes, in hypoxic tumor environments can also significantly promote the local accumulation of lactate, thereby exacerbating tumor microenvironment acidification. This lactate-enriched microenvironment can weaken anti-tumor immune responses through various pathways, promoting tumor progression and immune evasion. Recent studies have shown that elevated lactate levels in the tumor microenvironment can downregulate the expression of PPARγ in invariant natural killer T (iNKT) cells, thereby inhibiting cholesterol biosynthesis and IFN-γ production, which weakens their anti-tumor immune effects (105). Additionally, delivery systems targeting the metabolism of symbiotic anaerobic bacteria under hypoxic conditions can reduce lactate accumulation within tumors, reversing lactate-mediated immunosuppression and chemotherapy resistance (106). These findings suggest that microbially-mediated metabolic alterations may serve as significant functional hubs linking immunosuppression, tumor progression, and therapeutic resistance. In addition to single metabolomics studies, an increasing number of multi-omics integration analyses have further revealed the systematic associations among tumor-associated microorganisms, metabolic remodeling, and immune heterogeneity (10, 96, 107–109). This research trend emphasizes that metabolic changes are not isolated events, but rather key regulatory layers embedded within the tumor immune ecosystem. Although most current studies still focus on correlation analysis, the introduction of metabolomics has significantly enhanced the depth of immunological interpretations, providing new theoretical support for understanding how microorganisms regulate anti-tumor immunity. In the future, systematic research combining metabolomics, immune phenotype analysis, and functional experiments will contribute to further clarifying the causal role of microbially-mediated metabolic changes in tumor immune regulation, advancing its clinical translation toward predictive immunotherapy and combined intervention strategies.
4.5 Advances and challenges in detection technologies
The extremely low abundance of intratumoral microorganisms has led to ongoing debates regarding detection methods. In recent years, emerging technologies such as simplified metagenomic sequencing (2bRAD-M), upgraded 16S sequencing, spatial transcriptomics, and SLACS have significantly enhanced the ability to analyze low-abundance, spatial heterogeneity, and functional distribution (21, 110, 111). Additionally, the application of artificial intelligence and big data algorithms has made it possible to identify intratumoral microorganisms across different cancer types (20). However, challenges such as contamination control, sample standardization, and data consistency remain pressing issues in this field. A research team from Johns Hopkins University (112) reanalyzed whole-genome sequencing (WGS) data of 5,734 samples covering 25 cancer types from The Cancer Genome Atlas (TCGA) project, revealing that the presence of microorganisms in tumors is far less than previously reported, with sequencing read errors potentially reaching tens of thousands. This has cast even greater doubt on the actual existence and abundance assessment of intratumoral microorganisms. Effectively distinguishing genuine low-biomass microbial colonization from exogenous or environmental contaminants introduced during experimental procedures has become a core methodological challenge in the field of intratumoral microbiology. Without stringent quality control, microbial signals can easily be obscured by exogenous contamination. Therefore, the core principle guiding experimentation is that contamination control must be treated as a paramount consideration at every step—from experimental design and sample processing to data analysis—otherwise, microbial signals are highly susceptible to being masked by contamination. Additionally, there is an urgent need to establish unified detection and analytical protocols across different centers. This includes standardized protocols for sample collection and preservation, the standardized use of core reagents, and consistent bioinformatic analysis workflows and parameters.
4.6 Future perspectives
Future research directions can be deepened from the following aspects: 1. The first and most important point is: The lack of consistency across studies is particularly problematic in the context of intratumoral microbiomes, where microbial abundance is extremely low and sequencing artifacts can dominate the signal. Inadequate contamination control and heterogeneous analytical pipelines may lead to overestimation or misclassification of microbial presence, underscoring the urgent need for standardized methodologies and rigorously curated reference datasets. 2. Mechanistic studies should move beyond descriptive associations to clarify how intratumoral microorganisms functionally interact with distinct cellular components of the tumor microenvironment. In particular, the causal relationships between intratumoral microbes, immune cell subsets, cancer-associated fibroblasts, and local metabolic alterations remain poorly defined. Elucidating how these interactions evolve during tumor progression and treatment will be essential for distinguishing passenger microbes from functionally relevant regulators. 3. Greater emphasis should be placed on multi-omics approaches. While single-omics studies have provided important insights, they are often insufficient to capture the spatial heterogeneity of intratumoral microorganisms and their molecular effects. Combining spatial transcriptomics, metabolomics, and microbiome profiling at the tissue level may enable more accurate identification of microbe-associated molecular and immune phenotypes that are clinically relevant. 4. Current research remains heavily bacteria-centered, which likely underestimates the complexity of the intratumoral microecosystem. Emerging evidence suggests that fungi, viruses, and bacteriophages, as well as their interactions with bacteria, may influence tumor biology and immune regulation. Expanding research beyond bacteria alone will be necessary to construct a more complete and biologically meaningful map of the intratumoral microbiome. 5. Clinical translation and novel interventions: “Microbiota-targeted therapy” is gradually emerging as a potential clinical intervention strategy, which involves selectively modulating tumor-associated microorganisms and their functional states to improve the tumor immune microenvironment and therapeutic responses. For instance, the precise elimination of tumor-promoting or immunosuppressive bacterial strains using agents such as sterilizing vaccines and nanocarriers can alleviate immune suppression and reduce the risk of resistance; engineered bacteria can be utilized for localized delivery to produce relevant metabolites that modulate the immune microenvironment, thereby enhancing the efficacy of immunotherapy. But their efficacy, controllability, and long-term safety require further clinical validation.
Lastly, this study also has certain limitations. First, bibliometric analysis relies on databases and search strategies, which may result in the omission of some non-indexed or non-English literature. Second, bibliometric indicators themselves cannot directly reflect research quality or clinical value and should be supplemented by systematic reviews and mechanistic experiments. Additionally, given that research on intratumoral microbiomes is still rapidly evolving, the analysis of trends and hotspots may shift over time; therefore, conclusions need to be updated dynamically. In addition, when analyzing the status and trends of preclinical and clinical trial research on intratumoral microbiomes, several limitations should be acknowledged. First, database searches can be broadened to capture a more comprehensive set of sources. Second, the field is rapidly evolving, so trends identified at one time may shift with new technologies and approaches, necessitating periodic updates. Third, the methodological quality of included studies varies, introducing risk of bias in summaries and conclusions.
5 Conclusion
This study analyzed the current research status of intratumoral microbiomes based on bibliometric methods. Current literature identifies several stable hotspot areas. Immuno-oncology remains the core field, with intratumoral microorganisms implicated in the modulation of anti-tumor immunity, and their effects on responses to immune checkpoint inhibitors and the tumor microenvironment vary across different cancer contexts. Advances in intratumoral microbial mechanisms are driven by multi-omics integration, with particular emphasis on the connections between microbial metabolites, host immunometabolism pathways, and the tumor microenvironment. The gut–tumor axis and colonization by gut-derived microbes have become particularly active research directions, spurring investigations into determinants of colonization and potential regulatory strategies to support therapy. Therapeutic strategies, including nano-delivery systems, probiotics, and engineered bacteria—are increasingly represented, but most remain at the preclinical stage, with unresolved questions regarding safety, durability, and translational feasibility. Diagnostic and prognostic biomarker research is evident but remains exploratory, requiring validation across cohorts. Collectively, these hotspots suggest the field is moving toward mechanistic understanding and translational exploration, while emphasizing the need for robust validation and cross-institutional collaboration to translate findings into safe and effective clinical interventions.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
XT: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. CM: Data curation, Methodology, Project administration, Software, Writing – original draft, Writing – review & editing. ZL: Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. DG: Data curation, Project administration, Resources, Visualization, Writing – original draft, Writing – review & editing. YF: Data curation, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declared that financial support was received for this work and/or its publication. This work was supported partially by grants from: 1) Key project fund of Jiangsu Provincial HealthCommission (ZD2022052, K2023016); 2) Suqian science and technology support project fund (KY202203); 3) Social Development Fund of Zhenjiang (SH2024002,SH2024075); 4).Zhenjiang Special Fund Project for Basic Research (JC2024031).
Acknowledgments
The authors are grateful to all participants and clinicians involved in this study.
Conflict of interest
The author(s) declared that this work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declared that generative AI was not used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2026.1745842/full#supplementary-material
References
1
Catalano V Turdo A Di Franco S Dieli F Todaro M Stassi G et al . Tumor and its microenvironment: a synergistic interplay. Semin Cancer Biol. (2013) 23:522–32. doi: 10.1016/j.semcancer.2013.08.007
2
Kennel K Greten F . The immune microenvironment of colorectal cancer. Nat Rev Cancer. (2025) 41(6):1152–69. doi: 10.1038/s41568-025-00872-1
3
Deng X Huang S . Microbiome-macrophage crosstalk in the tumor microenvironment: implications for oral squamous cell carcinoma progression and therapy. Front Immunol. (2025) 16:1651837. doi: 10.3389/fimmu.2025.1651837
4
Nejman D Livyatan I Fuks G Gavert N Zwang Y Geller LT et al . The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. (2020) 368:973. doi: 10.1126/science.aay9189
5
Dzutsev A Trinchieri G . A New Dimension in Tumor Heterogeneity: Intratumoral Bacteria Impact Multifocal Hepatocellular Carcinoma Development. Cancer Discov. (2025) 15:1540–2. doi: 10.1158/2159-8290.Cd-25-0899
6
May MS Park H Moallem DH Seeram D Dajiang S Hibshoosh H et al . Low Bacterial Biomass in Human Pancreatic Cancer and Adjacent Normal Tissue. Int J Mol Sci. (2024) 26. doi: 10.3390/ijms26010140
7
Lu YQ Qiao H Tan XR Liu N . Broadening oncological boundaries: the intratumoral microbiota. Trends Microbiol. (2024) 32:807–22. doi: 10.1016/j.tim.2024.01.007
8
Huang QF Wei X Li WY Ma YB Chen GX Zhao L et al . Endogenous Propionibacterium acnes Promotes Ovarian Cancer Progression via Regulating Hedgehog Signalling Pathway. Cancers. (2022) 14. doi: 10.3390/cancers14215178
9
Jiang L Duan BF Jia P Zhang Y Yan X . The Role of Intratumor Microbiomes in Cervical Cancer Metastasis. Cancers. (2023) 15. doi: 10.3390/cancers15020509
10
Zhang B Liu JF Li H Huang BQ Zhang BL Song BY et al . Integrated multi-omics identified the novel intratumor microbiome-derived subtypes and signature to predict the outcome, tumor microenvironment heterogeneity, and immunotherapy response for pancreatic cancer patients. Front Pharmacol. (2023) 14:1244752. doi: 10.3389/fphar.2023.1244752
11
Gurizzan C Licata AG Lorini L Piazza C Mattavelli D Paderno A et al . The interplay of hypoxia, inflammation, and microbiota as indicators of malignant transformation in oral potentially malignant disorders. Oral Oncol. (2025) 168:107583. doi: 10.1016/j.oraloncology.2025.107583
12
Zhang J You Z Li X Hu J Li J Jing Z et al . Harnessing intratumoral microbiota: new horizons in immune microenvironment and immunotherapy. J Transl Med. (2025) 23:897. doi: 10.1186/s12967-025-06916-2
13
Tan Z Huang Y Fu S Li H Gong C Lv D et al . Construction of a gene-metabolite-microbiome regulatory network reveals novel therapeutic targets in bladder cancer through multi-omics analysis. Ann Med. (2025) 57:2553220. doi: 10.1080/07853890.2025.2553220
14
Liao K Wen J Liu Z Zhang B Zhang X Fu Y et al . The role of intratumoral microbiome in the occurrence, proliferation, metastasis of colorectal cancer and its underlying therapeutic strategies. Ageing Res Rev. (2025) 111:102820. doi: 10.1016/j.arr.2025.102820
15
Gnanasekar A Castaneda G Iyangar A Magesh S Perez D Chakladar J et al . The intratumor microbiome predicts prognosis across gender and subtypes in papillary thyroid carcinoma. Comput Struct Biotechnol J. (2021) 19:1986–97. doi: 10.1016/j.csbj.2021.03.032
16
John D Yalamarty R Barakchi A Chen T Chakladar J Li WT et al . Transcriptomic Analysis Reveals Dysregulation of the Mycobiome and Archaeome and Distinct Oncogenic Characteristics according to Subtype and Gender in Papillary Thyroid Carcinoma. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms24043148
17
Parida S Siddharth S Xia Y Sharma D . Concomitant analyses of intratumoral microbiota and genomic features reveal distinct racial differences in breast cancer. NPJ Breast Cancer. (2023) 9:4. doi: 10.1038/s41523-023-00505-6
18
Chakladar J John D Magesh S Uzelac M Li WT Dereschuk K et al . The Intratumor Bacterial and Fungal Microbiome Is Characterized by HPV, Smoking, and Alcohol Consumption in Head and Neck Squamous Cell Carcinoma. Int J Mol Sci. (2022) 23. doi: 10.3390/ijms232113250
19
Sun Z Huang S Zhu P Tzehau L Zhao H Lv J et al . Species-resolved sequencing of low-biomass or degraded microbiomes using 2bRAD-M. Genome Biol. (2022) 23:36. doi: 10.1186/s13059-021-02576-9
20
Wang C Ma A Li Y McNutt ME Zhang S Zhu J et al . A Bioinformatics Tool for Identifying Intratumoral Microbes from the ORIEN Dataset. Cancer Res Commun. (2024) 4:293–302. doi: 10.1158/2767-9764.Crc-23-0213
21
Lee S Kim G Lee J Lee AC Kwon S . Mapping cancer biology in space: applications and perspectives on spatial omics for oncology. Mol Cancer. (2024) 23:26. doi: 10.1186/s12943-024-01941-z
22
Kwon SY Ngo HTT Son J Hong Y Min JJ . Exploiting bacteria for cancer immunotherapy. Nat Rev Clin Oncol. (2024) 21:569–89. doi: 10.1038/s41571-024-00908-9
23
Wang YB Fu XM Lin MJ Cai RD Zhu Y Wu TT et al . An intratumor bacteria-targeted DNA nanocarrier for multifaceted tumor microenvironment intervention. Mater Today Bio. (2024) 27. doi: 10.1016/j.mtbio.2024.101144
24
Shiao SL Kershaw KM Limon JJ You SY Yoon J Ko EY et al . Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell. (2021) 39:1202. doi: 10.1016/j.ccell.2021.07.002
25
Inamura K Hamada T Bullman S Ugai T Yachida S Ogino S et al . Cancer as microenvironmental, systemic and environmental diseases: opportunity for transdisciplinary microbiomics science. Gut. (2022) 71:2107–22. doi: 10.1136/gutjnl-2022-327209
26
Geng S Guo P Li X Shi Y Wang J Cao M et al . Biomimetic Nanovehicle-Enabled Targeted Depletion of Intratumoral Fusobacterium nucleatum Synergizes with PD-L1 Blockade against Breast Cancer. ACS Nano. (2024) 18:8971–87. doi: 10.1021/acsnano.3c12687
27
Liu X Sun M Pu F Ren J Qu X . Transforming Intratumor Bacteria into Immunopotentiators to Reverse Cold Tumors for Enhanced Immuno-chemodynamic Therapy of Triple-Negative Breast Cancer. J Am Chem Soc. (2023) 145:26296–307. doi: 10.1021/jacs.3c09472
28
Wang S Wu K Zhang Z Xu Z Wu J Xu S et al . Mapping theme trends and recognizing research hot spots in the use of ultrasound in orthopaedics: a bibliometric analysis of global research. Am J Transl Res. (2021) 13:9892–911.
29
Galeano Niño JL Wu H LaCourse KD Kempchinsky AG Baryiames A Barber B et al . Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. (2022) 611:810–7. doi: 10.1038/s41586-022-05435-0
30
Arnone AA Tsai YT Cline JM Wilson AS Westwood B Seger ME et al . Endocrine-targeting therapies shift the breast microbiome to reduce estrogen receptor-a breast cancer risk. Cell Rep Med. (2025) 6. doi: 10.1016/j.xcrm.2024.101880
31
Goto Y Iwata S Miyahara M Miyako E . Discovery of Intratumoral Oncolytic Bacteria Toward Targeted Anticancer Theranostics. Adv Sci. (2023) 10. doi: 10.1002/advs.202301679
32
Murphy C Rettedal E Lehouritis P Devoy C Tangney M . Intratumoural production of TNFα by bacteria mediates cancer therapy. PloS One. (2017) 12:e0180034. doi: 10.1371/journal.pone.0180034
33
Qin G Shao X Liu X Xu J Wang X Wang W et al . A signaling molecule from intratumor bacteria promotes trastuzumab resistance in breast cancer cells. Proc Natl Acad Sci U.S.A. (2025) 122:e2421710122. doi: 10.1073/pnas.2421710122
34
Shen HS Zhang CY Li SJ Liang YM Lee LT Aggarwal N et al . Prodrug-conjugated tumor-seeking commensals for targeted cancer therapy. Nat Commun. (2024) 15. doi: 10.1038/s41467-024-48661-y
35
Wang Y Han Y Yang C Bai T Zhang C Wang Z et al . Long-term relapse-free survival enabled by integrating targeted antibacteria in antitumor treatment. Nat Commun. (2024) 15:4194. doi: 10.1038/s41467-024-48662-x
36
Zhao Y Li M Guo Y Jin J Pei F Wang W et al . Neutrophil hitchhiking nanoparticles enhance bacteria-mediated cancer therapy via NETosis reprogramming. J Ctrl Rel. (2024) 367:661–75. doi: 10.1016/j.jconrel.2024.01.068
37
Zhu Z Cai J Hou W Xu K Wu X Song Y et al . Microbiome and spatially resolved metabolomics analysis reveal the anticancer role of gut Akkermansia muciniphila by crosstalk with intratumoral microbiota and reprogramming tumoral metabolism in mice. Gut Microbes. (2023) 15:2166700. doi: 10.1080/19490976.2023.2166700
38
Song WF Zheng D Zeng SM Zeng X Zhang XZ . Targeting to Tumor-Harbored Bacteria for Precision Tumor Therapy. ACS Nano. (2022) 16:17402–13. doi: 10.1021/acsnano.2c08555
39
Deng H Muthupalani S Erdman S Liu H Niu Z Wang TC et al . Translocation of Helicobacter hepaticus synergizes with myeloid-derived suppressor cells and contributes to breast carcinogenesis. Oncoimmunology. (2022) 11:2057399. doi: 10.1080/2162402x.2022.2057399
40
Kamarajan P Ateia I Shin JM Fenno JC Le C Zhan L et al . Periodontal pathogens promote cancer aggressivity via TLR/MyD88 triggered activation of Integrin/FAK signaling that is therapeutically reversible by a probiotic bacteriocin. PloS Pathog. (2020) 16:e1008881. doi: 10.1371/journal.ppat.1008881
41
Chen LF Kang ZY Shen JJ Zhao R Miao Y Zhang L et al . An emerging antibacterial nanovaccine for enhanced chemotherapy by selectively eliminating tumor-colonizing bacteria. Sci Bull. (2024) 69:2565–79. doi: 10.1016/j.scib.2024.06.016
42
Chen LF Shen JJ Kang ZY Zhang ZM Zheng ZX Zhang L et al . Fusobacterium nucleatum-mimicking nanovehicles to overcome chemoresistance for breast cancer treatment by eliminating tumor-colonizing bacteria. Chem. (2024) 10:1783–755. doi: 10.1016/j.chempr.2024.01.030
43
Chen Q Zhu YF Wang KR Wang XY Zhang YY Zhou MT et al . Clearing intratumoral bacteria to augment tumor-therapeutic response by engineering antimicrobial dendritic mesoporous silica nanomedicine. Nano Today. (2023) 52. doi: 10.1016/j.nantod.2023.101994
44
Gao C Wang X Yang B Yuan W Huang W Wu G et al . Synergistic Target of Intratumoral Microbiome and Tumor by Metronidazole-Fluorouridine Nanoparticles. ACS Nano. (2023) 17:7335–51. doi: 10.1021/acsnano.2c11305
45
Gao JF Tang L Fu C Cao YQ Liu HN Yin Y et al . A Nano-Strategy for Advanced Triple-Negative Breast Cancer Therapy by Regulating Intratumoral Microbiota. Nano Lett. (2025) 25:6134–44. doi: 10.1021/acs.nanolett.5c00298
46
Hu J Ran S Huang Z Liu Y Hu H Zhou Y et al . Antibacterial tellurium-containing polycarbonate drug carriers to eliminate intratumor bacteria for synergetic chemotherapy against colorectal cancer. Acta Biomater. (2024) 185:323–35. doi: 10.1016/j.actbio.2024.06.042
47
Kang XX Bu FQ Feng WL Liu F Yang XK Li HF et al . Dual-Cascade Responsive Nanoparticles Enhance Pancreatic Cancer Therapy by Eliminating Tumor-Resident Intracellular Bacteria. Adv Mater. (2022) 34. doi: 10.1002/adma.202206765
48
Kim JS Park JE Choi SH Kang SW Lee JH Lee JS et al . ECM-targeting bacteria enhance chemotherapeutic drug efficacy by lowering IFP in tumor mouse models. J Ctrl Rel. (2023) 355:199–210. doi: 10.1016/j.jconrel.2023.02.001
49
LaCourse KD Zepeda-Rivera M Kempchinsky AG Baryiames A Minot SS Johnston CD et al . The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep. (2022) 41:111625. doi: 10.1016/j.celrep.2022.111625
50
Li X Ma Y Xin Y Ma F Gao H . Tumor-Targeting Nanoassembly for Enhanced Colorectal Cancer Therapy by Eliminating Intratumoral Fusobacterium nucleatum. ACS Appl Mater Interfaces. (2023) 15:14164–72. doi: 10.1021/acsami.3c01210
51
Liang M Liu Y Zhang Z Yang H Dai N Zhang N et al . Fusobacterium nucleatum induces MDSCs enrichment via activation the NLRP3 inflammosome in ESCC cells, leading to cisplatin resistance. Ann Med. (2022) 54:989–1003. doi: 10.1080/07853890.2022.2061045
52
Niu Y Zhao X Li Y Ma X Yang W Ma J et al . Neutrophil-Mimicking Nanomedicine Eliminates Tumor Intracellular Bacteria and Enhances Chemotherapy on Liver Metastasis of Colorectal Cancer. Adv Sci (Weinh). (2025) 12:e04188. doi: 10.1002/advs.202504188
53
Wang S Chan SY Deng Y Khoo BL Chua SL . Oxidative stress induced by Etoposide anti-cancer chemotherapy drives the emergence of tumor-associated bacteria resistance to fluoroquinolones. J Adv Res. (2024) 55:33–44. doi: 10.1016/j.jare.2023.02.011
54
Wu T Jin D Wu M Xin Y Liu H Li N et al . Blocking Fusobacterium nucleatum-host cell interactions with responsive supramolecular GalNAc-derived nanoplatform for enhanced chemotherapy in colorectal cancer. Nano Today. (2024) 56. doi: 10.1016/j.nantod.2024.102288
55
Yeung YWS Ma Y Deng Y Khoo BL Chua SL . Bacterial Iron Siderophore Drives Tumor Survival and Ferroptosis Resistance in a Biofilm-Tumor Spheroid Coculture Model. Adv Sci (Weinh). (2024) 11:e2404467. doi: 10.1002/advs.202404467
56
Yan X Chen Q An J Liu DE Huang Y Yang R et al . Hyaluronic acid/PEGylated amphiphilic nanoparticles for pursuit of selective intracellular doxorubicin release. J Mater Chem B. (2019) 7:95–102. doi: 10.1039/c8tb02370k
57
Colbert LE El Alam MB Wang R Karpinets T Lo D Lynn EJ et al . Tumor-resident Lactobacillus iners confer chemoradiation resistance through lactate-induced metabolic rewiring. Cancer Cell. (2023) 41:1945–62.e11. doi: 10.1016/j.ccell.2023.09.012
58
Geller LT Barzily-Rokni M Danino T Jonas OH Shental N Nejman D et al . Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. (2017) 357:1156–60. doi: 10.1126/science.aah5043
59
Li J Zheng H Leung S . Potential of bacteriophage therapy in managing Staphylococcus aureus infections during chemotherapy for lung cancer patients. Sci Rep. (2023) 13:9534. doi: 10.1038/s41598-023-36749-2
60
Brennan CA Nakatsu G Gallini Comeau CA Drew DA Glickman JN Schoen RE et al . Aspirin Modulation of the Colorectal Cancer-Associated Microbe Fusobacterium nucleatum. mBio. (2021) 12:e00547-21. doi: 10.1128/mBio.00547-21
61
Chen L Shen Y Shi H Chen Y Yang T Cao W et al . Engineered Bio-Heterojunction for Effective Treatment of Colorectal Cancer Through Intratumoral Bacteria Scavenging and Autophagy Regulation. Adv Funct Mater. (2025) 35. doi: 10.1002/adfm.202421288
62
Kang Z Chen L Li P Zheng Z Shen J Xiao Z et al . A polyvalent vaccine for selectively killing tumor-associated bacteria to prevent cancer metastasis. Sci Adv. (2025) 11:eadt0341. doi: 10.1126/sciadv.adt0341
63
Kong F Fang C Zhang Y Duan L Du D Xu G et al . Abundance and Metabolism Disruptions of Intratumoral Microbiota by Chemical and Physical Actions Unfreeze Tumor Treatment Resistance. Adv Sci (Weinh). (2022) 9:e2105523. doi: 10.1002/advs.202105523
64
Li FY Zheng BZ Chen JF Yan QL Lu ZJ Fang C et al . Regulating the Tumor Microbiome through Near-Infrared-III Light-Excited Photosynthesis. ACS Nano. (2025) 19:14107–20. doi: 10.1021/acsnano.4c18954
65
Mehta RS Nishihara R Cao Y Song M Mima K Qian ZR et al . Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol. (2017) 3:921–7. doi: 10.1001/jamaoncol.2016.6374
66
Wang X Chen Q Zhu Y Wang K Chang Y Wu X et al . Destroying pathogen-tumor symbionts synergizing with catalytic therapy of colorectal cancer by biomimetic protein-supported single-atom nanozyme. Signal Transduct Targ Ther. (2023) 8:277. doi: 10.1038/s41392-023-01491-8
67
Soto-Pantoja DR Gaber M Arnone AA Bronson SM Cruz-Diaz N Wilson AS et al . Diet Alters Entero-Mammary Signaling to Regulate the Breast Microbiome and Tumorigenesis. Cancer Res. (2021) 81:3890–904. doi: 10.1158/0008-5472.Can-20-2983
68
Zhang S Zhu H Peng X Zhou Y Huang R Wang YF et al . Efficient Artificial T Cell Therapy for Bacterial-Colonized Tumors. Adv Mater. (2025) 37:e2503087. doi: 10.1002/adma.202503087
69
Duncan JL Ahmad RN Danesi H Slade DJ Davalos RV Verbridge SS et al . Electro-antibacterial therapy (EAT) to enhance intracellular bacteria clearance in pancreatic cancer cells. Bioelectrochemistry. (2024) 157:108669. doi: 10.1016/j.bioelechem.2024.108669
70
Fu A Yao B Dong T Chen Y Yao J Liu Y et al . Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. (2022) 185:1356–72.e26. doi: 10.1016/j.cell.2022.02.027
71
Khan ZA Song SS Xu HQ Ahmad M Wang AT Abdullah A et al . Elimination of intracellular microbes using drug combination therapy and unveiling survival mechanism of host cells upon microbial invasion. Int J Antimicrob Agents. (2025) 65. doi: 10.1016/j.ijantimicag.2025.107471
72
Xie YL Wang JR Li L Wang M Sun JK Chang JY et al . A Metal Chelation Therapy to Effectively Eliminate Breast Cancer and Intratumor Bacteria While Suppressing Tumor Metastasis by Copper Depletion and Zinc Ions Surge. Angew Chem-Int Ed. (2025) 64. doi: 10.1002/anie.202417592
73
Qu X Yin F Pei M Chen Q Zhang Y Lu S et al . Modulation of Intratumoral Fusobacterium nucleatum to Enhance Sonodynamic Therapy for Colorectal Cancer with Reduced Phototoxic Skin Injury. ACS Nano. (2023) 17:11466–80. doi: 10.1021/acsnano.3c01308
74
Alam A Levanduski E Denz P Villavicencio HS Bhatta M Alhorebi L et al . Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. (2022) 40:153. doi: 10.1016/j.ccell.2022.01.003
75
Bender MJ McPherson AC Phelps CM Pandey SP Laughlin CR Shapira JH et al . Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell. (2023) 186:1846–62.e26. doi: 10.1016/j.cell.2023.03.011
76
Carroll CSE Andrew ER Malik L Elliott KF Brennan M Meyer J et al . Simple and effective bacterial-based intratumoral cancer immunotherapy. J Immunother Cancer. (2021) 9. doi: 10.1136/jitc-2021-002688
77
Gurbatri CR Radford GA Vrbanac L Im J Thomas EM Coker C et al . Engineering tumor-colonizing E. coli Nissle 1917 for detection and treatment of colorectal neoplasia. Nat Commun. (2024) 15:646. doi: 10.1038/s41467-024-44776-4
78
Han ZY Fu ZJ Wang YZ Zhang C Chen QW An JX et al . Probiotics functionalized with a gallium-polyphenol network modulate the intratumor microbiota and promote anti-tumor immune responses in pancreatic cancer. Nat Commun. (2024) 15. doi: 10.1038/s41467-024-51534-z
79
Li L He SH Liao BY Wang MC Lin HM Hu B et al . Orally Administrated Hydrogel Harnessing Intratumoral Microbiome and Microbiota-Related Immune Responses for Potentiated Colorectal Cancer Treatment. Research. (2024) 7. doi: 10.34133/research.0364
80
Li Y Xing S Chen F Li Q Dou S Huang Y et al . Intracellular Fusobacterium nucleatum infection attenuates antitumor immunity in esophageal squamous cell carcinoma. Nat Commun. (2023) 14:5788. doi: 10.1038/s41467-023-40987-3
81
Pan BL Zhang XX Ye DJ Yao YX Zhang Z Luo Y et al . Intratumoral Brevibacillus parabrevis enhances antitumor immunity by inhibiting NK cell ferroptosis in hepatocellular carcinoma. Cell Death Dis. (2025) 16. doi: 10.1038/s41419-025-07733-7
82
Shi YY Zheng WX Yang KT Harris KG Ni KY Xue L et al . Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med. (2020) 217. doi: 10.1084/jem.20192282
83
Wang XL Fang Y Liang W Wong CC Qin HL Gao YH et al . Fusobacterium nucleatum facilitates anti-PD-1 therapy in microsatellite stable colorectal cancer. Cancer Cell. (2024) 42:895 1729-46.e8. doi: 10.1016/j.ccell.2024.08.019
84
Wu H Leng X Liu Q Mao T Jiang T Liu Y et al . Intratumoral Microbiota Composition Regulates Chemoimmunotherapy Response in Esophageal Squamous Cell Carcinoma. Cancer Res. (2023) 83:3131–44. doi: 10.1158/0008-5472.Can-22-2593
85
Michikawa C Gopalakrishnan V Harrandah AM Karpinets TV Garg RR Chu RA et al . Fusobacterium is enriched in oral cancer and promotes induction of programmed death-ligand 1 (PD-L1). Neoplasia. (2022) 31:100813. doi: 10.1016/j.neo.2022.100813
86
Zhang J Wan S Zhou H Du J Li Y Zhu H et al . Programmed Nanocloak of Commensal Bacteria-Derived Nanovesicles Amplify Strong Immunoreactivity against Tumor Growth and Metastatic Progression. ACS Nano. (2024) 18:9613–26. doi: 10.1021/acsnano.3c13194
87
Zhang Q Wang DJ Zhuo GZ Wang SL Yuan YF Wang LP et al . Intratumoral Stenotrophomonas Maltophilia in Breast Cancer: Unraveling the Interplay with Hormone Receptors and Impact on Tumor Immunity. Int J Biol Sci. (2025) 21:974–88. doi: 10.7150/ijbs.98260
88
Jiang H Guo YD Yu ZG Hu P Shi JL . Nanocatalytic bacteria disintegration reverses immunosuppression of colorectal cancer. Natl Sci Rev. (2022) 9. doi: 10.1093/nsr/nwac169
89
Pushalkar S Hundeyin M Daley D Zambirinis CP Kurz E Mishra A et al . The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. (2018) 8:403–16. doi: 10.1158/2159-8290.Cd-17-1134
90
Zheng DW Deng WW Song WF Wu CC Liu J Hong S et al . Biomaterial-mediated modulation of oral microbiota synergizes with PD-1 blockade in mice with oral squamous cell carcinoma. Nat BioMed Eng. (2022) 6:32–43. doi: 10.1038/s41551-021-00807-9
91
Zhang X Chen X Guo Y Gao G Wang D Wu Y et al . Dual Gate-Controlled Therapeutics for Overcoming Bacterium-Induced Drug Resistance and Potentiating Cancer Immunotherapy. Angew Chem Int Ed Engl. (2021) 60:14013–21. doi: 10.1002/anie.202102059
92
Chen X Sun J Ye T Li F . Neutrophil-Camouflaged Stealth Liposomes for Photothermal-Induced Tumor Immunotherapy Through Intratumoral Bacterial Activation. Pharmaceutics. (2025) 17. doi: 10.3390/pharmaceutics17050614
93
Xiao C Feng X Zhao T Ding G Gao Y . Research trends and hotspots of intratumor microbiome: A comprehensive visualization and bibliometric study from 2012 to 2024. Chin Med J (Engl). (2025) 138:376–8. doi: 10.1097/cm9.0000000000003333
94
Wang G Wang H Ji X Wang T Zhang Y Jiang W et al . Intratumoral microbiome is associated with gastric cancer prognosis and therapy efficacy. Gut Microbes. (2024) 16:2369336. doi: 10.1080/19490976.2024.2369336
95
Zepeda-Rivera M Minot SS Bouzek H Wu H Blanco-Míguez A Manghi P et al . A distinct Fusobacterium nucleatum clade dominates the colorectal cancer niche. Nature. (2024) 628:424–32. doi: 10.1038/s41586-024-07182-w
96
Zhao F An R Ma Y Yu S Gao Y Wang Y et al . Integrated spatial multi-omics profiling of Fusobacterium nucleatum in breast cancer unveils its role in tumour microenvironment modulation and cancer progression. Clin Transl Med. (2025) 15:e70273. doi: 10.1002/ctm2.70273
97
Wei Z Gao G He Q Kwok LY Sun Z . The gut-tumor connection: the role of microbiota in cancer progression and treatment strategies. J Adv Res. (2025). doi: 10.1016/j.jare.2025.08.038
98
Roesel R Strati F Basso C Epistolio S Spina P Djordjevic J et al . Combined tumor-associated microbiome and immune gene expression profiling predict response to neoadjuvant chemo-radiotherapy in locally advanced rectal cancer. Oncoimmunology. (2025) 14:2465015. doi: 10.1080/2162402x.2025.2465015
99
Schorr L Mathies M Elinav E Puschhof J . Intracellular bacteria in cancer-prospects and debates. NPJ Biofilms Microbiomes. (2023) 9. doi: 10.1038/s41522-023-00446-9
100
Chen Y Liu S Tan S Zheng Y Chen Y Yang C et al . KRAS mutations promote the intratumoral colonization of enterotoxigenic bacteroides fragilis in colorectal cancer through the regulation of the miRNA3655/SURF6/IRF7/IFNβ axis. Gut Microbes. (2024) 16:2423043. doi: 10.1080/19490976.2024.2423043
101
Yu J Lu W Chen Y Li Y Yang J Cai Z et al . Butyrate derived from intratumoral microbiome facilitates pulmonary metastasis in osteosarcoma. Pharmacol Res. (2025) 219:107897. doi: 10.1016/j.phrs.2025.107897
102
Li X Wang Y Wu Y Wang J Qi Y Bi X et al . Intratumor microbiome-derived butyrate enhances progesterone sensitivity by inducing ferroptosis in endometrial cancer. Pharmacol Res. (2025) 219:107902. doi: 10.1016/j.phrs.2025.107902
103
Ma Y Chen H Li H Zheng M Zuo X Wang W et al . Intratumor microbiome-derived butyrate promotes lung cancer metastasis. Cell Rep Med. (2024) 5:101488. doi: 10.1016/j.xcrm.2024.101488
104
Zhu X Li K Liu G Wu R Zhang Y Wang S et al . Microbial metabolite butyrate promotes anti-PD-1 antitumor efficacy by modulating T cell receptor signaling of cytotoxic CD8 T cell. Gut Microbes. (2023) 15:2249143. doi: 10.1080/19490976.2023.2249143
105
Fu S He K Tian C Sun H Zhu C Bai S et al . Impaired lipid biosynthesis hinders anti-tumor efficacy of intratumoral iNKT cells. Nat Commun. (2020) 11:438. doi: 10.1038/s41467-020-14332-x
106
Lan X Zhang F Cui G Hu B Lai Y Lin H et al . Microbiota-directed lactic acid depleting nanoenzyme reactivates antitumor immunity and chemosensitivity in hypoxic tumor. Mater Today Bio. (2025) 35:102493. doi: 10.1016/j.mtbio.2025.102493
107
Wu Z Liu Z Wang Y Teng G Li X Lu T et al . A comprehensive analysis of the tryptophan metabolism-related gene signature to predict the prognosis of esophageal squamous cell carcinoma based on multi-omics. Front Mol Biosci. (2025) 12:1613539. doi: 10.3389/fmolb.2025.1613539
108
Luo D Chen Q Li Y Yang J Tao Y Ji L et al . Microbiome-metabolome interplay in pancreatic cancer progression: insights from multi-omics analysis. Mol Cancer. (2025) 24:240. doi: 10.1186/s12943-025-02458-9
109
Li T Zhao Z Peng M Zhang L Wang C Luo F et al . Multi-omics analysis reveals the interplay between intratumoral bacteria and glioma. mSystems. (2025) 10:e0045724. doi: 10.1128/msystems.00457-24
110
Wang Y Qu D Zhang Y Jin Y Feng Y Zhang H et al . Intra-tumoral microbial community profiling and associated metabolites alterations of TNBC. Front Oncol. (2023) 13:1143163. doi: 10.3389/fonc.2023.1143163
111
Bullman S . INVADEseq to study the intratumoural microbiota at host single-cell resolution. Nat Rev Cancer. (2023) 23:189. doi: 10.1038/s41568-023-00553-x
112
Ge Y Lu J Puiu D Revsine M Salzberg SL . Comprehensive analysis of microbial content in whole-genome sequencing samples from The Cancer Genome Atlas project. Sci Transl Med. (2025) 17:eads6335. doi: 10.1126/scitranslmed.ads6335
Summary
Keywords
bibliometric analysis, cancer therapy, Fusobacterium nucleatum, immune microenvironment, intratumoral microbiota
Citation
Tang X, Man C, Liu Z, Gong D and Fan Y (2026) Research on microbial communities in tumor microenvironments: cutting-edge dynamics and future trends from a bibliometric perspective. Front. Immunol. 17:1745842. doi: 10.3389/fimmu.2026.1745842
Received
13 November 2025
Revised
19 January 2026
Accepted
20 January 2026
Published
16 February 2026
Volume
17 - 2026
Edited by
Francisco Jose Roig, Universidad San Jorge, Spain
Reviewed by
Yimtar L. Pongen, SRM Institute of Science and Technology, India
Mohini Verma, VSB-Technical University of Ostrava, Czechia
Updates
Copyright
© 2026 Tang, Man, Liu, Gong and Fan.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Fan, yuf12345@ujs.edu.cn
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.