- 1Alimentary Pharmabiotic Centre, Biosciences Institute, University College Cork, Cork, Ireland
- 2School of Microbiology, University College Cork, Cork, Ireland
- 3Cardiff School of Biosciences, Cardiff University, Cardiff, UK
- 4Department of Surgery and Cancer, Centre for Digestive and Gut Health, Imperial College London, London, UK
- 5Department of Biological Sciences, Cork Institute of Technology, Cork, Ireland
In the current study, a number of salt-tolerant clones previously isolated from a human gut metagenomic library were screened using Phenotype MicroArray (PM) technology to assess their functional capacity. PM's can be used to study gene function, pathogenicity, metabolic capacity and identify drug targets using a series of specialized microtitre plate assays, where each well of the microtitre plate contains a different set of conditions and tests a different phenotype. Cellular respiration is monitored colorimetrically by the reduction of a tetrazolium dye. One clone, SMG 9, was found to be positive for utilization/transport of L-carnitine (a well-characterized osmoprotectant) in the presence of 6% w/v sodium chloride (NaCl). Subsequent experiments revealed a significant growth advantage in minimal media containing NaCl and L-carnitine. Fosmid sequencing revealed putative candidate genes responsible for the phenotype. Subsequent cloning of two genes did not replicate the L-carnitine-associated phenotype, although one of the genes, a σ54-dependent transcriptional regulator, did confer salt tolerance to Escherichia coli when expressed in isolation. The original clone, SMG 9, was subsequently found to have lost the original observed phenotype upon further investigation. Nevertheless, this study demonstrates the usefulness of a phenomic approach to assign a functional role to metagenome-derived clones.
Introduction
The ability to adapt to and tolerate increases in extracellular osmolarity is an important characteristic that enables bacteria to survive in stressful environments. Increased osmolarity, caused by sodium chloride (NaCl) for example, initiates a phased response in bacteria. Firstly during the primary response, potassium ions are rapidly accumulated within the bacterial cell to offset the detrimental effects of water loss and influx of toxic sodium and chloride ions (Sleator and Hill, 2002; Epstein, 2003). Once the cell has been stabilized, the secondary response begins and involves the synthesis or, more often the more energetically favorable, uptake of osmoprotectant compounds (also termed compatible solutes or osmolytes). Osmoprotectants are compatible with cellular functions and can accumulate to very high concentrations within the cell and function to protect proteins and to restore cell volume and thus, turgor pressure (Kempf and Bremer, 1998; Sleator and Hill, 2002; Kunte, 2006). Osmoprotectants can be grouped broadly as amino acids, polyols, sugars, trimethyl- and quaternary-ammonium compounds and their derivatives (Kempf and Bremer, 1998). The most widely utilized and best characterized osmoprotectants are glycine betaine, carnitine, proline, and ectoine. Numerous studies have shown carnitine to be important not only for salt tolerance, but also for survival in vivo and pathogenesis of infection (Sleator et al., 2001; Wemekamp-Kamphuis et al., 2004). Carnitine is also found abundantly in animal tissues and red meat and is an important compound in the host environment; for the human pathogen Listeria monocytogenes, carnitine and its uptake system OpuC are critical for infection in mice (Sleator et al., 2003). In addition to its osmoprotective properties, carnitine may also be catabolised as a carbon or nitrogen source to generate energy (Wargo and Hogan, 2009).
The emergence of “omics” technologies, an umbrella term to include analyses such as genomics, metagenomics, transcriptomics, proteomics, metabolomics, and phenomics to name a few, have been used to gain valuable information about the functions and interactions of various biological systems as a whole and can provide more information than more traditional and reductive approaches to biological problems. Sequence-based and functional metagenomic approaches have led to the discovery of many novel and diverse genes (Beja et al., 2000; Gillespie et al., 2002; Banik and Brady, 2008; Culligan et al., 2013; Yoon et al., 2013). While identifying clones which display a specific phenotype through functional metagenomic screening yields worthwhile results, characterizing the functional mechanisms responsible for the observed phenotype can be sometimes difficult owing to the fact that a large proportion of metagenome derived genes will be annotated as hypothetical proteins or have no known function or homology to existing proteins (Bork, 2000; Qin et al., 2010). Phenomic approaches can be used to study hundreds of phenotypic profiles of different bacterial strains concurrently. Comparing phenomic profiles of wild-type and mutant derivatives or host strains and clones identified through metagenomic screening can reveal differences between strains relating to gene function, pathogenicity and metabolism for example (Bochner et al., 2001; Bochner, 2009).
With this in mind, we have utilized combined metagenomic and phenomic approaches in this study to characterize a salt tolerant clone identified from a human gut microbiome metagenomic fosmid library. An overview of the study design using this combined approach can be seen in Figure 1.The BIOLOG phenotype microarray (PM) system was used to compare phenotypes between metagenomic clones and cloning host (carrying empty fosmid vector). PM plates measure cellular respiration colorimetrically via reduction of a tetrazolium dye with electrons from NADH generated during the process of respiration. Strongly metabolized substrates generate a more intense purple color which is recorded with a camera on the Omnilog instrument. If desired, thousands of phenotypes may be monitored simultaneously using the different available PM plates which can be grouped as those that measure carbon, nitrogen, phosphorous and sulfur metabolism, response to different pH conditions, osmolytes and chemicals such as anitbiotics. The full range of PM plates and their constituents, for investigation of bacterial phenotypes can be found at: http://www.biolog.com/products-static/phenotype_microbial_cells_use.php
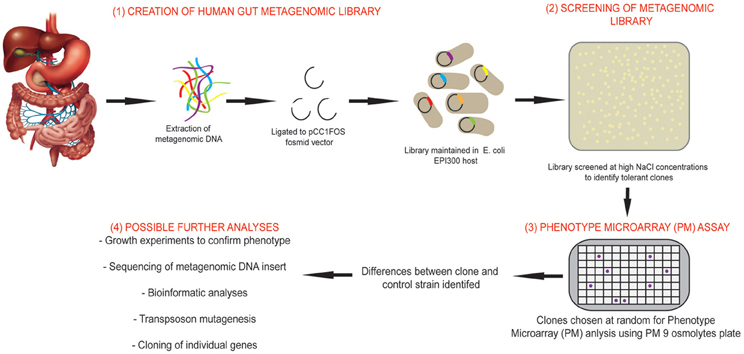
Figure 1. Overview of the study design using combined functional metagenomic and phenomic approaches. An overview of the study design and experimental process incorporating metagenomic library creation and screening, identification of salt tolerant clones, comparison of clone to control strain using Phenotypic Microarray (PM) assay and further analyses that may be performed to identify novel genes.
As our primary interest is salt tolerance we used the PM osmolyte plate (PM9) for analysis. The PM screen indicated that clone SMG 9 was positive for L-carnitine utilization in the presence of 6% w/v NaCl. Sequencing of the fosmid insert and cloning of two genes identified a novel salt tolerance gene, but did not replicate the carnitine-associated phenotype originally observed. Subsequent investigation revealed SMG 9 had lost this phenotype. Notwithstanding this phenomenon, this study demonstrates the usefulness of a phenomic approach to assign a functional role to metagenomic library-derived clones.
Materials and Methods
Bacterial Strains and Growth Conditions
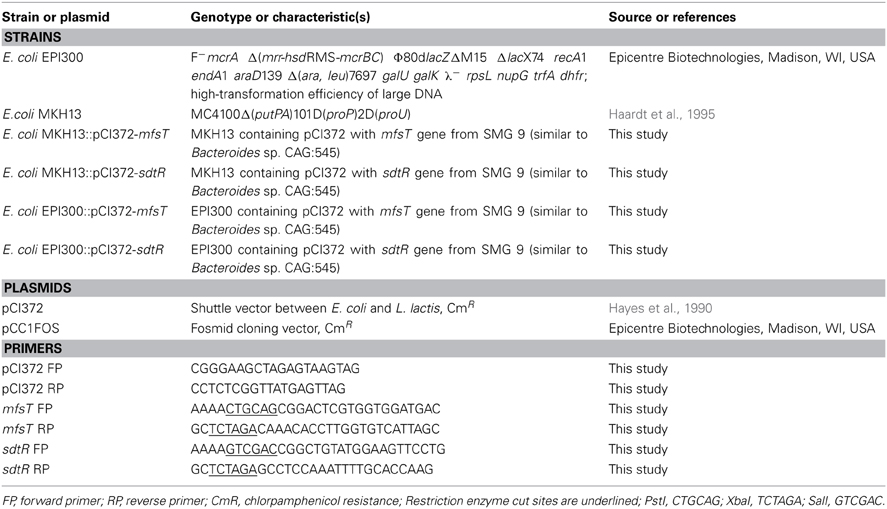
Bacterial strains, plasmids, and oligonucleotide primers (Eurofins, MWG Operon, Germany) used in this study are listed in Table 1. Escherichia coli EPI300::pCC1FOS (Epicentre Biotechnologies, Madison, WI, USA) was grown in Luria-Bertani (LB) medium containing 12.5 μg/ml chloramphenicol (Cm). E. coli MKH13 was grown in LB medium and in LB medium supplemented with 20 μg/ml Cm for strains transformed with the plasmid pCI372. LB media was supplemented with 1.5% w/v agar when required. All overnight cultures were grown at 37°C with shaking.
Metagenomic Library Construction and Screening
A previously constructed fosmid clone library (Jones and Marchesi, 2007; Jones et al., 2008), created from metagenomic DNA isolated from a fecal sample from a healthy 26 year old Caucasian male was used to screen for salt-tolerant clones. The library was screened as outlined previously (Culligan et al., 2012). Briefly, a total of 23,040 clones from the library were screened on LB agar supplemented with 6.5% (w/v) NaCl (a concentration which inhibits the growth of the cloning host, E. coli EPI300) and 12.5 μg/ml Cm using a Genetix QPix 2 XT colony picking/gridding robotics platform to identify clones with an increased salt tolerance phenotype compared to the cloning host (E. coli EPI300) carrying an empty fosmid vector (pCC1FOS). Identification of any salt tolerant clones will therefore most likely be due to a gene (or genes) present on the metagenomic insert from the human gut microbiota. Clones were gridded onto Q-trays (Genetix) using the robotics platform. Q-trays were incubated at 37°C for 3 days and checked twice daily for growth of likely salt-tolerant clones. Salt-tolerant clones were subsequently replica plated onto LB agar containing 12.5 μg/ml Cm and 6.5% NaCl and onto LB containing 12.5 μg/ml Cm, but without 6.5% NaCl (which represented a positive control plate). Each salt tolerant clone identified was streaked on LB agar + 12.5 μg/ml Cm to ensure a pure culture and all clones were maintained as glycerol stocks at −80°C.
Phenotype Microarray (PM) Assay
The PM9 osmolytes microplate was used to compare the cellular phenotypes of SMG 9 and the cloning host, E. coli EPI300::pCC1FOS (containing an empty fosmid vector) under 96 different conditions. Preparation of the different IF (Inoculating Fluids; proprietary formulation supplied by BIOLOG) solutions and inoculation of the PM plates was performed according to the BIOLOG PM protocol for E. coli and other Gram negative bacteria. Briefly, IF-0 solution was prepared by adding 25 ml of sterile water to 125 ml of 1.2× IF-0. IF-0 + dye mix A solution was prepared by adding 1.8 ml of dye mix A and 23.2 ml of sterile water to another bottle containing 125 ml of 1.2× IF-0. IF-10 solution was prepared by adding 1.5 ml of dye mix A and 23.5 ml of sterile water to a bottle containing 125 ml of 1.2× IF-10. E. coli strains were grown overnight on LB agar at 37°C by streaking from a frozen stock. Cells were sub-cultured by streaking again on LB agar and grown overnight again. Isolated colonies were removed from the agar plate using a sterile swab and added to a tube containing 16 ml of IF-0 solution until a cell suspension of 42% T (transmittance) was achieved using the BIOLOG Turbidimeter. 15 ml of this 42% T solution was diluted in 75 ml of IF-0 + dye mix A to achieve 85% T. 600 μl of the 85% T cell suspension solution was added to 120 ml of IF-10 + dye mix A. Finally, 100 μl of the final cell suspension was inoculated to each well of the PM 9 microplate. Plates were incubated at 37°C for 24 h in the Omnilog plate reader (BIOLOG).
Fosmid Sequencing and Analysis
Fosmid DNA was isolated from SMG 9 as described above to a concentration >200 ng/μl (approximately 5 μg in total). Sequencing of the full fosmid insert of SMG 9 was performed by GATC Biotech (Konstanz, Germany) using 454-pyrosequencing on a titanium mini-run of the Roche GS-FLX platform, achieving approximately 65-fold coverage. Sequencing reads were assembled into a single contig by GATC Biotech. The retrieved sequence was analyzed using the FGENESB software program (Softberry) to identify putative open reading frames and translated nucleotide sequences were subjected to BLASTP analysis to assign putative functions to the encoded proteins and identify homologous sequences from the National Centre for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). The full fosmid insert sequence of SMG 9 was submitted to GenBank and assigned the following accession number; KJ524644.
DNA Manipulations and Cloning of mfsT and sdtR Genes
Extraction of fosmids containing metagenomic DNA: 5 ml of bacterial culture was grown overnight with 12.5 μg/ml Cm. One millilitre of culture was used to inoculate 4 ml of fresh LB broth. To this, 5 μl of 1000× Copy Control Induction solution (Epicentre Biotechnologies) and 12.5 μg/ml Cm were added. The mixture was incubated at 37°C for 5 h with vigorous shaking (200–250 rpm) to ensure maximum aeration. Cells were harvested from the whole 5 ml of induced culture by centrifuging at 2100 × g for 12 min. Qiagen QIAprep Spin mini-prep kit was used to extract fosmids as per manufacturer's instructions. PCR products were purified with a Qiagen PCR purification kit and digested with XbaI and PstI (Roche Applied Science) for mfsT and with SalI and XbaI for sdtR, followed by ligation using the FastLink DNA ligase kit (Epicentre Biotechnologies) to similarly digested plasmid pCI372. Electrocompetent E. coli MKH13 and E. coli EPI300 were transformed with the ligation mixture and plated on LB agar plates containing 20 μg/ml Cm for selection. Colony PCR was performed on all resistant transformants using primers across the multiple cloning site (MCS) of pCI372 to confirm the presence and size of the insert.
Confirmation Tests for Observed Phenotype
Growth experiments were performed in defined M9 minimal media (M9MM) (Fluka) to confirm the observed phenotype. Single isolated colonies of SMG 9 and EPI300::pCC1FOS were grown overnight in M9MM (containing final concentrations of; D-glucose (0.4%), Bacto™ casamino acids (w/v 0.2%) (Becton, Dickinson and Co, Sparks, MD, USA), magnesium sulfate (MgSO4) (2 mM), calcium chloride (CaCl2) (0.1 mM) and 12.5 μg/ml Cm). Reagents were purchased from Sigma Aldrich (St. Louis, MO, USA) unless otherwise stated. Cells were harvested by centrifugation, washed in ¼ strength Ringers solution and resuspended in fresh M9MM. A 2% v/v inoculum was sub-cultured in fresh M9MM containing various concentrations of sodium chloride (0–8% w/v NaCl) and 1 mM of L-carnitine when required. Triplicate wells of a 96-well micro-titre plate were inoculated with 200 μl of the appropriate cell suspension. Plates were incubated at 37°C for 24–48 h in an automated spectrophotometer (Tecan Genios) which recorded the optical density at 595 nm (OD595 nm) every hour. After 48 h the data was retrieved and analyzed using the Magellan 3 software program and graphs were created with Sigma Plot 10.0 (Systat Software Inc, London, UK). Results are presented as the average of triplicate experiments, with error bars being representative of the standard error of the mean (SEM).
Results
Screening of Metagenomic Library
Approximately 23,000 clones from a metagenomic fosmid library from the human gut microbiome were screened previously and resulted in the identification of 53 salt tolerant clones which could grow on LB agar supplemented with 6.5% NaCl (a concentration which inhibits growth of the cloning host, E. coli EPI300::pCC1FOS) (Culligan et al., 2012). The salt tolerant clones identified were annotated as SMG 1-53 (Salt MetaGenome 1–53).
PM Osmolyte Plate Assay
A number of clones initially identified from the metagenomic library were chosen at random and phenotypically screened using PM 9 osmolytes plate from BIOLOG. The layout and contents of PM9 can be viewed at http://www.biolog.com/pdf/pm_lit/PM1-PM10.pdf. One clone, SMG 9, gave a positive result under one of the 96 different conditions tested. It was found that SMG 9 had an increased metabolic response (causing a reduction of the tetrazolium dye to generate a purple color, Figure 2A) compared to the cloning host containing an empty fosmid vector, E. coli EPI300::pCC1FOS (Figure 2B) in 6% w/v NaCl supplemented with L-carnitine. The color formation within each well was measured by BIOLOG's Omnilog machine, which produces a color-coded graph. A comparison of the kinetic data output from SMG 9 and EPI300::pCC1FOS can be seen in Figure 3.
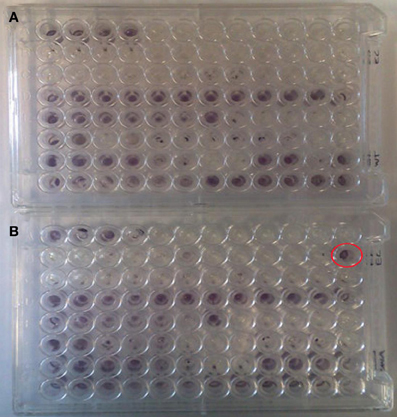
Figure 2. Appearance of PM 9 plates after incubation for 24 hours at 37°C. (A) Control EPI300::pCC1FOS and (B) SMG 9. PM plates measure cellular respiration colorimetrically via reduction of a tetrazolium dye with electrons from NADH generated during the process of respiration. Strongly metabolized substrates generate a more intense purple color. Development of a strong purple color can be seen in well B12 in Figure 2B (circled in red), which was inoculated with SMG 9, while no color development is visible in B12 of the control plate. This indicates SMG 9 has a greater ability to transport and utilise L-carnitine compared to the EPI300::pCC1FOS host strain.
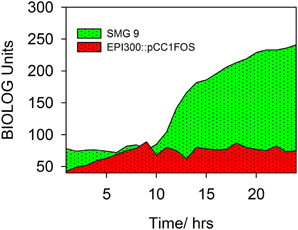
Figure 3. Kinetic data measured by BIOLOG Omnilog system. Color formation within each well was measured by BIOLOG's Omnilog machine, which produces a color coded graph. Kinetic data from two clones can be compared. EPI300::pCC1FOS is shown in red and that from SMG 9 is shown in green. The green color indicates more rapid metabolism by SMG 9 under the conditions in the well (6% NaCl + L-carnitine).
Confirmatory Experimentation of PM Analysis
In an attempt to confirm and replicate the result from the PM analysis, SMG 9 and EPI300::pCC1FOS were grown in M9MM containing various concentrations of NaCl (0–6% w/v) and supplemented with 1 mM L-carnitine when appropriate. Figure 4A shows growth of both clones in M9MM in the presence and absence of 1 mM L-carnitine. In the presence of L-carnitine, SMG 9 displays a growth defect, while growth is similar under all other conditions. The growth defect is alleviated when grown at 4% w/v NaCl + 1 mM L-carnitine and there is no difference in growth between clones either in the presence or absence of L-carnitine (Figure 4B). At 5% w/v NaCl however, SMG 9 has a significant growth advantage compared to EPI300::pCC1FOS both in the presence and absence of 1 mM L-carnitine (Figure 4C). The positive effect of L-carnitine on the growth of SMG 9 is evident with cells entering logarithmic phase growth sooner and reaching a much higher final optical density (OD595 nm). A similar, positive effect for L-carnitine on growth of SMG 9 is also seen at 6% w/v NaCl (Figure 4D).
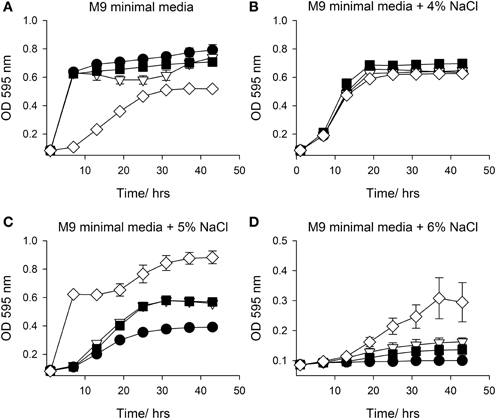
Figure 4. Growth in M9 minimal media with NaCl +/− 1mM L-carnitine. Growth of E. coli EPI300::pCC1FOS and SMG 9 in (A) M9 minimal media, (B) M9 minimal media + 4% NaCl, (C) M9 minimal media + 5% NaCl and (D) M9 minimal media + 6% NaCl. Legend: E. coli EPI300::pCC1FOS (• closed circle); SMG 9 (▽ open triangle); E. coli EPI300::pCC1FOS + 1mM L-carnitine (■ closed square); SMG 9 + 1 mM L-carnitine, (◊ open diamond).
Sequencing of SMG 9 Fosmid Insert and Analysis
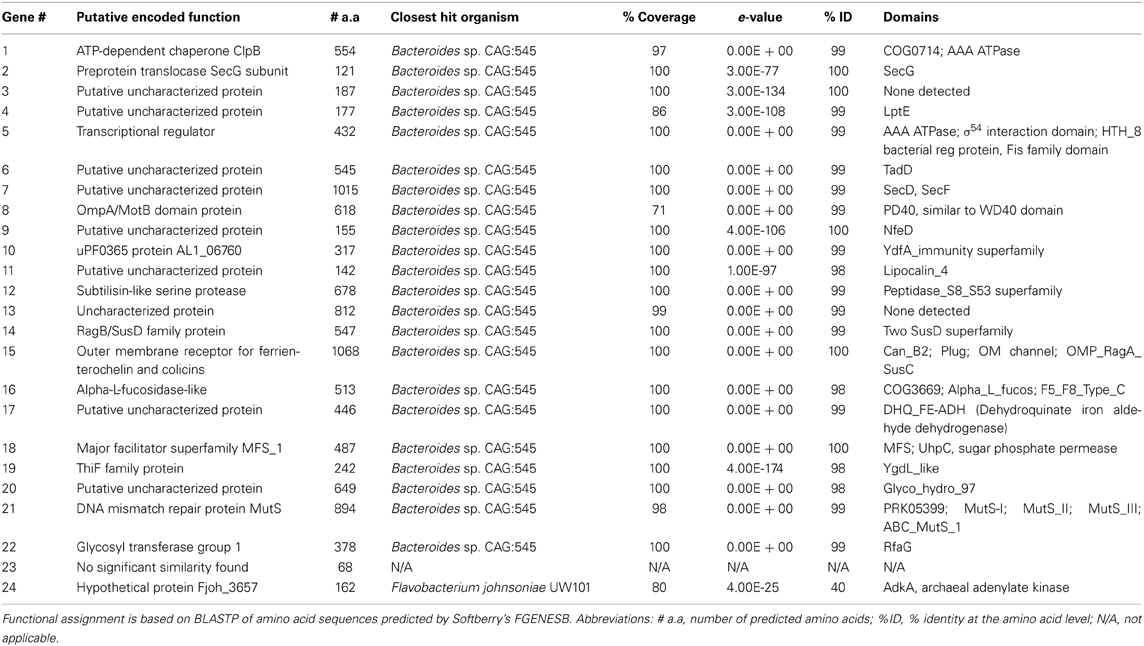
Fosmid SMG 9 was fully sequenced (454-pyrosequencing) and assembled by GATC Biotech. This generated a total of 2.3 × 106 base pairs of sequence data in 6939 sequencing reads. The average read length was 334 base pairs and coverage of 64.5× was achieved. Following vector trimming the length of the insert was approximately 36.5 kb and the %G+C content was 49.43%. Twenty-four putative open reading frames were predicted using Softberry's FGENESB, bacterial operon and gene prediction software (www.softberry.com) (Mavromatis et al., 2007). Translated nucleotide sequences were functionally annotated by homology searches using the BLASTP program to identify homologous sequences and determine their taxonomic origin. All but two of the genes encoded proteins with high identity (98–100% at the amino acid level) to Bacteroides sp. CAG:545. A list of the genes on SMG 9, their encoded functions and putative domains are presented in Table 2. The full fosmid insert sequence of SMG 9 has been submitted to GenBank and assigned the accession number, KJ524644.
Cloning of mfsT and sdtR Genes
Following initial inspection of the encoded proteins on SMG 9, the presence of an L-carnitine or general osmoprotectant transporter, nor indeed any protein with a functional link to carnitine metabolism was not immediately obvious. Transposon mutagenesis was attempted in order to create a knock-out mutant; this however, proved unsuccessful. Two genes (gene 5 and gene 18), which we felt may be likely to have a possible role in L-carnitine utilization based on bioinformatic analysis were cloned in isolation to further examine the phenotype. Genes 5 and 18 were annotated sdtR for sigma-dependent transcriptional regulator and mfsT for major facilitator superfamily transporter, respectively.
Both mfsT and sdtR were cloned in the vector pCI372 and expressed in both E. coli EPI300 and the osmosensitive strain E. coli MKH13 (Haardt et al., 1995). The effect of each gene on the growth of each strain under salt stress in the presence and absence of L-carnitine was assessed. The mfsT gene had no effect on growth under any of the conditions tested (data not shown). The sdtR gene on the other hand conferred a significant salt tolerance phenotype to E. coli MKH13 when grown in media supplemented with both 3 and 4% w/v NaCl (Figures 5C,D, respectively), while growth was similar in media lacking NaCl and in media supplemented with 2% w/v NaCl (Figures 5A,B, respectively). Cloning and expression of sdtR in EPI300 resulted in an increase in salt tolerance compare to wild-type EPI300 carrying an empty copy of the plasmid pCI372. Addition of 1 mM L-carnitine increased the growth rate and final optical density of both strains, but its effect on the sdtR+ strain was not significant relative to the EPI300::pCI372 control (Figures 6A,B).
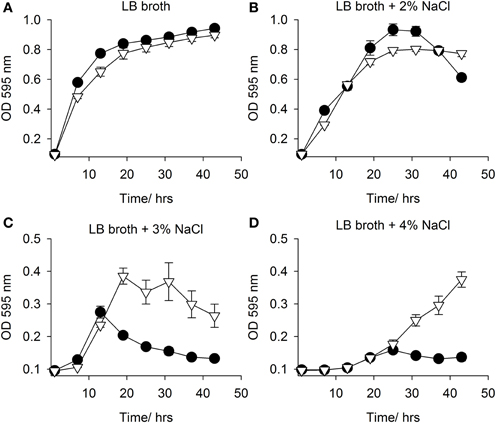
Figure 5. Growth in LB broth with NaCl. Growth in of E. coli MKH13::pCI372 (• black circle) and MKH13::pCI372-sdtR (▽ open triangle) in (A) LB broth and LB broth supplemented with (B) 2% NaCl, (C) 3% NaCl and (D) 4% NaCl.
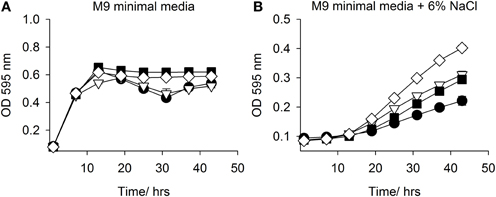
Figure 6. Growth in M9 minimal media with NaCl +/− L-carnitine. Growth of EPI300::pCI372 and EPI300::pCI372-sdtR in (A) M9 minimal media and (B) M9 minimal media + 6% NaCl. Legend: E. coli EPI300::pCI372 (• black circle); EPI300::pCI372 + 1 mM L-carnitine (▽ open triangle); EPI300::pCI372-sdtR (■ closed square); EPI300::pCI372-sdtR + 1mM L-carnitine, (◊ open diamond).
Although sdtR did confer a salt tolerance phenotype, neither of the cloned genes replicated the original phenotype related to L-carnitine. We re-examined clone SMG 9 and carried out further studies, however these revealed that SMG 9 had lost the carnitine-associated phenotype seen originally and unfortunately, it was therefore not possible to identify the gene(s) responsible.
Discussion
In the present study we have identified a novel salt tolerance gene from a metagenomic library clone from the human gut microbiome using a combined functional metagenomic and PM approach. The clone, SMG 9, was identified from a previous library screen to identify salt tolerant clones (Culligan et al., 2012) and was further characterized in this study using PM osmolyte plates. From the PM screen, SMG 9 showed an increased metabolic profile in the presence of 6% NaCl + 1 mM L-carnitine compared to the control strain carrying an empty fosmid vector (EPI300::pCC1FOS), indicating this clone could utilize or transport L-carnitine. Experiments to confirm the findings of the PM assay showed that SMG 9 displayed an increased growth profile at 5% and 6% NaCl in the presence of 1 mM L-carnitine compared to controls, similar to observations in the PM assay. Transposon mutagenesis was attempted to create phenotypic knock out mutants, using the EZTn5 system (Epicentre Biotechnologies; Goryshin and Reznikoff, 1998) but this proved unsuccessful. This may be due to the presence of a gene encoding a DNA repair protein MutS on the fosmid insert, which has been associated with transposon excision (specifically Tn5 and Tn10) (Lundblad and Kleckner, 1985).
Next generation sequencing of the full fosmid insert of SMG 9 and functional assignment of the encoded proteins using BLASTP revealed sequences shared highest genetic identity to Bacteroides sp. CAG:545. Species of Bacteroides are commonly found in the human gut, where the resident microbiota is largely composed of species from two dominant phyla, the Bacteroidetes and Firmicutes (Qin et al., 2010). The %G+C content of the SMG 9 insert was 49.44% which close to the reported range of 40–48% for genomes of species of Bacteroides (Shah, 1992). Sequencing and subsequent functional annotation did not reveal any obvious genes related to known L-carnitine transport or utilization systems, suggesting a novel mechanism may be involved. We conducted further bioinformatic analysis of the encoded proteins in an attempt to identify any link to salt tolerance or osmoprotectant transport. Gene 5 (sdtR) is predicted to encode a σ54-dependent transcriptional regulator, which contains a number of domains (see Table 2), including a helix-turn-helix 8 (HTH_8), Fis-family protein domain, while gene 18 (mfsT) is predicted to encode a major facilitator superfamily (MFS) protein with an UhpC sugar phosphate permease domain. Both genes were chosen for further study and cloning as Fis is a regulatory protein involved the regulation of proline (another important osmoprotectant) uptake (Xu and Johnson, 1995, 1997; Typas et al., 2007), while we further reasoned that sdtR could be regulating host EPI300 genes, contributing the L-carnitine-associated phenotype, while MFS transporters also play a role in osmoprotectant uptake (Culham et al., 1993; Haardt et al., 1995; Pao et al., 1998). Despite the presence of a sugar phosphate permease domain, indicating sugar transport, MFS transporters are known to have a diverse substrate range (Pao et al., 1998; Saier, 2000; Law et al., 2008).
The mfsT gene did not confer a salt tolerance or the L-carnitine associated phenotype to transformed cells (data not shown). The sdtR gene also did not confer the L-carnitine associated phenotype to E. coli, but did however confer an increased salt tolerance phenotype. sdtR therefore represents a novel salt tolerance gene and most likely functions by influencing expression (either positively or negatively) of host E. coli genes, although further work, comprising expression studies and microarray analysis, will be required to elucidate the genes involved as transcriptional regulators can influence a wide variety of genes. Transcriptional regulators are commonly involved in the response different stresses in bacteria (Hengge-Aronis et al., 1991; Cheville et al., 1996; Battesti et al., 2011; Hoffmann et al., 2013), while σ54 (RpoN) has been shown to play a role in osmotolerance in Listeria monocytogenes (Okada et al., 2006).
The loss of the carnitine-associated phenotype of SMG 9 prevented further characterization of this clone and ultimately the identification of the gene(s) responsible. It is difficult to pinpoint the cause of this phenotypic reversion, but a clue may be evident from Figure 4A, where a growth defect for SMG 9 is apparent when grown in M9MM + 1 mM L-carnitine. This indicates L-carnitine may be increasing the metabolic load on the cell and this metabolic stress is only relieved in the presence of NaCl, when L-carnitine may be utilized efficiently in an osmoprotective capacity. If the gene is constitutively expressed, a mutation may have occurred to counteract this phenomenon. The presence of a gene encoding MutS may also be relevant as mutations to MutS can result in a mutator phenotype in E. coli cells (Wu and Marinus, 1994). Furthermore, it is possible the original SMG 9 clone acquired a mutation on the fosmid insert that conferred the carnitine-associated phenotype and a subsequent suppressor mutation occurred to silence this mutation, returning the clone to its original phenotype.
In conclusion, we have identified a novel salt tolerance gene from the human gut microbiome using a combined functional metagenomic and PM approach. The gene originates from a species of Bacteroides and encodes a putative transcriptional regulator. Overall this study demonstrates the utility of functional metagenomics and phenomics for novel gene discovery and functional characterization of metagenome-derived clones.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Alimentary Pharmabiotic Centre is a research center funded by Science Foundation Ireland (SFI grant number 07/CE/B1368). We acknowledge the continued financial assistance of the Alimentary Pharmabiotic Centre, funded by Science Foundation Ireland. Julian R. Marchesi acknowledges funding from The Royal Society which supports the bioinformatic cluster (Hive) at Cardiff University, School of Biosciences. Roy D. Sleator is an ESCMID Research Fellow and Coordinator of ClouDx-i an EU FP7-PEOPLE-2012-IAPP project
References
Banik, J. J., and Brady, S. F. (2008). Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc. Natl. Acad. Sci. U.S.A. 105, 17273–17277. doi: 10.1073/pnas.0807564105.
Battesti, A., Majdalani, N., and Gottesman, S. (2011). The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213. doi: 10.1146/annurev-micro-090110-102946
Beja, O., Aravind, L., Koonin, E. V., Suzuki, M. T., Hadd, A., Nguyen, L. P., et al. (2000). Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902–1906. doi: 10.1126/science.289.5486.1902
Bochner, B. R. (2009). Global phenotypic characterization of bacteria. FEMS Microbiol. Rev. 33, 191–205. doi: 10.1111/j.1574-6976.2008.00149.x
Bochner, B. R., Gadzinski, P., and Panomitros, E. (2001). Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11, 1246–1255. doi: 10.1101/gr.186501
Bork, P. (2000). Powers and pitfalls in sequence analysis: the 70% hurdle. Genome Res. 10, 398–400. doi: 10.1101/gr.10.4.398
Cheville, A. M., Arnold, K. W., Buchrieser, C., Cheng, C. M., and Kaspar, C. W. (1996). rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 62, 1822–1824.
Culham, D. E., Lasby, B., Marangoni, A. G., Milner, J. L., Steer, B. A., Van Nues, R. W., et al. (1993). Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J. Mol. Biol. 229, 268–276. doi: 10.1006/jmbi.1993.1030
Culligan, E. P., Sleator, R. D., Marchesi, J. R., and Hill, C. (2012). Functional metagenomics reveals novel salt tolerance loci from the human gut microbiome. ISME J. 6, 1916–1925. doi: 10.1038/ismej.2012.38
Culligan, E. P., Sleator, R. D., Marchesi, J. R., and Hill, C. (2013). Functional environmental screening of a metagenomic library identifies stlA; a unique salt tolerance locus from the human gut microbiome. PLoS ONE 8:e82985. doi:10.1371/journal.pone.0082985
Epstein, W. (2003). The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 75, 293–320. doi.org/10.1016/S0079-6603(03)75008-9
Gillespie, D. E., Brady, S. F., Bettermann, A. D., Cianciotto, N. P., Liles, M. R., Rondon, M. R., et al. (2002). Isolation of antibiotics turbomycin a and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 68, 4301–4306. doi: 10.1128/AEM.68.9.4301-4306.2002
Goryshin, I. Y., and Reznikoff, W. S. (1998). Tn5 in vitro transposition. J. Biol. Chem. 273, 7367–7374.
Haardt, M., Kempf, B., Faatz, E., and Bremer, E. (1995). The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol. Gen. Genet. 246, 783–786.
Hayes, F., Daly, C., and Fitzgerald, G. F. (1990). Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 Plasmid pCI305. Appl. Environ. Microbiol. 56, 202–209.
Hengge-Aronis, R., Klein, W., Lange, R., Rimmele, M., and Boos, W. (1991). Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173, 7918–7924.
Hoffmann, R. F., Mclernon, S., Feeney, A., Hill, C., and Sleator, R. D. (2013). A single point mutation in the listerial betL sigma (A) -dependent promoter leads to improved osmo- and chill-tolerance and a morphological shift at elevated osmolarity. Bioengineered 4, 401–407. doi: 10.4161/bioe.24094
Jones, B. V., Begley, M., Hill, C., Gahan, C. G., and Marchesi, J. R. (2008). Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 105, 13580–13585. doi: 10.1073/pnas.0804437105
Jones, B. V., and Marchesi, J. R. (2007). Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat. Methods 4, 55–61. doi: 10.1038/nmeth964
Kempf, B., and Bremer, E. (1998). Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170, 319–330.
Kunte, H. J. (2006). Osmoregulation in bacteria: compatible solute accumulation and osmosensing. Environ. Chem. 3, 94–99. doi: 10.1071/EN06016
Law, C. J., Maloney, P. C., and Wang, D. N. (2008). Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62, 289–305. doi: 10.1146/annurev.micro.61.080706.093329
Lundblad, V., and Kleckner, N. (1985). Mismatch repair mutations of Escherichia coli K12 enhance transposon excision. Genetics 109, 3–19.
Mavromatis, K., Ivanova, N., Barry, K., Shapiro, H., Goltsman, E., Mchardy, A. C., et al. (2007). Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat. Methods 4, 495–500. doi: 10.1038/nmeth1043
Okada, Y., Okada, N., Makino, S., Asakura, H., Yamamoto, S., and Igimi, S. (2006). The sigma factor RpoN (sigma54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol. Lett. 263, 54–60. doi: 10.1111/j.1574-6968.2006.00405.x
Pao, S. S., Paulsen, I. T., and Saier, M. H. Jr. (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34.
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Saier, M. H. Jr. (2000). Families of transmembrane sugar transport proteins. Mol. Microbiol. 35, 699–710. doi: 10.1046/j.1365-2958.2000.01759.x
Shah, H. (1992). “The genus bacteroides and related taxa,” in The Prokaryotes, eds A. Balows, H. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (New York, NY: Springer), 3593–3607.
Sleator, R. D., Francis, G. A., O'Beirne, D., Gahan, C. G., and Hill, C. (2003). Betaine and carnitine uptake systems in Listeria monocytogenes affect growth and survival in foods and during infection. J. Appl. Microbiol. 95, 839–846. doi: 10.1046/j.1365-2672.2003.02056.x
Sleator, R. D., and Hill, C. (2002). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26, 49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x
Sleator, R. D., Wouters, J., Gahan, C. G., Abee, T., and Hill, C. (2001). Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl. Environ. Microbiol. 67, 2692–2698. doi: 10.1128/AEM.67.6.2692-2698.2001
Typas, A., Stella, S., Johnson, R. C., and Hengge, R. (2007). The -35 sequence location and the Fis-sigma factor interface determine sigmas selectivity of the proP (P2) promoter in Escherichia coli. Mol. Microbiol. 63, 780–796. doi: 10.1111/j.1365-2958.2006.05560.x
Wargo, M. J., and Hogan, D. A. (2009). Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology 155, 2411–2419. doi: 10.1099/mic.0.028787-0
Wemekamp-Kamphuis, H. H., Sleator, R. D., Wouters, J. A., Hill, C., and Abee, T. (2004). Molecular and physiological analysis of the role of osmolyte transporters BetL, Gbu, and OpuC in growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 70, 2912–2918. doi: 10.1128/AEM.70.5.2912-2918.2004
Wu, T. H., and Marinus, M. G. (1994). Dominant negative mutator mutations in the mutS gene of Escherichia coli. J. Bacteriol. 176, 5393–5400.
Xu, J., and Johnson, R. C. (1995). Fis activates the RpoS-dependent stationary-phase expression of proP in Escherichia coli. J. Bacteriol. 177, 5222–5231.
Xu, J., and Johnson, R. C. (1997). Activation of RpoS-dependent proP P2 transcription by the Fis protein in vitro. J. Mol. Biol. 270, 346–359. doi: 10.1006/jmbi.1997.1133
Keywords: metagenomics, functional metagenomics, gut microbiome, microbiota, salt tolerance, BIOLOG, phenotype microarray, transcriptional regulator
Citation: Culligan EP, Marchesi JR, Hill C and Sleator RD (2014) Combined metagenomic and phenomic approaches identify a novel salt tolerance gene from the human gut microbiome. Front. Microbiol. 5:189. doi: 10.3389/fmicb.2014.00189
Received: 26 March 2014; Paper pending published: 05 April 2014;
Accepted: 07 April 2014; Published online: 29 April 2014.
Edited by:
Anton G. Kutikhin, Kemerovo State Medical Academy, RussiaReviewed by:
Anton G. Kutikhin, Kemerovo State Medical Academy, RussiaArseniy E. Yuzhalin, University of Oxford, UK
Copyright © 2014 Culligan, Marchesi, Hill and Sleator. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julian R. Marchesi, School of Biosciences, Cardiff University, Museum Avenue, Cardiff CF10 3AX, UK e-mail:bWFyY2hlc2lqckBjYXJkaWZmLmFjLnVr;
Colin Hill, Alimentary Pharmabiotic Centre, School of Microbiology, University College Cork, Cork, Ireland e-mail:Yy5oaWxsQHVjYy5pZQ==;
Roy D. Sleator, Department of Biological Sciences, Cork Institute of Technology, Rossa Avenue, Bishopstown, Cork, Ireland e-mail:cm95LnNsZWF0b3JAY2l0Lmll
 Eamonn P. Culligan
Eamonn P. Culligan Julian R. Marchesi
Julian R. Marchesi Colin Hill
Colin Hill Roy D. Sleator
Roy D. Sleator