- 1School of Paediatrics and Child Health, The University of Western Australia, Perth, WA, Australia
- 2Centre for Vaccine and Infectious Disease Research, Telethon Kids Institute, The University of Western Australia, Perth, WA, Australia
Non-typeable Haemophilus influenzae (NTHi) and Haemophilus haemolyticus are closely related bacteria that reside in the upper respiratory tract. NTHi is associated with respiratory tract infections that frequently result in antibiotic prescription whilst H. haemolyticus is rarely associated with disease. NTHi and H. haemolyticus can be indistinguishable by traditional culture methods and molecular differentiation has proven difficult. This current review chronologically summarizes the molecular approaches that have been developed for differentiation of NTHi from H. haemolyticus, highlighting the advantages and disadvantages of each target and/or technique. We also provide suggestions for the development of new tools that would be suitable for clinical and research laboratories.
Introduction
Identification and taxonomic classification of Haemophilus species can be challenging (Norskov-Lauritsen, 2014). This is particularly true for Haemophilus haemolyticus, which is often misidentified as non-typeable H. influenzae (NTHi) despite significant differences in pathogenicity. In a landmark study in 2007, Murphy et al. identified NTHi isolates with altered culture phenotypes from patients with chronic obstructive pulmonary disease (COPD) (Murphy et al., 2007). To test the hypothesis that the phenotypically different NTHi isolates were also genetically different, the authors analyzed 490 culture-defined NTHi isolates. Using a combination of genetic and immunological techniques, they found that the variant isolates were actually non-hemolytic H. haemolyticus, a closely related respiratory tract commensal that appears similar to NTHi via culture. The NTHi misidentification rate was significant, with 27% (12/44) of nasopharyngeal isolates and 40% (102/258) of sputum isolates misidentified as NTHi by culture. Analysis of 130 culture-defined laboratory NTHi isolates from middle ear effusion found that none were H. haemolyticus. Further analysis of 58 invasive isolates from the United States national collection identified 4 H. haemolyticus isolates that were previously characterized as a cryptic genospecies. This study reaffirmed H. haemolyticus as a respiratory tract commensal that is rarely cultured from sterile sites and highlighted the issue of NTHi misidentification by culture to the scientific community. Since 2007, retrospective analysis of phenotypic NTHi isolates from other studies have identified similar rates of misidentification (Chang et al., 2010; Kirkham et al., 2010; Hare et al., 2012; Pickering et al., 2014b).
The impact of NTHi misidentification is far-reaching given that NTHi-positive cultures are used to diagnose chronic suppurative otitis media and exacerbations of COPD, prescribe antibiotics, and estimate the efficacy of treatments and preventative strategies for NTHi disease including vaccines. Furthermore, the misidentification of H. haemolyticus as NTHi has potentially impacted on estimations of the proportion of antibiotic resistant NTHi (Witherden et al., 2013).
X (hemin) and/or V (β-nicotinamide adenine dinucleotide) factor requirement is routinely used in diagnostic laboratories to distinguish between Haemophilus species, however NTHi and H. haemolyticus require both X and V factors. The principal phenotypic difference between NTHi and H. haemolyticus is the production of a hemolysin by H. haemolyticus allowing species differentiation on blood agar plates (Kilian, 1976b). However, this difference is often unreliable as H. haemolyticus can lose the defining hemolytic phenotype upon passage (Sandstedt et al., 2008), or the hemolytic phenotype may be absent from the outset. Earlier, it was suggested that H. haemolyticus is a hemolytic variant of H. influenzae (Broom and Sneath, 1981). However, modern phylogenetic studies have identified clear species differences (McCrea et al., 2008; Norskov-Lauritsen, 2011). It is now widely accepted that culture alone cannot reliably distinguish NTHi from H. haemolyticus.
An ideal molecular tool for NTHi and H. haemolyticus differentiation is one that is rapid, robust, inexpensive, requiring standard laboratory equipment, and limited technical expertise. A superior tool would be one that unambiguously determines whether an isolate is NTHi or H. haemolyticus in a single reaction to reduce the time and cost for identification. However, the development of such tools for NTHi and H. haemolyticus differentiation has been difficult due to extensive genetic similarities between these species. Over the last decade, considerable research effort has focused on identifying molecular targets and suitable methodologies to differentiate NTHi from H. haemolyticus. A chronological review of each potential target and discussion of the advantages and disadvantages of the methodologies is given below and summarized in Table 1.
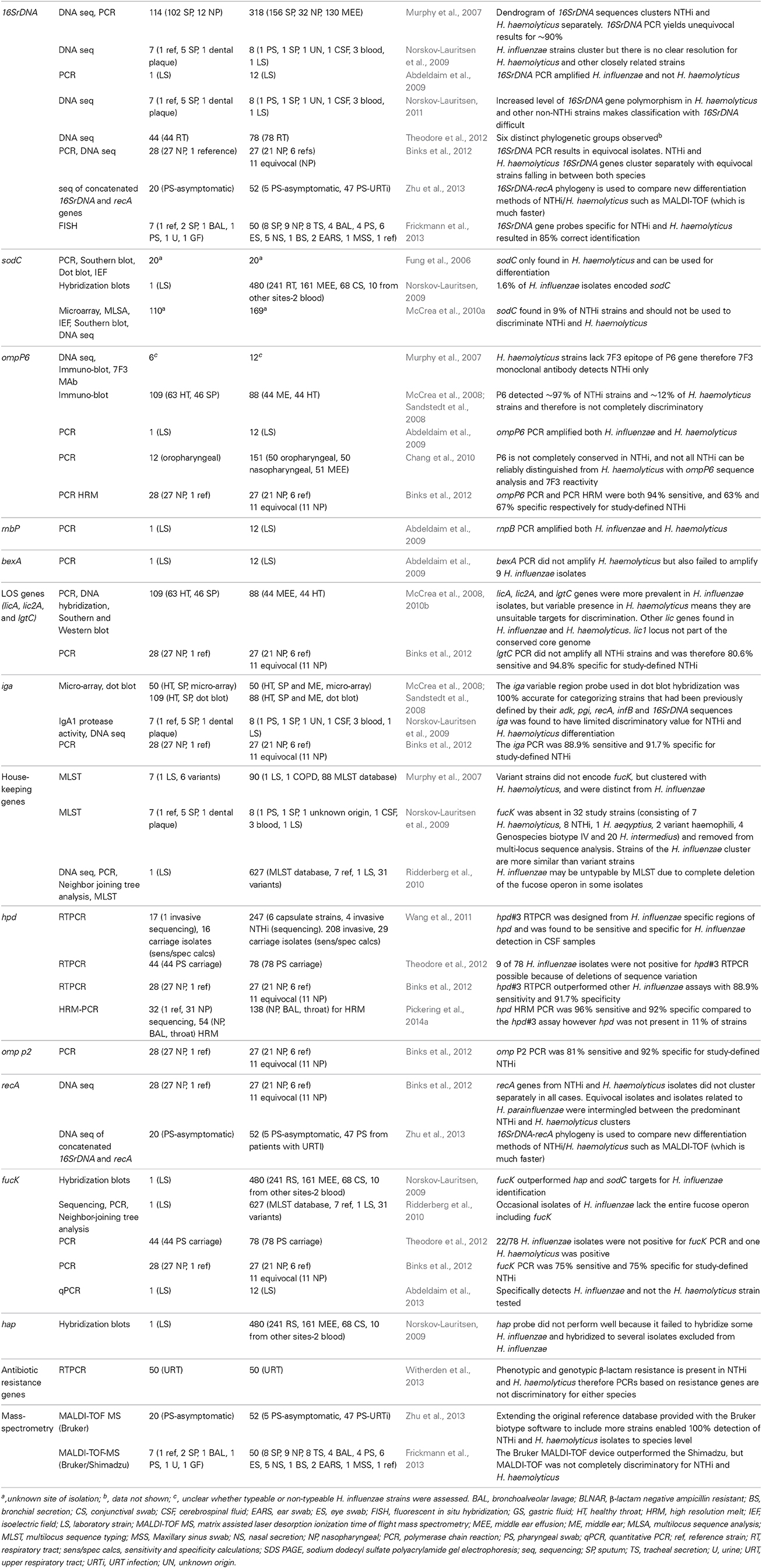
Table 1. Summary of gene targets and methodologies used to discriminate H. influenzae from H. haemolyticus.
Genetic Targets Investigated for Discrimination of NTHi from H. haemolyticus
The original discriminatory method used to distinguish NTHi from H. haemolyticus was a combination of 16SrDNA PCR, a monoclonal antibody targeting an epitope of the outer membrane protein (OMP) P6 of NTHi known as 7F3, and multilocus sequence analysis (MLSA) (Murphy et al., 2007). 16SrDNA PCR permitted easy identification of NTHi and H. haemolyticus, but only for 90% of strains. Recognition of the limitation of 16SrDNA PCR as a discriminatory tool for NTHi and H. haemolyticus is now widely accepted (Norskov-Lauritsen, 2011; Binks et al., 2012). In 2011, Norksov-Lauritsen further investigated the apparent low resolution of classification schemes based on 16SrDNA (Norskov-Lauritsen, 2011). 16SrDNA genes are historically recognized as being universally distributed and therefore appropriate targets for assessing lineages. However, further investigation into the NTHi/H. haemolyticus species border found high numbers of polymorphic nucleotide positions due to intragenomic 16SrDNA gene heterogeneity in isolates that were not NTHi. The increased level of 16SrDNA gene polymorphism in commensal taxa (not including pathogenic H. influenzae) could not be explained but did provide a reason for the difficulties of Haemophilus speciation using 16SrDNA gene-based classification. The 7F3 monoclonal antibody was found to be NTHi-specific and had the best differentiation capability, however its limited availability meant that widespread use of this method was unfeasible. Moreover, due to the cost and time, immunoblotting is not ideal for species identification in clinical diagnostic settings and subsequent studies have found that the 7F3 antibody does not identify all NTHi strains (McCrea et al., 2008). Multilocus sequence typing (MLST) is a standardized sequence-based profiling system that has been used to investigate NTHi diversity (Kaur et al., 2011; Schumacher et al., 2012; Puig et al., 2013) but, as discussed later, is not suitable for discrimination of NTHi from H. haemolyticus. MLSA is an extension of MLST that involves application of mathematical algorithms to assemble consensus trees (Tateno et al., 1994). In the Murphy study, MLSA identified that H. haemolyticus strains clustered separately to NTHi strains. Although MLSA is useful for understanding species boundaries, it requires a high level of technical expertise and is time consuming and therefore not ideal for routine diagnostics.
Another molecular target with the potential ability to completely differentiate NTHi from H. haemolyticus was simultaneously described by Fung et al. (2006). The sodC gene, which encodes the copper- and zinc-containing superoxide dismutase CuZnSOD, was found to be present in 20 H. haemolyticus isolates and absent in 20 NTHi isolates. Initial PCR results were confirmed by Southern and Western blotting. However, subsequent application of the sodC PCR to a larger collection of isolates in 2010 (110 H. haemolyticus and 169 NTHi) revealed that 9% of NTHi also possessed the sodC gene (McCrea et al., 2010a), demonstrating that the sodC gene was not a suitable target for complete discrimination of NTHi from H. haemolyticus.
In 2008, McCrea et al. thoroughly investigated the relationship of NTHi to hemolytic and non-hemolytic H. haemolyticus strains (McCrea et al., 2008). Taxonomic traits, MLSA and the presence of NTHi virulence-associated genes encoding lipooligosaccharide (licA, lic2A, lgtC), and IgA protease were compared. Eighty-eight capsulated and non-typeable H. influenzae (breakdown not given), and 109 culture-defined H. haemolyticus isolates were examined. The 109 H. haemolyticus isolates were not bound by iga hybridization probes, and this was the only target that differentiated all H. influenzae and H. haemolyticus isolates in the study. Whilst taxonomic traits such as H2S and indole production, urease and ornithine decarboxylase activity and hemolysis (Kilian, 1976b,a) were found to correlate with species identification, no trait completely differentiated NTHi from H. haemolyticus (McCrea et al., 2008). A main finding was that although hemolytic and non-hemolytic H. haemolyticus strains did not cluster as two separate subspecies, some NTHi genes (licA) and traits (urease activity) were more common in hemolytic strains compared with non-hemolytic strains. The authors remarked that no rapid, clinically useful marker was available to differentiate NTHi and H. haemolyticus, however X and V factor testing was sufficient for distinguishing H. influenzae from other haemophili that infect normally sterile sites and cause serious disease. The authors proposed that precise taxonomic division of these species is elusive, particularly with the high potential for genetic recombination between NTHi and H. haemolyticus that has since been demonstrated in vitro (Sondergaard et al., 2014). At this stage H. haemolyticus had not been associated with disease: there were two rare cases of H. haemolyticus causing endocarditis in 1923 (De Santo and White, 1933) and 1933 (Miller and Branch, 1923). The recent retrospective molecular analysis of NTHi culture-defined isolates has revealed additional cases in which H. haemolyticus was the apparent cause of bacteremia and septic arthritis (Anderson et al., 2012; Morton et al., 2012). Although H. haemolyticus infection is still rare, these cases reiterate the need for specific identification tools.
In 2008, Sandstedt et al. compared the generation of minimum evolution trees from concatenated sequences of 5 housekeeping genes (adk, pgi, recA, infB, and 16SrDNA), which had previously been shown to be the best method for distinguishing NTHi from H. haemolyticus (Norskov-Lauritsen et al., 2005; McCrea et al., 2008), with rapid and more cost-effective methods (Sandstedt et al., 2008). The three methods evaluated were DNA hybridization-based microarrays (targeting conserved and variable iga regions), genomic dot blot hybridization (also targeting conserved and variable iga regions), and dot blot immunoassays for OMP P6 with monoclonal antibody 7F3. Genomic dot blots targeting the iga variable region correlated most closely with the minimum evolution trees, whereas microarray detection of the variable iga region was favored for being high-throughput. Methods utilizing the conserved portion of the iga gene did not discriminate NTHi from H. haemolyticus. The authors recognized that the adoption of phylogenetic or molecular methods for NTHi/H. haemolyticus differentiation is dependent on the number of strains being analyzed and the purpose of doing so, which varies from laboratory to laboratory.
In 2009, the sodC, fucK (encodes fuculokinase), and hap (haemophilus adhesion protein) genes were investigated for their combined suitability to selectively identify NTHi (Norskov-Lauritsen, 2009). H. influenzae isolates (typeable and non-typeable) were expected to be sodC−, fucK+, and hap+. The fucK PCR gave the best discrimination between the 480 isolates investigated. It was suggested that phenotypic H. influenzae isolates lacking fucK were not H. influenzae and development of a fucK- based molecular discriminatory tool was proposed. Soon after this publication, the same group published a more detailed investigation into the delineation of H. influenzae by phenotype, multilocus sequence phylogeny and detection of marker genes (16SrDNA, hap, fucK, sodC, and virulence-associated genes hia, hmw1A, hmwC, hif, iga, lic2B) (Norskov-Lauritsen et al., 2009). In this study, the species borders for H. influenzae with (1) H. haemolyticus, (2) cryptic genospecies biotype IV, and (3) the then un-validated species “H. intermedius” were interrogated with MLSA for 6 of the 7 MLST housekeeping genes: adk, atpG, frdB, mdh, pgi, and recA (fucK was excluded from the MLST due to its absence in 42 strains). Individually, 16SrDNA, hap, fucK, and sodC genes correlated with the concatenated multilocus sequence phylogeny, but iga was found to have limited discriminatory value for NTHi and H. haemolyticus differentiation. This contrasted with previous studies detailing the discriminatory power of iga for H. influenzae detection (McCrea et al., 2008; Sandstedt et al., 2008). The virulence associated markers hia, hmw1a, hmwC, and hif were variably expressed in H. influenzae and therefore not discriminatory. Multilocus sequence phylogeny of H. haemolyticus strains produced separate lineages that also included H. intermedius and the cryptic genospecies biotype IV. This finding emphasized the difficulty of defining taxonomic boundaries within Haemophili. The authors observed that sequence analysis did not align with taxonomy, and suggested that different strains of H. haemolyticus may not share a common ancestor.
By 2010, it became apparent that not all strains of H. influenzae encode the fucK house-keeping gene, with some strains missing the entire fucose operon (Ridderberg et al., 2010). Therefore, the standardized MLST assay for H. influenzae is not suitable for all strains. In the same year, another study reported on variations in the OMP P6 (omp P6) gene of NTHi that obscures NTHi and H. haemolyticus differentiation (Chang et al., 2010). At this stage, none of the previously characterized targets remained attractive candidates for accurate identification of H. influenzae or H. haemolyticus.
Real-time (RT) PCR assays targeting 16SrDNA (Abdeldaim et al., 2009), omp P6 (Nelson et al., 1991; Abdeldaim et al., 2009), bexA (Corless et al., 2001), rnpB (Abdeldaim et al., 2009), and fucK (Abdeldaim et al., 2013) genes were developed for diagnostic detection of H. influenzae, however each was limited by poor specificity and/or sensitivity. For example, the rnpB RTPCR amplifies both NTHi and H. haemolyticus (Abdeldaim et al., 2009), whereas the bexA PCR does not amplify H. haemolyticus, but also failed to amplify all H. influenzae strains (Corless et al., 2001). In 2011, a quantitative RTPCR (hpd#3) based on the protein D gene (hpd) was developed that was sensitive and appeared to be specific for H. influenzae identification (Wang et al., 2011). Sixteen H. haemolyticus isolates were tested with the hpd#3 RTPCR and none were positive. A major advantage of the hpd#3 RTPCR was that it could be used directly on clinical samples, reducing cost and preparation time for H. influenzae identification. In 2012, the hpd#3 RTPCR assay was further investigated in a collection of 60 culture-defined NTHi from the nasopharynx of children with and without recurrent acute otitis media (Binks et al., 2012). 16SrDNA PCR had previously identified that only 37% (22/60) of the isolates were true NTHi, 27 were H. haemolyticus and the remaining 18% (11/60) could neither be defined as NTHi nor H. haemolyticus and were termed equivocal (Kirkham et al., 2010). Sequencing and concatenation of 16SrDNA and recA genes in this collection of isolates provided insight into the previously ambiguous equivocal isolates. Whilst 16SrDNA PCR-defined H. haemolyticus and NTHi strains were clearly separated from one another on the phylogenetic tree, the equivocal strains sat in the middle and were considered to either be divergent H. haemolyticus strains becoming NTHi or vice versa. An evolutionary continuum between the two species was suggested. This collection of isolates was considered to be ideal to test the limitations of existing discriminatory assays in their ability to identify the NTHi and H. haemolyticus. Seven of the most promising PCR targets (hpd, omp P2, omp P6, lgtC, 16SrDNA, fucK, and iga) were assessed (Binks et al., 2012). The study conceded that NTHi and H. haemolyticus could not be completely differentiated with any single gene target, however the hpd#3 RTPCR was superior for differentiating closely related strains. A subsequent study suggested conducting 3 tests: hpd#3 RTPCR, fucK PCR and then 16SrDNA sequencing for NTHi and H. haemolyticus differentiation (Theodore et al., 2012). However, the identification of strains lacking fucK remains an issue for its broad application, and conducting 2 PCRs followed by sequencing increases the cost and time for identification. Such lengthy and expensive tests are not ideal for clinical diagnostics or large-scale surveillance studies.
Recently, we developed a HRM (high resolution melt)-PCR to further investigate the potential use of the hpd gene to detect and differentiate NTHi and H. haemolyticus (Pickering et al., 2014a). The advantage of the PCR-HRM is low cost, speed and that only one reaction is required for differentiation of the two species. However, application of this assay to 180 clinical isolates revealed that even hpd, a host colonization-associated gene (Johnson et al., 2011) that was previously reported to be highly conserved (Song et al., 1995), was not present in 11% (19/180) of the isolates tested. Absence or variability of the hpd gene in NTHi has since been confirmed (Zhu et al., 2013; Smith-Vaughan et al., 2014) and suggests that the hpd gene is less conserved than originally thought. The hpd HRM-PCR is limited like all other single-target tools tested to date. Summarizing all H. influenzae/H. haemolyticus differentiation studies, it appears that single gene target approaches for discrimination are not ideal and that rapid tests incorporating multiple targets are required.
Proteomic and Whole Genome Approaches to Discrimination of NTHi and H. haemolyticus
Matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) analysis is revolutionizing the diagnostic laboratory. MALDI-TOF compares the spectral profiles of bacterial colonies to a database of species with known spectral profiles. The first assessment of the utility of MALDI-TOF to differentiate H. influenzae from H. haemolyticus reported 100% differentiation of 52 NTHi and 20 H. haemolyticus strains with the adaptation of a new reference database on the Bruker MALDI-TOF platform (Zhu et al., 2013). A second study compared two MALDI-TOF platforms (Shimadzu and Bruker) with fluorescent in situ hybridization (FISH) to identify 50 H. influenzae, 25 H. parainfluenzae, 7 H. haemolyticus, and 2 H. parahaemolyticus isolates (Frickmann et al., 2013). FISH failed to identify strains from all Haemophilus species tested including a high proportion (14%) of H. influenzae isolates. Neither MALDI-TOF platform correctly identified any H. haemolyticus isolates but addition of an H. haemolyticus reference spectrum to the Bruker database resulted in identification of all seven H. haemolyticus isolates. This demonstrates that the discriminatory power of MALDI-TOF is highly dependent on the comprehensiveness of species databases, which varies between laboratories. Improvement and global standardization of reference databases for H. influenzae and H. haemolyticus may permit the use of MALDI-TOF for high-throughput speciation of H. influenzae and H. haemolyticus in diagnostic laboratories in the near future.
An alternative approach to identifying and developing tools for multiple discriminatory targets of H. influenzae and H. haemolyticus is the use of whole genome sequencing and comparative genomics. There are several large-scale Haemophilus whole genome sequencing projects underway that will assist in development of such methods. Recently, comparison of 97 NTHi genomes revealed an NTHi population structure of 6 distinct clades. This high-resolution study is the first to identify a clonal-based evolution of NTHi (De Chiara et al., 2014). H. haemolyticus strains were not included in this study.
In summary, although the need for molecular identification is acknowledged, no single target or current methodology has been identified that can accurately identify all H. influenzae or H. haemolyticus strains. This is further complicated by the genetic relatedness of these species and the demonstration that inter-species horizontal gene transfer occurs. When new discriminatory tests are developed they must be validated on a large and diverse collection of strains. Future large-scale comparative genomic studies that compare H. influenzae core and accessory genes with H. haemolyticus have the potential to reveal new discriminatory targets and provide greater definition of species borders. This in turn will improve the accuracy of H. influenzae and H. haemolyticus identification for improved disease diagnosis and surveillance.
Author Contributions
Janessa Pickering prepared the manuscript, Peter C. Richmond and Lea-Ann S. Kirkham critically reviewed the manuscript. The authors do not have any competing interests.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Janessa Pickering is a recipient of an National Health and Medical Research Council (NHMRC) postgraduate award and Princess Margaret Hospital Foundation scholarship. Lea-Ann S. Kirkham is recipient of an NHMRC career development fellowship #1061428.
References
Abdeldaim, G. M., Stralin, K., Kirsebom, L. A., Olcen, P., Blomberg, J., and Herrmann, B. (2009). Detection of Haemophilus influenzae in respiratory secretions from pneumonia patients by quantitative real-time polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 64, 366–373. doi: 10.1016/j.diagmicrobio.2009.03.030
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Abdeldaim, G. M., Stralin, K., Olcen, P., Blomberg, J., Molling, P., and Herrmann, B. (2013). Quantitative fucK gene polymerase chain reaction on sputum and nasopharyngeal secretions to detect Haemophilus influenzae pneumonia. Diagn. Microbiol. Infect. Dis. 76, 141–146. doi: 10.1016/j.diagmicrobio.2013.02.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anderson, R., Wang, X., Briere, E. C., Katz, L. S., Cohn, A. C., Clark, T. A., et al. (2012). Haemophilus haemolyticus isolates causing clinical disease. J. Clin. Microbiol. 50, 2462–2465. doi: 10.1128/JCM.06575-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Binks, M. J., Temple, B., Kirkham, L. A., Wiertsema, S. P., Dunne, E. M., Richmond, P. C., et al. (2012). Molecular surveillance of true nontypeable Haemophilus influenzae: an evaluation of PCR screening assays. PLoS ONE 7:e34083. doi: 10.1371/journal.pone.0034083
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Broom, A., and Sneath, P. H. (1981). Numerical taxonomy of Haemophilus. J. Gen. Microbiol. 126, 123–149.
Chang, A., Adlowitz, D. G., Yellamatty, E., and Pichichero, M. (2010). Haemophilus influenzae outer membrane protein P6 molecular characterization may not differentiate all strains of H. Influenzae from H. haemolyticus. J. Clin. Microbiol. 48, 3756–3757. doi: 10.1128/JCM.01255-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Corless, C. E., Guiver, M., Borrow, R., Edwards-Jones, V., Fox, A. J., and Kaczmarski, E. B. (2001). Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39, 1553–1558. doi: 10.1128/JCM.39.4.1553-1558.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Chiara, M., Hood, D., Muzzi, A., Pickard, D. J., Perkins, T., Pizza, M., et al. (2014). Genome sequencing of disease and carriage isolates of nontypeable Haemophilus influenzae identifies discrete population structure. Proc. Natl. Acad. Sci. U.S.A. 111, 5439–5444. doi: 10.1073/pnas.1403353111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Santo, D. A., and White, M. (1933). Hemophilus hemolyticus endocarditis. Am. J. Pathol. 9, 381–392.
Frickmann, H., Christner, M., Donat, M., Berger, A., Essig, A., Podbielski, A., et al. (2013). Rapid discrimination of Haemophilus influenzae, H. parainfluenzae, and H. haemolyticus by fluorescence in situ hybridization (FISH) and two matrix-assisted laser-desorption-ionization time-of-flight mass spectrometry (MALDI-TOF-MS) platforms. PLoS ONE 8:e63222. doi: 10.1371/journal.pone.0063222
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fung, W. W., O'dwyer, C. A., Sinha, S., Brauer, A. L., Murphy, T. F., Kroll, J. S., et al. (2006). Presence of copper- and zinc-containing superoxide dismutase in commensal Haemophilus haemolyticus isolates can be used as a marker to discriminate them from nontypeable H. influenzae isolates. J. Clin. Microbiol. 44, 4222–4226. doi: 10.1128/JCM.01376-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hare, K. M., Binks, M. J., Grimwood, K., Chang, A. B., Leach, A. J., and Smith-Vaughan, H. (2012). Culture and PCR detection of Haemophilus influenzae and Haemophilus haemolyticus in Australian Indigenous children with bronchiectasis. J. Clin. Microbiol. 50, 2444–2445. doi: 10.1128/JCM.00566-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Johnson, R. W., McGillivary, G., Denoel, P., Poolman, J., and Bakaletz, L. O. (2011). Abrogation of nontypeable Haemophilus influenzae protein D function reduces phosphorylcholine decoration, adherence to airway epithelial cells, and fitness in a chinchilla model of otitis media. Vaccine 29, 1211–1221. doi: 10.1016/j.vaccine.2010.12.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaur, R., Chang, A., Xu, Q., Casey, J. R., and Pichichero, M. E. (2011). Phylogenetic relatedness and diversity of non-typable Haemophilus influenzae in the nasopharynx and middle ear fluid of children with acute otitis media. J. Med. Microbiol. 60, 1841–1848. doi: 10.1099/jmm.0.034041-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kilian, M. (1976a). The haemolytic activity of Haemophilus species. Acta Pathol. Microbiol. Scand. B 84B, 339–341.
Kilian, M. (1976b). A taxonomic study of the genus Haemophilus, with the proposal of a new species. J. Gen. Microbiol. 93, 9–62. doi: 10.1099/00221287-93-1-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kirkham, L. A., Wiertsema, S. P., Mowe, E. N., Bowman, J. M., Riley, T. V., and Richmond, P. C. (2010). Nasopharyngeal carriage of Haemophilus haemolyticus in otitis-prone and healthy children. J. Clin. Microbiol. 48, 2557–2559. doi: 10.1128/JCM.00069-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCrea, K. W., Wang, M. L., Xie, J., Sandstedt, S. A., Davis, G. S., Lee, J. H., et al. (2010a). Prevalence of the sodC gene in nontypeable Haemophilus influenzae and Haemophilus haemolyticus by microarray-based hybridization. J. Clin. Microbiol. 48, 714–719. doi: 10.1128/JCM.01416-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCrea, K. W., Xie, J., Lacross, N., Patel, M., Mukundan, D., Murphy, T. F., et al. (2008). Relationships of nontypeable Haemophilus influenzae strains to hemolytic and nonhemolytic Haemophilus haemolyticus strains. J. Clin. Microbiol. 46, 406–416. doi: 10.1128/JCM.01832-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCrea, K. W., Xie, J., Marrs, C. F., and Gilsdorf, J. R. (2010b). Prevalence of genetic differences in phosphorylcholine expression between nontypeable Haemophilus influenzae and Haemophilus haemolyticus. BMC Microbiol. 10:286. doi: 10.1186/1471-2180-10-286
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, C., and Branch, A. (1923). Subacute bacterial endocarditis due to a hemolytic hemophilic bacillus. Arch. Intern. Med. 32, 911–923. doi: 10.1001/archinte.1923.00110240104006
Morton, D. J., Hempel, R. J., Whitby, P. W., Seale, T. W., and Stull, T. L. (2012). An invasive Haemophilus haemolyticus isolate. J. Clin. Microbiol. 50, 1502–1503. doi: 10.1128/JCM.06688-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Murphy, T. F., Brauer, A. L., Sethi, S., Kilian, M., Cai, X., and Lesse, A. J. (2007). Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 195, 81–89. doi: 10.1086/509824
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nelson, M. B., Munson, R. S. Jr., Apicella, M. A., Sikkema, D. J., Molleston, J. P., and Murphy, T. F. (1991). Molecular conservation of the P6 outer membrane protein among strains of Haemophilus influenzae: analysis of antigenic determinants, gene sequences, and restriction fragment length polymorphisms. Infect. Immun. 59, 2658–2663.
Norskov-Lauritsen, N. (2009). Detection of cryptic genospecies misidentified as Haemophilus influenzae in routine clinical samples by assessment of marker genes fucK, hap, and sodC. J. Clin. Microbiol. 47, 2590–2592. doi: 10.1128/JCM.00013-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Norskov-Lauritsen, N. (2011). Increased level of intragenomic 16S rRNA gene heterogeneity in commensal strains closely related to Haemophilus influenzae. Microbiology 157, 1050–1055. doi: 10.1099/mic.0.047233-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Norskov-Lauritsen, N. (2014). Classification, identification, and clinical significance of haemophilus and aggregatibacter species with host specificity for humans. Clin. Microbiol. Rev. 27, 214–240. doi: 10.1128/CMR.00103-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Norskov-Lauritsen, N., Bruun, B., and Kilian, M. (2005). Multilocus sequence phylogenetic study of the genus Haemophilus with description of Haemophilus pittmaniae sp. nov. Int. J. Syst. Evol. Microbiol. 55, 449–456. doi: 10.1099/ijs.0.63325-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Norskov-Lauritsen, N., Overballe, M. D., and Kilian, M. (2009). Delineation of the species Haemophilus influenzae by phenotype, multilocus sequence phylogeny, and detection of marker genes. J. Bacteriol. 191, 822–831. doi: 10.1128/JB.00782-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pickering, J., Binks, M. J., Beissbarth, J., Hare, K. M., Kirkham, L. A., and Smith-Vaughan, H. (2014a). A PCR-high-resolution melt assay for rapid differentiation of nontypeable Haemophilus influenzae and Haemophilus haemolyticus. J. Clin. Microbiol. 52, 663–667. doi: 10.1128/JCM.02191-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pickering, J., Smith-Vaughan, H., Beissbarth, J., Bowman, J. M., Wiertsema, S., Riley, T. V., et al. (2014b). Diversity of nontypeable Haemophilus influenzae strains colonizing Australian Aboriginal and non-Aboriginal children. J. Clin. Microbiol. 52, 1352–1357. doi: 10.1128/JCM.03448-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Puig, C., Calatayud, L., Marti, S., Tubau, F., Garcia-Vidal, C., Carratala, J., et al. (2013). Molecular epidemiology of nontypeable Haemophilus influenzae causing community-acquired pneumonia in adults. PLoS ONE 8:e82515. doi: 10.1371/journal.pone.0082515
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ridderberg, W., Fenger, M. G., and Norskov-Lauritsen, N. (2010). Haemophilus influenzae may be untypable by the multilocus sequence typing scheme due to a complete deletion of the fucose operon. J. Med. Microbiol. 59, 740–742. doi: 10.1099/jmm.0.018424-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sandstedt, S. A., Zhang, L., Patel, M., McCrea, K. W., Qin, Z., Marrs, C. F., et al. (2008). Comparison of laboratory-based and phylogenetic methods to distinguish between Haemophilus influenzae and H. haemolyticus. J. Microbiol. Methods 75, 369–371. doi: 10.1016/j.mimet.2008.06.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schumacher, S. K., Marchant, C. D., Loughlin, A. M., Bouchet, V., Stevenson, A., and Pelton, S. I. (2012). Prevalence and genetic diversity of nontypeable Haemophilus influenzae in the respiratory tract of infants and primary caregivers. Pediatr. Infect. Dis. J. 31, 145–149. doi: 10.1097/INF.0b013e31823aaeb3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith-Vaughan, H. C., Chang, A. B., Sarovich, D. S., Marsh, R. L., Grimwood, K., Leach, A. J., et al. (2014). Absence of an important vaccine and diagnostic target in carriage- and disease-related nontypeable Haemophilus influenzae. Clin. Vaccine Immunol. 21, 250–252. doi: 10.1128/CVI.00632-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sondergaard, A., Witherden, E. A., Norskov-Lauritsen, L., and Tristram, S. (2014). “Horizontal transfer of chromosomally-encoded resistance to beta-lactam antibiotics between Haemophilus influenzae and Haemophilus haemolyticus,” in International Pasteurellaceae Conference 2014 (Prato).
Song, X. M., Forsgren, A., and Janson, H. (1995). The gene encoding protein D (hpd) is highly conserved among Haemophilus influenzae type b and nontypeable strains. Infect. Immun. 63, 696–699.
Tateno, Y., Takezaki, N., and Nei, M. (1994). Relative efficiencies of the maximum-likelihood, neighbor-joining, and maximum-parsimony methods when substitution rate varies with site. Mol. Biol. Evol. 11, 261–277.
Theodore, M. J., Anderson, R. D., Wang, X., Katz, L. S., Vuong, J. T., Bell, M. E., et al. (2012). Evaluation of new biomarker genes for differentiating Haemophilus influenzae from Haemophilus haemolyticus. J. Clin. Microbiol. 50, 1422–1424. doi: 10.1128/JCM.06702-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X., Mair, R., Hatcher, C., Theodore, M. J., Edmond, K., Wu, H. M., et al. (2011). Detection of bacterial pathogens in Mongolia meningitis surveillance with a new real-time PCR assay to detect Haemophilus influenzae. Int. J. Med. Microbiol. 301, 303–309. doi: 10.1016/j.ijmm.2010.11.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Witherden, E. A., Kunde, D., and Tristram, S. G. (2013). PCR screening for the N526K substitution in isolates of Haemophilus influenzae and Haemophilus haemolyticus. J. Antimicrob. Chemother. 68, 2255–2258. doi: 10.1093/jac/dkt189
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhu, B., Xiao, D., Zhang, H., Zhang, Y., Gao, Y., Xu, L., et al. (2013). MALDI-TOF MS distinctly differentiates nontypable Haemophilus influenzae from Haemophilus haemolyticus. PLoS ONE 8:e56139. doi: 10.1371/journal.pone.0056139
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Haemophilus haemolyticus, NTHi, identification, culture, molecular differentiation
Citation: Pickering J, Richmond PC and Kirkham L-AS (2014) Molecular tools for differentiation of non-typeable Haemophilus influenzae from Haemophilus haemolyticus. Front. Microbiol. 5:664. doi: 10.3389/fmicb.2014.00664
Received: 13 October 2014; Paper pending published: 08 November 2014;
Accepted: 15 November 2014; Published online: 02 December 2014.
Edited by:
Christina Maria Joseph Elisabeth Vandenbroucke-Grauls, VU University Medical Center, NetherlandsCopyright © 2014 Pickering, Richmond and Kirkham. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lea-Ann S. Kirkham, School of Paediatrics and Child Health, The University of Western Australia, Administration Building, Princess Margaret Hospital, Roberts Road, Perth, WA 6008, Australia e-mail:bGVhLWFubi5raXJraGFtQHV3YS5lZHUuYXU=
 Janessa Pickering
Janessa Pickering Peter C. Richmond
Peter C. Richmond Lea-Ann S. Kirkham
Lea-Ann S. Kirkham