- 1Environmental Engineering Laboratory, Department of Civil Engineering, Yeungnam University, Gyeongsan, South Korea
- 2Disasters Prevention Research Institute, Yeungnam University, Gyeongsan, South Korea
- 3Department of Microbiology, Bose Institute, Kolkata, India
Polycyclic aromatic hydrocarbons (PAHs) include a group of organic priority pollutants of critical environmental and public health concern due to their toxic, genotoxic, mutagenic and/or carcinogenic properties and their ubiquitous occurrence as well as recalcitrance. The increased awareness of their various adverse effects on ecosystem and human health has led to a dramatic increase in research aimed toward removing PAHs from the environment. PAHs may undergo adsorption, volatilization, photolysis, and chemical oxidation, although transformation by microorganisms is the major neutralization process of PAH-contaminated sites in an ecologically accepted manner. Microbial degradation of PAHs depends on various environmental conditions, such as nutrients, number and kind of the microorganisms, nature as well as chemical property of the PAH being degraded. A wide variety of bacterial, fungal and algal species have the potential to degrade/transform PAHs, among which bacteria and fungi mediated degradation has been studied most extensively. In last few decades microbial community analysis, biochemical pathway for PAHs degradation, gene organization, enzyme system, genetic regulation for PAH degradation have been explored in great detail. Although, xenobiotic-degrading microorganisms have incredible potential to restore contaminated environments inexpensively yet effectively, but new advancements are required to make such microbes effective and more powerful in removing those compounds, which were once thought to be recalcitrant. Recent analytical chemistry and genetic engineering tools might help to improve the efficiency of degradation of PAHs by microorganisms, and minimize uncertainties of successful bioremediation. However, appropriate implementation of the potential of naturally occurring microorganisms for field bioremediation could be considerably enhanced by optimizing certain factors such as bioavailability, adsorption and mass transfer of PAHs. The main purpose of this review is to provide an overview of current knowledge of bacteria, halophilic archaea, fungi and algae mediated degradation/transformation of PAHs. In addition, factors affecting PAHs degradation in the environment, recent advancement in genetic, genomic, proteomic and metabolomic techniques are also highlighted with an aim to facilitate the development of a new insight into the bioremediation of PAH in the environment.
Introduction
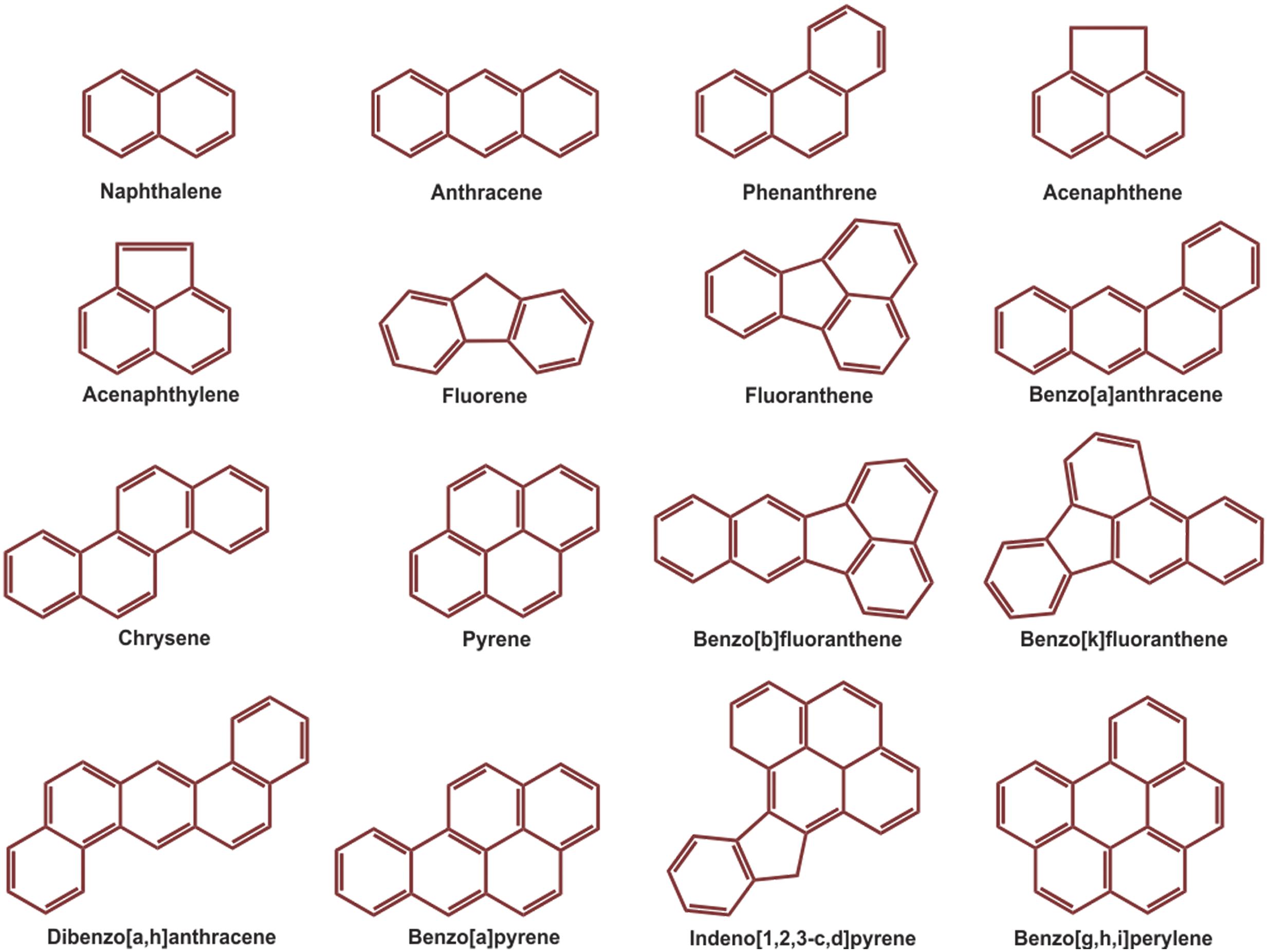
Over the last few decades, with an increasing global awareness about the potential adverse effects of pollutants on public health and environment, remediation and renovation of environment contaminated with hazardous materials have received increasing attention. Among others, polycyclic aromatic hydrocarbons (PAHs) include a group of priority organic pollutants of significant concern due to their toxic, genotoxic, mutagenic and/or carcinogenic properties (WHO, 1983; Cerniglia, 1992; Mastrangelo et al., 1996; Schützendübel et al., 1999). PAHs are composed of fused aromatic rings in linear, angular, or cluster arrangements. Generally, the electrochemical stability, persistency, resistance toward biodegradation and carcinogenic index of PAHs increase with an increase in the number of aromatic rings, structural angularity, and hydrophobicity, while volatility tends to decrease with increasing molecular weight (Mackay and Callcott, 1998; Marston et al., 2001). The PAHs have a natural potential for bioaccumulation in various food chains, which make their presence in the environment quite alarming (Morehead et al., 1986; Xue and Warshawsky, 2005), and are therefore being considered as substances of potential human health hazards (Mastrangelo et al., 1996; Binkova et al., 2000; Marston et al., 2001; Xue and Warshawsky, 2005). On the basis of abundance and toxicity, 16 PAHs are already enlisted as priority environmental pollutants by the United States Environmental Protection Agency (US EPA), (Agency for Toxic Substances and Disease Registry [ATSDR], 1990; Liu et al., 2001) which are depicted in Figure 1. Various physical-chemical properties and some relevant information of 16 PAHs enlisted as priority pollutants by US EPA are depicted in Table 1.
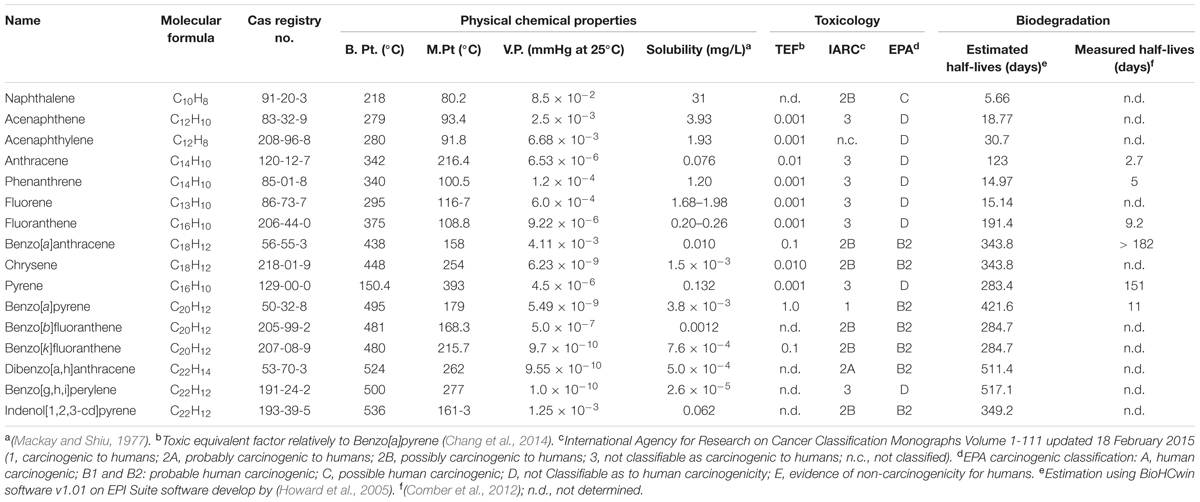
TABLE 1. Physical-chemical properties and some relevant information of 16 PAHs enlisted as priority pollutants by US EPA.
In their pure chemical form, PAHs generally exist as colorless, white, or pale yellow-green solids having a faint, pleasant odor. They are basically non-polar organic compounds, characteristically composed of carbon and hydrogen atoms. Mostly incomplete combustion of organic materials like coal, tar, oil and gas, automobile exhaust, tobacco or smoked food, either during industrial and other human activities or during geothermal reactions associated with the production of fossil-fuels and minerals, result in PAH formation. In nature, they are formed during forest fires, volcanic eruptions, or by plant and bacterial reactions (Blumer, 1976; Wilson and Jones, 1993). Diverse types of combustion yield different distributions of PAHs in both relative amounts of individual PAHs as well as their isomers. In nature, they are formed during forest fires, volcanic eruptions, or by plant and bacterial reactions (Blumer, 1976; Wilson and Jones, 1993). Nevertheless, the anthropogenic input of PAHs to the environment far exceeds the natural sources (National Academy of Sceinces [NAS], 1971).
Polycyclic aromatic hydrocarbons are formed whenever organic substances are exposed to high temperatures (pyrolysis), and the composition of the products thus formed depends largely on the nature of the starting material as well as the transformation temperature (Blumer, 1976; Cerniglia, 1992). Fossil fuels also contain huge amounts of PAHs which are released into the environment during incomplete combustion or by accidental discharge during transport, use, or disposal of petroleum products or as a result of uncontrolled emissions (Cerniglia, 1992; Wilson and Jones, 1993; Johansson and van Bavel, 2003). PAHs are widely present as contaminants in air, soil, aquatic environments, sediments, surface water as well as in ground water (Huntley et al., 1993; Van Brummelen et al., 1996; Boxall and Maltby, 1997; Holman et al., 1999; Lim et al., 1999; Ohkouchi et al., 1999). Natural and anthropogenic sources of PAHs, in combination with global transport phenomena, result in their worldwide distribution and consequently, PAHs get dispersed from the atmosphere to vegetation, ultimately leading to bioaccumulation in various food chains (Edwards, 1983; Morehead et al., 1986; Wagrowski and Hites, 1997). Apart from biodegradation, the fate of PAH in nature varies depending on the environment, for example, in air, PAH can undergo photo-oxidation, whereas in the case of soil and water, they can undergo both photo-oxidation and chemical oxidation while some PAHs like naphthalene and alkyl naphthalene are partly lost by volatilization (Cerniglia, 1992).
The toxicity of PAH was first recognized in 1761 by John Hill, a physician who documented a high incidence of nasal cancer in tobacco snuff consumers (Cerniglia, 1984). The low-molecular-weight (LMW) PAHs (containing two or three aromatic rings) are acutely toxic while the high-molecular-weight (HMW) PAHs (containing four or more rings) are largely considered as genotoxic (Cerniglia, 1992; Mueller et al., 1996; Abdel-Shafy and Mansour, 2016). It is a known fact that PAH can covalently bind to DNA, RNA and proteins, but it is the amount of covalent interaction between PAHs and DNA that correlates best with carcinogenicity (Marston et al., 2001; Santarelli et al., 2008). In addition, the transformation products of some PAHs are more toxic than parent PAHs and can lead to critical cellular effects (Schnitz et al., 1993). In humans cytochrome P450 monooxygenase group of enzymes oxidize PAHs to epoxides, some of which are highly reactive (such as “bay-region” diol epoxides) and known as ultimate carcinogens. They can bind to DNA and have the ability to transform normal cells to malignant one (Milo and Casto, 1992; Schnitz et al., 1993; Marston et al., 2001). Studies have shown that processing food at high temperatures, like grilling or barbecuing result in high levels of PAHs in cooked meat and smoked fish (Morehead et al., 1986). The concern associated with PAHs further increased due to their ability to interfere with hormone metabolizing enzymes of the thyroid glands, and their adverse effects on reproductive as well as immune system (Veraldi et al., 2006; Oostingh et al., 2008).
Consequently, cleaning of such polluted places has been thought to be one of the most essential alternatives for restoring environmental damage. Several physical and chemical treatment methods including incineration, base-catalyzed de-chlorination, UV oxidation, fixation, solvent extraction etc. are already in practice (Norris et al., 1993; Gan et al., 2009), but have several drawbacks including cost, complexity, regulatory burden etc. Moreover, these conventional techniques, in many cases, do not destroy the contaminating compounds completely, but instead transfer them from one environment or form to another. In order to solve this burning problem, researchers have devised an efficient and eco-friendly clean-up technique known as bioremediation, which is being progressively refined to fight pollution. This technique utilizes and manipulates the detoxification abilities of living organisms to convert hazardous organic wastes including xenobiotics into harmless products, often carbon dioxide and water (Cerniglia and Heitkamp, 1989; Mueller et al., 1996; Bamforth and Singleton, 2005; Johnsen et al., 2005). Bioremediation addresses the limitations associated with most of the physicochemical processes by destroying many organic contaminants at reduced cost, under ambient conditions and thus, has now become a popular remedial alternative for pollutant removal including PAHs (Young and Cerniglia, 1996; Juhasz and Naidu, 2000; Kastner, 2000; Lovley, 2001; Andreoni and Gianfreda, 2007; Jorgensen, 2007; Megharaj et al., 2011; Abdel-Shafy and Mansour, 2016).
In addition, many natural habitats (e.g., aquifers, aquatic sediments) contaminated with a huge amount of aromatic pollutants are often anoxic. In these environments, the anaerobic degradation of aromatic compounds by microorganisms plays a major role in the removal of contaminants, recycling of carbon and sustainable development of the ecosystem. Reports on anaerobic biodegradation of PAHs are relatively recent, and only a limited number of preliminary studies have demonstrated the anaerobic degradation of PAHs including naphthalene, anthracene, phenanthrene, fluorene, acenaphthene and fluoranthene (Foght, 2008; Carmona et al., 2009; Mallick et al., 2011). However, detailed information on anaerobic degradation of PAHs under sulfate-reducing and nitrate-reducing conditions is scarce and very little is known about their degradation pathways, catabolic genes/enzymes and/or regulatory mechanisms, but this emerging field is ready to blossom in various aspects of biotechnological applications.
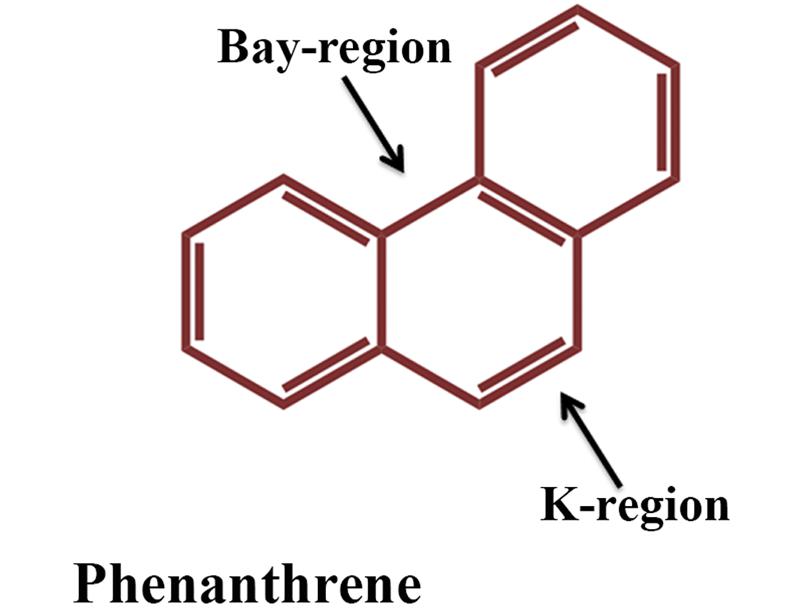
Compared to HMW PAHs, LMW PAHs are reasonably more volatile and more soluble in water and consequently more susceptible to biodegradation (Pannu et al., 2003). LMW PAHs like naphthalene, anthracene and phenanthrene are widely present throughout the environment and designated as prototypic PAHs and serve as signature compounds to detect PAH contamination. Naphthalene represents the simplest PAH whereas the chemical structures of anthracene and phenanthrene are found in many carcinogenic PAHs (such as in benzo[a]pyrene, benz[a]anthracene etc.), and phenanthrene represents the smallest PAH to have both the bay and K regions (Figure 2). Thus, they are often used as a model substrate for studies on the metabolism of carcinogenic PAHs (Mallick et al., 2011) (and the references therein). In line with the advances in the knowledge of bacterial diversity within an ecosystem, many unique metabolic pathways on degradation of the PAHs are reported and scattered in literatures, which would collectively deepen the understanding on the range of catabolism as well as the biochemical and genetic diversities. Timely collective update of literature is most important to address current status of subjects related to our life and surrounding environment where the ongoing threat due to PAH-mediated pollution is no exception rather an issue that deserves top priority. The present review provides an overview of the current knowledge of microbial degradation/transformation of PAHs. Moreover, factors affecting biodegradation of PAHs, recent advancement in genetic, genomic, proteomic and metabolomic techniques and their application in bioremediation of PAHs have also been described.
PAH Degradation by Bacteria and Halophilic Archaea
Bacterial Catabolism of PAHs
Bacteria, which have evolved more than three billion years ago, have developed strategies for obtaining energy from virtually every compound and have been considered as nature’s ultimate scavengers. Because of their quick adaptability, bacteria have largely been used to degrade or remediate environmental hazards. Various bacteria have been found to degrade PAHs, in which degradation of naphthalene and phenanthrene has been most widely studied. Numerous unique metabolic pathways for the bacterial degradation of PAHs have been well documented in a number of excellent review articles (Cerniglia, 1992; Peng et al., 2008; Seo et al., 2009; Mallick et al., 2011). So, the biochemical pathways for bacterial degradation of PAHs will not be discussed in details in this communication. Nevertheless, there are two major strategies to degrade PAHs depending on the presence or absence of oxygen. In the aerobic catabolism of aromatics, the oxygen is not only the final electron acceptor but also a co-substrate for the hydroxylation and oxygenolytic ring cleavage of the aromatic ring. In contrast, the anaerobic catabolism of aromatic compounds uses a completely different strategy to attack the aromatic ring, primarily based on reductive reactions (Foght, 2008; Carmona et al., 2009). While the aerobic catabolism of aromatic compounds has been studied for several decades, the anaerobic degradation of aromatic compounds is a more recently discovered microbial capacity that still awaits a deeper understanding. However, anoxic conditions dominate in many natural habitats and contaminated sites (e.g., aquifers, aquatic sediments and submerged soils) where biodegradation is carried out by anaerobes using alternative final electron acceptors such as nitrate, sulfate or ferric ions (Foght, 2008; Carmona et al., 2009).
Principally, bacteria favor aerobic conditions for degradation of PAHs via oxygenase-mediated metabolism (involving either monooxygenase or dioxygenase enzymes). Usually, the first step in the aerobic bacterial degradation of PAHs is the hydroxylation of an aromatic ring via a dioxygenase, with the formation of a cis-dihydrodiol, which gets rearomatized to a diol intermediate by the action of a dehydrogenase. These diol intermediates may then be cleaved by intradiol or extradiol ring-cleaving dioxygenases through either an ortho-cleavage or meta-cleavage pathway, leading to intermediates such as catechols that are ultimately converted to TCA cycle intermediates (Evans et al., 1965; Cerniglia, 1992; Eaton and Chapman, 1992; Mueller et al., 1996; Mallick et al., 2011). Dioxygenase is a multicomponent enzyme generally consisting of reductase, ferredoxin, and terminal oxygenase subunits (Mallick et al., 2011). Bacteria can also degrade PAH via the cytochrome P450-mediated pathway, with the production of trans-dihydrodiols (Sutherland et al., 1990; Moody et al., 2004) or under anaerobic conditions, e.g., under nitrate-reducing conditions (Foght, 2008; Carmona et al., 2009).
Naphthalene degrading bacteria are ubiquitous in nature and there are enormous numbers of reports documenting the bacterial degradation of naphthalene including the elucidation of the biochemical pathways, enzymatic mechanisms and genetic regulations (Cerniglia, 1992; Peng et al., 2008; Seo et al., 2009; Lu et al., 2011; Mallick et al., 2011). The naphthalene catabolic genes present in the plasmid NAH7 in Pseudomonas putida G7 are well characterized (Simon et al., 1993). In the plasmid NAH7, the naphthalene catabolic genes (nah) are organized into two operons: the nal operon containing the genes for the upper pathway enzymes involved in conversion of naphthalene to salicylate, and the sal operon containing the genes for the lower pathway enzymes involved in the conversion of salicylate to pyruvate and acetaldehyde (Simon et al., 1993). The operons are positively regulated by a common regulator NahR, a LysR type of positive transcriptional regulator and are widely dispersed in bacteria. NahR is induced in presence of salicylate leading to high-level expression of the nah genes in bacteria (Yen and Gunsalus, 1985; Peng et al., 2008). There are several reports on the nucleotide sequences of genes encoding the upper pathway enzymes in different Pseudomonas strains, and the genes are more than 90% identical (Menn et al., 1993; Simon et al., 1993; Yang et al., 1994; Bosch et al., 2000; Peng et al., 2008). Moreover, in Ralstonia sp. U2, the naphthalene catabolic operon (nag) contains all of the upper pathway genes similar to that of the classical nah genes of Pseudomonas strains in the same order, with the exception of two extra genes named nagG and nagH, which are structural subunits of salicylate-5-hydroxylase enzyme required for the conversion of naphthalene to gentisate (Zhou et al., 2001). Additionally, in Comamonas testeroni strain GZ42, the naphthalene catabolic genes are similar to that found in Ralstonia sp. U2 (Goyal and Zylstra, 1997). The lower pathway genes for the naphthalene catabolism are also similar in different Pseudomonas strains like P. putida G7, NCIB9816-4, ND6, and P. stutzeri AN10 (Habe and Omori, 2003; Peng et al., 2008). In those organisms, lower pathway of naphthalene operon contains 11 genes present in the order nahGTHINLOMKJY, in which nahY represent a naphthalene chemotaxis gene. However, in AN10 and ND6 strains another salicylate hydroxylase gene (nahW) has been found to be present outside, but near to the sal operon.
Along with Pseudomonas, a high proportion of the PAH-degrading isolates belong to the sphingomonads, which comprise a physiologically versatile group within the Alphaproteobacteria: mainly Sphingomonas, Sphingobium and Novosphingobium that are frequently found as aromatic degraders. Species belonging to these genera show great catabolic versatility, capable of degrading a wide range of natural and xenobiotic compounds including HMW PAHs (Basta et al., 2005; Peng et al., 2008; Stolz, 2009; Vila et al., 2015). In a few reports, it has been shown that the catabolic versatility of sphingomonads relies on the large plasmids present in those organisms (Basta et al., 2005; Peng et al., 2008). However, the plasmid-encoded degradative genes have been found to be largely scattered, or they are not organized in coordinately regulated operons (Romine et al., 1999). It may be possible that this kind of ‘flexible’ gene organization i.e., different combinations of conserved gene clusters allows sphingomonads to adapt easily and proficiently to degrade various aromatic compounds including xenobiotics (Basta et al., 2005; Peng et al., 2008). Recently an illustrative report on the catabolic versatility of sphingomonads has shown that strain PNB, acting on diverse monoaromatic and polyaromatic compounds, contains seven sets of ring-hydroxylating oxygenases (RHO) with different substrate specificities (Khara et al., 2014). In general, it has been seen that mobile genetic elements (MGEs) like plasmids and transposons play a crucial role in the biodegradation of organic pollutants like PAHs. The presence of foreign compounds can often lead to the selection of mutant bacteria that are capable of metabolizing them. Apart from vertical gene transfer, aromatics catabolic genes are often harbored by MGEs that successfully disseminate the catabolic traits to phylogenetically diverse bacteria by horizontal gene transfer (Top and Springael, 2003; Nojiri et al., 2004).
Among PAHs degrading bacteria the genus Rhodococcus is very unique, having an enormous catabolic versatility. In contrast to Pseudomonas and other Gram-negative bacteria whose naphthalene catabolic genes are usually clustered, the Gram-positive Rhodococcus strains usually exhibit only three structural genes required for naphthalene degradation (narAa, narAb and narB) (Kulakov et al., 2005; Larkin et al., 2005). It has been seen that the nar region is not arranged into a single operon, and there are several homologous transcription units in different Rhodococcus strains separated by non-homologous sequences containing direct and inverted repeats (Larkin et al., 2005). The narAa and narAb genes codes for the α- and β- subunit of the naphthalene dioxygenase (NDO). Both subunits of NDO in Rhodococcus sp. strain NCIMB12038 showed only 30% amino acid identity to the corresponding P. putida NDO subunits. In addition, there is no gene in Rhodococcus strains which is similar to the genes encoding the electron transport components reductase and ferredoxin of NDO in Pseudomonas strains (Kulakov et al., 2005; Larkin et al., 2005). Moreover, the narA and narB genes are transcribed as a single unit through different start sites, and their transcription is induced in the presence of naphthalene in contrast to salicylate-inducible naphthalene catabolic genes in Pseudomonas species. There are two putative regulatory genes (narR1 and narR2, GntR-like transcriptional regulator) which are shown to be transcribed as a single mRNA in naphthalene-induced cells (Kulakov et al., 2005; Larkin et al., 2005). Figure 3 represent the gene organization of the nar gene cluster of various naphthalene degrading Rhodococcus sp.
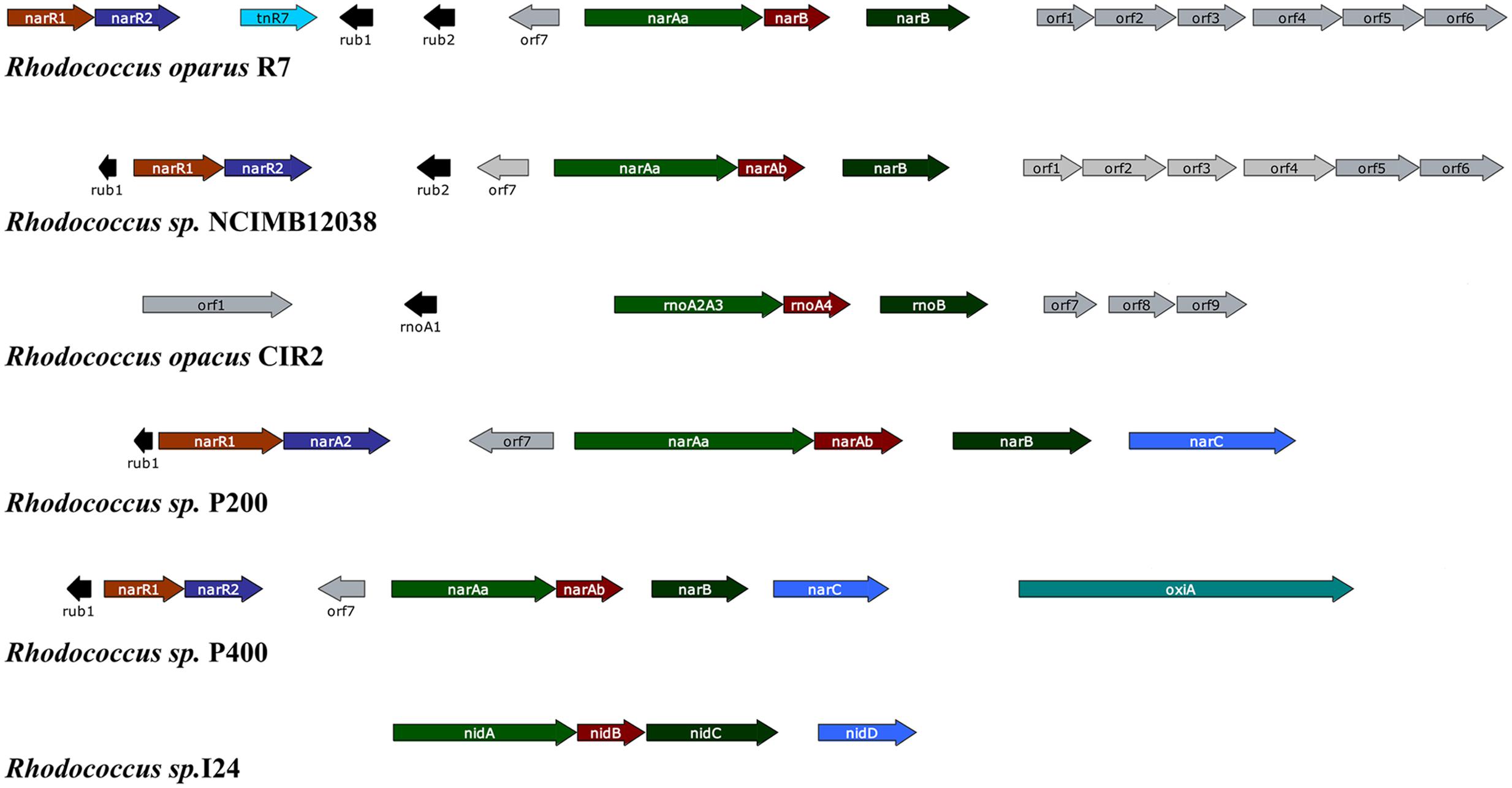
FIGURE 3. Gene organization of the nar gene cluster of various naphthalene degrading Rhodococcus sp. The respective Rhodococcus sp. along with the accession numbers of their corresponding naphthalene degrading gene cluster: Rhodococcus opacus R7 (accession no. DQ846881), Rhodococcus sp. NCIMB12038 (accession no. AF082663), Rhodococcus opacus CIR2 (accession no. AB024936), Rhodococcus sp. P200 (accession no. AY392424), Rhodococcus sp. P400 (accession no. AY392423), Rhodococcus sp. I24 (accession no. AF121905). The genes and their corresponding function of the gene products: narR1- GntR-like regulator protein; narR2- XylR-like regulator protein; rub1 and 2- Rubredoxin; narAa- Naphthalene dioxygenase large subunit; narAb- Naphthalene dioxygenase small subunit; narB- Naphthalene dihydrodiol dehydrogenase (adapted from Di Gennaro et al., 2010).
Along with naphthalene, a number of reports on phenanthrene degradation by various Gram negative and Gram positive bacterial species have been reported (Peng et al., 2008; Seo et al., 2009; Mallick et al., 2011). In a study, Mallick et al. (2007) reported the degradation of phenanthrene by Staphylococcus sp. strain PN/Y, by initiating the dioxygenation specifically at the 1,2-position followed by meta-cleavage of phenanthrene-1,2-diol, leading to the formation of 2-hydroxy-1-naphthoic acid as the metabolic intermediate; while the ortho-cleavage could yield the naphthalene-1,2-dicarboxylic acid. Authors also reported that 2-hydroxy-1-naphthoic acid was metabolized by a meta-cleavage enzyme 2-hydroxy-1-naphthoate dioxygenase leading to the formation of trans-2,3-dioxo-5-(2′-hydroxyphenyl)-pent-4-enoic acid, a novel metabolite in the phenanthrene degradation pathway, and was subsequently degraded via salicylic acid and catechol (Mallick et al., 2007). Later on, Ghosal et al. (2010) reported the assimilation of phenanthrene by Ochrobactrum sp. strain PWTJD, isolated from municipal waste contaminated soil sample using phenanthrene as a sole source of carbon and energy. The strain PWTJD could also degrade phenanthrene via 2-hydroxy-1-naphthoic acid, salicylic acid and catechol. The strain PWTJD was found to utilize 2-hydroxy-1-naphthoic acid and salicylic acid as sole carbon sources, while the former was metabolized by a ferric-dependent meta-cleavage dioxygenase. In the lower pathway, salicylic acid was metabolized to catechol and was further degraded by catechol 2,3-dioxygenase to 2-hydroxymuconoaldehyde acid, ultimately leading to TCA cycle intermediates. The metabolic pathway involved in the degradation of phenanthrene by Ochrobactrum sp. strain PWTJD is shown in Figure 4 (Ghosal et al., 2010). This was the first report of the complete degradation of a PAH molecule by Gram-negative Ochrobactrum sp. describing the involvement of the meta-cleavage pathway of 2-hydroxy-1-naphthoic acid in phenanthrene assimilation.
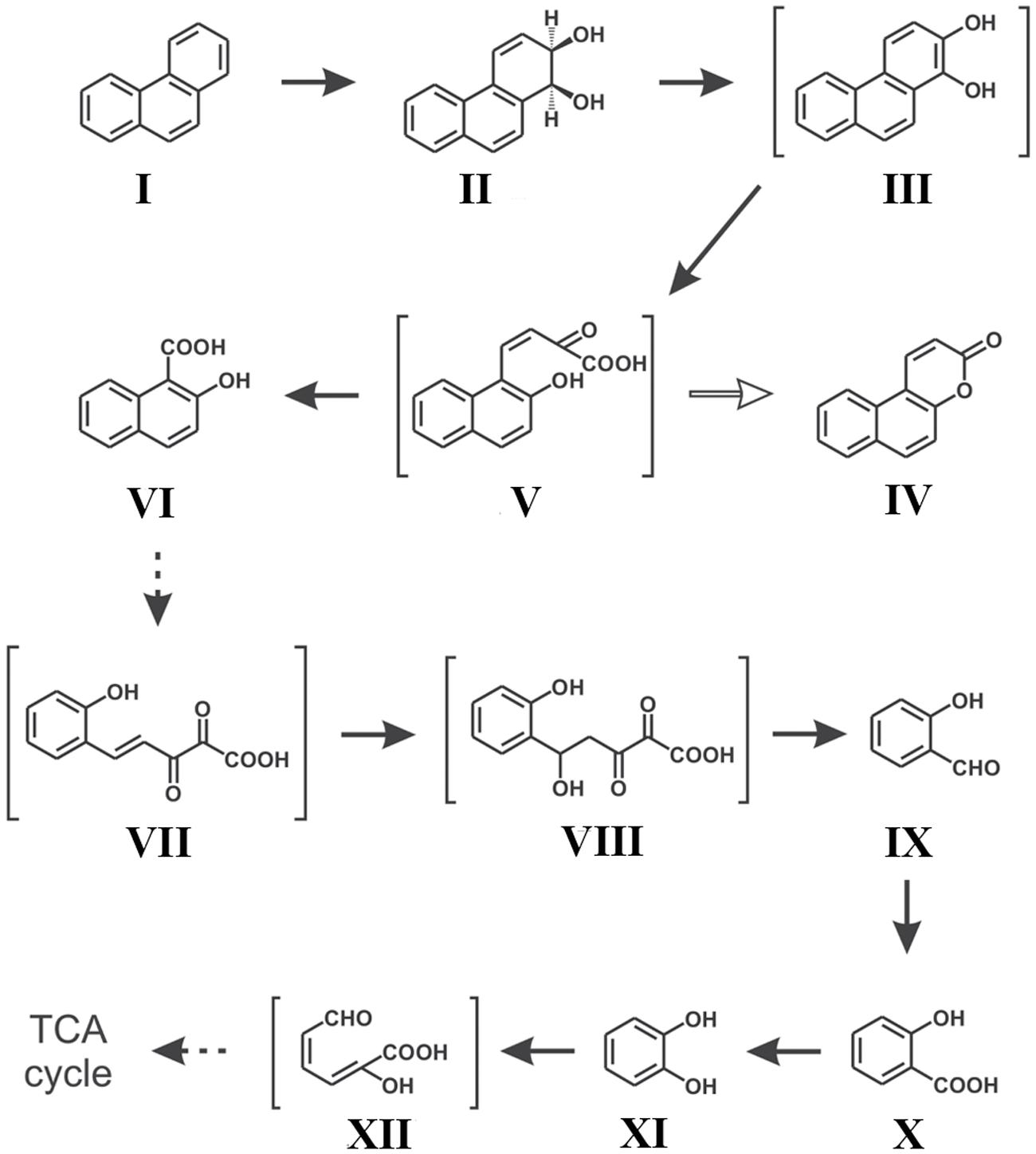
FIGURE 4. Proposed pathway for the degradation of phenanthrene by Ochrobactrum sp. strain PWTJD. The transient intermediates, which were not detected, are shown in parentheses. Filled arrows indicate mineralization; open arrows indicate a dead-end metabolite; and dotted arrows indicate multiple steps. Chemical designations: (I) phenanthrene; (II) cis-1,2-phenanthrenedihydrodiol; (III) 1,2-dihydroxyphenanthrene; (IV) 5,6-benzocoumarin; (V) cis-2-oxo-4-(20-hydroxynaphthyl)-but-3-enoic acid; (VI) 2-hydroxy-1-naphthoic acid; (VII) trans-2,3-dioxo-5-(20-hydroxyphenyl)- pent-4-enoic acid; (VIII) 2,3-dioxo-5-hydroxy-5-(20-hydroxyphenyl)-pentanoic acid; (XI) salicylaldehyde; (X) salicylic acid; (XI) catechol; (XII) 2-hydroxymuconaldehyde acid. Adapted from Ghosal et al. (2010).
Other LMW PAHs like anthracene, fluorene, acenaphthene and acenaphthylene are also found in high quantities in PAH-polluted sites and various bacterial species have the capability to utilize these compounds individually as sole carbon and energy sources (Moody et al., 2001; Peng et al., 2008; Seo et al., 2009; Mallick et al., 2011). Lately, Ghosal et al. (2013) reported assimilation of acenaphthene and acenaphthylene by the Acinetobacter sp. strain AGAT-W, isolated from municipal waste contaminated soil sample using acenaphthene as a sole source of carbon and energy. The strain AGAT-W could degrade acenaphthene via 1-acenaphthenol, naphthalene-1,8-dicarboxylic acid, 1-naphthoic acid, salicylic acid and cis, cis muconic acid ultimately leading to TCA cycle intermediates. The metabolic pathways involved in the degradation of acenaphthene and acenaphthylene by Acinetobacter sp. strain AGAT-W is shown in Figure 5 (Ghosal et al., 2013). This was the first report on the complete degradation of acenaphthene and acenaphthylene individually by strain AGAT-W belonging to the genus Acinetobacter.
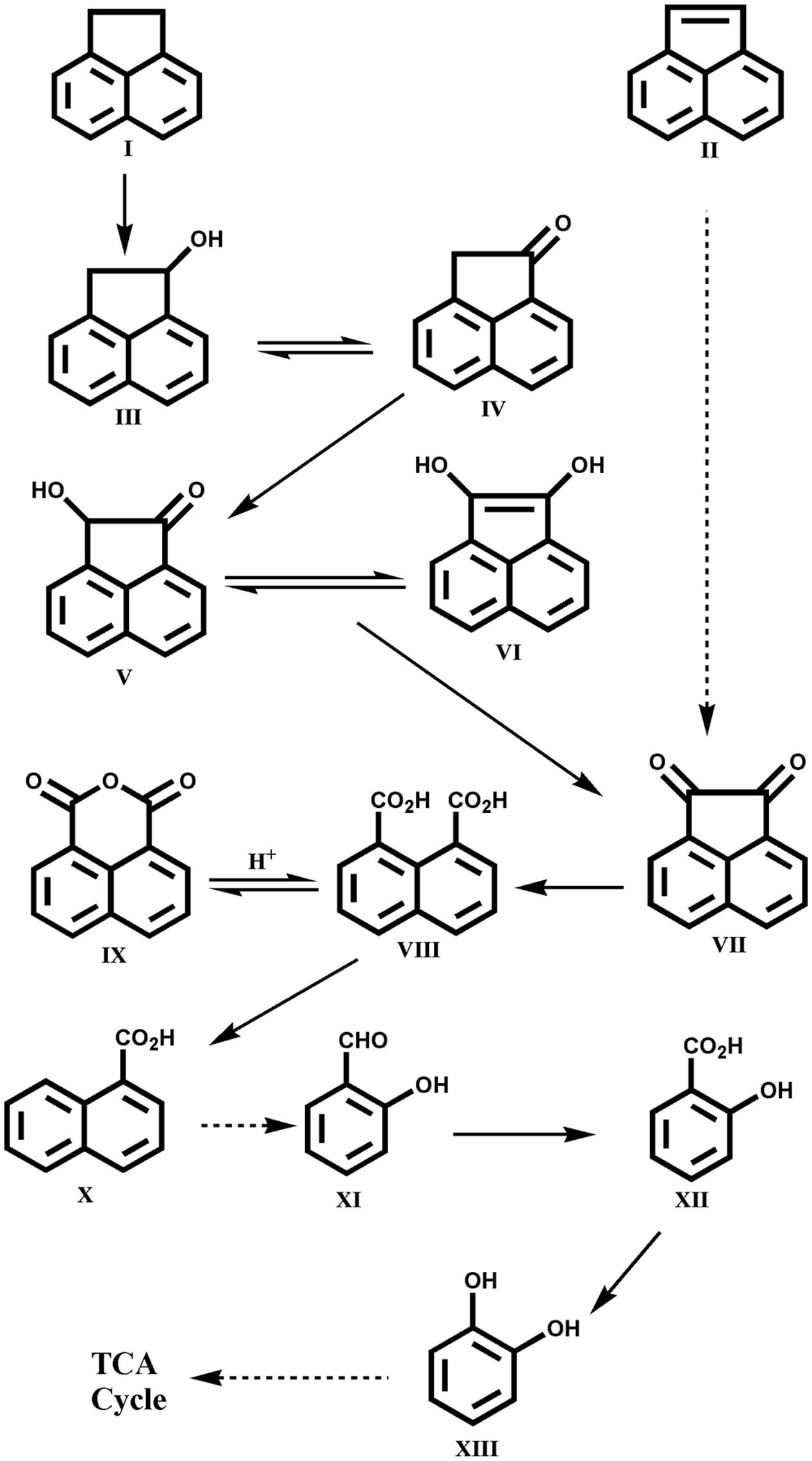
FIGURE 5. Proposed pathway for the degradation of acenaphthene by Acinetobacter sp. strain AGAT-W. Chemical designations: (I) acenaphthene; (II) acenaphthylene; (III) 1-acenaphthenol; (IV) 1-acenaphthenone; (V) 1-hydroxy-2-ketoacenaphthene; (VI) 1,2-dihydroxyacenaphthylene; (VII) acenaphthenequinone; (VIII) naphthalene-1,8-dicarboxylic acid; (IX) 1,8-naphthalic anhydride; (X) 1-naphthoic acid; (XI) salicylaldehyde; (XII) salicylic acid; (XIII) catechol. Adapted from Ghosal et al. (2013).
Polycyclic aromatic hydrocarbons with more than three rings viz. pyrene, benzo[a] pyrene (Bap), are generally referred to as HMW PAHs (Figure 1), which are of significant environmental concern due to their long persistence and high toxicity as well as their mutagenic and/or carcinogenic properties (Cerniglia, 1992; Kanaly and Harayama, 2000; Peng et al., 2008; Seo et al., 2009). In the last few decades research on microbial degradation of HMW PAHs has advanced significantly and a number of HMW PAH-degrading isolates have been reported (Cerniglia, 1992; Kanaly and Harayama, 2000, 2010; Peng et al., 2008; Seo et al., 2009). Biodegradation of HMW PAHs by microorganisms is discussed adequately in many excellent reviews and the pathways for their degradation have also been depicted (Kanaly and Harayama, 2000; Peng et al., 2008; Seo et al., 2009). However, further investigations are prerequisite in several areas of HMW PAH biodegradation, namely, research related to the regulatory mechanisms of HMW PAH biodegradation, biodegradation of HMW PAH associated with other hydrocarbons in mixtures; and the interactions of complex microbial community during HMW PAH degradation can be exploited in more details. These will enrich our understanding on the microbial ecology of HMW PAH-degrading communities and the mechanisms by which HMW PAH biodegradation occur. In addition, the outcome will also help in predicting the environmental fate of these recalcitrant compounds and aid for the development of convenient as well as cost-effective bioremediation strategies in near future.
Degradation of PAHs by Halophilic/Halotolerant Bacteria and Archaea
Environmental pollution due to anthropogenic activity has spread to all types of ecosystems and marine ecosystem is not excluded from this list. Hypersaline environments are regularly being polluted with organic pollutants including PAHs, through industrial and municipal effluents. Industrial effluents, specifically, petroleum industry where one of the main sources of contaminants in the waste waters are aromatic hydrocarbons including PAHs. Contamination and biodegradation in extreme environments like high salinity has been receiving increased attention in recent times (Oren et al., 1992; Margesin and Schinner, 2001a,b; Le Borgne et al., 2008; Bose et al., 2013; Cravo-Laureau and Duran, 2014; Elango et al., 2014; Fathepure, 2014; Genovese et al., 2014; Kappell et al., 2014; Kostka et al., 2014; Thomas et al., 2014; Torlapati and Boufadel, 2014). In addition, a survey of relevant literatures indicates that along with numerous biotechnological applications, halophilic microorganisms have more extended catabolic versatility than previously thought about (Margesin and Schinner, 2001b; Garcia et al., 2005; Smith et al., 2013; Kappell et al., 2014; Lamendella et al., 2014; Roling and Bodegom, 2014; Scott et al., 2014; Singh et al., 2014).
The biological treatment of industrial hypersaline waste waters and the bioremediation of polluted hypersaline environments are not possible with conventional microorganisms because they are incapable to function efficiently at salinities that of seawater or above. Thus, halophilic microorganisms are the best alternatives to overcome this problem (Oren et al., 1992; Garcia et al., 2005; Fathepure, 2014; Kostka et al., 2014). In the last decade there has been an increasing interest in the development and optimization of bioremediation processes via halophiles to deal with hypersaline environments that are contaminated with organic pollutants (Margesin and Schinner, 2001a; Mellado and Ventosa, 2003; Peyton et al., 2004; Garcia et al., 2005; Cui et al., 2008; Mnif et al., 2009; Zhao et al., 2009; Bonfa et al., 2011; Smith et al., 2013; Fathepure, 2014; Kappell et al., 2014; Lamendella et al., 2014; Singh et al., 2014; Thomas et al., 2014). Some halophiles that have shown PAHs degrading property are depicted in Supplementary Table S1. However, studies concerning the ability of this group of microbes to degrade PAHs are still in their infancy. Although in the last few years, a few studies on the degradation of PAHs by halophiles have been published. In this context, Gauthier et al. (1992) described the isolation of Marinobacter hydrocarbonoclasticus which can degrade various organic compounds including naphthalene (Gauthier et al., 1992). The presence of naphthalene dioxygenase, similar to those from Pseudomonas and Burkholderia spp. was reported in a naphthalene-degrading Marinobacter sp. strain NCE312 (Hedlund et al., 2001). Again, based on the analysis of the shoreline sand and rocks (Costa da Morte, northwestern Spain) affected by the Prestige oil spill, unveiled the high relative abundances of Sphingomonadaceae and Mycobacterium that could be associated with PAH degradation (Alonso-Gutierrez et al., 2009). In a similar study, Vila et al. (2010) reported the isolation of a marine microbial consortium from a beach polluted with the Prestige oil spill which is efficient in removing different hydrocarbon present in heavy fuel oil including three to five-ring PAHs (for example anthracene, fluoranthene, pyrene, benzo(a)anthracene, chrysene, and BaP) (Vila et al., 2010). In addition based on community dynamics analysis, it has been contemplated that Alphaproteobacteria (Maricaulis and Roseovarius), could be associated with the degradation of LMW PAHs, whereas Gammaproteobacteria (Marinobacter and Methylophaga), could be associated with the degradation of HMW PAHs. A similar bacterial community associated with the degradation of various organic compounds was found in a beach affected by the Deepwater Horizon oil spill (Kostka et al., 2011; Lamendella et al., 2014). Recently, Gallego et al. (2014) reported the isolation of a marine pyrene-degrading microbial consortium from a beach polluted by an oil spill and found that an uncultured Gordonia sp. is the key pyrene degrader in the consortium based on community structure and PAH ring-hydroxylating genes analyses (Gallego et al., 2014). Nevertheless, detailed information on the bioremediation of PAHs under high salinity by halophilic/halotolerant bacteria and archaea is still in its initial stage of exploration, but it is expected that this emerging field is ready to flourish in near future.
Occurrence of PAH-Degrading Machinery in Diverse Bacterial/Halophilic Archaeal Genera
The community analysis of indigenous microorganisms capable of degrading various aromatic compounds in diverse environments has been of great interest in the recent years (Watanabe et al., 2002; Brakstad and Lodeng, 2005; Gerdes et al., 2005; Yakimov et al., 2005; Coulon et al., 2007; Cui et al., 2008) and different strategies have been developed for the study of associated microbial communities (Widada et al., 2002). Previously it was thought that PAH degradation capabilities may be associated with certain genera or groups of bacteria independent of the origin of the source sample (Kastner et al., 1994; Mueller et al., 1997). But consequently, with time it has been reported that PAH-degrading bacteria are far more diverse (Widada et al., 2002; Andreoni et al., 2004; Cui et al., 2008; Hilyard et al., 2008). Currently, there are enormous reports of various bacterial as well as some archaeal genera capable of degrading PAHs. Supplementary Table S1 represents the bacterial/archaeal (halophilic) strains involved in the degradation of various PAHs. Figure 6 illustrates the 16S rRNA gene-based phylogenetic analysis of all the bacterial/archaeal strains cited in Supplementary Table S1. It has been seen that bacterial/archaeal strains having PAHs catabolic property are distributed mainly in six major groups: Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Actinomycetes, Firmicutes and Archaea (Halophiles). It may be mentioned here that many strains listed in Supplementary Table S1 have not been cultured in laboratory. Some PAHs degrading isolates represent novel strains which are isolated from various geographically diverse habitats. Thus, all this information illustrate that PAH-degrading machinery is not confined to a few particular genus reported so far but is more likely to be distributed widely in the prokaryotic kingdom found in diverse geographical niches.
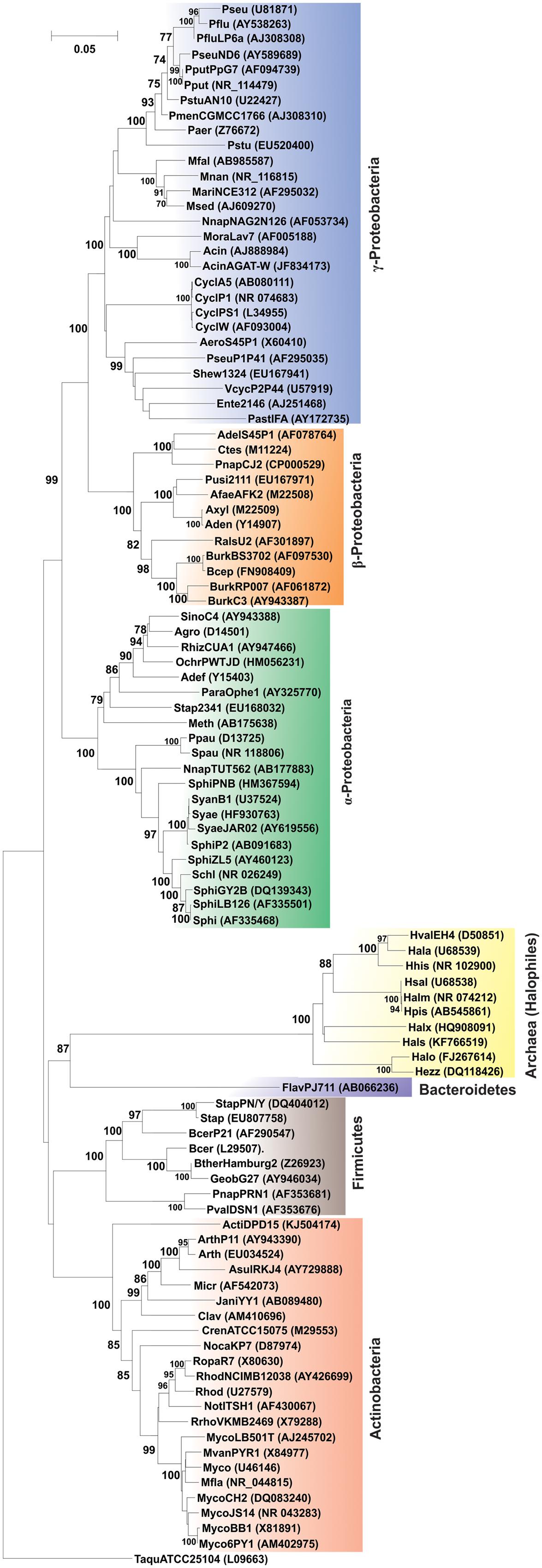
FIGURE 6. Phylogenetic tree based on 16S rRNA gene sequence of organisms enlisted in Supplementary Table S1. Organism designations are according to those given in Supplementary Table S1. However, for organisms devoid of 16S rRNA gene sequence information, sequence of representative type strain has been considered. Accession numbers are given within parentheses. 16S rRNA gene sequence of Thermus aquaticus strain ATCC 25104 was used as outgroup. Numbers at nodes indicate levels of bootstrap support based on neighbor-joining analysis of 100 resampled datasets. Bootstrap values below 70% are not shown. Bar represents 0.05 substitutions per nucleotide position.
Fungal Degradation of PAHs
The biodegradation of PAHs by fungi has been studied extensively in last several years and numerous fungal species have been reported to metabolize different PAHs (Cerniglia, 1992; Cerniglia and Sutherland, 2010). Most fungi cannot use PAHs as sole sources of carbon and energy; however, they may co-metabolize PAHs to a wide variety of oxidized products and sometimes to CO2. Bacterial PAHs degradation mainly involves dioxygenase enzymes and partially monooxygenase mediated reactions and the same is valid for algae. For example, the extent of dioxygenase vis-à-vis monooxygenase catalyzed transformation of naphthalene by Mycobacterium sp. was found to be in the ratio of around 25:1 (Kelley et al., 1990). On the other hand, fungal PAHs degradation mainly involves monooxygenase enzymes (Cerniglia and Sutherland, 2010) (and the references therein). However, the transformation of PAHs by fungi involves several enzymatic pathways and depends on the particular species and growth conditions. The fungi involved in PAHs biodegradation are mainly of two types- ligninolytic fungi or white-rot fungi (they have the ability to produce enzymes including lignin peroxidase (LiP), manganese peroxidase (MnP) and laccases to degrade the lignin in wood) and non-ligninolytic fungi (those who do not produce peroxidases or laccases but instead produce cytochrome P450 monooxygenase like enzymes) (Hofrichter, 2002; Tortella et al., 2005; Cerniglia and Sutherland, 2010; Li et al., 2010). Although, various ligninolytic fungi such as Phanerochaete chrysosporium and Pleurotus ostreatus can secrete both ligninolytic and non-ligninolytic type of enzymes, but it is uncertain to what extent each enzyme participates in the degradation of the PAHs (Bezalel et al., 1997). On the other hand, another type of ligninolytic fungi, known as brown-rot fungi, mainly produces hydrogen peroxide for degrading hemicelluloses and cellulose. Although, only limited data is available about PAH metabolism by brown rot fungi, some brown-rot fungi such as Laetiporus sulphureus and Flammulina velutipes have been shown to metabolize PAHs like phenanthrene, fluoranthene and fluorine (Sack et al., 1997; Cerniglia and Sutherland, 2010) (and the references therein).
Catabolism of PAHs by Non-ligninolytic Fungi
Polycyclic aromatic hydrocarbons metabolic pathway of non-ligninolytic fungi is similar to those formed by mammalian enzymes. In this case, the predominant pathway of initial oxidation of PAHs by non-ligninolytic fungi involves the activity of the cytochrome P450 monooxygenase enzymes. These enzymes catalyze a ring epoxidation to form an unstable arene oxide, which is further transformed to trans-dihydrodiol via an epoxide-hydrolase catalyzed reaction (Jerina, 1983; Sutherland et al., 1995). Non-ligninolytic fungi such as Cunninghamella elegans and ligninolytic fungi such as Pleurotus ostreatus, metabolize PAHs via this pathway (Bezalel et al., 1996; Tortella et al., 2005). For example, the transformation of fluoranthene by C. elegans produce fluoranthene trans-2,3-dihydrodiol, 8- and 9-hydroxyfluoranthene trans-2,3-dihydrodiols (Tortella et al., 2005). Similarly, P. ostreatus metabolizes pyrene into pyrene trans-4,5-dihydrodiol (Bezalel et al., 1996). Arene oxide produced by cytochrome P450 can also be rearranged to phenol derivatives by non-enzymatic reactions and are subsequently conjugated with sulfate, xylose, glucoronic acid, or glucose (Mueller et al., 1996; Pothuluri et al., 1996). In some fungi, cytochrome P450 monooxygenases oxidize PAHs to epoxides and dihydrodiols which are potent carcinogens and more toxic than the respective parent PAHs, while on the other hand peroxidase-mediated oxidation of PAHs produces quinines which are less toxic than the parent PAHs (Cerniglia and Sutherland, 2010). This is why oxidation of PAHs by ligninolytic enzymes could be a more logical strategy for the detoxification of PAHs contaminated environment.
Catabolism of PAHs by Ligninolytic Fungi
White-rot fungi are ubiquitous in nature and can produce ligninolytic enzymes which are secreted extracellularly. These enzymes can degrade lignin present in wood and other organic substances. Ligninolytic enzymes are mainly of two types, peroxidases and laccases. On the basis of reducing substrate types, peroxidase enzyme can be classified again into two types, lignin peroxidase and manganese peroxidase. Both types of peroxidases have the ability to oxidize PAHs (Hammel, 1995; Cerniglia and Sutherland, 2010). Laccases, the phenol oxidase enzymes, also have the ability to oxidize PAHs (Cerniglia and Sutherland, 2010). Compared to bacterial PAHs degrading enzymes, ligninolytic enzymes are not induced in the presence of PAHs or by its degradation products (Verdin et al., 2004). As ligninolytic enzymes are secreted extracellularly, they are able to diffuse toward the immobile PAHs, and that is why they are more useful than bacterial intracellular enzymes in making initial attack on PAHs in soil. In addition compared to bacterial PAH-degrading enzymes, ligninolytic enzymes have broad substrate specificity and are therefore able to transform a wide range of substrates, even those which are most recalcitrant (Tortella et al., 2005; Cerniglia and Sutherland, 2010). Ligninolytic enzymes can transform PAHs by producing hydroxyl free radicals by the donation of one electron, which oxidizes the PAH ring (Sutherland et al., 1995). As a result, PAH-quinones and acids are formed instead of dihydrodiols. It has been seen that ligninolytic fungi mineralize PAHs by a combination of ligninolytic enzymes, cytochrome P450 monooxygenases, and epoxide hydrolases (Bezalel et al., 1997). Numerous reports have been documented on the degradation of PAHs by white-rot fungi (Tortella et al., 2005; Cerniglia and Sutherland, 2010). The metabolic pathway for the degradation of phenanthrene by the ligninolytic fungus Pleurotus ostreatus is illustrated in Figure 7 (Bezalel et al., 1997). PAHs degradative potential of wood-rotting fungi Plrurotus ostreatus and Antrodia vaillantii was also examined in soil, artificially contaminated with fluorene, phenanthrene, pyrene, and benz[a]anthracene (Andersson et al., 2003). It has been reported that although P. ostreatus significantly increased the degradation of PAHs in soil, but in the process, accumulated toxic PAH metabolites. It has also been seen that this white-rot fungus inhibits the indigenous microbial populations in the soil, which may have prohibited the complete mineralization of the PAHs. Conversely, the brown-rot fungus A. vaillantii did not generate any dead-end metabolites, although its PAHs degradation rate was similar to that of P. ostreatus (Andersson et al., 2003). Oxidation of pyrene, anthracene, fluorene and BaP to the corresponding quinines by lignin peroxidase and manganese peroxidase was reported by the white-rot fungus Phanerochaete chrysosporium (Bogan et al., 1996a,b). In another study, complete mineralization of BaP (HMW PAHs) by the white-rot fungus P. chrysosporium was reported in a two-stage pilot scale reactor (May et al., 1997). Clemente et al. (2001) studied PAHs degradation by thirteen ligninolytic fungal strains and reported that the rate of degradation varies with a variation of lignolytic enzymes (Clemente et al., 2001). In this study, highest naphthalene degradation (69%) was reported by the strain 984 which have Mn-peroxidase activity, followed by strain 870 (17%) having lignin peroxidase and laccase activities. The ability of soil fungi to degrade PAHs that produce ligninolytic enzymes was also studied under microaerobic and very-low-oxygen conditions (Silva et al., 2009). It was reported that Aspergillus sp., Trichocladium canadense, and Fusarium oxysporum can degrade LMW PAHs (2–3 ring compounds) most extensively, whereas highest degradation of HMW PAHs (4–7 rings) was observed in T. canadense, Aspergillus sp., Verticillium sp., and Achremonium sp. These results suggest that, along with bacteria, fungi can be exploited as a valuable endeavor for the bioremediation of PAH contaminated sites.
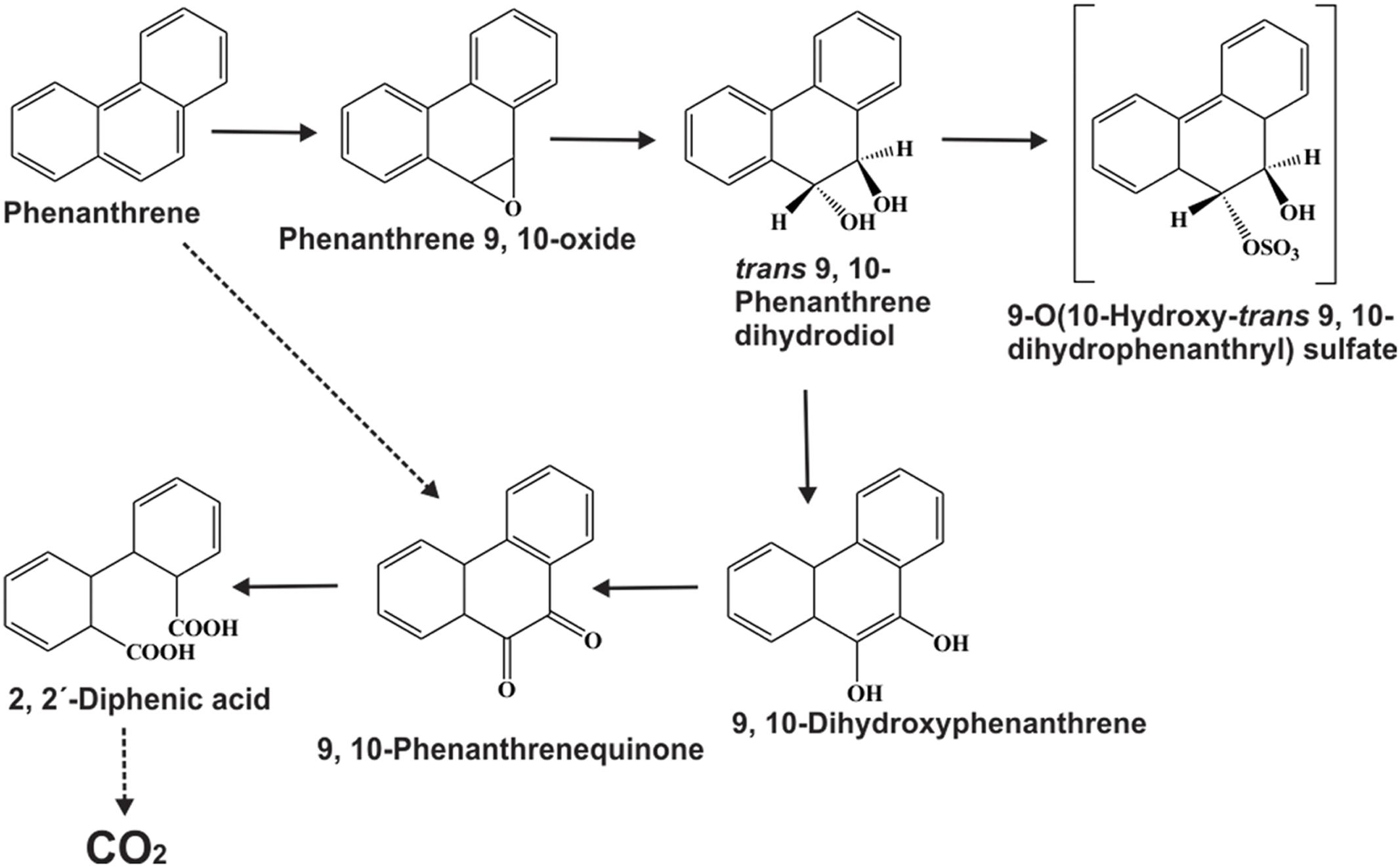
FIGURE 7. Proposed phenanthrene degradation pathway by the ligninolytic fungus Pleurotus ostreatus (adapted from Bezalel et al., 1997).
Microalgal Degradation of PAHs
Compared to bacteria and fungi, relatively little attention has been paid to the biodegradation of PAHs by microalgae (cyanobacteria, diatoms etc.). Microalgae are one of the major primary producers in aquatic ecosystems, and play vital roles in the fate of PAHs in those environments. Several strains of microalgae are known to metabolize/transform naphthalene, phenanthrene, anthracene, BaP and other PAHs (Cerniglia et al., 1979, 1980a,b; Narro et al., 1992a,b; Safonova et al., 2005; Chan et al., 2006; El-Sheekh et al., 2012). Supplementary Table S2 represents different microalgal strains involved in the bioremediation of various PAHs. Figure 8 illustrates the rubisco large subunit gene-based phylogenetic analysis of all the organisms enlisted in Supplementary Table S2. Figure 9 shows the biotransformation pathway of naphthalene by microalgae Oscillatoria sp., strain JCM (Cerniglia et al., 1980a). Under photoautotrophic growth conditions, strain JCM has been reported to oxidize naphthalene to 1-naphthol, whereas marine cyanobacterium Agmenellum quadruplicatum strain PR-6 can convert phenanthrene to phenanthrene trans-9,10-dihydrodiol and 1-methoxyphenanthrene (Narro et al., 1992a,b). In a separate study, oxidation of BaP by the microalgae Selanastum capricornutum has been evaluated. Transformation of BaP resulted in the formation of cis-4,5-, 7,8-, 9,10- and 11,12-BaP-dihydrodiols, involving a dioxygenase system, similar to bacterial PAH degradation systems but unlike those of eukaryotic organisms (like fungi) which involve monooxygenase systems (Warshawsky et al., 1988). In another study, the effects of gold, white or UV-A fluorescent lights for the biotransformation of BaP and phototoxicity of carcinogenic PAHs in different algal systems was determined. It has been found that algae like S. capricornutum, Scenedesmus acutus and Ankistrodesmus braunii are able to degrade BaP to dihydrodiols, and the degradation varies with the kind and intensity of light sources (Warshawsky et al., 1995). The removal efficiency for either fluoranthene or pyrene, or a mixture of fluoranthene and pyrene were also determined using Chlorella vulgaris, Scenedesmus platydiscus, Scenedesmus quadricauda, and Selenastrum capricornutum microalgal species (Lei et al., 2007). After 7 days of treatment PAHs removal by S. capricornutum and C. vulgaris was 78 and 48% respectively. The removal rate of fluoranthene and pyrene in a mixture was found to be similar, or higher than the respective single compound, which indicates that the presence of one PAH acts synergistically in the removal of the other PAH (Lei et al., 2007). In another study, microalgal strain Prototheca zopfii immobilized in polyurethane foam has been reported to accumulate a mixture of PAHs in the matrix, when incubated with mixture of aliphatic hydrocarbons and PAHs as substrates. Though the immobilized organism can degrade n-alkanes but it’s PAH degradation rate is very less, however, PAHs accumulation did not impair the degradation of PAHs; whereas in case of free-living cells, the organisms can reduce the concentration of both PAHs and n-alkanes satisfactorily (Ueno et al., 2006, 2008). Hong et al. (2008) have examined the accumulation and biodegradation of phenanthrene (Phn) and fluoranthene (Fla) by the two diatoms Skeletonema costatum and Nitzschia sp., enriched from a mangrove aquatic ecosystem. It was seen that the strains were capable of accumulating and degrading phenanthrene and fluoranthene simultaneously and the PAHs accumulation and degradation capability of Nitzschia sp. were higher than those of S. costatum. Further, it had been observed that the degradation of fluoranthene by the two diatoms was slower, compared to phenanthrene. The strains also showed similar or higher efficiency in the removal of the Phn–Fla mixture than Phn or Fla alone (Hong et al., 2008). In another study, the efficiency of seven microalgal species to remove pyrene from solution was reported (Lei et al., 2002). In a recent study removal of benzo(a)pyrene (BaP) by sorption and degradation was determined by two microalgal species Selenastrum capricornutum and Scenedesmus acutus (Garcia de Llasera et al., 2016). It has been seen that S. capricornutum can remove 99% of BaP after 15 h of exposure, whereas S. acutus can remove 95% after 72 h of exposure. In a separate study, the effects of metals on biosorption and biodegradation of fluorene, phenanthrene, fluoranthrene, pyrene and benzo[a]pyrene by Selenastrum capricornutum were investigated (Ke et al., 2010). It has been shown that both metal dosage and exposure time yielded a significant effect on the ability of removal of low molecular weight PAHs like fluorene and phenanthrene by the alga, whereas for high molecular weight PAHs like fluoranthrene, pyrene and BaP, the removal efficiency was not affected by the presence of metals. Patel et al. (2016) recently reported the biodegradation of anthracene and pyrene by Anabaena fertilissima (Patel et al., 2016), while Takáčová et al. (2014) reported the biodegradation of BaP by the microalgae Chlorella kessleri. Removal and transformation of seven high molecular weight PAHs in water was reported by live and dead cells of a freshwater microalga, Selenastrum capricornutum under gold and white light irradiation. The removal efficiency of PAHs, and the effectiveness of live and dead cells, was found to be predominantly PAH dependent (Ke et al., 2010; Luo L. et al., 2014; Luo S. et al., 2014). The first study on the potential of algal–bacterial microcosms was reported for the biodegradation of aromatic pollutants comprising salicylate, phenol and phenanthrene in a one-stage treatment (Borde et al., 2003). The green alga Chlorella sorokiniana was grown with those three aromatics at different concentrations, showing increasing inhibitory effects in the order salicylate < phenol < phenanthrene. However, a satisfactory removal (>85%) was achieved only in the system having both bacteria and algae, incubated under continuous lighting, indicating the synergistic relationship between the algal–bacterial microcosms in the removal of organic pollutants. An algal–bacterial consortium consisting of Chlorella sorokiniana and Pseudomonas migulae was also reported for the degradation of phenanthrene under photosynthetic conditions and without an external source of oxygen (Munoz et al., 2003). In another study, accelerated pyrene degradation by a bacterial-algal consortium was reported under photosynthetic condition (Luo L. et al., 2014; Luo S. et al., 2014). These studies indicate that microalgae can be used singly or along with bacteria as a potential candidate for biodegradation of PAHs. However, more research is needed for optimizing their efficiency to apply them successfully in the bioremediation of PAH-contaminated environment.
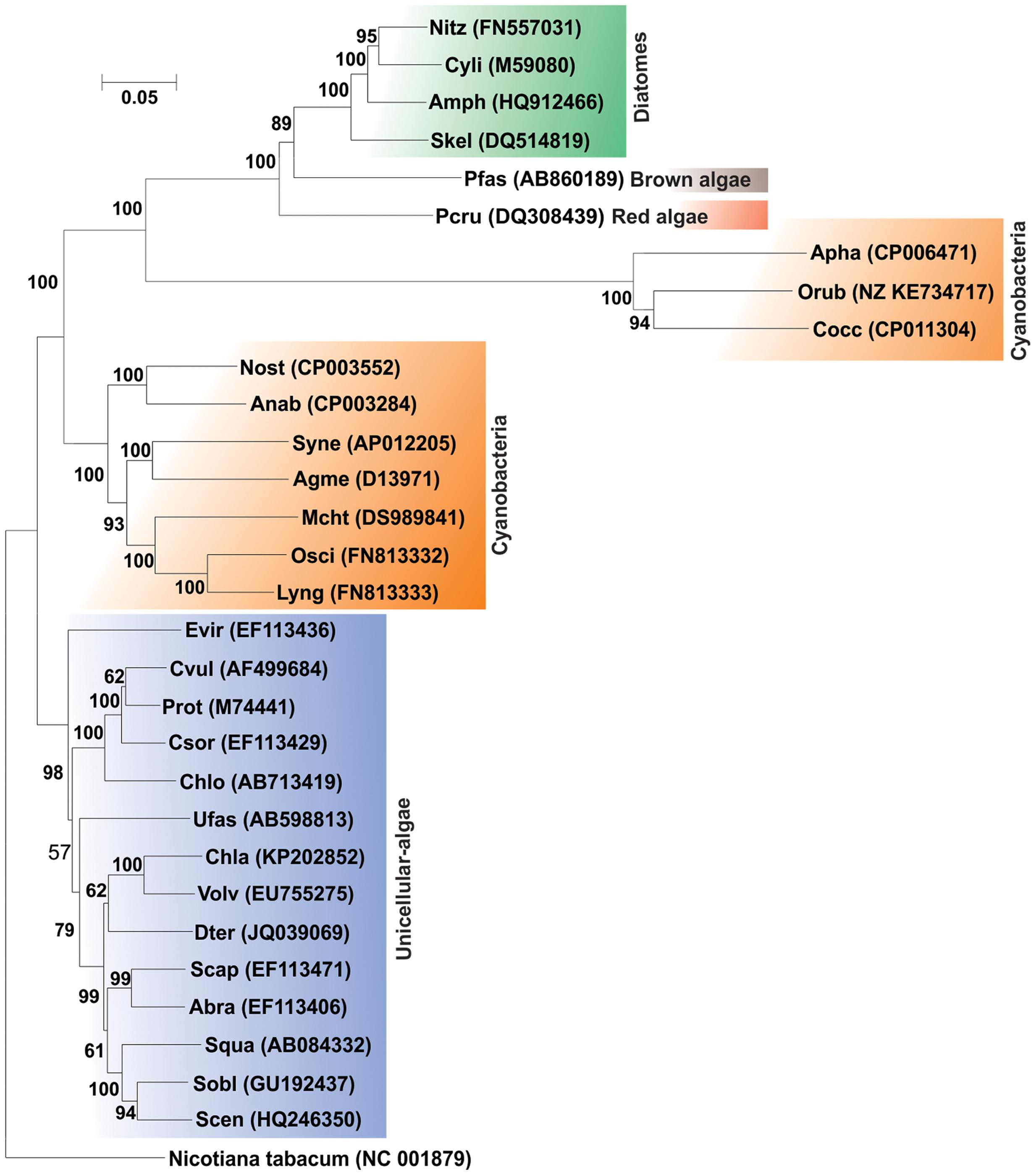
FIGURE 8. Phylogenetic tree based on ribulose 1, 5-bisphosphate carboxylase (rubisco) gene sequence (large subunit) of organisms enlisted in Supplementary Table S2. Organism designations are according to those given in Supplementary Table S2. rubisco gene sequences are taken from representative strains present in NCBI and the respective accession numbers are given within parentheses. rubisco gene sequence (large subunit) of organism Nicotiana tabacum was used as outgroup. Numbers at nodes indicate levels of bootstrap support based on neighbor-joining analysis of 100 resampled datasets. Bootstrap values below 60% are not shown. Bar represents 0.05 substitutions per nucleotide position.
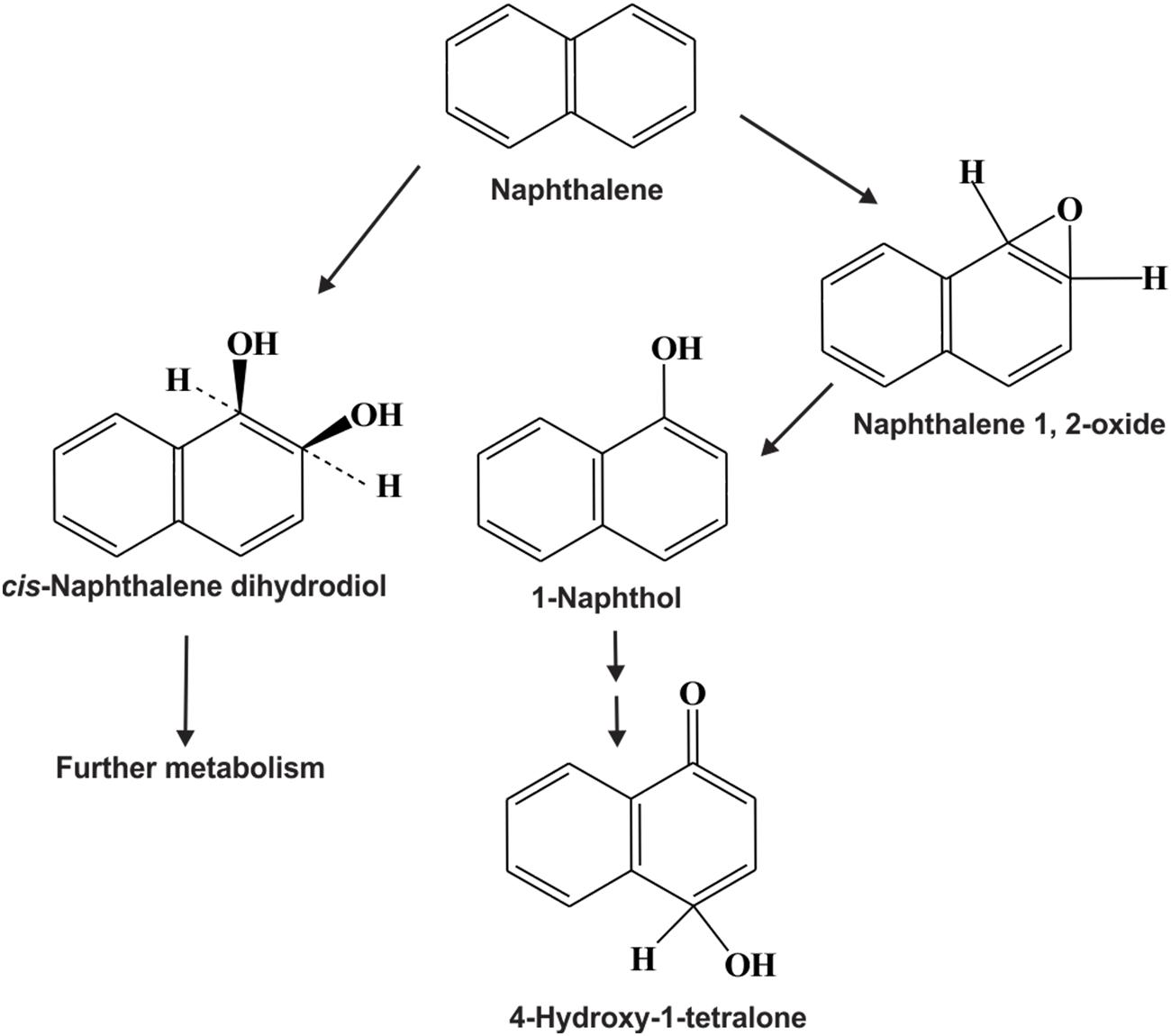
FIGURE 9. Proposed naphthalene transformation pathway by the cyanobacteria, Oscillatoria sp. strain JCM (adapted from Cerniglia et al., 1980a).
Degradation of PAHs by Microbial Consortia and Co-Metabolism
Occasionally it has been observed that a particular microorganism does not have all the genes required for the complete mineralization of a particular organic pollutant like PAH. Therefore, researchers are developing microbial consortia for complete degradation of such pollutants. In those consortia, each microorganism has specialized role in certain degradation steps where, intermediates produced by certain microorganisms are utilized by other members. On the other hand, cometabolism is a phenomenon by which a recalcitrant compound is degraded in the presence of an analogous degradable compound. Several microorganisms can co-metabolize PAHs, and it is a very complex phenomenon. As PAHs in the environment are present as a mixture, co-metabolism plays a very crucial role for bioremediation of PAHs contaminated sites. Co-metabolism of one PAH could have a synergistic effect on the degradation of other PAHs, specifically for the degradation of HMW PAHs (van Herwijnen et al., 2003). In a report, Rhodococcus sp. strain S1 when grown on anthracene has shown to cometabolize phenanthrene to phenanthrene trans-9,10-dihydrodiol (Tongpim and Pickard, 1999). The cometabolic degradation of phenanthrene, fluoranthene, anthracene and dibenzothiophene was reported by the fluorene grown Sphingomonas sp. LB126 (van Herwijnen et al., 2003). In a separate study, cometabolic degradation of a creosote-PAHs mixture including phenanthrene, fluoranthene, and pyrene, by the pyrene-degrading strain Mycobacterium sp. AP1 was reported (Lopez et al., 2008). However, it has been observed that the rate of degradation of individual PAHs was related to their aqueous solubility, for example, the biodegradation rate of fluoranthene and pyrene are significantly lower than that of phenanthrene (Lopez et al., 2008). In another study, cometabolism of acenaphthene and acenaphthylene by the succinate grown Beijerinckia sp. and one of its mutant strain, Beijerinckia sp. strain B8/36 was reported (Schocken and Gibson, 1984). Both the wild type and the mutant strains cometabolize acenaphthene to 1-acenaphthenol, 1-acenaphthenone, 1,2-acenaphthenediol, acenaphthenequinone, and 1,2-dihydroxyacenaphthylene. Furthermore, Sphingobium sp. strain PNB was observed to co-metabolize fluoranthene, acenaphthene, benz[a]anthracene, pyrene and benzo[a]pyrene, in presence of phenanthrene indicating metabolic robustness of the strain (Roy et al., 2013).
Various white-rot fungi can metabolize PAHs along with bacteria in consortium by improving bioavailability of target compounds. Due to lack of suitable enzymes, generally fungi cannot degrade HMW PAHs completely, but can transform them into comparatively polar metabolite(s) with their extracellular enzymes, which can further be degraded by bacteria and other microbes (Sutherland, 1992). Meulenberg et al. (1997) reported that the degradation product of anthracene by white-rot fungi can be mineralized by indigenous mixed bacterial cultures (e.g., activated sludge or soil) more quickly than anthracene itself (Meulenberg et al., 1997). It has been seen that, inoculation of fungal–bacterial co-cultures into the soils contaminated with PAHs, can significantly enhance degradation of HMW PAHs, like chrysene, benzo-[a]anthracene and dibenzo[a,h]anthracene (Boonchan et al., 2000). Consequently, it is assumed that PAH degradation in nature is a result of coordinate steps mediated by fungi and bacteria, with the fungi playing the initial oxidation step (Meulenberg et al., 1997; Boonchan et al., 2000; Cerniglia and Sutherland, 2010). Along with bacteria and fungi, various microalgal strains have been used for the degradation of PAHs in a consortium. Microalgae can be exploited as a potential candidate for degradation of PAHs specifically in aquatic environments. Metabolic competition is another feature of biodegradation when a combinations of two individually degradable PAHs are present in the medium (Bouchez et al., 1995). For example, when LMW PAHs such as phenanthrene and fluorene present in the medium, they can inhibit the degradation of fluoranthene and pyrene (Dean-Ross et al., 2002). Nevertheless, another cause of inhibition is due to the formation of dead-end products that result from co-metabolic degradation of non-growth substrates (Stringfellow and Aitken, 1995).
Factors Affecting the Bioremediation of PAHs
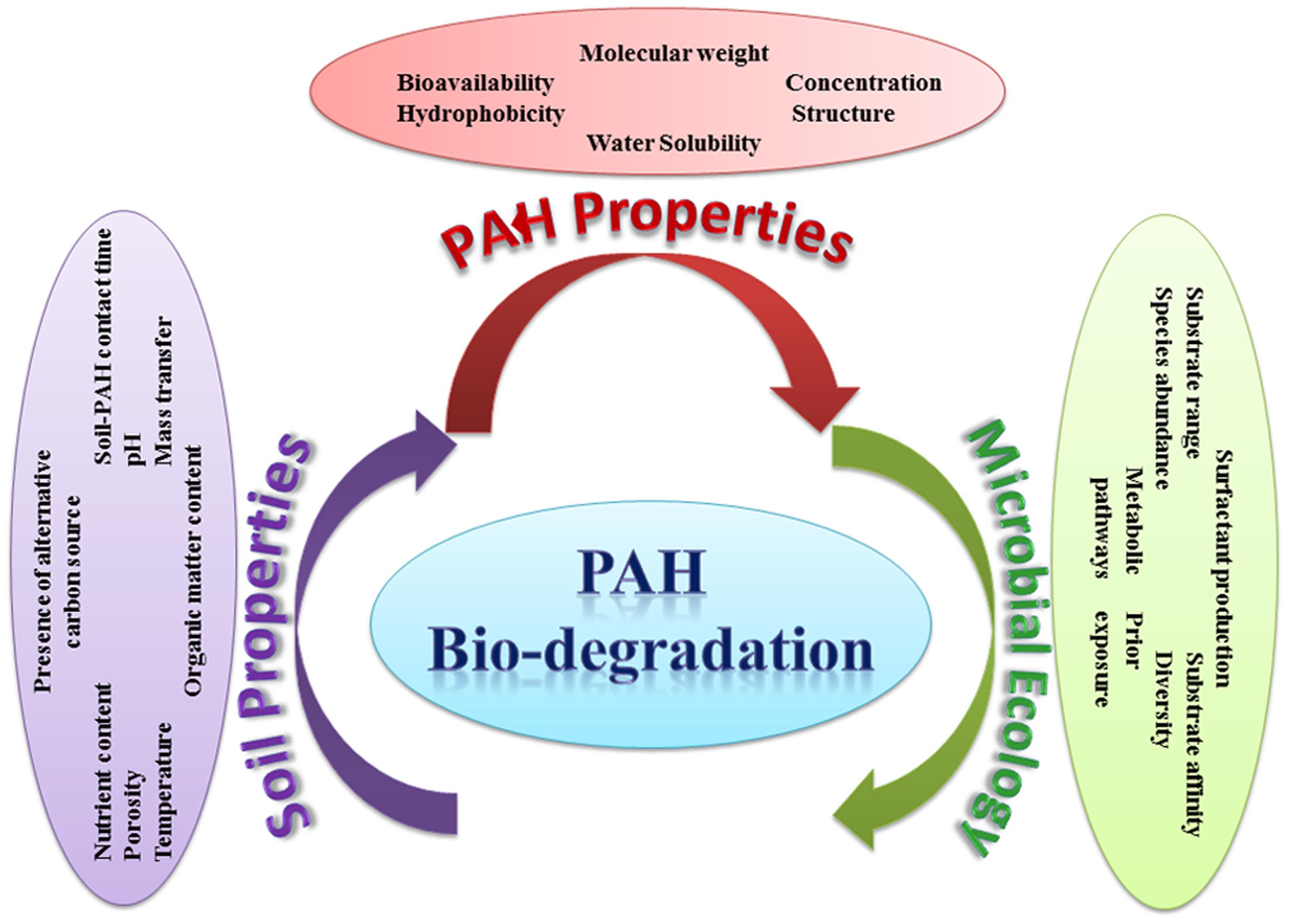
The effectiveness of bioremediation has been mainly investigated under ideal laboratory conditions, having a circum-neutral pH and ambient mesophilic temperature. However, in the real situation, bioremediation can be effective only at sites where environmental conditions permit microbial growth and express associated enzyme activity so that microorganisms can enzymatically attack the pollutants converting them to harmless products. Numerous abiotic and biotic factors (such as pH, nutrient availability and the bioavailability of the pollutants) can apparently differ from site to site, which in turn can influence the process of bioremediation in those environments either by inhibiting or accelerating the growth of the pollutant-degrading microorganisms. Figure 10 illustrates the various abiotic and biotic factors influencing PAHs degradation in soil. The main environmental factors that could affect the rate of biodegradation of PAHs in the environment are summarized below.
Temperature
Temperature has a profound effect on the biodegradation of PAHs in contaminated sites since those places are not always at ambient temperature for activity of the inhabitant microorganisms. When temperature increases, the solubility of PAHs also increases, which in turn increases the bioavailability of PAH molecules. On the other hand, with increasing temperature, dissolved oxygen level decreases, as a result of which the metabolic activity of aerobic mesophilic microorganisms reduces. There are some microorganisms called psychrophiles, which can operate at low temperature, and are in contrast to thermophiles that function at high temperature. At high temperature, some pollutants can get transformed into a new compound and often the daughter compound appears to be more toxic than the parent compound, which in turn inhibits its biodegradation rate (Müller et al., 1998). Generally in most of the studies biodegradation of PAHs have been examined under moderate temperatures, however, there are reports on PAHs biodegradation under extreme temperatures. For example, biodegradation of petroleum hydrocarbons including PAHs was reported in seawater at low temperatures (0–5°C) (Brakstad and Bonaunet, 2006) while the biodegradation of PAHs along with long chain alkanes was documented at very high temperature (60–70°C) by Thermus and Bacillus spp (Feitkenhauer et al., 2003).
pH
pH also plays a significant role in biodegradation processes including that of PAHs. Usually, microorganisms are pH-sensitive and near neutral conditions (6.5–7.5) are favored by most of them for their normal activity. However, it has been seen that in many PAHs contaminated sites, pH is very far from neutral settings. For example, some abandoned gasworks sites frequently contain a huge amount of demolition waste like concrete and brick. Leaching of these materials can result in the increase of pH of the native soil. Moreover, the leaching of some material like coal spoil can generate an acidic environment due to the oxidation of sulfides. These acidic or alkaline conditions can create adverse conditions for microbial activities, which in turn decrease the biodegradation of PAHs in those sites. Hence, it is recommended to adjust the pH at those sites by adding chemicals: basic soils can be treated with ammonium sulfate or ammonium nitrate while acidic soils can be treated by liming with calcium or magnesium carbonate (Bowlen and Kosson, 1995) to generate an environment for effective biodegradation.
Oxygen
Biodegradation of organic contaminants including PAHs can proceed under both aerobic and anaerobic conditions; however, most of the studies focus bioremediation of PAHs under aerobic condition, where oxygen acts as a co-substrate and a rate limiting factor. During aerobic biodegradation of PAHs, oxygen is required for the action of both mono- and dioxygenase enzymes in the initial oxidation of the aromatic rings. For in situ bioremediation of contaminants, sometimes oxygen is added from an external source. The addition of oxygen externally may be achieved by drainage, tilling, and addition of chemicals that release oxygen or by injection of air into the contaminated site (Pardieck et al., 1992; Atlas and Unterman, 1999; Bewley and Webb, 2001). In a study, enhanced intrinsic bioremediation of BTEX compounds in aquifer using an oxygen releasing compound (magnesium peroxide; ORC®) was reported (Odencrantz et al., 1996). Again, in a separate study, in situ bioremediation of an aquifer contaminated with various pollutants, including phenols, BTEX compounds and PAHs was reported, in which sodium nitrate was circulated through the aquifer as the oxygen source, via a series of injection and abstraction wells (Bewley and Webb, 2001).
Nutrients
Availability of nutrients is another rate-limiting factor for successful bioremediation of PAHs contaminated environments. Along with easily metabolized carbon source, microorganisms require various minerals like nitrogen, phosphorus, potassium, and iron for normal metabolism and growth. Thus, supplementation of nutrients, in the poor nutrient content pollutant contaminated sites is required to stimulate the growth of autochthonous microorganisms, to enhance bioremediation of pollutants (Atagana et al., 2003). In marine environments, poor biodegradation of petroleum hydrocarbon is due to a low level of nitrogen and phosphorous (Floodgate, 1984). On the other hand, excessive nutrient availability can also inhibit the bioremediation of pollutants (Chaillan et al., 2006). There are several reports on the negative effects of high nutrient levels on the biodegradation of organic pollutants especially PAHs (Carmichael and Pfaender, 1997; Chaîneau et al., 2005).
Bioavailability
In case of a biological system, the term bioavailability is defined as the fraction of a chemical that can be taken up or transformed by living organisms during the course of the experiment. This fraction can vary under the influence of mass transfer parameters, which include physicochemical processes governing dissolution, desorption and diffusion, hydrological processes like mixing and, finally, biological processes, such as uptake and metabolism (Bosma et al., 1997; Semple et al., 2003). Bioavailability is regarded as one of the most crucial factors in bioremediation of pollutants. Biodegradation of PAHs in the environment is often limited, due to their low aqueous solubility and their strong tendency to sorbs to mineral surfaces (like clays) and organic matters (like black carbon, coal tar humic acids) in the soil matrix, processes which reduce bioavailability. There are numerous reports on poor or unsuccessful remediation of PAHs contaminated sites due to absorption of PAHs in black carbon, coal-tar substances which decreases bioavailability of PAHs (Volkering et al., 1992; Luthy et al., 1993, 1994; Nelson et al., 1996; Northcott and Jones, 2001; Hong et al., 2003; Bucheli et al., 2004; Cornelissen et al., 2005; Liu et al., 2009; Benhabib et al., 2010; Rein et al., 2016). Again, the aqueous solubility of PAHs decreases with increasing molecular weight, which in turn reduces the bioavailability of PAHs. It has been seen that longer any hydrophobic organic contaminants (like PAHs) is in contact with soil, the more irreversible the sorption, and lesser is the chemical and biological extractability of the contaminants, which is known as ‘aging’ of the contaminant (Alexander, 2000; Luo et al., 2012). It has been reported that extractability and bioavailability of PAHs decrease significantly with time in aging processes which can be significantly rate-limiting for in situ bioremediation (Alexander, 2000; Megharaj et al., 2002; Luo et al., 2012; Abdel-Shafy and Mansour, 2016).
Inhibition Due to Excess Substrate or End Product
The major goal of bioremediation is the removal or detoxification of contaminants from a given environment. However, sometimes it is possible that the contaminant gets transformed into a more toxic dead end product. This is why it is essential to confirm that the pollutant is completely mineralized at the end of the treatment (Mendonca and Picado, 2002; Lundstedt et al., 2003). A study using a bioreactor to treat PAH-contaminated gasworks soil using in situ bioremediation monitored both the removal of PAHs and the accumulation of oxy-PAHs, such as PAH-ketones, quinines and coumarins as dead end products (Lundstedt et al., 2003). As oxy-PAHs are more toxic than the parent PAHs, this study highlights the significance of monitoring the metabolites during bioremediation, specifically for toxic dead-end products, and determining the toxicity of the metabolites both before and after treatment (Lundstedt et al., 2003).
Since PAHs in the environment are present as mixtures, the effect of substrate interaction during biodegradation is crucial in determining the fate of PAHs in nature. It has been reported that, when present as a mixture, the high molecular weight PAHs are degraded followed by the degradation of low molecular weight PAHs, by the bacterial community (Mueller et al., 1989). It has been also observed that a high concentration of naphthalene have inhibitory effect on the degradation of other PAHs, by a defined bacterial coculture (Bouchez et al., 1995). Similarly, a competitive inhibition of phenanthrene degradation by naphthalene, methylnaphthalene and fluorene in binary mixtures using two pure cultures was reported (Stringfellow and Aitken, 1995). Thus during in situ bioremediation of PAHs, concentration of PAHs in the contaminated sites, and a chance for the formation of the toxic dead end product(s) need to be considered for effective process development.
Recent Advancement in Molecular Techniques in Understanding Bacterial Degradation of PAHs and Future Directions
Nowadays, bioremediation has become an intensive area of research. However, more advancement has to be made in developing practically efficient microbial bioremediation processes. Although microorganisms have the capability to use numerous organic pollutants as their carbon and energy sources, their proficiency at eliminating such pollutants might not be of top class level for cleaning up present-day pollution. In fact, microorganisms have evolved in the direction of ecological fitness rather than biotechnological efficacy (Schmid et al., 2001). To enhance the catabolic efficiency of a microorganism for bioremediation, bioengineering is a prerequisite. Recent advancement in genetic, genomic, proteomic and metabolomic technologies, which are applied to study bioremediation of organic pollutants, have contributed considerably to enrich our knowledge on various aspects of the physiology, ecology, biochemistry and regulatory mechanisms of the microbial catabolic pathways. Hence, practical utilization of that knowledge is essential to manipulate or reconstruct as well as enhance the natural processes, thereby making strategies for the development of more competent biocatalysts for various biotechnological applications including bioremediation of culprit organic pollutants in the contaminated sites and transformation of toxic chemical into harmless product or other specialty chemicals (Schmid et al., 2001; Carmona et al., 2009).
A considerable advancement in microbial ecology was achieved based on the identification of conserved sequences present in a particular group of microorganisms, most notably the 16S rRNA or 18S rRNA genes which could provide a phylogenetic characterization of the microbial population present in a particular habitat (Pace et al., 1986; Amann et al., 1995). This approach is a significant achievement in the field of bioremediation because by determining the microbiota in a polluted environment it is possible to identify particular microorganisms inhabiting those environments, thereby predicting possible bioremediation potential. With the advent of Denaturing Gradient Gel Electrophoresis (DGGE), it is now possible to analyze the microbial community structure and dynamics of a particular habitat, more precisely (Gallego et al., 2014; Vila et al., 2015). Development of fluorescence in situ hybridization (FISH), which is another technique is quite useful in this field and has often been practiced (Amann et al., 2001).
Generally, there is a positive link between the relative abundance of the genes associated with pollutant removal and the efficiency of bioremediation. However, sometimes it is possible that the genes associated with pollutant removal can be present but not expressed. So, there is an increasing interest in quantifying the mRNA for key catabolic genes via real-time PCR (Schneegurt and Kulpa, 1998; Debruyn and Sayler, 2009). In addition transcriptomics, DNA-based stable isotope probing, single cell genomics and DNA microarray techniques are also emerging for application in the field of bioremediation (Wilson et al., 1999; Denef et al., 2005, 2006; Chain et al., 2006; Parnell et al., 2006; Macaulay and Voet, 2014; Mason et al., 2014; Mishamandani et al., 2014). DNA microarray which is a high-throughput version of DNA hybridization technique can detect an enormous number of genes in a single test. One of the recent applications of microarray in the PAHs biodegradation is the construction of GeoChip (He et al., 2010; Nostrand et al., 2012).
The advancement in genome sequencing technology practically revolutionized the field of bioremediation. By whole genome sequencing, it is now possible to study the physiology of microorganisms associated with pollutant removal in more details (Nierman and Nelson, 2002; Buermans and den Dunnen, 2014). Presently many complete or nearly complete genome sequences of cultivable microorganisms are available which have potential catabolic activity. However, unfortunately by using modern microbiological and molecular techniques over the past 40 years so far only an extremely small fraction (~1%) of the total microbial diversity has been cultivated from all habitats investigated (Hugenholtz and Pace, 1996; Hugenholtz et al., 1998). While culturing novel environmental microorganisms is indispensable (Zinder, 2002), perhaps the greatest promise to date comes from metagenomic approaches, which are the only ones currently available to adequately utilize the tremendous microbial diversity- one of the richest sources on earth (Handelsman et al., 1998). Although there are some associated challenges in metagenome-based approaches, scientists already overcame many of those problems (Liles et al., 2008; Mareckova et al., 2008). The progressively decreasing cost and increasing speed of DNA sequencing has made it possible to sequence billions of bases of metagenomic DNA. Culturable aromatics degrading bacteria are being isolated mostly on the basis of their ability to utilize aromatic compound as their sole carbon and energy sources and by genome sequencing, scientist can identify all the genes necessary to completely degrade the compound. Therefore, inclusive knowledge of the catabolic potential of a contaminated site virtually remains unknown by using culture-dependent techniques. High throughput metagenomics reduces that bottleneck. However, at present, relatively little resources have been spent for the sequencing of soil or marine metagenomes contaminated with aromatic pollutants, compared to those committed to the human microbiome (Blow, 2008). That is why there is a scarcity of information about the xenobiotics degrading novel bacteria that are difficult to culture in the laboratory. Sites contaminated with toxic chemicals have become biotechnological gold mines because the indigenous microorganisms may have evolved the necessary enzymes, to degrade or transform those toxic contaminants to an inanimate one, quite different from those found in common cultivable microorganisms (Handelsman et al., 1998; Galvao et al., 2005; Boubakri et al., 2006). In addition, sequencing of the soil or aquatic metagenome will also provide insights into the ecology of microorganisms which in turn will help to identify who the dominant and rare community members are and what are their probable roles in the degradation of recalcitrant molecules like PAHs and other related xenobiotics.
Recently another two techniques metaproteomics and metabolomics have been utilized to unfold various aspects of environmental microbiology and have shown their promise in the field of bioremediation (Nesatyy and Suter, 2007; Keum et al., 2008). Proteomics is an efficient technology to recognize proteins and their roles associated with catabolism of PAHs while metabolomics can be exploited to identify metabolites produced during PAHs biodegradation. Figure 11 illustrates a summarized representation of the molecular techniques involved in studying microbial degradation of PAH. In near future, functional metagenomics, metaproteomics, metabolomics, metatranscriptomics and DNA microarrays will become crucial tools to elucidate the mechanisms of biodegradation of PAHs in the environment, and will offer more information on as-yet-uncultured organisms associated with PAHs bioremediation (Wilson et al., 1999; Amann et al., 2001; Fromin et al., 2002; Liang, 2002; Zhou and Thompson, 2002; Baldwin et al., 2003; Ginige et al., 2004). Moreover, in silico biology is being progressively applied in different aspects in the field of bioremediation (Kweon et al., 2010; Chakraborty et al., 2012; Khara et al., 2014). Thus, it is expected that emerging molecular biological, analytical and computational methods which can predict the activity of microorganisms involved in biodegradation, should transform the bioremediation field from a largely empirical practice into a branch of modern science.
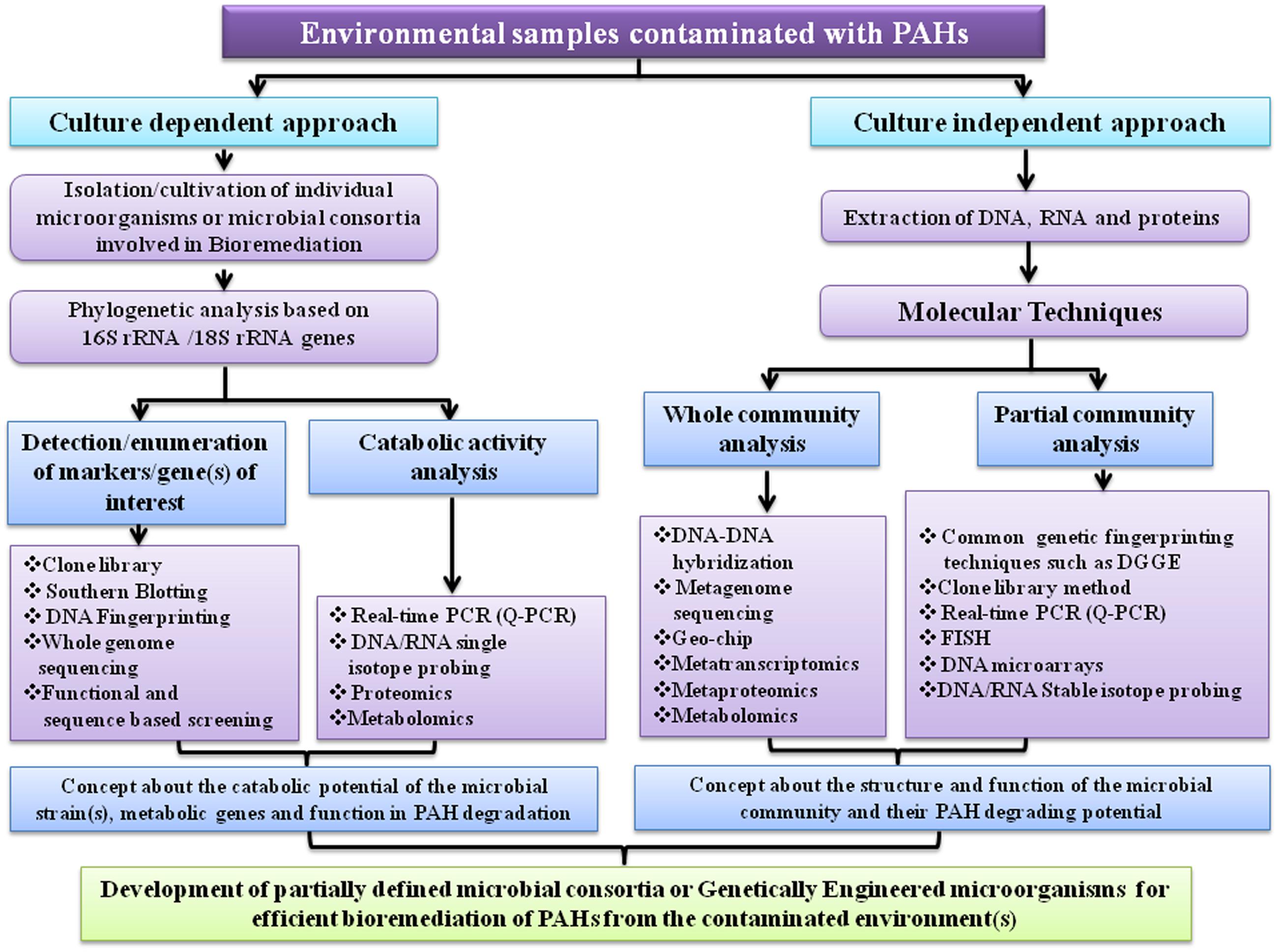
FIGURE 11. Schematic diagram summarizing different molecular techniques as future directions for better understanding of the microbial PAH degradation and ecologically sustainable bioremediation of sites contaminated with PAHs.
Approaches to the Bioremediation of PAH-Contaminated Environments
Various types of bioremediation technologies can be employed for contaminant removal. Intrinsic bioremediation (also called natural attenuation or ‘in situ’ bioremediation) depends on the bioremediation potential of the indigenous micro flora dwelling at the site contaminated with pollutants. These techniques provide the treatment at the site itself avoiding excavation and transport of contaminants, which makes them the most desirable options due to lower cost and fewer disturbances. On the other hand extrinsic bioremediation (also called ‘ex-situ’ bioremediation) mainly involves the physical removal of the contaminated material to another location for treatment (Carberry and Wik, 2001). While in the case of “biostimulation” process, it is generally possible to stimulate the indigenous microorganisms to use the contaminants as a food source at a much greater rate by compensating limiting parameters, for example, by introducing additional oxygen or nutrients to the indigenous population (Straube et al., 2003). Another method, “bioaugmentation,” involves introduction of exogenous microorganisms into the contaminated environment which are capable of degrading the target pollutants, either with or without additional nutrients (Straube et al., 2003). Whereas, “humification” is a process by which strongly persistent substances in the polluted environment are incorporated into the humic substances mostly by means of enzymatic reactions (Megharaj et al., 2011). In addition, “phytoremediation” involves removal of environmental contaminants using plants (Haritash and Kaushik, 2009; Megharaj et al., 2011). Another bioremediation technology called ‘landfarming,’ is used to stimulate indigenous biodegradative microorganisms by providing nutrients, water and oxygen, in order to facilitate aerobic degradation of contaminants (Megharaj et al., 2011). While ‘composting’ involves the degradation of pollutant along with other agricultural wastes such as manure etc. (Semple et al., 2001). Hence, it becomes clear that regardless of the method chosen, an ideal bioremediation strategy can be designed only on the basis of knowledge about the microorganisms and the contaminant present within the polluted environments, along with their catabolic potential and response to changes in the environmental conditions.
Treatment of Soils and Sediments
There are some reports on the treatment of PAH contaminated soils and sediments by in situ or ex situ bioremediation methods. Straube et al. (2003) reported that a pilot-scale landfarming treatment of PAH-contaminated soil from a wood treatment facility was achieved by biostimulation of the soil with water, ground rice hulls as a bulking agent, and palletized dried blood as a nitrogen source and bioaugmentation of the microbial community with an inoculum of Pseudomonas aeruginosa strain 64 (Straube et al., 2003). It has been seen that within 1 year of treatment ~86% of total PAHs were removed from the soil including a moderate reduction in HMW PAHs such as pyrene, BaP etc. In a separate study on the treatment of an aged gasworks soil contaminated with aromatic compounds including PAHs, it has been observed that LMW PAHs and heterocyclic compounds were degraded more quickly than the HMW counterparts (Lundstedt et al., 2003). In addition, the unsubstituted PAHs were degraded faster than the related alkyl-PAHs as well as nitrogen-containing heterocyclics. Treatment of soil at a tar contaminated site via composting along with conventional land treatment process revealed that composting led to more extensive PAH removal than did by two different land treatment processes (Guerin, 2000). Sasek et al. (2003) reported the remediation of a manufactured-gas plant soil contaminated with PAHs via composting. In this study, treatment of soil was performed in a thermally insulated chamber using mushroom compost containing wheat straw, chicken manure and gypsum, where at the end of 54 days, removal of ~20–60% of individual PAHs was reported, while ~37–80% of individual PAHs degradation was observed after another 100 days of composting (Sasek et al., 2003). Moreover, studies on the removal of PAHs present in contaminated soil using the associated bacterial communities in two aerobic, lab-scale, slurry-phase bioreactors, which were run semi-continuously and fed either on weekly or monthly basis, showed that most of the PAHs, including HMW PAHs were biodegraded to a greater extent in the weekly fed bioreactor for up to 76% (Singleton et al., 2011).
Treatment of Waters
Polycyclic aromatic hydrocarbons-polluted groundwater can also be bioremediated through both in situ and ex situ bioremediation procedures. However, due to the high cost associated with the extraction and shipment, ex situ treatment of contaminated groundwater is not generally performed. Few reports are documented on the treatment of PAH contaminated water. Bewley and Webb (2001) reported the in situ bioremediation of an aquifer which was contaminated with diverse pollutants, including phenols, BTEX compounds and PAHs. The site was bioremediated using a procedure involving a combination of bioaugmentation and biostimulation. Nutrients (a commercial mixture of urea and diammonium phosphate), a commercially available phenol-degrading mixed bacterial inoculum (PHENOBAC, Microbac Ltd, Durham), and sodium nitrate (an oxygen source) were circulated through the aquifer via a series of injection and abstraction wells. After treatment for over two and half years, the mean concentration of PAHs was reduced to 0.9 μgL-1 from 11 μgL-1 along with reduction of other contaminant (Bewley and Webb, 2001). Apart from that, groundwater remediation technology of petroleum-derived compounds (PDCs) including PAHs based on enhanced solubility of PDCs in humic acid was also reported (Van Stempvoort et al., 2002). Zein et al. (2006) reported the treatment of groundwater contaminated with PAHs, gasoline hydrocarbons, and methyl tert-butyl ether using an ex situ aerobic biotreatment system and it was observed that after 10 months of treatment, the concentration of PAHs was reduced to >99% along with other pollutants (Zein et al., 2006).
Treatment with Genetically Engineered Microorganisms (GEMs)
The bioremediation of PAHs contaminated site is generally very slow because there are a number of biotic and abiotic factors responsible for successful bioremediation. In addition, the incomplete success rates might be due to the fact that some places are heavily contaminated, and hence, the microorganisms are incapable to grow and degrade the contaminant at the same rate at which they are introduced into the environment. Although there is very few information on the use of GEMs in bioremediation, they can be a promising candidate for such processes (Ang et al., 2005; Singh et al., 2008; Megharaj et al., 2011). Using genetic engineering it is possible to enhance the activity or broad substrate specificity of certain enzymes associated with PAH-degrading pathways, which in turn will improve the mineralization of those pollutants in the environment (Timmis et al., 1994; Timmis and Pieper, 1999; Ang et al., 2005; Singh et al., 2008). In recent times, various molecular biology tools such as gene conversion, gene duplication, and transposon or plasmids mediated gene delivery are available, which might play vital roles to boost up the biodegrading potential of naturally occurring microorganisms (Timmis et al., 1994; Timmis and Pieper, 1999; Ang et al., 2005; Singh et al., 2008; Megharaj et al., 2011). However, it is crucial to ensure the stability of GEMs prior to their field application since the catabolic activity of released GEM is associated largely with the stability of the recombinant plasmid introduced into the organism (Brunel et al., 1988; Shaw et al., 1992; Samanta et al., 2002). As GEMs that are released into a contaminated site can spread to other places and multiply under favorable conditions, so before releasing into the environment there should be a clear understanding of the possible side effects associated with GEMs, which would possibly guide the restriction of their application in pollutant abatement (Samanta et al., 2002; Ang et al., 2005; Singh et al., 2008; Megharaj et al., 2011).
Conclusion
In last few decades, there has been a great deal of progress in the study of the bioremediation of PAHs. Numerous microorganisms have been isolated and characterized having PAHs catabolic potentials. In addition, many unique enzymes with different catabolic efficiency associated with PAHs degradation have been purified and different novel biochemical pathways for PAHs degradation have been elucidated. Moreover, many PAH catabolic operons have been sequenced, and their regulatory mechanism for PAH degradation has been determined. The advancement in genetic, genomic, proteomic and metabolomic approaches, which are employed to study catabolism of organic pollutants have contributed remarkably in understanding the physiology, ecology, biochemistry of PAHs degrading microorganisms. However, more detail research is a prerequisite to determine exactly what is going on in PAH-contaminated environment. In addition, there are still various aspects of bioremediation of PAHs that remain unknown or otherwise have insufficient information, which requires future attention. There is very scarce knowledge on genes, enzymes as well as the molecular mechanism of PAHs degradation in high salinity environments, or anaerobic environments. Also, there is very little information on the transmembrane trafficking of PAHs and their metabolites. Various transporters have been assumed to be participating in the transport of PAHs into microorganisms, but till date, none has been characterized.
There are various factors which may affect bioremediation of PAHs in a contaminated environment. Adjustment of oxygen concentration, pH, temperature, nutrient availability and improvement of bioavailability may increase PAHs degradation. It is seen that in contaminated soils and sediments, PAHs get entrapped in coal tar or black carbon particles, which results in unsuccessful remediation due to decrease in PAHs bioavailability (Bucheli et al., 2004). This phenomenon is a major bottleneck for successful remediation of PAHs contaminated environments (Ortega-Calvo and Alexander, 1994). Some microorganisms are known to excrete biosurfactants which enhance the bioavailability of organic pollutants. Many microorganisms exhibit chemotaxis toward pollutants. These strategies lead to enhanced degradation of organic pollutants. The addition of small amount of biosurfactant, which increases the bioavailability of PAHs, or some merely toxic chemicals, like salicylic acid, which induce PAHs catabolic operons may enhance biodegradation of PAHs in the environment. It has been seen that organic amendments influence the indigenous microbial community as well as efficiency of bioremediation of PAHs in contaminated soil (Gandolfi et al., 2010; Loick et al., 2012; Lukić et al., 2016; Rein et al., 2016). So adjustment of organic matter content of a polluted site with exogenous addition of compost or other substances like buffalo manure, food and vegetables waste may enhance the bioremediation efficiency of PAH polluted site. In addition, instead of treating with a single microorganism, a defined microbial consortium may give a better result in some cases for successful remediation of a contaminated site. So, more research is required on the catabolic capabilities of PAHs degrading microbial consortia, which will further deepen our understanding on the microbial consortia-mediated remediation of contaminated environments, and will help to develop potential microbial consortium having robust pollutant degrading potential. It is also possible to treat the contaminated site sequentially with fungi, bacteria and algae or in combination for more efficient pollutant removal.
In addition, genetic engineering can be employed to boost the catabolic efficiency of microorganisms used in bioremediation. Today, scientists are capable to create unique PAHs metabolic pathways by recombining different catabolic genes from different organisms in a single host cell. In this way it is possible to enhance the substrate specificity of a catabolic pathway to degrade new substrates, complete partial pathways and also to construct novel pathways not found in nature, which may permit the mineralization of highly recalcitrant compounds and avoid the accumulation of toxic dead end product. However, before releasing such GMOs into the environment, it is necessary to check thoroughly the possibility of any unwanted side effects produced by those GMOs. In addition, authorities should be convinced that the GMOs are safer, cheaper, and more efficient than the present alternatives. Thus, it is anticipated that the compiled information present in this manuscript will open up new avenues to the researchers, and the field of bioremediation will revolutionize in near future.
Author Contributions
DG translated the concept and wrote the manuscript. DG and SG analyzed the data, edited and formatted the manuscript. TD and YA generated the concept and ideas, critically revised the manuscript and approved the final version for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Yeungnam University Research Grant (2015).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01369
References
Abdel-Shafy, H. I., and Mansour, M. S. M. (2016). A review on polycyclic aromatic hydrocarbons: source, environmental impact, effect on human health and remediation. Egypt. J. Petrol. 25, 107–123. doi: 10.1016/j.ejpe.2015.03.011
Agency for Toxic Substances and Disease Registry [ATSDR] (1990). Toxicological profile for polycyclic aromatic hydrocarbons. Acenaphthene, Acenaphthylene, Anthracene, Benzo[a]anthracene, Benzo[a]pyrene, Benzo[b]fluoranthene, Benzo[g,i,h]perylene, Benzo[k]fluoranthene, Chrysene, Dibenzo[a,h]anthracene, Fluoranthene, Fluorene, Indeno[1,2,3-c,d]pyrene, Phenanthrene, Pyrene. Atlanta, GA: Agency for Toxic Substances and Disease Registry.
Alexander, M. (2000). Aging bioavailability, and overestimation of risk from environmental pollutants. Environ. Sci. Technol. 34, 4259–4265.
Alonso-Gutierrez, J., Figueras, A., Albaiges, J., Jimenez, N., Vinas, M., Solanas, A. M., et al. (2009). Bacterial communities from shoreline environments (costa da morte, northwestern Spain) affected by the prestige oil spill. Appl. Environ. Microbiol. 75, 3407–3418. doi: 10.1128/AEM.01776-08
Amann, R., Fuchs, B. M., and Behrens, S. (2001). The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12, 231–236. doi: 10.1016/S0958-1669(00)00204-4
Amann, R. I., Ludwig, W., and Schleifer, K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169.
Andersson, B. E., Lundstedt, S., Tornberg, K., Schnurer, Y., Oberg, L. G., and Mattiasson, B. (2003). Incomplete degradation of polycyclic aromatic hydrocarbons in soil inoculated with wood-rotting fungi and their effect on the indigenous soil bacteria. Environ. Toxicol. Chem. 22, 1238–1243. doi: 10.1002/etc.5620220608
Andreoni, V., Cavalca, L., Rao, M. A., Nocerino, G., Bernasconi, S., Dell’Amico, E., et al. (2004). Bacterial communities and enzyme activities of PAHs polluted soils. Chemosphere 57, 401–412. doi: 10.1016/j.chemosphere.2004.06.013
Andreoni, V., and Gianfreda, L. (2007). Bioremediation and monitoring of aromatic-polluted habitats. Appl. Microbiol. Biotechnol. 76, 287–308. doi: 10.1007/s00253-007-1018-5
Ang, J. L., Zhao, H., and Obbard, J. P. (2005). Recent advances in the bioremediation of persistent organic pollutants via biomolecular engineering. Enzyme Microb. Technol 37, 487–496. doi: 10.1016/j.enzmictec.2004.07.024
Atagana, H. I., Haynes, R. J., and Wallis, F. M. (2003). Optimization of soil physical and chemical conditions for the bioremediation of creosote-contaminated soil. Biodegradation 14, 297–307. doi: 10.1023/A:1024730722751
Atlas, R. M., and Unterman, R. (1999). “Bioremediation,” in Manual of Industrial Microbiology and Biotechnology, 2nd Edn, eds A. C. Demain and J. E. Davies (Washington, DC: ASM Press), 666–681.
Baldwin, B. R., Nakatsu, C. H., and Nies, L. (2003). Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl. Environ. Microbiol. 69, 3350–3358. doi: 10.1128/AEM.69.6.3350-3358.2003
Bamforth, S. M., and Singleton, I. (2005). Bioremediation of polycyclic aromatic hydrocarbons: current knowledge and future directions. J. Chem. Technol. Biotechnol. 80, 723–736. doi: 10.1002/jctb.1276
Basta, T., Buerger, S., and Stolz, A. (2005). Structural and replicative diversity of large plasmids from sphingomonads that degrade polycyclic aromatic compounds and xenobiotics. Microbiology 151(Pt 6), 2025–2037. doi: 10.1099/mic.0.27965-0
Benhabib, K., Faure, P., Sardin, M., and Simonnot, M. O. (2010). Characteristics of a solid coal tar sampled from a contaminated soil and of the organics transferred into water. Fuel 89, 352–359. doi: 10.1016/j.fuel.2009.06.009
Bewley, R. J. F., and Webb, G. (2001). In situ bioremediation of groundwater contaminated with phenols, BTEX and PAHs using nitrate as electron acceptor. Land Contam. Reclam. 9, 335–347.
Bezalel, L., Hadar, Y., and Cerniglia, C. E. (1997). Enzymatic mechanisms involved in phenanthrene degradation by the white rot fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 63, 2495–2501.
Bezalel, L., Hadar, Y., Fu, P. P., Freeman, J. P., and Cerniglia, C. E. (1996). Initial oxidation products in the metabolism of pyrene, anthracene, fluorene, and dibenzothiophene by the white rot fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 62, 2554–2559.
Binkova, B., Giguere, Y., Rossner, P. Jr., Dostal, M., and Sram, R. J. (2000). The effect of dibenzo[a,1]pyrene and benzo[a]pyrene on human diploid lung fibroblasts: the induction of DNA adducts, expression of p53 and p21(WAF1) proteins and cell cycle distribution. Mutat. Res. 471, 57–70. doi: 10.1016/S1383-5718(00)00111-X
Blow, N. (2008). Metagenomics: exploring unseen communities. Nature 453, 687–690. doi: 10.1038/453687a
Blumer, M. (1976). Polycyclic aromatic compounds in nature. Sci. Am. 234, 35–45. doi: 10.1038/scientificamerican0376-34
Bogan, B. W., Lamar, R. T., and Hammel, K. E. (1996a). Fluorene oxidation in vivo by phanerochaete chrysosporium and in vitro during manganese peroxidase-dependent lipid peroxidation. Appl. Environ. Microbiol. 62, 1788–1792.
Bogan, B. W., Schoenike, B., Lamar, R. T., and Cullen, D. (1996b). Expression of lip genes during growth in soil and oxidation of anthracene by Phanerochaete chrysosporium. Appl. Environ. Microbiol. 62, 3697–3703.
Bonfa, M. R., Grossman, M. J., Mellado, E., and Durrant, L. R. (2011). Biodegradation of aromatic hydrocarbons by Haloarchaea and their use for the reduction of the chemical oxygen demand of hypersaline petroleum produced water. Chemosphere 84, 1671–1676. doi: 10.1016/j.chemosphere.2011.05.005
Boonchan, S., Britz, M. L., and Stanley, G. A. (2000). Degradation and mineralization of high-molecular-weight polycyclic aromatic hydrocarbons by defined fungal-bacterial cocultures. Appl. Environ. Microbiol. 66, 1007–1019. doi: 10.1128/AEM.66.3.1007-1019.2000
Borde, X., Guieysse, B., Delgado, O., Munoz, R., Hatti-Kaul, R., Nugier-Chauvin, C., et al. (2003). Synergistic relationships in algal-bacterial microcosms for the treatment of aromatic pollutants. Bioresour. Technol. 86, 293–300. doi: 10.1016/S0960-8524(02)00074-3
Bosch, R., Garcia-Valdes, E., and Moore, E. R. (2000). Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene 245, 65–74. doi: 10.1016/S0378-1119(00)00038-X
Bose, A., Rogers, D. R., Adams, M. M., Joye, S. B., and Girguis, P. R. (2013). Geomicrobiological linkages between short-chain alkane consumption and sulfate reduction rates in seep sediments. Front. Microbiol. 4:386. doi: 10.3389/fmicb.2013.00386
Bosma, T. N. P., Middeldorp, P. J. M., Schraa, G., and Zender, A. J. B. (1997). Mass transfer limitation of biotransformation: quantifying bioavailability. Environ. Sci. Technol. 31, 248–252. doi: 10.1080/08927014.2012.662641
Boubakri, H., Beuf, M., Simonet, P., and Vogel, T. M. (2006). Development of metagenomic DNA shuffling for the construction of a xenobiotic gene. Gene 375, 87–94. doi: 10.1016/j.gene.2006.02.027
Bouchez, M., Blanchet, D., and Vandecasteele, J. P. (1995). Degradation of polycyclic aromatic hydrocarbons by pure strains and by defined strain associations: inhibition phenomena and cometabolism. Appl. Microbiol. Biotechnol. 43, 156–164. doi: 10.1007/BF00170638
Bowlen, G. F., and Kosson, D. S. (1995). “In situ processes for bioremediation of BTEX and petroleum fuel products,” in Microbial Transformation and Degradation of Toxic Organic Chemicals, eds L. Y. Young and C. E. Cerniglia (New York, NY: Wiley-Liss), 514–544.
Boxall, A. B., and Maltby, L. (1997). The effects of motorway runoff on freshwater ecosystems: 3. Toxicant confirmation. Arch. Environ. Contam. Toxicol. 33, 9–16. doi: 10.1007/s002449900216
Brakstad, O. G., and Bonaunet, K. (2006). Biodegradation of petroleum hydrocarbons in seawater at low temperatures (0-5 degrees C) and bacterial communities associated with degradation. Biodegradation 17, 71–82. doi: 10.1007/s10532-005-3342-8
Brakstad, O. G., and Lodeng, A. G. (2005). Microbial diversity during biodegradation of crude oil in seawater from the North Sea. Microb. Ecol. 49, 94–103. doi: 10.1007/s00248-003-0225-6
Brunel, B., Cleyet-Marel, J. C., Normand, P., and Bardin, R. (1988). Stability of Bradyrhizobium japonicum inoculants after introduction into soil. Appl. Environ. Microbiol. 54, 2636–2642.
Bucheli, T. D., Blum, F., Desaules, A., and Gustafsson, O. (2004). Polycyclic aromatic hydrocarbons, black carbon, and molecular markers in soils of Switzerland. Chemosphere 56, 1061–1076. doi: 10.1016/j.chemosphere.2004.06.002
Buermans, H. P., and den Dunnen, J. T. (2014). Next generation sequencing technology: advances and applications. Biochim. Biophys. Acta 1842, 1932–1941. doi: 10.1016/j.bbadis.2014.06.015
Carberry, J. B., and Wik, J. (2001). Comparison of ex situ and in situ bioremediation of unsaturated soils contaminated by petroleum. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 36, 1491–1503. doi: 10.1081/ESE-100105726
Carmichael, L. M., and Pfaender, F. K. (1997). The effect of inorganic and organic supplements on the microbial degradation of phenanthrene and pyrene in soils. Biodegradation 8, 1–13. doi: 10.1023/A:1008258720649
Carmona, M., Zamarro, M. T., Blazquez, B., Durante-Rodriguez, G., Juarez, J. F., Valderrama, J. A., et al. (2009). Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol. Mol. Biol. Rev. 73, 71–133. doi: 10.1128/MMBR.00021-08
Cerniglia, C. E. (1984). Microbial metabolism of polycyclic aromatic hydrocarbons. Adv. Appl. Microbiol. 30, 31–71. doi: 10.1016/S0065-2164(08)70052-2
Cerniglia, C. E. (1992). Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3, 351–368. doi: 10.1007/BF00129093
Cerniglia, C. E., Gibson, D. T., and Van Baalen, C. (1979). Algal oxidation of aromatic hydrocarbons: formation of 1-naphthol from naphthalene by Agmenellum quadruplicatum, strain PR-6. Biochem. Biophys. Res. Commun. 88, 50–58. doi: 10.1016/0006-291X(79)91695-4
Cerniglia, C. E., Baalen, C. V., and Gibson, D. T. (1980a). Metabolism of naphthalene by cyanobacterium Oscillatoria sp. Strain JCM. J. Gen. Microbiol. 116, 485–494.
Cerniglia, C. E., Gibson, D. T., and Van Baalen, C. (1980b). Oxidation of naphthalene by cyanobacteria and microalgae. J. Gen. Microbiol. 116, 495–500.
Cerniglia, C. E., and Heitkamp, M. A. (1989). “Microbial degradation of polycyclic aromatic hydrocarbons (PAH) in the aquatic environment,” in Metabolism of polycyclic aromatic hydrocarbons in the aquatic environment, ed. U. Varanasi (Boca Raton, FL: CRC Press).
Cerniglia, C. E., and Sutherland, J. B. (2010). “Degradation of polycyclic aromatic hydrocarbons by fungi,” in Handbook of Hydrocarbon and Lipid Microbiology, eds K. N. Timmis, T. J. McGenity, J. R. van der Meer, and V. de Lorenzo (Berlin: Springer), 2080–2110.
Chaillan, F., Chaineau, C. H., Point, V., Saliot, A., and Oudot, J. (2006). Factors inhibiting bioremediation of soil contaminated with weathered oils and drill cuttings. Environ. Pollut. 144, 255–265. doi: 10.1016/j.envpol.2005.12.016
Chain, P. S., Denef, V. J., Konstantinidis, K. T., Vergez, L. M., Agullo, L., Reyes, V. L., et al. (2006). Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. U.S.A. 103, 15280–15287. doi: 10.1073/pnas.0606924103
Chakraborty, J., Ghosal, D., Dutta, A., and Dutta, T. K. (2012). An insight into the origin and functional evolution of bacterial aromatic ring-hydroxylating oxygenases. J. Biomol. Struct. Dyn. 30, 419–436. doi: 10.1080/07391102.2012.682208
Chan, S. M., Luan, T., Wong, M. H., and Tam, N. F. (2006). Removal and biodegradation of polycyclic aromatic hydrocarbons by Selenastrum capricornutum. Environ. Toxicol. Chem. 7, 1772–1779. doi: 10.1897/05-354R.1
Chang, W. T., Fang, M. D., Lee, C. L., and Brimblecombe, P. (2014). Measuring bioavailable PAHs in estuarine water using semipermeable membrane devices with performance reference compounds. Mar. Pollut. Bull. 89, 376–383. doi: 10.1016/j.marpolbul.2014.09.031
Chaîneau, C. H., Rougeux, G., Yéprémian, C., and Oudot, J. (2005). Effects of nutrient concentration on the biodegradation of crude oil and associated microbial populations in the soil. Soil Biol. Biochem. 37, 1490–1497. doi: 10.1016/j.soilbio.2005.01.012
Clemente, A. R., Anazawa, T. A., and Durrant, L. R. (2001). Biodegradation of polycyclic aromatic hydrocarbons by soil fungi. Braz. J. Microbiol. 32, 255–261. doi: 10.1590/S1517-83822001000400001
Comber, M. I. H., den Haan, K. H., Djemel, N., Eadsforth, C. V., King, D., Paumen, M. L., et al. (2012). Primary Biodegradation of Petroleum Hydrocarbons in Seawater. Concawe Report, No.10/12. Brussels: Concawe.
Cornelissen, G., Gustafsson, O., Bucheli, T. D., Jonker, M. T., Koelmans, A. A., and van Noort, P. C. (2005). Extensive sorption of organic compounds to black carbon, coal, and kerogen in sediments and soils: mechanisms and consequences for distribution, bioaccumulation, and biodegradation. Environ. Sci. Technol. 39, 6881–6895. doi: 10.1021/es050191b
Coulon, F., McKew, B. A., Osborn, A. M., McGenity, T. J., and Timmis, K. N. (2007). Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ. Microbiol. 9, 177–186. doi: 10.1111/j.1462-2920.2006.01126.x
Cravo-Laureau, C., and Duran, R. (2014). Marine coastal sediments microbial hydrocarbon degradation processes: contribution of experimental ecology in the omics’ era. Front. Microbiol. 5:39. doi: 10.3389/fmicb.2014.00039
Cui, Z., Lai, Q., Dong, C., and Shao, Z. (2008). Biodiversity of polycyclic aromatic hydrocarbon-degrading bacteria from deep sea sediments of the Middle Atlantic Ridge. Environ. Microbiol. 10, 2138–2149. doi: 10.1111/j.1462-2920.2008.01637.x
Dean-Ross, D., Moody, J., and Cerniglia, C. E. (2002). Utilization of mixtures of polycyclic aromatic hydrocarbons by bacteria isolated from contaminated sediment. FEMS Microbiol. Ecol. 41, 1–7. doi: 10.1111/j.1574-6941.2002.tb00960.x
Debruyn, J. M., and Sayler, G. S. (2009). Microbial community structure and biodegradation activity of particle-associated bacteria in a coal tar contaminated creek. Environ. Sci. Technol. 43, 3047–3053. doi: 10.1021/es803373y
Denef, V. J., Klappenbach, J. A., Patrauchan, M. A., Florizone, C., Rodrigues, J. L., Tsoi, T. V., et al. (2006). Genetic and genomic insights into the role of benzoate-catabolic pathway redundancy in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 72, 585–595. doi: 10.1128/AEM.72.1.585-595.2006
Denef, V. J., Patrauchan, M. A., Florizone, C., Park, J., Tsoi, T. V., Verstraete, W., et al. (2005). Growth substrate- and phase-specific expression of biphenyl, benzoate, and C1 metabolic pathways in Burkholderia xenovorans LB400. J. Bacteriol. 187, 7996–8005. doi: 10.1128/JB.187.23.7996-8005.2005
Di Gennaro, P., Terreni, P., Masi, G., Botti, S., De Ferra, F., and Bestetti, G. (2010). Identification and characterization of genes involved in naphthalene degradation in Rhodococcus opacus R7. Appl. Microbiol. Biotechnol. 87, 297–308. doi: 10.1007/s00253-010-2497-3
Eaton, R. W., and Chapman, P. J. (1992). Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions. J. Bacteriol. 174, 7542–7554.
Edwards, N. T. (1983). Polycyclic aromatic hydrocarbons (PAHs) in the terrestrial environment—a review. J. Environ. Qual. 12, 427–441. doi: 10.2134/jeq1983.124427x
Elango, V., Urbano, M., Lemelle, K. R., and Pardue, J. H. (2014). Biodegradation of MC252 oil in oil: sand aggregates in a coastal headland beach environment. Front. Microbiol. 5:161. doi: 10.3389/fmicb.2014.00161
El-Sheekh, M. M., Ghareib, M. M., and Abou-El-Souod, G. W. (2012). Biodegradation of phenolic and polycyclic aromatic compounds by some algae and cyanobacteria. J. Bioremediat. Biodegrad. 3:133. doi: 10.4172/2155-6199.1000133
Evans, W. C., Fernley, H. N., and Griffiths, E. (1965). Oxidative metabolism of phenanthrene and anthracene by soil pseudomonads. The ring-fission mechanism. Biochem. J. 95, 819–831. doi: 10.1042/bj0950819
Fathepure, B. Z. (2014). Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments. Front. Microbiol. 5:173. doi: 10.3389/fmicb.2014.00173
Feitkenhauer, H., Muller, R., and Markl, H. (2003). Degradation of polycyclic aromatic hydrocarbons and long chain alkanes at 60-70 degrees C by Thermus and Bacillus spp [corrected]. Biodegradation 14, 367–372. doi: 10.1023/A:1027357615649
Floodgate, G. (1984). “The fate of petroleum in marine ecosystems,” in Petroleum Microbiology, ed. R. M. Atlas (New York, NY: Macmillion), 355–398.
Foght, J. (2008). Anaerobic biodegradation of aromatic hydrocarbons: pathways and prospects. J. Mol. Microbiol. Biotechnol. 15, 93–120. doi: 10.1159/000121324
Fromin, N., Hamelin, J., Tarnawski, S., Roesti, D., Jourdain-Miserez, K., Forestier, N., et al. (2002). Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4, 634–643. doi: 10.1046/j.1462-2920.2002.00358.x
Gallego, S., Vila, J., Tauler, M., Nieto, J. M., Breugelmans, P., Springael, D., et al. (2014). Community structure and PAH ring-hydroxylating dioxygenase genes of a marine pyrene-degrading microbial consortium. Biodegradation 25, 543–556. doi: 10.1007/s10532-013-9680-z
Galvao, T. C., Mohn, W. W., and de Lorenzo, V. (2005). Exploring the microbial biodegradation and biotransformation gene pool. Trends Biotechnol. 23, 497–506. doi: 10.1016/j.tibtech.2005.08.002
Gan, S., Lau, E. V., and Ng, H. K. (2009). Remediation of soils contaminated with polycyclic aromatic hydrocarbons (PAHs). J. Hazard. Mater. 172, 532–549. doi: 10.1016/j.jhazmat.2009.07.118
Gandolfi, I., Sicolo, M., Franzetti, A., Fontanarosa, E., Santagostino, A., and Bestetti, G. (2010). Influence of compost amendment on microbial community and ecotoxicity of hydrocarbon-contaminated soils. Bioresour. Technol. 101, 568–575. doi: 10.1016/j.biortech.2009.08.095
Garcia, M. T., Ventosa, A., and Mellado, E. (2005). Catabolic versatility of aromatic compound-degrading halophilic bacteria. FEMS Microbiol. Ecol. 54, 97–109. doi: 10.1016/j.femsec.2005.03.009
Garcia de Llasera, M. P., Olmos-Espejel Jde, J., Diaz-Flores, G., and Montano-Montiel, A. (2016). Biodegradation of benzo(a)pyrene by two freshwater microalgae Selenastrum capricornutum and Scenedesmus acutus: a comparative study useful for bioremediation. Environ. Sci. Pollut. Res. Int. 23, 3365–3375. doi: 10.1007/s11356-015-5576-2
Gauthier, M. J., Lafay, B., Christen, R., Fernandez, L., Acquaviva, M., Bonin, P., et al. (1992). Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Bacteriol. 42, 568–576. doi: 10.1099/00207713-42-4-568
Genovese, M., Crisafi, F., Denaro, R., Cappello, S., Russo, D., and Calogero, R. (2014). Effective bioremediation strategy for rapid in situ cleanup of anoxic marine sediments in mesocosm oils pill simulation. Front. Microbiol. 5:162. doi: 10.3389/fmicb.2014.00162
Gerdes, B., Brinkmeyer, R., Dieckmann, G., and Helmke, E. (2005). Influence of crude oil on changes of bacterial communities in Arctic sea-ice. FEMS Microbiol. Ecol. 53, 129–139. doi: 10.1016/j.femsec.2004.11.010
Ghosal, D., Chakraborty, J., Khara, P., and Dutta, T. K. (2010). Degradation of phenanthrene via meta-cleavage of 2-hydroxy-1-naphthoic acid by Ochrobactrum sp. strain PWTJD. FEMS Microbiol. Lett. 313, 103–110. doi: 10.1111/j.1574-6968.2010.02129.x
Ghosal, D., Dutta, A., Chakraborty, J., Basu, S., and Dutta, T. K. (2013). Characterization of the metabolic pathway involved in assimilation of acenaphthene in Acinetobacter sp. strain AGAT-W. Res. Microbiol. 164, 155–163. doi: 10.1016/j.resmic.2012.11.003
Ginige, M. P., Hugenholtz, P., Daims, H., Wagner, M., Keller, J., and Blackall, L. L. (2004). Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microbiol. 70, 588–596. doi: 10.1128/AEM.70.1.588-596.2004
Goyal, A. K., and Zylstra, G. J. (1997). Genetics of naphthalene and phenanthrene degradation by Comamonas testosteroni. J. Ind. Microbiol. Biotechnol. 19, 401–407. doi: 10.1038/sj.jim.2900476
Guerin, T. E. (2000). The differential removal of aged polycyclic aromatic hydrocarbons from soil during bioremediation. Environ. Sci. Pollut. Res. Int. 7, 19–26. doi: 10.1065/espr199910.004
Habe, H., and Omori, T. (2003). Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 67, 225–243. doi: 10.1271/bbb.67.225
Hammel, K. E. (1995). “Organopollutant degradation by ligninolytic fungi,” in Microbial Transformation and Degradation of Toxic Organic Chemicals, eds L. Y. Young and K. E. Cerniglia (New York: Wiley-Liss Inc.), 331–346.
Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J., and Goodman, R. M. (1998). Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5, R245–R249. doi: 10.1016/S1074-5521(98)90108-9
Haritash, A. K., and Kaushik, C. P. (2009). Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J. Hazard. Mater. 169, 1–15. doi: 10.1016/j.jhazmat.2009.03.137
He, Z., Deng, Y., Van Nostrand, J. D., Tu, Q., Xu, M., Hemme, C. L., et al. (2010). GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 4, 1167–1179. doi: 10.1038/ismej.2010.46
Hedlund, B. P., Geiselbrecht, A. D., and Staley, J. T. (2001). Marinobacter strain NCE312 has a Pseudomonas-like naphthalene dioxygenase. FEMS Microbiol. Lett. 201, 47–51. doi: 10.1111/j.1574-6968.2001.tb10731.x
Hilyard, E. J., Jones-Meehan, J. M., Spargo, B. J., and Hill, R. T. (2008). Enrichment, isolation, and phylogenetic identification of polycyclic aromatic hydrocarbon-degrading bacteria from Elizabeth River sediments. Appl. Environ. Microbiol. 74, 1176–1182. doi: 10.1128/AEM.01518-07
Hofrichter, M. (2002). Review: lignin conversion by manganese peroxidase (MnP). Enz. Microbiol. Technol. 30, 454–466. doi: 10.1016/S0141-0229(01)00528-2
Holman, H. Y. N., Tsang, Y. W., and Holman, W. R. (1999). Mineralization of sparsely water-soluble polycyclic aromatic hydrocarbons in a water table fluctuation zone. Environ. Sci. Technol. 33, 1819–1824. doi: 10.1021/es980701k
Hong, L., Ghosh, U., Mahajan, T., Zare, R. N., and Luthy, R. G. (2003). PAH sorption mechanism and partitioning behavior in lampblack-impacted soils from former oil-gas plant sites. Environ. Sci. Technol. 37, 3625–3634. doi: 10.1021/es0262683
Hong, Y. W., Yuan, D. X., Lin, Q. M., and Yang, T. L. (2008). Accumulation and biodegradation of phenanthrene and fluoranthene by the algae enriched from a mangrove aquatic ecosystem. Mar. Pollut. Bull. 56, 1400–1405. doi: 10.1016/j.marpolbul.2008.05.003
Howard, P., Meylan, W., Aronson, D., Stiteler, W., Tunkel, J., Comber, M., et al. (2005). A new biodegradation prediction model specific to petroleum hydrocarbons. Environ. Toxicol. Chem. 24, 1847–1860. doi: 10.1897/04-453R.1
Hugenholtz, P., Goebel, B. M., and Pace, N. R. (1998). Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180, 4765–4774.
Hugenholtz, P., and Pace, N. R. (1996). Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 14, 190–197. doi: 10.1016/0167-7799(96)10025-1
Huntley, S. L., Bonnevie, N. L., Wenning, R. J., and Bedbury, H. (1993). Distribution of polycyclic aromatic hydrocarbons (PAHs) in three northern New Jersey waterways. Bull. Environ. Contam. Toxicol. 51, 865–872. doi: 10.1007/BF00198283
Jerina, D. M. (1983). Metabolism of aromatic hydrocarbons by the cytochrome P450 system and epoxide hydrolase. Drug Metab. Dispos. 11, 1–4.
Johansson, I., and van Bavel, B. (2003). Levels and patterns of polycyclic aromatic hydrocarbons in incineration ashes. Sci. Total Environ. 311, 221–231. doi: 10.1016/S0048-9697(03)00168-2
Johnsen, A. R., Wick, L. Y., and Harms, H. (2005). Principles of microbial PAH-degradation in soil. Environ. Pollut. 133, 71–84. doi: 10.1016/j.envpol.2004.04.015
Jorgensen, K. S. (2007). In situ bioremediation. Adv. Appl. Microbiol. 61, 285–305. doi: 10.1016/S0065-2164(06)61008-3
Juhasz, A. L., and Naidu, R. (2000). Bioremediation of high molecular weight polycyclic aromatic hydrocarbons: a review of the microbial degradation of benzo[a] pyrene. Int. Biodeterior. Biodegradation 45, 57–88. doi: 10.1016/S0964-8305(00)00052-4
Kanaly, R. A., and Harayama, S. (2000). Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182, 2059–2067. doi: 10.1128/JB.182.8.2059-2067.2000
Kanaly, R. A., and Harayama, S. (2010). Advances in the field of high-molecular-weight polycyclic aromatic hydrocarbon biodegradation by bacteria. Microb. Biotechnol. 3, 136–164. doi: 10.1111/j.1751-7915.2009.00130.x
Kappell, A. D., Wei, Y., Newton, R. J., Van Nostrand, J. D., Zhou, J., McLellan, S. L., et al. (2014). The polycyclic aromatic hydrocarbon degradation potential of Gulf of Mexico native coastal microbial communities after the Deepwater Horizon oil spill. Front. Microbiol. 5:205. doi: 10.3389/fmicb.2014.00205
Kastner, M. (2000). “Degradation of aromatic and polyaromatic compounds,” in Biotechnology, 2nd Edn. Environmental Processes, Vol. 11(b), eds H.-J. Rehm, G. Reed, A. Pühler, and P. Stadler (Weinheim: Wiley-VCH), 211–239.
Kastner, M., Jammali, B. M., and Mahro, B. (1994). Enumeration and characterization of the soil microflora from hydrocarbon contaminated soil sites able to mineralize polycyclic aromatic hydrocarbons (PAH). Appl. Microbiol. Biotechnol. 41, 267–273. doi: 10.1007/BF00186971
Ke, L., Luo, L., Wang, P., Luan, T., and Tam, N. F. (2010). Effects of metals on biosorption and biodegradation of mixed polycyclic aromatic hydrocarbons by a freshwater green alga Selenastrum capricornutum. Bioresour. Technol. 101, 6961–6972. doi: 10.1016/j.biortech.2010.04.011
Kelley, I., Freeman, J. P., and Cerniglia, C. E. (1990). Identification of metabolites from degradation of naphthalene by a Mycobacterium sp. Biodegradation 1, 283–290. doi: 10.1007/BF00119765
Keum, Y. S., Seo, J. S., Li, Q. X., and Kim, J. H. (2008). Comparative metabolomic analysis of Sinorhizobium sp. C4 during the degradation of phenanthrene. Appl. Microbiol. Biotechnol. 80, 863–872. doi: 10.1007/s00253-008-1581-4
Khara, P., Roy, M., Chakraborty, J., Ghosal, D., and Dutta, T. K. (2014). Functional characterization of diverse ring-hydroxylating oxygenases and induction of complex aromatic catabolic gene clusters in Sphingobium sp. PNB. FEBS Open Bio. 4, 290–300. doi: 10.1016/j.fob.2014.03.001
Kostka, J. E., Prakash, O., Overholt, W. A., Green, S. J., Freyer, G., Canion, A., et al. (2011). Hydrocarbon-degrading bacteria and the bacterial community response in gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 77, 7962–7974. doi: 10.1128/AEM.05402-11
Kostka, J. E., Teske, A. P., Joye, S. B., and Head, I. M. (2014). The metabolic pathways and environmental controls of hydrocarbon biodegradation in marine ecosystems. Front. Microbiol. 5:471. doi: 10.3389/fmicb.2014.00471
Kulakov, L. A., Chen, S., Allen, C. C., and Larkin, M. J. (2005). Web-type evolution of rhodococcus gene clusters associated with utilization of naphthalene. Appl. Environ. Microbiol. 71, 1754–1764. doi: 10.1128/AEM.71.4.1754-1764.2005
Kweon, O., Kim, S. J., Freeman, J. P., Song, J., Baek, S., and Cerniglia, C. E. (2010). Substrate specificity and structural characteristics of the novel Rieske nonheme iron aromatic ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1. MBio 1:e00135-10. doi: 10.1128/mBio.00135-10
Lamendella, R., Strutt, S., Borglin, S., Chakraborty, R., Tas, N., Mason, O. U., et al. (2014). Assessment of the Deepwater Horizon oil spill impact on Gulf coast microbial communities. Front. Microbiol. 5:130. doi: 10.3389/fmicb.2014.00130
Larkin, M. J., Kulakov, L. A., and Allen, C. C. (2005). Biodegradation and Rhodococcus–masters of catabolic versatility. Curr. Opin. Biotechnol. 16, 282–290. doi: 10.1016/j.copbio.2005.04.007
Le Borgne, S., Paniagua, D., and Vazquez-Duhalt, R. (2008). Biodegradation of organic pollutants by halophilic bacteria and archaea. J. Mol. Microbiol. Biotechnol. 15, 74–92. doi: 10.1159/000121323
Lei, A. P., Hu, Z. L., Wong, Y. S., and Tam, N. F. (2007). Removal of fluoranthene and pyrene by different microalgal species. Bioresour. Technol. 98, 273–280. doi: 10.1016/j.biortech.2006.01.012
Lei, A. P., Wong, Y. S., and Tam, N. F. (2002). Removal of pyrene by different microalgal species. Water Sci. Technol. 46, 195–201.
Li, X., Lin, X., Zhang, J., Wu, Y., Yin, R., Feng, Y., et al. (2010). Degradation of polycyclic aromatic hydrocarbons by crude extracts from spent mushroom substrate and its possible mechanisms. Curr. Microbiol. 60, 336–342. doi: 10.1007/s00284-009-9546-0
Liles, R. M., Williamson, L. L., Rodbumrer, J., Torsvik, V., Goodman, R. M., and Handelsman, J. (2008). Recovery, purification, and cloning of high-molecular-weight DNA from soil microorganisms. Appl. Environ. Microbiol. 74, 3302–3305. doi: 10.1128/AEM.02630-07
Lim, L. H., Harrison, R. M., and Harrad, S. (1999). The contribution of traffic to atmospheric concentrations of polycyclic aromatic hydrocarbons. Environ. Sci. Technol. 33, 3538–3542. doi: 10.1021/es990392d
Liu, K., Han, W., Pan, W. P., and Riley, J. T. (2001). Polycyclic aromatic hydrocarbon (PAH) emissions from a coal-fired pilot FBC system. J. Hazard. Mater. 84, 175–188. doi: 10.1016/S0304-3894(01)00196-0
Liu, L., Endo, S., Eberhardt, C., Grathwohl, P., and Schmidt, T. C. (2009). Partition behavior of polycyclic aromatic hydrocarbons between aged coal tar and water. Environ. Toxicol. Chem 28, 1578–1584. doi: 10.1897/08-276.1
Loick, N., Hobbs, P. J., Hale, M. C. D., and Jones, D. L. (2012). Bioremediation of poly-aromatic hyrdocarbon (PAH)-contaminated soil by composting. Crit. Rev. Environ. Sci. Technol. 39, 271–332. doi: 10.1080/10643380701413682
Lopez, Z., Vila, J., Ortega-Calvo, J. J., and Grifoll, M. (2008). Simultaneous biodegradation of creosote-polycyclic aromatic hydrocarbons by a pyrene-degrading Mycobacterium. Appl. Microbiol. Biotechnol. 78, 165–172. doi: 10.1007/s00253-007-1284-2
Lovley, D. R. (2001). Bioremediation. Anaerobes to the rescue. Science 293, 1444–1446. doi: 10.1126/science.1063294
Lu, X. Y., Zhang, T., and Fang, H. H. (2011). Bacteria-mediated PAH degradation in soil and sediment. Appl. Microbiol. Biotechnol. 89, 1357–1371. doi: 10.1007/s00253-010-3072-7
Lukić, B., Huguenot, D., Panico, A., Fabbricino, M., van Hullebusch, E. D., Esposito, G., et al. (2016). Importance of organic amendment characteristics on bioremediation of PAH-contaminated soil. Environ. Sci. Pollut. Res. Int. 23, 15041–15052. doi: 10.1007/s11356-016-6635-z
Lundstedt, S., Haglund, P., and Oberg, L. (2003). Degradation and formation of polycyclic aromatic compounds during bioslurry treatment of an aged gasworks soil. Environ. Toxicol. Chem. 22, 1413–1420. doi: 10.1002/etc.5620220701
Luo, L., Lin, S., Huan, H., and Zhang, S. (2012). Relationships between aging of PAHs and soil properties. Environ. Pollut. 170, 177–182. doi: 10.1016/j.envpol.2012.07.003
Luo, L., Wang, P., Lin, L., Luan, T., Ke, L., and Tam, N. F. Y. (2014). Removal and transformation of high molecular weight polycyclic aromatic hydrocarbons in water by live and dead microalgae. Process Biochem. 49, 1723–1732. doi: 10.1016/j.procbio.2014.06.026
Luo, S., Chen, B., Lin, L., Wang, X., Tam, N. F., and Luan, T. (2014). Pyrene degradation accelerated by constructed consortium of bacterium and microalga: effects of degradation products on the microalgal growth. Environ. Sci. Technol. 48, 13917–13924. doi: 10.1021/es503761j
Luthy, R. G., Dzombak, D. A., Peters, C. A., Roy, S. B., Ramaswami, A., Nakles, D. V., et al. (1994). Remediating tar-contaminated soils at manufactured gas plant sites. Environ. Sci. Technol. 28, 266–276. doi: 10.1021/es00055a718
Luthy, R. G., Ramaswami, A., and Ghosal, S. (1993). Interfacial films in coal tar nonaqueous-phase liquid-water systems. Environ. Sci. Technol. 27, 2914–2918. doi: 10.1021/es00049a035
Macaulay, I. C., and Voet, T. (2014). Single cell genomics: advances and future perspectives. PLoS Genet. 10:e1004126. doi: 10.1371/journal.pgen.1004126
Mackay, D., and Callcott, D. (1998). “Partitioning and physical chemical properties of PAHs,” in The Handbook of Environmental Chemistry, ed. A. H. Neilson (Berlin: Springer-Verlag), 325–346.
Mackay, D., and Shiu, W. Y. (1977). Aqueous solubility of polynuclear aromatic hydrocarbons. J. Chem. Eng. 22, 399–402.
Mallick, S., Chakraborty, J., and Dutta, T. K. (2011). Role of oxygenases in guiding diverse metabolic pathways in the bacterial degradation of low-molecular-weight polycyclic aromatic hydrocarbons: a review. Crit. Rev. Microbiol. 37, 64–90. doi: 10.3109/1040841X.2010.512268
Mallick, S., Chatterjee, S., and Dutta, T. K. (2007). A novel degradation pathway in the assimilation of phenanthrene by Staphylococcus sp. strain PN/Y via meta-cleavage of 2-hydroxy-1-naphthoic acid: formation of trans-2,3-dioxo-5-(2′-hydroxyphenyl)-pent-4-enoic acid. Microbiology 153(Pt. 7), 2104–2115. doi: 10.1099/mic.0.2006/004218-0
Mareckova, M. S., Cermak, L., Novotna, J., Plhackova, K., Forstova, J., and Kopecky, J. (2008). Innovative methods for soil DNA purification tested in soils with widely differing characteristics. Appl. Environ. Microbiol. 74, 2902–2907. doi: 10.1128/AEM.02161-07
Margesin, R., and Schinner, F. (2001a). Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl. Microbiol. Biotechnol. 56, 650–663. doi: 10.1007/s002530100701
Margesin, R., and Schinner, F. (2001b). Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 5, 73–83. doi: 10.1007/s007920100184
Marston, C. P., Pereira, C., Ferguson, J., Fischer, K., Hedstrom, O., Dashwood, W. M., et al. (2001). Effect of a complex environmental mixture from coal tar containing polycyclic aromatic hydrocarbons (PAH) on the tumor initiation, PAH-DNA binding and metabolic activation of carcinogenic PAH in mouse epidermis. Carcinogenesis 22, 1077–1086. doi: 10.1093/carcin/22.7.1077
Mason, O. U., Han, J., Woyke, T., and Jansson, J. K. (2014). Single-cell genomics reveals features of a Colwellia species that was dominant during the Deepwater Horizon oil spill. Front. Microbiol. 5:332. doi: 10.3389/fmicb.2014.00332
Mastrangelo, G., Fadda, E., and Marzia, V. (1996). Polycyclic aromatic hydrocarbons and cancer in man. Environ. Health Perspect. 104, 1166–1170. doi: 10.1289/ehp.961041166
May, R., Schroder, P., and Sandermann, H. J. (1997). Ex-situ process for treating PAH contaminated soil with Phanerochaete chrysosporium. Environ. Sci. Technol. 31, 2626–2633. doi: 10.1021/es9700414
Megharaj, M., Ramakrishnan, B., Venkateswarlu, K., Sethunathan, N., and Naidu, R. (2011). Bioremediation approaches for organic pollutants: a critical perspective. Environ. Int. 37, 1362–1375. doi: 10.1016/j.envint.2011.06.003
Megharaj, M., Wittich, R. W., Blanco, E., Pieper, D. H., and Timmis, K. N. (2002). Superior survival and degradation of dibenzo-p-dioxin and dibenzofuran in soil by soil-adapted Sphingomonas sp. strain RW1. Appl. Microbiol. Biotechnol. 48, 109–114. doi: 10.1007/s002530051024
Mellado, E., and Ventosa, A. (2003). “Biotechnological potential of moderately and extremely halophilic microorganisms,” in Microorganisms for Health Care, Food and Enzyme Production, ed. J. L. Barredo (Kerala: Research Signpost), 233–256.
Mendonca, E., and Picado, A. (2002). Ecotoxicological monitoring of remediation in a coke oven soil. Environ. Toxicol. 17, 74–79. doi: 10.1002/tox.10034
Menn, F. M., Applegate, B. M., and Sayler, G. S. (1993). NAH plasmid-mediated catabolism of anthracene and phenanthrene to naphthoic acids. Appl. Environ. Microbiol. 59, 1938–1942.
Meulenberg, R., Rijnaarts, H. H. M., Doddema, H. J., and Field, J. A. (1997). Partially oxidized polycyclic aromatic hydrocarbons show an increased bioavailability and biodegradability. FEMS Microbiol. Lett. 152, 45–49. doi: 10.1111/j.1574-6968.1997.tb10407.x
Milo, G. E., and Casto, B. C. (1992). “Events of tumor progression associated with carcinogen treatment of epithelium and fibroblast compared with mutagenic events,” in Transformation of Human Epithelial Cells: Molecular and Oncogenetic Mechanisms, eds G. E. Milo, B. C. Casto, and C. F. Shuler (Boca Raton, FL: CRC Press), 261–284.
Mishamandani, S., Gutierrez, T., and Aitken, M. D. (2014). DNA-based stable isotope probing coupled with cultivation methods implicates Methylophaga in hydrocarbon degradation. Front. Microbiol. 5:76. doi: 10.3389/fmicb.2014.00076
Mnif, S., Chamkha, M., and Sayadi, S. (2009). Isolation and characterization of Halomonas sp. strain C2SS100, a hydrocarbon-degrading bacterium under hypersaline conditions. J. Appl. Microbiol. 107, 785–794. doi: 10.1111/j.1365-2672.2009.04251.x
Moody, J. D., Freeman, J. P., Doerge, D. R., and Cerniglia, C. E. (2001). Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67, 1476–1483. doi: 10.1128/AEM.67.4.1476-1483.2001
Moody, J. D., Freeman, J. P., Fu, P. P., and Cerniglia, C. E. (2004). Degradation of benzo[a]pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 70, 340–345. doi: 10.1128/AEM.70.1.340-345.2004
Morehead, N. R., Eadie, B. J., Lake, B., Landrum, P. D., and Berner, D. (1986). The sorption of PAH onto dissolved organic matter in Lake Michigan waters. Chemosphere 15, 403–412. doi: 10.1016/0045-6535(86)90534-5
Mueller, J. G., Cerniglia, C. E., and Pritchard, P. H. (1996). “Bioremediation of environments contaminated by polycyclic aromatic hydrocarbons,” in Bioremediation: Principles and Applications, eds R. L. Crawford and L. D. Crawford (Cambridge: Cambridge University Press), 125–194.
Mueller, J. G., Chapman, P. J., and Pritchard, P. H. (1989). Action of a fluoranthene-utilizing bacterial community on polycyclic aromatic hydrocarbon components of creosote. Appl. Environ. Microbiol. 55, 3085–3090.
Mueller, J. G., Devereux, R., Santavy, D. L., Lantz, S. E., Willis, S. G., and Pritchard, P. H. (1997). Phylogenetic and physiological comparisons of PAH-degrading bacteria from geographically diverse soils. Antonie Van Leeuwenhoek 71, 329–343. doi: 10.1023/A:1000277008064
Müller, R., Antranikian, G., Maloney, S., and Sharp, R. (1998). “Thermophilic degradation of environmental pollutants,” in Biotechnology of Extremophiles. Advances in Biochemical Engineering/Bio-technology, Vol. 61, ed. G. Antranikian (Berlin: Springer), 155–169.
Munoz, R., Guieysse, B., and Mattiasson, B. (2003). Phenanthrene biodegradation by an algal-bacterial consortium in two-phase partitioning bioreactors. Appl. Microbiol. Biotechnol. 61, 261–267. doi: 10.1007/s00253-003-1231-9
Narro, M. L., Cerniglia, C. E., Van Baalen, C., and Gibson, D. T. (1992a). Evidence for an NIH shift in oxidation of naphthalene by the marine cyanobacterium Oscillatoria sp. strain JCM. Appl. Environ. Microbiol. 58, 1360–1363.
Narro, M. L., Cerniglia, C. E., Van Baalen, C., and Gibson, D. T. (1992b). Metabolism of phenanthrene by the marine cyanobacterium Agmenellum quadruplicatum PR-6. Appl. Environ. Microbiol. 58, 1351–1359.
National Academy of Sceinces [NAS] (1971). Particulate Polycyclic Aromatic Matter. Washington, DC: National Academy of Sceinces.
Nelson, E. C., Ghoshal, S., Edwards, J. C., Marsh, G. X., and Luthy, R. G. (1996). Chemical characterization of coal tar-water interfacial films. Environ. Sci. Technol. 30, 1014–1022. doi: 10.1021/es950482s
Nesatyy, V. J., and Suter, M. J. (2007). Proteomics for the analysis of environmental stress responses in organisms. Environ. Sci. Technol. 41, 6891–6900. doi: 10.1021/es070561r
Nierman, W. C., and Nelson, K. E. (2002). Genomics for applied microbiology. Adv. Appl. Microbiol. 51, 201–245. doi: 10.1016/S0065-2164(02)51007-8
Nojiri, H., Shintani, M., and Omori, T. (2004). Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl. Microbiol. Biotechnol. 64, 154–174. doi: 10.1007/s00253-003-1509-y
Norris, R. D., Hinchee, R. E., Brown, R., McCarty, P. L., Semprini, L., Wilson, J. T., et al. (1993). Handbook of Bioremediation. Boca Raton, FL: Lewis Publishers.
Northcott, G. L., and Jones, K. C. (2001). Partitioning, extractability, and formation of nonextractable PAH residues in soil. 1. Compound differences in aging and sequestration. Environ. Sci. Technol. 35, 1103–1110. doi: 10.1021/es000071y
Nostrand, J. D., He, Z., and Zhou, J. (2012). Use of functional gene arrays for elucidating in situ biodegradation. Front. Microbiol. 3:339. doi: 10.3389/fmicb.2012.00339
Odencrantz, J. E., Johnson, J. G., and Koenigsberg, S. S. (1996). Enhanced intrinsic bioremediation of hydrocarbons using an oxygen-releasing compound. Autumn (Fall) 6, 99–114.
Ohkouchi, N., Kawamura, K., and Kawahata, H. (1999). Distributions of three- to seven-ring polynuclear aromatic hydrocarbons on the deep sea floor in the central Pacific. Environ. Sci. Technol. 33, 3086–3090. doi: 10.1021/es981181w
Oostingh, G. J., Schmittner, M., Ehart, A. K., Tischler, U., and Duschl, A. (2008). A high-throughput screening method based on stably transformed human cells was used to determine the immunotoxic effects of fluoranthene and other PAHs. Toxicol. In Vitro 22, 1301–1310. doi: 10.1016/j.tiv.2008.03.003
Oren, A., Gurevich, P., Azachi, M., and Henis, Y. (1992). Microbial degradation of pollutants at high salt concentrations. Biodegradation 3, 387–398. doi: 10.1007/BF00129095
Ortega-Calvo, J. J., and Alexander, M. (1994). Roles of bacterial attachment and spontaneous partitioning in the biodegradation of naphthalene initially present in nonaqueous-phase liquids. Appl. Environ. Microbiol. 60, 2643–2646.
Pace, N. R., Stahl, D. A., Lane, D. J., and Olsen, G. J. (1986). The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9, 1–55. doi: 10.1007/978-1-4757-0611-6_1
Pannu, J. K., Singh, A., and Ward, O. P. (2003). Influence of peanut oil on microbial degradation of polycyclic aromatic hydrocarbons. Can. J. Microbiol. 49, 508–513. doi: 10.1139/w03-068
Pardieck, D. L., Bouwer, E. J., and Stone, A. T. (1992). Hydrogen-peroxide use to increase oxidant capacity for in situ bioremediation of contaminated soils and aquifers—a review. J. Contam. Hydrol. 9, 221–242. doi: 10.1016/0169-7722(92)90006-Z
Parnell, J. J., Park, J., Denef, V., Tsoi, T., Hashsham, S., Quensen, J., et al. (2006). Coping with polychlorinated biphenyl (PCB) toxicity: physiological and genome-wide responses of Burkholderia xenovorans LB400 to PCB-mediated stress. Appl. Environ. Microbiol. 72, 6607–6614. doi: 10.1128/AEM.01129-06
Patel, J. G., Nirmal Kumar, J. I., Kumar, R. N., and Khan, S. R. (2016). Biodegradation capability and enzymatic variation of potentially hazardous polycyclic aromatic hydrocarbons—anthracene and pyrene by Anabaena fertilissima. Polycycl. Aromat. Compd. 36, 72–87. doi: 10.1080/10406638.2015.1039656
Peng, R. H., Xiong, A. S., Xue, Y., Fu, X. Y., Gao, F., Zhao, W., et al. (2008). Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 32, 927–955. doi: 10.1111/j.1574-6976.2008.00127.x
Peyton, B. M., Mormille, M. R., Alva, V., Oie, C., Roberto, F., Apel, W. A., et al. (2004). “Biotransformation of toxic organic and inorganic contaminants by halophilic bacteria,” in Halophilic Microorganisms, ed. A. Ventosa (Berlin: Springer), 315–331.
Pothuluri, J. V., Evans, F. E., Heinze, T. M., and Cerniglia, C. E. (1996). Formation of sulfate and glucoside conjugates of benzo[e]pyrene by Cunninghamella elegans. Appl. Microbiol. Biotechnol. 45, 677–683. doi: 10.1007/s002530050747
Rein, A., Adam, I. K., Miltner, A., Brumme, K., Kastner, M., and Trapp, S. (2016). Impact of bacterial activity on turnover of insoluble hydrophobic substrates (phenanthrene and pyrene)-Model simulations for prediction of bioremediation success. J. Hazard. Mater. 306, 105–114. doi: 10.1016/j.jhazmat.2015.12.005
Roling, W. F., and Bodegom, P. M. (2014). Toward quantitative understanding on microbial community structure and functioning: a modeling-centered approach using degradation of marine oil spills as example. Front Microbiol. 5:125. doi: 10.3389/fmicb.2014.00125
Romine, M. F., Stillwell, L. C., Wong, K. K., Thurston, S. J., Sisk, E. C., Sensen, C., et al. (1999). Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181, 1585–1602.
Roy, M., Khara, P., Basu, S., and Dutta, T. K. (2013). Catabolic versatility of Sphingobium sp. strain PNB capable of degrading structurally diverse aromatic compounds. J. Bioremediat. Biodegrad. 4, 1–6. doi: 10.4172/2155-6199.1000173
Sack, U., Heinze, T. M., Deck, J., Cerniglia, C. E., Martens, R., Zadrazil, F., et al. (1997). Comparison of phenanthrene and pyrene degradation by different wood-decaying fungi. Appl. Environ. Microbiol. 63, 3919–3925.
Safonova, E., Kvitko, K., Kuschk, P., Möder, M., and Reisser, W. (2005). Biodegradation of phenanthrene by the green alga Scenedesmus obliquus ES-55. Eng. Life Sci. 5, 234–239. doi: 10.1002/elsc.200520077
Samanta, S. K., Singh, O. V., and Jain, R. K. (2002). Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol. 20, 243–248. doi: 10.1016/S0167-7799(02)01943-1
Santarelli, R. L., Pierre, F., and Corpet, D. E. (2008). Processed meat and colorectal cancer: a review of epidemiologic and experimental evidence. Nutr. Cancer 60, 131–144. doi: 10.1080/01635580701684872
Sasek, V., Bhatt, M., Cajthaml, T., Malachova, K., and Lednicka, D. (2003). Compost-mediated removal of polycyclic aromatic hydrocarbons from contaminated soil. Arch. Environ. Contam. Toxicol. 44, 336–342. doi: 10.1007/s00244-002-2037-y
Schmid, A., Dordick, J. S., Hauer, B., Kiener, A., Wubbolts, M., and Witholt, B. (2001). Industrial biocatalysis today and tomorrow. Nature 409, 258–268. doi: 10.1038/35051736
Schneegurt, M. A., and Kulpa, C. F. J. (1998). The application of molecular techniques in environmental biotechnology for monitoring microbial systems. Biotechnol. Appl. Biochem. 27, 73–79. doi: 10.1111/j.1470-8744.1998.tb01377.x
Schnitz, A. R., Squibb, K. S., and O’Connor, J. M. (1993). Time-varying conjugation of 7,12-dimethylbenz[a]anthracene metabolites in rainbow trout (Oncorhynchus mykiss). Toxicol. Appl. Pharmacol. 121, 58–70. doi: 10.1006/taap.1993.1129
Schocken, M. J., and Gibson, D. T. (1984). Bacterial oxidation of the polycyclic aromatic hydrocarbons acenaphthene and acenaphthylene. Appl. Environ. Microbiol. 48, 10–16.
Schützendübel, A., Majcherczyk, A., Johannes, C., and Hüttermann, A. (1999). Degradation of fluorene, anthracene, phenanthrene, fluoranthene, and pyrene lacks connection to the production of extracellular enzymes by Pleurotus ostreatus and Bjerkandera adusta. Int. Biodeterior. Biodegradation 43, 93–100. doi: 10.1016/S0964-8305(99)00035-9
Scott, N. M., Hess, M., Bouskill, N. J., Mason, O. U., Jansson, J. K., and Gilbert, J. A. (2014). The microbial nitrogen cycling potential is impacted by polyaromatic hydrocarbon pollution of marine sediments. Front. Microbiol. 5:108. doi: 10.3389/fmicb.2014.00108
Semple, K. T., Morriss, A. W. J., and Paton, G. I. (2003). Bioavailability of hydrophobic organic contaminants in soils: fundamental concepts and techniques for analysis. Eur. J. Soil Sci. 54, 809–818. doi: 10.1046/j.1351-0754.2003.0564.x
Semple, K. T., Reid, B. J., and Fermor, T. R. (2001). Impact of composting strategies on the treatment of soils contaminated with organic pollutants. Environ. Pollut. 112, 269–283. doi: 10.1016/S0269-7491(00)00099-3
Seo, J. S., Keum, Y. S., and Li, Q. X. (2009). Bacterial degradation of aromatic compounds. Int. J. Environ. Res. Public Health 6, 278–309. doi: 10.3390/ijerph6010278
Shaw, J. J., Dane, F., Geiger, D., and Kloepper, J. W. (1992). Use of bioluminescence for detection of genetically engineered microorganisms released into the environment. Appl. Environ. Microbiol. 58, 267–273.
Silva, I. S., Grossman, M., and Durrant, L. R. (2009). Degradation of polycyclic aromatic hydrocarbons (2–7 rings) under microaerobic and very-low-oxygen conditions by soil fungi. Int. Biodeterior. Biodegradation 2, 224–229. doi: 10.1016/j.ibiod.2008.09.008
Simon, M. J., Osslund, T. D., Saunders, R., Ensley, B. D., Suggs, S., Harcourt, A., et al. (1993). Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene 127, 31–37. doi: 10.1016/0378-1119(93)90613-8
Singh, A. K., Sherry, A., Gray, N. D., Jones, D. M., Bowler, B. F., and Head, I. M. (2014). Kinetic parameters for nutrient enhanced crude oil biodegradation in intertidal marine sediments. Front. Microbiol. 5:160. doi: 10.3389/fmicb.2014.00160
Singh, S., Kang, S. H., Mulchandani, A., and Chen, W. (2008). Bioremediation: environmental clean–up through pathway engineering. Curr. Opin. Biotechnol. 19, 437–444. doi: 10.1016/j.copbio.2008.07.012
Singleton, D. R., Richardson, S. D., and Aitken, M. D. (2011). Pyrosequence analysis of bacterial communities in aerobic bioreactors treating polycyclic aromatic hydrocarbon-contaminated soil. Biodegradation 22, 1061–1073. doi: 10.1007/s10532-011-9463-3
Smith, C. B., Tolar, B. B., Hollibaugh, J. T., and King, G. M. (2013). Alkane hydroxylase gene (alkB) phylotype composition and diversity in northern Gulf of Mexico bacterioplankton. Front. Microbiol. 4:370. doi: 10.3389/fmicb.2013.00370
Stolz, A. (2009). Molecular characteristics of xenobiotic-degrading sphingomonads. Appl. Microbiol. Biotechnol. 81, 793–811. doi: 10.1007/s00253-008-1752-3
Straube, W. L., Nestler, C. C., Hansen, L. D., Ringleberg, D., Pritchard, P. H., and Meehan, J. J. (2003). Remediation of polyaromatic hydrocarbons (PAHs) through landfarming with biostimulation and bioaugmentation. Acta Biotechnol. 23, 179–196. doi: 10.1002/abio.200390025
Stringfellow, W. T., and Aitken, M. D. (1995). Competitive metabolism of naphthalene, methylnaphthalenes, and fluorene by phenanthrene-degrading pseudomonads. Appl. Environ. Microbiol. 61, 357–362.
Sutherland, J. B. (1992). Detoxification of polycyclic aromatic hydrocarbons by fungi. J. Ind. Microbiol. Biotechnol. 9, 53–62.
Sutherland, J. B., Freeman, J. P., Selby, A. L., Fu, P. P., Miller, D. W., and Cerniglia, C. E. (1990). Stereoselective formation of a K-region dihydrodiol from phenanthrene by Streptomyces flavovirens. Arch. Microbiol. 154, 260–266. doi: 10.1007/BF00248965
Sutherland, J. B., Rafii, F., Khan, A. A., and Cerniglia, C. E. (1995). “Mechanisms of polycyclic aromatic hydrocarbon degradation,” in Microbial Transformation and Degradation of Toxic Organic Chemicals, eds L. Y. Young and C. E. Cerniglia (New York, NY: Wiley-Liss), 269–306.
Takáčová, A., Smolinská, M., Ryba, J., Mackul’ak, T., Jokrllová, J., Hronec, P., et al. (2014). Biodegradation of Benzo[a]Pyrene through the use of algae. Cent. Eur. J. Chem. 12, 1133–1143. doi: 10.2478/s11532-014-0567-6
Thomas, J. C. I., Wafula, D., Chauhan, A., Green, S. J., Gragg, R., and Jagoe, C. (2014). A survey of deep water horizon (DWH) oil-degrading bacteria from the eastern oyster biome and its surrounding environment. Front. Microbiol. 5:149. doi: 10.3389/fmicb.2014.00149
Timmis, K. N., and Pieper, D. H. (1999). Bacteria designed for bioremediation. Trends Biotechnol. 17, 200–204. doi: 10.1016/S0167-7799(98)01295-5
Timmis, K. N., Steffan, R. J., and Unterman, R. (1994). Designing microorganisms for the treatment of toxic wastes. Annu. Rev. Microbiol. 48, 525–557. doi: 10.1146/annurev.mi.48.100194.002521
Tongpim, S., and Pickard, M. A. (1999). Cometabolic oxidation of phenanthrene to phenanthrene trans-9,10-dihydrodiol by Mycobacterium strain S1 growing on anthracene in the presence of phenanthrene. Can. J. Microbiol. 45, 369–376. doi: 10.1139/cjm-45-5-369
Top, E. M., and Springael, D. (2003). The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14, 262–269. doi: 10.1016/S0958-1669(03)00066-1
Torlapati, J., and Boufadel, M. C. (2014). Evaluation of the biodegradation of Alaska North Slope oil in microcosms using the biodegradation model BIOB. Front. Microbiol. 5:212. doi: 10.3389/fmicb.2014.00212
Tortella, G. R., Diez, M. C., and Duran, N. (2005). Fungal diversity and use in decomposition of environmental pollutants. Crit. Rev. Microbiol. 31, 197–212. doi: 10.1080/10408410500304066
Ueno, R., Ureno, N., and Wada, S. (2006). Synergistic effect of cell immobilization in polyurethane foam and use of thermo tolerant strain on mixed hydrocarbon substrate by Prototheca zopfii. Fish. Sci. 72, 1027–1033. doi: 10.1111/j.1444-2906.2006.01252.x
Ueno, R., Wada, S., and Urano, N. (2008). Repeated batch cultivation of the hydrocarbon-degrading, micro-algal strain Prototheca zopfii RND16 immobilized in polyurethane foam. Can. J. Microbiol. 54, 66–70. doi: 10.1139/w07-112
Van Brummelen, T. C., Verweij, R. A., Wedzinga, S., and Van Gestel, C. A. M. (1996). Enrichment of polycyclic aromatic hydrocarbons in forest soils near a blast furnace plant. Chemosphere 32, 293–314. doi: 10.1016/0045-6535(95)00339-8
van Herwijnen, R., Wattiau, P., Bastiaens, L., Daal, L., Jonker, L., Springael, D., et al. (2003). Elucidation of the metabolic pathway of fluorene and cometabolic pathways of phenanthrene, fluoranthene, anthracene and dibenzothiophene by Sphingomonas sp. LB126. Res. Microbiol. 154, 199–206. doi: 10.1016/S0923-2508(03)00039-1
Van Stempvoort, D. R., Lesage, S., Novakowski, K. S., Millar, K., Brown, S., and Lawrence, J. R. (2002). Humic acid enhanced remediation of an emplaced diesel source in groundwater. 1. Laboratory-based pilot scale test. J. Contam. Hydrol. 54, 249–276. doi: 10.1016/S0169-7722(01)00182-6
Veraldi, A., Costantini, A. S., Bolejack, V., Miligi, L., Vineis, P., and van Loveren, H. (2006). Immunotoxic effects of chemicals: a matrix for occupational and environmental epidemiological studies. Am. J. Ind. Med. 49, 1046–1055. doi: 10.1002/ajim.20364
Verdin, A., Sahraoui, A. L. H., and Durand, R. (2004). Degradation of benzo[a]pyrene by mitosporic fungi and extracellular oxidative enzymes. Int. Biodeterior. Biodegradation 53, 65–70. doi: 10.1016/j.ibiod.2003.12.001
Vila, J., María Nieto, J., Mertens, J., Springael, D., and Grifoll, M. (2010). Microbial community structure of a heavy fuel oil-degrading marine consortium: linking microbial dynamics with polycyclic aromatic hydrocarbon utilization. FEMS Microbiol. Ecol. 73, 349–362. doi: 10.1111/j.1574-6941.2010.00902.x
Vila, J., Tauler, M., and Grifoll, M. (2015). Bacterial PAH degradation in marine and terrestrial habitats. Curr. Opin. Biotechnol. 33, 95–102. doi: 10.1016/j.copbio.2015.01.006
Volkering, F., Breure, A. M., Sterkenburg, A., and van Andel, J. G. (1992). Microbial degradation of polycyclic aromatic hydrocarbons: effect of substrate availability on bacterial growth kinetics. Appl. Microbiol. Biotechnol. 36, 548–552. doi: 10.1007/BF00170201
Wagrowski, D. M., and Hites, R. A. (1997). Polycyclic aromatic hydrocarbon accumulation in urban, suburban, and rural vegetation. Environ. Sci. Technol. 31, 279–282. doi: 10.1021/es960419i
Warshawsky, D., Cody, T., Radike, M., Reilman, R., Schumann, B., LaDow, K., et al. (1995). Biotransformation of benzo[a]pyrene and other polycyclic aromatic hydrocarbons and heterocyclic analogs by several green algae and other algal species under gold and white light. Chem. Biol. Interact. 97, 131–148. doi: 10.1016/0009-2797(95)03610-X
Warshawsky, D., Radike, M., Jayasimhulu, K., and Cody, T. (1988). Metabolism of benzo(a)pyrene by a dioxygenase enzyme system of the freshwater green alga Selenastrum capricornutum. Biochem. Biophys. Res. Commun. 152, 540–544. doi: 10.1016/S0006-291X(88)80071-8
Watanabe, K., Futamata, H., and Harayama, S. (2002). Understanding the diversity in catabolic potential of microorganisms for the development of bioremediation strategies. Antonie Van Leeuwenhoek 81, 655–663. doi: 10.1023/A:1020534328100
WHO (1983). Polynuclear aromatic compounds, Part 1, chemical, environmental and experimental data. IARC Monogr. Eval. Carcinog. Risk Chem. Hum. 32, 1–453.
Widada, J., Nojiri, H., Kasuga, K., Yoshida, T., Habe, H., and Omori, T. (2002). Molecular detection and diversity of polycyclic aromatic hydrocarbon-degrading bacteria isolated from geographically diverse sites. Appl. Microbiol. Biotechnol. 58, 202–209. doi: 10.1007/s00253-001-0880-9
Wilson, M. S., Bakermans, C., and Madsen, E. L. (1999). In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl. Environ. Microbiol. 65, 80–87.
Wilson, S. C., and Jones, K. C. (1993). Bioremediation of soil contaminated with polynuclear aromatic hydrocarbons (PAHs): a review. Environ. Pollut. 81, 229–249. doi: 10.1016/0269-7491(93)90206-4
Xue, W., and Warshawsky, D. (2005). Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol. Appl. Pharmacol. 206, 73–93. doi: 10.1016/j.taap.2004.11.006
Yakimov, M. M., Denaro, R., Genovese, M., Cappello, S., D’Auria, G., Chernikova, T. N., et al. (2005). Natural microbial diversity in superficial sediments of Milazzo Harbor (Sicily) and community successions during microcosm enrichment with various hydrocarbons. Environ. Microbiol. 7, 1426–1441. doi: 10.1111/j.1462-5822.2005.00829.x
Yang, Y., Chen, R. F., and Shiaris, M. P. (1994). Metabolism of naphthalene, fluorene, and phenanthrene: preliminary characterization of a cloned gene cluster from Pseudomonas putida NCIB 9816. J. Bacteriol. 176, 2158–2164.
Yen, K. M., and Gunsalus, I. C. (1985). Regulation of naphthalene catabolic genes of plasmid NAH7. J. Bacteriol. 162, 1008–1013.
Young, L. Y., and Cerniglia, C. E. (1996). Microbial Transformation and Degradation of Toxic Organic Chemicals. Hoboken, NJ: Wiley.
Zein, M. M., Pinto, P. X., Garcia-Blanco, S., Suidan, M. T., and Venosa, A. D. (2006). Treatment of groundwater contaminated with PAHs, gasoline hydrocarbons, and methyl tert-butyl ether in a laboratory biomass-retaining bioreactor. Biodegradation 17, 57–69. doi: 10.1007/s10532-005-3049-x
Zhao, B., Wang, H., Mao, X., and Li, R. (2009). Biodegradation of phenanthrene by a halophilic bacterial consortium under aerobic conditions. Curr. Microbiol. 58, 205–210. doi: 10.1007/s00284-008-9309-3
Zhou, J., and Thompson, D. K. (2002). Challenges in applying microarrays to environmental studies. Curr. Opin. Biotechnol. 13, 204–207. doi: 10.1016/S0958-1669(02)00319-1
Zhou, N. Y., Fuenmayor, S. L., and Williams, P. A. (2001). nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183, 700–708. doi: 10.1128/JB.183.2.700-708.2001
Keywords: biodegradation, polycyclic aromatic hydrocarbons (PAHs), bacteria, fungi, algae
Citation: Ghosal D, Ghosh S, Dutta TK and Ahn Y (2016) Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. Front. Microbiol. 7:1369. doi: 10.3389/fmicb.2016.01369
Received: 02 May 2016; Accepted: 18 August 2016;
Published: 31 August 2016.
Edited by:
Pankaj Kumar Arora, M. J. P. Rohilkhand University, IndiaReviewed by:
Matthias E. Kaestner, Helmholtz Centre for Environmental Research – UFZ, GermanyEric D. Van Hullebusch, University of Paris-Est, France
Copyright © 2016 Ghosal, Ghosh, Dutta and Ahn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tapan K. Dutta, dGFwYW5AamNib3NlLmFjLmlu Youngho Ahn, eWhhaG5AeW51LmFjLmty
 Debajyoti Ghosal1
Debajyoti Ghosal1 Youngho Ahn
Youngho Ahn