- Molecular Genetics Group, Department of Biology, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia
by Logue, C. M., Wannemuehler, Y., Nicholson, B. A., Doetkott, C., Barbieri, N. L., and Nolan, L. K. (2017). Front. Microbiol. 8:283. doi: 10.3389/fmicb.2017.00283
Escherichia coli (E. coli) is mostly a commensal bacterium, part of the intestinal microbiota of a variety of animals, including humans (Bélanger et al., 2011; Vila et al., 2016). However, some E. coli strains can be pathogenic and depending on the spectrum of encoded virulence factors E. coli can cause either intestinal or extraintestinal infections (Kaper et al., 2004). It is known that the species E. coli has an extensive genetic substructure (Chaudhuri and Henderson, 2012) and that the substructure of E. coli populations differs among distinct geographical regions (Freitag et al., 2005; Walk et al., 2009) and bacterial hosts (Vadnov et al., 2017).
Initially, four different phylogenetic groups of E. coli were defined, A, B1, B2, and D (Chaudhuri and Henderson, 2012). Clermont et al. (2000) established a PCR-method, the so called triplex PCR, for assigning E. coli strains into these four phylogenetic groups, a method that was widely used to type and subtype commensal and pathogenic E. coli. Phylogenetic classification has been extensively used to compare with serogroup, virulence and resistance traits as well as distribution among various hosts. However, subsequently, on the basis of multi-locus sequence typing and complete genome data, additional E. coli phylogenetic groups were recognized (Walk et al., 2009; Luo et al., 2011). The number of defined phylogenetic groups thus rose to 8 (A, B1, B2, C, D, E, F that belong to E. coli sensu stricto, and the eighth—the Escherichia cryptic clade I). Thus, Clermont et al. (2013) proposed a revised, so called extended quadruplex method for assigning E. coli strains to phylogenetic groups that is now replacing the triplex method. The authors validated the extended quadruplex method on a set of 234 strains, which included the ECOR strains (Clermont et al., 2011) and 133 strains from Australia (Gordon et al., 2008). In addition, Clermont et al. (2013) used the new extended quadruplex method for phylogroup assignment of 293 human fecal E. coli strains from France and 373 human fecal E. coli strains from Australia (Clermont et al., 2013). The authors reported that 12.8% of the tested strains belonged to the new phylogroups C, E, F, and clade I and that strains previously assigned, with the triplex method, to the A and D group should be retested with the new extended quadruplex method. None of the investigated strains were not typeable (NT). Recently, Logue et al. (2017) performed a comparative analysis of phylogenetic assignment of human and avian extraintestinal pathogenic (ExPEC) and fecal commensal E. coli (FEC) strains and showed that in total 13.05% of studied human E. coli strains and 40.49% of avian E. coli strains had to be reclassified. The majority of reassignments among the human E. coli strains involved changes from phylogroup D to F (45 out of 139 reclassifications), A to C (29 out of 139 reclassifications) and D to B2 (26 out of 139 reclassifications), while among the avian E. coli strains, the majority were reclassified from phylogroup A to C (162 out of 377 reclassifications), D to F (139 out of 377 reclassifications), and D to E (26 out of 377 reclassifications) (Logue et al., 2017). Here, we compared phylogroup classification of our strain collections: E. coli from skin and soft tissue infections (Petkovšek et al., 2009), fecal E. coli strains from healthy humans (Starčič Erjavec et al., 2010), and avian fecal strains (Salmič and Stele, 2012), with both PCR methods and with the results presented in Logue et al. (2017) (Table 1). Compared to the latter study (Logue et al., 2017), among our strain collections, more human (27.60% of human) and less avian (23.33% of avian) strains had to be reclassified. Further, among our human strains, the majority involved reclassification from the D to E phylogroup (19 out of 53 reclassifications), and D to F (9 out of 53 reclassifications). On the other hand, only 4 out of 53 involved reclassification from A to C as well as from B1 to E, with the latter not reported by Logue et al. (2017). Further, among our avian fecal strains, the majority of reclassifications were from the D to NT (7 out of 21 reclassifications), D to E (5 out of 21 reclassifications) and A to NT (3 out of 21 reclassifications). Our results thus showed that among distinct E. coli populations, reclassifications to different groups occurred with different prevalences. This is also evident from our data obtained on our collection of 86 fecal E. coli strains from brown bears (Vadnov et al., 2017), where the most prevalent reclassification was from group D to Clade III/IV/V with 25 out of 61 reclassifications, followed by reclassification from D to E (10 out of 61 reclassifications) (Table 1). Further, the high number of reclassifications to NT observed among our avian fecal strains is striking, especially as Logue et al. (2017) reported only a small number for reclassifications to the NT, in total from A to NT only 5 out of 516 reclassifications, while from B1 to NT and from B2 to NT only 1 out of 516 reclassifications. A survey on published studies using the extended quadruplex Clermont method for assigning the phylogenetic group showed even higher numbers of NT E. coli strains: 16 E. coli strains (7.92%) from 202 E. coli strains isolated from in- and outpatients in Burkina Faso (Ouedraogo et al., 2016), 8 E. coli strains (24.24%) from 33 vaginal E. coli strains isolated from pregnant women in Mozambique (Sáez-López et al., 2016a), 9 E. coli strains (6.21%) from 145 vaginal and obstetric infection E. coli strains from women in Barcelona (Sáez-López et al., 2016b), 12 E. coli strains (3.05%) from 393 E. coli strains from mallard ducks in Germany (Rödiger et al., 2015), 38 E. coli strains (27.14%) from 140 uropathogenic E. coli strains in Iran (Iranpour et al., 2015).
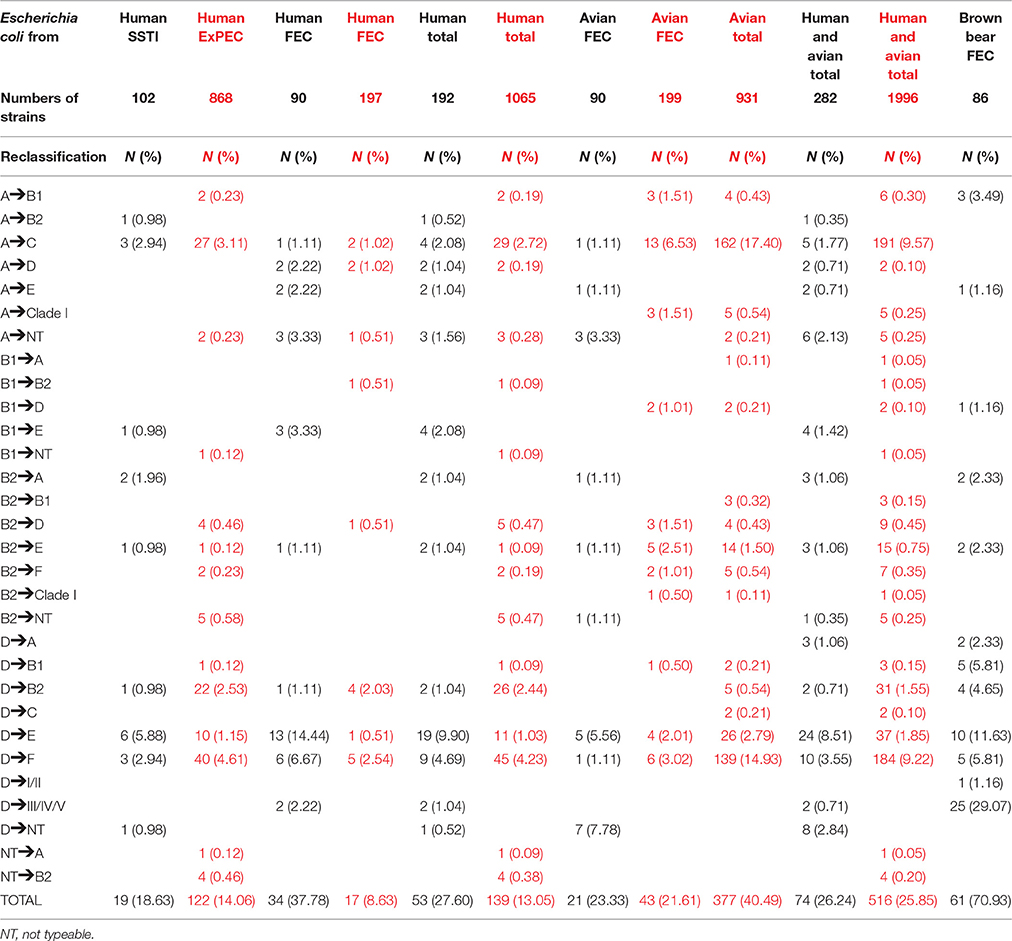
Table 1. Changes in phylogenetic group assignment based on application of the revised extended quadruplex Clermont method to assignment as determined by the original triplex Clermont method [our data in black, data from Logue et al. (2017) in red].
To conclude, Logue et al. (2017) stated that the new extended quadruplex method had a significant impact on avian pathogenic E. coli classification and definition of some human uropathogenic strains, a statement we fully support. However, we would like to emphasize that the extent of reclassifications into different groups differ among distinct E. coli populations and that, as with the new extended quadruplex method many strains are NT, the question arises whether there is a need for a revised revised phylogenetic typing method? It is well known that E. coli is a highly diverse bacterial species therefore, it is not unexpected that all strains from novel environments were not phylotyped with the new extended quadruplex method. We believe that there is a clear need to search for NT E. coli strains from novel environments (new hosts in not yet explored geographic regions). Whole genome sequence analysis of such strains should be performed in the search for markers that can be incorporated into a new rapid PCR phylotyping method.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication. MSE led the submission process.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was financed by Grant P1-0198 from the Slovenian Research Agency (ARRS).
References
Bélanger, L., Garenaux, A., Harel, J., Boulianne, M., Nadeau, E., and Dozois, C. M. (2011). Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol. Med. Microbiol. 62, 1–10. doi: 10.1111/j.1574-695X.2011.00797.x
Chaudhuri, R. R., and Henderson, I. R. (2012). The evolution of the Escherichia coli phylogeny. Infect. Genet. Evol. 12, 214–226. doi: 10.1016/j.meegid.2012.01.005
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Clermont, O., Olier, M., Hoede, C., Diancourt, L., Brisse, S., Keroudean, M., et al. (2011). Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11, 654–662. doi: 10.1016/j.meegid.2011.02.005
Freitag, T., Squires, R. A., Schmid, J., and Elliott, J. (2005). Feline uropathogenic Escherichia coli from Great Britain and New Zealand have dissimilar virulence factor genotypes. Vet. Microbiol. 106, 79–86. doi: 10.1016/j.vetmic.2004.11.014
Gordon, D. M., Clermont, O., Tolley, H., and Denamur, E. (2008). Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing vs. the PCR triplex method. Environ. Microbiol. 10, 2484–2496. doi: 10.1111/j.1462-2920.2008.01669.x
Iranpour, D., Hassanpour, M., Ansari, H., Tajbakhsh, S., Khamisipour, G., and Najafi, A. (2015). Phylogenetic groups of Escherichia coli strains from patients with urinary tract infection in Iran based on the new Clermont phylotyping method. Biomed. Res. Int. 2015:846219. doi: 10.1155/2015/846219
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Logue, C. M., Wannemuehler, Y., Nicholson, B. A., Doetkott, C., Barbieri, N. L., and Nolan, L. K. (2017). Comparative analysis of phylogenetic assignment of human and avian ExPEC and fecal commensal Escherichia coli using the (previous and revised) Clermont phylogenetic typing methods and its impact on avian pathogenic Escherichia coli (APEC) classification. Front. Microbiol. 8:283. doi: 10.3389/fmicb.2017.00283
Luo, C., Walk, S. T., Gordon, D. M., Feldgarden, M., Tiedje, J. M., and Konstantinidis, K. T. (2011). Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc. Natl. Acad. Sci. U.S.A. 108, 7200–7205. doi: 10.1073/pnas.1015622108
Ouedraogo, A. S., Sanou, M., Kissou, A., Sanou, S., Solaré, H., Kaboré, F., et al. (2016). High prevalence of extended-spectrum ß-lactamase producing enterobacteriaceae among clinical isolates in Burkina Faso. BMC Infect. Dis. 16:326. doi: 10.1186/s12879-016-1655-3
Petkovšek, Ž., Eleršič, K., Gubina, M., Žgur-Bertok, D., and Starčič Erjavec, M. (2009). Virulence potential of Escherichia coli isolates from skin and soft tissue infections. J. Clin. Microbiol. 47, 1811–1817. doi: 10.1128/JCM.01421-08
Rödiger, S., Kramer, T., Frömmel, U., Weinreich, J., Roggenbuck, D., Guenther, S., et al. (2015). Intestinal Escherichia coli colonization in a mallard duck population over four consecutive winter seasons. Environ. Microbiol. 17, 3352–3361. doi: 10.1111/1462-2920.12807
Sáez-López, E., Cossa, A., Benmessaoud, R., Madrid, L., Moraleda, C., Villanueva, S., et al. (2016a). Characterization of vaginal Escherichia coli isolated from pregnant women in two different African sites. PLoS ONE 11:e0158695. doi: 10.1371/journal.pone.0158695
Sáez-López, E., Guiral, E., Fernández-Orth, D., Villanueva, S., Goncé, A., López, M., et al. (2016b). Vaginal vs. obstetric infection Escherichia coli isolates among pregnant women: antimicrobial resistance and genetic virulence profile. PLoS ONE 11:e0146531. doi: 10.1371/journal.pone.0146531
Salmič, J., and Stele, F. (2012). Filogenetska analiza ptičjih izolatov bakterije Escherichia coli z metodo verižne reakcije s polimerazo: Raziskovalna naloga [Phylogenetic Analysis of Escherichia coli Avian Isolates with the Polymerase Chain Reaction: Research Project]. Ljubljana: Gimnazija Poljane.
Starčič Erjavec, M., Jesenko, B., Petkovšek, Ž., and Žgur-Bertok, D. (2010). Prevalence and associations of tcpC, a gene encoding a toll/interleukin-1 receptor domain-containing protein, among Escherichia coli urinary tract infection, skin and soft tissue infection, and commensal isolates. J. Clin. Microbiol. 48, 966–968. doi: 10.1128/JCM.01227-09
Vadnov, M., Barbič, D., Žgur-Bertok, D., and Starčič Erjavec, M. (2017). Escherichia coli isolated from faeces of brown bears (Ursus arctos) have a lower prevalence of human extraintestinal pathogenic E. coli virulence-associated genes. Can. J. Vet. Res. 81, 59–63.
Vila, J., Sáez-López, E., Johnson, J. R., Römling, U., Dobrindt, U., Cantón, R., et al. (2016). Escherichia coli: an old friend with new tidings. FEMS Microbiol. Rev. 40, 437–463. doi: 10.1093/femsre/fuw005
Keywords: Escherichia coli, phylogroup, avian, human, phylogenetic typing, fecal E. coli (FEC), extraintestinal pathogenic E. coli (ExPEC)
Citation: Starčič Erjavec M, Predojević L and Žgur-Bertok D (2017) Commentary: Comparative Analysis of Phylogenetic Assignment of Human and Avian ExPEC and Fecal Commensal Escherichia coli Using the (Previous and Revised) Clermont Phylogenetic Typing Methods and its Impact on Avian Pathogenic Escherichia coli (APEC) Classification. Front. Microbiol. 8:1904. doi: 10.3389/fmicb.2017.01904
Received: 08 August 2017; Accepted: 19 September 2017;
Published: 12 October 2017.
Edited by:
John W. A. Rossen, University Medical Center Groningen, NetherlandsReviewed by:
Eelco Franz, Centre for Infectious Disease Control, NetherlandsCopyright © 2017 Starčič Erjavec, Predojević and Žgur-Bertok. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marjanca Starčič Erjavec, bWFyamFuY2Euc3RhcmNpYy5lcmphdmVjQGJmLnVuaS1sai5zaQ==
 Marjanca Starčič Erjavec
Marjanca Starčič Erjavec Luka Predojević
Luka Predojević Darja Žgur-Bertok
Darja Žgur-Bertok