- Laboratory for Bioenergy and Biochemicals, School of Agricultural, Forest and Food Sciences, Bern University of Applied Sciences, Bern, Switzerland
The constraints and advantages in cellulolytic enzymes production by fungal biofilms for a consolidated bioconversion process were investigated during this study. The biofilm cultivations were carried out in reactors designed for consolidated bioprocessing Multispecies Biofilm Membrane reactors, (MBM) where an aerobic fungal biofilm produces the lignocellulolytic enzymes while a fermenting microorganism forms the fermentation product at anaerobic conditions. It was shown that although mycelial growth was limited in the MBM reactors compared to submerged cultivations, the secretion of cellulolytic enzymes per cell dry weight was higher. When Trichoderma reesei was used as the sole enzyme producer, cellobiose accumulated in the liquid medium as the result of the deficiency of β-glucosidase in the fungal secretome. To enhance β-glucosidase activity, T. reesei was co-cultivated with A. phoenicis which is a β-glucosidase overproducer. The two fungi formed a multispecies biofilm which produced a balanced cellulolytic cocktail for the saccharification of plant biomass. The mixed biofilm reached a 2.5 fold increase in β-glucosidase production, compared to the single T. reesei biofilm. The enzymatic systems of single and mixed biofilms were evaluated regarding their efficiency on cellulosic substrates degradation. Washed solids from steam pretreated beechwood, as well as microcrystalline cellulose were used as the substrates. The enzymatic system of the multispecies biofilm released four times more glucose than the enzymatic system of T. reesei alone from both substrates and hydrolyzed 78 and 60% of the cellulose content of washed solids from beechwood and microcrystalline cellulose, respectively.
Introduction
Biological decay, bioremediation and biodegradation of plant biomass are performed by the combined action of diverse microorganisms or by complex microbial communities. A paradigm of cellulolytic multispecies biofilms from nature is the cattle rumen where cellulose degradation is mainly performed by such multispecies biofilms which are formed on the lignocellulosic particles and on the rumen walls (McAllister et al., 1994). The successful degradation of cellulose by these microbes is based on the dynamic equilibrium among the different species: Rumen microorganisms benefit from each other's metabolism, in order to effectively access and consume their preferred substrates (Prescott et al., 2002; Nagaraja, 2016). These interactions create distinguished ecological niches, making the rumen suitable for many different microorganisms.
Microbial biofilms and microbial consortia have been used in various bioconversion processes of lignocellulosic biomass (Villena and Gutierrez-Correa, 2007; Wang and Chen, 2009; Kalyani et al., 2013). The formation of multispecies biofilm structures is beneficial for the bioconversion when the involved microorganisms require different conditions and/or when vicinity satisfies their (not always mutual) needs. Wang and Chen foresaw the possibility of a highly consolidated bioprocess (HCBP), which would incorporate delignification, saccharification and sugars co-fermentation in a single reactor inhabited by a multispecies biofilm (Wang and Chen, 2009). The consolidated bioconversion bioprocess integrates as many conversion steps as possible in one module and/or in one microbial host. Many different approaches of consolidated bioprocessing have been developed over the last decades, using different cultivation techniques, mainly in the framework of bio-ethanol research projects. The aim was to make the production of lignocellulosic ethanol economically attractive, by decreasing the cost of enzymes production and transport as well as the number of unit operations (Jouzani and Taherzadeh, 2015).
Brethauer and Studer (2014) developed such a CBP reactor for direct conversion of plant biomass to liquid fuels and chemicals, named Multispecies Biofilm Membrane (MBM) reactor. In this system, the simultaneous achievement and separation of aerobic and anaerobic conditions in the reactor allows fungal cellulolytic enzyme production and alcoholic yeast fermentation of the hydrolysis-derived sugars at the same time. After inoculation, a fungal biofilm is formed on a membrane, as the oxygen required for fungal growth and lignocellulolytic enzyme production is introduced to the cultivation only by diffusion through this membrane. Therefore, the fungal cells attach to the membrane; they grow and form a biofilm on it. The fungal biofilm itself is an oxygen sink, creating anaerobic conditions on the side of the liquid medium where the fermenting microorganism(s) may form an anaerobic biofilm or grow in the liquid, as the released fungal enzymes degrade lignocellulose to monomeric sugars (Brethauer and Studer, 2014).
The enzymatic hydrolysis of cellulose and hemicellulose is the slow step of most biomass bioconversion processes (Revin et al., 2016). Various enzymatic activities should act in synergy to degrade these polymers to free sugars, which will then be converted to the final products by the fermenting microorganisms. Cellobiohydrolases (CBHs), β-Glucosidases (BGs), endoglucanases (EGs), lytic polysaccharide monooxygenases (LPMOs) cleave different bonds on the cellulosic molecule. The lack of one of those slows down the hydrolytic process not only due to inability of the enzymatic system to cleave certain bonds, but also because the accumulation of intermediate products has inhibitory effects on the enzymatic activities. The presence of β-Glucosidase activity in sufficient quantities in the enzymatic cocktail is crucial in order to efficiently degrade cellobiose, which is a strong inhibitor of most cellulolytic enzyme activities (Philippidis et al., 1993; Singhania et al., 2013). Solid state cultivation (SSC) has been widely used for lignocellulolytic enzymes production, based on the well-known ability of fungi to grow on solid surfaces in nature. SSC studies on Aspergillus phoenicis which is a β-Glucosidase over producer have shown that co-cultures with other cellulolytic fungi enhance the cellulolytic efficiency of the produced enzymatic cocktail. SSC co-cultivations with Trichoderma reesei produced a very balanced enzymatic profile and showed about four times increase in β-Glucosidase production, in comparison with T. reesei alone (Wen et al., 2005).
The surface adhesion ability of fungi has been applied in biofilm fermentations which differ from SSC regarding the quantity of liquid being abundant (and free) in the culture environment (Gutiérrez-Correa et al., 2012). Fungal biofilms have been studied regarding their biotechnological applications (Villena and Gutierrez-Correa, 2007; Wang and Chen, 2009; Gamarra et al., 2010; Gutiérrez-Correa et al., 2012; Borghi et al., 2015) including lignocellulolytic enzyme production by Aspergillus strains (Villena and Gutierrez-Correa, 2007, 2012; Gamarra et al., 2010). A. phoenicis is known to be able to form biofilms in certain conditions; however its lignocellulolytic enzymes production, as well as biomass conversion applications of this fungus have not been studied in such cultivations.
In this study we evaluated the fungal biofilms as enzyme producing cultivations in comparison with the submerged cultivations. During T. reesei cultivations, the accumulation of cellobiose due to the lack of β-Glucosidase was shown. It was demonstrated that the MBM process can host more than one aerobic fungus in order to maximize the cellulolytic efficiency of the system. The cellulolytic efficiency of a single fungus biofilm (T. reesei) was compared with the multispecies biofilm (A. phoenicis and T. reesei) in the MBM system. A. phoenicis was selected, based on its secretome profile and its optimal growth conditions. We evaluated and compared the produced enzymatic extracts during enzymatic hydrolysis reactions using Avicel and washed solids from hydrothermally pretreated beechwood.
Materials and Methods
Strains and Chemicals
T. reesei RUT C 30 and A. phoenicis, both acquired from VTT culture collection (VTT Numbers: D-86271and D-76019, respectively) were used in this study. The fungi were received in lyophilized form and were reactivated according to the instructions by the provider. The fungi were grown on potato-dextrose-agar (PDA) slants for 5 days at 28°C. The slants were maintained as stock cultures at 4°C and renewed every 60 days. All chemicals were purchased from Sigma, Switzerland and VWR, Switzerland.
Pretreatment, Composition, and Preparation of Beech Wood Material for Enzymatic Hydrolysis
Beech wood (Fagus sylvatica) chips were milled to a final particle size below 1.5 mm. Steam explosion pretreatment was applied. A custom-built steam gun system (Industrieanlagen Planungsgesellschaft m.b.H., Austria) was used (Pielhop et al., 2016). Two hundred and fifty gram of wood with a moisture content of 6% w/w were inserted into the reactor chamber and saturated steam was injected to pretreat the material. Various experiments on beech wood steam explosion pretreatment revealed that different pretreatment conditions lead to an optimal xylan yield (180°C for 24.8 min) and to a maximum glucan yield (230°C for 14.9 min). Thus, beech wood were pretreated at 230°C for 14.9 min. After pretreatment, the slurry was collected, filtered and washed three times with an overall volume of de-ionized H2O equal to three times the volume of the slurry. The washed solids were then dried at 55°C until constant weight. The dry matter content was measured using a moisture analyzer (Mettler Toledo HB 43 -S, Switzerland). The cellulose and lignin contents in the pretreated beech wood washed solids (PBWS) were estimated using the NREL protocol for the determination of Structural Carbohydrates and Lignin in Biomass (Sluiter et al., 2008). The material was grounded in a Ball mill (Retsch MM 400, Haan, Germany) before analysis. Analyses were performed in triplicate. The same material was used in hydrolysis reactions.
Inoculum Preparation
For inoculum preparation, 6 mL deionized sterile water, containing 1% v/v Tween 80, were added to a PDA slant of the stock culture and aliquots (2 mL) of the mixture were used to inoculate 250 mL Erlenmeyer flasks (sterilized at 121°C for 20 min and cooled prior to inoculation) containing 50 mL of Mandels medium (containing the following ingredients (in g L−1): KH2PO4, 2; (NH4)2SO4, 1.4; MgSO4·7H2O, 0.3; CaCl2·6H2O, 0.4; urea, 0.3; peptone, 0.75; yeast extract, 0.25; and 1 mL L−1 trace element stock). The trace element stock contained (in g · L−1): FeSO4·7H2O, 5; MnSO4·H2O, 1.6; ZnSO4·7H2O, 1.4; CoCl2·6 H2O, 3.7 and 10 mL · L−1 concentrated hydrochloric acid and was sterile filtered. To avoid precipitation, 100 × CaCl2 and 100 × MgSO4 solutions were autoclaved separately). Avicel and glucose were used as the carbon sources during inoculum preparations for T. reesei and A. phoenicis, respectively.
Cultivations in MBM Reactors
Biofilm cultivations were performed in 32 mL MBM reactors set up as previously described (Brethauer and Studer, 2014). The reactors were filled with 31.5 mL of liquid medium. Avicel and washed solids from pretreated beech wood were used as the carbon sources at 2% w/w DM loading. The reactors were inoculated with 1.5 mL (~5% v/v) fungal inoculum (Brethauer and Studer, 2014) and incubated at 27°C. Wherever a co-cultivation of two fungi was performed, the second fungus was inoculated 48 h after the first inoculation (unless otherwise stated). Just before the second inoculation, the reactor was left to settle without stirring for 5 min and 1.5 mL of liquid (equal to the volume of the second inoculum) was removed from the reactor. All cultivations were performed in duplicate. Sampling was performed by harvesting a whole reactor.
Sampling and Fractionation of MBM Cultivations
Liquid Fraction and Solid Residue
After harvesting, the liquid fraction (including the residual solid substrate) was centrifuged at 20,000×g, at 4°C (Scanspeed 1580 R, Labogene, Denmark). An aliquot from the clear supernatant was used for the determination of enzymatic activities in the liquid fraction. The residue was washed as following: 10 mL citrate buffer pH 5, 50 mM, supplemented with 0.5% v/v Tween 80 were added to the pellet and the suspension was vortexed at room temperature for 1 h. The suspension was then centrifuged at 20,000×g, at 4°C. The washing procedure was performed twice. The two supernatants were pooled and were used for the determination of the enzymatic activities adsorbed on the solid substrate. The pellet after the second centrifugation was washed with 15 mL deionized H2O and checked for existence of fungal cells. A schematic representation is provided in Supplementary Figure 2, explaining the sampling and culture fractionation in the MBM system.
Biofilm Fraction
The fungal biofilm was removed from the membrane, suspended in 10 mL citrate buffer pH 5, 50 mM supplemented with 0.5% v/v Tween 80 and homogenized for 2 × 10 s in homogenizer (DT-50-M-gamma Tube with stirring device, Ultra Turrax, IKA, Germany). The suspension was vortexed at room temperature for 1 h and then centrifuged at 20,000×g, at 4°C. The pellet was resuspended in 10 mL citrate buffer pH 5, 50 mM supplemented with 0.5% v/v Tween 80 and the pooled supernatants were used for the determination of enzymatic activities bound to the biofilm. The pellet was then suspended in EDTA disodium salt (5 g · L−1), (Merck, Germany) and centrifuged again at 20,000×g, at 4°C, for 15 min to remove the remaining extracellular polymeric substances (EPS) and then washed with distilled H2O and centrifuged at 20,000×g, at 4°C. The washed pellet was used for fungal growth estimation (Supplementary Figure 2).
Fungal Growth Estimation
The fungal growth estimation was done by measuring the glucosamine (GlcN) content of the fungal cell wall. The method was based on previous published reports (Ride and Drysdale, 1972; Scotti et al., 2001). The samples were diluted in distilled H2O in order to ensure measurements in the linear range of the glucosamine calibration curve and a volume of 0.3 mL was used in the assay. Samples were placed in Pyrex screw capped tubes and 0.3 mL HCl (4M) was used to hydrolyze the fungal biomass. Samples were flushed with nitrogen after HCl addition, and hydrolyzed for 2 h at 121°C. Samples were cooled down and neutralized with 0.4 mL 2M Na2CO3. After neutralization, 0.5 mL of freshly prepared 2% v/v acetyl acetone in 1.5M Na2CO3 were added. Samples were heated in a boiling water bath for 20 min and then 1 ml absolute ethanol was added. 0.5 mL of the Ehrlich's reagent [2 g p-dimethylaminobenzaldehyde in 30 mL EtOH and 30 mL concentrated HCl (32% w/w)] were used for the colorimetric determination of the glucosamine content. The color formation was measured at 530 nm. A calibration curve using GlcN was constructed to determine the linear range of the measurements. Different calibration curves correlated the cell dry weight (CDW) for each fungus (grown on glucose) with their GlcN content. The influence of the substrates (Avicel and PBWS) on glucosamine measurements was also evaluated within the range of substrate concentrations used in the experiments.
Enzyme Assays
One international unit (IU) of enzyme activity was defined as the amount of enzyme required to liberate 1 μmol of product per min, at assay conditions.EG, CBH and BG activities were expressed as IU · mL−1 or as IU · g−1 cell dry weight (CDW), or as total IU produced in the reactor. Total units (produced in the reactor) were used to describe enzyme production and also comparisons among biofilm cultivations, because of the localization of the activities in different fractions of the MBM reactor. All enzyme assays were performed in duplicate at pH 5. Filter Paper Activity (FPA) was assayed as described by Wood and Bhat (1988) and expressed as Filter Paper Units (FPU). BG activity was measured using 1 mM pnp-G (pH 5) as the substrate (0.05 mL sample in a final volume 0.5 mL). The assay was carried out at 50°C for 10 min. The reaction was stopped by the addition of 0.1 mL Na2CO3 (15% w/v) and the absorbance was measured at 410 nm. EG and CBH assays were carried out as described previously (Xiros et al., 2008) using CMC (2% w/w) and Avicel (1% w/w) as the substrates, respectively, at pH 5 and 50°C.
Ultrafiltration of Enzyme Extracts and Enzymatic Hydrolysis Reactions
After harvesting the culture, 0.5% v/v of Tween 80 was added and homogenized for 2 × 10 s (DT-50-M-gamma Tube with stirring device, Ultra Turrax, IKA, Germany). The suspensions were then vortexed at room temperature for 1 h and then centrifuged at 20,000×g, at 4°C. The enzymatic extracts used in the reactions were generated after ultrafiltration of those supernatants using spin filters with a molecular weight cut off of 10 kDalton (Vivaspin®20, Sartorius, Switzerland). The enzymatic profiles of the enzymatic extracts before and after ultrafiltration are shown in Table 1. All enzymatic reactions took place in a thermomixer C (Eppendorf, Hamburg, Germany) at agitation speed 1,400 rpm, in a final volume of 1.5 mL. All reactions were performed in triplicate at pH 5 (phosphate buffer, 50 mM) and T = 30°C, unless otherwise is stated. Dry washed solids from steam pretreated beechwood (PBWS) as well as Avicel were used as substrates (2% w/w).
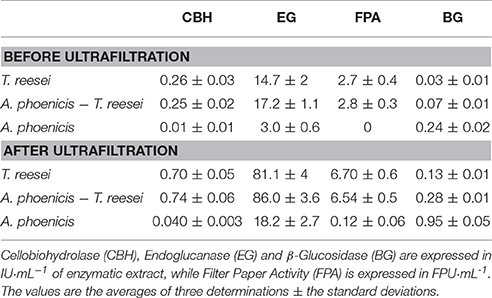
Table 1. Enzymatic profiles of the enzyme extracts produced by biofilm cultivations and used in hydrolysis experiments.
Evaluation of Inhibition Constants for Cellobiose
The effect of cellobiose on cellulose hydrolysis was assessed during hydrolysis reactions of Avicel and PBWS (2 % w/w cellulose in both cases) in citrate buffer pH 5, 50 mM, at 30°C. The reactions were performed in the presence of cellobiose in a range of concentrations from 0 to 30 g · L−1. The enzyme loading was 40 FPU g−1 of cellulose, in order to assure a significant amount of cellobiose release in a short time and to prevent erroneous results due to the cellobiose added in the reaction. δ-Gluconolactone (3 g · L−1) was added to the reaction mixtures to prevent the action of BG; however, the small amounts of glucose formed during the reaction were taken into account (Philippidis et al., 1993). The reaction time was 30 min. The inhibition constants for cellobiose were estimated with nonlinear regression (SigmaPlot v. 12.5) following the equation:
Where, r, is the initial reaction rate of cellulose hydrolysis to cellobiose; ro is the initial reaction rate in the absence of cellobiose; C, is the concentration of cellobiose; and Ki is the inhibition constant of cellobiose.
In Situ Soluble Sugars Removal
Enzymatic reactions were performed with an enzyme loading of 25 FPU g−1 and Avicel initial concentration of 2% w/w, at pH 5 and T = 30 °C. After 24 h of reaction soluble sugars were removed using microtube ultrafilitration membranes (cut off 10 kDalton) under centrifugation at 3,200 rpm at 8°C. After centrifugation, fresh buffer was added and the reaction continued in the initial reaction volume, without adding any fresh enzyme.
Reducing Sugars, Glucose, and Cellobiose Quantification
Reducing sugars were quantified using the DNS method (Miller, 1959). Glucose and cellobiose concentrations were quantified using high performance liquid chromatography (Waters 2695 Separation Module, Waters Corporation, Milford, MA, USA) using an Aminex HPX-87H column (Bio-Rad, Hercules, CA, USA) at 60°C, with 5 mM H2SO4 as the mobile phase (0.6 mL min−1) and a refractive index detector (Waters 410).
Statistical Evaluation of the Results
Significant differences between different experimental conditions were evaluated using T-test. A two sided unpaired t-test (for independent samples) was applied. Normal distribution was assumed. The T-test was applied for equal variance and 2 degrees of freedom for Figures 1–3, 8 and 4 degrees of freedom for Figures 6, 7 (n1 + n2 − 2). The “t critical” was calculated for DF = 2 or DF = 4 and probability = 0.05. The null hypothesis was that the groups compared were not different. The null hypothesis was rejected when the absolute t stat value was higher than the t critical value. In the figures the error bars represent the range of the two independent values in case of two replicates; while the standard deviation is shown in the cases where triplicate measurements were performed. For cellobiose inhibition (Figure 5), the significant difference of the constants calculated by the models was evaluated for 80% confidence intervals.
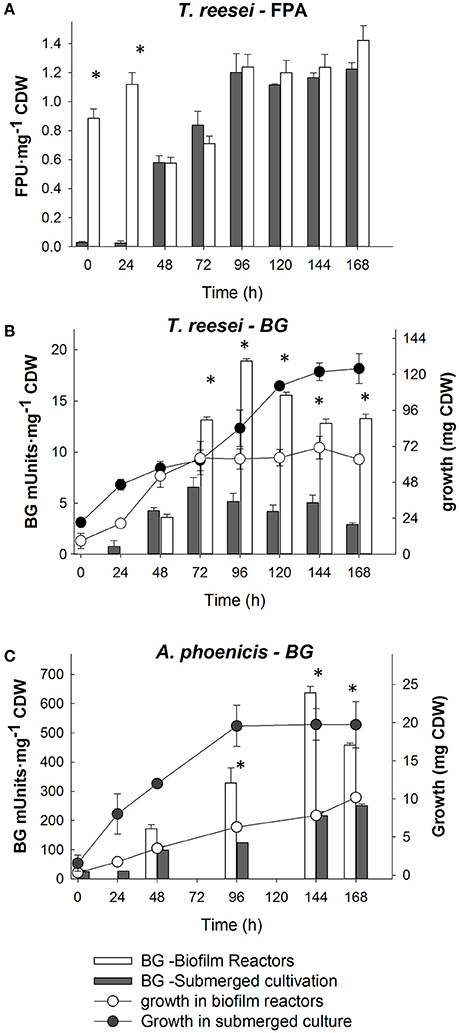
Figure 1. Comparison of cultivation techniques for fungal growth and cellulolytic enzymes production by T. reesei and A. phoenicis. The comparisons are based on specific enzyme activities (Units per mg of CDW). (A) Comparison of Filter paper activity production by T. reesei in submerged and biofilm cultivations. (B) Comparison of BG activity and growth by T. reesei between submerged and biofilm cultivations. (C) Comparison of BG activity and growth by A: phoenicis between submerged and biofilm cultivations. In all comparisons, equal inoculums and equal culture volumes were used. All cultivations were performed twice, and assays were performed in duplicate. Error bars represent the range between replicates. White color represents biofilm cultures, while gray represents submerged ones. The circles connected with lines show growth, while bars show specific enzyme activities. Only at the time points marked with asterisks, the activities measured were statistically different between submerged and biofilm cultivations.
Results
Cellulolytic Enzymes Production by Fungal Biofilms
Comparisons between Submerged and Biofilm Cultivations Regarding Growth and Enzyme Production
T. reesei RUT C-30, a filamentous fungus widely used for cellulolytic enzymes production, was grown at submerged and biofilm cultivations, using Avicel (2% w/w) as the carbon source. The fungal growth and also the cellulolytic enzymes production were compared between the two cultivation methods. The comparison was based on fungal growth and extracellular FPA and BG productions. Total FPU (for FPA) and total IU (for BG) for equal culture volumes were calculated. At submerged conditions, higher enzyme activities per culture volume were measured (data not shown). However, as shown in Figures 1A,B, where the enzyme activities were normalized based on fungal CDW, the higher activities at submerged conditions were mainly due to the significantly higher fungal growth at these conditions.
The comparison between the two cultivation techniques indicates that when T. reesei grew in a biofilm, the cells showed an enhanced metabolic activity. The production of both FPA and BG practically stopped after 96 or 120 h of cultivation in all cases. In the biofilm membrane reactors growth already stopped after 72 h. However, BG activity remained at very low levels (below 45 mU mL−1) in both cultivations. The low BG productivity was also reflected on the cellobiose levels in the liquid medium, measured during the cultivation in the membrane reactor. As shown in Figure 2A, cellobiose accumulated in the MBM reactors after 96 h of cultivation.
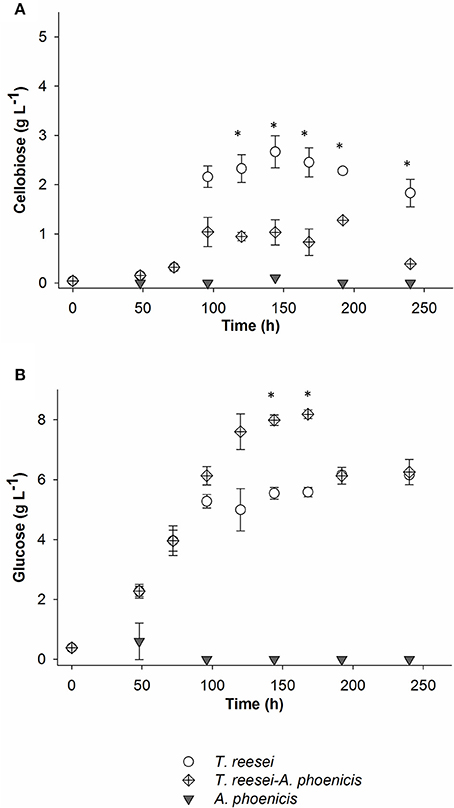
Figure 2. Cellobiose (A) and glucose (B) concentrations in the liquid fraction of the biofilm cultivations. All cultivations were performed twice. Error bars represent the range between replicates. In the case of the mixed culture, Time = 0 h, corresponds to the inoculation of T. reesei. A. phoenicis was inoculated 2 days later. The time points marked with asterisks indicate the concentrations that were statistically different between mixed and single (T. reesei) cultivations.
During the experiments with A. phoenicis as the sole microorganism, the formation of biofilm by A. phoenicis in the MBM system was confirmed only when glucose was used as the carbon source. In these experiments glucose and Avicel were tested as carbon sources. In the case of Avicel, the biofilm formation was very slow and not so clear. In fact, a very thin and not well formed biofilm was observed only after 5 or 6 days of cultivation. These observations reflected and confirmed the weak growth of A. phoenicis on Avicel (Figure 1C). A. phoenicis could not efficiently hydrolyze Avicel and grow on it, as indicated by the overall cellulase activity (expressed as FPA) measurements, which were close to the detection limits of the method used (Table 1 and Figure 3). As also shown in Table 1, there was a lack of CBH activity in A. phoenicis enzyme extract which explained the inability of the fungus to grow well on Avicel. Fungal growth in the membrane reactors was half of that in submerged cultivation, but as in the case of T. reesei, cells were metabolically very active as shown by the enhanced specific BG production compared to the submerged cultivation. As shown in Figure 1C, a three-fold difference in BG activity normalized on fungal growth (CDW) was observed after 4 days of A. phoenicis cultivation. BG activity after 6 days of cultivation was higher in the case of membrane reactors even when activities per culture volume were measured.
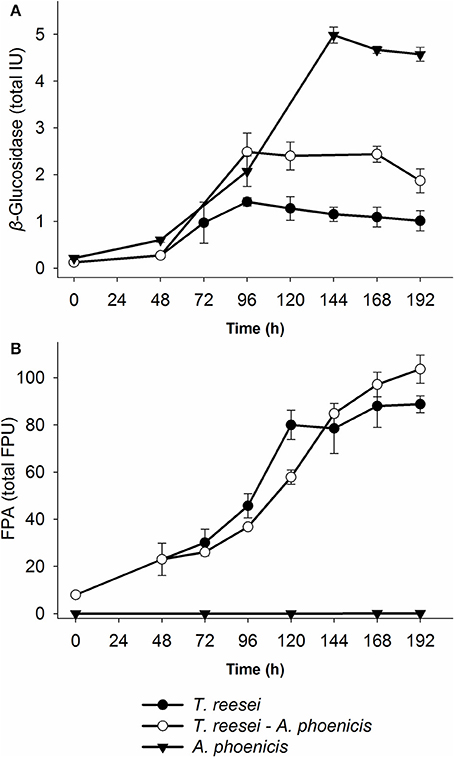
Figure 3. Time course of enzyme production by the single and mixed biofilm cultivations in the biofilm membrane reactors. (A) β-Glucosidase activity, and (B) Filter Paper Activity. In the case of the mixed cultivation, time zero corresponds to the inoculation of T. reesei, while A. phoenicis was added 48 h later. The activities shown are the sum of all three fractions (Liquid, Biofilm, cellulosic residue) for each time point. Therefore, total units were calculated for the whole MBM reactor. All cultivations were performed twice and enzyme assays were performed in duplicate. Error bars represent the range between replicates. BG activities between the mixed (A. phoenicis – T. reesei) and the single (T. reesei) cultivations measured after 120 h were statistically different. On the contrary, FPA was not found to show statistical differences between the same cultivations. Details on the statistical tests are given in materials and methods section.
Localization of Enzymatic Activities in Biofilm Membrane Reactors
Culture supernatants have been used for enzymatic activities estimation during experiments at submerged conditions in numerous studies. In SSCs of fungi, an extraction step is necessary to obtain and measure the produced (extracellular) activities (Xiros et al., 2008). Fungal biofilm cultivations have been considered as an intermediate between solid and submerged cultures: Fungi grow on a solid surface but completely covered by the liquid medium. In the present study, the activities measured in the liquid supernatants from the cultivations represented only a part of the cellulolytic enzyme production. The measurements of enzymatic activities in all three fractions (solid residue, biofilm, liquid medium) of the cultures revealed the true amounts of cellulolytic activities produced (Figure 4). The distribution of activities among these fractions did not remain constant, but depended on the activity measured as well as on the cultivation time. In Supplementary Figure 1 the localization of Filter Paper and β-Glucosidase activities over time is shown for T. reesei cultivations. As shown, the distribution was different for BG and FPA. BG activity increased over time in the biofilm fraction while it decreased in the liquid. Taking into account all three fractions, it can be observed from Figure 1 that BG production stopped 1 day after fungal growth reached its maximum value. BG activity increased until 96 h and changed only slightly thereafter. On the contrary, the general cellulolytic activity (FPA) increased in the liquid, as the cultivation time passed, possibly also due to cell lysis during the late stage of cultivation. However, FPA does represent a mixture of cellulolytic activities, and thus conclusions regarding its relation with growth are difficult.
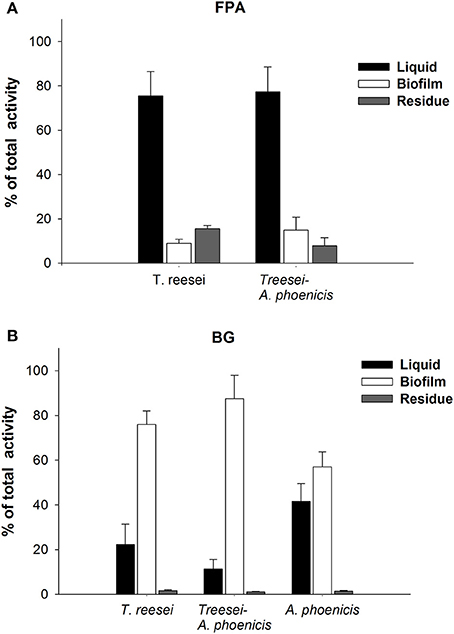
Figure 4. Localization of enzymatic activities in the biofilm membrane reactors during single and mixed cultivations. The activities are given as percentages of the total units produced in the whole reactor. (A) FPA, (B) BG. All cultivations were performed twice, and assays were performed in duplicate. Error bars represent the range between replicates.
Enhancement of β-Glucosidase Production by Multispecies Biofilm Formation in Membrane Reactors
Setup of T. reesei and A. phoenicis Co-cultivation
In preliminary experiments, three different inoculation procedures were tested: (a) Subsequent inoculation with T. reesei as the first microorganism, (b) subsequent inoculation with A. phoenicis as the first microorganism, and (c) simultaneous inoculation of the two fungi. The three setups were evaluated with regard to the cellulolytic activities produced. When T. reesei was the first microorganism inoculated and A. phoenicis was added 2 days afterwards, the best results in terms of enzyme production were obtained. This was expected as in this case, A. phoenicis, due to its enhanced β-Glucosidase production, could grow on the cellobiose which had been already released by T. reesei enzymes in the medium. Microscopic observations of the biofilms were performed for all different setups. However, both T. reesei and A. phoenicis in the biofilm could microscopically be observed when T. reesei was the first microorganism and A. phoenicis was added afterwards.
In single microorganism experiments, T. reesei formed a distinct biofilm after 48 h. On the contrary, A. phoenicis barely grew on Avicel due to the inability of this fungus to grow on cellulose as it lacks a complete cellulolytic enzymatic mixture. When both fungi were simultaneously inoculated in the MBM system, A. phoenicis hyphae could not be observed in the biofilm during microscopic observation and no differences were observed in BG production. The inoculation of A. phoenicis 2 days after T. reesei was found to be a much better procedure for the co-cultivation. However, a more detailed investigation would probably indicate more precisely the optimum inoculation time of A. phoenicis and would further enhance the BG production.
Enzyme Production in Multispecies Biofilm Membrane Reactors
In Figure 3, the enzymatic activities produced by the multispecies biofilm cultivations are compared with the single biofilm ones. As shown, there was a significant enhancement of BG activity produced by the mixed culture, compared to T. reesei alone: An increase by a factor of 2.5 was achieved after 2 days of common cultivation (corresponding to 4 days from T. reesei inoculation). The maximum BG activity was 2.5 IU for the whole reactor (Figure 3), corresponding to 0.084 IU·mL−1. However, BG activity did not reach the levels of the A. phoenicis single culture (5 IU). The increased BG activity was also reflected to the cellobiose concentrations during the cultivation. Cellobiose concentration decreased by 50 to 70% and remained at low levels (about 1g·L−1) throughout the experiment (Figure 2). FPA activity was only slightly affected by the addition of A. phoenicis and reached about 85% of its maximum value after 4 days of co-cultivation (Figure 3).
The microscopic observation of the mixed biofilms showed that there were distinct regions where the presence of one fungus was more intense and regions where the distribution of the two fungi was more balanced. Although a quantitative analysis of the microscopic images was not possible, it was clear that the distribution of the two fungi in the biofilm was changing overtime. The activities measured in the biofilm fraction (Figure 4 and Supplementary Figure 1) also showed differences related not only to the microorganisms involved but also related to the cultivation time. FPA was mainly found in the liquid fraction, while the amount adsorbed on the residual solids (Avicel) was changing overtime, and on the 7th day of the cultivation it only corresponded to 16 and 8% of the total FPA measured in all fractions (Figure 4), for T. reesei and the mixed culture (A. phoenicis – T. reesei), respectively. As also shown in the same figure, after 168 h of growth, BG activity was found mainly in the biofilm, except for the case of A. phoenicis single culture, where almost half of the activity was measured in the liquid fraction. However, this was expected, as in this case the biofilm was not well formed.
Hydrolytic Efficiency of Enzymes Secreted by Single- and Multi-Species Biofilms
Inhibition of Cellulases by Cellobiose
The effect of cellobiose on cellulose hydrolysis was studied for both cellulolytic systems (produced by T. reesei and by the mixed culture A. phoenicis – T. reesei) and for two types of cellulosic substrates (Avicel and PBWS). As can been seen in Figure 5, the regressions successfully described the effect of cellobiose on cellulases, with R2 for all cases ranging between 0.89 and 0.97. In the case of T. reesei enzymes, the estimated values for the inhibition constants varied between the two substrates (2.64 g · L−1 for Avicel and 9 g · L−1 for PBWS). The difference between the two constants was statistically significant for confidence intervals 80%. No significant difference between the substrates was observed in the case of the A. phoenicis – T. reesei enzymes.
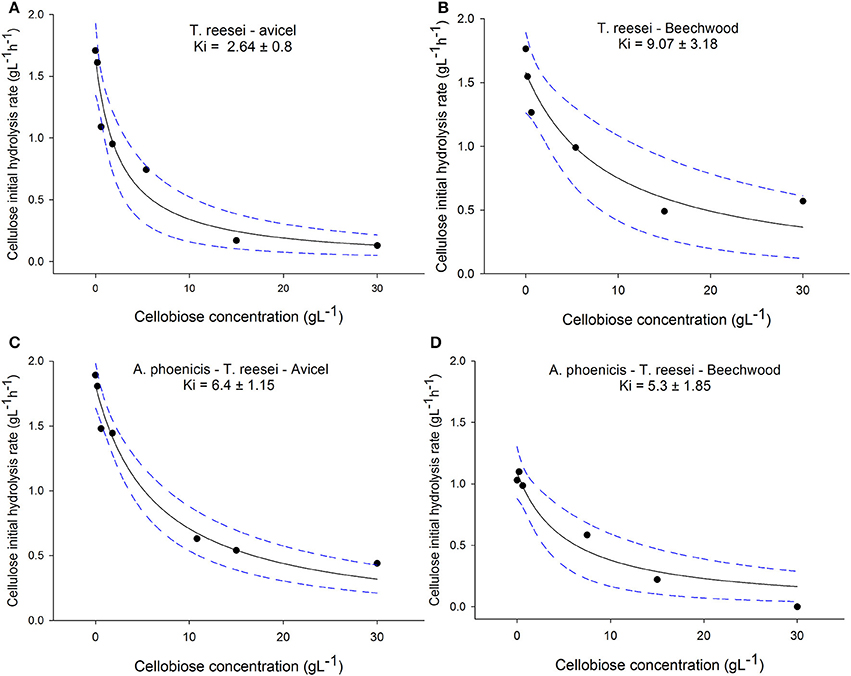
Figure 5. Effect of cellobiose on cellulose initial hydrolysis rates. All reactions were performed in duplicate, with 40 FPU g−1 of cellulose, at 30°C and at pH 5. The cellulose loading was 2% w/w in all cases. The R2 values for the regressions were 0.94, 0.89, for the enzymatic system of T. reesei on Avicel (A) and PBWS (B), respectively, and for the enzymatic system of the mixed culture (A. phoenicis − T. reesei) the R2 values were 0.97 (C) and 0.93 (D). The dashed lines show the 95% confidence band. The estimated inhibition constants for the enzymatic system of T. reesei were found statistically different between the two substrates for a confidence interval of 80%. No statistical differences between different substrates were found for the enzymatic system of the mixed culture.
Hydrolysis of Different Cellulosic Substrates Using Different Enzyme Dosages
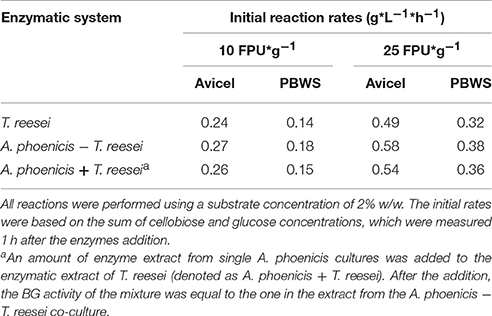
Hydrolysis reactions were designed and performed to evaluate the hydrolytic efficiency of the enzymatic systems produced by single and mixed cultures. The reactions were performed at the cultivation conditions (30°C, pH 5) in order to be able to derive lessons for a consolidated bioprocess based on a microbial consortium where fungi grow and secrete enzymes to release sugars fed to the fermenting microorganism in the anaerobic part of the reactor (Brethauer and Studer, 2014). Apart from Avicel, PBWS were also used as substrate. Analysis of PBWS for cellulose and acid insoluble lignin contents, gave mass fractions of 60.5% w/w (±5.9%) and 36.7 % w/w (±1.4%), respectively, on a dry weight basis. All hydrolysis reactions were performed with a substrate concentration of 2% w/w.
During Avicel hydrolysis, 10 FPU·g−1 of T. reesei enzymes hydrolyzed 17% of the substrate in 72 h, while 6.4 g·L−1 of glucose and cellobiose cumulatively were released after 150 h of reaction corresponding to a hydrolysis yield of 29%. However, cellobiose concentration reached 4.7 g·L−1, which was almost three times higher than the glucose released (1.7 g·L−1) at the same reaction time (Figure 6). The picture of released sugars was similar during hydrolysis of PBWS using the same amount of T. reesei enzymes, where 41% of total cellulose was hydrolyzed after 150 h, releasing 5.5 g·L−1 of glucose and cellobiose cumulatively, while glucose concentration reached 1.6 g·L−1 corresponding to less than half of the cellobiose concentration (3.9 g·L−1) (Figure 7).
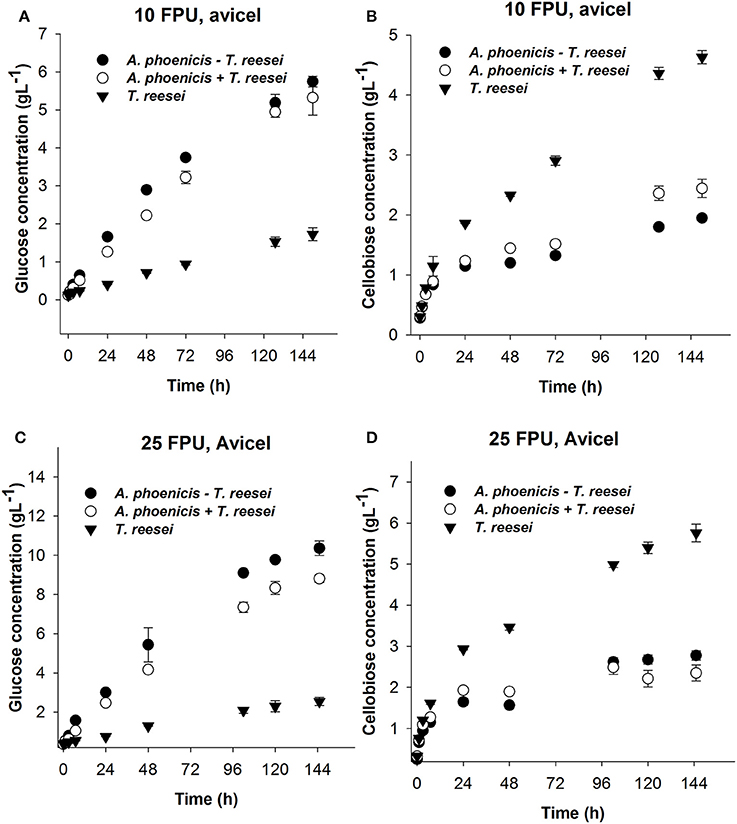
Figure 6. Glucose (A,C) and cellobiose (B,D) release during enzymatic hydrolysis of Avicel using 10 FPU (A,B) and 25 FPU (C,D) of enzymatic extract per g of dry substrate. The hydrolytic performance of the extract from the single culture (T. reesei) is compared with that from the mixed culture (A. phoenicis − T. reesei). In the case of T. reesei + A. phoenicis, the enzymatic extracts of single cultures were mixed to achieve a BG activity equal to that of the mixed culture. All reactions were performed at 30°C, pH 5, in triplicate. Error bars represent standard deviations. Cellobiose and glucose release were compared between the T. reesei enzymes and the enzymes of the mixed cultivation. In all cases the differences were found statistically significant. Details on the statistical tests are given in materials and methods section.
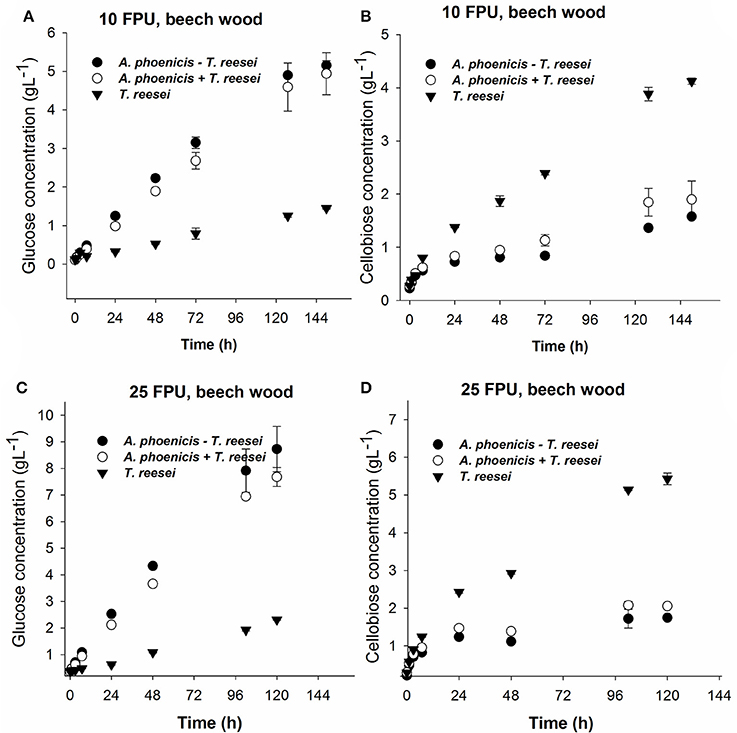
Figure 7. Glucose (A,C) and cellobiose (B,D) release during enzymatic hydrolysis of PBWS (washed solids from pretreated beechwood) using 10 FPU (A,B) and 25 FPU (C,D) of enzymatic extract per g of dry substrate. The hydrolytic performance of the extract from the single culture (T. reesei) is compared with that from the mixed culture (A. phoenicis − T. reesei). In the case of T. reesei + A. phoenicis, the enzymatic extracts of single cultures were mixed to achieve a BG activity equal to that of the mixed culture. All reactions were performed at 30°C, pH 5, in triplicate. Error bars represent standard deviations. Cellobiose and glucose release were compared between the T. reesei enzymes and the enzymes of the mixed cultivation. In all cases the differences were found statistically significant. Details on the statistical tests are given in materials and methods section.
The enzymatic system produced by the mixed biofilm of A. phoenicis and T. reesei was more efficient in hydrolyzing both Avicel and PBWS. Using 10 FPU of that enzyme extract, 7.7 g·L−1and 6.7 g·L−1 of glucose and cellobiose were cumulatively measured after 150 h hydrolysis of Avicel and PBWS, respectively (Figures 6, 7). In both cases and due to the enhanced BG activity, the released glucose was dramatically increased compared to the glucose release by only T. reesei enzymes: 5.7 g·L−1 and 5.2 g·L−1 of glucose was released from Avicel and PBWS, respectively, corresponding to 80–90% of total sugars released.
As expected, the use of higher enzyme dosage increased the initial hydrolysis rates (Table 2), and also the final hydrolysis yields. When 25 FPU/g solids were used in the reactions, the initial rates were at least doubled in all cases. The initial reaction rates also revealed the difference in digestibility between Avicel and PBWS. In all cases initial reaction rates of PBWS hydrolysis were significantly lower than those of Avicel hydrolysis. The released glucose from Avicel reached 10.4 g·L−1 and 2.5 g·L−1 for the enzymatic extracts from the mixed (A. phoenicis − T. reesei) biofilm and the single one (T. reesei), respectively. Comparing with the hydrolysis reactions with 10 FPU·g−1, the increase in glucose release was much higher in the case of the extract from the mixed biofilm, confirming that in the case of T. reesei extract, the crucial weakness is the lack of BG activity and not the enzyme dosage. The sum of glucose and cellobiose after 120 h of hydrolysis was 13.1 g·L−1 for the enzymes from mixed culture, and 8.3 g·L−1 for those from the single culture. In the case of PBWS, the sum of these two sugars reached 10.5 g·L−1 corresponding to 79% of cellulose content (Figures 6, 7).
Effect of in Situ Sugar Removal during Enzymatic Hydrolysis
In a consolidated bioprocess using the MBM reactor, the sugars released due to the action of fungal enzymes, are consumed mainly by the fermenting microorganism, resulting in the formation of the final products (Brethauer and Studer, 2014). Thus, like in simultaneous saccharification and fermentation, the sugars quickly disappear from the cultivation medium. To evaluate the hydrolytic efficiency of the different enzymatic systems at such conditions, hydrolysis reactions with in situ sugars removal were conducted. As shown in Table 3, a significant enhancement in reaction rates was observed right after the sugars removal as a result of decreased end product inhibitory effects on hydrolytic enzymes. The reactions rates decreased rapidly during the following 24–48 h of reactions. However, compared to the reactions where no in situ sugars removal was performed, the final hydrolysis yields increased 9 and 16% for the reactions with enzymes from mixed biofilm (A. phoenicis − T. reesei) and from the single one (T. reesei), respectively (Figure 8).
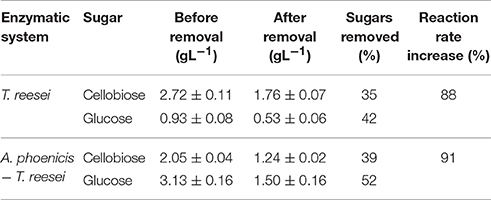
Table 3. In situ sugars removal during hydrolysis of Avicel by the enzymatic systems produced by single (T. reesei) and mixed cultures (A. phoenicis − T. reesei).
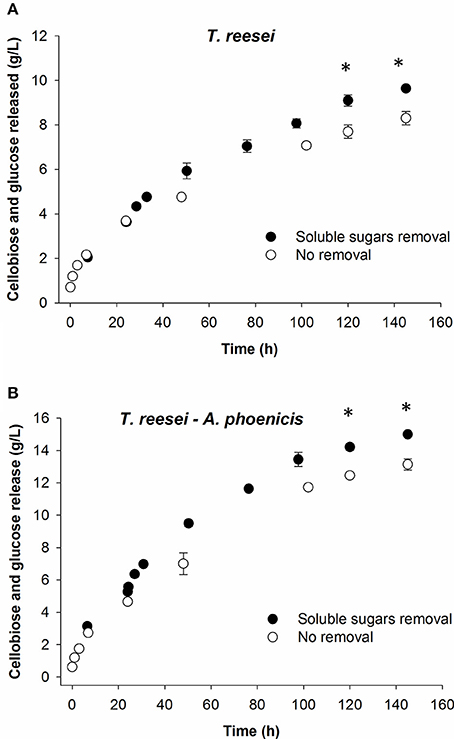
Figure 8. Effect of in situ hydrolysate removal on hydrolysis of Avicel by the enzymatic extracts of mixed (A) and single cultivations (B). In all cases, the sum of cellobiose and glucose is presented. For the black circles (hydrolysate removal), the calculated released sugars are shown (sugars concentrations measured plus those removed). Reactions were performed at 30°C, pH 5, in duplicate. Error bars represent range between replicates. Sugar release was compared between the removal and no-removal reactions. The statistical significant differences are marked with an asterisk. Details on the statistical tests are given in materials and methods section.
Discussion
Evaluation of T. reesei and A. phoenicis Biofilms for Cellulolytic Enzymes Production
As shown in Figure 1, both fungi grew better in submerged cultures than in biofilm membrane reactors. Although the enzymatic activities produced per volume of culture were significantly higher in the case of the submerged cultures, the FP and BG specific activities (activities normalized per CDW) were higher in the case of the MBM system (biofilm growth). Due to the biofilm membrane reactor design, growth is closely related to the available membrane area. Increase in the area (and more specifically increase in the ratio of membrane area to reactor volume) would result in increased fungal growth in terms of CDW and therefore it would further increase enzyme production. However, it is difficult to predict how it would affect specific enzymatic activities as activity is not linearly correlated with concentration. These findings are in accordance with previous studies showing that the cultivation technique influences the fungal morphology. The morphology in turn, affects fungal productivities and metabolism (Wösten et al., 1991; Quintanilla et al., 2015). Villena and Gutierez-Correa compared the specific enzyme activities produced by another Aspergillus species (A. niger) in submerged and biofilm cultivations, and showed that biofilm cultures achieved much higher specific activities (Villena and Gutierrez-Correa, 2007). In that case, as also in the present study, the different morphology of fungi at different cultivation conditions reflected differences in metabolism, which in turn, resulted in enhanced specific enzymatic productivities during biofilm growth. Thus, growth (rate and yield) and cultivation technique are two crucial parameters for the optimization of enzyme production by fungi.
It is well known from literature that the production of cellulases by fungi is associated with their growth. In batch cultivation, most cellulolytic activities are produced during the exponential or the early stationary phase (Xiros et al., 2008; Xiros and Christakopoulos, 2009). This is reasonable, since the fungus needs to degrade the substrate to gain the necessary carbon, and to do so, it needs to produce lignocellulolytic enzymes. This was partially observed during this study, as shown in Figures 1, 3, for both fungi and both enzyme activities measured over time: Only a slight increase in measured activities was observed after 100 (BG) or 120 h (FPA) of cultivation, while fungal growth reached highest levels 24 h or 48 h before that. Besides, the lack of major cellulolytic activities in the secretome of A. phoenicis (Table 1) resulted in very limited growth of this fungus on Avicel (Figure 1). This observation is in accordance with previous reports showing the inability of this fungus to produce a complete cellulolytic enzymatic mixture, capable of hydrolyzing cellulosic substrates (Wen et al., 2005). Therefore, A. phoenicis has rarely been studied as the sole microorganism in cultures using cellulose. Instead, lignocellulosic substrates also containing small amounts of free sugars like sugar beet pulp are more suitable for its growth (Deschamps and Huet, 1984).
Enhancement of BG Activity
Many Aspergillus species, including A. phoenicis have been used in co-cultures in order to enhance the production of BG activity (Castillo et al., 1994; Wen et al., 2005; Brijwani et al., 2010). However, a co-cultivation of A. phoenicis with T. reesei in biofilms had never been tried before. As also previously implied by Brethauer and Studer (2014) the low BG activity produced by T. reesei in the MBM system would lower the efficiency of the consolidated bioconversion system for any cellulose containing plant biomass. Therefore, a multispecies approach was applied during this study in order to enhance the BG productivity in the system. A. phoenicis a β-Glucosidase overproducer (Woodward and Wiseman, 1982) was selected to form a multispecies biofilm with T. reesei. The two fungi have similar optimal values for growth pH and T and also both microorganisms have been successfully co-cultivated before, in Mandel's medium (Woodward and Wiseman, 1982; Wen et al., 2005).
The progressive accumulation of glucose in the liquid medium was a strong indication that fungal anabolism slowed down and thus glucose was not assimilated by the fungi (Figure 2B). The fact that this happened not only in the case of T. reesei, but also in the case of the mixed cultivation where cellobiose levels were low, may imply that the cellulolytic rates were higher than the sugar uptake rates for this strain at these conditions. The enhancement of BG activity during the co-cultivation did not significantly affect FPA, which is in accordance with previous studies (Wen et al., 2005).
Hydrolytic Efficiency of the Enzymatic Systems
The enhancement of BG in enzymatic extract from the mixed culture affected sugars release from both substrates used, during the enzymatic reactions performed. The effects on glucose release were spectacular and these were reflected to a smaller extent on total sugars release. The effects of BG enhancement on the initial reaction rates were more visible when 25 FPU g−1 were applied as enzyme loading. In this case, the enzymatic extract of the mixed culture achieved 16% higher reaction rates than the one from T. reesei for both Avicel and PBWS. As observed, the reaction rates decreased rapidly after the first 10–20 h, but only slightly thereafter. The initial decrease in the rates was more evident in the case of PBWS. This implies structural unevenness in this substrate compared with the structurally more homogeneous Avicel (Xiros et al., 2011). The reactions took place at 30°C in order to simulate the conditions during the CBP process in the MBM reactor. At this temperature, the enzymes remained active for quite a long time, but the relatively low reaction rates would be a bottleneck for a commercial process. It is clear, that the use of thermotolerant fungi and fermenting microorganisms would boost the productivity of the system, allowing hydrolysis to take place in higher temperature. A higher reaction temperature would increase the initial rates and shorten significantly the reaction time, but could also result in a faster deactivation of the enzymes.
The initial reaction rates when 25 FPU·g−1 were used as the enzyme loading, are roughly double than the rates with 10 FPU·g−1 (Table 2) but not proportional to the enzyme dosage increase. This not proportional relationship (A Langmuir-like dependence) reflects the functional adsorption of enzymes on the cellulose. Beyond a certain enzyme loading, no further significant increase in the initial rates can be observed due to the saturation of the substrate with enzymes (Xiros et al., 2011). As shown in Table 2, initial reaction rates were much higher for Avicel hydrolysis than for PBWS. This was probably due to the adsorption (not functional binding) of enzymes on lignin in the case of PBWS leading to decreased amounts of active enzyme molecules. The differences in initial reaction rates between PBWS and Avicel hydrolysis reactions would decrease as enzyme dosage would increase, because lignin (in PBWS) would become more and more saturated with enzymes. The enzyme loading of 25 FPU·g−1 is somewhat less than half of the FPA in the biofilm reactors. Of course, a comparison between the two processes (hydrolysis and cultivation) is not possible, due to the different localization of the enzymes in the two systems and also due to the existence of active cells in the second case which metabolize the released sugars.
The substrate used in the cultivations is also of importance, since cellobiose inhibition on cellulases depends not only on the present enzymatic activities but also on the kind of cellulose that is hydrolyzed. Gruno et al. (2004) reported that cellobiose had much stronger inhibitory effects on enzymes that act on crystalline regions (CBHs) than on enzymes that act on amorphous cellulose (EGs). This is also reflected to the differences in the values of cellobiose inhibition constants on cellulases that had been reported for different substrates, and vary from 0.5 g·L−1 to 5.5 g·L−1 (Philippidis et al., 1993; Gruno et al., 2004). However, the Ki values in the literature are strongly depended on the reaction conditions and therefore they can hardly be compared. During the present study the significance of the substrate for cellobiose inhibition was confirmed in the case of T. reesei enzymes: When Avicel was used as the substrate, the Ki for cellobiose was almost four times lower than during PBWS hydrolysis (Statistically confirmed in 80% confidence intervals). Indeed, Avicel is a very crystalline form of cellulose, and its hydrolysis is very much depended on the CBH activity of the cellulolytic system. PBWS on the other hand was generated during steam pretreatment from beechwood, and contains amorphous regions which are hydrolyzed mainly by EGs. In the case of Avicel hydrolysis by T. reesei cellulases, the Ki value (2.64 g·L−1) showed that the cellobiose concentrations measured after 96 h of cultivation in MBM reactors (using Avicel as the carbon source), decreased the cellulolytic rate about 50%. On the other hand, BG production by the mixed biofilm (A. phoenicis − T. reesei) limited cellobiose accumulation up to 1.3 g·L−1. At these levels cellobiose could have decreased hydrolysis rates only by 20% (Ki = 6.4 g·L−1).
In situ sugars removal almost doubled the reaction rates. End-product inhibition of lignocellulolytic enzymes has been regarded as a major bottleneck of many bioconversion processes. However, during the present study, only slightly over one third of the cellobiose was removed. The cellobiose concentration in the reaction mixtures increased again quickly and contributed to a rapid decrease in reaction rates. This could be one reason for the moderate effect that the hydrolysate removal had on the final saccharification yields in both cases. Moreover, it should be kept in mind that inhibition by cellobiose does not give the whole picture of the inhibition, as various hydrolysis products have inhibitory effects on the hydrolytic enzymes including oligosacharides, which were not quantified before and after the removal (Kim et al., 2011; Xiros et al., 2011).
Conclusion
This study showed the efficiency of fungal biofilms as cellulolytic enzyme producers using two filamentous fungi T. reesei and A. phoenicis. The accumulation of cellobiose when T. reesei was the sole enzyme producer showed that there was a lack of BG activity in the system. A multispecies approach was applied using A. phoenicis as the BG producer microorganism. Although A. phoenicis could not grow on Avicel as the sole microorganism, due to the lack of CBH activity in its secretome, it was possible to form a multi-species biofilm with T. reesei in the MBM reactors, proving the versatility of the reactor design to host multi-species fungal biofilms. It was shown that fungal biofilms are effective systems for cellulolytic enzymes production compared to other cultivation techniques. The two fungi co-formed a biofilm, which produced a balanced cellulolytic system containing all main activities (CBHs, EGs, BGs) for cellulose hydrolysis. From the results of the enzymatic hydrolysis reactions on Avicel and PBWS by T. reesei enzymes, it was shown that the inhibition of cellulases by cellobiose was substrate depended. It was also concluded that the enhancement of BG activity (in the secretome of the multispecies biofilm) compared to T. reesei enzymatic system, not only was important for the glucose yields but also could positively affect the initial hydrolysis rates.
Author Contributions
CX conceived the study, conducted the experiments, and drafted the manuscript. CX and MS performed the experimental design and the analysis of results. MS and CX critically revised the manuscript and both authors approved the final version of the article.
Funding
This work was supported by the Swiss National Science Foundation in the framework of the National Research Programme “Energy Turnaround” (NRP 70) [grant number 407040_153868] as well as by the Commission for Technology and Innovation (CTI), Federal Department of Economic Affairs, Education and Research, Swiss Confederation, through SCCER biosweet [contract number KTI.2014.0116].
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CX and MS are grateful to Robert Shahab for all his technical help and also to Robert Balan for his help on the steam pretreatment of beech wood.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.01930/full#supplementary-material
Abbreviations
CDW, Cell Dry Weight; BG, β-Glucosidase; DM, Dry matter; EG, Endoglucanase; CBH, Cellobiohydrolase; FPA, Filter Paper Activity; PBWS, Washed Solids from Pretreated Beechwood; MBM (reactors), Multispecies Biofilm Membrane reactors; SSC, Solid State Cultivation; CBP, Consolidated bioprocess; HCBP, Highly Consolidated Bioprocess; NREL, National Renewable Energy Laboratory; DM, Dry matter; PDA, Photodiode array.
References
Borghi, E., Morace, G., Borgo, F., Rajendran, R., Sherry, L., Nile, C., et al. (2015). New strategic insights into managing fungal biofilms. Front. Microbiol. 6:1077. doi: 10.3389/fmicb.2015.01077
Brethauer, S., and Studer, M. H. (2014). Consolidated bioprocessing of lignocellulose by a microbial consortium. Energy Environ. Sci. 7, 1446–1453. doi: 10.1039/c3ee41753k
Brijwani, K., Oberoi, H. S., and Vadlani, P. V. (2010). Production of a cellulolytic enzyme system in mixed-culture solid-state fermentation of soybean hulls supplemented with wheat bran. Process. Biochem. 45, 120–128. doi: 10.1016/j.procbio.2009.08.015
Castillo, M. R., Gutierrez-Correa, M., Linden, J. C., and Tengerdy, R. P. (1994). Mixed culture solid substrate fermentation for cellulolytic enzyme production. Biotechnol. Lett. 16, 967–972. doi: 10.1007/BF00128635
Deschamps, F., and Huet, M. C. (1984). β-Glucosidase production by Aspergillus phoenicis in solid state fermentation. Biotechnol. Lett. 6, 55–60. doi: 10.1007/BF00128230
Gamarra, N. N., Villena, G. K., and Gutierrez-correa, M. (2010). Cellulase production by Aspergillus niger in biofilm, solid-state, and submerged fermentations. Appl. Microbiol. Biotechnol. 87, 545–551. doi: 10.1007/s00253-010-2540-4
Gruno, M., Valjamae, P., Pettersson, G., and Johansson, G. (2004). Inhibition of the Trichoderma reesei cellulases by cellobiose is strongly dependent on the nature of the substrate. Biotechnol. Bioeng. 86, 503–511. doi: 10.1002/bit.10838
Gutiérrez-Correa, M., Lude-a, Y., Ramage, G., and Villena, G. K. (2012). Recent advances on filamentous fungal biofilms for industrial uses. Appl. Biochem. Biotechnol. 167, 1235–1253. doi: 10.1007/s12010-012-9555-5
Jouzani, G. S., and Taherzadeh, M. J. (2015). Advances in consolidated bioprocessing systems for bioethanol and butanol production from biomass: a comprehensive review. Biofuel Res. J. 2, 152–195. doi: 10.18331/BRJ2015.2.1.4
Kalyani, D., Lee, K.-M., Kim, T.-S., Li, J., Dhiman, S. S., Kang, Y. C., et al. (2013). Microbial consortia for saccharification of woody biomass and ethanol fermentation. Fuel 107, 815–822. doi: 10.1016/j.fuel.2013.01.037
Kim, Y., Ximenes, E., Mosier, N. S., and Ladisch, M. R. (2011). Soluble inhibitors/deactivators of cellulase enzymes from lignocellulosic biomass. Enzyme Microb. Technol. 48, 408–415. doi: 10.1016/j.enzmictec.2011.01.007
McAllister, T. A., Bae, H. D., Jones, G. A., and Cheng, K. J. (1994). Microbial attachment and feed digestion in the rumen. J. Anim. Sci. 72, 3004–3018. doi: 10.2527/1994.72113004x
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428. doi: 10.1021/ac60147a030
Nagaraja, T. G. (2016). “Microbiology of the Rumen,” in Rumenology, eds D. D. Millen, M. de Beni Arrigoni, and R. D. Lauritano Pacheco (Cham: Springer International Publishing), 39–61.
Philippidis, G. P., Smith, T. K., and Wyman, C. E. (1993). Study of the enzymatic hydrolysis of cellulose for production of fuel ethanol by the simultaneous saccharification and fermentation process. Biotechnol. Bioeng. 41, 846–853. doi: 10.1002/bit.260410903
Pielhop, T., Amgarten, J., Rohr, P. R., and von Studer, M. H. (2016). Steam explosion pretreatment of softwood: the effect of the explosive decompression on enzymatic digestibility. Biotechnol. Biofuels 9:152. doi: 10.1186/s13068-016-0567-1
Quintanilla, D., Hagemann, T., Hansen, K., and Gernaey, K. V. (2015). “Fungal morphology in industrial enzyme production–modelling and monitoring,” in Filaments in Bioprocesses, eds R. Krull and T. Bley (Cham: Springer International Publishing), 29–54.
Revin, V., Atykyan, N., and Zakharkin, D. (2016). Enzymatic hydrolysis and fermentation of ultradispersed wood particles after ultrasonic pretreatment. Electron. J. Biotechnol. 20, 14–19. doi: 10.1016/j.ejbt.2015.11.007
Ride, J. P., and Drysdale, R. B. (1972). A rapid method for the chemical estimation of filamentous fungi in plant tissue. Physiol. Plant Pathol. 2, 7–15. doi: 10.1016/0048-4059(72)90043-4
Scotti, C., Vergoignan, C., Feron, G., and Durand, A. (2001). Glucosamine measurement as indirect method for biomass estimation of Cunninghamella elegans grown in solid state cultivation conditions. Biochem. Eng. J. 7, 1–5. doi: 10.1016/S1369-703X(00)00090-5
Singhania, R. R., Patel, A. K., Sukumaran, R. K., Larroche, C., and Pandey, A. (2013). Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour. Technol. 127, 500–507. doi: 10.1016/j.biortech.2012.09.012
Sluiter, A., Hames, B., Ruiz, R., Scarlata, C., Sluiter, J., Templeton, D., et al. (2008). Determination of Structural Carbohydrates and Lignin in Biomass. Laboratory Analytical Procedure 1617, NREL/TP-510-42618. Golden, CO: National Renewable Energy Laboratory.
Villena, G. K., and Gutierrez-Correa, M. (2007). Morphological patterns of Aspergillus niger biofilms and pellets related to lignocellulolytic enzyme productivities. Lett. Appl. Microbiol. 45, 231–237. doi: 10.1111/j.1472-765X.2007.02183.x
Villena, G. K., and Gutierrez-Correa, M. (2012). Kinetic analysis of Aspergillus niger cellulase and xylanase production in biofilm and submerged fermentation. J. Microbiol. Biotechnol. Res. 2, 805–814. Available online at: https://www.jmbronline.com/index.php/JMBR/article/view/197/197
Wang, Z.-W., and Chen, S. (2009). Potential of biofilm-based biofuel production. Appl. Microbiol. Biotechnol. 83, 1–18. doi: 10.1007/s00253-009-1940-9
Wen, Z., Liao, W., and Chen, S. (2005). Production of cellulase/β-glucosidase by the mixed fungi culture Trichoderma reesei and Aspergillus phoenicis on dairy manure. Process Biochem. 40, 3087–3094. doi: 10.1016/j.procbio.2005.03.044
Wood, T. M., and Bhat, K. (1988). “Methods for measuring cellulase activities,” in Cellulose and Hemicellulose, ed W. A. Wood (San Diego, CA: Academic Press), 87–112.
Woodward, J., and Wiseman, A. (1982). Fungal and other β-D-glucosidases -their properties and applications. Enzyme Microb. Techol. 4, 73–79. doi: 10.1016/0141-0229(82)90084-9
Wösten, H. A. B., Moukha, S. M., Sietsma, J. H., and Wessels, J. G. H. (1991). Localization of growth and secretion of proteins in Aspergillus niger. Microbiology 137, 2017–2023. doi: 10.1099/00221287-137-8-2017
Xiros, C., and Christakopoulos, P. (2009). Enhanced ethanol production from brewer's spent grain by a Fusarium oxysporum consolidated system. Biotechnol. Biofuels 2:4. doi: 10.1186/1754-6834-2-4
Xiros, C., Katapodis, P., and Christakopoulos, P. (2011). Factors affecting cellulose and hemicellulose hydrolysis of alkali treated brewers spent grain by Fusarium oxysporum enzyme extract. Bioresour. Technol. 102, 1688–1696. doi: 10.1016/j.biortech.2010.09.108
Keywords: multispecies biofilm membrane reactors, Trichoderma reesei, Aspergillus phoenicis, lignocellulose, cellulases, β-glucosidase, fungal biofilms
Citation: Xiros C and Studer MH (2017) A Multispecies Fungal Biofilm Approach to Enhance the Celluloyltic Efficiency of Membrane Reactors for Consolidated Bioprocessing of Plant Biomass. Front. Microbiol. 8:1930. doi: 10.3389/fmicb.2017.01930
Received: 04 July 2017; Accepted: 21 September 2017;
Published: 10 October 2017.
Edited by:
Qiang Wang, Institute of Hydrobiology (CAS), ChinaReviewed by:
Tania Pozzo, University of California, Davis, United StatesHans C. Bernstein, Pacific Northwest National Laboratory (DOE), United States
Copyright © 2017 Xiros and Studer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael H. Studer, bWljaGFlbC5zdHVkZXIxQGJmaC5jaA==
 Charilaos Xiros
Charilaos Xiros Michael H. Studer
Michael H. Studer