- 1Laboratorio de Biotecnología, Instituto de Nutrición y Tecnología de los Alimentos, Universidad de Chile, Santiago, Chile
- 2Departamento de Agroindustria y Enología, Facultad de Ciencias Agronómicas, Universidad de Chile, Santiago, Chile
- 3Helios Innovation, Valparaíso, Chile
The presence and load of species of LAB at the end of the malolactic fermentation (MLF) were investigated in 16 wineries from the different Chilean valleys (Limarí, Casablanca, Maipo, Rapel, and Maule Valleys) during 2012 and 2013, using PCR-RFLP and qPCR. Oenococcus oeni was observed in 80% of the samples collected. Dominance of O. oeni was reflected in the bacterial load (O. oeni/total bacteria) measured by qPCR, corresponding to >85% in most of the samples. A total of 178 LAB isolates were identified after sequencing molecular markers, 95 of them corresponded to O. oeni. Further genetic analyses were performed using MLST (7 genes) including 10 commercial strains; the results indicated that commercial strains were grouped together, while autochthonous strains distributed among different genetic clusters. To pre-select some autochthonous O. oeni, these isolates were also characterized based on technological tests such as ethanol tolerance (12 and 15%), SO2 resistance (0 and 80 mg l−1), and pH (3.1 and 3.6) and malic acid transformation (1.5 and 4 g l−1). For comparison purposes, commercial strain VP41 was also tested. Based on their technological performance, only 3 isolates were selected for further examination (genome analysis) and they were able to reduce malic acid concentration, to grow at low pH 3.1, 15% ethanol and 80 mg l−1 SO2. The genome analyses of three selected isolates were examined and compared to PSU-1 and VP41 strains to study their potential contribution to the organoleptic properties of the final product. The presence and homology of genes potentially related to aromatic profile were compared among those strains. The results indicated high conservation of malolactic enzyme (>99%) and the absence of some genes related to odor such as phenolic acid decarboxylase, in autochthonous strains. Genomic analysis also revealed that these strains shared 470 genes with VP41 and PSU-1 and that autochthonous strains harbor an interesting number of unique genes (>21). Altogether these results reveal the presence of local strains distinguishable from commercial strains at the genetic/genomic level and also having genomic traits that enforce their potential use as starter cultures.
Introduction
Malolactic fermentation (MLF) is a process performed by lactic acid bacteria (LAB) that transforms malic acid into lactic acid and CO2, which causes a decrease in the total acidity and improvement of the taste, flavor, and microbial stability of wine (Henick-Kling, 1995; Capozzi et al., 2010). Those bacteria are naturally present in grapes, musts and wines. The predominant genera are Leuconostoc, Pediococcus, Lactobacillus, and Oenococcus (Lonvaud-Funel, 1995).
Previous studies investigating autochthonous LAB in winemaking have reported the presence of Oenococcus oeni strains in spontaneous MLF (Marques et al., 2011; Nisiotou et al., 2015; Cafaro et al., 2016). The genetic diversity of O. oeni has been shown in studies from different winemaking regions worldwide. Bartowsky et al. (2003) determined that O. oeni strains that originated from the same winery in Australia were either indistinguishable or closely related to each other. In Castilla-La Mancha, Spain, Cañas et al. (2009) showed that MLF was dominated by a variable number of O. oeni genotypes. The same observation of the genetic diversity in these bacteria was reported in La Rioja, Spain, and Apulia, Italy (González-Arenzana et al., 2012; Garofalo et al., 2015).
Bacterial dynamics during MLF have been studied using culture-dependent techniques. The major drawback of this strategy is the impossibility of correctly obtaining the diversity and dynamics of LAB during MLF (Spano et al., 2007). However, culture-independent analysis methods have been developed and are commonly used to detect and identify microorganisms directly from wine by analyzing their DNA. Ilabaca et al. (2014) designed a 16S rRNA Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (16S rRNA PCR-RFLP) culture-independent strategy that was a reliable tool for the identification and differentiation of winemaking LAB strains isolated during the MLF process. González-Arenzana et al. (2012) studied LAB populations in red wine (La Rioja, Spain) and compared two strategies (culture-dependent and culture-independent methods). Both methods were complementary during MLF. However, the culture-independent methods allowed the detection of a vaster number of species than the culture-dependent methods. Therefore, O. oeni was the most frequently detected bacterium during MLF. The conclusions drawn from these molecular studies indicate that LAB populations are diverse during the early stages of MLF; however, O. oeni subsequently becomes the most dominant bacterial population during the MLF process. This result is consistent with the previous observations derived using culture-dependent approaches (Rodas et al., 2003; López et al., 2007).
The genetic characterization of O. oeni has permitted the evaluation of differences between O. oeni isolates from diverse winemaking locations using several molecular strategies, including multilocus sequence typing (MLST) (de las Rivas et al., 2004; Bilhère et al., 2009; Bridier et al., 2010; Bordas et al., 2013; Wang et al., 2015). Several studies reported a high level of allelic diversity and a combination of alleles among O. oeni isolates (Bilhère et al., 2009; Bon et al., 2009). However, genetic differences between O. oeni strains must be studied and understood because they might affect the quality of the wine, especially in terms of organoleptic properties. Recently, the genome sequences of O. oeni strains have been made available from different enological locations (Mills et al., 2005; Borneman et al., 2010; Capozzi et al., 2014; Lamontanara et al., 2014; Jara and Romero, 2015). Jara and Romero (2015) suggested that genomic analyses might provide insights into the adaptation of strains to wine-hostile conditions and their contributions to the organoleptic properties of the final product. Cappello et al. (2017) proposed an association between inter/intra-species diversity and bacterial metabolic traits that impacted the wine's organoleptic characteristics. Additionally, these authors showed evidence of the importance of the enzymatic potential of LAB to enrich the wine aroma.
In Chile, most MLF processes are conducted spontaneously by the resident LAB microbiota in the cellars. However, spontaneous MLF has drawbacks, including stuck fermentation and contamination by microorganisms, which risk altering the wine quality. To solve this problem, the use of microbial starters has been introduced with commercial strains isolated from other wine-producing countries. However, in Chile some of these starters have produced poor results due to their insufficient imposition during MLF (Ilabaca et al., 2014). Among the many factors that impede the development of O. oeni, the most important is the presence of inhibiting factors, such as a low pH, high ethanol content, and low malic acid content.
This study reports the first genetic and technological characterization of O. oeni strains retrieved from spontaneous MLF in different Chilean valleys. Additionally, the genomes of selected isolates were examined and compared them to PSU-1 and VP41 strains to study their potential contributions to the organoleptic properties of the final product. These characteristics could be the basis for obtaining autochthonous isolates to serve as starters capable of improving the typicity of Chilean wines.
Materials and Methods
Samples
Spontaneous MLF samples (58) were collected in 2012 and 2013 from 16 wineries, including cultivars (cvs.) Cabernet Sauvignon and Carménère, located in four Chilean valleys: Limarí (30°38′S–71°24′W), Maipo (33°45′S–70°46′W), Rapel (34°15′S–72°00′W), and Maule (35°58′S–72°19′W), sampling four wineries per valley. All the tested wineries carried out spontaneous MFL without commercial starter; sampling was performed at the end of the MLF. The winemaking process was initiated with healthy grapes harvested from March to May, followed by the traditional vinification practices of each winery. As a general rule of each winery alcoholic fermentation (AF) using commercial freeze-dried yeast was performed stainless steel tanks at 22–25°C. Spontaneous MLF was carried out immediately after AF in stainless steel tanks at 18–22°C for 30–40 days. Samples were aseptically collected at the end of MLF, where the wines showed average chemical parameters: ethanol (14.1%v/v) and pH (3.7). The criterion for defining the end of MLF in each winery is the reduction in the content of the L-malic acid (<0.3 g/L) in the wines determined using an enzymatic test (Boehringer Manheim; Mannheim, Germany). Samples were stored at 4°C until being processed.
Bacterial Isolation
The bacterial isolation was carried out using medium for Leuconosctoc oenos (MLO), following indications by Blasco et al. (2003). This medium was supplemented with 2 ml L−1 sodium azida (Winkler, Chile) and 3 ml L−1 cyclohexamide (Sigma-Aldrich) to eliminate yeasts and acid acetic bacteria, respectively (Ruiz et al., 2008). Serial dilutions were plated onto the MLO media and incubated for 5–7 days at 28°C, under anaerobic conditions. After count colonies (CFU mL1) 10 colonies per sample were randomly chosen. This selection was realized according to the phenotypic characterization (Mesas et al., 2011). Each selected colony was transferred and purified through two rounds of streak plating onto fresh agar plates. The isolates were maintained in a cryobank at −80°C.
Reference and Commercial Strains
Lactic acid bacteria commercial strains for comparison in genetic diversity study were used. This included Lallemand (Lalvin VP41®, Lalvin 31®, uvaferm Alpha®, uvaferm Beta®, Lalvin Elios®, PN4®, Lalvin MTO®); Lamothe Abiet (Oeno 1, Oeno 2); Laffort (Lactoenos B28 PreAc®). Lalvin VP41® was included in the technological evaluation.
DNA Extraction from Wine
The initial step for our culture independent approach was the extraction of DNA directly from wine with MLF, according to Jara et al. (2008) and Ilabaca et al. (2014). This DNA was used to quantify bacterial load (see below).
DNA Extraction from Bacterial Isolates
In the case of LAB isolates, each of the colonies selected was suspended in 200 mL PBS, with vigorous agitation, followed by centrifugation at 5,000 × g for 5 min. Subsequently, 20 μL aliquot of 20 mg mL−1 Lysozyme (Sigma) was added to the pellets, which were subsequently incubated at 37°C for 30 min. The Wizard Genomic DNA Purification Kit (Promega) was used for DNA extraction according to the protocol for isolating genomic DNA from Gram-positive bacteria the manufacturer's instructions. DNA obtained was frozen at −20°C until processed. The identification de LAB from samples MLF wine was done by 16S rRNA PCR-RFLP, according to Ilabaca et al. (2014).
Amplification rpoB gen
To distinguish O. oeni isolates, RNA polymerase ß subunit (rpoB) were PCR amplified using methodology described by Bridier et al. (2010). DNA sequencing was performed by Macrogen (USA) and the analyses were done by BLAST (basic Alignment Search Tool). From the rpoB sequence results of autochthonous O. oeni isolates and commercial strain (VP41), were analyzed with its differences about nucleotides among them by used BioEdit version 7.1.9, generating signature groups (Drancourt and Raoult, 2002). The signature sequences corresponded to part of a coding sequence of a gene which, because it is shared by different isolates, is thought to be evolutionarily conserved and therefore can serve to trace taxonomic relationships among these isolates (Gupta, 1998).
Quantitative PCR (qPCR) and Total Bacteria and O. oeni
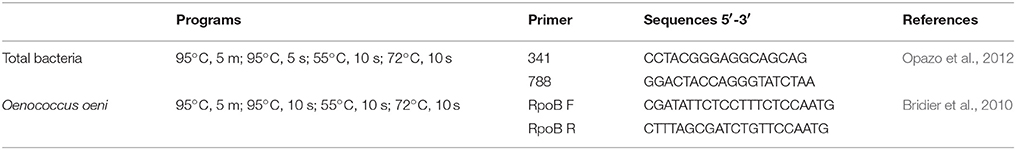
Both total bacteria and load of O. oeni were quantified by qPCR reactions based on detection of SYBR fluorescence. The qPCR reactions were performed using an AriaMx real-time PCR System (Agilent Technologies), using primers and programs described in Table 1. PCR amplification was performed in 10 μL of mix containing 1 μL of DNA 0.5 pmol/mL of each respective primer 5 μL of LightCycler 480 SYBR Green I Master (Roche) and 3.5 μL of Milli-Q sterile H2O. All samples were analyzed in triplicate. The statistical analyses among bacterial loads valleys were determined by ANOVA using R Development Core Team (2008), and the post-hoc test was performed by pairwise.t.test.
Multilocus Sequence Typing (MLST) Analysis
Based on rpoB gen analysis (signature groups), the autochthonous O. oeni isolates were selected about two criteria. First, the different rpoB sequences to obtain maximal diversity. Second, rpoB sequences isolates from different Chilean Valleys.
To analysis of MLST seven housekeeping genes for this study were selected. These genes were gyrB (Gyrase Beta subunit), g6pd (Glucose-6-phosphate dehydrogenase), mleA (Malolactic enzyme), pgm (Phosphoglucomutase), dnaE (DNA polymerase III, alfa subunit), pgm (Phosphorybosylaminoimidazole), purK (Phosphoribosylamino-imidazole carboxylase), rpoB (RNA polymerase, Beta subunit). The recP gene was not included in our analysis because it was not present in our isolates. After the examination of a subset of 114 O. oeni genomes originated from diverse geographic locations, only 40 strains harbored this gene (Supplementary Table S1). The PCR mixes were performed according to de las Rivas et al. (2004) and Bridier et al. (2010).
The PCR products were sequenced by Macrogen (USA). The analysis of sequences obtained by MLST was realized using BioEdit version 7.1.9 software and a dendrogram was constructed by UPGMA (unweighted pair-group method with arithmetic mean) method. The evolutionary distances were computed using the Maximum Composite Likelihood method, using the Mega 6 (version 6.0) software from the website (Tamura et al., 2013).
The sequences of each gen of autochthonous O. oeni isolates selected and commercial strain (VP41), were analyzed with its differences about nucleotides among them by used BioEdit version 7.1.9. The base pair of each gen analyzed were rpoB gen 579 bp; dnaE gen 641 bp; g6pd gen 591 bp; gyrB gen 544 bp; mleA gen 355 bp; pmg gen 580 bp; purK gen 493 bp.
Evaluation of Technological Properties of O. oeni Isolates
Based on the MLST analysis, some strains were included in the technological tests. For the different technological tests, autochthonous O. oeni isolates were grown in MLO broth to early stationary phase. An inoculum of 1*108 cells mL−1 of each O. oeni isolates was used to inoculate wine-like medium (Bordas et al., 2015). Our criterion to select the isolates was a first test; wine-like medium was supplemented with two ethanol concentrations (12% v/v and 15% v/v), the isolates natives were incubated at 25°C for 10 days. Then, the best isolates were submitted to wine-like medium supplemented with malic acid 1.5 and 4g L−1 at 25°C for 10 days. Wine-like medium either at pH 3.1 and 3.6 and were incubated at 25°C for 24 h and 5 days. Finally, the isolates were incubated into this medium, utilizing potassium metabisulfite concentrations (0 and 80 ppm free SO2) at 25°C for 7 days. All bacterial growth was per triplicate and monitored by culture dependent analyses. The significant differences among isolates of each tests were determined by Kruskal–Wallis test using R Development Core Team (2008), and the post-hoc test was performed by posthoc.kruskal.dunn.test belonging to the PMCMR package.
Genomic Analyses
Based on technological characteristics, the draft genome of three autochthonous O. oeni isolates were obtained and analyzed. Genome characteristics and the accession numbers about those isolates have been previously reported (Jara and Romero, 2015). Using the online tool at bioinformatics.psb.ugent.be, a Venn diagram was generated to compared genome composition of O. oeni autochthonous isolates with 2 reference strains: VP41 (ACSE00000000) and PSU-1 (NC_008528).
Three approaches to calculate average nucleotide identity from genome sequences of O. oeni autochthonous isolates and 2 reference strains: VP41 (ACSE00000000) and PSU-1 (NC_008528) were used. Those were: DNA–DNA hybridization (DDH), Average Nucleotide Identity (ANI) and Orthology (OrthoANI). The similarity obtained by DNA–DNA hybridization (DDH) to genome-sequence-based similarity according to Meier-Kolthoff et al. (2013) was determined. Second approach utilized was average nucleotide identity (ANI) according to Rosselló-Mora (2006). Finally, third approach was average nucleotide identity by orthology (OrthoANI) according to Lee et al. (2016).
The presence and homology of 21 (abf, arcA, alsS, alsD, arcB, arcC, bgl, citD, citE, citF, estA, estB, estC, gshR, maeP, metB, metC, metK, mleA, pad, prtP) genes potentially related to aromatic profile were analyzed from O. oeni, autochthonous isolates and reference strains VP41 and PSU-1 genome sequences. These genes were taken from literature (Sumby et al., 2009, 2013; Mtshali et al., 2012; Cappello et al., 2014). The orthologous clustering analysis were performed using Inparanoid (Fouts et al., 2012; Sonnhammer and Östlund, 2015). To analyze enzymes groups (glycosidases, esterases, proteases, citrate metabolism, and peptidases), that may play a role in the wine organoleptic properties into proteomes of O. oeni, autochthonous isolates and reference strains VP41 and PSU-1 genome sequences were realized using coding sequence for protein (CDS) by PfamScan (Finn et al., 2016).
Results
Lactic Acid Bacteria in Different Chilean Valleys
A total of 60 wine samples from four Chilean valleys were used to characterize the bacterial population dynamics at the end of spontaneous MLF with 16S rRNA PCR-RFLP. O. oeni was observed in 80% of the wine samples. Lactobacillus and Leuconostoc were detected in 4.5% and 2.3% of the samples, respectively. Furthermore, Oenococcus/Pediococcus and Oenococcus/Leuconoctoc were found together at frequencies of 4.5 and 2.3%, respectively.
Both the total bacterial and O. oeni loads were explored by qPCR. Figure 1 shows the number of microorganisms (log10 scale) represented by each valley ordered from north to south (Limarí, Maipo, Rapel, and Maule). Limarí and Rapel valleys showed the highest total bacterial load with 107 total bacteria per mL of wine. Significant differences were found in total bacterial loads among valleys; Limarí and Maipo valleys (p-value: 0.034); Maipo and Rapel valleys (p-value: 0.000013); Rapel and Maule valleys (p-value: 0.0014). On the other hand, Rapel and Maule valleys, showed the highest O. oeni loads, with 106 O. oeni per mL of wine. Significant differences were found in O. oeni load among valleys, Maipo and Maule valleys (p-value: 0.00048) and Maipo and Rapel valleys (p-value: 0.00012). In summary, the O. oeni loads and the total bacterial loads indicated a dominance of O. oeni at the end of MLF.
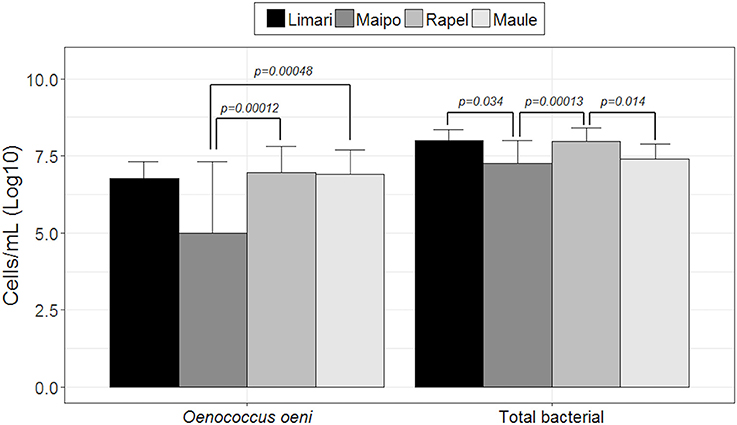
Figure 1. Total bacterial and O. oeni load quantified by qPCR in wine from spontaneous MLF different valleys examined (log10 scale).
Identification and Characterization of LAB Isolates
A total of 158 autochthonous LAB isolates were retrieved from red wine collected at the end of spontaneous MLF in wineries located in the Limarí, Maipo, Rapel, and Maule Valleys and characterized using phenotypic tests (catalase and Gram staining). Among them, 75 strains were identified as O. oeni by PCR-rpoB sequencing. The analysis of a 579-bp rpoB sequence generated signature groups according to the different nucleotides and positions using the rpoB sequence from the VP41 reference strain. Based on this analysis, 46 autochthonous O. oeni isolates were found to differ from VP41, and 7 signature groups were observed. The signature groups are described in detail in Table 2, including the signature, number of isolates and origin. Based on these signature groups, 10 autochthonous O. oeni isolates for analysis using MLST typing were selected. The selection was based on two inclusion criteria of at least 1 isolate from each signature group and two strains per valley. Additionally, 10 O. oeni commercial strains were included in the MLST genetic analysis for comparison. Concatenation of the sequences for each of the seven genes formed a 3,783-bp sequence, which was examined using the MEGA software. The resulting dendrogram (Figure 2) showed two major genetic groups of O. oeni (M and A). Group M included all of the commercial strains and some autochthonous O. oeni isolates (13, 399, 565, and 74). Furthermore, the commercial strain MTO presented a transposon in purK from 423 to 1,282 bp. In contrast, group A was only composed of autochthonous O. oeni. Group M was composed of two subgroups that showed different distributions of autochthonous isolates based on the signature groups. Interestingly, two isolates obtained from geographically separated valleys (500 kilometers) grouped together in A.
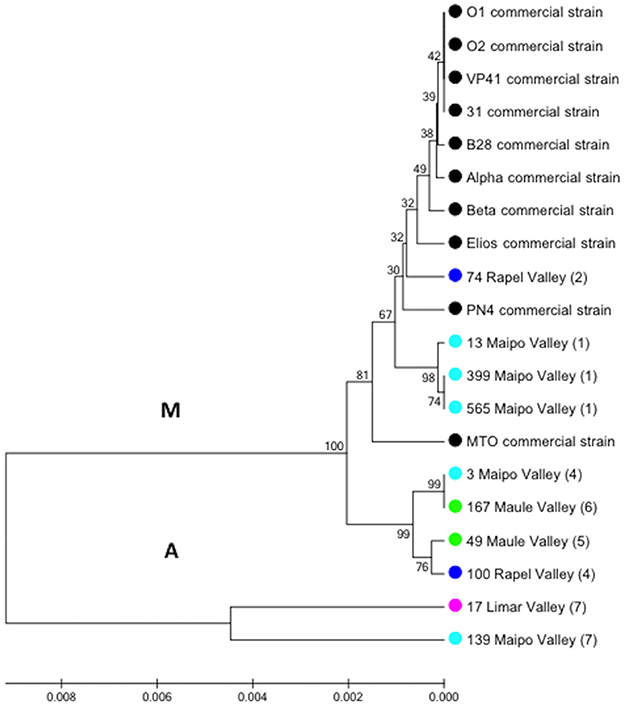
Figure 2. Phylogenetic reconstruction based on the seven gene markers MLST scheme. Analysis included O. oeni commercial strains and Chilean autochthonous O. oeni isolates, which were distributed in two genetic groups indicated as M and A. Colored circles indicate the origin of each isolate (Chilean valley or commercial). Numbers correspond to signature group described in Table 2.
Technological Properties of Autochthonous O. oeni Isolates
The ability of autochthonous strains to resist wine-like medium supplement with ethanol (12% v/v) was compared with strain VP41 (LABc). Three isolates were discarded because they were unable to survive in this medium. Therefore, only seven isolates were examined to assess their technological properties, including the kinetics of the transformation of malic acid, ethanol resistance, pH resistance, and SO2 resistance. Supplementary Figures S1A,B shows the ability of the isolates to degrade malic acid at two initial concentrations. The strains 139 and 565 showed the best reduction of this acid. Similarly, all strains grew in the presence of 12% ethanol, but 139 showed the best survival in 12 and 15% ethanol (Supplementary Figures S1C,D). To study the association between bacterial growth and pH tolerance, autochthonous strains were examined in media with different pH values (3.1 and 3.6). Supplementary Figures S1E,F, showed that strain 17 and 74 did not survive these conditions; in contrast, strain 139 presented the highest potential for growth in restrictive pH media. Similarly, the effect of sulfur oxide (0 and 80 ppm) was examined (Supplementary Figures S1G,H). Sulfur oxide was deleterious for strains 17, 74, and 167 whereas isolate 139 presented a high viable count over the 10-day period. In summary, the best strains according to their technological properties were 139 and 565.
Genome-Based Phylogeny and Genome Comparison
To examine the relationships among the autochthonous O. oeni strains (565, 399, and 139) and the reference strains (VP41 and PSU-1), genome analyses were performed. First, the number of common genes shared by these O. oeni strains was evaluated; the results are shown in a Venn diagram in Figure 3. This figure revealed 470 common genes, of which 63% were uncharacterized proteins and 9% were ribosomal genes. Each strain presented unique genes, indicating that the autochthonous strains harbored an interesting number of unique genes. Strain 139 presented 28 unique genes, strain 565 presented 27 unique genes and strain 399 presented 21 unique genes. These genes encoded ABC transporters; galactose metabolism; phosphostransferase system (PTS); pentose phosphatase pathway; starch and sucrose metabolism; two component system; nicotinate and nicotinamide metabolism that might improve bacterial performance in the wine environment. Then, the autochthonous O. oeni genomes were compared to the reference strains using 3 methodologies (ANIb, DDH, and OrthoANI) (Table 3). The three methods showed that strain 139 had the lowest similarity to VP41 and PSU-1.
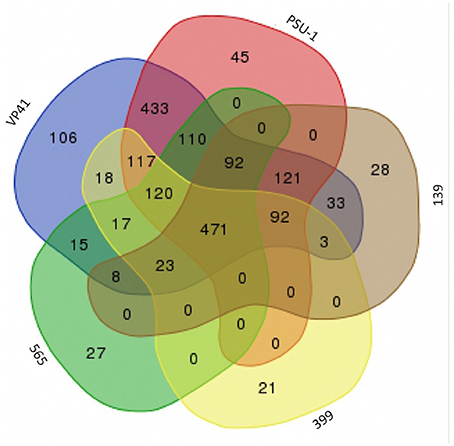
Figure 3. Venn diagram between genome of Chilean autochthonous O. oeni isolates compared with two reference strains VP41 and PSU-1.
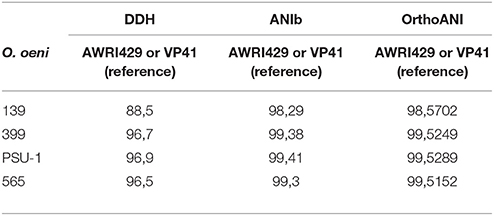
Table 3. Results of DDH, ANIb, and OrthoANI algorithms of autochthonous O. oeni genomes compared to reference strain VP41.
Genes that Contribute to Organoleptic Properties
To study the potential contributions to the organoleptic properties of the final product, genes related to the improvement of the wine's aromatic profile were analyzed. The autochthonous O. oeni genomes were compared to reference strains in terms of families and domains of interest for the enzymes that contributed to the organoleptic properties, such as esterases, glycosidases, enzymes involved in citrate metabolism, peptidases and proteases (Table 4). Our results showed that strain 139 presented a higher number of enzymes than strains 399 and 565.
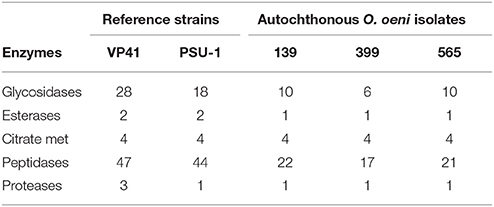
Table 4. Occurrence of esterases, glycosidases, citrate metabolism, peptidases, and proteases enzymes of autochthonous O. oeni genomes compared with reference strains (VP41 and PSU-1).
The genome analysis screened for the presence and identity of 21 genes encoding enzymes potentially related to the aromatic profile of the wine as previously described (Mtshali et al., 2010; Cappello et al., 2014). Table 5 showed that strain 139 harbored 14 of the 21 screened genes, which was higher than the numbers found for strains 565 and 399. Furthermore, the phenolic acid carboxylase gene (pad), carbamate kinase gene (arcC) and protease ptrP gene were absent in the autochthonous O. oeni genomes.
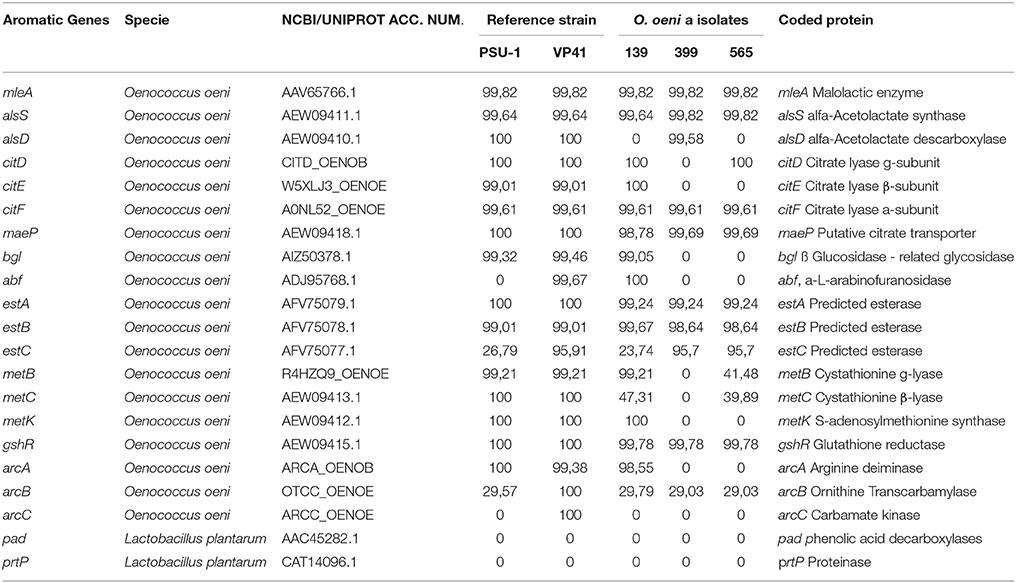
Table 5. Identities of aromatic genes found between autochthonous O. oeni genomes compared with reference strains (VP41 and PSU-1).
Genes linked to increased esters and ethyl esters that contributed to the wine's fruity aromas (estA, estB, estC, and metB) were present in the autochthonous O. oeni genomes. Genes linked to dyacetil, acetoin, butanediol, and acetate via citrate metabolism (citE, citF, citD maeP, and alsA) were present in isolate 139, but some of these genes were absent in the other autochthonous O. oeni genomes (565 and 399). Genes related to odorless non-volatile glycosides and glycosidase activities that contributed to the wine aroma (bgl and abf) were present in strain 139.
Discussion
One of the main objectives of this study was to analyze the load and diversity of LAB in wine produced with spontaneous MLF in Chilean valleys and to pre-select future starter cultures. This study covered an extensive Chilean enological region from 30°39′S to 35°50′S latitude and analyzed the total and lactic acid bacterial loads using culture-independent methodology. Our LAB screening results showed the highest prevalence O. oeni, which was coincident with reports from other countries (González-Arenzana et al., 2012; Pramateftaki et al., 2012; Cappello et al., 2017). Additionally, combinations of LAB, such as O. oeni/Leuconostoc and O. oeni/Pediococcus were found, which were coincident with the findings of Renouf et al. (2007), Pérez-Martín et al. (2015) and Miranda-Castilleja et al. (2016). However, the existence of Pediococcus in wine samples needs consideration, because these bacteria have been associated with ropiness of wine (Dols-Lafargue et al., 2008).
Furthermore, we found a predominance of the O. oeni load compared to the total bacterial load in all samples by qPCR analysis of the rpoB gen. This gene has been used for the description of LAB in fermentative environments (i.e., Renouf et al., 2006; Miranda-Castilleja et al., 2016 used the rpoB gene to study the dynamics and diversity of LAB in different cellars). Therefore, quantification of the bacterial DNA using the rpoB gen showed that this gene could be used as a marker of the O. oeni load and thus might be useful for monitoring MLF. However, the total bacterial and O. oeni loads were similar among the Limarí and Maule Valleys, which were located ~760 km apart. The influence of the local bacterial diversity on wine elaboration and the peculiar characteristics provide a local product fingerprint, as reported by Bokulich et al. (2014). Furthermore, rpoB was useful to obtain a prior genetic screening of the strains, since the sequencing of this gene allow us to distinguish between autochthonous and commercial O. oeni strains. This approach has been reported previously in other Gram positive bacteria such as Staphylococcus (Drancourt and Raoult, 2002).
Genetic typing examined using MLST evidenced the existence of two major phylogenetic clusters. Interestingly, half of the autochthonous isolates could be distinguishable from the commercial isolates, whereas the other half grouped together with the commercial strains. Within group M, the commercial strain MTO presented insertion of a transposon element in purK, which is one of the most interesting loci to analyze the genetic variability of the O. oeni strains (González-Arenzana et al., 2013). This insertion was reported in 7 O. oeni strains from Champagne, Burgundy, and Jura, France, 1 strain from Italy (Bridier et al., 2010) and 2 strains from Pineau and Jura, France (Bilhère et al., 2009). According to the MLST results, low genetic diversity among the autochthonous O. oeni isolates were found, which might be related to the use of housekeeping genes that could be under restricted variation. Other explanations are the exchange of DNA among O. oeni strains, as proposed by de las Rivas et al. (2004), where a favorable environment for horizontal gene transfer could be created in the fermentation tank/barrel. Dicks (1994) and Zúñiga et al. (1996) showed that O. oeni was able to receive foreign DNA by transformation in vitro and by conjugation. Interestingly, Campbell-Sills et al. (2015) reported that O. oeni isolated from MLF grouped together in a phylogenomic analysis and that strains outside this genetic group were absent during MLF.
Spontaneous MLF has drawbacks, including stuck fermentation and contamination by microorganisms, which risk altering the wine quality. To solve this problem, the use of microbial starters has been introduced with commercial strains isolated from other wine-producing countries. However, starter strains selected from the wine native microbiota of each region have better natural adaptation to the wine and maintain regional typicity (Zapparoli et al., 2003; Izquierdo et al., 2004). Ethanol and acidic environments are determinant factors for the growth of O. oeni in wine (Liu et al., 2014). In this context, strains able to tolerate 15% v/v ethanol and a pH of 3.1 were obtained. These results were different from the reports of Capozzi et al. (2010); Solieri et al. (2010) and Lerm et al. (2011), which showed that O. oeni strains were unable to survive in high ethanol concentrations (13% v/v). Strain 139 had high growth in 15% v/v ethanol and at a pH of 3.1 and exhibited high malolactic activity; these results suggested that this strain (139) adapted better to the wine environment than the other two strains (565 and 139). Hence, strain 139 may be proposed as the best candidate for use as a starter in MLF.
O. oeni strains have a compact genome of 1.8 Mb and several metabolic pathways related to growth in enological environments, including MLF and aroma production (Mills et al., 2005; Makarova et al., 2006; Makarova and Koonin, 2007). Furthermore, its compact genome most likely reflects a high level of organization and simplicity (Jara and Romero, 2015; Sternes and Borneman, 2016). This genomic organization may be the basis for its adaptation to the wine environment (Zé-Zé et al., 1998, 2000; Mills et al., 2005). Interestingly, the analyses of these genomes using ANIb, DDH and OrthoANI revealed 139 consistent differences from the autochthonous strains when the distance between genomes was calculated by aligning the whole sequences. According to Thompson et al. (2013), these isolates and reference strains (VP41 and PSU-1) shared more than 95% ANIb and hence could be considered the same species. A similar observation including more than 30 O. oeni genomes was also reported by Campbell-Sills et al. (2015).
A report by Borneman et al. (2010) compared the genomes of three O. oeni strains (PSU-1, BAA1163, and AWRIB429). These strains shared conserved genes corresponding to 52% of the observed ORFs. These authors claimed that this conserved region could be considered the core genome. A similar result was reported by Campbell-Sills et al. (2015) and Sternes and Borneman (2016), but these studies included more CDSs due to differences in the orthologous calculation. Borneman et al. (2010) posited that unique ORFs associated with bacteriophage-derived sequences or glycosyl hydrolases might be key from a winemaking perspective, because these ORFs might contribute to aromatic compound formation through the cleavage of the sugar moiety from the non-volatile (and therefore aroma-less) glycosidic precursors present in grape juice. These analyses suggest that genomic variation may be the key to ascertaining the phenotypic differences between O. oeni strains. In this context, our data showed that the Chilean strain contained 21–28 unique genes per strain related to metabolism and transport, some of which possibly explained some of the technological properties of the bacteria. Similarly, Campbell-Sills et al. (2015) reported that O. oeni isolates from Champagne showed 27 unique genes that might be related to technical properties. Taken together, the technical properties, unique characteristics and capacity for local adaptation of some LAB could provide the basis for obtaining suitable strains to serve as inocula in future products and contribute to the typicity of Chilean wines.
Several enzymes from O. oeni may contribute to the aromatic profile of the wine during MLF, including β-glucosidase, citrate lyase, esterases, proteases, and peptidases (Mtshali et al., 2010; Michlmayr et al., 2012; Cappello et al., 2017). The activity of some of these enzymes may be modulated by enological parameters, such as the pH, temperature, ethanol, or glucose and fructose concentrations (Grimaldi et al., 2005). Furthermore, Olguín et al. (2011) demonstrated that the expression of the β-glucosidase gene (bgl) in O. oeni might be induced by a moderate ethanol concentration.
Citrate metabolism is involved in the production of compounds, such as diacetyl, acetoin, butanediol and acetate, which are important for the wine aroma (Olguín et al., 2009). Diacetyl is the most important aroma compound during MLF (Cappello et al., 2017). Additionally, some genes involved in citrate metabolism have been shown to provide metabolic traits to different strains (Olguín et al., 2009). The inter-strain comparison of the transcriptional levels of genes involved in citrate metabolism (ackA and alsD) revealed that the strains had different metabolic features.
Esters are a key group of volatile compounds that can contribute to the wine aromatic profile (Swiegers et al., 2005). These compounds depend on the activity of esterases (Cappello et al., 2017). Sumby et al. (2013) showed that two purified O. oeni esterases (EstA2 and EstB28) had two activities (synthesis and hydrolysis) that suggested the contribution of O. oeni to the wine aroma profile.
The contribution of specific O. oeni strain to the organoleptic properties of wine may affect flavor formation depending on the wine parameters (Cappello et al., 2017). The selection of bacterial strains for MLF should consider the potential to improve the wine typicity. In this context, Chilean isolates showed different contents of genes encoding enzymes contributing to the aromatic profile; among them, strain 139 presented a higher number of glucosidases and promising enological properties and thus might be proposed as the best candidate for use as a starter in MLF.
To the best of our knowledge, this study is the first report to focus on both the genetic and technological characterization of O. oeni strains in Chile. This study reported that genetic (MLST) and genomic tools (ANI) might reveal the differences between commercial and autochthonous O. oeni strains. Similarly, autochthonous O. oeni strains showed some advantages in terms of technological properties. Thus, future studies should focus on determining the potential relationships between the phylogenetic and phenotypic characteristics of O. oeni strains; these results could help identify the effect of environmental conditions on the genetic content and evolution of the species. Furthermore, these analyses may provide useful information for the selection of strains with better industrial performances.
Author Contributions
JR and CJ: designed of the work; CI: data collection, data analysis, and interpretation; MR: Genetic and genomic analyses; CJ: drafting the article; JR: critical revision and edition of the article; CJ, CI, JR: Final approval of the version to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This was funded by grants from FONDEF Idea CA12I10123, FONDECYT 11121329 from CONICYT; INNOVA 12IDL2-13145 from Corfo. The authors also thank La Reserva de Caliboro winery. Work by CI was partially supported by the Stekel scholarship from INTA-Nestlé.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00090/full#supplementary-material
References
Bartowsky, E. J., McCarthy, J. M., and Henschke, P. (2003). Differentiation of Australian wine isolates of Oenococcus oeni using random amplified polymorphic DNA (RAPD). Aust. J. Grape Wine Res. 9, 122–126. doi: 10.1111/j.1755-0238.2003.tb00262.x
Bilhère, E., Lucas, P. M., Claisse, O., and Lonvaud-Funel, A. (2009). Multilocus sequence typing of Oenococcus oeni: detection of two subpopulations shaped by intergenic recombination. Appl. Environ. Microbiol. 75, 1291–1300. doi: 10.1128/AEM.02563-08
Blasco, L., Ferrer, S., and Pardo, I. (2003). Development of specific fluorescent oligonucleotide probes for in situ identification of wine lactic acid bacteria. FEMS Microbiol. Lett. 225, 115–123. doi: 10.1016/S0378-1097(03)00501-9
Bokulich, N. A., Thorngate, J. H., Richardson, P. M., and Mills, D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111, 139–148. doi: 10.1073/pnas.1317377110
Bon, E., Delaherche, A., Bilhère, E., De Daruvar, A., Lonvaud-Funel, A., and Le Marrec, C. (2009). Oenococcus oeni genome plasticity is associated with fitness. Appl. Environ. Microbiol. 75, 2079–2090. doi: 10.1128/AEM.02194-08
Bordas, M., Araque, I., Alegret, J. O., El Khoury, M., Lucas, P., Rozès, N., et al. (2013). Isolation, selection, and characterization of highly etanol tolerant strains of Oenococcus oeni. Int. Microbiol. 16, 113–123. doi: 10.2436/20.1501.01.186
Bordas, M., Araque, I., Bordons, A., and Reguant, C. (2015). Differential expression of selected Oenococcus oeni genes for adaptation in wine-like media and red wine. Ann. Microbiol. 65, 2277–2285. doi: 10.1007/s13213-015-1069-2
Borneman, A. R., Bartowsky, E. J., and McCarthy, J. (2010). Genotypic diversity in Oenococcus oeni by high-density microarray comparative genome hybridization and whole genome sequencing. Appl. Microbiol. Biotechnol. 86, 681–691. doi: 10.1007/s00253-009-2425-6
Bridier, J., Claisse, O., Coton, M., Coton, E., and Lonvaud-Funel, A. (2010). Evidence of distinct populations and specific subpopulations within the species Oenococcus oeni. Appl. Environ. Microbiol. 76, 7754–7764. doi: 10.1128/AEM.01544-10
Cafaro, C., Bonomo, M. G., Guerrieri, A., Crispo, F., Ciriello, R., and Salzano, G. (2016). Assessment of the genetic polymorphism and physiological characterization of indigenous Oenococcus oeni strains isolated from Aglianico del Vulture red wine. Folia Microbiol. 61, 1–10. doi: 10.1007/s12223-015-0402-2
Cañas, P. M., Pérez, P. R., Prieto, S. S., and Herreros, M. L. (2009). Ecological study of lactic acid microbiota isolated from Tempranillo wines of Castilla-La Mancha. J. Biosci. Bioeng. 108, 220–224. doi: 10.1016/j.jbiosc.2009.04.001
Campbell-Sills, H., El Khoury, M., Favier, M., Romano, A., Biasioli, F., Spano, G., et al. (2015). Phylogenomic analysis of Oenococcus oeni reveals specific domestication of strains to cider and wines. Genome Biol. Evol. 7, 1506–1518. doi: 10.1093/gbe/evv084
Capozzi, V., Russo, P., Beneduce, L., Weidmann, S., Grieco, F., Guzzo, J., et al. (2010). Technological properties of Oenococcus oeni strains isolated from typical southern Italian wines. Lett. Appl. Microbiol. 50, 327–334. doi: 10.1111/j.1472-765XX.2010.02795.x
Capozzi, V., Russo, P., Lamontanara, A., Orr,ù, L., Cattivelli, L., and Spano, G. (2014). Genome sequences of five Oenococcus oeni strains isolated from Nero di Troia Wine in Apulia, Southern Italy. Genome Announc. 2, 1–4. doi: 10.1128/genomeA.01077-14
Cappello, M. S., De Domenico, S., Logrieco, A., and Zapparoli, G. (2014). Bio-molecular characterization of indigenous Oenococcus oeni strains from Negroamaro wine. Food Microbiol. 42, 142–148. doi: 10.1016/j.fm.2014.02.004
Cappello, M. S., Zapparoli, G., Logrieco, A., and Bartowsky, E. J. (2017). Linking wine lactic acid bacteria diversity with wine aroma and flavor. Int. J. Food Microbiol. 243, 16–27. doi: 10.1016/j.ijfoodmicro.2016.11.025
de las Rivas, B., Marcobal, Á., and Muñoz, R. (2004). Allelic diversity and population structure in Oenococcus oeni as determined from sequence analysis of housekeeping genes. Appl. Environ. Microbiol. 70, 7210–7219. doi: 10.1128/AEM.70.12.7210-7219.2004
Dicks, L. M. T. (1994). Transformation of Leuconostoc oenos by electroporation. Biotechnol. Tech. 8, 901–904. doi: 10.1007/BF02447736
Dols-Lafargue, M., Lee, H. Y., Le Marrec, C., Heyraud, A., Chambat, G., and Lonvaud-Funel, A. (2008). Characterization of gtf, a glucosyltransferase gene in the genomes of Pediococcus parvulus and Oenococcus oeni, two bacterial species commonly found in wine. Appl. Environ. Microbiol. 74, 4079–4090. doi: 10.1128/AEM.00673-08
Drancourt, M., and Raoult, D. (2002). rpoB gene sequence-based identification of Staphylococcus species. J. Clin. Microbiol 40, 1333–1338. doi: 10.1128/JCM.40.4.1333-1338.2002
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Fouts, D. E., Brinkac, L., Beck, E., Inman, J., and Sutton, G. (2012). PanOCT: automated clustering of orthologs using conserved gene neighborhood for Pan-genomic analysis of bacterial strains and closely related species. Nucleic Acids Res. 40:e172. doi: 10.1093/nar/gks757
Garofalo, C., El Khoury, M., Lucas, P., Bely, M., Russo, P., Spano, G., et al. (2015). Autochthonous starter cultures and indigenous grape variety for regional wine production. J. Appl. Microbiol. 118, 1395–1408. doi: 10.1111/jam.12789
González-Arenzana, L., López, R., Santamaría, P., and López-Alfaro, I. (2013). Dynamics of lactic acid bacteria populations in Rioja wines by PCR-DGGE, comparison with culture-dependent methods. Appl. Microbiol. Biotechnol. 97, 6931–6941. doi: 10.1007/s00253-013-4974-y
González-Arenzana, L., López, R., Santamaría, P., Tenorio, C., and López-Alfaro, I. (2012). Dynamics of indigenous lactic acid bacteria populations in wine fermentations from La Rioja (Spain) during three vintages. Microb. Ecol. 62, 12–19. doi: 10.1007/s00248-011-9911-y
Grimaldi, A., Bartowsky, E., and Jiranek, V. (2005). A survey of glycosidase activities of commercial wine strains of Oenococcus oeni. Int. J. Food Microbiol. 105, 233–244. doi: 10.1016/j.ijfoodmicro.2005.04.011
Henick-Kling, T. (1995). Control of malolactic fermentation in wine: energetics, flavour modification and methods of starter culture preparation. J. Appl. Bacteriol. Symp. Suppl. 79, 29S–37S.
Ilabaca, C., Jara, C., and Romero, J. (2014). The rapid identification of lactic acid bacteria present in Chilean winemaking processes using culture-independent analysis. Ann. Microbiol. 64, 1857–1859. doi: 10.1007/s13213-014-0810-6
Izquierdo, P., García, E., Martínez, J., and Chacón, J. (2004). Selection of lactic bacteria to induce malolactic fermentation in red wine of cv. Cencibel. Vitis 43, 149–153.
Jara, C., and Romero, J. (2015). Genome sequences of three Oenococcus oeni strains isolated from Maipo Valley, Chile. Genome Announc. 3:e00866-15. doi: 10.1128/genomeA.00866-15
Jara, C., Mateo, E., Guillamón, J. M., Torija, M. J., and Mas, A. (2008). Analysis of several methods for the extraction of high quality DNA from acetic acid bacteria in wine and vinegar for characterization by PCR-based methods. Int. J. Food Microbiol. 128, 336–341. doi: 10.1016/j.ijfoodmicro.2008.09.008
Lamontanara, A., Orrù, L., Cattivelli, L., Russo, P., Spano, G., and Capozzi, V. (2014). Genome Sequence of Oenococcus oeni OM27, the first fully assembled genome of a strain isolated from an Italian wine. Genome Announc. 2, 1–4. doi: 10.1128/genomeA.00658-14
Lee, I., Ouk Kim, Y., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Lerm, E., Engelbrecht, L., and du Toit, M. (2011). Selection and characterisation of Oenococcus oeni and Lactobacillus plantarum South African wine isolates for use as malolactic fermentation starter cultures. S. Afr. J. Enol. Viticult. 32, 280–295. doi: 10.21548/32-2-1388
Liu, K., Gao, D., Liu, Z., Li, N., Bian, Y., Zheng, R., et al. (2014). Screening of lactic acid bacteria with high activities malolactic enzyme and analysis of indigenous flora in red wine. J. Chem. Pharm. Res. 6. 136–144.
Lonvaud-Funel, A. (1995). Microbiology of the malolactic fermentation: molecular aspects. FEMS Microbiol. Lett. 126, 209–214. doi: 10.1111/j.1574-6968.1995.tb07420.x
López, I., Carmen, T., Zarazaga, M., Dizy, M., Torres, C., and Ruiz-Larrea, F. (2007). Evidence of mixed wild populations of Oenococcus oeni strains during wine spontaneous malolactic fermentations. Eur. Food Res. Technol. 226, 215–223. doi: 10.1007/s00217-006-0529-0
Makarova, K. S., and Koonin, E. V. (2007). Evolutionary genomics of lactic acid bacteria. J. Bacteriol. 189, 1199–1208. doi: 10.1128/JB.01351-06
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., et al. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U.S.A. 42, 15611–15616. doi: 10.1073/pnas.0607117103
Marques, A. P., Duarte, A. J., Chambel, L., Teixeira, M. F., San Romão, M. V., and Tenreiro, R. (2011). Genomic diversity of Oenococcus oeni from different winemaking regions of Portugal. Int. Microbiol. 14, 155–162. doi: 10.2436/20.1501.01.144
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Mesas, J. M., Rodríguez, M. C., and Alegre, M. T. (2011). Characterization of lactic acid bacteria from musts and wines of three consecutive vintages of Ribeira Sacra. Lett. Appl. Microbiol. 52, 258–268. doi: 10.1111/j.1472-765X.2010.02991.x
Michlmayr, H., Nauer, S., Brandes, W., Schümann, C., Kulbe, K. D., and del Hierro, A. M. (2012). Release of wine monoterpenes from natural precursors by glycosidases from Oenococcus oeni. Food Chem. 135, 80–87. doi: 10.1016/j.foodchem.2012.04.099
Mills, D. A., Rawsthorne, H., Parker, C., Tamir, D., and Makarova, K. (2005). Genomic analysis of Oenococcus oeni PSU-1 and its relevance to winemaking. FEMS Microbiol. Rev. 29, 465–475. doi: 10.1016/j.femsre.2005.04.011
Miranda-Castilleja, D. E., Martínez-Peniche, R. A., Aldrete-Tapia, J. A., Soto-Muñoz, L., Iturriaga, M. H., Pacheco-Aguilar, J. R., et al. (2016). Distribution of native lactic acid bacteria in Wineries of Queretaro, Mexico and their resistance to wine-like conditions. Front. Microbiol. 7:1769. doi: 10.3389/fmicb.2016.01769
Mtshali, P. S., Divol, B., and du Toit, M. (2012). PCR detection of enzyme-encoding genes in Leuconostoc mesenteroides strains of wine origin. World J. Microbiol. Biotechnol. 28, 1443–1449. doi: 10.1007/s11274-011-0944-7
Mtshali, P. S., Divol, B., van Rensburg, P., and du Toit, M. (2010). Genetic screening of wine-related enzymes in Lactobacillus species isolated from South African wines. J. Appl. Microbiol. 108, 1389–1397. doi: 10.1111/j.1365-2672.2009.04535.x
Nisiotou, A. A., Dourou, D., Filippousi, M. E., Diamantea, E., Fragkoulis, P., Fragkoulis, P., et al. (2015). Genetic and technological characterization of vineyard- and winery-associated lactic acid bacteria. Biomed. Res. Int. 2015, 9–17. doi: 10.1155/2015/508254
Olguín, N., Bordons, A., and Reguant, C. (2009). Influence of ethanol and pH on the gene expression of the citrate pathway in Oenococcus oeni. Food Microbiol. 26, 197–203. doi: 10.1016/j.fm.2008.09.004
Olguín, N., Oriol Alegret, J., Bordons, A., and Reguant, C. (2011). β-Glucosidase activity and bgl gene expression of Oenococcus oeni strains in model media and Cabernet Sauvignon wine. Am. J. Enol. Vitic. 62, 99–105. doi: 10.5344/ajev.2010.10044
Opazo, R., Ortúzar, F., Navarrete, P., Espejo, R., and Romero, J. (2012). Reduction of soybean meal non-starch polysaccharides and alpha-galactosides by solid-state fermentation using cellulolytic bacteria obtained from different environments. PLoS ONE 7:e44783. doi: 10.1371/journal.pone.0044783
Pérez-Martín, F., Seseña, S., and Palop, M. L. (2015). Inventory of lactic acid bacteria populations in red wine varieties from Appellation of Origin Méntrida. Eur. Food Res. Technol. 240, 725–733. doi: 10.1007/s00217-014-2377-7
Pramateftaki, P. V., Matafa, M., Karapetrous, G., and Marmaras, G. (2012). Assessment of the genetic polymorphism and biogenic amine production of indigenous Oenococcus oeni strains isolated from Greek red wines. Food Microbiol. 29, 113–120. doi: 10.1016/j.fm.2011.09.007
R Development Core Team (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org.
Renouf, V., Claisse, O., and Lonvaud-Funel, A. (2007). Inventory and monitoring of wine microbial consortia. Appl. Microb. Cell Physiol. 75, 149–164. doi: 10.1007/s00253-006-0798-3
Renouf, V., Claisse, O., Miot-Sertier, C., and Lonvaud-Funel, A. (2006). Lactic acid bacteria evolution during winemaking: use of rpoB gene as a target for PCR-DGGE analysis. Food Microbiol. 23, 136–145. doi: 10.1016/j.fm.2005.01.019
Rodas, A. M., Ferrer, S., and Pardo, I. (2003). 16S-ARDRA, a tool for identification of lactic acid bacteria isolated from grape must and wine. Syst. Appl. Microbiol. 26, 412–422. doi: 10.1078/072320203322497446
Rosselló-Mora, R. (2006). “DNA-DNA reassociation methods applied to microbial taxonomy and their critical evaluation,” in Molecular Identification, Systematics, and Population Structure of Prokaryotes, ed E. Stackebrandt (Berlin: Springer), 23–50.
Ruiz, P., Izquierdo, P. M., Seseña, S., and Palop, M. L. (2008). Intraspecific genetic diversity of lactic acid bacteria from malolactic fermentation of Cencibel wines as derived from combined analysis of RAPD-PCR and PFGE patterns. Food Microbiol. 25, 942–948. doi: 10.1016/j.fm.2008.06.007
Solieri, L., Genova, F., De Paola, M., and Giudici, P. (2010). Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: a frame work for selection of new starter cultures. J. Appl. Microbiol. 108, 285–298. doi: 10.1111/j.1365-2672.2009.04428.x
Sonnhammer, E. L., and Östlund, G. (2015). InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 43, D234–D239. doi: 10.1093/nar/gku1203
Spano, G., Lonvaud-Funel, A., Claisse, O., and Massa, S. (2007). In vivo PCR-DGGE analysis of Lactobacillus plantarum and Oenococcus oeni populations in red wine. Curr. Microbiol. 54, 9–13. doi: 10.1007/s00284-006-0136-0
Sternes, P. R., and Borneman, A. R. (2016). Consensus pan-genome assembly of the specialized wine bacterium Oenococcus oeni. BMC Genomics 17:308. doi: 10.1186/s12864-016-2604-7
Sumby, K. M., Grbin, P. R., and Jiranek, V. (2013). Characterization of EstCOo8 and EstC34, intracellular esterases, from the wine-associated lactic acid bacteria Oenococcus oeni and Lactobacillus hilgardii. J. Appl. Microbiol. 114, 413–422. doi: 10.1111/jam.12060
Sumby, K. M., Matthews, A. H., Grbin, P. R., and Jiranek, V. (2009). Cloning and characterization of an intracellular esterase from the wine-associated lactic acid bacterium Oenococcus oeni. Appl. Environ. Microbiol. 75, 6729–6735. doi: 10.1128/AEM.01563-09
Swiegers, J. H., Bartowsky, E. J., Henschke, P. A., and Pretorius, I. S. (2005). Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 11, 139–173. doi: 10.1111/j.1755-0238.2005.tb00285.x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mo.l Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, C. C., Chimetto, L., Edwards, R. A., Swings, J., Stackebrandt, E., and Thompson, F. L. (2013). Microbial genomic taxonomy. BMC Genomics 14:913. doi: 10.1186/1471-2164-14-913
Wang, T., Li, H., Wang, H., and Su, J. (2015). Multilocus sequence typing and pulsed-field gel electrophoresis analysis of Oenococcus oeni from different wine-producing regions of China. Int. J. Food Microbiol. 16, 47–53. doi: 10.1016/j.ijfoodmicro.2015.01.006
Zapparoli, G., Spinelli, P., Torriani, S., and Dellaglio, F. (2003). The spontaneous malolactic fermentation in wines of the Valtellina area: a study of the kinetic parameters and of the dominant Oenococcus oeni strains. Riv. Vitic. Enol. 2, 47–56.
Zé-Zé, L., Teneiro, R., and Paveia, H. (2000). The Oenococcus oeni genome: physical and genetic map of strain GM and comparison with the genome of a ‘divergent’ strain, PSU 1. Microbiol. 146, 3195–3204. doi: 10.1099/00221287-146-12-3195
Zé-Zé, L., Teneiro, R., Brito, L., Santos, M. A., and Paveia, H. (1998). Physical map of the genome of Oenococcus oeni PSU-1 and localization of genetic markers. Microbiology 144, 1145–1156. doi: 10.1099/00221287-144-5-1145
Keywords: wine, malolactic fermentation, malolactic bacteria, Oenococcus oeni, terroir, genome, bacterial
Citation: Romero J, Ilabaca C, Ruiz M and Jara C (2018) Oenococcus oeni in Chilean Red Wines: Technological and Genomic Characterization. Front. Microbiol. 9:90. doi: 10.3389/fmicb.2018.00090
Received: 29 July 2017; Accepted: 15 January 2018;
Published: 14 February 2018.
Edited by:
Gustavo Cordero-Bueso, University of Cádiz, SpainReviewed by:
Giuseppe Spano, University of Foggia, ItalyLucía González-Arenzana, Instituto de Ciencias de la Vid y del Vino (ICVV), Spain
Copyright © 2018 Romero, Ilabaca, Ruiz and Jara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carla Jara, Y2FybGFqYXJhQHUudWNoaWxlLmNs
 Jaime Romero
Jaime Romero Carolina Ilabaca
Carolina Ilabaca Mauricio Ruiz
Mauricio Ruiz Carla Jara
Carla Jara