- Department of Food Science, Center for Food Safety, University of Arkansas, Fayetteville, AR, United States
The objective of the present study was to evaluate the prebiotic ability of Arkansas (AR) LaKast rice bran cultivars as a feed supplement to reduce Salmonella Typhimurium and other gut microbiota. An in vitro mixed anaerobic culture system was used to simulate conditions in the chicken ceca. Anaerobic cultures contained feed, cecal contents collected from 2, 4, and 6 weeks of chicken broilers, and with/without AR rice bran (pureline and hybrid). After 24 h pre-incubation, S. Typhimurium was inoculated into the anaerobic cultures and surviving S. Typhimurium were enumerated during anaerobic incubation up to 48 h. Samples were also collected after 0, 6, 12, 24, 48 h incubation for microbiome analysis with an Illumina MiSeq platform to investigate the changes in bacterial composition. Both pure and hybrid LaKast rice exhibited significant inhibitory effects in all experiments using 2, 4, and 6 weeks ceca but greater bactericidal effects by LaKast rice were observed at 6 weeks compared to 2- and 4-week ceca samples. For samples containing 6-week chicken ceca, the pureline and hybrid rice bran resulted in no viable S. Typhimurium and 6.58 log CFU/ml reduction after 48 h, respectively. Adding rice bran also led to changes in the cecal microbiota. LaKast rice bran resulted in more diverse bacterial population than control groups without any rice bran. The lowest abundance of Proteobacteria (at phylum level) and Enterobacteriaceae (at family and genus level) was exhibited in LaKast pure treated groups followed by LaKast hybrid and control. This may be attributed to a significant reduction of S. Typhimurium of the Enterobacteriaceae family and Proteobacteria phylum. This study suggests the beneficial functionality of LaKast rice brans as biological supplements in feed. The use of rice bran is favorable for both the consumer and the rice industry because of the perception of rice bran as a naturally occurring substance. As an abundant by-product of rice production, its use as a prebiotic in chicken feed may add economic value benefiting both the rice and poultry industries.
Introduction
Salmonella is a prevalent foodborne pathogen causing bacterial gastroenteritis in humans throughout the world and the organism is commonly isolated from various foods and environments, such as poultry products (Hope et al., 2002; Centers for Disease Control Prevention, 2009; Scallan et al., 2011). Pathogenic Salmonella is of great concern to the poultry industry thus various attempts to control Salmonella and prevent potential foodborne disease have been studied (Foley et al., 2011, 2013; Finstad et al., 2012; Howard et al., 2012). Prebiotics are defined as “a non-digestible ingredient which beneficially influences the host by selectively stimulating the growth and/or activity of one or a limited number of bacteria in the colon” and using prebiotics in feed is an emerging pathogen control method in the poultry industry (Gibson and Roberfroid, 1995; Patterson and Burkholder, 2003; Roberfroid, 2007; Ricke, 2015).
Rice bran is the hard outer layer of rice and an underutilized product of rice milling (Zullaikah et al., 2005). Rice bran is the main by-product of rice and is produced when the bran layer is removed from the rice kernel during the milling, whitening, or polishing process. Rice bran contains a variety of nutrients such as complex carbohydrates, proteins, amino acids, minerals, and vitamins as well as various components that exhibit prebiotic effects by modulating microbiota in the intestine (Ryan et al., 2011; Sheflin et al., 2015). Several studies have revealed potential prebiotic properties by demonstrating reduction of S. Typhimurium colonization in the gastrointestinal tract when rice bran was added in the diet (Kim et al., 2012, 2013; Kumar et al., 2012). Our previous study investigated the ability of three rice brans including Calrose, Jasmine, and Red Wells to control S. Typhimurium, and reported that Calrose rice bran resulted in a rapid inhibition of S. Typhimurium as well as significant changes in microbial populations and metabolites (Rubinelli et al., 2017).
Rice production in Arkansas (AR) ranks as the highest in the United States, accounting for 46.5% of the total production, making it a major crop in the state (Talbert and Burgos, 2007; Wilson et al., 2007). Further assessment of AR LaKast rice components could be beneficial to both the rice and the poultry industries prevalent in AR. However, only limited information is available on the effects of LaKast rice bran as a potential prebiotic. In the present study, the ability of LaKast rice bran to reduce S. Typhimurium was investigated in a controlled in vitro anaerobic mixed cecal culture to determine the potential of LaKast rice brans as prebiotics. Microbiome analysis was employed to evaluate bacterial compositional changes of LaKast rice bran on microbiota within the chicken ceca.
Materials and Methods
Bacterial Strains
A nalidixic acid-resistant (NAR) marker strain of Salmonella Typhimurium ST97 was kindly provided by Dr. Billy Hargis, University of Arkansas, Fayetteville, AR, United States. A second NAR marker strain used was derived from strain UK-1 (gift of Dr. Young Min Kwon, University of Arkansas) and passaged four times in chicks to increase virulence. The bacterial strains were cultured in sterile tubes containing Luria-Bertani (LB) medium supplemented with 20 μg/ml nalidixic acid at 37°C for 16 h with agitation at 250 rpm.
Arkansas Rice Bran Cultivars
The LaKast pureline and hybrid rice brans were kindly provided by Dr. Griffiths Atungulu, Department of Food Science, University of Arkansas, Fayetteville, AR, United States. These were derived from rice obtained from local rice producers in Burdette, AR, United States.
Cecal Contents Preparation
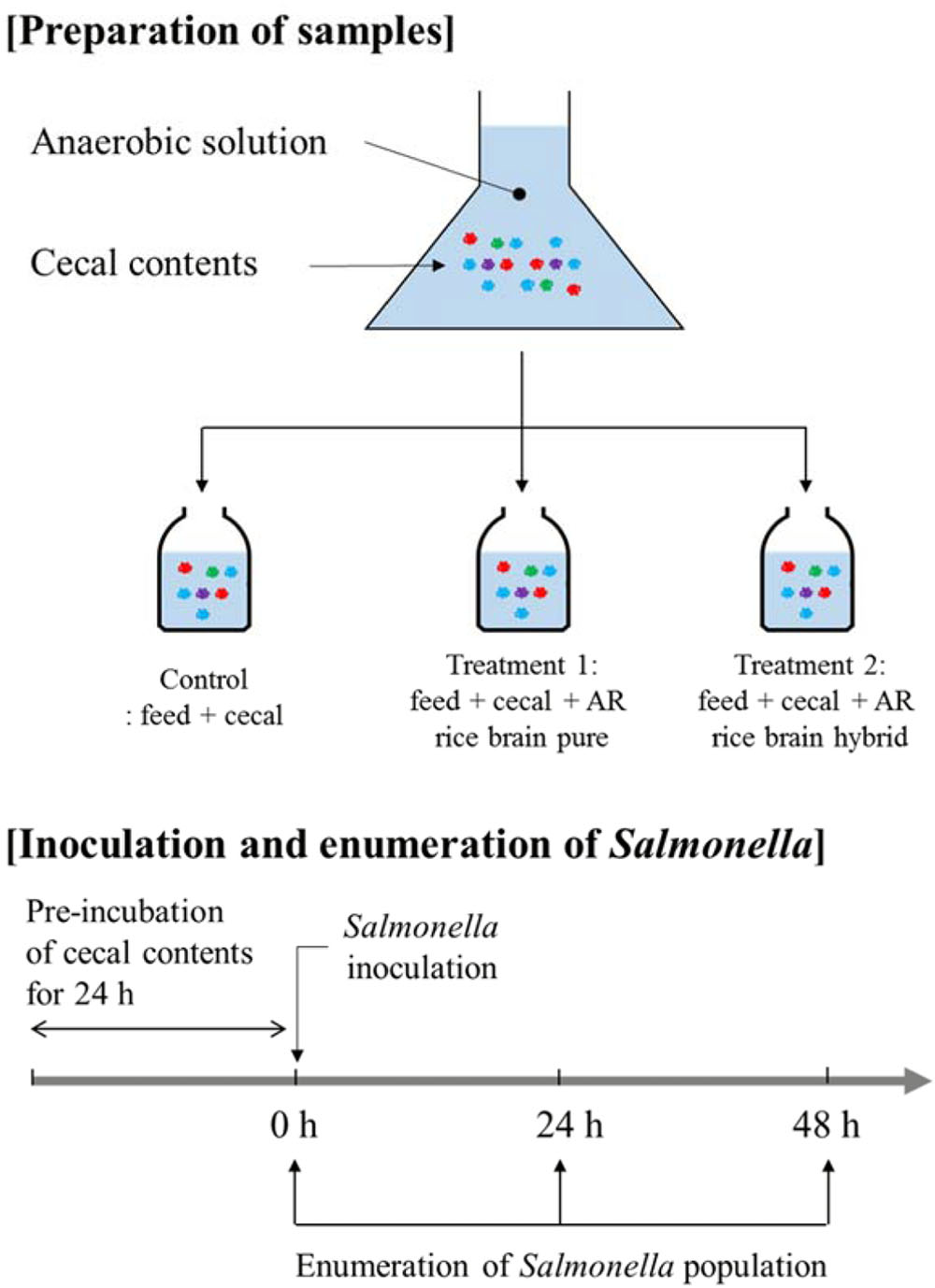
The overall experimental design used in the present study is shown in Figure 1. Cecal materials used in the present study were obtained from 14-, 28-, and 42-day-old Cobb male broiler chickens (Cobb-Vantress, Inc., Siloam Springs, AR, United States). Chickens were killed by CO2 asphyxiation using an approved Institutional Animal Care and Use Committee protocol (IACUC). Ceca were removed aseptically and immediately placed in a sterile sample bag in a portable anaerobic box (Mitsubishi Gas Chemical Co., Japan) containing oxygen-scrubbing sachets. These were subsequently transferred to an anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, MI, United States) containing an atmosphere of 90% nitrogen/5% CO2/5% hydrogen.
Anaerobic in Vitro Mixed Cultures
Anaerobic dilution solution was prepared fresh and consisted of 0.45 g/L K2HPO4, 0.45 g/L KH2PO4, 0.45 g/L (NH4)2SO4, 0.9 g/L NaCl, 0.1875 g/L MgSO4-7H2O, 0.12 g/L CaCl2-2H2O, 1 ml/L 0.1% resazurin, 0.05% cysteine-HCl, and 0.4% CO2-saturated sodium carbonate, with the sodium carbonate added last. This medium was used along with chicken feed and chemical fractions of rice bran to culture the anaerobic microorganisms of cecal contents, based on the methods described in previous research (Fan et al., 1995; Shermer et al., 1998; Saengkerdsub et al., 2006; Donalson et al., 2007, 2008; Dunkley et al., 2007; Rubinelli et al., 2016, 2017). Briefly, the anaerobic dilution solution was de-oxygenated using anaerobic gas mixture (90% nitrogen/5% carbon dioxide/5% hydrogen) in an anaerobic chamber for 30 min using an aquarium pump and airstone and subsequently autoclaved. The absence of S. Typhimurium in the cecal content was confirmed prior to the experiment by enrichment with tetrathionate (TT) broth (BD Biosciences, San Jose, CA, United States) followed by streaking onto brilliant green agar plates (BD Biosciences).
The cecal content was diluted 1:3000 by addition of 0.1 gram cecal content to 300 ml sterile anaerobic dilution solution. The 20 ml of diluted solution (the anaerobic mixed cultures) was transferred to sterile serum bottles containing 0.25 gram chicken feed as previously described (Fan et al., 1995; Shermer et al., 1998; Saengkerdsub et al., 2006; Donalson et al., 2007, 2008; Dunkley et al., 2007; Rubinelli et al., 2016, 2017). The control group contained only cecal content and feed (no rice bran), and the treatment group contained cecal content, feed, and LaKast rice bran (pure or hybrid) or rice bran fractions were added at a concentration of 1% (w/v). The feed was added to the same concentration used in previous research (Donalson et al., 2007). The anaerobic mixed cultures containing cecal, feed, and/or LaKast rice bran cultivar was pre-incubated at 37°C for 24 h (Model G25, New Brunswick, Inc.,) at 150 rpm. After pre-incubation, culture of S. Typhimurium was added to 20 ml culture (approximately 6 log CFU/ml) and incubated at 37°C.
Salmonella Inoculation and Enumeration of Survivors
After 0, 24, and 48 h, an aliquot of each culture was subjected to enumerate S. Typhimurium survivors by dilution with sterile phosphate buffer and spread plating onto 2 brilliant green agar plates supplemented with 20 μg/ml NA. If there were no S. Typhimurium at a particular time point in the undiluted culture, that culture was enriched with TT and streaked on brilliant green agar plates in order to confirm that no S. Typhimurium survived.
Microbiome Analysis
For the microbiome analysis, the in vitro anaerobic mixed cultures containing purline LaKast rice bran, hybrid LaKast rice bran, and “no rice bran” control (feed + cecal control) were collected at various time points (0, 6, 12, 24, and 48 h). Three biological replicate cecal contents were sampled from chickens of ages 2, 4, and 6 weeks old. These were sequenced as 3 technical replicates, for a total of 135 sequenced samples. Genomic DNA was extracted from the cecal contents via a QIAamp Fast DNA Stool Mini Kit following the manufacturer’s instructions (Qiagen, Valencia, CA, United States). Microbiome sequencing was conducted using an Illumina MiSeq platform (Illumina, San Diego, CA, United States), targeting the V4 hypervariable region of 16S rRNA and the resulting sequence reads were analyzed with the Quantitative Insights into Microbial Ecology (QIIME) pipeline (version 1.9.0) as described previously (Park et al., 2016; Kim et al., 2017a).
Statistical Analysis
The log CFU/ml was determined by averaging all biological replicates and evaluated the general linear model procedure in SAS (version 9.13; SAS Institute Inc., Cary, NC, United States). If analysis of variance (ANOVA) detected a significant result (P < 0.05), the mean values were tested using Tukey’s multiple-range test.
Results and Discussion
Recently, our laboratory reported on the activity of rice bran from different cultivars to reduce Salmonella using an in vitro mixed anaerobic culture system (Rubinelli et al., 2017). The ceca are the main site of Salmonella colonization and it contains a wide range of anaerobic bacteria (Salanitro et al., 1974; Hudault et al., 1985; Fan et al., 1995; Ricke and Pillai, 1999). However, when we collected cecal contents from different ages of broilers and employed an anaerobic mixed culture system to evaluate prebiotic-like effects of a commercial prebiotic, we observed differences in responses related to age of the bird (Rubinelli et al., 2016; Park et al., 2017). The previous experimental approach provides insight on cecal microbial development as a function of bird age (Park et al., 2017) thus the same incremental ages of birds were also employed in this study as sources of inocula.
In our previous study, two different experimental designs were employed; unadapted and adapted incubation (Rubinelli et al., 2017). In the unadapted incubation, the bacteria were inoculated at the beginning of the incubation along with feed, agents, and cecal bacteria. For the adapted incubation, the bacteria were added after pre-incubation for 24 h of the cecal bacteria. Park et al. (2017) observed that the inhibitory effects of Calrose rice bran against S. Typhimurium were significantly higher in the adapted condition; suggesting cecal microbiota play an important role to the reduction of S. Typhimurium. Further, the adapted condition simulates exposure of pathogenic bacteria after cecal bacteria are metabolically active, similar to the in vivo conditions. Thus, the adapted experimental condition was used exclusively in the present study.
Salmonella Typhimurium survival in the presence and absence of pureline and hybrid LaKast rice bran using cecal contents from 2-, 4-, and 6-week chickens are shown in Figures 2A,B,C, respectively. There were significant differences in S. Typhimurium populations between initial and 48 h samples (P < 0.05). Moreover, LaKast rice bran pure and hybrid resulted in less bacterial survival compared to the control.
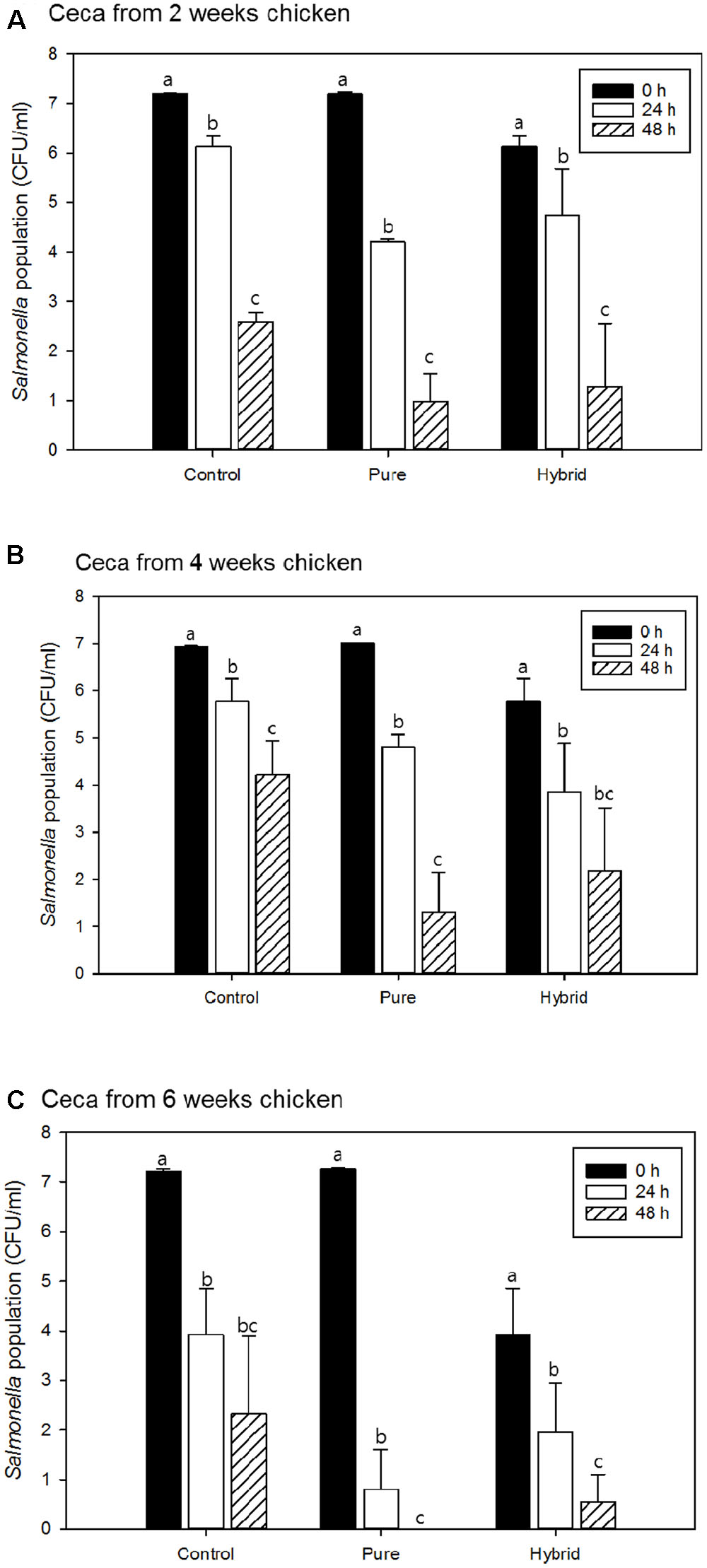
FIGURE 2. Salmonella Typhimurium population of control and treatments with pure and hybrid Arkansas rice bran with cecal from (A) 2 weeks, (B) 4 weeks, and (C) 6 weeks chicken. a-cMean values denoted by the different letters within each treatment were significantly different at P < 0.05.
When ceca from 2-week-old chickens were used in these experiments, S. Typhimurium populations were reduced by 1.07 log CFU/ml in control sample after 24 h while pureline and hybrid LaKast rice bran reduced S. Typhimurium by 3.00 and 2.54 log CFU/ml, respectively (Figure 2A). After 48 h, the control treatment had a 2.58 log CFU/ml reduction while the LaKast pureline and hybrid rice bran resulted in greater reductions in S. Typhimurium with 6.22 and 6.01 log CFU/ml reductions (P < 0.05), respectively. For samples containing 4-week-old chicken ceca, the control yielded 1.15 and 2.70 log CFU/ml reductions after 24 and 48 h incubation, respectively, while LaKast pureline and hybrid resulted in 2.22 (24 h) and 5.72 (48 h) log CFU/ml reduction and 3.10 (24 h) and 4.78 (48 h) log CFU/ml reduction (P < 0.05), respectively. For samples containing 6-week-old chicken ceca, greater bacterial reduction activities were observed by LaKast pureline and hybrid compared to 2- and 4-week ceca. In the control sample, the S. Typhimurium counts were reduced to 3.92 and 2.32 log CFU/ml (3.29 and 4.90 log reduction) after 24 and 48 h, respectively (Figure 2C). The initial cell density of S. Typhimurium was 7.27 log CFU/ml and LaKast pureline bran resulted in a 6.47 log reduction after 24 h and no viable S. Typhimurium after 48 h (P < 0.05). When samples were treated with LaKast hybrid bran, a 5.17 and 6.58 log CFU/ml reduction was exhibited after 24 and 48 h, respectively (P < 0.05).
We next examined the bacterial composition, via microbiome sequencing, in anaerobic mixed culture samples containing 2-, 4-, and 6-week-old chicken ceca exposed to LaKast pureline and hybrid bran as well as control at various culture time points (0, 6, 12, 24, and 48 h incubation starting from the initial inoculation with cecal contents). During sequencing data analysis with QIIME, some data groups were removed due to low read numbers.
Figure 3 represents the major bacteria among groups identified in anaerobic cultures from phylum to genus level when ceca of 2-week-old chickens were used in the experiment. At the phylum level, Firmicutes and Proteobacteria were predominant, accounting for 60.76 and 35.37% of the entire phyla, respectively (Figure 3A). Along with incubation time, abundance of Firmicutes was increased in the control (33.13% at 0 h to 76.24% at 24 h) as well as LaKast pureline (21.12% at 0 h to 63.91% at 48 h) and hybrid (56.31% at 0 h to 85.09% at 24 h) while that of Proteobacteria was decreased [control: 65.73–22.52%, LaKast pureline (8.54–34.07%), and hybrid (42.85–14.31%)]. In the 24 h incubation samples, LaKast pureline yielded the lowest relative abundance of Proteobacteria with 9.71%, followed by LaKast hybrid with 14.31% and control with 22.56%. At the family level, Enterobacteriaceae abundance decreased with the incubation time; from 64.65% (initial) to 21.31% (24 h) in control, 66.96 % (initial) to 34.10% (48 h) in the LaKast pureline, and 34.01% (initial) to 14.25% (24 h) in the LaKast hybrid (Figure 3D). When 24 h samples of groups were compared, Enterobacteriaceae and Lactobacillaceae were predominant in the control group with 23.31 and 22.61%, respectively, while Lachnospiraceae and Clostridiaceae were predominant in the LaKast pureline incubation (31.34 and 22.34%) and hybrid (30.96 and 25.98%) groups. The genus level microbiome data showed a decrease of Enterobacteriaceae; the combined abundance of Enterobacteriaceae (family level) and Other was 63.94, 66.67, and 41.75% in the control, LaKast pureline, and LaKast hybrid, respectively, at the initial time point but they were reduced to 19.52, 9.53, and 14.05% after 24 h incubation (Figure 3E).
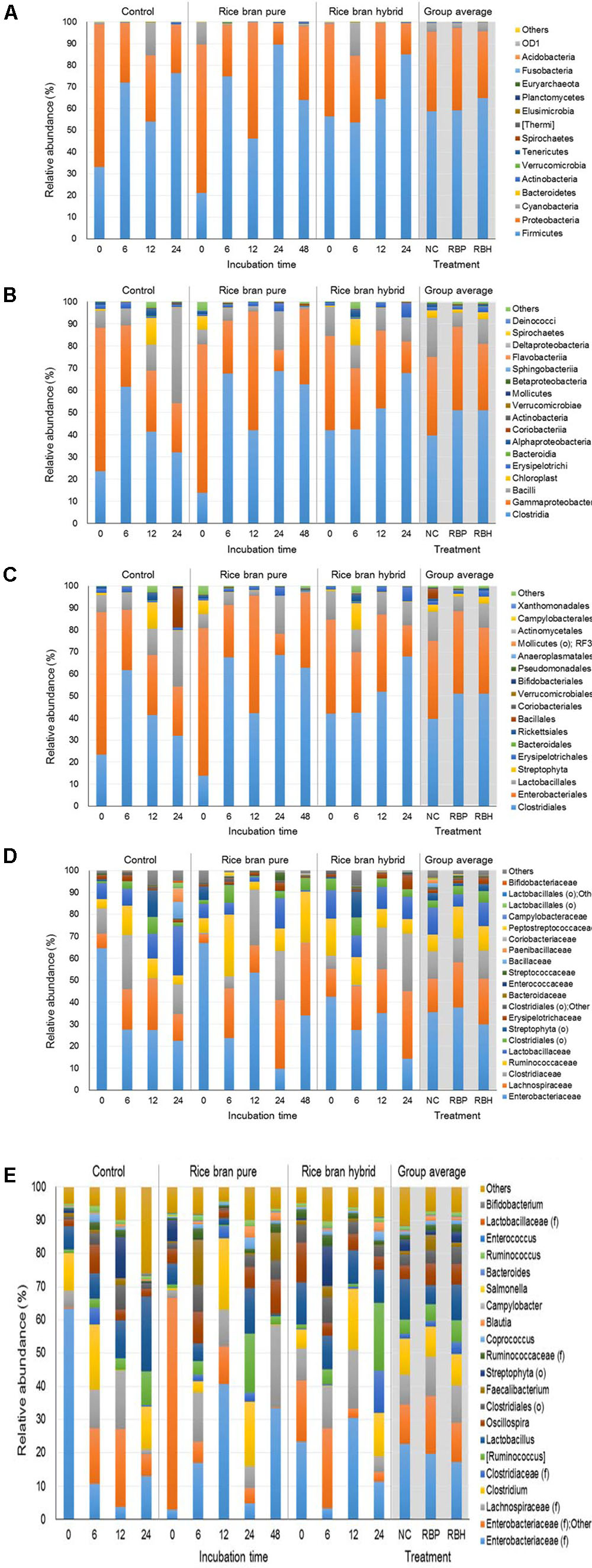
FIGURE 3. Relative abundance of major bacteria among different groups using ceca contents from 2 weeks of chicken broilers at a phylum (A), class (B), order (C), family (D), and genus level (E).
Figure 4 represents the major bacteria among groups identified in anaerobic cultures of control and LaKast treatments containing ceca of 4-week-old chickens from phylum to genus level. Similar to results from the experiment using 2-week-old chicken ceca, anaerobic cultures harbored the greatest proportion of Firmicutes constituting 63.82% of all phyla followed by Proteobacteria with 35.44% (Figure 4A). The Proteobacteria phylum includes a variety of pathogens such as Campylobacter, Salmonella, E. coli, Vibrio, and Yersinia (Nakagawa et al., 2007; Stopforth et al., 2007). In general, the proportion of Proteobacteria was reduced in the LaKast pureline from 44.65% (after 12 h) to 12.92% (after 48 h) while that of the control and the LaKast hybrid fluctuated during incubation times. Enterobacteriaceae, a large family of Gram-negative bacteria that contains pathogens such as Salmonella, E. coli, Yersinia, and Shigella, belongs to the Proteobacteria, and it exhibited a similar pattern at the family level with Proteobacteria at the phylum level (Figure 4D). The abundance of Enterobacteriaceae in LaKast pureline was reduced from 44.23% (12 h) to 15.88% (48 h). Comparing the bacterial abundance in 48 h samples at genus level, the lowest abundance of Enterobacteriaceae (family level) with Other was in LaKast pureline sample 15.60% while the control was 33.42% and LaKast hybrid was 45.13% (Figure 4E).
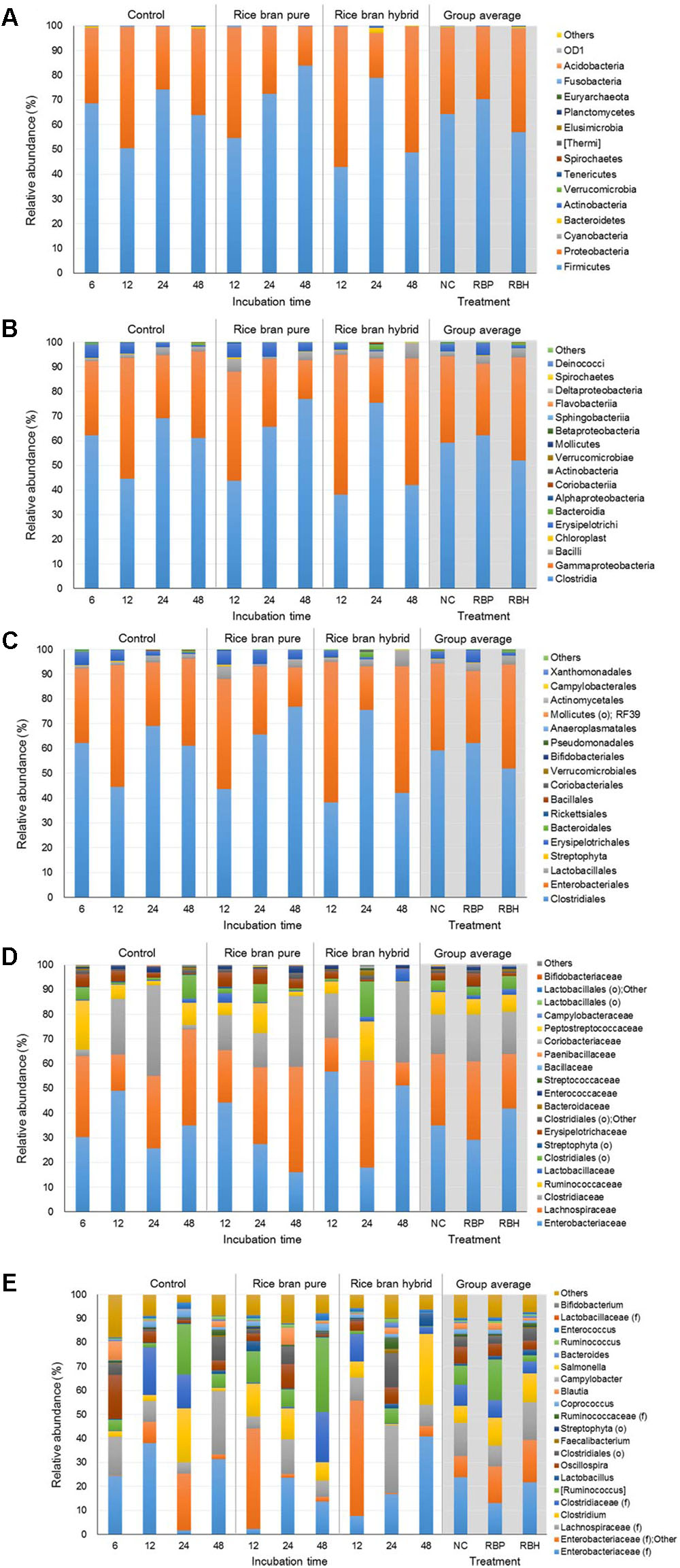
FIGURE 4. Relative abundance of major bacteria among different groups using ceca contents from 4 weeks of chicken broilers at a phylum (A), class (B), order (C), family (D), and genus level (E).
Figure 5 represents the major bacteria among groups identified in anaerobic cultures from phylum to genus level when ceca of 6-week-old chickens were used in the experiment. Similar to experiments using 2- and 4-week chickens, these incubations showed similar composition at the phylum level. The most predominant bacterial group was Firmicutes with 54.23% and Proteobacteria (39.03%; Figure 5A). In general, the proportion of Proteobacteria was reduced during incubation and LaKast pureline exhibited the lowest abundance (28.37%) of Proteobacteria at the end of the incubation (48 h) followed by LaKast hybrid (30.53%) and the control group (57.59%). This trend was also observed at other levels. LaKast pureline resulted in the lowest abundance of Enterobacteriaceae (28.30%, Figure 5D) at the family level and Enterobacteriaceae (family level) + Other (28.25%, Figure 5E) at the genus level while the control exhibited the highest proportion (55.53 and 55.39%, respectively).
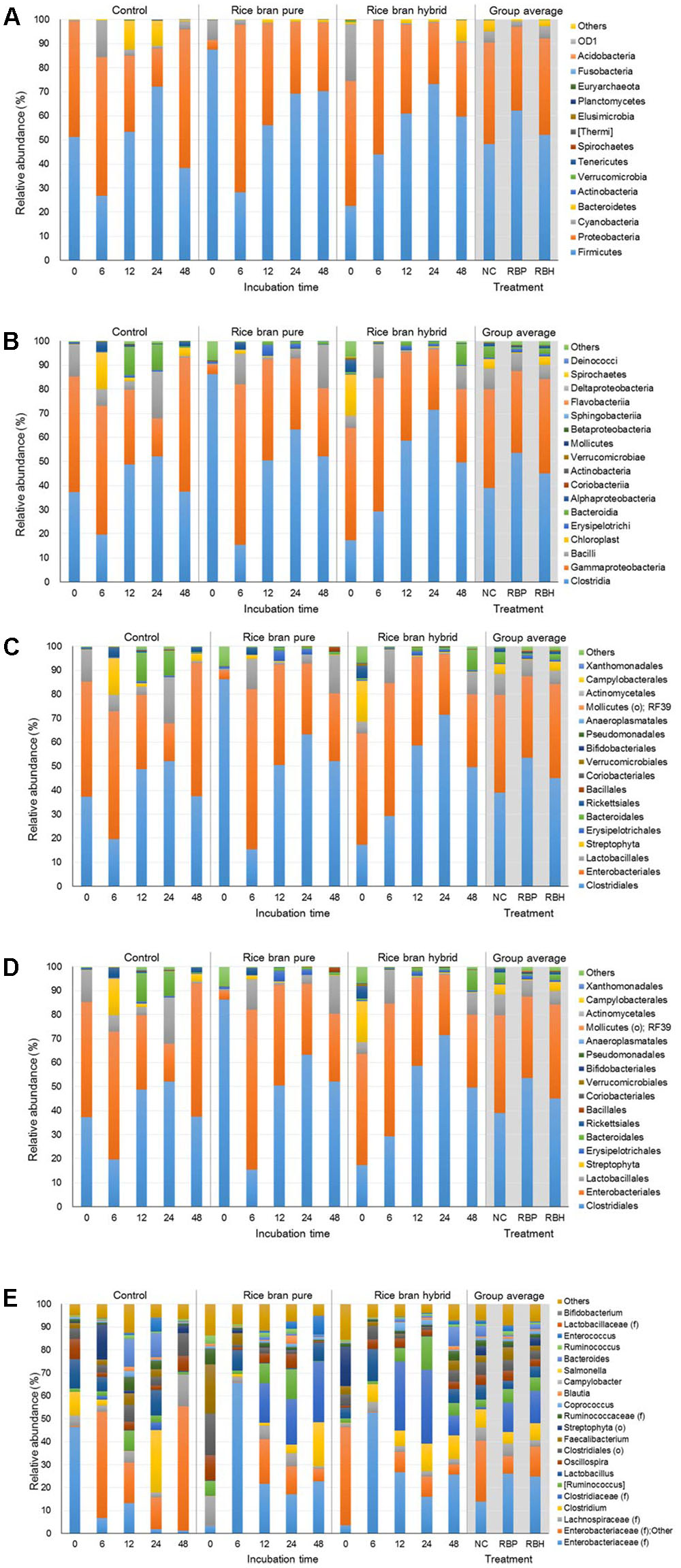
FIGURE 5. Relative abundance of major bacteria among different groups using ceca contents from 6 weeks of chicken broilers at a phylum (A), class (B), order (C), family (D), and genus level (E).
In this study, Salmonella was artificially inoculated in anaerobic culture to examine the prebiotic effects of LaKast rice. Salmonella belong to the Enterobacteriaceae family and the Proteobacteria phylum. In 16s rRNA gene sequencing, a ≥ 97% sequence identity is considered a positive identification (Janda and Abbott, 2007; Reller et al., 2007). Indeed, Salmonella artificially inoculated in chicken carcass rinsates were identified as Enterobacteriaceae at the genus level (Kim et al., 2017b). Our previous study reported that chicken ceca without any prebiotic treatment were dominated by Bacteriodetes (52.21%) and Firmicutes (34.35%; Park et al., 2016) while abundance of Proteobacteria was relatively small with 5.91%. However, the present study indicates that Proteobacteria are the second most predominant bacteria in anaerobic cultures at the phylum level and this could be attributed to the Salmonella spiked into these experiments.
The Chao 1 rarefaction plot which was employed to estimate species richness is presented in Figure 6. Anaerobic cultures containing cecal contents from older broilers exhibited more diverse microbiota. Sample groups with 6-week ceca showed the highest rarefaction value followed by groups with 4- and 2-week ceca (Figure 6A). Samples treated with LaKast rice bran exhibited greater diversity compared to the control groups, indicating that LaKast rice brans (both pureline and hybrid) resulted in richer microbiota than the control (Figure 6B). Gut microbiota play an important role to host health and commensal microbiota in the intestine can help to prevent colonization of invading pathogenic bacteria by interacting with pathogens directly and enhancement of defense mechanisms; thus diverse microbiota in this study would be expected to be more resistant to pathogens such as Salmonella (Kamada et al., 2013).
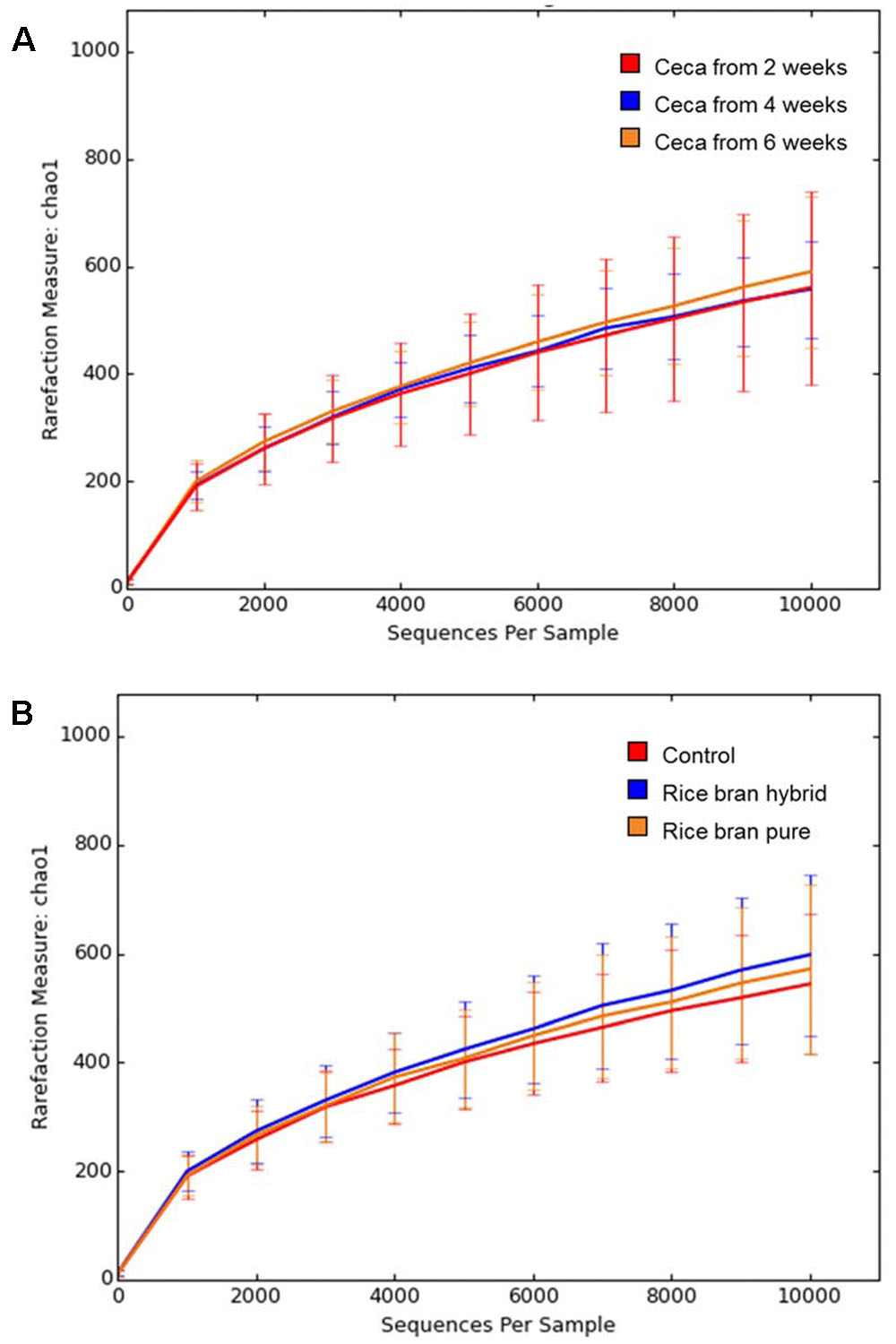
FIGURE 6. Rarefaction curves of Chao 1 in each group determined by (A) weeks of used ceca and (B) treatment (control or LaKast rice treated) from alpha diversity analysis.
In our plating experiments, the population of Salmonella was approximately 7 log CFU/ml in the anaerobic culture at the initial time point, indicating that Salmonella was dominant in samples. However, no Salmonella was identified in the microbiome analysis. These results suggest that the spiked Salmonella may be identified as Proteobacteria at the phylum level and Enterobacteriaceae (family level) or “Other” in the microbiome analysis at the genus level. Therefore, lower abundance of Proteobacteria and Enterobacteriaceae in LaKast rice samples compared to control in all experiments using ceca from different chicken ages may be due to the significant reductions of Salmonella as shown in plating experiment results.
Arkansas is the top producer of rice in the United States. Thus, tremendous quantities of rice bran as a by-product are available (Talbert and Burgos, 2007; Wilson et al., 2007). The present study clearly demonstrated the beneficial functionality of LaKast rice bran to inhibit pathogenic bacteria in both plating and microbiome analysis. In addition, with reduction of Salmonella-related bacteria, LaKast rice treated samples exhibited more diverse microbiota. Rice bran is a natural product derived from rice and thus the use of rice bran or fractions of rice bran could be acceptable and favorable to both the consumer and the animal feed industry. The results suggest that adding rice bran or bran fractions to animal feed as a biological supplement can contribute to ensure poultry safety, especially when antibiotics in feed are likely to be prohibited in the future. In conclusion, the present study demonstrates that AR LaKast rice bran or fractions thereof can add economic value to rice bran, benefiting both the rice and the poultry feed industries in post-antibiotic feed agriculture.
Author Contributions
SK, PR, SP, and SR designed the experiment. SK, PR, and SP performed the experiment and analyzed the data. SK wrote the manuscript, and PR, SP, and SR revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer GS and handling Editor declared their shared affiliation.
References
Centers for Disease Control Prevention (2009). Multistate outbreaks of Salmonella infections associated with live poultry-United States, 2007. MMWR Morb. Mortal. Wkly. Rep. 58, 25–29.
Donalson, L. M., Kim, W. K., Chalova, V. I., Herrera, P., Woodward, C. L., McReynolds, J. L., et al. (2007). In vitro anaerobic incubation of Salmonella enterica serotype Typhimurium and laying hen cecal bacteria in poultry feed substrates and a fructooligosaccharide prebiotic. Anaerobe 13, 208–214. doi: 10.1016/j.anaerobe.2007.05.001
Donalson, L. M., McReynolds, J. L., Kim, W. K., Chalova, V. I., Woodward, C. L., Kubena, L. F., et al. (2008). The influence of a fructooligosaccharide prebiotic combined with alfalfa molt diets on the gastrointestinal tract fermentation, Salmonella Enteritidis infection, and intestinal shedding in laying hens. Poult. Sci. 87, 1253–1262. doi: 10.3382/ps.2007-00166
Dunkley, K. D., Dunkley, C. S., Njongmeta, N. L., Callaway, T. R., Hume, M. E., Kubena, L. F., et al. (2007). Comparison of in vitro fermentation and molecular microbial profiles of high-fiber feed substrates incubated with chicken cecal inocula. Poult. Sci. 86, 801–810. doi: 10.1093/ps/86.5.801
Fan, Y. Y., Ricke, S. C., Scanlan, C. M., Nisbet, D. J., Vargas-Moskola, A. A., Corrier, D. E., et al. (1995). Use of differential rumen fluid-based carbohydrate agar media for culturing lactose-selected cecal bacteria from chickens. J. Food Prot. 58, 361–367. doi: 10.4315/0362-028X-58.4.361
Finstad, S., O’Bryan, C. A., Marcy, J. A., Crandall, P. G., and Ricke, S. C. (2012). Salmonella and broiler processing in the United States: relationship to foodborne salmonellosis. Food Res. Int. 45, 789–794. doi: 10.1016/j.foodres.2011.03.057
Foley, S. L., Johnson, T. J., Ricke, S. C., Nayak, R., and Danzeisen, J. (2013). Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol. Mol. Biol. Rev. 77, 582–607. doi: 10.1128/MMBR.00015-13
Foley, S. L., Nayak, R., Hanning, I. B., Johnson, T. J., Han, J., and Ricke, S. C. (2011). Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl. Environ. Microbiol. 77, 4273–4279. doi: 10.1128/aem.00598-11
Gibson, G. R., and Roberfroid, M. B. (1995). Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125, 1401–1412.
Hope, B. K., Baker, A. R., Edel, E. D., Hogue, A. T., Schlosser, W. D., Whiting, R., et al. (2002). An overview of the Salmonella Enteritidis risk assessment for shell eggs and egg products. Risk Anal. 22, 203–218. doi: 10.1111/0272-4332.00023
Howard, Z. R., O’Bryan, C. A., Crandall, P. G., and Ricke, S. C. (2012). Salmonella Enteritidis in shell eggs: current issues and prospects for control. Food Res. Int. 45, 755–764. doi: 10.1016/j.foodres.2011.04.030
Hudault, S., Bewa, H., Bridonneau, C., and Raibaud, P. (1985). Efficiency of various bacterial suspensions derived from cecal floras of conventional chickens in reducing the population level of Salmonella typhimurium in gnotobiotic mice and chicken intestines. Can. J. Microbiol. 31, 832–838. doi: 10.1139/m85-155
Janda, J. M., and Abbott, S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764. doi: 10.1128/JCM.01228-07
Kamada, N., Chen, G. Y., Inohara, N., and Núñez, G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690. doi: 10.1038/ni.2608
Kim, S. A., Park, S. H., Lee, S. I., Owens, C. M., and Ricke, S. C. (2017a). Assessment of chicken carcass microbiome responses during processing in the presence of commercial antimicrobials using a next generation sequencing approach. Sci. Rep. 7:43354. doi: 10.1038/srep43354
Kim, S. A., Park, S. H., Lee, S. I., and Ricke, S. C. (2017b). Development of a rapid method to quantify Salmonella Typhimurium using a combination of MPN with qPCR and a shortened time incubation. Food Microbiol. 65, 7–18. doi: 10.1016/j.fm.2017.01.013
Kim, S. P., Kang, M. Y., Park, J. C., Nam, S. H., and Friedman, M. (2012). Rice hull smoke extract inactivates Salmonella Typhimurium in laboratory media and protects infected mice against mortality. J. Food Sci. 71, M80–M85. doi: 10.1111/j.1750-3841.2011.02478.x
Kim, S. P., Park, S. O., Lee, S. J., Nam, S. H., and Friedman, M. (2013). A polysaccharide isolated from the liquid culture of Lentinus edodes (Shiitake) mushroom mycelia containing black rice bran protects mice against a Salmonella lipopolysaccharide-induced endotoxemia. J. Agric. Food Chem. 61, 10987–10994. doi: 10.1021/jf403173k
Kumar, A., Henderson, A., Forster, G., Goodyear, A., Weir, T., Leach, J., et al. (2012). Dietary rice bran promotes resistance to Salmonella enterica serovar Typhimurium colonization in mice. BMC Microbiol. 12:71. doi: 10.1186/1471-2180-12-71
Nakagawa, S., Takaki, Y., Shimamura, S., Reysenbach, A.-L., Takai, K., and Horikoshi, K. (2007). Deep-sea vent ε-proteobacterial genomes provide insights into emergence of pathogens. Proc. Natl. Acad. Sci. U.S.A. 104, 12146–12150. doi: 10.1073/pnas.0700687104
Park, S. H., Kim, S. A., Lee, S. I., Rubinelli, P. M., Roto, S. M., Pavlidis, H. O., et al. (2017). Original XPCTM effect on Salmonella Typhimurium and cecal microbiota from three different ages of broiler chickens when incubated in an anaerobic in vitro culture system. Front. Microbiol. 8:1070. doi: 10.3389/fmicb.2017.01070
Park, S. H., Lee, S. I., and Ricke, S. C. (2016). Microbial populations in naked neck chicken ceca raised on pasture flock fed with commercial yeast cell wall prebiotics via an Illumina MiSeq platform. PLOS ONE 11:e0151944. doi: 10.1371/journal.pone.0151944
Patterson, J. A., and Burkholder, K. M. (2003). Application of prebiotics and probiotics in poultry production. Poult. Sci. 82, 627–631. doi: 10.1093/ps/82.4.627
Reller, L. B., Weinstein, M. P., and Petti, C. A. (2007). Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 44, 1108–1114. doi: 10.1086/512818
Ricke, S. C. (2015). Potential of fructooligosaccharide prebiotics in alternative and nonconventional poultry production systems. Poult. Sci. 94, 1411–1418. doi: 10.3382/ps/pev049
Ricke, S. C., and Pillai, S. D. (1999). Conventional and molecular methods for understanding probiotic bacteria functionality in gastrointestinal tracts. Crit. Rev. Microbiol. 25, 19–38. doi: 10.1080/10408419991299176
Roberfroid, M. (2007). Prebiotics: the concept revisited. J. Nutr. 137, 830S–837S. doi: 10.1093/jn/137.3.830S
Rubinelli, P., Roto, S., Kim, S. A., Park, S. H., Pavlidis, H. O., McIntyre, D., et al. (2016). Reduction of Salmonella Typhimurium by fermentation metabolites of Diamond V Original XPC in an in vitro anaerobic mixed chicken cecal culture. Front. Vet. Sci. 3:83. doi: 10.3389/fvets.2016.00083
Rubinelli, P. M., Kim, S. A., Park, S. H., Roto, S. M., Nealon, N. J., Ryan, E. P., et al. (2017). Differential effects of rice bran cultivars to limit Salmonella Typhimurium in chicken cecal in vitro incubations and impact on the cecal microbiome and metabolome. PLOS ONE 12:e0185002. doi: 10.1186/1471-2180-12-71
Ryan, E. P., Heuberger, A. L., Weir, T. L., Barnett, B., Broeckling, C. D., and Prenni, J. E. (2011). Rice bran fermented with Saccharomyces boulardii generates novel metabolite profiles with bioactivity. J. Agric. Food Chem. 59, 1862–1870. doi: 10.1021/jf1038103
Saengkerdsub, S., Kim, W. K., Anderson, R. C., Nisbet, D. J., and Ricke, S. C. (2006). Effects of nitrocompounds and feedstuffs on in vitro methane production in chicken cecal contents and rumen fluid. Anaerobe 12, 85–92. doi: 10.1016/j.anaerobe.2005.11.006
Salanitro, J., Blake, I., and Muirhead, P. (1974). Studies on the cecal microflora of commercial broiler chickens. Appl. Microbiol. 28, 439–447.
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M.-A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States-major pathogens. Emerging Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Sheflin, A. M., Borresen, E. C., Wdowik, M. J., Rao, S., Brown, R. J., Heuberger, A. L., et al. (2015). Pilot dietary intervention with heat-stabilized rice bran modulates stool microbiota and metabolites in healthy adults. Nutrients 7, 1282–1300. doi: 10.3390/nu7021282
Shermer, C. L., Maciorowski, K. G., Bailey, C. A., Byers, F. M., and Ricke, S. C. (1998). Caecal metabolites and microbial populations in chickens consuming diets containing a mined humate compound. J. Sci. Food Agric. 77, 479–486. doi: 10.1002/(SICI)1097-0010(199808)77:4<479::AID-JSFA607>3.0.CO;2-L
Stopforth, J., O’connor, R., Lopes, M., Kottapalli, B., Hill, W., and Samadpour, M. (2007). Validation of individual and multiple-sequential interventions for reduction of microbial populations during processing of poultry carcasses and parts. J. Food Prot. 70, 1393–1401. doi: 10.4315/0362-028X-70.6.1393
Talbert, R. E., and Burgos, N. R. (2007). History and management of herbicide-resistant barnyardgrass (Echinochloa crus-galli) in Arkansas rice. Weed Technol. 21, 324–331. doi: 10.1614/WT-06-084.1
Wilson, C. Jr., Runsick, S., and Mazzanti, R. (2007). Trends in Arkansas rice production. BR Wells Rice Res. Stud. 560, 11–20. doi: 10.1007/s10886-013-0246-7
Keywords: Salmonella, poultry, reduction, inhibition, cecum, Arkansas rice, microbiome
Citation: Kim SA, Rubinelli PM, Park SH and Ricke SC (2018) Ability of Arkansas LaKast and LaKast Hybrid Rice Bran to Reduce Salmonella Typhimurium in Chicken Cecal Incubations and Effects on Cecal Microbiota. Front. Microbiol. 9:134. doi: 10.3389/fmicb.2018.00134
Received: 29 September 2017; Accepted: 19 January 2018;
Published: 06 February 2018.
Edited by:
Aldo Corsetti, Università degli Studi di Teramo, ItalyReviewed by:
Robin Anderson, Agricultural Research Service (USDA), United StatesGiovanna Suzzi, Università degli Studi di Teramo, Italy
Copyright © 2018 Kim, Rubinelli, Park and Ricke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven C. Ricke, c3JpY2tlQHVhcmsuZWR1
†Present address: Si Hong Park, Department of Food Science and Technology, Oregon State University, Corvallis, OR, United States Sun Ae Kim, Department of Food Science and Engineering, Ewha Womans University, Seoul, South Korea
 Sun Ae Kim
Sun Ae Kim Peter M. Rubinelli
Peter M. Rubinelli Si Hong Park
Si Hong Park Steven C. Ricke
Steven C. Ricke