- 1Institute of Pharmaceutical Biotechnology, Zhejiang University, Hangzhou, China
- 2Zhejiang Provincial Key Laboratory for Microbial Biochemistry and Metabolic Engineering, Hangzhou, China
- 3College of Life Sciences, Zhejiang University, Hangzhou, China
AdpA, an AraC/XylS family protein, had been proved as a key regulator for secondary metabolism and morphological differentiation in Streptomyces griseus. Here, we identify AdpAch, an ortholog of AdpA, as a “higher level” pleiotropic regulator of natamycin biosynthesis with bidirectional regulatory ability in Streptomyces chattanoogensis L10. DNase I footprinting revealed six AdpAch-binding sites in the scnRI–scnRII intergenic region. Further analysis using the xylE reporter gene fused to the scnRI–scnRII intergenic region of mutated binding sites demonstrated that the expression of scnRI and scnRII was under the control of AdpAch. AdpAch showed a bi-stable regulatory ability where it firstly binds to the Site C and Site D to activate the transcription of the two pathway-specific genes, scnRI and scnRII, and then binds to other sites where it acts as an inhibitor. When Site A and Site F were mutated in vivo, the production of natamycin was increased by 21% and 25%, respectively. These findings indicated an autoregulatory mechanism where AdpAch serves as a master switch with bidirectional regulation for natamycin biosynthesis.
Introduction
The secondary metabolic process in Streptomyces is regulated by a complex regulatory network involving pathway-specific, pleiotropic, and global regulators which respond to a variety of physiological and environmental condition alterations (van Wezel and McDowall, 2011; Liu et al., 2013). The best characterized is the A-factor regulatory cascade in which AdpA is the most important transcriptional factor for the secondary metabolism (Horinouchi, 2002; Ohnishi et al., 2005). In early culture stages, the transcription of adpA in Streptomyces griseus is repressed by ArpA, the receptor protein for A-factor (Onaka and Horinouchi, 1997). When A-factor reaches a critical concentration, it binds to ArpA and confers the conformational change of ArpA (Ohnishi et al., 1999). This results in dissociation of ArpA from the adpA promoter, in turn switching on the expression of adpA (Ohnishi et al., 1999). The induced AdpA then activates the transcription of various genes related to secondary metabolism such as strR, the pathway-specific regulatory genes for streptomycin in S. griseus (Retzlaff and Distler, 1995; Tomono et al., 2005).
AdpA is a member of the AraC/XylS family proteins (Gallegos et al., 1997). It has been suggested to form a dimer through the N-terminal portion which belong to the ThiJ/PfpI/DJ-1 family (Yamazaki et al., 2004; Ohnishi et al., 2005). To date, a number of AdpA orthologs have been described as having essential roles in the secondary metabolism in many Streptomyces species, such as Streptomyces lividans (Guyet et al., 2013), Streptomyces coelicolor A3(2) (Takano et al., 2001; Nguyen et al., 2003), Streptomyces ansochromogenes (Pan et al., 2009), Streptomyces avermitilis (Komatsu et al., 2010), Streptomyces hygroscopicus 5008 (Tan et al., 2015), and Streptomyces clavuligerus (López-García et al., 2010).
Typically, AdpA is regarded as an activator for downstream regulated genes, except itself which is proved to be negatively auto-regulated by binding to its own promoter region (Kato et al., 2005b; Hara et al., 2009). The molecular mechanism of transcriptional activation begins as a dimer of AdpA binds to the target sites with consensus sequences which then recruit RNA polymerase to the promoter for transcriptional initiation (Yamazaki et al., 2004; Kato et al., 2005a). For different target genes, AdpA showed a different number of binding sites in the promoter regions. For example, there are two AdpA-binding sites in the promoter of strR (Tomono et al., 2005), whereas there are three AdpA-binding sites for regulation of ssgA (Yamazaki et al., 2003a). However, the precise regulation mechanism how the AdpA binds to multiple sites to activate transcription has not been experimentally determined. Based on the importance of AdpA in the biosynthesis of the secondary metabolism, it is necessary to elucidate details of its regulatory mechanisms.
Natamycin, an antifungal polyene macrolide antibiotic, is synthesized by a type I polyketide synthase gene cluster. Previous analysis of the gene cluster of natamycin in Streptomyces chattanoogensis L10 revealed the existence of 17 open-reading frames, including two pathway-specific genes, scnRI and scnRII (Du et al., 2011a). These two genes showed high sequence identity to pimR and pimM of Streptomyces natalensis, respectively (Antón et al., 2007; Santos-Aberturas et al., 2012). Gene disruption of scnRI resulted in a large decrease in the expression of biosynthetic genes, indicating its role as a pivotal activator for the biosynthesis of natamycin (Du et al., 2011a). scnRII, adjacent but divergently transcribed transcriptional regulatory genes, was shown to act as a second positive regulator for natamycin production (Du et al., 2009). We also had proved that AdpAch controls the production of natamycin, but the detailed relationship among AdpAch, ScnRI, and ScnRII had not been well characterized (Du et al., 2011a).
Here, we reveal the sophisticated regulatory characteristics of AdpAch in the natamycin biosynthesis of S. chattanoogensis L10. AdpAch acts as a “higher level” pleiotropic regulator for transcription of the two divergently transcribed pathway-specific genes, scnRI and scnRII. In this regulatory process, AdpAch shows a bi-stable regulatory ability, where it firstly acts as an activator, then a repressor. Moreover, natamycin production was enhanced by mutating the AdpAch-binding sites which had an inhibitory effect. This work not only advances the understanding of detailed regulatory mechanism of AdpA, but also provides a potential target for the enhancement of other antibiotic production levels by manipulating the regulatory network.
Results
AdpAch Identified as a “Higher Level” Pleiotropic Regulator for Natamycin Biosynthesis
In our previous study, the biosynthetic gene cluster of natamycin has been cloned and characterized in S. chattanoogensis L10. Within this there are two divergently transcribed genes, scnRI and scnRII, encoding proteins that resemble pathway-specific regulators (Du et al., 2009, 2011a). Although the functions of these two regulators have been well characterized, an important question remains as to whether there are multiple levels of control in the biosynthesis of natamycin. Based on our previous study that AdpAch affected the transcription of these two pathway-specific genes (Du et al., 2011a), we speculated that AdpAch may act as a “higher level” pleiotropic regulator for regulating the natamycin biosynthesis.
To test this hypothesis, electrophoretic mobility shift assays (EMSAs) were applied. As shown in Figure 1, retardation was readily detected upon the addition of 50 pM AdpAch with the probe RI–RII, while the addition of 50- to 100-fold excess of unlabeled specific PCR product reduced the proportion of the labeled promoter-containing fragment (Figure 1). These data clearly demonstrate that AdpAch could specifically bind to the scnRI–scnRII intergenic region and could control the expression of these two pathway-specific genes.
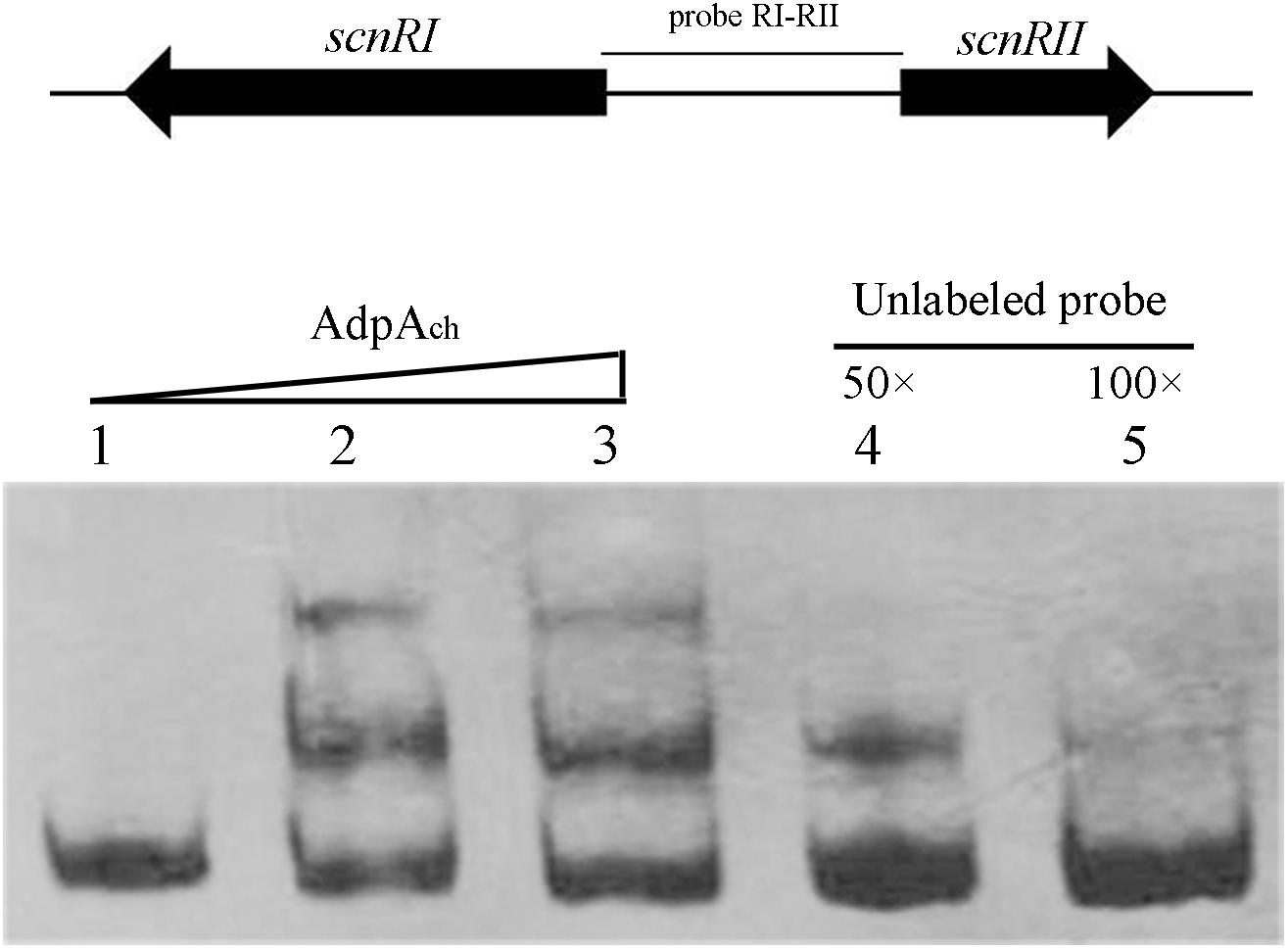
FIGURE 1. AdpAch binds to the DNA sequence of the intergenic promoter region between scnRI and scnRII. Lanes 1–3, DNA probe with AdpAch protein 0, 50, and 100 pM, respectively. Lanes 4 and 5, 50- and 100-fold excess of unlabeled specific PCR product was added into binding reactions.
DNase I Footprinting Assay Reveals Six AdpAch-Binding Sites in the scnRI–scnRII Intergenic Region
To identify the exact DNA sequences that AdpAch protected in the scnRI–scnRII intergenic region, DNase I footprinting assays, in absence or presence of purified recombinant AdpAch, were performed. In our previous studies, we had determined the transcription start site (TSS) of the two pathway-specific genes, scnRI and scnRII (Du et al., 2011a). As seen in Figure 2A, at a lower AdpAch protein concentration of 100 pM, the DNA strands of the scnRI–scnRII intergenic region showed two protected regions, Site C and Site D, extending from positions -69 to -44 and -106 to -74 relative to the TSS of scnRI. When increasing the protein concentration to 500 pM, another four protected regions (Sites A, B, E, and F) were observed. With respect to the scnRI TSS, the AdpAch-binding Site A locates at positions +8 to +54, Site B at positions -20 to +2, Site E at positions -161 to -114, and Site F at positions -283 to -259 (Figure 2B). The six AdpAch-binding sites were spread over the scnRI–scnRII intergenic region. Notably, Site A was located downstream of the scnRI TSS, while Site B overlapped the -10 region of the scnRI promoter. Site F was located downstream of the scnRII TSS, and Site E overlapped the -35 region of the scnRII promoter. This data suggest that AdpAch might have a negative regulatory ability for the expression of these two pathway-specific genes. Additionally, the results from the DNase I footprinting assay also reveal that AdpAch may have higher affinity to Site C and Site D than to the others.
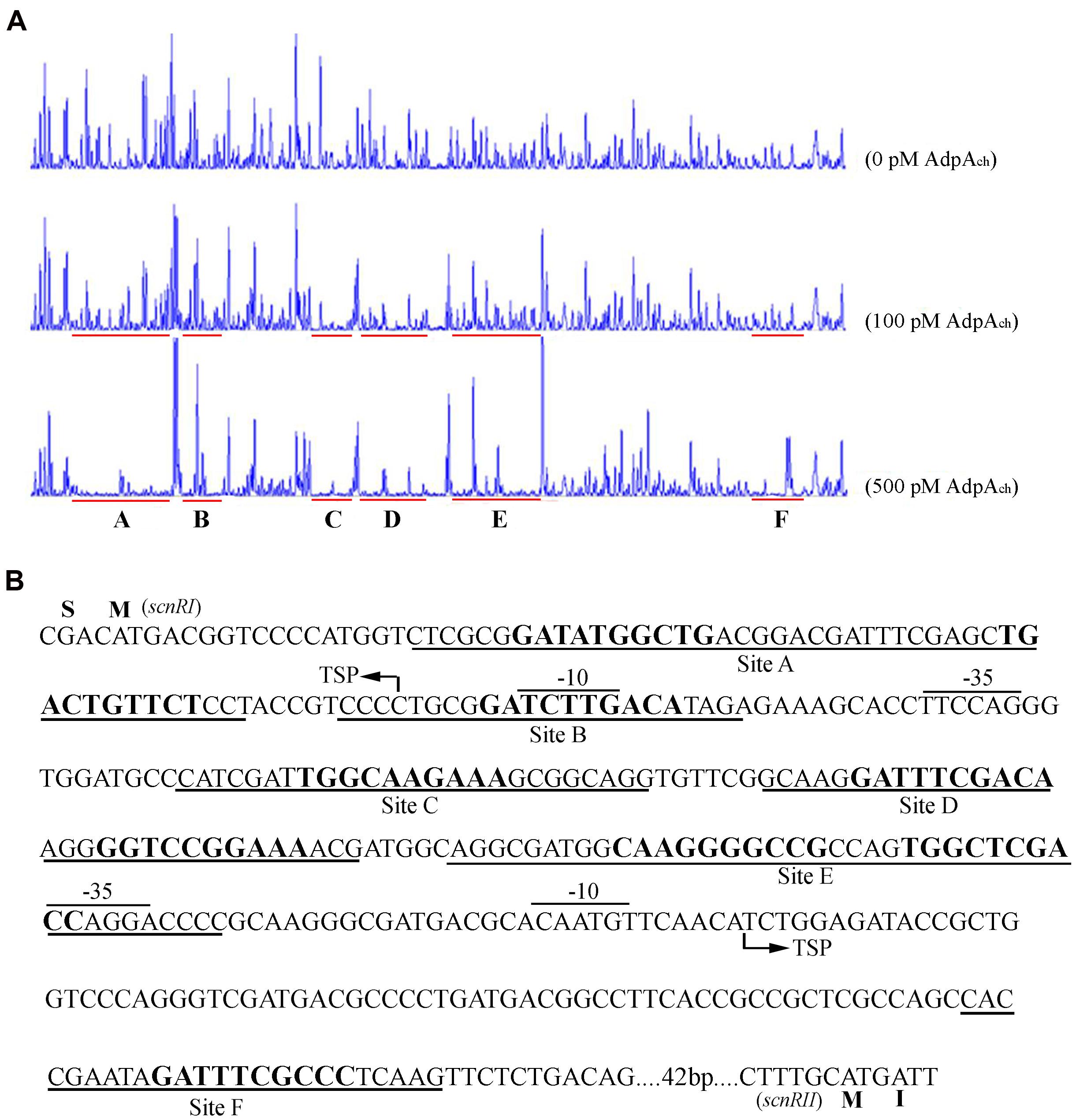
FIGURE 2. DNase I footprinting assay for determination of the AdpAch-binding sites. (A) A 5′-FAM-labeled probe pRI-RII was used in the DNase I footprinting assay with 0, 100, and 500 pM purified AdpAch, respectively. The protected regions are underlined. (B) Nucleotide sequences of the scnRI–scnRII intergenic region showing the predicted AdpAch-binding sites. The TSS is marked by a bent arrow, the AdpAch-binding sites are underlined, and the –10 and –35 regions are overlined.
The Consensus AdpAch-Binding Sequence in the AdpAch-Binding Sites
The orthologs of AdpAch identified in S. griseus and S. coelicolor have been reported to have the consensus binding sequence, 5-TGGCSNGWWY-3 (S: G or C; W: A or T; Y: T or C; N: any nucleotide) (Yamazaki et al., 2004). After alignment of these six protected regions, we also found that there were highly conserved AdpAch-binding sequences in each binding site (Figure 3A). To further study the roles of these consensus sequences in the AdpAch-binding ability, EMSAs were carried out using the probes containing either the sequences of wild-type (wt) binding sites or the mutated sites (Figure 3A). As shown in Figure 3B, no binding shift was detected for the mutated sites A–F when compared with their corresponding wt targets. Taken together, these data demonstrated that AdpAch indeed has six binding sites in the scnRI–scnRII intergenic region and the consensus sequence is essential for the binding activity of AdpAch.
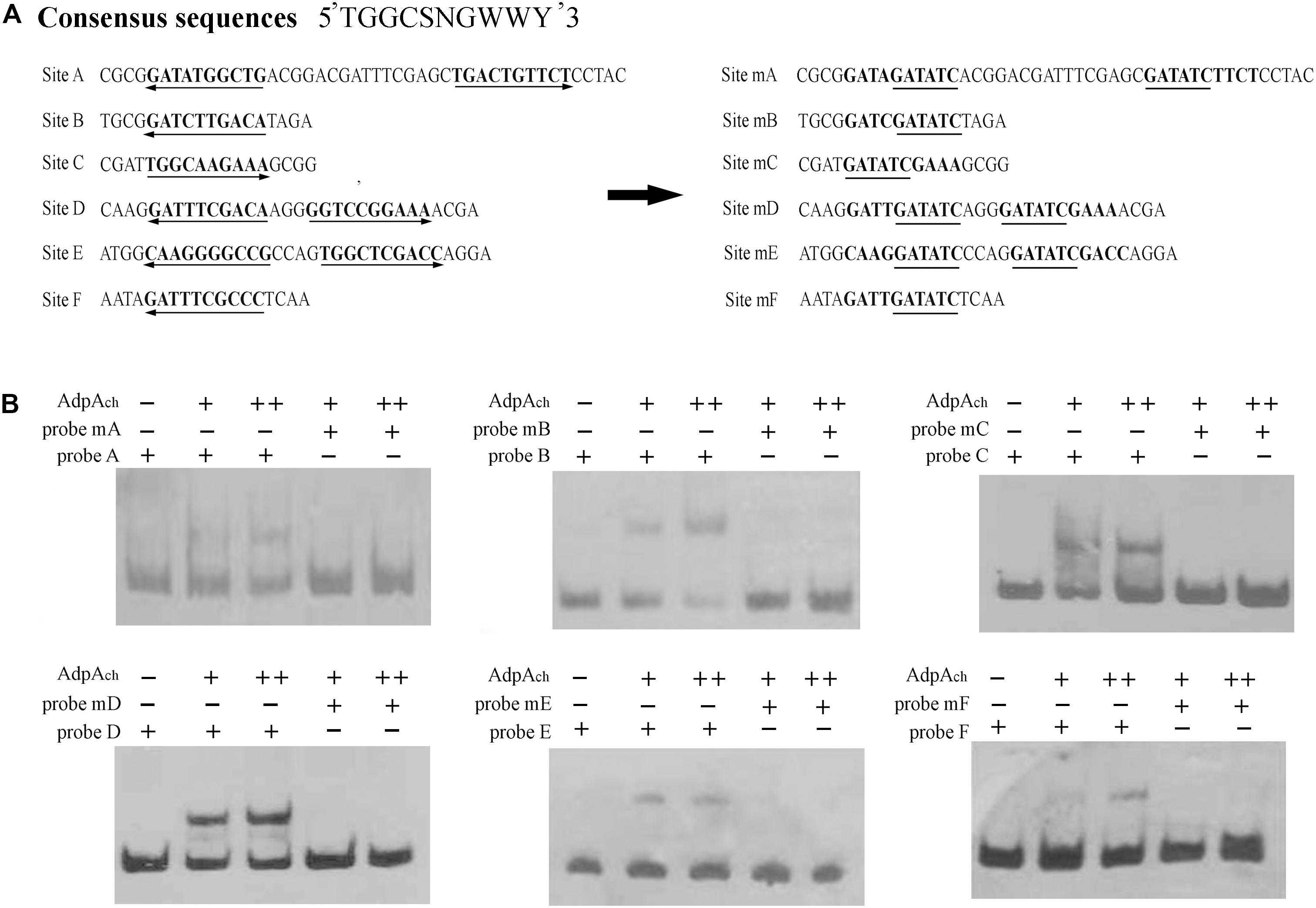
FIGURE 3. Mutational analysis of the AdpAch-binding sites. (A) Mutations introduced in the six putative AdpAch-binding sites. The predicted AdpAch-binding consensus sequences are in bold, and these consensus sequences are changed with an EcoRI site indicated with underlines. (B) EMSAs for determination of AdpAch binding to mutated sequences. Probes A–F contained the fragment of Sites A–F as shown in A, respectively. Probes mA–mF contained the fragment of corresponding mutated sites. The amounts of AdpAch protein used were 50 and 100 pM.
AdpAch Has Differing Affinities for Different Binding Sites
In the DNase I footprinting analysis, Site C and Site D were occupied with a lower concentration of AdpAch than the other sites. This suggests that there may be affinity differences for AdpAch between the six binding sites. To test this possibility, competitive EMSAs with 50- to 100-fold excess of unlabeled fragments of six AdpAch-binding sites were used to compete with each labeled fragment. As shown in Figure 4A, 100-fold excess of unlabeled SB′ (Site B) and SF′ (Site F) could not completely abolish AdpAch complex formation with the labeled probe SA (Site A). However, the same amount of unlabeled SC′ (Site C), SD′ (Site D), and SE′ (Site E) outcompeted the labeled probe SA. This result indicated that AdpAch binds to Site A more tightly than Site B and Site F, but less tightly than Site C, Site D, and Site E. Following this way, we could conclude that Site B has less affinity for AdpAch than others, except for Site F (Figure 4B), which was the weakest affinity among the six binding sites (Figure 4F), and Site D was the strongest affinity of these six sites (Figure 4D). The affinity of Site E for AdpAch was between that of Site C and Site A (Figures 4A,C,E). Therefore, we determined the affinity of AdpAch to different binding sites in the following order: Site D > Site C > Site E > Site A > Site B > Site F.
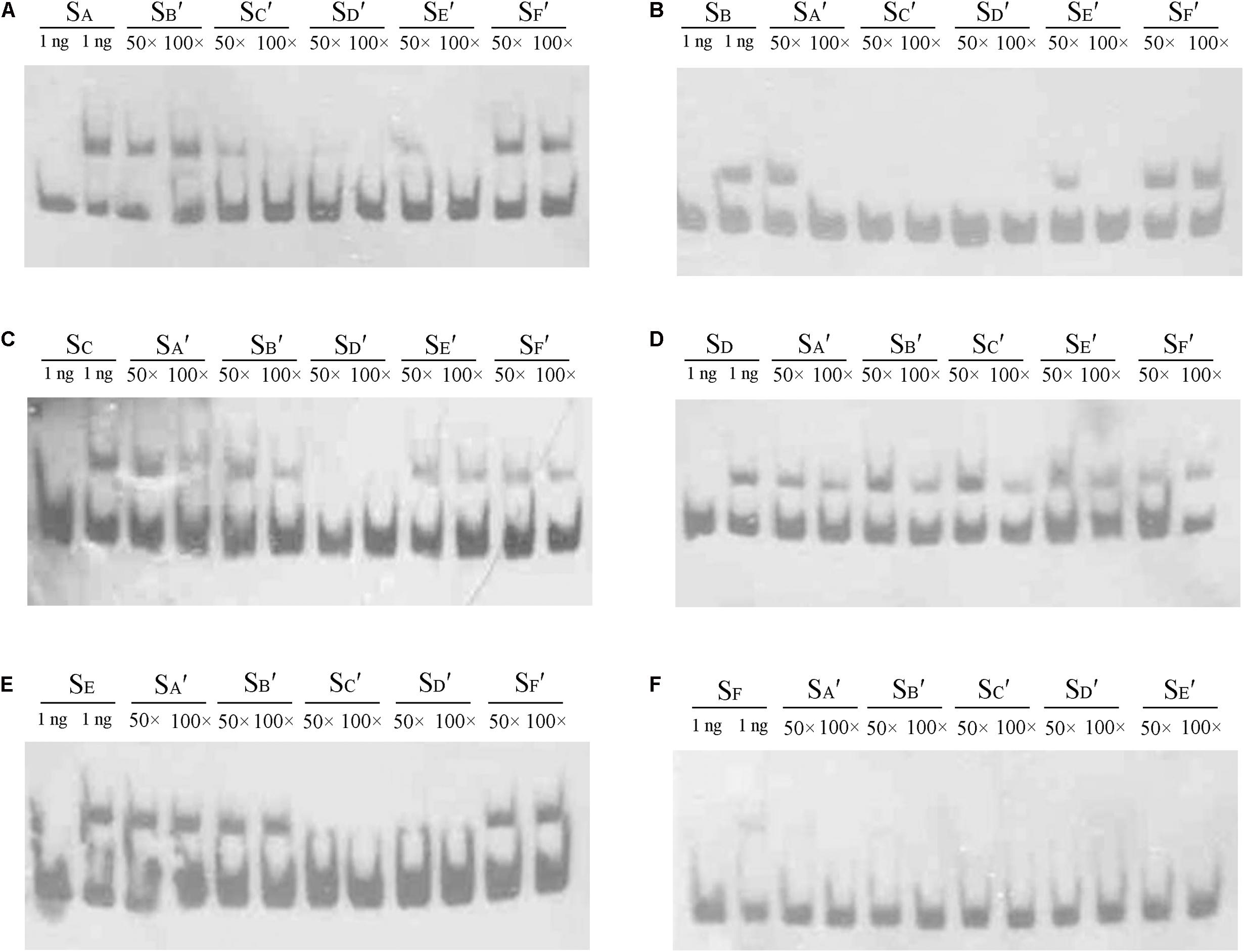
FIGURE 4. Comparison of the relative affinity of AdpAch with different binding sites. Labeled probes SA, SB, SC, SD, SE, and SF contained the fragment of Sites A–F as shown in Figure 3A, respectively. Probes SA′, SB′, SC′, SD′, SE′, and SF′ also contained the fragment of Sites A–F as shown in Figure 3A, respectively, but they are unlabeled. The amount of AdpAch protein used was 100 pM.
Promoter-Probe Assays of the AdpAch-Binding Sites in the scnRI–scnRII Intergenic Region
The binding sites of AdpAch in the scnRI–scnRII intergenic region were adjacent to either the scnRI or the scnRII start codon. This raised the possibility that this intergenic region might harbor a bidirectional promoter allowing AdpAch to regulate transcriptions of the divergently transcribed flanking genes, scnRI and scnRII (Figure 2B). To investigate the promoter activities of the two pathway-specific genes with each of the AdpAch-binding sites, we used the promoter-probe plasmid pIJ8601 carrying the xylE gene, encoding catechol 2,3-dioxygenase, as the reporter. As shown in Figure 5A, the transcriptional profiles of scnRI were severely decreased when the AdpAch-binding Site C and Site D were mutated. Conversely, its transcriptional activity was increased when Site A and Site B were mutated and remained almost unchanged when Site E and Site F were mutated. For the promoter activity of scnRII, we did not detect any consistent differences when Sites A, B, and C were mutated, but mutation in the Sites D and E resulted in a large decreases of up to 70 and 40%, respectively, compared to those of the wt. The mutation in Site F resulted in a statistically significant increase (Figure 5B). These findings indicated that expressions of scnRI and scnRII are both under the control of AdpAch, which has a completely different regulatory ability (activation or inhibition) when binding to different binding sites.
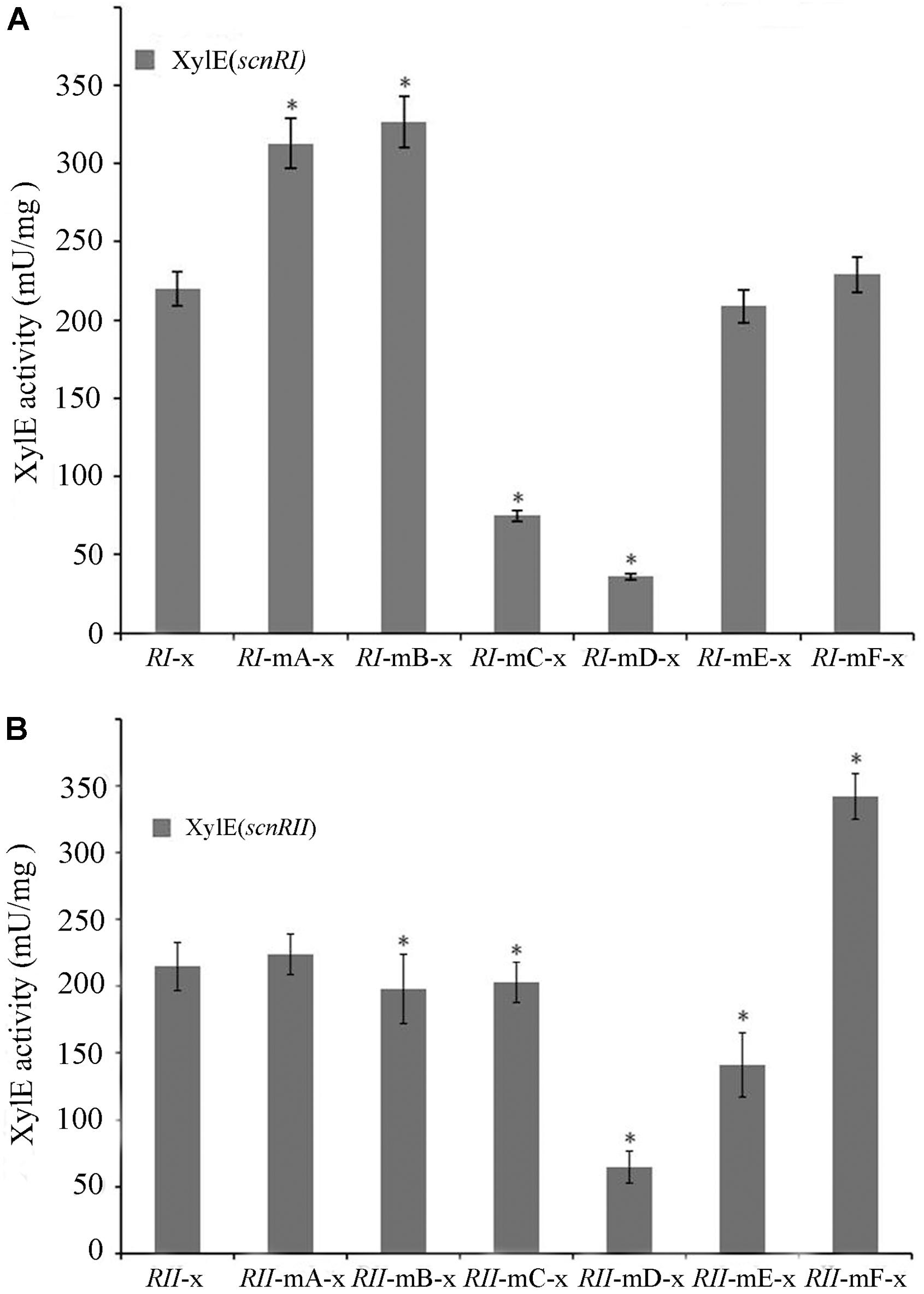
FIGURE 5. Promoter activities of scnRI (A) and scnRII (B) with the effect of mutations in the AdpAch-binding sites. The strains were grown in YEME medium for 24 h, and catechol dioxygenase activity was calculated as the change of catechol quantity (mmol) per minute. Error bars correspond to the standard error of the mean of four culture replicates. ∗ Indicates significant differences between promoter mutants and promoter wt (P < 0.05).
Effect of Mutated AdpAch-Binding Sites On Natamycin Production in Vivo
There have been some reports where effects upon DNA-binding sites were found in vitro that failed to be exhibited in vivo. In order to test this possibility and reveal the function of the six AdpAch-binding sites in natamycin biosynthesis in vivo, a series of mutants were constructed as described in Experimental procedures. As shown in Figure 6A, compared to the WT strain, the level of natamycin production in the R-mA (mutation in Site A) and R-mF (mutation in Site F) had increased by 21 and 25%, respectively. However, the constructed strains of R-mC (mutation in Site C), R-mD (mutation in Site D), and R-mE (mutation in Site E) showed up to 31, 42 and 15% reductions, respectively. The natamycin production of R-mB (mutation in Site B) mutant exhibited almost no change. This finding indicated that the AdpAch-binding Sites A and F play negative roles for natamycin biosynthesis, while the functions of the Sites C, D, and E were positive. Quantitative real-time PCR (qRT-PCR) analysis showed that the promoting effect of site mutation on natamycin production was due to alteration of the pathway-specific genes at the transcriptional level (Figure 6B).
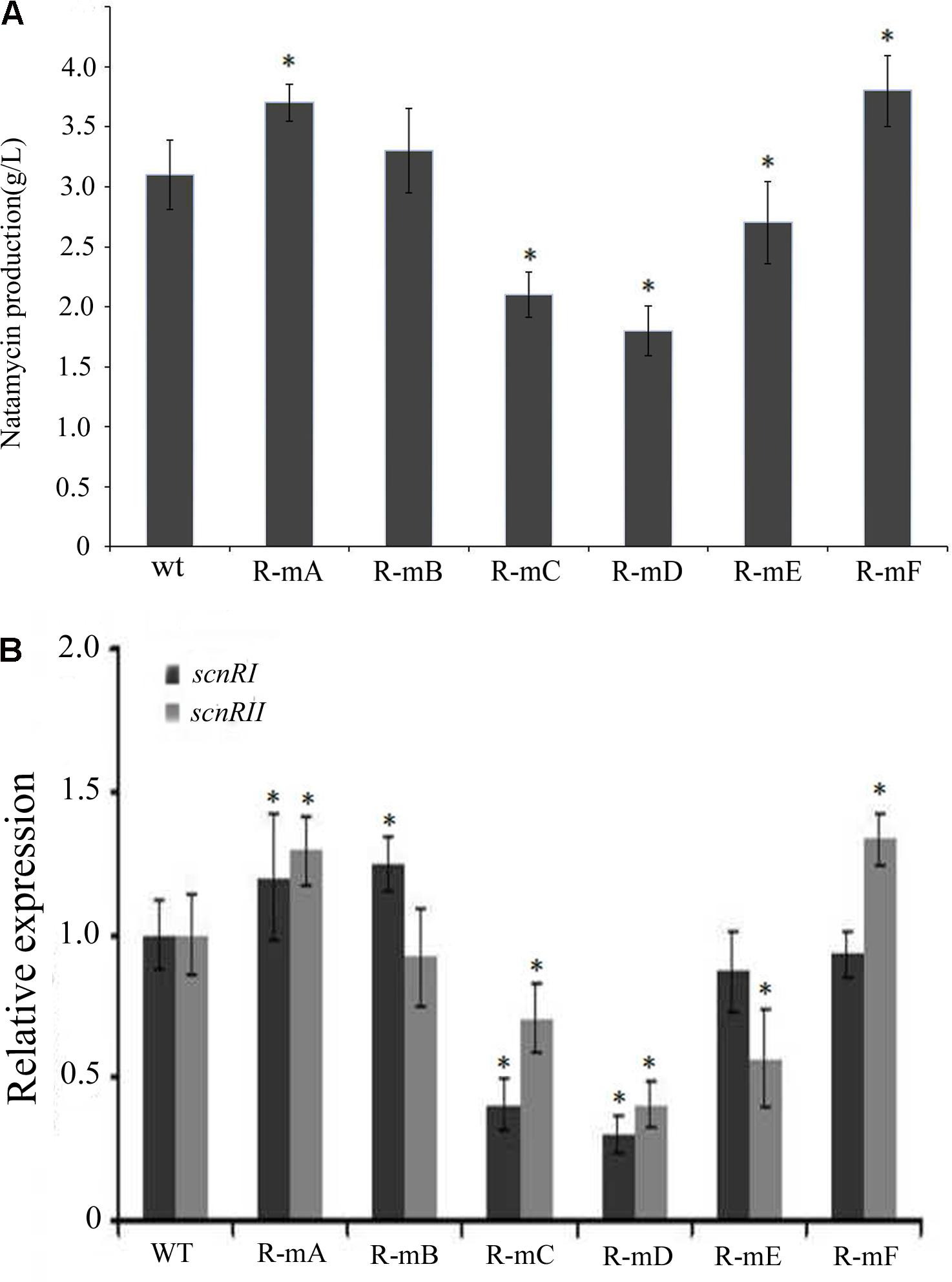
FIGURE 6. (A) The effect of mutated AdpAch-binding sites on the natamycin production in vivo. The strains were grown in YEME medium for 96 h. Vertical error bars correspond to the standard error of the mean of four replicated cultures. (B) Real-time RT-PCR analysis of the scnRI and scnRII transcript in the wt strain and mutated AdpAch-binding sites strain. The expression level of scnRI and scnRII is presented relative to the wt sample from 24 h, which was arbitrarily assigned a value of 1. The transcription of hrdB was assayed as an internal control. Error bars were calculated by measuring the standard deviation among three replicates of each sample. ∗ Indicates significant differences between AdpAch-binding site mutants and wt (P < 0.05).
Discussion
Streptomyces spp. have developed complicated mechanisms to adapt to altered circumstances (Santos-Beneit et al., 2009; Yu et al., 2012). Among these mechanisms, the multiple levels of regulation in controlling the expression of the genes responsible for the formation of the secondary metabolism are drawing increased attention. In this study, we focused on the regulatory network of natamycin biosynthesis in S. chattanoogensis L10, an industrial strain for natamycin production. In our previous study, we determined that gamma-butyrolactones (GBLs) serve as quorum-sensing signaling molecules for activating natamycin production in S. chattanoogensis L10 (Du et al., 2011b), and ScnRII acts as a positive regulator by directly binding to the promoters of natamycin biosynthetic genes (Du et al., 2009) where ScnRI acts as a positive regulator for the transcription of scnRII (Du et al., 2011a). However, the deletion of scnRI did not result in a complete halt of the transcription of scnRII (our unpublished data). This is quite different from the function of PimR in S. natalensis where the deletion of pimR almost completely destroys the transcription of pimM (Antón et al., 2004; Santos-Aberturas et al., 2012). As the regulation of antibiotic biosynthesis involves numerous transcription factors (McKenzie and Nodwell, 2007; van Wezel and McDowall, 2011), participation of other regulator(s) is possible, in the regulation of scnRII.
With AdpAch being able to regulate the expression of both of the pathway-specific genes, scnRI and scnRII, it provides a possible explanation that there is a coordinate regulation in controlling expression of scnRII by AdpAch and ScnRI. This regulatory pattern may occur in following steps. Firstly, AdpAch binds to the scnRI–scnRII intergenic region and activates both transcription of scnRI and scnRII. Then ScnRI also binds to the scnRI–scnRII intergenic region which, in turn, promotes the transcriptional level of scnRII. However, these two genes were not completely controlled by AdpAch. Trace expression of scnRI was observed in the adpAch mutant, and then ScnRI would promote the transcription of scnRII (Du et al., 2011a). Notably, a certain amount of AdpAch is required for binding to the scnRI–scnRII intergenic region (∼50 pM). This is why we did not detect the shifted band with low concentration AdpAch (∼1 pM) in the binding reaction of our previous study (Du et al., 2011a).
In most cases, AdpA acts as an activator for the target genes, except for itself where it exhibits an autorepression (Kato et al., 2005b). In this study, we concluded from promoter-probe assays in vivo that AdpAch could not only regulate both pathway-specific genes, but also displayed completely opposite regulatory abilities in control of them. The AdpAch-binding Site C and Site D were involved in activating the transcription of scnRI, while AdpAch binding to Sites A and B resulted in repression. For the promoter activity of scnRII, mutation in the Site C and Site D resulted in a decrease of transcriptional profiles, while a mutation in the Site F led to a statistically significant increase. A similar phenotype was observed in S. ansochromogenes where transcription of sanG decreased when Site I and Site V were mutated but increased when other three AdpA-L-binding sites were mutated (Pan et al., 2009). However, when combinations of binding site mutations were carried out, the promoter activities were not in accordance with our predictions. For example, mutations in both Sites E and F reduced the transcriptional level of scnRII (data not shown). Based on the short distances between the AdpAch-binding sites which are spread over the scnRI–scnRII intergenic region, there may be complicated interactions between different AdpAch dimmers to explain this.
With further analysis using competitive gel shift assays, we could conclude that AdpAch binds to Sites A–F with the following affinities: Site D > Site C > Site E > Site A > Site B > Site F (Figure 4). These data are consistent with the footprinting assay where the regions of Site C and Site D were previously protected at a lower AdpAch protein concentration (Figure 2A). This gives a hint that the regulatory ability of AdpAch may occur in a growth phase-dependent manner. In the early stage, AdpAch firstly binds to the Site C and D to recruit RNA polymerase to the promoter and initiates the transcription of scnRI and scnRII. This in turn triggers natamycin production (Figure 7). When AdpAch is accumulated to a certain critical level, it will bind to other binding sites located near the TSS. A DNA loop may be formed via the interaction between different AdpAch dimers, thus preventing RNA polymerase from access to the promoter of the pathway-specific genes (Figure 7). Reduced transcription of the pathway-specific genes will result in a low rate of natamycin production.
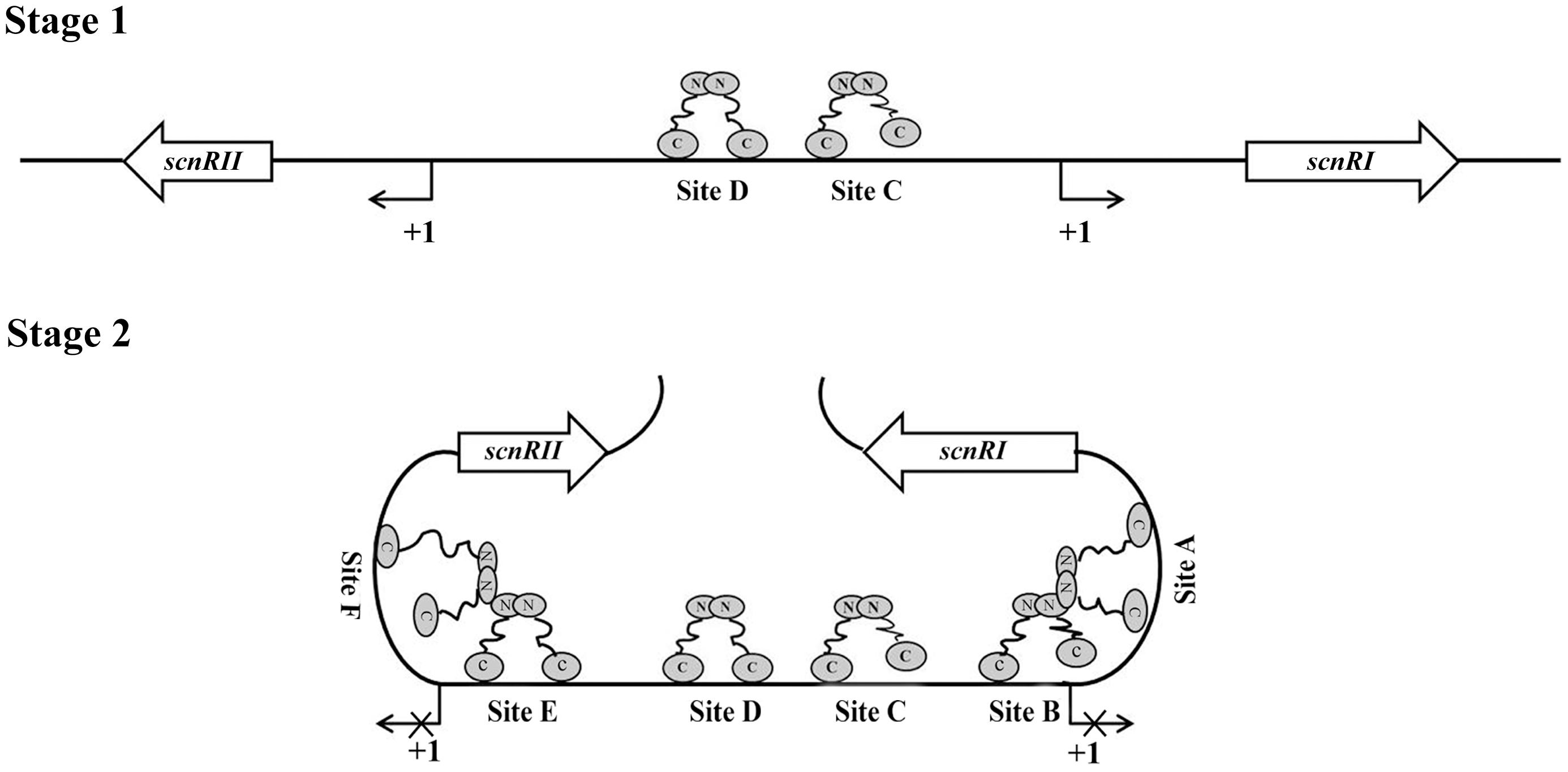
FIGURE 7. Proposed model of AdpAch regulation for scnRI and scnRII transcription. The AdpAch-binding sites are spread over the scnRI–scnRII intergenic region, and the presumptive manner of AdpAch-binding forms a dimmer. Stage 1: AdpAch binds to the Site C and Site D when the concentration of AdpAch is low in the early growth stage. Stage 2: When the concentration AdpAch reaches a high level, AdpAch binds to Site A, Site B, Site E, and Site F, most of which located downstream of the -35 sequences of the corresponding genes. A DNA loop may be formed via interaction between different AdpAch dimmers, thus preventing RNA polymerase from access to the promoter.
The discovery of this bidirectional regulation of AdpAch in the control of natamycin biosynthesis reveals an artful adaptive mechanism in microbial cells. Microorganisms produce molecules with antibiotic activity and expel them into the environment, presumably enhancing their ability to compete with their neighbors (Berdy, 2005; Hopwood, 2007). However, most of these molecules are toxic to the producer (Mak et al., 2014; Moody, 2014). Mechanisms must exist to ensure that antibiotic production reaches a reasonable level. The proposed model of AdpAch in Figure 7 may provide a fresh mechanistic insight into how S. chattanoogensis controls the production level of natamycin via AdpAch. However, further work will be needed to prove the proposed model and the detailed mechanism of how AdpAch responds to the signal of natamycin. In all, the complicated regulatory network involving AdpAch, ScnRI, and ScnRII helps advance our understanding of the molecular regulation mechanisms of antibiotic biosynthesis and provides an effective strategy to help improve yields in industrial strains.
Materials and Methods
Media, Plasmids, Strains, and Growth Conditions
All plasmids and bacterial strains used in this study are listed in Table 1. General techniques for the manipulation of nucleic acids and bacterial growth were carried out according to the standard protocols as previously described (Kieser et al., 2000). Escherichia coli DH5α was the general cloning host. Vectors used were pSET152, pIJ8660, pTA2. S. chattanoogensis L10 strains were grown at 28°C on YMG agar for sporulation and at 30°C in YEME medium (3 g/l yeast extract, 3 g/l malt extract, 5 g/l tryptone, 10 g/l glucose) for natamycin production.
Electrophoretic Mobility-Shift Assays (EMSAs)
His-AdpAch, histidine-tagged protein was purified from the soluble fractions of E. coli BL21 (DE3) harboring the plasmids pET32a-adpAch, as previously described (Du et al., 2011a). The Bradford reagent (Bio-Rad) was used to determine the protein concentration. For probe preparation, all primers used in this study are listed in Supplementary Table S1. The EMSA DNA probe RI–RII (517 bp) spanning the entire scnRI–scnRII intergenic region was amplified by PCR using primer pair RI–RII-F and RI–RII-R. The PCR product was then cloned into a pTA2-vector (TOYOBO) to generate the plasmid pT-RI–RII. The biotin-labeled probe RI–RII was made with 5′-biotin-labeled M13 universal primer pair using pT-RI–RII as a template by PCR amplification. The probes A (295 bp), B (281 bp), C (294 bp), D (282 bp), E (288 bp), F (284 bp), mA (295 bp), mB (281 bp), mC (294 bp), mD (282 bp), mE (288 bp), and mF (284 bp) were prepared following the above-mentioned method. In the EMSAs assay, 1 ng of the probe was incubated with varying quantities of AdpAch, at 25°C for 30 min in the buffer (20 mM Tris, pH 7.5, 5% glycerol, 0.01% BSA, 50 μg ml-1 sheared sperm DNA). For the competition assay, 100 times of excessive un-labeled probes and non-specific DNA were added to the reaction buffer, respectively. Reactions were displayed on 5% acrylamide gels for separation in 0.5× TBE buffer. EMSA gels were then electro-blotted onto the nylon membrane and UV-fixed by UV crosslinker. Labeled DNA was detected with streptavidin-HRP and BeyoECL plus (Beyotime, China) as described by the manufacturer.
DNase I Footprinting Assay
DNase I footprinting assay was performed as previously described (Mao et al., 2009). Firstly, AdpAch protein was ultra-filtered with YM-10 (Millipore) for 10 kD cut-off and eluted in 20 mM Tris buffer, pH 7.5. Then, FAM-labeled probe was amplified using 5′-(6-FAM)-labeled M13 universal primers from plasmid pT-RI-RII, followed by gel recovery. About 50 ng of fluorescently labeled probe was added to the reaction mixture to a final volume of 50 μl. After binding of the AdpAch protein to 5′-(6-FAM)-labeled probe (30°c, 30 min), 0.01 U of DNase I (Promega) was added for 1 min at 30°C, followed with equal volume of 100 mM EDTA to stop the reactions and extracted by phenol/chloroform. After precipitation with 40 μg of glycogen, 0.75 M ammonium acetate (NH4Ac), and ethanol, the digested DNA mixture was loaded into ABI 3130 DNA sequencer with Liz-500 DNA marker (MCLAB). DNA sequencing ladder was prepared according to Thermo Sequenase Dye Primer Manual Cycle Sequencing Kit (USB).
Alterations of the Consensus Sequence for AdpAch-Binding Sites
The consensus sequence of AdpAch-binding sites A–F was replaced by the sequence of EcoRV restriction sequence sites using overlapping primers (Supplementary Table S1). The PCR product was then cloned into a pTA2-vector (TOYOBO). The resulted plasmids were used as template for PCR to amplify mutated probes using 5′-biotin-labeled M13 universal primers, and the binding ability was measured by EMSAs.
Construction and Analysis of Transcriptional Fusions to the xylE Reporter Gene
For xylE fusions, the xylE gene was PCR amplified with the primers xylE-F and xylE-R. This fragment was digested with NdeI and NotI, and introduced into the likewise-digested pIJ8660 (Sun et al., 1999) to construct pIJ8601. To probe scnRIp and scnRIIp activities with the mutation of AdpAch-binding sites, the wt and mutated promoter regions were amplified by PCR using upstream primers carrying a BamHI site listed in Supplementary Table S1. These promoter fragments were cloned into BamHI-cut pIJ8601 and transferred by conjugation into S. chattanoogensis L10. Plasmid-containing strains were grown on YEME medium for 24 h. Cell pellets from 1 ml culture samples were kept on ice and measured immediately. Assays of catechol 2,3-dioxygenase were performed as previously described (Kieser et al., 2000).
Mutational Analysis of the AdpAch-Binding Sites On Natamycin Biosynthesis
The 1.8 kb DNA fragment containing the sequence of scnRI–scnRII intergenic region was amplified by PCR using primers scnRI-F and scnRII-R. The resulted 1.8 kb sequence was used as template to amplify the DNA fragment for construction of mutated AdpAch-binding sites in vivo using overlapping primers (Supplementary Table S1), then PCR product was purified and ligated into pKC1139. The resulting plasmids containing DNA fragment of mutated sites was conjugated by E. coli ET12567/pUZ8002 into S. chattanoogensis L10. The mutants were selected by replica plating for apramycin-sensitive colonies and they were used as template for PCR with primer pairs RI-RII-F and RI-RII-R. The amplified sequences were digested with EcoRV to confirm the mutants.
Determination of Natamycin Production by HPLC Analysis
Natamycin production was confirmed by HPLC analysis with the Agilent 1100 HPLC system. HC-C18 column (5 μm, 4.6 by 250 mm) was used with UV detector set at 303 nm. Mobile phase and gradient elution process were as described previously (Du et al., 2009).
Author Contributions
PY, Q-TB, and Y-LT performed the experiments. X-MM assisted with the primary data analysis. Y-QL supervised the project and revised the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by the National Key Research and Development Program (2016YFD0400805), the National Natural Science Foundation of China (Nos. 31520103901 and 31470212), and the China Postdoctoral Science Foundation (2016M601953).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully thank Dr. Chris Wood, a native English biologist for his critical reading of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00316/full#supplementary-material
References
Antón, N., Santos-Aberturas, J., Mendes, M. V., Guerra, S. M., Martín, J. F., and Aparicio, J. F. (2007). PimM, a PAS domain positive regulator of pimaricin biosynthesis in Streptomyces natalensis. Microbiology 153, 3174–3183. doi: 10.1099/mic.0.2007/009126-0
Antón, N., Vendes, M. V., Martin, J. F., and Aparicio, J. F. (2004). Identification of PimR as a positive regulator of pimaricin biosynthesis in Streptomyces natalensis. J. Bacteriol. 186, 2567–2575. doi: 10.1128/JB.186.9.2567-2575.2004
Berdy, J. (2005). Bioactive microbial metabolites : a personal view. J. Antibiot. 58, 1–26. doi: 10.1038/ja.2005.1
Du, Y. L., Chen, S. F., Cheng, L. Y., Shen, X. L., Tian, Y., and Li, Y. Q. (2009). Identification of a novel Streptomyces chattanoogensis L10 and enhancing its natamycin production by overexpressing positive regulator ScnRII. J. Microbiol. 47, 506–513. doi: 10.1007/s12275-009-0014-0
Du, Y. L., Li, S. Z., Zhou, Z., Chen, S. F., Fan, W. M., and Li, Y. Q. (2011a). The pleiotropic regulator AdpAch is required for natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis. Microbiology 157, 1300–1311. doi: 10.1099/mic.0.046607-0
Du, Y. L., Shen, X. L., Yu, P., Bai, L. Q., and Li, Y. Q. (2011b). Gamma-butyrolactone regulatory system of Streptomyces chattanoogensis links nutrient utilization, metabolism and developmental programme. Appl. Environ. Microbiol. 77, 8415–8426. doi: 10.1128/AEM.05898-11
Gallegos, M. T., Schleif, R., Bairoch, A., Hofmann, K., and Ramos, J. L. (1997). Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61, 393–410.
Gust, B., Challis, G. L., Fowler, K., Kieser, T., and Chater, K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 100, 1541–1546. doi: 10.1073/pnas.0337542100
Guyet, A., Gominet, M., Benaroudj, N., and Mazodier, P. (2013). Regulation of the clpP1clpP2 operon by the pleiotropic regulator AdpA in Streptomyces lividans. Arch. Microbiol. 195, 831–841. doi: 10.1007/s00203-013-0918-2
Hara, H., Ohnishi, Y., and Horinouchi, S. (2009). DNA microarray analysis of global gene regulation by A-factor in Streptomyces griseus. Microbiology 155, 2197–2210. doi: 10.1099/mic.0.027862-0
Hopwood, D. A. (2007). Streptomyces in Nature and Medicine. The Antibiotic Makers. New York, NY: Oxford University Press Inc.
Horinouchi, S. (2002). A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7, d2045–d2057.
Kato, J. Y., Chi, W. J., Ohnishi, Y., Hong, S. K., and Horinouchi, S. (2005a). Transcriptional control by A-factor of two trypsin genes in streptomyces griseus. J. Bacteriol. 187, 286–295.
Kato, J. Y., Ohnishi, Y., and Horinouchi, S. (2005b). Autorepression of AdpA of the AraC/XylS family, a key transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. J. Mol. Biol. 350, 12–26.
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation.
Komatsu, M., Uchiyama, T., Omura, S., Cane, D. E., and Ikeda, H. (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 2646–2651. doi: 10.1073/pnas.0914833107
Liu, G., Chater, K. F., Chandra, G., Niu, G., and Tan, H. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. doi: 10.1128/MMBR.00054-12
López-García, M. T., Santamarta, I., and Liras, P. (2010). Morphological differentiation and clavulanic acid formation are affected in a Streptomyces clavuligerus adpA-deleted mutant. Microbiology 156, 2354–2365. doi: 10.1099/mic.0.035956-0
Macneil, D. J., and Klapko, L. M. (1987). Transformation of Streptomyces avermitilis by plasmid DNA. J. Ind. Microbiol. 2, 209–218. doi: 10.1007/BF01569542
Mak, S., Xu, Y., and Nodwell, J. R. (2014). The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol. Microbiol. 93, 391–402. doi: 10.1111/mmi.12689
Mao, X. M., Zhou, Z., Cheng, L. Y., Hou, X. P., Guan, W. J., and Li, Y. Q. (2009). Involvement of SigT and RstA in the differentiation of Streptomyces coelicolor. FEBS. Lett. 583, 3145–3150. doi: 10.1016/j.febslet.2009.09.025
McKenzie, N. L., and Nodwell, J. R. (2007). Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J. Bacteriol. 189, 5284–5292. doi: 10.1128/JB.00305-07
Moody, S. C. (2014). Microbial co-culture: harnessing intermicrobial signaling for the production of novel antimicrobials. Future Microbiol. 9, 575–578. doi: 10.2217/fmb.14.25
Nguyen, K. T., Tenor, J., Stettler, H., Nguyen, L. T., Nguyen, L. D., and Thompson, C. J. (2003). Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 185, 7291–7296. doi: 10.1128/JB.185.24.7291-7296.2003
Ohnishi, Y., Kameyama, S., Onaka, H., and Horinouchi, S. (1999). The A-factor regulatory cascade leading to streptomycin production in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34, 102–111. doi: 10.1046/j.1365-2958.1999.01579.x
Ohnishi, Y., Yamazaki, H., Kato, J. Y., Tomono, A., and Horinouchi, S. (2005). AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69, 431–439. doi: 10.1271/bbb.69.431
Onaka, H., and Horinouchi, S. (1997). DNA-binding activity of the A-factor receptor protein and its recognition DNA sequences. Mol. Microbiol. 24, 991–1000. doi: 10.1046/j.1365-2958.1997.4081772.x
Pan, Y., Liu, G., Yang, H., Tian, Y., and Tan, H. (2009). The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol. Microbiol. 72, 710–723. doi: 10.1111/j.1365-2958.2009.06681.x
Retzlaff, L., and Distler, J. (1995). The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol. Microbiol. 18, 151–162. doi: 10.1111/j.1365-2958.1995.mmi_18010151.x
Santos-Aberturas, J., Vicente, C. M., Payero, T. D., Martín-Sánchez, L., Cañibano, C., Martín, J. F., et al. (2012). Hierarchical control on polyene macrolide biosynthesis: PimR modulates pimaricin production via the PAS-LuxR transcriptional activator PimM. PLoS One 7:e38536. doi: 10.1371/journal.pone.0038536
Santos-Beneit, F., Rodriguez-Garcia, A., Sola-Landa, A., and Martin, J. F. (2009). Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol. Microbiol. 72, 53–68. doi: 10.1111/j.1365-2958.2009.06624.x
Sun, J., Kelemen, G. H., Fernandez-Abalos, J. M., and Bibb, M. J. (1999). Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145, 2221–2227. doi: 10.1099/00221287-145-9-2221
Takano, E., Chakraburtty, R., Nihira, T., Yamada, Y., and Bibb, M. J. (2001). A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41, 1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x
Tan, G. Y., Peng, Y., Lu, C., Bai, L., and Zhong, J. J. (2015). Engineering validamycin production by tandem deletion of γ-butyrolactone receptor genes in Streptomyces hygroscopicus 5008. Metab. Eng. 28, 74–81. doi: 10.1016/j.ymben.2014.12.003
Tomono, A., Tsai, Y., Yamazaki, H., Ohnishi, Y., and Horinouchi, S. (2005). Transcriptional control by A-factor of strR, the pathway-specific transcriptional activator for streptomycin biosynthesis in Streptomyces griseus. J. Bacteriol. 187, 5595–5604. doi: 10.1128/JB.187.16.5595-5604.2005
van Wezel, G. P., and McDowall, K. J. (2011). The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat. Prod. Rep. 28, 1311–1333. doi: 10.1039/c1np00003a
Yamazaki, H., Ohnishi, Y., and Horinouchi, S. (2003a). Transcriptional switch on of ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus. J. Bacteriol. 185, 1273–1283.
Yamazaki, H., Tomono, A., Ohnishi, Y., and Horinouchi, S. (2004). DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53, 555–572. doi: 10.1111/j.1365-2958.2004.04153.x
Keywords: bidirectional regulation, AdpA, natamycin biosynthesis, Streptomyces chattanoogensis L10, pathway-specific gene
Citation: Yu P, Bu Q-T, Tang Y-L, Mao X-M and Li Y-Q (2018) Bidirectional Regulation of AdpAch in Controlling the Expression of scnRI and scnRII in the Natamycin Biosynthesis of Streptomyces chattanoogensis L10. Front. Microbiol. 9:316. doi: 10.3389/fmicb.2018.00316
Received: 18 December 2017; Accepted: 09 February 2018;
Published: 02 March 2018.
Edited by:
Eung-Soo Kim, Inha University, South KoreaReviewed by:
Yinhua Lu, Shanghai Institutes for Biological Sciences (CAS), ChinaYasuo Ohnishi, The University of Tokyo, Japan
Copyright © 2018 Yu, Bu, Tang, Mao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Quan Li, bHlxQHpqdS5lZHUuY24=
 Pin Yu
Pin Yu Qing-Ting Bu
Qing-Ting Bu Yi-Li Tang
Yi-Li Tang Xu-Ming Mao
Xu-Ming Mao Yong-Quan Li
Yong-Quan Li