- 1State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 2School of Life Sciences, Northeast Agricultural University, Harbin, China
Milbemycins, a group of 16-membered macrolide antibiotics, are used widely as insecticides and anthelmintics. Previously, a limited understanding of the transcriptional regulation of milbemycin biosynthesis has hampered efforts to enhance antibiotic production by engineering of regulatory genes. Here, a novel ArpA/AfsA-type system, SbbR/SbbA (SBI_08928/SBI_08929), has been identified to be involved in regulating milbemycin biosynthesis in the industrial strain S. bingchenggensis BC04. Inactivation of sbbR in BC04 resulted in markedly decreased production of milbemycin, while deletion of sbbA enhanced milbemycin production. Electrophoresis mobility shift assays (EMSAs) and DNase I footprinting studies showed that SbbR has a specific DNA-binding activity for the promoters of milR (the cluster-situated activator gene for milbemycin production) and the bidirectionally organized genes sbbR and sbbA. Transcriptional analysis suggested that SbbR directly activates the transcription of milR, while represses its own transcription and that of sbbA. Moreover, 11 novel targets of SbbR were additionally found, including seven regulatory genes located in secondary metabolite biosynthetic gene clusters (e.g., sbi_08420, sbi_08432, sbi_09158, sbi_00827, sbi_01376, sbi_09325, and sig24sbh) and four well-known global regulatory genes (e.g., glnRsbh, wblAsbh, atrAsbh, and mtrA/Bsbh). These data suggest that SbbR is not only a direct activator of milbemycin production, but also a pleiotropic regulator that controls the expression of other cluster-situated regulatory genes and global regulatory genes. Overall, this study reveals the upper-layer regulatory system that controls milbemycin biosynthesis, which will not only expand our understanding of the complex regulation in milbemycin biosynthesis, but also provide a basis for an approach to improve milbemycin production via genetic manipulation of SbbR/SbbA system.
Introduction
Streptomyces species are important sources for commercially available antibiotics (Kitani et al., 2011; Niu et al., 2016). The production of these antibiotics is typically specified by large gene clusters that usually include cluster-situated regulators (CSRs). In many cases, transcription of antibiotic biosynthetic gene clusters is under the direct regulation of CSRs, which are in turn controlled by a plethora of higher-level regulatory systems that respond to various environmental and physiological signals (Bibb, 2005; van Wezel and McDowall, 2011; Liu et al., 2013).
Milbemycins, a group of 16-membered macrolide antibiotics, were isolated from the culture broths of several Streptomyces species, including Streptomyces hygroscopicus subsp. aureolacrimosus, Streptomyces thermoarchaensis, and Streptomyces bingchenggensis (Rugg et al., 2005; Xiang et al., 2007). Similar to avermectins, milbemycins possess a lactone ring and are potent anthelmintic and insecticidal agents that are widely used in veterinary, agricultural, and medical fields (Shoop et al., 1995; Danaher et al., 2012; Jacobs and Scholtz, 2015). In particular, milbemycins have been used to control avermectin and organophosphorus pesticides-resistant mites, Liriomyza, aphidoidea, and aleyrodidae on more than two dozen plant species. Milbemectin, a mixture of milbemycin A3 and A4, shows high acaricidal activity and has been widely used against agricultural mites since 1990 (Pluschkell et al., 1999). The semisynthetic derivative of milbemycin A3/A4, milbemycin oxime, has been used to treat pests against filarid and nematode (Shoop et al., 1995; Danaher et al., 2012). Moreover, other milbemycin A3/A4 derivatives, such as lepimectin and latidectin, have also been commercialized and widely used in agricultural field (Ikari, 2013; Kim et al., 2016).
The species S. bingchenggensis, isolated in 2007 by our group, has been used for the industrial production of milbemycin (Gao et al., 2007; Wang et al., 2010). Given the importance of milbemycin A3/A4 and their derivatives, many efforts have been made to increase the titer of milbemycin. In S. bingchenggensis, a variety of mutagenic methods, including treatment with N-methyl-N′-nitro-N-nitrosoguanidine (NTG) (Wang et al., 2009), ultraviolet mutagenesis (Wang et al., 2009), as well as room temperature plasma (ARTP) mutations have been adopted to increase the titer of milbemycin A3/A4 (Wang et al., 2014). Moreover, in silico-based metabolic engineering strategies such as deletion of milD (encoding C5-O-methyltransferase) to remove undesired C5-O-methylmilbemycins and deletion of the nanchangmycin gene cluster, which putatively competes for the same precursors, such as malonyl-CoA and methylmalonyl-CoA, have been performed to enhance the production of milbemycin A3/A4 in S. bingchenggensis (Zhang J. et al., 2013). However, a limited understanding of the transcriptional regulation of milbemycin production has impeded the efforts to further improve antibiotic titers via manipulation of regulatory genes. Identification and characterization of novel transcriptional regulators involved in milbemycin biosynthesis are essential for the elucidation of the underlying regulatory mechanism, which in turn will benefit the genetic engineering of S. bingchenggensis to generate new hyper-producer strains.
In 2010, the milbemycin biosynthesis gene cluster (mil) was identified from the genome of S. bingchenggensis (Wang et al., 2010). The mil cluster (sbi_00726–sbi_00790) comprised of 10 genes, including one regulatory gene (milR) and nine structural genes located in six operons (i.e., milA2-C, milA4-E, milR-A3, milA1-D, milF, and orf1), and a large (62 kb) insertion fragment between milR and milA1 (Wang et al., 2010; Zhang et al., 2016). milR, the only CSR gene in the mil cluster, encodes a large ATP-binding regulator of the LuxR (LAL) family. Previously, MilR was found to be essential for milbemycin production, and activates the expression of milA4-E and milF directly (Zhang et al., 2016). However, the higher-level transcriptional regulators controlling milR expression and, thus, the detailed regulatory networks of milbemycin production, remain elusive.
In Streptomyces, antibiotic production is often regulated by low-molecular-weight compounds, such as γ-butyrolactone signals (GBLs), avenolides, and furans (Bibb, 2005; O’Rourke et al., 2009; Kitani et al., 2011; Niu et al., 2016). GBLs, the largest group of these compounds, can elicit antibiotic biosynthesis at nanomolar concentrations by binding to cognate receptor proteins (Kitani et al., 2011). They share a characteristic 2,3-disubstituted-γ-butyrolactone core but differ in the C2 side chains that determine signaling specificity (Takano, 2006). In the process of cell growth, a small increase in expression of the GBL synthase, leads to gradually accumulated GBLs, which in turn binds to the receptor protein and releases its interaction with target genes (Mehra et al., 2008). To date, the well-studied GBL systems include A-factor-mediated ArpA/AfsA system in Streptomyces griseus (Khokhlov et al., 1967), SCBs-mediated ScbR/ScbA system in Streptomyces coelicolor (Cuthbertson and Nodwell, 2013), VBs-mediated BarA/BarX system in Streptomyces virginiae (Kinoshita et al., 1997), IM-2-mediated FarA/FarX system in Streptomyces lavendulae (Waki et al., 1997), and SVB1-mediated JadR3/JadW1 system in Streptomyces venezuelae (Zou et al., 2014). Given the large number of more than 900 Streptomyces species described and the very few GBL-mediated ArpA/AfsA-like systems known, further research on the ArpA/AfsA-like regulatory cascades is required (Niu et al., 2016).
In this report, we describe a putative ArpA/AfsA-like system, SBI_08928/SBI_08929, which is designated as SbbR/SbbA in S. bingchenggensis. SbbR is a homolog of GBL receptors, while SbbA resembles proteins involved in the production of GBL signals. We demonstrate that the SbbR/SbbA system plays an important role in the production of milbemycin. SbbR controls milbemycin production by directly activating the transcription of milR, while SbbA appears to have a repressive effect on milbemycin production; both SbbR and SbbA have positive effects on cell growth. Interestingly, SbbR is also a pleiotropic regulator that controls the transcription of other CSR genes and global regulatory genes. This work deepens our understanding of an upper-level of regulation in milbemycin biosynthesis, and provides an effective approach to improve milbemycin production via genetic manipulation of SbbR/SbbA system in S. bingchenggensis.
Materials and Methods
Bacterial Strains, Plasmids, Primers, and Growth Conditions
Bacterial strains and plasmids used in this study are summarized in Table 1; primers are listed in Supplementary Table S1. S. bingchenggensis BC04, the producer of milbemycin A3/A4, is derived from S. bingchenggensis CGMCC 1734 which is deposited at China General Microbiological Culture Collection (CGMCC). S. bingchenggensis BC04 and its derivatives were grown at 28°C on SKYM (0.4% sucrose, 0.1% skimmed milk powder, 0.2% yeast extract, and 0.5% malt extract pH 7.2) agar for spore collection. Besides, SKYM medium together with minimal medium (MM) (0.1% L-asparagine, 0.1% K2HPO4, 0.04% MgSO4⋅7H2O, 0.002% FeSO4⋅7H2O, 2% glucose, and 2% agar) was used to analyze morphological differentiation. The media and methods used for milbemycin production were the same as previously reported (Zhang et al., 2016).
Construction of the Phylogenetic Tree
The amino acid sequences of SbbR with its 36 homologs were individually aligned with multiple sequences obtained from the GenBank/EMBL/DDBJ databases using CLUSTAL X 1.83 software. The phylogenetic tree was reconstructed with the neighbor-joining algorithms (Saitou and Nei, 1987) using MEGA software version 6.06 (Tamura et al., 2013). Phylogenetic distances were calculated with the Jones-Taylor-Thornton (JTT) model (Wang et al., 2008). The stability of the clades in the trees was appraised using a bootstrap procedure with 1000 repeats (Felsenstein, 1985). All positions containing gaps and missing data were eliminated from the dataset (complete deletion option).
Construction of sbbR and sbbA Disruption Mutants and Their Complementation
The sbbR and sbbA gene disruption were performed by the method of homologous recombination. To construct the sbbR disruption mutant, two fragments (sbbR-L: 1.9 kb, sbbR-R: 2.0 kb) flanking sbbR were amplified with the S. bingchenggensis BC04 genomic DNA as template by polymerase chain reaction (PCR) using the primer pairs sbbRLF/R and sbbRRF/R (Supplementary Table S1). The fragment sbbR-L was cut with HindIII and XbaI, and the fragment sbbR-R was cut with KpnI and EcoRI. The plasmid pUC119::neo was used to obtain the kanamycin-resistance gene (neo) after HindIII and KpnI digestion. The three resultant DNA segments were ligated into the sites of EcoRI and HindIII in pKC1139 to give pKC1139::sbbR::neo, in which a 519-bp fragment of sbbR was replaced by neo. pKC1139::sbbR::neo was subsequently introduced into S. bingchenggensis BC04 via conjugation (Kieser et al., 2000). The exconjugants were selected on MS agar containing apramycin (Ahn et al., 2012). After incubation on SKYM solid medium at 37°C for 9 days, double cross-over recombinant strains were obtained by selection for kanamycin-resistant (KanR) and apramycin-sensitive (AprS) clones. The obtained sbbR disrupted strain, designated as ΔsbbR, was confirmed by PCR using primer VsbbR-F and VsbbR-R followed by DNA sequencing.
Complementation was performed with pSET152 carrying sbbR expressed from its own promoter. The primers CsbbR-F and CsbbR-R were used to amplify the 1301-bp DNA fragment containing the sbbR ORF and its upstream region from genomic DNA of BC04 (Supplementary Table S1). The obtained DNA fragment was inserted into EcoRI and XbaI sites of pSET152 to create sbbR complementation vector pSET152::sbbR, which was introduced into ΔsbbR by intergenic conjugation to obtain the complemented strain ΔsbbR/sbbR (Kuhstoss et al., 1991).
To construct a sbbA deletion mutant, two fragments (sbbA-L: 1.5 kb and sbbA-R: 2.0 kb) flanking sbbA were amplified by primers sbbALF/R and sbbARF/R using the genomic DNA of BC04 as template, respectively. The fragment sbbA-L was digested with HindIII/XbaI, while the sbbA-R fragment was digested with KpnI/EcoRI. These two fragments together with neo digested with HindIII/KpnI were inserted into the site of EcoRI and HindIII in pKC1139 to generate pKC1139::sbbA::neo, in which a 385-bp fragment of sbbA was replaced by neo. The sbbA mutants were obtained with the same method as described previously, and were confirmed by PCR with VsbbA-F and VsbbA-R as primers, followed by DNA sequencing. The sbbA disruption mutant was designated as ΔsbbA.
For complementation of ΔsbbA, a 1643-bp fragment containing the intact sbbA ORF and its putative promoter region was amplified by PCR using primers CsbbA-F and CsbbA-R. The XbaI-EcoRV digested DNA fragment was ligated into pSET152 to give the sbbA complementation vector pSET152::sbbA, which was introduced into ΔsbbR by conjugation to obtain the complemented strain ΔsbbA/sbbA.
The plasmid pSET152 was introduced into BC04 to generate control strain BC04/pSET152.
HPLC Analysis of Milbemycin A3/A4
Milbemycin A3/A4 was analyzed by the method previously reported (Zhang et al., 2016). HPLC was performed by Agilent 1100 HPLC (Zorbax, SB-C18 column, 4.6 mm × 250 mm, 5 μm) at a flow rate of 1.0 ml/min with gradient from 0 to 100% of solvent B in 15 min (Solvent A: CH3CN:H2O:CH3OH = 7:1:2, v/v/v; Solvent B: CH3OH) and detected at 242 nm.
Overexpression and Purification of SbbR-His6 Protein
To prepare the SbbR-His6 protein, primers proSbbR-F and proSbbR-R were used to amplify the sbbR gene (the stop codon was excluded) with the BC04 genomic DNA. The amplified PCR products were digested with NdeI/XhoI and cloned between the NdeI/XhoI sites of pET23b (+) to generate the overexpression plasmid pET23b::sbbR, which was verified by nucleotide sequencing and then introduced into E. coli BL21 (DE3) for protein expression. IPTG (final concentration 0.1 mM) was used to induce SbbR-His6 expression. The nickel-nitrilotriacetic acid (Ni-NTA) agarose chromatography was used to purify the SbbR-His6 based on the protocol of the manufacturer (Novagen). The Amicon Ultra 0.5-ml centrifugal filters (Millipore membrane, 3 kDa cut-off size) (Millipore) were used to concentrate the protein based on the protocol provided by the manufacturer. Concentration of the total recovered protein was determined by BCA protein assay Kit (Novagen) with a standard of Bovine Serum Albumin (BSA). The purified protein was stored in 5% glycerol at -70°C.
Electrophoretic Mobility Shift Assays (EMSAs)
Electrophoresis mobility shift assays were carried out as described previously (Wei et al., 2014; Li et al., 2016). In brief, two fragments [PmilR (413-bp) and PR-A (464-bp)] corresponding to the upstream region of milR and the intergenic region of sbbR and sbbA were generated by PCR from the genomic DNA of S. bingchenggensis BC04 with primer pairs pmilR-F/R and psbbR-A-F/R, respectively. Primers phrdB-F and phrdB-R were used to amplify the control probe PhrdB (443-bp). The other 79 primer pairs used to amplify promoter region probes are listed in Supplementary Table S1. The DNA probe (10 ng) was incubated with various concentrations of SbbR-His6 in 20 μl reaction mixture containing 20 mM Tris base, 2 mM dithiothreitol (DTT), 5 mM MgCl2, 0.5 μg calf BSA, and 5% (v/v) glycerol, 500 ng poly[d(I-C)] was added when necessary. After incubation at 25°C for 25 min, DNA-protein complexes and free DNA were separated by electrophoresis on non-denaturing 4% (wt/vol) polyacrylamide gel at 4°C with 0.5× TBE as the running buffer. After electrophoresis, DNA in the gel was stained with SYBR Gold nucleic acid gel stain for 30 min in 1.0× TBE and photographed under ultraviolet transillumination using Quantity One.
DNase I Footprinting
In order to determine the binding sites of SbbR on PmilR and PR-A, DNase I footprinting assays were carried out as described previously (Li et al., 2016). DNA probes were prepared by PCR using 5′ FAM fluorescence-labeled primers listed in Supplementary Table S1. After being purified from agarose gel, labeled DNA probe (200 ng) and protein with different concentrations of SbbR-His6 were added to a final reaction volume of 50 μl, and incubated for 25 min at 25°C. DNase I (Promega) digestions were carried out for 75 s at 25°C and stopped with 20 mM EGTA. After ethanol extraction and precipitation, the purified samples were loaded in an Applied Biosystems 3730 DNA genetic analyzer together with the internal-lane size standard ROX-500 (Applied Biosystems). The dye primer-based sequencing kit (Thermo) was used to further precisely determine the sequences after aligning the capillary electrophoresis results of reactions. The results were then processed with GeneMarker v2.2.0 software.
Site-Directed Mutagenesis of SbbR Binding Sequences
To assess the specificity of SbbR on its binding sequence, site-directed mutagenesis was performed. Two fragments PmilR and PR-A were ligated into the EcoRV-digested plasmid pBluescript KS (+) to generate plasmids pBmilR and pBsbbA, respectively. A fragment containing mutated PmilR was amplified by primer pair mupmilR-F/R using the plasmid pBmilR as template. Similarly, the fragment containing mutated PR-A was amplified by primer pair mupsbbA-F/R using pBsbbA as template. These two fragments were self-ligated, generating mutant plasmids mupBmilR and mupBsbbA, respectively. The mutated sequences at the binding sites were further verified by DNA sequencing. Then mupBmilR and mupBsbbA were digested with HindIII/BamHI to generate mutant probes PmilR∗ and PR-A∗, respectively. The binding activity of SbbR-His6 to these probes was subsequently determined by EMSAs.
RNA Isolation and Quantitative Real-Time PCR
Transcription levels of genes were compared between S. bingchenggensis BC04 and ΔsbbR by quantitative real-time PCR (qRT-PCR) analysis. For this purpose, total RNAs were isolated from cultures of S. bingchenggensis BC04 grown at 28°C at various time points (1, 2, 3, 5, and 9 days). RNA extraction was performed as described previously (Zhang Y. et al., 2013). The RNA sample was treated with RQ1 RNase-free DNase I (Promega) to remove genomic DNA. UV spectroscopy and agarose gel electrophoresis were used to examine the RNA quality and quantity. Synthesis of cDNA and subsequent qRT-PCR were essentially the same as described previously (Zhang Y. et al., 2013).
Determination of Transcriptional Start Points of milR, sbbR, and sbbA
The transcriptional start points (tsp) of milR, sbbR and sbbA were determined by using a 5′ RLM-RACE Kit (Thermo) according to the manufacturer’s instructions. Briefly, total RNAs were prepared from a 3 days culture of S. bingchenggensis BC04 (for milR and sbbR) and a 2 days culture of ΔsbbR (for sbbA). First, total RNAs (10 μg) were successively treated with Calf Intestinal Alkaline Phosphatase (CIAP) and tobacco acid pyrophosphatase (TAP) and ligated with the 5′ RLM-RACE adapter using T4 RNA ligase. Reverse transcription was preceded by using M-MLV Reverse Transcriptase with random decamers supplied. A first round of PCR was performed with the cDNAs derived from RT Reactions using 5′ RLM-RACE outer primer and another gene specific-primer listed in Supplementary Table S1. To obtain a single specific band, a second round of PCR was performed with the original PCR product (25-fold dilution) as template, by using 5′ RLM-RACE inner primer and another nested primer listed in Supplementary Table S1. The final PCR product was inserted into pBluescript (KS+) and sent for sequencing.
GFP Reporter Assay in E. coli
For construction of the reporter plasmid, the original plasmid pSET152 was cut with BamHI and XbaI, and the promoters PmilR and PsbbA were amplified with two primer pairs pmilRGFP-F/R and psbbAGFP-F/R, respectively. The green fluorescence gene (gfp) was amplified with primers GFP-F and GFP-R from pTAC (Yin et al., 2015). Prior to PCR amplification, primers pmilRGFP-R, psbbAGFP-R, and GFP-F were phosphorylated with T4 polynucleotide kinase to facilitate subsequent ligation reactions. PmilR and PsbbA were digested with BamHI and the gfp coding region was digested with XbaI. The PmilR and PsbbA promoters and the gfp coding region were ligated into pSET152 which was cut with BamHI/XbaI in a three-piece ligation reaction to generate pSET152::PmilRgfp and pSET152::PsbbAgfp, respectively. To evaluate the regulatory effects of SbbR on PmilR and PsbbA promoters, the strong constitutive promoter SF14 and the coding region of sbbR were amplified from plasmid pGusTSF14 and genomic DNA of BC04 with primers pSF14-F/pSF14-R and psbbRGFP-F/psbbRGFP-R, respectively (Yin et al., 2015). The plasmids pSET152::PmilRgfp and pSET152::PsbbAgfp were digested with NheI and assembled with the SF14 promoter and the sbbR coding region to obtain the corresponding reporter plasmids pSET152::PmilRgfp::SF14sbbR and pSET152::PsbbAgfp::SF14sbbR, respectively, which include, both SF14 promoter controlled sbbR and a gfp controlled by PmilR or PsbbA, respectively. sbbR and gfp were placed in opposite orientations. These four plasmids together with the empty vector pSET152, which was used as a control, were each transferred to DH5α to detect green fluorescence (excitation at 485 nm; emission at 535 nm, Synergy H4 Multi-Mode Reader). All fluorescence values were normalized to growth rates (OD600). The average and standard deviation of three experimental replicates are presented by each value and error bar, respectively.
Preparation of GBL Crude Extracts
The procedure used for preparation of GBL crude extracts was as described previously with slight modification (Kitani et al., 2011; Zou et al., 2014). A total of 100 ml culture broth of BC04 or ΔsbbA grown in seed medium for 2 days was extracted with an equal volume of ethyl acetate. The organic phase was dried in a vacuum evaporator and re-dissolved in 1 ml DMSO.
Determination of Cell Dry Weight
Two-milliliter cell cultures were collected by vacuum filtration and dried at 55°C to a constant weight.
Statistical Analysis
All experiments mentioned above were carried out at least three times, independently. The mean values were presented as ± SD (standard deviation). The Student’s t-test was used to analyze the data. P < 0.05 is used as a standard criterion of statistical significance.
Results
Identification of the ArpA/AfsA System SbbR/SbbA in S. bingchenggensis
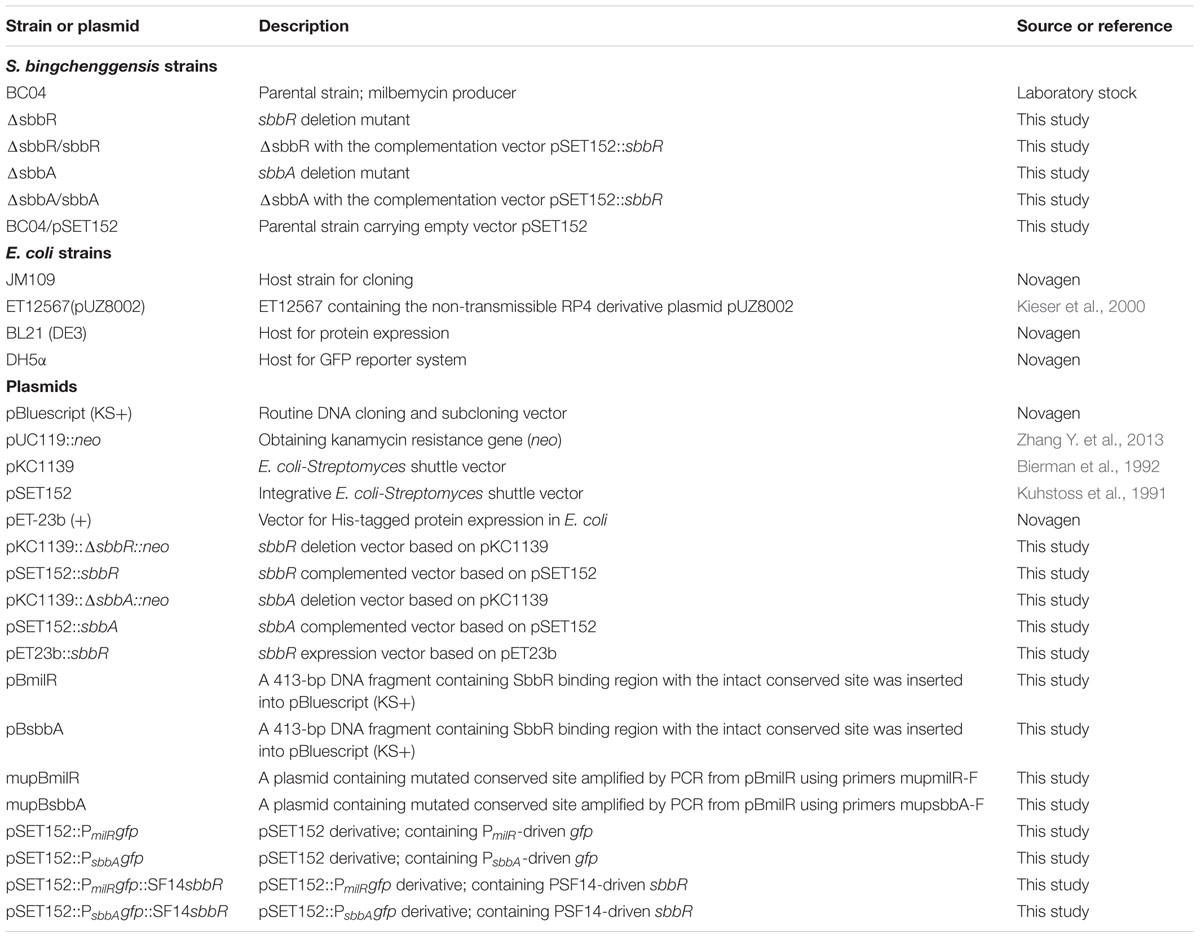
Genome analysis of S. bingchenggensis revealed an arpA/afsA-type system, sbbR/sbbA (sbi_08928/sbi_08929), which is ∼104 kb far downstream of the mil cluster (sbi_00726–sbi_00790) (Figure 1A). An accurate prediction of open reading frame (ORF) is important for the study of gene function. After careful examination, we did not find a putative ribosome binding site (RBS) in the upstream region of the annotated sbbR or sbbA translation start codon; therefore, we re-annotated the coding sequences of sbbR and sbbA. The true sbbR coding region may span from nucleotide (nt) 10567682 to 10567062 on the reverse strand of the S. bingchenggensis chromosome consisting of 621 nucleotides, and sbbA’s real ORF may span from nt 10567842 to 10568729 on the forward strand of the chromosome consisting of 888 nucleotides.
FIGURE 1. Bioinformatic analysis of SbbR in S. bingchenggensis BC04. (A) Schematic representation of the relative positions of sbbR, sbbA, and the mil cluster. Each arrow indicates a separate open reading frame (ORF) and orientation of transcription. (B) Phylogenetic analyses of SbbR and its homologs. SbbR of S. bingchenggensis BC04 is highlighted in bold. The CLUSTALW program was used for performing sequence alignment. The phylogenetic tree was constructed by applying the neighbor-joining algorithm. Bootstrap values >50% (based on 1000 replications) are shown at branch points. Bar, 0.02 substitutions per amino acid position for the phylogenetic tree of SbbR.
sbbR encodes a TetR (Tetracycline Repressor) family regulator that shows similarity to GBL receptors such as Gbr from Streptomyces tacrolimicus (53% identity) (Salehi-Najafabadi et al., 2011), AvaR1 from Streptomyces avermitilis (31% identity) (Kitani et al., 2011), and JadR3 from S. venezuelae (30% identity) (Zou et al., 2014). sbbA, which is located upstream of the oppositely oriented sbbR gene, encodes a putative GBL biosynthesis protein that shows 32% identity with FarX from S. lavendulae (Waki et al., 1997), 31% identity with SrrX from Streptomyces rochei (Arakawa et al., 2007), 27% identity with AfsA from S. griseus (Khokhlov et al., 1967), and 25% identity with JadW1 from S. venezuelae (Zou et al., 2014). Previously, Nishida et al. (2007) constructed a phylogenetic tree for the GBL receptor homologs. These homologs were classified into two groups; one consists of canonical or “true” GBL receptors that bind GBLs as ligands, whereas the others are “pseudo” GBL receptors that do not bind GBLs. To get an indication of SbbR function, we reconstructed the phylogenetic tree for SbbR homologs. Phylogenetic analysis showed that SbbR belongs to the group of “true” GBL receptors represented by BarA (Kinoshita et al., 1997), ArpA (Khokhlov et al., 1967), and FarA (Waki et al., 1997) (Figure 1B). Furthermore, true GBL receptors and “pseudo” GBL receptors can be distinguished by their pI values: the true receptors appear to have acidic or neutral pI values, while the “pseudo” ones show basic pI values (Kitani et al., 2008). As SbbR has a pI value of 5.81, we hypothesize it belongs to the group of true GBL receptors, but this requires further experimental confirmation. In screening of the whole genome of S. bingchenggensis, no other homologs of SbbR or SbbA were discovered. This indicates that there is likely only one ArpA/AfsA-type system present in S. bingchenggensis.
Effects of sbbR and sbbA Disruption on Phenotype and Milbemycin Production
To determine the roles of SbbR and SbbA in morphological differentiation and milbemycin production, sbbR and sbbA were disrupted individually. In ΔsbbR, a 519-bp fragment internal to sbbR ORF, containing the complete coding region of the HTH-type DNA-binding motif (a.a. 12–58) essential for TetR function, was replaced by a kanamycin resistance gene neo via homologous recombination (Supplementary Figure S1A). Similarly, the sbbA deletion mutant ΔsbbA was constructed with a 385-bp fragment covering much of the A-factor biosynthesis hotdog domain (a.a. 21–159) of sbbA replaced by neo (Supplementary Figure S1B). The resulting mutants ΔsbbR and ΔsbbA were confirmed by PCR (Supplementary Figures S1C,D) and DNA sequencing (data not shown).
Compared with the parental strain S. bingchenggensis BC04 and BC04/pSET152 controls, ΔsbbR formed spores earlier and the spores were darker pigmented on SKYM (a rich medium for sporulation) and MM; while on MM, ΔsbbR presented an obvious weak growth of aerial mycelium (Figure 2A). ΔsbbA grew a little sparse but sporulated normally on these two media (Figure 2A). In complementation experiments, pSET152::sbbR was introduced into ΔsbbR to generate ΔsbbR/sbbR and pSET152::sbbA was transferred to ΔsbbA to generate ΔsbbA/sbbA. As expected, the complementation strains gave full complementation (Figure 2A). These results suggested that sbbR and sbbA are involved in the regulation of morphological development.
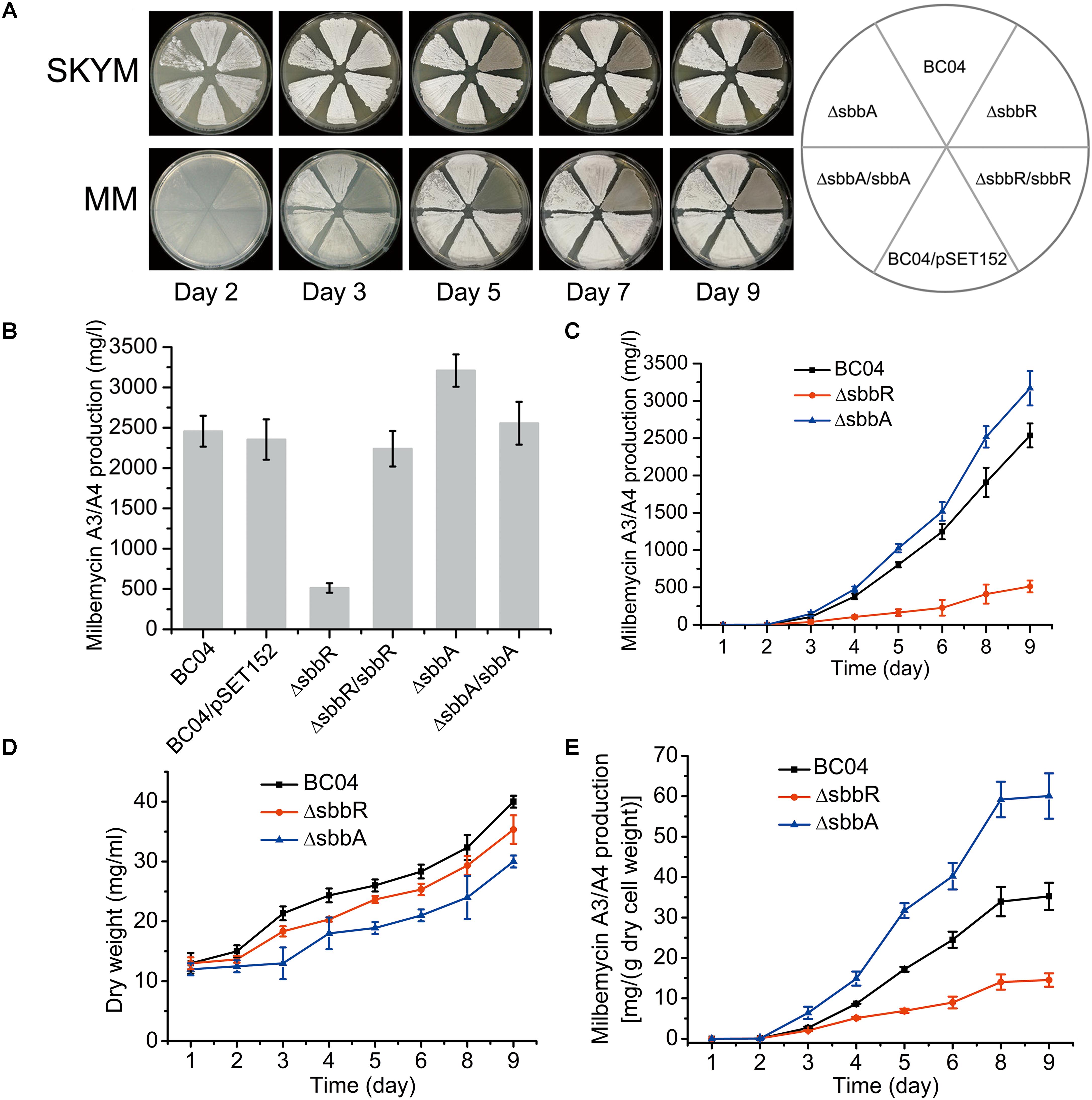
FIGURE 2. Effects of sbbR and sbbA on sporulation, milbemycin production, and cell growth. (A) Effects of sbbR and sbbA deletion on morphological differentiation. (B) Comparative milbemycin A3/A4 production in BC04, BC04/pSET152, ΔsbbR, ΔsbbR/sbbR, ΔsbbA, and ΔsbbA/sbbA cultured in fermentation medium. (C) Quantitative milbemycin A3/A4 production of BC04, ΔsbbR, and ΔsbbA cultured in fermentation medium. (D) Growth curves of BC04, ΔsbbR, and ΔsbbA strains cultured in fermentation medium. Biomass is expressed as dry cell weight. (E) Milbemycin A3/A4 yield per g dry cell weight of BC04, ΔsbbR, and ΔsbbA strains. Data are presented as the averages of the results of three independent experiments. Error bars show standard deviations.
In comparison with the parental strain S. bingchenggensis BC04, production of milbemycin A3/A4 in ΔsbbR was significantly reduced (Figure 2B and Supplementary Figure S2); whereas the titer of milbemycin A3/A4 in ΔsbbA was increased (Figure 2B and Supplementary Figure S2). To verify that the opposite phenotypic changes were due to the separate disruption of sbbR and sbbA, complementation strains ΔsbbR/sbbR and ΔsbbA/sbbA were also tested for milbemycin production. The results showed that ΔsbbR/sbbR and ΔsbbA/sbbA produced similar levels of milbemycin A3/A4 as those of BC04 and BC04/pSET152 controls, indicating that SbbR is an activator of milbemycin production and SbbA has a repressive effect on milbemycin production (Figure 2B and Supplementary Figure S2). Next, milbemycin production (Figure 2C), the cell growth (Figure 2D), and milbemycin yield per gram dry cell weight (Figure 2E) of ΔsbbR, ΔsbbA and the parental strain BC04 were quantitatively compared in fermentation medium. Compared with the parental strain BC04, deletion of sbbR decreased milbemycin production dramatically by 80% (from 2537 to 514 mg/l) and reduced cell growth; the disruption of sbbA increased milbemycin production by 25% (from 2537 to 3169 mg/l) but reduced biomass accumulation throughout the entire time-course (Figures 2C,D). These data revealed that sbbR plays a positive regulatory role in milbemycin production and cell growth; meanwhile, sbbA plays a negative role in milbemycin production and is also important for cell growth.
SbbR Binds to the Upstream Regions of milR, sbbR, and sbbA
To assess whether the SbbR/SbbA system directly regulates milbemycin biosynthesis, the ArpA-like protein SbbR was expressed as a C-terminally His6-tagged protein and tested in EMSAs with all potential promoter regions in the mil cluster, including the putative promoters of milA2, milA4, milE, orf1, milF, milR, milA3, and milA1 (Figure 3A and Supplementary Table S2). The intergenic region of sbbR-sbbA (PR-A) was also included for EMSA, because sbbR and sbbA are adjacent on the chromosome, transcribed divergently, and regulation of a neighboring oppositely arranged gene is a common feature of TetR-family regulators (Ahn et al., 2012; Yuan et al., 2016). Results from EMSAs showed that SbbR only formed complexes with PmilR and PR-A in a concentration-dependent manner (Figures 3C,D), while 500-fold excess of non-specific poly (dI-dC) could not dissociate SbbR from PmilR (DNA region upstream of milR) or PR-A (Supplementary Figure S3). Negative control PhrdB also showed no band-shift when SbbR concentration reaches 0.4 μM (Figure 3E). These results suggested that SbbR binds specifically to PmilR and PR-A.
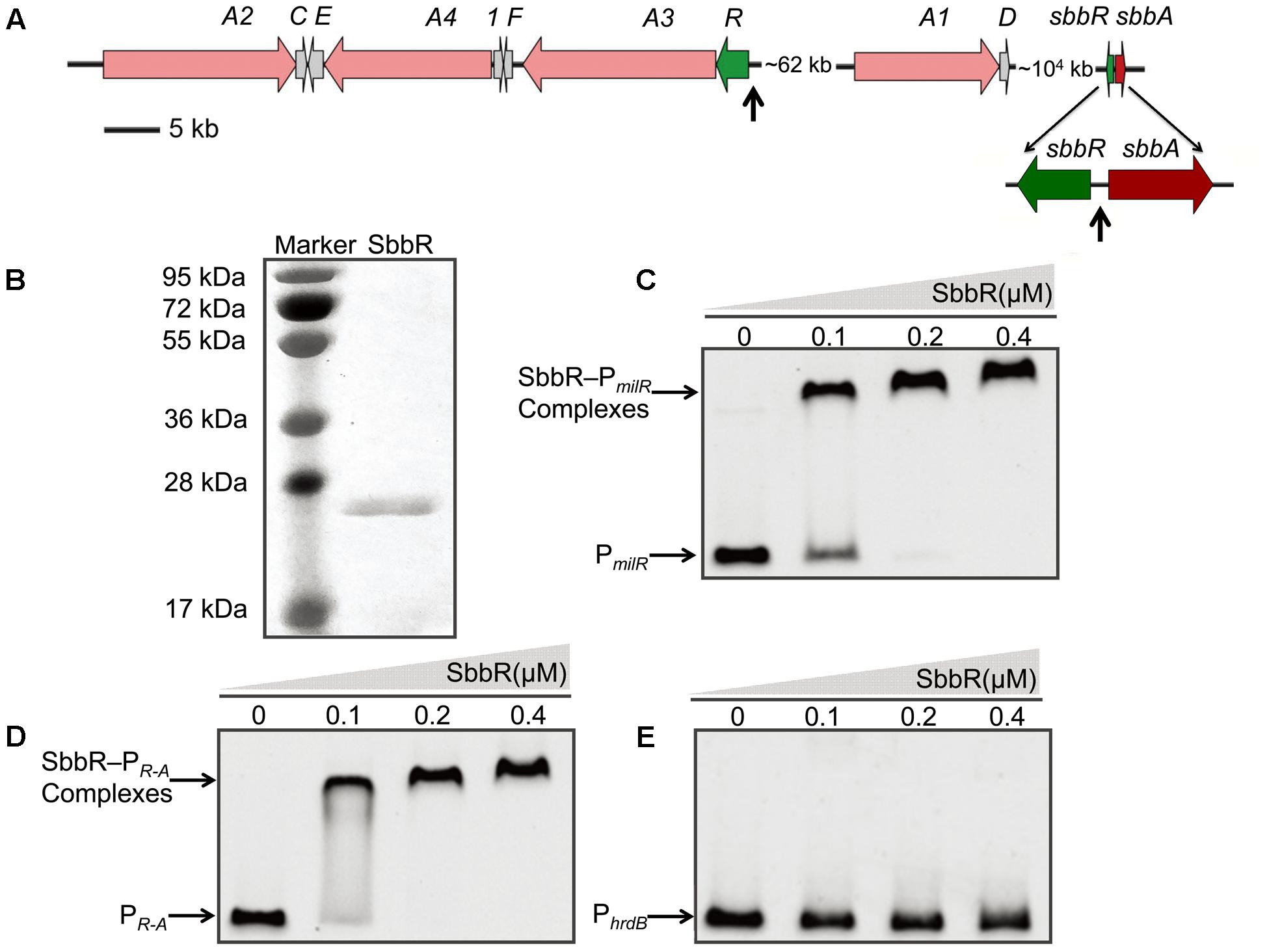
FIGURE 3. Binding of purified SbbR to the promoter regions of milR, sbbR, and sbbA. (A) Schematic representation of the relative positions of sbbR, sbbA, and the mil cluster. The vertical arrows indicate the binding region of SbbR. (B) SDS-PAGE analysis of the purified SbbR-His6 (23.6 kDa). (C–E) EMSAs of SbbR binding to the promoters PmilR, PR-A, and PhrdB. PmilR: the promoter region of milR. PR-A: the intergenic region of sbbR and sbbA. PhrdB: the promoter region of hrdB. Each lane contains 10 ng of DNA probes. The promoter regions of milR, the bidirectional sbbR/sbbA and hrdB were prepared as 413, 464, and 443 bp probes, respectively. DNA-protein complexes and free probes are indicated by arrows.
To further define the accurate binding sequences of SbbR in the upstream region of milR and the intergenic region of sbbR-sbbA, the transcription start points (tsps) of milR, sbbR, and sbbA were determined by 5′ RACE. For milR, the tsp was identified at a C, 39 nt upstream of the milR translation start codon (tsc) (Supplementary Figure S4A). For sbbR, the tsp was identified at a C, 15 nt upstream of the sbbR tsc (Supplementary Figure S4B). For sbbA, the tsp was identified at a G, 43 nt upstream of the sbbA tsc (Supplementary Figure S4C). DNase I footprinting experiments with 5′ FAM labeled probes were carried out to define the protected sites of SbbR on its target promoters. As shown in Figure 4, SbbR protected a region spanning from nt -279 to -252 relative to the milR tsp (Figures 4A,C), far away from the -35 and -10 elements of milR promoter. The footprint of the sbbR promoter region covered a region from nt -106 to -71 (Figures 4B,D), and this region corresponds to positions -30 to +5 relative to the tsp of sbbA (Figures 4B,E). Analysis of these two protected sequences interacting with SbbR showed no consensus motif. However, after careful examination, a 6 nt imperfect inverted repeat (CCGAYG [Y=C or T]) with 8 bp spacing within the binding region of SbbR on PmilR, and another different 6 nt inverted repeat (CCATHA [H=C or A]) with 11 bp spacing within the protected sequences of SbbR on PR-A were defined, respectively.
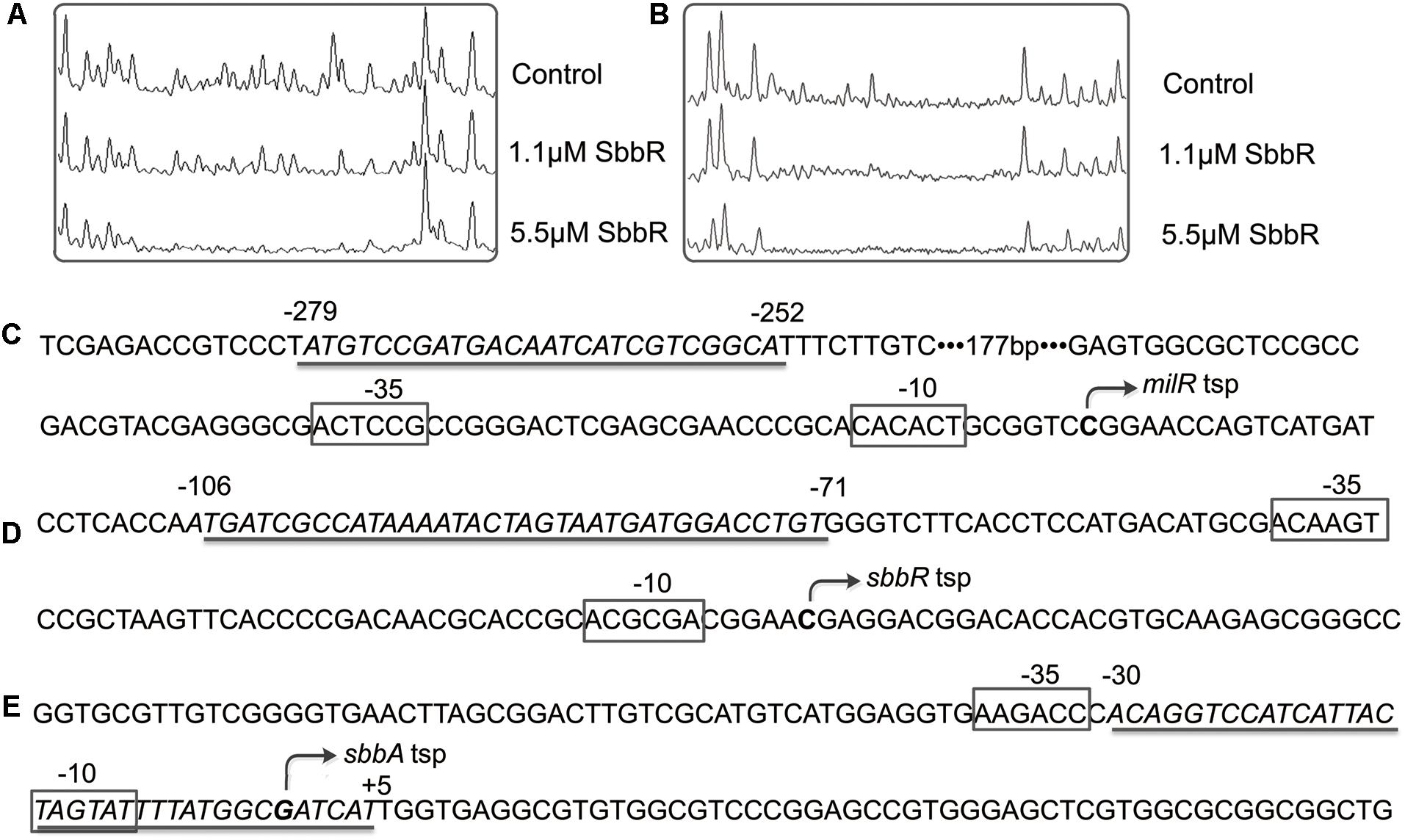
FIGURE 4. Binding sequences of SbbR on the promoter regions of milR, sbbR, and sbbA. (A) DNase I footprinting assay of SbbR binding site on milR promoter region (PmilR). (B) DNase I footprinting assay of SbbR binding site on sbbR (sbbA) promoter region PR-A. Upper fluorograms: control reaction with 10 μM BSA. Protection fluorograms were obtained with increasing amounts of SbbR-His6. (C–E) Nucleotide sequences of the promoter regions of milR, sbbR, and sbbA. The tsp is marked by a bent arrow. The SbbR-binding sequences are underlined, and the putative –10 and –35 regions are marked by boxes. Numbers indicate distance (nt) from respective tsp.
To determine the actual contributions of these two different inverted repeats to the binding activity of SbbR. PmilR and PR-A were separately mutated to PmilR∗ and PR-A∗. To obtain PmilR∗, the 6 nt imperfect inverted repeat of PmilR was replaced by EcoRI and SpeI restriction sites (Figure 5A); similarly, the 6 nt imperfect inverted repeat of PR-A was also replaced by EcoRI and SpeI restriction sites, generating PR-A∗ (Figure 5C). PmilR∗ and PR-A∗ were then tested in EMSAs with SbbR. As shown in Figure 5, compared with the wild-type DNA probes, only a small amounts of complexes were formed for PmilR∗ or PR-A∗ (Figures 5B,D). Thus, these two inverted repeats were critical in the binding of SbbR.
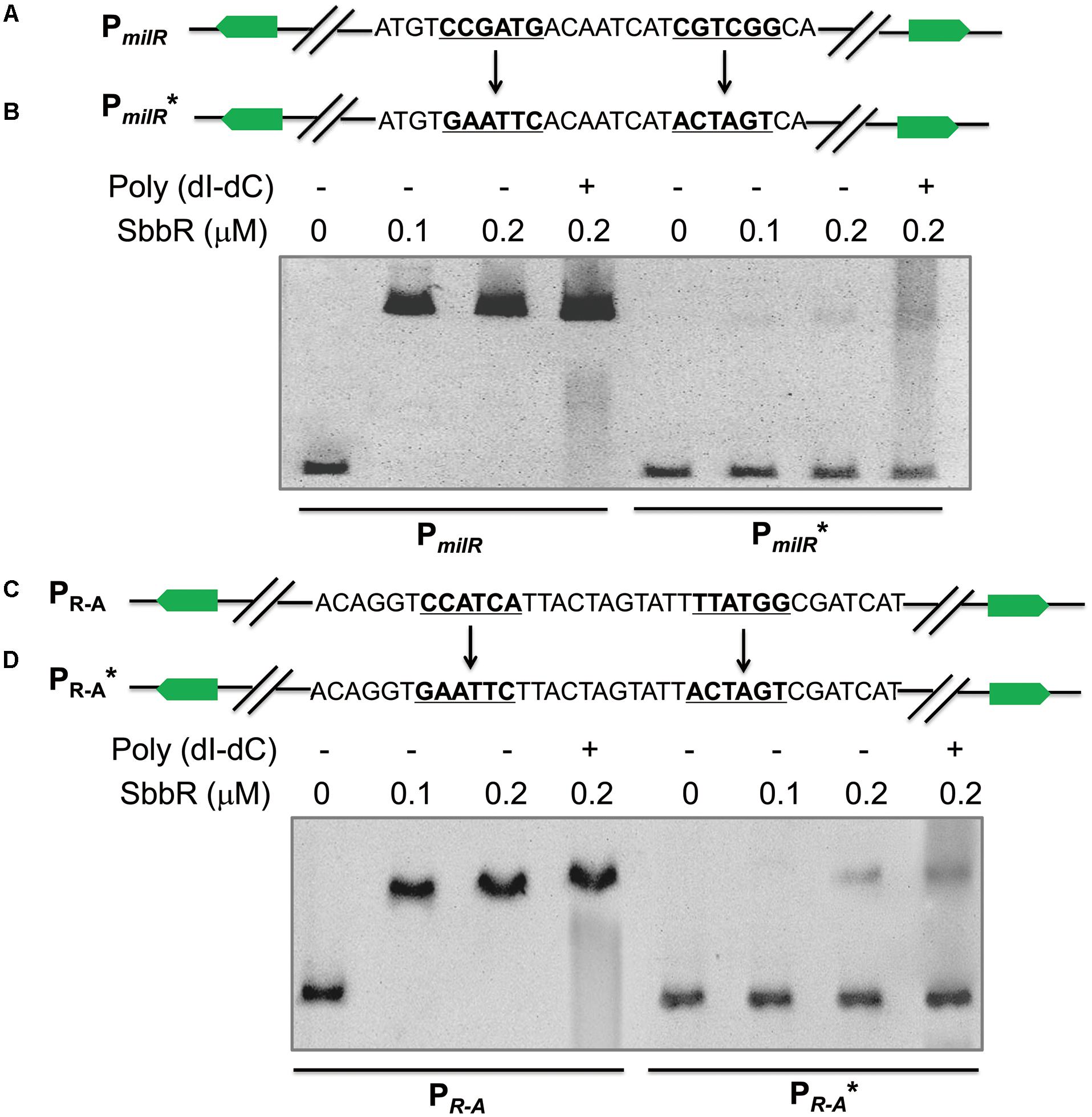
FIGURE 5. Mutational analysis of SbbR binding sites on PmilR and PR-A. (A) Mutations introduced into the 6 nt inverted repeat of PmilR. The changed nucleotides are underlined. (B) EMSAs of the wild-type probe PmilR and mutated probe PmilR∗. Each lane contains 10 ng of DNA probes. The concentrations of SbbR used in lanes 1 to 4 were 0, 0.1, 0.2, and 0.2 μM, respectively. Line 4 contains 500-fold poly(dI-dC). (C) Mutations introduced into the 6 nt inverted repeat of PR-A. The changed nucleotides are underlined. (D) EMSAs of the wild-type probe PR-A and mutated probe PR-A∗.
SbbR Activates the Transcription of milR but Represses sbbR and sbbA
To examine the effect of SbbR on the transcription of its targets, RNA samples were extracted from the mycelia of BC04 and ΔsbbR cultivated for various days (1, 2, 3, 5, and 9 days). The transcription levels of milR, sbbR (the ORF of sbbR in ΔsbbR was partially left at its 5′ end) and sbbA were then assessed by quantitative real-time RT-PCR (qRT-PCR). The results showed that transcription of milR decreased significantly in ΔsbbR at all tested time points, whereas the transcript levels of sbbR and sbbA were shown to be higher in ΔsbbR than in BC04 (Figure 6). The transcription profiles of sbbR and sbbA were similar, reached the highest level at about 3 days of cultivation when milbemycin production was initiated, and then the transcripts decreased gradually (Figure 6). Since milA3 is located in the same operon with milR (the milR-A3 operon), and MilR activates the expression of the milA4-E operon and milF directly (Zhang et al., 2016); thus, transcription of milA3, milA4, and milF was also measured. Transcripts of these three genes were also decreased dramatically (Figure 6). These results suggested that SbbR regulates milbemycin biosynthesis by activating the transcription of milR directly, and SbbR is a direct repressor of its own transcription and that of sbbA.
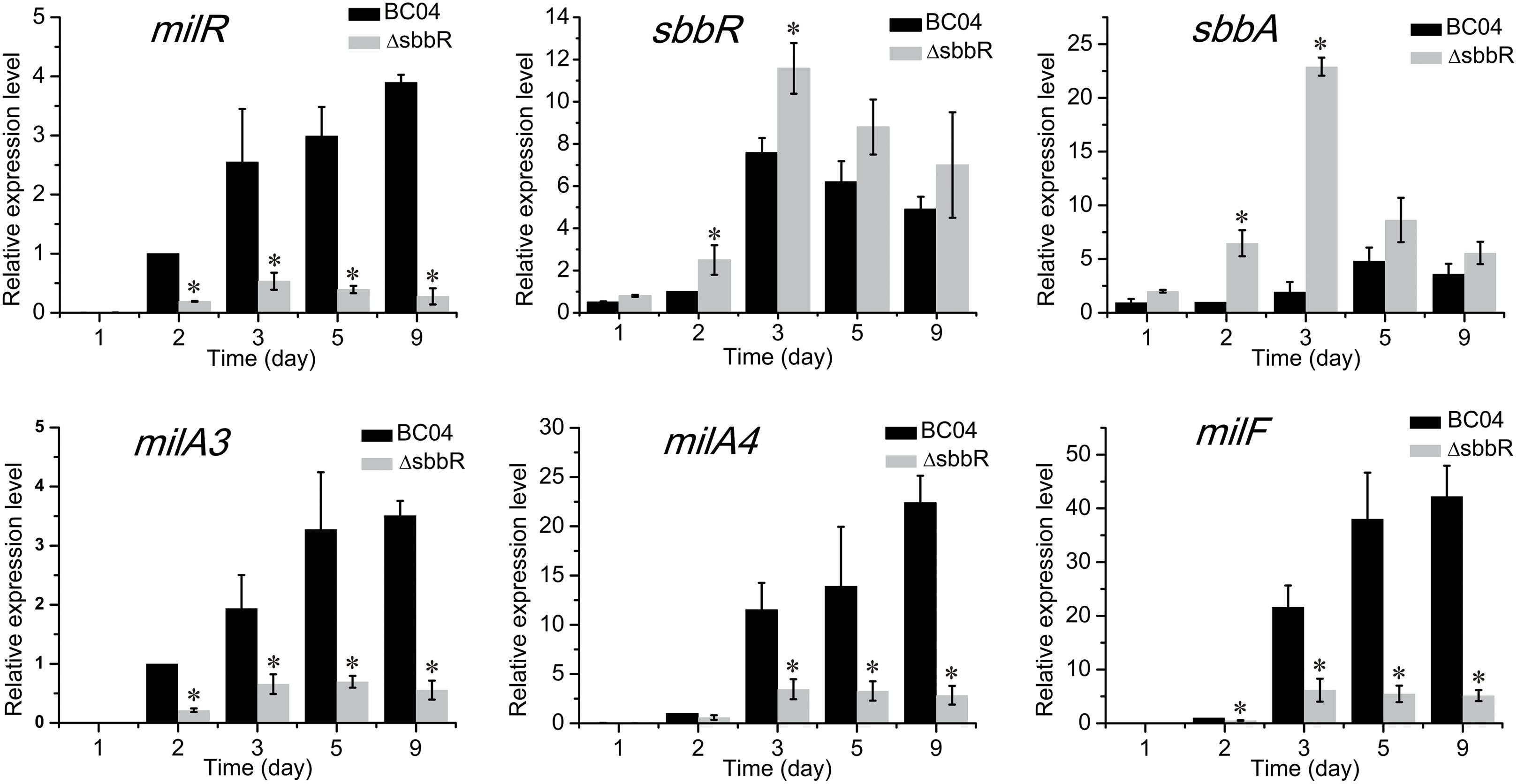
FIGURE 6. Quantitative real-time RT-PCR analysis of milR, sbbR, sbbA, milA3, milA4, and milF in BC04 and ΔsbbR. All RNA samples were isolated from 1, 2, 3, 5, and 9 days cultures. The transcriptional levels of related genes are presented relative to that of BC04 collected after fermentation for 24 h, which was arbitrarily assigned a value of 1. Data are presented as the averages of three independent experiments conducted in triplicate. 16S rRNA transcription was monitored and used as the internal control. Error bars show standard deviations. P-values were determined by Student’s t-test. ∗P < 0.05.
The regulatory relationships between SbbR and its target genes (milR and sbbA) were further verified using a heterologous promoter-reporter system in Escherichia coli (Yin et al., 2015). As shown in Figure 7A, the promoters of milR and sbbA were cloned separately upstream of gfp [encoding green fluorescent protein (GFP)] to generate pSET152::PmilRgfp and pSET152::PsbbAgfp, respectively. The resulting plasmids were then each transferred to E. coli. Afterwards, sbbR controlled by SF14 was assembled into the above two recombinant vectors to generate pSET152::PmilRgfp::SF14sbbR and pSET152::PsbbAgfp::SF14sbbR, respectively, which were also introduced into E. coli. Then the four strains constructed above together with the control strain harboring pSET152 were detected for green fluorescence. Fluorescence in the strain harboring pSET152::PmilRgfp::SF14sbbR was enhanced 2.4-fold compared to that in the strain harboring pSET152::PmilRgfp, while fluorescence in pSET152::PsbbAgfp::SF14sbbR-containing strain exhibited a decrease of 87% compared to that of pSET152::PsbbAgfp strain (Figure 7B). These results agreed with the data obtained from transcriptional profiles in S. bingchenggensis BC04, and provided convincing evidence that SbbR activates the expression of milR and represses the transcription of sbbA directly.
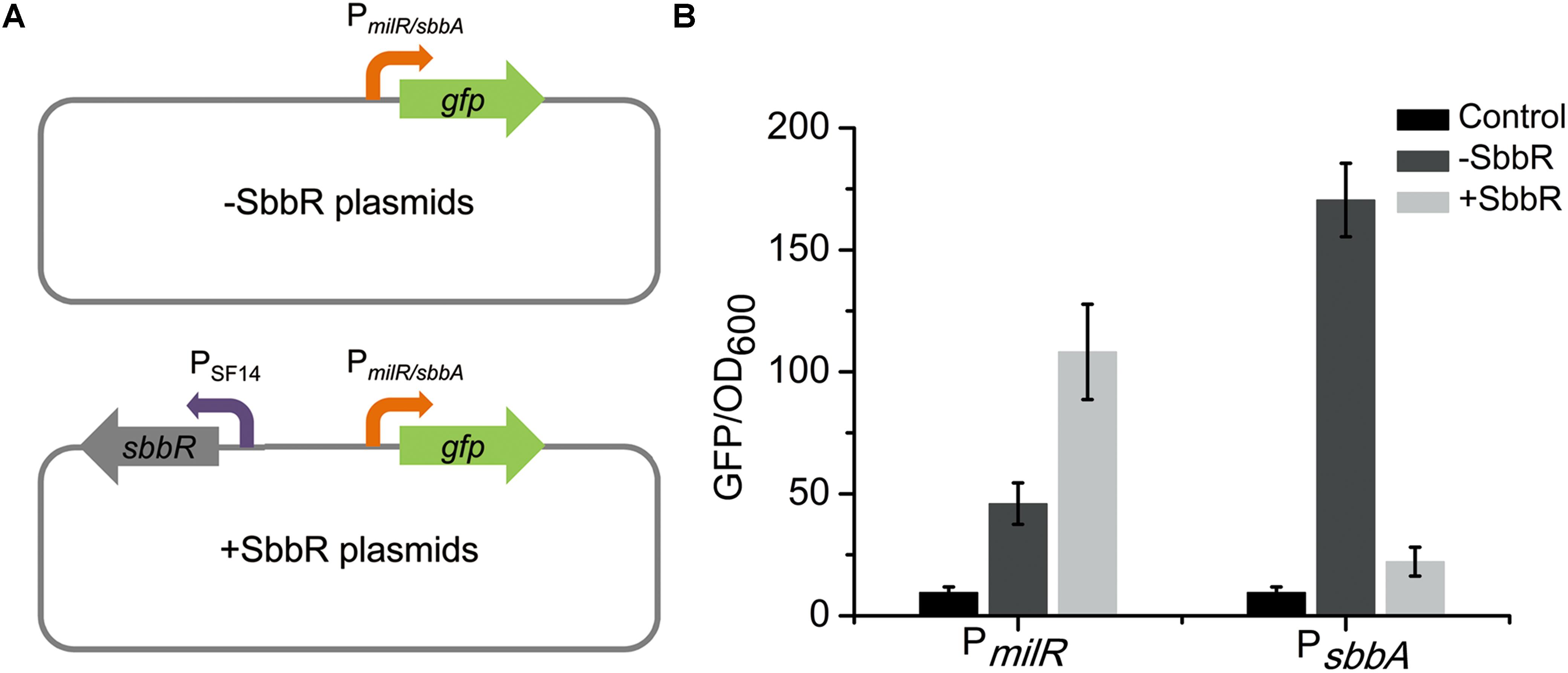
FIGURE 7. Analyses of regulatory relationships between SbbR and its target genes (milR and sbbA) in E. coli. (A) An illustration of the reporter plasmids. (B) Determination of the regulatory relationships between SbbR and its target promoters (PmilR and PsbbA). All values are in relative fluorescence unit (GFP/OD600) and represent the averages of at least three independent readings.
Identification of New SbbR Targets That Encode Transcriptional Regulatory Proteins
When analyzing the function of SbbR/SbbA system, it is possible that SbbR/SbbA may be involved in a more comprehensive regulation of diverse cellular processes directly or via regulatory cascades. As shown in Figures 3, 4, the promoter of milR and the intergenic region of sbbR and sbbA were the primary targets of SbbR; however, no highly conserved recognition sequence could be extracted from the two binding sites. The relatively unconserved binding sites for SbbR make it difficult to predict novel targets through in silico screening of the S. bingchenggensis genome sequence using MAST/MEME tool. Therefore, to further explore the SbbR/SbbA regulon, we selectively searched for binding targets of SbbR that encode transcriptional regulators. A total of 43 genes located in putative secondary metabolism clusters and 29 genes that encodes homologs of global or pleiotropic regulators were selected for EMSAs (Supplementary Table S2). The probes containing the upstream regions of the 72 genes were amplified (Supplementary Table S1). Subsequent EMSAs showed that SbbR has strong affinity for 11 DNA probes (Figure 8), and the deduced products of the putative target genes were as follows: two SARP family CSRs (SBI_08420 and SBI_08432) of nanchangmycin biosynthetic gene cluster (Wang et al., 2013), one SARP family CSR (SBI_09158) of actagardine biosynthetic gene cluster (Wang et al., 2013), one LAL family CSR (SBI_00827) of a cluster encoding non-ribosomal peptide synthases (Wang et al., 2013), one LAL family CSR (SBI_01376) of lasalocid biosynthetic gene cluster (Wang et al., 2013), one putative LAL family regulator (SBI_09325) of melanin biosynthetic gene cluster (Wang et al., 2013), one putative Sig24 homologous regulator (SBI_06451, Sig24sbh) of desferrioxamine biosynthetic gene cluster (Wang et al., 2013), one GlnR homolog (SBI_05051, GlnRsbh), one WblA-like protein (SBI_05811, WblAsbh), one AtrA homolog protein (SBI_05779, AtrAsbh), and one MtrA/B-type two-component system (TCS: SBI_06494/06495, MtrA/Bsbh).
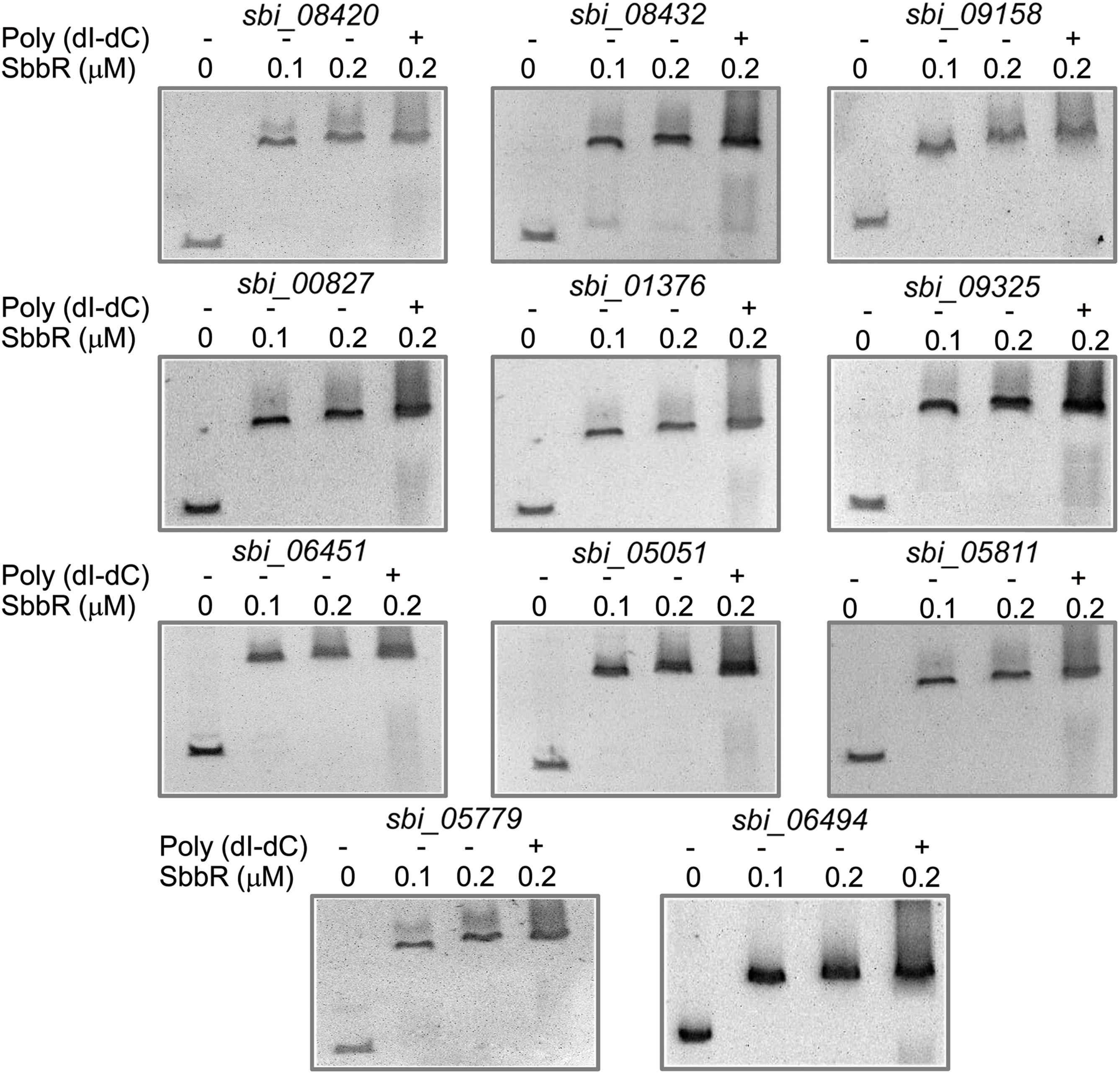
FIGURE 8. Electrophoresis mobility shift assays (EMSAs) of SbbR binding to the upstream regions of 11 novel targets. The amounts of SbbR (μM) used were as indicated and 10 ng of DNA probes was added to each EMSA reaction. Lane 2–4 contains 0.1, 0.2, and 0.2 μM SbbR and Line 4 contains 500-fold non-specific poly (dI-dC).
To investigate the regulatory role of SbbR in the transcription of these new target genes, qRT-PCR analysis was carried out to assess the transcription of related genes. As shown in Figure 9, SbbR repressed the transcription of two SARP-family regulatory genes (sbi_08432 and sbi_09158) and one LAL-family regulatory gene sbi_09325, as the transcriptional levels of these genes were increased in ΔsbbR compared with BC04 at day 3 or both days (days 3 and 5). In contrast, transcription of the other eight genes was lower in ΔsbbR than in BC04 at day 5 or both days, indicating that SbbR is an activator of these genes (Figure 9). These data suggested that SbbR plays a differential regulatory role toward different target genes. It should be noted that these 11 novel targets of SbbR are identified only from the above 72 transcriptional regulatory genes that act as putative CSRs or pleiotropic regulators. Genome-wide identification of SbbR targets via chromatin immunoprecipitation followed by sequencing (Chip-seq) will be carried out in our future works.
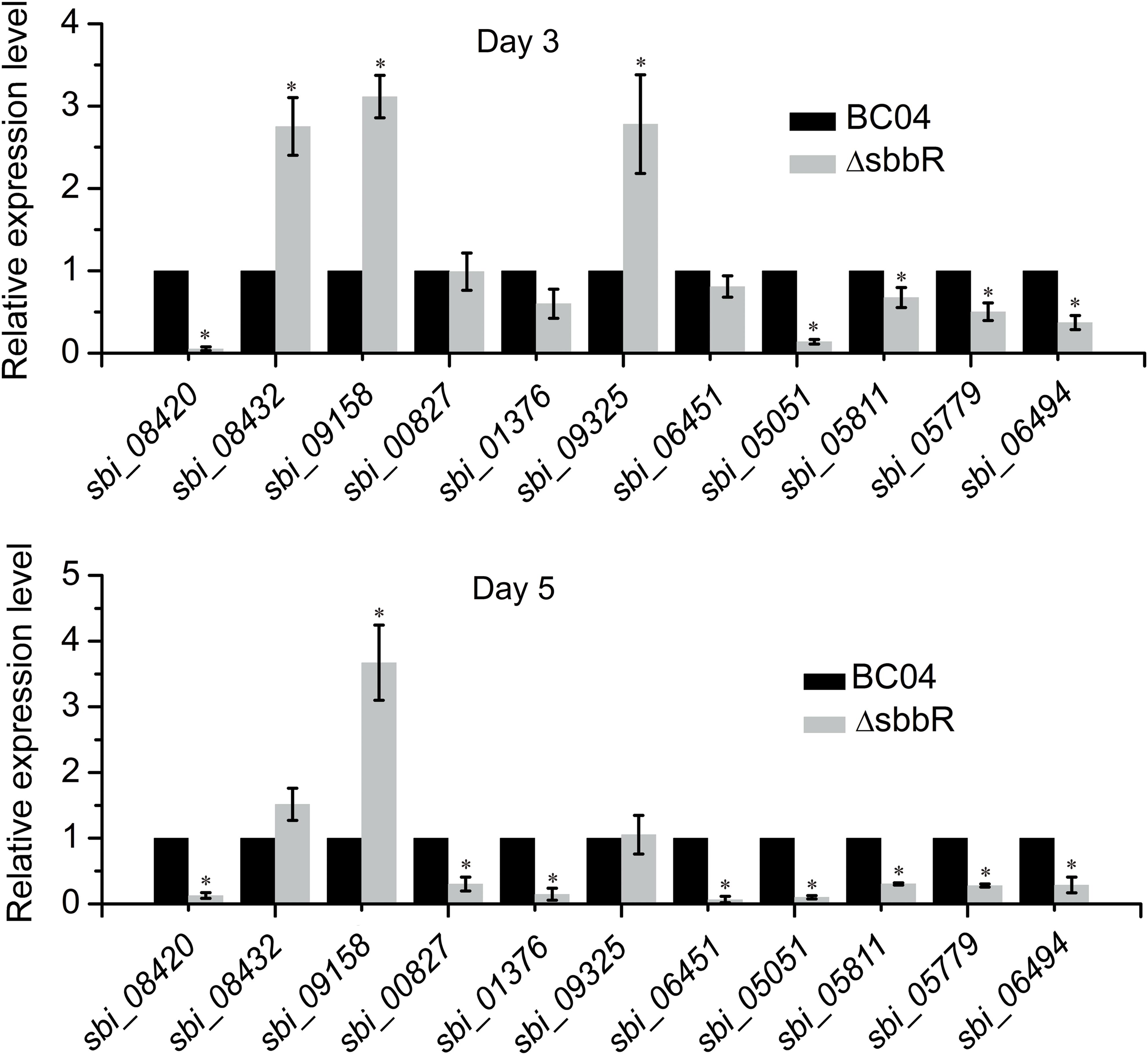
FIGURE 9. Quantitative real-time PCR (qRT-PCR) analysis examining the effects of sbbR deletion on the transcription of new SbbR target genes. RNA samples were isolated from 3 and 5 days cultures. The transcription of genes in BC04 was assigned a value of 1. 16S rRNA transcription was monitored and used as the internal control. Data are presented as the averages of three independent experiments conducted in triplicate. Error bars show standard deviations. ∗P < 0.05.
Effect of Culture Extracts From S. bingchenggensis on the Binding Activity of SbbR
In S. bingchenggensis, SbbR and SbbA constitute the quorum-sensing system, in which SbbR can respond to the likely GBL-type signaling molecules generated by SbbA. Thus, to determine whether SbbR is able to respond to small signaling molecules produced by S. bingchenggensis, ethyl acetate extracts of strains BC04 and ΔsbbA were prepared and assayed for their effect on the binding activity of SbbR to PmilR. As expected, extracts from BC04 could dissociate SbbR from SbbR-PmilR complexes, leading to about forty thousand inhibitory units/l (1 inhibitory units is defined as the activity of dissociating about half of 10 ng of PmilR from SbbR) (Supplementary Figure S5). Unexpectedly, when added at a high concentration, extracts from ΔsbbA could also dissociate SbbR from SbbR-PmilR complexes (Supplementary Figure S5). This indicated that SbbR could bind the factor determined by the GBL biosynthesis protein SbbA; it could also bind to some unknown ligands produced in ΔsbbA. It is interesting to probe the putative chemicals that bind to SbbR produced by ΔsbbA, which will be the focus of our future work. These results suggested that SbbR may be a genuine signaling molecular receptor protein, and it is possible that the molecules produced by SbbA regulate the biosynthesis of milbemycin via interactions with SbbR, but the further structural analysis is impeded by the fact that they are normally produced in very small quantities (Niu et al., 2016).
Discussion
In the Gram-positive Streptomyces, ArpA/AfsA systems are important for antibiotic production and morphological differentiation (Tan et al., 2013; Zou et al., 2014). In the present study, genome analysis of sequenced S. bingchenggensis revealed a potential ArpA/AfsA-type system annotated as SbbR/SbbA. sbbR and sbbA are adjacent and divergently transcribed, but they locate far from the mil cluster. We demonstrate that SbbR, a GBL receptor homolog, activates the biosynthesis of milbemycin by directly activating the transcription of milR, the CSR of the mil cluster; while the GBL synthase homolog, SbbA, has a repressive effect on milbemycin production. Interestingly, both SbbR and SbbA have positive effects on cell growth, and SbbR also acts pleiotropically by controlling the expression of many other CSR genes and global regulatory genes.
Generally, it is thought that GBLs produced by the AfsA homologs are essential for antibiotic production, and mutation of the corresponding homologous genes lead to impaired or reduced production of the respective antibiotics, while disruption of the cognate receptor genes leads to the opposite or no effect (Takano, 2006; Zou et al., 2014). For example, in S. hygroscopicus 5008, inactivation of afsA homologs (shbA1, shbA2, or shbA3) dramatically decreased biosynthesis of validamycin; whereas deletion of arpA homologs (shbR1 or shbR3) increased validamycin production by 26% (ΔshbR1) and 20% (ΔshbR3), and deletion of shbR2 had no significant effects on validamycin production (Tan et al., 2015). However, in S. bingchenggensis, the SbbR/SbbA system presented an opposite regulatory pattern: inactivation of the GBL synthase gene (sbbA) led to an increase of antibiotic production instead of the anticipated reduction or abolishment, while mutation of sbbR resulted in reduced milbemycin production. Virtually, titer improvement of milbemycin in sbbA mutant strain is not surprising. In S. bingchenggensis BC04, SbbR activates milR expression by binding to the upstream region of milR; expressed milR can activate the transcription of biosynthetic structural genes milA4-E and milF. The putative compound produced by SbbA could directly bind to SbbR, leading to its dissociation from the milR promoter. In ΔsbbA, production of the compound might be abolished, and SbbR could bind milR promoter persistently and activates its expression, thus leading to an increased milbemycin production. Previously, titer improvements of antibiotic in GBL synthase gene deficiency strains have been reported. In the model strain S. coelicolor, deletion of scbA, a homolog of afsA, led to overproduction of actinorhodin and undecylprodigiosins (Takano et al., 2001; Li et al., 2015). In S. lividans, deletion of scbA also improved the titers of actinorhodin and undecylprodigiosins (Butler et al., 2003). The mechanisms underlying the role of ScbA in increasing actinorhodin and undecylprodigiosins production in these two strains are unclear, but are supposed to be the molecular events that are closely related with the complex regulatory cascade mediated by GBLs. In this work, inactivation of sbbA obviously improved the production of milbemycin by 25% (from 2537 to 3169 mg/l); thus the sbbA mutant can be used as a new parental strain in the future genetic engineering to obtain milbemycin high-producing strains.
In this work, SbbR played differential regulatory roles toward its targets: it represses the expression of its own gene and the divergently transcribed sbbA, but positively regulates milbemycin biosynthesis by directly activating the transcription of milR. The promoter sites covered by SbbR were from nt -106 to -71, nt -30 to +5, and nt -279 to -252 relative to the tsps of sbbR, sbbA, and milR. The auto-repression of SbbR on its own is a common feature exerted by TetR family regulators (Ahn et al., 2012). The mechanism whereby SbbR represses sbbA is easy to understand, the protected site overlaps the -35 and -10 elements, SbbR may sterically block the access of RNA polymerase to sbbA promoter. While as an activator of milR, SbbR locates far upstream of the -35 element and may recruit RNA polymerase to the promoter via direct protein–protein interaction (Rodionov, 2007). GBL receptor homologs that bind (far) upstream of promoter region and act as the direct activator of antibiotic CSRs have been reported previously. For instance, in jadomycin biosynthesis in S. venezuelae, the GBL (SVB1) receptor, JadR3, can activate the expression of jadR1, the cluster-situated activator of jadomycin biosynthesis. JadR3 has three binding sites in the promoter of jadR1. Among these sites, AREII, a region from nt -252 to -219 relative to the jadR1 tsp, is essential for the activation function of JadR3 (Zou et al., 2014). In S. ansochromogenes, SabR binds to the sequences (from nt -64 to -29 relative to the tsp of sanG) adjacent to or overlapping partially with the -35 element and positively regulates sanG transcription (He et al., 2010). There are also exceptions, AvaR1, the ArpA homolog from S. avermitilis, protects two regions (regions from nt -262 to -233 and nt -223 to -193) far upstream of AveR (the cluster-situated activator of avermectins) and acts as a repressor, but not an activator (Zhu et al., 2017); one of the two binding sites (nt -222 to -244 and nt -3 to -35 relative to the cpkO tsp) of ScbR from S. coelicolor is also far away from cpkO promoter and ScbR is a repressor of cpkO (Takano et al., 2005). These examples complicate the regulatory mechanisms exerted by GBL receptor homologs. Nevertheless, our study and the observations reported previously lead to the conclusion that, in addition to being a repressor of its own expression and that of other targets, GBL receptor homologs could also interact with the upstream regions of cluster-situated activator genes to promote gene expression, suggesting that a GBL receptor homolog-mediated activation of antibiotic biosynthesis might exist in many streptomycetes.
In the search of SbbR/SbbA regulon, 11 genes that encode seven putative secondary metabolism CSRs and four well-known pleiotropic regulatory homologs have been identified. Of these novel targets, the SARP-family regulators (SBI_08420, SBI_08432, and SBI_09158), the LAL-family regulators (SBI_00827, SBI_01376, and SBI_09325) and Sig24sbh are the potential cluster-situated activators, e.g., SBI_08420, designated as NanR2, has recently been reported to be an essential activator of nanchangmycin production (Yu et al., 2012), suggesting the importance of SbbR/SbbA in the regulation of secondary metabolism. Moreover, homologs of the four pleiotropic regulators GlnRsbh, WblAsbh, AtrAsbh, and MtrA/Bsbh have been reported to be involved in the development of Streptomyces as well as in the production of various antibiotics in streptomycetes (Rabyk et al., 2011; Lee et al., 2013). Many detailed studies concerning the downstream regulatory networks of these four regulators have been reported (Liu et al., 2013), but little is known about the regulatory proteins that function upstream of these pleiotropic regulators, although AdpAch from Streptomyces chattanoogensis L10 has been confirmed to be a direct activator of wblAch (Yu et al., 2014), and AdpA homolog from Streptomyces roseosporus directly regulate the atrA homolog (Mao et al., 2015). Our finding provides an interesting clue that SbbR, as a GBL receptor homolog, could be a direct controller of these important regulators that act pleiotropically in Streptomyces. The molecular regulatory mechanism of SbbR as the upper regulator of these pleiotropic regulators needs further investigation.
SbbR could regulate milR directly, however, it is doubted that this is the only regulatory pattern that is exerted by SbbR/SbbA system to regulate milbemycin production. Note that both deletion of sbbR and sbbA decreased cell biomass in fermentation medium, suggesting the importance of sbbR and sbbA in cell growth (Figure 2D); moreover, SbbR has positive effects on the four well-known pleiotropic regulators (i.e., GlnRsbh, WblAsbh, AtrAsbh, and MtrA/Bsbh). Therefore, it is possible that SbbR/SbbA system may exert pleiotropic effects on primary and secondary metabolism, and the changes of milbemycin production in ΔsbbR and ΔsbbA is not only due to the changed expression levels of biosynthetic genes, but also the changed supplies of precursors and cofactors. Further studies are required to investigate the regulatory targets of SbbR/SbbA system through comparative transcriptomic analysis among BC04, ΔsbbR, and ΔsbbA, and Chip-seq of SbbR, and thereby gain a more comprehensive overview of the roles of SbbR/SbbA system.
Author Contributions
HH, YZ, and WX designed the research. HH and YZ wrote the manuscript. HH performed the experiments and LY determined the dry weight and production curves. HW, CL, XG, and XW helped to modify the article. All authors reviewed and approved the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31572070 and 31401814).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Doctor Nyssa Cullin (Oregon Health and Science University, Portland, OR, United States) for critical reading in preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01064/full#supplementary-material
References
Ahn, S. K., Cuthbertson, L., and Nodwell, J. R. (2012). Genome context as a predictive tool for identifying regulatory targets of the TetR family transcriptional regulators. PLoS One 7:e50562. doi: 10.1371/journal.pone.0050562
Arakawa, K., Mochizuki, S., Yamada, K., Noma, T., and Kinashi, H. (2007). gamma-Butyrolactone autoregulator-receptor system involved in lankacidin and lankamycin production and morphological differentiation in Streptomyces rochei. Microbiology 153(Pt 6), 1817–1827. doi: 10.1099/mic.0.2006/002170-0
Bibb, M. J. (2005). Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8, 208–215. doi: 10.1016/j.mib.2005.02.016
Bierman, M., Logan, R., O’Brien, K., Seno, E. T., Rao, R. N., and Schoner, B. E. (1992). Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49. doi: 10.1016/0378-1119(92)90627-2
Butler, M. J., Takano, E., Bruheim, P., Jovetic, S., Marinelli, F., and Bibb, M. J. (2003). Deletion of scbA enhances antibiotic production in Streptomyces lividans. Appl. Microbiol. Biotechnol. 61, 512–516. doi: 10.1007/s00253-003-1277-8
Cuthbertson, L., and Nodwell, J. R. (2013). The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77, 440–475. doi: 10.1128/MMBR.00018-13
Danaher, M., Radeck, W., Kolar, L., Keegan, J., and Cerkvenik-Flajs, V. (2012). Recent developments in the analysis of avermectin and milbemycin residues in food safety and the environment. Curr. Pharm. Biotechnol. 13, 936–951. doi: 10.2174/138920112800399068
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Gao, A. L., Wang, X. J., Xiang, W. S., Gao, J. G., Wang, Q., Jiang, B., et al. (2007). Selection and Identification of a New Streptomyces hygroscopicus Strain. Faisalabad: Journal of Northeast Agricultural University, 3, 361–364. doi: 10.3969/j.issn.1005-9369.2007.03.016
He, X., Li, R., Pan, Y., Liu, G., and Tan, H. (2010). SanG, a transcriptional activator, controls nikkomycin biosynthesis through binding to the sanN-sanO intergenic region in Streptomyces ansochromogenes. Microbiology 156(Pt 3), 828–837. doi: 10.1099/mic.0.033605-0
Ikari, K. (2013). Pest control composition. U.S. Patent No EP, EP2,601,836. Washington, DC: U.S. Patent and Trademark Office.
Jacobs, C. T., and Scholtz, C. H. (2015). A review on the effect of macrocyclic lactones on dung-dwelling insects: toxicity of macrocyclic lactones to dung beetles. Onderstepoort J. Vet. Res. 82:858. doi: 10.4102/ojvr.v82i1.858
Khokhlov, A. S., Tovarova, I. I., Borisova, L. N., Pliner, S. A., Shevchenko, L. N., and Kornitskaia, E. (1967). [The A-factor, responsible for streptomycin biosynthesis by mutant strains of Actinomyces streptomycini]. Dokl Akad Nauk SSSR 177, 232–235.
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces genetics: A Laboratory Manual. Norwich: John Innes Foundation.
Kim, S. W., Rahman, M. M., Abd El-Aty, A. M., Truong, L. T., Choi, J. H., Park, J. S., et al. (2016). Residue level and dissipation pattern of lepimectin in shallots using high-performance liquid chromatography coupled with photodiode array detection. Biomed. Chromatogr. 30, 1835–1842. doi: 10.1002/bmc.3759
Kinoshita, H., Ipposhi, H., Okamoto, S., Nakano, H., Nihira, T., and Yamada, Y. (1997). Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae. J. Bacteriol. 179, 6986–6993. doi: 10.1128/jb.179.22.6986-6993.1997
Kitani, S., Hoshika, M., and Nihira, T. (2008). Disruption of sscR encoding a gamma-butyrolactone autoregulator receptor in Streptomyces scabies NBRC 12914 affects production of secondary metabolites. Folia Microbiol. 53, 115–124. doi: 10.1007/s12223-008-0017-y
Kitani, S., Miyamoto, K. T., Takamatsu, S., Herawati, E., Iguchi, H., Nishitomi, K., et al. (2011). Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc. Natl. Acad. Sci. U.S.A. 108, 16410–16415. doi: 10.1073/pnas.1113908108
Kuhstoss, S., Richardson, M. A., and Rao, R. N. (1991). Plasmid cloning vectors that integrate site-specifically in Streptomyces spp. Gene 97, 143–146. doi: 10.1016/0378-1119(91)90022-4
Lee, H. N., Kim, J. S., Kim, P., Lee, H. S., and Kim, E. S. (2013). Repression of antibiotic downregulator WblA by AdpA in Streptomyces coelicolor. Appl. Environ. Microbiol. 79, 4159–4163. doi: 10.1128/aem.00546-13
Li, X., Wang, J., Li, S., Ji, J., Wang, W., and Yang, K. (2015). ScbR- and ScbR2-mediated signal transduction networks coordinate complex physiological responses in Streptomyces coelicolor. Sci. Rep. 5:14831. doi: 10.1038/srep14831
Li, Y., Li, J., Tian, Z., Xu, Y., Zhang, J., Liu, W., et al. (2016). Coordinative modulation of chlorothricin biosynthesis by binding of the glycosylated intermediates and end product to a responsive regulator ChlF1. J. Biol. Chem. 291, 5406–5417. doi: 10.1074/jbc.M115.695874
Liu, G., Chater, K. F., Chandra, G., Niu, G., and Tan, H. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. doi: 10.1128/mmbr.00054-12
Mao, X. M., Luo, S., Zhou, R. C., Wang, F., Yu, P., Sun, N., et al. (2015). Transcriptional regulation of the daptomycin gene cluster in Streptomyces roseosporus by an autoregulator. AtrA. J. Biol. Chem. 290, 7992–8001. doi: 10.1074/jbc.M114.608273
Mehra, S., Charaniya, S., Takano, E., and Hu, W. S. (2008). A bistable gene switch for antibiotic biosynthesis: the butyrolactone regulon in Streptomyces coelicolor. PLoS One 3:e2724. doi: 10.1371/journal.pone.0002724
Nishida, H., Ohnishi, Y., Beppu, T., and Horinouchi, S. (2007). Evolution of γ-butyrolactone synthases and receptors in Streptomyces. Environ. Microbiol. 9, 1986–1994. doi: 10.1111/j.1462-2920.2007.01314.x
Niu, G., Chater, K. F., Tian, Y., Zhang, J., and Tan, H. (2016). Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 40, 554–573. doi: 10.1093/femsre/fuw012
O’Rourke, S., Wietzorrek, A., Fowler, K., Corre, C., Challis, G. L., and Chater, K. F. (2009). Extracellular signalling, translational control, two repressors and an activator all contribute to the regulation of methylenomycin production in Streptomyces coelicolor. Mol. Microbiol. 71, 763–778. doi: 10.1111/j.1365-2958.2008.06560.x
Pluschkell, U., Horowitz, A. R., and Ishaaya, I. (1999). Effect of milbemectin on the sweetpotato whitefly, Bemisia tabad. Phytoparasitica 27, 183–191. doi: 10.1007/BF02981457
Rabyk, M., Ostash, B., Rebets, Y., Walker, S., and Fedorenko, V. (2011). Streptomyces ghanaensis pleiotropic regulatory gene wblA(gh) influences morphogenesis and moenomycin production. Biotechnol. Lett. 33, 2481–2486. doi: 10.1007/s10529-011-0728-z
Rodionov, D. A. (2007). Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 107, 3467–3497. doi: 10.1021/cr068309
Rugg, D., Buckingham, S. D., Sattelle, D. B., and Jansson, R. K. (2005). “The insecticidal macrocyclic lactones,” in Comprehensive Molecular Insect Science 3. Pharmacology, eds I. Lawrence, K. I. Gilbert, and S. Sarjeet Gill (Cambridge, MA: Academic Press Elsevier), 69–96. doi: 10.1016/B0-44-451924-6/00065-X
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Salehi-Najafabadi, Z., Barreiro, C., Martinez-Castro, M., Solera, E., and Martin, J. F. (2011). Characterisation of a gamma-butyrolactone receptor of Streptomyces tacrolimicus: effect on sporulation and tacrolimus biosynthesis. Appl. Microbiol. Biotechnol. 92, 971–984. doi: 10.1007/s00253-011-3466-1
Shoop, W. L., Mrozik, H., and Fisher, M. H. (1995). Structure and activity of avermectins and milbemycins in animal health. Vet. Parasitol. 59, 139–156. doi: 10.1016/0304-4017(94)00743-V
Takano, E. (2006). Gamma-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9, 287–294. doi: 10.1016/j.mib.2006.04.003
Takano, E., Chakraburtty, R., Nihira, T., Yamada, Y., and Bibb, M. J. (2001). A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41, 1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x
Takano, E., Kinoshita, H., Mersinias, V., Bucca, G., Hotchkiss, G., Nihira, T., et al. (2005). A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol. 56, 465–479. doi: 10.1111/j.1365-2958.2005.04543.x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, G. Y., Bai, L., and Zhong, J. J. (2013). Exogenous 1,4-butyrolactone stimulates A-factor-like cascade and validamycin biosynthesis in Streptomyces hygroscopicus 5008. Biotechnol. Bioeng. 110, 2984–2993. doi: 10.1002/bit.24965
Tan, G. Y., Peng, Y., Lu, C., Bai, L., and Zhong, J. J. (2015). Engineering validamycin production by tandem deletion of gamma-butyrolactone receptor genes in Streptomyces hygroscopicus 5008. Metab. Eng. 28, 74–81. doi: 10.1016/j.ymben.2014.12.003
van Wezel, G. P., and McDowall, K. J. (2011). The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat. Prod. Rep. 28, 1311–1333. doi: 10.1039/c1np00003a
Waki, M., Nihira, T., and Yamada, Y. (1997). Cloning and characterization of the gene (farA) encoding the receptor for an extracellular regulatory factor (IM-2) from Streptomyces sp. strain FRI-5. J. Bacteriol. 179, 5131–5137. doi: 10.1128/jb.179.16.5131-5137.1997
Wang, H. C., Li, K., Susko, E., and Roger, A. J. (2008). A class frequency mixture model that adjusts for site-specific amino acid frequencies and improves inference of protein phylogeny. BMC Evol. Biol. 8:331. doi: 10.1186/1471-2148-8-331
Wang, H. Y., Zhang, J., Zhang, Y. J., Zhang, B., Liu, C. X., He, H. R., et al. (2014). Combined application of plasma mutagenesis and gene engineering leads to 5-oxomilbemycins A3/A4 as main components from Streptomyces bingchenggensis. Appl. Microbiol. Biotechnol. 98, 9703–9712. doi: 10.1007/s00253-014-5970-6
Wang, X. J., Wang, X. C., and Xiang, W. S. (2009). Improvement of milbemycin-producing Streptomyces bingchenggensis by rational screening of ultraviolet- and chemically induced mutants. World J. Microbiol. Biotechnol. 25, 1051–1056. doi: 10.1007/s11274-009-9986-5
Wang, X. J., Yan, Y. J., Zhang, B., An, J., Wang, J. J., Tian, J., et al. (2010). Genome sequence of the milbemycin-producing bacterium Streptomyces bingchenggensis. J. Bacteriol. 192, 4526–4527. doi: 10.1128/JB.00596-10
Wang, X. J., Zhang, B., Yan, Y. J., An, J., Zhang, J., Liu, C. X., et al. (2013). Characterization and analysis of an industrial strain of Streptomyces bingchenggensis by genome sequencing and gene microarray. Genome 56, 677–689. doi: 10.1139/gen-2013-0098
Wei, J. H., Tian, Y. Q., Niu, G. Q., and Tan, H. R. (2014). GouR, a TetR family transcriptional regulator, coordinates the biosynthesis and export of gougerotin in Streptomyces graminearus. Appl. Environ. Microbiol. 80, 714–722. doi: 10.1128/AEM.03003-13
Xiang, W. S., Wang, J. D., Wang, X. J., and Zhang, J. (2007). Two new beta-class milbemycins from Streptomyces bingchenggensis: fermentation, isolation, structure elucidation and biological properties. J. Antibiot. 60, 351–356. doi: 10.1038/ja.2007.47
Yin, S., Wang, W., Wang, X., Zhu, Y., Jia, X., Li, S., et al. (2015). Identification of a cluster-situated activator of oxytetracycline biosynthesis and manipulation of its expression for improved oxytetracycline production in Streptomyces rimosus. Microb. Cell Fact. 14:46. doi: 10.1186/s12934-015-0231-7
Yu, P., Liu, S. P., Bu, Q. T., Zhou, Z. X., Zhu, Z. H., Huang, F. L., et al. (2014). WblAch, a pivotal activator of natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis L10, is positively regulated by AdpAch. Appl. Environ. Microbiol. 80, 6879–6887. doi: 10.1128/aem.01849-14
Yu, Q., Du, A., Liu, T., Deng, Z., and He, X. (2012). The biosynthesis of the polyether antibiotic nanchangmycin is controlled by two pathway-specific transcriptional activators. Arch. Microbiol. 194, 415–426. doi: 10.1007/s00203-011-0768-8
Yuan, P. H., Zhou, R. C., Chen, X., Luo, S., Wang, F., Mao, X. M., et al. (2016). DepR1, a TetR Family transcriptional regulator, positively regulates daptomycin production in an industrial producer, Streptomyces roseosporus SW0702. Appl. Environ. Microbiol. 82, 1898–1905. doi: 10.1128/AEM.03002-15
Zhang, J., An, J., Wang, J. J., Yan, Y. J., He, H. R., Wang, X. J., et al. (2013). Genetic engineering of Streptomyces bingchenggensis to produce milbemycins A3/A4 as main components and eliminate the biosynthesis of nanchangmycin. Appl. Microbiol. Biotechnol. 97, 10091–10101. doi: 10.1007/s00253-013-5255-5
Zhang, Y., Pan, G., Zou, Z., Fan, K., Yang, K., and Tan, H. (2013). JadR∗-mediated feed-forward regulation of cofactor supply in jadomycin biosynthesis. Mol. Microbiol. 90, 884–897. doi: 10.1111/mmi.12406
Zhang, Y., He, H., Liu, H., Wang, H., Wang, X., and Xiang, W. (2016). Characterization of a pathway-specific activator of milbemycin biosynthesis and improved milbemycin production by its overexpression in Streptomyces bingchenggensis. Microb. Cell Fact. 15:152. doi: 10.1186/s12934-016-0552-1
Zhu, J., Chen, Z., Li, J., and Wen, Y. (2017). AvaR1, a butenolide-type autoregulator receptor in Streptomyces avermitilis, directly represses avenolide and avermectin biosynthesis and multiple physiological responses. Front. Microbiol. 8:2577. doi: 10.3389/fmicb.2017.02577
Keywords: Streptomyces bingchenggensis, milbemycins, SbbR, SbbA, transcriptional regulation
Citation: He H, Ye L, Li C, Wang H, Guo X, Wang X, Zhang Y and Xiang W (2018) SbbR/SbbA, an Important ArpA/AfsA-Like System, Regulates Milbemycin Production in Streptomyces bingchenggensis. Front. Microbiol. 9:1064. doi: 10.3389/fmicb.2018.01064
Received: 05 March 2018; Accepted: 04 May 2018;
Published: 23 May 2018.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Cláudia Vicente, Institut National de la Recherche Agronomique (INRA), FranceYvonne Mast, Universität Tübingen, Germany
Copyright © 2018 He, Ye, Li, Wang, Guo, Wang, Zhang and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanyan Zhang, eXl6aGFuZ0BpcHBjYWFzLmNu Wensheng Xiang, eGlhbmd3ZW5zaGVuZ0BuZWF1LmVkdS5jbg==
 Hairong He
Hairong He Lan Ye
Lan Ye Chuang Li1,2
Chuang Li1,2 Wensheng Xiang
Wensheng Xiang