- Laboratory of Molecular Environmental Microbiology, Department of Environmental Science and Ecological Engineering, Korea University, Seoul, South Korea
Many petroleum-polluted areas are considered as extreme environments because of co-occurrence of low and high temperatures, high salt, and acidic and anaerobic conditions. Alkanes, which are major constituents of crude oils, can be degraded under extreme conditions, both aerobically and anaerobically by bacteria and archaea of different phyla. Alkane degraders possess exclusive metabolic pathways and survival strategies, which involve the use of protein and RNA chaperones, compatible solutes, biosurfactants, and exopolysaccharide production for self-protection during harsh environmental conditions such as oxidative and osmotic stress, and ionic nutrient-shortage. Recent findings suggest that the thermophilic sulfate-reducing archaeon Archaeoglobus fulgidus uses a novel alkylsuccinate synthase for long-chain alkane degradation, and the thermophilic Candidatus Syntrophoarchaeum butanivorans anaerobically oxidizes butane via alkyl-coenzyme M formation. In addition, gene expression data suggest that extremophiles produce energy via the glyoxylate shunt and the Pta-AckA pathway when grown on a diverse range of alkanes under stress conditions. Alkane degraders possess biotechnological potential for bioremediation because of their unusual characteristics. This review will provide genomic and molecular insights on alkane degraders under extreme conditions.
Background
Extremophiles are microorganisms that can not only survive but also prefer to grow under extremely harsh conditions (Rampelotto, 2013). Based on their habitat, extremophiles are classified as acidophiles (pH 0.5–5), alkaliphiles (pH 9–12), barophiles (approximately 0.1 MPa), psychrophiles (0°C–20°C), thermophiles (50°C–70°C), halophiles (5–20% salt). Extremophiles have evolved to possess special features for adapting to extreme conditions. For example, psychrophilic bacteria synthesize cold-shock proteins, which function as transcriptional enhancers and RNA-binding proteins, to protect RNA from numerous stressors (Wouters et al., 1999). In addition, many oligotrophs increase their surface area by synthesizing appendages for easy nutrient absorption and utilization of a wide range of substrates (Huyop and Cooper, 2012). Halophilic bacteria assimilate glycine betaine from the environment and produce osmoprotectants, such as trehalose or N-acetylglutaminylglutamine amide (Sagot et al., 2010).
n-alkanes, the major components of petroleum, can be metabolized aerobically or anaerobically by numerous microorganisms. Several alkane oxidation mechanisms have been documented till date, namely, terminal oxidation, subterminal oxidation, biterminal oxidation, and the Finnerty pathway. Alkane oxidation by aerobic bacteria produces fatty acids, which are subsequently metabolized via β-oxidation to generate acetyl-CoA (Figure 1). On the contrary, anaerobic alkane degradation is initiated by the addition of fumarate onto the terminal or subterminal carbon of alkane molecules, yielding alkylsuccinate derivatives as intermediates (Figure 1). Anaerobic alkane degradation was first proposed by So and Young, who demonstrated anaerobic alkane metabolism by fumarate- addition in the sulfate-reducing Desulfatibacillum alkenivorans AK-01 (So and Young, 1999; Rojo, 2009). Furthermore, recent studies also suggested that initial hydrocarbon activation by fumarate-addition occurs in syntrophic bacteria which produces acetate, CO2, and/or H2 that are utilized by methanogens (Berdugo-Clavijo and Gieg, 2014). Anaerobic hydrocarbon-degrading syntrophic bacteria possess genes encoding an alkylsuccinate (assA) or a benzylsuccinate synthase (bssA) as well as genes for syntrophic processes, such as H2 and acetate production (Washer and Edwards, 2007; Kato S. et al., 2009; Walker et al., 2009; Fowler et al., 2012; Aitken et al., 2013; Cheng et al., 2013). However, several questions regarding the mechanism underlying anaerobic alkane degradation persist, such as the nature of all key enzymes and metabolic pathways for methanogenic alkane degradation by syntrophic bacteria and methanogen (Tan et al., 2015a).
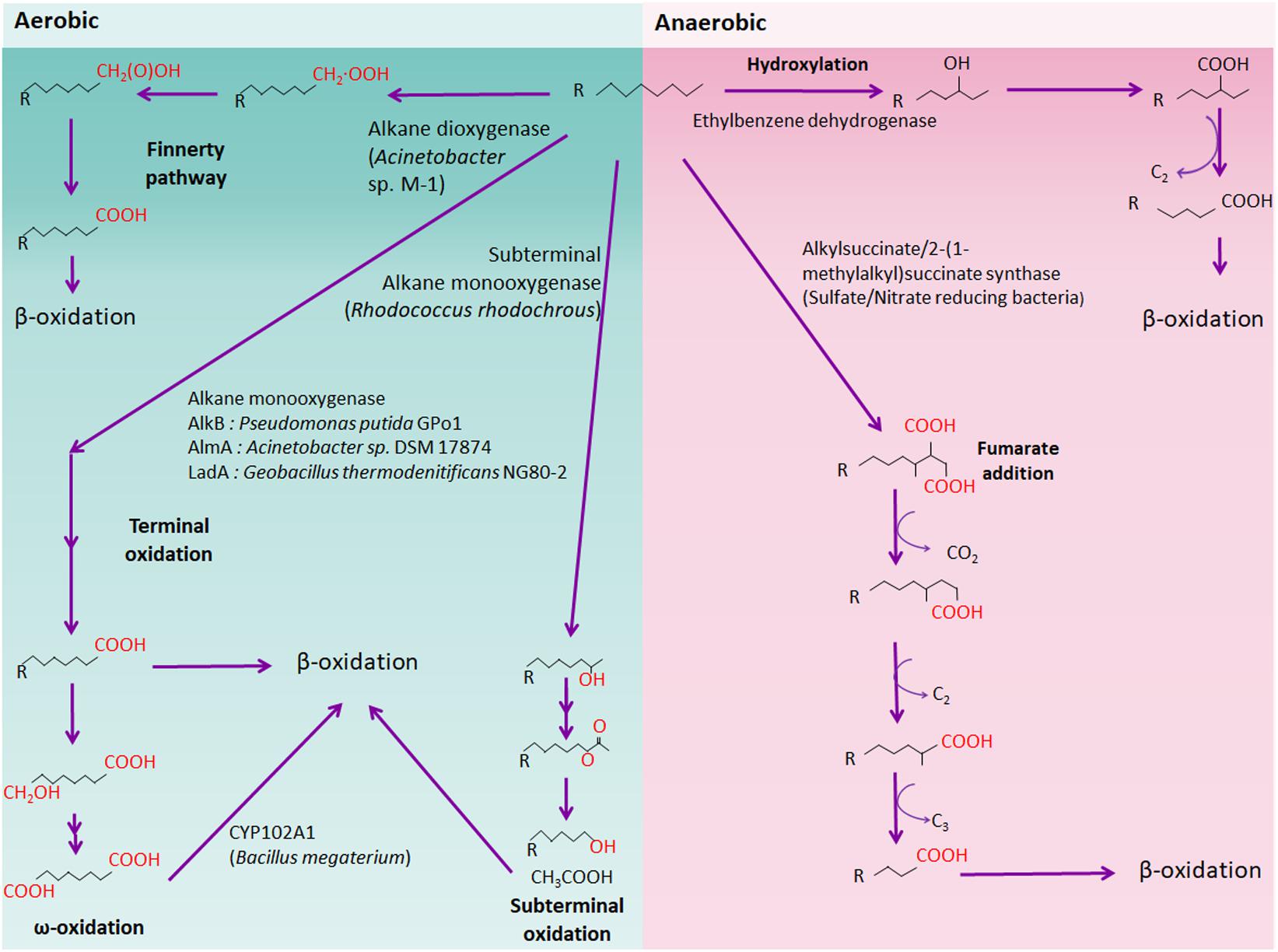
FIGURE 1. Schematic aerobic and anaerobic alkane degradation pathway. Aerobic (green background) and anaerobic (red background) alkane degradation pathways are shown. Each alkane oxidizer (e.g., alkane monooxygenase) is shown and representative bacteria are written in brackets.
Alkane monooxygenases have unique substrate ranges, which are classified into three categories based on the alkane chain length (van Beilen and Funhoff, 2007). Methane (C1) to butane (C4) conversion is catalyzed by methane monooxygenase (MMO)-like enzymes such as soluble MMO, particulate MMO, propane monooxygenase (PMO), and butane monooxygenase (BMO). In addition, integral membrane non-heme iron (AlkB) or cytochrome P450 enzymes (CYP153) oxidize pentane (C5) to hexadecane (C16), and alkanes longer than heptadecane are metabolized by a novel alkane dioxygenase, putative flavin-binding monooxygenase (AlmA), or long-chain alkane monooxygenase (LadA) (Maeng et al., 1996; Feng et al., 2007; Throne-Holst et al., 2007). However, a recent study showed that a novel Rieske-type alkane monooxygenase degrades pentane (C5) to tetracosane (C24) using NADH as a cofactor (Li et al., 2013). In addition, many bacteria possess multiple alkane hydroxylases (AH), and therefore, it is possible that a wide range of alkanes can be degraded by one microorganism (Whyte et al., 1998; Rozhkova-Novosad et al., 2007; Amouric et al., 2010; Liu et al., 2011; Park et al., 2017).
Biodegradation (transformation or mineralization) of a wide range of hydrocarbons, including aliphatic, aromatic, halogenated, and nitrated compounds, occurs in various extreme habitats. In addition, biodegradation of petroleum hydrocarbons by extremophiles has also been demonstrated under several conditions. For example, Rhodococcus sp. strain Q15, a psychrophile, showed short- and medium-alkane (C10 to C21) degradation at 5 °C. In addition, solid-phase microextraction–gas chromatography-mass spectrometry showed the co-appearance of 1-hexadecanol and 2-hexadecanol when strain Q15 utilized hexadecane, suggesting that Q15 assimilate alkanes via both the terminal and subterminal oxidation pathways (Whyte et al., 1998). A novel halophilic species, Amycolicicoccus subflavus, retains defensive genes against high salinity, osmotic stress, and poor nutrient availability, resulting in growth in the presence of 1–12% NaCl. Furthermore, genome analysis revealed that Amycolicicoccus subflavus possesses four AH (AlkB, CYP153, LadA, and PMO), which allows growth on C10 to C36 alkanes and propane (Nie et al., 2013). Thermophilic Geobacillus and Aeribacillus species, isolated from petroleum reservoirs and a hot spring, respectively, grew from 38°C to 68–70°Ñ, with maximum growth at 60°Ñ, and utilized C10–C30 alkanes as carbon source. Both strains are proven to have an alkB-type alkane monooxygenase. Two other Geobacillus strains (G. toebii B-1024 and 1017) also possess genes homologous to ladA and show activity similar to that of G. toebii B-1027 during long-chain alkane metabolism (Tourova et al., 2016).
Alkane-degrading extremophiles can be used in bioremediation of diverse oil-contaminated environments because of their special capabilities under extreme conditions. Cold-adapted hydrocarbon degraders have been applied to oil-polluted cold soil (Margesin et al., 2003; Aislabie et al., 2004; Wang J. et al., 2015) and wastewater (Margesin and Schinner, 1998; Gratia et al., 2009). In addition, psychrophiles from alpine habitats showed the highest degradation rate at 10°C within 8 days (40–60%) and at 4°C after 8 days (20–40%), showing high biodegradation efficiency (Margesin et al., 2000; Das and Chandran, 2011). Another well-studied example of bioremediation is the utilization of halophiles. Hydrocarbon biodegradation in the presence of high salinity is substantially valuable, not only from the economical aspect, but also in ecological and scientific studies. Recently, four Pseudomonas aeruginosa strains isolated from oil-contaminated saline soil were used to treat 10 g kg-1 of crude oil with more than 40% biodegradation efficiency in the presence of 300 mM NaCl after 120 days (Ebadi et al., 2017). Marinobacter hydrocarbonoclasticus showed high biodegradation activities of n-alkanes such as hexadecane (100%), eicosane (91%), and heneicosane (84%) in the presence of 4.6–20% NaCl (Gauthier et al., 1992). In another study, artificially weathered crude oil was proven to be degraded by microorganisms in a sandy salt marsh with a nitrogen–phosphorus–potassium (NPK) fertilizer (Lin et al., 1999). Several studies have demonstrated the feasibility of applying extremophiles to bioremediation and provided useful information for optimization of biodegradation efficiency.
However, despite the versatile potential of extremophiles in biodegradation, the molecular mechanisms and biological characteristics of these microorganisms have not been well-studied. This review discusses recent studies on alkane monooxygenase and 16S rRNA-based phylogenetic analysis of extremophiles, genotypic and phenotypic characteristics of pollutant-degrading extremophiles, and the ecological/advanced aspects of bioremediation. Overall, this review presents an updated overview of petroleum hydrocarbon degradation by microorganisms in different ecosystems.
Phylogenetic Affiliation and Distribution of Alkane-Utilizing Extremophiles
Extremophiles Possessing AlkB and CYP153 Family Alkane Hydroxylases
The genetic diversity of alkane hydroxylases has been extensively investigated under several conditions, including various oil-polluted and non-polluted environments. In particular, AlkB and members of the CYP153 family are widely distributed and have been extensively investigated in soil, marine environments, and groundwater (Sotsky et al., 1994; Knaebel and Crawford, 1995; Stapleton and Sayler, 1998; Luz et al., 2004; Kuhn et al., 2009).
Psychrophiles
Biodegradation of several petroleum components by indigenous cold-adapted microbial populations has been observed at low temperatures in hydrocarbon-contaminated Arctic and Antarctic soils. A large population of Pseudomonas species was observed in Arctic soil after diesel contamination. Furthermore, gene expression analysis confirmed that Pseudomonas and Rhodococcus species induce hydrocarbon degradation genes in Arctic biopile soils during bioremediation, indicating the importance of both species in oil-contaminated Arctic soil (Yergeau et al., 2012). DNA-based study showed that rhodococcal alkB genotypes (Rh alkB) are generally distributed in both contaminated and pristine Antarctic soils, whereas alkB from Pseudomonas putida (Pp alkB) was detected more in contaminated soils than in non-contaminated soils, implying that Rhodococcus spp. may be the predominant alkane-degradative bacteria in both polar soils, but Pseudomonas spp. may be enriched after contamination (Whyte et al., 2002). Populations of γ-proteobacteria such as Colwellia, Marinomonas, and Glaciecola appeared to be predominated in oil-contaminated Arctic fjord ice cores, (Brakstad et al., 2008). The 16S rRNA gene libraries of both non-contaminated and crude oil-contaminated seawater from sub-Antarctic areas showed the predominance of Roseobacter, Sulfitobacter, Staleya, Glaciecola, Colwellia, Marinomonas, Cytophaga, and Cellulophaga. However, non α- and γ- proteobacteria (𝜀-proteobacteria and Bacteroidetes) such as Arcobacter, Polaribacter, Ulvibacter, and Tenacibaculum started to appear when seawater was contaminated with hydrocarbons (Prabagaran et al., 2007). Therefore, α-, and γ-proteobacteria contribute maximally to alkane degradation in cold marine environments, whereas non α- and γ- proteobacteria have minor contribution.
Halophiles
The metagenomic database of a seawater sample from the Sargasso Sea (Venter et al., 2004) revealed the presence of alkB and cyp153 (van Beilen and Funhoff, 2007), implying wide distribution of halophilic and halotolerant alkane degraders in the ocean environment. Other PCR-based study examining the Atlantic Ocean surface seawater revealed that both alkB and cyp153 genes coexist in Alcanivorax and Salinisphaera species, whereas all Parvibaculum species possess only cyp153 and discovered new culturable alkane-degraders belonging to Brachybacterium, Idiomarina, Leifsonia, Martelella, Kordiimonas, Parvibaculum, and Tistrella (Wang W. et al., 2010; Supplementary Table S1). Characterization of culturable Alcanivorax strains, Marinobacter, Nocardioides, Parvibaculum strains, originating from deep-sea hydrothermal vents, also revealed that only Parvibaculum strain possesses an alkane-oxidizing cytochrome P450 (CYP)-like protein to degrade alkanes (Bertrand et al., 2013). In subtropical seawater, alkB was detected in Gallaecimonas, Castellaniella, Paracoccus, and Leucobacter species, which shows a completely different bacterial community compared to the pelagic area (Wang L. et al., 2010), indicating that the distribution of alkane degraders varies with ocean conditions and geographical location. The number of α- and γ-proteobacteria (dominated by Sphingomonadaceae, Rhodobacteraceae, and Chromatiales) increased when crude oil was spilled in the supratidal and intertidal zones, on the other hand, γ- and δ-proteobacteria were more abundant in subtidal zones. The phylum Actinobacteria, particularly the genus Rhodococcus, was a key player in microbial response to the spillage especially in the supratidal sediment (Acosta-González et al., 2015). Likewise, no alkB sequence from Actinobacteria was detected in the marine metagenome despite a high proportion of alkB sequences from Actinobacteria in terrestrial and freshwater metagenomes (Nie et al., 2014).
Acidophiles
Although information on acidophilic alkane degraders is limited, some studies on alkane degradation at low pH have indicated the existence of hydrocarbon-degrading acidophiles. 16S rRNA library analysis of acidic soil from natural hydrocarbon seeps (pH 2.8–3.8) showed that majority of sequences were from heterotrophic acidophilic bacteria, including Acidisphaera and Acidiphilium species of α-proteobacteria, as well as the iron-and sulfur-oxidizing chemolithotroph Acidithiobacillus strain. A novel Acidisphaera-related strain C197 possesses alkB homologs (92.5% similarity with an alkB fragment from Xanthobacter flavus) and performs hexadecane metabolism (Hamamura et al., 2005). Mycobacterium is also a predominant bacterial genus found in extreme acidic sulfur block environments and alkane degradation of strain AGS10, an acidophilic Mycobacterium, was demonstrated. Interestingly strain AGS10 has both alkB and cyp153 genes, implying the possibility of two alkane hydroxylase families in many acidophilic Mycobacteria strains (Ivanova et al., 2014).
Extremophiles Possessing AlmA and LadA Family Alkane Hydroxylases
Flavin-binding family alkane hydroxylase (AlmA) and long-chain alkane monooxygenase (LadA) have recently been investigated; however, little is known about the distribution of either gene in the environment. Studies on functional diversity of alkane hydroxylase genes, including almA and ladA homologs, provided evidence that some psychrophiles possess AH for long-chain alkane degradation. Alignments with combined Pfam data sets showed the presence of almA homologs in Paraglaciecola psychrophila 170 (five copies) and Psychrobacter cryohalolentis K5 (one copy). On the contrary, a ladA candidate appears in the genomes of Octadecabacter antarcticus 307, Octadecabacter arcticus 238, and Terriglobus saanensis SP1PR4 (Bowman and Deming, 2014). These results hint at the occurrence of long-chain alkane metabolism at low temperatures, although the functionality of almA and ladA genes has not been shown experimentally. The diversity of almA in marine bacteria and oil-enrichment bacterial communities was investigated in three surface seawater samples from the South China Sea, Indian Ocean, and Atlantic Ocean (Wang and Shao, 2012). The acquired DNA sequences suggested two clades of almA gene. Class I almA contained a few sequences represented by the Salinisphaera and Parvibaculum genera, whereas class II was larger and more diverse and had many proteobacteria-related sequences, mainly from Alcanivorax and Marinobacter. However, metagenomic studies for detecting ladA in marine or saline conditions have not been conducted so far, although several marine hydrocarbonoclastic bacteria and halophilic hydrocarbon-utilizing bacteria harboring either AlmA- or LadA-type homologs, or both, have been reported (Liu et al., 2011; Nie et al., 2013; Meier et al., 2016). Thus, further investigation of long-chain alkane metabolism, especially the involvement of almA and ladA, under conditions of high salinity, are warranted.
Plasmid-born ladA gene was first investigated in Geobacillus thermodenitrificans NG80-2, indicating high possibility of horizontal gene transfer (HGT) of the ladA gene (Feng et al., 2007; Li et al., 2008). However, other member of Geobacillus, G. thermoleovorans B23 harbors three genes encoding LadA-type alkane hydroxylases (ladAαB23, ladAβB23, and ladBB23, collectively named the ladAB region) on its chromosome. Comparative genome analysis of relevant Geobacillus strains confirmed the absence of the ladA gene on the chromosome of G. thermodenitrificans NG80-2, but the presence of the ladAB gene island in many other Geobacillus strains (Boonmak et al., 2014).
We retrieved whole genome data of extremophiles from the National Center for Bio-technology Information (NCBI)1 and Integrated Microbial Genomes (IMG)2 databases. Total 66 identified genomes were aligned and analyzed (Supplementary Table S1). alkB genes are widely distributed in aerobic alkane-degrading extremophiles (psychrophiles, halophiles, acidophiles, and thermophiles), although their association with HGT was not clear; nonetheless, the different evolution patterns of cyp and 16S rRNA, shown in Figure 2, indicate the occurrence of HGT. Interestingly, alkB, cyp, and almA were mostly detected in psychrophiles, whereas halophiles and thermophiles possess ladA (Figure 3). In addition, acidophilic alkane degraders possess only alkB, which is not present in the genome of thermophilic alkane degraders (Figure 2 and Supplementary Table S1).
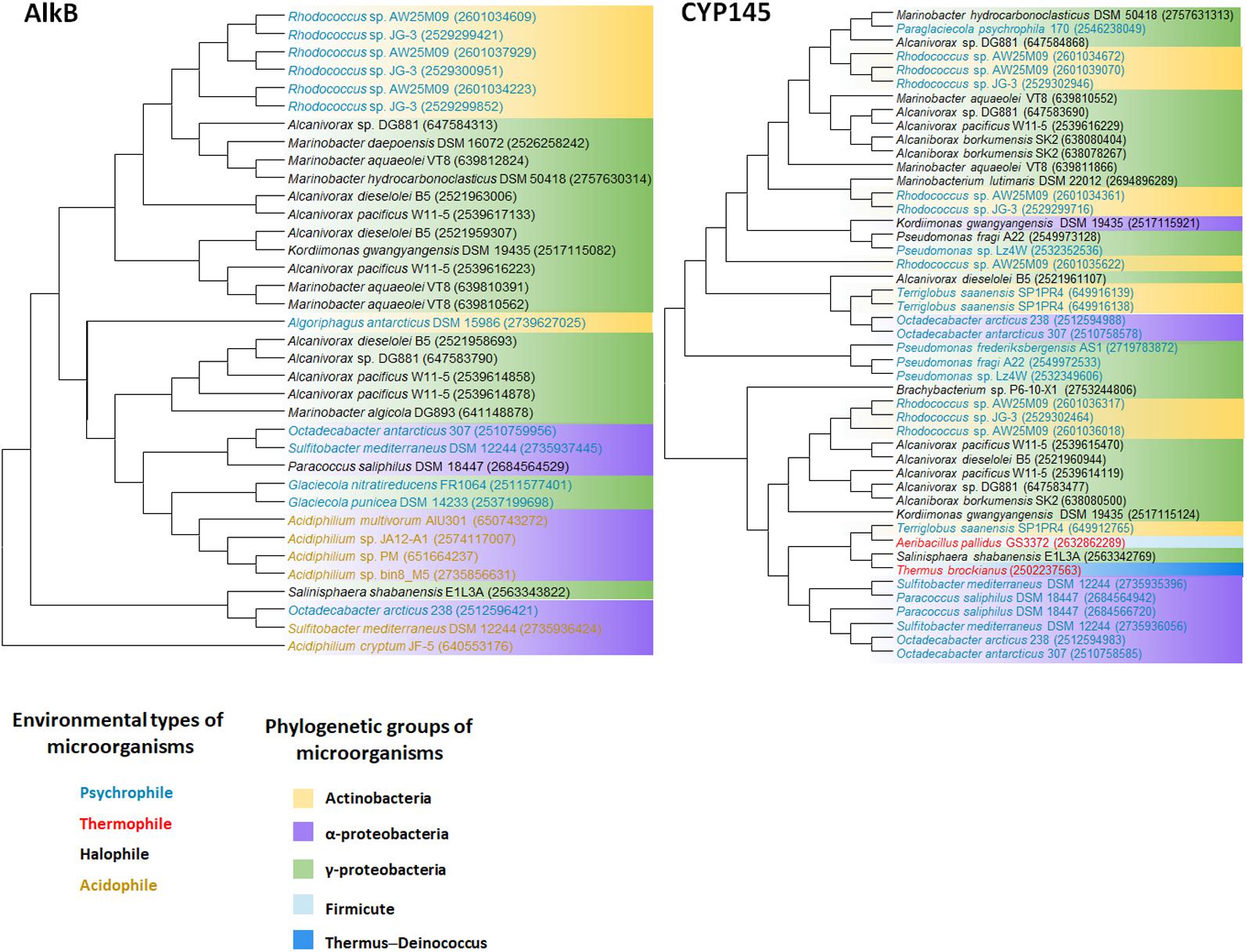
FIGURE 2. AlkB and Cytochrome P450 (CYP145) phylogenetic tree of extremophiles. Collected sequences were aligned and phylogenetic tree was drawn using the MEGA 6 program. Colored words indicate the following environmental types of microorganisms: blue, psychrophile; red, thermophile; black, halophile; yellow, acidophile. Colored backgrounds indicate the following phylogenetic groups: yellow, Actinobacteria; purple, Alphaproteobacteria; green, Gammaproteobacteria; light blue, Firmicute; dark blue, Thermus–Deinococcus.
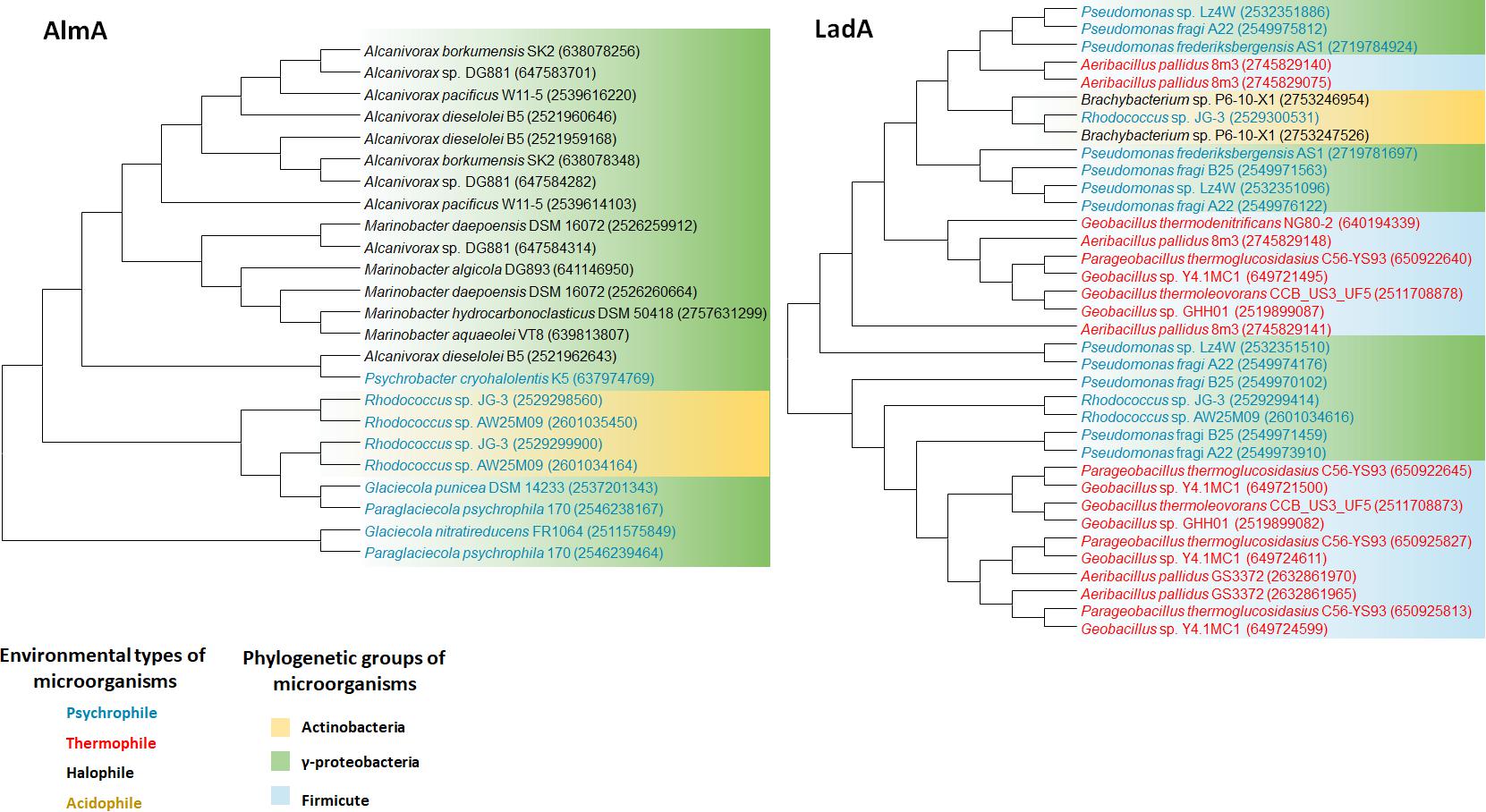
FIGURE 3. AlmA and LadA phylogenetic tree of extremophiles. Collected sequences were aligned and phylogenetic tree was drawn using the MEGA 6 program. Colored words indicate the following environmental types of microorganisms: blue, psychrophile; red, thermophile; black, halophile; yellow, acidophile. Colored backgrounds indicate the following phylogenetic groups: yellow, Actinobacteria; purple, Alphaproteobacteria; green, Gammaproteobacteria.
Methanotrophic and Anaerobic Extremophiles
Anaerobic methane oxidation (AOM) via reverse methanogenesis commonly occurs in anaerobic methane-oxidizing archaea (ANME) under anoxic environments, and was first identified in marine sediments where AOM was coupled to sulfate reduction. Therefore, ANME usually formed a community with sulfate-reducing bacteria (SRB) of δ-proteobacteria. ANME are generally classified into three categories: ANME-1 (sub-clusters a, and b) related to Methanomicrobiales and Methanosarcinales, ANME-2 (sub-clusters a, b, and c) related to cultivated members of Methanosarcinales, and ANME-3 related to Methanococcoides spp. (Timmers et al., 2017). However, all mcr genes (encoding methyl-coenzyme M reductase) in the class Methanomicrobia, such as Methanoculleus, Methanoregula, and Methanosaeta showed over 60% identity each other, which is indicative of its high conservation (Figure 4). Compared to samples containing only methane, samples from natural freshwater gas sources containing methane and sulfate, harbored ANME-2a/b such as Methanolobus, and AOM-associated archaea such as Candidatus Methanoperedens nitroreducens, as determined by archaeal 16S rDNA analysis (Timmers et al., 2016).

FIGURE 4. AssA and Mcr phylogenetic tree of extremophiles. Collected sequences were aligned and phylogenetic tree was drawn using the MEGA 6 program. Colored backgrounds indicate the following phylogenetic groups: pink, Deltaproteobacteria; purple, Metanomicrobia (archaea); green, Archaeoglobi (archaea).
Methanogenic hydrocarbon-degrading δ-proteobacteria (Syntrophus/Smithella, Desulfosarcina/Desulfococcus), Firmicutes (Desulfotomaculum, Desulfosporosinus, and Pelotomaculum), and relatively few bacteria belonging to Spirochaetes, Bacteroidetes, Chloroflexi, and β-proteobacteria have been identified in hydrocarbon-contaminated aquifers (Tan et al., 2015a). In addition, previous reports showed that the γ-proteobacterial genus Marinobacter, and δ-proteobacterial genera Syntrophus and Smithella, are the predominant hydrocarbon degraders in samples from estuarine sand and oil sand tailings under methanogenic conditions (Gray et al., 2011; Siddique et al., 2012). Hydrocarbon degradation in marine environments is predominantly conducted by SRB, and on-site stable isotope probing revealed that Desulfosarcina/Desulfococcus (DSS) of δ-proteobacteria actively oxidize short- (butane) and long-chain (dodecane) alkanes. Although few SRB alkane degraders have been identified, several culture-dependent and independent studies at oil-contaminated seeps have indicated the presence of more diverse species than the currently known SRB (Kleindienst et al., 2014). In experimental condition, cultures enriched from oil sands tailings ponds included predominant δ-proteobacteria, such as Desulfoglaeba, Desulfobacteraceae, and Geobacter when short-chain alkane-degrading culture with a mixture of C6, C7, C8, and C10 n-alkanes as predominant organic carbon sources was transferred into sulfate-supplemented medium (Tan et al., 2015b).
Survival Strategies of Extremophiles Under Harsh Condition
Psychrophiles
Many DNA-binding proteins are present in extremophiles such as psychrophiles. For example, cold-shock proteins (Csp) play essential roles in DNA or RNA stabilization and membrane rigidity by regulating unsaturated fatty-acid synthesis (D’Amico et al., 2006). Interestingly, Methanogenium frigidum, a stenopsychrophilic archaea, possesses csp family of genes, which are generally not detected in archaeal (such as thermophilic and hyperthermophilic) genomes (Giaquinto et al., 2007). The Csp sequences of M. frigidum and Escherichia coli, are highly similar, and complementation of csp-knockout E. coli strain with csp gene from M. frigidum, results in growth recovery at low temperature, showing compatibility between archaeal and bacterial Csps. We observed that Pseudomonas fragi strains A22 and B25 (Supplementary Table S1) have the most abundant csp genes (6 genes) among putative alkane-degrading psychrophiles. However, six csp genes were also present in the genome of the mesophilic alkane degrader Acinetobacter oleivorans DR1. Thus, it is assumed that other strategies facilitate cold adaptation in psychrophiles regardless of the number of csp genes.
Most cold-adapted proteins in psychrophiles also show other characteristics, including high structure flexibility, low proportion of acyl chains in the cell membrane, and increased protein volume (Siddiqui and Cavicchioli, 2006). Chemical and biological reactions are slower at low temperatures, and therefore, the synthesis of cold-active enzymes with higher activity than mesophilic enzymes are required for maintaining an appropriate rate for essential metabolic reactions. To achieve high enzyme activity in cold conditions, the catalytic center must be flexible and unstable. Therefore, cold-adapted enzymes are heat labile and possess lower Gibbs free energy (ΔG) than mesophilic or thermophilic enzymes (Feller and Gerday, 2003; Siddiqui and Cavicchioli, 2006). Comparative sequence alignments of putative alkane hydroxylases from mesophiles and psychrophiles revealed that substitution by specific amino acids, such as alanine or glycine, contributes to increased flexibility of loops, bends, and α-helical structures of psychrophilic alkane hydroxylases. Furthermore, reports show that the flexible loops, bends, and helical regions of P450 from Glaciecola psychrophila are located in the center of three methionine residues, which is known to act as an alternative heme-binding site at low-temperatures (Bowman and Deming, 2014).
Trehalose production plays significant roles in resistance to freezing in cold environments. Observations from neutron diffraction, Raman spectroscopy, and inelastic neutron scattering revealed that trehalose disrupts the tetrahedral intermolecular network of water formed at low temperatures and thereby protects from ice damage. Image analysis based on field-emission transmission electron microscopy showed that trehalose was better than sucrose in preventing ice formation (Magazù et al., 2012). Currently, only psychrophilic Rhodococcus is known to produce trehalose lipid as a biosurfactant during alkane degradation (Rapp and Gabriel-Jürgens, 2003). However, there is a high possibility of trehalose lipid production by psychrophilic alkane degraders as trehalose synthesis can be induced by low temperature (Phadtare, 2004). Taken together, trehalose can be utilized as a cryoprotectant or biosurfactant by psychrophiles under cold conditions.
Halophiles
Many halophilic microorganisms develop strategies for surviving in highly saline environments, such as transportation of inorganic ions (mainly potassium) for stabilizing external osmotic pressure or synthesis of metabolism-compatible osmolytes. Genome and expression analyses of the moderately halophilic alkane degrader, Amycolicicoccus subflavus DQS3-9A1T showed upregulation of the KdpD/KdpE two component system (high-affinity K+ uptake system), Trk-type K+ transporter and Na+/K+ antiporter, MtrA/MtrB two component system (BetP regulator), and glycine/betaine transporter (BetP) in response to high salt stress (Nie et al., 2013). Trk-type K+ transporter, one of main K+ transporters, requires three genes for cellular K+ uptake, namely, trkA (1,374 bp, encoding cytoplasmic NAD+/NADH binding protein), trkH (1,449 bp, encoding Trk transporter), and trkI (1,479 bp, encoding Trk transporter). Genome data show that several halophilic alkane degraders, such as Paracoccus saliphilus, Marinobacterium lutimaris, Salinisphaera, and Marinobacter hydrocarbonoclasticus, possess multiple Trk potassium uptake proteins, implying that Trk transporter provides osmolarity balance to halophiles during alkane degradation under high salinity conditions.
Several classes of organic compatible solutes, such as amino acids, alcohols, sugars, and derivatives act as organic osmoprotectants, which can be classified into three chemical categories: zwitterionic, non-polar, and anionic solutes (Roberts, 2005). These organic osmoprotectants are regulated by extracellular salt concentration. The synthesis rate of trehalose in Haladaptatus paucihalophilus decreases with increase in salinity; on the contrary, the intracellular concentration of glycine betaine increases due to import at high salinities, suggesting dynamic regulation of two compatible solutes for osmoadaptation (Youssef et al., 2014). The obligate hydrocarbon bacterium, Alcanivorax borkumensis SK2, synthesizes and accumulates ectoine to maintain osmolality under hyperosmosis, which promotes active metabolism and maintains cell integrity albeit low hydrocarbon degrading performance, thereby protecting cells from damage due to hyperosmosis (Scoma and Boon, 2016).
Undesirable interactions that destroy internal microbial proteins by dehydration can be avoided by controlling net charge. Compared to non-halophilic proteins, halophilic proteins contain several glutamate and aspartate residues on their surfaces, which provides high water solubility in the presence of salinity (Edbeib et al., 2016). Although comparative sequence alignment and structural studies of halophilic alkane hydroxylases have not been performed so far, we speculate that large numbers of glutamate and aspartate on their surfaces distributed to the stabilization of alkane hydroxylases.
Acidophiles
Specific adaptation to and metabolism of alkane degradation have not been extensively studied in acidophiles. Acidophiles possess several systems for surviving in highly acidic conditions (Baker-Austin and Dopson, 2007), such as a cytoplasmic buffering, DNA protection and repair, blocking of extracellular proton uptake, and organic acid utilization (Babu et al., 2015). Buffer capacity in the cytoplasm is determined mainly by amino acids, such as glutamate and arginine, which sequester protons. An acid-resistant E. coli strain showed that decarboxylated amino acids which require intracellular proton consumption enhanced internal pH, resulting in maintenance of pH homeostasis. However, the buffering capacities of acidophilic bacteria Bacillus acidocaldarius and other neutrophilic bacilli were not significantly different, suggesting that the survival mechanism encompasses several ways via which acidophilic bacteria adapt to low pH (Richard and Foster, 2004).
A large number of DNA and protein repair genes in acidophiles contribute to preservation of pH homeostasis (Dopson et al., 2005). A DNA-binding protein in starved cells (Dps), which protects DNA from reactive oxygen species (ROS) generated by the Fenton reaction, plays a significant role when cells are exposed to oxidative and nutritional stress. Studies showed that the E. coli O157:H7 wild-type strain survives better than dps::nptI mutants in the initial 1 h of exposure to acidic conditions. Furthermore, complementation of wild type dps gene to dps::nptI mutants restored survival (Choi et al., 2000). However, available genome data showed that Dps is not present in Acidiphilium, suggesting that Dps synthesis does not occur by all alkane-degrading acidophiles for surviving under acidic conditions.
Upregulation of an outer membrane porin (Omp40) in Acidithiobacillus ferrooxidans was observed when pH was shifted from 3.5 to 1.5 (Amaro et al., 1991). Further studies illustrated that the positively charged L3 loop of the porin protein controls pore size and ion selectivity at pH 2.5 (Guiliani and Jerez, 2000). In addition, the decrease in membrane fluidity of E. coli O157:H7 may confer resistance to protons under acidic conditions (Yuk and Marshall, 2004). When lipids of acidophilic strains were adapted to acidic condition, palmitic acid (C16:0) concentrations increased, whereas that of cis-vaccenic acid (C18:1ω7c) decreased, resulting in reduced membrane fluidity, which regulated porin pore size. Low membrane fluidity confers resistance to toxicity from hydrocarbons and acidic condition, but compromises alkane uptake.
Thermophiles
Most proteins with catalytic and regulatory functions in thermophiles are thermostable and are structurally more compact than their mesophilic counterparts. Hyperthermostable proteins in Thermococcus onnurineus NA1, hydperthermophilic archaea, incubated at 100°C include intracellular protease I, thioredoxin reductase, triosephosphate isomerase, putative hydroperoxide reductase, proteasome, and translation initiation factors (Yun et al., 2011). Furthermore, transcriptomic and proteomic analyses of thermophiles revealed that many thermostable proteins of certain categories [heat stable proteins, chaperonin, metalloenzyme, putative ribosomal associated proteins, and superoxide dismutase (SOD)] are translated at high temperatures, implying that they are essential for thermophile survival at those temperatures (Li et al., 2010; Shih and Pan, 2011; Wang W. et al., 2014). These proteins have large numbers of disulfide bonds and positively charged amino acids, such as lysine and arginine, which are critical for their stability (Brocchieri, 2004; Beeby et al., 2005; Ma et al., 2010). Additionally, in silico studies illustrated that disulfide bonds are ubiquitous among thermophiles. BLAST revealed the presence of nine genes encoding FAD-dependent pyridine nucleotide-disulfide oxidoreductase, a potential key enzyme in thermophilic intracellular disulfide-bond formation, in the thermophilic alkane degrader Geobacillus sp. Y4.1MC1 (Beeby et al., 2005). Increase in the number of small residues such as Gly, Ala, Ser, and Val, and decrease in the number of Cys and polar residues (Asp, Asn, Glu, Gln, and Arg) were observed in membrane proteins of many thermophiles, which increases stringent membrane hydrophobicity at high temperatures (Meruelo et al., 2012). The amino acid preference might occur in membrane proteins of thermophilic alkane degraders.
Proteome analyses in most thermophiles has demonstrated the presence of genes encoding highly thermostable proteins such as antioxidants, antitoxins, heat-shock proteins (Hsps), and enzymes involved in carbon metabolism pathways, including glycolysis (Wang Q. et al., 2015). Several antioxidants and antitoxins, such as the VapBC complex, are translated in response to high-temperature stress (Shih and Pan, 2011). Global transcriptomic analysis demonstrated that VapBC proteins, the virulence-associated proteins in the hyperthermophilic crenarchaeon Sulfolobus solfataricus, are induced at elevated temperatures ranging from 80–90°C. Furthermore, a ΔvapBC S. solfataricus knockout strain showed a highly altered transcriptomic profile and susceptibility to heat shock (Cooper et al., 2009). However, S. solfataricus VapBC homologues do not exist in any thermophilic alkane degrading bacteria (Geobacillus species, Thermus brockianus, and Aeribacillus pallidus) and archaea (Archaeoglobus fulgidus DSM 4304), indicating that the VapBC system is not a ubiquitous heat-shock defense mechanism. Small Hsps (sHSPs) bind to proteins and confer protection from denaturation under high temperatures (Li et al., 2012). Unlike Csp proteins, Hsp proteins are abundant in many thermophilic alkane degraders. Five Hsp-encoding genes are present in the Geobacillus thermodenitrificans NG 80-2 genome, and most Geobacillus species possess more than four genes. Thus, Hsp proteins in alkane-degrading thermophiles may play an important role in survival at high temperatures.
Energy Production of Extremophiles During Alkane Metabolism
Most alkane-degrading extremophiles have ubiquitous aerobic or anaerobic alkane oxidation pathways represented by ω-oxidation or fumarate addition, respectively (Figure 1). It is assumed that most aerobic psychrophiles, halophiles, acidophiles, and thermophiles degrade alkanes through general alkane oxidation pathways, as demonstrated by the possession of AlkB, CYP, AlmA, and LadA-type alkane hydroxylases (Bertrand et al., 2013; Nie et al., 2013; Ivanova et al., 2014). However, some extremophiles oxidize alkanes via a unique pathway and produce energy via alternative pathways to avoid stress under harsh conditions.
Novel Aerobic Alkane Degradation Systems in Extremophiles
The aerobic alkane degrader, Dietzia DQ12-45-1b, grown at 4–45°C, pH 6.0–12.0, and 0–20% (w/v) NaCl (Wang et al., 2011), possesses an alkW1-alkX system, which is co-expressed and induced by fatty acids (Liang et al., 2016). The proposed mechanism is that alkX, a TetR family regulator, competes with DNA polymerase at the alkW1 promoter region and represses the expression of alkW1, an alkB-type alkane hydroxylase-rubredoxin fusion gene, in the absence of alkanes. However, fatty acids produced by alkane oxidation directly bind to AlkX, removing it from the alkW1 promoter when alkanes are present, indicating that this system is energy-cost effective. Furthermore, phylogenetic and gene alignment analyses showed that alkW1-alkX may be ubiquitous in Actinobacteria such as Rhodococcus and Mycobacterium. Thus, alkW-alkX system confer benefits to Actinobacteria living in harsh conditions characterized by high pH, osmotic stress, and lack of nutrients (Liang et al., 2016).
Recent genome sequencing analysis revealed that the moderate halophile, Amycolicicoccus subflavus DQS3-9A1T possesses four different alkane hydroxylation systems (propane monooxygenase, AlkB, CYP, and LadA) (Nie et al., 2013). A previously described halophile, Alcanivorax dieselolei B-5, harbors multiple copies of alkane hydroxylases (two AlkB-, one CYP153-, and AlmA-type alkane hydroxylase) and showed broad range of alkane-degrading capability (C5–C36) (Liu et al., 2011). Furthermore, the available complete genome sequence of Marinobacter aquaeolei VT8 shows the existence of three AlkB-, two CYP153-, and one AlmA-like alkane hydroxylase enzymes in the VT8 genome. Although the evolutionary reasons for possession of multiple alkane hydroxylase systems are not yet clear, it probably facilitates utilization of diverse hydrocarbon sources by halophilic bacteria under high saline environments. Illumination and addition of casamino acid as an organic fertilizer were tested to measure biodegradation efficiency of the extremely halophilic archaeal strains, Haloferax, Halobacterium, and Halococcus in highly saline soil (>22% salinity) and pond water (>16% salinity), which resulted in significantly enhanced biodegradation by halophilic archaea in the presence of casamino acid and illumination (Al-Mailem et al., 2012). It was hypothesized that archaea in hypersaline environment can synthesize ATP using a red pigment, perhaps a bacteriorhodopsin-like system, under low oxygen tension. Casamino acid also confers better growth and degradation of halophilic archaea, indicating promotion of biodegradation efficiency by addition of organic nitrogen source. Further studies are required to understand enhanced hydrocarbon degradation by haloarchaea in the presence of light and N source.
Alternative Pathways Producing Energy in Extremophiles
The glyoxylate shunt in the mesophilic alkane-degrading bacteria, Acinetobacter oleivorans DR1, is activated to generate energy during triacontane metabolism (Park et al., 2017). The glyoxylate shunt requires isocitrate lyase (encoded by aceA) and malate synthase (encoded by aceB or glcB), which are activated under oxidative stress or in the presence of C2 carbon sources (Ahn et al., 2016). Among psychrophiles, upregulation of glyoxylate cycle-related genes in Nesterenkonia sp. AN1, isolated from Antarctic soil, was observed at 5°C. Furthermore, a large number of glyoxylate shunt-related genes belonging to the psychrophilic bacteria Colwellia and Neptuniibacter were observed in samples from the area surrounding the Deepwater Horizon oil spill (Rivers et al., 2013). In Colwellia maris, the expression level of icl was higher at 0°C than at 15°C (Watanabe et al., 2002), implying that Colwellia possess the advantage of surviving in cold environment by activating the glyoxylate shunt during alkane degradation.
Little is known about complete energy producing metabolism of alkane-degrading psychrophiles. The presence of methylglyoxal synthase in psychrophilic bacteria, Exiguobacterium sibiricum, was proposed as an important alternative catabolic pathway for glyceraldehyde phosphate (Rodrigues et al., 2008). In addition, the glyoxalase family proteins in Planococcus halocryophilus Or1 have been emphasized as key enzymes for utilization of carbon sources under cold environments. In addition, repression of energy metabolism at low temperature (-15°C) in Planococcus halocryophilus Or1, but increased expression of succinic semialdehyde dehydrogenase, alcohol dehydrogenase, and several oxidoreductases indicate maintenance of energy metabolism and ATP levels (Mykytczuk et al., 2013). Similarly, transcriptomic analysis of Pseudomonas extremaustralis showed activation of the ethanol oxidation pathway, involving a pyrroloquinoline quinone (PQQ)-dependent ethanol dehydrogenase, cytochrome c550, and an aldehyde dehydrogenase (encoded by exaA, exaB, and exaC, respectively) although genes associated with tricarboxylic acid (TCA) and cytochrome synthesis are repressed under cold conditions (Tribelli et al., 2018). Proteomics study suggested that enzymes participating in the fatty acid degradation pathway (β-oxidation) were more abundant at low temperatures (4–10°C) in cold adapting marine bacterium, Sphingopyxis alaskensis under artificial seawater medium (Ting et al., 2010). Cold adaptation strategy which uses fatty acid metabolism was also observed in Pseudomonas putida KT2440 at 10°C in Luria-Bertani (LB) medium. Analyses of proteomics and RNA sequencing data in KT2440 strain showed upregulation of the 2-methylcitrate and branched amino acid degradation pathways under cold temperature, implying conversion of propionate or propionyl-CoA to succinate and pyruvate via 2-methyl citrate under cold condition (Fonseca et al., 2011).
Proteomic analysis of the halophilic bacteria, Alcanivorax borkumensis SK2, also showed upregulation of AceA (ABO_2741) and GlcB (ABO_1267) and downregulation of TCA cycle enzymes, including isocitrate dehydrogenase (icd, ABO_1281) and 2-oxoglutarate dehydrogenase (lpdG, ABO_1494), when bacteria were grown in hexadecane-supplemented media, suggesting that the glyoxylate shunt is the main pathway for hexadecane metabolism (Sabirova et al., 2006). Similarly, another strategy for adapting to high temperatures involves a shift in the carbohydrate metabolism pathways. Transcriptomic and proteomic analysis of Thermus filiformis at 63°C, 70°C, or 77°C revealed that oxidative stress generated by high temperature induced genes related to the pentose phosphate (PP) pathway, whereas genes participating in glycolysis and the TCA cycle were downregulated (Mandelli et al., 2017), indicating evasion of ROS generation. In addition, metabolomic analysis revealed accumulation of ROS scavengers such as oxaloacetate and α-ketoglutarate, indicating preservation of ROS homeostasis. It has been reported that the PP pathway, glyoxylate cycle, and ROS defense systems are essential for detoxifying petroleum hydrocarbons (Feng et al., 2007; Kato T. et al., 2009; Shih and Pan, 2011). Thus, exposure to heat shock probably assists the growth of thermophilic alkane degraders at high temperatures in the presence of alkanes. The same pattern was observed in the genomic and proteomic analysis of thermophilic bacteria Geobacillus thermodenitrificans NG80-2, which showed activated glyoxylate shunt instead of the TCA cycle in the presence of hexadecane (Feng et al., 2007).
The interface between alkane degraders and hydrocarbon compounds may form microoxic or anoxic niches because of hydrophobicity (Suter et al., 2017). In addition, alkane oxidation generates excessive acetyl-CoA (Sabirova et al., 2006; Schönfeld and Wojtczak, 2016; Park et al., 2017), creating appropriate conditions for aerobic fermentation, which is also called the Crabtree effect (Zhou et al., 2017). Acetate assimilation via the Pta-AckA pathway occurs in the presence of long-chain alkanes (triacontane), producing ATP (Park et al., 2017). Although studies on the Pta-Ack pathway during alkane metabolism are limited, evidence shows that extremophiles can synthesize ATP via the Pta-AckA pathway. Phenotypic analysis of Psychrobacter arcticus 273-4 shows that acetic acid can be diffused into cells without energy consumption, whereas energy-consuming glucose could not be utilized at low temperatures (Bergholz et al., 2009). In addition, transcriptomic and proteomic analysis of Psychrobacter sp. PAMC 21119, which was grown under cold conditions, revealed upregulation of acetyl-CoA metabolism, although the central energy production and conversion pathways were downregulated (Koh et al., 2017). Geobacillus thermodenitrificans NG80-2 harbors pta and ackA, which are indicative of possible methods of ATP synthesis during alkane assimilation (Feng et al., 2007). Methanogenic archaea and bacteria also possess pta and ackA genes. Furthermore, methanogenic and halophilic archaea have the same ancestral Pta lineage (Barnhart et al., 2015), implying the possibility of acetate production via the Pta-AckA pathway during alkane metabolism at high salinity.
Novel Alkane Degradation Pathways Under Anaerobic Conditions
Although many alkane hydroxylase genes involved in aerobic alkane degradation were detected in extremophiles, anaerobic alkane metabolism may have advantages, particularly because extreme environments confer oxidative stress on bacteria. The thermophilic sulfate-reducing archaeon, Archaeoglobus fulgidus strain VC-16, which degrades C10–C21 alkanes with thiosulfate or sulfate as a terminal electron acceptor, possess pyruvate formate lyase (encoded by pflD), which is homologous to alkylsuccinate synthase (encoded by assA). Phylogenetic analysis showed that homologs of PflD and PflC mainly belong to bacteria such as Firmicutes, δ-proteobacteria, γ-proteobacteria, and Actinobacteria, indicating that these genes are acquired from bacteria via HGT. Furthermore, three-dimensional simulation suggested that PflD might be a potential alkylsuccinate synthase, which is supported by the upregulation of pflD gene expression in the presence of hexadecane but not fatty acid (Khelifi et al., 2014). An anaerobic hyperthermophile, Geoglobus acetivorans, isolated from deep-sea hydrothermal vents, requires ferric iron (Fe (III)) as an electron acceptor during alkane degradation. Genome analysis revealed that this archaeon also harbors close homologs of Archaeoglobus fulgidus PflC and PflD (69 and 57% identity, respectively). However, Gace_0240 (homolog to PflC in Archaeoglobus fulgidus) shares features with alkyl and benzylsuccinate synthases, rather than with pyruvate formate lyase-activating enzymes (Mardanov et al., 2014).
A methanogenic culture, enriched from freshwater hydrocarbon-contaminated aquifer sediments, consumed octacosane (C28) and generated methane. Smithella was predominant and assA was abundant in the presence of octacosane, furthermore, phylogenetic analysis showed that assA copies in the culture were closely related to assA of Smithella, indicating that fumarate addition in octacosane is mainly performed by Smithella. Interestingly, the detection of α,ω-dicarboxylic acids in the octacosane-only media implies simultaneous activation by fumarate addition, although the mechanism remains unclear (Oberding and Gieg, 2017). Another alkane degradation pathway has been suggested in Verrucomicrobia methylotrophs, which grow optimally at pH 2.0–2.5 and consume methane. Interestingly, analysis of the draft Verrucomicrobia genome detected genes encoding pMMO, which is homologous to the pMMO in methanotrophic proteobacteria. However, genes for subsequent methanol and formaldehyde oxidation were incomplete or missing, suggesting that the bacterium uses novel methylotrophic pathways (Dunfield et al., 2007). Further analysis showed that three pmoA genes in Verrucomicrobia were located in a different clade of proteobacterial pmo homologs, indicating divergent evolution with proteobacterial methanotrophs. Recent metatranscriptome, metaproteome, and metagenome analyses of Candidatus Syntrophoarchaeum butanivorans showed that genes encoding β-oxidation enzymes, carbon monoxide dehydrogenase, and reversible C1 methanogenesis enzymes [reverse methyl coenzyme M (Mcr)] are also expressed, indicating that enzymes for complete butane metabolism exist in Ca. Syntrophoarchaeum butanivorans. Furthermore, ultra-high resolution mass spectrometry data showed that Ca. Syntrophoarchaeum butanivorans generates alkyl-coenzyme M. Phylogenetic analysis showed that MCR was present in the draft genomes of Bathyarchaeota and pan-genome of uncultivated Hadesarchaea, indicating that uncultivated alkane degraders may perform non-methane alkane degradation (Laso-Pérez et al., 2016).
Biotechnological Application of Extremophiles in Bioremediation
In situ bioremediation of petroleum, mainly applied to soil and groundwater, includes biostimulation and bioventing. Biostimulation was recently performed in desert soil near the Arabian Gulf region (Al-Mailem et al., 2015). Community analysis revealed that 19 thermophilic hydrocarbonoclastic bacterial taxa were present in the soil sample. Furthermore, predominant species belonged to Amycolatopsis, Chelativorans, Isoptericola, Nocardia, Aeribacillus, Aneurinibacillus, Brevibacillus, Geobacillus, Kocuria, Marinobacter, and Paenibacillus, which showed better growth and hydrocarbon consumption rate at 50°C than at 30°C. However, enhanced growth and hydrocarbon consumption were observed after supplementation with Ca2+ (∼2.5 M CaSO4) or addition of 8% w/v dipicolinic acid (DPA), implying that Ca2+ and DPA-mediated increased heat resistance of alkane degraders may stimulate biodegradation at high temperatures (Radwan et al., 2017). A bioelectrochemical system was recently applied to marine sediments for hydrocarbon biodegradation, wherein a graphite rod (snorkel) is placed between the anoxic contaminated sediment and the O2-containing water. The snorkel acts as an electron acceptor and shuttles electrons generated by anaerobic hydrocarbon oxidation using reducing elements, such as S2 and Fe2+. Electrons move to the water phase along the graphite and fuse with oxygen and protons to form water (Viggi et al., 2015). This device serves as a sustainable respiratory electron acceptor in anaerobic oxidation of hydrocarbons, and can stimulate hydrocarbon degradation by SRB by scavenging toxic sulfide (Mapelli et al., 2017). Psychrophilic bacteria can potentially be used to degrade organic pollutants from soil and water at low temperatures, mainly, psychrophilic petroleum degraders such as Acinetobacter, Arthrobacter, Colwellia, Cytophaga, Halomonas, Marinobacter, Marinomonas, Pseudoalteromonas, Oleispira, Rhodococcus, and Shewanella (Brakstad et al., 2008). Acinetobacter spp. isolated from oil-polluted soils of King George Island were inoculated into a microcosm of contaminated soil, resulting in reduction of contaminant concentration by 75% after 50 days, compared to NP addition, which had no significant influence (Ruberto et al., 2003). Rhoder, composed of hydrocarbon-degrading bacteria Rhodococcus ruber and R. erythrococcus, oil, and gas, was used for removing oil pollution (Murygina et al., 2000). Over 99% bioremediation rate was obtained in open aquatic surfaces with light contamination (<20 g L-1); however, primary mechanical collection of oil and triple Rhoder treatment resulted in a 94% bioremediation rate in heavily polluted aquatic systems (thickness of oil film > 3 mm) and wetland (2,000 m2) in Urai, indicating that complex bioremediation strategies for effective oil-degradation are required.
Biopile bioremediation involves excavation of polluted soil, followed by supply of nutrients and air for increasing microbial hydrocarbon degradation and promoting bioremediation. Biopile remediation has recently been applied to polluted extreme environments such as cold regions (Gomez and Sartaj, 2014; Dias et al., 2015; Whelan et al., 2015; Martínez Álvarez et al., 2017). In a recent study, a previously suggested nutrient ratio (C:N:P ratio of 100:17.6:1.73) for biological activity (as FDA levels) was used to treat 0.4-ton geomembrane-covered biopiles from diesel fuel storage tanks of Antarctic expedition (Martínez Álvarez et al., 2017). Results showed a higher hydrocarbon removal rate (75.9%) in biostimulated soil within 40 days than in the control biopile (49.5%). In another study, an optimized biopile condition was obtained using surface response methodology (RSM) - a combination of 3 mL m-3 microbial consortia and 5% mature compost. After 94 days, 90.7% petroleum hydrocarbon reduction was observed, whereas hydrocarbon was reduced by only 56% in control soil (excavated from Val-des-Bois, in the Outaouais region of Quebec, Canada) between 1 and 8°C, indicating synergistic interaction between bioaugmentation and biostimulation for bioremediation enhancement. Additionally, biostimulation of Antarctic soils with fishmeal reduced 71% of total hydrocarbon and facilitated a bacterial community shift, increasing Actinobacteria over the 50-day study period (Dias et al., 2015). Landfarming has also been demonstrated as an effective bioremediation method in polar regions, which involves aggressive soil tilling and selective fertilization with a mixture of fertilizers (ammonium phosphate and urea) over a span of 70 days (McCarthy et al., 2004). However, soil concentrations of diesel range organics in northeast (380 mg kg-1) and southwest soil (430 mg kg-1) were already reduced to the target concentrations (500 mg kg-1) at 31 and 55 days, respectively.
Concluding Remarks
Although many alkane degraders living in extreme environment has been isolated and characterized, there is a great number of uncharacterized extremophiles that appear to be alkane-degraders, such as Brachybacterium, Idiomarina, and Leifsonia. Thus, further investigation of such novel alkane-degrading extremophiles will provide new metabolic pathways and survival strategies under harsh environments. Recent studies on discovery of new alkane metabolic systems in aerobic-, anaerobic extremophiles, such as AlkW1-AlkX system, Pfl homologues, and alkyl-coenzyme M indicate that there are several questions to be solved regarding alkane degradation metabolism. The evidences demonstrated the activation of several alternative pathways, such as glyoxylate shunt, Pta-AckA, or alcohol/fatty acid metabolism, instead of TCA cycle when extremophiles survive under extreme condition, implying that they are beneficial during alkane degradation. Because of high adaptability, strong stress resistance, and unique biodegradation capability, extremophiles are promising bioremediation-reagents to clean up polluted environments with low cost and high efficiency.
Author Contributions
CP and WP designed and coordinated the study. CP collected the data. CP wrote the first complete draft of the manuscript. WP provided substantial modifications. All authors contributed to and approved the final version of the manuscript.
Funding
This work was supported by a National Research Foundation of Korea (NRF) grant to WP, funded by the Korea Government (MSIP) (No. NRF-2017R1A2B4005838).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01081/full#supplementary-material
Footnotes
References
Acosta-González, A., Martirani-von Abercron, S. M., Rosselló-Móra, R., Wittich, R. M., and Marqués, S. (2015). The effect of oil spills on the bacterial diversity and catabolic function in coastal sediments: a case study on the Prestige oil spill. Environ. Sci. Pollut. Res. Int. 22, 15200–15214. doi: 10.1007/s11356-015-4458-y
Ahn, S., Jung, J., Jang, I. A., Madsen, E. L., and Park, W. (2016). Role of glyoxylate shunt in oxidative stress response. J. Biol. Chem. 291, 11928–11938. doi: 10.1074/jbc.M115.708149
Aislabie, J. M., Balks, M. R., Foght, J. M., and Waterhouse, E. J. (2004). Hydrocarbon spills on Antarctic soils: effects and management. Environ. Sci. Technol. 38, 1265–1274. doi: 10.1021/es0305149
Aitken, C. M., Jones, D. M., Maguire, M. J., Gray, N. D., Sherry, A., and Bowler, B. F. J. (2013). Evidence that crude oil alkane activation proceeds by different mechanisms under sulfate-reducing and methanogenic conditions. Geochim. Cosmochim. Acta 109, 162–174. doi: 10.1016/j.gca.2013.01.031
Al-Mailem, D. M., Eliyas, M., and Radwan, S. S. (2012). Enhanced haloarchaeal oil removal in hypersaline environments via organic nitrogen fertilization and illumination. Extremophiles 16, 751–758. doi: 10.1007/s00792-012-0471-y
Al-Mailem, D. M., Kansour, M. K., and Radwan, S. S. (2015). Moderately thermophilic, hydrocarbonoclastic bacterial communities in Kuwaiti desert soil: enhanced activity via Ca2+ and dipicolinic acid amendment. Extremophiles 19, 573–583. doi: 10.1007/s00792-015-0739-0
Amaro, A. M., Chamorro, D., Seeger, M., Arredondo, R., Pierano, I., and Jerez, C. A. (1991). Effect of external pH perturbations on in vivo protein synthesis by the acidophilic bacterium Thiobacillus ferrooxidans. J. Bacteriol. 173, 910–915. doi: 10.1128/jb.173.2.910-915.1991
Amouric, A., Quéméneur, M., Grossi, V., Liebgott, P. P., Auria, R., and Casalot, L. (2010). Identification of different alkane hydroxylase systems in Rhodococcus ruber strain SP2B, a hexane-degrading actinomycete. J. Appl. Microbiol. 108, 1903–1916. doi: 10.1111/j.1365-2672.2009.04592.x
Babu, P., Chandel, A. K., and Singh, O. V. (2015). Extremophiles and their Applications in Medical Processes. New York, NY: Springer International Publishing. doi: 10.1007/978-3-319-12808-5
Baker-Austin, C., and Dopson, M. (2007). Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 15, 165–171. doi: 10.1016/j.tim.2007.02.005
Barnhart, E. P., McClure, M. A., Johnson, K., Cleveland, S., Hunt, K. A., and Fields, M. W. (2015). Potential role of acetyl-coa synthetase (acs) and malate dehydrogenase (mae) in the evolution of the acetate switch in bacteria and archaea. Sci. Rep. 5:12498. doi: 10.1038/srep12498
Beeby, M., O’Connor, B. D., Ryttersgaard, C., Boutz, D. R., Perry, L. J., and Yeates, T. O. (2005). The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 3:e309. doi: 10.1371/journal.pbio.0030309
Berdugo-Clavijo, C., and Gieg, L. M. (2014). Conversion of crude oil to methane by a microbial consortium enriched from oil reservoir production waters. Front. Microbiol. 5:197. doi: 10.3389/fmicb.2014.00197
Bergholz, P. W., Bakermans, C., and Tiedje, J. M. (2009). Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J. Bacteriol. 191, 2340–2352. doi: 10.1128/JB.01377-08
Bertrand, E. M., Keddis, R., Groves, J. T., Vetriani, C., and Austin, R. N. (2013). Identity and mechanisms of alkane-oxidizing metalloenzymes from deep-sea hydrothermal vents. Front. Microbiol. 4:109. doi: 10.3389/fmicb.2013.00109
Boonmak, C., Takahashi, Y., and Morikawa, M. (2014). Cloning and expression of three ladA-type alkane monooxygenase genes from an extremely thermophilic alkane-degrading bacterium Geobacillus thermoleovorans B23. Extremophiles 18, 515–523. doi: 10.1007/s00792-014-0636-y
Bowman, J. S., and Deming, J. W. (2014). Alkane hydroxylase genes in psychrophile genomes and the potential for cold active catalysis. BMC Genomics 15:1120. doi: 10.1186/1471-2164-15-1120
Brakstad, O. G., Nonstad, I., Faksness, L. G., and Brandvik, P. J. (2008). Responses of microbial communities in Arctic sea ice after contamination by crude petroleum oil. Microb. Ecol. 55, 540–552. doi: 10.1007/s00248-007-9299-x
Brocchieri, L. (2004). Environmental signatures in proteome properties. Proc. Natl. Acad. Sci. U.S.A. 101, 8257–8258. doi: 10.1073/pnas.0402797101
Cheng, L., Ding, C., Li, Q., He, Q., Dai, L. R., and Zhang, H. (2013). DNA-SIP reveals that Syntrophaceae play an important role in methanogenic hexadecane degradation. PLoS One 8:e66784. doi: 10.1371/journal.pone.0066784
Choi, S. H., Baumler, D. J., and Kasper, C. W. (2000). Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 66, 3911–3916. doi: 10.1128/AEM.66.9.3911-3916.2000
Cooper, C. R., Daugherty, A. J., Tachdjian, S., Blum, P. H., and Kelly, R. M. (2009). Role of vapBC toxin-antitoxin loci in the thermal stress response of Sulfolobus solfataricus. Biochem. Soc. Trans. 37, 123–126. doi: 10.1042/BST0370123
D’Amico, S., Collins, T., Marx, J. C., Feller, G., and Gerday, C. (2006). Psychrophilic microorganisms: challenges for life. EMBO Rep. 7, 385–389. doi: 10.1038/sj.embor.7400662
Das, N., and Chandran, P. (2011). Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnol. Res. Int. 2011:941810. doi: 10.4061/2011/941810
Dias, R. L., Ruberto, L., Calabró, A., Balbo, A. L., Del Panno, M. T., and Mac Cormack, W. P. (2015). Hydrocarbon removal and bacterial community structure in on-site biostimulated biopile systems designed for bioremediation of diesel-contaminated Antarctic soil. Polar Biol. 5, 677–687. doi: 10.1007/s00300-014-1630-7
Dopson, M., Baker-Austin, C., and Bond, P. L. (2005). Analysis of differential protein expression during growth states of Ferroplasma strains and insights into electron transport for iron oxidation. Microbiology 151, 4127–4137. doi: 10.1099/mic.0.28362-0
Dunfield, P. F., Yuryev, A., Senin, P., Smirnova, A. V., Stott, M. B., Hou, S., et al. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450, 879–882. doi: 10.1038/nature06411
Ebadi, A., Khoshkholgh Sima, N. A., Olamaee, M., Hashemi, M., and Ghorbani Nasrabadi, R. (2017). Effective bioremediation of a petroleum-polluted saline soil by a surfactant-producing Pseudomonas aeruginosa consortium. J. Adv. Res. 8, 627–633. doi: 10.1016/j.jare.2017.06.008
Edbeib, M. F., Wahab, R. A., and Huyop, F. (2016). Halophiles: biology, adaptation, and their role in decontamination of hypersaline environments. World J. Microbiol. Biotechnol. 32:135. doi: 10.1007/s11274-016-2081-9
Feller, G., and Gerday, C. (2003). Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1, 200–208. doi: 10.1038/nrmicro773
Feng, L., Wang, W., Cheng, J., Ren, Y., Zhao, G., Gao, C., et al. (2007). Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Natl. Acad. Sci. U.S.A. 104, 5602–5607. doi: 10.1073/pnas.0609650104
Fonseca, P., Moreno, R., and Rojo, F. (2011). Growth of Pseudomonas putida at low temperature: global transcriptomic and proteomic analyses. Environ. Microbiol. Rep. 3, 329–339. doi: 10.1111/j.1758-2229.2010.00229.x
Fowler, S. J., Dong, X. L., Sensen, C. W., Suflita, J. M., and Gieg, L. M. (2012). Methanogenic toluene metabolism: community structure and intermediates. Environ. Microbiol. 14, 754–764. doi: 10.1111/j.1462-2920.2011.02631.x
Gauthier, M. J., Lafay, B., Christen, R., Fernandez, L., Acquaviva, M., Bonin, P., et al. (1992). Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Bacteriol. 42, 568–576. doi: 10.1099/00207713-42-4-568
Giaquinto, L., Curmi, P. M., Siddiqui, K. S., Poljak, A., DeLong, E., DasSarma, S., et al. (2007). Structure and function of cold shock proteins in archaea. J. Bacteriol. 189, 5738–5748. doi: 10.1128/JB.00395-07
Gomez, F., and Sartaj, M. (2014). Optimization of field scale biopiles for bioremediation of petroleum hydrocarbon contaminated soil at low temperature conditions by response surface methodology (RSM). Int. Biodeterior. Biodegradation 89, 103–109. doi: 10.1016/j.ibiod.2014.01.010
Gratia, E., Weekers, F., Margesin, R., D’Amico, S., Thonart, P., and Feller, G. (2009). Selection of a cold-adapted bacterium for bioremediation of wastewater at low temperatures. Extremophiles 13, 763–768. doi: 10.1007/s00792-009-0264-0
Gray, N. D., Sherry, A., Grant, R. J., Rowan, A. K., Hubert, C. R., Callbeck, C. M., et al. (2011). The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ. Microbiol. 13, 2957–2975. doi: 10.1111/j.1462-2920.2011.02570.x
Guiliani, N., and Jerez, C. A. (2000). Molecular cloning, sequencing, and expression of Omp-40, the gene coding for the major outer membrane protein from the acidophilic bacterium Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 66, 2318–2324. doi: 10.1128/AEM.66.6.2318-2324.2000
Hamamura, N., Olson, S. H., Ward, D. M., and Inkskeep, W. P. (2005). Diversity and functional analysis of bacterial community associated with natural hydrocarbon seeps in acidic soils at Rainbow Springs, Yellowstone National Park. Appl. Environ. Microbiol. 71, 5943–5950. doi: 10.1128/AEM.71.10.5943-5950.2005
Huyop, F., and Cooper, R. (2012). Degradation of millimolar concentration of the herbicide dalapon (2,2-dichloropropionic Acid) by Rhizobium sp. isolated from soil. Biotechnol. Biotechnol. Equip. 26, 3106–3112. doi: 10.5504/BBEQ.2012.0058
Ivanova, A. E., Sukhacheva, M. V., Kanat’eva, A. Y., Kravchenko, I. K., and Kurganov, A. A. (2014). Hydrocarbon-oxidizing potential and the genes for n-alkane biodegradation in a new acidophilic mycobacterial association from sulfur blocks. Microbiology 83, 764–772. doi: 10.1134/S0026261714060095
Kato, S., Kosaka, T., and Watanabe, K. (2009). Substrate-dependent transcriptomic shifts in Pelotomaculum thermopropionicum grown in syntrophic co-culture with Methanothermobacter thermautotrophicus. Microb. Biotechnol. 2, 575–584. doi: 10.1111/j.1751-7915.2009.00102.x
Kato, T., Miyanaga, A., Kanaya, S., and Morikawa, M. (2009). Alkane inducible proteins in Geobacillus thermoleovorans B23. BMC Microbiol. 9:60. doi: 10.1186/1471-2180-9-60
Khelifi, N., Amin Ali, O., Roche, P., Grossi, V., Brochier-Armanet, C., Valette, O., et al. (2014). Anaerobic oxidation of long-chain n-alkanes by the hyperthermophilic sulfate-reducing archaeon, Archaeoglobus fulgidus. ISME J. 8, 2153–2166. doi: 10.1038/ismej.2014.58
Kleindienst, S., Herbst, F. A., Stagars, M., Von Netzer, F., Von Bergen, M., Seifert, J., et al. (2014). Diverse sulfate-reducing bacteria of the Desulfosarcina/Desulfococcus clade are the key alkane degraders at marine seeps. ISME J. 8, 2029–2044. doi: 10.1038/ismej.2014.51
Knaebel, D. B., and Crawford, R. L. (1995). Extraction and purification of microbial DNA from petroleum-contaminated soils and detection of low numbers of toluene, octane and pesticide degraders by multiplex polymerase chain reaction and Southern analysis. Mol. Ecol. 4, 579–591. doi: 10.1111/j.1365-294X.1995.tb00258.x
Koh, H. Y., Park, H., Lee, J. H., Han, S. J., Sohn, Y. C., and Lee, S. G. (2017). Proteomic and transcriptomic investigations on cold-responsive properties of the psychrophilic Antarctic bacterium Psychrobacter sp. PAMC 21119 at subzero temperatures. Environ. Microbiol. 19, 628–644. doi: 10.1111/1462-2920.13578
Kuhn, E., Bellicanta, G. S., and Pellizari, V. H. (2009). New alk genes detected in Antarctic marine sediments. Environ. Microbiol. 11, 669–673. doi: 10.1111/j.1462-2920.2008.01843.x
Laso-Pérez, R., Wegener, G., Knittel, K., Widdel, F., Harding, K. J., Krukenberg, V., et al. (2016). Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature 539, 396–401. doi: 10.1038/nature20152
Li, D. C., Yang, F., Lu, B., Chen, D. F., and Yang, W. J. (2012). Thermotolerance and molecular chaperone function of the small heat shock protein HSP20 from hyperthermophilic archaeon, Sulfolobus solfataricus P2. Cell Stress Chaperones 17, 103–108. doi: 10.1007/s12192-011-0289-z
Li, H., Ji, X., Zhou, Z., Wang, Y., and Zhang, X. (2010). Thermus thermophilus proteins that are differentially expressed in response to growth temperature and their implication in thermoadaptation. J. Proteome Res. 9, 855–864. doi: 10.1021/pr900754y
Li, L., Liu, X., Yang, W., Xu, F., Wang, W., Feng, L., et al. (2008). Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase. J. Mol. Biol. 376, 453–465. doi: 10.1016/j.jmb.2007.11.069
Li, P., Wang, L., and Feng, L. (2013). Characterization of a novel Rieske-type alkane monooxygenase system in Pusillimonas sp. strain T7-7. J. Bacteriol. 195, 1892–1901. doi: 10.1128/JB.02107-12
Liang, J. L., Nie, Y., Wang, M., Xiong, G., Wang, Y. P., Maser, E., et al. (2016). Regulation of alkane degradation pathway by a TetR family repressor via an autoregulation positive feedback mechanism in a Gram-positive Dietzia bacterium. Mol. Microbiol. 99, 338–359. doi: 10.1111/mmi.13232
Lin, Q., Mendelssohn, I. A., Henry, C. B., Roberts, P. O., Walsh, M. M., Overton, E. B., et al. (1999). Effects of bioremediation agents on oil degradation in mineral and sandy salt marsh sediments. Environ. Technol. 20, 825–837. doi: 10.1080/09593332008616878
Liu, C., Wang, W., Wu, Y., Zhou, Z., Lai, Q., and Shao, Z. (2011). Multiple alkane hydroxylase systems in a marine alkane degrader, Alcanivorax dieselolei B-5. Environ. Microbiol. 13, 1168–1178. doi: 10.1111/j.1462-2920.2010.02416.x
Luz, A. P., Pellizari, V. H., Whyte, L. G., and Greer, C. W. (2004). A survey of indigenous microbial hydrocarbon degradation genes in soils from Antarctica and Brazil. Can. J. Microbiol. 50, 323–333. doi: 10.1139/w04-008
Ma, B. G., Goncearenco, A., and Berezovsky, I. N. (2010). Thermophilic adaptation of protein complexes inferred from proteomic homology modeling. Structure 18, 819–828. doi: 10.1016/j.str.2010.04.004
Maeng, J. H., Sakai, Y., Tani, Y., and Kato, N. (1996). Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J. Bacteriol. 178, 3695–3700. doi: 10.1128/jb.178.13.3695-3700.1996
Magazù, S., Migliardo, F., Benedetto, A., La Torre, R., and Hennet, L. (2012). Bio-protective effects of homologous disaccharides on biological macromolecules. Eur. Biophys. J. 41, 361–367. doi: 10.1007/s00249-011-0760-x
Mandelli, F., Couger, M. B., Paixão, D. A. A., Machado, C. B., Carnielli, C. M., Aricetti, J. A., et al. (2017). Thermal adaptation strategies of the extremophile bacterium Thermus filiformis based on multi-omics analysis. Extremophiles 21, 775–788. doi: 10.1007/s00792-017-0942-2
Mapelli, F., Scoma, A., Michoud, G., Aulenta, F., Boon, N., Borin, S., et al. (2017). Biotechnologies for marine oil spill cleanup: indissoluble ties with microorganisms. Trends Biotechnol. 35, 860–870. doi: 10.1016/j.tibtech.2017.04.003
Mardanov, A. V., Slododkinab, G. B., Slobodkinb, A. I., Beletskya, A. V., Gavrilovb, S. N., Kublanovb, I. V., et al. (2014). The Geoglobus acetivorans genome: Fe(III) reduction, acetate utilization, autotrophic growth, and degradation of aromatic compounds in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 81, 1003–1012. doi: 10.1128/AEM.02705-14
Margesin, R., Gander, S., Zacke, G., Gounot, A. M., and Schinner, F. (2003). Hydrocarbon degradation and enzyme activities of cold-adapted bacteria and yeasts. Extremophiles 7, 451–458. doi: 10.1007/s00792-003-0347-2
Margesin, R., and Schinner, F. (1998). Low-temperature bioremediation of a waste water contaminated with anionic surfactants and fuel oil. Appl. Microbiol. Biotechnol. 49, 482–486. doi: 10.1007/s002530051202
Margesin, R., Zimmerbauer, A., and Schinner, F. (2000). Monitoring of bioremediation by soil biological activities. Chemosphere 40, 339–346. doi: 10.1016/S0045-6535(99)00218-0
Martínez Álvarez, L. M., Ruberto, L., Lo Balbo, A., and Mac Cormack, W. P. (2017). Bioremediation of hydrocarbon-contaminated soils in cold regions: development of a pre-optimized biostimulation biopile-scale field assay in Antarctica. Sci. Total Environ. 590–591, 194–203. doi: 10.1016/j.scitotenv.2017.02.204
McCarthy, K., Walker, L., Vigoren, L., and Bartel, J. (2004). Remediation of spilled petroleum hydrocarbons by in situ landfarming at an arctic site. Cold Reg. Sci. Technol. 40, 31–39. doi: 10.1016/j.coldregions.2004.05.001
Meier, D. V., Bach, W., Girguis, P. R., Gruber-Vodicka, H. R., Reeves, E. P., Richter, M., et al. (2016). Heterotrophic Proteobacteria in the vicinity of diffuse hydrothermal venting. Environ. Microbiol. 18, 4348–4368. doi: 10.1111/1462-2920.13304
Meruelo, A. D., Han, S. K., Kim, S., and Bowie, J. U. (2012). Structural differences between thermophilic and mesophilic membrane proteins. Protein Sci. 21, 1746–1753. doi: 10.1002/pro.2157
Murygina, V., Arinbasarov, M., and Kalyuzhnyi, S. (2000). Bioremediation of oil polluted aquatic systems and soils with novel preparation ‘Rhoder’. Biodegradation 11, 385–389. doi: 10.1023/A:1011680703911
Mykytczuk, N. C., Foote, S. J., Omelon, C. R., Southam, G., Greer, C. W., and Whyte, L. G. (2013). Bacterial growth at -15°C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J. 7, 1211–1226. doi: 10.1038/ismej.2013.8
Nie, Y., Chi, C. Q., Fang, H., Liang, J. L., Lu, S. L., Lai, G. L., et al. (2014). Diverse alkane hydroxylase genes in microorganisms and environments. Sci. Rep. 4:4968. doi: 10.1038/srep04968
Nie, Y., Fang, H., Li, Y., Chi, C. Q., Tang, Y. Q., and Wu, X. L. (2013). The genome of the moderate halophile Amycolicicoccus subflavus DQS3-9A1(T) reveals four alkane hydroxylation systems and provides some clues on the genetic basis for its adaptation to a petroleum environment. PLoS One 8:e70986. doi: 10.1371/journal.pone.0070986
Oberding, L. K., and Gieg, L. M. (2017). Methanogenic paraffin biodegradation: alkylsuccinate synthase gene quantification and dicarboxylic acid production. Appl. Environ. Microbiol. 84:e01773-17. doi: 10.1128/AEM.01773-17
Park, C., Shin, B., Jung, J., Lee, Y., and Park, W. (2017). Metabolic and stress responses of Acinetobacter oleivorans DR1 during long-chain alkane degradation. Microb. Biotechnol. 10, 1809–1823. doi: 10.1111/1751-7915.12852
Phadtare, S. (2004). Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6, 125–136.
Prabagaran, S. R., Manorama, R., Delille, D., and Shivaji, S. (2007). Predominance of Roseobacter, Sulfitobacter, Glaciecola and Psychrobacter in seawater collected off Ushuaia, Argentina, Sub-Antarctica. FEMS Microbiol. Ecol. 59, 342–355. doi: 10.1111/j.1574-6941.2006.00213.x
Radwan, S. R., Al-Mailem, D. M., and Kansour, M. K. (2017). Calcium (II) - and dipicolinic acid mediated-biostimulation of oil-bioremediation under multiple stresses by heat, oil and heavy metals. Sci. Rep. 7:9534. doi: 10.1038/s41598-017-10121-7
Rampelotto, P. H. (2013). Extremophiles and extreme environments. Life 3, 482–485. doi: 10.3390/life3030482
Rapp, P., and Gabriel-Jürgens, L. H. (2003). Degradation of alkanes and highly chlorinated benzenes, and production of biosurfactants, by a psychrophilic Rhodococcus sp. and genetic characterization of its chlorobenzene dioxygenase. Microbiology 149, 2879–2890. doi: 10.1099/mic.0.26188-0
Richard, H., and Foster, J. W. (2004). Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 186, 6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004
Rivers, A. R., Sharma, S., Tringe, S. G., Martin, J., Joye, S. B., and Moran, M. A. (2013). Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 7, 2315–2329. doi: 10.1038/ismej.2013.129
Roberts, M. F. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Systems 1:5. doi: 10.1186/1746-1448-1-5
Rodrigues, D. F., Ivanova, N., He, Z., Huebner, M., Zhou, J., and Tiedje, J. M. (2008). Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million year old permafrost: a genome and transcriptome approach. BMC Genomics 9:547. doi: 10.1186/1471-2164-9-547
Rojo, F. (2009). Degradation of alkanes by bacteria. Environ. Microbiol. 11, 2477–2490. doi: 10.1111/j.1462-2920.2009.01948.x
Rozhkova-Novosad, E. A., Chae, J. C., Zylstra, G. J., Bertrand, E. M., Alexander-Ozinskas, M., Deng, D., et al. (2007). Profiling mechanisms of alkane hydroxylase activity in vivo using the diagnostic substrate norcarane. Chem. Biol. 14, 165–172. doi: 10.1016/j.chembiol.2006.12.007
Ruberto, L., Vazquez, S. C., and Mac Cormack, W. P. (2003). Effectiveness of the natural bacterial flora, biostimulation and bioaugmentation on the bioremediation of a hydrocarbon contaminated Antarctic soil. Int. Biodeterior. Biodegradation 52, 115–125. doi: 10.1016/S0964-8305(03)00048-9
Sabirova, J. S., Ferrer, M., Regenhardt, D., Timmis, K. N., and Golyshin, P. N. (2006). Proteomic insights into metabolic adaptations in Alcanivorax borkumensis induced by alkane utilization. J. Bacteriol. 188, 3763–3773. doi: 10.1128/JB.00072-06
Sagot, B., Gaysinski, M., Mehiri, M., Guigonis, J. M., Le Rudulier, D., and Alloing, G. (2010). Osmotically induced synthesis of the dipeptide N-acetylglutaminylglutamine amide is mediated by a new pathway conserved among bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 12652–12657. doi: 10.1073/pnas.1003063107
Schönfeld, P., and Wojtczak, L. (2016). Short- and medium-chain fatty acids in the energy metabolism–the cellular perspective. J. Lipid Res. 57, 943–954. doi: 10.1194/jlr.R067629
Scoma, A., and Boon, N. (2016). Osmotic stress confers enhanced cell integrity to hydrostatic pressure but impairs growth in Alcanivorax borkumensis SK2. Front. Microbiol. 7:729. doi: 10.3389/fmicb.2016.00729
Shih, T. W., and Pan, T. M. (2011). Stress responses of thermophilic Geobacillus sp. NTU 03 caused by heat and heat-induced stress. Microbiol. Res. 166, 346–359. doi: 10.1016/j.micres.2010.08.001
Siddique, T., Penner, T., Klassen, J., Nesbø, C., and Foght, J. M. (2012). Microbial communities involved in methane production from hydrocarbons in oil sands tailings. Environ. Sci. Technol. 46, 9802–9810. doi: 10.1021/es302202c
Siddiqui, K. S., and Cavicchioli, R. (2006). Cold-adapted enzymes. Annu. Rev. Biochem. 75, 403–433. doi: 10.1146/annurev.biochem.75.103004.142723
So, C. M., and Young, L. Y. (1999). Initial reactions in anaerobic alkane degradation by a sulfate reducer, strain AK-01. Appl. Environ. Microbiol. 65, 5532–5540.
Sotsky, J. B., Greer, C. W., and Atlas, R. M. (1994). Frequency of genes in aromatic and aliphatic hydrocarbon biodegradation pathways within bacterial populations from Alaskan sediments. Can. J. Microbiol. 40, 981–985. doi: 10.1139/m94-157
Stapleton, R. D., and Sayler, G. S. (1998). Assessment of the microbiological potential for the natural attenuation of petroleum hydrocarbons in a shallow aquifer system. Microb. Ecol. 36, 349–361. doi: 10.1007/s002489900121
Suter, E. A., Pachiadaki, M., Taylor, G. T., Astor, Y., and Edgcomb, V. P. (2017). Free-living chemoautotrophic and particle-attached heterotrophic prokaryotes dominate microbial assemblages along a pelagic redox gradient. Environ. Microbiol. 20, 693–712. doi: 10.1111/1462-2920.13997
Tan, B., Fowler, S. J., Abu Laban, N., Dong, X., Sensen, C. W., Foght, J., et al. (2015a). Comparative analysis of metagenomes from three methanogenic hydrocarbon-degrading enrichment cultures with 41 environmental samples. ISME J. 9, 2028–2045. doi: 10.1038/ismej.2015.22
Tan, B., Semple, K., and Foght, J. (2015b). Anaerobic alkane biodegradation by cultures enriched from oil sands tailings ponds involves multiple species capable of fumarate addition. FEMS Microbiol. Ecol. 91:fiv042. doi: 10.1093/femsec/fiv042
Throne-Holst, M., Wentzel, A., Ellingsen, T. E., Kotlar, H. K., and Zotchev, S. B. (2007). Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl. Environ. Microbiol. 73, 3327–3332. doi: 10.1128/AEM.00064-07
Timmers, P. H. A., Suarez-Zuluaga, D. A., van Rossem, M., Diender, M., Stams, A. J., and Plugge, C. M. (2016). Anaerobic oxidation of methane associated with sulfate reduction in a natural freshwater gas source. ISME J. 10, 1400–1412. doi: 10.1038/ismej.2015.213
Timmers, P. H. A., Welte, C. U., Koehorst, J. J., Plugge, C. M., Jetten, M. S. M., and Stams, A. J. M. (2017). Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1654237. doi: 10.1155/2017/1654237
Ting, L., Williams, T. J., Cowley, M. J., Lauro, F. M., Guilhaus, M., Raftery, M. J., et al. (2010). Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ. Microbiol. 12, 2658–2676. doi: 10.1111/j.1462-2920.2010.02235.x
Tourova, T. P., Sokolova, D. S., Semenova, E. M., Shumkova, E. S., Korshunova, A. V., Babich, T. L., et al. (2016). Detection of n-alkane biodegradation genes alkB and ladA in thermophilic hydrocarbon-oxidizing bacteria of the genera Aeribacillus and Geobacillus. Microbiology 85, 693–707. doi: 10.1134/S0026261716060199
Tribelli, P. M., Rossi, L., Ricardi, M. M., Gomez-Lozano, M., Molin, S., Raiger Iustman, L. J., et al. (2018). Microaerophilic alkane degradation in Pseudomonas extremaustralis: a transcriptomic and physiological approach. J. Ind. Microbiol. Biotechnol. 45, 15–23. doi: 10.1007/s10295-017-1987-z
van Beilen, J. B., and Funhoff, E. G. (2007). Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 74, 13–21. doi: 10.1007/s00253-006-0748-0
Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D., and Eisen, J. A. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74. doi: 10.1126/science.1093857
Viggi, C. C., Presta, E., Bellagamba, M., Kaciulis, S., Balijepalli, S. K., Zanaroli, G., et al. (2015). The “Oil-Spill Snorkel”: an innovative bioelectrochemical approach to accelerate hydrocarbons biodegradation in marine sediments. Front. Microbiol. 6:881. doi: 10.3389/fmicb.2015.00881
Walker, C. B., He, Z. L., Yang, Z. K., Ringbauer, J. A., He, Q., and Zhou, J. H. (2009). The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J. Bacteriol. 191, 5793–5801. doi: 10.1128/JB.00356-09
Wang, J., Wang, J., Zhang, Z., Li, Y., Zhang, B., Zhang, Z., et al. (2015). Cold-adapted bacteria for bioremediation of crude oil-contaminated soil. J. Chem. Technol. Biotechnol. 91, 2286–2297. doi: 10.1002/jctb.4814
Wang, L., Wang, W., Lai, Q., and Shao, Z. (2010). Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ. Microbiol. 12, 1230–1242. doi: 10.1111/j.1462-2920.2010.02165.x
Wang, Q., Cen, Z., and Zhao, J. (2015). The survival mechanisms of thermophiles at high temperatures: an angle of omics. Physiology 30, 97–106. doi: 10.1152/physiol.00066.2013
Wang, W., Ma, T., Zhang, B., Yao, N., Li, M., Cui, L., et al. (2014). Novel mechanism of protein thermostability: a unique N-terminal domain confers heat resistance to Fe/Mn-SODs. Sci. Rep. 4:7284. doi: 10.1038/srep07284
Wang, W., Wang, L., and Shao, Z. (2010). Diversity and abundance of oil-degrading bacteria and alkane hydroxylase (alkB) genes in the subtropical seawater of Xiamen Island. Microb. Ecol. 60, 429–439. doi: 10.1007/s00248-010-9724-4
Wang, W., and Shao, Z. (2012). Diversity of flavin-binding monooxygenase genes (almA) in marine bacteria capable of degradation long-chain alkanes. FEMS Microbiol. Ecol. 80, 523–533. doi: 10.1111/j.1574-6941.2012.01322.x
Wang, X. B., Chi, C. Q., Nie, Y., Tang, Y. Q., Tan, Y., Wu, G., et al. (2011). Degradation of petroleum hydrocarbons (C6–C40) and crude oil by a novel Dietzia strain. Bioresour. Technol. 17, 7755–7761. doi: 10.1016/j.biortech.2011.06.009
Washer, C. E., and Edwards, E. A. (2007). Identification and expression of benzylsuccinate synthase genes in a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 73, 1367–1369. doi: 10.1128/AEM.01904-06
Watanabe, S., Yamaoka, N., Takada, Y., and Fukunaga, N. (2002). The cold-inducible icl gene encoding thermolabile isocitrate lyase of a psychrophilic bacterium, Colwellia maris. Microbiology 148, 2579–2589. doi: 10.1099/00221287-148-8-2579
Whelan, M. J., Coulon, F., Hince, G., Rayner, J., McWatters, R., Spedding, T., et al. (2015). Fate and transport of petroleum hydrocarbons in engineered biopiles in polar regions. Chemosphere 131, 232–240. doi: 10.1016/j.chemosphere.2014.10.088
Whyte, L. G., Hawari, J., Zhou, E., Bourbonnière, L., Inniss, W. E., and Greer, C. W. (1998). Biodegradation of variable-chain-length alkanes at low temperatures by a psychrotrophic Rhodococcus sp. Appl. Environ. Microbiol. 64, 2578–2584.
Whyte, L. G., Schultz, A., Beilen, J. B., Luz, A. P., Pellizari, V., Labbé, D., et al. (2002). Prevalence of alkane monooxygenase genes in Arctic and Antarctic hydrocarbon-contaminated and pristine soils. FEMS Microbiol. Ecol. 41, 141–150. doi: 10.1111/j.1574-6941.2002.tb00975.x
Wouters, J. A., Rombouts, F. M., de Vos, W. M., Kuipers, O. P., and Abee, T. (1999). Cold shock proteins and low-temperature response of Streptococcus thermophilus CNRZ302. Appl. Environ. Microbiol. 65, 4436–4442.
Yergeau, E., Sanschagrin, S., Beaumier, D., and Greer, C. W. (2012). Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high arctic soils. PLoS One 7:e30058. doi: 10.1371/journal.pone.0030058
Youssef, N. H., Savage-Ashlock, K. N., McCully, A. L., Luedtke, B., Shaw, E. I., Hoff, W. D., et al. (2014). Trehalose/2-sulfotrehalose biosynthesis and glycine-betaine uptake are widely spread mechanisms for osmoadaptation in the Halobacteriales. ISME J. 8, 636–649. doi: 10.1038/ismej.2013.165
Yuk, H. G., and Marshall, D. L. (2004). Adaptation of Escherichia coli O157:H7 to pH alters membrane lipid composition, verotoxin secretion, and resistance to simulated gastric fluid acid. Appl. Environ. Microbiol. 70, 3500–3505. doi: 10.1128/AEM.70.6.3500-3505.2004
Yun, S. H., Choi, C. W., Kwon, S. O., Lee, Y. G., Chung, Y. H., Jung, H. J., et al. (2011). Enrichment and proteome analysis of a hyperthermostable protein set of archaeon Thermococcus onnurineus NA1. Extremophiles 15, 451–461. doi: 10.1007/s00792-011-0376-1
Keywords: extremophiles, alkane oxidizer, survival strategies, energy production, bioremediation
Citation: Park C and Park W (2018) Survival and Energy Producing Strategies of Alkane Degraders Under Extreme Conditions and Their Biotechnological Potential. Front. Microbiol. 9:1081. doi: 10.3389/fmicb.2018.01081
Received: 30 December 2017; Accepted: 07 May 2018;
Published: 25 May 2018.
Edited by:
Kian Mau Goh, Universiti Teknologi Malaysia, MalaysiaReviewed by:
Rachel Narehood Austin, Columbia University, United StatesNatsuko Hamamura, Kyushu University, Japan
Copyright © 2018 Park and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woojun Park, d3BhcmtAa29yZWEuYWMua3I=
 Chulwoo Park
Chulwoo Park Woojun Park
Woojun Park