- Department of Biology, Reed College, Portland, OR, United States
Enteropathogenic Escherichia coli (EPEC) is a significant cause of infant morbidity and mortality in developing regions of the world. Horizontally acquired genetic elements encode virulence structures, effectors, and regulators that promote bacterial colonization and disease. One such genetic element, the locus of enterocyte effacement (LEE), encodes the type three secretion system (T3SS) which acts as a bridge between bacterial and host cells to pass effector molecules that exert changes on the host. Due to its importance in EPEC virulence, regulation of the LEE has been of high priority and its investigation has elucidated many virulence regulators, including master regulator of the LEE Ler, H-NS, other nucleoid-associated proteins, GrlA, and PerC. Media type, environmental signals, sRNA signaling, metabolic processes, and stress responses have profound, strain-specific effects on regulators and LEE expression, and thus T3SS formation. Here we review virulence gene regulation in EPEC, which includes approaches for lessening disease by exploiting the elucidated regulatory pathways.
Introduction
Diarrheal disease is a leading cause of infant death worldwide, according to the World Health Organization. Enteropathogenic Escherichia coli (EPEC) is a major player in diarrheal disease—a study in Nairobi, Kenya of 207 clinical isolates from children under five with diarrhea found that the majority (19.3%) of isolates were EPEC (Makobe et al., 2012). Additionally, a Global Enteric Multicenter Study of children in sub-Saharan Africa and south Asia found that EPEC is associated with increased risk of death in children ages 0–11 months (Kotloff et al., 2013). EPEC shares many genetic and phenotypic similarities with enterohemorrhagic E. coli (EHEC), a zoonotic pathogen that infects children and adults, with a major reservoir being livestock (Hartland and Leong, 2013). EPEC and EHEC have been implicated in numerous acute diarrheal outbreaks worldwide (Majowicz et al., 2014; Hu and Torres, 2015). EPEC’s burden on children in developing countries and its relatedness to an infectious strain with a broader host range, namely EHEC, has incited bacteriologists to extensively study its disease mechanism in a collective, ongoing investigation.
Enteropathogenic Escherichia coli belongs to an over-arching group of infectious E. coli, referred to as attaching and effacing E. coli (AEEC). Attaching and effacing (A/E) pathogens are referred to as such because they intimately adhere to the host cell surface, efface brush border microvilli, and form characteristic actin-rich pedestals. Strains in this group are defined by the presence of a 35–43 kb horizontally acquired pathogenicity island named the locus of enterocyte effacement (LEE), which has inserted into various E. coli strains in many independent instances (Elliott et al., 1998; Hazen et al., 2013). The EPEC strain E2348/69 LEE contains 41 open reading frames with an average GC-content of 38.3% compared to the genome average of 50.6% (Elliott et al., 1998; Iguchi et al., 2009). Operons LEE1, LEE2, and LEE3 encode the type three secretion system (T3SS) inner and outer membrane components including SepD and SepQ, as well as the outer membrane porin EscC and the ATPase EscN (Elliott et al., 1998). LEE4 encodes the filament structure protein EspA and translocator proteins EspD and EspB along with other structural proteins and SepL (Elliott et al., 1998). LEE5 encodes intimin and translocated intimin receptor (Tir), which facilitate intimate attachment of EPEC cells to the epithelium, as well as the Tir chaperone CesT (Sánchez-SanMartín et al., 2001). Other genes of the LEE include espG, grlRA, cesF, map, and escD (Iguchi et al., 2009). The LEE1 operon encodes a master regulator of the LEE PAI named the LEE-encoded regulator (Ler) (Mellies et al., 1999; Elliott et al., 2000; Sperandio et al., 2000; Bustamante et al., 2001; Sánchez-SanMartín et al., 2001; Haack et al., 2003; Li et al., 2004; Bingle et al., 2014). Cloning the EPEC LEE pathogenicity island from the archetypal E2348/69 strain into an E. coli K-12 strain confers the A/E phenotype on cultured epithelial cells (McDaniel and Kaper, 1997). Mutations in the eae gene encoding intimin show decreased virulence- diminished colonic hyperplasia and CFU per gram of tissue using the A/E pathogen Citrobacter rodentium in a mouse model of infection (Reece et al., 2002), and Tir is essential for virulence in mice (Deng et al., 2003). Deletion of ler results in loss of LEE expression, effector secretion, pedestal formation, and virulence in the mouse model (Deng et al., 2004). Use of the A/E pathogen C. rodentium, as an infection model of LEE-associated virulence in mice has been reviewed previously (Mundy et al., 2005; Yang et al., 2010; Collins et al., 2014). Similarly, in rabbit EPEC (rEPEC), a ler deletion mutant is well tolerated by the animals whereas the wt parent strain causes severe diarrhea (Zhu et al., 2006). The LEE is the common genetic element of AEEC.
Though EPEC acquired the LEE, they are distinct from other AEEC in certain ways. EPEC lack the EHEC-containing Shiga-toxin (stx) that results in painful, bloody stool and can cause the complication known as hemolytic uremic syndrome in about 15% of cases (Tarr et al., 2005). Typical EPEC (tEPEC) contain the LEE and 14 genes that encode the bundle-forming pilus (BFP), which is most often encoded on the 70- to 95-kb (without or with tra transfer genes, respectively) E. coli adherence factor plasmid (pEAF). Atypical EPEC (aEPEC) contain the LEE and, and by definition, lack bfp (and the pEAF in most cases) (Brinkley et al., 2006; Hazen et al., 2013, 2015b). The pEAF additionally contains the plasmid encoded regulators, perABC, also termed bfpTVW (Tobe et al., 1996; Okeke et al., 2001; Brinkley et al., 2006). EPEC is parsed into six lineages, of which two are typical EPEC and the remaining are atypical (Hazen et al., 2013). Infection with typical EPEC can be asymptomatic to causing severe, acute watery diarrhea (Ahmed et al., 2014; Hu and Torres, 2015), which can become lethal. Some data may indicate that aEPEC is less virulent than typical strains containing the pEAF plasmid and the bundle-forming pilus (Levine et al., 1985), but clinical data indicate that aEPEC cause prolonged diarrhea. In a study by Nguyen et al. (2006), the mean number of days with aEPEC diarrhea was 12.1 (Nguyen et al., 2006). In many parts of the world, aEPEC is becoming more prevalent than tEPEC (Hu and Torres, 2015).
Enteropathogenic Escherichia coli spreads via the ‘fecal-oral route,’ which necessitates its ability to withstand not only the environment exterior to the host, but also the acidic environment of the stomach and the viscous mucous layer atop the epithelium. In colonization of the distal small intestine, tEPEC uses flagella to navigate the lumen and mucous layer, and utilizes both flagella and BFP to attach to the apical epithelial lining and accumulate together in micro-colonies of five to 200 bacteria, forming the ‘localized adherence’ (LA) phenotype (Tobe and Sasakawa, 2001; Giron et al., 2002; Cleary et al., 2004). Although none are as well characterized as the LA phenotype, aEPEC strains attach to the epithelial lining by various mechanisms such as ‘LA-like’ attachment, diffuse adherence, or aggregative adherence (Scaletsky et al., 2009, 2010).
Upon attachment, EPEC forms the T3SS that provides a bridge between the bacterial and the host cell cytoplasms, through which effector molecules pass from bacteria to host. Upon attachment of the T3SS to the host cell, the Tir protein is translocated into the cytoplasm where it then embeds into the membrane and is phosphorylated by host cell tyrosine kinases (Kenny et al., 1997b; Kenny, 1999). Actin recruitment occurs, forming pedestals on which the bacterium sits, with tight attachment resulting when the bacterial outer membrane protein intimin binds to Tir (reviewed in Kaper et al., 2004; Frankel and Phillips, 2008; Wong et al., 2011; Pearson et al., 2016). Strain E2348/69 has 7 LEE-encoded translocated effectors (Tir, Map, EspF, EspG, EspH, SepZ, and EspB) and fourteen non-LEE encoded (nle) effectors on prophages and integrative elements (Garmendia et al., 2005; Iguchi et al., 2009). Effector molecules work to further stabilize bacterial attachment to the apical enterocyte membrane, disrupt tight junctions between enterocytes, efface brush border microvilli, and cause actin polymerization and accumulation beneath the adherent bacterium, creating the characteristic cup-like pedestal in what is termed the attaching and effacing (A/E) lesion (Knutton et al., 1993). Infection by EPEC causes persistent secretory diarrhea, low fever, anorexia, and can cause death within a few days (Arenas-Hernández et al., 2012). Secretory diarrhea is characterized by increased chloride secretion, decreased sodium absorption, and increased mucosal permeability (Arenas-Hernández et al., 2012). Effector molecule deployment through the LEE-encoded T3SS is thought to allow EPEC to evade the host intestinal immune system by inhibiting phagocytosis, protein secretion, inflammation, and apoptotic cell death, which is reviewed in depth elsewhere (Santos and Finlay, 2015; Pearson et al., 2016).
Connections between environmental signals and regulation of the LEE and other aspects of virulence have shown that many stimuli can have profound effects on EPEC virulence. Specific, tightly regulated control of virulence gene expression ensure that EPEC deploys colonization tactics once in its ideal niche. Connolly et al. (2015) reviewed the effects of environment on EHEC LEE expression, thus this review aims to summarize the environmental effects on EPEC virulence gene regulation. Here, we discuss the mechanisms of the key regulators PerC, GrlA, and Ler, further regulation of the LEE, and manners in which LEE regulation and other virulence mechanisms are controlled in response to environmental stimuli.
Lee Regulation
Regulation of the LEE is controlled by many factors, including a range of nucleoid-associated proteins (Table 1). Master regulator Ler is encoded on the first open reading frame of LEE1, and is a transcriptional activator of LEE2, LEE3, LEE4, and LEE5, as well as extra-LEE genes including nleI/G to nleF, espC, and an espG homolog (Mellies et al., 1999; Elliott et al., 2000; Sperandio et al., 2000; Bustamante et al., 2001; Sánchez-SanMartín et al., 2001; Haack et al., 2003; Li et al., 2004; Bingle et al., 2014) (Figure 1). Ler represses expression of its own operon (Bhat et al., 2014). The importance of Ler in greater regulation of the LEE has driven investigation of the regulators and environmental signals affecting LEE1. The LEE1 operon has two promoters, LEE1 P1A and P1B, of which P1B is dominant in transcripts and in translated products (Jeong et al., 2012). Transcription off of the P1B promoter, but not the P1A promoter, is dependent on integration host factor (IHF), which binds to the promoter and induces expression (Friedberg et al., 1999; Jeong et al., 2012). Another DNA-binding protein, factor for inversion stimulation (Fis), increases transcription of the LEE1 by bending DNA (Pan et al., 1996; Goldberg et al., 2001). Hha binds to the LEE1 promoter and represses transcription in EHEC (Sharma and Zuerner, 2004). Non-nucleoid associated factors that positively regulate LEE1 include BipA and the quorum sensing regulator QseA (Figure 1). The ribosome-binding GTPase BipA decreases transcription of ler in a Per, IHF, and H-NS independent manner (Grant et al., 2003). In EHEC, QseA directly binds the LEE1 promoter and activates transcription (Sharp and Sperandio, 2007). The effect of these factors on transcription of the LEE, particularly LEE1 encoding Ler, is important for virulence (Deng et al., 2004; Zhu et al., 2006) (Figure 1).
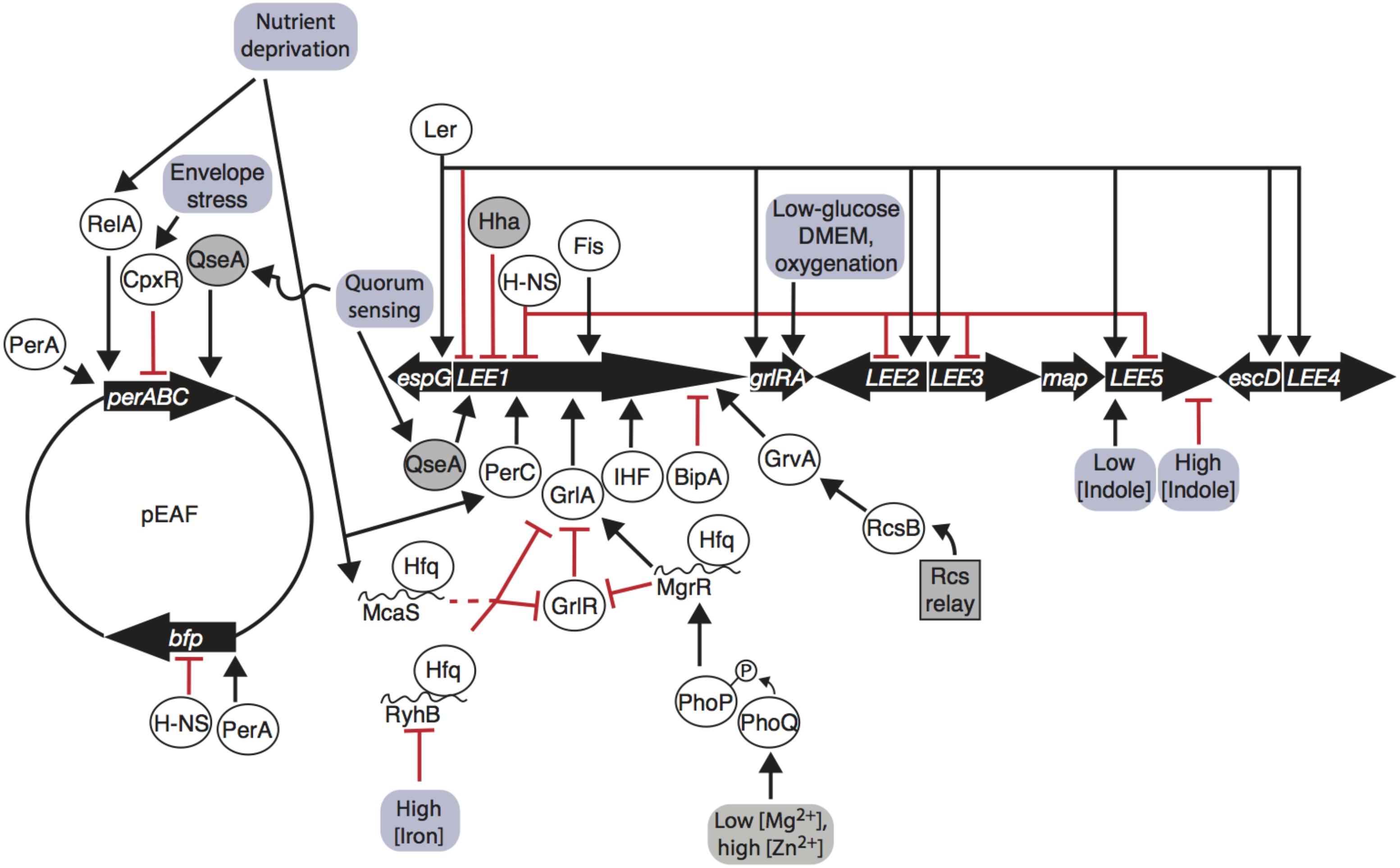
FIGURE 1. Enteropathogenic Escherichia coli (EPEC) LEE and pEAF gene regulation is influenced by environmental inputs. The LEE1 operon encodes Ler, which promotes transcription of all operons and auto-represses its own promoter. LEE1 is regulated by a number of nucleoid associated proteins (Ler, H-NS, Hha, Fis, and IHF), pEAF-encoded regulator PerC, LEE-encoded GrlA/GrlR, quorum-sensing factor QseA, BipA, and GrvA. RelA, CpxR, and QseA regulate the per operon on the pEAF, PerA promotes bfp transcription and auto-activates per. Environmental signals are transmitted through phosphorelay systems, sRNAs, stringent responses, quorum-sensing responses, and envelope stress responses to affect transcription of the LEE and pEAF. Arrows indicate transcription/translation promotion, blunt arrows indicate transcription/translation repression; proteins are in ovals; sRNAs are indicated by curvy lines; genetic elements are indicated by thick black arrows; environmental inputs are in red boxes; dashed line indicates indirect regulation.
Ler and H-Ns
The mechanisms of action of Ler and H-NS at LEE operons have been investigated extensively. Data indicate Ler increases transcription by displacing the silencer H-NS and other nucleoid-associated proteins, which permits transcription (Friedberg et al., 1999; Sánchez-SanMartín et al., 2001; Mellies et al., 2011). Recently, Shin (2017) demonstrated using lacZ reporter gene fusions that Ler only activates the LEE5 promoter in the presence of H-NS. This result was recapitulated by in vitro transcription—the relative number of transcripts off of the LEE5 promoter were the same with and without Ler in the absence of H-NS (Shin, 2017). However, there is some evidence that Ler regulates LEE5 in both H-NS-dependent and H-NS-independent manners (Sánchez-SanMartín et al., 2001; Umanski et al., 2002). Predictably, the dissociation constants of Ler to PLEE5 (Kd = 4.59 × 10-8) and that of H-NS to PLEE5 (Kd = 1.43 × 10-7) by surface plasmon resonance indicate that Ler has a greater binding affinity to the promoter than H-NS (Shin, 2017). The C-terminal regions of the proteins are the most conserved (Sperandio et al., 2000). Mutations in this conserved region abolishes Ler and H-NS binding to DNA (Yerushalmi et al., 2008; Sette et al., 2009).
The structures of Ler and H-NS, and how the proteins interact with DNA have also been investigated. A Ler 70–116 peptide binds to the minor groove of AT-rich DNA in the LEE2/LEE3 regulatory region, where Ler was shown to bind by DNA footprinting experiments (Sperandio et al., 2000; Cordeiro et al., 2011). For both proteins, A Q/RGR loop motif in the C-terminus mediates binding (Gordon et al., 2011). H-NS forms head-to-head, tail-to-tail multimers, bridging DNA and forming plectonemic structures (Arold et al., 2010), while Ler wraps DNA, forming toroids (Mellies et al., 2011). At LEE5, Ler binds over an extended region between -190 and -70 (Haack et al., 2003), while at most promoters H-NS binds upstream and downstream of the transcriptional start site. +1. In addition, data indicate that H-NS binds cooperatively, while Ler binds non-cooperatively to DNA (Winardhi et al., 2014). Ler has a shorter N-terminal coiled-coil domain than H-NS, containing only two α-helices and is “missing” the H-NS residues 59–68, which in H-NS are key in stabilizing dimerization (Arold et al., 2010), suggesting that Ler does not oligomerize in the same fashion as H-NS. Thus, Ler and H-NS recognize similar or identical motifs, but the manner in which they oligomerize and alter DNA structure differs (Mellies et al., 2011; Leh et al., 2017). Investigating the signaling affecting LEE1 transcription, and the molecular mechanisms of Ler-mediated control of LEE expression is paramount for understanding EPEC virulence.
Global Regulator of Ler (GrlA)
The Global regulator of Ler (GrlA) activates transcription of LEE1 in EPEC (Figure 1) and EHEC, controls transcription of a hemolysin and its secretion machinery encoded by ehxCABD and the flagellar regulator genes flhDC in EHEC (Padavannil et al., 2013). grlA is transcribed with its repressor grlR in an operon within the LEE PAI (Barba et al., 2005). GrlA activates Ler independently of PerC and IHF, and Ler increases GrlRA expression in a positive feedback loop (Barba et al., 2005). GrlA binds to the LEE1 promoter between -54 and the transcriptional start site (Padavannil et al., 2013). GrlR binds to the GrlA, outcompeting the LEE1, ehxCABD, and flhDC promoters and diminishing transcription (Jimenez et al., 2010; Padavannil et al., 2013). GrlR dimerizes and interacts with the HTH domain and C-terminus of a GrlA monomer, blocking the DNA-binding region GrlA (Padavannil et al., 2013). Thus, GrlA binds to DNA to positively regulate hemolysis, motility, and the LEE through master virulence regulator ler, and GrlA and the plasmid-encoded regulator perC activate the LEE1 operon in parallel.
The per Operon
Plasmid-encoded genes have been implicated in aiding bacteria to survive in specific local environments (Eberhard, 1989). The pEAF encodes two major operons, bfp and per, that advantage tEPEC in colonizing the human small intestine by production of type IV pili and promoting transcription of the LEE, respectively (Figure 1). Expressed from the perABC operon, PerA induces bfp transcription and auto-activates per (Gomez-Duarte and Kaper, 1995; Porter et al., 2004). PerA is part of the AraC/XylS family of transcriptional regulators, a group of over 1,500 proteins known to regulate adaptation to environmental changes and stress responses, carbon and nitrogen metabolism, and virulence (Ibarra et al., 2013). Regulators of this family have a DNA-binding domain in the C-terminus and two non-conserved HTH structures in the N-terminus, typically for dimerization and effector molecule-binding (Ibarra et al., 2013). The C- and N-termini in PerA contain key residues that allow PerA to bind to DNA, likely as a monomer (Porter et al., 2004; Ibarra et al., 2013). PerA binds to an AT-rich 29-bp sequence upstream of the -35 positions of the transcriptional start sites of bfp and per and increases transcription of both operons (Porter et al., 2004; Ibarra et al., 2013). The function of PerB is largely uncharacterized, but it is thought that PerB aids in activating bfp (Gomez-Duarte and Kaper, 1995; Tobe et al., 1996). Recent findings indicate that PerA and PerB are involved in greater control of the EPEC transcriptome.
An initial observation of smaller and larger colony morphologies of EPEC when cultured in low-glucose DMEM and then plated on agar led to the finding that bimodal per expression is correlated with bimodal colony morphologies controlled by PerA and PerB (Leh et al., 2017; Ronin et al., 2017). Small colonies are associated with high per expression and large with weak per expression (Leh et al., 2017; Ronin et al., 2017). It is argued that per expression dictates distinct transcriptomes among subpopulations, which advantages EPEC in unpredictable milieus (Leh et al., 2017; Ronin et al., 2017). Remarkably, bimodal colony morphology can be induced in E. coli K-12 strain MG1655 by expression of perAB, indicating that the role of per on colony morphology bimodality is not solely dependent on its transcription of the LEE PAI (Ronin et al., 2017). The small colonies showed increased aggregation and microcolony formation on cultured epithelial cells, but it is unknown how these different morphologies might affect virulence in an in vivo infection model, or contribute to disease (Ronin et al., 2017). However, it is becoming increasingly evident that per has an important role in allocation of transcriptional resources in response to nutrient availability in addition to encoding a virulence gene activator.
The third per-encoded protein, PerC activates the LEE1 promoter which encodes Ler, thereby stimulating virulence gene expression encoded in the LEE PAI (Mellies et al., 1999; Bustamante et al., 2011). Along with stimulating virulence gene expression, PerC regulates many genes. RNA-sequencing revealed that PerC regulates, either directly or indirectly, transcription of the fim operon, various metabolism genes, ribosomal RNAs, tRNAs, sRNAs, and others (Mellies et al., 2017). PerC’s mechanism of action has remained largely elusive, but some observations are worth examining. It is known that PerC requires DNA from positions -54 to +216 in relation to the start site of LEE1 to activate transcription (Bustamante et al., 2011). It has been hypothesized that PerC homologs (Pch) in EHEC bind to DNA and de-repress transcriptional activity by dislodging H-NS in a manner similar to Ler because of the following findings: PchA binds preferentially to AT-rich DNA or DNA complexes, Pch reduces the amount of H-NS bound to DNA, Pch and Ler preferentially bind in H-NS binding regions, and Pch is dependent on Hha and H-NS for activation of LEE1 in EHEC (Fukui et al., 2016). In contradiction, Porter et al. (2005) was unable to detect EPEC LEE1 promoter DNA-PerC complexes in vitro by electrophoretic mobility shift assay to a protein concentration of 1 μM, and thus concluded that PerC either does not bind to DNA, or only binds in the presence of other factors (Porter et al., 2005). Additionally, PerC in EPEC does not require H-NS to activate transcription—an IHF and H-NS double mutant maintains PerC-dependent activation of LEE1, indicating that PerC activation is not IHF or H-NS dependent, rather the two nucleoid-associated proteins act antagonistically at the LEE1 promoter (Porter et al., 2005). Taken together, it is undetermined whether PerC binds to DNA in the presence of other proteins or acts by another mechanism in which it does not bind to DNA. Thus, PerC regulates beyond LEE1 activation by an unknown mechanism.
Per and Ler regulation are intimately linked (Figure 1). A microarray study revealed that Ler represses per expression roughly threefold, indicating a negative feedback loop (Bingle et al., 2014). The intertwined regulation of these important players has implications in interpretation of their observed regulons. For instance, an increase in bfp transcripts in a ler mutant is likely due to decreased repression of perA, a known positive regulator of bfp (Bustamante et al., 2011; Bingle et al., 2014; Mellies et al., 2017). Conversely, the fim operon was found to be increased in expression in both perC- and ler-deletion mutants. Down-regulation of fim by these regulators could possibly be explained by decreased fimE recombinase transcription, inverting the fimS element to the OFF orientation due to Ler in both cases (Bingle et al., 2014; Mellies et al., 2017). The negative feedback loop between per and Ler dictates controlled regulation of the LEE, which is also coordinated by environmental signals.
Environmental Signals
Nutritional stress has a profound effect on virulence gene expression. Initially, it was observed that low-glucose DMEM, which has 5.5 mM D-glucose, or 0.1% (while high-glucose DMEM has 25 mM D-glucose, or 0.45%), provides increased expression of virulence genes compared to LB when grown at 37°C to an OD600 of 0.2 to 0.5 (Puente et al., 1996; Rosenshine et al., 1996; Leverton and Kaper, 2005; Bustamante et al., 2011; Hazen et al., 2015a). per, bfpA, and grlA transcription are all increased more than 4 log2 fold compared to LB at an OD600 of 0.5 in strain E2348/69, and similarly ler is expressed over 2 log2 fold in this strain (Hazen et al., 2015a). This pattern of regulation is not upheld in all tEPEC strains tested by Hazen et al. (2015a)—in fact, perC has decreased expression in tEPEC strain C581-05 under these conditions, and ler and grlA expression was unchanged (Hazen et al., 2015a). Growth conditions further dictate LEE expression—in a ΔperC mutant, ler transcription is 5.5-fold less than WT in DMEM with 0.45% glucose and grown statically with 5% CO2, but unchanged in shaking conditions without added CO2 (Bustamante et al., 2011). This regulation has downstream effects, as effector secretion of EspA, EspB, EspC, and EspD is decreased under static conditions, but not shaken conditions (Bustamante et al., 2011). Additionally, butyrate, which is abundant in the colon where EHEC colonizes, has been shown to increase pchA expression in EHEC (Nakanishi et al., 2009). Growth conditions affect GrlA-mediated control of the LEE as well. In grlA mutant, ler transcription was decreased fivefold when shaken in DMEM with 0.45% glucose and without CO2, but transcription was not decreased in static growth conditions with CO2 (Barba et al., 2005; Bustamante et al., 2011). Importantly, environmental signals have a significant influence on regulators, and ultimately affect timing of virulence gene expression.
The per operon is implicated in affecting timing of virulence expression. per is transcribed in response to environmental stimuli and in turn modulates expression of the LEE PAI and other genes that are predicted to advantage tEPEC in its ileal niche (Nakanishi et al., 2006; Spira et al., 2014; Leh et al., 2017; Mellies et al., 2017; Ronin et al., 2017). per gene expression occurs maximally in early exponential growth and has been implicated in the temporal control of colonization (Bustamante et al., 2011; Bueris et al., 2015). Surprisingly, aEPEC can be induced to colonize earlier when per is introduced in trans (Bueris et al., 2015). These data indicate the larger role in timing of virulence gene expression in response to beneficial environments for colonization.
These initial observations of the effect of media type on virulence gene transcription led to the finding that it is the transition from nutrient-dense LB to low-glucose DMEM that induces virulence gene expression in EHEC (Nakanishi et al., 2006). This transition induces the stringent response, in which relA and spoT encoding synthetases of guanosine (penta-) tetra-phosphate (herein collectively referred to as ppGpp), and DksA are necessary for the growth-phase dependent increase in LEE1 expression in EHEC (Nakanishi et al., 2006). RelA overexpression increases EspB and Tir secretion in the presence of functional pchA and ler genes in EHEC (Nakanishi et al., 2006). Intriguingly, in EPEC, RelA induces per expression, is necessary for bfp expression at WT levels, and plays a role in adherence to HEp-2 cells (Spira et al., 2014). Additionally, ppGpp over production depresses Fis expression, and to a lesser extent that of H-NS (Brandi et al., 2016). Fis also increases transcription of many genes involved in primary metabolism, such as tRNAs and rRNAs, likely because those are the genes that are being expressed in the early exponential growth phase when Fis is expressed (Ross et al., 1990; González-Gil et al., 1996; Goldberg et al., 2001). Nutrient deprivation is sensed and relayed through the stringent response to control per and virulence gene expression. Beyond growth conditions, other virulence determinants have been linked to environmental signal sensing, including calcium-mediated secretion system assembly, sRNA global regulation and metabolism, and envelope stress induced mediation of virulence factors.
Type Three Secretion
Calcium-Mediated Secretion
Calcium-deplete conditions have been observed to reduce secretion of the earlier translocators and cause “hypersecretion” of the later secreted effectors through the T3SS, likely by signaling that is transmitted through the gatekeeper protein SepL and/or through the “ruler” protein EscP (Kenny et al., 1997a; Ide et al., 2003; Deng et al., 2004; Gaytán et al., 2017; Shaulov et al., 2017). The membrane-associated translocator receptor SepL has been implicated in modulating the switch from early substrates to translocators and from translocators to effectors by interacting with different proteins in various mechanisms (Portaliou et al., 2017; Shaulov et al., 2017). It’s suggested that effector secretion is blocked by an interaction of SepL and EscP, and in calcium-deplete conditions, SepL and EscP dissociate and effectors are secreted (Shaulov et al., 2017). Shaulov et al. (2017) suggest that the calcium-rich extracellular milieu leaks in to the base of the basal body during T3SS formation, and upon completion of the T3SS and closed connection of bacterial and host cells, the calcium concentration drops to intracellular levels, causing a dissociation of SepL and EscP (Monjarás Feria et al., 2012; Shaulov et al., 2017). Many studies have observed calcium-dependent altered secretion, and this is one possible mechanism for these observations.
Another mechanism provides a calcium-independent explanation for control of secretion by SepL. A stepwise theory in which early substrates, then translocators, and finally effectors are secreted necessitates that SepL, SepD, and EscV interact in a single mechanism to control both translocator and effector secretion (Portaliou et al., 2017). SepL interacts with SepD in a calcium-independent manner, in which SepD mediates the crosstalk between SepL and the major T3SS component EscV, activating SepL and permitting translocator targeted secretion while blocking effector secretion (Gaytán et al., 2017; Portaliou et al., 2017). SepD releases, uncoupling EscV and SepL crosstalk and permitting both translocator and effector secretion (Portaliou et al., 2017). Then, the translocator receptor SepL releases, freeing up space for effectors to bind EscV and to be secreted (Portaliou et al., 2017). By these two studies, it’s suggested that SepL is involved in controlling the timing of secretion in parallel, calcium-dependent and -independent mechanisms.
Disruption of Host Homeostasis
Enteropathogenic Escherichia coli infection, by T3SS-mediated mechanisms, disrupts a number of host metabolic and physiologic processes. Live EPEC with a functional T3SS, but not non-pathogenic E. coli, dead EPEC, nor EPEC supernatant, decreases vitamin B1 (thiamin) uptake of Caco-2 cells by affecting the thiamin uptake receptors hTHTR-1 and hTHTR-2 (Ashokkumar et al., 2009). Effectors espF and espH at least partially depress thiamin uptake (Ashokkumar et al., 2009). EPEC, in a T3SS-dependent manner, induces activity of Na+/H+ transporters NHE1 and NHE2, thus increasing Na+ cation uptake, disrupting electrolyte homeostasis (Hecht et al., 2004). In fact, EPEC infection causes an increase in epithelial short circuit current that is partially dependent on an induction of chloride secretion (Collington et al., 1998). EPEC infection causes an increase in protein tyrosine phosphatase (PTPase), specifically by activation of Src-homology-2 domain containing PTPase, which decreases tyrosine phosphorylation of the serotonin transporter, decreasing transporter function and thus deregulation of serotonin uptake (Singhal et al., 2017). The EPEC T3SS is necessary for many instances of host homeostasis disruption.
sRNA Signaling and Bacterial Metabolism
Small RNAs and Their Chaperones
Small RNAs (sRNAs) are regulators of many metabolic processes, motility, virulence, and other cell functions. The majority of discovery of pathogenic E. coli sRNA regulation has been done in EHEC. Bhatt et al. (2016) comprehensively reviewed sRNA control in EPEC and EHEC, thus discoveries in this area are mentioned here to outline recent discovery and to highlight their importance in linking metabolism and virulence in EPEC. sRNAs respond rapidly to various environmental changes and are energetically advantageous as regulators because they need not be translated into functional protein to exert regulatory control (Bhatt et al., 2016). sRNAs non-specifically regulate a wide variety of genes with breadth of functions, such as metabolism and virulence genes.
sRNAs are 50 to 500 nt molecules that most often act by post-transcriptionally modulating expression from messenger RNAs (mRNAs) by base-pairing complementary sequences, blocking transcription elongation, translation, and/or mRNA stability (Bhatt et al., 2016). The majority of sRNAs in E. coli are trans-encoded, or remotely encoded with respect to its target subject, thus create short and often discontinuous complementary segments of 6–25 bp (Bhatt et al., 2016). Trans-encoded sRNAs require a chaperone, which is most often Hfq in E. coli (Bhatt et al., 2016). The extensive role of Hfq and Hfq-dependent sRNAs in EHEC indicates that there is likely a large role of Hfq-dependent sRNA regulation in EPEC that has not been explored.
Enteropathogenic Escherichia coli Hfq-dependent sRNAs MgrR, RyhB, and McaS affect grlRA translation (Bhatt et al., 2017) (Figure 1). MgrR represses grlR directly by binding to the upstream leader region of the mRNA, while activating grlA (Bhatt et al., 2017). RyhB directly represses grlRA translation by base-pairing to a shorter RNA sequence to repress translation, and McaS likely acts indirectly to repress grlRA (Bhatt et al., 2017). These three sRNAs respond to different environmental conditions. Expression of MgrR, which increases expression of the LEE via more stable grlA transcripts, increases in the case of low magnesium cation concentrations likely via PhoQ-PhoP phosphorelay (Bhatt et al., 2017). High abundance of iron cations bound to iron regulatory protein Fur repress RyhB, in which case RyhB cannot decrease concentration of the LEE through grlRA, thus LEE is activated (Bhatt et al., 2017) (Figure 1). The discovery of these three Hfq-dependent sRNAs that specifically regulate EPEC LEE expression is the tip of the iceberg of many sRNA interactions tying environmental sensing and virulence that have yet to be characterized.
Another RNA-binding protein, CsrA (and its homolog RsmA) co-regulates metabolite sensing, motility, stringent response, and virulence via a relaxed sequence specificity that enables base-pairing and regulation of a large number of operons that control many aspects of cell function. CsrA is part of the carbon storage regulation system, which encodes RNAs CsrB and CsrC, as well as protein CsrD that targets these RNAs for RNase E degradation (Bhatt et al., 2009). CsrA is intimately connected to the stringent response, involving DksA and ppGpp. CsrA acts by a number of different mechanisms—some operons are affected when CsrA is in high concentration and creates large homo-multimers, and other operons are affected at lower concentrations (Bhatt et al., 2009). CsrA positively regulates by binding distantly from the Shine–Dalgarno sequence and creating mRNA stability, and negative regulates by binding in proximity to this sequence, which prevents the 30S ribosomal subunit from binding and inhibiting translation (Bhatt et al., 2009). The flagellar operon flhDC is likely regulated by CsrA in EPEC because there is a conserved CsrA binding sequence identified in E. coli K-12 (Bhatt et al., 2009). It is proposed that CsrA could be involved in differential regulation of that operon in response to glucose concentrations, because the K-12 strain is not motile in 10 mM concentrations of glucose, whereas EPEC is motile (Bhatt et al., 2009).
CsrA regulates virulence by a number of factors, one of which is by increasing the sepL transcript, the product of which facilitates secretion of translocators together with SepD (Bhatt et al., 2009). CsrA represses grlA at high concentrations of the regulator, which has downstream negative regulatory effect on LEE expression, possibly in an advantageous effect of conserving resources when glucose is abundant (Bhatt et al., 2009). Importantly, an EPEC strain with csrA deleted is unable to create pedestals nor depolarize the enterocyte membrane in vitro, in part because the mutant no longer encourages tryptophan metabolism via tnaA (Bhatt et al., 2009). CsrA indirectly promotes transcription of tnaA (Figure 2), which encodes the enzyme tryptophanase of the tri-cistronic operon tnaCAB that is similarly tied to both metabolism and virulence (Bhatt et al., 2009). Additionally, CesT protein binds to CsrA, antagonizing the global regulator (Katsowich et al., 2017; Ye et al., 2018). Wide genomic control via sRNAs and their chaperone proteins is coming to light as determinately important in tying environmental stimulation to metabolism and virulence control in strain-specific manners.
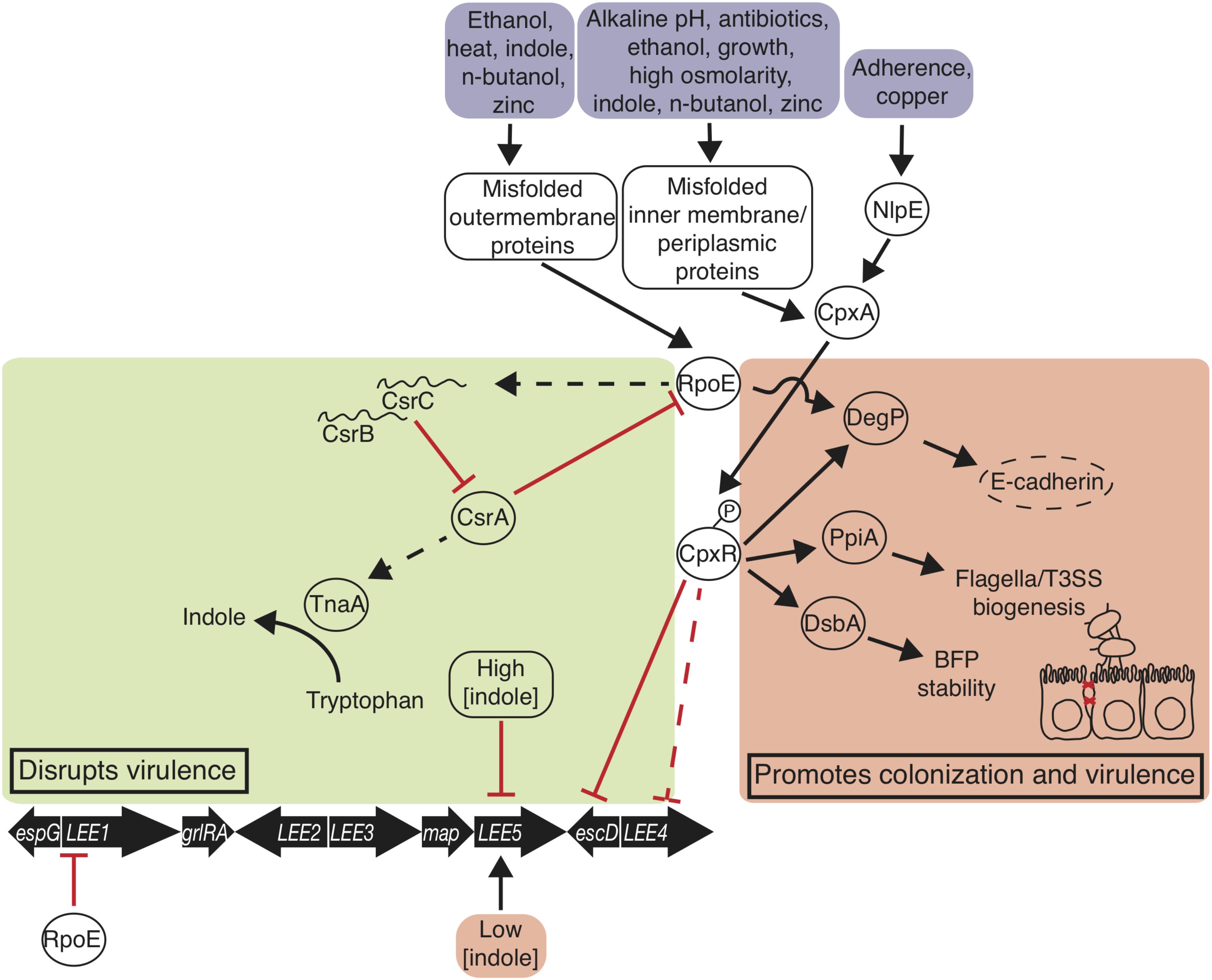
FIGURE 2. Envelope stress responses disrupt and promote EPEC virulence and colonization. The σE and Cpx responses are triggered when EPEC encounters environmental signals. σE promotes degP transcription, which degrades host E-cadherin disrupting adherens junctions, and represses LEE1 transcription. σE disrupts virulence by decreasing csrA transcription. Phosphorylated CpxR promotes degP, ppiA, and dsbA transcription, and represses perC transcription. Indole at 1.5 mM represses LEE5 transcription and at 50 μM promotes LEE5 transcription. Thin straight arrows indicate transcription promotion; blunt arrows indicate transcription repression; curved arrow indicates enzymatic reaction; bold arrows indicate genetic elements; proteins are in ovals; environmental signals are in boxes.
Tryptophan Metabolism and Virulence
The presence of tryptophan has a positive effect on virulence expression in EPEC (Figure 2). Overall, tryptophan and tnaCAB expression causes an increase in expression of LEE1, LEE4, and secreted factors that affect Caenorhabditis elegans survival (Bhatt et al., 2011). The tnaCAB operon, which encodes a cis-acting regulator (TnaC), the catalytic enzyme tryptophanase that cleaves tryptophan into indole, pyruvate, and ammonia (TnaA), and the primary permease important for tryptophan uptake into the bacterium (TnaB), is essential for EPEC-mediated killing of C. elegans (Bhatt et al., 2011). At low concentrations, indole promotes tir transcription. However, at high concentrations, indole acts as a competitive inhibitor of tryptophanase by binding to the catalytic site which retards LEE transcription (Bhatt et al., 2011) (Figure 2). This direct tie between a metabolic gene function and virulence expression is an indication that these cellular processes are delicately intertwined. The exploitation of these connections has already begun in the role of indole and indole-derivatives in suppressing pathogenesis (Bommarius et al., 2013).
Indole and the two indole-derivatives, indole-3-carboxyaldehyde (ICA) and indole-3-acetic acid (IAA) are potential therapeutics because of their repressive effect on LEE expression (Bommarius et al., 2013). Indole is a stationary phase quorum-sensing and signaling molecule produced not only by EPEC, but by resident, non-pathogenic E. coli, which potentially explains part of the contribution of resident intestinal microbiota in protecting against pathogenic infection (Bommarius et al., 2013). At the low concentration of 50 μM, indole and ICA induce expression of tir, but at higher than 1 mM levels, indole, ICA, and IAA each inhibit expression of LEE genes (Bommarius et al., 2013). ICA most effectively represses LEE expression—at 1.5 mM, ICA reduces pedestals formed by EPEC on cultured cells by sevenfold (Bommarius et al., 2013). When Citrobacter rodentium is administered to MyD88-/- mice, a model for EPEC infection in humans, in a lethal dose, oral administration of ICA reduces the CFU in the colon 50-fold (Bommarius et al., 2013). Indole and ICA could be promising therapeutics against EPEC.
Envelope Stress Responses
Envelope stress responses are linked to virulence attenuation in EPEC (Figure 2). Of many stress responses, the alternative sigma factor RpoE (σE) stress response and the two-component system CpxRA are deployed in response to a number of stresses as well as misfolded inner and outer membrane proteins and periplasmic proteins. σE is one of six alternative sigma factors in E. coli (Gruber and Gross, 2003), although the protypical laboratory strain EPEC E2348/69 houses a deleterious mutation in σS (Bleibtreu et al., 2014). In the σE stress response, heat (Erickson and Gross, 1989), n-butanol (Rutherford et al., 2010), zinc (Yamamoto and Ishihama, 2005), indole (Yitzhaki et al., 2012), and other factors cause outer membrane protein misfolding that leads to a proteolytic cascade that ultimately shears anti-σ factor RseA, releasing σE (Mecsas et al., 1993) (Figure 2). Growth-phase dependent activation of σE is ppGpp-dependent (Costanzo and Ades, 2006), and DksA and ppGpp directly activate σE-dependent transcription (Costanzo et al., 2008). σE associates with RNA polymerase and guides it to promoters to drive expression of chaperone, protease, and outer membrane biogenesis factors (Raivio et al., 2013). CsrA represses σE translation, and σE indirectly activates transcription of csrB and csrC, which bind and sequester CsrA, with potential effects on virulence as stated previously (Yakhnin et al., 2017) (Figure 2).
In E. coli, the two-component system CpxRA is stimulated by a number of signals, including indole (Raffa and Raivio, 2002), high osmolarity (Jubelin et al., 2005), ethanol (Bury-Moné et al., 2009), n-butanol (Rutherford et al., 2010), copper (Yamamoto and Ishihama, 2006), alkaline pH (Danese and Silhavy, 1998), amino-glycoside antibiotics (Kashyap et al., 2011), adhesion (Otto and Silhavy, 2002), growth (DiGiuseppe and Silhavy, 2003; Wolfe et al., 2008), and a number of membrane/periplasmic protein and phospholipid disruptions (Jones et al., 1997; Wang et al., 2010; Itou et al., 2012; Raivio et al., 2013). Zinc does not induce CpxRA induction (Yamamoto and Ishihama, 2005). Outer membrane lipoprotein NlpE activates CpxA in response to abiotic surface adherence (Otto and Silhavy, 2002), and cpxP transcription is transcribed at high levels in the response and blocks phosphorylation of CpxA in a negative feedback loop (Raivio et al., 1999). Inner transmembrane sensory histidine kinase CpxA directly senses stressors, autophosphorylates its cytoplasmic sensory domain, then phosphorylates CpxR, which goes on to bind DNA sequences to regulate gene expression (Grabowicz and Silhavy, 2017) (Figure 2). The activation of the Cpx response regulates the LEE and other key proteins that promote bacterial colonization.
Key Cpx response proteins DsbA, PpiA, and DegP are activated by the Cpx response and promote virulence regulation. DsbA is a disulfide oxidase that facilitates stability of the major BFP subunit bundlin (Zhang and Donnenberg, 1996; Heras et al., 2009; Vogt et al., 2010). PpiA is a peptidyl-prolyl-isomerase and may facilitate biogenesis of the type three secretion system and flagella (Justice et al., 2005; MacRitchie et al., 2012). As well as in response to Cpx, protease/chaperone-encoding degP is transcribed in response to σE activation (MacRitchie et al., 2012; Abfalter et al., 2016). DegP degrades misfolded periplasmic envelope proteins and cleaves human E-cadherin, disrupting cell-cell adherens junctions (MacRitchie et al., 2012; Abfalter et al., 2016). α-bundalin of BFP binds to N-acetyllactosamine glycan receptors on human enterocytes, and BFP is consequently degraded by DegP (Hyland et al., 2008; Humphries et al., 2010). In addition to these auxiliary factors that contribute to virulence, Cpx has been implicated in direct repression of the LEE.
The Cpx envelope stress response has a negative effect on LEE expression, especially on operons LEE4 and LEE5 (MacRitchie et al., 2008) (Figure 2). A CpxR binding site at LEE5 suggests that CpxR may be acting directly at that operon (MacRitchie et al., 2008; Vogt et al., 2010). BFP, Intimin, Tir, and translocator proteins EspA, EspB, EspD, and the needle complex EscF and inner and outer membrane components EscJ and EscC are diminished as consequence (MacRitchie et al., 2008; Vogt et al., 2010; MacRitchie et al., 2012). Despite the positive effect on pathogenesis through BFP and flagella promotion and E-cadherin degradation by some Cpx response proteins (Figure 2), the overwhelming effect of envelope stress is a decrease in virulence of EPEC, demonstrated by zinc administration.
Heavy metals induce the envelope stress responses and while most are not feasible as therapies because they are toxic, the World Health Organization has been supplementing with zinc to aid in treating acute diarrheal infections since 2008. Zinc is administered along with rehydration methods to help treat acute diarrheal infections because it has been observed to diminish the severity and duration of disease (Sazawal et al., 1995). Specifically, zinc causes cell envelope stress and indirectly suppresses virulence, possibly through multiple stress mechanisms. For instance, zinc increases transcription of rpoE (Xue et al., 2015). Ultimately, there is an observable decrease in expression of perC, bfpA, ler, and espA likely due to indirect downregulation which results in decreased adherence to epithelial cells (Crane et al., 2007; Xue et al., 2015). Additionally, EPEC growth rates decrease and membrane perturbation and permeability increases (Xue et al., 2015). Zinc as a virulence inhibitor has been and continues to be deployed in response to EPEC and other enteric pathogens.
Conclusion
The study of EPEC virulence has identified the T3SS, many regulators, effector molecules, and environmental stresses that are tied to sensory responses and metabolism, all necessary for pathogenesis. Here we present our current, overall understanding of the complex regulatory networks that promote colonization and virulence, and also those that disrupt this process. Beyond commonly administered antibiotics, the disruption of tryptophan metabolism with indole derivatives, or the now commonly given zinc dietary supplements are two alternate therapy approaches. While ICA and IAA are yet to be used clinically, inexpensive zinc supplements have been given with oral rehydration therapy to children and infants with diarrhea for nearly a decade. Such targeted therapies require an understanding of conserved regulatory pathways. While the majority of studies have been conducted with archetypal EPEC strains, strain-specific regulatory control of virulence genes in response to environmental inputs, like those observed by Hazen et al. (2015a), illustrate the importance of expanding our investigations to encompass a wider variety of related strains (Platenkamp and Mellies, 2017). Research on virulence gene regulation as it relates to environmental cues, stress responses, metabolism, and host-associated signaling will add to the growing body of knowledge necessary for minimizing disease caused by EPEC and related pathogens.
Author Contributions
JM and AP co-authored and edited the manuscript. AP drew the figures, and compiled the table.
Funding
This work was supported by NIH grant 1R21AI115193-01 awarded to JM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abfalter, C. M., Schubert, M., Götz, C., Schmidt, T. P., Posselt, G., and Wessler, S. (2016). HtrA-mediated E-cadherin cleavage is limited to DegP and DegQ homologs expressed by gram-negative pathogens. Cell Commun. Signal. 14:30. doi: 10.1186/s12964-016-0153-y
Ahmed, S. F., Shaheen, H. I., Abdel-Messih, I. A., Mostafa, M., Putnam, S. D., Kamal, K. A., et al. (2014). The epidemiological and clinical characteristics of diarrhea associated with enteropathogenic, enteroaggregative and diffuse-adherent Escherichia coli in egyptian children. J. Trop. Pediatr. 60, 397–400. doi: 10.1093/tropej/fmu034
Arenas-Hernández, M. M., Martínez-Laguna, Y., and Torres, A. G. (2012). Clinical implications of enteroadherent Escherichia coli. Curr. Gastroenterol. Rep. 14, 386–394. doi: 10.1007/s11894-012-0277-1
Arold, S. T., Leonard, P. G., Parkinson, G. N., and Ladbury, J. E. (2010). H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl. Acad. Sci. U.S.A. 107, 15728–15732. doi: 10.1073/pnas.1006966107
Ashokkumar, B., Kumar, J. S., Hecht, G. A., and Said, H. M. (2009). Enteropathogenic Escherichia coli inhibits intestinal vitamin B1 (thiamin) uptake: studies with human-derived intestinal epithelial Caco-2 cells. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G825–G833. doi: 10.1152/ajpgi.00250.2009
Barba, J., Bustamante, V. H., Flores-Valdez, M. A., Deng, W. Y., Finlay, B. B., and Puente, J. L. (2005). A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 187, 7918–7930. doi: 10.1128/JB.187.23.7918-7930.2005
Bhat, A., Shin, M., Jeong, J. H., Kim, H. J., Lim, H. J., Rhee, J. H., et al. (2014). DNA looping-dependent autorepression of LEE1 P1 promoters by Ler in enteropathogenic Escherichia coli (EPEC). Proc. Natl. Acad. Sci. U.S.A. 111, E2586–2595. doi: 10.1073/pnas.1322033111
Bhatt, S., Anyanful, A., and Kalman, D. (2011). CsrA and TnaB coregulate tryptophanase activity to promote exotoxin-induced killing of Caenorhabditis elegans by enteropathogenic Escherichia coli. J. Bacteriol. 193, 4516–4522. doi: 10.1128/JB.05197-11
Bhatt, S., Edwards, A. N., Nguyen, H. T. T., Merlin, D., Romeo, T., and Kalman, D. (2009). The RNA binding protein csra is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect. Immun. 77, 3552–3568. doi: 10.1128/IAI.00418-09
Bhatt, S., Egan, M., Jenkins, V., Muche, S., and El-Fenej, J. (2016). The tip of the iceberg: on the roles of regulatory small rnas in the virulence of enterohemorrhagic and enteropathogenic Escherichia coli. Front. Cell. Infect. Microbiol. 6:105. doi: 10.3389/fcimb.2016.00105
Bhatt, S., Egan, M., Ramirez, J., Xander, C., Jenkins, V., Muche, S., et al. (2017). Hfq and three Hfq-dependent small regulatory RNAs-MgrR, RyhB and McaS-coregulate the locus of enterocyte effacement in enteropathogenic Escherichia coli. Pathog. Dis. 75:ftw113. doi: 10.1093/femspd/ftw113
Bingle, L. E., Constantinidou, C., Shaw, R. K., Islam, M. S., Patel, M., Snyder, L. A., et al. (2014). Microarray analysis of the Ler regulon in enteropathogenic and enterohaemorrhagic Escherichia coli strains. PLoS One 9:e80160. doi: 10.1371/journal.pone.0080160
Bleibtreu, A., Clermont, O., Darlu, P., Glodt, J., Branger, C., Picard, B., et al. (2014). The rpoS gene is predominantly inactivated during laboratory storage and undergoes source-sink evolution in Escherichia coli species. J. Bacteriol. 196, 4276–4284. doi: 10.1128/JB.01972-14
Bommarius, B., Anyanful, A., Izrayelit, Y., Bhatt, S., Cartwright, E., Wang, W., et al. (2013). A family of indoles regulate virulence and Shiga toxin production in pathogenic E. coli. PLoS One 8:e54456. doi: 10.1371/journal.pone.0054456
Brandi, A., Giangrossi, M., Giuliodori, A. M., and Falconi, M. (2016). An interplay among fis, h-ns, and guanosine tetraphosphate modulates transcription of the Escherichia coli cspA gene under physiological growth conditions. Front. Mol. Biosci. 3:19. doi: 10.3389/fmolb.2016.00019
Brinkley, C., Burland, V., Keller, R., Rose, D. J., Boutin, A. T., and Klink, S. A., et al. (2006). Nucleotide sequence analysis of the enteropathogenic Escherichia coli adherence factor plasmid pMAR7. Infect. Immun. 74, 5408–5413. doi: 10.1128/IAI.01840-05
Bueris, V., Huerta-Cantillo, J., Navarro-Garcia, F., Ruiz, R. M., Cianciarullo, A. M., and Elias, W. P. (2015). Late establishment of the attaching and effacing lesion caused by atypical enteropathogenic Escherichia coli depends on protein expression regulated by Per. Infect. Immun. 83, 379–388. doi: 10.1128/IAI.02563-14
Bury-Moné, S., Nomane, Y., Reymond, N., Barbet, R., Jacquet, E., Imbeaud, S., et al. (2009). Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 5:e1000651. doi: 10.1371/journal.pgen.1000651
Bustamante, V. H., Santana, F. J., Calva, E., and Puente, J. L. (2001). Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39, 664–678. doi: 10.1046/j.1365-2958.2001.02209.x
Bustamante, V. H., Villalba, M. I., Garcia-Angulo, V. A., Vazquez, A., Martinez, L. C., Jimenez, R., et al. (2011). PerC and GrlA independently regulate Ler expression in enteropathogenic Escherichia coli. Mol. Microbiol. 82, 398–415. doi: 10.1111/j.1365-2958.2011.07819.x
Cleary, J., Lai, L. C., Shaw, R. K., Straatman-Iwanowska, A., Donnenberg, M. S., Frankel, G., et al. (2004). Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 150, 527–538. doi: 10.1099/mic.0.26740-0
Collington, G. K., Booth, I. W., and Knutton, S. (1998). Rapid modulation of electrolyte transport in Caco-2 cell monolayers by enteropathogenic Escherichia coli (EPEC) infection. Gut 42, 200–207. doi: 10.1136/gut.42.2.200
Collins, J. W., Keeney, K. M., Crepin, V. F., Rathinam, V. A., Fitzgerald, K. A., Finlay, B. B., et al. (2014). Citrobacter rodentium: infection, inflammation and the microbiota. Nat. Rev. Microbiol. 12, 612–623. doi: 10.1038/nrmicro3315
Connolly, J. P. R., Finlay, B. B., and Roe, A. J. (2015). From ingestion to colonization: the influence of the host environment on regulation of the LEE encoded type III secretion system in enterohaemorrhagic Escherichia coli. Front. Microbiol. 6:568. doi: 10.3389/fmicb.2015.00568
Cordeiro, T. N., Schmidt, H., Madrid, C., Juarez, A., Bernado, P., Griesinger, C., et al. (2011). Indirect DNA readout by an H-NS related protein: structure of the DNA complex of the C-terminal domain of Ler. PLoS Pathog. 7:e1002380. doi: 10.1371/journal.ppat.1002380
Costanzo, A., and Ades, S. E. (2006). Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3’,5’-bispyrophosphate (ppGpp). J. Bacteriol. 188, 4627–4634. doi: 10.1128/JB.01981-05
Costanzo, A., Nicoloff, H., Barchinger, S. E., Banta, A. B., Gourse, R. L., and Ades, S. E. (2008). ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 67, 619–632. doi: 10.1111/j.1365-2958.2007.06072.x
Crane, J. K., Naeher, T. M., Shulgina, I., Zhu, C. R., and Boedeker, E. C. (2007). Effect of zinc in enteropathogenic Escherichia coli infection. Infect. Immun. 75, 5974–5984. doi: 10.1128/IAI.00750-07
Danese, P. N., and Silhavy, T. J. (1998). CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180, 831–839.
Deng, W., Puente, J. L., Gruenheid, S., Li, Y., Vallance, B. A., Vázquez, A., et al. (2004). Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U.S.A. 101, 3597–3602. doi: 10.1073/pnas.0400326101
Deng, W., Vallance, B. A., Li, Y., Puente, J. L., and Finlay, B. B. (2003). Citrobacter rodentium translocated intimin receptor (Tir) is an essential virulence factor needed for actin condensation, intestinal colonization and colonic hyperplasia in mice. Mol. Microbiol. 48, 95–115. doi: 10.1046/j.1365-2958.2003.03429.x
DiGiuseppe, P. A., and Silhavy, T. J. (2003). Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185, 2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003
Eberhard, W. G. (1989). Why do bacterial plasmids carry some genes and not others? Plasmid 21, 167–174. doi: 10.1016/0147-619X(89)90040-1
Edwards, A. N., Patterson-Fortin, L. M., Vakulskas, C. A., Mercante, J. W., Potrykus, K., Vinella, D. et al. (2011). Circuitry linking the Csr and stringent response global regulatory systems. Mol. Microbiol. 80, 1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x
Elliott, S. J., Sperandio, V., Giron, J. A., Shin, S., Mellies, J. L., Wainwright, L., et al. (2000). The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68, 6115–6126. doi: 10.1128/IAI.68.11.6115-6126.2000
Elliott, S. J., Wainwright, L. A., McDaniel, T. K., Jarvis, K. G., Deng, Y. K., Lai, L. C., et al. (1998). The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28, 1–4. doi: 10.1046/j.1365-2958.1998.00783.x
Erickson, J. W., and Gross, C. A. (1989). Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 3, 1462–1471. doi: 10.1101/gad.3.9.1462
Frankel, G., and Phillips, A. D. (2008). Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cell. Microbiol. 10, 549–556. doi: 10.1111/j.1462-5822.2007.01103.x
Friedberg, D., Umanski, T., Fang, Y., and Rosenshine, I. (1999). Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34, 941–952. doi: 10.1046/j.1365-2958.1999.01655.x
Fukui, N., Oshima, T., Ueda, T., Ogasawara, N., and Tobe, T. (2016). Gene activation through the modulation of nucleoid structures by a horizontally transferred regulator, pch, in enterohemorrhagic Escherichia coli. PLoS One 11:e0149718. doi: 10.1371/journal.pone.0149718
Garmendia, J., Frankel, G., and Crepin, V. F. (2005). Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 73, 2573–2585. doi: 10.1128/IAI.73.5.2573-2585.2005
Gaytán, M. O., Monjarás Feria, J., Soto, E., Espinosa, N., Benítez, J. M., Georgellis, D., et al. (2017). Novel insights into the mechanism of SepL-mediated control of effector secretion in enteropathogenic Escherichia coli. Microbiologyopen. 7:e00571. doi: 10.1002/mbo3.571
Giron, J. A., Torres, A. G., Freer, E., and Kaper, J. B. (2002). The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44, 361–379. doi: 10.1046/j.1365-2958.2002.02899.x
Goldberg, M. D., Johnson, M., Hinton, J. C., and Williams, P. H. (2001). Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41, 549–559. doi: 10.1046/j.1365-2958.2001.02526.x
Gomez-Duarte, O. G., and Kaper, J. B. (1995). A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63, 1767–1776.
González-Gil, G., Bringmann, P., and Kahmann, R. (1996). FIS is a regulator of metabolism in Escherichia coli. Mol. Microbiol. 22, 21–29. doi: 10.1111/j.1365-2958.1996.tb02652.x
Gordon, B. R., Li, Y., Cote, A., Weirauch, M. T., Ding, P., Hughes, T. R., et al. (2011). Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 10690–10695. doi: 10.1073/pnas.1102544108
Grabowicz, M., and Silhavy, T. J. (2017). Envelope stress responses: an interconnected safety net. Trends Biochem. Sci. 42, 232–242. doi: 10.1016/j.tibs.2016.10.002
Grant, A. J., Farris, M., Alefounder, P., Williams, P. H., Woodward, M. J., and O’Connor, C. D. (2003). Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48, 507–521. doi: 10.1046/j.1365-2958.2003.t01-1-03447.x
Gruber, T. M., and Gross, C. A. (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466. doi: 10.1146/annurev.micro.57.030502.090913
Haack, K. R., Robinson, C. L., Miller, K. J., Fowlkes, J. W., and Mellies, J. L. (2003). Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71, 384–392. doi: 10.1128/IAI.71.1.384-392.2003
Hartland, E. L., and Leong, J. M. (2013). Enteropathogenic and enterohemorrhagic E. coli: ecology, pathogenesis, and evolution. Front. Cell. Infect. Microbiol. 3:15. doi: 10.3389/fcimb.2013.00015
Hazen, T. H., Daugherty, S. C., Shetty, A., Mahurkar, A. A., White, O., Kaper, J. B., et al. (2015a). RNA-Seq analysis of isolate- and growth phase-specific differences in the global transcriptomes of enteropathogenic Escherichia coli prototype isolates. Front. Microbiol. 6:569. doi: 10.3389/fmicb.2015.00569
Hazen, T. H., Kaper, J. B., Nataro, J. P., and Rasko, D. A. (2015b). Comparative genomics provides insight into the diversity of the attaching and effacing Escherichia coli virulence plasmids. Infect. Immun. 83, 4103–4117. doi: 10.1128/IAI.00769-15
Hazen, T. H., Sahl, J. W., Fraser, C. M., Donnenberg, M. S., Scheutz, F., and Rasko, D. A. (2013). Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 12810–12815. doi: 10.1073/pnas.1306836110
Hecht, G., Hodges, K., Gill, R. K., Kear, F., Tyagi, S., Malakooti, J., et al. (2004). Differential regulation of Na+/H+ exchange isoform activities by enteropathogenic E. coli in human intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G370–G388. doi: 10.1152/ajpgi.00432.2003
Heras, B., Shouldice, S. R., Totsika, M., Scanlon, M. J., Schembri, M. A., and Martin, J. L. (2009). DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 7, 215–225. doi: 10.1038/nrmicro2087
Hu, J., and Torres, A. G. (2015). Enteropathogenic Escherichia coli: foe or innocent bystander. Clin. Microbiol. Infect. 21, 729–734. doi: 10.1016/j.cmi.2015.01.015
Humphries, R. M., Griener, T. P., Vogt, S. L., Mulvey, G. L., Raivio, T., Donnenberg, M. S., et al. (2010). N-acetyllactosamine-induced retraction of bundle-forming pili regulates virulence-associated gene expression in enteropathogenic Escherichia coli. Mol. Microbiol. 76, 1111–1126. doi: 10.1111/j.1365-2958.2010.07192.x
Hyland, R. M., Sun, J., Griener, T. P., Mulvey, G. L., Klassen, J. S., Donnenberg, M. S., et al. (2008). The bundlin pilin protein of enteropathogenic Escherichia coli is an N-acetyllactosamine-specific lectin. Cell. Microbiol. 10, 177–187.
Ibarra, J. A., Garcia-Zacarias, C. M., Lara-Ochoa, C., Carabarin-Lima, A., Tecpanecatl-Xihuitl, J. S., Perez-Rueda, E., et al. (2013). Further characterization of functional domains of pera, role of amino and carboxy terminal domains in DNA binding. PLoS One 8:e56977. doi: 10.1371/journal.pone.0056977
Ide, T., Michgehl, S., Knappstein, S., Heusipp, G., and Schmidt, M. A. (2003). Differential modulation by Ca2+ of type III secretion of diffusely adhering enteropathogenic Escherichia coli. Infect. Immun. 71, 1725–1732. doi: 10.1128/IAI.71.4.1725-1732.2003
Iguchi, A., Thomson, N. R., Ogura, Y., Saunders, D., Ooka, T., Henderson, I. R., et al. (2009). Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191, 347–354. doi: 10.1128/JB.01238-08
Itou, A., Matsumoto, K., and Hara, H. (2012). Activation of the Cpx phosphorelay signal transduction system in acidic phospholipid-deficient pgsA mutant cells of Escherichia coli. Biochem. Biophys. Res. Commun. 421, 296–300. doi: 10.1016/j.bbrc.2012.04.003
Jeong, J. H., Kim, H. J., Kim, K. H., Shin, M., Hong, Y., Rhee, J. H., et al. (2012). An unusual feature associated with LEE1 P1 promoters in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 83, 612–622. doi: 10.1111/j.1365-2958.2011.07956.x
Jimenez, R., Cruz-Migoni, S. B., Huerta-Saquero, A., Bustamante, V. H., and Puente, J. L. (2010). Molecular characterization of GrlA, a specific positive regulator of ler expression in enteropathogenic Escherichia coli. J. Bacteriol. 192, 4627–4642. doi: 10.1128/JB.00307-10
Jones, C. H., Danese, P. N., Pinkner, J. S., Silhavy, T. J., and Hultgren, S. J. (1997). The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16, 6394–6406. doi: 10.1093/emboj/16.21.6394
Jubelin, G., Vianney, A., Beloin, C., Ghigo, J. M., Lazzaroni, J. C., and Lejeune, P., et al. (2005). CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187, 2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005
Justice, S. S., Hunstad, D. A., Harper, J. R., Duguay, A. R., Pinkner, J. S., Bann, J., et al. (2005). Periplasmic peptidyl prolyl cis-trans isomerases are not essential for viability, but SurA is required for pilus biogenesis in Escherichia coli. J. Bacteriol. 187, 7680–7686. doi: 10.1128/JB.187.22.7680-7686.2005
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Kashyap, D. R., Wang, M., Liu, L. H., Boons, G. J., Gupta, D., and Dziarski, R. (2011). Peptidoglycan recognition proteins kill bacteria by activating protein-sensing two-component systems. Nat. Med. 17, 676–683. doi: 10.1038/nm.2357
Katsowich, N., Elbaz, N., Pal, R. R., Mills, E., Kobi, S., Kahan, T., et al. (2017). Host cell attachment elicits posttranscriptional regulation in infecting enteropathogenic bacteria. Science 355, 735–739. doi: 10.1126/science.aah4886
Kenny, B. (1999). Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol. Microbiol. 31, 1229–1241. doi: 10.1046/j.1365-2958.1999.01265.x
Kenny, B., Abe, A., Stein, M., and Finlay, B. B. (1997a). Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect. Immun. 65, 2606–2612.
Kenny, B., DeVinney, R., Stein, M., Reinscheid, D. J., Frey, E. A., and Finlay, B. B. (1997b). Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520. doi: 10.1016/S0092-8674(00)80437-7
Kim, S., Song, I., Eom, G., Kim, S. (2018). A small periplasmic protein with a hydrophobic C-terminal residue enhances DegP proteolysis as a suicide activator. J. Bacteriol. 200:e00519-17. doi: 10.1128/JB.00519-17
Knutton, S., Baldwin, T., Williams, P., Manjarrezhernandez, A., and Aitken, A. (1993). The attaching and effacing virulence property of enteropathogenic Escherichia-coli. Zentralbl. Bakteriol. 278, 209–217. doi: 10.1016/S0934-8840(11)80838-8
Kotloff, K. L., Nataro, J. P., Blackwelder, W. C., Nasrin, D., Farag, T. H., Panchalingam, S., et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222. doi: 10.1016/S0140-6736(13)60844-2
Lee, L. J., Barrett, J. A., and Poole, R. K. (2005). Genome-wide transcriptional response of chemostat-cultured Escherichia coli to zinc. J. Bacteriol. 187, 1124–1134. doi: 10.1128/JB.187.3.1124-1134.2005
Leh, H., Khodr, A., Bouger, M. C., Sclavi, B., Rimsky, S., and Bury-Mone, S. (2017). Bacterial-chromatin structural proteins regulate the bimodal expression of the locus of enterocyte effacement (LEE) pathogenicity island in enteropathogenic Escherichia coli. mBio 8:e00773-17. doi: 10.1128/mBio.00773-17
Leverton, L. Q., and Kaper, J. B. (2005). Temporal expression of enteropathogenic Escherichia coli virulence genes in an in vitro model of infection. Infect. Immun. 73, 1034–1043. doi: 10.1128/IAI.73.2.1034-1043.2005
Levine, M. M., Nataro, J. P., Karch, H., Baldini, M. M., Kaper, J. B., Black, R. E., et al. (1985). The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152, 550–559. doi: 10.1093/infdis/152.3.550
Li, M., Rosenshine, I., Tung, S. L., Wang, X. H., Friedberg, D., Hew, C. L., et al. (2004). Comparative proteomic analysis of extracellular proteins of enterohemorrhagic and enteropathogenic Escherichia coli strains and their ihf and ler mutants. Appl. Environ. Microbiol. 70, 5274–5282. doi: 10.1128/AEM.70.9.5274-5282.2004
MacRitchie, D. M., Acosta, N., and Raivio, T. L. (2012). DegP is involved in Cpx-mediated posttranscriptional regulation of the type III secretion apparatus in enteropathogenic Escherichia coli. Infect. Immun. 80, 1766–1772. doi: 10.1128/IAI.05679-11
MacRitchie, D. M., Ward, J. D., Nevesinjac, A. Z., and Raivio, T. L. (2008). Activation of the Cpx envelope stress down-regulates expression of several locus of enterocyte-efacement-encoded genes in enteronathogenic response Eschetichia coli. Infect. Immun. 76, 1465–1475. doi: 10.1128/IAI.01265-07
Majowicz, S. E., Scallan, E., Jones-Bitton, A., Sargeant, J. M., Stapleton, J., Angulo, F. J., et al. (2014). Global incidence of human Shiga toxin-producing Escherichia coli infections and deaths: a systematic review and knowledge synthesis. Foodborne Pathog. Dis. 11, 447–455. doi: 10.1089/fpd.2013.1704
Makobe, C. K., Sang, W. K., Kikuvi, G., and Kariuki, S. (2012). Molecular characterization of virulence factors in diarrhoeagenic Escherichia coli isolates from children in Nairobi, Kenya. J. Infect. Dev. Ctries. 6, 598–604. doi: 10.3855/jidc.2082
McDaniel, T. K., and Kaper, J. B. (1997). A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E-coli K-12. Mol. Microbiol. 23, 399–407. doi: 10.1046/j.1365-2958.1997.2311591.x
Mecsas, J., Rouviere, P. E., Erickson, J. W., Donohue, T. J., and Gross, C. A. (1993). The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7, 2618–2628. doi: 10.1101/gad.7.12b.2618
Mellies, J. L., Benison, G., McNitt, W., Mavor, D., Boniface, C., and Larabee, F. J. (2011). Ler of pathogenic Escherichia coli forms toroidal protein-DNA complexes. Microbiology 157, 1123–1133. doi: 10.1099/mic.0.046094-0
Mellies, J. L., Elliott, S. J., Sperandio, V., Donnenberg, M. S., and Kaper, J. B. (1999). The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33, 296–306. doi: 10.1046/j.1365-2958.1999.01473.x
Mellies, J. L., Platenkamp, A., Osborn, J., and Ben-Avi, L. (2017). PerC manipulates metabolism and surface antigens in enteropathogenic Escherichia coli. Front. Cell. Infect. Microbiol. 7:32. doi: 10.3389/fcimb.2017.00032
Monjarás Feria, J., García-Gómez, E., Espinosa, N., Minamino, T., Namba, K., and González-Pedrajo, B. (2012). Role of EscP (Orf16) in injectisome biogenesis and regulation of type III protein secretion in enteropathogenic Escherichia coli. J. Bacteriol. 194, 6029–6045. doi: 10.1128/JB.01215-12
Mundy, R., MacDonald, T. T., Dougan, G., Frankel, G., and Wiles, S. (2005). Citrobacter rodentium of mice and man. Cell. Microbiol. 7, 1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x
Nakanishi, N., Abe, H., Ogura, Y., Hayashi, T., Tashiro, K., Kuhara, S., et al. (2006). ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 61, 194–205. doi: 10.1111/j.1365-2958.2006.05217.x
Nakanishi, N., Tashiro, K., Kuhara, S., Hayashi, T., Sugimoto, N., and Tobe, T. (2009). Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology 155, 521–530. doi: 10.1099/mic.0.023499-0
Nguyen, R. N., Taylor, L. S., Tauschek, M., and Robins-Browne, R. M. (2006). Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg. Infect. Dis. 12, 597–603. doi: 10.3201/eid1204.051112
Okeke, I. N., Borneman, J. A., Shin, S., Mellies, J. L., Quinn, L. E., and Kaper, J. B. (2001). Comparative sequence analysis of the plasmid-encoded regulator of enteropathogenic Escherichia coli Strains. Infect. Immun. 69, 5553–5564. doi: 10.1128/IAI.69.9.5553-5564.2001
Otto, K., and Silhavy, T. J. (2002). Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 2287–2292. doi: 10.1073/pnas.042521699
Padavannil, A., Jobichen, C., Mills, E., Velazquez-Campoy, A., Li, M., and Leung, K. Y., et al. (2013). Structure of GrlR-GrlA complex that prevents GrlA activation of virulence genes. Nat. Commun. 4:2546. doi: 10.1038/ncomms3546.
Pan, C. Q., Finkel, S. E., Cramton, S. E., Feng, J. A., Sigman, D. S., and Johnson, R. C. (1996). Variable structures of Fis-DNA complexes determined by flanking DNA-protein contacts. J. Mol. Biol. 264, 675–695. doi: 10.1006/jmbi.1996.0669
Pearson, J. S., Giogha, C., Wong Fok Lung, T., and Hartland, E. L. (2016). The Genetics of Enteropathogenic Escherichia coli Virulence. Annu. Rev. Genet. 50, 493–513. doi: 10.1146/annurev-genet-120215-035138
Portaliou, A. G., Tsolis, K. C., Loos, M. S., Balabanidou, V., Rayo, J., Tsirigotaki, A., et al. (2017). Hierarchical protein targeting and secretion is controlled by an affinity switch in the type III secretion system of enteropathogenic. EMBO J. 36, 3517–3531. doi: 10.15252/embj.201797515
Porter, M. E., Mitchell, P., Free, A., Smith, D. G., and Gally, D. L. (2005). The LEE1 promoters from both enteropathogenic and enterohemorrhagic Escherichia coli can be activated by PerC-like proteins from either organism. J. Bacteriol. 187, 458–472. doi: 10.1128/JB.187.2.458-472.2005
Porter, M. E., Mitchell, P., Roe, A. J., Free, A., Smith, D. G. E., and Gally, D. L. (2004). Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol. Microbiol. 54, 1117–1133. doi: 10.1111/j.1365-2958.2004.04333.x
Puente, J. L., Bieber, D., Ramer, S. W., Murray, W., and Schoolnik, G. K. (1996). The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20, 87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x
Raffa, R. G., and Raivio, T. L. (2002). A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45, 1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x
Raivio, T. L., Leblanc, S. K., and Price, N. L. (2013). The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J. Bacteriol. 195, 2755–2767. doi: 10.1128/JB.00105-13
Raivio, T. L., Popkin, D. L., and Silhavy, T. J. (1999). The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 181, 5263–5272.
Reece, S., Simmons, C. P., Fitzhenry, R. J., Batchelor, M., Hale, C., Matthews, S., et al. (2002). Mutagenesis of conserved tryptophan residues within the receptor-binding domain of intimin: influence on binding activity and virulence. Microbiology 148, 657–665. doi: 10.1099/00221287-148-3-657
Ronin, I., Katsowich, N., Rosenshine, I., and Balaban, N. Q. (2017). A long-term epigenetic memory switch controls bacterial virulence bimodality. eLife 6:e19599 doi: 10.7554/eLife.19599
Rosenshine, I., Ruschkowski, S., and Finlay, B. B. (1996). Expression of attaching/effacing activity by enteropathogenic Escherichia coli depends on growth phase, temperature, and protein synthesis upon contact with epithelial cells. Infect. Immun. 64, 966–973.
Ross, W., Thompson, J. F., Newlands, J. T., and Gourse, R. L. (1990). E.coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 9, 3733–3742.
Rutherford, B. J., Dahl, R. H., Price, R. E., Szmidt, H. L., Benke, P. I., Mukhopadhyay, A., et al. (2010). Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl. Environ. Microbiol. 76, 1935–1945. doi: 10.1128/AEM.02323-09
Sánchez-SanMartín, C., Bustamante, V. H., Calva, E., and Puente, J. L. (2001). Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183, 2823–2833. doi: 10.1128/JB.183.9.2823-2833.2001
Santos, A. S., and Finlay, B. B. (2015). Bringing down the host: enteropathogenic and enterohaemorrhagic Escherichia coli effector-mediated subversion of host innate immune pathways. Cell. Microbiol. 17, 318–332. doi: 10.1111/cmi.12412
Sazawal, S., Black, R. E., Bhan, M. K., Bhandari, N., Sinha, A., and Jalla, S. (1995). Zinc supplementation in young children with acute diarrhea in India. N. Engl. J. Med. 333, 839–844. doi: 10.1056/NEJM199509283331304
Scaletsky, I. C. A., Aranda, K. R. S., Souza, T. B., and Silva, N. P. (2010). Adherence factors in atypical enteropathogenic Escherichia coli strains expressing the localized adherence-like pattern in HEp-2 cells. J. Clin. Microbiol. 48, 302–306. doi: 10.1128/JCM.01980-09
Scaletsky, I. C. A., Aranda, K. R. S., Souza, T. B., Silva, N. P., and Morais, M. B. (2009). Evidence of pathogenic subgroups among atypical enteropathogenic Escherichia coli Strains. J. Clin. Microbiol. 47, 3756–3759. doi: 10.1128/JCM.01599-09
Sette, M., Spurio, R., Trotta, E., Brandizi, C., Brandi, A., Pon, C. L., et al. (2009). Sequence-specific recognition of DNA by the C-terminal domain of nucleoid-associated protein H-NS. J. Biol. Chem. 284, 30453–30462. doi: 10.1074/jbc.M109.044313
Sharma, V. K., and Zuerner, R. L. (2004). Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186, 7290–7301. doi: 10.1128/JB.186.21.7290-7301.2004
Sharp, F. C., and Sperandio, V. (2007). QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect. Immun. 75, 2432–2440. doi: 10.1128/IAI.02003-06
Shaulov, L., Gershberg, J., Deng, W., Finlay, B. B., and Sal-Man, N. (2017). The ruler protein escp of the enteropathogenic escherichia coli type III secretion system is involved in calcium sensing and secretion hierarchy regulation by interacting with the gatekeeper protein SepL. mBio 8:e01733-16 doi: 10.1128/mBio.01733-16
Shin, M. S. (2017). The mechanism underlying Ler-mediated alleviation of gene repression by H-NS. Biochem. Biophys. Res. Commun. 483, 392–396. doi: 10.1016/j.bbrc.2016.12.132
Singhal, M., Manzella, C., Soni, V., Alrefai, W. A., Saksena, S., Hecht, G. A., et al. (2017). Role of SHP2 protein tyrosine phosphatase in SERT inhibition by enteropathogenic. Am. J. Physiol. Gastrointest. Liver Physiol. 312, G443–G449. doi: 10.1152/ajpgi.00011.2017
Sperandio, V., Mellies, J. L., Delahay, R. M., Frankel, G., Crawford, J. A., Nguyen, W., et al. (2000). Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38, 781–793. doi: 10.1046/j.1365-2958.2000.02168.x
Spira, B., Ferreira, G. M., and de Almeida, L. G. (2014). relA enhances the adherence of enteropathogenic Escherichia coli. PLoS One 9:e91703. doi: 10.1371/journal.pone.0091703
Tarr, P. I., Gordon, C. A., and Chandler, W. L. (2005). Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365, 1073–1086. doi: 10.1016/S0140-6736(05)71144-2
Tobe, T., and Sasakawa, C. (2001). Role of bundle-forming pilus of enteropathogenic Escherichia coli in host cell adherence and in microcolony development. Cell. Microbiol. 3, 579–585. doi: 10.1046/j.1462-5822.2001.00136.x
Tobe, T., Schoolnik, G. K., Sohel, I., Bustamante, V. H., and Puente, J. L. (1996). Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21, 963–975. doi: 10.1046/j.1365-2958.1996.531415.x
Umanski, T., Rosenshine, I., and Friedberg, D. (2002). Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148, 2735–2744. doi: 10.1099/00221287-148-9-2735
Vogt, S. L., Nevesinjac, A. Z., Humphries, R. M., Donnenberg, M. S., Armstrong, G. D., and Raivio, T. L. (2010). The Cpx envelope stress response both facilitates and inhibits elaboration of the enteropathogenic Escherichia coli bundle-forming pilus. Mol. Microbiol. 76, 1095–1110. doi: 10.1111/j.1365-2958.2010.07145.x
Wang, P., Kuhn, A., and Dalbey, R. E. (2010). Global change of gene expression and cell physiology in YidC-depleted Escherichia coli. J. Bacteriol. 192, 2193–2209. doi: 10.1128/JB.00484-09
Wendrich, T. M., Blaha, G., Wilson, D. N., Marahiel, M. A., and Nierhaus, K. H. (2002). Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10, 779–788. doi: 10.1016/S1097-2765(02)00656-1
Winardhi, R. S., Gulvady, R., Mellies, J. L., and Yan, J. (2014). Locus of enterocyte effacement-encoded regulator (Ler) of pathogenic Escherichia coli competes off histone-like nucleoid-structuring protein (H-NS) through noncooperative DNA binding. J. Biol. Chem. 289, 13739–13750. doi: 10.1074/jbc.M113.545954
Wolfe, A. J., Parikh, N., Lima, B. P., and Zemaitaitis, B. (2008). Signal integration by the two-component signal transduction response regulator CpxR. J. Bacteriol. 190, 2314–2322. doi: 10.1128/JB.01906-07
Wong, A. R., Pearson, J. S., Bright, M. D., Munera, D., Robinson, K. S., Lee, S. F., et al. (2011). Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80, 1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x
Xue, Y., Osborn, J., Panchal, A., and Mellies, J. L. (2015). The RpoE stress response pathway mediates reduction of the virulence of enteropathogenic Escherichia coli by zinc. Appl. Environ. Microbiol. 81, 3766–3774. doi: 10.1128/AEM.00507-15
Yakhnin, H., Aichele, R., Ades, S. E., Romeo, T., and Babitzke, P. (2017). Circuitry linking the global Csr and σ dependent cell envelope stress response systems. J. Bacteriol. doi: 10.1128/JB.00484-17 [Epub ahead of print].
Yamamoto, K., and Ishihama, A. (2005). Transcriptional response of Escherichia coli to external zinc. J. Bacteriol. 187, 6333–6340. doi: 10.1128/JB.187.18.6333-6340.2005
Yamamoto, K., and Ishihama, A. (2006). Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci. Biotechnol. Biochem. 70, 1688-1695. doi: 10.1271/bbb.60024
Yang, J., Tauschek, M., Hart, E., Hartland, E. L., and Robins-Browne, R. M. (2010). Virulence regulation in Citrobacter rodentium: the art of timing. Microb. Biotechnol. 3, 259–268. doi: 10.1111/j.1751-7915.2009.00114.x
Ye, F., Yang, F., Yu, R., Lin, X., Qi, J., Chen, Z., et al. (2018). Molecular basis of binding between the global post-transcriptional regulator CsrA and the T3SS chaperone CesT. Nat. Commun. 9:1196. doi: 10.1038/s41467-018-03625-x
Yerushalmi, G., Nadler, C., Berdichevski, T., and Rosenshine, I. (2008). Mutational analysis of the locus of enterocyte effacement-encoded regulator (Ler) of enteropathogenic Escherichia coli. J. Bacteriol. 190, 7808–7818. doi: 10.1128/JB.00663-08
Yitzhaki, S., Rostron, J. E., Xu, Y., Rideout, M. C., Authement, R. N., Barlow, S. B., et al. (2012). Similarities between exogenously- and endogenously-induced envelope stress: the effects of a new antibacterial molecule, TPI1609-10. PLoS One 7:e44896. doi: 10.1371/journal.pone.0044896
Zhang, H. Z., and Donnenberg, M. S. (1996). DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol. Microbiol. 21, 787–797. doi: 10.1046/j.1365-2958.1996.431403.x
Zhu, C., Feng, S., Thate, T. E., Kaper, J. B., and Boedeker, E. C. (2006). Towards a vaccine for attaching/effacing Escherichia coli: a LEE encoded regulator (ler) mutant of rabbit enteropathogenic Escherichia coli is attenuated, immunogenic, and protects rabbits from lethal challenge with the wild-type virulent strain. Vaccine 24, 3845–3855. doi: 10.1016/j.vaccine.2005.07.019
Keywords: EPEC, LEE, environment, virulence, sRNA, metabolism, envelope stress
Citation: Platenkamp A and Mellies JL (2018) Environment Controls LEE Regulation in Enteropathogenic Escherichia coli. Front. Microbiol. 9:1694. doi: 10.3389/fmicb.2018.01694
Received: 20 April 2018; Accepted: 09 July 2018;
Published: 27 July 2018.
Edited by:
Xihui Shen, Northwest A&F University, ChinaReviewed by:
Emily Mallick, Fluid-Screen, United StatesChristian Rueter, Universitätsklinikum Münster, Germany
Copyright © 2018 Platenkamp and Mellies. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jay L. Mellies, amF5Lm1lbGxpZXNAcmVlZC5lZHU=
 Amy Platenkamp
Amy Platenkamp Jay L. Mellies
Jay L. Mellies