Abstract
The objective of this study was to evaluate the presence of Cronobacter sakazakii and microbiological parameters in dairy products associated with a food alert. Ninety dairy product samples were analyzed, including seven commercial brands and two product types (liquid and powdered) from four countries. Aerobic plate count (APC) and Enterobacteriaceae count were performed according to Chilean standards. Cronobacter spp. and C. sakazakii were identified by polymerase chain reaction real time amplification of rpoB and cgcA genes and the genotype by multilocus sequence typing. Eighty-eight percent of dairy products showed APC higher than the detection limit. Fifty percent of liquid commercial brand samples contained APC: 2.6, 2.3, 1.1, and 2.9 CFU/mL in brands A, C, E, and G, respectively. Results for powdered commercial brands were 3.0, 3.6, and 5.7 CFU/g in brands B, D, and F, respectively. Maximum count (5.7 CFU/g) occurred in brand F dairy product manufactured in Chile. Enterobacteriaceae were found in 55% of the samples, 64% in liquid and 51% in powdered commercial brands. In 50% of brands B, D, and E, samples contained 2.9, 2.8, and 2.7 log CFU/g, respectively. Only liquid commercial brands from the United States had Enterobacteriaceae values between 0.1 and 4.5 CFU/mL. Seventeen suspicious strains were isolated and nine were identified as Enterobacter spp. Only eight suspicious strains from four powdered commercial brands (Chile and Singapore) were confirmed as C. sakazakii by rpoB and cgcA gene amplification and fusA sequencing. C. sakazakii prevalence in the analyzed samples was 8.8%. There were 11% of powdered milk brands that contained APC between 4.0 and 4.7 log CFU/g and 55% of the samples contained Enterobacteriaceae. C. sakazakii was found in dairy products manufactured in Chile and Singapore. On the basis of this information, the Chilean Ministry of Health (RSA) decreed a national and international food alert and recalled all the product batches that resulted positive in the present study from supermarkets and pharmacies.
Introduction
On June 2, 2017 the Chilean Ministry of Health issued a national and international food alert as a result of the presence of Cronobacter sakazakii in two powdered formula samples intended for children under 10. Researchers from the Universidad del Bío-Bío conducted a study which led to the food alert. This preventive measure was adopted because of the risk of disease associated with Cronobacter spp. and C. sakazakii in hypersensitive groups of the population (Food and Agriculture Organization and World Health Organization [FAO/WHO], 2008; Jason, 2015).
Cronobacter spp. was initially defined as the new bacterial species Enterobacter sakazakii by Farmer et al. (1980); it was later classified by Iversen et al. (2008) and Joseph et al. (2012a,b) as Cronobacter spp. and included seven species: C. sakazakii, C. malonaticus, C. universalis, C. turicensis, C. muytjensii, C. dublinensis, and C. condimenti.
Cronobacter spp. is considered as an emerging pathogen that is especially aggressive in hypersensitive individuals, such as children and the elderly (Fanning and Forsythe, 2008; Hunter and Bean, 2013). Newborns and the elderly are the population groups that are most affected by C. sakazakii, although the highest incidence and severity occurs in preterm infants (Hariri et al., 2013). Outbreaks have generally been the most common cause of infection (Jason, 2012). Clinical symptoms are mainly found in meningitis, septicemia, or necrotizing enteritis in infants (Block et al., 2002; Bowen and Braden, 2006), but diarrhea, urinary tract infection, and septicemia have also been observed. Mortality rates are associated with general infection (42–80%) and neonatal meningitis and septicemia (15–25%) (Holý and Forsythe, 2014).
The disease is associated with the consumption of rehydrated milk as a carrier of the pathogen, as well as the eventual involvement of utensils and equipment as reservoirs (Food and Agriculture Organization and World Health Organization [FAO/WHO], 2008; Kalyantanda et al., 2015). Since it is widespread, it can be isolated in powdered infant formula (PIF), rehydrated milk (R-PIF), infant cereals, various foods, water, surfaces, homes, and hospitals (Baumgartner et al., 2009). Even when the source of primary contamination is unclear (Norberg et al., 2012), some researchers suggest that the natural habitat is PIF manufacturing plants. This situation has been reported by Jacobs et al. (2011) and Li et al. (2014), who identified Cronobacter spp. in different parts of PIF plants in both China and Australia. It has also been strongly associated with by-products used in its formulation, which are also probable carriers (Jongenburger et al., 2011). The control of this pathogen in the first stages of PIF production is the most important step to reduce its incidence in the final product (Yan et al., 2012; Fei et al., 2015) because viable Cronobacter spp. strains have been found 2 years after the product was packaged (Caubilla-Barron and Forsythe, 2007).
Although updated detection and identification techniques are being used, there are still cases of disease and mortality every year (Norberg et al., 2012). It is therefore necessary to improve hygiene and the production process to reduce the impact of C. sakazakii. Biochemical tests (API 20E, RAPID, BIOLOG microarray), molecular confirmation of the Cronobacter spp. genus by polymerase chain reaction (PCR) (Lehner et al., 2004; Cetinkaya et al., 2012), and especially multilocus sequence typing (MLST) have been used to complement its identification. These techniques have allowed advances in correctly identifying it, and thus decreasing the possibility of false negatives (Baldwin et al., 2009; Joseph and Forsythe, 2011; Yan et al., 2015; Ogrodzki and Forsythe, 2017). Several primers have been generated to detect Cronobacter spp. by amplifying specific sequences of variable and conserved regions of 16S ribosomal rDNA of the bacterium (Lehner et al., 2004; Hassan et al., 2007), OmpA (Mohan-Nair and Venkitanarayanan, 2006), as well as others that are more specific, which detect C. sakazakii by rpoB (Stoop et al., 2009; Lehner et al., 2012; Li et al., 2016) and cgcA gene amplification (Carter et al., 2013).
Studies of Cronobacter spp. incidence in powdered milk have demonstrated a positivity range between 3 and 30% (Chap et al., 2009; Siqueira-Santos et al., 2013; Fei et al., 2017). Sáez et al. (2012) found 5% positivity of Cronobacter spp. in 80 PIF samples from a dairy processing plant in the Los Lagos Region in Chile. Parra-Flores et al. (2015a) found an incidence of 9.5% C. sakazakii in an exploratory study with a limited number of samples using MLST in PIF manufactured in Chile in 2014. If PIF samples manufactured in other countries are considered, incidence was 2.7% in all the analyzed PIF samples.
Given the need to ensure safety in PIFs, the FAO/WHO have held expert meetings to study cases of diseases related to its consumption, whether epidemiologically or microbiologically. Three categories of microorganisms were identified based on the soundness of evidence of a causal relationship between their presence in food and the disease: (A) microorganisms with clear causality evidence, enteric Salmonella, and Cronobacter spp. (E. sakazakii); (B) microorganisms in which causality is possible but has not yet been demonstrated, primarily from the Enterobacteriaceae family; and (C) microorganisms in which causality is less probable or has not yet been demonstrated, and have not been identified in PIF (Food and Agriculture Organization and World Health Organization [FAO/WHO], 2006; Jackson et al., 2015). The WHO therefore recommended the absence of Cronobacter spp., Salmonella, and Enterobacteriaceae in dairy products (Food and Agriculture Organization and World Health Organization [FAO/WHO], 2004, 2006).
Given that PIFs are not sterile foods, the determination of microbial indicators, such as aerobic plate count (APC) and Enterobacteriaceae (ENT), provides useful information about the hygienic conditions of their preparation or post-process contamination (Friedemann, 2009; Parra-Flores et al., 2015a; Heperkan et al., 2017).
Cronobacter spp. was not considered in the Chilean Food Sanitary Regulations (Reglamento Sanitario de los Alimentos, RSA) when the Chilean Ministry of Health decreed the food alert. The decision was taken due to the risk of disease associated with the pathogen described in the scientific literature (Jason, 2015), factors that affect PIF contamination (Parra-Flores et al., 2015b), and the variability in its cellular response (Parra-Flores et al., 2016). The PIFs associated with this alert are not only commercialized in Chile but throughout the Americas.
Therefore, the objective of this study was to evaluate the presence of C. sakazakii and the microbiological parameters of APC and ENT in dairy products associated with a food alert in Chile in June 2017.
Materials and Methods
Food Samples
Sampling was conducted from August 2016 to May 2017. Ninety samples were collected in four countries (United States, Singapore, Chile, and Holland), from three manufacturers (1, 2, and 3), seven commercial dairy brands (A, B, C, D, E, F, G) of which B, D, and F were powdered and A,C, E, and G were liquid products sold in supermarkets and pharmacies in Chile. All the analyses were performed in duplicate. The sampling criteria used as a reference were the standards of the Chilean RSA and CAC/RPC 66 of the Codex Alimentarius.
Microbiological Quantification
The APC of the mesophilic microorganisms and ENT count were used. Quantification of both microbial groups and identification of isolated enterobacteria (including suspicious strains of Salmonella or Escherichia coli) were performed in the Accredited Food Testing and Certification Laboratory (LECYCA-UBB) and the Molecular Epidemiology and Microbiology Laboratory of the Universidad del Bío-Bío. References are NCh 2659 (2002) for AMC, NCh 2676 (2002) for ENT, NCh 2636 (2002) for E. coli, and NCh 2675 (2002) to isolate Salmonella.
Isolation of Cronobacter spp.
The technique described by Parra-Flores et al. (2015a) was applied. For each sample, 225 mL of buffered peptone water (BPW) were added to 25 g of powdered infant formula (PIF) or dairy product (DP) and then homogenized in a stomacher at a mean velocity for 60 s. Liquid products in their original container were directly incubated at 37°C. Then 10 mL of each sample was inoculated after incubation at 37°C for 24 h in 90 mL Enterobacteriaceae enrichment broth (BD Difco, Sparks, MD, United States). A loop was extracted from the culture suspension and striated in Brilliance Chromogenic Agar CM 1035 (Oxoid Thermo Fisher, United Kingdom) at 37°C for 20 h. Five strains, presumed to be colonies of Cronobacter spp. (green or blue), were striated in trypticase soy agar (BD Difco, Sparks, MD, United States) to verify their purity prior to future analyses. The isolated strains were maintained in a strain collection and stored at -80°C.
Identification of Cronobacter spp. and Cronobacter sakazakii
Genomic DNA of the suspicious strains was extracted and purified with the Ultra Clean® Microbial DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, United States). The strains were confirmed as Cronobacter spp. by OmpA gene amplification (Mohan-Nair and Venkitanarayanan, 2006) and later identified as C. sakazakii by qPCR amplification of rpoB and cgcA genes (Stoop et al., 2009; Carter et al., 2013) (Table 1) in the Stratagene Mx3000P qPCR System equipment (Agilent Technologies).
Table 1
| Identification | Gene | Primer sequence | Reference | Annealing temperature |
|---|---|---|---|---|
| Cronobacter spp. | 16S | F: GCT YTG CTG ACG AGT GGC GGR: ATC TCT GCA GGA TTC TCT GG | Lehner et al., 2004 | 60°C |
| Cronobacter spp. | OmpA | F: GGATTTAACCGTGAACTTTTCCR: CGCCAGCGATGTTAGAAGA | Mohan-Nair and Venkitanarayanan, 2006 | 60°C |
| Cronobacter sakazakii | rpoB | F: ACG CCA AGC CTA TCT CCG CGR: ACG GTT GGC GTC ATC GTG | Stoop et al., 2009 | 67°C |
| Cronobacter sakazakii | cgcA | F: GTGGCSGGGTATGACAAAGACR: GGCGGACGAAGCCTCAGAGAGT | Carter et al., 2013 | 62°C |
| Cronobacter sakazakii | fusA | F: GAAACCGTATGGCGTCAG’R: AGAACCGAAGTGCAGACG | Baldwin et al., 2009. | 58°C |
Polymerase chain reaction (PCR) primers used in the study.
Sequencing of fusA Gene to Identify Species of Cronobacter spp.
The methodology described by Baldwin et al. (2009) was followed using PCR CORE Kit QIAGEN (Cat No. 201225) solutions. Amplified products were sent to MACROGEN in Korea for sequencing. Identification was performed with the free access online database https://pubmlst.org/cronobacter/ and BLASTn (NCBI).
Bioinformatic and Statistical Analyses
The sequenced products were analyzed with the Gentle software and later aligned with the ClustalW software. A phylogenetic tree was constructed using the maximum likelihood method with the MEGA7 software. Statistical description included measures of central tendency, dispersion, and position for quantitative variables, while absolute frequencies and percentages were used for qualitative variables. The Mann–Whitney and Kruskal–Wallis tests were used for comparison purposes with the STATA 14 software at the significance level α = 0.05.
Results
Of the 90 analyzed Chilean and foreign samples, 79 had APC. When analyzing APC for each DP commercial brand, no significant statistical differences were found (p > 0.05). However, half of the liquid DP commercial brands contained 2.6 log CFU/mL, 2.3 log CFU/mL, 1.1 log CFU/mL, and 2.9 log CFU/mL for brands A, C, E, and G, respectively. The powdered DP (including PIF) commercial brands had values of 3.0 log CFU/g, 3.6 log CFU/g, and 5.7 log CFU/g for brands B, D, and F, respectively. Positivity for ENT was found in all the evaluated brands. In the liquid DP brands A, C, E, and G, the sample means were 1.8, 0.8, <0.1, and <0.1 log UFC/mL, respectively. In half of powdered DP brands B, D, and E, samples contained 2.9, 2.8, and 2.7 log CFU/g, respectively. Statistical differences were only found in the ENT counts (p = 0.012). For DP manufacturers, company 2 had the highest count with 5.9 log CFU/g. Enterobacteriaceae counts were found in 55% of the total analyzed samples. Company 1 obtained the highest counts with 3.5 log CFU/g (Table 2).
Table 2
| Counts | ||
|---|---|---|
| APC log CFU/g Median (minimum–maximum) | ENT log CFU/g Median (minimum–maximum) | |
| Commercial brand | ||
| A | 2.6 (1.7–5.7) | 1.8 (1.6–4.5) |
| B | 3.0 (2.3–4.1) | 2.9 (1.0–3.6) |
| C | 2.3 (2.0–5.8) | 0.85 (0–2.9) |
| D | 3.6 (1.3–5.9) | 2.8 (1.0–4.4) |
| E | 1.1 (0.9–5.8) | <0.1 (0.1–4.5) |
| F | 5.7 (1.3–6.7) | 2.7 (1.0–3.0) |
| G | 2.9 (2.6–5.7) | <0.1 |
| p-value∗ | ns | 0.0121 |
| Manufacturer | ||
| 1 | 3.0 (1.3–5.9) | 2.6 (0–4.5) |
| 2 | 5.7 (0.9–6.8) | 2.7 (1.0–3.0) |
| 3 | 2.9 (2.6–5.7) | <0.1 |
| p-value∗ | ns | 0.0107 |
Aerobic plate count (APC) and Enterobacteriaceae (ENT) count for each commercial brand and manufacturer.
∗p-value according to Kruskal–Wallis test. Brands A, C, E, and G: liquid dairy products; Brands B, D, and F: powdered milk products; ns: non-significant.
Regarding the country of origin (Table 3), Chile exhibited the highest APC count means with 5.0 log CFU/g and the United States showed the lowest count with 2.1 log CFU/g. The DP produced in Holland had the highest ENT counts followed by Singapore with 3.6 and 3.2 log CFU/g, respectively. The lowest count was obtained in the US with 0.1 log CFU/g. No significant differences existed in the APC and ENT counts for country of origin (p > 0.05). As for the type of DP, 100% of the liquid DP brands contained APC and 55% ENT counts. Only the liquid DP brands produced in the US had ENT counts with values between 0 and 4.5 log CFU/mL. The highest count in powdered DPs was obtained in Holland with 4.4 log CFU/g.
Table 3
| Counts | ||
|---|---|---|
| APC log CFU Median (minimum–maximum) | ENT log CFU Median (minimum–maximum) | |
| Country | ||
| Chile | 5.0 (1.3–6.8) | 2.7 (1.0–3.0) |
| Holland | 3.6 (1.3–5.9) | 2.8 (1.0–4.4) |
| Singapore | 3.0 (2.3–4.1) | 2.9 (1.0–3.6) |
| United States | 2.1 (0.9–5.8) | 1.7 (0–4.5) |
| p-value∗ | ns | ns |
Aerobic plate count (APC) and Enterobacteriaceae (ENT) count for each country.
∗p-value according to Kruskal–Wallis test; ∗∗p-value according to Mann–Whitney test. ns, non-significant.
Of the total analyzed samples, 17 suspicious strains were isolated from the chromogenic agar. Nine were identified as E. cloacae, Klebsiella pneumoniae, E. hormaechei, and Enterobacter spp., whereas Salmonella spp. was not isolated in any of the samples. However, E. coli was identified in one powdered milk (PM) product manufactured in Chile.
Only eight suspicious strains from the PM from Chile and Singapore were confirmed as Cronobacter spp. by amplifying the ompA gene. These strains were subsequently confirmed as C. sakazakii through the amplification of the gene products for rpoB and cgcA by PCR in real time. One of the PM products in which C. sakazakii was isolated was intended for consumption by infants under 2 years (CH84), and another from Singapore was intended for consumption by children older than 1 year (CH65).
Furthermore, six more strains were confirmed, which were not part of the food alert because of their expiry date and were PM products manufactured in Chile (CH42, CH43, CH44, CH45, CH50, and CH85) (Table 4). Two more samples were also detected by real-time PCR with Cronobacter spp. from PIFs manufactured in Holland, but it was not possible to recover the pathogen from the samples.
Table 4
| Strain | Gene amplification | Identification | |||||
|---|---|---|---|---|---|---|---|
| 16S | OmpA | rpoB | cgcA | fusA sequencing allelle | MLST database | BLAST | |
| CH42 | + | + | + | + | 1 | Cronobacter sakazakii | Cronobacter sakazakii |
| CH43 | + | + | + | + | 1 | Cronobacter sakazakii | Cronobacter sakazakii |
| CH44 | + | + | + | + | 1 | Cronobacter sakazakii | Cronobacter sakazakii |
| CH45 | + | + | + | + | 16 | Cronobacter sakazakii | Cronobacter sakazakii |
| CH50 | + | + | + | + | 16 | Cronobacter sakazakii | Cronobacter sakazakii |
| CH65 | + | + | + | + | 1 | Cronobacter sakazakii | Cronobacter sakazakii |
| CH84 | + | + | + | + | 71 | Cronobacter spp. | Cronobacter sakazakii |
| CH85 | + | + | + | + | 71 | Cronobacter spp. | Cronobacter sakazakii |
Identification of Cronobacter spp. and Cronobacter sakazakii by molecular amplification.
Cronobacter sakazakii incidence in the total evaluated samples was 8.8% (Table 5).
Table 5
| Positivity of Cronobacter sakazakii in dairy product (DP) samples | |||||
|---|---|---|---|---|---|
| Country | n | Powdered infant formula | Liquid DP | Total | |
| (+) | (+) | (+) | % | ||
| Chile | 20 | 7 | 0 | 7 | 35 |
| United States | 25 | NE | 0 | 0 | 0 |
| Holland | 35 | 0 | NE | 0 | 0 |
| Singapore | 10 | 1 | NE | 1 | 10 |
| Total | 90 | 8 | 0 | 8 | 8.8 |
Positivity of Cronobacter sakazakii for country of origin and product type.
NE, not evaluated.
All the C. sakazakii strains were genotyped by sequencing the fusA gene using MLST in the database https://pubmlst.org/cronobacter/ and BLASTn (NCBI). The information of the sequences was later used to construct a phylogenetic tree (Figure 1).
FIGURE 1
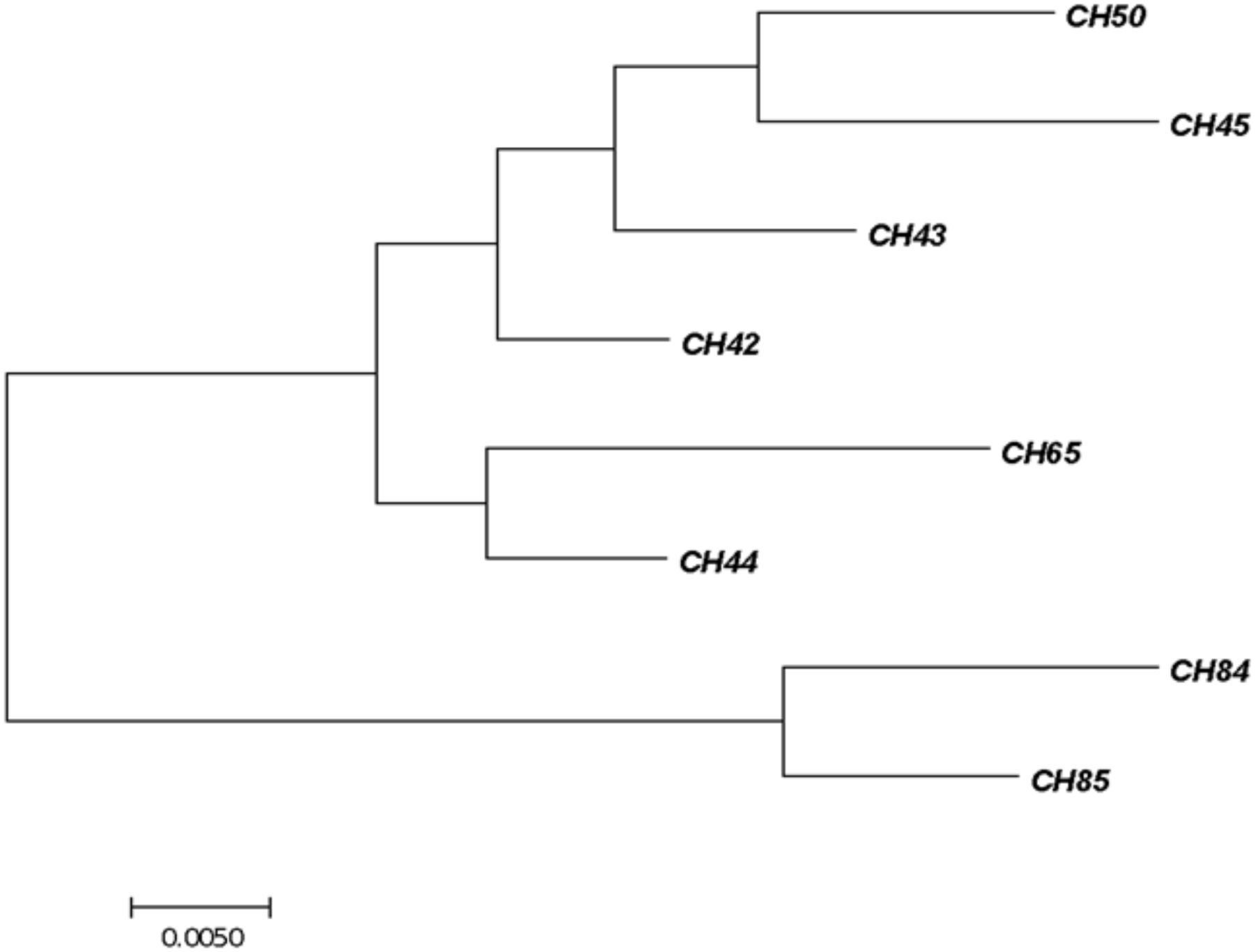
Phylogenetic tree of fusA sequencing identified as Cronobacter sakazakii. The tree with the highest log likelihood (–1450.2262) is shown. The initial tree for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms. The analysis involved 24 nucleotide sequences. There were 401 positions in the final dataset. Evolutionary analyses were conducted with the MEGA7 software program.
Discussion
The PIFs analyzed in the present study are commercialized throughout the Americas. Therefore, evaluating their microbiological quality allows determining aspects such as the hygienic conditions in which they were prepared, as well as identifying microbial hazards from probable recontamination occurring when they are supplemented with nutrients after pasteurization (Kent et al., 2015).
For APC, 72% of all analyzed powdered and liquid DP samples contained less than 3 log CFU. There were 11% of PM brands that contained between 4.0 and 4.7 log CFU/g originating only from the United States and Singapore; two products manufactured in Chile had values of 6.7 log CFU/g. Liquid DPs revealed five samples that ranged between 5.6 and 5.8 log CFU/g, four of which were produced in the United States and one in Chile. These APC values are within ranges reported by other authors (Iversen et al., 2004; Kim et al., 2011; Parra-Flores et al., 2015a). However, it was a concern to find counts greater than 5 log CFU/g in PM; these values were very high compared to results reported by Chap et al. (2009), who found 2% in this range. Heperkan et al. (2017) found values between 1.7 log for PIF and 4.9 log CFU/g for PM in a study of 80 PM samples; counts were similar to those determined in the present study. There is an evident need to control the contamination sources of DM products due to the wide range of microorganisms present in the APC and the higher susceptibility of infection in children of different ages who consume DPs (Chap et al., 2009).
Positivity was found for ENT in 55% of all analyzed samples, and there was a significant statistical relationship in the counts for commercial brand (p = 0.012) and manufacturer (p = 0.010). Eight PM and two liquid DPs obtained count means of 2 log CFU and 11 samples had values from 3.4 to 4.5 log CFU. Muytjens et al. (1988) encountered ENT in 52% of 141 evaluated formulas from 35 countries. On the other hand, ENT was found in 47% of PIF manufactured in Indonesia and Malaysia (Estuningsih et al., 2006), 22.5% in Ivory Coast (Yao et al., 2012), and 100% in Chile (Parra-Flores et al., 2015a). All these ENT counts are much higher than the values permitted according to the international standard of Codex Alimentarius Commission (2008), which requires the absence of this indicator in 10 g. In the present study, the high ENT positivity is compatible with the presence of several opportunistic microorganisms and pathogens associated with disease in infants reported in different publications (Food and Agriculture Organization and World Health Organization [FAO/WHO], 2006; Kent et al., 2015). Therefore, these findings should be analyzed in terms of risk associated with the consumption of PM by infants, the lack of control by manufacturers, and health authorities responsible for inspection in Chile. Although the association between the risk of falling ill with the consumption of ENT-contaminated PM has not yet been established with certainty, its absence in PM provides additional protection for newborns, especially the preterm, immunocompromised, and those with low (<2,500 g) and very low (<1,500 g) birth weight during the preparation, storage, and administration of infant feeding (Abdullah Sani et al., 2013).
Other microorganisms belonging to this family were also identified in the present study. E. cloacae, K. pneumoniae, and E. hormaechei were found. This situation does not seem altogether exceptional considering that other authors have also found these microorganisms of the ENT family in PIF (Iversen et al., 2004; Giammanco et al., 2011; Kim et al., 2011; Abdullah Sani et al., 2013), and especially for the risk associated with its ability to maintain itself for at least 8 months under desiccation conditions (Caubilla-Barron and Forsythe, 2007; Juma et al., 2016). Furthermore, the Food and Agriculture Organization and World Health Organization [FAO/WHO] (2004, 2006) also recognized that other ENT can be recovered from PIF and could present a risk to infants, although no reported cases had been confirmed at that time. Jackson et al. (2015) re-evaluated a reported C. sakazakii outbreak through the consumption of contaminated reconstituted PIF in Mexico. Using DNA sequencing, they demonstrated that the causative agents were misidentified strains of E. hormaechei and Enterobacter spp. Meanwhile, E. hormaechei has been shown to have clinical significance with the report of several outbreaks of sepsis in neonatal intensive care units in Brazil and the United States (Wenger et al., 1997; Campos et al., 2007; Townsend et al., 2008).
The high APC and ENT values can indicate a non-strict adherence to hygienic practices recommended for the preparation of PIF, which has been mentioned by other authors (Mullane et al., 2007; Jongenburger et al., 2011). This can imply a permanent risk for populations that usually consume this product.
The C. sakazakii incidence was 8.8% in the total of evaluated samples, particularly in 10 and 35% of samples produced in Singapore and Chile, respectively. This high positivity should give rise to greater control by the manufacturers and health authorities because of its high lethality, related neurological sequela, and risk of falling ill by C. sakazakii (Lai, 2001; Holý and Forsythe, 2014). An infection rate of 1 in 100,000 newborns has been estimated in the United States; this rate increases to 8.7 in 100,000 in infants weighing less than 1500 g, and 1 in 10,660 preterm infants with low birth weight (Hunter and Bean, 2013). In Holland, Cronobacter spp. causes from 0.5 to 0.7% of all the cases of meningitis in infants, with a probable range of infection of 0.00062 to 0.62 cases per year. When this probability is adjusted with all the cases that have occurred in the last 30 years, the projected probability is 0.53 cases of infection per year with a rate of 1 in 100,000 infants (Reij et al., 2009). There is no doubt that these values are greatly underestimated (Kucerova et al., 2011; Jason, 2015). Patrick et al. (2014) stated that the median age in adults is 59 for disease caused by Cronobacter spp., this value has been widely referred to by the lay press and the representatives of formula manufacturers. Parra-Flores et al. (2016) evaluated cell response variability of C. sakazakii after mild heat treatments using stochastic approaches and reported that these can better describe microbial single cell response than deterministic models. They found that the mean probability of illness from the initial inoculum size of 1 cell was less than 0.2 in all cases, while the mean probability of illness was greater than 0.7 in most cases for the inoculum size of 50 cells.
A principal aspect of our study was the correct identification of Cronobacter spp. and C. sakazakii species by several methods described in the literature (Table 1). When comparing the methods, a very good correlation was found for these methods by using different primers with fusA gene sequencing, which today enables the most accurate speciation because it follows the whole genome phylogeny and adjusts to taxonomic changes (Forsythe et al., 2014; Jackson et al., 2014; Xu et al., 2014; Alsonosi et al., 2015; Vojkovska et al., 2016). Therefore, the information generated when using molecular techniques can improve the confidence level in the identification and confirmation of presumptive strains even when molecular tests provide the best identification and phenotyping methods (Joseph et al., 2013; Jackson and Forsythe, 2016).
The robustness of the results was a primary facet in our decision to declare the national and international food alert and the massive recall of the products involved. There was another important point health authorities needed to consider, that is, the 2007 WHO recommendation to use water at >70°C to rehydrate PM to limit the risk of infection by Cronobacter spp. (World Health Organization [WHO], 2007). It was also recommended that rehydrated PM for children be administered within 2 h of its preparation or conservation under refrigeration at <4°C. This was the main idea of the publicity campaign launched by the authorities for the Chilean population.
Unfortunately, there are situations that warn us that microbiological control cannot be relaxed in food products consumed by hypersensitive populations, such as children and the elderly. For example, the case of the recall of C. sakazakii-contaminated PM destined for children in Argentina in 2015, and the recent contamination of Lactalis milk with Salmonella spp., which affected 83 European countries. However, recent studies demonstrate that the microbiological quality of these products is still inadequate even when we know that milk and DPs for child feeding are not sterile (Parra-Flores et al., 2015a).
In summary, an inadequate microbiological quality of powdered and liquid PM consumed by children under 10 was found in the present study. The presence of ENT and C. sakazakii was also identified, which is a wake-up call to manufacturers and public health regulatory authorities in Chile and throughout the Americas. It is therefore necessary to establish greater control of hygienic conditions in PM production and microbiological vigilance to prevent unnecessary risks for the child population that massively consumes these products (Koletzko et al., 2012). Disease caused in children as a consequence of any pathogen present in the PM they consume, requires manufacturers and health authorities to ensure the highest possible level of food safety (Food and Agriculture Organization and World Health Organization [FAO/WHO], 2006; Jason, 2015; Kent et al., 2015).
Conclusion
Powdered infant milk formulas (PIF) are not sterile products; according to the specifications established by the Codex Alimentarius, this type of product should be treated as a possible food safety issue for high risk populations, such infants and neonates, due to the presence of the C. sakazakii pathogen. A total of 11% of the powdered milk brands contained APCs between 4.0 and 4.7 log CFU/g, which is considered as the rejection level by the updated Chilean Food Sanitary Regulations (RSA). Of all the samples, 55% contained Enterobacteriaceae; E. cloacae, E. hormaechei, and K. pneumoniae were identified. The overall incidence of C. sakazakii was 8.8%, which was found in samples produced either in Chile or Singapore.
Based on this information, the Chilean Ministry of Health decreed a national and international food alert and recalled all the product batches from supermarkets and pharmacies that tested positive in the study. After the first survey conducted for PIF contaminated with Cronobacter spp., it was pointed out that this microorganism was present and represented a risk that was not considered in the Chilean food safety standards. The RSA therefore included a new regulation for Cronobacter spp. in PIF in November 2017 because of social media pressure and the scientific results provided by our team.
Statements
Author contributions
JP-F conceived the experiments. JP-F, FC-L, and JA designed the experiments. NV-R, AC, and JP-F conducted the laboratory work. JP-F, NV-R, AR, and FC-L drafted the manuscript. All authors reviewed and approved the final manuscript.
Funding
This study was provided by the Research Directorate of the Universidad del Bío-Bío, Projects 161720 3/R, 091824/R, and GI 171220/EF.
Acknowledgments
We wish to thank the Chilean Ministry of Health, the Chilean Institute of Public Health, and the Research Directorate of the Universidad del Bío-Bío.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Abdullah SaniN.HartantyoS.ForsytheS. (2013). Microbiological assessment and evaluation of rehydration instructions on powdered infant formulas, follow-up formulas and infant foods in Malaysia.J. Dairy Sci.961–8. 10.3168/jds.2012-5409
2
AlsonosiA.HaririS.KajsikM.OrieskovaM.HanulikV.RoderovaM.et al (2015). The speciation and genotyping of Cronobacter isolates from hospitalised patients.Eur. J. Clin. Microbiol. Infect. Dis.341979–1988. 10.1007/s10096-015-2440-8
3
BaldwinA.LoughlinM.Caubilla-BarronJ.KucerovaE.ManningG.DowsonC.et al (2009). Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes.BMC Microbiol.9:223. 10.1186/1471-2180-9-223
4
BaumgartnerA.GrandM.LinigerM.IversenC. (2009). Detection and frequency of Cronobacter spp. (Enterobacter sakazakii) in different categories of ready-to-eat foods other than infant formula.Int. J. Food Microbiol.136189–192. 10.1016/j.ijfoodmicro.2009.04.009
5
BlockC.PelegO.MinsterN.Bar-OzB.SimhonA.AradI.et al (2002). Cluster of neonatal infections in Jerusalem due to unusual biochemical variant of Enterobacter sakazakii.Eur. J. Clin. Microbiol. Infect. Dis.21613–616. 10.1007/s10096-002-0774-5
6
BowenA.BradenC. (2006). Invasive Enterobacter sakazakii disease in infants.Emerg. Infect. Dis.121185–1189. 10.3201/eid1208.051509
7
CamposL.LobiancoL.SekiL.SantosR.AsensiM. (2007). Outbreak of Enterobacter hormaechei septicaemia in newborns caused by contaminated parenteral nutrition in Brazil.J. Hosp. Infect.6695–97. 10.1016/j.jhin.2007.02.013
8
CarterL.LindseyL. A.GrimC. J.SathyamoorthyV.JarvisK. G.GopinathG.et al (2013). Multiplex PCR assay targeting a diguanylate cyclase-encoding gene, cgcA, to differentiate species within the genus Cronobacter.Appl. Environ. Microbiol.79734–737. 10.1128/AEM.02898-12
9
Caubilla-BarronJ.ForsytheS. J. (2007). Dry stress and survival time of Enterobacter sakazakii and other Enterobacteriaceae in dehydrated powdered infant formula.J. Food Prot.702111–2117. 10.4315/0362-028X-70.9.2111
10
CetinkayaE.JosephS.AyhanK.ForsytheS. (2012). Comparison of methods for the microbiological identification and profiling of Cronobacter species from ingredients used in the preparation of infant formula.Mol. Cell. Probes2760–64. 10.1016/j.mcp.2012.10.003
11
ChapJ.JacksonP.SiqueiraR.GasparN.QuintasC.ParkJ.et al (2009). International survey of Cronobacter sakazakii and other Cronobacter spp. in follow up formulas and infant foods.Int. J. Food Microbiol.136185–188. 10.1016/j.ijfoodmicro.2009.08.005
12
Codex Alimentarius Commission (2008). CAC/RCP 66 – 2008. Code of Hygienic Practice for Powdered Formulae for Infants and Young Children. Codex Web site. Available at: www.fao.org/input/download/standards/11026/CXP_066e.pdf
13
EstuningsihS.KressC.HassanA.AkinedenO.SchneiderE.UsleberE. (2006). Enterobacteriaceae in dehydrated powdered infant formula manufactured in Indonesia and Malaysia.J. Food Prot.693013–3017. 10.4315/0362-028X-69.12.3013
14
FanningS.ForsytheS. (2008). “Isolation and identification of Enterobacter sakazakii,” inEnterobacter sakazakii. Emerging Issues in Food Safety SeriesedsFarberJ. M.ForsytheS. J. (Washington, DC: ASM Press), 27–59.
15
FarmerJ. J.AsburyM. A.HickmanF.BrennerD.The Enterobacteriaceae Study Group (1980). Enterobacter sakazakii, new species of Enterobacteriaceae isolated from clinical specimens.Int. J. Syst. Bacteriol.30569–584. 10.1099/00207713-30-3-569
16
FeiP.JiangY.FengJ.ForsytheS. J.LiR.ZhouY.et al (2017). Antibiotic and desiccation resistance of Cronobacter sakazakii and C. malonaticus isolates from powdered infant formula and processing environments.Front. Microbiol.8:316. 10.3389/fmicb.2017.00316
17
FeiP.ManC.LouB.ForsytheS.ChaiY.LiR.et al (2015). Genotyping and source tracking of the Cronobacter sakazakii and C. malonaticus isolated from powdered infant formula and an infant formula production factory in China.Appl. Environ. Microbiol.815430–5439. 10.1128/AEM.01390-15
18
Food and Agriculture Organization and World Health Organization [FAO/WHO] (2004). “Enterobacter sakazakii and Other Microorganisms in Powdered Infant Formula: Meeting Report. Microbiological Risk Assessment. Series No. 6. Geneva: WHO.
19
Food and Agriculture Organization and World Health Organization [FAO/WHO] (2006). Enterobacter sakazakii and Salmonella Powdered Infant Formula. Microbiological Risk Assessment. Series No. 10. Rome: FAO.
20
Food and Agriculture Organization and World Health Organization [FAO/WHO] (2008). Enterobacter sakazakii (Cronobacter spp.) in Powdered Follow-Up Formulae. Microbiological Risk Assessment. Series No. 15. Rome: FAO.
21
ForsytheS. J.DickinsB.JolleyK. A. (2014). Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics15:1121. 10.1186/1471-2164-15-1121
22
FriedemannM. (2009). Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections.Eur. J. Clin. Microbiol. Infect. Dis.281297–1304. 10.1007/s10096-009-0779-4
23
GiammancoG.AleoA.GuidaI.MamminaC. (2011). Molecular epidemiological survey of Citrobacter freundii misidentified as Cronobacter spp. (Enterobacter sakazakii) and Enterobacter hormaechei isolated from powdered infant milk formula.Foodborne Pathog. Dis.8517–525. 10.1089/fpd.2010.0719
24
HaririS.JosephS.ForsytheS. J. (2013). Cronobacter sakazakii ST4 strains and neonatal meningitis, United States.Emerg. Infect. Dis.19175–177. 10.3201/eid1901.120649
25
HassanA.AkinedenA.KressC.EstuningsihS.SchneiderE.UsleberE. (2007). Characterization of the gene encoding the 16S rRNA of Enterobacter sakazakii and development of a species specific PCR method.Int. J. Food Microbiol.116214–220. 10.1016/j.ijfoodmicro.2006.12.011
26
HeperkanD.Dalkilic-KayaG.JunejaV. (2017). Cronobacter sakazakii in baby foods and baby food ingredients of dairy origin and microbiological profile of positive samples.LWT Food Sci. Technol.75402–407. 10.1016/j.lwt.2016.09.013
27
HolýO.ForsytheS. J. (2014). Cronobacter species as emerging causes of healthcare-associated infection.J. Hosp. Infect.86169–177. 10.1016/j.jhin.2013.09.011
28
HunterC. J.BeanJ. F. (2013). Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis.J. Perinatol.33581–585. 10.1038/jp.2013.26
29
IversenC.LaneM.ForsytheS. J. (2004). The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula milk.Lett. Appl. Microbiol.38378–382. 10.1111/j.1472-765X.2004.01507.x
30
IversenC.MullaneN.Mc CardellB.TallB. D.LehnerA.FanningS.et al (2008). Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov. comb. nov., C. malonaticus sp. nov., C. turicensis sp. nov., C. muytjensii sp. nov., C. dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, C. dublinensis sp. nov. subsp. dublinensis subsp. nov., C. dublinensis sp. nov. subsp. lausannensis subsp. nov., and C. dublinensis sp. nov. subsp. lactaridi subsp. nov.Int. J. Syst. Evol. Microbiol.581442–1447. 10.1099/ijs.0.65577-0
31
JacksonE.ForsytheS. J. (2016). Comparative study of Cronobacter identification according to phenotyping methods.BMC Microbiol.16:146. 10.1186/s12866-016-0768-6
32
JacksonE.Parra-FloresJ.Fernandez-EscartinE.ForsytheS. J. (2015). Reevaluation of a suspected Cronobacter sakazakii outbreak in Mexico.J. Food Prot. 2015 781191–1196. 10.4315/0362-028X.JFP-14-563
33
JacksonE.SonbolH.MasoodN.ForsytheS. (2014). Genotypic and phenotypic characteristics of Cronobacter species, with particular attention to the newly reclassified species Cronobacter helveticus, Cronobacter pulveris, and Cronobacter zurichensis.Food Microbiol.44226–235. 10.1016/j.fm.2014.06.013
34
JacobsC.BraunP.HammerP. (2011). Reservoir and routes of transmission of Enterobacter sakazakii (Cronobacter spp.) in a milk powder-producing plant.J. Dairy Sci.943801–3810. 10.3168/jds.2011-4318
35
JasonJ. (2012). Prevention of invasive Cronobacter infection in young infants fed powdered infant formulas.Pediatrics1301076–1084. 10.1542/peds.2011-3855
36
JasonJ. (2015). The roles of epidemiologists, laboratorians, and public health agencies in preventing invasive Cronobacter infection.Front. Pediatr.3:110. 10.3389/fped.2015.00110
37
JongenburgerI.ReijM.BoerE.GorrisL.ZwieteringM. (2011). Actual distribution of Cronobacter spp in industrial batches of powdered infant formula and consequences for performance of sampling strategies.Int. J. Food Microbiol.15162–69. 10.1016/j.ijfoodmicro.2011.08.003
38
JosephS.CetinkayaE.DrahovskaH.LevicanA.FiguerasM. J.ForsytheS. J. (2012a). Cronobacter condimenti sp. Nov., isolated from spiced meat, and Cronobacter universalis sp. Nov., a species designation for Cronobacter sp. Geneomoespecies 1, recovered from a leg infection, water and food ingredients.Int. J. Syst. Evol. Microbiol.621277–1283. 10.1099/ijs.0.032292-0
39
JosephS.ForsytheS. (2011). Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections.Emerg. Infect. Dis.171713–1715. 10.3201/eid1709.110260
40
JosephS.HaririS.ForsytheS. (2013). Lack of continuity between Cronobacter biotypes and species as determined using multilocus sequence typing.Mol. Cell. Probes27137–139. 10.1016/j.mcp.2013.02.002
41
JosephS.SonbolH.HaririS.DesaiP.McClellandM.ForsytheS. (2012b). Diversity of the Cronobacter genus as revealed by multilocus sequence typing.J. Clin. Microbiol.503031–3039. 10.1128/JCM.00905-12
42
JumaN.ManningG.ForsytheS. (2016). Desiccation survival of Acinetobacter spp. in infant formula.Food Control68162–166. 10.1016/j.foodcont.2016.03.043
43
KalyantandaG.ShumyakL.ArchibaldL. K. (2015). Cronobacter species contamination of powdered infant formula and the implications for neonatal health.Front. Pediatr.3:56. 10.3389/fped.2015.00056
44
KentR.FitzgeraldG.HillC.StantonC.RossR. (2015). Novel approaches to improve the intrinsic microbiological safety of powdered infant milk formula.Nutrients71217–1244. 10.3390/nu7021217
45
KimS.OhS.LeeY.ImmJ.HwangI.KangD.et al (2011). Microbial contamination of food products consumed by infants and babies in Korea.Lett. Appl. Microbiol.53532–538. 10.1111/j.1472-765X.2011.03142.x
46
KoletzkoB.ShamirR.AshwellM. (2012). Quality and safety aspects of infant nutrition.Ann. Nutr. Metab.60179–184. 10.1159/000338803
47
KucerovaE.JosephS.ForsytheS. (2011). The Cronobacter genus: ubiquity and diversity.Qual. Assur. Safety Crops Foods3104–122. 10.1111/j.1757-837X.2011.00104.x
48
LaiK. K. (2001). Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and review of the literature.Medicine80113–122. 10.1097/00005792-200103000-00004
49
LehnerA.Fricker-FeerC.StephanR. (2012). Identification of the recently described Cronobacter condimenti by an rpoB-gene-based PCR system.J. Med. Microbiol.611034–1035. 10.1099/jmm.0.042903-0
50
LehnerA.TasaraT.StephanR. (2004). 16S rRNA gene based analysis of Enterobacter sakazakii strains from different sources and development of a PCR assay for identification.BMC Microbiol.4:43. 10.1186/1471-2180-4-43
51
LiY.ChenQ.ZhaoJ.LuF.BieX.LuZ. (2014). Isolation, identification and antimicrobial resistance of Cronobacter spp. iolated from various foods in China.Food Control37109–114. 10.1016/j.foodcont.2013.09.017
52
LiZ.GeW.LiK.GanJ.ZhangY.ZhangQ.et al (2016). Prevalence and Characterization of Cronobacter sakazakii in retail milk-based infant and baby foods in Shaanxi, China.Foodborne Pathog. Dis.131–7. 10.1089/fpd.2015.2074
53
Mohan-NairM. K.VenkitanarayananK. S. (2006). Cloning and sequencing of the ompA gene of Enterobacter sakazakii and development of an ompA-targeted PCR for rapid detection of Enterobacter sakazakii in infant formula.Appl. Environ. Microbiol.722539–2546. 10.1128/AEM.72.4.2539-2546.2006
54
MullaneN.WhyteP.WallP.QuinnT.FanningS. (2007). Application of pulsed-field gel electrophoresis to characterise and trace the prevalence of Enterobacter sakazakii in an infant formula processing facility.Int. J. Food Microbiol.11673–81. 10.1016/j.ijfoodmicro.2006.12.036
55
MuytjensH. L.ZanenH. C.SonderkampH. J.KolleeL. A.WachsmuthI. K.FarmerJ. J. (1988). Analysis of eight cases of neonatal meningitis and sepsis due to Enterobacter sakazakii.J. Clin. Microbiol.18115–120.
56
NorbergS.StantonC.RossR.HillC.FitzgeraldG.CotterP. (2012). Cronobacter spp. in powdered infant formula.J. Food Prot.75607–620. 10.4315/0362-028X.JFP-11-285
57
OgrodzkiP.ForsytheS. J. (2017). DNA-sequence based typing of the Cronobacter genus using MLST, CRISPR-cas array and capsular profiling.Front. Microbiol.8:1875. 10.3389/fmicb.2017.01875
58
Parra-FloresJ.JunejaV.Garcia de FernandoG.AguirreJ. (2016). Variability in cell response of Cronobacter sakazakii after mild-heat treatments and its impact on food safety.Front. Microbiol.7:535. 10.3389/fmicb.00535
59
Parra-FloresJ.OliverasL.RodriguezA.RiffoF.JacksonE.ForsytheS. (2015a). Risk of Cronobacter sakazakii contamination in powdered milk for infant nutrition.Rev. Chil. Nutr.4283–89. 10.4067/S0717-75182015000100011
60
Parra-FloresJ.RodriguezA.RiffoF.Arvizu-MedranoS. M.Arias-RiosE. V.AguirreJ. (2015b). Investigation on the factors affecting Cronobacter sakazakii contamination levels in reconstituted powdered infant formula.Front. Pediatr.3:72. 10.3389/fped.2015.00072
61
PatrickM.MahonB.GreeneS.RoundsJ.ConquistA.WymoreK.et al (2014). Incidence of Cronobacter spp. infections, United States, 2003–2009.Emerg. Infect. Dis.201520–1523. 10.3201/eid2009.140545
62
ReijM.JongerburgerI.GkogkaE.GorrisL.ZwieteringM. (2009). Perspective on the risk to infants in the Netherlands associated with Cronobacter spp. occurring in powdered infant formula.Int. J. Food Microbiol.136232–237. 10.1016/j.ijfoodmicro.2009.07.011
63
SáezM.LlanosS.TamayoR. (2012). Primer aislamiento de Cronobacter spp. (Enterobacter sakazakii) en fórmula láctea en polvo producida en Chile.Rev. Chil. Salud Púb.1611–15. 10.5354/0719-5281.2012.18607
64
Siqueira-SantosR. F. S.da SilvaN.JunqueiraV. C. A.KajsikM.ForsytheS.PereiraJ. L. (2013). Screening for Cronobacter species in powdered and reconstituted infant formulas and from equipment used in formula preparation in maternity hospitals.Ann. Nutr. Metab.6362–68. 10.1159/000353137
65
StoopB.LenherA.IversenC.FanningS. (2009). Development and evaluation of rpoB based PCR systems to differentiate the six proposed species within the genus Cronobacter.Int. J. Food Microbiol.136165–168. 10.1016/j.ijfoodmicro.2009.04.023
66
TownsendS.HurrellE.ForsytheS. (2008). Virulence studies of Enterobacter sakazakii isolates associated with a neonatal intensive care unit outbreak.BMC Microbiol8:64. 10.1186/1471-2180-8-64
67
VojkovskaH.KarpiskovaR.OrieskovaM.DrahovskaH. (2016). Characterization of Cronobacter spp. isolated from food of plant origin and environmental samples collected from farms and from supermarkets in the Czech Republic.Int. J. Food Microbiol.217130–136. 10.1016/j.ijfoodmicro.2015.10.017
68
WengerP.TokarsJ. I.BrennanP.SamelC.BlandL.MillerM.et al (1997). An Outbreak of Enterobacter hormaechei infection and colonization in an intensive care nursery.Clin. Infect. Dis.241243–1244. 10.1086/513650
69
World Health Organization [WHO] (2007). Safe Preparation, Storage and Handling of Powdered Infant Formula: Guidelines.Geneva: WHO.
70
XuX.QingpingW.JumeiZ.YingwangY.XiaojuanY.XiaohuiD. (2014). Occurrence and characterization of Cronobacter spp. in powdered formula from Chinese retail markets.Foodborne Pathog. Dis.11307–312. 10.1089/fpd.2013.1657
71
YanQ.CondellO.PowerK.ButtlerF.TallB.FanningS. (2012). Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: a review of our current understanding of the biology of this bacterium.J. Appl. Microbiol.1131–15. 10.1111/j.1365-2672.2012.05281.x
72
YanQ.WangJ.GangiredlaJ.CaoY.MartinsM.GopinathG.et al (2015). Comparative genotypic and phenotypic analysis of Cronobacter species cultured from four powdered infant formula production facilities: indication of pathoadaptation along the food chain.Appl. Environ. Microbiol.814388–4402. 10.1128/AEM.00359-15
73
YaoK.ZinzendorfN.BohouaG.KouassiK.KouaA.KouadioL.et al (2012). Occurrence of Enterobacter sakazakii (Cronobacter) and other Enterobacteriaceae in commercial powdered infant formula in Abidjan, Ivory Coast.Food Nutr. Sci.3822–826. 10.4236/fns.2012.36110
Summary
Keywords
Cronobacter sakazakii, food alert, microbiological parameters, powdered infant formula, liquid dairy formula
Citation
Parra-Flores J, Cerda-Leal F, Contreras A, Valenzuela-Riffo N, Rodríguez A and Aguirre J (2018) Cronobacter sakazakii and Microbiological Parameters in Dairy Formulas Associated With a Food Alert in Chile. Front. Microbiol. 9:1708. doi: 10.3389/fmicb.2018.01708
Received
16 April 2018
Accepted
09 July 2018
Published
31 July 2018
Volume
9 - 2018
Edited by
Giovanna Suzzi, Università di Teramo, Italy
Reviewed by
Chiara Montanari, Università degli Studi di Bologna, Italy; Antonio Martínez, Consejo Superior de Investigaciones Científicas (CSIC), Spain
Updates
Copyright
© 2018 Parra-Flores, Cerda-Leal, Contreras, Valenzuela-Riffo, Rodríguez and Aguirre.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julio Parra-Flores, juparra@ubiobio.cl; juparraf@gmail.com
This article was submitted to Food Microbiology, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.