- 1Department of Marine and Coastal Sciences, Rutgers University, New Brunswick, NJ, United States
- 2Geophysical Laboratory, Carnegie Institution of Washington, Washington, DC, United States
- 3Institute of Marine Science, National Research Council, Ancona, Italy
- 4Earth-Life Science Institute, Tokyo Institute of Technology, Tokyo, Japan
- 5Institute of Marine Sciences, Middle East Technical University, Mersin, Turkey
- 6Department of Biochemistry and Microbiology, Rutgers University, New Brunswick, NJ, United States
In this study, we integrated geochemical measurements, microbial diversity surveys and physiological characterization of laboratory strains to investigate substrate-attached filamentous microbial biofilms at Tor Caldara, a shallow-water gas vent in the Tyrrhenian Sea. At this site, the venting gases are mainly composed of CO2 and H2S and the temperature at the emissions is the same as that of the surrounding water. To investigate the composition of the total and active fraction of the Tor Caldara biofilm communities, we collected established and newly formed filaments and we sequenced the 16S rRNA genes (DNA) and the 16S rRNA transcripts (cDNA). Chemoautotrophic sulfur-oxidizing members of the Gammaproteobacteria (predominantly Thiotrichales) dominate the active fraction of the established microbial filaments, while Epsilonproteobacteria (predominantly Sulfurovum spp.) are more prevalent in the young filaments. This indicates a succession of the two communities, possibly in response to age, sulfide and oxygen concentrations. Growth experiments with representative laboratory strains in sulfide gradient medium revealed that Sulfurovum riftiae (Epsilonproteobacteria) grew closer to the sulfide source than Thiomicrospira sp. (Gammaproteobacteria, Thiotrichales). Overall, our findings show that sulfur-oxidizing Epsilonproteobacteria are the dominant pioneer colonizers of the Tor Caldara biofilm communities and that Gammaproteobacteria become prevalent once the community is established. This succession pattern appears to be driven - among other factors - by the adaptation of Epsilon- and Gammaproteobacteria to different sulfide concentrations.
Introduction
Hydrothermal and gas vents are manifestations of volcanism on earth. Both environments are characterized by venting of gas and/or hydrothermal fluids, enriched in reduced, inorganic chemical species, into the oxygenated water column. This creates a redox disequilibrium, which is then harnessed by the prokaryotic communities to convert chemical energy into ATP. Both types of vents have been known to occur at varying depths, from intertidal regions to abyssal depths (Tarasov et al., 2005). Vents that occur at depths ≤ 200 meters were traditionally classified as “shallow-water” hydrothermal vents, and the ones that occur at depths greater than 200 meters were classified as “deep-sea” hydrothermal vents (Tarasov et al., 2005). Shallow-water vents (both hydrothermal and gas) have been discovered at seamounts, submarine volcanoes, volcanic arcs and back arcs (Price and Giovannelli, 2017). Common features of both shallow-water and deep-sea vent ecosystems include high concentrations of reduced compounds, gases (e.g., H2, CH4, H2S) and heavy metals. However, the temperature of hydrothermal fluids at shallow-water vents isn’t as high as that of the deep-sea vents, where it can exceed 400°C (Fornari et al., 1998). Shallow vents are also affected by input of meteoric water and terrigenous organic carbon. At most shallow-water vents, it has been observed that H2 content of the venting fluid-gas is exceedingly low as compared to deeper vents (Tarasov et al., 2005). Along with geochemical energy, the availability of light energy leads to co-occurrence of photoautotrophy and chemoautotrophy, setting these shallow systems apart from their deep-sea counterparts. In the broader context, coastal geothermal environments can serve as model systems to study the effects of ocean acidification and increasing temperatures in the oceans. Further, high levels of potentially toxic heavy metals have been associated with shallow-water vents, making them probable sources of coastal pollution leading to bioaccumulation and biotransformation of these metals in the organisms living in the vicinity (Price and Giovannelli, 2017).
The collision of the African and European tectonic plates leads to the subduction of the oceanic part of the African plate below Europe, which causes hydrothermalism in the Mediterranean Sea (Dando et al., 1999). The resulting volcanic arcs and back arcs are found in the Aegean and Tyrrhenian Seas (e.g., the Aeolian Islands arc). Hydrothermal venting in volcanic arc systems is characterized by the release of volcanic gases. Volcanic arc venting is a combination of degassing from subducted slab and mantle as well as decomposition of the carbonaceous sediment on top (Dando et al., 1999). While the terrestrial volcanic systems of the Mediterranean region have been well studied because of their impact on local population, the submarine component started receiving attention only in the 1990s (Dando et al., 1999). The Tyrrhenian margin of Southern and Central Italy is a volcanic geothermal region with a thinned continental crust. It is characterized by heat flows and degassing of CO2 of deep magmatic/mantle origin due to the alkali-potassic volcanoes present in the region from the Quaternary period (Carapezza et al., 2012). One of the relatively recent volcanoes is the Colli Albani, whose eruptive activity began 600 ka years ago and went on till about 5.8 ka (Holocene). Although the age of its most recent eruption is controversial, there is unanimity about its quiescent status (Carapezza et al., 2012; Marra et al., 2016). Due to the infiltration of cold meteoric water, there aren’t any thermal springs or fumaroles in the immediate vicinity of the Colli Albani volcano. However, there are several gas emission sites associated with the flanks of this volcanic system near the coast of the Tyrrhenian sea, Tor Caldara being one of them (Carapezza et al., 2012). The helium isotopic composition of the gases released from Colli Albani suggests a likely origin from a deep magma source affected by crustal contamination during its primary generation in a subduction process (Carapezza and Tarchini, 2007).
Till date, the microbiology of shallow-water hydrothermal vents has been investigated at the Aeolian Islands and Milos in the Mediterranean Sea, at the Azores and Iceland in the Atlantic Ocean, and at White Point, New Caledonia, Papua New Guinea, Kueishan and Taketomi islands in the Pacific Ocean (Sievert et al., 1999, 2000a,b; Marteinsson et al., 2001; Kalanetra et al., 2004; Hirayama et al., 2007; Giovannelli et al., 2013; Meyer-Dombard et al., 2013; Quéméneur et al., 2014; Gugliandolo et al., 2015; Wang et al., 2017). Previous shallow-water vent microbial diversity studies in the Tyrrhenian Sea have mostly been focused on vents off the coast of the Aeolian Islands. However, none of these studies investigated the active fraction of the microbial communities and very few carried out physiological studies of bacteria isolated from these vents. These previous studies have revealed that, within the Proteobacteria, thermophilic as well as mesophilic chemoautotrophic members belonging to the Gammaproteobacteria and Epsilonproteobacteria dominate these sites, whereas members of the Deltaproteobacteria class are abundant in the sediments. Occurrence of phototrophic members belonging to Alphaproteobacteria, Cyanobacteria, Chlorobi and Chlorofexi were recorded in addition to thermophilic heterotrophs belonging to the Firmicutes phylum and members of the Euryarchaeota and Crenarchaeota (Gugliandolo and Maugeri, 1993, 1998; Gugliandolo et al., 2006, 2015; Maugeri et al., 2008, 2010, 2013; Lentini et al., 2014).
In the present study, we used next generation 16S rRNA amplicon sequencing, geochemical analyses and physiological studies of pure cultures to investigate the abundance, diversity and adaptation to sulfide of the filamentous microbial biofilms as well as the gas composition of the shallow-water gas vents at Tor Caldara.
Materials and Methods
Study Site
Tor Caldara is a natural reserve located near the town of Anzio, around 60 km south of Rome and 40 km south-west of the Colli Albani volcano. It sits on a buried carbonate basement that has been hypothesized as one of the sources of CO2 at this site (Carapezza et al., 2012). The study site (41°29′ 9″ N 12°35′ 23″ E) is a submarine gas vent located at a water depth of approximately 3 m off the coast of the natural reserve. Along the coastline, hydrothermal alterations of the rocks and soil are visible in correspondence with the study site. A significant amount of degassing can be observed underwater and at the surface. The rocks occurring on the sandy seabed in the vicinity of the gas discharges are colonized by filamentous microbial communities (Supplementary Figure 1a).
Collection of Biological Samples
Samples were collected by a SCUBA diver in the summer (month of August) over a course of 5 years from 2012 to 2016. Sampling was unsuccessful in 2014 due to inclement weather conditions. Samples of established filaments (EF from here on) growing on rocks in the vicinity of the gas emissions were collected in all 4 years (2012, 2013, 2015, and 2016). A total of four samples (one per each year) were used for downstream applications. In 2012, filament samples were collected by scraping them off the rocky substrate; in the following years (2013, 2015, and 2016) filament samples were collected using a 60 ml sterile syringe. Briefly, the syringe plunger was retracted to generate vacuum and draw the filaments into the barrel. Once the sampling was completed, the syringe was inverted to let the filament deposit onto the plunger seal and the excess seawater was slowly pushed out of the barrel. The remaining filaments were pushed into a sterile tube containing RNA Later. To investigate the early stage of substrate colonization, in 2016 we performed a biofilm colonization experiment by mounting several sterile glass slides very close to each other on an aluminum rod at the venting site for 4 days (Supplementary Figure 1b). The young filaments (YF from here on) growing on these slides were then collected for further analyses. All samples were stored in ambient seawater at 4°C to be used as inocula for cultivation work and in RNA Later at −80°C for further nucleic acid extraction. The EF and YF were also stored in 2% formaldehyde at 4°C for microscopy.
Collection of Gas Samples
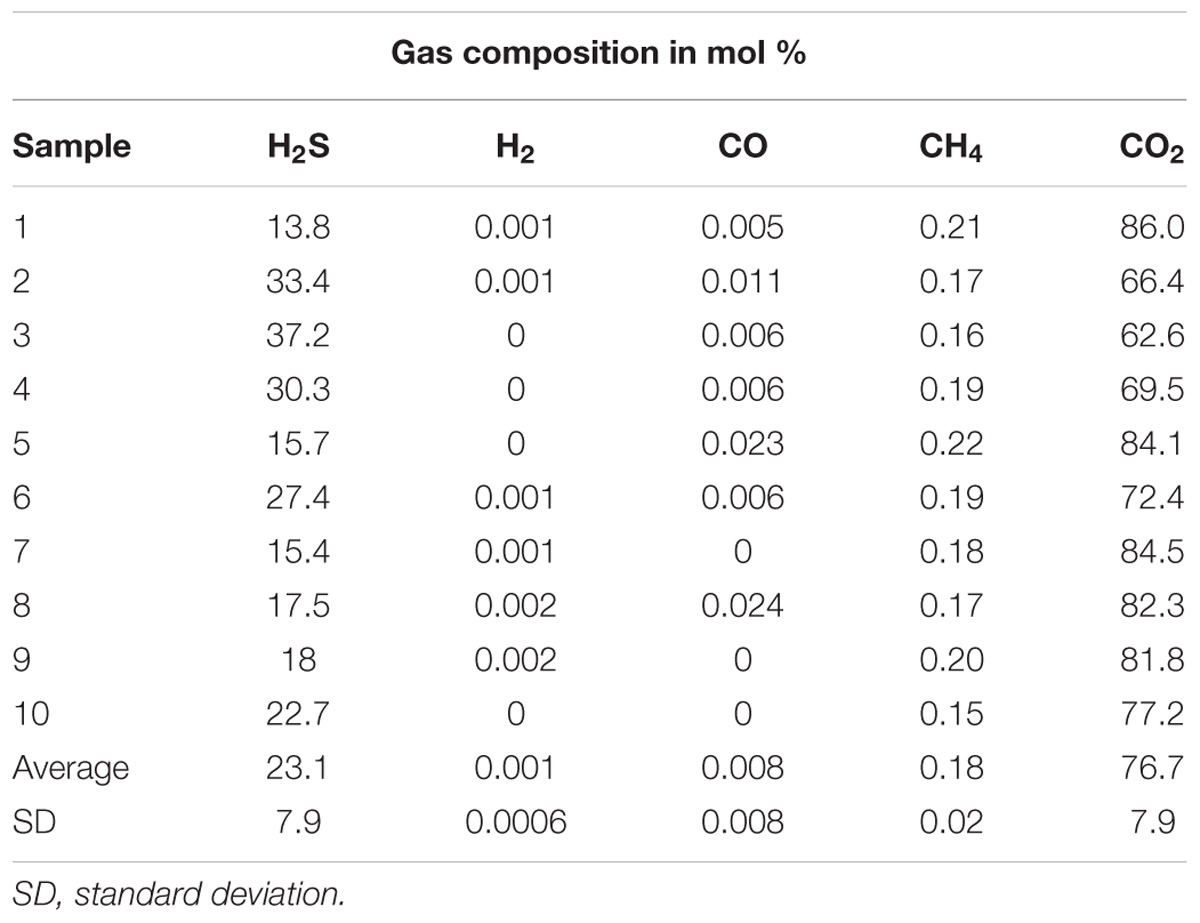
In 2015, we built an in-house apparatus for collecting gas samples (10 replicates) at Tor Caldara. A weighted inverted funnel was deployed over the gas discharges. The gases trapped inside the inverted funnel were conveyed via connected tubing to a gas-tight valve, which was finally connected to a syringe. Twenty milliliters of gas were collected and injected into 25 ml stoppered borosilicate vials containing 4 ml of 6% (w/v) AgNO3, 25% (w/v) H3PO4 and 20 ml Argon grade zero. The vials were pretreated by following protocols discussed elsewhere (Foustoukos, 2012). The precipitated Ag2S and gases in the headspace was measured and analyzed. Concentrations of H2(g), CO(g), CO2(g), and CH4(g) were measured by a SRI 8610C gas chromatograph equipped with TCD/FID detector and a Carboxen-1010 Plot/Silica-Gel column. The detection limit for these volatiles is 1–5 μmol/kg (<0.001 mol%) (Foustoukos et al., 2011). Analytical errors are within 1–15% (2σ) and reflect the larger values of errors estimated either by instrument calibration and/or by duplicate analysis of individual samples. The precipitated Ag2S in the solution was filtered on to a 0.2 μm polycarbonate and weighed. Temperature and pH (using an Oakton waterproof pH/temperature probe) and salinity (using a Spartan A 366 ATC refractometer) measurements were also taken in the venting area as well as 100 m away from venting in 2015.
Microscopy and Elemental Composition of the Filament Communities
Biofilm samples, as well as manually teased individual filaments stored in 2% formaldehyde, were mounted on aluminum pin stubs using double sided carbon adhesives for microscopy. Scanning electron micrographs were obtained for EF and YF using a Phenom ProX table top scanning electron microsope (SEM) at a voltage of 10 kV. Spot analysis using the Elemental Mapping & Line Scan software was performed at 15 kV using the same instrument to examine the elemental composition of the sample.
Nucleic Acid Extraction
Both DNA and RNA were extracted from samples stored in RNA Later following a phenol:chloroform extraction protocol to study the total and active communities, respectively. Samples were centrifuged at 10,000 rmp for 3 min to remove the RNA Later supernatant. 0.8 g (wet weight) of the pelleted filaments was added to 850 μl extraction buffer (50 mM Tris-HCl, 20 mM EDTA, 100 mM NaCl; pH 8.0) supplemented with 100 μl of lysozyme (100 mg/ml) and incubated at 37°C for 30 min. This mix was then supplemented with 5 μl of proteinase K (20 mg/ml) and incubated at 37°C for 30 min followed by the addition of 50 μl SDS (20%) and further incubated at 65°C for 1 h. Nucleic acids were extracted by performing a series of phenol:chloroform: isoamylalcohol (25:24:1) and chloroform:isoamyl alcohol (24:1) extractions. Phenol:chloroform:isoamylalcohol (25:24:1) at pH 4.3 was used for RNA extractions. Multiple replicates of the same sample were co-extracted to reduce potential bias. The final supernatant was precipitated in 3 M sodium-acetate and isopropanol, washed twice with 70% ice cold ethanol and resuspended in ultra-pure water. For RNA preparation, carryover DNA was removed by treating the RNA with the TURBO DNase kit (Invitrogen, Carlsbad, CA, United States), according to the manufacturer’s specifications. This DNase treated RNA was reverse transcribed into cDNA using the Invitrogen cDNA synthesis kit (Invitrogen, Carlsbad, CA, United States), according to the manufacturer’s specifications. The quality of the genomic DNA and cDNA was assessed by polymerase chain reaction amplification of the 16S rRNA gene using primers Bact 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and Univ 519R (5′-ATTACCGCGGCTGCTGG-3′). PCR positive (DNA template known to produce an amplicon) and negative (no DNA template) controls were included, but not sequenced.
16S rRNA Gene Amplification and Sequencing
The DNA and cDNA from the Tor Caldara microbial communities were used as templates to amplify the variable 4 (V4) region of the 16S rRNA gene and transcript, respectively, using the prokaryotic universal primers (515F 5′-GTG CCA GCM GCC GCG GTA A-3′ and 806R 5′-GGA CTA CVS GGG TAT CTA AT-3′ (Caporaso et al., 2011). Amplicons were sequenced using the PGM Ion Torrent platform at the Molecular Research LP, Shallowater, TX, United States. Multiple PCR reactions for each sample were combined to reduce potential bias. Sequences are available through the NCBI Short Read Archive database with accession number SRP167116. Sequences were first depleted of barcodes and primers, then sequences < 150 bp, those with ambiguous base calls and with homopolymer runs exceeding 6 bp were removed. The amplicon size obtained was around 300 bp. The total number of reads for 16 samples after the chimera check was 1,311,749. The average number of reads per sample was 81,984.
Bioinformatic Analyses
The 16S rRNA gene sequences were analyzed using QIIME 1.9 (Caporaso et al., 2010). Chimeric sequences were removed using ChimeraSlayer (Haas et al., 2011). Operational Taxonomic Units (OTUs) were picked at 97% similarity using the “pick_open_reference_otus.py” which clusters reads against the Greengenes v13_8 database (DeSantis et al., 2006). Reads that did not hit the database were subsequently clustered de novo. This method was chosen based on the assumption that the study site might harbor communities that are not well represented by the reference database. The OTUs were then taxonomically classified using the Ribosomal Database Project Classifier against the 2013 Greengenes database (Wang et al., 2007). OTUs that were not resolved up to the genus level were manually checked using NCBI BLAST (Altschul et al., 1990).
Phylogenetic Analyses
Sequences of the V4 region of 16S rRNA genes of OTUs, full length 16S rRNA sequences of their closest cultured relatives (obtained by searching the NCBI non-redundant database by nucleotide BLAST search), and full length 16S rRNA sequences of cultured representatives from this study were aligned using Clustal Omega (Sievers et al., 2011) within SEAVIEW (Galtier et al., 1996; Gouy et al., 2010). The resulting alignment was used to reconstruct phylogenetic trees using the Maximum Likelihood algorithm with PhyML (Guindon and Gascuel, 2003) using the general time reversible (GTR) model, aLRT scores calculated.
Statistical Analyses
The contribution of Gammaproteobacteria and Epsilonproteobacteria in each biofilm sample was further investigated using a Gammaproteobacteria to Epsilonproteobacteria ratio (GE ratio) calculated using the relative abundance of each class as follows: (%Gammaproteobacteria – %Epsilonproteobacteria)/ (%Gammaproteobacteria + % Epsilonproteobacteria). The obtained index is constrained between 1 (only Gammaproteobacteria present; Epsilonproteobacteria absent) and −1 (only Epsilonproteobacteria present), with a value of 0 representing equal relative amount of Gamma- and Epsiloproteobacteria (Supplementary Table 1). Since the majority of Gamma- and Epsiloproteobacteria were sulfur-oxidizing bacteria, this GE ratio was used to understand their shift in the YF and EF communities. The GE ratio was calculated for all the biofilm samples and differences among groups were tested by means of two-way analysis of variance (ANOVA) using the R statistical software (R: The R Project for Statistical Computing, 2018). Where ANOVA assumptions were rejected, a more conservative level of p was chosen. Both the biofilm age (established vs. young) and metabolic state (active vs. total communities) were used as factors in the analysis as well as the interaction term (age by metabolic state). In case of significant differences, a HSD Tukey post hoc test was performed.
Isolation of Microorganisms
All primary enrichment cultures was set up by inoculating a slurry containing filament samples (stored at 4°C) re-suspended in 1 ml of artificial seawater into 10 ml of liquid medium or/and in 25 ml of solidified medium. A variety of different media were used, selecting for chemoautotrophic and heterotrophic microorganisms. Primary enrichments in liquid media were diluted to extinction and incubated at 30°C. Aliquots from the last dilution tube with growth were inoculated into fresh medium and pure cultures were obtained by performing three consecutive series of dilutions to extinction. During the isolation procedures, the cultures were incubated at 30°C.
Isolation of Heterotrophic Bacteria
Isolation of heterotrophs was targeted using full strength (FS) and low strength (LS) artificial seawater medium (ASW). FS ASW medium contained (per liter): 24 g NaCl, 0.7 g KCl, 7.0 g MgCl2, 3 g yeast extract, and 2.5 g peptone. LS ASW had the same amounts of salts per liter but lower amount of yeast extract (0.15 g) and peptone (0.125 g). For solid FS ASW and LS ASW, medium was supplemented with 15 g of agar per liter. Sulfide-oxidizing heterotrophs were isolated in sulfide gradient agar plugs (S-plugs) with yeast extract. S-plugs were prepared in 20 ml stoppered Hungate tubes (Bellco Glass). The S-plug medium contained a 3 ml solid (1.8% agar) sulfide reservoir (6.6 mM Na2S) on the bottom with 7 ml of solid (5% agar) THST medium (Podgorsek et al., 2004) with 0.05% yeast extract added on top.
Isolation of Chemolithoautotrophic Bacteria
Isolation of sulfur-oxidizing chemolithoautotrophs was carried out using liquid 1011 medium (Inagaki, 2004), which contained (l−1): 30 g NaCl, 0.14 g K2HPO4, 0.14 g CaCl2 ⋅ 2H2O, 3.4 g MgSO4 ⋅ 7H2O, 4.18 g MgCl2 ⋅ 6H2O, 0.33 g KCl, 0.5 mg NiCl2 ⋅ 6H2O, 0.5 mg Na2SeO3 ⋅ 5H2O, 0.01 g Fe(NH4)2(SO4)2 ⋅ 6H2O, 0.25 g NH4Cl, 1.5 g NaHCO3, 1.5 g Na2S2O3 ⋅ 5H2O, 10 ml trace mineral solution and 1 ml of trace vitamins. The medium was supplemented with a gas phase of CO2/O2 (95:5; 200kPa), and 10% (w/v) potassium nitrate under a N2/CO2 gas phase (80:20; 200 kPa), for isolating aerobic and anaerobic sulfur-oxidizing chemoautotrophs, respectively. Sulfide-oxidizing chemolithoautotrophs were isolated using S-plugs (as described in the previous paragraph) without yeast extract. The THST medium of the S-plugs was amended with 100 μl/10 ml of potassium nitrate (500 mM) and ammonium ferric citrate (500 mM) to enrich for anaerobic sulfide-oxidizers.
The isolation of Thiothrix-like bacteria was attempted (unsuccessfully) using Thiothrix growth medium (DSMZ medium 573), which contained (per liter): 0.20 g NH4Cl, 0.01 g K2HPO4, 0.01 g MgSO4, 20 ml CaSO4, 5 ml trace element solution from DSMZ medium 155, 0.10 g of Na-acetate, 12 g agar and 0.3 g Na2S.
Growth of Sulfurovum riftiae and Thiomicrospira spp. in Sulfide Gradients and Generation of Sulfide Concentration Profiles
S-plugs for growth of Thiomicrospira sp. strain EPR 85 (Houghton et al., 2016) and Sulfurovum riftiae DSM 101780 (Giovannelli et al., 2016) were prepared as described above. However, the THST medium was replaced with medium 1011 without sodium thiosulfate (Inagaki, 2004). Replicate S-plug tubes were inoculated with cultures individually. The inoculated tubes along with un-inoculated controls were incubated at 30°C. Thiomicrospira sp. strain EPR 85 and S. riftiae were grown under a gas phase of CO2/O2 (95:5; 200 kPa) and N2/CO2 (80:20; 200 kPa), respectively. The upward diffusion of sulfide from the reservoir into the medium created a gradient of decreasing sulfide concentrations, thus providing micro-niches for selective growth of the two strains. A voltammetric microsensor was used to profile the concentration of total dissolved sulfide (HS- + H2S) throughout uninoculated S-plug tubes. To this end, dissolved sulfide (total of HS− and H2S) in the S-plug medium was measured using cyclic voltammetry with the gold amalgam (Au/Hg) working microelectrode (Luther et al., 2003; Yücel, 2013). The three-electrode voltammetric sensor was calibrated for S(-II) before the first profile using standard additions of a stock solution prepared with Na2S and deoxygenated distilled water. In order to profile the sulfide concentration throughout the medium, the Au/Hg glass working electrode was attached to a micromanipulator with counter and reference electrodes placed in the overlying fluid of the plug. The working electrode was then vertically maneuvered with a minimum step of 1 mm. The scans were taken in cyclic voltammetry form, between −0.1 V and −1.8 V at a rate of 1000 mV sec−1. Before each scan, the electrode was electrochemically conditioned at −0.9 V for 10 s to remove any adsorbed species (Yücel, 2013). Detection limit was 0.2 μM for S(-II). Voltammograms were recorded from the electrodes using a bench-top potentiostat (PalmSens) and the data were processed with the software provided by the manufacturer.
Results
Composition of the Gas Emissions at Tor Caldara
The temperature measured during the summer in the water surrounding the Tor Caldara gas vents varied between 22 and 26°C, with an average salinity of 40 ppt (the avg. salinity of the Tyrrhenian Sea varies between 36 and 39 ppt) (Kallel et al., 1997). The pH (7.48) in the area of venting was slightly lower than the pH (7.9) away from venting. The venting gases were mainly composed of CO2 (avg. of 76.7 mol%) and H2S (avg. of 23.1 mol%) with minor contribution of CH4 (avg. of 0.18 mol%) and traces of CO (avg. of 0.008 mol%) and H2 (avg. of 0.001 mol%) (Table 1). Since the gas venting at Tor Caldara is a peripheral manifestation of the quiescent Colli Albani volcanic complex, whose activity began 600 ka ago (Carapezza et al., 2012), the system is thought to be very stable over time in comparison to the more dynamic venting associated with active hydrothermal vents.
Identification of Culturable Bacteria From Tor Caldara
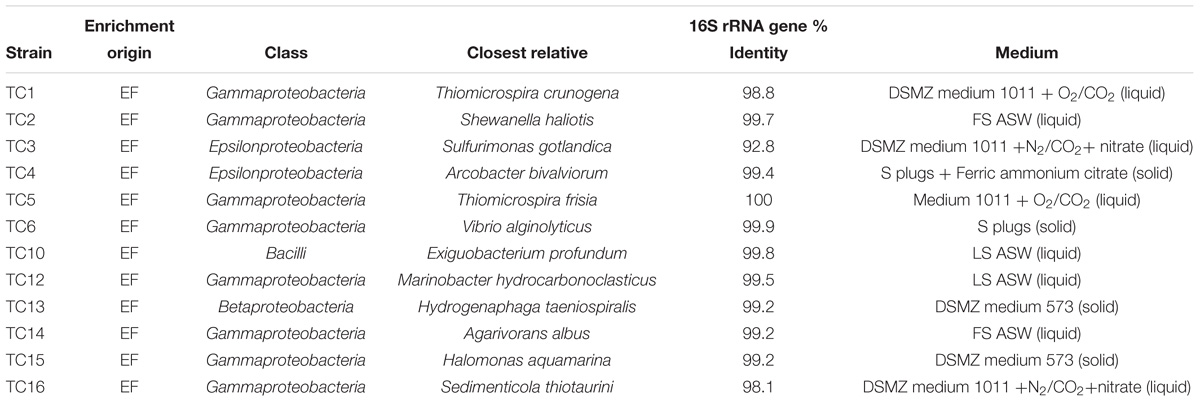
EF samples were used as inoculum to obtain primary enrichments in a variety of heterotrophic and autotrophic media. Subsequently, pure cultures were obtained after undergoing three consecutive series of end-point dilutions. A total of 12 cultures were obtained from all the enrichments (Table 2). Among the isolates, four were sulfur-oxidizing chemolithotrophs: strain TC1 (Figure 1), whose 16S rRNA gene was 98.8% identical to that of Thiomicrospira crunogena (Jannasch et al., 1985), strain TC3 (92.8% identical to Sulfurimonas gotlandica Figure 2) (Labrenz et al., 2013), strains TC5 (100% identical to Thiomicrospira frisia; Figure 1) (Brinkhoff et al., 1999) and TC16 (98.1% similarity to Sedimenticola thiotaurini) (Flood et al., 2015). Phylogenetic analysis showed that the novel epsilonproteobacterial strain TC3 clustered together with two uncultured 16S rRNA gene clones recovered from deep-sea vents (Figure 2).
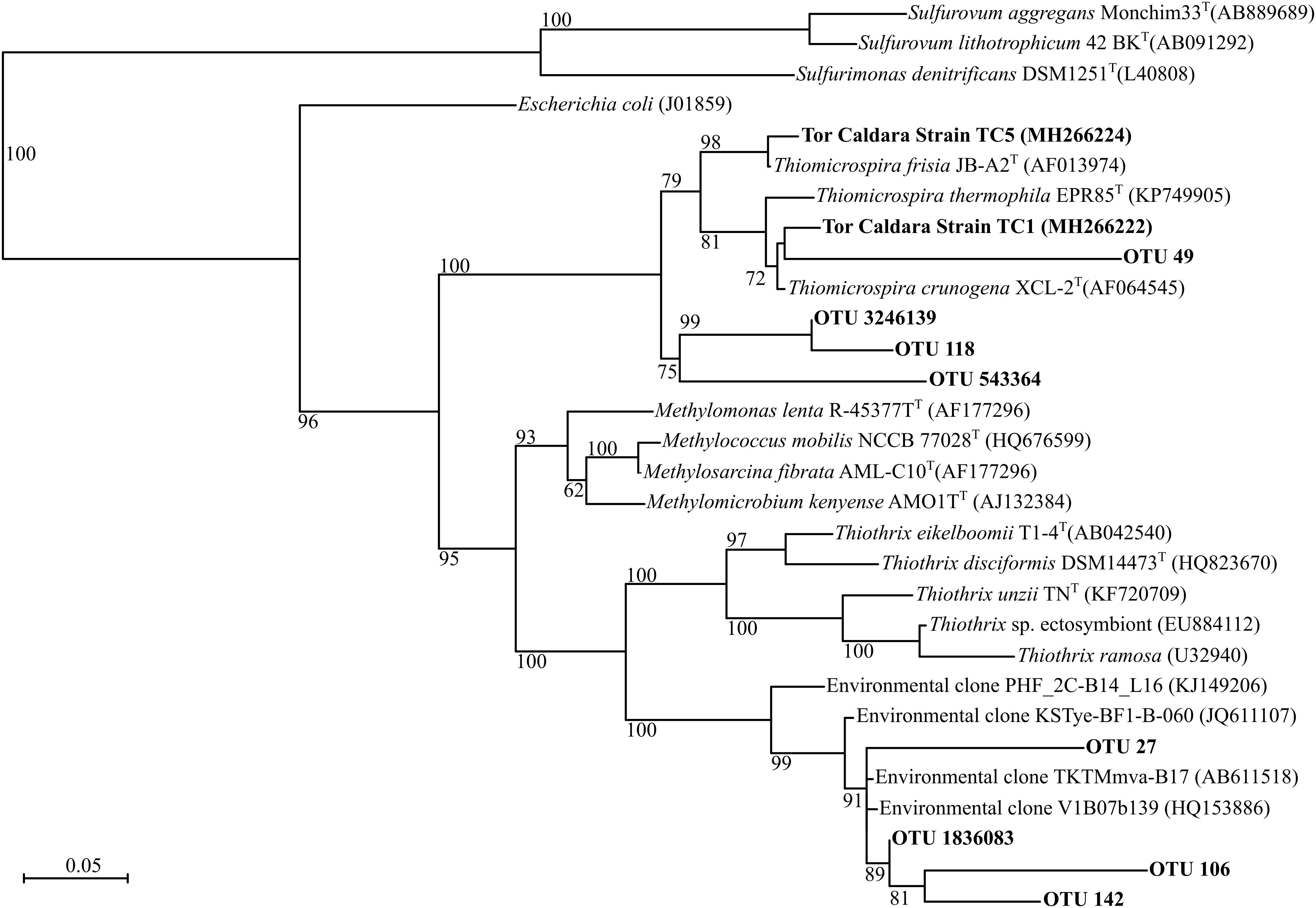
Figure 1. Maximum-likelihood phylogenetic tree derived from V4 regions and full length 16S rRNA gene sequences showing the positions of three Thiothrix-like CF-26 OTUs, three Thiomicrospira OTUs and Thiomicrospira strains TC1 and TC5 within the Gammaproteobacteria. aLRT branch support values higher than 50% were based on 1000 replicates and are shown at each node. Bar, 0.05% substitutions per position. Sequences belonging to the class Epsilonproteobacteria were used as outgroup.
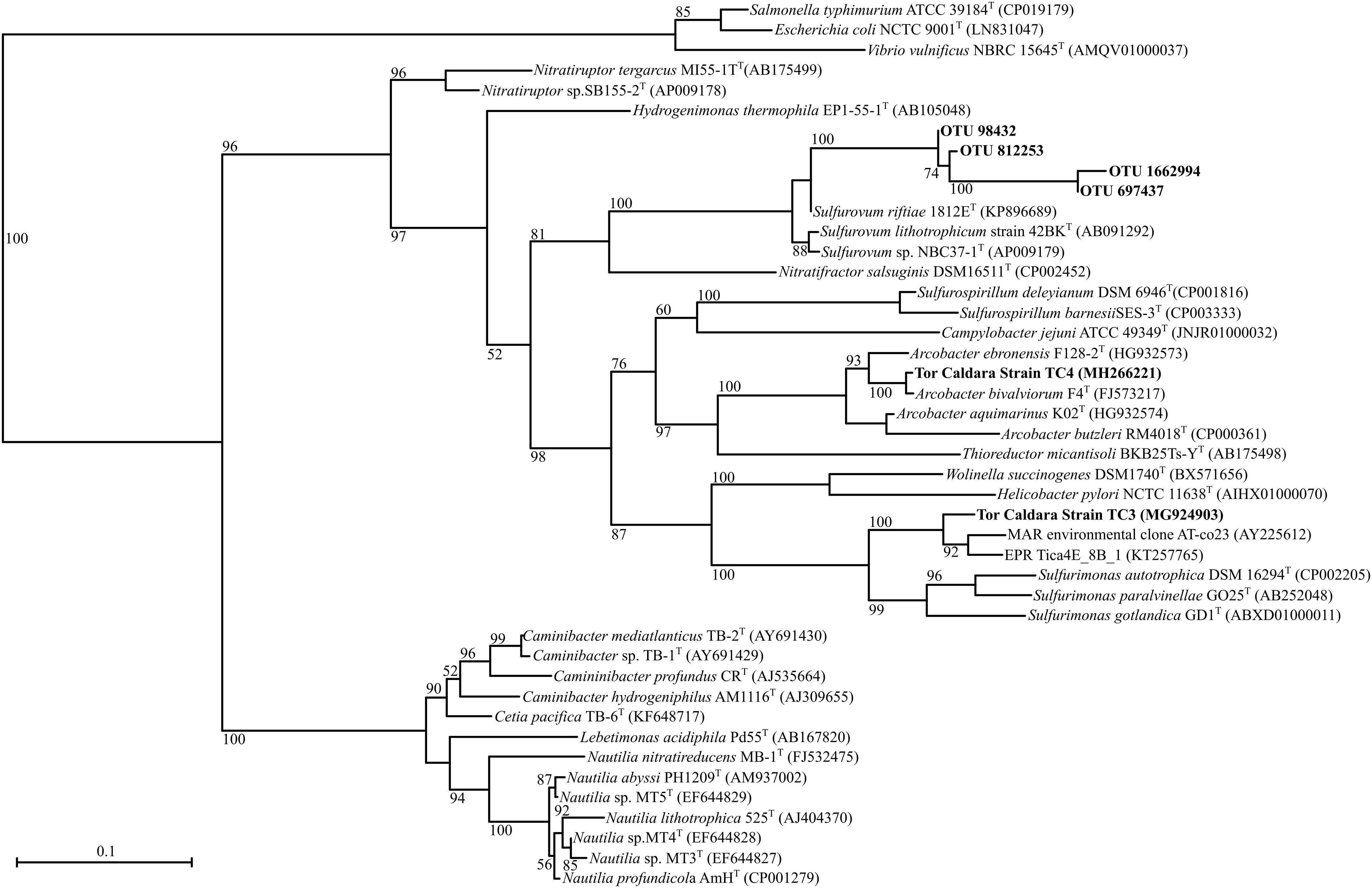
Figure 2. Maximum-likelihood phylogenetic tree derived from V4 regions and full length 16S rRNA gene sequences showing the position of OTUs classified as Sulfurovum within the Epsilonproteobacteria and strains TC3 and TC4. aLRT branch support values higher than 50% were based on 1000 replicates and are shown at each node. Bar, 0.1% substitutions per position. Sequences belonging to the class Gammaproteobacteria were used as outgroup.
The remaining eight heterotrophic strains included: strain TC2 (99.7% identical to Shewanella haliotis) (Kim et al., 2007), strain TC4 (99.4% identity to Arcobacter bivalviorum; Figure 2) (Levican et al., 2012), strain TC6 (99.9 % identity to Vibrio alginolyticus) (Sakazaki, 1968), strain TC10 (99.8% identity to Exiguobacterium profundum) (Crapart et al., 2007), strain TC12 (99.5% identity to Marinobacter hydrocarbonoclasticus) (Gauthier et al., 1992), strain TC13 (99.2% identity to Hydrogenophaga taeniospiralis) (Lalucat et al., 1982; Willems et al., 1989), strain TC14 (99.2% identity to Agarivorans albus) (Kurahashi and Yokota, 2004) and strain TC15 (99.2% identity to Halomonas aquamarina; Dobson and Franzmann, 1996). Strain TC13 grew either chemolithoautrophically using sodium sulfide or heterotrophically using acetate.
Microscopy
Scanning electron micrographs of the EF showed at least three types of different filaments: thin filaments with sulfur inclusions (Supplementary Figure 2a), thicker filaments with an outer sheath and segmented cells (Supplementary Figure 2b) and Thiothrix-like sheathed cells with visible septa (Supplementary Figure 2c). The YF filaments appeared to be embedded in a matrix containing sulfur crystals (Supplementary Figure 2d). The crystals were identified by using the elemental analyzer in conjunction with the SEM.
Composition of Tor Caldara Microbial Communities
A total of 1,311,749 sequences were generated, which clustered in 19,128 unique OTUs at 97% similarity across the 16 samples. Out of these, 19,100 OTUs were bacterial and only 28 OTUs were archaeal. Archaeal sequences contributed to less than 0.01% of the total sequences. Rarefaction curve analysis showed that, on average, EF community was more diverse than the YF (Supplementary Figure 3). The total EF community was more diverse than its active fraction (avg. chao1 of 5412 vs. 3909; Supplementary Table 2), while the total and active fraction of YF showed comparable diversity (avg. chao1 of 4111 and 4621, respectively; Supplementary Table 2).
Established and Young Filamentous Microbial Communities
Overall, 15 phyla were abundant (≥0.1%) in the EF community, as opposed to 8 phyla in the YF community (Supplementary Table 3).
The total and active fractions of the EF community were numerically dominated by Proteobacteria (75.3 and 91.2% on average, respectively), followed by Bacteroidetes (9.2 and 4.1%, respectively) and Cyanobacteria (2.3% each). The 2012 filament sample (EF12) was an exception, with Firmicutes making up 45.2 and 8.3% of the total (EF12T) and active filament (EF12A) community. The total and active fractions of the YF community were numerically dominated by Proteobacteria (93.7 and 94% on average, respectively), followed by Bacteroidetes (3.5% each) and candidate phylum GN02, later renamed as Gracilibacteria (Rinke et al., 2013) (1.5 and 1.4%, respectively). Within the Proteobacteria, the classes Gammaproteobacteria and Epsilonproteobacteria dominated both the EF and the YF communities, respectively (Figure 3).
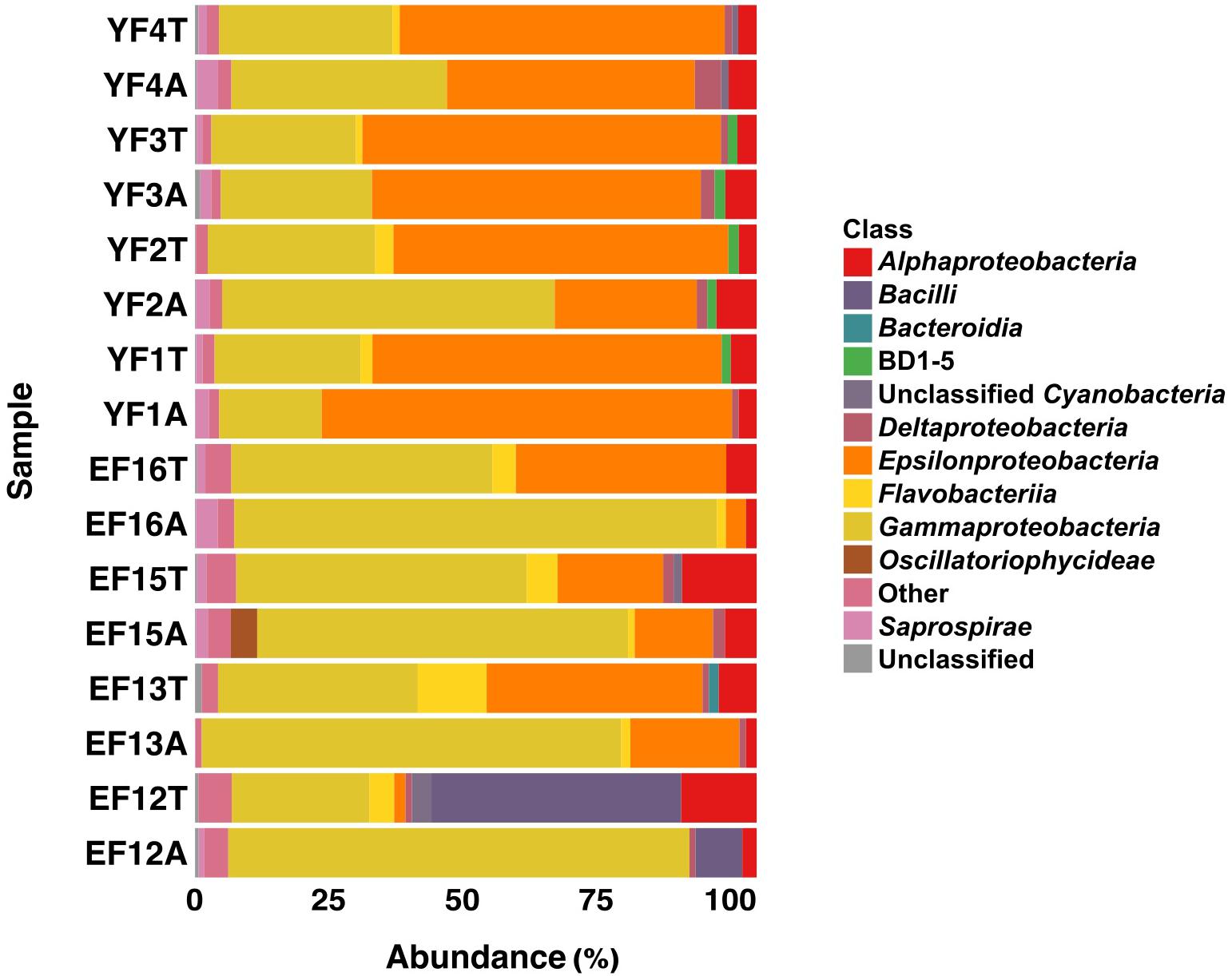
Figure 3. Class level distribution of 16S rRNA gene and transcript sequences recovered from EF and YF. Phylogenetic categories representing classes that account for at least 1% of overall abundance in all samples are shown. YF, Young Filaments; EF, Established Filaments; T, Total community (rRNA genes); A, Active Community (rRNA transcripts); the number for YF denotes the slide replicate, the number for EF denotes the sampling year.
On average, Gammaproteobacteria constituted 39.5% of the total EF community and 77.1% of its active fraction, whereas Epsilonproteobacteria constituted 24.2% of the total EF community and 12.3% of its active fraction. A reverse trend was observed in the YF community where, on average, Epsilonproteobacteria constituted 60.9% of the total filament community and 50.3% of its active fraction, whereas Gammaproteobacteria constituted 28.1% of the total filament community and 35.7% of its active fraction. The Gammaproteobacteria to Epsilonproteobacteria ratio (GE ratio) shows statistically significant differences between the established vs. young filaments (ANOVA p < 0.001; Figure 4A) with a predominance of Gammaproteobacteria in the established filaments (GE ratio of 0.57 ± 0.38) and a predominance of Epsilonproteobacteria in the young filaments (GE ratio of −0.26 ± 0.31). This difference is not evident when the GE ratio of the active vs. total community is calculated (ANOVA p > 0.05; Figure 4B), suggesting that the shift in the community is controlled predominantly by the age rather than by the metabolic state of the communities. Further, both the active community and the total community are statistically different between the established and young filaments (Tukey HSD post hoc test p < 0.01 for the active and p < 0.05 for the total community respectively; Figure 4C).
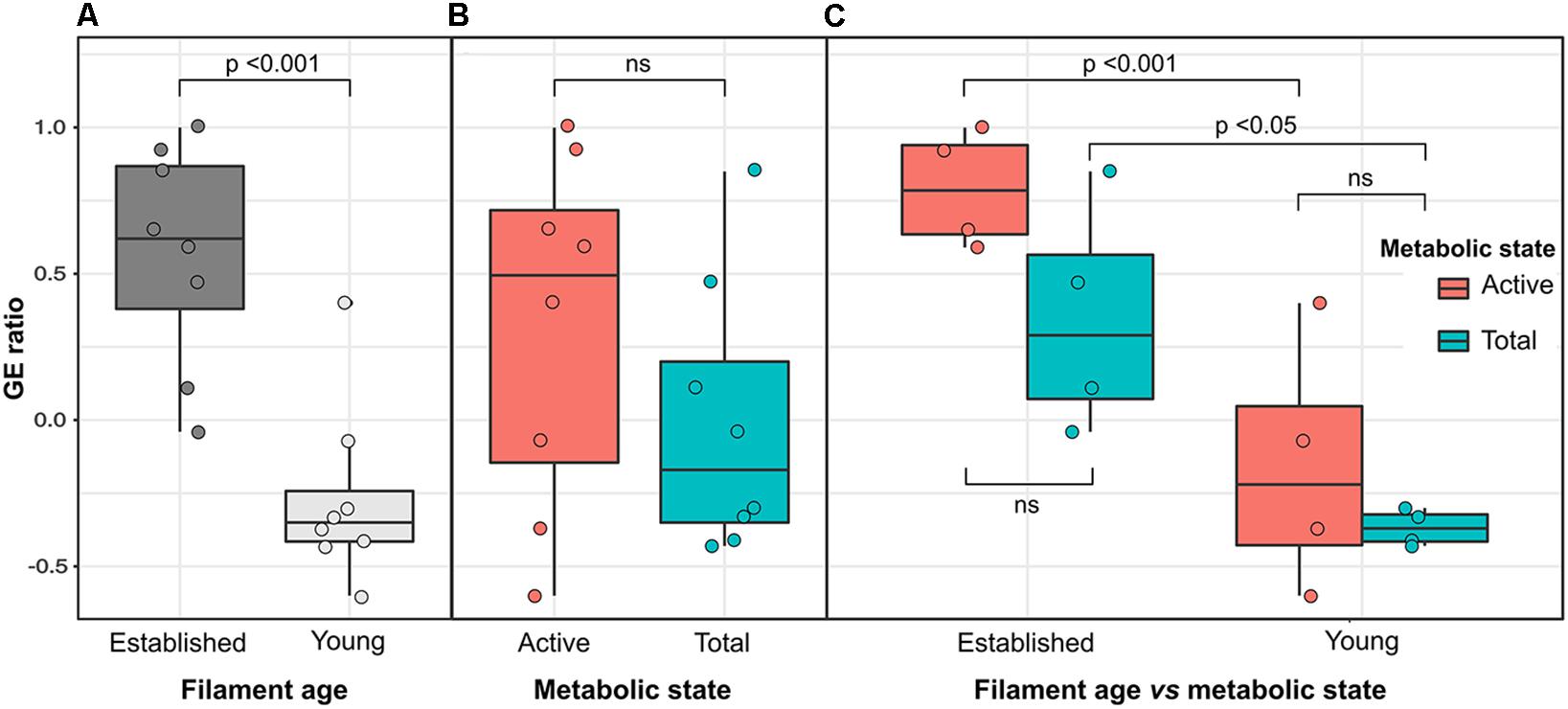
Figure 4. Variation of the Gamma- to Epsilonproteobacteria ratio (GE ratio) between the established (EF) and young filament (YF) communities. (A) Total plus active established vs. young filaments ratio; (B) Active vs. total ratio for established and young filaments combined; (C) Active and total community ratio calculated independently for the established and young filament communities.
The other abundant groups in the total and active fractions of the EF community were Alphaproteobacteria (9.7 and 3%, respectively), Flavobacteria (6.6 and 1.5 %, respectively), Saprospirae (2.3 and 1.5%, respectively) and Deltaproteobacteria (1.4 and 1.5%, respectively) (Figure 3). The remaining fractions of the total and active YF community were Alphaproteobacteria (3.6 and 5.2%, respectively), Flavobacteriia (1.9% and undetectable, respectively), Saprospirae (1.2 and 2.6%, respectively) and Deltaproteobacteria (1.2 and 2.5%, respectively) except for the BD1-5 group (1.7 and 1.7%, respectively) within the candidate phylum GN02/Gracilibacteria, which was unique to the YF community.
At the genus level, the most abundant members of the total EF community were related to the sulfur-oxidizing Gammaproteobacteria, Thiomicrospira, Thiothrix, Cocleimonas, Thiothrix-related CF-26, and to the Epsilonproteobacteria, Sulfurovum, and Sulfurimonas (Figures 1, 2, 5). Many sequences within the Gammaproteopbacteria that were not resolved at the genus level were manually analyzed and were closely related (94–96% similarity) to Thioprofundiculum, Thiothrix, Thiohalomonas, Thiomicrospira, and Galenea. The Alphaproteobacteria-related sequences were mainly affiliated with the Rhodobacteraceae order. Sequences in the Bacteriodetes phylum were mainly related to the Lutibacter, Crocinitomix, and Saprospira genera (Figure 5). In spite of this diversity in the total community, Thiomicrospira and Thiothrix-related CF-26 heavily dominated the active EF community, with the latter accounting for more than 60% of the sequences in three out of the four samples (Figure 5). In the total YF community, Sulfurovum, Thiomicrospira, Thiothrix-related CF-26 and Sulfurimonas were the most abundant genera, with Sulfurovum (53.7% on average) dominating the community (Figure 5). Sequences related to Crocinitomix and Saprospira within the Bacteriodetes phylum were also recovered in addition to unresolved sequences belonging to Deltaproteobacteria and the order Rhodobacteraceae. Sulfurovum (57.6%) also dominated the active fractions of all, but one of the YF communities.
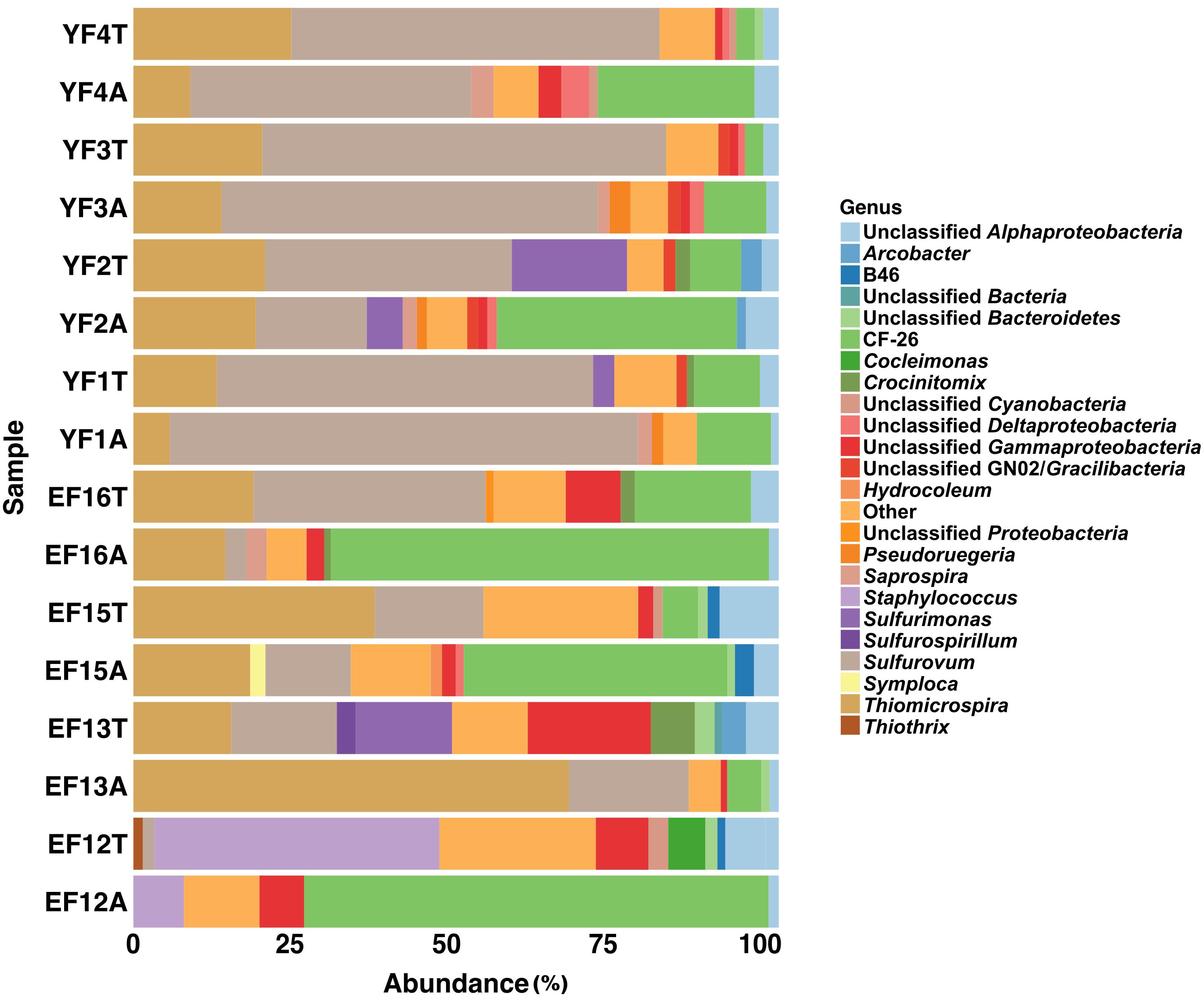
Figure 5. Genus level distribution of 16S rRNA gene and transcript sequences recovered from EF and YF samples. Phylogenetic categories representing genera that account for at least 1% of overall abundance in all samples are shown. YF, Young Filaments; EF, Established Filaments; T, Total community (rRNA genes); A, Active Community (rRNA transcripts); the number for YF denotes the slide replicate, the number for EF denotes the sampling year.
Adaptation of Sufurovum and Thimicrospira spp. to Different Sulfide Concentrations
Since phylotypes related to sulfur-oxidizing Sulfurovum (Epsilonproteobacteria) dominated the YF, while Thiomicrospira and Thiothrix (Gammaproteobacteria) dominated the EF (Figure 5), we posited that the observed shift might be related, among other factors, to a different adaptation to sulfide of the two groups of bacteria. To test this hypothesis, we grew Sulfurovum riftiae (Epsilonproteobacteria; Figure 2) and Thiomicrospira sp. EPR 85 (Gammaproteobacteria; Figure 1) from the laboratory culture collection in S-plug medium. These two strains were chosen for the following reasons: (1) both bacteria are closely related to strains or phylotypes obtained from the Tor Caldara biofilms (Figures 1, 2); (2) the Thiomicrospira strains isolated from Tor Caldara were not characterized beyond the 16S rRNA gene sequence, and generally much harder to maintain in culture; (3) in contrast, Thiomicrospira sp. strain EPR 85 and Sulfurovum riftiae are well characterized physiologically and their genomes have been sequenced; (4) to date, no Sulfurovum spp. have been isolated from shallow-water marine geothermal systems. The sulfide gradient that forms in the S-plug tubes allows the bacteria to position themselves at their optimal sulfide concentration. Subsequently, we used cyclic voltammetry to profile the sulfide concentration throughout the medium and we evaluated the growth of the two strains relative to the concentration of sulfide. Thiomicrospira sp. EPR 85 grew over a relatively broad range of sulfide concentrations: while most of the biomass was visible at about 20–25 mm from the top of the agar, where the sulfide concentration varied between 5 and 52.5 μM (Figures 6a,b), some growth could be detected down to 32 mm, where the sulfide concentration was 77.5 μM (Figures 6a,b). In contrast, S. riftiae showed a much more restricted growth pattern with a band at about 35 mm from the top of the agar, where the sulfide concentration was about 100 μM (Figures 6a,b).
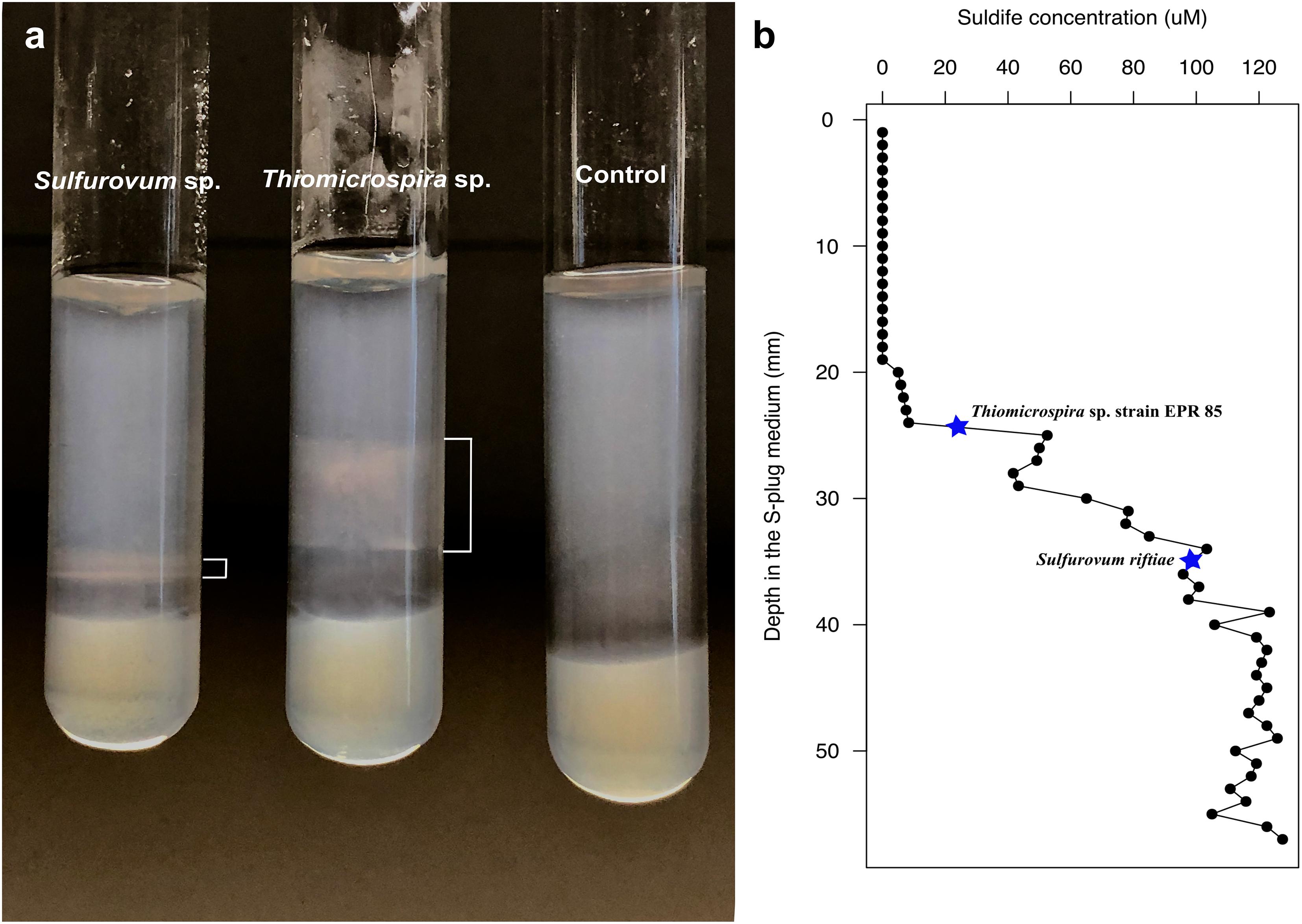
Figure 6. Growth of Sulfurovum riftiae and Thiomicrospira sp. strain 85 in sulfide gradient medium; (a) growth pattern in S-plug tubes; (b) Total sulfide concentration along the gradient. Stars indicate the total sulfide concentration for optimal growth of the two strains.
Discussion
The aim of this study was to explore the microbial diversity and gas composition of the newly discovered shallow-water gas vents located at Tor Caldara, Italy. The venting gas is mainly composed of CO2 (avg. 77 mol%) and H2S (avg. 23 mol%), with trace amounts of CH4 (0.18 mol%). This is in accordance with observations at other shallow-water vents in the Tyrrhenian Sea and elsewhere (Dando et al., 1999; Gugliandolo et al., 1999; Tarasov et al., 2005; Maugeri et al., 2008). However, the Tor Caldara gas is more enriched in H2S as compared to other shallow-water vent sites. This could be attributed to the fact that, unlike other shallow-water vent sites, the temperature at Tor Caldara is not affected by geothermal processes and H2S is a product of low temperature hydrothermal activity in the perivolcanic outgassing areas (Pizzino et al., 2002; Boatta et al., 2013). Salinity of the Mediterranean is known to be high (between 36 ppt near Gibraltar to 39 ppt in the eastern basin) because the evaporation processes are more dominant than fresh water input from precipitation and river discharge (Kallel et al., 1997). At 40 ppt, salinity at Tor Caldara was found to be marginally higher that the average salinity in the Mediterranean Sea. The pH was slightly lower in the vicinity of the venting due to the high input of CO2 from the gas emissions.
In the course of several summers of sampling the Tor Caldara gas vents, we noted that the density of the sessile filamentous biofilms was substantially lower following intense wave motion due to high winds. This was predictable, as shallow-water vents are dynamic environments, strongly influenced by factors such as tides, winds and waves (Yücel et al., 2013; Price and Giovannelli, 2017), and implies that these biofilm communities must be experiencing intermittent disruption, resetting the clock for new colonization.
We investigated the total and active fractions of the microbial biofilm communities using 16S rRNA gene and transcript amplicon analysis. Hierarchical clustering (Supplementary Figure 4) and nMDS (Supplementary Figure 5) analyses carried on the microbial biofilm communities showed that, with few exceptions, EF and YF communities clustered separately. One such exception is EF12T, in which Firmicutes comprised 45.2% of the OTUs (Figures 3, 5 and Supplementary Figure 4). We tend to rule out a possible laboratory contamination for two reasons: (1) The no-template control associated with this sample did not show amplification; (2) the abundance of the Firmicutes in the corresponding active fraction (EF12A sample) was much lower and showed that the main active bacteria were related, similarly to the other established filaments, to the Thiothrix-related CF-26 group (Figures 3, 5). One possible explanation for the abundance of Firmicutes in EF12T is that the rock substrate and sand associated with the filaments were co-sampled in 2012. This was the first year of sampling and, instead of collecting the filaments with a syringe, we scraped them from the substrate, with possible carryover of sedimentary debris. The presence of Firmicutes in the Tor Caldara sediments is not surprising, as these bacteria have been found to be relatively abundant in coastal sediments associated with gas venting (Kerfahi et al., 2014).
Succession in Microbial Communities of the Young and Established Filaments
Microbial biofilms have been studied at cold seeps (Ristova et al., 2015; Paul et al., 2017), sulfidic caves (Engel et al., 2004; Macalady et al., 2006), thermal springs (Coman et al., 2013; Beam et al., 2016), mud volcanoes (Heijs et al. (2005); Omoregie et al., 2008), as well as deep-sea (Lopez-Garcia et al., 2003; Alain et al., 2004; Crépeau et al., 2011; Dahle et al., 2013; Gulmann et al., 2015; O’Brien et al., 2015; Stokke et al., 2015; Teske et al., 2016) and shallow-water vents (Gugliandolo et al., 2006; Miranda et al., 2016). Many of these microbial communities are dominated by members of the Gammaproteobacteria and Epsilonproteobacteria (Macalady et al., 2006; Crépeau et al., 2011; O’Brien et al., 2015). However, there is a paucity of such studies in the Mediterranean Sea, where previous work has focused more on sedimentary environments (Gugliandolo and Maugeri, 1993; Maugeri et al., 2013; Kerfahi et al., 2014).
Previous field observations of the distribution of microorganisms in biofilms from geothermal habitats indicated that Epsilon- and Gammaproteobacteria are spatially segregated, with the former dominating higher sulfide habitats and the latter being more prevalent in lower sulfide, possibly more oxic habitats (Engel et al., 2004; Reigstad et al., 2011; Giovannelli et al., 2013; O’Brien et al., 2015; Miranda et al., 2016). Based on these observations, and on the high concentrations of sulfide measured in the gases at Tor Caldara (Table 1), we had predicted that the EF biofilms would be dominated by sulfide-oxidizing Epsilonproteobacteria. However, in the initial stages of this study, we detected a prevalence of Gammaproteobacteria related to the genera Thiothtrix and Thimicrospira in the EF biofilms (Figures 1, 5). Following these initial observations, we hypothesized that sulfide-oxidizing Epsilonproteobacteria might play a role in the early stages of biofilm formation at Tor Caldara, and that a colonization pattern might occur on a temporal, rather than spatial, scale. To test this hypothesis, in 2016 we conducted a biofilm colonization experiment by deploying sterile glass slides in the same area for 4 days, till visible biofilm (YF) formation was observed (Supplementary Figure 1b). Then we analyzed the taxonomic composition of the YF and we compared that with that of the EF biofilms (Supplementary Figure 1a). Both types of filaments showed similar diversity at the phylum level, except for Cyanobacteria and GN02/Gracilibacteria, which were unique to the EF and YF, respectively. Members of candidate phylum GN02/Gracilibacteria were first reported from a high sulfide concentration zone of a hyper saline microbial mat in Mexico and, later, single amplified genomes (SAGs) from this phylum were recovered from the East Pacific Rise hydrothermal vent system (Ley et al., 2006; Rinke et al., 2013). The presence of Cyanobacteria-related sequences in both the total and active fraction of established filaments at Tor Caldara might indicate a more permissible (e.g., less sulfidic and more oxic) environment as compared to the newly formed community (Ley et al., 2006).
The white and feathery EF resembled those seen at the oxic/anoxic boundary of sulfidic environments. Members of the Thiotrichales, especially those closely related to Thiothrix, have been shown to dominate these white filaments (Engel et al., 2004; Gugliandolo et al., 2006; Macalady et al., 2006; Reigstad et al., 2011; O’Brien et al., 2015). We observed sequences related to Thiothrix in our EF samples, where Thiomicrospira and Thiothrix-related CF-26 dominated the total and active fraction of these communities. Two species of Thiomicrospira, T. frisia and T. crunogena were also isolated from EF in pure culture under strict sulfur-oxidizing chemolithoautotrophic conditions, further establishing their abundance and importance. Thiothrix-related CF-26 is an uncultured genus first described as a clone recovered from the flocculent mats associated with submarine volcanoes along the Kermadec Arc. The OTUs annotated as CF-26 show high 16S rRNA sequence identity (>97%) to sequences recovered from Hatoma Knoll deep-sea vent site (GenBank accession number AB611518) (Yoshida-Takashima et al., 2012), microbial mats at shallow-vents from South Tonga Arc (GenBank accession number HQ153886) (Murdock et al., 2010) and Kuieshan Island shallow water vents (GenBank accession number JQ611107) (unpublished) with Thiothrix as the closest genus (16S rRNA gene sequence identity: 88–90%; Figure 1). However, the scanning electron micrographs of the filaments didn’t show rosette-like formations that are characteristic of Thiotrix, while sheathed cells with visible septa as described by Williams et al. (1987) were observed along with other types of filaments (Supplementary Figure 2). Members of the genus Thiothrix are sulfur-oxidizing chemoautotrophic, heterotrophic as well as mixotrophic bacteria (Howarth et al., 1999). Similar to the other chemoautotrophic members of the Gammaproteobacteria, Thiotrix species fix CO2 via the CBB cycle (Odintsova et al., 1993). Unfortunately, our attempts to cultivate Thiotrix-related bacteria in selective medium were unsuccessful. Thiomicrospira are also sulfur oxidizing obligate chemolithoautotrophs that fix CO2 using the CBB cycle, with mixotrophy observed in some species (Boden et al., 2017). Sequences related to the genus B46 within Gammaproteobacteria with 95% similarity to Methylomolas and Methylococcaceae were also recovered. Similar sequences have been recovered from the Tica vent on the East Pacific Rise deep-sea hydrothermal vent system (GenBank accession number KT257824) (Gulmann et al., 2015) as well as microbial mats at White Point, California (GenBank accession number KX422153) (Miranda et al., 2016) indicating potential occurrence of methylotrophy in addition to chemoautotrophy. Successful isolation of Sedimenticola thiotaurini, another sulfur-oxidizing chemolithotroph belonging to Gammaproteobacteria, further confirmed their dominance in the EF. The isolation of several non-sulfur-oxidizing heterotrophic bacterial strains suggests that the EF is a complex community that also harbors heterotrophs, in addition to primary producers. The enrichment and isolation approach provided an initial culture collection from the Tor Caldara gas vents for future physiological and genomic characterization (Patwardhan and Vetriani, 2016).
Following a 4-day deployment, the glass slides (YF) were predominantly colonized by Sulfurovum species. Sequences related to the genus Sulfurospirillum were also recovered. Both genera are sulfur-oxidizing chemolithoautotrophic members of Epsilonproteobacteria that are abundant in both shallow and deep-sea vents environments (Sievert and Vetriani, 2012; Giovannelli et al., 2013; Vetriani et al., 2014; O’Brien et al., 2015; Miranda et al., 2016). We also isolated a pure culture from Tor Caldara that had ∼93% 16S rRNA gene sequence similarity to Sulfurimonas gotlandica, which is another sulfur-oxidizing chemolithoautotrophic Epsilonproteobacterium.
Adaptation to Different Sulfide Concentrations May Drive Ecological Succession of Epsilon- and Gammaproteobacteria in Young and Established Biofilms
The dominance of Epsilonproteobacteria related to Sulfurovum spp. in the YF active community indicates that these bacteria were the early colonizers in the Tor Caldara filamentous biofilms and that Gammaproteobacteria related to Thiomicrospira and Thiothrix spp. became prevalent as the biofilm matured (Figures 3, 5). Therefore, we posited that the metabolic activities of Sulfurovum sp. during the early stages of colonization may be critical to condition the environment for the settlement of subsequent species. Previous studies have observed that Epsilon- and Gammaproteobacteria occupy spatially separated niches based on their adaptation to sulfide concentrations (Engel et al., 2004; Reigstad et al., 2011; O’Brien et al., 2015; Miranda et al., 2016; Meier et al., 2017). In particular, in a 16S rRNA gene- and metagenomic-based study of the distribution of microorganisms at deep-sea hydrothermal vents in the Manus Basin, off Papua New Guinea, Meier et al. (2017) proposed that sulfide and oxygen concentrations are determining factors for niche partitioning between sulfur-oxidizing Epsilon- and Gammaproteobacteria. However, to our knowledge this hypothesis had not been experimentally tested prior to this study.
We tested this hypothesis experimentally by evaluating the growth pattern of an Epsilonproteobacterium (S. riftiae) and a Gammaproteobacterium (Thiomicrospira sp. EPR 85) relative to the concentration of total dissolved sulfide throughout S-plug tubes. Our findings show that S. riftiae grew at higher sulfide concentration (100 μM) than Thiomicrospira sp. EPR 85 (5 – 52.5 μM; Figures 6a,b). These sulfide concentrations are consistent with those measured in situ at deep-sea hydrothermal vents. For instance, the sulfide concentrations measured by O’Brien et al. (2015) in Epsilonproteobacteria-dominated diffuse flow deep-sea vents were up to 67 mM, while it ranged between 0.3–2.9 mM at Gammaproteobacteria-dominated adjacent control sites. Similarly, Gulmann et al. (2015) reported maximum dissolved sulfide concentrations of 100 μM in fluids from a diffuse flow vent at the same site. It is worth noting that Meier et al. (2017) inferred that the distribution of SUP05-related Gammaproteobacteria at deep-sea vents correlated with sulfide concentrations between 0 and 100 μM, while Sulfurovum and Sulfurimonas-related Epsilonproteobacteria correlated with sulfide concentrations between about 100 and 750 μM. While some of the latter concentrations are higher than those measured in the S-plug experiment, it should be noted that S. riftiae grew near the highest sulfide concentration attainable in the S-plug tube. Hence, it is possible that its observed narrow growth range could be limited by sulfide availability and that the bacterium is able to tolerate higher sulfide concentrations than those observed in this experiment.
Based on these observations, it is reasonable to infer that, at the early stages of formation of the Tor Caldara biofilms, Sulfurovum-related spp. experience and cope with the high sulfide (and possibly low oxygen) concentration in the gas emissions. Laboratory studies of pure cultures of Sulfurovum sp. revealed that these bacteria conserve energy by the oxidation of reduced sulfur species to sulfate (Yamamoto et al., 2010). Hence, at Tor Caldara, respiration by Sulfurovum spp. and other sulfur-oxidizing Epsilonproteobacteria may contribute to the decrease in sulfide concentration within the biofilm structure during the early stages of colonization, effectively conditioning the habitat via sulfide detoxification for the subsequent colonization by Gammaproteobacteria related to Thiomicrospira and Thiothrix spp. This is consistent with the observation that the less sulfide-tolerant, aerobic Gammaproteobacteria become dominant in the EF community (Figures 3, 5). Interestingly, the observed temporal succession from Epsilon- to Gammaprotebacteria dominance in the Tor Caldara biofilms appears to be driven by ecosystem conditioning rather than by the spatial segregation often observed at deep-sea hydrothermal vents (Engel et al., 2004; Reigstad et al., 2011; O’Brien et al., 2015; Miranda et al., 2016; Meier et al., 2017).
Conclusion
Our findings show that the gas composition of the Tor Caldara shallow-water vents is composed mainly by CO2 and H2S. These gases support dense sessile filamentous microbial biofilms prevalently composed of sulfur-oxidizing Epsilon- and Gammaproteobacteria. We observed a temporal succession in the biofilm species composition that appears to be driven, among other factors, by the different adaptation of some of the most abundant Epsilon- and Gammaproteobacteria (Sulfurovum and Thiomicrospira spp.) to sulfide concentration. Future studies investigating the metabolic potential and function of these biofilm communities will be important to further elucidate their ecophysiological role.
Author Contributions
SP performed the experiments, analyzed the data, and wrote the manuscript. DF analyzed the data and participated in the writing of the manuscript. DG collected samples, analyzed the data, and participated in the writing of the manuscript. MY analyzed the data and participated in the writing of the manuscript. CV directed and supervised the research, collected samples, and wrote the manuscript.
Funding
This work was partially supported by NSF grants OCE 11-24141 and OCE 11-36451 to CV, NSF grant MCB 15-17567 to CV and DG, NSF grants OCE-1155246 and MCB-1517560 to DF, and NASA grant NNX15AM18G to CV. This paper is C-DEBI contribution 447.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the crew of the R/V Antares for their support during the sampling operations.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02970/full#supplementary-material
References
Alain, K., Zbinden, M., Le Bris, N., Lesongeur, F., Quérellou, J., Gaill, F., et al. (2004). Early steps in microbial colonization processes at deep-sea hydrothermal vents. Environ. Microbiol. 6, 227–241. doi: 10.1111/j.1462-2920.2003.00557
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Beam, J. P., Bernstein, H. C., Jay, Z. J., Kozubal, M. A., Jennings, R. D., Tringe, S. G., et al. (2016). Assembly and succession of iron oxide microbial mat communities in acidic geothermal springs. Front. Microbiol. 7:25. doi: 10.3389/fmicb.2016.00025
Boatta, F., D’Alessandro, W., Gagliano, A. L., Liotta, M., Milazzo, M., Rodolfo-Metalpa, R., et al. (2013). Geochemical survey of Levante Bay, Vulcano Island (Italy), a natural laboratory for the study of ocean acidification. Mar. Pollut. Bull. 73, 485–494. doi: 10.1016/j.marpolbul.2013.01.029
Boden, R., Scott, K. M., Williams, J., Russel, S., Antonen, K., Rae, A. W., et al. (2017). An evaluation of Thiomicrospira, Hydrogenovibrio and Thioalkalimicrobium: reclassification of four species of Thiomicrospira to each Thiomicrorhabdus gen. nov. and Hydrogenovibrio, and reclassification of all four species of Thioalkalimicrobium to Thiomicrospira. Int. J. Syst. Evol. Microbiol. 67, 1140–1151. doi: 10.1099/ijsem.0.001855
Brinkhoff, T., Muyzer, G., Wirsen, C. O., and Kuever, J. (1999). Thiomicrospira kuenenii sp. nov. and Thiomicrospira frisia sp. nov., two mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacteria isolated from an intertidal mud flat. Int. J. Syst. Bacteriol. 49, 385–392. doi: 10.1099/00207713-49-2-385
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carapezza, M. L., Barberi, F., Ranaldi, M., Ricci, T., Tarchini, L., Barrancos, J., et al. (2012). Hazardous gas emissions from the flanks of the quiescent Colli Albani volcano (Rome, Italy). Appl. Geochem. 27, 1767–1782. doi: 10.1016/j.apgeochem.2012.02.012
Carapezza, M. L., and Tarchini, L. (2007). Accidental gas emission from shallow pressurized aquifers at Alban Hills volcano (Rome, Italy): geochemical evidence of magmatic degassing? J. Volcanol. Geotherm. Res. 165, 5–16. doi: 10.1016/j.jvolgeores.2007.04.008
Coman, C., Drugã, B., Hegedus, A., Sicora, C., and Dragoş, N. (2013). Archaeal and bacterial diversity in two hot spring microbial mats from a geothermal region in Romania. Extremophiles 17, 523–534. doi: 10.1007/s00792-013-0537-5
Crapart, S., Fardeau, M.-L., Cayol, J.-L., Thomas, P., Sery, C., Ollivier, B., et al. (2007). Exiguobacterium profundum sp. nov., a moderately thermophilic, lactic acid-producing bacterium isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 57, 287–292. doi: 10.1099/ijs.0.64639-0
Crépeau, V., Cambon Bonavita, M.-A., Lesongeur, F., Randrianalivelo, H., Sarradin, P.-M., Sarrazin, J., et al. (2011). Diversity and function in microbial mats from the Lucky Strike hydrothermal vent field. FEMS Microbiol. Ecol. 76, 524–540. doi: 10.1111/j.1574-6941.2011.01070
Dahle, H., Roalkvam, I., Thorseth, I. H., Pedersen, R. B., and Steen, I. H. (2013). The versatile in situ gene expression of an Epsilonproteobacteria-dominated biofilm from a hydrothermal chimney. Environ. Microbiol. Rep. 5, 282–290. doi: 10.1111/1758-2229.12016
Dando, P. R., Stüben, D., and Varnavas, S. P. (1999). Hydrothermalism in the Mediterranean Sea. Prog. Oceanogr. 44, 333–367. doi: 10.1016/S0079-6611(99)00032-4
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Dobson, S. J., and Franzmann, P. D. (1996). Unification of the genera Deleya (Baumann et al. 1983), Halomonas (Vreeland et al. 1980), and Halovibrio (Fendrich 1988) and the Species Paracoccus halodenitrificans (Robinson and Gibbons 1952) into a Single Genus, Halomonas, and Placement of the Genus Zymobacter in the Family Halomonadaceae. Int. J. Syst. Evol. Microbiol. 46, 550–558. doi: 10.1099/00207713-46-2-550
Engel, A. S., Porter, M. L., Stern, L. A., Quinlan, S., and Bennett, P. C. (2004). Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria.”. FEMS Microbiol. Ecol. 51, 31–53. doi: 10.1016/j.femsec.2004.07.004
Flood, B. E., Jones, D. S., and Bailey, J. V. (2015). Sedimenticola thiotaurini sp. nov., a sulfur-oxidizing bacterium isolated from salt marsh sediments, and emended descriptions of the genus Sedimenticola and Sedimenticola selenatireducens. Int. J. Syst. Evol. Microbiol. 65, 2522–2530. doi: 10.1099/ijs.0.000295
Fornari, D. J., Shank, T., Von Damm, K. L., Gregg, T. K. P., Lilley, M., Levai, G., et al. (1998). Time-series temperature measurements at high-temperature hydrothermal vents, East Pacific Rise 9°49′–51′N: evidence for monitoring a crustal cracking event. Earth Planet. Sci. Lett. 160, 419–431. doi: 10.1016/S0012-821X(98)00101-0
Foustoukos, D. I. (2012). Metastable equilibrium in the C-H-O system: graphite deposition in crustal fluids. Am. Mineral. 97, 1373–1380. doi: 10.2138/am.2012.4086
Foustoukos, D. I., Houghton, J. L., Seyfried, W. E., Sievert, S. M., and Cody, G. D. (2011). Kinetics of H2–O2–H2O redox equilibria and formation of metastable H2O2 under low temperature hydrothermal conditions. Geochim. Cosmochim. Acta 75, 1594–1607. doi: 10.1016/j.gca.2010.12.020
Galtier, N., Gouy, M., and Gautier, C. (1996). SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12, 543–548. doi: 10.1093/bioinformatics/12.6.543
Gauthier, M. J., Lafay, B., Christen, R., Fernandez, L., Acquaviva, M., Bonin, P., et al. (1992). Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Evol. Microbiol. 42, 568–576. doi: 10.1099/00207713-42-4-568
Giovannelli, D., Chung, M., Staley, J., Starovoytov, V., Le Bris, N., and Vetriani, C. (2016). Sulfurovum riftiae sp. nov., a mesophilic, thiosulfate-oxidizing, nitrate-reducing chemolithoautotrophic epsilonproteobacterium isolated from the tube of the deep-sea hydrothermal vent polychaete Riftia pachyptila. Int. J. Syst. Evol. Microbiol. 66, 2697–2701. doi: 10.1099/ijsem.0.001106
Giovannelli, D., d’Errico, G., Manini, E., Yakimov, M. M., and Vetriani, C. (2013). Diversity and phylogenetic analyses of bacteria from a shallow-water hydrothermal vent in Milos island (Greece). Front. Extreme Microbiol. 4:184. doi: 10.3389/fmicb.2013.00184
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Gugliandolo, C., Italiano, F., and Maugeri, T. (2006). The submarine hydrothermal system of Panarea (Southern Italy): biogeochemical processes at the thermal fluids - sea bottom interface. Ann. Geophys. 49, 783–792. doi: 10.4401/ag-3139
Gugliandolo, C., Italiano, F., Maugeri, T. L., Inguaggiato, S., and Caccamo, D. (1999). Submarine hydrothermal vents of the Aeolian Islands: relationship between microbial communities and thermal fluids. Geomicrobiol. J. 16, 105–117. doi: 10.1080/014904599270794
Gugliandolo, C., Lentini, V., Bunk, B., Overmann, J., Italiano, F., and Maugeri, T. L. (2015). Changes in prokaryotic community composition accompanying a pronounced temperature shift of a shallow marine thermal brine pool (Panarea Island, Italy). Extremophiles 19, 547–559. doi: 10.1007/s00792-015-0737-2
Gugliandolo, C., and Maugeri, T. L. (1993). Chemolithotrophic, sulfur-oxidizing bacteria from a marine, shallow hydrothermal vent of Vulcano (Italy). Geomicrobiol. J. 11, 109–120. doi: 10.1080/01490459309377939
Gugliandolo, C., and Maugeri, T. L. (1998). Temporal variations in heterotrophic mesophilic bacteria from a marine shallow hydrothermal vent off the island of Vulcano (Eolian Islands, Italy). Microb. Ecol. 36, 13–22. doi: 10.1007/s002489900088
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Gulmann, L. K., Beaulieu, S. E., Shank, T. M., Ding, K., Seyfried, W. E., and Sievert, S. M. (2015). Bacterial diversity and successional patterns during biofilm formation on freshly exposed basalt surfaces at diffuse-flow deep-sea vents. Front. Microbiol. 6:901. doi: 10.3389/fmicb.2015.00901
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Heijs, S. K., Damsté, J. S., and Forney, L. J. (2005). Characterization of a deep-sea microbial mat from an active cold seep at the Milano mud volcano in the Eastern Mediterranean Sea. FEMS Microbiol. Ecol. 54, 47–56. doi: 10.1016/j.femsec.2005.02.007
Hirayama, H., Sunamura, M., Takai, K., Nunoura, T., Noguchi, T., Oida, H., et al. (2007). Culture-dependent and -independent characterization of microbial communities associated with a shallow submarine hydrothermal system occurring within a coral reef off Taketomi Island, Japan. Appl. Environ. Microbiol. 73, 7642–7656. doi: 10.1128/AEM.01258-07
Houghton, J. L., Foustoukos, D. I., Flynn, T. M., Vetriani, C., Bradley, A. S., and Fike, D. A. (2016). Thiosulfate oxidation by Thiomicrospira thermophila: metabolic flexibility in response to ambient geochemistry. Environ. Microbiol. 18, 3057–3072. doi: 10.1111/1462-2920.13232
Howarth, R., Unz, R. F., Seviour, E. M., Seviour, R. J., Blackall, L. L., Pickup, R. W., et al. (1999). Phylogenetic relationships of filamentous sulfur bacteria (Thiothrix spp. and Eikelboom type 021N bacteria) isolated from wastewater-treatment plants and description of Thiothrix eikelboomii sp. nov., Thiothrix unzii sp. nov., Thiothrix fructosivorans sp. nov. and Thiothrix defluvii sp. nov. Int. J. Syst. Evol. Microbiol. 49, 1817–1827. doi: 10.1099/00207713-49-4-1817
Inagaki, F. (2004). Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the ε-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 54, 1477–1482. doi: 10.1099/ijs.0.03042-0
Jannasch, H. W., Wirsen, C. O., Nelson, D. C., and Robertson, L. A. (1985). Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 35, 422–424. doi: 10.1099/00207713-35-4-422
Kalanetra, K. M., Huston, S. L., and Nelson, D. C. (2004). Novel, attached, sulfur-oxidizing bacteria at shallow hydrothermal vents possess vacuoles not involved in respiratory nitrate accumulation. Appl. Environ. Microbiol. 70, 7487–7496. doi: 10.1128/AEM.70.12.7487-7496.2004
Kallel, N., Paterne, M., Labeyrie, L., Duplessy, J.-C., and Arnold, M. (1997). Temperature and salinity records of the Tyrrhenian Sea during the last 18,000 years. Palaeogeogr. Palaeoclimatol. Palaeoecol. 135, 97–108. doi: 10.1016/S0031-0182(97)00021-7
Kerfahi, D., Hall-Spencer, J. M., Tripathi, B. M., Milazzo, M., Lee, J., and Adams, J. M. (2014). Shallow water marine sediment bacterial community shifts along a natural CO2 gradient in the Mediterranean sea off Vulcano, Italy. Microb. Ecol. 67, 819–828. doi: 10.1007/s00248-014-0368-7
Kim, D., Baik, K. S., Kim, M. S., Jung, B.-M., Shin, T.-S., Chung, G.-H., et al. (2007). Shewanella haliotis sp. nov., isolated from the gut microflora of abalone, Haliotis discus hannai. Int. J. Syst. Evol. Microbiol. 57, 2926–2931. doi: 10.1099/ijs.0.65257-0
Kurahashi, M., and Yokota, A. (2004). Agarivorans albus gen. nov., sp. nov., a γ-proteobacterium isolated from marine animals. Int. J. Syst. Evol. Microbiol. 54, 693–697. doi: 10.1099/ijs.0.02778-0
Labrenz, M., Grote, J., Mammitzsch, K., Boschker, H. T. S., Laue, M., Jost, G., et al. (2013). Sulfurimonas gotlandica sp. nov., a chemoautotrophic and psychrotolerant epsilonproteobacterium isolated from a pelagic redoxcline, and an emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 63, 4141–4148. doi: 10.1099/ijs.0.048827-0
Lalucat, J., Pares, R., and Schlegel, H. G. (1982). Pseudomonas taeniospiralis sp. nov., an R-body-containing hydrogen bacterium. Int. J. Syst. Evol. Microbiol. 32, 332–338. doi: 10.1099/00207713-32-3-332
Lentini, V., Gugliandolo, C., Bunk, B., Overmann, J., and Maugeri, T. L. (2014). Diversity of prokaryotic community at a shallow marine hydrothermal site elucidated by Illumina sequencing technology. Curr. Microbiol. 69, 457–466. doi: 10.1007/s00284-014-0609-5
Levican, A., Collado, L., Aguilar, C., Yustes, C., Diéguez, A. L., Romalde, J. L., et al. (2012). Arcobacter bivalviorum sp. nov. and Arcobacter venerupis sp. nov., new species isolated from shellfish. Syst. Appl. Microbiol. 35, 133–138. doi: 10.1016/j.syapm.2012.01.002
Ley, R. E., Harris, J. K., Wilcox, J., Spear, J. R., Miller, S. R., Bebout, B. M., et al. (2006). Unexpected diversity and complexity of the guerrero negro hypersaline microbial mat. Appl. Environ. Microbiol. 72, 3685–3695. doi: 10.1128/AEM.72.5.3685-3695.2006
Lopez-Garcia, P., Duperron, S., Philippot, P., Foriel, J., Susini, J., and Moreira, D. (2003). Bacterial diversity in hydrothermal sediment and Epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ. Microbiol. 5, 961–976. doi: 10.1046/j.1462-2920.2003.00495
Luther, G. W., Brendel, P. J., Lewis, B. L., Sundby, B., Lefrançois, L., Silverberg, N., et al. (2003). Simultaneous measurement of O2, Mn, Fe, I-, and S(—II) in marine pore waters with a solid-state voltammetric microelectrode. Limnol. Oceanogr. 43, 325–333. doi: 10.4319/lo.1998.43.2.0325
Macalady, J. L., Lyon, E. H., Koffman, B., Albertson, L. K., Meyer, K., Galdenzi, S., et al. (2006). Dominant microbial populations in limestone-corroding stream biofilms, Frasassi cave system, Italy. Appl. Environ. Microbiol. 72, 5596–5609. doi: 10.1128/AEM.00715-06
Marra, F., Gaeta, M., Giaccio, B., Jicha, B. R., Palladino, D. M., Polcari, M., et al. (2016). Assessing the volcanic hazard for Rome: 40Ar/39Ar and In-SAR constraints on the most recent eruptive activity and present-day uplift at Colli Albani Volcanic District. Geophys. Res. Lett. 43, 6898–6906. doi: 10.1002/2016GL069518
Marteinsson, V. T., Kristjánsson, J. K., Kristmannsdóttir, H., Dahlkvist, M., Sæmundsson, K., Hannington, M., et al. (2001). Discovery and description of giant submarine smectite cones on the seafloor in Eyjafjordur, northern Iceland, and a novel thermal microbial habitat. Appl. Environ. Microbiol. 67, 827–833. doi: 10.1128/AEM.67.2.827-833.2001
Maugeri, T. L., Lentini, V., Gugliandolo, C., Cousin, S., and Stackebrandt, E. (2010). Microbial diversity at a hot, shallow-sea hydrothermal vent in the Southern Tyrrhenian Sea (Italy). Geomicrobiol. J. 27, 380–390. doi: 10.1080/01490450903451518
Maugeri, T. L., Lentini, V., Gugliandolo, C., Italiano, F., Cousin, S., and Stackebrandt, E. (2008). Bacterial and Archaeal populations at two shallow hydrothermal vents off Panarea Island (Eolian Islands, Italy). Extremophiles 13, 199–212. doi: 10.1007/s00792-008-0210-6
Maugeri, T. L., Lentini, V., Spanò, A., and Gugliandolo, C. (2013). Abundance and diversity of picocyanobacteria in shallow hydrothermal vents of Panarea Island (Italy). Geomicrobiol. J. 30, 93–99. doi: 10.1080/01490451.2011.653088
Meier, D. V., Pjevac, P., Bach, W., Hourdez, S., Girguis, P. R., Vidoudez, C., et al. (2017). Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 11, 1545–1558. doi: 10.1038/ismej.2017.37
Meyer-Dombard, D. R., Amend, J. P., and Osburn, M. R. (2013). Microbial diversity and potential for arsenic and iron biogeochemical cycling at an arsenic rich, shallow-sea hydrothermal vent (Tutum Bay, Papua New Guinea). Chem. Geol. 348, 37–47. doi: 10.1016/j.chemgeo.2012.02.024
Miranda, P. J., McLain, N. K., Hatzenpichler, R., Orphan, V. J., and Dillon, J. G. (2016). Characterization of chemosynthetic microbial mats associated with intertidal hydrothermal sulfur vents in white point, San Pedro, CA, USA. Front. Microbiol. 7:1163. doi: 10.3389/fmicb.2016.01163
Murdock, S., Johnson, H., Forget, N., and Juniper, S. K. (2010). Composition and diversity of microbial mats at shallow hydrothermal vents on Volcano 1, South Tonga Arc. 7. Cah. Biol. Mar. 51, 407–413.
O’Brien, C. E., Giovannelli, D., Govenar, B., Luther, G. W., Lutz, R. A., Shank, T. M., et al. (2015). Microbial biofilms associated with fluid chemistry and megafaunal colonization at post-eruptive deep-sea hydrothermal vents. Deep Sea Res. Part II Top. Stud. Oceanogr. 121, 31–40. doi: 10.1016/j.dsr2.2015.07.020
Odintsova, E. V., Wood, A. P., and Kelly, D. P. (1993). Chemolithoutotrophic growth of Thiothrix ramosa. Arch. Microbiol. 160, 152–157. doi: 10.1007/BF00288718
Omoregie, E. O., Mastalerz, V., de Lange, G., Straub, K. L., Kappler, A., Roy, H., et al. (2008). Biogeochemistry and community composition of iron- and sulfur-precipitating microbial mats at the chefren mud volcano (Nile Deep Sea Fan, Eastern Mediterranean). Appl. Environ. Microbiol. 74, 3198–3215. doi: 10.1128/AEM.01751-07
Patwardhan, S., and Vetriani, C. (2016). Varunaivibrio sulfuroxidans gen. nov., sp. nov., a facultatively chemolithoautotrophic, mesophilic alphaproteobacterium from a shallow-water gas vent at Tor Caldara, Tyrrhenian Sea. Int. J. Syst. Evol. Microbiol. 66, 3579–3584. doi: 10.1099/ijsem.0.001235
Paul, B. G., Ding, H., Bagby, S. C., Kellermann, M. Y., Redmond, M. C., Andersen, G. L., et al. (2017). Methane-oxidizing bacteria shunt carbon to microbial mats at a marine hydrocarbon seep. Front. Microbiol. 8:186. doi: 10.3389/fmicb.2017.00186
Pizzino, L., Galli, G., Mancini, C., Quattrocchi, F., and Scarlato, P. (2002). Natural gas hazard (CO2, 222Rn) within a quiescent volcanic region and its relations with tectonics: the case of the Ciampino-Marino area, Alban Hills Volcano, Italy. Nat. Hazards 27, 257–287. doi: 10.1023/A:1020398128649
Podgorsek, L., Petri, R., and Imhoff, J. F. (2004). Cultured and genetic diversity, and activities of sulfur-oxidizing bacteria in low-temperature hydrothermal fluids of the North Fiji Basin. Mar. Ecol. Prog. Ser. 266, 65–76. doi: 10.3354/meps266065
Price, R. E., and Giovannelli, D. (eds). (2017). “A review of the geochemistry and microbiology of marine shallow-water hydrothermal vents,” in Reference Module in Earth Systems and Environmental Sciences, (Berlin: Elsevier). doi: 10.1016/B978-0-12-409548-9.09523-3
Quéméneur, M., Bes, M., Postec, A., Mei, N., Hamelin, J., Monnin, C., et al. (2014). Spatial distribution of microbial communities in the shallow submarine alkaline hydrothermal field of the Prony Bay, New Caledonia. Environ. Microbiol. Rep. 6, 665–674. doi: 10.1111/1758-2229.12184
R: The R Project for Statistical Computing (2018). R: The R Project for Statistical Computing. Available at: https://www.r-project.org/ [accessed May 17, 2018].
Reigstad, L. J., Jorgensen, S. L., Lauritzen, S.-E., Schleper, C., and Urich, T. (2011). Sulfur-oxidizing chemolithotrophic proteobacteria dominate the microbiota in high arctic thermal springs on svalbard. Astrobiology 11, 665–678. doi: 10.1089/ast.2010.0551
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J.-F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Ristova, P. P., Wenzhöfer, F., Ramette, A., Felden, J., and Boetius, A. (2015). Spatial scales of bacterial community diversity at cold seeps (Eastern Mediterranean Sea). ISME J. 9, 1306–1318. doi: 10.1038/ismej.2014.217
Sakazaki, R. (1968). Proposal of Vibrio alginolyticus for the biotype 2 of Vibrio parahaemolyticus. Jpn. J. Med. Sci. Biol. 21, 359–362. doi: 10.7883/yoken1952.21.359
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Sievert, S., and Vetriani, C. (2012). Chemoautotrophy at deep-sea vents: past, present, and future. Oceanography 25, 218–233. doi: 10.5670/oceanog.2012.21
Sievert, S. M., Brinkhoff, T., Muyzer, G., Ziebis, W., and Kuever, J. (1999). Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 65, 3834–3842.
Sievert, S. M., Kuever, J., and Muyzer, G. (2000a). Identification of 16S ribosomal DNA-defined bacterial populations at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 66, 3102–3109.
Sievert, S. M., Ziebis, W., Kuever, J., and Sahm, K. (2000b). Relative abundance of Archaea and Bacteria along a thermal gradient of a shallow-water hydrothermal vent quantified by rRNA slot-blot hybridization. Microbiology 146, 1287–1293.
Stokke, R., Dahle, H., Roalkvam, I., Wissuwa, J., Daae, F. L., Tooming-Klunderud, A., et al. (2015). Functional interactions among filamentous Epsilonproteobacteria and Bacteroidetes in a deep-sea hydrothermal vent biofilm. Environ. Microbiol. 17, 4063–4077. doi: 10.1111/1462-2920.12970
Tarasov, V. G., Gebruk, A. V., Mironov, A. N., and Moskalev, L. I. (2005). Deep-sea and shallow-water hydrothermal vent communities: two different phenomena? Chem. Geol. 224, 5–39. doi: 10.1016/j.chemgeo.2005.07.021
Teske, A., de Beer, D., McKay, L. J., Tivey, M. K., Biddle, J. F., Hoer, D., et al. (2016). The Guaymas basin hiking guide to hydrothermal mounds, chimneys, and microbial mats: complex seafloor expressions of subsurface hydrothermal circulation. Front. Microbiol. 7:75. doi: 10.3389/fmicb.2016.00075
Vetriani, C., Voordeckers, J. W., Crespo-Medina, M., O’Brien, C. E., Giovannelli, D., and Lutz, R. A. (2014). Deep-sea hydrothermal vent Epsilonproteobacteria encode a conserved and widespread nitrate reduction pathway (Nap). ISME J. 8, 1510–1521. doi: 10.1038/ismej.2013.246
Wang, L., Cheung, M. K., Liu, R., Wong, C. K., Kwan, H. S., and Hwang, J.-S. (2017). Diversity of total bacterial communities and chemoautotrophic populations in sulfur-rich sediments of shallow-water hydrothermal vents off Kueishan Island, Taiwan. Microb. Ecol. 73, 571–582. doi: 10.1007/s00248-016-0898-2
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Willems, A., Busse, J., Goor, M., Pot, B., Falsen, E., Jantzen, E., et al. (1989). Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophaga flava comb. nov. (Formerly Pseudomonas flava), Hydrogenophaga palleronii (Formerly Pseudomonas palleronii), Hydrogenophaga pseudoflava (Formerly Pseudomonas pseudoflava and “Pseudomonas carboxydoflava”), and Hydrogenophaga taeniospiralis (Formerly Pseudomonas taeniospiralis). Int. J. Syst. Evol. Microbiol. 39, 319–333. doi: 10.1099/00207713-39-3-319
Williams, T. M., Unz, R. F., and Doman, J. T. (1987). Ultrastructure of Thiothrix spp. and “Type 021N” Bacteria. Appl. Environ. Microbiol. 53, 1560–1570.
Yamamoto, M., Nakagawa, S., Shimamura, S., Takai, K., and Horikoshi, K. (2010). Molecular characterization of inorganic sulfur-compound metabolism in the deep-sea epsilonproteobacterium Sulfurovum sp. NBC37-1. Environ. Microbiol. 12, 1144–1153. doi: 10.1111/j.1462-2920.2010.02155
Yoshida-Takashima, Y., Nunoura, T., Kazama, H., Noguchi, T., Inoue, K., Akashi, H., et al. (2012). Spatial distribution of viruses associated with planktonic and attached microbial communities in hydrothermal environments. Appl. Environ. Microbiol. 78, 1311–1320. doi: 10.1128/AEM.06491-11
Yücel, M. (2013). Down the thermodynamic ladder: a comparative study of marine redox gradients across diverse sedimentary environments. Estuar. Coast. Shelf Sci. 131, 83–92. doi: 10.1016/j.ecss.2013.07.013
Keywords: shallow-water gas vent, Tyrrhenian Sea, Tor Caldara, microbial biofilms, active microbial communities, Epsilonproteobacteria, geothermal
Citation: Patwardhan S, Foustoukos DI, Giovannelli D, Yücel M and Vetriani C (2018) Ecological Succession of Sulfur-Oxidizing Epsilon- and Gammaproteobacteria During Colonization of a Shallow-Water Gas Vent. Front. Microbiol. 9:2970. doi: 10.3389/fmicb.2018.02970
Received: 18 August 2018; Accepted: 18 November 2018;
Published: 06 December 2018.
Edited by:
Anna-Louise Reysenbach, Portland State University, United StatesReviewed by:
Julie L. Meyer, University of Florida, United StatesIda Helene Steen, University of Bergen, Norway
Copyright © 2018 Patwardhan, Foustoukos, Giovannelli, Yücel and Vetriani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Costantino Vetriani, dmV0cmlhbmlAbWFyaW5lLnJ1dGdlcnMuZWR1
 Sushmita Patwardhan
Sushmita Patwardhan Dionysis I. Foustoukos
Dionysis I. Foustoukos Donato Giovannelli
Donato Giovannelli Mustafa Yücel
Mustafa Yücel Costantino Vetriani
Costantino Vetriani