- 1Department of Biological Sciences, The University of Southern Mississippi, Hattiesburg, MS, United States
- 2U.S. Army Engineer Research and Development Center, Environmental Laboratory, Vicksburg, MS, United States
- 3Gulf Coast Research Laboratory, The University of Southern Mississippi, Hattiesburg, MS, United States
The U. S. Gulf of Mexico is experiencing a dramatic increase in tidal marsh restoration actions, which involves planting coastal areas with smooth cordgrass (Spartina alterniflora) and black needlerush (Juncus roemerianus) for erosion control and to provide habitat for fish and wildlife. It can take decades for sedimentary cycles in restored marshes to approach reference conditions, and the contribution of the sediment microbial communities to these processes is poorly elucidated. In this study, we addressed this gap by comparing rhizosphere microbiomes of S. alterniflora and J. roemerianus from two restored marshes and a natural reference marsh located at Deer Island, MS. Our results revealed that plants from the restored and reference areas supported similar microbial diversity indicating the rapid colonization of planted grasses with indigenous soil microbiota. Although close in composition, the microbial communities from the three studied sites differed significantly in the relative abundance of specific taxa. The observed differences are likely driven by the host plant identity and properties of sediment material used for the creation of restored marshes. Some of the differentially distributed groups of bacteria include taxa involved in the cycling of carbon, nitrogen, and sulfur, and may influence the succession of vegetation at the restored sites to climax condition. We also demonstrated that plants from the restored and reference sites vary in the frequency of culturable rhizobacteria that exhibit traits commonly associated with the promotion of plant growth and suppression of phytopathogenic fungi. Our findings will contribute to the establishment of benchmarks for the assessment of the outcome of coastal restoration projects in the Gulf of Mexico and better define factors that affect the long-term resiliency of tidal marshes and their vulnerability to climate change.
Introduction
Coastal marshes are highly productive ecosystems, exceeding primary production estimates of species rich ecosystems like tropical rainforests or coral reefs (Bertness, 2006). These wetlands provide key ecosystem benefits such as reducing fertilizer and pollutant run-off into the ocean, protection from storm surge, and habitat and shelter for many 100s of species of fish, birds, and other marine animals (Costanza et al., 1997). Despite the great faunal diversity, coastal marshes are dominated by a limited number of key species of perennial grasses, rushes, and sedges that have adapted to tolerate stresses associated with salinity changes, flooding, and temperature fluctuations. The most common of these plants in the northern Gulf of Mexico are black needlerush (Juncus roemerianus) and smooth cordgrass (Spartina alterniflora), which both form rhizomes and can appear in genetically identical tall and short forms (Eleuterius and McDaniel, 1978). The presence of these two marsh plant species represents a key component in the successful establishment and succession of coastal salt marshes. In the northern Gulf, marsh plant communities exhibit clear patterns of zonation, with S. alterniflora dominant in the low intertidal, and J. roemerianus dominant in the mid-marsh elevations, as a result of a host of biogeochemical factors (Craft et al., 2003; Herbert et al., 2015).
In Mississippi, coastal marshes have come under increasing pressure from both natural and anthropogenic sources. Increases in hurricane and storm activity combined with industrial and suburban development have resulted in significant habitat loss (Eleuterius, 1973). Such environmental issues can be best illustrated at Deer Island (GPS 30 21.99′–88 49.56′), which is located in the Mississippi Sound near the mouth of Biloxi Bay and the City of Biloxi, Mississippi. The 4.5-mile-long rod-shaped Deer Island originates from the late Pleistocene beach ridges and is not a true barrier island, but is a remnant of the mainland (Schmid and Otvos, 2003). Historical records indicate that Deer Island has suffered continuous erosion, resulting in the loss of approximately 300 acres over the last 150 years, or about one third of its 1850 footprint (Gerhardt Smith et al., 2015). While the island provides a diverse habitat for flora and fauna, certain portions of Deer Island’s shoreline are eroding rapidly, leading to a loss of sediment stabilizing vegetation. Increased exposure to wave energy, coupled with gradual sea level rise, has hastened erosion of wetlands along both Deer Island as well as the mainland of Mississippi. As a result, the island has been a pilot study site for wetland restoration by reestablishing S. alterniflora and J. roemerianus using beneficial use (BU) dredged material with the long-term goals of providing habitat for native species of the Gulf Coast and to aid the city of Biloxi, MS with protection during storm events (Gerhardt Smith et al., 2015).
The Mississippi Department of Marine Resources (MDMR) and the U.S. Army Corps of Engineers (USACE) began a multiyear restoration project of the eroded northeastern end of Deer Island in 2003 – Deer Island Multiyear Restoration (DIMR1) (Figure 1). Dredged material from the maintenance of the Biloxi Ship Channel was used to create approximately 30 acres of tidal marsh on the north shore of the east end of the island. After dewatering and stabilization of this fill, planting of S. alterniflora, J. roemerianus, and Spartina patens (salt marsh hay) along the sand-berm and selected areas of dredge fill was completed in spring 2005. Additional replanting of selected areas has been ongoing since then to stabilize the eroding sand-berm and new fill that has been added to the site since 2009 (Lang, 2012). A second round of berm construction and fill placement at DIMR1 was completed in 2013. In addition, a new phase of construction began in 2014 to approximately double the area of created marsh. This new area, DIMR2, was partially planted in spring of 2016 with approximately 36,000 plants of S. alterniflora, J. roemerianus, S. patens, Uniola paniculata (sea oats), and Panicum amarum (beach panic grass).
A post-construction assessment at Deer Island of the older constructed wetland (DIMR1) and an adjacent reference site showed the site dominated by S. alterniflora, S. patens, Distichlis spicata (salt grass), and J. roemerianus with no significant differences in abundance or diversity values across the sites (Lang, 2012). However, further monitoring of the newly constructed marsh site (DIMR2) at Deer Island, including more detailed microbial analysis of the sediments, is needed for a clearer understanding of the role of sedimentary ecosystem processes facilitated by the microbial community and the long-term stability of the site. Toward this objective we compared the rhizosphere microbial communities of the two dominant late successional plant species, S. alterniflora and J. roemerianus, from restoration sites DIMR1 and DIMR2 and a natural marsh reference site via high-throughput sequencing of 16S rRNA amplicons. We also isolated and characterized rhizobacteria that exhibit traits associated with the capacity to control phytopathogens and promote plant growth. In particular, we identified several species capable of antagonizing Fusarium spp., a group of plant pathogens which infect stressed S. alterniflora plants and play a significant secondary role in the ecological disruption of salt marshes during outbreaks of sudden vegetation dieback (SVD) (Elmer and Marra, 2011; Elmer, 2014). Collectively these findings reveal how microbial communities compare between natural marsh and restored sites as a function of construction practices and restoration age and help define potential factors that may affect the long-term restoration success and resiliency of coastal marshes.
Materials and Methods
Sample Collection and Processing
Three study sites, DIMR1, DIMR2, and natural marsh, were sampled over the course of this study (Figure 1). DIMR1 is a restoration site on the northeast of Deer Island, periodically planted between 2004 and 2015. DIMR2 was constructed to the west of DIMR1, with partial planting completed in 2015–2016. The natural marsh is a virgin, Juncus-dominated reference site located in the southeast portion of the island. The natural vegetation of the island exists in distinct elevation zones; with typical marsh zones dominated either by S. alternifora (low marsh), J. roemerianus (mid marsh), or a mixture of upland salt marsh vegetation including S. patens, Baccharis halimifolia (sea myrtle), and D. spicata (high marsh). At each site, the vegetation was surveyed by establishing a 100-m long transect perpendicular to the shoreline. The starting point of transect was within <3 m of a georeferenced permanent marker. Transects were surveyed on November 10, 2016, which represents the end of the growing season in the northern Gulf of Mexico and coincides with maximal live above-ground biomass resulting in decomposition and remineralization of the growing season organic biomass. Transect orientation was set to capture changes in plant zonation as a function of elevation change. Within each site and near the transect, several clumps each of S. alterniflora and J. roemerianus were selected randomly every few meters, dug with a shovel to below the sediment surface (about 30 cm) and placed in large plastic bags. Global positioning coordinates (GPSs) of collected plants from all sites were recorded (Table 1). Sediment samples (four per site) were collected from the same general locations using a 2′′ diameter soil core sampler (model no. 404.50, AMS, American Falls, ID, United States) for sediment geochemistry analysis on August 8, 2017. All collected plants and sediment cores were transported on ice to the laboratory at the Hattiesburg USM campus. In the laboratory, the plant material was kept at 4°C and processed within 24 h, while sediment samples were frozen at -20°C. Analysis of sediment samples was conducted by the Agricultural and Environmental Services Laboratories (AESL) at the University of Georgia (Athens, GA, United States) for total carbon and nitrogen, total organic carbon (TOC), ammonium, nitrate, nitrite, sulfate, and soil texture.
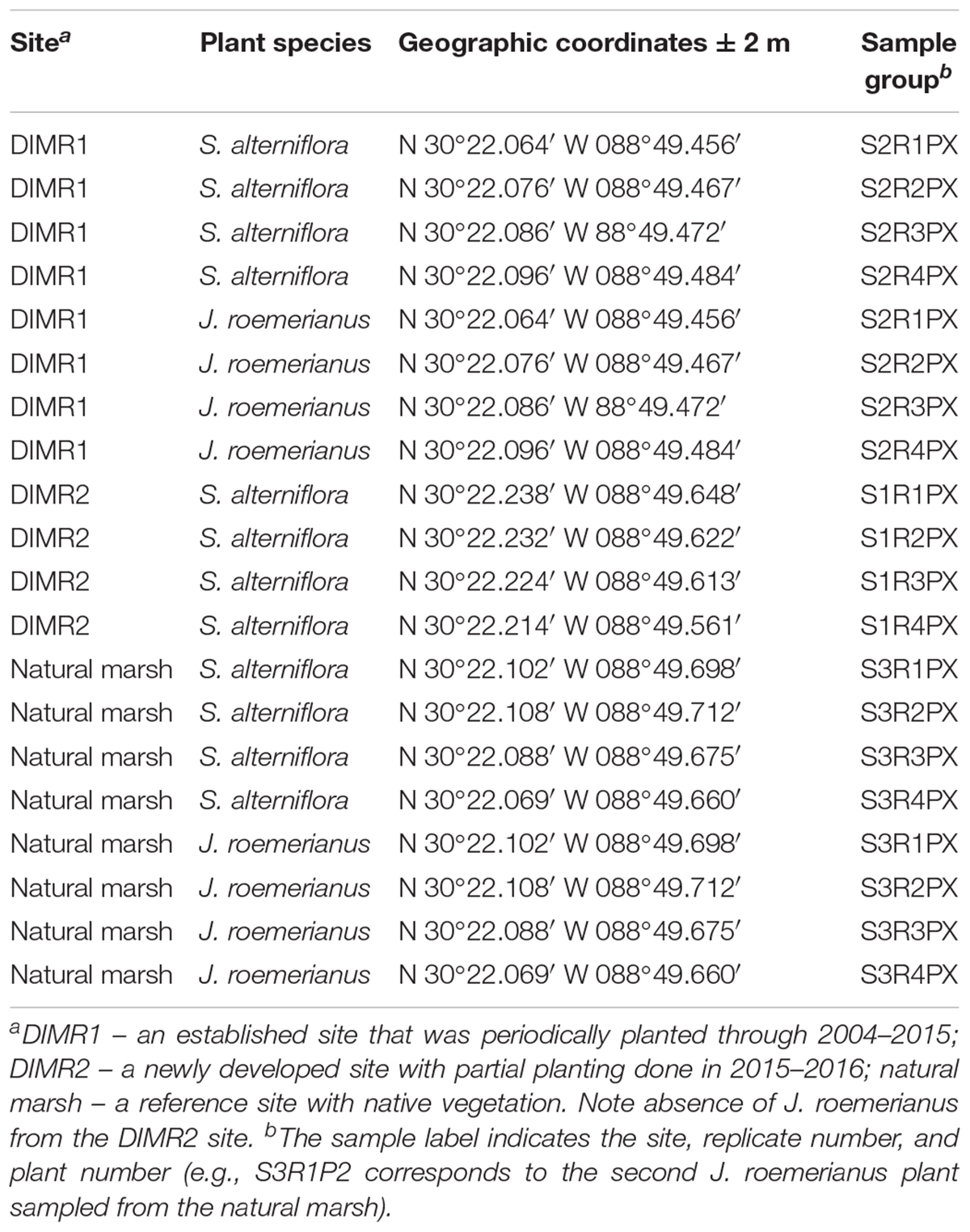
Table 1. Description of samples of S. alterniflora and J. roemerianus collected from the two DIMR restoration sites and natural marsh on Deer Island.
Isolation of Soil DNA and High-Throughput Sequencing of 16S rRNA Amplicons
To isolate soil DNA partial root systems were excised from larger clumps, excess sediment was carefully removed, and root systems with the adhering rhizosphere soil were placed into 50 mL Falcon tubes. Twenty milliliters of sterile saline were added to each tube and root-associated bacteria were dislodged by vortexing and treatment in a sonicating bath (1 min each). Rhizosphere soil DNA was extracted from 1.5 mL aliquots of root washes using a Power Soil DNA Isolation kit (MO BIO Laboratories, Carlsbad, CA, United States). The quality of the extracted DNA was verified by gel electrophoresis and PCR with primers 8F and 1492R that target eubacterial 16S rRNA (Weisburg et al., 1991). The amplifications were carried out in 50-μL reaction mixtures containing 1× DreamTaq DNA polymerase buffer, 200 μM each of dATP, dTTP, dGTP, and dCTP, 20 pmol of each primer, and 0.06 U of DreamTaq DNA polymerase (Thermo Fisher Scientific, Waltham, MA, United States). The amplifications were performed with a T100 gradient thermal cycler (Bio-Rad Laboratories, Hercules, CA, United States) and the cycling program consisting of initial denaturation at 94°C for 2 min followed by 30 cycles of 94°C for 20 s, 55°C for 15 s, and 72°C for 1.5 min, and a final extension at 72°C for 3 min. 16S rRNA amplicons for high-throughput sequencing were generated using the primer pair 515F-806R with each reverse primer containing a barcode following the protocol of Costello et al. (2009). Amplicons were then purified using a Promega Wizard PCR Cleanup kit (Madison, WI, United States), quantified, and sequenced on an Illumina MiSeq sequencer (Illumina, San Diego, CA, United States) using the Illumina MiSeq Reagent Kit v2 (300 cycles) following the manufacturer’s instructions. The 16S rRNA sequencing data from MiSeq runs were trimmed, demultiplexed and quality filtered using the default settings of the Illumina MiSeq Reporter software (v 2.6).
Microbial Community Analysis
To compare bacterial microbiomes associated with roots of S. alterniflora and J. roemerianus collected at the three sites microbial community analysis was performed. From the 16S rRNA sequence data, OTU assignments were performed with the open reference OTU clustering package implemented in the QIIME (Quantitative Insights Into Microbial Ecology) bioinformatics pipeline (Caporaso et al., 2010) which performs an initial closed reference clustering against the Greengenes Database with a 97% sequence similarity threshold. Taxonomy assignments were performed using the naïve Bayesian Ribosomal Database Project (RDP) Classifier (Wang et al., 2007) as implemented in the MICCA (MICrobial Community Analysis) software pipeline (Albanese et al., 2015). Bar chart comparisons at different taxonomy levels were performed using a 1% relative abundance threshold to remove rare OTUs. Differences between sites were determined at the order level using a Kruskal–Wallis test with a significance threshold of P < 0.05. Order level was selected to provide the highest possible resolution of community composition between sites. Statistical comparisons of significant differences between individual phyla were performed using ANOVA. Analysis of similarities (ANOSIM) was performed using the compare_categories.py script in QIIME. Comparisons were made between S. alterniflora collected from DIMR1 and DIMR2 and natural marsh samples and between J. roemerianus collected from DIMR1 and natural marsh (this species was not found at DIMR2). Comparisons were also performed between S. alterniflora and J. roemerianus samples from the natural marsh site only to identify differences in community structure that were due to the type of vegetation present. Non-metric multidimensional scaling (NMDS) with Bray–Curtis distances was also performed in R Core Team (2014) using the Phyloseq package (Mcmurdie and Holmes, 2013). Samples were first filtered to remove rare OTUs (appearing less than five times in at least 50% of the samples). Following the filtering step, a variance-stabilizing transformation (VST) was applied. VST is a transformation method which removes the dependence of the variance on the mean (Anders and Huber, 2010). The resulting data set was pruned to the nine most abundant phyla for generating the NMDS plot.
Isolation of Culturable Bacteria and Estimation of Bacterial Populations
Population levels of total culturable heterotrophic bacteria in the rhizosphere of Spartina and Juncus were estimated using the terminal dilution endpoint assay (Mavrodi et al., 2012). Root washes were serially diluted in microplates and cultured in one-third-strength King’s Medium B (1/3 KMB) (King et al., 1954) and 1-10th-strength Tryptic Soy broth (1/10 TSB) (Martin, 1975) supplemented with 15% artificial saltwater SW30 (Dyall-Smith, 2009) and 100 μg mL-1 of cycloheximide. The bacterial growth was estimated after 72 h of growth at 27°C by measuring optical density at 600 nm (OD600) with a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT, United States). An OD600 of >0.1 was considered positive for bacterial growth. Population densities of bacteria were calculated based on the final dilution with positive growth. The detection limit of this technique was log 3.2 CFU g-1 root fresh weight. We also assembled a collection of rhizobacterial isolates by spread-plating the root washes on 1/3 KMB agar and 1-10th-strength Tryptic Soy agar (1/10 TSA) amended with 15% SW30 and cycloheximide to suppress fungi. The inoculated plates were incubated at 27°C for 72 h and distinct bacterial morphotypes were transferred onto fresh 1/3 KMB or 1/10 TSA and serially passaged to obtain pure cultures. All isolates were stored at -80°C in 1/3 KMB or 1/10 TSB supplemented with 30% glycerol for later screening. Bacterial population densities were converted to log CFU g-1 of fresh root weight. Differences among treatments in S. alterniflora were determined by the standard one-way analysis of variance (ANOVA) with mean comparisons among treatments performed by the Fisher’s protected least significant difference (LSD) test (P = 0.05) or by the Kruskal–Wallis test (P = 0.05). Bacterial populations in J. roemerianus were compared by the two-sample t-test (P = 0.05) or Wilcoxon Rank Sum test (P = 0.05). All statistical analyses were conducted with Statistix 10 (Analytical Software, Saint Paul, MN, United States).
Screening of Rhizosphere Isolates for Plant Beneficial Traits
A subset of the rhizosphere isolate collection was screened for traits commonly associated with the capacity to proliferate in the rhizosphere (siderophores, motility), antagonize plant pathogens (production of exoprotease, biosurfactants, and HCN), and promote plant growth [production of ACC deaminase and indoleacetic acid (IAA)]. A total of 287 isolates (n = 55–61 per sampled plant species per site) from roots of Spartina and Juncus were randomly selected and cultured for 2 days at 27°C in 96-well microplates filled with 1/5 TSB supplemented with SW30. These microplates served as a source of inoculum for individual assays and allowed for the rapid transfer of bacteria with the help of a 96-pin replicator. The secretion of protease was assessed by observing a clearing zone surrounding bacterial growth on skim milk agar after 3 days of incubation at 27°C (Sacherer et al., 1994). The siderophore production was determined by pre-growing bacteria on 1/5 TSA with SW30 for 2 days at 27°C and then overlaying plates with 30 mL of soft O-CAS agar (Perez-Miranda et al., 2007). The overlayed plates were incubated at 27°C and the appearance of yellow/orange halo around bacterial colonies was scored at 4, 8, and 24 h. The production of hydrogen cyanide was monitored by spotting bacteria on TSA supplemented with 4.4 g L-1 glycine (Bakker and Schippers, 1987). The inoculated Petri dishes were sealed with Parafilm, and the production of HCN was detected by placing on the lid a piece of indicator filter paper impregnated with 0.5% picric acid and 2.0% sodium carbonate. The change in the color of the indicator paper from yellow to bright orange after 2–5 days of growth at 27°C indicated the production of HCN. The production of biosurfactants was assayed by the droplet collapse assay via spotting 10 μL of a culture grown in 1/5 TSB with SW30 onto a hydrophobic surface (Parafilm). The presence of biosurfactants in the culture medium causes the collapse of the droplet due to the reduction in the surface tension. The motility of bacteria was assessed by spotting bacteria on the semi-soft 1/5 TSA and scoring the zone of spread after the first and second day of incubation at 27°C. The growth on ACC as a sole nitrogen source was determined by a modified protocol of Penrose and Glick (2003). The bacteria were pre-grown for 2 days with shaking (190 rpm) at 27°C in deep-well microtiter plates filled with 1/5 TSB with SW30. The cells were washed twice and replica-plated onto DF agar without N source (control), or the DF agar supplemented with (NH4)2SO4 or 3.0 mM ACC. Plates were scored for growth after 2–4 days of incubation at 27°C. All isolates that were positive for growth on ACC were confirmed for the capacity to use this compound as a N-source in liquid DF medium. The production of IAA was determined by growing isolates at 27°C and 190 rpm in deep-well microplates filled with 1/5 TSB + SW30 with and without tryptophan (500 μg mL-1). After 2 days of incubation, the optical density at 600 nm of bacteria grown in two media was measured, and cells were removed by centrifugation at 5000 rpm for 10 min at 10°C. The presence of IAA was detected by mixing 40 μL of supernatant with 160 μL of Salkowski’s reagent (0.5 M FeCl3 and 35% perchloric acid) and measuring the absorbance at 540 nm after the 25-min incubation at room temperature (Gordon and Weber, 1951). All assays were repeated at least twice with two replicates per experimental condition. Frequencies of rhizobacteria with beneficial traits at DIMR1, DIMR2, and the natural marsh were compared using chi-square test (P = 0.05) with Statistix 10.
Antagonism of Rhizosphere Isolates Toward Plant-Pathogenic Fusarium
The capacity of rhizosphere isolates to antagonize the growth of Fusarium pseudograminearum and F. culmorum was assayed on 1/5 TSA. A 5-mm-diameter agar plug of a 5-day-old culture of Fusarium grown on potato dextrose agar was transferred to the center of a 1/5 TSA plate. Next, 10 μL of an exponentially growing bacterial culture adjusted to an OD600 of 0.1 was spotted 1 cm from the edge the agar plate. The inoculated plates were incubated in the dark at 27°C for 5 days, after which the distance from the center of the plug to the leading edge of the fungus (x) and the distance from the edge of the bacterial growth to the growing edge of the fungus (y) were measured and the inhibition index (i) was calculated as i = y/(x + y) × 100%. The model biocontrol strain Pseudomonas protegens Pf-5 was used as a positive control while negative control plates contained only fungus. All assays were repeated twice with 3–6 replicates per isolate. The most active antagonists were identified by amplifying 16S rRNA with primers 8F and 1492R, end-sequencing the resultant amplicons, and analyzing sequences with the Sequence Match tool of the Ribosomal Database Project (RDP-II)1.
Screening of Soil DNA and Bacterial Isolates for the Presence of Genes Involved in the Production of Phenazines and 2,4-Diacetlphloroglucinol
Samples of DNA extracted from plant rhizosphere or selected bacterial isolates were screened by PCR for the presence of genes involved in the synthesis of phenazines and 2,4-diacetlphloroglucinol (2,4-DAPG), since both of these antimicrobial compounds were implicated in the biological control of phytopathogenic fungi (including Fusarium spp.) by certain groups of beneficial rhizobacteria (Weller et al., 2007; Mazurier et al., 2009). The amplifications were performed in 25-μL reaction mixtures containing PCR reagents, 1 μL of template (10-fold diluted soil DNA) and primers targeting the phenazine biosynthesis gene phzF (primers Ps_up1 and Ps_low1) (Mavrodi et al., 2010), or the 2,4-DAPG biosynthesis gene phlD (primers BPR4 and B2BF) (Gardener et al., 2001). The cycling programs for the amplification of phzF and phlD loci were as described by Gardener et al. (2001) and Mavrodi et al. (2010), respectively. Pseudomonas brassicacearum Q8r1-96 was used as a positive control for the amplification of 2,4-DAPG genes, while P. synxantha 2-79 served as a positive control for the presence of phenazine biosynthesis genes.
Results
Site Descriptions at the Sampling Time
At the time of sampling in late 2016, field observations showed that all three transects were primarily in S. alterniflora dominated intertidal marsh zone. Vegetation percent cover was high, at least 50%, and increased with age of the site. Both S. alterniflora and J. roemerianus were present at the natural marsh site (control) occupying the mid marsh elevation, but S. alterniflora was dominant in contrast to J. roemerianus which grew as isolated plants or dense clumps. Sediment was muddy but well bound by extensive root mat, and it was mostly easy to traverse except in unvegetated areas. Vegetation cover was 70–90%, canopy height was typically 60 cm or less and 1.2–1.5 m for S. alterniflora and J. roemerianus, respectively. During the sampling at the DIMR1 site, we observed some S. alterniflora toward an interior lagoon in the low- to mid-marsh zone, but no J. roemerianus, which has remained mostly absent for more than 10 years since the original plantings in April 2005. Much of the central part of the transect was in high marsh habitat with sandy sediments that were dominated by S. patens. Vegetation percent cover varied from >90 to <10% in sandy areas with vegetation height of less than 60 cm. Other plants observed included Ipomoea pes-caprae (bayhops), Hydrocotyle bonariensis (largeleaf pennywort), and D. spicata. The DIMR2 site was recently planted in the spring of 2016 as part of the ongoing restoration and during sampling had only S. alterniflora present in both the low- and mid-marsh elevations. The canopies of the plants were tall for this species ranging between 1.2 and 1.5 m in height, and the vegetation percent cover ranged from 50 to 80% over the site. Sediments were muddy clay, with surface biofilm indicating regular submergence. Most areas were compact enough to walk without difficulty. The analysis of sediments revealed substantial differences between the three sampled areas. Sediment cores collected at the natural marsh had the highest overall levels (although not significantly different from DIMR2) of carbon, nitrogen, TOC, and sulfate (Supplementary Table 1). In contrast, samples from DIMR1 were low on C, N, TOC, ammonium, and sulfate. The DIMR1 sediments also contained a significantly higher amount of sand and less silt and clay compared to the natural marsh and DIMR2.
Microbial Community Analysis
We compared bacterial microbiomes associated with roots of S. alterniflora and J. roemerianus collected at the three sites. The removal of low-quality reads yielded 5 million sequences spanning the V4 region of the 16S rRNA gene (n = 4 per sampled plant species per site) with a median read count per sample of 83,000. The high-quality reads were binned at >97% sequence identity into OTUs which were utilized for subsequent analyses (Supplementary Figure 1). The rarefaction analysis [phylogenetic diversity (PD) whole tree] at a depth of 10,000 reads per sample indicated sufficient sampling depth to represent the OTU richness of the studied microbial communities (data not shown). The alpha diversity calculated using Faith’s PD metric showed that differences in alpha diversity between the two restored sites and the natural marsh were not significant (non-parametric t-test; P > 0.5) (Figure 2). The core rhizosphere microbiomes of S. alterniflora and J. roemerianus included members of the phyla Proteobacteria, Bacteroidetes, and Acidobacteria as the most numerous taxa represented by OTU numbers. At the class level, samples were dominated in descending order by Gamma-, Delta-, Alpha-proteobacteria, Bacteroidia, and Ignavibacteria and had a high abundance of rare or poorly defined taxa (Figure 3).
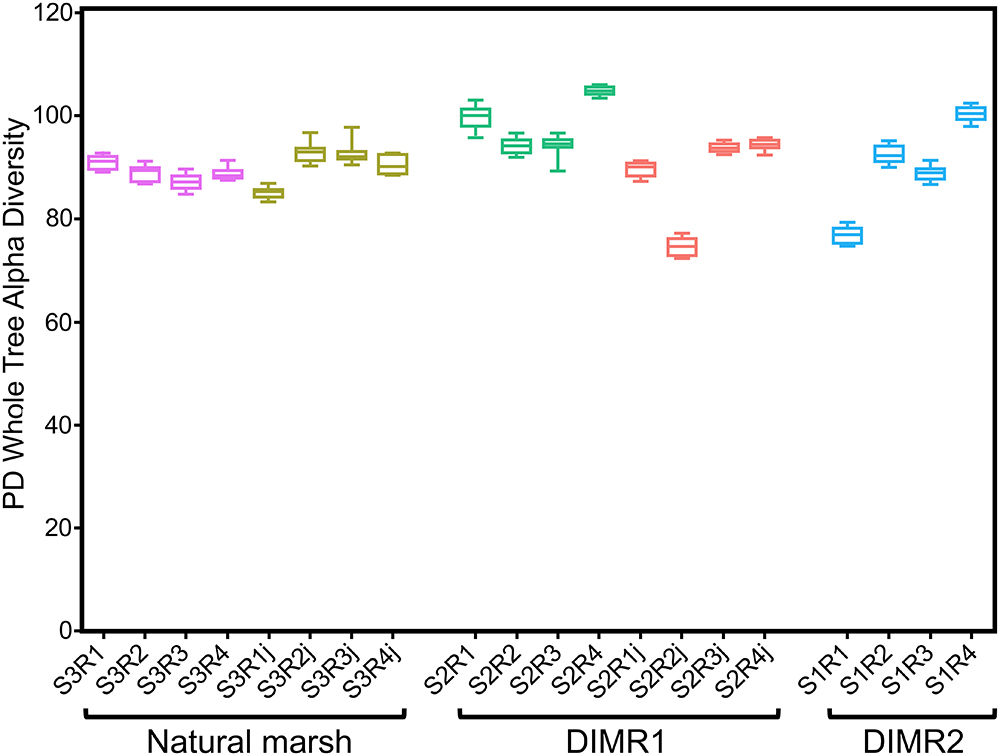
Figure 2. Alpha diversity [Faith’s phylogenetic diversity (PD) whole tree metrics] by plant hosts Spartina alterniflora and Juncus roemerianus (series labels followed by “j”) at the natural marsh (S3R series) and restored DIMR1 (S2R series) and DIMR2 (S1R series) sites. Samples were rarified to 10,000 reads per sample and compared using a non-parametric t-test. No significant differences were found between any of the samples (P < 0.5).
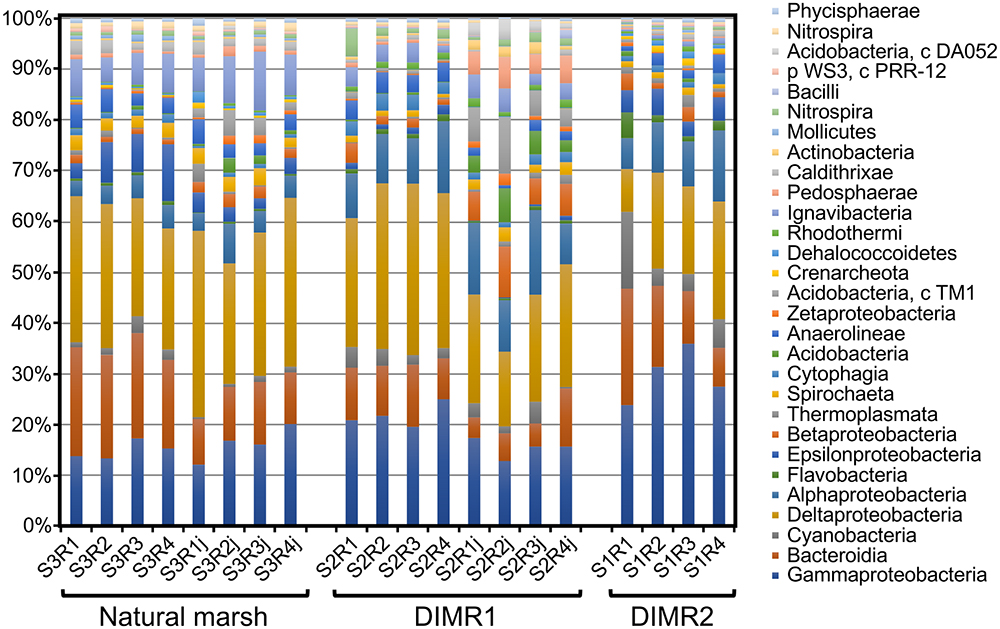
Figure 3. Relative abundance at the class level of the dominant bacterial taxa in microbial communities associated with roots of S. alterniflora and J. roemerianus (series labels followed by “j”) collected at the natural tidal marsh (S3R series) and restored DIMR1 (S2R series) and DIMR2 (S1R series) sites on Deer Island, MS.
Non-metric multidimensional scaling plots based on the Bray–Curtis dissimilarities matrix showed a separation between samples from the natural reference marsh, DIMR1, and DIMR2 (Figure 4). Similar pattern was observed when samples were subjected to the principal coordinate analysis (PCoA) using weighted UniFrac distances (Supplementary Figure 2). These differences in overall microbial community structure between the three sites were significant at a 95% confidence interval. The relative abundance of microbial taxa was then compared at the order level to infer key microbial phylotypes associated with the rhizosphere of plants from the restored and natural marshes. Multivariate analysis identified several taxa that appeared to drive the separation between S. alterniflora and J. roemerianus and between the natural marsh and restored sites DIMR1 and DIMR2 (Figure 4). Non-parametric multivariate analysis of variance indicated these differences were significant (F = 12.34, P < 0.001). In particular, Acidobacteria and Verrucomicrobia appeared to be more closely linked with S. alterniflora from DIMR1 while Chlorobi seemed to be a driver in the separation of J. roemerianus at DIMR1. Furthermore, Cyanobacteria were linked with Spartina at DIMR2, whereas Bacteroidetes, Nitrospirae, and Chloroflexi were associated with Juncus from the natural marsh (Figure 4).
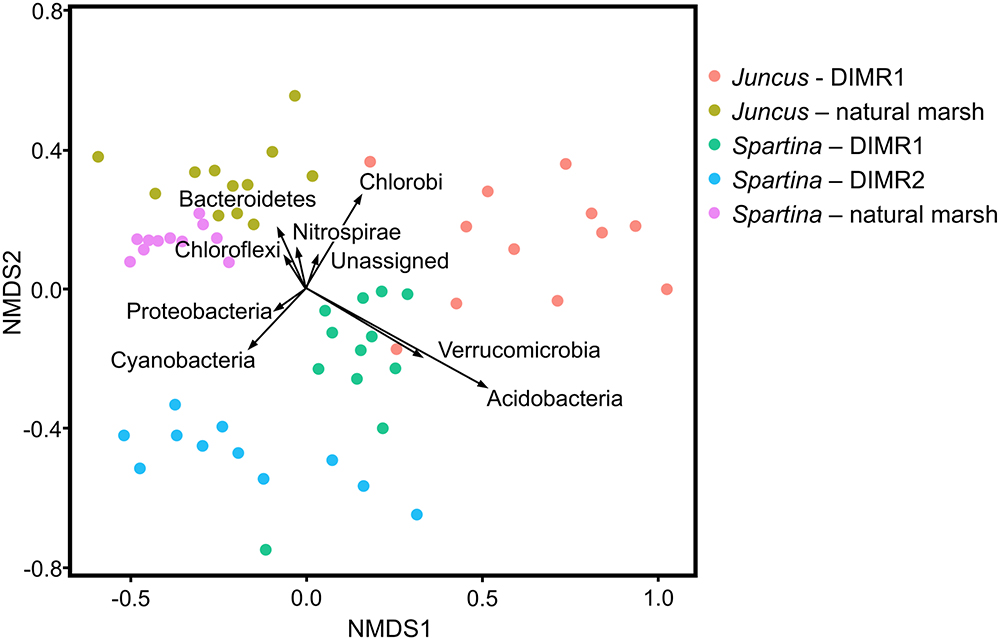
Figure 4. Two-dimensional non-metric multidimensional scaling (NMDS) plot with vectors displaying most abundant bacteria phyla identified in samples from S. alterniflora and J. roemerianus. The length of the vector is correlated to relative abundance of each of the dominant phyla. Samples are color-coded based on the plant species and sampling site. Controls are natural marsh samples.
Differences in the relative abundance of dominant phyla between the sampled communities were also determined at the phylum level using a Kruskal–Wallis test (P < 0.001). Overall, Proteobacteria represented a dominant group in all analyzed Spartina samples although their levels were significantly higher in both DIMR sites (60% relative abundance) than in the natural marsh (50% relative abundance) (Supplementary Figure 3). In contrast, Bacteriodetes were significantly higher (P < 0.001) in the reference marsh with a relative abundance of 20% compared to around only 14% in the two restored sites. All other intersite differences were in agreement with results of the NMDS analysis described above (Supplementary Figure 4). We also used samples collected at the natural marsh and DIMR1 for intrasite comparisons of microbial communities of Juncus and Spartina. In the reference site, the two plant species had overall similar microbiomes that differed only in the abundance of Bacteriodetes (P < 0.001) (Supplementary Figure 5). These results were further supported by NMDS analysis which showed close clustering of samples from the natural marsh regardless of plant species (Figure 4). The examination of communities from DIMR1 revealed significantly higher levels (P < 0.001) of Proteobacteria and Bacteriodetes in the rhizosphere of S. alterniflora, while Acidobacteria and Verrucomicrobia were more abundant (P < 0.001) on roots of J. roemerianus (Supplementary Figure 6). The observed differences were most significant with Acidobacteria that were >5-fold higher abundance in Juncus as compared to Spartina.
Analysis of Total Culturable Heterotrophic Rhizobacteria in the Rhizosphere of S. alterniflora and J. roemerianus
The population levels of total culturable heterotrophic bacteria in the rhizosphere of Spartina and Juncus ranged between log 6.4 and 7.7 CFU g-1 root fresh weight, with slightly higher overall populations observed in S. alterniflora (Table 2). On 1/3 KMB, the populations of rhizobacteria were significantly higher on S. alterniflora at the DIMR1 site in comparison with DIMR2 and the natural marsh. On 1/10 TSB, the levels of bacteria recovered from the DIMR1 and DIMR2 were similar but significantly higher than those in the natural marsh. In J. roemerianus, the populations of culturable rhizobacteria were highest on 1/3 KMB in the natural marsh samples, whereas no difference between sites was observed when bacteria were plated on 1/10 TSB. The actual number of isolates per site ranged from 108 to 190 with a broader range of distinct morphotypes (i.e., colony diameter, shape, elevation, margin, pigmentation, opacity, and texture) observed on 1/10 TSB in comparison with 1/3 KMB (Supplementary Figure 7). A total of 751 bacterial isolates were recovered from the rhizosphere of S. alterniflora, and J. roemerianus collected at the three surveyed Deer Island sites (Table 2). At each site, we observed 20 to 30 different morphotypes, and it is likely that these morphotypes were represented in the final collection by multiple isolates.
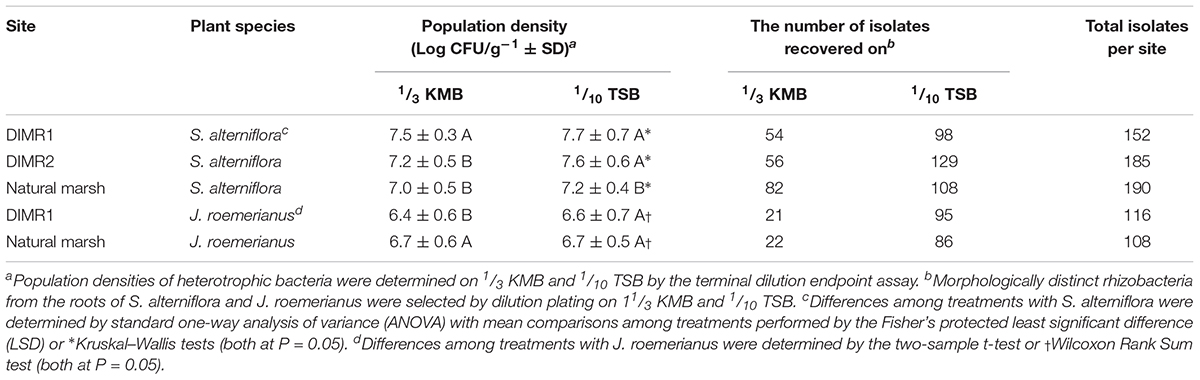
Table 2. Population densities of culturable aerobic heterotrophic bacteria and the number of isolated bacteria from rhizospheres of S. alterniflora and J. roemerianus collected on Deer Island.
Plant Growth Promotion and Biocontrol Traits of Bacterial Isolates
Results of the screening indicate that a total of 72 (25.1%) strains were motile with approximately equal distribution between the plant hosts and sites (Table 3). The secretion of siderophores was detected in 100 (34.8%) strains based on their ability to chelate iron from the dye complex and change the color of the indicator O-CAS medium from blue to yellow. The production of siderophores was evaluated using a modified overlay technique that minimized the inhibition of Gram-positive bacteria by hexadecyltrimethylammonium bromide (HDTMA), which is used as an indicator in CAS medium (Perez-Miranda et al., 2007). The production of 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase and IAA by rhizobacteria are important determinants in beneficial plant-bacterial interactions because the release of these compounds results in the formation of longer and more robust root systems (Penrose and Glick, 2003). Our screening identified 10 strains (7.9%) that grew in the defined DF medium supplemented with ACC as a sole nitrogen source, and 21 strains (7.3%) that produced IAA in low (3.9–16.2 fg cell-1), medium (32.3–54.3 fg cell-1), or high (176.2–215.5 fg cell-1) quantities (data not shown). The production of antimicrobial biosurfactants and hydrogen cyanide was detected in 41 (14.3%), and 6 strains, respectively, while the secretion of the protease was noted in a total of 68 (44.7%) strains. Not all isolates grew in culture media used for the detection of ACC-deaminase (DF medium), exoprotease (milk agar), or hydrogen cyanide (TSA medium supplemented with glycine) (Supplementary Figure 8). Hence, the percent of positive strains in each category was calculated based on the number of isolates capable of growing in the corresponding semi-selective or indicator medium. Overall, the frequencies of bacterial isolates with beneficial traits did not differ significantly between the natural marsh and DIMR1 (χ2 = 6.9, P = 0.3305, d.f. = 6 for Juncus, and χ2 = 8.05, P = 0.0897, d.f. = 4 for Spartina). In contrast, we observed a significant difference (χ2 = 19.41, P = 0.0016, d.f. = 5 for Spartina) in the frequencies of beneficial isolates between the natural marsh and recently established DIMR2 site.
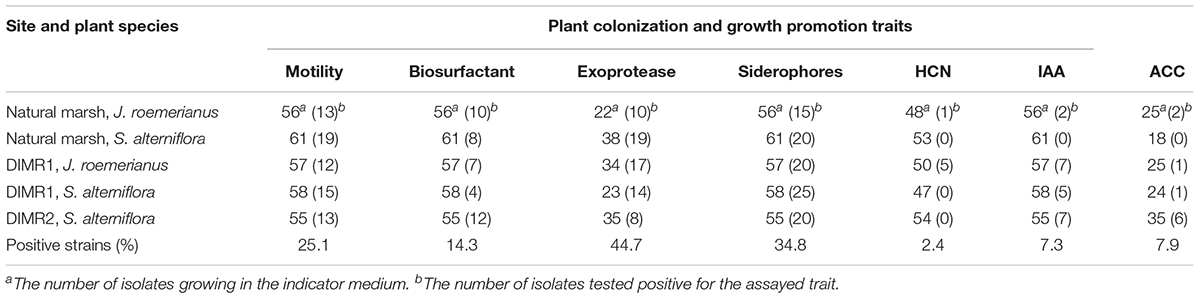
Table 3. Screening of bacterial isolates from J. roemerianus and S. alterniflora for traits associated with the competitive rhizosphere colonization and plant growth promotion.
In vitro Inhibition of Fusarium by Rhizobacterial Strains
Previous studies reported that S. alterniflora harbors different species of Fusarium, some of which were common in salt marshes affected by SVD (Elmer and Marra, 2011; Elmer, 2014). Among these, F. palustre was strongly associated with S. alterniflora in the parasite/host relationship manner and was capable of inciting disease in pathogenicity trials. We could not obtain an isolate of F. palustre and instead used the plant pathogens F. pseudograminearum and F. culmorum. Although not identical, the three species are closely related and belong to the same Fusarium species complex (i.e., F. sambucinum) (Aoki et al., 2014). Screening for the antifungal activity yielded six strains that actively inhibited the mycelial growth of F. pseudograminearum and F. culmorum. Only one of these strains was isolated from J. roemerianus (collected in the natural marsh), while the rest originated from the roots of S. alterniflora collected at both DIMR2 and the natural marsh (Table 4). All six strains inhibited F. pseudograminearum, but only four out of six antagonized F. culmorum. Partial sequencing of 16S rDNA identified strains that were most inhibitory toward Fusarium (isolates 461, 449, 577) as members of Gynuella sunshinyii (Table 4 and Supplementary Figure 9). Two strains with slightly lower antifungal activity (isolates 9 and 619) were identified as Bacillus pumilus that originated from J. roemerianus and S. alterniflora collected at the natural marsh site. Interestingly, the antagonism toward to Fusarium was species-specific, and both B. pumilus strains inhibited only F. pseudograminearum but not F. culmorum (Table 4 and Supplementary Figure 9). The last antagonistic strain, isolate 294, was identified as Tenacibaculum discolor and exhibited activity against both species of Fusarium. We screened genomic DNA of all antagonistic strains by PCR with primers targeting genes for the synthesis of antifungal metabolites phenazine and 2,4-DAPG. We also used the same PCR assay to screen all samples of soil DNA that were extracted from the rhizosphere of S. alterniflora and J. roemerianus. The quality of extracted soil DNA was assessed by PCR with universal eubacterial 16S primers 8F and 1492R and revealed that all samples were of high quality and free of PCR inhibitors. The positive controls also worked as expected and produced the 629- and 427-bp amplicons that matched, respectively, the amplified portions of phlD and phzF genes. In contrast, no amplification of phlD or phzF occurred in PCR reactions containing rhizosphere soil DNA or DNA extracted from the six strains that actively inhibited Fusarium (data not shown).
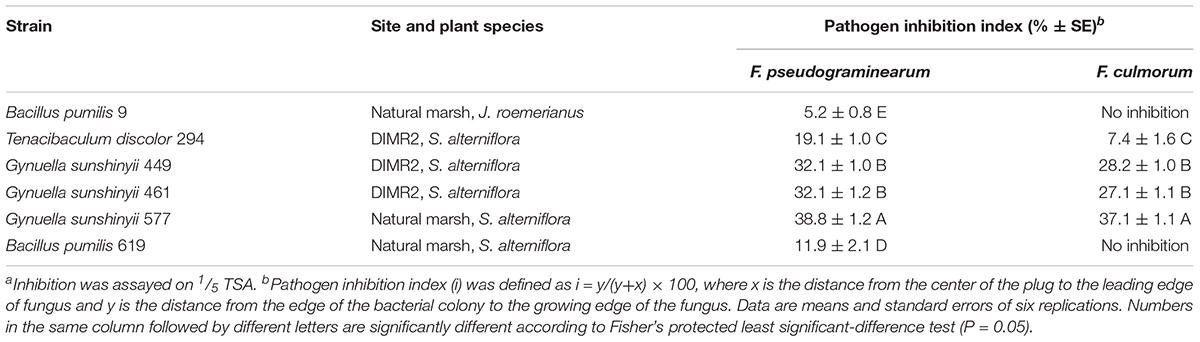
Table 4. Inhibition of Fusarium pseudograminearum and F. culmorum by strains isolated from the rhizosphere of S. alterniflora and J. roemerianusa.
Discussion
To gain a clearer understanding of the sedimentary microbial processes occurring during plant succession after initial restoration, and thereby better inform the potential for long-term stability of restored marsh habitat via beneficial use of dredged material, additional microbial analysis was conducted at Deer Island restoration sites DIMR1 and DIMR2 and compared to the results from a nearby virgin natural marsh. Previously a post-construction assessment showed the site dominated by S. alterniflora, S. patens, D. spicata, and J. roemerianus with no significant differences in plant abundance or diversity values between the reference site and the DIMR1 restored site (Lang, 2012). Similarly, in this study analysis of rhizosphere associated microbial communities for S. alterniflora and J. roemerianus from the natural and two different aged restored sites showed that the two restoration sites (DIMR1 and DIMR2) had similar soil microbial communities when compared to the natural marsh. Collectively, these findings suggest that native rhizosphere-associated microbial communities established on S. alterniflora planted at the restored areas over a short period of time (within 6 month of planting).
Natural tidal marshes are highly productive coastal ecosystems characterized by sediments rich in organic matter resulting from the decay of plant material and root exudation (Bertness, 2006). Because of the frequent waterlogging, the abundant residual organic matter is actively decomposed via anaerobic microbial respiration through sulfate reduction. The sulfate reducers produce hydrogen sulfide and other reduced sulfur compounds that serve as electron donors for sulfur-oxidizing microorganisms, which complete the sulfur cycle (Wasmund et al., 2017). All rhizosphere samples collected across the three study sites showed an abundance of sulfate-reducing and sulfur-oxidizing organisms, which supports the importance of sulfur cycling and is consistent with the anaerobic nature of saturated tidal marsh sediments. The sulfate-reducing group was dominated by Desulfobacterales, which are metabolically flexible and especially well-adapted to tidal marshes due to the ability to tolerate oxidative stress from oxygen penetration in shallow sediments (Wasmund et al., 2017). We also identified the presence of Nitrospirae, which include the thermophilic sulfate-reducing bacteria of the genus Thermodesulfovibrio. Analysis of the community data showed that members of the family Thermodesulfovibrionaceae were present but represented <1% total relative abundance and differences in abundance between restored and control sites were not statistically significant (P < 0.05). The most abundant group of sulfur oxidizers was represented by Chromatiales that use sulfides as a reducing agent, which is further oxidized to elemental sulfur. The roots of Spartina and Juncus also supported diverse Bacteroidia, which are known to carry multiple sulfatase genes and specialize in degrading sulfated organics in anoxic subsurface sediments (Baker et al., 2015).
The presence and oxidation state of sulfur can influence vegetation patterns and contribute to the natural zonation of wetland plant species (Lamers et al., 2013). In the sites studied, S. alterniflora is typically the dominant species in the low- to mid-marsh zone, where abiotic stress including inundation and anaerobic soil conditions can lead to high sulfide concentrations. Sulfide is toxic to plant metabolism, and it is thought that marsh grasses benefit from the interaction with sulfur-oxidizing microorganisms that help to lower hydrogen sulfide levels in the rhizosphere. Results of the intersite comparisons on Deer Island identified Bacteroidetes and Nitrospirae among taxa associated with J. roemerianus from the natural marsh, while Chlorobi (a group known to contain sulfur-oxidizers) were a driver in the separation of Juncus microbiome at DIMR1. Such fluctuations in the abundance of taxa involved in the transformation of different sulfur compounds may be of importance for the plant fitness and ecosystem functioning in newly created marshes. Juncus roemerianus, which typically dominates in mid-marsh elevations in the northern Gulf of Mexico, was frequently found to occur in low abundance (DIMR1) or be entirely absent (DIMR2) from this zone in the two restored sites, potentially related to site-specific sediment microbial processes including sulfate reduction. Other significant differences in the microbial communities were observed between the rhizospheres of S. alterniflora and J. roemerianus. Proteobacteria and Bacteriodetes were significantly higher (P < 0.001) in samples recovered from roots of Spartina while Acidobacteria and Verrucomicrobia were significantly higher (P < 0.001) in the rhizosphere of Juncus. The difference was greatest with Acidobacteria which were fivefold higher in Juncus dominated areas. Although the significance of these differences for the establishment of restored marshes is unclear, both Acidobacteria and Verrucomicrobia represent diverse lineages of bacteria that are particularly abundant in soil habitats and are involved in the subsurface cycling of carbon and nitrogen (Schlesner et al., 2006; Kielak et al., 2016). Previously, an analysis of vegetated sediments in Louisiana saltmarshes indicated that S. alterniflora microbial sediment communities had greater proportions of Bacteroidetes and Lentisphaerae compared to J. roemerianus, while J. roemerianus sediments contained higher proportions of α-Proteobacteria and β-Proteobacteria (Rietl et al., 2016). The predominance of Proteobacteria and Bacteriodetes in some of our samples is consistent with results of that study.
It is likely that the observed intersite variations in the composition of rhizosphere microbial communities are partially driven by differences in the properties of the dredged material that was used for the establishment of artificial marshes at the two DIMR sites. Results of our analyses revealed that sediments from DIMR2 had more silt and clay and more closely resembled samples from the natural marsh (Supplementary Table 1). In contrast, sediments from DIMR1 contained significantly less sulfate, nitrate, and organic matter, and had higher amounts of sand as a result of regular rebuilding of the berm area to replace losses from ongoing erosion. The coarse sediments at DIMR1 are probably more susceptible to oxygen exposure and tidal flushing of porewater nutrients in comparison to finer sediments at DIMR2 and the reference site. Higher levels of dissolved O2 and redox potentials were previously reported in restored marshes constructed with sandy sediments (Craft et al., 1991).
Plants have developed various biochemical and physiological mechanisms to respond and adapt to stress conditions. One of them is the fostering of root-associated microbial communities that positively influence plant fitness in response to abiotic stressors (e.g., drought, salt, temperature, and soil pollution) (Dimkpa et al., 2009). The capacity of rhizobacteria to alleviate the detrimental effect of abiotic stress is a multifactorial phenotype. Beneficial rhizobacteria improve the nutritional status of the host plant by fixing atmospheric nitrogen, solubilizing inorganic phosphate, and secreting iron-chelating siderophores (Kim et al., 2011). Many root-colonizing microorganisms stimulate plant organ development through the production of phytohormones such as auxins, cytokinins or gibberellins, which stimulate the formation of root hairs, root growth, and branching, and can improve mineral and nutrient uptake (Kim et al., 2011). Certain beneficial rhizobacteria are capable of modulating levels of the plant hormone ethylene, which leads to the formation of longer roots and enhanced growth following environmental stress. Such bacteria produce the enzyme 1-aminocyclopropane-1-carboxylate (ACC) deaminase, which removes amine group from the immediate precursor of ethylene (Gamalero and Glick, 2015). In comparison to terrestrial plants, the diversity of plant growth-promoting rhizobacteria in tidal marshes and their effect on plant health remains poorly understood. In this study, we surveyed roots of S. alterniflora and J. roemerianus for culturable rhizobacteria with traits that are commonly associated with competitive rhizosphere colonization and plant growth promotion. Our results demonstrated the presence of bacteria capable of secreting iron-chelating siderophores, antimicrobial metabolites (exoprotease, biosurfactants, and HCN), and the plant growth-promoting metabolites ACC deaminase and IAA (Table 3). The 16S rRNA-based identification of selected isolates revealed that these organisms were taxonomically diverse and included Gram-positive Firmicutes (Bacillus), as well as Gram-negative Flavobacteria (Tenacibaculum), Alphaproteobacteria (Marimonas), and Gammaproteobacteria (Gallaecimonas, Gynuella, Halomonas, Photobacterium, Vibrio) (Supplementary Table 2). Although the 16S based community analysis revealed that all these taxa were present at the on relative abundance of <1% (Vibrio spp. were the most abundant at 0.8%), their population levels estimated by dilution plating were between 106 and 107 CFU g-1 root fresh weight. These values are on par with population levels reported for some well-characterized groups of beneficial rhizobacteria in terrestrial agroecosystems (Weller et al., 2007; Kinkel et al., 2012). Some taxa characterized in this study were also recovered previously from small cordgrass (Spartina maritima) and denseflower cordgrass (Spartina densiflora) collected in salt marshes of the south Atlantic Spanish coast (Mateos-Naranjo et al., 2015; Paredes-Paliz et al., 2016), and treatment of Spartina with these bacteria improved plant growth in soil polluted with heavy metals (Mesa et al., 2015). Interestingly, the overall frequencies of isolates with beneficial traits differed between Spartina from the natural marsh and the recently established DIMR2, potentially suggesting that the natural disturbance may affect the dynamics of beneficial microorganisms in the rhizosphere of marsh plants.
Previous culture-based and culture-independent surveys of S. alterniflora and J. roemerianus from tidal marshes revealed the presence of diverse fungal communities (Gillevet et al., 2009; Walker and Campbell, 2010; Neori and Agami, 2017). While most of these fungi are considered harmless endophytes, surface saprophytes or even mutualistic symbionts (mycorrhizae), some species can act as opportunistic pathogens when host plants are stressed by the environment. Under certain conditions, populations of S. alterniflora and J. roemerianus in the Gulf Coast marshes undergo a rapid and progressive decline known as saltwater marsh dieback (SMD). One of the most severe SMD events occurred in 2000 and affected 100,000 hectares of salt marsh along Louisiana’s coast (Elmer et al., 2013). Interactive climate conditions and sea level anomalies are thought to be the primary cause of the sudden marsh dieback. Biotic factors associated with SMD are poorly understood, but independent assessments of the possible role of pathogens during dieback events in Louisiana and New England suggested the involvement of soilborne fungi of the genus Fusarium (Elmer and Marra, 2011). It has been proposed that Fusarium normally has an endophytic association with Spartina but may turn pathogenic in plants predisposed by abiotic stressors. We tested a subset of bacterial isolates from the rhizosphere of S. alterniflora and J. roemerianus for their capacity to inhibit plant pathogenic fungi F. pseudograminearum and F. culmorum. Our screening identified several Gram-positive and Gram-negative organisms that effectively antagonized one or both species of Fusarium, with the best antifungal activity observed in strains 441, 461, and 577 of G. sunshinyii (Table 4). Interestingly, Gynuella is a poorly characterized halophilic bacterium that was recently discovered in the rhizosphere of a salt-marsh sedge (Carex scabrifolia) and proved to inhibit a broad range of plant pathogenic fungi and oomycetes, including Botrytis, Colletotrichum, Pythium, Rhizoctonia, Sclerotinia, and Phytophthora (Chung et al., 2015). In terrestrial ecosystems, beneficial root-colonizing (rhizosphere) bacteria play a significant role in defending plants against soilborne pathogens (Mendes et al., 2011; Philippot et al., 2013; Raaijmakers and Mazzola, 2016). The suppression of pathogens by beneficial rhizobacteria is a complex phenomenon and often results from the synergistic interaction of different types of biocontrol mechanisms. Most often the biocontrol results from antagonism, which involves the destruction of the pathogen by antibiotics, toxins, and lytic enzymes, or parasitism/predation of the pathogen by the biocontrol agent. Alternatively, antagonism may result from indirect interactions where the beneficial microorganism suppresses the pathogen by competing for essential nutrients or space. It is tempting to speculate that species like Gynuella represent a specific group of beneficial microorganisms that are associated with roots of marsh plants and may contribute to their protection against different fungal pathogens. As a follow up to this study, we sequenced genomes of G. sunshinyii 449, G. sunshinyii 577, M. ostereistagnii 398, and M. spartinae 468 and are currently in the process of analyzing them for the potential plant growth promotion and antibiotic biosynthesis genes.
The analysis of recently sequenced microbial genomes has revealed that most beneficial rhizobacteria can produce multiple antibiotics and antagonistic metabolites, often with broad and overlapping spectra of activity. To date, however, only a small proportion of such antibiotics have been studied, as is evidenced by the recent flood of genomic-based discoveries of new antimicrobial and insecticidal compounds. In biocontrol strains, the biochemistry and genetics of antibiotic production are best studied in members of Bacillus, Streptomyces, and Pseudomonas (Gross and Loper, 2009; Raaijmakers et al., 2010; Barka et al., 2016). The types of antibiotics synthesized by these biocontrol agents are very diverse and include peptides and polyketides and hybrids thereof, cyclic lipopeptides, derivatives of chorismate and amino acids, macrolides and aminoglycosides. In terrestrial agroecosystems, rhizosphere-dwelling bacteria that produce 2,4-diacetylphloroglucinol, phenazines, and various polyketides have been implicated as key antagonistic components of microbial communities suppressive to diseases caused by Fusarium and other phytopathogenic fungi (Weller et al., 2007; Mazurier et al., 2009; Mendes et al., 2011; Kinkel et al., 2012; Cha et al., 2016). We screened all bacterial isolates that antagonized Fusarium, as well as samples of soil DNA extracted from the rhizosphere of S. alterniflora and J. roemerianus by PCR targeting genes for the synthesis of 2,4-diacetylphloroglucinol and phenazines. Results of these screens were negative suggesting that the antagonism was likely due to the presence of other types of antimicrobials.
It can take decades for sedimentary cycles in restored marshes to approach reference condition and the contribution of the sediment microbial communities to these processes is still poorly elucidated. In this study, we addressed this gap by comparing rhizosphere microbiomes of S. alterniflora and J. roemerianus from two restored tidal marshes and the natural reference marsh located at Deer Island, MS. Our results revealed that plants from the restored and reference areas supported similar microbial diversity indicating the rapid colonization of planted grasses with indigenous soil microbiota. Although close in composition, the microbial communities from the three studied sites differed significantly in the relative abundance of specific taxa. The observed differences are likely driven by the host plant identity and properties of sediment material used for the creation of restored marshes. Some of the differentially distributed groups of bacteria include taxa involved in the cycling of carbon, nitrogen, and sulfur, and may influence the succession of vegetation at the restored sited to climax condition. We also demonstrated that plants from the restored and reference sites vary in the frequency of culturable rhizobacteria that exhibit traits commonly associated with the promotion of plant growth and suppression of phytopathogenic fungi. Our findings will contribute to the establishment of benchmarks for the assessment of the outcome of coastal restoration projects in the Gulf of Mexico and better define factors that affect the long-term resiliency of tidal marshes and their vulnerability to climate change.
Author Contributions
DM, OM, PB, and KI conceived the research project. OM, DM, SH, and PB collected field samples. OM, SN, and SH extracted soil DNA and isolated and characterized culturable rhizobacteria. CJ and JE performed Illumina sequencing and conducted the microbiome analysis. DM, OM, CJ, JE, PB, and KI wrote the manuscript. All authors contributed to the manuscript revision.
Funding
This study was funded by cooperative agreement W912HZ-16-2-0020 from the U.S. Army Corps of Engineers Research and Development Center (ERDC) under the terms of the Gulf Coast Cooperative Ecosystems Studies Unit (CESU). Views, opinions, and/or findings contained herein are those of the authors and should not be construed as an Official Department of the Army position or decision unless so designated by other official documentation. The authors also acknowledge support from the Mississippi INBRE, funded by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant P20GM103476.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. Carl Qualls (USM Department of Biological Sciences) for the help with statistical analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03049/full#supplementary-material
Footnote
References
Albanese, D., Fontana, P., De Filippo, C., Cavalieri, D., and Donati, C. (2015). MICCA: a complete and accurate software for taxonomic profiling of metagenomic data. Sci. Rep. 5:9743. doi: 10.1038/srep09743
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Aoki, T., O’Donnell, K., and Geiser, D. M. (2014). Systematics of key phytopathogenic Fusarium species: current status and future challenges. J. Gen. Plant Pathol. 80, 189–201. doi: 10.1007/s10327-014-0509-3
Baker, B. J., Lazar, C. S., Teske, A. P., and Dick, G. J. (2015). Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3, 1–12. doi: 10.1186/s40168-015-0077-6
Bakker, A. W., and Schippers, B. (1987). Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp. mediated plant growth stimulation. Soil Biol. Biochem. 19, 451–457. doi: 10.1016/0038-0717(87)90037-X
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Meier-Kolthoff, J. P., et al. (2016). Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Bertness, M. (2006). Atlantic Shorelines: Natural History and Ecology. Woodstock: Princeton University Press.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cha, J. Y., Han, S., Hong, H. J., Cho, H., Kim, D., Kwon, Y., et al. (2016). Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 10, 119–129. doi: 10.1038/ismej.2015.95
Chung, E. J., Park, J. A., Jeon, C. O., and Chung, Y. R. (2015). Gynuella sunshinyii gen. nov., sp nov., an antifungal rhizobacterium isolated from a halophyte, Carex scabrifolia steud. Int. J. Syst. Evol. Microbiol. 65, 1038–1043. doi: 10.1099/ijs.0.000060
Costanza, R., Darge, R., Degroot, R., Farber, S., Grasso, M., Hannon, B., et al. (1997). The value of the world’s ecosystem services and natural capital. Nature 387, 253–260. doi: 10.1038/387253a0
Costello, E. K., Lauber, C. L., Hamady, M., Fierer, N., Gordon, J. I., and Knight, R. (2009). Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697. doi: 10.1126/science.1177486
Craft, C., Megonigal, P., Broome, S., Stevenson, J., Freese, R., Cornell, J., et al. (2003). The pace of ecosystem development of constructed Spartina alterniflora marshes. Ecol. Appl. 13, 1417–1432. doi: 10.1890/02-5086
Craft, C. B., Seneca, E. D., and Broome, S. W. (1991). Porewater chemistry of natural and created marsh soils. J. Exp. Mar. Biol. Ecol. 152, 187–200. doi: 10.1016/0022-0981(91)90214-H
Dimkpa, C., Weinand, T., and Asch, F. (2009). Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 32, 1682–1694. doi: 10.1111/j.1365-3040.2009.02028.x
Dyall-Smith, M. (2009). The Halohandbook: Protocols for Haloarchaeal Genetics. Available at: https://haloarchaea.com/halohandbook/
Eleuterius, L. N. (1973). “The marshes of Mississippi,” in Cooperative Gulf of Mexico Estuarine Inventory and Study (GMEI), ed. J. Y. Christmas (Ocean Springs, MS: Gulf Coast Research Laboratory), 147–190.
Eleuterius, L. N., and McDaniel, S. (1978). The salt marsh flora of Mississippi. Castanea 43, 86–95.
Elmer, W. H. (2014). A tripartite interaction between Spartina alterniflora, Fusarium palustre, and the purple marsh crab (Sesarma reticulatum) contributes to sudden vegetation dieback of salt marshes in New England. Phytopathology 104, 1070–1077. doi: 10.1094/PHYTO-08-13-0219-R
Elmer, W. H., Lamondia, J. A., Useman, S., Mendelssohn, I. A., Schneider, R. W., Jimenez-Gasco, M. M., et al. (2013). Sudden vegetation dieback in Atlantic and gulf coast salt marshes. Plant Dis. 97, 436–445. doi: 10.1094/PDIS-09-12-0871-FE
Elmer, W. H., and Marra, R. E. (2011). New species of Fusarium associated with dieback of Spartina alterniflora in Atlantic salt marshes. Mycologia 103, 806–819. doi: 10.3852/10-155
Gamalero, E., and Glick, B. R. (2015). Bacterial modulation of plant ethylene levels. Plant Physiol. 169, 13–22. doi: 10.1104/pp.15.00284
Gardener, B. B. M., Mavrodi, D. V., Thomashow, L. S., and Weller, D. M. (2001). A rapid polymerase chain reaction-based assay characterizing rhizosphere populations of 2,4-diacetylphloroglucinol-producing bacteria. Phytopathology 91, 44–54. doi: 10.1094/PHYTO.2001.91.1.44
Gerhardt Smith, J., Mcdonald, J., Rees, S. I., and Lovelace, N. (2015). Deer Island Aquatic Ecosystem Restoration Project. Available at: http://www.dtic.mil/dtic/tr/fulltext/u2/a621493.pdf
Gillevet, P. M., Sikaroodi, M., and Torzilli, A. P. (2009). Analyzing salt-marsh fungal diversity: comparing ARISA fingerprinting with clone sequencing and pyrosequencing. Fungal Ecol. 2, 160–167. doi: 10.1016/j.funeco.2009.04.001
Gordon, S. A., and Weber, R. P. (1951). Colorimetric estimation of indoleacetic acid. Plant Physiol. 26, 192–195. doi: 10.1104/pp.26.1.192
Gross, H., and Loper, J. E. (2009). Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 26, 1408–1446. doi: 10.1039/b817075b
Herbert, E. R., Marton, J. M., and Craft, C. B. (2015). “Tidal wetland restoration,” in Wetland Soils: Genesis, Hydrology, Landscapes, and Classification, 2nd Edn, eds M. J. Vepraskas and C. B. Craft (New York, NY: CRC Press), 447–468. doi: 10.1201/b18996-22
Kielak, A. M., Barreto, C. C., Kowalchuk, G. A., Van Veen, J. A., and Kuramae, E. E. (2016). The ecology of Acidobacteria: moving beyond genes and genomes. Front. Microbiol. 7:744. doi: 10.3389/fmicb.2016.00744
Kim, Y. C., Leveau, J., Mcspadden Gardener, B. B., Pierson, E. A., Pierson, L. S., and Ryu, C. M. (2011). The multifactorial basis for plant health promotion by plant-associated bacteria. Appl. Environ. Microbiol. 77, 1548–1555. doi: 10.1128/AEM.01867-10
King, E. O., Ward, M. K., and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44, 301–307.
Kinkel, L. L., Schlatter, D. C., Bakker, M. G., and Arenz, B. E. (2012). Streptomyces competition and co-evolution in relation to plant disease suppression. Res. Microbiol. 163, 490–499. doi: 10.1016/j.resmic.2012.07.005
Lamers, L. P. M., Govers, L. L., Janssen, I. C. J. M., Geurts, J. J. M., Van Der Welle, M. E. W., Van Katwijk, M. M., et al. (2013). Sulfide as a soil phytotoxin—a review. Front. Plant. Sci. 4:268. doi: 10.3389/fpls.2013.00268
Lang, M. J. (2012). Post-Construction Assessment of Saltmarsh Habitat on Deer Island, Biloxi, Mississippi. Master’s thesis, University of South Alabama, Mobile, AL.
Martin, J. K. (1975). Comparison of agar media for counts of viable soil bacteria. Soil Biol. Biochem. 7, 401–402. doi: 10.1016/0038-0717(75)90057-7
Mateos-Naranjo, E., Mesa, J., Pajuelo, E., Perez-Martin, A., Caviedes, M. A., and Rodriguez-Llorente, I. D. (2015). Deciphering the role of plant growth-promoting rhizobacteria in the tolerance of the invasive cordgrass Spartina densiflora to physicochemical properties of salt-marsh soils. Plant Soil 394, 45–55. doi: 10.1007/s11104-015-2504-7
Mavrodi, D. V., Mavrodi, O. V., Parejko, J. A., Bonsall, R. F., Kwak, Y. S., Paulitz, T. C., et al. (2012). Accumulation of the antibiotic phenazine-1-carboxylic acid in the rhizosphere of dryland cereals. Appl. Environ. Microbiol. 78, 804–812. doi: 10.1128/AEM.06784-11
Mavrodi, D. V., Peever, T. L., Mavrodi, O. V., Parejko, J. A., Raaijmakers, J. M., Lemanceau, P., et al. (2010). Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. Microbiol. 76, 866–879. doi: 10.1128/AEM.02009-09
Mazurier, S., Corberand, T., Lemanceau, P., and Raaijmakers, J. M. (2009). Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J. 3, 977–991. doi: 10.1038/ismej.2009.33
Mcmurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Mendes, R., Kruijt, M., De Bruijn, I., Dekkers, E., Van Der Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Mesa, J., Mateos-Naranjo, E., Caviedes, M. A., Redondo-Gomez, S., Pajuelo, E., and Rodriguez-Llorente, I. D. (2015). Endophytic cultivable bacteria of the metal bioaccumulator Spartina maritima improve plant growth but not metal uptake in polluted marshes soils. Front. Microbiol. 6:1450. doi: 10.3389/fmicb.2015.01450
Neori, A., and Agami, M. (2017). The functioning of rhizosphere biota in wetlands – a review. Wetlands 37, 615–633. doi: 10.1093/femsre/fuw040
Paredes-Paliz, K. I., Caviedes, M. A., Doukkali, B., Mateos-Naranjo, E., Rodriguez-Llorente, I. D., and Pajuelo, E. (2016). Screening beneficial rhizobacteria from Spartina maritima for phytoremediation of metal polluted salt marshes: comparison of gram-positive and gram-negative strains. Environ. Sci. Pollut. Res. Int. 23, 19825–19837. doi: 10.1007/s11356-016-7184-1
Penrose, D. M., and Glick, B. R. (2003). Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 118, 10–15. doi: 10.1034/j.1399-3054.2003.00086.x
Perez-Miranda, S., Cabirol, N., George-Tellez, R., Zamudio-Rivera, L. S., and Fernandez, F. J. (2007). O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 70, 127–131. doi: 10.1016/j.mimet.2007.03.023
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and Van Der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
R Core Team (2014). R: A Language and Environment for Statistical Computing. Available at: http://www.R-project.org/
Raaijmakers, J. M., De Bruijn, I., Nybroe, O., and Ongena, M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x
Raaijmakers, J. M., and Mazzola, M. (2016). Soil immune responses. Science 352, 1392–1393. doi: 10.1126/science.aaf3252
Rietl, A. J., Overlander, M. E., Nyman, A. J., and Jackson, C. R. (2016). Microbial community composition and extracellular enzyme activities associated with Juncus roemerianus and Spartina alterniflora vegetated sediments in Louisiana saltmarshes. Microb. Ecol. 71, 290–303. doi: 10.1007/s00248-015-0651-2
Sacherer, P., Defago, G., and Haas, D. (1994). Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 116, 155–160. doi: 10.1111/j.1574-6968.1994.tb06694.x
Schlesner, H., Jenkins, C., and Staley, J. (2006). “The phylum verrucomicrobia: a phylogenetically heterogeneous bacterial group,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 881–896.
Schmid, K., and Otvos, E. O. (2003). Deer island, coastal mississippi – a geological and historical story. J. Mississippi Acad. Sci. 48:45.
Walker, A. K., and Campbell, J. (2010). Marine fungal diversity: a comparison of natural and created salt marshes of the north-central Gulf of Mexico. Mycologia 102, 513–521. doi: 10.3852/09-132
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wasmund, K., Mussmann, M., and Loy, A. (2017). The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ. Microbiol. Rep. 9, 323–344. doi: 10.1111/1758-2229.12538
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Keywords: Spartina altemiflora, Juncus roemarianus, rhizosphere microbiome, coastal marshes, coastal restoration
Citation: Mavrodi OV, Jung CM, Eberly JO, Hendry SV, Namjilsuren S, Biber PD, Indest KJ and Mavrodi DV (2018) Rhizosphere Microbial Communities of Spartina alterniflora and Juncus roemerianus From Restored and Natural Tidal Marshes on Deer Island, Mississippi. Front. Microbiol. 9:3049. doi: 10.3389/fmicb.2018.03049
Received: 25 June 2018; Accepted: 27 November 2018;
Published: 11 December 2018.
Edited by:
Pilar Martínez-Hidalgo, University of Salamanca, SpainReviewed by:
Brian Raymond Murphy, Trinity College Dublin, IrelandAngelica Cibrian-Jaramillo, Centro de Investigación y de Estudios Avanzados (CINVESTAV), Mexico
Copyright © 2018 Mavrodi, Jung, Eberly, Hendry, Namjilsuren, Biber, Indest and Mavrodi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karl J. Indest, S2FybC5KLkluZGVzdEB1c2FjZS5hcm15Lm1pbA== Dmitri V. Mavrodi, ZG1pdHJpLm1hdnJvZGlAdXNtLmVkdQ==
†These authors have contributed equally to this work
‡Present address: Jed O. Eberly, Central Agricultural Research Center, Montana State University, Moccasin, MT, United States
 Olga V. Mavrodi
Olga V. Mavrodi Carina M. Jung
Carina M. Jung Jed O. Eberly
Jed O. Eberly Samuel V. Hendry
Samuel V. Hendry Sanchirmaa Namjilsuren
Sanchirmaa Namjilsuren Patrick D. Biber
Patrick D. Biber Karl J. Indest
Karl J. Indest Dmitri V. Mavrodi
Dmitri V. Mavrodi