- 1Department of Plant Protection, Faculty of Agriculture, University of Tabriz, Tabriz, Iran
- 2Department of Sustainable Agro-Ecosystems and Bioresources, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige, Italy
- 3Center Agriculture Food Environment (C3A), University of Trento, San Michele all'Adige, Italy
Strawberry anthracnose caused by Colletotrichum spp. is considered one of the most serious and destructive disease of strawberry worldwide. Weeds, as possible hosts of the pathogen, could have a role as potential inoculum reservoir. To prove this hypothesis, symptomless weeds were collected in strawberry fields showing anthracnose symptoms in Iran. Ten isolates with Colletotrichum-like colonies were recovered from symptomless Amaranthus viridis L., Convolvulus arvensis L., Fumaria officinalis L., Lactuca serriola L., and Sonchus oleraceus L. plants. The isolates were identified as C. nymphaeae, based on a combination of morphological and sequence data of TUB and GADPH genes. This identification was further validated using Rep-PCR fingerprinting analysis, which produces species-specific DNA fingerprints and unveils inter and intra variation of the species examined in this study. Moreover, rep-PCR marker was used to reveal accurate taxonomic position of Colletorichum spp. causing strawberry anthracnose belonging to the C. acutatum complex, including C. acutatum sensu stricto, C. fiorinae, C. godetiae, C. nymphaeae, C. salicis, and C. simmondsii. The C. nymphaeae isolates originating from symptomless weeds confirmed their pathogenicity on detached strawberry, proving that weeds in strawberry field may have a role as reservoir of inoculum. However, further studies are necessary to quantify their actual contribution to anthracnose epidemics in strawberry fields.
Introduction
Colletotrichum spp. are among the most widespread and destructive phytopathogenic fungi worldwide. The inclusion of these species among the top 10 fungal pathogens indicates their economic importance and also justifies the interest of scientists (Dean et al., 2012). Most Colletotrichum spp., viz. C. acutatum, C. gloeosporioides, C. dematium, C. destructivum, C. gigasporum, C. boninense and C. orbiculare, are known as complex species comprising different cryptic species (Damm et al., 2009, 2012a,b, 2013, 2014; Weir et al., 2012; Liu et al., 2014). Nearly all crops worldwide are susceptible to one or more species of Colletotrichum (Dean et al., 2012). Strawberry is an important and valuable crop and it is frequently infected by Colletotrichum spp. (Freeman and Katan, 1997; Karimi et al., 2017). Anthracnose, the main disease symptom caused by these fungal pathogens, was initially used to name the destructive disease of strawberry caused by Colletotrichum fragariae Brooks (Howard et al., 1992). Three species C. acutatum, C. gloeosporioides and C. fragariae, are commonly indicated as the causal agents of anthracnose on strawberry (Howard et al., 1992; Freeman and Katan, 1997). However, the advent of new molecular approaches, such as multi-gene approach in fungal taxonomy, combined with traditional diagnostic methods (polyphasic approach), revealed that C. acutatum and C. gloeosporioides are complex species, comprising several species causing anthracnose symptoms on strawberry (Damm et al., 2012a; Weir et al., 2012; Karimi et al., 2017). Nowadays, multi-gene technique is mainly used as the most confident approach to reveal cryptic species among complex species of fungi, particularly Colletotrichum spp. (Damm et al., 2009, 2012a, 2014). However, this method is expensive and not accessible for many mycologists all over the world. Therefore, alternative PCR-based genomic fingerprinting, such as rep-PCR, can be very useful to resolve cryptic species. Rep-PCR fingerprinting was originally developed to amplify sequences located between repetitive elements in prokaryotic genomes. However, various studies proved that this molecular marker is capable to provide the fingerprinting of eukaryotic genomes, including fungi (Mehta et al., 2002; Godoy et al., 2004; Alves et al., 2007).
Colletotrichum nymphaeae, one of the members of C. acutatum complex species, seems to be the most common and devastating Colletotrichum sp. on strawberry worldwide, including Iran (Damm et al., 2012a; Baroncelli et al., 2015; Karimi et al., 2017). Besides strawberry, this pathogen can cause anthracnose on other important crops, namely apple, pepper, grapevine and celery (Velho et al., 2014; Yamagishi et al., 2015; Liu et al., 2016; Nasehi et al., 2016). Colletotrichum nymphaeae has become a serious problem, threatening strawberry industry in Iran, since its first outbreak in 2013 (Karimi et al., 2017). Different conventional methods, including chemical fungicides, crop sanitation, cultural methods, and resistant cultivars, are used to control the disease (Smith, 2013). It was originally assumed that the primary infection in strawberry fields starts from infected plants growing near the strawberry field (Smith, 2013), since most of the Colletotrichum spp. such as C. nymphaeae may infect many other hosts than strawberry, including common weeds. Therefore, if this hypothesis is true, the identification and eradication of alternative hosts harboring the pathogen in order to remove pathogen inoculum could become a crucial agronomic practice to prevent infections of strawberry plants. The fact that many alternative hosts, including weeds, may not show any typical symptoms of anthracnose disease due to endophytic nature of Colletotrichum spp., could let growers to underestimate the risk. Limited knowledge is available on the potential role of weeds in the epidemiology of Colletotrichum acutatum sensu lato in strawberry anthracnose (Berrie and Burgess, 2003). In particular, (Berrie and Burgess, 2003) noticed that the pathogen can overwinter on weeds and survive on infected crop debris in absence of strawberry.
In our study, we isolated a Colletotrichum sp. from symptomless common weeds across the anthracnose-infected strawberry fields in Iran (Kurdistan province) and we compared them with type strains of C. nymphaeae, C. acutatum sensu stricto, C. godetiae, C. fioriniae, C. salicis and C. simmondsii, belonging to the C. acutatum seneu lato, in terms of morphological, molecular and pathogenicity.
Materials and Methods
Sampling and Isolation
The strawberry fields across Sanandaj and Kamyaran counties (Supplementary Figure 1) (two main areas of strawberry production in Kurdistan province, Iran), were inspected from May to June 2017. In this study, anthracnose-infected fields were identified and weeds were collected inside and around those fields with focusing mostly on broad-leaved weeds, because it has been previously determined that C. acutatum sensu lato can be mainly found on those plants (Berrie and Burgess, 2003). Symptomless common weeds across anthracnose-infected strawberry fields were collected and transferred to the lab inside facial bags. In the lab, different plant parts of each sample (leaf, stem, and root) were superficially disinfected by immersion in 90 % ethanol for 1 min, 1 % solution of sodium hypochlorite for 4 min and rinsed three times in sterile distilled water. All plant parts were left to dry on sterile Whatman paper under laminar flow hood. The plant parts were then cultured on potato dextrose agar (PDA; Sigma Aldrich, UK) and monitored for fungal growth on a daily base. Single spore cultures were established on water agar as described by Arzanlou et al. (2015) from colonies growing on PDA. Purified isolates were long term stored on home-made potato carrot agar (20 gr potatoes, 20 gr carrots, 20 gr technical agar per liter) at 4°C for further investigations. Fungal type strains of C. nymphaeae (CBS 515.78), C. acutatum sensu stricto (CBS 112996), C. godetiae (CBS 133.44), C. fioriniae (CBS 128517), C. salicis (CBS 607.94), and C. simmondsii (CBS 122122) were purchased from Westerdijk Fungal Biodiversity Institute, The Netherlands.
Morphological Description
Macro morphological features of fungal isolates including colony characterization and growth rate were studied on PDA, malt extracts agar (MEA; Sigma Aldrich, UK) and oat meal agar (OA; Sigma Aldrich, UK) as described by Karimi et al. (2017). Color charts of Rayner (1970) was used to rate the color of colonies.
DNA Extraction, Amplification, And Sequencing
Total genomic DNA of fungal isolates was extracted using a Fast DNA spin kit for soil (MP Biomedical, USA), according to the manufacturer's instructions. DNA quantification was determined using NanoDrop 8000 spectrophotometer (Thermo Scientific, USA) and proportional dilutions were made. The genomic regions of beta-tubulin (TUB) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were amplified with PCR, using the primer sets T1/Bt-2b (Glass and Donaldson, 1995; O'Donnell and Cigelnik, 1997) and GDF1/GDR1 (Guerber et al., 2003), respectively. The reaction mixture of the PCR was prepared in a total volume of 25 μl by mixing 1 μl (50 ng/ μl) of extracted DNA, 1 μl (0.2 μM) of forward and reverse primers, 9.5 μl of nuclease free sterile water and 12.5 μl of DreamTaq Green PCR Master Mix (Fermentase). PCR for TUB and GAPDH was carried out in a Biometra T Professional thermal cycler, as described by Karimi et al. (2016): an initial denaturation at 94°C for 5 min, followed by 40 cycles of 30 s at 95°C, 30 s at 52°C and 30 s at 72°C and final extension at 72°C for 7 min. Amplified products were visualized on 1% agarose gel stained with ethidium bromide. The products were then purified with Exo-Sap enzyme (Euroclone S.p.a., Italy), according to the manufacturer's instructions and sequenced using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA).
Phylogenetic Analysis
Consensus sequences were established from raw trace files of forward and reverse primers using Staden package program ver. 2.0.0b9 (Staden, 1996). The assembled sequences were subjected to BLAST searches of the GenBank database hosted by NCBI as queries. Two alignments were made for each of the two markers used in this study, by comparing downloaded sequences with the highest similarity and sequences of type species obtained from relevant literature for C. acutatum complex (Damm et al., 2012a). Sequences were then aligned with the Muscle software (Edgar, 2004) implemented in Mega6 (Tamura et al., 2013) and manually checked when necessary. Multi-gene analyses were performed using concatenated datasets of TUB and GAPDH made by Mesquite software (Maddison and Maddison, 2018).
For each alignment, the best evolutionary model was calculated using the software MrModelTest v. 2.3 (Nylander, 2004). All analyses were performed using MrBayes v. 3.2.1 (Ronquist and Huelsenbeck, 2003), with heating parameter set at 0.15, and four MCMC running up to 1000000 generations and sampling trees every 1,000 generations. Satisfactory convergence was assessed using the standard deviation of split frequency. The first 25% of saved trees were discarded as the burn-in, and consensus trees and their posterior probabilities (PP) were determined from the remaining trees. The generated phylogenetic trees were inspected and printed using FigTree v. 1.3.1 (Rambaut, 2009). Two strains of C. orchidophilum (CBS 128556 and CBS 632.80) were used as out-group taxa during phylogenetic analysis (Damm et al., 2012a). Sequences derived from this study were deposited at NCBI's GenBank nucleotide database (http://www.ncbi.nlm.nih.gov; Table 1).
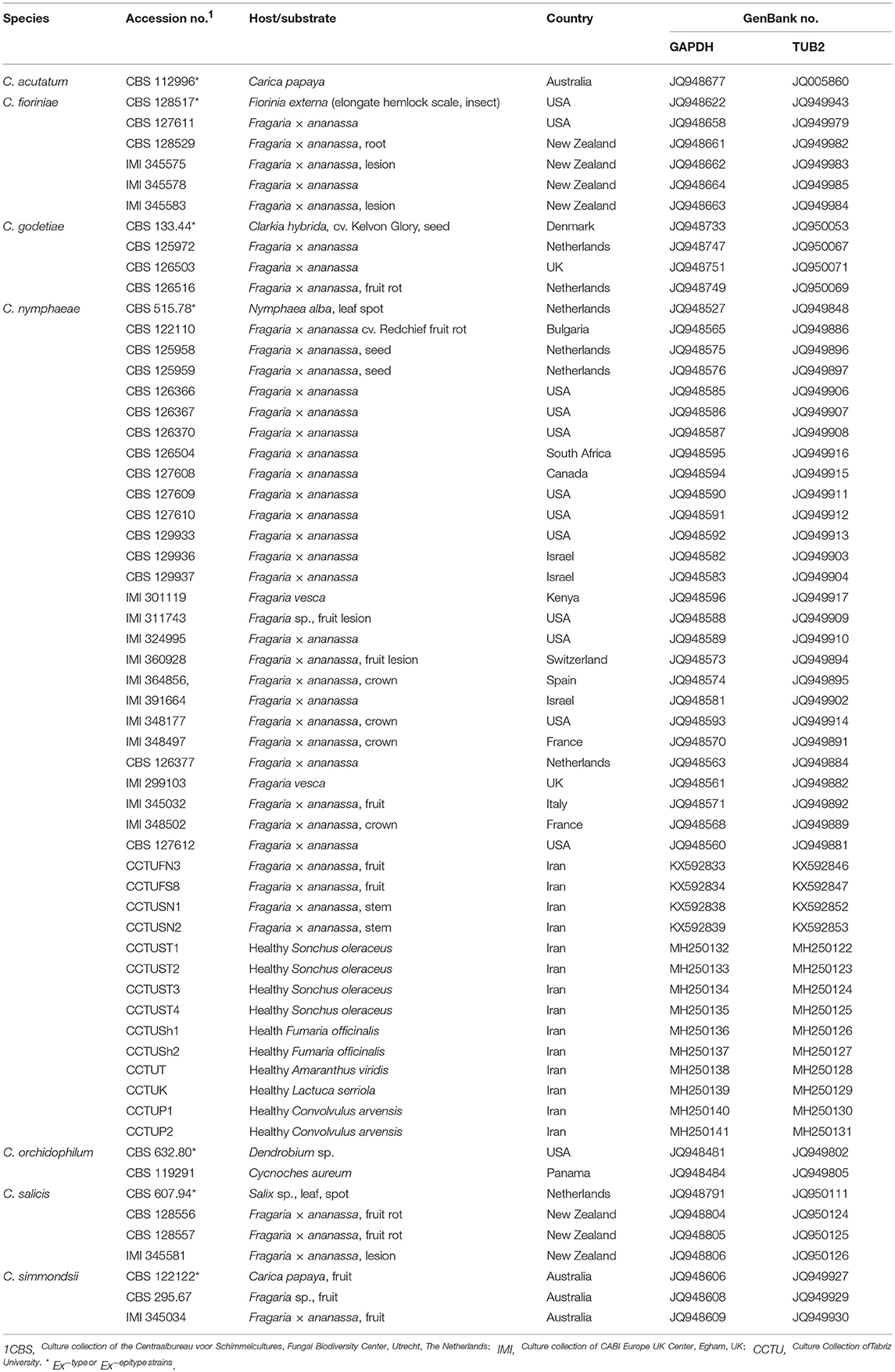
Table 1. Colletotrichum spp. associated with strawberry anthracnose used in multi-gene analysis in this study, host/substrate, and country of isolation and GenBank accessions.
Pathogenicity Test on Detached Leaves of Strawberry
To fulfill Koch's postulates, a pathogenicity test on detached untreated leaves of Fragaria × ananassa cv. Elsanta (a susceptible cultivar to strawberry anthracnose). The test was performed to evaluate the pathogenicity of the ten fungal isolates recovered in this study, in comparison to the type strains of C. nymphaeae, C. acutatum sensu stricto, C. godetiae, C. fioriniae, C. salicis, C. simmondsii and C. nymphaeae. Colletotrichum nymphaeae strain Cch32 was used as positive control due to its high aggressiveness on strawberry fruits of Fragaria × ananassa cv. Elsanta in previous study (Karimi et al., 2017). Detached leaves were first surface-sterilized by dipping in 1 % sodium hypochlorite for 30 s and ethanol 70% followed by rinsing three times in sterile distilled water. The leaves were left to dry on sterile paper under a laminar hood before inoculation. Inoculation was carried out according to Saha et al. (2010), with some modifications. Based on the leaf size, four to 12 wounds were initially made on the adaxial surface of each leaf using a sterile sharp needle. For inoculation, a plug of 5 mm of seven-day-old fungal cultures was placed on the wounds of each leaf. After inoculation, leaves were placed in plastic containers with moistened filter papers at 23°C to maintain high relative humidity (>95%). Five detached leaves were inoculated for each fungal strain and pathogen-free PDA plugs were used as control. After 8 days, the severity of anthracnose disease were recorded and calculated for each strawberry leaf using imageJ v. 1.51i software (https://imagej.nih.gov/ij/index.html).
Rep-PCR Fingerprinting
A polymorphic analysis based on repetitive-sequence-based PCR (rep-PCR) genomic fingerprinting was carried out for 17 fungal strains in this study using the primers of BOXA1R, REP1R-1/REP2-1, and ERIC1R/ERIC2 (Versalovic et al., 1991, 1994). PCR reactions were repeated twice (independent reactions) to confirm reproducibility of the results. PCR reaction mixtures were prepared in 25 ml volumes as follows: 12.5 μl 1X Dream Taq Green PCR Mastermix (Fer-mentas) 9.5 μl sterile distilled water, 1 μl each of each primer (0.2 μmol/μl stock) and 1 μl of template DNA (50 ng). DNA-free water (nuclease free) was used as a control. The PCR cycles were carried out as follows: an initial denaturation at 96 °C for 5 min, followed by 30 cycles at 94°C for 60 s, 40°C (REP primers) or 51°C (ERIC/BOX primers) for 1 min and 65°C for 8 min and a final extension at 65°C for 8 min. An aliquot of 5 μl PCR product of each isolate was loaded onto 1.5 % agarose gel 1 × Tris-Acetate-EDTA (TAE) buffer at 100V for 2.5 h and then stained with ethidium bromide. Amplicons size was determined in comparison to the GeneRulerTM 1kb DNA ladder. Unambiguous bands were scored as “0” and “1” to make a matrix data according to their presence or absence. UPGMA analysis was carried out for each marker using a simple matching coefficient with NTSYS-PC v2.2 and finally a consensus dendrogram was built.
Statistical Analyses
One-way ANOVA was performed to analyze results of pathogenicity test using SAS software package (SAS® software v. 9.1.3 SP4 portable). Least significant difference (LSD) was used to compare the means.
Results
Isolation
Of the various weeds sampled in the strawberry fields, 10 fungal isolates with a colony morphology resembling the one of Colletotrichum were recovered. These isolates were obtained from the leaves of asymptomatic plants of Amaranthus viridis L. (one isolate), Convolvulus arvensis L. (two isolates), Fumaria officinalis L. (two isolates), Lactuca serriola L. (one isolate) and Sonchus oleraceus L. (four isolates).
Morphological Description
Macro morphological features of type strains and isolates recovered from common weeds of strawberry fields are represented in Table 2. All isolates obtained from weeds were morphologically similar to each other and to C. nymphaeae strain CCTUCch32 reported from strawberry (Table 2, Figures 2c,d,g,h,k,l). On the contrary, colony morphology and growth rate of these strains were almost different compared to the type strain of C. nymphaeae (CBS 515.78) (Table 2, Figures 2b–d,f–h,j–l). Other fungal strains of Colletotrichum spp. in this study shared some similar characteristics (colony morphology and growth rate) compared to isolates recovered from weeds in this study (Table 2, Figures 1, 2).
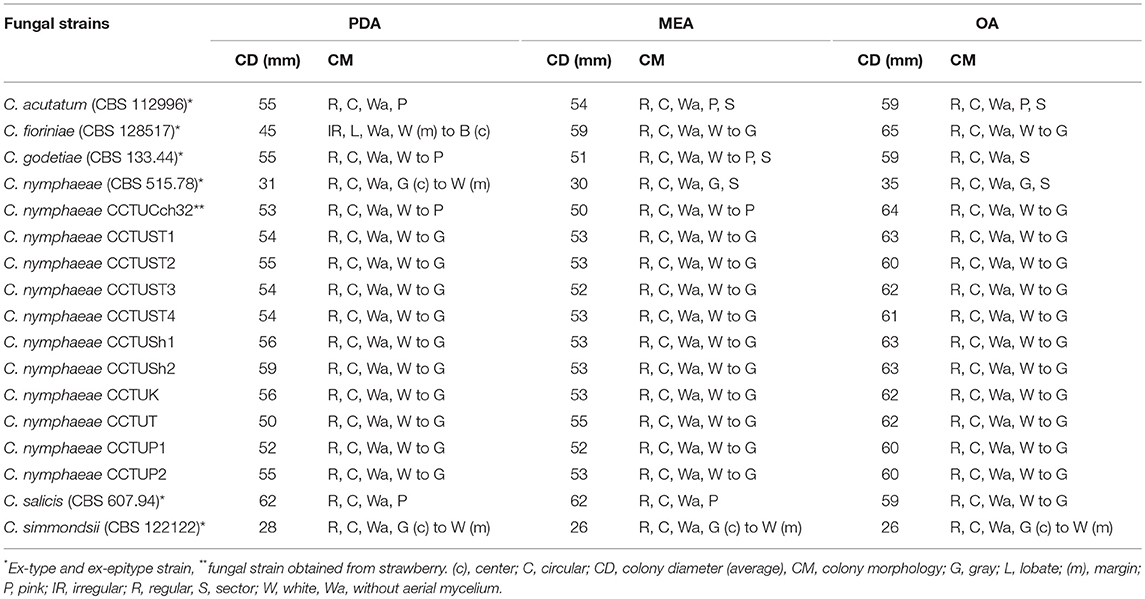
Table 2. Colony morphology and morphometry of fungal strains recovered from weeds and type strains belonging to C. acutatum complex.

Figure 1. Colony morphology of four fungal strains of C. acutatum (CBS 112996), C. salicis (CBS 607.94), C. simmondsii (CBS 122122) and C. fioriniae (CBS 128517) on PDA (a,b,c,d), MEA (e,f,g,h), and OA (i,j,k,l), respectively plus their pathogenicity on strawberry leaves of Fragaria × ananassa cv. Elsanta (m,n,o,p) respectively.

Figure 2. Colony morphology of four fungal strains of C. godetiae (CBS 133.44), C. nymphaeae (CBS 515.78), C. nymphaeae CCTUCch32 and C. nymphaeae CCTUK on PDA (a,b,c,d), MEA (e,f,g,h), and OA (i,j,k,l), respectively plus their pathogenicity on strawberry leaves of Fragaria × ananassa cv. Elsanta (m,n,o,p), respectively.
Phylogenetic Analysis
The final aligned TUB and GADPH datasets contained 74 in-group taxa with a total of 757 (gene boundaries, GAPDH: 1–265, TUB: 266–757) characteristics containing 109 and 94 unique site patterns, respectively. MrModeltest v. 2.3 found GTR+G+I to be the most fitting replacement models for both concatenated datasets. Bayesian inference of both concatenated datasets placed all the isolates sequenced in this study in the C. nymphaeae clade with the highest posterior probability (Figure 3). The C. nymphaeae clade with high posterior probability contained five subclades. All the isolates generated in this study were placed in subclade S5 containing the strains reported from strawberry plants besides one isolate from Mahonia sp. in Italy.
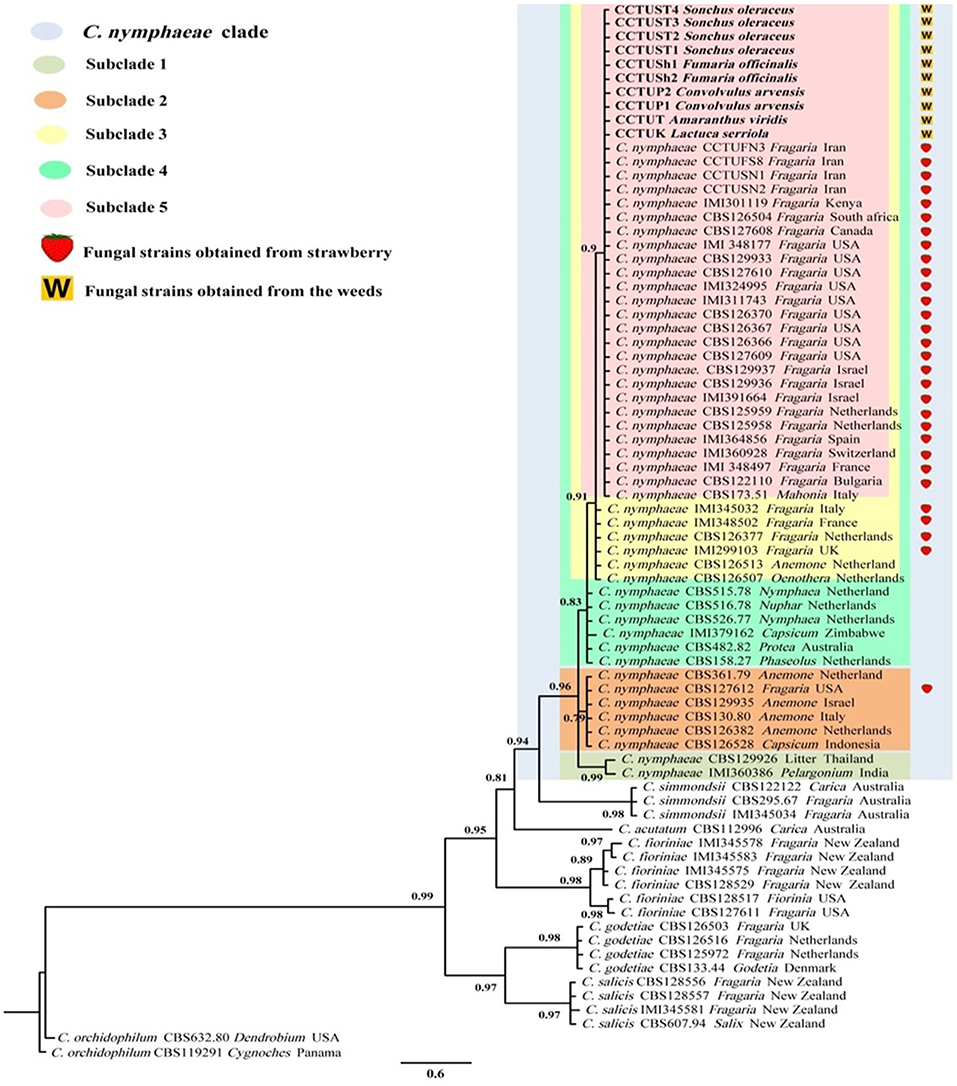
Figure 3. Bayesian inference phylogenetic tree of the Colletotrichum spp. reported from strawberry belonging to C. acutatum species complex. The tree was built using concatenated sequences of GAPDH (glyceraldehyde 3-phosphate dehydrogenase) and TUB2 (beta-tubulin), with the GTR+G+I model. Two strains of C. orchidophilum (CBS 632.80, CBS 119291) were used as out-group. The scale bar shows 0.6 expected changes per site. Strawberry isolates are labeled with a strawberry and the isolates recovered from the weeds in this study are marked with a W.
Pathogenicity Test
Pathogenicity test revealed that all isolates obtained from weeds in this study and other type strains of Colletotrichum spp. were able to induce anthracnose symptoms on strawberry leaves of Fragaria × ananassa cv. Elsanta (Figures 1, 2). However, there was a significant difference between fungal strains in terms of disease severity (df = 17, F = 6.95, p = < 0.0001). Colletotrichum nymphaeae CCTUSh1 showed the highest pathogenicity, although it had no significant difference with most strains such as the type strains of C. acutatum, C. salicis and C. fioriniae (Figures 1, 2, 4). The type strain of C. nymphaeae had significantly the lowest virulence compared to other C. nymphaeae strains in this study besides C. nymphaeae CCTUK (Figure 4). Moreover, C. nymphaeae strain CCTUCch32 after four strains of CCTUSh1, CCTUST3, CCTUT, and CCTUP1 had the highest pathogenicity compared to others (Figure 4). Type strains of C. godetiae and C. simmondsii showed the lowest pathogenicity and had no significant difference with healthy control (Figure 4).
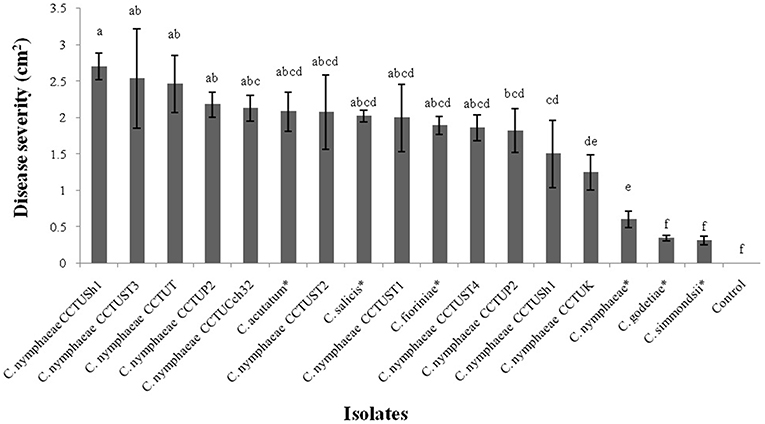
Figure 4. Disease severity of anthracnose on strawberry leaves of Fragaria × ananassa cv. Elsanta caused by the Colletotrichum spp. in this study. The mean with the common letters have no significant differences at level of p < 0.01. Error bars show standard error of the means. *Ex-type or ex-epitype strains.
Repetitive-Sequence-Based PCR
No amplification was observed for primer set REP1R-1/REP2-1 based on repetitive-sequence-based PCR analysis after several attempts while two primer sets, BOXA1R and ERIC1R/ERIC2, amplified the genomic DNA with reproducible DNA fingerprints (Figure 5). The size of amplified amplicons for each primer set ranged from 400 to 5,000 bp and from 200 to 6,000 bp, respectively (Figure 5). UPGMA clustering analysis of individual and combined data matrix generated with the amplicon patterns of BOXA1R and ERIC1R/ERIC2 placed all strains of this study at level of species showing the efficacy of these primers to generate species-specific DNA fingerprints even in the case of cryptic species (Figures 6–8). Individual and consensus dendrograms resided the type strain of C. nymphaeae as subclade compared to other C. nymphaeae strains obtained from weed plants and strawberry (Figures 6–8).
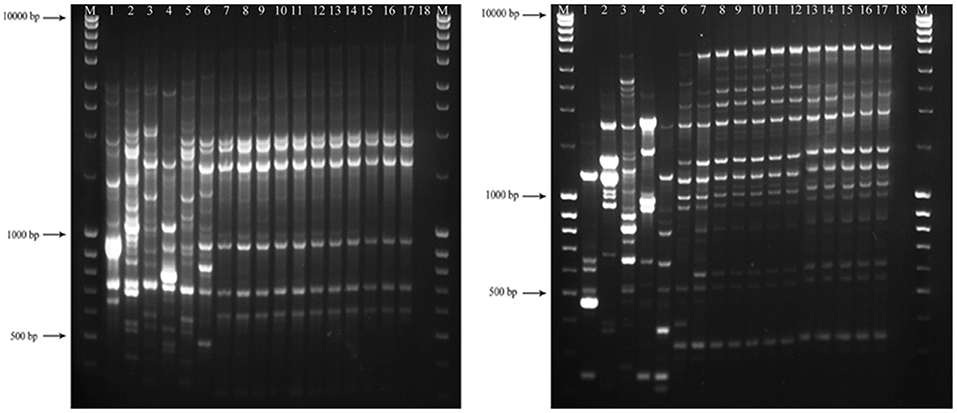
Figure 5. Amplified profiles of genomic DNA belong to Colletotrichum spp. generated with BOX (left-hand image) and ERIC1R/ERIC2 (right-hand image) primers. The first and the last columns (M) show GeneRuler Ladder bands (#SM0403). The lanes with consecutive numbers from one to seventeen correspond to C. acutatum (CBS 112996), C. salicis (CBS 607.94), C. fioriniae (CBS 128517), C. godetiae (CBS 133.44), C. simmondsii (CBS 122122), C. nymphaeae (CBS 515.78), C. nymphaeae CCTUCch32, C. nymphaeae CCTUST1, C. nymphaeae CCTUST2, C. nymphaeae CCTUST3, C. nymphaeae CCTUST4, C. nymphaeae CCTUK, C. nymphaeae CCTUT, C. nymphaeae P1, C. nymphaeae CCTUP2, C. nymphaeae CCTUSh1 and C. nymphaeae CCTUSh2. The eighteenth well represents negative control.
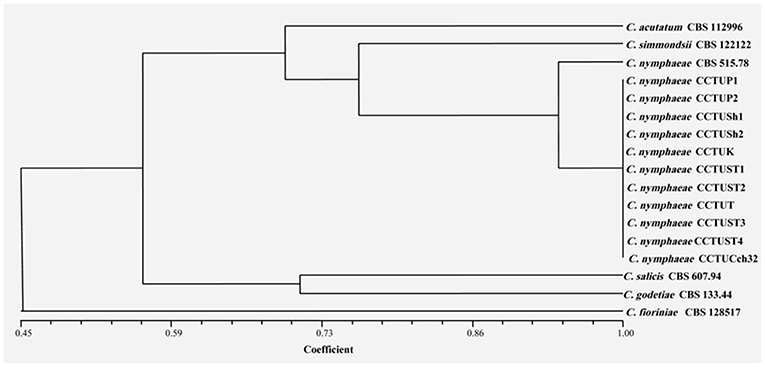
Figure 6. Dendrogram generated with DNA banding patterns of BOX primer using UPGMA cluster analysis. Similarity between the patterns was calculated with simple matching coefficient. Ex-type and Ex-epitype cultures are represented with CBS mark.
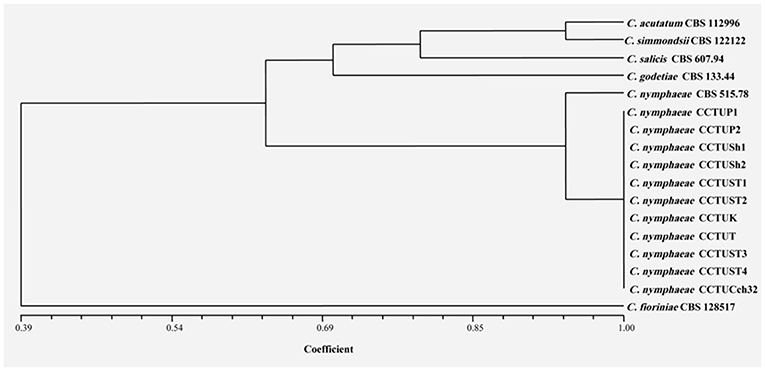
Figure 7. Dendrogram generated with DNA banding patterns of ERIC1R/ERIC2 primer using UPGMA cluster analysis. Similarity between the patterns was calculated with simple matching coefficient. Ex-type and Ex-epitype cultures are represented with CBS mark.
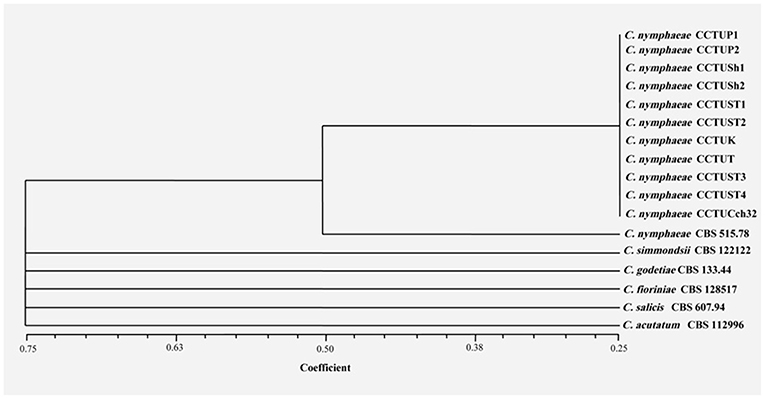
Figure 8. Consensus dendrogram generated with combined DNA banding patterns of BOX and ERIC1R/ERIC2 primers using UPGMA cluster analysis. Similarity between the patterns was calculated with simple matching coefficient. Ex-type and Ex-epitype cultures are represented with CBS mark.
Discussion
This study confirmed the hypothesis that weeds present in strawberry fields can host C. nymphaeae as a potential inoculum reservoir. The characterization of the Colletotrichum isolates recovered from the common weeds across the strawberry fields in Iran, indicates that weed isolates shared similar macro morphological characteristics (Table 2, Figure 2) and were identical to C. nymphaeae CCTUCch32 isolated from symptomatic strawberry (Karimi et al., 2017). On the other hand, in terms of colony features and growth rate, the 10 isolates showed some discrepancies with type strain of C. nymphaeae from Nymphaea alba (Table 2, Figures 1, 2). Moreover, they shared some features with other type strains including C. acutatum, C. salicis, C. simmondsii, C. fioriniae and C. godetiae (Table 2, Figures 1, 2), indicating the variability and unreliability of macro-morphological characterization for species delimitation. However, Bayesian inference of concatenated alignments of TUB and GAPDH clustered our isolates in C. nymphaeae clade inside subclade 5, containing other C. nymphaeae strains reported from strawberry worldwide including Iran such as C. nymphaeae FN3, FS8, SN1, and SN2 (Karimi et al., 2017). Both TUB and GAPDH genes have been recommended as appropriate barcode genes to reveal cryptic species of C. acutatum complex (Damm et al., 2012a). Base on that, using a combination of molecular and morphological characterizations, the 10 Colletotrichum sp. isolates from symptomless plants of A. viridis, C. arvensis, F. officinalis, L serriola, and S. oleraceus were identified as C. nymphaeae (Figures 2, 3). This result was also in line with multi-gene and fingerprinting analyses where type strains of C. nymphaeae resided in a different subclade (Figures 3, 6–8). Furthermore, in the pathogenicity test, type strain of C. nymphaeae showed the lowest virulence on strawberry leaves compared to other C. nymphaea strains, except for C. nymphaeae CCTUK (Figure 3). Overall, these results further confirm our previous findings (Karimi et al., 2017) that subclade 5 containing strains recovered from strawberry are evolutionary different and constitute a distinct subpopulation of C. nymphaeae with host-preference on strawberry. Moreover, this variability in the aggressiveness of a species within C. acutatum complex could be associated with the capability of different populations of these species to infect a wider range of hosts and the susceptibility to various environmental conditions (Baroncelli et al., 2015).
Molecular typing based on rep-PCR revealed that this technique can generate species-specific DNA fingerprints, at least for Colletotrichum spp. causing strawberry anthracnose belonging to C. acutatum complex (Figures 5–8). It is worth noting that dendrograms resulted from BOX, REP1R-1/REP2-1 and their combination showed high similarity with multi-gene tree generated with both TUB and GAPDH genes particularly one resulted from BOX (Figures 3, 6–8). Accordingly, amplified patterns generated by rep-PCR fingerprinting analysis revealed that all isolates recovered from weeds were highly similar to C. nymphaeae CCTUCch32 isolated from strawberry, while they showed some discrepancies with the type strain of C. nymphaeae (Figure 5). Similarly to multi-gene analysis, the type strain of C. nymphaeae resides in a distinct subclade (Figures 6–8). Based on the dendrograms of this study, rep-PCR confirmed to be an appropriate molecular marker to reveal intra and inter species variation of Colletotrichum spp. causing anthracnose on strawberry. It seems that this molecular technique can reveal all cryptic species among C. acutatum sensu lato, however to further confirm this hypothesis, more type and epi-type fungal strains belong to C. acutatum sensu lato should be investigated. Although rep-PCR fingerprinting analysis has been originally used for molecular typing and identification of bacterial species (Versalovic et al., 1991, 1994), the efficacy of this molecular typing technique in unveiling of fungal cryptic species belonging to various fungal complex species was shown in some studies (Alves et al., 2007; Faedda et al., 2011). However, this is the first time that rep-PCR is used as authentic molecular typing technique for unveiling Colletorichum spp. causing strawberry anthracnose belonging to the C. acutatum sensu lato. This result is consistent with a previous study (Faedda et al., 2011) where dendrograms obtained with RAPD-PCR analysis, a molecular technique very similar to Rep-PCR (Gillings and Holley, 1997), were showed to be almost identical, and even more informative of intraspecific variability, compared to ITS- and TUB-based UPGMA phylogetic trees of isolates of the C. acutatum species complex from various host plants, including strawberry.
To the best of our knowledge, this is the first time that asymptomatic plants of A. viridis, C. arvensis, F. officinalis, L. serriola, and S. oleraceus are reported as possible inoculum reservoirs of C. nymphaeae. In this study, fungal isolates were isolated using the protocol which is commonly used for the isolation of endophytic fungi, highlighting their either endophytic or long lasting biotrophic nature inside those plants. A broad diversity of approaches including biotrophs, nectrotrophs, hemibiotrophs to endophytes are employed by Colletotrichum species to colonize and obtain nutrients from their hosts (Baroncelli et al., 2017). Previously, only asymptomatic plants of the weed genera Vicia and Conyza were reported as inoculum reservoirs of C. acutatum sensu lato (Freeman et al., 2001) and the pathogenicity of C. acutatum sensu lato was demonstrated on some other weeds in strawberry fields in UK, including Malva sylvestris, Ranunchulus repens, Sinapis arvensis, Rumex obtusifolius, Geranium dissectum, Polygonatum aviculare, and Plantago lanceolata (Berrie and Burgess, 2003). It is noteworthy that the aggressiveness of four C. nymphaeae strains recovered from weeds including C. nymphaeae CCTUSh1, CCTUST3, CCTUT, and CCTUP2 was higher than C. nymphaeae CCTUCch32, which was the most aggressive strain in our previous studies (Karimi et al., 2017). These findings highlight that the infected weeds close to strawberry fields can possibly act as a potential reservoirs of pathogen inoculum during crop season, as highlighted by Berrie and Burgess (2003). In view of the fact that no specific resting structure, which can let it to survive in buried plant residues, has so far been reported for C. acutatum sensu lato (Ureña-Padilla et al., 2001; Freeman, 2008), weed plants identified in this study could allow/contribute to the survival of the pathogen between seasons. Therefore, the removal of weeds that could act as potential pathogen reservoirs should be considered to optimize disease management.
Conclusion
The presence of this pathogen on common weeds growing close to or within strawberry fields indicates that they may serve as a reservoir of inoculum and calls for further practical investigations to assess the advantage of their removal to reduce pathogen inoculum and improve the efficacy of anthracnose disease control on strawberry. Moreover, our study demonstrates that, when the type strains of each species are available, rep-PCR is able to reveal intra and inter species variation of fungal species belonging to the C. acutatum complex, even in case of cryptic species.
Author Contributions
KK designed and performed the experiments, analyzed and discussed the results, and wrote the manuscript. MA and IP discussed the results and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Iran National Science Foundation (INSF) and the University of Tabriz for the grant for the post-doc to KK. The research reported here is also partially funded by the Autonomous Province of Trento.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00129/full#supplementary-material
References
Alves, A., Phillips, A. J., Henriques, I., and Correia, A. (2007). Rapid differentiation of species of Botryosphaeriaceae by PCR fingerprinting. Res. Microbiol. 158, 112–121. doi: 10.1016/j.resmic.2006.10.003
Arzanlou, M., Bakhshi, M., Karimi, K., and Torbati, M. (2015). Multigene phylogeny reveals three new records of Colletotrichum spp. and several new host records for the mycobiota of Iran J. Plant Prot. Res. 55, 198–211. doi: 10.1515/jppr-2015-0027
Baroncelli, R., Talhinhas, P., Pensec, F., Sukno, S. A., Le Floch, G., and Thon, M. R. (2017). The Colletotrichum acutatum species complex as a model system to study evolution and host specialization in plant pathogens. Front. Microbiol. 8:2001. doi: 10.3389/fmicb.2017.02001
Baroncelli, R., Zapparata, A., Sarrocco, S., Sukno, S. A., Lane, C. R., Thon, M. R., et al. (2015). Molecular diversity of anthracnose pathogen populations associated with UK strawberry production suggests multiple introductions of three different Colletotrichum species. PLOS ONE 10:e0129140. doi: 10.1371/journal.pone.0129140
Berrie, A. M., and Burgess, C. M. (2003). A review of research on epidemiology and control of blackspot of strawberry (Colletotrichum acutatum) with special reference to weeds as alternative hosts. IOBC/wrps Bull. 26, 163–168.
Damm, U., Cannon, P. F., Liu, F., Barreto, R. W., Guatimosim, E., and Crous, P. W. (2013). The Colletotrichum orbiculare species complex: important pathogens of field crops and weeds. Fungal. Divers. 61, 29–59. doi: 10.1007/s13225-013-0255-4
Damm, U., Cannon, P. F., Woudenberg, J. H., and Crous, P. W. (2012a). The Colletotrichum acutatum species complex. Stud. Mycol. 73, 37–113. doi: 10.3114/sim0010
Damm, U., Cannon, P. F., Woudenberg, J. H., Johnston, P. R., Weir, B. S., Tan, Y. P., et al. (2012b). The Colletotrichum boninense species complex. Stud. Mycol. 73, 1–36. doi: 10.3114/sim0002
Damm, U., O'Connell, R. J., Groenewald, J. Z., and Crous, P. W. (2014). The Colletotrichum destructivum species complex–hemibiotrophic pathogens of forage and field crops. Stud. Mycol. 79, 49–84. doi: 10.1016/j.simyco.2014.09.003
Damm, U., Woudenberg, J. H., Cannon, P. F., and Crous, P. W. (2009). Colletotrichum species with curved conidia from herbaceous hosts. Fungal. Divers. 18, 39–45.
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. plant pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic. Acids. Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Faedda, R., Agosteo, G. E., Schena, L., Mosca, S., Frisullo, S., Di San Lio, G. M., et al. (2011). Colletotrichum clavatum sp. nov. identified as the causal agent of olive anthracnose in Italy. Phytopathol. Mediterr. 50, 283–302. Available online at: https://www.jstor.org/stable/26458702
Freeman, S. (2008). Management, survival strategies, and host range of Colletotrichum acutatum on strawberry. Hortscience 43, 66–68.
Freeman, S., Horowitz, S., and Sharon, A. (2001). Pathogenic and nonpathogenic lifestyles in Colletotrichum acutatum from strawberry and other plants. Phytopathol 91, 986–992. doi: 10.1094/PHYTO.2001.91.10.986
Freeman, S., and Katan, T. (1997). Identification of Colletotrichum species responsible for anthracnose and root necrosis of strawberry in Israel. Phytopathol 87, 516–521. doi: 10.1094/PHYTO.1997.87.5.516
Gillings, M., and Holley, M. (1997). Repetitive element PCR fingerprinting (rep-PCR) using enterobacterial repetitive intergenic consensus (ERIC) primers is not necessarily directed at ERIC elements. Lett. Appl. Microbiol. 25, 17–21. doi: 10.1046/j.1472-765X.1997.00162.x
Glass, N. L., and Donaldson, G. (1995). Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330.
Godoy, P., Cano, J., Gené, J., Guarro, J., Höfling-Lima, A. L., and Lopes Colombo, A. (2004). Genotyping of 44 isolates of Fusarium solani, the main agent of fungal keratitis in Brazil. J. Clin. Microbiol. 42, 4494–4497. doi: 10.1128/JCM.42.10.4494-4497.2004
Guerber, J. C., Liu, B., Correll, J. C., and Johnston, P. R. (2003). Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95, 872–895. doi: 10.1080/15572536.2004.11833047
Howard, C. M., Maas, J. L., Chandler, C. K., and Afbregts, E. E. (1992). Anthracnose of strawberry caused by the Colletotrichum complex in Florida. Plant Dis. 76, 976–981. doi: 10.1094/PD-76-0976
Karimi, K., Ahari, A. B., Arzanlou, M., Amini, J., Pertot, I., and Rota-Stabelli, O. (2017). Application of the consolidated species concept to identify the causal agent of strawberry anthracnose in Iran and initial molecular dating of the Colletotrichum acutatum species complex. Eur. J. Plant Pathol. 147, 375–387. doi: 10.1007/s10658-016-1009-4
Karimi, K., Khodaei, S., Rota-Stabelli, O., Arzanlou, M., and Pertot, I. (2016). Identification and characterization of two new fungal pathogens of Polygonatum odoratum (Angular Solomon's seal) in Italy. J. Phytopathol. 164, 1075–1084. doi: 10.1111/jph.12528
Liu, F., Cai, L., Crous, P. W., and Damm, U. (2014). The Colletotrichum gigasporum species complex. Persoonia 33, 83–97. doi: 10.3767/003158514X684447
Liu, M., Zhang, W., Zhou, Y., Liu, Y., Yan, J. Y., Li, X. H., et al. (2016). First report of twig anthracnose on grapevine caused by Colletotrichum nymphaeae in China. Plant Dis. 100, 2530–2530. doi: 10.1094/PDIS-05-16-0632-PDN
Maddison, W. P., and Maddison, D. R. (2018). Mesquite: a Molecular System for Evolutionary Analysis. version 3.40. Available online at: http://mesquiteproject.org
Mehta, Y. R., Mehta, A., and Rosato, Y. B. (2002). ERIC and REP-PCR banding patterns and sequence analysis of the internal transcribed spacer of rDNA of Stemphylium solani isolates from cotton. Curr. Microbiol. 44, 323–328. doi: 10.1007/s00284-001-0026-4
Nasehi, A., Kadir, J., Rashid, T. S., Awla, H. K., Golkhandan, E., and Mahmodi, F. (2016). Occurrence of anthracnose fruit rot caused by Colletotrichum nymphaeae on pepper (Capsicum annuum) in Malaysia. Plant Dis. 100, 1244–1244. doi: 10.1094/PDIS-08-15-0876-PDN
Nylander, J. A. A. (2004). MrModeltest v2. Program Distributed by the Author. Sweden: Evolutionary Biology Centre, Uppsala University.
O'Donnell, K., and Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are non orthologous. Mol. Phylogenet. Evol. 7, 103–116.
Rambaut, A. (2009). FigTree v1.3.1. Available online at: http://tree.bio.ed.ac.uk/software/figtree/
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Saha, A., Isha, M., Dasgupta, S., and Saha, D. (2010). Pathogenicity of Colletotrichum gloeosporioides (Penz.) Sacc. causal agent of anthracnose in different varieties of eggplant (Solanum melongena L.) determined by levels of cross-reactive antigens shared by host and pathogen. Arch. Phytopathology. Plant Protect. 43, 1781–1795. doi: 10.1080/03235400902753667
Smith, B. J. (2013). Strawberry anthracnose: progress toward control through science. Int. J. Fruit Sci. 13, 91–102. doi: 10.1080/15538362.2012.697025
Staden, R. (1996). The staden sequence analysis package. Mol. Biotech. 5:233. doi: 10.1007/BF02900361
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Ureña-Padilla, A. R., Mitchell, D. J., and Legard, D. E. (2001). Oversummer survival of inoculum for Colletotrichum crown rot in buried strawberry crown tissue. Plant Dis. 85, 750–754. doi: 10.1094/PDIS.2001.85.7.750
Velho, A. C., Stadnik, M. J., Casanova, L., Mondino, P., and Alaniz, S. (2014). First report of Colletotrichum nymphaeae causing apple bitter rot in southern Brazil. Plant Dis. Note. 98:567. doi: 10.1094/PDIS-06-13-0671-PDN
Versalovic, J., Koeuth, T., and Lupski, J. R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic. Acid. Res. 19, 6823–6831. doi: 10.1093/nar/19.24.6823
Versalovic, J., Schneider, M., de Bruijn, F. J., and Lupski, J. R. (1994). Genomic fingerprinting of bacteria using repetitive sequence based polymerase chain reaction. Methods. Mol. Cell. Biol. 5, 25–40.
Weir, B. S., Johnston, P. R., and Damm, U. (2012). The Colletotrichum gloeosporioides species complex. Stud. Mycol. 73, 115–180. doi: 10.3114/sim0011
Keywords: Fragaria × ananassa, anthracnose, asymptomatic plants, inoculum reservoir, molecular identification, kurdistan province
Citation: Karimi K, Arzanlou M and Pertot I (2019) Weeds as Potential Inoculum Reservoir for Colletotrichum nymphaeae Causing Strawberry Anthracnose in Iran and Rep-PCR Fingerprinting as Useful Marker to Differentiate C. acutatum Complex on Strawberry. Front. Microbiol. 10:129. doi: 10.3389/fmicb.2019.00129
Received: 11 April 2018; Accepted: 21 January 2019;
Published: 12 February 2019.
Edited by:
Gustavo Henrique Goldman, University of São Paulo, BrazilReviewed by:
Soner Soylu, Mustafa Kemal University, TurkeySanta Olga Cacciola, Università degli Studi di Catania, Italy
Copyright © 2019 Karimi, Arzanlou and Pertot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaivan Karimi, S2VpdmFua2FyaW1pQHRhYnJpenUuYWMuaXI=; S2FpdmFuLmthcmltaUBndWVzdHMuZm1hY2guaXQ=
 Kaivan Karimi
Kaivan Karimi Mahdi Arzanlou
Mahdi Arzanlou Ilaria Pertot
Ilaria Pertot