- 1State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
- 2Department of Clinical Laboratory, The 307th Hospital of the Chinese People’s Liberation Army, Beijing, China
- 3Zhejiang Provincial Center for Disease Control and Prevention, Hangzhou, China
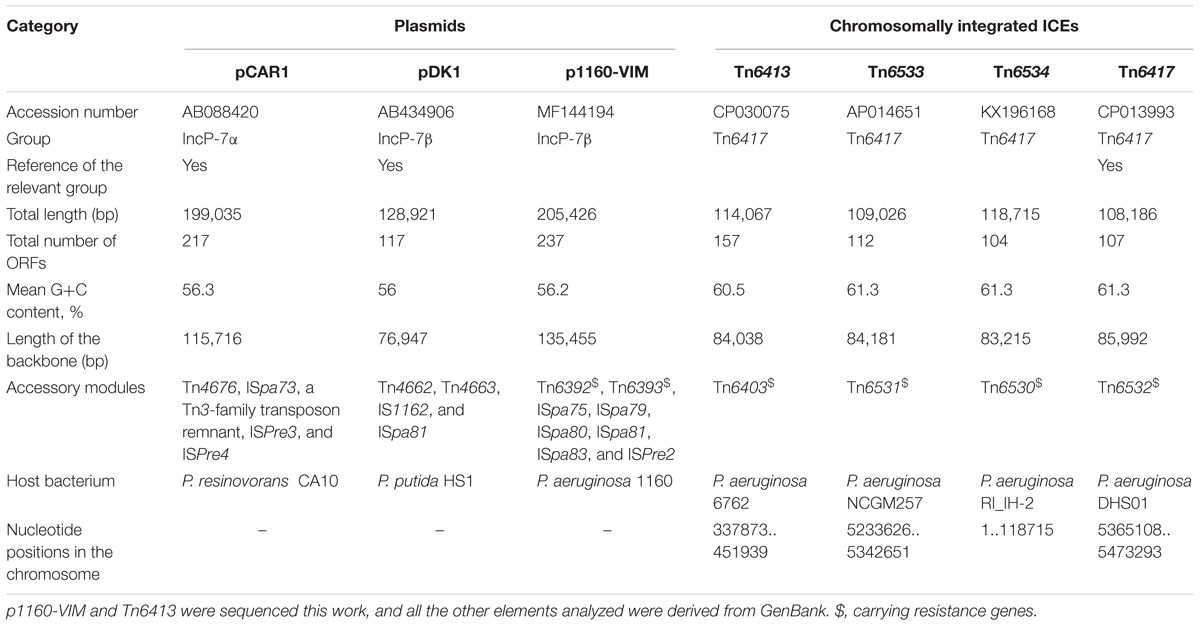
This study presents three novel integrons In1394, In1395, and In1443, three novel unit transposons Tn6392, Tn6393, and Tn6403, one novel conjugative element (ICE) Tn6413, and the first sequenced IncP-7 resistance plasmid p1160-VIM from clinical Pseudomonas aeruginosa. Detailed sequence comparison of p1160-VIM (carrying Tn6392 and Tn6393) and Tn6413 (carrying Tn6403) with related elements were performed. Tn6392, Tn6393, and Tn6403 were generated from integration of In1394 (carrying blaVIM–24), In1395 and In1443 (carrying blaVIM–4) into prototype Tn3-family unit transposons Tn5563, Tn1403, and Tn6346, respectively. To the best of our knowledge, this is the first report of a blaVIM–24-carrying P. aeruginosa isolate.
Introduction
Plasmids of thirteen incompatibility groups in Pseudomonas (IncP-1 to IncP-7 and IncP-9 to IncP-14) have been recognized, varying in genetic structure, size and host range. IncP-7 plasmids, with a narrow host range, are of particular interest in environmental biodegradative potentials. Most sequenced members of this group, such as pCAR1 (Maeda et al., 2003), pND6_1 (Li et al., 2004), pWW53 (Pickup and Williams, 1985), pDK1 (Kunz and Chapman, 1981), and pHE24 (Supplementary Table S1), belong to toluene catabolic or degradation plasmids (D-plasmids) rather than resistance plasmids (R-plasmids).
Integrative and conjugative elements (ICEs), also known as conjugative transposons, are typically found integrated into host bacterial chromosomes and encode integrase (Int), excisionase (Xis) and type IV secretion system responsible for integration, excision, interbacterial transfer, respectively. ICEs confer antibiotic resistance (such as Tn916) (Franke and Clewell, 1981), heavy metal resistance (such as R391) (Peters et al., 1991), and carbon utilization (such as ICEclc) (Gaillard et al., 2006).
Verona integron-encoded metallo-β-lactamase (VIM) is one of the most predominant families among class B carbapenemases and can hydrolyze nearly all β-lactams including carbapenems, except aztreonam (Queenan and Bush, 2007). This study dealt with a detailed genetic characterization of a novel blaVIM–24-carrying IncP-7β plasmid p1160-VIM and a novel blaVIM−4-carrying ICE Tn6413 recovered from two different clinical P. aeruginosa isolates.
Materials and Methods
Bacterial Isolates
Pseudomonas aeruginosa 1160 was isolated in 2015 from a sputum specimen of an elderly patient in a teaching hospital in Hebei Province, China. P. aeruginosa 6762 was recovered in 2016 from a sputum specimen of an elderly patient in a public hospital in Lanzhou Province, China. Bacterial species was identified by 16S rRNA gene sequencing and PCR detection of P. aeruginosa-specific oafA gene (Choi et al., 2013).
Conjugal Transfer
Conjugal transfer experiments were carried out with rifampin-resistant P. aeruginosa PAO1 used as recipients and each of the blaVIM-positive 1160 or 6762 isolate as donor. Three milliliters of overnight cultures of each of donor and recipient bacteria were mixed together, harvested and resuspended in 80 μl of Brain Heart Infusion (BHI) broth (BD Biosciences). The mixture was spotted on a 1 cm2 hydrophilic nylon membrane filter with a 0.45 μm pore size (Millipore) that was placed on BHI agar (BD Biosciences) plate and then incubated for mating at 30°C for 12 to 18 h. Bacteria were washed from filter membrane and spread on Muller-Hinton (MH) agar (BD Biosciences) plates containing 1000 μg/ml rifampin together with 2 μg/ml meropenem for selecting an P. aeruginosa transconjugant carrying blaVIM.
Sequencing and Annotation
The genomic DNA of strain 6762 or the plasmid DNA of strain 1160 was isolated using an UltraClean Microbial Kit or a Large Construct Kit (Qiagen, NW, Germany), respectively, and then sequenced from a mate-pair library with average insert size of 5 kb (ranged from 2 to 10 kb) using a MiSeq sequencer (Illumina, CA, United States). DNA contigs were assembled based on their contig coverages using Newbler 2.6 (Nederbragt, 2014). Open reading frames and pseudogenes were predicted using RAST 2.0 (Brettin et al., 2015) combined with BLASTP/BLASTN (Boratyn et al., 2013) searches against the UniProtKB/Swiss-Prot database (Boutet et al., 2016) and the RefSeq database (O’Leary et al., 2016). Annotation of resistance genes, mobile elements, and other features was carried out using the online databases including CARD (Liang et al., 2017), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and the Tn Number Registry (Roberts et al., 2008). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 0.48.11.
Phylogenetic Analysis
The nucleotide sequences of repA coding regions of indicative plasmids were aligned using MUSCLE 3.8.31 (Edgar, 2004). The unrooted neighbor-joining trees were generated from the aligned repA sequences using MEGA7 (Kumar et al., 2016), and evolutionary distances were estimated using the maximum composite likelihood method, with a bootstrap iteration of 1000.
Phenotypic Assays
Activity of Ambler class A/B/D carbapenemases in bacterial cell extracts was determined by a modified CarbaNP test (Wei et al., 2016). Bacterial antimicrobial susceptibility was tested by BioMérieux VITEK 2 and interpreted as per the 2017 Clinical and Laboratory Standards Institute (CLSI) guidelines (Wayne, 2017).
Nucleotide Sequence Accession Numbers
The sequence of p1160-VIM and that of the 6762 chromosome were submitted to GenBank under accession numbers MF144194 and CP030075, respectively.
Results and Discussion
Overview of Sequenced p1160-VIM and Tn6413
Two blaVIM-positive P. aeruginosa isolates, designated 1160 and 6762, were subjected to high-throughput genome sequencing. The 1160 isolate harbored a blaVIM–24-carrying plasmid p1160-VIM, which had a circular DNA sequence of 205.4 kb in length, with an average G+C content of 56.3%. p1160-VIM belonged to the IncP-7 group because it had a IncP-7 repA gene responsible for plasmid replication initiation.
A 114.1-kb blaVIM–4-harboring ICE Tn6413 was found to integrate into tRNAGly gene in the 6762 chromosome. The modular structure of each of p1160-VIM and Tn6413 was divided into the backbone (responsible for replication, maintenance and conjugal transfer) and separate accessory modules (defined as acquired DNA regions associated with mobile elements) integrated at different sites of the backbone (Supplementary Figures S1, S2 and Table 1).
p1160-VIM could be transferred from the 1160 isolate into P. aeruginosa PAO1 through conjugation, generating the transconjugant 1160-VIM-PAO1. The self-transmissible nature of p1160-VIM was consistent with the presence of complete conjugal transfer regions in this plasmid. Strains 1160 and 1160-VIM-PAO1 had class B carbapenemase activity, and they were resistant to cefuroxime, ceftazidime, ceftriaxone and cefepime (with minimal inhibitory concentration values ≥ 64), and imipenem and meropenem (with minimal inhibitory concentration values ≥ 4), which were resulted from production of VIM enzymes in these strains. Repeated conjugation attempts failed to transfer Tn6413 from the 6762 isolate to PAO1.
Subgrouping of IncP-7 Plasmids Including p1160-VIM
A group of ten completely or partially sequenced plasmids (Supplementary Table S1; including p1160-VIM) with IncP-7 repA genes (≥ 95% nucleotide identity to that of p1160-VIM), were collected, and two phylogenetic trees (Figure 1) were constructed based on repA nucleotide and amino acid sequences, respectively. These ten plasmids could be divided into two separately clustering subgroups designated IncP-7α and IncP-7β. As shown by pairwise comparison of repA nucleotide sequences, plasmids within each of these two subgroups showed ≥ 99% nucleotide identity, while plasmids from these two different subgroups displayed ≤ 96% nucleotide identity (Supplementary Table S2a). Considerable genetic diversity was found between the repA genes of IncP-7α and IncP-7β, representing two separated lineages.
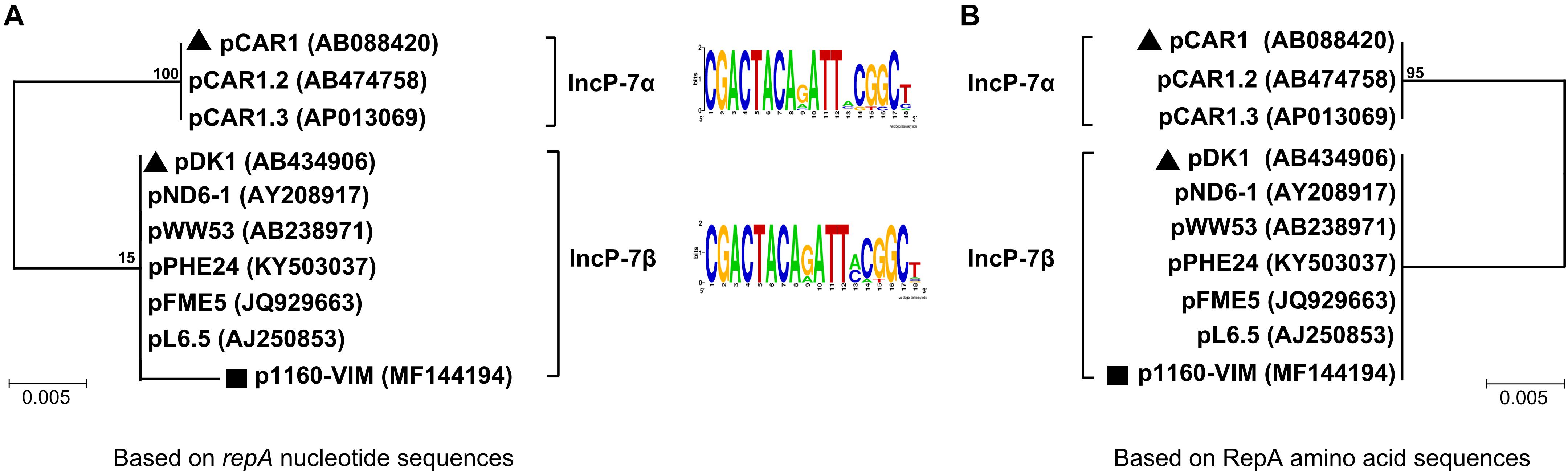
Figure 1. Neighbor-joining phylogenetic trees of sequenced IncP-7 plasmids. Phylogenetic trees are constructed based on repA nucleotide (A) and amino acid sequences (B), respectively. Degree of support (percentage) for each cluster of associated taxa, as determined by bootstrap analysis, is shown next to each branch. Triangles indicate IncP-7α and IncP-7β reference plasmids, while squares denote p1160-VIM sequenced in this work.
Predicted iterons (RepA-binding sites) were found within the oriV region downstream of repA, and plasmids from both subgroups shared a conserved iteron motif and an identical iteron copy number (Figure 1 and Supplementary Table S1).
pCAR1 (Maeda et al., 2003) and pDK1 (Kunz and Chapman, 1981) were identified as IncP-7α and IncP-7β reference plasmids, respectively, because they were the first sequenced plasmids harboring complete conjugal transfer regions. In the phylogenetic tree based on nucleotide sequences, p1160-VIM displayed a long branch, which resulted from presence of five single nucleotide polymorphisms (SNPs) in p1160-VIM, while all other plasmids had identical repA sequences (Supplementary Figure S3). Notably, these five SNPs did not lead to mutations of RepA amino acid sequences.
Comparison of p1160-VIM With pCAR1 and pDK1
pCAR1, pDK1 and p1160-VIM were included in a genomic comparison. These three plasmids had > 92% nucleotide identity across > 52% of their backbone sequences (Supplementary Table S2b), and their conserved backbone was composed of gene or gene loci responsible for replication initiation (repA), partitioning (parABCW), and conjugal transfer (rlx, cpl, tivF3, and tivF6). There were three major modular differences within their backbones (Figure 2): (i) a terABC region could be found in p1160-VIM and pCAR1 rather than pDK1; (ii) a 23.9-kb orf324–to–orf891 region was found in only p1160-VIM; and (iii) the endA gene was intact in pCAR1 but was interrupted or truncated in p1160-VIM and pDK1. All these modular differences were resulted from integration of relevant accessory modules.
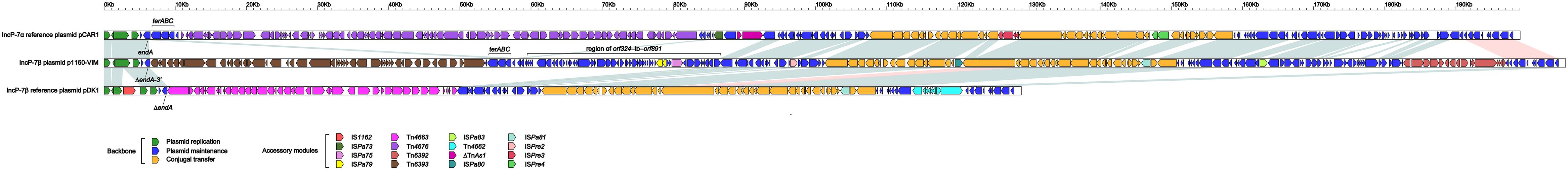
Figure 2. Linear comparison of p1160-VIM with related plasmids. A linear comparison is carried out for complete DNA sequences of pCAR1 (accession number AB088420), p1160-VIM (this study), and pDK1 (accession number AB434906). Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Blue shading denotes regions of homology (> 95% nucleotide identity), and pink shading denotes regions of homology (nucleotide identity between 90 and 95%).
pCAR1, pDK1, and p1160-VIM carried totally different profiles of accessory modules (Table 1), which were composed of 10 distinct IS elements (ISPre2, ISPre3, ISPre4, IS1162, ISpa73, ISpa75, ISpa79, ISpa80, ISpa81, and ISpa83), 5 different intact Tn3-family unit transposons (Tn4676 from pCAR1, Tn4662, and Tn4663 from pDK1, and Tn6392 and Tn6393 from p1160-VIM; a typical unit transposon encodes a transposase and a site-specific recombinase or resolvase as core transposition determinants, and also carries one or several accessory genes), and one Tn3-family transposon remnant. Only Tn6392 and Tn6393 of the above accessory modules (Table 2). Tn4676 (Supplementary Figure S4a) carried core transposition genes (tnpAC and tnpST) genetically related to Tn4651 (Maeda et al., 2003), and also an ant (two-component anthranilate 1,2-dioxygenase) operon (Urata et al., 2004) interrupted by insertion of ISPre1 and a car (carbazole/dioxin degradation) operon (Nojiri et al., 2001). Tn4662 encoded a RelBE toxin-antitoxin system involved in plasmid maintenance. Tn4663 (Supplementary Figure S4b) was derived from Tn4659 (Yano et al., 2007) and harbored a toluene-catabolic xyl gene cluster (Yano et al., 2010).
Comparison of Tn6392 With Tn5563
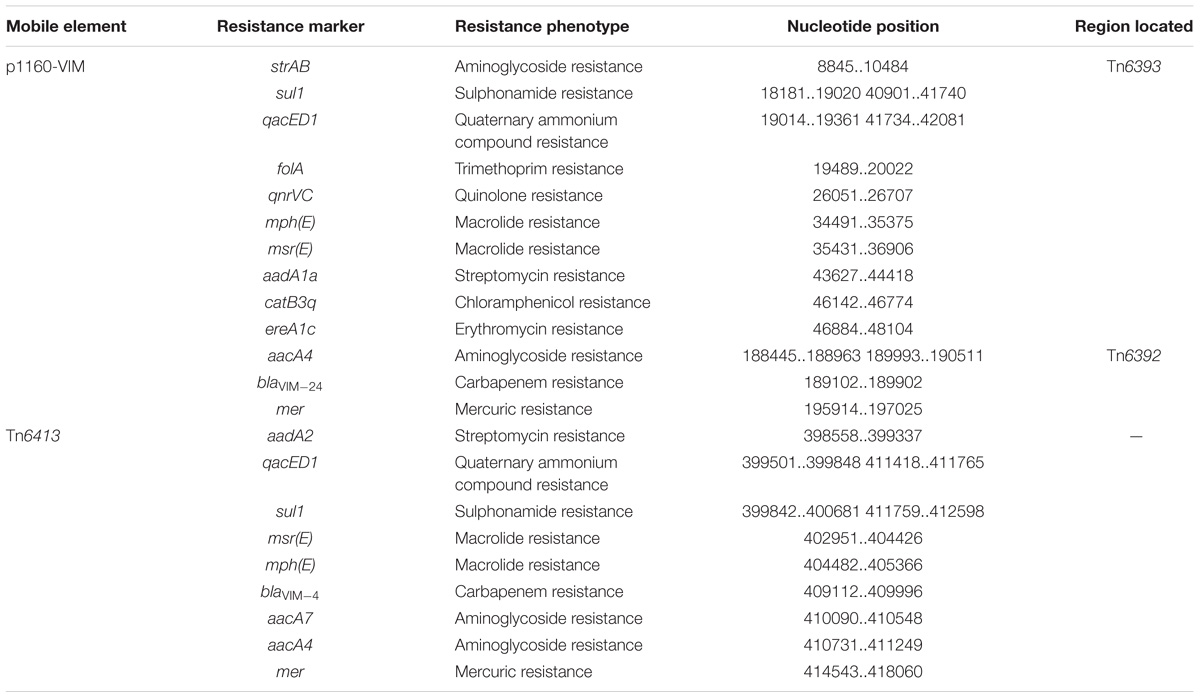
Tn6392 (Figure 3) from p1160-VIM was a novel derivative of Tn5563, which was originally characterized in P. alcaligenes and had the structure IRL (inverted repeat left)–tnpR (resolvase)–res (resolution site)–orf2 (hypothetical protein)–pliT (pilT domain-containing protein)–tnpA (transposase)–mer (mercuric resistance gene locus)–IRR (inverted repeat right), bracketed by 5-bp or 7-bp direct repeats (DRs; target site duplication signals) at both ends (Yeo et al., 1998). Tn6392 differed from Tn5563 by insertion of a novel class 1 integron In1394 into res. The prototype Tn402-associated class 1 integron was typically organized as IRi (inverted repeat at the integrase end), 5’-CS [5’-conserved segment: intI1 (integrase)-attI1 (a specific recombination site)], GCA (gene cassette array), 3’-CS [3’-conserved segment: qacED1–sul1–orf5–orf6], a Tn402 tni module [tniA (transposase)–tniB (ATP-binding protein)–tniQ (transposition auxiliary protein)–res–tniR (serine resolvase)], and IRt (inverted repeat at the tni end) (Gillings et al., 2008). In1394, bracketed by 5 bp DRs at both ends, contained all the above core integron structures except 3’-CS. The GCA of In1394 consisted of a blaVIM–24 gene and two copies of aacA4.
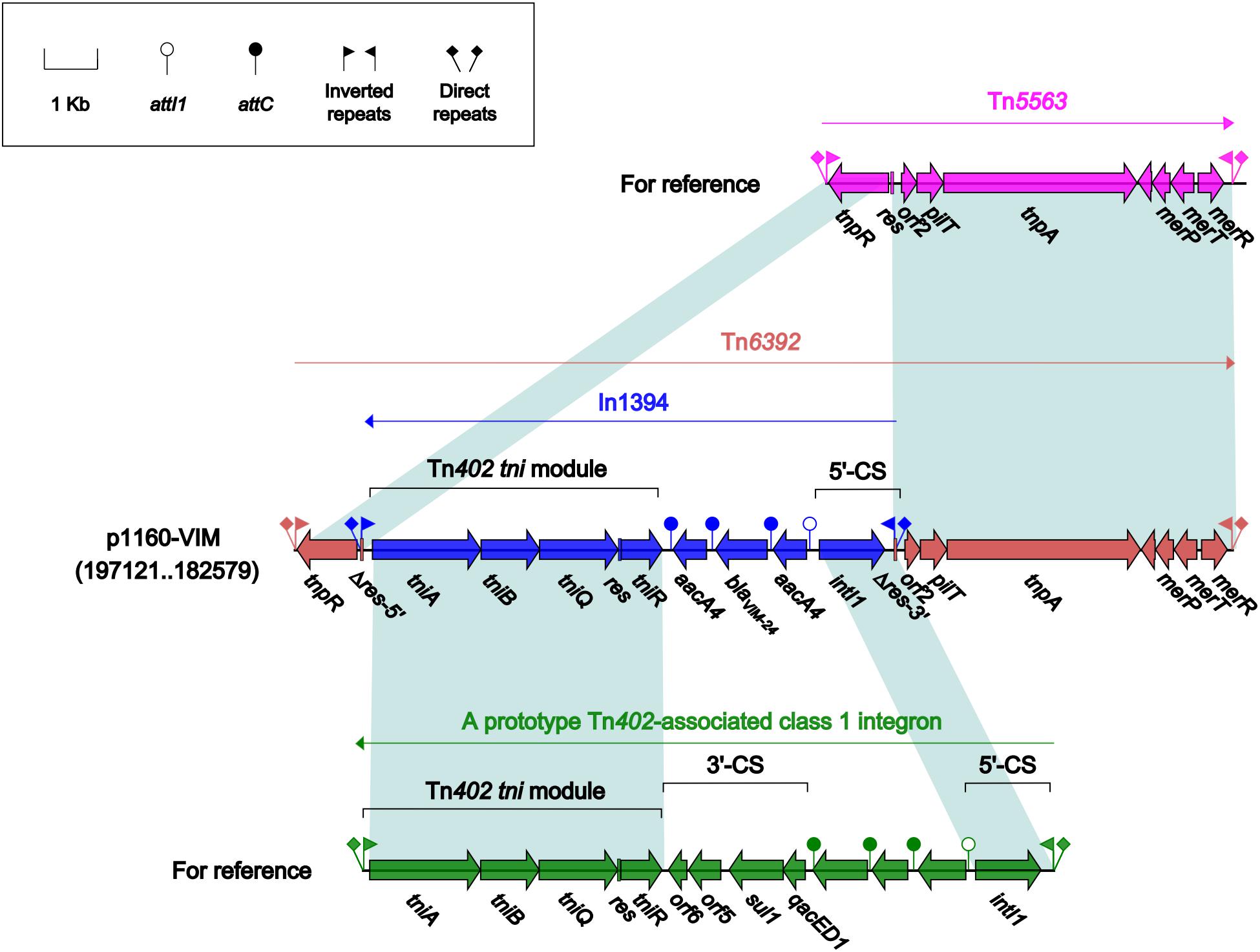
Figure 3. Organization of Tn6392 and comparison to related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity > 95%). Numbers in brackets indicate nucleotide positions within corresponding plasmid sequences. Accession number of Tn5563 for reference is U88088.
Comparison of Tn6393 With Tn1403
Tn6393 (Figure 4) was a novel derivative from Tn1403 after insertion of a novel class 1 integron In1395 instead of In28 at the same position within res. Tn1403 was initially identified in P. aeruginosa and displayed a backbone structure IRL–tnpAR–res–sup–uspA–dksA–yjiK–IRR, with integration of accessory modules In28 and Tn5393c into res and dksA, respectively (Stokes et al., 2007). In1395 belonged to complex class 1 integron, which was typically organized as IRi–5’-CS–VR1 (variable region 1)–3’-CS1 (the first copy of 3’-CS1: qacED1–sul1)–ISCR1 (comment region)–VR2 (variable region 2)–3’-CS2 (a second 3’-CS: qacED1–sul1–orf5–orf6)–tni–IRt. In1395, bracketed by 5-bp DRs at both ends, was composed of IRi, 5’-CS, VR1 [GCA: gcu104–aacA1–catB3q:ISpa62– gcu161–ereA1c:ISpa62], 3’-CS1, ISCR1 (further interrupted by ΔTn4662b–ΔIScs605–msr(E)–mph(E)–Tn4662b), VR2 [containing qnr, ΔISCR1, folA and other genes], 3’-CS2, IS6100 (replacing tni) and IRt.
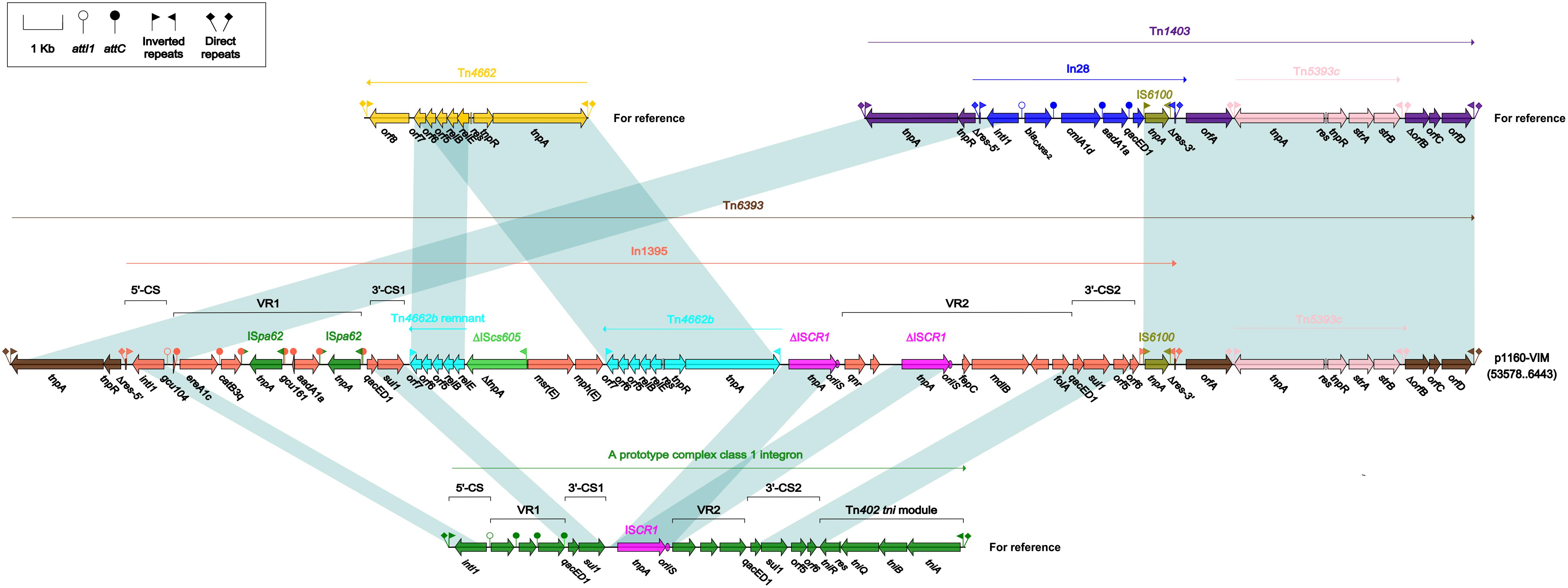
Figure 4. Organization of Tn6393 and comparison to related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity > 95%). Numbers in brackets indicate nucleotide positions within corresponding plasmid sequences. Accession number of Tn1403 for reference is AF313472.
Comparison of Tn6413 With Tn6534, Tn6533, and Tn6417
Tn6413 (Supplementary Figure S2) was a novel ICE that could be divided into a single 30-kb accessory module Tn6403 (Figure 6) and the remaining backbone regions. Tn6413 belonged to a collection of 31 ICE or ICE-like sequences (Supplementary Table S3, including Tn6417, Tn6534, and Tn6533) with > 95% nucleotide identity across > 59% of Tn6413 backbone. Tn6417 was the first sequenced one and identified as the reference of these 31 Tn6417-family ICE sequences. A genomic comparison (Figure 5) was subjected to Tn6413, Tn6534, Tn6533, and Tn6417 because they shared mostly highly similar backbones with 99% nucleotide identity and > 94% query coverage. These four Tn6417-family ICEs, which genetically differed from the two existing ICE families in P. aeruginosa (Kung et al., 2010) shared conserved DNA processing and conjugation genes. Three major modular differences were fund within the backbones of these four ICEs: (i) presence of orf348 in only Tn6417; (ii) presence of orf645 in only Tn6417; and iii) 3’-terminal regions (orf432–to–orf1188, orf693–to–orf866, orf798–to–orf468, and orf693–to–orf1068 from Tn6413, Tn6534, Tn6533 and Tn6417, respectively) differed from one another.
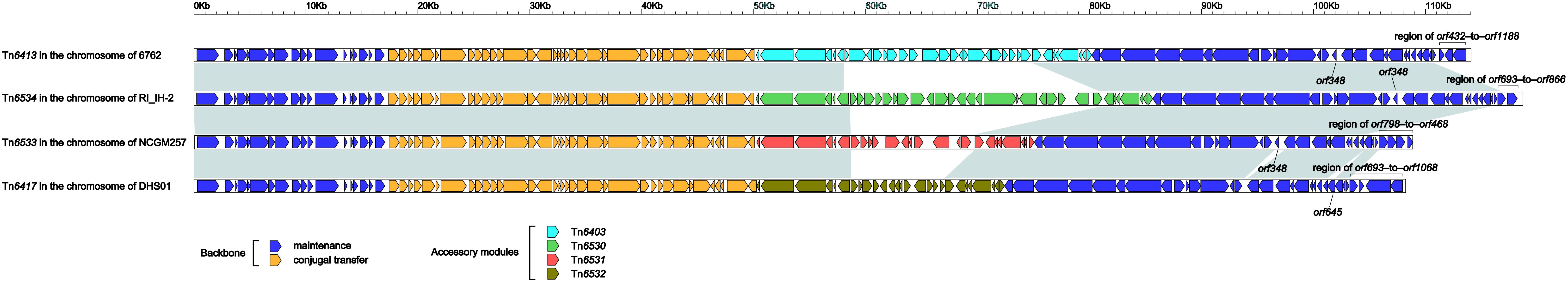
Figure 5. Linear comparison of Tn6413 with related ICEs. A linear comparison is carried out for DNA sequences of Tn6413 (this study), Tn6534 (accession number KX196168), Tn6533 (accession number AP014651), and Tn6417 (accession number CP013993). Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (> 95% nucleotide identity).
Each of these four Tn6417-family ICEs carried a single accessory module: Tn6403, Tn6531, Tn6530, and Tn6532 (Figure 6) from Tn6413, Tn6533, Tn6534, and Tn6417, respectively; all these accessory modules were integrated at the same site of the ICE backbones and identified as Tn6346 derivatives. The Tn3-family unit transposon Tn6346, originally found in heavy metal-tolerant Achromobacter spp., was a hybrid of the core transposition module tnpAR–res of Tn5051 and the mer region of Tn501 (Ng et al., 2009). Tn6403, Tn6531, Tn6530, and Tn6532 differed from Tn6346 by (i) interruption of original tnpATn6346 due to insertion of IS1071, and (ii) insertion of four different class 1 integrons at the same position within the urf2 gene of mer. Tn6403, Tn6530 and Tn6532, rather than Tn6531, were bracketed by 5-bp DRs.
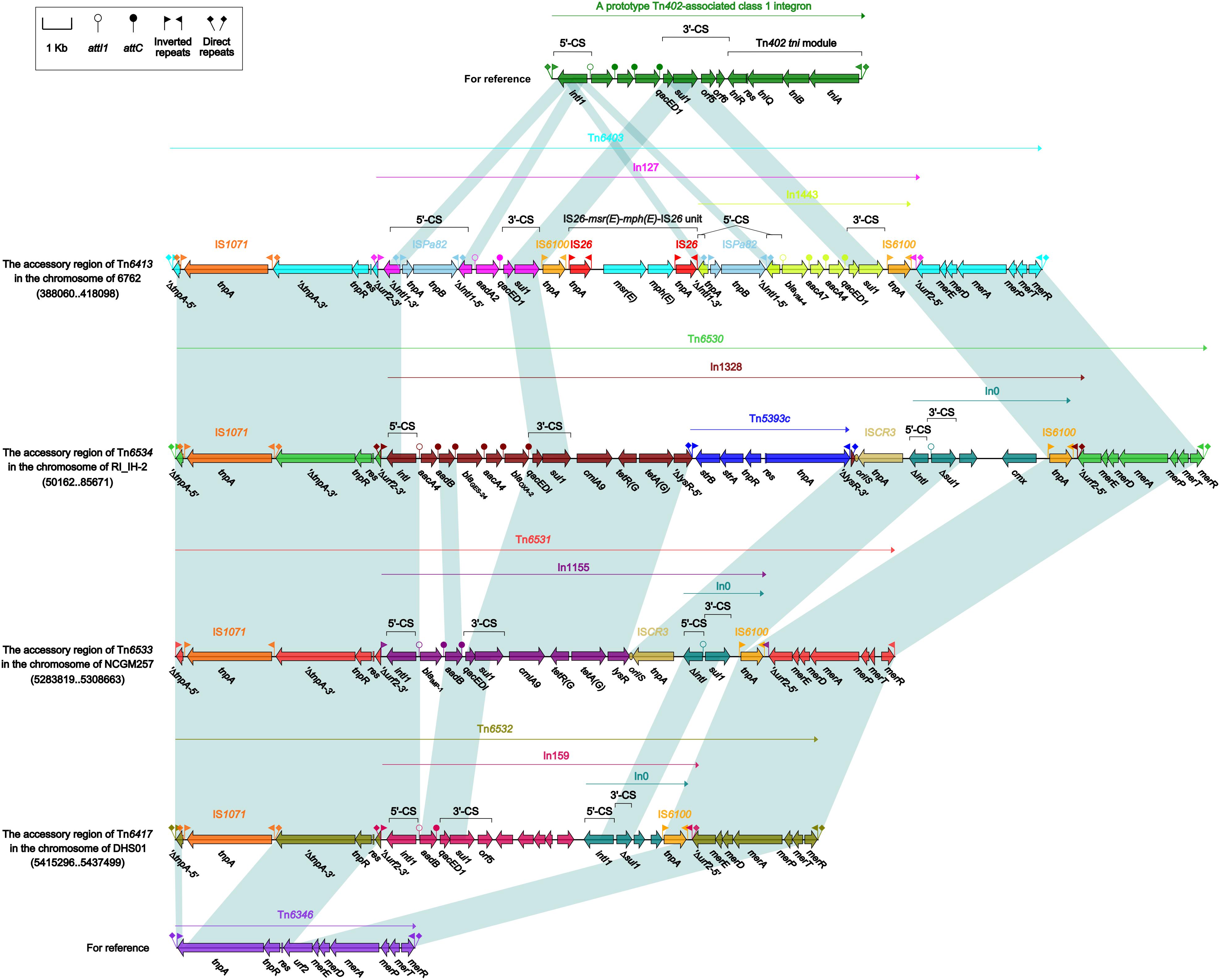
Figure 6. Organization of Tn6403 and comparison with related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity > 95%). Numbers in brackets indicate nucleotide positions within corresponding chromosome sequences. Accession number of Tn6346 for reference is EU696790.
In127, In1328, In1155, and In159 found in Tn6403, Tn6531, Tn6530, and Tn6532, respectively, were intact integrons because all of them had paired terminal 25-bp repeats. Except In1155, all the other three were bracketed by 5-bp DRs. Notably, these integrons captured additional elements beside GCAs: IS26–msr(E)–mph(E)–IS26 unit and a novel blaVIM–4-carrying class 1 integron In1443, cmlA9–tetRA(G)–Tn5393c–ISCR3 and cmx-carrying In0, cmlA9–tetRA(G)–ISCR3 and empty In0, and empty In0 in In127, In1328, In1155, and In159, respectively. In1443 was organized as IRi–5’-CS (interrupted by insertion of ISPa82)–GCA (blaVIM–4–aadA7–aadA4)–Δ3’-CS–IS6100 (replacing tni)–IRt.
Conclusion
IncP-7 R-plasmids are not commonly found in natural isolates, and p1160-VIM represents the first fully sequenced IncP-7 R-plasmid. Based on repA sequences, IncP-7 plasmids can be further divided into two separately clustering subgroups IncP-7α and IncP-7β. The two novel blaVIM-carrying transposons Tn6392 and Tn6413, which are integrated into the IncP-7β plasmid p1160-VIM and the P. aeruginosa chromosome, respectively, represent two different categories of transposons: Tn3-family unit transposon and Tn6417-family ICE. Tn6392 and Tn6413 contain novel class 1 integrons In1394 and In1443, which harbor the two GCAs aacA4–blaVIM–24–aacA4 and blaVIM–4-aadA7–aadA4, respectively. The blaVIM–24 gene was initially discovered from a Klebsiella pneumoniae isolate in Colombia in 2011 (Montealegre et al., 2011). This study presents the first report of a blaVIM–24-carrying P. aeruginosa isolate and a blaVIM-carrying IncP-7 plasmid. Both p1160-VIM and Tn6413 are conjugative (self-transmissible) mobile elements, promoting horizontal transfer of resistance genes carried. Presence of IRi/IRt and a complete tni module would ensure In1394 self-transferable, while replacement of tni by IS6100 would impair mobility of In1443. Class 1 integrons (e.g., In1394 and In1443) could be integrated into a transposon (e.g., Tn6392 and Tn6413) to restore or enhance their mobility.
Ethics Statement
The use of human specimens and all related experimental protocols were approved by the Committee on Human Research of the First Affiliated Hospital of Hebei North University and that of the General Hospital of Xinjiang Military Region, and carried out in accordance with the approved guidelines. The research involving biohazards and all related procedures were approved by the Biosafety Committee of the Beijing Institute of Microbiology and Epidemiology.
Author Contributions
DZ and ZY conceived the study and designed experimental procedures. LZ, ZZ, LH, XJ, and YJZ performed the experiments. LZ, ZZ, YJZ, JF, BG, YEZ, and WY analyzed the data. LZ, ZZ, and HY contributed reagents and materials. DZ, ZY, LZ, and ZZ wrote this manuscript.
Funding
This work was supported by the National Key R&D Program (2018YFC1200100) of China.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00213/full#supplementary-material
FIGURE S1 | Plasmid schematic maps. Three plasmids pCAR1, pDK1 and p1160-VIM are included. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and color, respectively. Innermost circle presents GC-skew [(G–C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. Next-to-innermost circle presents GC content.
FIGURE S2 | ICE schematic maps. Four ICEs Tn6413, Tn6534, Tn6533, and Tn6417 are included. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and color, respectively. Innermost circle presents GC-skew [(G–C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. Next-to-innermost circle presents GC content.
FIGURE S3 | Alignment of repA nucleotide sequences. Red-labeled nucleotides indicate SNP sites. Sequence of IncP-7β reference plasmid pDK1 was bolded.
FIGURE S4 | Organization of Tn4676 or Tn4663 comparison with related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Blue shading denotes regions of homology (nucleotide identity > 95%), and pink shading denotes regions of homology (average nucleotide identity 82%). Numbers in brackets indicate nucleotide positions within corresponding plasmid sequences. Accession number of Tn4651 for reference is AJ344068.
TABLE S1 | Collection of repA and iteron sequences of IncP-7 plasmids.
TABLE S2 | Pairwise comparison of repA and backbone sequences.
TABLE S3 | List of the Tn6417-related sequences.
Footnotes
References
Boratyn, G. M., Christiam, C., Cooper, P. S., George, C., Amelia, F., Ning, M., et al. (2013). Blast: a more efficient report with usability improvements. Nucleic Acids Res. 41, 29–33. doi: 10.1093/nar/gkt282
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-prot, the manually annotated section of the uniprot knowledgebase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Choi, H. J., Kim, M. H., Cho, M. S., Kim, B. K., Kim, J. Y., Kim, C., et al. (2013). Improved PCR for identification of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 97, 3643–3651. doi: 10.1007/s00253-013-4709-0
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Franke, A. E., and Clewell, D. B. (1981). Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of ”conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494.
Gaillard, M., Vallaeys, T., Vorholter, F. J., Minoia, M., Werlen, C., Sentchilo, V., et al. (2006). The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188, 1999–2013. doi: 10.1128/JB.188.5.1999-2013.2006
Gillings, M., Boucher, Y., Labbate, M., Holmes, A., Krishnan, S., Holley, M., et al. (2008). The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 190, 5095–5100. doi: 10.1128/JB.00152-08
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kung, V. L., Ozer, E. A., and Hauser, A. R. (2010). The accessory genome of Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 74, 621–641. doi: 10.1128/MMBR.00027-10
Kunz, D. A., and Chapman, P. J. (1981). Isolation and characterization of spontaneously occurring TOL plasmid mutants of Pseudomonas putida HS1. J. Bacteriol. 146, 952–964.
Li, W., Shi, J., Wang, X., Han, Y., Tong, W., Ma, L., et al. (2004). Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 336, 231–240. doi: 10.1016/j.gene.2004.03.027
Liang, Q., Zhe, Y., Zhao, Y., Liang, L., Jiao, F., Zhe, Z., et al. (2017). Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int. J. Antimicrob. Agents 49, 709–718. doi: 10.1016/j.ijantimicag.2017.01.021
Maeda, K., Nojiri, H., Shintani, M., Yoshida, T., Habe, H., and Omori, T. (2003). Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J. Mol. Biol. 326, 21–33. doi: 10.1016/s0022-2836(02)01400-6
Montealegre, M. C., Correa, A., Briceno, D. F., Rosas, N. C., De La Cadena, E., Ruiz, S. J., et al. (2011). Novel VIM metallo-beta-lactamase variant, VIM-24, from a Klebsiella pneumoniae isolate from Colombia. Antimicrob. Agents Chemother. 55, 2428–2430. doi: 10.1128/AAC.01208-10
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Nederbragt, A. J. (2014). On the middle ground between open source and commercial software - the case of the Newbler program. Genome Biol. 15:113. doi: 10.1186/gb4173
Ng, S. P., Davis, B., Palombo, E. A., and Bhave, M. (2009). A Tn5051-like mer-containing transposon identified in a heavy metal tolerant strain Achromobacter sp. AO22. BMC Res. Notes 2:38. doi: 10.1186/1756-0500-2-38
Nojiri, H., Sekiguchi, H., Maeda, K., Urata, M., Nakai, S. I., Yoshida, T., et al. (2001). Genetic characterization and evolutionary implications of a car gene cluster in the carbazole degrader Pseudomonas sp. Strain CA10. J. Bacteriol. 183, 3663–3679. doi: 10.1128/JB.183.12.3663-3679.2001
O’Leary, N. A., Wright, M. W., Rodney, B. J., Stacy, C., Diana, H., Rich, M. V., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44:D733. doi: 10.1093/nar/gkv1189
Peters, S. E., Hobman, J. L., Strike, P., and Ritchie, D. A. (1991). Novel mercury resistance determinants carried by IncJ plasmids pMERPH and R391. Mol. Gen. Genet. 228, 294–299. doi: 10.1007/BF00282479
Pickup, R. W., and Williams, P. (1985). Evolutionary conservation ofgenescoding formeta pathway enzymeswithin TOL Plasmids pWWO andpWW53. J. Bacteriol. 164, 887–895.
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile -lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/cmr.00001-07
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, 32–36. doi: 10.1093/nar/gkj014
Stokes, H. W., Elbourne, L. D., and Hall, R. M. (2007). Tn1403, a multiple-antibiotic resistance transposon made up of three distinct transposons. Antimicrob. Agents Chemother. 51, 1827–1829. doi: 10.1128/AAC.01279-06
Urata, M., Miyakoshi, M., Kai, S., Maeda, K., Habe, H., Omori, T., et al. (2004). Transcriptional regulation of the ant operon, encoding two-component anthranilate 1,2-dioxygenase, on the carbazole-degradative plasmid pCAR1 of Pseudomonas resinovorans strain CA10. J. Bacteriol. 186, 6815–6823. doi: 10.1128/JB.186.20.6815-6823.2004
Wayne, P. A. (2017). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Informational Supplement M100-S27. Wayne, PA: CLSI.
Wei, F., Zhou, D., Qian, W., Luo, W., Zhang, D., Qiang, S., et al. (2016). Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci. Rep. 6:33419. doi: 10.1038/srep33419
Yano, H., Garruto, C. E., Sota, M., Ohtsubo, Y., Nagata, Y., Zylstra, G. J., et al. (2007). Complete sequence determination combined with analysis of transposition/site-specific recombination events to explain genetic organization of IncP-7 TOL plasmid pWW53 and related mobile genetic elements. J. Mol. Biol. 369, 11–26. doi: 10.1016/j.jmb.2007.02.098
Yano, H., Miyakoshi, M., Ohshima, K., Tabata, M., Nagata, Y., Hattori, M., et al. (2010). Complete nucleotide sequence of TOL plasmid pDK1 provides evidence for evolutionary history of IncP-7 catabolic plasmids. J. Bacteriol. 192, 4337–4347. doi: 10.1128/JB.00359-10
Yeo, C. C., Tham, J. M., Kwong, S. M., Yiin, S., and Poh, C. L. (1998). Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. FEMS Microbiol. Lett. 165, 253–260. doi: 10.1111/j.1574-6968.1998.tb13154.x
Keywords: IncP-7 plasmid, unit transposon, integrative and conjugative element, blaVIM, Pseudomonas aeruginosa
Citation: Zeng L, Zhan Z, Hu L, Jiang X, Zhang Y, Feng J, Gao B, Zhao Y, Yang W, Yang H, Yin Z and Zhou D (2019) Genetic Characterization of a blaVIM–24-Carrying IncP-7β Plasmid p1160-VIM and a blaVIM–4-Harboring Integrative and Conjugative Element Tn6413 From Clinical Pseudomonas aeruginosa. Front. Microbiol. 10:213. doi: 10.3389/fmicb.2019.00213
Received: 11 September 2018; Accepted: 24 January 2019;
Published: 26 February 2019.
Edited by:
Charles W. Knapp, University of Strathclyde, United KingdomReviewed by:
Hong-Yu Ou, Shanghai Jiao Tong University, ChinaMariagrazia Perilli, University of L’Aquila, Italy
Copyright © 2019 Zeng, Zhan, Hu, Jiang, Zhang, Feng, Gao, Zhao, Yang, Yang, Yin and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhe Yin, amVycnk5eWluQDE2My5jb20= Dongsheng Zhou, ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t
†These authors have contributed equally to this work
 Lijun Zeng1,2†
Lijun Zeng1,2† Yanjun Zhang
Yanjun Zhang Jiao Feng
Jiao Feng Wenhui Yang
Wenhui Yang Zhe Yin
Zhe Yin Dongsheng Zhou
Dongsheng Zhou