- 1Department of Entolomology, Cornell AgriTech, Cornell University, Geneva, NY, United States
- 2School of Integrative Plant Science, Cornell University, Ithaca, NY, United States
- 3Department of Natural Resources and the Environment, University of New Hampshire, Durham, NH, United States
- 4Department of Natural Resources, Cornell University, Ithaca, NY, United States
Despite decades of research, our understanding of the importance of invertebrates for soil biogeochemical processes remains incomplete. This is especially true when considering soil invertebrate effects mediated through their interactions with soil microbes. The aim of this study was to elucidate how soil macroinvertebrates affect soil microbial community composition and function within the root zone of a managed grass system. We conducted a 2-year field mesocosm study in which soil macroinvertebrate communities were manipulated through size-based exclusion and tracked changes in microbial community composition, diversity, biomass and activity to quantify macroinvertebrate-driven effects on microbial communities and their functions within the rhizosphere. The presence of soil macroinvertebrates created distinct microbial communities and altered both microbial biomass and function. Soil macroinvertebrates increased bacterial diversity and fungal biomass, as well as increased phenol oxidase and glucosidase activities, which are important in the degradation of organic matter. Macroinvertebrates also caused distinct shifts in the relative abundance of different bacterial phyla. Our findings indicate that within the rhizosphere, macroinvertebrates have a stimulatory effect on microbial communities and processes, possibly due to low-intensity microbial grazing or through the dispersal of microbial cells and spores by mobile invertebrates. Our results suggest that macroinvertebrate activity can be an important control on microbially-mediated processes in the rhizosphere such as nitrogen mineralization and soil organic matter formation.
Introduction
Soil microbes are recognized as the main drivers of soil organic matter (SOM) formation and decomposition. However, soil invertebrates can also directly impact SOM processes, particularly through litter decomposition (Coleman, 2008). Soil macroinvertebrates, including earthworms, soil-dwelling insects, myriapods and isopods, are categorized by their larger size (>2 mm) and are known for their roles as ecosystem engineers where they modify soil habitat as well as resource distribution (Jones et al., 1996). This invertebrate group is functionally distinct from other soil biota such as mesofauna (0.1–2 mm) and microfauna (<0.1 mm) (Swift et al., 1979; Beare et al., 1995; Brussaard, 1997; Coleman et al., 2004; Bradford et al., 2007) and through the transport and breakdown of plant litter they are capable of accelerating the incorporation of plant residues into soil (Bradford et al., 2002; Wall et al., 2008; Garcia-Palacios et al., 2013).
While the direct impact of soil macroinvertebrates on soil carbon cycling is notable, it has been proposed that the greatest contribution invertebrates make to soil processes is through their interactions with the soil microbial community (Grandy et al., 2016; Trap et al., 2016). Soil invertebrate-microbe interactions can take diverse direct and indirect forms. For instance, grazing, by macroinvertebrates, such as isopods and millipedes, can reduce fungal biomass (Crowther et al., 2011a), alter fungal community composition (Crowther et al., 2011b) and increase fungal extracellular enzymatic activity (Crowther et al., 2011c). Microbial grazing by macroinvertebrates has also been shown to increase bacterial biomass (Lussenhop, 1992; Dempsey et al., 2013). In addition to affecting microbial communities directly, macroinvertebrates also impact soil microbes indirectly by altering the composition and distribution of microbial resources. For instance, through litter fragmentation and translocation between litter and soil layers, earthworms and millipedes can alter microbial biomass (Maraun and Scheu, 1996; Chang et al., 2017) and community composition (Dempsey et al., 2011, 2013). Furthermore, macroinvertebrates can alter litter chemistry through ingestion and gut passage, changing resource availability and quality for microbes (Filley et al., 2008). These examples illustrate that macroinvertebrates can alter microbial communities through diverse channels and suggest that such alterations may have important consequences for key belowground SOM processes.
There has been a long-standing interest in quantifying the effects of macroinvertebrates on soil microbial dynamics. To date, studies have focused primarily on microbial responses to single species of invertebrates, with the majority of studies focusing on earthworms, isopods or millipedes (Maraun and Scheu, 1996; Groffman et al., 2004; McLean et al., 2006; Crowther et al., 2011a,b,c, 2015; Dempsey et al., 2011). Studies attempting to link macroinvertebrate-driven changes in microbial community composition to shifts in microbial function have highlighted that macroinvertebrates induce strong changes in microbial biomass and can elicit contrasting responses in fungi and bacteria (Dempsey et al., 2013; Chang et al., 2017). However, under natural conditions, microbial communities are exposed simultaneously to diverse macroinvertebrate taxa, each with its own potential to impact microbial communities through distinct pathways. Thus, in order to fully understand the role of macroinvertebrates in shaping soil microbial dynamics in natural settings it will be important to quantify microbial responses to mixed macroinvertebrate communities, which are likely have multiple interactive effects on microbes.
Previous efforts to quantify the impact of soil invertebrates on soil microbial community composition and function have also focused almost exclusively on leaf litter and other surface-confined plant residues as the dominant microbial resource input (e.g., Bradford et al., 2002; Bardgett, 2005; Hättenschwiler et al., 2005), and much less is known about how invertebrates influence microbial interactions with root-derived organic matter. Root-derived carbon differs greatly from foliar litter in both quantity and chemistry (Gale et al., 2000; Jones et al., 2009; Bradford et al., 2012) and it is acknowledged as a major resource for soil food webs (Albers et al., 2006; Pollierer et al., 2007; Elfstrand et al., 2008). Recent studies have also demonstrated that root-derived inputs constitute the dominant source of SOM (Austin et al., 2017; Shi et al., 2018; Sokol et al., 2019). Given the growing awareness of the importance root-derived inputs for SOM formation, and the acknowledged role of soil microbes in driving SOM formation (Cotrufo et al., 2013; Wieder et al., 2014), it is equally important to quantify the effects of soil animals on rhizosphere microbial communities.
The goal of this study was to better understand the impacts of macroinvertebrates on microbial community composition and function in the rhizosphere. We selected an urban grass ecosystem where rhizosphere inputs are important for soil biological communities (De Deyn et al., 2003; Ostle et al., 2007), where soil invertebrates are abundant and diverse (Rochefort et al., 2006; Pouyat et al., 2010) and where SOM cycling and accrual are of particular interest for soil carbon management (Qian and Follett, 2012; Shi et al., 2012). In order to quantify the importance of soil macroinvertebrates as mediators of soil microbial function, we carried out an exclusion-based mesocosm study within the root zone of lawn-type grasses and tracked changes in microbial community composition and activity over 2 years. Because macroinvertebrates are known to graze intensively on fungal biomass, we hypothesized that their inclusion would (1) decrease fungal biomass but stimulate bacterial biomass; (2) decrease fungal diversity and increase bacterial diversity, and overall, stimulate the activity of extracellular enzymes and subsequently C and N mineralization rates. Finally, we anticipated that the above responses to macroinvertebrates would change over the course of the 2- year experiment.
Materials and Methods
Research Site and Experimental Design
This experiment was conducted at the Bluegrass Lane Turf and Landscape Research Center (Ithaca, NY, United States) from 2015 to 2017 in an area dominated by turf-type tall fescue (Festuca arundinacea) and regularly mowed to a height of approximately 7.5 centimeters. Soils at the site are classified as Arkport fine sandy loam (mesic Lamellic Hapludalfs) with a pH of 6.5. Total annual precipitation at the site was 80.8 cm in 2016 and 108.1 cm in 2017 (Northeast Regional Climate Center, 2016–2017). In the fall of 2015, soil mesocosms were installed in 16 replicate 1 m2 plots for the purpose of manipulating soil faunal communities. Thirty-two mesocosms (10 cm height × 10 cm in diameter) were created using stainless steel mesh: 16 with 5 mm openings to allow for colonization by both soil macroinvertebrates and mesoinvertebrates (“macro-mesh”) and 16 with 1 mm openings to restrict colonization by macroinvertebrates (“meso-mesh”). Both mesocosm types allowed for ingrowth of grass roots. All mesocosms were filled with defaunated soils that were collected as one intact core per mesocosm (10 cm depth × 10 cm in diameter) and each core was kept separate throughout the defaunation process. These soils were subsequently sieved (4 mm) then subjected to two heating and freezing cycles (24 h at +80°C, 24 h at -20°C) to eliminate soil fauna (adapted from Huhta et al., 1989; Bardgett et al., 1998). Soils were then adjusted to field moist conditions (based on gravimetric moisture content, recorded at the time of soil collection) and added to mesocosms at about field bulk density. The mesocosms then installed to 10 × 10 cm core holes and buried under an actively growing grass layer (2–3 cm thick) by replacing a live turfgrass plug directly above each newly installed mesocosm. The placement of the live turfgrass plug was done to ensure root growth directly into the mesocosms to mirror the extensive root growth in the grass system. Two mesocosms were placed in adjacent 1 m2 plot in November 2015 and all mesocosms were in a uniform 24 m2 area. In November 2016, 1 year after burial, 16 mesocosms (8 macro-mesh and 8 meso-mesh) were harvested and the remaining mesocosms were harvested in November 2017, 2 years after burial.
Soils were carefully removed from the mesocosms in the lab and divided for soil fauna extractions, soil microbial community analyses and analysis of physical and chemical properties. Soils for microbial analysis were sieved (4 mm). Soils were subsampled for extracellular enzyme activity, microbial biomass through phospholipid fatty acids (PLFAs), bacterial and fungal community composition and diversity and stored at -20°C. Soils for analysis of physical and chemical properties were air-dried and stored at ambient temperature. Ingrown roots within each mesocosm from the turf layer directly above were carefully separated, dried and weighed. Soil samples were also collected directly adjacent to the mesocosms prior to each mesocosm harvest and handled and analyzed in the same way to assess mesocosm effects.
Soil Invertebrates
All macroinvertebrates from harvested mesocosms were collected by hand, counted and identified to major taxonomic groups. A subsample (approximately 150 g) of soil was placed onto modified Berlese funnels for heat extraction of soil mesofauna. Extraction temperature began at 30°C and was increased by 10°C over a 3 day period, to a final extraction temperature of 50°C. Soil invertebrates were identified to major taxonomic groups using Borror and Delong Key for Insects (Triplehorn and Johnson, 2005) for insects and collembola and following the taxonomy in the Key to Major Mite Taxa (Walter, 2005) for mites. Abundances are reported as number of individuals kg-1 dry soil.
Microbial Community Composition and Diversity
DNA was isolated from a 250 mg soil subsample using the PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, United States). For 16S rRNA amplifications, 1 μl of each bacterial primer and 8 μl of 5 PRIME HotMaster Mix (5 PRIME, Inc., Gaithersburg, MD, United States) were added to 1:10 diluted DNA solutions, yielding 20 μL reaction volumes. The universal bacterial primers 341F (5′-195 CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) (Herlemann et al., 2011) were used with overhangs included for index attachment. The PCR protocol for 16S rRNA gene amplifications was as follows: 94°C for 2 min; 25 cycles of 94°C for 20 s, 55°C for 20 s and 72°C for 30 s; with a final elongation at 72°C for 5 min, using a Bio-Rad C1000 Thermal Cycler (Bio-Rad, Hercules, CA, United States). For fungal internal transcriber spacer (ITS) amplifications, 0.5 μl of each primer, 8 μl of 5 prime HotMaster mix, and 1 μl DMSO were added to 1:10 diluted DNA solutions. For ITS amplifications, we used the primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and 58A2R (5′-CTGCGTTCTTCATCGAT-3′) (Gardes and Bruns, 1993; Martin and Rygiewicz, 2005) with the required adaptors attached. The PCR settings for ITS were as follows: 94°C for 3 min; 35 cycles of 94°C for 20 s, 45°C for 30 s and 72°C for 45 s; with a final elongation at 72°C for 5 min, and were conducted on the Bio-Rad C1000 Thermal Cycler.
The amplicons were cleaned with MagBio HighPrep PCR beads (MagBio Genomics, Gaithersburg MD, United States) in clear 96-well plates. The cleaned amplicons received attachments of unique two-barcode index combinations through combination of the following into each well of a 96-well plate: 5 μL of sample, 2.5 μL of forward and reverse primers containing designated barcodes that target the attached overhangs, 2.5 μL of water, and 12.5 μL of Q5 High Fidelity 2X Master Mix (New England Biolabs, Inc., Ipswich, MA, United States). The PCR conditions for index attachment were set as: 98°C for 1 min; 8 cycles of 98°C for 15 s, 55°C for 30 s and 72°C for 20 s; with a final elongation at 72°C for 3 min. The resulting barcoded amplicons were normalized by using the SequalPrep Normalization Kit (Thermo Fisher Scientific, Waltham, MA, United States). A 5 μL aliquot of each normalized sample was added into separate pools for 16S rRNA gene and ITS amplicons. The mixture was concentrated and run on 1.2% agarose gels, and bands of the expected size were excised and processed using the Promega Wizard SV Gel and PCR Clean-Up System (Promega, Middleton, WI, United States) to a final concentration of 30 μL. Samples were sequenced on the Illumina MiSeq at the Cornell Genomics Facility (Ithaca, NY, United States), using a 500-cycle MiSeq Reagent Kit v.2 for the ITS pool, and a 600-cycle MiSeq Reagent Kit v.3 for the 16S rRNA gene pool.
Initial sequence processing was based on the Brazilian Microbiome Project Pipeline (Pylro et al., 2014), with some modifications (see Howard et al., 2017). In mothur v.1.35.1 (Schloss et al., 2009), paired-end sequences were merged (make.contigs), primers trimmed (trim.seqs, pdiffs = 2, maxambig = 0), and singleton sequences removed (unique.seqs - > split.abund, cutoff = 1). In QIIME (Caporaso et al., 2010), clustering of 97% OTUs and chimera removal (RDP Gold and UNITE databases provided by http://www.brmicrobiome.org/) were performed using VSEARCH v.2.82 (Rognes et al., 2016). In mothur, representative OTU sequences were classified (classify.seqs, cutoff = 80) using the GreenGenes v. 13.8 database for 16S rRNA gene sequences and UNITE v.7 database for ITS sequences, and OTUs that were suspected to not be of fungal or bacterial origin were removed (remove.lineage). OTU tables were formatted by QIIME. The number of 16S rRNA gene sequences and ITS sequences were rarefied based off of the minimum available reads per soil sample to normalize inter-sample comparisons for downstream analyses in R. All raw sequencing files were submitted to the NCBI SRA database (SRA accession PRJNA508306).
Fatty Acid Methyl Ester Analysis and Microbial Biomass
Phospholipid-derived fatty acids (PLFAs) were extracted from 1 g of sieved, root-free, freeze-dried soil. We used a modified Bligh and Dyer (1959) extraction procedure (White et al., 1979; Guckert et al., 1985) where a single-phase solvent system (chloroform) was modified to include a phosphate buffer. This initially extracts lipids from only viable microorganisms captured at the time of sampling. Lipid extracts were then fractionated on silicic acid columns into neutral, glyco- and polar lipids. Polar lipids were collected and then methylated with 0.2 M methanolic KOH to form fatty acid methyl esters (FAMEs). Purified FAMEs were brought to volume with hexane before injection onto a Varian 3800 FID GC. FAME identification and quantification of each peak was based on retention time data with known standards from Matreya, LLC®. The polyenoic unsaturated fatty acids, 18:2ω6 and 18:1ω9c, were considered as fungal biomarkers (Bardgett et al., 1996; Bååth, 2003). Bacterial markers included saturated Gram-positive fatty acids (i15:0, a15:0, i16:0, i17:0, and a17:0), monoenoic and cyclopropane unsaturated Gram-negative fatty acids (18:1ω7c and cy19:0), and general bacterial markers (15:0, 16:1ω7c, and 16:1ω7t) and fungal markers included non arbuscular mycorrhizal fungi (18:2ω6,9 and 18:1ω9) and arbuscular mycorrhizal fungi (AMF)(16:1ω5) (Ekelund et al., 2003; Leckie et al., 2004).
Enzyme Activities
Potential soil microbial extracellular enzyme activity was assessed using protocols outlined by Saiya-Cork et al. (2002); Grandy et al. (2008), and Wickings and Grandy (2011). The activities of three hydrolytic enzymes, N-acetyl-β-D-glucosaminidase (NAG), β-glucosidase (BG) and acid phosphatase (PHOS) and two oxidative enzymes, phenol oxidase (POX) and peroxidase (PER), were measured. Soil slurries were created from a 1 g soil subsample from each mesocosm and 120 mL sodium acetate buffer (pH 6.5). Hydrolytic enzyme activities were measured on black 96 well plates receiving one of three different substrates and the fluorescent compound methylumbelliferone (MUB). Oxidative enzymes were measured using clear 96 well plates, receiving L-3,4-dihydroxyphenylalanine (L-DOPA) alone for phenol oxidase or L-DOPA plus hydrogen peroxide for peroxidase. Hydrolytic enzyme plates were incubated for 3–4 h and oxidative enzyme plates were incubated for 22–24 h. Hydrolytic enzyme plates were then run at 360 nm excitation and 460 nm emission wavelengths and oxidative enzyme plates at 450 nm absorbance wavelength using a microplate reader (Synergy, BioTek Instruments, Winooski, VT, United States). Potential enzyme activity for each substrate was calculated as nmol of substrate h-1 g-1 dry soil.
Carbon and Nitrogen Mineralization
Soils from mesocosms collected in the second year (2 years of burial) were used for estimating potential carbon mineralization. Soils were air-dried the day of collection and remoistened 5 days later to 60% water holding capacity. Soil CO2 flux was measured using 30 g of soil in 90 mL serum vials. The serum vials were capped and two headspace measurements were taken, one immediately after capping and one after a pre-determined time to assess accumulation of CO2. The incubation time between the initial and second gas measurement was increased progressively from 2 to 24 h over 30 days. Measurements were taken daily for 15 days then at 2–3 day intervals from days 15 to 30. CO2 concentrations were determined using a LiCor 820 infrared gas analyzer (LiCor, Lincoln, NE, United States). CO2 flux was calculated using the increase in CO2 from the initial to the second measurement and by converting ppm CO2 to μg C-CO2 g-1 soil day-1. Cumulative CO2 over the 30-day incubation period was calculated by consecutively adding the daily flux measurements.
Soil samples collected at the start and at the end of the 30-day incubation period for C mineralization (above) were extracted with 0.05 M K2SO4 to quantify nitrogen mineralization and net nitrification. Ammonium and nitrate were measured using colorimetric assays in clear 96-well plates, modified from Doane and Horwath (2003) and Sinsabaugh (2010). To obtain potential nitrogen mineralization and nitrification values, the start of incubation values were subtracted from the end of incubation values. Accumulation of ammonium, nitrate and net nitrogen mineralization (ammonium + nitrate) results were reported in μg g-1 dry soil day-1.
Statistical Analyses
The overall experimental design was a randomized complete block design with 16 replicate plots and in the end 29 total mesocosms divided across two collection dates. Linear mixed effect models were used to analyze soil invertebrates (macroinvertebrates and mesofauna), microbial biomass through PLFAs and extracellular enzyme activities within the mesocosms. Mesocosm mesh size was treated as the fixed effect and plot and mesocosm were treated as random effects. Measurements after 1 year of burial and 2 years of burial were analyzed separately for these measurements. Root biomass was analyzed separately with a linear mixed effect model with mesocosm mesh size and number of years of burial as fixed effects and plot and mesocosm as random effects. Data normality was determined graphically through histograms and Q–Q plots for residuals. To identify significant mesh size effects, analysis of variance (ANOVA) was used with Satterthwaite’s method to calculate degrees of freedom.
The effects of mesocosm mesh size and burial time on soil bacterial and fungal communities were analyzed via permutational multivariate analysis of variance (PERMANOVA). Bray–Curtis dissimilarities between samples were calculated and non-metric multidimensional scaling (NMDS) was used to visualize differences in the composition of the microbial communities. Invertebrate effects (densities of macroinvertebrates and mesofauna) on the microbial community were visualized using vectors plotting correlations to the ordination. Significance values for the vectors were generated with 999 permutations. The differences in Shannon diversity and relative abundance of each phylum were assessed using linear mixed effect models, where mesocosm mesh size was treated as a fixed effect and plot and mesocosm were treated as random effects. The 2 years of the study were analyzed separately for Shannon diversity and relative abundance of each phylum.
For carbon mineralization, the cumulative and daily measurements were analyzed using repeated measures ANOVA, with mesocosm mesh size and measurement date as between-subject effects and individual mesocosms as within-subject effects. Nitrogen mineralization values were also analyzed using a linear mixed effect model with mesocosm mesh size as a fixed effect and plot and mesocosm as random effects.
Data on soils collected from outside the mesocosms were also analyzed using linear mixed effects models with a fixed effect that included the two mesh sizes and outside as treatments and plot and mesocosm as random effects. This analysis was done in order to distinguish mesocosm effects on soil biota and biological traits with differences for undisturbed soils.
All statistical analyses were performed in R (R Core Team, 2018). The lme4 and lmerTest packages were used for the linear mixed effects models. The phyloseq package was used to aggregate OTU tables. The vegan package was used microbial community analyses (PERMANOVA, Bray–Curtis distances, NDMS, and Shannon diversity). All P-values less than 0.05 were considered significant.
Results
Soil Fauna Within and Outside Mesocosms
Soil macroinvertebrates included earthworms (Lumbricidae), centipedes (Chilopoda), millipedes (Diplopoda), and beetle larvae (Elateridae and Scarabaeidae) (Table 1). Immature lumbricid earthworms were the dominant macroinvertebrate group. Macroinvertebrate densities were significantly different between treatments as macroinvertebrates were absent from the meso-mesh for both years of the study (1 year: F1,14 = 48.8, P < 0.000001, 2 years: F1,10 = 8.75, P < 0.02; Table 1), confirming that the small mesh size excluded macroinvertebrates. Soil mesofauna were comprised mainly of collembola and mites, with no significant differences in mesofaunal densities between treatments in either year (1 year: F1,10 = 1.78, P = 0.21, 2 years: F1,10 = 0.894, P = 0.37; Table 1), confirming that colonization of the mesocosm soils by mesofauna communities was not impacted by mesh size.
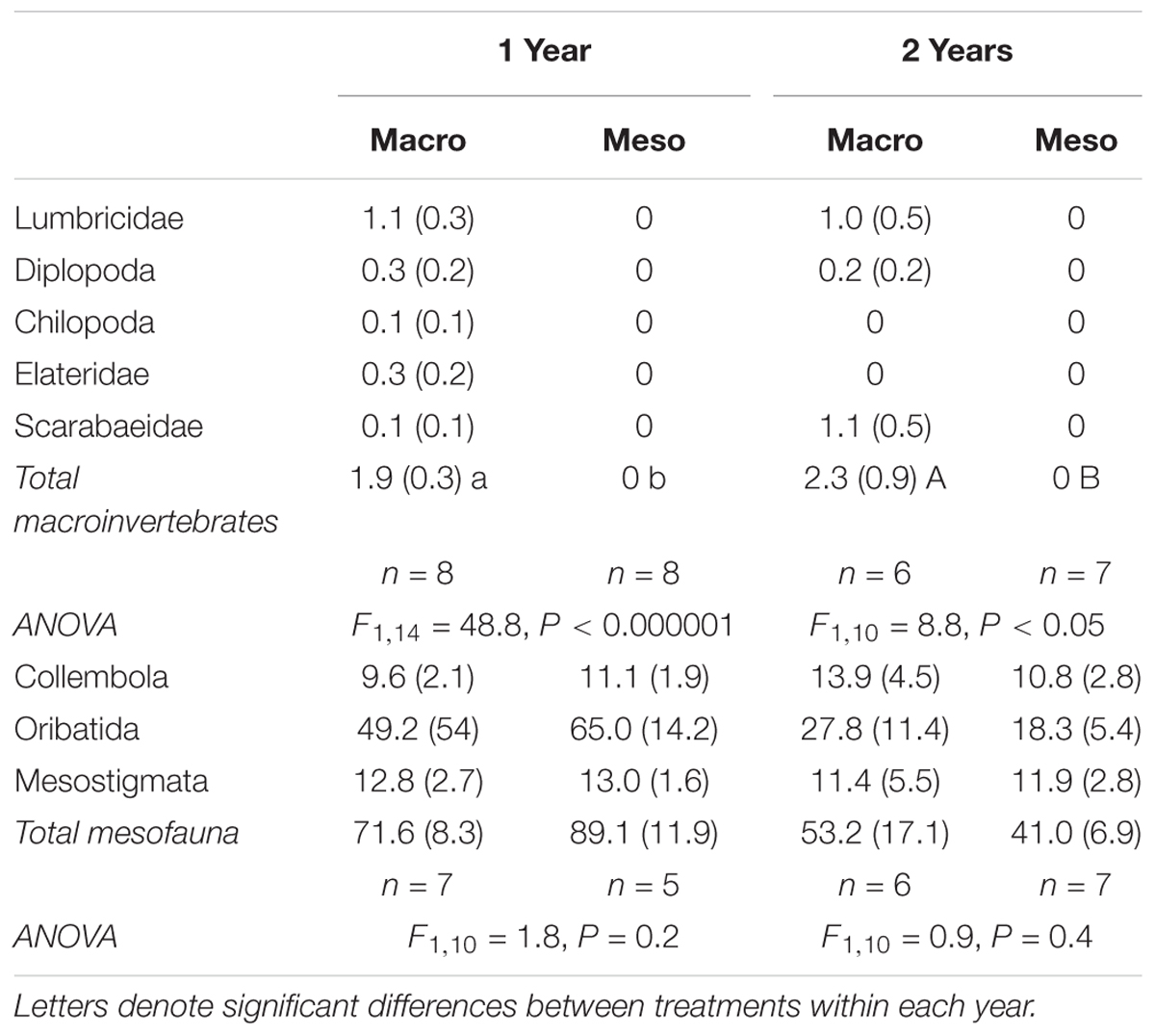
Table 1. Fauna densities (number individuals kg-1 dry soil, average ± standard error) for mesocosms with macroinvertebrates (macro) and mesocosms excluding macroinvertebrates (meso) and associated ANOVA results.
Macroinvertebrate densities inside the mesocosms were significantly lower than outside the mesocosms (1 year: F2,25 = 18.7, P < 0.0001, 2 years: F2,22 = 21.9, P < 0.00001; Supplementary Table 1) where outside mesocosms had 56–62% more macroinvertebrates compared to inside the mesocosms. Similarly, mesofauna densities inside the mesocosms were significantly lower than densities outside the mesocosms (1 year: F2,20 = 28.5, P < 0.00001, 2 years: F2,17.4 = 21.9, P < 0.001; Supplementary Table 1), where outside mesocosm soils had 83–84% more mesofauna. More specifically, for both years of the study, oribatid mites were the least successful colonizers of the mesocosms relative to densities assessed outside the mesocosms (88–90% decrease in oribatid mites inside compared to outside mesocosm) but were the most abundant animal group collected within the mesocosms (Table 1).
Root Biomass Within Mesocosms
After 1 year of burial, average total root biomass within the mesocosms was 0.81 ± 0.36 or 1027 ± 329 g cubic meter soil-1 for macro-mesh and 0.41 ± 0.11 or 522 ± 136 g cubic meter soil-1 for meso-mesh mesocosms. After 2 years of burial, average total root biomass within the mesocosms was 0.62 ± 0.17 or 787 ± 219 g cubic meter soil-1 for macro-mesh and 0.40 ± 0.050 or 511 ± 63.5 g cubic meter soil-1 for meso-mesh mesocosms, respectively. Root biomass within the mesocosms was not significantly different between treatments (F1,24 = 1.85, P = 0.19) or between years (F1,24 = 0.891, P = 0.35) and the treatment-year interaction was also not significant (F1,24 = 0.526, P = 0.48; Table 2). The turfgrass roots growing into the mesocosms extended to the full depth or almost full depth of the mesocosms (10 cm).
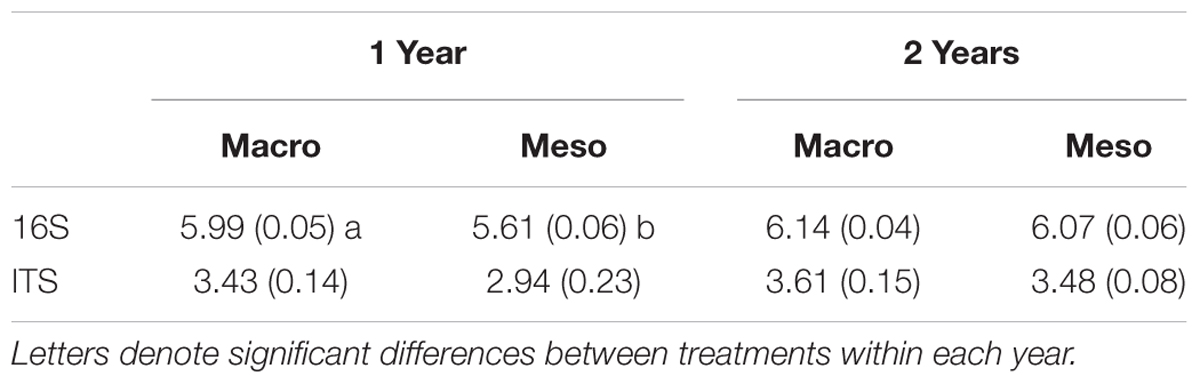
Table 2. Shannon diversity index (average ± standard error) for bacterial diversity and fungal diversity for mesocosms with macroinvertebrates (macro) and mesocosms excluding macroinvertebrates (meso).
Microbial Community Composition and Diversity
Both bacterial and fungal communities (bacterial 16S rRNA gene and fungal ITS) within mesocosms differed significantly between treatments (P < 0.01; Figures 1, 2). Though the differences in the communities were not significant between years (16S: P = 0.63, ITS: P = 0.81), the treatment by time interaction was significant for the bacterial community (16S: P < 0.05, ITS: P = 0.11). NMDS ordination also revealed relationships between soil invertebrates and microbial community structure. For the bacterial community ordination, the stress value was 0.0724 and the non-metric fit R2 was 0.995. For the fungal community ordination, the stress value was 0.134 and the non-metric R2 0.982. Invertebrate community correlations to the bacterial community ordination were significant for total macroinvertebrate density, earthworm density, total mesofauna density and oribatid mite densities (P < 0.05, Figure 1). Invertebrate community correlations to the fungal community ordination were significant for total macroinvertebrate densities and earthworm densities (P < 0.01, Figure 2).
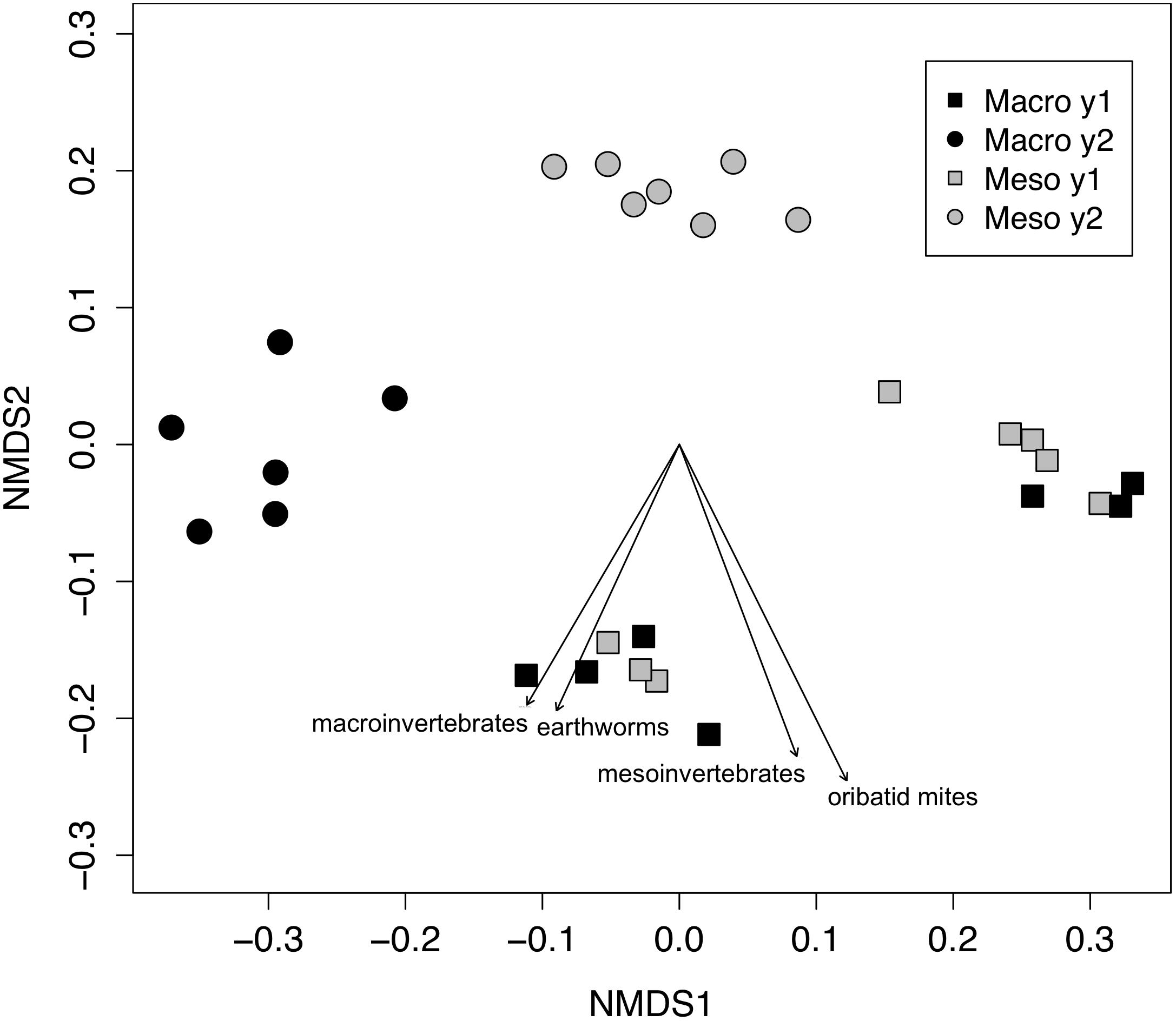
Figure 1. Non-metric multidimensional scaling (NMDS) of bacterial community from 16SrRNA (Bray–Curtis dissimilarities). Macroinvertebrate manipulations resulted in significant shifts in bacterial taxa (PERMANOVA, P < 0.01). Vectors indicate significant correlations (P < 0.05) between invertebrate densities and microbial community ordination scores for ordination axis two. Vectors include total macroinvertebrate density (macroinvertebrates), earthworm (earthworms), total mesofauna density (mesoinvertebrates) and oribatid mite density (oribatid mites). Non-significant vectors are not shown. Black symbols denote microbial communities from soils permitting macroinvertebrates. Gray symbols denote microbial communities from soils excluding macroinvertebrates. Squares indicate communities from year one and circles from year two.
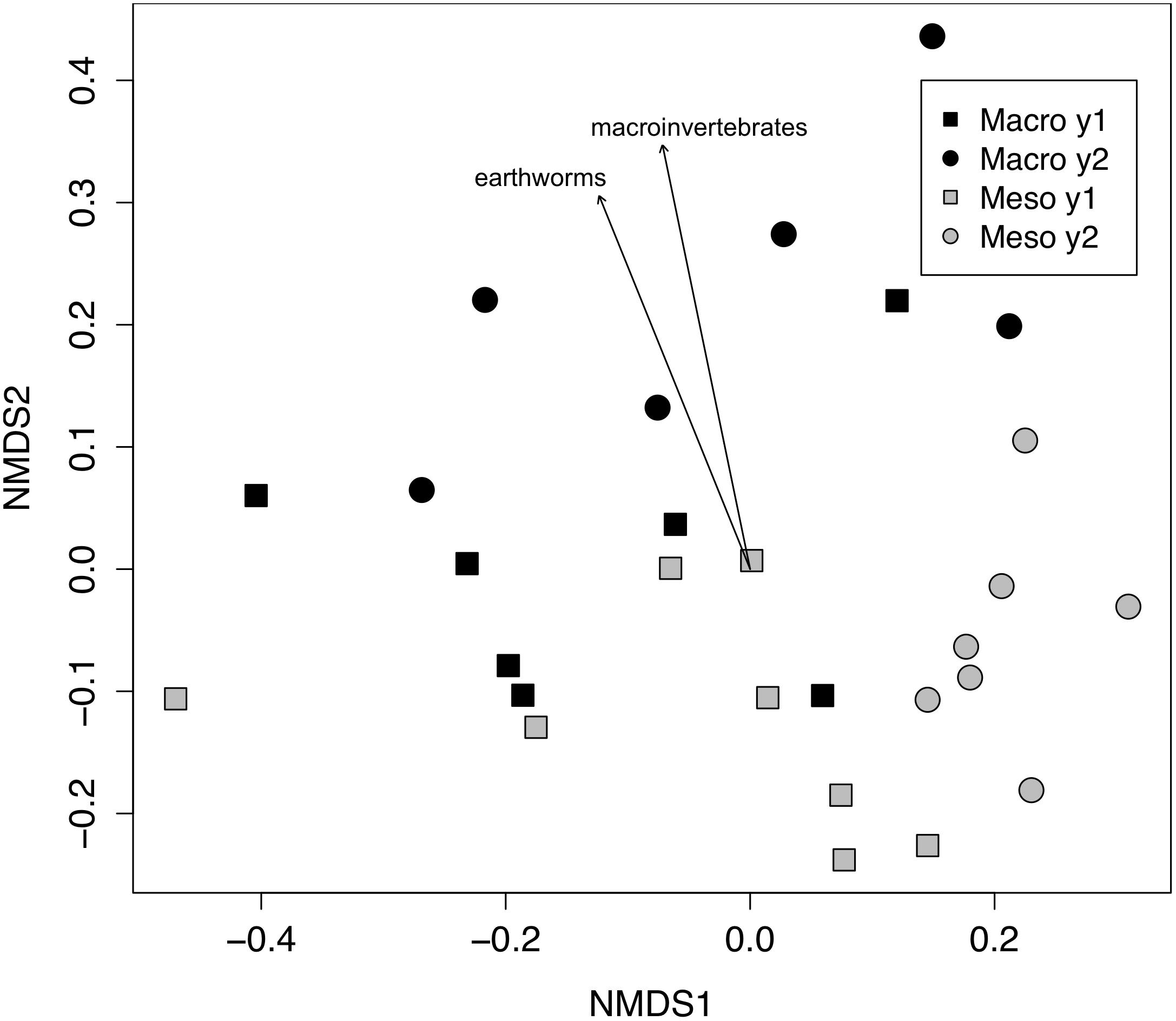
Figure 2. Non-metric multidimensional scaling (NMDS) of fungal community from ITS (Bray–Curtis dissimilarities). Macroinvertebrate manipulations resulted in significant shifts in fungal taxa (PERMANOVA, P < 0.01). Vectors indicate significant correlations (P < 0.01) between invertebrate densities and microbial community ordination scores for ordination axis two. Vectors include total macroinvertebrate density (macroinvertebrates) and earthworm density (earthworms). Non-significant vectors are not shown. Black symbols denote microbial communities from soils permitting macroinvertebrates. Gray symbols denote microbial communities from soils excluding macroinvertebrates. Squares indicate communities from year one and circles from year two.
Shannon diversity of the bacterial community was significantly higher when macroinvertebrates were present after 1 year of burial (F1,5 = 39.7, P < 0.01; Table 2) but this difference was not significant after 2 years of burial (F1,10 = 1.21, P = 0.30, Table 2). Diversity of the fungal community was not significantly altered by treatment in either year (1 year: F1,12 = 3.13, P = 0.10; 2 years: F1,10 = 0.596, P = 0.46; Table 2). Microbial community composition measurements from outside the mesocosms are included in the supplement (Supplementary Figures 1, 2 and Supplementary Table 2).
The most abundant bacterial phyla identified were Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, Planctomycetes, Proteobacteria, and Verrucomicrobia. Relative abundance of Actinobacteria (macro: 15.9 ± 1.6, meso: 23.3 ± 1.6) and Firmicutes (macro: 1.0 ± 0.1, meso: 3.7 ± 0.3) was significantly lower with macroinvertebrates after 1 year of burial (F1,13 = 10.4, P < 0.01; F1,13 = 70.0, P < 0.00001; Supplementary Table 2), whereas the relative abundance of Bacteroidetes (macro: 12.7 ± 1.7, meso: 6.4 ± 0.4) and Verruomicrobia (macro: 8.7 ± 0.4, meso: 6.7 ± 0.4) was significantly higher (F1,13 = 19.5, P < 0.001; F1,13 = 11.9, P < 0.01; Supplementary Table 2). After 2 years of burial, the pattern for the relative abundance of Actinobacteria (macro: 9.0 ± 0.9, meso: 13.0 ± 1.2) and Firmicutes (macro: 1.4 ± 0.3, meso: 3.5 ± 0.5) (F1,10 = 12.9, P < 0.01; F1,10 = 19.8, P < 0.01; Supplementary Table 2) remained the same and was also evident in Planctomycetes (macro: 6.6 ± 0.2, meso: 4.4 ± 0.4) (F1,10 = 21.4, P < 0.001; Supplementary Table 2).
The most abundant fungal phyla were Ascomycota, Basidiomycota, and Zygomycota. There were no significant differences in relative abundance after 1 year of burial; however, the relative abundance of Zygomycota (macro: 17.3 ± 1.5, meso: 32.3 ± 3.5) was significantly lower with macroinvertebrates after 2 years of burial (F1,10 = 13.3, P < 0.01; Supplementary Table 2).
Microbial Biomass (PLFAs)
Total bacterial biomass based upon PLFAs was not significantly different between treatments for either year (1 year: F1,6 = 5.44, P = 0.56; 2 years: F1,7 = 1.09, P = 0.33; Figure 3), although there was a trend toward higher bacterial biomass in year 1 with macroinvertebrates present (Figure 3). Total non-AMF fungal biomass from PLFAs was significantly higher when macroinvertebrates were present for year 1 (+68%) (F1,6 = 8.671, P < 0.05; Figure 3) but not for year 2 (F1,7 = 0.34, P = 0.58). The opposite pattern was observed in AMF biomass, which was significantly higher in mesocosms with macroinvertebrates after 2 years (+48%) (F1,8 = 5.61,1, P < 0.05; Figure 3). Total bacterial and non-AMF fungal biomass from PLFAs was significantly higher when macroinvertebrates were present for year 1 (+82%) (F1,6 = 6.35, P < 0.05; Figure 3) but not for year 2 (F1,8 = 0.81, P = 0.40). Macroinvertebrates did not significantly affect the fungal to bacterial ratio in either year (1 year: F1,7 = 0.0053, P = 0.94; year 2: F1,8 = 51.64, P = 0.24). Complete lipid marker profiles are summarized in Table 3.
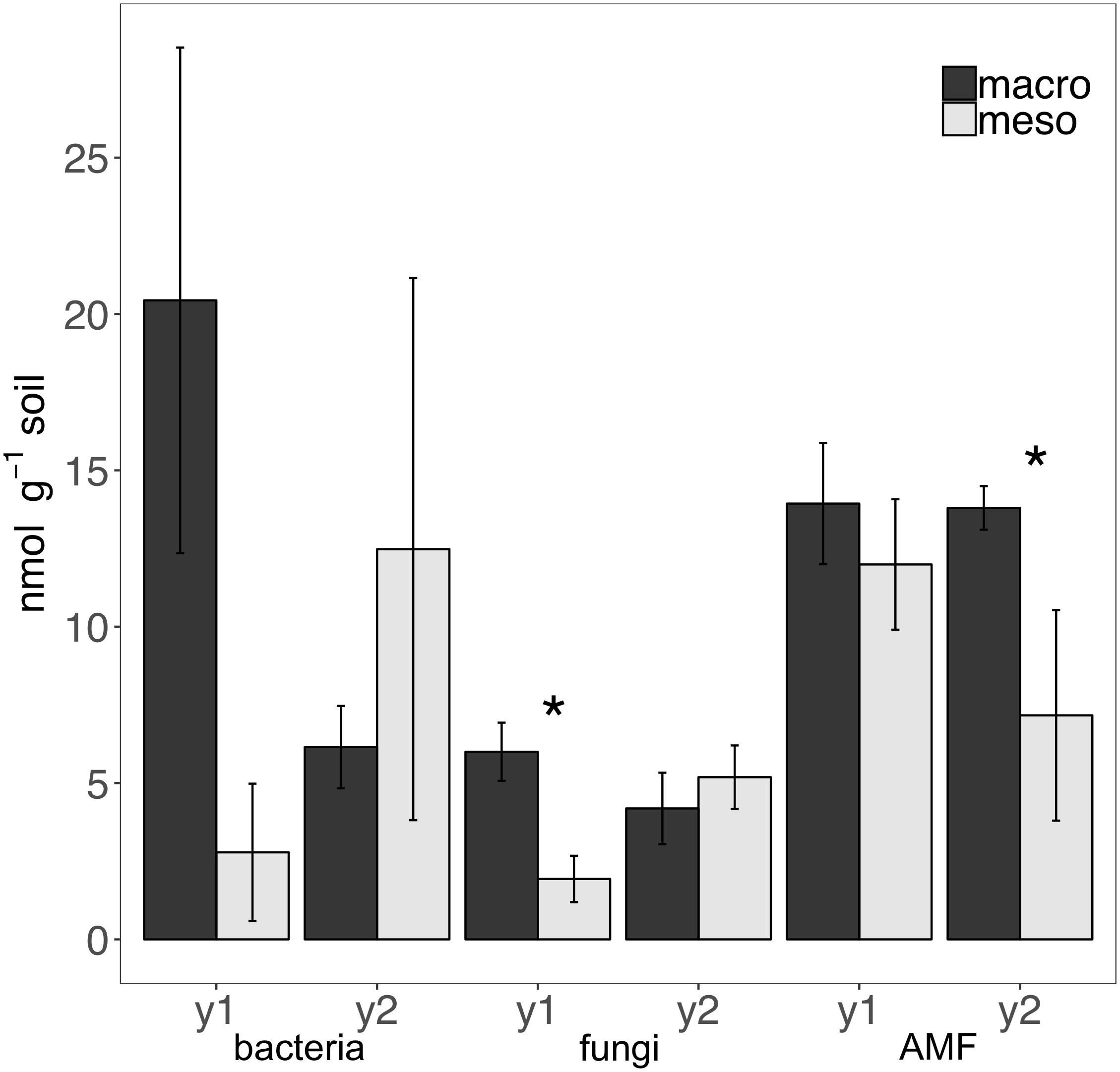
Figure 3. Microbial biomass based on phospholipid fatty acids (PLFAs) (nmol g-1 soil, average ± standard error) in years 1 (y1) and 2 (y2) for total bacteria, total non-AMF fungi (fungi) and AMF. Mesocosms with macroinvertebrates (macro) are represented in dark gray and mesocosms excluding macroinvertebrates (meso) are in light gray. Asterisks denote significant differences between mesocosm treatments within years (P < 0.05).
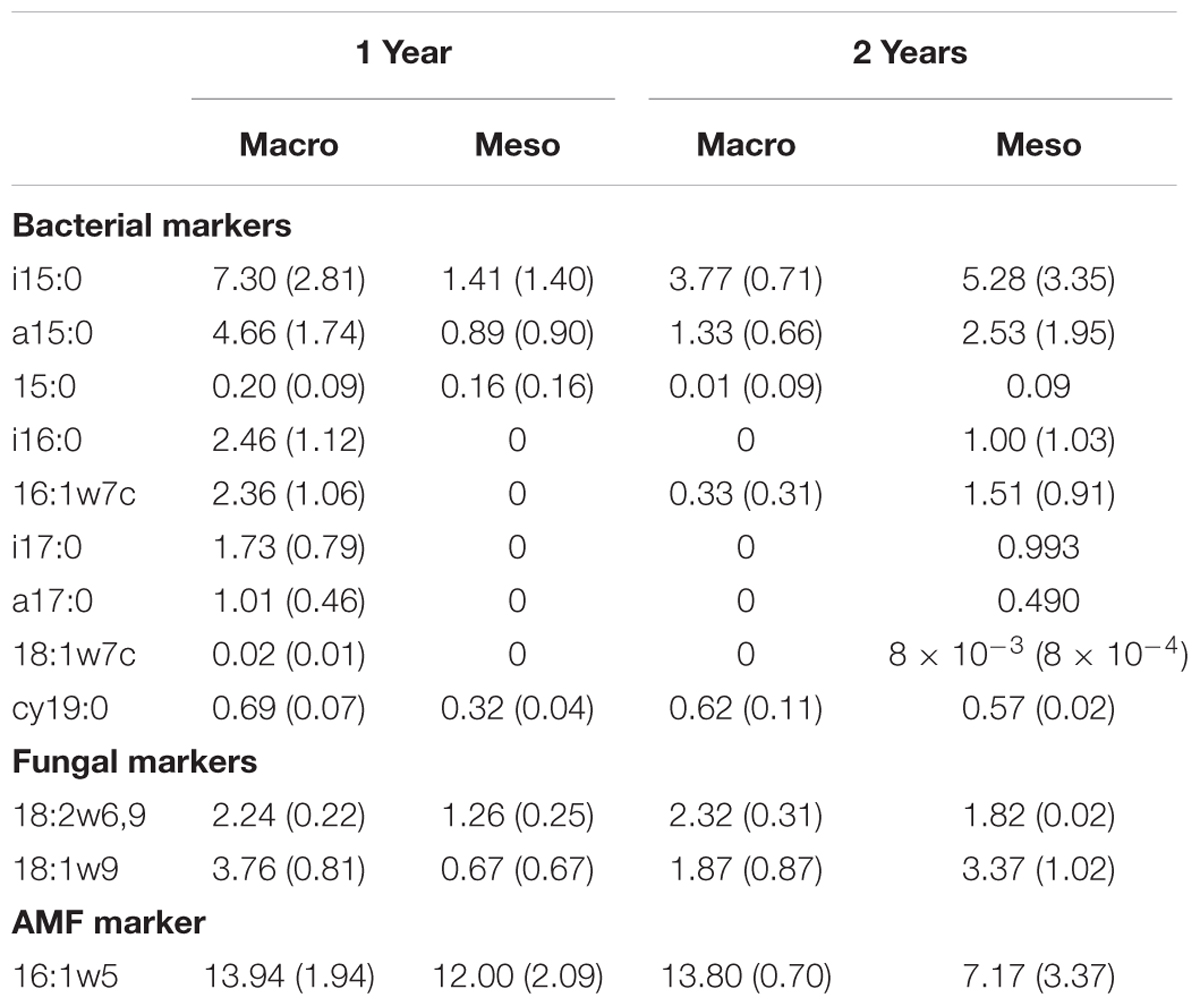
Table 3. Phospholipid fatty acid (PLFA) lipid biomarkers (nmol g-1 soil, average ± standard error) for bacteria, fungi, and AM fungi across both years for mesocosms with macroinvertebrates (macro) and mesocosms excluding macroinvertebrates (meso).
Enzymes
The presence of macroinvertebrates increased the activity of phenol oxidase (POX) by 37.5% (F1,13 = 5.38, P < 0.05; Figure 4) after 1 year of burial and β-glucosidase (BG) by 35.3% (F1,12 = 6.82, P < 0.05; Figure 5) after 2 years. Other measured enzymes, NAG, PHOS, and PER showed no differences across treatments in either year of the study (averages for each year reported in Supplementary Table 1).
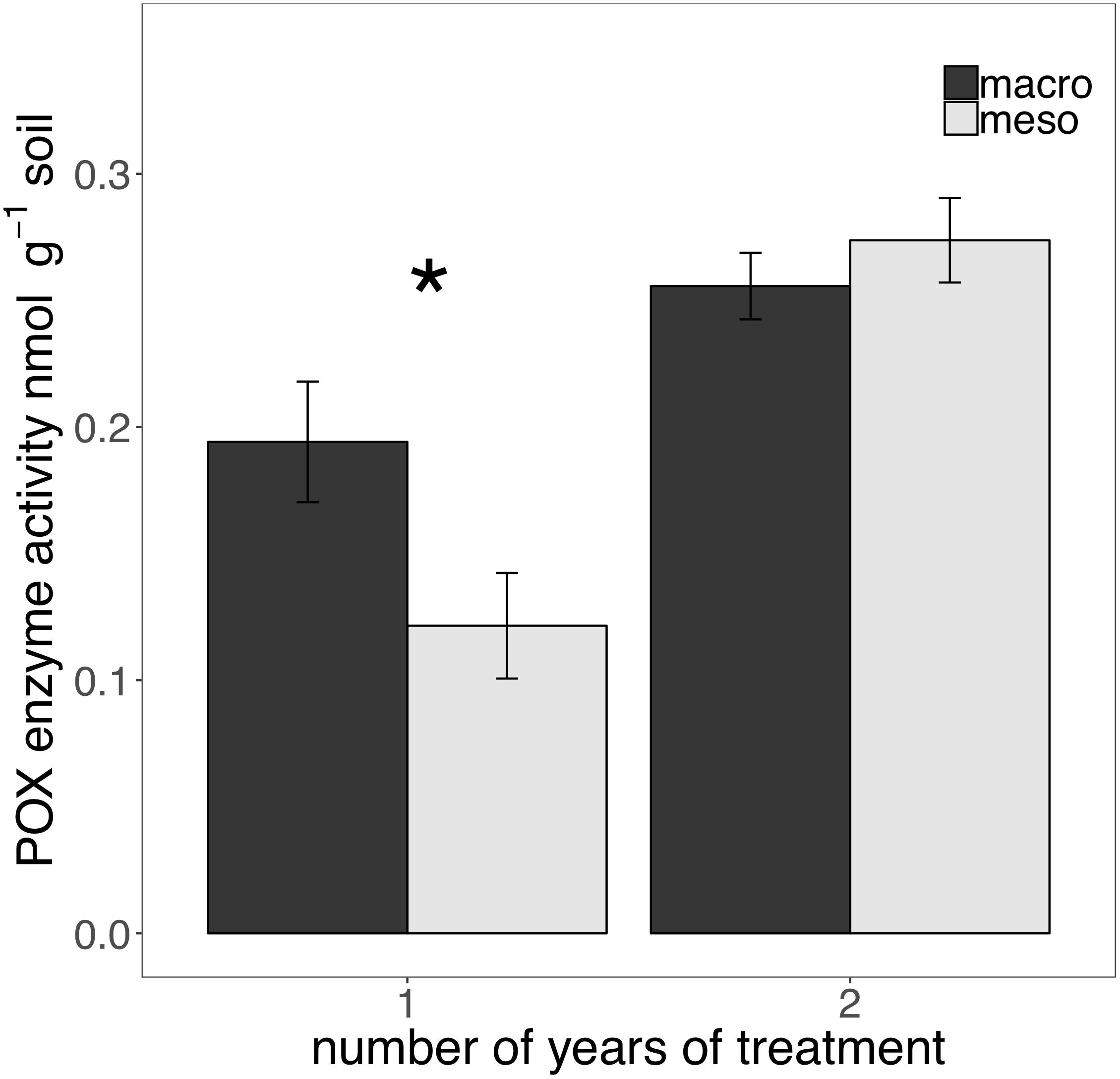
Figure 4. Potential extracellular phenol oxidase (POX) activity (nmol g-1 soil, average ± standard error) in years 1 and 2. Mesocosms with macroinvertebrates (macro) are represented in dark gray and mesocosms excluding macroinvertebrates (meso) are in light gray. Asterisks denote significant differences between mesocosm treatments within years (P < 0.05).
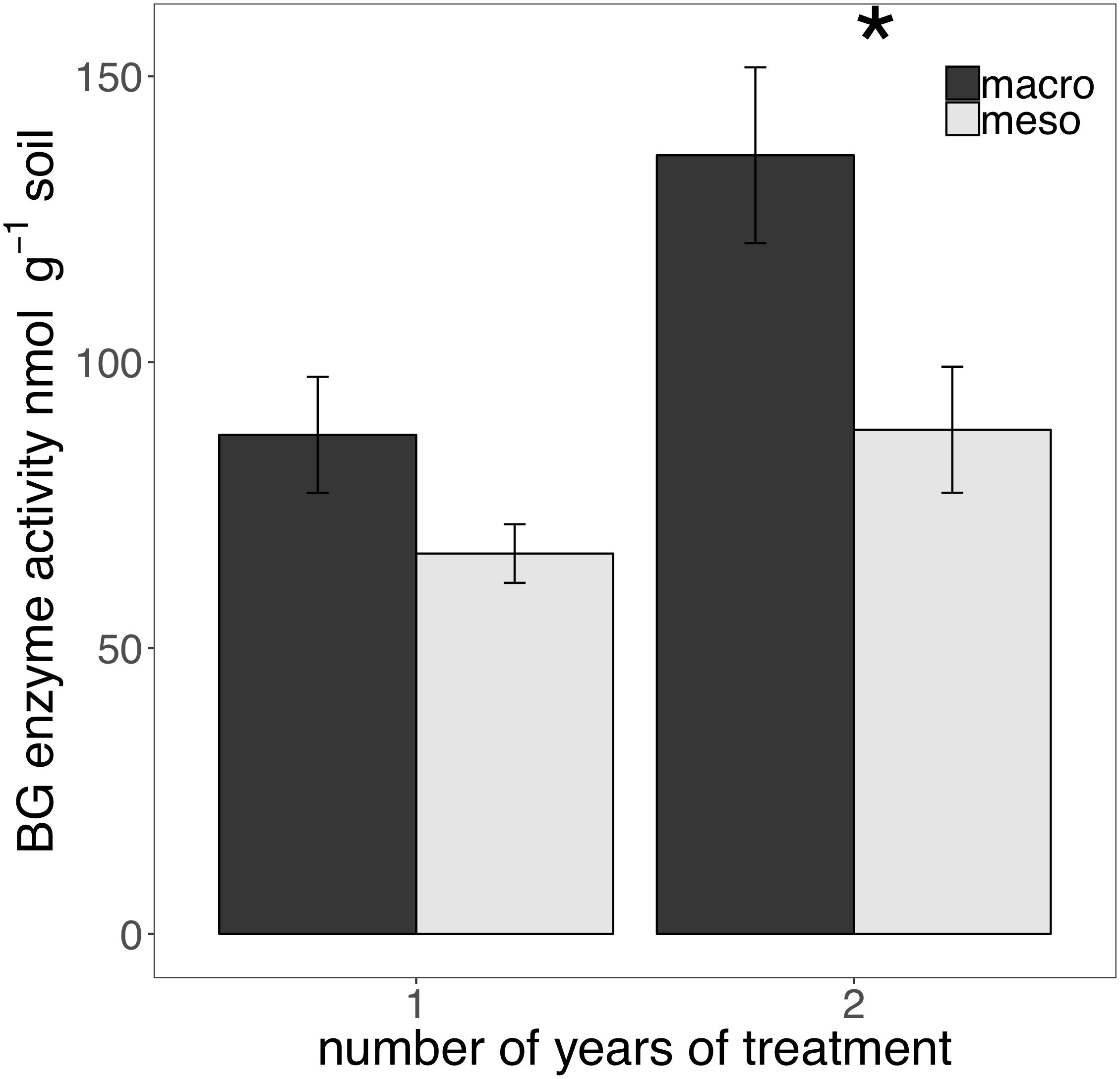
Figure 5. Potential extracellular β-glucosidase (BG) activity (nmol g-1 soil, average ± standard error) in years 1 and 2. Mesocosms with macroinvertebrates (macro) are represented in dark gray and mesocosms excluding macroinvertebrates (meso) are in light gray. Asterisks denote significant differences between mesocosm treatments within years (P < 0.05).
Carbon and Nitrogen Mineralization
Cumulative carbon mineralization potential measured after 2 years differed between the two treatments (F1,19 = 10.9, P < 0.01; Figure 6 and Supplementary Table 3) and was significantly higher when macroinvertebrates were included (+19%). Net ammonium and net nitrate were both affected by mesocosm treatment: net ammonium accumulation was lower (-150%) (F1,7 = 24.4, P < 0.01; Figure 7 and Supplementary Table 3) and net nitrification was higher (+30%) (F1,11 = 25.1, P < 0.01; Figure 7 and Supplementary Table 3) when macroinvertebrates were included. However, potential net nitrogen mineralization (ammonium plus nitrate) was not significantly between the two fauna treatments 2 years after burial (F1,6 = 0.181, P = 0.7).
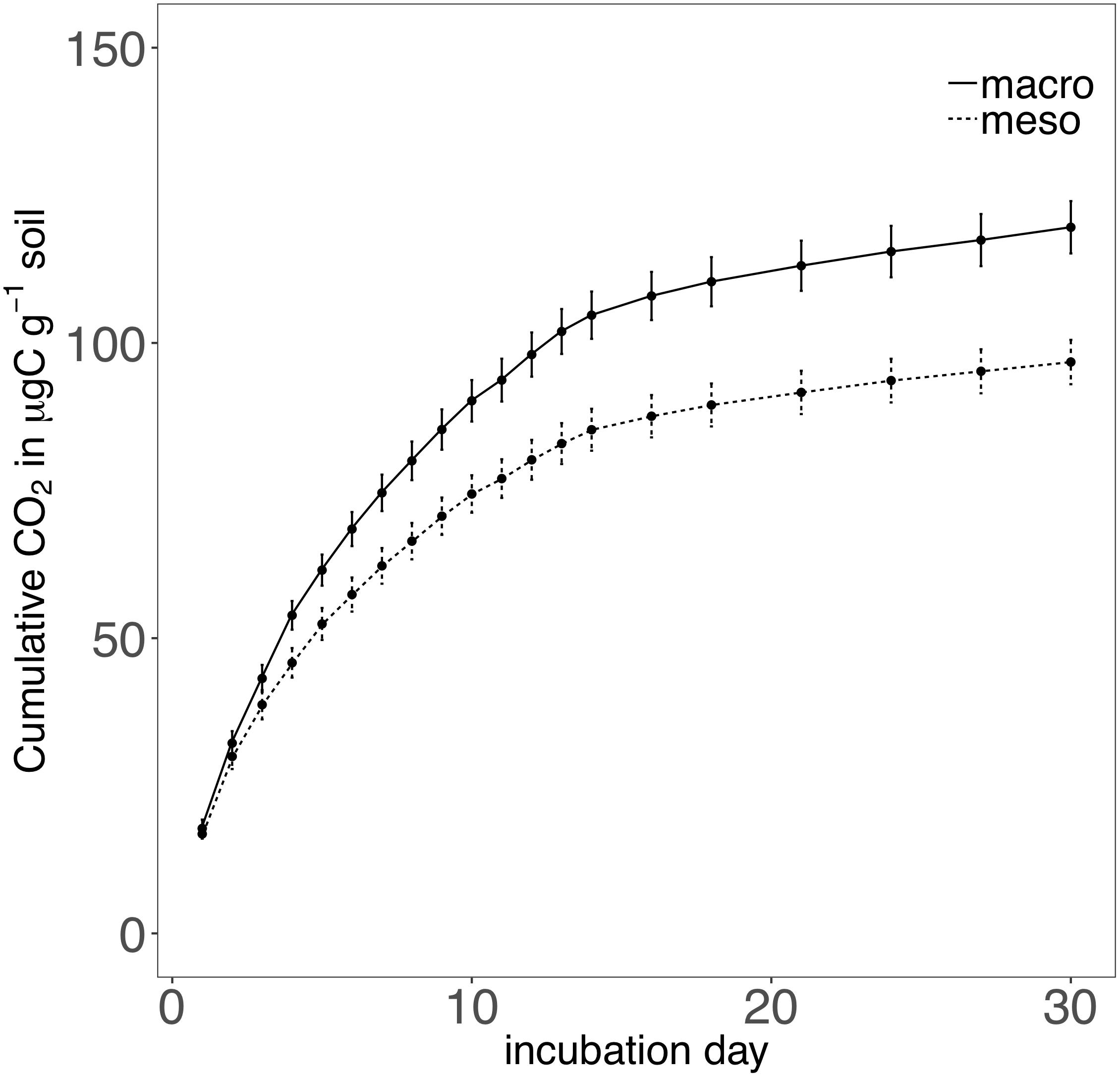
Figure 6. Cumulative CO2 (μg carbon g-1 soil) over a 30 day incubation from mesocosm soils recovered following 2 years of burial. Mesocosms with macroinvertebrates (macro) are represented with a solid line and mesocosms excluding macroinvertebrates (meso) are represented with a dashed line. Asterisks denote significant differences between mesocosm treatments (P < 0.05).
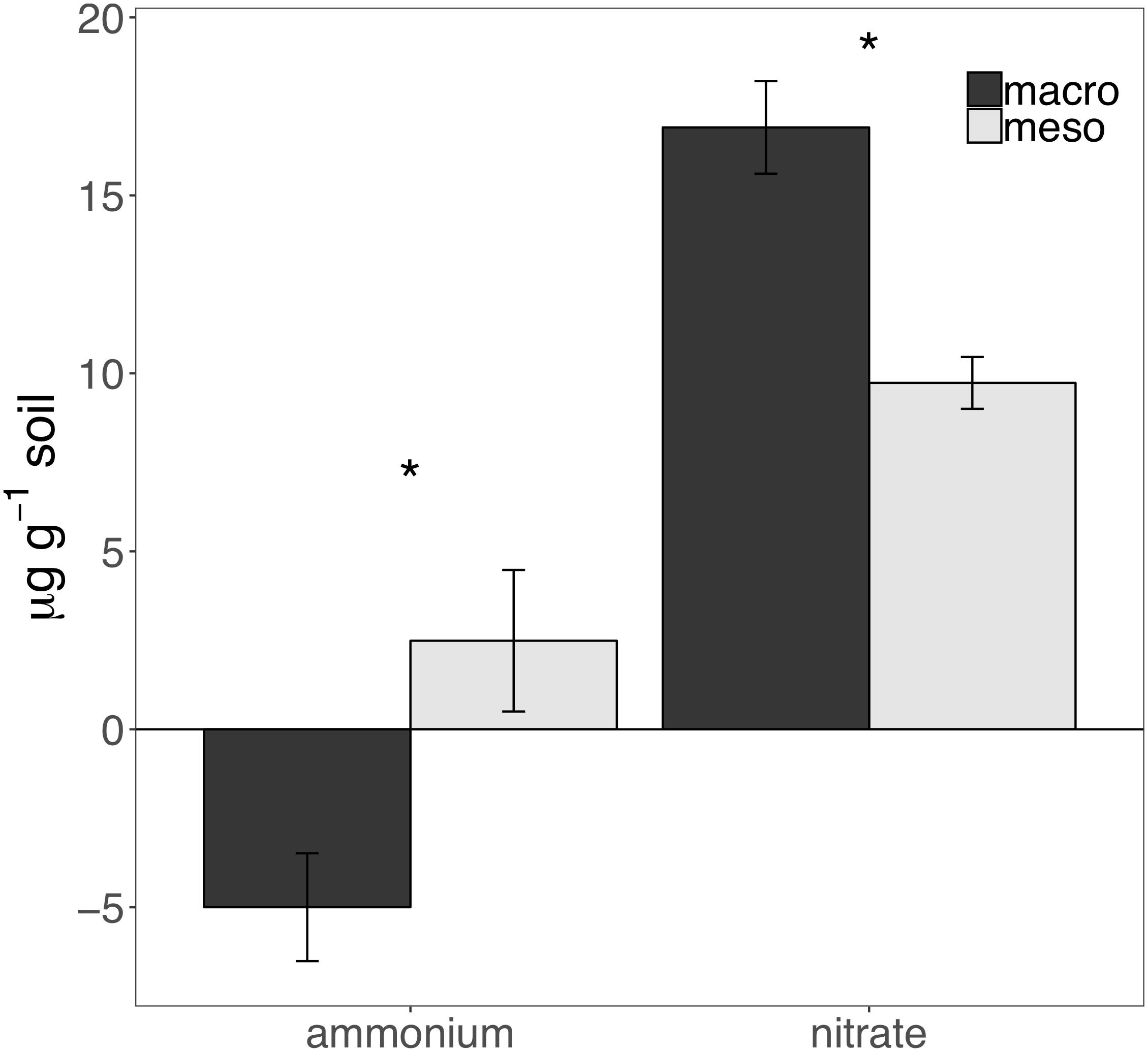
Figure 7. Ammonium and nitrate mineralization (μg g-1 soil, average ± standard error) following 30 days of incubation of mesocosm soils collected after 2 years of burial. Mesocosms with macroinvertebrates (macro) are represented in dark gray and mesocosms excluding macroinvertebrates (meso) are in light gray. Asterisks denote significant differences between mesocosm treatments (P < 0.05).
Discussion
Our findings indicate that macroinvertebrates can significantly affect microbial community composition and that they stimulate microbial biomass and activity within the rhizosphere. Soil macroinvertebrates increased bacterial diversity and stimulated microbial biomass, phenol oxidase and glucosidase activities along with potential carbon mineralization and nitrification. However, contrary to our hypothesis, macroinvertebrates did not cause a decrease in the fungal to bacterial ratio. Instead, fungal biomass increased significantly at least after the first year of the study in the presence of macroinvertebrates. Moreover, many of the observed treatment responses that were significant after the first year were not observed after 2 years.
Some of the differences in treatment responses between the first and second year are likely a result of changing physical and chemical conditions within the mesocosms. The sieving and defaunation process used to eliminate soil animals prior to the study likely resulted in a pulse of labile organic matter as pre-existing macroaggregates were disturbed or destroyed (Six et al., 2000; Denef et al., 2001); however, such an effect on organic matter availability to decomposers should be relatively short-lived and probably minimal after a full year of incubation. The introduction of new grass roots and colonization by soil invertebrates and microbial communities would be expected to result in transient changes in biotic communities and activity during the first growing season and may explain some of the treatments effects observed in year 1. Additionally, soil invertebrate densities within the mesocosms were lower compared to outside the mesocosms. The length of the experiment (2 years) highlights how slowly invertebrates recolonize soils. This is supported by previous work showing both slow recovery of soil by invertebrates following disturbance (Adl et al., 2006) along with taxon- and system-dependent differences in soil animal colonization (Cole et al., 2006). Our findings provide insight into the role of soil animals in shaping microbial processes as both microbes and invertebrates colonize new habitats or existing soil following disturbances. We suggest that these initial responses to the presence of macroinvertebrates represent a reorganization phase of soil ecosystem development, and were perhaps more apparent during the first year of the study due to the initial colonization of the soil by roots and invertebrates and reestablishment of microbial communities.
Our mesocosm soils were primarily populated by a mixture of earthworms, herbivorous beetle larvae and predatory and detritivorous myriapods. Previous studies have shown that macroinvertebrates elicit mixed microbial responses that can vary even at the species level. For example, earthworms were found to increase microbial biomass, whereas millipedes decreased biomass and their co-occurrence led to an increase in biomass (Seeber et al., 2006). Additionally, different species of native and invasive earthworms had dissimilar effects on microbial biomass (Zhang et al., 2000; Scheu et al., 2002; Chang et al., 2017). The abundance of earthworms in our macro-mesh mesocosms was moderately high (approximately 180 individuals per m2 on average across both years) compared to other grasslands (e.g., Gastine et al., 2003; Xu et al., 2013) and nearby forested areas (e.g., Dempsey et al., 2011), although lower (56–62%) compared to undisturbed soils adjacent to the mesocosms. The variability in microbial response observed among studies is likely due to differences in behavior and ecological function such as SOM mixing, microbial grazing intensity and patterns of fecal/waste deposition among different macroinvertebrate species (Ineson and Anderson, 1985; Hassall et al., 1987; Devliegher and Verstraete, 1995, 1997; Brown et al., 2000). Thus, the impact of macroinvertebrates on microbial biomass likely depends upon macroinvertebrate abundance and community composition. Our findings indicate that despite large differences in ecological function and potential interactions types among the macroinvertebrates occurring under natural conditions within our mesocosms their presence had an overall stimulatory effect on microbial biomass.
The observed increase in microbial biomass was driven primarily by an increase in fungal biomass, specifically non-AMF fungi in year 1 and AMF in year 2. This finding was surprising given that many previous studies have shown that macroinvertebrates decrease fungal biomass (Crowther et al., 2011a; Dempsey et al., 2011 but see Dempsey et al., 2013). Fungi are an important food source for earthworms and other macroinvertebrates (Bonkowski et al., 2000; Brown et al., 2004; Pollierer et al., 2007) and therefore it is logical that macroinvertebrate activity would result in a decline in fungal biomass. However, effects on fungal biomass have been shown to vary with grazer identity and grazing intensity. For example, Crowther and A’Bear (2012) concluded that high intensity grazing by larger invertebrates decreases microbial biomass, whereas low intensity grazing by smaller mesoinvertebrates and other mesofauna can increase microbial biomass. Increases in biomass due to invertebrate grazing have also been attributed to compensatory growth (Lussenhop, 1992). The density of macroinvertebrates in our mesocosms was lower compared to the typical densities observed outside of our mesocosms, suggesting that grazing intensity by macroinvertebrates in our soils may have been relatively low, resulting in stimulation rather than suppression of fungal biomass. The presence of earthworms has also been shown to increase AMF colonization rates (Zarea et al., 2009). Our observed increased in AMF in year 2 in the presence of macroinvertebrates supports this finding; however, we were unable to confirm whether the increase in AMF in soil was associated with an increase in AMF root colonization.
Under natural conditions, grazing is only one of the many macroinvertebrate activities that fungi are exposed to. For instance, bioengineering and litter-soil mixing by macroinvertebrates can improve soil habitat and resource distribution for microbes (Lavelle and Spain, 2001; Coleman et al., 2004). Similarly, many macroinvertebrates are capable of fungal spore dispersal through gut passage and fecal deposition, or by the passive transport of fungal spores on their exoskeletons or cuticles (McIlveen and Cole, 1976; Rabatin and Stinner, 1985; Gange, 1993; Moody et al., 1996; Lilleskov and Bruns, 2005). Our results suggest that fungal grazing either had a stimulatory or minor impact on fungal biomass and that its effects on the microbial community were likely tempered or outweighed by other factors such as improved resource distribution or fungal spore dispersal. Furthermore, the increase in non-AMF fungal biomass was only observed in year 1 and may be due to the introduction of fungi to the mesocosms by macroinvertebrates during the initial colonization phase after 1 year of burial. The disappearance of this effect in year 2 was driven not by a decrease in fungal biomass in the presence of macroinvertebrates, but instead by an increase in their absence. This supports evidence that soil animals do in fact play an important role in fungal spore dispersal (McIlveen and Cole, 1976; Rabatin and Stinner, 1985; Gange, 1993; Moody et al., 1996; Lilleskov and Bruns, 2005) but our results further indicate that fauna across all size classes are capable of contributing to this process over different timescales. Thus the rate of fungal colonization of new resource inputs to soil may depend upon soil animal community composition.
One of the most striking findings of our study was the strong disconnect between microbial biomass and community composition responses to macroinvertebrates. For instance, the increase in fungal biomass observed in year 1 was not accompanied by a change in the relative abundance of any individual fungal phyla, indicating that the initial fungal response to macroinvertebrates was community-wide. However, it is also possible that the fungal taxa responsible for the observed shifts in fungal biomass were not captured by the primers used for ITS sequencing as some fungal taxa are excluded (e.g., Glomeromycota, Schoch et al., 2012). In contrast, we found that macroinvertebrates caused an increase in bacterial diversity and shifts in the relative abundance of the phyla Bacteriodetes, Verrucomicrobia, Actinobacteria, and Firmicutes. Similarly, macroinvertebrates caused a relative decrease in the fungal phylum Zygomycota in year 2. Neither of these taxon-level shifts in bacteria or fungi was associated with notable changes in total bacterial or fungal biomass. One mechanism that may explain this response is the modification of microbial communities upon gut passage and fecal material deposition, which has been shown to alter bacterial community composition (Nechitaylo et al., 2010). For instance, Bacteroidetes is recognized as an important phylum in the digestive tracts of invertebrates (Van Borm et al., 2002; Egert et al., 2003; Schmitt-Wagner et al., 2003; Nechitaylo et al., 2010) and has been shown to increase in soils in response to earthworm additions (Bernard et al., 2012).
Fungus-grazing invertebrates may also be capable of altering fungal community composition through selective grazing on distinct fungal taxa (Klironomos and Kendrick, 1996; Maraun et al., 2003), which may explain the decline in only Zygomycota in year 2 of the study. However, the community-wide increase in total fungal biomass observed in year 1 may reflect the finding that macroinvertebrates show lower selectivity than microinvertebrates, such as nematodes and protozoans, when grazing on fungi (Maraun et al., 2003). Despite these and other studies investigating species-level interactions between microbes and soil invertebrates, most studies on microbial responses to soil animals under field conditions have employed coarse levels of microbial taxonomic resolution. Our study is one of only a handful studies to use next generation sequencing to investigate the impacts of macroinvertebrates on soil microbial communities and demonstrates that microbial responses can emerge at different taxonomic levels (Crowther et al., 2015): some effects (e.g. dispersal of fungal spores) manifest uniformly across taxa, while others (e.g., intensive fungivory) may occur at finer taxonomic levels and can only be captured using sequencing and other molecular approaches.
Changes in microbial biomass and community composition are commonly associated with distinct shifts in microbial activity (Maraun and Scheu, 1996; Groffman et al., 2004; Hedde et al., 2007) and our findings indicate that macrofauna modify both taxonomic and functional aspects of soil microbial communities within the rhizosphere. Specifically, macrofauna stimulated phenol oxidase activity in year 1 and glucosidase activity in year 2. Beta glucosidase is important for the breakdown of labile carbon compounds and is often used as a general indicator for SOM cycling (Stott et al., 2010), while phenol oxidase plays a role in lignin degradation and is responsive to shifts in plant and microbial communities (Sinsabaugh, 2010). This finding is supported by previous work showing that invertebrates often stimulate microbial C and N cycling activities (Ineson and Anderson, 1985; Hassall et al., 1987; Wickings and Grandy, 2011). The increase in phenol oxidase activity in year 1 may have been driven by the community-wide increase in fungal biomass, as fungi are known to produce lignin-degrading enzymes (Fioretto et al., 2000; Romani et al., 2006; Harner et al., 2009; Sinsabaugh, 2010). Phenol oxidase activity may have also been stimulated by the consumption and gut passage of fine roots by macroinvertebrates during the first year of the study (Fisk et al., 2004). The functional consequences of phylum-level changes in the microbial community are more difficult to interpret. However, there is some evidence that suggests that Bacteroidetes, classified as copiotrophic (Fierer et al., 2007), are associated with increased carbon availability and carbon mineralization rates. This explanation aligns with the stimulation of glucosidase activity and carbon mineralization observed in year 2 of the study. The sieving and defaunation process used to establish our treatments likely created a pulse of labile organic matter within our mesocosm soils. We would expect this to elicit stimulatory responses in hydrolytic microbial enzyme activities (Fontaine et al., 2003) in the first year of the experiment, yet we observed an increase in phenol oxidase activity only. The pulse of labile organic matter was likely not captured in sampling 1 year after burial of the mesocosms and potentially could have been measured if samples had been taken immediately after the installation of the mesocosms and in the weeks and months following. Macroinvertebrate activity also caused notable responses in nitrogen cycling by microbes: increased immobilization of ammonium and stimulation of net nitrification, which ultimately led to no net effect on net nitrogen mineralization. Higher nitrification could reflect an increased abundance of nitrifier species and nitrification rates in casts and burrows of lumbricid earthworms, as has been observed in agricultural soils by Parkin and Berry (1994, 1999), however, our sampling approach prevented us from distinguishing species-level responses in the microbial communities.
Conclusion
Our findings highlight the potentially important role of macroinvertebrate communities in shaping the composition and activity soil microbial communities within the rhizosphere. Despite evidence in previous studies for both positive and negative effects of macrofauna on microbial biomass and activity, our findings suggest that under natural conditions, with diverse invertebrate communities, macroinvertebrates stimulate microbial biomass and processes. Such stimulatory responses may stem from diverse animal-microbe interaction types such as low-intensity grazing, microbial dispersal and changes in microbial resource quality or availability. We also observed macroinvertebrate-driven shifts in microbial community composition that extend out to impact carbon and nutrient cycling. We remain cautious in interpreting our findings beyond the 2-year scope of our study and acknowledge that the treatment effects observed likely reflect a community establishment and colonization phase for both microbes and invertebrates. Longer-term experiments will be required to fully capture the role of mixed macroinvertebrate communities in shaping microbial dynamics under natural conditions. Future studies on fauna-microbe interactions should also continue to incorporate sequencing and other molecular approaches to fully elucidate both community-wide and taxon-specific changes in the microbial communities and their consequences for soil ecosystem processes such as SOM formation in response to soil invertebrates.
Author Contributions
NB and KW designed the experiments and wrote the first draft of the manuscript. NB performed the experiments under the supervision of KW and JK-K, except for PLFA analyses which were performed under the direction of SF. NB analyzed the data. All authors revised the text of the manuscript.
Funding
This work was supported by an NSF IGERT cross-scale biogeochemistry and climate program small grant, a Cornell University Department of Entomology Grace H. Griswold grant and a Cornell University College of Agricultural and Life Sciences Andrew W. Mellon grant.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Abigail Wentworth, Grant Thompson, Maria Gannett, Martin Ward and Brett Welch for their help in the lab and in the field, Melissa Knorr and Amber Kittle for their help with PLFA analysis, Dr. Erika Mudrak for her help with statistical analysis, and Liang Cheng and Joshua Garcia for their help with the sequence processing and analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00256/full#supplementary-material
References
Adl, S. M., Coleman, D. C., and Read, F. (2006). Slow recovery of soil biodiversity in sandy loam soils of Georgia after 25 years of no-tillage management. Agric. Ecosyst. Environ. 114, 323–334. doi: 10.1016/j.agee.2005.11.019
Albers, D., Schaefer, M., and Scheu, S. (2006). Incorporation of plant carbon into the soil animal food web of an arable system. Ecology 87, 235–245. doi: 10.1890/04-1728
Austin, E. E., Wickings, K., McDaniel, M. D., Robertson, G. P., and Grandy, A. S. (2017). Cover crop root contributions to soil carbon in a no-till corn bioenergy cropping system. Gcb Bioenergy 9, 1252–1263. doi: 10.1111/gcbb.12428
Bååth, E. (2003). The use of neutral lipid fatty acids to indicate the physiological conditions of soil fungi. Microb. Ecol. 45, 373–383. doi: 10.1007/s00248-003-2002-y
Bardgett, R. (2005). The Biology of Soil: A Community and Ecosystem Approach. Oxford: Oxford University Press. doi: 10.1093/acprof:oso/9780198525035.001.0001
Bardgett, R. D., Hobbs, P. J., and Frostegård, Å. (1996). Changes in soil fungal: bacterial biomass ratios following reductions in the intensity of management of an upland grassland. Biol. Fertil. Soils 22, 261–264. doi: 10.1007/BF00382522
Bardgett, R. D., Keiller, S., Cook, R., and Gilburn, A. S. (1998). Dynamic interactions between soil animals and microorganisms in upland grassland soils amended with sheep dung: a microcosm experiment. Soil Biol. Biochem. 30, 531–539. doi: 10.1016/S0038-0717(97)00146-6
Beare, M. H., Coleman, D. C., Crossley, D. A. Jr., Hendrix, P. F., and Odum, E. P. (1995). “A hierarchical approach to evaluating the significance of soil biodiversity to biogeochemical cycling,” in The Significance and Regulation of Soil Biodiversity, eds H. P. Collins, G. Philip Robertson, and M. J. Klug (Berlin: Springer), 5–22.
Bernard, L., Chapuis-Lardy, L., Razafimbelo, T., Razafindrakoto, M., Pablo, A. -L., Legname, E., et al. (2012). Endogeic earthworms shape bacterial functional communities and affect organic matter mineralization in a tropical soil. ISME J. 6:213. doi: 10.1038/ismej.2011.87
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/y59-099
Bonkowski, M., Griffiths, B. S., and Ritz, K. (2000). Food preferences of earthworms for soil fungi. Pedobiologia (Jena). 44, 666–676. doi: 10.1078/S0031-4056(04)70080-3
Bradford, M. A., Jones, T. H., Bardgett, R. D., Black, H. I. J., Boag, B., Bonkowski, M., et al. (2002). Impacts of soil faunal community composition on model grassland ecosystems. Science (80-.) 298, 615–618. doi: 10.1126/science.1075805
Bradford, M. A., Strickland, M. S., DeVore, J. L., and Maerz, J. C. (2012). Root carbon flow from an invasive plant to belowground foodwebs. Plant Soil 359, 233–244. doi: 10.1007/s11104-012-1210-y
Bradford, M. A., Tordoff, G. M., Black, H. I. J., Cook, R., Eggers, T., Garnett, M. H., et al. (2007). Carbon dynamics in a model grassland with functionally different soil communities. Funct. Ecol. 21, 690–697. doi: 10.1111/j.1365-2435.2007.01268.x
Brown, G. G., Barois, I., and Lavelle, P. (2000). Regulation of soil organic matter dynamics and microbial activityin the drilosphere and the role of interactionswith other edaphic functional domains. Eur. J. Soil Biol. 36, 177–198. doi: 10.1016/S1164-5563(00)01062-1
Brown, G. G., Doube, B. M., and Edwards, C. A. (2004). Functional interactions between earthworms, microorganisms, organic matter, and plants. Earthworm Ecol. 2, 213–239.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335. doi: 10.1038/nmeth.f.303
Chang, C. -H., Szlavecz, K., and Buyer, J. S. (2017). Amynthas agrestis invasion increases microbial biomass in Mid-Atlantic deciduous forests. Soil Biol. Biochem. 114, 189–199. doi: 10.1016/j.soilbio.2017.07.018
Cole, L., Bradford, M. A., Shaw, P. J. A., and Bardgett, R. D. (2006). The abundance, richness and functional role of soil meso-and macrofauna in temperate grassland: a case study. Appl. Soil Ecol. 33, 186–198. doi: 10.1016/j.apsoil.2005.11.003
Coleman, D. C. (2008). From peds to paradoxes: linkages between soil biota and their influences on ecological processes. Soil Biol. Biochem. 40, 271–289. doi: 10.1016/j.soilbio.2007.08.005
Coleman, D. C., Crossley, D. A., and Hendrix, P. F. (2004). Fundamentals of Soil Ecology. Cambridge: Academic press.
Cotrufo, M. F., Wallenstein, M. D., Boot, C. M., Denef, K., and Paul, E. (2013). The M icrobial E fficiency-M atrix S tabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: do labile plant inputs form stable soil organic matter? Glob. Change Biol. 19, 988–995. doi: 10.1111/gcb.12113
Crowther, T. W., and A’Bear, A. D. (2012). Impacts of grazing soil fauna on decomposer fungi are species-specific and density-dependent. Fungal Ecol. 5, 277–281. doi: 10.1016/j.funeco.2011.07.006.
Crowther, T. W., Boddy, L., and Jones, T. H. (2011a). Species-specific effects of soil fauna on fungal foraging and decomposition. Oecologia 167, 535–545. doi: 10.1007/s00442-011-2005-1
Crowther, T. W., Boddy, L., and Jones, T. H. (2011b). Outcomes of fungal interactions are determined by soil invertebrate grazers. Ecol. Lett. 14, 1134–1142. doi: 10.1111/j.1461-0248.2011.01682.x
Crowther, T. W., Jones, T. H., Boddy, L., and Baldrian, P. (2011c). Invertebrate grazing determines enzyme production by basidiomycete fungi. Soil Biol. Biochem. 43, 2060–2068. doi: 10.1016/j.soilbio.2011.06.003
Crowther, T. W., Thomas, S. M., Maynard, D. S., Baldrian, P., Covey, K., Frey, S. D., et al. (2015). Biotic interactions mediate soil microbial feedbacks to climate change. Proc. Natl. Acad. Sci. U.S.A. 112, 7033–7038. doi: 10.1073/pnas.1502956112
De Deyn, G. B., Raaijmakers, C. E., Zoomer, H. R., Berg, M. P., de Ruiter, P. C., Verhoef, H. A., et al. (2003). Soil invertebrate fauna enhances grassland succession and diversity. Nature 422:711. doi: 10.1038/nature01548
Dempsey, M. A., Fisk, M. C., and Fahey, T. J. (2011). Earthworms increase the ratio of bacteria to fungi in northern hardwood forest soils, primarily by eliminating the organic horizon. Soil Biol. Biochem. 43, 2135–2141. doi: 10.1016/j.soilbio.2011.06.017
Dempsey, M. A., Fisk, M. C., Yavitt, J. B., Fahey, T. J., and Balser, T. C. (2013). Exotic earthworms alter soil microbial community composition and function. Soil Biol. Biochem. 67, 263–270. doi: 10.1016/j.soilbio.2013.09.009
Denef, K., Six, J., Paustian, K., and Merckx, R. (2001). Importance of macroaggregate dynamics in controlling soil carbon stabilization: short-term effects of physical disturbance induced by dry–wet cycles. Soil Biol. Biochem. 33, 2145–2153. doi: 10.1016/S0038-0717(01)00153-5
Devliegher, W., and Verstraete, W. (1995). Lumbricus terrestris in a soil core experiment: nutrient-enrichment processes (NEP) and gut-associated processes (GAP) and their effect on microbial biomass and microbial activity. Soil Biol. Biochem. 27, 1573–1580. doi: 10.1016/0038-0717(95)00090-2
Devliegher, W., and Verstraete, W. (1997). Microorganisms and soil physico-chemical conditions in the drilosphere of Lumbricus terrestris. Soil Biol. Biochem. 29, 1721–1729. doi: 10.1016/S0038-0717(97)00068-0
Doane, T. A., and Horwath, W. R. (2003). Spectrophotometric determination of nitrate with a single reagent. Anal. Lett. 36, 2713–2722. doi: 10.1081/AL-120024647
Egert, M., Wagner, B., Lemke, T., Brune, A., and Friedrich, M. W. (2003). Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69, 6659–6668. doi: 10.1128/AEM.69.11.6659-6668.2003
Ekelund, F., Olsson, S., and Johansen, A. (2003). Changes in the succession and diversity of protozoan and microbial populations in soil spiked with a range of copper concentrations. Soil Biol. Biochem. 35, 1507–1516. doi: 10.1016/S0038-0717(03)00249-9
Elfstrand, S., Lagerlöf, J., Hedlund, K., and Mårtensson, A. (2008). Carbon routes from decomposing plant residues and living roots into soil food webs assessed with 13 C labelling. Soil Biol. Biochem. 40, 2530–2539. doi: 10.1016/j.soilbio.2008.06.013
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Filley, T. R., McCormick, M. K., Crow, S. E., Szlavecz, K., Whigham, D. F., Johnston, C. T., et al. (2008). Comparison of the chemical alteration trajectory of Liriodendron tulipifera L. leaf litter among forests with different earthworm abundance. J. Geophys. Res. Biogeosci. 113:G01027.
Fioretto, A., Papa, S., Curcio, E., Sorrentino, G., and Fuggi, A. (2000). Enzyme dynamics on decomposing leaf litter of Cistus incanus and Myrtus communis in a Mediterranean ecosystem. Soil Biol. Biochem. 32, 1847–1855. doi: 10.1016/S0038-0717(00)00158-9
Fisk, M. C., Fahey, T. J., Groffman, P. M., and Bohlen, P. J. (2004). Earthworm invasion, fine-root distributions, and soil respiration in north temperate forests. Ecosystems 7, 55–62. doi: 10.1007/s10021-003-0130-3
Fontaine, S., Mariotti, A., and Abbadie, L. (2003). The priming effect of organic matter: a question of microbial competition? Soil Biol. Biochem. 35, 837–843. doi: 10.1016/S0038-0717(03)00123-8
Gale, W. J., Cambardella, C. A., and Bailey, T. B. (2000). Root-derived carbon and the formation and stabilization of aggregates. Soil Sci. Soc. Am. J. 64, 201–207. doi: 10.2136/sssaj2000.641201x
Gange, A. C. (1993). Translocation of mycorrhizal fungi by earthworms during early succession. Soil Biol. Biochem. 25, 1021–1026. doi: 10.1016/0038-0717(93)90149-6
Garcia-Palacios, P., Maestre, F. T., Kattge, J., and Wall, D. H. (2013). Climate and litter quality differently modulate the effects of soil fauna on litter decomposition across biomes. Ecol. Lett. 16, 1045–1053. doi: 10.1111/ele.12137
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Gastine, A., Scherer-Lorenzen, M., and Leadley, P. W. (2003). No consistent effects of plant diversity on root biomass, soil biota and soil abiotic conditions in temperate grassland communities. Appl. Soil Ecol. 24, 101–111. doi: 10.1016/S0929-1393(02)00137-3
Grandy, A. S., Sinsabaugh, R. L., Neff, J. C., Stursova, M., and Zak, D. R. (2008). Nitrogen deposition effects on soil organic matter chemistry are linked to variation in enzymes, ecosystems and size fractions. Biogeochemistry 91, 37–49. doi: 10.1007/s10533-008-9257-9
Grandy, A. S., Wieder, W. R., Wickings, K., and Kyker-Snowman, E. (2016). Beyond microbes: are fauna the next frontier in soil biogeochemical models? Soil Biol. Biochem. 102, 40–44. doi: 10.1016/j.soilbio.2016.08.008
Groffman, P. M., Bohlen, P. J., Fisk, M. C., and Fahey, T. J. (2004). Exotic earthworm invasion and microbial biomass in temperate forest soils. Ecosystems 7, 45–54. doi: 10.1007/s10021-003-0129-9
Guckert, J. B., Antworth, C. P., Nichols, P. D., and White, D. C. (1985). Phospholipid, ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol. Lett. 31, 147–158. doi: 10.1111/j.1574-6968.1985.tb01143.x
Harner, M. J., Crenshaw, C. L., Abelho, M., Stursova, M., Follstad Shah, J. J., and Sinsabaugh, R. L. (2009). Decomposition of cottonwood and Russian olive litter in riparian habitats of a semiarid region. Ecol. Appl. 19, 1135–1146. doi: 10.1890/08-0294.1
Hassall, M., Turner, J. G., and Rands, M. R. W. (1987). Effects of terrestrial isopods on the decomposition of woodland leaf litter. Oecologia 72, 597–604. doi: 10.1007/BF00378988
Hättenschwiler, S., Tiunov, A. V., and Scheu, S. (2005). Biodiversity and litter decomposition in terrestrial ecosystems. Annu. Rev. Ecol. Evol. Syst. 36, 191–218. doi: 10.1146/annurev.ecolsys.36.112904.151932.
Hedde, M., Bureau, F., Akpa-Vinceslas, M., Aubert, M., and Decaëns, T. (2007). Beech leaf degradation in laboratory experiments: effects of eight detritivorous invertebrate species. Appl. Soil Ecol. 35, 291–301. doi: 10.1016/j.apsoil.2006.08.002
Herlemann, D. P. R., Labrenz, M., Jürgens, K., Bertilsson, S., Waniek, J. J., and Andersson, A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5:1571. doi: 10.1038/ismej.2011.41
Howard, M. M., Bell, T. H., and Kao-Kniffin, J. (2017). Soil microbiome transfer method affects microbiome composition, including dominant microorganisms, in a novel environment. FEMS Microbiol. Lett. 364:092.
Huhta, V., Wright, D. H., and Coleman, D. C. (1989). Characteristics of defaunated soil, 1: a comparison of three techniques applied to two different forest soils. Pedobiologia (German DR) 33, 417–426.
Ineson, P., and Anderson, J. M. (1985). Aerobically isolated bacteria associated with the gut and faeces of the litter feeding macroarthropods Oniscus asellus and Glomeris marginata. Soil Biol. Biochem. 17, 843–849. doi: 10.1016/0038-0717(85)90145-2
Jones, C. G., Lawton, J. H., and Shachak, M. (1996). “Organisms as ecosystem engineers,” in Ecosystem Management: Selected Readings, eds F. B. Samson and F. L. Knopf (New York, NY: Springer), 130–147.
Jones, D. L., Nguyen, C., and Finlay, R. D. (2009). Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321, 5–33. doi: 10.1007/s11104-009-9925-0
Klironomos, J. N., and Kendrick, W. B. (1996). Palatability of microfungi to soil arthropods in relation to the functioning of arbuscular mycorrhizae. Biol. Fertil. Soils 21, 43–52. doi: 10.1007/BF00335992
Lavelle, P., and Spain, A. V. (2001). Soil Ecology. Dordrecht: Kluwer Academic Publishers. doi: 10.1007/978-94-017-5279-4
Leckie, S. E., Prescott, C. E., Grayston, S. J., Neufeld, J. D., and Mohn, W. W. (2004). Comparison of chloroform fumigation-extraction, phospholipid fatty acid, and DNA methods to determine microbial biomass in forest humus. Soil Biol. Biochem. 36, 529–532. doi: 10.1016/j.soilbio.2003.10.014
Lilleskov, E. A., and Bruns, T. D. (2005). Spore dispersal of a resupinate ectomycorrhizal fungus, Tomentella sublilacina, via soil food webs. Mycologia 97, 762–769. doi: 10.1080/15572536.2006.11832767
Lussenhop, J. (1992). Mechanisms of microarthropod-microbial interactions in soil. Adv. Ecol. Res. 23, 1–33. doi: 10.1016/S0065-2504(08)60145-2
Maraun, M., Martens, H., Migge, S., Theenhaus, A., and Scheu, S. (2003). Adding to the enigma of soil animal diversity: fungal feeders and saprophagous soil invertebrates prefer similar food substrates. Eur. J. Soil Biol. 39, 85–95. doi: 10.1016/S1164-5563(03)00006-2
Maraun, M., and Scheu, S. (1996). Changes in microbial biomass, respiration and nutrient status of beech (Fagus sylvatica) leaf litter processed by millipedes (Glomeris marginata). Oecologia 107, 131–140. doi: 10.1007/BF00582243
Martin, K. J., and Rygiewicz, P. T. (2005). Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5:28. doi: 10.1186/1471-2180-5-28
McIlveen, W. D., and Cole, H. Jr (1976). Spore dispersal of Endogonaceae by worms, ants, wasps, and birds. Can. J. Bot. 54, 1486–1489. doi: 10.1139/b76-161
McLean, M. A., Migge-Kleian, S., and Parkinson, D. (2006). Earthworm invasions of ecosystems devoid of earthworms: effects on soil microbes. Biol. Invasions 8, 1257–1273. doi: 10.1007/s10530-006-9020-x
Moody, S. A., Piearce, T. G., and Dighton, J. (1996). Fate of some fungal spores associated with wheat straw decomposition on passage through the guts of Lumbricus terrestris and Aporrectodea longa. Soil Biol. Biochem. 28, 533–537. doi: 10.1016/0038-0717(95)00172-7
Nechitaylo, T. Y., Yakimov, M. M., Godinho, M., Timmis, K. N., Belogolova, E., Byzov, B. A., et al. (2010). Effect of the earthworms Lumbricus terrestris and Aporrectodea caliginosa on bacterial diversity in soil. Microb. Ecol. 59, 574–587. doi: 10.1007/s00248-009-9604-y
Northeast Regional Climate Center (2016–2017). Ithaca Climate Page. Available at: http://www.nrcc.cornell.edu/wxstation/ithaca/ithaca.html
Ostle, N., Briones, M. J. I., Ineson, P., Cole, L., Staddon, P., and Sleep, D. (2007). Isotopic detection of recent photosynthate carbon flow into grassland rhizosphere fauna. Soil Biol. Biochem. 39, 768–777. doi: 10.1016/j.soilbio.2006.09.025
Parkin, T. B., and Berry, E. C. (1994). Nitrogen transformations associated with earthworm casts. Soil Biol. Biochem. 26, 1233–1238. doi: 10.1016/0038-0717(94)90148-1
Parkin, T. B., and Berry, E. C. (1999). Microbial nitrogen transformations in earthworm burrows. Soil Biol. Biochem. 31, 1765–1771. doi: 10.1016/S0038-0717(99)00085-1
Pollierer, M. M., Langel, R., Körner, C., Maraun, M., and Scheu, S. (2007). The underestimated importance of belowground carbon input for forest soil animal food webs. Ecol. Lett. 10, 729–736. doi: 10.1111/j.1461-0248.2007.01064.x
Pouyat, R. V, Szlavecz, K., Yesilonis, I. D., Groffman, P. M., and Schwarz, K. (2010). “Chemical, physical, and biological characteristics of urban soils,” in Urban Ecosystem Ecology, eds J. Aitkenhead-Peterson and A. Volder (Madison, WI: American Society of Agronomy), 119–152.
Pylro, V. S., Roesch, L. F. W., Morais, D. K., Clark, I. M., Hirsch, P. R., and Tótola, M. R. (2014). Data analysis for 16S microbial profiling from different benchtop sequencing platforms. J. Microbiol. Methods 107, 30–37. doi: 10.1016/j.mimet.2014.08.018
Qian, Y., and Follett, R. (2012). Carbon dynamics and sequestration in urban turfgrass ecosystems, in Carbon Sequestration in Urban Ecosystems (Berlin: Springer), 161–172. doi: 10.1007/978-94-007-2366-5_8
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available at: https://www.R-project.org/.
Rabatin, S. C., and Stinner, B. R. (1985). Arthropods as consumers of vesicular-arbuscular mycorrhizal fungi. Mycologia 77, 320–322. doi: 10.1080/00275514.1985.12025103
Rochefort, S., Therrien, F., Shetlar, D. J., and Brodeur, J. (2006). Species diversity and seasonal abundance of Collembola in turfgrass ecosystems of North America. Pedobiologia (Jena) 50, 61–68. doi: 10.1016/j.pedobi.2005.10.007
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Romani, A. M., Fischer, H., Mille-Lindblom, C., and Tranvik, L. J. (2006). Interactions of bacteria and fungi on decomposing litter: differential extracellular enzyme activities. Ecology 87, 2559–2569. doi: 10.1890/0012-9658(2006)87[2559:IOBAFO]2.0.CO;2
Saiya-Cork, K. R., Sinsabaugh, R. L., and Zak, D. R. (2002). The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 34, 1309–1315. doi: 10.1016/S0038-0717(02)00074-3
Scheu, S., Schlitt, N., Tiunov, A. V, Newington, J. E., and Jones, H. T. (2002). Effects of the presence and community composition of earthworms on microbial community functioning. Oecologia 133, 254–260. doi: 10.1007/s00442-002-1023-4
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schmitt-Wagner, D., Friedrich, M. W., Wagner, B., and Brune, A. (2003). Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69, 6007–6017. doi: 10.1128/AEM.69.10.6007-6017.2003
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U.S.A. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Seeber, J., Scheu, S., and Meyer, E. (2006). Effects of macro-decomposers on litter decomposition and soil properties in alpine pastureland: a mesocosm experiment. Appl. Soil Ecol. 34, 168–175. doi: 10.1016/j.apsoil.2006.02.004
Shi, S., Herman, D. J., He, Z., Pett-Ridge, J., Wu, L., Zhou, J., et al. (2018). Plant roots alter microbial functional genes supporting root litter decomposition. Soil Biol. Biochem. 127, 90–99. doi: 10.1016/j.soilbio.2018.09.013
Shi, W., Bowman, D., and Rufty, T. (2012). “Microbial control of soil carbon accumulation in turfgrass systems,” in Carbon Sequestration in Urban Ecosystems, eds R. Lal and B. Augustin (Berlin: Springer), 215–231. doi: 10.1007/978-94-007-2366-5_11
Sinsabaugh, R. L. (2010). Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 42, 391–404. doi: 10.1016/j.soilbio.2009.10.014
Six, J., Paustian, K., Elliott, E. T., and Combrink, C. (2000). Soil structure and organic matter I. Distribution of aggregate-size classes and aggregate-associated carbon. Soil Sci. Soc. Am. J. 64, 681–689. doi: 10.2136/sssaj2000.642681x
Sokol, N. W., Kuebbing, S. E., Karlsen-Ayala, E., and Bradford, M. A. (2019). Evidence for the primacy of living root inputs, not root or shoot litter, in forming soil organic carbon. New Phytol. 221, 233–246.
Stott, D. E., Andrews, S. S., Liebig, M. A., Wienhold, B. J., and Karlen, D. L. (2010). Evaluation of β-glucosidase activity as a soil quality indicator for the soil management assessment framework. Soil Sci. Soc. Am. J. 74, 107–119. doi: 10.2136/sssaj2009.0029
Swift, M. J., Heal, O. W., Anderson, J. M., and Anderson, J. M. (1979). Decomposition in Terrestrial Ecosystems. Berkeley, CA: Univ of California Press.
Trap, J., Bonkowski, M., Plassard, C., Villenave, C., and Blanchart, E. (2016). Ecological importance of soil bacterivores for ecosystem functions. Plant Soil 398, 1–24. doi: 10.1007/s11104-015-2671-6
Triplehorn, C. A., and Johnson, N. F. (2005). Borror and Delong’s Introduction to the Study of Insects. Dallas, TX: Thompson Brooks/Cole.
Van Borm, S., Buschinger, A., Boomsma, J. J., and Billen, J. (2002). Tetraponera ants have gut symbionts related to nitrogen–fixing root–nodule bacteria. Proc. R. Soc. Lond. B Biol. Sci. 269, 2023–2027. doi: 10.1098/rspb.2002.2101
Wall, D. H., Bradford, M. A., Trofymow, J. A., Behan-Pelletier, V., Bignell, D. E., et al. (2008). Global decomposition experiment shows soil animal impacts on decomposition are climate-dependent. Glob. Chang. Biol. 14, 2661–2677. doi: 10.1111/j.1365-2486.2008.01672.x
Walter, D. E. (2005). Major Mite Taxa. Available at: http://idtools.org/id/mites/invasive_mite/Invasive_Mite_Identification/key/Major_Mite_taxa/Media/Html/Home_Major_Mite_Taxa.html
White, D. C., Davis, W. M., Nickels, J. S., King, J. D., and Bobbie, R. J. (1979). Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia 40, 51–62. doi: 10.1007/BF00388810
Wickings, K., and Grandy, A. S. (2011). The oribatid mite Scheloribates moestus (Acari: Oribatida) alters litter chemistry and nutrient cycling during decomposition. Soil Biol. Biochem. 43, 351–358. doi: 10.1016/j.soilbio.2010.10.023
Wieder, W. R., Grandy, A. S., Kallenbach, C. M., and Bonan, G. B. (2014). Integrating microbial physiology and physio-chemical principles in soils with the microbial-mineral carbon stabilization (MIMICS) model. Biogeosciences 11, 3899–3917. doi: 10.5194/bg-11-3899-2014
Xu, S., Johnson-Maynard, J. L., and Prather, T. S. (2013). Earthworm density and biomass in relation to plant diversity and soil properties in a Palouse prairie remnant. Appl. Soil Ecol. 72, 119–127. doi: 10.1016/j.apsoil.2013.06.006
Zarea, M. J., Ghalavand, A., Goltapeh, E. M., Rejali, F., and Zamaniyan, M. (2009). Effects of mixed cropping, earthworms (Pheretima sp.), and arbuscular mycorrhizal fungi (Glomus mosseae) on plant yield, mycorrhizal colonization rate, soil microbial biomass, and nitrogenase activity of free-living rhizosphere bacteria. Pedobiologia (Jena) 52, 223–235. doi: 10.1016/j.pedobi.2008.10.004
Keywords: soil macroinvertebrates, soil microbial communities, rhizosphere, mesocosms, microbial activity
Citation: Bray N, Kao-Kniffin J, Frey SD, Fahey T and Wickings K (2019) Soil Macroinvertebrate Presence Alters Microbial Community Composition and Activity in the Rhizosphere. Front. Microbiol. 10:256. doi: 10.3389/fmicb.2019.00256
Received: 19 September 2018; Accepted: 30 January 2019;
Published: 22 February 2019.
Edited by:
Eoin L. Brodie, Lawrence Berkeley National Laboratory (DOE), United StatesReviewed by:
Jennifer Pett-Ridge, Lawrence Berkeley National Laboratory (DOE), United StatesJincai Ma, Jilin University, China
Copyright © 2019 Bray, Kao-Kniffin, Frey, Fahey and Wickings. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyle Wickings, a2d3MzdAY29ybmVsbC5lZHU=
 Natalie Bray
Natalie Bray Jenny Kao-Kniffin
Jenny Kao-Kniffin Serita D. Frey
Serita D. Frey Timothy Fahey4
Timothy Fahey4 Kyle Wickings
Kyle Wickings