- Department of Life and Environmental Sciences, Marche Polytechnic University, Ancona, Italy
The spoilage yeasts belonging to the genus Dekkera (anamorph Brettanomyces) are associated with the fermentation process and can be responsible for off-flavors in wine. Brettanomyces bruxellensis is difficult to isolate from natural environments because of its low diffusion, low presence on the grape surface and low competition capacity, slow growth, and VBNC (viable but not culturable) state, even when selective media are used. In this study, to investigate the origins and occurrence of B. bruxellensis in winemaking, a total of 62 samples from grapes, winery environment, and fermenting musts were taken through direct isolation with a selective medium. B. bruxellensis was not directly detected in the grape samples but was instead widely isolated from the winery environment samples. However, using a combination of enrichment and selective media, eight of fifteen grape samples were positive for B. bruxellensis. Analysis of the genetic traits of the isolates indicated a strict relationship among the strains from the vineyard and the winery. Isolates from the vineyard and the winery were both part of the more common and dominant biotypes suggesting that the vineyard may be the contamination source of B. bruxellensis in the winery environment. For this, grapes may represent the possible primary origin source from which a flow toward the winery environment originates. On the other hand, the wide occurrence of B. bruxellensis in winery indicates that this environment can be considered as the favorable ecological niche for colonization and diffusion of these yeast.
Introduction
Brettanomyces bruxellensis is a yeast that has been isolated from several natural ecological niches, and it is intrinsically linked with various industrial fermented products, such as wine, beer, cider, kombucha tea, and bioethanol (Martens et al., 1997; Greenwalt et al., 2000; Morrissey et al., 2004; Silva et al., 2004; Teoh et al., 2004; Smith and Divol, 2016). The role of B. bruxellensis in these fermentation processes is often ambiguous. In some industrial processes, as beer production, the presence of B. bruxellensis can be considered favorable, as is produces aromas contributing to the specific style of some specialty beers (Gilliland, 1961; Spitaels et al., 2014). However, the same aroma compounds are considered as off-flavors in the production of other beer styles and of wines (Verachtert, 1992; Wedral et al., 2010; Steensels et al., 2015). In winemaking, the main factor that affects the sensory properties that are imparted by B. bruxellensis is the production of 4-ethylphenol, 4-ethylguaiacol, and tetrahydropiridine, which are considered unpleasant aromas and are defined as the “Brett character” (Heresztyn, 1986; Chatonnet et al., 1992; Dias et al., 2003; Ciani and Comitini, 2014; Valdetara et al., 2017).
Brettanomyces bruxellensis is mainly associated with wine aging in barrels, and with stuck or sluggish fermentation (Suárez et al., 2007). This is possible due to the physiological traits of B. bruxellensis, as this yeast shows high ethanol tolerance, SO2 resistance and growth at low nutrient availability (Loureiro and Malfeito-Ferreira, 2003; Barata et al., 2008; Coulon et al., 2011; Steensels et al., 2015; Avramova et al., 2018). Compared with white wines, red wines are particularly susceptible to B. bruxellensis contamination due to their higher pH, lower SO2 addition, higher polyphenols content, and barrel aging (i.e., oxygen availability) (Benito et al., 2009). Due to this major role for Dekkera/Brettanomyces yeast in industrial fermentation processes, their key phenotypic characteristics have been extensively studied, and especially for B. bruxellensis. Although many of these Dekkera/Brettanomyces strains occupy similar ecological niches to Saccharomyces cerevisiae, their general physiology and phenotypic traits show remarkable differences (Nardi et al., 2010; Steensels et al., 2015).
Since the 1980’s, numerous studies have speculated on the occurrence and diffusion of Brettanomyces spp. in winemaking (Pretorius, 2000; Suárez et al., 2007; Agnolucci et al., 2009; Campolongo et al., 2010; Di Toro et al., 2015), although their origin is not well defined yet (Aguilar-Uscanga et al., 2000; Curtin and Pretorius, 2014; Avramova et al., 2018). Many studies have associated this spoilage yeast with poor hygienic practices in the cellar, presence of cellobiose and micro-oxygenation related to the use of wooden barrel, together with high ethanol content (Chatonnet et al., 1999; Renouf and Lonvaud-Funel, 2007; Boulton et al., 2013). It was only in 2007 that the presence of B. bruxellensis on the grape berry surface was clearly demonstrated, using an enrichment medium (Renouf and Lonvaud-Funel, 2007). The use of this formulated medium represents a useful tool to detect B. bruxellensis, as this yeast appears to have low presence and to lack competitive ability in ecological niches such as the grape berry surface. Moreover, B. bruxellensis could entry in VBNC state (Capozzi et al., 2016). Even using more sensitive recent techniques as transcriptome approach/new generation sequencing (NGS) B. bruxellensis was not isolated (Bokulich et al., 2014; Pinto et al., 2014; Kecskeméti et al., 2016; Morgan et al., 2017). Albertin et al. (2014) compared B. bruxellensis strains detected from grape and wine by microsatellite analysis. This study shows that there is not strict genetic pattern depending on the substrate. Namely, strains isolated from grape clustered together with strains coming from wine, suggesting a strong connection between grapes and the cellar.
The present study investigated the occurrence of B. bruxellensis. The presence of this yeast was monitored on the grape berry surface and in the winery during the winemaking process, with the isolated strains characterized by molecular typing.
Materials and Methods
Sampling
The 12 sites selected for this B. bruxellensis isolation and characterization study are part of a farm of over 100 ha of vineyards situated in the hills to the south-west of Florence, near the town of Vicarello, at 50 km from the winery in Montespertoli (FI), thus avoiding possible cross-contamination (Goddard et al., 2010). Indeed, the farm is only held the cultivation and harvesting grapes. The grapes mechanical harvesting, were loaded into wagons and transported to the cellar.
In this situation any anthropogenic contamination from the farm to the winery and vice versa was excluded, since there were different workers involved in the winery activity and in the vineyard management.
The sampling campaign was performed during the 2014 harvest, as grape samples collected from 12 locations in the vineyard that contained four different grapevine varieties: “Syrah,” “Merlot,” “Cabernet Franc,” and “Cabernet Sauvignon.” Each vineyard sampling site had very similar characteristics in terms of age, pruning system, canopy management, and sun exposure, and they were also characterized by similar climatic conditions (September: mean air temperature, 28°C; rainfall, 70.5 mm).
The vineyard sampling sites were almost 800 m from each other, and they were randomly selected, although taking into account the position of the grapes harvested (i.e., beginning, middle, and end of grapevines rows). A total of 26 different grape samples of 2–3 kg each were analyzed. Samples were also collected from the winery environment: from the transport trailer (100 mL of grape juice from the bottom of the grape bin), the destemmer (1 Kg of grapes), the washing water of the valves between the destemmers and the tanks (50 mL of residue liquid), the air (using an SAS-Super 180 high microbiological air sampler; Bioscience International, Rockville, MD, United States). During the fermentation seven vats of 450 hL were monitored at the start, middle, and end of the fermentation process (one Syrah, two Merlot, two Cabernet Franc, and two Cabernet Sauvignon).
All of the samples were collected aseptically and stored at 4°C until arrival in the laboratory (within 6 h of collection), and then they were processed immediately.
Detection and Isolation of B. bruxellensis From Grapes and the Winery Environment
The refrigerated bags that contained the harvested grape berries were opened in the laboratory under sterile conditions, and 45 berries per sample were taken and placed into a 250 mL sterile flask containing 150 mL of the enrichment B. bruxellensis (EBB) medium (Renouf and Lonvaud-Funel, 2007). The EBB medium had the following composition: 200 mL/L red grape juice (commercial grape juice; Folicello, Modena, Italia with 19.1% sugar, nitrogen content YAN 115 mgN/L and pH 3.4), 40 mL/L ethanol (VWR International, Milan, Italy), 1.5 g/L malt extract (Sigma-Aldrich, Milan, Italy), 1.5 g/L yeast extract, 0.5 g/L (NH4)2SO4 (Sigma-Aldrich, Milan, Italy), 0.2 g/L MgSO4, and 0.5 mL/L Tween 80 (Merck, Hohenbrunn, Germany). The pH was adjusted to 5.0 with sodium hydroxide. The EBB medium was also supplemented with 0.2 g/L (w/v) biphenyl (Fluka, Steinheim, Switzerland) and 0.05 g/L (w/v) chloramphenicol (Sigma-Aldrich, Milan, Italy), to limit mold and bacteria development, respectively. The prepared flasks were incubated at 25°C with gentle agitation (120 rpm), and after 2 h the first sample was taken for analysis on Petri dishes with the Dekkera/Brettanomyces differential medium (DBDM) (Rodrigues et al., 2001). Subsequently, to monitor the evolution of the B. bruxellensis population, further sampling of the EBB medium cultures was carried out on days 10, 40, and 80. These determinations of viable and culturable cells of Dekkera/Brettanomyces were carried out semi quantitatively, as absent (-) or present (+). To detect Brettanomyces yeasts in the winery, microbial sampling (1 mL) was performed at the following steps: (i) from the trailer, before load discharge into the destemmer; (ii) during the pressing and destemming of the grapes; (iii) during grape juice fermentation (at the start, middle, and end of process). The following grape varieties were monitored: “Syrah,” “Merlot,” “Cabernet Franc,” and “Cabernet Sauvignon.”
Fermentation was carried out in stainless steel fermentation tanks (450 hL) at 22 to 27°C. All fermentations were inoculated with S. cerevisiae commercial starter strain (Zymaflore F15 – Laffort). Samples for B. bruxellensis analysis were also taken from the connecting fermenter valves, as 10 mL of the residual washing water, with 1 mL from each solution used on Petri dishes with the DBDM medium. Finally, analysis of the air was performed using a microbiological air sampler that can suck in multiple aliquots of air (according to need), and project it directly onto a Petri dish with DBDM medium. This test was performed at the beginning and in the middle of the harvesting campaign. Three samples were also taken from sites near the exhaust (in the winery square) and near the destemmer, at the start and in the middle of the harvesting.
Molecular Identification and Biotyping of B. bruxellensis
Preliminary molecular identification of 250 presumptive B. bruxellensis strains was carried out to confirm the genus and species of these yeast. For this, all of the isolated strains underwent DNA extraction (Stringini et al., 2009) and sequence analysis of the 26S-D1/D2 region of rDNA, which identified the specific nucleotide sequences of the individual species (Kurtzman and Robnett, 1998).
The DNA extracted from the B. bruxellensis strains underwent biotyping using the RAPD-PCR primers M13 (5′-GAG GGT GGCGGT TCT-3′); M14 (5′-GAGGGT GGG GCC GTT-3′); OPC20 (5′-ACTTCGCCAC-3′); OPK03 (5′-CCAGCTTAGG-3′) and the minisatellite primers PIR1 (forward: 5′-GCCACTACTGCTTCCTCCAA-3′; reverse: 5′-TGGACCAACCAGCAGCATAG-3′) and PIR3 (forward: 5′-TCCTCCGTCGCCTCATCTAA-3′; reverse: 5′-GGCACTGAGAACCAATGTGC-3′). For molecular typing the type strain coming from University of Perugia collection (DBVPG 6706) was included. The conditions used for the amplification protocols were as described by Crauwels et al. (2014) and Canonico et al. (2015).
Statistical Analysis
The genotypic analysis of the isolates into biotype clusters was performed using JMP version 11 (JMP SAS-Genomics, United States). The cluster analysis was carried out according to the Ward’s minimum variance method, and it is represented as a constellation plot. This diagram, arranges the samples as endpoints and each cluster join as a new point. The lines represent membership in a cluster. The length of a line between cluster joins approximates the distances between the cluster that were joined.
Results
Occurrence of B. bruxellensis on Grape Surfaces and in the Winery
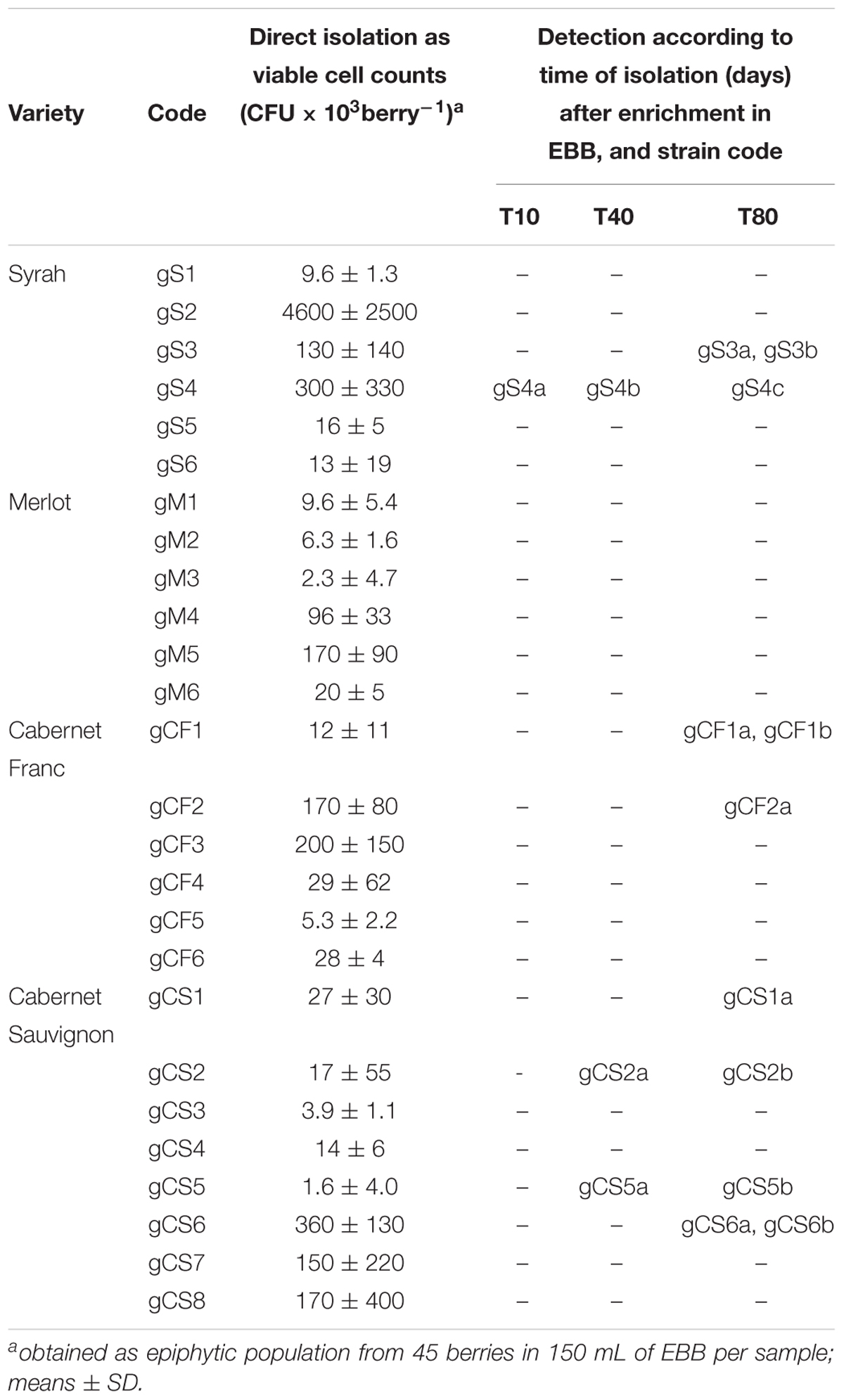
At harvest (day 0), all of the grape samples showed a quantitative yeast population of about 104 CFU/mL, without any significant variations among these, with exception of a sample of the “Syrah” variety (code: gS2), with a yeast population two orders of magnitude greater. The main representative yeast belonged to the Hanseniaspora and Pichia genera, and grew on Wallerstein laboratory (WL) agar medium (data not shown). However, no B. bruxellensis strains were detected, even using the EBB medium. After 10 days of incubation of the grapes in EBB medium, B. bruxellensis was isolated from only one Syrah sample (code: gS4), while after 40 days, a “Cabernet Sauvignon” variety was positive for Brettanomyces strains in two samples (codes: gCS2, gCS5) (Table 1). The last sampling was carried out on day 80, with B. bruxellensis isolated from eight different grape samples (Table 1). None of the “Merlot” variety grape samples showed any B. bruxellensis throughout these sampling periods. After the further cultivation in DBDM medium, three or four colonies per sample were collected, and the eight positive samples yielded 15 B. bruxellensis isolates, which were identified and stored for further molecular characterization.
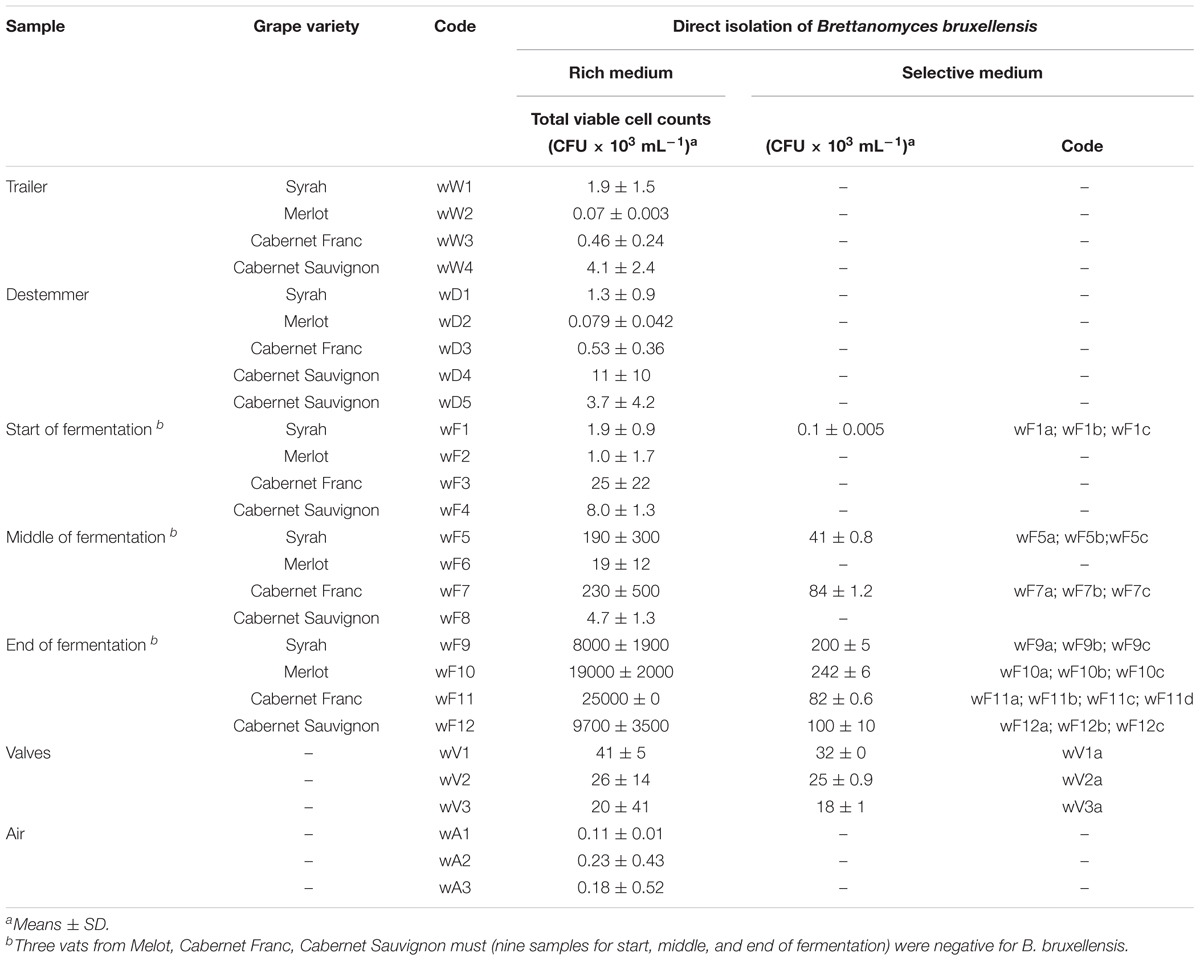
In the samples collected from the winery environment, B. bruxellensis was not detected from the trailers, the destemming process, or the air samples. The only positive samples were collected from two of the three valves (order of magnitude, 104 CFU/mL) used as connections for the transfer of the grape must from one vat to another. During the fermentation conducted under maceration conditions, only one must sample (code: wF1) was positive for B. bruxellensis (order of magnitude, 102 CFU/mL) at the initial fermentation stage, while there was wide occurrence of this B. bruxellensis spoilage yeast at the middle (two positive samples out of seven) and the end (four positive samples out of seven) of the process, where ∼105 CFU/mL was achieved (Table 2). Therefore, B. bruxellensis was isolated from four of the seven tanks monitored. Following these winery sampling procedures, a total of 24 B. bruxellensis strains were isolated, identified and stored for further molecular characterization.
B. bruxellensis From the Grapes and the Winery: Genetic Relationships
The characterization at strain level carried out using the M13, M14, OPC20 and OPK03 RAPD and the PIR1 and PIR3 minisatellite primers was reported in Figure 1. The use of the OPK03 primer showed the higher discrimination power at strain level with resulting electrophoretic profiles grouped in seven different biotypes (Figure 1D). OPC20 primer grouped the isolated into six biotypes, while the other two RAPD primers, M14 and M13 divided the strains into four and five biotypes, respectively.
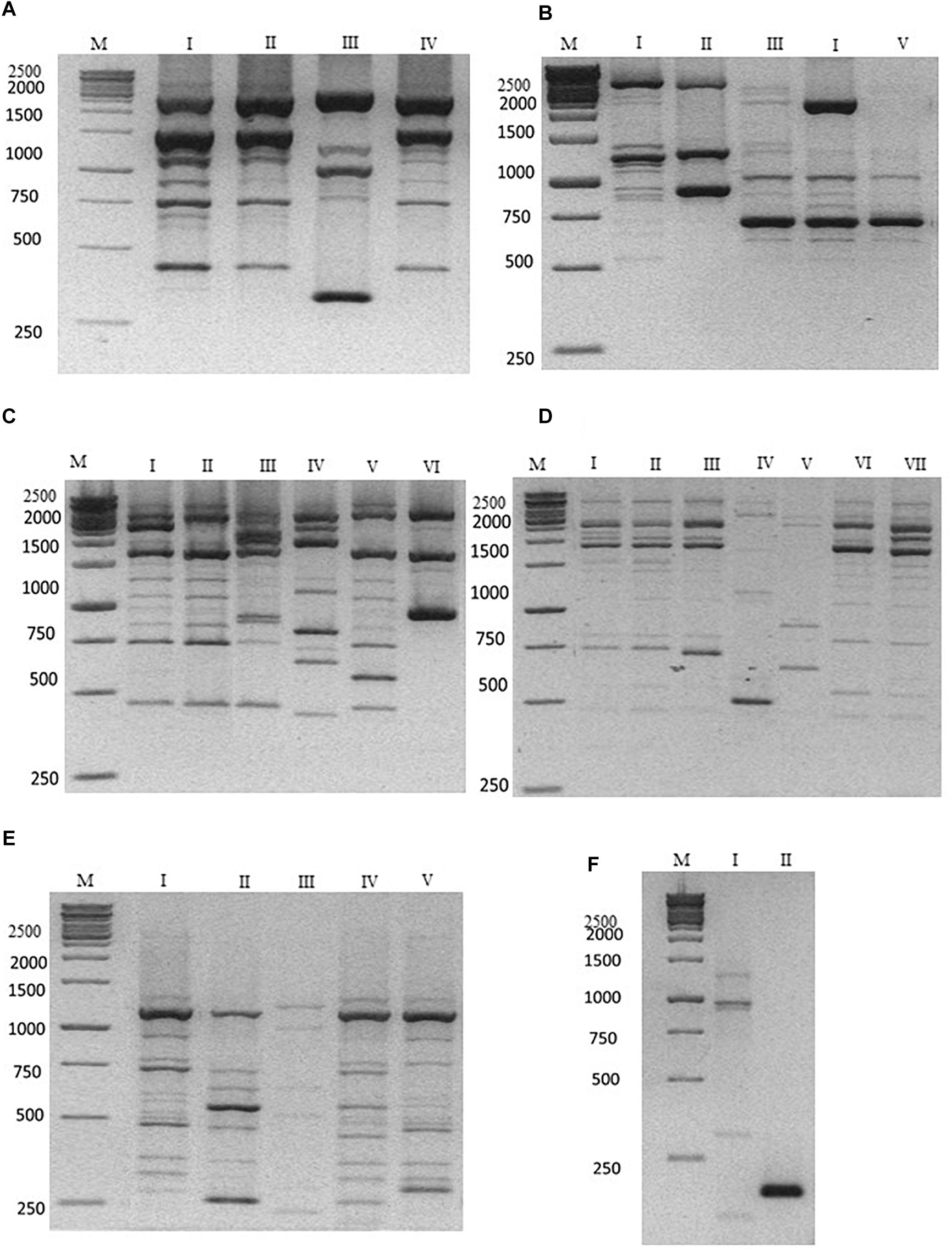
Figure 1. Representative electrophoresis analysis after PCR amplification using (A) M14; (B) M13; (C) OPC20; (D) OPK03 RAPD primers; (E) PIR1; and (F) PIR3 minisatellite primers. Lane M: Gene Ruler 1kb (Fermentas) as indicated on the left of each gel. Lane I–VII (A–F) indicate the representative biotypes, as extensively reported in Table 3.
The discriminatory power was compared with other PCR-fingerprinting based on minisatellites. In particular, PIR1 and PIR3 showed five and two different electrophoretic profiles, respectively. These last results highlighted a lower discrimination power of PIR3 primer in comparison with RAPD primers. The data obtained are summarized according to the biotype profiles in Table 3.

Table 3. The discriminatory power of primers on the B. bruxellenisis isolates (see Figure 1).
The overall typing using the combination of above mentioned primers showed 22 different biotypes as reported in the constellation plot using Ward clustering (Figure 2) and in tree with Bootstrap analysis (Figure 1 and Supplementary Material). The plot showed three main biotypes, containing isolates coming both from grapes and winery. One of these biotypes contained the isolates gCF1a and gCF1b (grape from Cabernet Franc); gS4b and gS3b (grape from Syrah); wF9c, wF10c and wF11c (end of fermentation, winery); wV3a (from valve, winery). This result showed that the isolates coming from winery and grapes are grouped together and could be considered clones of the same genotype. The second largest biotype includes two isolates gCS1a and gCS2b (grape from Cabernet Sauvignon) and three isolates wF11b, wF9b, wF12a (end fermentation, from winery). The last largest biotype is formed by isolates coming from winery environment (middle fermentation and valve) (wF7b, wV1a, and wV2a), and one isolate from Syrah grape (gS4a). The Figure 2 showed also three small biotypes: one includes two isolates coming from two different winery samples at the end fermentation (wF11c, wF12b); the second one is composed by wF5c, isolate coming from winery sample at middle fermentation and gCS6b (grape from Cabernet Sauvignon). The last one contains two strains from the Cabernet Sauvignon (gCS5a and gCS5b). Finally, 15 isolates (five from grapes and 10 from winery), showed unique biotype.
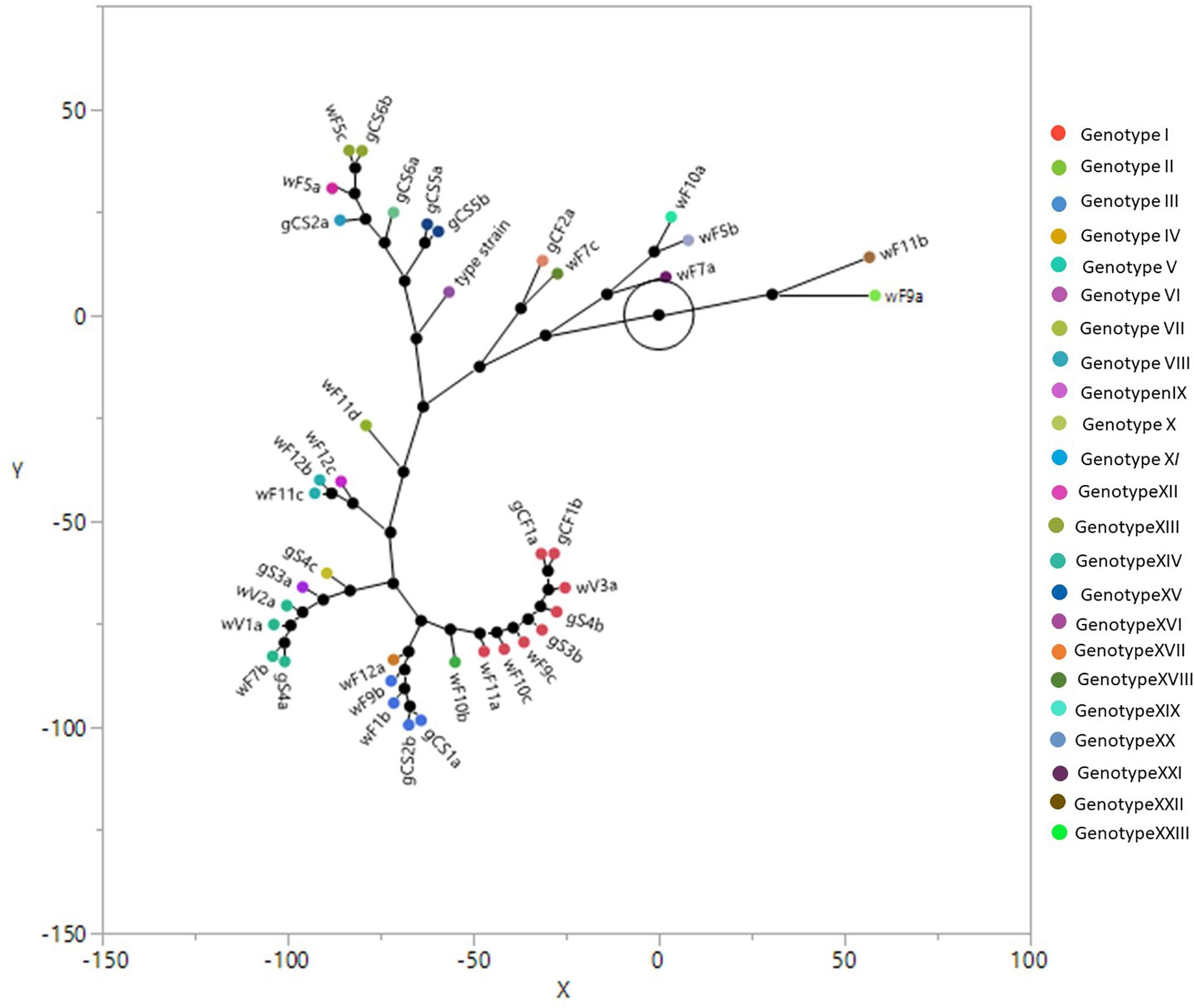
Figure 2. Ward clustering and constellation plot of the Brettanomyces bruxellensis biotypes using M13, M14, OPC20 and OPK03 RAPD primers and the PIR1 and PIR3 minisatellite primers. For yeast strain codes, see Tables 1, 2.
Discussion
Nowadays, the issue of the B. bruxellensis origins in the winemaking field remains under debate and has never been clearly demonstrated. The origins of these spoilage yeast in the vineyard has been long neglected in favor of the winery (Ibeas et al., 1996). This has probably been due to their poor detection on grapes, because of their low growth and competition ability, poor surface adhesion, the use of chemical treatments, possible entrance in a VBNC state and possible negative interactions with other microorganisms (Arvik and Henick-Kling, 2002; Schifferdecker et al., 2014; Coton et al., 2017). Indeed, isolation of B. bruxellensis from a complex yeast community generally requires differential medium formulations that include cycloheximide, to eliminate the sensitive fastest growing species (Rodrigues et al., 2001; Morneau et al., 2011). However, B. bruxellensis was probably not isolated from grapes due to the lack of specific cultivation methods and due their presence in very low numbers. The combined use of the specific enrichment liquid medium (i.e., EBB) and the selective DBDM agar medium (the Dekkera/ Brettanomyces differential medium) (Rodrigues et al., 2001; Renouf and Lonvaud-Funel, 2007; Garijo et al., 2015) has now allowed the isolation of B. bruxellensis strains from grapes.
The present study has confirmed the previous failure to directly isolate B. bruxellensis from the grape surface, through our use of the EBB selective medium. Thus, we isolated B. bruxellensis from eight samples out of 26 using this combination of enrichment and selective media. In contrast, during the must fermentation, B. bruxellensis was directly detected with quantitative levels from 102 CFU/mL to 105 CFU/mL, in seven of the total of 21 samples. Indeed, although B. bruxellensis isolates are usually present as a minority species in the winery environment, they can increase in number during the more nutritionally advantageous conditions, while exploiting their slow metabolism (Fugelsang et al., 1993). This can occur in particular once the alcoholic fermentation is almost complete, when the trace levels of residual sugars could allow B. bruxellensis to proliferate (Oelofse et al., 2008). Indeed, wine will represent a suitable ecological niche for some B. bruxellensis strains that can exploit its chemical characteristics, such as the presence of oxygen (i.e., pumping over; a wooden barrel), the sufficient nitrogen levels, the ethanol presence, the low pH, and the presence of various vitamins (Smith and Divol, 2016).
However, in the present study in the winery environment, the spread of B. bruxellensis was limited. Indeed, no B. bruxellensis were detected in the air, or in the trailer or the destemmer. This agrees with previous studies (Donnelly, 1977), although it is in contrast with (Stratford, 2006), who reported Brettanomyces/Dekkera in the air samples taken near to barrels located in an area that communicated with the outdoors. Other studies have reported the presence of B. bruxellensis in wineries using a combination of culture-based methods and direct detection with molecular probes, and particularly for equipments that are difficult to sanitize, such as cement storage vessels and oak barrels (Fugelsang, 1997; Curtin et al., 2015). In agreement with their results, we isolated B. bruxellensis from the valves, where must and wine can remain during the transfer operations from one fermenter to another. Although these connections are washed, some B. bruxellensis cells might survive, to find favorable conditions for their colonization and multiplication, which creates a dangerous and uncontrolled transit system in the winery.
Regarding the uncertain origins of B. bruxellensis wine strains, in the present study some aspects of the ecology of these yeasts were elaborated suggesting the grape surfaces as primary sources. The clustering of the biotypes did not show any clear separation between the vineyard and winery strains, but rather the presence of the same biotypes on the grapes and in the winery. Indeed, analyzing all B. bruxellensis molecular profiles after characterization procedure, it can be noted that biotypes include several isolates coming from vineyard and winery, without a clear distinction between the two environments. In this specific condition (the large distance 50 Km, the exclusion of any human flux from winery to vineyard) the grape surface could represents the primary source of these B. bruxellensis even if other contamination ways such as insects or birds could be not theoretically excluded. In this regard, to support this hypothesis further investigations involving more years and/or other vineyards and wineries needed, also in the light of the patterns depicted studying the diversity of the model organism S. cerevisiae on a comparable scale (Knight and Goddard, 2015). On the other hand, the wine and some of the winery equipments are more suitable ecological niche for B. bruxellensis colonization, growth and diffusion. Indeed, almost all of B. bruxellensis strains that were isolated from the grapes showed the same biotypes as those recovered in the winery (i.e., from the valves and the final stages of grape must fermentation). Furthermore, the absence of B. bruxellensis strains from the “Merlot” grapevine variety from the same environment indicates that the grape variety can influence the presence of certain yeast, in agreement with Raspor et al. (2006) and Cordero-Bueso et al. (2011), who reported strong correlations among grape varieties and yeast biota.
In conclusion, the results of the present study show that B. bruxellensis can be isolated (with great difficulty) from grapes. We did not find distinct populations between strains from grapes and strains from winery, but the presence of the same biotypes in both environments was observed. In the condition tested, this statement suggests a B. bruxellensis flux from the grapes to the winery even if more investigations (subsequent vintages and/or more vineyards and wineries) are necessary to generalize this evidence. B. bruxellensis reveals a great metabolic plasticity, as its appear to quickly adapt to the winery environmental. The differences between grape varieties might play a role for the presence and diffusion B. bruxellensis in the winemaking environment, although further investigations are needed to confirm this hypothesis.
Author Contributions
LO, LC, VM, MC, and FC contributed equally to this manuscript. All authors participated in the design and discussion of the research. LO and VM carried out the experimental part of the work. LC, LO, MC, and FC carried out the analysis of the data and wrote the manuscript. All authors have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Marchesi De’ Frescobaldi Società Agricola s.r.l. for the support of the research, and two reviewers who helped to improve the quality of the final manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00415/full#supplementary-material
References
Agnolucci, M., Vigentini, I., Capurso, G., Merico, A., Tirelli, A., Compagno, C., et al. (2009). Genetic diversity and physiological traits of Brettanomyces bruxellensis strains isolated from Tuscan Sangiovese wines. Int. J. Food Microbiol. 130, 238–244. doi: 10.1016/j.ijfoodmicro.2009.01.025
Aguilar-Uscanga, G. M., Delia, M. L., and Strehaiano, P. (2000). Nutritional requirements of Brettanomyces bruxellensis: growth and physiology in batch and chemostat cultures. Can. J. Microbiol. 46, 1046–1050. doi: 10.1139/w00-089
Albertin, W., Panfili, A., Miot-Sertier, C., Goulielmakis, A., Delcamp, A., Salin, F., et al. (2014). Development of microsatellite markers for the rapid and reliable genotyping of Brettanomyces bruxellensis at strain level. Food Microbiol. 42, 188–195. doi: 10.1016/j.fm.2014.03.012
Arvik, T. J., and Henick-Kling, T. (2002). “Overview of Brettanomyces, its occurrence growth and effect on wine flavors,” in Proceeding of 31st Annual New York Wine Industry Workshop, New York, NY.
Avramova, M., Cibrario, A., Peltier, E., Coton, M., Coton, E., Schacherer, J., et al. (2018). Brettanomyces bruxellensis population survey reveals a diploid-triploid complex structured according to substrate of isolation and geographical distribution. Sci. Rep. 8:4136. doi: 10.1038/s41598-018-22580-7
Barata, A., Caldeira, J., Botelheiro, R., Pagliara, D., Malfeito-Ferreira, M., and Loureiro, V. (2008). Survival patterns of Dekkera bruxellensis in wines and inhibitory effect of sulphur dioxide. Int. J. Food Microbiol. 121, 201–207. doi: 10.1016/j.ijfoodmicro.2007.11.020
Benito, S., Palomero, F., Morata, A., Calderón, F., and Suárez-Lepe, J. A. (2009). Factors affecting the hydroxycinnamate decarboxylase/vinylphenol reductase activity of Dekkera/Brettanomyces: application for Dekkera/Brettanomyces control in red wine making. J. Food Sci. 74, M15–M22. doi: 10.1111/j.1750-3841.2008.00977.x
Bokulich, N. A., Thorngate, J. H., Richardson, P. M., and Mills, D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111, E139–E148. doi: 10.1073/pnas.1317377110
Boulton, R. B., Singleton, V. L., Bisson, L. F., and Kunkee, R. E. (2013). Principles and Practices of Winemaking. Berlin: Springer Science & Business Media.
Campolongo, S., Rantsiou, K., Giordano, M., Gerbi, V., and Cocolin, L. (2010). Prevalence and biodiversity of Brettanomyces bruxellensis in wine from Northwestern Italy. Am. J. Enol. Vitic. 61, 486–491. doi: 10.5344/ajev.2010.10034
Canonico, L., Comitini, F., and Ciani, M. (2015). TdPIR minisatellite fingerprinting as a useful new tool for Torulaspora delbrueckii molecular typing. Int. J. Food Microbiol. 200, 47–51. doi: 10.1016/j.ijfoodmicro.2015.01.020
Capozzi, V., Di Toro, M. R., Grieco, F., Michelotti, V., Salma, M., Lamontanara, A., et al. (2016). Viable But Not Culturable (VBNC) state of Brettanomyces bruxellensis in wine: new insights on molecular basis of VBNC behaviour using a transcriptomic approach. Food Microbiol. 59, 196–204. doi: 10.1016/j.fm.2016.06.007
Chatonnet, P., Dubourdie, D., Boidron, J. N., and Pons, M. (1992). The origin of ethylphenols in wines. J. Sci. Food Agric. 60, 165–178. doi: 10.1111/j.1574-6968.2008.01192.x
Chatonnet, P., Masneuf, I., Gubbiotti, M. C., and Dubourdieu, D. (1999). Prévention et détection des contaminations par Brettanomyces au cours de la vinification et de l’élevage des vins. Rev. Fr. Oenol. 179, 20–24.
Ciani, M., and Comitini, F. (2014). “Brettanomyces,” in Encyclopedia of Food Microbiology, eds C. A. Batt and M. L. Tortorello (Amsterdam: Elsevier), 316–323. doi: 10.1016/B978-0-12-384730-0.00046-X
Cordero-Bueso, G., Arroyo, T., Serrano, A., Tello, J., Aporta, I., Vélez, M. D., et al. (2011). Influence of grape farming system and vine variety on yeast communities associated with grape berries. Int. J. Food Microbiol. 145, 132–139. doi: 10.1016/j.ijfoodmicro.2010.11.0
Coton, M., Pawtowski, A., Taminiau, B., Burgaud, G., Deniel, F., Coulloumme-Labarthe, L., et al. (2017). Unraveling microbial ecology of industrial-scale Kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiol. Ecol. 93, 1–16. doi: 10.1093/femsec/fix048
Coulon, J., Raffestin, V., Bellan, M., and Lonvaud-Funel, A. (2011). “Molecular SO2 effect on Brettanomyces bruxellensis populations during a one-year storage,” in Proceedings of the 9th International Symposium of Oenology, Bordeaux.
Crauwels, S., Zhu, B., Steensels, J., Busschaert, P., De Samblanx, G., Marchal, K., et al. (2014). Assessing genetic diversity among Brettanomyces yeasts by DNA fingerprinting and whole-genome sequencing. Appl. Environ. Microbiol. 80, 4398–4413. doi: 10.1128/AEM.00601-14
Curtin, C., Varela, C., and Borneman, A. (2015). Harnessing improved understanding of Brettanomyces bruxellensis biology to mitigate the risk of wine spoilage. Aust. J. Grape Wine R. 21, 680–692. doi: 10.1111/ajgw.12200
Curtin, C. D., and Pretorius, I. S. (2014). Genomic insights into the evolution of industrial yeast species Brettanomyces bruxellensis. FEMS Yeast Res. 14, 997–1005. doi: 10.1111/1567-1364.12198
Di Toro, M. R., Capozzi, V., Beneduce, L., Alexandre, H., Tristezza, M., Durante, M., et al. (2015). Intraspecific biodiversity and ‘spoilage potential’of Brettanomyces bruxellensis in Apulian wines. LWT Food Sci. Technol. 60, 102–108. doi: 10.1016/j.lwt.2014.06.059
Dias, L., Pereira-da-Silva, S., Tavares, M., Malfeito-Ferreira, M., and Loureiro, V. (2003). Factors affecting the production of 4-ethylphenol by the yeast Dekkera bruxellensis in enological conditions. Food Microbiol. 20, 377–384. doi: 10.1016/S0740-0020(03)00023-6
Donnelly, D. M. (1977). Airborne microbial contamination in a winery bottling room. Am. J. Enol. Vitic. 28, 176–181.
Fugelsang, K. C. (1997). Wine Microbiology. New York: Chapman & Hall. doi: 10.1007/978-1-4757-6970-8
Fugelsang, K. C., Osborn, M. M., and Muller, C. J. (1993). “Brettanomyces and Dekkera. Implications in winemaking,” in Beer and Wine Production: Analysis, Characterization and Technological Advances, ed. B. H. Gump (Washington, DC: American Chemical Society), 110–131. doi: 10.1021/bk-1993-0536.ch007
Garijo, P., Gonzalez-Arenzana, L., Lopez-Alfaro, I., Garde-Cerdan, T., Lopez, R., Santamaria, P., et al. (2015). Analysis of grapes and the first stages of the vinification process in wine contamination with Brettanomyces bruxellensis. Eur. Food Res. Technol. 240, 525–532. doi: 10.1007/s00217-014-2351-4
Gilliland, R. B. (1961). Brettanomyces. I. Occurrence, characteristics, and effects on beer flavour. J. Inst. Brew. 67, 257–261. doi: 10.1002/j.2050-0416.1961.tb01791.x
Goddard, M. R., Anfang, N., Tang, R., Gardner, R. C., and Jun, C. (2010). A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ. Microbiol. 12, 63–73. doi: 10.1111/j.1462-2920.2009.02035.x
Greenwalt, C. J., Steinkraus, K. H., and Ledford, R. A. (2000). Kombucha, the fermented tea: microbiology, composition, and claimed health effects. J. Food Protect. 63, 976–981. doi: 10.4315/0362-028X-63.7.976
Heresztyn, T. (1986). Formation of substituted tetrahydropyridines by species of Brettanomyces and Lactobacillus isolated from mousy wines. Am. J. Enol. Vitic. 37, 127–132.
Ibeas, J. I., Lozano, I., Perdigones, F., and Jimenez, J. (1996). Detection of Dekkera/Brettanomyces strains in sherry by a nested PCR method. Appl. Environ. Microbiol. 62, 998–1003.
Kecskeméti, E., Berkelmann-Löhnertz, B., and Reineke, A. (2016). Are epiphytic microbial communities in the carposphere of ripening grape clusters (Vitis vinifera L.) different between conventional, organic, and biodynamic grapes? PLoS One 11:e0160852. doi: 10.1371/journal.pone.0160852
Knight, S., and Goddard, M. R. (2015). Quantifying separation and similarity in a Saccharomyces cerevisiae metapopulation. ISME J. 9, 361–370. doi: 10.1038/ismej.2014.132
Kurtzman, C. P., and Robnett, C. J. (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73, 331–371. doi: 10.1023/A:1001761008817
Loureiro, V., and Malfeito-Ferreira, M. (2003). Spoilage yeasts in the wine industry. Int. J. Food Microbiol. 86, 23–50. doi: 10.1016/S0168-1605(03)00246-0
Martens, H., Iserentant, D., and Verachtert, H. (1997). Microbiological aspects of a mixed yeast bacterial fermentation in the production of a special Belgian acidic ale. J. Inst. Brew. 103, 85–91. doi: 10.1002/j.2050-0416.1997.tb00939.x
Morgan, H. H., du Toit, M., and Setati, M. E. (2017). The grapevine and wine microbiome: insights from high-throughput amplicon sequencing. Front. Microbiol. 8:820. doi: 10.3389/fmicb.2017.00820
Morneau, A. D., Zuehlke, J. M., and Edwards, C. G. (2011). Comparison of media formulations used to selectively cultivate Dekkera/Brettanomyces. Lett. Appl. Microbiol. 53, 460–465. doi: 10.1111/j.1472-765X.2011.03133.x
Morrissey, W., Davenport, B., Querol, A., and Dobson, A. (2004). The role of indigenous yeasts in traditional Irish cider fermentations. J. Appl. Microbiol. 97, 647–655. doi: 10.1111/j.1365-2672.2004.02354.x
Nardi, T., Remize, F., and Alexandre, H. (2010). Adaptation of yeasts Saccharomyces cerevisiae and Brettanomyces bruxellensis to winemaking conditions: a comparative study of stress genes expression. Appl. Microbiol. Biotechnol. 88, 925–937. doi: 10.1007/s00253-010-2786-x
Oelofse, A., Pretorius, I. S., and du Toit, M. (2008). Significance of Brettanomyces and Dekkera during winemaking: a synoptic review. S. Afr. J. Enol. Vitic. 29, 128–144.
Pinto, C., Pinho, D., Sousa, S., Pinheiro, M., Egas, C., and Gomes, A. C. (2014). Unravelling the diversity of grapevine microbiome. PLoS One 9:e85622. doi: 10.1371/journal.pone.0085622
Pretorius, I. S. (2000). Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16, 675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B
Raspor, P., Milek, D. M., Polanc, J., Mozina, S. S., and Cadez, N. (2006). Yeasts isolated from three varieties of grapes cultivated in different locations of the Dolenjska wine-growing region, Slovenia. Int. J. Food Microbiol. 109, 97–102. doi: 10.1016/j.ijfoodmicro.2006.01.017
Renouf, V., and Lonvaud-Funel, A. (2007). Development of an enrichment medium to detect Dekkera/Brettanomyces bruxellensis, a spoilage wine yeast, on the surface of grape berries. Microbiol. Res. 162, 154–167. doi: 10.1016/j.micres.2006.02.006
Rodrigues, N., Gonçalves, G., Pereira-da-Silva, S., Malfeito-Ferreira, M., and Loureiro, V. (2001). Development and use of a new medium to detect yeast of the genera Dekkera/Brettanomyces sp. J. Appl. Microbiol. 90, 588–599. doi: 10.1046/j.1365-2672.2001.01275.x
Schifferdecker, A. J., Dashko, S., Ishchuk, O. P., and Piškur, J. (2014). The wine and beer yeast Dekkera bruxellensis. Yeast 31, 323–332. doi: 10.1002/yea.3023
Silva, P., Cardoso, H., and Géros, H. (2004). Studies on the wine spoilage capacity of Brettanomyces/Dekkera spp. Am. J. Enol. Vitic. 55, 65–72. doi: 10.1590/S1517-83822013005000010
Smith, B. D., and Divol, B. (2016). Brettanomyces bruxellensis, a survivalist prepared for the wine apocalypse and other beverages. Food Microbiol. 59, 161–175. doi: 10.1016/j.fm.2016.06.008
Spitaels, F., Wieme, A. D., Janssens, M., Aerts, M., Daniel, H. M., Van Landschoot, A., et al. (2014). The microbial diversity of traditional spontaneously fermented lambic beer. PLoS one 9:e95384. doi: 10.1371/journal.pone.0095384
Steensels, J., Daenen, L., Malcorps, P., Derdelinckx, G., Verachtert, H., and Verstrepen, K. J. (2015). Brettanomyces yeasts - from spoilage organisms to valuable contributors to industrial fermentations. Int. J. Food Microbiol. 206, 24–38. doi: 10.1016/j.ijfoodmicro.2015.04.005
Stratford, M. (2006). “Food and beverage spoilage yeasts,” in Yeasts in Food and Beverages, eds H. Querol and G. Fleet (Berlin: Springer-Verlag), 335–379. doi: 10.1007/978-3-540-28398-0_11
Stringini, M., Comitini, F., Taccari, M., and Ciani, M. (2009). Yeast diversity during tapping and fermentation of palm wine from Cameroon. Food Microbiol. 26, 415–420. doi: 10.1016/j.fm.2009.02.006
Suárez, R., Suárez-Lepe, J. A., Morata, A., and Calderón, F. (2007). The production of ethylphenols in wine by yeasts of the genera Brettanomyces and Dekkera. A review. Food Chem. 102, 10–21. doi: 10.1016/j.foodchem.2006.03.030
Teoh, A. L., Heard, G., and Cox, J. (2004). Yeast ecology of Kombucha fermentation. Int. J. Food Microbiol. 95, 119–126. doi: 10.1016/j.ijfoodmicro.2003.12.020
Valdetara, F., Fracassetti, D., Campanello, A., Costa, C., Foschino, R., Compagno, C., et al. (2017). A response surface methodology approach to investigate the effect of sulfur dioxide, pH, and ethanol on DbCD and DbVPR gene expression and on the volatile phenol production in Dekkera/Brettanomyces bruxellensis CBS2499. Front. Microbiol. 8:1727. doi: 10.3389/fmicb.2017.01727
Verachtert, H. (1992). Lambic and gueuze brewing: mixed cultures in action. Microbial. Contam. 7, 243–262.
Keywords: Brettanomyces bruxellensis, grape berry, winery, molecular characterization, ecological distribution
Citation: Oro L, Canonico L, Marinelli V, Ciani M and Comitini F (2019) Occurrence of Brettanomyces bruxellensis on Grape Berries and in Related Winemaking Cellar. Front. Microbiol. 10:415. doi: 10.3389/fmicb.2019.00415
Received: 19 September 2018; Accepted: 18 February 2019;
Published: 07 March 2019.
Edited by:
Vittorio Capozzi, University of Foggia, ItalyReviewed by:
Lucía González-Arenzana, Instituto de Ciencias de la Vid y del Vino (ICVV), SpainFrancesco Grieco, Istituto Scienze delle Produzioni Alimentari (CNR), Italy
Copyright © 2019 Oro, Canonico, Marinelli, Ciani and Comitini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesca Comitini, Zi5jb21pdGluaUB1bml2cG0uaXQ=
 Lucia Oro
Lucia Oro Laura Canonico
Laura Canonico Valentina Marinelli
Valentina Marinelli Maurizio Ciani
Maurizio Ciani Francesca Comitini
Francesca Comitini