- 1Research Unit on Communicable Diseases, Laboratory of Research in Applied Biology, Polytechnic School of Abomey-Calavi, University of Abomey-Calavi, Abomey-Calavi, Benin
- 2Microbial Resources Division, Institute of Bioresources and Sustainable Development (IBSD), Takyelpat Institutional Area, Imphal, India
- 3Central Livestock Laboratory, Niamey, Niger
Traditional Wagashi cheese and fermented cow milk are among the most popular dairy products appreciated by people from Benin, Niger, and the neighboring region. These products are the main source of protein in the diet of the low-income population in the region. The fermented milk is prepared by spontaneous fermentation without back-slopping. Whereas, the leaf extract of Calotropis procera is used for curdling the milk to prepare the soft Wagashi cheese. The present study aims to provide in-depth analysis of yeast communities associated with these traditional milk products by high-throughput Illumina MiSeq amplicon sequencing of internal transcribed spacer (ITS) region of fungal rRNA genes. A total of 60 samples, 20 samples of fermented milk each from Benin and Niger, and 20 samples of Wagashi cheese from Benin were used for analysis. The metagenomic investigation revealed that Kluyveromyces marxianus, Saccharomyces cerevisiae, Candida parapsilosis, and Sagenomella keratitidis were the predominant yeast species present in the traditional milk products. Furthermore, we noticed a high presence of K. marxianus (61.1% relative abundance) in the Wagashi cheese and S. cerevisiae (28.4% relative abundance) in the fermented milk of Niger. The presence of potential pathogenic yeast C. parapsilosis and S. keratitidis in these African milk products calls for further investigation to assess their safety. The predominant yeast K. marxianus and S. cerevisiae, recognized with generally regarded as safe (GRAS) status, could be further selected as starter culture along with lactic acid bacteria for developing controlled fermentation processes with enhanced product quality and safety.
Introduction
In most of Sub-Sahara Africa countries, milk production occupies a prominent place due to its economic value, as well as its nutritional, social, and cultural importance (Mattiello et al., 2018). Several projects in the region have aimed at improving the reproductive potential and milk production potential of the local breeds. Republics of Niger and Benin are considered as significant producers of milk in the region with annual cow milk productions of 596,968 and 109,660 tons, respectively (Gagara et al., 2017; Ministry of Agriculture Livestock and Fisheries [MAEP], 2017). Because of its high perishability, the raw milk produced in Benin and Niger is transformed into various derived products by traditional processes. These milk products do not meet regulatory quality requirements and consumer preferences because of unhygienic conditions for milking, uncontrolled processes, and storage practices. Among these, spontaneously fermented milk and Wagashi cheese are considered as the major products and most appreciated by the consumers in Benin and Niger (Boko et al., 2016; Dossou et al., 2016; Gagara et al., 2017; Okry et al., 2017). The naturally fermented milk produced in the two countries is obtained from cow milk by incubation of unpasteurized milk for 1 day at room temperature to allow spontaneous fermentation without back-slopping (Figure 1). Fermented milk is commonly used as a dessert or refreshment by people in these countries (Boko et al., 2016). These by-products are characterized by high variability from producer to producer due to uncontrolled spontaneous fermentation (Mattiello et al., 2018). Wagashi (also called as Gassire in the local Fulfulde language) is a soft fresh cheese produced from cow milk. For its preparation, milk is cleaned from impurities and boiled for about 5 min. Each liter of boiled milk is mixed with about 0.5 L of fresh unheated milk with the extract of Calotropis procera leaves (approximately 15 g). The mixture is kept at a warm temperature (60–70°C) until coagulation is achieved. After coagulation, the heating is stopped following the separation of curd and whey. The heated curd is then drained and molded without being pressed and incubated at room temperature (Dossou et al., 2006; Tossou, 2018). Wagashi cheese is a source of protein and other essential nutrients, especially for low-income populations and commonly used as a replacement of meat or fish in different dishes (Ministry of Agriculture Livestock and Fisheries [MAEP], 2017). Despite the importance of these dairy products in the diet of the Benin and Niger people, very limited information is currently available on the microbial species associated with these indigenous products. Moreover, understanding the microbial and biochemical changes in these traditional products is the first step toward developing technologies for their safety and quality improvement. Besides bacteria, yeasts frequently occur in dairy products and represent an important component of many cheese varieties and fermented milk products (Fasoli et al., 2015). As a part of starter cultures together with bacteria, yeasts especially relevant in fermented products contribute to the ripening of cheese, enhancing flavor development and accelerating maturation (Garcia et al., 2004; Legras et al., 2018). Thus, special attention is given to the role of yeasts in the complex microbial interactions prevailing in several dairy products and to control fermentation, maturation, and important product characteristics (Jakobsen and Narvhus, 1996). To our knowledge, previous studies conducted on fungal ecology of Wagashi cheese and naturally fermented milk from Benin and Niger are limited by cultivation on conventional media (Sessou, 2013). As it is well known that the cultivability of the microbiota is still a limiting factor in understanding the microbial ecology of natural food fermentation, cultivation-independent in-depth metagenomic analysis by amplicon sequencing of the rRNA gene is used to get a realistic view of the microbial community associated with traditionally fermented food products. Most of the next-generation sequencing-based metagenomics studies on fermented milk products described bacterial diversity in the natural milk fermentation (Parker et al., 2018; Shangpliang et al., 2018). There is a need to know the yeast ecology of these milk products to understand their role in the fermentation, maturation, and quality. We aim to use Illumina MiSeq amplicon sequencing of ITS region of rRNA gene amplified from the food metagenome to get a realistic view of yeast community associated with the naturally fermented milk and cheese products of Benin and Niger. The present study is the first cultivation-independent report on yeast community structure present in the naturally fermented milk products from African countries.
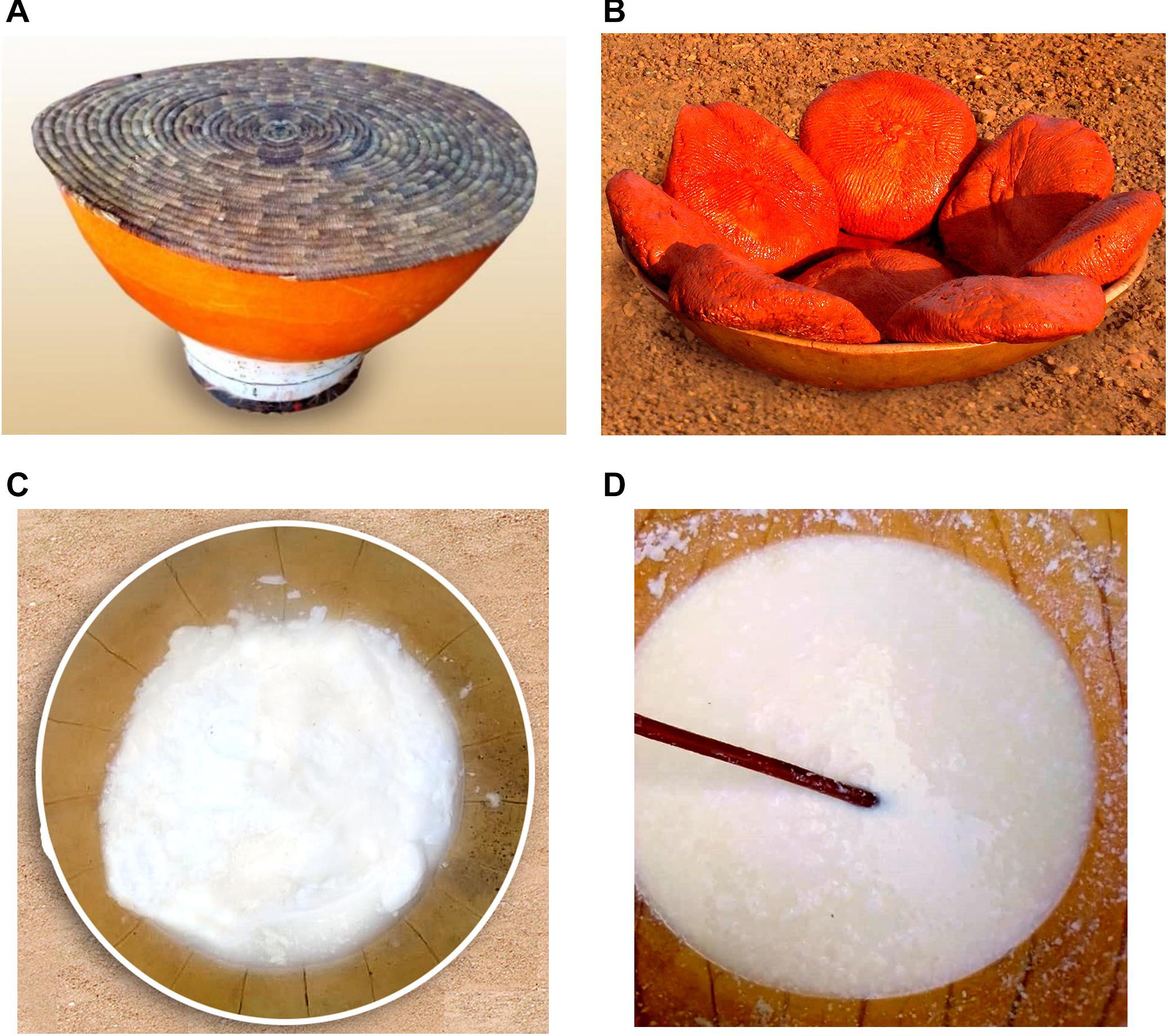
Figure 1. Pictures of the studied dairy products of Benin and Niger. (A) The traditional container used for milk fermentation in Niger, (B) the pink colored Wagashi cheese marketed in Benin, and the naturally fermented milk product of Benin (C) and Niger (D) are shown here. The colored Wagashi cheese is prepared by soaking into a millet leaf brine for the pink color formation.
Materials and Methods
Sampling
Twenty samples of naturally fermented milk were collected from three regions, namely, Niamey, Tillaberi, and Dosso of Niger, and 20 samples of each milk product, namely, Wagashi cheese and naturally fermented milk were collected from different geographical regions, namely, Djougou, Parakou, Pehunco, Agouna, and Houeyogbe of Benin. The fermented milk samples were aseptically collected from the producers in Fulani camps whereas Wagashi cheese samples were collected from retailers in the markets of targeted areas, transported in an ice-box, and stored in the laboratory at −80°C.
Metagenomic DNA Extraction
DNA was extracted as per the method described in Keisam et al. (2016) with some minor modifications. Briefly, 2 g of Wagashi cheese or 2 mL of fermented milk samples were homogenized with 98 mL 2% sodium citrate buffer in a Stomacher® 400 Circulator (Seward, United Kingdom) at 200 rpm for 2 min. After homogenization, the big debris was allowed to settle down and the clear homogenate was used for DNA extraction; 1.5 mL of the homogenate was transferred to a sterile 2 mL screw-cap tube containing 0.5 g of zirconia/silica beads (0.1 mm) (BioSpec Products, Inc., United States) and four glass beads (2 mm) and centrifuged for 10 min at 18,000 × g. To the pellet, 400 μL of TES buffer (50 mM Tris, 1 mM EDTA, 8.7% sucrose, pH 8), 50 KU lysozyme, 25 U mutanolysin, and 20 U lyticase (Sigma-Aldrich, United States) were added and incubated at 37°C for 1 h. After incubation, 25 μL Proteinase-K (25 mg/mL) (Himedia, India) was added to the mixture and further incubated at 65°C for 1 h. This was followed by the addition of 300 μL pre-warmed (65°C) NTS buffer (0.2 M NaCl, 0.1 M Tris, 2% SDS) (Promega, United States) and further incubation at 65°C for 10 min. The DNA was then purified once with phenol and twice with chloroform: isoamyl alcohol mixture (24:1) (Merck, India) each time with centrifugation at 4°C for 15 min at 15,000 × g. DNA was precipitated with isopropanol and the DNA pellet dissolved in 50 μL TE buffer (10 mM Tris, 1 mM EDTA) (Sigma-Aldrich, United States). The quality (A260/280 and A260/230) and quantity of the extracted DNA were measured using spectrophotometer (NanoDrop ND-1000, United States). DNA was stored at −20°C until required.
Barcoded Illumina MiSeq Sequencing
For in-depth yeast community analyses, barcoded Illumina MiSeq amplicon sequencing targeting the variable internal transcribed spacer (ITS) region was performed using a forward primer (ITS1-F: 5′-CTTGGTCATTTAGAGGAAGTAA-3′) and reverse primer (ITS2: 5′-GCTGCGTTCTTCATCGATGC-3′) (White et al., 1990; Motooka et al., 2017). The 5′ end of the reverse primers was barcoded with a 12-bp error correcting Golay barcodes (Caporaso et al., 2012) to enable sample multiplexing. The complete list of the forward and barcoded reverse primers used in the present study is listed in Supplementary Table S1. Each PCR was performed in a 25 μL reaction volume containing 1X high-fidelity reaction buffer, 1.0 mM MgCl2, 0.1 μM forward primer, 0.1 μM reverse primer (IDT, United States), 0.5 U Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA, United States) and nuclease-free water, with the following conditions: 94°C for 1 min; 94°C for 30 s, 52°C for 30 s and 68°C for 30 s for 35 cycles and 68°C for 7 min. A template-free reaction was used as the control. The PCR products were separated in a 2.0% agarose gel (w/v) and purified using QIAquick gel extraction kit (Qiagen, New Delhi, India) as per the manufacturer’s instructions. The purified DNA was quantified using Qubit dsDNA BR Assay Kit (Invitrogen, United States) in a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, United States) and the individual samples were pooled in equimolar proportions. The final DNA pool was sent to the NGS facility in Xcelris Genomics (Ahmedabad, India) for paired-end Illumina MiSeq sequencing.
Data Processing
The raw sequence reads obtained was analyzed using QIIME2 version 2019.11 bioinformatics pipeline (Caporaso et al., 2012). Briefly, removal of adaptor sequences, generation of paired-end reads and sample demultiplexing was performed by QIIME2 scripts2 in a Linux platform following the procedure used by Romi et al. (2015). The resulting sequences were quality-filtered based on the number of ambiguous bases and Phred quality scores using the default options. Chimera checking and removal of chimeric sequences were performed using the ChimeraSlayer algorithm. The filtered sequences were clustered into operational taxonomic units (OTUs) at 97% identity threshold and picking up a representative sequence in QIIME2 was done by using the furthest-neighbor algorithm. The representative sequences of OTUs were taxonomically annotated using the UNITE fungal ITS database release version 7.1 (Nilsson et al., 20183) using QIIME2 pipeline. The unassigned OTUs were further blasted with UNITE database4 and the closest relatives with % of sequence similarity were obtained. Multiple OTUs with closer similarity to a particular yeast species were merged together during the relative abundance estimation. Bacteria-specific and unassigned OTUs were filtered from the final OTU table before performing yeast community statistical analyzes. To establish the phylogenetic relationship of two OTUs of Saccharomyces cerevisiae present in the fermented milk samples, the nucleotide sequences (481 bases) of ITS of 26S large subunit ribosomal RNA gene retrieved from the fermented milk metagenome were aligned with the ITS sequences of S. cerevisiae cultures of milk and wine origin available in the GenBank by using ClustalW. A neighbor-joining phylogenetic tree based on the Kimura-2 parameter evolutionary distance matrix with 1000 bootstrap replications was constructed from the aligned sequences using MEGA6 software (Tamura et al., 2013). The ITS sequence of Saccharomyces bayanus CBS8715 was maintained as outgroup.
Statistical Analysis
The difference in the overall yeast community structure was analyzed by an unsupervised principal component analysis (PCA) clustering using Canoco software v4.52 (Wageningen University, Netherlands). The relative abundance of the filtered yeast species-level OTUs were normalized by log transformation (log xi + 1) before performing the PCA analysis. The principal coordinate shows a significant difference was visualized further with horizontal boxplot using BoxPlotR5. The significant difference in the yeast community structure of different food types or regions was calculated by PERMANOVA test with 9999 permutations using Bray–Curtis distances in PAST v3.08 (Hammer et al., 2001). ANOSIM also calculated to support the significant difference. The main diversity indices used for ecological application, namely, Chao-1, Shannon, Simpson, Evenness, Dominance, Fisher alpha, Berger–Parker indices (Morris et al., 2014) were analyzed to show the difference in yeast diversity between the food types and regions. Whittaker index was calculated to show the beta diversity. The diversity indices were calculated using the species level OTUs using PAST. ANOVA was performed to show the statistical difference in yeast diversity. Any significant difference in the abundance of specific taxa or diversity between the food groups or the region was calculated as “Bonferroni” corrected p-value using R (v3.1.3). The relative abundance (%) of yeast taxa was used for generating the bar chart (Figure 2A). We tested the yeast taxa that differently abundant across the sample groups (country, food type, and region of sample collection) with ANCOM analysis (Mandal et al., 2015) by using QIIME2. The OTU table file along with sample metadata (country, food type, and region of sample collection) was used for ANCOM visualization. The statistical significance of the difference in the abundance of yeast taxa between the groups (food types or regions) was also obtained by Wilcoxon test with Bonferroni corrected p-value (q-value) using “svDialogs” in R package (v3.1.3). The yeast taxa showed significant differential abundance between the food types were visualized with boxplots using BoxPlotR.
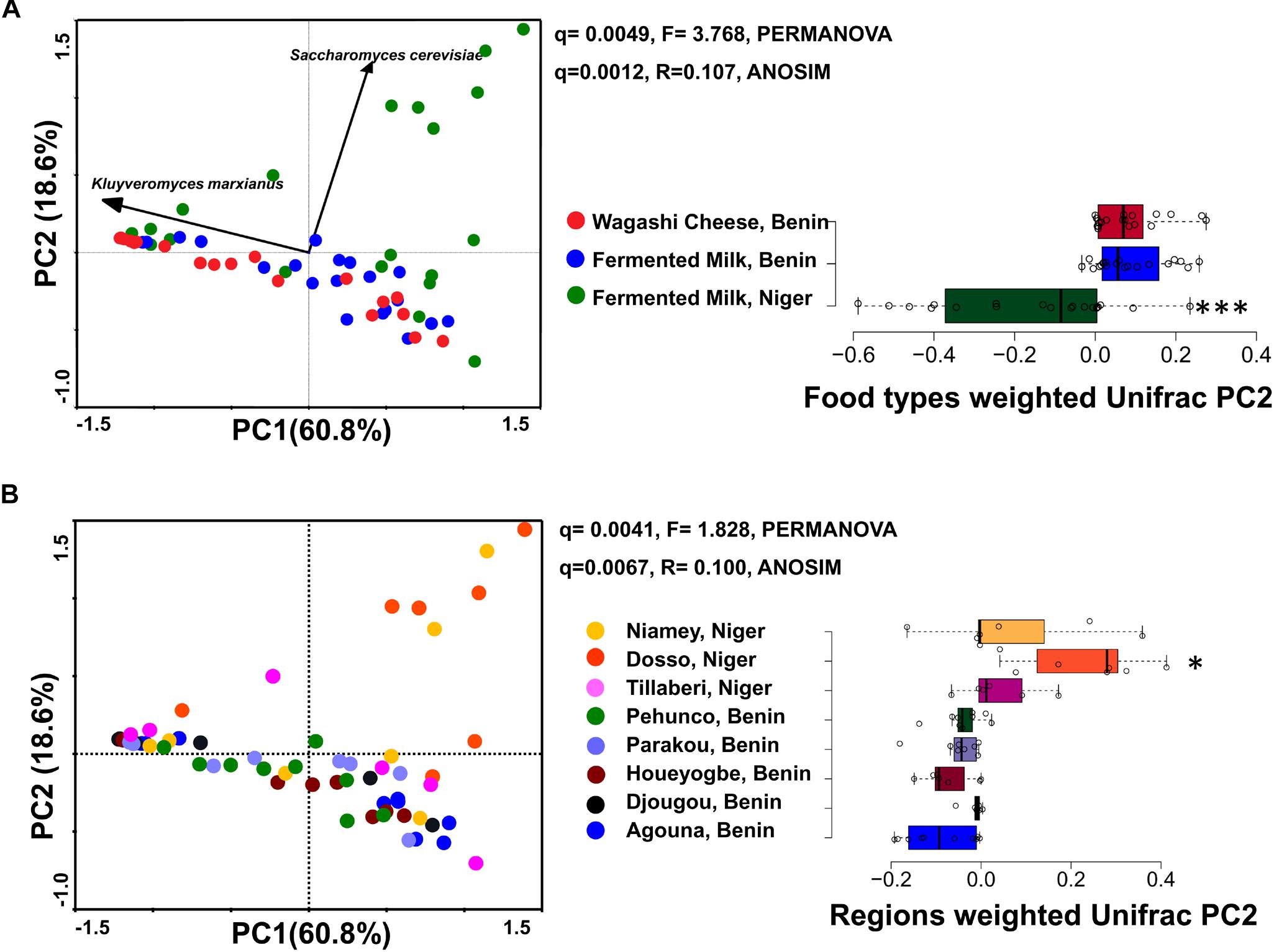
Figure 2. PCA plots show the difference in the yeast community structure among the fermented milk products of Niger and Benin (A) and the regions of sample collection (B). The direction of yeast species with significant association is indicated with arrows. PERMANOVA and ANOSIM used to visualize the separation among the groups using Bray–Curtis distance and the significance in difference was expressed as Bonferroni corrected q-values. The horizontal box plot shows the principal coordinates of weighted Unifrac that significantly differ among the groups (indicated as ∗q < 0.05, ∗∗∗q < 0.001).
Sensory Analysis
The texture of the traditionally fermented milk products of Niger and Benin were analyzed by consistency grading as per the International Dairy Federation (IDF) (Karagül-Yüceer and Drake, 2007). Twenty samples each of marketed fermented milk from Benin and Niger were analyzed for the consistency grades of setting, lumps or flakes, dripping, gritty, sticky, too thick, too fluid, ropy/stringy, dried, brittle, and gelatinous nature.
Data Accession
The yeast ITS sequences generated in the present study was deposited in NCBI SRA with the accession number PRJNA506750 (Supplementary Data Sheets S1, S2).
Results
The yeast communities of the naturally fermented milk from Niger and Benin and the Wagashi cheese from Benin were analyzed for the first time by MiSeq amplicon sequencing of fungal ITS region. This in-depth analysis resulted in a total of 13,51,858 quality-filtered sequences of 200–550 bp read length with an average reads of 22,530 ± 8464 per sample. The taxonomic assignment of the sequences at 97% identity in UNITE database resulted with 1548 yeast OTUs. The yeast community structure of the fermented milk products was compared based on the yeast species-level OTUs relative abundance profiles by a PCA using Bray–Curtis distance metrics. The yeast community structure in fermented milk products of Niger and Benin significantly differed (q = 0.001, F = 3.4, PERMANOVA) (Figure 2A). The principal coordinate-2 that showed a distinct separation between the two countries was visualized with a boxplot. Pair-wise ANOVA comparison showed that the yeast community structure of fermented milk of Niger differed significantly with fermented milk of Benin (q = 0.015, Bonferroni corrected) and Wagashi cheese of Benin (q = 0.0069, Bonferroni corrected). Further analysis to see the yeast community difference across the sample collection regions also showed a significant difference (q = 0.0041, PERMANOVA) (Figure 2B). ANOVA pair-wise comparison showed a significant difference in the yeast community of fermented milk samples collected between the Dosso region of Niger, and Houeyogbe (q = 0.042, Bonferroni corrected) and Parakou (q = 0.042, Bonferroni corrected) regions of Benin. We also noticed a significant difference in the diversity indices between the fermented milk products of Niger and Benin (Table 1). The fermented milk of Benin had a high yeast species richness (Chao-1) and high yeast diversity (Shannon index) than the other two milk products (q < 0.05, ANOVA). The samples collected from Pehunco and Djougou regions of Benin had low yeast richness and significantly differed with the samples from Niamey and Tillaberi regions of Niger (q < 0.05, Bonferroni corrected). Moreover, beta diversity analysis resulted with Whittaker index value of 20.42. Our results supported the association of different yeast community structure in the fermented milk products of Benin and Niger.
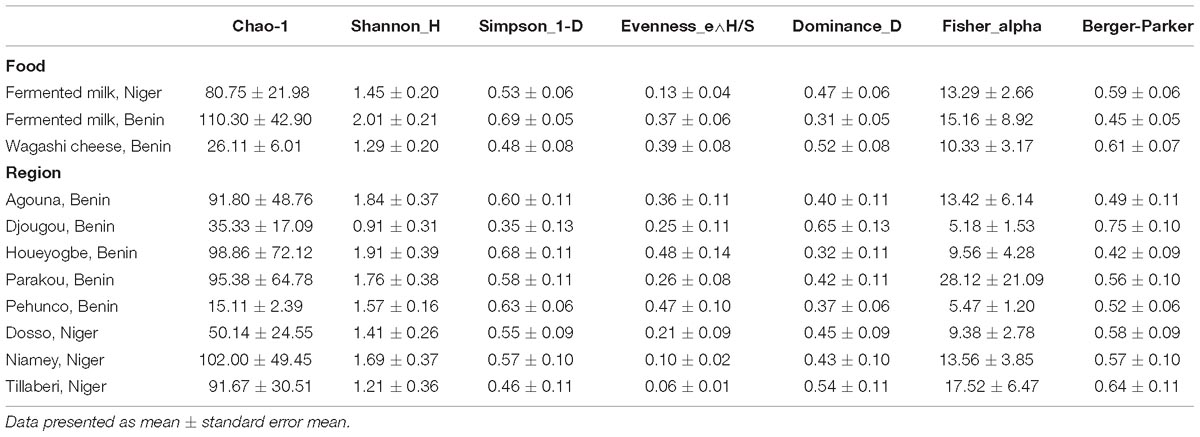
Table 1. Yeast diversity in fermented milk products collected from different regions of Benin and Niger.
The predominant yeast taxa associated with the fermented milk products of Benin and Niger are shown with a relative abundance graph (Figure 3A). Kluyveromyces marxianus (39.42%), S. cerevisiae (17.28%), Candida parapsilosis (7.23%), and Sagenomella keratitidis (5.62%) were predominant in the naturally fermented milk of Benin and Niger. Regarding the cheese samples, K. marxianus (61.13%), S. keratitidis (6.44%), C. parapsilosis (4.93%), and S. cerevisiae (4.65%) were prevalent. Further analysis showed that S. cerevisiae as the key differentiating taxa (q < 0.05, Wilcoxon test, Bonferroni corrected) with a high presence in the fermented milk products of Niger (Figure 3B). We used ANCOM differential abundance analysis using QIIME2 to visualize the yeast taxa that significantly differed among the samples by group (country, food type, and the region of sample collection). ANCOM visualization (Figure 4) showed that two OTUs of S. cerevisiae (OTU965 and OTU240) significantly differed between Benin and Niger. The W-value for the two OTUs is much higher (>1500) than other yeast taxa. Whereas OTU240 was exclusively present in the fermented milk of Niger (Figures 5A,B). Table 2 shows the ANCOM percentile abundance of features by the group for the significantly differing yeast taxa. We did not see any yeast taxa that significantly differed between the regions of sample collection within two countries. The neighbor-joining phylogenetic tree (Figure 5C) generated based on the nucleotide sequence similarity of ITS region of 26S rRNA gene showed the closest relative of the two OTUs of S. cerevisiae present in the fermented milk products. OTU965 showed a closer similarity with the S. cerevisiae strains of wine origin. Whereas, OTU240 showed a divergent with S. cerevisiae strains of milk origin. We observed a high presence of K. marxianus in the Wagashi cheese of Benin (Figure 3C). Also noticed a denser texture in the K. marxianus predominated fermented milk of Benin, whereas lighter texture in the S. cerevisiae predominated fermented milk of Niger. The other way around, the low presence of S. cerevisiae associated with the denser structure and low presence of K. marxianus associated with the lighter texture. Consistency analysis by IDF grading (Table 3) showed that most of the samples (16/20) from Benin showed thick texture with lumps and flakes. Whereas, the samples of Niger (18/20) showed with too fluid with dripping texture.
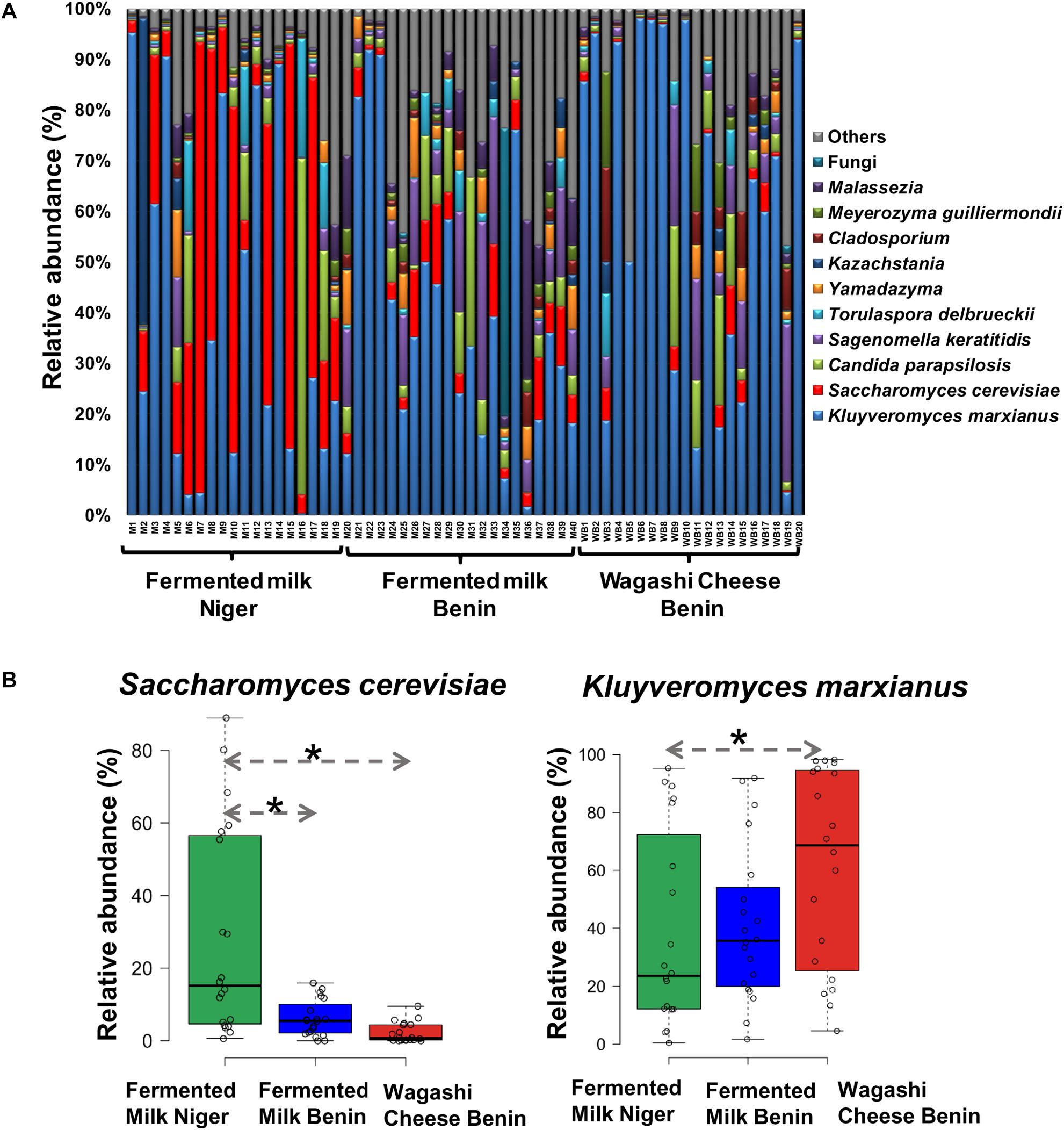
Figure 3. (A) Yeast taxon plot shows the relative abundance of predominant yeast present in the naturally fermented milk products of Benin and Niger. Each column represents the relative abundance (%) of yeast taxa investigated by using Illumina MiSeq amplicon sequencing of ITS region. OTUs with similar species-level identity at 97% similarity in the UNITE database are merged here. Taxa with less than 1% mean relative abundance across the samples studied are combined and shown as others. (B) Boxplot shows the differential abundance of Saccharomyces cerevisiae and Kluyveromyces marxianus among the fermented milk products of Niger and Benin. The significance in difference was calculated by Wilcoxon test with Bonferroni correction and indicated as ∗q < 0.05.
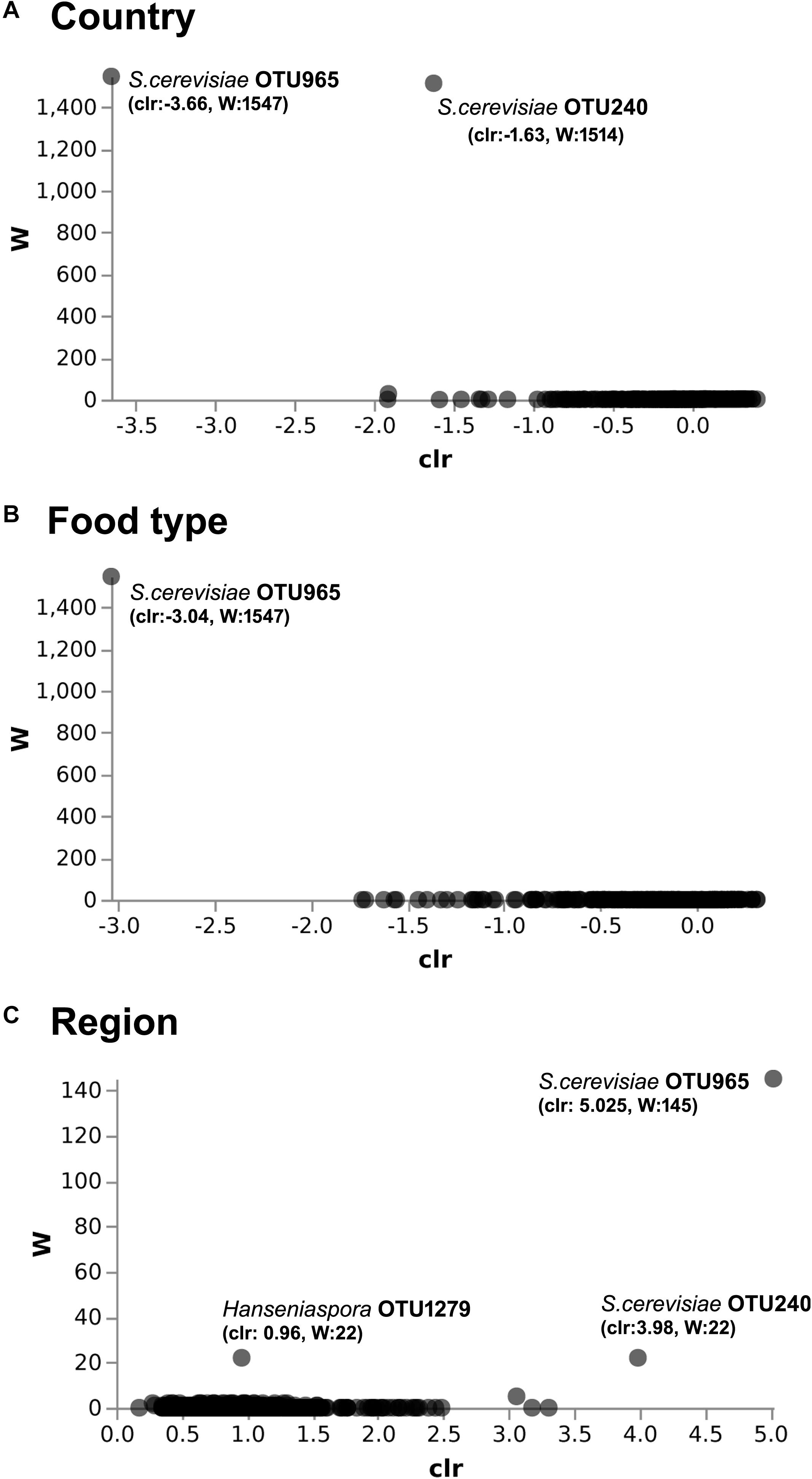
Figure 4. ANCOM differential abundance testing shows the yeast taxa significantly differ across the (A) country (Benin and Niger) and (B) food type (fermented milk and cheese). (C) No significant difference observed between the regions of sample collection. QIIME2 version 2019.1 was used for ANCOM analysis. The percentile abundances of features by the group of significantly differing yeast taxa are shown in Table 2.
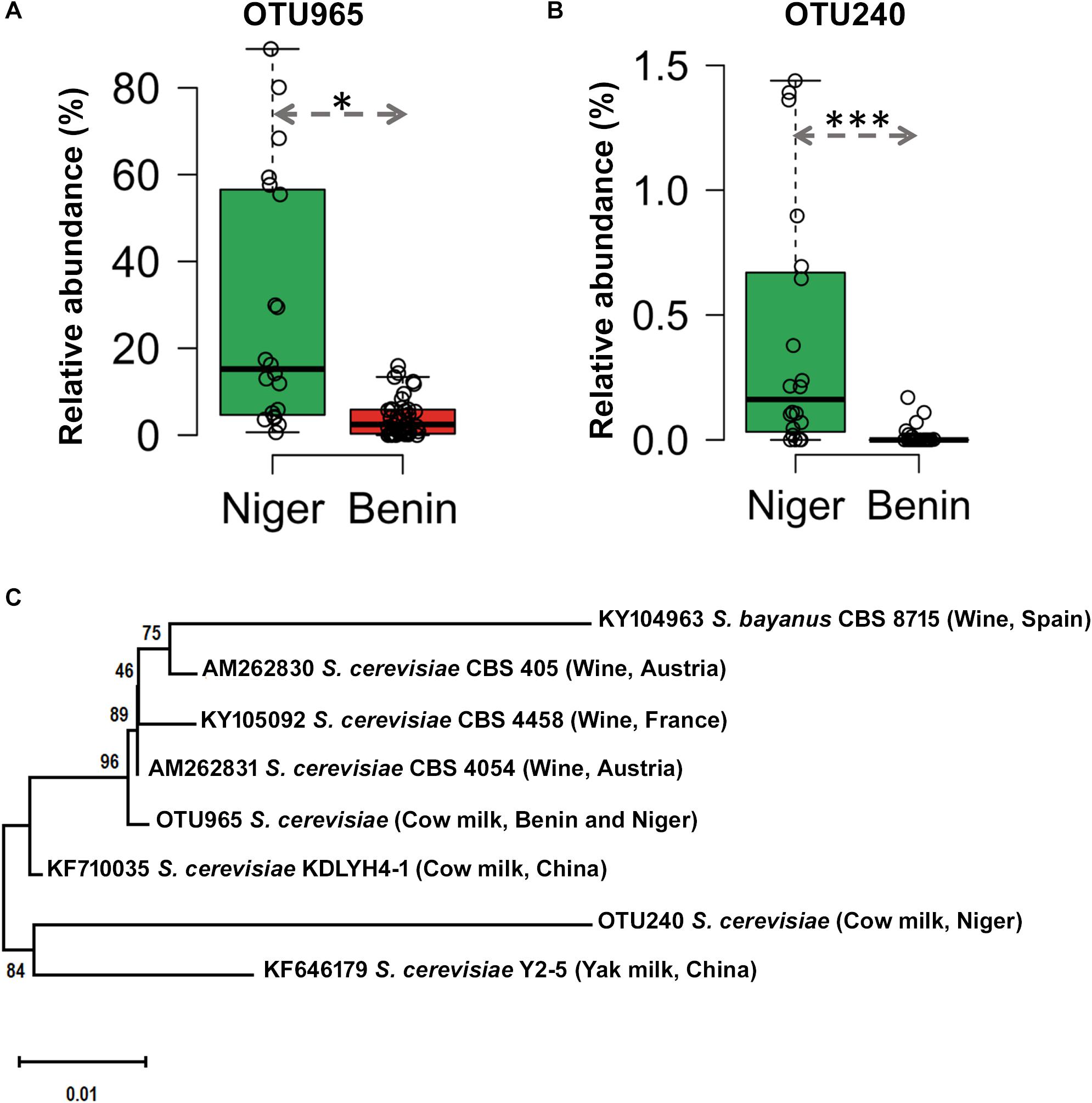
Figure 5. Difference in the occurrence and phylogenetic affiliation of two OTUs of Saccharomyces cerevisiae retrieved from the metagenome of fermented milk products of Niger and Benin. (A,B) Box plots shows the difference in the occurrence of OTU965 and OTU240 in the fermented milk products of Niger and Benin. The significance in difference was calculated by Wilcoxon test with Bonferroni correction and indicated as ∗q < 0.05, ∗∗∗q < 0.001. (C) The neighbor-joining phylogenetic tree generated based on the sequences of the internal transcribed spacer (ITS) region of 26S large subunit ribosomal RNA gene of the two OTUs of S. cerevisiae in comparison with the ITS sequence of earlier characterized cultures of S. cerevisiae shows its close relatives of wine and milk origins. Saccharomyces bayanus CBS8715 was maintained as an out-group. The evolutionary distances were computed using the Kimura two-parameter method and the scale indicates the units of the number of base substitutions per site.
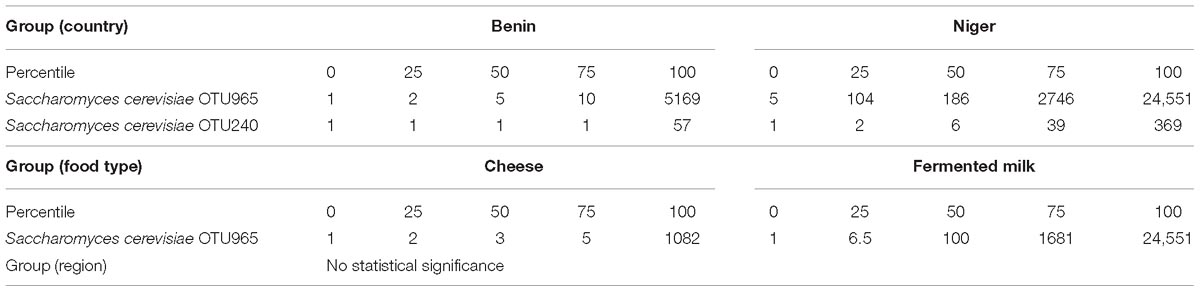
Table 2. ANCOM differential abundance testing resulted significantly differing yeast taxa and their percentile abundance of samples by group.
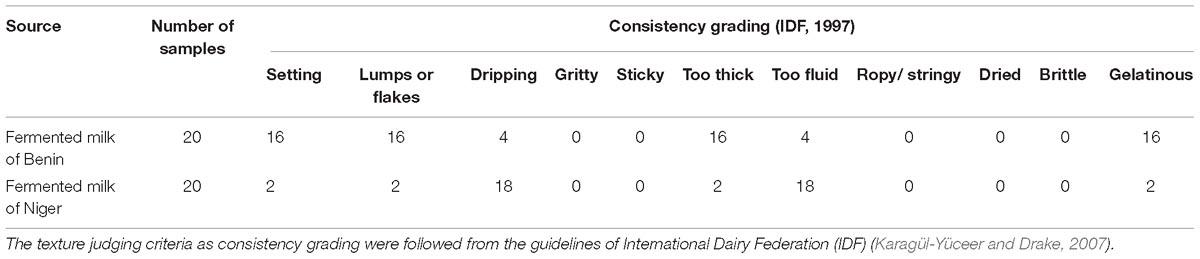
Table 3. Difference in the consistency of the naturally fermented milk products marketed in Benin and Niger.
Discussion
Yeast and lactic acid bacteria together contribute to the characteristic taste, texture, and flavor of naturally fermented milk products. The difference in their composition contributes to the differences in chemical composition and led to a variety of milk products. The present study investigated the yeast ecology of naturally fermented milk products of Benin and Niger and showed that the yeast composition differed between the countries. The difference in the yeast composition in the studied products may be linked to the difference in indigenous production processes, geographical location, different climatic conditions, and grassland ecosystems of the two countries. Two yeast species, K. marxianus and S. cerevisiae, dominated the fermented milk products of Benin and Niger. Earlier studies observed S. cerevisiae, K. marxianus, Kluyveromyces lactis, Debaromyces hansenii, and Yarrowia lipolytica (Rai and Jeyaram, 2017; Hittinger et al., 2018) as the main yeast species present in the similar type of naturally fermented milk products. Similar to the Wagashi cheese of Benin, K. marxianus dominantly present in several naturally fermented milk products, namely, Indian hard cheese prepared from yak milk “churpi” (Rai et al., 2016) and traditional Mongolian fermented mare’s milk “airag” (Watanabe et al., 2008). The dominance of S. cerevisiae in natural milk fermentation also reported from African milk products, “nyarmie” of Ghana (Obodai and Dodd, 2006) and “amabere amaruranu” of Kenya (Nyambane et al., 2014). Pichia kudriavzevii along with S. cerevisiae dominated in a West African fermented yogurt-like milk product – “nunu” (Owusu-Kwarteng et al., 2017). Among the other yeast species C. parapsilosis, S. keratitidis, Torulaspora delbrueckii, Yamadazyma, Kazachstania, Cladosporium, Meyerozyma guilliermondii, and Malassezia were also relatively abundant in the studied milk products. Whereas earlier studies showed the presence of Galactomyces geotrichum, Issatchenkia orientalis, Pichia mandshurica, Pichia fermentans, P. kudriavzevii, Candida lusitaniae, Candida rugosa, Candida tropicalis in several dairy products (Shangpliang et al., 2018). In addition, the predominance of Galctomyces spp. (Murugesan et al., 2018) and Debaryomyces spp. (Dos Santos et al., 2017) reported in several cheese varieties. The presence of diverse yeast species and their compositional difference is linked with the unique sensory properties of the different naturally fermented milk products.
The dominance of K. marxianus in the investigated milk products is a good trait as this food-grade thermotolerant yeast produces a wide array of volatile molecules and contributes to the flavor development of different fermented beverages (Morrissey et al., 2015). It is a lactose-fermenting yeast conventionally recognized as Candida kefyr and phylogenetically related to S. cerevisiae, which possesses biotechnological and probiotic potential including the ability to survive in the gastric and intestinal environments (Gethins et al., 2016). Several studies have recognized K. marxianus as an emerging probiotic, fermentation starter with generally regarded as safe (GRAS) status. K. marxianus is used as a starter culture along with Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus to enhance the flavor (Zhang et al., 2017), taste (Fasoli et al., 2016), and denser texture (Gethins et al., 2016). The denser texture observed in the fermented milk of Benin with high abundance of K. marxianus or low abundance of S. cerevisiae, and lighter texture in the fermented milk of Niger with high abundance of S. cerevisiae or low abundance of K. marxianus is worthy to study further on developing a product with a difference in texture. Besides K. marxianus, non-lactose-fermenting S. cerevisiae is also recognized as a potential starter culture for fermented milk production with improved qualities (FAO/WHO/CODEX ALIMENTARIUS, 2018). Further studies on selective isolation of the strain of S. cerevisiae OTU240 exclusively present in the fermented milk of Niger and characterizing in the angle of its association with milk products is worthwhile. The dominance of K. marxianus and S. cerevisiae could significantly contribute to the organoleptic profile of these dairy products by producing CO2 as well as aroma compounds (Aponte et al., 2010), thus provide an advantage of improving texture and flavor. Further studies on chemical profiling by mass spectrometry will give an insight into the role of yeast in the biotransformation of milk fermentation.
The identified yeast species may contribute significantly to many desirable effects in the fermented milk products in one hand and potential pathogens in the other hand. The presence of S. keratitidis and C. parapsilosis in the investigated products may be of risk for the consumers. This is the first report on the predominance of S. keratitidis in the naturally fermented milk products of Africa. S. keratitidis is a potential agent causing keratitis, inflammation of cornea and eyes. Moreover, S. keratitidis found to be an emerging agent of nail infection (Tsang, 2017). Systemic fungal infection by Sagenomella lead to Juvenile idiopathic arthritis has been also reported (Ried and Fakler, 2018). Indeed, C. parapsilosis is recognized as an emerging major human pathogen that causes invasive candidal disease (Trofa et al., 2008). Recently, C. parapsilosis has been recognized as the most frequent non-albicans Candida (NAC) species of invasive candidiasis in neonates (Garzillo et al., 2017). However, several reports observed the presence of C. parapsilosis in traditional food fermentation (Chaves-López et al., 2014). Similarly, M. guilliermondiii regarded as an emerging infectious yeast of the NAC observed in the studied milk products also predominately involved in the natural bamboo shoot fermentation (Romi et al., 2014). The presence of potential yeast pathogens in the uncontrolled natural milk fermentation and its impact on health particularly on children to be studied further to confirm its safety. As S. keratitidis infect the nails, unhygienic milking and further spread through marketing of fermented milk products require an epidemiological study. Further studies on understanding the biochemical changes by different yeast species in a controlled milk fermentation will improve our understanding on the role of yeast in milk fermentation.
Conclusion
This study provides a baseline understanding of the yeast ecology of traditional fermented milk products of the two African countries Benin and Niger. The results showed that K. marxianus and S. cerevisiae were the prevalent yeast species which may have an important role during natural milk fermentation and Wagashi cheese production. The composition difference of these two yeast species might contribute to the differences in the physical and chemical character of the studied products. Further studies are necessary to selectively isolate the predominant indigenous yeasts K. marxianus and S. cerevisiae and screen them for desirable fermentative and functional properties with aim of developing a controlled milk fermentation for producing safe and quality milk products. The presence of potential pathogenic yeasts S. keratitidis and C. parapsilosis in these traditional foods is to be further investigated to assess safety. The study offers tremendous opportunities to predict and control the growth and survival of desirable or undesirable microorganisms in the studied products.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
PS, MG, and KJ conceived and designed the research. PS and SK acquired the data, interpreted the results, and wrote the manuscript. NT assisted with UNITE database analysis. KJ and SF critically revised the manuscript. All authors read and approved the final manuscript.
Funding
This project was funded by the Centre for Science and Technology of the Non-Aligned and Other Developing Countries (NAM S&T Centre), New Delhi (India), through the award of a Research Training Fellowship-Developing Country Scientists to PS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express our heartfelt gratitude to NAM Centre and Institute of Bioresources and Sustainable Development for facilities. We acknowledge the help of Rajendra Kumar Labala for ANCOM analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00594/full#supplementary-material
Footnotes
- ^ https://docs.qiime2.org/2019.1/
- ^ https://docs.qiime2.org/2019.1/tutorials/
- ^ http://unite.ut.ee
- ^ https://unite.ut.ee/analysis.php
- ^ http://shiny.chemgrid.org/boxplotr/
References
Aponte, M., Pepe, O., and Blalotta, G. (2010). Identification and technological characterization of yeast strains isolated from samples of water buffalo Mozzarella cheese. J. Dairy Sci. 93, 2358–2361. doi: 10.3168/jds.2009-2948
Boko, C., Adje, Y., Atindogbe, G., Hounmanou, M. G. Y., Dossa, F., Sessou, P., et al. (2016). Evaluation of the production technologies and the microbial and physico-chemical qualities of curdled milk produced in Benin. J. Appl. Biosci. 104, 10019–10033. doi: 10.4314/jab.v104i1.14
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Chaves-López, C., Serio, A., Grande-Tovar, C. D., Cuervo-Mulet, R., Delgado-Ospina, J., and Paparella, A. (2014). Traditional fermented foods and beverages from a microbiological and nutritional perspective: the Colombian heritage. Compr. Rev. Food Sci. F. 13, 1031–1048. doi: 10.1111/1541-4337.12098
Dos Santos, M. T. P. G., Benito, M. J., de Guía Córdoba, M., Alvarenga, N., and de Herrera, S. R. M. S. (2017). Yeast community in traditional Portuguese Serpa cheese by culture-dependent and-independent DNA approaches. Int. J. Food Microbiol. 262, 63–70. doi: 10.1016/j.ijfoodmicro.2017.09.013
Dossou, S., and Soule, H. (2006). Production et Transformation du Lait Frais en Fromage Peulh au Benin – Guide de Bonnes Pratiques. Available at: www.repol.info/IMG/pdf/Fiche_wagashi_VF.pdf (accessed October 20, 2018).
Dossou, J., Montcho, J. K., Londji, S., Atchouke, G. D. L., and Odjo, S. (2016). Procédé amélioré de conservation et de stabilisation du fromage peuhl wagashi par l’effet combiné du traitement thermique et du conditionnement sous vide. Eur. Sci. J. 12, 189–209. doi: 10.19044/esj.2016.v12n36p189
FAO/WHO/CODEX ALIMENTARIUS (2018). Standards for Fermented Milk CXS 243-2003. Adopted in 2003. Revised in 2008, 2010, 2018. Available at: http://www.fao.org/fao-who-codexalimentarius/sh-proxy/en/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FStandards%252FCODEX%2BSTAN%2B243-2003%252FCXS_243e.pdf (accessed October 20, 2018).
Fasoli, G., Barrio, E., Tofalo, R., Suzzi, G., and Belloch, C. (2016). Multilocus analysis reveals large genetic diversity in Kluyveromyces marxianus strains isolated from Parmigiano Reggiano and Pecorino di Farindola cheeses. Int. J. Food Microbiol. 233, 1–10. doi: 10.1016/j.ijfoodmicro.2016.05.028
Fasoli, G., Tofalo, R., Lanciotti, R., Schirone, M., Patrignani, F., Grazia, L., et al. (2015). Chromosome arrangement, differentiation of growth kinetics and volatile molecule profiles in Kluyveromyces marxianus strains from Italian cheeses. Int. J. Food Microbiol. 214, 151–158. doi: 10.1016/j.ijfoodmicro.2015.08.001
Gagara, M., Sessou, P., Ahounou, S., Azokpota, P., Youssao, I., Farougou, S., et al. (2017). Analyses of constraints related to milk production in Liptako Gourma in Niger. Afr. J. Agric. Res. 12, 1949–1958. doi: 10.5897/AJAR2016.12029
Garcia, T., Mayoral, B., Gonzales, I., Lopez, C., Sanz, A., Hernandez, P. E., et al. (2004). Enumeration of yeasts in dairy products: a comparison of immunological and genetic techniques. J. Food Protect. 67, 354–363.
Garzillo, C., Bagattini, M., Bogdanović, L., Di Popolo, A., Iula, V. D., Catania, M. R., et al. (2017). Risk factors for Candida parapsilosis bloodstream infection in a neonatal intensive care unit: a case-control study. Ital. J. Pediatr. 43:10. doi: 10.1186/s13052-017-0332-5
Gethins, L., Rea, M. C., Stanton, C., Ross, R. P., Kilcawley, K., O’Sullivan, M., et al. (2016). Acquisition of the yeast Kluyveromyces marxianus from unpasteurised milk by a kefir grain enhances kefir quality. FEMS Microbiol. Lett. 363:165. doi: 10.1093/femsle/fnw165
Hammer, Ø, Harper, D., and Ryan, P. (2001). Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9.
Hittinger, C. T., Steele, J. L., and Ryder, D. S. (2018). Diverse yeasts for diverse fermented beverages and foods. Curr. Opin. Biotechnol. 49, 199–206. doi: 10.1016/j.copbio.2017.10.004
Jakobsen, M., and Narvhus, J. (1996). Yeasts and their possible beneficial and negative effects on the quality of dairy products. Int. Dairy J. 6, 755–768. doi: 10.1016/0958-6946(95)00071-2
Karagül-Yüceer, Y.,and Drake, M. (2007). “Sensory analysis of yogurt,” in Manufacturing Yogurt and Fermented Milks, ed. R. C. Chandan (Hoboken, NJ: Blackwell Publishing).
Keisam, S., Romi, W., Ahmed, G., and Jeyaram, K. (2016). Quantifying the biases in metagenome mining for realistic assessment of microbial ecology of naturally fermented foods. Sci. Rep. 6:34155. doi: 10.1038/srep34155
Legras, J. L., Galeote, V., Bigey, F., Camarasa, C., Marsit, S., Nidelet, T., et al. (2018). Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 35, 1712–1727. doi: 10.1093/molbev/msy066
Mandal, S., Van Treuren, W., White, R. A., Eggesbø, M., Knight, R., and Peddada, S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. doi: 10.3402/mehd.v26.27663
Mattiello, S., Caroprese, M., Matteo, C. G., Fortina, R., Martini, A., Martini, M., et al. (2018). Typical dairy products in Africa from local animal resources. Ital. J. Anim. Sci. 17, 740–754. doi: 10.1080/1828051x.2017.1401910
Ministry of Agriculture Livestock and Fisheries [MAEP] (2017). Strategic Plan for Agricultural Sector Development (PSDSA) 2025 and National Plan for Agricultural Investments and Food Security and Nutrition PNIASAN 2017 – 2021. Available at: http://www.agriculture.gouv.bj/IMG/pdf/synthese_psdsa_2025_pniasan_2021_cadre_prog_cadre_insti_promo_fil.pdf (accessed July 22, 2018).
Morris, E. K., Caruso, T., Buscot, F., Fischer, M., Hancock, C., Maier, T. S., et al. (2014). Choosing and using diversity indices: insights for ecological applications from the German biodiversity exploratories. Ecol. Evol. 4, 3514–3524. doi: 10.1002/ece3.1155
Morrissey, P. J., Etschmann, W. M. M., Schrader, J., and de Billerbeck, M. G. (2015). Cell factory applications of the yeast Kluyveromyces marxianus for the biotechnological production of natural flavour and fragrance molecules. Yeast 32, 3–16. doi: 10.1002/yea.3054
Motooka, D., Fujimoto, K., Tanaka, R., Yaguchi, T., Gotoh, K., Maeda, Y., et al. (2017). Fungal ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut mycobiota of healthy Japanese individuals. Front. Microbiol. 8:238. doi: 10.3389/fmicb.2017.00238
Murugesan, S., Reyes-Mata, M. P., Nirmalkar, K., Chavez-Carbajal, A., Juárez-Hernández, J. I., Torres-Gómez, R. E., et al. (2018). Profiling of bacterial and fungal communities of Mexican cheeses by high throughput DNA sequencing. Food Res. Int. 113, 371–381. doi: 10.1016/j.foodres.2018.07.023
Nilsson, R. H., Larsson, K.-H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2018). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Nyambane, B., Thari, W. M., Wangoh, J., and Njage, P. M. K. (2014). Lactic acid bacteria and yeasts involved in the fermentation of amabere amaruranu, a Kenyan fermented milk. Food Sci. Nutr. 2, 692–699. doi: 10.1002/fsn3.162
Obodai, M., and Dodd, C. E. R. (2006). Characterization of dominant microbiota of a Ghanaian fermented milk product, nyarmie, by culture- and nonculture-based methods. J. Appl. Microbiol. 100, 1355–1363. doi: 10.1111/j.1365-2672.2006.02895.x
Okry, F., Kpenavoun Chogou, S., Fanou-Fogny, N., Moumouni, I., and Hounhouigan, J. (2017). Local governance of a dairy sector in Benin Republic: actors, roles, power relationships and perception of interdependency. Asian J. Agric. Ext. Econ. Sociol. 21, 1–14. doi: 10.9734/ajaees/2017/38050
Owusu-Kwarteng, J., Akabanda, F., Johansen, P., Jespersen, L., and Nielsen, D. S. (2017). Chapter 15 - Nunu, A West African Fermented Yogurt-Like Milk Product. Yogurt in Health and Disease Prevention. Cambridge, MA: Academic Press, 275–283.
Parker, M., Zobrist, S., Donahue, C., Edick, C., Mansen, K., Nadjari, M. H. Z., et al. (2018). Naturally fermented milk from Northern Senegal: bacterial community composition and probiotic enrichment with Lactobacillus rhamnosus. Front. Microbiol. 9:2218. doi: 10.3389/fmicb.2018.02218
Rai, A. K., and Jeyaram, K. (2017). “Role of yeasts in food fermentation,” in Yeast Diversity in Human Welfare, eds T. Satyanarayana and G. Kunze (Singapore: Springer).
Rai, A. K., Kumari, R., Sanjukta, S., and Sahoo, D. (2016). Production of bioactive protein hydrolysate using the yeasts isolated from soft chhurpi. Bioresour. Technol. 219, 239–245. doi: 10.1016/j.biortech.2016.07.129
Ried, K., and Fakler, P. (2018). Rare fungal infection linked to a case of juvenile arthritis. Cureus 10:e3229. doi: 10.7759/cureus.3229
Romi, W., Ahmed, G., and Jeyaram, K. (2015). Three-phase succession of autochthonous lactic acid bacteria to reach a stable ecosystem within 7 days of natural bamboo shoot fermentation as revealed by different molecular approaches. Mol. Ecol. 24, 3372–3389. doi: 10.1111/mec.13237
Romi, W., Keisam, S., Ahmed, G., and Jeyaram, K. (2014). Reliable differentiation of Meyerozyma guilliermondii from Meyerozyma caribbica by internal transcribed spacer restriction fingerprinting. BMC Microbiol. 14:52. doi: 10.1186/1471-2180-14-52
Sessou, P. (2013). Promotion of Fulani Cheese “Wagashi” in Benin: Endogenous Conservation Practices, Screening of Contamination Mold and Perspectives of Biological Fight. Ph.D. thesis, University of Abomey-Calavi, Abomey Calavi.
Shangpliang, H. N. J., Rai, R., Keisam, S., Jeyaram, K., and Tamang, J. P. (2018). Bacterial community in naturally fermented milk products of Arunachal Pradesh and Sikkim of India analysed by high-throughput amplicon sequencing. Sci. Rep. 8:1532. doi: 10.1038/s41598-018-19524-6
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tossou, M. L. (2018). Nutritional Quality and Safety of Processed Wagashi Cheese Using Calotropis procera Found in Benin and Kenya. Available at: http://ir.jkuat.ac.ke/bitstream/handle/123456789/4695/Thesis%20M.%20L.%20TOSSOU.pdf?sequence=1&isAllowed=y (accessed October 20, 2018).
Trofa, D., Gácser, A., and Nosanchuk, J. D. (2008). Candida parapsilosis, an emerging fungal pathogen. Clin. Microbiol. Rev. 21, 606–625. doi: 10.1128/cmr.00013-08
Tsang, C. (2017). Diversity of Novel and Emerging Pathogenic Fungi in Hong Kong. Ph.D. thesis, University of Hong Kong, Pokfulam.
Watanabe, K., Fujimoto, J., Sasamoto, M., Dugersuren, J., Tumursuh, T., and Demberel, S. (2008). Diversity of lactic acid bacteria and yeasts in Airag and Tarag, traditional fermented milk products of Mongolia. World J. Microbiol. Biotechnol. 24:1313. doi: 10.1007/s11274-007-9604-3
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, NY: Academic Press Inc.), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Keywords: Wagashi, Calotropis, MiSeq amplicon sequencing, internal transcribed spacer, Kluyveromyces, Saccharomyces, Sagenomella
Citation: Sessou P, Keisam S, Tuikhar N, Gagara M, Farougou S and Jeyaram K (2019) High-Throughput Illumina MiSeq Amplicon Sequencing of Yeast Communities Associated With Indigenous Dairy Products From Republics of Benin and Niger. Front. Microbiol. 10:594. doi: 10.3389/fmicb.2019.00594
Received: 27 November 2018; Accepted: 08 March 2019;
Published: 03 April 2019.
Edited by:
Eugenia Bezirtzoglou, Democritus University of Thrace, GreeceReviewed by:
Kristina Marie Feye, University of Arkansas, United StatesAlex Galanis, Democritus University of Thrace, Greece
Copyright © 2019 Sessou, Keisam, Tuikhar, Gagara, Farougou and Jeyaram. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philippe Sessou, cGhpbGlwcGUuc2Vzc291QGVwYWMudWFjLmJq Kumaraswamy Jeyaram, amV5YXJhbS5pYnNkQG5pYy5pbg==
 Philippe Sessou
Philippe Sessou Santosh Keisam
Santosh Keisam Ngangyola Tuikhar2
Ngangyola Tuikhar2 Souaïbou Farougou
Souaïbou Farougou Kumaraswamy Jeyaram
Kumaraswamy Jeyaram