- Department of Microbiology and Biotechnology, College of Life Sciences, Northeast Agricultural University, Harbin, China
In the cation diffusion facilitator (CDF) family, the transported substrates are confined to divalent metal ions, such as Zn2+, Fe2+, and Mn2+. However, this study identifies a novel CDF member designated MceT from the moderate halophile Planococcus dechangensis. MceT functions as a Na+(Li+, K+)/H+ antiporter, together with its capability of facilitated Zn2+ diffusion into cells, which have not been reported in any identified CDF transporters as yet. MceT is proposed to represent a novel CDF group, Na-CDF, which shares significantly distant phylogenetic relationship with three known CDF groups including Mn-CDF, Fe/Zn-CDF, and Zn-CDF. Variation of key function-related residues to “Y44-S48-Q150” in two structural motifs explains a significant discrimination in cation selectivity between Na-CDF group and three major known CDF groups. Functional analysis via site-directed mutagenesis confirms that MceT employs Q150, S158, and D184 for the function of MceT as a Na+(Li+, K+)/H+ antiporter, and retains D41, D154, and D184 for its facilitated Zn2+ diffusion into cells. These presented findings imply that MceT has evolved from its native CDF family function to a Na+/H+ antiporter in an evolutionary strategy of the substitution of key conserved residues to “Q150-S158-D184” motif. More importantly, the discovery of MceT contributes to a typical transporter model of CDF family with the unique structural motifs, which will be utilized to explore the cation-selective mechanisms of secondary transporters.
Introduction
Cation diffusion facilitator (CDF) family proteins are ubiquitous secondary transmembrane transporters in all three kingdoms of living organisms including bacteria, archaea and eukaryotes, which play an important role in the homeostasis of divalent metal cations (Me2+) including Zn2+, Cd2+, Co2+, Fe2+, Ni2+, Mn2+ and possibly Cu2+ and Pb2+ (Paulsen and Saier, 1997; Haney et al., 2005; Montanini et al., 2007). Most members of this family function as Me2+/H+ antiporters, which utilize the proton motive force for the transport of divalent metal ions from the cytoplasm to the outside of cells or into subcellular compartments. In addition, Escherichia coli ZitB and Bacillus subtilis CzcD can exchange Me2+ for H+ and also K+ (Guffanti et al., 2002; Lee et al., 2002). Based on the phylogenetic analysis, CDF transporters are classified into three major groups with different metal ion specificity: (i) Mn-CDF with the sole substrate of Mn2+; (ii) Fe/Zn CDF with the substrates of Fe2+ and Zn2+ and also other metal ions; (iii) Zn-CDF with substrates of Zn2+ and other metal ions but not including Fe2+ or Mn2+ (Montanini et al., 2007). Most CDF members possess a cation-transporting transmembrane domain (TMD) composed of six transmembrane helices (TMHs), followed by a ∼100 aa long cytoplasmic regulatory C-terminal domain (CTD) folded into two α helices and three β strands (Paulsen and Saier, 1997; Kolaj-Robin et al., 2015).
X-ray structure of E. coli YiiP in complex with Zn2+ provides an archetypal 3D model for CDF members (Lu and Fu, 2007; Lu et al., 2009). Of three Zn2+-binding sites (A-C) of YiiP, the major binding site A is located in the center of TMD and consists of four coordinating residues: D45 and D49 (DD) in TMH2 and H153 and D157 (HD) in TMH5, whereas other two binding sites (B and C) are located in the cytoplasmic Loop 2,3 and CTD domain, respectively. Six TMHs are grouped into two bundles with four (TMH1-TMH2-TMH4-TMH5) and two (TMH3-TMH6) helices (Coudray et al., 2013). So far, DD, ND, or HD motifs in TMH2 and HD motif in TMH5 have been widely accepted to be responsible for ionic selectivity between divalent metal ions (Montanini et al., 2007; Kolaj-Robin et al., 2015; Martin and Giedroc, 2016). For example, residue swapping of HD to DD of human ZnT5 or ZnT8 abolished their ionic selectivity against Cd2+, but had no effect on Zn2+ transport (Hoch et al., 2012). Mutation of DD to HD in TMH2 resulted in the Zn2+and Cd2+ specificity of E. coli YiiP to its preferred Zn2+ (Hoch et al., 2012). An H90D mutation in TMH2 of rice CDF OsMTP1 abolished Zn2+ transport but improved Fe2+ transport (Menguer et al., 2013). Mutation of HD to ND in TMH2 of human ZnT1 resulted in the loss of its native Zn2+ transport activity and the conversion into a Mn2+ efflux transporter, as human ZnT10 with ND motif in its TMH2 (Nishito et al., 2016).
Na+/H+ antiporters are a category of secondary transmembrane transporters that catalyze the exchange of Na+ for H+, which play a major role in maintaining intracellular pH and Na+ homeostasis (Padan and Landau, 2016). Under high saline-alkaline stress, halophiles should have been driven to evolve a larger number of Na+/H+ antiporters to stabilize their intracellular osmotic and ionic state. This has been strongly supported by our recent reports on several novel Na+/H+ antiporters such as UPF0118, UmpAB, RDD, and MdrP from the slight or moderate halophiles (Dong et al., 2017; Meng et al., 2017; Abdel-Motaal et al., 2018; Shao et al., 2018). Secondary transporters are proposed to share similar structures but different transported substrates (Shi, 2013; Yan, 2013), and thus a transporter model may alter the substrate selectivity by changing conserved functional residues. Therefore, some proteins from halophiles may have evolved from its native family functions to Na+ efflux transporters by changing conserved functional residues, but remaining the homologies and similar structures.
In this study, a moderate halophile, Planococcus dechangensis NEAU-ST10-9T (Wang et al., 2015) was used as a research object for gene mining, in order to screen Na+/H+ antiporters especially novel ones, and even to discover new functions or structure models of members within known ionic transporter families. Consequently, a novel CDF transporter, MceT, was obtained and identified to function as a Na+(Li+, K+)/H+ antiporter, together with the capability of facilitated Zn2+ diffusion into cells, which has not been reported in identified CDF members as yet. The function-related structural motifs for Na+ efflux were identified through site-directed mutagenesis of conserved residues. These presented findings imply that MceT has evolved from a Zn2+-efflux model of CDF members to a novel Na+-efflux model of Na+/H+ antiporters through the substitution of key conserved residues in its structural motifs.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Strains and plasmids are presented in Supplementary Table 1. P. dechangensis NEAU-ST10-9T was grown in a modified S-G liquid medium as previously described (Wang et al., 2015). A Na+/H+ antiporter-deficient mutant E. coli KNabc (ΔnhaAΔnhaBΔchaA) was used for NaCl complementation, growth tests and Na+/H+ antiport activity assays. A Zn2+-sensitive E. coli mutant KZAB04 (ΔzntAΔzitB) was used for ZnCl2 resistance experiments and intracellular Zn2+ accumulation analysis. This mutant was constructed with E. coli DH5α as an original strain by inserting a kanamycin resistance gene from pBBR1MCS-2 into zntA and a gentamicin resistance gene from pBBR1MCS-5 into zitB, via homologous recombination with the aid of pKD46 containing a λ Red recombinase system (Datsenko and Wanner, 2000). E. coli strains were cultured to OD600 nm at 1.0 in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast, 1% NaCl), LBO broth (LB without the addition of NaCl) or LBK broth (LB with the addition of 86 mM KCl instead of NaCl), followed by the inoculation into the same fresh media and incubation at 37°C within 24 h. In the physiological experiments, NaCl (0–0.4 M), LiCl (0–10 mM), or ZnCl2 (0–0.75 mM) was added into the indicated media. pH was adjusted by the addition of 10 mM Tris-HCl buffer (7.0–8.5) at the final concentration to the tested media supplemented by 50 mM NaCl, which is essential for the alkaline pH resistance of Na+/H+ antiporters as previously described (Krulwich et al., 2011; Quinn et al., 2012; Meng et al., 2014; Padan, 2014).
Isolation and Subcloning of a Na+/H+ Antiporter Gene Candidate
The pUC18 vector, which was digested by BamHI and dephosphorylated by a bacterial alkaline phosphatase, was ligated with Sau3AI-partially-digested genomic DNA fragments from strain NEAU-ST10-9T as described in our previous study (Meng et al., 2017). After electroporation into E. coli KNabc cells, the recombinant plasmid designated pUC-S5 was separated by functional complementation with E. coli KNabc on LBK medium plates containing 0.2 M NaCl. The 2712 bp nucleotide sequence was submitted to the GenBank database with the accession No. MH845411. For the subcloning of mceT gene, expression vector pTrcHisB-mceT was constructed through the fusion of the ORF sequence of mceT gene in frame with an N-terminal 6xHis tag followed by an enterokinase cleavage site into an expression vector, pTrcHisB. The gene czcD encoding an identified CDF member from B. subtilis subsp. subtilis strain 168 (Guffanti et al., 2002), was cloned as the positive control of a Zn2+ efflux transporter via the same cloning strategy as mceT. The resultant constructs, pTrcHisB-mceT and pTrcHisB-czcD were verified by sequencing. Primers are listed in Supplementary Table 2.
Preparation of Everted Membrane Vesicles
Everted membrane vesicles were prepared as previously described (Rosen and Tsuchiya, 1979) with a minorly-modified buffer containing 10 mM Tris-HCl (pH7.5), 140 mM choline chloride, 0.5 mM dithiothreitol (DTT), 1 mM phenylmethanesulfonyl fluoride (PMSF) and 250 mM sucrose. Cells of E. coli KNabc carrying pTrcHisB-mceT or its variants, as well as the empty vector pTrcHisB, were harvested and everted by one passage through a JG-1A French Press (NingBo Scientz Biotechnology Co., Ltd., China) at 1000 psi system pressure, and cell debris was removed by centrifugation at 12,000 g for 15 min. The resultant membrane vesicles were collected by centrifugation at 100,000 g for 60 min. The above procedures were performed at 4 °C. The pellets were resuspended in the same buffer, and stored at −80°C. During this course, cell extract and cytoplasmic fraction membrane fraction existing as everted membrane vesicles were sampled, respectively, for the determination of expression and localization of MceT or its variants, as described in our recent studies (Abdel-Motaal et al., 2018; Shao et al., 2018).
Western Blot
Protein samples with 30 μg for each were separated on 12% SDS-polyacrylamide gels and blotted (Bio-Rad Laboratories, Inc., China) onto polyvinylidene difluoride membranes. Western blot detection was performed by using a polyclonal mouse anti-6xHis tag antibody (Beyotime Biotechnology Co. Ltd., Shanghai, China) and an HRP-labeled goat anti-mouse IgG(H+L) (Nachuan Biotechnology Co., Ltd., Changchun, China). The blots were visualized using a BeyoECL Star kit (Beyotime Biotechnology Co. Ltd., Shanghai, China) and recorded by a Tanon-5200 imaging system (Tanon Co. Ltd., China).
Measurements of Na+(Li+, K+)/H+ Antiport Activity by Fluorescence
Na+(Li+, K+)/H+ antiport activity assays were performed as previously described (Bassilana et al., 1984; Nakamura et al., 1986; Goldberg et al., 1987). Everted vesicles containing approximately 100 μg of total membrane protein were added into a 2 ml reaction buffer containing 140 mM choline chloride, 250 mM sucrose, 1 μM acridine orange and 10 mM BTP/HCl adjusted to the indicated pH. Respiration-dependent formation of ΔpH was initiated by the addition of 10 mM Tris-D-lactate, which resulted in the quenching of acridine orange fluorescence. Antiport activity was estimated from the dequenching percentage after the addition of NaCl, LiCl or KCl at the final concentration of 5 mM. Fluorescence was measured with a fluorescence spectrophotometer F-7000 (Hitachi High-Technologies, Japan) with excitation at 490 nm and emission at 530 nm. The apparent affinity of the antiporter for the cations was estimated through the calculation of K0.5 values, which were obtained by fitting the antiport activity as the functions of corresponding cation concentrations followed by non-linear regression analysis using the software Prism 7.0.
Analysis of Intracellular Zn2+ Accumulation
Overnight-grown cultures were diluted 100-fold into 200 ml of fresh LBK broth and continued to grow till OD600nm reached 1.0. Cells were harvested and washed two times with a buffer containing 140 mM choline chloride, 0.2% glucose, 10 mM Tris-HCl at pH 7.5 and then resuspended and adjusted to the same total protein concentration in 2 ml of the same buffer at 4°C. Four moicroliter of cell suspensions were transferred into 96 μl of the same buffer and incubated at 25°C for 10 min before the reaction. In order to start the reaction, appropriate amounts of ZnCl2 were added to the indicated final concentrations. After incubation at 25°C, the reaction was terminated by the addition of 4 ml of the same ice-pre-cooled buffer and immediately filtered through a polyethersulfone (PES) membrane (0.45 μm). Ten microliter of the same ice-pre-cooled buffer was passed through the PES membrane to wash the cells. Finally, 2.5 ml of 5% trichloroacetic acid (TCA) was passed five times through the PES membrane to lyse the cells and dissolve the intracellular Zn2+. Zn2+ contents were determined using an atomic absorption spectrophotometer AA-6650 (Shimadzu, Kyoto, Japan).
Site-Directed Mutagenesis
Site-directed mutagenesis was performed with a Fast Mutagenesis System Kit (Transgen Biotech, Beijing, China), according to the manufacturer’s instructions. Variants were generated by PCR using pTrcHisB-mceT as a template and mutagenic primers (Supplementary Table 2), and verified for the sequence accuracy by sequencing.
Protein Concentration
Protein concentration was determined by the Bradford protein assay with bovine serum albumin as the standard (Bradford, 1976).
Bioinformatic Analysis
DNA sequencing was carried out by Beijing Genomics Institute (Beijing, China). ORFs were deduced by DNAMAN 8.0 and aligned using BlastP at the NCBI website https://blast.ncbi.nlm.nih.gov/Blast.cgi. Promoters were predicted at the website http://www.fruitfly.org/seq_tools/promoter.html (Reese, 2001). Topological analysis was performed at the PredictProtein website https://www.predictprotein.org. CDF members were downloaded from the TCDB database (Saier et al., 2016) at the website http://www.tcdb.org. Amino acid sequence logos were created by submitting the multiple sequence alignment to the WebLogo 3 website http://weblogo.threeplusone.com/. The taxonomy of the hosts was recognized in the UniProtKB/Swiss-Prot database at the website https://www.uniprot.org/taxonomy/. Protein sequences were aligned with the software ClustalX 1.83, followed by the construction of a neighbor-joining phylogenetic tree via the software MEGA 5.0 on the basis of a bootstrap analysis on the clustering stability (1000 replications) (Saitou and Nei, 1986). The modeled structure of MceT was constructed using the Swiss-Model server at the website https://www.swissmodel.expasy.org/interactive. Structure assessment enclosed in Swiss-Model server was used to verify the reliability of the modeled structure of MceT.
Results
Screening for a Na+/H+ Antiporter Gene Candidate
In this study, a Na+/H+ antiporter-deficient E. coli mutant KNabc (ΔnhaA, ΔnhaB, ΔchaA) (Nozaki et al., 1996) was employed to screen Na+/H+ antiporter gene from strain NEAU-ST10-9T by functional complementation on LBK medium plates containing 0.2 M NaCl, which is the upper limit for the growth of E. coli KNabc and routinely selected as the growth condition for the screening of Na+/H+ antiporter genes. As a result, a recombinant plasmid designated pUC-S5 with a 2712 bp digestion fragment succeeded in complementing with E. coli KNabc. Sequence analysis showed three open reading frames (ORFs 1–3) in the fragment (Supplementary Figure 1). ORF1 shares the highest identity (48%) with a hypothetical protein (accession version No. AQU78343.1) from Planococcus faecalis, ORF2 shares the highest identity (69%) with a TetR/AcrR family transcriptional regulator (accession version WP_052144530.1) from Bacillus okhensis, and ORF3 shares the highest identity (63%) with a CDF transporter (accession version No. WP_084309370.1) from Bacillus okhensis. Each ORF is preceded by a predicted promoter and a Shine-Dalgarno (SD) sequence and also ORF3 is followed by one possible terminator (Supplementary Figure 1). It seems that each of them can be a Na+/H+ antiporter gene candidate.
However, each of three ORFs shares no identity with identified single-gene Na+/H+ antiporters or proteins reported to exhibit Na+/H+ antiport activity, the subunit of double-gene or multiple-gene Na+/H+ antiporters, and even predicted Na+/H+ antiporters. Topological analysis showed that ORF3 is the sole transmembrane protein consisting of six TMHs including TMH1 (6–33), TMH2 (35–62), TMH3 (74–104), TMH4 (113–135), TMH5 (142–173), and TMH6 (180–211), and two α helices including α1 (217–229) and α2 (264–281), and also three β stands including β1 (236–245), β2 (250–258) and β3 (288-295) (Supplementary Figure 2), which is a typical topological characteristic of CDF members (Lu and Fu, 2007; Coudray et al., 2013). Therefore, ORF3 may be the real Na+/H+ antiporter gene candidate. For the convenience of the following description, ORF3 was designated MceT on the basis of its main function as a monovalent cation efflux transporter.
Alignment of MceT With Its Homologs
To confirm whether MceT belongs to CDF family, MceT was aligned with its putative homologs using BlastP at the NCBI website. Ten representative homologs were selected from the different species widely distributed in eight phyla (Supplementary Table 3) to show the multiple alignment with MceT (Supplementary Figure 2). In addition to the putative CDF member from B. okhensis, MceT shares 28–45% identities with the selected CDF members (Supplementary Table 3). Also, MceT and its homologs contain the signature sequence between TMH2 and TMH3, as is almost consistent with those of reported CDF members (Montanini et al., 2007). Moreover, MceT shares four relatively highly conserved motifs (Motifs 1–4) with its putative homologs (Supplementary Figure 2).
Because the above-mentioned homologs were predicted on the basis of sequence homology, all the identified and several putative CDF members were also downloaded from the TCDB database, and aligned with MceT using BlastP at the NCBI website. MceT shares ≤24% identities with eleven identified CDF members within the query cover range from 54 to 90% (Supplementary Table 3). These eleven proteins were also aligned with MceT to find out their homology and difference with MceT (Supplementary Figure 3). Interestingly, two motifs, Motif 2 and Motif 4, located in the beginning of TMH3 and TMH6 share almost the same sequence similarity whereas the other two motifs, Motif 1 and Motif 3, located in TMH2 and TMH5, respectively, are significantly varied at the corresponding positions to Y44, S48, and Q150 of MceT (Supplementary Figure 3), compared with those of identified CDF members (Montanini et al., 2007). Based on the above analysis, MceT may be different from identified CDF members, although it should belong to CDF family.
Growth Tests for Salt Tolerance and Alkaline pH Resistance
For the functional analysis of MceT, the mceT gene was fused in frame with an N-terminal 6 × His tag of an expression vector pTrcHisB. As a preliminary test, the growth tests were carried out for salt tolerance and alkaline pH resistance using the resultant construct designated pTrcHisB-mceT and the empty vector pTrcHisB as a negative control (Supplementary Table 1). Expression of MceT in E. coli KNabc led to the increase of Na+ and Li+ tolerance of the host from 5 mM LiCl and 0.2 M NaCl to 0.3 M (Figure 1A) and 7.5 mM (Figure 1B), respectively. Also, expression of MceT significantly enhanced the host growth under alkaline pH conditions, especially at pH 8.0, in contrast to the empty vector (Figure 1C). Therefore, MceT is likely to function as a Na+/H+ antiporter.
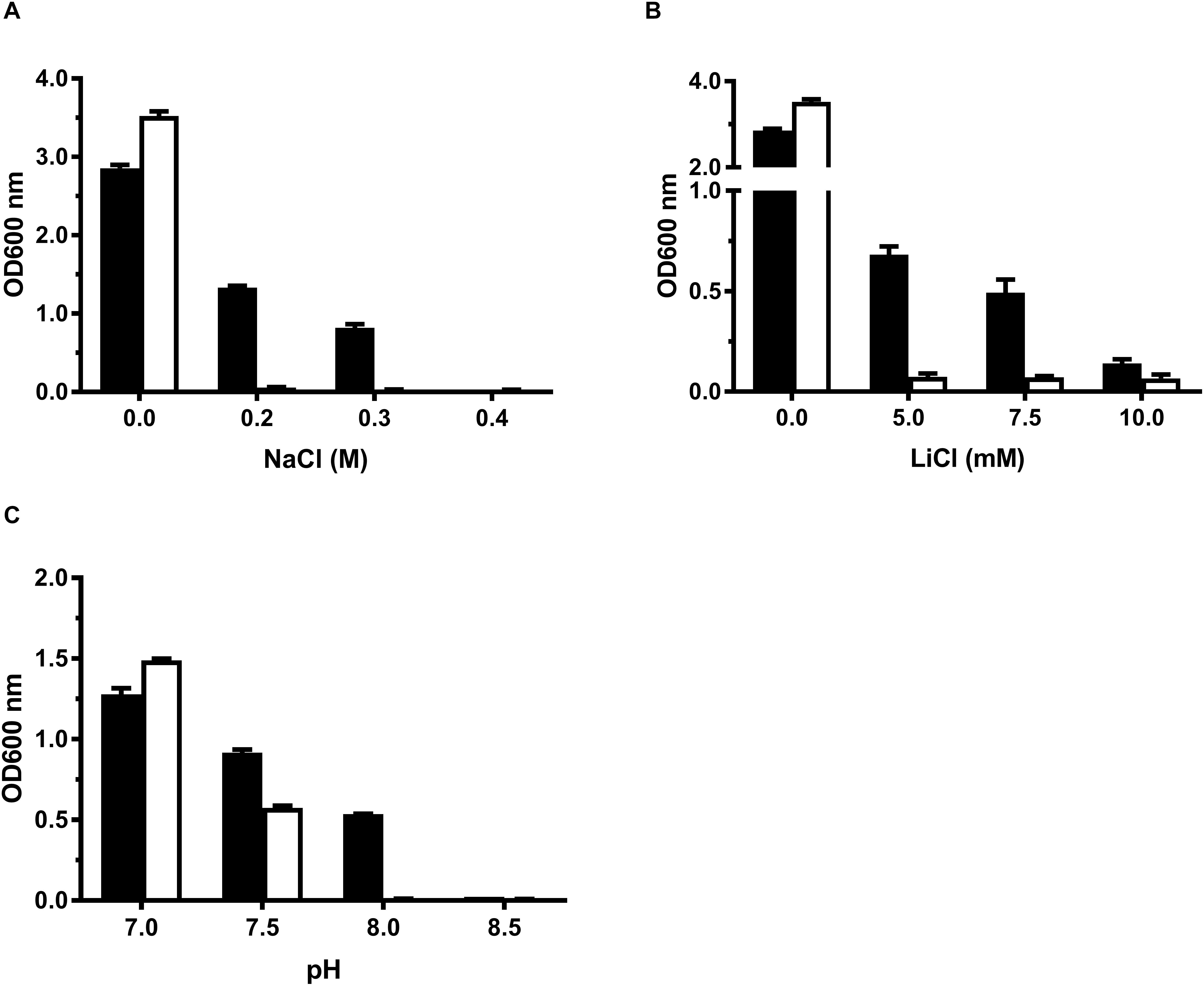
Figure 1. Growth tests for E. coli KNabc transformants under saline or alkaline conditions. For the growth tests under saline or alkaline conditions, E. coli KNabc/pTrcHisB-mceT (black column) and KNabc/pTrcHisB (white column) were grown in LBK broths containing 0–0.4 M NaCl (A) or 0–10 mM LiCl (B), or at the pH values from 7.0 to 8.5 supplemented by 50 mM NaCl (C). The pre-cultures of E. coli KNabc transformant cells were grown to OD600 nm at 1.0 in LBK broths at pH 7.0 at 37°C. The above-mentioned cell growth was ended after 24 h and the values for OD600 nm then evaluated. Each data point represents Mean ± SD of three independent cultures.
Establishment of MceT as a Transmembrane Protein by Western Blot
To establish the expression of MceT, cell extract, cytoplasmic fraction and membrane fraction were sampled during the preparation of everted membrane vesicles from E. coli KNabc carrying pTrcHisB-mceT and the empty vector pTrcHisB as a negative control, followed by the running in the SDS-PAGE (Supplementary Figure 4A) and detection by western blot (Supplementary Figure 4B). His tag-labeled MceT was detected in both cell extract and membrane fraction from E. coli KNabc/pTrcHisB-mceT, but not in those of KNabc/pTrcHisB (Supplementary Figure 4B). Also, no positive signal was detected in cytoplasmic fraction from E. coli KNabc/pTrcHisB-mceT or KNabc/pTrcHisB. These results reveal that MceT is indeed located in the cytoplasmic membrane of E. coli KNabc.
Function of MceT as a Na+(Li+, K+)/H+ Antiporter
Na+/H+ antiporters simultaneously possess Na+/H+ and Li+/H+ antiport activity (Krulwich et al., 2011; Padan, 2014), and also some of them exhibit K+/H+ antiport activity (Fujisawa et al., 2007; Meng et al., 2017; Abdel-Motaal et al., 2018; Shao et al., 2018). To identify the function of MceT, everted membrane vesicles were prepared from E. coli KNabc/pTrcHisB-mceT or KNabc/pTrcHisB and Na+ (Li+, K+)/H+ antiport activity was measured using a conventional fluorescence dequenching method with the acridine orange as a pH indicator. As a result, MceT exhibited Na+(Li+)/H+ (Figures 2A,B) antiport activity, and also K+/H+ (Figure 2C) antiport activity, which were detected within the pH range from 7.0 to 9.5 with the optimal for Na+(Li+)/H+ antiport activity at pH 8.0 and K+/H+ antiport activity at pH 8.0–8.5 (Figure 2D). K0.5 values of MceT were analyzed for Na+, K+, and Li+ in order to assess the apparent affinity of MceT for the cations, and calculated each of them to be 0.21 ± 0.03 mM (Figure 3A), 0.33 ± 0.07 mM (Figure 3B), and 0.34 ± 0.06 mM (Figure 3C), respectively. Therefore, MceT can function as a Na+(Li+, K+)/H+ antiporter.
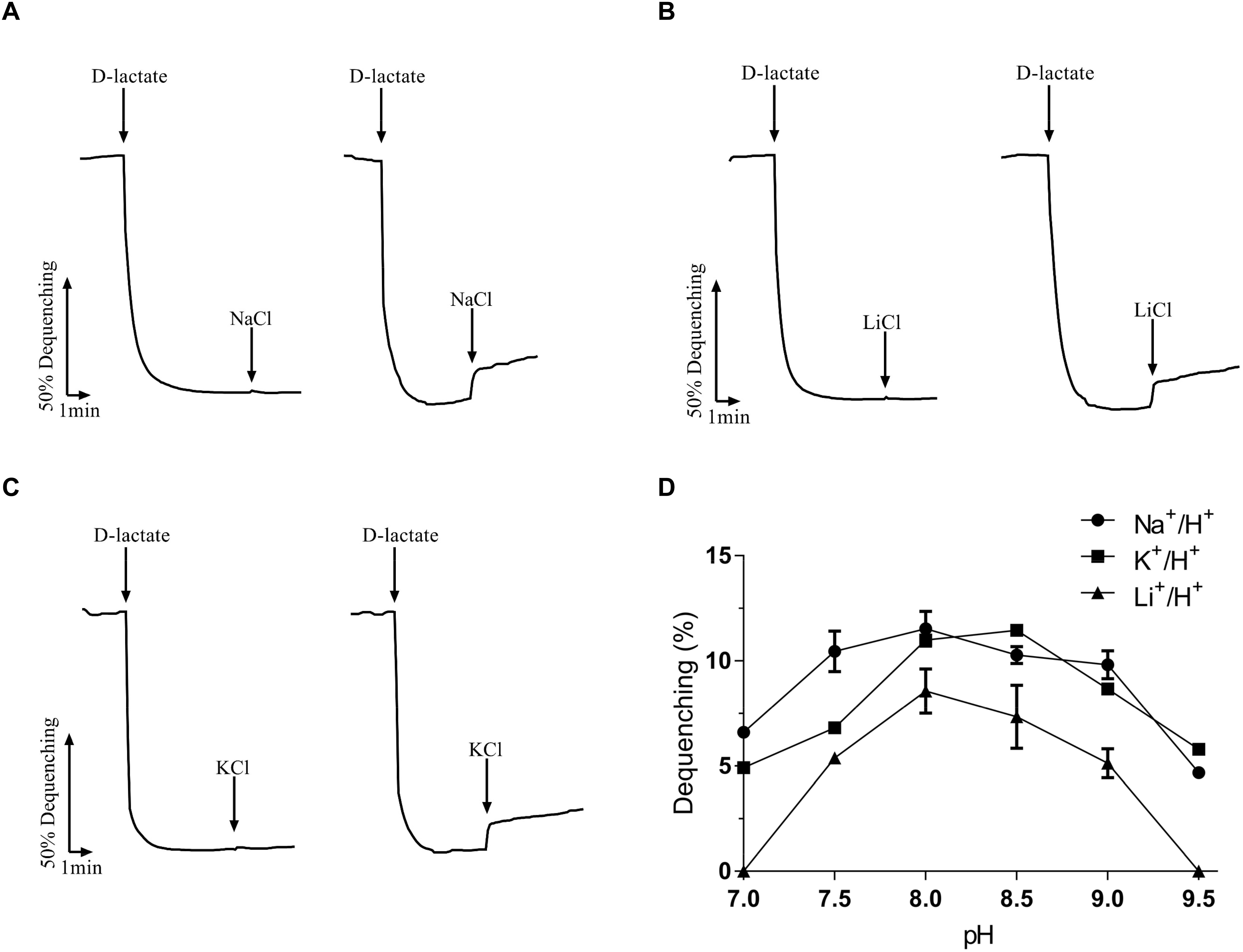
Figure 2. Na+/H+ antiporter activity measured by the acridine orange fluorescence quenching method. | Everted membrane vesicles (100 μg of total membrane protein) prepared from cells of E. coli KNabc/pTrcHisB-mceT (right) or KNabc/pTrcHisB (left) were added in a 2 ml reaction mixture. At the time points indicated, Tris-D-lactate was added at the final concentration of 10 mM to initiate fluorescence quenching due to the H+ transport caused by respiration. Then NaCl, LiCl or KCl was added at the final concentration of 5 mM to the assay mixture at the indicated time points. The transport activity at different pH values was determined by calculating the percentage of dequenching to total quenching (Dequenching %). The antiport activity at pH 8.0 for Na+/H+ (A), Li+/H+ (B), or at pH 8.5 for K+/H+ (C) was shown as the representatives. Also, the antiport activity pH profile (D) for Na+/H+ (filled circle), Li+/H+ (filled triangle) and K+/H+ (filled square) was also plotted at the indicated pH values. Each data point represents Mean ± SD of three independent experiments.
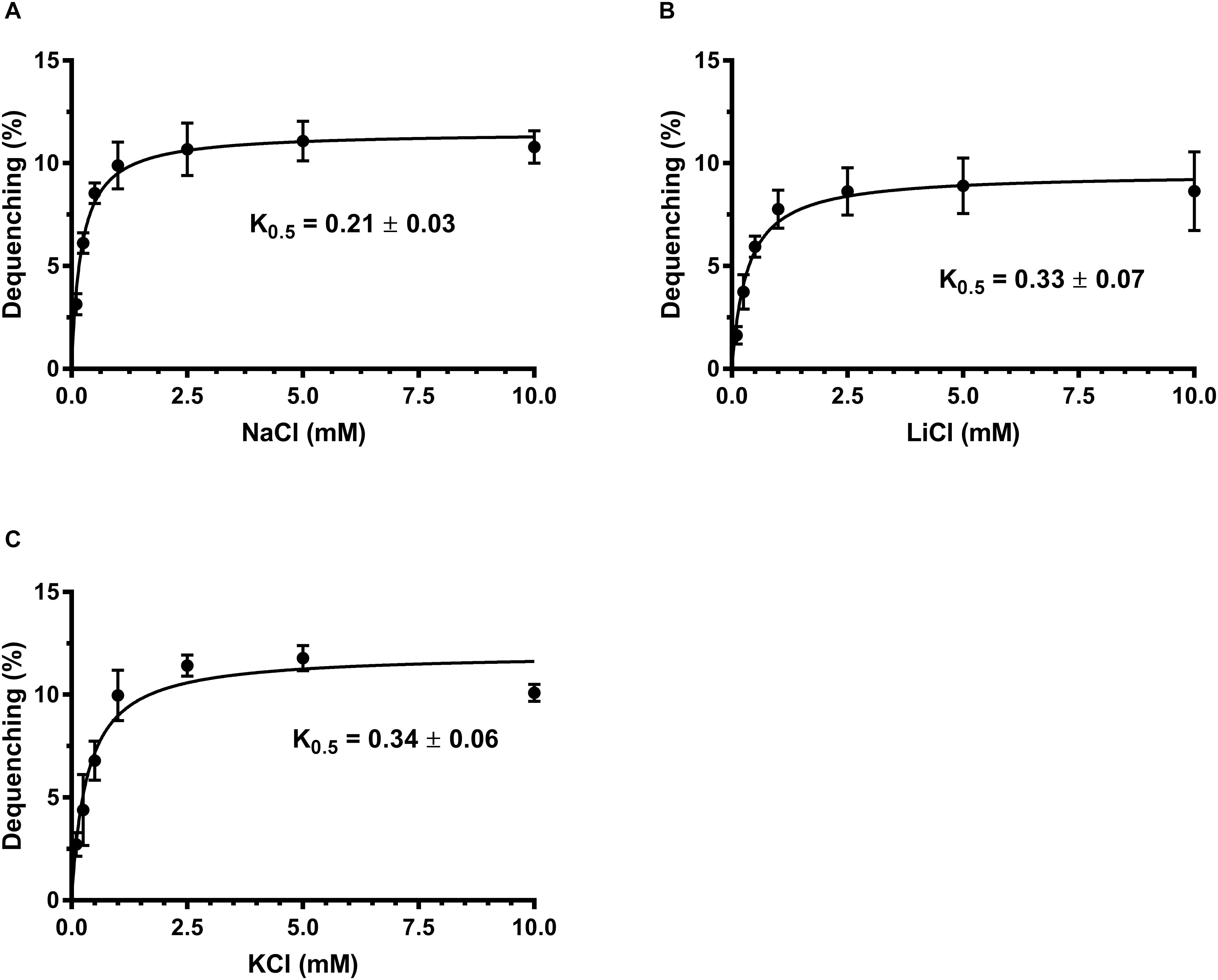
Figure 3. Calculation of K0.5 values of MceT for Na+, Li+, and K+. The antiport activity was determined at pH 8.0 for Na+/H+ (A), Li+/H+ (B), and K+/H+ (C) at the cation concentrations varied from 0 to 10 mM. K0.5 values of MceT for the tested cations were calculated by fitting the antiport activity as the functions of the corresponding cation concentrations followed by non-linear regression analysis with the software Prism 7.0. Each data point represents Mean ± SD of three independent experiments.
Zn2+ Sensitivity of E. coli KZAB04 Exacerbated by MceT
Among identified CDF members, MceT shares relatively higher cover range and identity with Zn-CDF members such as ZnT2 from Rattus norvegicus, ZnT4 from R. norvegicus and Homo sapiens (Supplementary Table 3 and Supplementary Figure 3). Therefore, Zn2+ was chosen as its potential substrate to test whether MceT can function as a Zn2+ efflux transporter. For this purpose, a Zn2+-sensitive E. coli mutant KZAB04 was constructed through the disruption of two major Zn2+ efflux transporters, ZntA (Beard et al., 1997; Rensing et al., 1997), and ZitB (Grass et al., 2001). Also, B. subtilis CzcD (Guffanti et al., 2002) was constructed as the positive control of a prokaryotic Zn2+ efflux transporter into an expression vector pTrcHisB (Supplementary Table 1). At first, growth tests were carried out in LB broths containing ZnCl2 concentrations varied from 0 to 0.75 mM (Figure 4A). Compared with E. coli KZAB04/pTrcHisB, the growth of E. coli KZAB04/pTrcHisB-czcD was significantly enhanced as ZnCl2 concentrations increased from 0.30 to 0.75 mM while that of KZAB04/pTrcHisB-mceT was slightly reduced the growth of E. coli in the presence of ZnCl2, even completely abolished at 0.75 mM ZnCl2 (Figure 5A). K+ was reported to be one of the coupling substrates of CzcD for exchange with the extracellular Zn2+ (Guffanti et al., 2002). Therefore, the growth of the above transformants was tested in LBK broths at the same tested ZnCl2 concentrations (Figure 4B). The growth trend in LBK broths was similar to that in LB broths, except that all the transformants showed higher growth in the former than in the latter. These results reveal that MceT can’t function as a Zn2+ efflux transporter, and also the growth failure of E. coli KZAB04/pTrcHisB-mceT at 0.75 mM ZnCl2 (Figures 4A,B) implies that Zn2+ sensitivity of E. coli KZAB04 should be exacerbated by MceT.
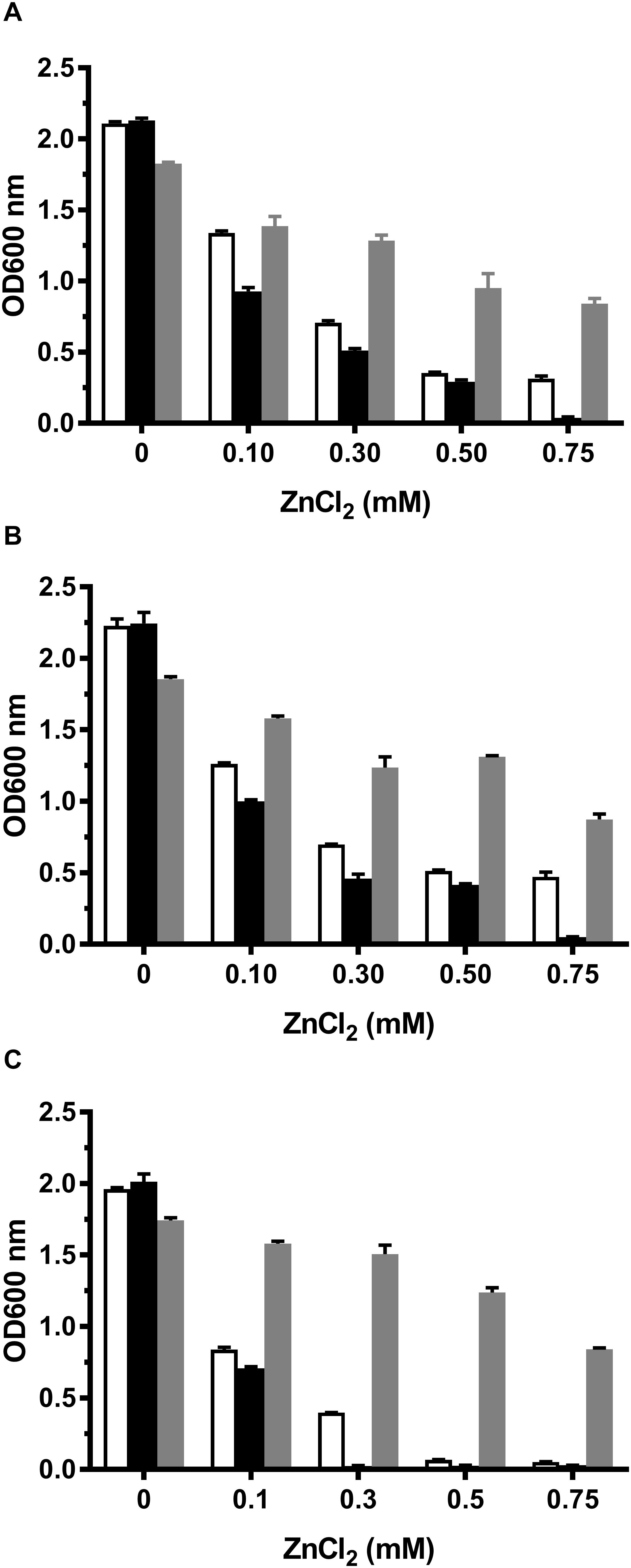
Figure 4. Growth tests for E. coli KZAB04 transformants in LB, LBK, and LBO broths containing different ZnCl2 concentrations. To test whether MceT functions as a Zn2+ efflux transporter, E. coli KZAB04/pTrcHisB (white column), KZAB04/pTrcHisB-mceT (black column) and KZAB04/pTrcHisB-czcD (gray column) as the positive control of a Zn2+ efflux transporter were grown in LB (A) or LBK (B) broths containing 0–0.75 mM ZnCl2. Also, to test the effect of the presence of Na+ or K+ on Zn2+ diffusion facilitated by MceT, the above-mentioned E. coli KZAB04 transformants were grown in LBO (C) broths containing 0–0.75 mM ZnCl2. The pre-cultures of E. coli KZAB04 transformant cells were grown to OD600 nm at 1.0 in LBO broth at pH 7.0 at 37°C. The above-mentioned cell growth was ended after 24 h and the values for OD600 nm then evaluated. Each data point represents Mean ± SD of three independent cultures.
Na+/K+ Independence of Zn2+ Sensitivity by MceT
To test whether the presence of Na+ or K+ may affect Zn2+ sensitivity by MceT, the growth tests were also performed in LBO broths with no addition of NaCl or KCl. In contrast to the empty vector pTrcHisB, expression of CzcD rendered the significant ZnCl2 resistance of E. coli KZAB04 in LBO broths (Figure 4C), which was similarly found in LB or LBK broths (Figures 4A,B). However, the major difference is that E. coli KZAB04 carrying pTrcHisB-mceT or pTrcHisB failed to grow in LBO broths containing 0.50 mM ZnCl2 and above (Figure 4C). That may be attributed to that the absence of Na+ or K+ inhibited the host growth, to some extent. Importantly, E. coli KZAB04/pTrcHisB could grow at 0.30 mM ZnCl2 while KZAB04/pTrcHisB-mceT lost the growth under the same stress (Figure 4C). These results not only confirm that Zn2+ sensitivity of E. coli KZAB04 was exactly exacerbated by MceT but also reveal that this phenomenon was not affected by the presence of Na+ or K+.
Facilitated Diffusion of Zn2+ Into Cells by MceT
Considering Zn2+ sensitivity of E. coli KZAB04 was exacerbated by MceT, MceT may lose Zn2+ efflux activity but retain the Zn2+-binding ability and therefore facilitate the diffusion of Zn2+ into cells. To test this hypothesis, the difference in intracellular Zn2+ accumulation was compared between E. coli KZAB04/pTrcHisB-mceT and KZAB04/pTrcHisB under the stress of high ZnCl2 concentrations. As expected, E. coli KZAB04 cells expressing MceT accumulated significantly higher intracellular Zn2+ concentrations in the presence of 0.75 mM ZnCl2 than those with the empty vector, as the incubation time increased from 0 to 30 min (Figure 5A). Also, the similar difference was found between E. coli KZAB04/pTrcHisB-mceT and KZAB04/pTrcHisB within 20 min when ZnCl2 concentrations were varied from 0 to 1.50 mM (Figure 5B). These results reveal that expression of MceT indeed facilitate the diffusion of extracellular Zn2+ into the cells of E. coli KZAB04.
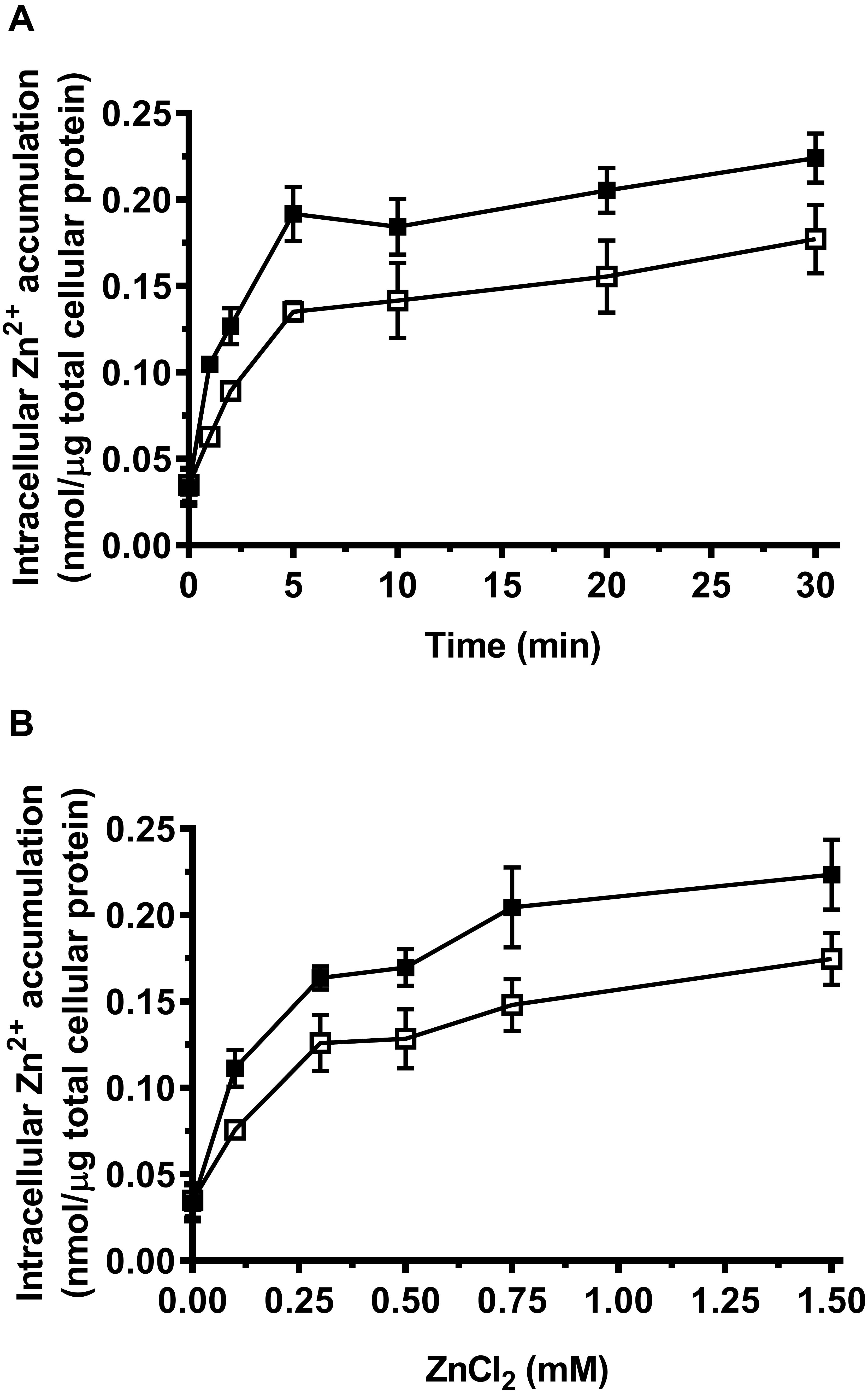
Figure 5. Comparison of intracellular Zn2+ accumulation between E. coli KZAB04/pTrcHisB-mceT and KZAB04/pTrcHisB. | E. coli KZAB04/pTrcHisB-mceT (filled square) and KZAB04/pTrcHisB as a negative control (open square) were grown to OD600 nm at 1.0, respectively. Aliquots of prepared cells with the equal total protein were incubated at 0.75 mM ZnCl2 from 0 to 30 min (A) for the plotting of a time curve of intracellular accumulated Zn2+ contents and also incubated within 20 min at the different ZnCl2 concentrations ranging from 0 to 1.50 mM (B) for the comparison of intracellular accumulated Zn2+ contents between E. coli KZAB04/pTrcHisB-mceT and KZAB04/pTrcHisB. Each data point represents Mean ± SD of three independent experiments.
A Representative of MceT as a Novel CDF Group
On the basis of the above results, MceT may represent a novel class of CDF members. To establish this hypothesis, the phylogenetic relationship was analyzed using Neighbor-Joining (NJ) method between MceT and the representatives of identified or putative CDF members. For the accuracy and representativeness of the phylogenetic tree, the number of target sequences were set to the maximum numerical of 20,000 in the setting of BlastP algorithm parameters. Finally, 62 putative homologs with the identity range of 28–63% were selected and guaranteed to widely distribute in different species or strains from eight phyla as possible (Supplementary Table 3). All the identified or putative CDF members (Supplementary Table 3) were also downloaded from the TCDB database. To avoid biased group distribution, long gaps on extended N or C termini typical of some CDF members were extruded. As shown in Figure 6, CDF members from the TCDB database are clustered into three known CDF groups including Zn-CDF, Fe/Zn-CDF, and Mn-CDF, respectively, except for three exceptional Zn-CDF members such as H. sapiens ZnT9, Saccharomyces cerevisiae Zrg17 and Streptococcus pneumoniae CzcD and one exceptional Mn-CDF member, Sinorhizobium meliloti YiiP. This result reflects the reliability of the constructed phylogenetic tree. Interestingly, MceT exactly clustered with its 62 putative homologs with a bootstrap value of 73% (Figure 6). This suggests that MceT, together with its putative homologs, may constitute a novel CDF group designated Na-CDF with Na+ as the preferred substrate, which is significantly distant with three known CDF groups.
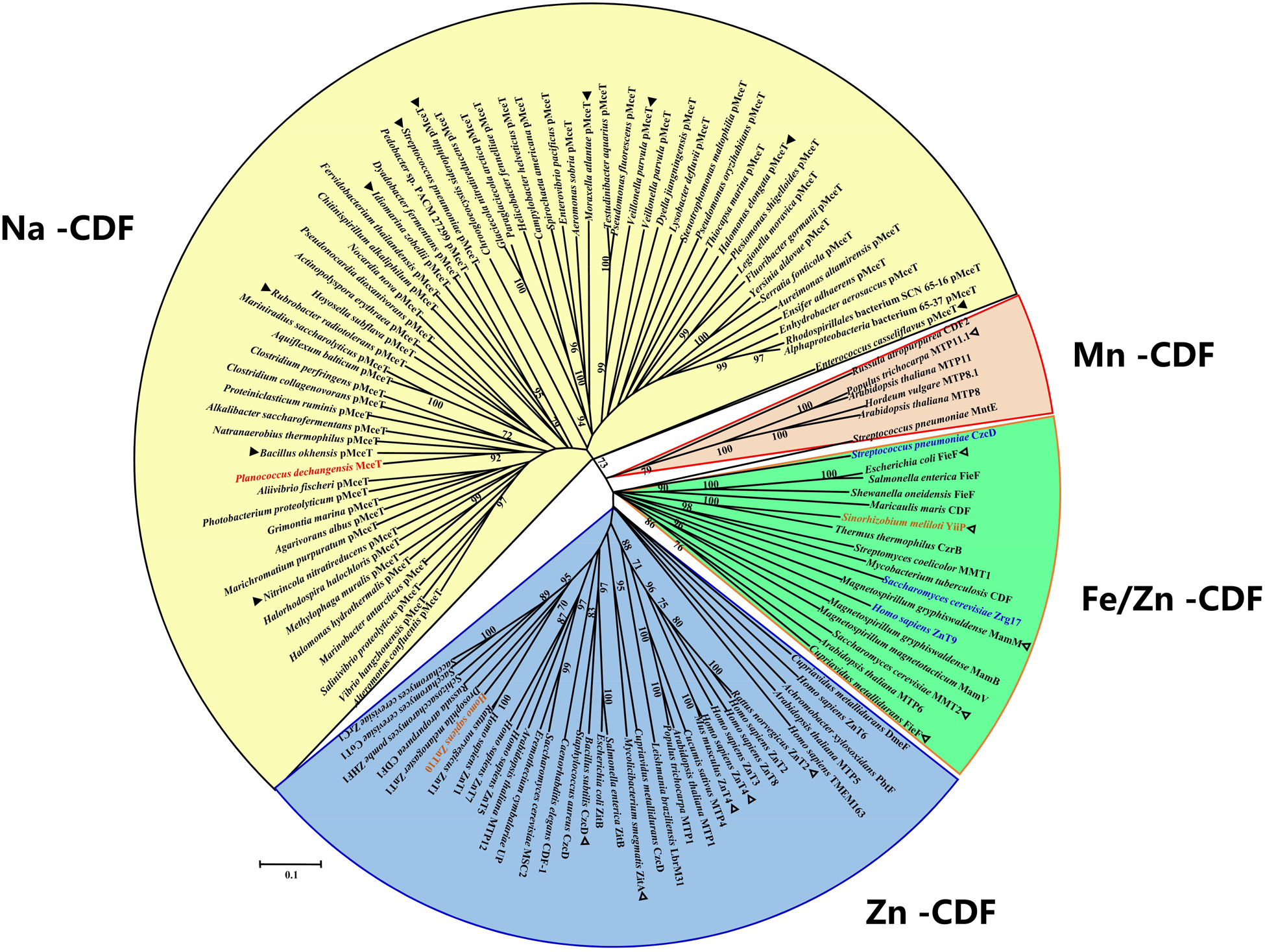
Figure 6. Neighbor-joining phylogenetic tree of MceT with CDF members. For the construction of phylogenetic tree, 62 putative MceT (pMceT) homologs were selected through BlastP at the NCBI website using the deduced amino acid sequence of MceT, and all the identified or putative CDF members collected in the TCDB database were also selected as the respective representatives of three known CDF groups including Zn-CDF (in a blue fan-shaped sector or three exceptional members in blue), Fe/Zn-CDF (in a green fan-shaped sector), and Mn-CDF (in a light red fan-shaped sector or one exceptional member in light red). MceT (in red) and its putative homologs are highlighted in a yellow fan-shaped sector. Ten homologs (filled triangle) were selected as the respective representatives of ten different clusters or clades and aligned with MceT, as shown in Supplementary Figure 2. Eleven identified CDF members (open triangle) with the query cover range above 50% were used for the alignment with MceT, as shown in Supplementary Figure 3. Accession version numbers of selected proteins were listed in the Supplementary Table 3. Bootstrap values ≥70% (based on 1000 replications) are shown at branch points. Bar, 0.1 substitutions per amino acid residue position.
Significantly Different Conserved Residues Located Within Motif 1 and Motif 3 of Na-CDF Group
To establish whether MceT represents a novel CDF group, the web-based amino acid sequence logos were created to analyze the difference in conserved residues between Na-CDF group members and three known CDF groups. The polar or aromatic residues including Y44, S45, Q150, W151, and Y182 located within Motif 1 and Motif 3 were found to be highly conserved within Na-CDF group, which are different or non-conserved at the corresponding positions within three known CDF groups (Figure 7). Also, other negatively-charged, polar or aromatic residues such as E8, S15, F40, C93, C127, E147, and F165 unlocated within the four conserved motifs are highly conserved within Na-CDF group, which are not conserved at the corresponding positions within three known CDF groups (Figure 7). Moreover, negatively-charged or polar residues including S35 unlocated within the four conserved motifs, and D41, E78, D154, S158, and D184 located within the four conserved motifs are highly conserved between Na-CDF group and three known CDF groups. Exceptionally, S48 located within Motif 1 is not highly conserved between Na-CDF group members (Figure 7), although MceT shows significant difference at this position from eleven identified CDF members (Supplementary Figure 3). The most important finding is that “D41-Y44-S45” within Motif 1 of MceT and “Q150-D154” within Motif 3 of MceT are different from “D47-H50-D54” within Motif 1 of CzcD, and “H154-D158” within Motif 3 of CzcD, which supports that MceT, together with its putative homologs, should constitute a novel CDF group, Na-CDF.

Figure 7. Comparison of web-based amino acid sequence logos between MceT homologs and identified CDF members. Amino acid sequence logos were created by submitting the alignment of MceT with its selected 62 homologs within the phylogenetic tree (Figure 6) and the alignment of B. subtilis CzcD with all the identified and putative CDF members downloaded from the TCDB database to the WebLogo 3 website http://weblogo.threeplusone.com/, respectively. The detailed information of proteins included in the phylogenetic tree is shown in Supplementary Table 3. The positions of residues correspond to those of CzcD and MceT, respectively. The heights of amino acid symbols stand for their conservation in the multiple alignment. Six predicted TMHs (TMH 1-6) of MceT (yellow filled cylinder) or CzcD (blue filled cylinder) were shown above the logos, respectively. Four conserved motifs within Na-CDF group and three known CDF groups are labeled and highlighted in the open rectangles. The highly conserved negatively-charged, polar or aromatic residues distributed within TMHs of MceT are marked with the black filled downward triangles above the logo of MceT and its homologs.
Functional Analysis of Conserved Residues Located Within TMHs of MceT
Conserved residues located within TMHs play a critical role in transporting activity of CDF transporters (Montanini et al., 2007; Hoch et al., 2012; Kolaj-Robin et al., 2015; Martin and Giedroc, 2016; Nishito et al., 2016) or Na+/H+ antiporters (Inoue et al., 1995; Habibian et al., 2005; Jiang et al., 2013). Based on the predicted topological model of MceT (Figure 8), 17 conserved residues excluding C93 and S48 were replaced to test their importance for Na+ efflux or facilitated Zn2+ diffusion. C127 was selected as a representative of C93 and C127, and also non-conserved S48 was not selected (Figure 7). Moreover, CzcD was chosen as the positive control of a Zn2+ efflux transporter and the empty vector as a negative control. For the functional analysis, E. coli KNabc and KZAB04 transformants expressing wild-type MceT or each of its variants were grown in LBK broth containing 0.2 M NaCl (Figure 9A, left panel) and LBK broth containing 0.75 mM ZnCl2 (Figure 9A, right panel), respectively. Expression of wild-type MceT and variants was verified in E. coli KNabc by western blot. In contrast to wild-type MceT, all its variants were identified to be expressed at the similar or higher level, with the sole exception of non-expressed S35A (Figure 9B). Two variants, Q150A and S158A, recovered no growth of E. coli KNabc in the presence of 0.2 M NaCl (Figure 9A, left panel), while still could inhibit the growth of E. coli KZAB04 in the presence of 0.75 mM ZnCl2 (Figure 9A, right panel). The activity assay showed that Q150A and S158A absolutely abolished Na+/H+ antiport activity (Figure 9C, left panel), indicating that Q150 and S158 are key residues of MceT only for Na+ efflux. Two variants, D41A and D154A, rendered Na+ tolerance of E. coli KNabc (Figure 9A, left panel) but completely or largely restored ZnCl2 resistance of E. coli KZAB04 (Figure 9A, right panel). Also, D41A and D154A lost the capability of facilitated Zn2+ diffusion (Figure 9C, right panel), indicating that D41 and D154 are involved in Zn2+ diffusion facilitated by MceT. Notably, D184A restored not only entire NaCl sensitivity of E. coli KNabc but also partial ZnCl2 resistance of E. coli KZAB04, implying D184 is the prerequisite for both Na+ and Zn2+ transport. This is supported by the results of activity assays (Figure 9C). Moreover, E. coli KNabc expressing F40A, S45A or E78A showed weak growth (Figure 9A, left panel). This reveals that these three residues are related, to some extent, to Na+ efflux by MceT. The non-expressed S35A (Figure 9B) had no effect on NaCl sensitivity of E. coli KNabc or ZnCl2 resistance of E. coli KZAB04 (Figure 9A). The possible reason can’t be speculated for non-expression of S35A. However, this finding confirms that the expression of wild-type MceT indeed can exacerbate ZnCl2 sensitivity of E. coli KZAB04 and even inhibit the host growth.
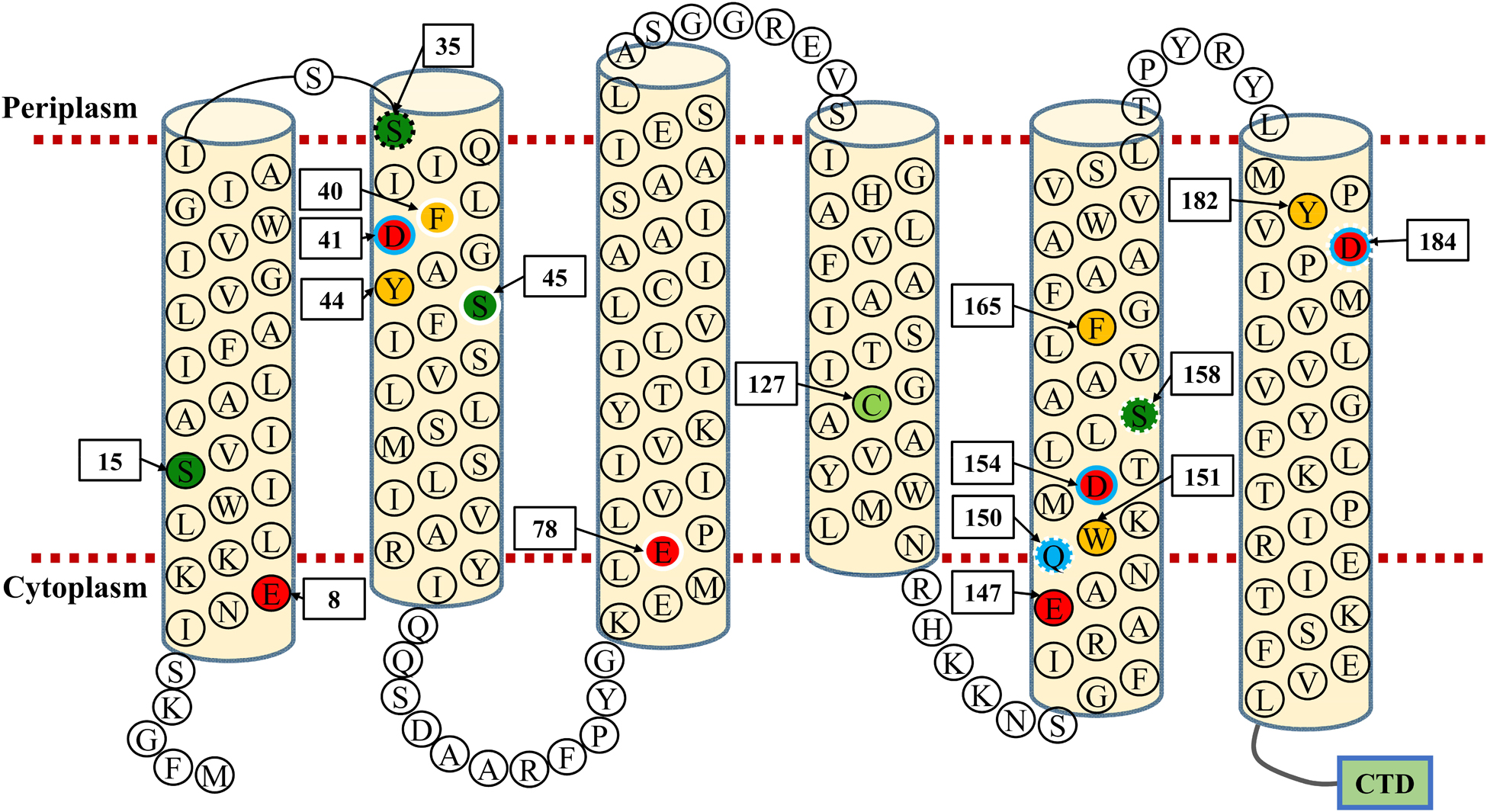
Figure 8. Topological model of MceT. This model is constructed based on the predicted topology using the deduced amino acid sequence of MceT. Six predicted TMHs (TMH 1–6) are marked with light yellow filled cylinders. Conserved negatively-charged residues (red filled circle), conserved aromatic residues (yellow filled circle), conserved serine residues (deep green filled circle), one conserved cysteine residue (light green filled circle) and one conserved glutamine residue (blue filled circle) are highlighted, respectively. S35, whose variant substituted by alanine showed no expression in the cytoplasmic membrane, is shown in the black dotted-line circle; three residues (F40, S45, E78), whose variants substituted by alanine significantly reduced the complementation capacity with E. coli KNabc, are shown in white solid-line circles; three residues (Q150, S158, D184), whose variants substituted by alanine lost the complementation capacity with E. coli KNabc, are shown in white dotted-line circles; three residues (D41, D154, D184), whose variants substituted by alanine rescued the growth of E. coli KZAB04, are shown in blue solid-line circles. CTD (green filled rectangle) stands for the C-terminal domain of MceT composed of 95 aa.
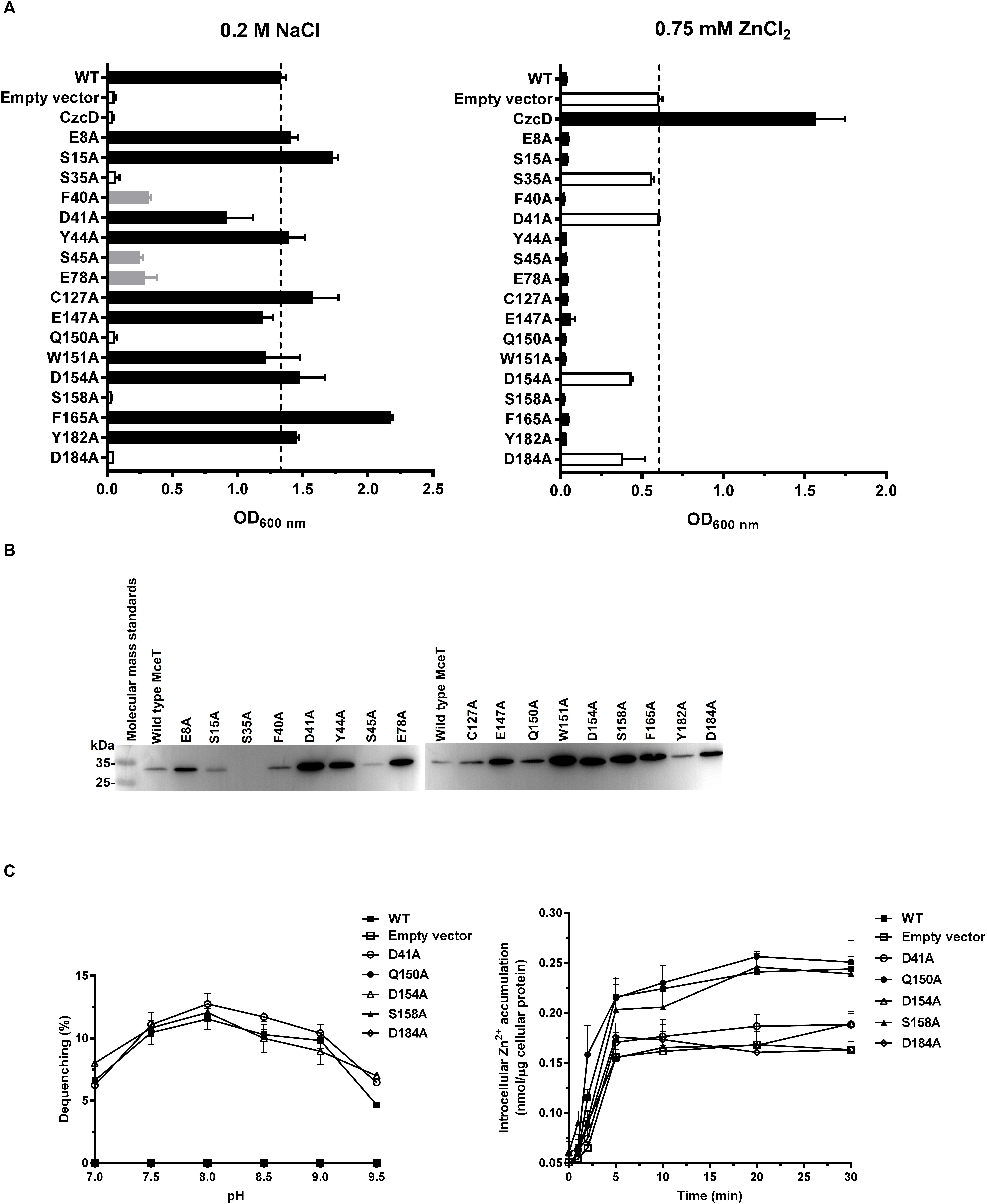
Figure 9. Functional analysis of MceT variants. To analyze the effect of MceT variants by site-directed mutagenesis on the Na+/H+ antiport activity or facilitated Zn2+ diffusion, E. coli KNabcor KZAB04 transformants (A) with the empty vector (negative control), and expressing CzcD (positive control as a Zn2+ efflux transporter), MceT or its variants substituted by alanine were grown in LBK broths containing 0.2 M NaCl or 0.75 mM ZnCl2 within 24 h, respectively, followed by the measurements of OD600 nm. To analyze the expression of MceT variants, western blot detection (B) was also carried out using everted membrane vesicles prepared from E. coli KNabc transformants of WT and its variants. The activity for Na+/H+ antiport and facilitated Zn2+ diffusion of MceT variants including Q150A, S158A, D41A, D154A, D184A was assayed (C).
Predicted Na+ and Zn2+ Transport Models for MceT
In the predicted topological model of MceT (Figure 8), D41 in TMH2 and D184 in TMH6 are adjacent to the periplasmic side. Also, Q150, D154, and S158 are located in the center of TMH5, of which Q150 is closer to the cytoplasmic side and S158 closer to the periplasmic side. The modeled structure of MceT (Figure 10) with the template of 3D structure of Shewanella oneidensis YiiP (PDB ID: 5vrf) verifies the reliability of its predicted topological model and better shows the location of the above five key residues. Based on the current results, the Na+ and Zn2+ transport models for MceT were predicted, respectively (Figure 10). In these two models, MceT may employ the same transmembrane channel for Na+ and Zn2+ translocation (Figure 10). However, Q150, S158, and D184 are responsible for Na+ efflux and selectivity, and may transport Na+ outside the cells in a relay mode of Na+/H+ antiporters. In contrast, D41, D154 and D184 can still retain the capability of Zn2+ binding. D41 and D184 may form a metal ion coordinating site adjacent to the periplasmic side and thus can bind with the extracellular Zn2+. After their coordination of with Zn2+, D154 may act as a vital relay site and thus facilitate Zn2+ diffusion into cells under the stress of high concentrations of extracellular Zn2+.
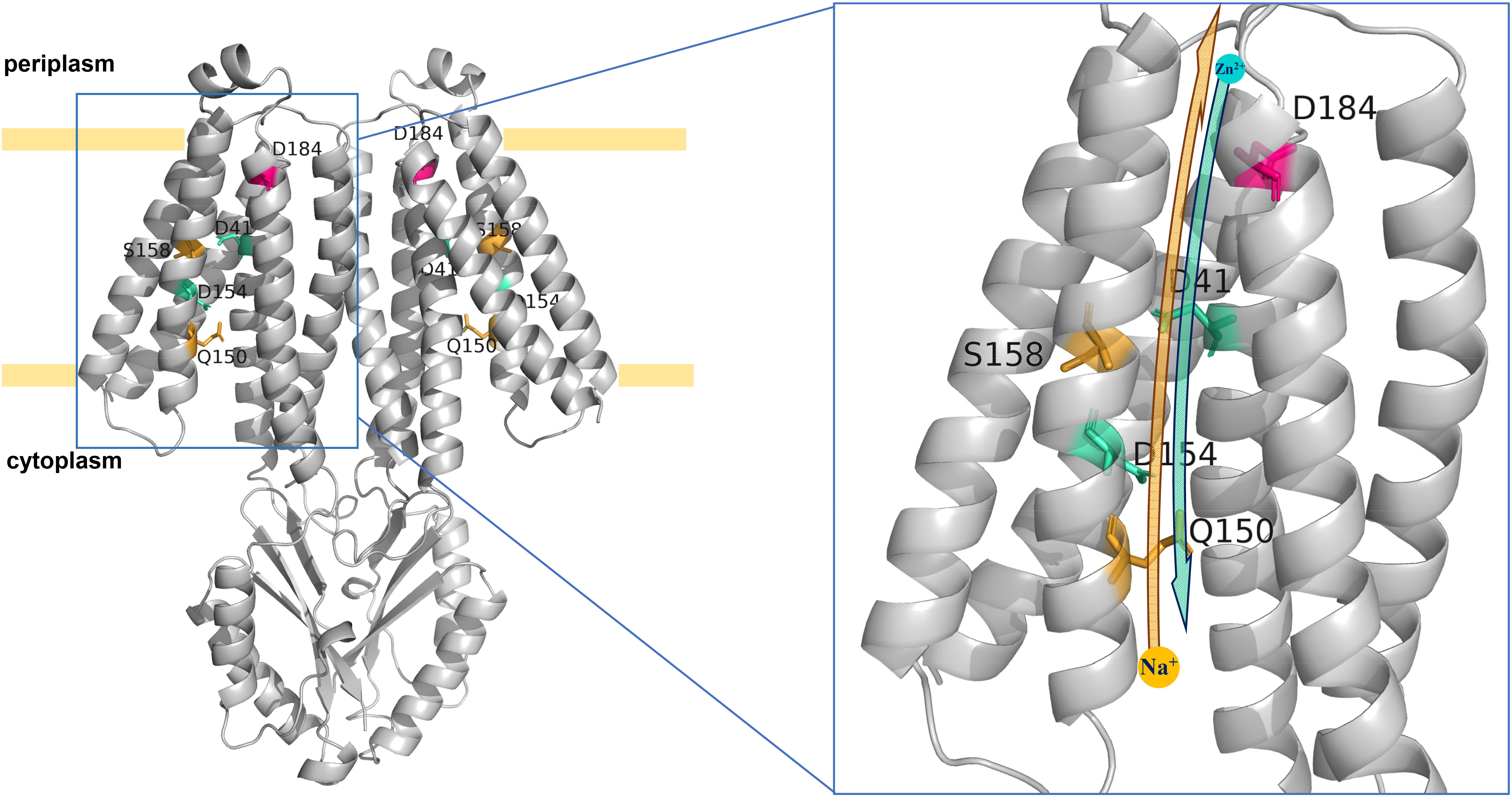
Figure 10. Predicted Na+ and Zn2+ transport models for MceT. The Na+ and Zn2+ transport models were predicted on the basis of the modeled structure of MceT with the template of 3D structure of S. oneidensis YiiP (PDB ID: 5vrf). Na+ and Zn2+ are drawn as a yellow filled circle and a cyan filled circle, respectively. Key residues including D41, Q150, D154, S158, and D184 are labeled and colored. The yellow and cyan arrows stand for the transport orientations of Na+ and Zn2+, respectively. In two models of MceT, MceT may employ the same ionic translocation channel for Na+ efflux and facilitated Zn2+ diffusion. In the presence of extracellular Na+, MceT can extrude Na+ in exchange for external H+ in a relay mode of three key Na+/H+-binding residues (Q150 > S158 > D184). Under the stress of high concentrations of extracellular Zn2+, MceT can facilitate the diffusion of Zn2+ into cells in a relay mode of three possible key Zn2+-transporting residues (D184 and D41 > D154).
Discussion
This study identifies a novel CDF transporter, MceT, from the moderate halophile P. dechangenesis NEAU-ST10-9T. This transporter functions as a Na+(Li+, K+)/H+ antiporter, together with its facilitated Zn2+ diffusion into cells, which is significantly different from all identified CDF members (Paulsen and Saier, 1997; Haney et al., 2005; Montanini et al., 2007). MceT is proposed to represent a novel CDF group, Na-CDF, which shares relatively distant phylogenetic relationship with three known CDF groups including Mn-CDF, Fe/Zn-CDF, and Zn-CDF (Montanini et al., 2007). This can be strongly supported by a significant discrimination in key conserved residues of two function-related structural motifs between Na-CDF group and three known CDF groups. Site-directed mutagenesis implies that this discrimination leads to the evolution of MceT from a Zn2+-efflux transporter to a Na+(Li+, K+)/H+ antiporter. These presented findings provide the important evolutionary implications that CDF transporters can change the ionic selectivity from divalent cations to monovalent ones through the substitution of key conserved residues in their major structural motifs. More importantly, the discovery of MceT contributes to a typical transporter model of CDF family with the unique structural motifs, which will be utilized to explore the cation-selective mechanisms of secondary transporters.
In identified CDF transporters, four conserved coordinating residues in TMH2 and TMH5 are proposed as a major metal ion binding site (Coudray et al., 2013), in which the structural motifs such as DD-HD, HD-HD and ND-HD are responsible for the selection specificity between divalent cations including Zn2+, Cd2+, Fe2+, or Mn2+ (Montanini et al., 2007; Hoch et al., 2012; Kolaj-Robin et al., 2015; Martin and Giedroc, 2016; Nishito et al., 2016). At the corresponding positions of MceT, the residues in Motif 1 are varied to Y44 and S48 (YS) whereas the residues in Motif 3 are varied to Q150 and D154 (QD). This remarkable discrimination can elucidate the loss of Zn2+ efflux activity by MceT. Notably, YS-QD motifs in MceT remain a conserved aspartic acid residue (D154), which is consistent with the corresponding one of identified CDF members (Montanini et al., 2007; Hoch et al., 2012; Kolaj-Robin et al., 2015; Martin and Giedroc, 2016; Nishito et al., 2016). Mutation of D154A partially abolished the facilitated extracellular Zn2+ diffusion of MceT in cells, which suggests the Zn2+ coordinating ability of D154. The consistent conservation of D41 in Motif 1 and D184 in Motif 4 exists between Na-CDF and three known CDF groups. Mutations in either of two corresponding aspartic acid residues in two CDF members, E. coli ZitB and Populus trichocarpa ×Populus deltoides MTP1, completely abolished their resistance to Zn2+ (Blaudez et al., 2003; Anton et al., 2004; Montanini et al., 2007). Therefore, the corresponding residues of D41 and D184 may form an additional metal ion coordinating site in CDF transporters. In CDF family transporters, the metal ion coordinating ability of these two residues has not attracted enough attention. This may be because that their roles are masked by the existence of four coordinating residues in the shared structural motifs (Lu and Fu, 2007; Montanini et al., 2007; Lu et al., 2009; Hoch et al., 2012; Gupta et al., 2014; Kolaj-Robin et al., 2015; Martin and Giedroc, 2016; Nishito et al., 2016). However, varied motifs of MceT lack three key residues in the shared structural motifs of identified CDF transporters and thus highlight the significant roles of D41 and D184 in Zn2+ transport. Therefore, we propose to improve D-D (H, N)-D in TMH2, H-D in TMH5 and D in TMH6 as the common structural motifs of CDF members, which are responsible for the metal ion binding and/or selectivity.
It’s very interesting why and how MceT selects Na+ as the preferred substrate. 3D struture of E. coli NhaA provides a Na+-transporting model of Na+/H+ antiporters, in which D133, D163 and D164 constitute a core region for Na+ efflux in exchange with external H+ (Hunte et al., 2005). D184 is indispensable for both Na+ and Zn2+ transport of MceT, indicating that the aspartic acid residue at this position plays a decisive role in the cation coordination. Polar residues also play critical roles in the antiport activity of many Na+/H+ antiporters (Inoue et al., 1995; Habibian et al., 2005; Jiang et al., 2013). Mutations of Q150A and S158A led to the loss of Na+ tolerance, but had no effect on Zn2+ transport. The activity assay reveals that these two residues are closely involved in the Na+/H+ antiport activity of MceT, although it’s unclear whether they can bind with Na+ or H+. The negatively-charged aspartic acid and glutamic acid residues play predominant roles in the Na+ binding or protonation of Na+/H+ antiporters (Inoue et al., 1995; Habibian et al., 2005; Hunte et al., 2005; Jiang et al., 2013). However, a key glutamine residue sets Na+ pump rhodopsins from marine bacteria apart from H+ pump rhodopsins from marine bacteria, which may determine the Na+ coupling of Na+ pump rhodopsins (Inoue et al., 2015). Therefore, Q150 may act as a determinant for the selectivity of MceT for Na+.
Halophiles, which can grow at a wide range of 0.5–32.5% (w/v) NaCl, have to confront the challenges to the presence of molar concentrations of salt ions or high alkaline pH conditions and therefore may be driven to evolve their proteins for the adaptation to the extremely saline-alkaline habitats (Ventosa et al., 1998; Oren, 1999). Na+/H+ antiporters play a predominant role in maintaining intracellular pH and Na+ homeostasis (Padan and Landau, 2016). In the phylogenetic analysis, MceT and its homologs form an independent group, which is significantly distant with three known CDF groups. Combined with the functional characterization of MceT, this implies that MceT may have evolved from a Zn2+-efflux transporter to a Na+/H+ antiporter. MceT shares the homologies and similar structures with identified CDF members, but the function-related structural motifs are varied in key residues. Therefore, MceT is very likely to have been modified from its native CDF family function to a Na+/H+ antiporter in an evolutionary strategy of the substitution of key conserved residues in its function-related structural motifs. During the evolution, MceT selected YS-QD motifs in TMH1 and TMH3 and thus lost the Zn2+ efflux activity, and also employs Q150 for the selectivity of Na+ at this important cation-selective site. Also, it may retain D184 in TMH6 as the Na+-binding site and S158 in TMH3 as an assistant for the ionic transport, since they are conserved within CDF members (Paulsen and Saier, 1997; Haney et al., 2005; Montanini et al., 2007) and also can be utilized for the participation in the Na+/H+ antiport (Inoue et al., 1995; Habibian et al., 2005; Hunte et al., 2005). Moreover, MceT is not essential to modify D41 In TMH1 or D154 in TMH3, due to their no effect on the Na+/H+ antiport. This also explains why MceT can facilitate extracellular Zn2+ diffusion into cells. MceT retains D41, D154, and D184, and thus can still employ them to bind with Zn2+. However, without the aid of key residues for Zn2+ efflux, MceT is forced to facilitate extracellular Zn2+ diffusion into cells under the stress of high concentrations of extracellular Zn2+.
Although the Na+ and Zn2+ transport models were predicted for MceT, the exact roles of function-related residues remain to be clarified. In the future study, it needs to be confirmed whether D184 can bind with Na+, Zn2+, or H+ and whether D41 and D154 can bind with Zn2+. Also, it is very worthy of being explored whether Q150 or even S158 can bind directly with Na+ and thus cause MceT to select Q150 and S158 for the Na+/H+ antiport. Therefore, we have over-expressed and purified MceT and its variants for the isothermal titration calorimetry (ITC) test in order to analyze the cation-binding ability of the above-mentioned residues. To better answer the above questions, we also plan to discover its structure using purified MceT on the basis of X-ray crystallography. Furthermore, it needs to be confirmed whether variation in structural motifs of MceT leads to the loss of Zn2+ efflux activity. We have been modifying the residues of MceT through the site-directed mutagenesis based on the Zn2+-transporting model of CzcD and hope to reconstruct MceT as a Zn2+-efflux transporter, as the functions of identified CDF members.
Data Availability
The datasets generated for this study can be found in GenBank, No. MH845411.
Author Contributions
TX and JJ contributed to the study design, analyzed the data, and revised the manuscript. TX performed the construction of expression vector, zinc-sensitive E. coli mutant, and protein variants. SH performed the cloning of mceT. TX, HC, JL, LS, XZ, QZ, YW, and SG contributed to the cultures, preparation of everted membrane vesicles, and transporting activity determination. TX drafted the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 31770051).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Prof. Terry A. Krulwich (Department of Pharmacology and Systems Therapeutics, Mount Sinai School of Medicine, New York, United States) for friendly donation of E. coli strain KNabc, Dr. W. Todd Lowther (School of Medicine, Wake Forest University, North Carolina, United States) for friendly donation of the plasmid pKD46, and Prof. Jose Enrique Ruiz-Sainz (Department of Microbiology, University of Seville, Spain) for friendly donation of the plasmids pBBR1MCS-2 and pBBR1MCS-5.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00607/full#supplementary-material
References
Abdel-Motaal, H., Meng, L., Zhang, Z., Abdelazez, A. H., Shao, L., Xu, T., et al. (2018). An uncharacterized major facilitator superfamily transporter from Planococcus maritimus exhibits dual functions as a Na+(Li+, K+)/H+ antiporter and a multidrug efflux pump. Front. Microbiol. 9:1601. doi: 10.3389/fmicb.2018.01601
Anton, A., Weltrowski, A., Haney, C. J., Franke, S., Grass, G., Rensing, C., et al. (2004). Characteristics of zinc transport by two bacterial cation diffusion facilitators from Ralstonia metallidurans CH34 and Escherichia coli. J. Bacteriol. 186, 7499–7507. doi: 10.1128/JB.186.22.7499-7507.2004
Bassilana, M., Damiano, E., and Leblanc, G. (1984). Relationships between the Na+-H+ antiport activity and the components of the electrochemical proton gradient in Escherichia coli membrane vesicles. Biochemistry 23, 1015–1022. doi: 10.1021/bi00300a033
Beard, S., Hashim, R., Membrillo-Hernandez, J., Hughes, M., and Poole, R. K. (1997). Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol. Microbiol. 25, 883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x
Blaudez, D., Kohler, A., Martin, F., Sanders, D., and Chalot, M. (2003). Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 15, 2911–2928. doi: 10.1105/tpc.017541
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Coudray, N., Valvo, S., Hu, M., Lasala, R., Kim, C., Vink, M., et al. (2013). Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 110, 2140–2145. doi: 10.1073/pnas.1215455110
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Dong, P., Wang, L., Song, N., Yang, L., Chen, J., Yan, M., et al. (2017). A UPF0118 family protein with uncharacterized function from the moderate halophile Halobacillus andaensis represents a novel class of Na+(Li+)/H+ antiporter. Sci. Rep. 7:45936. doi: 10.1038/srep45936
Fujisawa, M., Ito, M., and Krulwich, T. A. (2007). Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity. Proc. Natl. Acad. Sci. U. S. A. 104, 13289–13294. doi: 10.1073/pnas.0703709104
Goldberg, E. B., Arbel, T., Chen, J., Karpel, R., Mackie, G. A., Schuldiner, S., et al. (1987). Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 84, 2615–2619. doi: 10.1073/pnas.84.9.2615
Grass, G., Fan, B., Rosen, B. P., Franke, S., Nies, D. H., and Rensing, C. (2001). ZitB (YbgR), a member of the cation diffusion facilitator family, is an additional zinc transporter in Escherichia coli. J. Bacteriol. 183, 4664–4667. doi: 10.1128/JB.183.15.4664-4667.2001
Guffanti, A. A., Wei, Y., Rood, S. V., and Krulwich, T. A. (2002). An antiport mechanism for a member of the cation diffusion facilitator family: divalent cations efflux in exchange for K+ and H+. Mol. Microbiol. 45, 145–153. doi: 10.1046/j.1365-2958.2002.02998.x
Gupta, S., Chai, J., Cheng, J., D’Mello, R., Chance, M. R., and Fu, D. (2014). Visualizing the kinetic power stroke that drives proton-coupled zinc(II) transport. Nature 512, 101–104. doi: 10.1038/nature13382
Habibian, R., Dzioba, J., Barrett, J., Galperin, M. Y., Loewen, P. C., and Dibrov, P. (2005). Functional analysis of conserved polar residues in Vc-NhaD, Na +/H+ antiporter of Vibrio cholerae. J. Biol. Chem. 280, 39637–39643. doi: 10.1074/jbc.M509328200
Haney, C. J., Grass, G., Franke, S., and Rensing, C. (2005). New developments in the understanding of the cation diffusion facilitator family. J. Ind. Microbiol. Biotechnol. 32, 215–226. doi: 10.1007/s10295-005-0224-3
Hoch, E., Lin, W., Chai, J., Hershfinkel, M., Fu, D., and Sekler, I. (2012). Histidine pairing at the metal transport site of mammalian ZnT transporters controls Zn2+ over Cd2+ selectivity. Proc. Natl. Acad. Sci. 109, 7202–7207. doi: 10.1073/pnas.1200362109
Hunte, C., Screpanti, E., Venturi, M., Rimon, A., Padan, E., and Michel, H. (2005). Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202. doi: 10.1038/nature03692
Inoue, H., Noumi, T., Tsuchiya, T., and Kanazawa, H. (1995). Essential aspartic acid residues, Asp-133, Asp-163 and Asp-164, in the transmembrane helices of a Na+/H+ antiporter (NhaA) from Escherichia coli. FEBS Lett. 363, 264–268. doi: 10.1016/0014-5793(95)00331-3
Inoue, K., Konno, M., Abe-yoshizumi, R., and Kandori, H. (2015). The role of the NDQ motif in sodium-pumping rhodopsins. Angew.Chem. Int. Ed. 54, 11536–11539. doi: 10.1002/anie.201504549
Jiang, J., Wang, L., Zou, Y., Lu, W., Zhao, B., Zhang, B., et al. (2013). Identification of important charged residues for alkali cation exchange or pH regulation of NhaH, a Na+/H+antiporter of Halobacillus dabanensis. Biochim. Biophys. Acta 1828, 997–1003. doi: 10.1016/j.bbamem.2012.11.015
Kolaj-Robin, O., Russell, D., Hayes, K. A., Pembroke, J. T., and Soulimane, T. (2015). Cation diffusion facilitator family: structure and function. FEBS Lett. 589, 1283–1295. doi: 10.1016/j.febslet.2015.04.007
Krulwich, T. A., Sachs, G., and Padan, E. (2011). Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343. doi: 10.1038/nrmicro2549
Lee, S. M., Grass, G., Haney, C. J., Fan, B., Rosen, B. P., Anton, A., et al. (2002). Functional analysis of the Escherichia coli zinc transporter ZitB. FEMS Microbiol. Lett. 215, 273–278. doi: 10.1186/1471-2164-8-107
Lu, M., Chai, J., and Fu, D. (2009). Structural basis for autoregulation of the zinc transporter YiiP. Nat. Struct. Mol. Biol. 16, 1063–1067. doi: 10.1038/nsmb.1662
Lu, M., and Fu, D. (2007). Structure of the zinc transporter YiiP. Science 317, 1746–1748. doi: 10.1126/science.1143748
Martin, J. E., and Giedroc, D. P. (2016). Functional determinants of metal ion transport and selectivity in paralogous cation diffusion facilitator transporters CzcD and MntE in Streptococcus pneumoniae. J. Bacteriol. 198, 1066–1076. doi: 10.1128/JB.00975-15
Meng, L., Hong, S., Liu, H., Huang, H., Sun, H., Xu, T., et al. (2014). Cloning and identification of Group 1 mrp operon encoding a novel monovalent cation/proton antiporter system from the moderate halophile Halomonas zhaodongensis. Extremophiles 18, 963–972. doi: 10.1007/s00792-014-0666-5
Meng, L., Meng, F., Zhang, R., Zhang, Z., Dong, P., Sun, K., et al. (2017). Characterization of a novel two-component Na+(Li+, K+)/H+ antiporter from Halomonas zhaodongensis. Sci. Rep. 7:4221. doi: 10.1038/s41598-017-04236-0
Menguer, P. K., Farthing, E., Peaston, K. A., Ricachenevsky, F. K., Fett, J. P., and Williams, L. E. (2013). Functional analysis of the rice vacuolar zinc transporter OsMTP1. J. Exp. Bot. 64, 2871–2883. doi: 10.1093/jxb/ert136
Montanini, B., Blaudez, D., Jeandroz, S., Sanders, D., and Chalot, M. (2007). Phylogenetic and functional analysis of the cation diffusion facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8:107. doi: 10.1186/1471-2164-8-107
Nakamura, T., Hsu, C., and Rosen, B. P. (1986). Cation/proton antiport systems in Escherichia coli. Solubilization and reconstitution of delta pH-driven sodium/proton and calcium/proton antiporters. J. Biol. Chem. 261, 678–683. doi: 10.1016/0006-291X(78)91403-1
Nishito, Y., Tsuji, N., Fujishiro, H., Takeda, T. A., Yamazaki, T., Teranishi, F., et al. (2016). Direct comparison of manganese detoxification/efflux proteins and molecular characterization of ZnT10 protein as a manganese transporter. J. Biol. Chem. 291, 14773–14787. doi: 10.1074/jbc.M116.728014
Nozaki, K., Inaba, K., Kuroda, T., Tsuda, M., and Tsuchiya, T. (1996). Cloning and sequencing of the gene for Na+/H+Antiporter of Vibrio parahaemolyticus. Biochem. Biophys. Res. Commun. 222, 774–779. doi: 10.1006/bbrc.1996.0820
Padan, E. (2014). Functional and structural dynamics of NhaA, a prototype for Na+and H+antiporters, which are responsible for Na+and H+homeostasis in cells. Biochim. Biophys. Acta Bioenerg. 1837, 1047–1062. doi: 10.1016/j.bbabio.2013.12.007
Padan, E., and Landau, M. (2016). Sodium-proton (Na(+)/H(+)) antiporters: properties and roles in health and disease. Met. Ions Life Sci. 16, 391–458. doi: 10.1007/978-3-319-21756-7-12
Paulsen, I. T., and Saier, M. H. (1997). A novel family of ubiquitous heavy metal ion transport proteins. J. Membr. Biol. 156, 99–103. doi: 10.1007/s002329900192
Quinn, M. J., Resch, C. T., Sun, J., Lind, E. J., Dibrov, P., and Häse, C. C. (2012). NhaP1 is a K +(Na +)/H + antiporter required for growth and internal pH homeostasis of Vibrio cholerae at low extracellular pH. Microbiology 158, 1094–1105. doi: 10.1099/mic.0.056119-0
Reese, M. G. (2001). Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26, 51–56. doi: 10.1016/S0097-8485(01)00099-7
Rensing, C., Mitra, B., and Rosen, B. P. (1997). The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 94, 14326–14331. doi: 10.1073/pnas.94.26.14326
Rosen, B. P., and Tsuchiya, T. (1979). Preparation of everted membrane vesicles from Escherichia coli for the measurement of calcium transport. Methods Enzymol. 56, 233–241. doi: 10.1016/0076-6879(79)56026-1
Saier, M. H., Reddy, V. S., Tsu, B. V., Ahmed, M. S., Li, C., and Moreno-Hagelsieb, G. (2016). The transporter classification database (TCDB): recent advances. Nucleic Acids Res. 44, D372–D379. doi: 10.1093/nar/gkv1103
Saitou, N., and Nei, M. (1986). The number of nucleotides required to determine the branching order of three species, with special reference to the human-chimpanzee-gorilla divergence. J. Mol. Evol. 24, 189–204. doi: 10.1007/BF02099966
Shao, L., Abdel-Motaal, H., Chen, J., Chen, H., Xu, T., Meng, L., et al. (2018). Characterization of a functionally unknown arginine-aspartate-aspartate family protein from Halobacillus andaensis and functional analysis of its conserved arginine/aspartate residues. Front. Microbiol. 9:807. doi: 10.3389/fmicb.2018.00807
Shi, Y. (2013). Common folds and transport mechanisms of secondary active transporters. Annu. Rev. Biophys. 42, 51–72. doi: 10.1146/annurev-biophys-083012-130429
Ventosa, A., Márquez, M. C., Garabito, M. J., and Arahal, D. R. (1998). Moderately halophilic gram-positive bacterial diversity in hypersaline environments. Extremophiles 2, 297–304. doi: 10.1007/s007920050072
Wang, K., Zhang, L., Li, J., Pan, Y., Meng, L., Xu, T., et al. (2015). Planococcus dechangensis sp. nov., a moderately halophilic bacterium isolated from saline and alkaline soils in Dechang Township, Zhaodong City, China. Int. J. Gen. Mol. Microbiol. 107, 1075–1083. doi: 10.1007/s10482-015-0399-1
Keywords: cation selectivity, cation diffusion facilitator family, Na+(Li+, K+)/H+ antiporter, facilitated Zn2+ diffusion, secondary transporter, moderate halophile
Citation: Xu T, Chen H, Li J, Hong S, Shao L, Zheng X, Zou Q, Wang Y, Guo S and Jiang J (2019) Implications for Cation Selectivity and Evolution by a Novel Cation Diffusion Facilitator Family Member From the Moderate Halophile Planococcus dechangensis. Front. Microbiol. 10:607. doi: 10.3389/fmicb.2019.00607
Received: 09 January 2019; Accepted: 11 March 2019;
Published: 22 March 2019.
Edited by:
Masahiro Ito, Toyo University, JapanReviewed by:
Jun Liu, Tianjin Institute of Industrial Biotechnology (CAS), ChinaHeven Sze, University of Maryland, College Park, United States
Copyright © 2019 Xu, Chen, Li, Hong, Shao, Zheng, Zou, Wang, Guo and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juquan Jiang, ampxZGFpbnR5QDE2My5jb20=
 Tong Xu
Tong Xu Huiwen Chen
Huiwen Chen Li Shao
Li Shao Juquan Jiang
Juquan Jiang