- 1Key Laboratory of Coastal Biology and Bioresource Utilization, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
- 2College of Resources and Environment, University of Chinese Academy of Sciences, Beijing, China
- 3Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 4Shandong Huibang Bohai Agriculture Development Limited Company, Dongying, China
Cbp1, a chemoreceptor containing a PilZ domain was identified in Azorhizobium caulinodans ORS571, a nitrogen-fixing free-living soil bacterium that induces nodule formation in both the roots and stems of the host legume Sesbania rostrata. Chemoreceptors are responsible for sensing signals in the chemotaxis pathway, which guides motile bacteria to beneficial niches and plays an important role in the establishment of rhizobia-legume symbiosis. PilZ domain proteins are known to bind the second messenger c-di-GMP, an important regulator of motility, biofilm formation and virulence. Cbp1 was shown to bind c-di-GMP through the conserved RxxxR motif of its PilZ domain. A mutant strain carrying a cbp1 deletion was impaired in chemotaxis, a feature that could be restored by genetic complementation. Compared with the wild type strain, the Δcbp1 mutant displayed enhanced aggregation and biofilm formation. The Δcbp1 mutant induced functional nodules when inoculated individually. However, the Δcbp1 mutant was less competitive than the wild type in competitive root colonization and nodulation. These data are in agreement with the hypothesis that the c-di-GMP binding chemoreceptor Cbp1 in A. caulinodans is involved in chemotaxis and nodulation.
Introduction
C-di-GMP (bis-(3′–5′)-cyclic dimeric guanosine monophosphate) is a second messenger commonly found in the bacterial kingdom that regulates multiple cellular functions including motility, extracellular polysaccharide production and biofilm formation (Hengge, 2009). Generally, high intracellular c-di-GMP concentrations down regulate motility and promote surface attachment, which correlates with the transition from a motile to sessile lifestyle (Povolotsky and Hengge, 2012; Purcell et al., 2012; Roy et al., 2012). C-di-GMP turnover is controlled by diguanylate cyclases (DGCs) and c-di-GMP specific phosphodiesterases (PDEs) (Chan et al., 2004; Rao et al., 2008). C-di-GMP is synthesized from two molecules of GTP by DGCs containing a GGDEF domain with a conserved GG(D/E)EF motif and hydrolyzed into pGpG by PDEs containing an EAL domain or into two molecules of GMP by PDEs containing a HD-GYP domain (Christen et al., 2005; Ryjenkov et al., 2005; Ryan et al., 2006).
C-di-GMP exerts its biological effects by binding to various effectors including riboswitches and proteins (Lee et al., 2007; Newell et al., 2009; Bordeleau et al., 2015). Among these, proteins with PilZ domains have been extensively studied (Ryan et al., 2012). The PilZ domain is characterized by conserved RxxxR and D/NxSxxG motifs responsible for c-di-GMP binding (Benach et al., 2007). PilZ domain proteins display a broad range of c-di-GMP binding affinities and are involved in the control of different bacterial behaviors. In Escherichia coli, the PilZ domain protein YcgR inhibits motility at high c-di-GMP concentrations by interacting with the flagellar proteins FliG and FliM (Paul et al., 2010). In Pseudomonas aeruginosa, the PilZ domain protein HapZ regulates a two-component signaling pathway by interacting with the histidine kinase SagS (Xu et al., 2016a), while another PilZ domain protein Alg44 is required for alginate biosynthesis (Merighi et al., 2007). In Xanthomonas oryzae pv. oryzae, PilZ domain proteins were shown to regulate virulence (Yang et al., 2015).
Chemotaxis enables motile bacteria to detect environmental changes, such as chemical gradients, and respond by moving to beneficial niches that support optimal growth. Chemoreceptors (also known as methyl-accepting chemotaxis proteins, MCPs) are signaling proteins that transduce chemotactic signals via interaction with both CheA (histidine kinase) and CheW (coupling protein) of the chemotaxis pathway (Wadhams and Armitage, 2004). As a histidine kinase, CheA can be autophosphorylation with ATP, and CheA-P phosphorylates its response regulator CheY. CheY-P then binds to the flagellar rotary motor to bring about a reversal in rotational direction (Tindall et al., 2012). Chemoreceptor binding of a chemotactic signal molecule regulates the kinase activity of CheA, thus controls the probability of directional changes (Lai et al., 2006). The signal ligand binding affinity of chemoreceptors depends on their methylation state (Li and Weis, 2000). There are chemotaxis adaptation proteins CheB (methylesterase) and CheR (methyltransferase) present in the chemotaxis pathway. CheB is also phosphorylated by CheA-P, and CheB-P competes with CheR to regulate the methylation state of chemoreceptors, thus resetting their signal sensitivity (Wuichet et al., 2007). Azorhizobium caulinodans ORS571 is a versatile nitrogen fixer that can fix nitrogen both in the symbiotic and free-living states under microaerobic conditions. It is a symbiont of the legume Sesbania rostrata and can form nitrogen fixing nodules both on roots and stems (Dreyfus et al., 1988). Chemotaxis of A. caulinodans ORS571 is controlled by a single chemotaxis system that contains five chemotaxis proteins: CheA, CheW, CheY1, CheB, and CheR (Liu et al., 2018). Chemotaxis toward chemoattractants in root exudates promotes attachment between rhizobia and legume roots and subsequent infection, thus benefiting the establishment of rhizobia-legume symbiosis (Greer-Phillips et al., 2004; Scharf et al., 2016).
A considerable number of c-di-GMP-related genes were identified in rhizobia (Gao et al., 2014). The c-di-GMP-related genes have been investigated in Sinorhizobium meliloti and a single PilZ domain protein, McrA, was identified as a c-di-GMP binding effector able to repress motility at elevated c-di-GMP concentrations (Schäper et al., 2016). The PilZ domain containing chemoreceptor Tlp1 from Azospirillum brasiencse has been shown to be involved in the chemotactic signaling pathway (Russell et al., 2013). The integration of c-di-GMP into the chemotactic signaling pathway prompted us to investigate the potential interplay between c-di-GMP and chemotaxis in A. caulinodans ORS571. The whole genome of ORS571 has been sequenced (Lee et al., 2008), and there are 43 chemoreceptors as shown in the Pfam database (Jiang et al., 2016). In the present study, we report the characterization of a PilZ domain containing chemoreceptor that we named Cbp1 (c-di-GMP binding protein 1) and show the involvement of Cbp1 in chemotaxis and symbiosis with legume host S. rostrata.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
Strains and plasmids used in this study are listed in Table 1. A. caulinodans ORS571 and mutant strain derivatives were grown at 37°C in TY medium and L3 minimal medium with the corresponding antibiotics (Jiang et al., 2016). E. coli strains used for cloning and expression were routinely grown at 37°C in LB medium. Solid media plates contained 1.5% agar. Antibiotics were used at the following concentrations: ampicillin 100 μg/mL, nalidixic acid 25 μg/mL, kanamycin 25 μg/mL, gentamycin 50 μg/mL, and tetracycline 10 μg/mL.
Protein Expression and Purification
Plasmid pBAD/M with a C-terminal 6×His tag was used to overexpress Cbp1 in E. coli Top10 (Sun et al., 2015). A DNA fragment encoding Cbp1 was amplified from A. caulinodans ORS571 genomic DNA with the primer pair Cbp1-NdeI-F/HindIII-R and cloned into the NdeI/HindIII site of pBAD/M to create the recombinant expression plasmid. The resulting plasmid was then transformed into E. coli Top10. The Cbp1R320A mutant was generated using the QuikChange®Site-Directed Mutagenesis Kit (Stratagene) with the primer pair Cbp1R320A-F/R. Protein expression was induced with 0.1% L-arabinose and incubation at 16°C for 24 h. Protein purification was performed as described previously (Sun et al., 2015). Proteins were analyzed by SDS-PAGE and their concentration was measured using the standard BCA protein assay.
Protein Thermal Shift Assay
The protein thermal shift assay was performed as described previously with some modifications (Huynh and Partch, 2015). The StepOne Plus Quantitative Real-time PCR system (Thermo Fisher Scientific) was used to determine the thermal shift of purified proteins in the presence of SYPRO orange dye, which binds hydrophobic regions of proteins. The reaction buffer was 25 mM Tris–HCl (pH7.9) with 10 mM MgCl2 and the final assay concentration of SYPRO orange dye was 1× (stock solution was 5000×). For measurement of protein thermal shift without a ligand, 5 μM of purified Cbp1 or Cbp1R320A protein was added. For measurement of protein thermal shift with a ligand, 5 μM of the purified protein was added with 5 μM c-di-GMP. 50 μL of reaction buffer with 1 × SYPRO orange was used as a negative control (No Protein Control, NPC). Before being measured in the qRT-PCR instrument, all reaction mixtures were incubated for 1 h at 37°C. The experimental parameters were selected as described by Huynh and Partch (2015). The experimental data was analyzed using Protein Thermal Shift Software 1.3 (Thermo Fisher Scientific).
Isothermal Titration Calorimetry (ITC)
Isothermal titration calorimetry measurements were performed as described previously with some modifications (Huang et al., 2016). A NANO ITC 2G (TA Instruments) was used to detect c-di-GMP binding to Cbp1 and Cbp1R320A. Both the Cbp1 and Cbp1R320A proteins and the ligand c-di-GMP were dissolved in buffer (25 mM Tris–HCl, 10 mM MgCl2, pH7.9). 20 μM of the purified protein was titrated with aliquots of 300 μM c-di-GMP. The experimental data was analyzed using NanoAnalyze software 3.5.0 (TA Instruments).
Construction of Mutant and Complemented Strains
The Δcbp1 mutant was constructed using the allelic exchange vectors pCM351 and pCM157, a pair of vectors carrying the cre/lox system (Marx and Lidstrom, 2002). 702-bp upstream and 690-bp downstream fragments of the cbp1 gene were amplified from A. caulinodans ORS571 genomic DNA with the primer pairs Cbp1Up-XbaI-F/NdeI-R and Cbp1Down-AgeI-F/SacI-R. The fragments were then digested and inserted into the plasmid vector pCM351. The resulting recombinant plasmid (pCM351:Up:Down) was introduced into A. caulinodans ORS571 by tri-parental conjugation with the helper plasmid pRK2013 (Figurski and Helinski, 1979). Allelic exchange between the recombinant plasmid pCM351:Up:Down and the ORS571 chromosome led to the replacement of cbp1 with a gentamicin gene cassette and the cassette was further deleted following the introduction of plasmid pCM157. To complement the Δcbp1 mutant, full-length cbp1 and 300-bp upstream encompassing the predicted promoter region was amplified by PCR using the primer pair Cbp1P-KpnI-F/SacI-R. The amplified fragment was then cloned into the KpnI/SacI site of the broad-host-range vector pBBR1MCS-2 (Kovach et al., 1995) to create the plasmid pBBR-cbp1. The plasmid pBBR-cbp1R320A was then constructed using the site-directed mutagenesis protocol mentioned above with primer pair Cbp1R320A-F/R. The complementation plasmids were introduced into the Δcbp1 mutant by tri-parental conjugation with the helper plasmid pRK2013. The empty plasmid pBBR1MCS-2 was also introduced into wild type ORS571 and Δcbp1 mutant as control. All primers used in mutant construction are listed in Table 2.
Growth Curve Assay
Growth of the strains were measured as previously described (Liu et al., 2017). ORS571 and the mutants were grown overnight in TY medium at 37°C. The overnight-grown cultures were collected by centrifugation and washed three times with L3+N liquid medium. The cells were then inoculated into 50 mL TY medium or L3+N medium (with succinate as a carbon source) at an initial concentration of OD600 0.02 in sterile 250 mL conical flasks. Cells were grown at 150 rpm and 37°C and cell density was monitored every 2 h (0–12 h), 3 h (12–24 h), and 6 h (24–48 h).
Competitive Capillary Chemotaxis Assay
The competitive capillary chemotaxis assay was performed as previously described with modifications (Liu et al., 2018). Overnight grown cells of ORS571 and mutants were collected by centrifugation, washed three times with PBS buffer, and the suspensions were then adjusted with a final concentration of OD600 0.01. The wild type and each mutant were then mixed at a 1:1 ratio. The 1:1 ratio was re-confirmed by counting CFUs of each strain through serial dilution. 200 μL aliquots of the mixed cell suspensions were placed into the wells of a 96-well plate. Capillaries were heat-sealed at one end using a flame and the open end was then inserted into the PBS buffer and 10 mM succinate solution for 5 min. Solution-filled capillaries were then removed to wells containing bacterial mixtures. The 96-well plates were incubated at 37°C for 1 h. The sealed ends of then capillaries were broken and the contents were then emptied into 1 mL PBS buffer using a rubber adaptor. The ratio of wild type to mutant cells was calculated by counting CFUs of each strain entering the capillaries.
Aggregation Assay
Overnight grown cells of ORS571 and the Δcbp1 mutant were collected by centrifugation and washed three times with L3+N liquid medium. Cells were then inoculated into 15 mL fresh L3+N medium (with succinate as a carbon source) to an initial concentration of OD600 0.02 in sterile 50 mL conical centrifuge tubes. The tubes were then incubated in an orbital shaker and shaken at 220 rpm for 24 and 48 h at 37°C. The tubes were taken out and left standing for a further 20 min. When the aggregated cells settled to the bottom of the tubes, the OD600 of the upper cell suspension was measured. The aggregated cells were then vortexed and the total OD600 of the cell cultures was measured. The aggregation ratio of each strain was then calculated using the formula: aggregation ratio % = 100∗(ODtotal-ODsuspension)/ODtotal.
Quantification of Biofilm Formation
Overnight grown cells of wild type and the Δcbp1 mutant were collected by centrifugation and washed with L3+N liquid medium. 1.5 mL of resuspended cells with a final concentration of OD600 0.1 was added to a sterile glass tube. The strains were then incubated at 37°C for 3 days. For crystal violet staining, the cells were removed and the glass tubes were washed carefully with sterile water. 2 mL of 1% crystal violet was added to each tube. After 20 min of staining, crystal violet was removed and 1 mL of 30% acetic acid was added to dissolve the crystal violet-stained biofilms. The OD595 of solution from each tube was then measured for biofilm quantification.
Plant Growth, Root Colonization and Nodule Occupancy Competition Assays
Sesbania rostrata seeds were treated with concentrated sulfuric acid for 30 min and washed thoroughly with sterile water and then soaked in sterile water in the dark at 37°C for 48 h, as previously described (Jiang et al., 2016). For the root colonization assay, overnight grown cells of wild type and the Δcbp1 mutant were harvested and resuspended in sterile L3-N liquid medium. Normalized cell suspensions of wild type and the Δcbp1 mutant were mixed in a 1:1 ratio and added into sterile vermiculite. The 1:1 ratio was re-confirmed by counting CFUs of each strain. The germinated seedlings were then transferred into vermiculite for 4 and 24 h. After that, the seedlings were removed, washed and then mashed to recover bacteria from the root surface. Serial dilutions were then plated on TY plates. After growth at 37°C, at least 200 single colonies were analyzed by PCR with the primer pair Cbp1Up-XbaI-F /Cbp1Down-SacI-R to calculate the colonization occupation ratio. For nodulation analysis, germinated seedlings were planted in vermiculite in Leonard jars. Nodulation on roots was performed by adding cell cultures into vermiculite, and nodulation on stems was performed by inoculation of stem-root primordia with sterile cotton wool moistened with a bacterial suspension as previously described (Liu et al., 2018). For individual nodulation, cells of a single strain were used. For competitive nodulation, cells of wild type and the Δcbp1 mutant were mixed in a 1:1 ratio (re-confirmed by counting CFUs of each strain). All plants were grown at 26°C with a daylight illumination period of 12 h in a greenhouse. The root and stem nodules were harvested 4 weeks after inoculation. Bacteria were then re-isolated from crushed nodules and colonies were tested by PCR as above to determine the nodulation occupation ratio.
Acetylene Reduction Activity (ARA) Assays
Acetylene reduction activity of the nodules was determined using gas chromatography (Agilent Technologies, 7890A). The ARA was expressed as μmol of C2H4 production/h/g of fresh nodule, as described by Liu et al. (2017). Each measurement was repeated at least three times.
Results
The Chemoreceptor Cbp1 Binds c-di-GMP Through the Conserved RxxxR Motif
Of the 43 chemoreceptors in A. caulinodans ORS571 (Jiang et al., 2016), the only chemoreceptor containing a PilZ domain is encoded by the gene AZC_3349, here named Cbp1. Cbp1 is an internal soluble protein without a transmembrane region consisting of an N-terminal MA domain (methyl-accepting chemotaxis-like domain) and a C-terminal PilZ domain (Supplementary Figure S1A). All chemoreceptors were identified by the presence of a MA domain, a highly conserved cytoplasmic signaling domain that interacts with the CheA histidine kinase and CheW coupling protein (Wadhams and Armitage, 2004).
Multiple alignment of Cbp1 PilZ domain with other proteins showed that the characteristic c-di-GMP binding motifs RxxxR and D/NxSxxG are fully conserved in the Cpb1 PilZ domain (Supplementary Figure S1C). Conservation of the binding motif suggested that Cbp1 may bind c-di-GMP. To determine whether Cbp1 can bind c-di-GMP, Cbp1 was heterologously expressed in E. coli Top10 to obtain purified protein (Supplementary Figure S1B). Protein thermal shift assays (also known as different scanning fluorimetry or ThermoFluor) have been widely used to identify stabilizing solution conditions and small-molecule ligands for purified proteins (Huynh and Partch, 2015). The protein thermal shift assay was applied here to analyze the c-di-GMP binding ability of Cbp1. As shown in Figure 1, the denaturation profile of Cbp1 (green line) indicated that Cbp1 had exposed hydrophobic regions in the native state (SYPRO orange interacts with hydrophobic regions in proteins). The red line (Cbp1 with c-di-GMP) indicates a denaturation profile of a well-folded protein: low fluorescence emission at room temperature and then emission increases with increasing temperature. The difference between these lines suggested that Cbp1 could specifically bind c-di-GMP and c-di-GMP binding led to a significant conformational change in Cbp1. In order to better characterize the binding of c-di-GMP to Cbp1, isothermal titration calorimetry (ITC) was performed. Purified Cbp1 was titrated with c-di-GMP and the result of ITC showed that Cbp1 binds c-di-GMP with a KD of 14.94 ± 1.6 μM (Figure 2A). A previous study on A. brasilense Tlp1 has shown that mutation of Arg563, the first R in RxxxR motif of the PilZ domain, led to complete loss of c-di-GMP binding ability of Tlp1 (Russell et al., 2013). In Cbp1, the corresponding Arg residue is at position 320. Thus, the Arg320 residue of Cbp1 PilZ domain was replaced by Ala. The Cbp1R320A variant was then analyzed for c-di-GMP binding. As expected, there was no obvious change in the denaturation profile of Cbp1R320A with c-di-GMP, which suggested that Cbp1R320A failed to bind c-di-GMP (Figure 1). The ITC analysis of Cbp1R320A further confirmed that it cannot bind c-di-GMP (Figure 2B). Together, the results showed that Cbp1 is a c-di-GMP binding chemoreceptor and the PilZ domain is responsible for c-di-GMP binding.
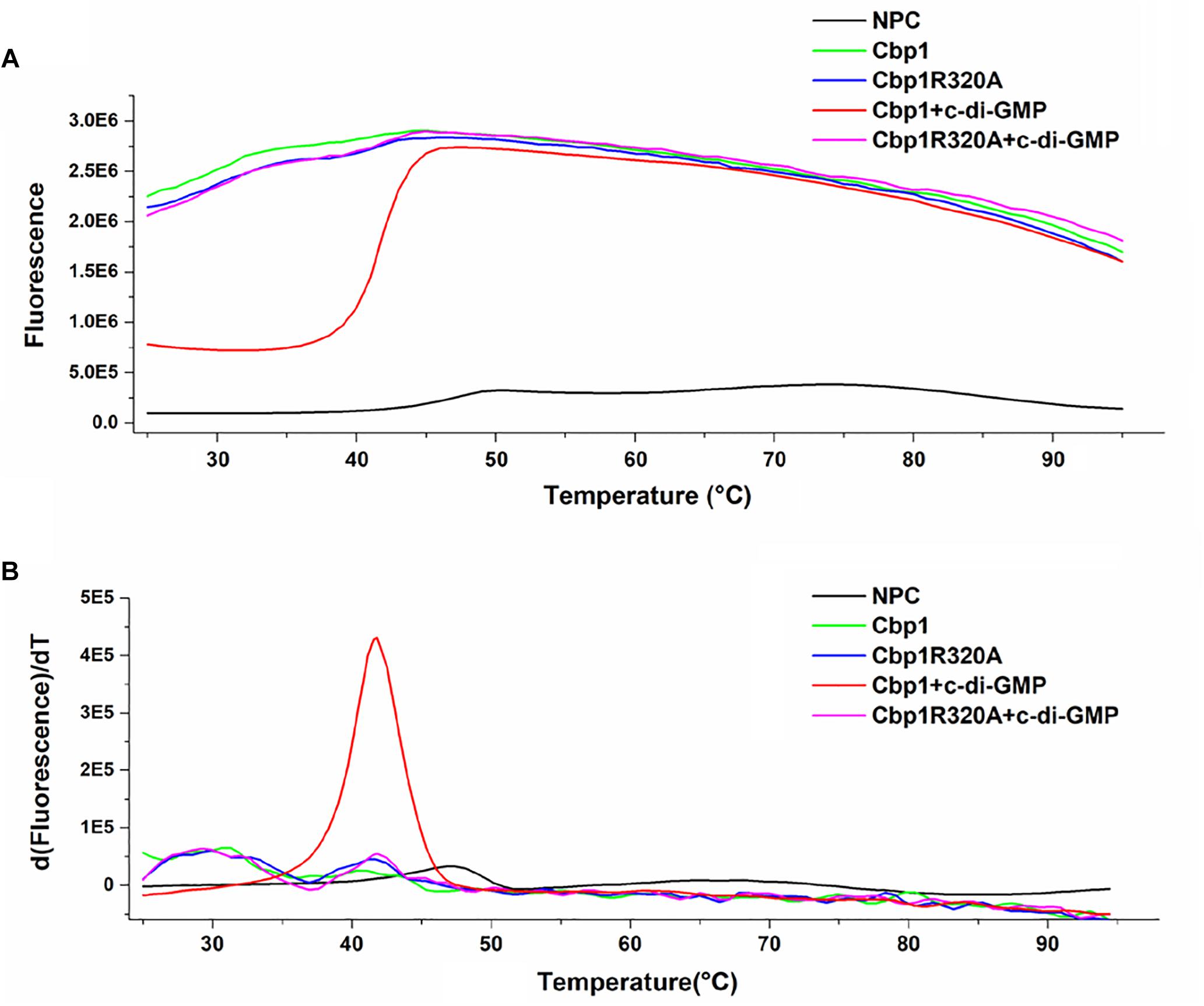
Figure 1. Protein thermal shift assay of Cbp1 and Cbp1R320A. (A) The thermal denaturation profile of proteins. (B) The profile of the derivative of the fluorescence emission as a function of temperature (dF/dT). NPC (no protein control, black line), Cbp1 (green line), Cbp1R320A (blue line), Cbp1 with c-di-GMP (red line), Cbp1R320A with c-di-GMP (magenta line).
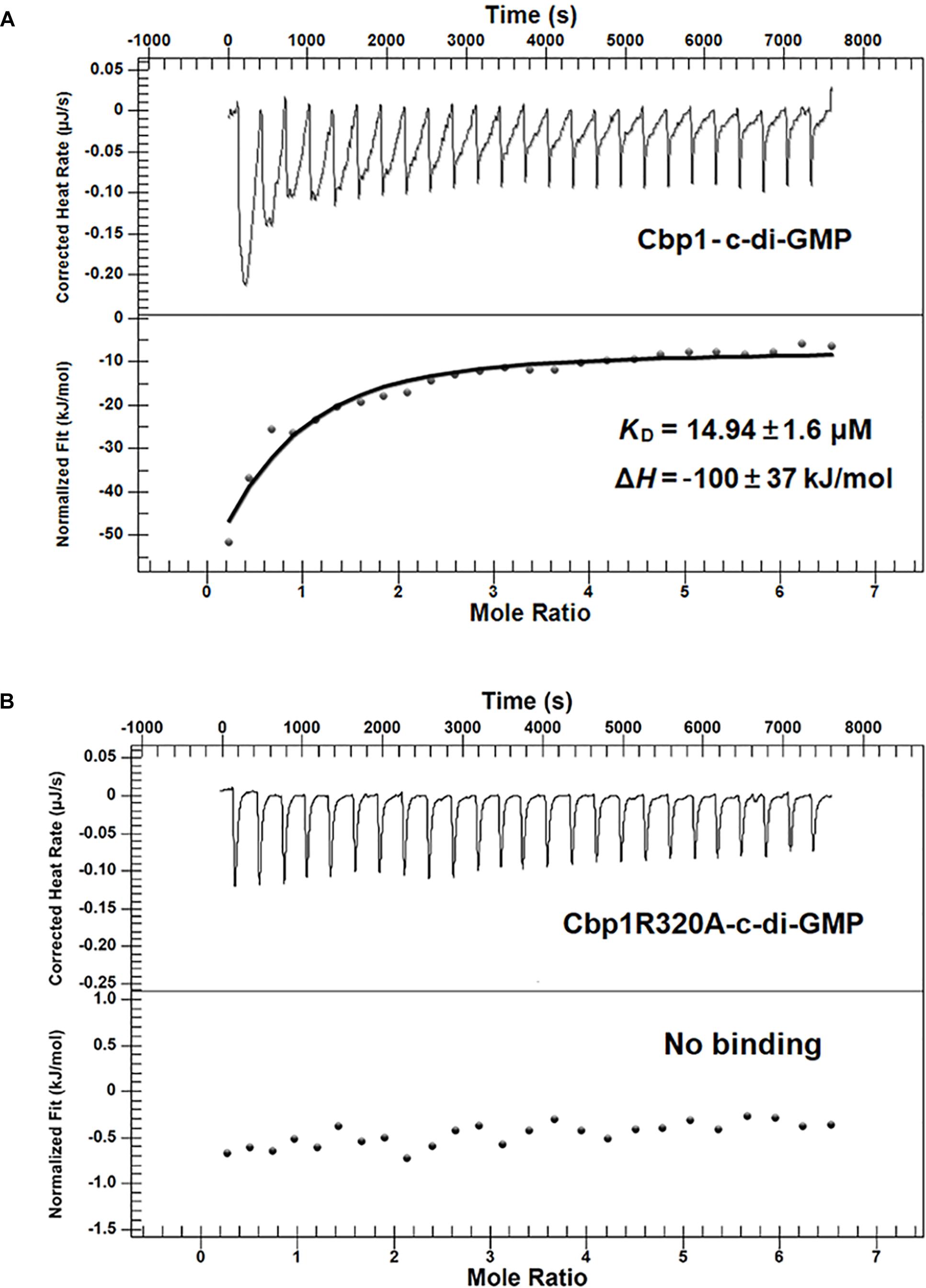
Figure 2. Measurement of the c-di-GMP binding affinity of Cbp1 and Cbp1R320A by ITC. For titration, 20 μM of the purified proteins (Cbp1 and Cbp1R320A) and 300 μM c-di-GMP were used. (A) Cbp1 binds c-di-GMP with a KD of 14.94 ± 1.6 μM, and the ΔH is –100 ± 37 KJ/mol. (B) Cbp1R320A cannot bind c-di-GMP.
The Δcbp1 Mutant Is Impaired in Chemotaxis and c-di-GMP Binding Ability of Cbp1 Is Essential for Normal Chemotaxis
To determine the role of Cbp1 in chemotaxis, a Cbp1 deletion mutant (Δcbp1) was constructed. The growth curves of the strains were first determined and the results suggested that the growth rate of the Δcbp1 mutant had no obvious difference compared with that of the wild type in both rich (TY) and minimal media (L3+N) with succinate as a carbon source (Supplementary Figure S2). The competitive capillary chemotaxis assay was performed to characterize the chemotactic behavior of wild type and mutants. For the competitive capillary assay, wild type and the Δcbp1 mutant were mixed in a 1:1 ratio and capillaries containing PBS buffer or the chemoattractant succinate were immersed in bacterial suspensions. As shown in Figure 3A, the number of Δcbp1 mutant cells entering the capillary containing PBS buffer was about the same as the wild type (50% vs. 50%), which suggested that the swimming ability of the Δcbp1 mutant was not affected. However, cells of the Δcbp1 mutant entering the capillary containing the chemoattractant succinate were greatly reduced compared to the wild type (23.5% vs. 76.5%), confirming that the chemotaxis ability of the Δcbp1 mutant was impaired.
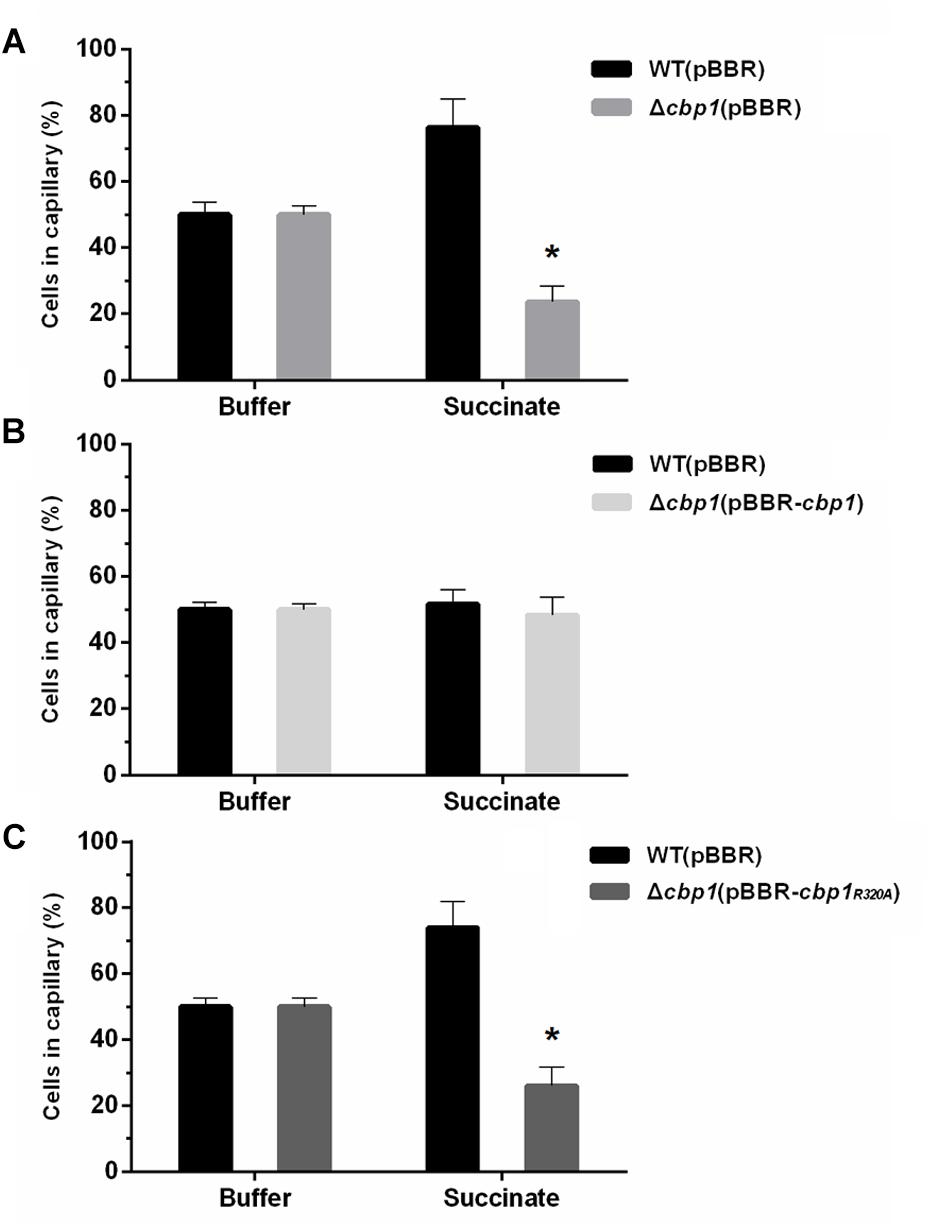
Figure 3. Chemotactic behavior of wild-type ORS571 and mutants. (A) Competitive chemotactic responses of wild type and Δcbp1 mutant using the quantitative capillary assay. (B) Competitive chemotactic responses of wild type and Δcbp1(pBBR-cbp1) mutant using the quantitative capillary assay. (C) Competitive chemotactic responses of wild type and Δcbp1(pBBR-cbp1R320A) mutant using the quantitative capillary assay. Wild type and each mutant were mixed in a 1:1 ratio. The capillary contained either PBS buffer as control or 10 mM succinate solution as a chemoattractant. Error bars indicate standard deviations for three independent biological replicates and asterisks represent significant differences (∗P < 0.01).
Since Cbp1 binds c-di-GMP, the role of c-di-GMP in chemotaxis may be of great importance. To investigate any potential interaction between c-di-GMP and chemotaxis, genetic complementation experiments were performed using the Δcbp1 mutant and broad-host-range plasmids expressing wild-type cbp1 or cbp1R320A under the control of their native promoters. As shown in Figure 3B, cells of the complemented strain Δcbp1 (pBBR-cbp1) entering the capillary containing chemoattractant were about the same as the wild type, suggesting that the chemotaxis defect of the Δcbp1 mutant was completely restored by the expression of wild-type cbp1. However, the plasmid pBBR-cbp1R320A failed to restore the chemotaxis defect of the Δcbp1 mutant (Figure 3C). The result suggested that loss of c-di-GMP binding ability significantly impaired chemotaxis and the c-di-GMP binding ability of Cbp1 was essential for normal chemotaxis.
The Δcbp1 Mutant Has Enhanced Aggregation Ratio
As previously reported, A. caulinodans ORS571 can aggregate and form flocs in minimal medium when incubated with high rotational speed (Nakajima et al., 2012). Chemotaxis has been implicated in the regulation of cell aggregation (Alexandre, 2015). To determine whether Cbp1 played a role in aggregation, wild type and Δcbp1 mutant cells were analyzed for their ability to aggregate. As shown in Figure 4, the aggregation ratios of the wild type and Δcbp1 mutant were similar after 24 h of incubation. Aggregation and floc formation enhanced over time for both strains. However, after 48 h of incubation, the Δcbp1 mutant showed elevated aggregation ratio compared with the wild type (31% vs. 22%) and the flocs formed by the Δcbp1 mutant were also bigger than those of the wild type.
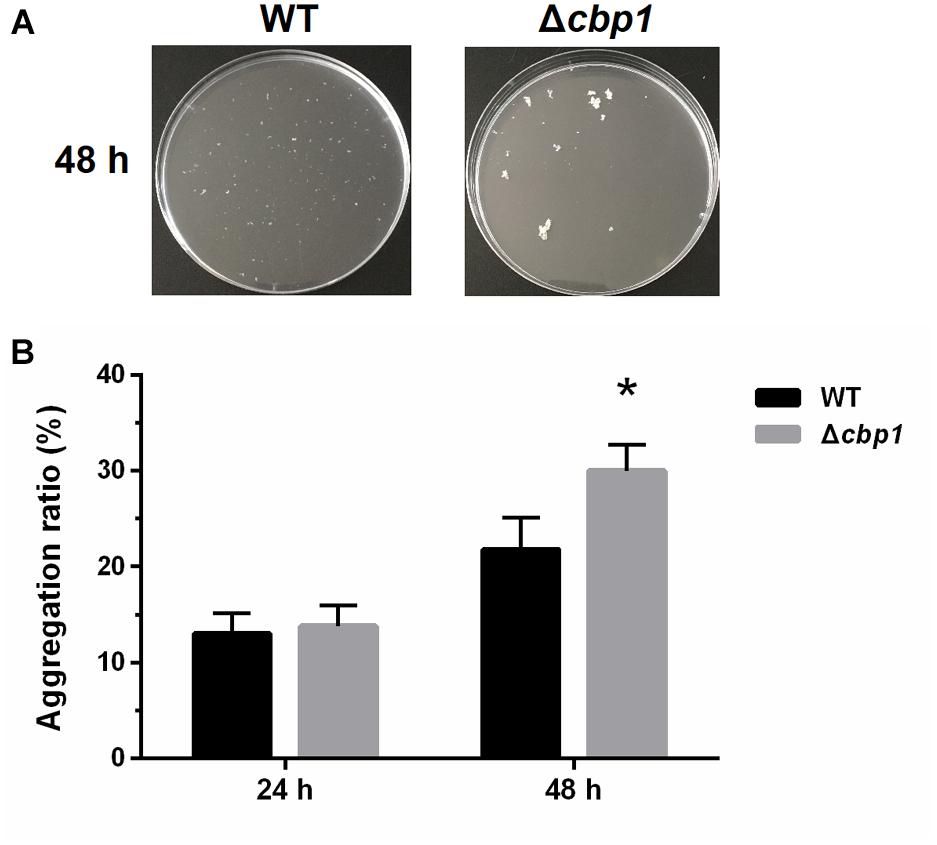
Figure 4. Aggregation of wild-type ORS571 and the Δcbp1 mutant. (A) The aggregation morphologies of wild type and Δcbp1 mutant in L3+N liquid medium after 48 h of incubation. (B) The aggregation ratios of wild type and Δcbp1 mutant. Error bars indicate standard deviations for three independent biological replicates and asterisk represent significant difference (∗P < 0.01).
The Δcbp1 Mutant Has Enhanced Biofilm Formation
Besides cell aggregation, chemotaxis also has a pronounced effect on biofilm formation under static conditions (Liu et al., 2018). Thus, biofilm formation of the Δcbp1 mutant on abiotic surfaces was measured using crystal violet staining. After 3 days of incubation, the biofilm formed at the air-liquid interface by the Δcbp1 mutant appeared to be thicker than that of the wild type (Figure 5A), which was confirmed by quantitative determination (Figure 5B). This indicates that the chemoreceptor Cbp1 may modulate biofilm formation.
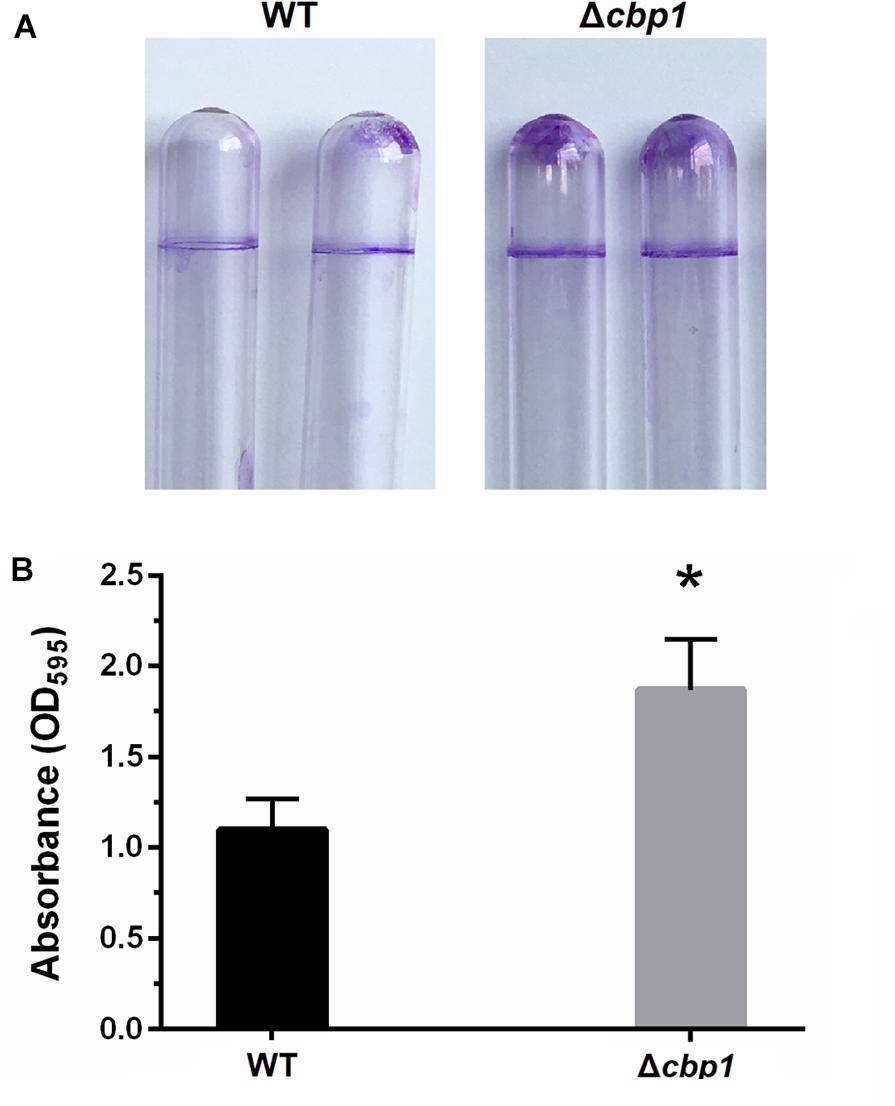
Figure 5. Biofilm formation of wild-type ORS571 and the Δcbp1 mutant. (A) The biofilm morphologies of wild type and Δcbp1 mutant following crystal violet staining. (B) Quantification of crystal violet-stained biofilms. Error bars indicate standard deviations for three independent biological replicates and asterisk represent significant difference (∗P < 0.01).
The Δcbp1 Mutant Is Impaired in Competitive Root Colonization and Nodulation
Establishment of the rhizobia-legume symbiosis depends on colonization of the root system, which itself involves bacterial chemotaxis toward roots (Greer-Phillips et al., 2004). Since the Δcbp1 mutant was impaired in chemotaxis, we expected that it would also be impaired in root colonization. Colonization abilities of both strains on the root surface were assessed. Seedlings were placed in vermiculite containing a mixture of both strains (1:1 ratio) for 4 and 24 h. The colonized strains were then re-isolated and quantified. As shown in Figure 6A, the wild type performed better than the Δcbp1 mutant (69% of wild type vs. 31% of Δcbp1 mutant) in root colonization after 4 h of incubation. The result suggested that the Δcbp1 mutant was less competitive than the wild type in root surface colonization and the defect in chemotaxis affected the initial colonization of rhizobia on legume roots. After 24 h of incubation, the occupation ratio of the wild type and the Δcbp1 mutant was 60%: 40%. The slightly elevated occupation ratio of the Δcbp1 mutant may suggest that the lower colonization efficiency caused by the chemotaxis defect could be partially compensated with a longer incubation time.
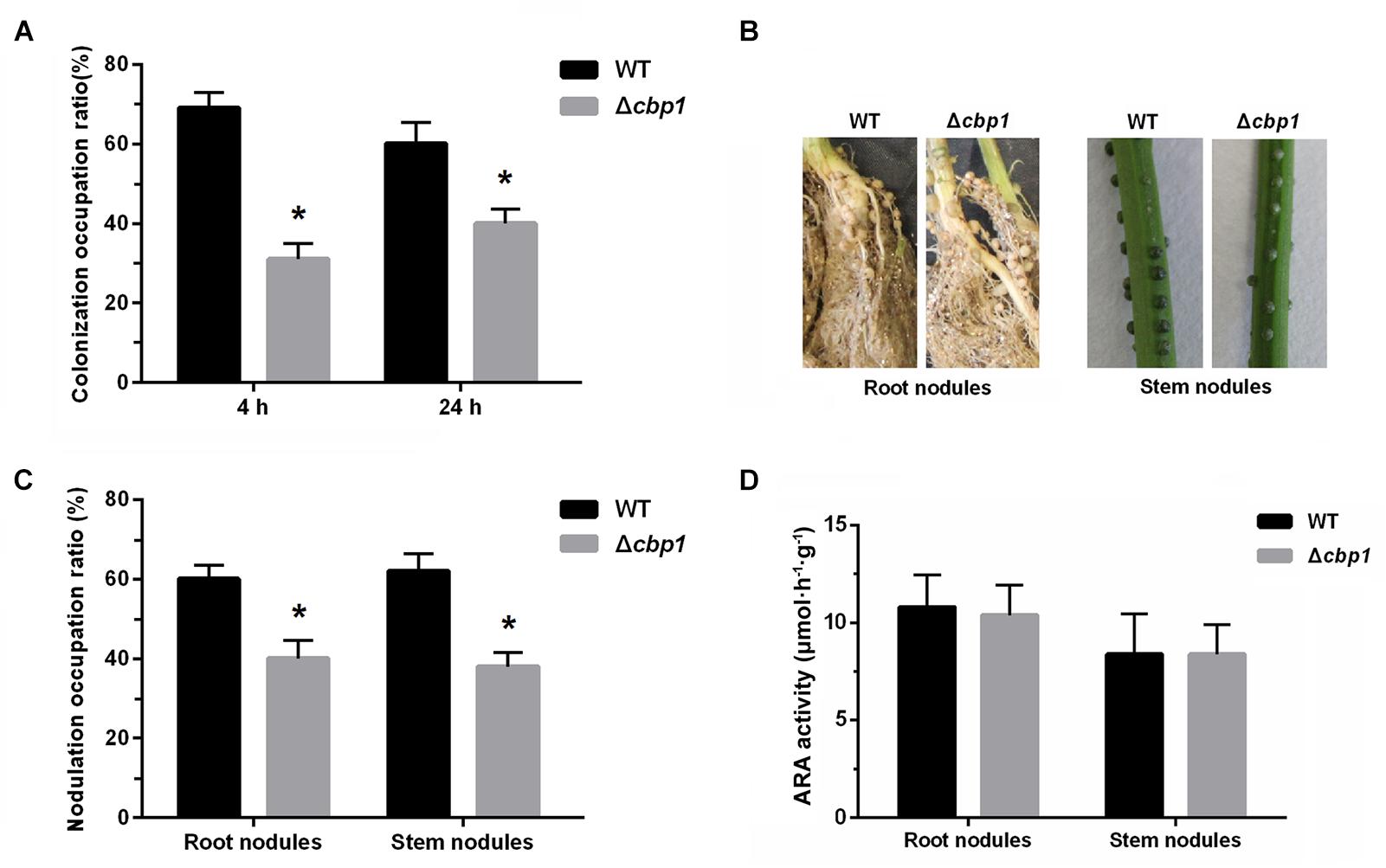
Figure 6. Root colonization, nodulation, and nitrogen fixation of wild-type ORS571 and the Δcbp1 mutant. (A) The competitive root colonization ratio of wild type and Δcbp1 mutant. (B) The morphologies of root nodules and stem nodules induced by wild type and Δcbp1 mutant. (C) The competitive root and stem nodulation ratio of wild type and Δcbp1 mutant. (D) The nitrogen fixation efficiencies of wild type and Δcbp1 mutant. Error bars indicate standard deviations for three independent biological replicates and asterisks represent significant differences (∗P < 0.01).
Nodulation of wild type and the Δcbp1 mutant were then further tested. Figure 6B shows that, similar to the wild type, the Δcbp1 mutant successfully induced nodules on both roots and stems. However, when competitive experiments were performed, using wild type and the Δcbp1 mutant at a 1: 1 ratio, the Δcbp1 mutant was impaired in competitive nodulation on both roots and stems. The number of root nodules formed by the wild type was about 1.5-fold higher than that of the Δcbp1 mutant and the ratio for stem nodules was about the same (Figure 6C). To test whether the Δcbp1 mutant induced functional nodules, the ARA of nodules was also determined. The results showed that the ARA of nodules formed by the Δcbp1 mutant was similar as that of wild type (Figure 6D). Together, these data suggested that Cbp1 was involved in competitive colonization and nodulation, but the nitrogen fixation of ORS571 was not affected by a lack of Cbp1.
Discussion
In this study, the chemoreceptor Cbp1 was shown to bind c-di-GMP and a mutation in the conserved RxxxR motif caused complete loss of c-di-GMP binding. In other systems, the role of the five key residues of the RxxxR and D/NxSxxG motifs of the PilZ domain in c-di-GMP binding has been extensively analyzed. For YcgR from E. coli, substitution of second Arg (from the RxxxR motif) with Asp completely abolished c-di-GMP binding, while the role of Ser residue (from the D/NxSxxG) in c-di-GMP binding was not critical (Ryjenkov et al., 2006). For DgrA from Caulobacter crescentus, the first Arg residue of the RxxxR motif was shown to be essential for c-di-GMP binding (Christen et al., 2007). In this study, replacement of the first Arg residue of the RxxxR motif by Ala also resulted in the complete loss of c-di-GMP binding ability of Cbp1. Thus, we have confirmed the essential role of the RxxxR motif in c-di-GMP binding.
In this study, we have determined that Cbp1 binds c-di-GMP with a KD of 14.94 ± 1.6 μM. C-di-GMP binding affinities of PilZ domain proteins span a sizable range of c-di-GMP concentrations. In Salmonella typhimurium, the KD of YcgR for c-di-GMP is 0.19 ± 0.018 μM, while BcsA binds c-di-GMP with a KD of 8.24 ± 2.4 μM (Pultz et al., 2012). In P. aeruginosa, MapZ and Alg44 bind c-di-GMP with KD of 8.8 ± 1.2 and 12.74 ± 1.68 μM, respectively (Pultz et al., 2012; Xu et al., 2016b). The PilZ domain containing chemoreceptor Tlp1 from A. brasiencse binds c-di-GMP with a moderate KD of 6.9 ± 1.2 μM (Russell et al., 2013). The intracellular concentration of c-di-GMP is speculated to be in the submicromolar to low micromolar range, but local concentrations may be considerably higher (Hengge, 2009). The range of c-di-GMP binding affinities of PilZ domain proteins correlates well with that of intracellular c-di-GMP concentrations.
There are 43 chemoreceptors and a single chemotaxis pathway (CheAWY1BR) in A. caulinodans ORS571 (Jiang et al., 2016; Liu et al., 2018). Chemoreceptors are responsible for sensing signals that are then transmitted to the coupling protein CheW and the histidine kinase CheA. Next, CheY, the response regulator of CheA, regulates the directional change of flagellar rotation (Wuichet et al., 2007). Intracellular concentrations of c-di-GMP respond to a variety of external signals (Schirmer, 2016). As a c-di-GMP binding chemoreceptor, Cbp1 may be responsible for sensing changes in intracellular c-di-GMP concentrations and transmitting the signal to downstream chemotaxis proteins. It is known that chemoreceptor interacts with downstream CheA/CheW, and the initiation of signal transduction is the conformational change of chemoreceptor upon ligand binding (Lai et al., 2006). The result of thermal shift assay showed that binding of c-di-GMP caused significant conformational change of Cbp1. The chemotaxis defect caused by lacking of c-di-GMP binding ability of Cbp1 suggested that the c-di-GMP-induced conformational change may be essential for Cbp1-mediated chemotaxis.
As adaptation proteins present in the chemotaxis pathway, methylesterase CheB and methyltransferase CheR compete with each other to regulate methylation state and ligand binding affinity of chemoreceptors to reset chemotaxis sensitivity (Li and Hazelbauer, 2005). The PilZ domain protein MapZ from P. aeruginosa has been shown to interact with CheR1 (Xu et al., 2016b). C-di-GMP binding to MapZ induces dramatic conformational changes in MapZ. The conformational changes are crucial for MapZ-CheR1 binding, which regulates the activity of CheR1 and the methylation state of chemoreceptor PctA (Yan et al., 2018). In ORS571, there is another possibility that Cbp1 interacts with the adaptation protein CheR via its PilZ domain at the presence of c-di-GMP. The potential interaction between Cbp1 and CheR may regulate the activity of CheR and chemotaxis sensitivity. The impaired chemotaxis of the Δcbp1 mutant may result from lacking of functional adaptational modification of other chemoreceptors. However, the underlying mechanistic function of Cbp1 remains to be further investigated. An analysis of 20681 chemoreceptors has shown that 52 of them contained a C-terminal PilZ domain, which suggests that the integration of c-di-GMP into chemotaxis is not unique to only one or two species (Russell et al., 2013).
Chemotaxis allows motile bacteria to rapidly move toward beneficial niches or away from detrimental conditions, thus affecting the transient cell-cell contact and the probability of cell clumping, which leads to aggregation (Alexandre, 2015). It has been reported that A. brasilense mutants with defective chemotaxis ability had elevated aggregation ratios compared with wild type (Bible et al., 2012). In this study, the Δcbp1 mutant aggregated more than the wild-type ORS571 after long incubation. The defect in chemotaxis caused by loss of the chemoreceptor Cbp1 may account for the enhanced aggregation of the Δcbp1 mutant. Besides cell-cell contact, chemotaxis also affects cell-surface interactions involved in biofilm formation. PilZ domain containing proteins have been implicated in the regulation of biofilm formation (Martinez-Granero et al., 2014). Because of the dual role of Cbp1 in chemotaxis and the c-di-GMP signal transduction network, biofilm formation by the Δcbp1 mutant may be regulated by these two mechanisms together.
Chemotaxis has been shown to play an important role in root surface colonization and nodulation. For example, Pseudomonas putida KT2440 colonized corn root surface by rapid chemotaxis, chemotaxis enhanced the root colonization of the PGPR (plant-growth-promoting rhizobacteria) strain Bacillus amyloliquefaciens SQR9 on cucumber roots, and the che1 chemotaxis gene cluster of Rhizobium leguminosarum was found to be essential for competitive nodulation (Espinosa-Urgel et al., 2002; Miller et al., 2007; Feng et al., 2018). As previously reported, an A. brasilense Sp7 mutant lacking the energy-sensing chemoreceptor Tlp1 was severely impaired in chemotaxis and root surface colonization of wheat (Greer-Phillips et al., 2004). Another study in our lab showed that an A. caulinodans ORS571 mutant ΔtlpA1 with a chemotaxis defect was also compromised in competitive root surface colonization and nodulation of S. rostrata (Liu et al., 2017). Consistent with the former findings, the competitive root colonization efficiency of the Δcbp1 mutant was lower than that of the wild-type ORS571. The results further showed the vital role of chemotaxis in competitive colonization and nodulation.
Conclusion
In conclusion, we identified a c-di-GMP binding chemoreceptor Cbp1 in A. caulinodans ORS571, showing that the Δcbp1 mutant was impaired in chemotaxis and competitive root colonization and nodulation of the host legume S. rostrata. Our research provides an example of the interplay between chemotaxis and c-di-GMP, which may benefit our understanding of bacterial signal transduction networks.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
YS and ZX conceived and designed the experiments and wrote the manuscript. YS, FS, and XL carried out the experiments. YS analyzed data and prepared figures and tables. WC helped with the improvement and revision of the manuscript. All authors approved the submission for publication.
Funding
This work was financed by the NSFC-Shandong Joint Fund Key Projects (U1806206), the National Natural Science Foundation of China (31570063 and 31870020), the Mechanisms of Marginal Land Productivity Expansion and Technologies of Storing Grain in Land (KFZD-SW-112), Shandong Key Research and Development Program (2017GSF17129), Shandong Key Scientific and Technological Innovation Program (2017CXGC0303).
Conflict of Interest Statement
WC was employed by the company Shandong Huibang Bohai Agriculture Development Limited Company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Claudine Elmerich for helpful and insightful comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00638/full#supplementary-material
References
Alexandre, G. (2015). Chemotaxis control of transient cell aggregation. J. Bacteriol. 197, 3230–3237. doi: 10.1128/JB.00121-15
Benach, J., Swaminathan, S., Tamayo, R., Handelman, S., Folta-Stogniew, E., Ramos, J., et al. (2007). The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 26, 5153–5166. doi: 10.1038/sj.emboj.7601918
Bible, A., Russell, M. H., and Alexandre, G. (2012). The Azospirillum brasilense Che1 chemotaxis pathway controls swimming velocity, which affects transient cell-to-cell clumping. J. Bacteriol. 194, 3343–3355. doi: 10.1128/JB.00310-12
Bordeleau, E., Purcell, E. B., Lafontaine, D. A., Fortier, L. C., Tamayo, R., and Burrus, V. (2015). Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. J. Bacteriol. 197, 819–832. doi: 10.1128/JB.02340-14
Chan, C., Paul, R., Samoray, D., Amiot, N. C., Giese, B., Jenal, U., et al. (2004). Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 101, 17084–17089. doi: 10.1073/pnas.0406134101
Christen, M., Christen, B., Allan, M. G., Folcher, M., Jeno, P., Grzesiek, S., et al. (2007). DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. U.S.A. 104, 4112–4117. doi: 10.1073/pnas.0607738104
Christen, M., Christen, B., Folcher, M., Schauerte, A., and Jenal, U. (2005). Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280, 30829–30837. doi: 10.1074/jbc.M504429200
Dreyfus, B., Garcia, J. L., and Gillis, M. (1988). Characterization of Azorhizobium caulinodans gen. nov., sp. nov., a stem nodulating nitrogen-fixing bacterium isolated from Sesbania rostrata. Int. J. Syst. Bacteriol. 38, 89–98. doi: 10.1099/00207713-38-1-89
Espinosa-Urgel, M., Kolter, R., and Ramos, J. (2002). Root colonization by Pseudomonas putida: love at first sight. Microbiology 148, 341–343. doi: 10.1099/00221287-148-2-341
Feng, H., Zhang, N., Du, W., Zhang, H., Liu, Y., Fu, R., et al. (2018). Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol. Plant Microbe Interact. 31, 995–1005. doi: 10.1094/MPMI-01-18-0003-R
Figurski, D. H., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
Gao, S., Romdhane, S. B., Beullens, S., Kaever, V., Lambrichts, I., Fauvart, M., et al. (2014). Genomic analysis of cyclic-di-GMP-related genes in rhizobial type strains and functional analysis in Rhizobium etli. Appl. Microbiol. Biotechnol. 98, 4589–4602. doi: 10.1007/s00253-014-5722-7
Greer-Phillips, S. E., Stephens, B. B., and Alexandre, G. (2004). An energy taxis transducer promotes root colonization by Azospirillum brasilense. J. Bacteriol. 186, 6595–6604. doi: 10.1128/JB.186.19.6595-6604.2004
Hengge, R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273. doi: 10.1038/nrmicro2109
Huang, Z., Ni, B., Jiang, C. Y., Wu, Y. F., He, Y. Z., Parales, R. E., et al. (2016). Direct sensing and signal transduction during bacterial chemotaxis toward aromatic compounds in Comamonas testosteroni. Mol. Microbiol. 101, 224–237. doi: 10.1111/mmi.13385
Huynh, K., and Partch, C. L. (2015). Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 79, 28.9.1–28.9.14. doi: 10.1002/0471140864.ps2809s79
Jiang, N., Liu, W., Li, Y., Wu, H., Zhang, Z., Alexandre, G., et al. (2016). A chemotaxis receptor modulates nodulation during the Azorhizobium caulinodans-Sesbania rostrata symbiosis. Appl. Environ. Microbiol. 82, 3174–3184. doi: 10.1128/AEM.00230-16
Kovach, M., Elzer, P., Hill, D., Robertson, G., Farris, M., Martin Roop, R. II, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Lai, W. C., Beel, B. D., and Hazelbauer, G. L. (2006). Adaptational modification and ligand occupancy have oppsite effects on positioning of the transmembrane signalling helix of a chemoreceptor. Mol. Microbiol. 61, 1081–1090. doi: 10.1111/j.1365-2958.2006.05296.x
Lee, K. B., De Backer, P., Aono, T., Liu, C. T., Suzuki, S., Suzuki, T., et al. (2008). The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9:271. doi: 10.1186/1471-2164-9-271
Lee, V. T., Matewish, J. M., Kessler, J. L., Hyodo, M., Hayakawa, Y., and Lory, S. (2007). A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x
Li, G., and Weis, R. M. (2000). Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100, 357–365. doi: 10.1016/S0092-8674(00)80671-6
Li, M., and Hazelbauer, G. L. (2005). Adaptational assistance in clusters of bacterial chemoreceptors. Mol. Microbiol. 56, 1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x
Liu, W., Sun, Y., Shen, R., Dang, X., Liu, X., Sui, F., et al. (2018). A chemotaxis-like pathway of Azorhizobium caulinodans controls flagella-driven motility, which regulates biofilm formation, exopolysaccharide biosynthesis, and competitive nodulation. Mol. Plant Microbe Interact. 31, 737–749. doi: 10.1094/MPMI-12-17-0290-R
Liu, W., Yang, J., Sun, Y., Liu, X., Li, Y., Zhang, Z., et al. (2017). Azorhizobium caulinodans transmembrane chemoreceptor TlpA1 involved in host colonization and nodulation on roots and stems. Front. Microbiol. 8:1327. doi: 10.3389/fmicb.2017.01327
Martinez-Granero, F., Navazo, A., Barahona, E., Redondo-Nieto, M., Gonzalez de Heredia, E., Baena, I., et al. (2014). Identification of flgZ as a flagellar gene encoding a PilZ domain protein that regulates swimming motility and biofilm formation in Pseudomonas. PLoS One 9:e87608. doi: 10.1371/journal.pone.0087608
Marx, C. J., and Lidstrom, M. E. (2002). Broad-host-range cre-lox system for antibiotic marker recycling in Gram-negative bacteria. Biotechniques 22, 1062–1067. doi: 10.2144/02335rr01
Merighi, M., Lee, V. T., Hyodo, M., Hayakawa, Y., and Lory, S. (2007). The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65, 876–895. doi: 10.1111/j.1365-2958.2007.05817.x
Miller, L. D., Yost, C. K., Hynes, M. F., and Alexandre, G. (2007). The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol. Microbiol. 63, 348–362. doi: 10.1111/j.1365-2958.2006.05515.x
Nakajima, A., Aono, T., Tsukada, S., Siarot, L., Ogawa, T., and Oyaizu, H. (2012). Lon protease of Azorhizobium caulinodans ORS571 is required for suppression of reb gene expression. Appl. Environ. Microbiol. 78, 6251–6261. doi: 10.1128/AEM.01039-12
Newell, P. D., Monds, R. D., and O’Toole, G. A. (2009). LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U.S.A. 106, 3461–3466. doi: 10.1073/pnas.0808933106
Paul, K., Nieto, V., Carlquist, W. C., Blair, D. F., and Harshey, R. M. (2010). The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38, 128–139. doi: 10.1016/j.molcel.2010.03.001
Povolotsky, T. L., and Hengge, R. (2012). ‘Life-style’ control networks in Escherichia coli: signaling by the second messenger c-di-GMP. J. Biotechnol. 160, 10–16. doi: 10.1016/j.jbiotec.2011.12.024
Pultz, I. S., Christen, M., Kulasekara, H. D., Kennard, A., Kulasekara, B., and Miller, S. I. (2012). The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol. Microbiol. 86, 1424–1440. doi: 10.1111/mmi.12066
Purcell, E. B., McKee, R. W., McBride, S. M., Waters, C. M., and Tamayo, R. (2012). Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 194, 3307–3316. doi: 10.1128/JB.00100-12
Rao, F., Yang, Y., Qi, Y., and Liang, Z. X. (2008). Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190, 3622–3631. doi: 10.1128/JB.00165-08
Roy, A. B., Petrova, O. E., and Sauer, K. (2012). The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194, 2904–2915. doi: 10.1128/JB.05346-11
Russell, M. H., Bible, A. N., Fang, X., Gooding, J. R., Campagna, S. R., Gomelsky, M., et al. (2013). Integration of the second messenger c-di-GMP into the chemotactic signaling pathway. mBio 4:e00001-13. doi: 10.1128/mBio.00001-13
Ryan, R. P., Fouhy, Y., Lucey, J. F., Crossman, L. C., Spiro, S., He, Y. W., et al. (2006). Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U.S.A. 103, 6712–6717. doi: 10.1073/pnas.0600345103
Ryan, R. P., Tolker-Nielsen, T., and Dow, J. M. (2012). When the PilZ don’t work: effectors for cyclic di-GMP action in bacteria. Trends Microbiol. 20, 235–242. doi: 10.1016/j.tim.2012.02.008
Ryjenkov, D. A., Simm, R., Romling, U., and Gomelsky, M. (2006). The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281, 30310–30314. doi: 10.1074/jbc.C600179200
Ryjenkov, D. A., Tarutina, M., Moskvin, O. V., and Gomelsky, M. (2005). Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187, 1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005
Schäper, S., Krol, E., Skotnicka, D., Kaever, V., Hilker, R., Sogaard-Andersen, L., et al. (2016). Cyclic di-GMP regulates multiple cellular functions in the symbiotic alphaproteobacterium Sinorhizobium meliloti. J. Bacteriol. 198, 521–535. doi: 10.1128/JB.00795-15
Scharf, B. E., Hynes, M. F., and Alexandre, G. M. (2016). Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol. Biol. 90, 549–559. doi: 10.1007/s11103-016-0432-4
Schirmer, T. (2016). C-di-GMP Synthesis: structural aspects of evolution, catalysis and regulation. J. Mol. Biol. 428, 3683–3701. doi: 10.1016/j.jmb.2016.07.023
Sun, Y., Zhao, H., Wang, J., Zhu, J., and Wu, S. (2015). Identification and regulation of the catalytic promiscuity of (-)-gamma-lactamase from Microbacterium hydrocarbonoxydans. Appl. Microbiol. Biotechnol. 99, 7559–7568. doi: 10.1007/s00253-015-6503-7
Tindall, M. J., Gaffney, E. A., Maini, P. K., and Armitage, J. P. (2012). Theoretical insights into bacterial chemotaxis. Wiley Interdiscip. Rev. Syst. Biol. Med. 4, 247–259. doi: 10.1002/wsbm.1168
Wadhams, G. H., and Armitage, J. P. (2004). Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5, 1024–1037. doi: 10.1038/nrm1524
Wuichet, K., Alexander, R. P., and Zhulin, I. B. (2007). Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 422, 3–31. doi: 10.1016/S0076-6879(06)22001-9
Xu, L., Venkataramani, P., Ding, Y., Liu, Y., Deng, Y., Yong, G. L., et al. (2016a). A cyclic di-GMP-binding adaptor protein interacts with histidine kinase to regulate two-component signaling. J. Biol. Chem. 291, 16112–16123. doi: 10.1074/jbc.M116.730887
Xu, L., Xin, L., Zeng, Y., Yam, J., Ding, Y., Venkataramani, P., et al. (2016b). A cyclic di-GMP-binding adaptor protein interacts with a chemotaxis methyltransferase to control flagellar motor switching. Sci. Signal. 9:ra102.
Yan, X. F., Xin, L., Yen, J. T., Zeng, Y., Jin, S., Cheang, Q. W., et al. (2018). Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein. J. Biol. Chem. 293, 100–111. doi: 10.1074/jbc.M117.815704
Keywords: c-di-GMP, PilZ domain, chemoreceptor, chemotaxis, nodulation
Citation: Sun Y, Xie Z, Sui F, Liu X and Cheng W (2019) Identification of Cbp1, a c-di-GMP Binding Chemoreceptor in Azorhizobium caulinodans ORS571 Involved in Chemotaxis and Nodulation of the Host Plant. Front. Microbiol. 10:638. doi: 10.3389/fmicb.2019.00638
Received: 15 November 2018; Accepted: 13 March 2019;
Published: 02 April 2019.
Edited by:
Marta Martín, Autonomous University of Madrid, SpainReviewed by:
Matias Castro, Fundación Ciencia & Vida, ChileMichael Göttfert, Dresden University of Technology, Germany
Copyright © 2019 Sun, Xie, Sui, Liu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihong Xie, emh4aWVAeWljLmFjLmNu
 Yu Sun
Yu Sun Zhihong Xie
Zhihong Xie Fu Sui1,2
Fu Sui1,2 Xiaolin Liu
Xiaolin Liu
