- 1Guangdong Institute of Microbiology, State Key Laboratory of Applied Microbiology Southern China, Guangdong Open Laboratory of Applied Microbiology, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangzhou, China
- 2College of Food Science, South China Agricultural University, Guangzhou, China
- 3Department of Food Science and Technology, Jinan University, Guangzhou, China
Listeria monocytogenes is a globally notorious foodborne pathogen. This study aimed to qualitatively and quantitatively detect L. monocytogenes from meat and meat products in China and to establish their virulence profiles and population diversity. From 1212 meat and meat product samples, 362 (29.9%) were positive for L. monocytogenes. Of these positive samples, 90.6% (328/362) had less than 10 MPN/g, 5.5% (20/364) samples had 10–110 MPN/g, and 3.9% (14/362) of the positive samples had over 110 MPN/g. Serogroup analysis showed that the most prevalent serogroup of L. monocytogenes was I.1 (1/2a-3a), which accounted for 45.0% (123/458) of the total, followed by serogroup I.2 (1/2c-3c) that comprised 26.9%, serogroup II.1 (4b-4d-4e) that comprised 4.8%, and serogroup II.2 (1/2b-3b-7) that comprised 23.3%. A total of 458 isolates were grouped into 35 sequence types (STs) that belonged to 25 clonal complexes (CCs) and one singleton (ST619) by multi-locus sequence typing. The most prevalent ST was ST9 (26.9%), followed by ST8 (17.9%), ST87 (15.3%), ST155 (9.4%), and ST121 (7.6%). Thirty-seven isolates harbored the llsX gene (representing LIPI-3), and they belonged to ST1/CC1, ST3/CC3, ST288/CC288, ST323/CC288, ST330/CC288, ST515/CC1, and ST619, among which ST323/CC288, ST330/CC288, and ST515/CC1 were newly reported to carry LIPI-3. Seventy-five isolates carried ptsA, and they belonged to ST87/CC87, ST88/CC88, and ST619, indicating that consumers may be exposed to potential hypervirulent L. monocytogenes. Antibiotics susceptibility tests revealed that over 90% of the isolates were susceptible to 11 antibiotics; however, 40.0% of the isolates exhibited resistance against ampicillin and 11.8% against tetracycline; further, 45.0 and 4.6% were intermediate resistant and resistant to ciprofloxacin, respectively. The rise of antibiotic resistance in L. monocytogenes suggests that stricter regulations should be formulated to restrict the use of antibiotic agents in human listeriosis treatment and livestock breeding.
Introduction
Listeria monocytogenes is a facultative Gram-positive foodborne pathogen responsible for life-threatening listeriosis diseases, including septicaemia, meningitis, encephalitis, and miscarriage (Drevets and Bronze, 2008). The most susceptible groups are pregnant women, fetuses, elderly people, and immunocompromised individuals, who show a considerably high mortality rate (10–40%) (Hof, 2004). This pathogen can resist stressful conditions in foods and associated environments. It can grow in high salinity (10%), low temperatures (4°C), low water activity (<0.9), and a wide pH range (4.1–9.6) (Guenther et al., 2009), resulting in a wide range of habitats during different stages of the food processing. Listeriosis cases occur mainly due to the consumption of L. monocytogenes-contaminated foods.
Previous studies report 0.27 and 0.39 reported cases of listeriosis annually per 100,000 individuals in the United States and France, respectively (Goulet et al., 2012; Silk et al., 2012). In China, 147 clinical cases and 82 outbreak-related cases were reported from 28 provinces between 1964 and 2010 (Feng et al., 2013). In recent years, the occurrence of listeriosis diseases have been increasing in China, especially in developed cities (Wang et al., 2015, 2018). A total of 253 invasive listeriosis cases were reported between 2011 and 2016 in 19 provinces, with a fatality rate of 25.7% (Li et al., 2018b). Listeriosis has therefore become a severe public health concern to consumers in China. Although a national human listeriosis pilot surveillance was started in 2013, a risk assessment on the prevalence and characteristics of L. monocytogenes in foods is an urgent necessity. Although several studies have reported the presence of L. monocytogenes in various foods, including ready-to-eat products, mushrooms, aquatic products, meat and meat products, and frozen foods, most such studies are regionally focused (Chen et al., 2014, 2015; Liu et al., 2017; Yang et al., 2017). A comprehensive surveillance of L. monocytogenes in foods throughout China is of crucial importance.
Among 13 serotypes of L. monocytogenes, serotypes 4b, 1/2a, 1/2b, and 1/2c account for over 95% of the isolates recovered from foods and clinical cases (Orsi et al., 2011). The pathogenicity of L. monocytogenes may depend on the presence of virulence genes. Several virulence genes and their encoded proteins have been described in previous studies. Listeria pathogenicity island-1 (LIPI-1), along with inlA and inlB, participate in the L. monocytogenes infection cycle in host cells (Kreft and Vazquez-Boland, 2001). The llsX gene (listeriolysin S, representing LIPI-3), which encodes a haemolytic cytotoxic factor associated with the destruction of gut microbiota during infection, is mainly present in a subset of lineage I (Cotter et al., 2008; Quereda et al., 2017a,b). LIPI-3 has been detected in several sequence types (STs) of L. monocytogenes strains, including ST1, ST3, ST4, ST6, ST77, ST79, ST191, ST213, ST217, ST224, ST288, ST363, ST380, ST382, ST389, ST489, ST554, ST581, ST619, ST778, ST999, ST1000, and ST1001 (Chen et al., 2018b; Kim et al., 2018; Wang et al., 2018). Furthermore, a cellobiose-family phosphotransferase system with a cluster of six genes was recently identified as LIPI-4 (Maury et al., 2016). LIPI-4, which is strongly associated with neural and placental infection, was first identified as a clonal complex (CC) 4-specific virulence factor. Several STs (ST87, ST213, ST217, ST363, ST382, ST388, ST663, ST1002, ST1166, and ST619) harboring the ptsA gene (representing LIPI-4) have been reported in recent studies (Chen et al., 2018a; Kim et al., 2018). The pathogenic potential of L. monocytogenes may differ though the presence of LIPI-1, inlA and inlB gene. However, LIPI-3 (llsX) and LIPI-4 (ptsA) are strongly associated with the L. monocytogenes infection.
The objectives of the present study were to (i) determine the qualitative and quantitative data on L. monocytogenes in meat and meat products; (ii) evaluate the potential virulence and antimicrobial resistance profiles of L. monocytogenes isolates; and (iii) characterize the molecular serotype and genetic diversity of L. monocytogenes isolates recovered from the Chinese retail aquatic system. This data will be invaluable for future risk assessments.
Materials and Methods
Samples
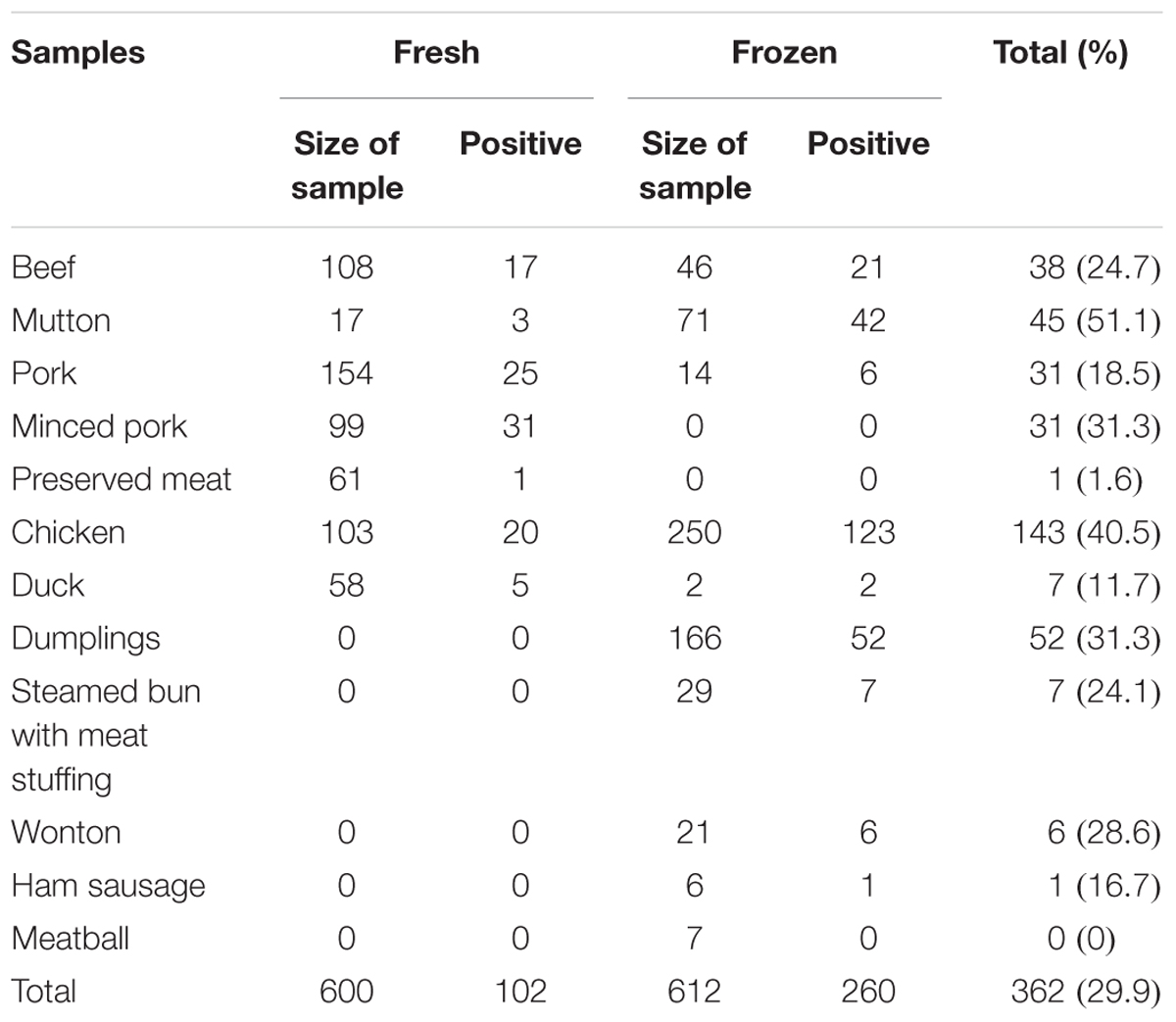
Between July 2012 and April 2016, 1212 retail raw meat and meat product samples were collected from 43 cities of China, including beef (fresh = 108 samples, frozen = 46 samples), mutton (fresh = 17, frozen = 71), pork (fresh = 154, frozen = 14), minced meat (n = 99), preserved pork (n = 61), chicken (fresh = 103, frozen = 250), duck (fresh = 58, frozen = 2), dumplings (n = 166), steamed bun with meat (n = 29), wonton (n = 21), ham sausage (n = 6), and meat balls (n = 7). All samples were placed in insulated shipping coolers with frozen gel packs placed on the sides, middle, and top of the samples. All samples were kept below 4°C during transportation, and testing was initiated within 4 h after receiving the samples.
Qualitative and Quantitative Analysis
Qualitative detection of L. monocytogenes was performed based on the National Food Safety Standard of China (4789.30-2010) (Anonymous, 2010), with minor adaptations. Briefly, 25 g of homogenized samples were added to 225 mL Listeria enrichment broth 1 (LB1) (Guangdong Huankai Co., Ltd., Guangzhou, China). The cultures in LB1 media were incubated at 30°C for 24 h. After incubation, 100°μL of the LB1 enrichment culture was transferred to 10 mL Listeria enrichment broth 2 (LB2) and incubated at 30°C for 24 h. A loopful (about 10°μL) of the LB2 enrichment culture was streaked onto Listeria selective agar plates (Guangdong Huankai Co., Ltd.) and incubated at 37°C for 48 h. Three to five (when possible) presumptive colonies were selected for the identification of L. monocytogenes using the Microgen ID Listeria identification system (Microgen, Camberley, United Kingdom), according to the manufacturer’s instructions.
For quantitative detection, a nine-tube most probable number (MPN) method was used, based on a previous study (Gombas et al., 2003). Briefly, nine tubes were divided into three sets of three tubes each. Homogenized samples (25 g) were added to 225 mL half frasher broth. The first set of tubes contained 10 mL of the sample homogenate in 225 mL half Frasher broth, while the second and third sets contained 10 mL of half Fraser broth (Guangdong Huankai Co., Ltd.) inoculated with 1 and 0.1 mL of the homogenate, respectively. These different volumes (10, 1, and 0.1 mL) of the sample homogenate represented 1.0, 0.1, and 0.01 g of the original sample, respectively. The nine tubes were incubated at 30 ± 2°C for 24 ± 2 h. The darkened Fraser tubes were streaked onto Listeria Chromagar plates. If a Fraser broth tube did not darken, it was examined again after an additional 26 ± 2 h of incubation. The presumptive pure colonies were streaked onto TSA plates and identified using the Microgen ID Listeria identification system. The MPN value was determined based on the number of positive tube(s) in each of the three sets and the MPN table (United States Department of Agriculture, 1998).
Serotyping and Virulotype Determination
Multiplex PCR was used for identifying the serotypes of the 458 isolates, as described previously (Doumith et al., 2004) (Supplementary Table S1). PCR was performed in a thermal cycler (Biometra, Gottingen, Germany) with the following conditions: an initial denaturation at 94°C for 3 min; followed by 35 cycles of 94°C for 35 s, 53°C for 50 s, and 72°C for 60 s; and a final cycle of 72°C for 7 min. Two additional PCRs were performed to detect the llsX and ptsA genes (representing LIPI-3 and LIPI-4, respectively) in the L. monocytogenes isolates (Clayton et al., 2011; Maury et al., 2016). The PCR primers used are shown in Supplementary Table S1. The amplicons were separated on 1.5% agarose gels in TAE buffer and visualized by Goldview® staining (0.005%, v/v).
Antimicrobial Susceptibility Test
The antibiotic susceptibility of the L. monocytogenes isolates was determined using the KB method, according to the breakpoints for Staphylococci spp., as recommended by the Clinical Laboratory Standards Institute (Clinical, and Laboratory Standards Institute [CLSI], 2014) for Staphylococcus. The breakpoints of ampicillin and penicillin G for specific Listeria have been defined (M45-A2 Vol. 30 No. 18). The following 17 common antibiotic agents (disk load), including those used to treat human listeriosis, were tested: kanamycin (30°μg), gentamicin (10°μg), ciprofloxacin (5°μg), levofloxacin (5°μg), ofloxacin (5°μg), sulfamethoxazole with trimethoprim (23.75/1.25°μg), streptomycin (10°μg), rifampin (5°μg), doxycycline (30°μg), chloramphenicol (30°μg), erythromycin (15°μg), tetracycline (30°μg), meropenem (10°μg), vancomycin (30°μg), linezolid (30°μg), amoxycillin/clavulanic acid (10°μg), and sulbactam/ampicillin (10/10°μg) (Oxoid, Basingstoke, United Kingdom). Briefly, pure cultures were transferred to brain heart infusion (BHI) broth and incubated at 37°C overnight. A cell suspension was adjusted to 0.5 MacFarland standards by 0.85% NaCl (w/v). The suspension was spread onto the surface of Mueller-Hinton agar (Huankai Co., Ltd., Guangzhou). The diameters of the inhibition zones were measured using precision calipers after 24°h incubation. Staphylococcus aureus ATCC 25923 and Escherichia coli ATCC 25922 were used as quality control strains. Isolates exhibiting resistance to at least three classes of the tested antimicrobial agents were considered multidrug-resistant (Magiorakos et al., 2012).
Multi-Locus Sequence Typing
Multi-locus sequence typing (MLST) analysis of L. monocytogenes was performed according to a previously published method (Ragon et al., 2008), which was based on seven house-keeping genes (abcZ, bglA, cat, dapE, dat, ldh, and lhkA) (Supplementary Table S2). A detailed protocol of MLST analysis, including primers, PCR conditions, STs and CCs assignments were performed according to the recommendation of Pasteur Institute website. PCR products were sequenced (Thermo Fisher Co., Ltd., Shanghai, China), and an allele number was assigned based on each variant locus of each housekeeping gene; STs and CCs were assigned via the Listeria MLST database at the Pasteur Institute website1. A minimum spanning tree (MST) was constructed to analyze the relationships between the isolates using the BioNumerics software Version 7.6 (Applied Maths, Belgium).
Results
Occurrence and Contamination Levels of L. monocytogenes
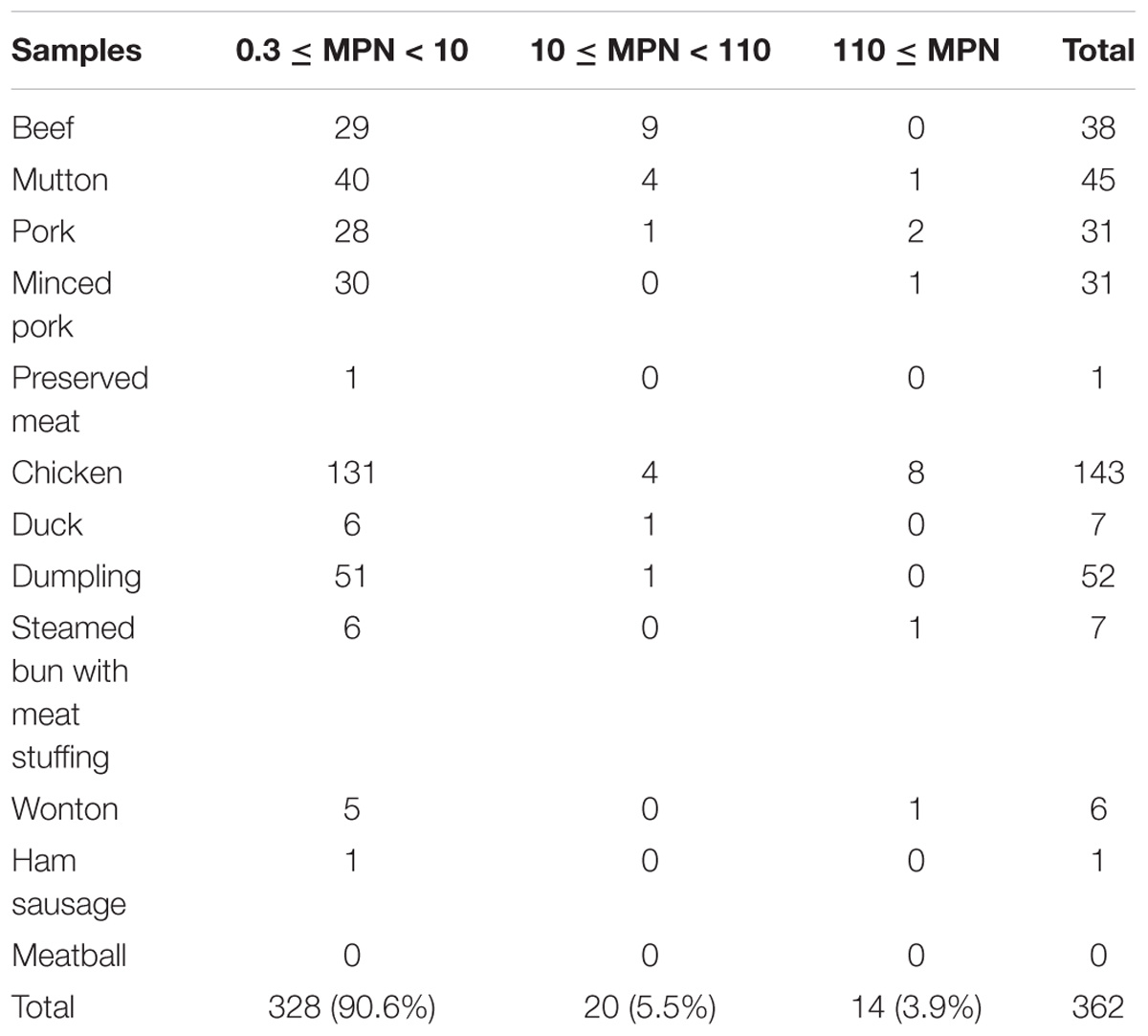
A total of 1212 meat and meat products (12 types) were tested in this study. As shown in Table 1, the overall prevalence of L. monocytogenes in meat and meat products was 29.9% (362/1212); it was detected in 45 (51.1%) mutton samples, 143 (40.5%) chicken samples, 31.3% of both minced pork (31/99) and dumpling samples (52/166), 38 (24.7%) beef samples, 6 (28.6%) wonton samples, 7 (24.1%) steamed bun with meat stuffing samples, 7 (11.7%) duck samples, 31 (18.5%) pork samples, and 1 (16.7%) ham sausage sample. Only one preserved meat sample showed the presence of L. monocytogenes, while no positive samples were found in meatball samples. In addition, as risk identification required quantitative data to estimate the impact of L. monocytogenes on consumer health, the level of contamination was also determined in the meat and meat product samples. Most samples, 90.6% (328/362) had less than 10 MPN/g, 5.5% (20/364) samples had 10-110 MPN/g, and 3.9% (14/362) of the positive samples had over 110 MPN/g, 8 of which were from chicken samples (Table 2).
Serogroups
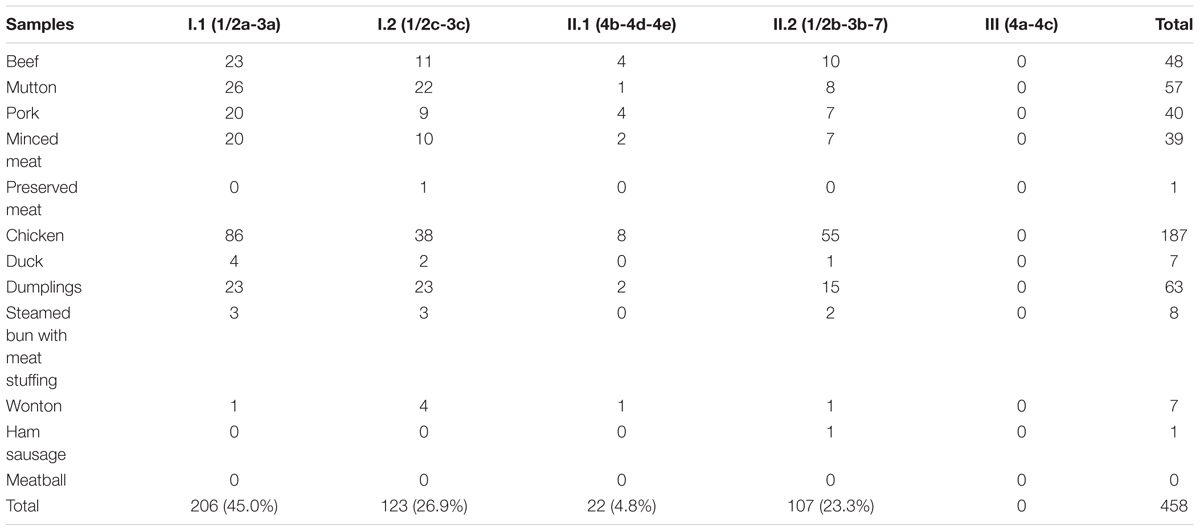
Molecular serogrouping was performed by multiplex PCR on 458 L. monocytogenes strains isolated from all 364 positive samples. As shown in Table 3, serogroup I.1 (1/2a-3a) was the most prevalent (45.0%). As for the other serogroups, 26.9% (123/458) of the samples were in serogroup I.2 (1/2c-3c), 4.8% (22/458) were in serogroup II.1 (4b-4d-4e), 23.3% (107/458) were in serogroup II.2 (1/2b-3b-7), and none were in serogroup III (4a-4c).
Antibiotic Susceptibility Test
All L. monocytogenes isolates recovered from meat and meat products were susceptible to vancomycin and amoxicillin/clavulanic acid. Over 90% of the isolates were susceptible to kanamycin, gentamicin, ofloxacin, sulfamethoxazole with trimethoprim, doxycycline, meropenem, linezolid, sulbactam/ampicillin, and penicillin. However, to some extent, L. monocytogenes isolates were resistant to some antibiotics, including ciprofloxacin, levofloxacin, streptomycin, rifampin, tetracycline, and ampicillin. Approximately 40.0% of the isolates exhibited resistance to ampicillin, while 8.1% were resistant to penicillin. Fifty-four (11.8%) isolates were resistant to tetracycline, and 11 exhibited intermediate resistance. In addition, 45.0 and 4.6% of the isolates were intermediate-resistant or resistant to ciprofloxacin, respectively; further, 20.3% were also intermediate-resistant to levofloxacin (Table 4). In total, 27 multidrug-resistant strains were counted.
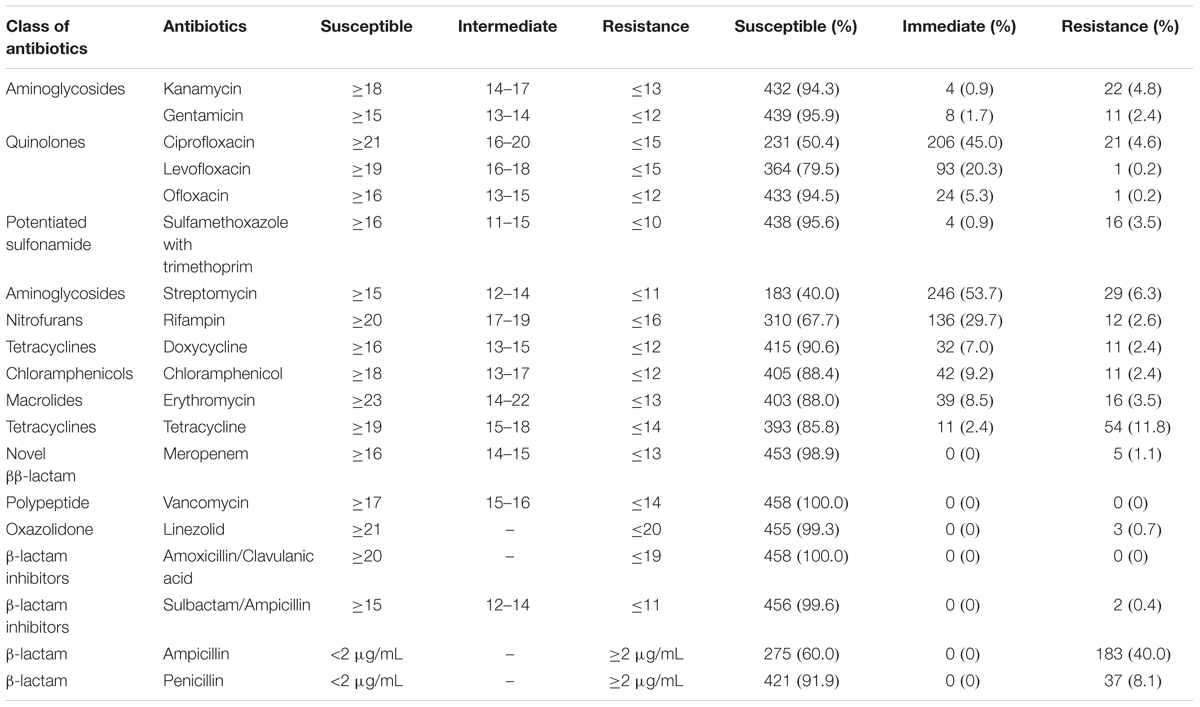
Table 4. Antibiotic susceptibilities of Listeria monocytogenes isolates from meat and meat products.
MLST Analysis
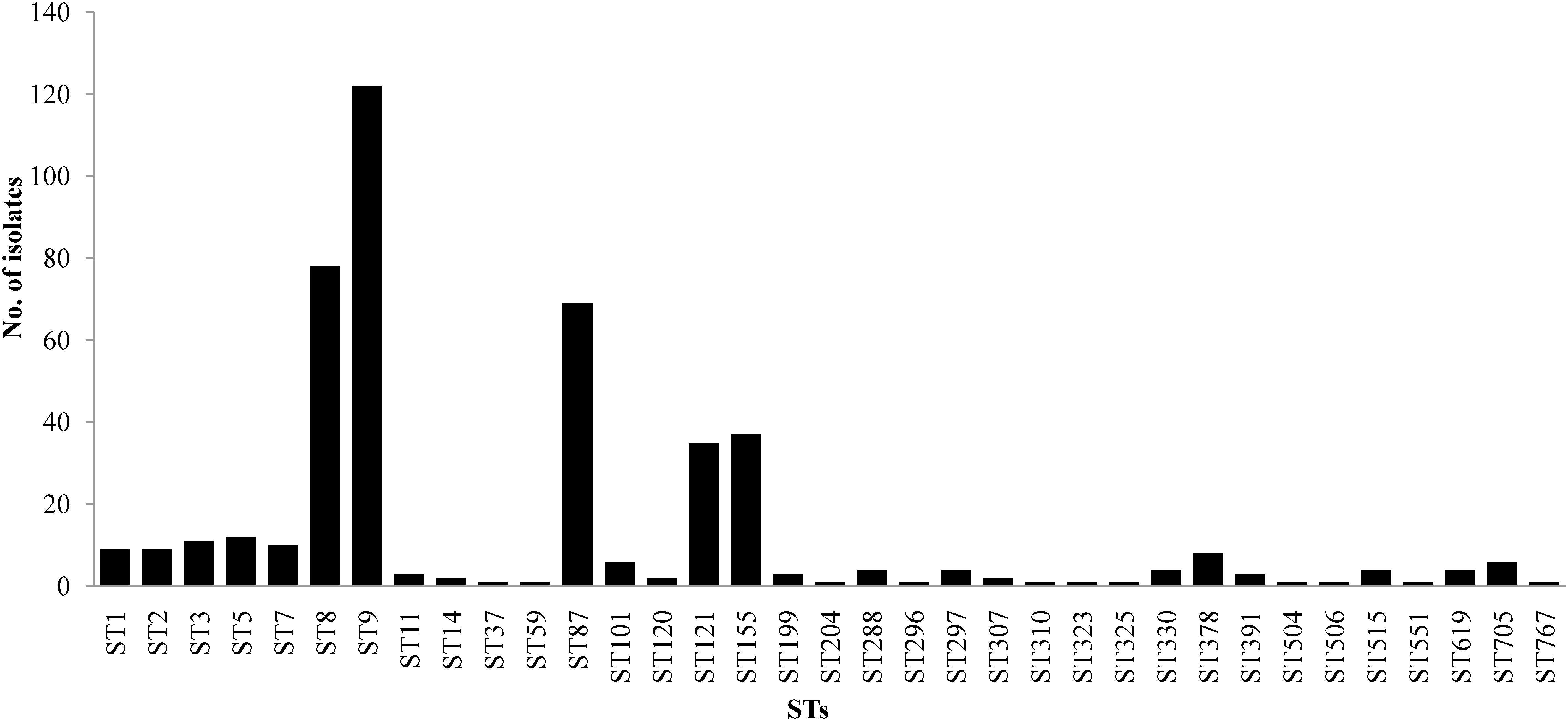
The 458 L. monocytogenes isolates were grouped into 35 different STs belonging to 25 CCs and one singleton (ST619) by MLST analysis (Figures 1, 2). Thirteen STs (35.1% of all STs) were represented by single isolates. Five CCs were the most prevalent: CC9 (n = 123 isolates; 26.9%) and CC8 (n = 82; 17.9%); followed by CC87 (n = 70; 15.3%), CC155 (n = 43, 9.4%), and CC121 (n = 35; 7.6%) (Figure 2). The remaining 20 CCs and one singleton (n = 105, 22.9%) were sporadically distributed (Figure 2). In addition, the presence of the llsX (representing LIPI-3) and ptsA genes (representing in LIPI-4) were also determined in the L. monocytogenes isolates. Thirty-seven isolates harbored llsX, and they belonged to ST1/CC1, ST3/CC3, ST288/CC288, ST323/CC288, ST330/CC288 ST515/CC1, and ST619; seventy-five isolates harbored ptsA, and they belonged to ST87/CC87, ST88/CC88, and ST619. Interestingly, isolates belonging to ST619 carried both llsX and ptsA.
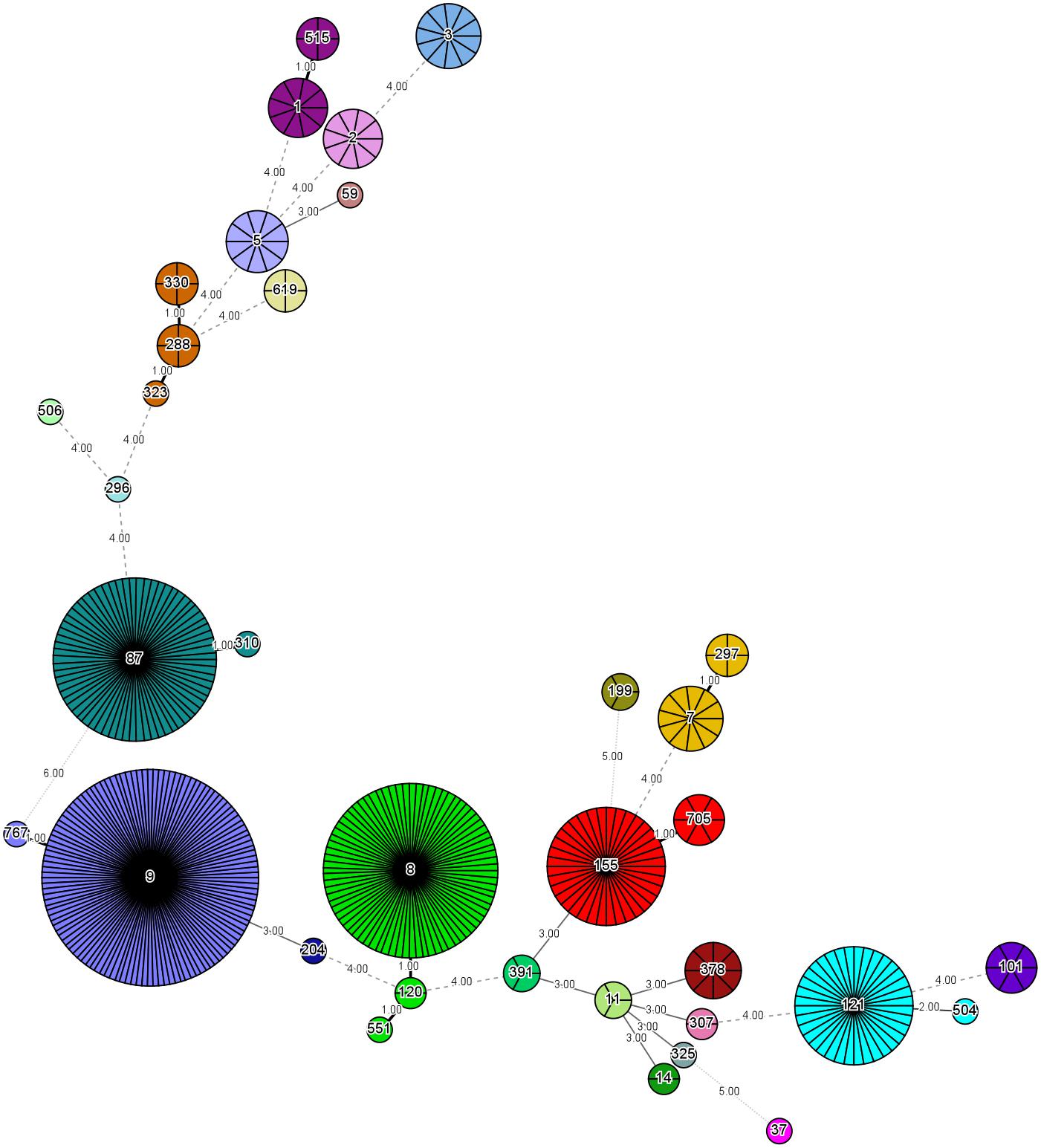
Figure 2. Minimum spanning tree of multi-locus sequence typing data for 458 Listeria monocytogenes isolates. Each circle represents one ST, and each fragment of the pie chart corresponds to a single isolate. The size of the circle is proportional to the number of isolates of that ST.
Discussion
Listeriosis is a major public health concern worldwide, with a high morbidity rate. Surveillance for L. monocytogenes in food items is of utmost importance for risk assessment. In this study, the contamination levels of L. monocytogenes in different meat and meat products in China were determined, and the phenotypic and genotypic characteristics of isolates were analyzed by serotype, antibiotic resistance, and genetic diversity. The overall prevalence of L. monocytogenes in meat and meat products in China was 29.9% (362/1212), similar result (26.6%) was observed conducted in Beijing city by Ma (2015) and the results from Addis Ababa, Ethiopia (Derra et al., 2013); while the different contamination rate were reported in Changchun city by Zhu et al. (2016) (43.3%) and in Liaoning province by Wen et al. (2015) (8.88%). However, other countries have been reported to have different levels of L. monocytogenes prevalence in meat and meat products (Ndahi et al., 2014; Ristori et al., 2014). This variation maybe attributed to differences in sample size, sample constitution, or geographical location. Among the 12 types of meat and meat products analyzed, the highest prevalence was found in mutton (51.1%), followed by chicken (40.5%) and beef (24.7%), which was consistent with the contamination reported in retail-level beef meat in Poland (Wieczorek et al., 2012). The occurrence of L. monocytogenes in pork (18.5%) was higher than that reported in previous studies in several countries (2.6–12.8%) (Pesavento et al., 2010; Derra et al., 2013; Li et al., 2016; Hamidiyan et al., 2018). The high prevalence of L. monocytogenes in meat and meat products in some countries but not others suggest meat and meat products contaminated with L. monocytogenes may occur at processing level, which maybe associate with the hygiene conditions of retail environments of these products. Hoelzer et al. (2012) reported that transfer probabilities of L. monocytogenes may be from cutting boards, scales, deli cases, deli preparation sinks to product, floor drains, walk-in cooler floors, and knife racks to food contact surfaces. In addition, it should pay attention to the persistence of L. monocytogenes in food production process. Simmons et al. (2014) reported that one or more PFGE types were isolated on at least three separate occasions, suggesting that the persistence of a given L. monocytogenes subtype in the delis. The highest prevalence of persistent predominant genotypes of L. monocytogenes was also observed on the Finish dairy farm with the poorest production hygiene, such as feeding surfaces, water troughs, and floors (Castro et al., 2018). Due to the high prevalence of L. monocytogenes in meat and meat products, some sanitization measures and regulations should be formulated to reduce the prevalence of contamination at the processing level.
Quantitative data are invaluable for estimating the impact of L. monocytogenes on consumer health. In the present study, the level of contamination in meat and meat products was also assessed using the MPN method. Most positive samples (90.5%) had less than 10 MPN/g, and only 3.9% of the samples had above 110 MPN/g, which were mainly chicken samples. These results were consistent with those of studies in other countries such as Poland (Modzelewska-Kapitula and Maj-Sobotka, 2014), Brazil (Ristori et al., 2014), and Ireland (Khen et al., 2015); similar results were also reported in China for other food items (Chen et al., 2018a,b), suggesting low levels of L. monocytogenes contamination in fresh food products. The low number of L. monocytogenes in most samples could still be problematic; being psychrotrophic, it may grow during the storage period. It was reported that an initial contamination by only 10 CFU/g of L. monocytogenes can make the food unsafe within 8 days (Salvat and Fravalo, 2004). These results demonstrate the need for further processing of meat and meat products after purchase. Additionally, cross-contamination of food items should be carefully avoided during storage to ensure food safety.
Serotyping is a classical method for L. monocytogenes subtyping. Our results showed that the most prevalent serotypes were serogroups I.1 (1/2a-3a), I.2 (1/2c-3c), and II.2 (1/2b-3b-7), which was consistent with the results of previous studies on L. monocytogenes isolated from food items (Korsak et al., 2012; Shen et al., 2013; Martin et al., 2014; Vallim et al., 2015). Interestingly, serogroup II.1 (4b-4d-4e), which we found to be scattered in all meat and meat products analyzed, has been reported to be the most predominant serogroup in ready-to-eat foods in China, including cooked meat (Chen et al., 2014; Wu et al., 2016). Serotypes 4b, 1/2b, and 1/2a have been shown to be predominant in human listeriosis cases (Orsi et al., 2011), suggesting that these isolates may exhibit pathogenicity against consumers. In fact, L. monocytogenes exhibit variable pathogenicity at the species-level, even though each isolate carries inlA, inlB, and virulence genes of LIPI-1.
Our MLST analysis results showed that five CCs were the most prevalent: CC9 (n = 123 isolates; 26.9%) and CC8 (n = 82; 17.9%), followed by CC87 (n = 70; 15.3%), CC155 (n = 43, 9.4%), and CC121 (n = 35; 7.6%). These results were consistent with those of previous studies in China (Wang et al., 2012; Li et al., 2018a), France (Felix et al., 2018), the European Union (Rychli et al., 2018), and Spain (Martin et al., 2014). The most prevalent ST was ST9 (26.9%), followed by ST8 (17.9%), ST87 (15.3%), ST155 (9.4%), and ST121 (7.6%) in this study. Thus, ST9 and ST121 may be dominant in food and food processing environments globally. In addition, ST87 has been found to be prevalent in other kinds of foods in China, including edible mushrooms and aquatic products (Chen et al., 2018a,b). ST2 and ST87 were reported to be persistent in prepacked smoked salmon in Singapore (Chau et al., 2017). To the best of our knowledge, ST87 strains are rarely reported in western countries, indicating that ST87 isolates may have a geographically associated distribution in Asia. Further surveillance should be performed for the presence of ST87 strains in food, food processing environments, and clinic cases.
llsX (belonging to LIPI-3) encodes a bacteriocin-like haemolytic and cytotoxic virulence factor, which plays a role in the destruction of the gut microbiota. It is critical for the establishment of infection and for the survival of the pathogen in polymorphonuclear neutrophils (Cotter et al., 2008; Quereda et al., 2016, 2017a,b). We found that 37 isolates harbored llsX, and they belonged to serogroups II.1 (4b-4d-4e) and II.2 (1/2b-3b-7). These isolates were present in ST1/CC1, ST3/CC3, ST288/CC288, ST323/CC288, ST330/CC288, ST515/CC1, and ST619, implying that these lineage-I isolates carrying llsX may be responsible for epidemic listeriosis outbreaks. ST323, ST330, and ST515 were newly reported to be llsX-carrying isolates. Furthermore, LIPI-4, a cluster of six genes called the cellobiose-family phosphotransferase system, was mainly involved in neural and placental infection; it was first found only in CC4 strains (Maury et al., 2016). In our results, 75 isolates carried ptsA, and they belonged to ST87/CC87, ST88/CC88, and ST619. Interestingly, hypervirulent CC4 strains carrying both llsX and ptsA are known to be overrepresented in human isolates (Maury et al., 2016). ST619 isolates also carried both llsX and ptsA, suggesting that they may pose a hyper-pathogenic risk to public health. In addition, one of the predominant strains found in this study, ST87, is a known epidemiological hypervirulent ST in China (Li et al., 2018b; Wang et al., 2018). Thus, potential hypervirulent isolates are present in meat and meat products.
It was reported that 162,000 tons of antibiotics was used in China in 2013, of which 84,240 tons was used for livestock breeding and cultivation (Zhang et al., 2015). The extensive use of antibiotics has facilitated the emergence of antibiotic resistance in L. monocytogenes (Wilson et al., 2018). The first antibiotic-resistant L. monocytogenes strain was isolated in 1988. An increasing number of antibiotic-resistant L. monocytogenes strains are being reported worldwide. Ampicillin, amoxicillin with or without gentamicin, and trimethoprim-sulfamethoxazole are the first-line therapeutic antibiotics used for listeriosis treatment (Alonso-Hernando et al., 2012). Approximately 40.0% of the strains isolated in this study were ampicillin-resistant, indicating the necessity for regular surveillances for resistance against one of the oldest antibiotics used in livestock breeding and patient treatment. In addition, 11.8% of the isolates in this study were tetracycline-resistant; this result was consistent with previous studies, in which tetracycline-resistant isolates were frequently recovered from foods (Bertrand et al., 2016; Akrami-Mohajeri et al., 2018; Noll et al., 2018). A total of 1,540 tons of tetracycline was used for humans and livestock in 2013 in China (Zhang et al., 2015); the widespread resistance may be attributed to this excessive use of tetracycline. Fluoroquinolone antibiotics are also extensively used in both human and livestock; in 2013, 25,500 tons of these antibiotics were used in China (Zhang et al., 2015). The results of this study showed that 45.0 and 4.6% of the isolates were intermediate-resistant or resistant to ciprofloxacin, respectively; further, 20.3% of the isolates were intermediate-resistant to levofloxacin (Table 4). These results were consistent with the results of previous studies (Chen et al., 2015; Wieczorek and Osek, 2017; Wilson et al., 2018). It has to be noted that intermediate-resistant strains could develop into completely resistant strains under certain circumstances (Ruiz-Bolivar et al., 2011). Several molecular mechanisms for ciprofloxacin-resistance have been documented, including gene mutations (Godreuil et al., 2003), efflux pump (Godreuil et al., 2003; Guerin et al., 2014; Bertrand et al., 2016), and plasmid-mediated resistance (Wang et al., 2003; Jacoby et al., 2006; Jiang et al., 2008). In recent years, the high prevalence of ciprofloxacin, tetracycline, and streptomycin of L. monocytogenes isolated from foods were reported in China (Yan et al., 2010, Yan et al., 2014), suggesting that the abuse of these antibiotics may accelerate the emergence of antibiotic resistance in L. monocytogenes. Although we did detect multidrug-resistant L. monocytogenes strains, the majority of isolates were sensitive to antibiotics commonly used in listeriosis treatment. Over 90% of the isolates were susceptible to 11 antibiotics, including kanamycin, gentamicin, ofloxacin, sulfamethoxazole with trimethoprim, doxycycline, meropenem, linezolid, sulbactam/ampicillin, penicillin, vancomycin, and amoxicillin/claulanic acid, which are commonly used to treat human listeriosis (Olaimat et al., 2018). However, the emerging threat of antibiotic resistance highlights the necessity for continuous surveillance and elucidation of molecular mechanisms behind antibiotics resistance in L. monocytogenes from foods, environment, and clinical cases.
In conclusion, 458 L. monocytogenes strains were isolated from 1212 meat and meat product samples. These strains were characterized based on serogroup, antibiotic susceptibility, and MLST. Five STs (ST8, ST9, ST87, ST155, and ST121) were predominant in meat and meat products. Several isolates carried llsX and/or pstA virulence factors, which play an important role in human listeriosis diseases, posing a potential public health concern for consumers. In addition, the rising trend of antibiotics resistance in L. monocytogenes suggests that strict regulations to restrict the abuse of antibiotics should be formulated urgently.
Author Contributions
QW, JZ, and MC conceived and designed the experiments. MC, JC, and YC performed the experiments. LX, HZ, SW, RP, and HW conducted the bioinformatics analyses. MC, QW, SZ, TL, and XW drafted the manuscript. QW, YZ, and YD reviewed the manuscript. All authors read and approved the final manuscript.
Funding
We would like to acknowledge the financial support from the National Natural Science Foundation of China (31701718 and 31501580), the Natural Science Foundation of Guangdong Province, China (2017A030313173), the Pearl River S&T Nova Program of Guangzhou (201710010018), and the GDAS’ Special Project of Science and Technology Development (2017GDASCX-0201).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JZ declared a shared affiliation, with no collaboration, with one of the authors, YC, to the handling editor at the time of review.
Acknowledgments
The authors would like to thank the team of curators of the Institute Pasteur MLST databases for curating the data and making them publicly available at http://bigsdb.pasteur.fr/.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00946/full#supplementary-material
Footnotes
References
Akrami-Mohajeri, F., Derakhshan, Z., Ferrante, M., Hamidiyan, N., Soleymani, M., Conti, G. O., et al. (2018). The prevalence and antimicrobial resistance of Listeria spp in raw milk and traditional dairy products delivered in Yazd, central Iran (2016). Food Chem. Toxicol. 114, 141–144. doi: 10.1016/j.fct.2018.02.006
Alonso-Hernando, A., Prieto, M., Garcia-Fernandez, C., Alonso-Calleja, C., and Capita, R. (2012). Increase over time in the prevalence of multiple antibiotic resistance among isolates of Listeria monocytogenes from poultry in Spain. Food Control 23, 37–41. doi: 10.1016/j.foodcont.2011.06.006
Anonymous (2010). National Food Safety Standard Food Microbiological Examination: Listeria monocytogenes (GB4789.30-2010). Beijing: Ministry of Health of People’s Republic of China.
Bertrand, S., Ceyssens, P. J., Yde, M., Dierick, K., Boyen, F., Vanderpas, J., et al. (2016). Diversity of Listeria monocytogenes strains of clinical and food chain origins in Belgium between 1985 and 2014. PLoS One 11:e0164283. doi: 10.1371/journal.pone.0164283
Castro, H., Jaakkonen, A., Hakkinen, M., Korkeala, H., and Lindström, M. (2018). Occurrence, persistence, and contamination routes of Listeria monocytogenes genotypes on three Finnish dairy cattle farms: a longitudinal study. Appl. Environ. Microbiol. 84, e2000–e2017. doi: 10.1128/AEM.02000-17
Chau, M. L., Aung, K. T., Hapuarachchi, H. C., Lee, P. S., Lim, P. Y., Kang, J. S., et al. (2017). Microbial survey of ready-to-eat salad ingredients sold at retail reveals the occurrence and the persistence of Listeria monocytogenes sequence types 2 and 87 in pre-packed smoked salmon. BMC Microbiol. 17:46. doi: 10.1186/s12866-017-0956-z
Chen, M., Cheng, J., Wu, Q., Zhang, J., Chen, Y., Xue, L., et al. (2018a). Occurrence, antibiotic resistance, and population diversity of Listeria monocytogenes isolated from fresh aquatic products in China. Front. Microbiol. 9:2215. doi: 10.3389/fmicb.2018.02215
Chen, M., Cheng, J., Wu, Q., Zhang, J., Chen, Y., Zeng, H., et al. (2018b). Prevalence, potential virulence, and genetic diversity of Listeria monocytogenes isolates from edible mushrooms in chinese markets. Front. Microbiol. 9:1711. doi: 10.3389/fmicb.2018.01711
Chen, M., Wu, Q., Zhang, J., Wu, S., and Guo, W. (2015). Prevalence, enumeration, and pheno- and genotypic characteristics of Listeria monocytogenes isolated from raw foods in South China. Front. Microbiol. 6:1026. doi: 10.3389/fmicb.2015.01026
Chen, M. T., Wu, Q. P., Zhang, J. M., Yan, Z. A., and Wang, J. (2014). Prevalence and characterization of Listeria monocytogenes isolated from retail-level ready-to-eat foods in South China. Food Control 38, 1–7. doi: 10.1016/j.foodcont.2013.09.061
Clayton, E. M., Hill, C., Cotter, P. D., and Ross, R. P. (2011). Real-time PCR assay to differentiate listeriolysin s-positive and -negative strains of Listeria monocytogenes. Appl. Environ. Microbiol. 77, 163–171. doi: 10.1128/AEM.01673-10
Clinical, and Laboratory Standards Institute [CLSI] (2014). Performance Standards for Antimicrobial Susceptibility Testing: 24th Informational Supplement(M100- S24). Wayne, PA: Clinical and Laboratory Standards Institute.
Cotter, P. D., Draper, L. A., Lawton, E. M., Daly, K. M., Groeger, D. S., Casey, P. G., et al. (2008). Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 4:e1000144. doi: 10.1371/journal.ppat.1000144
Derra, F. A., Karlsmose, S., Monga, D. P., Mache, A., Svendsen, C. A., Felix, B., et al. (2013). Occurrence of Listeria spp. in retail meat and dairy products in the area of Addis Ababa, Ethiopia. Foodborne Pathog. Dis. 10, 577–579. doi: 10.1089/fpd.2012.1361
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi: 10.1128/jcm.42.8.3819-3822.2004
Drevets, D. A., and Bronze, M. S. (2008). Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol. Med. Microbiol. 53, 151–165. doi: 10.1111/j.1574-695X.2008.00404.x
Felix, B., Feurer, C., Maillet, A., Guillier, L., Boscher, E., Kerouanton, A., et al. (2018). Population genetic structure of Listeria monocytogenes strains isolated from the pig and pork production chain in France. Front. Microbiol. 9:684. doi: 10.3389/fmicb.2018.00684
Feng, Y. F., Wu, S. Y., Varma, J. K., Klena, J. D., Angulo, F. J., and Ran, L. (2013). Systematic review of human listeriosis in China, 1964-2010. Trop. Med. Int. Health 18, 1248–1256. doi: 10.1111/tmi.12173
Godreuil, S., Galimand, M., Gerbaud, G., Jacquet, C., and Courvalin, P. (2003). Efflux pump lde is associated with fluoroquinolone resistance in Listeria monocytogenes. Antimicrob. Agents Chemother. 47, 704–708. doi: 10.1128/Aac.47.2.704-708.2003
Gombas, D. E., Chen, Y., Clavero, R. S., and Scott, V. N. (2003). Survey of Listeria monocytogenes in ready-to-eat foods. J. Food Prot. 66, 559–569.
Goulet, V., Hebert, M., Hedberg, C., Laurent, E., Vaillant, V., De Valk, H., et al. (2012). Incidence of listeriosis and related mortality among groups at risk of acquiring listeriosis. Clin. Infect. Dis. 54, 652–660. doi: 10.1093/cid/cir902
Guenther, S., Huwyler, D., Richard, S., and Loessner, M. J. (2009). Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 75, 93–100. doi: 10.1128/AEM.01711-08
Guerin, F., Galimand, M., Tuambilangana, F., Courvalin, P., and Cattoir, V. (2014). Overexpression of the novel MATE fluoroquinolone efflux pump FepA in Listeria monocytogenes Is driven by inactivation of its local repressor FepR. PLoS One 9:e106340. doi: 10.1371/journal.pone.0106340
Hamidiyan, N., Salehi-Abargouei, A., Rezaei, Z., Dehghani-Tafti, R., and Akrami-Mohajeri, F. (2018). The prevalence of Listeria spp. food contamination in Iran: a systematic review and meta-analysis. Food Res. Int. 107, 437–450. doi: 10.1016/j.foodres.2018.02.038
Hoelzer, K., Oliver, H. F., Kohl, L. R., Hollingsworth, J., Wells, M. T., and Wiedmann, M. (2012). Structured expert elicitation about Listeria monocytogenes cross-contamination in the environment of retail deli operations in the United States. Risk Anal. 2, 1139–1156. doi: 10.1111/j.1539-6924.2011.01729.x
Hof, H. (2004). An update on the medical management of listeriosis. Exp. Opin. Pharmacother. 5, 1727–1735. doi: 10.1517/14656566.5.8.1727
Jacoby, G. A., Walsh, K. E., Mills, D. M., Walker, V. J., Oh, H., Robicsek, A., et al. (2006). qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50, 1178–1182. doi: 10.1128/Aac.50.4.1178-1182.2006
Jiang, Y., Zhou, Z. H., Qian, Y., Wei, Z. Q., Yu, Y. S., Hu, S. N., et al. (2008). Plasmid-mediated quinolone resistance determinants qnr and aac(6’)-Ib-cr in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 61, 1003–1006. doi: 10.1093/jac/dkn063
Khen, B. K., Lynch, O. A., Carroll, J., McDowell, D. A., and Duffy, G. (2015). Occurrence, antibiotic resistance and molecular characterization of Listeria monocytogenes in the beef Chain in the Republic of Ireland. Zoonoses Public Health 62, 11–17. doi: 10.1111/zph.12106
Kim, S. W., Haendiges, J., Keller, E. N., Myers, R., Kim, A., Lombard, J. E., et al. (2018). Genetic diversity and virulence profiles of Listeria monocytogenes recovered from bulk tank milk, milk filters, and milking equipment from dairies in the United States (2002 to 2014). PLoS One 13:e0197053. doi: 10.1371/journal.pone.0197053
Korsak, D., Borek, A., Daniluk, S., Grabowska, A., and Pappelbaum, K. (2012). Antimicrobial susceptibilities of Listeria monocytogenes strains isolated from food and food processing environment in Poland. Int. J. Food Microbiol. 158, 203–208. doi: 10.1016/j.ijfoodmicro.2012.07.016
Kreft, J., and Vazquez-Boland, J. A. (2001). Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291, 145–157. doi: 10.1078/1438-4221-00111
Li, H., Wang, P., Lan, R., Luo, L., Cao, X., Wang, Y., et al. (2018a). Risk factors and level of Listeria monocytogenes contamination of raw pork in retail markets in China. Front. Microbiol. 9:1090. doi: 10.3389/fmicb.2018.01090
Li, L., Olsen, R. H., Ye, L., Wang, W., Shi, L., Yan, H., et al. (2016). Characterization of antimicrobial resistance of Listeria monocytogenes strains isolated from a pork processing plant and its respective meat markets in southern China. Foodborne Pathog. Dis. 13, 262–268. doi: 10.1089/fpd.2015.2087
Li, W., Bai, L., Fu, P., Han, H., Liu, J., and Guo, Y. (2018b). The epidemiology of Listeria monocytogenes in China. Foodborne Pathog. Dis. 15, 459–466. doi: 10.1089/fpd.2017.2409
Liu, H. X., Xu, Y., Yang, Z. S., Tang, X. Z., Zou, Y. Q. S., Guo, Y. D., et al. (2017). Contamination of Listeria monocytogenes in cooked meat products and catering food in Yunnan in 2010-2016. J. Food Saf. Qual. 8, 3768–3772.
Ma, A. (2015). Isolation and Characterization of Listeria monocytogenes Isolated from Raw Meat and Aquatic Product Samples. Beijing: Chiense Center for Disease Control and Prevention.
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Martin, B., Perich, A., Gomez, D., Yanguela, J., Rodriguez, A., Garriga, M., et al. (2014). Diversity and distribution of Listeria monocytogenes in meat processing plants. Food Microbiol. 44, 119–127. doi: 10.1016/j.fm.2014.05.014
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
Modzelewska-Kapitula, M., and Maj-Sobotka, K. (2014). The microbial safety of ready-to-eat raw and cooked sausages in Poland: Listeria monocytogenes and Salmonella spp. occurrence. Food Control 36, 212–216. doi: 10.1016/j.foodcont.2013.08.035
Ndahi, M. D., Kwaga, J. K., Bello, M., Kabir, J., Umoh, V. J., Yakubu, S. E., et al. (2014). Prevalence and antimicrobial susceptibility of Listeria monocytogenes and methicillin-resistant Staphylococcus aureus strains from raw meat and meat products in Zaria, Nigeria. Lett. Appl. Microbiol. 58, 262–269. doi: 10.1111/lam.12183
Noll, M., Kleta, S., and Al Dahouk, S. (2018). Antibiotic susceptibility of 259 Listeria monocytogenes strains isolated from food, food-processing plants and human samples in Germany. J. Infect. Public Health 11, 572–577. doi: 10.1016/j.jiph.2017.12.007
Olaimat, A. N., Al-Holy, M. A., Shahbaz, H. M., Al-Nabulsi, A. A., Abu Ghoush, M. H., Osaili, T. M., et al. (2018). Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: a comprehensive review. Compar. Rev. Food Sci. Food Saf. 17, 1277–1292. doi: 10.1111/1541-4337.12387
Orsi, R. H., Bakker, H. C. D., and Wiedmann, M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi: 10.1016/j.ijmm.2010.05.002
Pesavento, G., Ducci, B., Nieri, D., Comodo, N., and Lo Nostro, A. (2010). Prevalence and antibiotic susceptibility of Listeria spp. isolated from raw meat and retail foods. Food Control 21, 708–713. doi: 10.1016/j.foodcont.2009.10.012
Quereda, J. J., Dussurget, O., Nahori, M. A., Ghozlane, A., Volant, S., Dillies, M. A., et al. (2016). Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. U.S.A. 113, 5706–5711. doi: 10.1073/pnas.1523899113
Quereda, J. J., Meza-Torres, J., Cossart, P., and Pizarro-Cerda, J. (2017a). Listeriolysin S: a bacteriocin from epidemic Listeria monocytogenes strains that targets the gut microbiota. Gut Microbes 8, 384–391. doi: 10.1080/19490976.2017.1290759
Quereda, J. J., Nahori, M. A., Meza-Torres, J., Sachse, M., Titos-Jimenez, P., Gomez-Laguna, J., et al. (2017b). Listeriolysin S is a streptolysin s-like virulence factor that targets exclusively prokaryotic cells in vivo. Mbio 8:e259-17. doi: 10.1128/mBio.00259-17
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. doi: 10.1371/journal.ppat.1000146
Ristori, C. A., Rowlands, R. E., Martins, C. G., Barbosa, M. L., Yoshida, J. T., and Franco, B. D. (2014). Prevalence and populations of Listeria monocytogenes in meat products retailed in São Paulo, Brazil. Foodborne Pathog Dis. 11, 969–973. doi: 10.1089/fpd.2014.1809
Ruiz-Bolivar, Z., Neuque-Rico, M. C., Poutou-Pinales, R. A., Carrascal-Camacho, A. K., and Mattar, S. (2011). Antimicrobial susceptibility of Listeria monocytogenes food isolates from different cities in Colombia. Foodborne Pathog. Dis. 8, 913–919. doi: 10.1089/fpd.2010.0813
Rychli, K., Stessl, B., Szakmary-Brandle, K., Strauss, A., Wagner, M., and Schoder, D. (2018). Listeria monocytogenes isolated from illegally imported food products into the european union harbor different virulence factor variants. Genes (Basel.) 9:E428. doi: 10.3390/genes9090428
Salvat, G., and Fravalo, P. (2004). Risk assessment strategies for Europe: integrated safety strategy or final product control: example of Listeria monocytogenes in processed products from pork meat industry. Dtsch Tierarztl Wochenschr 111, 331–334.
Shen, J., Rump, L., Zhang, Y., Chen, Y., Wang, X., and Meng, J. (2013). Molecular subtyping and virulence gene analysis of Listeria monocytogenes isolates from food. Food Microbiol. 35, 58–64. doi: 10.1016/j.fm.2013.02.014
Silk, B. J., Date, K. A., Jackson, K. A., Pouillot, R., Holt, K. G., Graves, L. M., et al. (2012). Invasive listeriosis in the foodborne diseases active surveillance network (FoodNet), 2004-2009: further targeted prevention needed for higher-risk groups. Clin. Infect. Dis. 54(Suppl. 5), S396–S404. doi: 10.1093/cid/cis268
Simmons, C., Stasiewicz, M. J., Wright, E., Warchocki, S., Roof, S., Kause, J. R., et al. (2014). Listeria monocytogenes and Listeria spp. contamination patterns in retail delicatessen establishments in three U.S. states. J. Food Prot. 77, 1929–1939. doi: 10.4315/0362-028X.JFP-14-183
United States Department of Agriculture. (1998). “Isolation and identification of Listeria monocytogenes from meat, poultry and egg products,” in Microbiology Guide Book, 3rd Edn, Chap.8, eds B. P. Dey and C. P. Lattuada (Washington, DC: Government Printing Office).
Vallim, D. C., Hofer, C. B., Lisboa, R. D., Barbosa, A. V., Rusak, L. A., dos Reis, C. M. F., et al. (2015). Twenty Years of Listeria in Brazil: occurrence of Listeria species and Listeria monocytogenes serovars in food samples in Brazil between 1990 and 2012. Biomed. Res. Int. 2015:540204. doi: 10.1155/2015/540204
Wang, H., Luo, L., Zhang, Z., Deng, J., Wang, Y., Miao, Y., et al. (2018). Prevalence and molecular characteristics of Listeria monocytogenes in cooked products and its comparison with isolates from listeriosis cases. Front. Med. 12:104–112. doi: 10.1007/s11684-017-0593-9
Wang, M. G., Tran, J. H., Jacoby, G. A., Zhang, Y. Y., Wang, F., and Hooper, D. C. (2003). Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47, 2242–2248. doi: 10.1128/Aac.47.7.2242-2248.2003
Wang, Y., Jiao, Y., Lan, R., Xu, X., Liu, G., Wang, X., et al. (2015). Characterization of Listeria monocytogenes isolated from human Listeriosis cases in China. Emerg. Microbes Infect. 4:e50. doi: 10.1038/emi.2015.50
Wang, Y., Zhao, A., Zhu, R., Lan, R., Jin, D., Cui, Z., et al. (2012). Genetic diversity and molecular typing of Listeria monocytogenes in China. BMC Microbiol. 12:119. doi: 10.1186/1471-2180-12-119
Wen, T., Wang, W. S., Sun, W., and Li, X. (2015). Contamination of Listeria monocytogenes in foods in Liaoning province,2010-2014. Chin. J. Public Health 31, 1475–1477. doi: 10.11847/zgggws2015-31-11-33
Wieczorek, K., Dmowska, K., and Osek, J. (2012). Characterization and antimicrobial resistance of Listeria monocytogenes isolated from retail beef meat in Poland. Foodborne Pathog. Dis. 9, 681–685. doi: 10.1089/fpd.2012.1137
Wieczorek, K., and Osek, J. (2017). Prevalence, genetic diversity and antimicrobial resistance of Listeria monocytogenes isolated from fresh and smoked fish in Poland. Food Microbiol. 64, 164–171. doi: 10.1016/j.fm.2016.12.022
Wilson, A., Gray, J., Chandry, P. S., and Fox, E. M. (2018). Phenotypic and genotypic analysis of antimicrobial resistance among Listeria monocytogenes isolated from australian food production Chains. Genes 9, E80. doi: 10.3390/genes9020080
Wu, S., Wu, Q., Zhang, J., Chen, M., and Guo, W. (2016). Analysis of multilocus sequence typing and virulence characterization of Listeria monocytogenes isolates from chinese retail ready-to-eat food. Front. Microbiol. 7:168. doi: 10.3389/fmicb.2016.00168
Yan, H., Neogi, S. B., Mo, Z., Guan, W., Shen, Z., Zhang, S., et al. (2010). Prevalence and characterization of antimicrobial resistance of foodborne Listeria monocytogenes isolates in Hebei province of Northern China, 2005-2007. Int. J. Food Microbiol. 144, 310–316. doi: 10.1016/j.ijfoodmicro.2010.10.015
Yan, S., Yang, D., Yu, D., Gan, X., Wang, W., Hu, Y., et al. (2014). Antibiotic resistance and MLST analysis of foodborne pathogenic Listeria monocytogenes in China in 2012. Chin. J. Food Hyg. 2014, 537–542.
Yang, X. J., Zhao, W., Liu, G. H., Huang, X., Wang, H., Zhang, W. Y., et al. (2017). Analysis on the monitoring data of Listeria monocytogenes in food of Jilin Province in 2011∼2015. J. Food Saf. Qual. 8, 105–110.
Zhang, Q. Q., Ying, G. G., Pan, C. G., Liu, Y. S., and Zhao, J. L. (2015). Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 49, 6772–6782. doi: 10.1021/acs.est.5b00729
Keywords: Listeria monocytogenes, animal-derived food products, LIPI-3, LIPI-4, multi-locus sequence typing, antibiotic resistance
Citation: Chen M, Cheng J, Zhang J, Chen Y, Zeng H, Xue L, Lei T, Pang R, Wu S, Wu H, Zhang S, Wei X, Zhang Y, Ding Y and Wu Q (2019) Isolation, Potential Virulence, and Population Diversity of Listeria monocytogenes From Meat and Meat Products in China. Front. Microbiol. 10:946. doi: 10.3389/fmicb.2019.00946
Received: 04 December 2018; Accepted: 15 April 2019;
Published: 07 May 2019.
Edited by:
Learn-Han Lee, Monash University Malaysia, MalaysiaReviewed by:
Jianmin Zhang, South China Agricultural University, ChinaPeter Bergholz, North Dakota State University, United States
Copyright © 2019 Chen, Cheng, Zhang, Chen, Zeng, Xue, Lei, Pang, Wu, Wu, Zhang, Wei, Zhang, Ding and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, d3VxcDIwM0AxNjMuY29t
†Co-first authors
 Moutong Chen
Moutong Chen Jianheng Cheng
Jianheng Cheng Jumei Zhang
Jumei Zhang Yuetao Chen1,2
Yuetao Chen1,2 Haiyan Zeng
Haiyan Zeng Liang Xue
Liang Xue Tao Lei
Tao Lei Rui Pang
Rui Pang Shi Wu
Shi Wu Haoming Wu
Haoming Wu Yu Ding
Yu Ding Qingping Wu
Qingping Wu