- 1College of Animal Science and Technology, Northwest A&F University, Xianyang, China
- 2Department of Animal Science, McGill University, Montreal, QC, Canada
Two-component systems (TCSs) are dominant regulating components in bacteria for responding to environmental stimuli. However, little information is available on how TCSs in Enterococcus faecium respond to bile salts – an important environmental stimulus for intestinal bacteria. In this study, the gene expression of 2 TCSs, BsrXRS and LiaFSR, was positively correlated with survival rates of different E. faecium isolates during exposure to ox gall. Moreover, gene disruptions of bsrR, bsrS, liaS, and liaR significantly reduced the survival rates of E. faecium in the presence of ox gall. Finally, EMSA results indicated that BsrR functioned as a transcription regulator for expression of its own gene as well as lipoate-protein ligase A (lplA). Additional 27 potential target genes by BsrR were revealed through in silico analyses. These findings suggest that BsrXRS and LiaFSR systems play important roles in bile salt resistance in E. faecium.
Introduction
Bile salts are known as a natural detergent that emulsifies and solubilizes lipids, thereby playing a crucial role in fat digestion and absorption. At the same time, the amphipathic property of bile salts may induce membrane damage and impair membrane functions of intestinal bacteria. In addition to attacking cell membrane, bile salts have numerous other toxic effects on bacterial cells including disturbing macromolecule stability and inducing oxidative stress, DNA damage and protein misfolding in bacterial cells (Ruiz et al., 2013). Thus, the ability to adapt and respond to bile salts is essential for the survival and persistence of intestinal bacteria in gastrointestinal tracts.
Two-component systems (TCSs) are a primary means by which bacteria sense and respond to their environmental changes (Capra and Laub, 2012; Groisman, 2016). Although exact mechanisms for bacteria to respond to bile salts have not been completely elucidated, it is likely to be associated with TCSs. A typical TCS consists of two proteins: a signal-sensing histidine protein kinase (HK) in the cell membrane that senses an environmental change, and a cytoplasmic cognate response regulator (RR) that generates an appropriate response, usually regulating expression of target genes.
Several studies have shown that several types of bacteria can up-regulate expression of TCS genes and proteins in the presence of bile salts. For example, exposure to bile salts significantly increased expression of baeS-baeR, phoP3-phoR3, and vraR-vraS in Lactobacillus rhamnosus GG (Koskenniemi et al., 2011). A whole-genome microarray analysis of Lactobacillus acidophilus NCFM found a significant up-regulation of a TCS operon (LBA1425 to LBA1432) in response to bile salts and mutations in the HK (LBA1430) and the RR (LBA1431) of this operon decreased the survival rates (Pfeiler et al., 2007). Similarly, basR and basS genes were up-regulated after exposure to bile salts in Escherichia coli O157:H7 (Kus et al., 2011). E. coli Nissle 1917 became more resistant to bile salts after introduction of a mutated envZ (envZP41L) from E. coli MG1655 (the envZ gene encodes a histidine kinase and induces a higher level of phosphorylated OmpR) (Adediran et al., 2014). In Propionibacterium freudenreichii SI41, addition of bile salts was found to induce expression of a putative RR (ORF0001) protein at two-dimensional electrophoresis (Leverrier et al., 2003). In another study, a RR (BL1000) was detected when exposure to ox bile in Bifidobacterium longum NCIMB 8809 (Sanchez et al., 2005). Disruption of cbrR in Campylobacter jejuni F38011 rendered the strain more sensitive to ox bile (Raphael et al., 2005). Mutation of the cpxAR in Klebsiella pneumoniae NTUH-K2044 led to low survival rates during exposure to bile at the concentrations higher than 0.5% (Srinivasan et al., 2012). Le Breton et al. (2003) found that mRNA amount of ehk10-err10 was increased after incubation with 0.08% bile salts in Enterococcus faecalis JH2-2. However, the expression of ehk10-err10 TCS was not significantly increased in a transcriptional analysis of E. faecalis V583 response to ox gall (Solheim et al., 2007). Collectively, these findings suggest that TCSs are essential for resistance to bile salts in various bacterial species. However, there is no information available on involvement of TCSs in the bile resistance in E. faecium.
Enterococcus faecium is a common commensal organism in the gastrointestinal tract of a wide variety of hosts. At the same time, it is also an important cause of multidrug-resistant hospital-acquired infection (Lebreton et al., 2013). Moreover, E. faecium strains have been considered as potential bio-preservatives due to the strong antimicrobial activity against Listeria monocytogenes in fermented food production (Pingitore et al., 2012; Favaro et al., 2014). As an important group of lactic acid bacteria (LAB), E. faecium has been used as probiotics to confer health benefit to the host (Veljovic et al., 2017). E. faecium harbors more than 15 TCSs in the chromosome and in plasmids (Ortet et al., 2015). Four TCSs in E. faecium, CroRS, ChtRS, LiaFSR, and VanSR, have been characterized for antibiotic resistance. The CroRS system responds to cell wall stress and is critical for the resistance against cell wall-targeting antibiotics in E. faecium. Mutation of croRS in the E. faecium 1,141,733 strain remarkably reduced the resistance to cefepime and ceftriaxone (Kellogg et al., 2017). Guzman Prieto et al. (2017) revealed that the ChtRS system contributed to chlorhexidine tolerance. Mutants of chtR and chtS exhibited a slower exponential growth rate than the wild-type E. faecium E1162 in the presence of chlorhexidine. The LiaFSR system is an important regulator of the cell envelope stress response to the membrane-targeting agents (Tran et al., 2016). This system consists of a sensor histidine kinase (LiaS), a response regulator (LiaR) and a membrane-anchored protein (LiaF) that negatively affect the function of LiaS. The role of LiaFSR in enterococci has been described recently in resistance to daptomycin (Tran et al., 2016). Deletion of liaR in E. faecium R446F and R497 that resistant to daptomycin fully reversed resistance (Panesso et al., 2015). The VanSR two-component system regulates the downstream van gene cluster, which alters the cell wall precursors to allow bacteria to resist vancomycin (Hong et al., 2008).
The objective of this study was to identify potential TCSs in the resistance of E. faecium to bile salts. Our results show that the gene expression of liaS, liaR, bsrR, and bsrS was positively correlated with survival rate of different E. faecium isolates after exposure to bile salts. Moreover, gene disruptions of bsrR, bsrS, liaS, and liaR significantly reduced the survival rates of E. faecium after exposure to different concentrations of bile salts. In addition, BsrR could function as a transcription regulator for its own expression and expression of lplA. The in silico analyses also revealed 27 other target genes for BsrR. These findings provide a first glance at the molecular mechanisms for bile salt resistance in E. faecium.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
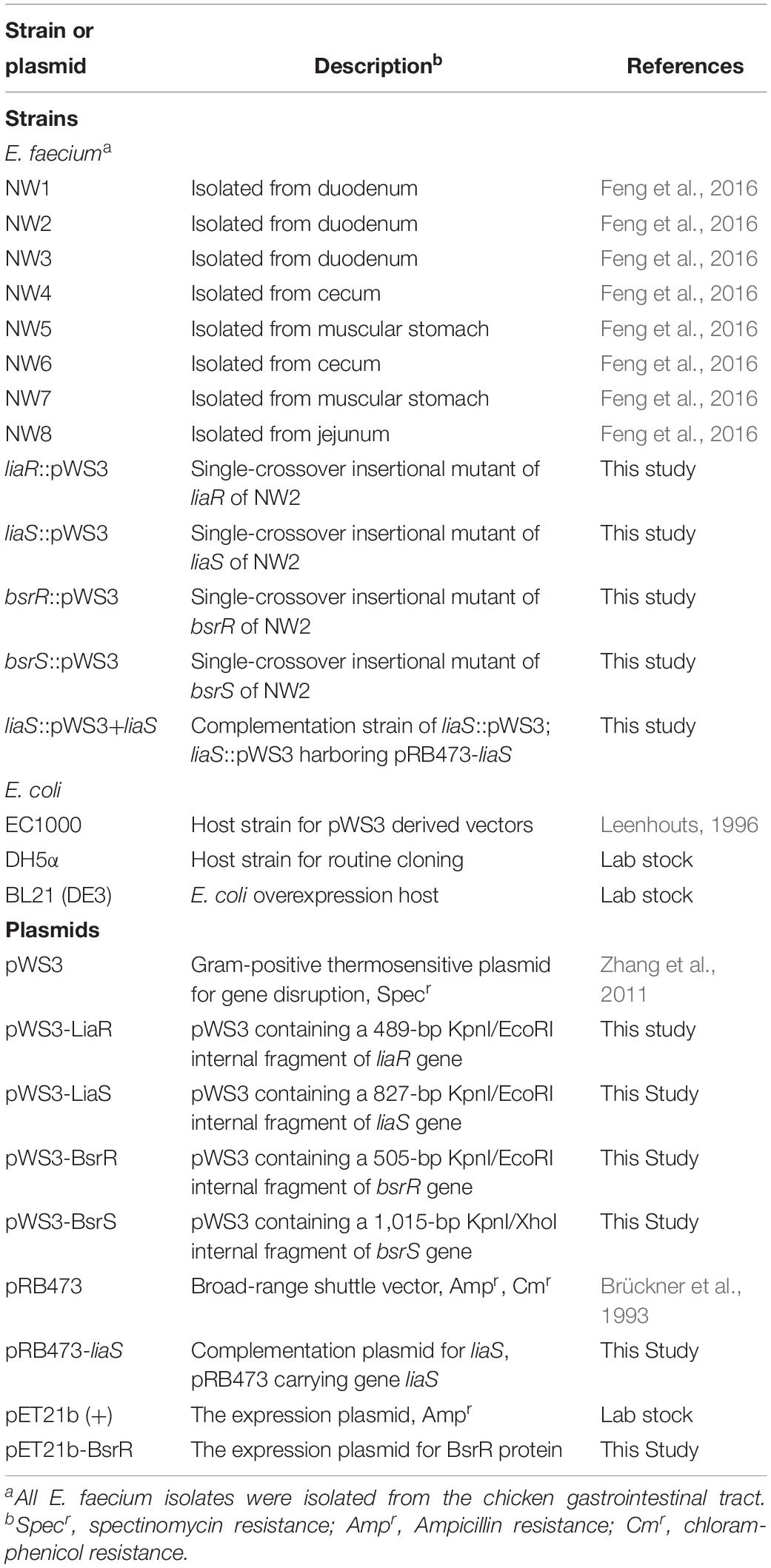
Enterococcus faecium isolates, E. coli strains and plasmids used in this study are listed in Table 1. E. faecium were isolated from a previous study (Feng et al., 2016). E. faecium isolates were identified by 16s rRNA gene sequencing and subsequent blasting of the sequences, using the Basic Local Alignment Search Tool (BLAST) program. The genomic DNA was extracted from overnight culture using an EasyPure Genomic DNA kit (TransGen Biotech, Beijing, China) as per manufacturer’s protocol. A PCR reaction was performed to amplify the 16S rRNA genes using universal primers 27F and 1492R (Supplementary Table S1). PCR products were then sequenced using the Sanger sequencing. E. faecium isolates were routinely grown in the de Man, Rogosa, and Sharpe (MRS) broth or on the agar at 37°C. E. coli EC1000 (Leenhouts, 1996) was used for plasmid construction and was grown in the Luria-Bertani (LB) broth at 30°C. Where necessary, antibiotics were used at the following concentrations: chloramphenicol 5 μg/ml for E. faecium, spectinomycin 300 μg/ml for E. faecium, and 100 μg/ml for E. coli, ampicillin 100 μg/ml for E. coli. All antibiotics were obtained from Sigma (St. Louis, MO, United States).
Resistance of E. faecium to Bile Salts
To determine the resistant abilities of 8 E. faecium isolates to bile salts, overnight cultures were diluted 100-fold in 20 ml of the fresh MSR medium. When bacterial cells were grown to the mid-log-phase (OD600 of ∼0.4), 1 ml aliquots of the cultures were collected as control samples (t = 0 min). Another 1 ml aliquots of the cultures were collected and centrifuged at 12,000 × g for 2 min, then re-suspended in the same volume of the fresh medium supplemented with different concentrations of 0.5, 1, 2, and 5% bile salts (Ox gall, Sigma, United States). After incubation for 30 min at 37°C (t = 30 min), the numbers of colony-forming units per milliliter (CFU/ml) of each sample were determined by plating a serial of 10-fold dilutions on MRS agar and then incubated at 37°C for 48 h. The percentage survival of E. faecium in bile salts was calculated by comparing CFU at 30 min to CFU at time zero. The assays were performed in three independent experiments.
RNA Preparation, RT-PCR, and RT-qPCR
Eight TCSs in E. faecium genomes were selected to study the effect of TCSs on resistance to bile salts. Among them, 5 TCSs are homologous to TCSs in other species known to be involved in bile resistance (Supplementary Table S2), while ChtRS, CroRS, and LiaFSR were selected because they respond to cell envelope-targeting antimicrobial agents. To quantify gene expression, overnight cultures were diluted 100-fold in 20 ml of the fresh MSR medium. When bacterial cells were grown to the mid-log-phase (OD600 of ∼0.4), 5 ml aliquots of the culture were centrifuged for 2 min at 12,000 × g at 4°C and pellets were flash frozen in liquid N2 prior to RNA extraction. This sample served as the control (t = 0 min) prior to the addition of bile salts. Bile salts (final concentration 0.5%, w/v) were added into the remaining culture. After 30 min of incubation at 37°C, 5 ml aliquots of the culture were centrifuged and flash frozen as described above. Total RNA was extracted by using the E.Z.N.A. Bacterial RNA Kit (Omega, United States). One μg of extracted RNA was converted into cDNA using a PrimeScriptTM RT reagent Kit (Takara, Dalian, China) according to the manufacturer’s instructions. To identify the transcript structure, reverse transcription-PCR (RT-PCR) was performed on cDNA synthesized from wild-type E. faecium NW2 RNA with primers listed in Supplementary Table S1. RNA without RT and genomic DNA were used as the negative control and positive control, respectively. The levels of expression of genes were determined by quantitative real-time RT-PCR (RT-qPCR) using a real-time PCR kit (Takara, Dalian, China) and specific primers (Supplementary Table S1). The transcript levels were determined by the 2–ΔΔCt method using the adk gene as an endogenous control gene.
Generation of Insertional Mutants and in trans Complementation
To verify whether the LiaFSR and BsrXRS systems were responsible for resistance to bile salts, gene disruption mutants were constructed in E. faecium NW2 as previously described Zhang et al. (2011). The internal DNA fragments of liaR, liaS, bsrR, and bsrS genes were amplified using primers LiaR-mut-F/LiaR-mut-R, LiaS-mut-F/LiaS-mut-R, BsrR-mut-F/BsrR-mut-R, and BsrS-mut-F/BsrS-mut-R, respectively (Supplementary Table S1), and cloned to a Gram-positive thermosensitive vector pWS3. The resulting constructs were transformed into E. coli EC1000 for propagation and grown on LB plates containing 100 μg/ml spectinomycin at 30°C. The inserts from each construct were sequenced using the primers M13-F and SK-R to ensure that no mutations arose during cloning. The correct vectors (Table 1) were then electrotransformed into E. faecium NW2 using Gene Pulser XcellTM apparatus (Bio-Rad), operating at 2,500 V, 25 mF capacitance, 200 Ω resistance, and 2-mm cuvettes that contained 2 μg of plasmid DNA plus 100 μl of electrocompetent cells. The electrocompetent cells were obtained as previously described (Zhang et al., 2012). After electrotransformation, the cells were allowed to recover for 2 h at the permissive temperature of 30∘C, after which the cells were plated on BHI plates supplemented with 300 μg/ml spectinomycin at 30°C to select for transformants. Spectinomycin-resistant colonies were picked and grown overnight in 2 ml of BHI broth supplemented with 300 μg/ml spectinomycin at an elevated temperature (37°C) to cure the plasmid. Following serial passages at 37°C, the cells were then plated on BHI agar plates with 300 μg/ml spectinomycin at 37°C. Single-cross-over integration into liaR, liaS, bsrR, or bsrS was verified by PCR with pWS3-specific primer SK-R and gene-specific primers LiaR-d-check, LiaS-d-check, BsrR-d-check, BsrS-d-check, respectively (Supplementary Table S1).
The liaS mutant was complemented in trans by cloning liaS gene into the shuttle plasmid pRB473 (Brückner et al., 1993). The gene lias fragment was generated from the NW2 genomic DNA using primers LiaS-comp-F and LiaS-comp-R (Supplementary Table S1). The PCR product was then ligated into pRB473, generating the complementation plasmid pRB473-liaS. Sequencing of the full insert was performed to verify the absence of errors. The correct plasmid was introduced into the liaS mutant strain by electroporation as described above. Unfortunately, despite repeated attempts, we were unable to obtain complemented strains for liaR, bsrR, and bsrS genes.
Search of the Binding Sites and Target Genes of BsrR in E. faecium in silico
Available complete genome sequence of the E. faecium DO was downloaded from the NCBI RefSeq database in GenBank (.gbk) format. The amino acid sequence of BsrR orthologs from 7 bacterial species (including E. faecium DO) in the Enterococcus genus were downloaded from OrtholugeDB (Whiteside et al., 2013). The upstream DNA sequences of BsrXRS system from 7 bacterial species were obtained from the NCBI GenBank database. Then these DNA sequences were used to search the putative binding sites for BsrR using the motif finding in DMINDA 2.0 web server with default parameters (Yang et al., 2017). As for searching target genes, all promoter sequences in the E. faecium DO genome were downloaded from DOOR 2.0 (Mao et al., 2014) and submitted to DMINDA 2.0. Multiple sequence alignments of DNA and protein sequences were performed using the CLUSTAL W (Thompson et al., 1994).
Overexpression and Purification of BsrR
To further examine if the predicted binding sites are directly bound by BsrR, we used the electrophoretic mobility shift assays (EMSAs) to detect the protein–DNA interactions. The bsrR gene was amplified from the NW2 genome using primers BsrRc-F/BsrRc-R and cloned into the pET21b vector, then was transformed into E. coli strain BL21 (DE3) for overexpressing C-terminally six-His-tagged BsrR. Successful transformants were inoculated into LB broth containing ampicillin (100 μg/ml) and incubated at 37°C with vigorous shaking (200 rpm) to achieve an OD600 of 0.55. Once the correct OD600 obtained, cultures were kept on ice for 10 min followed by the addition of 0.1 mM (final concentration) isopropyl-β-d-1-thiogalactopyranoside (IPTG) to induce BsrR expression. The cultures were further incubated at 18°C for overnight, and the cells were harvested by centrifugation at 12,000 × g for 5 min at 4°C. To purify the recombinant BsrR, the cell pellet was resuspended in binding buffer [15 mM imidazole, 50 mM Tris-HCl, 0.5 mM EDTA, 50 mM NaCl and 10% glycerol (v/v), pH = 7.5] and disrupted by sonication, and the cell debris was removed by centrifugation at 12,000 × g for 15 min at 4°C. This soluble fraction, which contained the BsrR protein, was placed in a HisTrap prepacked column (GE healthcare). The same binding buffer was used for equilibration of the column. Whole-cell lysate was allowed to bind to the resin. The column was then washed five times with binding buffer. Bound BsrR were eluted using elution buffer containing 250 mM imidazole [250 mM imidazole, 50 mM Tris-HCl, 0.5 mM EDTA, 50 mM NaCl, 10% glycerol (v/v), pH = 7.5]. The eluted samples were dialyzed in the storage buffer [10 mM Tris, 50 mM KCl, 1 mM DTT, 10% glycerol (v/v), pH 7.5] at 4°C. The concentration of BsrR protein was determined by the Bradford assay (Bio-Rad) using the bovine serum albumin (BSA) as the standard.
Electrophoretic Mobility Shift Assays (EMSAs)
The intergenic regions upstream of bsrX (143 bp) and lplA (162 bp) were amplified from the E. faecium NW2 genomic DNA using the primers pBsrRS-F/pBsrRS-R and pLplA-F/pLplA-R (Supplementary Table S1), respectively. A DNA fragment as a negative control was amplified from the coding region of housekeeping gene ddlA using the primer pNC-F/pNC-R, which does not share sequence identity with the upstream of bsrX and lplA. The EMSA experiments were performed using the LightShift EMSA Optimization and Control Kit according to the manufacturer’s instructions (Pierce, Thermo Fisher Scientific, Waltham, MA, United States). Briefly, the 5′-end biotin labeled DNA probes (20 fmol) were incubated with various concentrations of recombinant BsrR protein (0, 0.1, 0.2, and 0.3 nM) in binding reaction (20 μl) containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 0.5 mM DTT, 1 mM EDTA, 7.5% glycerol, 1 mg of BSA per ml, and 1 μg of poly (dI-dC). To verify the specificity of BsrR-probe interaction, 200-fold excess amounts of unlabeled non-specific DNA (negative control) or each unlabeled specific probe were first incubated with BsrR protein for 20 min, followed by the addition of each labeled specific probe. Reactions were incubated for 40 min at 25°C, and then were loaded in 6% non-denaturing polyacrylamide gels containing 5% glycerol and electrophoresed in the 0.5 × Tris-borate-EDTA (TBE) for 90 min at 120 V and 4°C. Biotin-labeled DNA-protein complexes were detected using the Chemiluminescent Nucleic Acid Detection Module Kit follow according to the manufacturer’s instructions (Pierce, Thermo Fisher Scientific).
Statistical Analysis
One-way analysis of variance (ANOVA) was used to analyze data using the R software (Version 3.4.3) (R Core Team, 2017). The differences between treatment means were calculated using the Duncan’s new multiple range test in the Agricolae Packages (Version 1.2-8) (Mendiburu, 2017). Data are expressed as mean ± standard deviation (SD) and P < 0.05 was considered significantly different.
Results
Resistance of E. faecium to Ox Gall
To assay the resistant abilities to ox gall of 8 E. faecium isolates, the survival rates were measured in the MRS broth supplemented with different concentrations of ox gall. The resistant activities to bile salts varied widely among the isolates. After exposure to 1% ox gall, more than 20% of E. faecium NW1, 2, and 3 remained viable, whereas the survival rate of NW7 and 8 fell below 1% (Figure 1A). After exposure to 2% ox gall, E. faecium NW1, 2, 3 still had more than 1% survival rates, whereas survival rates of E. faecium NW7 and 8 were lower than 0.1% (Figure 1B). When the concentration of ox gall was increased to 5%, the survival of all E. faecium isolates except NW1 was less than 0.1% (Figure 1C). On the other hand, three isolates (E. faecium NW1, 2, and 3) showed more than 70% survival rates after exposure to 0.5% ox gall (Figure 2A). The 0.5% ox gall was selected for further studies, unless indicated otherwise.
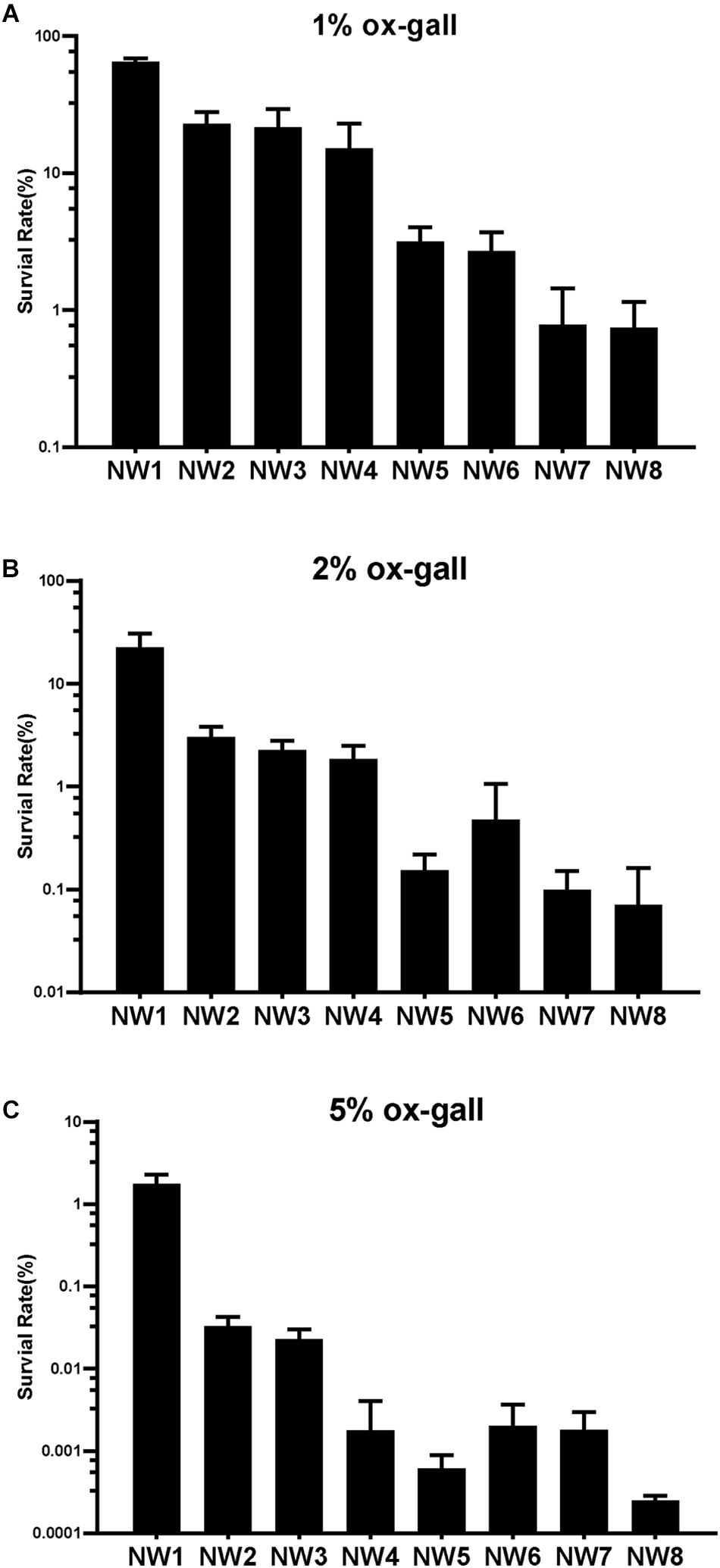
Figure 1. Resistance of Enterococcus faecium to bile salts. Survival rates of E. faecium isolates after 30 min exposure to 1 (A), 2 (B), and 5% (C) ox gall. Data are shown as mean ± SD from three independent biological replicates.
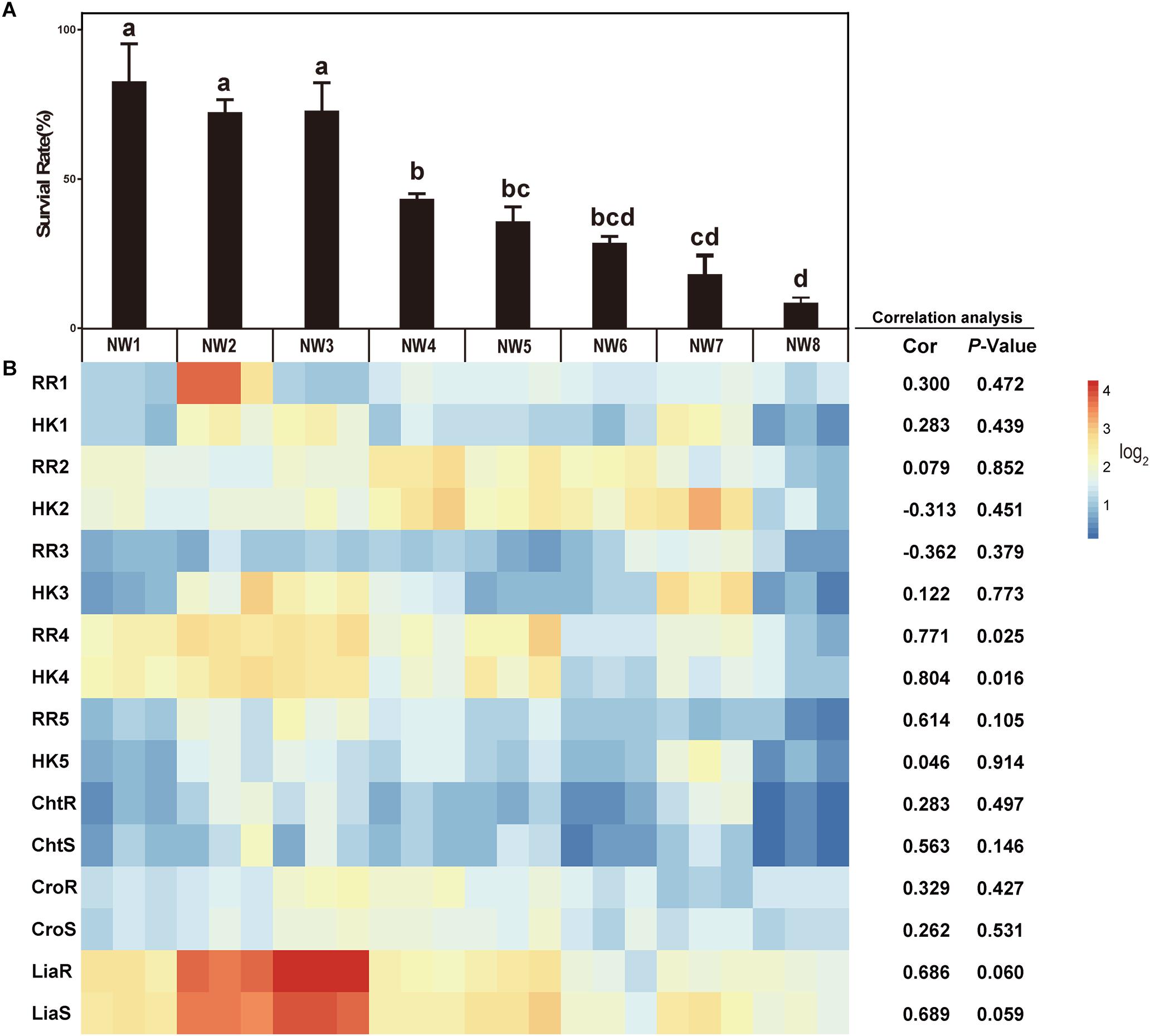
Figure 2. The correlations between the gene expressions of TCSs and bile salt resistance. (A) Survival rates of E. faecium isolates after 30 min exposure to 0.5% ox gall. Data are shown as mean ± SD from three independent biological replicates. Different letters above bars indicate the survival rates are significant differences among E. faecium isolates (P < 0.05). (B) The heatmap of expression of TCS genes after 0.5% ox gall addition compared with the time point preceding the bile addition (B, left panel). Red indicates up-regulated gene expression and blue denotes down-regulated gene expression. The color bar right indicates the expression scales [represented as log2 (fold change)]. Correlations between the survival rates in 0.5% ox gall and the expressions of TCS genes in E. faecium were performed using Pearson’s product-moment correlation in R software. The Cor value represents the correlation coefficient and P-value < 0.05 indicates that the correlation is significantly different.
The Correlations Between the Gene Expression of Putative TCSs and Bile Salt Resistance
To determine whether TCSs were involved in the resistance of E. faecium to bile salts, 8 TCSs were selected for measuring the expression of these genes in response to 0.5% ox gall. As shown in Figure 2B, there was a significantly positive correlation between the survival rate and the expressions of RR4 (0.771, P = 0.025) and HK4 (0.804, P = 0.016). The positive correlation between the survival rate and the expressions of liaR (0.686, P = 0.060) and liaS (0.689, P = 0.059) tended to be significant. These results suggested that liaR, liaS, RR4, and HK4 genes may contribute to bile salt resistance in E. faecium. The correlations between the gene expression of the remaining putative or known TCSs and bile salt resistance were not significant.
The Effect of liaR, liaS, bsrR, and bsrS Mutants on the Bile Salt Resistance in E. faecium NW2
Considering the importance of RR4 and HK4 genes in bile salt resistance, these putative and previously unstudied genes were renamed as bsrR and bsrS, for b ile s alt r esistance r esponse regulator and b ile s alt r esistance s ensor histidine kinase, respectively.
To confirm the involvement of liaR, liaS, bsrR, and bsrS genes in resistance to bile salts, mutants of gene disruption were constructed using E. faecium NW2 due to its high bile resistant ability and up-regulation of liaR, liaS, bsrR, and bsrS genes after exposure to ox gall. All mutants had similar growth curves as the parental strain NW2 in MRS broth in the absence of ox gall (data not shown). As showed in Figure 3, disruption of liaR, liaS, bsrR, and bsrS significantly decreased the survival rate after exposure to 0.5, 1, and 2% ox gall compared to the E. faecium NW2 wild type. Complementation of the liaS gene resulted in restoration of survival in ox gall to levels of wild-type NW2 (Figure 3). However, the liaR, bsrR, and bsrS complemented strains were not obtained after several attempts. These results indicated that liaR, liaS, bsrR, and bsrS were important for bile salt resistance in E. faecium.
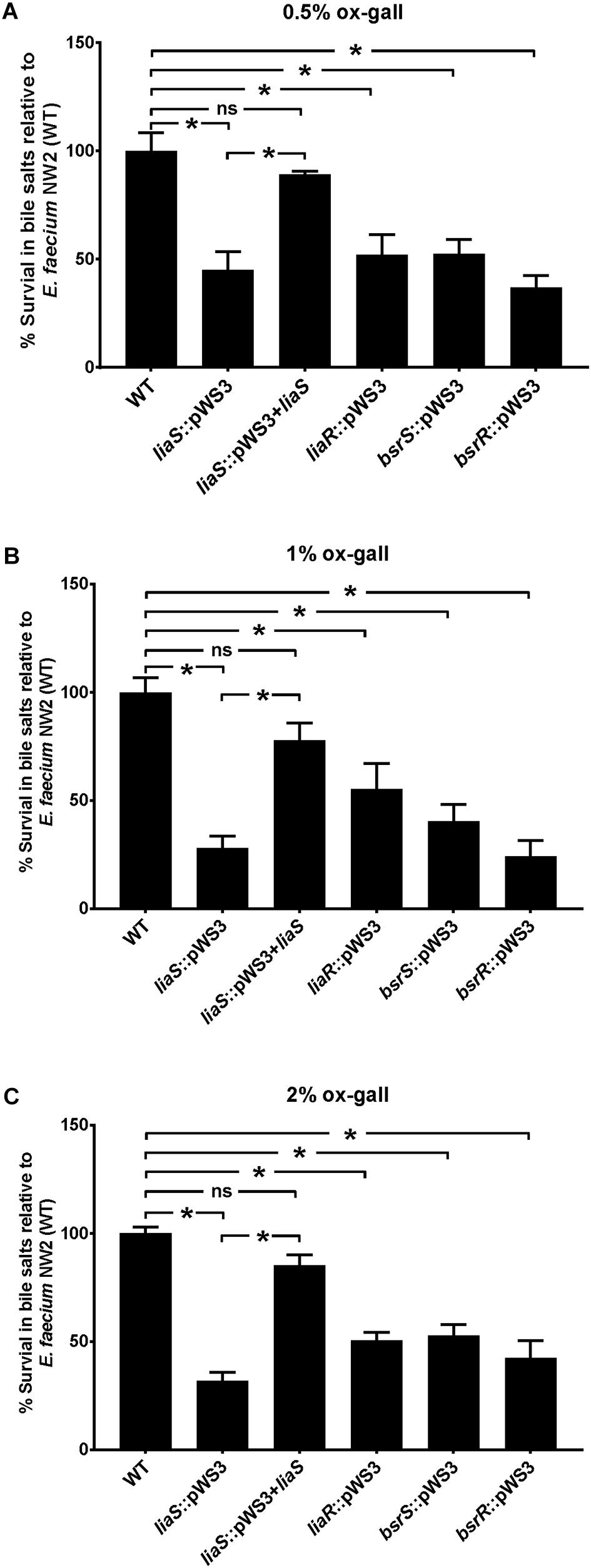
Figure 3. The effect of mutation of liaR, liaS, bsrR, and bsrS genes on resistance to ox gall by E. faecium NW2. Survival rates of E. faecium NW2 wild type (WT) and its mutants with insertional mutagenesis of liaS (liaS::pWS3), liaR (liaR::pWS3), bsrR (bsrR::pWS3), and bsrS (bsrS::pWS3) genes and the complementation strain of liaS (liaS::pWS3 + liaS) after 30 min exposure to 0.5 (A), 1 (B), 2% ox gall (C). Data are mean ± SD of three independent experiments. Asterisk above bars indicate the survival rates are significant differences between WT and mutant strains or complementation strain (P < 0.05).
Characterization of the BsrXRS System in E. faecium
In analysis of the genomic location of bsrR and bsrS, it was found that the 5′-end of the bsrR overlaps with 20 bases of the 3′-end of a 351-bp gene encoding a hypothetical protein (HMPREF0351_11749, renamed as bsrX) and the 5′-end of the bsrS overlaps with a base of the 3′-end of bsrR, which suggest that bsrX, bsrR, and bsrS may exist in an operon (Figure 4A). We validated that these 3 genes were co-transcribed, forming a single operon by RT-PCR (Figure 4B).
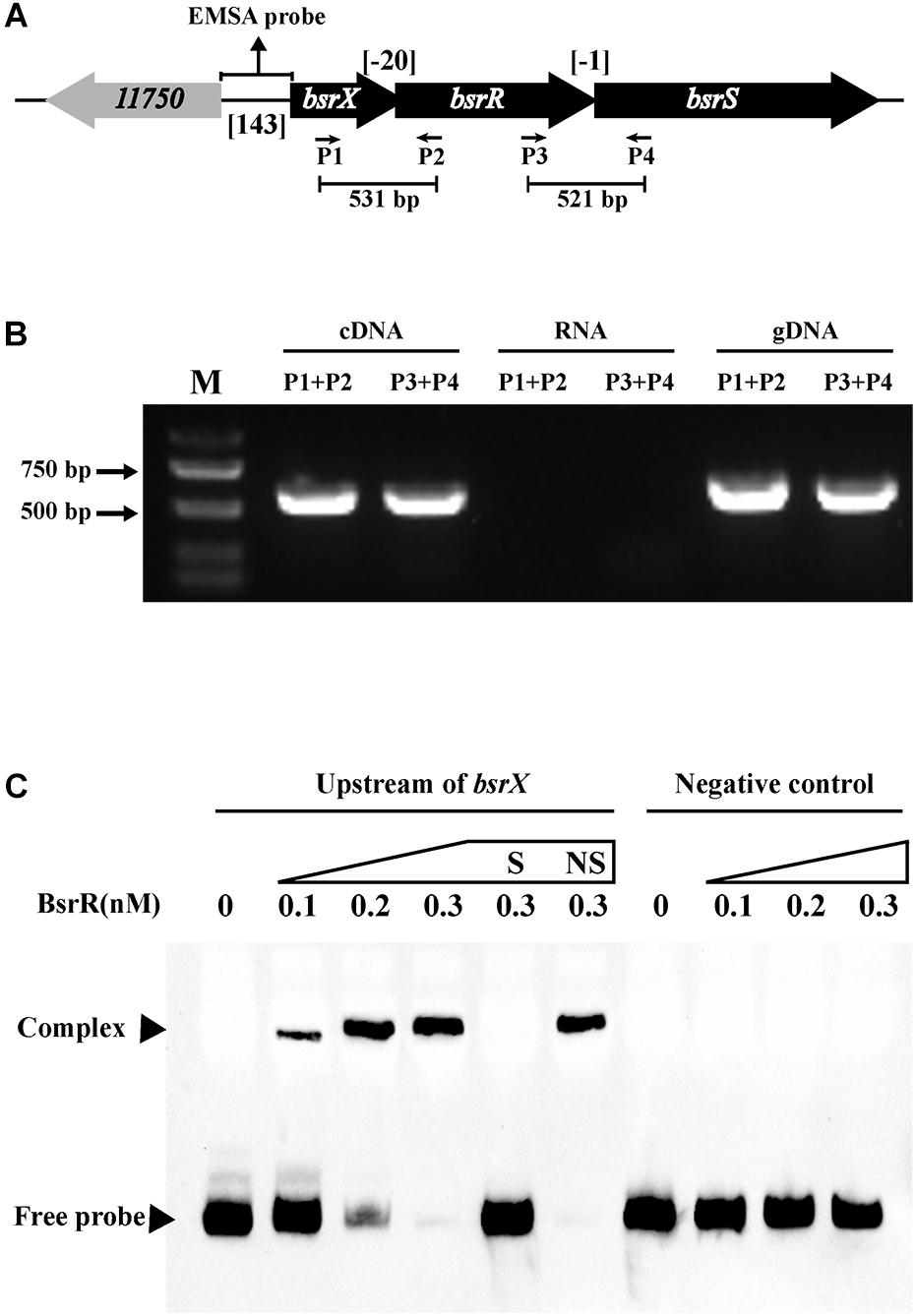
Figure 4. Characterization of the BsrXRS system in E. faecium. (A) Schematic representation of the BsrXRS system in E. faecium. (B) RT-PCR was performed for studying co-transcription using cDNA, RNA (negative control) and genomic DNA (positive control). Location of each intergenic primer pair for RT-PCR is shown in (A). (C) EMSA using biotin-labeled probes containing the intergenic region upstream of bsrX (143-bp) or negative control (150 bp) incubated with indicated amounts of BsrR protein. The negative control was amplified from the coding region of housekeeping gene ddlA, which does not share sequence identity with the intergenic region upstream of bsrX. EMSAs in the presence of 200-fold unlabeled specific probe (S, upstream of bsrX) or non-specific (NS, negative control) competitor DNA were performed as controls. Free DNA fragments and BsrR-DNA complexes are labeled.
The sequence analysis of BsrR revealed that the C-terminal of BsrR harbors a DNA-binding domain, suggesting that BsrR may function as a transcription regulator. To examine whether BsrR autoregulates directly, we performed EMSA with purified BsrR protein. The BsrR was showed to bind to a 143-bp intergenic region directly upstream of bsrX, thus suggesting BsrR to directly regulate the transcription of its own operon (Figure 4C).
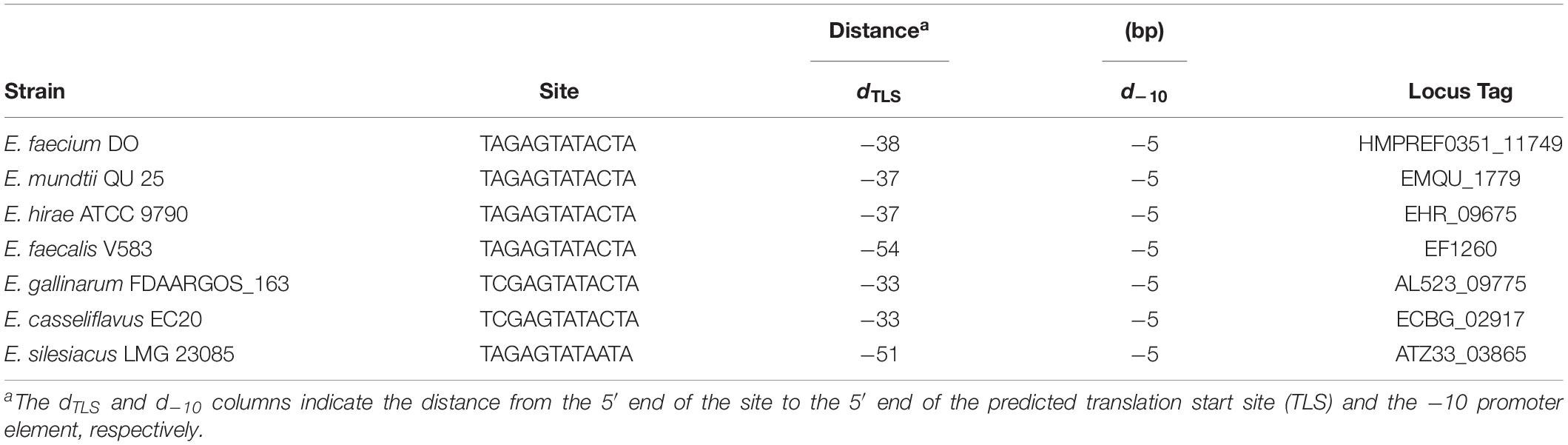
To further determine whether BsrR in the Enterococcus genus can function as a transcription regulator, protein sequences of the BsrR from 7 strains including E. faecium DO were aligned. There were 86% identities and a conserved DNA-binding domain (Thr-164, Ser-183, Arg-184, Thr-202, Ala-205, Lys-213, Ile-214, Thr-224, and Gly-229) (Supplementary Figure S1), implying that these BsrR homologs might have a conserved DNA-binding site. By submitting the DNA sequences of upstream of the BsrXRS system from 7 bacterial species to the DMINDA 2.0 web server (Yang et al., 2017), a putative BsrR-binding site, the consensus sequence TMGAGTATAMTA, was found in the Enterococcus genus bacteria (Table 2).
Characterization of the Potential Target Genes for the BsrXRS System
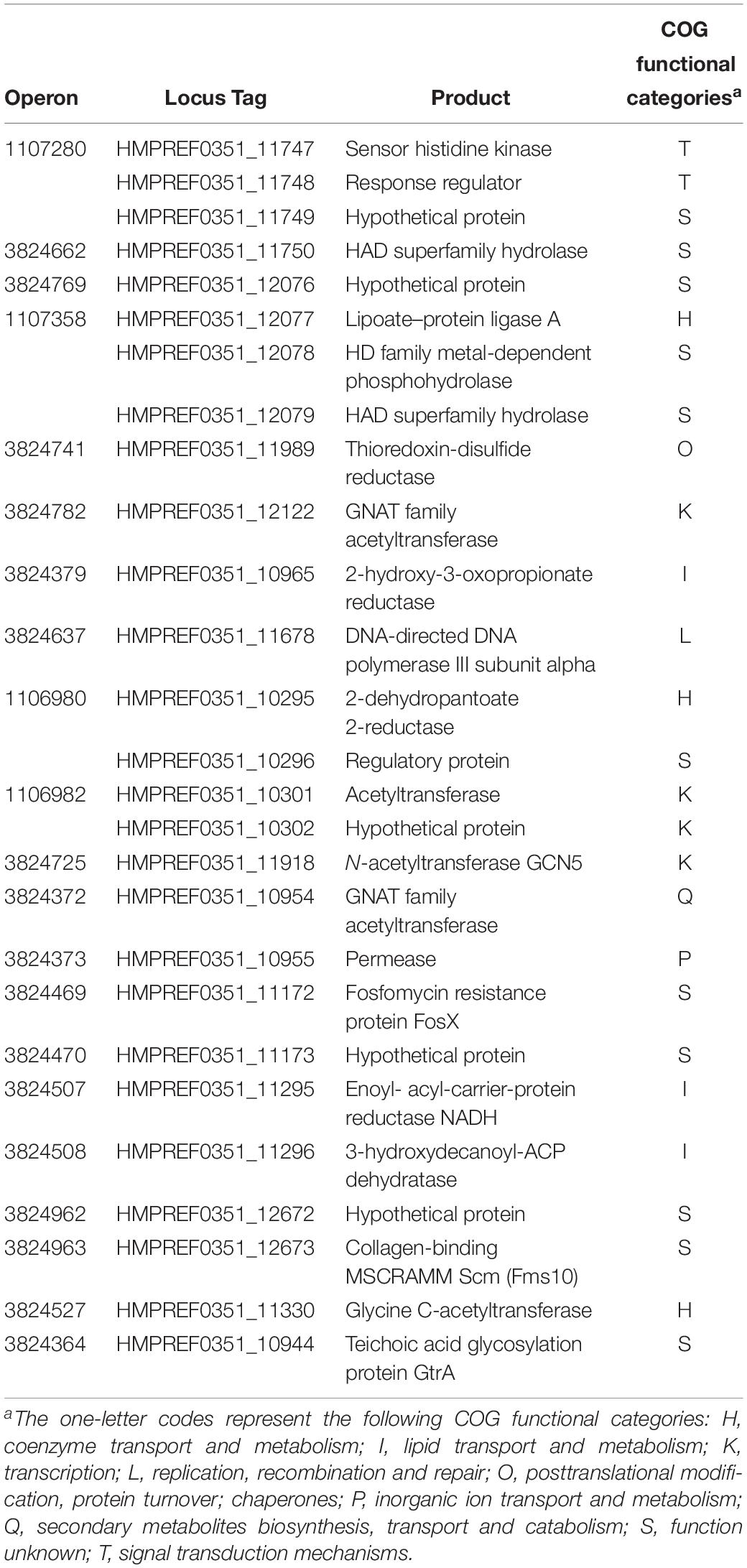
To determine whether BsrR in the Enterococcus genus can also function as a transcription regulator for expression of other genes, an in silico genome-wide search was performed for putative BsrR target genes using the identified consensus sequence TMGAGTATAMTA as the BsrR-binding motif. As shown in Table 3, 27 putative target genes were identified.
Expression of 25 putative target genes was measured by RT-qPCR, while expression of HMPREF035_10301 and 10302 was not measured due to lack of suitable specific primers. Among 25 tested genes, 16 genes were differentially expressed between the E. faecium NW2 wild-type and the bsrR mutant after exposure to 0.5% ox gall (Figure 5). Three genes, bsrX, bsrR, and bsrS, exist in an operon bsrXRS as described above. HMPREF0351_12077 is annotated as encoding a lipoate-protein ligase A (lplA). The lplA, 12078 and 12079 were co-transcribed and belong to a same operon (Figures 6A,B). As shown in Figure 6C, BsrR was able to bind the upstream of lplA gene, indicating that BsrR directly regulate the lplA operon. Seven genes are putatively involved in amino acid metabolism (HMPREF0351_11989 and 10965), fatty acid biosynthesis (HMPREF0351_11295), secondary metabolites biosynthesis (HMPREF0351_10954), inorganic ion transport and metabolism (HMPREF0351_10955), cell wall maintenance (HMPREF0351_10944), and DNA mismatch repair (HMPREF0351_11678). The other target genes encode hypothetical proteins of unknown functions (HMPREF0351_11172, 11173, 12076, and 12672).
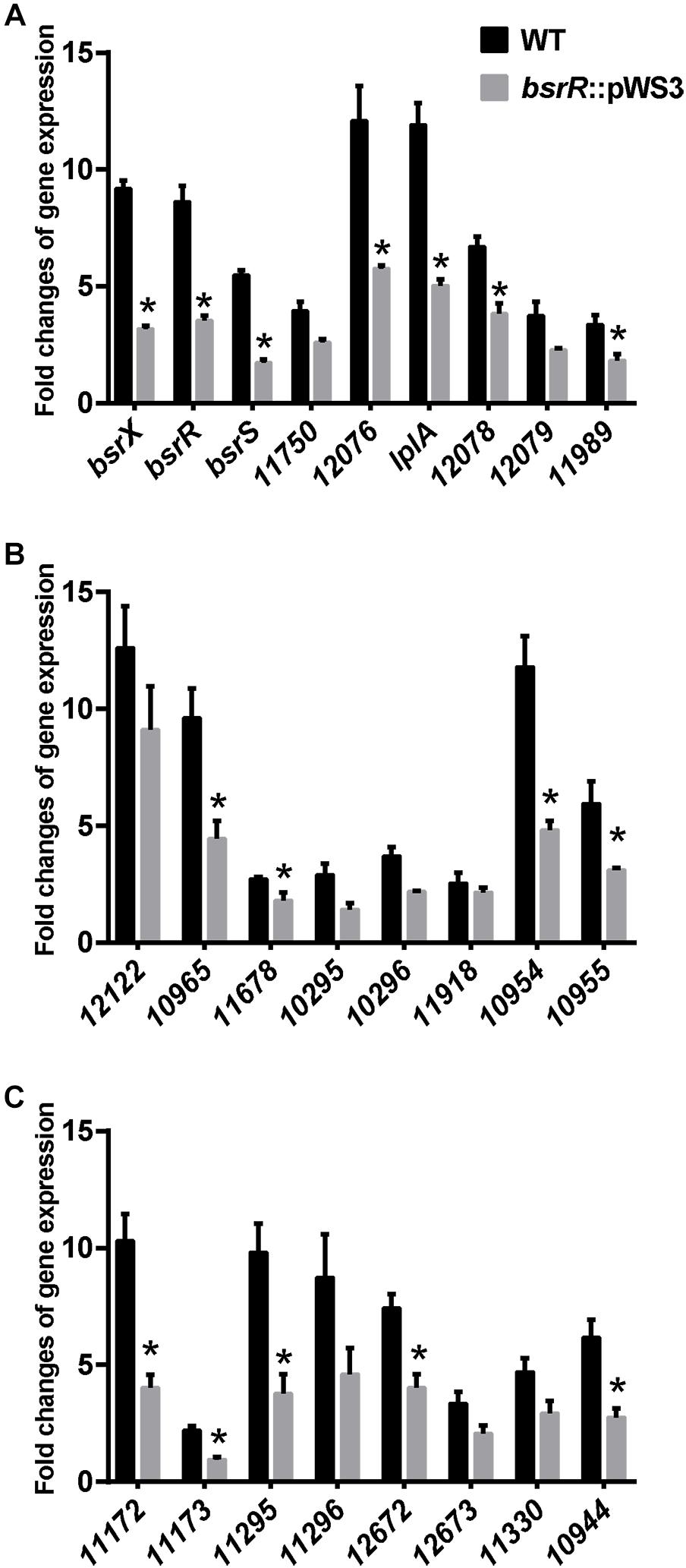
Figure 5. Theexpression of target genes of BsrR in response to bile salts. (A–C) Relative expression of target genes of BsrR in response to 0.5% ox-gall was determined by RT-qPCR. Fold changes of gene expression of wild-type (WT) and the bsrR mutant (bsrR::pWS3) of E. faecium NW2 after exposure to 0.5% ox-gall for 30 min, were calculated relative to WT and bsrR::pWS3 strains before the bile salts addition, respectively. Data are shown as mean ± SD from three independent biological replicates. Asterisk above bars indicate significant difference between WT and the bsrR mutant (P < 0.05). The target genes, bsrX (HMPREF0351_11749), bsrR (HMPREF0351_11748), bsrS (HMPREF0351_11747), 11750 (HMPREF0351_11750), 12076 (HMPREF0351_12076), lplA (HMPREF0351_12077), 12078 (HMPREF0351_12078), 12079 (HMPREF0351_12079), 11989 (HMPREF0351_11989), 12122 (HMPREF0351_12122), 10965 (HMPREF0351_10965), 11678 (HMPREF0351_11678), 10295 (HMPREF0351_10295), 10296 (HMPREF0351_10296), 11918 (HMPREF0351_11918), 10954 (HMPREF0351_10954), 10955 (HMPREF0351_10955), 11172 (HMPREF0351_11172), 11173 (HMPREF0351_11173), 11295 (HMPREF0351_11295), 11296 (HMPREF0351_11296), 12672 (HMPREF0351_12672), 12673 (HMPREF0351_12673), 11330 (HMPREF0351_11330), 10944 (HMPREF0351_10944), were predicted with putative BsrR-binding motif in genome of E. faecium DO.
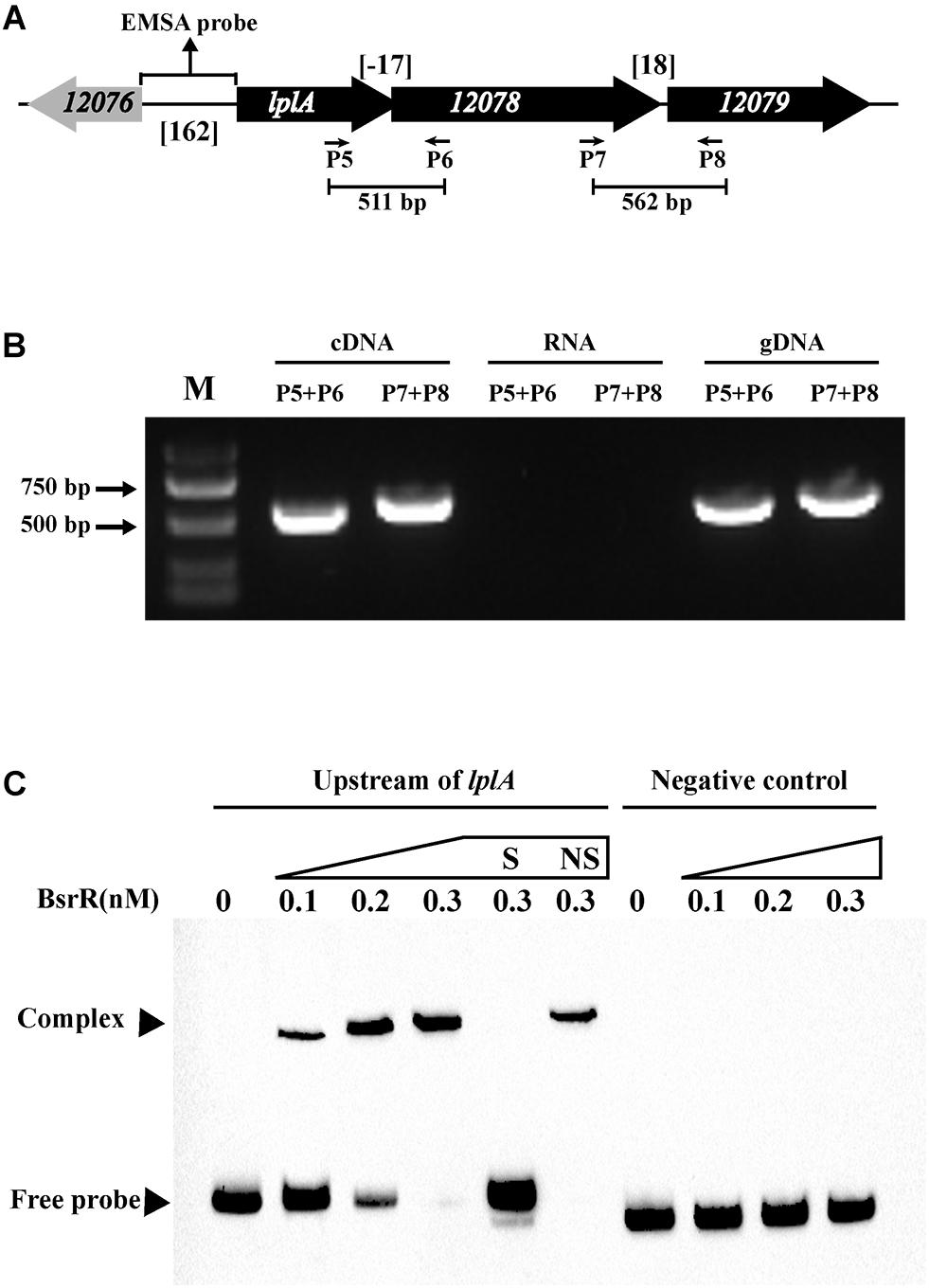
Figure 6. Characterization of the lplA operon that regulated by BsrR. (A) Schematic representation of lplA operon in E. faecium. (B) RT-PCR was performed for studying co-transcription using cDNA, RNA (negative control) and genomic DNA (positive control). Location of each intergenic primer pair for RT-PCR is shown in (A). (C) EMSA using biotin-labeled probes containing the intergenic region upstream of lplA (162-bp) or negative control (150 bp) incubated with indicated amounts of BsrR protein. The negative control was amplified from the coding region of housekeeping gene ddlA, which does not share sequence identity with the intergenic region upstream of bsrX. EMSAs in the presence of 200-fold unlabeled specific probe (S, upstream of lplA) or non-specific (NS, negative control) competitor DNA were performed as controls. Free DNA fragments and BsrR-DNA complexes are labeled.
Discussion
Bile salts execute antimicrobial effects through disturbing cell membrane and macromolecule stability, and inducing oxidative stress, DNA damage and protein mis-folding in bacterial cells (Ruiz et al., 2013). Thus, the ability to adapt and respond to bile salts is essential for the survival and persistence of intestinal bacteria in gastrointestinal tracts. Two-component systems (TCSs) are dominant regulated mechanisms of bacteria in response to environmental stimuli (Groisman, 2016). Several studies have shown that TCSs are involved in resistance to bile salts in various bacterial species. However, the involvement of TCSs in E. faecium’s resistance to bile salts has not been reported. As a successful colonizer in the gastrointestinal tract of various hosts including humans and animals, E. faecium must have evolved mechanisms to sense, respond to and resist bile salts. The results from the current study indicate that LiaFSR and BsrXRS contributed to bile salt resistance of E. faecium.
LiaFSR was shown to be involved in bile resistance in E. faecium isolates. The positive correlation between the survival rate in 0.5% ox gall and the expressions of liaSR genes tended to be significant. More importantly, disruption of liaR or liaS significantly reduced the survival rate of mutants in different concentrations of bile salts compared to the E. faecium wild type. Our results for the LiaFSR system in E. faecium were supported by a previous study in E. faecalis in which the liaR mutant had a low survival rate after exposure to bile (Harp et al., 2016). The LiaFSR system was first identified in the response against lipid II interfering antibiotics (bacitracin, nisin, ramoplanin, and vancomycin) in Bacillus subtilis (Mascher et al., 2004). The role of LiaFSR in E. faecium and E. faecalis has mainly been described recently in the context of daptomycin resistance (Reyes et al., 2015). Given that cell membrane is a major attack target of bile salts, our finding expands the role of LiaFSR in dealing with cell membrane stress in E. faecium.
Cross-resistance against bile salts and antibiotics may exist. Saito et al. (2014) found the supplementing cultures with 0.2% bile significantly enhanced survival of E. faecalis OG1RF when exposed to daptomycin through incorporation of exogenous fatty acids and altering the fatty acid composition of cell membrane. In addition, LiaR-deficient strain of E. faecalis can incorporate exogenous fatty acids into its membrane, then increase the survival rate when exposed to a variety of membrane damage agents, such bile, SDS and daptomycin (Harp et al., 2016).
The functions of BsrXRS have not been explored before. In this study, expression of bsrR and bsrS was significantly correlated with the survival rate of E. faecium in 0.5% ox gall. There were 4 amino acid polymorphisms (F for NW1, 2, 3, 5, and 6 and V for NW4, 7, and 8 at position 176; K for NW2, 3, 4, 7, and 8 and R for NW 1, 5, and 6 at position 243; A for NW1, 2, 3, 5, and 6 and T for NW4, 7, and 8 at position 471; K for NW1, 2, 3, 5, and 6 and E for NW4, 7, and 8 at position 481) in the BsrS protein, while no difference was found in the BsrR protein. The amino acid at position 176 is located in a predicted trans-membrane domain in the sensor histidine kinase BsrS, which is essential for signal transmission from extracellular to cytoplasm (Zschiedrich et al., 2016). The other two amino acid substitutions at positions 471 and 481 are located in C-terminal ATP-binding catalytic domain, which is responsible for autophosphorylation of HK. These amino acid substitutions may affect the signal transduction and activation of BsrR and change the transcription of target genes, resulting in different bile salt resistance in E. faecium strains. The sequence analysis of BsrX using revealed that BsrX harbors 3 transmembrane helices, suggesting BsrX may be a small membrane protein (116 aa). Despite repeated attempts, we were unable to obtain a mutant strain of bsrX gene. Therefore, the function of bsrX gene was not validated in the present study.
Twenty-seven potential target genes for BsrR were revealed and these target genes are involved in multiple cellular functions. Sixteen genes were found as differentially expressed between E. faecium NW2 wild-type and bsrR mutant after exposure to 0.5% ox gall. Of the 16 differentially expressed genes, 7 genes (bsrX, bsrR, bsrS, lplA, HMPREF0351_10944, 11173, 12672) were upregulated after exposure to 0.5% ox gall. Our result was supported by a previous study in E. faecium E1162 in which these 7 genes were also upregulated after addition of bile salts to the culture medium (Zhang et al., 2013). However, none of the 7 genes showed significant differential expression in E. faecalis V583 in the presence of ox gall (Solheim et al., 2007). On the other hand, a TCS CroRS system was induced in E. faecalis V583 after exposure to ox gall (Solheim et al., 2007). However, expression of the CroRS system was not significant in E. faecium NW2 in our study and also in E. faecium E1162 (Zhang et al., 2013). Although E. faecium is a close relative of E. faecalis, these findings imply that different mechanisms are involved in bile resistance in E. faecium and E. faecalis.
Among these 16 differentially expressed target genes of BsrR, 3 genes (bsrX, bsrR, and bsrS) constitute an operon bsrXRS as discussed above. Another operon targeted by BsrR consists of 3 genes, lplA encoding a lipoate–protein ligase A (HMPREF0351_12077), a gene encoding HD family metal-dependent phosphohydrolase (HMPREF0351_12078) and a gene encoding HAD superfamily hydrolase (HMPREF0351_12079). Whether involvement of the HD family metal-dependent phosphohydrolase and HAD superfamily hydrolase in bile resistance is unknown, lplA gene may play an important role in bile salt resistance. This was supported by the observation that mutation of lplA induced a 2-log reduction of viability in comparison with the wild type L. monocytogenes after 8 h exposure to porcine gallbladder bile (Dowd et al., 2011). LplA catalyzes the formation of lipoyl-AMP from lipoate and ATP and transfers the lipoyl moiety to a specific lysine residue on the acyltransferase subunit of α-ketoacid dehydrogenase complexes. In particular, the branched-chain-ketoacid dehydrogenase (BCDH) participates in the degradation of branched chain amino acids to generate branched-chain CoA (BC-CoA) molecules. In many Gram-positive bacteria, these short BC-CoA molecules are used chiefly as primers for generating longer branched-chain fatty acids that can modulate membrane fluidity. This change of membrane fluidity may promote E. faecium to resist bile salts toxicity. However, further study is required to corroborate this hypothesis.
Bacterial cell membrane and cell wall are main targets of bile salts. Thus changes in their composition appear to be the most prevalent actions to resist bile salt in bacteria (Ruiz et al., 2013). Studies have shown that resistance to bile salt in E. faecalis involved changes in fatty acid biosynthesis (Solheim et al., 2007) and in membrane composition or cell wall synthesis (Rincé et al., 2003). In addition, DNA mismatch repair proteins repair the DNA damage caused by bile salts and are important for bacterial bile resistance (Rincé et al., 2003). In this study, the target genes of BsrR from functional annotation are involved in amino acid metabolism (HMPREF0351_11989 and 10965), fatty acid biosynthesis (HMPREF0351_11295), secondary metabolites biosynthesis (HMPREF0351_10954), inorganic ion transport and metabolism (HMPREF0351_10955), cell wall maintenance (HMPREF0351_10944), and DNA mismatch repair (HMPREF0351_11678). These target genes may contribute to bile salt resistance in E. faecium.
The Enterococcus genus is an ancient genus of microbes that are highly adapted to living in complex environments and surviving harsh conditions. The BsrXRS system is highly conserved in Enterococcus genus, indicating that BsrXRS may play a prominent role in responding to environmental changes among bacteria in the Enterococcus genus.
Conclusion
In summary, our data demonstrate that E. faecium can improve the resistance to bile salts by up-regulating LiaFSR and BsrXRS systems. The BsrXRS system up-regulates gene expression of target genes that involving in change of membrane fluidity, fatty acid biosynthesis, cell wall maintenance, DNA mismatch repair. These findings provide potential molecular mechanisms for resistance to bile salts by E. faecium.
Author Contributions
LZ and XZ contributed to the design of the experiments and prepared the manuscript of this publication. LZ, LW, PT, and TB performed the experiments. LZ and LL analyzed the data. All authors read and approved the final manuscript.
Funding
This work was supported by funding from National Natural Science Foundation of China (31672445) and a University Scientific Research Fund project (Z109021718).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Axel Hartke (University of Caen, France), Willem van Schaik (University of Birmingham, United Kingdom), and Reinhold Brückner (University of Kaiserslautern, Germany) for generously providing the E. coli EC1000 strain and plasmid pWS3 and pRB473.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01048/full#supplementary-material
References
Adediran, J., Leatham-Jensen, M. P., Mokszycki, M. E., Frimodt-Moller, J., Krogfelt, K. A., Kazmierczak, K., et al. (2014). An Escherichia coli Nissle 1917 missense mutant colonizes the streptomycin-treated mouse intestine better than the wild type but is not a better probiotic. Infect. Immun. 82, 670–682. doi: 10.1128/IAI.01149-13
Brückner, R., Wagner, E., and Götz, F. (1993). Characterization of a sucrase gene from Staphylococcus xylosus. J. Bacteriol. 175, 851–857. doi: 10.1128/jb.175.3.851-857.1993
Capra, E. J., and Laub, M. T. (2012). Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347. doi: 10.1146/annurev-micro-092611-150039
Dowd, G. C., Joyce, S. A., Hill, C., and Gahan, C. G. (2011). Investigation of the mechanisms by which Listeria monocytogenes grows in porcine gallbladder bile. Infect. Immun. 79, 369–379. doi: 10.1128/IAI.00330-10
Favaro, L., Basaglia, M., Casella, S., Hue, I., Dousset, X., Gombossy de Melo Franco, B. D., et al. (2014). Bacteriocinogenic potential and safety evaluation of non-starter Enterococcus faecium strains isolated from home made white brine cheese. Food Microbiol. 38, 228–239. doi: 10.1016/j.fm.2013.09.008
Feng, J., Wang, L., Zhou, L., Yang, X., and Zhao, X. (2016). Using in vitro immunomodulatory properties of lactic acid bacteria for selection of probiotics against Salmonella infection in broiler chicks. PLoS One 11:e0147630. doi: 10.1371/journal.pone.0147630
Groisman, E. A. (2016). Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 70, 103–124. doi: 10.1146/annurev-micro-102215-095331
Guzman Prieto, A. M., Wijngaarden, J., Braat, J. C., Rogers, M. R. C., Majoor, E., Brouwer, E. C., et al. (2017). The two-component system ChtRS contributes to chlorhexidine tolerance in Enterococcus faecium. Antimicrob. Agents Chemother. 61:e2122–16. doi: 10.1128/AAC.02122-16
Harp, J. R., Saito, H. E., Bourdon, A. K., Reyes, J., Arias, C. A., Campagna, S. R., et al. (2016). Exogenous fatty acids protect Enterococcus faecalis from daptomycin-induced membrane stress independently of the response regulator LiaR. Appl. Environ. Microbiol. 82, 4410–4420. doi: 10.1128/AEM.00933-16
Hong, H. J., Hutchings, M. I., and Buttner, M. J. (2008). “Vancomycin resistance VanS/VanR two-component systems,” in Bacterial Signal Transduction: Networks and drug Targets. Advances in Experimental Medicine and Biology, Vol. 631, ed. R. Utsumi (New York, NY: Springer), 200–213. doi: 10.1007/978-0-387-78885-2_14
Kellogg, S. L., Little, J. L., Hoff, J. S., and Kristich, C. J. (2017). Requirement of the CroRS two-component system for resistance to cell wall-targeting antimicrobials in Enterococcus faecium. Antimicrob. Agents Chemother. 61:e02461–16. doi: 10.1128/AAC.02461-16
Koskenniemi, K., Laakso, K., Koponen, J., Kankainen, M., Greco, D., Auvinen, P., et al. (2011). Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol. Cell Proteomics 10:M110.002741. doi: 10.1074/mcp.M110.002741
Kus, J. V., Gebremedhin, A., Dang, V., Tran, S. L., Serbanescu, A., and Barnett Foster, D. (2011). Bile salts induce resistance to polymyxin in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 193, 4509–4515. doi: 10.1128/JB.00200-11
Le Breton, Y., Boel, G., Benachour, A., Prevost, H., Auffray, Y., and Rince, A. (2003). Molecular characterization of Enterococcus faecalis two-component signal transduction pathways related to environmental stresses. Environ. Microbiol. 5, 329–337. doi: 10.1046/j.1462-2920.2003.00405.x
Lebreton, F., van Schaik, W., McGuire, A. M., Godfrey, P., Griggs, A., Mazumdar, V., et al. (2013). Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534–13. doi: 10.1128/mBio.00534-13
Leenhouts, K. (1996). A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253, 217–224. doi: 10.1007/s004380050315
Leverrier, P., Dimova, D., Pichereau, V., Auffray, Y., Boyaval, P., and Jan, G. (2003). Susceptibility and adaptive response to bile salts in Propionibacterium freudenreichii: physiological and proteomic analysis. Appl. Environ. Microbiol. 69, 3809–3818. doi: 10.1128/aem.69.7.3809-3818.2003
Mao, X., Ma, Q., Zhou, C., Chen, X., Zhang, H., Yang, J., et al. (2014). DOOR 2.0: presenting operons and their functions through dynamic and integrated views. Nucleic Acids Res. 42, D654–D659. doi: 10.1093/nar/gkt1048
Mascher, T., Zimmer, S. L., Smith, T. A., and Helmann, J. D. (2004). Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48, 2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004
Mendiburu, F. D. (2017). Agricolae: Statistical Procedures for Agricultural Research (Version 1.2-8). Available at: https://CRAN.R-project.org/package=agricolae (accessed September 12, 2017).
Ortet, P., Whitworth, D. E., Santaella, C., Achouak, W., and Barakat, M. (2015). P2CS: updates of the prokaryotic two-component systems database. Nucleic Acids Res. 43, D536–D541. doi: 10.1093/nar/gku968
Panesso, D., Reyes, J., Gaston, E. P., Deal, M., Londono, A., Nigo, M., et al. (2015). Deletion of liaR reverses daptomycin resistance in Enterococcus faecium independent of the genetic background. Antimicrob. Agents Chemother. 59, 7327–7334. doi: 10.1128/AAC.01073-15
Pfeiler, E. A., Azcarate-Peril, M. A., and Klaenhammer, T. R. (2007). Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 189, 4624–4634. doi: 10.1128/JB.00337-07
Pingitore, E. V., Todorov, S. D., Sesma, F., and Franco, B. D. (2012). Application of bacteriocinogenic Enterococcus mundtii CRL35 and Enterococcus faecium ST88Ch in the control of Listeria monocytogenes in fresh Minas cheese. Food Microbiol. 32, 38–47. doi: 10.1016/j.fm.2012.04.005
R Core Team (2017). R: A Language and Environment for Statistical Computing (Version 3.4.3). Available at: https://www.R-project.org (accessed November 30, 2017).
Raphael, B. H., Pereira, S., Flom, G. A., Zhang, Q., Ketley, J. M., and Konkel, M. E. (2005). The Campylobacter jejuni response regulator, CbrR, modulates sodium deoxycholate resistance and chicken colonization. J. Bacteriol. 187, 3662–3670. doi: 10.1128/JB.187.11.3662-3670.2005
Reyes, J., Panesso, D., Tran, T. T., Mishra, N. N., Cruz, M. R., Munita, J. M., et al. (2015). A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J. Infect. Dis. 211, 1317–1325. doi: 10.1093/infdis/jiu602
Rincé, A., Le Breton, Y., Verneuil, N., Giard, J.-C., Hartke, A., and Auffray, Y. (2003). Physiological and molecular aspects of bile salt response in Enterococcus faecalis. Int. J. Food Microbiol. 88, 207–213. doi: 10.1016/s0168-1605(03)00182-x
Ruiz, L., Margolles, A., and Sanchez, B. (2013). Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 4:396. doi: 10.3389/fmicb.2013.00396
Saito, H. E., Harp, J. R., and Fozo, E. M. (2014). Incorporation of exogenous fatty acids protects Enterococcus faecalis from membrane-damaging agents. Appl. Environ. Microbiol. 80, 6527–6538. doi: 10.1128/AEM.02044-14
Sanchez, B., Champomier-Verges, M. C., Anglade, P., Baraige, F., de Los Reyes-Gavilan, C. G., Margolles, A., et al. (2005). Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187, 5799–5808. doi: 10.1128/JB.187.16.5799-5808.2005
Solheim, M., Aakra, A., Vebo, H., Snipen, L., and Nes, I. F. (2007). Transcriptional responses of Enterococcus faecalis V583 to bovine bile and sodium dodecyl sulfate. Appl. Environ. Microbiol. 73, 5767–5774. doi: 10.1128/AEM.00651-07
Srinivasan, V. B., Vaidyanathan, V., Mondal, A., and Rajamohan, G. (2012). Role of the two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. PLoS One 7:e33777. doi: 10.1371/journal.pone.0033777
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tran, T. T., Miller, W. R., Shamoo, Y., and Arias, C. A. (2016). Targeting cell membrane adaptation as a novel antimicrobial strategy. Curr. Opin. Microbiol. 33, 91–96. doi: 10.1016/j.mib.2016.07.002
Veljovic, K., Popovic, N., Miljkovic, M., Tolinacki, M., Terzic-Vidojevic, A., and Kojic, M. (2017). Novel aggregation promoting factor AggE contributes to the probiotic properties of Enterococcus faecium BGGO9-28. Front. Microbiol. 8:1843. doi: 10.3389/fmicb.2017.01843
Whiteside, M. D., Winsor, G. L., Laird, M. R., and Brinkman, F. S. (2013). OrtholugeDB: a bacterial and archaeal orthology resource for improved comparative genomic analysis. Nucleic Acids Res. 41, D366–D376. doi: 10.1093/nar/gks1241
Yang, J., Chen, X., McDermaid, A., and Ma, Q. (2017). DMINDA 2.0: integrated and systematic views of regulatory DNA motif identification and analyses. Bioinformatics 33, 2586–2588. doi: 10.1093/bioinformatics/btx223
Zhang, X., Bierschenk, D., Top, J., Anastasiou, I., Bonten, M. J., Willems, R. J., et al. (2013). Functional genomic analysis of bile salt resistance in Enterococcus faecium. BMC Genomics 14:299. doi: 10.1186/1471-2164-14-299
Zhang, X., Paganelli, F. L., Bierschenk, D., Kuipers, A., Bonten, M. J., Willems, R. J., et al. (2012). Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet. 8:e1002804. doi: 10.1371/journal.pgen.1002804
Zhang, X., Vrijenhoek, J. E., Bonten, M. J., Willems, R. J., and van Schaik, W. (2011). A genetic element present on megaplasmids allows Enterococcus faecium to use raffinose as carbon source. Environ. Microbiol. 13, 518–528. doi: 10.1111/j.1462-2920.2010.02355.x
Keywords: two-component systems, Enterococcus faecium, bile salts, LiaFSR, BsrXRS
Citation: Zhou L, Wang L, Tian P, Bao T, Li L and Zhao X (2019) The LiaFSR and BsrXRS Systems Contribute to Bile Salt Resistance in Enterococcus faecium Isolates. Front. Microbiol. 10:1048. doi: 10.3389/fmicb.2019.01048
Received: 15 January 2019; Accepted: 25 April 2019;
Published: 10 May 2019.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Jean-Christophe Giard, University of Caen Normandy, FranceCécile Muller, University of Caen Normandy, France
Copyright © 2019 Zhou, Wang, Tian, Bao, Li and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Zhao, eGluLnpoYW9AbWNnaWxsLmNh
 Luoxiong Zhou1
Luoxiong Zhou1 Lianbin Li
Lianbin Li Xin Zhao
Xin Zhao