- 1Jiangxi Key Laboratory of Bioprocess Engineering and Co-Innovation Center for In-vitro Diagnostic Reagents and Devices of Jiangxi Province, College of Life Sciences, Jiangxi Science and Technology Normal University, Nanchang, China
- 2National Key Laboratory of Enzyme – Protein Technology, VNU University of Science, Hanoi, Vietnam
- 3Faculty of Biology, VNU University of Science, Hanoi, Vietnam
Mevalonate diphosphate decarboxylase (MVD; EC 4.1.1.33) is a key enzyme of the mevalonic acid (MVA) pathway. In fungi, the MVA pathway functions as upstream of ergosterol biosynthesis, and MVD is also known as Erg19. Previously, we have identified Aoerg19 in Aspergillus oryzae using bioinformatic analysis. In this study, we showed that AoErg19 function is conserved using phylogenetic analysis and yeast complementation assay. Quantitative reverse transcription–PCR (qRT-PCR) indicated that Aoerg19 expression changed in different growth stages and under different forms of abiotic stress. Subcellular localization analysis showed that AoErg19 was located in the vacuole. Overexpression of Aoerg19 decreased the ergosterol content in A. oryzae, which may due to the feedback-mediated downregulation of Aoerg8. Consistent with the decrease in ergosterol content, both Aoerg19 overexpression and RNAi strains of A. oryzae are sensitive to abiotic stressors, including ergosterol biosynthesis inhibitor, temperature, salt and ethanol. Thus, we have identified the function of AoErg19 in A. oryzae, which may assist in genetic modification of MVA and the ergosterol biosynthesis pathway.
Introduction
Mevalonate diphosphate decarboxylase (MVD; EC 4.1.1.33) is a key enzyme of the mevalonic acid (MVA) pathway (Krepkiy and Miziorko, 2004). It catalyzes the decarboxylation of the six-carbon mevalonate 5-diphosphate (MVAPP) to the five-carbon isopentenyl diphosphate (IPP) (Krepkiy and Miziorko, 2004), the basic structure required for the biosynthesis of isoprenoids, an important cellular intermediate (Bergès et al., 1997). MVD plays important roles in different organisms, including microbes, plants, and animals (Abbassi et al., 2015). In fungi, IPP is the precursor required for ergosterol biosynthesis and MVD is a crucial enzyme required for cell viability because the mutant of the corresponding gene was lethal at 35°C (Bergès et al., 1997). In higher plants, IPP is the basic skeleton required for the biosynthesis of isoprenoids, which is the precursor for many plant-specific molecules such as growth regulators (e.g., gibberellins and abscisic acid), photosynthetic pigments, phytotoxins, phytoalexins, and other compounds required for plant defense, as well as aromatic terpenoids and natural rubbers (Chappell, 1995; Shi et al., 2012; Abbassi et al., 2016). In rat, the reduced activity of MVD has been associated with lower serum cholesterol levels and subsequently severe hypertension and cerebral hemorrhage (Michihara et al., 1998).
Studies have shown that the function of MVD is conserved among many organisms as the genes encoding MVDs in Candida albicans, human, Arabidopsis, and Ganoderma lucidum can complement the temperature-sensitive phenotype of the Saccharomyces cerevisiae mvd mutant (Cordier et al., 1999; Dassanayake et al., 2002; Voynova et al., 2008; Shi et al., 2012). The enzymatic properties of MVD from several organisms have been identified. The decarboxylation catalyzed by MVD requires one molecule of ATP and the participation of Mg2+ (Bergès et al., 1997). The yeast two-hybrid assay showed that in human, rat, yeast, and Arabidopsis, MVD forms homodimers in vivo; it also revealed that MVD heterodimers can be formed between S. cerevisiae and Arabidopsis proteins, which further supported the evolutionarily conserved function of MVD (Cordier et al., 1999). The subcellular localization of MVD in different organisms has also been investigated. In the past decade, MVD was reported to be localized in peroxisome in human and rat cells (Kovacs et al., 2002). However, Hogenboom et al. (2004) showed that both endogenous human MVD and overexpressed MVD were localized in cytosol. Consistent with this result (Michihara et al., 2008) also showed that MVD is predominantly located in the cytosol of both B16 and B16F10 cells in mouse. Unlike animals, MVD has been reported to be localized in peroxisomes in plants (Clastre et al., 2011; Simkin et al., 2011). However, studies on the subcellular localization of MVD in fungi are limited.
In fungi, the MVA pathway acts as the upstream of ergosterol biosynthesis. Therefore, the MVD gene was also known as erg19. Ergosterol is a specific cell membrane component and has been widely used as a marker to assess fungal biomass (Beni et al., 2014). It is not only an important cell membrane component that affects fluidity and permeability, which plays important roles in fungal growth, reproduction and stress tolerance (Kodedova and Sychrova, 2015), but also an important pharmaceutical raw material that can be used as a precursor of vitamin D2, cortisone, and progesterone (Görög, 2011). Studies on yeast and other fungi have revealed that the ergosterol biosynthesis pathway involves 18 reactions and 24 enzymes in S. cerevisiae (Hu et al., 2017). It has been reported that erg19 is one of the key genes in the ergosterol biosynthesis pathway (Bergès et al., 1997). The function of erg19 in S. cerevisiae, C. albicans and other fungi has been studied. In erg19 mutants, ergosterol biosynthesis was blocked, and the cell membrane composition was altered, which rendered the mutant temperature-sensitive (Cordier et al., 1999).
Till date, erg19 in filamentous fungi has been scarcely studied. Aspergillus oryzae, one of the most important filamentous fungi with industrial applications, is a FDA and WHO identified safe production species. In addition to its long history in traditional food fermentation, and condiment and brewing industries in China, A. oryzae is increasingly being used in modern biotechnology industries such as enzyme and recombinant protein production (Yamada et al., 2014). Previously, we identified only one paralog of erg19 in A. oryzae (named Aoerg19), as most of the other genes in the ergosterol biosynthesis pathway have multiple paralogs (Hu et al., 2019). Therefore, erg19 may act as the key gene in ergosterol biosynthesis in A. oryzae. In this study, we determined the function, expression pattern, and subcellular localization of AoErg19 and observed that both overexpression and RNAi of Aoerg19 decreased ergosterol content in A. oryzae and changed the response of A. oryzae to abiotic stress agents.
Materials and Methods
Strains and Growth Conditions
Aspergillus oryzae 3.042 (CICC 40092) was obtained from the China Center of Industry Culture Collection (Beijing, China) and used as the wild type strain. The A. oryzae 3.042 aaapyrG was constructed using Agrobacterium tumefaciens-mediated transformation in our laboratory (Sun et al., 2019). A. oryzae was cultured on (Czapek-Dox Agar) medium (2% sucrose, 0.2% NaNO3, 0.1% KH2PO4, 0.05% MgSO4, 0.05% KCl, 0.05% NaCl, 0.002% FeSO4, 1.6% agar, pH 5.5) at 30°C. Spores were harvested after 3 days of culture by scraping the agar surface with a sterile glass spreader under a laminar flow hood and suspending the spores in sterile water containing 0.05% Tween-80. The concentration of spores was determined using a hemocytometer. Escherichia coli DH5α (Grant et al., 1990) was used for plasmid construction and A. tumefaciens AGL1 (Lazo et al., 1991) was used for genetic transformation of A. oryzae. Both E. coli and A. tumefaciens were cultured in LB medium supplemented with suitable antibiotics at 37°C and 28°C, respectively.
DNA and RNA Extraction
The medium was covered with cellophane for DNA and RNA extraction. The mycelia were harvested and immediately frozen in liquid nitrogen and pulverized. DNA extraction was performed according to the instructions of the fungal DNA midi kit (Beyotime Institute of Biotechnology, Nantong, China). Total RNA was extracted from 0.5 g mycelia using a fungal RNA kit (Omega Bio-tek, Norcross, GA, United States) according to the manufacturer’s instructions, with the addition of an RNase-free DNase I treatment (Omega). The quality and concentration of the DNA and RNA samples were determined using NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, United States).
Expression Analysis of MVD
Total RNA was isolated from different growth stages or different stress conditions. The cDNAs were synthesized from 1 μg of total RNA using the Prime ScriptTM RT reagent kit (Perfect Real Time; Takara). All quantitative reverse transcription–PCR (qRT–PCR) measurements were performed using a CFX96 real-time PCR detection system (Bio-Rad, CA, United States) with SYBR Premix Ex Taq (Takara, Japan), according to the manufacturer’s instructions. The house-keeping gene encoding histone H4 was used as a normalization control (Maruyama et al., 2002). The relative expression was calculated using the formula 2-ΔΔCt. The primer sequences for qRT–PCR are provided in Supplementary Table S1.
Functional Complementation of Aoerg19 in Yeast
The erg19 (Y41208) mutant was purchased from EUROSCARF1 and the yeast strain BY4741 was the wild-type control. Plasmid pYES2-Aoerg19 contains Aoerg19 full-length coding sequence (CDS) under the control of the GAL1 promoter. To construct pYES2-Aoerg19, the CDS of Aoerg19 was amplified using PCR (the primer sequences are listed in Supplementary Table S2) and HindIII and EcoRI restriction sites were added to facilitate cloning of the fragments into the pYES2 vector (Invitrogen, Shanghai, China). The recombinant plasmid was sequenced. Then, the pYES2-Aoerg19 construct was transformed into the yeast mutant. The control and transformants were grown on YPD (1% yeast extract, 2% Bacto-Peptone, 2% glucose), and YPG (1% yeast extract, 2% Bacto-Peptone, 2% galactose) to assess the phenotypes.
Overexpression and RNAi of AoErg19
To construct the overexpression vector, the CDS of Aoerg19 was fused to DsRed at the C- and N-terminal of AoErg19 using fusion PCR, and the primers are listed in Supplementary Table S2. The binary vector pEX2B (Nguyen et al., 2017), was linearized with XhoI and BamHI, followed by cloning of the fusion DNA fragments into pEX2B using a one-step cloning kit (Vazyme Biotech Co., Ltd., China) to construct pEX2B-AoErg19-DsRed and pEX2B-DsRed-AoErg19 vectors. For the RNAi of Aoerg19, the 500 bp sequence of 5′ Aoerg19 mRNA was cloned to construct the hairpin structure, and the 79 bp intron of α-amylase was used as the loop of the hairpin structure by three times PCR using RNAi-forward R1, R2, and R3. The primer sequences for RNAi are listed in Supplementary Table S2. All the constructed vectors were transformed into A. tumefaciens AGL1. Then, the vectors were transformed into A. oryzae 3.042 aaapyrG according to the procedure published by our laboratory (Sun et al., 2019). Single spores of the transformants were cultured for further analysis.
Subcellular Localization of AoErg19
The constructed pEX2B-Aoerg19-DsRed and pEX2B-DsRed-Aoerg19 plasmids were transformed into A. oryzae 3.042 aaapyrG to determine the subcellular localization of the fusion protein. The pEX2B vector was transformed as a control. The peroxisome targeted signal (PTS1), SRL amino acid, was fused to the C-terminal of GFP using PCR to visualize peroxisomes (Escano et al., 2009). Then, the GFP sequence of pEX1-ptrA (Nguyen et al., 2016) was replaced with GFP-PTS1 to construct pEX1-ptrA-GFP-PTS1 (Sun et al., 2019). The first 72 amino acids of AoCit1 protein was used as the mitochondria targeted signal (MTS) (Takaya et al., 2009). The MTS encoded sequence was fused at the N-terminal of GFP by PCR (primers: MTS-F and MTS-R) to visualize mitochondria and the corresponding construct was named pEX1-ptrA-MTS-GFP. Aspergillus nidulans vacuolar carboxypeptidase Y (CPY) and A. oryzae vacuolar protein AoVam3 were used as vacuole maker by fusion the GFP at the C-terminal and N-terminal, respectively (Ohneda et al., 2002; Shoji et al., 2006). The CPY encoding sequences was synthetized by Sangon Biotech (Shanghai, China) and the corresponding construct was named pEX1-ptrA-CPY-GFP. The Aovam3 sequence was amplified from the cDNA of A. oryzae using AoVam3-F and AoVam3-R primers and the corresponding vector was named pEX1-ptrA-GFP-AoVam3. For co-localization, these different GFP localization vectors were transformed into A. oryzae 3.042 aaapyrG, respectively. Then, pEX2B-Aoerg19-DsRed or pEX2B-DsRed-Aoerg19 was transformed into the transformants. The fluorescence was observed using confocal microscopy (Olympus FV1000MPE2). A 488 nm and 554 argon ion laser line was used for excitation of GFP and DsRed, while 505–515 nm and 585–595 nm emission filters were used for simultaneously capturing GFP and DsRed fluorescence, respectively, using the Olympus FV1000MPE2 confocal microscope. Primer sequences used for plastid construction were provided in Supplementary Table S2. For mitochondrial staining, mycelia grown on the slides were transferred into a medium containing Mito-tracker green (Beyotime Institute of Biotechnology, Nantong, China) and incubated for 30 min at 30°C (Tanabe et al., 2011). The mycelia were then washed twice with distilled water and observed using fluorescence microscopy.
Measurement of Ergosterol, Mevalonic-5-PP and Isopentenyl-PP Content
Ergosterol extraction and quantification was performed according to Fowler et al. (2011). Briefly, the 48 h-old A. oryzae mycelium was harvested and vacuum freeze-dried to a constant weight. Then, the mycelium was pulverized into powder for the measurement of ergosterol, mevalonic-5-PP and isopentenyl-PP contents. Three milliliters of alcoholic KOH (25 g KOH plus 35 ml ddH2O, with 100% ethanol added to a total volume of 100 ml) was added to the 50 mg dry A. oryzae powder and mixed by vortexing for 1 min. The suspensions were then transferred to glass tubes and incubated at 85°C in a water bath for 1 h, after which the tubes were allowed to cool to ambient temperature. Three milliliters of n-heptane (Sigma-Aldrich, St Louis, MO, United States) was added to the tube, followed by vigorous vortexing for 3 min to extract ergosterol. The upper layer (the n-heptane layer) was isolated and stored at -20°C for 24 h prior to high-performance liquid chromatographic (HPLC) analysis. HPLC was conducted on Waters Alliance e2695 HPLC (Milford, MA) using a UV detector set at 282 nm with a Zorbax SB-C18 column. Methanol/water (95:5, v/v) was used as the mobile phase, and the elution rate was 1.5 ml min-1. Ergosterol (Sigma-Aldrich) was used to obtain a calibration curve. The measurement of mevalonic-5-PP and isopentenyl-PP content was performed by using ELISA kits (Qingdao Kechuang, China) according to the manufacturer’s instructions.
Phylogenetic Analysis of MVD
The protein sequences of different organisms were collected from NCBI2. The IDs of the sequences are as follows: Zea mays-MVD1 (GI: 670375213), Zea mays-MVD2 (GI: 1142711841), Oryza sativa-MVD1(GI: 1443059899), Oryza sativa-MDV2 (GI: 1002245003), Danio rerio (GI: 55925435), Mus musculus (GI: 256985114), Homo sapiens (GI: 568815582), Arabidopsis thaliana (GI: 240254678), S. cerevisiae (GI: 330443715), A. niger (GI: 966767527), A. oryzae (GI: 391869300), A. flavus (GI: 238491808), E. coli (GI: 1205338453), Lactobacillus composti (GI: 737476048), and Staphylococcus aureus (GI: 739736044). Phylogenetic analysis was performed using the MEGA 5 software (Tamura et al., 2011). Briefly, the full-length amino acid sequences of these homologous genes were aligned using Muscle and a maximum likelihood tree was constructed using the alignment sequence with bootstrap value of 1000. Protein sequences were aligned using the DNAMAN software.
Results
AoErg19 Is Evolutionarily Conserved in Different Organisms
To investigate the function of MVD in A. oryzae, we performed phylogenetic analysis of MVD in different organisms. As shown in Figure 1, all the analyzed organisms contain at least one copy of MVD. The evolutionary relationships of the MVD genes are consistent with species evolution. For example, based on the origin of the MVD genes, the phylogenetic tree can be divided into plants, animals, fungi, and bacterial branches; in the fungi branch, the evolutionary distance of Aspergillus species (A. niger, A. oryzae, and A. flavus) is nearer than that of S. cerevisiae. However, MVD evolved two different paralogs in dicotyledons such as Zea mays and Oryza sativa (Figure 1). Protein alignment showed that MVD is conserved across plants, animals, fungi, and bacteria (Supplementary Figure S1), and the identity is 48%. MVD in the filamentous fungus A. oryzae is also known as erg19 homolog in the yeast S. cerevisiae; therefore we named the homologous gene in A. oryzae as Aoerg19. These results indicate that Aoerg19 is evolutionarily conserved across species.
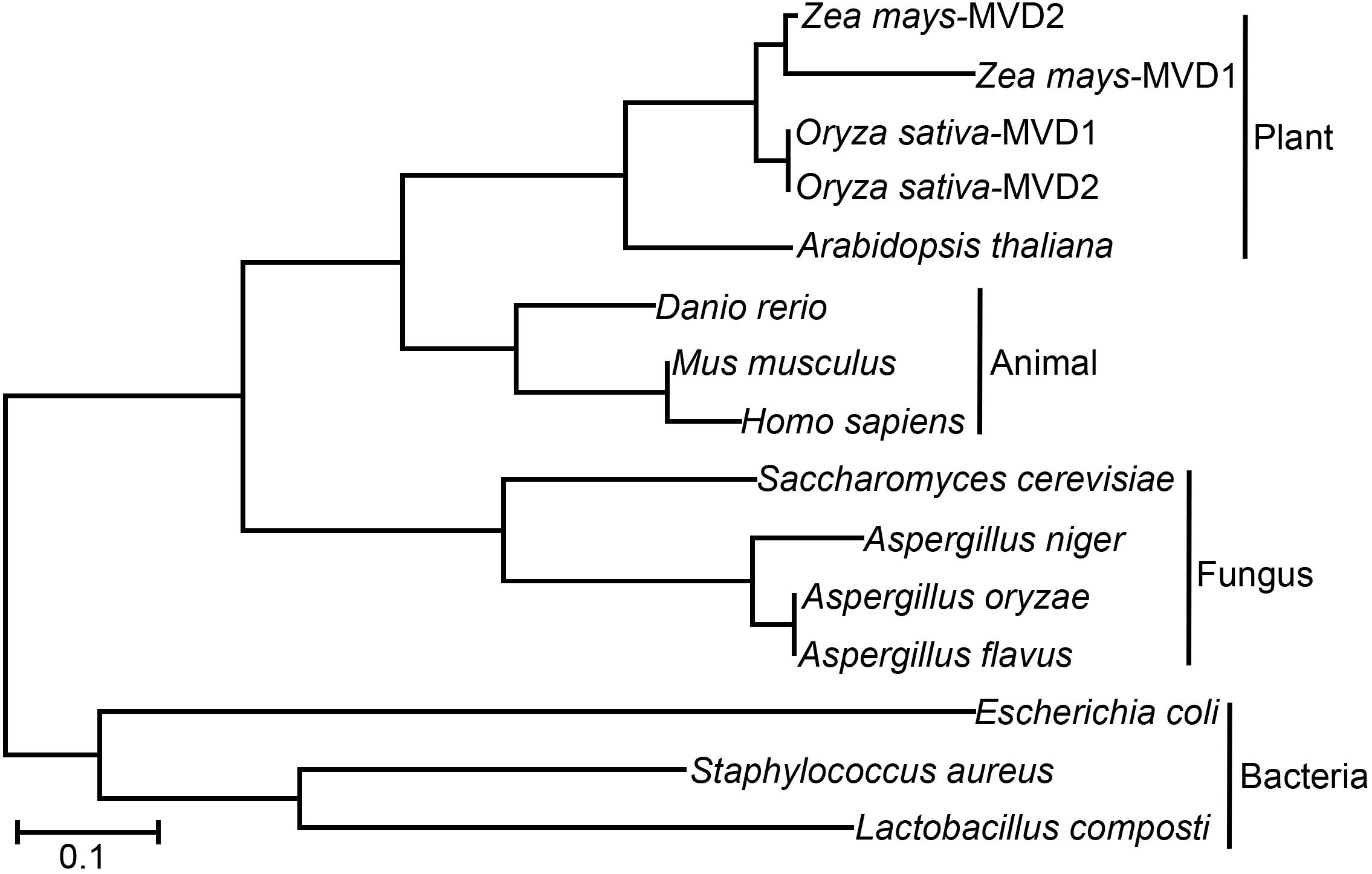
Figure 1. Phylogenetic analysis and amino acid sequence alignment of MVD in different species. Unrooted phylogenetic tree of MVD and homologous proteins in Zea Mays, Oryza sativa, Danio rerio, Mus musculus, Homo sapiens, Arabidopsis thaliana, S. cerevisiae, A. niger, A. oryzae, A. flavus, E. coli, Lactobacillus composti, and Staphylococcus aureus.
Aoerg19 Recovers the Phenotypes of the erg19 Mutant of S. cerevisiae
Reports showed that the S. cerevisiae erg19 (Y41208) mutant is temperature-sensitive (Dassanayake et al., 2002). To identify whether the function of Aoerg19 is conserved, the full length CDS of Aoerg19 was cloned into the yeast expression vector pYES2 and transformed into the erg19 mutant. As the CDS of Aoerg19 was under the GAL1 promoter in the pYES2 vector, the transformants were grown on the YPD (with glucose) and YPG (with galactose as inducer) media at 30°C and 37°C to determine whether the temperature-sensitive phenotype of erg19 was restored. Results showed that the erg19 mutation was lethal at 37°C on YPD and YPG; Aoerg19/erg19 can restore the lethal phenotype of the yeast on YPG (Figures 2A,B). Next, we detected the ergosterol content of these strains. Results showed that the Aoerg19/erg19 transformants can recover the ergosterol deficiency of the erg19 mutant (Figure 2C). Therefore, the function of AoErg19 is conserved between S. cerevisiae and A. oryzae.
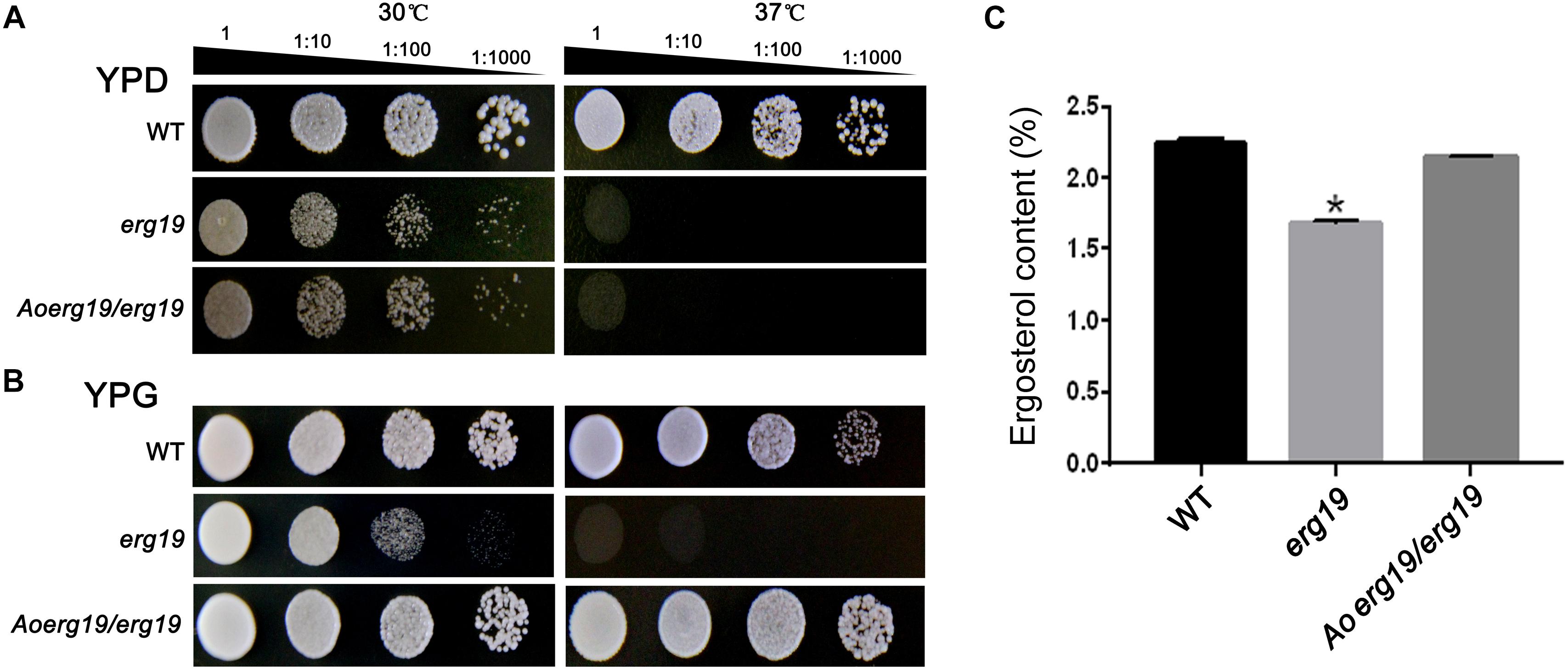
Figure 2. Phenotypes and ergosterol contents of S. cerevisiae with heterologous expression of AoErg19. (A,B) Growth of wild type S. cerevisiae, erg19 mutant, and AoErg19/erg19 transformant on YPD and YPG media. (C) Ergosterol content in wild type S. cerevisiae, erg19 mutant, and AoErg19/erg19 transformant. Asterisks denote significant differences compared to the control (P < 0.05).
Aoerg19 Expression Differed at Different Growth Stages and Stress Environments
To investigate the role of Aoerg19 during A. oryzae growth, we determined the expression patterns of Aoerg19 at different developmental stages using qRT-PCR. As shown in Figure 3A, the transcript level of Aoerg19 increased during A. oryzae, and it was maximum in the 72 h-old mycelium. The colony morphologies of A. oryzae after 24, 48, and 72 h cultivation was shown as Supplementary Figure S2. Ergosterol is reported to be involved in stress responses in S. cerevisiae (Kodedova and Sychrova, 2015), therefore we also investigated the expression of Aoerg19 under different stress conditions. A. oryzae was subjected to salt, temperature, and ethanol stress. Results showed that the Aoerg19 transcript level decreased under these abiotic stress conditions; the transcript level of Aoerg19 decreased with increase in salt and ethanol concentration and temperature (Figures 3B–D). Conclusively, the expression level of Aoerg19 changed at different growth stages and under different forms of abiotic stress.
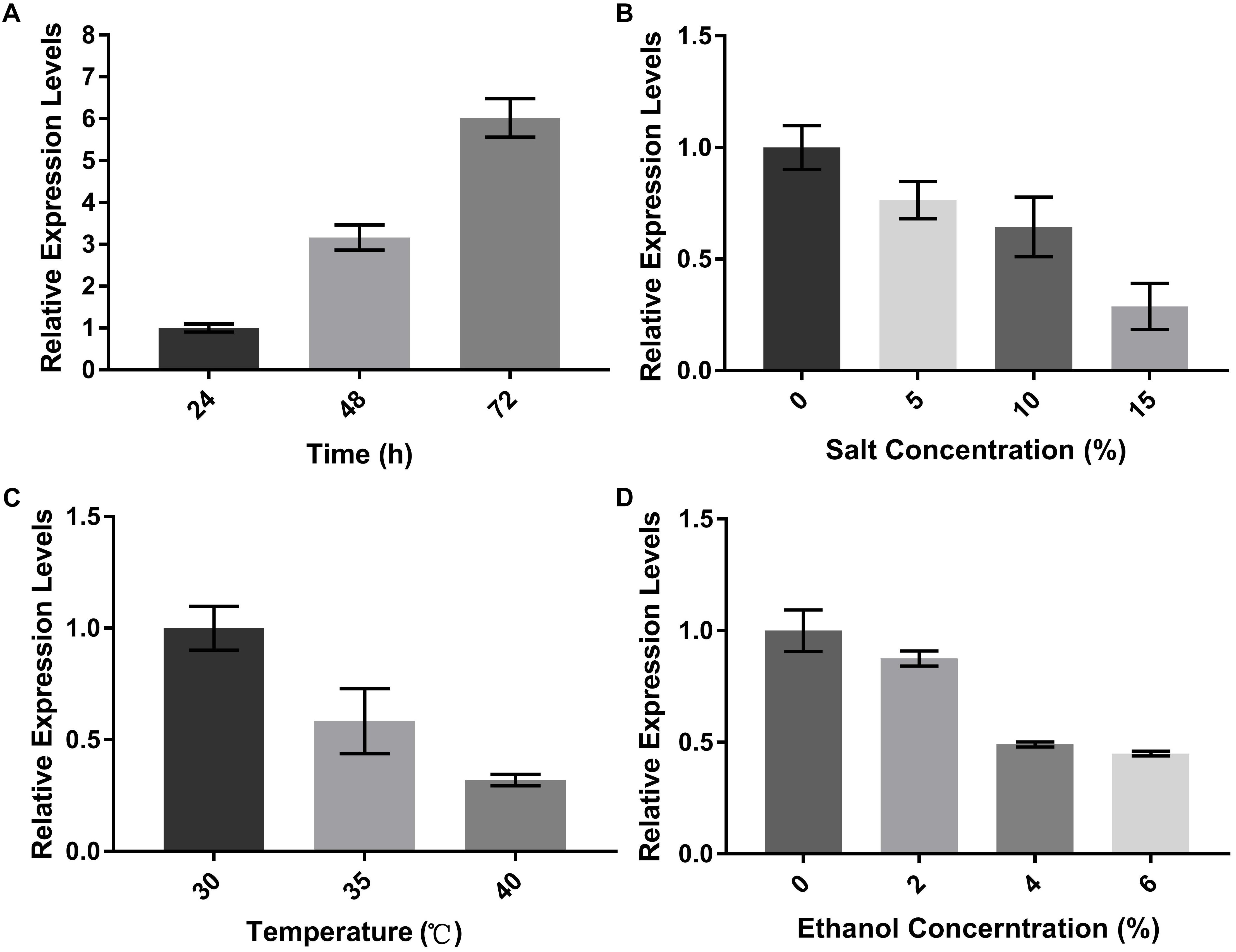
Figure 3. Expression levels of AoErg19 at different developmental stages and under different abiotic stress. (A) Expression of AoErg19 at 24, 48, and 72 h of growth; (B–D) Expression of AoErg19 under salt, temperature, and ethanol stress conditions. The wild-type A. oryzae spore suspension was plated onto CD agar medium or CD agar medium supplied with NaCl or ethanol and incubated at 30°C (except temperature stress). For the determination of AoErg19 mRNA levels in different growth stages, mycelia were harvested at 24, 48, and 72 h; for other tests, the mycelia were harvested at 48 h. Values represent the mean ± SD of three independent experiments.
AoErg19 Was Localized in the Vacuole
Mevalonate diphosphate decarboxylase has been reported to localize in cytosol or peroxisomes in plants or animals (Kovacs et al., 2002; Hogenboom et al., 2004; Michihara et al., 2008; Clastre et al., 2011; Simkin et al., 2011). However, studies on the subcellular localization of Erg19 homologs in filamentous fungi are limited. Bioinformatic analysis showed that there were no specific targeting signal sequences in the amino acid sequence of AoErg19 (data not shown). Therefore, we used DsRed as the reporter gene to investigate the subcellular localization of AoErg19 in A. oryzae. First, we fused the DsRed in the C-terminal and N-terminal region of AoErg19 to construct pYES2-DsRed-Aoerg19 and pYES2-Aoerg19-DsRed vectors. Both vectors were transformed in the S. cerevisiae erg19 mutant. Results showed that both DsRed-AoErg19 and AoErg19-DsRed can recover the temperature-sensitive phenotype and ergosterol deficiency of erg19 mutant, indicating that the DsRed tag does not affect the function of AoErg19 (Supplementary Figure S3). Then, we transferred AoErg19-DsRed into pEX2B to construct A. oryzae transformation vectors. Both vectors were transformed into A. oryzae for the determination of AoErg19 subcellular localization. We observed that DsRed fluorescence displayed punctate structure with different sizes in the mycelium (Figure 4A). To further determine the organelle of AoErg19 localization, we investigated whether AoErg19 co-localized with peroxisomes, mitochondria or vacuole. The construct containing GFP-PTS1, MTS-GFP, CPY-GFP or GFP-AoVam3 was co-transformed with AoErg19-DsRed into A. oryzae, respectively. As shown in Figures 4B,C, the DsRed fluorescence was not coincident with GFP-PTS1 or MTS-GFP fluorescence, indicating that AoErg19 was not localized in peroxisomes or mitochondria. Mitochondria staining also confirmed that AoErg19 was not co-localized with mitochondria (Supplementary Figure S4). However, the DsRed fluorescence was coincident with both the two vacuole-localized GFP fluorescence (Figure 4D). We also performed the co-localization by co-transformation DsRed-AoErg19 with these different localized GFP vectors and find that AoErg19-DsRed was also co-localized with vacuole-localized GFP fluorescence (Supplementary Figure S5). Therefore, we concluded that AoErg19 was located in the vacuole.
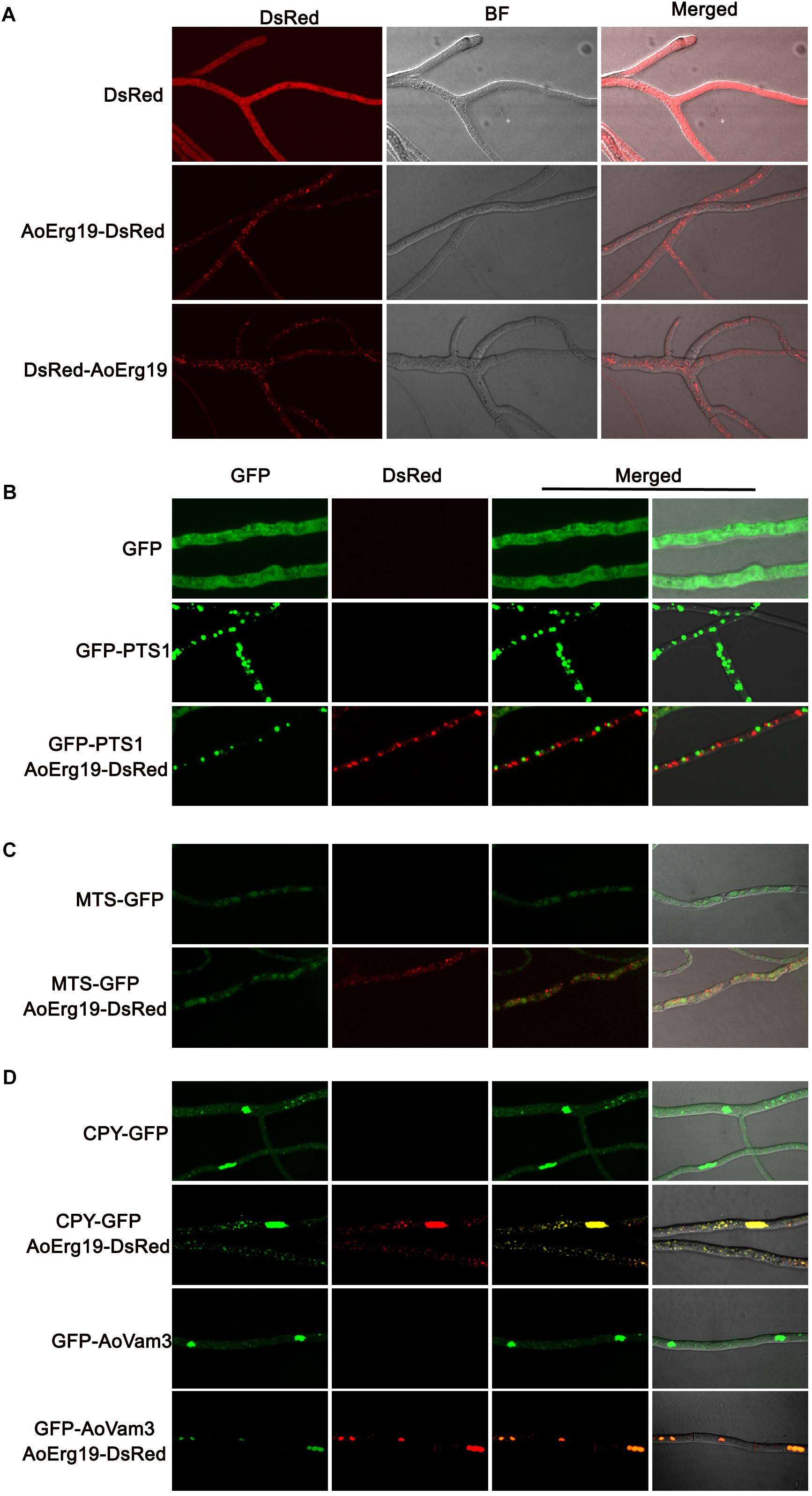
Figure 4. Subcellular localization of AoErg19. (A) The mycelium of A. oryzae 3.042 aaapyrG transformed with DsRed, AoErg19-DsRed, and DsRed-AoErg19 vectors. Left to right: fluorescent image of DsRed, bright field, and merged image of DsRed and bright field. (B) Co-localization of AoErg19-DsRed and peroxisome. The mycelium of A. oryzae 3.042 ΔpyrG transformed with GFP, GFP-PTS1, and co-transformed with GFP-PTS1 and AoErg19-DsRed. (C) Co-localization of AoErg19-DsRed and mitochondria. The mycelium of A. oryzae 3.042 ΔpyrG transformed with MTS-GFP and AoErg19-DsRed. (D) Co-localization of AoErg19-DsRed and vacuole. The mycelium of A. oryzae 3.042 aaapyrG transformed with CPY-GFP and GFP-AoVam3, and co-transformed with CPY-GFP or GFP-AoVam3 with AoErg19-DsRed, respectively. In (B–D), left to right: fluorescent image of GFP, DsRed, merged image of GFP and DsRed, and merged image of GFP, DsRed and bright field (data not shown).
Overexpression and RNAi of Aoerg19 Affects the Sporulation and Ergosterol Content of A. oryzae
We took advantage of the overexpression vector and the decrease of the target gene expression by RNAi to determine the function of Aoerg19 in A. oryzae. As shown in Figure 5, the colony diameters of AoErg19 overexpression strains were not significantly different from that of the control; however, the RNAi strains were smaller (Figures 5A–C). The mRNA levels of Aoerg19 in three overexpression strains and five RNAi strains were detected to confirm the effect of overexpression and RNAi. We observed that both overexpression and RNAi worked efficiently (Figures 5D,E). Then, the AoOE1# and AoRNAi3# were selected for further identification. The growth rate of AoOE1# colony was not significantly different from that of the control; however, the growth rate of AoRNAi3# was significantly lower (Figure 5F). Quantitation of the spore number and sporulation time of AoOE1# and AoRNAi3# showed that the spore number of both AoOE1# and AoRNAi3# decreased, whereas sporulation time of both increased (Figure 5G).
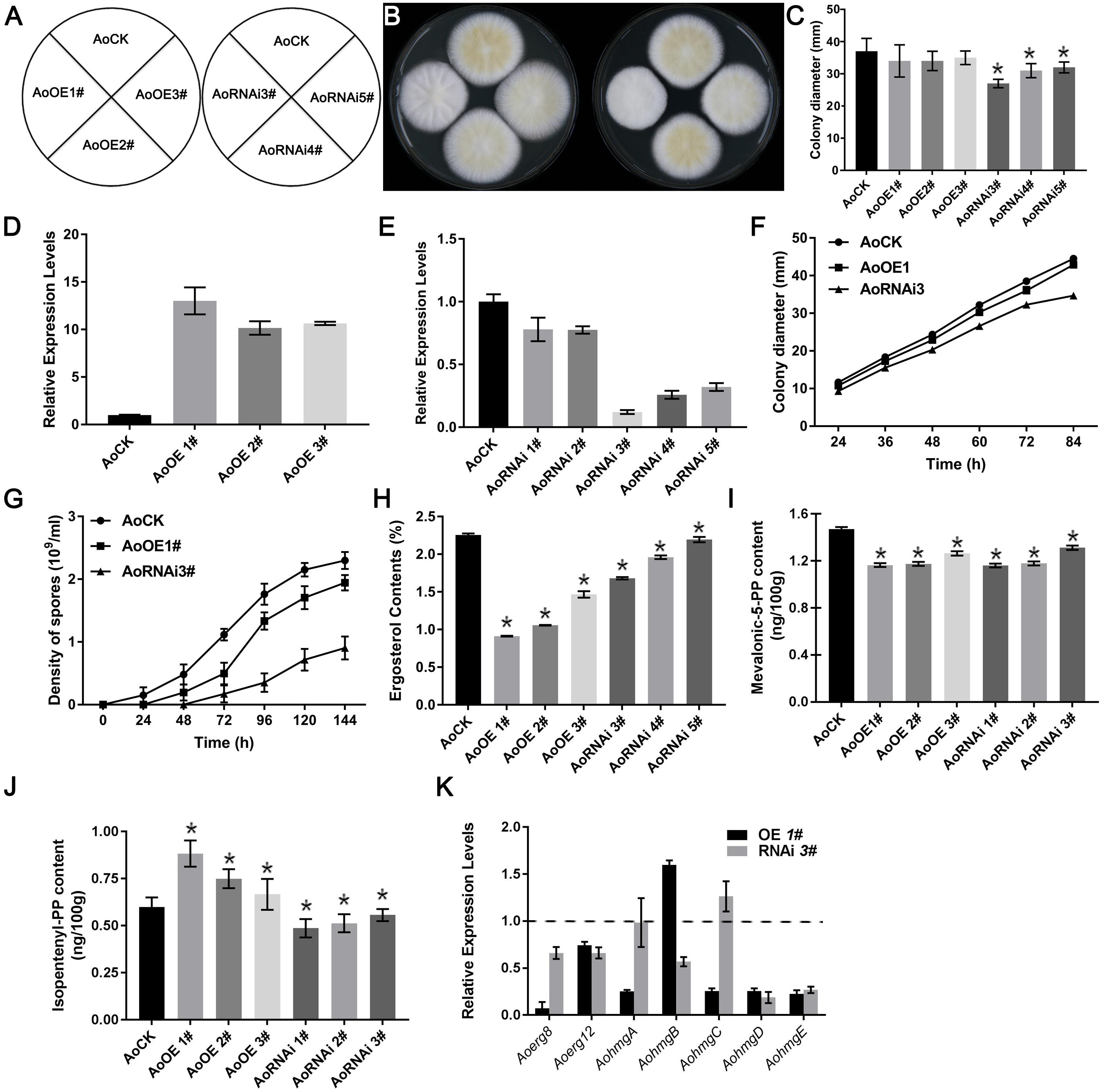
Figure 5. Ergosterol contents of AoErg19 overexpression and RNAi strains. (A) Scheme showing different transgenic strains. (B) Colony morphologies of the control (AoCK, wild-type A. oryzae), overexpression and RNAi strains on the DPY media after 72 h of incubation. Spore suspension of identical concentrations of different A. oryzae strains were plated onto DPY agar medium and incubated at 30°C for 72 h. (C) Diameter of different transgenic strain colonies. (D,E) Relative mRNA levels of AoErg19 in overexpression and RNAi strains. (F) The colony diameter of AoOE1# and AoRNAi3# at different growth time. (G) The spore number and sporulation time of AoOE1# and AoRNAi3#. (H) Ergosterol content in Aoerg19 overexpression and RNAi strains. (I) Mevalonic-5-PP contents in AoErg19 overexpression and RNAi strains. (J) Isopentenyl-PP contents in AoErg19 overexpression and RNAi strains. (K) Expression levels of Aoerg8, Aoerg12, and Aohmgs in AoOE1# and AoRNAi3#. The colony diameter was calculated using the crisscross method, in which three independent experiments were performed. For qRT-PCR, the mycelia were harvested at 48 h; values represent the mean ± SD of three independent experiments. Asterisks denote significant differences compared to the control (P < 0.05).
Because Aoerg19 acts as the upstream of the ergosterol biosynthesis pathway, impairment or overexpression of erg19 decreased ergosterol content in S. cerevisiae (Bergès et al., 1997). Hence, we detected the ergosterol contents of strains AoOE1#-AoOE3# and AoRNAi3#-AoRNAi5#. As expected, the ergosterol contents were reduced in the RNAi strains (Figure 5H). However, the ergosterol contents also decreased significantly in the overexpression strains, and were even lower than those of the RNAi strains (Figure 5H). We also detected the contents of the direct substrate (mevalonic-5-PP) and product (isopentenyl-PP) of AoErg19 in both OE and RNAi A. oryzae strains. The results showed that in both OE and RNAi strains, mevalonic-5-PP was decreased (Figure 5I); in OE strains, isopentenyl-PP contents increased while the isopentenyl-PP content was decreased in RNAi strains (Figure 5J). To investigate whether the decrease in ergosterol content in the overexpression strains was caused by feedback regulation, we used qRT-PCR to assess the expression of genes located in the upstream of Aoerg19 (Aoerg8, Aoerg12, and Aohmgs) in the ergosterol biosynthesis pathway. Results showed that with the exception of AohmgB, the expression of these genes was reduced in AoOE1#, and Aoerg8 showed the maximum reduction (Figure 5K), indicating that AoErg8 is the feedback target in the Aoerg19 overexpression strains; of the expression of these genes was lowered in AoRNAi3#, with the exception of AohmgC (Figure 5K). Taken together, these results showed that both up- and down-regulation of eAoerg19 mRNA levels in A. oryzae impair sporulation and decrease ergosterol content.
Aoerg19 Overexpression and RNAi Strains Are More Sensitive to Abiotic Stress
Ergosterol deficiency in S. cerevisiae rendered it more sensitive to ergosterol biosynthesis inhibitor (Zuzana et al., 2013), and overexpression of erg19 increased sensitivity to NaCl stress (Bhattacharya et al., 2018). Therefore, we investigated the growth of Aoerg19 overexpression and RNAi strains to different abiotic stresses, including ergosterol biosynthesis inhibitor, temperature, salt, and ethanol. The differences among the three colonies on the DPY media were not obvious at 37°C (Figure 6B). However, we observed that under liquid culture conditions, the hyphal pellets of the three strains showed different phenotypes. For example, at 30°C, all the three strains can form hyphal pellets, although their shapes were slightly different (Figure 6D); the hyphal pellets were smaller at 37°C; however, the hyphal pellets of AoOE1# and AoRNAi3# strains were considerably smaller than that of the control (Figure 6D). The AoOE1# and AoRNAi3# strains were more sensitive than the control to the ergosterol biosynthesis inhibitor, temperature, salt, and ethanol treatment (Figures 6E–K and Supplementary Figure S6). Interestingly, the color of the overexpression colony turned red under ethanol stress (Figure 6H) which may be because of the accumulation AoErg19-DsRed protein. Thus, overexpression and RNAi of Aoerg19 appear to increase sensitivity to different abiotic stress conditions.
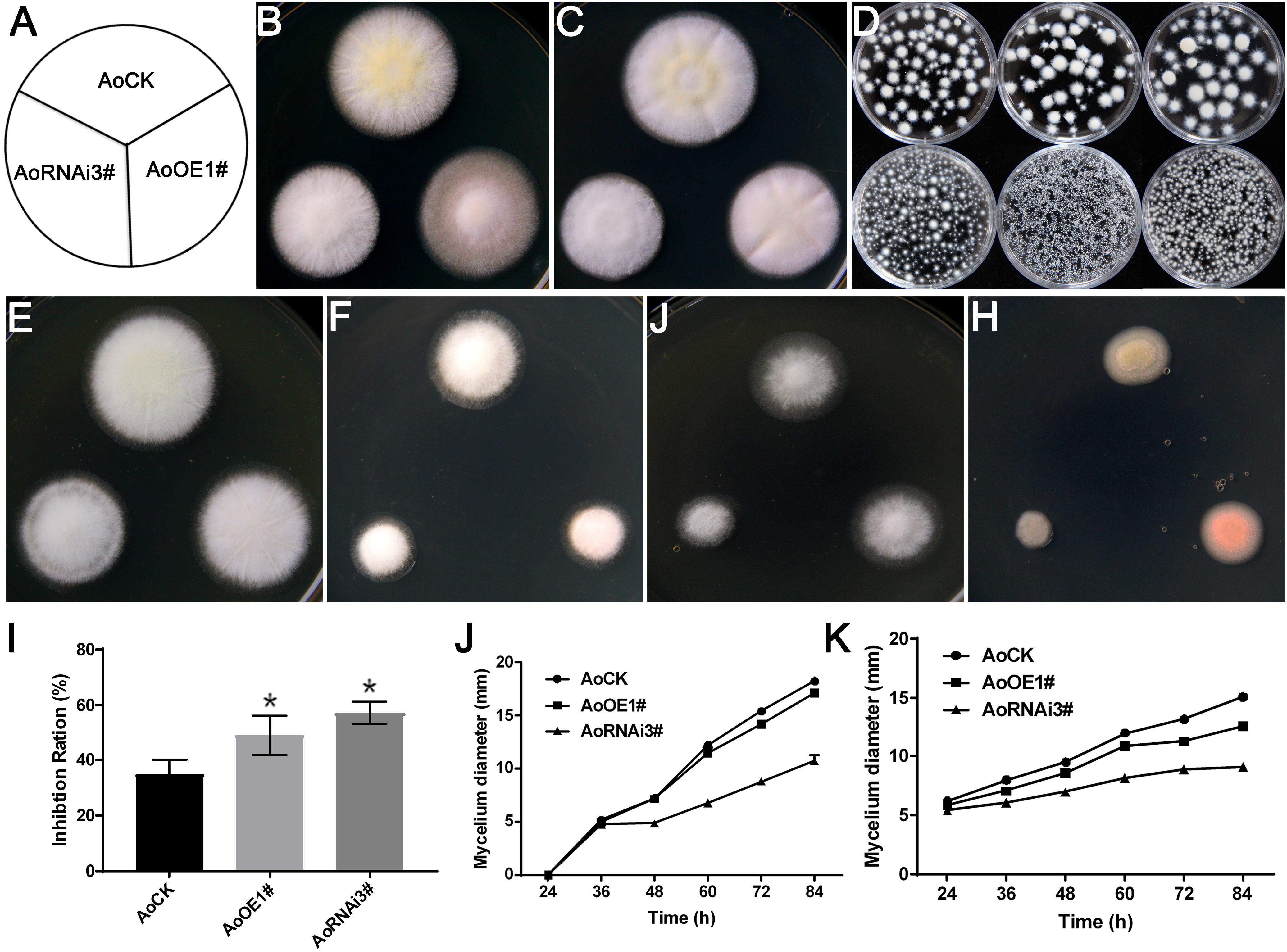
Figure 6. Colony morphologies of control, AoOE1#, and AoRNAi3# strains under different stress conditions. (A) Scheme showing the different A. oryzae strains in Figure 6. (B,C) The colony of control, overexpression, and RNAi strains cultured for 60 h on DPY media at 30 and 37°C. (D) Phenotypes of control, overexpression, and RNAi strains at 30°C (upper panel) and 37°C (lower panel) in liquid DPY media, from left to right: AoCK, AoOE1#, AoRNAi3#. (E–H) Phenotypes of control, AoOE1#, and RNAi3# strains cultured on DPY, DPY with 0.35 μg/L terbinafine, DPY with 10% NaCl, and DPY with 4% ethanol. (I) The inhibition ratio of terbinafine on the growth of control, AoOE1#, and RNAi3# stains. (J,K) The diameter of different strains cultured on 10% NaCl and 4% ethanol media at different time points. Asterisks denote significant differences compared to the control (P < 0.05).
Discussion
The identification of genes involved in MVA pathway is important for genetic modification of MVA pathway, which has potential application for producing isoprenoids, an important feed-stock for commercial production of synthetic rubber and a high energy biofuel (Ma et al., 2011; Miziorko, 2011). Bioisoprene is synthesized by isoprene synthase from dimethylallyl diphosphate (DMAPP), the direct biosynthesis product of MVD (Hu et al., 2017). Meanwhile, in fungi, the MVA pathway is the upstream of ergosterol biosynthesis pathway, which has been well studied in S. cerevisiae (Hu et al., 2019). However, there was little research about ergosterol biosynthesis pathway in A. oryzae. Our previous study revealed that the ergosterol biosynthesis pathway is conserved between S. cerevisiae and A. oryzae by bioinformatics analysis. However, the ergosterol biosynthesis genes in A. oryzae are more complicated as there are multiple paralogs encoding the same biosynthetic enzymes (Hu et al., 2019). The research of Aoerg19 does not only of significance in understanding MVA pathway but also in ergosterol biosynthesis pathway.
The Function of AoErg19 in A. oryzae
The MVA pathway was conserved among different species (Jonathan and David, 2011). The function of MVD (AoErg19) in A. oryzae has not been identified. In this study, phylogenetic analysis and yeast complementation assay suggested that MVD function was conserved across fungal species (Figures 1, 2). The Aoerg19 overexpression and RNAi strains of A. oryzae showed delayed sporulation and sensitive to abiotic stress, indicating that ergosterol is involved in sporulation and stress response. Besides, we have noticed that under ethanol stress, the color of the overexpression colony turns red, which may be caused by the accumulation the AoErg19-DsRed protein, indicating that ethanol stress can also affect the accumulation of AoErg19. The subcellular localization of MVD in different species has been studied and it was reported to vary in plants and animals. In plants MVD was located in peroxisomes, whereas in animals it was mainly located in the cytosol (Kovacs et al., 2002; Hogenboom et al., 2004; Michihara et al., 2008; Clastre et al., 2011; Simkin et al., 2011). However, the MVD of filamentous fungi have been scarcely studied. Here, we demonstrated that MVD in A. oryzae was located in the vacuole (Figure 4). The different subcellular localization of MVD among plants, animals, and fungi can provide some information for genetic modification of MVA pathway across different species.
Ergosterol Content in Aoerg19 Overexpression and RNAi Strains
Ergosterol is a specific cell membrane component that is not only an important cell membrane component that affects fluidity and permeability, but also plays important roles in fungal growth, reproduction, and stress tolerance (Kodedova and Sychrova, 2015). Usually, ergosterol content is strictly regulated to appropriate levels by feedback regulation of ergosterol synthesizing activities at the transcriptional, translational, and posttranslational levels (Espenshade and Hughes, 2007). In yeast, many rate-limiting reactions in ergosterol biosynthesis have been identified. For example, the HMG is the first rate-limiting enzyme; overexpression HMG in S. cerevisiae resulted in squalene accumulation, but did not affect the content of ergosterol (Polakowski et al., 1998). Other enzymes such as Erg3, Erg4, Erg11, and Erg27 are also reported to be the possible rate-limiting enzyme in S. cerevisiae (Lees et al., 1995; Arthingtonskaggs et al., 1996; Veen et al., 2003). In filamentous fungi, there are also some researches about ergosterol biosynthesis, and main of them focused on fungicide discovery or drug resistance by using ergosterol biosynthesis inhibitors (Alcazar-Fuoli and Mellado, 2012; Shao et al., 2016; Dhingra and Cramer, 2017). Little of them focused on the function identification of these genes.
In A. oryzae, the research about MVA or ergosterol pathway is of great significance because A. oryzae is the GRAS microorganism and has broadly applied in industry. Our previous studies showed that ergosterol biosynthesis in A. oryzae is more complicated than in S. cerevisiae (Hu et al., 2019). In this study, we revealed that overexpression of Aoerg19 resulting in the decreased ergosterol content by feedback regulation of Aoerg8 (Figure 5H). The intermediate, mevalonic-5-PP was decreased in both OE and RNAi A. oryzae strains which was possible the comprehensive results of the gene expression of Aoerg8 and Aoerg19 (Figure 5I). Consistent with the function of ergosterol in fungal growth, reproduction, and stress tolerance, the decrease in ergosterol content in both overexpression and RNAi strains rendered A. oryzae more sensitive to abiotic stress (Figure 6). We observed that although the ergosterol content of the overexpression strain was lower than that of the RNAi strain, the RNAi stains are more sensitive to the abiotic stress (Figure 6). One explanation is that the ergosterol content in the overexpression strains was reduced by feedback regulation, and under stress conditions, the feedback regulation adjusted the ergosterol level to a suitable level for survival; alternatively, the accumulation of IPP, geranyl diphosphate, or FPP in the overexpression strains can act as anti-stress agents that assist in adapting to stress conditions.
Conclusion
In summary, this study determined the function, expression pattern, and subcellular localization of Aoerg19. Overexpression of Aoerg19 decreased ergosterol content, suggesting Aoerg19 is the rate-limiting enzyme in the sterol biosynthesis pathway of A. oryzae. Further studies about the regulation network in ergosterol biosynthesis can be done to uncover the feedback regulation of MVA and ergosterol pathway. Thus, the gene function identification of Aoerg19 in A. oryzae may provide some useful information for the genetic modification of MVA pathway in A. oryzae.
Author Contributions
ZH and BZ designed the study and wrote the manuscript. YS and YN performed the main experiments. HH participated in the subcellular localization of AoErg19. BH and LM participated in the analysis of sterols. YT participated in the confocal microscopy. V-TT contributed to technical support of A. oryzae transformation and manuscript revision.
Funding
This study was supported by the National Natural Science Foundation of China (NSFC Grant Nos. 31700068 and 31460447), Natural Science Foundation of Jiangxi Province (Grant No. 20181BAB214001), and the Science and Technology Research Project of Jiangxi Provincial Department of Education (Grant No. GJJ180600).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01074/full#supplementary-material.
Footnotes
References
Abbassi, S., Patel, K., Khan, B., Bhosale, S., and Gaikwad, S. (2016). Functional and conformational transitions of mevalonate diphosphate decarboxylase from Bacopa monniera. Int. J. Biol. Macromol. 83, 160–170. doi: 10.1016/j.ijbiomac.2015.11.067
Abbassi, S. J., Vishwakarma, R. K., Patel, P., Kumari, U., and Khan, B. M. (2015). Bacopa monniera recombinant mevalonate diphosphate decarboxylase: biochemical characterization. Int. J. Biol. Macromol. 79, 661–668. doi: 10.1016/j.ijbiomac.2015.05.041
Alcazar-Fuoli, L., and Mellado, E. (2012). Ergosterol biosynthesis in aspergillus fumigatus: its relevance as an antifungal target and role in antifungal drug resistance. Front. Microbiol. 3:439. doi: 10.3389/fmicb.2012.00439
Arthingtonskaggs, B. A., Crowell, D. N., Yang, H., Sturley, S. L., and Bard, M. (1996). Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. Febs Lett. 392, 161–165. doi: 10.1016/0014-5793(96)00807-1
Beni, A., Soki, E., Lajtha, K., and Fekete, I. (2014). An optimized HPLC method for soil fungal biomass determination and its application to a detritus manipulation study. J. Microbiol. Meth. 103, 124–130. doi: 10.1016/j.mimet.2014.05.022
Bergès, T., Guyonnet, D., and Karst, F. (1997). The Saccharomyces cerevisiae mevalonate diphosphate decarboxylase is essential for viability, and a single Leu-to-Pro mutation in a conserved sequence leads to thermosensitivity. J. Bacteriol. 179, 4664–4670. doi: 10.1128/jb.179.15.4664-4670.1997
Bhattacharya, S., Esquivel, B. D., and White, T. C. (2018). Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio 9:e1291–18. doi: 10.1128/mBio.01291-18
Chappell, J. (1995). Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu. Rev. Plant Phys. 46, 521–547. doi: 10.1146/annurev.pp.46.060195.002513
Clastre, M., Papon, N., Courdavault, V., Giglioli-Guivarc’H, N., St-Pierre, B., and Simkin, A. J. (2011). Subcellular evidence for the involvement of peroxisomes in plant isoprenoid biosynthesis. Plant Signal. Behav. 6, 2044–2046. doi: 10.4161/psb.6.12.18173
Cordier, H., Karst, F., and Bergès, T. (1999). Heterologous expression in Saccharomyces cerevisiae of an Arabidopsis thaliana cDNA encoding mevalonate diphosphate decarboxylase. Plant Mol. Biol. 39, 953–967. doi: 10.1023/A:1006181720100
Dassanayake, R. S., Cao, L., Samaranayake, L. P., and Berges, T. (2002). Characterization, heterologous expression and functional analysis of mevalonate diphosphate decarboxylase gene (MVD) of Candida albicans. Mol. Genet. Genomics 267, 281–290. doi: 10.1007/s00438-002-0648-7
Dhingra, S., and Cramer, R. A. (2017). Regulation of sterol biosynthesis in the human fungal pathogen aspergillus fumigatus: opportunities for therapeutic development. Front. Microbiol. 8:92. doi: 10.3389/fmicb.2017.00092
Escano, C. S., Juvvadi, P. R., Jin, F. J., Takahashi, T., Koyama, Y., Yamashita, S., et al. (2009). Disruption of the aopex11-1 gene involved in peroxisome proliferation leads to impaired woronin body formation in Aspergillus oryzae. Eukaryot Cell 8, 296–305. doi: 10.1128/EC.00197-08
Espenshade, P. J., and Hughes, A. L. (2007). Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 41, 401–427. doi: 10.1146/annurev.genet.41.110306.130315
Fowler, D. M., Cooper, S. J., Stephany, J. J., Hendon, N., Nelson, S., and Fields, S. (2011). Suppression of statin effectiveness by copper and zinc in yeast and human cells. Mol. Biosyst. 7, 533–544. doi: 10.1039/c0mb00166j
Görög, S. (2011). Advances in the analysis of steroid hormone drugs in pharmaceuticals and environmental samples (2004–2010). J. Pharm. Biomed. 55, 728–743. doi: 10.1016/j.jpba.2010.11.011
Grant, S. G., Jessee, J., Bloom, F. R., and Hanahan, D. (1990). Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U.S.A. 87, 4645–4649. doi: 10.1073/pnas.87.12.4645
Hogenboom, S., Tuyp, J. M., Koster, J., Wanders, R. J., and Waterham, H. R. (2004). Human mevalonate pyrophosphate decarboxylase is localized in the cytosol. Mol. Genet. Metab. 81, 216–224. doi: 10.1016/j.ymgme.2003.12.001
Hu, Z., He, B., Ma, L., Sun, Y., Niu, Y., and Zeng, B. (2017). Recent advances in ergosterol biosynthesis and regulation mechanisms in Saccharomyces cerevisiae. Indian J. Microbiol. 57, 270–277. doi: 10.1007/s12088-017-0657-1
Hu, Z., Li, G., Sun, Y., Niu, Y., Ma, L., He, B., et al. (2019). Gene transcription profiling of Aspergillus oryzae 3.042 treated with ergosterol biosynthesis inhibitors. Braz. J. Microbiol. 50, 43–52. doi: 10.1007/s42770-018-0026-1
Jonathan, L., and David, M. (2011). Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol. Biol. Evol. 28, 87–99. doi: 10.1093/molbev/msq177
Kodedova, M., and Sychrova, H. (2015). Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS One 10:e0139306. doi: 10.1371/journal.pone.0139306
Kovacs, W. J., Olivier, L. M., and Krisans, S. K. (2002). Central role of peroxisomes in isoprenoid biosynthesis. Prog. Lipid Res. 41, 369–391. doi: 10.1016/S0163-7827(02)00002-4
Krepkiy, D., and Miziorko, H. M. (2004). Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of phospho transferases. Protein Sci. 13, 1875–1881. doi: 10.1110/ps.04725204
Lazo, G. R., Stein, P. A., and Ludwig, R. A. (1991). A DNA transformation-competent arabidopsis genomic library in agrobacterium. Biotechnology 9, 963–967. doi: 10.1038/nbt1091-963
Lees, N. D., Skaggs, B., Kirsch, D. R., and Bard, M. (1995). Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae —a review. Lipids 30, 221–226. doi: 10.1007/BF02537824
Ma, S. M., Garcia, D. E., Reddingjohanson, A. M., Friedland, G. D., Chan, R., Batth, T. S., et al. (2011). Optimization of a heterologous mevalonate pathway through the use of variant HMG-CoA reductases. Metab. Eng. 13, 588–597. doi: 10.1016/j.ymben.2011.07.001
Maruyama, J., Nakajima, H., and Kitamoto, K. (2002). Observation of EGFP-visualized nuclei and distribution of vacuoles in Aspergillus oryzae arpA null mutant. FEMS Microbiol. Lett. 206, 57–61. doi: 10.1111/j.1574-6968.2002.tb10986.x
Michihara, A., Morita, S., Toda, K., Akasaki, K., and Tsuji, H. (2008). Mevalonate pyrophosphate decarboxylase is predominantly located in the cytosol of both B16 and B16F10 cells in mouse melanoma treated with lovastatin. J. Health Sci. 54, 216–223. doi: 10.1248/jhs.54.216
Michihara, A., Sawamura, M., Nara, Y., Ikeda, K., and Yamori, Y. (1998). Lower mevalonate pyrophosphate decarboxylase activity is caused by the reduced amount of enzyme in stroke-prone spontaneously hypertensive rat. J. Biochem. 124, 40–44. doi: 10.1093/oxfordjournals.jbchem.a022094
Miziorko, H. M. (2011). Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch. Biochem. Biophys. 505, 131–143. doi: 10.1016/j.abb.2010.09.028
Nguyen, K. T., Ho, Q. N., Do, L., Mai, L. T. D., Pham, D. N., Tran, H. T. T., et al. (2017). A new and efficient approach for construction of uridine/uracil auxotrophic mutants in the filamentous fungus Aspergillus oryzae using Agrobacterium tumefaciens-mediated transformation. World J. Microbiol. Biotechnol. 33:107. doi: 10.1007/s11274-017-2275-9
Nguyen, K. T., Ho, Q. N., Pham, T. H., Phan, T. N., and Tran, V. T. (2016). The construction and use of versatile binary vectors carrying pyrG auxotrophic marker and fluorescent reporter genes for agrobacterium-mediated transformation of Aspergillus oryzae. World J. Microbiol. Biotechnol. 32:204. doi: 10.1007/s11274-016-2168-3
Ohneda, M., Arioka, M., Nakajima, H., and Kitamoto, K. (2002). Visualization of vacuoles in Aspergillus oryzae by expression of CPY-EGFP. Fungal Genet. Biol. 37, 29–38. doi: 10.1016/s1087-1845(02)00033-6
Polakowski, T., Stahl, U., and Lang, C. (1998). Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl. Microbiol. Biotechnol. 49, 66–71. doi: 10.1007/s002530051138
Shao, J., Shi, G., Wang, T., Wu, D., and Wang, C. (2016). Antiproliferation of berberine in combination with fluconazole from the perspectives of reactive oxygen species, ergosterol and drug efflux in a fluconazole-resistant candida tropicalis isolate. Front. Microbiol. 7:1516. doi: 10.3389/fmicb.2016.01516
Shi, L., Qin, L., Xu, Y., Ren, A., Fang, X., Mu, D., et al. (2012). Molecular cloning, characterization, and function analysis of a mevalonate pyrophosphate decarboxylase gene from Ganoderma lucidum. Mol. Biol. Rep. 39, 6149–6159. doi: 10.1007/s11033-011-1431-9
Shoji, J. Y., Arioka, M., and Kitamoto, K. (2006). Vacuolar membrane dynamics in the filamentous fungus Aspergillus oryzae. Eukaryot Cell 5, 411–421. doi: 10.1128/ec.5.2.411-421.2006
Simkin, A. J., Guirimand, G., Papon, N., Courdavault, V., Thabet, I., Ginis, O., et al. (2011). Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta 234, 903–914. doi: 10.1007/s00425-011-1444-6
Sun, Y., Niu, Y., He, B., Ma, L., Li, G., Tran, V. T., et al. (2019). A dual selection marker transformation system using Agrobacterium tumefaciens for the industrial Aspergillus oryzae 3.042. J. Microbiol. Biotechnol. 29, 230–234. doi: 10.4014/jmb.1811.11027
Takaya, K., Higuchi, Y., Kitamoto, K., and Arioka, M. (2009). A cytosolic phospholipase A2-like protein in the filamentous fungus Aspergillus oryzae localizes to the intramembrane space of the mitochondria. FEMS Microbiol. Lett. 301, 201–209. doi: 10.1111/j.1574-6968.2009.01818.x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tanabe, Y., Maruyama, J., Yamaoka, S., Yahagi, D., Matsuo, I., Tsutsumi, N., et al. (2011). Peroxisomes are involved in biotin biosynthesis in aspergillus and arabidopsis. J. Biol. Chem. 286, 30455–30461. doi: 10.1074/jbc.M111.247338
Veen, M., Stahl, U., and Lang, C. (2003). Combined overexpression of genes of the ergosterol biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae. FEMS Yeast Res. 4, 87–95. doi: 10.1016/s1567-1356(03)00126-0
Voynova, N. E., Fu, Z., Battaile, K. P., Herdendorf, T. J., Kim, J. J. P., and Miziorko, H. M. (2008). Human mevalonate diphosphate decarboxylase: characterization, investigation of the mevalonate diphosphate binding site, and crystal structure. Arch. Biochem. Biophys. 480, 58–67. doi: 10.1016/j.abb.2008.08.024
Yamada, R., Yoshie, T., Wakai, S., Asai-Nakashima, N., Okazaki, F., Ogino, C., et al. (2014). Aspergillus oryzae-based cell factory for direct kojic acid production from cellulose. Microb. Cell Fact. 13:71. doi: 10.1186/1475-2859-13-71
Keywords: mevalonate diphosphate decarboxylase, Aspergillus oryzae, sporulation, stress tolerance, ergosterol biosynthesis
Citation: Sun Y, Niu Y, Huang H, He B, Ma L, Tu Y, Tran V-T, Zeng B and Hu Z (2019) Mevalonate Diphosphate Decarboxylase MVD/Erg19 Is Required for Ergosterol Biosynthesis, Growth, Sporulation and Stress Tolerance in Aspergillus oryzae. Front. Microbiol. 10:1074. doi: 10.3389/fmicb.2019.01074
Received: 03 February 2019; Accepted: 29 April 2019;
Published: 16 May 2019.
Edited by:
Maurizio Sanguinetti, Catholic University of the Sacred Heart, ItalyReviewed by:
Satoshi Suzuki, National Food Research Institute, JapanGang Liu, Chinese Academy of Sciences, China
Copyright © 2019 Sun, Niu, Huang, He, Ma, Tu, Tran, Zeng and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Zeng, WmVuZ3R4MDAxQGFsaXl1bi5jb20= Zhihong Hu, aHV6aGlob25nNDI2QDE2My5jb20=
 Yunlong Sun1
Yunlong Sun1 Van-Tuan Tran
Van-Tuan Tran Zhihong Hu
Zhihong Hu