- 1Produce Safety and Microbiology Research Unit, U.S. Department of Agriculture, Agricultural Research Services, Western Regional Research Center, Albany, CA, United States
- 2School of Food and Agriculture, University of Maine, Orono, ME, United States
- 3Department of Plant and Soil Science, Texas Tech University, Lubbock, TX, United States
- 4Department of Biological Science and Technology, National Chiao Tung University, Hsinchu, Taiwan
Optical biosensors for rapid detection of significant foodborne pathogens are steadily gaining popularity due to its simplicity and sensitivity. While nanomaterials such as gold nanoparticles (AuNPs) are commonly used as signal amplifiers for optical biosensors, AuNPs can also be utilized as a robust biosensing platform. Many reported optical biosensors were designed for individual pathogen detection in a single assay and have high detection limit (DL). Salmonella spp. is one of the major causative agents of foodborne sickness, hospitalization and deaths. Unfortunately, there are around 2,000 serotypes of Salmonella worldwide, and rapid and simultaneous detection of multiple strains in a single assay is lacking. In this study, a comprehensive and highly sensitive simultaneous colorimetric detection of nineteen (19) environmental and outbreak Salmonella spp. strains was achieved by a novel optical biosensing platform using oligonucleotide-functionalized AuNPs. A pair of newly designed single stranded oligonucleotides (30-mer) was displayed onto the surface of AuNPs (13 nm) as detection probes to hybridize with a conserved genomic region (192-bases) of ttrRSBCA found on a broad range of Salmonella spp. strains. The sandwich hybridization (30 min, 55°C) resulted in a structural formation of highly stable oligonucleotide/AuNPs-DNA complexes which remained undisturbed even after subjecting to an increased salt concentration (2 M, final), thus allowing a direct discrimination via color change of target (red color) from non-target (purplish-blue color) reaction mixtures by direct observation using the naked eye. In food matrices (blueberries and chicken meat), nineteen different Salmonella spp. strains were concentrated using immunomagnetic separation and then simultaneously detected in a 96-well microplate by oligonucleotide-functionalized AuNPs after DNA preparation. Successful oligonucleotide/AuNPs-DNA hybridization was confirmed by gel electrophoresis while AuNPs aggregation in non-target and control reaction mixtures was verified by both spectrophotometric analysis and TEM images. Results showed that the optical AuNP biosensing platform can simultaneously screen nineteen (19) viable Salmonella spp. strains tested with 100% specificity and a superior detection limit of <10 CFU/mL or g for both pure culture and complex matrices setups. The highly sensitive colorimetric detection system can significantly improve the screening and detection of viable Salmonella spp. strains present in complex food and environmental matrices, therefore reducing the risks of contamination and incidence of foodborne diseases.
Introduction
Nanomaterials are commonly incorporated in the development of optical biosensors to enhance its sensitivity (Pérez-López and Merkoçi, 2011). Nanomaterials such as gold nanoparticles (AuNPs), gold nanorods (GNRs), and quantum dots (QDs) are also utilized to increase capture efficiency of analytes, signal amplification and improvement of detection limit (DL). AuNPs within 13–20 nm diameter range possess excellent dispersity and biocompatibility with relative ease of functionalization and integration into colorimetric, electrochemical, scattering and fluorescence detection methods targeting bacterial cells, nucleic acids, toxins, viruses and other analytes of interests (Yola et al., 2013, 2015b; Chen et al., 2014; Quintela et al., 2015). After modification, functionalization or physico-chemical enhancement, the resulting AuNPs ultimately lead to usable morphological transformations such as color change via dispersion and aggregation (Chen et al., 2014).
AuNPs surface modification via ligand exchange is an excellent approach in the design and development of biosensors (Zhu et al., 2017). For various biosensing purposes, AuNPs have also facilitated enhancement of electrode conductivity and increased sensitivity (Yola and Atar, 2014). Successful detection of DNA hybridization using an impedimetric biosensor integrated with AuNPs has been reported (Gupta et al., 2013). In addition, a molecular imprinted electrochemical biosensor based on polyoxometalate and graphene oxide decorated-AuNPs has successfully detected chemicals such as triclosan (TCS) from environmental water samples (Yola et al., 2015b). For colorimetric assay, the bio functionality of AuNPs has been demonstrated in previous works such as by using quaternary ammonium ligands and β-galactosidase enzyme with cationic AuNPs in which its binding event with the target bacterium, Escherichia coli XL1, delivered a colorimetric read-out for sensing and quantification (Miranda et al., 2011). Similarly, cationic AuNPs have been non-covalently conjugated with anionic polyparaphenyleneethynylene (PPE) polymers for rapid and efficient bacterial identification in a microarray setup (Phillips et al., 2008). This sensor array relies on the displacement of the fluorescent PPE from the AuNPs quenchers after binding with the target bacteria. Various shapes (i.e., oval and popcorn-shaped) of AuNPs can also be tailored and functionalized for rapid colorimetric detection of significant pathogens (Khan et al., 2011; Afonso et al., 2013). An optical biosensor for Chlamydia trachomatis DNA detection that utilized GNRs as molecular probes has been previously reported (Parab et al., 2010). The principle behind the DNA detection is based on the simultaneous biorecognition-mediated hybridization of target DNA in a sandwich type manner with two different capture GNRs-DNA probe which leads to aggregation (Ko and Grant, 2006). It successfully detected PCR amplified C. trachomatis pathogen gene (concentration range, 0.25–20 nM). A fiber optic portable biosensor utilizing the principle of fluorescence resonance energy transfer (FRET) was developed for Salmonella Typhimurium detection in pork samples and had a detection limit of 105 colony forming unit or CFU/g (Ko and Grant, 2006). Though various molecular and immunological-based optical biosensors have been widely studied and designed for rapid detection and identification of foodborne bacterial pathogens, many of these methods are only capable of detecting a single pathogen with high detection limit (DL) approximately 104–105 CFU/mL. An aptamer-based colorimetric detection assay by Bayraç et al. (2017) was able to specifically screen the target strain, S. Enteriditis, however, when it was applied on real food sample (i.e., milk), the detection limit was only 103 CFU/mL. A colorimetric aptasensor-based platform based on the peroxidase-like activity of ZnFe2O4-reduced graphene oxide nanostructures with a sensitivity of 2 log CFU/mL (milk sample) has been reported by Wu et al. (2017); however, it was designed only for single pathogen (S. Typhimurium) detection. More so, an interesting paper was reported by Abdelhaseib et al. (2016) on BARDOT (bacterial rapid detection using optical scattering technology) that could facilitate the detection of S. Enteritidis and S. Typhimurium serovars using fiber optic sensor but with a detection limit of 103 CFU/mL from selectively enriched poultry products. Therefore, a more comprehensive system that can detect multiple target pathogens simultaneously in a single assay with superior DL is desired.
Salmonella spp. is a significant bacterial foodborne pathogen causing outbreaks and sporadic cases of gastroenteritis (Franklin et al., 2011). The nature and severity of diseases caused by Salmonella spp. is based on serotype, host species and presence of virulence factors such as Salmonella pathogenicity islands (SPIs), plasmids, toxins, fimbriae, and flagella (Zou et al., 2010). SPIs are small chromosomal regions that carry multiple Salmonella spp. virulence genes. ttrRSBCA is a locus near SPI-2 that encodes for tetrathionate reductase enzyme for anaerobic respiration. Specifically, Salmonella spp. utilizes a sulfur compound tetrathionate which acts as one of the terminal electron acceptors during anaerobic respiration (James et al., 2013).
In this work, we developed a novel approach for simultaneous optical detection of various Salmonella spp. strains in contaminated complex matrices (i.e., food and environmental samples) by utilizing oligonucleotide-functionalized AuNPs as a sensitive optical biosensing platform in combination with an efficient sample pooling and immunomagnetic separation (IMS) system that ensure detection of viable cells. It features a highly sensitive detection technology that specifically targets conserved region of ttrRSBCA locus by a sandwich hybridization approach. The development of the detection technology that simultaneously and directly detects a comprehensive outbreak bacterial strains in complex matrices can efficiently reduce the risks and incidence of foodborne diseases.
Materials and Methods
Reagents and Materials
Gold (III) chloride trihydrate (HAuCl4⋅3H2O) and sodium citrate (C6H5Na3O7) were purchased from Sigma-Aldrich (St. Louis, MO, United States). Sodium chloride (NaCl) and disodium hydrogen phosphate (Na2HPO4) were obtained from Fisher Scientific (Lawn Fair, NJ, United States). Culture media: Brain Heart Infusion (BHI) broth and Hektoen Enteric (HE) Agar, and MacConkey Agar with Sorbitol (SMAC) were purchased from Neogen (Lansing, MI, United States) while Buffered Peptone Water (BPW) was supplied by Becton, Dickinson and Company (Franklin Lakes, NJ, United States). Pathatrix 5 Pooling Salmonella spp. was purchased from Life Technologies (Grand Island, NY, United States) and DNeasy Blood & Tissue Kit for DNA extraction was obtained from Qiagen (Hilden, Germany). Asymmetric PCR (asPCR) primers were customized by Integrated DNA Technologies (Coralville, IA, United States): For (For-192-Sal) 5′-ACC CAC GCG TTT CAT CGG TT-3′ and Rev (Rev-192-Sal) 5′-GCC GGC AAT CCC TAT CAC CC-3′. Two sets of thiol-modified oligonucleotides labeled as Probe 1 and Probe 2 with sequences (P1-Sal 5′-AGC AAC TGG CGG GAG AAA GCG GTC TTG ACG-3) and (P2-192-Sal HS-(CH2)6 -5′-GCA GGA ACA CCC GAT TGA CTC GTC CGT CCC-3′), respectively, were designed from S. Typhimurium complete genome sequence (Gen Bank accession no. CP019649.1), synthesized and thiol-modified by Eurogentec (San Diego, CA, United States). All reagents used in this study were analytical grade and utilized without further treatment. Water was purified (>18.3 MΩ/cm) using a Barnstead Nanopure filtration system (Lake Balboa, CA, United States).
Bacterial Cultures and Genomic DNA Preparation
All bacterial strains used are listed in Table 1. All bacterial cultures were initially stored in cryogenic beads with Brucella broth and glycerol (CryoSavers; Hardy Diagnostics, Santa Maria, CA, United States) at -80°C before activating them overnight using BHI (Neogen) at 37°C. HE Agar (Neogen) and MacConkey Agar with Sorbitol (Neogen) were used for viable Salmonella spp. and STEC O157:H7 ATCC 35150 cell counts, respectively. Genomic bacterial DNA from all samples were extracted using DNeasy Blood & Tissue Kit (Qiagen) following manufacturer’s protocol. Asymmetric PCR (asPCR) was conducted with a highly specific pair of primers, For-192-Sal and Rev-192-Sal (10:1 ratio). All products were kept at -20°C until further use.
Designing and Synthesis of AuNPs-Oligonucleotide Probes
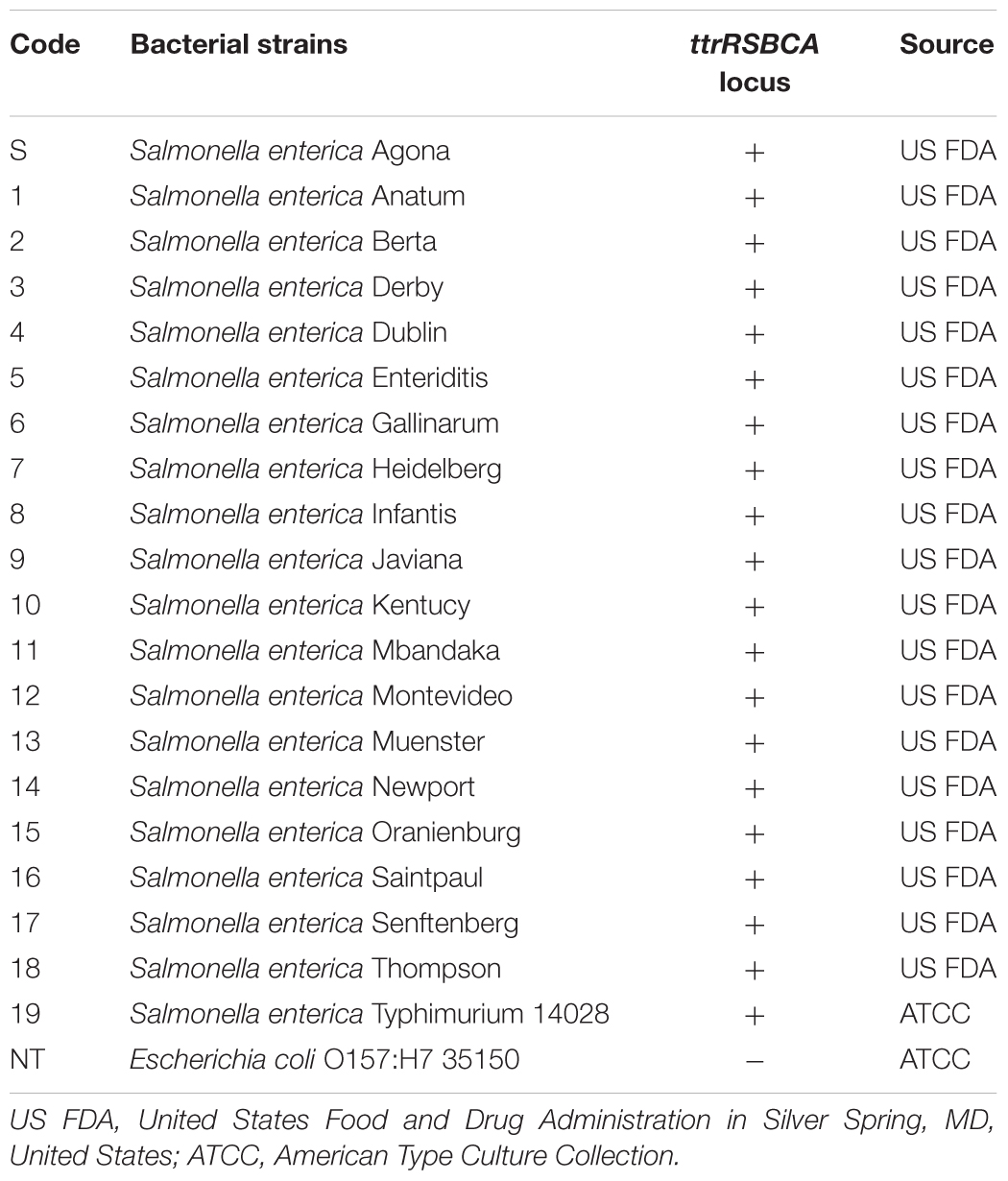
AuNPs were synthesized by using citrate reduction method as previously described (Lin et al., 2008). A microplate reader spectrophotometer (BioTek Power Wave XS, Winoskii, VT, United States) confirmed the average size of the newly synthesized AuNPs at 400–700 nm based on wavelength absorption spectra. Probes 1 and 2 (30-mer, 20 μM, 20 μL) were conjugated onto the surface of AuNPs (Supplementary Figure S1). In brief, each oligonucleotide probe was added to AuNPs solution (20 μM, 980 μL) and then subjected to increasing salt concentration (0.05–1.0 M NaCl in 10 mM Na2HPO4, pH 7.4) for 24 h in water bath at 37°C to maximize the oligonucleotide loading capacity. After incubation, unbound probes were discarded by centrifugation (19,530 ×g) for 30 min and the oily-like AuNPs which remained at the bottom of the tube was resuspended in salt buffer solution (NaCl in 10 mM Na2HPO4, pH 7.4). These freshly synthesized AuNPs-oligonucleotide probes were placed in dark storage at room temperature until further use. Shift in the absorbance peak of AuNPs was measured to confirm successful conjugation of oligonucleotide probes. The novel principles behind the simultaneous AuNPs optical biosensing and the schematic representation of DNA sandwich hybridization targeting ttrRSBCA locus which is found in a broad range of Salmonella spp. strains is presented in Scheme 1.
Scheme 1. The novel principles behind AuNPs optical biosensing and the schematic representation of DNA sandwich hybridization targeting ttrRSBCA locus of Salmonella spp. The high complementarity between the AuNPs-oligonucleotide probes (Probes 1 and 2) and ttrRSBCA regions allows the occurrence of DNA sandwich hybridization. Probe-target complexes form with high specificity and retain in the solution even after increasing its salt concentration. Positive reaction (ttrRSBCA locus) shows no AuNPs-oligonucleotide probes aggregation thus no color change (original red) is observed. Formation of probe-target complexes did not occur in blank or non-target DNA samples; therefore, the salt-induced AuNPs-oligonucleotide probes aggregation occurs that eventually changes the color of the reaction mixtures (from the original red to purplish blue). Color differentiation facilitates direct detection of positive and negative samples by observation using the naked eye. Size not to scale (reproduced with some modifications and with permissions from Quintela et al. (2015); copyright 2015, Royal Society of Chemistry).
AuNPs Optical Biosensing Assay on Pure Bacterial Cultures
In brief, asPCR products (50 μL) or blank was mixed with AuNPs-oligonucleotide probes (Probe 1 and Probe 2, 50 μL each) and incubated for 30 min at 55°C for DNA sandwich hybridization. All mixtures were then transferred to a 96-well microplate for color challenge test by adding a salt solution (final concentration, 2 M NaCl in 10 mM Na2HPO4, pH 7.4). The color changes in various reaction mixtures were simultaneously observed within 5 min. The rest of reaction mixtures retained its original red color. Pre- and post-color challenge test absorbance (400–700 nm) and ratio at two wavelengths, 625 nm and 525 nm (A625/525 nm), were also collected for spectrophotometric analysis. Gel electrophoresis (3% agarose gel and ethidium bromide) was performed to all pre-color challenge test reaction mixtures (50 μL) to verify the occurrence of a successful oligonucleotide/AuNP-DNA sandwich hybridization.
Sample Pooling and Detection of Live Salmonella spp. Strains in Complex Matrices
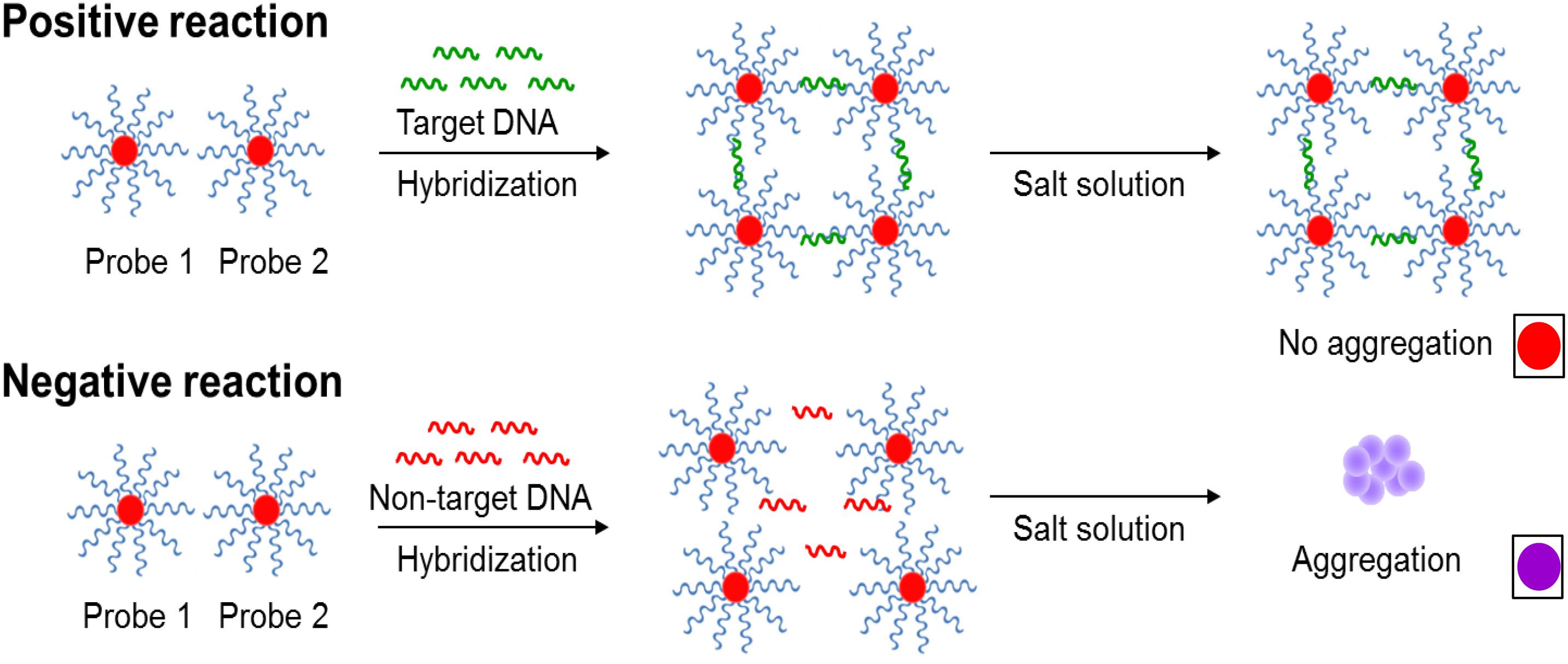
A robust sample pooling plan was designed to efficiently test post-enriched samples prior to the application of the IMS system, Pathatrix (Matrix MicroScience, Newmarket, United Kingdom) with Pathatrix 5 Pooling Salmonella spp. (Life Technologies). In Scheme 2, a workflow shows the rapid simultaneous detection of Salmonella spp. strains in complex matrices using the novel AuNPs optical biosensor. In brief, chicken meat with skin was purchased from a local retailer while unwashed frozen blueberries were supplied by a local farmer. Individual overnight cultures of Salmonella spp. as listed in Section “Bacterial Cultures and Genomic DNA Preparation” were randomly spiked at 0–9 bacterial cells/g (<10 CFU/g) inoculum levels to food samples (25 g × 25 samples) placed in individual stomacher bags (Fisher Scientific). After which, 225 mL of BPW (Becton, Dickinson and Company) was added, as recommended in the USDA/Food Safety and Inspection Service (FSIS) Microbiology Laboratory Guidebook (MLG) 4.08 for the isolation and identification of Salmonella spp. with minor modifications. Samples were homogenized using a Pulsifier (Microbiology International, Frederick, MD, United States) and then enriched for 6 h at 37°C.
Scheme 2. A workflow showing rapid Salmonella spp. strains detection in complex matrices using the novel AuNPs optical biosensor coupled with a robust sample pooling plan and an IMS system. (A) Twenty-five different individual enriched food or environmental samples are randomly pooled into five groups: Pools 1–5. (B) Each pool is circulated in the IMS system for capture and concentration of Salmonella spp. strains. Genomic DNA are extracted from the concentrated IMS samples and amplified (C) DNA sandwich hybridization reaction mixtures are transfered into a 96-well microplate for color challenge test by increasing its salt concentration. If pools change from red to purplish blue after adding salt, reactions are interpreted as negative for ttRSBCA; all of the individual subsamples are therefore ttrRSBCA-negative. If pools remain red after adding salt, reaction mixtures are interpreted as positive for ttrRSBCA and all of the individual subsamples are further tested. (D) AuNPs optical biosensing is conducted for all individual subsamples belonging to the ttrRSBCA-positive pools. S = Subsamples.
Five individual (50 mL each) samples of each kind, either chicken meat with skin or blueberry samples were pooled together. Pooled samples were concentrated (37°C, 30 min) following the manufacturer’s instructions (Pathatrix, Matrix Microscience). For environmental samples, five different sampling sites were randomly chosen for soil with chicken manures (25 g × 5 samples) samples collection. Genomic DNA was extracted as described in section “Bacterial Cultures and Genomic DNA Preparation”. Detection was conducted by applying the previous protocols used for AuNPs optical biosensing of pure cultures. AuNP optical biosensing was performed in parallel with selective plating (HE Agar) for direct comparison.
Transmission Electron Microscopy (TEM)
The morphological properties and behavior of AuNPs were analyzed using Philips CM10 TEM (Philips, Eindhoven, Netherlands) operating at 80 kV. Two microliters each of modified AuNPs and reaction mixtures were deposited directly onto 200 mesh carbon-coated copper grids then air-dried for 30–45 min at ambient temperature and pressure for TEM imaging.
Statistical Analysis
Three trials were performed per each experiment. To evaluate the reproducibility of the assay, all samples tested were in duplicates, and the mean of the data ± standard deviation (S.D.) was analyzed by JMP software using one-way ANOVA for significance (p < 0.05). Fisher’s least significance difference (LSD) was used for post hoc analysis to confirm the significant differences between groups.
Results and Discussion
Functionalization of AuNPs With Single-Stranded Thiolated-Oligonucleotides
AuNPs synthesis using citrate reduction method was based on HAuCl4 reduction and instantaneous stabilization in trisodium citrate. To limit the negative effects of ionic interactions of synthesized AuNPs, low concentration of reactants (25 mL, 2.5 mM CHLOROAURIC ACID, 3 ML 38.3 MM sodium citrate solution) was used before resuspending the final product in aqueous salt solution (Gittins and Caruso, 2001). The solution shifted from the initial bluish-gray color to purplish until it completely turned to ruby red solution which was one of the primary attributes of spherical AuNPs within 5–15 nm diameter range (Wuithschick et al., 2015). The controlled and morphologically consistent size of colloidal AuNPs has a direct correlation with the ratio of citrate to gold (Hinterwirth et al., 2013). Generally, AuNPs (≤20 nm diameter, 𝜀 = 108/cm M) in aqueous media has 520 nm plasmon band on the average which makes it very compatible for color differentiation-based optical biosensing (Chen et al., 2014).
To fully take advantage of the useful properties of nanomaterials, its functionalization is necessary which can also preserve its properties and biocompatibilities (DeLong et al., 2010). In this study, we designed highly specific oligonucleotides and conjugated it onto the surface of AuNPs via thiol linkage, HS-(CH2)6, which was initially introduced chemically to either 5′- or 3′- end of the oligonucleotide probes. The stability of AuNPs-probes is supported by a strong covalent bond that forms between thiol and gold (Au-S) and mediated by the sulfhydryl (SH) functional group (Li et al., 2013). Sulfhydryl compounds are small molecules that exert less steric hindrance which provide thiolated-oligonucleotide with superior accessibility to AuNPs by citrate ions replacement without altering its biological functions (Chegel et al., 2011). Specifically, functionalization of AuNPs involved “salt aging” method which requires excess amount of thiolated-DNA with periodic addition of salt solution until the final salt concentration and salt-stable nanoparticles are achieved. Herein, the final salt concentration in phosphate buffer was 1 M NaCl in 10 mM Na2HPO4, pH 7.4. Compared to other buffer solutions such as Tris, phosphate buffer has relatively higher DNA loading capacity which makes it suitable for AuNPs functionalization. Previous studies have shown that negatively charged DNA bind to positively charged amine groups that were present in Tris resulting to bigger sizes and eventually reducing the available space (approx. 16% less strands per particle at 1 M NaCl) as well as surface loading capacity of AuNPs (Hurst et al., 2006). It was noteworthy that there was no obvious color change between the pre- and post-AuNPs functionalized solutions. Ultimately, the absorbance peak of AuNPs shifted by 2–3 nm after immobilizing thiolated-oligonucleotide mainly due to increase in its refractive index which was a direct evidence of successful AuNPs surface modification (Supplementary Figure S2).
AuNPs Optical Biosensing of Salmonella spp. Strains in Pure Cultures
In this study, amplified ttrRSBCA of Salmonella spp. strains from inoculum levels 10–103 and 108 CFU/mL were tested along with STEC O157:H7 ATCC 35150 as non-target and nuclease-free water as blank. It was simultaneously observed by naked eye that the reaction mixture wells of various Salmonella spp. strains remained red in color whereas the wells containing STEC O157:H7 ATCC 35150 or blank changed to purplish-blue color after the addition of salt solution (2 M, final concentration).
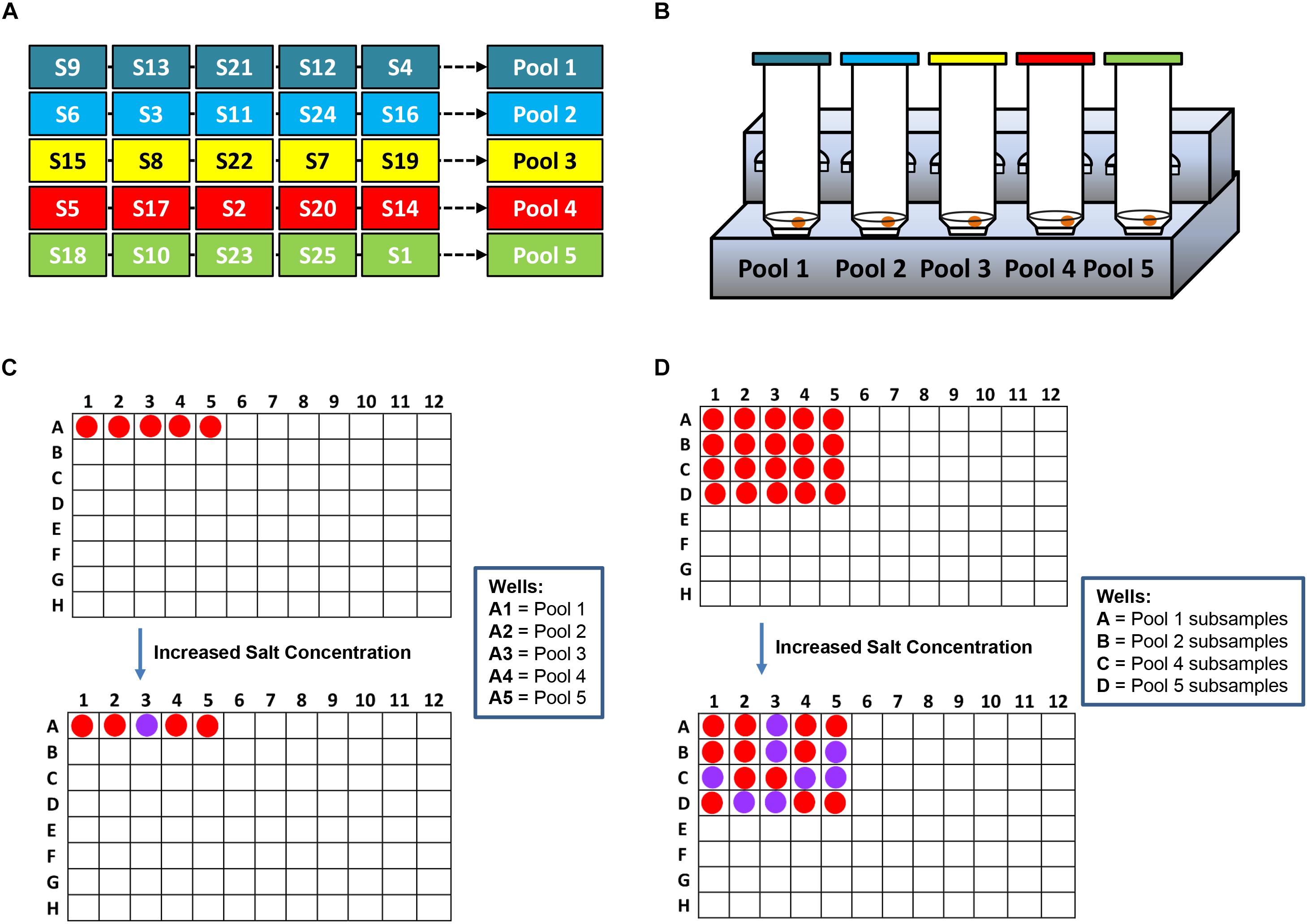
For research purpose, further confirmation was conducted by gel electrophoresis and spectrophotometric analysis. Specifically, as shown in Figure 1, S. Agona was selected as the representative strain for AuNPs optical biosensing at 108 CFU/mL inoculum level. In Figure 1A, the successful hybridization of oligonucleotide/AuNPs probes with its complementary region was observed as it retained its original red color after adding salt solution. This can be attributed to the formation of a network and complexes through DNA sandwich hybridization that stabilized AuNPs even in an increased salt concentration. The phosphate groups on the DNA backbone promote interparticle electrostatic repulsion which counteracted the elevated ionic strength of the salt solution as well as the van der Waals attraction force. The interactions between the complimentary probes and target regions may have led to a stronger ordered B-form structure, thus limiting co-localization of AuNPs and allowing the retention of original red color. Successful DNA sandwich hybridization between oligonucleotide/AuNPs and the ttrRSBCA of S. Agona was also confirmed by the formation of a gel band around the 200-bp region while non-target (STEC O157:H7 ATCC 35150) and blank (nuclease-free water) did not show any bands. The non-target and blank reaction mixtures shifted from red to purplish-blue color primarily due to absence of complementary strands that could have facilitated the network formation and rendered interparticle stability. The repulsion force exerted by AuNPs was weakened and neutralized by the sheer ionic strength of the salt solution that ultimately led to the aggregation of AuNPs.
Figure 1. AuNPs optical biosensing targeting ttrRSBCA region of S. Agona in pure culture setup. (A) AuNPs optical biosensing showing three reaction mixtures before and after adding salt solution (50 μL, 2 M NaCl–Na2HPO4 in final reaction). Both B and NT samples changed from red to bluish-purple color while no color change was observed in T reaction mixture. DNA sandwich hybridization between ttrRSBCA and AuNPs-probes was confirmed by a gel band (192-bp). (B) A625/525nm of the reaction mixtures after increasing its salt concentration showed significant differences at p < 0.05 between B, NT and T (C) TEM images confirmed salt induced AuNPs-probe aggregation in B or NT (left image with a bluish-purple solution as inserted) sample while no significant AuNPs aggregation was observed in S. Agona (right image with a red color solution as inserted). Both NT and T had 108 CFU/mL bacterial concentration. B = Blank, nuclease-free water; NT = non-target, STEC O157:H7 ATCC 35150; T = target, S. Agona. ∗Significant difference at p < 0.05.
As an aggregation parameter of AuNPs, empirical measurement was derived to quantify the variation of integrated absorbance of samples after adding salt solution through its absorbance ratio at 625 and 525 nm (A625/525nm) wavelengths. This was determined based on the average absorbance of target reaction mixtures which peaked at around 525 nm while blank and non-target reaction mixtures had both absorbance peak at a longer wavelength at 625 nm. The A625/525nm of S. Agona after adding salt solution in Figure 1B clearly showed a significant difference at p < 0.05 as compared to blank and non-target samples. TEM images in Figure 1C confirmed salt induced AuNPs-probe aggregation in blank or non-target sample while no significant aggregation of AuNPs was observed in S. Agona reaction mixture.
We next challenged the sensitivity of the AuNPs optical biosensing on various Salmonella spp. strains as shown Figure 2. At bacterial levels 10, 102, and 103 CFU/mL, S. Typhimurium ATCC 14028 showed positive results while negative for both bank (nuclease-free water) and non-target STEC O157:H7 ATCC 35150 as presented in Figure 2A. The absorbance (A625/525nm) in Figure 2B showed significant differences at p < 0.05 between target, blank and non-target. Similar results shown in Figure 2C were generated when the remaining strains of Salmonella spp. strains as listed in Section “Bacterial Cultures and Genomic DNA Preparation” were subjected to simultaneous AuNPs optical biosensing set at 10 CFU/mL; both blank and non-target changed to purplish blue while no color change was observed on the remaining target samples. Finally, the absorbance (A625/525nm) of various Salmonella spp. strains reaction mixtures (10 CFU/mL) after adding salt, as shown in Figure 2D, had significant differences at p < 0.05 between target and non-target or blank. Based on these results, the AuNPs optical biosensor has successfully and simultaneously detected Salmonella spp. strains via ttrRSBCA from 108 CFU/mL down to 10 CFU/mL. Based on the color changes differentiation and statistically valid results, the AuNPs optical biosensor has achieved 100% specificity with a DL of <10 CFU/mL. These results were also in agreement with the gel electrophoresis and TEM images (Supplementary Figure S3). DNA concentration measurements of Salmonella spp. strains and STEC O157:H7 (non-target) is presented in Supplementary Table S1.
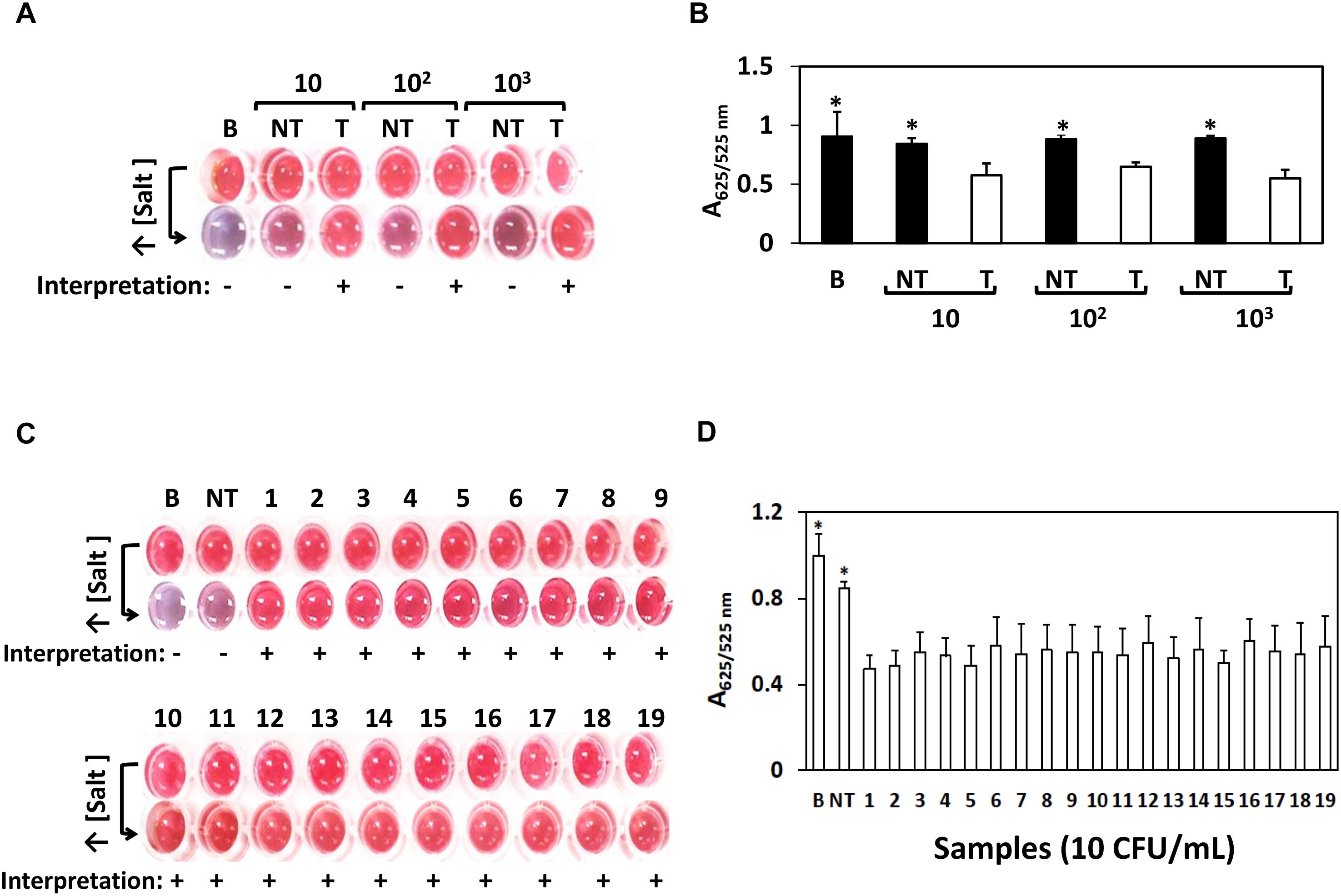
Figure 2. AuNPs optical biosensing – pure culture setup using various Salmonella spp. strains. (A) AuNPs optical biosensing a 10, 102 and 103 CFU/mL bacterial count levels, showing positive results for T and negative for both B and NT reaction mixtures after adding salt (B) A625/525nm of the reaction mixtures after increasing its salt concentration showing significant differences at p < 0.05 between B, NT and T (C) AuNPs optical biosensing of various Salmonella spp. strains at 10 CFU/mL bacterial counts. Reaction mixtures containing B and NT changed from red to purplish blue after adding salt solution while the original color of all target numbered samples remained unchanged. (D) A625/525nm of the reaction mixtures presented in panel (C) after increasing its salt concentration showing significant differences at p < 0.05 between B, NT and T. B = Blank, nuclease-free water; NT = non-target, STEC O157:H7 ATCC 35150, T = target, Salmonella spp. strains as numbered 1–19 in Table 1. ∗Significant difference at p < 0.05.
Taxonomically, all 19 Salmonella spp. strains that were used in this study belonged to S. enterica species and were also environmental and/or outbreak strains with the exception of S. Typhimurium ATCC 14028. Twelve of these strains are high on the top 20 list of the most prevalent pathogenic Salmonella serotypes in humans in the United States (CDC, 2014). Salmonella spp. utilizes tetrathionate as a terminal respiratory electron acceptor and the locus ttrRSBCA which is needed for tetrathionate respiration is located near the SPI-2 (Hensel et al., 1999; González-Escalona et al., 2012). SPIs in congruent with other virulence factors, facilitate host cell invasions and intracellular pathogenesis (Marcus et al., 2000). SPIs have been studied so far and each SPI influences the phenotypic characteristics of known Salmonella serotypes (Marcus et al., 2000; Morgan, 2007). SPI-2 has two main elements; 25 kb size portion which is only found in S. enterica and a 15 kb portion present in both S. enterica and S. bongori (Marcus et al., 2000; Hensel, 2004). The enzyme tetrathionate reductase is encoded by the structural genes, ttrA, ttrB and ttrC (Hensel et al., 1999). The 192-bp target region that was identified in this study and has high complementarity with the AuNPs-probes is located on the ttrA gene (3,063 bp). Since tetrathionate respiration is an important life cycle event of Salmonella spp. strains, these three structural genes are genetically stable which made ttrRSBCA an excellent choice as a marker for Salmonella spp. diagnostic detection. Based on our extensive literature review, there have been no optical biosensing assay reports that have simultaneously screened and detected Salmonella spp. as comprehensive as this study (i.e., nineteen different serovars; Ahn and Walt, 2005; Mazumdar et al., 2007; Valadez et al., 2009; Jokerst et al., 2012; Ohk and Bhunia, 2013; Yang et al., 2013; Viter et al., 2017). Additionally, Table 2 presents previously reported Salmonella spp. detection methods and detection limits achieved for selected serovars. Specifically, in comparison to the present study, these methods had longer enrichment periods and/or were only designed for single Salmonella spp. strain detection with inferior detection limits. Additionally, since Salmonella spp. strains are highly associated with poultry products that often come in a large volume of samples for testing, efficient processing methods and analysis that can save both time and cost without diminishing the sensitivity of the assay are necessary.
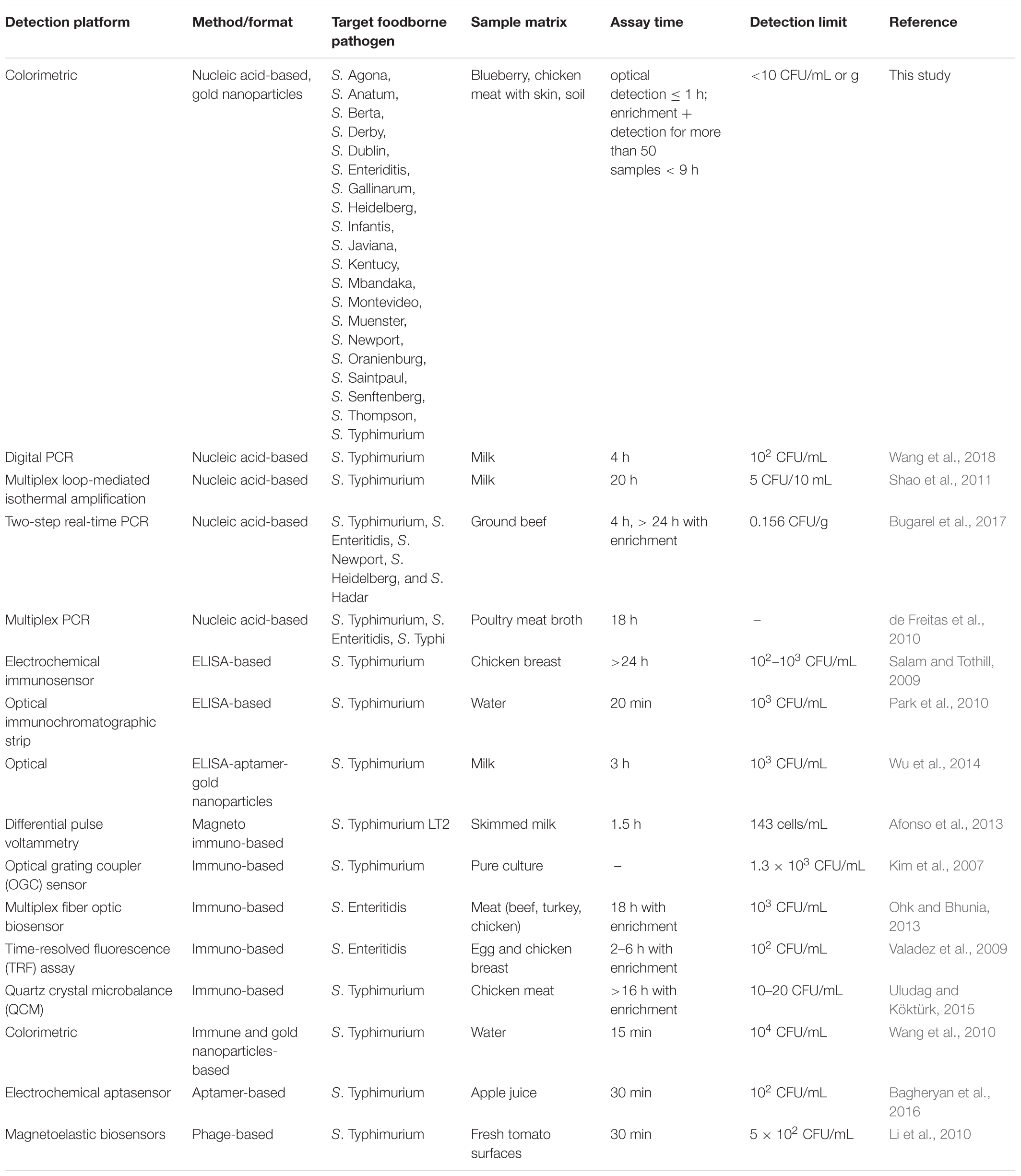
Table 2. Reported Salmonella spp. detection methods (alternative to culture methods) and detection limits achieved for selected serovars.
Application of the AuNPs Optical Biosensing in Food and Environmental Samples
We designed a pooling technique that allowed processing of a large number of samples (>50 samples) in a short period of time (<9 h) including both enrichment and detection of Salmonella spp. strains. Pooled ttrRSBCA-negative samples would mean that all the original individual subsamples were not contaminated with Salmonella spp. strains. However, individual subsamples were further tested when its original pooled sample source was positive for ttrRSBCA. After the short enrichment and sample preparation, the actual simultaneous AuNPs optical biosensing only took ≤1 h to complete.
In Figure 3A, results show that four out of five pooled enriched chicken meat with skin samples tested positive and subsequently in Figure 3B, eight of its 20 subsamples also tested positive for ttrRSBCA. For the blueberry setup in Figure 3C, 3 out of five pooled samples were positive and Figure 3D shows that 12 of its 15 subsamples were also positive for ttrRSBCA. For the natural and uninoculated environmental soil samples, Figure 3E shows that all samples tested negative for ttrRSBCA. A standard plating technique as recommended in the Bacteriological Analytical Manual (BAM) of the US FDA was performed in parallel with the newly developed optical biosensor. In this validation step, HE Agar was used and the results showed development of green colonies with black centers (presumptive positive) for all ttrRSBCA positive samples while yellow to salmon orange colonies (presumptive negative) for all ttrRSBCA negative samples were observed (Supplementary Figure S4).
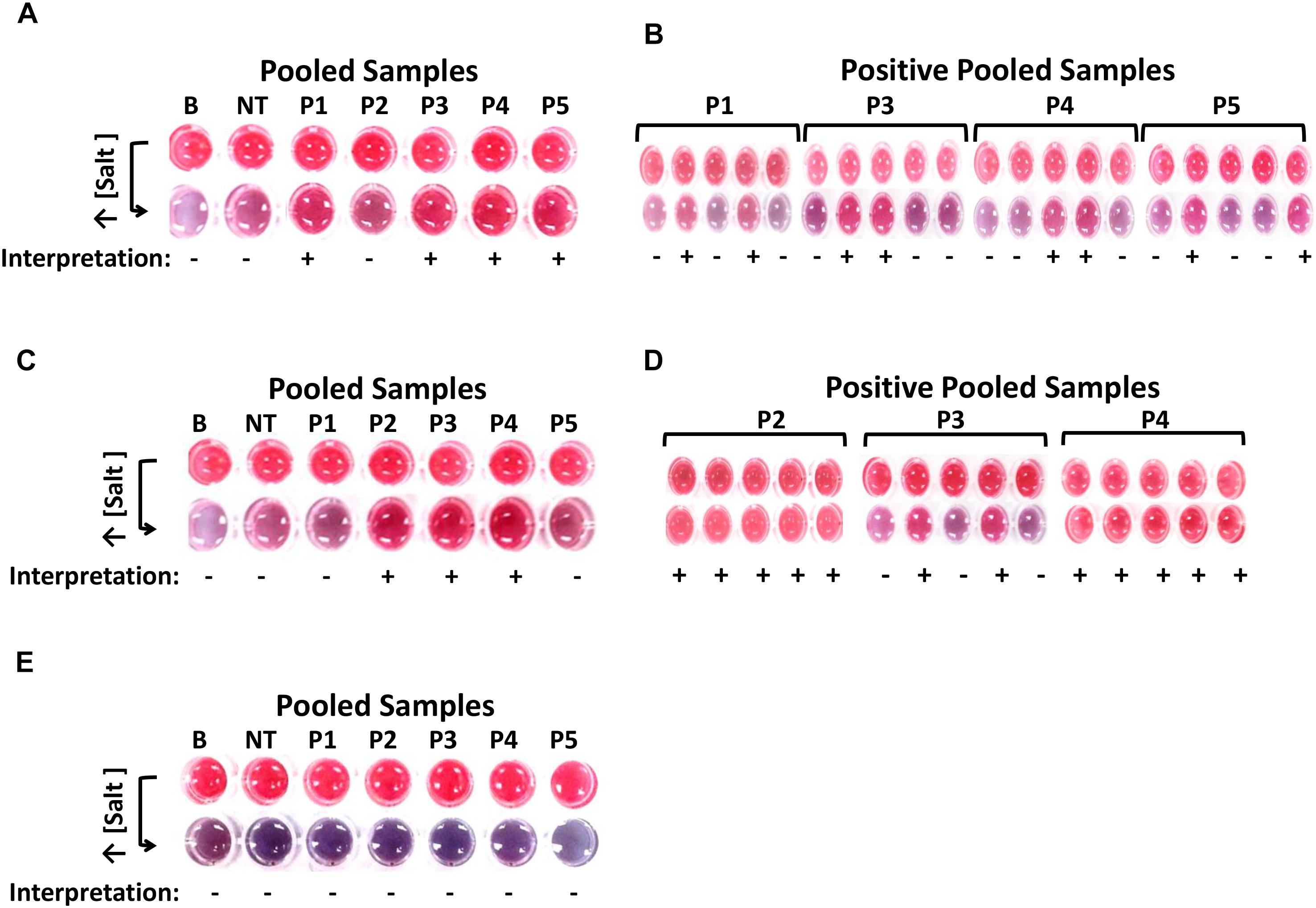
Figure 3. Detection of various Salmonella spp. strains in complex matrices. (A) After IMS system, five pools (P1-P5) of chicken meat with skin were tested for the presence of ttrRSBCA. Among these five pools, P2 tested negative so as its five subsamples. The other 4 pools were positive: P1, P3, P4 and P5. (B) From the four positive pools, a total of 8 individual subsamples tested positive for ttrRSBCA. (C) Three out of 5 blueberry pools: P2, P3 and P4 were positive for ttrRSBCA. (D) Individual subsamples from the three positive blueberry pools were tested and 12 out of 15 these individual subsamples showed positive for ttrRSBCA. (E) All five soil sample pools: P1-P5 were negative for ttrRSBCA. B = Blank, nuclease-free water; NT = non-target, STEC O157:H7 ATCC 35150.
In this study, the AuNPs optical biosensor shows efficient simultaneous detection of Salmonella spp. When addressing the detection of viable cells from complex matrices, which is an important aspect in the detection of foodborne pathogens in food and environmental samples, the AuNPs optical biosensing coupled with a shortened enrichment and strategic pooling with IMS could process and screen more than 50 samples in <9 h, as compared to the conventional method (>24 h). Sample pooling increases the efficiency of processing large volume of materials that need to be tested (Singer et al., 2006). By collecting naturally pooled fecal-contaminated materials such as from large number of birds, the actual chance of including fecal samples from infected chickens is increased as compared to collection of individual swabs or droppings (Carrique-Mas and Davies, 2008). However, pooling of poultry manures for Salmonella spp. detection has a critical dependency on the “dilution effect” in which mixing of negative and positive samples decreases the likelihood of detection (Arnold et al., 2011). Thus, IMS system was incorporated to eliminate the negative impacts of the dilution effect on the sensitivity of the Salmonella spp. detection that might arise during the development of the technology. Even if the dilution effects occurred while processing the samples, it would be very negligible and pooling of samples would still mean more efficient as compared to individual sampling for Salmonella spp. detection (Arnold and Cook, 2009). IMS is an efficient tool that separates and concentrates target bacteria from food and environmental samples. This separation of bacteria allows the exclusion of interfering substances that may contribute to non-specificity and cross-reactivity of the downstream assays. Physical separation, capture and concentration of target bacteria add more layers of selectivity from the competitive microbiota especially when dealing with complex matrices.
There have been increasing efforts toward developing and improving rapid detection of Salmonella spp. in poultry and poultry products. Salmonella outbreaks in the recent years encourage the need for reliable, cost-effective and high-throughput detection methods (Park et al., 2014). A multiplex PCR for the detection of Salmonella targeting STM4492 gene and the differentiation of S. Typhimurium and S. Enteriditis in chicken meat was previously reported with a DL of 105 CFU/g (Saeki et al., 2013). The specificity of the detection method in the present study is based on the highly conserved ttrRSBCA region that is present in Salmonella spp. strains, its complementary AuNPs-probes as well as the optical features of AuNPs. There was no non-specificity that was observed in this experiment even when the assay was applied in complex matrices, however, the color intensity of the reaction mixtures can still be optimized by further reducing the background noise to provide better resolution for ease of differentiation between target and non-target samples. Future studies may include further quantitative aspects to correlate color intensity with the actual viable cell counts, though additional verification steps may be needed aside from direct observation using the naked eye.
Conclusion
We have developed a sensitive optical detection platform coupled with an efficient sample pooling in conjunction with an IMS that allowed simultaneous optical biosensing of various viable Salmonella spp. strains on contaminated poultry, produce and natural environmental samples. AuNPs were synthesized and surface functionalized with short oligonucleotides that rendered high stability without compromising its biocompatibility. Sandwich hybridization between specific AuNPs-probes and highly conserved regions of ttrRSBCA locus provided the opportunity to optically differentiate target and non-target samples after increasing the salt concentration of the reaction mixtures. The results showed that the highly sensitive assay toward its target with a superior DL of <10 CFU/mL or g and 100% specificity. The total turn-around time with enrichment is <9 h for detection of viable Salmonella spp. in more than 50 food and environmental samples tested. This approach has significantly shortened the screening process for simultaneously detecting outbreak and environmental strains of Salmonella spp. as compared to the conventional method. The unique colorimetric properties of AuNPs have provided this assay a simple and stable platform at a very low cost, allowing rapid screening of a highly significant group of foodborne pathogens.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
VW and IQ designed the experiments and performed data analysis. IQ conducted the experiments including TEM imaging. IQ prepared the manuscript, VW, C-SL, and BdlR edited the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the United States Department of Agriculture NIFA-AFRI Competitive Grant (Award # 2011-68003-30420).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Darcy Hanes and Dr. Atin Datta of the U.S. FDA for providing the Salmonella spp. strains, Kelly Edwards of the University of Maine Electron Microscopy Laboratory for helping with setting up and calibrating the TEM and Ai Kitazumi for her assistance during the conduct of the experiment.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01138/full#supplementary-material
References
Abdelhaseib, M. U., Singh, A. K., Bailey, M., Singh, M., El-Khateib, T., and Bhunia, A. K. (2016). Fiber optic and light scattering sensors: complimentary approaches to rapid detection of Salmonella enterica in food samples. Food Control 61, 135–145.
Afonso, A. S., Pérez-López, B., Faria, R. C., Mattoso, L. H. C., Hernández-Herrero, M., Roig-Sagués, A. X., et al. (2013). Electrochemical detection of Salmonella using gold nanoparticles. Biosens. Bioelectron. 40, 121–126. doi: 10.1016/j.bios.2012.06.054
Ahn, S., and Walt, D. R. (2005). Detection of Salmonella spp. using microsphere-based, fiber-optic DNA microarrays. Anal. Chem. 77, 5041–5047.
Arnold, M., Carrique-Mas, J., Mclaren, I., and Davies, R. (2011). A comparison of pooled and individual bird sampling for detection of Salmonella in commercial egg laying flocks. Prev. Vet. Med. 99, 176–184. doi: 10.1016/j.prevetmed.2010.12.007
Arnold, M., and Cook, A. (2009). Estimation of sample sizes for pooled faecal sampling for detection of Salmonella in pigs. Epidemiol. Infect. 137, 1734–1741. doi: 10.1017/S0950268809002702
Bagheryan, Z., Raoof, J. B., Golabi, M., Turner, A. P., and Beni, V. (2016). Diazonium-based impedimetric aptasensor for the rapid label-free detection of Salmonella typhimurium in food sample. Biosens. Bioelectron. 80, 566–573. doi: 10.1016/j.bios.2016.02.024
Bayraç, C., Eyidoǧan, F., and Öktem, H. A. (2017). DNA aptamer-based colorimetric detection platform for Salmonella Enteritidis. Biosens. Bioelectron. 98, 22–28. doi: 10.1016/j.bios.2017.06.029
Bugarel, M., Tudor, A., Loneragan, G. H., and Nightingale, K. K. (2017). Molecular detection assay of five Salmonella serotypes of public interest: Typhimurium, Enteritidis, Newport, Heidelberg, and Hadar. J. Microbiol. Methods 134, 14–20. doi: 10.1016/j.mimet.2016.12.011
Carrique-Mas, J., and Davies, R. (2008). Sampling and bacteriological detection of Salmonella in poultry and poultry premises: a review. Rev. Sci. Tech. 27, 665–677.
CDC (2014). Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet Surveillance Report for 2012 (Final Report). Atlanta, GA: Centers for Disease Control and Prevention.
Chegel, V., Rachkov, O., Lopatynskyi, A., Ishihara, S., Yanchuk, I., Nemoto, Y., et al. (2011). Gold nanoparticles aggregation: drastic effect of cooperative functionalities in a single molecular conjugate. J. Phys. Chem. C 116, 2683–2690.
Chen, J., Shi, X., Gehring, A. G., and Paoli, G. C. (2014). Automated immunomagnetic separation for the detection of Escherichia coli O157:H7 from spinach. Int. J. Food Microbiol. 179, 33–37. doi: 10.1016/j.ijfoodmicro.2014.03.022
de Freitas, C. G., Santana, A. P., da Silva, P. H. C., Gonçalves, V. S. P., Barros Mde, A., Torres, F. A., et al. (2010). PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int. J. Food Microbiol. 139, 15–22. doi: 10.1016/j.ijfoodmicro.2010.02.007
DeLong, R. K., Reynolds, C. M., Malcolm, Y., Schaeffer, A., Severs, T., and Wanekaya, A. (2010). Functionalized gold nanoparticles for the binding, stabilization, and delivery of therapeutic DNA, RNA, and other biological macromolecules. Nanotechnol. Sci. Appl. 3, 53–63. doi: 10.2147/NSA.S8984
Franklin, K., Lingohr, E. J., Yoshida, C., Anjum, M., Bodrossy, L., Clark, C. G., et al. (2011). Rapid genoserotyping tool for classification of Salmonella serovars. J. Clin. Microbiol. 49, 2954–2965. doi: 10.1128/JCM.02347-10
Gittins, D. I., and Caruso, F. (2001). Spontaneous phase transfer of nanoparticulate metals from organic to aqueous media. Angew. Chem. Int. Ed. 40, 3001–3004. doi: 10.1002/1521-3773(20010817)40:16
González-Escalona, N., Brown, E. W., and Zhang, G. (2012). Development and evaluation of a multiplex real-time PCR (qPCR) assay targeting ttrRSBCA locus and invA gene for accurate detection of Salmonella spp. in fresh produce and eggs. Food Res. Int. 48, 202–208.
Gupta, V. K., Yola, M. L., Qureshi, M. S., Solak, A. O., Atar, N., and Üstündaǧ, Z. (2013). A novel impedimetric biosensor based on graphene oxide/gold nanoplatform for detection of DNA arrays. Sens. Actuators B Chem. 188, 1201–1211.
Hensel, M. (2004). Evolution of pathogenicity islands of Salmonella enterica. Int. J. Med. Microbiol. 294, 95–102.
Hensel, M., Hinsley, A. P., Nikolaus, T., Sawers, G., and Berks, B. C. (1999). The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32, 275–287.
Hinterwirth, H., Kappel, S., Waitz, T., Prohaska, T., Lindner, W., and LäMmerhofer, M. (2013). Quantifying thiol ligand density of self-assembled monolayers on gold nanoparticles by inductively coupled plasma–mass spectrometry. ACS Nano 7, 1129–1136.
Hurst, S. J., Lytton-Jean, A. K., and Mirkin, C. A. (2006). Maximizing DNA loading on a range of gold nanoparticle sizes. Anal. Chem. 78, 8313–8318.
James, M. J., Coulthurst, S. J., Palmer, T., and Sargent, F. (2013). Signal peptide etiquette during assembly of a complex respiratory enzyme. Mol. Microbiol. 90, 400–414. doi: 10.1111/mmi.12373
Jokerst, J. C., Adkins, J. A., Bisha, B., Mentele, M. M., Goodridge, L. D., and Henry, C. S. (2012). Development of a paper-based analytical device for colorimetric detection of select foodborne pathogens. Anal. Chem. 84, 2900–2907. doi: 10.1021/ac203466y
Khan, S. A., Singh, A. K., Senapati, D., Fan, Z., and Ray, P. C. (2011). Targeted highly sensitive detection of multi-drug resistant Salmonella DT104 using gold nanoparticles. Chem. Commun. 47, 9444–9446.
Kim, N., Park, I.-S., and Kim, W.-Y. (2007). Salmonella detection with a direct-binding optical grating coupler immunosensor. Sens. Actuators B Chem. 121, 606–615.
Ko, S., and Grant, S. A. (2006). A novel FRET-based optical fiber biosensor for rapid detection of Salmonella typhimurium. Biosens. Bioelectron. 21, 1283–1290.
Li, F., Zhang, H., Dever, B., Li, X.-F., and Le, X. C. (2013). Thermal stability of DNA functionalized gold nanoparticles. Bioconjug. Chem. 24, 1790–1797.
Li, S., Li, Y., Chen, H., Horikawa, S., Shen, W., Simonian, A., et al. (2010). Direct detection of Salmonella typhimurium on fresh produce using phage-based magnetoelastic biosensors. Biosens. Bioelectron. 26, 1313–1319. doi: 10.1016/j.bios.2010.07.029
Lin, Y.-H., Chen, S.-H., Chuang, Y.-C., Lu, Y.-C., Shen, T. Y., Chang, C. A., et al. (2008). Disposable amperometric immunosensing strips fabricated by Au nanoparticles-modified screen-printed carbon electrodes for the detection of foodborne pathogen Escherichia coli O157: H7. Biosens. Bioelectron. 23, 1832–1837. doi: 10.1016/j.bios.2008.02.030
Marcus, S. L., Brumell, J. H., Pfeifer, C. G., and Finlay, B. B. (2000). Salmonella pathogenicity islands: big virulence in small packages. Microb. Infect. 2, 145–156.
Mazumdar, S. D., Hartmann, M., Kämpfer, P., and Keusgen, M. (2007). Rapid method for detection of Salmonella in milk by surface plasmon resonance (SPR). Biosens. Bioelectron. 22, 2040–2046.
Miranda, O. R., Li, X., Garcia-Gonzalez, L., Zhu, Z.-J., Yan, B., Bunz, U. H. F., et al. (2011). Colorimetric bacteria sensing using a supramolecular enzyme-nanoparticle biosensor. J. Am. Chem. Soc. 133, 9650–9653. doi: 10.1021/ja2021729
Morgan, E. (2007). “Salmonella pathogenicity islands,” in Salmonella Molecular Biology and Pathogenesis, eds M. Rhen, D. Maskell, P. Mastroeni, and J. Threlfall (Wymondham: Horizon Bioscience), 67–87.
Ohk, S.-H., and Bhunia, A. K. (2013). Multiplex fiber optic biosensor for detection of Listeria monocytogenes, Escherichia coli O157: H7 and Salmonella enterica from ready-to-eat meat samples. Food Microbiol. 33, 166–171. doi: 10.1016/j.fm.2012.09.013
Parab, H. J., Jung, C., Lee, J. H., and Park, H. G. (2010). A gold nanorod-based optical DNA biosensor for the diagnosis of pathogens. Biosens. Bioelectron. 26, 667–673. doi: 10.1016/j.bios.2010.06.067
Park, S., Kim, Y. T., and Kim, Y.-K. (2010). Optical enzyme-linked immunosorbent assay on a strip for detection of Salmonella typhimurium. Biochip J. 4, 110–116.
Park, S. H., Aydin, M., Khatiwara, A., Dolan, M. C., Gilmore, D. F., Bouldin, J. L., et al. (2014). Current and emerging technologies for rapid detection and characterization of Salmonella in poultry and poultry products. Food Microbiol. 38, 250–262. doi: 10.1016/j.fm.2013.10.002
Pérez-López, B., and Merkoçi, A. (2011). Nanomaterials based biosensors for food analysis applications. Trends Food Sci. Technol. 22, 625–639.
Phillips, R. L., Miranda, O. R., You, C. C., Rotello, V. M., and Bunz, U. H. (2008). Rapid and efficient identification of bacteria using gold-nanoparticle–poly (para-phenyleneethynylene) constructs. Angew. Chem. Int. Ed. 47, 2590–2594.
Quintela, I. A., De Los Reyes, B. G., Lin, C.-S., and Wu, V. C. (2015). Simultaneous direct detection of Shiga-toxin producing Escherichia coli (STEC) strains by optical biosensing with oligonucleotide-functionalized gold nanoparticles. Nanoscale 7, 2417–2426. doi: 10.1039/c4nr05869k
Saeki, E. K., Alves, J., Bonfante, R. C., Hirooka, E. Y., and Oliveira, T. C. R. M. (2013). Multiplex PCR (mPCR) for the detection of Salmonella spp. and the differentiation of the typhimurium and enteritidis serovars in chicken meat. J. Food Saf. 33, 25–29. doi: 10.1089/fpd.2017.2290
Salam, F., and Tothill, I. E. (2009). Detection of Salmonella typhimurium using an electrochemical immunosensor. Biosens. Bioelectron. 24, 2630–2636.
Shao, Y., Zhu, S., Jin, C., and Chen, F. (2011). Development of multiplex loop-mediated isothermal amplification-RFLP (mLAMP-RFLP) to detect Salmonella spp. and Shigella spp. in milk. Int. J. Food Microbiol. 148, 75–79. doi: 10.1016/j.ijfoodmicro.2011.05.004
Singer, R. S., Cooke, C. L., Maddox, C. W., Isaacson, R. E., and Wallace, R. L. (2006). Use of pooled samples for the detection of Salmonella in feces by polymerase chain reaction. J. Vet. Diagn. Invest. 18, 319–325.
Uludag, Y., and Köktürk, G. (2015). Determination of prostate-specific antigen in serum samples using gold nanoparticle based amplification and lab-on-a-chip based amperometric detection. Microchim. Acta 182, 1685–1691.
Valadez, A. M., Lana, C. A., Tu, S.-I., Morgan, M. T., and Bhunia, A. K. (2009). Evanescent wave fiber optic biosensor for Salmonella detection in food. Sensors 9, 5810–5824. doi: 10.3390/s90705810
Viter, R., Tereshchenko, A., Smyntyna, V., Ogorodniichuk, J., Starodub, N., Yakimova, R., et al. (2017). Toward development of optical biosensors based on photoluminescence of TiO2 nanoparticles for the detection of Salmonella. Sens. Actuators B Chem. 252, 95–102.
Wang, M., Yang, J., Gai, Z., Huo, S., Zhu, J., Li, J., et al. (2018). Comparison between digital PCR and real-time PCR in detection of Salmonella typhimurium in milk. Int. J. Food Microbiol. 266, 251–256. doi: 10.1016/j.ijfoodmicro.2017.12.011
Wang, S., Singh, A. K., Senapati, D., Neely, A., Yu, H., and Ray, P. C. (2010). Rapid colorimetric identification and targeted photothermal lysis of Salmonella bacteria by using bioconjugated oval-shaped gold nanoparticles. Chemistry 16, 5600–5606. doi: 10.1002/chem.201000176
Wu, S., Duan, N., Qiu, Y., Li, J., and Wang, Z. (2017). Colorimetric aptasensor for the detection of Salmonella enterica serovar typhimurium using ZnFe2O4-reduced graphene oxide nanostructures as an effective peroxidase mimetics. Int. J. Food Microbiol. 261, 42–48. doi: 10.1016/j.ijfoodmicro.2017.09.002
Wu, W., Li, J., Pan, D., Li, J., Song, S., Rong, M., et al. (2014). Gold nanoparticle-based enzyme-linked antibody-aptamer sandwich assay for detection of Salmonella typhimurium. ACS Appl. Mater. Interfaces 6, 16974–16981. doi: 10.1021/am5045828
Wuithschick, M., Birnbaum, A., Witte, S., Sztucki, M., Vainio, U., Pinna, N., et al. (2015). Turkevich in new robes: key questions answered for the most common gold nanoparticle synthesis. ACS Nano 9, 7052–7071. doi: 10.1021/acsnano.5b01579
Yang, M., Peng, Z., Ning, Y., Chen, Y., Zhou, Q., and Deng, L. (2013). Highly specific and cost-efficient detection of Salmonella Paratyphi A combining aptamers with single-walled carbon nanotubes. Sensors 13, 6865–6881. doi: 10.3390/s130506865
Yola, M. L., and Atar, N. (2014). A novel voltammetric sensor based on gold nanoparticles involved in p-aminothiophenol functionalized multi-walled carbon nanotubes: application to the simultaneous determination of quercetin and rutin. Electrochim. Acta 119, 24–31.
Yola, M. L., Atar, N., Eren, T., Karimi-Maleh, H., and Wang, S. (2015a). Sensitive and selective determination of aqueous triclosan based on gold nanoparticles on polyoxometalate/reduced graphene oxide nanohybrid. RSC Adv. 5, 65953–65962.
Yola, M. L., Eren, T., and Atar, N. (2015b). A sensitive molecular imprinted electrochemical sensor based on gold nanoparticles decorated graphene oxide: application to selective determination of tyrosine in milk. Sens. Actuators B Chem. 210, 149–157.
Yola, M. L., Atar, N., Üstündaǧ, Z., and Solak, A. O. (2013). A novel voltammetric sensor based on p-aminothiophenol functionalized graphene oxide/gold nanoparticles for determining quercetin in the presence of ascorbic acid. J. Electroanal. Chem. 698, 9–16.
Zhu, X. L., Chen, Y. Y., Feng, C., Wang, W., Bo, B., Ren, R. X., et al. (2017). Assembly of self-cleaning electrode surface for the development of refreshable biosensors. Anal. Chem. 89, 4131–4138. doi: 10.1021/acs.analchem.6b05177
Keywords: gold nanoparticles, optical biosensor, ttrRSBCA, colorimetric, oligonucleotides, Salmonella
Citation: Quintela IA, de los Reyes BG, Lin C-S and Wu VCH (2019) Simultaneous Colorimetric Detection of a Variety of Salmonella spp. in Food and Environmental Samples by Optical Biosensing Using Oligonucleotide-Gold Nanoparticles. Front. Microbiol. 10:1138. doi: 10.3389/fmicb.2019.01138
Received: 13 March 2019; Accepted: 06 May 2019;
Published: 31 May 2019.
Edited by:
Lin Lin, Jiangsu University, ChinaReviewed by:
Wei-Lung Tseng, National Sun Yat-sen University, TaiwanDedi Futra, Riau University, Indonesia
Copyright © 2019 Quintela, de los Reyes, Lin and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vivian C. H. Wu, dml2aWFuLnd1QGFycy51c2RhLmdvdg==
 Irwin A. Quintela
Irwin A. Quintela Benildo G. de los Reyes3
Benildo G. de los Reyes3 Vivian C. H. Wu
Vivian C. H. Wu