- 1Institute of Physiology, University of Zurich, Zurich, Switzerland
- 2Department of Gastroenterology and Hepatology, University Hospital Zurich – University of Zurich, Zurich, Switzerland
- 3Laboratory of Food Biotechnology, Department of Health Sciences and Technology, ETH Zurich, Zurich, Switzerland
Milk oligosaccharides exert a prebiotic action that contributes to the development of the infant gut microbiota during lactation. Given that milk oligosaccharides remain intact after passage through stomach and small intestine, they can potentially influence the composition of the gut microbiota when ingested as dietary supplements after weaning. To address the regulatory effects of specific oligosaccharides in colitis linked to the microbiota composition, we have supplemented interleukin-10 null (Il10-/-) mice with four fucosylated and sialylated oligosaccharides. We found that oral supplementation with 2-fucosyllactose significantly decreased the severity of colitis as displayed by reduced inflammatory marker expression, histological and diarrhea scores, an increased epithelial integrity and less pronounced colon shortening. Oral supplementation with 2-fucosyllactose led to a marked expansion of the commensal Ruminococcus gnavus, which was accompanied by an enhanced cecal concentration of propionate. Decreased activation of immune cells by R. gnavus was confirmed by reconstitution of antibiotic-treated Il10-/- mice and by stimulation of dendritic cells in vitro. This study demonstrates that post-weaning administration of specific oligosaccharides can shift the composition of the gut microbiota to lessen chronic inflammation as observed in Il10-/- mice. The expansion of R. gnavus sets a positive microbial environment at the cost of pro-inflammatory Gram-negative bacteria, thereby lowering intestinal inflammation.
Introduction
The gastrointestinal tract is colonized by a complex ecosystem of microorganisms that are referred to as the intestinal microbiota (Lynch and Pedersen, 2016). It consists of trillions of microbes, mainly found in the colon, from hundreds of different species that live in a mutually beneficial interdependency with the host organism (Eckburg et al., 2005; Xu et al., 2007). Playing an important role in immune homeostasis, vitamin provision as well as nutrient utilization of indigestible components, the intestinal microbiota fulfills critical physiological roles within the colonized host (Mazmanian et al., 2005; LeBlanc et al., 2013; Ramakrishna, 2013; Honda and Littman, 2016). In the gut, energy for bacterial growth can be derived through processes such as fermentation of dietary and host carbohydrates (Chassard and Lacroix, 2013). Bacterial fermentation products, such as the short chain fatty acids (SCFA) propionate, acetate and especially butyrate have beneficial effects on host glucose homeostasis, protect from inflammation and act as important energy sources for colonocytes (Sun et al., 2017). Furthermore, an established intact microbial community also protects the host against pathogen colonization and expansion by competing for nutrients and space (Gill et al., 2011). In contrast to an intact microbiota, a destabilized microbial community is referred to as dysbiosis (Weiss and Hennet, 2017). Several pathological conditions such as obesity, diabetes and inflammatory bowel disease (IBD) correlate with dysbiosis (Turnbaugh et al., 2006; Maloy and Powrie, 2011; Winer et al., 2017). For instance, a reduced microbial diversity featuring reduced Firmicutes and outgrown Proteobacteria are typical hallmarks of dysbiosis in IBD (Swidsinski et al., 2002). Beside other factors including antibiotic treatment and bacterial toxins, the host diet contributes to microbial shifts, especially in the initial postnatal colonization of the gut (Hussey et al., 2011; Zeissig and Blumberg, 2014; Backhed et al., 2015). Accordingly, nutrients provided by breast milk, usually the primary diet in the first period of life, are critical for defining a healthy microbiota by supporting growth of beneficial bacterial strains (Pannaraj et al., 2017). Consequently, among the many beneficial effects that breastfeeding provides for infants, one is to lower the occurrence of Crohn’s disease or ulcerative colitis later in life (Xu et al., 2017). Representing a prominent component of breast milk, oligosaccharides are of particular interest in this regard (Hennet and Borsig, 2016).
In human breast milk, oligosaccharides represent a structurally diverse group of about 200 soluble carbohydrates that sum up to 5–10 g per liter (Zivkovic et al., 2011). The composition of milk oligosaccharides varies between individual mothers and over the course of lactation (Coppa et al., 1993; Marx et al., 2014). To date, the functional significance for this oligosaccharide diversity is still debated. In sharp contrast to the essential contribution of breast milk as a source of energy for the infant, milk oligosaccharides are indigestible to the newborn (Sela and Mills, 2010). In fact, mammals lack the enzymatic machinery necessary to release monosaccharides, such as fucose, N-acetylglucosamine and sialic acid, which are common building blocks of milk oligosaccharides (Engfer et al., 2000). Instead, milk oligosaccharides act as prebiotics, used as nutrients by selected bacterial groups, such as bifidobacteria (Davis et al., 2016). In addition to their prebiotic effect, milk oligosaccharides also affect host-microbe interactions, for example by acting as receptor decoys preventing pathogen binding to host epithelial cells (Newburg et al., 2005). Moreover, milk oligosaccharides affect gene expression in intestinal epithelial cells, influencing their differentiation, survival and surface glycan expression (Angeloni et al., 2005; Kuntz et al., 2008).
Previous studies have shown that prebiotic polysaccharides used as food additives, such as galacto-oligosaccharides or fructo-oligosaccharides of the inulin type, have a beneficial effect in Crohn’s disease and ulcerative colitis patients (Wilson and Whelan, 2017). The mechanisms underlying the positive effect of prebiotic carbohydrates on colitis and the role of the microbiota in these processes are largely unknown. Given the beneficial impact of milk oligosaccharides in newborns, these oligosaccharides may also exert positive effects on dysbiotic microbiota after lactation (Peterson et al., 2013). To date, little is known about the impact of milk oligosaccharides supplemented in children post-weaning and in adults (Elison et al., 2016). To clarify the role of naturally occurring oligosaccharides featuring fucosyl- and sialyl-residues on the course of colitis, we have investigated the effects of the orally supplemented oligosaccharides 2-fucosyllactose (2FL), 3-fucosyllactose (3FL), 3-sialyllactose (3SL), and 6-sialyllactose (6SL) on the course of intestinal inflammation in interleukin-10 null (Il10-/-) mice after weaning. These trisaccharides represent the simplest fucosylated and sialylated carbohydrates that remain uncleaved by host-encoded carbohydrate hydrolases.
Results
2FL Supplementation Decreased Inflammation in Il10-/- Mice
The lack of the immunosuppressive effects mediated by interleukin-10 leads to a progressive enterocolitis related to the continuous stimulation of the mucosal immune system by the microbiota (Kuhn et al., 1993). To assess the impact of naturally occurring prebiotic oligosaccharides on the course and severity of colitis, weaned wildtype (WT) and Il10-/- mice were supplemented with oligosaccharides in drinking water starting at 3 weeks of age for 4 weeks. A concentration of 5 mM of either lactose (Lac), 2FL, 3FL, 3SL, or 6SL was provided with the drinking water. This concentration was chosen, because it fails to induce any signs of discomfort such as soft stool, diarrhea, decreased weight gain and behavioral changes. Mice may display intolerance symptoms related to high lactose concentrations after down-regulation of lactase expression post-weaning (Fang et al., 2006).
The analysis of inflammatory markers, cytokines, and markers of epithelial integrity in the distal colon of mice after 4 week of oligosaccharide supplementation revealed that one oligosaccharide, 2FL, led to significantly decreased expression of the pro-inflammatory markers iNOS, IL-1β and IL-6 (Figure 1). At the same time, the expression of TGFβ, a factor involved in wound healing-associated tissue remodeling, as well as occludin, a tight junction protein associated with epithelial integrity, were increased in Il10-/- mice after 2FL supplementation. By contrast, the structurally related oligosaccharide 3FL and the sialylated oligosaccharide 6SL did not induce any changes in Il10-/- mice after oral supplementation. The other sialylated oligosaccharide, 3SL, decreased IL-1β and TNF expression, while it also decreased expression of occludin (Figure 1). The same trend was observed in proximal colon tissue of Il10-/- mice, which showed a lower degree of inflammation when compared to distal colon (Supplementary Figure S1A). None of the oligosaccharides tested changed the expression of inflammation markers in WT colonic tissue (Supplementary Figure S1B). These data indicated that specific oligosaccharides influenced the course of intestinal inflammation. Interestingly, the structurally related fucosylated oligosaccharides 2FL and 3FL exerted different effects on the expression of marker genes, thus underlining the importance of the glycosidic linkages for the biological roles of oligosaccharides.
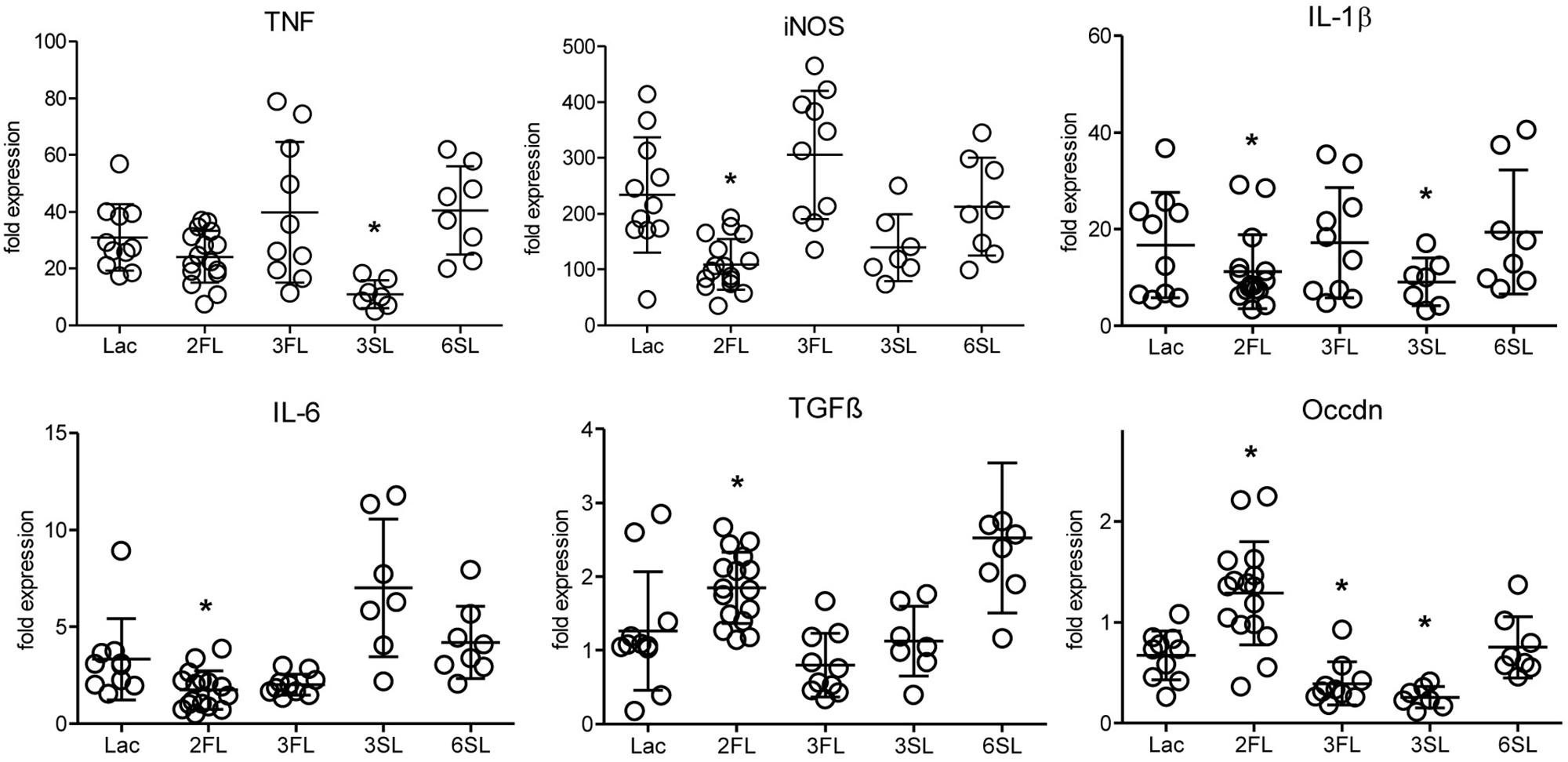
Figure 1. Influence of oligosaccharide supplementation on expression of inflammation markers in Il10-/- mice. Il10-/- mice (open circles) were supplemented with oligosaccharides for 4 weeks. Expression levels of inflammation markers were assessed by qPCR of distal colonic tissue at 7 weeks of age. Expression of the iNOS, the cytokines IL-1β, IL-6, TGFβ, TNF, and the tight junction protein occludin was determined. β-actin was used for normalization and the sample that represents median from Lac supplemented WT mice was used as reference sample according to the ΔΔCt method. Each group consisted of at least six animals (N = 6–12) from 2 (3SL, 6SL), 3 (3FL), or 4 (Lac, 2FL) independent supplementation experiments. Error bars indicate SD. P-values were determined by one-way ANOVA with Dunnett Post-test and are indicated by asterisk (∗) if significantly different from control (Lac) condition (p < 0.05).
The anti-inflammatory effect of 2FL supplementation on colitis in Il10-/- mice was confirmed by measuring the changes in colon length in oligosaccharide-supplemented WT and Il10-/- mice. Only 2FL supplementation maintained a normal colon length in Il10-/- mice, whereas the other oligosaccharides had no effect on the shortening of the colon occurring through inflammation (Figure 2A). Similarly, supplementation with 2FL decreased diarrhea (Figure 2B) and intestinal permeability (Figure 2C), in line with the observed increased expression of occludin in 2FL supplemented Il10-/- mice (Figure 1). Mucosal damage was examined histologically and scored in distal colon sections of Lac, 2FL, and 3FL supplemented Il10-/- mice. Whereas Lac and 3FL supplemented animals showed leukocyte infiltration, lamina propria swelling, cryptitis, goblet cell loss and epithelial damage, these traits were markedly decreased in 2FL supplemented mice (Figure 2D). Blind scoring of histological sections confirmed that supplementation with 2FL, but not 3FL reduced colitis in comparison to control treatment with Lac (Figure 2E).
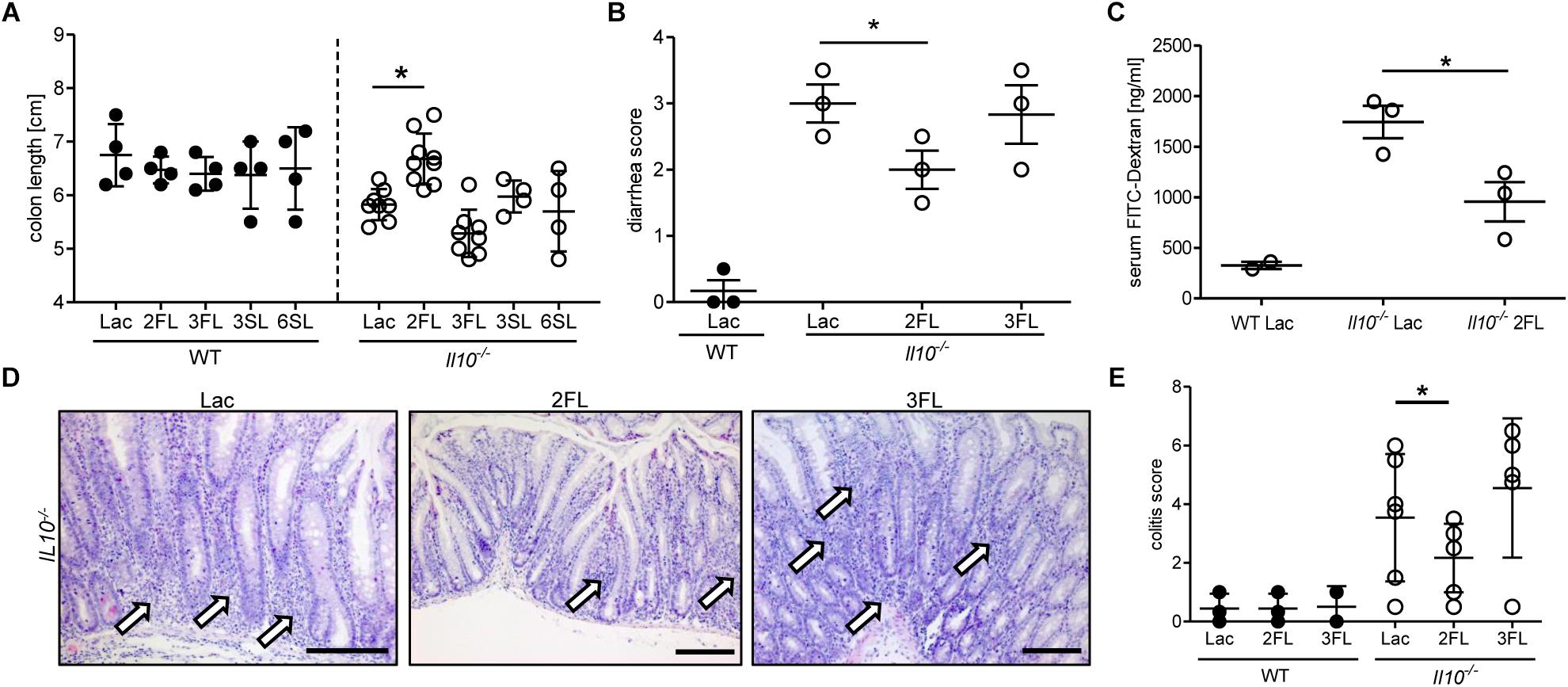
Figure 2. Signs of decreased colonic inflammation after 2FL supplementation. WT (filled circles) and Il10-/- (open circles) were supplemented with Lac, 2FL, or 3FL for 4 weeks. (A) Colon length and (B) diarrhea scores (Sakai et al., 2013). (C) Intestinal permeability of supplemented Il10-/- and WT mice as determined by serum FITC-dextran levels after gavage. (D) Representative images of H&E stained colonic tissue sections. Massive leukocyte infiltration sites are indicated by white arrows. Scale bar = 100 μm. (E) Colitis score of colonic tissue sections. Each group consisted of at least three animals (N = 3–7) from 2 independent supplementation experiments. Error bars indicate SD, P-values were determined by one-way ANOVA with Dunnett Post-test, and indicated by asterisk (∗) if significant (p > 0.05).
Oligosaccharide Supplementation Affected the Intestinal Microbiota in Il10-/- Mice
Considering the selective effects of 2FL against related oligosaccharides, we anticipated a similar variable impact of each oligosaccharide supplemented on the intestinal microbiota. Shifts in intestinal microbial community are likely to influence the local concentrations of bacterial fermentation products, such as SCFAs (Macfarlane and Macfarlane, 2003). Accordingly, we measured the cecal levels of SCFAs in oligosaccharide-supplemented WT and Il10-/- mice. Whereas most of the oligosaccharides did not change SCFA levels, 2FL supplementation in Il10-/- mice was paralleled by increased cecal concentrations of acetate, propionate, and valerate (Figure 3). To further document the shifts in intestinal microbial composition over the course of 2FL and 3FL supplementation, fecal pellets were collected on day 0, day 14, and day 28 of supplementation, and bacterial DNA diversity was assessed by 16S rRNA sequencing. Oligosaccharide supplementation did not appear to alter microbial species richness in neither WT nor Il10-/-, as shown by alpha diversity rarefaction curves (Supplementary Figure S2A). Microbial diversity was, however, lower in Il10-/- mice when compared to WT animals, which is in accordance to previous findings (Gong et al., 2016). Taxonomic evaluation of sequencing data at the family level revealed considerable differences in WT vs. Il10-/- mice (Supplementary Figure S2B), with a higher abundance of Porphyromonadaceae and a lower abundance of Bacteroidaceae and Lachnospiraceae in WT animals in comparison to Il10-/- mice (Supplementary Figure S2C). Notably, some operational taxonomic units (OTU) that were abundant in WT mice appeared to be almost completely absent in Il10-/- mice, such as S24-7, Prevotellaceae, and Desulfovibrionaceae.
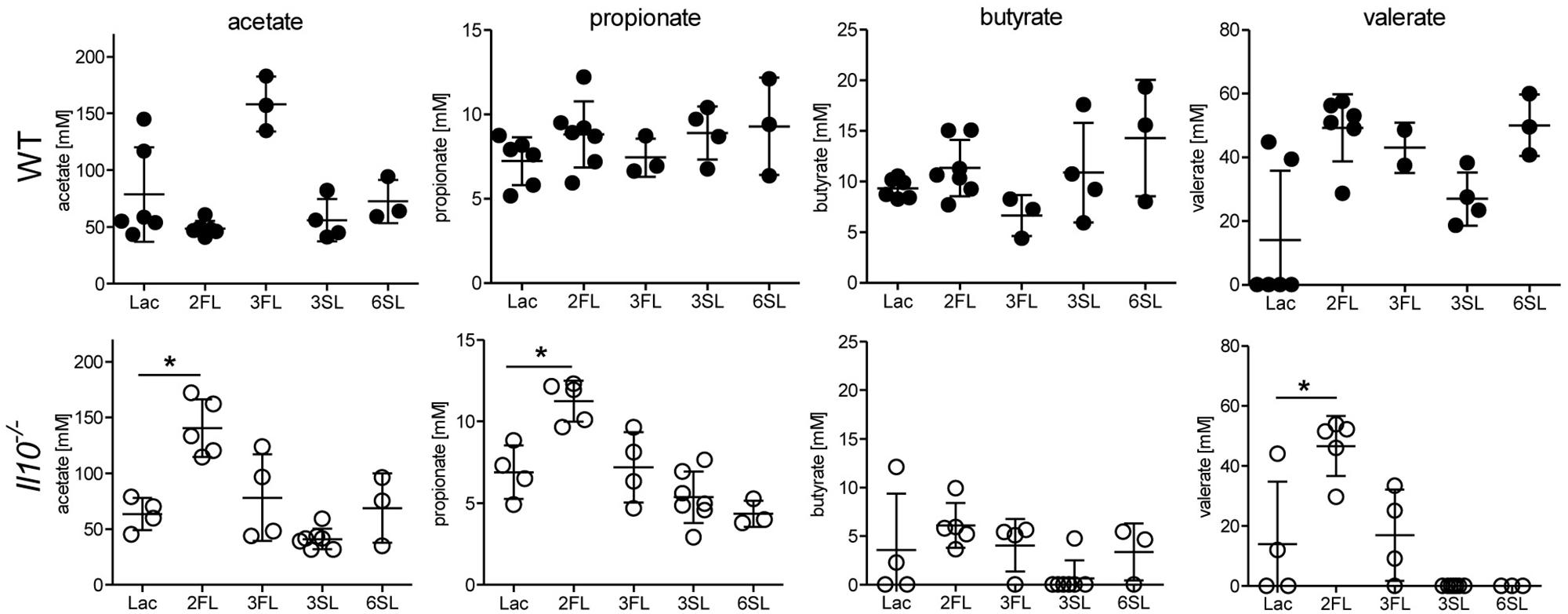
Figure 3. Oligosaccharide supplementation change short chain fatty acid concentration in cecal fluid. Concentrations of the SCFAs acetate, propionate, butyrate, and valerate were analyzed in cecal fluids of WT mice (upper row) and Il10-/- mice (lower row) after 4 weeks of oligosaccharide supplementation in drinking water. Each group consisted of at least three animals (N = 3–7) from 2 (1 for 3FL and 6SL) independent supplementation experiments. Error bars indicate SD, P-values were determined by one-way ANOVA with Dunnett Post-test and are indicated by asterisk (∗) if significant (p > 0.05).
Oral supplementation of Il10-/- mice with Lac did not affect the microbial composition significantly. By contrast, the ingestion of 2FL and 3FL induced a marked increase in Lachnospiraceae over 4 weeks of supplementation (Figure 4A). In 2FL-supplemented Il10-/- mice, the rise in Lachnospiraceae (from 10.6 ± 5.5% to 37.1 ± 12.1%) was matched by a pronounced decrease in Bacteroidaceae (from 51.1 ± 8.4% to 33.0 ± 7.8%). In 3FL-supplemented Il10-/- mice, the increase of Lachnospiraceae was accompanied by a decrease in Ruminococcaceae, while Bacteroidaceae remained unchanged. Further OTU assignment revealed that the increment of Lachnospiraceae in 2FL-supplemented animals was almost exclusively due to expansion of the species Ruminococcus gnavus (from 4.0 ± 3.2% to 31.1 ± 8.3%). On the other hand, the main representants of Bacteroidaceae consisted of Bacteroides acidifaciens and Bacteroides vulgatus, which decreased during 2FL supplementation from 26.3 ± 5.5% to 16.0 ± 3.4% and from 12.6 ± 3.5% to 7.4 ± 2.8%, respectively (Figure 4B). In 3FL-supplemented animals, expansion of R. gnavus (from 2.1 ± 1.5% to 18.7 ± 5.2%) also mainly accounted for the increase in Lachnospiraceae, which, however, yielded in a lower total abundance of this microbe after D28. In clear contrast to 2FL treatment, 3FL supplementation showed a tendency to induce an increase of B. vulgatus from 7.8 ± 4.5% to 15.1 ± 1.8%. Altogether, these data indicate that oral supplementation of Il10-/- mice with 2FL and 3FL led to increased abundance of the Lachnospiraceae R. gnavus especially upon ingestion of 2FL.
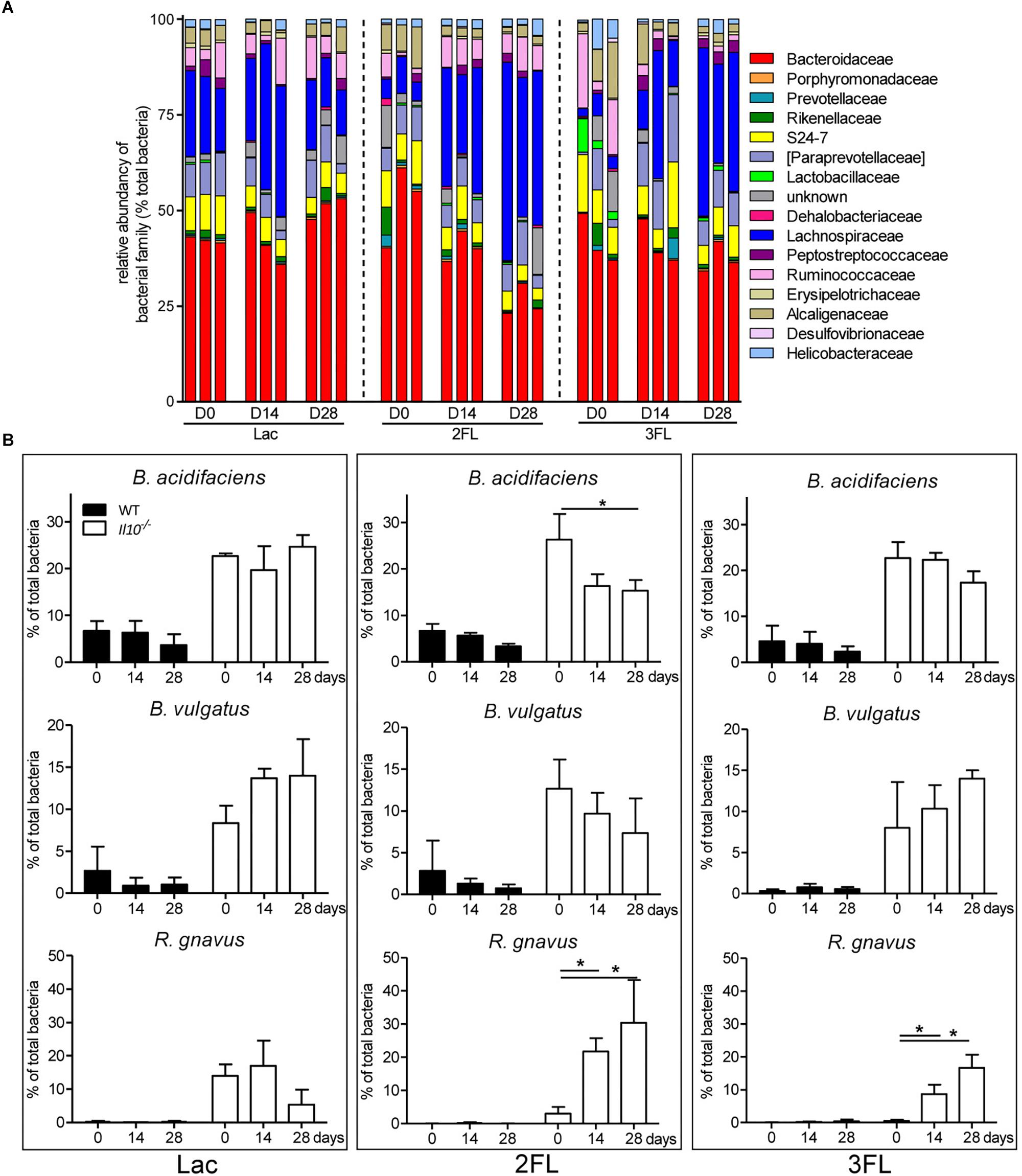
Figure 4. Shift in microbiota at family level after 2FL supplementation in IL-10-/- mice. Bacterial DNA was isolated from fecal pellets of 2FL, 3FL and Lac control supplemented mice on day 0 (D0), day 14 (D14), and day 28 (D28) in Il10-/- mice. Bacterial composition of each sample was assessed by 16S rRNA sequencing. Individual biological samples of one representative supplementation experiment (N = 3) are shown. Each taxa is shown as the percentage of total OTUs within the sample. Only the bacterial taxa representing at least 1% of total identified sequences are presented. (A) Bacterial families are shown as percentage of total OTUs. Each individual replicate is shown. (B) Effect of oligosaccharide supplementation on relative levels of B. acidifaciens, B. vulgatus, and R. gnavus. Mean values are shown, error bars indicate SD. P-values were determined by one-way ANOVA with Dunnett Post-test and are indicated by asterisk (∗) if significant (p > 0.05).
To confirm the growth promoting effect of 2FL on Lachnospiraceae and R. gnavus in particular, microbiota composition was additionally analyzed by quantitative real-time PCR in mice that were supplemented for 4 weeks after weaning with Lac or 2FL. The relative abundance of Bacteroidaceae, Lachnospiraceae, Prevotellaceae, and Porphyromonadaceae in fecal samples was determined by using specific primers for these groups. In line with the data obtained from 16S rRNA sequencing, the increase of Lachnospiraceae could be clearly confirmed (Figure 5). At the species level, the concomitant increase of R. gnavus upon 2FL supplementation was also evident. The analysis of additional mice, however, did not confirm the decrease in Bacteroidaceae levels to a significance threshold.
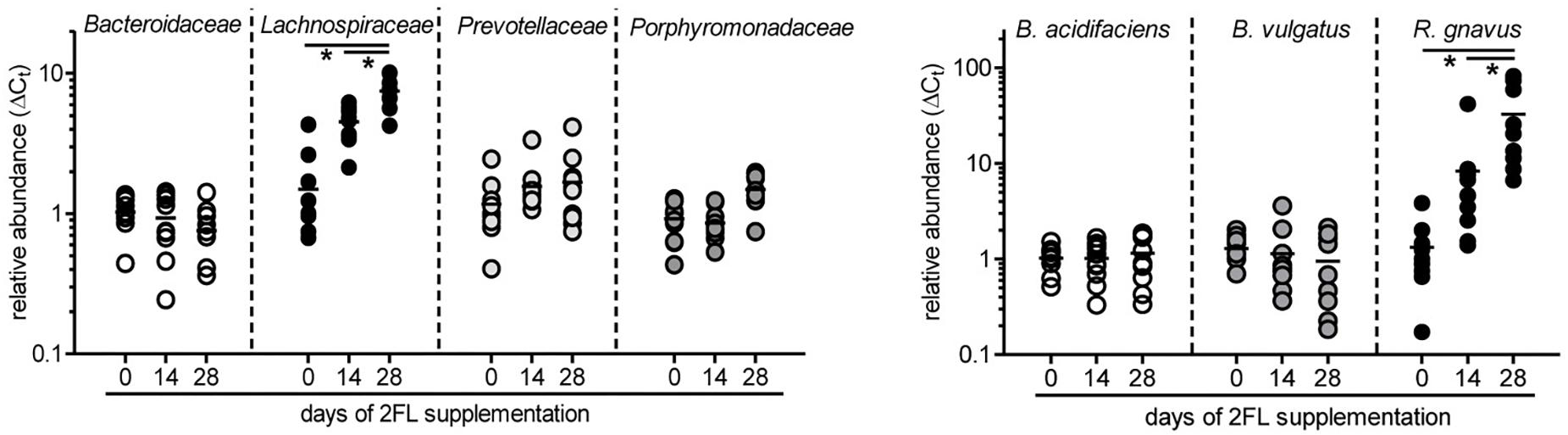
Figure 5. Relative expansion of bacterial taxa after 2FL supplementation. Bacterial DNA was isolated from fecal pellets of 2FL supplemented Il10-/- mice on day 0, 14, and 28. Bacterial composition of each sample was assessed by qPCR analysis. Bacterial taxa at family level (left) and species level (right). To obtain relative taxa expansion, 16S DNA was used for total bacteria normalization and samples were referenced to mean taxa abundance on a D0 sample that represents the median according to ΔΔCt method. Individual data points from 10 biological replicates of 3 independent supplementation experiments (N = 10). P-values were determined by one-way ANOVA with Dunnett Post-test and are indicated by asterisk (∗) if significant (p > 0.05).
Growth Promoting Effect of Fucosylated Oligosaccharides on R. gnavus
The expansion of R. gnavus during supplementation with fucosylated oligosaccharides likely reflected a growth advantage conferred by these carbohydrates on that bacterial species. R. gnavus has been previously shown to efficiently utilize mucin glycans including the oligosaccharides 2FL and 3FL as carbon source (Crost et al., 2013). We confirmed the growth of R. gnavus ATCC 29149 and compared it to the proliferation of B. acidifaciens JCM 10556 and B. vulgatus ATCC 29327 cultured in minimal medium supplemented with glucose, fucose, Lac, 2FL, and 3FL as unique carbohydrate sources. The three bacterial strains had comparable growth kinetics when glucose was provided as sole nutrient (Figure 6). Whereas B. vulgatus failed to efficiently utilize the other carbohydrates, the growth of B. acidifaciens was robust with glucose and Lac, and moderate with 2FL and 3FL, indicating that B. acidifaciens expressed α-fucosidase and β-galactosidase enzymes able to release galactose and glucose as nutrients. B. acidifaciens, however, only metabolized free fucose inefficiently. R. gnavus presented the opposite picture with a robust growth in presence of fucose (Figure 6). The onset of proliferation was delayed to 50 h post-seeding, indicating that growth was probably either conditional on the selection of mutants with fucose-metabolizing ability, or the expression of a fucose-utilization operon. 2FL, but not 3FL, initiated an early proliferation of R. gnavus, suggesting that the bacteria express a linkage-specific fucosidase conferring the preferential release of α1–2 linked fucose. This linkage specificity may be related to the stronger and earlier expansion of R. gnavus observed in 2FL- vs. 3FL-supplemented mice (Figure 4B).
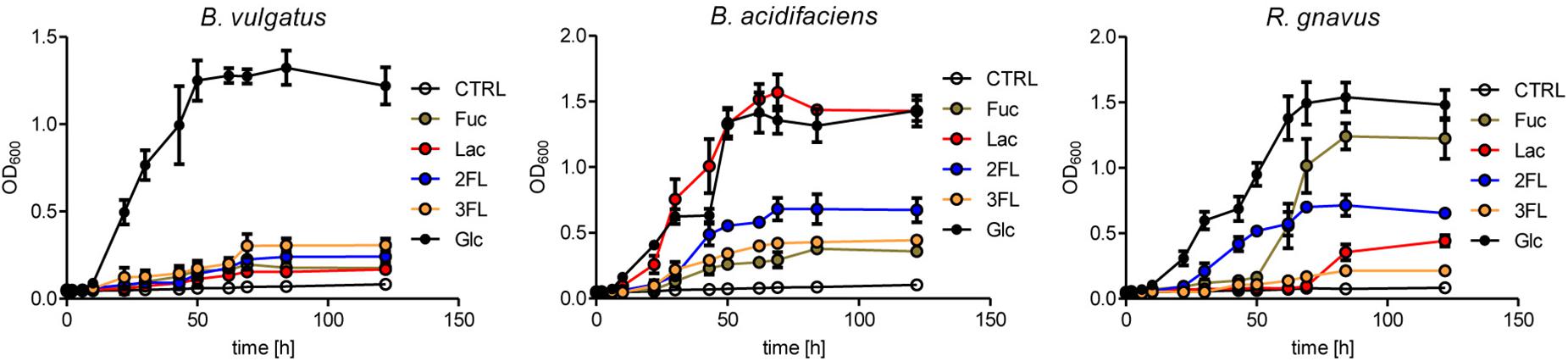
Figure 6. Mono- and oligosaccharide utilization by bacterial cultures. B. acidifaciens, B. vulgatus, and R. gnavus were cultured in chopped meat medium (CMM) to a final OD600 of 0.8. Then they were transferred to Hungate tubes with growth-limiting YCFA medium containing 20 mM of fucose (Fuc), glucose (Glc), 2-fucosyllactose (2FL), 3-fucosyllactose (3FL), or 10 mM of lactose (Lac) to an initial OD600 of 0.05. Negative control tubes (CTRL) contained YCFA medium without any carbohydrate substrate. Growth was monitored until stationary phase was reached. Mean values are shown from triplicates of 2 independent growth experiments. Error bars represent SD.
Because the growth of pure bacterial strains in vitro does not reflect the competition for nutrients prevailing in the gastrointestinal tract, we have assessed the ability of R. gnavus to repopulate the gut microbiota of Il10-/- mice after antibiotic treatment. In general, the induction of spontaneous colitis in Il10-/- mice is strongly associated with the gut microbial community (Sellon et al., 1998). Treatment of mice with broad spectrum antibiotics for 7 days drastically reduced the levels of intestinal bacteria, which readily returned to initial levels a week after cessation of the treatment (Figure 7A). This antibiotic treatment decreased intestinal inflammation for at least 14 days before restoration of the microbiota as assessed by diarrhea score (Figure 7B) and colon weight/length ratios (Figure 7C). Antibiotic-treated Il10-/- mice were reconstituted with 109 CFU of either B. acidifaciens, B. vulgatus, or R. gnavus to determine the effect of these bacterial strains on inflammation. In case of R. gnavus and B. acidifaciens, reconstitution yielded a pronounced increase of these bacteria, which was higher than the initial abundancies of the respective strains in Il10-/- mice. By contrast, B. vulgatus only expanded marginally after oral reconstitution (Supplementary Figure S3). The reconstitution of mice with Bacteroides did not decrease intestinal inflammation beyond the stage reached by natural recolonization after antibiotic treatment (Figure 7D). Reconstitution with R. gnavus, however, further alleviated inflammation as observed by the reduced expression of the inflammation markers iNOS, TNF and IL1β in distal colon tissue (Figure 7D). These results indicate that the expansion of R. gnavus mediated by 2FL supplementation contributed to decreased colitis in Il10-/- mice.
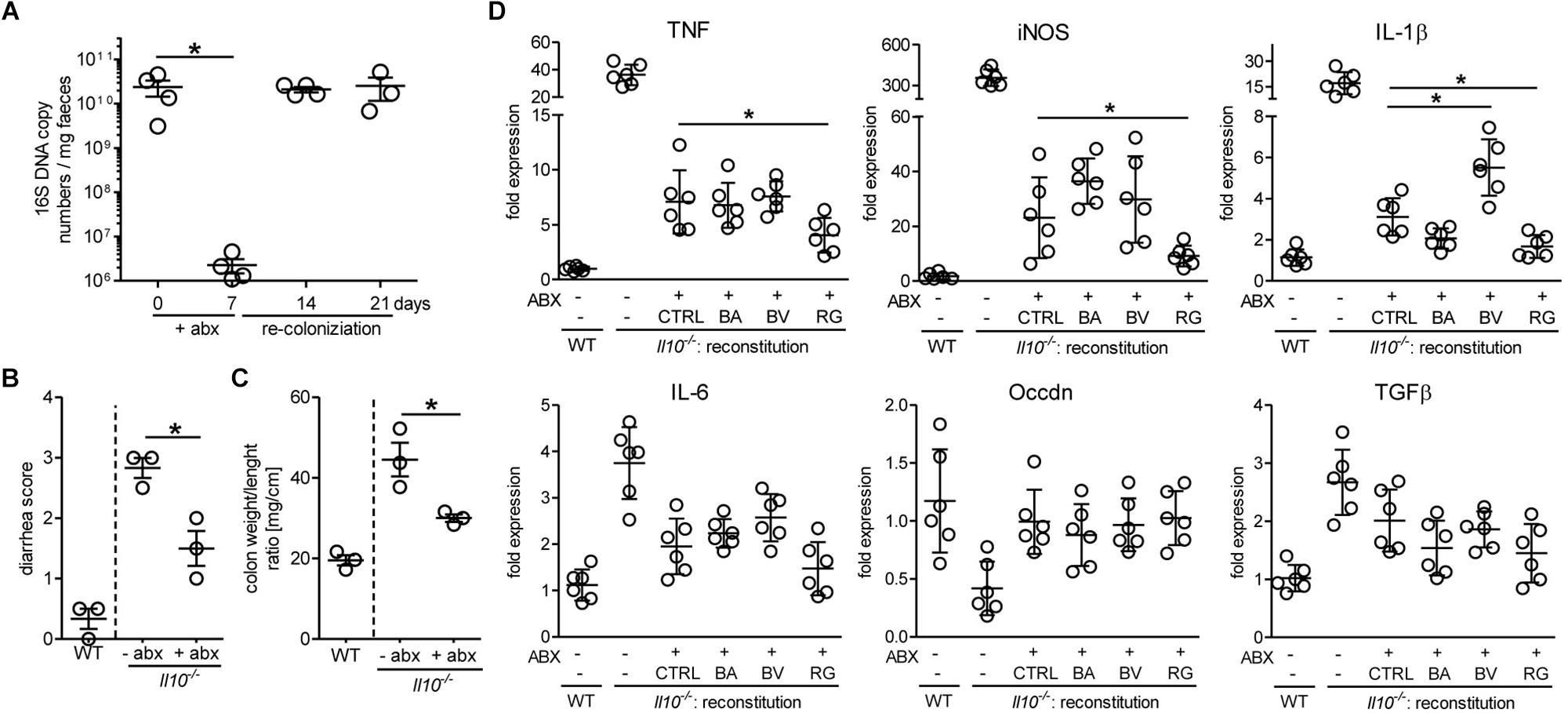
Figure 7. Antibiotic treatment and reconstitution of Il10-/- mice. Mice were treated with antibiotics for 7 days and microbiota allowed to re-colonize for 14 days. (A) Efficacy of antibiotic treatment was assessed by analyzing 16S rRNA copy numbers in DNA isolated from fecal pellets from Il10-/- mice by qPCR using the ΔΔCt method. (B) + (C) Colon weight/length ratio (B) and diarrhea score (C) were analyzed to determine colitis severity. (D) Inflammatory marker gene expression after reconstitution of antibiotic-treated Il10-/- mice with different bacterial strains or culture medium (CTRL). β-actin was used for normalization and the sample that represents median from untreated WT mice was used as reference sample according to the ΔΔCt method. Biological replicates are shown from 1 (A–C) or 2 (D) independent experiments (N = 3–6). Error bars indicate SD. P-values were determined by one-way ANOVA with Dunnett Post-test and are indicated by asterisk (∗) if significant (p > 0.05).
Differential Activation of Bone Marrow-Derived Dendritic Cells by Bacterial Strains
To evaluate the impact of R. gnavus on inflammation and immune reactions, we have compared the activation of dendritic cells mediated R. gnavus, B. acidifaciens, B. vulgatus, and Escherichia coli. Bone marrow-derived dendritic cells (BMDCs) from WT mice were incubated with fixed bacteria and cell activation was assessed by measuring the surface expression of the activation markers CD40, CD86, and MHC-II, and by analyzing cytokine profiles. Stimulation of BMDCs with E. coli and with pure lipopolysaccharide (LPS) strongly activated BMDC as evidenced by increased expression of the markers CD40, CD86, and MHC-II (Figure 8A). Stimulation with B. vulgatus also yielded a strong induction of activation markers, whereas B. acidifaciens and especially R. gnavus only yielded a moderate expression of CD40, CD86, and MHC-II (Figure 8B). The bacteria tested induced different cytokine responses in BMDCs. As expected, E. coli and LPS elicited a strong pro-inflammatory cytokine response characterized by elevated TNF, IL-1β, IFN-γ, IL-9, IL-12, and IL-27 production (Figure 8C). B. vulgatus also induced a potent pro-inflammatory cytokine response with elevated TNF, IFN-γ, IL-9, IL-12, and IL-27. The cytokine responses to B. acidifaciens and R. gnavus stimulation were more restricted and mitigated. Interestingly, R. gnavus induced a strong production of IL-17A, probably originating from T-cells present in the enriched BMDCs fractions. Although IL-17A production is increased in IBD (Fujino et al., 2003), the role of this cytokine in the etiology of the disease is still controversial. Neutralization of IL-17A was even shown to be ineffective at curing patients with moderate to severe Crohn’s disease (Hueber et al., 2012). Surprisingly, only the pro-inflammatory bacteria E. coli and B. vulgatus elicited the production of IL-10 (Figure 8C), which is essential in the maintenance of immune homeostasis in the gut. The low stimulatory effect of R. gnavus on BMDC is probably related to its surface molecules, which lacks LPS. The low stimulatory effect of R. gnavus on immune cells therefore confirms the beneficial impact on intestinal inflammation achieved through 2FL supplementation in Il10-/- mice.
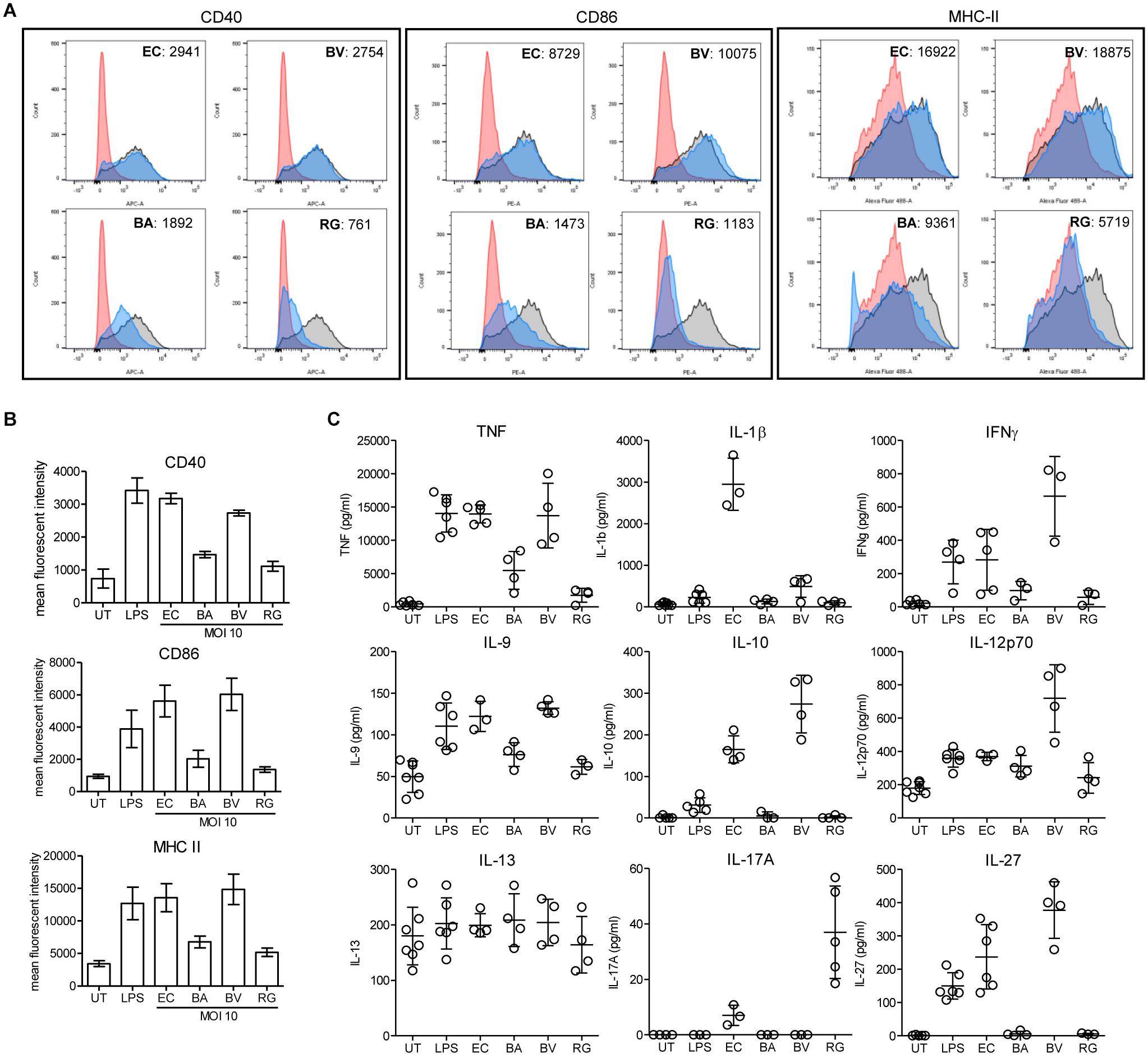
Figure 8. BMDC stimulation by fixed bacteria. Bone marrow cells were isolated from WT mice and in vitro-differentiated into BMDCs and stimulated with PBS (red histogram), 100 ng/ml LPS (black/gray histogram) or fixed cultures of E. coli (EC), B. acidifaciens (BA), B. vulgatus (BV), or R. gnavus (RG) at a MOI of 10 (blue histogram). Activation of BMDCs was determined by expression of activation markers or cytokine secretion. (A) Representative histograms of activation marker expression of CD40, CD86, and MHC-II. (B) Mean fluorescent intensity of activation markers in 3 independent experiments. (C) Cytokine secretion of stimulated BMDCs. Biological triplicates are shown (N = 3–5) from 3 independent experiments. Error bars indicate ± SD. P-values were determined by one-way ANOVA with Dunnett Post-test and are indicated by asterisk (∗) if significant (p > 0.05).
Discussion
Whereas monosaccharides are mainly resorbed in the small intestine, oligosaccharides transit and remain intact down to the microbe-rich colon. Our study in Il10-/- mice demonstrated that the oligosaccharide 2FL specifically promoted the expansion of the Lachnospiraceae R. gnavus, thereby decreasing the development of colitis in young animals after weaning. By this means, 2FL could have an onset-delaying effect in individuals with a colitis-prone predisposition even in the post-lactation period. This observed effect was definite for 2FL, while sialylated oligosaccharides such as 3SL or 6SL, and even the structurally similar 3FL failed to induce colitis protection, indicating that the mechanism involves a high linkage specificity. The colitis-reducing effect was accompanied by a shift in the microbiota that involved a significant increase of Lachnospiraceae. Interestingly, 2FL could also be shown to induce significant shifts of microbiota in a cohort of adult human individuals, showing that the impact of the bacterial community is not limited to early phase of life and opening new potential therapy options (Elison et al., 2016). A potential beneficial effect of 2FL was also seen in a different study using a dextran sulfate sodium model of colitis. However, the expansion of Lachnospiraceae was only transient and eventually overtaken by bacteria of the Barnesiella genus after 6 weeks (Weiss et al., 2014). Apart from IBD, 2FL supplementation has been shown to be beneficial in necrotizing enterocolitis models of newborn rats and pigs. However, this was not observed in combination with a change in microbiota and needs to be further evaluated with intervention on human newborns (Autran et al., 2016; Cilieborg et al., 2016).
Accumulating evidence suggests the involvement of certain bacterial taxa of the microbiota to pathological conditions, such as IBD (Zeng et al., 2017). Nevertheless, a substantial cause-impact relationship of specific bacterial taxa to disease phenotype is yet unidentified, probably due to the multifactorial pathological mechanism of IBD. However, bacteria in general are believed to be a key factor in IBD progression, whereas their role in the etiology of IBD is open to discussion. The results of our experiments with mice that were treated with antibiotics clearly show that microbiota plays a major role in the progression of colitis in the Il10-/- model. In our study, the 2FL-mediated reduction of colitis in the Il10-/- model was accompanied by the outgrowth of R. gnavus. In contrast to this finding, other studies and meta-analyses revealed an expansion of R. gnavus to be indicative for several pathological conditions, such as Crohn’s disease and allergic diseases in infants (Chua et al., 2018; Sokol et al., 2018). However, these findings need to be handled with caution because of their correlative nature. On the one hand, R. gnavus could just be a bystander as mucus-degrading bacteria, profiting from mucus changes in inflammatory conditions, such as increased mucus thickness and upregulation of Muc2 secretion, which have both been described for Crohn’s disease (Smirnova et al., 2001; Strugala et al., 2008; Crost et al., 2013). Intriguingly, R. gnavus abundance was significantly higher in Il10-/- mice compared to WT animals at steady state as shown in the present study. On the other hand, taxonomic profiling of stool samples from IBD patients revealed that blooming of R. gnavus is accountable to only one specific clade that is particularly adapted to oxidative stress given by the inflammatory environment (Hall et al., 2017). Moreover, Lachnospiraceae and especially R. gnavus were found to be highly frequent in early-life gut colonization (Sagheddu et al., 2016). Interestingly, a study showed that in 2-months old breast-fed infants Lachnospiraceae consisted exclusively of R. gnavus, while a cow’s-milk based formula resulted in a more diverse Lachnospiraceae community (Tannock et al., 2013).
While a colitis-protective effect was only seen with 2FL, outgrowth of R. gnavus was observed in 2FL as well as 3FL supplemented animals. One mechanism possibly explaining the anti-inflammatory effect of 2FL against 3FL may be the faster rate of expansion of R. gnavus and the higher relative abundance of this bacterium achieved with 2FL. Supplementation of 2FL might enable R. gnavus to contribute to a microbial environment that restricts the colonization with bacteria triggering robust inflammatory responses. By contrast, the slower expansion of R. gnavus mediated by 3FL was less efficient at dampening the expansion of other bacterial taxa to the same extent. The SCFA propionate has been shown to be protective for colonocytes by alleviating intestinal inflammation through promotion of regulatory T cell development (Arpaia et al., 2013; Smith et al., 2013). The increased proprionate production observed in 2FL-treated mice (see Figure 3) may thus contribute to the overall protective effects of the oligosaccharide treatment. Submucosal invasion of bacteria at local sites of epithelial disruption, which is frequently seen in the Il10-/- model, leads to activation of immune cells, release of pro-inflammatory cytokines, such as TNF, IL-6 and IFN-γ and ultimately even to more epithelial cell damage and increased leukocyte infiltration (Gunther et al., 2015). We could show that Bacteroidaceae strains, and especially B. vulgatus, are much more potent in the activation of dendritic cells and secretion of those cytokines when compared to R. gnavus. This might be a possible mode of action how 2FL supplementation and ensuing R. gnavus expansion leads to a reduction of Bacteroidaceae in the microbial community and therefore to a dampened inflammation response. A drastic reduction of Bacteroidaceae might also be the mechanism providing protection against colitis in antibiotic-treated Il10-/- mice. However, our study failed to provide any direct evidence for this mechanism given that the reduction of Bacteroides species such as B. vulgatus did not reach a significant level. Alternatively, the relative proportion of Bacteroidaceae measured by 16S rRNA sequencing may not represent changes in absolute amounts, but indirectly reflect the relative outgrowth of other bacterial taxa. Apart from that, we cannot exclude that 2FL also exerts a direct microbiota-independent effect on intestinal epithelial or submucosal immune cells. Some oligosaccharides have been shown to alter epithelial and immune cell responses, leading to differential gene expression that might influence TLR-mediated activation or tissue healing (Kuntz et al., 2009; He et al., 2014). However, at least for DCs recent studies have shown that 2FL lead to no direct modulation of differentiation or maturation by 2FL (Perdijk et al., 2018). A potential indirect mechanism for differential immune cell polarization might include 2FL-mediated R. gnavus outgrowth that resulted in a selective expression of the cytokine IL-17 in this study. Taken together, our data suggest that supplementation in the post-lactation period with the prebiotic 2FL induces a beneficial microbial community shift in terms of colitis. Hence, it might be an interesting and promising supplement for diet in IBD patients in terms of disease progression.
Materials and Methods
Mouse Models
All animal experiments conducted in this study were performed in compliance with the Swiss Animal Protection Ordinance and approved by the Veterinary Office of the Canton of Zurich, Switzerland. B6.129P2-Il10tm1Cgn/J mice, termed Il10-/- throughout this article, were obtained from Jackson Laboratories and bred on a C57BL/6J background (WT). All mice were held in light-cycled and climate-controlled housing conditions in an IVC facility in T2L cages, received laboratory chow diet (KLIBA extrudat #3436, Provimi Kliba, SA, Switzerland) and sterilized drinking water ad libitum, if not stated otherwise. For experimental conditions, 3–4 mice were held in a cage. At the end of the experiment mice were euthanized using CO2. Animals were not reused for multiple experiments. Biological material from the same mouse was subjected to different types of analyses.
Animal Experiments
Il10-/- mice were weaned 21 days after birth and supplemented with a 5 mM solution of either 2-fucosyllactose (2FL), 3-fucosyllactose (3FL), 3-sialyllactose (3SL), 6-sialyllactose (6SL) (Glycom A/S, Hørsholm, Denmark) or D-lactose (Sigma-Aldrich, Buchs, Switzerland) in sterile filtrated water ad libitum for 28 days. For depletion of intestinal microbiota, 4–6 weeks old mice were supplied with 0.5 g/L vancomycin (AppliChem), 1 g/L ampicillin (VWR), 1 g/L neomycin (Fisher Bioreagents) and 0.2% aspartame (Sigma) in drinking water for 6 days. Once per day, mice received intragastric gavage of 250 μL of a 1 g/L metronidazole (Sigma) solution. After antibiotic treatment, mice were treated with 10% PEG 3000 solution (Sigma) in drinking water to flush the remaining bacteria and antibiotic. Neo-colonization with specific bacterial strains was done by intragastric gavage of 250 μL containing 109 CFU of the respective strain in PBS. Fecal pellets were collected to monitor changes of the microbiota.
Bacterial Cultures
Anaerobic cultures of Ruminococcus gnavus [ATCC 29149, (Moore et al., 1976)], Bacteroides acidifaciens [JCM 10556, (Miyamoto and Itoh, 2000)] and Bacteroides vulgatus [ATCC 29327, (Eggerth and Gagnon, 1933)] were expanded in anoxic chopped meat medium (DSMZ, #78 including hemin and vitamin K1) in Hungate tubes at 37°C. To determine single carbon source substrate utilization, cultures were transferred to Hungate tubes containing YCFA medium (Duncan et al., 2002) supplemented with the respective carbon source as indicated and cultured at 37°C. Growth was monitored twice a day for at least 5 days by determination of OD600. The E. coli EHV2 strain that was isolated and characterized previously, was cultured in LB medium at 37°C under aerobic conditions (Huang et al., 2015).
Diarrhea Score
The most remarkable changes in individual consistency of feces are represented in the diarrhea score, assessed in a blinded manner and modified after Sakai et al. (2013). 0: normal, dry and solid feces; 1: swollen, moist feces; 2: soft and mucous feces; 3: wet and shapeless feces; 4: bloody diarrhea.
Trans-Epithelial Integrity
Mice were intragastrically gavaged with 200 μL of a 60 mg/ml FITC-Dextran (MW 3000-5000; Sigma) solution in ddH2O 90 min prior to euthanasia. Blood serum was collected after centrifugation at 1500 ×g for 15 min. Serum fluorescence intensity was measured using a multi-detection microplate reader (Tecan Infinite® M200 Pro, Switzerland) at an excitation wavelength of 485 nm and an emission wavelength of 535 nm. FITC concentration was calculated from a standard curve using serial dilutions of FITC–dextran.
Histology and Colitis Score
A tissue sample of 2 cm from distal colon was isolated, briefly washed with PBS, fixed in 10% neutral buffered formalin (Sigma) for at least 24 h and processed for paraffin embedding and sectioning. Histopathological analysis to determine colitis scores was performed on deparaffinized 5 μm Hematoxylin and Eosin (Sigma) stained tissue sections. Sections were scored individually by an independent investigator blinded to the type of treatment as described previously (Steidler et al., 2000).
RNA Isolation and qPCR
Tissue from proximal or distal colon was homogenized with a Precellys® 24 tissue homogenizer (Bertin Instruments) and RNA was isolated using Trizol reagent (Sigma) after manufacturer’s instructions. Total RNA (1 μg) was reverse transcribed using High-Capacity cDNA Reverse transcription Kit (Applied BioSystemsTM) following manufacturer’s instructions. For quantitative PCR reactions KAPA SYBR® Fast (Sigma) Master mix was used and amplified with a CFX95 TouchTM Real-Time System with each 50 nM of the following oligonucleotides: iNOS for: 5′-AGC CTT GCA TCC TCA TTG GG-3′; iNOS rev: 5′-CCT TTG AGC CCT TTG TGC TG-3′; TNF for: 5′-GGC CTC CCT CTC ATC AGT TC-3′; TNF rev: 5′-CAC TTG GTG GTT TGC TAC GAC-3′; IL-1b for: 5′-GCT GGA GAG TGT GGA TCC CAA G-3′; IL-1b rev: 5′-TGC TGA TGT ACC AGT TGG GG-3′; IL-6 for: 5′-CAC GGC CTT CCC TAC TTC AC-3′; IL-6 rev: 5′-GCC ATT GCA CAA CTC TTT TCT C-3′; TGFβ for: 5′-TGG AGC AAC ATG TGG AAC TC-3′; TGFβ rev: 5′-GTC AGC CGG TTA CCA-3′; Occdn for: 5′-CCC TGA CCA CTA TGA AAC AG-3′; Occdn rev: 5′-TTG ATC TGA AGT GAT AGG TG-3′. Cycling conditions were 50 cycles at 95°C for 10 s and 60°C for 30 s after an initial denaturation at 95°C for 3 min. All targets included an exon-exon junction-spanning oligonucleotide to reduce genomic DNA amplification. ΔΔct method was applied to analyze the relative gene expression with β-actin as an endogenous normalization reference and a sample representing the median from lactose treated WT mice used as control sample.
Isolation, Amplification and Sequencing of Fecal Pellet Bacterial DNA
Bacterial DNA was isolated from fecal pellets using the QIAamp DNA stool Mini Kit (QIAGEN) in compliance to manufacturer’s instructions. 20 ng of bacterial DNA were used to amplify the V3-V4 16S rRNA region, using oligonucleotides with Illumina adapters sequences: V3F 340-356: 5′-CTT TCC CTA CAC GAC GCT CTT CCG ATC TCC TAC GGR AGG CAG CAG-3′ and V4R 805-786: 5′-GGA GTT CAG ACG TGT GCT CTT CCG ATC TGG ACT ACH VGG GTW TCT AAT-3′. Amplicon sequencing was performed using an Illumina-MiSeq® system (Genotoul, France). The bioinformatics pipeline Quantitative Insight Into Microbial Ecology (QIIME) open source software package (Versions 1.8.0 and 1.9.0) was used for taxonomic classification of 16S rRNA gene sequencing (Caporaso et al., 2010). Raw datasets were processed by merging, trimming, chimeric reads purging and OTU construction following the UPARSE pipeline (Edgar, 2013). Assignment of taxonomic units was done using the Greengenes database (Version 13.8) as a reference database (DeSantis et al., 2006).
For determination of bacterial taxa by qPCR 50 nM of specific oligonucleotides were mixed with KAPA SYBR Fast (Sigma) Master mix and amplified with a CFX95 TouchTM Real-Time System. Cycling conditions were 50 cycles at 95°C for 10 s and 60°C for 30 s after an initial denaturation at 95°C for 3 min. The following primers were used: total bacteria 16S for (Eub338F): 5′- ACT CCT ACG GGA GGC AGC AG-3′; rev (Eub518R): 5′- ATT ACC GCG GCT GCT GG -3′ (Guo et al., 2008); R. gnavus for: 5′-CCA ATT ACG GAA AGC TGG AT-3′; rev: 5′-TCT GCT TTC CAT GTA TCT TCA CA-3′ (Crost et al., 2013); B. acidifaciens Rec A for: 5′-AAC CTG ATA ACG GTG AGC AGG CG-3′; rev: 5′-GCT GAC AGC CGA GGT CAA TTT G-3′; B. vulgatus for: -5′-GGA GGG GAA AGA CTT ATT TTG C-3′; rev: 5′-TTC CAC CAC TTC TGC CGA C-3′ (Huang et al., 2015). Lachnospiraceae for: 5′-CGG TAC CTG ACT AAG AAG C-3′; rev: 5′-AGT TTY ATT CTT GCG AAC G-3′ (Rinttila et al., 2004); Bacteroidaceae for: 5′-AAG GTC CCC CAC ATT GG-3′; rev: 5′-GAG CCG CAA ACT TTC ACA A-3′ (Hermann-Bank et al., 2013); Porphyromonadaceae for: 5′-ACG CGC GAA TCC CGA AAA CC-3′; rev: 5′- CTT CGT ACG CTC CTT GCG GTT G-3′ (Ramirez-Farias et al., 2009). The lack of significant sequence similarity of the selected primers with unrelated bacterial sequences was confirmed by BLAST analysis. ΔΔct method was applied to analyze the relative taxon abundance with universal bacterial 16S rRNA (Eub338F and Eub518R) as an endogenous normalization reference and a sample representing the median of samples from day 0 was used as a control sample.
Quantitative Analysis of Cecal SCFA
Mouse cecal contents were isolated and centrifuged at 4°C and 8,000 × g for 10 min. SCFA analysis was done by HPLC using a cation-H refill cartridge (30 × 4.6 mm) and an Aminex HPX-87H column at a flow rate of 0.4 ml.min-1 for 60 min and eluted with 10 mM sulphuric acid solution. Quantification was done by detection of refractive index. The concentration was calculated by integral area comparison with authentic standard solutions.
In vitro Differentiation and Stimulation of BMDCs
Bone marrow cells were isolated from tibia, femur and ilium of C57BL/6 WT mice as previously published by Matheu et al. (2008) with minor modifications. Briefly, red blood cell lysis was performed by incubation with ammonium-chloride-potassium lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) for 5 min on ice. Isolated bone marrow cells were cultured and differentiated in Iscove’s Modified Dulbecco’s Medium (Gibco), with 10% FCS, GlutaMAX® supplement (Gibco), antibiotic-antimycotic solution (Gibco), and 1 mM sodium pyruvate (Gibco). Cells were differentiated using 30 ng/mL GM-CSF (Peprotech) and 40 ng/mL IL-4 (Peprotech), replated after 6 days and stimulated after 11 days of isolation. For stimulation, E. coli EHV2, B. acidifaciens, B. vulgatus, and R. gnavus were fixed in 2% paraformaldehyde for 15 min at RT, washed with sterile PBS and reconstituted to a density of 5 × 108 CFU/mL. BMDCs were stimulated with either LPS (O111:B4; 100 ng/mL) or the respective fixed bacterial strains with a multiplicity of infection (MOI) of 10. After 18 h, cells were washed with PBS and harvested by scraping. Activation of BMDCs was determined by fluorescence-labeled antibodies (Biolegend): FITC α-mouse MHC-II (clone M5/114.15.2; #107696); APC α-mouse CD40 (clone 3/23; #124612); PE α-mouse CD86 (clone GL-1; #105008). Samples were acquired using a FACSCantoTM II flow cytometer (BD Biosciences).
BMDC Cytokine Secretion Analysis
Expression and secretion of cytokines after BMDC stimulation was determined by ProcartaPlexTM Multiplex immunoassay (Thermo Fisher Scientific) according to manufacturer’s instructions. Snap-frozen supernatant from BMDCs was diluted 1:2 and incubated with bead mixes B (Lot#140144000) and C (Lot#144194000). Absolute cytokine concentrations were assessed by a serial dilution of Standard Mix A (Lot#146651101).
Statistical Analysis
All multiple group comparisons were performed using one-way ANOVA with Dunnett post hoc test to compare all groups to a control indicated in the respective figure legend. Continuous variables are shown as ± SD and a p-value < 0.05 was considered significant. Throughout this study, all data points are shown, without excluding any outliers. All statistical analyses were performed using GraphPad Prism (Version 5.03).
Data Availability
The raw sequence data has been submitted to the European Nucleotide Archive (ENA) database and can be found under the project number PRJEB31580.
Ethics Statement
This study was carried out in accordance with the recommendations of the Swiss Animal Protection Ordinance. The protocol was approved by the Veterinary Office of the Canton of Zürich, Switzerland.
Author Contributions
TH designed and conceived the study and secured the funding. TG performed and analyzed the experiments and prepared the figures. JGG performed the BMDC FACS experiments. MH performed the histopathological scoring. AG and CL analyzed the 16S sequencing data. TG and TH wrote the manuscript.
Funding
This work was supported by the Swiss National Foundation grants CRSII5_180353 and 314730_172880 and by the Novartis FreeNovation Program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Glycom A/S (Hørsholm, Denmark) for providing oligosaccharides.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01385/full#supplementary-material
References
Angeloni, S., Ridet, J. L., Kusy, N., Gao, H., Crevoisier, F., Guinchard, S., et al. (2005). Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology 15, 31–41. doi: 10.1093/glycob/cwh143
Arpaia, N., Campbell, C., Fan, X., Dikiy, S., van der Veeken, J., deRoos, P., et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. doi: 10.1038/nature12726
Autran, C. A., Schoterman, M. H., Jantscher-Krenn, E., Kamerling, J. P., and Bode, L. (2016). Sialylated galacto-oligosaccharides and 2’-fucosyllactose reduce necrotising enterocolitis in neonatal rats. Br. J. Nutr. 116, 294–299. doi: 10.1017/s0007114516002038
Backhed, F., Roswall, J., Peng, Y., Feng, Q., Jia, H., Kovatcheva-Datchary, P., et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Chassard, C., and Lacroix, C. (2013). Carbohydrates and the human gut microbiota. Curr. Opin. Clin. Nutr. Metab. Care 16, 453–460. doi: 10.1097/mco.0b013e3283619e63
Chua, H. H., Chou, H. C., Tung, Y. L., Chiang, B. L., Liao, C. C., Liu, H. H., et al. (2018). Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology 154, 154–167. doi: 10.1053/j.gastro.2017.09.006
Cilieborg, M. S., Bering, S. B., Ostergaard, M. V., Jensen, M. L., Krych, L., Newburg, D. S., et al. (2016). Minimal short-term effect of dietary 2’-fucosyllactose on bacterial colonisation, intestinal function and necrotising enterocolitis in preterm pigs. Br. J. Nutr. 116, 834–841. doi: 10.1017/s0007114516002646
Coppa, G. V., Gabrielli, O., Pierani, P., Catassi, C., Carlucci, A., and Giorgi, P. L. (1993). Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics 91, 637–641.
Crost, E. H., Tailford, L. E., Le Gall, G., Fons, M., Henrissat, B., and Juge, N. (2013). Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One 8:e76341. doi: 10.1371/journal.pone.0076341
Davis, J. C., Totten, S. M., Huang, J. O., Nagshbandi, S., Kirmiz, N., Garrido, D. A., et al. (2016). Identification of oligosaccharides in feces of breast-fed infants and their correlation with the gut microbial community. Mol. Cell. Proteomics 15, 2987–3002. doi: 10.1074/mcp.m116.060665
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/aem.03006-05
Duncan, S. H., Hold, G. L., Harmsen, H. J. M., Stewart, C. S., and Flint, H. J. (2002). Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52, 2141–2146. doi: 10.1099/00207713-52-6-2141
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Eggerth, A. H., and Gagnon, B. H. (1933). The bacteroides of human feces. J. Bacteriol. 25, 389–413.
Elison, E., Vigsnaes, L. K., Rindom Krogsgaard, L., Rasmussen, J., Sorensen, N., McConnell, B., et al. (2016). Oral supplementation of healthy adults with 2′-O-fucosyllactose and lacto-N-neotetraose is well tolerated and shifts the intestinal microbiota. Br. J. Nutr. 116, 1356–1368. doi: 10.1017/s0007114516003354
Engfer, M. B., Stahl, B., Finke, B., Sawatzki, G., and Daniel, H. (2000). Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am. J. Clin. Nutr. 71, 1589–1596. doi: 10.1093/ajcn/71.6.1589
Fang, R., Olds, L. C., and Sibley, E. (2006). Spatio-temporal patterns of intestine-specific transcription factor expression during postnatal mouse gut development. Gene Expr. Patterns 6, 426–432. doi: 10.1016/j.modgep.2005.09.003
Fujino, S., Andoh, A., Bamba, S., Ogawa, A., Hata, K., Araki, Y., et al. (2003). Increased expression of interleukin 17 in inflammatory bowel disease. Gut 52, 65–70. doi: 10.1136/gut.52.1.65
Gill, N., Wlodarska, M., and Finlay, B. B. (2011). Roadblocks in the gut: barriers to enteric infection. Cell Microbiol. 13, 660–669. doi: 10.1111/j.1462-5822.2011.01578.x
Gong, D., Gong, X., Wang, L., Yu, X., and Dong, Q. (2016). Involvement of reduced microbial diversity in inflammatory bowel disease. Gastroenterol. Res. Pract. 2016:6951091.
Gunther, C., Buchen, B., He, G. W., Hornef, M., Torow, N., Neumann, H., et al. (2015). Caspase-8 controls the gut response to microbial challenges by Tnf-alpha-dependent and independent pathways. Gut 64, 601–610. doi: 10.1136/gutjnl-2014-307226
Guo, X., Xia, X., Tang, R., Zhou, J., Zhao, H., and Wang, K. (2008). Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 47, 367–373. doi: 10.1111/j.1472-765x.2008.02408.x
Hall, A. B., Yassour, M., Sauk, J., Garner, A., Jiang, X., Arthur, T., et al. (2017). A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9:103.
He, Y., Liu, S., Leone, S., and Newburg, D. S. (2014). Human colostrum oligosaccharides modulate major immunologic pathways of immature human intestine. Mucosal Immunol. 7, 1326–1339. doi: 10.1038/mi.2014.20
Hermann-Bank, M. L., Skovgaard, K., Stockmarr, A., Larsen, N., and Molbak, L. (2013). The gut microbiotassay: a high-throughput qPCR approach combinable with next generation sequencing to study gut microbial diversity. BMC Genomics 14:788. doi: 10.1186/1471-2164-14-788
Honda, K., and Littman, D. R. (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. doi: 10.1038/nature18848
Huang, Y. L., Chassard, C., Hausmann, M., von Itzstein, M., and Hennet, T. (2015). Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat. Commun. 6:8141.
Hueber, W., Sands, B. E., Lewitzky, S., Vandemeulebroecke, M., Reinisch, W., Higgins, P. D., et al. (2012). Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700. doi: 10.1136/gutjnl-2011-301668
Hussey, S., Wall, R., Gruffman, E., O’Sullivan, L., Ryan, C. A., Murphy, B., et al. (2011). Parenteral antibiotics reduce bifidobacteria colonization and diversity in neonates. Int. J. Microbiol. 2011:130574.
Kuhn, R., Lohler, J., Rennick, D., Rajewsky, K., and Muller, W. (1993). Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274. doi: 10.1016/0092-8674(93)80068-p
Kuntz, S., Kunz, C., and Rudloff, S. (2009). Oligosaccharides from human milk induce growth arrest via G2/M by influencing growth-related cell cycle genes in intestinal epithelial cells. Br. J. Nutr. 101, 1306–1315.
Kuntz, S., Rudloff, S., and Kunz, C. (2008). Oligosaccharides from human milk influence growth-related characteristics of intestinally transformed and non-transformed intestinal cells. Br. J. Nutr. 99, 462–471. doi: 10.1017/s0007114507824068
LeBlanc, J. G., Milani, C., de Giori, G. S., Sesma, F., van Sinderen, D., and Ventura, M. (2013). Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 24, 160–168. doi: 10.1016/j.copbio.2012.08.005
Lynch, S. V., and Pedersen, O. (2016). The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379. doi: 10.1056/nejmra1600266
Macfarlane, S., and Macfarlane, G. T. (2003). Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 62, 67–72. doi: 10.1079/pns2002207
Maloy, K. J., and Powrie, F. (2011). Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306. doi: 10.1038/nature10208
Marx, C., Bridge, R., Wolf, A. K., Rich, W., Kim, J. H., and Bode, L. (2014). Human milk oligosaccharide composition differs between donor milk and mother’s own milk in the NICU. J. Hum. Lact. 30, 54–61. doi: 10.1177/0890334413513923
Matheu, M. P., Sen, D., Cahalan, M. D., and Parker, I. (2008). Generation of bone marrow derived murine dendritic cells for use in 2-photon imaging. J. Vis. Exp. 17:773.
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118. doi: 10.1016/j.cell.2005.05.007
Miyamoto, Y., and Itoh, K. (2000). Bacteroides acidifaciens sp, nov., isolated from the caecum of mice. Int. J. Syst. Evol. Microbiol. 50, 145–148. doi: 10.1099/00207713-50-1-145
Moore, W. E. C., Johnson, J. L., and Holdeman, L. V. (1976). Emendation of Bacteroidaceae and Butyrivibrio and Descriptions of Desulfomonas Gen-Nov and 10 New Species in Genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium, and Ruminococcus. Int. J. Syst. Bacteriol. 26, 238–252.
Newburg, D. S., Ruiz-Palacios, G. M., and Morrow, A. L. (2005). Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 25, 37–58. doi: 10.1146/annurev.nutr.25.050304.092553
Pannaraj, P. S., Li, F., Cerini, C., Bender, J. M., Yang, S., Rollie, A., et al. (2017). Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 171, 647–654.
Perdijk, O., van Neerven, R. J. J., van den Brink, E., Savelkoul, H. F. J., and Brugman, S. (2018). The oligosaccharides 6’-sialyllactose, 2’-fucosyllactose or galactooligosaccharides do not directly modulate human dendritic cell differentiation or maturation. PLoS ONE 13:e0200356. doi: 10.1371/journal.pone.0200356
Peterson, R., Cheah, W. Y., Grinyer, J., and Packer, N. (2013). Glycoconjugates in human milk: protecting infants from disease. Glycobiology 23, 1425–1438. doi: 10.1093/glycob/cwt072
Ramakrishna, B. S. (2013). Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 28(Suppl. 4), 9–17. doi: 10.1111/jgh.12294
Ramirez-Farias, C., Slezak, K., Fuller, Z., Duncan, A., Holtrop, G., and Louis, P. (2009). Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 101, 541–550. doi: 10.1017/s0007114508019880
Rinttila, T., Kassinen, A., Malinen, E., Krogius, L., and Palva, A. (2004). Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97, 1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x
Sagheddu, V., Patrone, V., Miragoli, F., Puglisi, E., and Morelli, L. (2016). Infant early gut colonization by lachnospiraceae: high frequency of Ruminococcus gnavus. Front. Pediatr. 4:57. doi: 10.3389/fped.2016.00057
Sakai, H., Sagara, A., Matsumoto, K., Hasegawa, S., Sato, K., Nishizaki, M., et al. (2013). 5-Fluorouracil induces diarrhea with changes in the expression of inflammatory cytokines and aquaporins in mouse intestines. PLoS One 8:e54788. doi: 10.1371/journal.pone.0054788
Sela, D. A., and Mills, D. A. (2010). Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18, 298–307. doi: 10.1016/j.tim.2010.03.008
Sellon, R. K., Tonkonogy, S., Schultz, M., Dieleman, L. A., Grenther, W., Balish, E., et al. (1998). Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 66, 5224–5231.
Smirnova, M. G., Kiselev, S. L., Birchall, J. P., and Pearson, J. P. (2001). Up-regulation of mucin secretion in HT29-MTX cells by the pro-inflammatory cytokines tumor necrosis factor-alpha and interleukin-6. Eur. Cytokine Netw. 12, 119–125.
Smith, P. M., Howitt, M. R., Panikov, N., Michaud, M., Gallini, C. A., Bohlooly, Y. M., et al. (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. doi: 10.1126/science.1241165
Sokol, H., Jegou, S., McQuitty, C., Straub, M., Leducq, V., Landman, C., et al. (2018). Specificities of the intestinal microbiota in patients with inflammatory bowel disease and Clostridium difficile infection. Gut Microbes 9, 55–60. doi: 10.1080/19490976.2017.1361092
Steidler, L., Hans, W., Schotte, L., Neirynck, S., Obermeier, F., Falk, W., et al. (2000). Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289, 1352–1355. doi: 10.1126/science.289.5483.1352
Strugala, V., Dettmar, P. W., and Pearson, J. P. (2008). Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn’s disease. Int. J. Clin. Pract. 62, 762–769. doi: 10.1111/j.1742-1241.2007.01665.x
Sun, M., Wu, W., Liu, Z., and Cong, Y. (2017). Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 52, 1–8. doi: 10.1007/s00535-016-1242-9
Swidsinski, A., Ladhoff, A., Pernthaler, A., Swidsinski, S., Loening-Baucke, V., Ortner, M., et al. (2002). Mucosal flora in inflammatory bowel disease. Gastroenterology 122, 44–54.
Tannock, G. W., Lawley, B., Munro, K., Gowri Pathmanathan, S., Zhou, S. J., Makrides, M., et al. (2013). Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl. Environ. Microbiol. 79, 3040–3048. doi: 10.1128/aem.03910-12
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031.
Weiss, G. A., Chassard, C., and Hennet, T. (2014). Selective proliferation of intestinal Barnesiella under fucosyllactose supplementation in mice. Br. J. Nutr. 111, 1602–1610. doi: 10.1017/s0007114513004200
Weiss, G. A., and Hennet, T. (2017). Mechanisms and consequences of intestinal dysbiosis. Cell. Mol. Life Sci. 74, 2959–2977. doi: 10.1007/s00018-017-2509-x
Wilson, B., and Whelan, K. (2017). Prebiotic inulin-type fructans and galacto-oligosaccharides: definition, specificity, function, and application in gastrointestinal disorders. J. Gastroenterol. Hepatol. 32(Suppl. 1), 64–68. doi: 10.1111/jgh.13700
Winer, D. A., Winer, S., Dranse, H. J., and Lam, T. K. (2017). Immunologic impact of the intestine in metabolic disease. J. Clin. Invest. 127, 33–42. doi: 10.1172/jci88879
Xu, J., Mahowald, M. A., Ley, R. E., Lozupone, C. A., Hamady, M., Martens, E. C., et al. (2007). Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 5:e156. doi: 10.1371/journal.pbio.0050156
Xu, L., Lochhead, P., Ko, Y., Claggett, B., Leong, R. W., and Ananthakrishnan, A. N. (2017). Systematic review with meta-analysis: breastfeeding and the risk of Crohn’s disease and ulcerative colitis. Aliment. Pharmacol. Ther. 46, 780–789. doi: 10.1111/apt.14291
Zeissig, S., and Blumberg, R. S. (2014). Life at the beginning: perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat. Immunol. 15, 307–310. doi: 10.1038/ni.2847
Zeng, M. Y., Inohara, N., and Nunez, G. (2017). Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 10, 18–26. doi: 10.1038/mi.2016.75
Keywords: colitis, IBD, milk oligosaccharide, fucose, carbohydrates, microbiota, Lachnospiraceae, Ruminococcus
Citation: Grabinger T, Glaus Garzon JF, Hausmann M, Geirnaert A, Lacroix C and Hennet T (2019) Alleviation of Intestinal Inflammation by Oral Supplementation With 2-Fucosyllactose in Mice. Front. Microbiol. 10:1385. doi: 10.3389/fmicb.2019.01385
Received: 04 March 2019; Accepted: 03 June 2019;
Published: 19 June 2019.
Edited by:
Michael Gänzle, University of Alberta, CanadaReviewed by:
Jens Walter, University of Nebraska–Lincoln, United StatesLaure B. Bindels, Catholic University of Louvain, Belgium
Copyright © 2019 Grabinger, Glaus Garzon, Hausmann, Geirnaert, Lacroix and Hennet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thierry Hennet, dGhpZXJyeS5oZW5uZXRAdXpoLmNo
 Thomas Grabinger
Thomas Grabinger Jesus Francisco Glaus Garzon1
Jesus Francisco Glaus Garzon1 Martin Hausmann
Martin Hausmann Annelies Geirnaert
Annelies Geirnaert Christophe Lacroix
Christophe Lacroix Thierry Hennet
Thierry Hennet