- Lehrstuhl für Technische Mikrobiologie, Technische Universität München, Freising, Germany
Staphylococcus (S.) xylosus is a coagulase-negative Staphylococcus species naturally present in food of animal origin with a previously described potential for biofilm formation. In this study we characterized biofilm formation of five selected strains isolated from raw fermented dry sausages, upon different growth conditions. Four strains exhibited a biofilm positive phenotype with strain-dependent intensities. Biofilm formation of S. xylosus was influenced by the addition of glucose, sodium chloride and lactate to the growth medium, respectively. It was further dependent on strain-specific cell surface properties. Three strains exhibited hydrophobic and two hydrophilic cell surface properties. The biofilm positive hydrophilic strain TMW 2.1523 adhered significantly better to hydrophilic than to hydrophobic supports, whereas the differences in adherence to hydrophobic versus hydrophilic supports were not as distinct for the hydrophobic strains TMW 2.1023, TMW 2.1323, and TMW 2.1521. Comparative genomics enabled prediction of functional biofilm-related genes and link these to phenotypic variations. While a wide range of biofilm associated factors/genes previously described for S. aureus and S. epidermidis were absent in the genomes of the five strains analyzed, they all possess the gene encoding biofilm associated protein Bap. The only biofilm negative strain TMW 2.1602 showed a mutation in the bap sequence. This study demonstrates that Bap and surface hydrophobicity are important factors in S. xylosus biofilm formation with potential impact on the assertiveness of a starter strain against autochthonous staphylococci by competitive exclusion during raw sausage fermentation.
Introduction
Staphylococcus (S.) xylosus is a Gram-positive, coagulase negative species often found on mammal skin. S. xylosus is also widely used as starter organism in raw sausage fermentations (Vos et al., 2009) and has been described as biofilm producer in the past (Planchon et al., 2006; Xu et al., 2017). This ability can be positively associated with food fermentation processes, as adhesion and biofilm formation may increase the assertiveness of a starter organism against the autochthonous microbiota by concomitant induction of colonization resistance in a particular ecological niche. Additionally, biofilms offer a physical protection to bacteria against stress factors including antimicrobial substances (An and Friedman, 2010). In general, the lifecycle of a biofilm can be divided into the stages attachment, maturation and detachment (Otto, 2008). Thereby the first two stages are the main steps of the biofilm formation process, in which multiple factors are involved, and which is often dependent on environmental factors and availability of nutrients (Götz, 2002). Primarily, adherence to a certain support is mediated by nonspecific and/or specific adhesion factors. The latter are termed microbial surface components recognizing adhesive matrix molecules (MSCRAMMS), comprising adhesins on the cell surface of bacteria that bind specifically to extracellular matrix proteins, such as collagen, fibronectin or elastin (An and Friedman, 2010). Following initial adhesion, biofilm accumulation sets in with cells adhering to each other and producing a matrix in which they are embedded in. This extracellular matrix is usually composed of polysaccharides, proteins, and eDNA (Flemming and Wingender, 2010). The multifactorial mechanisms involved in biofilm formation of staphylococci have been described extensively for S. aureus and S. epidermidis in the past (Götz, 2002; Fey and Olson, 2010), often focusing on two important gene loci with functional redundancy, i.e., presence of either one correlates with strong biofilm production (Moretro et al., 2003; Cucarella et al., 2004; Tormo et al., 2005). The polysaccharide intercellular adhesin (PIA), which is synthesized by the products of the ica operon (Cramton et al., 1999) and the biofilm associated protein (Bap). Members of the Bap family are known to be involved in adhesion and biofilm forming processes (Latasa et al., 2006) and comprise among others Bhp, a surface protein often found in S. epidermidis (Tormo et al., 2005) and Esp, a surface protein found in Enterococcus faecalis (Shankar et al., 1999).
This study aimed to characterize phenotypic variations among different strains of S. xylosus regarding their ability to form biofilms, investigate factors influencing biofilm formation, and employed comparative genomic analysis to further comprehend primary adhesion and biofilm accumulation mechanisms in S. xylosus.
Materials and Methods
Bacterial Strains and Culture Conditions
Five S. xylosus strains from the strain collection of Technische Mikrobiologie Weihenstephan (TMW), which were originally isolated from raw fermented sausages, and S. epidermidis RP62A obtained from DSMZ were selected for all experiments. Unless otherwise indicated, strains were grown from cryocultures in tryptic soy broth (TSB, casein peptone 15 g/l, soy peptone 15 g/l, yeast extract 3 g/l) aerobically cultivated until stationary phase (approximately 18 h) at 37°C and shaken at 200 rpm until further use.
Congo Red Agar Assay
To screen for slime production, the congo red agar test was performed as described by Freeman et al. (1989). Briefly, cultures were cultivated on a mixture of 37 g/l brain heart infusion broth (Carl Roth, Germany), 10 g/l agar and 50 g/l sucrose. The medium was supplemented with a solution of separately autoclaved 0.8 g/l of Congo Red (Carl Roth, Germany). After incubation of the isolates on the plates for 24 h at 37°C and 12 h at room temperature, plates were screened for differences in colony morphology. Black and dry crystalline colonies reveal slime producer, while non-slime producer usually develop pink and smooth colonies. Pictorial examples for different kinds of phenotypes is given in Knobloch et al. (2002).
Quantitative Biofilm Formation Assay on Hydrophilic and Hydrophobic Support in Different Cultivation Media
Biofilm formation was tested according to Christensen et al. (1985), with some minor modifications. Basically, overnight cultures of the selected strains were washed and diluted to an OD590 of 0.05 in medium. 200 μl of the adjusted cultures were pipetted into the wells of a 96-well plate and statically incubated for 24 h. After incubation, OD590 was measured again to confirm adequate cell growth in all wells. The wells were carefully decanted and plates were washed twice with sterile phosphate buffered saline (PBS) (NaCl 9 g/l, Na2HPO4∗7H2O 0.795 g/l, KH2PO4 0.114 g/l, pH 7.2). For biofilm fixation, plates were dried in an inverted position in a heat chamber (60°C) for at least 1 h. Adherent biofilm was stained with 200 μl 0.1% safranin-O (Sigma Aldrich, United States) for 5 min. Unbound safranin was removed, and plates were washed again twice with PBS. After air drying of the plates, the stain was solubilized with ethanol (95%) and absorbance was quantified at 490 nm.
In order to test dependence of phenotypic variations and expression of a biofilm positive phenotype on the presence of certain substances, the biofilm assay was performed using different cultivation media (TSB, TSB + 1% glucose, TSB + 1% glucose + 3% sodium chloride, using lactic acid). Additionally, two different supports were used, polystyrene 96-well plates (Sarstedt, Germany) and NunclonTM delta surface 96-well plates (Thermo Fisher Scientific, United States) as hydrophobic and hydrophilic representatives, respectively.
Experiments were conducted in at least three independent biological replicates. Each biological replicate was performed in technical triplicates. Wells containing sterile medium only, served as a control in every experiment performed. S. epidermidis RP62A described as a strong biofilm producer and commonly used as model strain (Mack et al., 1992; Conlon et al., 2002) was included as a positive control for biofilm formation into the experiments.
Microbial Adhesion to Hydrocarbon (MATH)
For determining the surface hydrophobicity of cells, the adherence of bacteria to n-hexadecane was measured as described by Rosenberg (2006). Cells from overnight cultures were washed and resuspended in imidazole/PBS (KH2PO4 0.1 g/l, Na2HPO4∗2H2O 4.45 g/l, imidazole 1.7 g/l, pH 6.2) to an OD590 of 0.35 to 0.4 (AB). 5 ml of the cell suspension were overlaid with 0.4 ml n-hexadecane (Sigma Aldrich, United States) and incubated for 10 min at 37°C. Mixtures were then vortexted for 2 min and statically incubated for another 15 min at room temperature until phase separation was completed. The absorbance (AA) of the aqueous phase was measured and the affinity for n-hexadecane (%) determined by using the following formula:
If values were over 50%, strains were considered as highly hydrophobic, if values were under 20%, as hydrophilic. Each experiment was conducted in three independent runs.
DNA Isolation, Sequencing and Bioinformatics Analysis
For isolation of high-molecular-weight DNA from liquid (tryptic soy broth) bacterial overnight cultures, the E.Z.N.A®kit (Omega Bio-Tek Inc., United States) was used. Whole genome sequencing followed using SMRT (Single molecule real time) sequencing technology (PacBio RS II). The sequencing was carried out at GATC Biotech (Konstanz, Germany). For library creation an insert size of 8 to 12 kb was constructed, delivering at least 200 Mb of raw data from one to two SMRT cells (1 × 120-min movies), when P4-C2 chemistry is applied. SMRT Analysis version 2.2.0.p2 and the hierarchical genome assembly process (HGAP) were used for de novo assembly (Chin et al., 2013). Completion by manual processing according to PacBio instructions followed. Annotation of the genomes was based on the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) and the Rapid Annotations using Subsystems Technology (RAST) Server (Aziz et al., 2008; Tatusova et al., 2016). Bioinformatic analysis and comparative genomics were performed using CLC Main Workbench 8 software (CLC bio, Denmark). To determine strain diversity, average nucleotide identity (ANI) values were calculated using additionally available whole genome sequencing data of four other S. xylosus strains (C2A (LN554884), S170 (CP013922), HKUOPL8 (CP007208), and SMQ-121 (CP008724)). Therefore, the ANIb algorithm (Goris et al., 2007) which is implemented within JspeciesWS web service (Richter et al., 2016) was applied and a neighbor-joining distance tree was built using MEGA7 software.
Statistical Analysis
For statistical analysis, Shapiro–Wilk test was performed to assure normal distribution of data. Means of the technical triplicates were determined first, followed by calculating the means of the biological triplicates including error propagation, which were then used for subsequent statistical comparison of differences. Two-tailed Student’s t-tests assuming unequal variances were performed using SigmaPlot Version 12.5 (Systat Software GmbH, Germany). A difference of means was considered as being significant if p-values were less than 0.05 (P < 0.05). Student’s t-test were performed to compare biofilm intensities of the strains on hydrophilic vs. hydrophobic support and in TSB supplemented with glucose compared to TSB, TSB supplemented with 3% NaCl + 1% glucose compared to TSB + 1% glucose as well as TSB + 1% glucose + lactate (pH 6) compared to TSB + 1% glucose.
Results
Surface Hydrophobicity
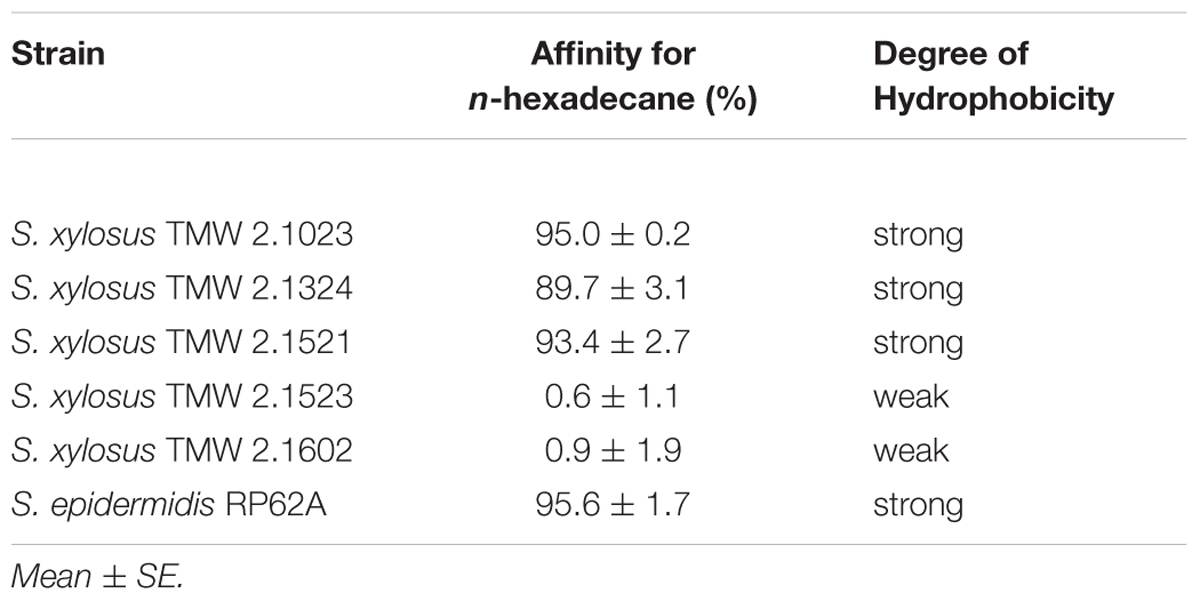
According to the MATH test, only two of the tested strains possess hydrophilic surface properties (TMW 2.1523, TMW 2.1602). All other strains expressed a decisive affinity for the hydrocarbon phase, thus can be considered as strongly hydrophobic (Table 1).
Behavior of Colonies in the Congo Red Agar Assay
All S. xylosus isolates were tested negative for slime production by the congo red agar test. Colonies were mostly smooth, shiny and pink. Yet, changes to a darker color in parts where colonies were in close proximity to each other were observed for TMW 2.1523. The colonies of TMW 2.1523 also showed a rough instead of a smooth surface and a lobate margin. A dry surface with a lobate margin was observed for TMW 2.1521 as well. However, the typical overall black and dry crystalline morphology of a slime producer couldn’t be detected for any of the S. xylosus strains. S. epidermidis RP62A served as positive control.
Influence of Support Hydrophobicity on Biofilm Formation
Adherence potential of S. xylosus to either hydrophobic or hydrophilic supports differed as shown in Figure 1. Among the strains that proved to be of hydrophobic nature, TMW 2.1023 and TMW 2.1521 weakly (A490 < 1.5) adhered to both supports, TMW 2.1324 adhered slightly better to hydrophobic than to hydrophilic support and S. epidermidis RP62A formed significantly more biofilm on hydrophilic than on hydrophobic support. Among the two hydrophilic strains, TMW 2.1602 adhered to neither of the supports (A490< 0.5), while TMW 2.1523 produced significantly more biofilm on hydrophilic compared to the hydrophobic support. In general, relations of biofilm formation on the two tested supports were similar in TSB and TSB + 1% glucose (compare Figures 1A,B), implicating that medium composition had no major influence on the adherence preference of the examined strains to either of the supports. Moreover, S. xylosus proved to be able to form comparable intensities of biofilm as the well characterized biofilm producer S. epidermidis RP62A.
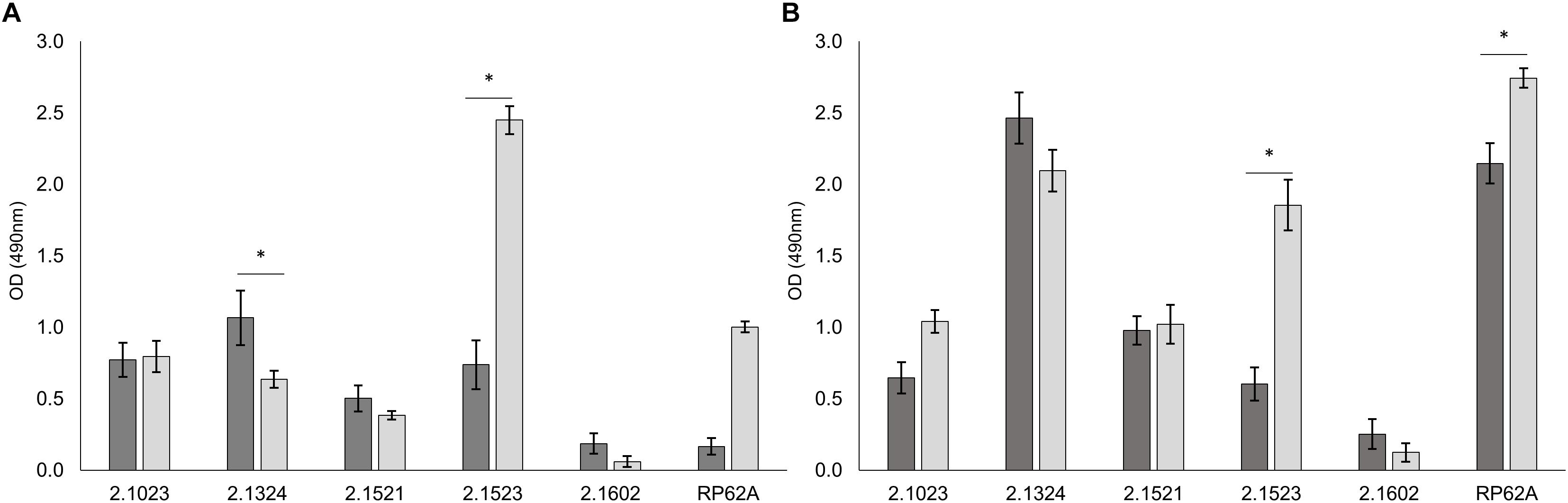
Figure 1. Biofilm formation is dependent on surface hydrophobicity of the support. Biofilm formation on hydrophobic ( ) and hydrophilic (
) and hydrophilic ( ) support in TSB (A) and TSB + 1% glucose (B). Significant differences of mean are marked by ∗. Mean ± SE.
) support in TSB (A) and TSB + 1% glucose (B). Significant differences of mean are marked by ∗. Mean ± SE.
Influence of Media Composition on Biofilm Formation
Media composition was found to influence adherence potential in a strain dependent matter (Figure 2). S. xylosus strain TMW 2.1602 proved again to be a non-biofilm producer regardless of which additive the media contained (A490 < 0.5).
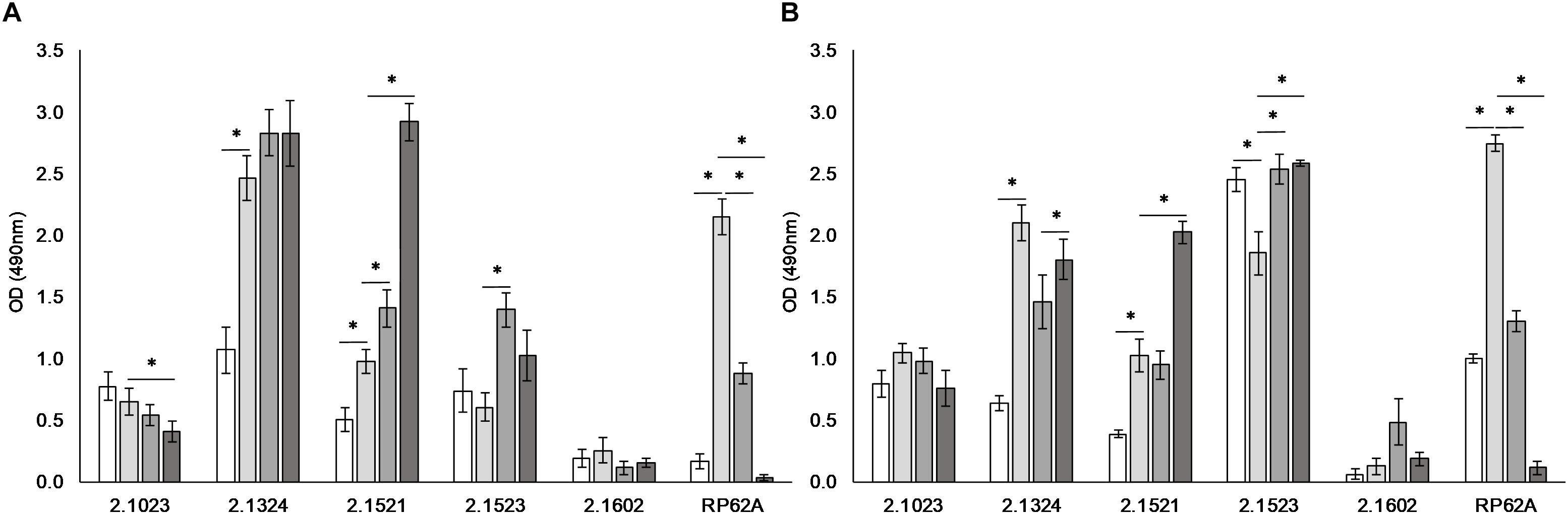
Figure 2. Biofilm formation is dependent on environmental conditions. Biofilm formation on hydrophobic (A) and hydrophilic (B) support when incubated in TSB ( ), TSB + 1% glucose (
), TSB + 1% glucose ( ), TSB + 1% glucose + 3% NaCl (
), TSB + 1% glucose + 3% NaCl ( ), and TSB + 1% glucose + lactate (pH 6) (
), and TSB + 1% glucose + lactate (pH 6) ( ). Significant differences of mean are marked by ∗. Mean ± SE.
). Significant differences of mean are marked by ∗. Mean ± SE.
S. xylosus TMW strains 2.1324 and 2.1521 as well as S. epidermidis RP62A displayed significantly enhanced biofilm formation on both supports tested upon the addition of 1% glucose to the culture medium. On the contrary, biofilm formation was significantly reduced by the presence of glucose in TMW 2.1523 on hydrophilic support. In weak biofilm producer TMW 2.1023, supplementation of glucose had no significant effect on adherence potential. Upon the addition of 3% NaCl to the culture medium, no clear pattern was identifiable for TMW strains 2.1023, 2.1324, and 2.1521. However, biofilm formation was significantly enhanced with NaCl present in TMW 2.1523 and significantly reduced in S. epidermidis RP62A on both supports, respectively. Acidification to pH 6 by lactate had a significantly enhancing effect on biofilm formation of TMW 2.1521 and 2.1523 while it significantly reduced biofilm formation of RP62A. The promoting effect of lactate on biofilm formation was especially distinct in TMW 2.1521, as the strain displayed a weak biofilm phenotype in TSB, TSB enriched with glucose and TSB enriched with a combination of glucose and NaCl. Using lactic acid, however, enhanced the strains biofilm formation to a degree that was comparable to the strong biofilm formers S. xylosus TMW 2.1324, 2.1523 and S. epidermidis RP62A (A490 > 2.0).
General Genome Features
Supplementary Table S1 summarizes the main genome features of the sequenced strains as well as the respective accession numbers. All five sequenced S. xylosus strains possess a single circular chromosome with sizes ranging from 2.8 to 2.9 Mbp, a GC content of 32.7 – 32.9 mol% and a strain-dependent plasmid quantity. The calculated ANI values (Supplementary Figure S1) confirmed genomic diversity among the isolates and revealed certain groups within the species S. xylosus. One comprising most of the TMW strains as well as S. xylosus C2A, originating from human skin (Götz et al., 1983) and S. xylosus SMQ-121, a starter used in the fermentation of processed meat (Labrie et al., 2014). Within this group, the strains TMW 2.1023 and TMW 2.1521 show the lowest genomic distance, while TMW 2.1523 seems to be considerably different from all the other S. xylosus strains. The second group comprised TMW 2.1602 and two additional S. xylosus strains that were both isolated in Asia, S170 from leaf vegetables (Hong and Roh, 2018) and HKUOPL8 from feces of healthy panda (Ma et al., 2014).
Genetic Screening for Adhesion and Biofilm Formation Related Factors
To further investigate the observed phenotypic differences, sequenced genomes of the five S. xylosus isolates were screened for the presence of genes that have been described to be associated with adhesion and biofilm formation processes of well characterized biofilm producers S. epidermidis and S. aureus (Table 2). S. xylosus carries only a small fraction of the described genes, among them autolysin atl/atlE, known to be involved in unspecific adhesion, MCSCRAMMs such as ebpS, eno, fnb as well as bap, a protein important in the biofilm accumulation process. Other genes, also associated with biofilm accumulation, such as aap and the ica-operon are lacking in all S. xylosus strains. Solely TMW 2.1602 carries parts of the ica operon, however, icaD is missing and only icaR, icaC, icaB, and icaA are present in the genome. Moreover, six out of eight genes of the ess cluster, encoding the ESAT-6 secretion system (ESS), were detected in TMW 2.1523 (esxA, esaA, essA, esaB, essB, essC, and A2I72_12780-12805). Compared to the ess cluster of S. aureus Newman (Burts et al., 2008), only esaC and esxB are missing in TMW 2.1523, both of which encode secreted polypeptides. All biofilm related genes analyzed in this study are located on the chromosome of the corresponding S. xylosus strains and not on their plasmids.
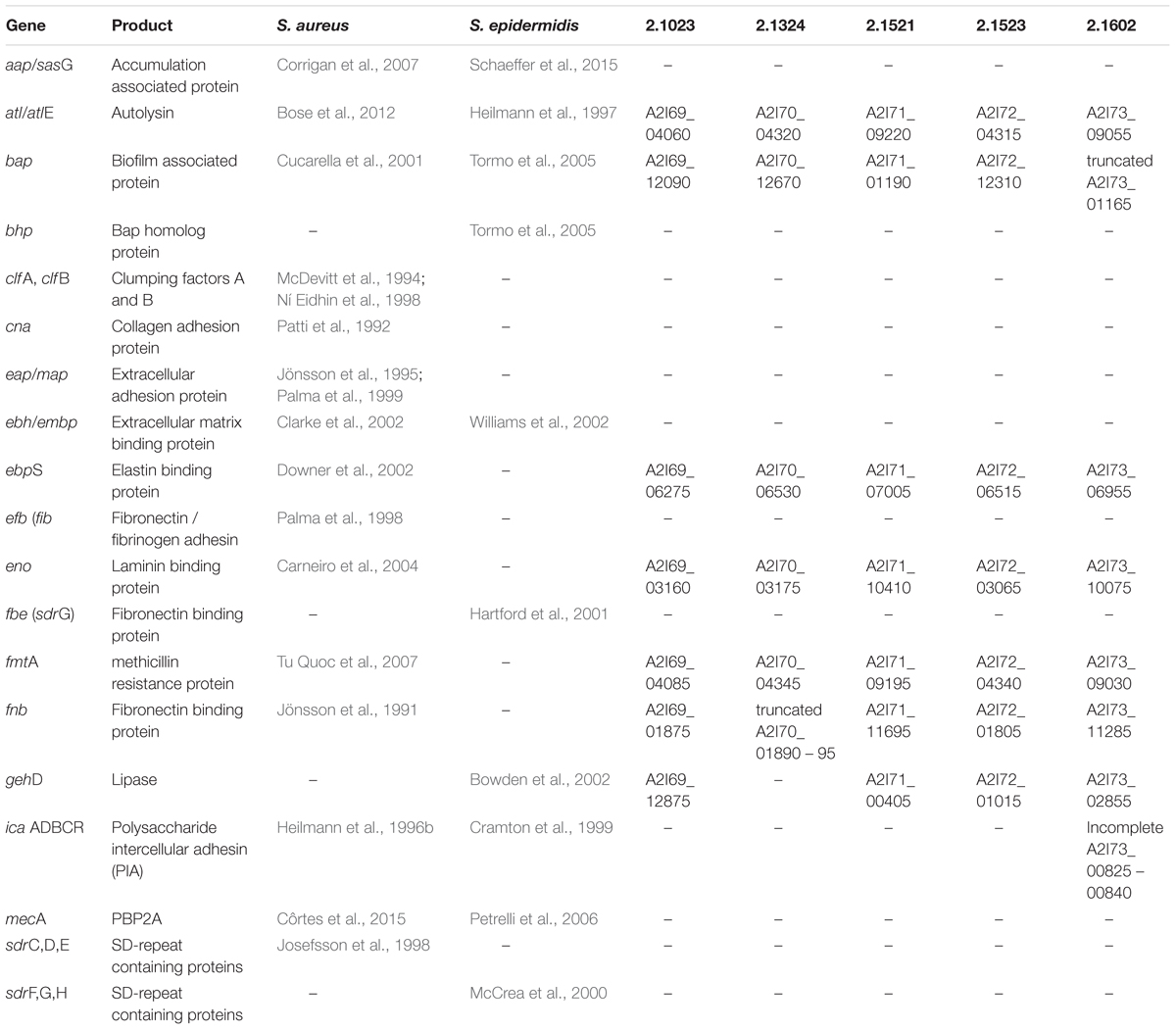
Table 2. Analysis of adhesion and biofilm associated genes, described for S. aureus and S. epidermidis regarding their presence in the sequenced genomes of S. xylosus TMW 2.1023, TMW 2.1324, TMW 2.1521, TMW 2.1523, and TMW 2.1602.
Two truncated genes related to biofilm formation were found in the investigated S. xylosus genomes. In TMW 2.1602 the bap gene encoding the biofilm associated protein carries a mutation. In TMW 2.1324, fnb, responsible for the synthesis of a fibronectin binding protein is truncated. TMW 2.1324 additionally lacks the gehD – lipase gene, which has been described for being involved in adhesion to collagen (Bowden et al., 2002).
Structural Analysis of the Biofilm Associated Protein (Bap) in S. xylosus
In ica-negative strains, Bap plays a major role in biofilm formation. Thus, a detailed in situ structural analysis of the Bap sequence was performed (Figure 3). General structural features were adapted from Cucarella et al. (2001), and Tormo et al. (2005), and Bap structure of S. aureus V329 (GenBank: AY220730.1) was included into the analysis. The bap gene is present in the genomes of all five S. xylosus isolates. However, the bap sequence of strain TMW 2.1602 contains a stop codon after 94 amino acids (aa) indicating an early termination during translation. All other Bap protein sequences show typical structural characteristics. At the N-terminal site of S. xylosus Bap, the YSIRK signal sequence (45 aa) for extracellular secretion is followed by region A (315 aa) which contains two short repeats of 5 aa. The signal sequence is missing in the NCBI-defined open reading frame (ORF) of strains TMW 2.1023 and TMW 2.1521, however, the missing sequence is present in the unprocessed consensus sequence indicating a false delimitation of the ORF. Region B (458 aa) possesses the most conserved part of the protein as it shows the highest sequence identity among the S. xylosus strains (protein identity 98.7 – 100%) as well as 80% identity to the B region of S. aureus V329 Bap. Region C starts with a short spacer region (48 aa) followed by a long core section which encompasses a varying number of Ig-like domain repeats (83 – 86 aa). The highest number of C repeats is present in the genome of S. xylosus TMW 2.1523 (13), followed by TMW strains 2.1324 (10), 2.1023 (7), and 2.1521 (7). The carboxy-terminal region D is characterized by differing numbers (12 – 17) of nearly identical 6 aa tandem repeats. Additionally, it contains an LPxTG motif, which is a well-known cell wall anchor sequence in Gram-positive bacteria. Regarding Bap of TMW 2.1602, not just the early stop codon indicates a truncation of the protein, also the B region misses 73 aa, the spacer region is much shorter and the sequence of the C and D repeats is different than in the other S. xylosus strains, where the repeating sequence was homolog and only the amount of repeats differed among the strains. Compared to S. aureus V329, the biggest difference in the organization of Bap in S. xylosus involves the number of C and D repeats, as size and amino acid sequence differ.
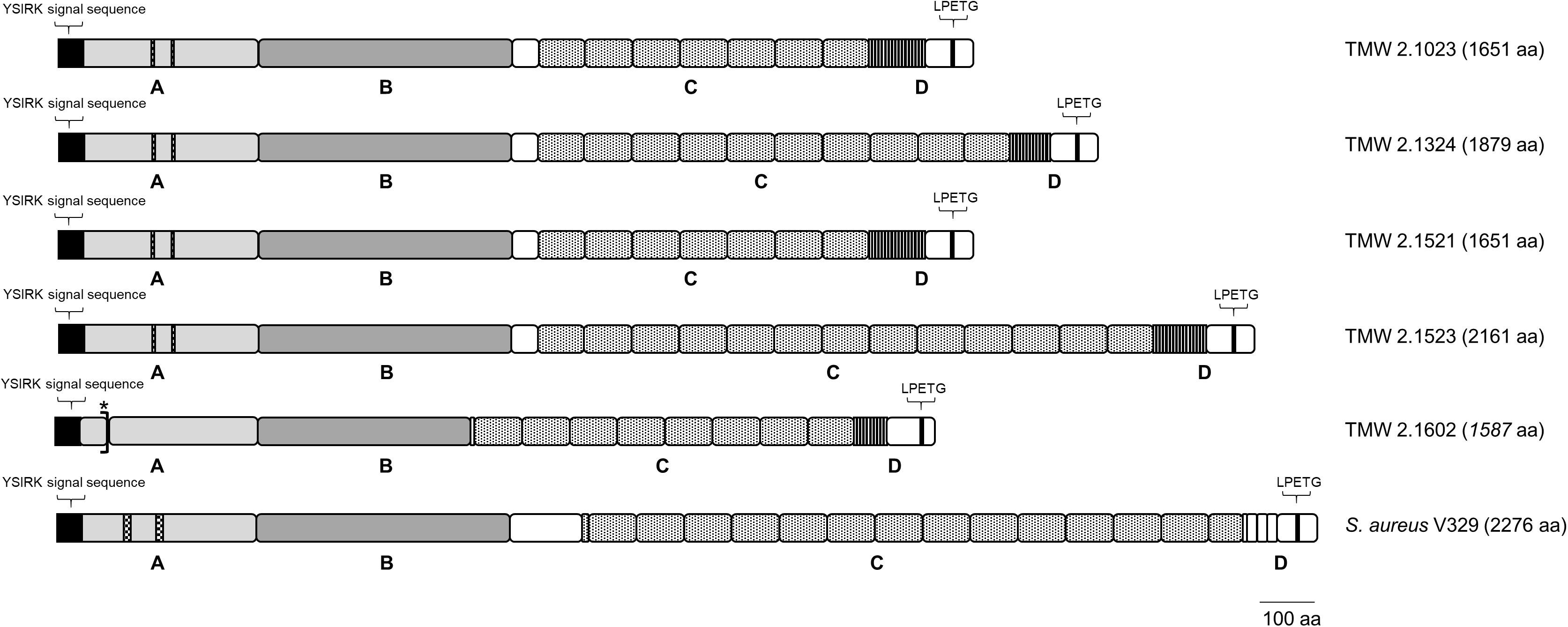
Figure 3. Bap structure of S. xylosus and S. aureus. The positions of the YSIRK signal sequence, the LPxTG cell wall anchor motif as well as the four domains (A–D) of the protein are shown; asterisk indicates early translation termination due to a stop codon in the aa sequence of TMW 2.1602.
Discussion
This study investigated variations in the biofilm forming capacity of five S. xylosus strains isolated from raw fermented sausages in dependence of different supports and media compositions. It was demonstrated that S. xylosus strains with hydrophobic surface properties (TMW 2.1023, TMW 2.1324, and TMW 2.1521) adhered equally well or with minor differences to the two supports tested (hydrophobic, hydrophilic). The only hydrophilic biofilm positive S. xylosus strain (TMW 2.1523) on the other hand adhered distinctly better to hydrophilic than to the hydrophobic support. This is in accordance with previous studies, which have proven that bacteria with hydrophobic surface properties adhere generally well to both kinds of supports while hydrophilic strains prefer hydrophilic supports (Heilmann et al., 1996a; Planchon et al., 2006). Hydrophobic interactions are an important factor for adhesion, and cell surface hydrophobicity is influenced by a combination of the activity of autolysins such as AtlE, teichoic acids, cell surface structures, and surface net charge as well as components of the growth medium (Heilmann et al., 1997; Gross et al., 2001; An and Friedman, 2010). In this study it was further proven that biofilm formation is affected by additives to the growth medium, i.e., glucose, NaCl and lactate. The tested additives had no general stimulating or inhibitory effect on biofilm formation of all strains, but rather displayed varying strain-dependent effects. The here reported controversial effect of glucose on biofilm formation of S. xylosus has been reported for other staphylococci in previous studies (Hennig et al., 2007; Potter et al., 2009). For certain strains, such as S. epidermidis RP62A, addition of 1% glucose is essential for biofilm formation (Mack et al., 1992), which could be confirmed in this study. A generally positive effect on biofilm formation by addition of sodium chloride, previously reported for S. epidermidis and S. aureus (Rachid et al., 2000; Moretro et al., 2003) was not as distinct in the investigated S. xylosus strains.
Generally, the impact of glucose, sodium chloride and lactate on biofilm formation of Staphylococcus spp. has been mainly associated with changes in physicochemical interactions between cell and surface (Planchon et al., 2006) as well as differential expression of the ica operon upon stress exposure (Rachid et al., 2000; Knobloch et al., 2001). Since S. xylosus is ica negative, biofilm formation should be differently regulated by environmental stimuli. Therefore, it seems more likely that the addition of glucose or sodium chloride to the culture medium or the change of pH by lactic acid influences the physiochemical surface properties such as the surface charge of the S. xylosus cells (Briandet et al., 1999). These changes, can impair the cell surface hydrophobicity, change electrostatic forces between support and cell, and therefore interfere with adhesion. Quorum sensing is another regulatory factor, often discussed in context with staphylococcal biofilm formation (Vuong et al., 2003). It appears that quorum sensing effects don’t account for differences in biofilm phenotypes in this study though, as growth rates did not differ significantly in the tested media among the five S. xylosus strains (Data not shown). This is in contrary to the growth enhancing effects of 20 g/l NaCl addition that Planchon et al. (2006) reported. We solely observed a significantly higher growth rate and ODmax in TSB + 1% glucose compared to TSB lacking glucose for S. epidermidis RP62A (Data not shown).
Staphylococci that are ica-positive and thus are able to synthesize PIA often display a slime-positive phenotype on congo red agar (Petrelli et al., 2006). In this study, none of the analyzed S. xylosus strains showed a positive phenotype in the CRA tests, which confirmed the in silico analysis of S. xylosus being ica negative. It also confirms the hypothesis that S. xylosus TMW 2.1602 is most likely not synthesizing PIA despite carrying some genes of the ica operon. However, as Götz (2002) has also reported, icaD is of importance for PIA expression and icaD is missing in TMW 2.1602. TMW 2.1523 showed some characteristics of a CRA-positive phenotype by part of the colonies turning dark, rough and undulated instead of remaining round and shiny. This might be related to congo red being able to not only interact with exopolysaccharides but also proteins (Cucarella et al., 2001). Thus, either the presence of the ess cluster in the genome of TMW 2.1523, which mediates the excretion of certain polypeptides (Burts et al., 2008) or extracellular Bap might cause the reported phenotypic change on CRA. In general, the impact of the ess cluster encoded ESAT-6 secretion system on biofilm formation of S. aureus has been questioned in the past (Wang et al., 2016), yet for Mycobacterium marinum a correlation between ESAT-6 and biofilm formation has been reported (Lai et al., 2018). Therefore, the secreted polypeptides might be part of the biofilm matrix of TMW 2.1523.
To address the question of biofilm intensity formed by S. xylosus, S. epidermidis RP62A, known for being a strong biofilm producer, was taken into account as a reference strain in this study. Hereby, it was shown that ica-negative S. xylosus strains are able to form similar intensities of biofilm as the ica-positive S. epidermidis RP62A strain does. In order to investigate the mechanism of S. xylosus biofilm formation, a comparative genomic analysis of the S. xylosus strains was performed and genomes were screened for presence or absence of genes, which have previously been identified as being involved in biofilm formation of S. aureus and S. epidermidis. Bap seems to be a major factor in S. xylosus biofilm formation, as other well-known biofilm accumulation factors such as the ica operon and aap were absent in the analyzed genomes. Additionally, the physiological data support the thesis that Bap plays a major role in S. xylosus biofilm formation, as the biofilm negative strain TMW 2.1602 carried a truncated bap sequence. The importance of Bap in ica-negative strains has been described for other staphylococci before, e.g., Tormo et al. (2005), have proven that ica negative strains lose their ability to form biofilm once the bap gene is disrupted. It is possible though, that other, yet unknown mechanisms can contribute to biofilm formation. Comparison of the Bap sequences in S. xylosus demonstrated variations in the number of C and D repeats of the protein. However, it has been assumed that at least a varying number of C repeats does not influence the functionality of Bap, as for instance Cucarella et al. (2004) could not identify a correlation between number of C repeats and bap-mediated biofilm formation of S. aureus isolates. Furthermore, Bap has been described as being carried on the pathogenicity island SaPIbov2 in S. aureus (Ubeda et al., 2003). Yet, for S. xylosus no indicators were found that the bap locus was carried on or within a mobile genetic element.
Biofilm formation may contribute to fitness and survival of starter cultures in a particular ecological niche. This assumption is based on the principle of colonization resistance, a phenomenon well known from the human intestine where the microbiota prevents inflammation by occupying all niches along the intestinal tract (Lawley and Walker, 2013). In the sausage matrix, starters with high adhesion and biofilm forming potential may occupy microniches within the meat matrix during fermentation and thus increase their assertiveness against autochthonous staphylococci. The knowledge obtained in this study can be used to explain strain-specific differences of assertiveness in raw sausage fermentation previously identified (Vogel et al., 2017). Screening for a defined set of marker genes derived from the reported comparative genomics results may support the choice of assertive biofilm formers among S. xylosus. Taken together, this study demonstrated variability in biofilm formation of different S. xylosus strains and analyzed for the first time, which adhesion and biofilm related genes are present and absent among different S. xylosus strains displaying distinct phenotypes.
Data Availability
The datasets generated for this study can be found in Genbank, CP015538, CP015539 – CP015541, CP015542 – CP015545, CP015546 – CP015551, and CP015555 – CP015556.
Author Contributions
CS conducted all the experiments, evaluated the data, generated the figures and tables, and wrote the first draft of the manuscript. MH helped in the bioinformatics analyses. ME supervised the work of CS and helped with biofilm tests. RV initiated the project, leaded the design of the study, and supervised CS. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
Part of this work was funded by the German Federal Ministry for Economic Affairs and Energy via the German Federation of Industrial Research Associations (AiF) and the Forschungskreis der Ernährungsindustrie E.V. (FEI), projects AiF17897N and 19690N. Publication of this work was supported by the Technische Universität München as part of the funding program Open Access Publishing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01387/full#supplementary-material
References
An, Y. H., and Friedman, R. J. (eds) (2010). Handbook of Bacterial Adhesion: Principles, Methods, and Applications. Totowa, NJ: Humana Press.
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bose, J. L., Lehman, M. K., Fey, P. D., and Bayles, K. W. (2012). Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One 7:e42244. doi: 10.1371/journal.pone.0042244
Bowden, M. G., Visai, L., Longshaw, C. M., Holland, K. T., Speziale, P., and Hook, M. (2002). Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J. Biol. Chem. 277, 43017–43023. doi: 10.1074/jbc.M207921200
Briandet, R., Meylheuc, T., Maher, C., and Bellon-Fontaine, M. N. (1999). Listeria monocytogenes scott a: cell surface charge, hydrophobicity, and electron donor and acceptor characteristics under different environmental growth conditions. Appl. Environ. Microbiol. 65, 5328–5333.
Burts, M. L., DeDent, A. C., and Missiakas, D. M. (2008). EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol. Microbiol. 69, 736–746. doi: 10.1111/j.1365-2958.2008.06324.x
Carneiro, C. R. W., Postol, E., Nomizo, R., Reis, L. F. L., and Brentani, R. R. (2004). Identification of enolase as a laminin-binding protein on the surface of Staphylococcus aureus. Microbes Infect. 6, 604–608. doi: 10.1016/j.micinf.2004.02.003
Chin, C.-S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Christensen, G. D., Simpson, W. A., Younger, J. J., Baddour, L. M., Barrett, F. F., Melton, D. M., et al. (1985). Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22, 996–1006.
Clarke, S. R., Harris, L. G., Richards, R. G., and Foster, S. J. (2002). Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect. Immun. 70, 6680–6687.
Conlon, K. M., Humphreys, H., and O’Gara, J. P. (2002). icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184, 4400–4408.
Corrigan, R. M., Rigby, D., Handley, P., and Foster, T. J. (2007). The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153, 2435–2446. doi: 10.1099/mic.0.2007/006676-0
Côrtes, M. F., Beltrame, C. O., Ramundo, M. S., Ferreira, F. A., and Figueiredo, A. M. S. (2015). The influence of different factors including fnbA and mecA expression on biofilm formed by MRSA clinical isolates with different genetic backgrounds. Int. J. Med. Microbiol. 305, 140–147. doi: 10.1016/j.ijmm.2014.11.011
Cramton, S. E., Gerke, C., Schnell, N. F., Nichols, W. W., and Götz, F. (1999). The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67, 5427–5433.
Cucarella, C., Solano, C., Valle, J., Amorena, B., Lasa, I., and Penadés, J. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183, 2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001
Cucarella, C., Tormo, M. A., Ubeda, C., Trotonda, M. P., Monzon, M., Peris, C., et al. (2004). Role of biofilm-associated protein Bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 72, 2177–2185. doi: 10.1128/IAI.72.4.2177-2185.2004
Downer, R., Roche, F., Park, P. W., Mecham, R. P., and Foster, T. J. (2002). The elastin-binding protein of Staphylococcus aureus (EbpS) is expressed at the cell surface as an integral membrane protein and not as a cell wall-associated protein. J. Biol. Chem. 277, 243–250. doi: 10.1074/jbc.M107621200
Fey, P. D., and Olson, M. E. (2010). Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 5, 917–933. doi: 10.2217/fmb.10.56
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Freeman, D. J., Falkiner, F. R., and Keane, C. T. (1989). New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 42, 872–874.
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Götz, F. (2002). Staphylococcus and biofilms. Mol. Microbiol. 43, 1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x
Götz, F., Zabielski, J., Philipson, L., and Lindberg, M. (1983). DNA homology between the arsenate resistance plasmid pSX267 from Staphylococcus xylosus and the penicillinase plasmid pI258 from Staphylococcus aureus. Plasmid 9, 126–137. doi: 10.1016/0147-619X(83)90015-X
Gross, M., Cramton, S. E., Götz, F., and Peschel, A. (2001). Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69, 3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001
Hartford, O., O’Brien, L., Schofield, K., Wells, J., and Foster, T. J. (2001). The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology 147, 2545–2552. doi: 10.1099/00221287-147-9-2545
Heilmann, C., Gerke, C., Perdreau-Remington, F., and Götz, F. (1996a). Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64, 277–282.
Heilmann, C., Schweitzer, O., Gerke, C., Vanittanakom, N., Mack, D., and Götz, F. (1996b). Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x
Heilmann, C., Hussain, M., Peters, G., and Götz, F. (1997). Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24, 1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x
Hennig, S., Nyunt Wai, S., and Ziebuhr, W. (2007). Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int. J. Med. Microbiol. 297, 117–122. doi: 10.1016/j.ijmm.2006.12.001
Hong, J., and Roh, E. (2018). Complete genome sequence of biofilm-producing strain Staphylococcus xylosus S170. Korean J. Micobiol. 54, 167–168. doi: 10.7845/kjm.2018.54.3.308
Jönsson, K., McDevitt, D., McGavin, M. H., Patti, J. M., and Höök, M. (1995). Staphylococcus aureus expresses a major histocompatibility complex class II analog. J. Biol. Chem. 270, 21457–21460.
Jönsson, K., Signas, C., Müller, H.-P., and Lindberg, M. (1991). Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 202, 1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x
Josefsson, E., McCrea, K. W., Ní Eidhin, D., O’Connell, D., Cox, J., Höök, M., et al. (1998). Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144(Pt 12), 3387–3395. doi: 10.1099/00221287-144-12-3387
Knobloch, J. K., Bartscht, K., Sabottke, A., Rohde, H., Feucht, H. H., and Mack, D. (2001). Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183, 2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001
Knobloch, J. K.-M., Horstkotte, M. A., Rohde, H., and Mack, D. (2002). Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med. Microbiol. Immunol. 191, 101–106. doi: 10.1007/s00430-002-0124-3
Labrie, S. J., El Haddad, L., Tremblay, D. M., Plante, P.-L., Wasserscheid, J., Dumaresq, J., et al. (2014). First complete genome sequence of Staphylococcus xylosus, a meat starter culture and a host to propagate Staphylococcus aureus phages. Genome Announc. 2:e0671-14. doi: 10.1128/genomeA.00671-14
Lai, L.-Y., Lin, T.-L., Chen, Y.-Y., Hsieh, P.-F., and Wang, J.-T. (2018). Role of the Mycobacterium marinum ESX-1 secretion system in sliding motility and biofilm formation. Front. Microbiol. 9:1160. doi: 10.3389/fmicb.2018.01160
Latasa, C., Solano, C., Penadés, J. R., and Lasa, I. (2006). Biofilm-associated proteins. C. R. Biol. 329, 849–857. doi: 10.1016/j.crvi.2006.07.008
Lawley, T. D., and Walker, A. W. (2013). Intestinal colonization resistance. Immunology 138, 1–11. doi: 10.1111/j.1365-2567.2012.03616.x
Ma, A. P. Y., Jiang, J., Tun, H. M., Mauroo, N. F., Yuen, C. S., and Leung, F. C.-C. (2014). Complete genome sequence of Staphylococcus xylosus HKUOPL8, a potential opportunistic pathogen of mammals. Genome Announc. 2:e0653-14. doi: 10.1128/genomeA.00653-14
Mack, D., Siemssen, N., and Laufs, R. (1992). Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60, 2048–2057.
McCrea, K. W., Hartford, O., Davis, S., Eidhin, D. N., Lina, G., Speziale, P., et al. (2000). The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology 146(Pt 7), 1535–1546. doi: 10.1099/00221287-146-7-1535
McDevitt, D., Francois, P., Vaudaux, P., and Foster, T. J. (1994). Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11, 237–248.
Moretro, T., Hermansen, L., Holck, A. L., Sidhu, M. S., Rudi, K., and Langsrud, S. (2003). Biofilm formation and the presence of the intercellular adhesion locus ica among staphylococci from food and food processing environments. Appl. Environ. Microbiol. 69, 5648–5655. doi: 10.1128/AEM.69.9.5648-5655.2003
Ní Eidhin, D., Perkins, S., Francois, P., Vaudaux, P., Höök, M., and Foster, T. J. (1998). Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30, 245–257. doi: 10.1046/j.1365-2958.1998.01050.x
Palma, M., Haggar, A., and Flock, J. I. (1999). Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding activity. J. Bacteriol. 181, 2840–2845.
Palma, M., Wade, D., Flock, M., and Flock, J. I. (1998). Multiple binding sites in the interaction between an extracellular fibrinogen-binding protein from Staphylococcus aureus and fibrinogen. J. Biol. Chem. 273, 13177–13181.
Patti, J. M., Jonsson, H., Guss, B., Switalski, L. M., Wiberg, K., Lindberg, M., et al. (1992). Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J. Biol. Chem. 267, 4766–4772.
Petrelli, D., Zampaloni, C., D’Ercole, S., Prenna, M., Ballarini, P., Ripa, S., et al. (2006). Analysis of different genetic traits and their association with biofilm formation in Staphylococcus epidermidis isolates from central venous catheter infections. Eur. J. Clin. Microbiol. Infect. Dis. 25, 773–781. doi: 10.1007/s10096-006-0226-8
Planchon, S., Gaillard-Martinie, B., Dordet-Frisoni, E., Bellon-Fontaine, M. N., Leroy, S., Labadie, J., et al. (2006). Formation of biofilm by Staphylococcus xylosus. Int. J. Food Microbiol. 109, 88–96. doi: 10.1016/j.ijfoodmicro.2006.01.016
Potter, A., Ceotto, H., Giambiagi-Demarval, M., Dos Santos, K. R. N., Nes, I. F., and Bastos Mdo, C. (2009). The gene bap, involved in biofilm production, is present in Staphylococcus spp. Strains from nosocomial infections. J. Microbiol. 47, 319–326. doi: 10.1007/s12275-009-0008-y
Rachid, S., Ohlsen, K., Wallner, U., Hacker, J., Hecker, M., and Ziebuhr, W. (2000). Alternative transcription factor sigma B is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182, 6824–6826. doi: 10.1128/JB.182.23.6824-6826.2000
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Rosenberg, M. (2006). Microbial adhesion to hydrocarbons: twenty-five years of doing MATH. FEMS Microbiol. Lett. 262, 129–134. doi: 10.1111/j.1574-6968.2006.00291.x
Schaeffer, C. R., Woods, K. M., Longo, G. M., Kiedrowski, M. R., Paharik, A. E., Büttner, H., et al. (2015). Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect. Immun. 83, 214–226. doi: 10.1128/IAI.02177-14
Shankar, V., Baghdayan, A. S., Huycke, M. M., Lindahl, G., and Gilmore, M. S. (1999). Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67, 193–200.
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tormo, M. A., Knecht, E., Götz, F., Lasa, I., and Penades, J. R. (2005). Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology 151, 2465–2475. doi: 10.1099/mic.0.27865-0
Tu Quoc, P. H., Genevaux, P., Pajunen, M., Savilahti, H., Georgopoulos, C., Schrenzel, J., et al. (2007). Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect. Immun. 75, 1079–1088. doi: 10.1128/IAI.01143-06
Ubeda, C., Tormo-Mas, M. A., Cucarella, C., Trotonda, P., Foster, T. J., Lasa, I., et al. (2003). Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol. Microbiol. 49, 193–210. doi: 10.1046/j.1365-2958.2003.03577.x
Vogel, R. F., Lechner, A., Ruhland, K., and Ehrmann, M. A. (2017). “Assertiveness of Staphylococcus carnosus and Staphylococcus xylosus in sausage fermentation,” in Proceedings of the 3rd International Symposium on Fermented Meats (France: Clermont-Ferrand), 15.
Vos, P., de Garrity, G. M., and Jones, D. (eds) (2009). Bergey’s Manual of Systematic Bacteriology: Volume 3: The Firmicutes. Dordrecht: Springer.
Vuong, C., Gerke, C., Somerville, G. A., Fischer, E. R., and Otto, M. (2003). Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188, 706–718. doi: 10.1086/377239
Wang, Y., Hu, M., Liu, Q., Qin, J., Dai, Y., He, L., et al. (2016). Role of the ESAT-6 secretion system in virulence of the emerging community-associated Staphylococcus aureus lineage ST398. Sci. Rep. 6:25163. doi: 10.1038/srep25163
Williams, R. J., Henderson, B., Sharp, L. J., and Nair, S. P. (2002). Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 70, 6805–6810. doi: 10.1128/IAI.70.12.6805-6810.2002
Xu, C.-G., Yang, Y.-B., Zhou, Y.-H., Hao, M.-Q., Ren, Y.-Z., Wang, X.-T., et al. (2017). Comparative proteomic analysis provides insight into the key proteins as possible targets involved in aspirin inhibiting biofilm formation of Staphylococcus xylosus. Front. Pharmacol. 8:543. doi: 10.3389/fphar.2017.00543
Keywords: Staphylococcus xylosus, biofilm, biofilm associated protein (Bap), surface hydrophobicity, coagulase negative staphylococci
Citation: Schiffer C, Hilgarth M, Ehrmann M and Vogel RF (2019) Bap and Cell Surface Hydrophobicity Are Important Factors in Staphylococcus xylosus Biofilm Formation. Front. Microbiol. 10:1387. doi: 10.3389/fmicb.2019.01387
Received: 26 April 2019; Accepted: 03 June 2019;
Published: 25 June 2019.
Edited by:
Rosanna Tofalo, University of Teramo, ItalyReviewed by:
Régine Talon, Institut National de la Recherche Agronomique (INRA), FranceKrzysztof Skowron, Nicolaus Copernicus University in Toruǹ, Poland
Copyright © 2019 Schiffer, Hilgarth, Ehrmann and Vogel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rudi F. Vogel, cnVkaS52b2dlbEB0dW0uZGU=; cnVkaS52b2dlbEB3encudHVtLmRl
 Carolin Schiffer
Carolin Schiffer Maik Hilgarth
Maik Hilgarth Matthias Ehrmann
Matthias Ehrmann Rudi F. Vogel
Rudi F. Vogel