Abstract
Bacterial communities are fundamental symbionts of corals. However, the process by which bacterial communities are acquired across the life history of corals, particularly in larval and early juvenile stages, is still poorly characterized. Here, transfer of bacteria of the Scleractinian coral Acropora digitifera from adults to spawned egg-sperm bundles was analyzed, as well as acquisition across early developmental stages (larvae and newly settled spat), and 6-month-old juveniles. Larvae were reared under manipulated environmental conditions to determine the source (maternal, seawater, or sediment) of bacteria likely to establish symbiotic relationships with the host using amplicon sequencing of the 16S rRNA gene. Maternal colonies directly transferred bacteria from the families Rhodobacteraceae, Cryomorphaceae, and Endozoicimonaceae to egg-sperm bundles. Furthermore, significant differences in the microbial community structure were identified across generations, yet the structure of the coral bacterial community across early life history stages was not impacted by different environmental rearing conditions. These data indicate that the uptake and structure of bacterial communities is developmentally, rather than environmentally, regulated. Both maternal coral colonies and ubiquitous bacteria found across environmental substrates represent a potential source of symbionts important in establishing the coral microbiome. Uniquely, we report the presence of variation with ontogeny of both the core and resident bacterial communities, supporting the hypothesis that microbial communities are likely to play specific roles within the distinct life history stages of the coral host.
Introduction
Corals host mutualistic relationships with a dense, dynamic and highly diverse consortium of microorganisms such as dinoflagellate unicellular algae in the family Symbiodiniaceae, bacteria, archaea, viruses and fungi. This complex system has been termed the “coral holobiont" (Rohwer et al., 2002). Although it is well recognized that Symbiodiniaceae are essential for the health of corals and for their success in oligotrophic waters (Fournier, 2013), other components of the holobiont (i.e., bacteria) might equally affect coral fitness. Coral bacterial symbionts contribute to various aspects of coral biology, including nutrition, defence, growth, survival and the general health status (Rosenberg et al., 2007; Bourne et al., 2016; Peixoto et al., 2017). A crucial step in the relationship of corals with their bacterial partners is the establishment, or onset, of the association, whereby bacteria are first acquired by the coral host (Apprill et al., 2009; Sharp et al., 2012; Lema et al., 2014). This critical first association between bacteria and coral early life stages has been attributed to both vertical (maternal) and horizontal (environmental) transmission modes. Vertical transmission of bacterial symbionts occurs in several invertebrates including shallow-water bivalves (Krueger et al., 1996), sponges (Sharp et al., 2007; Lee et al., 2009) and bryozoans (Haygood and Davidson, 1997). This mechanism offers potential for co-diversification and co-evolution of the coral holobiont enabling longevity of these relationships (Munson et al., 1991). Horizontal transmission of symbionts from the environment is also a commonly used mechanism and has been documented in invertebrates such as Hydra (Franzenburg et al., 2013), tubeworms (Kikuchi et al., 2007), the broad-headed bug Riptortus clavatus (Nussbaumer et al., 2006) and also in the bobtail squid Euprymna scolopes where the luminous bacterium Vibrio fischeri is acquired (Nyholm and McFall-Ngai, 2004). Both vertical and horizontal transmission modes are likely to be influenced by host reproductive strategies (Apprill et al., 2009, 2012; Sharp et al., 2010, 2012) and several studies suggest that the acquisition of microbial symbionts is consistent with reproductive mode (brooding vs. broadcast spawning) within corals (Hartmann et al., 2017). However, understanding of the mechanisms for acquiring bacterial associations, and how these evolve and are maintained through development stages, is poorly understood.
Recently, deep-sequencing methodologies have been applied to understand the diversity of the bacterial communities associated with specific early life stages of corals (Leite et al., 2017; Zhou et al., 2017). However, these have not yet been applied to document the diversity across development or to track the source (parental or environmental) of bacterial community acquisition. Among broadcast spawning coral only ∼20% acquire their algal symbionts vertically, while the remaining portion take them up from their immediate surrounding environment (Stat et al., 2006; Baird et al., 2009; Quigley et al., 2017). Given their benthic position on the reef, the most likely sources of bacterial partners for corals are the surrounding seawater and/or marine sediment. Seawater and sediment are both characterized by dynamic and diverse free-living bacterial communities, directly available for acquisition via horizontal transfer to the coral host. For corals that acquire Symbiodiniaceae from the environment, this occurs by the larvae, post-settlement and after metamorphosis to a juvenile polyp (Weis et al., 2001). Less information is available on horizontal transmission patterns of bacterial communities (Sharp et al., 2010). Early life stages contain a higher diversity of bacterial taxa in comparison to adult colonies, suggesting that the environment is a reservoir of bacterial symbionts during development (Apprill et al., 2009; Littman et al., 2009a,b; Lema et al., 2014). The main source of bacteria in coral early life history stages is thought to be the water column (Patten et al., 2008; Apprill et al., 2009, 2012). However, sediment bacterial communities are among the most taxonomically diverse within coral reef habitats (Schöttner et al., 2012) and coral mucus has a higher similarity of bacterial taxa with sediment than with the water column (Carlos et al., 2013), thus suggesting that sediments also act as an important source that seed coral microbiomes including symbionts that may play a fundamental role in coral ontogeny and establishment.
Scleractinian corals depend on their associated bacterial symbionts (Rosenberg et al., 2007), however, it is still relatively unclear how the composition and the structure of these associations change through life history stages and which taxa are likely to play a fundamental role. For instance, members of the genera Burkholderia, Pseudomonas, Acinetobacter, Ralstonia, Inquilinus, Bacillus, Marinobacter, Roseobacter and the orders Rhizobiales, Oceanospirellales, and Alteromonadales were ubiquitously found across various coral life stages and were proposed to be important in nitrogen fixation, degradation of the organic sulfur metabolite dimethyl-sulfoniopropionate (DMSP), pollution degradation and production of antibiotics (Nissimov et al., 2009; Raina et al., 2009; Sharp and Ritchie, 2012; Lema et al., 2014; Leite et al., 2017). Some of these taxa were regularly identified as members of the adult coral microbiome (Littman et al., 2009b; Raina et al., 2009; Bourne et al., 2013; Leite et al., 2017), thus suggesting that they may contribute to the formation of the core or resident microbiome (Leite et al., 2017). Another approach that can be applied to elucidate potential bacterial taxa that may play key functional roles is to identify those taxa that occur in very high frequency across individuals of the same host species. This community can be described as the core microbiome and is defined as those taxa that are present in all, or the vast majority, of individuals (Turnbaugh et al., 2007). In coral holobionts “core” and “resident” bacterial communities have been introduced, representing taxa found at high, medium and low frequencies, respectively (Hernandez-Agreda et al., 2018). Therefore, as shown in our study, characterizing bacterial communities based on their frequency and across various developmental stages, may help identify bacterial partners that are important in the development of the coral host.
Given the importance of bacteria in maintaining coral health, it is imperative to understand the establishment of coral-bacterial associations. In this study we documented the associations of bacteria across adult, egg-sperm bundles, larvae, spat and juveniles of the broadcast coral Acropora digitifera, one of the dominant species on Ningaloo Reef and widespread across the world (Aeby et al., 2014), using amplicon 16S rRNA gene sequencing. We specifically tracked the vertical transmission of bacteria to spawned bundles of five individual adult colonies. In addition, we experimentally manipulated the exposure of larvae to seawater and sediment to determine horizontal transmission.
Materials and Methods
Colonies of the Scleractinian coral A. digitifera were collected from Ningaloo Reef in Coral Bay, Western Australia (23°10.698′S, 113° 45.634′E) 3–5 days prior to predicted spawning. Eight healthy colonies (Supplementary Figure 1) were collected from the reef and transported to the Coral Bay jetty (Figure 1A). A few hours prior to spawning, colonies were placed into individual 60 L tubs containing seawater (Figure 1B and Supplementary Figure 1I). All colonies spawned at 2200–2300 h on 21 March 2017. Egg–sperm bundles were gently mixed in 20 L of seawater for fertilization (Figure 1C) and after 3 h, embryos were rinsed with 0.22 μm filtered seawater (FSW).
FIGURE 1
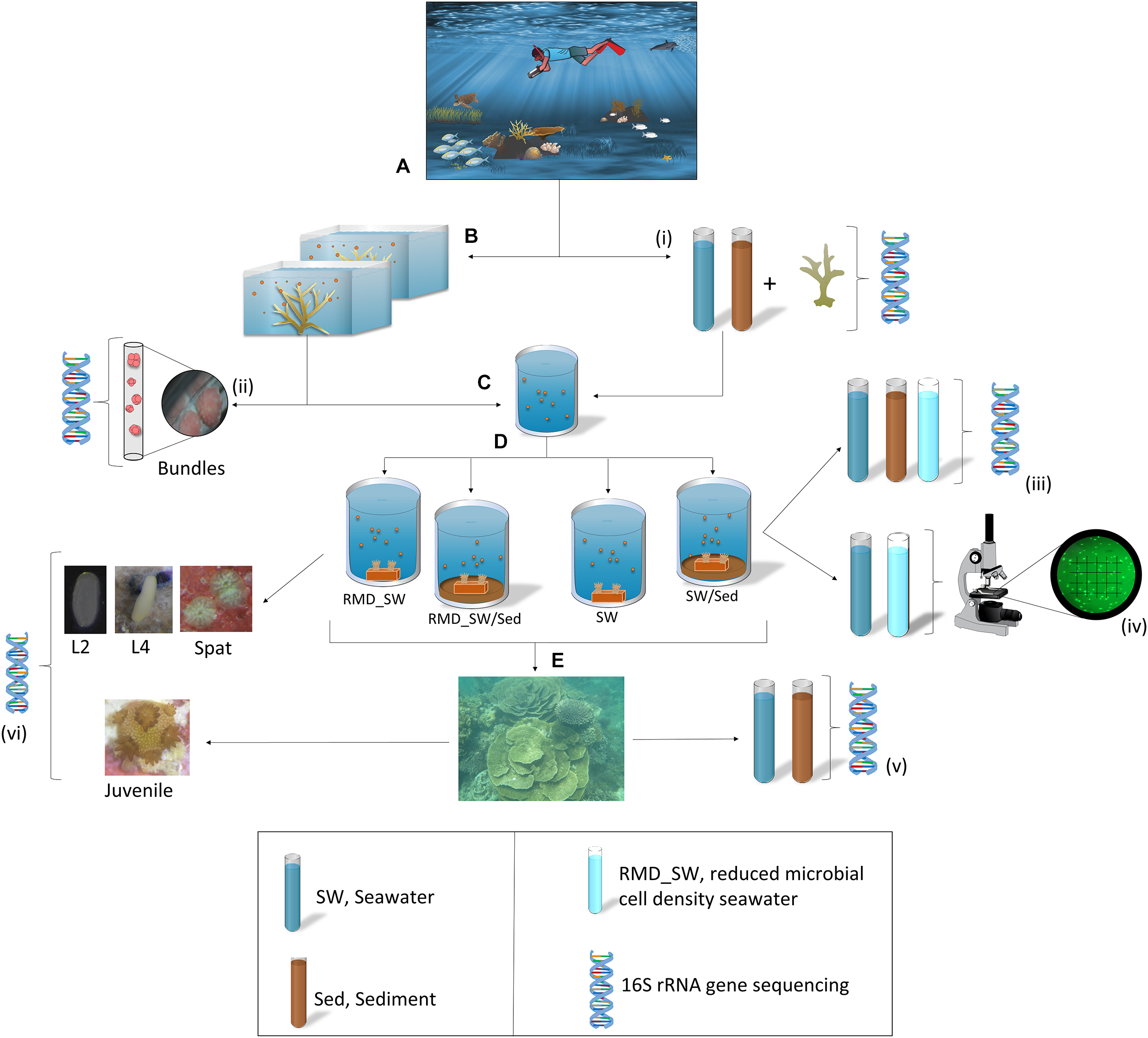
Sampling and experimental design. (A)Acropora digitifera adult coral colonies were relocated from the reef to the Coral Bay jetty. (B) A few hours prior to spawning colonies were placed into individual tubs containing seawater. (C) Egg–sperm bundles were collected for fertilization, and (D) 8 h old larvae were transferred to four experimental treatments for larval rearing: RMD_SW, reduced microbial cell density seawater; RMD_SW/Sed, reduced microbial cell density seawater and sediment; SW, seawater, SW/Sed, seawater and sediment. Seawater (SW) and sediment (Sed) samples were collected from the collection site and used in tanks during the manipulative experiment. (E) Once settled onto terracotta tiles, newly settled spat were deployed back onto the reef for 6 months. (i–iii,v,vi): DNA symbols indicates where various samples (n = 5 in all cases) were taken for 16S rRNA gene sequencing. (iv) Samples of SW and RMD_SW (n = 3) were preserved in 2% formalin at 4°C and bacterial density was determined via microscopy. Symbols are from the Integration and Application Network (ian.umces.edu/symbols/).
In order to characterize their bacterial communities, adults and bundles were collected and snap frozen in liquid nitrogen. For adults, fragments of each colony (three replicate tissue subsamples, ∼1 cm2 each) were collected prior to spawning (Figure 1i). Bundles were collected during spawning, from five coral colonies, placed in a cell strainer (40 μm nylon mash, Scientific Laboratory supplies), and rinsed in FSW prior to freezing (Figure 7ii).
Microbial Acquisition Experiment
Approximately 8 h old larvae were randomly assigned to one of four treatments (Figure 1D), each with five replicates (20 acid washed tanks in total). The treatments were designed to manipulate larval exposure to microbial communities from sediment and seawater and were: (1) reduced microbial cell density seawater (RMD_SW, filtered at 0.22 μm), (2) reduced microbial cell density seawater + sediment (RMD_SW/Sed), (3) seawater (SW) and, (4) seawater + sediment (SW/Sed). To establish the treatments, seawater was collected from the ocean surface while sediment was collected with 50 mL sterile tubes by scooping the oxic layer (∼1 cm depth) within 1 m from the coral colonies. Five replicate sediment (1.8 mL each), and seawater (2 L each, filtered on sterivex filters) samples were snap frozen in liquid nitrogen for bacterial analysis (Figure 1i).
For the experiment, the tanks each contained 2 L of seawater (either reduced or entire microbial density) and those with sediment had 50 mL of sediment thinly covering the bottom. Sediment was added to the tanks 7 h before the experiment started in order to allow it to completely settle. Sediment, seawater, and reduced microbial cell density seawater samples were snap frozen in liquid nitrogen for bacterial analysis (n = 5, Figure 1iii). The temperature was maintained at 25°C to match the daily mean local water temperature (25.1°C). After 24 h a gentle airflow was added to each tank. Larvae were maintained at ∼1 larva mL-1 until fully competent (Tebben et al., 2011), approximately 5 days after fertilization. One pre-conditioned terracotta tile was placed inside each tank to enable larval settlement (Figure 1D). Each tile was labeled, and then tiles were removed from tanks and randomly stacked with a ∼0.5 cm gap between tiles and deployed for 6 months onto the reef where adult coral colonies were collected (Figure 1E). Five replicate sediment and seawater samples from the deployment site were collected and snap frozen in liquid nitrogen for bacterial analysis (Figure 1v). Twenty larvae (days 2 and 4) and 20 spat (day 7) were collected from each tank and 10 juveniles (6 months) were collected from each tile (Figure 1vi). Samples were rinsed in FSW, and snap frozen in liquid nitrogen for 16S rRNA gene sequencing analysis. Samples of seawater in the SW and RMD_SW treatments (n = 3) were preserved in 2% formalin at 4°C and bacterial density was determined via microscopy. Cells were filtered onto 0.22 μm membranes, stained with SYBR Gold and enumerated under epifluorescence microscopy. Ten fields of view were counted for each sample (Figure 1iv).
Ethics Statement
Permission to conduct the fieldwork and collect coral samples was granted by the Western Australian Department of Parks and Wildlife (permit#SF010989) and Western Australian Department of Fisheries (permit#2895). No special animal ethics approval was needed.
Microbial Community Characterization: MiSeq Illumina Analysis of 16S rRNA Gene
Prokaryotic diversity for all samples (Figure 1) was assessed through 16S rRNA gene sequencing using an Illumina MiSeq platform (Illumina, San Diego, CA, United States) at Curtin University, Perth (Australia). Mixed genomic DNA was extracted from coral samples using the PowerPlant DNA Isolation Kit (MoBio, Solana Beach, CA, United States), following Sunagawa’s modifications for maximum yield (Sunagawa et al., 2010). Mucus and tissue were removed from the adult coral fragments with an air gun and 1X PBSE pH 7.4, while early life stages were added directly into the extraction tubes. Mixed genomic DNA was extracted from water and sediment samples with the PowerSoil DNA Isolation Kit (MoBio, Solana Beach, CA, United States), following the manufacturer’s instructions. Sediment samples where purified using AMPure XB Beads (Agencourt, Beckman Coulter, United States) following the manufacturer’s instructions.
Bacterial communities were sequenced using barcoded primers 515F (F: 5′-GTGCCAGCMGCCGCGGTAA G-3′) and 806R (R: 5′-GGACTACHVGGGTWTCTAAT-3′) for the V4 variable region of the 16S rRNA gene (Caporaso et al., 2012). Quantitative polymerase chain reaction (qPCR) was used to determine the optimal yield of DNA to be added to each PCR reaction (Murray et al., 2015). Amplicons were produced using a single round of PCR with fusion tag primers consisting of Illumina adaptor regions, MID tags unique to each sample and the 16S rRNA template specific primers. Reagents included 1× AmpliTaq Gold Buffer (Life Technologies, Carlsbad, CA, United States), 2 mM MgCl2, 0.25 μM deoxyribonucleotide triphosphate (dNTPs), 10 μg Bovine Serum Albumin (BSA), 5 pmol of each primer, 0.12 × SYBR Green (Life Technologies), 1 Unit AmpliTaq Gold DNA polymerase (Life Technologies), 2 μL of DNA and UltrapureTMDistilled Water (Life Technologies) to 25 μL. PCRs were run on Applied Biosystems StepOnePlus Real-Time PCR: initial denaturation at 95°C for 5 min, 35 cycles of 30 s at 95°C, 30 s at 50°C, and 45 s at 72°C, with a final extension for 10 min at 72°C. Prior to library pooling, duplicate PCR products from each sample were combined. Extraction controls containing no samples, and PCR negative controls, were added to sequencing runs to assess contamination, and taxa detected within them were excluded from analyses (Supplementary Data 1).
Libraries were prepared by pooling amplicons from each sample in equimolar ratios determined by quantification on a Labchip R GX Touch HT (Perkin Elmer, Waltham, MA, United States). Final libraries were size selected using a Pippin Prep (Sage Science, Beverly, MA, United States), purified using the Qiaquick PCR purification Kit (Qiagen, Venlo, Netherlands) and sequenced unidirectionally using Illumina 300 cycle MiSeq V2 Reagent Kits and standard flow cells on an Illumina MiSeq platform.
Bioinformatics Analysis of MiSeq Illumina Sequencing Outputs
Raw sequence data was extracted with Geneious version 8.1.71 (Kearse et al., 2012). Using 100% matches the adaptor, MID tags and primer sequences were removed. De-noising and quality filtering was performed using Quantitative Insights Into Microbial Ecology, Mac version 1.9.1 (MacQIIME) (Caporaso, 2011). Short sequences (<200 bp), sequences containing ambiguous base calls or homopolymer runs above 6 bp were discarded. Chimeras were removed using ChimeraSlayer (Haas et al., 2011). The remaining sequences were clustered into Operational Taxonomic Units (OTUs) based on an open-reference OTU picking method (Rideout et al., 2014) at 99% identity using UCLAST (Edgar, 2010). This method was used in order to keep sequences that are not perfectly matched to the GreenGenes reference database release 13_8 (DeSantis et al., 2006), therefore preventing the loss of novel OTUs. Alignment was done with PyNAST version 1.2.2 (Caporaso et al., 2010), and taxonomy was assigned to OTUs using the RDP classifier (Wang et al., 2007), against the GreenGenes database. Non-bacterial sequences (i.e., Archaea, Eukarya, chloroplast, and mitochondria) and absolute singletons (OTUs that occur only once in the dataset) were removed in a post-OTU picking step.
To avoid the analysis of rare sequences possibly generated by sequencing error, OTUs present in less than 0.01% of the total abundance in the table were excluded. This filter reduced the quantity of phylotypes from 18,051 to 973 OTUs. The resulting dataset contained 663,578 high quality sequences, ranging from 584 to 14,407 sequences per sample. To avoid biases generated by unequal sampling depth, sequences were rarefied to an even number (n = 831) which corresponded to the sequence depth present in the sample with the third lowest number of sequences, resulting in 962 OTUs (Supplementary Data 2) which were used in all downstream analyses.
MacQIIME and excel were used to explore the dynamics of core and resident bacteria associated with A. digitifera development stages. Bacterial phylotypes consistently reported in >80% of the samples within each development stage were considered the “core bacterial communities,” bacteria phylotypes present in 50–79% of the samples within each life stage were considered “resident,” following Ainsworth et al. (2015) and Hernandez-Agreda et al. (2016).
Alpha diversity calculations of the microbial communities, along with relative abundance summaries, were performed with MacQIIME using alpha_diversity.py, and summarize_taxa_through_plots.py as well as excel, while Venn Diagrams were built using MetaComet (Wang et al., 2016). Alpha diversity metrics included number of OTUs, Chao1 (Chao, 1984), and Shannon indices (Shannon, 1948) were each compared between early life stages (L2, L4, spat, and juvenile) using the non-parametric Kruskal–Wallis test in SPSS Version 25.0.
Data Availability
Sequence data were deposited and are publicly available in the NCBI Sequence Read Archive (SRA) under the BioProject ID PRJNA526947, accession numbers SAMN11097120 – SAMN11097248 and SAMN11603544-SAMN11603546.
Statistical Analysis
Sorenson (Bray-Curtis) distance was used on the final fourth root transformed dataset to construct a resemblance matrix and non-metric multidimensional scaling (nMDS) was used to visualize patterns in microbial community structure among treatments and development stages. PERMANOVA was used to examine the differences in microbial assemblages between treatments, and across development stages. Prior to PERMANOVA analyses data were tested to ensure equal dispersion using a resemblance-based permutation test (PERMDISP). As the PERMDISP assumptions were not met (p < 0.05), the PERMANOVA significance level was set to 0.01 as a precaution. A similarity percentage (SIMPER) was used to determine the key contributing OTUs responsible for the observed patterns of separation between groups and similarity within groups. Multivariate analyses and nMDS plots were completed using PRIMER-E 7 Ltd. (Clarke and Warwick, 2001).
Results
The data presented here are based on the final high confidence rarefied (n = 831) dataset (Supplementary Data 2). High-confidence non-rarefied data (Supplementary Figure 2A) provided comparable results (data not shown). In total, five adults and their respective bundles as well as a total of nineteen 2-day-old larvae (L2), eighteen 4-day-old larvae (L4), eighteen newly settled spat (Spat), and nineteen 6-month-old juveniles (Juvenile) were successfully sequenced at the 16S rRNA gene (Supplementary Data 2 and Supplementary Figures 2B, 3).
Source of Microbial Symbionts in Acropora digitifera Early Life Stages
Adult coral colonies hosted 125 bacterial OTUs. Of these, 15 belonged to the Endozoicimonaceae and contributed 76.61% of the mean relative abundance of sequence reads. Other taxa such as Alcanivoracaceae, Trueperaceae, and Streptococcaceae showed mean relative abundances between 7.20 and 0.024% (Supplementary Data 2). A subset of bacterial symbionts were present in both adult coral colonies and egg-sperm bundles (Figure 2A–G). While specific OTUs found in both adult colony and corresponding bundles were variable (Figure 2B–F and Supplementary Figure 4), taxa such as Cryomorphaceae, and Endozoicimonaceae were found in all adults and bundles (Figure 1G and Supplementary Figure 4), and Rhodobacteraceae were found in four out of five adults and associated bundles (Supplementary Figures 4B–E). Of the 15 OTUs belonging to the Endozoicimonaceae, OTU 4397109 was reported among all adult coral colonies (relative abundance between 28.28 and 77.62%) and 20 out of 22 bundle samples (relative abundance between 2.72 and 94.87%) (Supplementary Figure 5). Bundle samples with a lower abundance of OTU 4397109 were all released by the same coral colony (Adult_1) and were dominated by OTU 3041665 (Supplementary Figure 5). Five OTUs belonging to the family Endozoicimonaceae (OTUs 573686, 221108, 4397109, 831558, and 206565) were identified as part of the core bacterial community of adults (Figure 3A), and were also found within each egg-sperm bundle sample (Figure 2B–F and Supplementary Figure 4). Taxa such as Alteromonadaceae (OTUs 823476) and Pseudomonadaceae (OTU 646549) were found in association with four out of five adult coral colonies and their associated bundles (Supplementary Figures 4B–E).
FIGURE 2
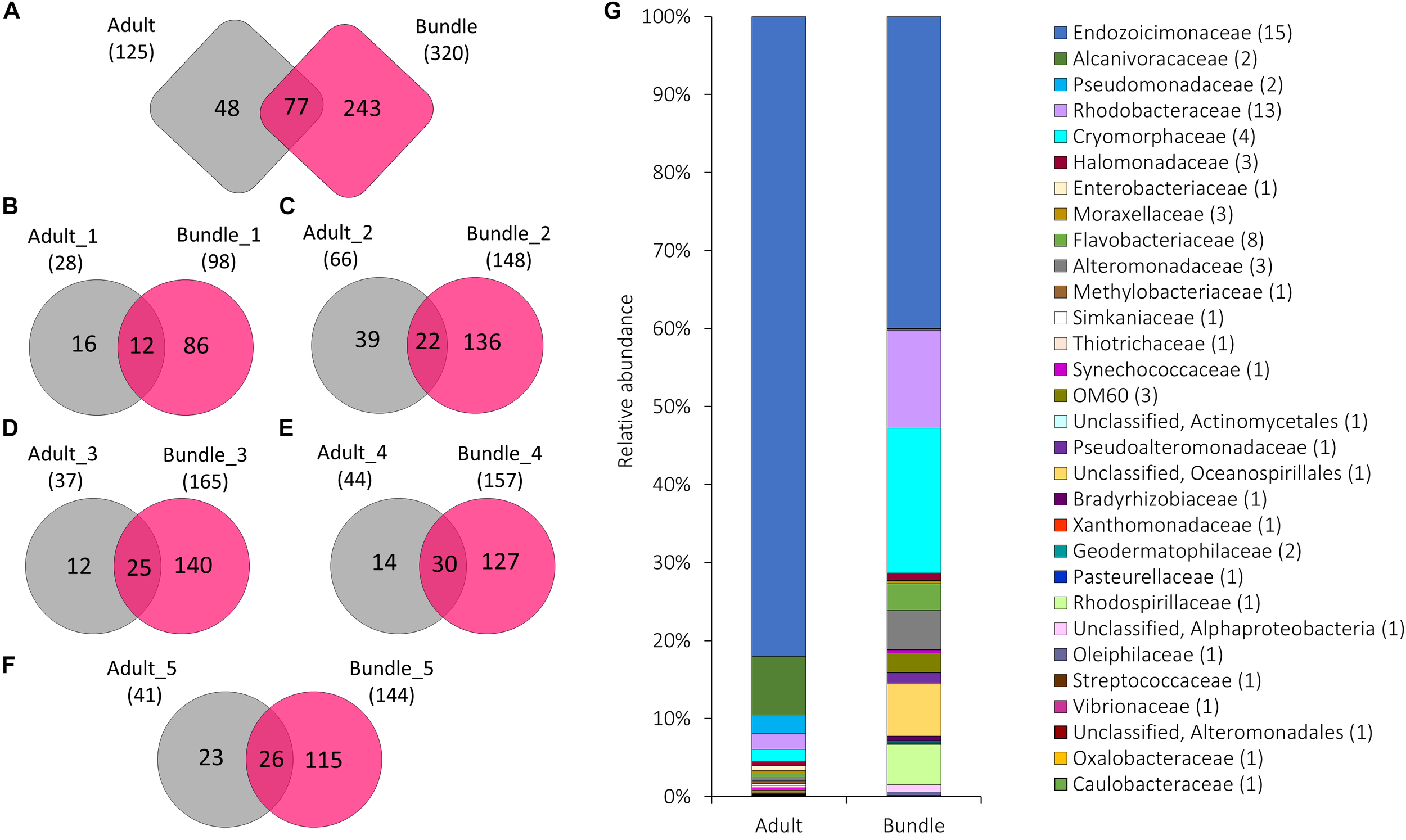
The number of OTUs unique to or shared between (A) all five maternal coral colonies (adult) and the released egg-sperm bundles (bundle), and between each individual coral colony and resulting bundles (B–F). The total number of OTUs associated with each group is represented in parentheses. Relative abundance of the 90 bacterial OTUs shared among adult colonies and egg-sperm bundles (G) (represented in A). Sequences were classified at family level where possible, with unclassified sequences indicated at higher taxonomic resolution. Numbers in parenthesis demark the number of different OTUs within the respective families/taxa.
FIGURE 3
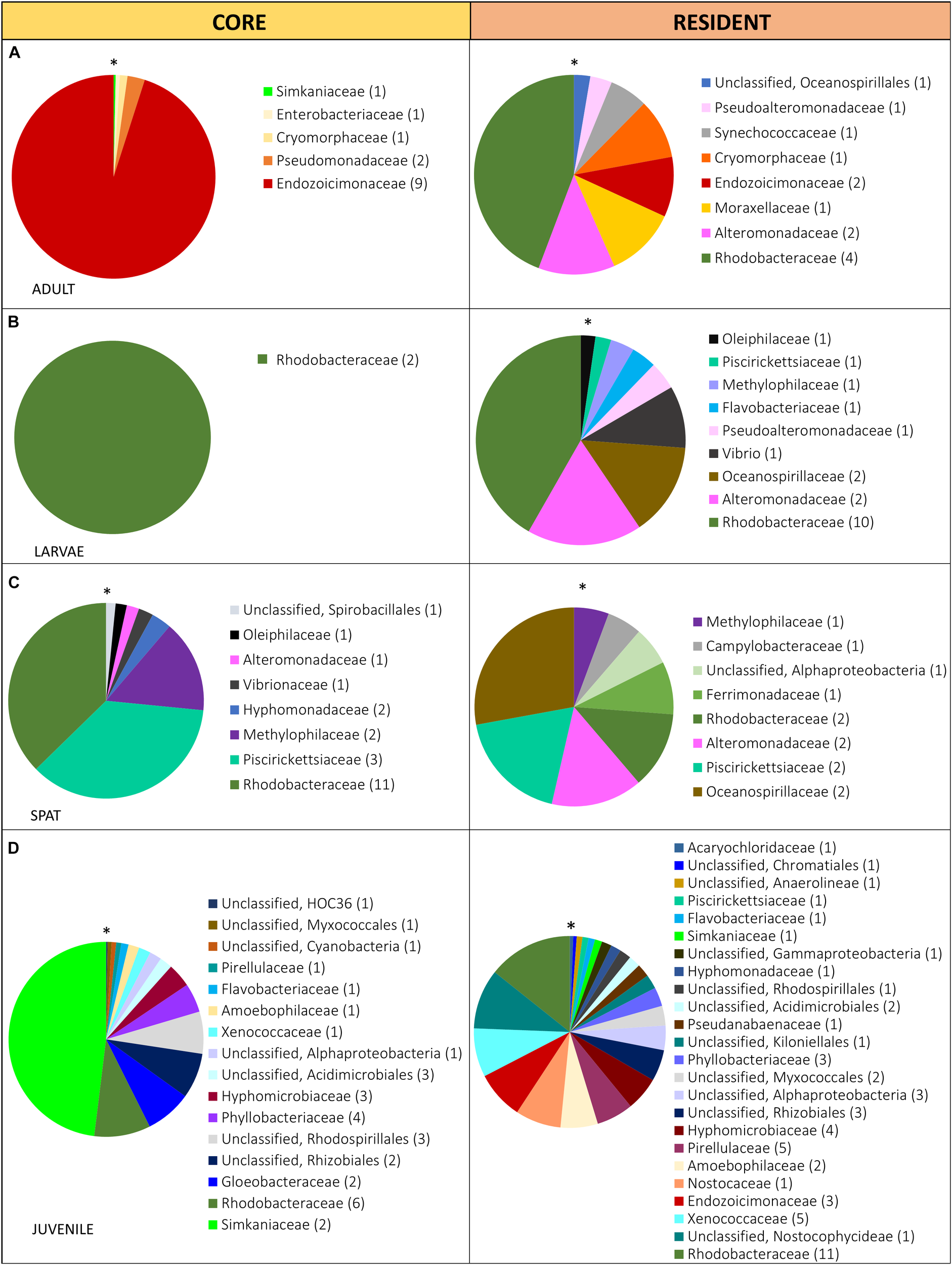
Relative abundance of the bacterial taxa within the core (present in 80% of the samples) and the resident (present in 50% of the samples) microbiome in: (A) Adult, adult colonies, (B) Larvae, 2- and 4-day-old larvae; (C) Spat, newly settled spat; and (D) Juvenile, 6-month-old juveniles. Sequences were classified at family level where possible, with unclassified species indicated at higher taxonomic resolution. Families within each pie chart are organized from the lowest (*) to the highest abundance (clockwise direction).
Filtering seawater reduced the bacterial density significantly (one-way ANOVA, p < 0.05) (Supplementary Table 1) to approximately 1 × 105 cells/L in comparison to raw seawater with average cell abundances of approximately 1 × 108/L (Supplementary Figure 6), but did not affect the overall composition of the bacterioplankton (PERMANOVA, p > 0.05; Supplementary Table 2). Consistent coral bacterial communities were found, regardless of exposure to seawater or sediment bacterial communities (PERMANOVA, p = 0.2427; Supplementary Table 3 and Figure 4, 5). Adults and 2-day-old larvae shared more OTUs with the surrounding water column than with sediment (Figure 4A,B) while 4-day-old larvae, spat and juveniles shared more OTUs with the sediment samples (Figure 4C–E). However, the investigation of the bacterial community structure showed that adult coral colonies, L2 and juveniles were more similar to the sediment samples (Supplementary Figures 6A–E,N,O), while L4 and spat had community structure that was similar to both sediment and the water column (Supplementary Figures 6F–M). The number of OTUs shared with both water and sediment increased from the larval stages (Figure 4B,C), to the spat (Figure 4D), and the juveniles (Figure 4E), but then dropped in adult coral colonies (Figure 4A).
FIGURE 4
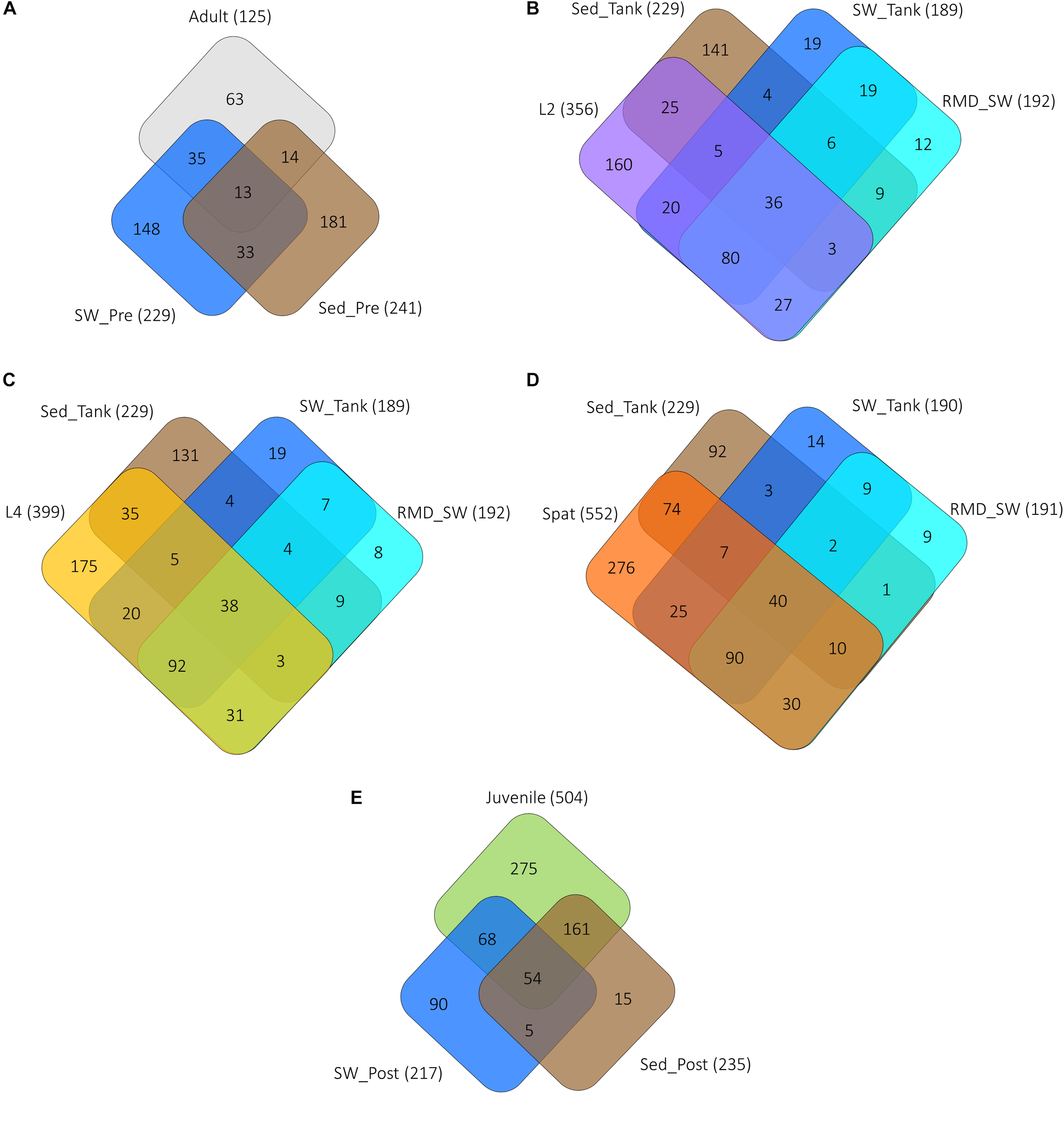
Unique and shared OTUs between the various life history stages and the surrounding environments of seawater and sediment. (A) Adult, adult coral colonies; (B) L2, 2-day-old larvae; (C) L4, 4-day-old larvae; (D) Spat, newly settled spat, and (E) Juvenile, 6-month-old juveniles. SW, seawater from the reef; SED, sediment from the reef, RMD_SW, reduced microbial density seawater (filtered 0.22 μm). The total number of OTUs associated with each sample type is represented in parentheses. Abbreviations are SW and Sed represent samples taken: “Pre” prior to spawning, “Tank” from the tank experiment, “Post” near juveniles on the reef.
FIGURE 5
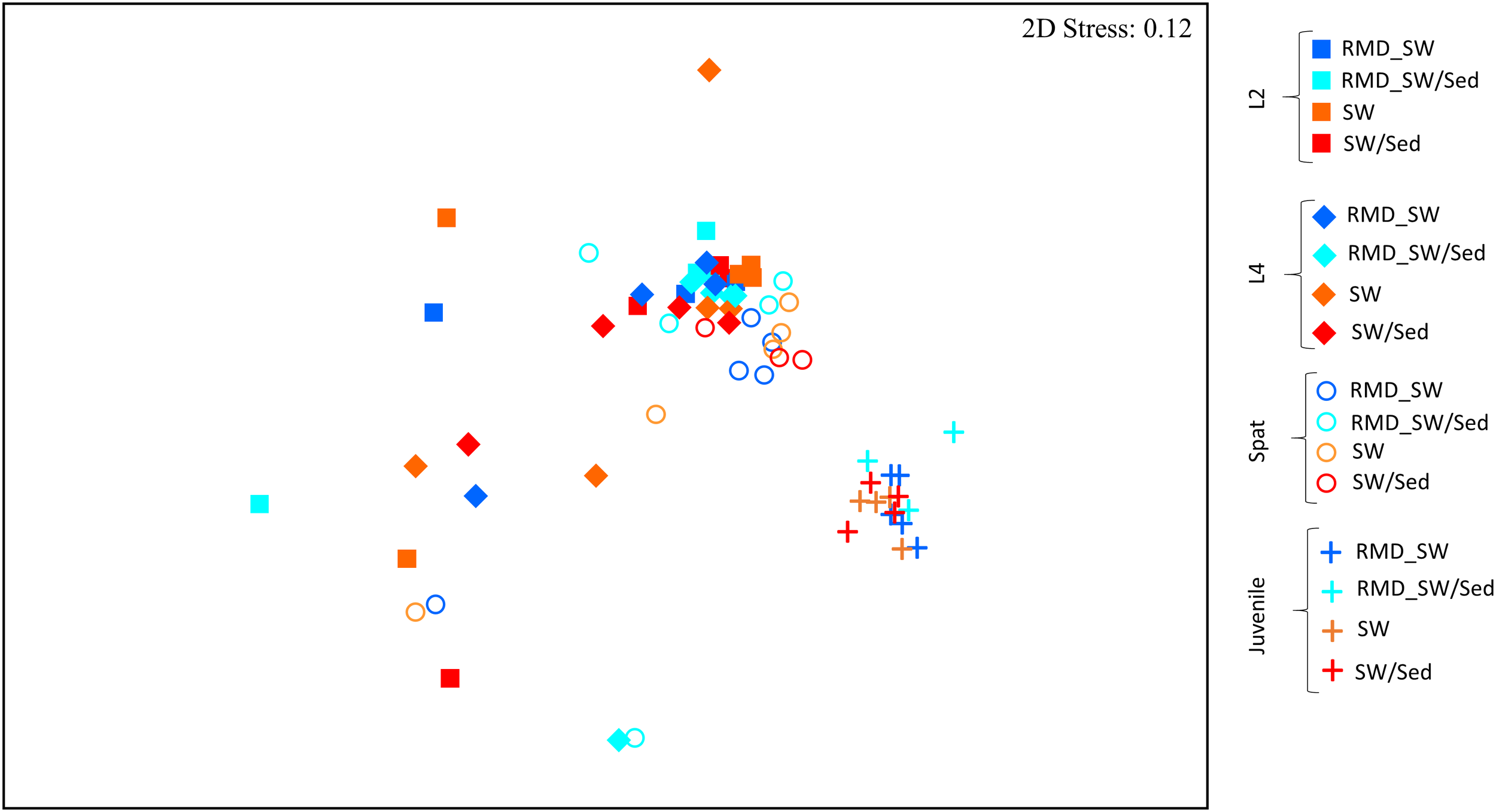
Non-metric multidimensional scaling (nMDS) of entire microbial communities associated with Acropora digitifera early life stages and reared among different environmental conditions. Development stages are indicated as: L2, 2-day-old larvae (squares); L4, 4-day-old larvae (diamonds); Spat, newly settled spat (circles); Juvenile, 6-month-old juveniles (cross). While the colors indicated the rearing environment: RMD_SW, reduced microbial cell density seawater (blue); RMD_SW/Sed, reduced microbial cell density seawater and sediment (light blue); SW, seawater (orange); SW/Sed, seawater and sediment (red).
Bacterial Diversity and Dynamics in Early Life Stages of Acropora digitifera
Each developmental stage of A. digitifera had a unique bacterial community (PERMANOVA, p < 0.01) (Figure 5 and Supplementary Table 3), with the exception of L2 and L4 (p = 0.1773) (Figure 5 and Supplementary Table 3). Bacterial communities also became increasingly complex with early development stages (Figure 6 and Supplementary Table 3).
FIGURE 6
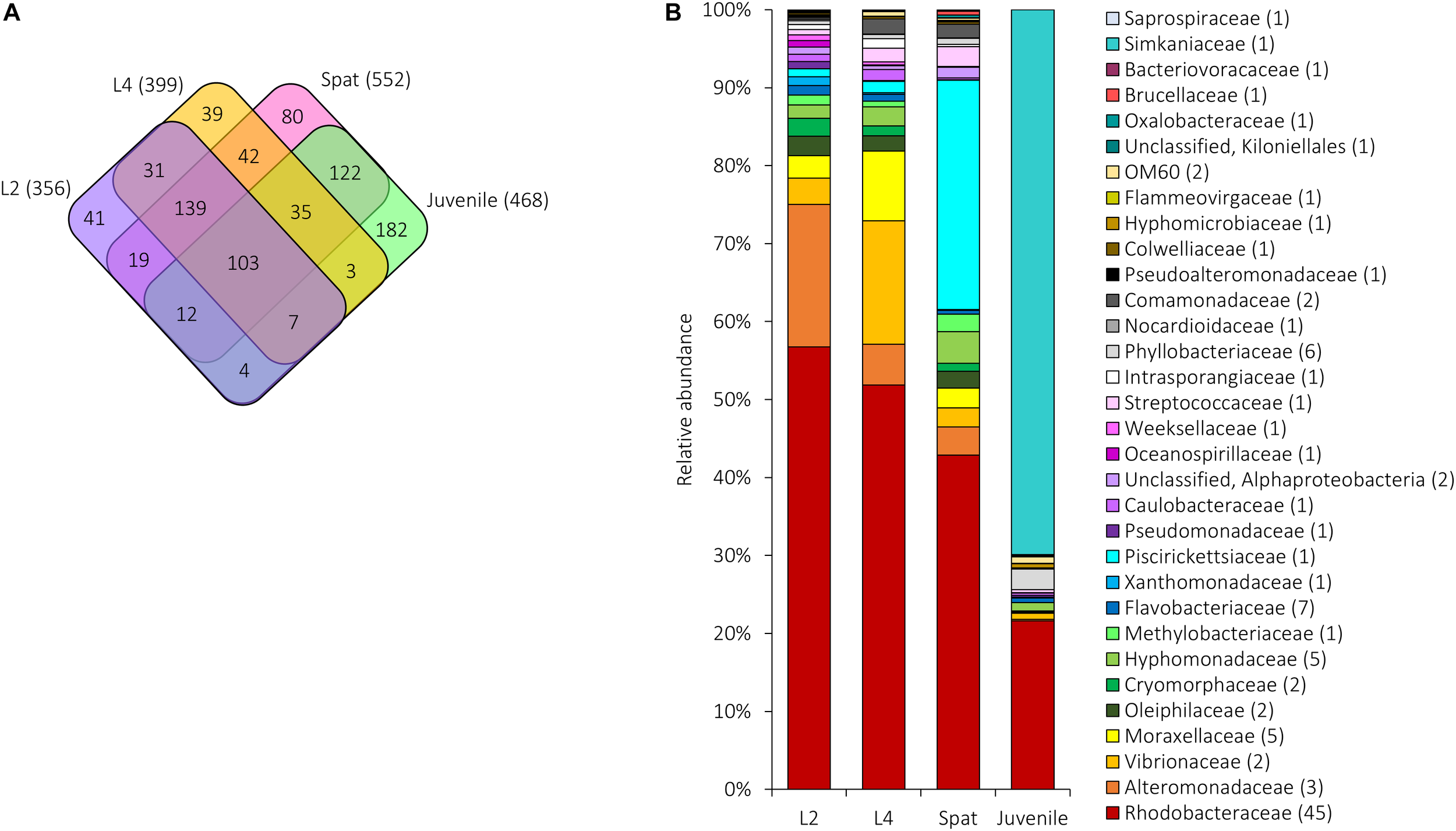
Shared and unique microbial communities associated with A. digitifera early life stages: L2, 2-day-old larvae; L4, 4-day-old larvae; Spat, newly settled spat; Juvenile, 6-month-old juveniles. (A) The number of OTUs unique to or shared between the development phases. The total number of OTUs associated with each stage is in parentheses. (B) Relative abundance of the 109 bacterial OTUs shared among early life stages. Sequences were classified at family level where possible, with unclassified sequences indicated at higher taxonomic resolution. Numbers in parenthesis demark the number of different OTUs within the respective families/taxa.
The microbial communities of 2- and 4-day-old larvae had, respectively, a mean of 76 (±3.07 SE) OTUs from 14 phyla and 75 OTUs (±3.30 SE) from 14 phyla. Both stages were dominated by Proteobacteria (L2 = 78.16%, L4 = 76.37%), Actinobacteria (L2 = 6.94%, L4 = 10.09%), Bacteroidetes (L2 = 10.55%, L4 = 8.19%), and Firmicutes (L2 = 4.14%, L4 = 3.35%). Within spat, 14 phyla were present with an average of 126 ( ± 5.46 SE) OTUs per sample, dominated by Proteobacteria (81.99%), Bacteroidetes (7.66%), Actinobacteria (4.79%), Firmicutes (3.59%), and lower abundance phyla such as Plancomycetia (0.11%). Fifteen phyla were identified within the juveniles based on an average of 173 ( ± 4.23 SE) OTUs, and juveniles were dominated by Proteobacteria (48.28%), Chlamydiae (25.26%), Cyanobacteria (14.49%), Bacteroidetes (6.35%), Planctomycetes (2.11%), and Actinobacteria (2.04%) (Supplementary Data 1). Overall, 859 OTUs were found within the early life stages, and significant differences in alpha diversity were identified between early life stages (Kruskal–Wallis H-test, p < 0.05; Supplementary Table 4A). Both larval stages and spat had a significantly lower number of OTUs and Chao1 index compared to juveniles (see Supplementary Tables 4B,C), while the Shannon index was significantly higher in juveniles compared to the larval stages (Supplementary Table 3D).
All development stages had 103 OTUs in common (Figure 6A). Of these, the relative abundance shifted across development (Figure 6B). Both larvae and spat were dominated by Proteobacteria (L2 = 94.02%, L4 = 94.32%, Spat = 95.40%) which declined to 28.98% in juveniles (Figure 6B). Within the Proteobacteria, members of the Rhodobacteraceae dominated all four stages (L2 = 56.74%, L4 = 51.88%, Spat = 42.87%, Juvenile = 21.63%). Alteromonadaceae reached maximum relative abundance in L2 (18.31%), and then declined in L4 (5.20%), spat (3.60%), and juveniles (0.19%). Xanthomonadaceae slightly increased their highest relative abundance in L2 (1.63%) and then declined in L4 (0.29%) and spat (0.27%), before increasing in juvenile (0.54%). Vibrionaceae (15.86%; OTUs 939811 and 837366) reached their maximum relative abundances within L4. Once larvae settled, the bacterial genus Methylophaga (family Piscirickettsiaceae) (OTU 823160) became dominant (29.40%), and then declined (Juveniles = 0.03%). After 6 months exposure to natural environmental conditions, the relative abundance of the genus Simkaniaceae (OTU 217851), increased to 69.85%. Other bacterial taxa such as Bacteroidetes, and Actinobacteria, and Firmicutes were present in lower abundance (0.03–4.24%) across all development stages (Figure 6B).
Bacterial community structure within both larval stages was highly dissimilar to spat [average dissimilarity of 79.76% (L2 vs. Spat) and 78.76% (L4 vs. Spat)]. Taxa such as Piscirickettsiaceae genus Methylophaga (OTUs 823160 and 278985) and Methylophilaceae genus Methylotenera (OTU 1091363) were predominantly associated with spat rather than larvae. Microbial communities within both larval stages were also dissimilar to juveniles [average dissimilarity = 92.68% (L2 vs. Juvenile) and 92.20% (L4 vs. Juveniles)], predominantly driven by the increased relative abundance of the Chlamydiiae-like OTU 217851 (family Simkaniaceae). Differences between microbial communities of newly settled spat and juveniles (average dissimilarity of 86.98%) were driven by the dominance of the family Simkaniaceae in juveniles, and the genus Methylophaga in spat (SIMPER analysis; see Supplementary Table 5).
In total, 898 OTUs occurred across coral generations and 22 occurred consistently within all stages (Figure 7A). Rhodobacteraceae were found within each generation and included genera such as Loktanella and Thalassobius (Figure 6B). Among the OTUs that were shared across all generations (Figure 7B) the Pseudomonadaceae species Pseudomonas veronii (OTU 646549) showed a decline in relative abundance from adults to early life history stages. Larvae were characterized by an increased abundance of Alteromonadaceae belonging to the genus Alteromonas and to the family Xanthomonadaceae (L2; Figure 7B).
FIGURE 7
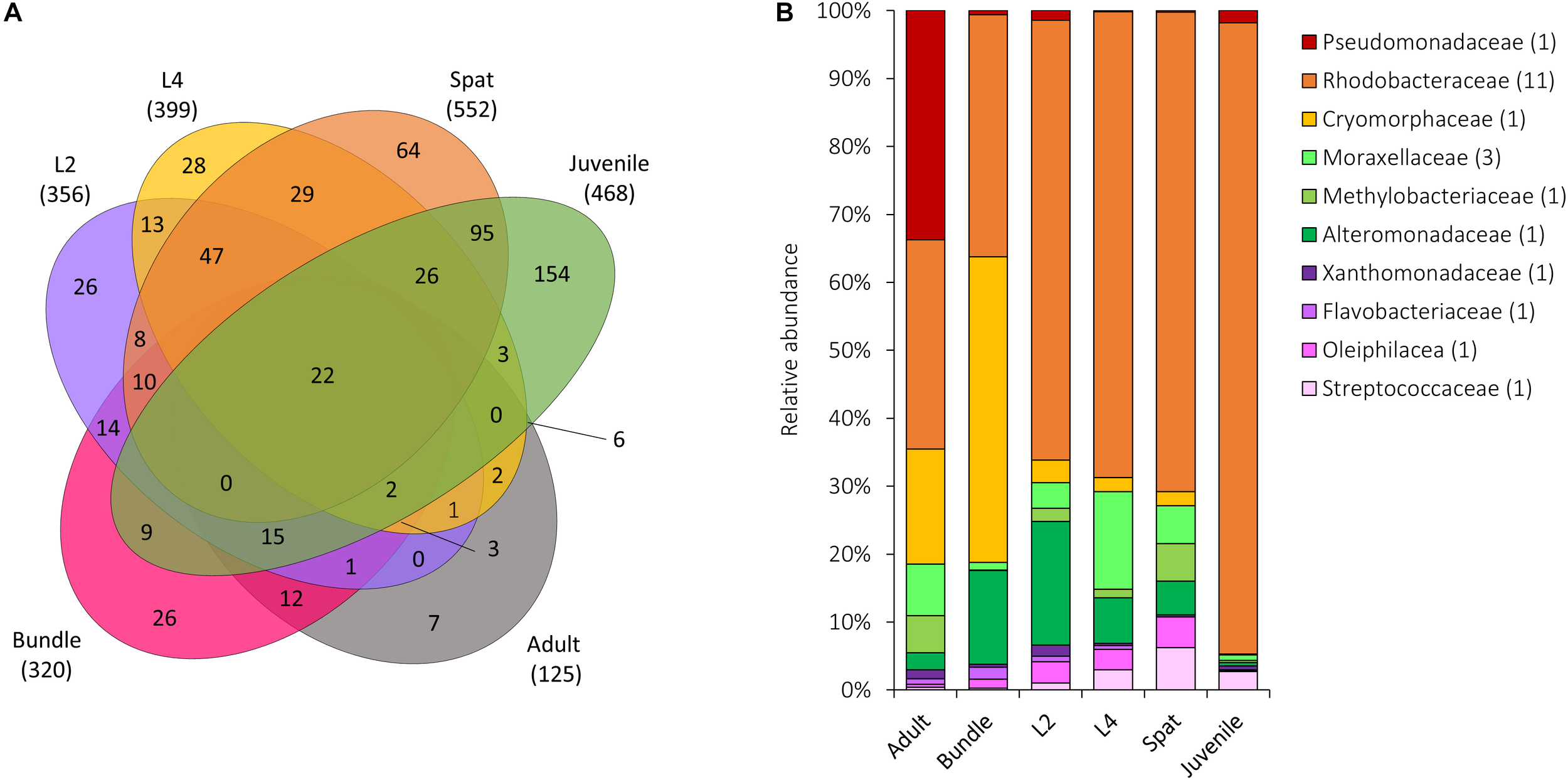
Microbial communities shared across all life history stages of Acropora digitifera. (A) The number of OTUs unique to or shared between stages, with the total number of OTUs associated with each group represented in parentheses. (B) Relative abundance of the 28 bacterial OTUs shared among all generations. Sequences were classified at a family level where possible, with unclassified species indicated at higher taxonomic resolution. Numbers in parenthesis demark the number of different OTUs within the respective families. Adult, adult colonies; Bundle, egg-sperm bundles; L2, 2-day-old larvae; L4, 4-day-old larvae; Spat, newly settled spat; Juvenile, 6-month-old juveniles.
Core and Resident Bacterial Communities Across Generations
The diversity of the core and the resident bacterial communities varied among development stages (Figure 3). In general, Rhodobacteraceae (i.e., genera Rhodovulum and Loktanella) were identified among both the core and resident bacterial communities. The dominant core members were characterized by: Endozoicimonaceae (adults, Figure 3A), Rhodobacteraceae (Larvae: L2 and L4; Figure 3B) and Piscirickettsiaceae genus Methylophaga (spat, Figure 3C), and Simkaniaceae (Juveniles, Figure 3D). The adult core bacteria Endozoicimonaceae (OTU 555869 and OTU 831558) were identified also in the juvenile resident microbiome (Figure 2A,D). During the adult, larval and spat stages the resident microbiome was characterized by the presence of Alteromonadaceae (Figure 2A–C), and adult and larval resident microbiomes also contained Pseudoalteromonadaceae (Figure 2A,B).
Discussion
Using 16S rDNA amplicon data, we provide evidence of both vertical and horizontal transmission modes in Acropora digitifera and thus suggest that the mechanisms by which bacteria are acquired in corals are more variable (Leite et al., 2017) than previously expected. Some members of the bacterial community within A. digitifera displayed a ubiquitous presence in all developmental stages including gametes, yet the core and resident bacterial communities were variable across the host generations. Altogether, this is likely reflective of stage-specific metabolic requirements of the coral host and associated symbionts. Larvae and juveniles harbored more diverse and flexible bacterial communities than adults, supporting the presence of an initial phase represented by a highly diverse and variable bacterial community, followed by a phase of selection (winnowing) and decreased diversity toward the establishment of the final adult consortium (Epstein et al., 2019; Littman et al., 2009a,b). Winnowing processes are known during the establishment host-Symbiodiniaceae partnerships in early life stages of broadcast spawning corals (Abrego et al., 2009; Cumbo et al., 2013), as well as in other holobiont models such as Hydra (Franzenburg et al., 2013) and the bobtail squid (Nyholm and McFall-Ngai, 2004). This mechanism of “selective colonization” is likely to be controlled by the host and/or other symbionts (i.e., Symbiodiniaceae or bacteria) for maximum benefit to the physiological requirements specific to each developmental stage and environmental conditions (Little et al., 2004; Nyholm and McFall-Ngai, 2004; Nissimov et al., 2009).
This study demonstrates that A. digitifera has the ability to exploit various strategies for the acquisition of bacterial symbionts, indicating flexibility in the mechanisms employed during establishment of symbiotic relationships. The majority of broadcast spawning corals acquire their bacterial symbionts from the surrounding environment as larvae or during the post-settlement and metamorphosis as a juvenile (Weis et al., 2001; Knowlton and Rohwer, 2003; Apprill et al., 2009; Sharp et al., 2010). However, there is some evidence of vertical transmission of Symbiodiniaceae and bacteria from adult broadcast corals to eggs released during spawning (Padilla-Gamiño et al., 2011; Leite et al., 2017). Mixed-mode strategies are common in nature (Russell, 2019) and have been documented in various organisms such as the brooding coral Seriatopora hystrix (Quigley et al., 2016) and Pocillopora damicronis (Epstein et al., 2019), marine sponges (Schmitt et al., 2008; Webster et al., 2010), solemyid bivalves (Russell et al., 2017), and humans (as reviewed in Ebert, 2013). Mixed strategies combine the best of the horizontal and vertical transmission modes and play an important role in the evolution and ecology of host-symbiont relationships (Bayer et al., 2013; Ebert, 2013; Russell et al., 2017). For instance, through vertical transmission the offspring benefit by not having to “search” for symbionts and transmission of beneficial partners (Byler et al., 2013). However, in environmental conditions different from those of the parents, this may confer a disadvantage if the transmitted symbionts are not optimal. In horizontal acquisition, the host can take up genetically diverse symbionts well suited to a specific environment but they may lack beneficial symbionts inherited by the parental colony (Byler et al., 2013). Therefore, corals showing both strategies are likely to be physiologically and evolutionarily advantaged compared to those strictly relying on one or the other.
We report the presence of bacterial taxa that were found in association with specific life stages, and ubiquitous taxa with either stable or fluctuating relative abundances, suggesting that proliferation of some microbial partners was promoted while others were suppressed (Little et al., 2004). These processes could represent a mechanism of “competitive exclusion” were microbes compete for the resources available (Hibbing et al., 2010). However, we hypothesize that changes in the relative abundance of bacterial phenotypes reflect variations in metabolic requirements at specific life stages. Clostridiaceae were reported within 4-day-old larvae and newly settled spat. Members of the Clostridiaceae, as well as Rhodobacteraceae and the genus Vibrio, contribute to carbon and/or nitrogen availability through the degradation of chitin produced by symbiotic zooxanthellae (Wainwright, 1963; Neulinger et al., 2008; Bourne et al., 2016). Larvae were dominated by the diazotrophic bacteria Alteromonas and Vibrio, which are expected to provide an additional source of fixed nitrogen to the host (Lema et al., 2014) and associated Symbiodiniaceae (Olson et al., 2009). Potential beneficial roles of Alteromonas include protection against pathogens (Gil-Turnes et al., 1989), and induction of larval settlement (Webster et al., 2004; Ceh et al., 2013). The protective and nutritional properties of these taxa could be fundamental during the first vulnerable life stages, where larvae face a gradual decline in metabolism rate expressed by oxygen consumption (Nakamura et al., 2011), but are still capable of swimming up to 200 days without feeding (Graham et al., 2008).
In the spat and juvenile phases we found the presence and increased relative abundance of bacterial taxa involved in the sulfur cycle (Raina et al., 2010). Specifically, the relative abundance of two common coral-associated dimethylsulfoniopropinate (DMSP) degrading bacteria (Raina et al., 2010; Leite et al., 2018), Desulfovibrionaceae and Desulfobacteraceae, increased in spat and juveniles. DMSP is likely to be synthesized by both the host and Symbiodiniaceae (Van Alstyne et al., 2006; Raina et al., 2013), and degradation of DMSP produces DMS, acrylate and dimethylsulfoxde (DMSO) which, due to their antioxidant properties, are involved in stress response (Raina et al., 2013, 2016; Deschaseaux et al., 2014) and antimicrobial activities (Slezak et al., 1994; Broadbent and Jones, 2004). Spat were also characterized by the presence of OTUs belonging to dimethylsulfide (DMS) degrading Methylophaga, while within juveniles this role may be played by other taxa such as Vibrio, Pseudomonas, and Alteromonas (Raina et al., 2010). Spat were also characterized by an increased relative abundance of a common coral-associated bacteria (Silveira et al., 2017), Janthinobacterium, known to produce antifungal metabolites (Brucker et al., 2008). The early establishment of these symbioses is likely to contribute beneficially to biological processes such as growth as well as protection at highly vulnerable life stages.
Spat and juveniles hosted an increased relative abundance of Rhizobiales, and juveniles hosted a higher relative abundance of Simkaniaceae. Members of the genus Simkaniaceae are among the most common bacterial taxa associated with both corals (Bayer et al., 2013; Ogawa, 2014; Wright et al., 2016) and octocorals (van de Water et al., 2018), however, their functional role is unknown. Rhizobiales are nitrogen fixing bacteria frequently reported in early life stages of corals (Sharp et al., 2012; Lema et al., 2014) and may be specifically associated with Symbiodiniaceae (Olson et al., 2009; Ainsworth et al., 2015). This could signify that they provide nutrients to Symbiodiniaceae which are in turn responsible for the production of chitin involved in the formation of the skeleton (Watanabe et al., 2003) which starts developing within the juvenile stage (Wainwright, 1963). The presence of diazotroph bacteria across early life stages highlights the possible functional importance of nitrogen fixing bacteria for the coral holobiont (Lema et al., 2014), due to their ability to provide additional source of fixed nitrogen in oligotrophic waters (Lesser et al., 2004, 2007; Olson et al., 2009; Lema et al., 2012, 2014). Early life stages were all characterized by the ubiquitous presence of members of the family Xanthomonadaceae which are known to promote plant growth and suppress pathogenic symbionts (Li et al., 2015) and establish symbiotic relationships with a wide range of organisms such as beetle larvae (Geib et al., 2009; Rizzi et al., 2013) as well as coral (Polson, 2007; Ceh et al., 2012; Godwin et al., 2012; Beatty et al., 2018).
Taxa such as Cryomorphaceae and the substantially dominant Endozoicimonaceae were ubiquitously found in bundles, while members of the Rhodobacteraceae, Alteromonadaceae, and Pseudomonadaceae were reported in four out of five adult/bundle individuals and across generations. Within the Endozoicimonaceae OTUs identified in adult and associated bundle OTU 4397109 matched in NCBI (100% similarity) with Endozoicimonas eunicola associated with the octocorals Eunicea fusca and Plexaura sp. (Accession number: NR_109684). Therefore, suggesting that vertical transmission ensures the generational transfer of beneficial microbial taxa likely to possess specific mechanisms to promote the coral host growth, development, nutrition and protection.
Here, both seawater and sediment surrounding the coral host were identified as potential reservoirs of microbes acquired throughout coral development. Methylophaga (family Piscirickettsiaceae; OTU 823160) and Pseudomonas (OTU 646549) were likely acquired during larval stages from sediment and seawater, respectively. Endozoicimonaceae, found in both adults and juveniles, were most likely taken up from the seawater at both larval and post-settlement stages. However, some of the bundles were associated with the same OTUs (i.e., Pseudomonas OTU 646549), therefore suggesting that they are also likely to be transmitted by the parental coral colony. The mixed transmission mode documented in this study may represent a functional compromise between horizontal and vertical strategies. Indeed, as suggested by Russell et al. (2017) horizontal transmission improves deleterious mutations and genome stasis through the facilitation of recombination and horizontal gene transfer. Vertical transmission supports the evolution of co-adapted host-symbiont genotypes, but also guarantees the colonization of each generation with symbionts. Thus, by combining both strategies, populations might be able to respond more rapidly to short and long term evolutionary pressures (Kaltz and Koella, 2003; Byler et al., 2013).
The core bacterial community has been proposed to be intimately linked with host requirements, while the variable community is thought to be influenced by surrounding environmental conditions (Hernandez-Agreda et al., 2016). We describe distinct core (>80% of the samples of each life stage) and resident bacterial communities (50–79% of each life stage samples) in the various life history stages, and propose that changes in communities reflect the ability of the host to adapt their microbiome to the distinct metabolic and/or environmental requirements of each stage. We also verified that the resident microbiome was characterized by a higher diversity than the core bacterial communities. The core bacterial communities of adults was dominated by the Endozoicimonaceae, as has previously been reported (Ainsworth et al., 2015; Hernandez-Agreda et al., 2018). Recently members of the Endozoicimonaceae have been described as being able to recognize, translocate, communicate with and modulate the coral host (Ding et al., 2016), as well as contribute to protein provision and cycling of carbohydrates (Neave et al., 2017). Interestingly, Endozoicimonaceae did not appear within the core or resident microbiome of larvae or spat, but subsequently became part of the juvenile resident microbiome. Similarly, members of the Simkaniaceae were dominant taxa within the juvenile core microbiome and part of the adult core microbiome. The simultaneous presence of Endozoicimonaceae and Simkaniaceae (Bayer et al., 2013; Pogoreutz, 2016; Goldsmith et al., 2018) could indicate that they play a critical functional role and/or that there is an interaction between the two. The core microbiome of spat and juveniles was dominated by Rhodobacteraceae, Piscirickettsiaceae, and Simkaniaceae, respectively. The presence of other taxa (i.e., Inquilinus), was previously reported as part of the core microbiome in early life stages (Leite et al., 2017) but this is the first report of core/resident bacterial communities changing through development. Temporal dynamicity of the core bacterial communities may enable the identification of consistent members across complex bacterial assemblages which are likely to facilitate the development, functioning and health of the host during each development stage.
While each stage hosted a distinct bacterial community, a notable component of both the core and resident bacterial communities of the various stages was the presence of the Rhodobacteraceae which has previously been identified (Williams et al., 2015) as ubiquitously distributed across coral life stages. The consistent presence of Rhodobacteraceae and Pseudomonas had been identified previously within the core bacterial communities of coral mucus within a broadcast spawning species (Leite et al., 2018), and were vertically transmitted (Leite et al., 2017). These studies and ours provide mounting evidence that components of the core bacterial communities can potentially be transmitted vertically across generations. The importance of Rhodobacteraceae might be due to their ability to obtain energy via sulfur oxidation, their ability to scavenge small amounts of organic material as a source of carbon (Neulinger et al., 2008), and their tolerance of anaerobic conditions such as those found in the mucus of tropical corals (Kellogg, 2004), and in polyps after feeding (Neulinger et al., 2008). We suggest that the core and resident coral bacterial communities are likely determined by the host, and possibly by the interaction with symbionts. However, the specific mechanisms that facilitate this selective process are still relatively unknown and require further investigation. It is noteworthy that some bacterial taxa found within these putatively key communities (i.e., Alteromonadaceae, Vibrionaceae, Simkaniaceae, Desulfobacteraceae, and Desulfovibrionaceae) include species commonly found within corals (Bernasconi et al., 2018) that can be beneficial as well as responsible for the outbreak of coral diseases (Bourne et al., 2009). As our specimens were healthy, these members may be considered part of the natural coral microbiome and opportunistic pathogens.
Conclusion
Our study shows that the mixed mode acquisition and subsequent winnowing of symbionts are likely to represent fundamental mechanisms in the ecology and evolution of corals. Particularly, in changing environmental conditions, mixed mode acquisition may represent a mechanism for the formation of new beneficial symbioses (Byler et al., 2013) and of new interactions with other symbionts (i.e., Symbiodiniaceae) within the coral host. Future investigations must focus on mechanisms behind the specific uptake of the microbial communities, including the roles played by other symbionts. As the condition of reefs decline worldwide (Paredes et al., 2003; Birkeland, 2019) coral taxa need to adapt to rapidly changing environmental conditions or risk extinction. We advocate that a better understanding of how microbes are acquired and utilized by corals is necessary to develop a more complete picture of coral resilience and adaptation.
Statements
Data availability statement
Sequence data were deposited and are publicly available in the NCBI Sequence Read Archive (SRA) under the BioProject ID PRJNA526947, accession numbers SAMN11097120 – SAMN11097248 and SAMN11603544-SAMN11603546.
Ethics statement
Permission to conduct the fieldwork and collect coral samples was granted by the Western Australian Department of Parks and Wildlife (permit#SF010989) and Western Australian Department of Fisheries (permit#2895). No special animal ethics approval was needed.
Author contributions
RB, MS, AK, and MH designed the experiment and interpreted the results. RB and MH collected the samples, ran the experiment, and wrote the manuscript. RB analyzed the data. MS and MB assisted RB with laboratory work and data analyses and AP with the bioinformatics analyses. MS, AK, AP, and MB edited the manuscript.
Funding
This work was made possible by financial support from the Holsworth Wildlife Research Endowment – Equity Trustees Charitable Foundation & the Ecological Society of Australia, the Woman Divers Hall of Fame, and the School of Science (Edith Cowan University).
Acknowledgments
We would like to thank Christopher Doropoulos and Russ Babcock from the CSIRO, Luis Alonso Gomez Lemos from the Griffith University, and Frazer McGregor from the Coral Bay Research Station for their assistance in the field. We also thank members of the TrEnD Lab, particularly Matthew Power for laboratory assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01529/full#supplementary-material
Footnotes
References
1
Abrego D. Van Oppen M. J. H. Willis B. L. (2009). Onset of algal endosymbiont specificity varies among closely related species of Acropora corals during early ontogeny.Mol. Ecol.183532–3543. 10.1111/j.1365-294X.2009.04276.x
2
Aeby E. R. Richards Z. T. Delbeek J. T. Reboton C. Bass D. (2014). Acropora digitifera.IUCN Red List Threat. Species2014:e.T133250A54223617.
3
Ainsworth T. D. Krause L. Bridge T. Torda G. Raina J. B. Zakrzewski M. et al (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts.ISME J.92261–2274. 10.1038/ismej.2015.39
4
Apprill A. Marlow H. Q. Martindale M. Q. Rappé M. S. (2009). The onset of microbial associations in the coral Pocillopora meandrina.ISME J.3685–699. 10.1038/ismej.2009.3
5
Apprill A. Marlow H. Q. Martindale M. Q. Rappé M. S. (2012). Specificity of associations between bacteria and the coral Pocillopora meandrina during early development.Appl. Environ. Microbiol.787467–7475. 10.1128/AEM.01232-12
6
Baird A. H. Guest J. R. Willis B. L. (2009). Systematic and biogeographical patterns in the reproductive biology of Scleractinian corals.Annu. Rev. Ecol. Evol. Syst.40551–571. 10.1146/annurev.ecolsys.110308.120220
7
Bayer T. Neave M. J. Alsheikh-Hussain A. Aranda M. Yum L. K. Mincer T. et al (2013). The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria.Appl. Environ. Microbiol.794759–4762. 10.1128/AEM.00695-13
8
Beatty D. S. Clements C. S. Stewart F. J. Hay M. E. (2018). Intergenerational effects of macroalgae on a reef coral: major declines in larval survival but subtle changes in microbiomes.Mar. Ecol. Prog. Ser.58997–114. 10.3354/meps12465
9
Bernasconi R. Stat M. Koenders A. Huggett M. J. (2018). Global Networks of Symbiodinium-bacteria within the coral holobiont.Microb. Ecol.77794–807. 10.1007/s00248-018-1255-4
10
Birkeland C. (ed.). (2019). “Global status of coral reefs,” inCombination, Disturbances and Stressors Become Ratchets, 2nd Edn, (Amsterdam: Elsevier Ltd.). 10.1016/B978-0-12-805052-1.00002-4
11
Bourne D. G. Dennis P. G. Uthicke S. Soo R. M. Tyson G. W. Webster N. (2013). Coral reef invertebrate microbiomes correlate with the presence of photosymbionts.ISME J.71452–1458. 10.1038/ismej.2012.172
12
Bourne D. G. Garren M. Work T. M. Rosenberg E. Smith G. W. Harvell C. D. (2009). Microbial disease and the coral holobiont.Trends Microbiol.17554–562. 10.1016/j.tim.2009.09.004
13
Bourne D. G. Morrow K. M. Webster N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems.Annu. Rev. Microbiol.70317–340. 10.1146/annurev-micro-102215-095440
14
Broadbent A. D. Jones G. B. (2004). DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef.Mar. Freshw. Res.55849–855. 10.1071/MF04114
15
Brucker R. M. Harris R. N. Schwantes C. R. Gallaher T. N. Flaherty D. C. Lam B. A. et al (2008). Amphibian chemical defense: antifungal metabolites of the microsymbiont Janthinobacterium lividum on the salamander Plethodon cinereus.J. Chem. Ecol.341422–1429. 10.1007/s10886-008-9555-7
16
Byler K. A. Carmi-Veal M. Fine M. Goulet T. L. (2013). Multiple symbiont acquisition strategies as an adaptive mechanism in the coral Stylophora pistillata.PLoS One8:e59596. 10.1371/journal.pone.0059596
17
Caporaso (2011). QIIME allows analysis of high-throughput community sequencing data.Nature91–15. 10.1038/nmeth.f.303.QIIME
18
Caporaso J. G. Bittinger K. Bushman F. D. Desantis T. Z. Andersen G. L. Knight R. (2010). PyNAST: a flexible tool for aligning sequences to a template alignment.Bioinformatics26266–267. 10.1093/bioinformatics/btp636
19
Caporaso J. G. Lauber C. L. Walters W. A. Berg-Lyons D. Huntley J. Fierer N. et al (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms.ISME J.61621–1624. 10.1038/ismej.2012.8
20
Carlos C. Torres T. T. Ottoboni L. M. M. (2013). Bacterial communities and species-specific associations with the mucus of Brazilian coral species.Sci. Rep.3:1624. 10.1038/srep01624
21
Ceh J. Raina J. B. Soo R. M. van Keulen M. Bourne D. G. (2012). Coral-bacterial communities before and after a coral mass spawning event on Ningaloo Reef.PLoS One7:e36920. 10.1371/journal.pone.0036920
22
Ceh J. van Keulen M. Bourne D. G. (2013). Intergenerational transfer of specific bacteria in corals and possible implications for offspring fitness.Microb. Ecol.65227–231. 10.1007/s00248-012-0105-z
23
Chao A. (1984). Nonparametric estimation of the number of classes in a population author.Scanad. J. Stat.11265–270. 10.1214/aoms/1177729949
24
Clarke K. R. Warwick R. M. (2001). Change in Marine Communities: an Approach to Statistical Analysis and Interpretation, 2nd Edn. Plymouth: PRIMER-E, 176.
25
Cumbo V. R. Baird A. H. van Oppen M. J. H. (2013). The promiscuous larvae: flexibility in the establishment of symbiosis in corals.Coral Reefs32111–120. 10.1007/s00338-012-0951-7
26
DeSantis T. Z. Hugenholtz P. Larsen N. Rojas M. Brodie E. L. Keller K. et al (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB.Appl. Environ. Microbiol.725069–5072. 10.1128/AEM.03006-05
27
Deschaseaux E. S. M. Jones G. B. Deseo M. A. Shepherd K. M. Kiene R. P. Swan H. B. et al (2014). Effects of environmental factors on dimethylated sulfur compounds and their potential role in the antioxidant system of the coral holobiont.Limnol. Oceanogr.59758–768. 10.4319/lo.2014.59.3.0758
28
Ding J. Y. Shiu J. H. Chen W. M. Chiang Y. R. Tang S. L. (2016). Genomic insight into the host-endosymbiont relationship of Endozoicomonas montiporae CL-33T with its coral host.Front. Microbiol.7:251. 10.3389/fmicb.2016.00251
29
Ebert D. (2013). The epidemiology and evolution of symbionts with mixed-mode transmission.Annu. Rev. Ecol. Evol. Syst.44623–643. 10.1146/annurev-ecolsys-032513-100555
30
Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST.Bioinformatics262460–2461. 10.1093/bioinformatics/btq461
31
Epstein H. E. Torda G. Munday P. L. van Oppen M. J. H. (2019). Parental and early life stage environments drive establishment of bacterial and dinoflagellate communities in a common coral.ISME J.131635–1638. 10.1038/s41396-019-0358-3
32
Fournier A. (2013). The story of symbiosis with zooxanthellae, or how they enable their host to thrive in a nutrient poor environment.Biosci. Master Rev.1–8. 10.1073/pnas.1121198109
33
Franzenburg S. Fraune S. Altrock P. M. Künzel S. Baines J. F. Traulsen A. et al (2013). Bacterial colonization of Hydra hatchlings follows a robust temporal pattern.ISME J.7781–790. 10.1038/ismej.2012.156
34
Geib S. M. del Mar Jimenez-Gasco M. Carlson J. E. Tien M. Jabbour R. Hoover K. (2009). Microbial community profiling to investigate transmission of bacteria between life stages of the wood-boring beetle, Anoplophora glabripennis.Microb. Ecol.58199–211. 10.1007/s00248-009-9501-4
35
Gil-Turnes M. S. Hay M. E. Fenical W. (1989). Symbiotic marine bacteria chemically defend crustacean embryos from a pathogenic fungus.Science246116–118. 10.1126/science.2781297
36
Godwin S. Bent E. Borneman J. Pereg L. (2012). The role of coral-associated bacterial communities in Australian subtropical white syndrome of Turbinaria mesenterina.PLoS One7:e44243. 10.1371/journal.pone.0044243
37
Goldsmith D. B. Kellogg C. A. Morrison C. L. Gray M. A. Stone R. P. Waller R. G. et al (2018). Comparison of microbiomes of cold-water corals Primnoa pacifica and Primnoa resedaeformis, with possible link between microbiome composition and host genotype.Sci. Rep.8:12383. 10.1038/s41598-018-30901-z
38
Graham E. M. Baird A. H. Connolly S. R. (2008). Survival dynamics of scleractinian coral larvae and implications for dispersal.Coral Reefs27529–539. 10.1007/s00338-008-0361-z
39
Haas B. J. Gevers D. Earl A. M. Feldgarden M. Ward D. V. Giannoukos G. et al (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons.Genome Res.21494–504. 10.1101/gr.112730.110.Freely
40
Hartmann A. C. Baird A. H. Knowlton N. Huang D. (2017). The paradox of environmental symbiont acquisition in obligate mutualisms.Curr. Biol.273711–3716.e3. 10.1016/j.cub.2017.10.036
41
Haygood M. G. Davidson S. K. (1997). Small-subunit rRNA genes and in situ hybridization with oligonucleotides specific for the bacterial symbionts in the larvae of the bryozoan.Microbiology634612–4616.
42
Hernandez-Agreda A. Leggat W. Bongaerts P. Ainsworth T. D. (2016). The microbial signature provides insight into the mechanistic basis of coral success across reef habitats.mBio7:e00560-16. 10.1128/mBio.00560-16
43
Hernandez-Agreda A. Leggat W. Bongaerts P. Herrera C. Ainsworth T. D. (2018). Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere.mBio9:e00812-18. 10.1128/mBio.00812-18
44
Hibbing M. E. Fuqua C. Parsek M. R. Peterson S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle.Natl. Rev. Microbiol.815–25. 10.1038/nrmicro2259.Bacterial
45
Kaltz O. Koella J. C. (2003). Host growth conditions regulate the plasticity of horizontal and vertical transmission in Holospora undulata, a bacterial parasite of the protozoan Paramecium caudatum.Evolution571535–1542. 10.1111/j.0014-3820.2003.tb00361.x
46
Kearse M. Moir R. Wilson A. Stones-Havas S. Cheung M. Sturrock S. et al (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Bioinformatics281647–1649. 10.1093/bioinformatics/bts199
47
Kellogg C. A. (2004). Tropical Archaea: diversity associated with the surface microlayer of corals.Mar. Ecol. Prog. Ser.27381–88. 10.3354/meps273081
48
Kikuchi Y. Hosokawa T. Fukatsu T. (2007). Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation.Appl. Environ. Microbiol.734308–4316. 10.1128/AEM.00067-07
49
Knowlton N. Rohwer F. (2003). Multispecies microbial mutualisms on coral reefs: the host as a habitat.Am. Nat.162S51–S62. 10.1086/378684
50
Krueger D. M. Gustafson R. G. Cavanaugh C. M. (1996). Vertical transmission of chemoautotrophic symbionts in the bivalve Solemya velum (Bivalvia: Protobranchia).Biol. Bull.190195–202. 10.2307/1542539
51
Lee O. O. Chiu P. Y. Wong Y. H. Pawlik J. R. Qian P. Y. (2009). Evidence for vertical transmission of bacterial symbionts from adult to embryo in the Caribbean sponge Svenzea zeai.Appl. Environ. Microbiol.756147–6156. 10.1128/AEM.00023-09
52
Leite D. C. A. Leão P. Garrido A. G. Lins U. Santos H. F. Pires D. O. et al (2017). Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core.Front. Microbiol.8:176. 10.3389/fmicb.2017.00176
53
Leite D. C. A. Salles J. F. Calderon E. N. van Elsas J. D. Peixoto R. S. (2018). Specific plasmid patterns and high rates of bacterial co-occurrence within the coral holobiont.Ecol. Evol.81818–1832. 10.1002/ece3.3717
54
Lema K. A. Bourne D. G. Willis B. L. (2014). Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora.Mol. Ecol.234682–4695. 10.1111/mec.12899
55
Lema K. A. Willis B. L. Bourneb D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria.Appl. Environ. Microbiol.783136–3144. 10.1128/AEM.07800-11
56
Lesser M. P. Falcón L. I. Rodríguez-Román A. Enríquez S. Hoegh-Guldberg O. Iglesias-Prieto R. (2007). Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa.Mar. Ecol. Prog. Ser.346143–152. 10.3354/meps07008
57
Lesser M. P. Mazel C. H. Gorbunov M. Y. Falkowski P. G. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals.Science305997–1000. 10.1126/science.1099128
58
Li X. Zhang Y. Ding C. Jia Z. He Z. Zhang T. et al (2015). Declined soil suppressiveness to Fusarium oxysporum by rhizosphere microflora of cotton in soil sickness.Biol. Fertil. Soils51935–946. 10.1007/s00374-015-1038-8
59
Little A. F. van Oppen M. J. H. Willis B. L. (2004). Flexibility in algal endosymbioses shapes growth in reef corals.Science3041492–1494. 10.1126/science.1095733
60
Littman R. A. Willis B. L. Bourne D. G. (2009a). Bacterial communities of juvenile corals infected with different Symbiodinium (dinoflagellate) clades.Mar. Ecol. Prog. Ser.38945–59. 10.3354/meps08180
61
Littman R. A. Willis B. L. Pfeffer C. Bourne D. G. (2009b). Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef.FEMS Microbiol. Ecol.68152–163. 10.1111/j.1574-6941.2009.00666.x
62
Munson M. A. Baumann P. Clark M. A. Baumann L. Moran N. A. Voegtlin D. J. et al (1991). Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families.J. Bacteriol.1736321–6324. 10.1128/jb.173.20.6321-6324.1991
63
Murray D. C. Coghlan M. L. Bunce M. (2015). From benchtop to desktop: important considerations when designing amplicon sequencing workflows.PLoS One10:e0124671. 10.1371/journal.pone.0124671
64
Nakamura M. Ohki S. Suzuki A. Sakai K. (2011). Coral larvae under ocean acidification: survival, metabolism, and metamorphosis.PLoS One6:e14521. 10.1371/journal.pone.0014521
65
Neave M. J. Michell C. T. Apprill A. Voolstra C. R. (2017). Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts.Sci. Rep.7:40579. 10.1038/srep40579
66
Neulinger S. C. Järnegren J. Ludvigsen M. Lochte K. Dullo W. C. (2008). Phenotype-specific bacterial communities in the cold-water coral Lophelia pertusa (Scleractinia) and their implications for the coral’s nutrition, health, and distribution.Appl. Environ. Microbiol.747272–7285. 10.1128/AEM.01777-08
67
Nissimov J. Rosenberg E. Munn C. B. (2009). Antimicrobial properties of resident coral mucus bacteria of Oculina patagonica.FEMS Microbiol. Lett.292210–215. 10.1111/j.1574-6968.2009.01490.x
68
Nussbaumer A. D. Fisher C. R. Bright M. (2006). Horizontal endosymbiont transmission in hydrothermal vent tubeworms.Nature441345–348. 10.1038/nature04793
69
Nyholm S. V. McFall-Ngai M. J. (2004). The winnowing: establishing the squid - Vibrios symbiosis.Nat. Rev. Microbiol.2632–642. 10.1038/nrmicro957
70
Ogawa D. (2014). The Effects of Climate Change on the Acropora aspera holobiont.Ph.D. thesis, James Cook University, Cairns. 10.3354/cr00733
71
Olson N. D. Ainsworth T. D. Gates R. D. Takabayashi M. (2009). Diazotrophic bacteria associated with Hawaiian Montipora corals: diversity and abundance in correlation with symbiotic dinoflagellates.J. Exp. Mar. Biol. Ecol.371140–146. 10.1016/j.jembe.2009.01.012
72
Padilla-Gamiño J. L. Weatherby T. M. Waller R. G. Gates R. D. (2011). Formation and structural organization of the egg-sperm bundle of the scleractinian coral Montipora capitata.Coral Reefs30371–380. 10.1007/s00338-010-0700-8
73
Paredes G. Warner R. R. Jackson J. B. C. Pandolfi J. M. Bradbury R. H. Sala E. et al (2003). Global trajectories of the long-term decline of coral reef ecosystems.Science301955–958. 10.1126/science.1085706
74
Patten N. L. Mitchell J. G. Middelboe M. Eyre B. D. Seuront L. Harrison P. L. et al (2008). Bacterial and viral dynamics during a mass coral spawning period on the Great Barrier Reef.Aquat. Microb. Ecol.50209–220. 10.3354/ame01179
75
Peixoto R. Rosado P. Leite D. Rosado A. S. Bourne D. (2017). Beneficial Microorganisms for Corals (BMC): proposed mechanisms for coral health and resilience.Front. Microbiol.8:341. 10.3389/fmicb.2017.00341
76
Pogoreutz C. (2016). Coral Holobiont Functioning Under Global Environmental Change.Ph.D. thesis, University of Bremen, Bremen.
77
Polson S. W. (2007). Comparative Analysis of Microbial Community Structure Associated with Acroporid Corals During a Disease Outbreak in the Florida Reef Tract.Ph.D. disseartion, Medical University of South Carolina, Charleston, SC. 10.13140/2.1.2852.4484
78
Quigley K. M. Willis B. L. Bay L. K. (2016). Maternal effects and Symbiodinium community composition drive differential patterns in juvenile survival in the coral Acropora tenuis.R. Soc. Open Sci.3:160471. 10.1098/rsos.160471
79
Quigley K. M. Willis B. L. Bay L. K. (2017). Heritability of the Symbiodinium community in vertically-and horizontally-transmitting broadcast spawning corals.Sci. Rep.7:100453. 10.1038/s41598-017-08179-4
80
Raina J. Tapiolas D. Motti C. A. Tebben J. Willis B. L. Bourne D. G. (2016). Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals.PeerJ4:e2275. 10.7717/peerj.2275
81
Raina J. B. Dinsdale E. A. Willis B. L. Bourne D. G. (2010). Do the organic sulfur compounds DMSP and DMS drive coral microbial associations?Trends Microbiol.18101–108. 10.1016/j.tim.2009.12.002
82
Raina J. B. Tapiolas D. Willis B. L. Bourne D. G. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur.Appl. Environ. Microbiol.753492–3501. 10.1128/AEM.02567-08
83
Raina J. B. Tapiolas D. M. Forêt S. Lutz A. Abrego D. Ceh J. et al (2013). DMSP biosynthesis by an animal and its role in coral thermal stress response.Nature502677–680. 10.1038/nature12677
84
Rideout J. R. He Y. Navas-Molina J. A. Walters W. A. Ursell L. K. Gibbons S. M. et al (2014). Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences.PeerJ2:e545. 10.7717/peerj.545
85
Rizzi A. Crotti E. Borruso L. Jucker C. Lupi D. Colombo M. et al (2013). Characterization of the bacterial community associated with larvae and adults of Anoplophora chinensis collected in Italy by culture and culture-independent methods.Biomed Res. Int.2013:420287. 10.1155/2013/420287
86
Rohwer F. Seguritan V. Azam F. Knowlton N. (2002). Diversity and distribution of coral-associated bacteria.Mar. Ecol. Prog. Ser.2431–10. 10.3354/meps243001
87
Rosenberg E. Koren O. Reshef L. Efrony R. Zilber-Rosenberg I. (2007). The role of microorganisms in coral health, disease and evolution.Nat. Rev. Microbiol.5355–362. 10.1038/nrmicro1635
88
Russell S. L. (2019). Transmission mode is associated with environment type and taxa across bacteria-eukaryote symbioses.FEMS Microbiol. Lett. 366 10.1101/412882
89
Russell S. L. Corbett-Detig R. B. Cavanaugh C. M. (2017). Mixed transmission modes and dynamic genome evolution in an obligate animal-bacterial symbiosis.ISME J.111359–1371. 10.1038/ismej.2017.10
90
Schmitt S. Angermeier H. Schiller R. Lindquist N. Hentschel U. (2008). Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts.Appl. Environ. Microbiol.747694–7708. 10.1128/AEM.00878-08
91
Schöttner S. Wild C. Hoffmann F. Boetius A. Ramette A. (2012). Spatial scales of bacterial diversity in cold-water coral reef ecosystems.PLoS One7:e32093. 10.1371/journal.pone.0032093
92
Shannon C. E. (1948). A mathematical theory of communication.Bell Syst. Tech. J.27379–423. 10.1002/j.1538-7305.1948.tb01338.x
93
Sharp K. H. Distel D. Paul V. J. (2012). Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides.ISME J.6790–801. 10.1038/ismej.2011.144
94
Sharp K. H. Eam B. John Faulkner D. Haygood M. G. (2007). Vertical transmission of diverse microbes in the tropical sponge Corticium sp.Appl. Environ. Microbiol.73622–629. 10.1128/AEM.01493-06
95
Sharp K. H. Ritchie K. B. (2012). Multi-partner interactions in corals in the face of climate change.Biol. Bull.22366–77. 10.1086/bblv223n1p66
96
Sharp K. H. Ritchie K. B. Schupp P. J. Ritson-Williams R. Paul V. J. (2010). Bacterial acquisition in juveniles of several broadcast spawning coral species.PLoS One5:e10898. 10.1371/journal.pone.0010898
97
Silveira C. B. Gregoracci G. B. Coutinho F. H. Silva G. G. Z. Haggerty J. M. de Oliveira L. S. et al (2017). Bacterial community associated with the reef coral Mussismilia braziliensis’s momentum boundary layer over a diel cycle.Front. Microbiol.8:784. 10.3389/fmicb.2017.00784
98
Slezak D. M. Puskaric S. Herndl G. J. (1994). Potential role of acrylic acid in bacterioplankton communities in the sea.Mar. Ecol. Prog. Ser.105191–198. 10.3354/meps105191
99
Stat M. Carter D. Hoegh-Guldberg O. (2006). The evolutionary history of Symbiodinium and scleractinian hosts-symbiosis, diversity, and the effect of climate change.Perspect. Plant Ecol. Evol. Syst.823–43. 10.1016/j.ppees.2006.04.001
100
Sunagawa S. Woodley C. M. Medina M. (2010). Threatened corals provide underexplored microbial habitats.PLoS One5:e9554. 10.1371/journal.pone.0009554
101
Tebben J. Tapiolas D. M. Motti C. A. Abrego D. Negri A. P. Blackall L. L. et al (2011). Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium.PLoS One6:e19082. 10.1371/journal.pone.0019082
102
Turnbaugh P. J. Ley R. E. Hamady M. Fraser-liggett C. Knight R. Gordon J. I. (2007). The human microbiome project: exploring the microbial part of ourselves in a changing world.Nature449804–810. 10.1038/nature06244.
103
Van Alstyne K. L. Schupp P. Slattery M. (2006). The distribution of dimethylsulfoniopropionate in tropical Pacific coral reef invertebrates.Coral Reefs25321–327. 10.1007/s00338-006-0114-9
104
van de Water J. A. J. M. Allemand D. Ferrier-Pagès C. (2018). Host-microbe interactions in octocoral holobionts - recent advances and perspectives.Microbiome6:64. 10.1186/s40168-018-0431-6
105
Wainwright S. A. (1963). Skeletal organization in the coral, Pocillopora damicornis.Q. J. Microsc. Sci.104169–183. 10.1016/j.molonc.2007.11.002
106
Wang Q. Garrity G. M. Tiedje J. M. Cole J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy.Appl. Environ. Microbiol.735261–5267. 10.1128/AEM.00062-07
107
Wang Y. Xu L. Gu Y. Q. Coleman-Derr D. (2016). MetaCoMET: a web platform for discovery and visualization of the core microbiome.Bioinformatics323469–3470. 10.1093/bioinformatics/btw507
108
Watanabe T. Fukuda I. China K. Isa Y. (2003). Molecular analyses of protein components of the organic matrix in the exoskeleton of two scleractinian coral species.Comp. Biochem. Physiol. B Biochem. Mol. Biol.136767–774. 10.1016/S1096-4959(03)00177-5
109
Webster N. S. Smith L. D. Heyward A. J. Watts J. E. M. Webb R. I. Blackall L. L. et al (2004). Metamorphosis of a Scleractinian coral in response to microbial biofilm.Appl. Environ. Microbiol.701213–1221. 10.1128/AEM.70.2.1213
110
Webster N. S. Taylor M. W. Behnam F. Lücker S. Rattei T. Whalan S. et al (2010). Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts.Environ. Microbiol.122070–2082. 10.1111/j.1462-2920.2009.02065.x
111
Weis V. M. Reynolds W. S. DeBoer M. D. Krupp D. A. (2001). Host-symbiont specificity during onset of symbiosis between the dinoflagellates Symbiodinium spp. and planula larvae of the scleractinian coral Fungia scutaria.Coral Reefs20301–308. 10.1007/s003380100179
112
Williams A. D. Brown B. E. Putchim L. Sweet M. J. (2015). Age-related shifts in bacterial diversity in a reef coral.PLoS One10:e0144902. 10.1371/journal.pone.0144902
113
Wright R. M. Kenkel C. D. Dunn C. E. Shilling E. N. Bay L. K. Mikhail V. M. (2016). Higher stress and immunity responses are associated with higher mortality in reef-building coral exposed to a bacterial challenge.bioRxiv [Preprint]. 10.1101/059444
114
Zhou G. Cai L. Yuan T. Tian R. Tong H. Zhang W. et al (2017). Microbiome dynamics in early life stages of the scleractinian coral Acropora gemmifera in response to elevated pCO2.Environ. Microbiol.193342–3352. 10.1111/1462-2920.13840
Summary
Keywords
core bacterial communities, symbiosis, 16S rRNA, metabarcoding, bacterial communities, seawater, sediment, transmission
Citation
Bernasconi R, Stat M, Koenders A, Paparini A, Bunce M and Huggett MJ (2019) Establishment of Coral-Bacteria Symbioses Reveal Changes in the Core Bacterial Community With Host Ontogeny. Front. Microbiol. 10:1529. doi: 10.3389/fmicb.2019.01529
Received
12 March 2019
Accepted
18 June 2019
Published
09 July 2019
Volume
10 - 2019
Edited by
Zhiyong Li, Shanghai Jiao Tong University, China
Reviewed by
Sen-Lin Tang, Biodiversity Research Center, Academia Sinica, Taiwan; Julie L. Meyer, University of Florida, United States
Updates
Copyright
© 2019 Bernasconi, Stat, Koenders, Paparini, Bunce and Huggett.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rachele Bernasconi, r.bernasconi@ecu.edu.au
This article was submitted to Microbial Symbioses, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.